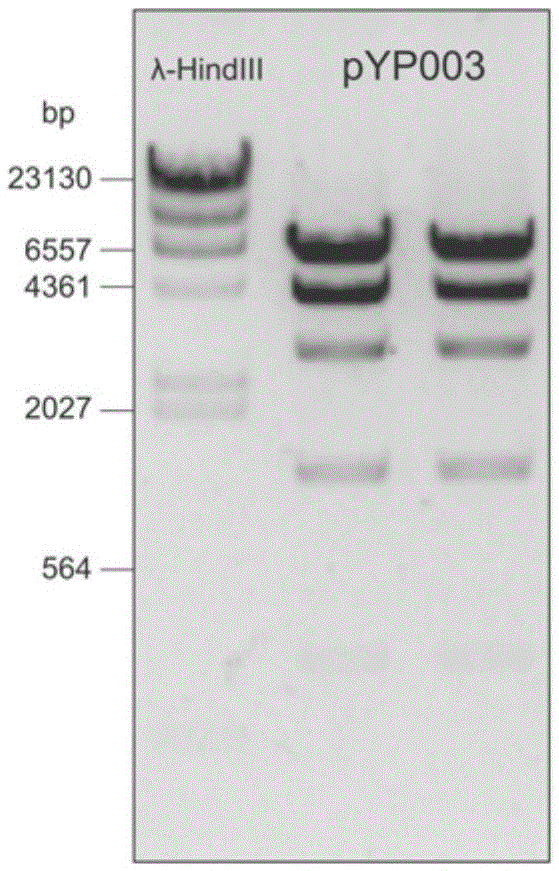
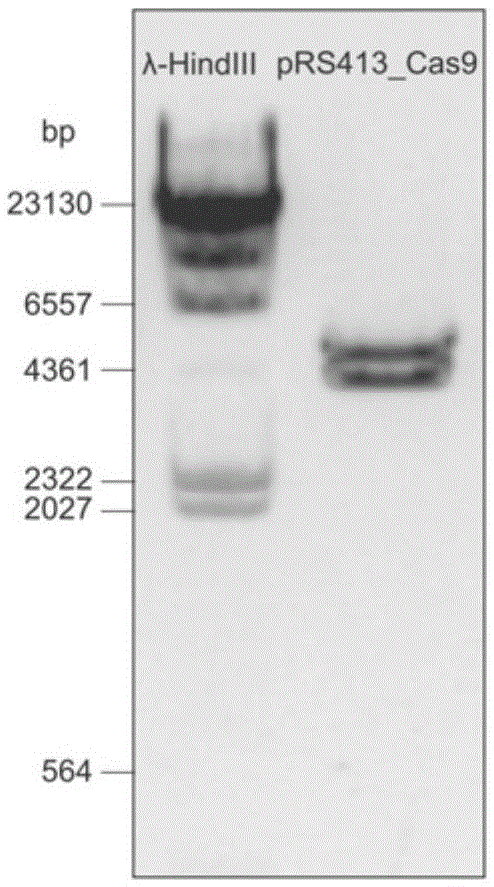
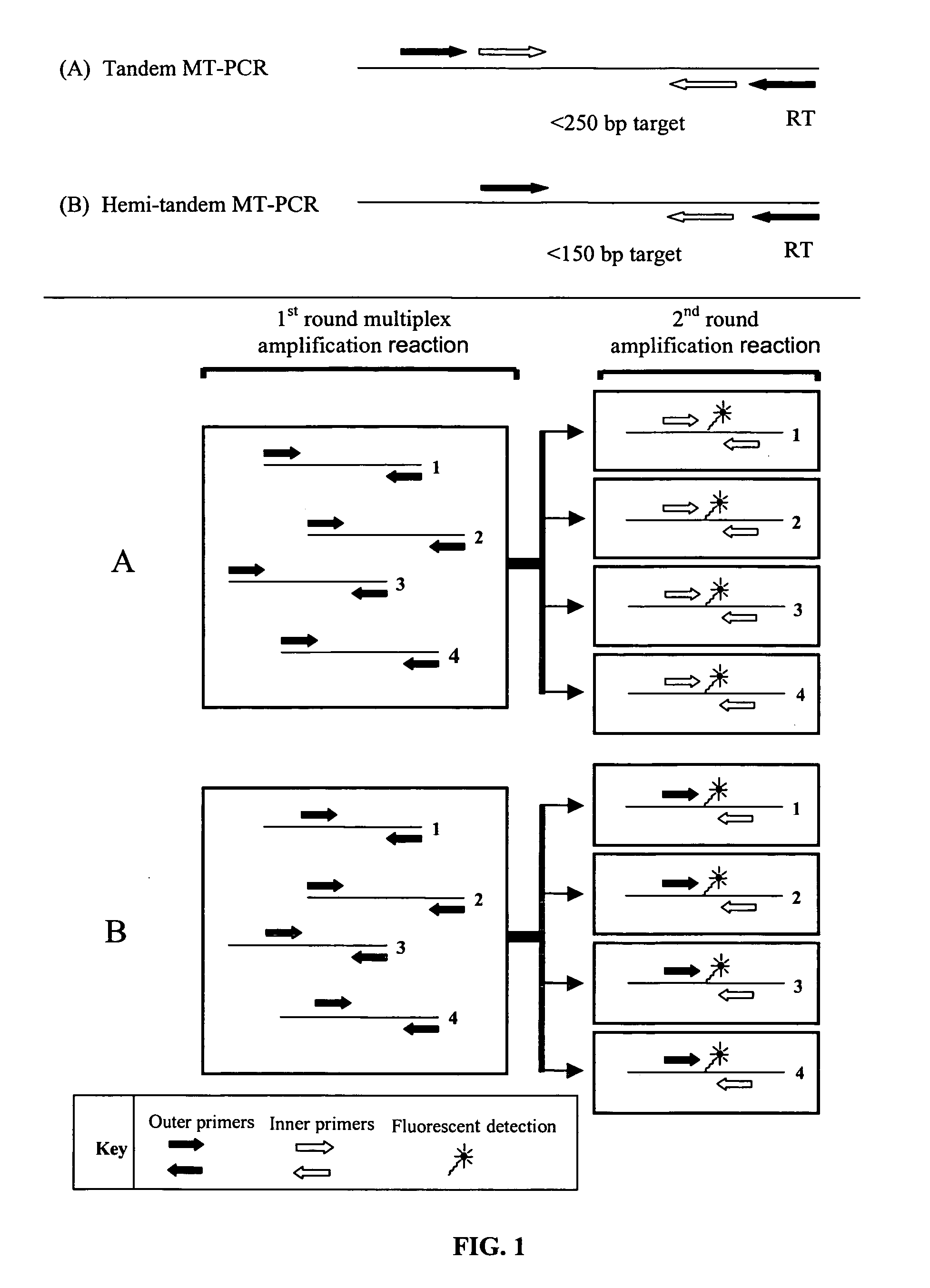
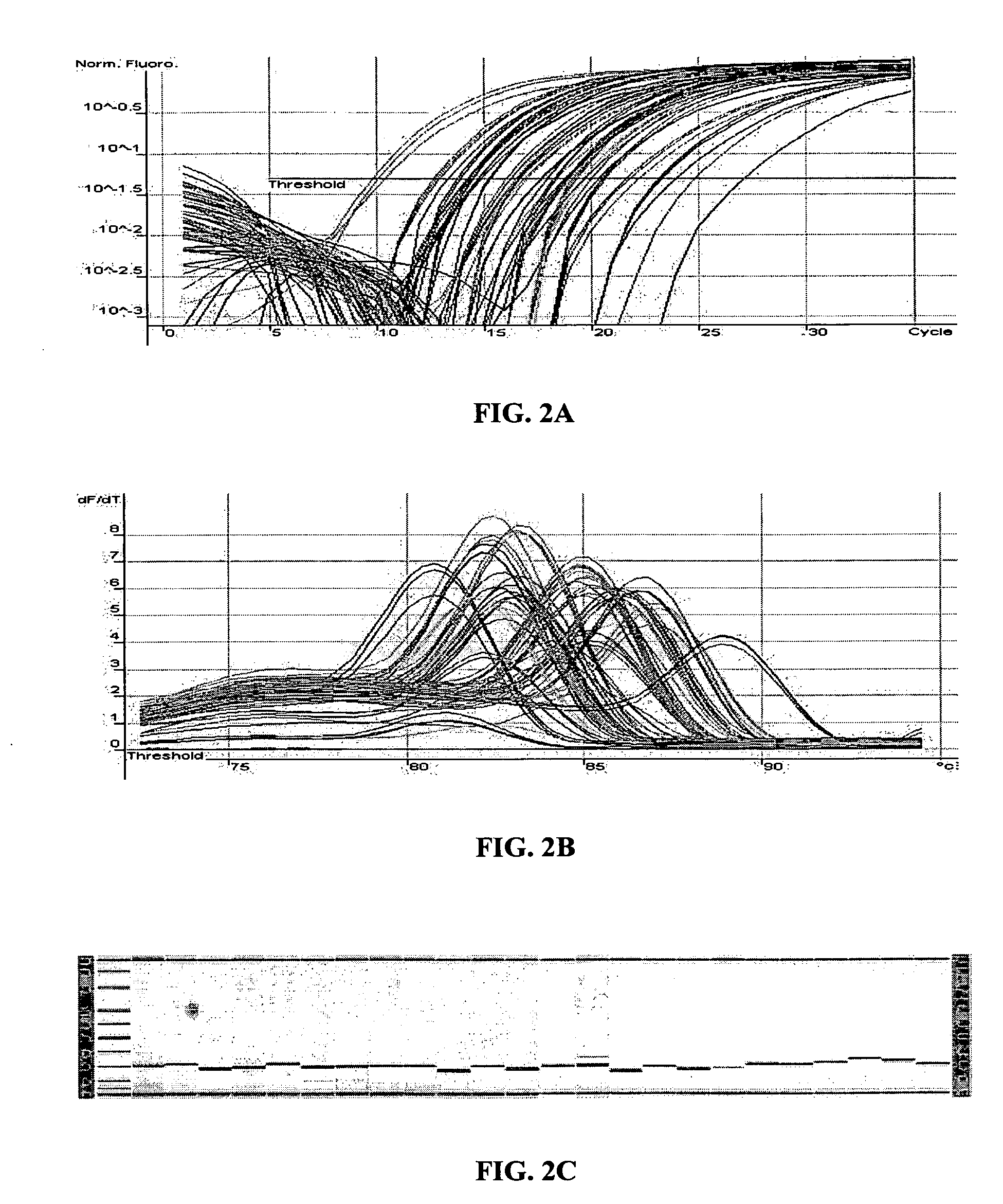
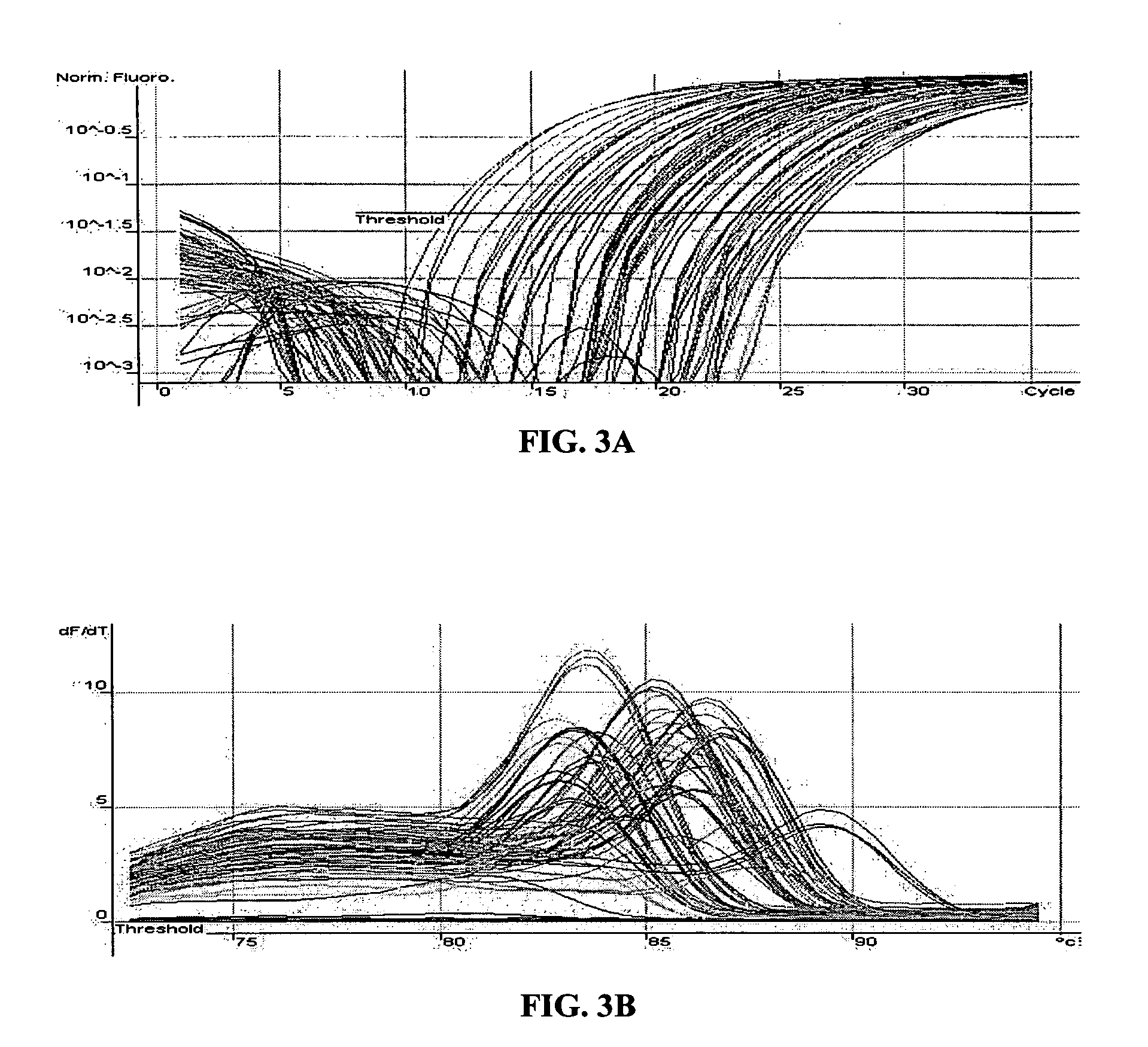
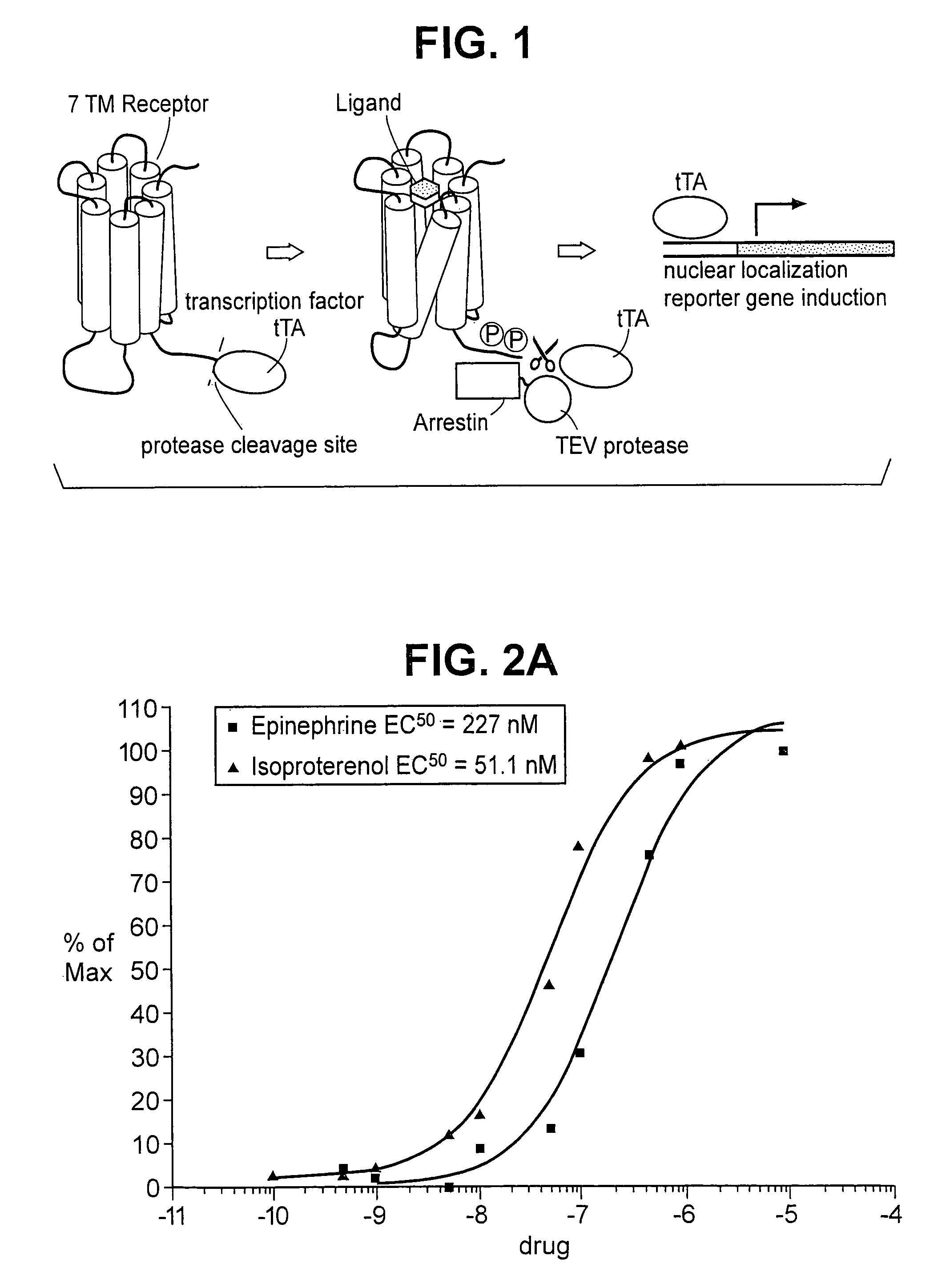
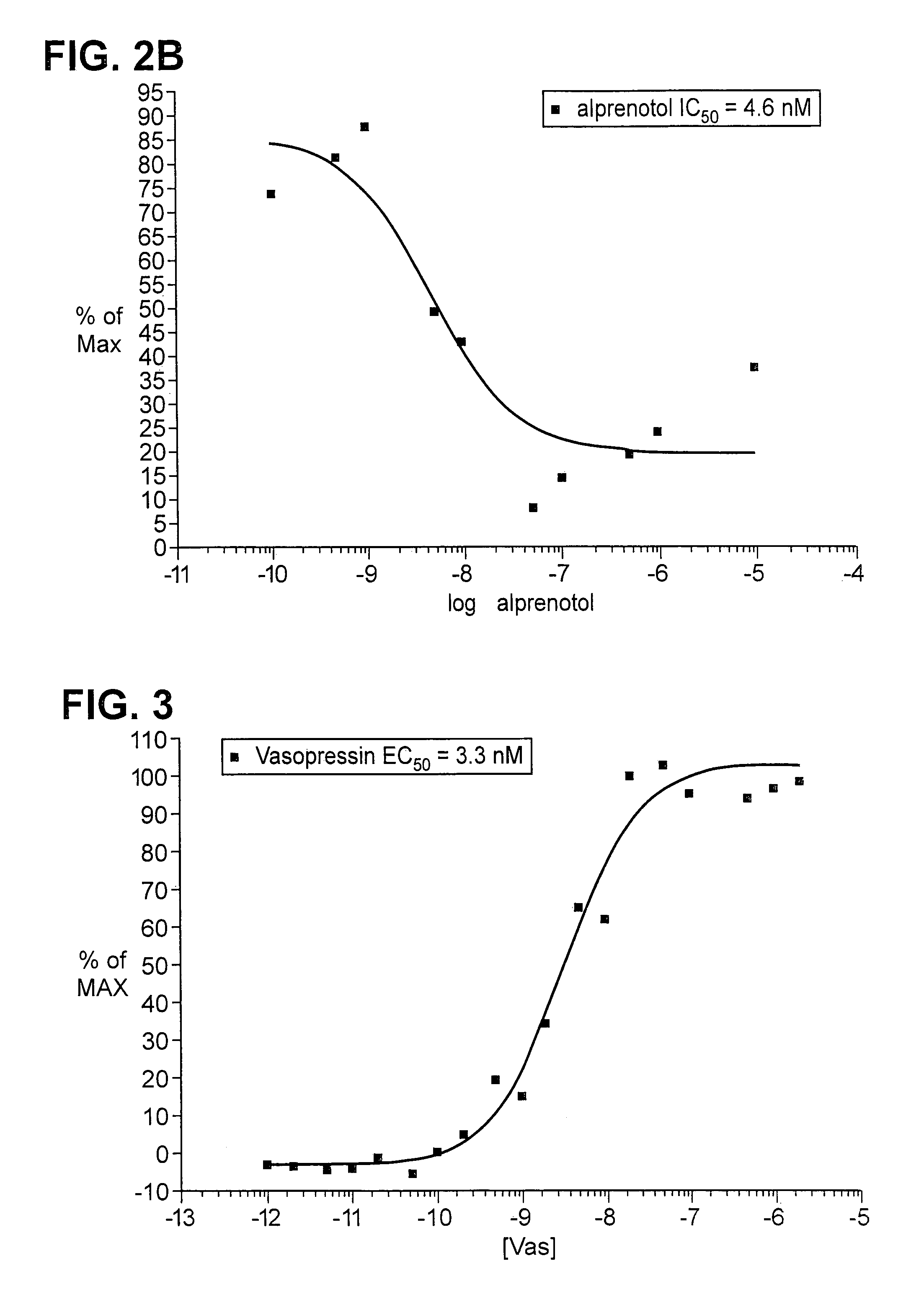
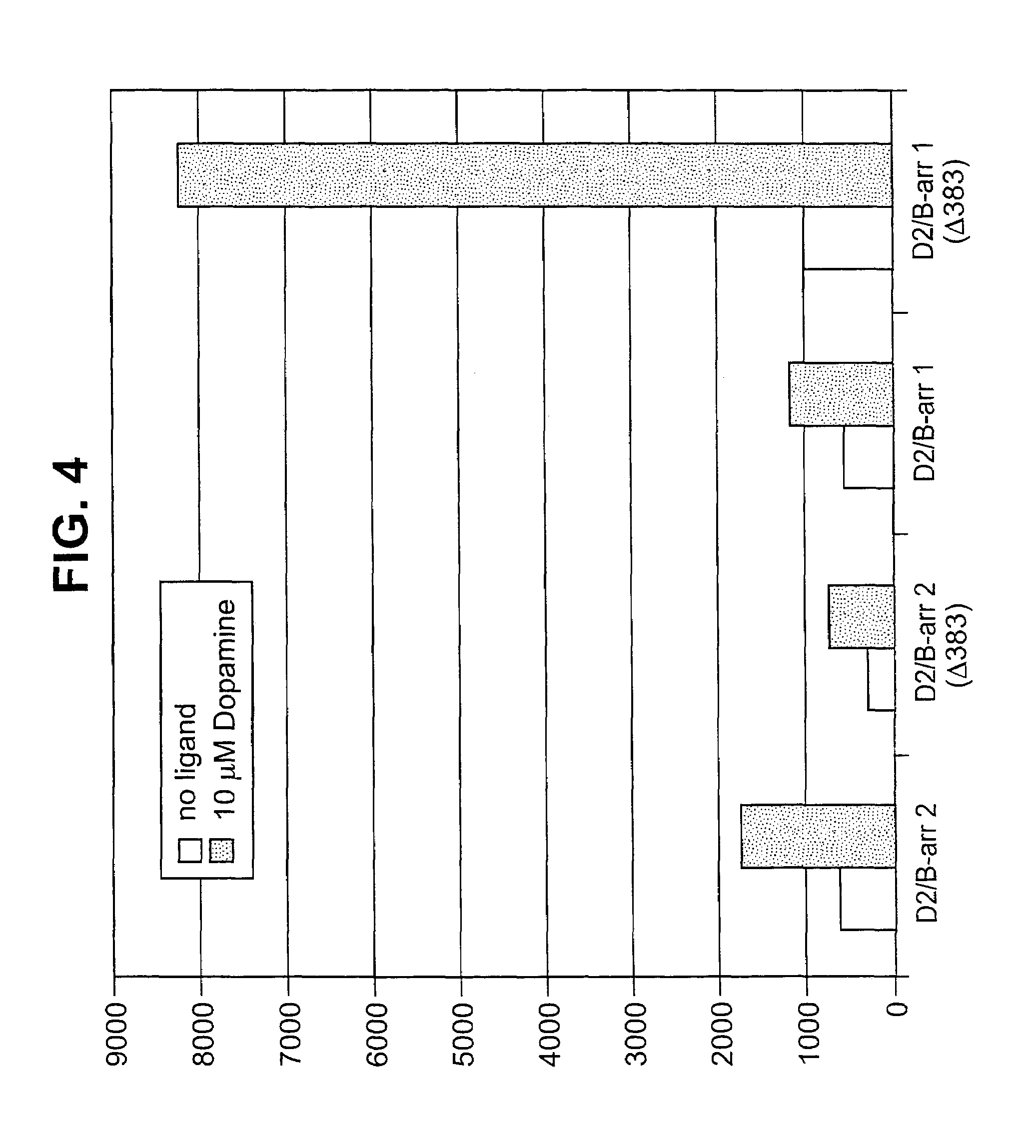
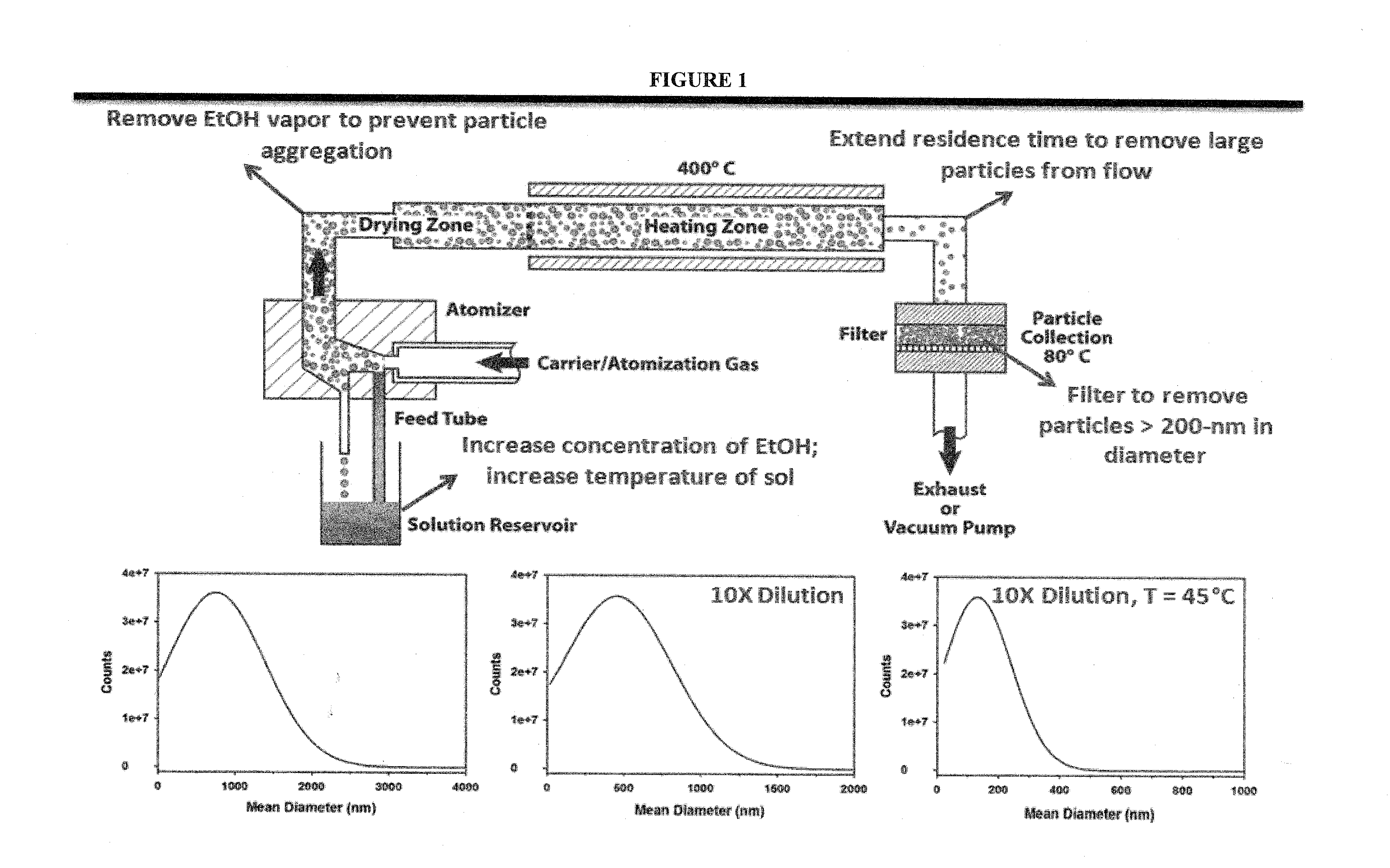
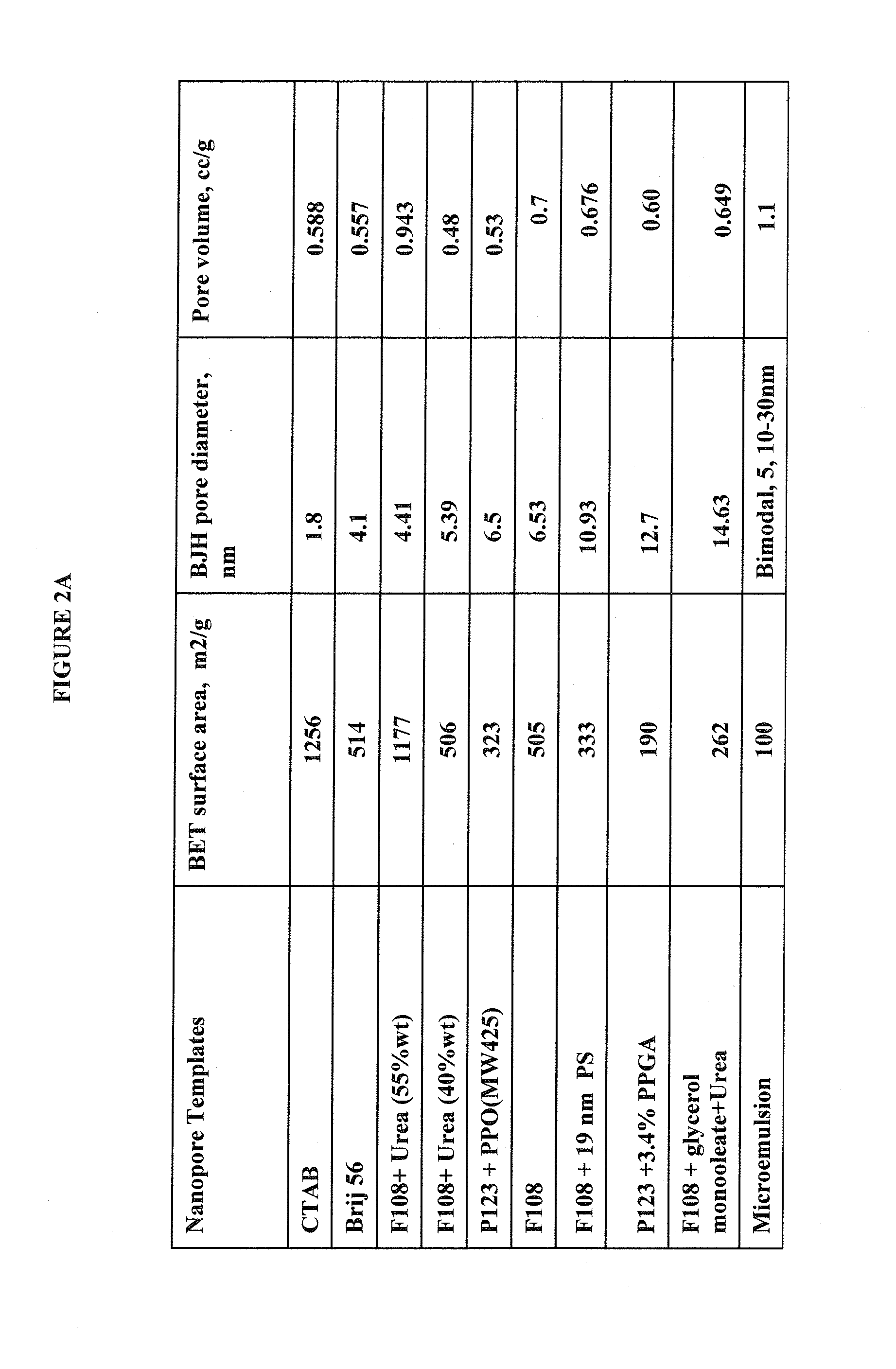
Patents
Literature
1554 results about "Reporter gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In molecular biology, a reporter gene (often simply reporter) is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Certain genes are chosen as reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population.
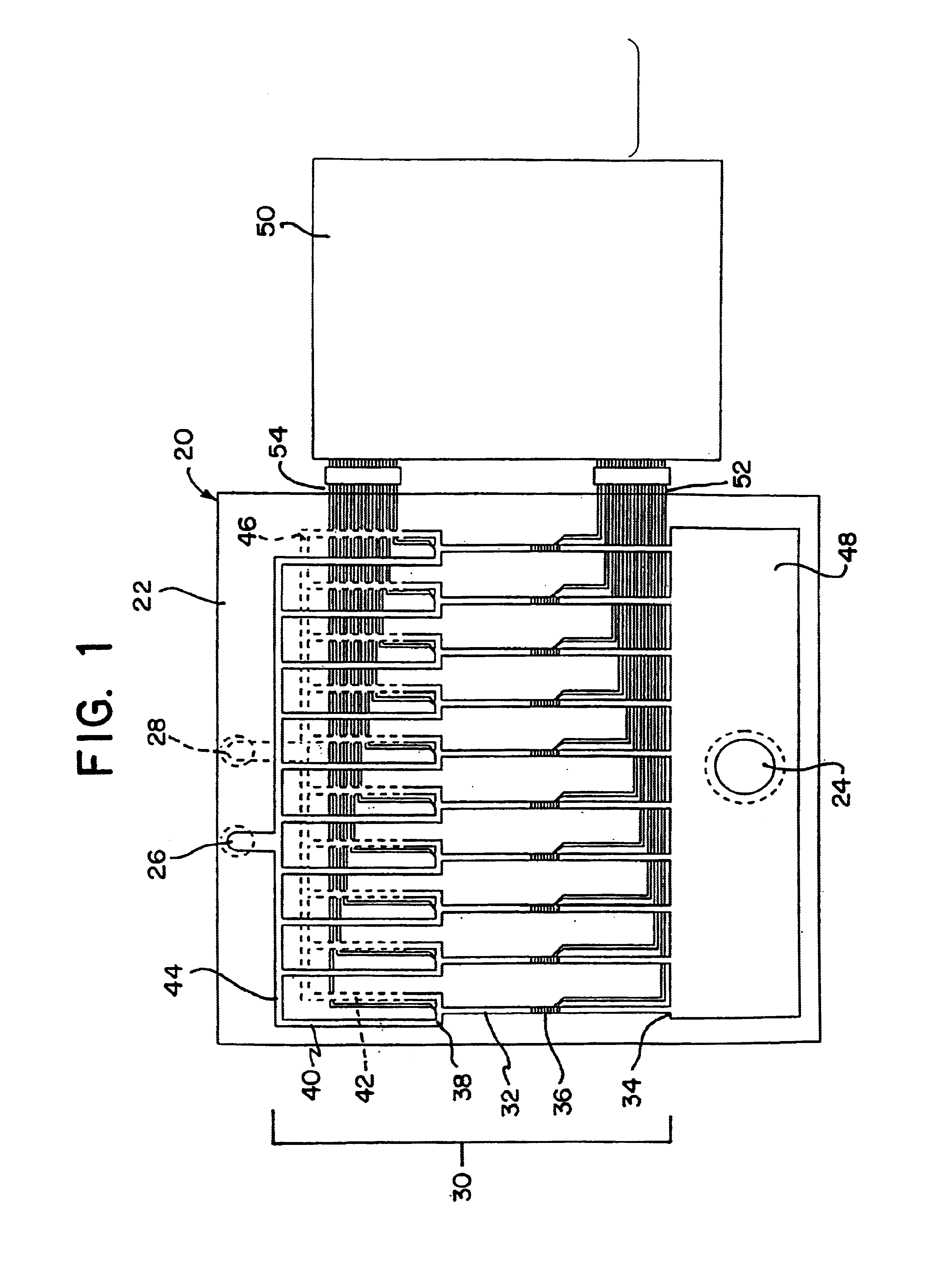
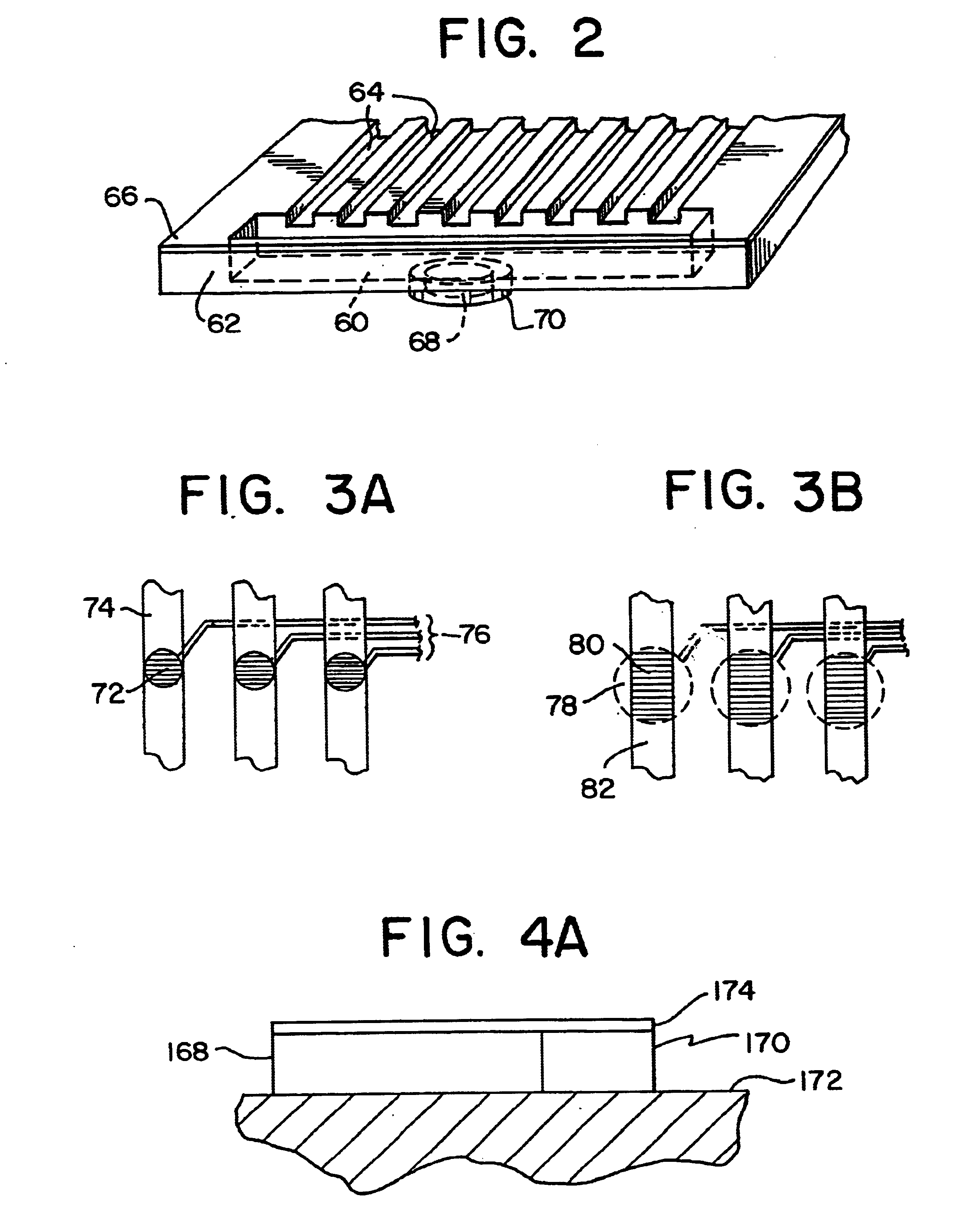
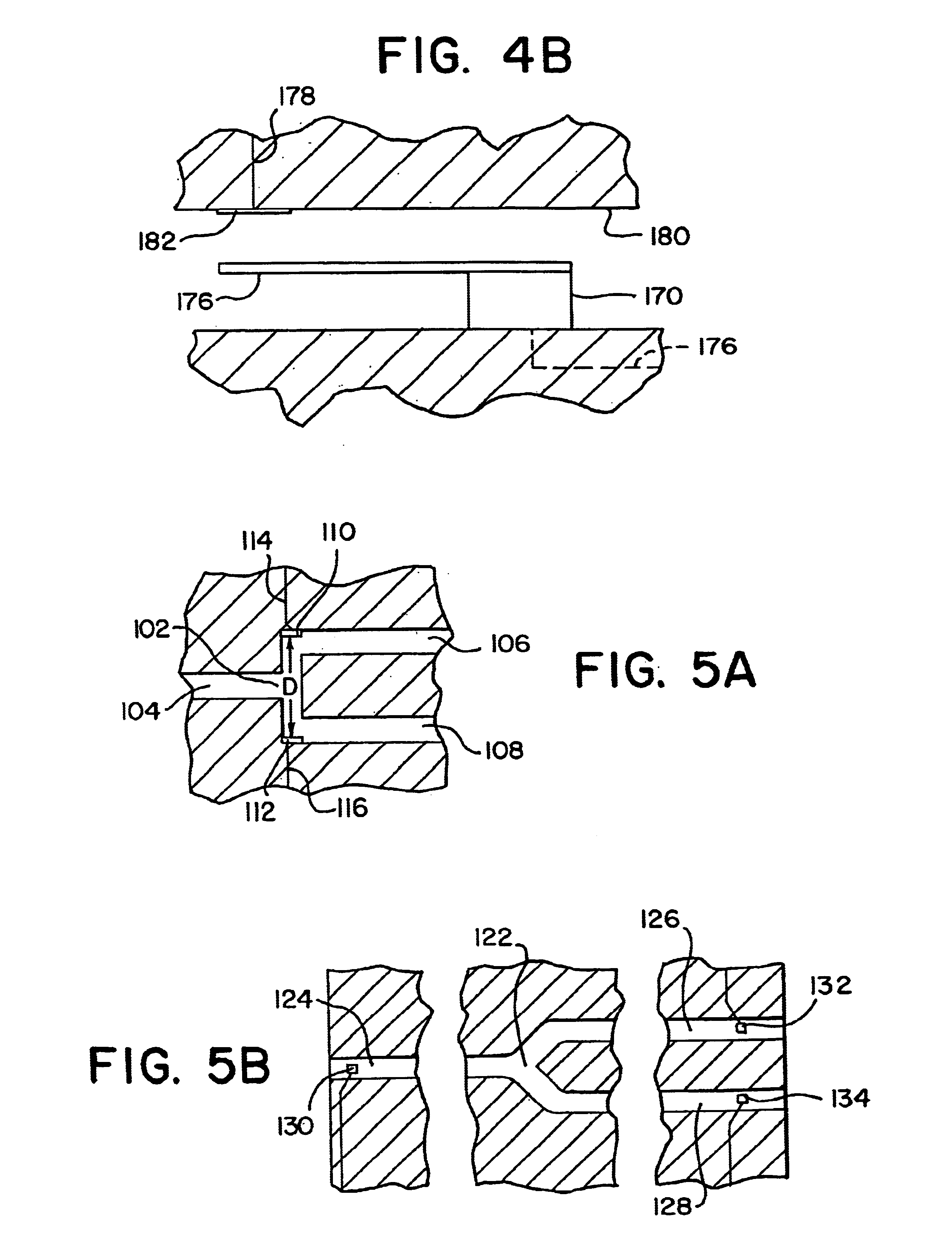
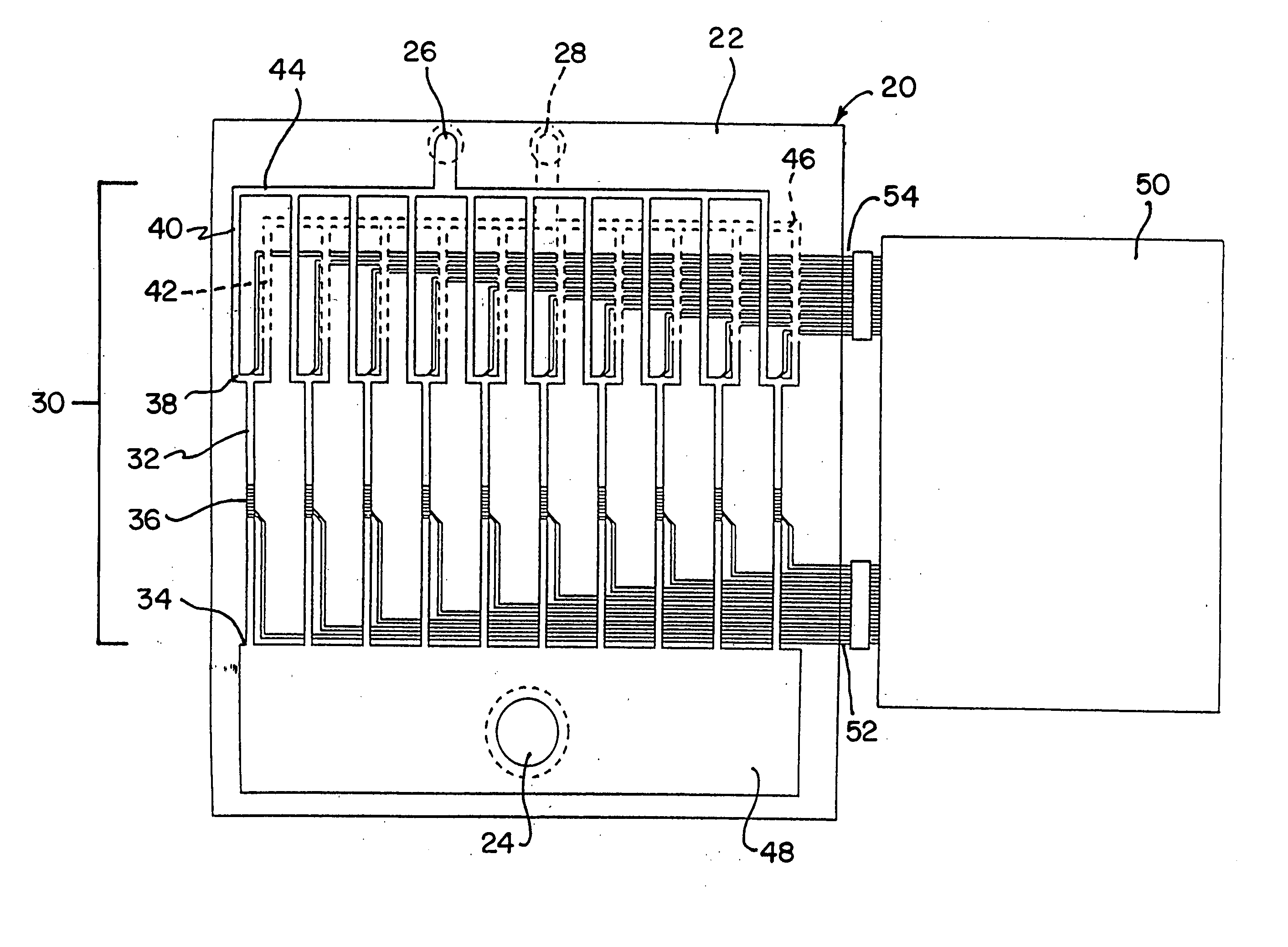
Integrated active flux microfluidic devices and methods
InactiveUS6767706B2Rapid and complete exposureQuick and accurate and inexpensive analysisBioreactor/fermenter combinationsFlow mixersAntigenHybridization probe
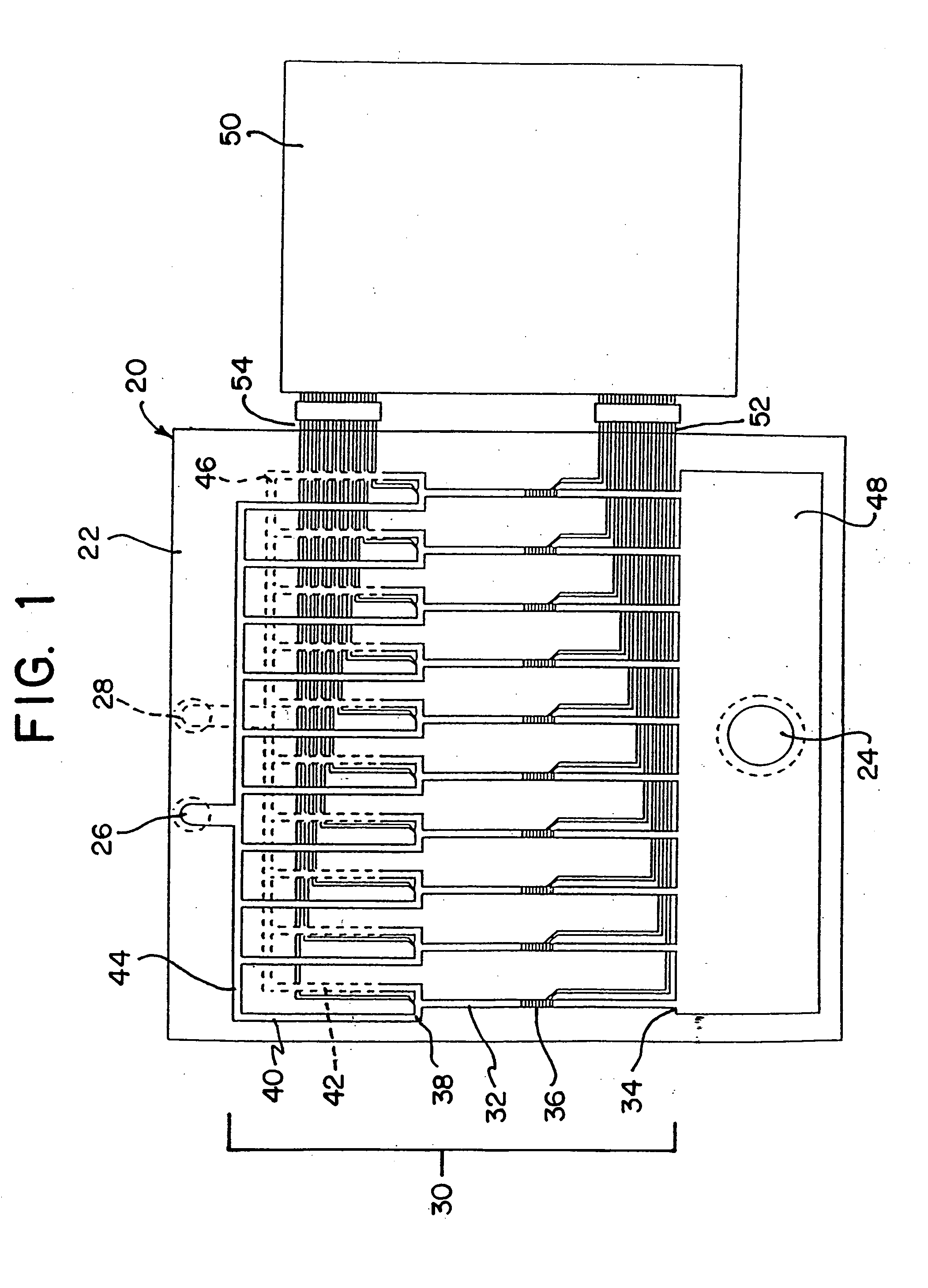
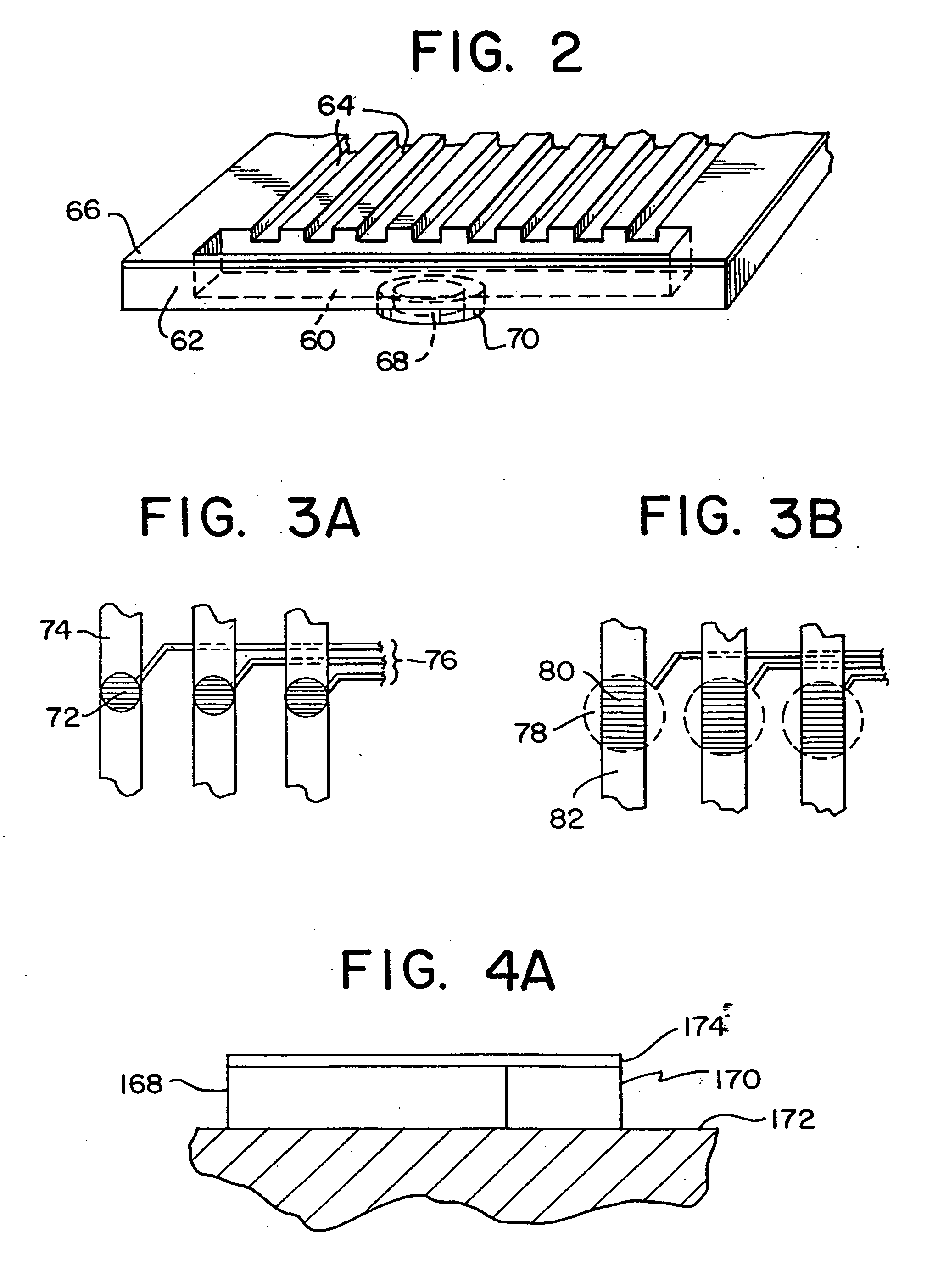
The invention relates to a microfabricated device for the rapid detection of DNA, proteins or other molecules associated with a particular disease. The devices and methods of the invention can be used for the simultaneous diagnosis of multiple diseases by detecting molecules (e.g. amounts of molecules), such as polynucleotides (e.g., DNA) or proteins (e.g., antibodies), by measuring the signal of a detectable reporter associated with hybridized polynucleotides or antigen / antibody complex. In the microfabricated device according to the invention, detection of the presence of molecules (i.e., polynucleotides, proteins, or antigen / antibody complexes) are correlated to a hybridization signal from an optically-detectable (e.g. fluorescent) reporter associated with the bound molecules. These hybridization signals can be detected by any suitable means, for example optical, and can be stored for example in a computer as a representation of the presence of a particular gene. Hybridization probes can be immobilized on a substrate that forms part of or is exposed to a channel or channels of the device that form a closed loop, for circulation of sample to actively contact complementary probes. Universal chips according to the invention can be fabricated not only with DNA but also with other molecules such as RNA, proteins, peptide nucleic acid (PNA) and polyamide molecules.
Owner:CALIFORNIA INST OF TECH
Integrated active flux microfluidic devices and methods
InactiveUS20040248167A1Increase speedImprove accuracyBioreactor/fermenter combinationsFlow mixersAntigenHybridization probe
The invention relates to a microfabricated device for the rapid detection of DNA, proteins or other molecules associated with a particular disease. The devices and methods of the invention can be used for the simultaneous diagnosis of multiple diseases by detecting molecules (e.g. amounts of molecules), such as polynucleotides (e.g., DNA) or proteins (e.g., antibodies), by measuring the signal of a detectable reporter associated with hybridized polynucleotides or antigen / antibody complex. In the microfabricated device according to the invention, detection of the presence of molecules (i.e., polynucleotides, proteins, or antigen / antibody complexes) are correlated to a hybridization signal from an optically-detectable (e.g. fluorescent) reporter associated with the bound molecules. These hybridization signals can be detected by any suitable means, for example optical, and can be stored for example in a computer as a representation of the presence of a particular gene. Hybridization probes can be immobilized on a substrate that forms part of or is exposed to a channel or channels of the device that form a closed loop, for circulation of sample to actively contact complementary probes. Universal chips according to the invention can be fabricated not only with DNA but also with other molecules such as RNA, proteins, peptide nucleic acid (PNA) and polyamide molecules.
Owner:CALIFORNIA INST OF TECH
Identification and comparison of protein-protein interactions that occur in populations and identification of inhibitors of these interactors
InactiveUS6057101AEfficient screeningLess experimentally significant and specific indicationMaterial nanotechnologyFungiDiseaseBinding site
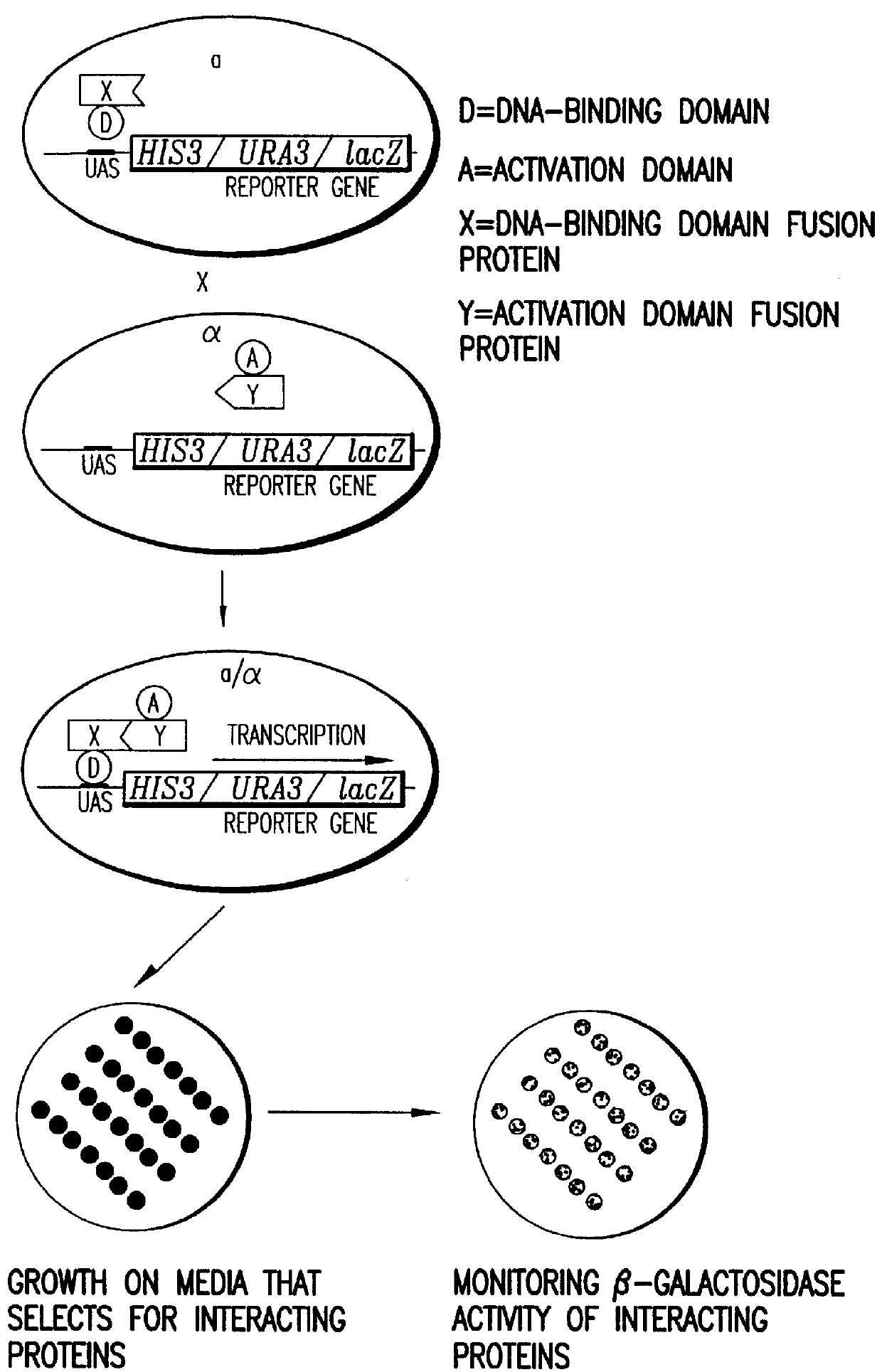
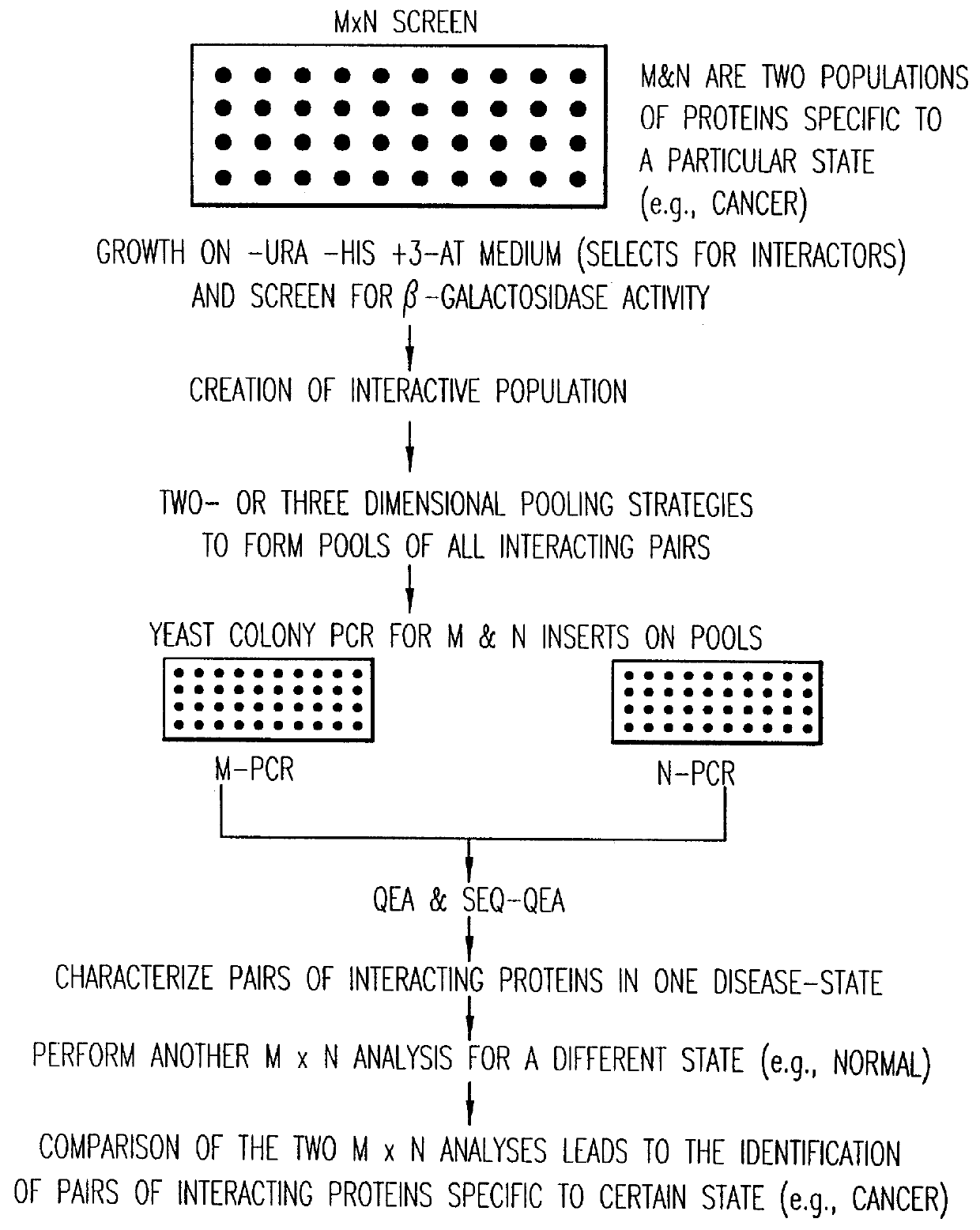
Methods are described for detecting protein-protein interactions, among two populations of proteins, each having a complexity of at least 1,000. For example, proteins are fused either to the DNA-binding domain of a transcriptional activator or to the activation domain of a transcriptional activator. Two yeast strains, of the opposite mating type and carrying one type each of the fusion proteins are mated together. Productive interactions between the two halves due to protein-protein interactions lead to the reconstitution of the transcriptional activator, which in turn leads to the activation of a reporter gene containing a binding site for the DNA-binding domain. This analysis can be carried out for two or more populations of proteins. The differences in the genes encoding the proteins involved in the protein-protein interactions are characterized, thus leading to the identification of specific protein-protein interactions, and the genes encoding the interacting proteins, relevant to a particular tissue, stage or disease. Furthermore, inhibitors that interfere with these protein-protein interactions are identified by their ability to inactivate a reporter gene. The screening for such inhibitors can be in a multiplexed format where a set of inhibitors will be screened against a library of interactors. Further, information-processing methods and systems are described. These methods and systems provide for identification of the genes coding for detected interacting proteins, for assembling a unified database of protein-protein interaction data, and for processing this unified database to obtain protein interaction domain and protein pathway information.
Owner:CURAGEN CORP
Screening for west nile virus antiviral therapy
InactiveUS20050058987A1Improve efficiencySsRNA viruses positive-senseVectorsHigh-Throughput Screening MethodsImmunogenicity
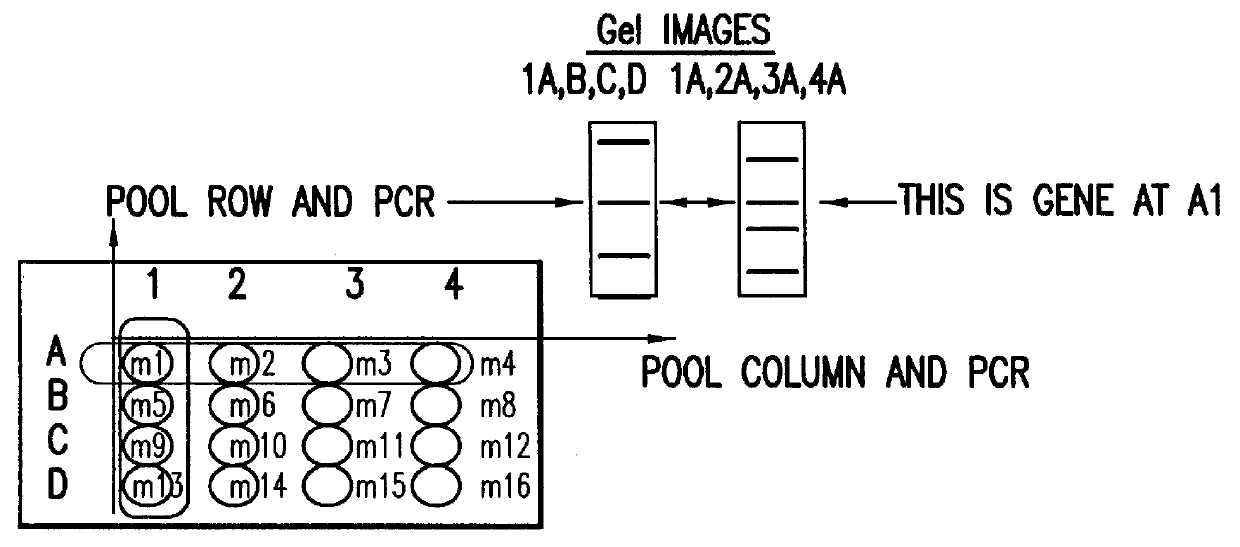
The instant invention provides stable and novel lineage I WNV reverse genetics systems, and methods for making the reverse genetics systems, specifically, a fully-infectious lineage I WNV cDNA or replicon system engineered with one or more nucleotide sequences each encoding a reporter gene to be used in high throughput cell-based screening assays for the identification of novel antiflaviviral chemotherapeutics and / or vaccines effective to treat and / or immunize against infections by WNV and other emerging flaviviruses, such as, for example, JEV, SLEV, AV, KV, JV, CV, YV, TBEV, DENV-1, DENV-2, DENV-3, DENV-4, YFV and MVEV. The present invention further provides methods of high throughput screening of antiflaviviral compounds or improved derivatives thereof using novel lineage I WNV reverse genetics systems and / or cell lines stably containing the reverse genetics systems. Also, the invention provides novel pharmaceutical compositions comprising an attenuated lineage I WNV that is less virulent but similarly immunogenic as the parent WNV and is capable of providing a protective immune response in a host.
Owner:HEALTH RES INC
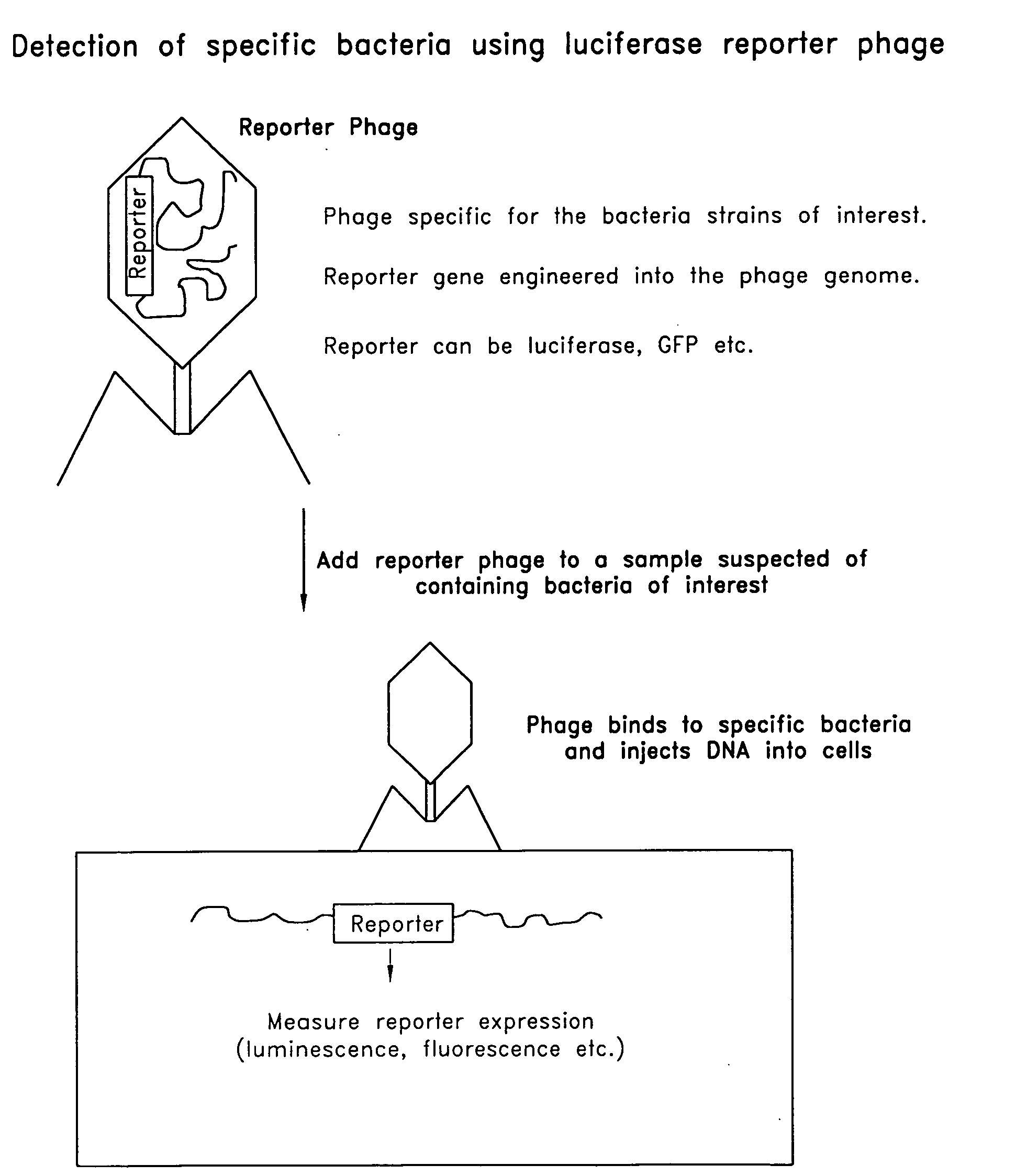
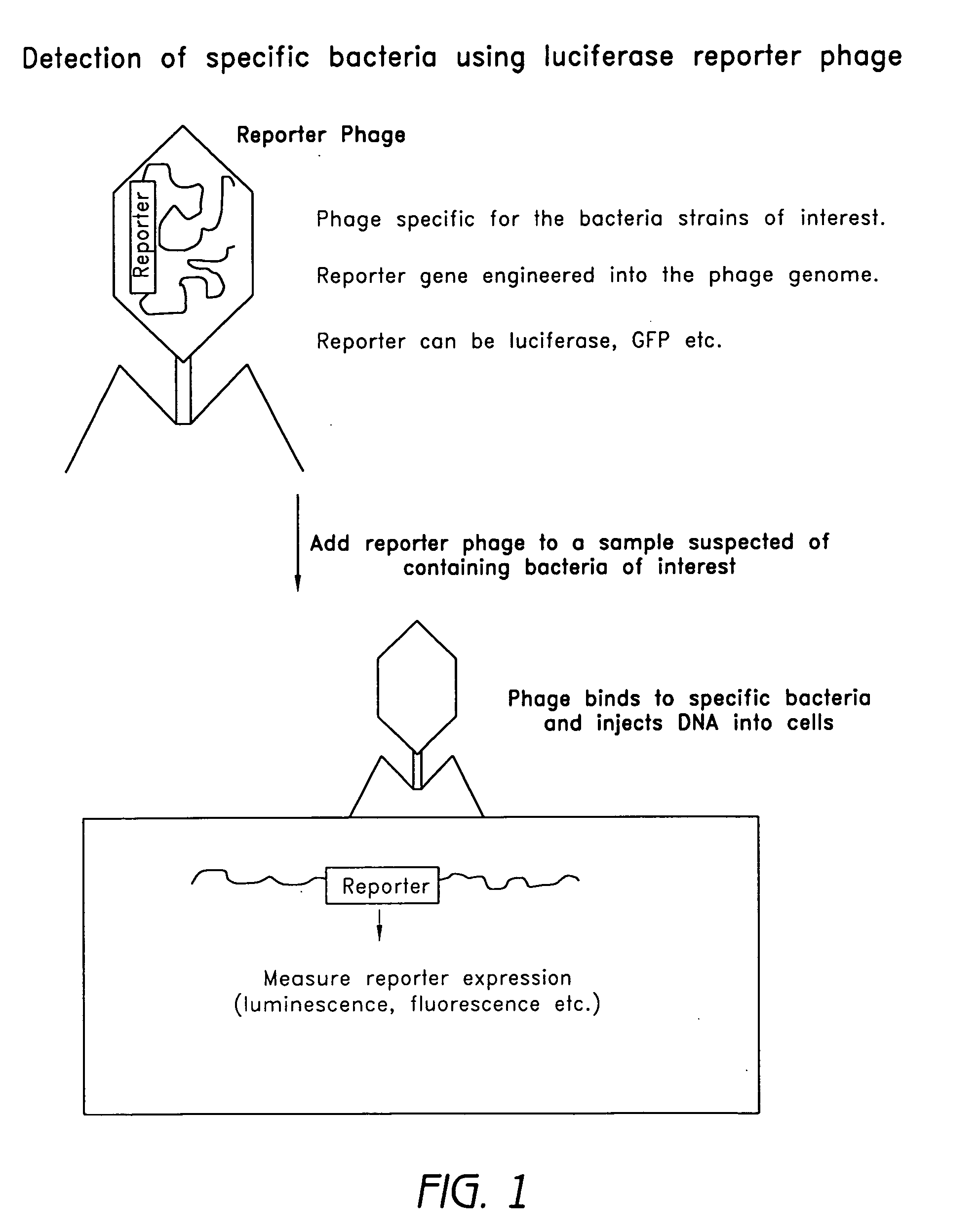
Reporter plasmid phage packaging system for detection of bacteria
InactiveUS20090155768A1Microbiological testing/measurementOther foreign material introduction processesBacteroidesOrigin of replication
The invention is related to a transducing particle that comprises a bacteriophage coat and a DNA core that comprises plasmid DNA comprising: a) a host-specific bacteriophage packaging site wherein the packaging site is substantially in isolation from sequences naturally occurring adjacent thereto in the bacteriophage genome, b) a reporter gene, c) a bacteria-specific promoter operably linked to said reporter gene, d) a bacteria-specific origin of replication, and optionally e) an antibiotic resistance gene. The invention includes phage transducing particles, methods of making transducing particles, and methods of using the transducing particles in bacterial detection.
Owner:UNITED STATES OF AMERICA
System and Method for Localization of Large Numbers of Fluorescent Markers in Biological Samples
ActiveUS20120193530A1Reduce respective collection solid angleExtensive collectionMaterial analysis using wave/particle radiationElectric discharge tubesFluorescenceQuantum dot
A method and system for the imaging and localization of fluorescent markers such as fluorescent proteins or quantum dots within biological samples is disclosed. The use of recombinant genetics technology to insert “reporter” genes into many species is well established. In particular, green fluorescent proteins (GFPs) and their genetically-modified variants ranging from blue to yellow, are easily spliced into many genomes at the sites of genes of interest (GoIs), where the GFPs are expressed with no apparent effect on the functioning of the proteins of interest (PoIs) coded for by the GoIs. One goal of biologists is more precise localization of PoIs within cells. The invention is a method and system for enabling more rapid and precise PoI localization using charged particle beam-induced damage to GFPs. Multiple embodiments of systems for implementing the method are presented, along with an image processing method relatively immune to high statistical noise levels.
Owner:FEI CO
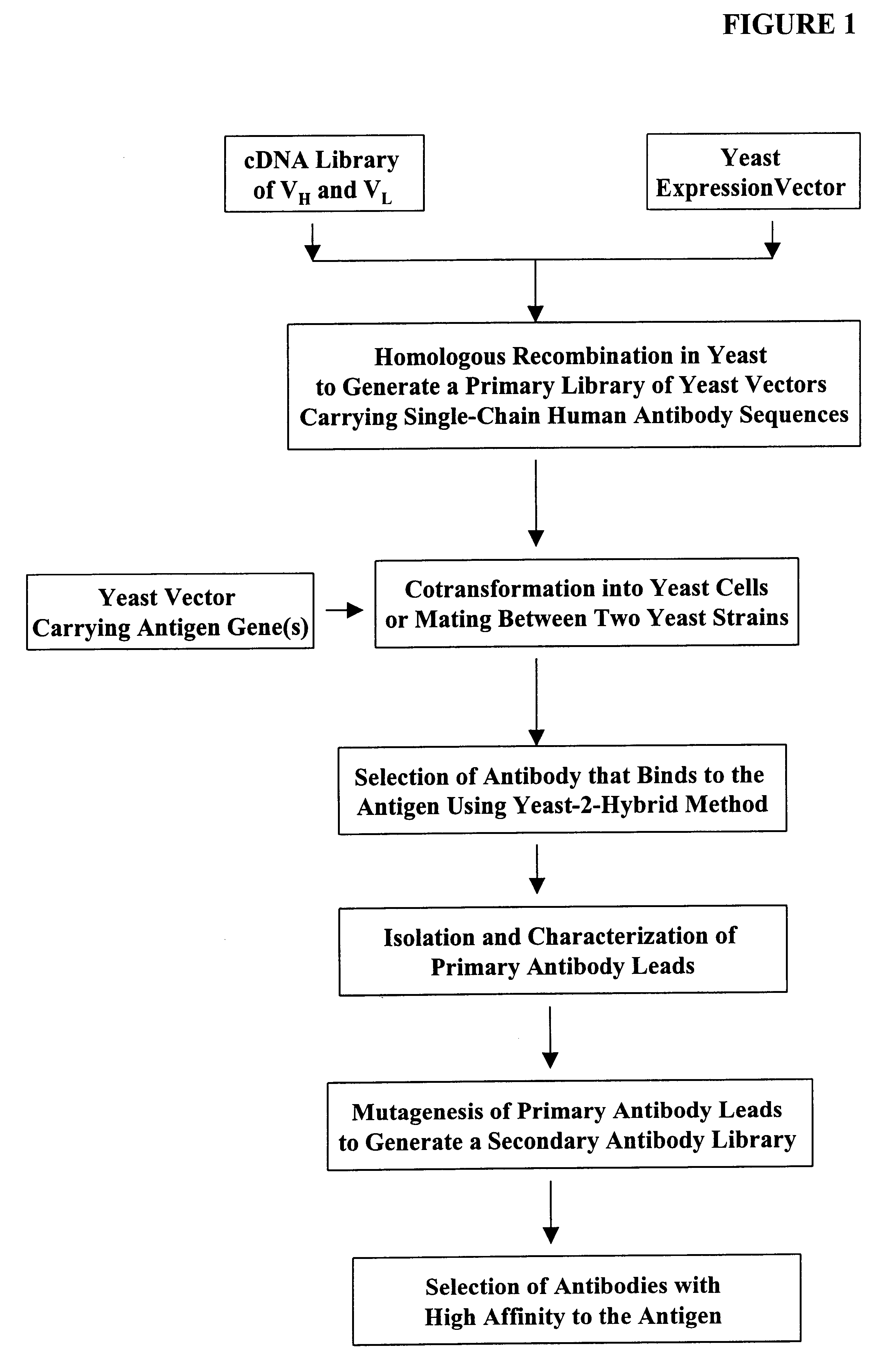
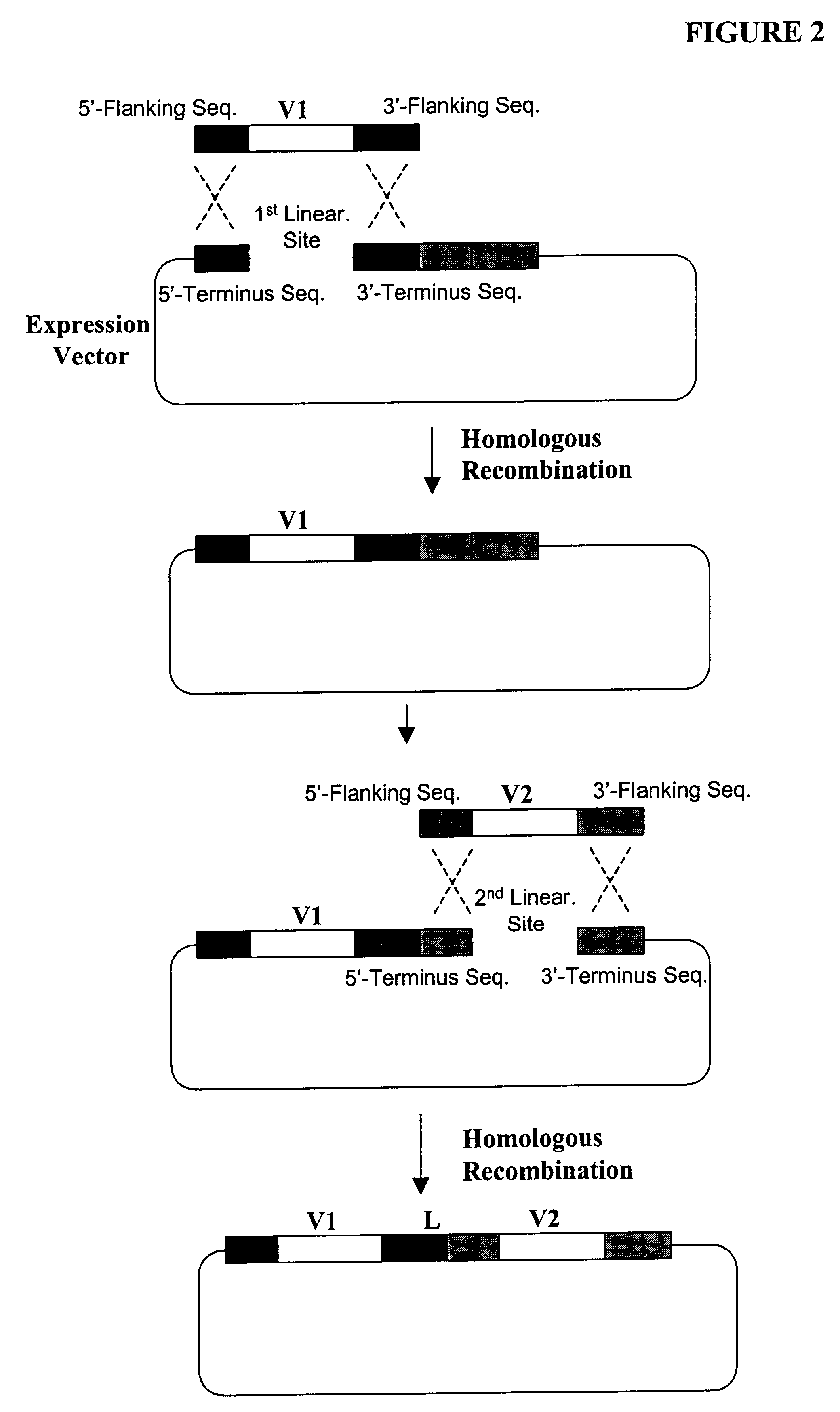
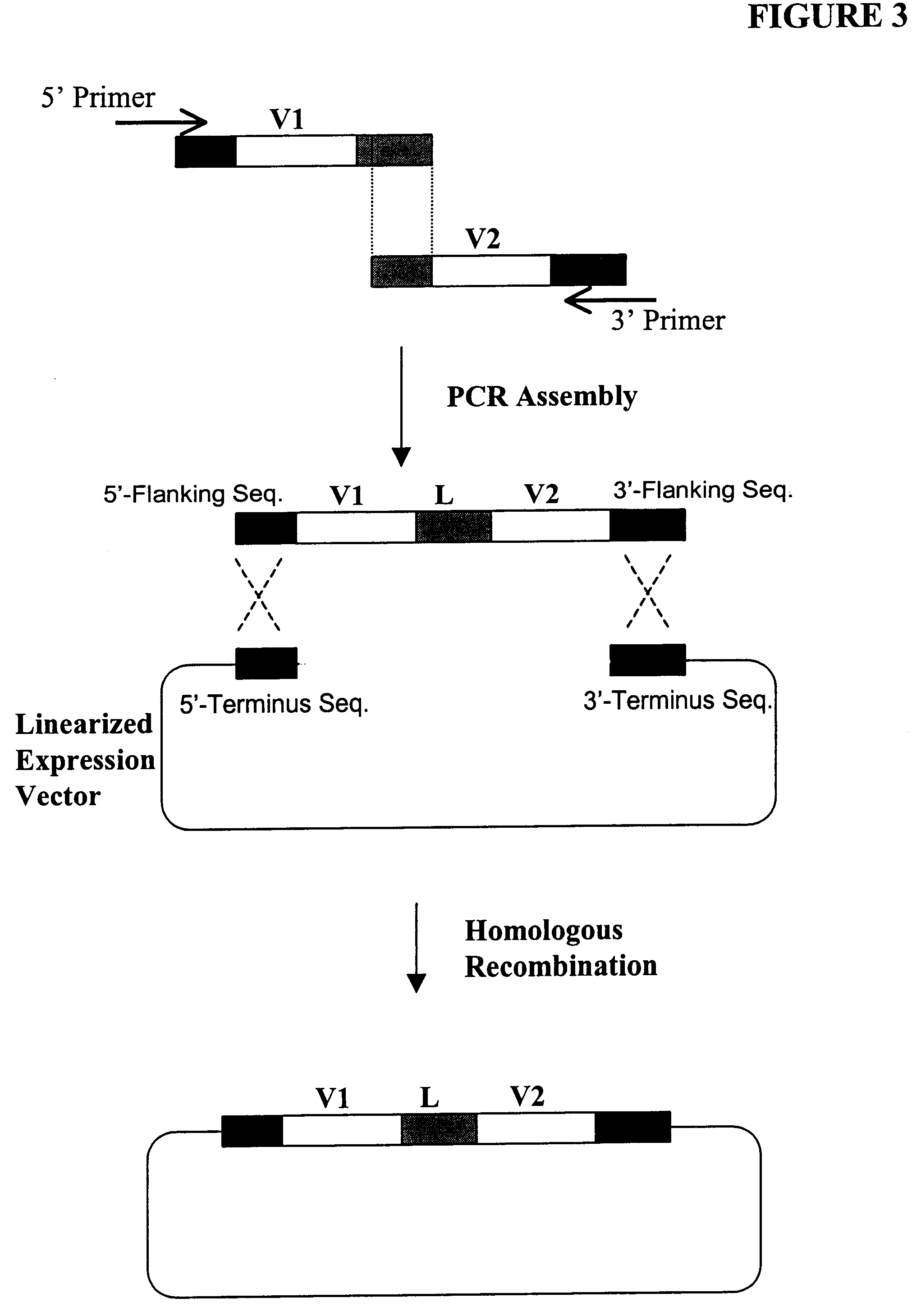
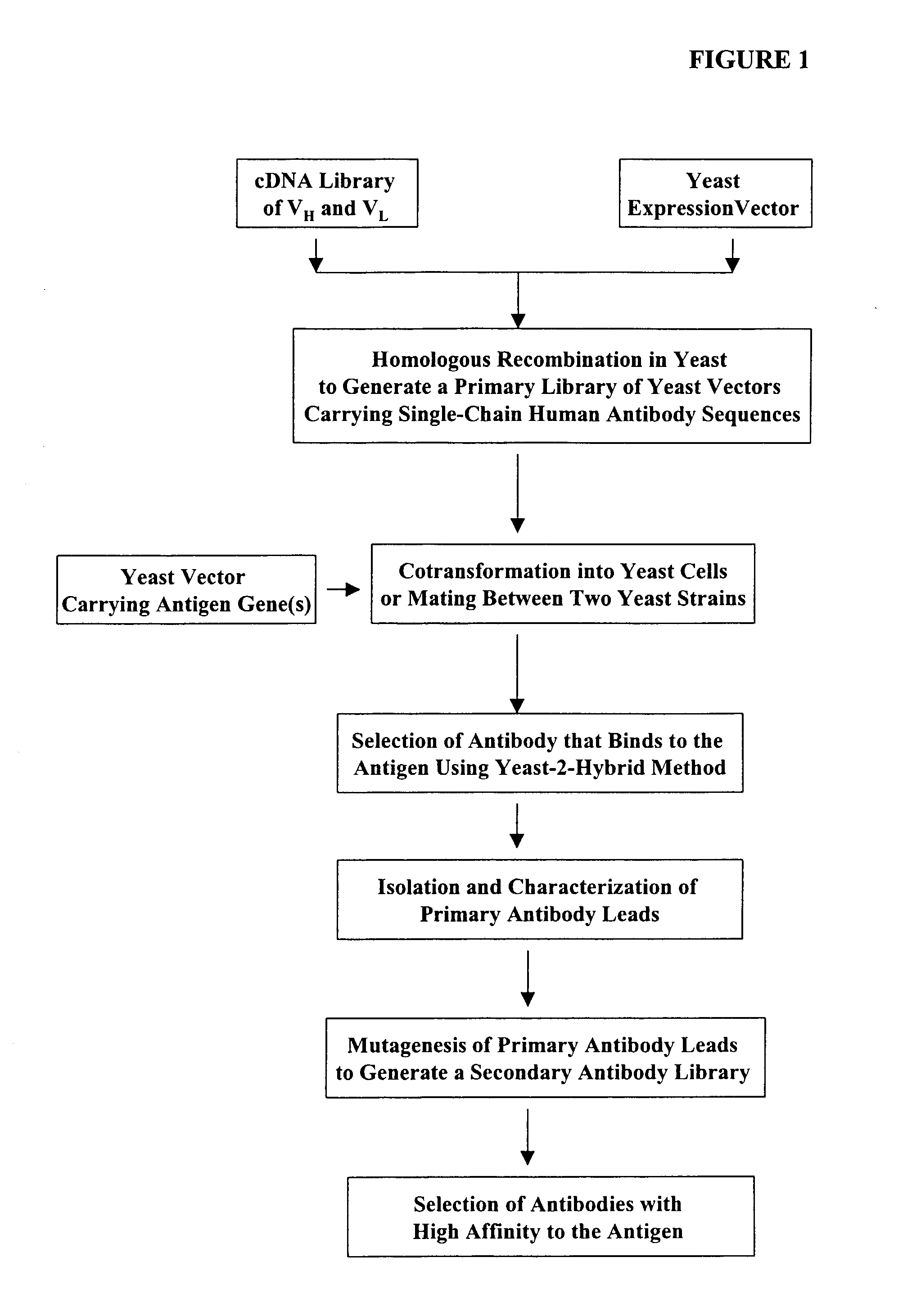
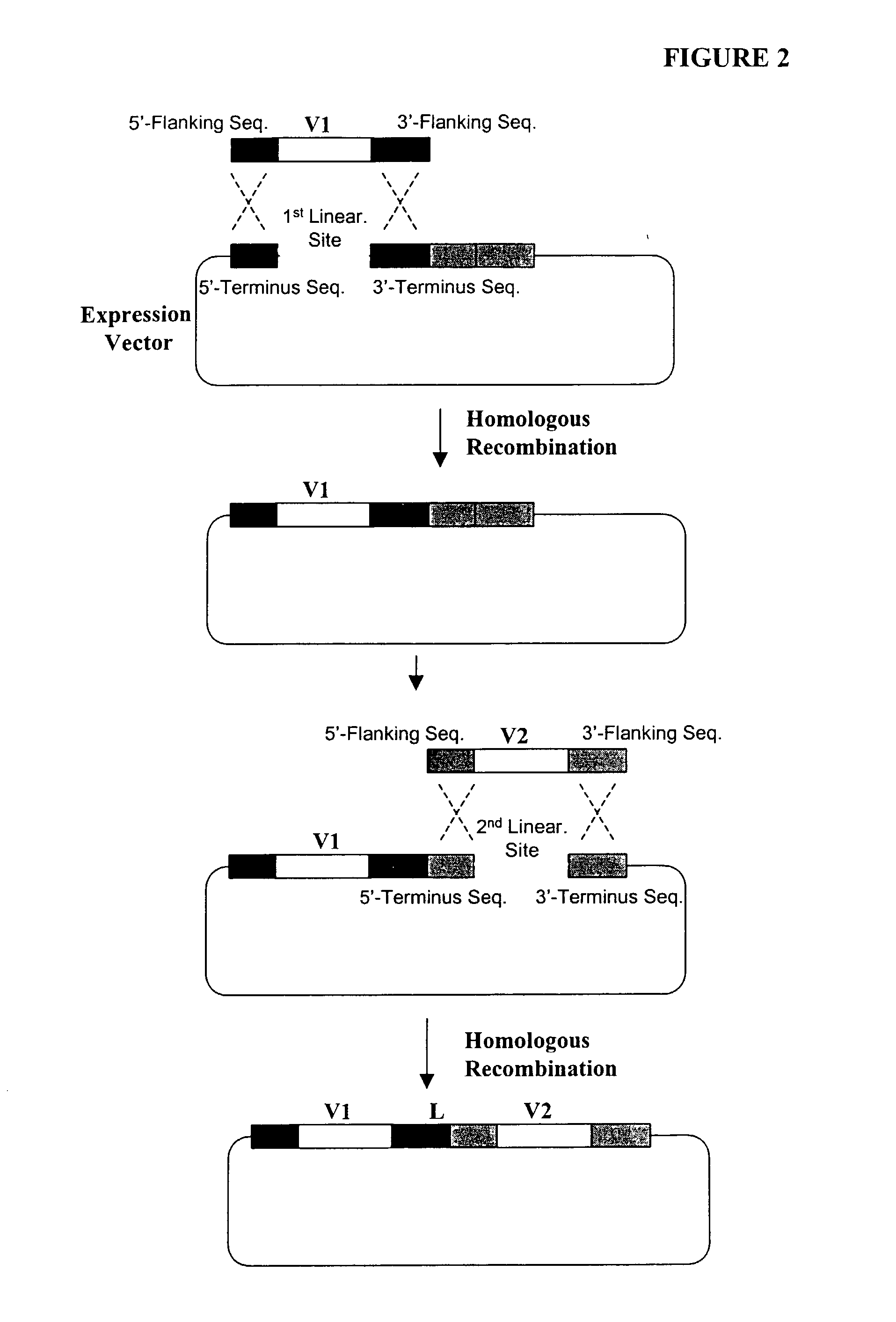
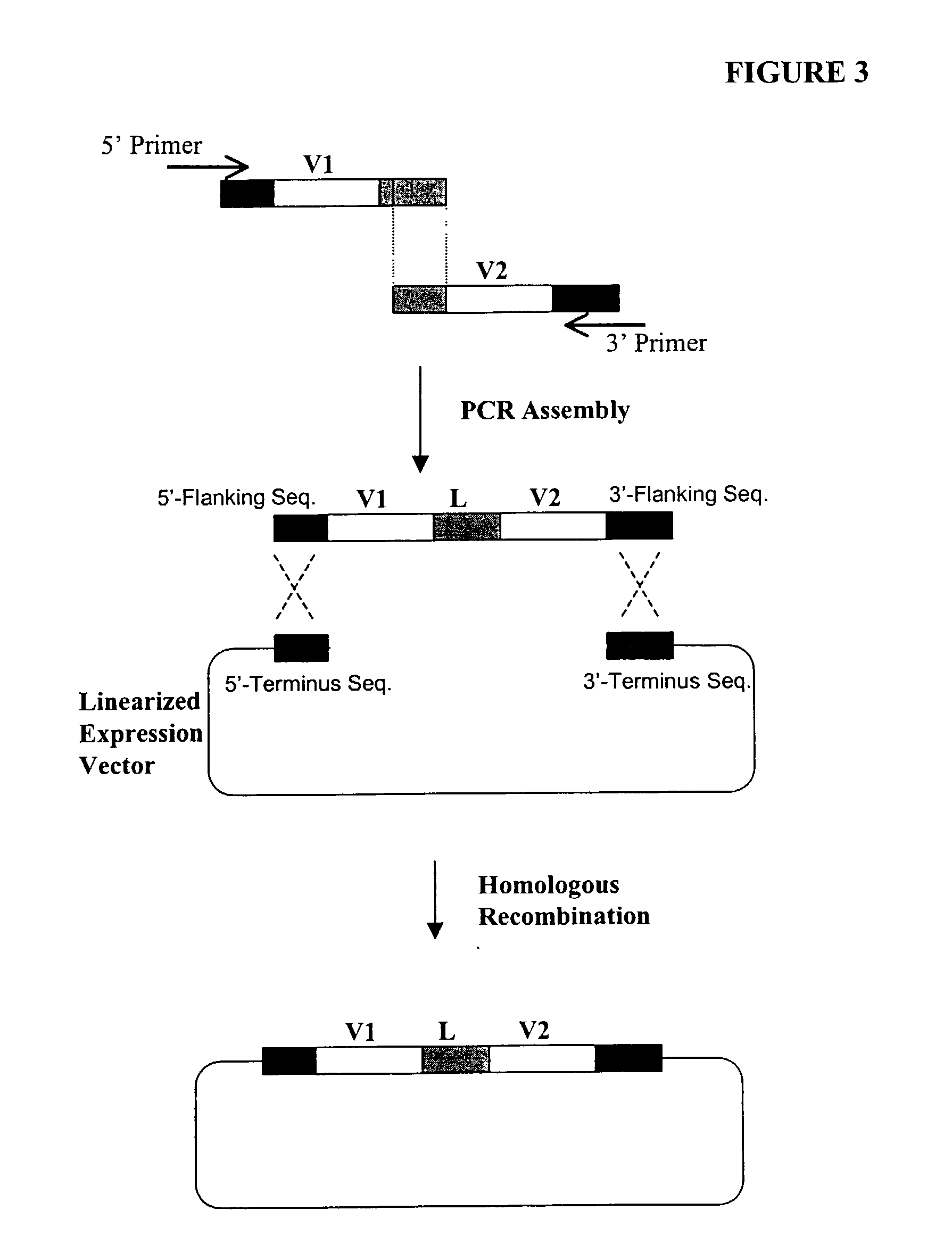
High throughput generation and screening of fully human antibody repertoire in yeast
InactiveUS6406863B1High affinityEasy to assembleMicrobiological testing/measurementImmunoglobulinsTarget peptideIn vivo
Compostions, kits and methods are provided for generating highly diverse libraries of proteins such as antibodies via homologous recombination in vivo, and screening these libraries against protein, peptide and nucleic acid targets using a two-hybrid method in yeast. The method for screening a library of tester proteins against a target protein or peptide comprises: expressing a library of tester proteins in yeast cells, each tester protein being a fusion protein comprised of a first polypeptide subunit whose sequence varies within the library, a second polypeptide subunit whose sequence varies within the library independently of the first polypeptide, and a linker peptide which links the first and second polypeptide subunits; expressing one or more target fusion proteins in the yeast cells expressing the tester proteins, each of the target fusion proteins comprising a target peptide or protein; and selecting those yeast cells in which a reporter gene is expressed, the expression of the reporter gene being activated by binding of the tester fusion protein to the target fusion protein.
Owner:GENETASTIX CORP
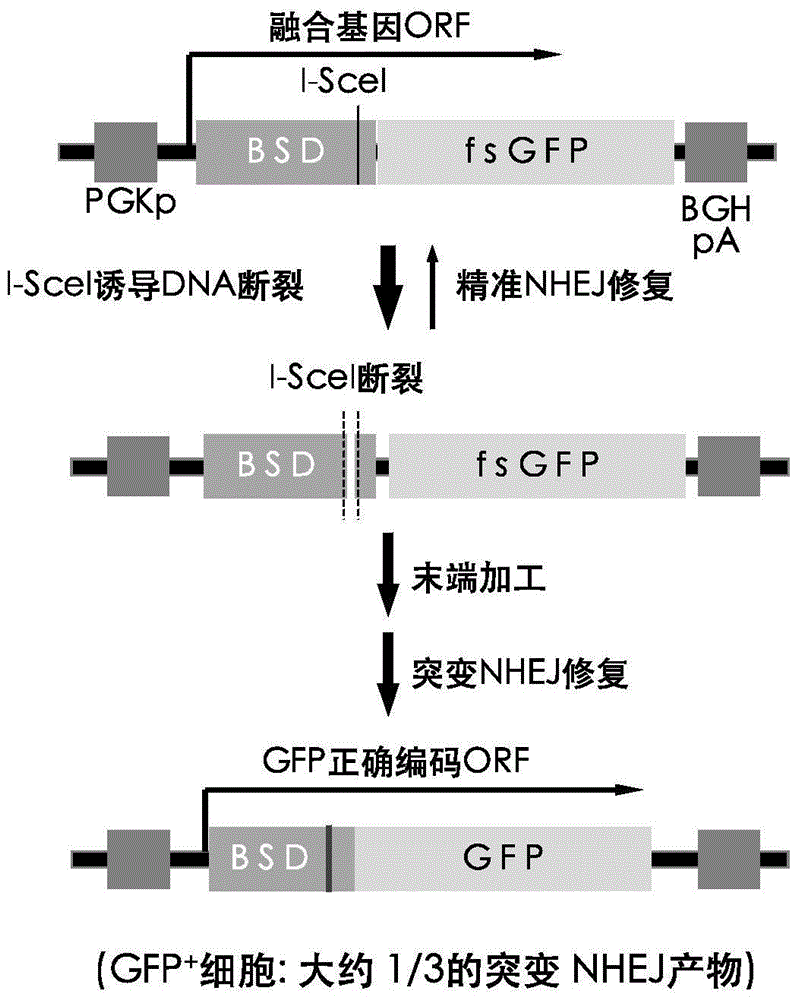
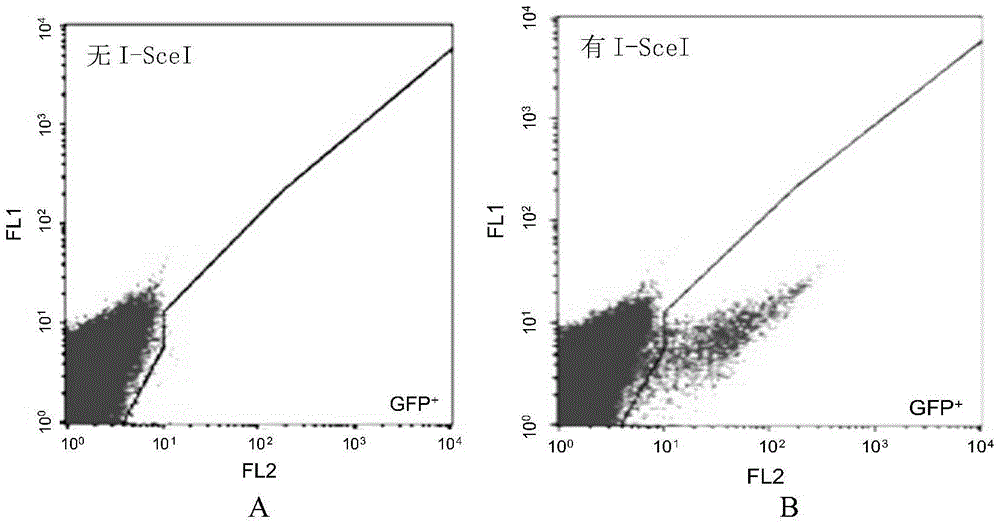
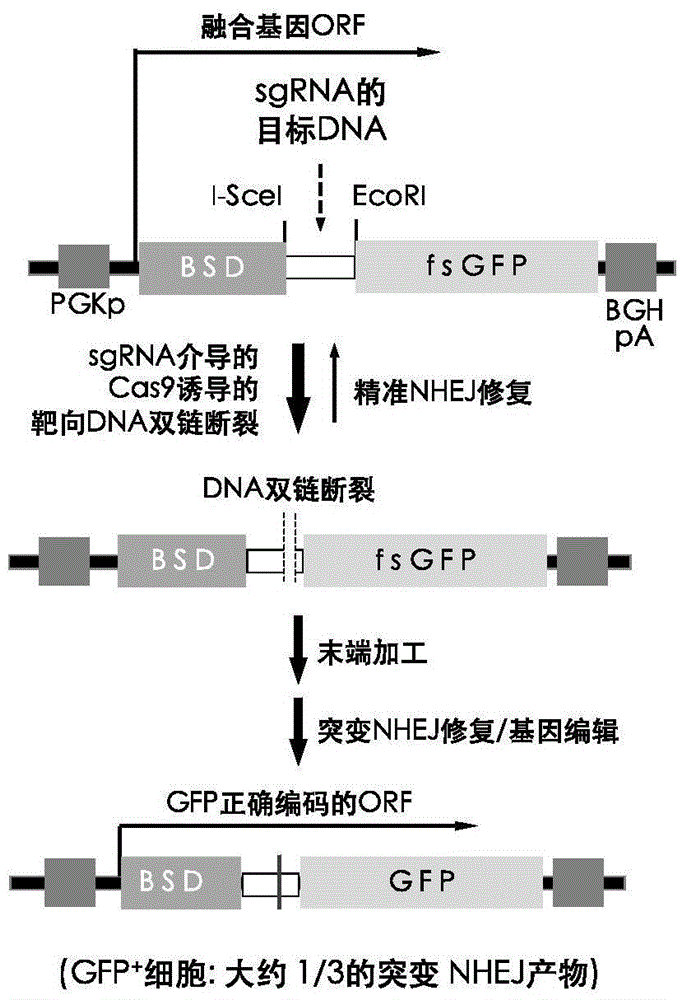
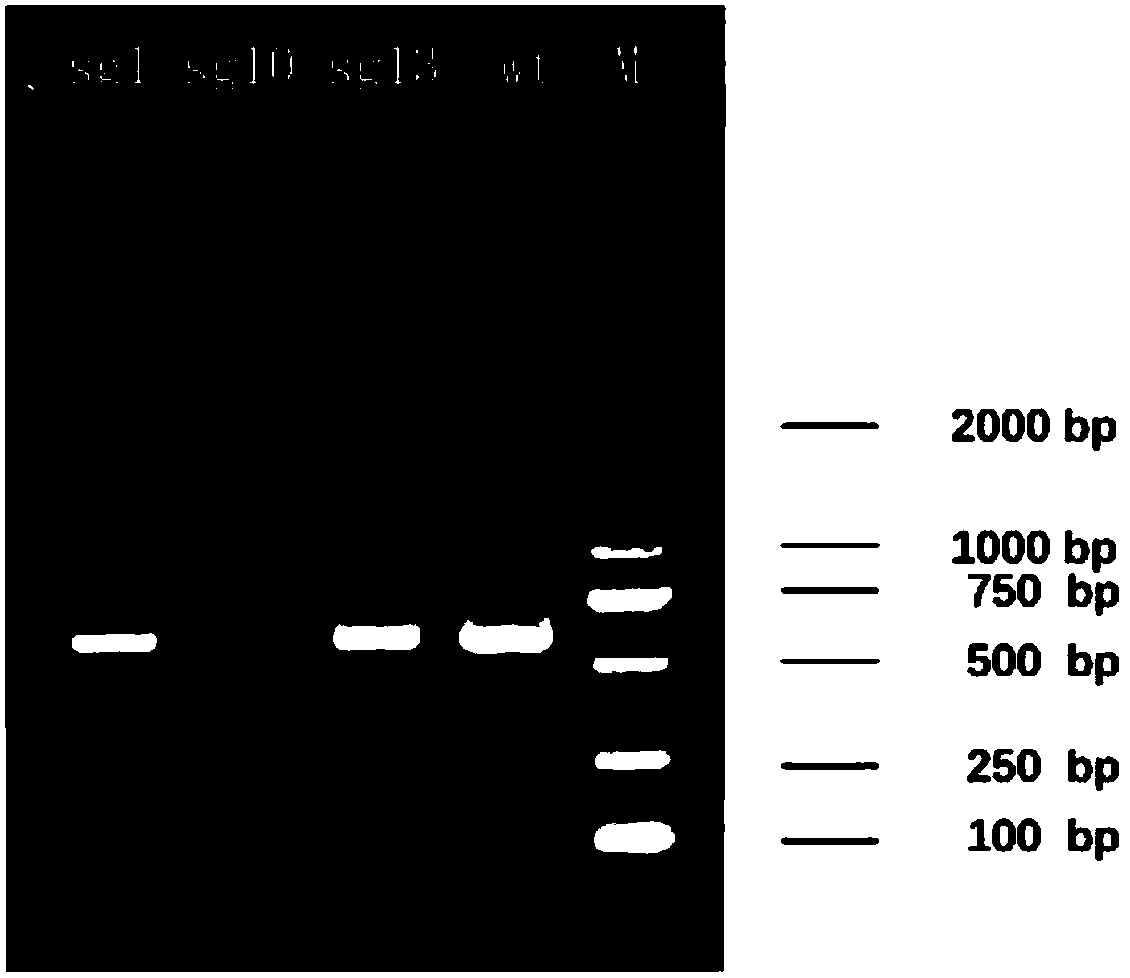
Fast CRISPR-Cas9 working efficiency testing system and application thereof
InactiveCN105647968ATest accurateTested and reliableMicrobiological testing/measurementPeptidesEnzyme digestionRapid testing
The invention discloses a fast CRISPR-Cas9 working efficiency testing system and application thereof. The testing system comprises a plasmid used for expressing sg RNA, a plasmid used for expressing Cas9 and a reporting system used for testing the CRISPR-Cas9 gene editing efficiency; the reporting system can splice the C-terminal of a nucleotide segment capable of coding effective protein and the N-terminal of a reporter gene and insert two restriction endonuclease enzyme digestion sites into the splicing position; before a special gene is edited (knocked out) through the CRISPR-Cas9 system, selection of a target sequence is vital, and the selection can influence the recognition efficiency of the sg RNA to target DNA, the binding efficiency of the sg RNA with the target DNA, the targeted cutting efficiency of the Cas9 and the NHEJ repairing efficiency. According to the system, the gene editing efficiency of different sgRNA-target DNA sequences can be quantitatively compared, the sgRNA with the best working effect can be determined within a short time, the actual knockout success rate is increased, therefore, the working cost can be lowered, the working efficiency can be improved, and the working progress can be promoted.
Owner:ZHEJIANG UNIV
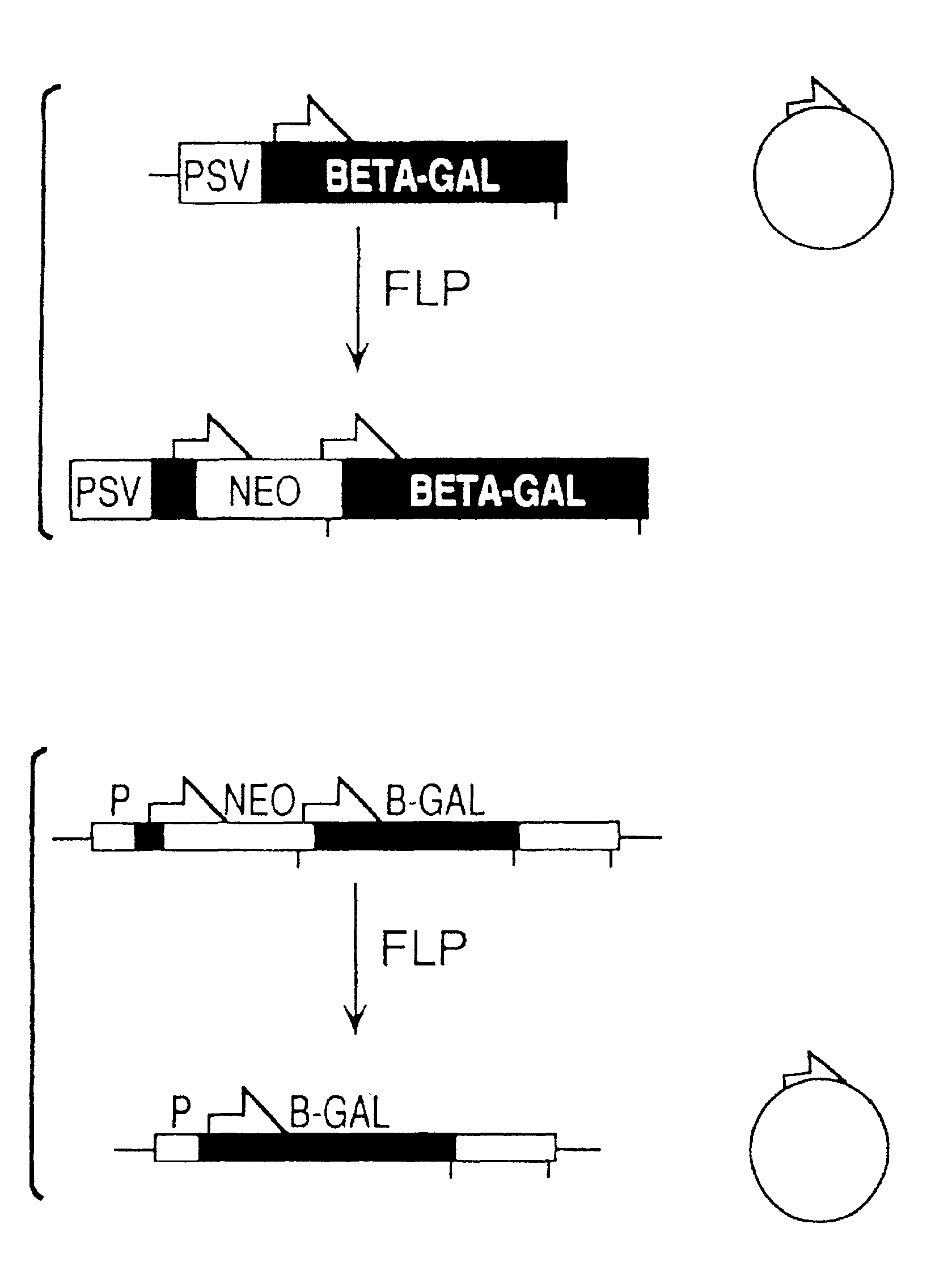
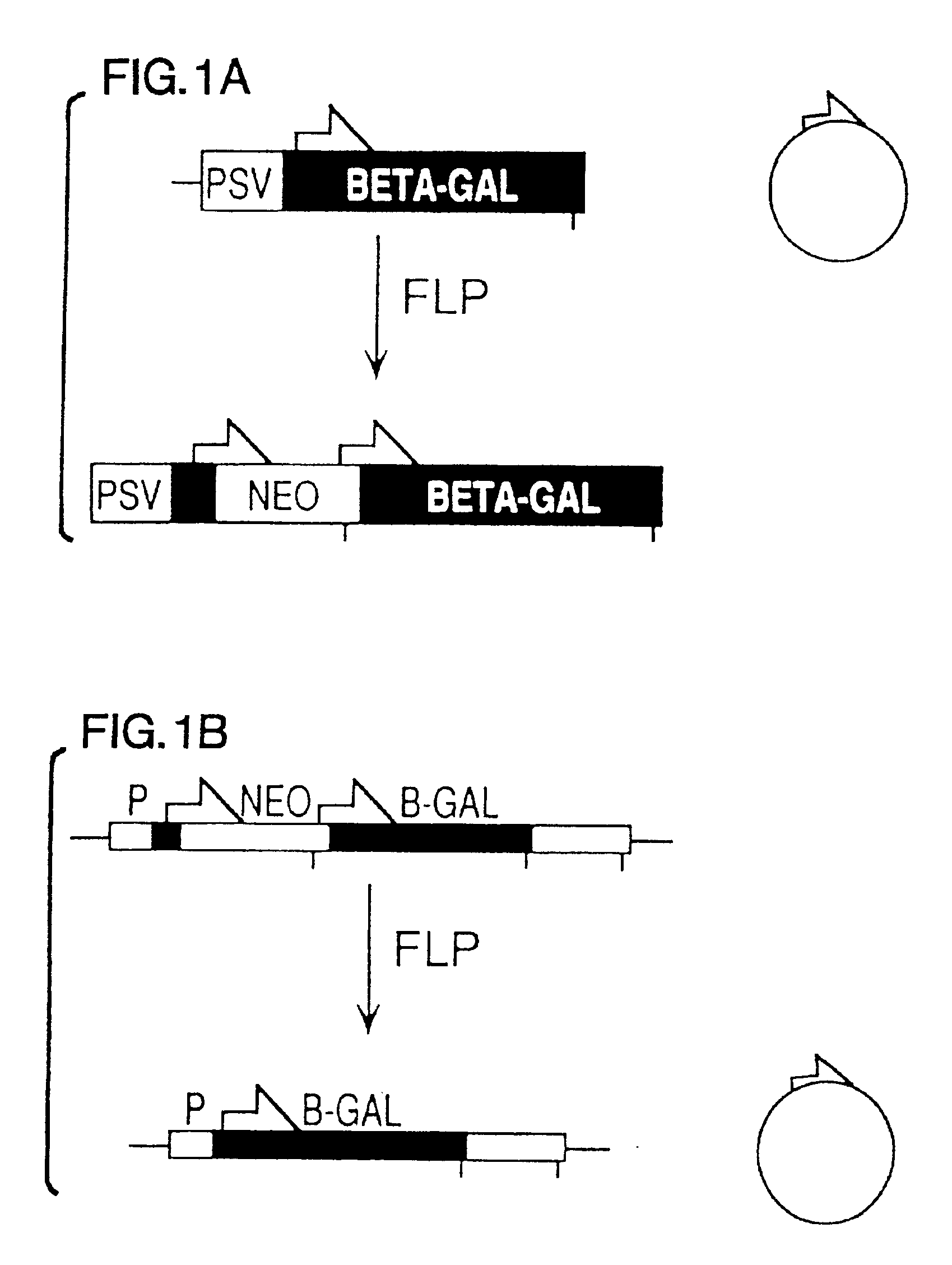
FLP-mediated gene modification in mammalian cells, and compositions and cells useful therefor
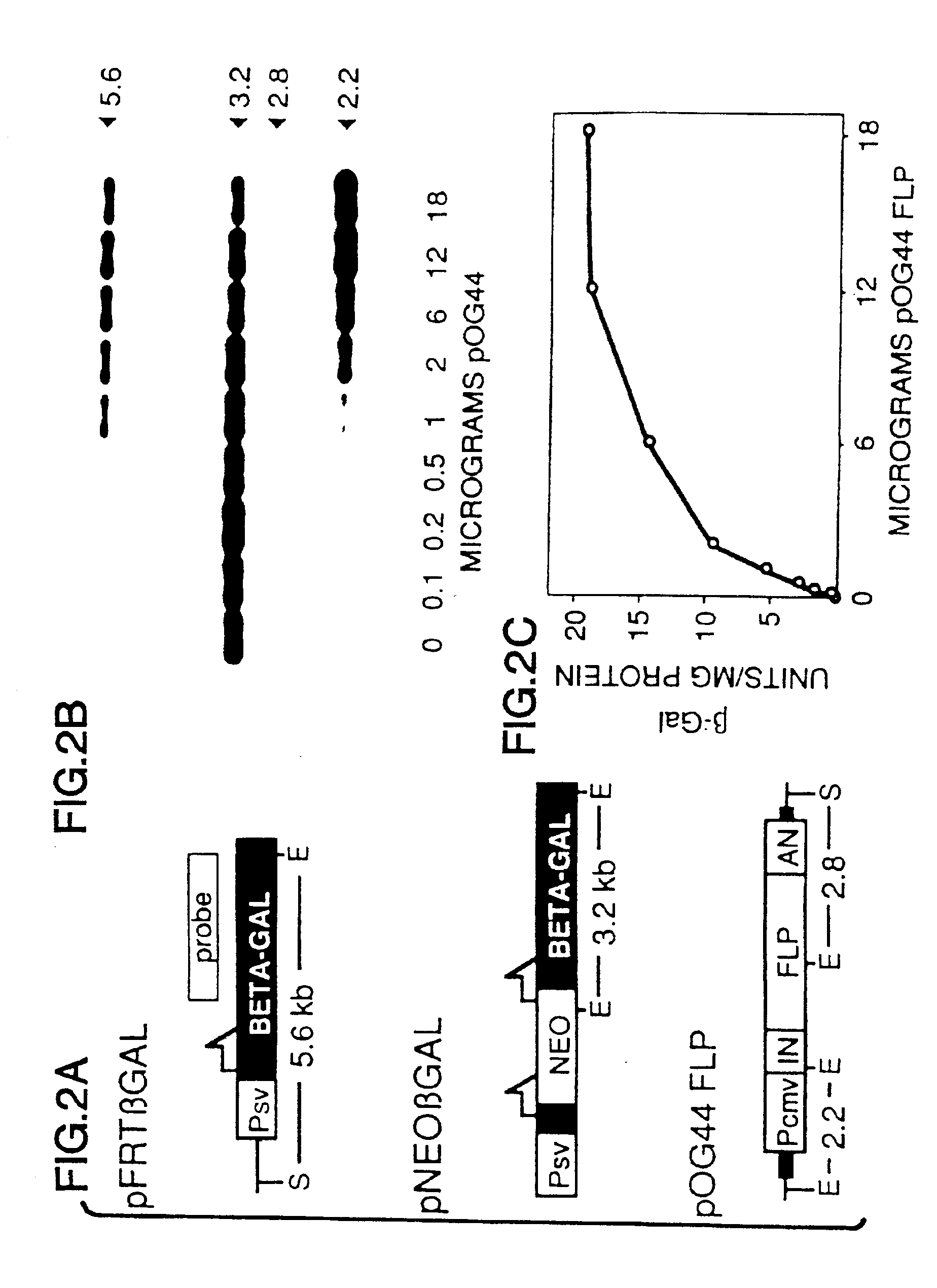
A gene activation / inactivation and site-specific integration system has been developed for mammalian cells. The invention system is based on the recombination of transfected sequences by FLP, a recombinase derived from Saccharomyces. In several cell lines, FLP has been shown to rapidly and precisely recombine copies of its specific target sequence. For example, a chromosomally integrated, silent β-galactosidase reporter gene was activated for expression by FLP-mediated removal of intervening sequences to generate clones of marked cells. Alternatively, the reverse reaction can be used to target transfected DNA to specific chromosomal sites. These results demonstrate that FLP can be used, for example, to mosaically activate or inactivate transgenes for a variety of therapeutic purposes, as well as for analysis of vertebrate development. The FLP recombination system of the present invention can be incorporated in transgenic, non-human mammals to achieve site-specific integration of transgenes, to construct functional genes or to disrupt existing genes.
Owner:CORN PROD DEV INC
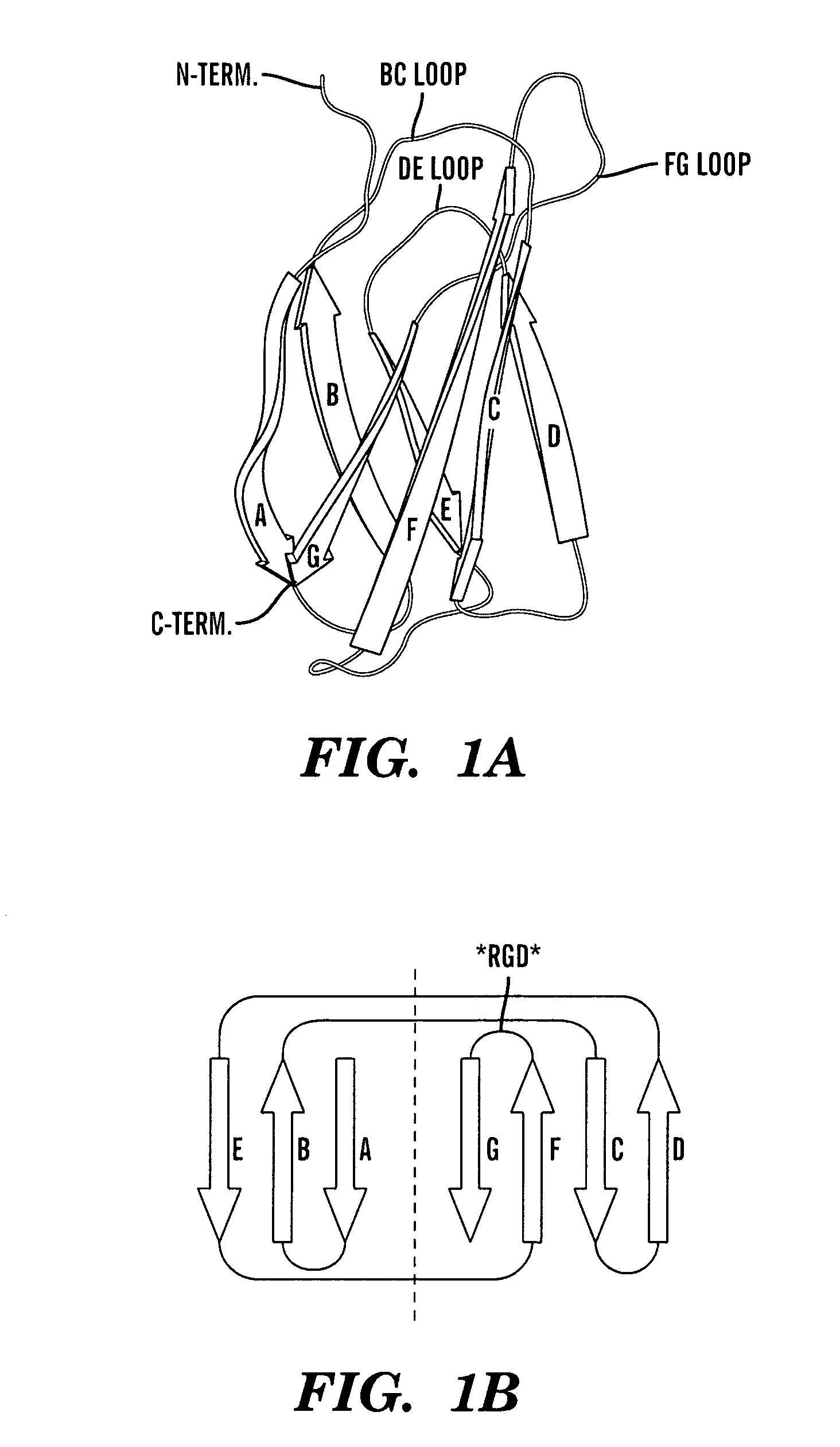
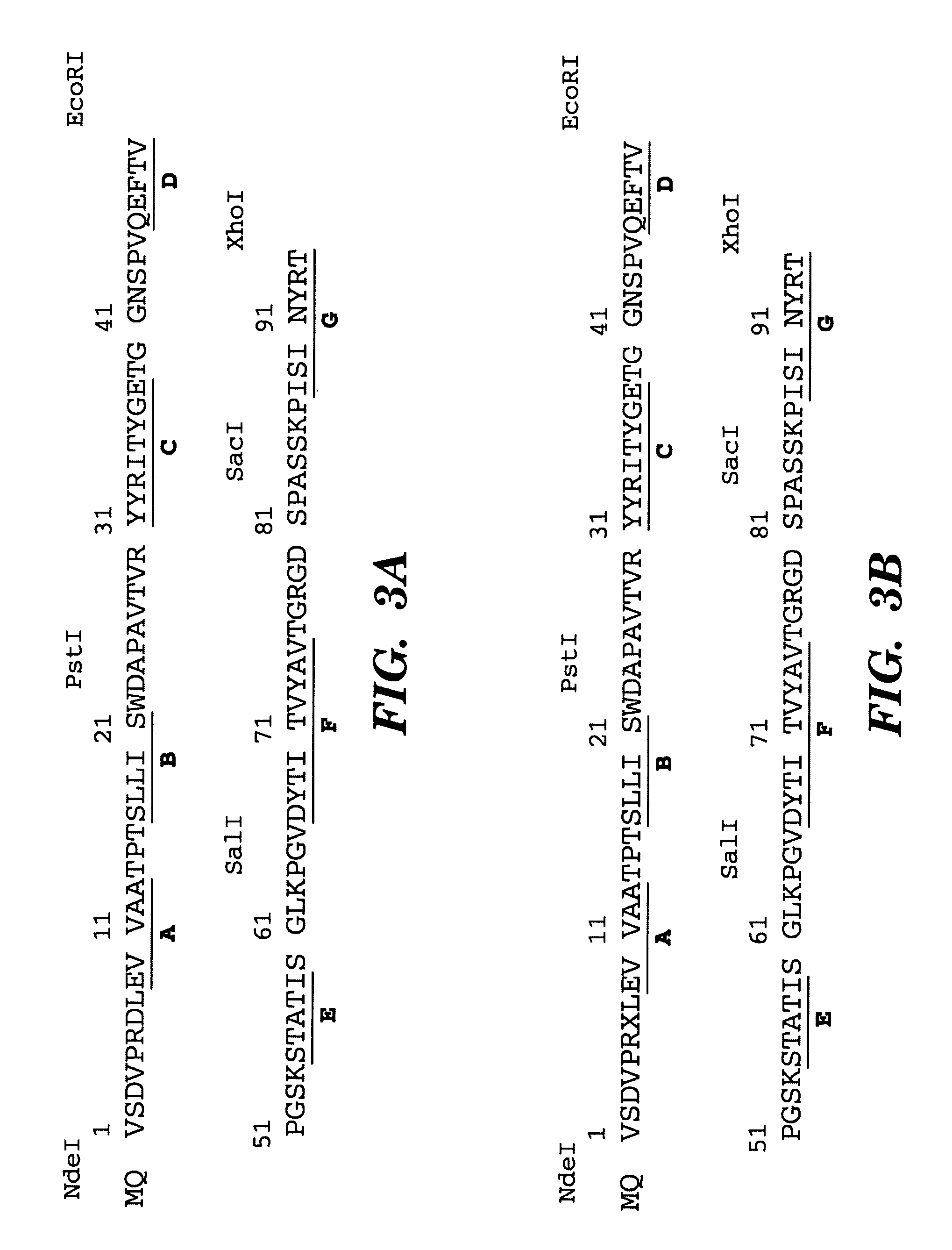
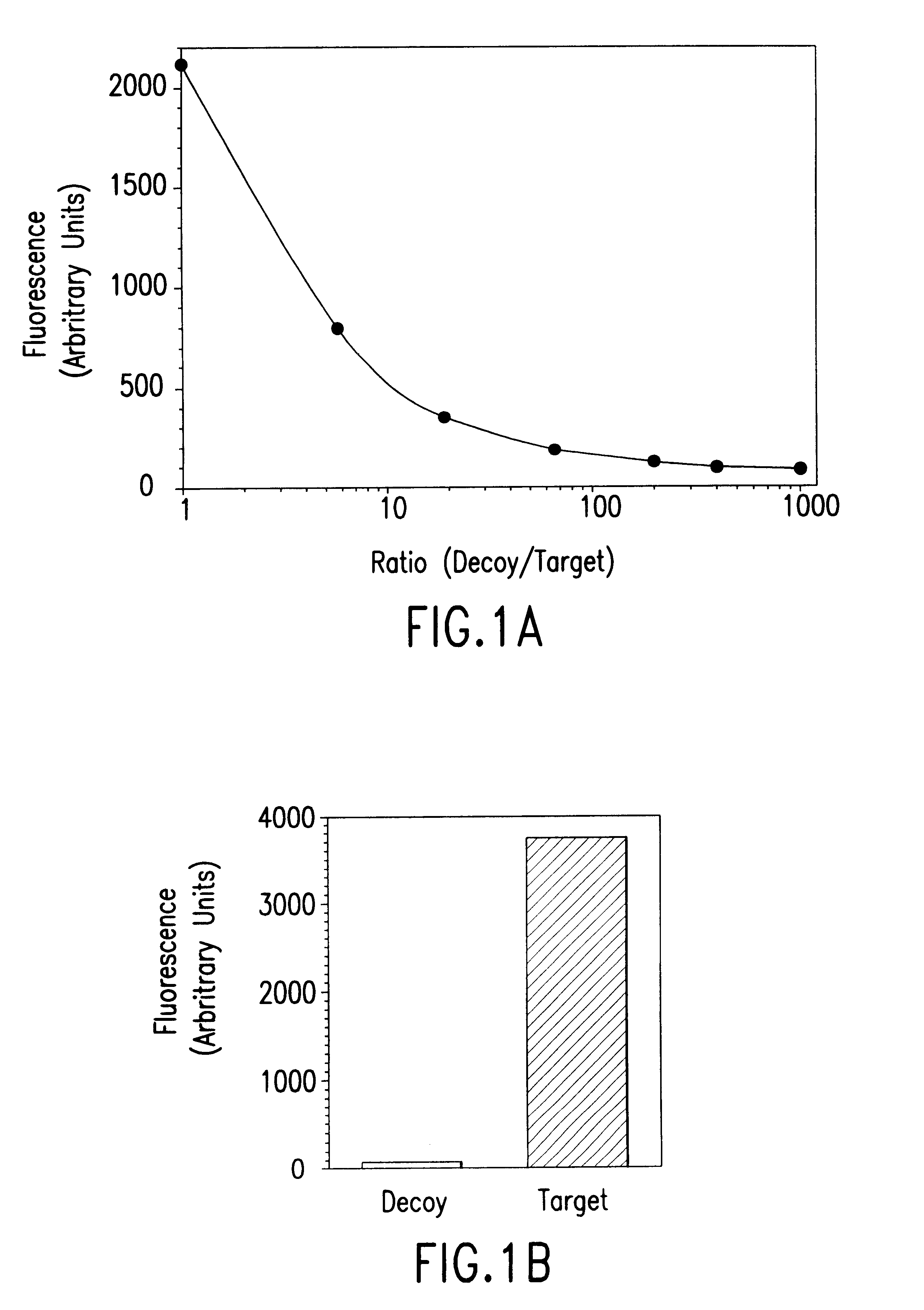
Method of identifying polypeptide monobodies which bind to target proteins and use thereof
InactiveUS7598352B2Peptide/protein ingredientsAntibody mimetics/scaffoldsDNA-binding domainProtein target
A method of identifying a polypeptide monobody having target protein binding activity, said method comprising: providing a host cell comprising (i) a reporter gene under control of a 5′ regulatory region operable in the host cell, (ii) a first chimeric gene which encodes a first fusion polypeptide comprising a target protein, or fragment thereof, fused to a C-terminus of a DNA-binding domain which binds to the 5′ regulatory region of the reporter gene, and (iii) a second chimeric gene which encodes a second fusion polypeptide comprising a polypeptide monobody fused to a transcriptional activation domain; and detecting expression of the reporter gene, which indicates binding of the polypeptide monobody of the second fusion polypeptide to the target protein such that the transcriptional activation domain of the second fusion polypeptide is in sufficient proximity to the DNA-binding domain of the first fusion polypeptide to allow expression of the reporter gene.
Owner:UNIVERSITY OF ROCHESTER
Targeted methods of drug screening using co-culture methods
The present invention provides methods of screening for a molecule that inhibits the expression or activity of a protein encoded by a target gene which affects the fitness of a cell. The methods are based on a co-culture assay, and entail culturing together two cell populations, each of which is a population of identical cells, of the same species that differs substantially only in the expression or activity of the gene to be targeted or its encoded protein and the presence or absence of a reporter gene. The screen can be applied to cultured cells, unicellular and multicellular organisms. Manipulating the expression or activity of the target gene sensitizes the host to a molecule which inhibits the target gene or its encoded protein such that the cell or organism comprising the manipulated target gene grows at a different rate from the cell or organism comprising the unmanipulated gene in response to exposure to the molecule. The methods of the invention can be used for identifying drugs, proteins or any other molecules that inhibit the function of proteins encoded by target genes.
Owner:ROSETTA INPHARMATICS LLC
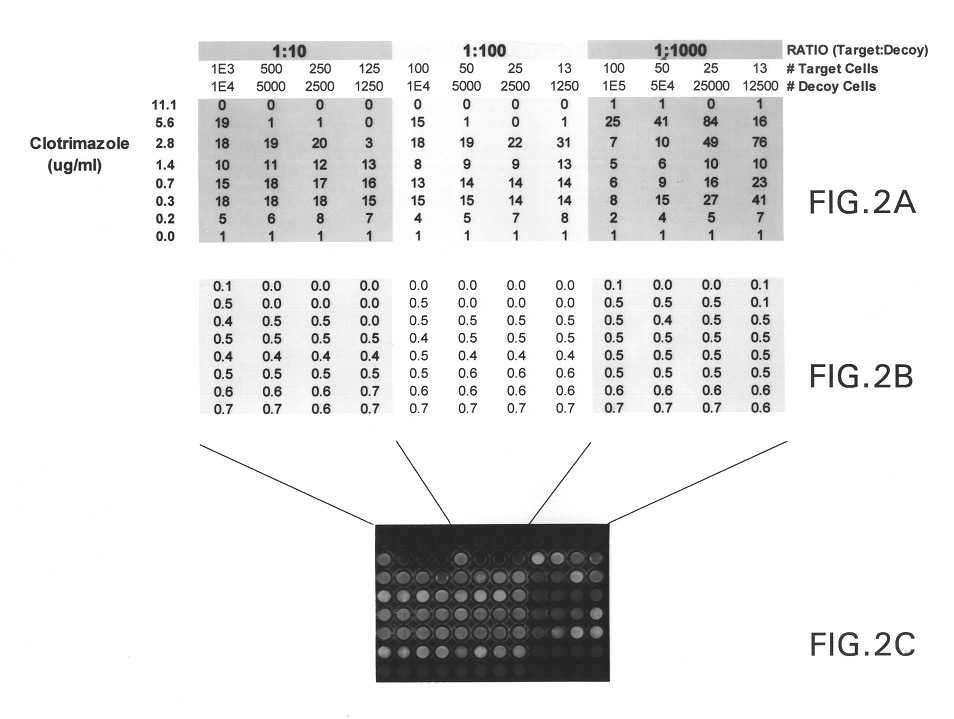
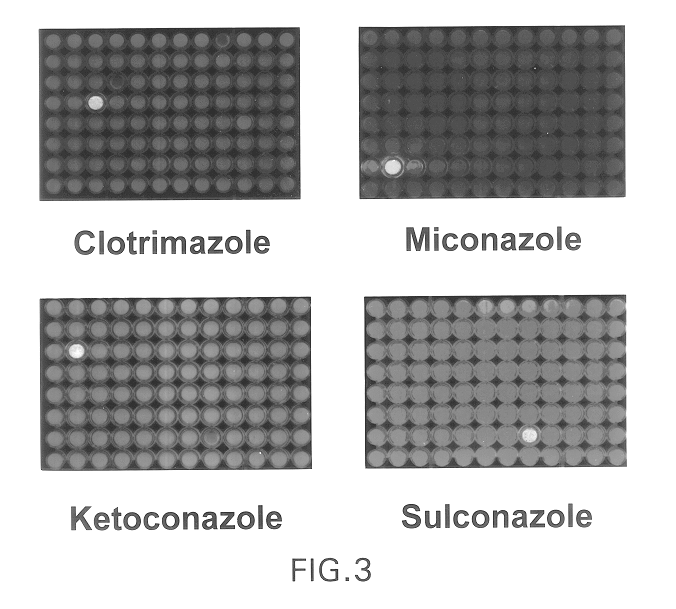
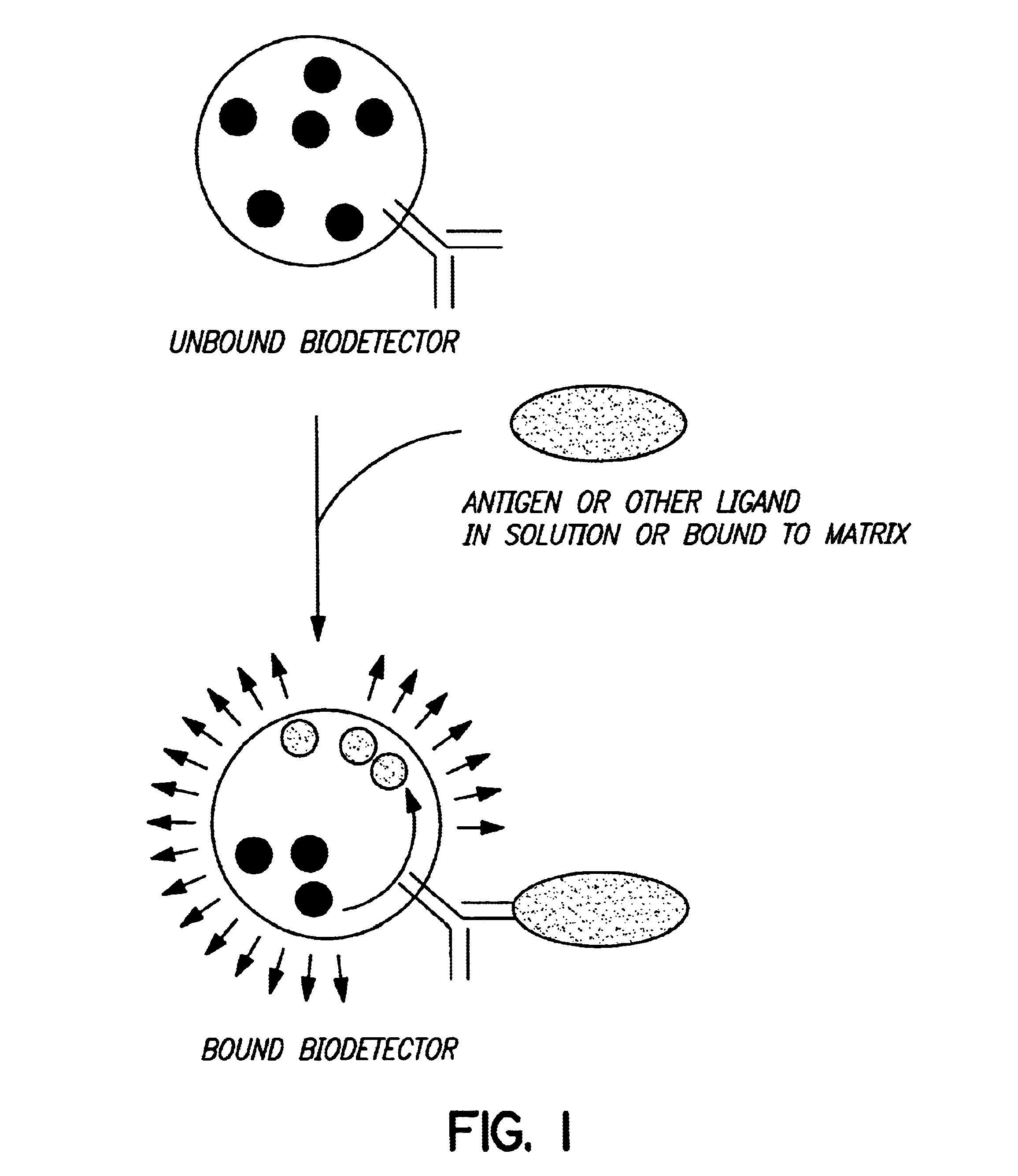
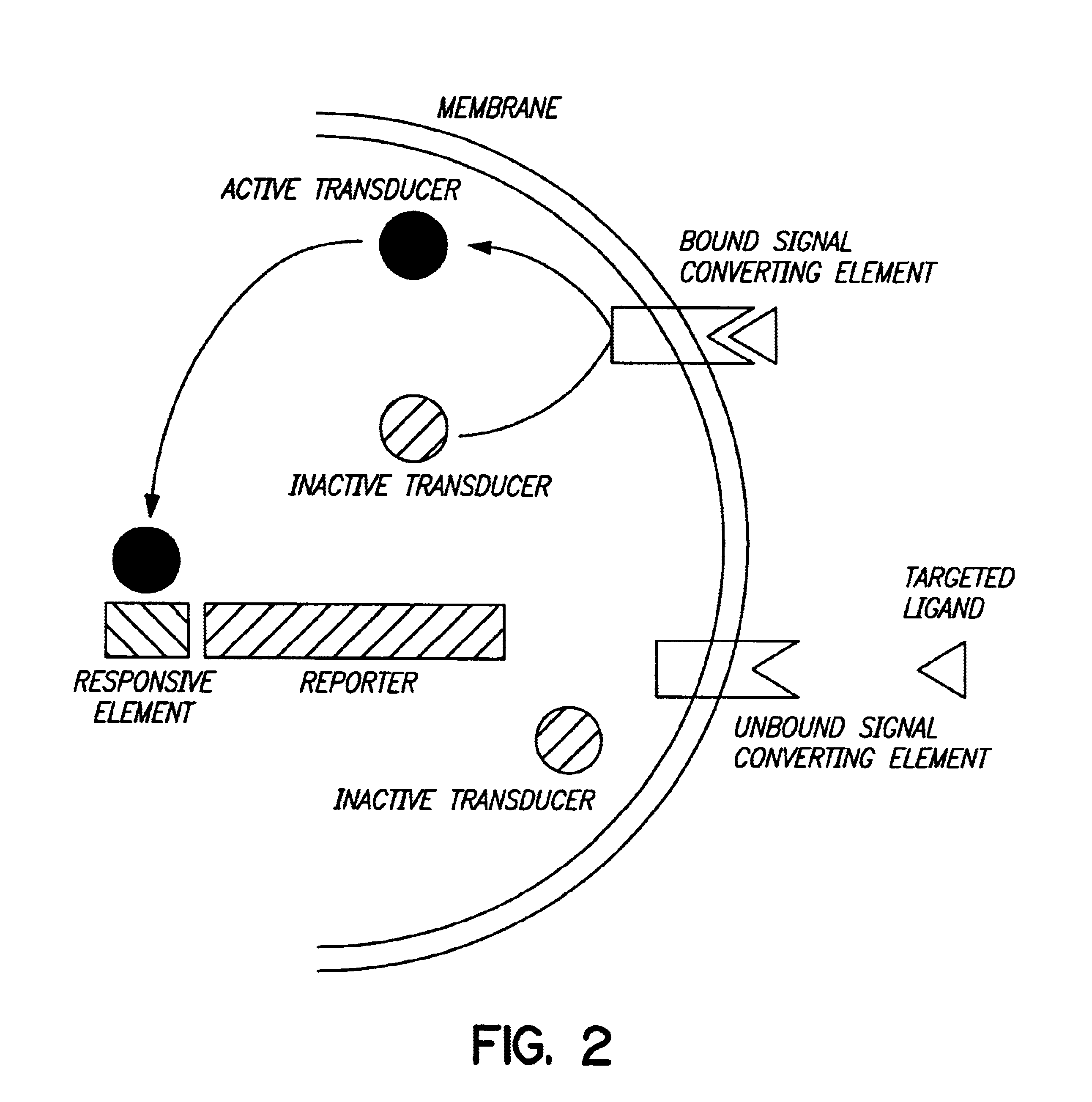
Biodetectors targeted to specific ligands
The present invention relates to biodetectors for detecting and quantifying molecules in liquid, gas, or matrices. More specifically, the present invention relates to biodetectors comprising a molecular switching mechanism to express a reporter gene upon interaction with target substances. The invention further relates to methods using such biodetectors for detecting and quantifying selected substances with high specificity and high sensitivity.
Owner:XENOGEN CORP
Method to measure the activation state of signaling pathways in cells
InactiveUS20020127604A1Microbiological testing/measurementWithdrawing sample devicesDiseaseSignalling pathways
The activity of multiple proteins in a single living cell, portion of a cell or in a group of cells is simultaneously measured by introducing reporter molecules. The reporter(s) is chemically modified by the enzyme of interest. In some cases the enzyme(s) is affected by the addition of a stimulus or a pharmaceutical compound to the cell. The reactions between the enzymes and the reporters are diminished or terminated, and the reporter and modified reporter are removed. The activity of the enzyme(s) is determined by measuring the amount of reporter remaining, the amount of altered reporter produced, or by comparing the amount of reporter to the amount of altered reporter. A database is compiled of the activities of the different proteins. By performing a series of experiments at different time points, conditions, and varieties of cell types, a database is developed for molecular cellular mechanisms in health and disease states. By exposing cells to a variety of compounds data for drug development and screening is provided.
Owner:RGT UNIV OF CALIFORNIA
Exogenous gene knocking-in and integrating system on basis of CRISPR/Cas9, method for establishing exogenous gene knocking-in and integrating system and application thereof
ActiveCN106191116ANucleic acid vectorVector-based foreign material introductionTarget geneIntegrated systems
The invention provides an exogenous gene knocking-in and integrating system on the basis of CRISPR / Cas9, a method for establishing the exogenous gene knocking-in and integrating system and application thereof. The exogenous gene knocking-in and integrating system comprises vectors with report / donor functions and Cas9 expression vectors. Each report / donor vector comprises two target gent homologous arms and an exogenous sequence fragment positioned between the two target gene homologous arms; homologous sequences, which are positioned on a target gene, of the two target gene homologous arms of each report / donor vector are respectively positioned on two sides of a target sequence of the target gene and are connected with the target sequence of the target gene; the exogenous sequence fragments comprise promoters, resistant genes, shorn peptide sequences, report genes and polyA tails which are sequentially arrayed, two SSA repair homologous sequences of each resistant gene are inserted into the resistant gene, and the target sequence of each target gene is inserted in a space between the two corresponding SSA repair homologous sequences. The exogenous gene knocking-in and integrating system, the method and the application have the advantages that exogenous genes can be integrated with endogenous gene sequences in an efficient site-directed and accurately targeted manner, and double-chromosome allelic gene double-knocking-in can be efficiently carried out.
Owner:成都中科奥格生物科技有限公司
Methods and systems for molecular fingerprinting
InactiveUS20020034748A1Fast and accurate determinationHigh resolutionBioreactor/fermenter combinationsBiological substance pretreatmentsSmall samplePolynucleotide
This invention relates in general to a method for molecular fingerprinting. The method can be used for forensic identification (e.g. DNA fingerprinting, especially by VNTR), bacterial typing, and human / animal pathogen diagnosis. More particularly, molecules such as polynucleotides (e.g. DNA) can be assessed or sorted by size in a microfabricated device that analyzes the polynucleotides according to restriction fragment length polymorphism. In a microfabricated device according to the invention, DNA fragments or other molecules can be rapidly and accurately typed using relatively small samples, by measuring for example the signal of an optically-detectable (e.g., fluorescent) reporter associated with the polynucleotide fragments.
Owner:CALIFORNIA INST OF TECH
Methods for identifying compounds that modulate untranslated region-dependent gene expression and methods of using same
InactiveUS20070111203A1Reduced stabilityReduces translation efficiencyCompound screeningPeptide librariesHigh-Throughput Screening MethodsHigh-throughput screening
The present invention relates to methods for identifying compounds that modulate untranslated region-dependent expression of a target gene. The invention particularly relates to using untranslated regions of a target gene or fragments thereof linked to a reporter gene to identify compounds that modulate untranslated region-dependent expression of a target gene. The methods of the present invention provide a simple, sensitive assay for high-throughput screening of libraries of compounds to identify pharmaceutical leads.
Owner:PTC THERAPEUTICS INC
Methods of treating asthma
InactiveUS20050164323A1Increase kinase activityDecrease kinase activityCompound screeningApoptosis detectionMast cellKinase activity
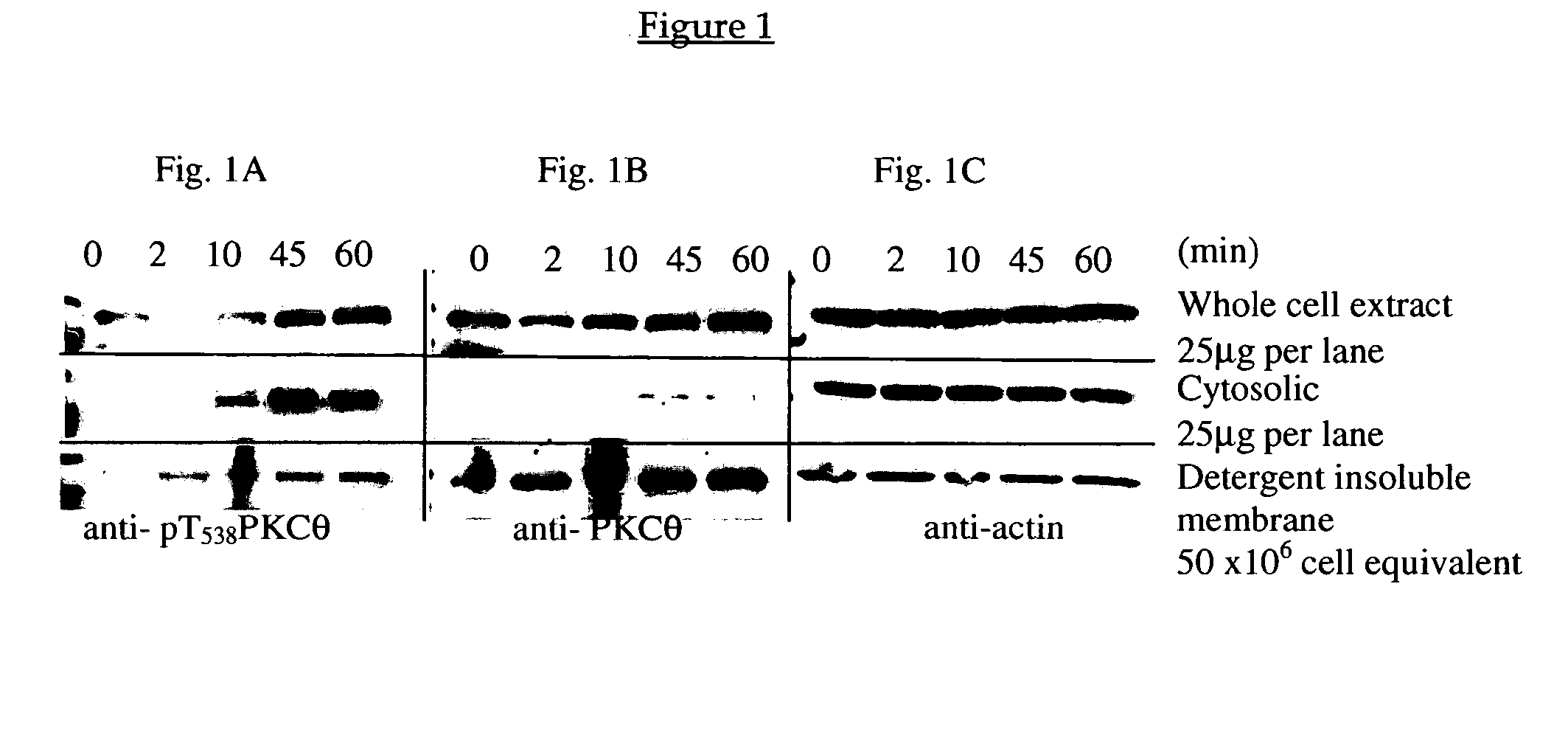
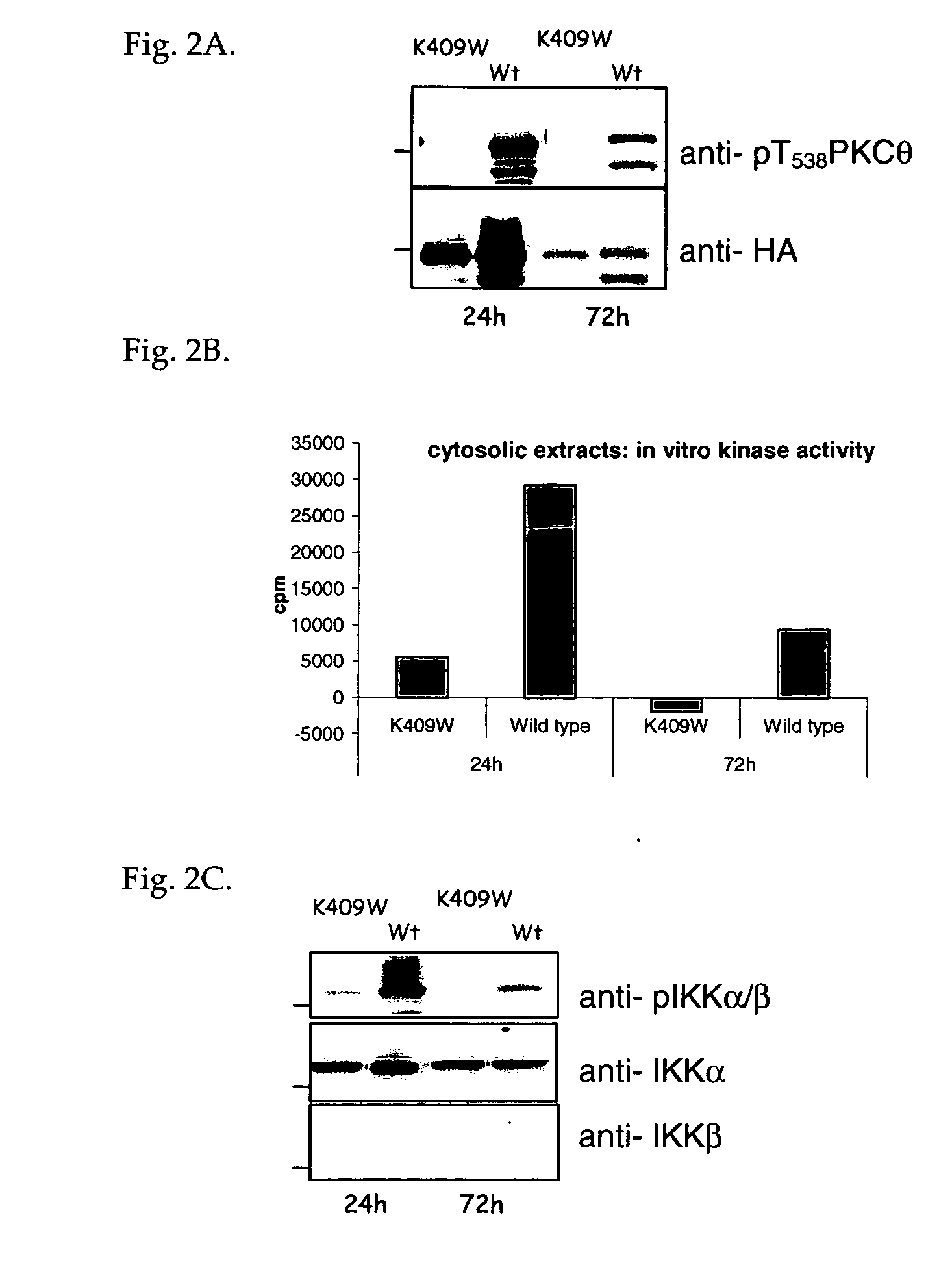
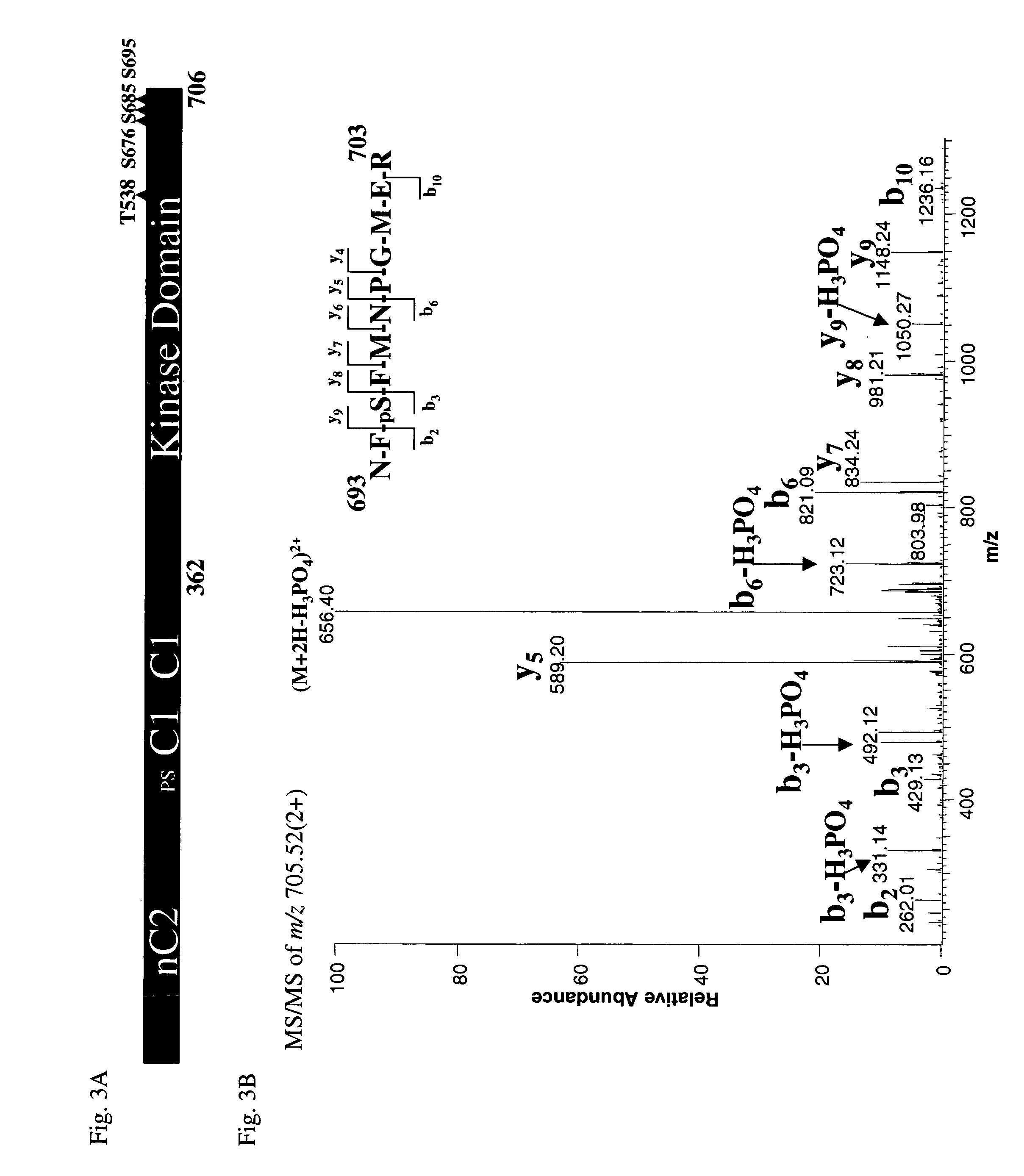
Methods for agents useful for treating asthma are disclosed. The methods include screening for agents that inhibit the production of a PKC-θ protein, as well as for agents that inhibit the kinase activity of a PKC-θ protein, or a functional fragment thereof, wherein such agents are useful for treating asthma. The methods also include screening for agents that inhibit the production of a reporter gene product encoded by a nucleic acid sequence operably linked to a PKC-θ promoter. Also disclosed are methods of treating asthma that include administering an agent that inhibits the production of a functional PKC-θ protein or the kinase activity of a PKC-θ protein or a functional fragment thereof. An isolated mast cell lacking expression of endogenous PKC-θ is also disclosed.
Owner:WYETH LLC
CRISPR-Cas9 system used for assembling DNA and DNA assembly method
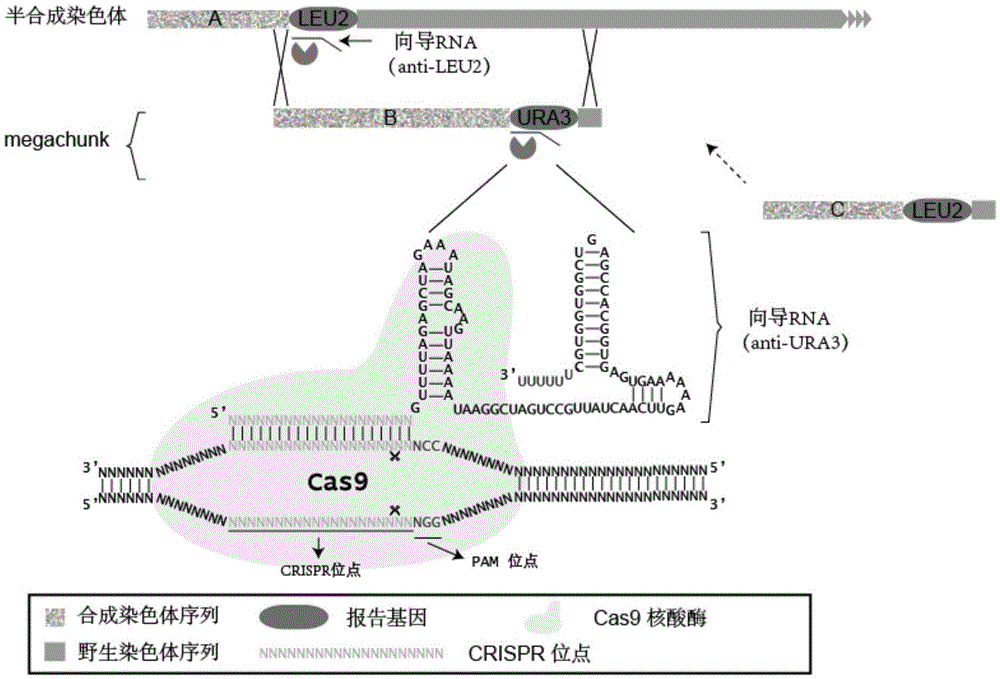
InactiveCN105821072ARepeatableSuccessfully synthesizedFungiMicroorganism based processesReporter geneGuide RNA
The invention discloses a CRISPR-Cas9 system used for assembling DNA and a DNA assembly method. The CRISPR-Cas9 system includes the following parts: a plasmid used for expressing Cas9 gene, a first guide RNA, and / or a plasmid used for expressing the first guide RNA, wherein the first guide RNA has a CRISPR site. The CRISPR site is combined with a first reporter gene carried by a semisynthetic chromosome used for assembling according to base complementation pairing rule. The CRISPR-Cas9 system has advantages of high replacement success rate of homologous recombination, less species of guide RNA which needs to design and use, less harmful effect subjected by the genomic sequence, and low off-target rate.
Owner:SHENZHEN HUADA GENE INST
Methods for the amplification, quantitation and identification of nucleic acids
ActiveUS20070190540A1Avoid pollutionPromote digestionMicrobiological testing/measurementFermentationNucleic acid sequencingFluorescent reporter
The invention relates to improved methods of amplifying and optionally quantifying and / or identifying a plurality of selected nucleic acid molecules from a pool of nucleic acid molecules. A first round of multiplex amplification used where the amplification reaction is allowed to proceed to a point prior to that at which significant competition between amplicons for reaction components has occurred. This is the followed by a second round of amplification that typically includes a fluorescent reporter to allow for each of the selected nucleic acid sequences to be quantified. The methods are useful for the amplification and quantification of nucleic acids from a variety of sources, such as gene expression products, whereby many such products may be amplified and quantified from very limited samples and from degraded archival samples.
Owner:AUSDIAGNOSTICS
Method for assaying protein—protein interaction
The invention relates to a method for determining if a test compound, or a mix of compounds, modulates the interaction between two proteins of interest. The determination is made possible via the use of two recombinant molecules, one of which contains the first protein a cleavage site for a proteolytic molecules, and an activator of a gene. The second recombinant molecule includes the second protein and the proteolytic molecule. If the test compound binds to the first protein, a reaction is initiated whereby the activator is cleaved, and activates a reporter gene.
Owner:LIFE TECH CORP +1
In-vitro evaluation cell model of skin sensitization of compound and construction method of cell model
ActiveCN108103098AEffective resolutionSensitization Study SpecificHydrolasesDrug screeningSkin sensitizationLuciferase Gene
The invention discloses an in-vitro evaluation cell model of skin sensitization of a compound and a construction method of the cell model. The construction method of the cell model comprises the following steps of: designing and constructing an sgRNA (small guide ribonucleic acid) expression vector with CRISPR / Cas9 (clustered regularly interspaced short palindromic repeats / CRISPR associated protein 9), designing and constructing a homologous recombinant vector capable of knocking a reporter gene connected with a spontaneous lysis peptide sequence into an expression cassette of an HMOX1 (heme oxygenase (decycling) 1) gene, and cotransfecting a cell with the homologous recombinant vector, hCas9 (humanized Cas9) plasmid and the sgRNA expression vector for monoclone enlarging culture to form the cell model. The HaCaT cell model with a luciferase gene knocked in before a termination codon of the HMOX1 gene by a combined CRISPR / CAS9 cell monoclone technology. The cell model achieves synchronous expression of the luciferase gene and the HMOX1 gene, so that a sensitization compound and a non-sensitization compound are effectively differentiated, and the more specific and sensitive cell model is provided for compound sensitization studies.
Owner:SOUTH CHINA UNIV OF TECH
Engraftable neural progenitor and stem cells for brain tumor therapy
One of the impediments to the treatment of some human brain tumors (e.g. gliomas) has been the degree to which they expand, migrate widely, and infiltrate normal tissue. We demonstrate that a clone of multipotent neural progenitor stem cells, when implanted into an experimental glioma, will migrate along with and distribute themselves throughout the tumor in juxtaposition to widely expanding and aggressively advancing tumor cells, while continuing to express a foreign reporter gene. Furthermore, drawn somewhat by the degenerative environment created just beyond the infiltrating tumor edge, the neural progenitor cells migrate slightly beyond and surround the invading tumor border. When implanted at a distant sight from the tumor bed (e.g., into normal tissue, into the contralateral hemisphere, into the lateral ventricles) the donor neural progenitor / stem cells will migrate through normal tissue and specifically target the tumor cells. These results suggest the adjunctive use of neural progenitor / stem cells as a novel, effective delivery vehicle for helping to target therapeutic genes and vectors to invasive brain tumors that have been refractory to treatment.
Owner:CHILDRENS MEDICAL CENT CORP +2
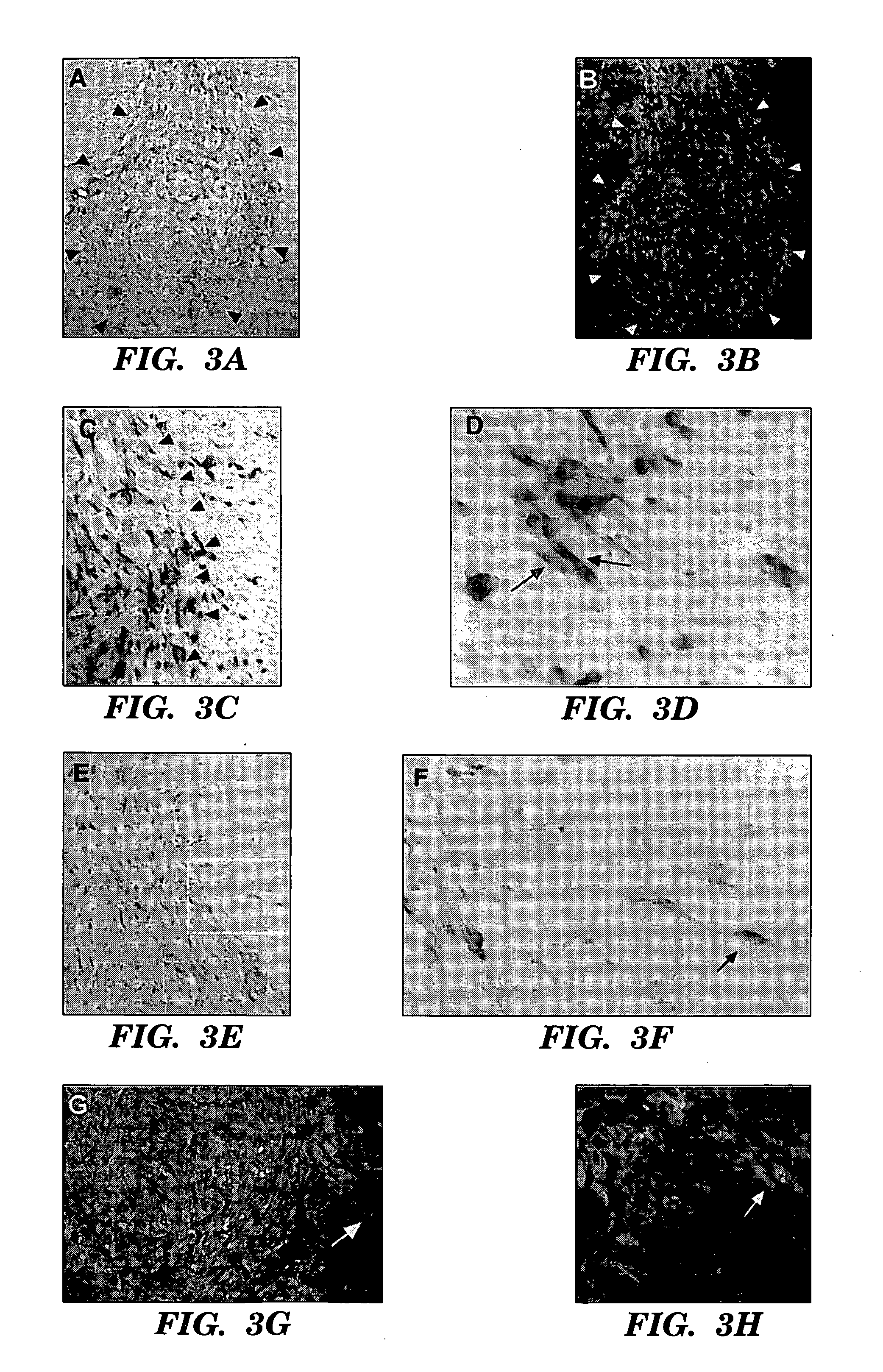
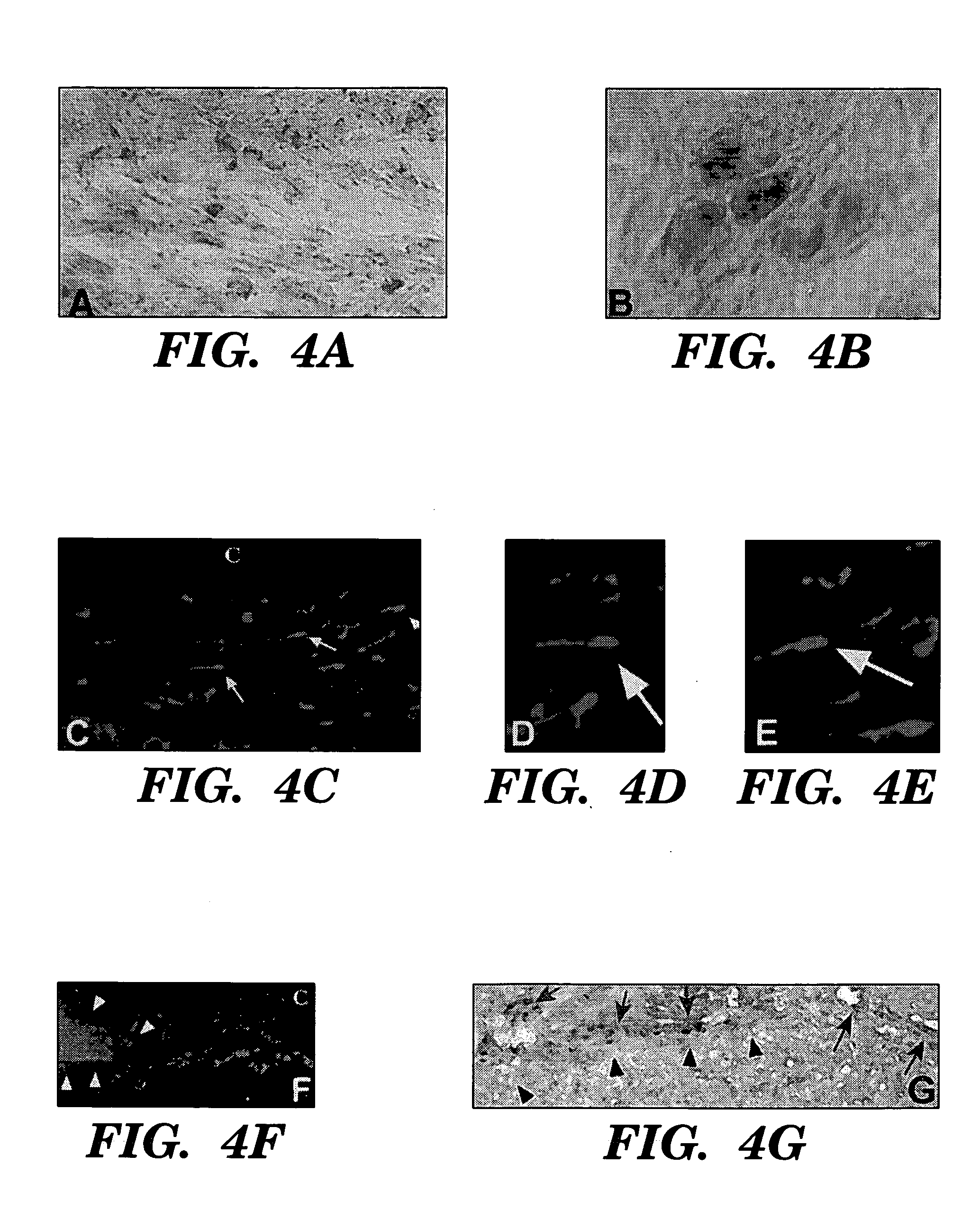
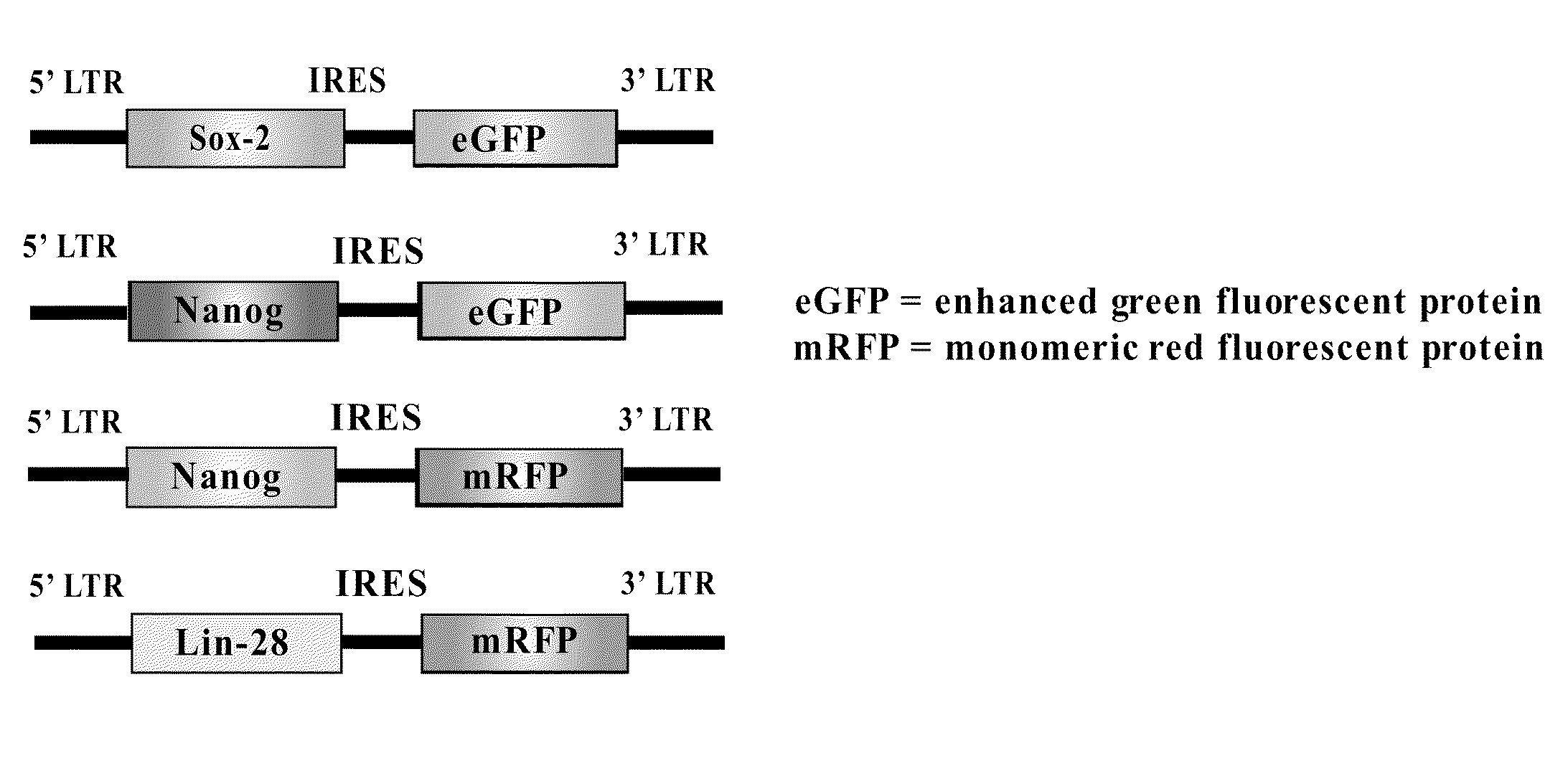
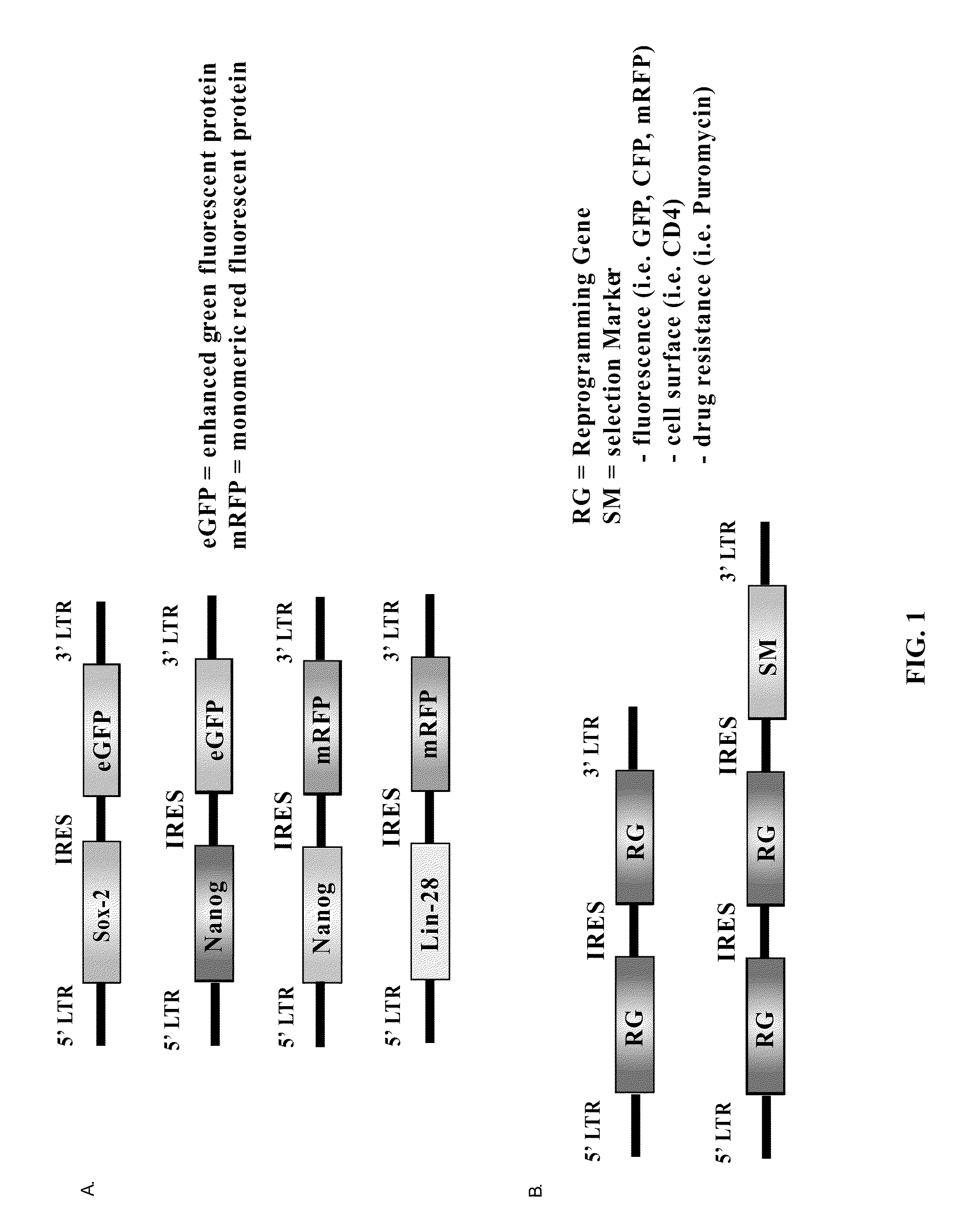
Methods for the production of ips cells
ActiveUS20100041054A1Improving transfectionReduce chanceVirusesGenetically modified cellsGeneHuman Induced Pluripotent Stem Cells
Methods and composition of induction of pluripotent stem cells are disclosed. For example, in certain aspects methods for generating induced pluripotent stem cells using reporter genes are described. Furthermore, the invention provides novel reprogramming vectors employing reporter genes.
Owner:FUJIFILM CELLULAR DYNAMICS INC
Methods and Compositions for Identification of Hydrocarbon Response, Transport and Biosynthesis Genes
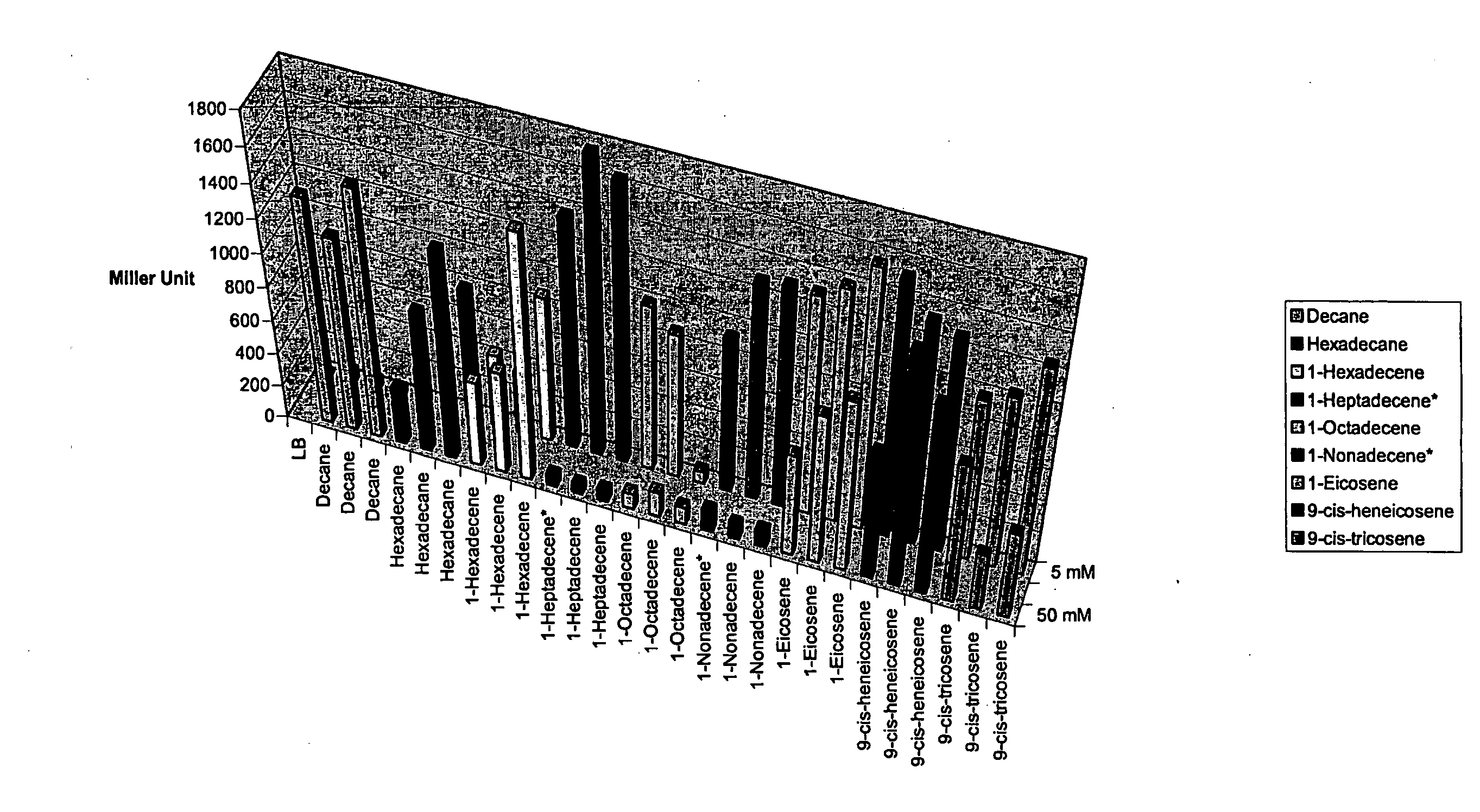
InactiveUS20080293060A1BacteriaMicrobiological testing/measurementBiosynthetic genesResponse element
Disclosed is a method using an alkane response element (ARE) from, e.g., Acinetobacter spp. to (i) identify and clone hydrocarbon biosynthesis genes, (ii) identify and clone hydrocarbon transporter genes (iii) identify and clone hydrocarbon response genes. Screening cells were developed that expressed a transcriptional activator, e.g., alkR, and included a reporter gene, e.g., GFP operatively linked to an ARE promoter, e.g., the alkM promoter. The cells were transformed with libraries from organisms capable of hydrocarbon biosynthesis. Transformed cells that expressed the reporter gene harbored library-derived genes involved in one or more of the above-mentioned processes; and these genes were isolated from the cells using standard molecular biology techniques. Additional systems were designed wherein screening cells also expressed a gene identified in the original screen, e.g., an additional hydrocarbon pathway gene, e.g., an enhancer.
Owner:LS9 INC +1
Haploid yeast cells transformed with a library of expression vectors encoding a fully human antibody repertoire
InactiveUS20030027213A1High affinityEasy to assembleFungiMicrobiological testing/measurementProtein targetTarget peptide
Compostions, kits and methods are provided for generating highly diverse libraries of proteins such as antibodies via homologous recombination in vivo, and screening these libraries against protein, peptide and nucleic acid targets using a two-hybrid method in yeast. The method for screening a library of tester proteins against a target protein or peptide comprises: expressing a library of tester proteins in yeast cells, each tester protein being a fusion protein comprised of a first polypeptide subunit whose sequence varies within the library, a second polypeptide subunit whose sequence varies within the library independently of the first polypeptide, and a linker peptide which links the first and second polypeptide subunits; expressing one or more target fusion proteins in the yeast cells expressing the tester proteins, each of the target fusion proteins comprising a target peptide or protein; and selecting those yeast cells in which a reporter gene is expressed, the expression of the reporter gene being activated by binding of the tester fusion protein to the target fusion protein.
Owner:GENETASTIX CORP
Porous nanoparticle-supported lipid bilayers (protocells) for targeted delivery including transdermal delivery of cargo and methods thereof
InactiveUS20150272885A1Large specific surface areaAmenable to high capacity loadingOrganic active ingredientsPowder deliveryCancers diagnosisBinding peptide
The present invention is directed to protocells for specific targeting of hepatocellular and other cancer cells which comprise a nanoporous silica core with a supported lipid bilayer; at least one agent which facilitates cancer cell death (such as a traditional small molecule, a macromolecular cargo (e.g. siRNA or a protein toxin such as ricin toxin A-chain or diphtheria toxin A-chain) and / or a histone-packaged plasmid DNA disposed within the nanoporous silica core (preferably supercoiled in order to more efficiently package the DNA into protocells) which is optionally modified with a nuclear localization sequence to assist in localizing protocells within the nucleus of the cancer cell and the ability to express peptides involved in therapy (apoptosis / cell death) of the cancer cell or as a reporter, a targeting peptide which targets cancer cells in tissue to be treated such that binding of the protocell to the targeted cells is specific and enhanced and a fusogenic peptide that promotes endosomal escape of protocells and encapsulated DNA. Protocells according to the present invention may be used to treat cancer, especially including hepatocellular (liver) cancer using novel binding peptides (c-MET peptides) which selectively bind to hepatocellular tissue or to function in diagnosis of cancer, including cancer treatment and drug discovery.
Owner:STC UNM +1
Non-invasive evaluation of physiological response in a transgenic mouse
Owner:XENOGEN CORP
Cell regulatory genes, encoded products, and uses related thereto
InactiveUS6946256B1Peptide/protein ingredientsImmunoglobulins against animals/humansSuppressorApoptosis
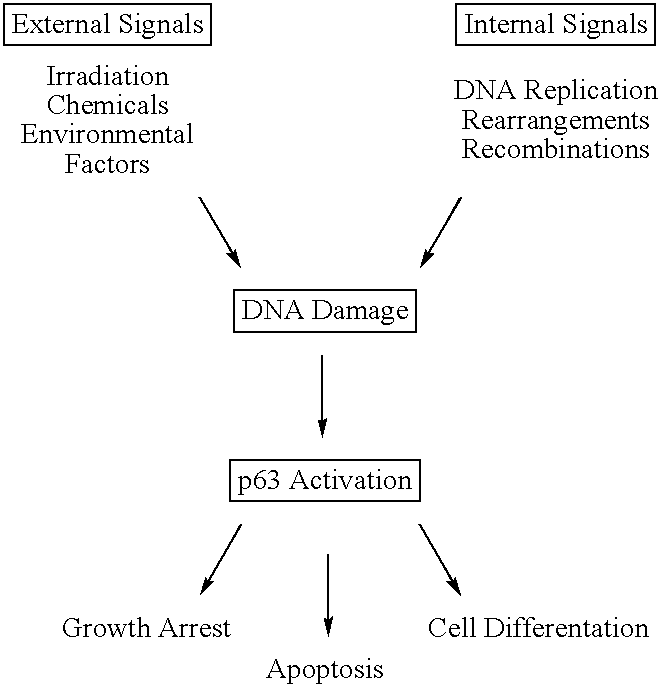
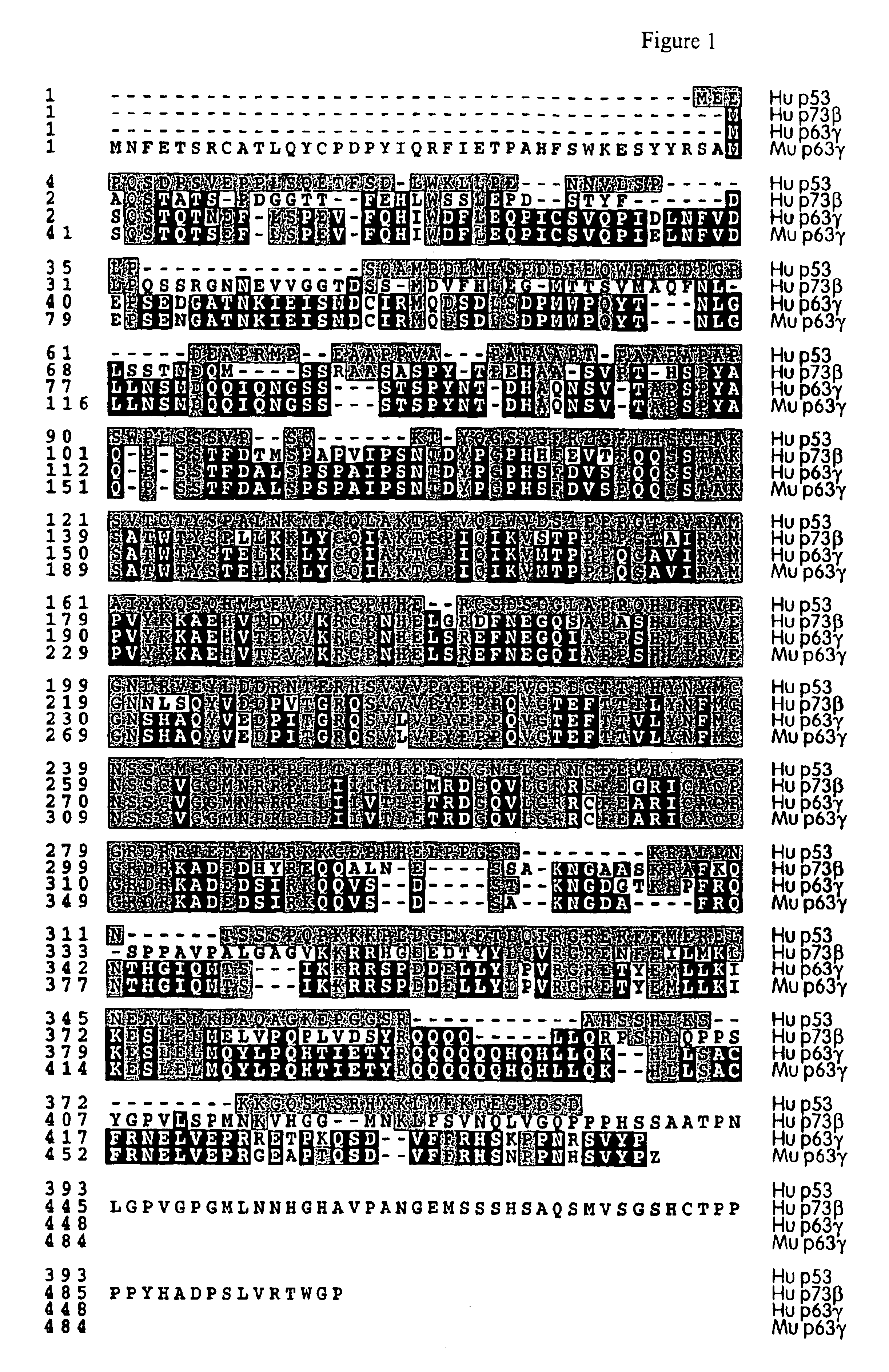
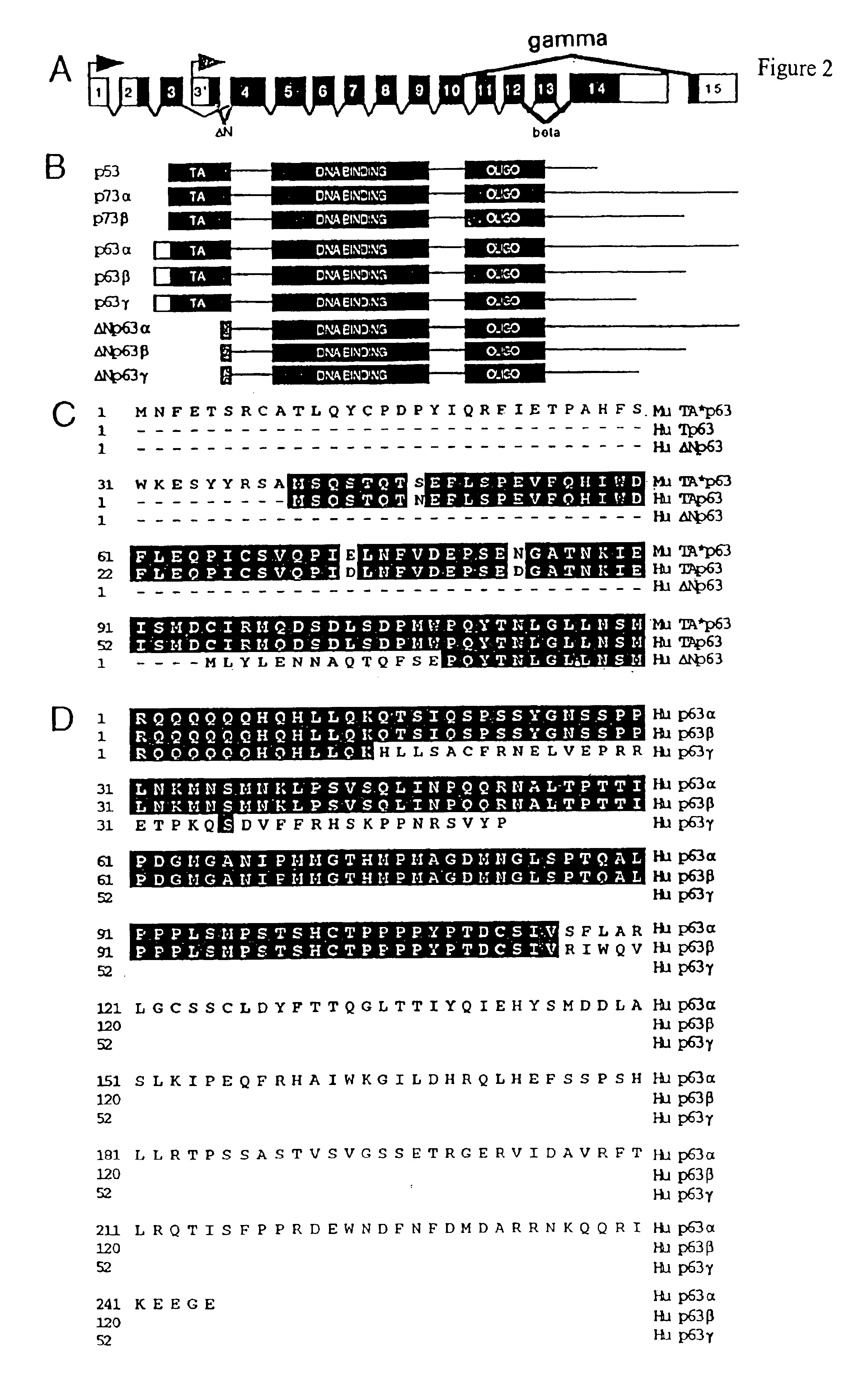
This application describes the cloning of p63, a gene at chromosome 3q27-29, that bears homology to the tumor suppressor p53. The p63 gene encodes at least six different isotypes. p63 was detected in a variety of human and mouse tissue and demonstrates remarkably divergent activities, such as the ability to transactivate p53 reporter genes and induce apoptosis. Isotopes of p63 lacking a transactivation domain act as dominant negatives towards the transactivation by p53 and p63.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC +2
Particle-based multiplex assay system with three or more assay reporters
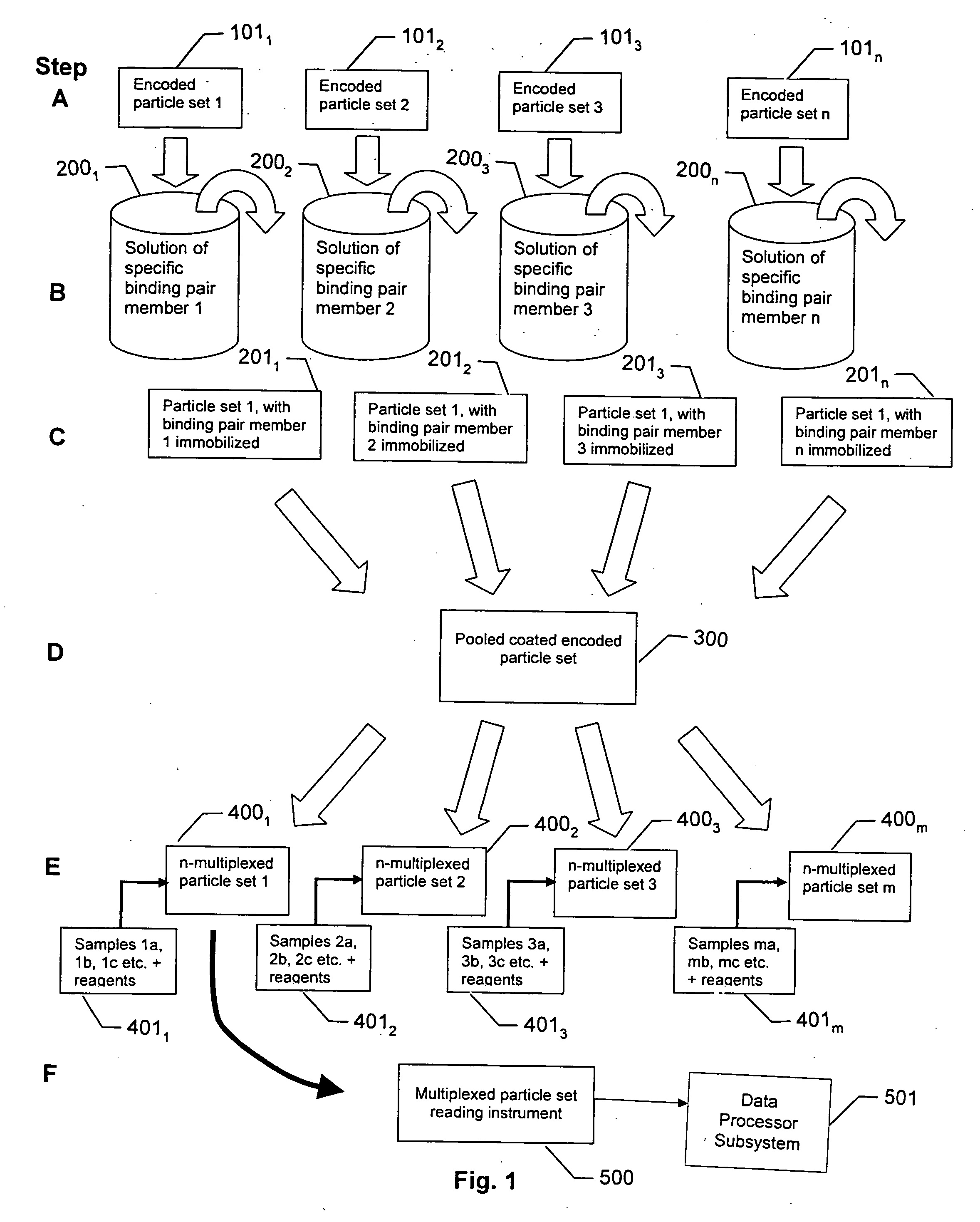
InactiveUS20060177850A1Increase the number ofImprove abilitiesBioreactor/fermenter combinationsSequential/parallel process reactionsReference sampleDiscriminator
A system and method for developing and utilizing particle-based n-multiplexed assays that include three or more reporters utilizes n particle sets that are associated with particle identification images or labels (IDs) that differ between sets. The encoded particles for a given set are coated with a specific binding member, or in the case of the sandwich assay with coupled capture and detector binding pair members, to form particle types. The sets of particle types are then pooled, and aliquots of the particle types are removed to assay vessels. Next, samples with three or four reporter molecules are supplied to the respective vessels. After one or more incubation periods, the particles are supplied to a reader system, which determines the particle IDs to identify the particle types and also detects the reporter signals. The reader system includes multiple excitation lasers that excite the various reporters in sequence or in parallel, to supply associated signals to one or more detectors. Emission filters and wavelength discriminators are included such that a given detector receives at a given time the signals associated with a single assay binding label. The system further develops greater capacity sandwich assays by assigning subsets of capture and detector antibody pairings to the three or four reporters, respectively. The system performs greater numbers of differential RNA expressions based on the use of the three or more reporters, with one or more reporters assigned to the reference sample and the other reporters assigned to respective test samples. The system and method are also capable of performing greater numbers of SNPs utilizing primer extension reactions, by assigning different color reporters to the respective nucleotides or terminators.
Owner:PERKINELMER HEALTH SCIENCES INC
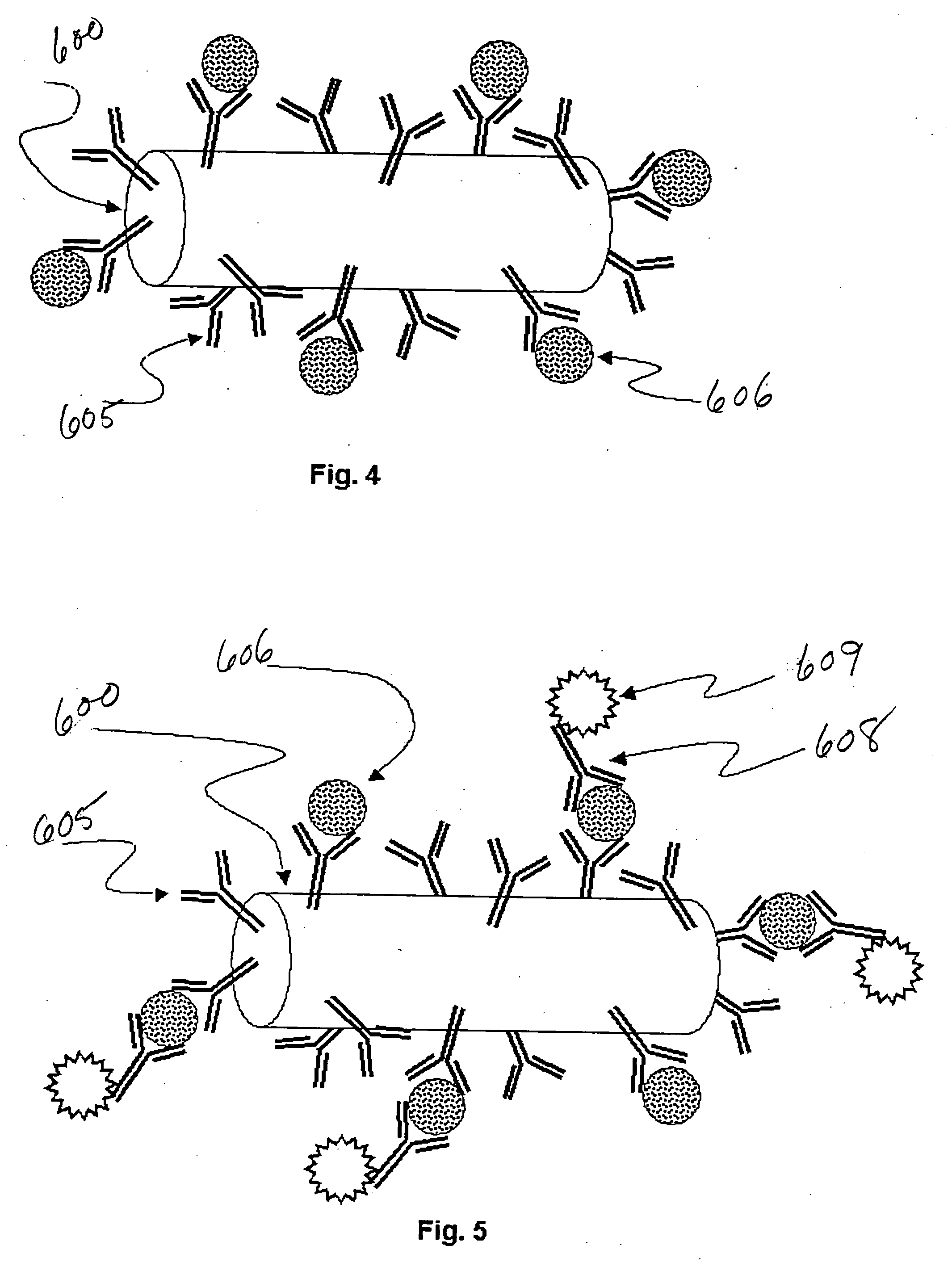
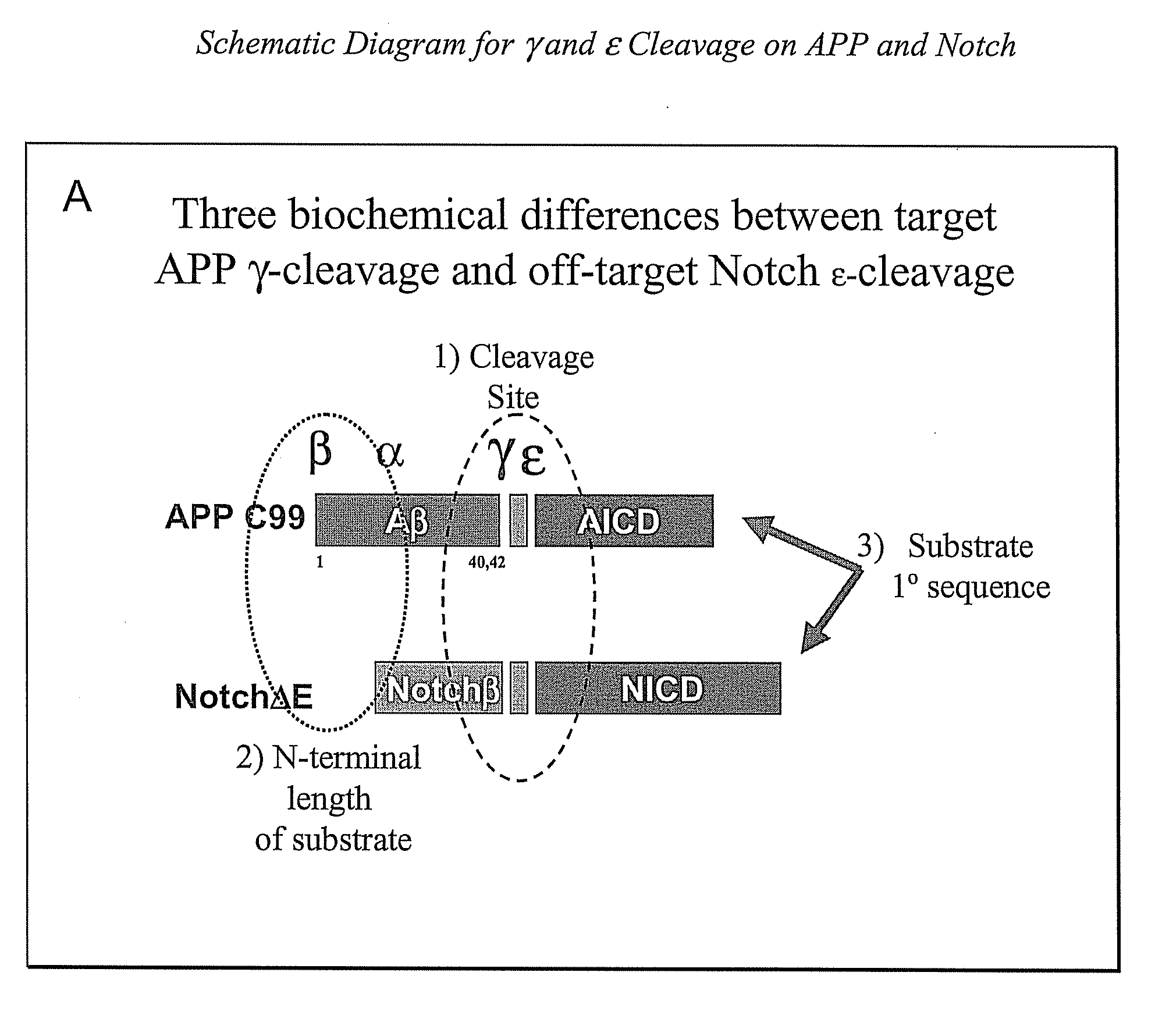
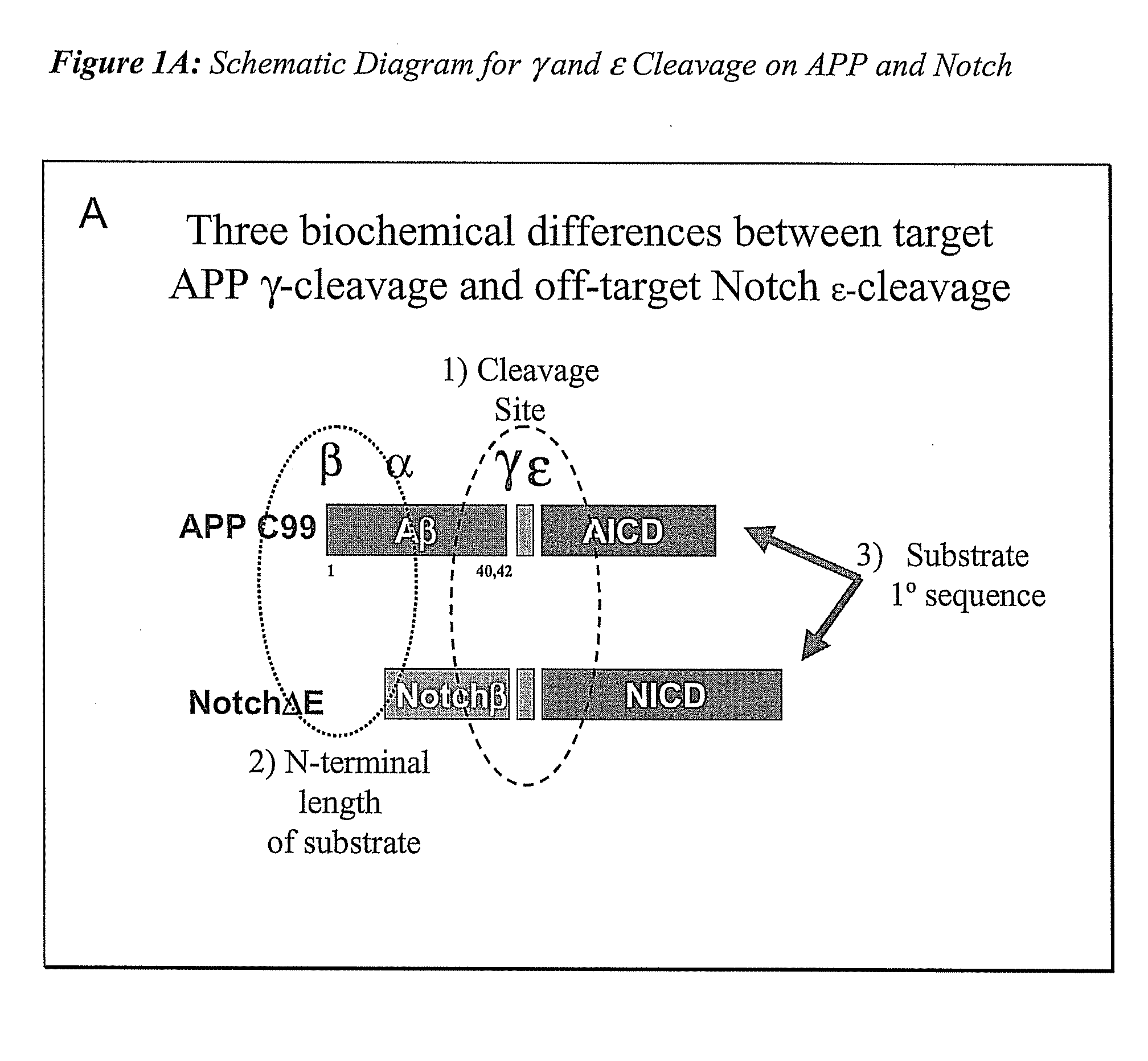
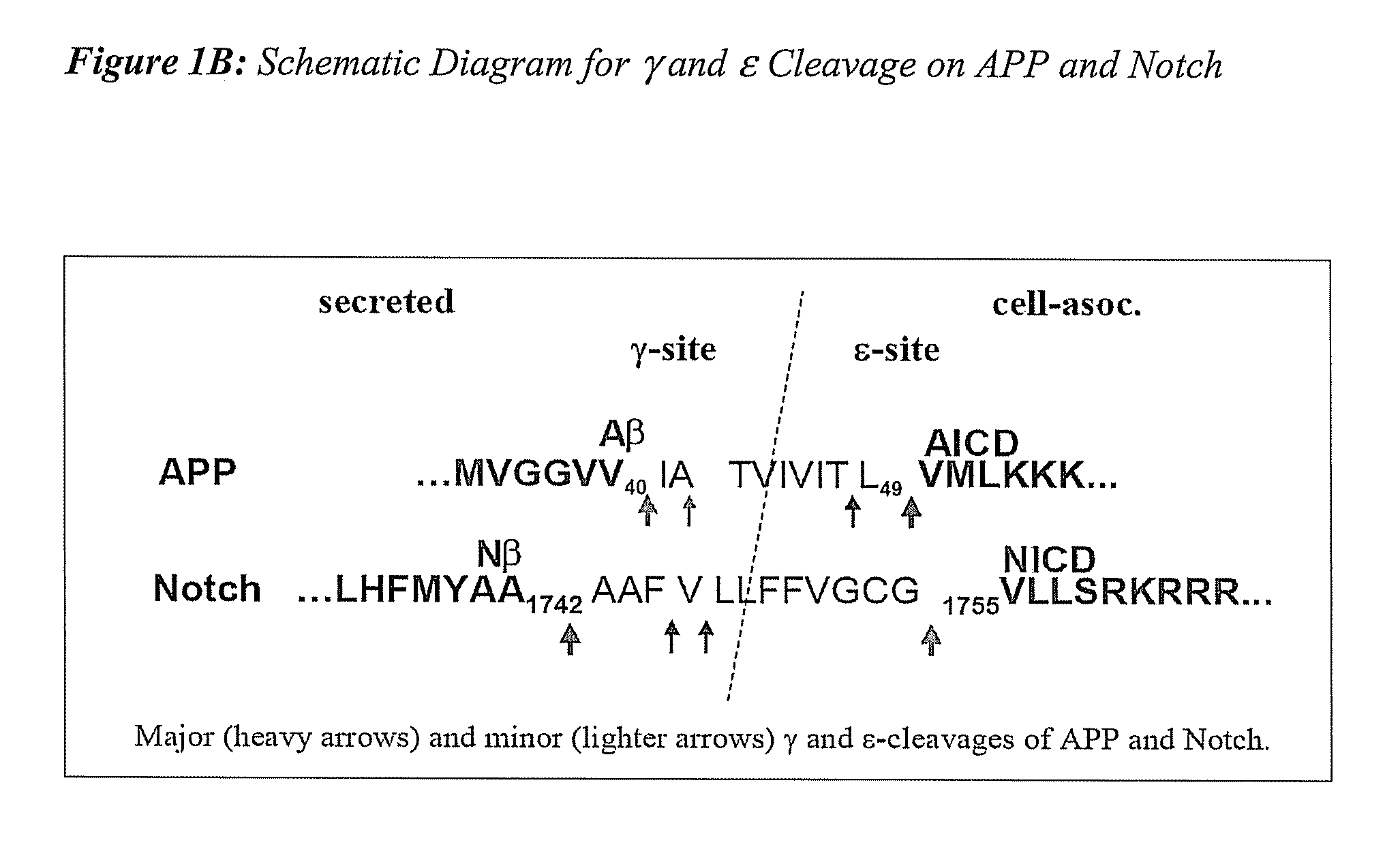
Triple Assay System for Identifying Substrate Selectivity of Gamma Secretase Inhibitors
The invention provides assays and methods for determining whether a compound inhibits gamma secretase in a substrate specific manner. The invention provides an isolated cell wherein the cell stably expresses APP and at least one gamma secretase substrate other than APP. The invention provides assays and methods comprising contacting a cell with gamma secretase and detecting production of Abeta, detecting production of intracellular domain (ICD), and detecting a signal from a reporter gene under transcriptional control of the ICD. The invention also provides compounds that inhibit gamma secretase, pharmaceutical compositions comprising such compounds, and methods of treating Alzheimer's disease using such compounds.
Owner:ELAN PHARM INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com