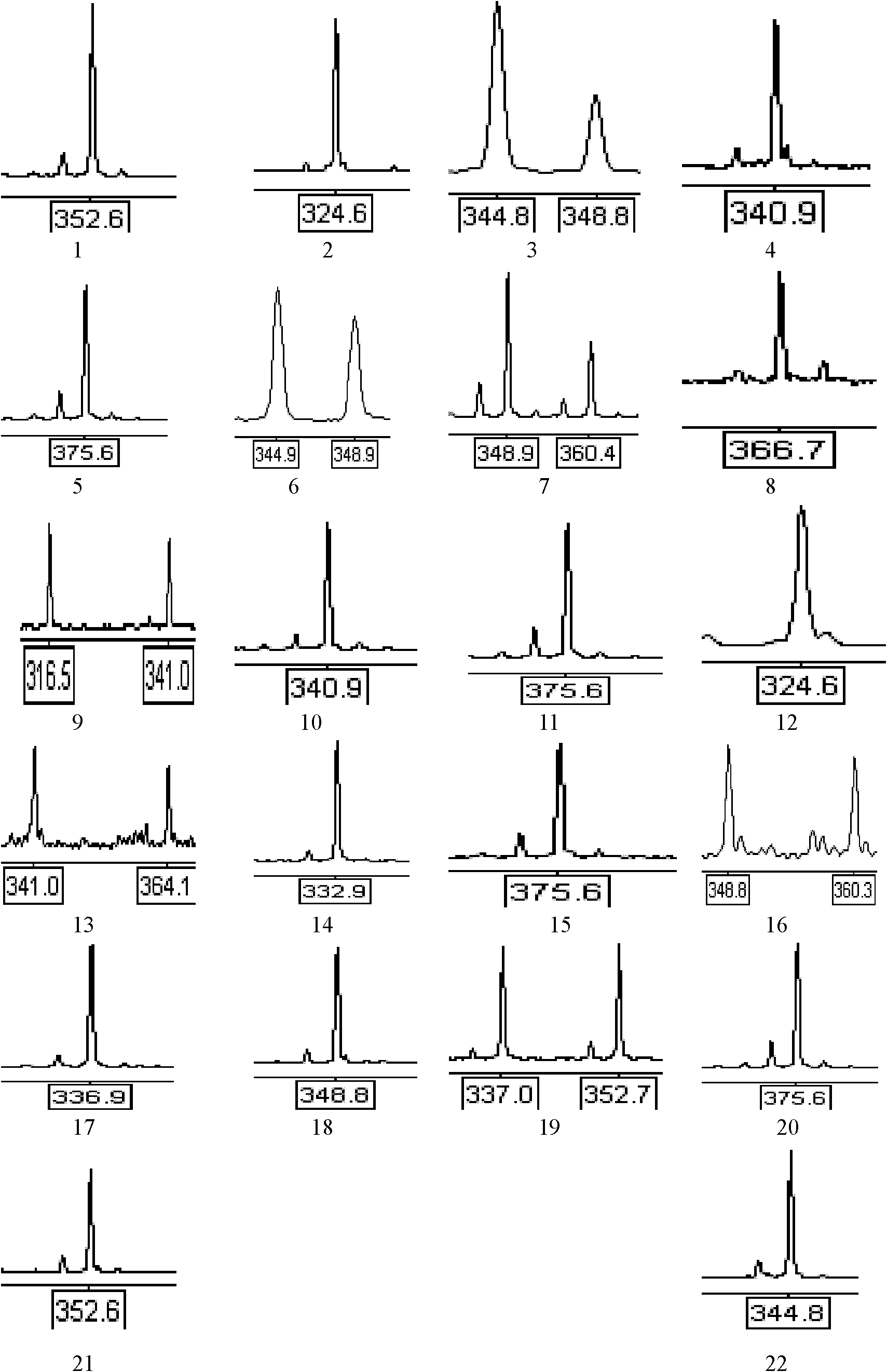
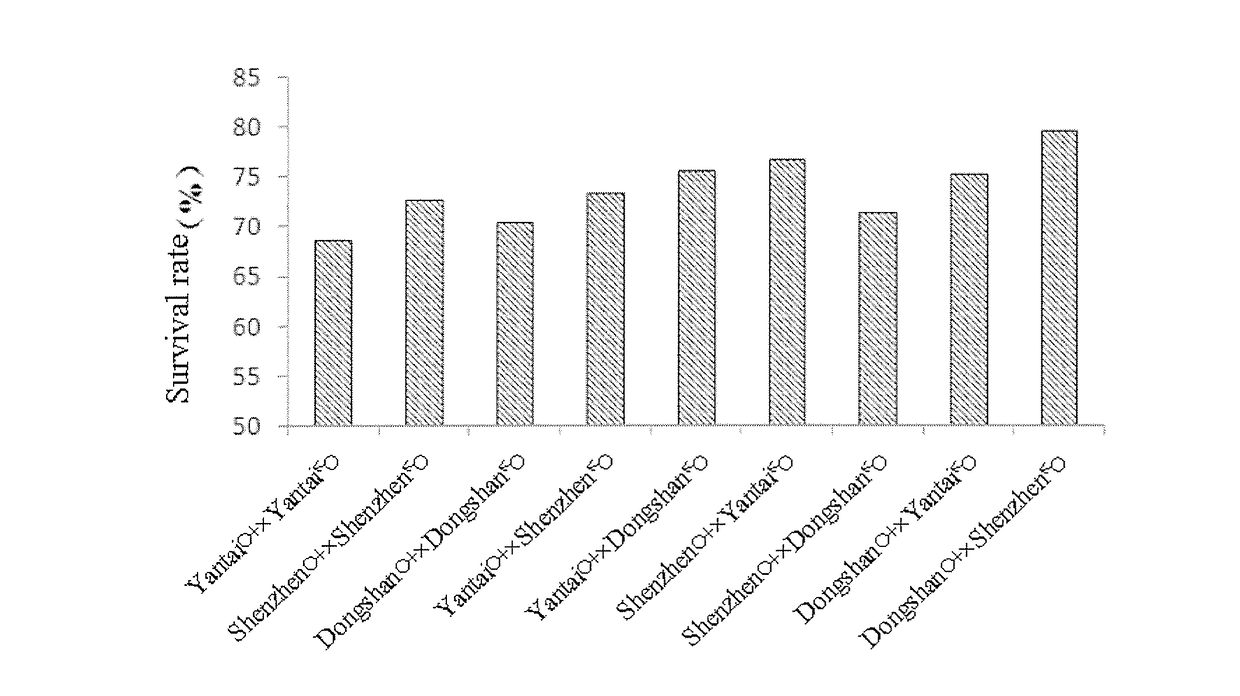
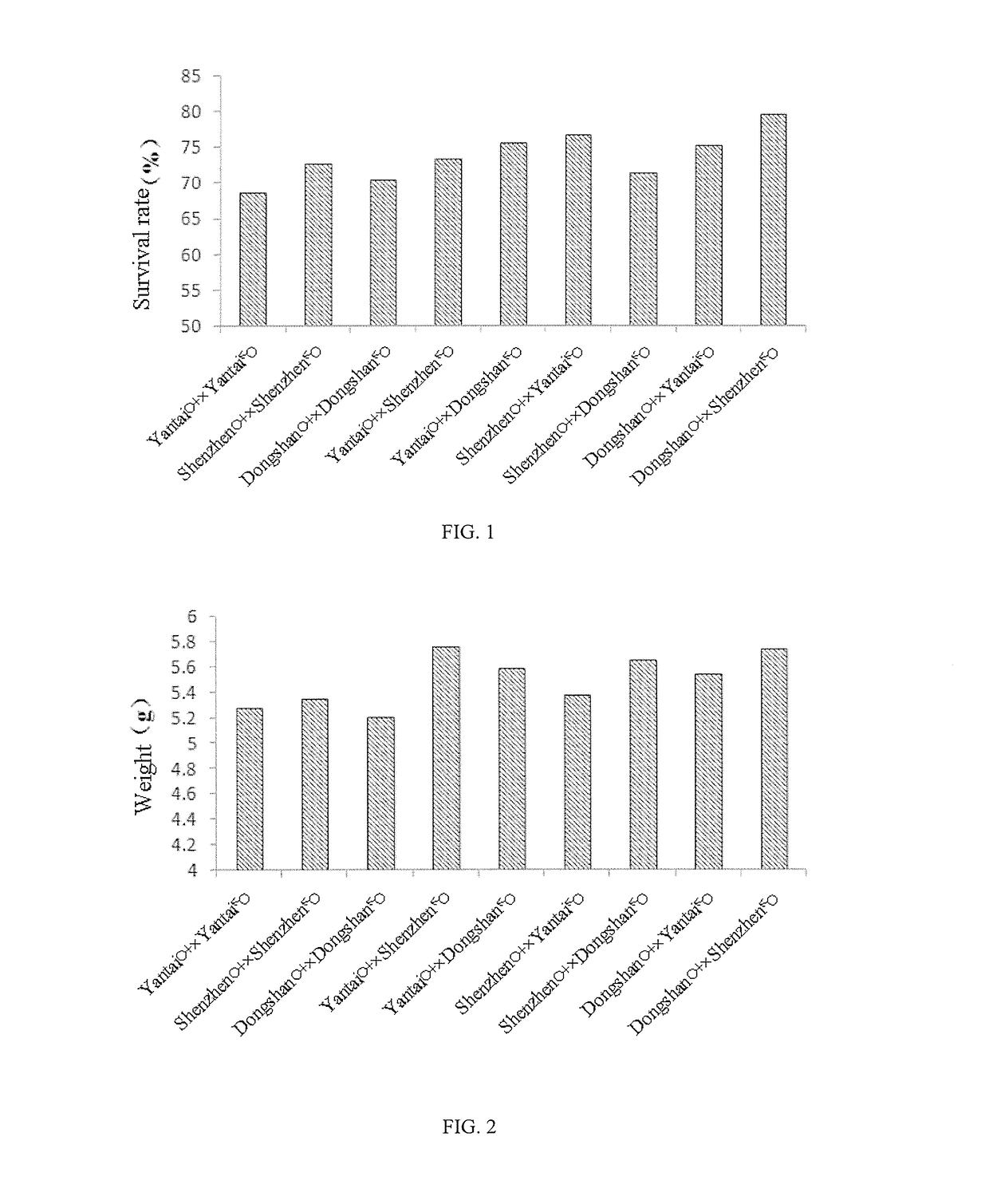
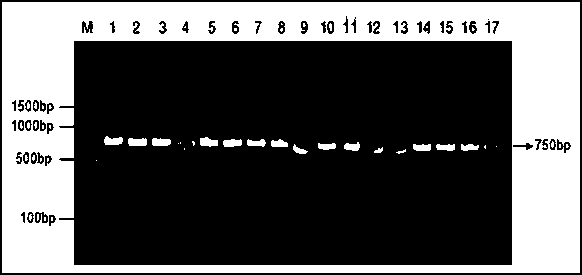
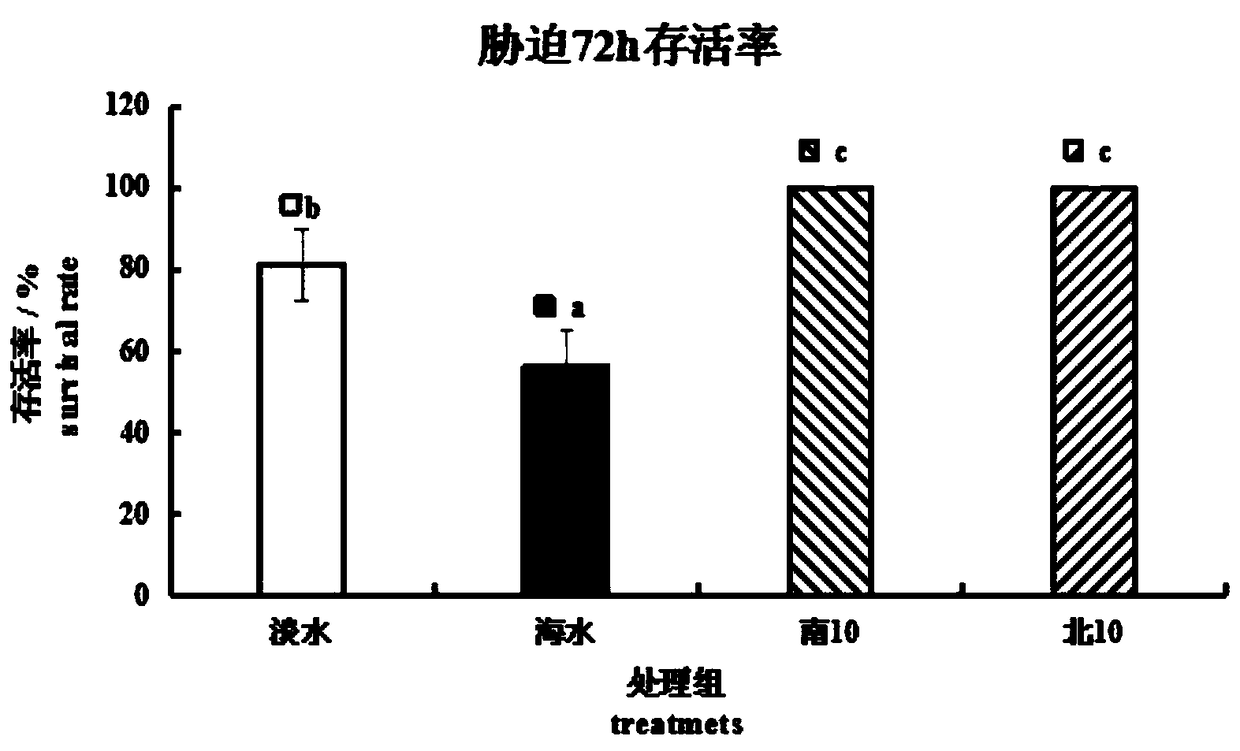
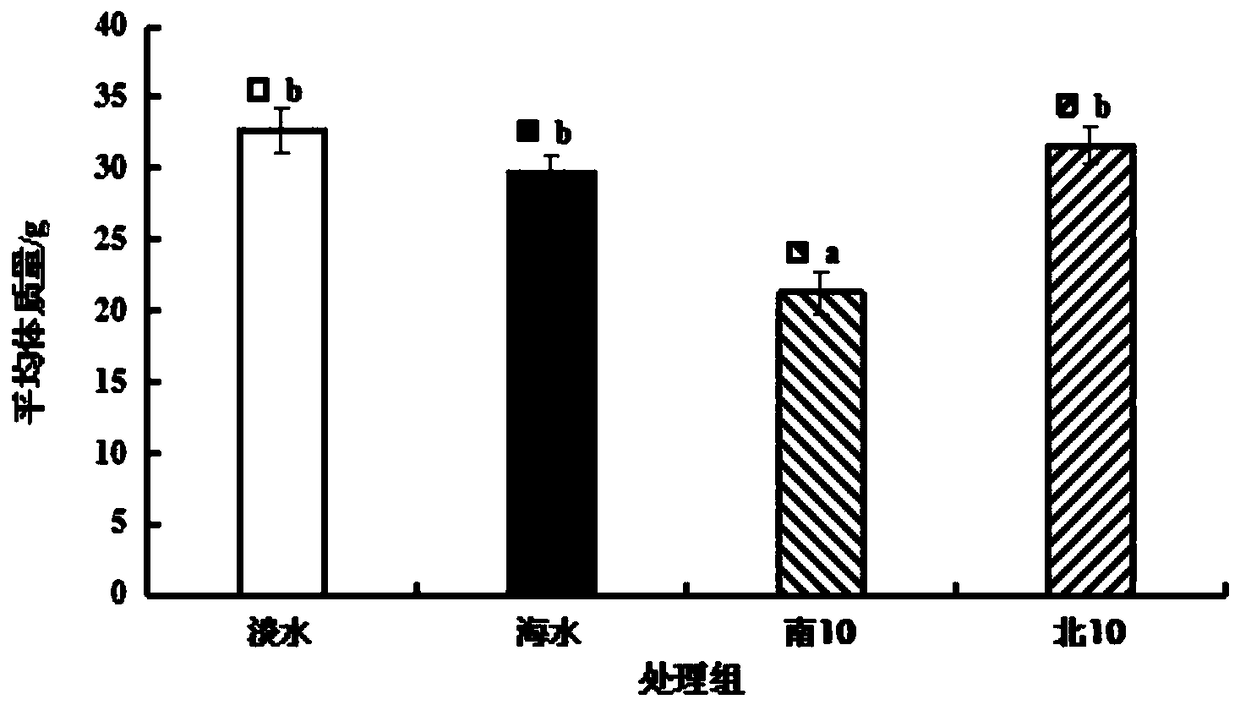
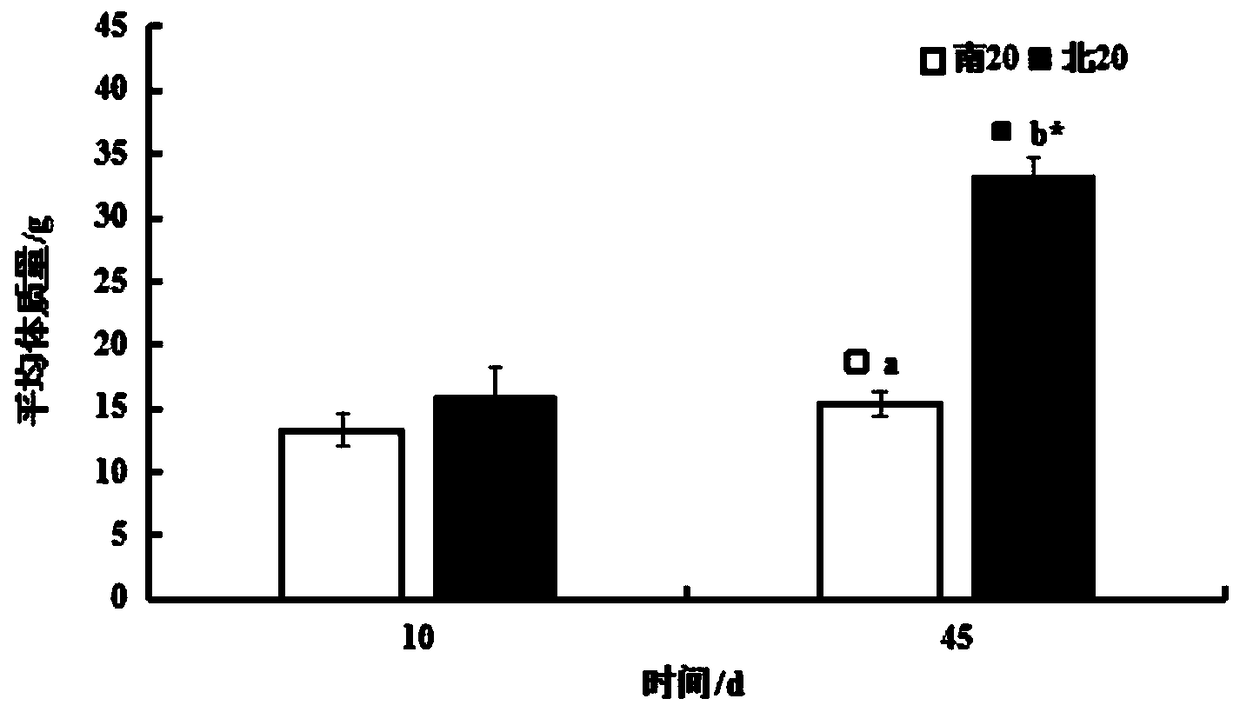
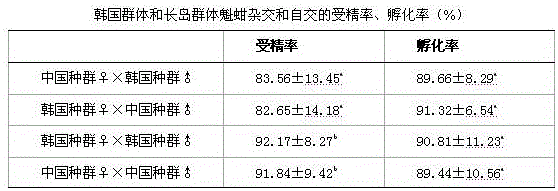
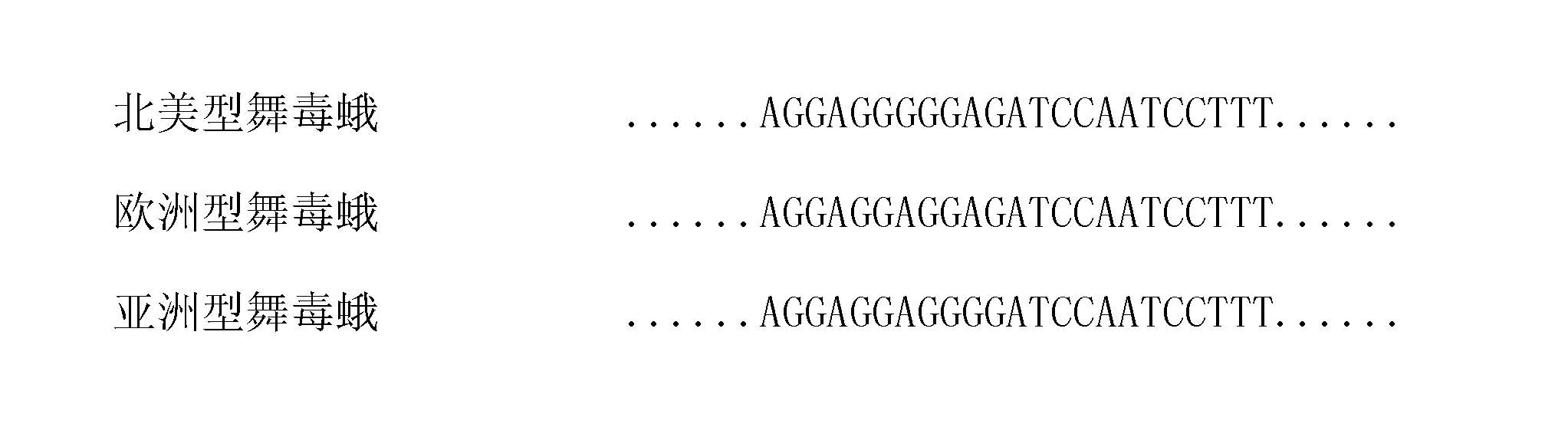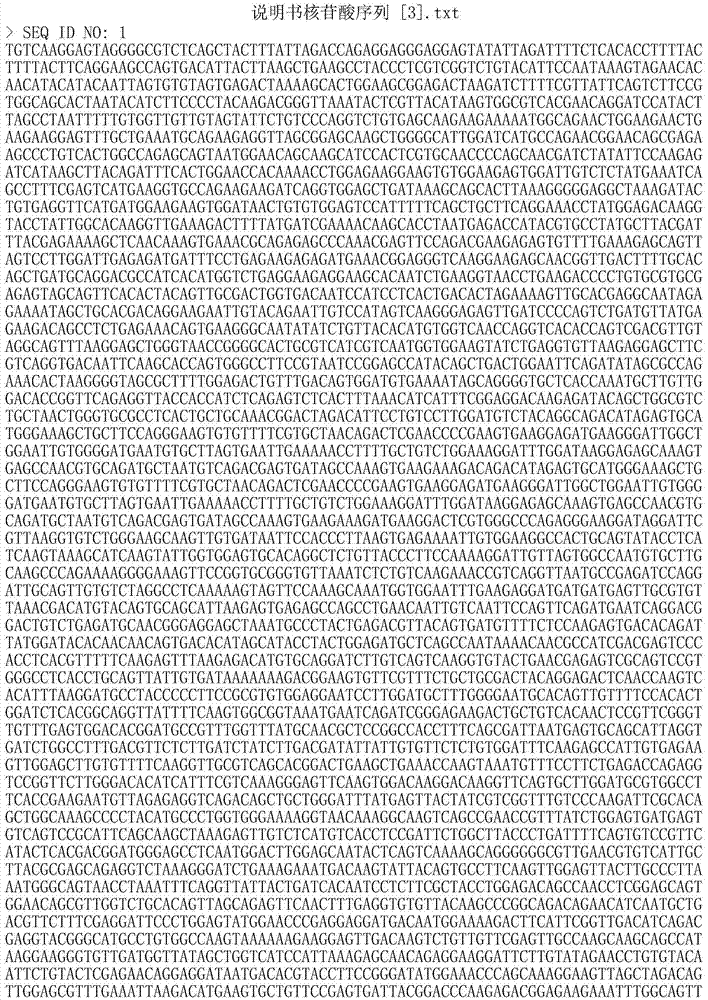Patents
Literature
47 results about "Geographical population" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
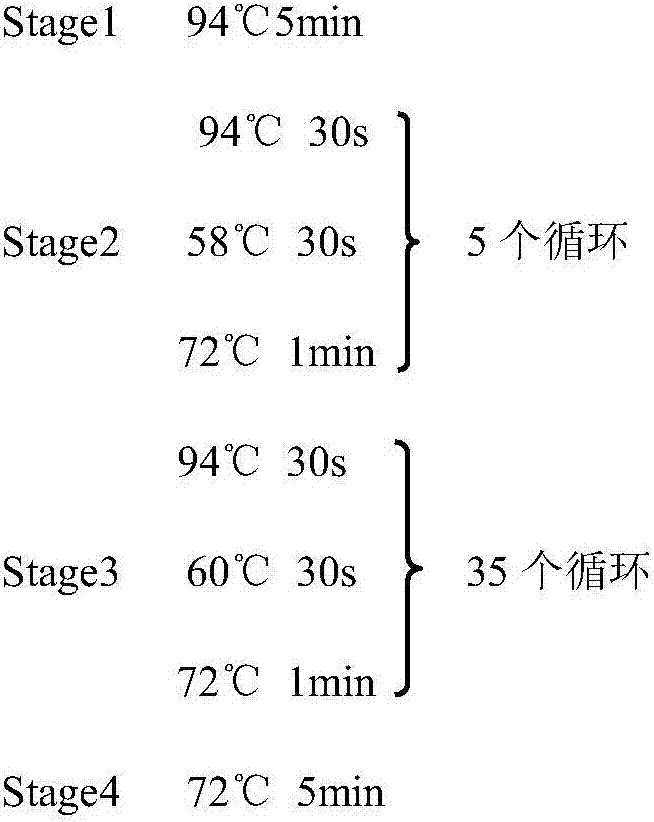
Amplification primer for grouper catostomidae mitochondrial genome complete sequence and design method thereof
InactiveCN101698842AEasy splicingNo mismatchMicrobiological testing/measurementDNA/RNA fragmentationGermplasmComplete sequence
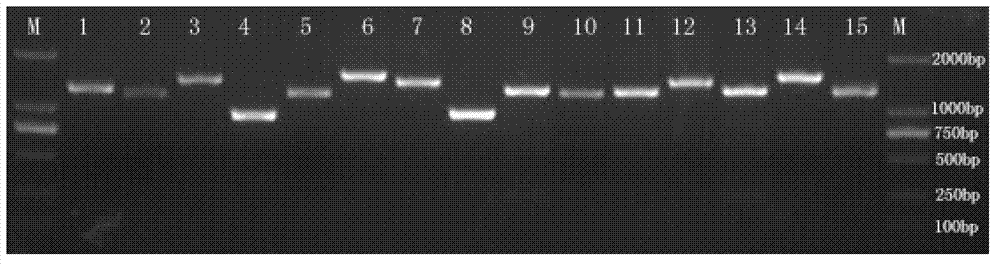
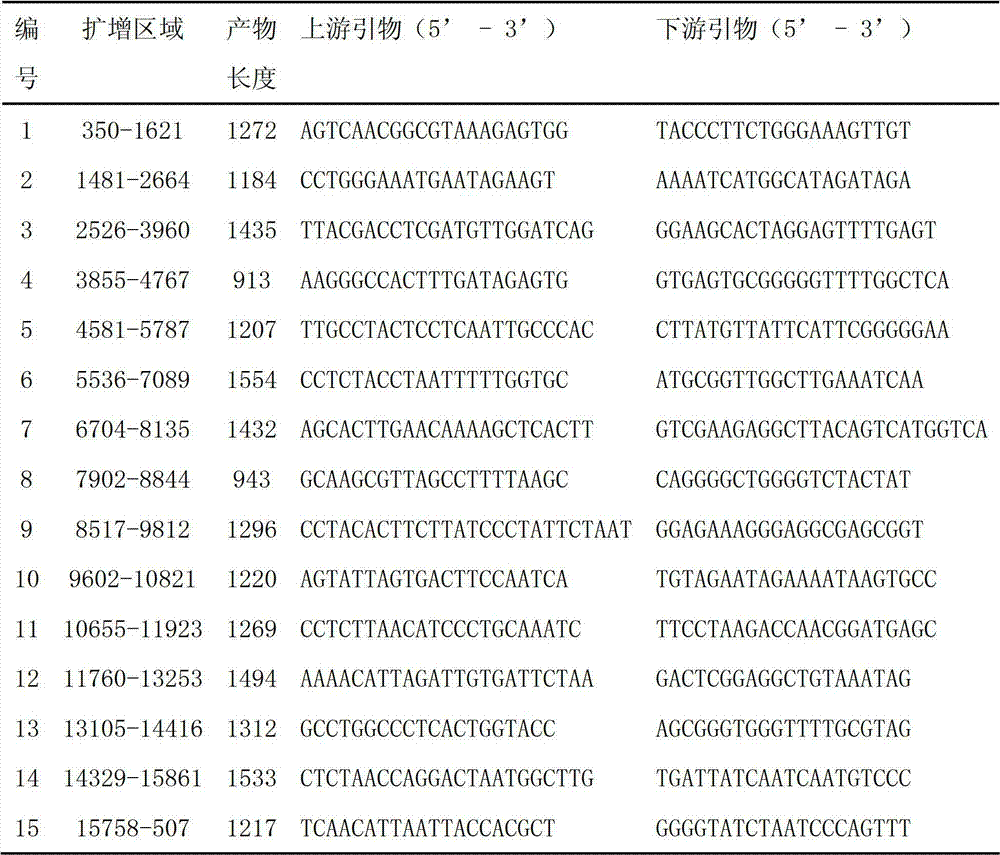
The invention discloses an amplification primer for a grouper catostomidae mitochondrial genome complete sequence and a design method thereof, relates to groupers, and particularly relates to 16 pairs of amplification primers mainly used for specifically amplifying most grouper catostomidae mitochondrial genome complete sequences and a design method thereof. The invention provides an amplification primer for a grouper catostomidae mitochondrial genome complete sequence, a design method thereof and application thereof. The amplification primer for the grouper catostomidae mitochondrial genome complete sequence consists of 16 pairs of single-chain oligonucleotides, wherein each pair consists of one light-chain primer and one heavy-chain primer. The amplification primer for the grouper catostomidae mitochondrial genome complete sequence can be used in the aspects of variety authentication, geographical population authentication, germplasm resource evaluation and the like.
Owner:XIAMEN UNIV
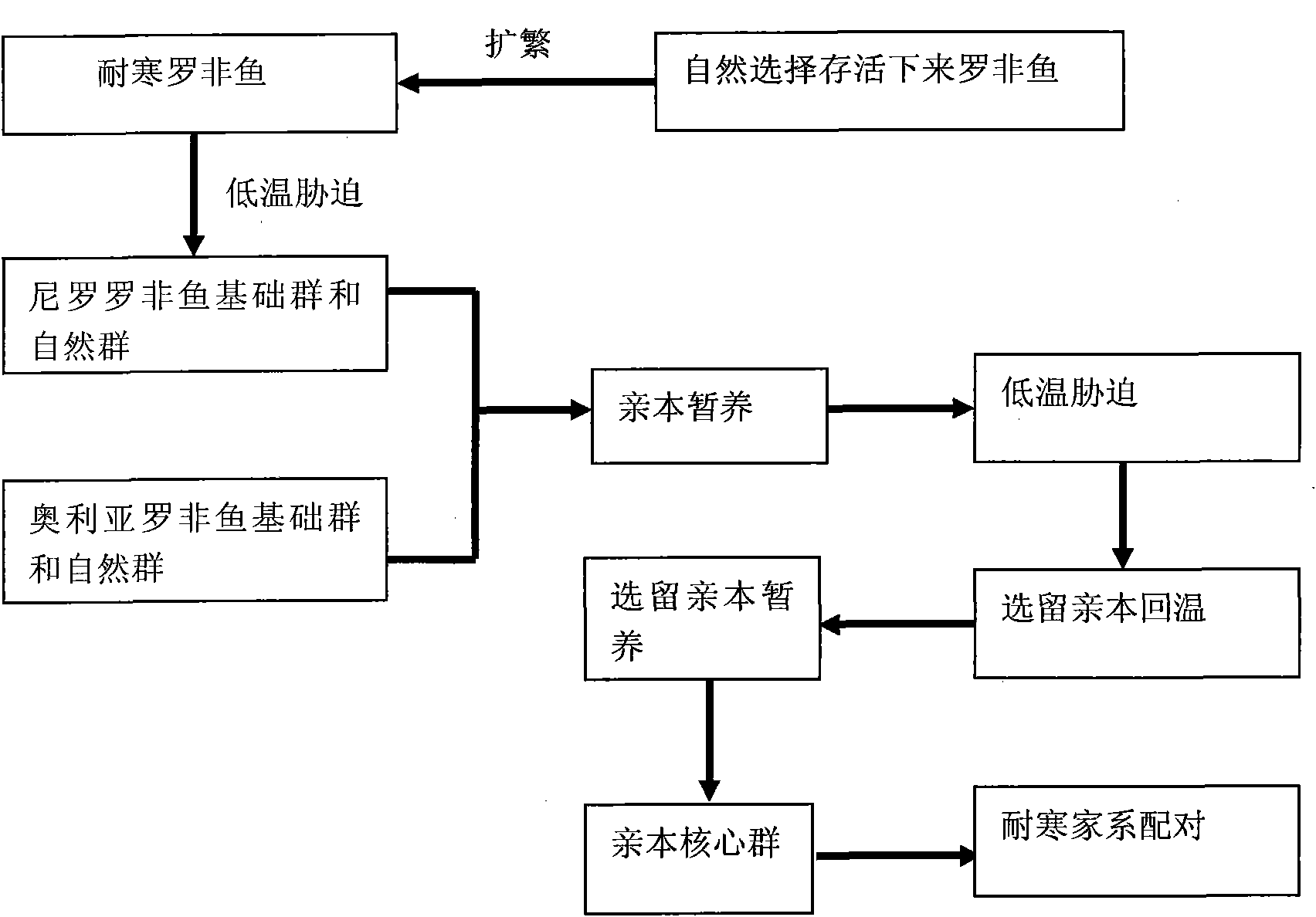
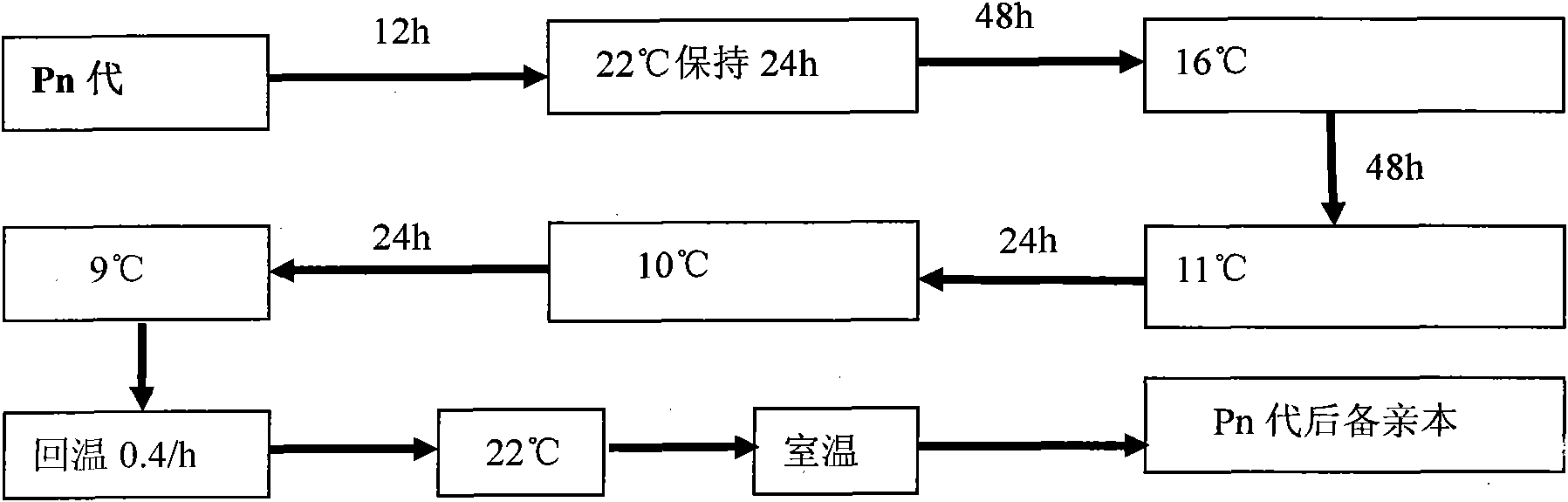
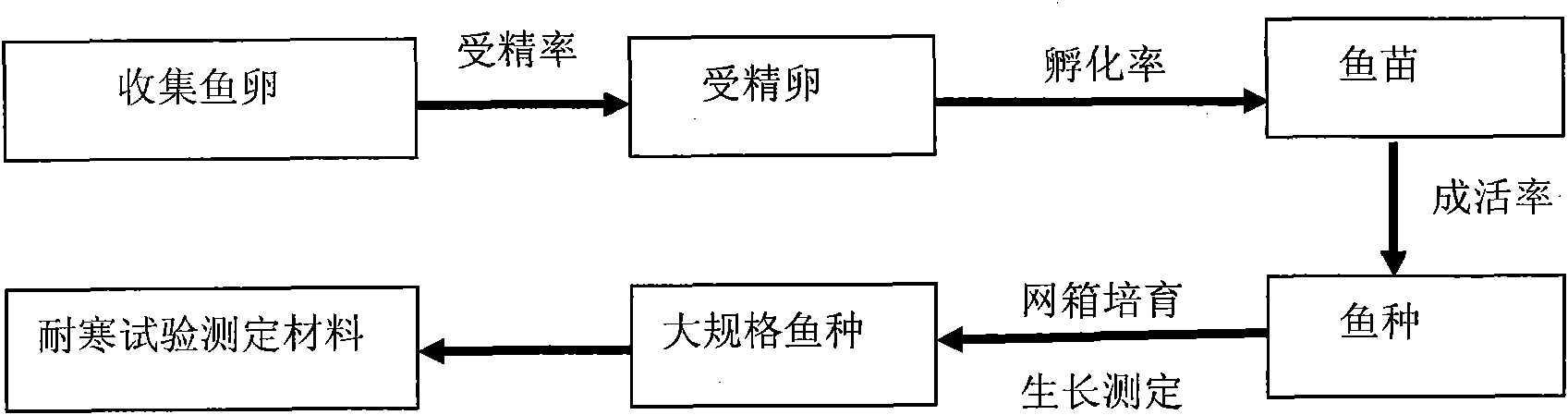
Selection method of new cold-resistant species of tilapia
InactiveCN101889561AStrong cold resistanceSelective methods are effectiveClimate change adaptationPisciculture and aquariaTemperature stressTilapia
The invention provides a selection method of new cold-resistant species of tilapia. The invention is characterized by determining the cold resistance of tilapia of different geographical population and screening the tilapia of different geographical population by adopting the technology such as colony continuous progeny selection, family selection, specialized line selection and the like and using natural screening and artificial low-temperature stress selection as the means, selecting the cold-resistant tilapia of specialized line and creating the selection method of new cold-resistant species of tilapia. The technology has the characteristics of strong pertinency, obvious effect and the like. The overwintering survival rate of the cold-resistant tilapia fries is over 80% and the low-temperature mortality of the tilapia is reduced by more than 15% than that of the common tilapia. The selection method opens up a new technical approach for breeding of cold-resistant species of fish, is suitable for being popularized and applied in fish selection and has great significance and application value in safe overwintering of the cultivated fish.
Owner:GUANGXI INST OF FISHERIES
Amplimer of large yellow croaker mitochondrion complete genome sequence and application thereof
ActiveCN102776183AEasy splicingIncrease success rateMicrobiological testing/measurementDNA preparationGerm plasmGene orders
The invention discloses an amplimer of a large yellow croaker mitochondrion complete genome sequence and an application thereof. The 15 pairs of amplimer are applied, a gene order is respectively shown as SEQ ID No: 1-30, the amplimer is further used for obtaining a Tai Qu family large yellow croaker mitochondrion complete genome sequence and a Min-Yue East family large yellow croaker mitochondrion complete genome sequence which are respectively shown as SEQ ID No: 31 and SEQ ID No: 32, and an amplimer of large yellow croakers in a high variation region is obtained. The amplimer and the application have the advantages that the Tai Qu family large yellow croaker and Min-Yue East family large yellow croaker mitochondrion complete genome sequences can be quickly and efficiently obtained respectively through the 15 pairs of amplimers; and the amplimer of the large yellow croakers in the high variation region provides support for the research of species identification, geographical population identification, germ plasm resources and genetic diversity of the large yellow croakers, and further provide the basis for developing an identification technology of Tai Qu family large yellow croakers and Min-Yue East family large yellow croakers.
Owner:NINGBO UNIV
Microsatellite DNA markers for Trionyx sinensis
The invention discloses microsatellite DNA markers for Trionyx sinensis, which is characterized in that an enriched library of microsatellites (CA)n, (AAC)n, (ACAG)n and (GATA)n from the Trionyx sinensis is constructed, and positive clones of microsatellite sequences are screened and sequenced to obtain 63 clones containing microsatellite repetitive sequences, thus determining 15 microsatellite markers with rich polymorphism, namely, PSW01, PSW02, PSW03, PSW04, PSW05, PSW06, PSW07, PSW08, PSW09, PSW10, PSW11, PSW12, PSW13, PSW14 and PSW15. The microsatellite DNA markers for the Trionyx sinensis of the invention can be applied to researching genetic diversity and paternity test for the Trionyx sinensis in different geographical populations, have the advantage of good repeatability, and are reliable and effective molecular markers.
Owner:ANHUI NORMAL UNIV
Breeding method of babylonia areolata cross-breeding seeds
ActiveCN104798709AProtect environmentReduce breeding diseasesClimate change adaptationPisciculture and aquariaEcological environmentAntibiotic Y
The invention provides a breeding method of babylonia areolata cross-breeding seeds. Male Thailand babylonia areolata and female Hainan babylonia areolata as well as male Hainan babylonia areolata and female Thailand babylonia areolata are respectively adopted for cross breeding, and through processes of parent babylonia areolata sex determination, parent babylonia areolata of different geographical population cross breeding pairing, parent babylonia areolata nutrition enhacnign culture, oocyst collection and hatching, larva culture, young babylonia areolata culture and the like, the cross-breeding seeds with high growth speed and strong disease resistance are cultured. When the method is used for breeding cross-breeding seeds, the culture diseases are reduced, and the occurrence of diseases such as turgescence diseases and shucking diseases can be avoided. The cross-breeding seed stocking is adopted, the yield is increased by 8.2 to 20.3 percent through being compared with that of the traditional breeding seeds, the production value of per square meter of culture pools is increased by 80 to 189.6 Yuan, and the culture economic benefits are improved; antibiotics are not used in the culture process, the product quality is improved, the cultured product conforms to the pollution-free food standard, positive effects are realized on marine ecological environment protection, and the stable healthy and continuous development of the babylonia areolata culture industry is promoted.
Owner:海南蓝泰邦生物技术有限公司
Artificial medium of trichogramma
ActiveCN103859210AImprove the breeding effectStable breeding effectFood processingAnimal feeding stuffTrichogramma dendrolimiHemolymph
The invention discloses an artificial medium of trichogramma. The artificial medium comprises the following ingredients in mass fraction: 19-40% of insect pupae hemolymph, 17-38% of egg yolk, 10-33% of milk, 4-19% of a Nissl salt solution, 0.8-8% of trehalose, 2-15% of a ganoderan solution with a mass fraction of 5% and 2-15% of a chitosan oligosaccharide solution with a mass fraction of 3.6%. By adopting the artificial medium of trichogramma, the breeding effect of trichogramma is obviously better than that of a traditional artificial medium through the trehalose, the ganoderan solution and the chitosan oligosaccharide solution via effective composition, and the content of insects can be reduced, so that the breeding cost of trichogramma is reduced. Through the artificial medium of trichogramma, trichogramma can be bred by subculturing, no degradation phenomenon exists, and the breeding effect is stable. By adopting the artificial medium of trichogramma, various kinds of trichogramma such as trichogramma dendrolimi, trichogramma confusum, Asian corn borer trichogramma, European corn borer trichogramma and geographical populations thereof can be bred, and good effects are realized.
Owner:INST OF ZOOLOGY GUANGDONG ACAD OF SCI
A breeding method for obtaining heterosis in lined seahorses
ActiveUS20170142941A1Improve survival rateHigh speedClimate change adaptationPisciculture and aquariaPregnancyGenetic diversity
The present invention discloses a breeding method for obtaining heterosis in lined seahorses, which comprises the following steps: S1, selecting parents of lined seahorse from populations with great differences in genetic background; S2, intensified rearing the parents before pregnancy; S3, matching and breeding the parents of lined seahorse from different geographical populations according to complete diallel cross method; S4, finely nursing pregnant seahorses; S5, respectively collecting all postlarvae (filial generations) hatched by each breeding group in one week; S6, rearing the filial generations; and S7, comparing survival rate and growth performance of filial generations. The present invention, via hybridization of different geographical populations to obtain lined seahorse, makes effective use of heterosis. Survival rate and growth rate of filial generations are apparently enhanced compared to that of those filial generations without hybridization. Such method enhances genetic diversity of lined seahorses and accelerates breeding of fine strain of lined seahorses.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Manila clam pedigree and breeding process thereof
InactiveUS20100162963A1Quick upgradeImprove stress resistanceClimate change adaptationPisciculture and aquariaHigh resistanceHigh stress
A process for breeding a Manila clam pedigree, includes: directively breeding present Manila clam pedigrees having different shell shapes to obtain a shell shape pedigree (A) having best phenotypic character; directively breeding the shell shape pedigrees (A) from different geographical populations to obtain a geographical population pedigree (B) having best phenotypic character; directively breeding the geographical population pedigrees (B) having different shell colors to obtain a growth shell color pedigree (C) of fastest growth and a resistance shell color pedigrees (E) of highest stress resistance; hybridizing the growth shell color pedigree (C) and the resistance shell color pedigree (E) to obtain a first hybridied pedigree (F1) of faster growth and higher stress resistance than the present Manila clam pedigrees; and directively breeding the first hybridied pedigree (F1) to obtain a second hybridied pedigree (F2) maintaining faster growth and higher stress resistance than the present Manila clam pedigrees.
Owner:DALIAN FISHERIES UNIVERSITY
Method for obtaining variety of Oplopanax elatus with heterosis
InactiveCN101278659AGood production traitsFast growthClimate change adaptationPisciculture and aquariaHeterosisUltraviolet radiation
A seed production method for obtaining trepang heterosis is characterized in that Chinese seed trepangs and Japanese seed trepangs are ripened synchronously firstly; then, artificial egg taking is processed under the conditions that the environment is dried in the shade and the temperature is heated and seawater is irradiated by ultraviolet radiation; the sperm and the egg of Chinese trepangs and Japanese trepangs are hybridized; finally, the hatch and the seed rearing of hybridized trepangs are processed by a normal method. The invention has the advantages as follows: Chinese trepangs and Japanese trepangs are ripened synchronously by temperature adjustment; two procreation-isolated and different geographical populations are hybridized to cultivate a hybridized trepang seedling with fast growth speed, strong stress resistance and high survival rate. The production character of the hybridized trepang seedling is obviously better than Chinese local selfing control population, which has very good industrialization application prospect.
Owner:OCEAN UNIV OF CHINA
Phomopsis RNA (ribonucleic acid) polymerase (RPB1) gene amplification primer, as well as design method and application thereof
InactiveCN104195262ARapid Genetic Background AnalysisAccurate identificationMicrobiological testing/measurementMicroorganism based processesPolymerase LPhomopsis
The invention discloses a phomopsis RNA polymerase (RPB1) gene amplification primer, as well as a design method and application thereof. The primer is designed to aim at a universal primer of phomopsis funguses. An upstream primer PhR1-F has 43 basic groups: CGCCAGGGTTTTCCCAGTCACGACTGCAGCAAGGTGTTGGCTG, and a downstream primer PhR1-R has 43 basic groups: AGCGGATAACAATTTCACACAGGAGCGATGTCGTTGTCCATGTA. The primer can rapidly perform large-scaled genetic background analysis on the phomopsis funguses, and provides an important tool for species identification, geographical population identification or inspection and quarantine works of phomopsis in the country of China.
Owner:ANIMAL & PLANT & FOOD INSPECTION CENT OF TIANJIN ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Molecular marker for distinguishing Heilongjiang megalobrama terminalis from southern megalobrama fishes, kit and distinguishing method
ActiveCN108277287AQuick distinctionEfficient distinctionMicrobiological testing/measurementDNA/RNA fragmentationMegalobrama terminalisElectrophoresis
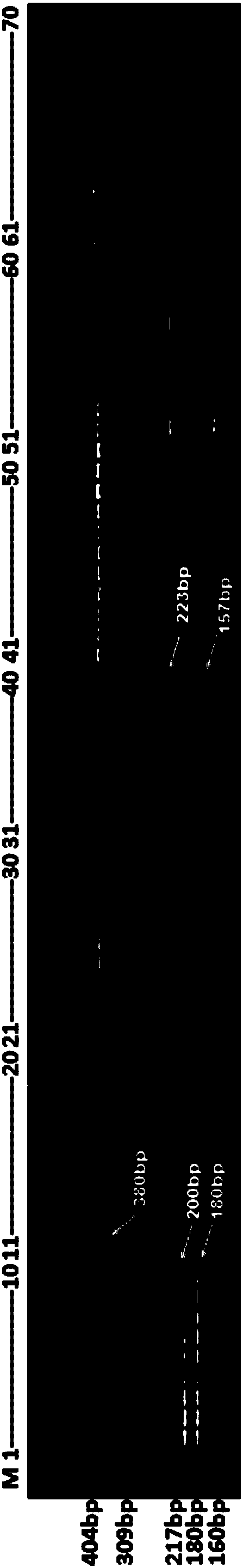
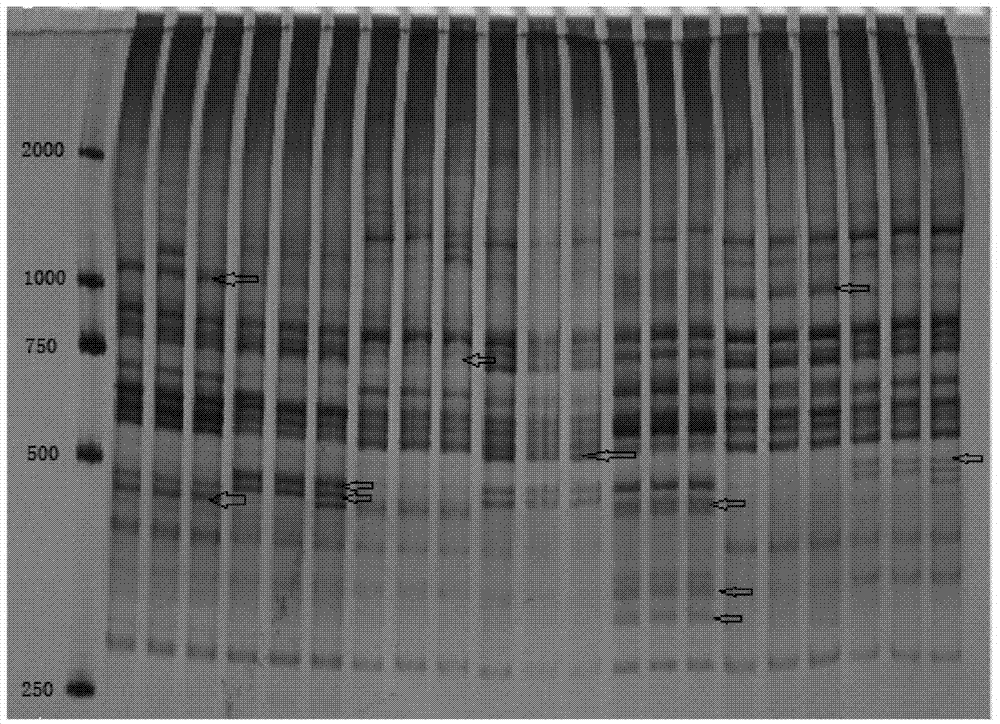
The invention discloses a molecular marker for distinguishing Heilongjiang megalobrama terminalis from southern megalobrama fishes, a kit and a distinguishing method, and relates to the molecular maker for distinguishing the megalobrama fishes and application of the molecular marker. A primer pair of the molecular marker can be HLJFLf1 and HLJFLr1. The kit comprises molecular marker primer pairs HLJFLf1 and HLJFLr1. The distinguishing method comprises the following steps: extracting fin rays DNA; 2, performing PCR amplification by using the HLJFLf1 and the HLJFLr1 as primers; 3, performing enzyme cutting on a PCR product by utilizing restriction endonuclease; 4, performing electrophoresis, wherein an electrophoresis result shows that corresponding individuals of double straps 200bp and 180bp are Heilongjiang megalobrama terminalis. According to the method disclosed by the invention, representative geographical population of the Heilongjiang megalobrama terminalis from the southern megalobrama fishes can be quickly, effectively and stably distinguished under a situation of no cross breeding.
Owner:HEILONGJIANG RIVER FISHERY RES INST CHINESE ACADEMY OF FISHERIES SCI
Specific primers for amplifying plecoptera insect mitochondrion COI (cytochrome c oxidase I) gene
InactiveCN104946645AAccurate identificationImprove accuracyFermentationPlant genotype modificationMetapopulationGene
Owner:YANGZHOU UNIV
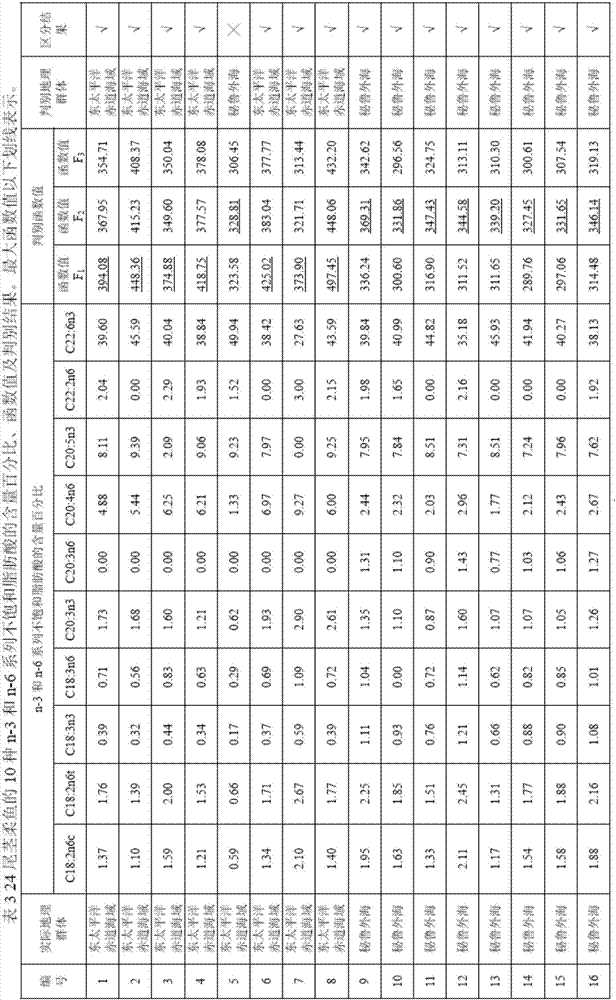
Method for distinguishing dosidicus gigas of multiple geographical populations with muscle essential fatty acid composition
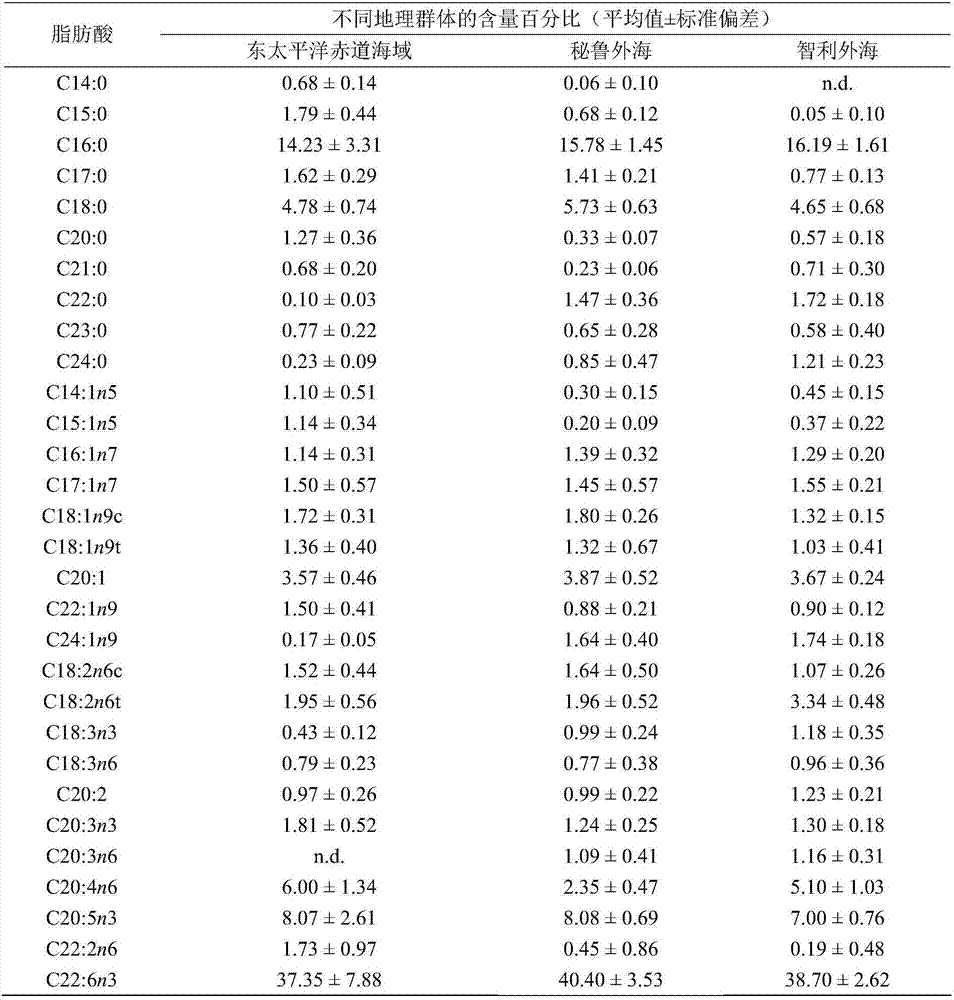
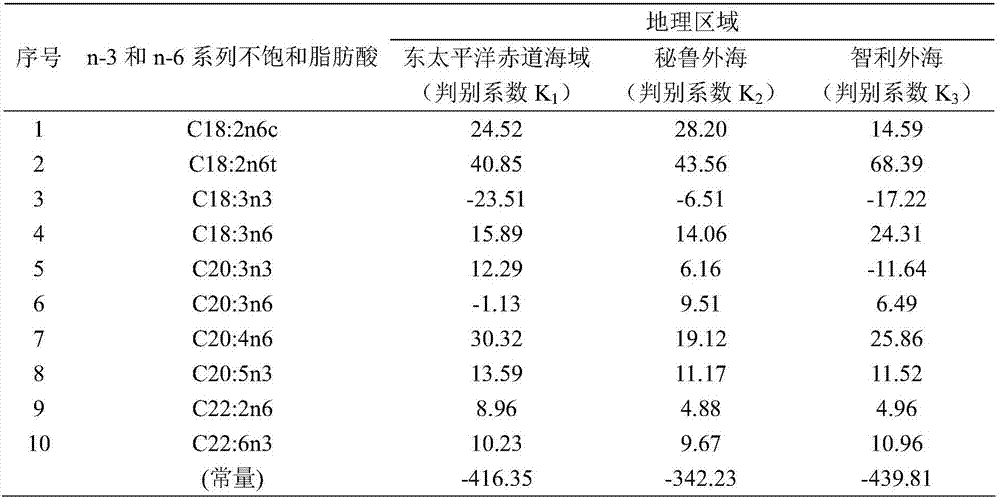
The invention relates to a method for distinguishing dosidicus gigas of multiple geographical populations with muscle essential fatty acid composition. The method specifically includes: (1) extracting dosidicus gigas muscle essential fatty acid; (2) determining fatty acid by GC-MS; (3) analyzing muscle fatty acid data; (4) calculating the n-3 and n-6 series unsaturated fatty acid discriminant coefficients and constants of dosidicus gigas of multiple geographical populations; and (5) performing discrimination: utilizing the to-be-discriminated dosidicus gigas muscle's n-3 and n-6 series unsaturated fatty acid percent content obtained by step (1)-(3) and the discriminant coefficients and constants obtained by step (4) to calculate a plurality of discriminant function values, and using a geographic area corresponding to a maximum function value to represent the belonging geographical population. The method provided by the invention has the advantages of rapidity and high accuracy.
Owner:SHANGHAI OCEAN UNIV
Efficient breeding method for families of Portunus trituberculatus
ActiveCN105918173AIncrease specificationImprove survival rateClimate change adaptationPisciculture and aquariaStage iiMetapopulation
The invention discloses an efficient breeding method for families of Portunus trituberculatus and belongs to the field of fishery breeding. According to the method, different wild geographical populations of the Portunus trituberculatus are collected, parent crab mating is completed with a manually controlled directional mating technology, after mating, large pincer muscles of male crabs are taken and stored at the ultralow temperature being subzero 80 DEG C, mated female crabs live through the winter and discharge the young, families are constructed, and after discharging of the young, large pincer muscles of the female crabs are taken and stored at the ultralow temperature being subzero 80 DEG C; when the young of the families develop to young crabs in the stage II, the young crabs are transferred outdoors for breeding, all the families are mixed in the equal proportion for releasing, test populations are established, and meanwhile, a conservation population is established for each family; at the 120-day age, growth traits of each individual of the test populations are measured, then family identification is performed on the individuals by the aid of screened micro-satellite primers, and statistics about survival rates and the growth traits of the families is performed; comprehensive indexes are calculated according to the survival rates and the growth traits of the families, breeding of the families is performed, reservation families are required, individual selection from the families is performed on conservation populations, parent crab mating is completed with an indoor manual directional mating technology, and accurate breeding of the families is realized.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Real-time fluorescent polymerase chain reaction (PCR) detection method for European Lymantria dispar, and primers and probe for detection
InactiveCN102534024AShorten detection timeHigh identification accuracyMicrobiological testing/measurementDNA/RNA fragmentationLymantria disparFluorescent pcr
The invention discloses a real-time fluorescent polymerase chain reaction (PCR) detection method for European Lymantria dispar, and primers and a probe for detection. The specific primers EU-F / R and the probe EU-TaqMan are designed according to sequences of partial genes in mtDNA of Lymantria dispar geographical populations such as Asian gypsy moth, European Lymantria dispar and North American Lymantria dispar, and the real-time fluorescent PCR detection method for the European Lymantria dispar is established, so that an aim of quickly and accurately detecting and identifying the European Lymantria dispar is fulfilled.
Owner:SHANGHAI ENTRY EXIT INSPECTION & QUARANTINE BUREAU OF P R C
Molecular marking method for carrying out genetic relationship and clustering analysis on citrus fruit flies
InactiveCN104087664AFast cluster analysisAccurate cluster analysisMicrobiological testing/measurementDendrogramSequence-related amplified polymorphism
The invention relates to a molecular marking method for carrying out genetic relationship and clustering analysis on citrus fruit flies and belongs to the technical field of molecular marking. The molecular marking method for carrying out genetic relationship and clustering analysis on the citrus fruit flies comprises the following steps: carrying out PCR (polymerase chain reaction) amplification on DNA of citrus fruit flies by utilizing seven RAPD (random amplification polymorphism DNA) primers and 10 pairs of SRAP (sequence-related amplified polymorphism) primer combinations, carrying out agarose gel electrophoresis on amplification products, taking pictures, recording results, and carrying out statistics on an amplification zone of a detected material; and analyzing with genetic analysis software to obtain genetic distance between individuals, and drawing a dendrogram of citrus fruit flies in different geographical populations for distinguishing different class groups. The molecular marking method for carrying out genetic relationship and clustering analysis on the citrus fruit flies has the advantages of stable reaction, clear amplified fragments and high polymorphism, a molecular reaction system for population genetic diversity of the citrus fruit flies is constructed, genetic diversity among populations can be shown, clustering analysis can be rapidly and accurately carried out on the citrus fruit flies, different populations can be distinguished, and the molecular marking method for carrying out genetic relationship and clustering analysis on the citrus fruit flies has an important significance on prevention and control of the citrus fruit flies.
Owner:CITRUS RES INST SOUTHWEST UNIV
Method for identifying common warehousing liposcelis quickly based on multiple PCR technology
InactiveCN102181563ARapid identificationImprove accuracyMicrobiological testing/measurementEnzyme digestionMale individual
The invention relates to a method for identifying common warehousing liposcelis quickly based on a multiple polymerase chain reaction (PCR) technology. The method comprises the following steps of: extracting deoxyribonucleic acid (DNA) of the genome of monomeric liposcelis or colonial liposcelis as a polymerase chain reaction (PCR) template, and designing specific primers to perform a multiple PCR reaction in a system and a program of an ordinary PCR kit, and determining liposcelis varieties by the detection of gel electrophoresis. The method is quick, efficient and high in accuracy and stability, 6 liposcelis varieties can be identified only by one PCR reaction, and regardless of colonial or monomeric liposcelis, female or male individuals, nymphae or imagoes, the reliable conclusion canbe obtained among the same geographical populations or different geographical populations; and compared with the enzyme digestion method, the method is simple, easy and economic, and general technicians can operate without complex technical steps and expensive reagents.
Owner:SOUTHWEST UNIVERSITY
Method for screening scylla paramamosain microsatellite molecular marker
InactiveCN102140449AEasy to operateAccurate operationMicrobiological testing/measurementDNA preparationSequence analysisGermplasm
The invention relates to a method for screening a scylla paramamosain microsatellite molecular marker. The method comprises the following steps of: extracting scylla paramamosain genome DNA (Deoxyribonucleic Acid) and diluting the DNA for later use; designing a 5' anchor primer and performing PCR (Polymerase Chain Reaction) amplification; cloning an amplification product and sequencing; performing sequence analysis and designing a microsatellite specific marker; performing PCR amplification on the genome DNA of different geographical populations of scylla paramamosain or individuals in the populations by using the primer; detecting a PCR amplification product by using 6 percent denaturing polyacrylamide gel electrophoresis; and determining the genotype of each individual according to different migration distances of the amplification product to obtain a polymorphic map of the genetic variance of scylla paramamosain. The method is easy to operate, is safe and has a high positive rate and a true and reliable result. By adopting the method, the polymorphic map of the genetic variance of scylla paramamosain can be obtained quickly. The method is mainly applied to genetic variance analysis of scylla paramamosain, protection and management of the germ plasm resource, establishment of a genetic linkage map and genetic breeding.
Owner:EAST CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Specific primer for amplifying COI genes of mitochondria of lepidoptera insects
ActiveCN104164433AEfficient amplificationGood sequencing profileDNA preparationDNA/RNA fragmentationCommunity evolutionForest insect
The invention relates to a primer pair for amplifying COI (cytochrome c oxidase I) genes of mitochondria of lepidoptera insects, wherein the sequence of the primer pair is shown in SEQ ID NO. 1 and SEQ ID NO. 2. By using the amplification primer disclosed by the invention, the COI genes of various lepidoptera species can be effectively amplified, a large-scale genetic background analysis is carried out on significant agriculture and forest insects in lepidoptera, some affinis species which are difficult to authenticate are accurately authenticated, and an important tool is provided for species authentication, phylogeny, population genetic structure, community evolution history and geographical population authentication for lepidoptera insects in China.
Owner:YANGZHOU UNIV
Primer special for D-loop region complete sequence sequencing of culter erythropterus and application
InactiveCN107475408AGuarantee personal safetyRapid precipitationMicrobiological testing/measurementDNA preparationCulter erythropterusComplete sequence
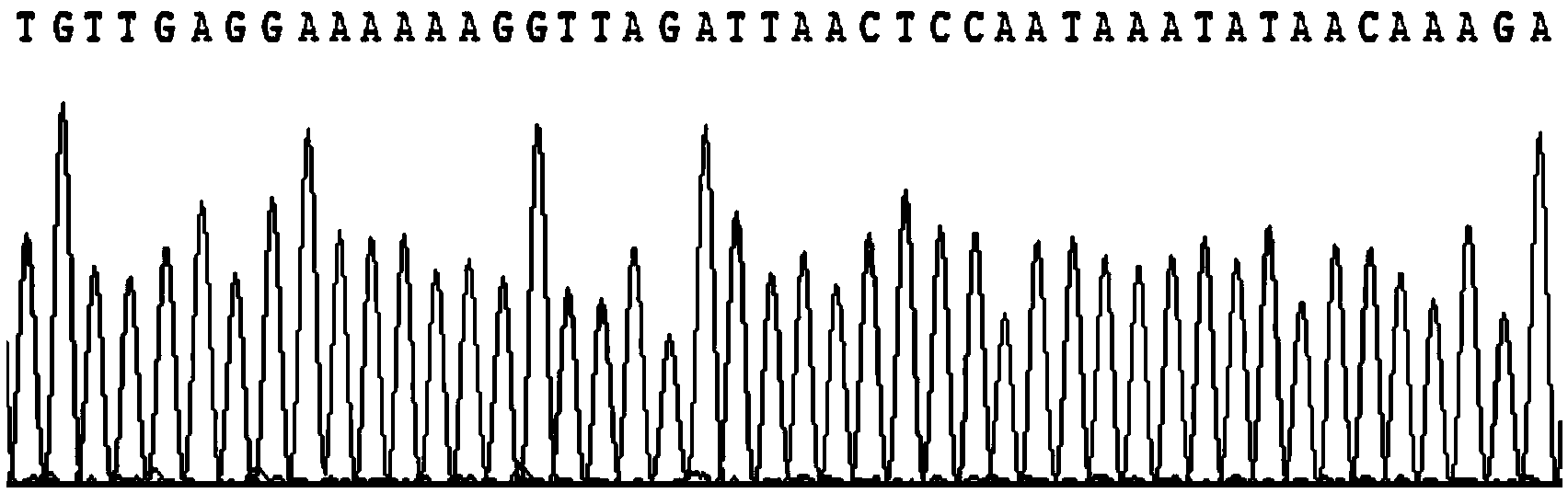
The invention relates to the field of biological genome research and specifically discloses a primer special for D-loop region complete sequence sequencing of culter erythropterus and an application. According to the invention, a degenerate primer is adopted for performing PCR amplification and then a special primer is used for sequencing. A D-loop complete sequence sequencing method provided by the invention has the advantages that cost is low, mastering is easy and the complete sequence can be acquired by once positively and negatively sequencing, so that an efficient experimental technical support is supplied for geographical population identification of culter erythropterus, research on molecular genetics and molecule system evolutionism and early resource identification and the enlightenment is supplied for the other animal experiments.
Owner:INST OF AQUATIC LIFE ACAD SINICA
Method for analyzing genetic diversity of Portunus trituberculatus by using DArT (diversity arrays technology) labelling technique
ActiveCN103710444ALow costMicrobiological testing/measurementLibrary creationGenetic diversityLabelling
The invention discloses a method for analyzing genetic diversity of Portunus trituberculatus by using a DArT (diversity arrays technology) labelling technique. The method is characterized by comprising the steps of: constructing a representative DNA (deoxyribonucleic acid) fragment library of Portunus trituberculatus genome; pointing a representative genome DNA chip and setting a series of positive and negative contrasts on the chip; respectively hybridizing DNAs of Portunus trituberculatus of different geographical populations with the representative genome DNA chip and cleaning; finally, scanning through a chip scanner after the hybridizing step, extracting data through LuxScan3.0 software, and calculating to obtain polymorphic efficiency, polymorphic information content and genetic profiles. The method has the advantages of being capable of obtaining stable and clear genetic profiles, the method can be applied in fields of genetic diversity detection of Portunus trituberculatus, wild population and family identification, creation of genetic maps and fingerprints, and the like, and the method has the advantages of being low in cost, efficient, stable and accurate.
Owner:NINGBO UNIV
Breeding method of pinctada martensi black-shell strain for sea pearl cultivation
ActiveCN103975883AIncrease productionIncreased survival rate during bead breeding periodClimate change adaptationPisciculture and aquariaReciprocal crossBase population
The invention relates to a breeding method of a pinctada martensi black-shell strain for sea pearl cultivation and belongs to the technical field of pearl breeding. The breeding method includes taking two geographical populations, namely the wild population of Lingshui of Hainan and the cultured population of Leizhou of Beibuwan as the breeding base populations, selecting high-quality individualities as crossing parents from the two populations , breeding progeny by means of reciprocal cross and selfing of different geographical populations, and selecting the progeny of the combination of 'leizhou *lingshui' with remarkable growth superiority from the progeny to perform enclosed breeding of population so as to obtain a certain amount of the pinctada martensi black-shell strain with obvious feature of black shells, good economic character and stable heritability. The breeding method has the advantages of feasible principle, simple processes and low production cost, can improve the survival rate, growth rate, pearl quality and output of the pinctada martensi greatly, and accordingly can improve sustainable development of the national sea pearl cultivation field.
Owner:GUANGDONG OCEAN UNIVERSITY
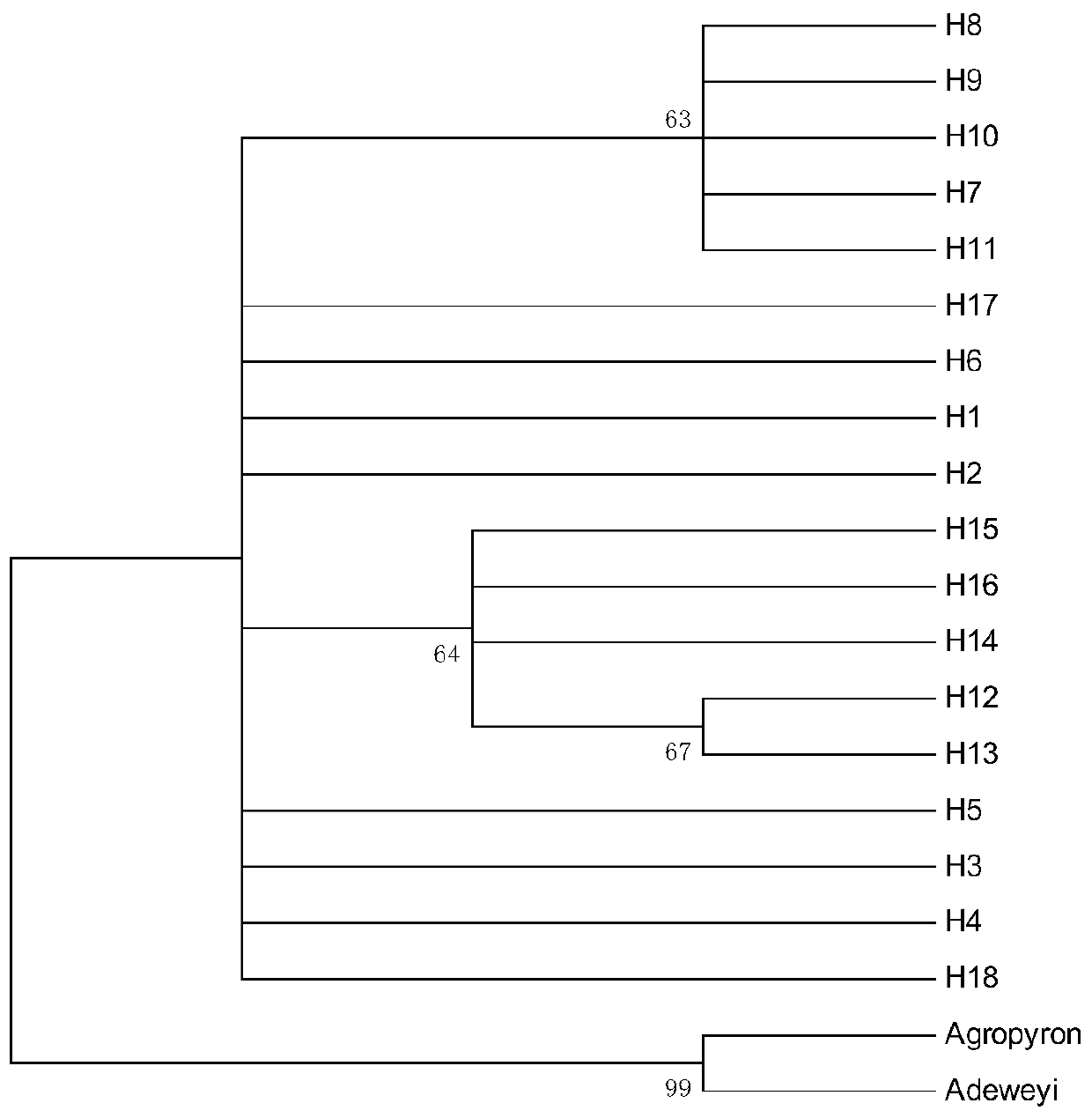
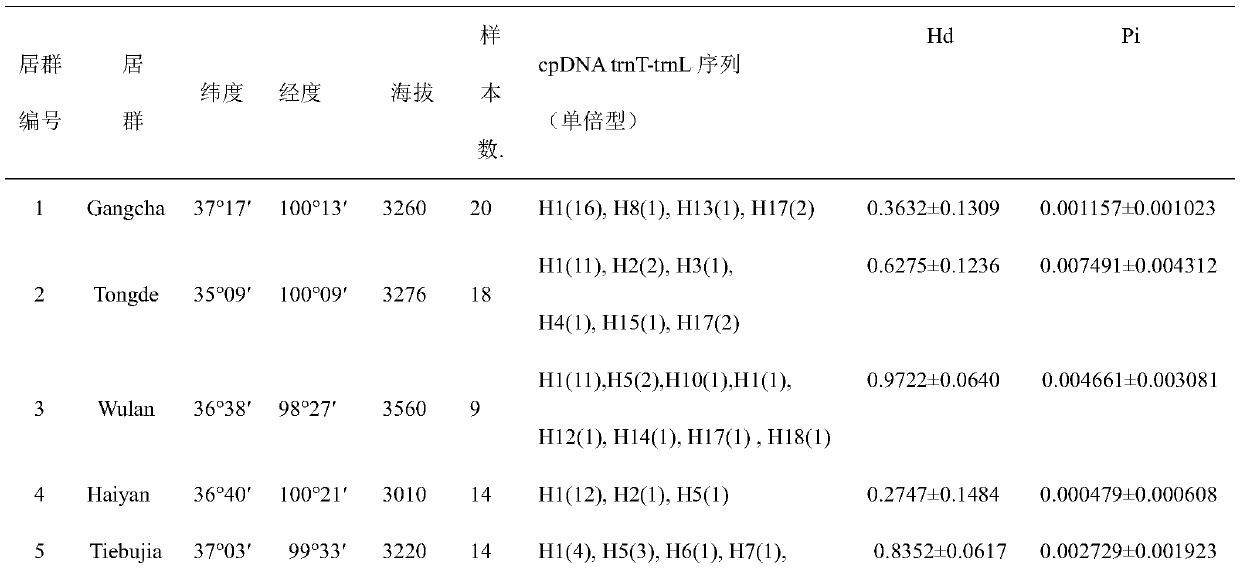
Method for researching polymorphism of different geographical populations cpDNatnT-trnL of Snowfox Crested wheatgrass
InactiveCN110060732AAccurately capture levels of genetic variationAccurately obtain population genetic informationMicrobiological testing/measurementBiostatisticsNucleotide sequencingSequence variation
The invention discloses a method for researching polymorphism of different geographical populations cpDNatnT-trnL of Snowfox Crested wheatgrass. The method includes the steps: collecting a plurality of tender leaves of the Snowfox Crested wheatgrass in a wheatgrass population; extracting the total DNA of each tender leaf of the Snowfox Crested wheatgrass; amplifying the trnT-trnL fragment of the cpDNA of each tender leaf of the Snowfox Crested wheatgrass to obtain a plurality of amplification products; carrying out sequencing to obtain a nucleotide sequence of a trnT-trnL amplified fragment ofthe cpDNA of each tender leaf of the Snowfox Crested wheatgrass; analyzing the sequence data by adopting software; and according to data analysis, obtaining cpDNAtrnT-trnL polymorphism information ofthe Snowfox Crested wheatgrass in different geographical populations, including sequence variation, genetic diversity, genetic structure of populations and phylogenetic relationship information amongthe populations. The method for researching polymorphism of different geographical populations cpDNatnT-trnL of Snowfox Crested wheatgrass can accurately obtain the polymorphism information of different geographical populations of the Snowfox Crested wheatgrass, and can provide a scientific basis for species protection.
Owner:青海省铁卜加草原改良试验站
Amplification primer of cardiidae shellfish mitochondria 16S rRNA (16small-subunit ribosomal RNA) gene
InactiveCN103103186AMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityGermplasm
According to the invention, 53 cardiidae 16S rRNA (16small-subunit ribosomal RNA) sequences amplified by the use of a universal primer are subjected to comparative analysis through CLUSTALW software, 3 pairs of primers are designed by primer designing software Primer Premier5 in a determined conserved region, 218 individuals of 24 species of cardiidae are repeatedly subjected to PCR reaction test in 10mu l system, and finally a specific amplification primer of cardiidae shellfish mitochondria 16S rRNA gene sequence is screened out through agarose gel electrophoresis detection. The amplification primer provided by the invention is characterized by NG16Sa5'-GCCTGTTTATCAAAAACATCTC-3'NG16Sb5'-TCTGAACTCAGATCACGTAAGGAT-3'. By adopting the cardiidae shellfish mitochondria 16S rRNA primer provided by the invention, the target fragments of different species of cardiidae shellfish can be quickly and efficiently amplified, and the amplification efficiency reaches 100%, thereby providing support to the identification of cardiidae species, identification of geographical population and research on germplasm resource and genetic diversity and laying a foundation for further developing artificial breeding of various important economic shellfishes of cardiidae.
Owner:OCEAN UNIV OF CHINA
Method for detecting Jassr131 microsatellite deoxyribonucleic acid (DNA) marker of Charybdis japonica
InactiveCN102162009AEasy to detectSimple methodMicrobiological testing/measurementCharybdis japonicaGenomic DNA
The invention discloses a method for detecting a Jassr131 microsatellite deoxyribonucleic acid (DNA) marker of Charybdis japonica. The method is characterized by comprising the following steps of: extracting genomic DNA of the Charybdis japonica and diluting for later use; designing a specific primer at both ends of a Jassr131 microsatellite core sequence in an enriched microsatellite library of the Charybdis japonica; performing polymerase chain reaction (PCR) amplification on genomic DNA of different geographical populations of the Charybdis japonica or genomic DNA of individuals in the populations by using the primer, and detecting a PCR product by using metamorphic polyacrylamide gel; and analyzing by utilizing stripes of the product, and determining a genotype of each individual so as to obtain a polymorphic map of high genetic variation of the Charybdis japonica in a Jassr131 core sequence region. Sequences of the specific primer of the Jassr131 microsatellite core sequence are that: the sequence of a plus strand is 5'-CCAGGGAATTGAAACACT-3', and the sequence of a minus strand is 3'-TATGAAGGCTCTGCGAAA-5'; and the annealing temperature is 50 DEG C. The method is simple and convenient; and the genotype of each individual of the Charybdis japonica at the site can be intuitively detected according to the obtained results.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Long-term safe breeding method for lateolabrax japonicus juvenile fish
InactiveCN108834955AHigh alkalinity toleranceClimate change adaptationPisciculture and aquariaJuvenile fishEuryhaline
The invention discloses a long-term safe breeding method for lateolabrax japonicus juvenile fish. Initial lateolabrax japonicus juvenile fish bodies with the mass of 14.11+ / -0.45 g in the early stageare selected, and the safe alkalinity of long-term breeding is 10 mmol / L. The method has the advantages that the setting of the safe alkalinity of the lateolabrax japonicus juvenile fish is firstly opened to the public according to the long-term breeding experimental result, the alkalinity is 10 mmol / L, early alkalinity adaptation can enhance the higher alkalinitytolerance of the lateolabrax japonicus juvenile fish, in actual production, data support is provided for the dividing of south and north groups of lateolabrax japonicas, and technical support is provided for salt and alkali water areapopularization and geographical population identification of early lateolabrax japonicas fry.
Owner:OCEAN UNIV OF CHINA +1
Method for synchronous development control of scapharca broughtonii in different geographical populations
InactiveCN106508759AIncrease food intakeSimultaneous maturity ensuresClimate change adaptationPisciculture and aquariaMicroscopic examAccessory gonad
The invention relates to a method for synchronous development control of scapharca broughtonii in different geographical populations. The method is characterized by including steps: acquiring scapharca broughtonii parents in geographical populations A and B, temporarily culturing in the sea, heating indoors for maturity promotion, and performing dissection and microscopic examination in a process of heating for maturity promotion to control synchronous development of scapharca broughtonii in different geographical populations. By collection for temporary culturing and heating for maturity promotion of the scapharca broughtonii parents in the different geographical populations and by temperature and bait regulation in the process of heating for maturity promotion, synchronous gonadal maturity of the scapharca broughtonii parents in the different geographical populations is guaranteed while spermiation and ovulation are realized, feasibility and easiness in operation of cross breeding are achieved, scapharca broughtonii hybridization breeding is technically supported, and development of the shellfish industry is promoted.
Owner:中国水产科学研究院长岛增殖实验站
Method for analyzing corn borer population genetic diversity by ISSR system
ActiveCN109576377AAmplification reaction is stableAmplified fragments are clearMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityGenetic architecture
The invention provides a method for analyzing the corn borer population genetic diversity by an ISSR system, and relates to a method for analyzing the corn borer population genetic diversity. The invention aims at solving the problems that in the prior art, no effective molecular marking method, particularly an ISSR molecular marking method is used for identifying the corn borer, particularly thegenetic structure and the affinity relationship of different geographical populations of asiatic corn borer, so that the migration change condition of the corn borer is specified. The invention provides the method for analyzing the corn borer population genetic diversity by the ISSR system. The method is a simple and easy-to-operate molecular marking technology; for 14 ISSR primer sequences selected by the method, the PCR amplification reaction in the corn borer is stable; the amplified fragments are clear; the polymorphism is high; the technical support is provided for scientific research such as corn borer resource identification and genetic diversity analysis. The method is applied to the field of the corn borer.
Owner:黑龙江省植检植保站
REMAP marker for evaluating genetic diversity of cyprinus carpio population
ActiveCN104498488AImprove identification efficiencyMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityGenetics
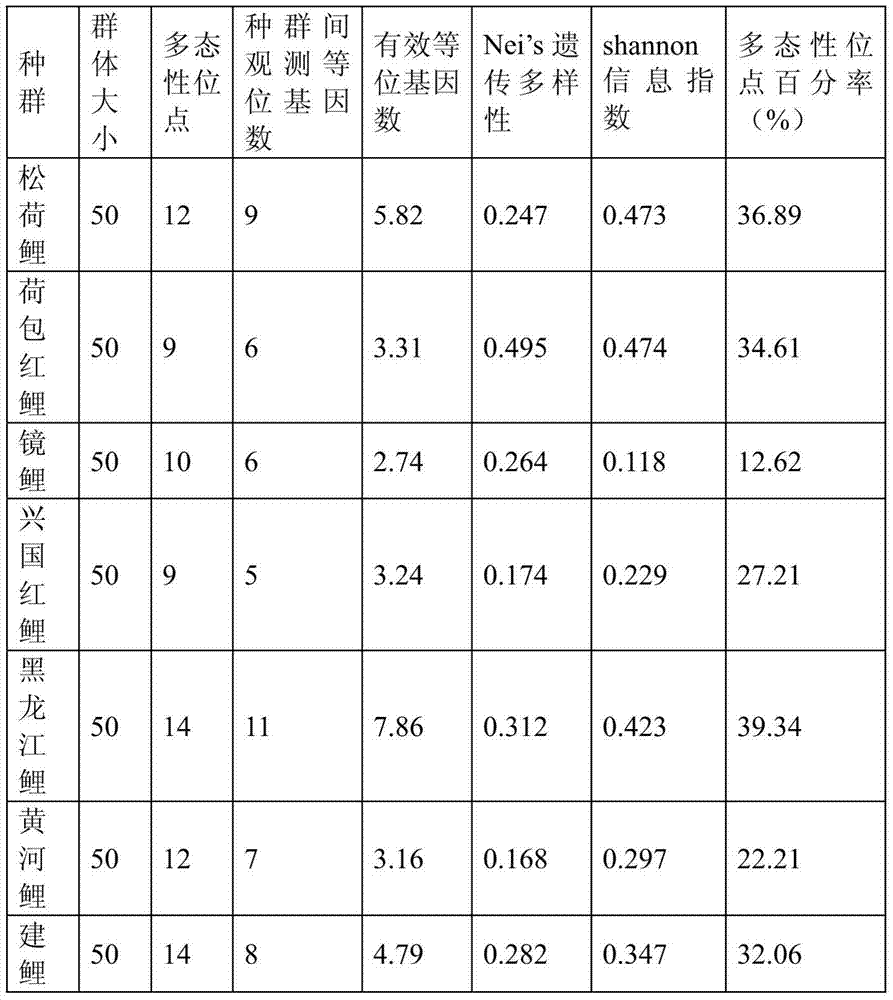
The invention discloses a cyprinus carpio retrotransposon marker. A high-polymorphism retrotransposon REMAP marker of cyprinus carpio is developed by predicting and identifying a cyprinus carpio retrotransposon sequence, screening a cyprinus carpio polymorphism REMAP marker and performing genetic polymorphism detection on a cyprinus carpio retrotransposon locus. The cyprinus carpio retrotransposon marker disclosed by the invention can be applied to the research on the population genetic structure, genetic variation level and the like of the cyprinus carpio population and the germplasm identification of the geographical population of cyprinus carpio.
Owner:CHINESE ACAD OF FISHERY SCI
Method suitable for hybridization of different geographical populations of escargots
ActiveCN109699539APromote growth and reproductionMeet growth requirementsClimate change adaptationPisciculture and aquariaNutritionEnvironment of Albania
The invention discloses a method suitable for hybridization of different geographical populations of escargots. The method mainly includes the steps of net cage preparation, isolated breeding, reserveparent selecting, pairing hybridization and the like. According to the method, the escargots with different geographical populations, different genders and different hybrid varieties are isolated indifferent net cages to be bred, the breeding water temperature and the dissolved oxygen amount are independently controlled within a proper range according to the respective growth characteristics ofthe escargots, the growth requirements of the escargots with different varieties are met, thus, hybridization of different geographical populations of the escargots can be accurately established, thesteps are simple, operation and popularization are easy, meanwhile, demersal bait biological films suitable for escargot breeding are cultured, high-nutrition bait can be provided for the escargots, and environmental pollution cannot be caused.
Owner:GUANGXI ACADEMY OF FISHERY SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com