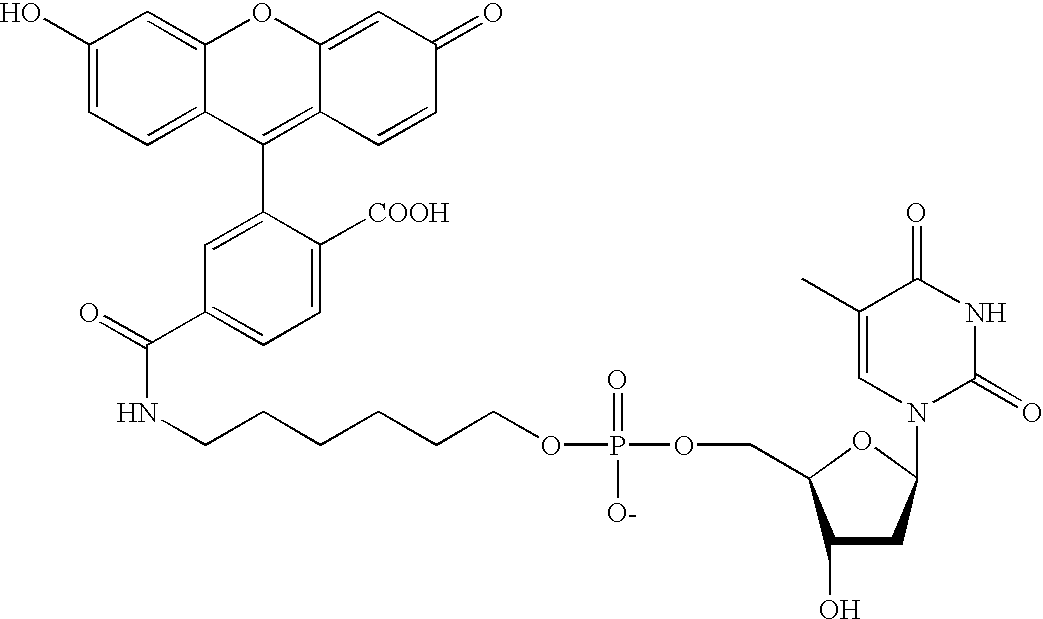
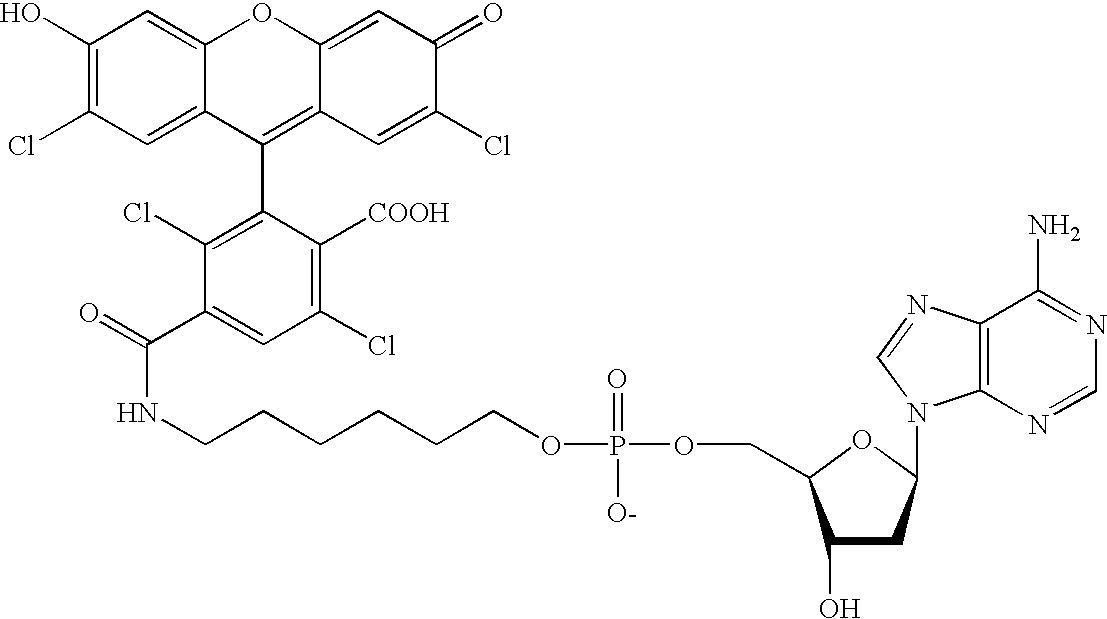
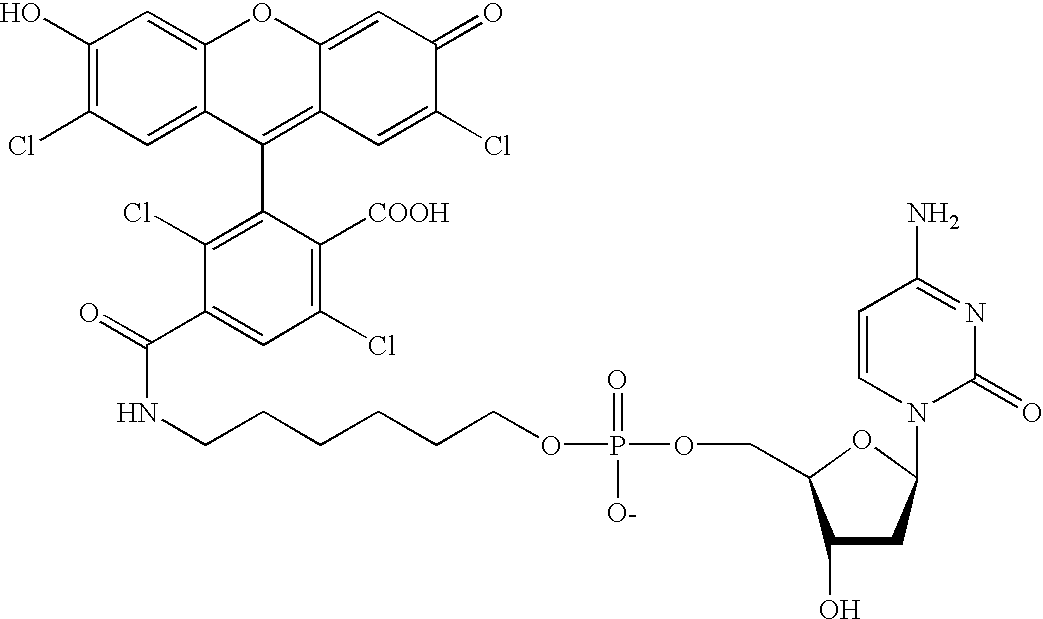
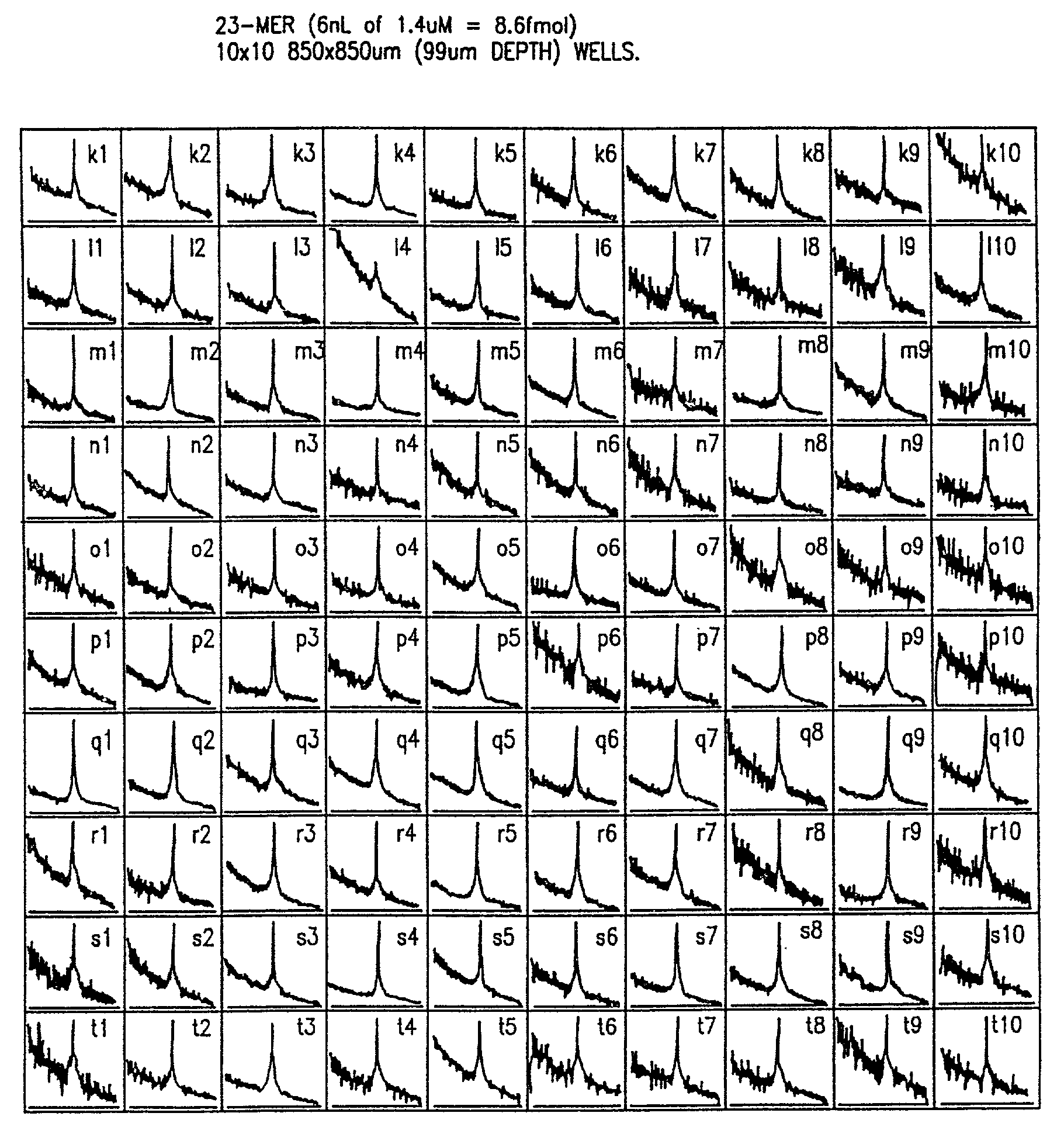
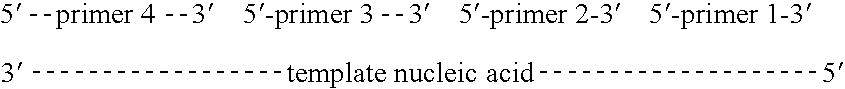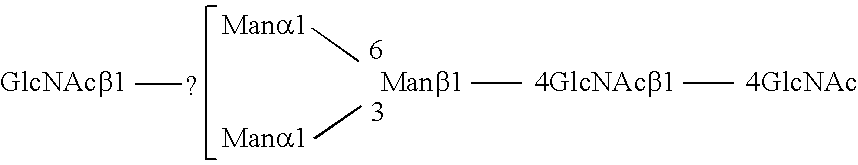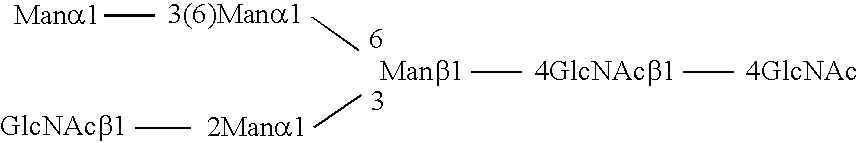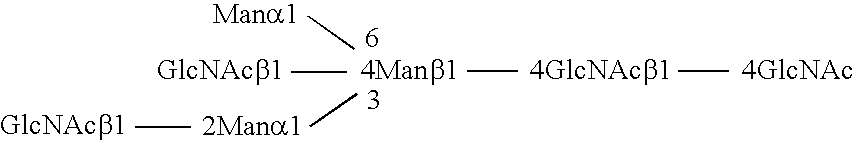Patents
Literature
1923results about "Peptide libraries" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
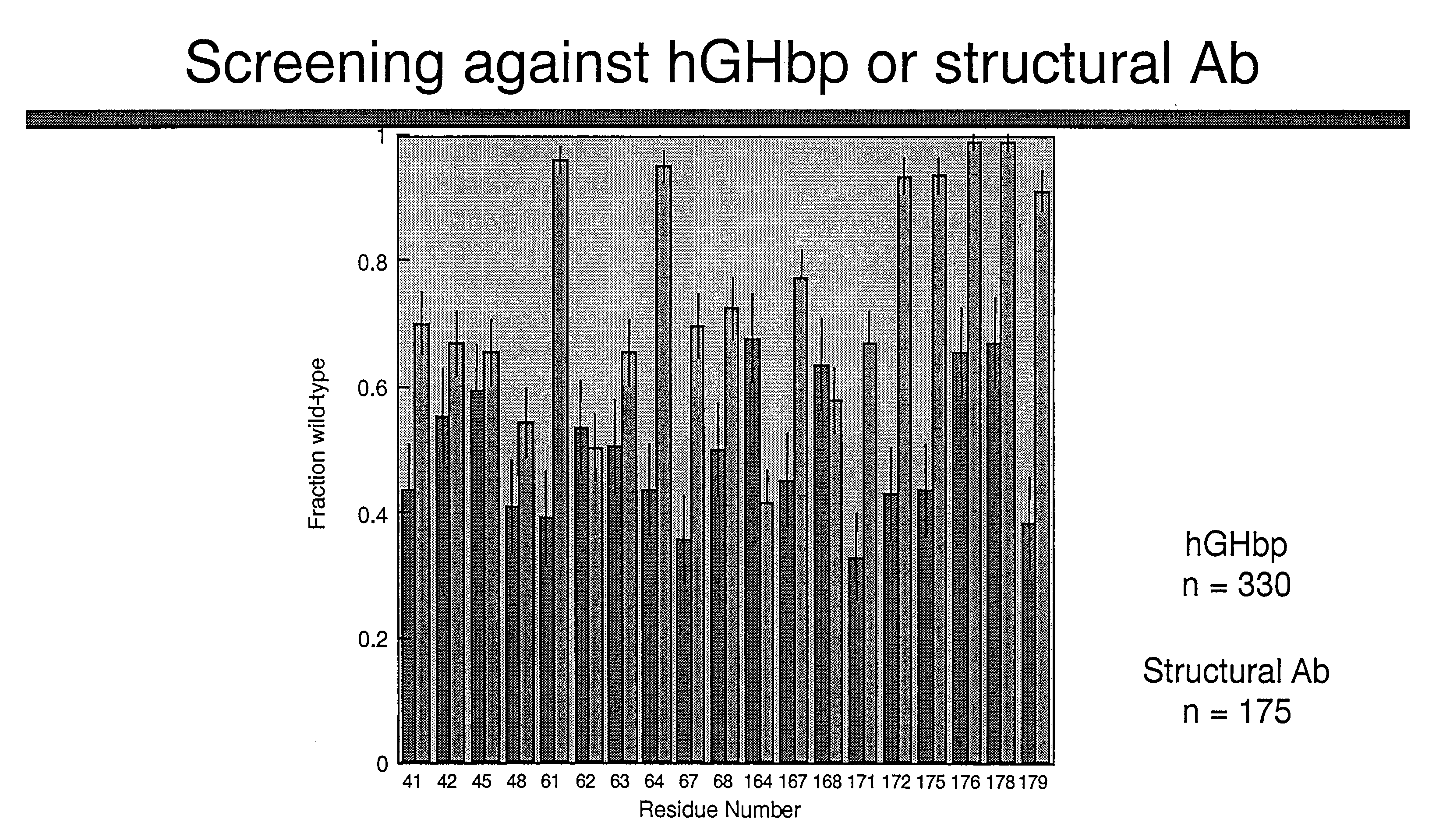
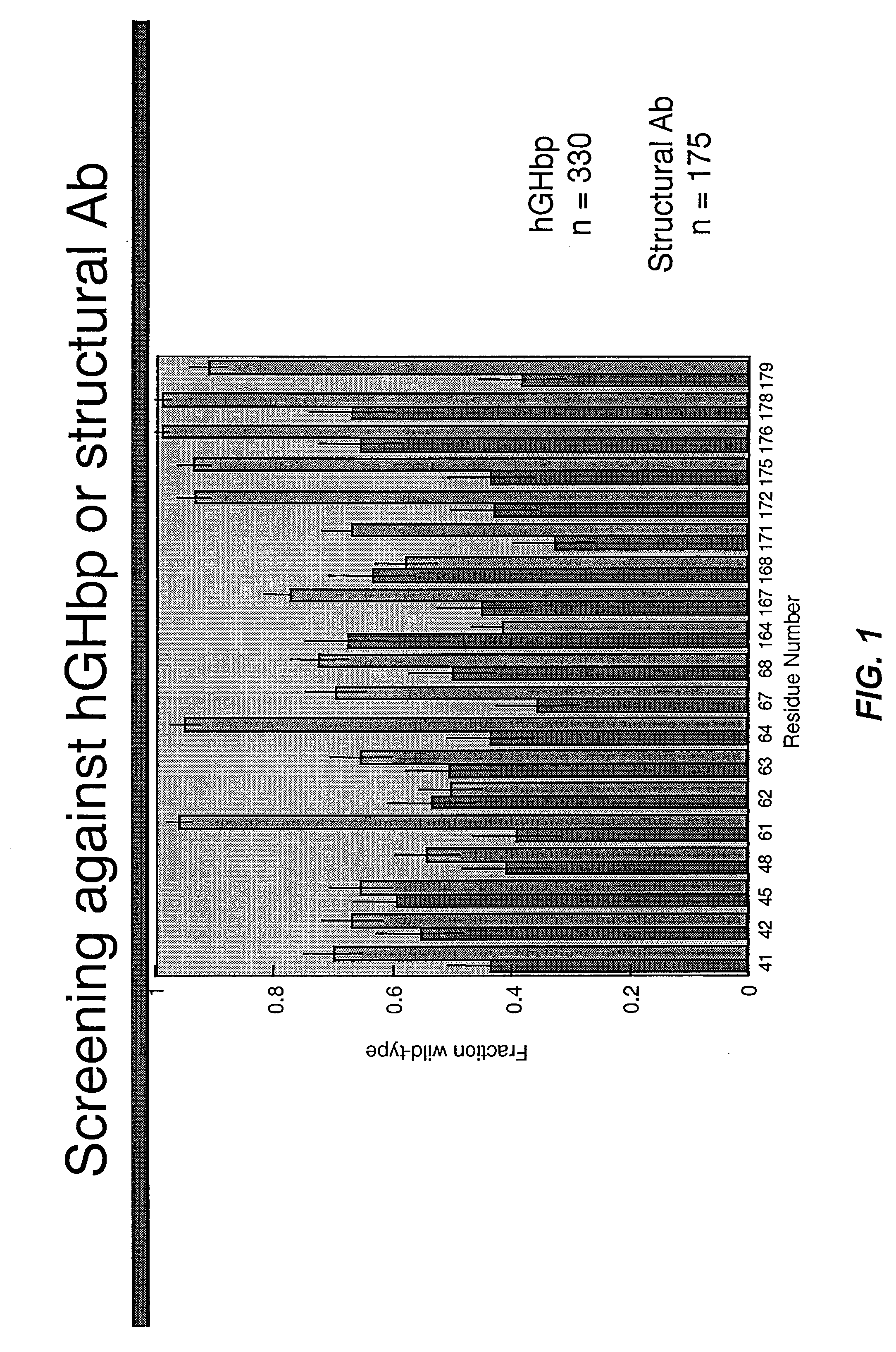
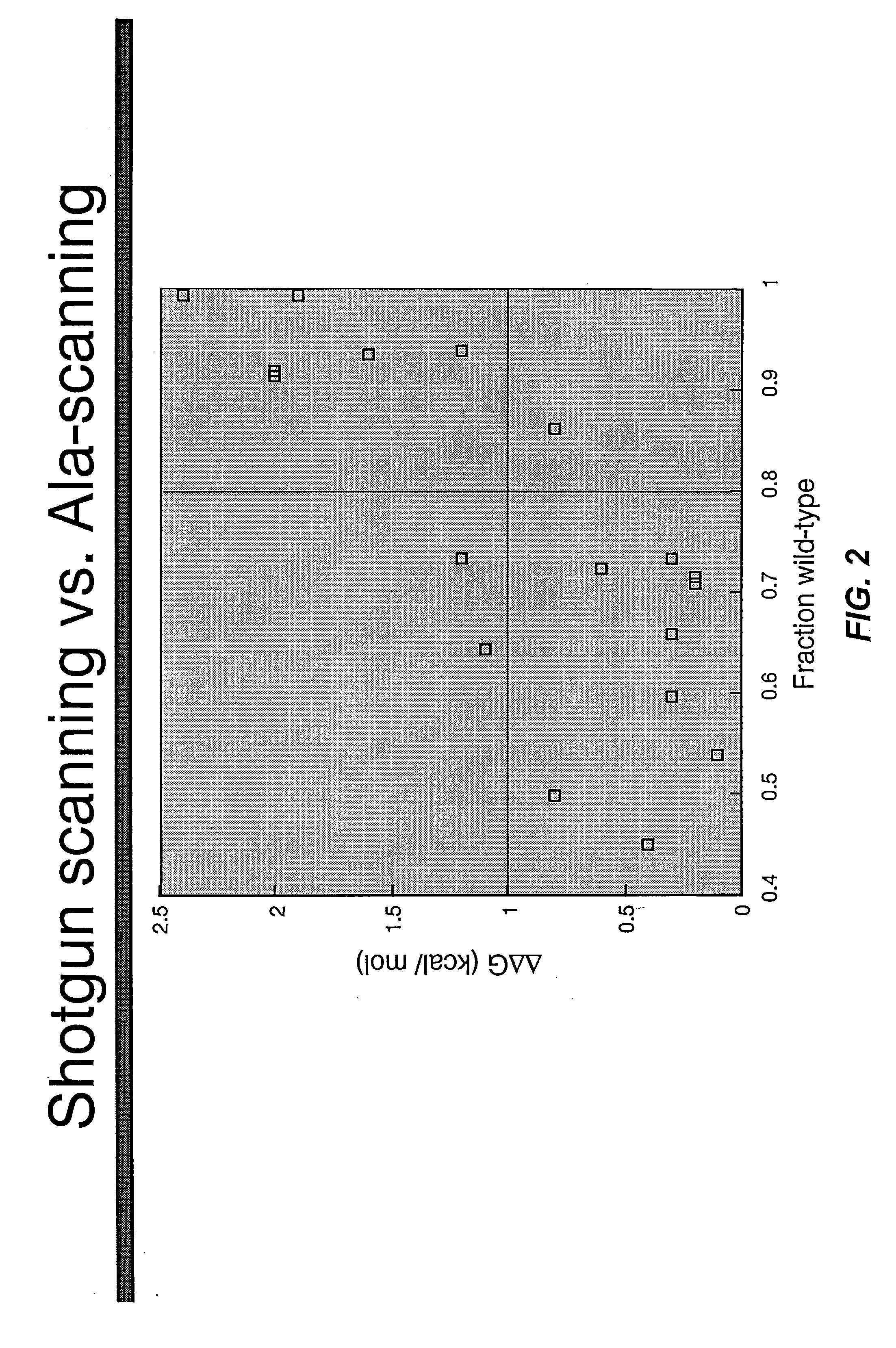
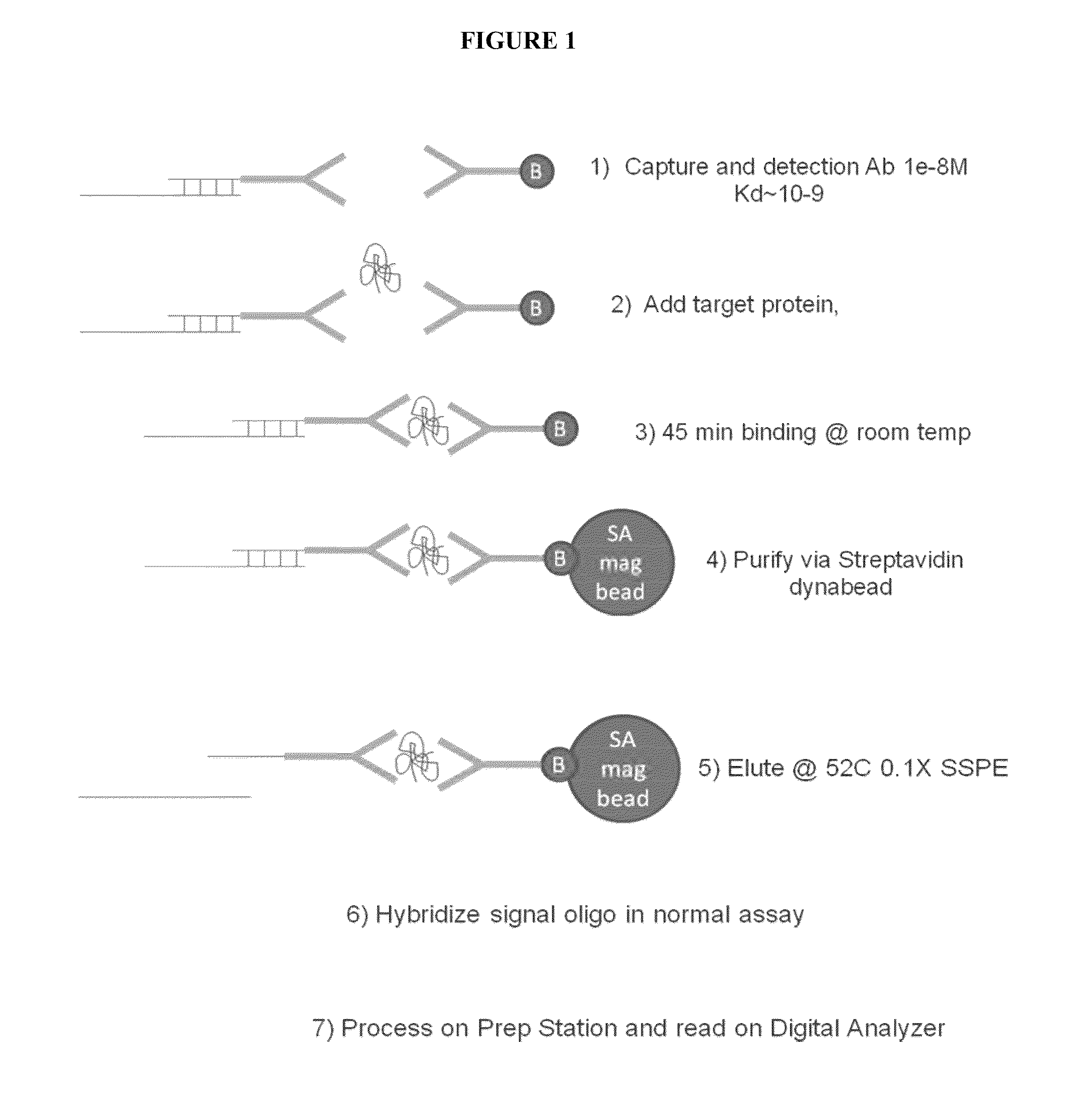
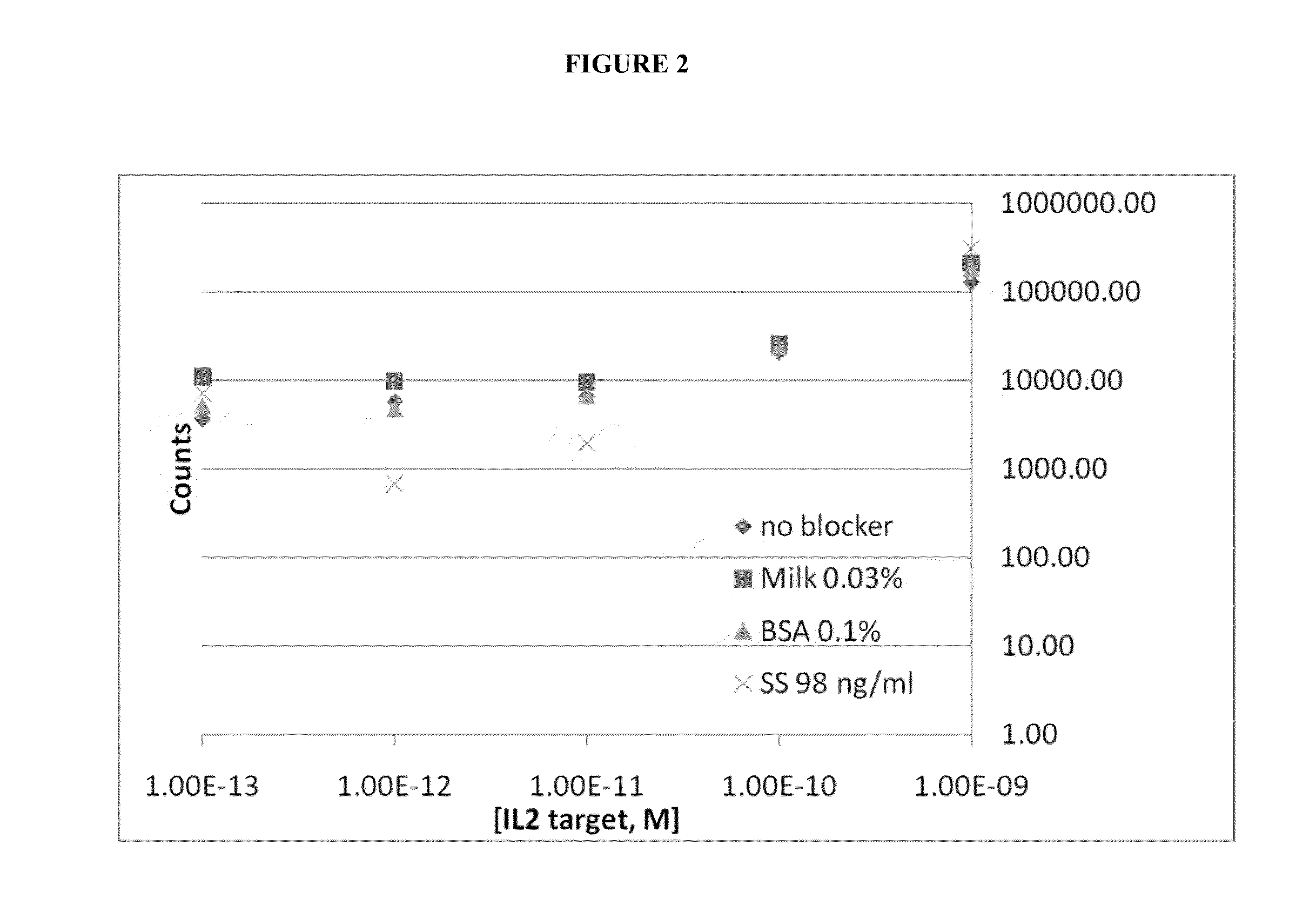
Shotgun scanning
A combinatorial method that uses statistics and DNA sequence analysis rapidly assesses the functional and structural importance of individual protein side chains to binding interactions. This general method, termed “shotgun scanning”, enables the rapid mapping of functional protein and peptide epitopes and is suitable for high throughput proteomics.
Owner:GENENTECH INC
Methods of amplifying and sequencing nucleic acids
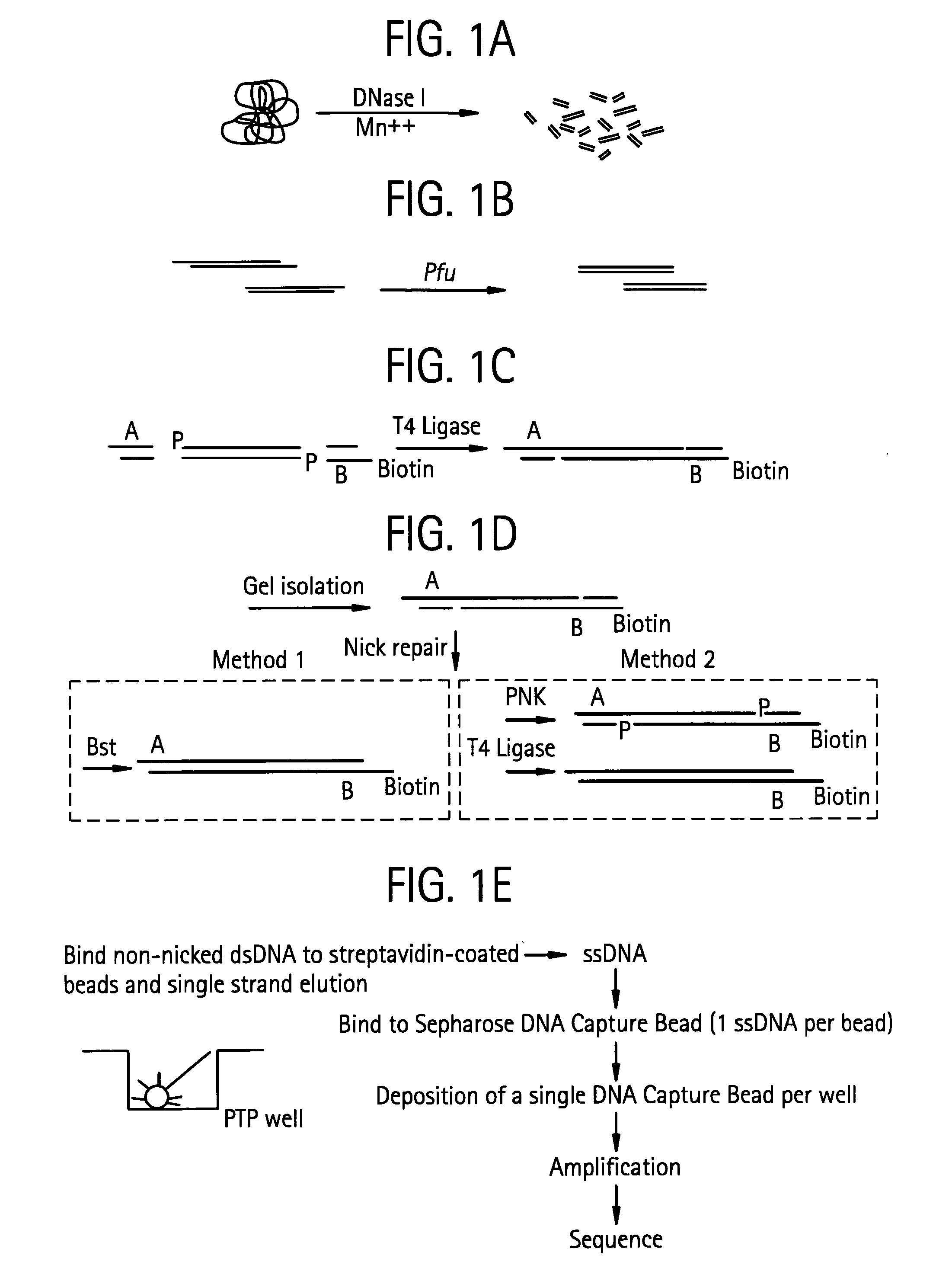
An apparatus and method for performing rapid DNA sequencing, such as genomic sequencing, is provided herein. The method includes the steps of preparing a sample DNA for genomic sequencing, amplifying the prepared DNA in a representative manner, and performing multiple sequencing reaction on the amplified DNA with only one primer hybridization step.
Owner:454 LIFE SCIENCES CORP
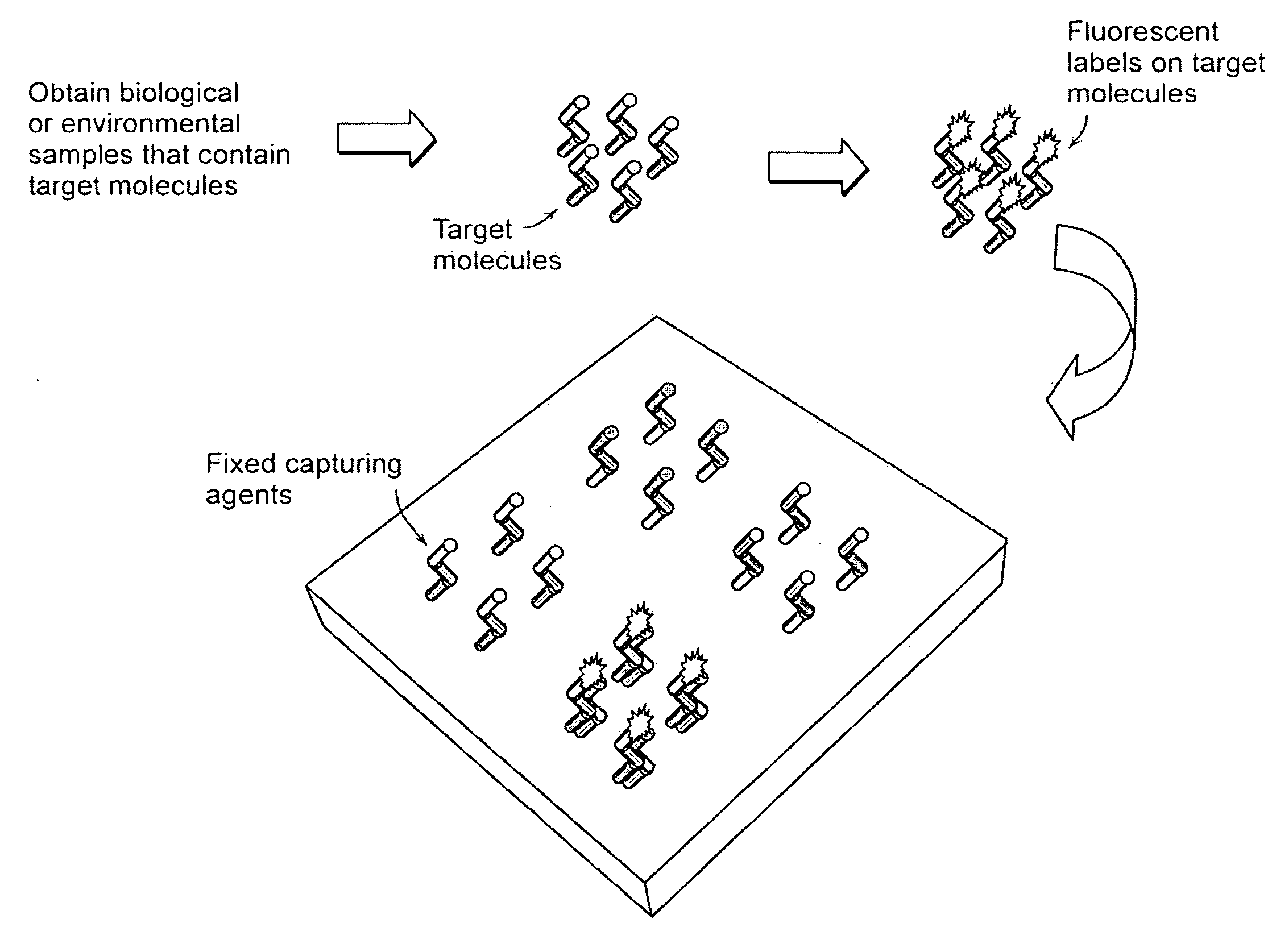
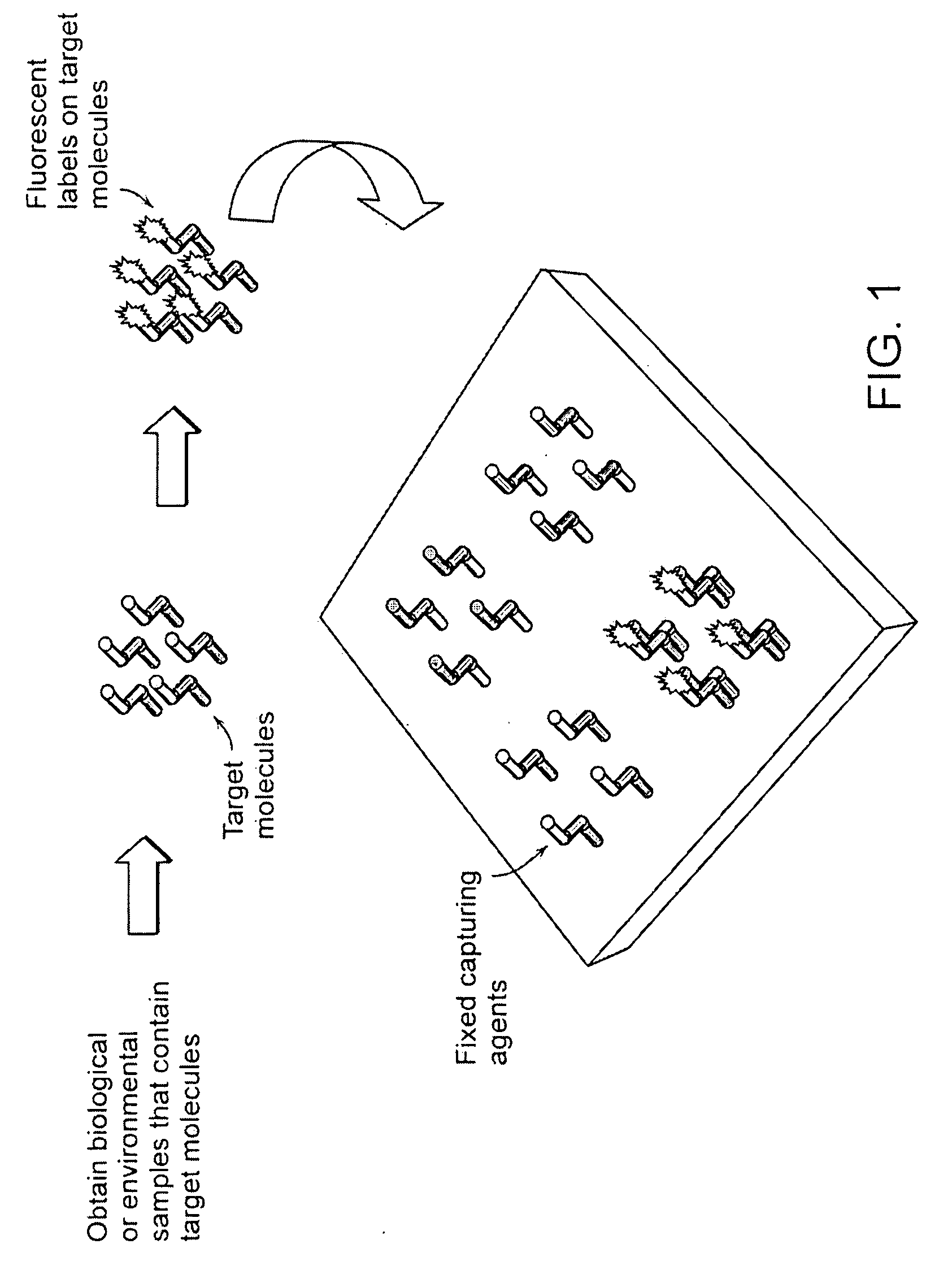
Target analyte sensors utilizing microspheres
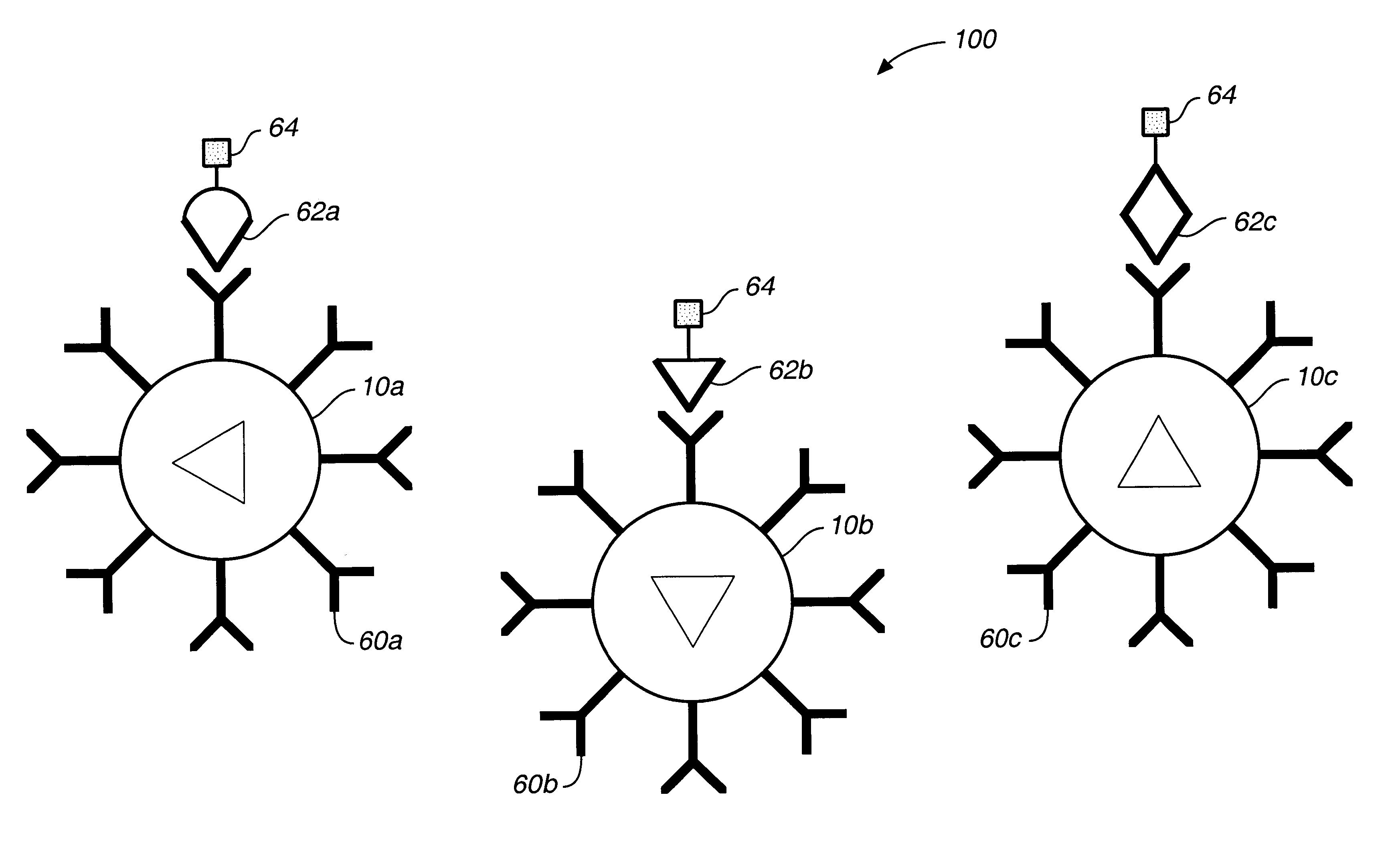
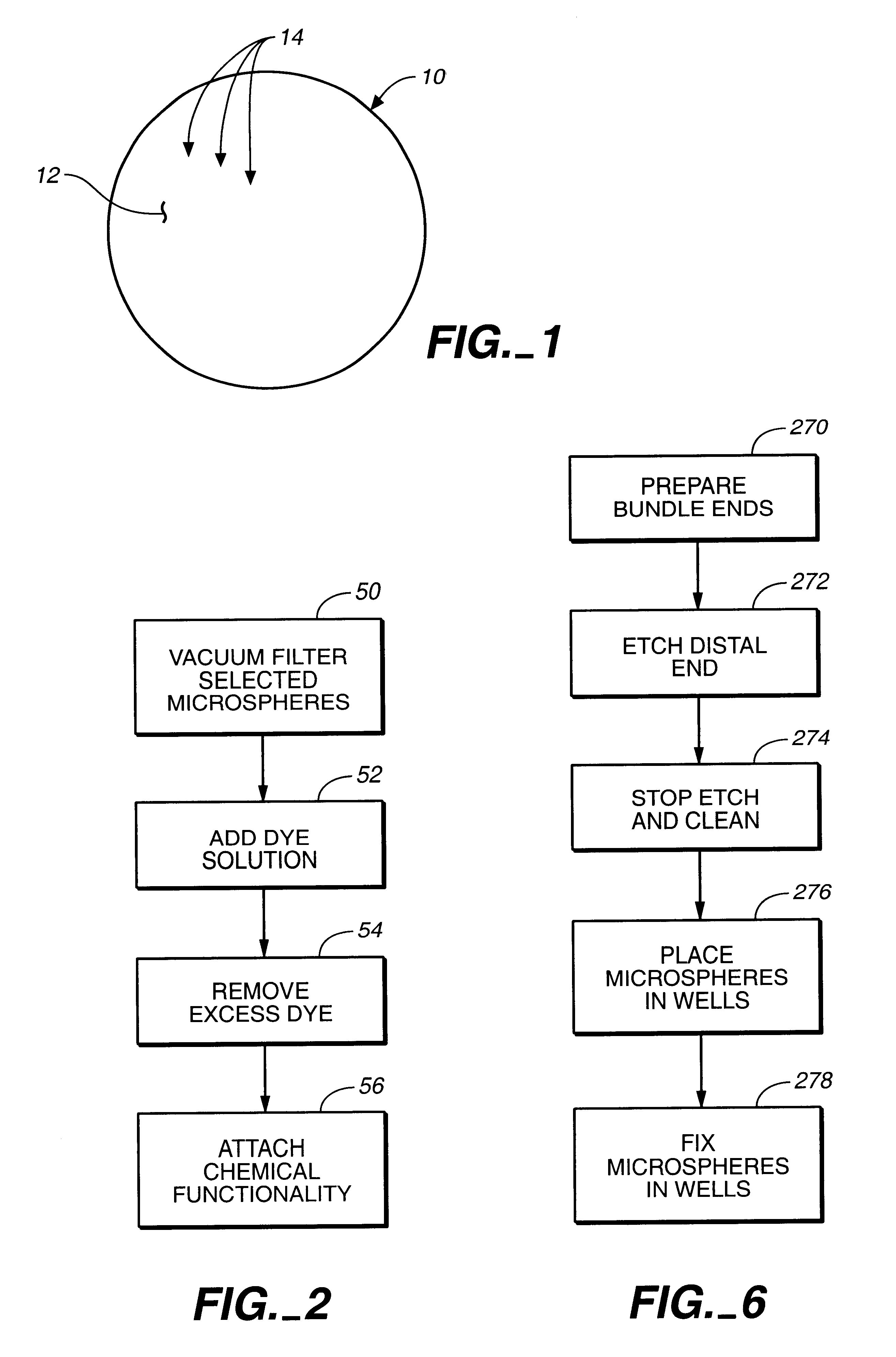
A microsphere-based analytic chemistry system and method for making the same is disclosed in which microspheres or particles carrying bioactive agents may be combined randomly or in ordered fashion and dispersed on a substrate to form an array while maintaining the ability to identify the location of bioactive agents and particles within the array using an optically interrogatable, optical signature encoding scheme. A wide variety of modified substrates may be employed which provide either discrete or non-discrete sites for accommodating the microspheres in either random or patterned distributions. The substrates may be constructed from a variety of materials to form either two-dimensional or three-dimensional configurations. In a preferred embodiment, a modified fiber optic bundle or array is employed as a substrate to produce a high density array. The disclosed system and method have utility for detecting target analytes and screening large libraries of bioactive agents.
Owner:TRUSTEES OF TUFTS COLLEGETHE
Novel proteins with targeted binding
InactiveUS20050089932A1Stimulate and inhibit activityEasy screeningPeptide librariesPeptide/protein ingredientsMonomerComputational biology
Methods for identifying discrete monomer domains and immuno-domains with a desired property are provided. Methods for generating multimers from two or more selected discrete monomer domains are also provided, along with methods for identifying multimers possessing a desired property. Presentation systems are also provided which present the discrete monomer and / or immuno-domains, selected monomer and / or immuno-domains, multimers and / or selected multimers to allow their selection. Compositions, libraries and cells that express one or more library member, along with kits and integrated systems, are also included in the present invention.
Owner:AMGEN MOUNTAIN VIEW
Droplet Libraries
Owner:BIO RAD LAB INC
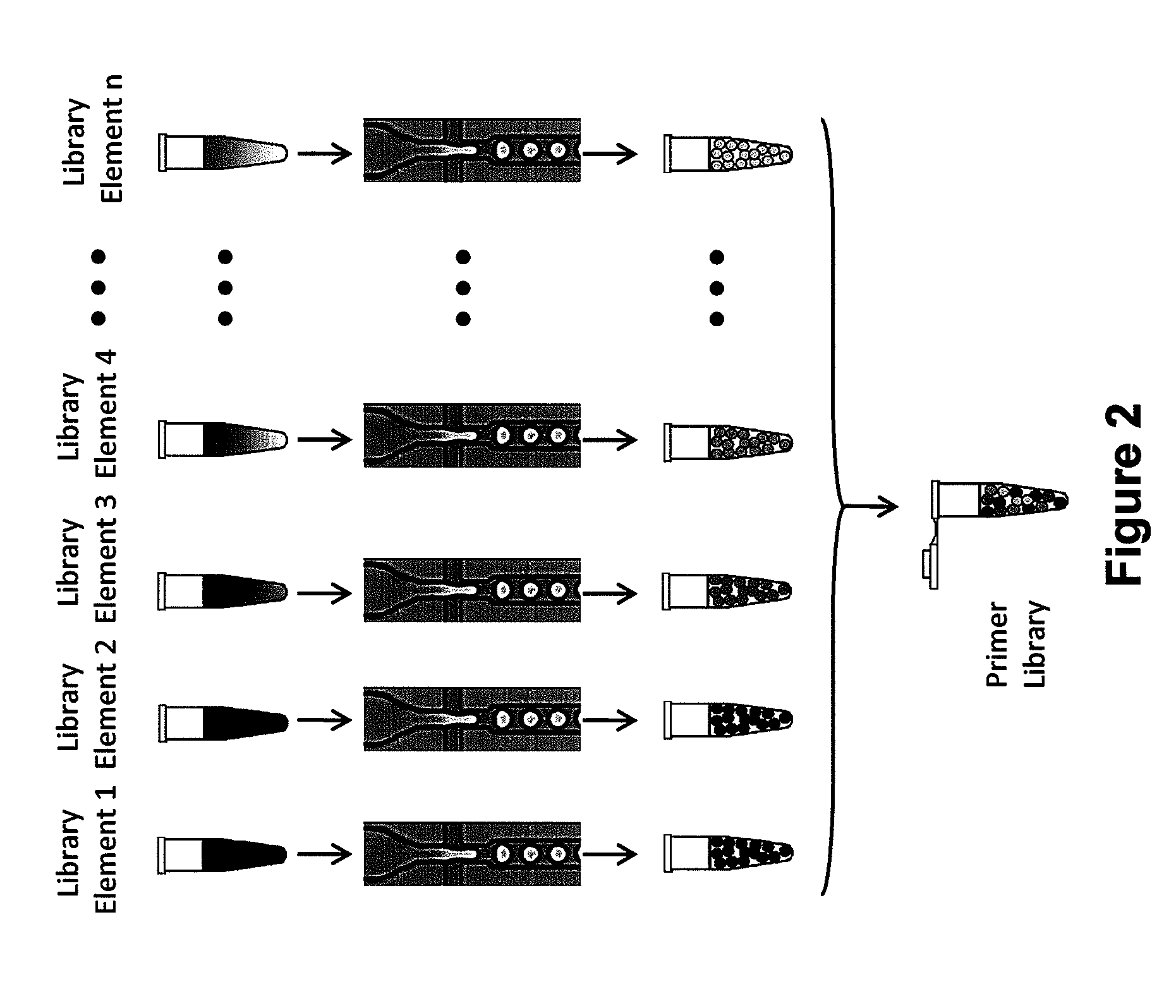
Combinatorial libraries of monomer domains
InactiveUS20050221384A1Specificity of bindingEasy screeningPeptide librariesMicrobiological testing/measurementMonomerChemistry
Methods for identifying discrete monomer domains and immuno-domains with a desired property are provided. Methods for generating multimers from two or more selected discrete monomer domains are also provided, along with methods for identifying multimers possessing a desired property. Presentation systems are also provided which present the discrete monomer and / or immuno-domains, selected monomer and / or immuno-domains, multimers and / or selected multimers to allow their selection. Compositions, libraries and cells that express one or more library member, along with kits and integrated systems, are also included in the present invention.
Owner:AMGEN MOUNTAIN VIEW
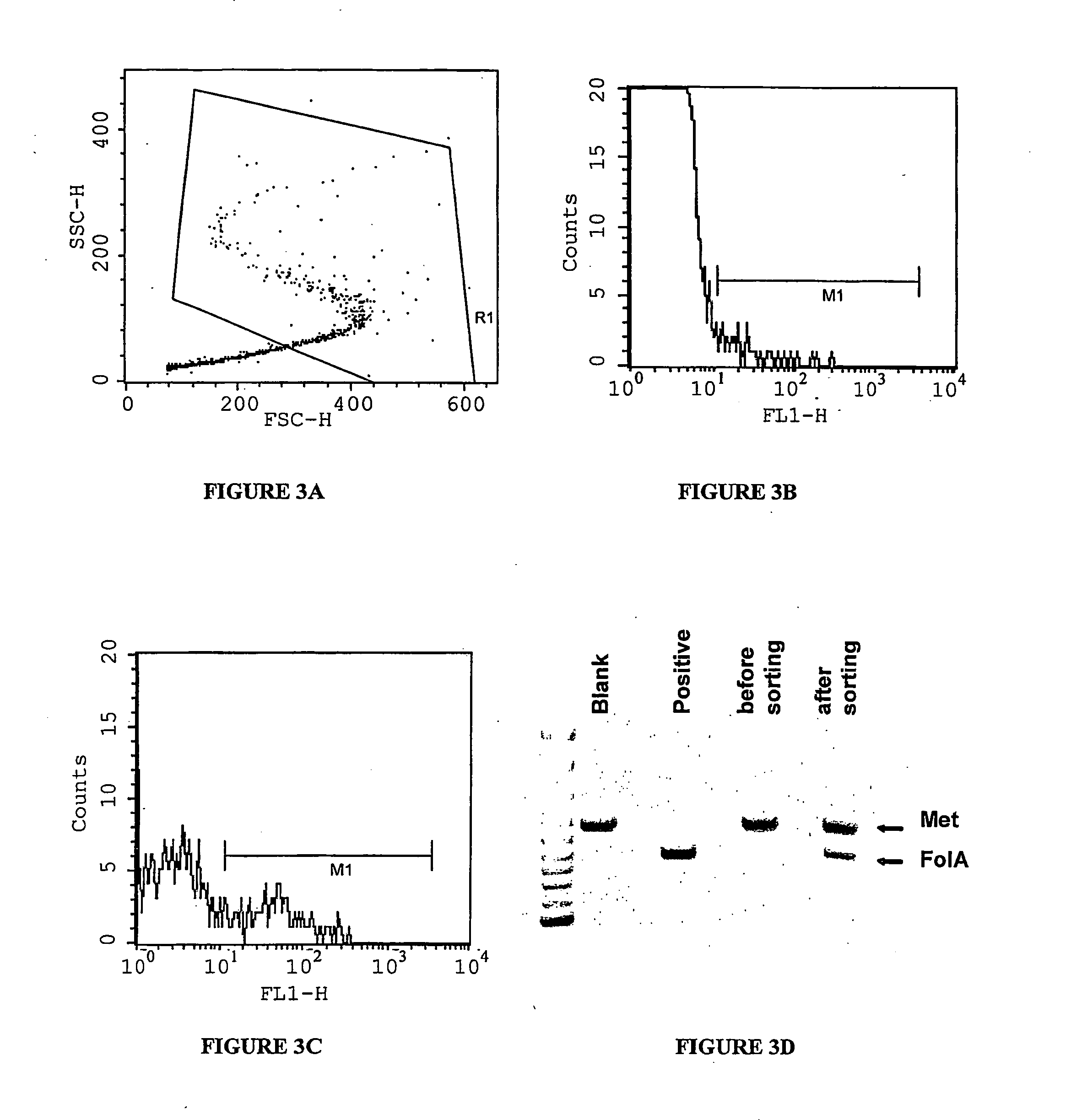
c-Met kinase binding proteins
InactiveUS20060008844A1Stimulate and inhibit activityCompound screeningPeptide librariesKinase bindingC-Met
Owner:AMGEN MOUNTAIN VIEW
Method to screen phage display libraries with different ligands
InactiveUS6846634B1Overcome inherent biasGuaranteed effective sizePeptide librariesLibrary screeningBinding siteBacteriophage
Owner:DORMANTIS LTD
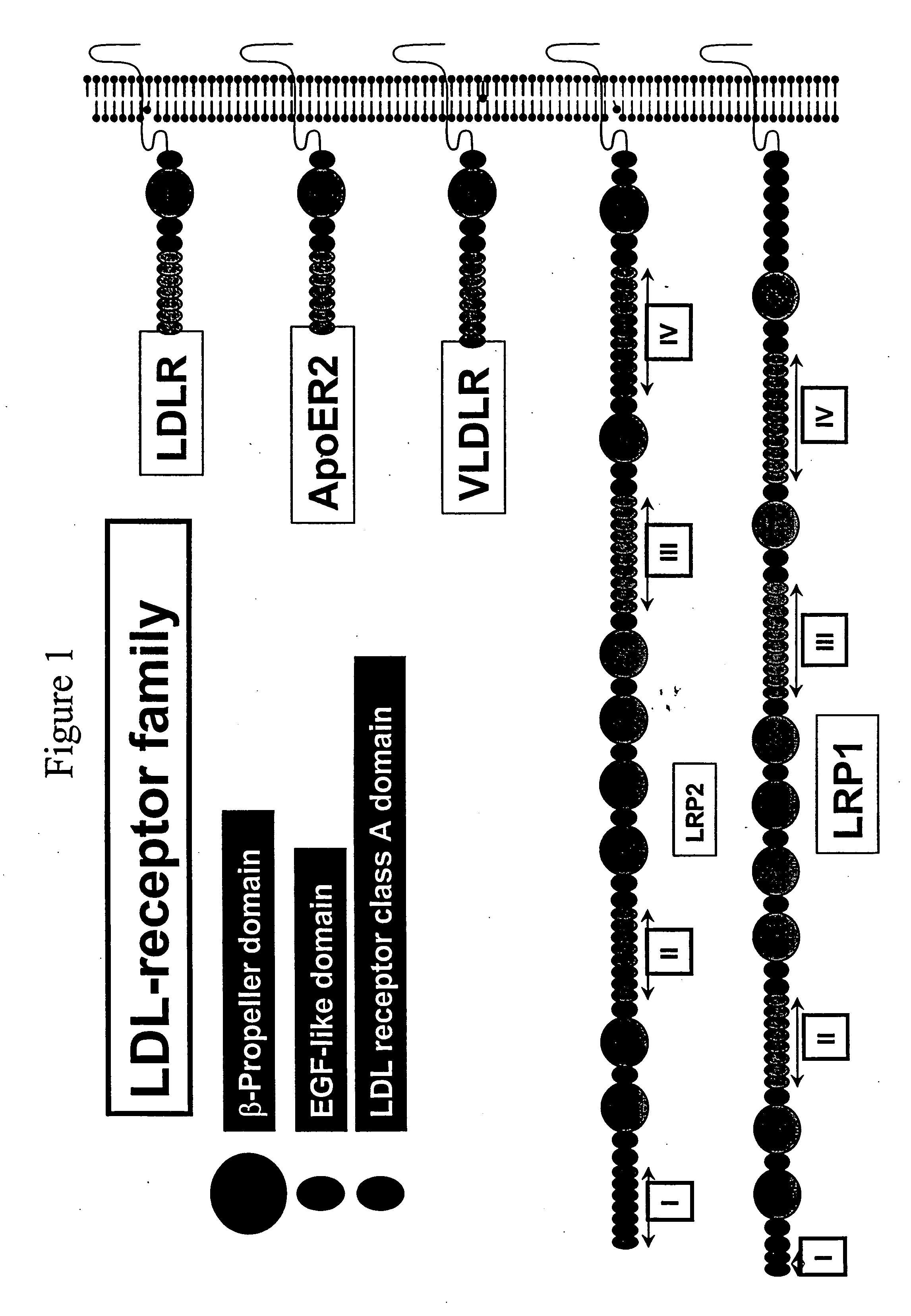
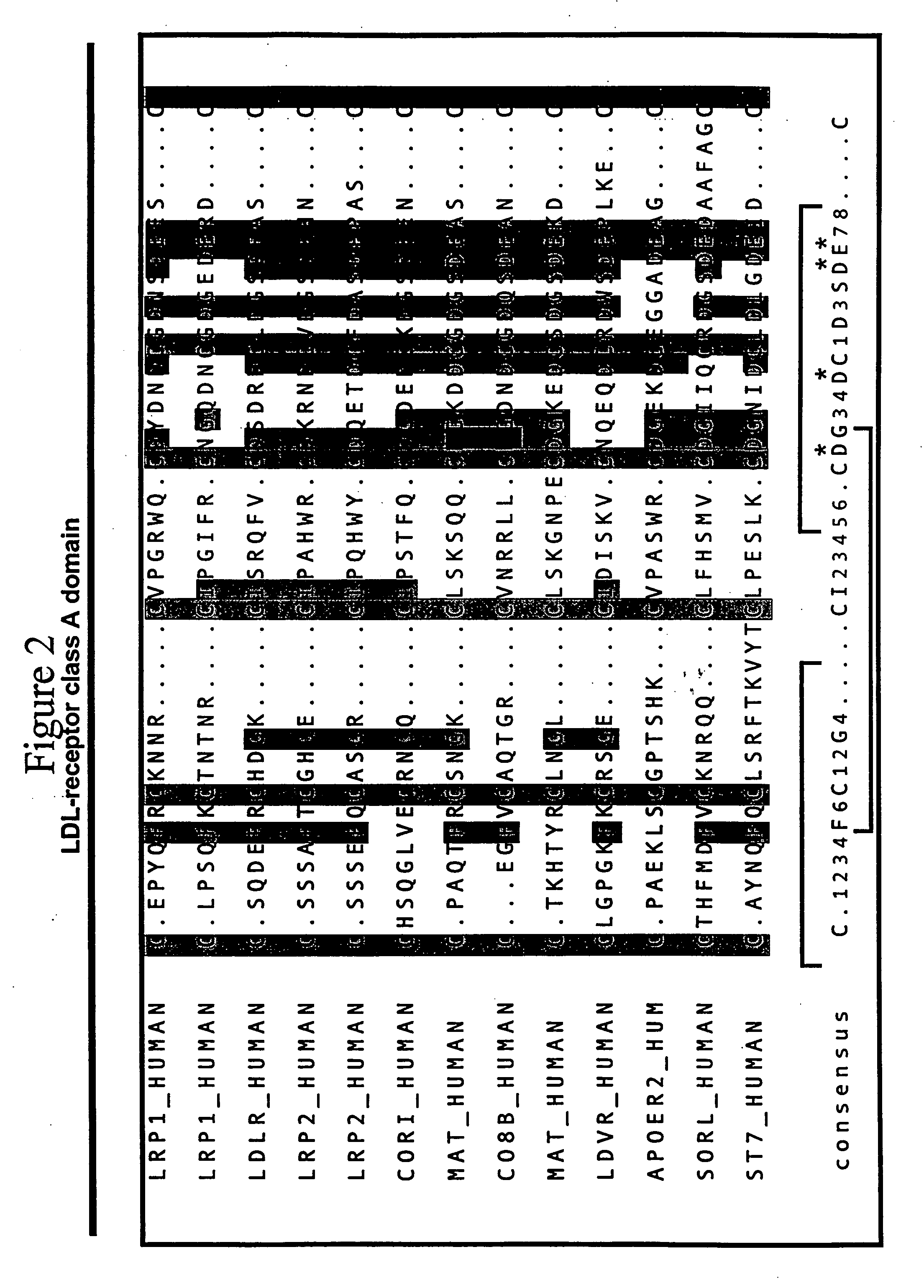
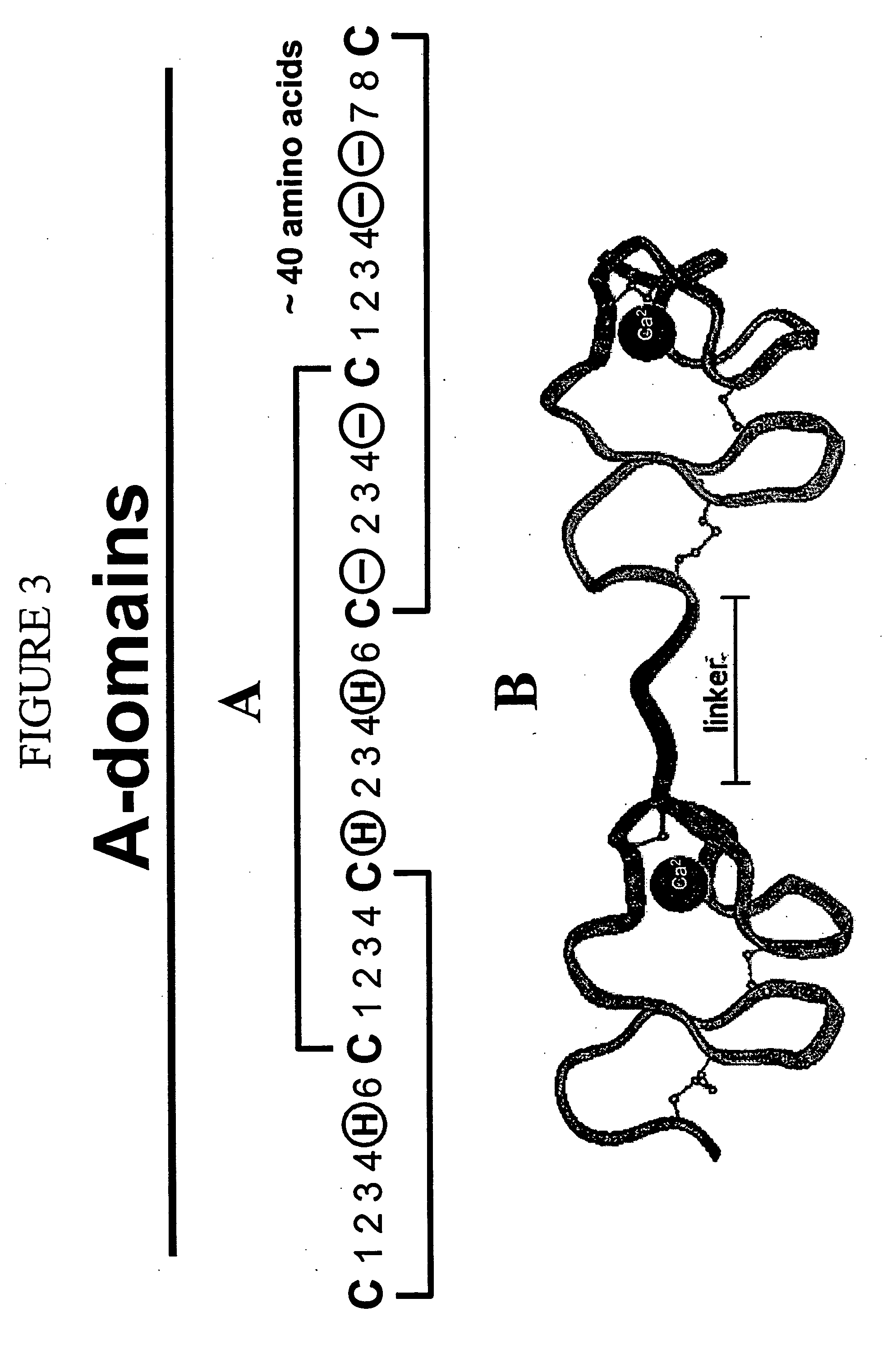
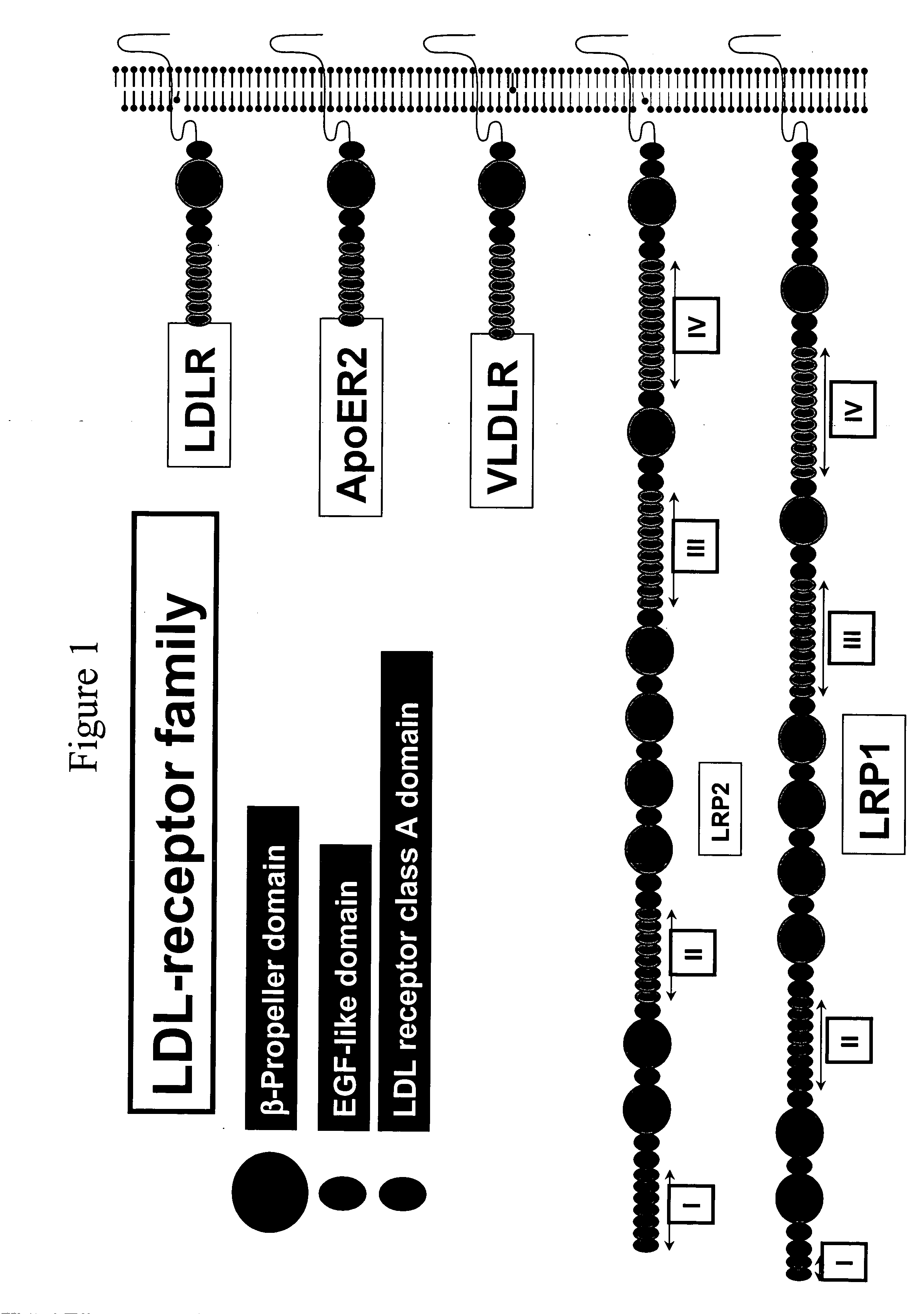
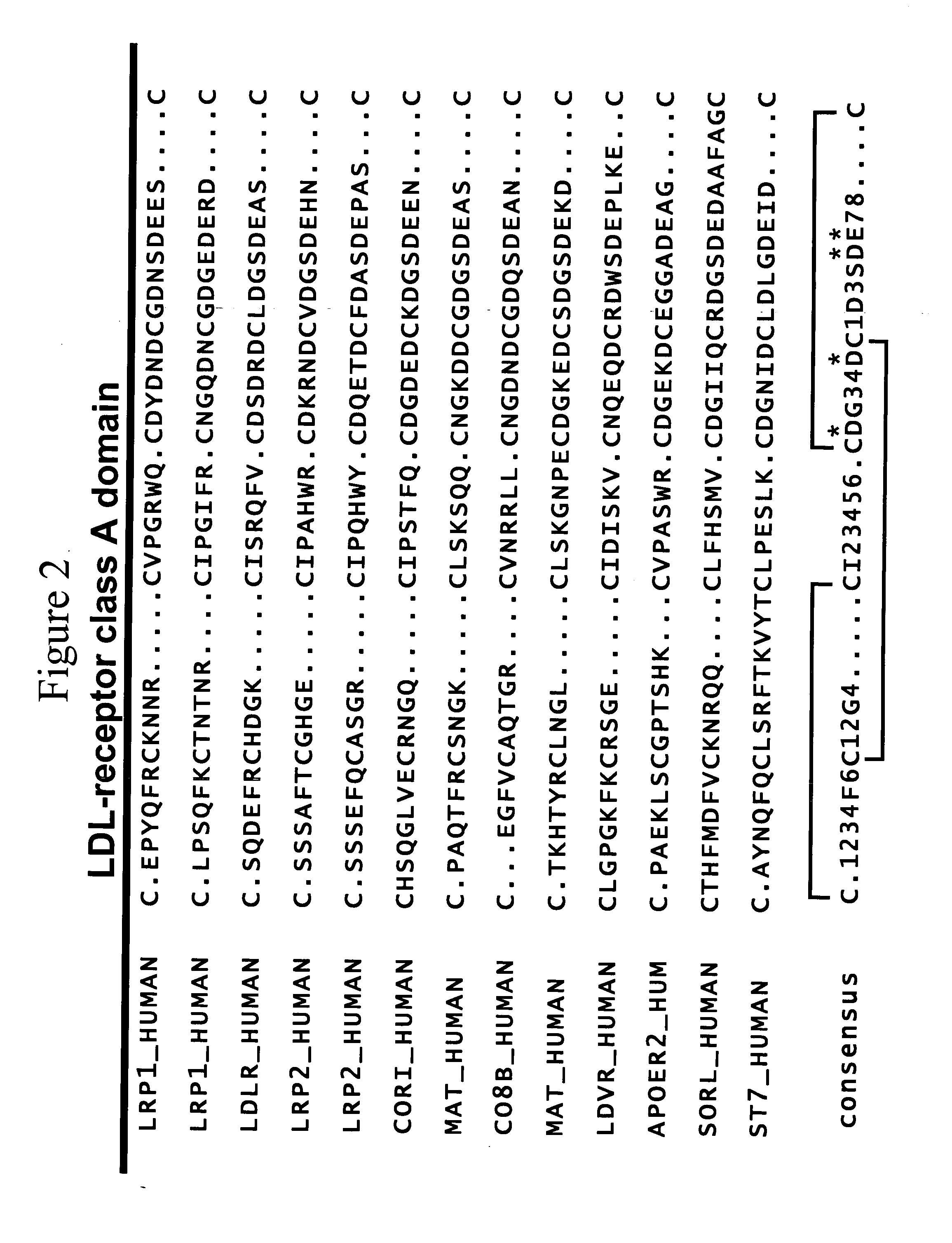
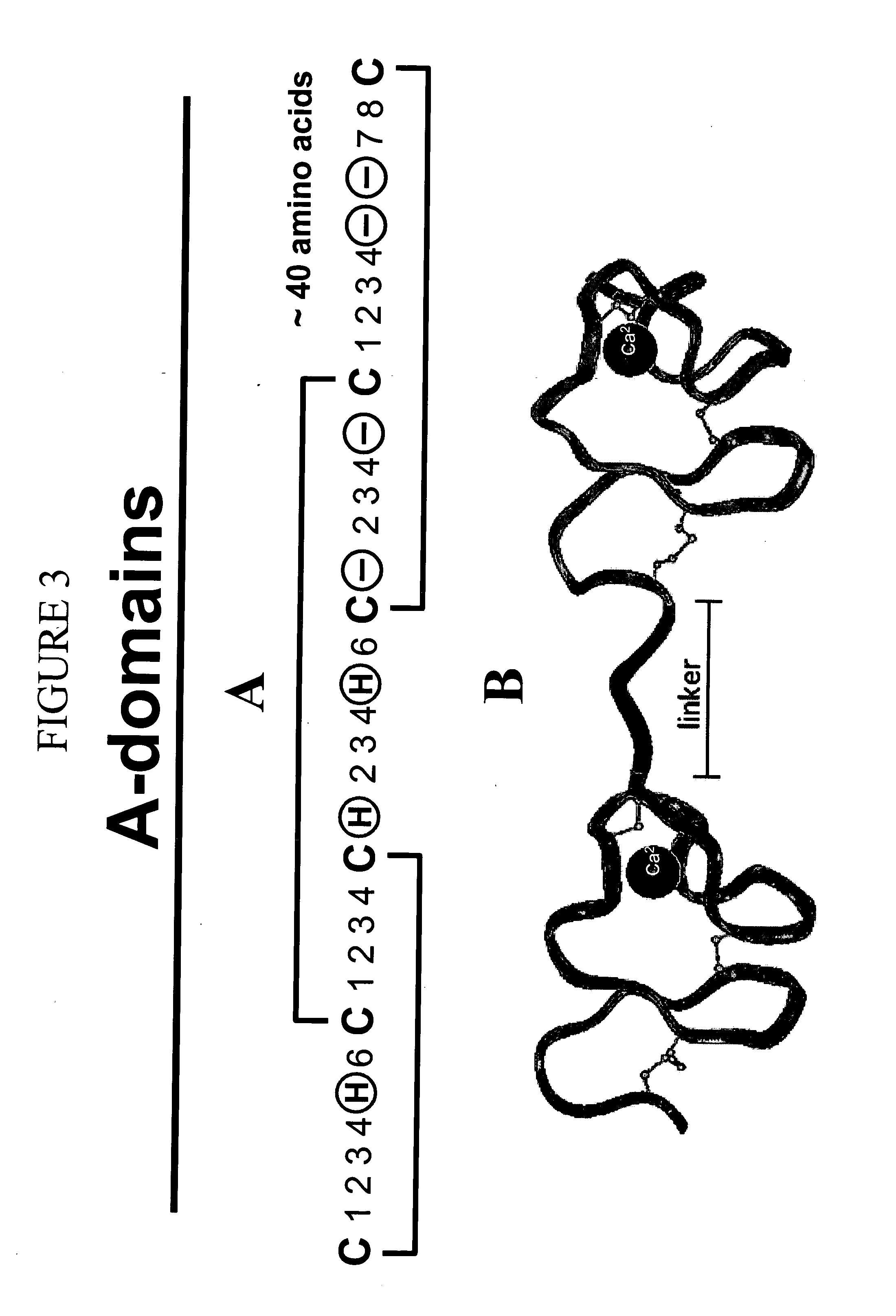
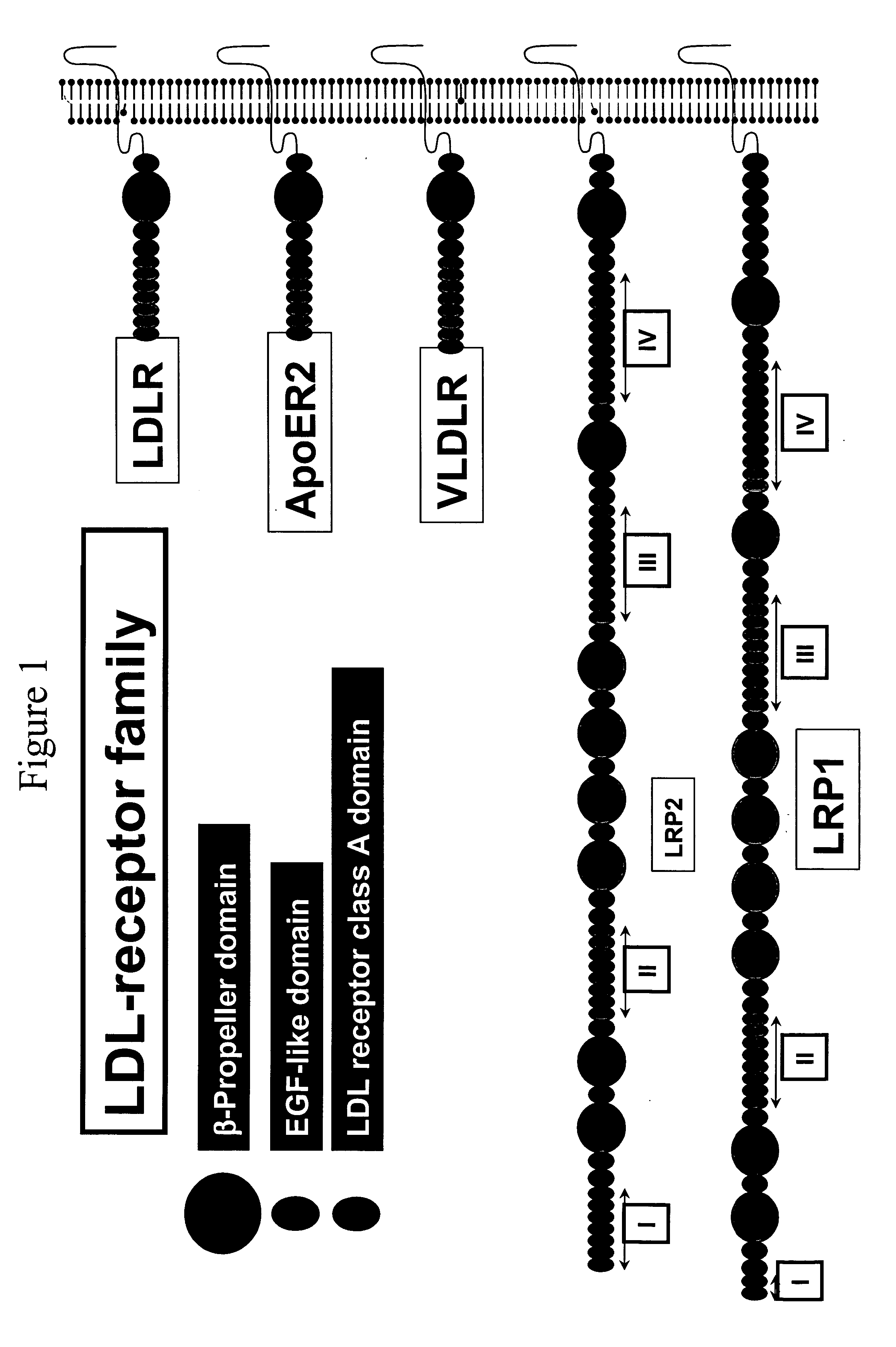
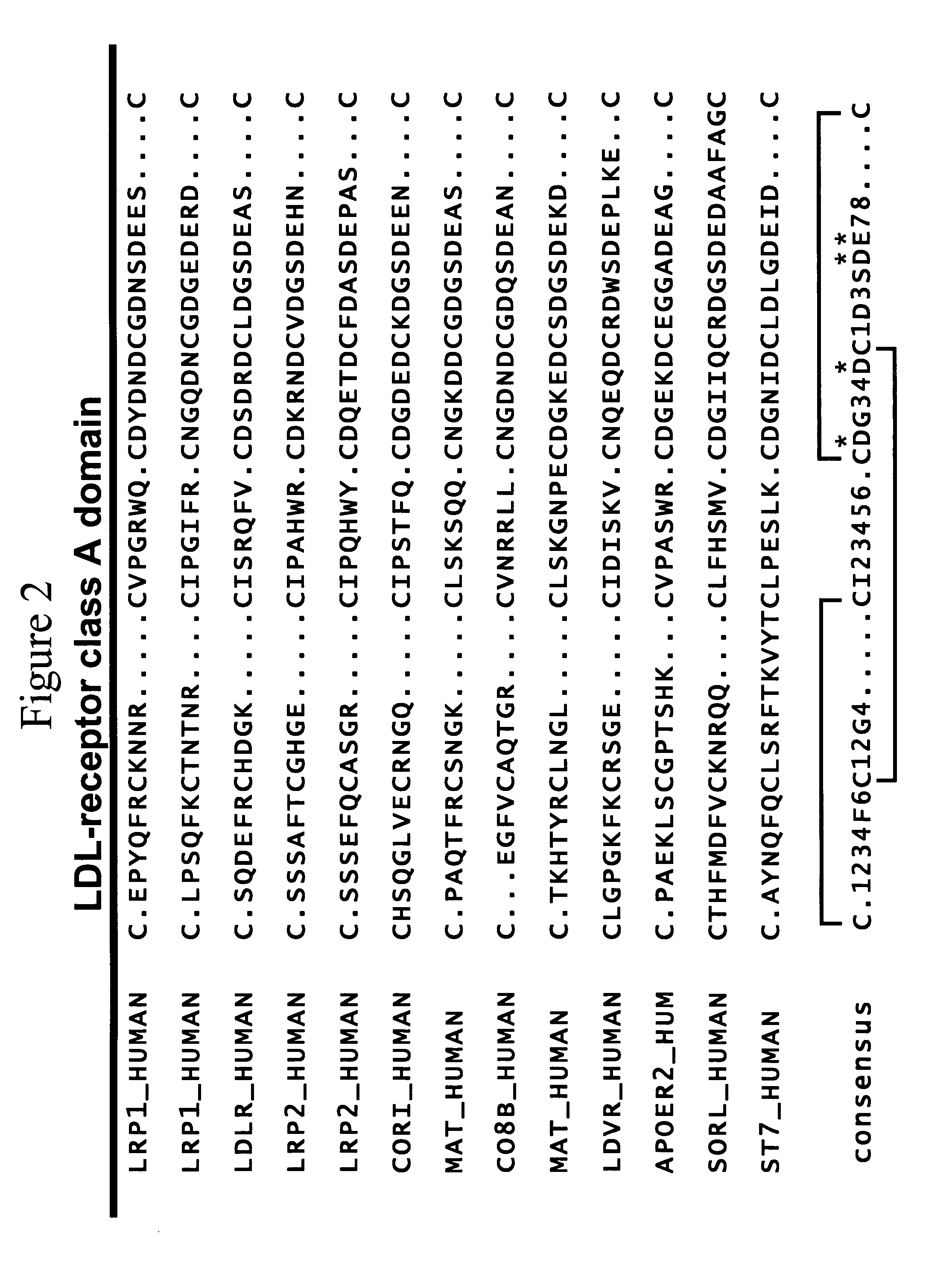
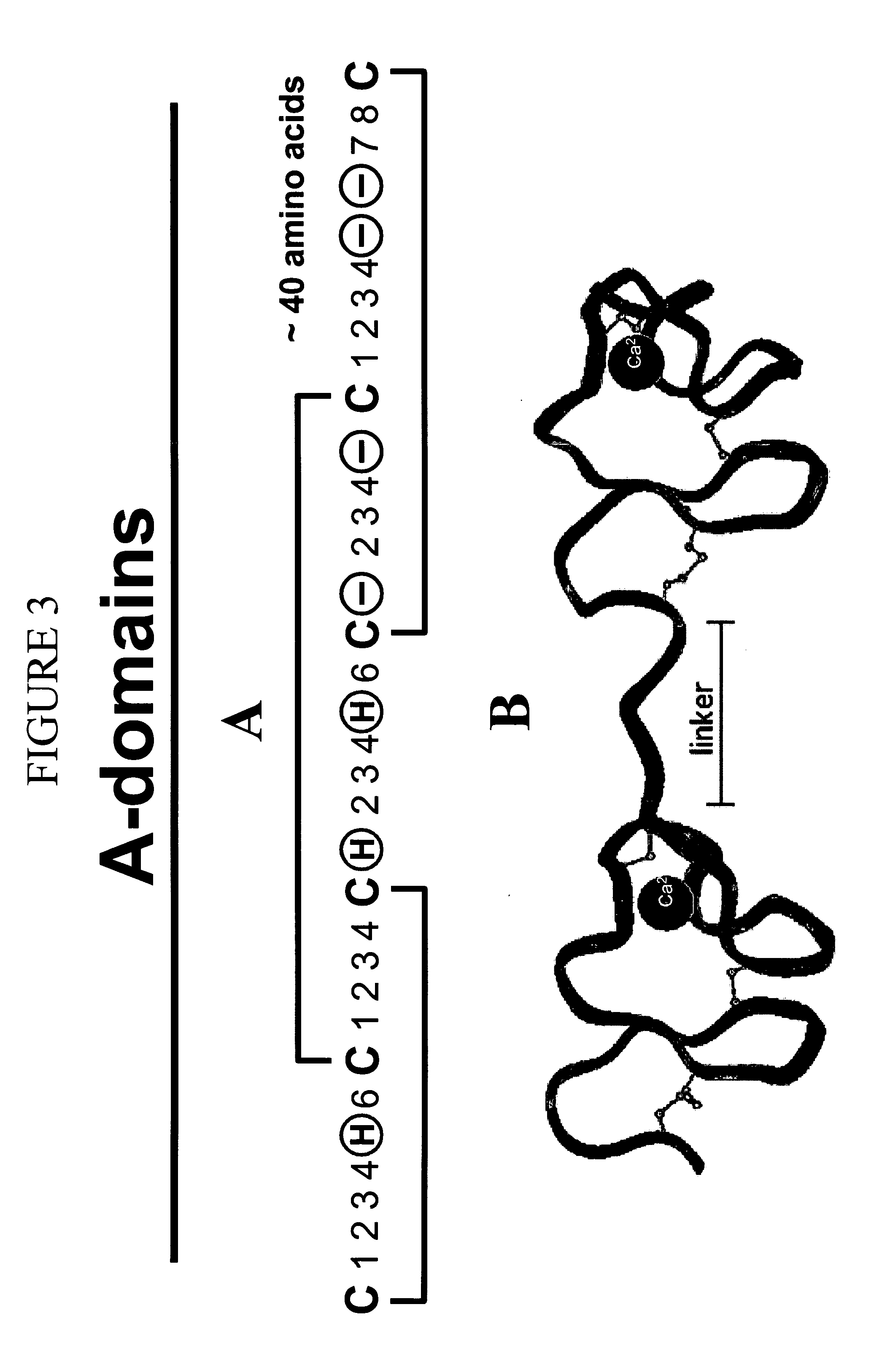
LDL receptor class A and EGF domain monomers and multimers
InactiveUS20050164301A1High affinityStimulate and inhibit activityPeptide librariesPeptide/protein ingredientsMonomerIntegrated systems
Specific monomer domains and multimers comprising the monomer domains are provided. Methods, compositions, libraries and cells that express one or more library member, along with kits and integrated systems, are also included in the present invention.
Owner:AMGEN MOUNTAIN VIEW
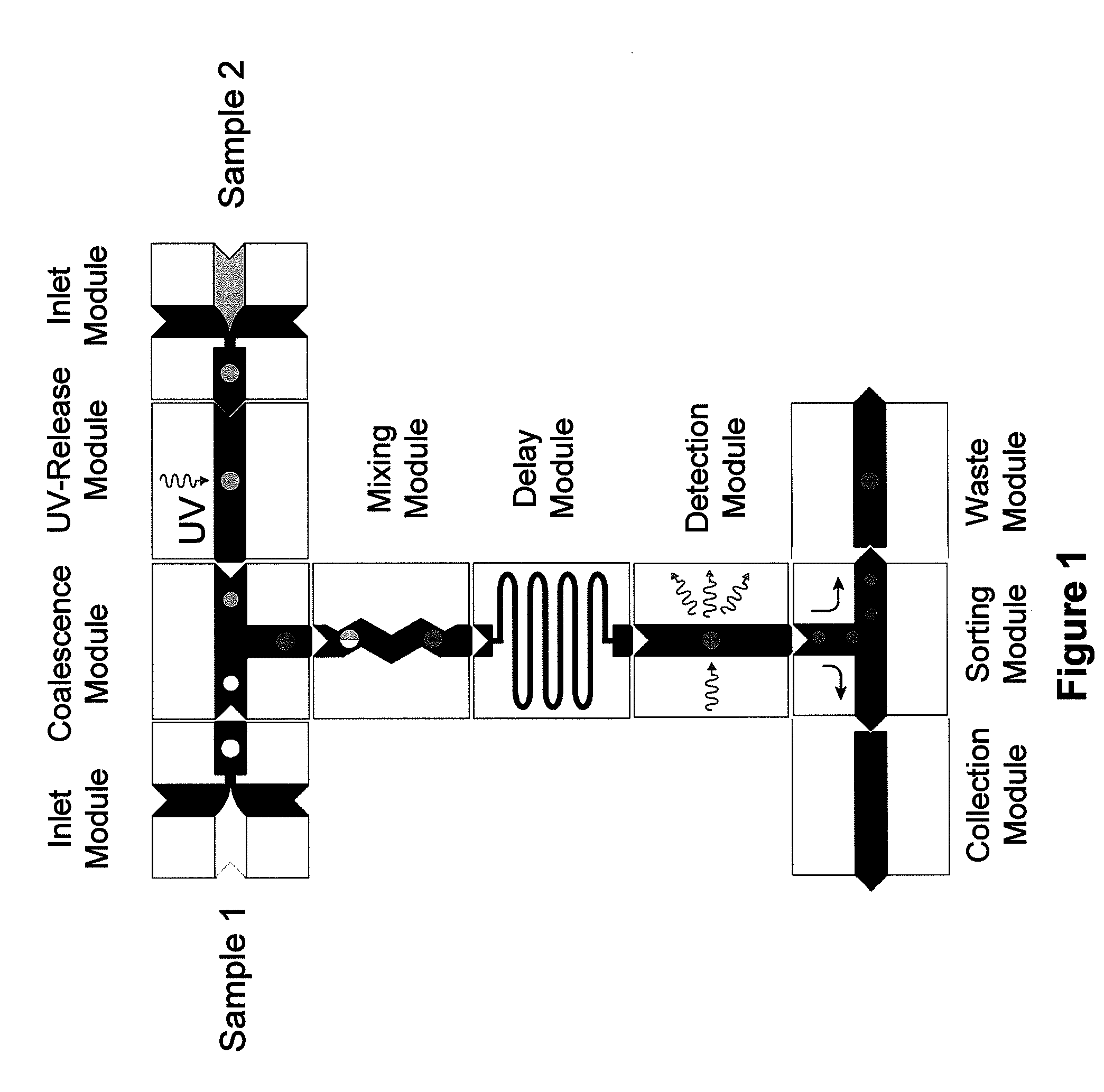
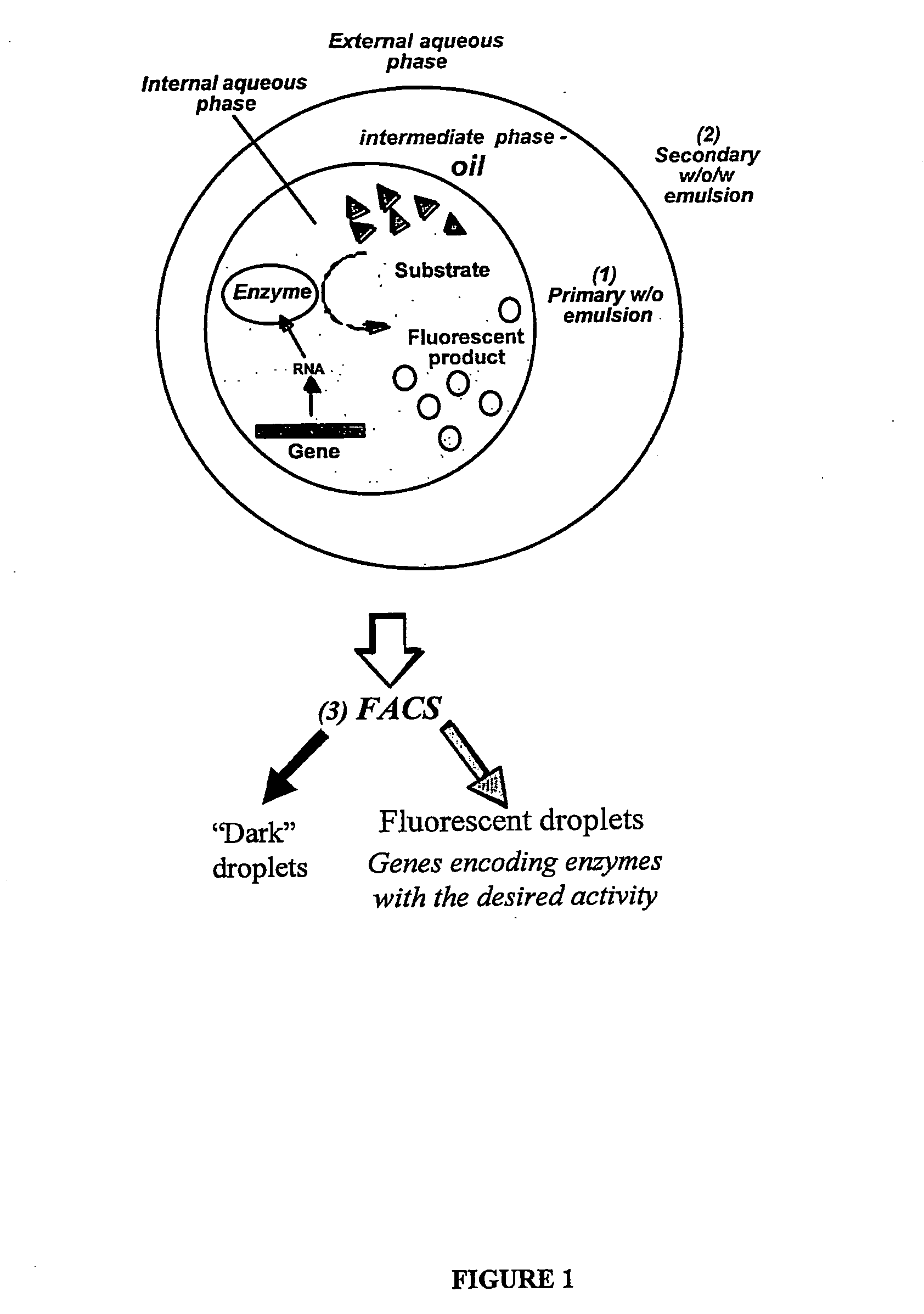
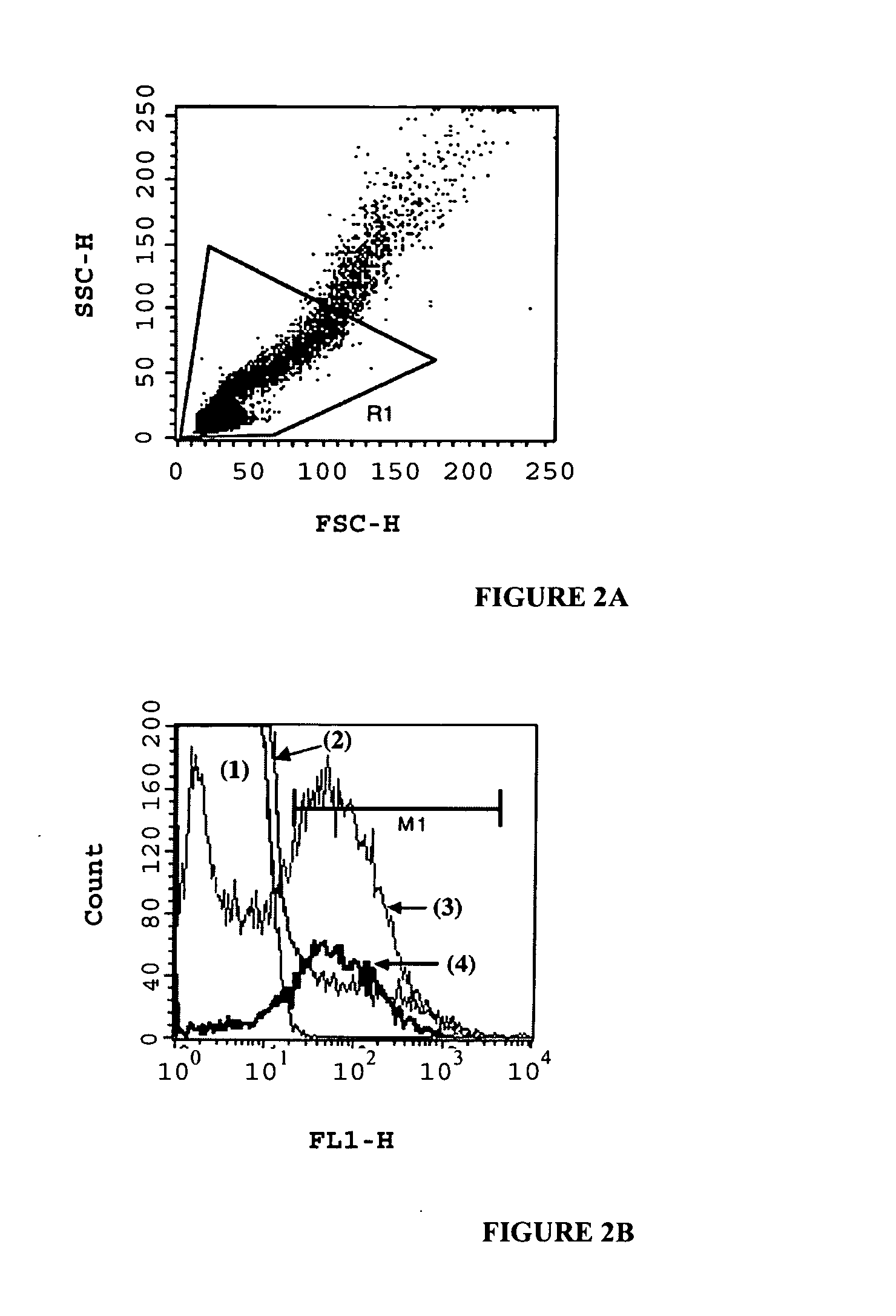
Compositions and methods for in vitro sorting of molecular and cellular libraries
InactiveUS20070077572A1Emulsion stabilizationWide potentialPeptide librariesMicrobiological testing/measurementCell biologyIn vitro system
The present invention provides an in vitro system for compartmentalization of molecular or cellular libraries and provides methods for selection and isolation of desired molecules or cells from the libraries. The library includes a plurality of distinct molecules or cells encapsulated within a water-in-oil-in-water emulsion. The emulsion includes a continuous external aqueous phase and a discontinuous dispersion of water-in-oil droplets. The internal aqueous phase of a plurality of such droplets comprises a specific molecule or cell that is within the plurality of distinct molecules or cells of the library.
Owner:MEDICAL RESEARCH COUNCIL +2
Method for isolation of independent, parallel chemical micro-reactions using a porous filter
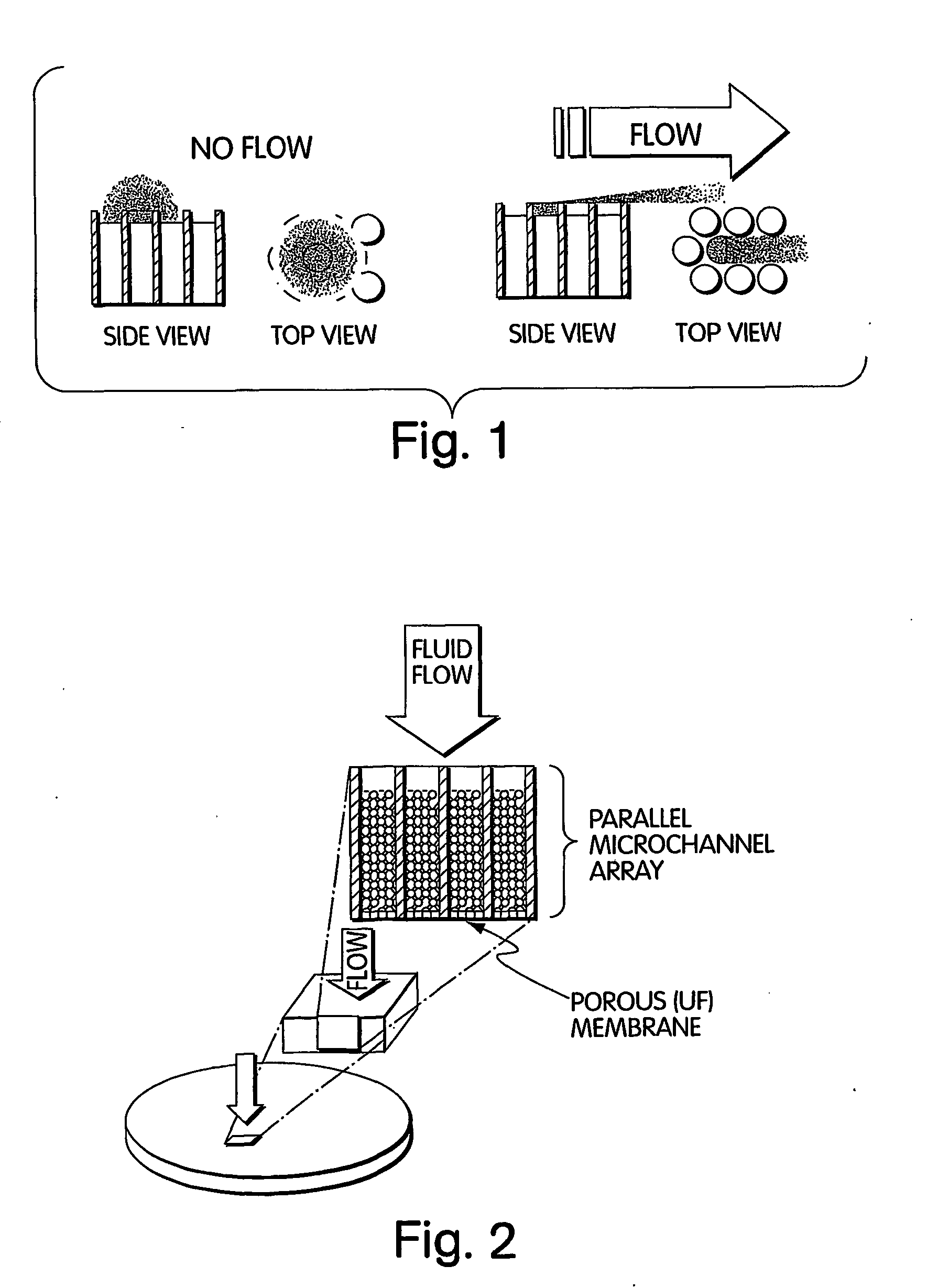
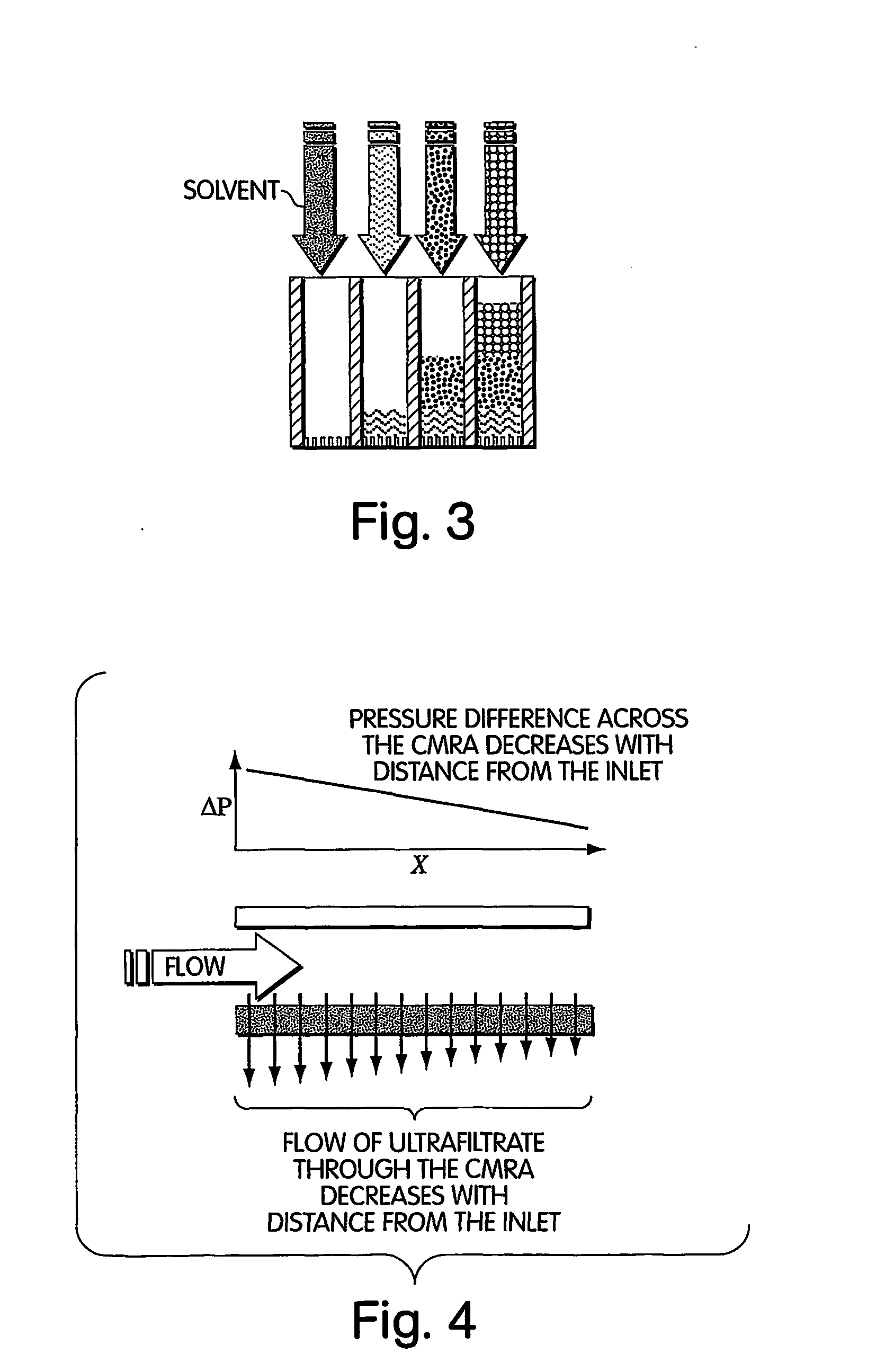
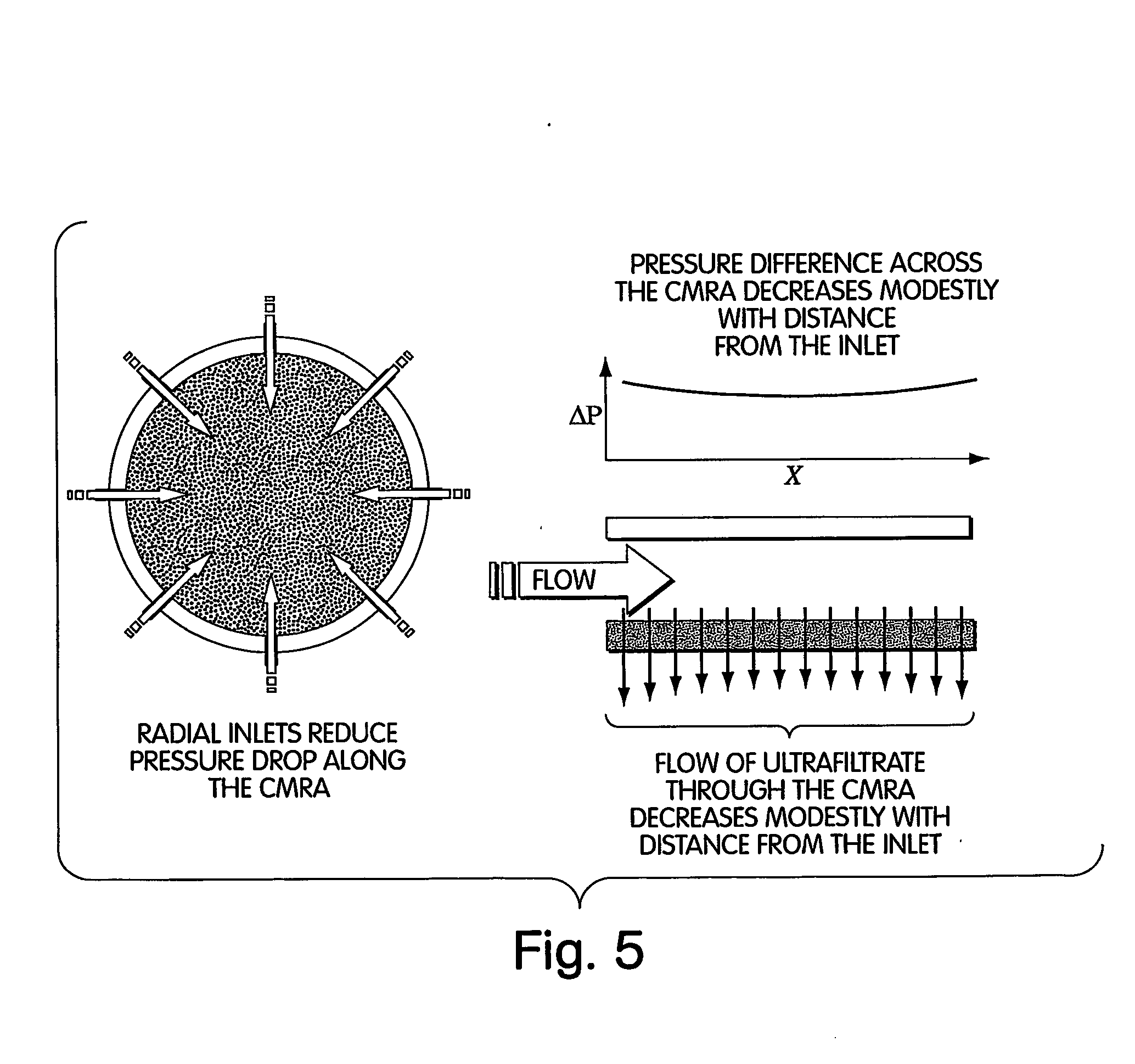
InactiveUS20050009022A1Fast deliveryFaster more complete removalBioreactor/fermenter combinationsPeptide librariesChemical reactionCompound (substance)
The present invention relates to the field of fluid dynamics. More specifically, this invention relates to methods and apparatus for conducting densely packed, independent chemical reactions in parallel in a substantially two-dimensional array. Accordingly, this invention also focuses on the use of this array for applications such as DNA sequencing, most preferably pyrosequencing, and DNA amplification.
Owner:WEINER MICHAEL P +4
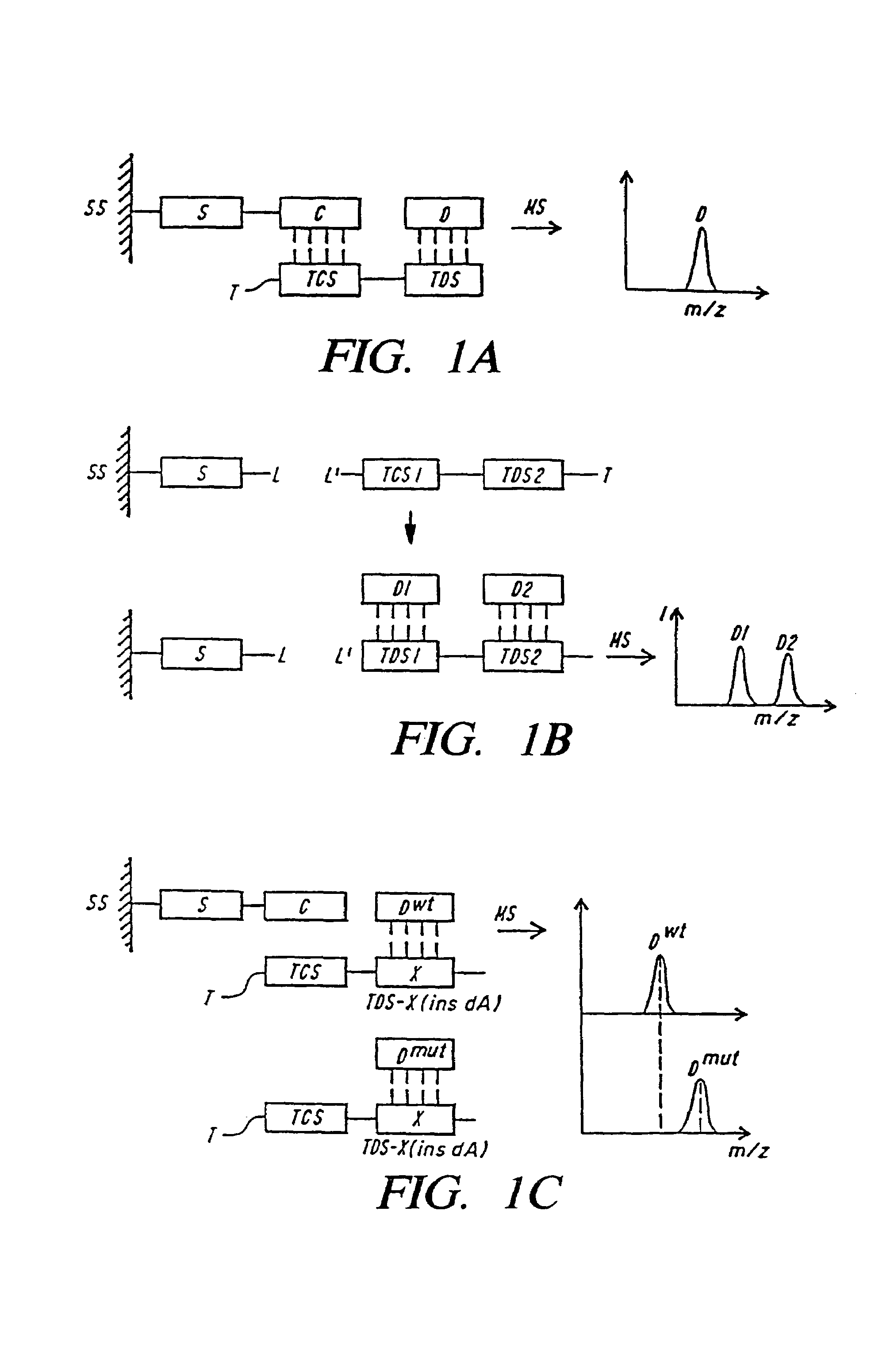
DNA diagnostics based on mass spectrometry
InactiveUS7198893B1Increase mass resolution massIncrease mass mass accuracyPeptide librariesSequential/parallel process reactionsMass spectrometry imagingOrganism
Fast and highly accurate mass spectrometry-based processes for detecting a particular nucleic acid sequence in a biological sample are provided. Depending on the sequence to be detected, the processes can be used, for example, to diagnose a genetic disease or chromosomal abnormality; a predisposition to a disease or condition, infection by a pathogenic organism, or for determining identity or heredity.
Owner:AGENA BIOSCI
Crystal growth devices and systems, and methods for using same
Owner:FLUIDIGM CORP
Protein scaffolds and uses thereof
InactiveUS20060223114A1Specificity of bindingEasy screeningPeptide librariesAntibody mimetics/scaffoldsChemistryIntegrated systems
The present invention provides thrombospondin, thyroglobulin and trfoil / PD monomer domains and multimers comprising the monomer domains are provided. Methods, compositions, libraries and cells that express one or more library member, along with kits and integrated systems, are also included in the present invention.
Owner:AMGEN MOUNTAIN VIEW
Assay cartridges and methods of using the same
Assay modules, preferably assay cartridges, are described as are reader apparatuses which may be used to control aspects of module operation. The modules preferably comprise a detection chamber with integrated electrodes that may be used for carrying out electrode induced luminescence measurements. Methods are described for immobilizing assay reagents in a controlled fashion on these electrodes and other surfaces. Assay modules and cartridges are also described that have a detection chamber, preferably having integrated electrodes, and other fluidic components which may include sample chambers, waste chambers, conduits, vents, bubble traps, reagent chambers, dry reagent pill zones and the like. In certain preferred embodiments, these modules are adapted to receive and analyze a sample collected on an applicator stick.
Owner:MESO SCALE TECH LLC
Proteinaceous pharmaceuticals and uses thereof
InactiveUS20070191272A1High disulfide densityReduce molecular weightPeptide librariesNervous disorderCysteine thiolateBiology
The present invention provides cysteine-containing scaffolds and / or proteins, expression vectors, host cell and display systems harboring and / or expressing such cysteine-containing products. The present invention also provides methods of designing libraries of such products, methods of screening such libraries to yield entities exhibiting binding specificities towards a target molecule. Further provided by the invention are pharmaceutical compositions comprising the cysteine-containing products of the present invention.
Owner:AMUNIX OPERATING INC
Methods and products related to the improved analysis of carbohydrates
The invention relates, in part, to the improved analysis of carbohydrates. In particular, the invention relates to the analysis of carbohydrates, such as N-glycans and O-glycans found on proteins and saccharides attached to lipids. Improved methods, therefore, for the study of glycosylation patterns on cells, tissue and body fluids are also provided. Information from the analysis of glycans, such as the glycosylation patterns on cells, tissues and in body fluids, can be used in diagnostic and treatment methods as well as for facilitating the study of the effects of glycosylation / altered glycosylation. Such methods are also provided. Methods are further provided to assess production processes, to assess the purity of samples containing glycoconjugates, and to select glycoconjugates with the desired glycosylation.
Owner:MASSACHUSETTS INST OF TECH +2
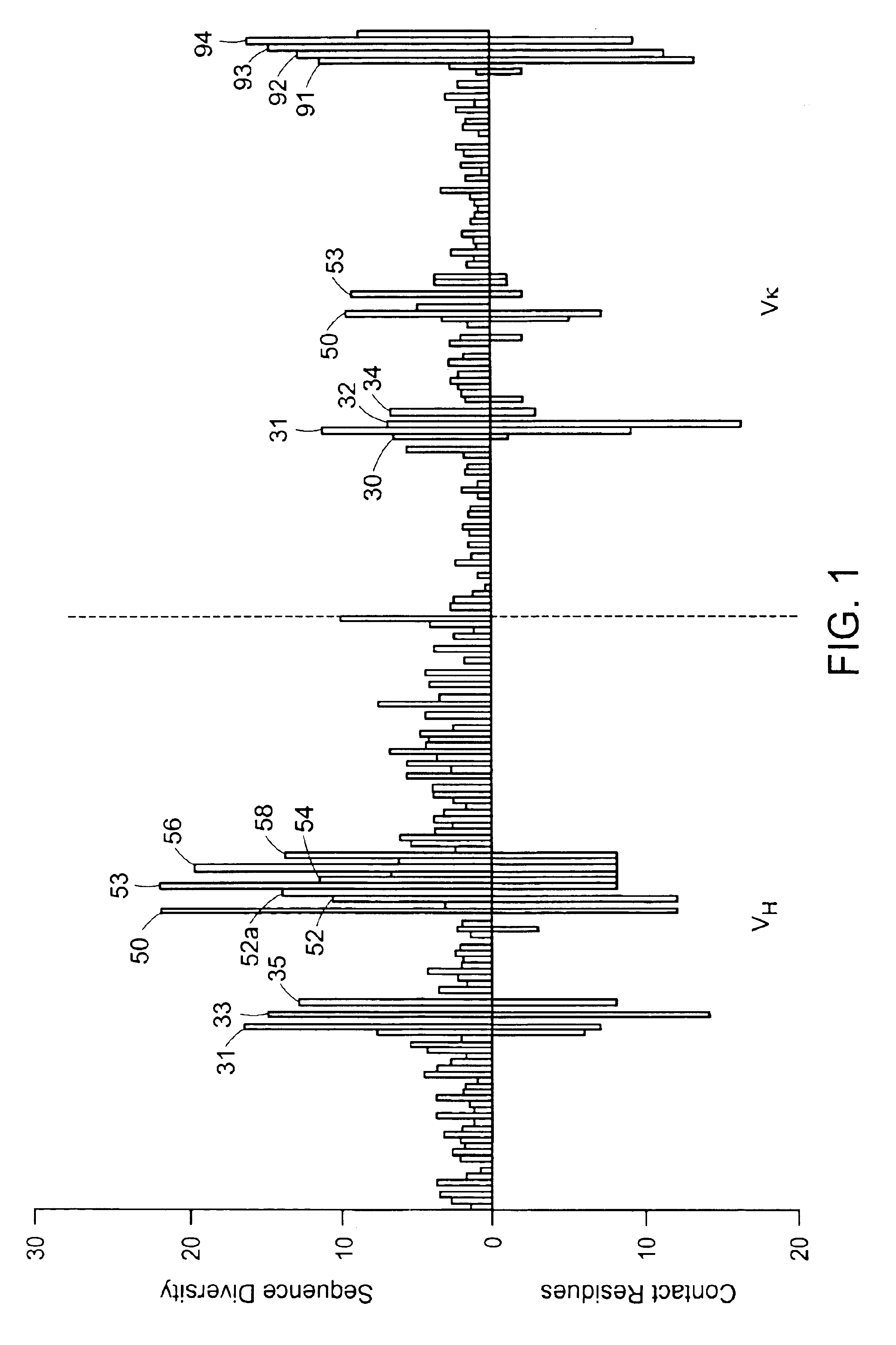
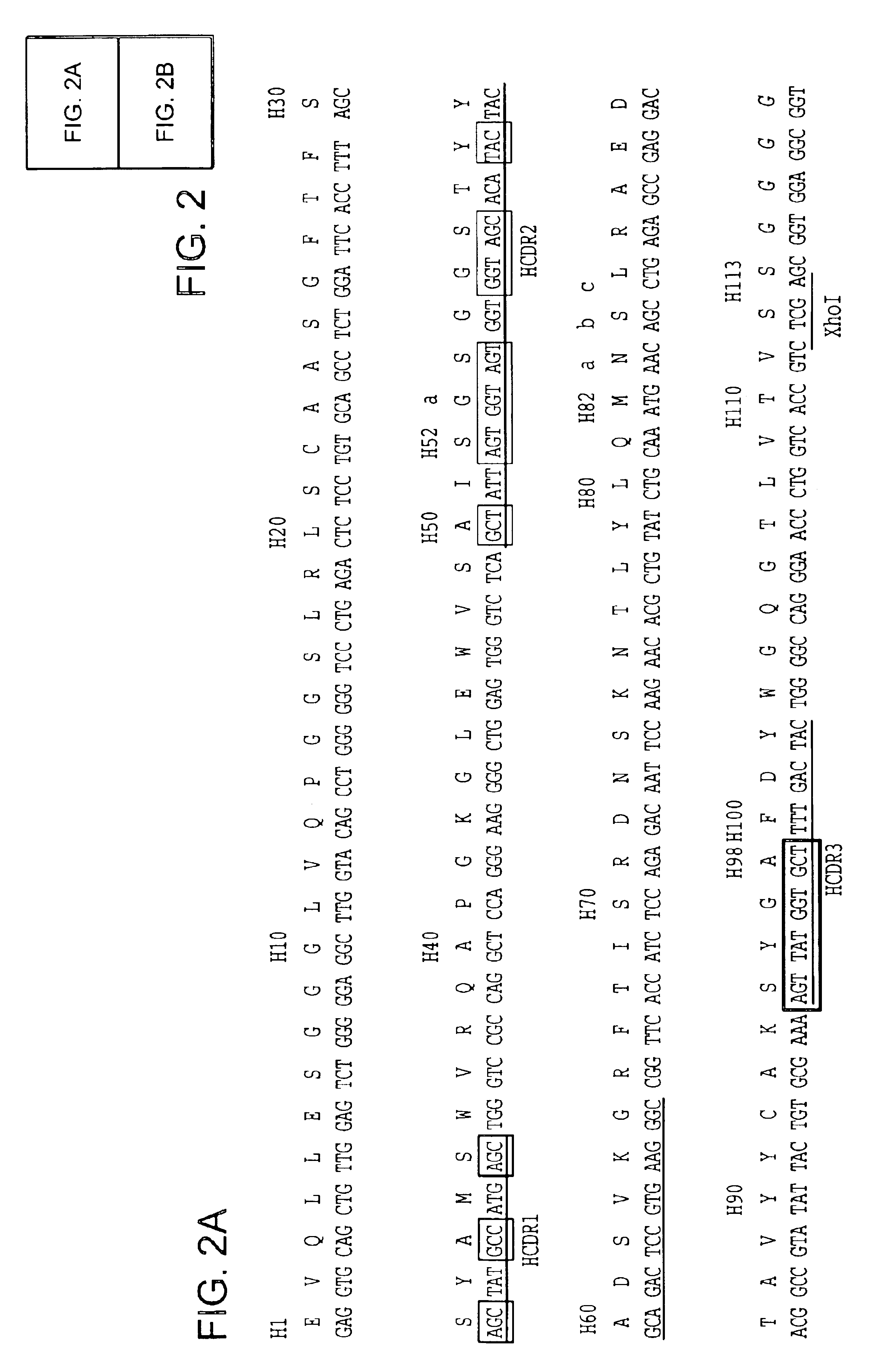
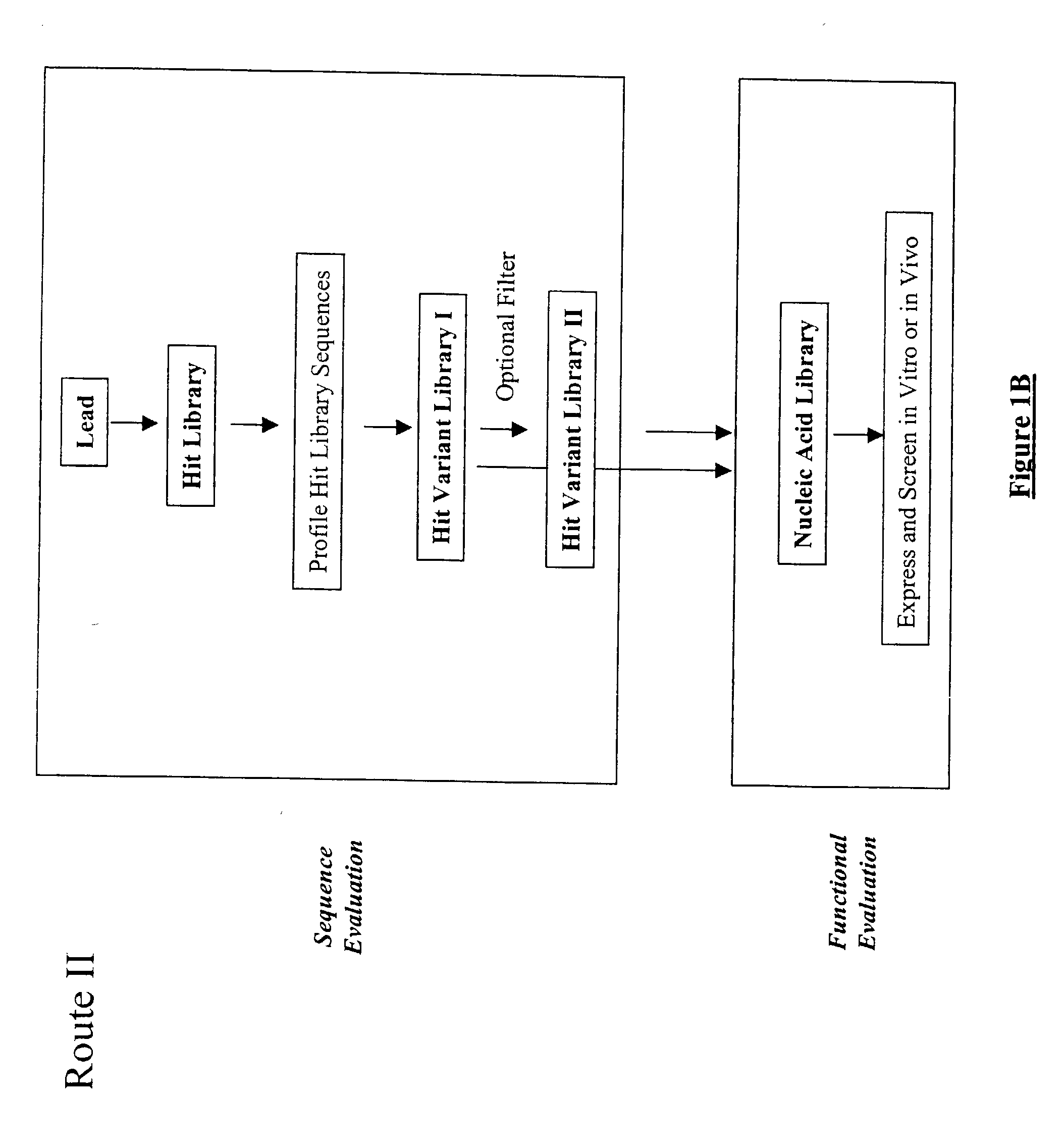
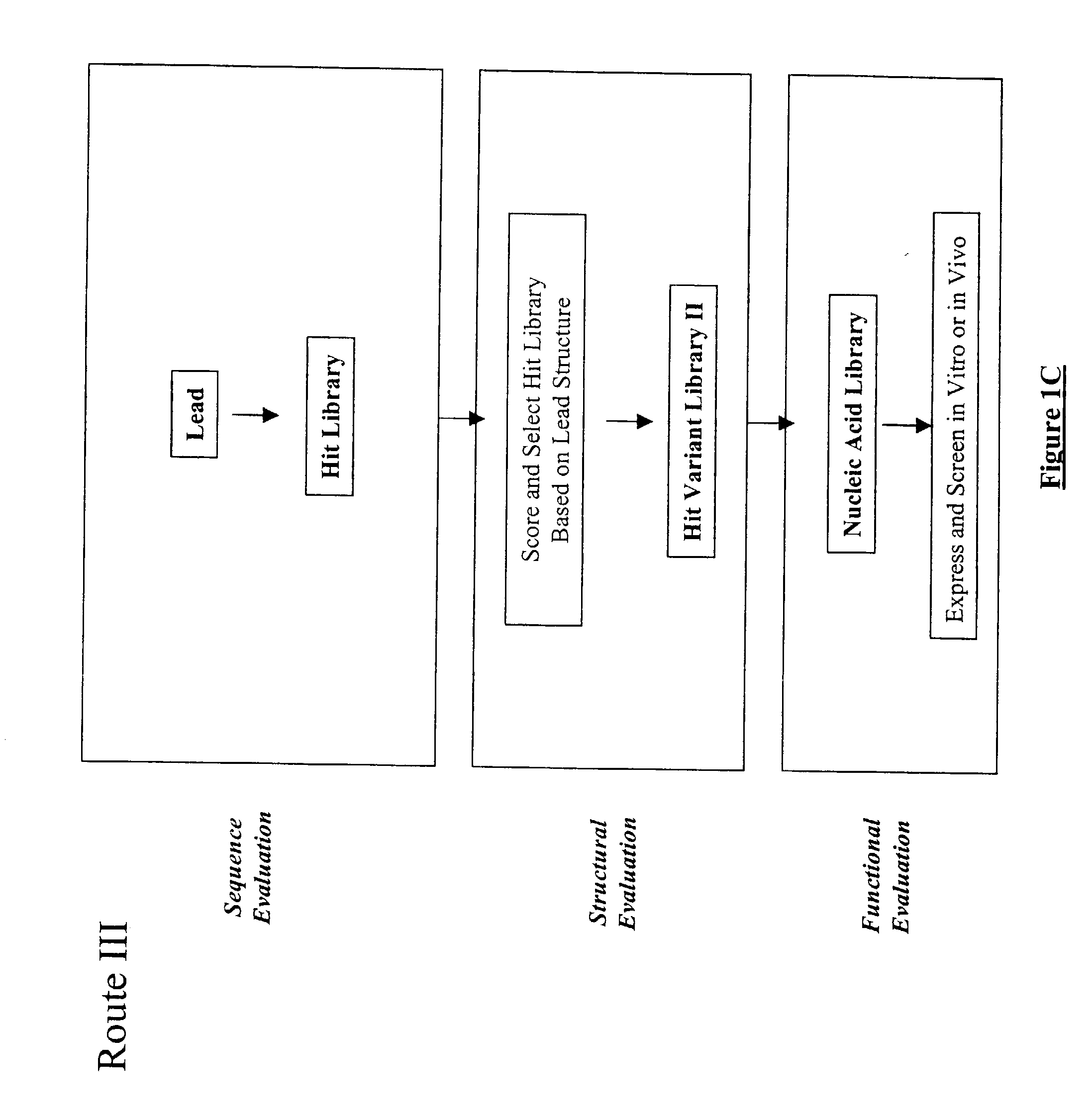
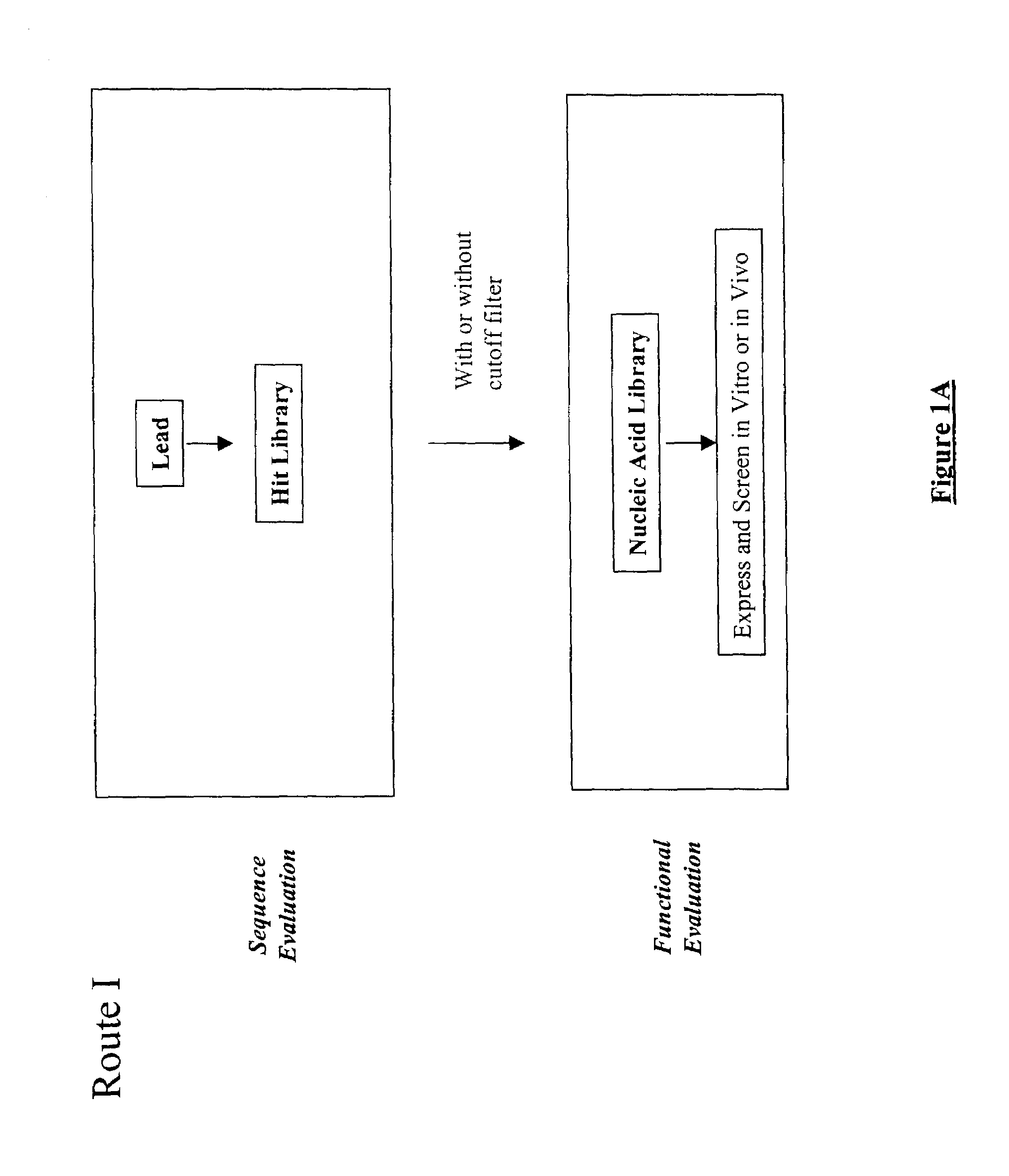
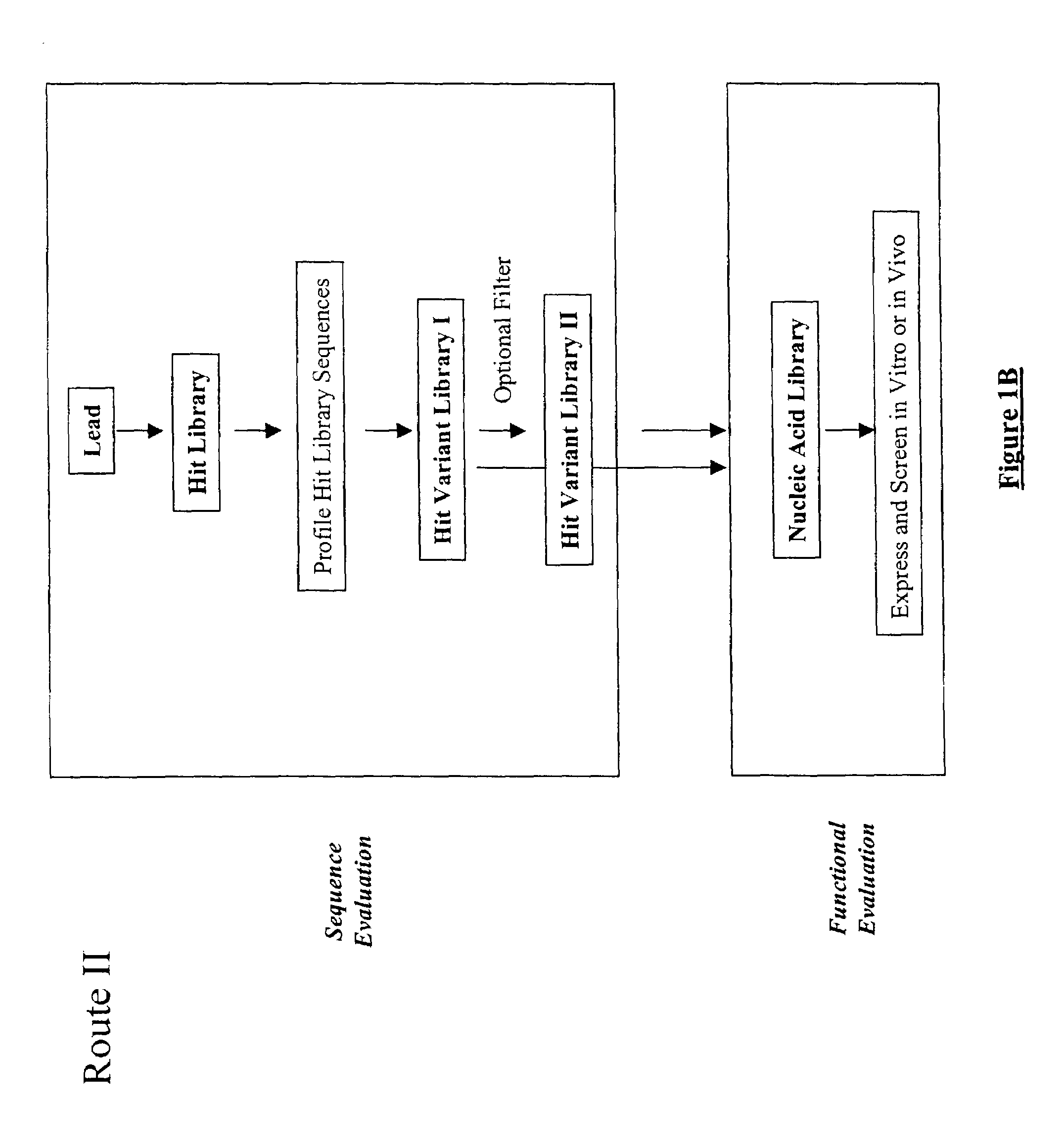
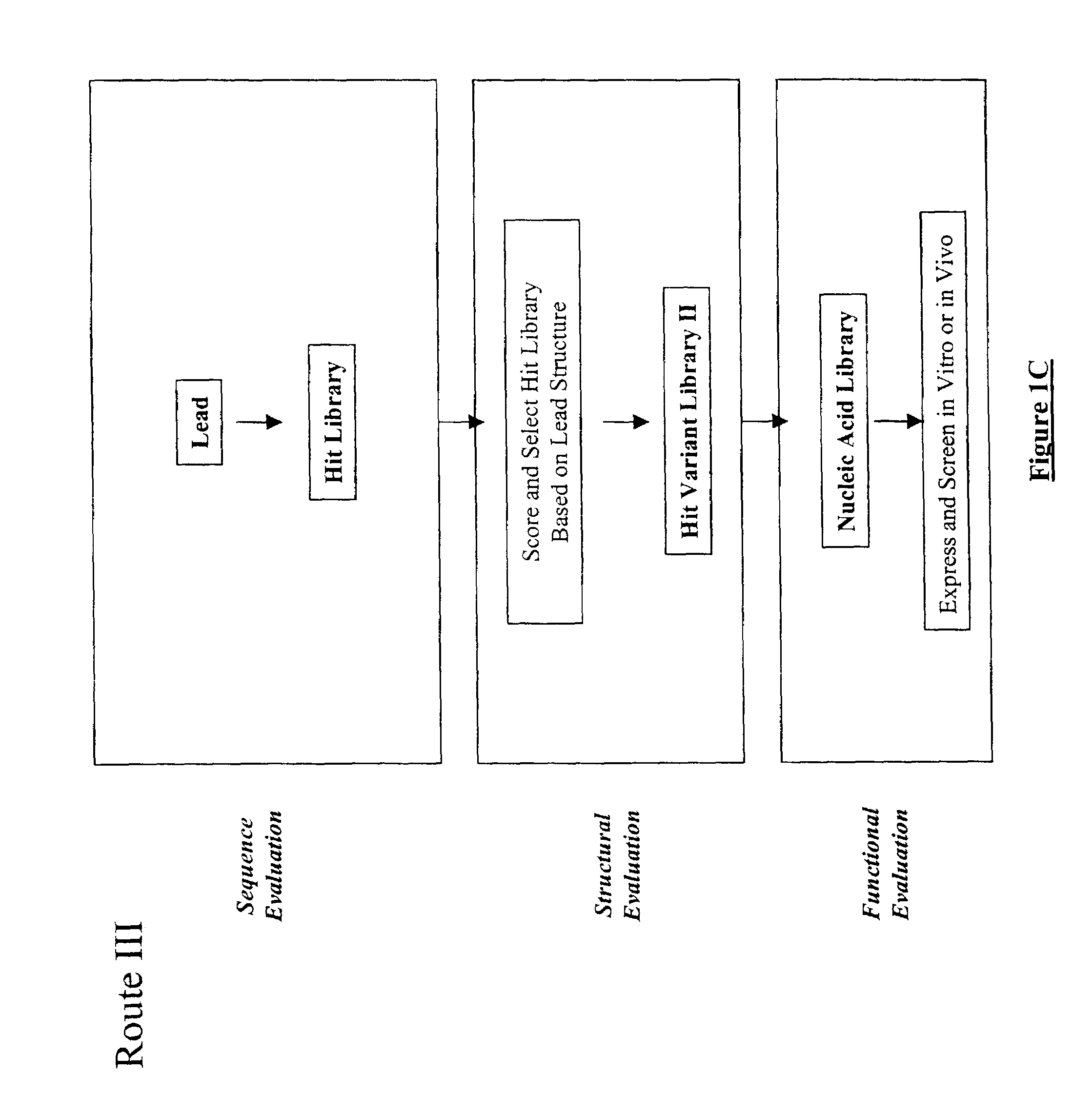
Structure-based selection and affinity maturation of antibody library
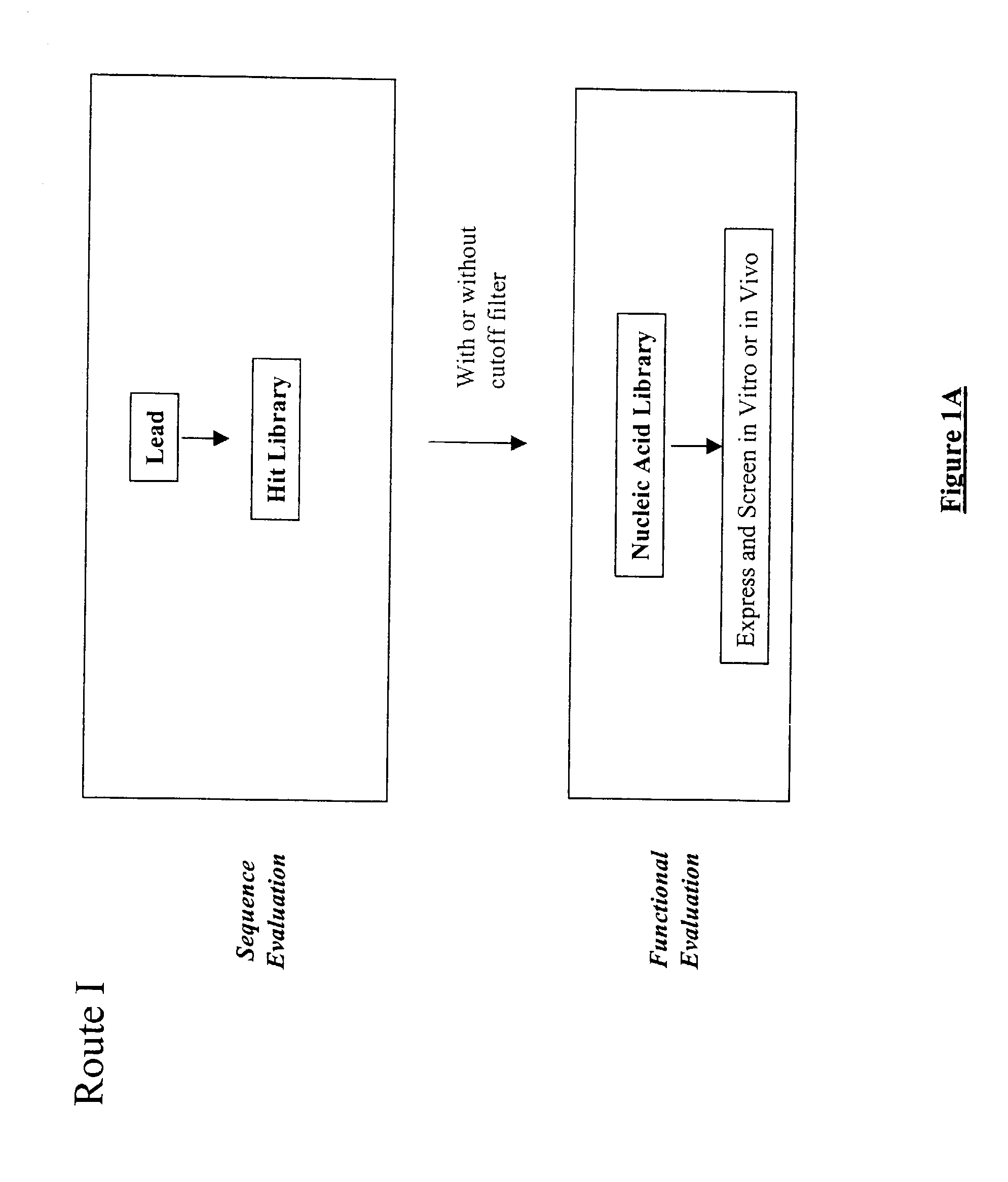
The present invention provides a structure-based methodology for efficiently generating and screening protein libraries for optimized proteins with desirable biological functions, such as antibodies with high binding affinity and low immunogenicity in humans. In one embodiment, a method is provided for constructing a library of antibody sequences based on a three dimensional structure of a lead antibody. The method comprises: providing an amino acid sequence of the variable region of the heavy chain (VH) or light chain (VL) of a lead antibody, the lead antibody having a known three dimensional structure which is defined as a lead structural template; identifying the amino acid sequences in the CDRs of the lead antibody; selecting one of the CDRs in the VH or VL region of the lead antibody; providing an amino acid sequence that comprises at least 3 consecutive amino acid residues in the selected CDR, the selected amino acid sequence being a lead sequence; comparing the lead sequence profile with a plurality of tester protein sequences; selecting from the plurality of tester protein sequences at least two peptide segments that have at least 10% sequence identity with lead sequence, the selected peptide segments forming a hit library; determining if a member of the hit library is structurally compatible with the lead structural template using a scoring function; and selecting the members of the hit library that score equal to or better than or equal to the lead sequence. The selected members of the hit library can be expressed in vitro or in vivo to produce a library of recombinant antibodies that can be screened for novel or improved function(s) over the lead antibody.
Owner:ABMAXIS
Structure-based selection and affinity maturation of antibody library
InactiveUS7117096B2High affinityImprove throughputPeptide librariesPeptide/protein ingredientsProtein insertionIn vivo
The present invention provides a structure-based methodology for efficiently generating and screening protein libraries for optimized proteins with desirable biological functions, such as antibodies with high binding affinity and low immunogenicity in humans. In one embodiment, a method is provided for constructing a library of antibody sequences based on a three dimensional structure of a lead antibody. The method comprises: providing an amino acid sequence of the variable region of the heavy chain (VH) or light chain (VL) of a lead antibody, the lead antibody having a known three dimensional structure which is defined as a lead structural template; identifying the amino acid sequences in the CDRs of the lead antibody; selecting one of the CDRs in the VH or VL region of the lead antibody; providing an amino acid sequence that comprises at least 3 consecutive amino acid residues in the selected CDR, the selected amino acid sequence being a lead sequence; comparing the lead sequence profile with a plurality of tester protein sequences; selecting from the plurality of tester protein sequences at least two peptide segments that have at least 10% sequence identity with lead sequence, the selected peptide segments forming a hit library; determining if a member of the hit library is structurally compatible with the lead structural template using a scoring function; and selecting the members of the hit library that score equal to or better than or equal to the lead sequence. The selected members of the hit library can be expressed in vitro or in vivo to produce a library of recombinant antibodies that can be screened for novel or improved function(s) over the lead antibody.
Owner:ABMAXIS
Rationally Designed, Synthetic Antibody Libraries and Uses Therefor
ActiveUS20090181855A1Peptide librariesImmunoglobulins against animals/humansPolynucleotideSynthetic antibody
The present invention overcomes the inadequacies inherent in the known methods for generating libraries of antibody-encoding polynucleotides by specifically designing the libraries with directed sequence and length diversity. The libraries are designed to reflect the preimmune repertoire naturally created by the human immune system and are based on rational design informed by examination of publicly available databases of human antibody sequences.
Owner:ADIMAB LLC
Multiplexed measurement of membrane protein populations
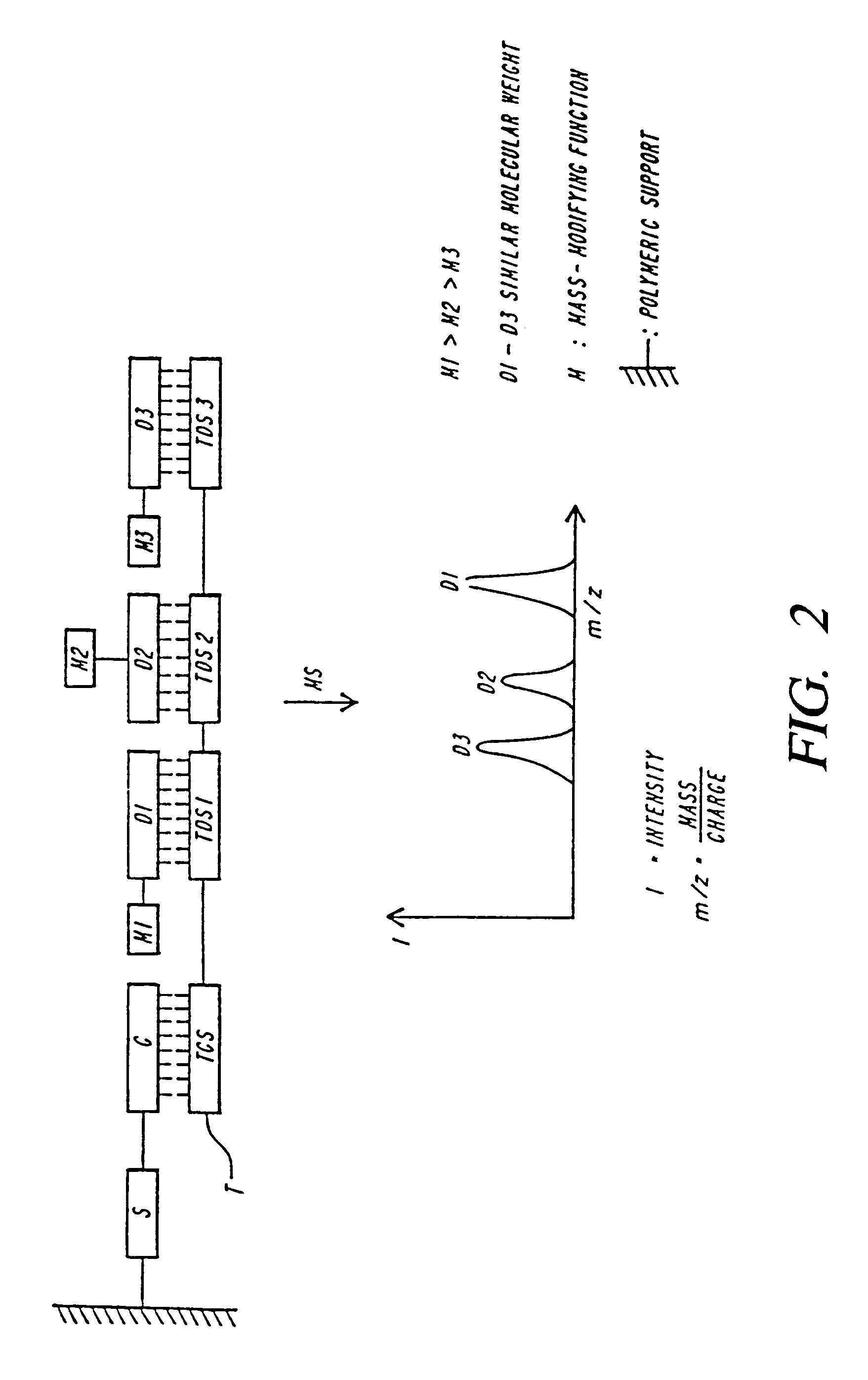
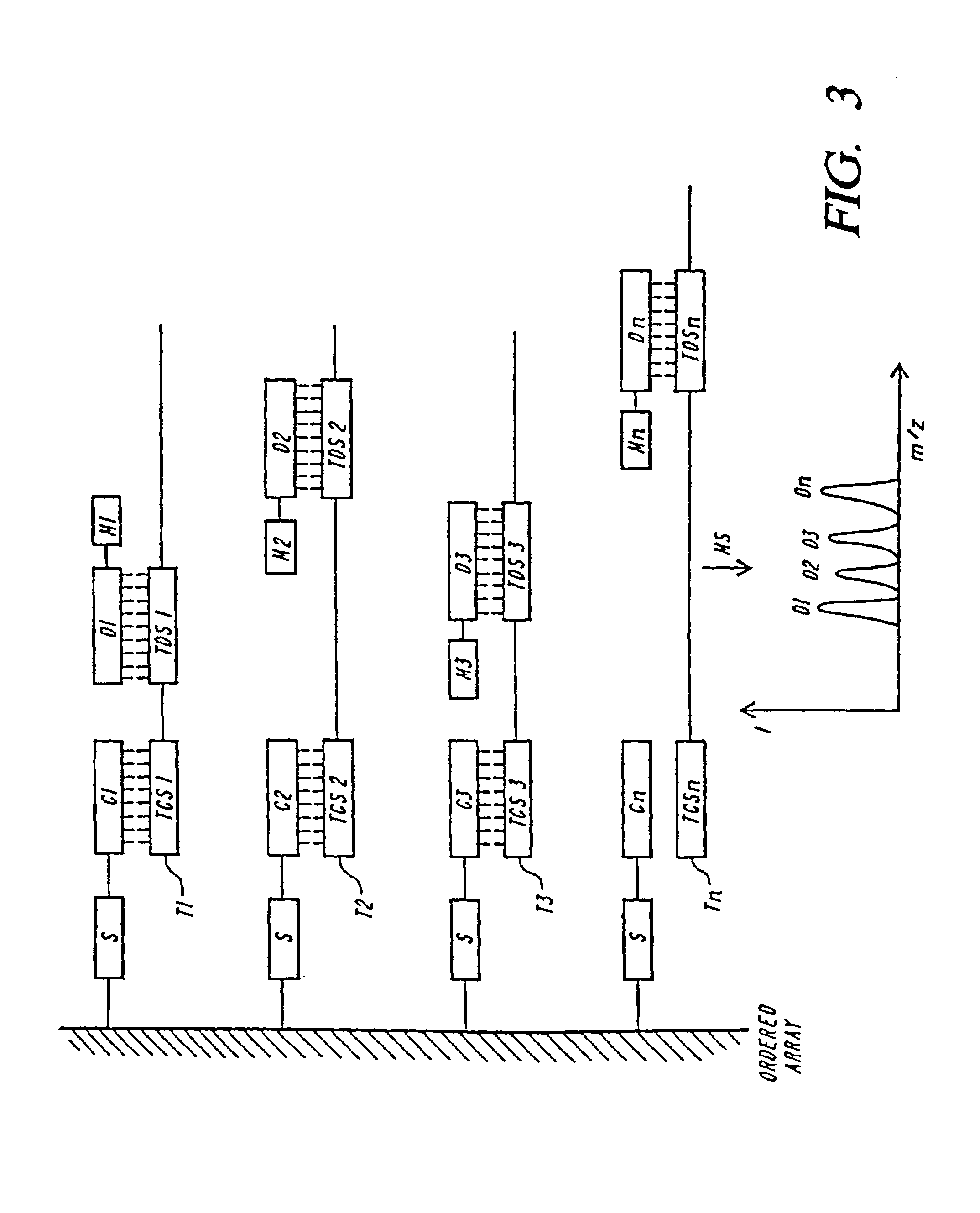
Families of compositions are provided as labels, referred to as eTag reporters for attaching to polymeric compounds and assaying based on release of the eTag reporters from the polymeric compound and separation and detection. For oligonucleotides, the eTag reporters are synthesized at the end of the oligonucleotide by using phosphite or phosphate chemistry, whereby mass-modifying regions, charge-modifying regions and detectable regions are added sequentially to produce the eTag labeled reporters. By using small building blocks and varying their combination large numbers of different eTag reporters can be readily produced attached to a binding compound specific for the target compound of interest for identification. Protocols are used that release the eTag reporter when the target compound is present in the sample.
Owner:MONOGRAM BIOSCIENCES
Systems and methods for preparing and analyzing low volume analyte array elements
InactiveUS7285422B1Rapid productionLess-expensive to employPeptide librariesSequential/parallel process reactionsAnalyteMass Spectrometry-Mass Spectrometry
The invention provides methods for dispensing tools that can be employed to generate multi-element arrays of sample material on a substrate surface. The substrates surfaces can be flat or geometrically altered to include wells of receiving material. The tool can dispense a spot of fluid to a substrate surface by spraying the fluid from the pin, contacting the substrate surface or forming a drop that touches against the substrate surface. The tool can form an array of sample material by dispensing sample material in a series of steps, while moving the pin to different locations above the substrate surface to form the sample array. The invention then passes the prepared sample arrays to a plate assembly that disposes the sample arrays for analysis by mass spectrometry. To this end, a mass spectrometer is provided that generates a set of spectra signal which can be understood as indicative of the composition of the sample material under analysis.
Owner:AGENA BIOSCI
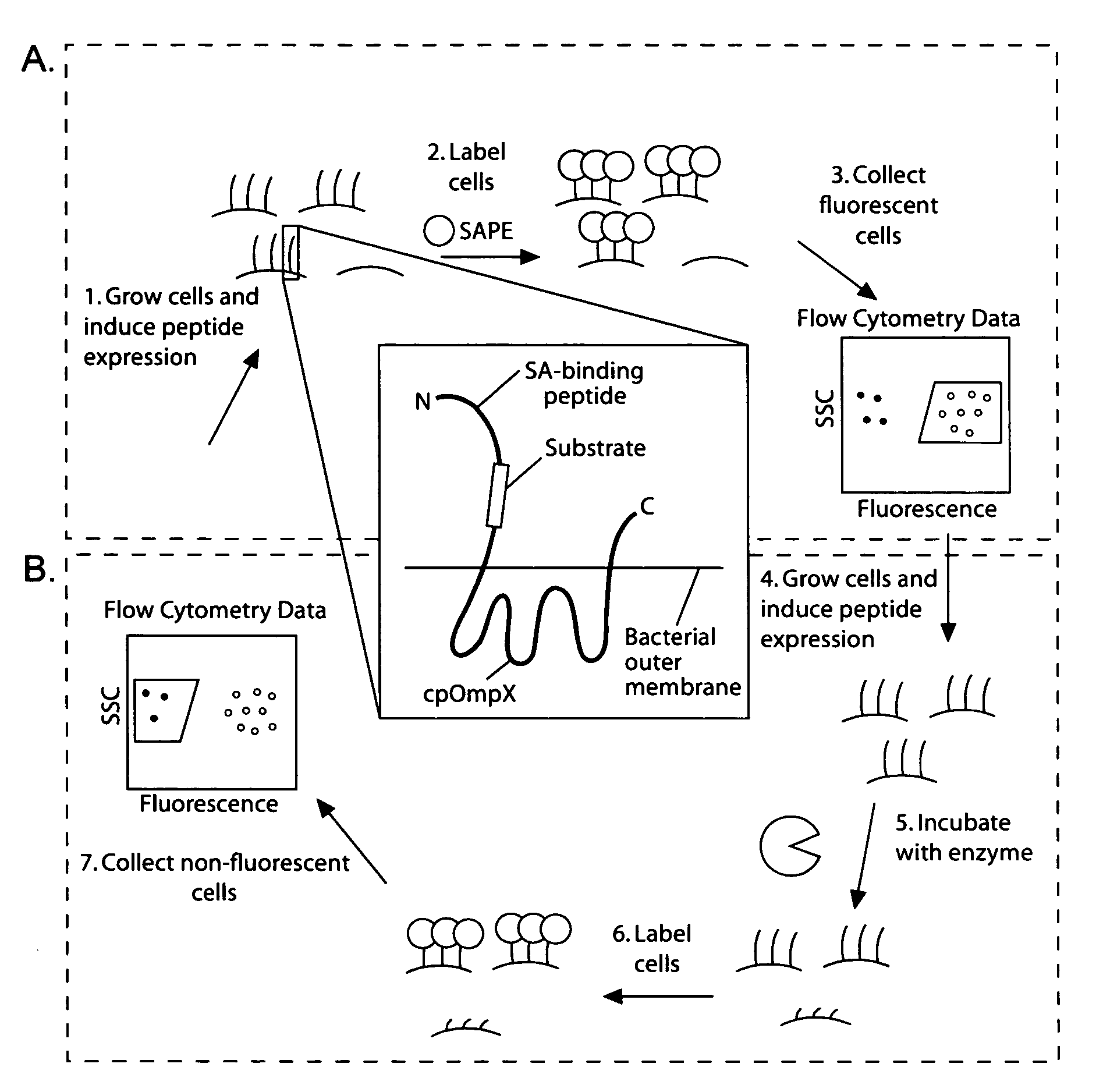
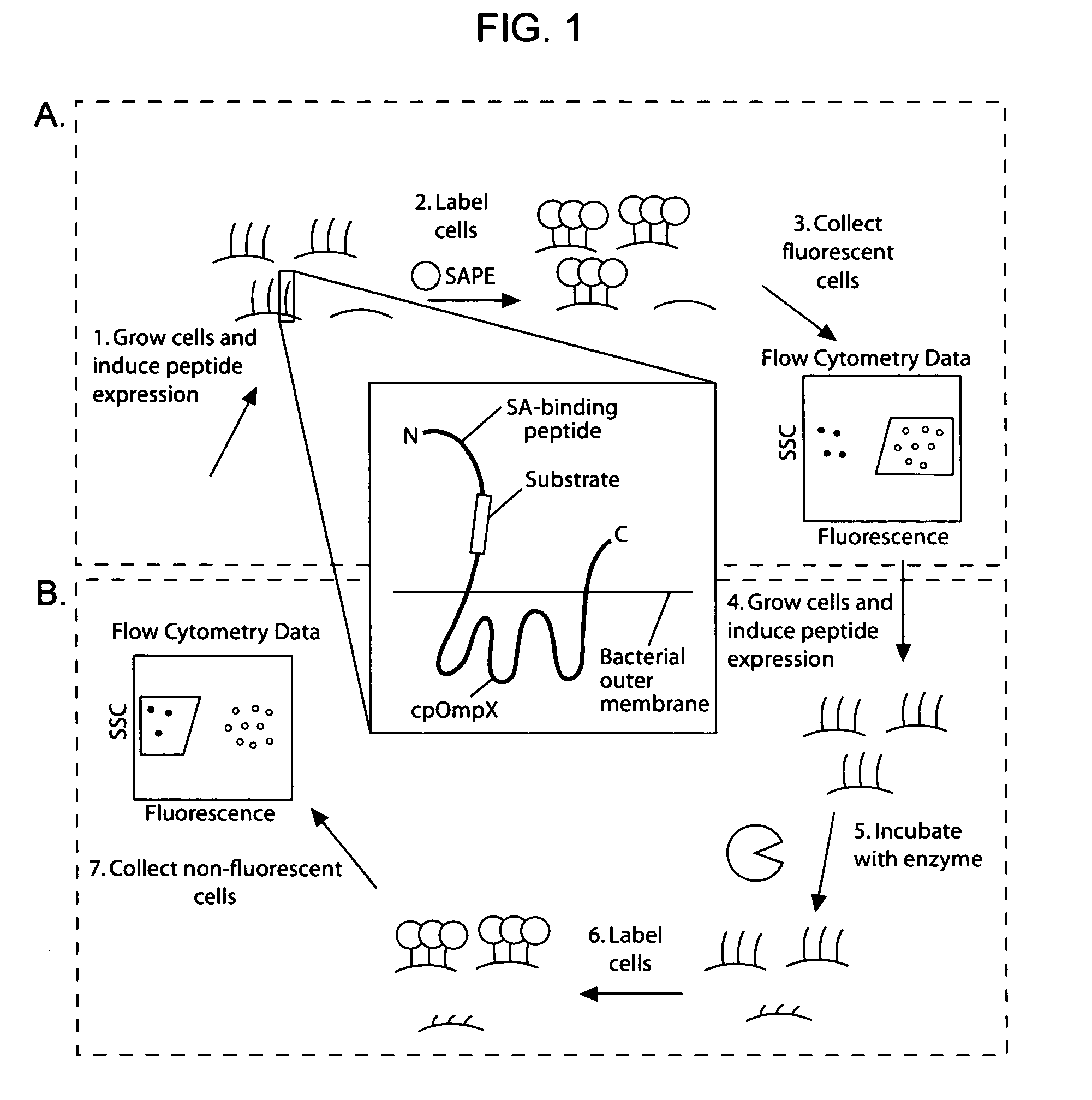
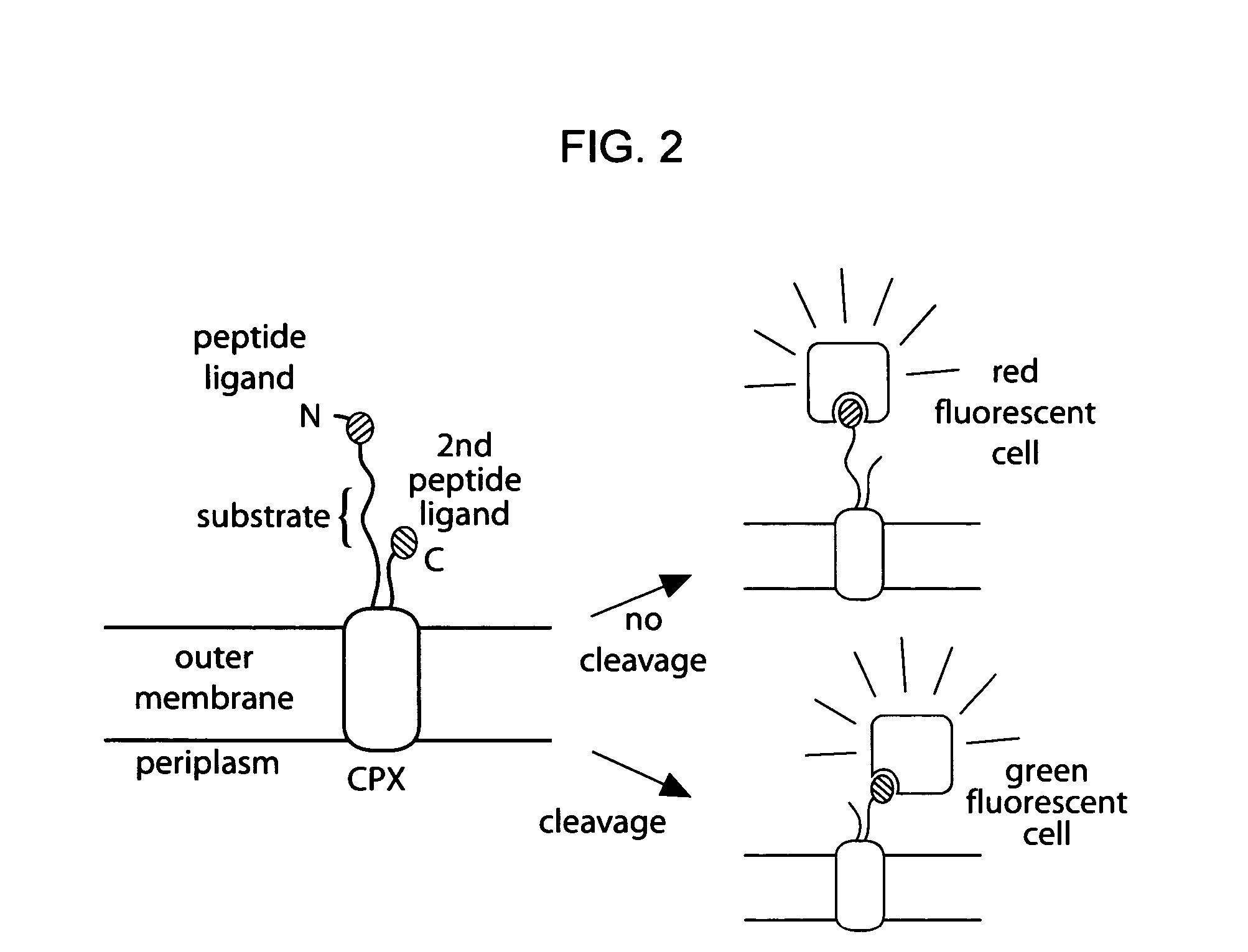
Cellular libraries of peptide sequences (CLiPS) and methods of using the same
The present invention provides compositions including peptide display scaffolds that present at least one candidate peptide and at least one detectable moiety in at least one of the N-terminal and C-terminal candidate peptide presenting domains that when expressed in a cell are accessible at a surface of the cell outermembrane. In addition, the present invention also provides kits and methods for screening a library of cells presenting the candidate peptides in peptide display scaffolds to identify a ligand for an enzyme.
Owner:RGT UNIV OF CALIFORNIA
Arrays of protein-capture agents and methods of use thereof
Owner:WAGNER PETER +3
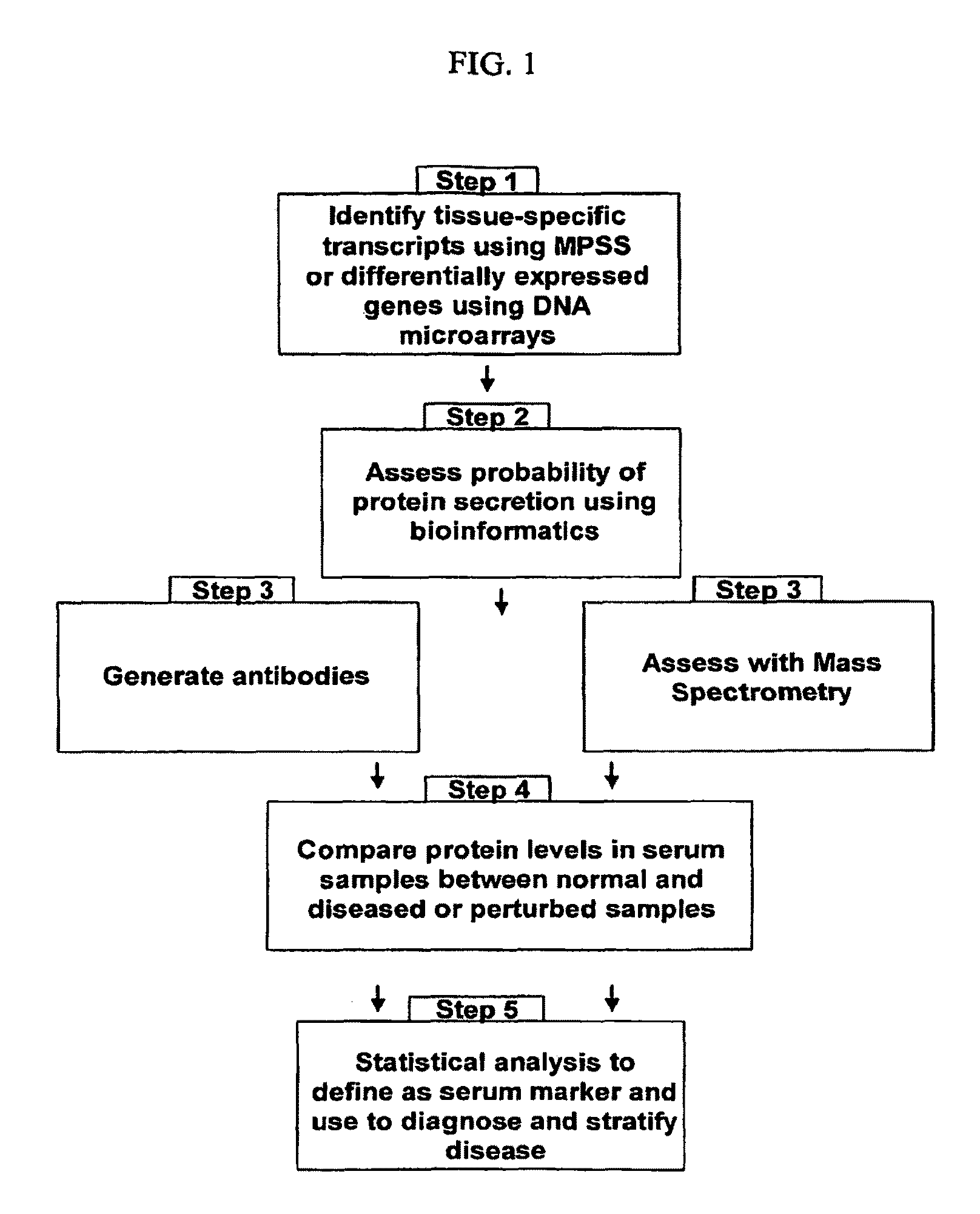
Organ-specific proteins and methods of their use
The present invention relates generally to methods for identifying and using organ-specific proteins and transcripts. The present invention further provides compositions comprising organ-specific proteins and transcripts encoding the same, detection reagents for detecting such proteins and transcripts, and diagnostic panels, kits and arrays for measuring organ-specific proteins / transcripts in blood, biological tissue or other biological fluid.
Owner:INSTITUTE FOR SYSTEMS BIOLOGY +1
Reaction plenum with magnetic separation and/or ultrasonic agitation
InactiveUS6277332B1Easy to runPeptide librariesSequential/parallel process reactionsSolid phasesChemical compound
The inventive apparatus is comprised of a means for producing cavitation in a complex reaction mixture to enhance the yield of the selected reaction product, a means for controlling the temperature of the complex reaction mixture, especially during cavitation, and a means for affecting magnetic separation of paramagnetic beads to which the selected reaction product is attached from the complex reaction mixture. In one embodiment, the compound(s) or molecule(s) is synthesized in situ, and isolated using the inventive apparatus. The apparatus finds use in the fields of solid phase organic synthesis, and for isolation and purification of a selected compound(s) or molecule (s), especially where automation is desired.
Owner:PROTANA
Systems and methods of conducting multiplexed experiments
A method for multiplexed detection and quantification of analytes by reacting them with probe molecules attached to specific and identifiable carriers. These carriers can be of different size, shape, color, and composition. Different probe molecules are attached to different types of carriers prior to analysis. After the reaction takes place, the carriers can be automatically analyzed. This invention obviates cumbersome instruments used for the deposition of probe molecules in geometrically defined arrays. In the present invention, the analytes are identified by their association with the defined carrier, and not (or not only) by their position. Moreover, the use of carriers provides a more homogenous and reproducible representation for probe molecules and reaction products than two-dimensional imprinted arrays or DNA chips.
Owner:VITRA BIOSCIENCES ASSIGNMENT FOR THE BENEFIT OF CREDITORS LLC +1
Protein Detection Via Nanoreporters
ActiveUS20110086774A1Peptide librariesMicrobiological testing/measurementProtein detectionBiochemistry
The invention provides methods, compositions, kits and devices for the detection of proteins. In some embodiments, the invention allows for multiplexed protein detection.
Owner:NANOSTRING TECH INC
Structured Substrates for Optical Surface Profiling
ActiveUS20090011948A1Increase heightQuick changePeptide librariesNucleotide librariesOptical surfaceBiology
This disclosure provides methods and devices for the label-free detection of target molecules of interest. The principles of the disclosure are particularly applicable to the detection of biological molecules (e.g., DNA, RNA, and protein) using standard SiO2-based microarray technology.
Owner:TRUSTEES OF BOSTON UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com