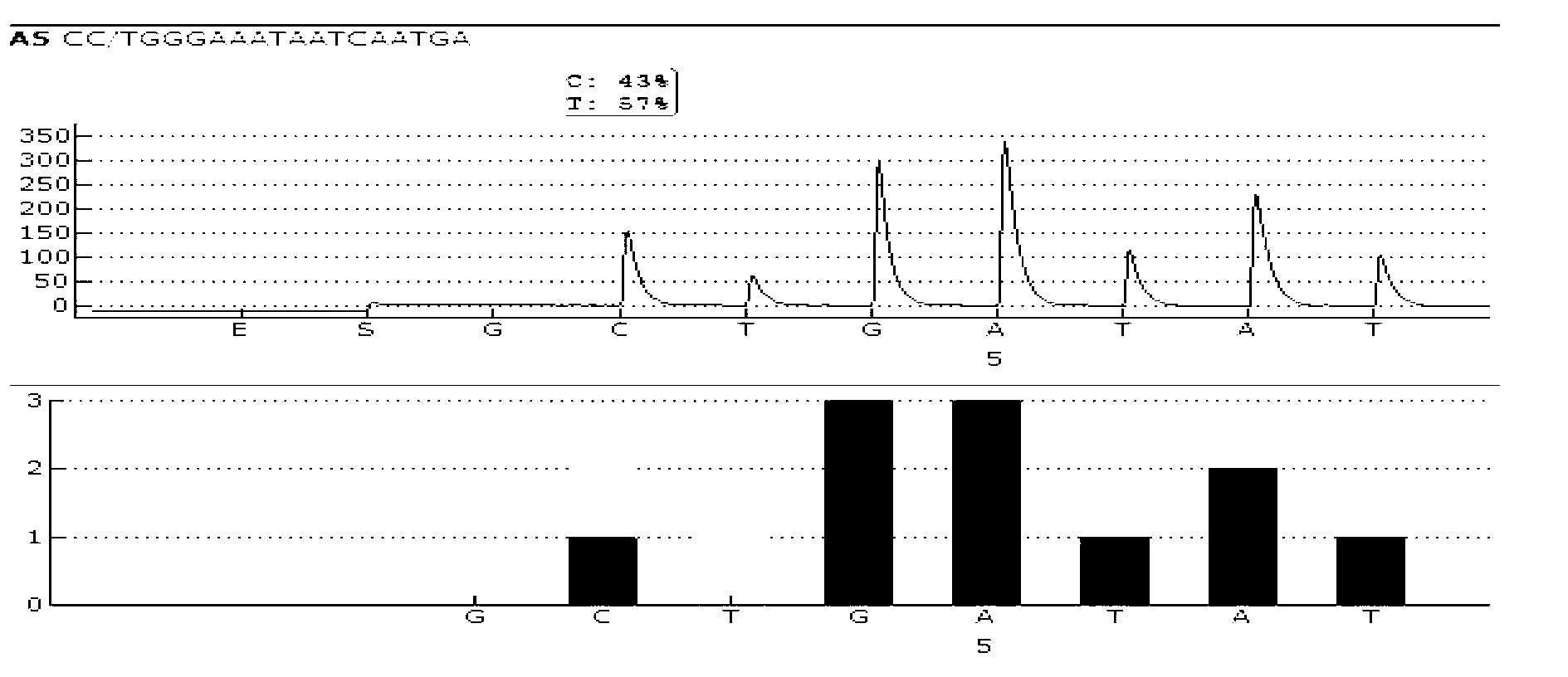
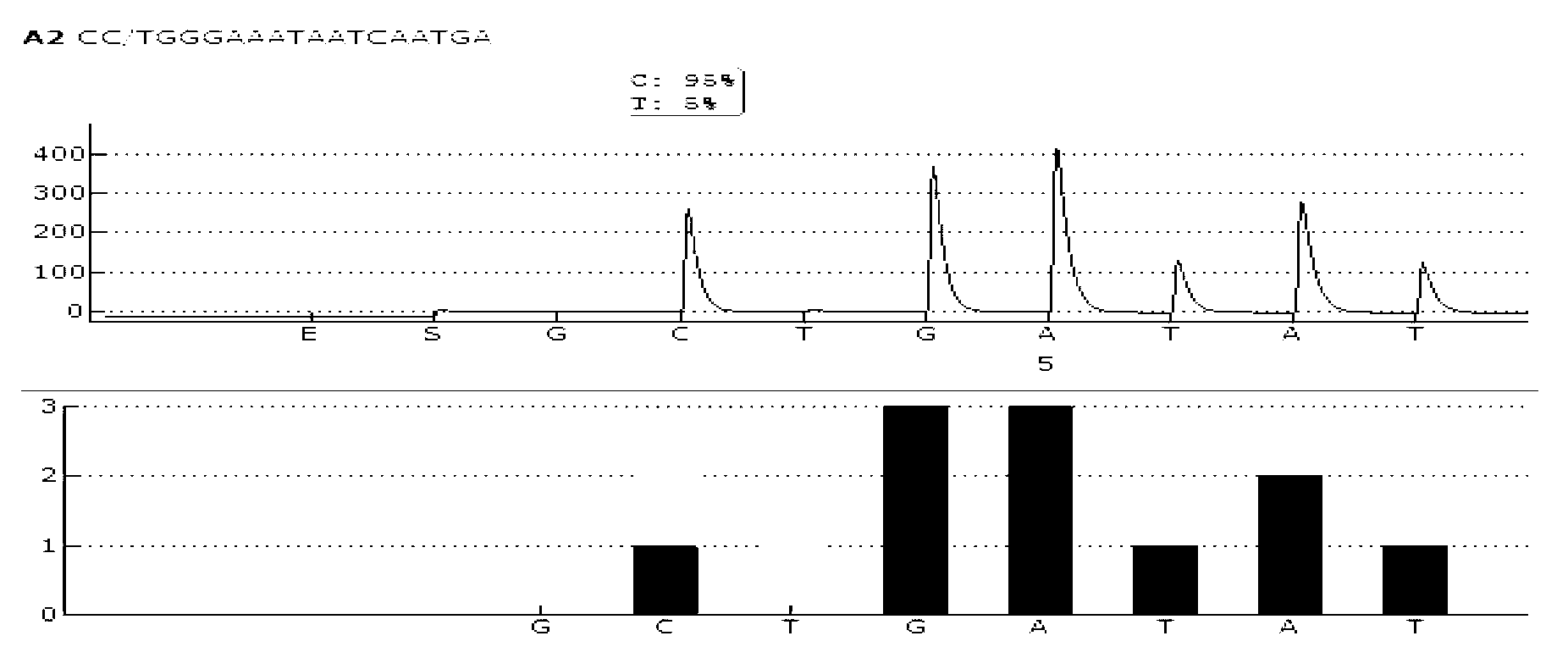
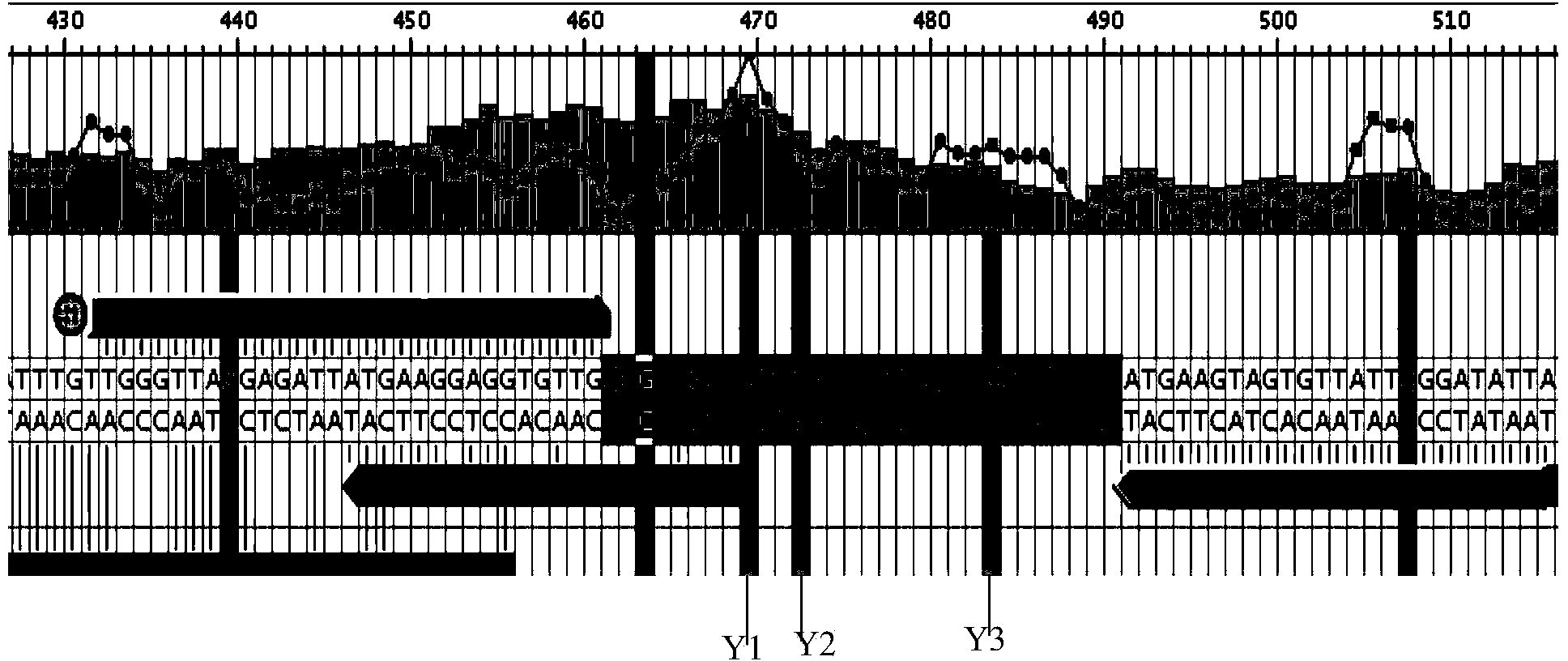
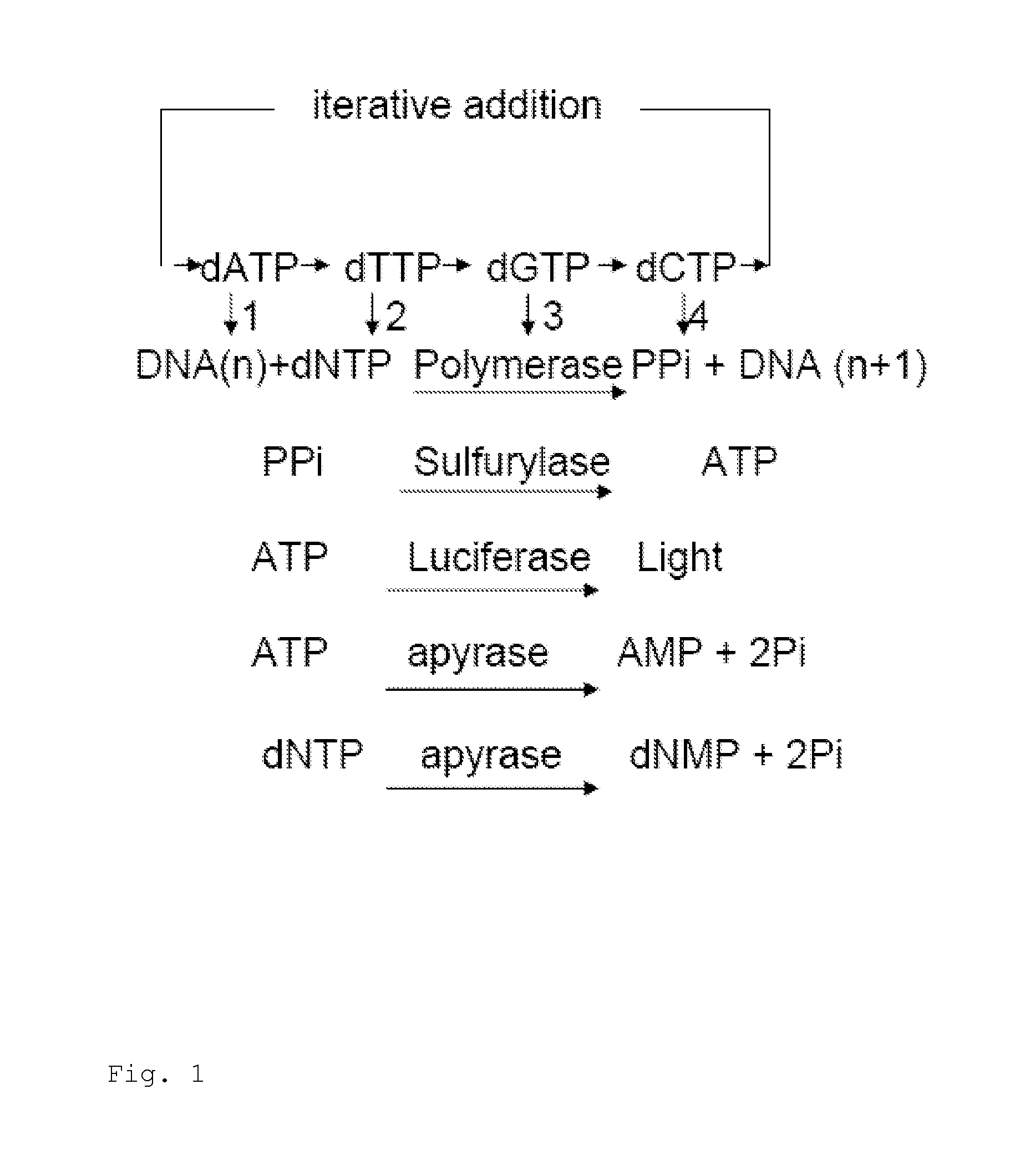
Patents
Literature
332 results about "Pyrosequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
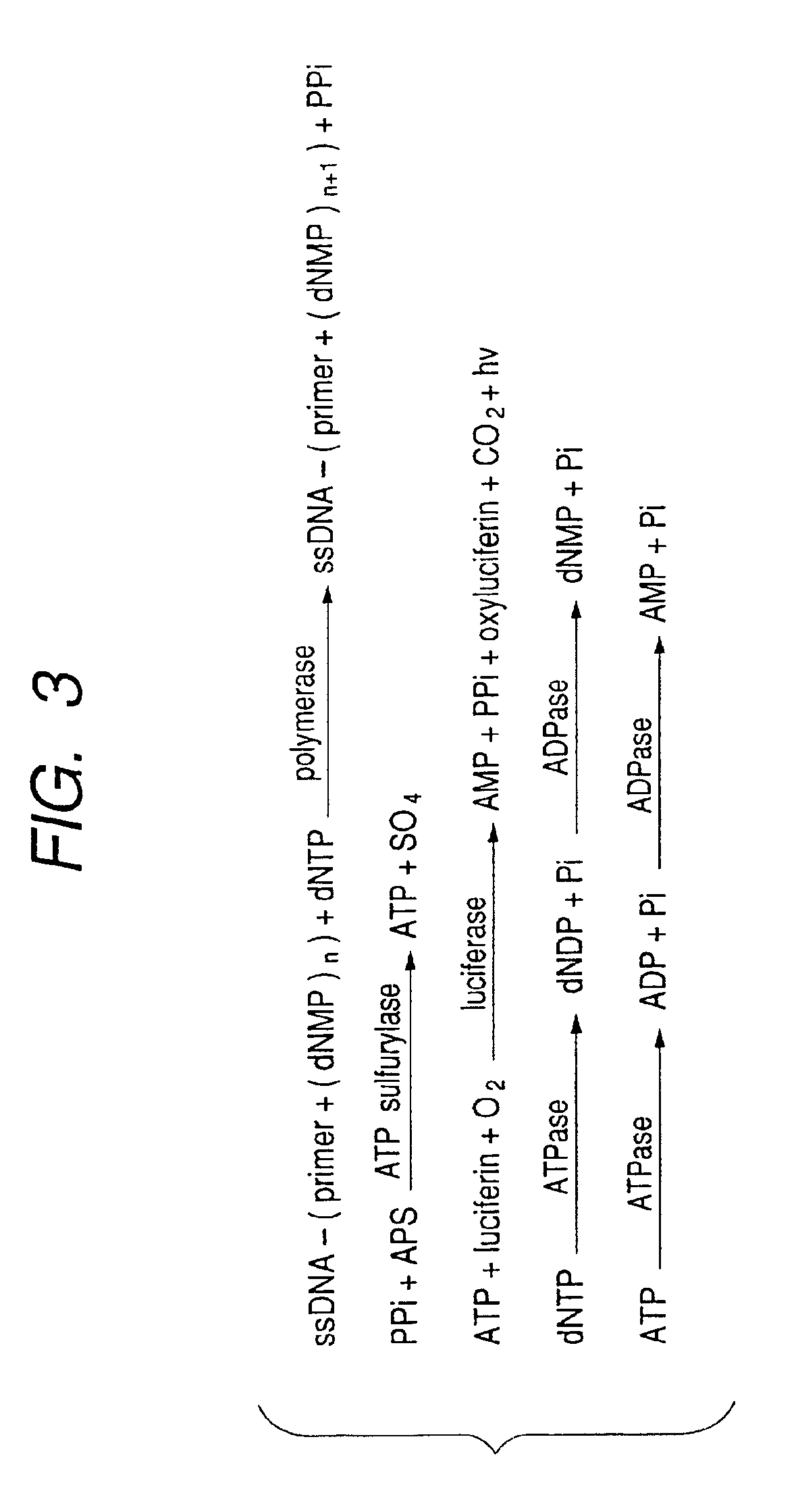
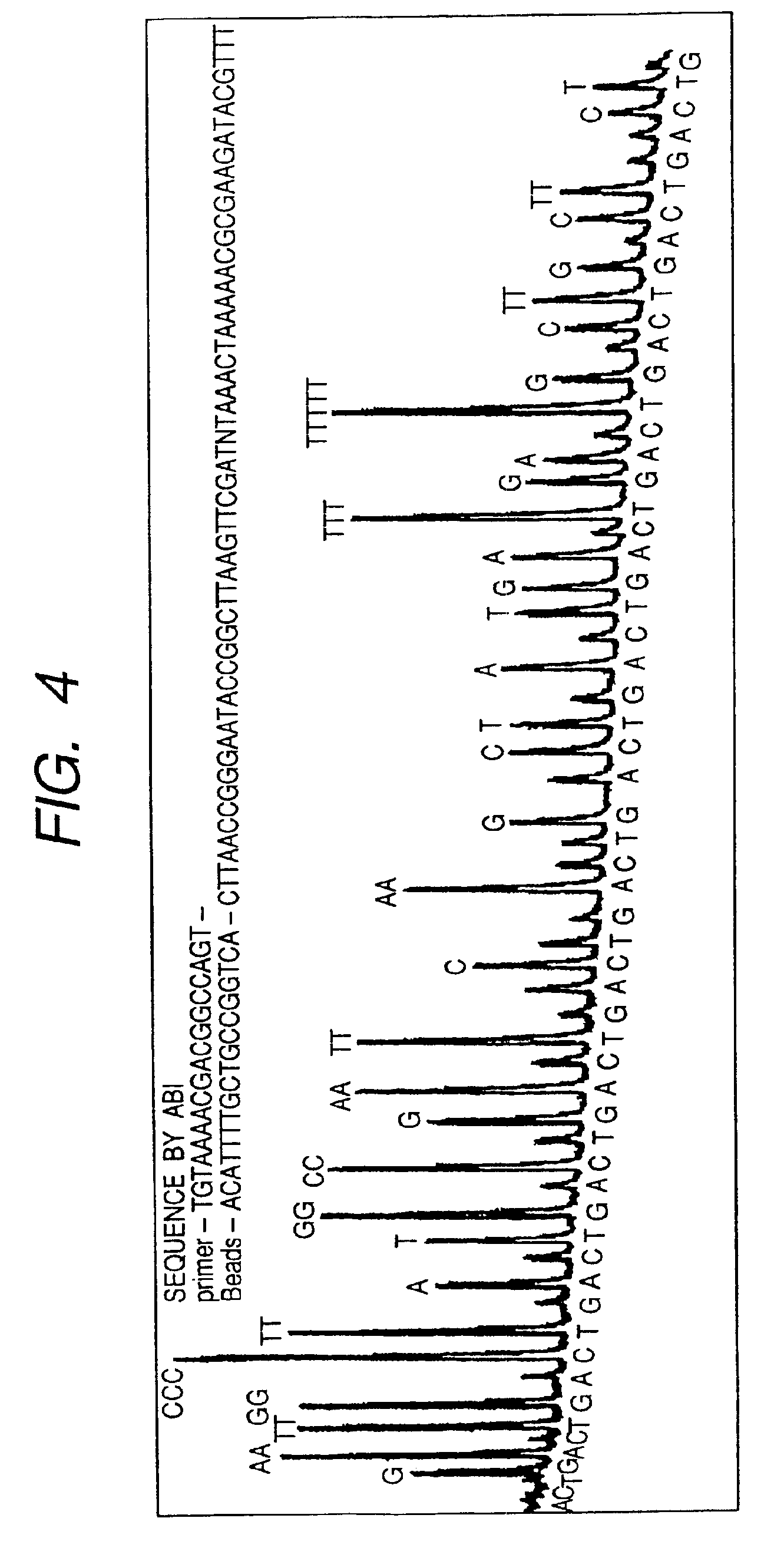
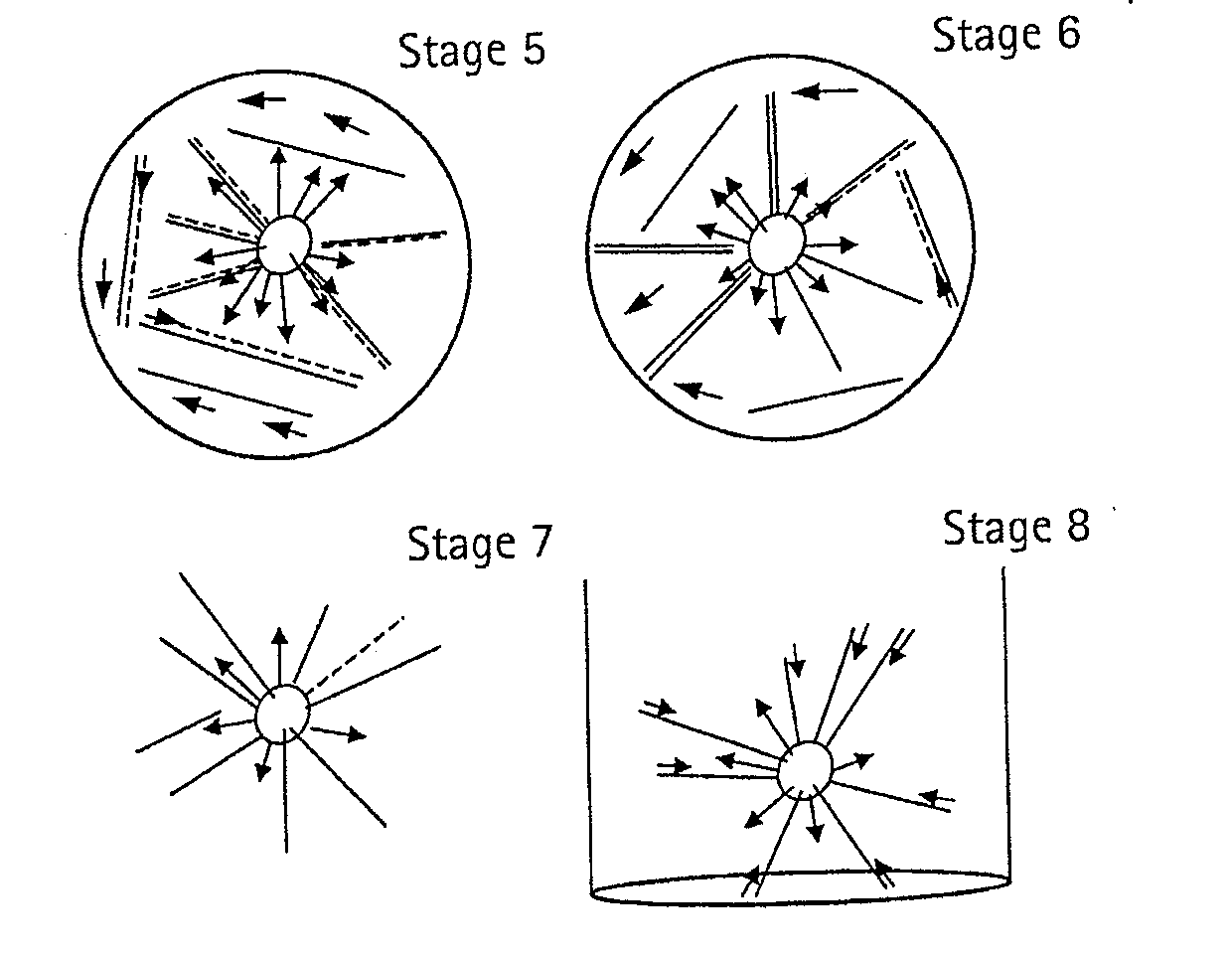
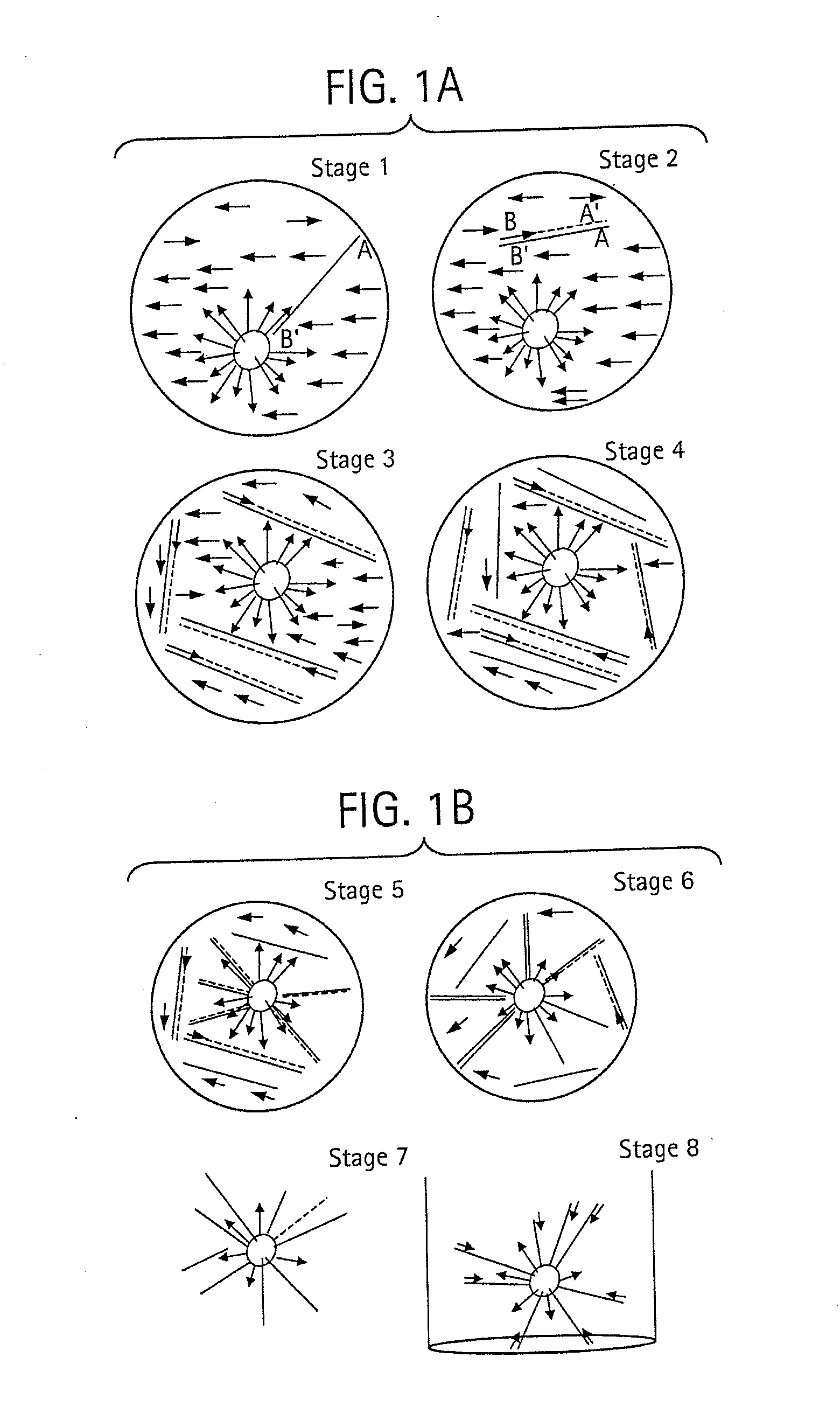
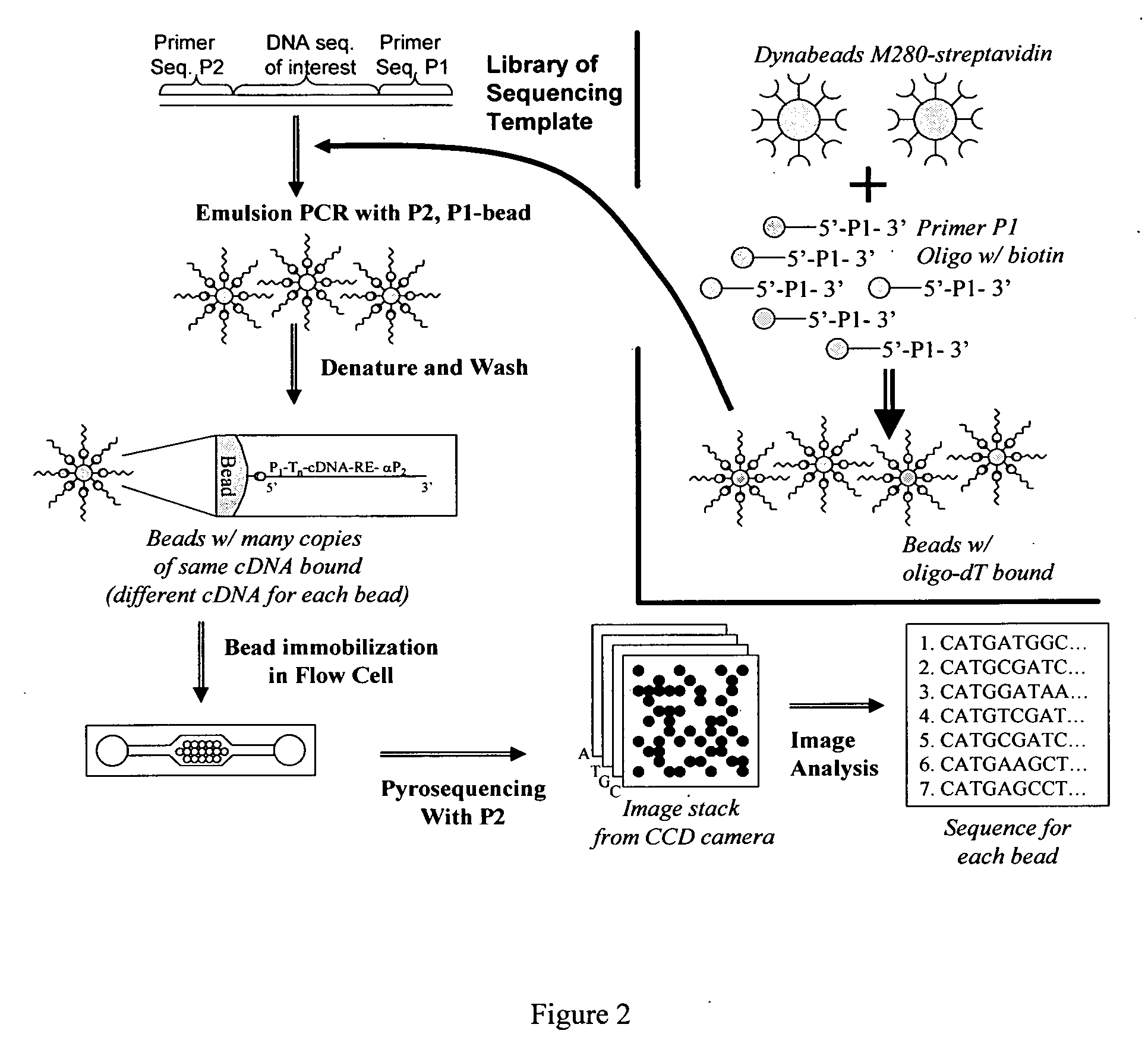
Pyrosequencing is a method of DNA sequencing (determining the order of nucleotides in DNA) based on the "sequencing by synthesis" principle, in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase. Pyrosequencing relies on light detection based on a chain reaction when pyrophosphate is released. Hence, the name pyrosequencing.
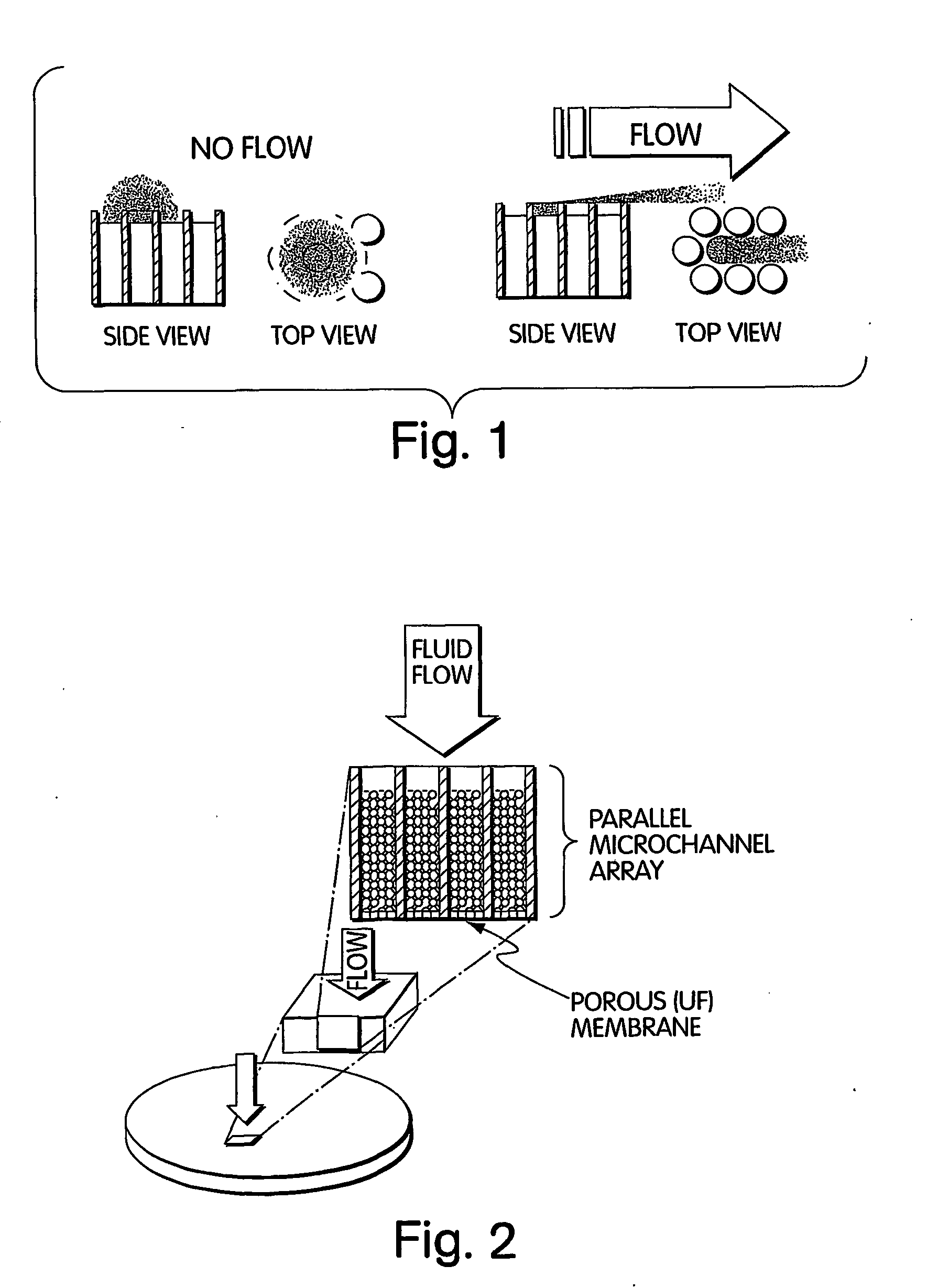
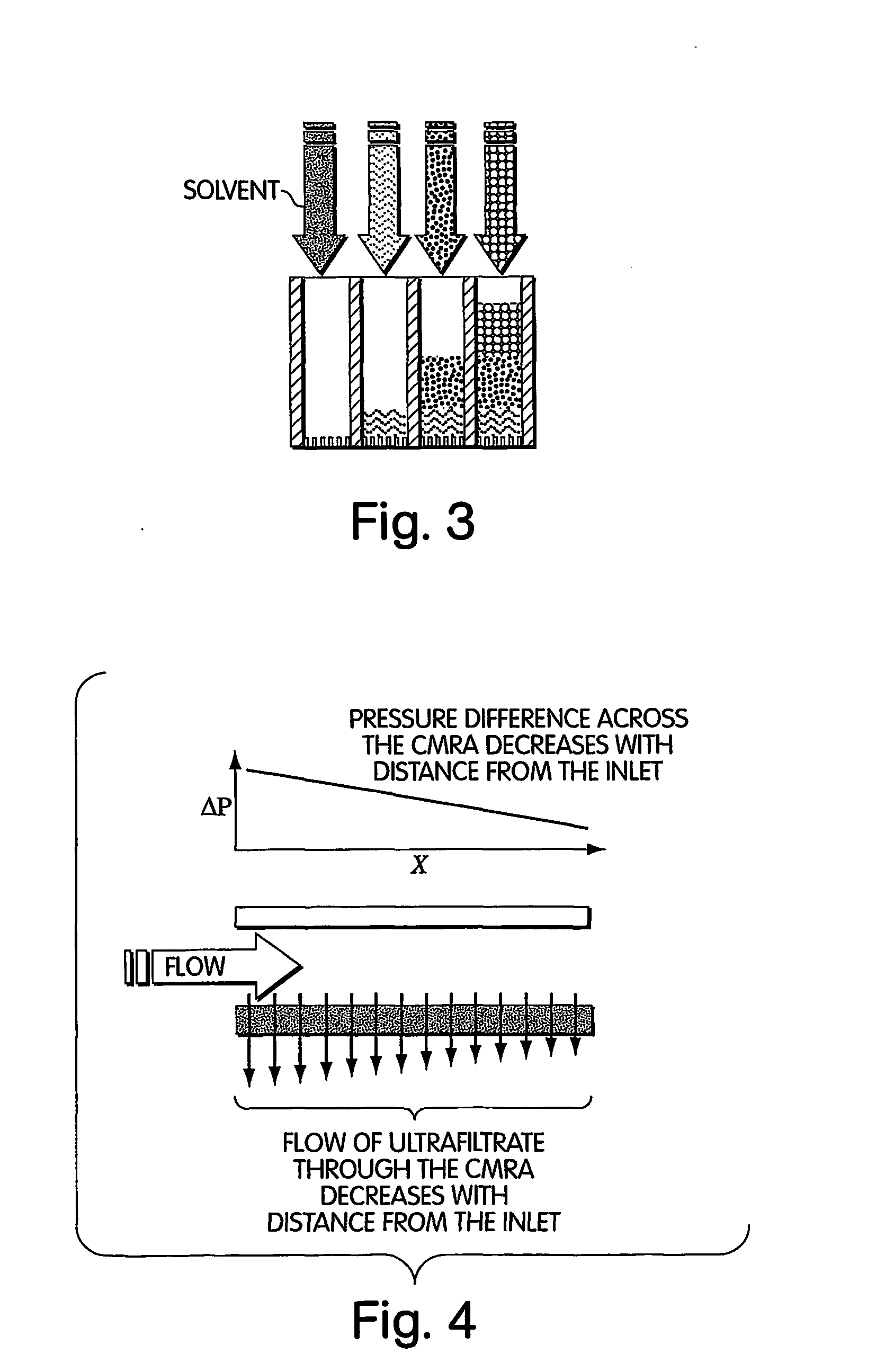
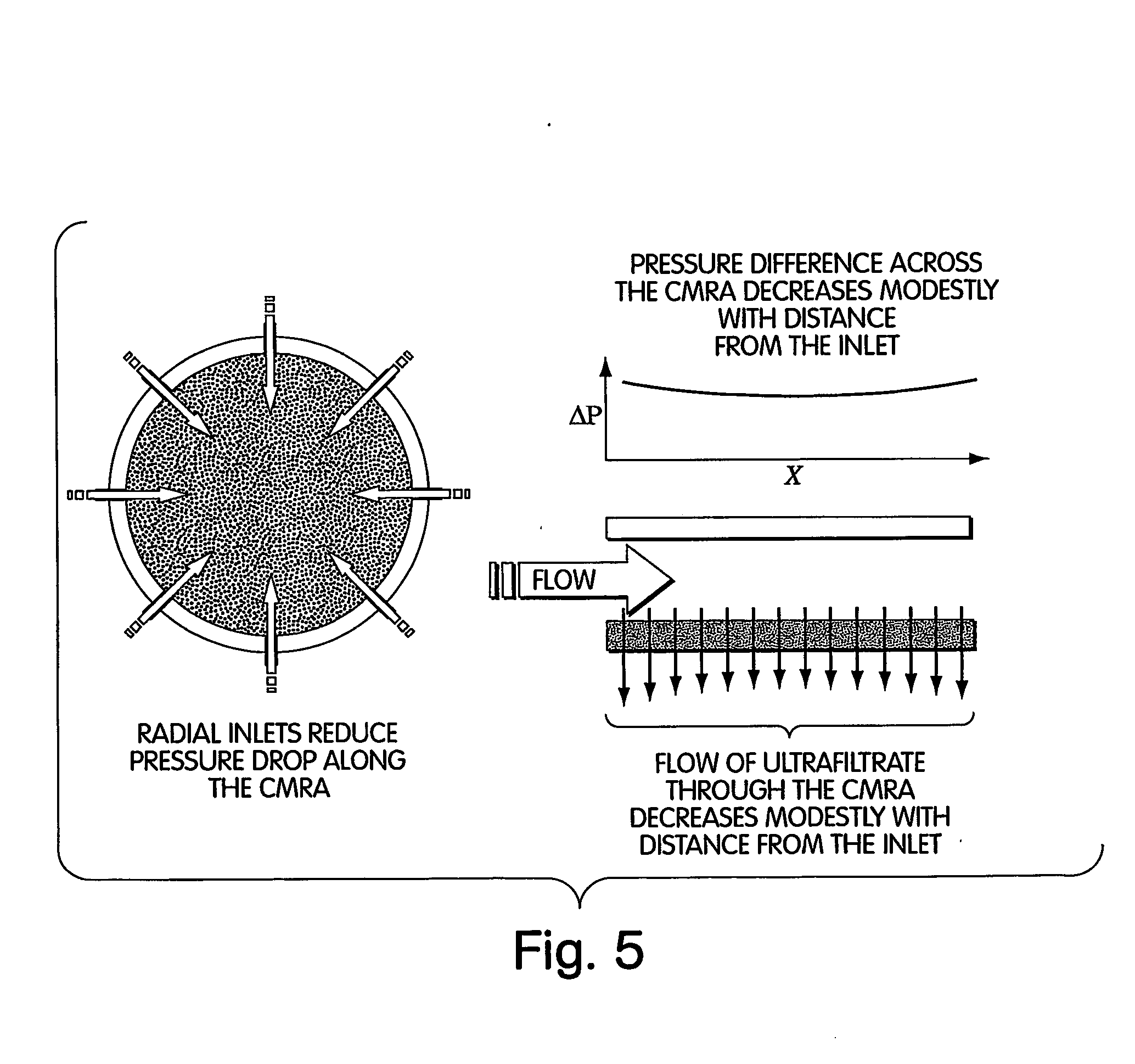
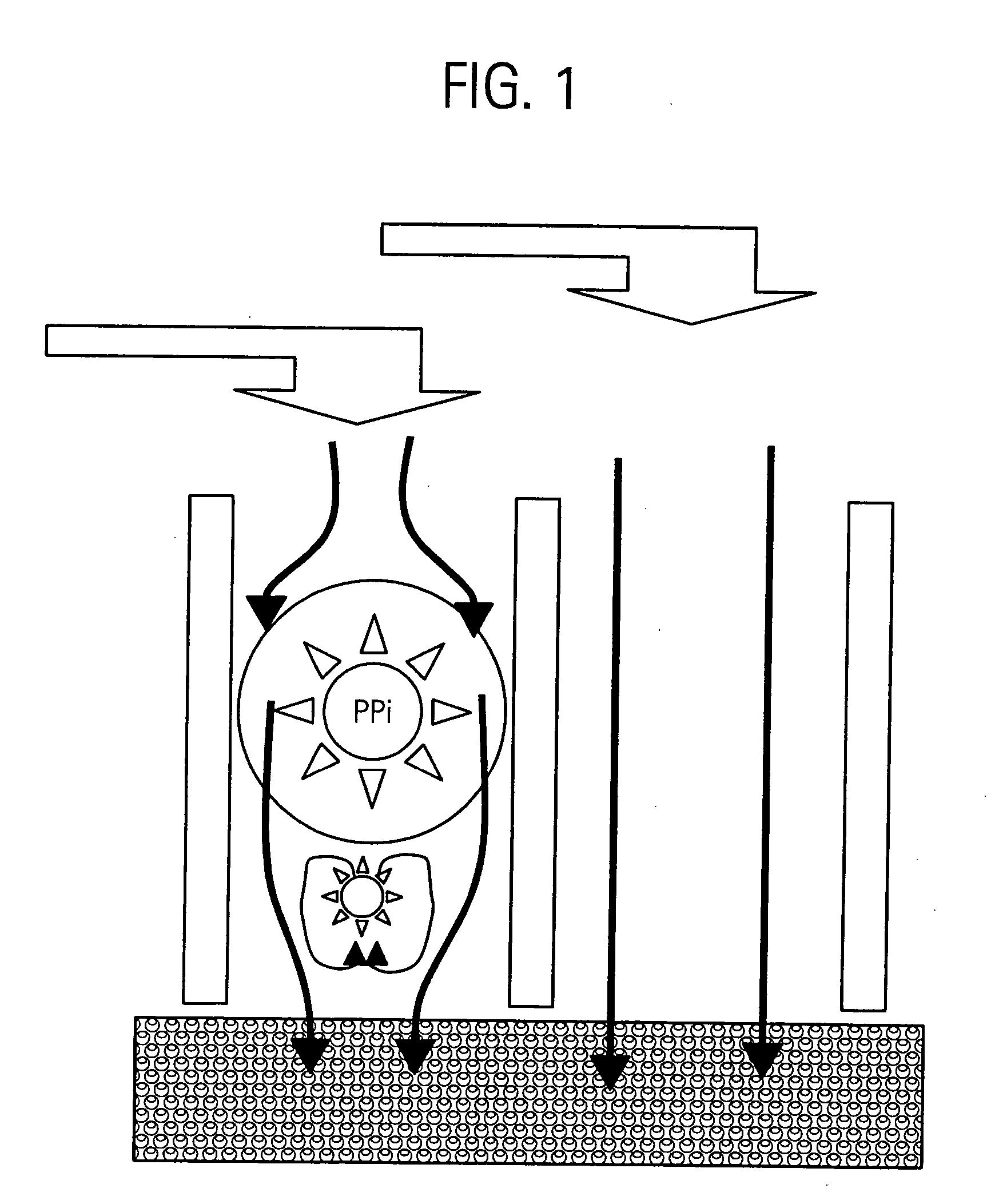
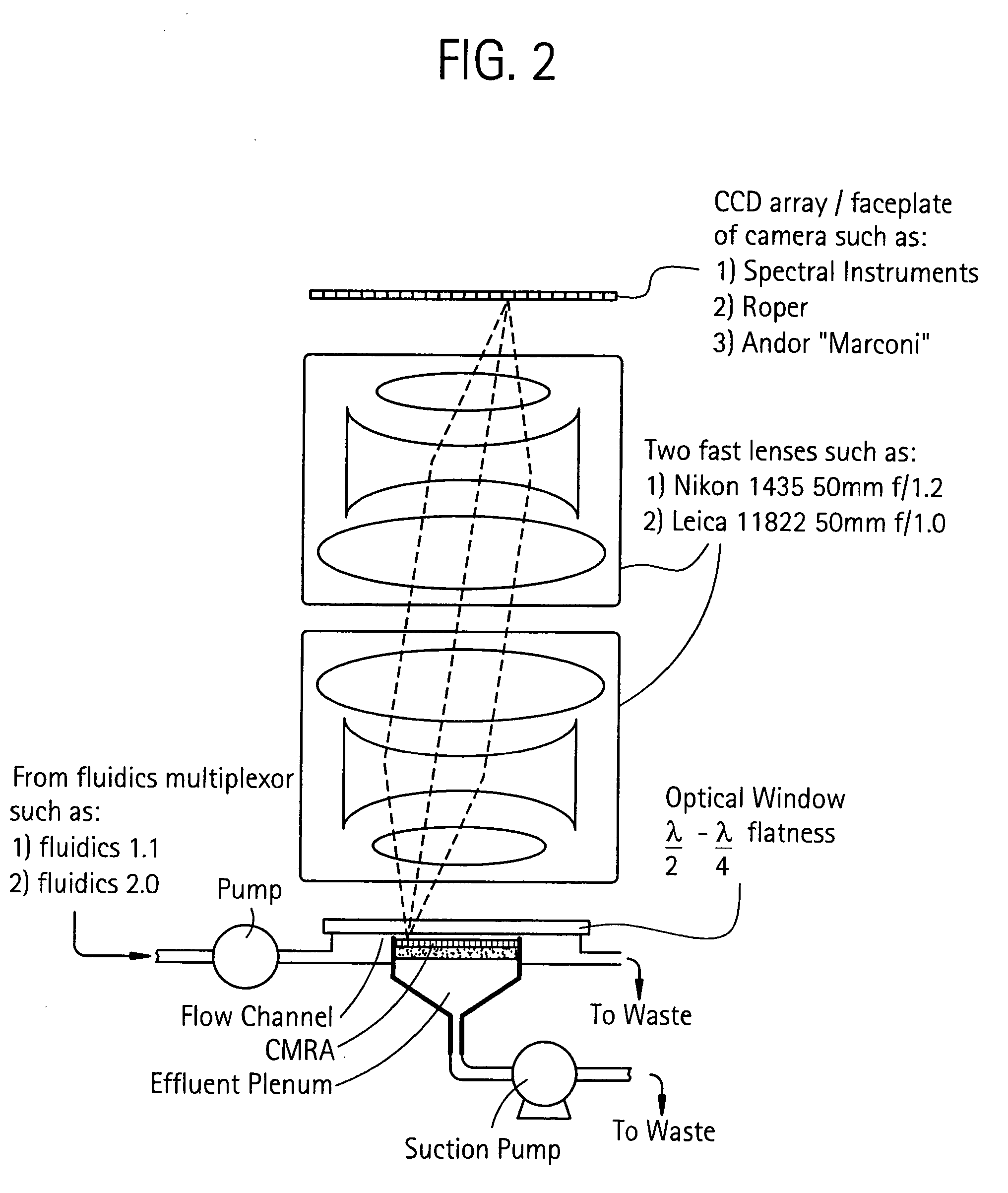
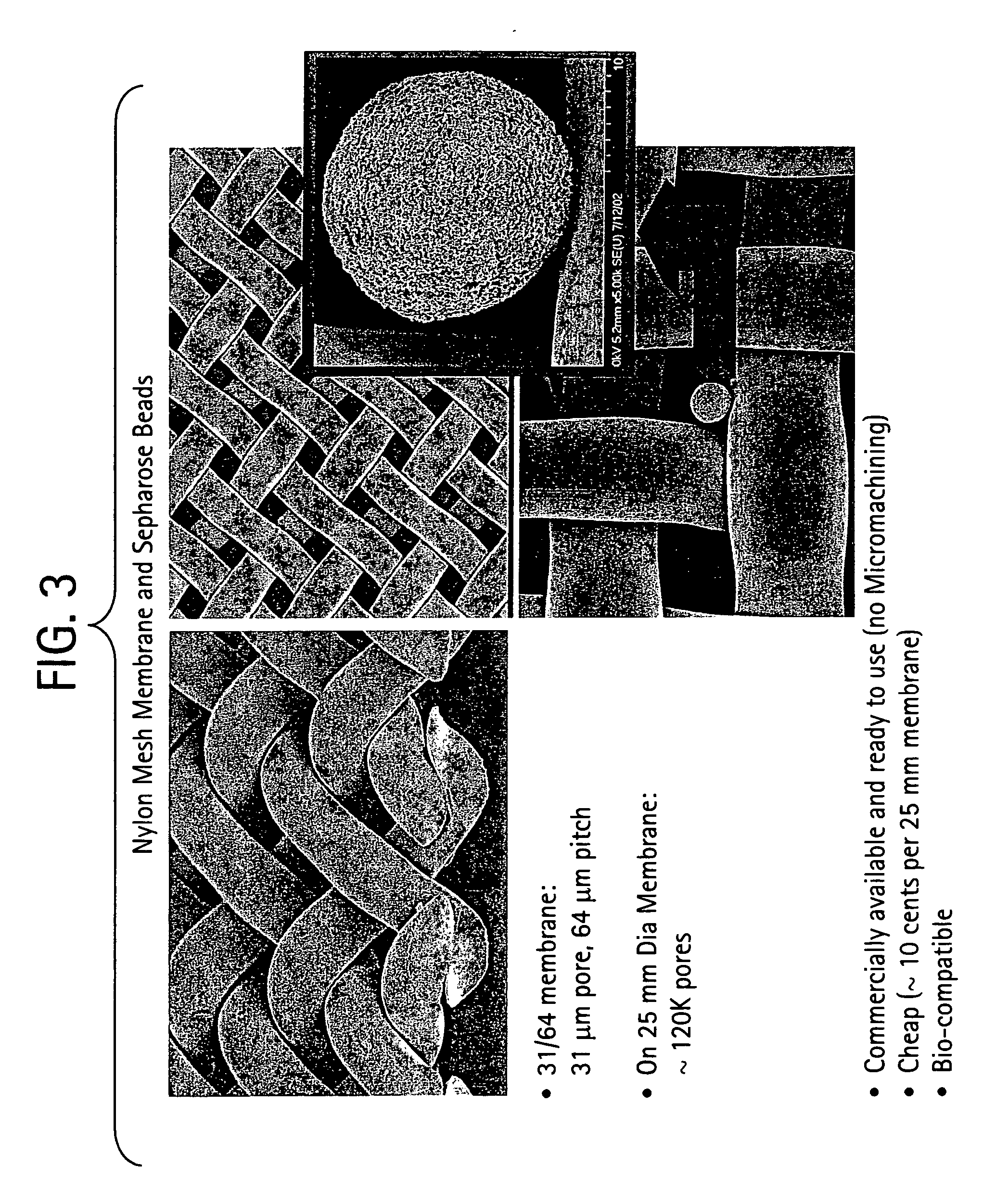
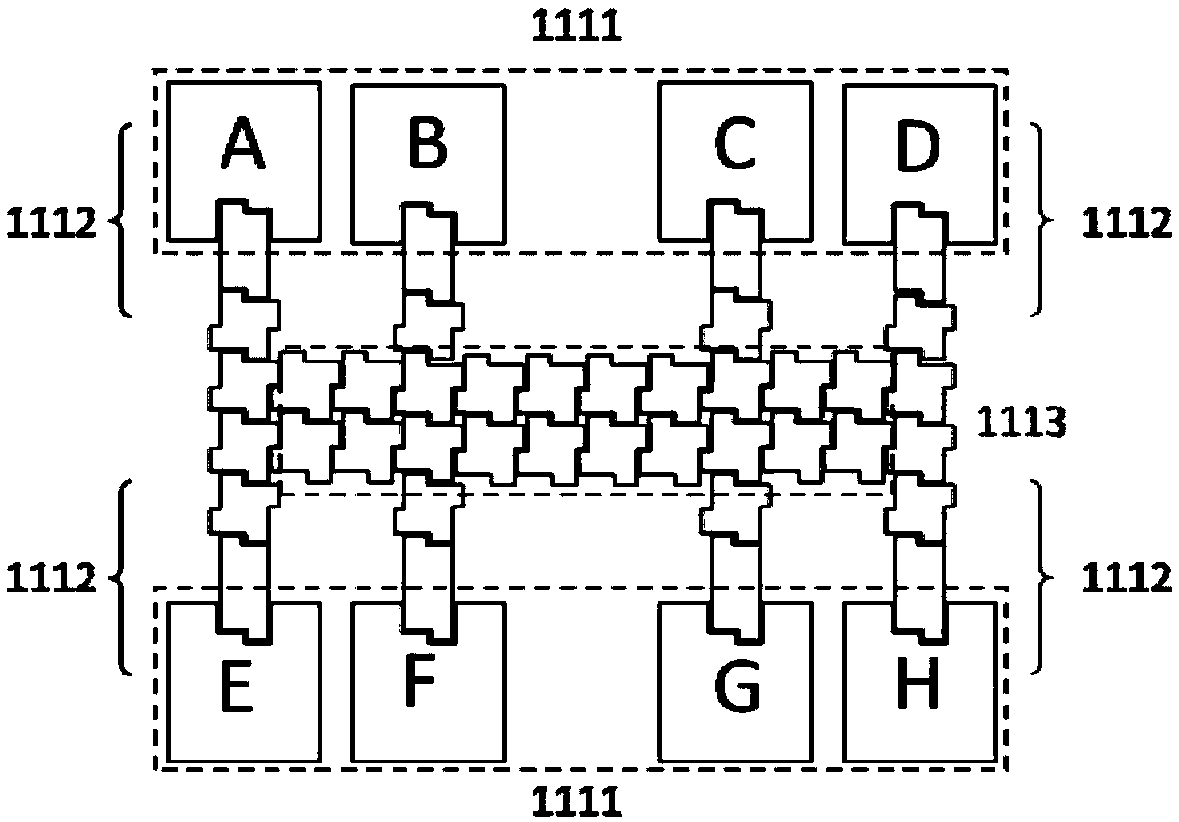
Method for isolation of independent, parallel chemical micro-reactions using a porous filter
InactiveUS20050009022A1Fast deliveryFaster more complete removalBioreactor/fermenter combinationsPeptide librariesChemical reactionCompound (substance)
The present invention relates to the field of fluid dynamics. More specifically, this invention relates to methods and apparatus for conducting densely packed, independent chemical reactions in parallel in a substantially two-dimensional array. Accordingly, this invention also focuses on the use of this array for applications such as DNA sequencing, most preferably pyrosequencing, and DNA amplification.
Owner:WEINER MICHAEL P +4
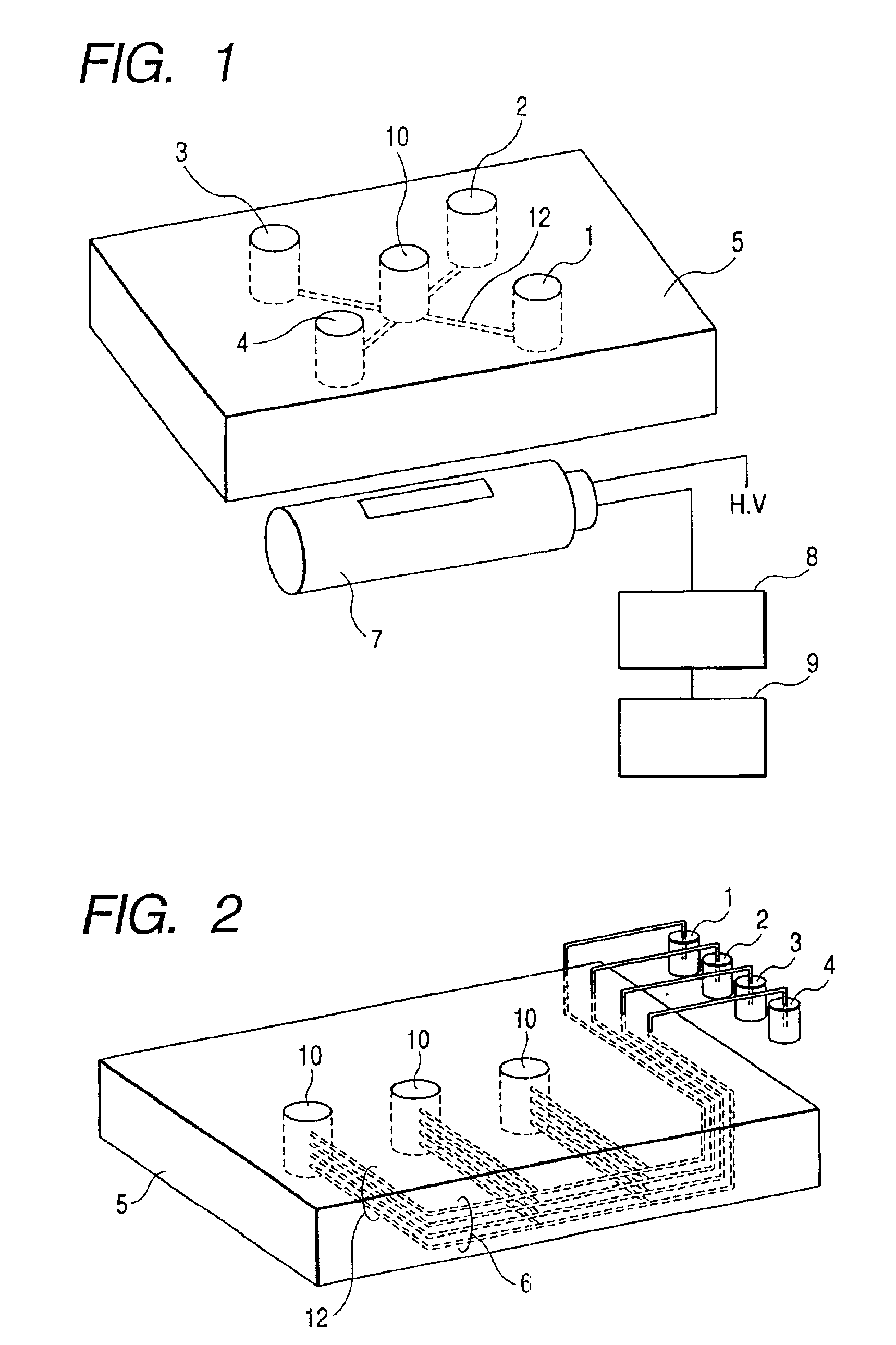
DNA base sequencing system
InactiveUS6841128B2Increase supplySize of apparatus be minimizedBioreactor/fermenter combinationsElectrolysis componentsA-DNAReagent
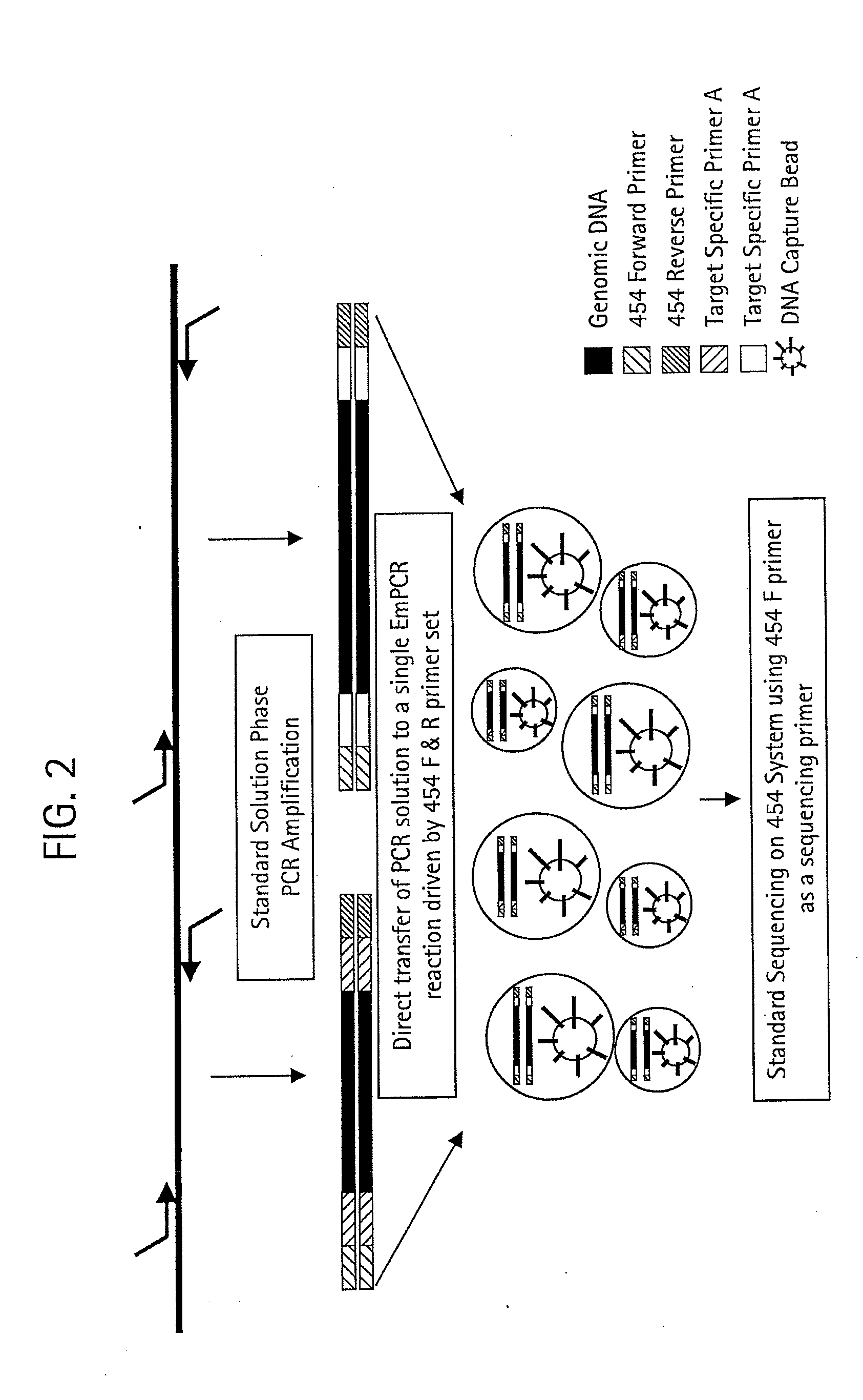
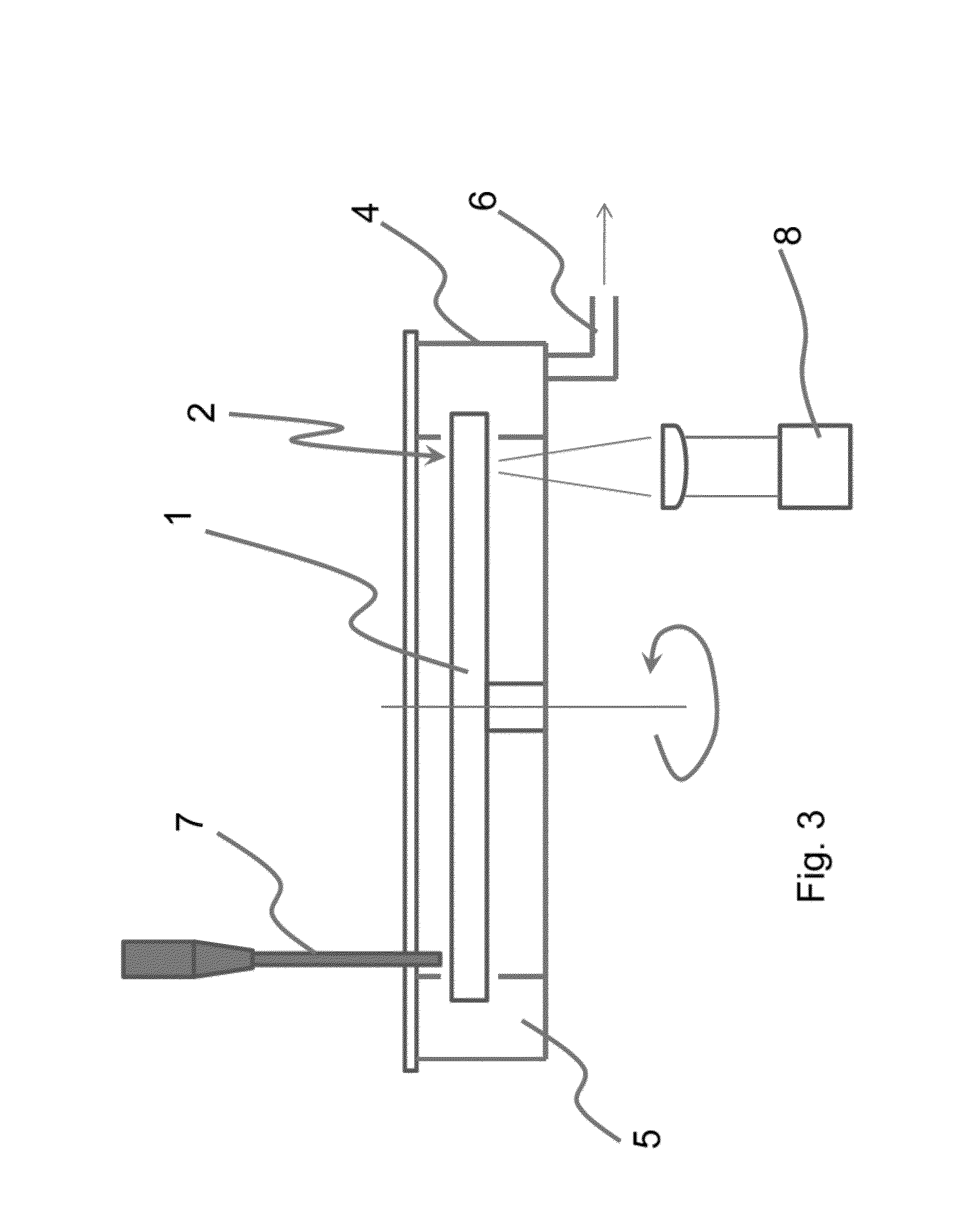
The present invention provides a DNA base sequencing system having a compact, simple and convenient structure.In one embodiment of the present invention, a reaction chamber module for pyrosequencing in which a multiple number of reaction vessels (reaction chambers) 10 and reagent-introducing narrow tubes 6 are integrated is formed in a device board 5. Reagents held in reagent reservoirs 1, 2, 3 and 4 mounted separately from this reaction chamber module are introduced into the reaction vessels 10 via reagent-introducing narrow tubes (capillaries) 6. The reagent-introducing narrow tubes (capillaries) 6 at the area of 2 cm from the reaction vessels 10 are structured with narrow capillaries having an inner diameter of about 0.1 mm and the conductance of these capillaries for the reagent solution determines the injection speed of the solution.Using the present invention, many kinds of DNAs can be analyzed simultaneously.
Owner:HITACHI LTD
Cancer screening method
InactiveUS20080311570A1Accurate screening resultEasy diagnosisMicrobiological testing/measurementFermentationScreening cancerScreening method
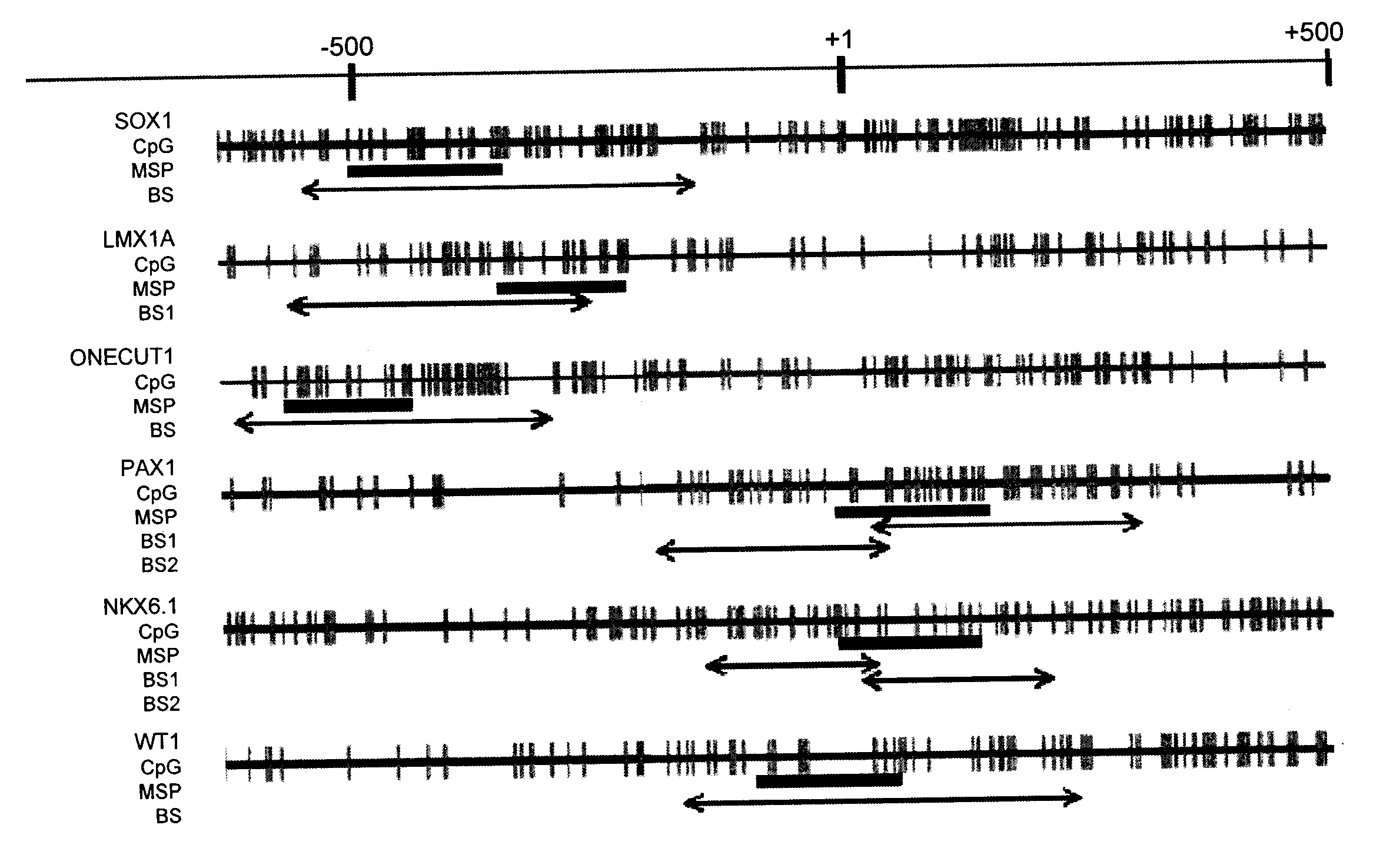
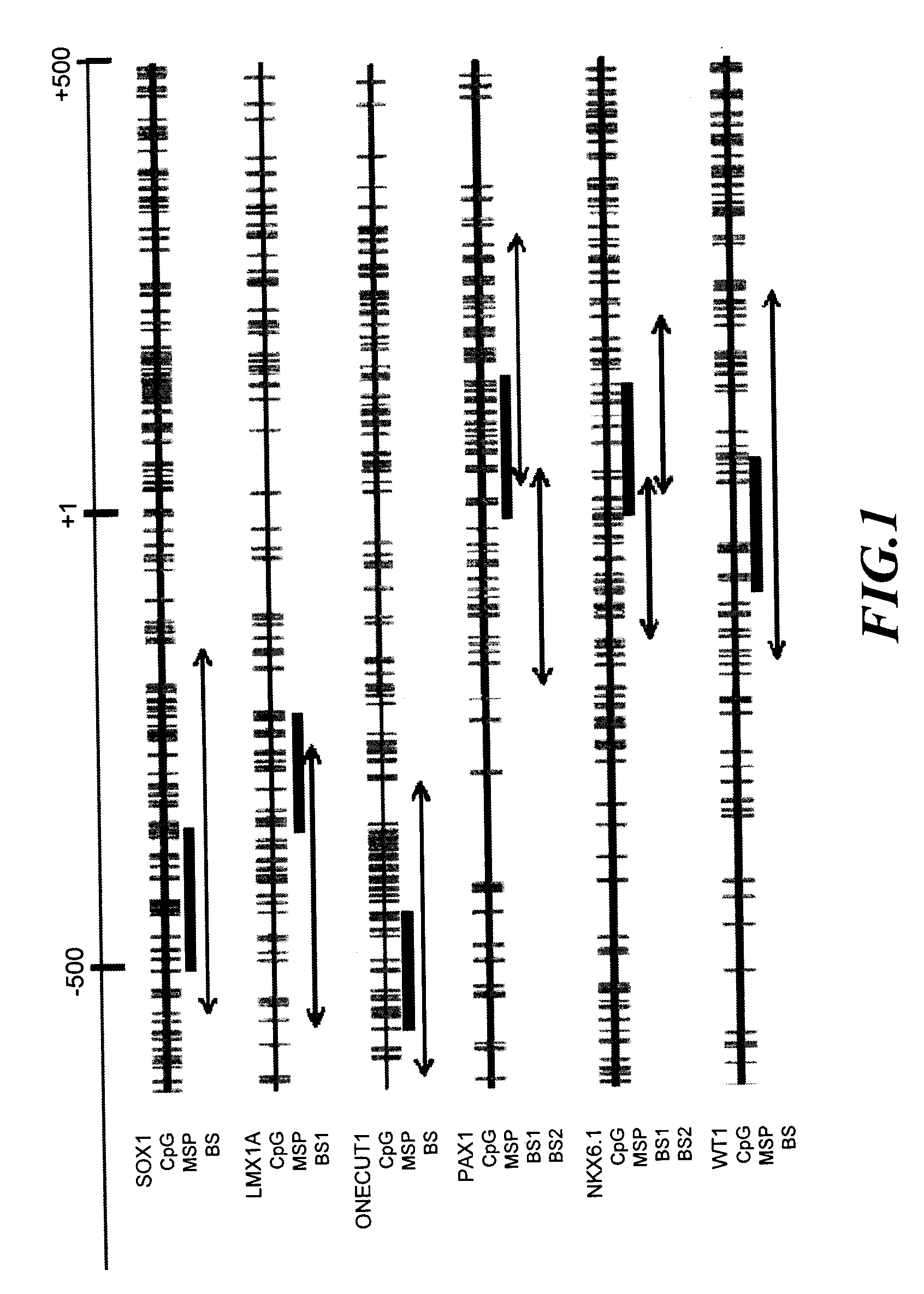
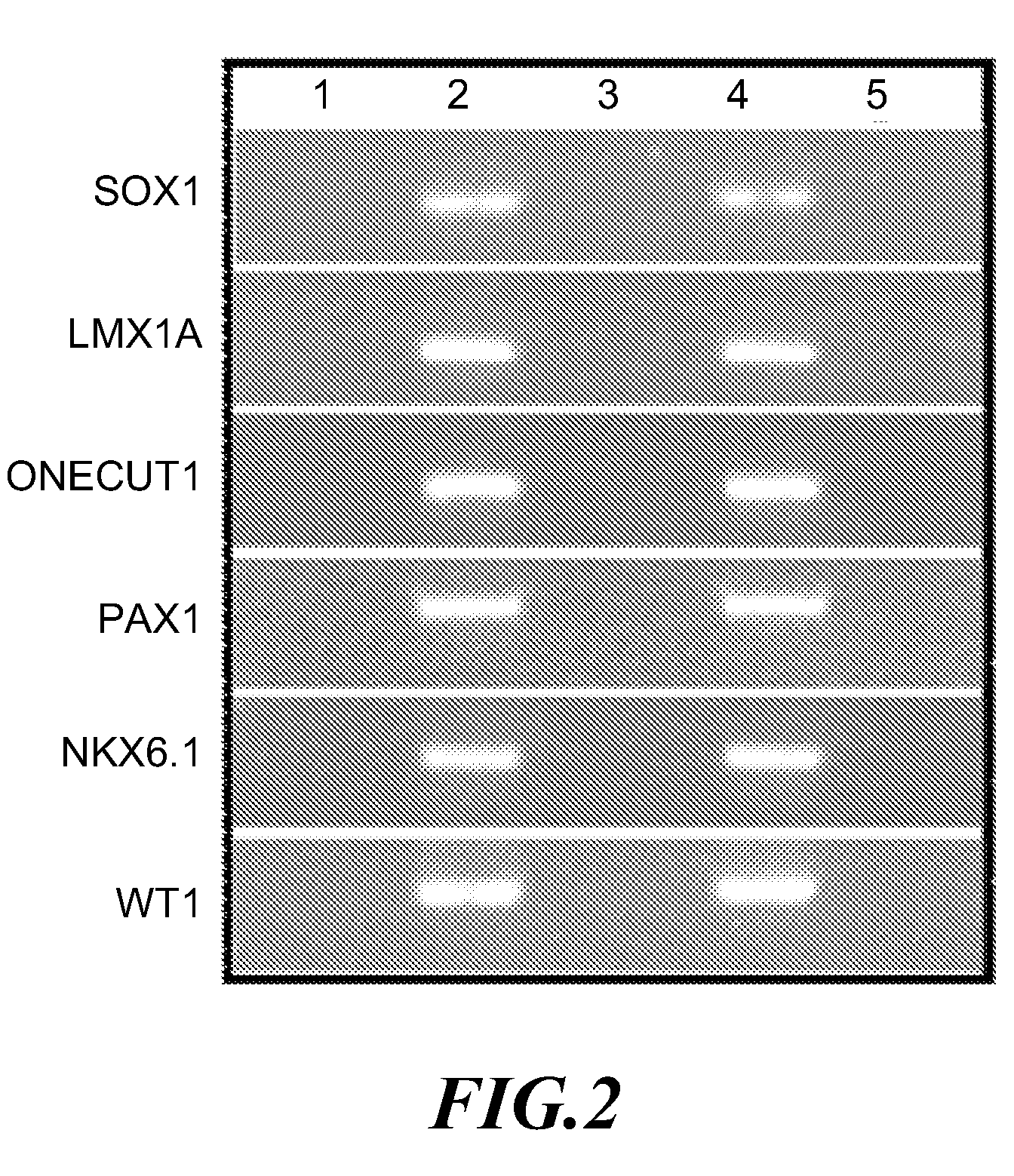
A method for screening cancer comprises the following steps: (1) providing a test specimen; (2) detecting the methylation state of the CpG sequence in at least one target gene within the genomic DNA of the test specimen, wherein the target genes is consisted of SOX1, PAX1, LMX1A, NKX6-1, WT1 and ONECUT1; and (3) determining whether there is cancer or cancerous pathological change in the specimen based on the presence or absence of the methylation state in the target gene; wherein method for detecting methylation state is methylation-specific PCR (MSP), quantitative methylation-specific PCR (QMSP), bisulfite sequencing (BS), microarrays, mass spectrometer, denaturing high-performance liquid chromatography (DHPLC), and pyrosequencing.
Owner:NAT DEFENSE MEDICAL CENT
Methods and compositions for sequencing nucleic acids using charge
ActiveUS20120052489A1Improve performanceMicrobiological testing/measurementNucleotideNucleotide sequencing
The invention provides methods and compositions, and systems for determining the identity of nucleic acids in nucleotide sequences, including sequences with one or more homopolymer regions. The methods of the invention include improvements so as to accurately identify sequences, including the difficult homopolymer sequences that are encountered during nucleotide sequencing, such as pyrosequencing.
Owner:ISOPLEXIS
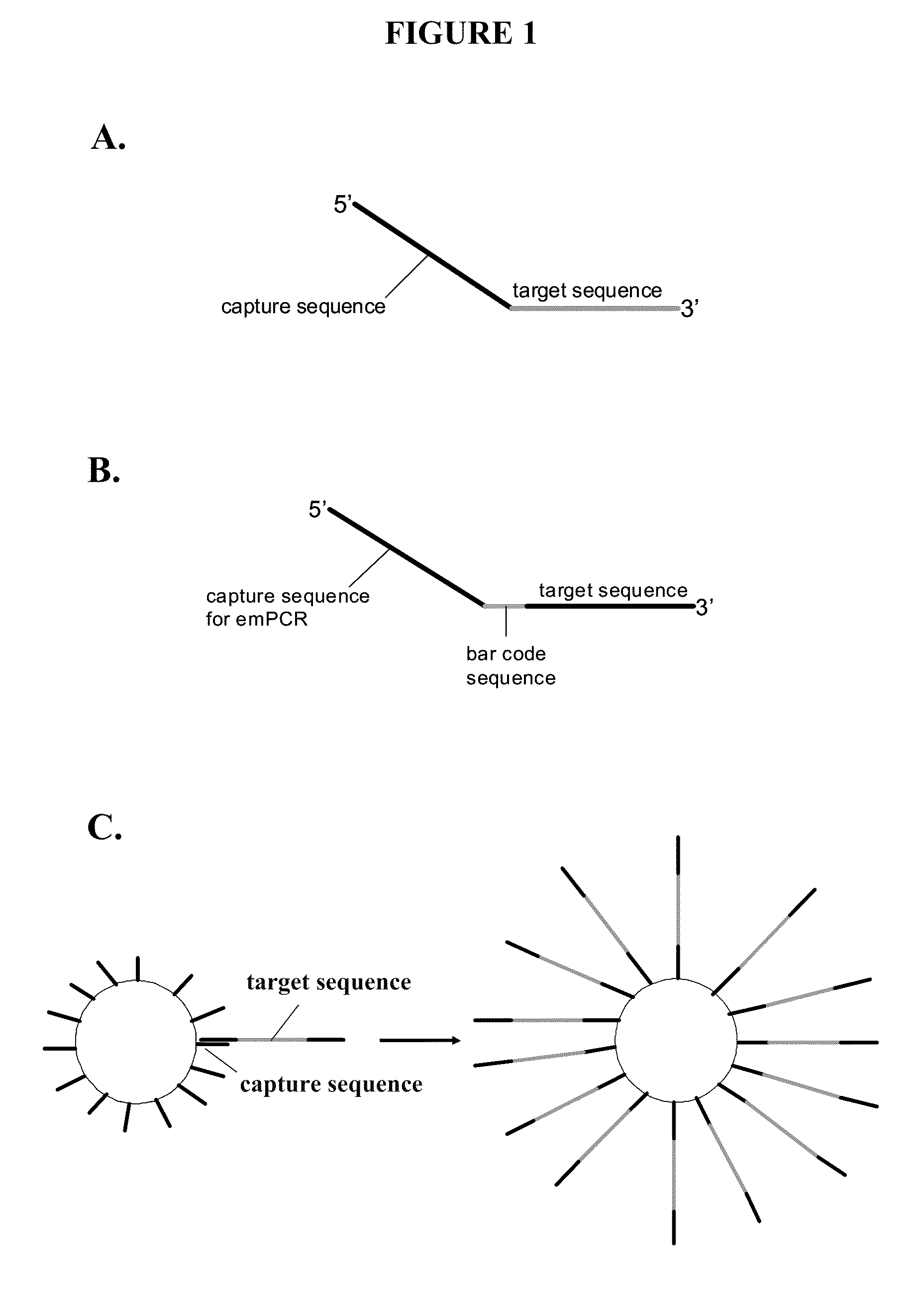
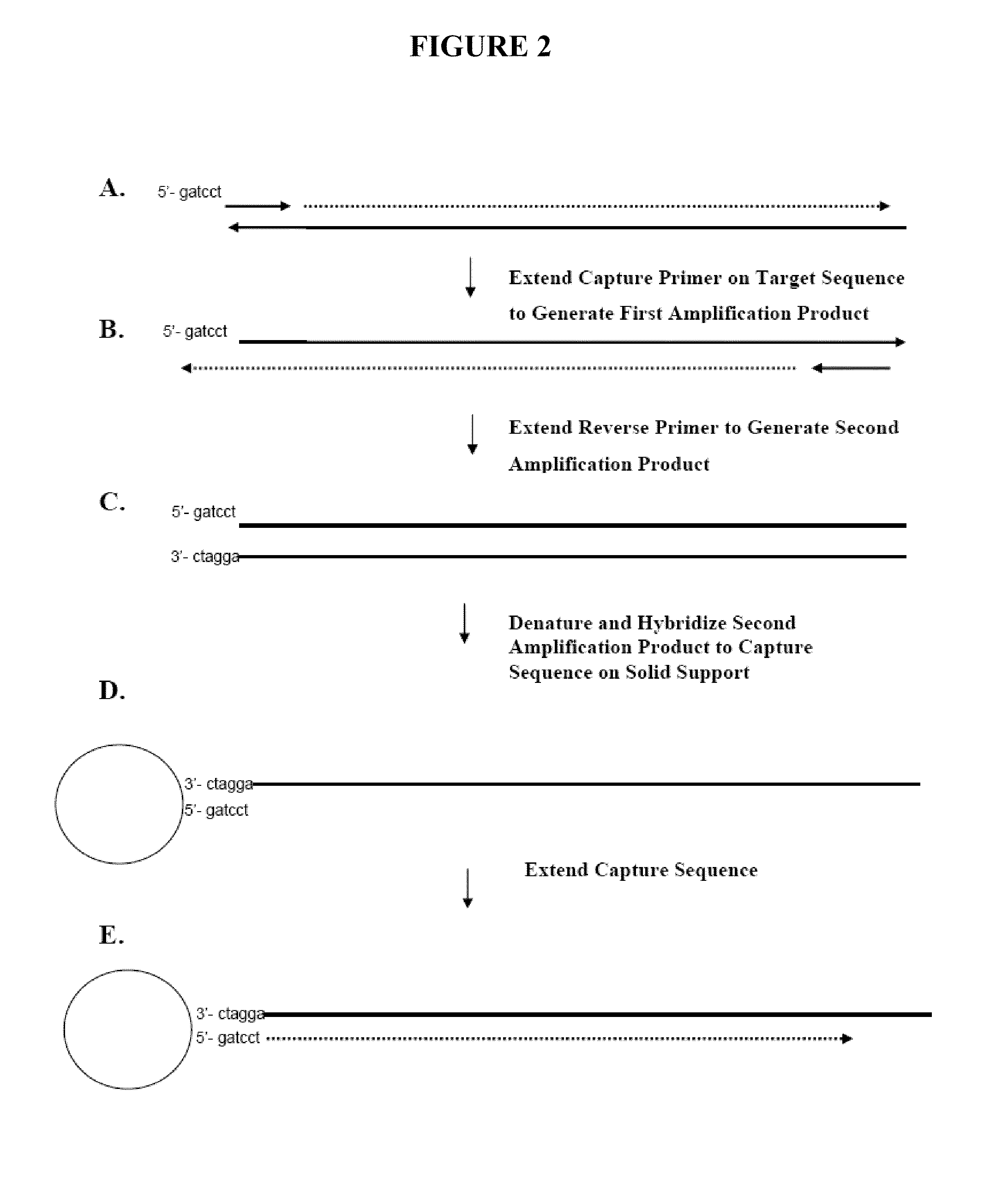
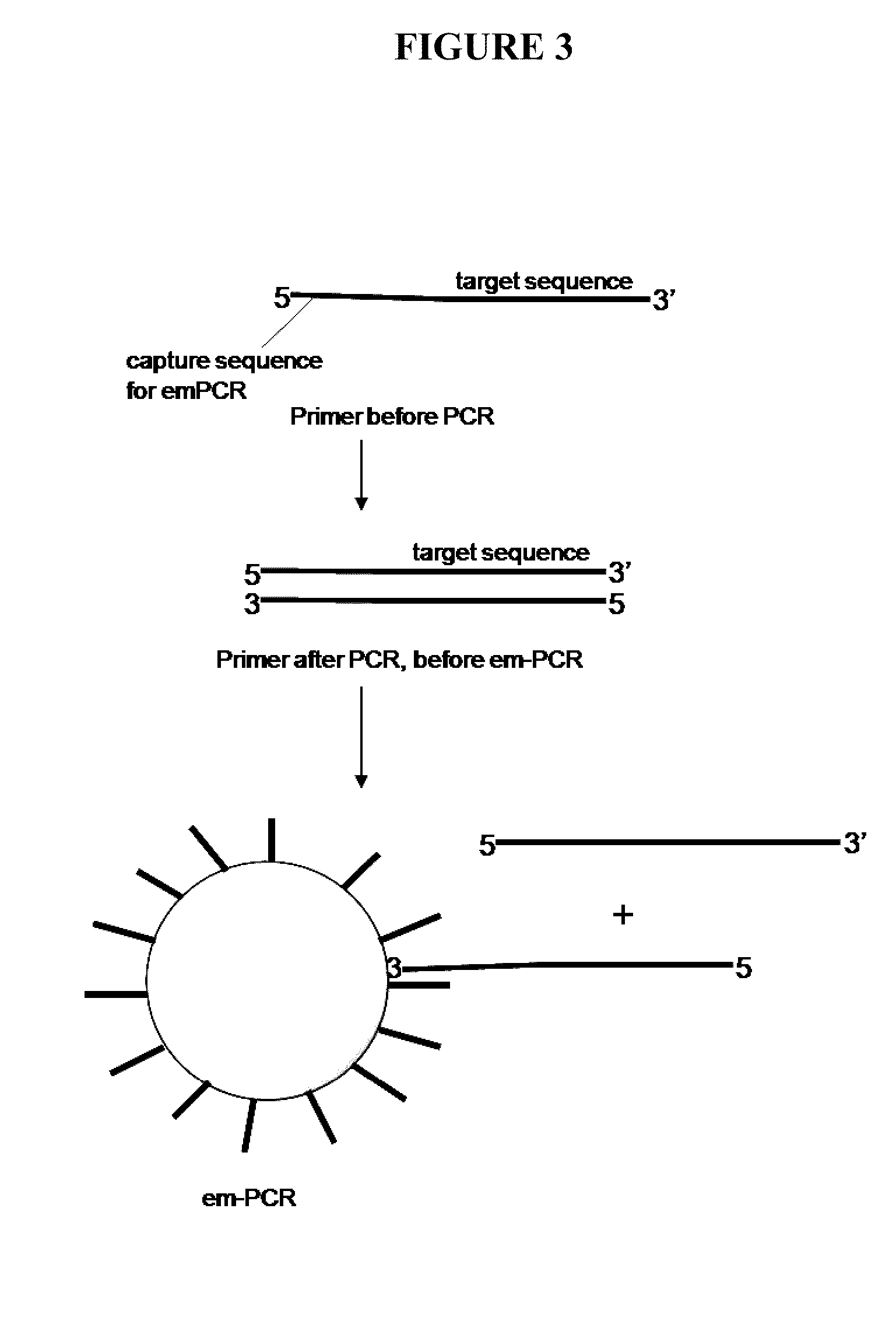
Capture primers and capture sequence linked solid supports for molecular diagnostic tests
InactiveUS9416409B2Microbiological testing/measurementSequence analysisMolecular diagnosticsMolecular Diagnostic Testing
The present invention provides systems, methods, and compositions for performing molecular tests. In particular, the present invention provides methods, compositions and systems for generating target sequence-linked solid supports (e.g., beads) using a solid support linked to a plurality of capture sequences and capture primers composed of a 3′ target-specific portion and a 5′ capture sequence portion. In certain embodiments, the target sequence linked solid support is used in sequencing methods (e.g., pyrosequencing, zero-mode waveguide type sequencing, nanopore sequencing, etc.) to determine the sequence of the target sequence (e.g., in order to detect the identity of a target nucleic acid in sample).
Owner:IBIS BIOSCI
Multiplex pyrosequencing using non-interfering noise cancelling polynucleotide identification tags
ActiveUS20140094376A1Sugar derivativesMicrobiological testing/measurementPyrosequencingPolynucleotide
The present disclosure generally pertains to a multiplex method for analyzing samples comprising using polynucleotide amplification to produce amplified products wherein one or more target sequences are tagged with a non-interfering, non-canceling target-specific polynucleotide identification tag, pyrosequencing the amplified products through the non-canceling target-specific polynucleotide identification tag sequence to detect the presence of one or more specific polynucleotide identification tags. The presence of a specific polynucleotide identification tag being correlated with the presence of a specific target sequence.
Owner:IREPERTOIRE INC
Methods for Determining Sequence Variants Using Ultra-Deep Sequencing
InactiveUS20120264632A1Reduce and effort and lossReduce and and product lossMicrobiological testing/measurementLibrary screeningUltra deep sequencingDirect sequencing
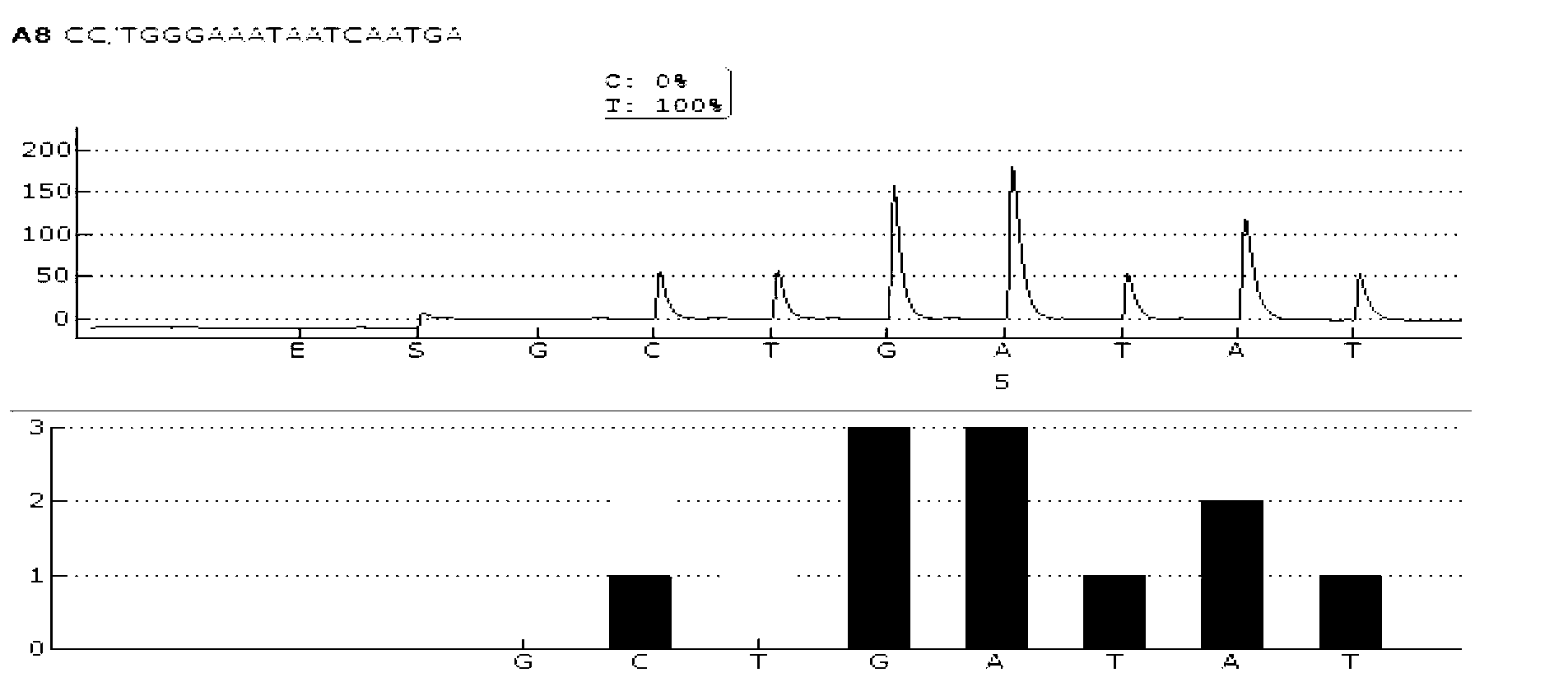
The claimed invention provides for new sample preparation methods enabling direct sequencing of PCR products using pyrophosphate sequencing techniques. The PCR products may be specific regions of a genome. The techniques provided in this disclosure allows for SNP (single nucleotide polymorphism) detection, classification, and assessment of individual allelic polymorphisms in one individual or a population of individuals. The results may be used for diagnostic and treatment of patients as well as assessment of viral and bacterial population identification.
Owner:454 LIFE SCIENCES CORP
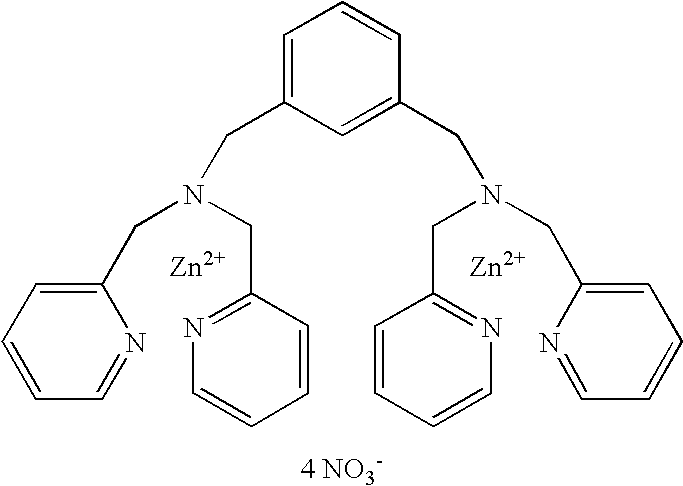
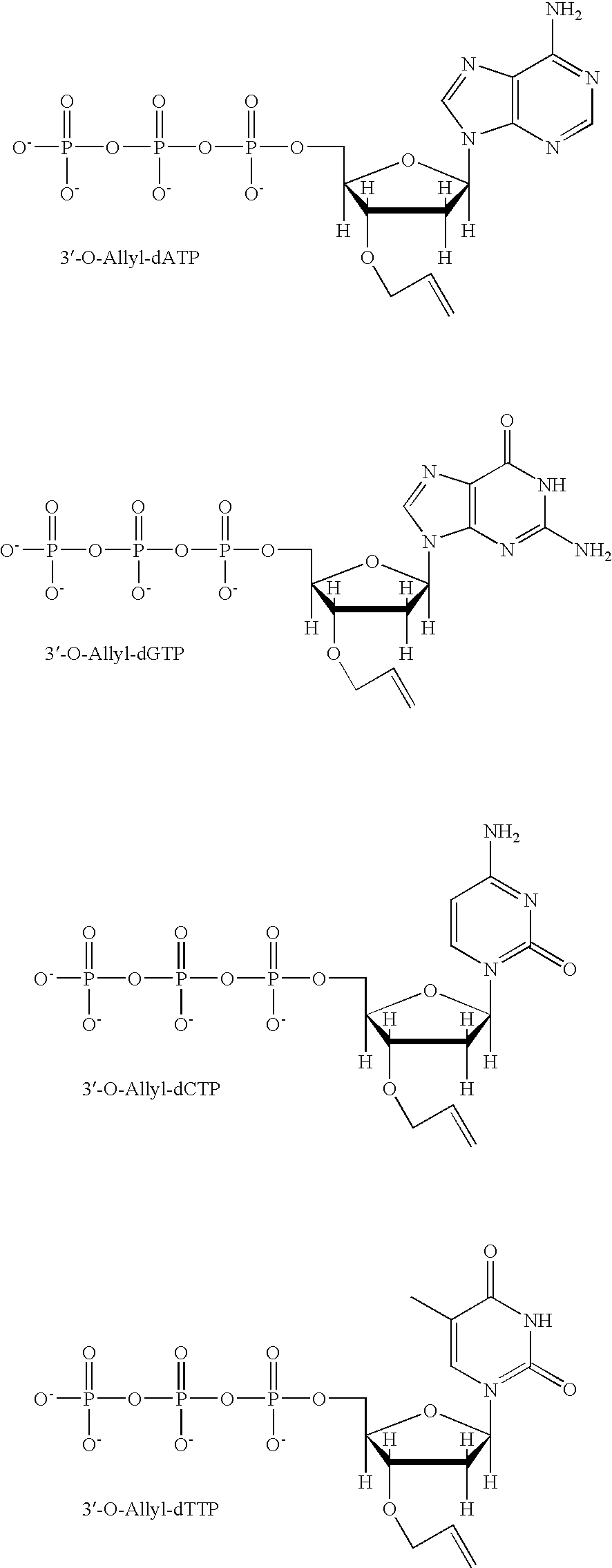
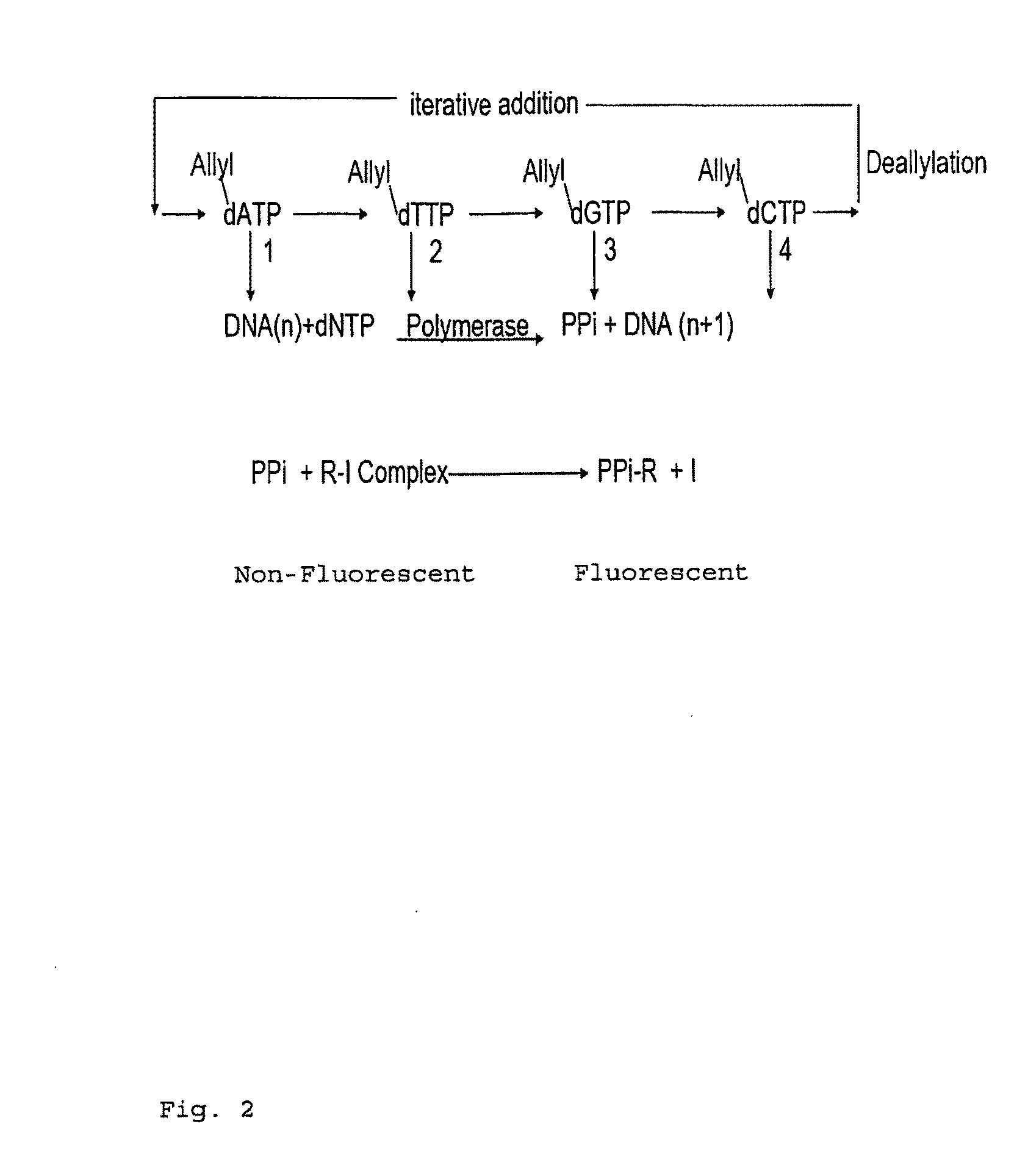
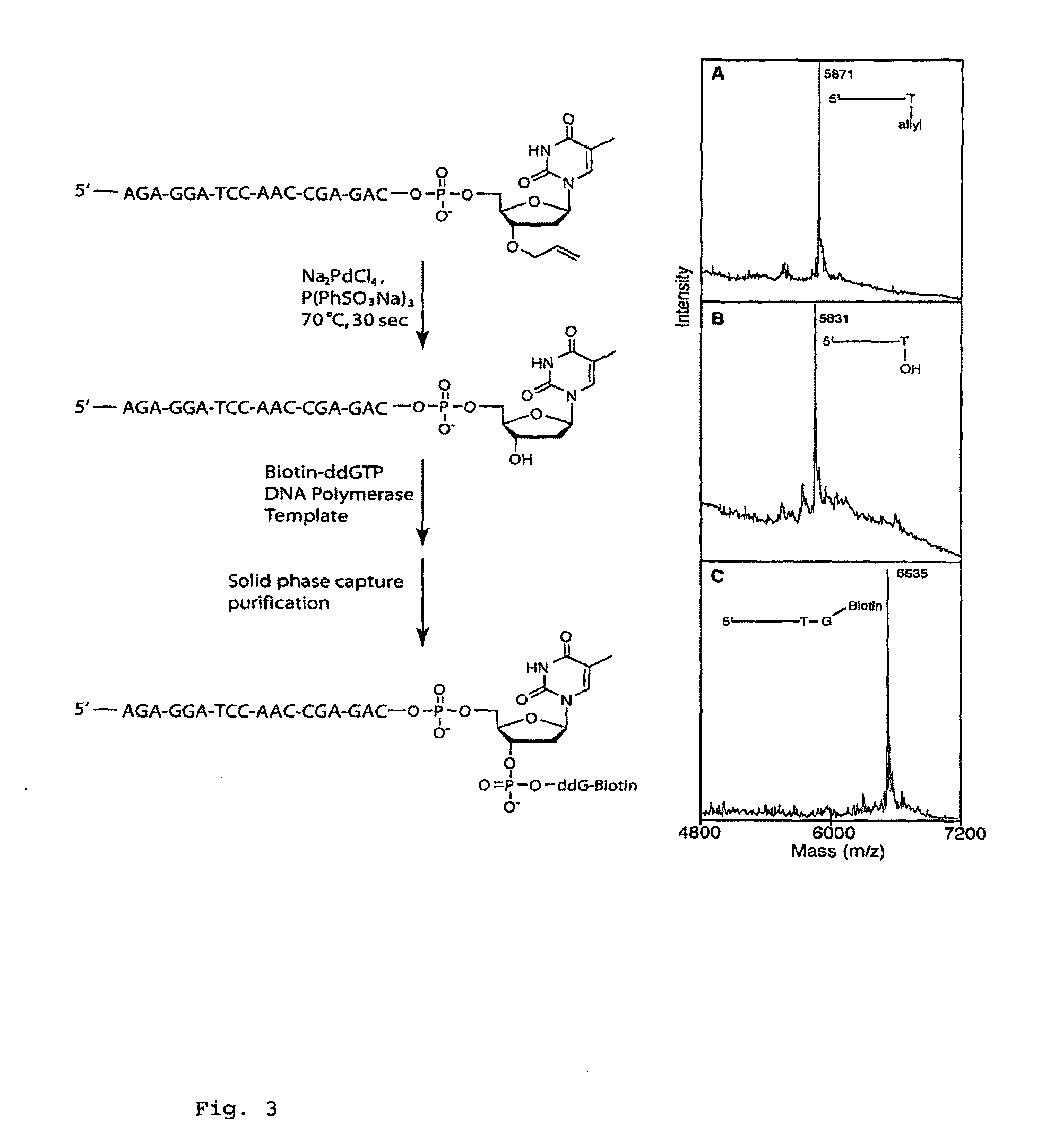
Pyrosequencing Methods and Related Compositions
This invention provides methods for pyrosequencing and compositions comprising 3′-O— modified deoxynucleoside triphosphates.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
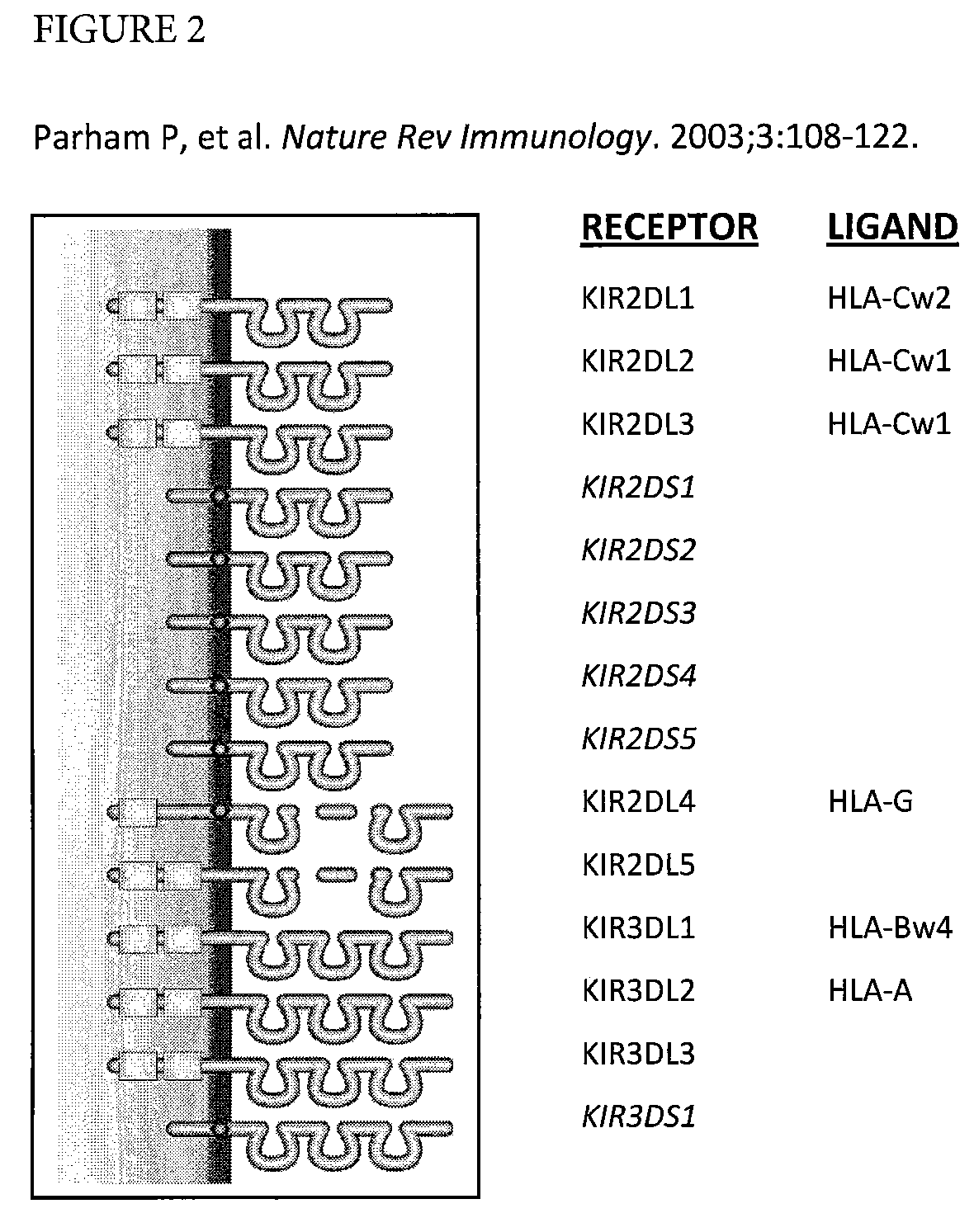
Determination of KIR haplotypes associated with disease
Disclosed is a method of determining KIR genotypes for one or more individuals in parallel, the method comprising: for each individual, amplifying the polymorphic exon sequences of the KIR genes, pooling the KIR amplicons, performing emulsion PCR followed by pyrosequencing in parallel to determine all the amplicon sequences present in the individual to determine which KIR alleles are present in the individual.
Owner:CHILDREN S HOSPITAL &RES CENT AT OAKLAN +2
Method for isolation of independent, parallel chemical micro-reactions using a porous filter
InactiveUS20060019264A1Prevent evaporationFast deliveryBioreactor/fermenter combinationsSequential/parallel process reactionsChemical reactionDna amplification
The present invention relates to methods and apparati for conducting densely packed, independent chemical reactions in parallel in a substantially two-dimensional array. Accordingly, this invention also focuses on the use of this array for applications such as DNA sequencing, most preferably pyrosequencing, and DNA amplification.
Owner:ATTIYA SAID +6
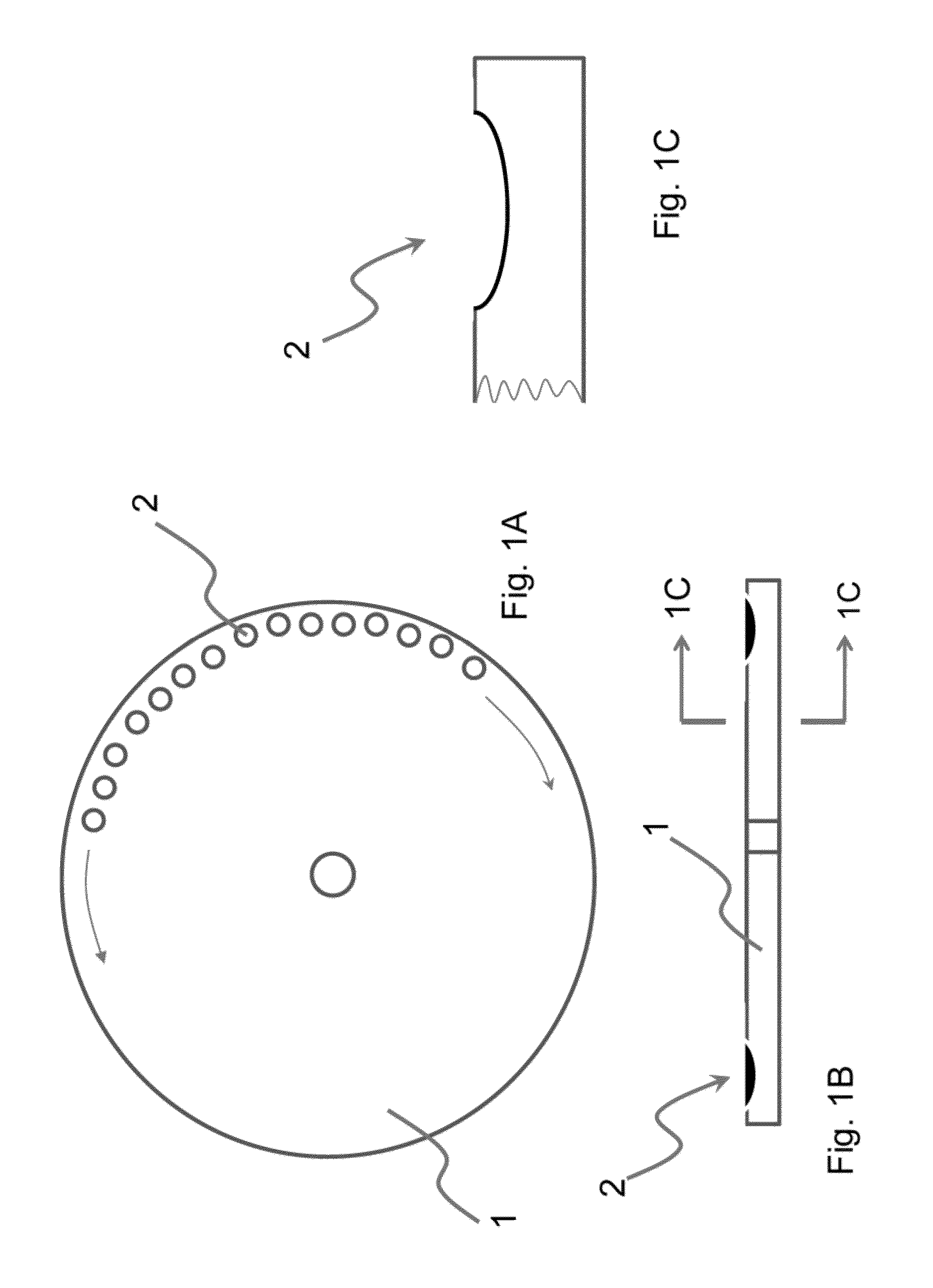
Method and apparatus for conducting an assay
ActiveUS8597882B2Simple stepsReduce wasteBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid sequencingBiology
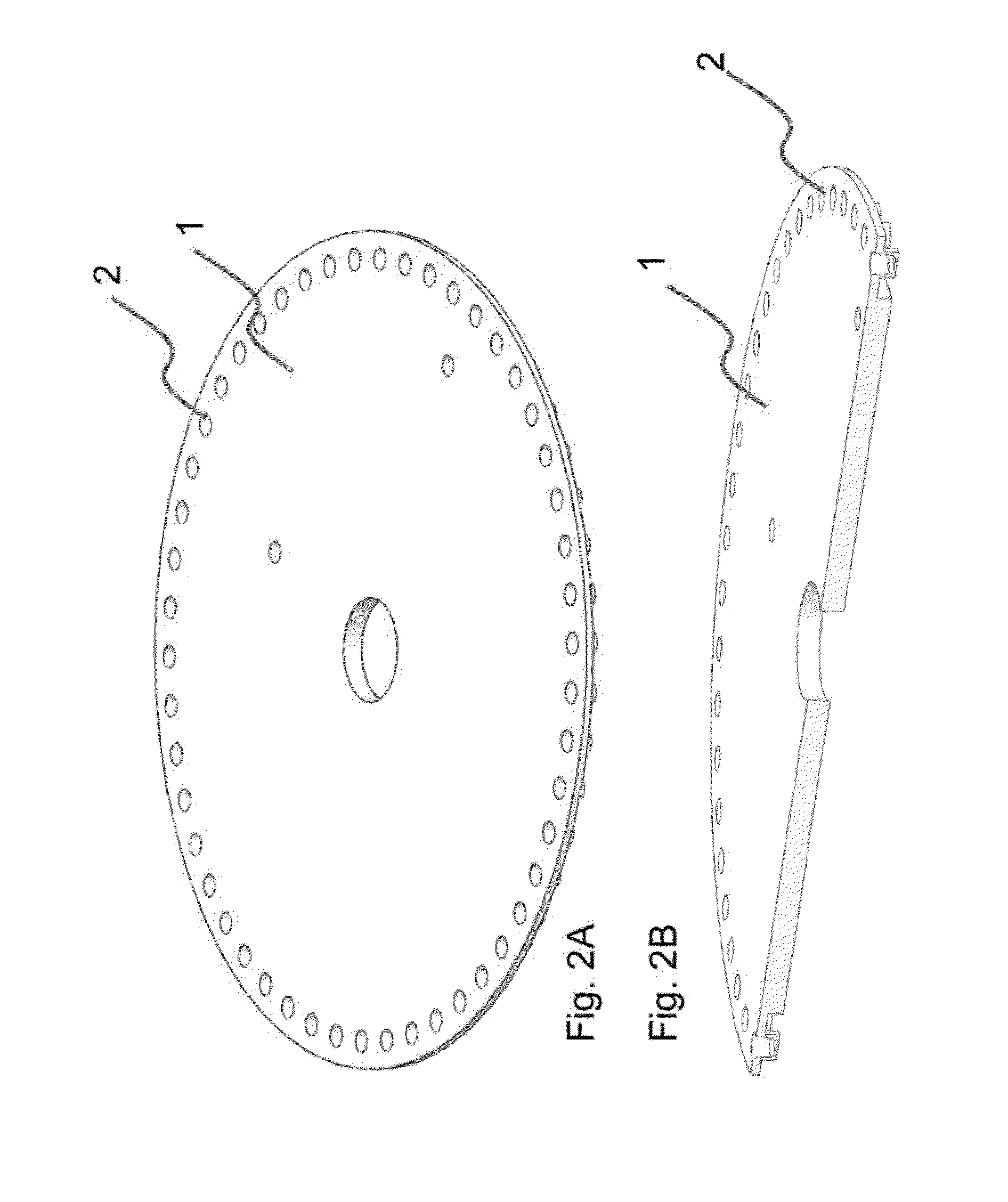
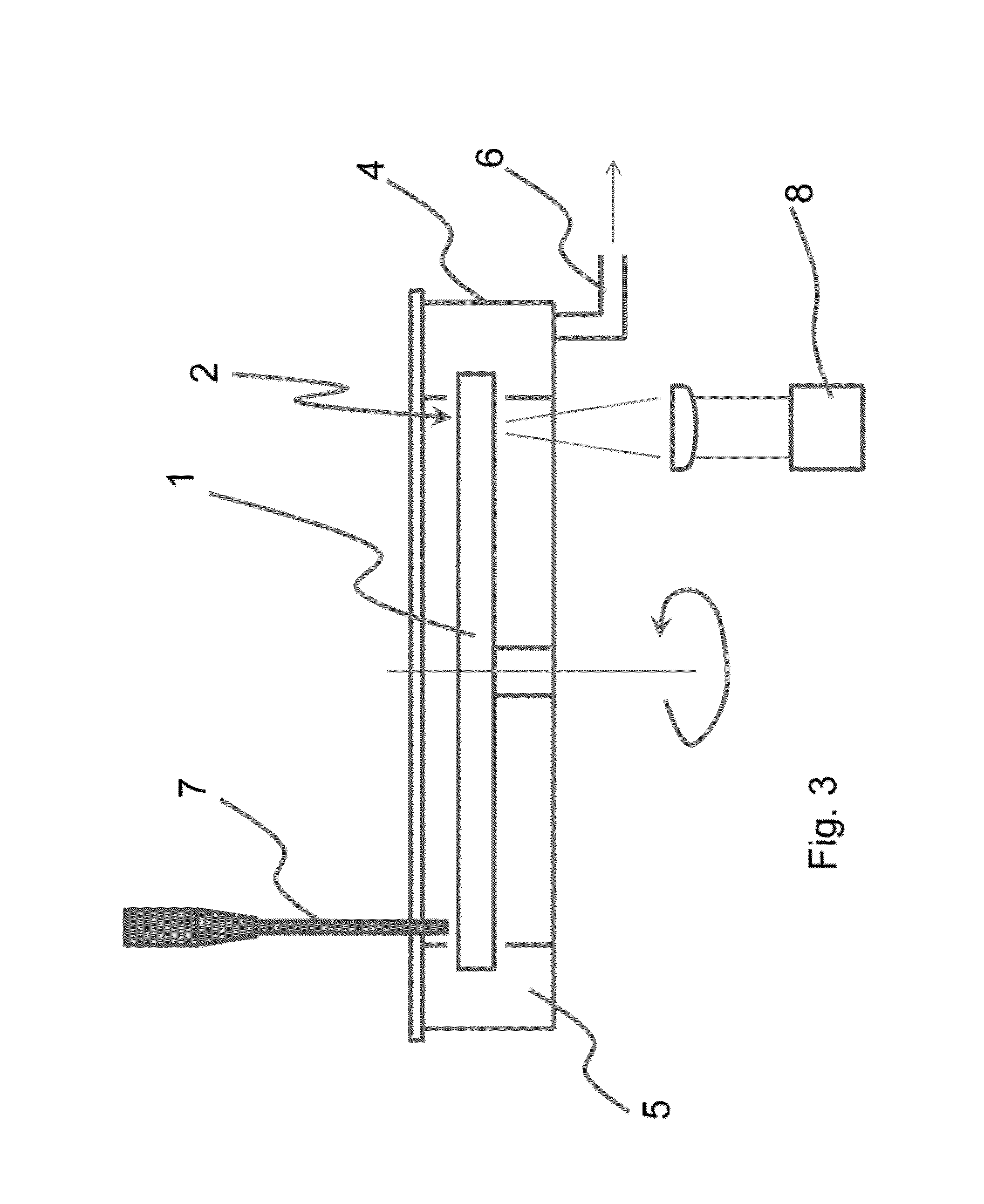
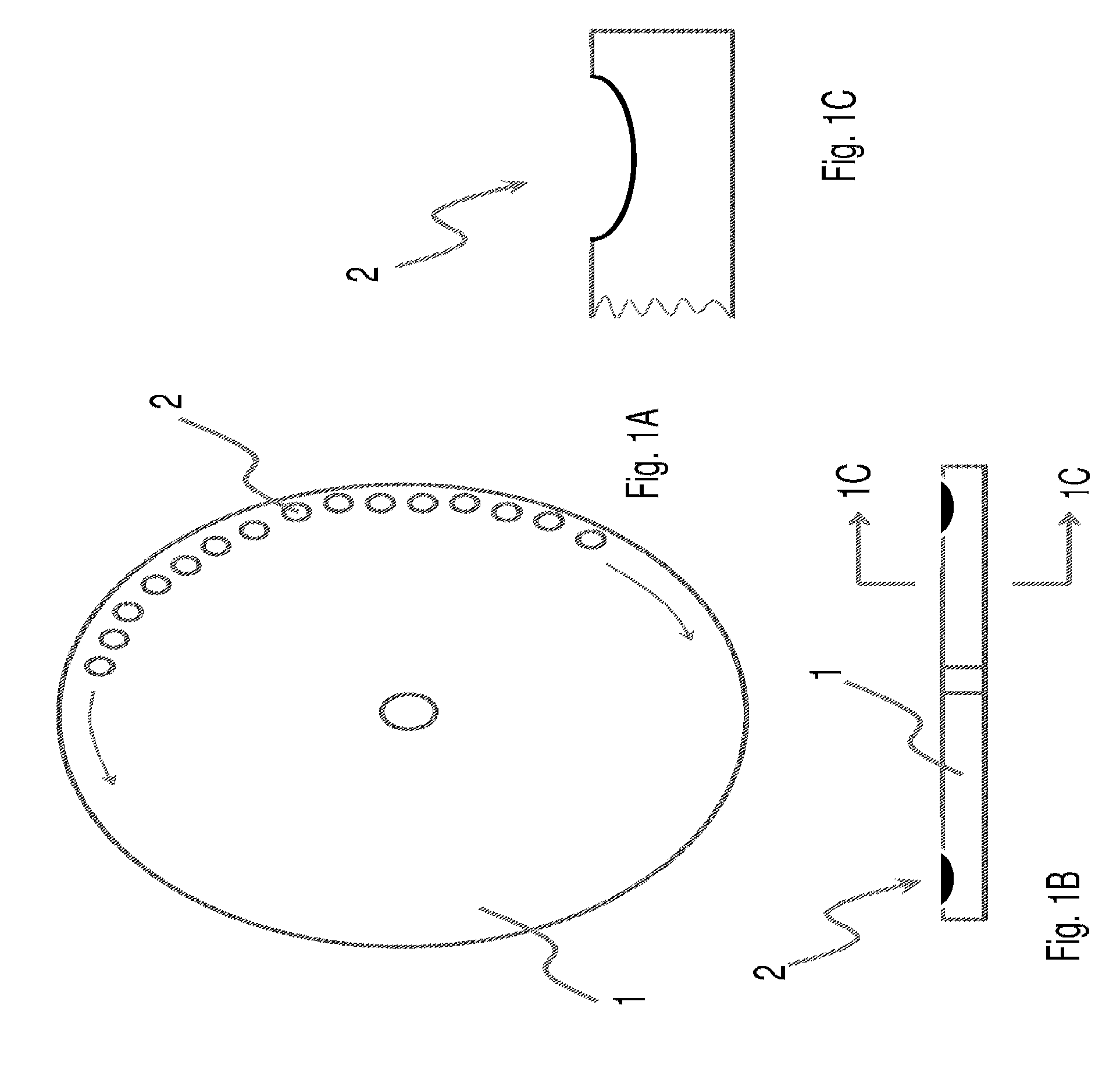
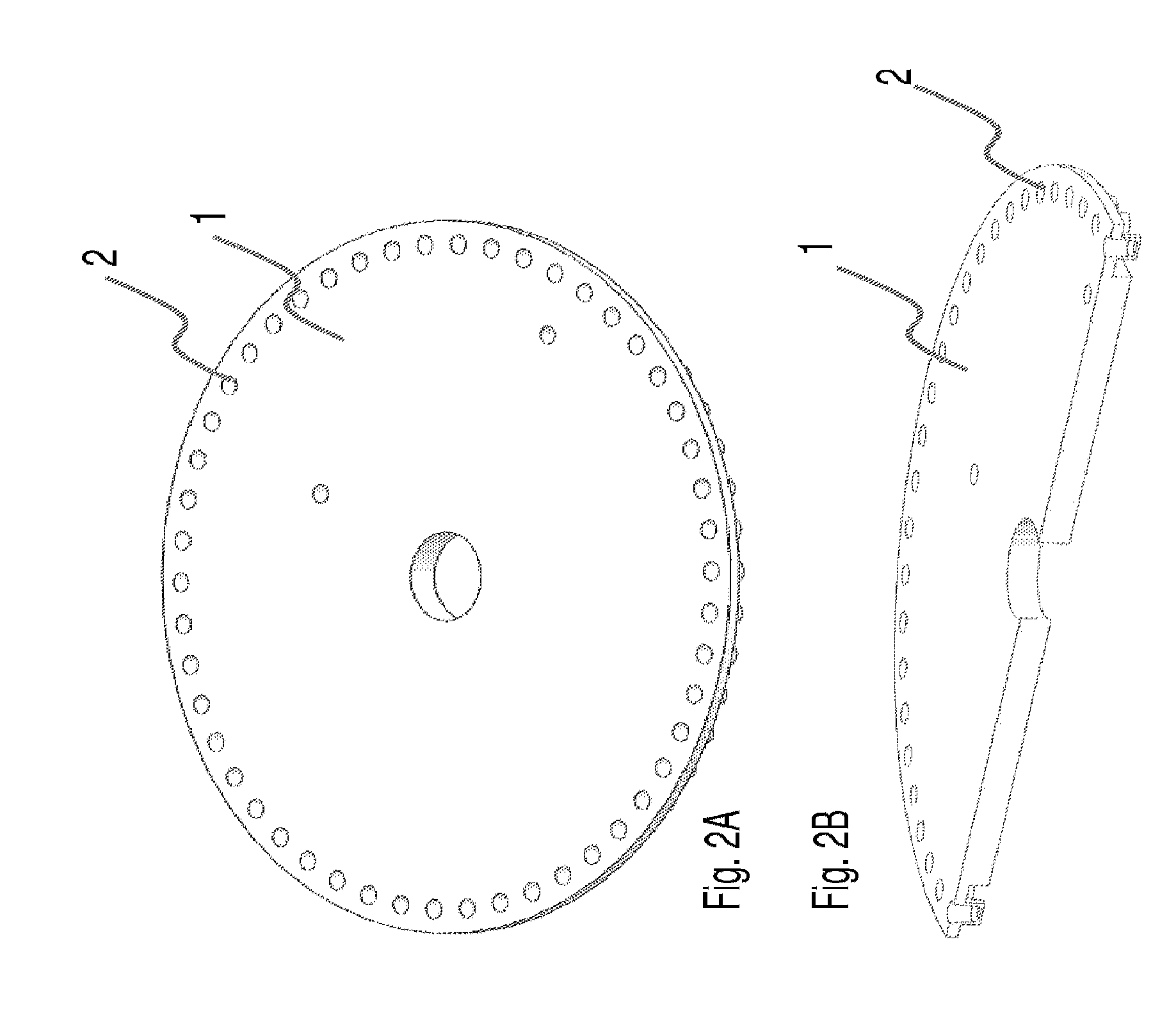
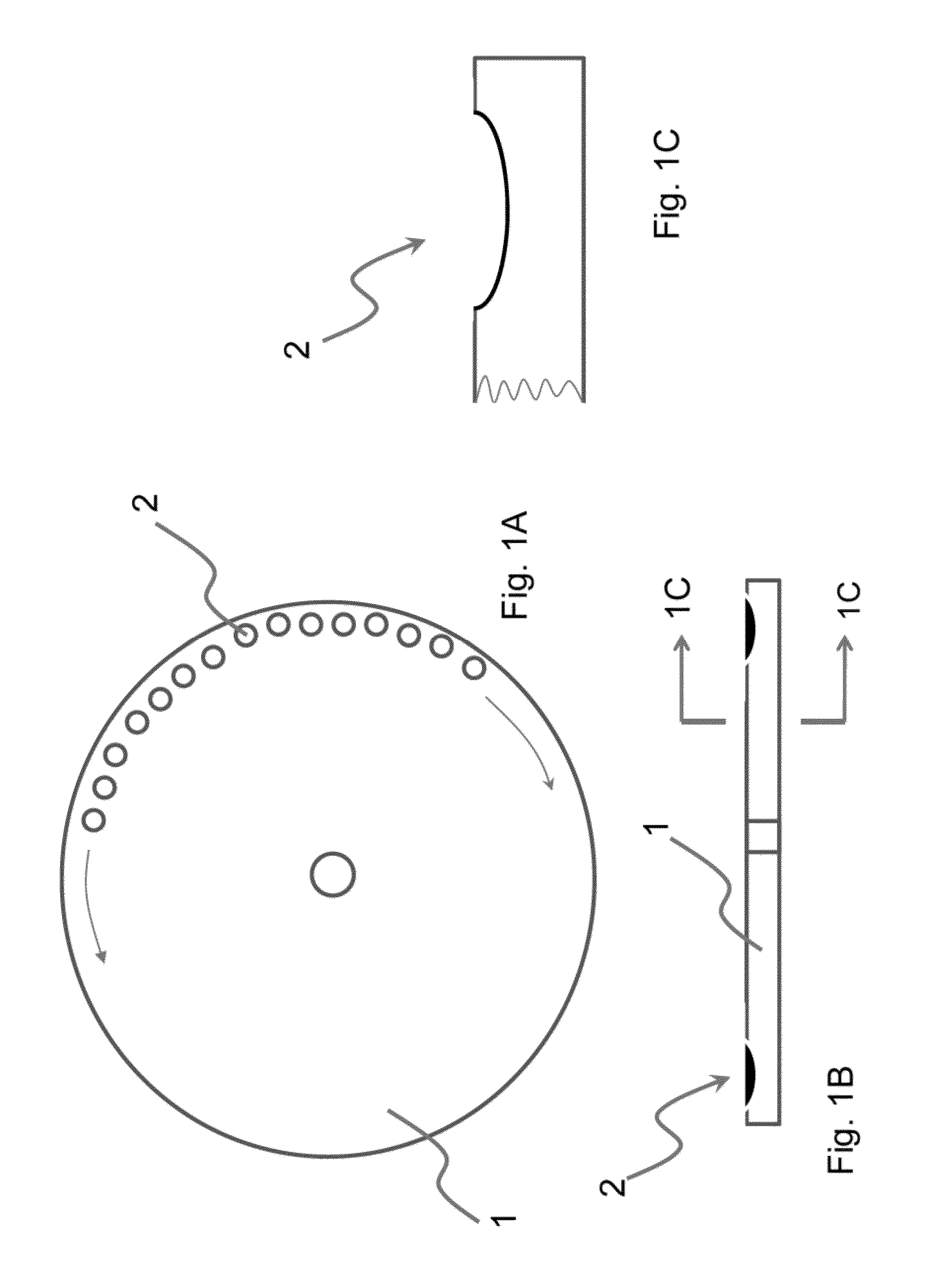
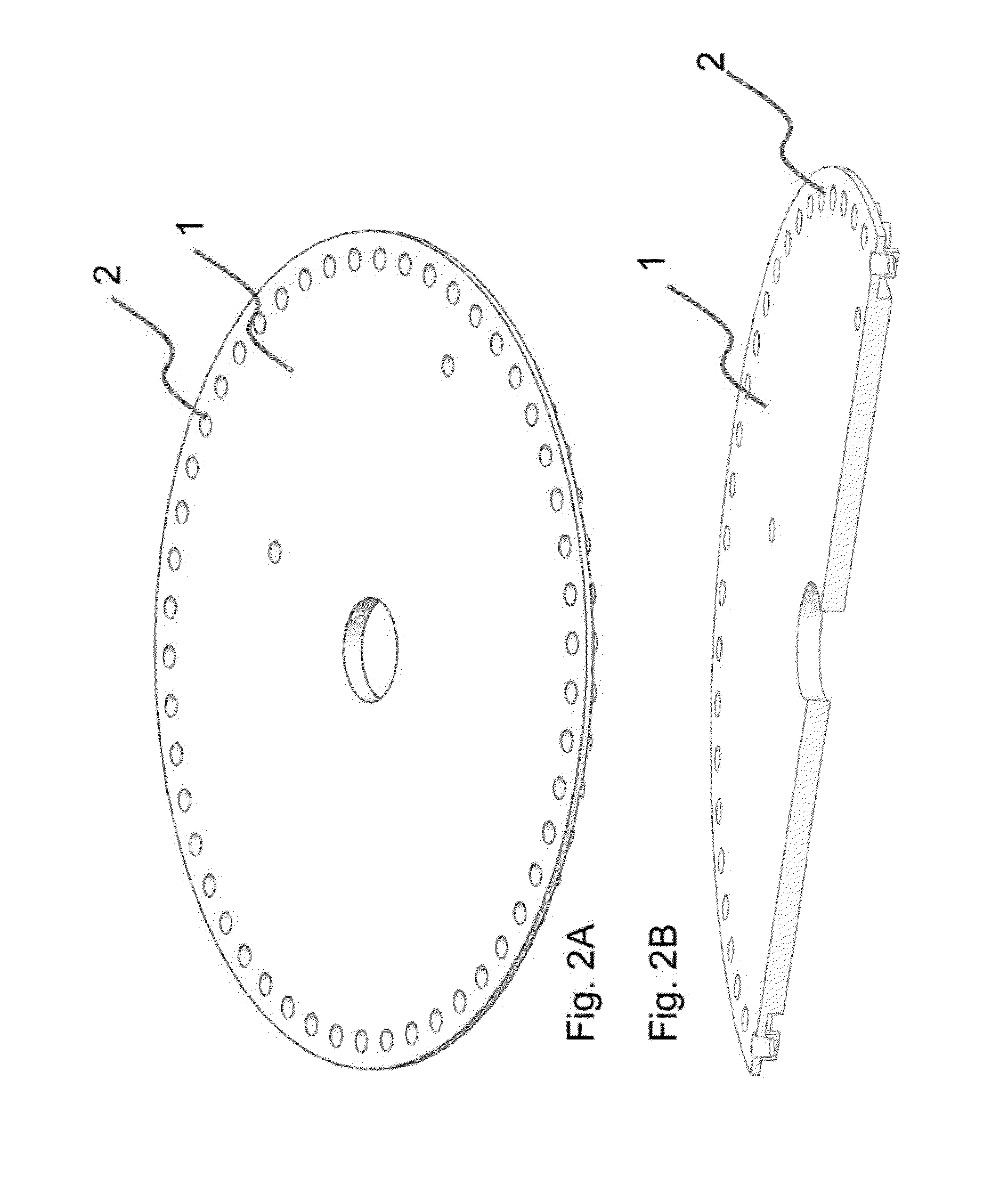
The present invention relates to methods and apparatus for conducting nucleic acid sequencing by pyrosequencing. The method comprises the steps of providing a platform having at least one well for containing at least one support surface, and providing at least one support surface within each well, wherein the support surface is adapted to immobilize a first binding partner or selectively immobilize a second binding partner. The method further comprises the steps of binding or immobilizing the first or second binding partner to the support surface and dispensing into each well from a point external of said platform a reagent, wherein after the dispensing step the platform is rotated sufficiently such that any residual or unreacted said reagent is substantially centrifugally removed from each well and / or each support surface, wherein during rotation each support surface is held within each well.
Owner:PYROBETT PTE
Method and Apparatus for Conducting an Assay
InactiveUS20120282708A1Reduce the amount requiredSimple stepsLaboratory glasswaresBiological testingEngineeringPyrosequencing
The present invention relates to methods and apparatus for conducting an assay. In particular, the present invention relates to a rotatable platform which can be used for conducting an assay, in particular multi-step assays. The present invention provides a rotatable platform adapted to immobilise a first binding partner in one or more discrete areas on a surface of said platform, or to selectively immobilise a second binding partner in one or more discrete areas on a surface of said platform. The invention also relates to methods, apparatus, a kit and the use of the rotatable platform for conducting an assay. In particular, the invention has been developed primarily for use in sequencing nucleic acid by pyrosequencing, however the invention is not limited to this field.
Owner:PYROBETT PTE
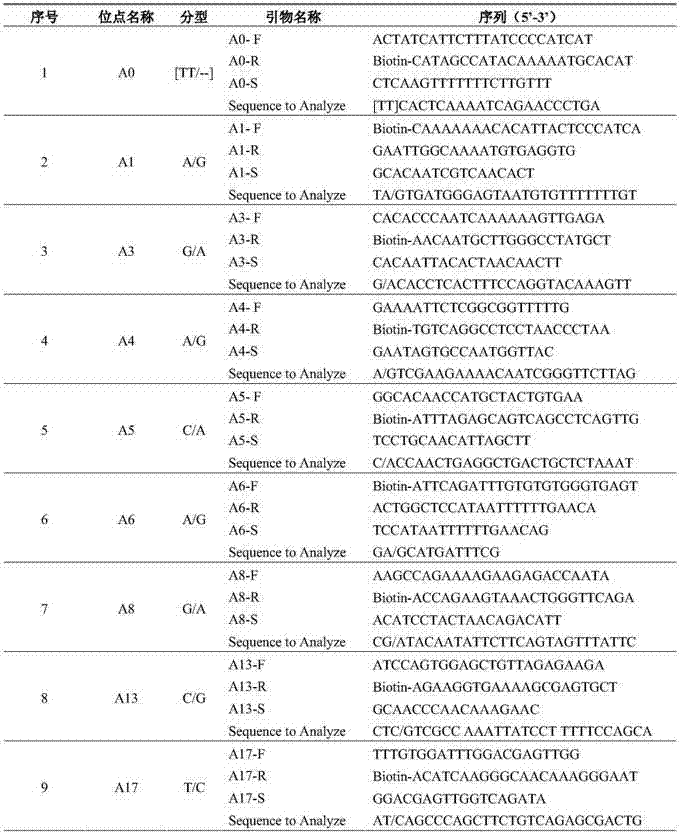
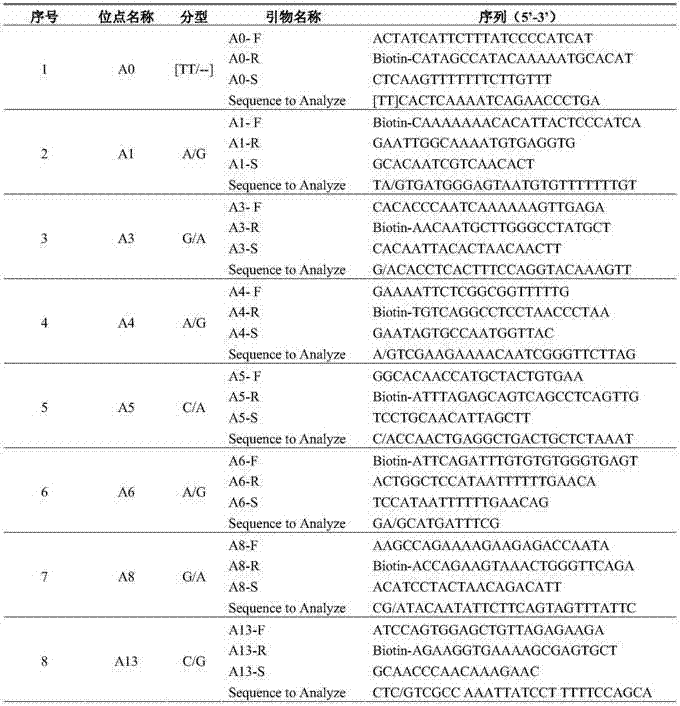
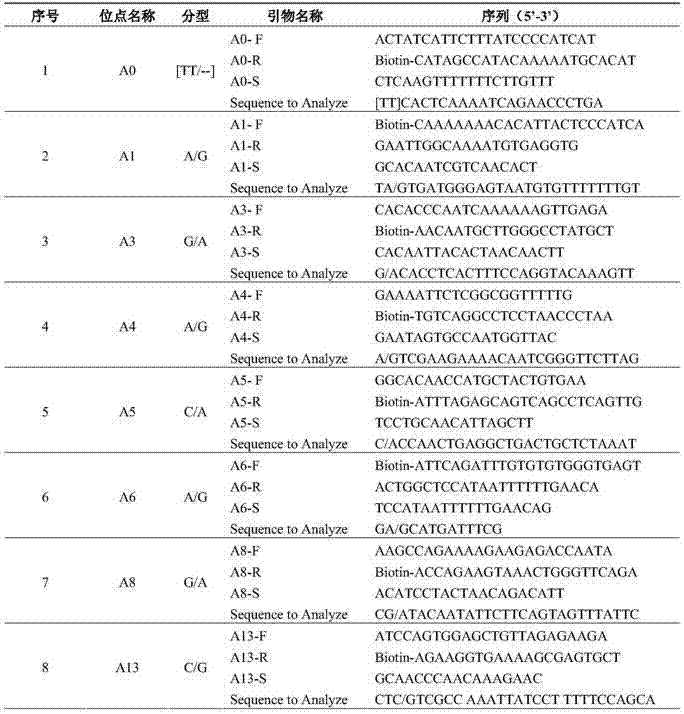
Multiplex PCR combined pyrosequencing kit used for guiding individualized medication of warfarin and detection method thereof
The invention provides a multiplex PCR combined pyrosequencing kit used for guiding individualized medication of warfarin and a detection method thereof, relating to warfarin. The kit comprises a whole blood genome DNA extraction reagent, multiplex PCR amplification primers, a multiplex PCR amplification reagent, a single-stranded DNA separating and purifying reagent, pyrosequencing primers, a pyrosequencing reagent and a kit body. The detection method comprises the following steps: extraction of human whole blood genome DNA; multiplex PCR amplification; separation and purification of a single-stranded DNA sample; and pyrosequencing and result analysis. According to the invention, main factors influencing difference of individual dosage of warfarin, i.e., important gene polymorphic sites of VKORC1 and CYP2C9, specifically being VKORC1-1639G>A, CYP2C9*2 and CYP2C9*3, are detected so as to guide individualized dosage of warfarin in treatment of patients needing to take warfarin, thereby achieving the purpose of reduction in side effects.
Owner:ZHONGSHAN HOSPITAL XIAMEN UNIV +1
Method and apparatus for conducting an assay
ActiveUS20130203049A1Reduce the amount requiredIncrease read lengthBioreactor/fermenter combinationsHeating or cooling apparatusNucleic acid sequencingBiology
The present invention relates to methods and apparatus for conducting nucleic acid sequencing. The method comprises the steps of providing a platform having at least one well for containing at least one support surface, and providing at least one support surface within each well, wherein the support surface is adapted to immobilise a first binding partner or selectively immobilise a second binding partner. The method further comprises the steps of binding or immobilising the first or second binding partner to the support surface and dispensing into each well from a point external of said platform a reagent, wherein after the dispensing step the platform is rotated sufficiently such that any residual or unreacted said reagent is substantially centrifugally removed from each well and / or each support surface, wherein during rotation each support surface is held within each well. The invention also relates to kits and uses of the kits for conducting nucleic acid sequencing. In particular, the invention has been developed primarily for use in sequencing nucleic acid by pyrosequencing, however the invention is not limited to this field.
Owner:PYROBETT PTE
Sequencing primer for qualitative detection of cytochrome oxidase CYP2C19 genetic typing and kit of sequencing primer
InactiveCN102912013AQualitatively accurateIncreased sensitivityMicrobiological testing/measurementDNA/RNA fragmentationPositive controlTaq polymerase
The invention provides a sequencing primer for qualitative detection of cytochrome oxidase CYP2C19 genetic typing and a kit of the sequencing primer and belongs to the field of external nucleic acid detection. The kit comprises uracil DNA (Deoxyribonucleic Acid) glycosylase, Taq polymerase, PCR (Polymerase Chain Reaction) reaction liquor, PCR amplification primers, pyrophosphoric acid sequencing primers and positive control products. The kit is high in sensitivity and good in specificity, PCR products can be simply treated for a pyrophosphoric acid sequenator for sequencing, operation is simple, reaction time is short, the sensitivity of the kit is higher than that of a golden standard-capillary electrophoresis sequencing, and the kit is suitable for use for mutation analysis.
Owner:CHANGSHA 3G BIOTECH
Primers, method and kit for detecting secreted frizzled-related protein (SFRP)2 gene CpG island methylation
ActiveCN103882119AStrong specificityImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationSFRP2 geneDNA fragmentation
The invention discloses primers, method and kit for detecting secreted frizzled-related protein (SFRP)2 gene CpG island methylation. The primers include specific primers for amplifying deoxyribonucleic acid (DNA) segments of three CG loci (related to intestinal cancer) on an SFRP2 gene CpG island from a sample nucleic acid, and primers for pyrosequencing the obtained nucleic acid segments. By the primers, the method and the kit disclosed by the invention, the frequency of the SFRP2 gene methylation related to colorectal cancer susceptibility can be detected by adopting a pyrosequencing technique, so as to screen out susceptible people with early colorectal cancer. The primers, the method and the kit are good in specificity, and high in accuracy, and can detect samples at high flux.
Owner:郑州艾迪康医学检验所(普通合伙)
Pyrosequencing methods and related compositions
ActiveUS9169510B2Bioreactor/fermenter combinationsAnalysis using chemical indicatorsPyrophosphoric acidBiology
This invention provides methods for pyrosequencing and compositions comprising 3′-O— modified deoxynucleoside triphosphates.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
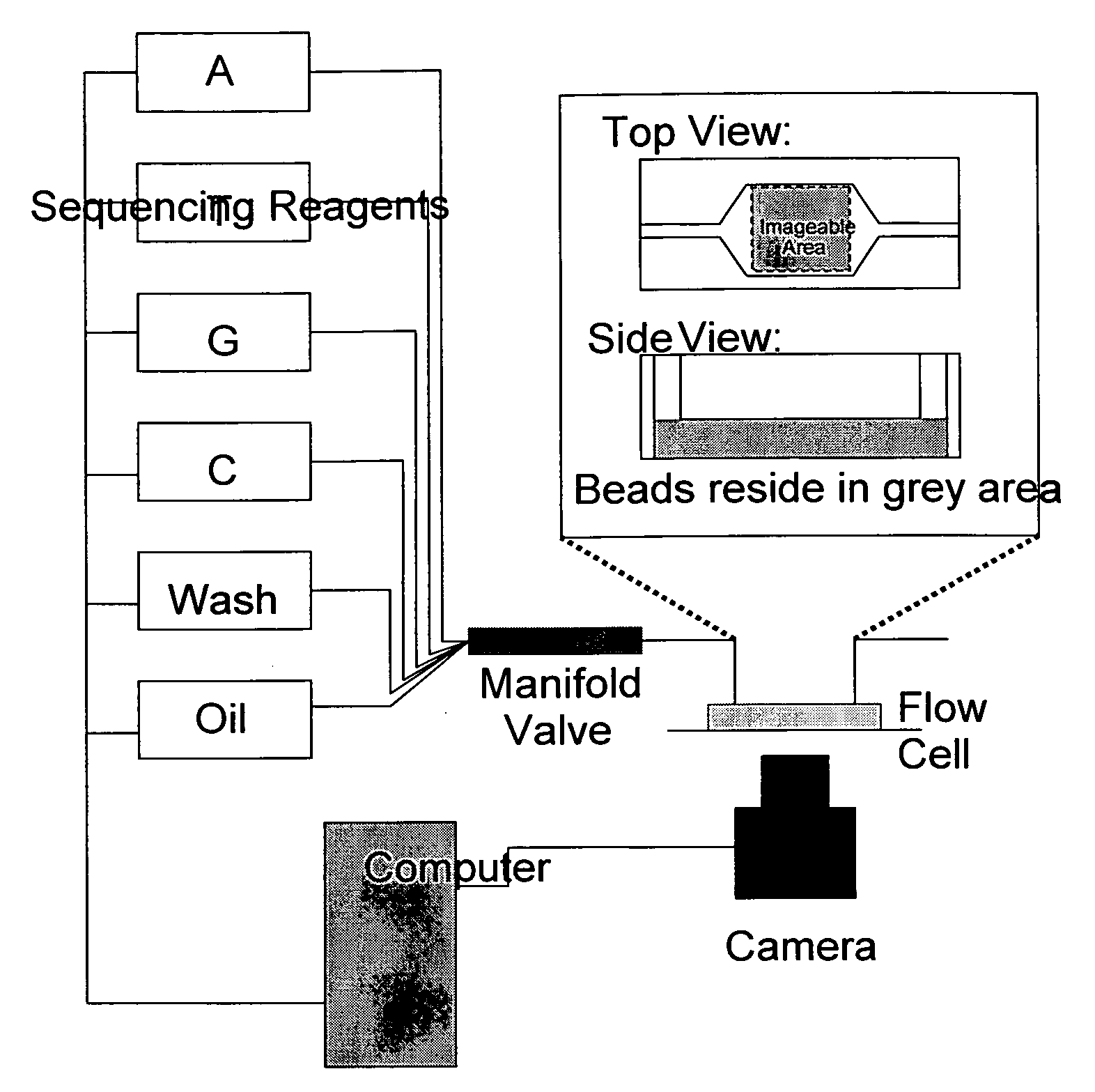
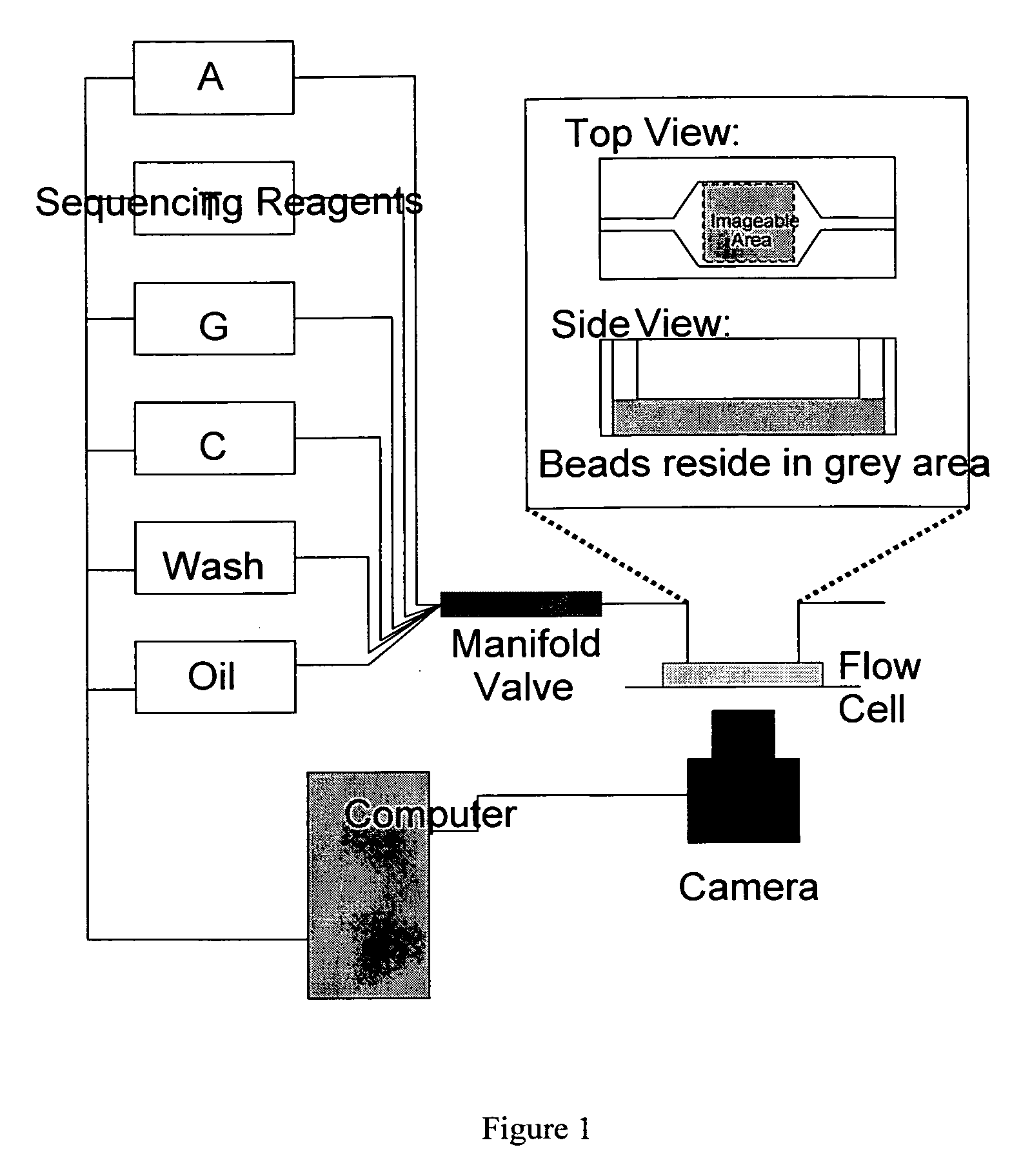
High throughput DNA sequencing method and apparatus
InactiveUS20100021915A1Quick sortingBioreactor/fermenter combinationsBiological substance pretreatmentsPyrophosphateNucleic acid sequencing
The present invention relates to a method for high throughput nucleic acid sequencing using a multi-bead flow cell and pyrophosphate sequencing, a sequencer capable of performing this method, and a kit of the pyrophosphate sequencing reagents.
Owner:UD TECH CORP +1
Cucumber variety identification method based on SNP markers
PendingCN104846062ASimple and fast operationSuitable for high-throughput operationsMicrobiological testing/measurementDNA/RNA fragmentationGenotype AnalysisMicrobiology
The present invention discloses a cucumber variety identification method based on single nucleotide polymorphism markers, wherein 12 single nucleotide polymorphism markers of cucumber are subjected to genotyping analysis by using a pyrosequencing technology so as to achieve authenticity identification on the cucumber variety. With the detection method of the present invention, the cucumber variety authenticity identification can be completed within 5 h, and characteristics of stability, high-throughput, accuracy and the like are provided.
Owner:TIANJIN INSTITUE OF QUALITY STANDARD & TESTING OF AGRICULTUAL PRODS
Kit and method for detecting gene polymorphism of irinotecan personalized medicine by pyrophosphoric acid sequencing method
InactiveCN102643906AQuick analysisAccurate analysisMicrobiological testing/measurementIrinotecanPyrophosphoric acid
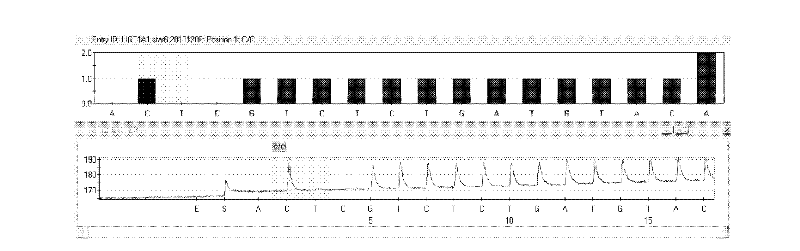
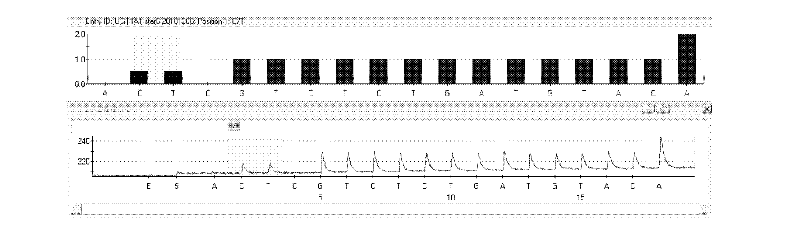
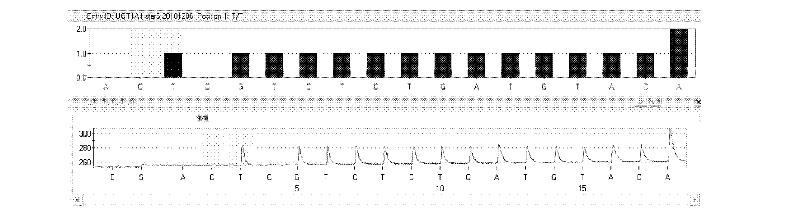
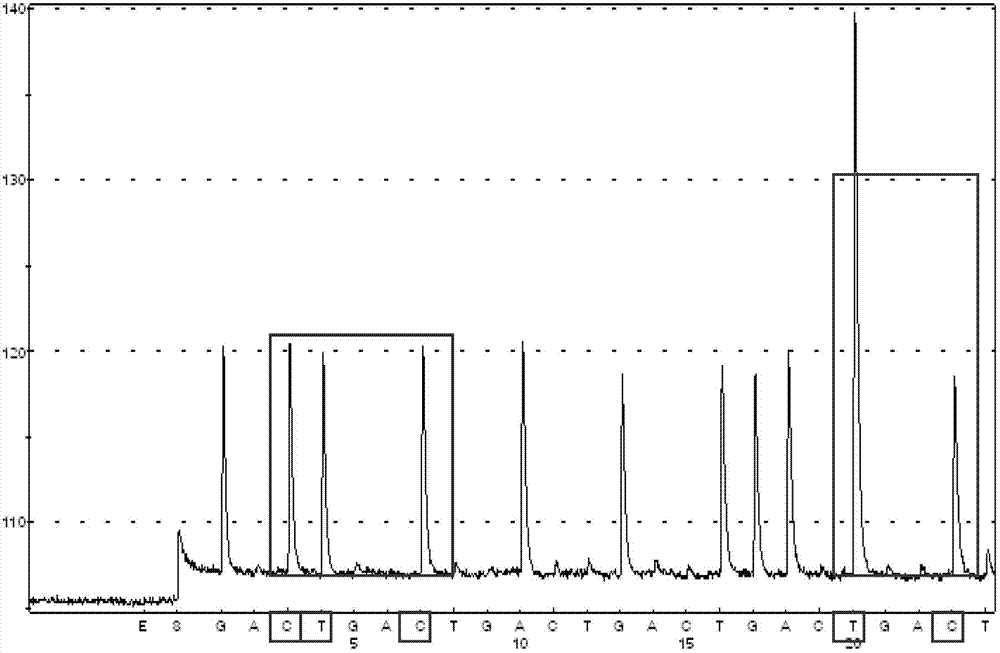
The invention discloses a kit and method for detecting gene polymorphism of an irinotecan personalized medicine by a pyrophosphoric acid sequencing method. The kit is used for parting an irinotecan medicine gene, particularly single nucleotide polymorphism of UGT1A1*6 (rs4148323) and UGT1A1*28 (rs3064744). The kit comprises primers shown as SEQ ID NO. 3-8. According to the kit, accurate, quick and high-flux detection of the UGT1A1*6 (rs4148323) and the UGT1A1*28 (rs3064744) can be realized, so that individual administration for realizing safe, rational and effective irinotecan medicine is achieved.
Owner:周宏灏
Kit for detecting hot spot mutation sites in colorectal cancer PIK3CA gene
InactiveCN102816839AStrong specificityImprove accuracyMicrobiological testing/measurementPositive controlBiotin
The invention discloses a kit detecting hot spot mutation sites in a colorectal cancer PIK3CA gene. The kit comprises an erythrocyte lysate, a whole blood DNA extraction reagent, an anhydrous ethanol, a PCR amplification reaction solution, a positive control, a negative control and a pyrosequencing reaction solution. The kit is characterized by comprising upstream and downstream primers 9F-Biotin and 9R for detecting exon 9 of the target gene and a sequencing primer 9RCX, and upstream and downstream primers 20F-Biotin and 20R for detecting exon 20 of the target gene and a sequencing primer 20RCX. The kit can be used to detect polymorphism of hot spot mutation sites (E542K, E545K and H1047R) in the colorectal cancer related PIK3CA gene, and has good specificity and high accuracy.
Owner:杭州艾迪康医学检验中心有限公司
Kit and method for determining mutation sites of genes of acetaldehyde dehydrogenase 2 and methylene tetrahydrofolic acid reductase by virtue of single tube at the same time
InactiveCN103834733AMicrobiological testing/measurementMultiplex pcrsMethylenetetrahydrofolate reductase
The invention provides a kit and method for determining mutation sites of genes of acetaldehyde dehydrogenase 2 and methylene tetrahydrofolic acid reductase by virtue of a single tube at the same time and belongs to the field of molecular biology test. The kit comprises a whole blood genome DNA (deoxyribonucleic acid) extraction reagent, a multiple PCR (polymerase chain reaction) amplification primer, a multiple PCR amplification reaction reagent, a single-strained DNA separation and purification reagent, a pyrosequencing primer, a pyrosequencing reagent and a kit body. The method comprises the following steps: extracting human whole blood genome DNA, carrying out multiple PCR amplification reaction, separating and purifying a single-strained DNA sample, pyrosequencing and analyzing results. The kit can be used by a person subjected to physical examination and hope to know individual ethyl alcohol and folic acid metabolic capability, can be used for predicting individual nitroglycerin metabolic capability and curative effect and guiding a patient suffering from myocardial infarction to reasonably select and prepare vasodilatation first-aid medicaments and can also be used for predicting the individual folic acid metabolic capability and guiding pregnant and lying-in women to supplement the right dosage of folic acid supplements.
Owner:ZHONGSHAN HOSPITAL XIAMEN UNIV +1
Palmtop pyrosequencing system based on digital microfluidic technology
InactiveCN107937265AAvoid dilutionReduce consumptionBioreactor/fermenter combinationsBiological substance pretreatmentsMiniaturizationComputer module
The invention discloses a palmtop pyrosequencing system based on digital microfluidic technology, and belongs to the technical field of digital microfluidics. A digital microfluidic chip, an electromagnetic module, and an integrated circuit are connected, and the integrated circuit and a chemiluminescent collection module are connected to a computer. According to the palmtop pyrosequencing systembased on digital microfluidic technology, a printed circuit board is used as a chip substrate, and based on an on-chip dielectric wetting principle, the electrifying sequence of electrodes that are located on the chip is regulated to achieve liquid drop generation, transportation, mixing and splitting in a sequencing process. The system is easy to operate and is miniaturized, and the produced optical signal can be fast rapidly read, so that single base on the chip is measured. The system is simple in structure, low in cost, easy to integrate, and high in detection sensitivity, the sequencing cost is greatly reduced, and the system can be widely applied to the field of pyrosequencing-based gene detection.
Owner:XIAMEN UNIV
Typing method for human papilloma virus gene
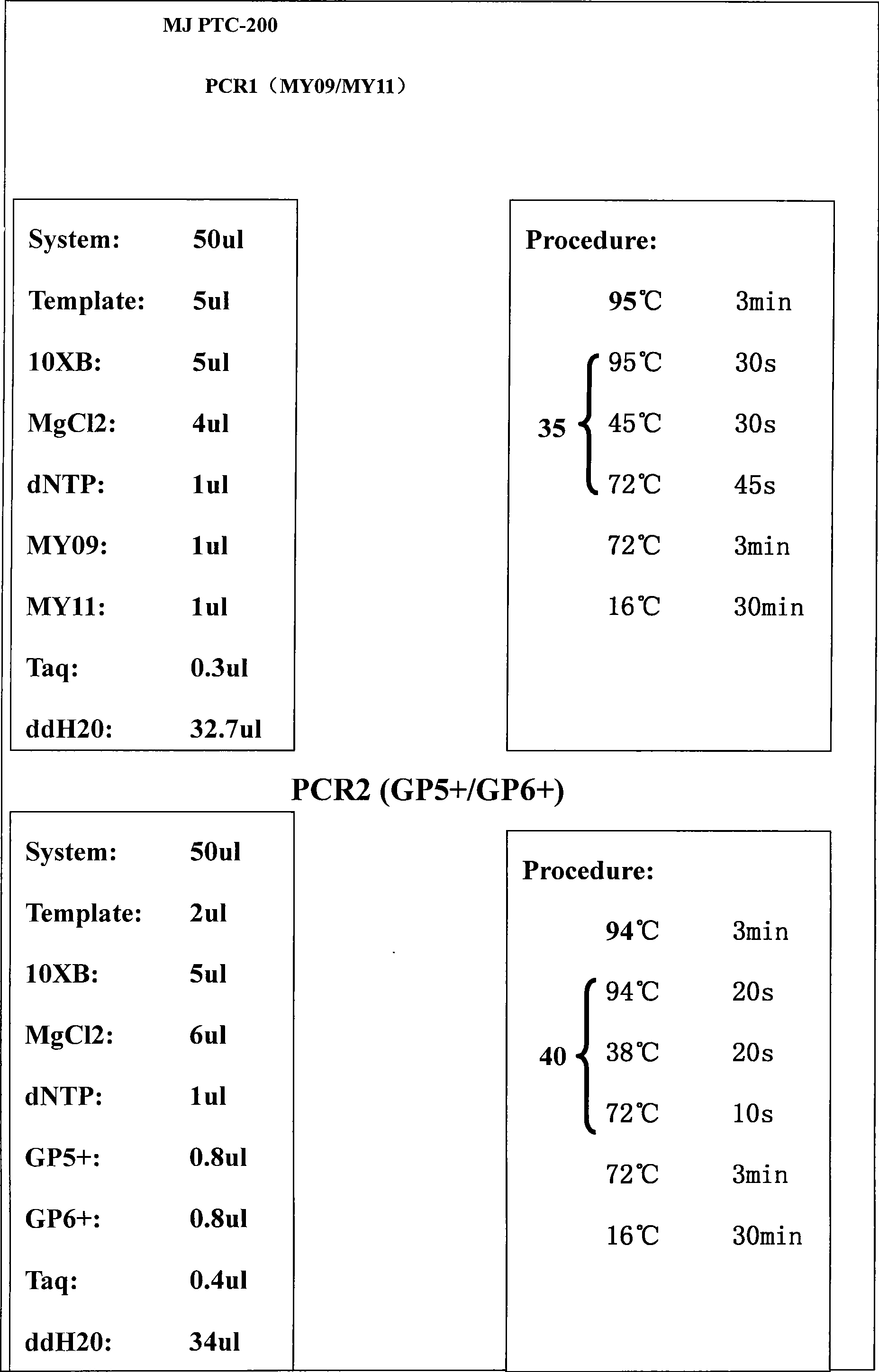
InactiveCN101397590ALow costEasy to operateMicrobiological testing/measurementSpecial data processing applicationsDesign softwareTyping methods
The invention discloses a genotyping method of human papilloma virus, which carries out permutation and combination on the sequences of 25bp behind GP5 of 40 common HPV subtypes by designed software, and the detailed steps are as follows: 1. the sequences of 25bp behind GP5 of a single type of the 40 common HPV subtypes; 2. mixed sequence mode of 25bp behind GP5 after arbitrary two types of the 40 common HPV subtypes are combined; 3. mixed sequence mode of 25bp behind GP5 after arbitrary three types of the 40 common HPV subtypes are combined; 4. mixed sequence mode of 25bp behind GP5 after arbitrary four types of the 40 common HPV subtypes are combined; 5. mixed sequence mode of 25bp behind GP5 after arbitrary five types of the 40 common HPV subtypes are combined; and 6. mixed sequence mode of 25bp behind GP5 after arbitrary six types of the 40 common HPV subtypes are combined. All the modes carry out pairing with the bases following GP5 by the ATCG sequence, and the pairing number is recorded, circulation is carried out in this way until the sequences of 25bp behind GP5 are all paired, then a pyrosequencing mode database infected by 1 to 6 types of different subtypes is created, the sample is carried out DNA extraction and MY09 / 11 amplification, the amplification product is taken as a template and then carried out amplification by GP5+ / 6+, then GP5 is taken as a sequencing primer, and the sequencing result is compared with the pyrosequencing mode database to determine the type. The genotyping method has the advantages of low cost, convenient operation, strong specificity and synchronously detecting a plurality of types of inflection.
Owner:HANGZHOU D A GENETIC ENG
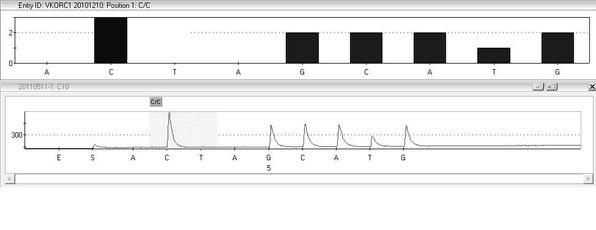
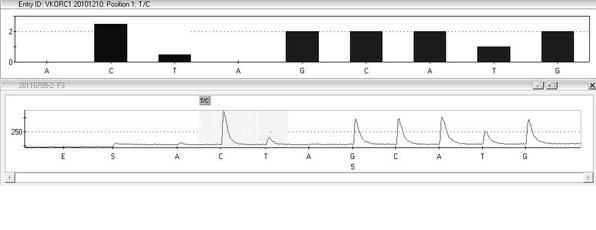
Kit and method for detecting gene polymorphism related to warfarin personalized medication by pyro sequencing method
InactiveCN102676666AQuick analysisAccurate analysisMicrobiological testing/measurementVKORC1Nucleotide
The invention discloses a kit and a method for detecting the gene polymorphism related to warfarin personalized medication by a pyro sequencing method. The kit is used for typing genes related to the warfarin medication, and the single nucleotide polymorphism of VKORC1-1639 G>A (rs9923231) and CYP2C9*31075A>C(rs1057910) is involved; and the kit comprises primers shown as SEQ ID NO.3-8. By the kit, VKORC1-1639G>A and CYP2C9*3 1075A>C can be detected accurately and quickly in high flux, so that the safe, reasonable and effective personalized administration of the warfarin medication is realized.
Owner:周宏灏
Primer pair and kit for detecting ALDH2 (Aldehyde Dehydrogenase 2) genotype with pyrosequencing method
InactiveCN105177159AHigh sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationGenotypeBiotin
The invention relates to a sequencing primer pair for detecting an ALDH2 (Aldehyde Dehydrogenase 2) genotype with a pyrosequencing method and a kit thereof, and belongs to the technical field of in-vitro nucleic acid detection. The primer pair comprises a positive amplification primer, a reverse amplification primer and a sequencing primer, wherein a 5' end of the positive amplifier primer is subjected to biotin labeling. The kit comprises the positive amplification primer, PCR (Polymerase Chain Reaction) reaction liquid containing the reverse amplification primer, the sequencing primer, uracil DNA (Deoxyribose Nucleic Acid) glycosylase and Taq polymerase. The kit provided by the invention has the advantages of accurate detection result, high specificity, short detection period, easiness in operation, and capability of effectively meeting clinical examination requirements. Moreover, the kit further has the advantages that a reaction process can be monitored in real time, the reaction time is short, a PCR product can be subjected to pyrosequencing by a pyrosequencing instrument and high-flux sample detection through simple treatment, and the sensitivity is higher compared with a golden standard method, namely, a capillary electrophoresis sequencing method.
Owner:CHANGSHA 3G BIOTECH
Method for detecting community structure and abundance of ammonia oxidizing bacteria in wastewater system
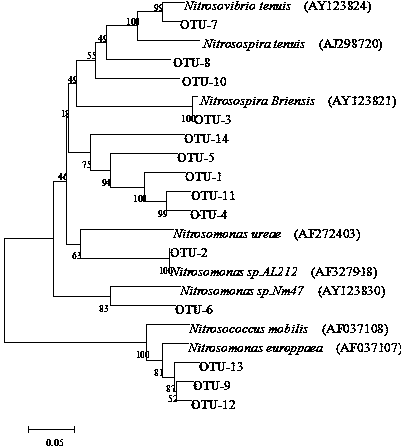
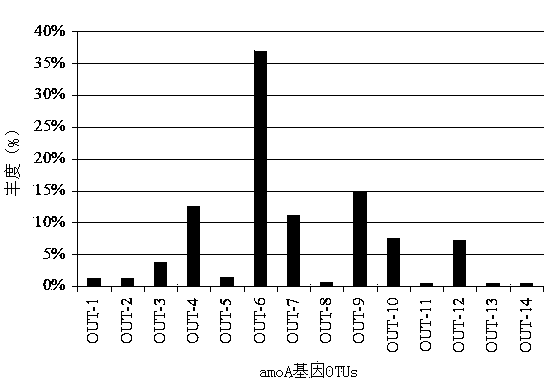
The invention discloses a method for detecting the community structure and abundance of ammonia oxidizing bacteria in a wastewater system. The method comprises the following specific steps: (A) extracting all genome DNA (Deoxyribonucleic Acid) of microorganisms in active sludge of the wastewater system; (B) performing PCR (polymerase chain reaction) amplification by taking all genome DNA as a template and taking amoAlF with a sequence number of SEQ ID No.1 and amoA2R with a sequence number of SEQ ID No.2 as primers; and (C) sequencing an amplification product by adopting a Roche 454 pyrosequencing process, dividing an operational taxonomic unit for a sequencing result by applying a Mothur program, performing online BLASTn comparison on representative sequences with the similarity of 99% in the operational taxonomic unit and sequences disclosed in a database GenBank, and dividing a system evolutionary tree according to a comparison result to obtain the community structure distribution and abundance of the ammonia oxidizing bacteria. The method is simple to perform, low in cost, quick in detection and high in sequence analysis accuracy, can be used for qualitatively analyzing different community structures of the ammonia oxidizing bacteria and quantitatively analyzing the abundance of different species of the ammonia oxidizing bacteria, can be used for simultaneously analyzing a plurality of samples, and is relatively high in practicality and easy to popularize and apply.
Owner:HIGH TECH RES INST NANJING UNIV LIANYUNGANG +1
Method for fast appraising purity of cucumber hybrid seed
InactiveCN101928766ASimple and fast operationHigh precisionMicrobiological testing/measurementFluorescence/phosphorescenceHybrid seedRoom temperature
The invention discloses a method for fast appraising the purity of cucumber hybrid seed, which comprises the following steps of: taking the genome DNA of a 'Jingyou 38' test seed of a new product of the room temperature cucumber as a template, informing to analyze the differentiation segment of parent DNA of the known 'Jingyou 38', and appraising the purity of the 'Jingyou 38' hybrid seed by applying a pyrophosphoric acid sequencing technology. The method can appraise the purity of the seed within 3h, has the characteristics of being simple and easy to operate, high in sensitivity, fast, low in cost, convenient for popularization and the like, and developing a wide foreground for appraising the purity of the seed.
Owner:CENT LAB TIANJIN ACADEMY OF AGRI SCI +1
Kit and method for detecting tamoxifen personalized medicine genetic polymorphism by use of pyrosequencing technique
InactiveCN102643905AQuick analysisAccurate analysisMicrobiological testing/measurementPersonalized medicinePyrosequencing
The invention discloses a kit and method for detecting the tamoxifen personalized medicine genetic polymorphism by use of the pyrosequencing technique. The genetic polymorphism specifically refers to the single nucleotide polymorphism of CYP2D6*10(rs1065852) and (SULT1A1*2) (rs9282861). The kit comprises the primer shown by SEQ ID NO.3-8. The kit disclosed by the invention can realize accurate, quick and high-flux detection on CYP2D6*10(rs1065852) and (SULT1A1*2) (rs9282861) so as to realize safe, reasonable and effective personalized administration of the tamoxifen medicine.
Owner:CENT SOUTH UNIV
Method of Assessing Soil Quality and Health
InactiveUS20120129706A1Microbiological testing/measurementLibrary screeningData setMultivariate statistical
The present invention relates to a method of assessing the soil quality and health of mined and un-mined lands. The present invention further relates to a method of assessing ecosystem health and soil restoration efforts. The method employs a multi-prong approach combining DNA fingerprinting and in-depth pyrosequencing of soil microorganisms that are responsible for maintaining soil fertility and productivity. The method incorporates assessing the indigenous bacterial community structure data derived from PCR-DGGE analysis and 165 rRNA gene clone libraries, and then further identification of those microorganisms that are known to promote soil health by soil metagenome analysis. Disparity between un-mined and post-mined soils are comprehensively studied via multivariate statistical analyses on the polyphasic dataset.
Owner:CHAUHAN ASHVINI +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com