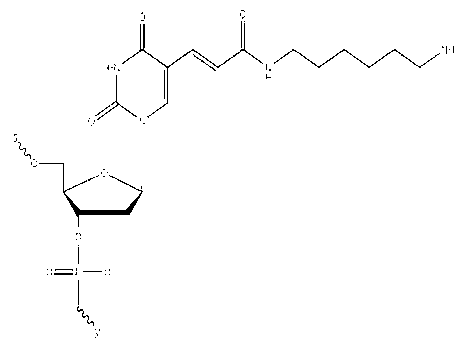
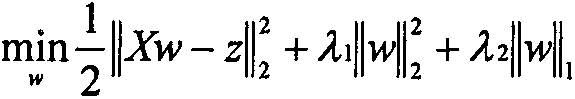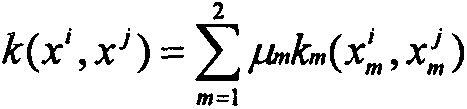Patents
Literature
128 results about "Genotype Analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
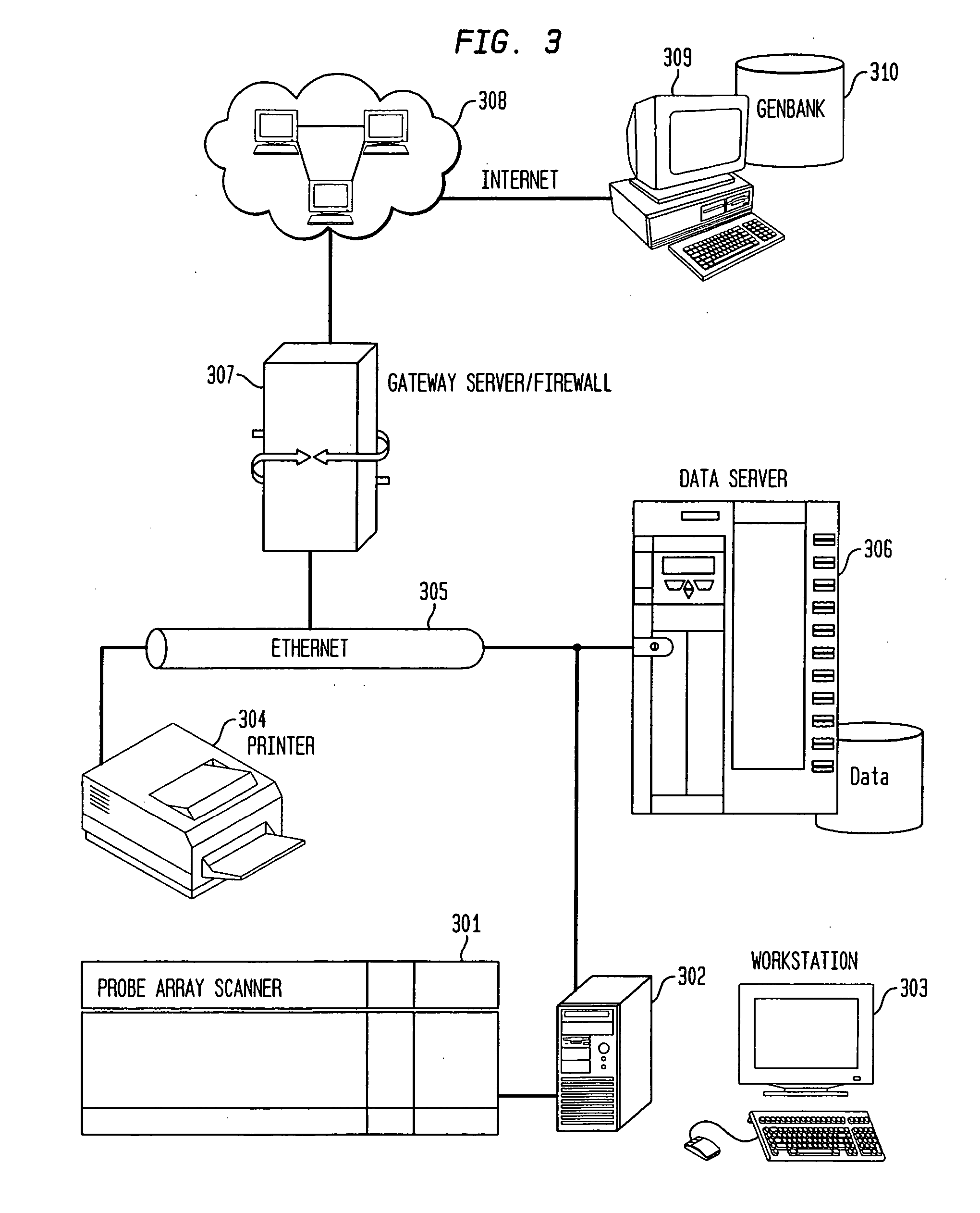
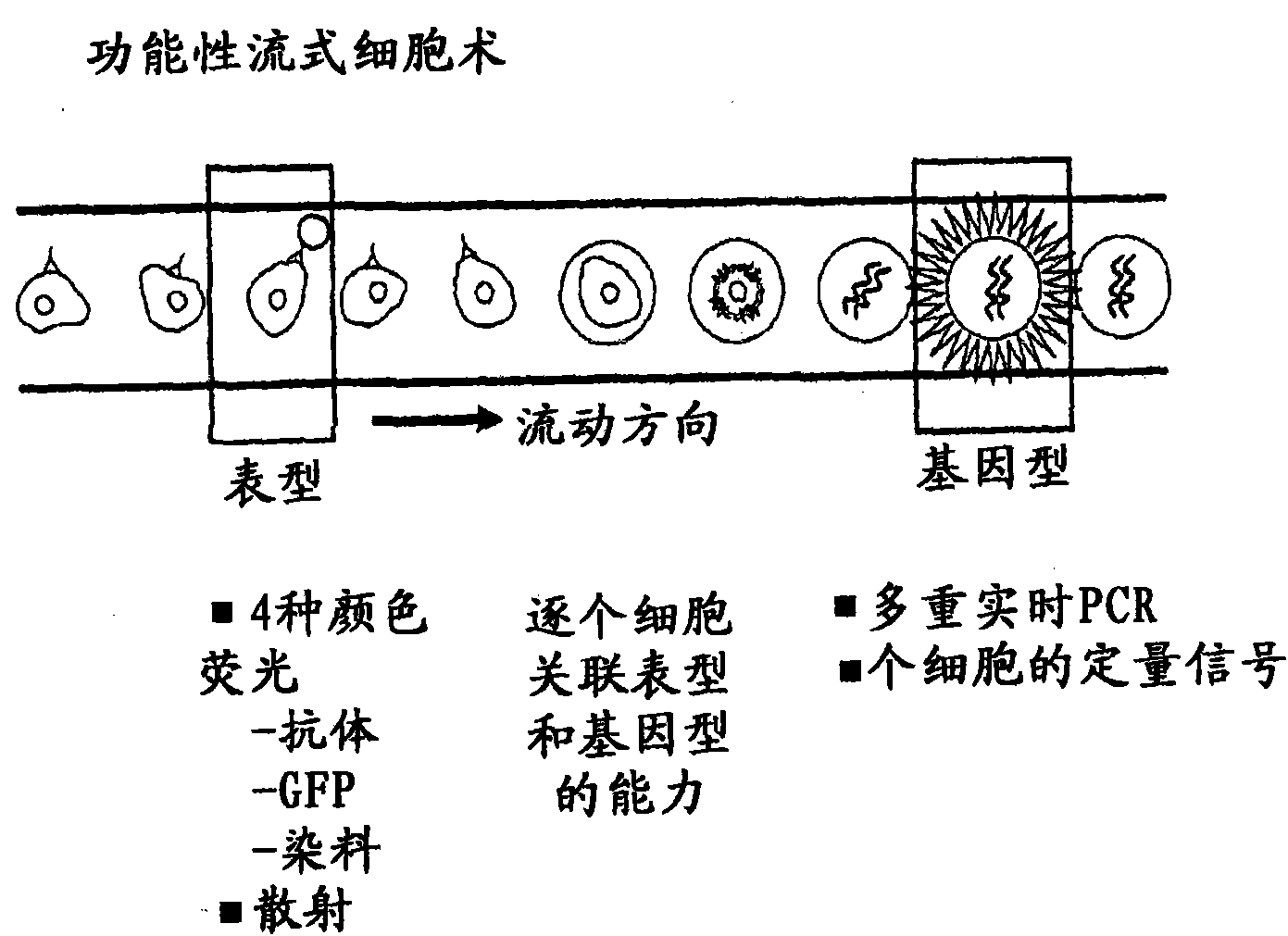
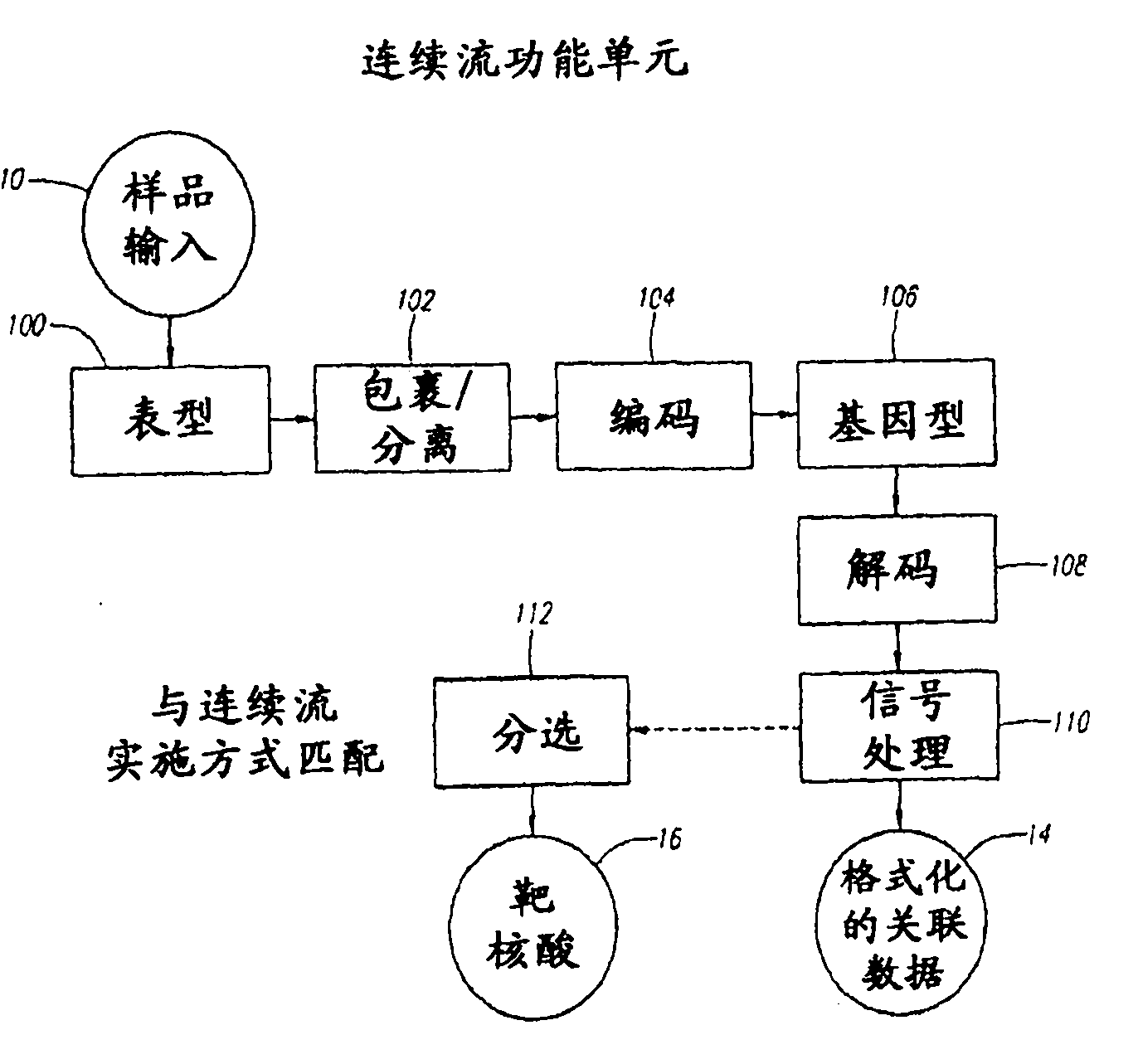
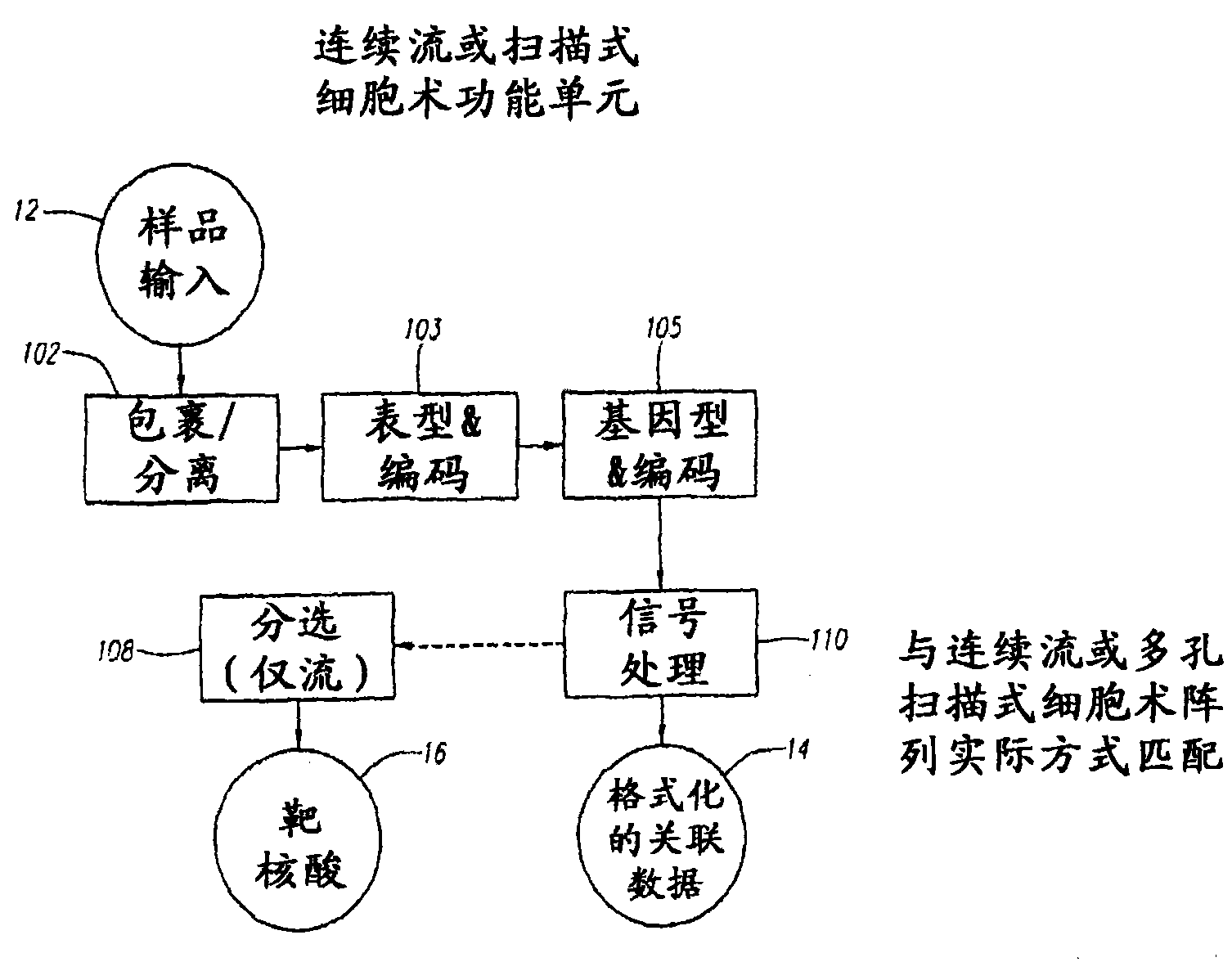
Methods and Devices for Correlated, Multi-Parameter Single Cell Measurements and Recovery of Remnant Biological Material
InactiveUS20090042737A1Reduces number of point of interrogationHeating or cooling apparatusLibrary screeningPhenotype genotypeGenotype Analysis
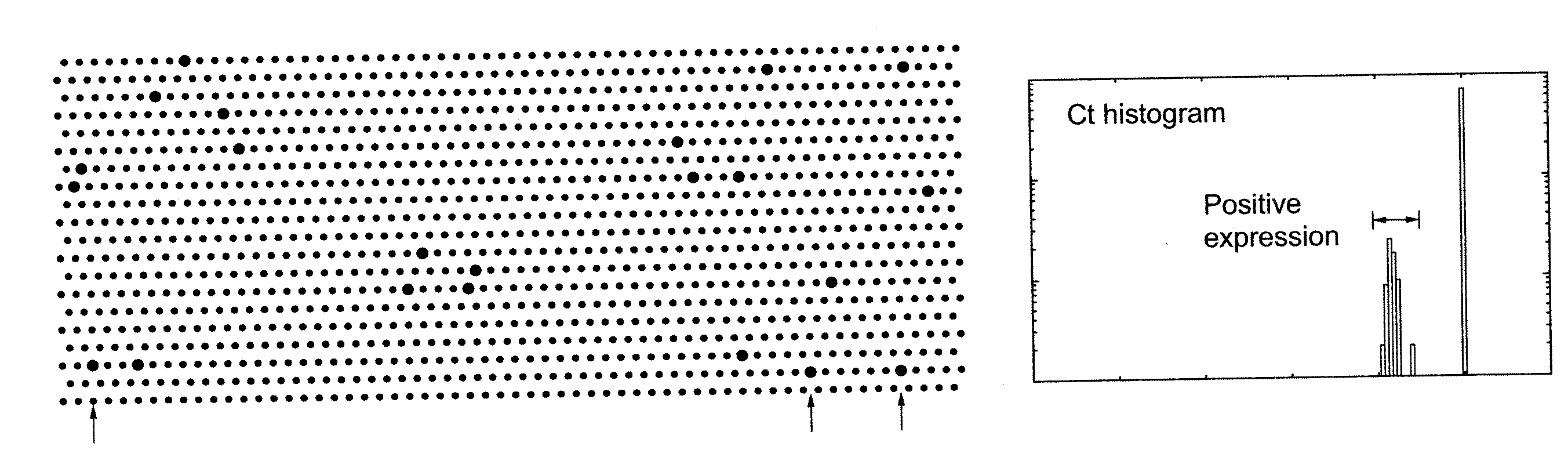
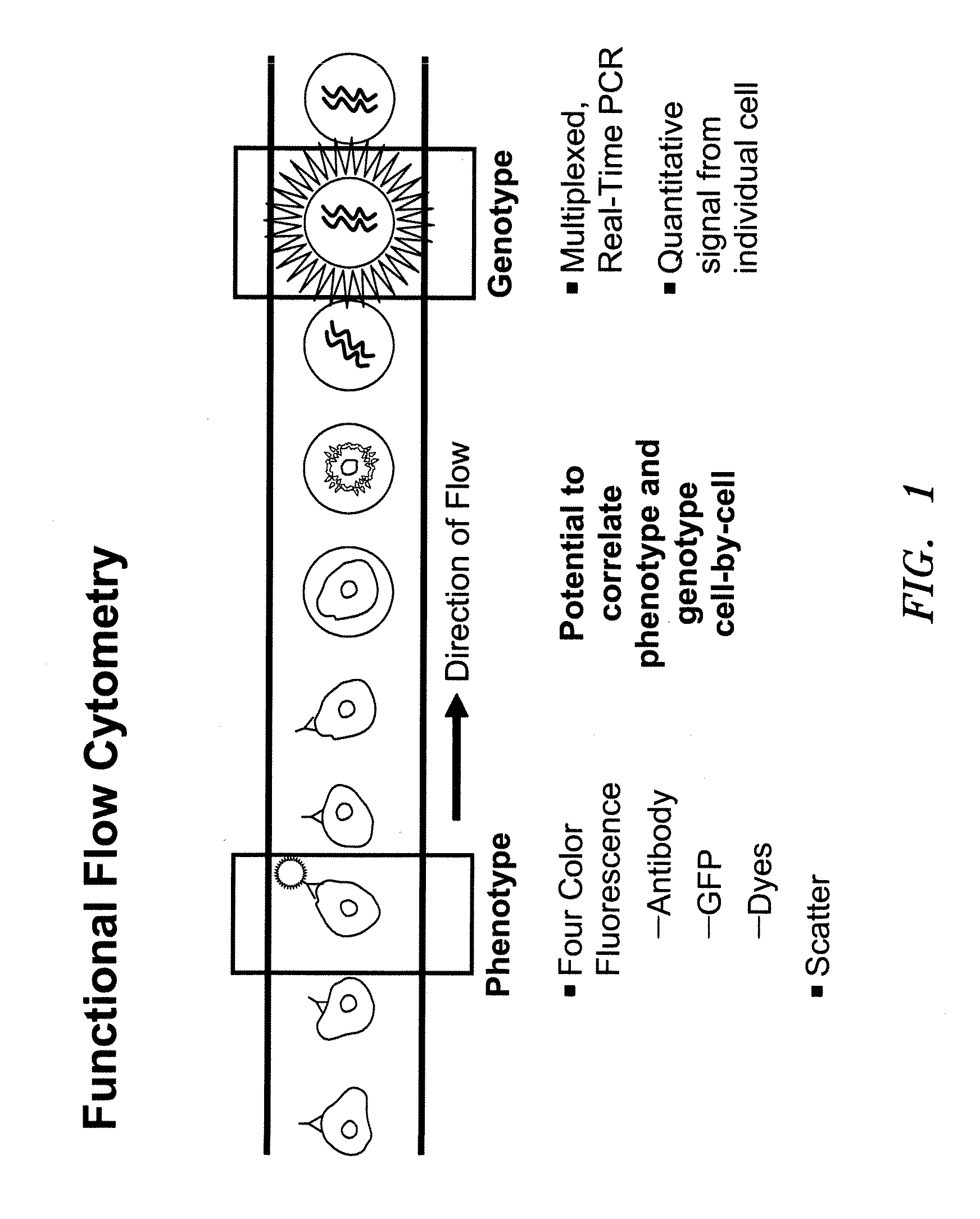
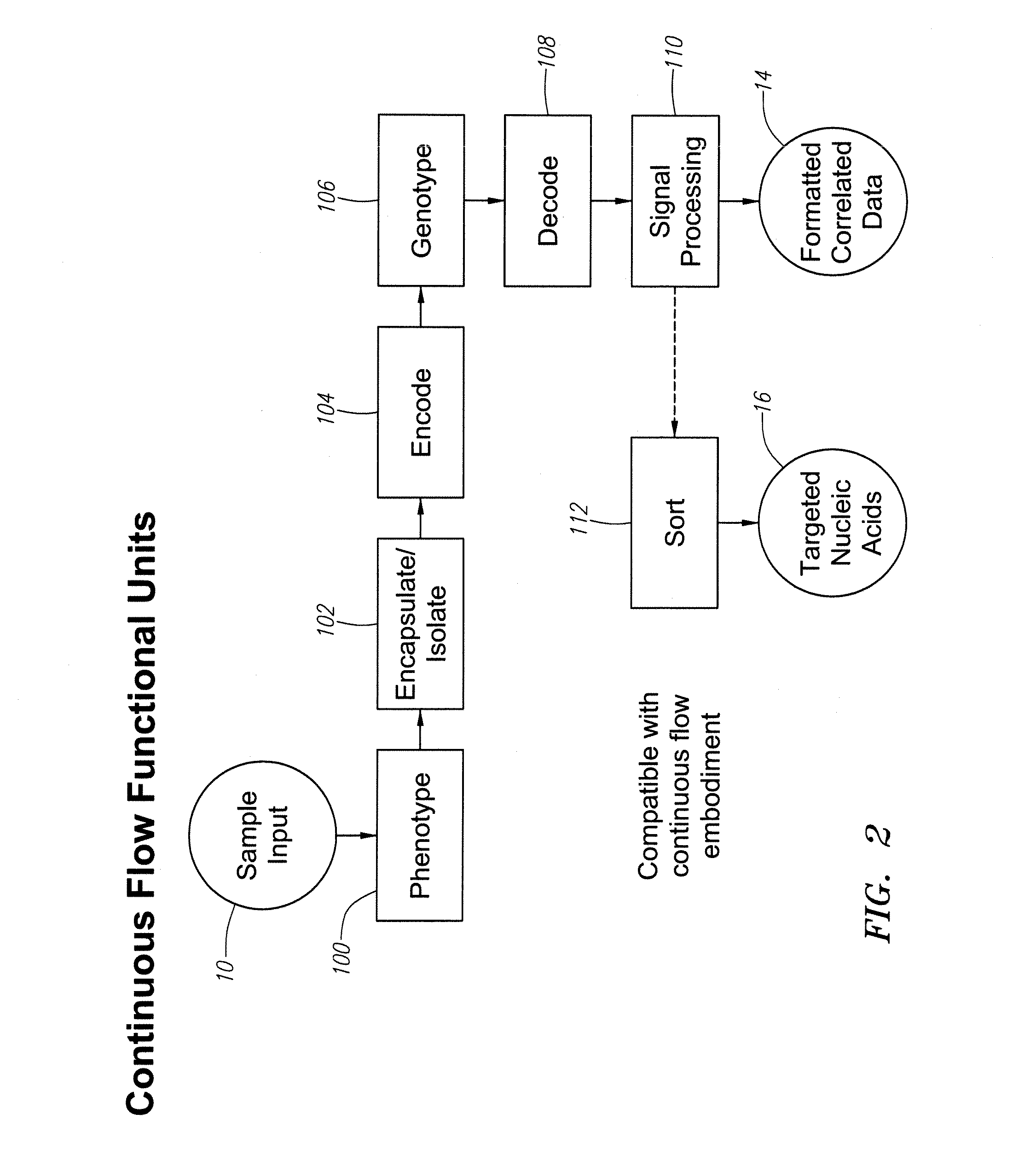
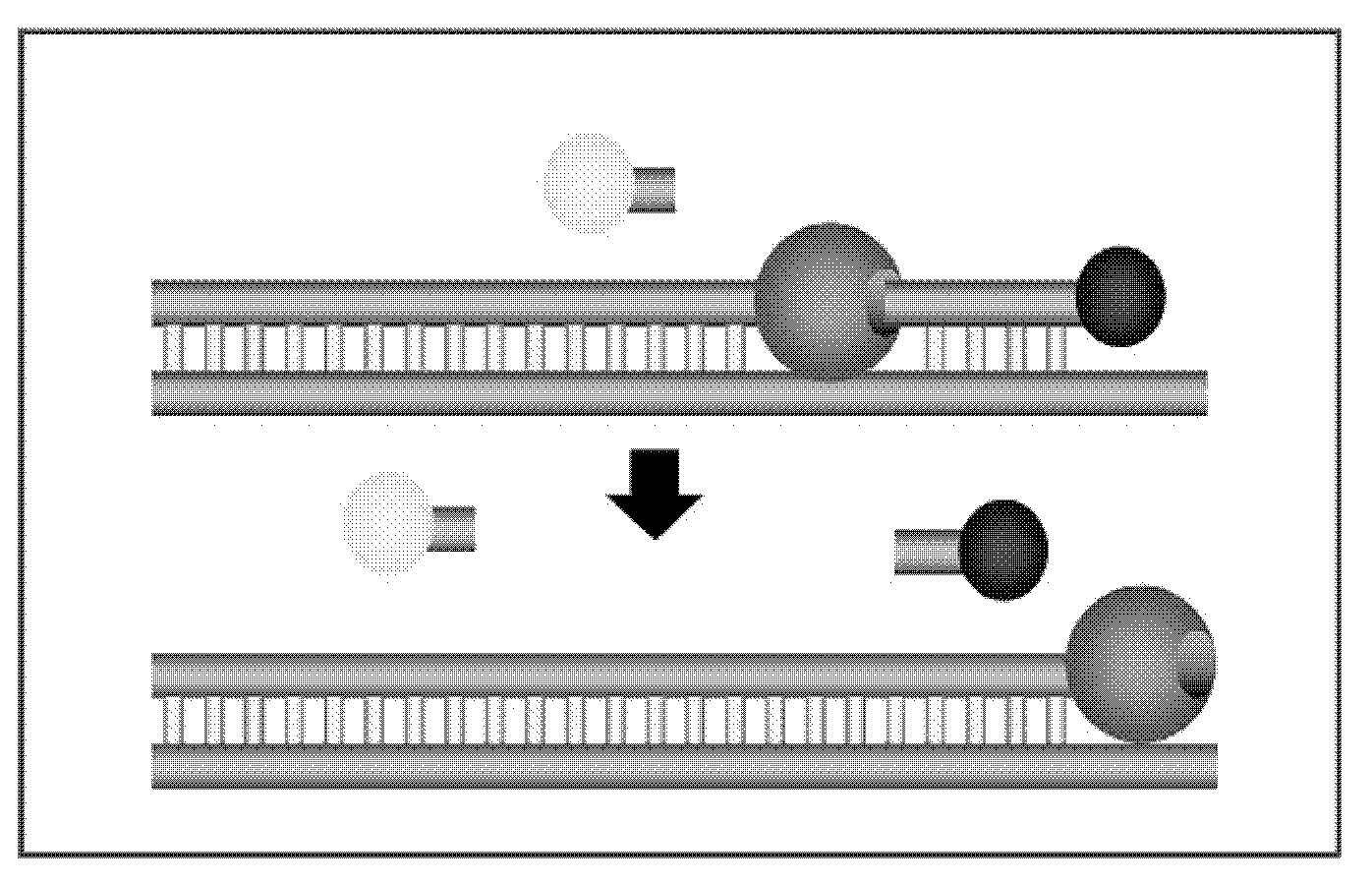
Methods and apparatus are provided for analysis and correlation of phenotypic and genotypic information for a high throughput sample on a cell by cell basis. Cells are isolated and sequentially analyzed for phenotypic information and genotypic information which is then correlated. Methods for correlating the phenotype-genotype information of a sample population can be performed on a continuous flow sample within a microfluidic channel network or alternatively on a sample preloaded into a nano-well array chip. The methods for performing the phenotype-genotype analysis and correlation are scalable for samples numbering in the hundreds of cells to thousands of cells up to the tens and hundreds of thousand cells.
Owner:PROGENITY INC
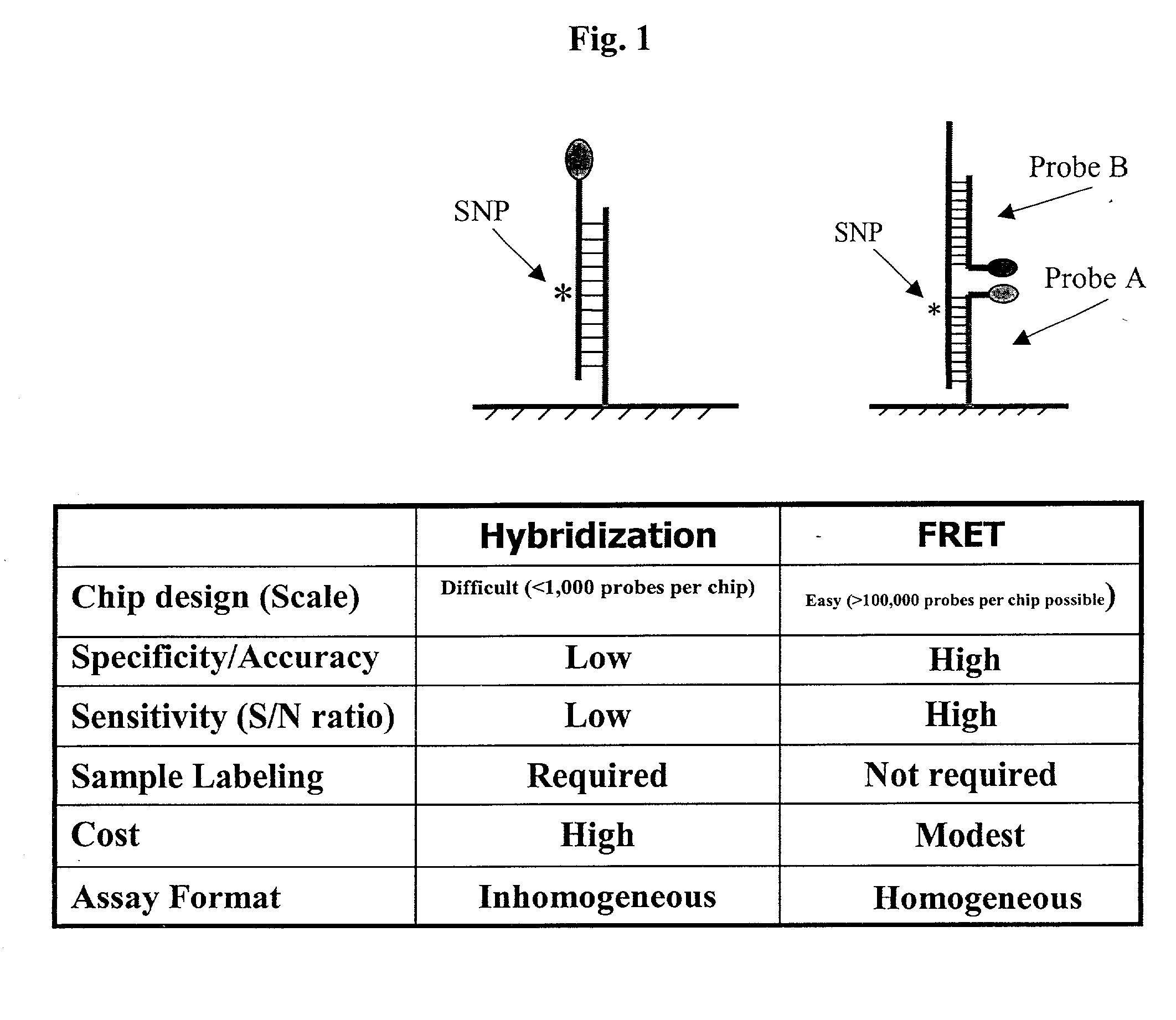
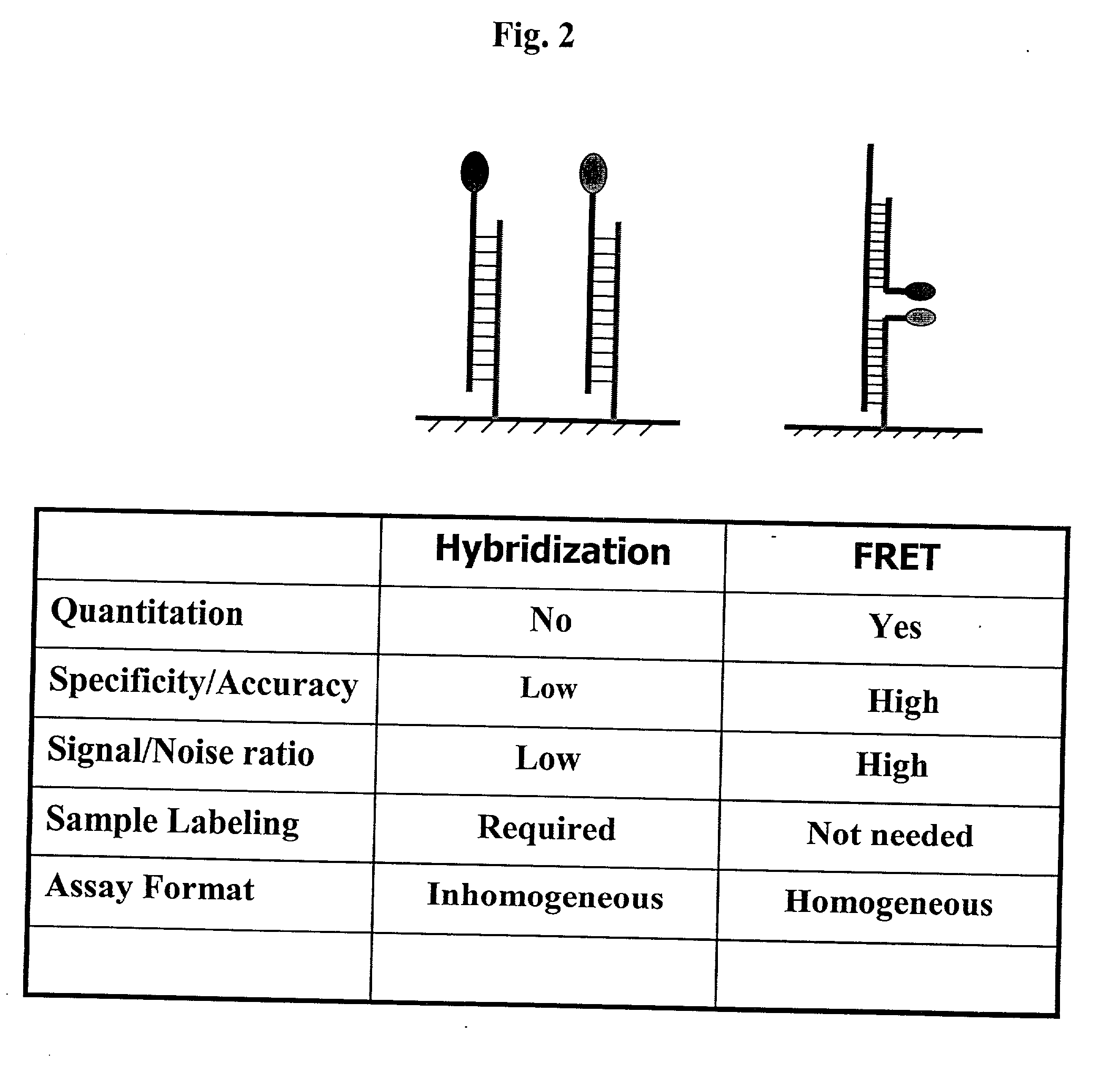
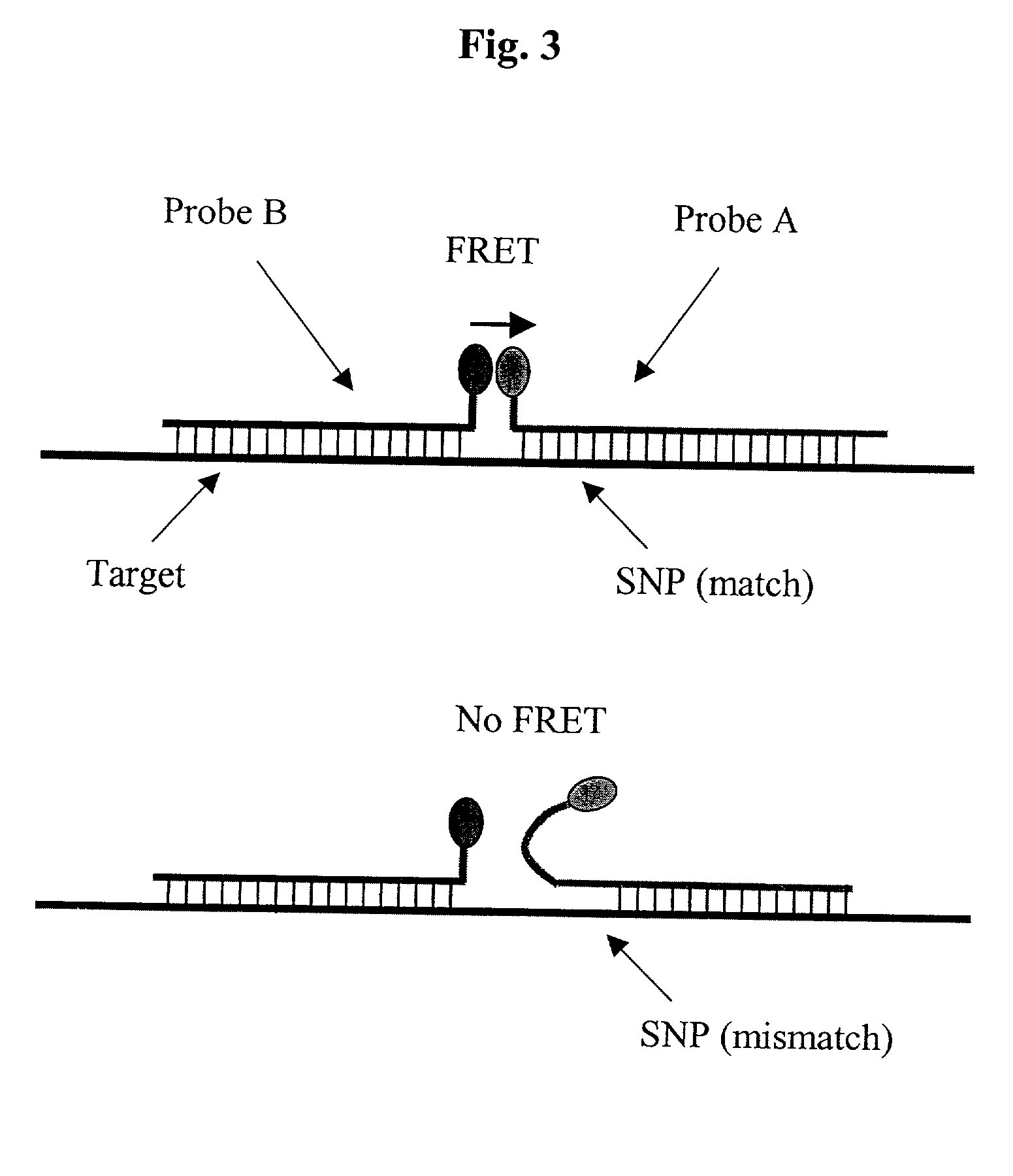
Pairs of nucleic acid probes with interactive signaling moieties and nucleic acid probes with enhanced hybridization efficiency and specificity
InactiveUS20020177157A1Improve efficiencyEnhanced hybridizationSugar derivativesMicrobiological testing/measurementNucleotideGenotype Analysis
The invention provides methods and kits for detecting and / or quantifying nucleic acid sequences of interest by using pairs of probes containing donor-acceptor moieties that when hybridized on a target polynucleotide, with one of the probes being hybridized to a sequence of interest in the target polynucleotide, places the donor-acceptor moieties in sufficiently close proximity such that a detectable signal is generated. Methods of the invention are particularly useful for genotyping analysis and gene expression profiling. Methods of the invention are easily adaptable to arrays and automation. The invention also provides probes with enhanced hybridization efficiency and / or specificity comprising a probe portion having a spacer element and / or a minor groove binder molecule, and methods and kits for using these probes.
Owner:GENOSPECTRA
Method for breeding tcf25 gene deletion type zebra fish through gene knockout
InactiveCN107988268AEfficient and More Precise SilenceEasy to makeHydrolasesMicroinjection basedDiseaseGenotype Analysis
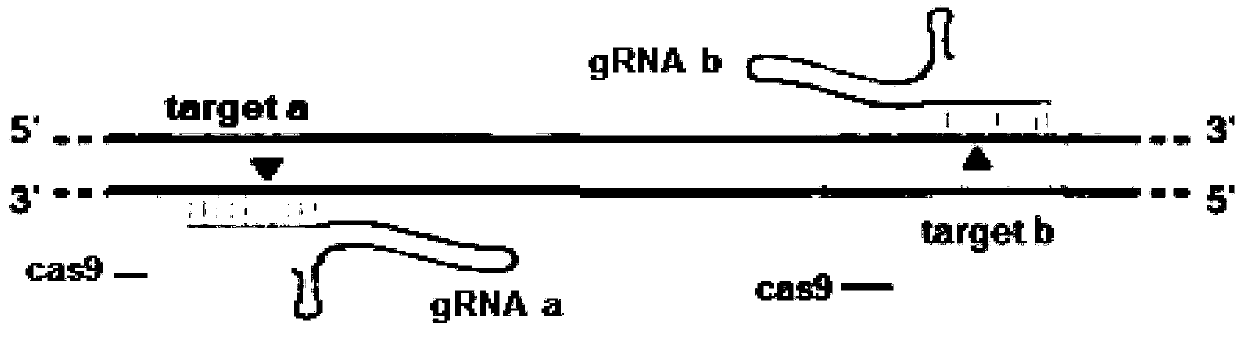
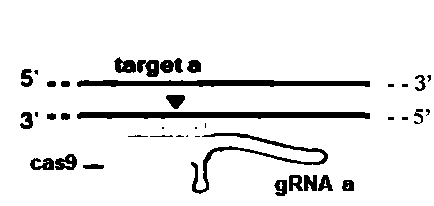
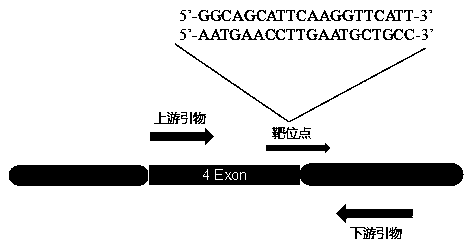
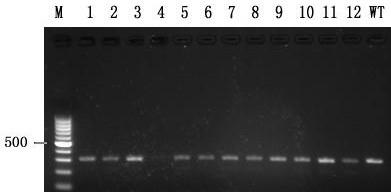
The invention relates to the technical field of gene knockout, and particularly discloses a method for breeding a tcf25 gene deletion type zebra fish through gene knockout. The method comprises the steps of through a CRISPR / Cas9 gene editing technology, designing an appropriate targeting gene locus on a tcf25 gene of the zebra fish, synthesizing in vitro to obtain specific sgRNA and Cas9-mRNA, microinjecting into a cell of the zebra fish, and after culturing an embryo for 60h, selecting the embryo for carrying out genotyping, so that the effectiveness of the selected locus is verified. The method provided by the invention is lower in off-target rate, removes the tcf25 gene through interference, is beneficial to further revealing the whole process of cardiac morphogenesis and a molecular mechanism regulating the process through researching functions through a genetics means, and is of great importance on understanding a cardiac disease pathology and researching and developing a new therapeutic schedule medically.
Owner:HUNAN NORMAL UNIVERSITY
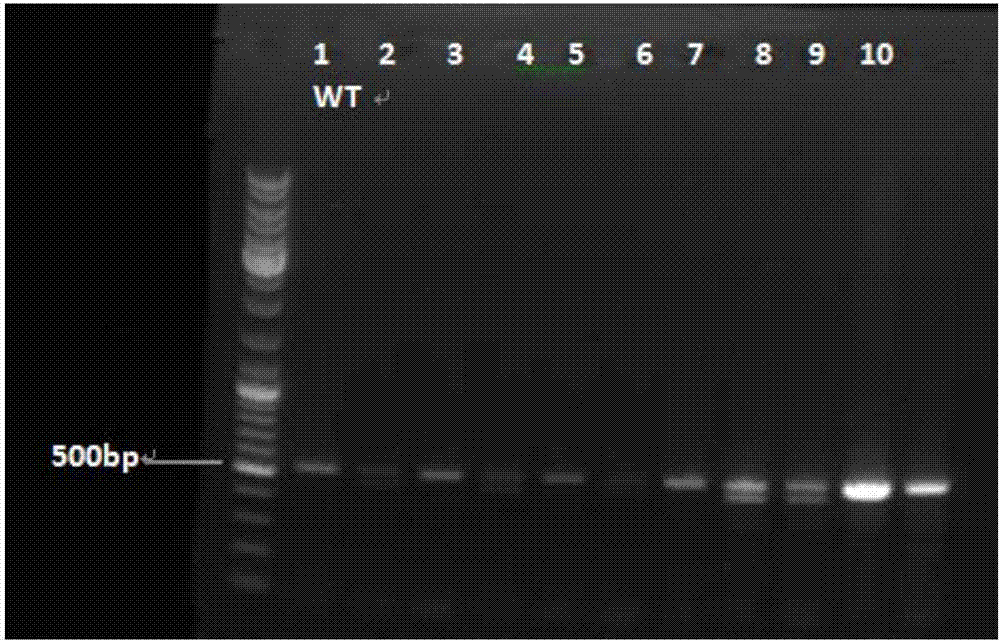
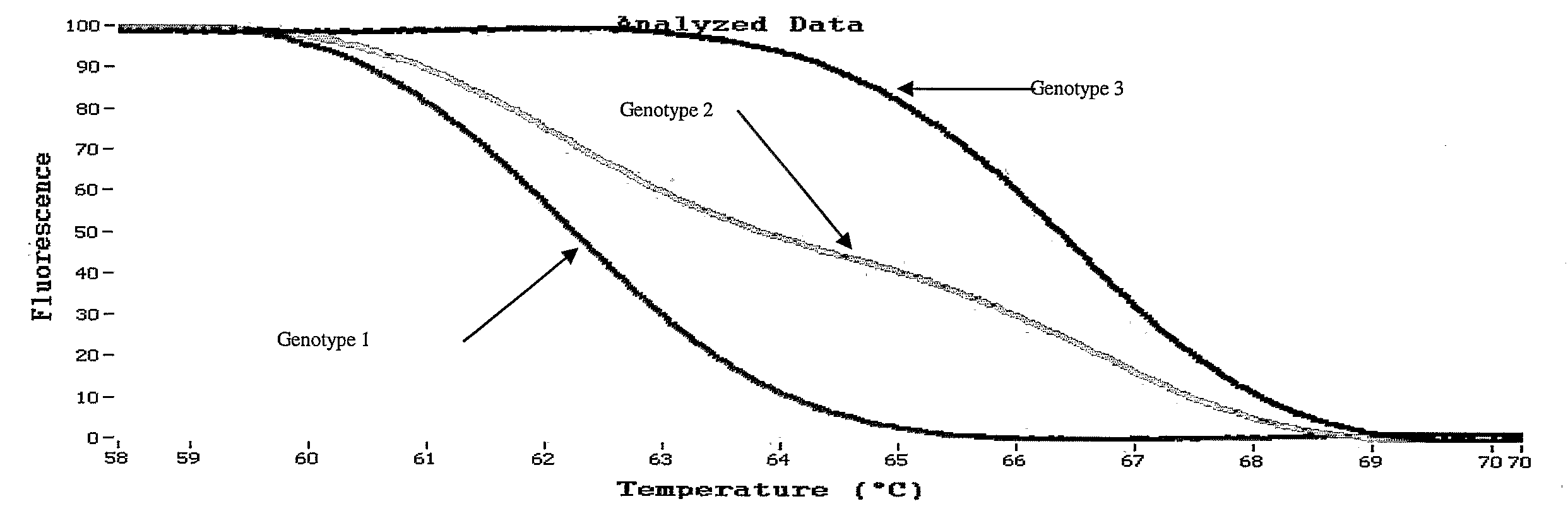
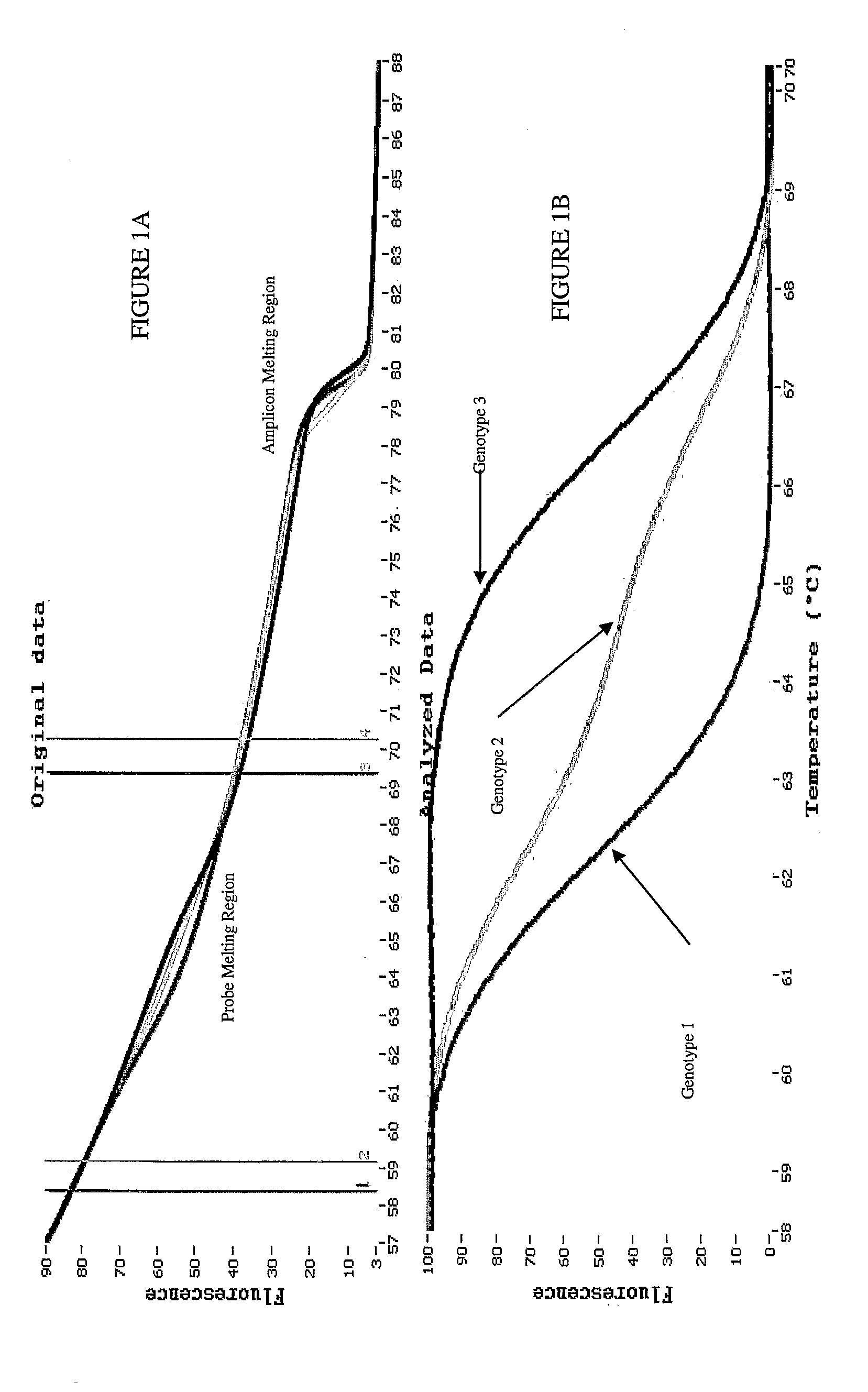
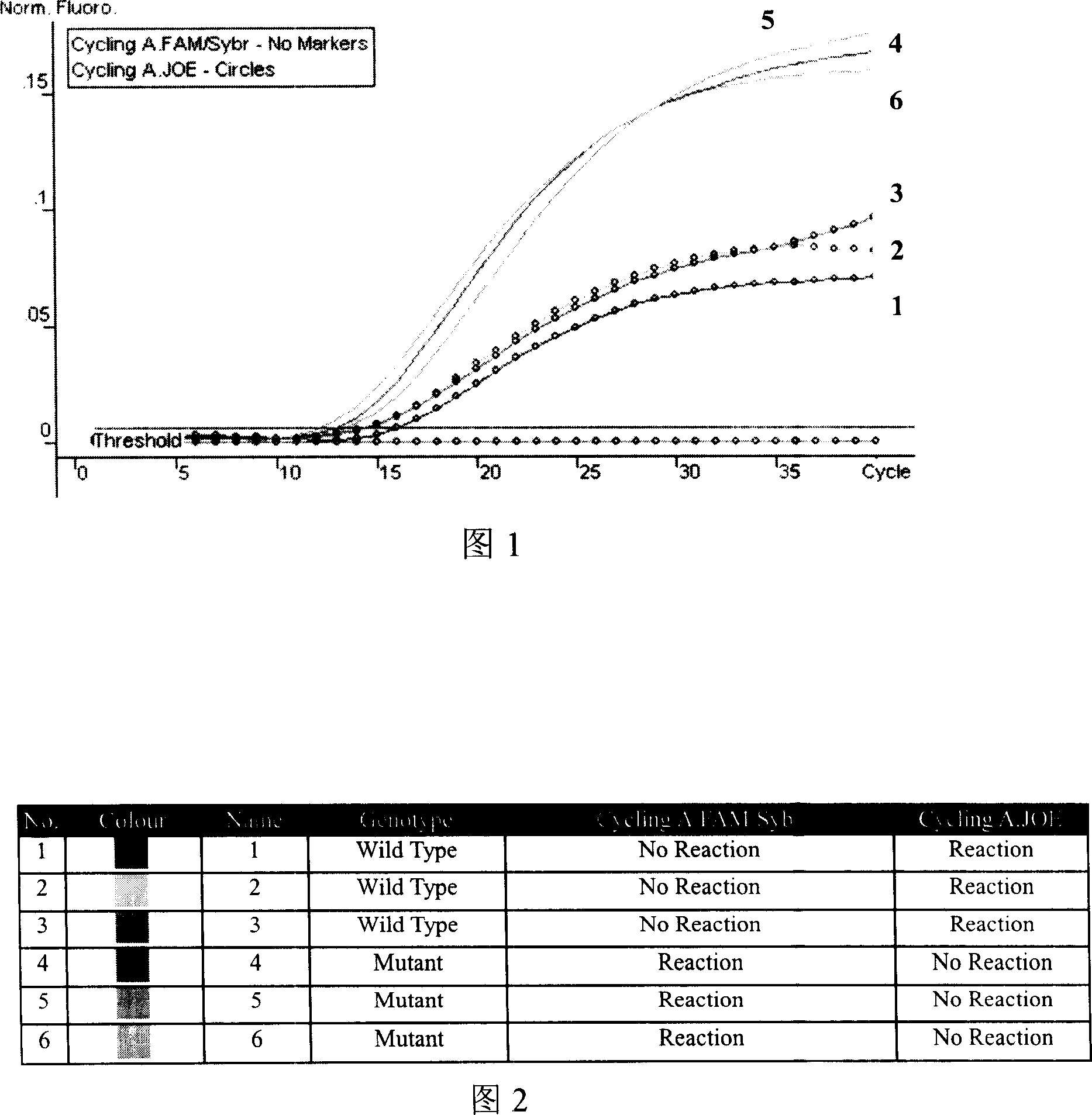
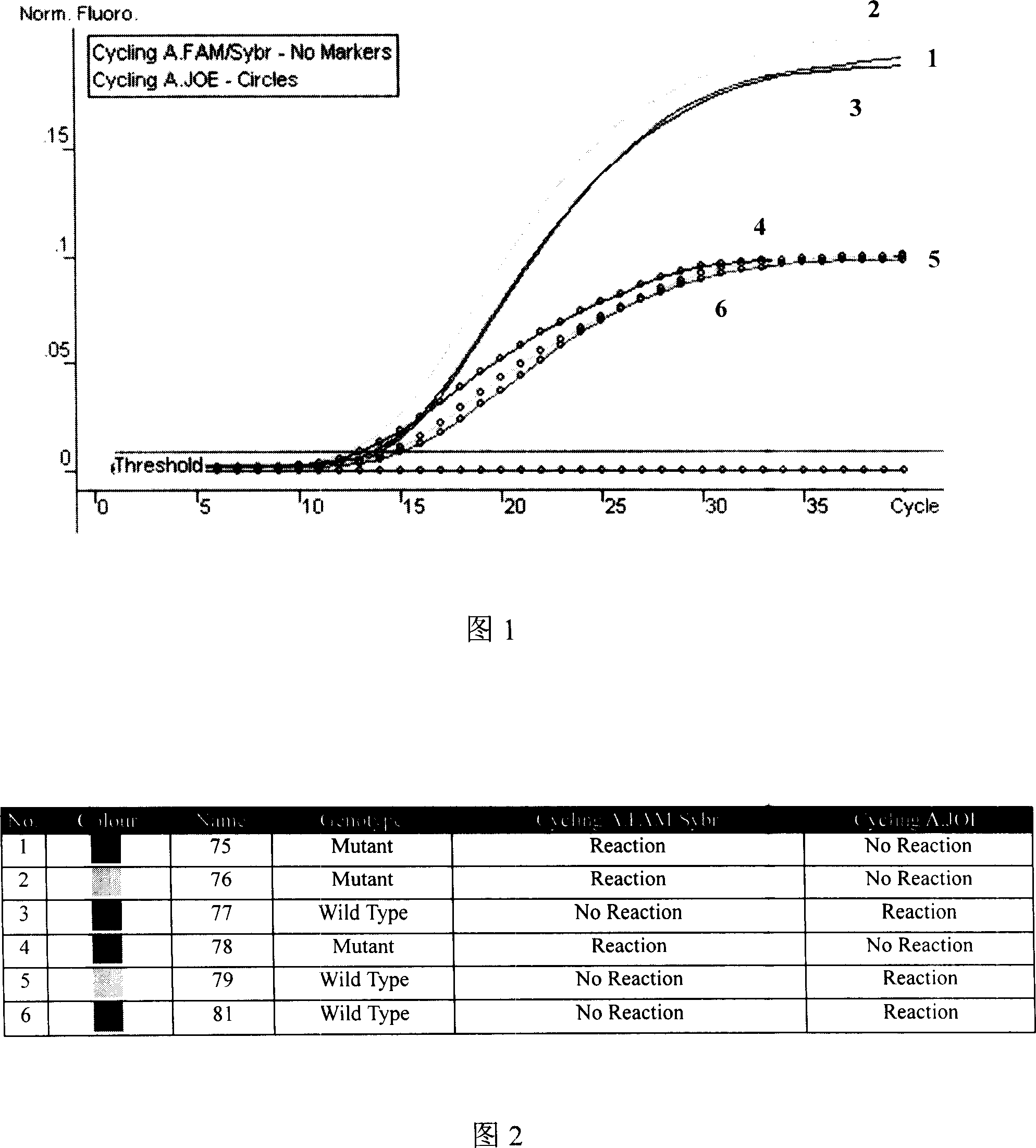
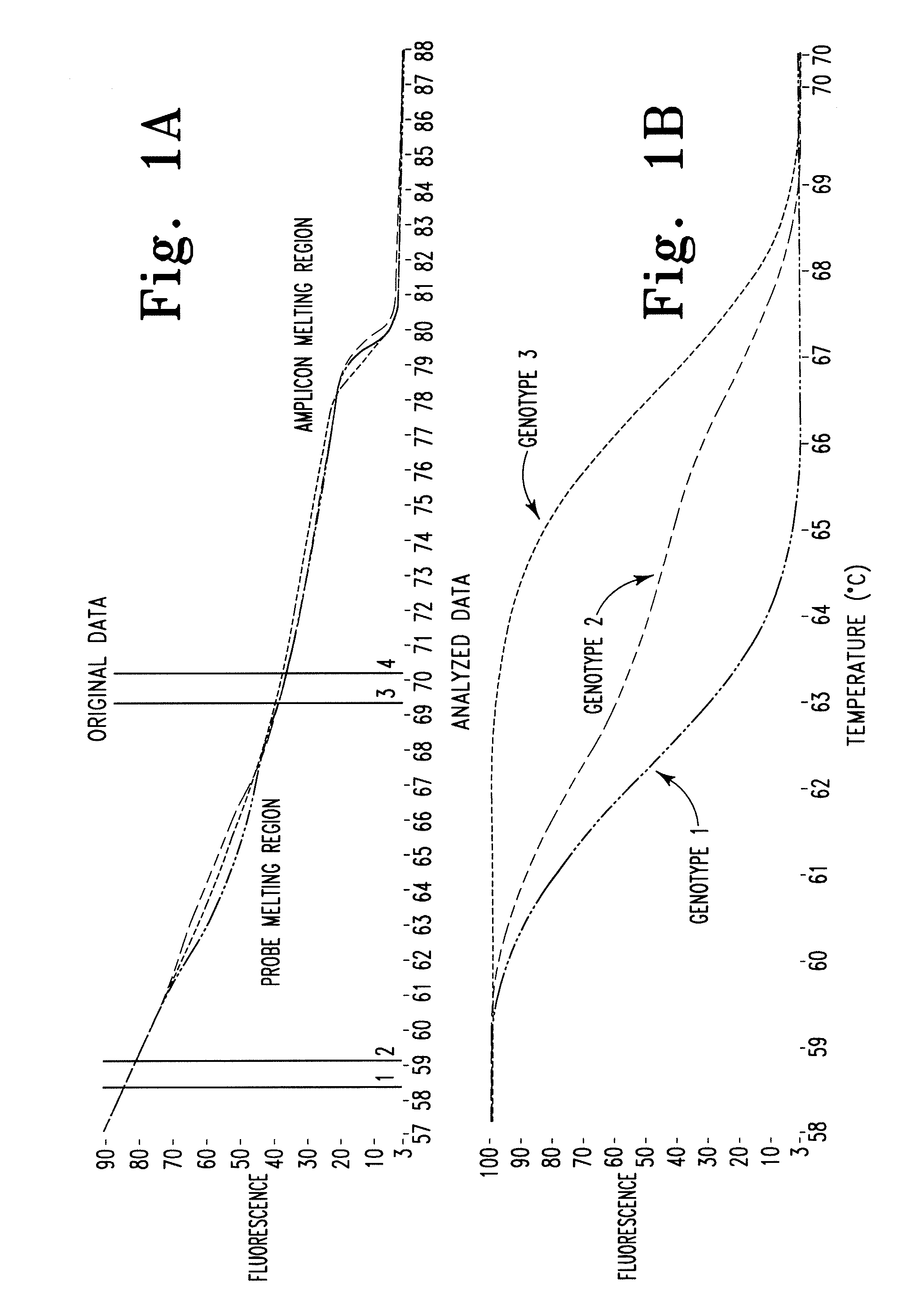
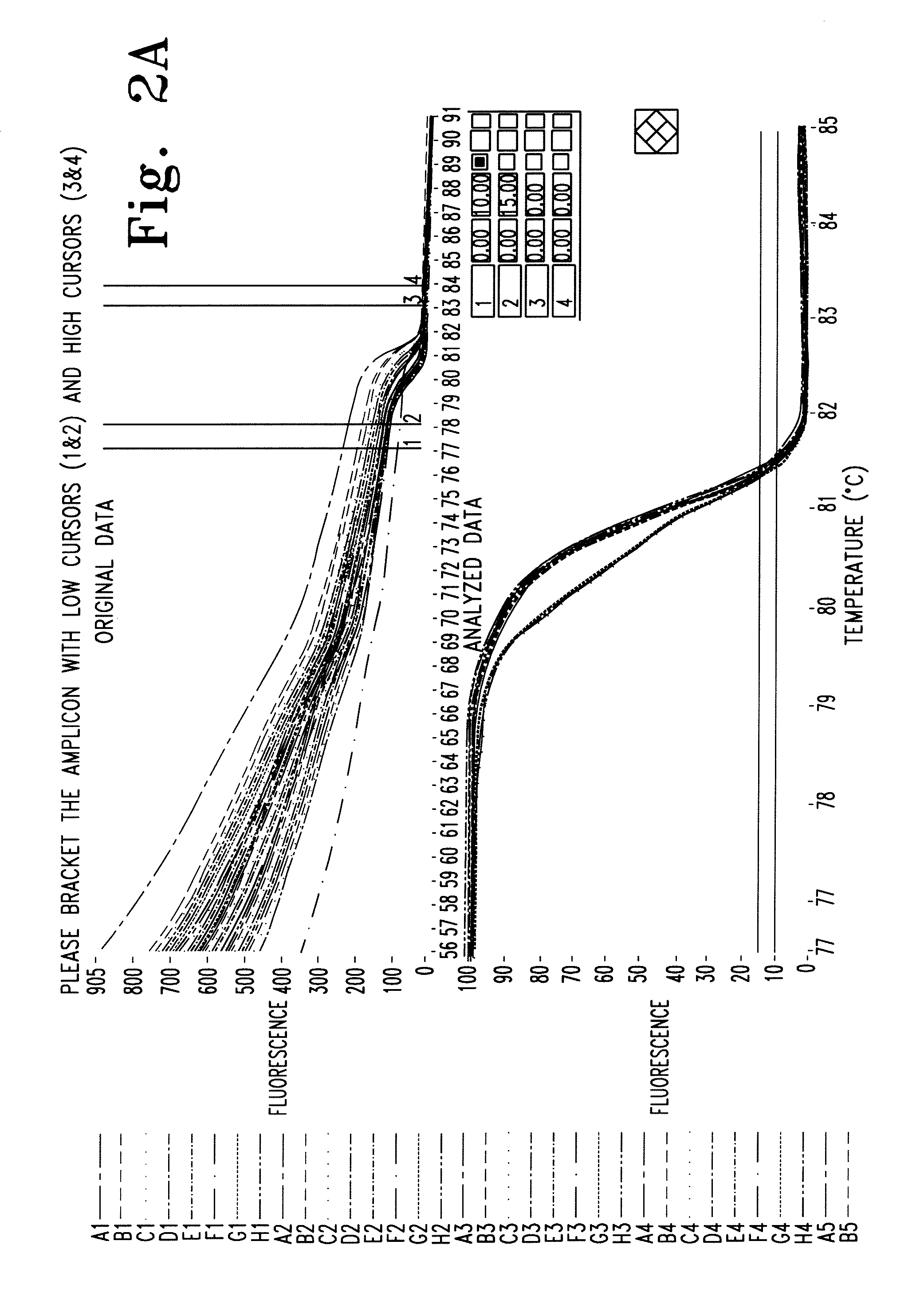
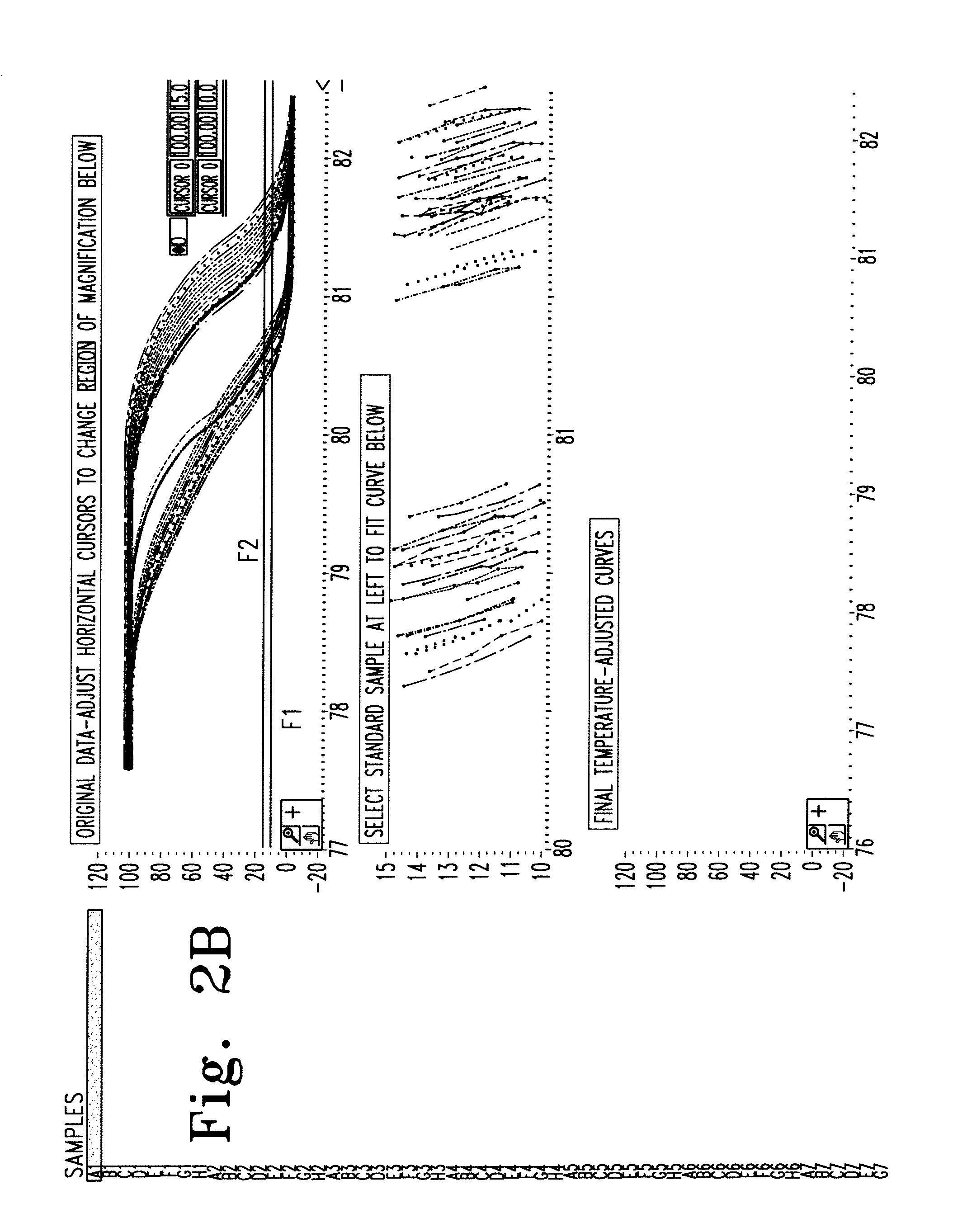
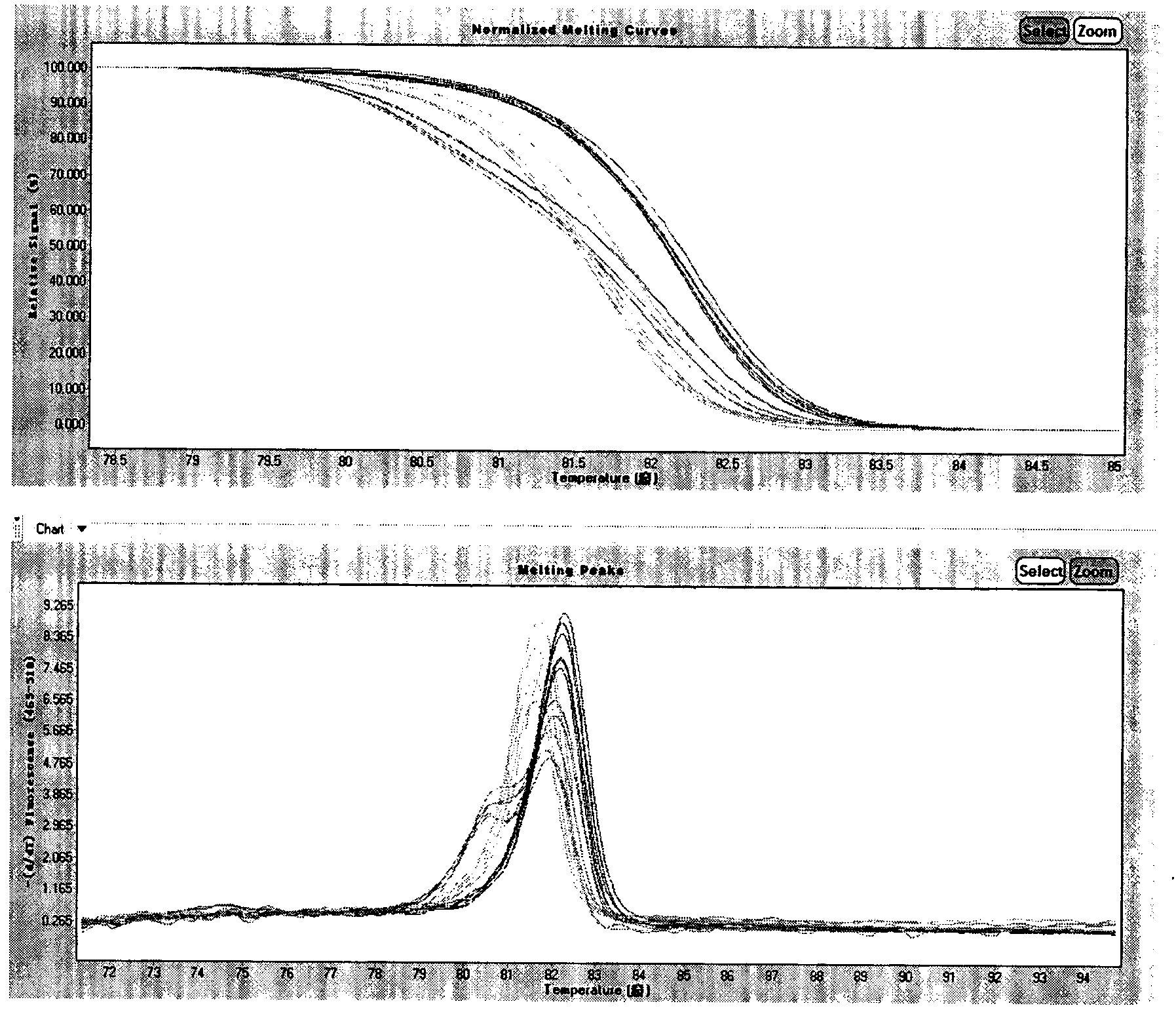
Melting Curve Analysis with Exponential Background Subtraction
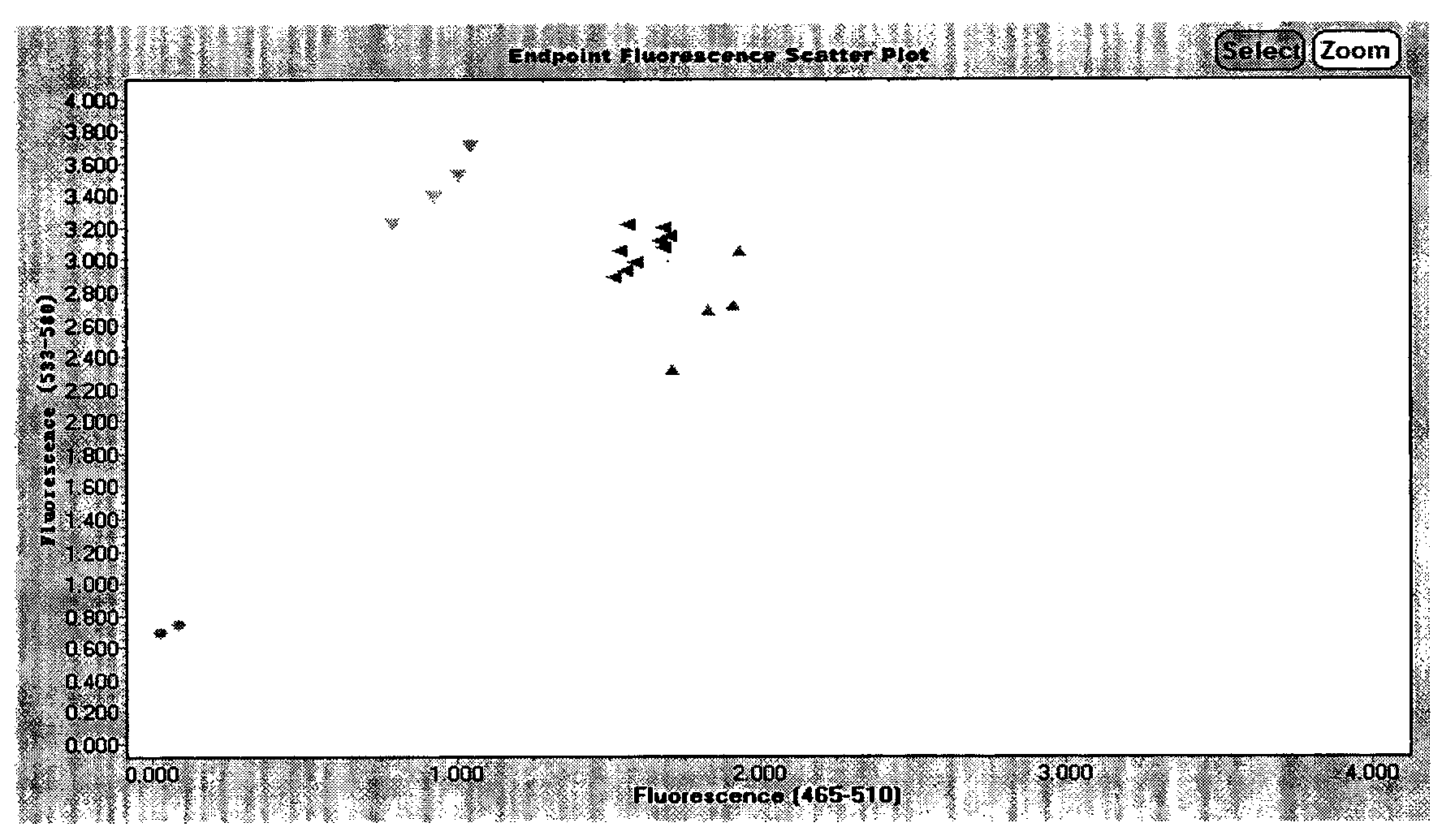
A system and methods are provided for melting curve genotyping analysis of nucleic acids. Melting curves are generated by plotting fluorescence of a sample as a function of temperature. In one illustrative example, an exponential algorithm is employed to remove the background from generated melting curves and thereby perform comparative analysis to other melting curves. Additional illustrative examples provide for measuring the differences between two or more melting curves and clustering the genotypes of the provided sample nucleic acids.
Owner:UNIV OF UTAH RES FOUND
Magnetic fluorescent microspheres and preparation method thereof
InactiveCN101787163AEasy to manufactureSimplify separabilityOrganic/organic-metallic materials magnetismMicroballoon preparationGenotype AnalysisMicroparticle
The invention relates to magnetic fluorescent microspheres and a preparation method thereof. The particle size of the magnetic fluorescent microspheres is 5-10 mu m, the fluorescence excitation wavelength range is 400-700nm, and the fluorescence intensity is not reduced within 24h. The prepration method comprises the following steps: adding a swelling agent into monodisperse carboxylated polystyrene microspheres, and adding magnetic nano microparticles into a swelling system; shaking on a decolorization shaker for 12-48h; using mixed solution of cyclohexane and ethanol for cleaning sediment, and sequentially carrying out ultrasonic dispersion and centrifugal separation till supernatant liquid is colorless under an ultraviolet lamp; and saving a final sediment product in 1ml of liquor. Compared with the existing magnetic fluorescent microspheres, the magnetic fluorescent microspheres have the advantages of uniform and controllable diameter, good fluorescence stability, simple preparation process, multiple types of fluorescence codes and the like, and can not only carry out fast separation and purification on reactants by being applied in the biological macromolecular detection, but also simultaneously detect a plurality of target molecules in a sample to be detected in a reaction tube and a hole, thereby being widely applied in the fields of immunoassay, nucleic acid hybridization, genotype analysis and the like.
Owner:TIANJIN UNIV
Taq ManMGB probe for detecting matrilinear inheritance chondriosome deafness gene C1494T mutation
ActiveCN1987463ASimplified electrophoresis detectionReduce the risk of contaminationMicrobiological testing/measurementBiological testingGenotype AnalysisWild type
Owner:金政策
Probe for detecting matrilinear inheritance chondriosome deafness gene A1555G and its use
ActiveCN1987462ASimplified electrophoresis detectionReduce the risk of contaminationMicrobiological testing/measurementBiological testingFluorescenceGenotype Analysis
The invention designs two pieces of Taqman mutant type and wild type probe and a pair of primer. Using Taqman probe method of real time fluorescence quantitation carries out genotype analysis for A1555G mutation of deaf gene of maternal inheritance mitochondria so as to diagnose genetic deaf disease of maternal mitochondria. The method is suitable to large-scale screening or preventative inspecting A1555G mutation of deaf mitochondria gene of maternal inheritance. Features are: simple, time saving, high specificity, high sensitivity, intuitive tested result, accurate and reliable.
Owner:金政策
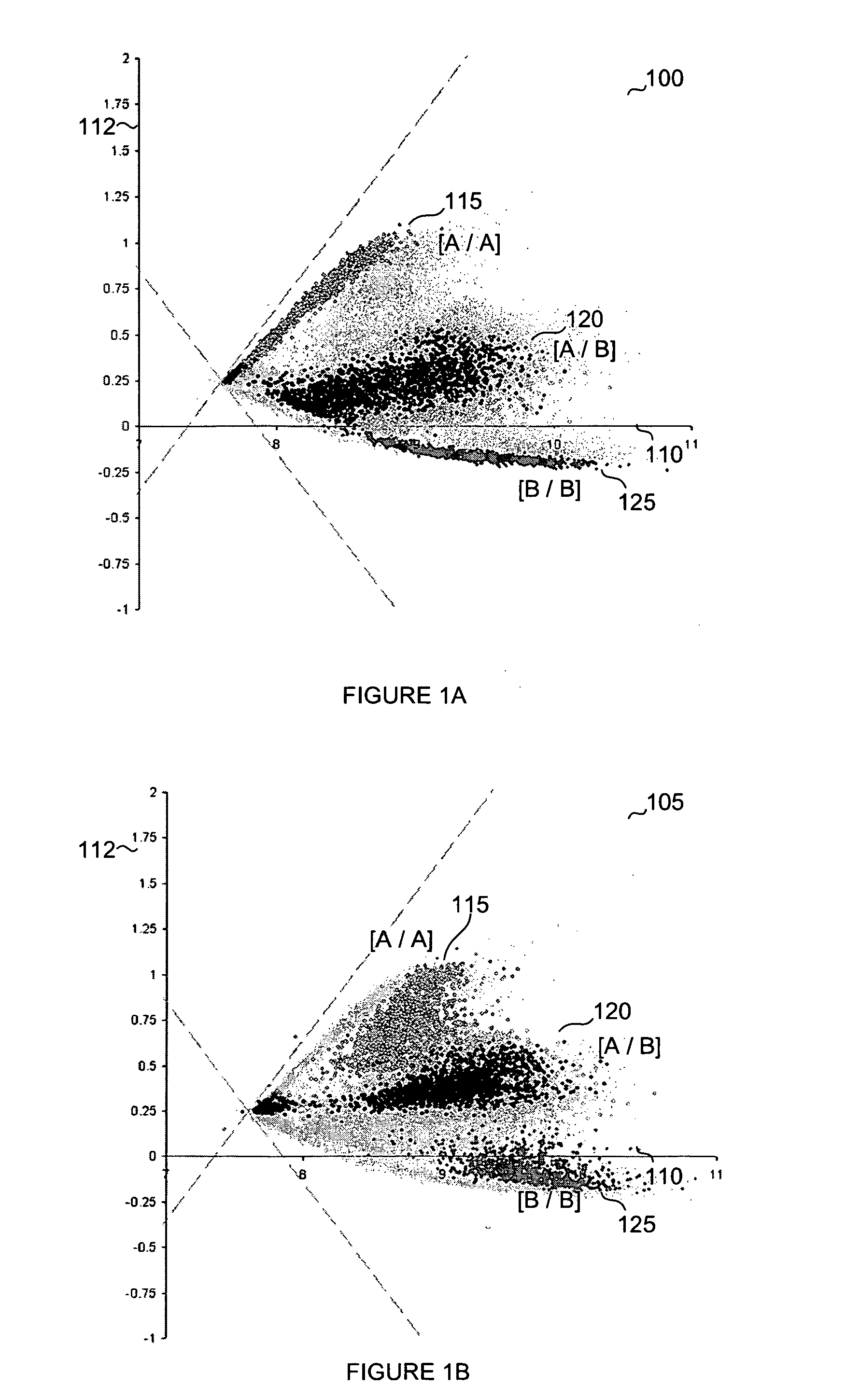
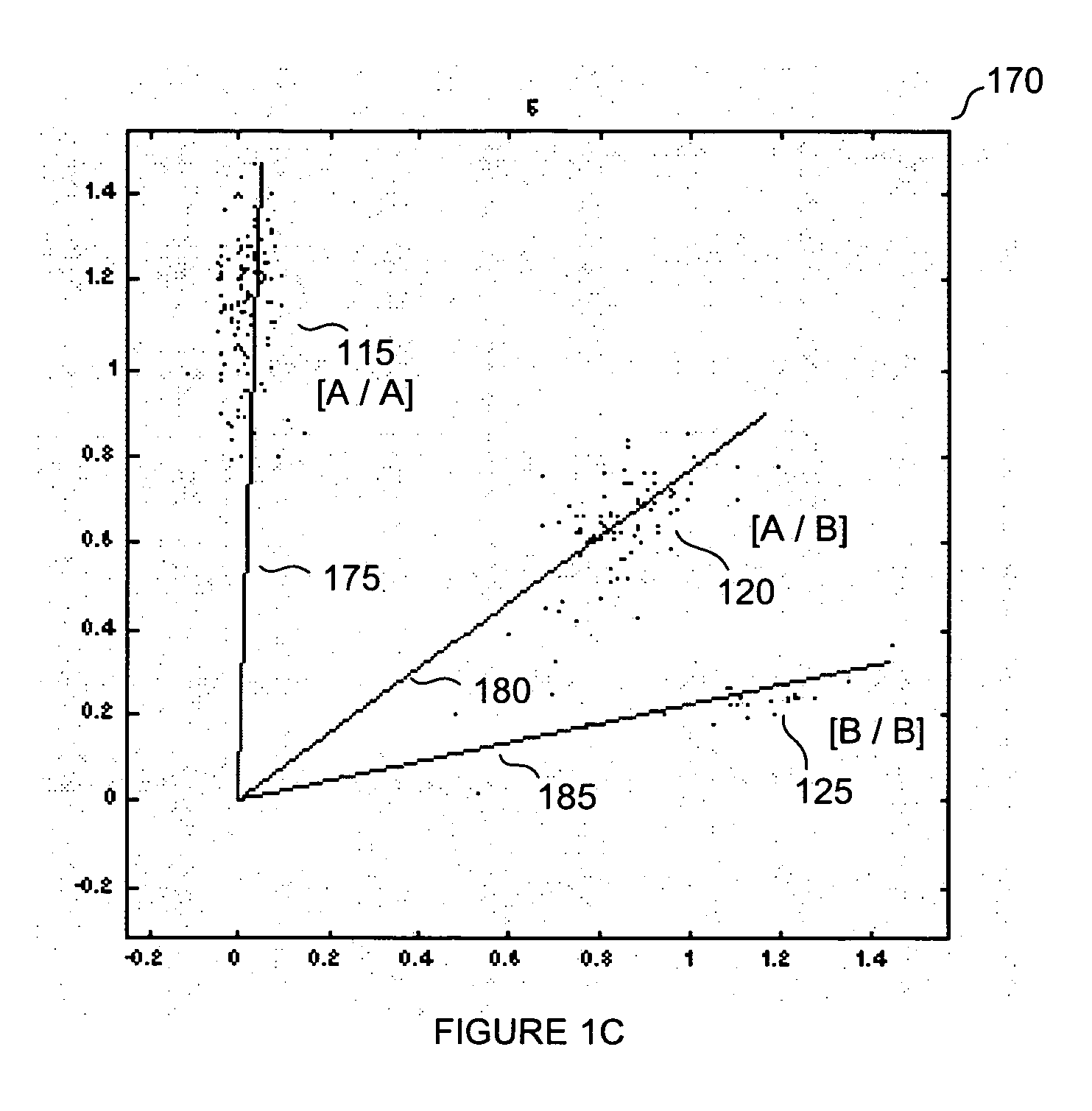
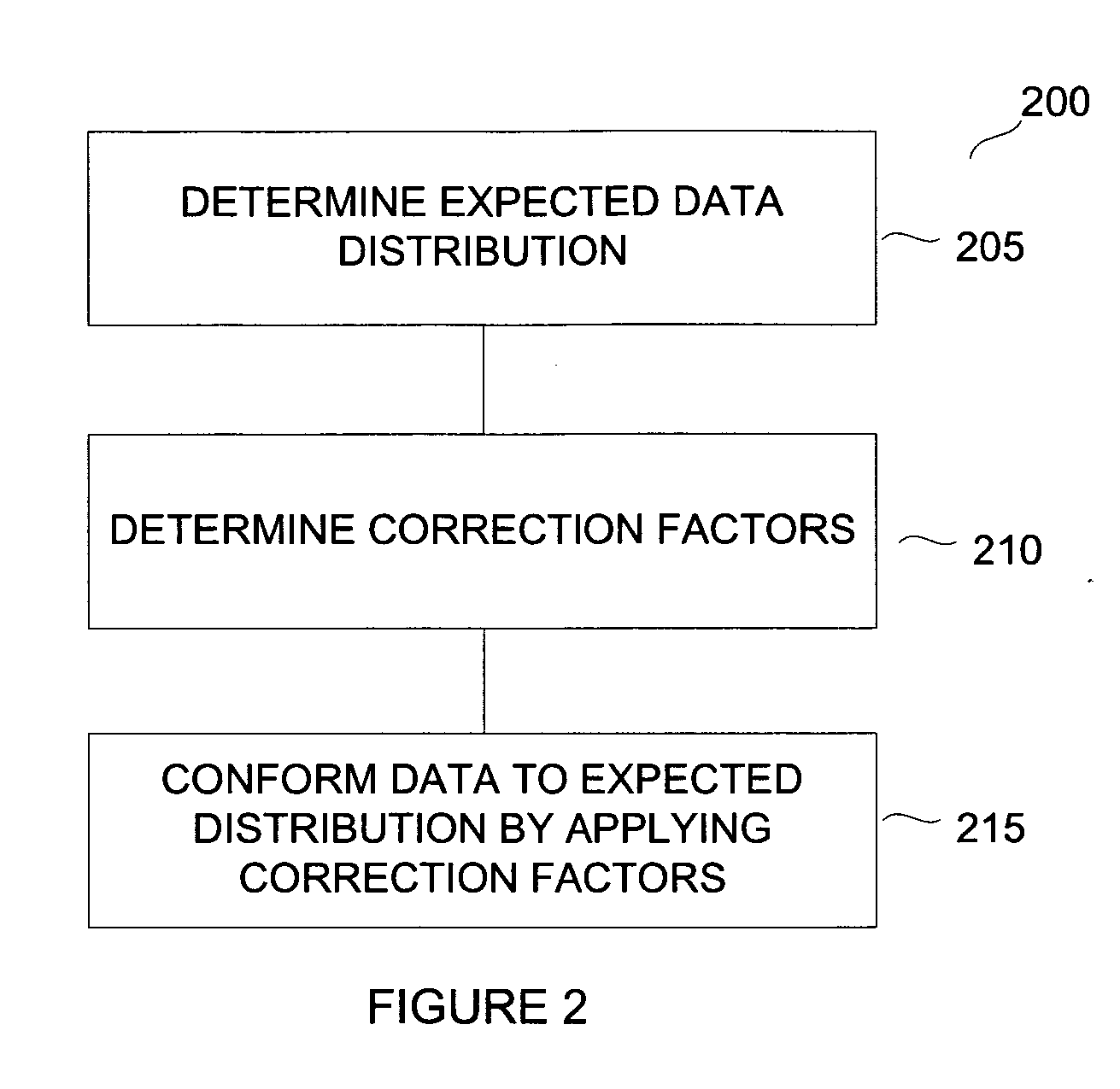
Normalization methods for genotyping analysis
InactiveUS20060178835A1Facilitate comparisonEasy to processBiostatisticsBiological testingData setHigh density
In arrays and other high density analysis platforms variabilities between data points and / or data sets may arise for a number of reasons. Disclosed are methods for addressing these variabilities and generating correction factors that may be used in conforming the data to expected or desired distributions. The methods may be adapted to operate with existing data analysis approaches and software applications to improve downstream analysis.
Owner:APPL BIOSYSTEMS INC
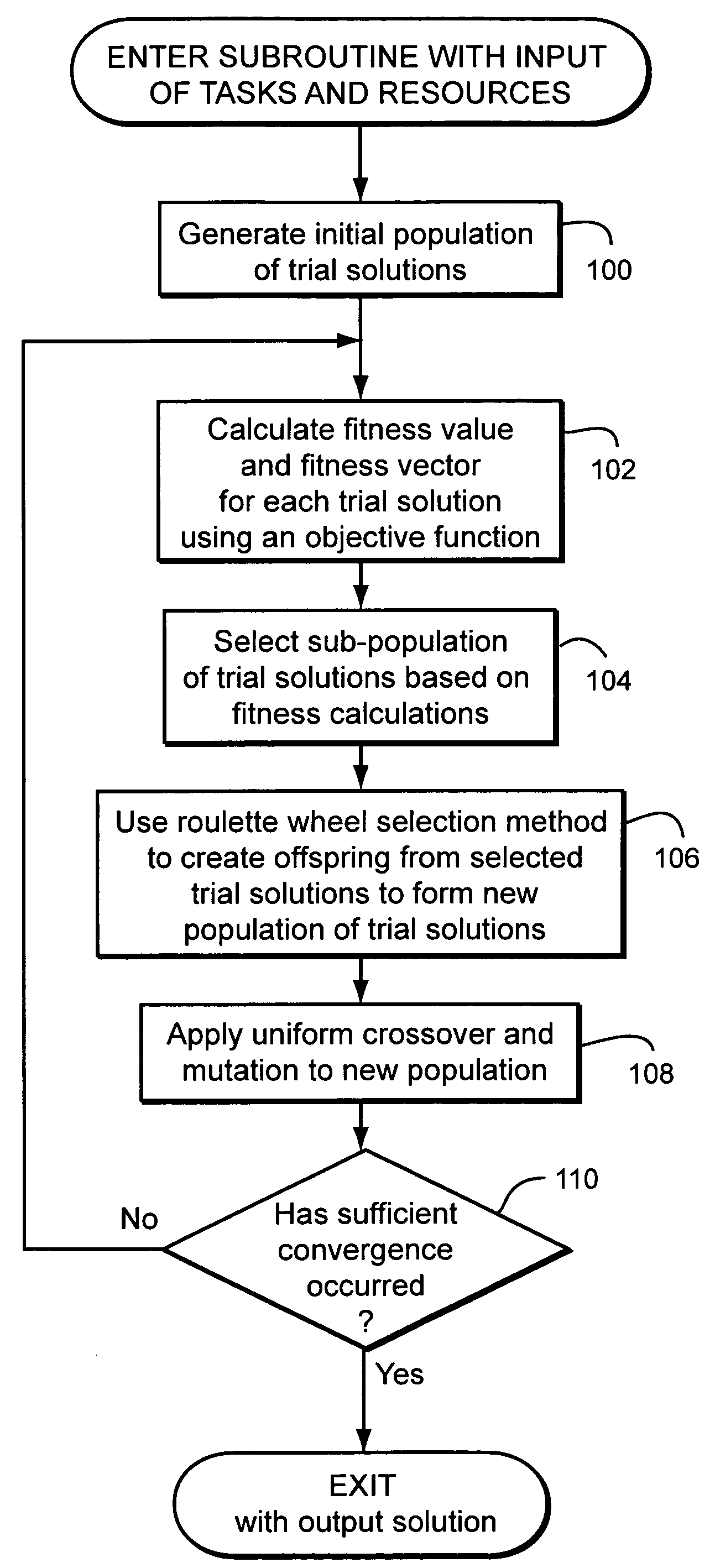
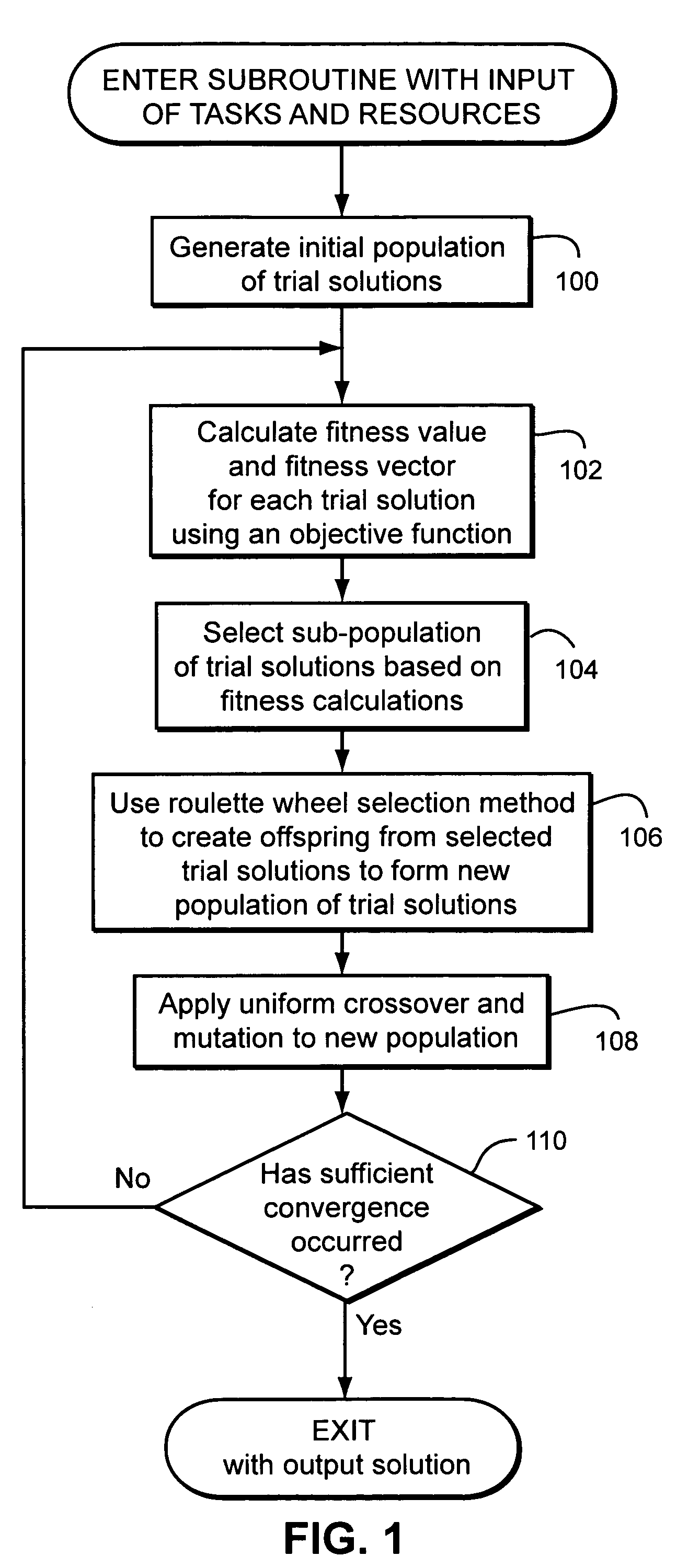
Method of combinatorial multimodal optimisation
InactiveUS7593905B2Reduce the impactDigital computer detailsMultiprogramming arrangementsGenotype AnalysisGenetic algorithm
A method of combinatorial multimodal optimization uses a genetic algorithm to find simultaneous global optimal solutions to combinatorial problems. Each individual within the population is associated not only with a fitness value but with a fitness vector, using which the persistence of all of the best individuals into the next generation can be guaranteed. Phenotype as well as genotype analysis is an integral part of the method.
Owner:BRITISH TELECOMM PLC
System, method, and computer software for genotyping analysis and identification of allelic imbalance
InactiveUS20050250151A1Microbiological testing/measurementData visualisationGenotype AnalysisGenotyping
Methods, systems and computer software products are provided for determining genotype of a sample using a plurality of probes. In one preferred embodiment, a tentative genotype call is made based upon the relative allele signals. Pattern recognition is then used to validate the tentative call.
Owner:AFFYMETRIX INC
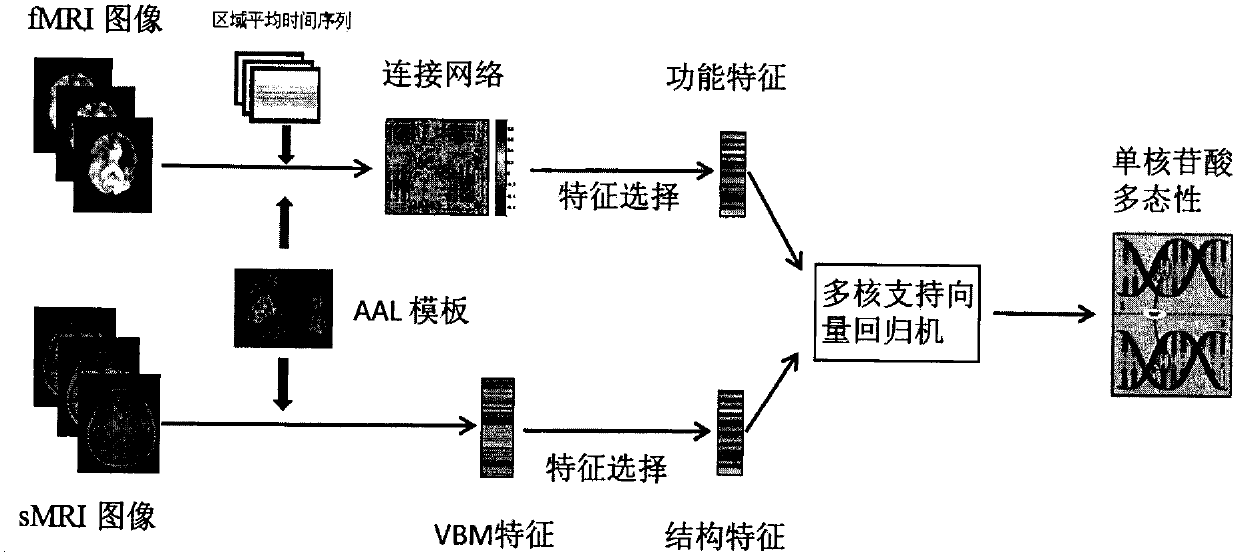
Novel genotype analysis method based on multimodal brain images
InactiveCN107507162AOptimize forecast resultsImage enhancementImage analysisDiagnostic Radiology ModalityNODAL
The invention discloses a novel genotype analysis method based on multimodal brain images. With utilization of information, including a structural image and a functional image, of two kinds of modes, regression on the rs429358 locus genotype linked to the Alzheimer's disease closely is realized. The method comprises: extracting voxel features from a brain structure images and extracting connecting network features from a brain function image; carrying out feature extraction on the two kinds of features by using an elastic network and keeping brain area and network connection with a high association degree; and then carrying out fusion and genotype regression on selected nodes and side attributes by using a multi-kernel support vector. The method has the following advantages: for the brain image with the complicated structure and functions and genotype data, a novel regression analysis method for a single locus genotype based on structural and functional multimodal brain images is put forward; the analysis effect is good; and complementary information of the two modes is utilized fully.
Owner:NANJING UNIV OF AERONAUTICS & ASTRONAUTICS
Methods and devices for correlated, multi-parameter single cell measurements and recovery of remnant biological material
ActiveCN101842159AHeating or cooling apparatusLaboratory glasswaresPhenotype genotypeGenotype Analysis
Methods and apparatus are provided for analysis and correlation of phenotypic and genotypic information for a high throughput sample on a cell by cell basis. Cells are isolated and sequentially analyzed for phenotypic information and genotypic information which is then correlated. Methods for correlating the phenotype-genotype information of a sample population can be performed on a continuous flow sample within a microfluidic channel network or alternatively on a sample preloaded into a nano-well array chip. The methods for performing the phenotype-genotype analysis and correlation are scalable for samples numbering in the hundreds of cells to thousands of cells up to the tens and hundreds of thousand cells.
Owner:CELULA CHINA MED TECH CO LTD
Melting curve analysis with exponential background subtraction
A system and methods are provided for melting curve genotyping analysis of nucleic acids. Melting curves are generated by plotting fluorescence of a sample as a function of temperature. In one illustrative example, an exponential algorithm is employed to remove the background from generated melting curves and thereby perform comparative analysis to other melting curves. Additional illustrative examples provide for measuring the differences between two or more melting curves and clustering the genotypes of the provided sample nucleic acids.
Owner:UNIV OF UTAH RES FOUND
Modified ear tags and method for removing tissue
Owner:IDNOSTICS
Molecular markers closely linked with multiple-effect major quantitative trait loci (QTL) of grain weight or silique length of rape, and application thereof
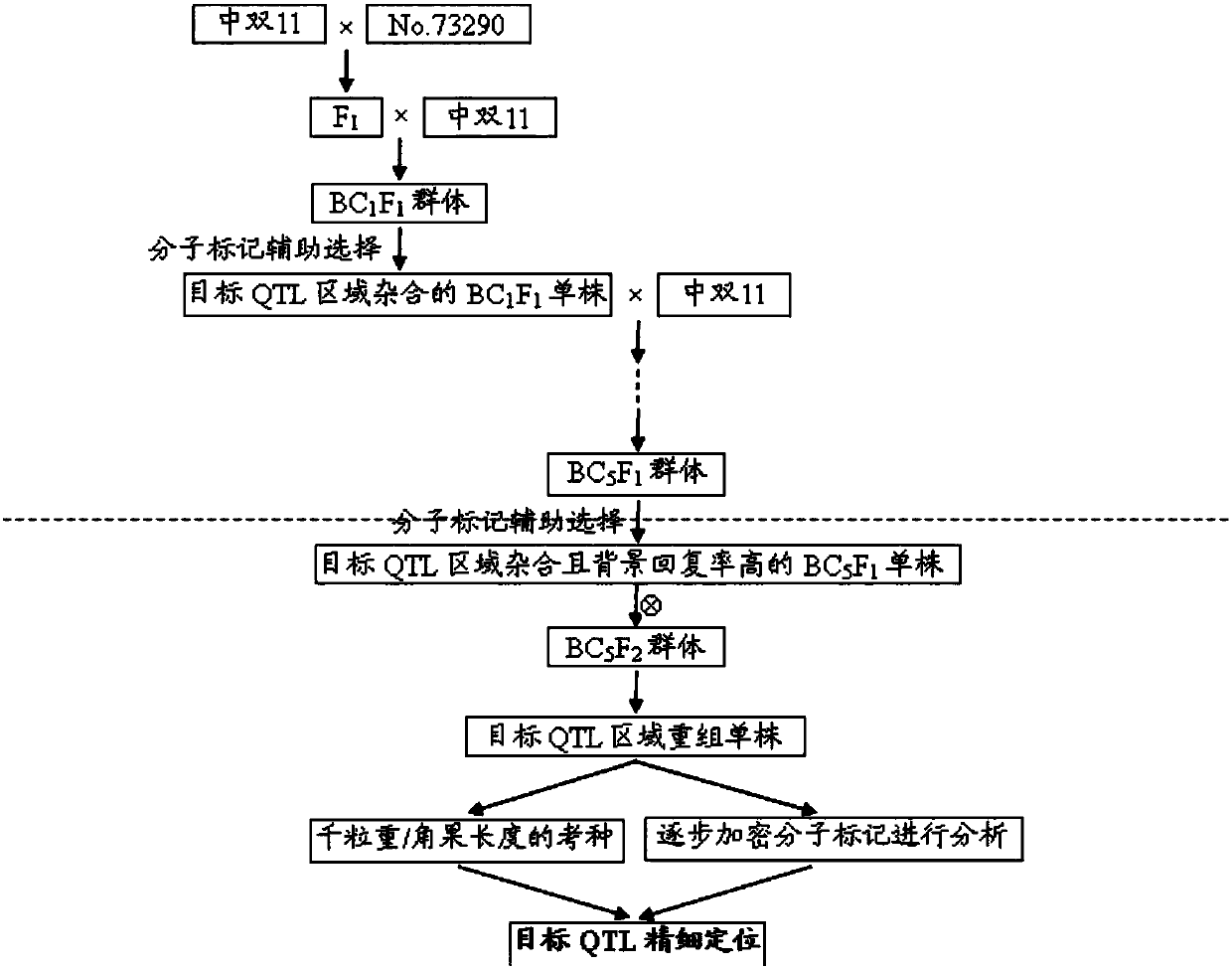
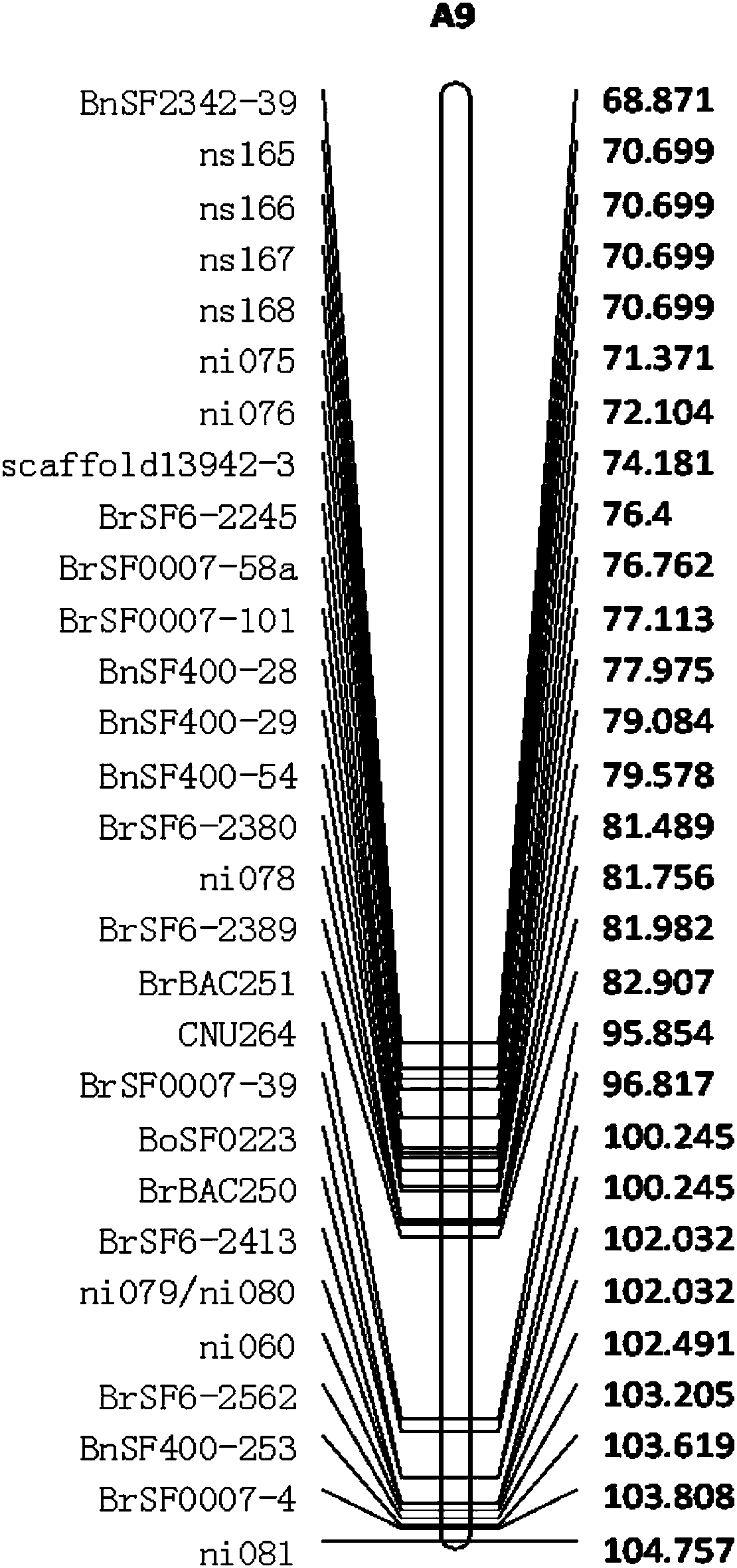
ActiveCN107630099ANot affectedClear locationMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionGenotype Analysis
The invention discloses molecular markers closely linked with multiple-effect major QTL of the grain weight or silique length of rape, and application thereof. The closely-linked molecular markers canbe used for molecular marker-assisted selection of rape and map-based cloning of the QTL. According to the invention, a hybrid F1 and parents are subjected to continuous backcrosses for multiple generations, and a hybrid near-isogenic line is constructed by cooperatively using a molecular marker-assisted selection method; individual plants with high background reversion rate are subjected to selfing so as to obtain a QTL-NIL segregation population; molecular markers are gradually encrypted in a target region so as to realize genotype analysis of the population and searching of crossover individual plants; and thus, the molecular markers BrSF7-101 and BrSF6-2389 are obtained, and the physical distance between the two molecular markers is only 134 kb. The two closely-linked molecular markers are used for genetic typing of linked populations, and it is found that two alleles are greatly different in grain weight and silique length; and thus, the molecular markers can be applied to molecular marker-assisted selection of rape, improve selection efficiency and accelerate breeding progress.
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
Gene knockout method for breeding selection of fhl1b gene-deletion type zebra fish
InactiveCN108048486AEfficient and More Precise SilenceEasy to makeHydrolasesMicroinjection basedDiseaseGenotype Analysis
The invention relates to the technical field of gene knockout and particularly discloses a gene knockout method for breeding selection of a fhl1b gene-deletion type zebra fish. According to the method, by means of a CRISPR / Cas9 gene editing technique, an appropriate target locus is designed on a fhl1b gene of the zebra fish, specific sgRNA and Cas9-mRNA which are synthesized outside the fish bodyare microscopically injected into one cell of the zebra fish, and 60 hours after embryo cultivation is carried out, by selecting embryos to conduct genotype analysis, the effectiveness of the selectedlocus is proved. The off-target rate is low, not only is the fhl1b gene removed by interference, but also more convenience is provided for further disclosing the whole process of generation of a heart shape and regulating and controlling a molecular mechanism of the process by adopting a genetic approach to studying the functions of the fhl1b gene, and the method has a significant meaning in understanding of medical pathology of heart diseases and research and development on new therapeutic schemes.
Owner:HUNAN NORMAL UNIVERSITY
Method for evaluating inheritance effect of SNP (Single Nucleotide Polymorphism) sites to traits and application thereof
InactiveCN103146821AImprove accuracyFast analysisMicrobiological testing/measurementSpecial data processing applicationsAnalysis aspectCalculation methods
The invention relates to a method for evaluating the inheritance effect of SNP (Single Nucleotide Polymorphism) sites to traits. The method comprises the following steps of: (1) obtaining gene fragments related to traits to be analyzed, and determining SNP sites related to the traits and genotypes of the SNP sites; and (2) analyzing the relevance between the SNP sites and the traits, wherein the specific calculation method is as follows: when the number of the SNP sites is one, the relevance between the SNP site and the traits is the direct influence of the SNP site, and when the number of the SNP sites is more than two, the relevance between the i-th SNP site and the traits is the sum of the direct influence and all indirect influence of the i-th SNP site. The method has the advantages that the problem in the prior art that blanks exist in the screening and evaluating of the inheritance effect of the SNP sites to the traits is solved; the method is simple, convenient and feasible, is high in accuracy and high in analyzing speed, so that many complicated tests are not needed; and the method is time-saving and labor-saving.
Owner:ANHUI AGRICULTURAL UNIVERSITY
Cucumber variety identification method based on SNP markers
PendingCN104846062ASimple and fast operationSuitable for high-throughput operationsMicrobiological testing/measurementDNA/RNA fragmentationGenotype AnalysisMicrobiology
The present invention discloses a cucumber variety identification method based on single nucleotide polymorphism markers, wherein 12 single nucleotide polymorphism markers of cucumber are subjected to genotyping analysis by using a pyrosequencing technology so as to achieve authenticity identification on the cucumber variety. With the detection method of the present invention, the cucumber variety authenticity identification can be completed within 5 h, and characteristics of stability, high-throughput, accuracy and the like are provided.
Owner:TIANJIN INSTITUE OF QUALITY STANDARD & TESTING OF AGRICULTUAL PRODS
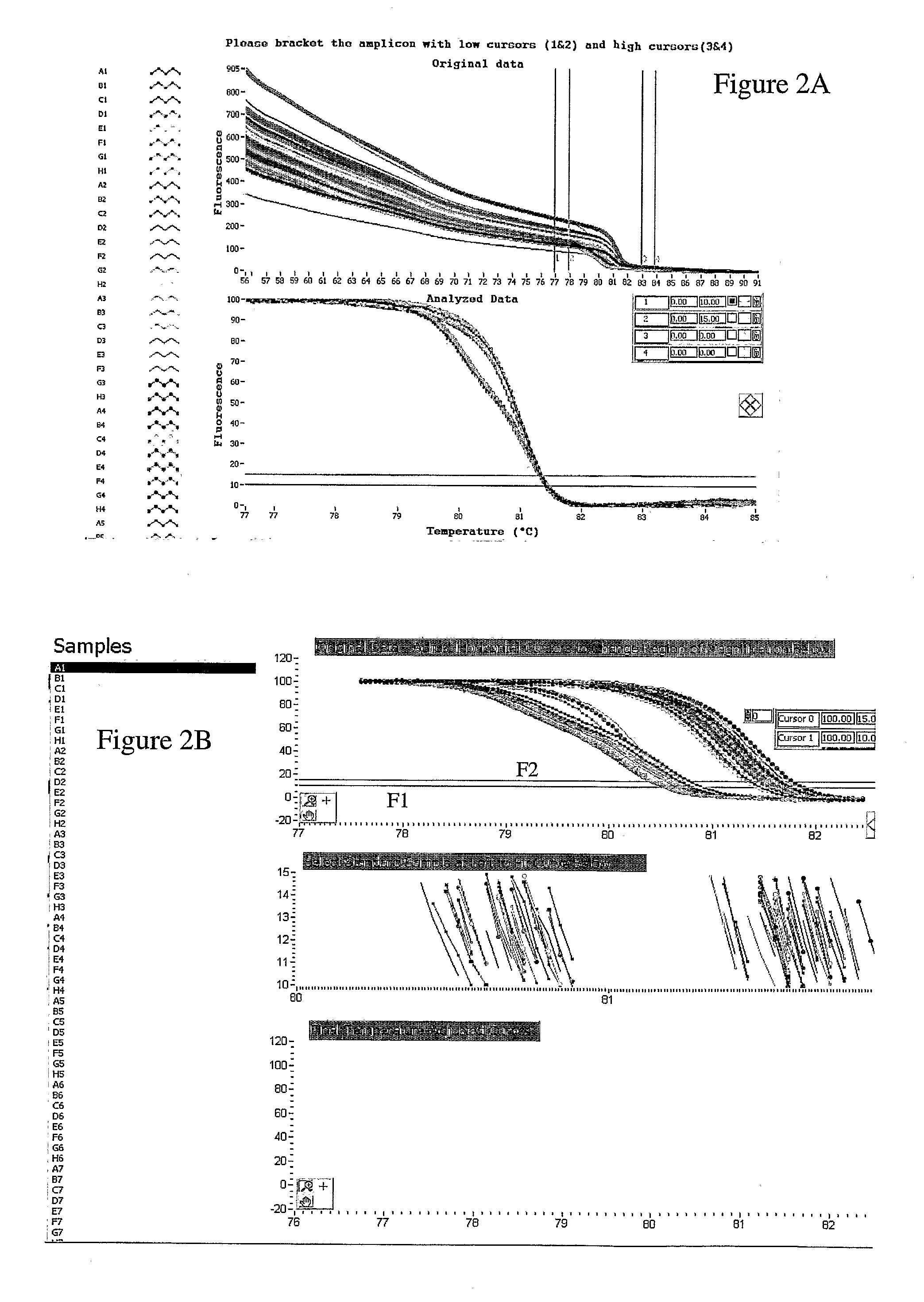
System and method for genotype analysis and enhanced monte carlo simulation method to estimate misclassification rate in automated genotyping
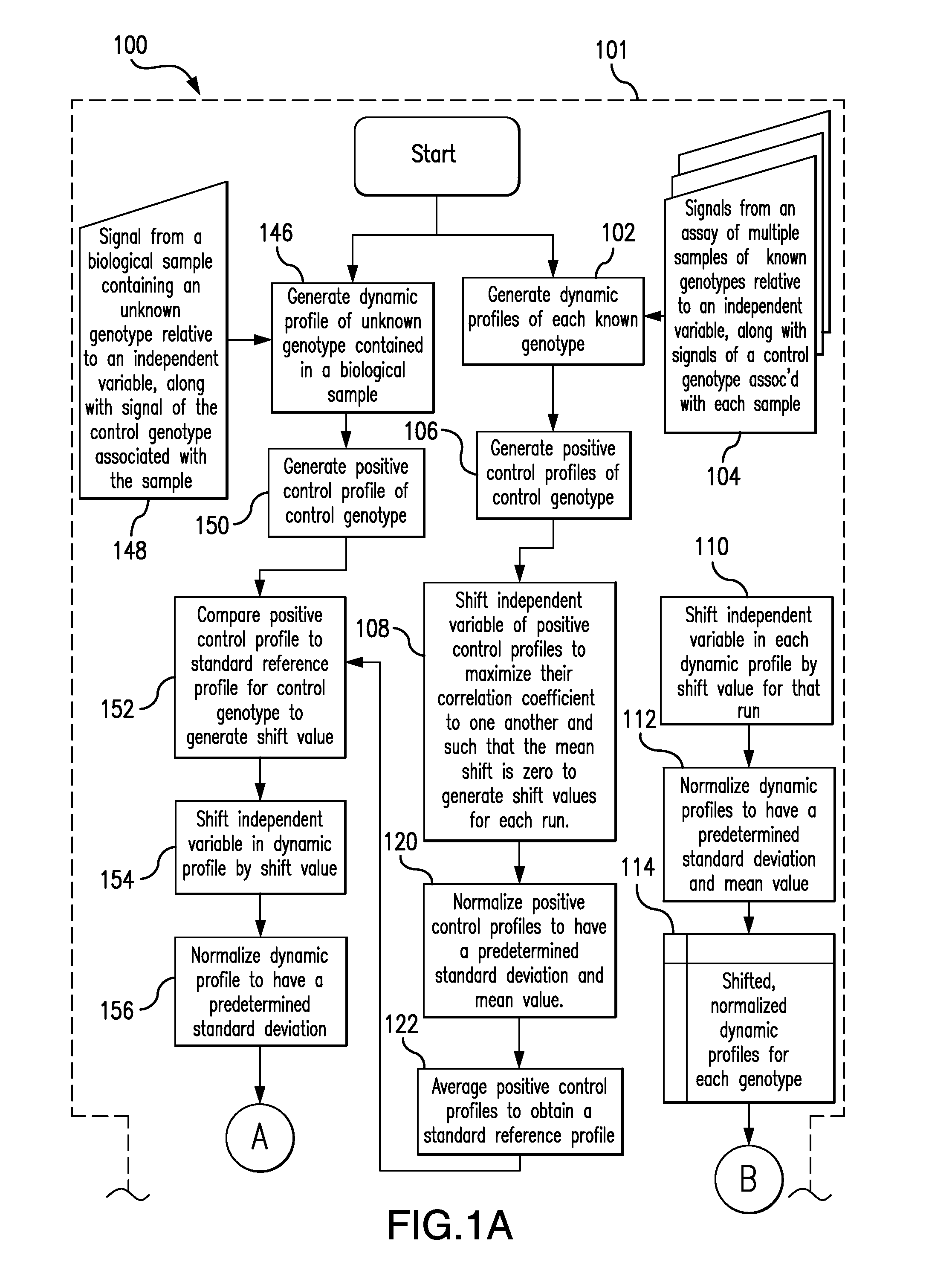
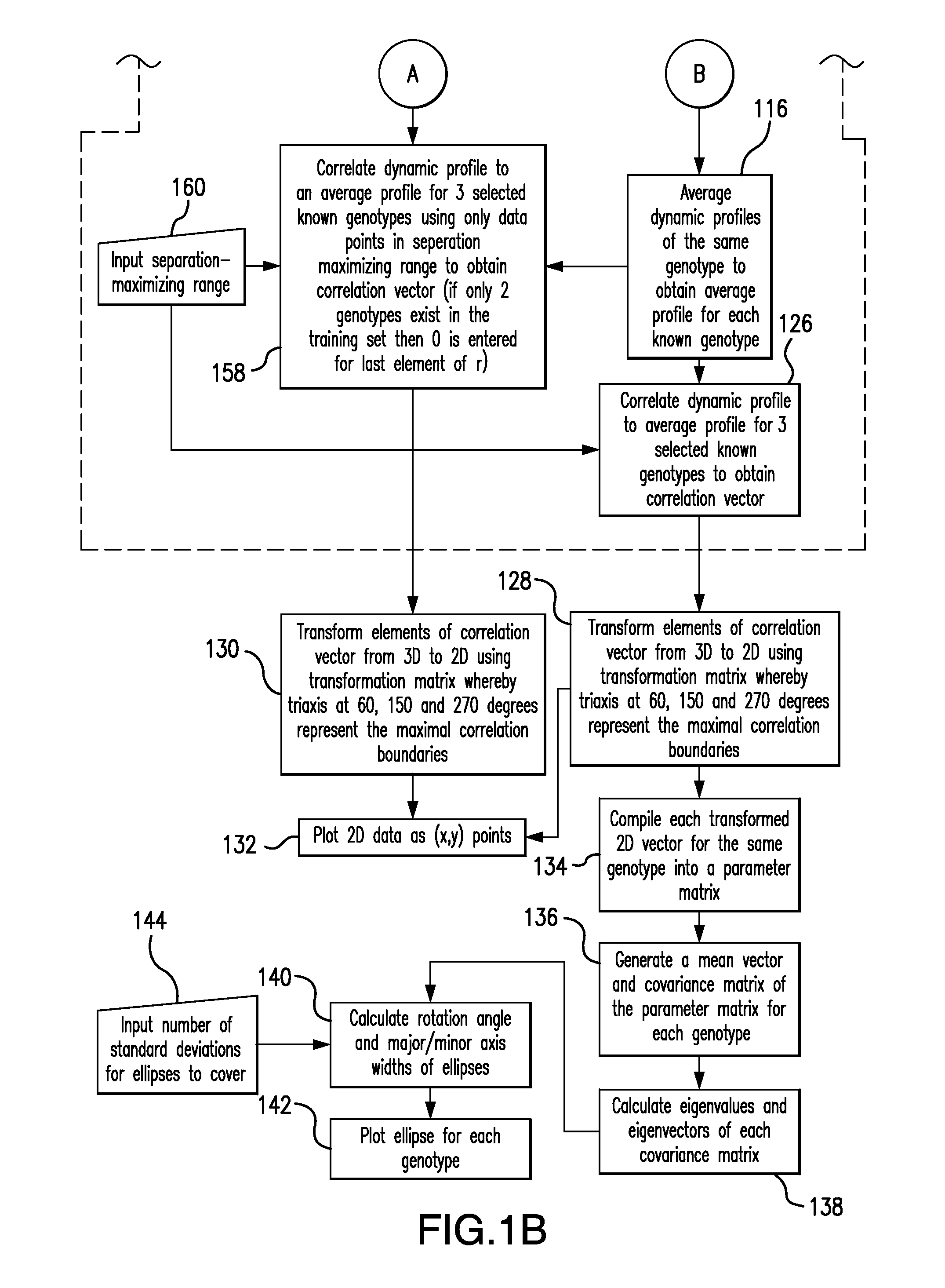
The present invention relates to methods and systems for the analysis of the dissociation behavior of nucleic acids. The present invention includes methods and systems for analyzing dynamic profiles of genotypes of nucleic acids, including the steps of using a computer, including a processor and a memory, to convert dynamic profiles of known genotypes of a nucleic acid to multi-dimensional data points, wherein the dynamic profiles each comprise measurements of a signal representing a physical change of a nucleic acid containing the known genotype relative to an independent variable; using the computer to reduce the multi-dimensional data points into reduced-dimensional data points; and generating a plot of the reduced-dimensional data points for each genotype. The present invention also relates to methods and systems for calculating error statistics for an assay for identifying a genotype in a biological sample using an enhanced Monte Carlo simulation method to generate a set of N random data points for each known genotype within a class of known genotypes, where each set of N random data points has the same mean data point and covariance matrix as a data set for each of the known genotypes.
Owner:CANON US LIFE SCIENCES INC
Kit and method for detecting porcine circovirus type-2 (PCV-2) and subtypes thereof
InactiveCN102206715AThe result is easy to judgeEasy to makeMicrobiological testing/measurementFluorescence/phosphorescenceGenotype AnalysisFluorescent pcr
The invention provides a kit for detecting porcine circovirus type-2 (PCV-2) and subtypes thereof, belonging to the field of biological detection and solving the technical problems of complex process and long period of a method for detecting the PCV-2 and the subtypes thereof. The kit internally comprises fluorescent PCR (Polymerase Chain Reaction) reaction liquid, a TaqMan specific probe a and a TaqMan specific probe b, wherein the fluorescent PCR reaction liquid contains specific primers, the sequence of the upstream primers is as shown in SEQIDNO: 3, and the sequence of the downstream primers is as shown in SEQIDNO: 4; the TaqMan specific probe a aims at a PCV-2a (Porcine Circovirus-2a); and the TaqMan specific probe b aims at a PCV-2b (Porcine Circovirus-2b). The invention also provides a method for detecting the PCV-2 and the subtypes thereof by adopting the kit. The bifluorescence PCR detection kit can fast and accurately detect the PCV-2 and also identify the subtypes of the PCV-2 and is suitable for the fast diagnosis of the PCV-2, epidemiological survey, risk assessment and genotyping analysis.
Owner:SHANGHAI ANIMAL EPIDEMIC PREVENTION & CONTROL CENT
Computer software for genotyping analysis using pattern recognition
Methods, systems and computer software products are provided for determining genotype of a sample using a plurality of probes. In one preferred embodiment, a tentative genotype call is made based upon the relative allele signals. Pattern recognition is then used to validate the tentative call.
Owner:AFFYMETRIX INC
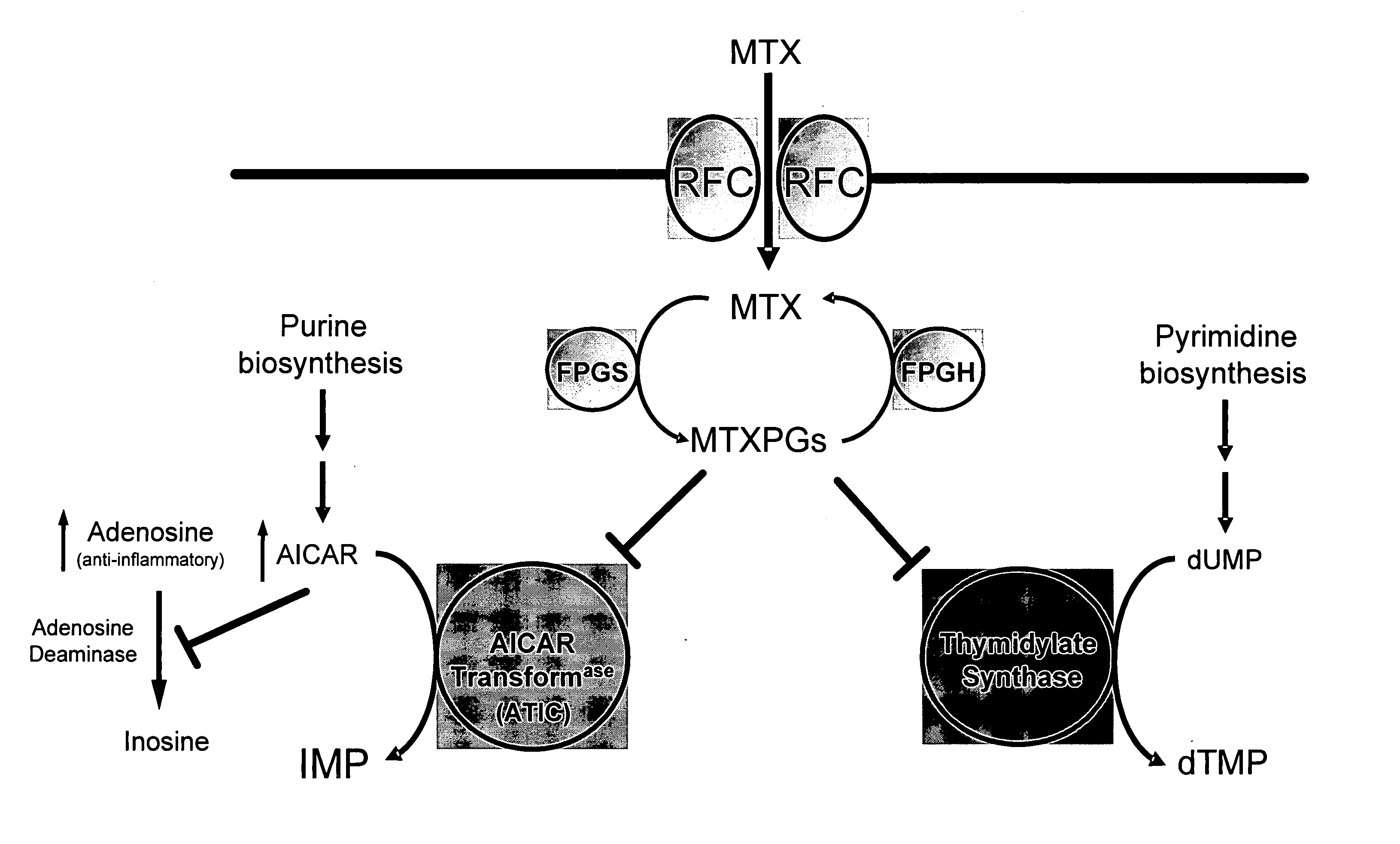
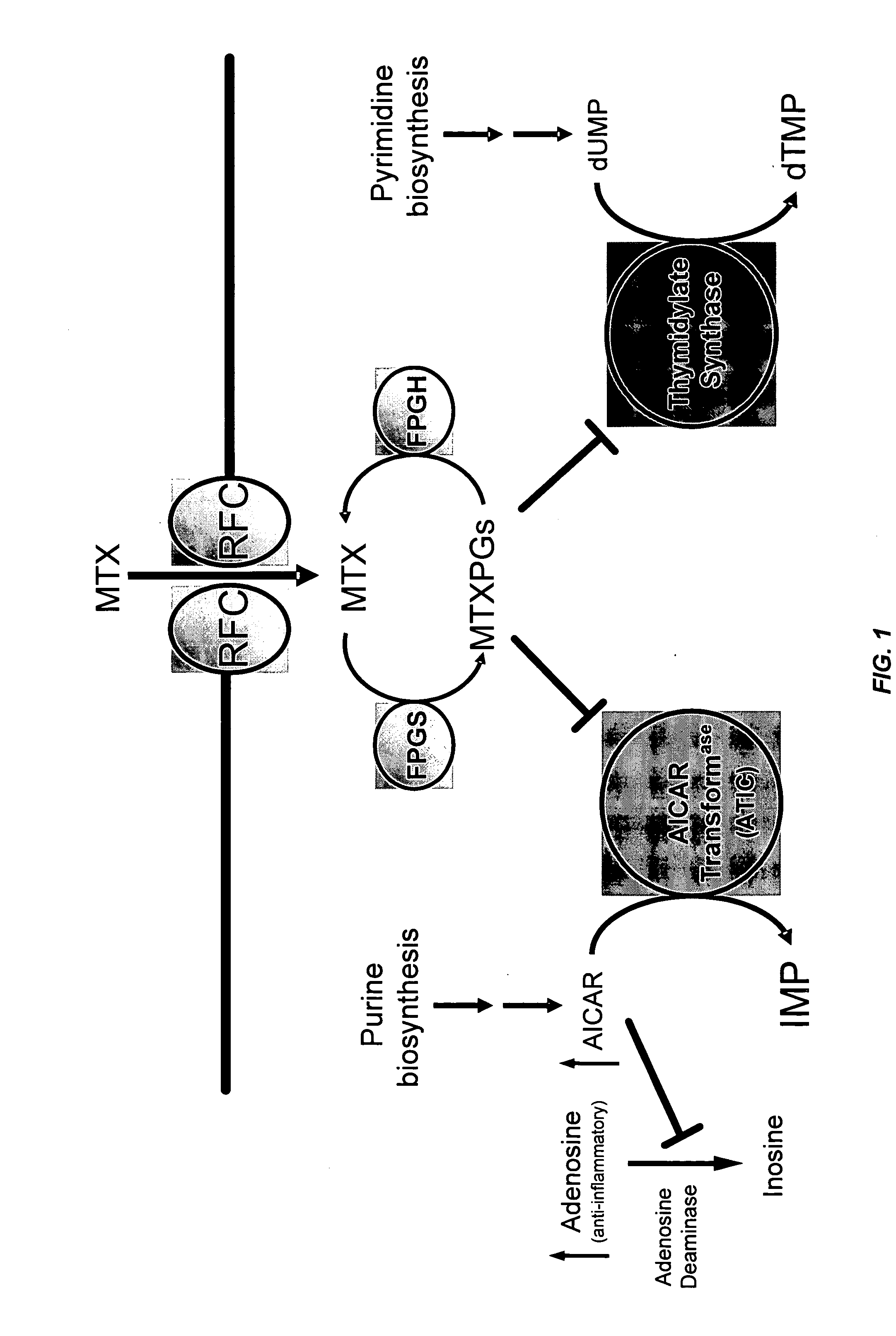
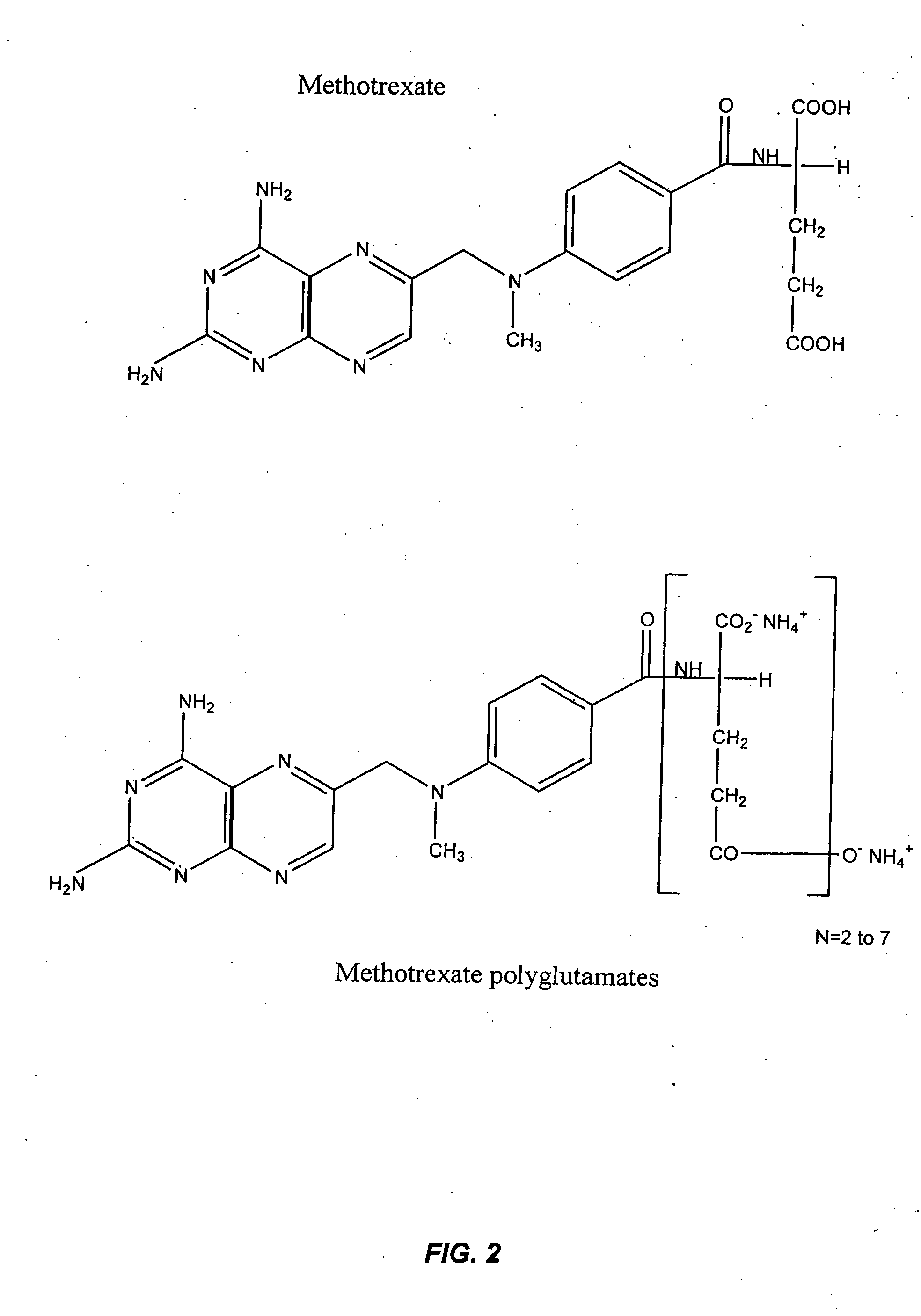
Methods for optimizing clinical responsiveness to methotrexate therapy using metabolite profiling and pharmacogenetics
The present invention provides methods for optimizing clinical responsiveness to chemotherapy in an individual through genotypic analysis of polymorphisms in at least one gene. The methods of the present invention may further comprise determining the level of at least one long-chain methotrexate polyglutamate (MTXPG) in a sample obtained from the individual. The present invention also provides methods for generating a pharmacogenetic index for predicting clinical responsiveness to chemotherapy in an individual through genotypic analysis of polymorphisms in at least one gene. In addition, the present invention provides methods for optimizing therapeutic efficacy of chemotherapy in an individual by calculating the level of at least one long-chain MTXPG in a sample obtained from the individual.
Owner:EXAGEN INC
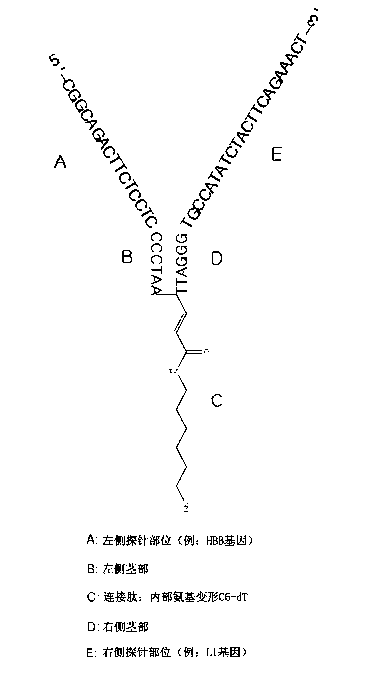
Y-probe and variation thereof, and DNA microarray, kit, and gene analysis method using the Y-probe and the variation thereof
ActiveCN103097533AAccurate diagnosisFast and Accurate DiagnosisSugar derivativesMicrobiological testing/measurementNucleotideGenotype Analysis
Owner:GOODGENE
Method for screening candidate bacterial strain from fish streptococcus agalactiae vaccine
ActiveCN102676683AHigh immune protection rateLarge range of protectionMicrobiological testing/measurementMicroorganism based processesGenotype AnalysisBacterial strain
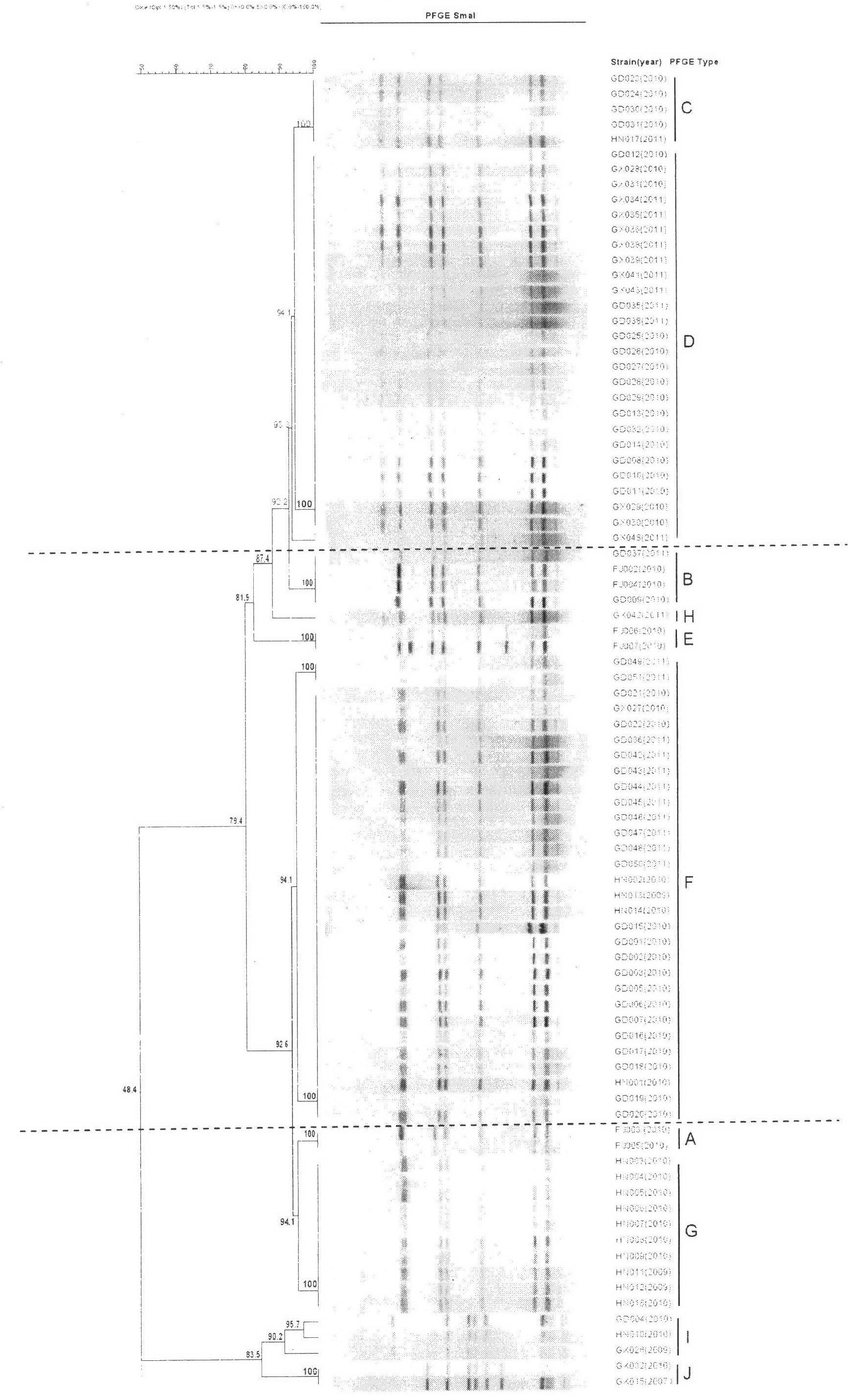
The invention discloses a method for screening candidate bacterial strains from fish streptococcus agalactiae vaccines. The method comprises the steps: separation and breed conservation of tilapia streptococcicosis agalactiae epidemic strains, identification of the epidemic strains and the establishing of a pathogenic library, PFGE genotype analysis of epidemic strains, toxicity determination of PFGE genotype representative strains, and immunogenicity and protection domain test of the PFGE genotype representative strains so as to obtain a candidate bacterial strains combination of a tilapia streptococcicosis agalactiae immunoprophylaxis vaccine in China. The technology has the characterizes of strong pertinence, high screening efficiency, remarkable effect and the like, and the screen candidate bacterial strains combination of the tilapia streptococcosis agalactiae immunoprophylaxis vaccine in China can protect 90% of genotypes and 96.47% of epidemic strains in the pathogenic library. The technology provides a new method for screening the candidate bacterial strains from the fish streptococcus agalactiae vaccines, is suitable for screening the candidate bacterial strains from the fish streptococcus agalactiae vaccines, and has significance and using value in immune prevention and control on fish streptococcicosis.
Owner:GUANGXI INST OF FISHERIES
System, method, and computer software for genotyping analysis and identification of allelic imbalance
InactiveUS6988040B2Microbiological testing/measurementData visualisationGenotype AnalysisComputer software
Owner:AFFYMETRIX INC
Gene knockout breeding method for tnni3k gene deletion zebrafish
PendingCN109266687AEfficient and More Precise SilenceEasy to makeHydrolasesMicrobiological testing/measurementGenotype AnalysisEmbryo
The invention relates to the technical field of gene knockout, in particular, the invention discloses a gene knockout breeding method for tnni3k gene deletion zebrafish, which adopts CRISPR / Cas9 geneediting technology to design a suitable target site on the tnni3k gene of zebrafish, and synthesizes specific gRNA and Cas9-protein was co-injected into zebrafish cells. After 48 hours of embryo culture, the embryos were selected for genotypic analysis, which confirmed the validity of the selected sites. The invention can more efficiently and more accurately silence a specific gene in the genome of an organism, and has the advantages of simple production, low cost, and can simultaneously cut a plurality of loci on a target gene and silence any number of single genes. At that same time, zebrafish embryo interfered with the tnni3k gene appear obvious developmental abnormalities.
Owner:HUNAN NORMAL UNIVERSITY
Detection method and kit for coronary heart disease susceptibility loci rs1801133
ActiveCN104059990AEasy to operateReduce use costMicrobiological testing/measurementGenotype AnalysisCoronary heart disease
The invention relates to a detection method and a kit for coronary heart disease susceptibility loci. The method comprises the following steps: (1) extracting a genome deoxyribonucleic acid (DNA) of a sample, and amplifying a region containing rs1801133 loci in a methylene tetrahydrofolate reductase (MTHFR) gene exon of the sample, so as to obtain an amplification product; (2) detecting the genetype of single nucleotide polymorphism (SNP) loci rs1801133 in the amplification product by using a high resolution melting (HRM) analysis technology. The genetype analysis is carried out on the region containing the rs1801133 loci in the amplified MTHFR gene by adopting the HRM analysis technology. The method is simple and quick to operate, low in use cost and accurate in result, and can achieve batch inspection; whether the tested people carry with coronary heart disease susceptibility genes can be clearly judged by using the kit, the coronary heart disease susceptibility people can be screened out from people, the poor living habit is improved, and the target of prevention is achieved.
Owner:光翰科技(上海)有限公司
Seed number per pod character major gene site of rape and application thereof
InactiveCN102226189AThe detection method is convenient and fastPrecise screeningMicrobiological testing/measurementFermentationField experimentGenotype Analysis
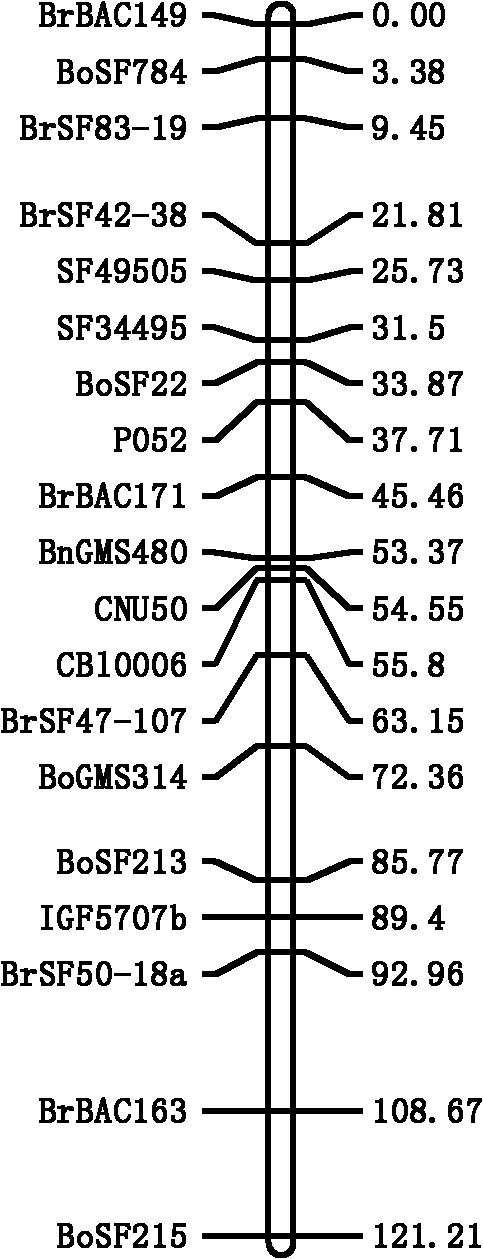
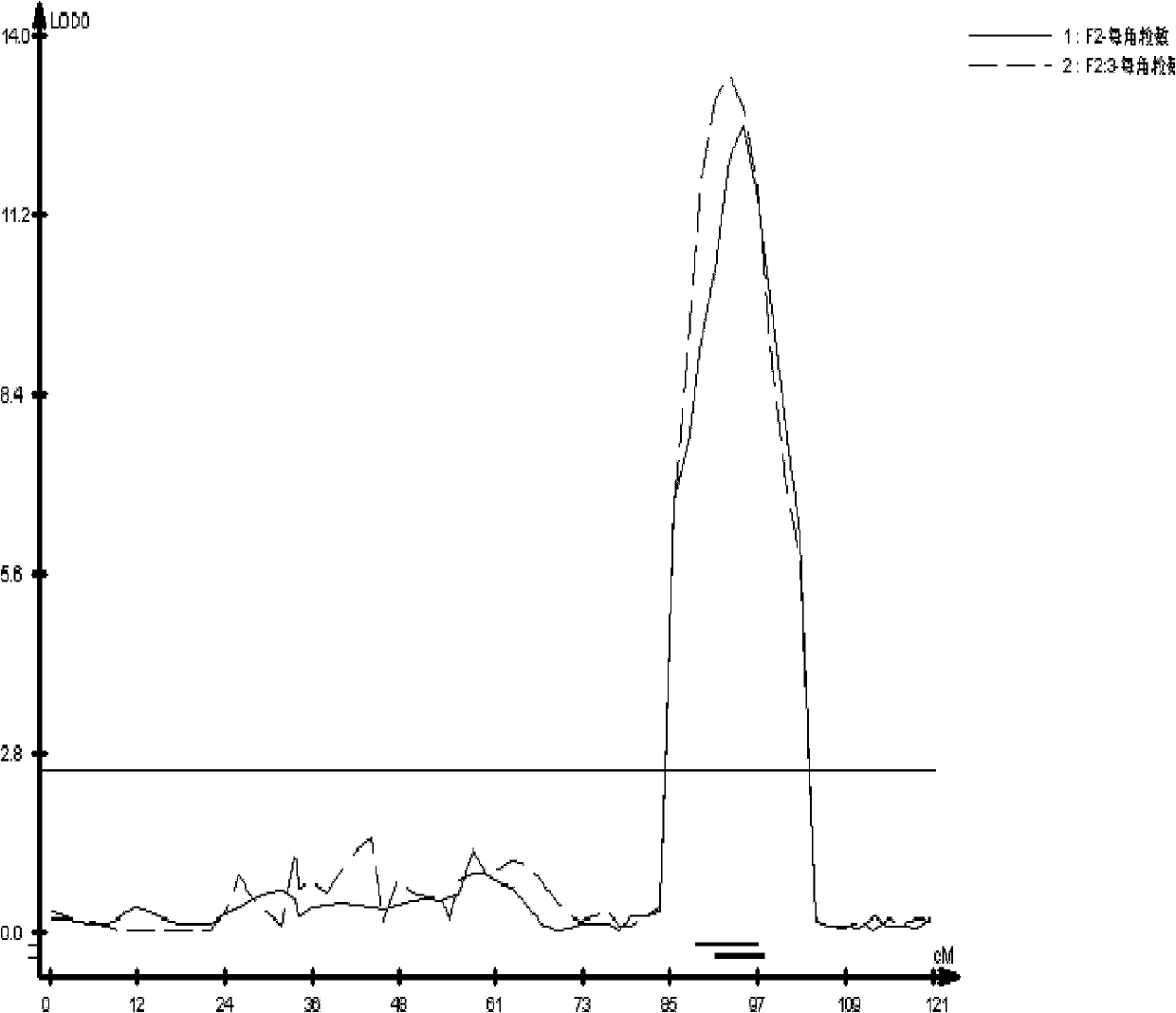
The invention discloses a seed number per pod character major gene site of rape and application thereof. The process comprises the following steps: (1) hybridizing by Brassica napus variety with obvious difference on each seed number per pod character; (2) carrying out polymorphism screening for parent DNA by public and developed SSR (Simple Sequence Repeat) and SNP (Single Nucleotide Polymorphism) primers, and building a genetic linkage map by molecular mark gene type analysis to an F2-generation segregation population; (3) obtaining the phenotype data of each seed number per pod character by field experiments and variety research to F2 and F2:3 segregation population; and (4) combining the gene type and the phenotype data of the segregation population to carry out QTL (Quantitative Trait Loci) detection. The major gene site qQN.A6 and the molecular marker BrSF50-18 for controlling seed number per pod of rape on an A6 linkage group are obtained. The F3 generation derived from two parents is analyzed by the marker, and an individual plant with the marker is kept. A variety research result shows that the ratio of the seed number per pod is 83.4% higher than the F3 individual plate of F2:F3 family mean value, and thus, the marker is used for assisted selection to greatly improve the selection efficiency of high-yield breeding.
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
Multiple PCR primer for expanding plasmodium falciparum drug resistance-related gene and method
ActiveCN101161822AImprove detection efficiencyReduce dosageMicrobiological testing/measurementFermentationGenotype AnalysisMultiplex pcrs
The invention relates to a plasmodium falciparum drug resistance related gene multiple PCR amplification method, wherein, plasmodium falciparum genome DNA is adopted as template; a multiple PCR primer set designed by the invention is adopted for monotube PCR reaction to realize amplification of five specific gene (plasmodium falciparum chloroquine resistance transport protein gene Pfcrt, plasmodium falciparum multidrug-resistance gene Pfmdrl, plasmodium falciparum dihydropteroic synthetase gene Pfdhps, plasmodium falciparum dihyrofolate reductase gene Pfdhfr and plasmodium falciparum adenosine triphosphatase Sixth subunit gene PfATPase6) target sequences; moreover, the amplification product covers the currently known 21 plasmodium falciparum drug-resistance related SNP sites. With fewer operational procedures, the invention is quick and convenient and saves cost; moreover, the invention which is particularly suitable for high-throughput genotype analysis is sure to accelerate the wide application of plasmodium falciparum resistance related SNP in field monitoring.
Owner:STATION OF VIRUS PREVENTION & CONTROL CHINA DISEASES PREVENTION & CONTROL CENT
Single nucleotide polymorphism sequence of dairy cattle FGF2 (Fibroblast Growth Factor 2) gene and detection method of single nucleotide polymorphism sequence
InactiveCN102533774AImprove survival rateMicrobiological testing/measurementFermentationInteinGenotype Analysis
The invention relates to a single nucleotide polymorphism sequence of a dairy cattle FGF2 (Fibroblast Growth Factor 2) gene and a detection method of the single nucleotide polymorphism sequence, belonging to the technical field of genetic engineering. The single nucleotide polymorphism sequence of the dairy cattle FGF2 gene comprises a single nucleotide polymorphism sequence of which the 11646th site of a first intron of the dairy cattle FGF2 gene is A or G. The detection method comprises the following steps of: with dairy cattle genome DNA as a template and a primer pair P as a primer, carrying out PCR (Polymerase Chain Reaction) amplification on the dairy cattle FGF2 gene; and sequencing a purified PCR product to determine that the single nucleotide polymorphism 11646th site of the dairy cattle FGF2 is A or G. According to the single nucleotide polymorphism sequence, the SNP (Single nucleotide Polymorphism) site of the dairy cattle FGF2 gene is related to butterfat production, butterfat percentage, somatic cell count and reproductive capacity; and found through genotyping an externally-fertilized embryo, the SNP site is related to the embryo survival rate; and through association analysis study on FGF2 genetic polymorphism and character, the SNP site can be used for dairy cattle molecular mark assisted breeding and increase of the externally-fertilized embryo survival rate.
Owner:NORTHWEST A & F UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com