Gene knockout method for breeding selection of fhl1b gene-deletion type zebra fish
A gene deletion and gene knockout technology, applied in the field of gene knockout, can solve the problems of high off-target rate and low efficiency of targeting technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
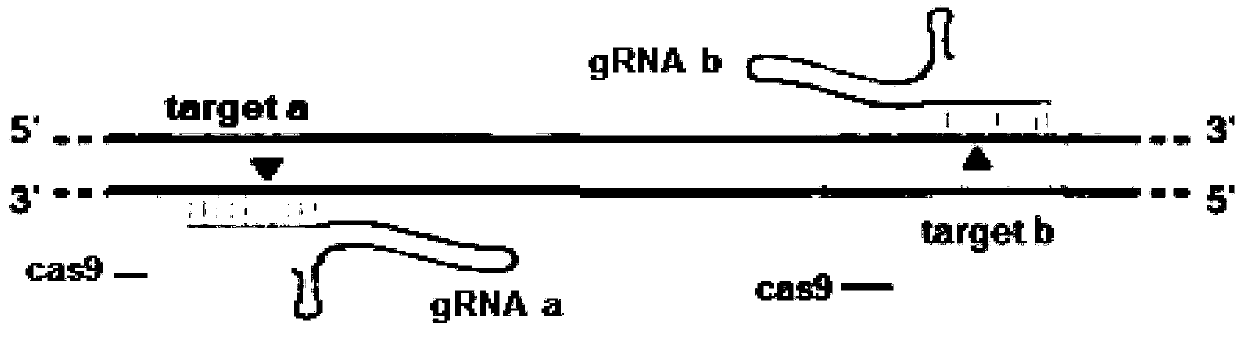
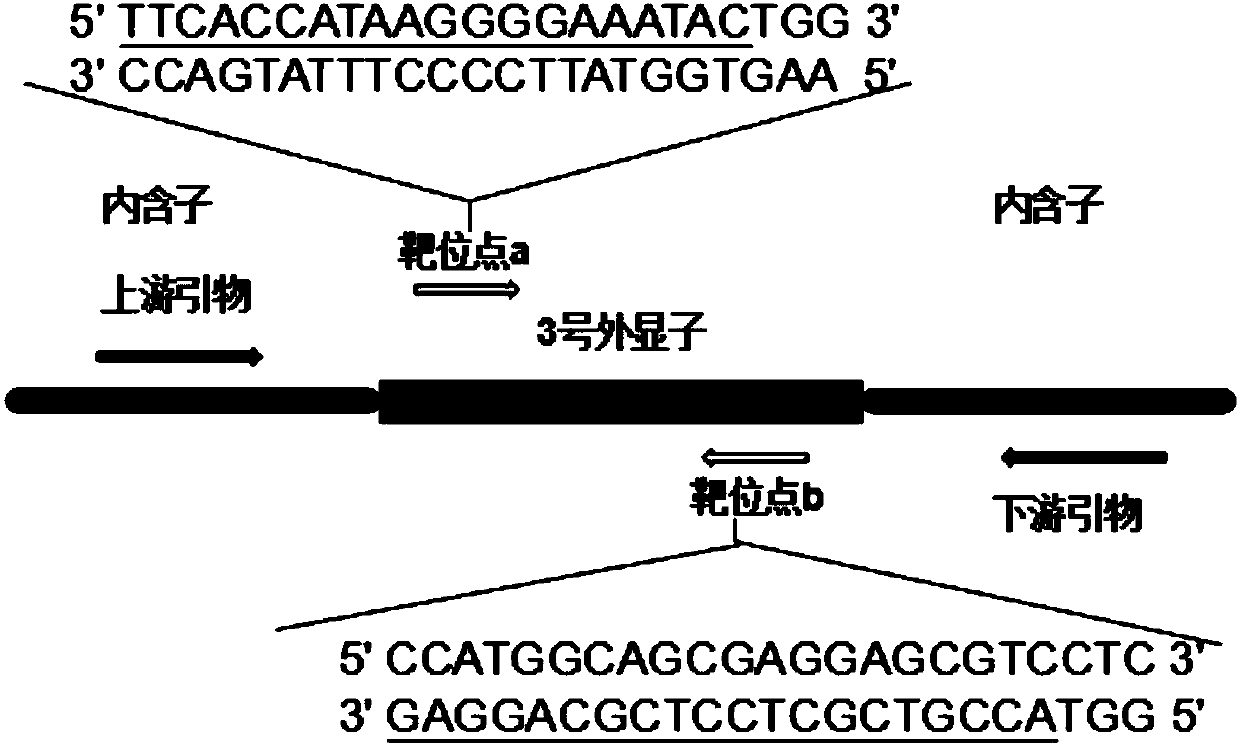
[0078] 1) Design CRISPR / Cas9 gene knockout target sites and detection primers
[0079] On the National Center for Biotechnology Information (NCBI), query the genomic DNA sequence of the zebrafish fhl1b gene, on the website SMART ( http: / / smart.embl-heidelberg.de / ) to analyze its functional domains, according to the principle of CRISPR / Cas knockout, on the website The ZiFiT Targeter ( http: / / zifit.partners.org / ZiFiT / ) to design the target site of fhl1b gene. The selection of targets must follow this standard: 5'-GG-(N)18-NGG-3'. The GG dinucleotide at the 5' end is part of the T7 promoter, and this restriction is not required when designing the target site, but it must be ensured that the 3' end of the target site is NGG. The selected position of the target point must be within the structural domain of the gene to ensure that the insertion or deletion of bases at the target site can affect the entire structural domain of the fhl1b gene, thereby changing the expression o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com