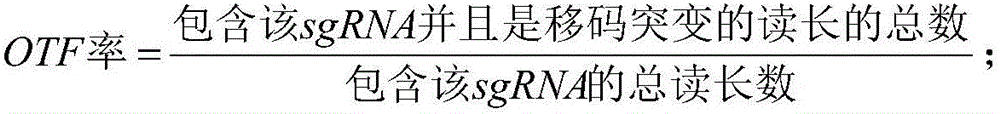Patents
Literature
2559 results about "Enzyme digestion" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Digestive enzymes are proteins that break down larger molecules like fats, proteins and carbs into smaller molecules that are easier to absorb across the small intestine. Without sufficient digestive enzymes, the body is unable to digest food particles properly, which may lead to food intolerances.
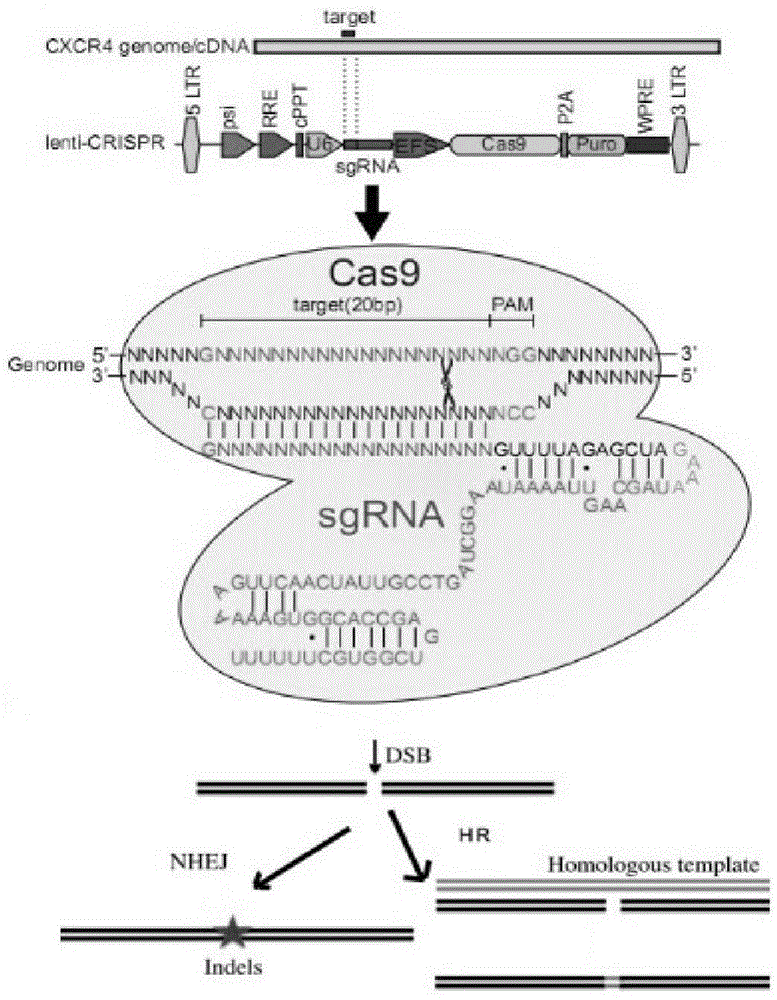
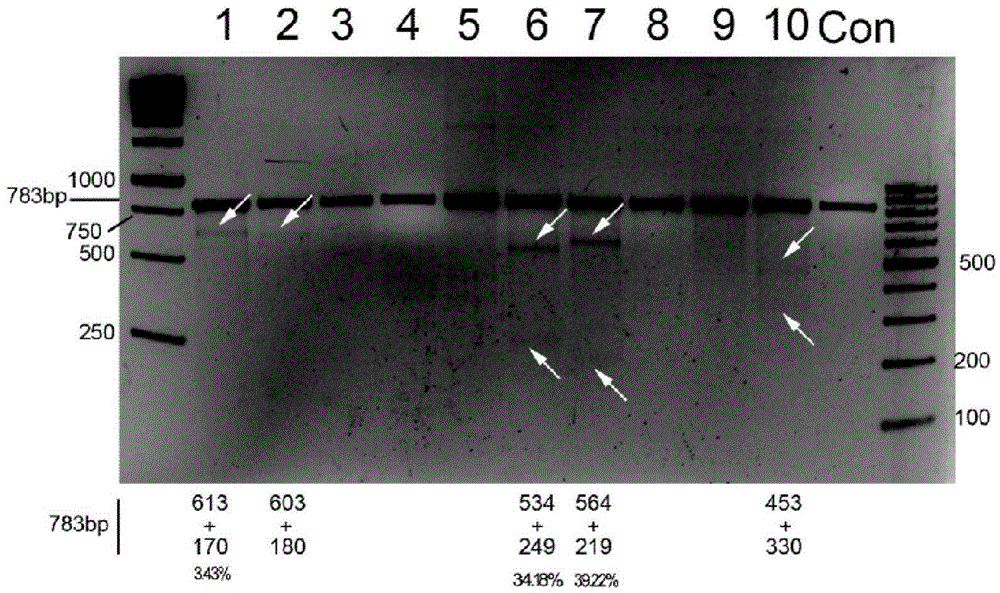
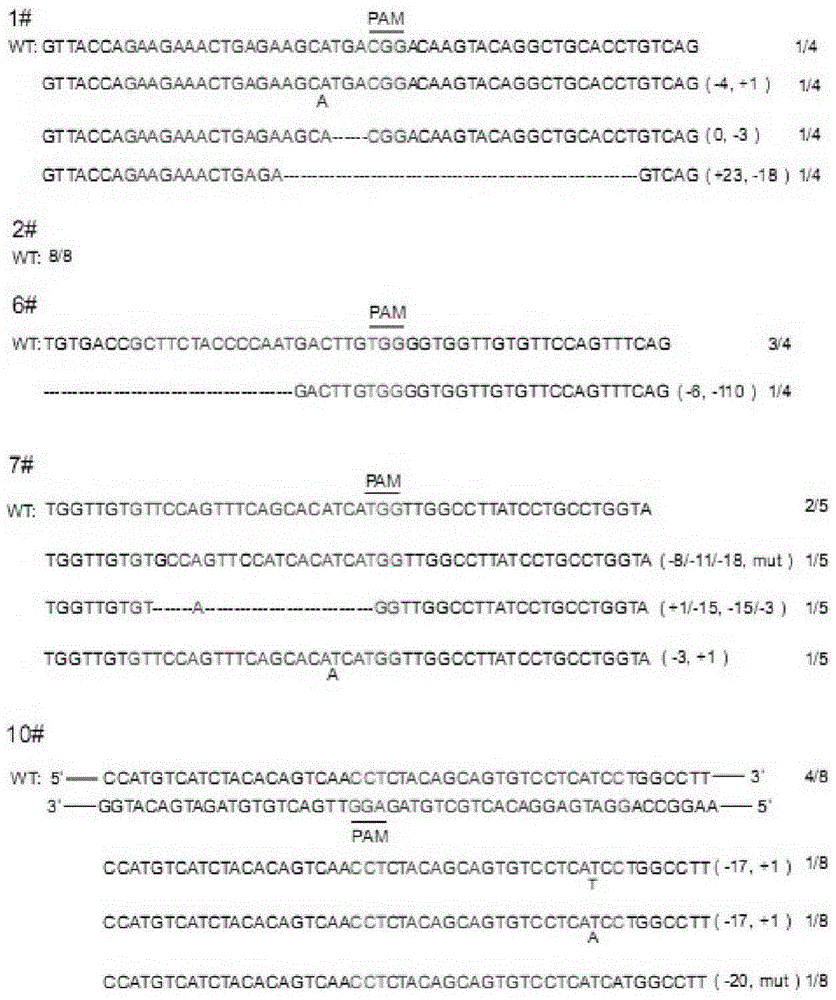
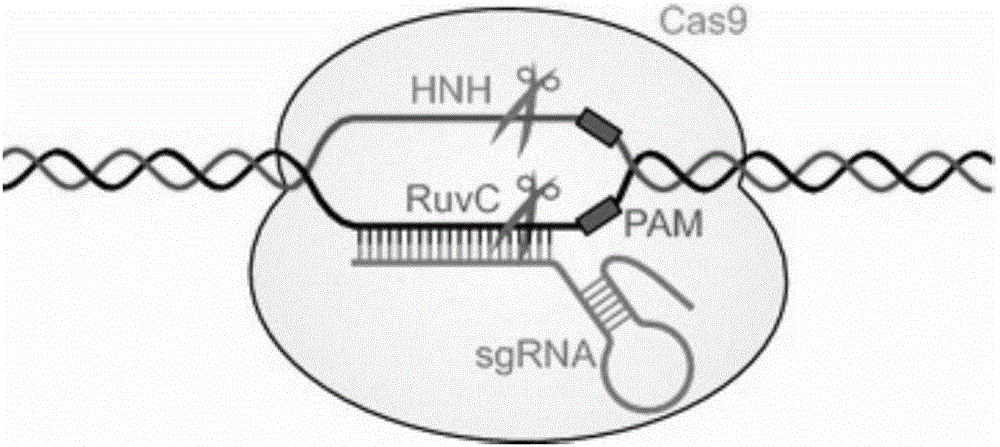
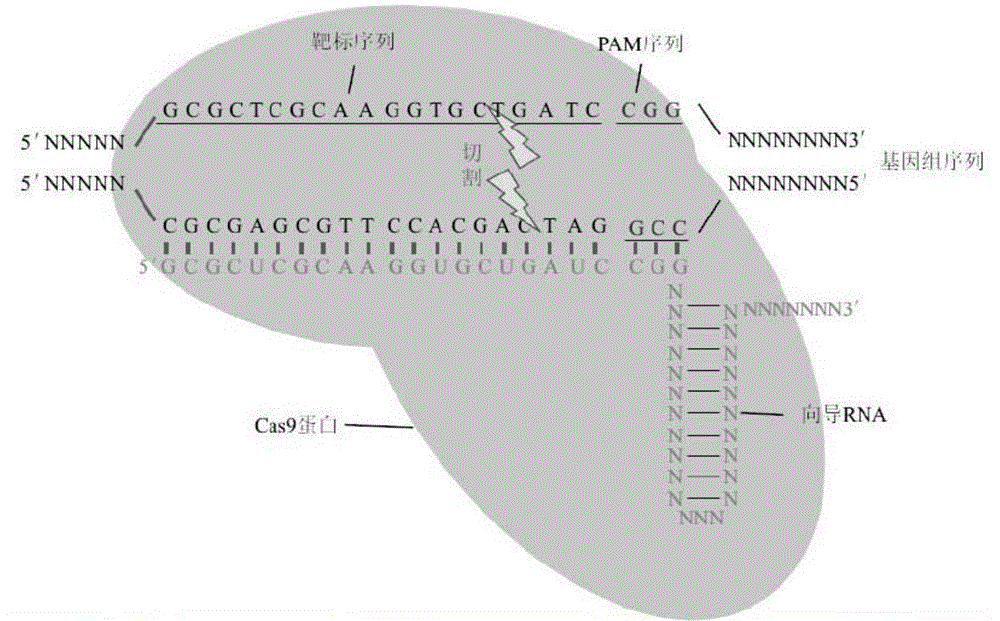
CRISPR/Cas9 recombinant lentiviral vector for human immunodeficiency virus gene therapy and lentivirus of CRISPR/Cas9 recombinant lentiviral vector
ActiveCN104480144AAvoid or delay intrusionInhibit the spread of infectionGenetic material ingredientsAntiviralsEnzyme digestionCXCR4
The invention belongs to the field of pharmaceutical and biological engineering, and relates to a CRISPR / Cas9 recombinant lentiviral vector for human immunodeficiency virus gene therapy and a lentivirus of the CRISPR / Cas9 recombinant lentiviral vector. The recombinant lentiviral vector is prepared by carrying out enzyme digestion on a lentiviral vectorlentiCRISPR by BsmBI and connecting into a BsmBI cohesive end-containing CXCR4 specific target sequence to recombine; the obtained CRISPR / Cas9 recombinant lentiviral vector is capable of mutating gene sequences at four different loci of ahuman immunodeficiency virusco-receptor CXCR4 and themutatuin rate is high and up to 25-75%. The cells transformed by the recombinant lentiviral vector cannot be infected by the human immunodeficiency virus. Compared with theRNAi-Knockdown, ZFN and TALEN technologies, the method has higher efficiency of suppressing the human immunodeficiency virus replication; the system is rapid to construct, simple and low in cost, is capable of preventing the invasion of the human immunodeficiency virus and is suitable for human immunodeficiency virus gene therapy.
Owner:WUHAN UNIV
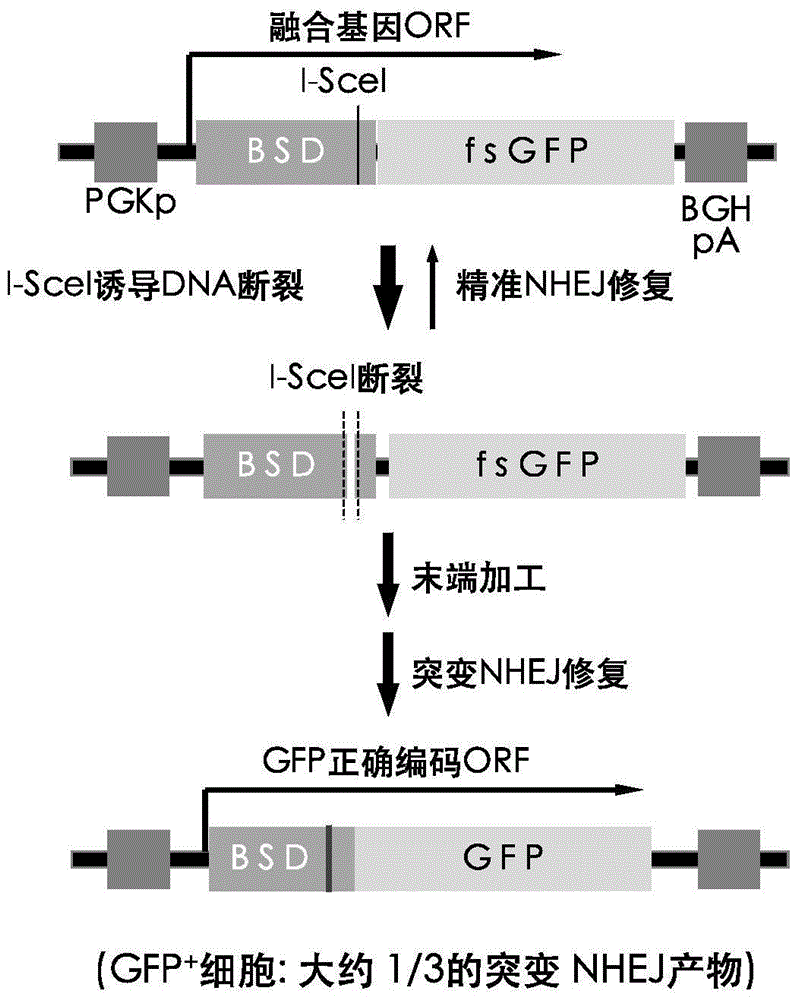
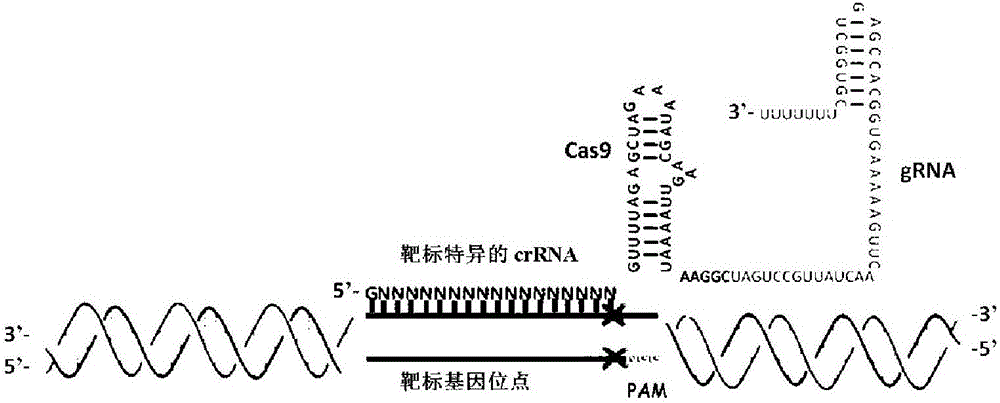
Fast CRISPR-Cas9 working efficiency testing system and application thereof
InactiveCN105647968ATest accurateTested and reliableMicrobiological testing/measurementPeptidesEnzyme digestionRapid testing
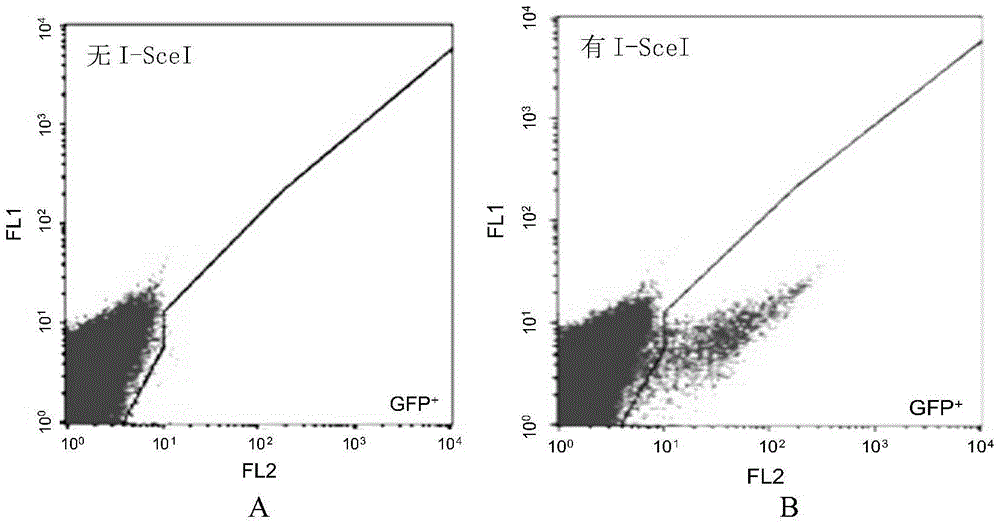
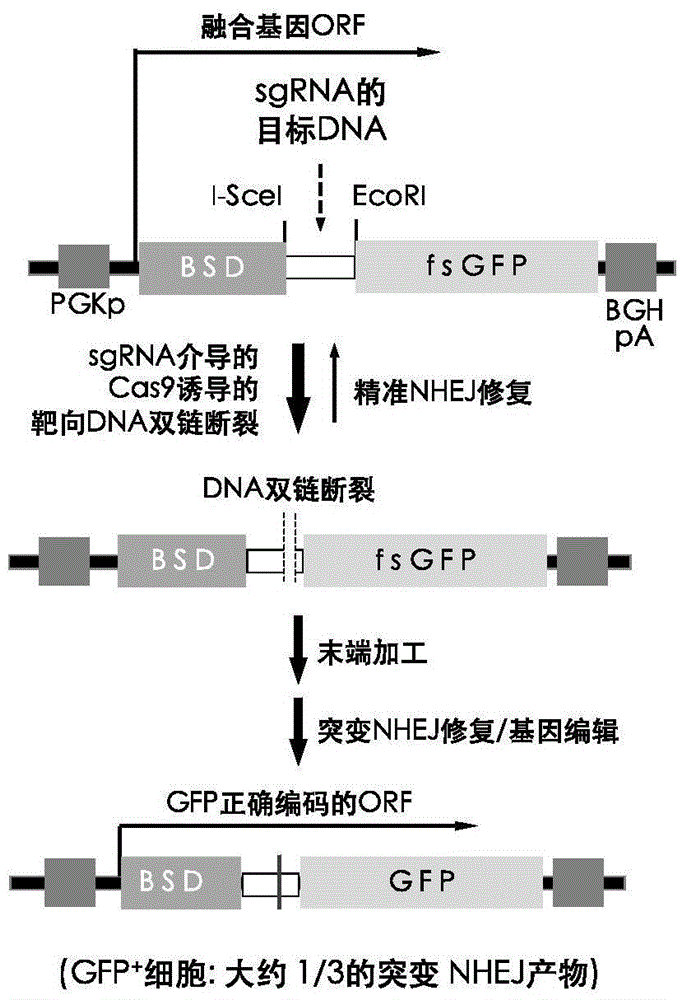
The invention discloses a fast CRISPR-Cas9 working efficiency testing system and application thereof. The testing system comprises a plasmid used for expressing sg RNA, a plasmid used for expressing Cas9 and a reporting system used for testing the CRISPR-Cas9 gene editing efficiency; the reporting system can splice the C-terminal of a nucleotide segment capable of coding effective protein and the N-terminal of a reporter gene and insert two restriction endonuclease enzyme digestion sites into the splicing position; before a special gene is edited (knocked out) through the CRISPR-Cas9 system, selection of a target sequence is vital, and the selection can influence the recognition efficiency of the sg RNA to target DNA, the binding efficiency of the sg RNA with the target DNA, the targeted cutting efficiency of the Cas9 and the NHEJ repairing efficiency. According to the system, the gene editing efficiency of different sgRNA-target DNA sequences can be quantitatively compared, the sgRNA with the best working effect can be determined within a short time, the actual knockout success rate is increased, therefore, the working cost can be lowered, the working efficiency can be improved, and the working progress can be promoted.
Owner:ZHEJIANG UNIV
Circular chromosome conformation capture (4C)
ActiveUS8642295B2High throughput analysisAccurate mappingSugar derivativesMicrobiological testing/measurementEnzyme digestionRestriction enzyme digestion
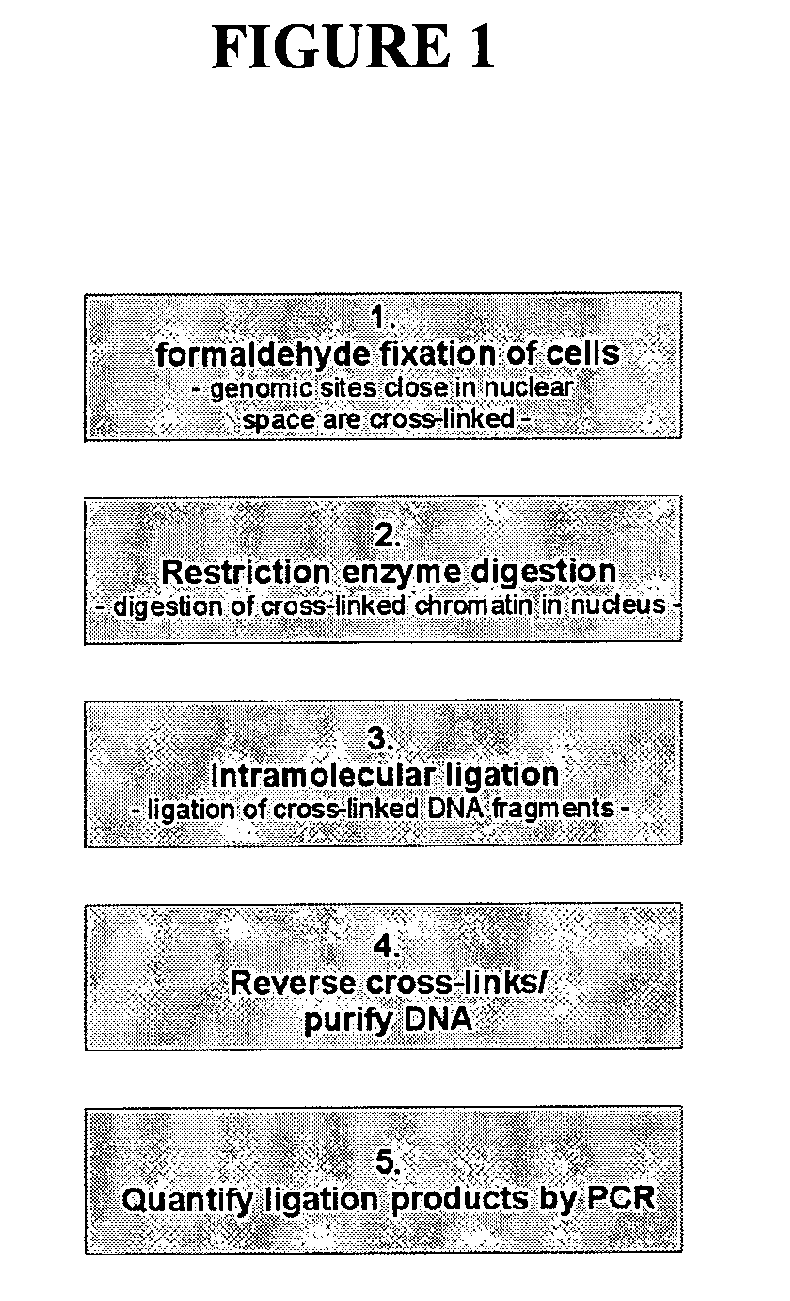
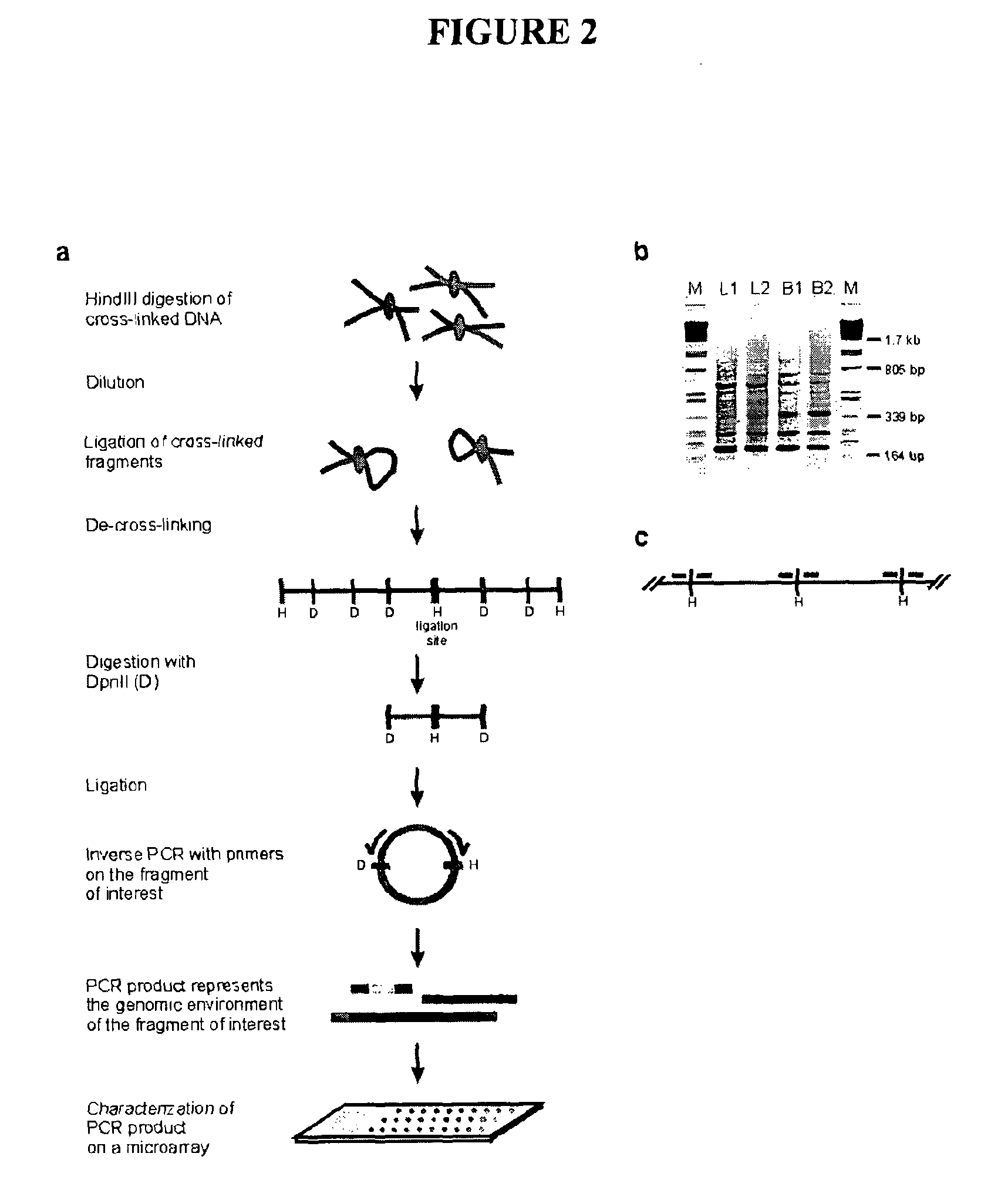
The present invention relates in one aspect to a method for analyzing the frequency of interaction of a target nucleotide sequence with one or more nucleotide sequences of interest (eg. one or more genomic loci) comprising the steps of: (a) providing a sample of cross-linked DNA; (b) digesting the cross-linked DNA with a primary restriction enzyme; (c) ligating the cross-linked nucleotide sequences; (d) reversing the cross linking; (e) optionally digesting the nucleotide sequences with a secondary restriction enzyme; (f) optionally ligating one or more DNA sequences of known nucleotide composition to the available secondary restriction enzyme digestion site(s) that flank the one or more nucleotide sequences of interest; (g) amplifying the one or more nucleotide sequences of interest using at least two oligonucleotide primers, wherein each primer hybridises to the DNA sequences that flank the nucleotide sequences of interest; (h) hybridising the amplified sequence(s) to an array; and (i) determining the frequency of interaction between the DNA sequences.
Owner:ERASMUS UNIV MEDICAL CENT ROTTERDAM ERASMUS MC
CRISPR/Cas9 system-containing targeted knockout vector and adenovirus and applications thereof
InactiveCN104004778AHigh knockout rateGenetic material ingredientsViruses/bacteriophagesBiotechnologyEnzyme digestion
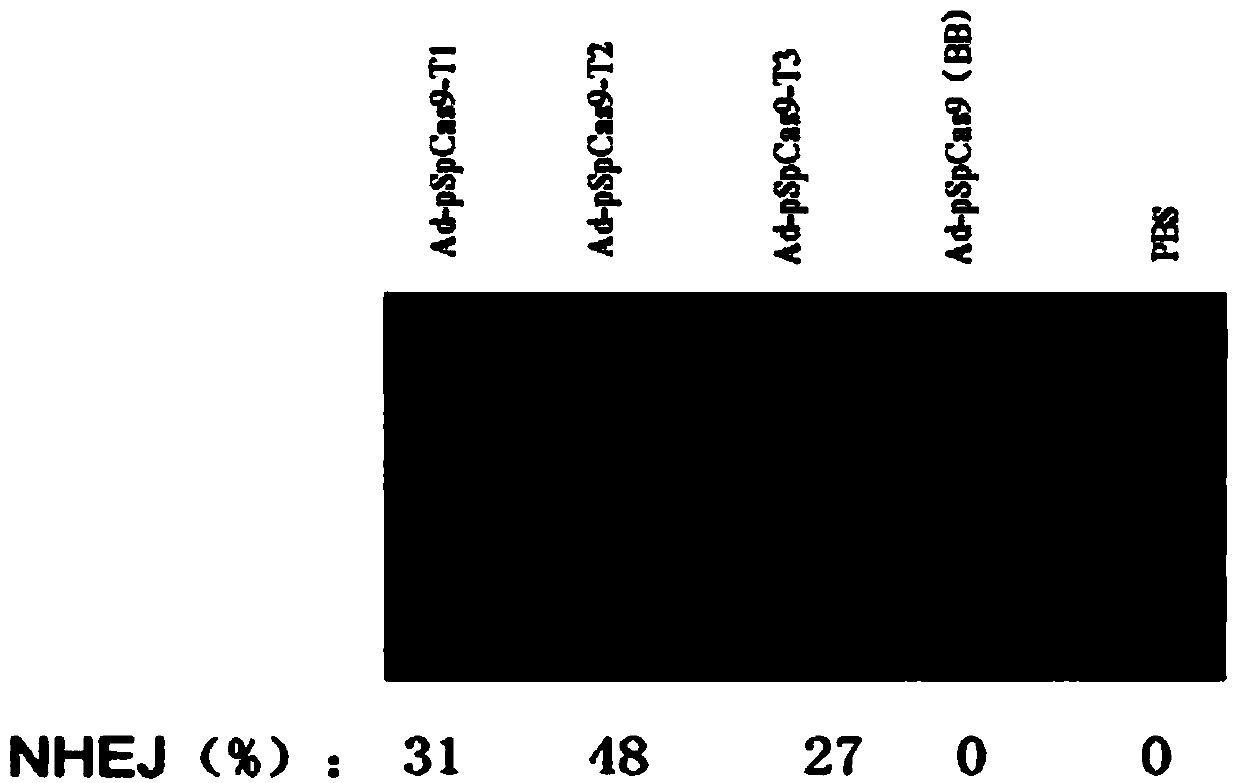
The invention discloses a CRISPR / Cas9 system-containing targeted knockout vector and adenovirus and applications thereof. The targeted knockout vector is prepared through the following steps: after a pX330 U6-Chimeric_BB-CBh-hSpCas9 plasmid is subjected to enzyme digestion and filling-in by using EcoRI and SacII, connecting the pX330 U6-Chimeric_BB-CBh-hSpCas9 plasmid with a pAdTrack-CMV plasmid subjected to enzyme digestion and filling-in by using BstXI; after the obtained product is linearized by using BbsI, connecting the obtained product to a specific target sequence of a desired gene; and after the obtained object is linearized by using PmeI and dephosphorylated by using CIAP alkaline phosphatase, recombining the obtained product with a pAdEasy-1 plasmid. The targeted knockout vector can mutate gene sequences in target sequence areas, and the mutation rate is high, up to 30.6-45.8%, therefore, the targeted knockout vector can be used for gene site-directed mutation, and lays a foundation for gene therapy.
Owner:重庆高圣生物医药有限责任公司
Gene editing method for knocking out rice MIRNA393b stem-loop sequences with application of CRISPR(clustered regulatory interspersed short palindromic repeat)-Cas9 system
InactiveCN105647962AKnockout worksNucleic acid vectorVector-based foreign material introductionEscherichia coliEnzyme digestion
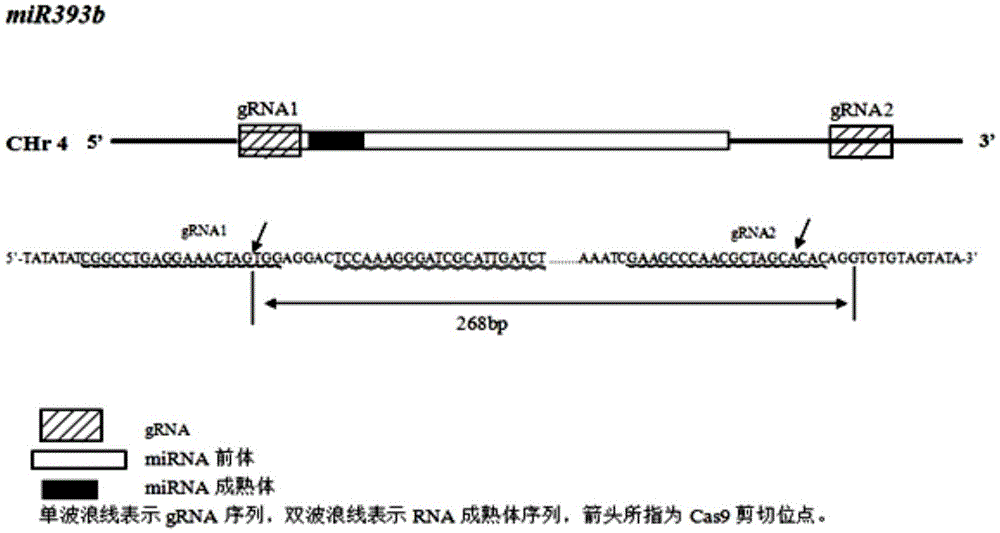
The invention relates to construction of rice transgenic materials and aims to provide a gene editing method for knocking out rice MIRNA393b stem-loop sequences with application of a CRISPR(clustered regulatory interspersed short palindromic repeat)-Cas9 system. The gene editing method comprises steps as follows: gRNA target sites are selected for cloning and GG linking, enzyme digestion is performed after amplification, and a product is linked with a pGREB 32 vector; escherichia coli competent cells are transformed; plasmids with a correct sequencing result are used for transforming agrobacteria, transgenic plants are obtained through mediated transformation of rice calli, and transgenic positive lines are obtained; the T0-generation mutant plant seeds are collected for seeding, and the T1-generation plants are subjected to homozygote screening; homozygous lines which are discovered to be negative through MIRNA393b expression are rice mutants completely losing the MIRNA393b stem-loop sequences and MIRNA393b stem-loop sequence expression. According to the gene editing method, MIRNA stem-loop sequences can be effectively knocked out, and loss-of-function mutants of different members in the same MIRNA family can be prepared; the mutant plant propagates to obtain a large number of seeds and is an ideal material for acquiring rice MIRNA393b gene functions successfully.
Owner:ZHEJIANG UNIV
CRISPR Cas9 system system for Bacillus subtilis genome edition and establishment method thereof
ActiveCN105671070AReduce the chance of infectionReduce chancePeptidesNucleic acid vectorEscherichia coliEnzyme digestion
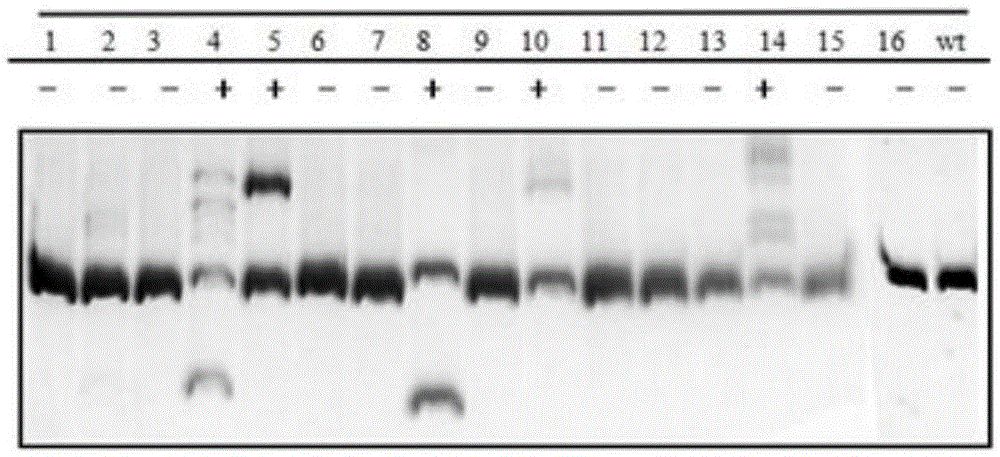
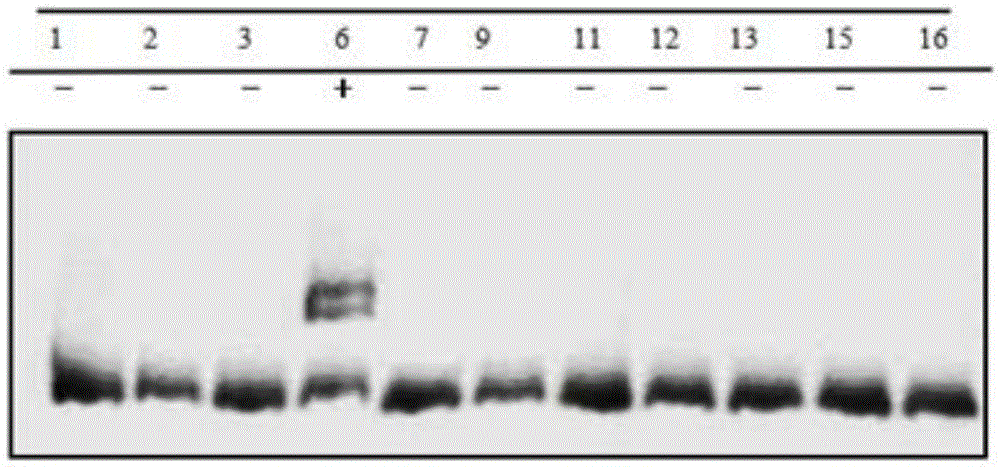
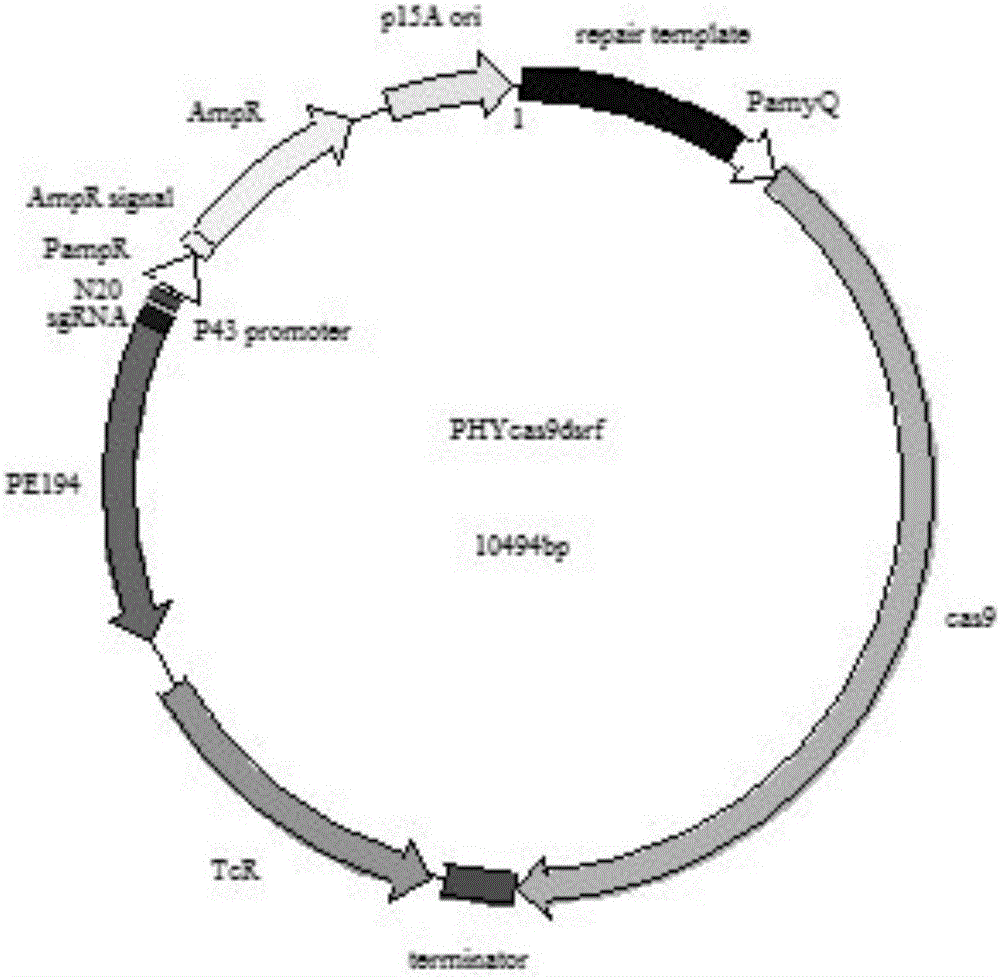
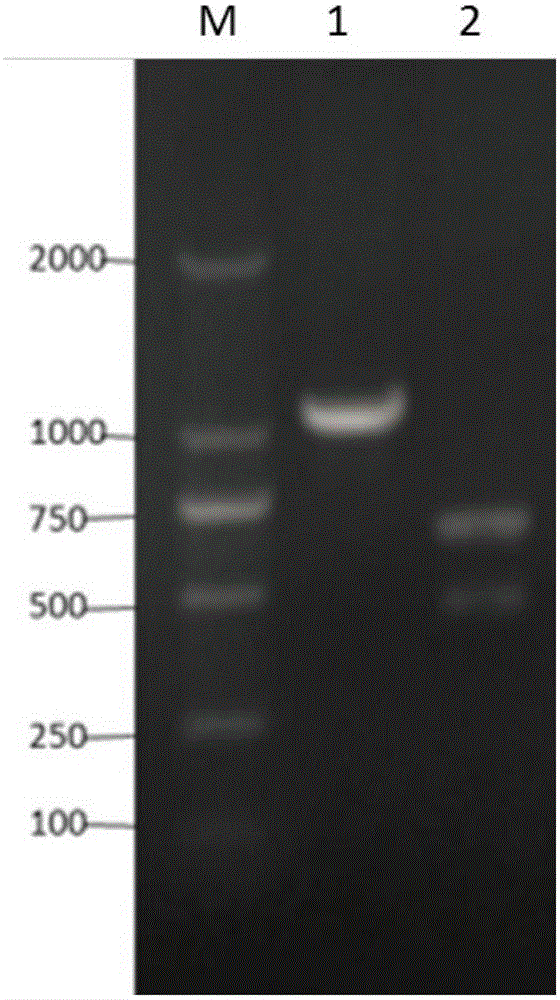
The invention discloses a CRISPR Cas9 system for Bacillus subtilis genome edition and an establishment method thereof, belonging to the technical field of gene engineering. A knock-out plasmid PHY300dsrf is established; and the plasmid comprises sgRNA and cas9 genes of the specific targeted target gene, a homologous restoration arm, and replication initial points and tolerance screening markers of Escherichia coli and Bacillus subtilis. The sgRNA of the specific targeted srfA-C gene can guide the cas9 protein to cut double strands at the specific site of the srfA-C gene, and can be used for accurate homologous recombination on the genome under the guide of the homologous restoration arm, thereby introducing the XhoI enzyme digestion site at the rupture. When the tank feed fermentation culture is carried out on the Bacillus subtilis of which the srfA-C gene is successfully knocked out, the foam is obviously reduced, which proves that the CRISPR / Cas9 gene editing system can effectively perform gene edition on the Bacillus subtilis genome.
Owner:JIANGNAN UNIV +1
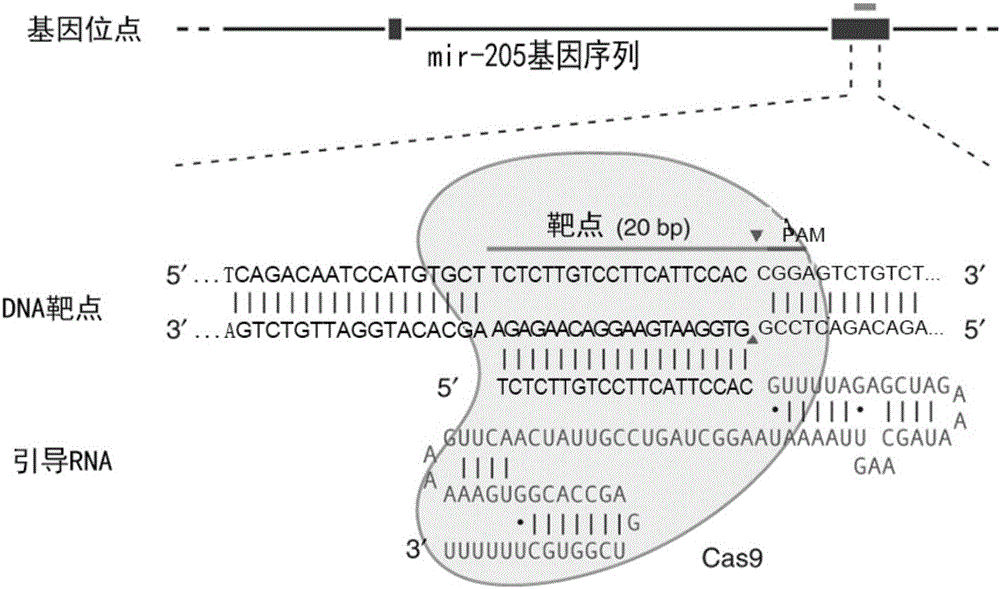
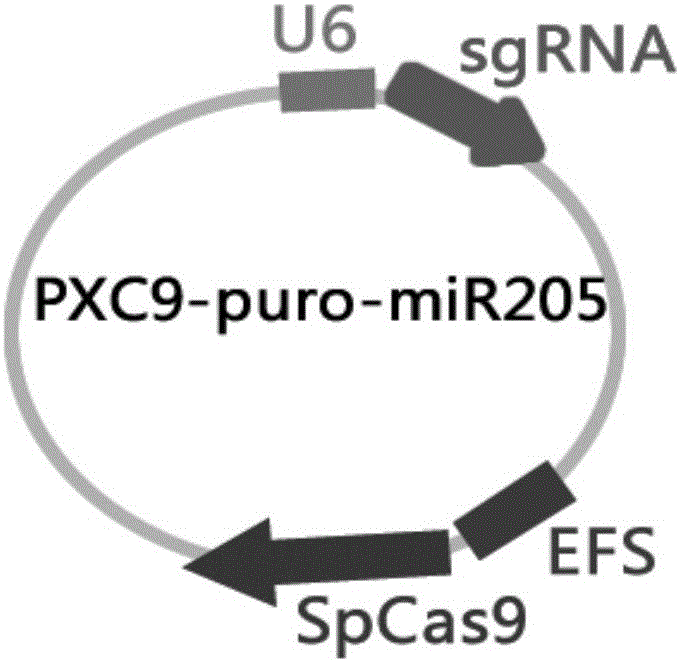
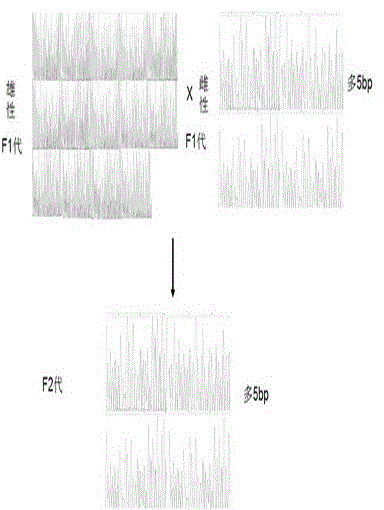
MiR-205 gene knockout kit based on CRISPR-Cas9 gene knockout technology
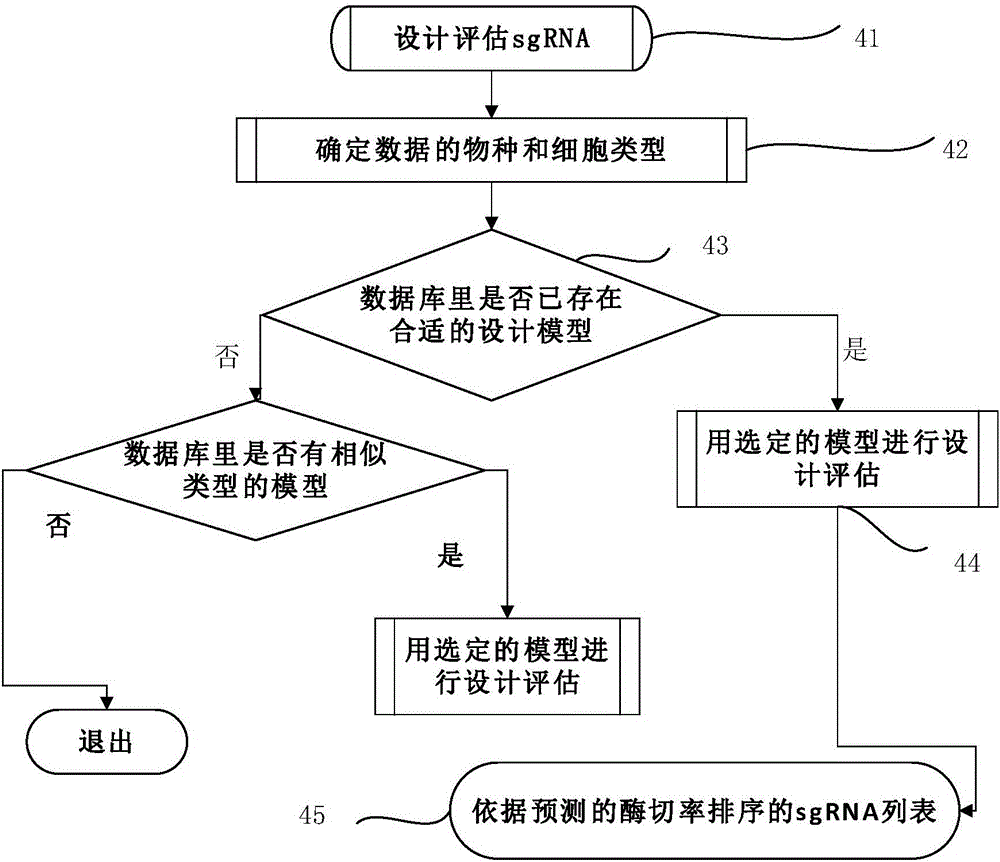
ActiveCN105112445AVector-based foreign material introductionDNA/RNA fragmentationEnzyme digestionFluorescence
The invention discloses a miR-205 gene knockout kit based on CRISPR-Cas9 gene knockout technology. According to the invention, an optimal CRISPR-Cas9 target sequence of a certain amount of miR-205 is obtained through target design software and an sgRNA single chain is synthesized in vitro; an insertion fragment is obtained through processing; then sgRNA is inoculated into a plasmid vector by using T4 ligase; and a miR-205 gene knockout cell strain is obtained through transfection of an LNCap cell strain, continuous drug screening and fluorescence detection. Heterogenous hybridization double strands are obtained by extracting DNAs of a cell, PCR amplification of miR-205, purification, denaturation of a PCR product and annealing; T7E1 enzyme digestion test is employed to determining shearing efficiency of a CRISPR-Cas9 system on miR-205; a verified optimized a miR-205 gene knockout CRISPR-Cas9 target sequence is obtained; and the kit is constructed on the basis of the target sequence and can be used for directional knockout of miR-205 genes. The kit has the characteristics of high gene knockout efficiency, fast speed, easiness and economic performance and has wide prospects in the aspects of construction of animal models and clinical research of medical science.
Owner:广州辉园苑医药科技有限公司
Method for obtaining bmp2a gene knocked-out zebra fish through CRISPR/Cas9
The invention discloses a method for obtaining bmp2a gene knocked-out zebra fish through CRISPR / Cas9. A novel gRNA sequence is designed between a first exon and an intron of BMP2a, the gRNA sequence is GGAGCCCATCACTAGACTCTTGG, and enzyme digestion is HinfI. Compared with a traditional gene knockout technology, the CRISPR / Cas9 technology has the advantages of low toxicity, high accuracy, high efficiency, short success cycle and the like. Therefore, the BMP2a gene can be theoretically knocked out faster.
Owner:WUXI NO 2 PEOPLES HOSPITAL
CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-Cas9) homing sequences and primers thereof, and transgenic expression vector and establishment method thereof
ActiveCN105907758AEasy and fast to buildHigh efficiency of editing functionNucleic acid vectorFermentationEnzyme digestionNucleotide
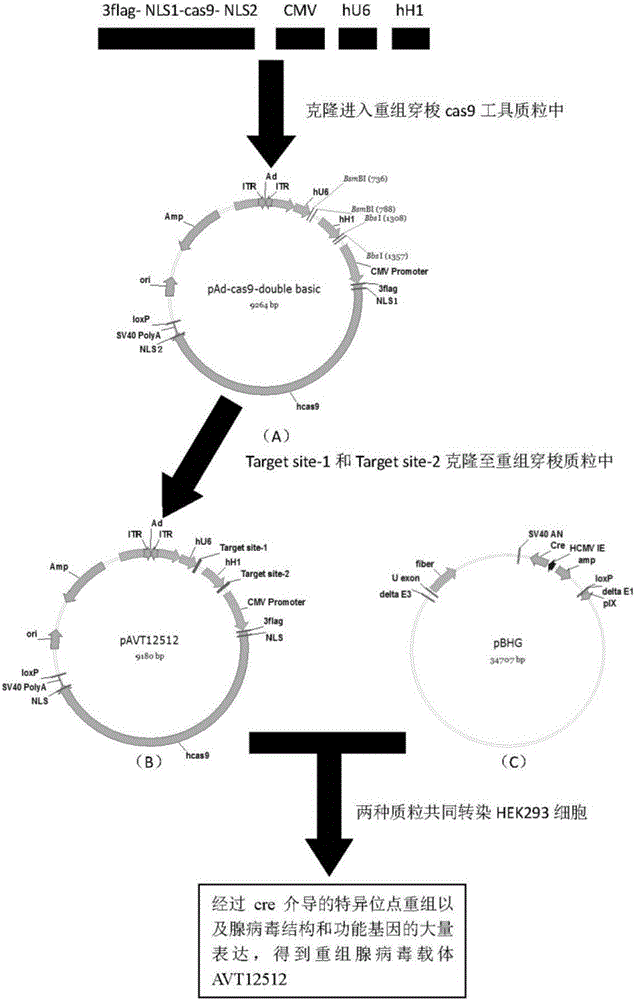
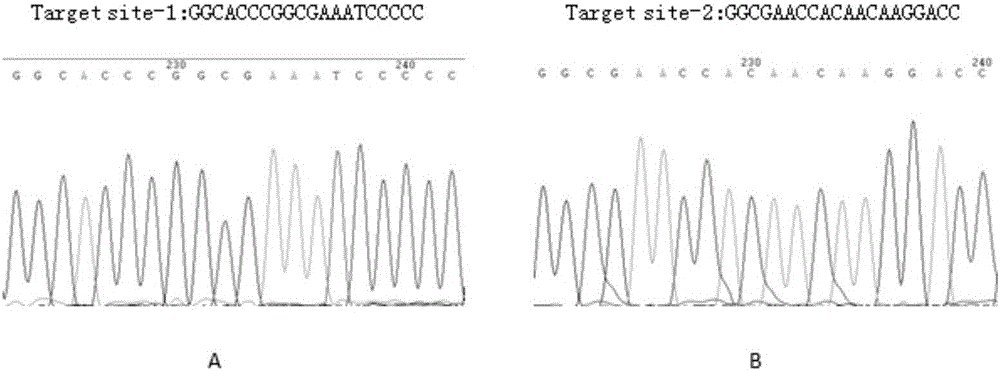
The invention belongs to the technical field of biomedicine, and particularly relates to CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-Cas9) homing sequences and primers thereof, and a transgenic expression vector and an establishment method thereof. The CRISPR-Cas9 homing sequence sgRNA comprises SIDT1-gene-specific Target site-1 of which the target is positioned on E1 exon, and SIDT1-gene-specific Target site-2 of which the target is positioned on E2 exon, wherein the nucleotide sequence of the Target site-1 is disclosed as SEQ ID NO.8, and the nucleotide sequence of the Target site-2 is disclosed as SEQ ID NO.9. The CRISPR-Cas9 transgenic expression vector is formed by connecting the Cas9 homing sequence Target site-1 and Target site-2 to the BsmB I and Bbs I enzyme digestion sites of a recombinant shuttle Cas9 tool plasmid. The recombinant shuttle Cas9 tool plasmid can be quickly assembled with homing sequences of the two target sites of the genome, thereby packaging the recombinant adenovirus vector specifically for the two target sites; and the recombinant shuttle Cas9 tool plasmid can independently complete the editing of the large-segment genome without dependence on the synergistic actions. The establishment process is simple and quick, and has high efficiency for performing the genome large-segment editing function.
Owner:世翱(上海)生物医药科技有限公司
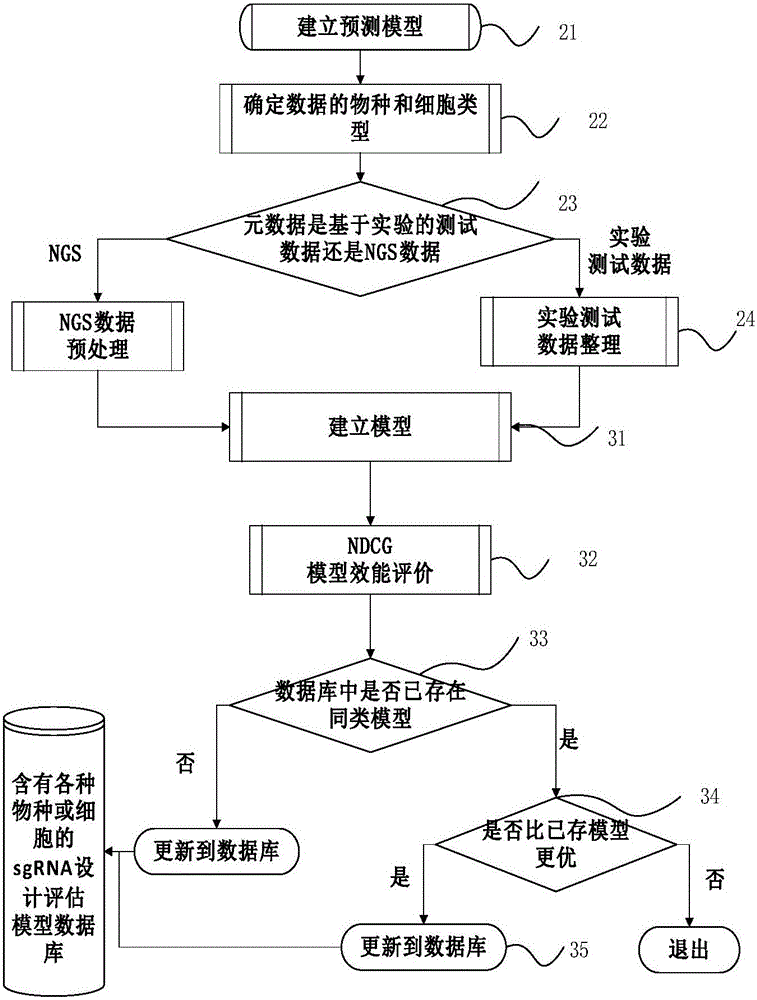
CRISPR/Cas9-based sgRNA design method
InactiveCN106446600AImprove assessment accuracyFully featuredSpecial data processing applicationsAnalysis dataEnzyme digestion
The invention relates to a CRISPR / Cas9-based sgRNA design method. The method is characterized by comprising the following steps of obtaining the value of enzyme digestion efficiency of sgRNA and corresponding Cas9; building a personalized sgRNA design model; measuring the quality of the built personalized sgRNA design model by applying an NDCG algorithm and updating a database; and designing sgRNA and giving out an assessment value of each sgRNA. Compared with the prior art, the method has the characteristics of high accuracy, complete characteristics, wide application range and wide data analysis range.
Owner:TONGJI UNIV
Establishment method of caffeine synthetase CRISPR/Cas9 genome editing vector
InactiveCN105821075ANucleic acid vectorVector-based foreign material introductionEnzyme digestionGolden Gate Cloning
The invention relates to an establishment method of a caffeine synthetase CRISPR / Cas9 genome editing vector. The method comprises the following steps: designing two target sequences T1 and T2 on the basis of tea tree caffeine synthetase genes; respectively establishing sgRNA expression cassettes of the target sequences T1 and T2; and carrying out enzyme digestion-coupling reaction on a double-element expression vector pYLCRISPR / Cas9P35S-H, the T1sgRNA expression cassette and the T2sgRNA expression cassette so that the T1sgRNA expression cassette and the T2sgRNA expression cassette are connected and inserted into the double-element expression vector, thereby obtaining the CRISPR / Cas9 genome editing vector. By taking the tea tree caffeine synthetase as the example, conventional PCR (polymerase chain reaction), Overlapping PCR and Golden Gate Cloning techniques are combined to establish the CRISPR / Cas9 gene editing vector comprising the tea tree caffeine synthetase double targets, thereby laying a solid foundation for application of the CRISPR / Cas9 gene editing technique in tea trees, and providing technical supports for the establishment of CRISPR / Cas9 gene editing vectors of other plants.
Owner:HUNAN AGRICULTURAL UNIV
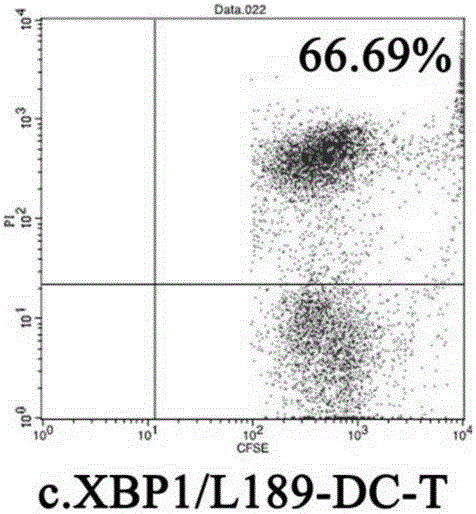
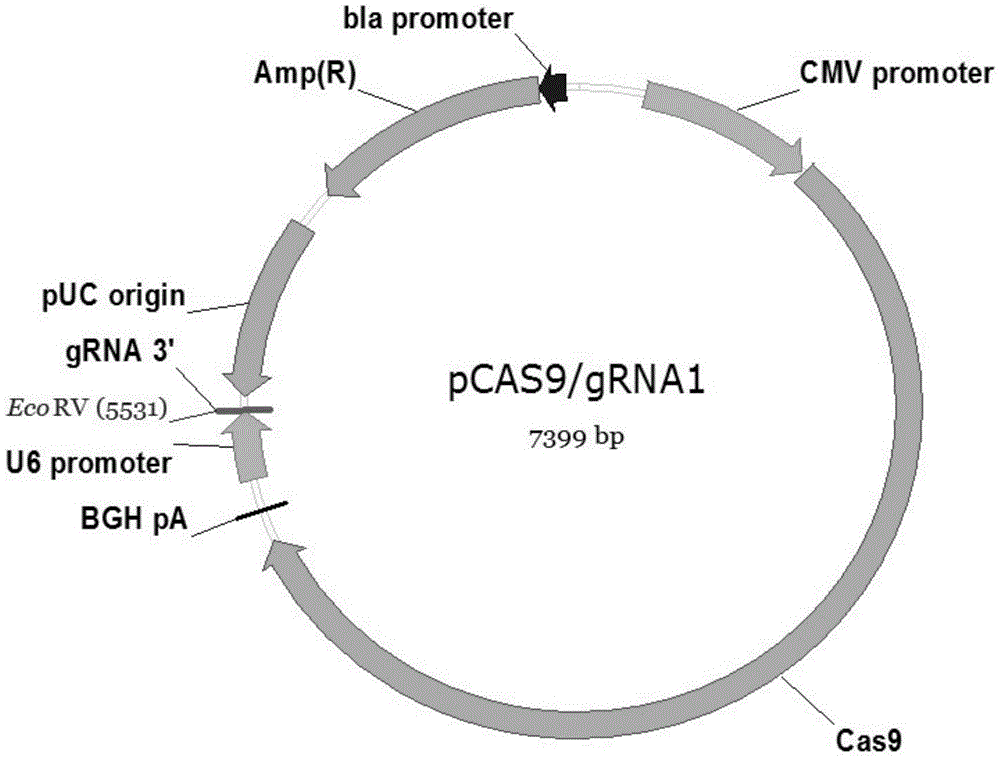
High-efficiency knockout method for XBP1 gene in DC cell
InactiveCN105602987AGuaranteed immune stimulationEffective anti-tumor immunityBlood/immune system cellsVector-based foreign material introductionCompetent cellXBP1
The invention provides a high-efficiency knockout method for the XBP1 gene in a DC cell. The high-efficiency knockout method for the XBP1 gene in the DC cell comprises the following steps: designing of a gene knockout target and oligonucleotide; annealing of oligonucleotide; enzyme digestion of a linearized vector; connection and reaction of the linearized vector with double-strand oligonucleotide; transformation of competent cells and knockout of the XBP1 gene in the DC cell; etc. According to the invention, the knockout method provided by the invention employs an improved CRISPR / pCas9 gene knockout system, uses the XBP1 as a target gene for designing of a CRISPR targeting sequence and preparation of pCas9 / gRNA1-XBP1 plasmid and further allows the plasmid to transfect the DC cell under the treatment action of an L189 drug; thus, the XBP1 gene in the DC cell can be effectively knocked out, the immunostimulation effect of the DC cells can be effectively guaranteed, and antineoplastic immunization effect of the DC cell is given to effective play.
Owner:SHENZHEN MORECELL BIOMEDICAL TECH DEV CO LTD
Human-STAT6-targerted CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 system and application thereof in treating allergic diseases
ActiveCN106222177AKnockout achievedLow efficiencyOrganic active ingredientsGenetic material ingredientsDiseaseEnzyme digestion
The invention provides a CRISPR (clustered regularly interspaced short palindromic repeats) knock-out system for carrying out gene knock-out on STAT6. According to the system, a plurality of sgRNAs (small guide ribonucleic acids) can be utilized to implement high-efficiency knock-out on the STAT6 gene. The sgRNAs capable of specific STAT6-gene-targeted knock-out in the CRISPR-Cas9 are designed and synthesized; and the sgRNA oligonucleotide vector and the enzyme digestion plasmid are utilized to respectively and successfully transfect cells, thereby implementing STAT6 gene knock-out. The invention provides a scheme for quickly, simply, efficiently and specifically knocking out STAT6 by using Cas9 / sgRNA. The efficiency is high, and the STAT6 knock-out efficiency reaches 80% or above. The system can be used for knocking out the single coded sequence or segment of STAT6. The sgRNAs can be massively produced only by synthesizing a small amount of polynucleotide segments, thereby saving the cost and enhancing the efficiency.
Owner:GEMPHARMATECH CO LTD
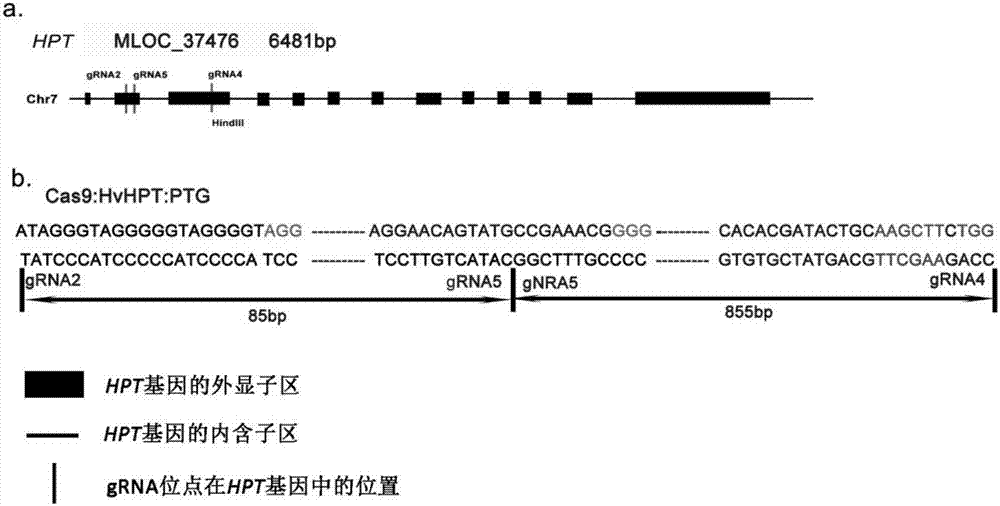
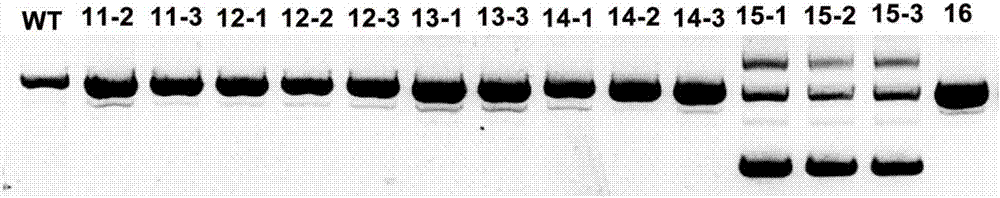
Method for applying CRISPR-Cas9 system to knockout of key gene HPT in VE synthesis pathway of barley
The invention relates to construction of a barley transgenic material and provides a method for applying a CRISPR-Cas9 system to knockout of the key gene HPT in the VE synthesis pathway of barley. The method comprises the following steps: selection of a gRNA target site; cloning of gRNA fragments; connection of GG(gRNA-gRNA) fragments; PCR amplification of a connection product; enzyme digestion of a purified product and a target vector; a ligation reaction between the purified product and the target vector; transformation of competent Escherichia coli cells with a ligation product; preparation of Agrobacterium-mediated transgenic barley plants; screening of positive transgenic plants; and sequencing of mutants. According to the invention, the advanced gene editing technology, i.e., the CRISPR-Cas9 system, is employed for target gene editing of the key gene (HPT) in the VE synthesis pathway of barley, and effective loss-of-function mutants are obtained, so conditions are created for research on biologically-active substances in barley.
Owner:ZHEJIANG UNIV
Goat TLR4 gene knock-out vector and construction method thereof
InactiveCN106755097AThe method is simple and fastHigh knockout efficiencyNucleic acid vectorVector-based foreign material introductionEscherichia coliCompetent cell
The invention discloses a goat TLR4 gene knock-out vector and a construction method thereof. The construction method comprises the following steps: firstly, designing an sgRNA fragment of the TLR4 gene by adopting a CRISPR / cas9 system, synthesizing an sgRNA nucleotide sequence, constructing and simultaneously expressing the sgRNA and plasmid PYSY-sgRNA of Cas9 D10A, connecting and transforming to an Escherichia coli DH5 alpha competent cell, and verifying the transformant; and judging and proving by enzyme digestion and sequencing that the TLR4 gene knock-out vector is constructed correctly. The invention adopts the CRISPR / cas9 for constructing the vector, and provides a theoretical basis for subsequently acquiring a goat TLR4 gene deletion type alveolar epithelial cell system, and studying the immune response molecular mechanism of mycoplasma pneumonia infection.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Construction method of homologous repair vector based on CRISPR/Cas9 system
PendingCN107937427ARealize fixed-point editing functionImprove assembly efficiencyVector-based foreign material introductionEnzyme digestionWild type
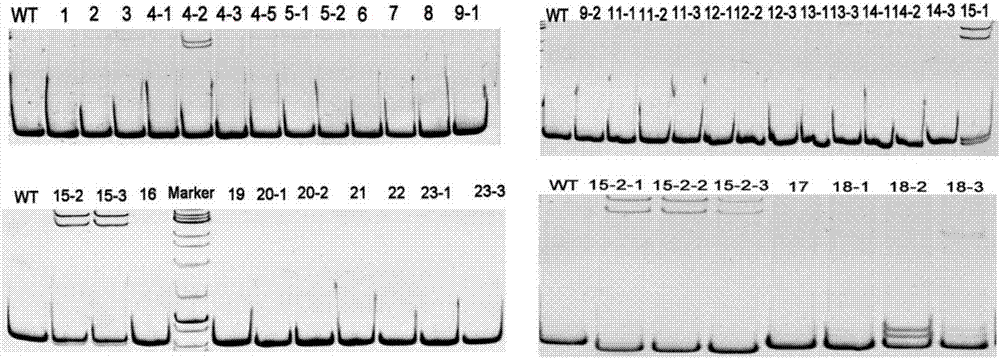
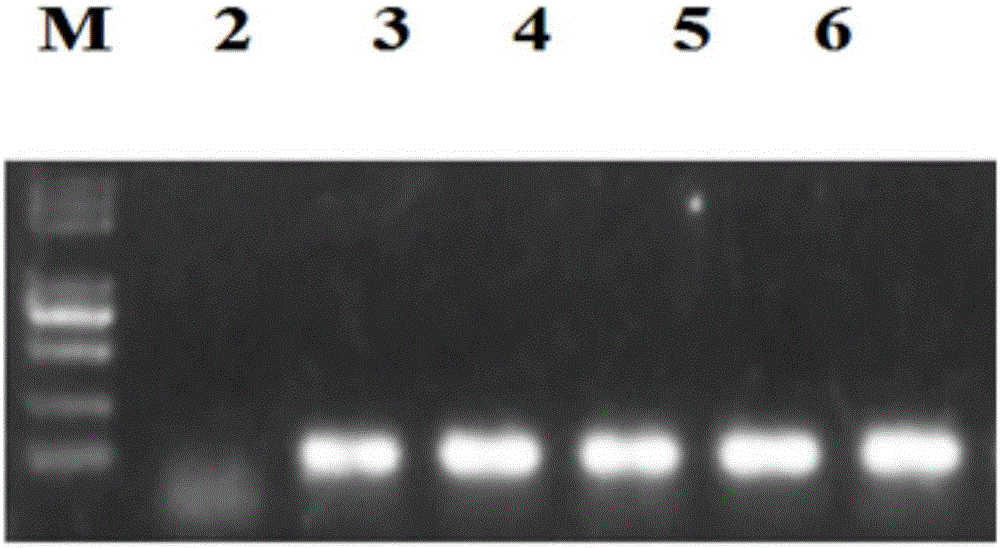
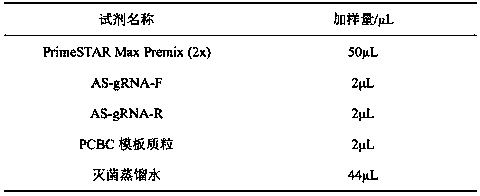
The invention relates to a construction method of a homologous repair vector based on a CRISPR / Cas9 system, and belongs to the technical field of genetic engineering. The construction method comprisesthe following steps: with a plasmid PCBC and wild-type arabidopsis genome as templates, amplifying a target band AS-gRNA containing a target sequence and an AS homologous repair template segment by virtue of PCR, implementing electrophoresis and gel cutting, and recovering the target band; implementing enzyme digestion on a plasmid PHDE-mCH by virtue of Bsa1; assembling and linking a PHDE-mCh vector, which undergoes complete enzyme digestion, with the ASgRNA by virtue of homologous recombinase, so that a recombinant plasmid PHDE-ASgRNA is formed, implementing transformation and sequencing identification, then linking the PHDE-ASgRNA plasmid, which is correct in sequencing and is subjected to enzyme digestion by virtue of EcoR1, with AS homologous repair template segment homologous recombinase, and implementing transformation and sequencing identification, so that the PHDE-ASgRNA-AS homologous repair vector is constructed. According to the technique provided by the invention, the method for constructing the CRISPR / Cas9 system which has a site-specific editing function on target biology genome is actually implemented; the vector is constructed just by conducting PCR by one step, sothat the method is simple and easy to implement; and the method, which is unnecessary to purify or recover a digestion vector and a PCR product, is high in assembly efficiency.
Owner:GUANGDONG UNIV OF PETROCHEMICAL TECH
Method for biosynthesis preparation of human GLP-1 polypeptide or analogue thereof
InactiveCN106434717AEasy to separateHigh expressionNucleic acid vectorFusion with protease siteEscherichia coliEnzyme digestion
The invention relates to a method for biosynthesis preparation of human glucagon-like peptide-1 (GLP-1) and an analogue thereof. With adopting of a gene engineering technology, a recombinant escherichia coli expressed GLP-1 fusion protein is constructed, and a protein enzyme digestion site is designed in the fusion protein; a fusion gene has a gene sequence with a form of A-B-C structure, wherein A is a chaperonin gene, B is a nucleotide sequence encoding a connection peptide containing the enzyme digestion site, and C is a gene encoding the GLP-1 or the analogue thereof. After recombinant engineering bacteria is subjected to induced expression, the fusion protein is purified and subjected to enzyme digestion, and then the GLP-1 and the analogue thereof are obtained and are detected to have biological activity. The preparation method of the GLP-1 and the analogue thereof provided by the invention is simple and quick, the production conditions are mild, the product is convenient to separate and extract, the process is simple, and the industrialization prospect is good.
Owner:HANGZHOU JIUYUAN GENE ENG +1
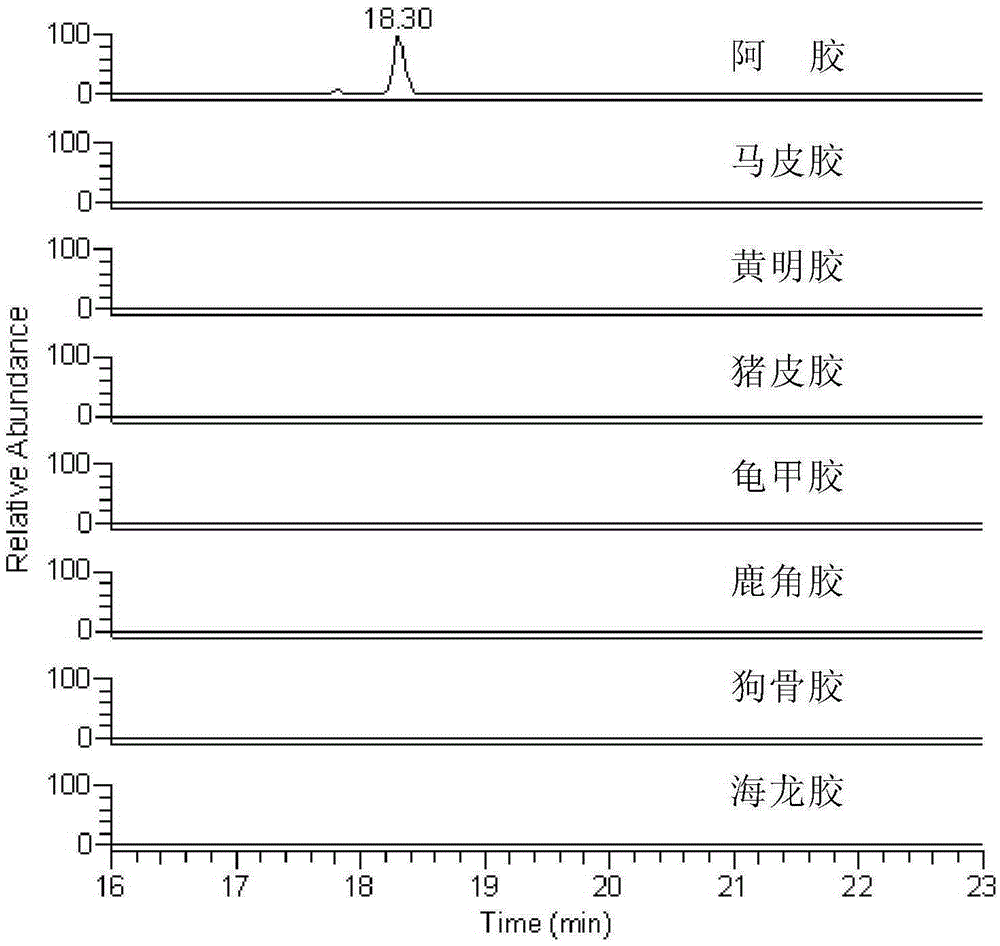
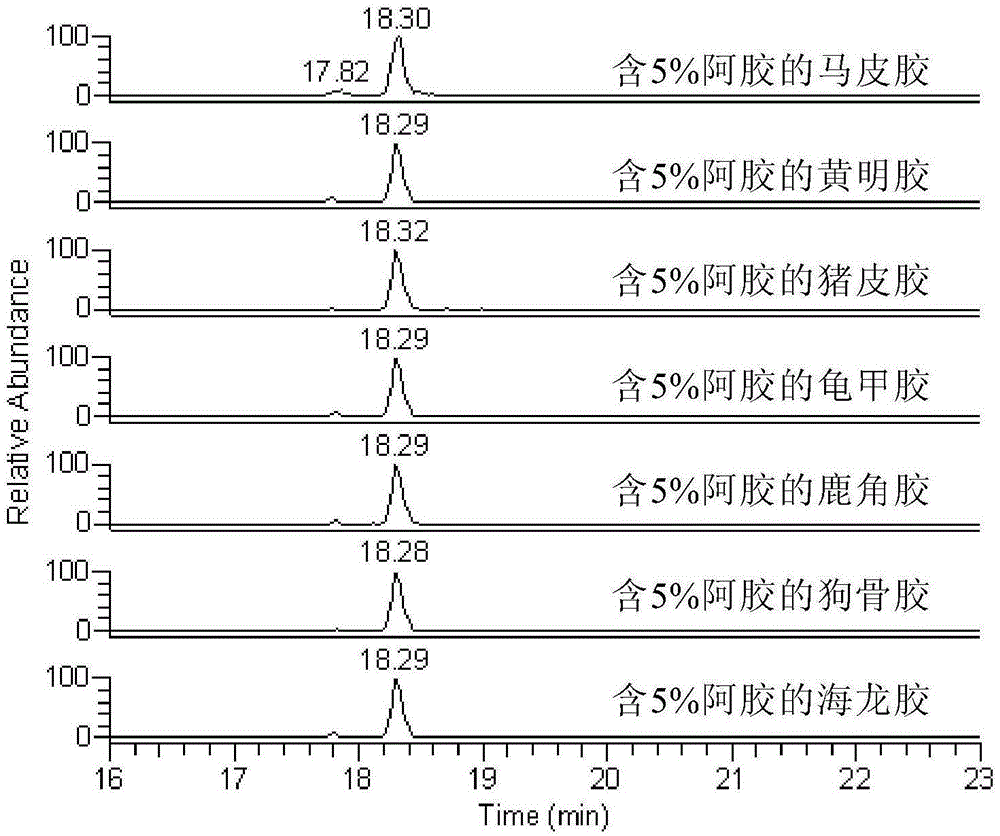
Donkey characteristic polypeptide and application thereof to detection on donkey skin derived ingredients
ActiveCN105301165AIncrease contentTo achieve the purpose of detectionComponent separationTandem mass spectrometryEnzyme digestion
The invention discloses donkey characteristic polypeptide and application thereof to detection on donkey skin derived ingredients. The amino acid sequence of the donkey characteristic polypeptide provided by the invention is shown as SEQ ID NO.1 and the polypeptide is specifically contained in collagen of a donkey. Moreover, the invention also provides a method for detecting donkey skin derived ingredients in glue traditional Chinese medicine, which comprises: carrying out enzyme digestion on the glue traditional Chinese medicine by adopting trypsin or arginine-C enzyme so as to enable the characteristic polypeptide to be dissociated from the collagen; then carrying out detection by adopting liquid chromatography-tandem mass spectrometry so as to judge whether the donkey skin derived ingredients are included in the glue traditional Chinese medicine. The method has the advantages of strong characteristic performance, high sensitivity, simplicity for operation and the like and can accurately authenticate whether the donkey skin derived ingredients are included in the glue traditional Chinese medicine.
Owner:SHAN DONG DONG E E JIAO
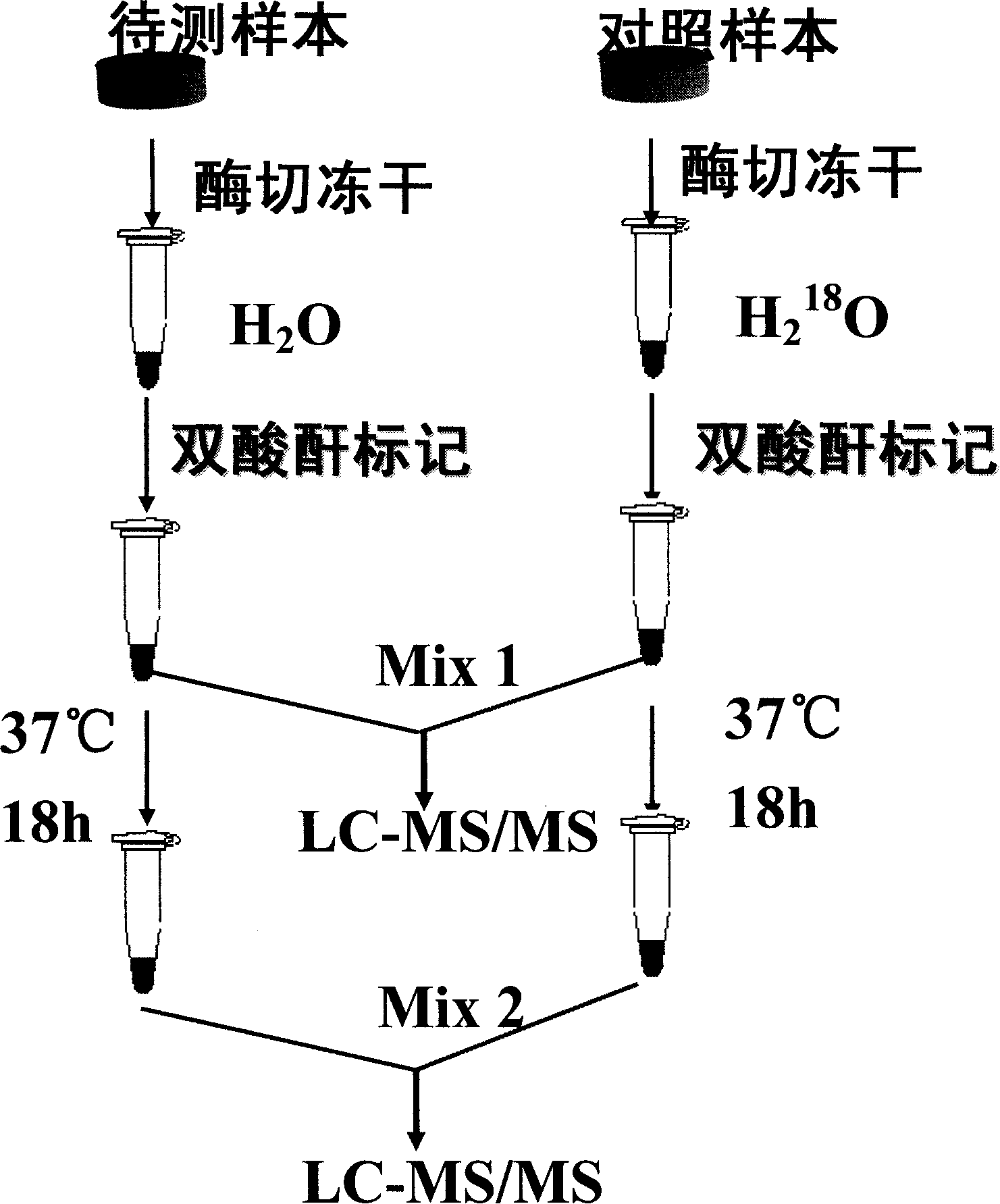
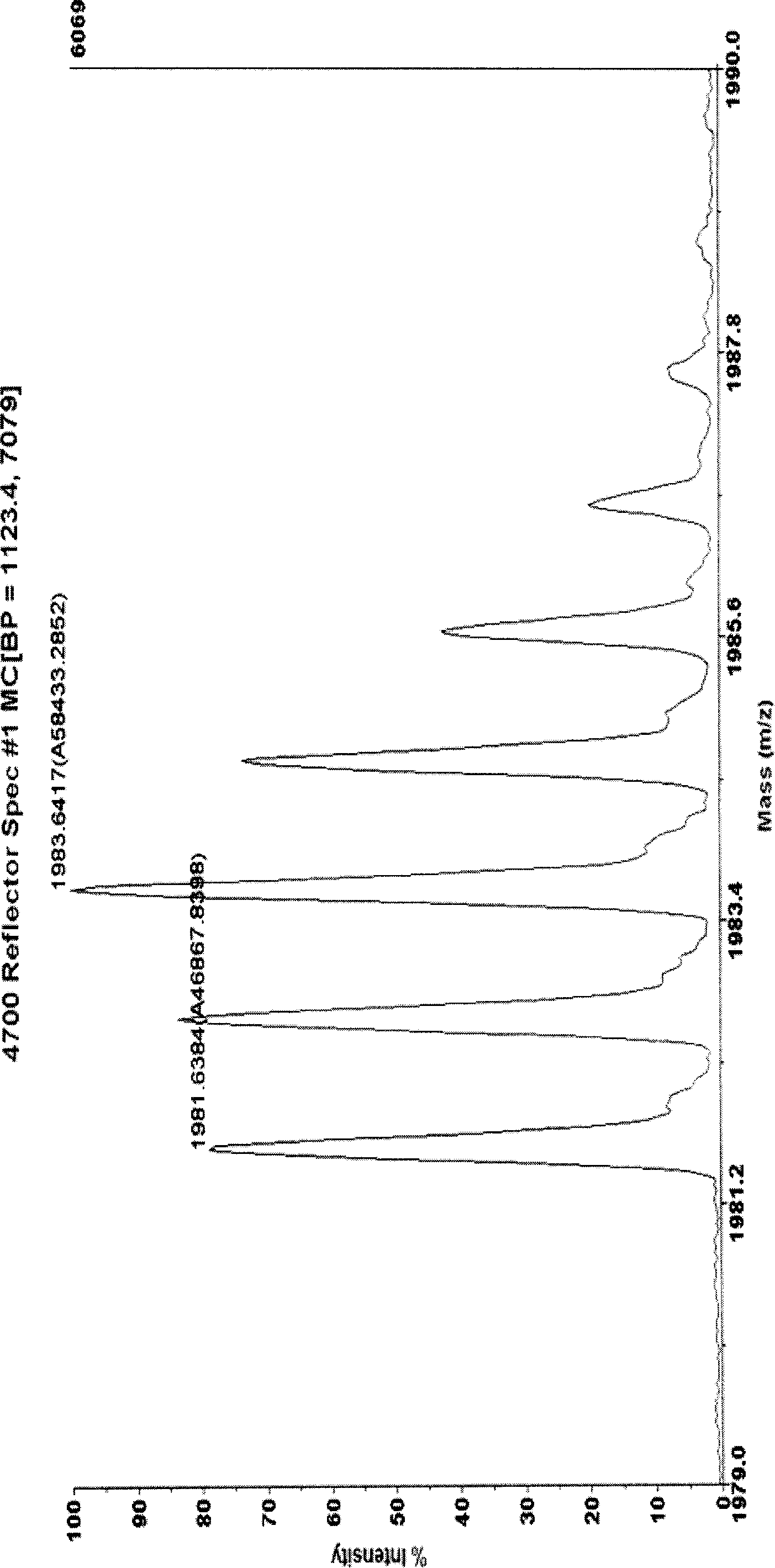
Dual quantification method and reagent kit for stable isotope 18 O marked proteome
InactiveCN101173926AFast wayEasy to useComponent separationBiological testingChemical labelingEnzyme digestion
The invention provides a new method for double quantifying the protein group marked on the basis of stable oxygen isotope <18>O, which is characterized in that: based on the fact that the N end peptide section and the C end peptide section are generally labeled, and by means of combining the <18>O chemical labeling and enzyme digestion, the quantifying strategy for two <18>O markings of the same sample can be accomplished. The method relates to the reductive alkylation and enzyme digestion of protein, dual-functional reagent modifying and <18>O marking of peptides, multi-dimensional liquid phase chromatogram separation and multistage Tandem Mass Spectrometry quantification and identification. A first mass spectrum peak area is used to quantify; and the identification of the protein is conducted through a second stage mass spectrum and by searching the database. The invention also provides a reagent box for facilitating the implementation of the method.
Owner:INST OF RADIATION MEDICINE ACAD OF MILITARY MEDICAL SCI OF THE PLA
Preparation method of sea cucumber intestine polypeptide capsule
InactiveCN101422205AIncrease the degree of hydrolysisIncrease profitAnimal proteins working-upFood preparationNutrientEnzyme digestion
The invention relates to a method for preparing a sea cucumber intestine opypeptide capsule with sea cucumber intestine. The sea cucumber intestine is taken as raw material, put into a high pressure sterilizing pot to be sterilized and softened under high pressure and low temperature, cut into small segments and then added with 5 times to 20 times of water; the mixture is pounded to paste and then proteolytic enzyme accounting for 1 percent to 3 percent of the weight of the sea cucumber intestine is added; at the temperature of 40 DEG C to 60 DEG C, enzyme digestion lasts for 3 hours to 5 hours to produce a sea cucumber intestine hydrolyzate; enzyme inactivation, precipitation removal, ultratemperature instant sterilization and hyperfiltration are conducted to obtain a sea cucumber intestine opypeptide liquid with the molecular weight below 3000 daltons; and then concentration and drying are carried out to prepare a sea cucumber intestine opypeptide powder which is filled into capsules to obtain the finished product. The sea cucumber intestine opypeptide capsule has the healthy functions of antifatigue, decreasing blood pressure, controlling blood fat, regulating body immunity and the like and is easy to digest and absorb and the utilization ratio of various nutriments is high. The preparation method is reasonable, easy and convenient to operate and has high operability, thus being easy to realize commercial process.
Owner:SHANDONG HOMEY AQUATIC DEV +1
Lamellar corneal stroma bracket as well as preparation method and application thereof
The invention discloses a lamellar corneal stroma bracket as well as a preparation method and application thereof. The method comprises the following steps: carrying out low-permeability swelling, repetitive freeze-thawing, enzyme digesting, drying and sterilizing treatment on a corneal stroma sheet of a fresh animal eyeball cut down under a sterile condition, wherein the enzyme digesting adopts buffer liquor containing DNA (deoxyribonucleic acid) enzyme and RNA (ribonucleic acid) enzyme to treat, ultrasonic treatment is carried out after the enzyme digestion treatment, and then drying and sterilizing treatment is carried out. The invention further provides application of the lamellar corneal stroma bracket as a corneal stroma substrate. The method disclosed by the invention can be used for reducing damages on a corneal stroma collagen structure to the greatest extent; collagenous fiber arrangement is orderly, holes are uniform and regular, cell residues are avoided, the lamellar structure is kept complete, and the biocompatibility of the bracket material is improved.
Owner:QINGDAO CHUNGHAO TISSUE ENG
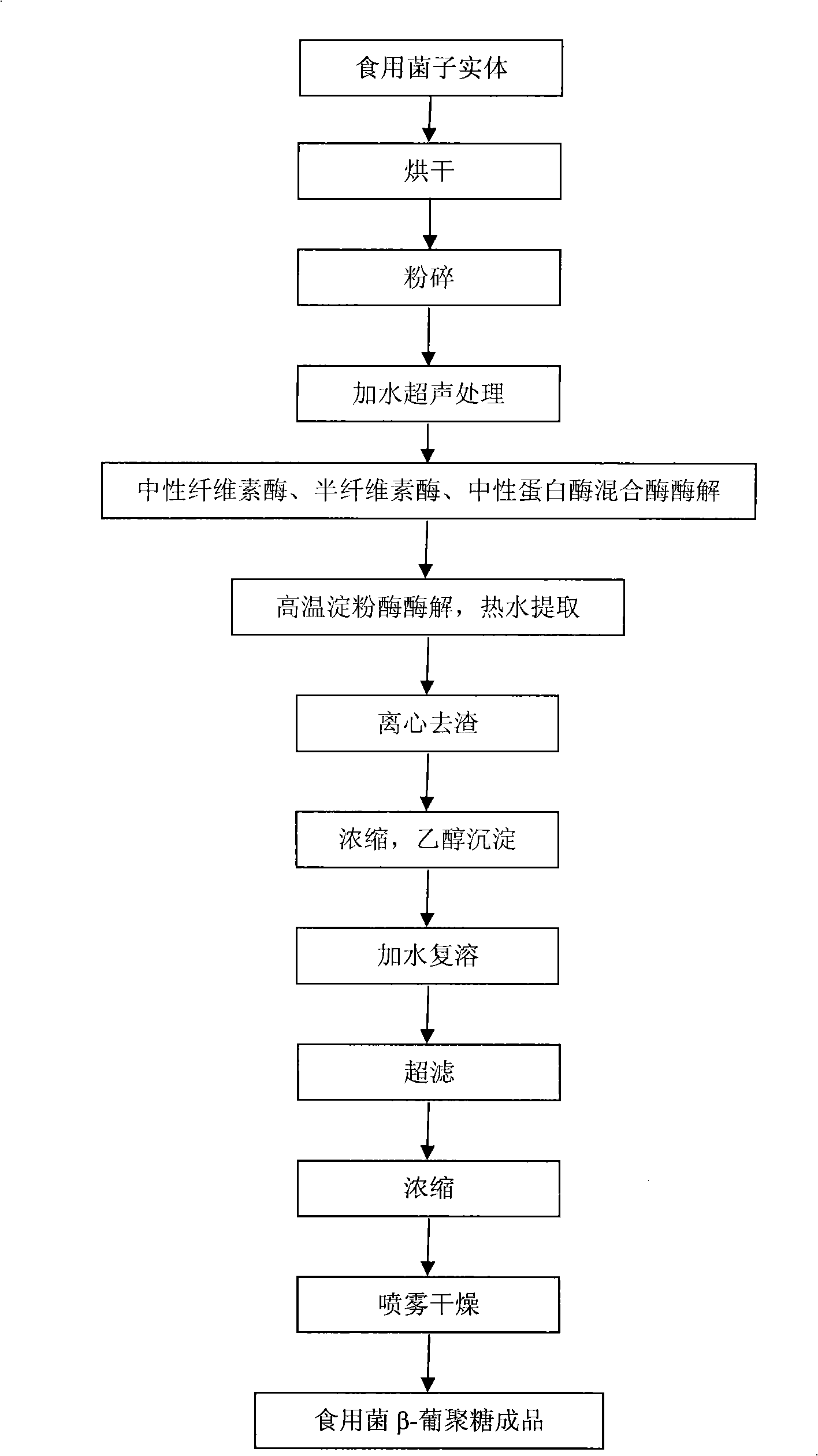
Preparation of edible fungus beta-dextran
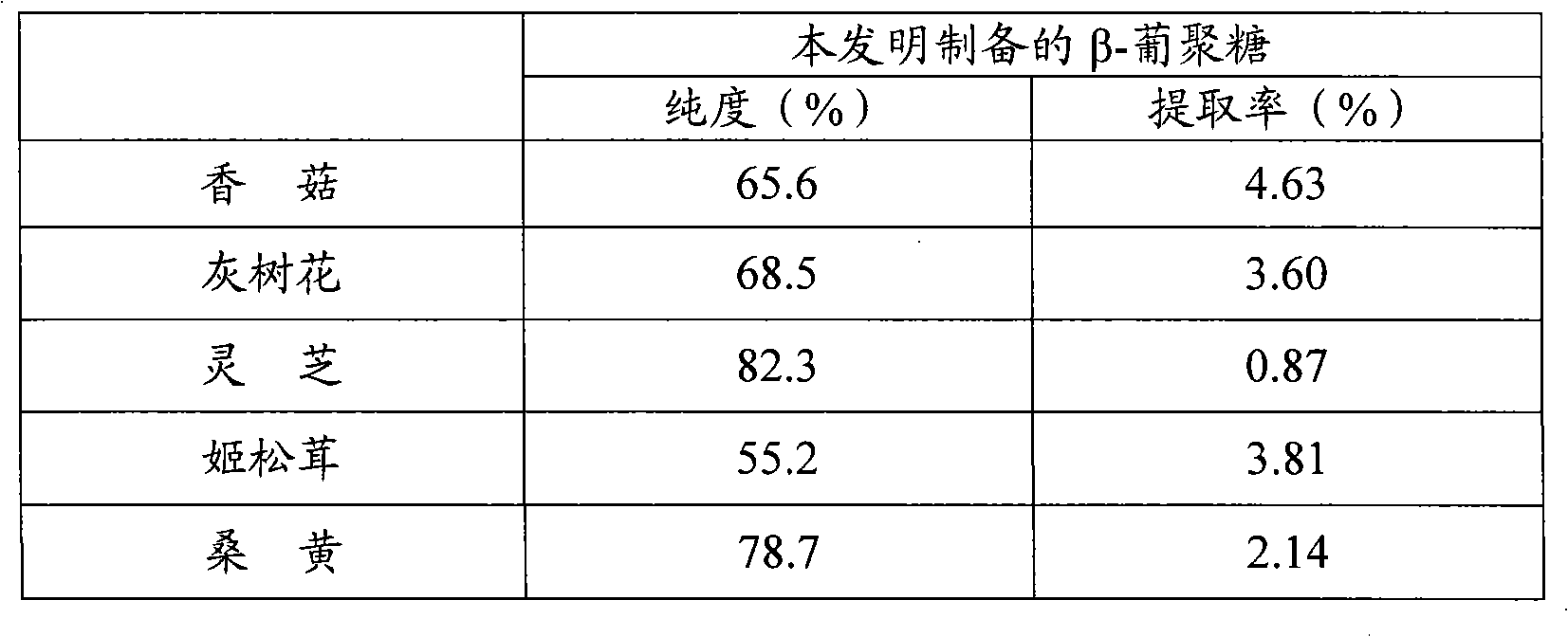
The invention provides a preparation method of edible fungi Beta-glucan. In the method, ultrasonic pretreatment, neutral cellulose, hemi-cellulase, the cellulase enzyme digested by a neutral protease mixed enzyme, hemicellulose (like xylan, gelose and a plurality of heteroglycans) and proteins (and peptides) are mainly adopted; a high-temperature amylase is added for the enzyme digestion of the amyloid simultaneously when hot water is extracted; and a target is obtained through a sequent ethanol deposition and the edible fungi Beta-glucan with better activity is further obtained through the hyperfiltration of a film with an interception of 10,000. The preparation method has the advantages that: the product purity of the edible fungi Beta-glucan by adopting the method is high (55.2 to 82.3 percent); the preparation process does not have the residues of poisonous and harmful chemical reagents; the product is safe; the technical condition is moderate and is easy to be operated; and the preparation method is suitable for preparing all the edible fungi Beta-glucan and is suitable for commercial production.
Owner:ZHEJIANG UNIV OF TECH
Gene-specific molecular marker Pi2SNP of rice blast-resistant gene Pi2 as well as preparation method and application thereof
ActiveCN103320437AStrong specificityHigh sequence identityMicrobiological testing/measurementDNA preparationAgricultural scienceEnzyme digestion
The invention discloses a gene-specific molecular marker Pi2SNP of a rice blast-resistant gene Pi2 as well as a preparation method and application thereof. The molecular marker Pi2SNP is a nucleotide fragment II in a specific banding pattern with the rice blast-resistant gene Pi2 obtained by amplifying F1 and R1 from the total DNA (Deoxyribose Nucleic Acid) of the rice blast-resistant variety carrying rice blast-resistant gene Pi2 by use of a primer to obtain a nucleotide fragment I and then performing enzyme digestion of the nucleotide fragment I by use of restriction endonuclease Pst I or Hinf I. The molecular marker is the first Pi2 gene-specific SNP marker developed for the sequence in the Pi2 gene, and has the advantages of high specificity and low cost and high flux in practical application, and can be widely applied to the colonies with different inheritance backgrounds. By adopting the molecular marker, the utilization efficiency of the gene in molecular marker-assisted selective breeding, gene pyramiding breeding and transgenic breeding can be improved.
Owner:PLANT PROTECTION RES INST OF GUANGDONG ACADEMY OF AGRI SCI
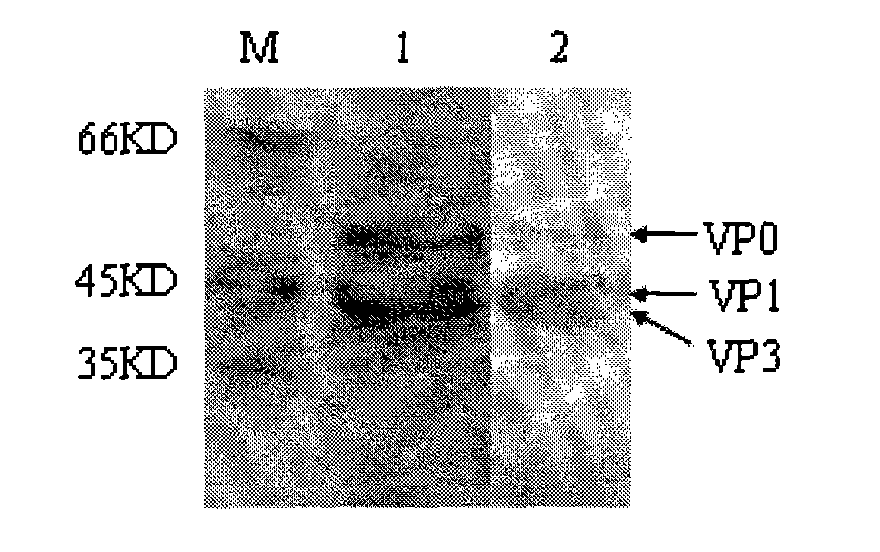
Foot and mouth disease virus-like particle, preparation method and application thereof
ActiveCN101914501AImproving immunogenicityImprove biological activityInactivation/attenuationAntiviralsEnzyme digestionStructural protein
The invention discloses an Asia I type foot and mouth disease virus-like particle, a preparation method and an application thereof. The Asian I type foot and mouth disease virus-like particle comprises structural proteins of VP0, VP3 and VP1 of Asia I type foot and mouth disease virus, wherein the gene sequence of VP0 is shown in SEQ 1, the gene sequence of VP3 is shown in SEQ 3, and the gene sequence of VP1 is shown in SEQ 2. The preparation method of the Asian I type foot and mouth disease virus-like particle has the following steps: performing amplification to obtain the VP0, VP3 and VP1, performing enzyme digestion to obtain a recombinant expression vector, performing enzyme digestion on fusion protein by small ubiquitin-like modifier (SUMO), and carrying out in-vitro assembling to obtain the foot and mouth disease virus-like particle.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
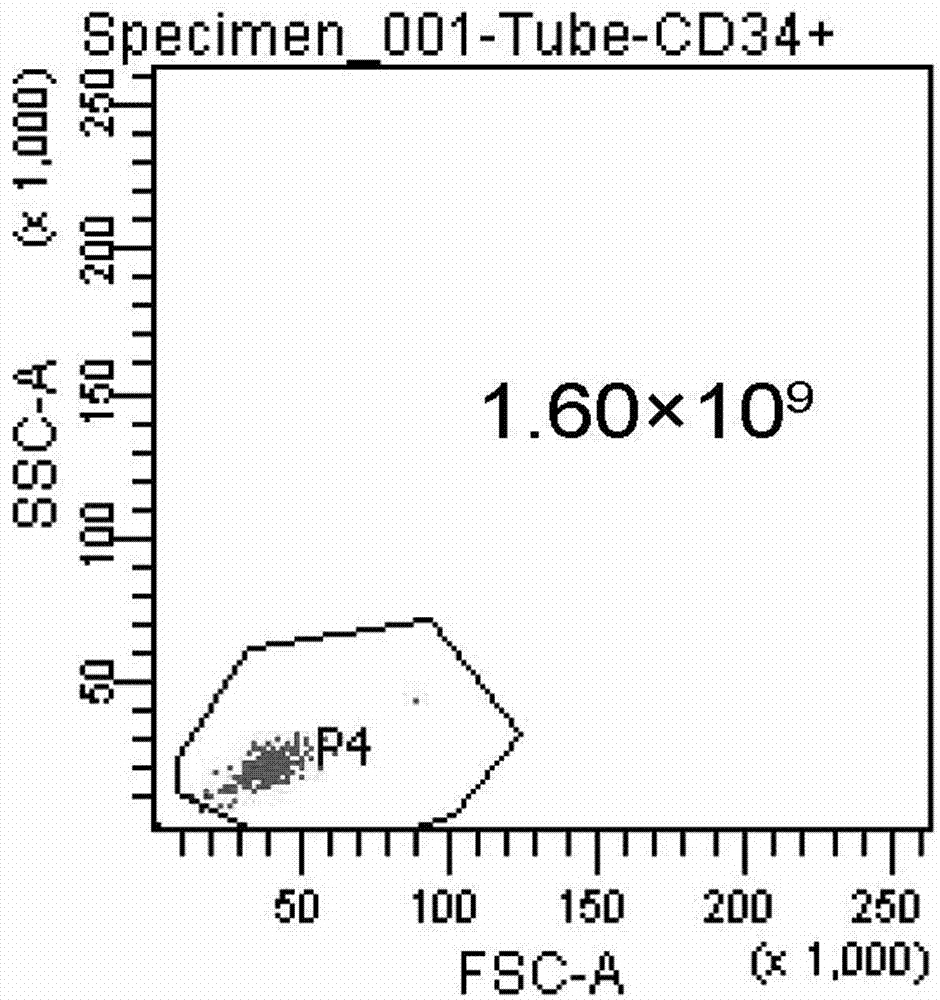
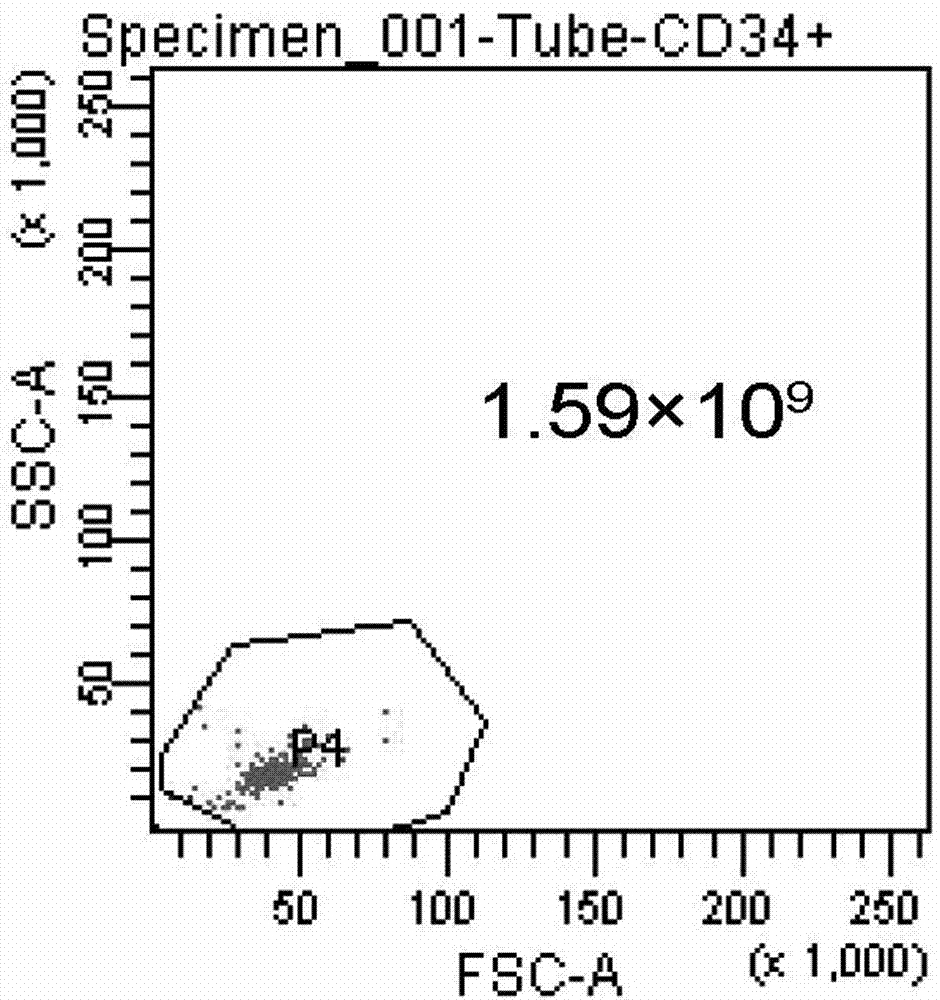
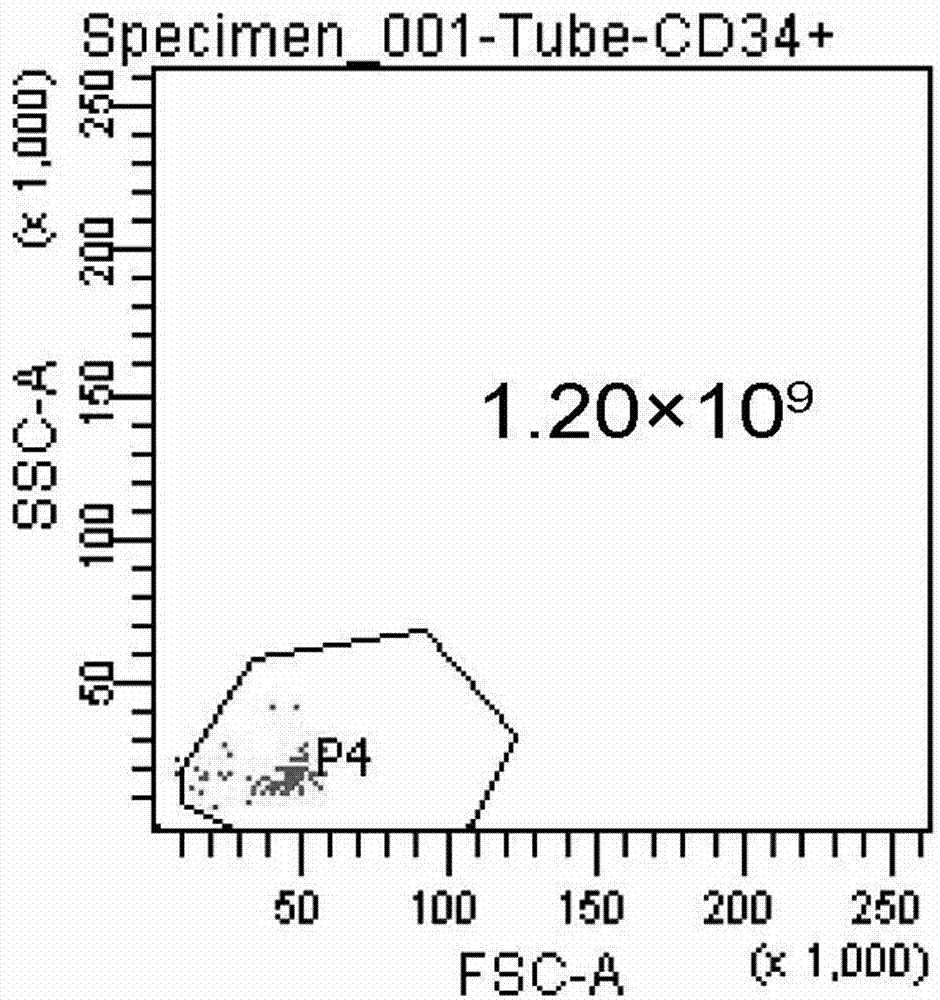
Preparation method and preservation method of clinical application-level placental hematopoietic stem cells
InactiveCN103789262AIncrease contentEnhance cell viabilityDead animal preservationBlood/immune system cellsHydroxyethyl starchEnzyme digestion
The invention provides a preparation method of clinical application-level placental hematopoietic stem cells. The preparation method comprises the steps of pretreating a fresh delivered placenta immediately and then separating out umbilical arteries and umbilical veins, pouring a cleaning fluid into the chorionic vessels of the placenta and colleting the cleaning fluid after pouring, pouring an enzyme digestion fluid into the chorionic vessels of the placenta and digesting at 37 DEG C for 5-20min, pouring the collected fluid into the placenta and collecting the liquid after pouring, centrifuging the obtained fluid and re-suspending cells after removing the supernatant, and separating the resuspended cell suspension through a two-step process with hydroxyethyl starch and a lymphocyte separation medium to obtain the clinical application-level placental hematopoietic stem cells. The method is capable of increasing the number and the activity of the prepared placental hematopoietic stem cells and the content of CD34 positive cells, and the prepared placental hematopoietic stem cells are at the clinical application level and free of potential pathogenic contamination, and moreover, the method is low in preparation cost.
Owner:台州恩源生物科技有限公司
Human-like collagen gene, homodirection tandem gene with different repeat numbers, recombinant plasmid having tandem gene and preparation method
ActiveCN101200718AEasy to operateConnective tissue peptidesFermentationEnzyme digestionMature technology
The present invention discloses a human-like collagen gene, a same direction serial gene thereof with different repeat numbers, a recombined plasmid containing the serial gene and a preparation method. The base sequence of the human-like collagen gene Gel of the present invention is shown as SEQ ID NO: 1 in a sequence list, and the serial gene with different repeat numbers and the recombined plasmid containing the serial gene with different repeat numbers are obtained by the external enzyme digestion and the joint of the gene. The present invention is a complete novel sequence, and the length is greatly less than natural human collagen gene, which realizes the operation simplification at the molecular level and is easier to realize the gene engineering bacteria fermentation production of the collagen. The human-like collagen gene Gel also contains the characteristic of the natural human collagen gene, and corresponding human-like collagen expressed by the human-like collagen gene Gel can be provided with the good characteristic of the natural human collagen gene. The preparation method is just the simple external enzyme digestion connection reaction and the prokaryotic transformation and sieving with mature technology, which realizes the series connection of the target gene with any repeat number.
Owner:JIANGSU JLAND BIOTECH CO LTD
Conditional gene knockout method constructed by using CRISPR/Cas9 system
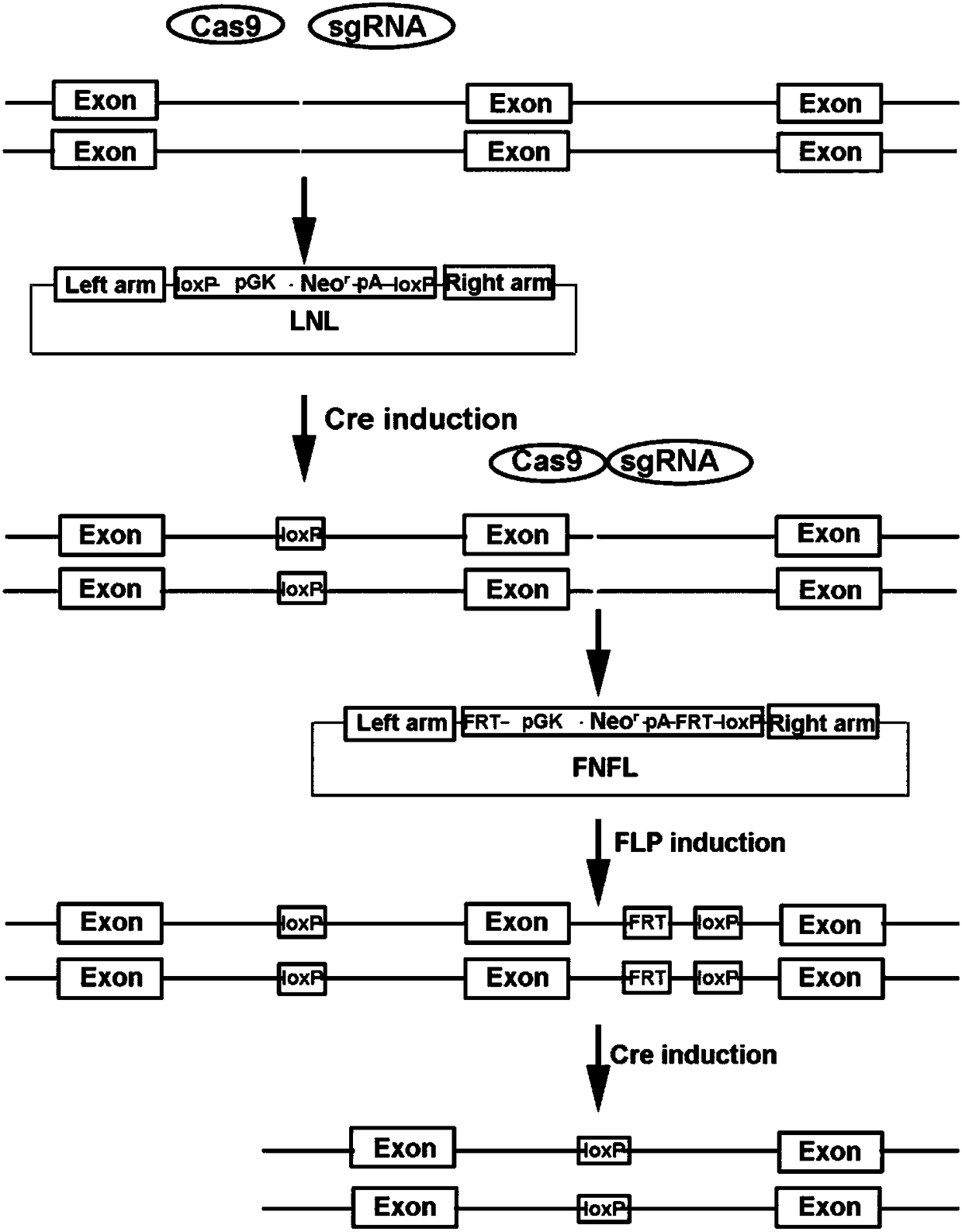
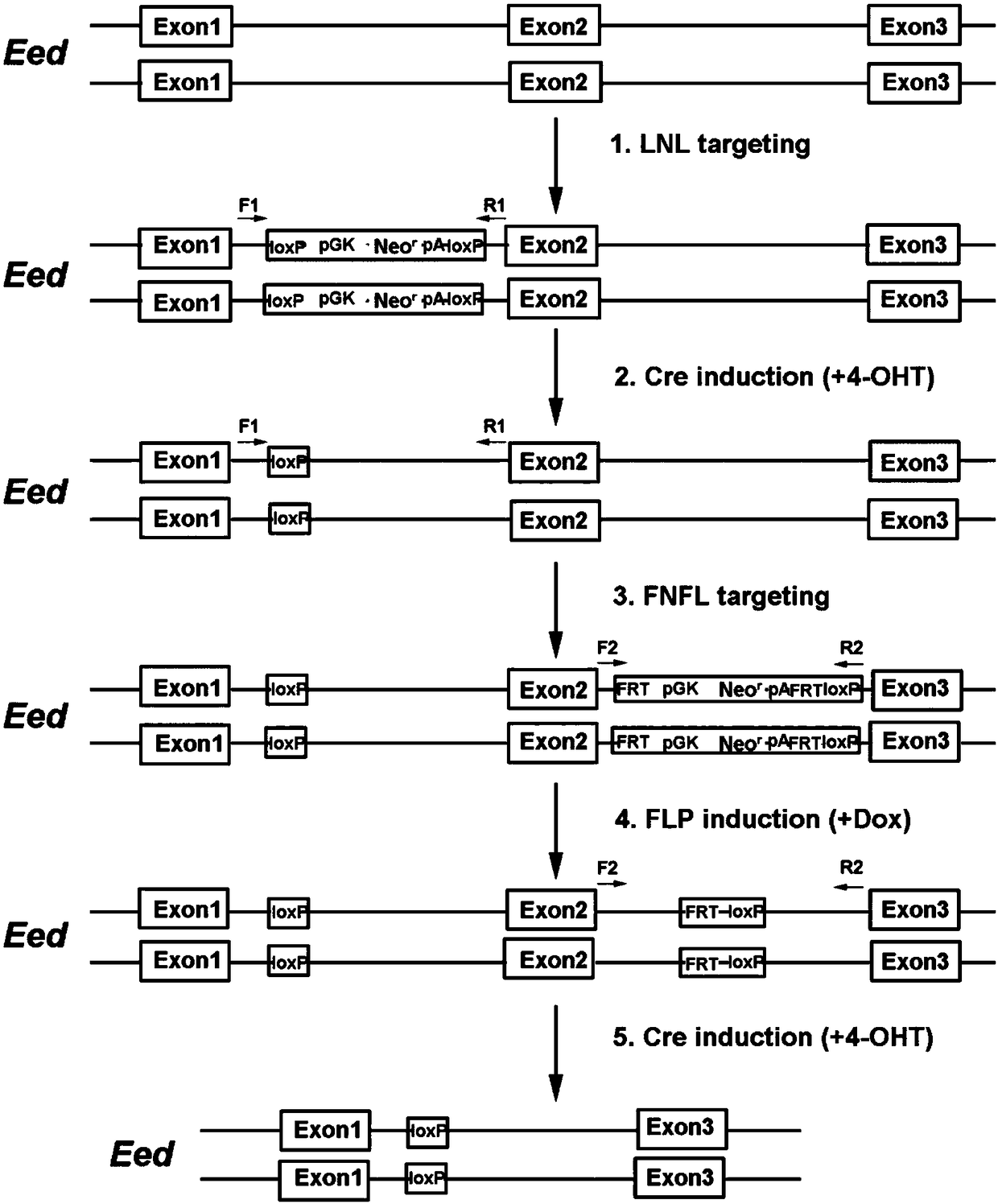
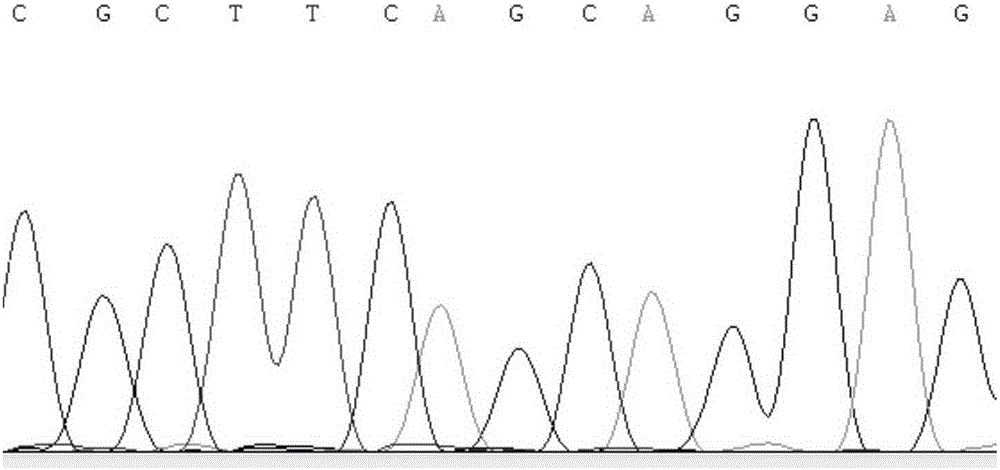
ActiveCN108441520AImprove recombination efficiencySave time and costHydrolasesStable introduction of DNAEnzyme digestionSequence Insertions
The invention relates to a conditional gene knockout method constructed by using a CRISPR / Cas9 system. LoxP-antibiotic-LoxP and FRT-antibiotic-FRT-LoxP sequence insertion sites are selected from leftand right sides of a to-be-knocked-out area respectively; left and right homologous arms are selected from upstream and downstream positions of to-be-inserted sites, amplification, purification, enzyme digestion, linking and conversion are performed, and left and right targeting plasmids are obtained; Cas9 plasmid, the left or right targeting plasmid of the knockout area and corresponding gRNA expression plasmids are sequentially or simultaneously introduced into cells, screening is performed with antibiotics after transfection, and target cells with left and right targeting sequences insertedare screened out simultaneously; 4-OHT and DOX are used for inducing Cre-LoxP and FLP-FRT medicated homologous recombination, so that loxp sites are introduced in left and right sides of the to-be-knocked-out area; 4-OHT is used for inducing Cre-LoxP medicated homologous recombination, and cloning of target fragment knockout is obtained.
Owner:SUZHOU UNIV
SNP (Single Nucleotide Polymorphism) molecular marker related with back fat thickness trait of pig and detection method of SNP molecular marker
ActiveCN105255870AEarly detectionQuick checkMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionNucleotide
The invention provides an SNP (Single Nucleotide Polymorphism) molecular marker related with a back fat thickness trait of a pig and a detection method of the SNP molecular marker. By detecting exon regions of MARK4 (Microtubule Affinity-Regulating Kinase) genes of different pig species, one SNP locus (G-A) is found to exist at 55th basic group (mRNA (Messenger Ribonucleic Acid) total-length 2581th base group) in 3 minute UTR (Untranslated Region) of the MARK4 gene, an allele A is positively correlated with low back fat trait, and an allele G is positively correlated with high back fat trait. The invention provides the method for detecting the SNP molecular marker. Pig genomic DNA (Deoxyribonucleic Acid) to be detected is amplified by using a primer pair with the nucleotide sequence shown in SEQ ID NO.1-2 and is subjected to enzyme digestion by using pstI; if an enzyme-digested product is 251bp, a base group at a mutation is A; if the enzyme-digested product is 218bp, a base group at the mutation is G. According to the method provided by the invention, the back fat thickness of the pig can be predicted early, quickly and effectively at low cost; the SNP molecular marker can be used as a useful molecular marker for genetic improvement of the pig species.
Owner:湘潭市家畜育种站
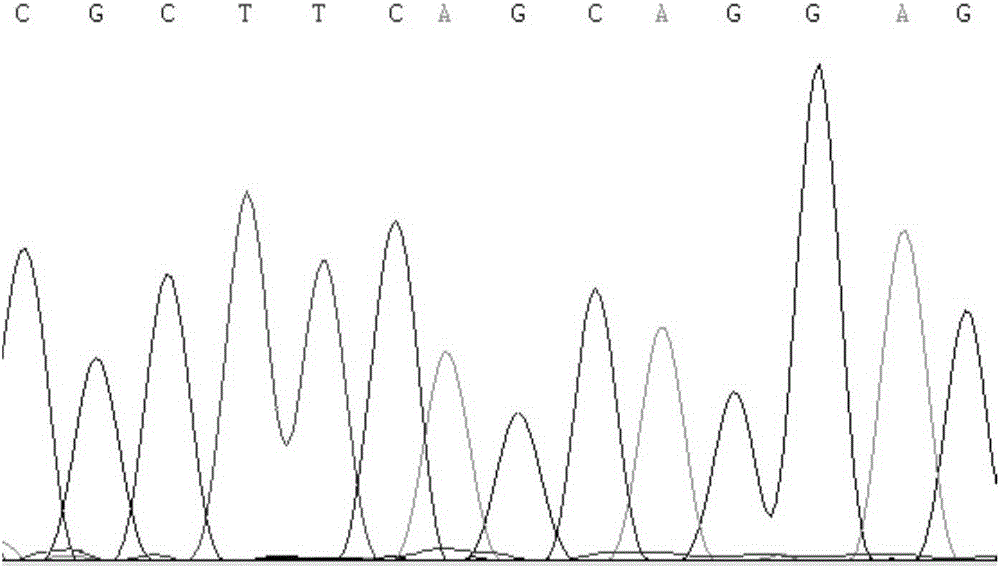
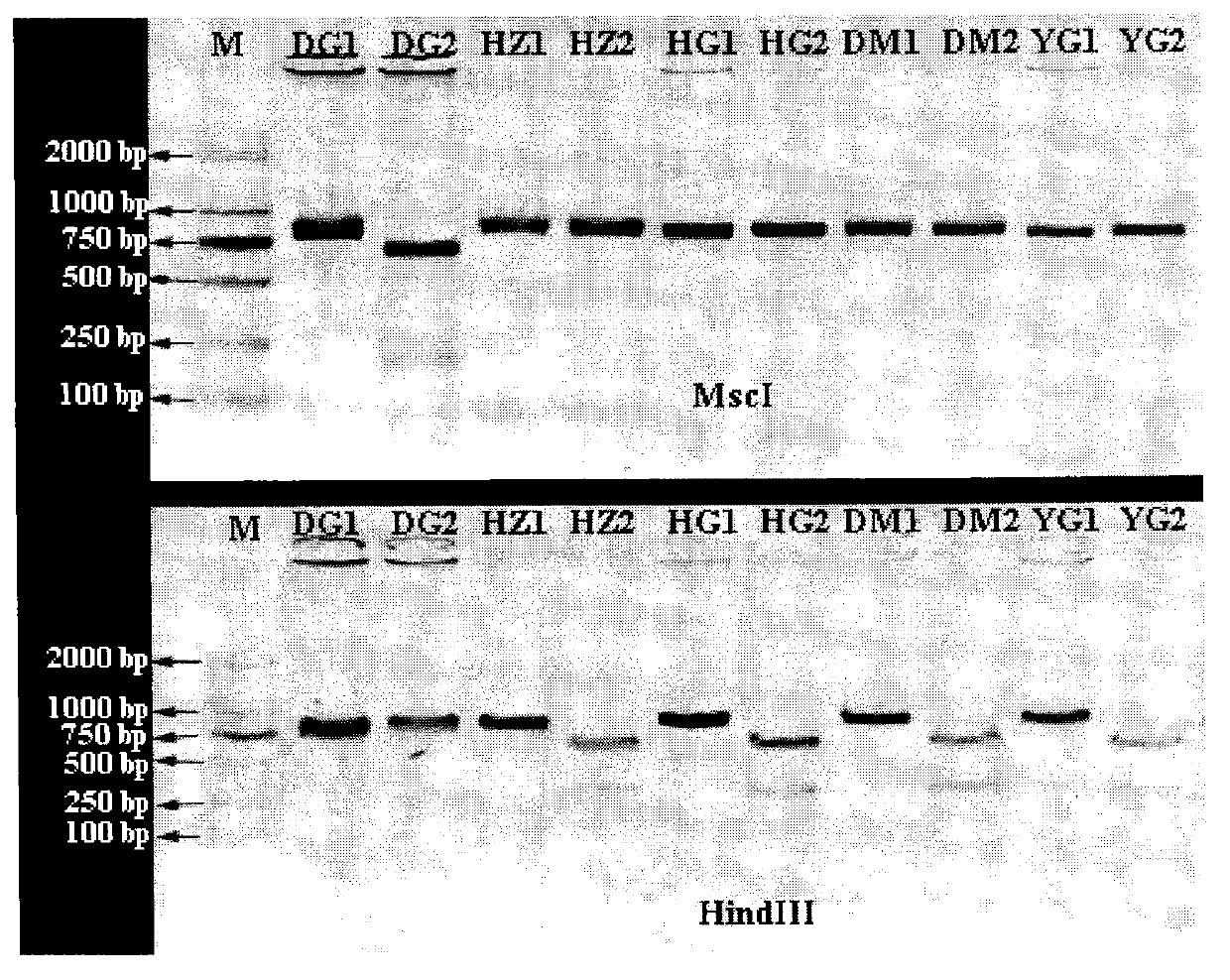
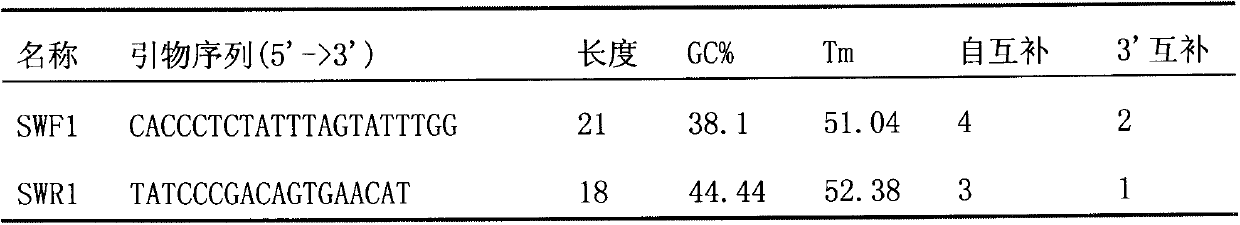
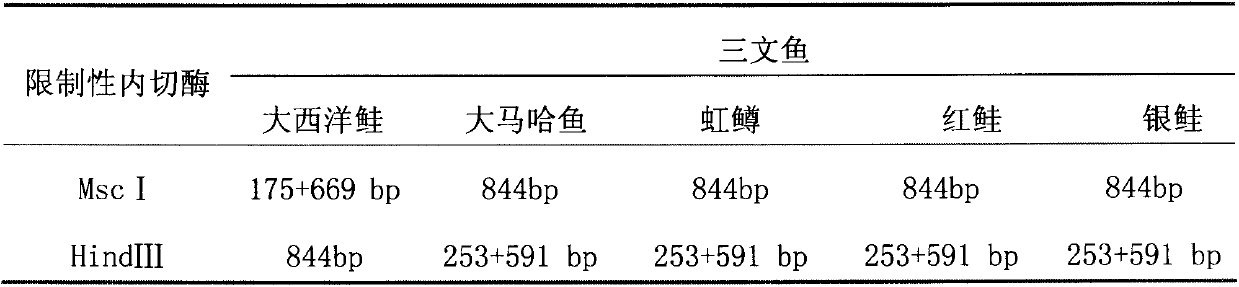
Molecular identification method of atlantic salmon
ActiveCN103122386ALower requirementEasy to operateMicrobiological testing/measurementMolecular identificationEnzyme digestion
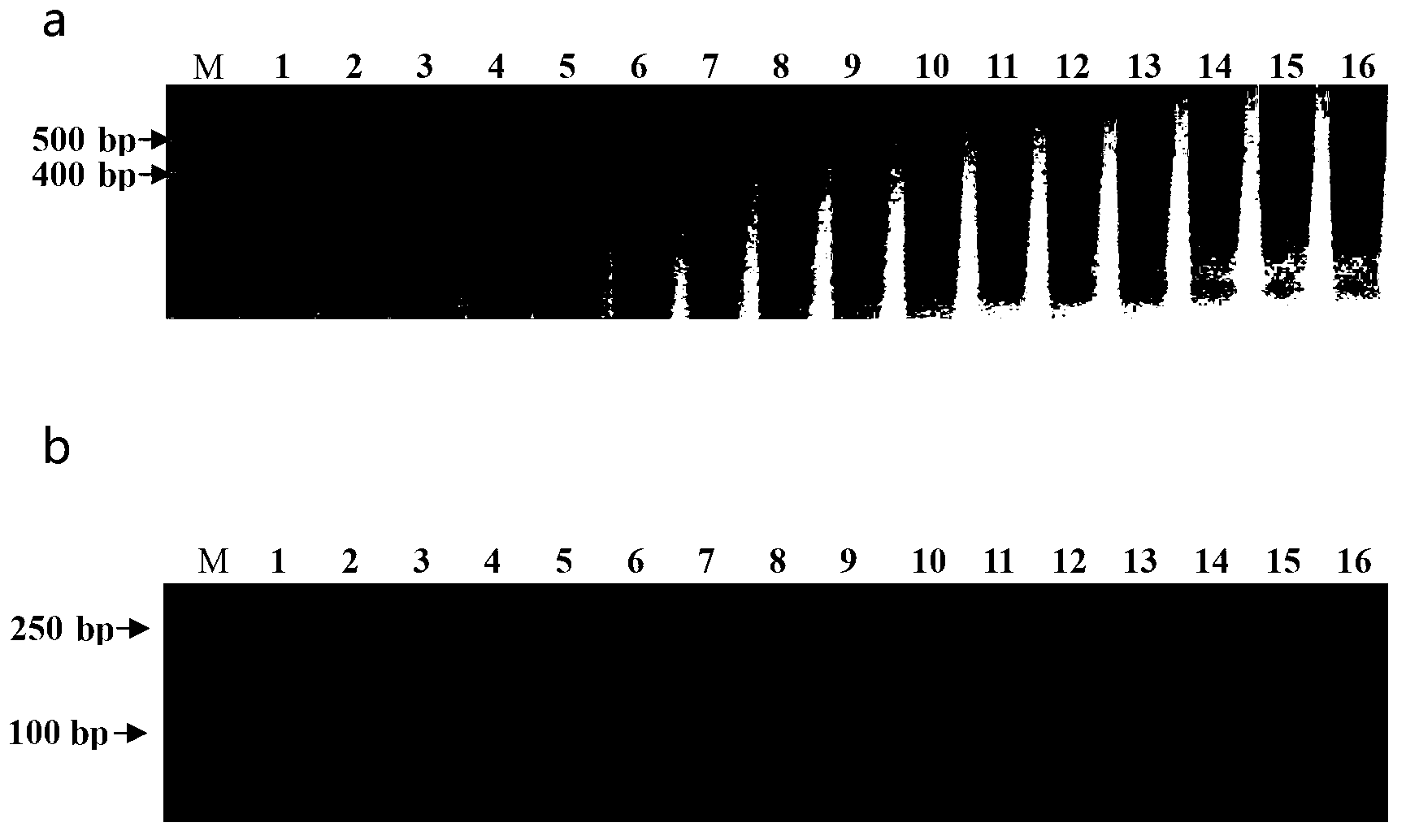
The invention discloses a molecular identification method of an atlantic salmon. The molecular identification method comprises the following steps of 1, extraction of fish DNA (deoxyribonucleic acid); 2, PCR (polymerase chain reaction) augmentation; 3, enzyme digestion and electrophoresis detection; and 4, identification. The atlantic salmon can be accurately determined in four steps. The molecular identification method of the atlantic salmon has the characteristics of being simple to operate, rapid and accurate, low in price, low in requirement on an instrument and equipment, and the like.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Polymerase chain reaction-sequence based typing (PCR-SBT) method for ABO blood type genotyping and reagent
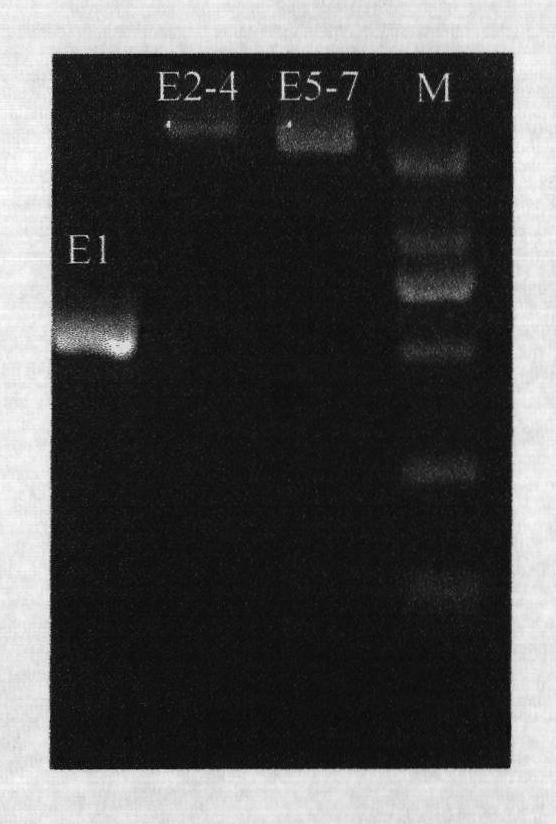
ActiveCN101921834ASolve the recombination phenomenonThe result is accurateMicrobiological testing/measurementSodium acetateHuman DNA sequencing
The invention provides a polymerase chain reaction-sequence based typing (PCR-SBT) method for ABO blood type genotyping. The method comprises the following steps of: preparing human genome DNA; amplifying segments of ABO gene exon 1, exons 2-4 and exons 5-7; performing double enzyme digestion purification on the obtained amplified products; performing a sequencing PCR reaction on the purified products; purifying the sequenced products by a sodium acetate-ethanol precipitation method and performing capillary electrophoresis sequencing; and analyzing the obtained sequences by using software to determine the genotype. The method has the advantages of solving the problems of identification of an ABO subtype, judgment of difficult blood types, discovery of a new mutational site, gene recombination among genes, genetic polymorphism detection and the like, exerting the characteristics of high flux and result accuracy of ABO genotyping operation by PCR-SBT, achieving great importance for the relative application in the fields of clinical transfusion medicinal research, genetics and the like and having important practical significance for medicinal research units, pharmic research and reagent development units.
Owner:浙江省血液中心
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com