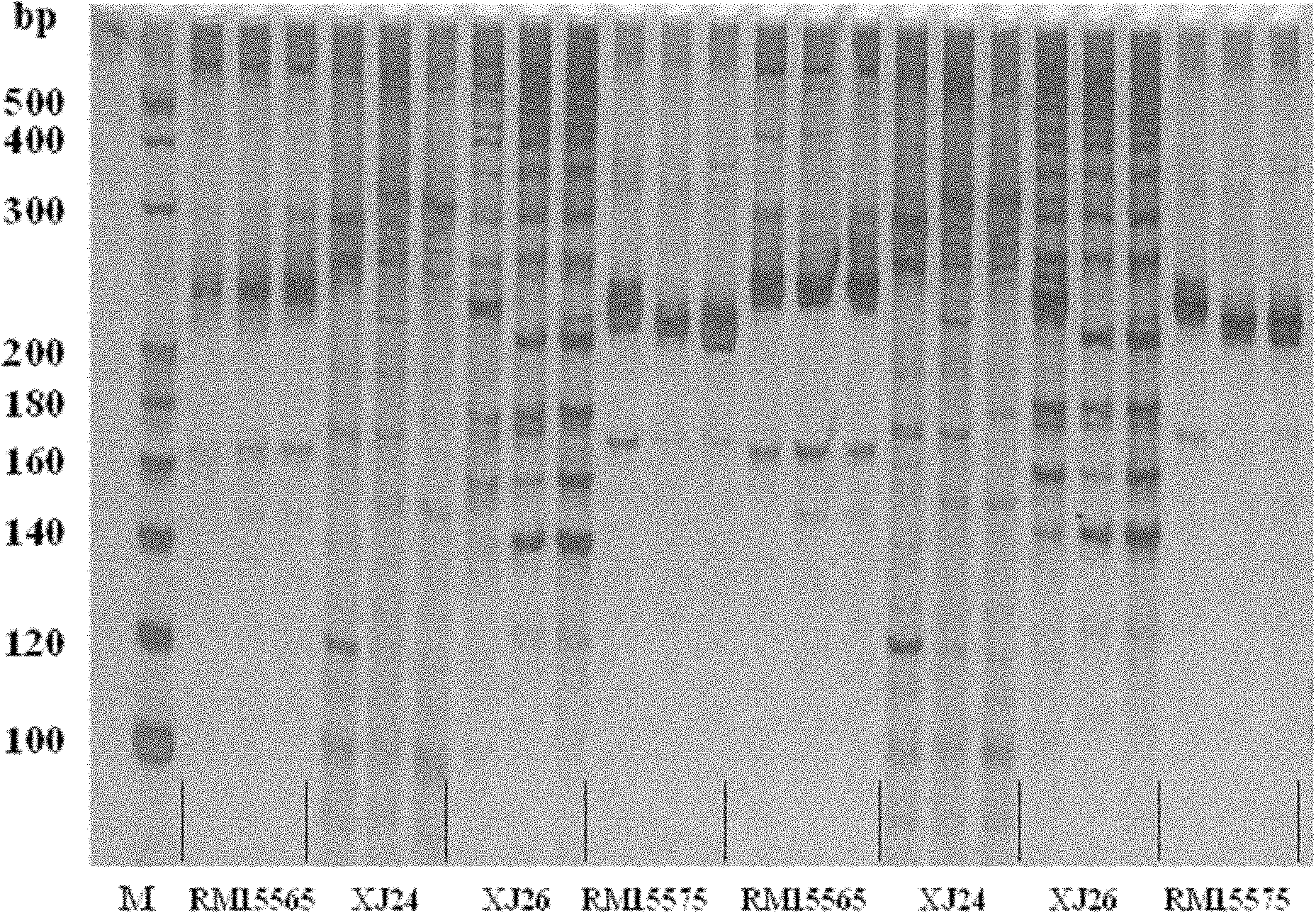Patents
Literature
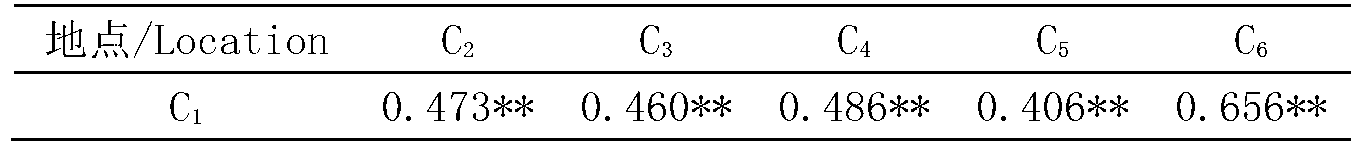
523 results about "Sequence repeat" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
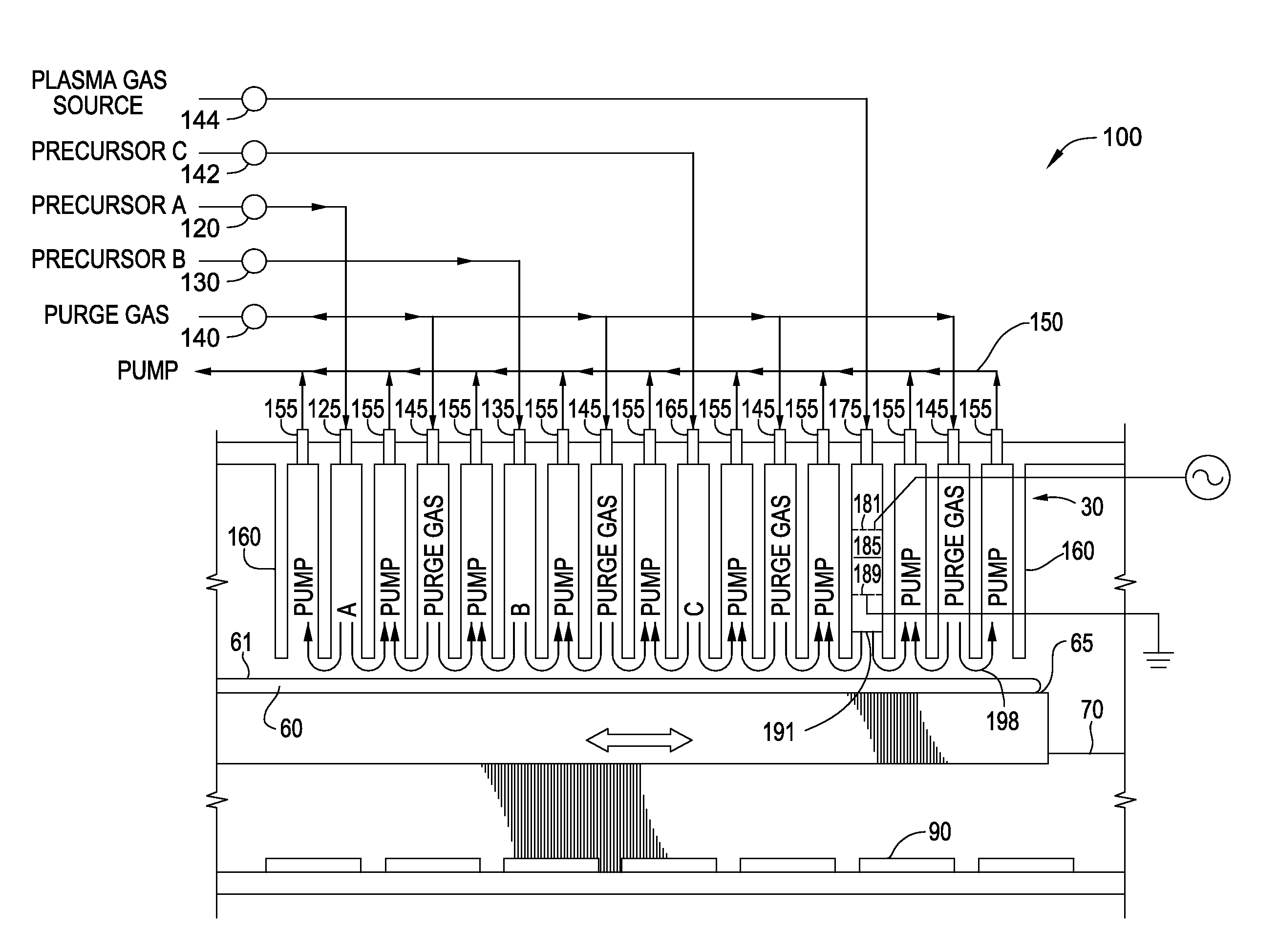
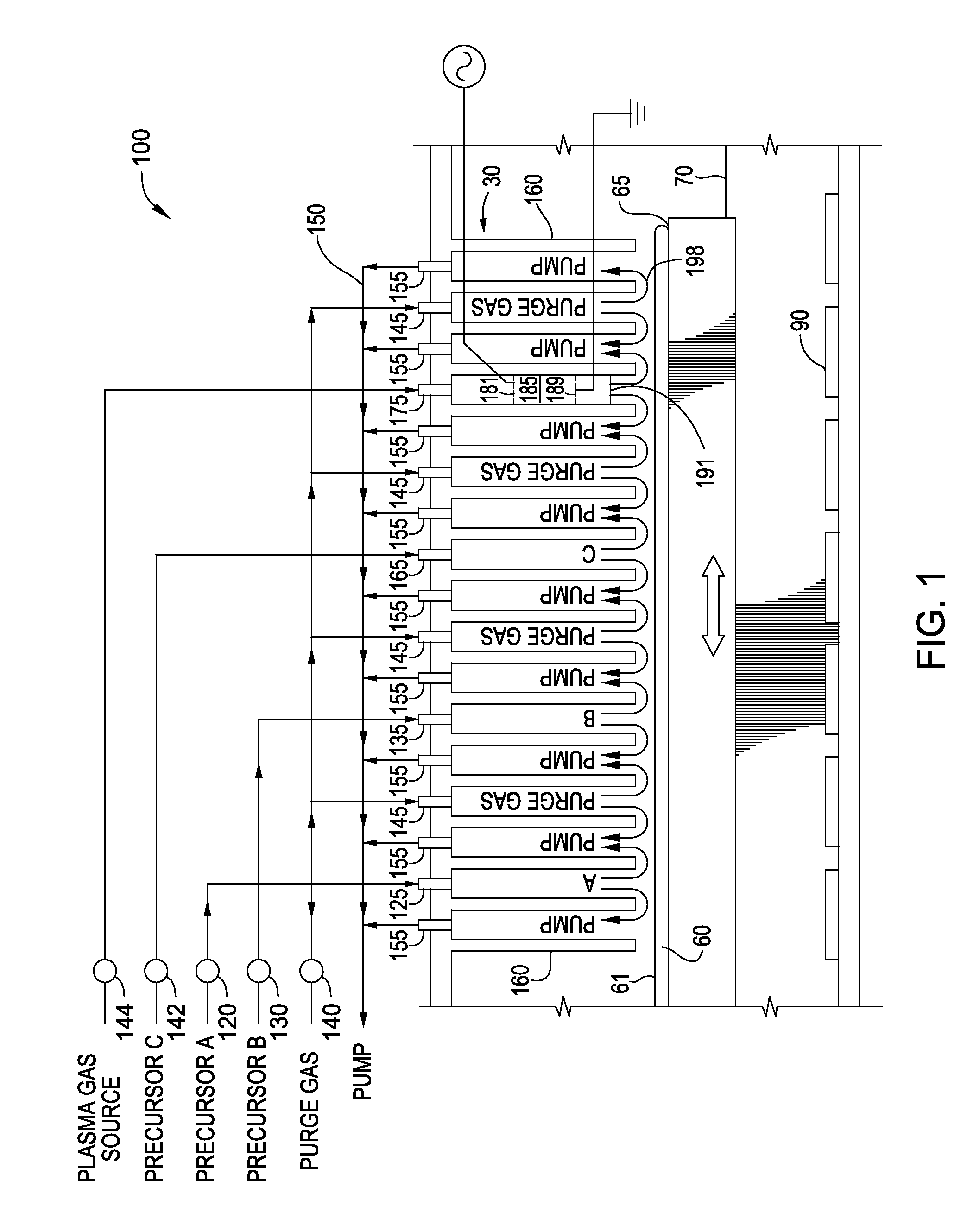
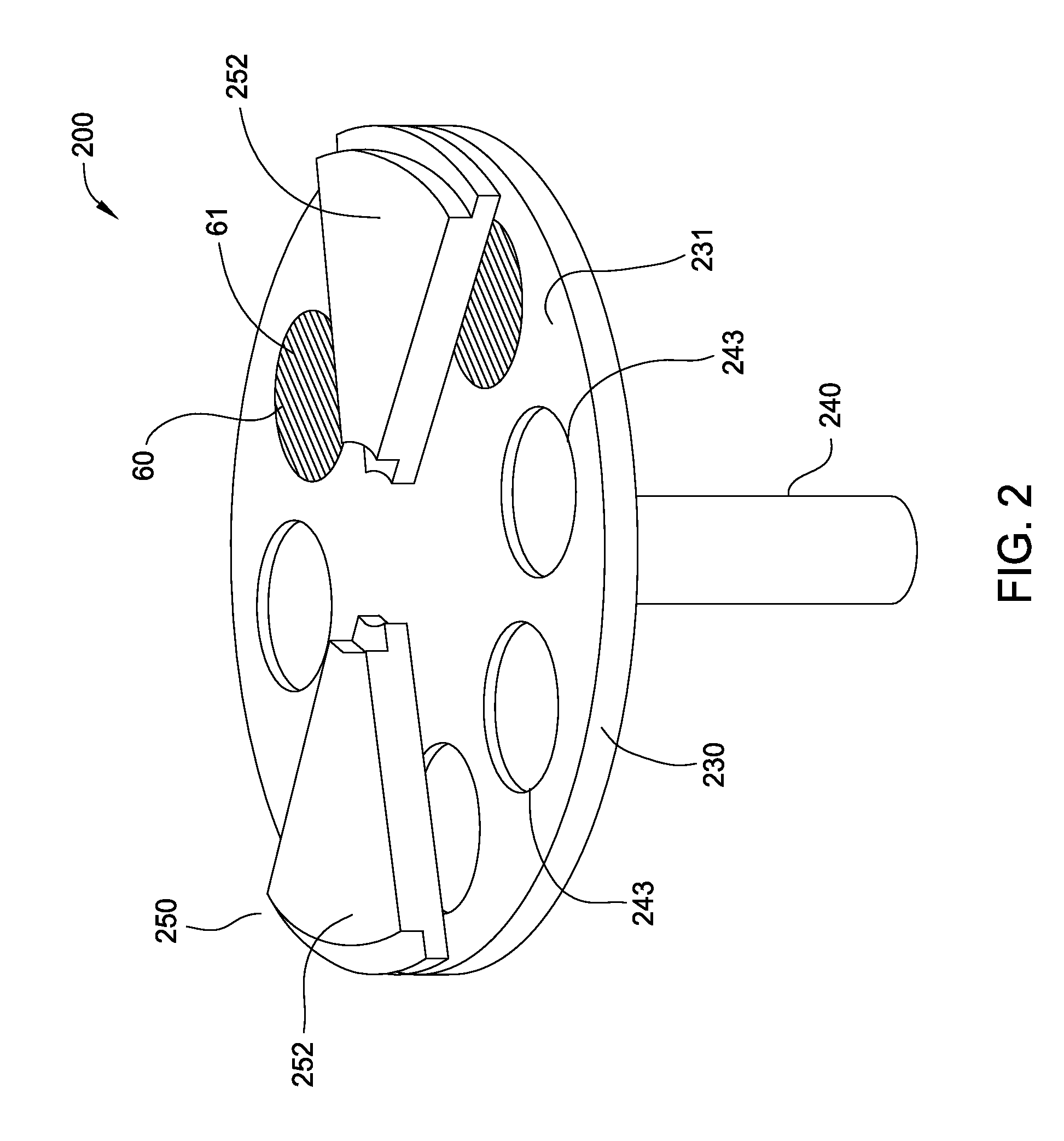
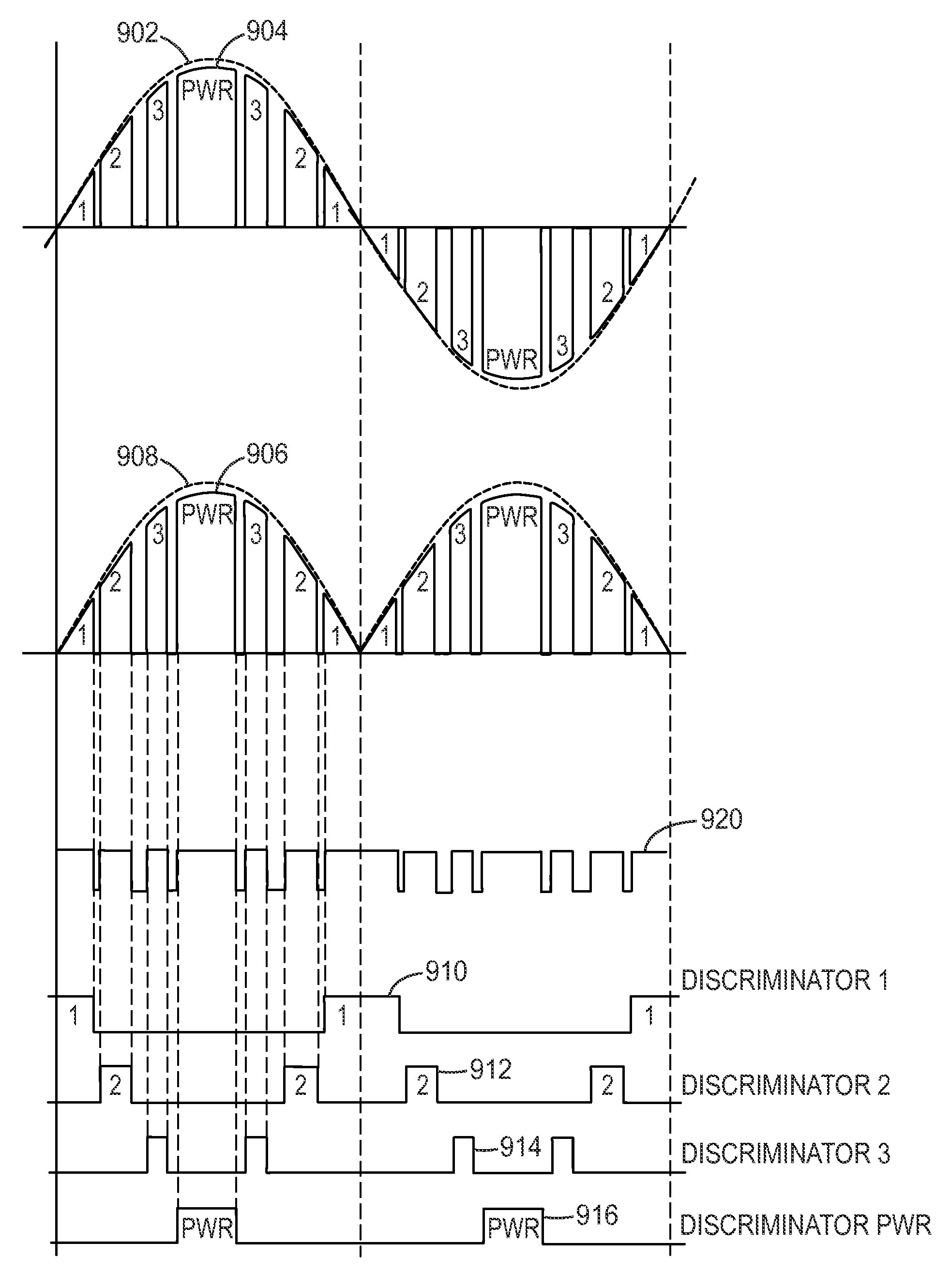
Seamless gap-fill with spatial atomic layer deposition
InactiveUS20150255324A1Semiconductor/solid-state device manufacturingChemical vapor deposition coatingDielectric layerAtomic layer deposition
Embodiments disclosed herein generally relate to forming dielectric materials in high aspect ratio features. In one embodiment, a method for filling high aspect ratio trenches in one processing chamber is disclosed. The method includes placing a substrate inside a processing chamber, where the substrate has a surface having a plurality of high aspect ratio trenches and the surface is facing a gas / plasma distribution assembly. The method further includes performing a sequence of depositing a layer of dielectric material on the surface of the substrate and inside each of the plurality of trenches, where the layer of dielectric material is on a bottom and side walls of each trench, and removing a portion of the layer of dielectric material disposed on the surface of the substrate, where an opening of each trench is widened. The sequence repeats until the trenches are filled seamlessly with the dielectric material.
Owner:APPLIED MATERIALS INC
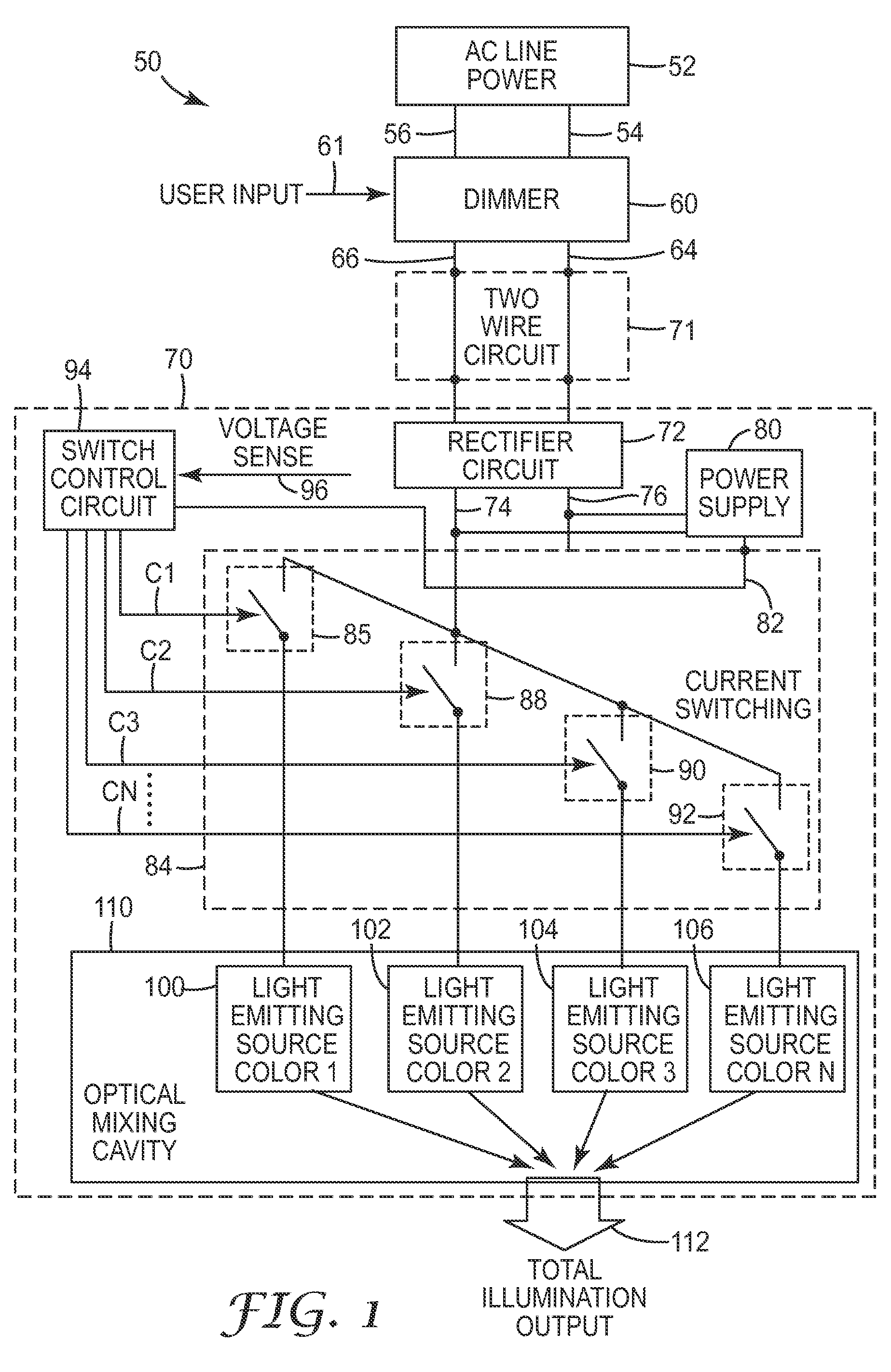
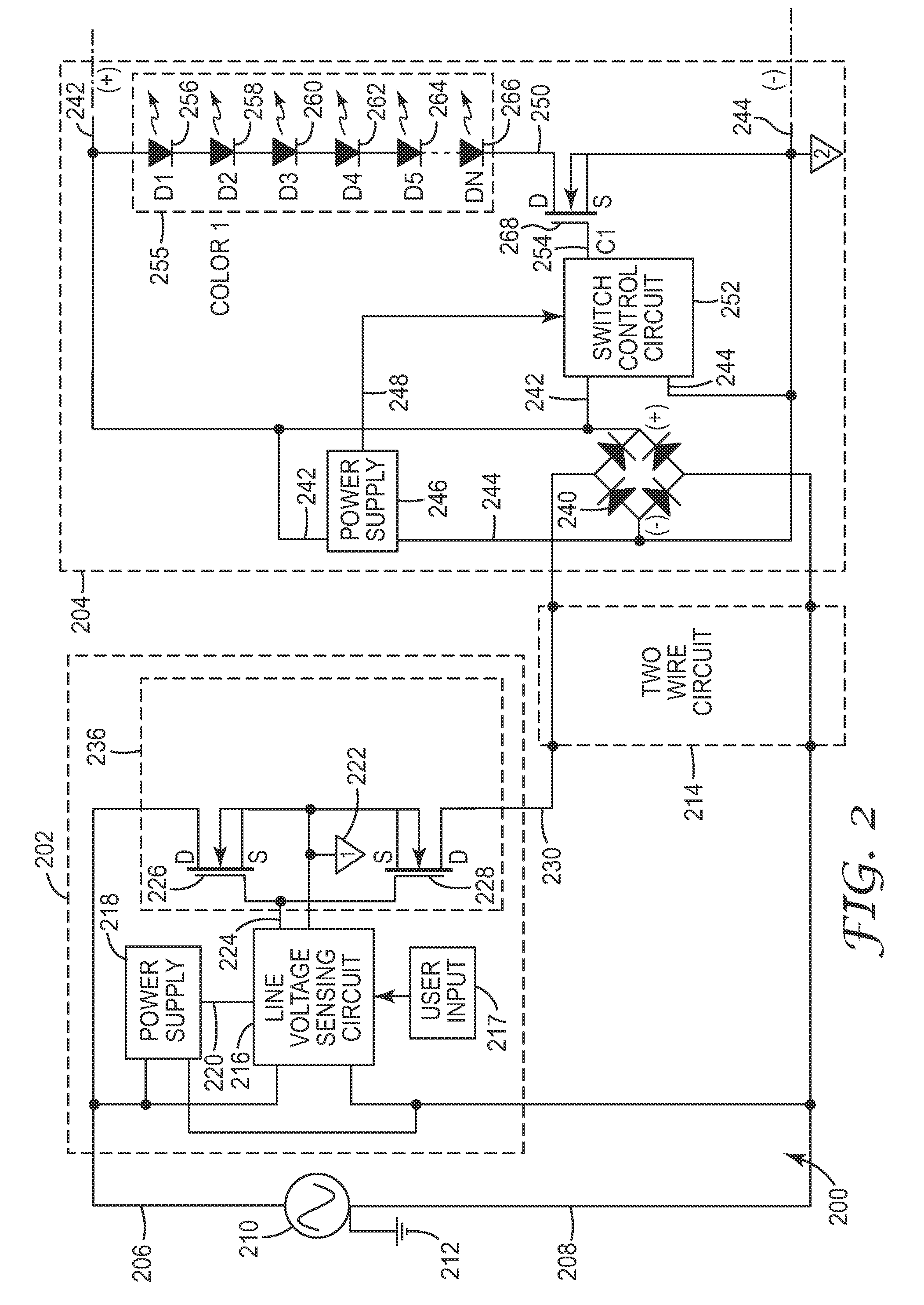
AC illumination apparatus with amplitude partitioning
Owner:3M INNOVATIVE PROPERTIES CO
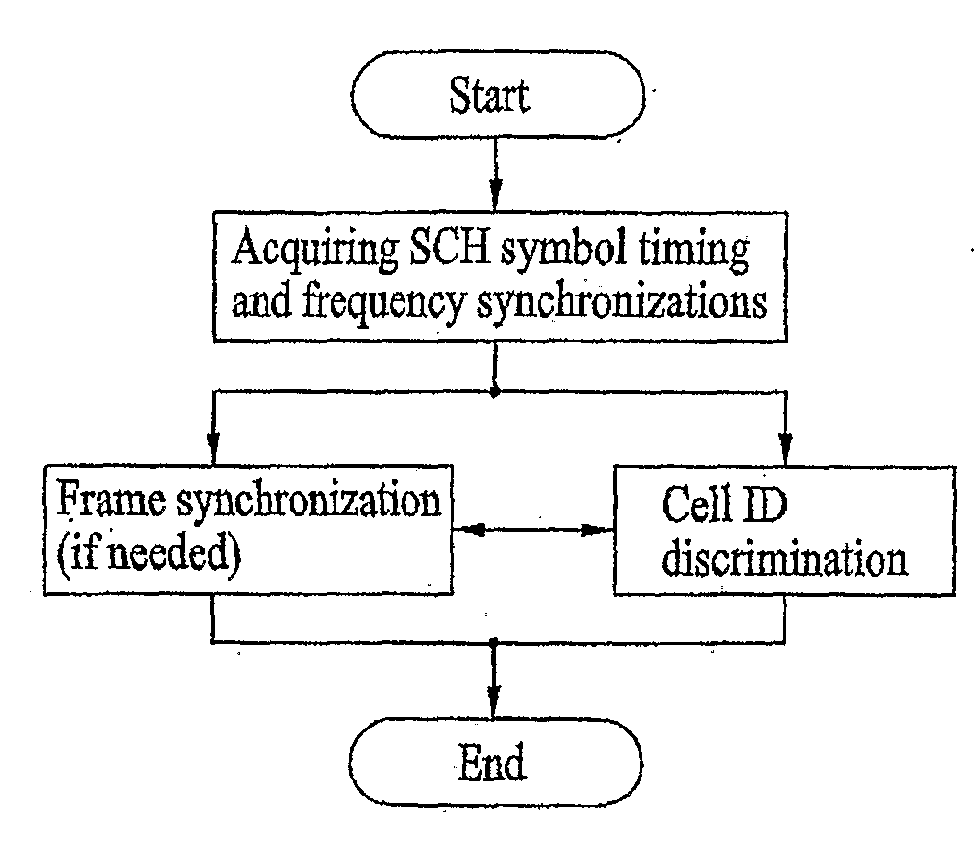
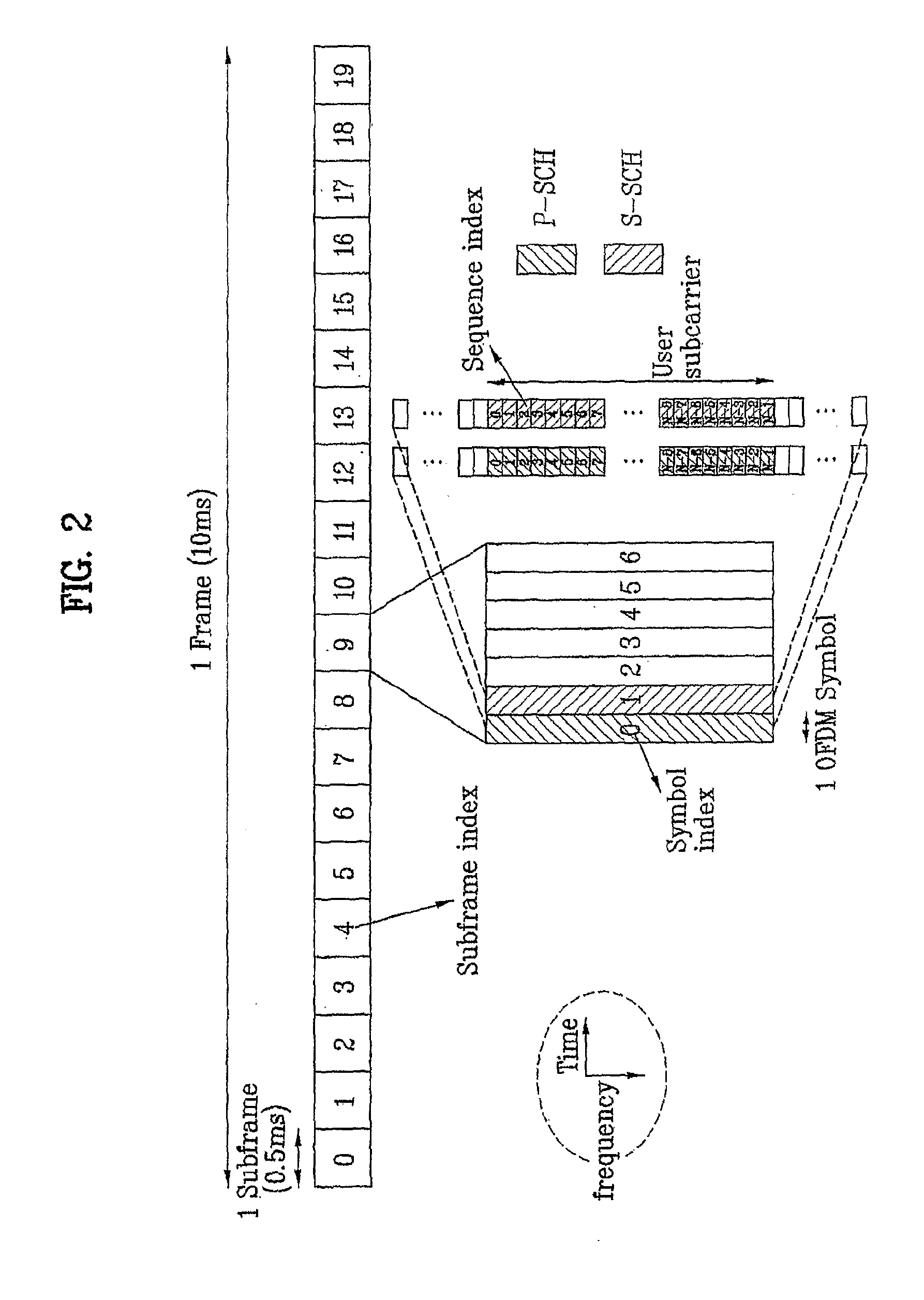
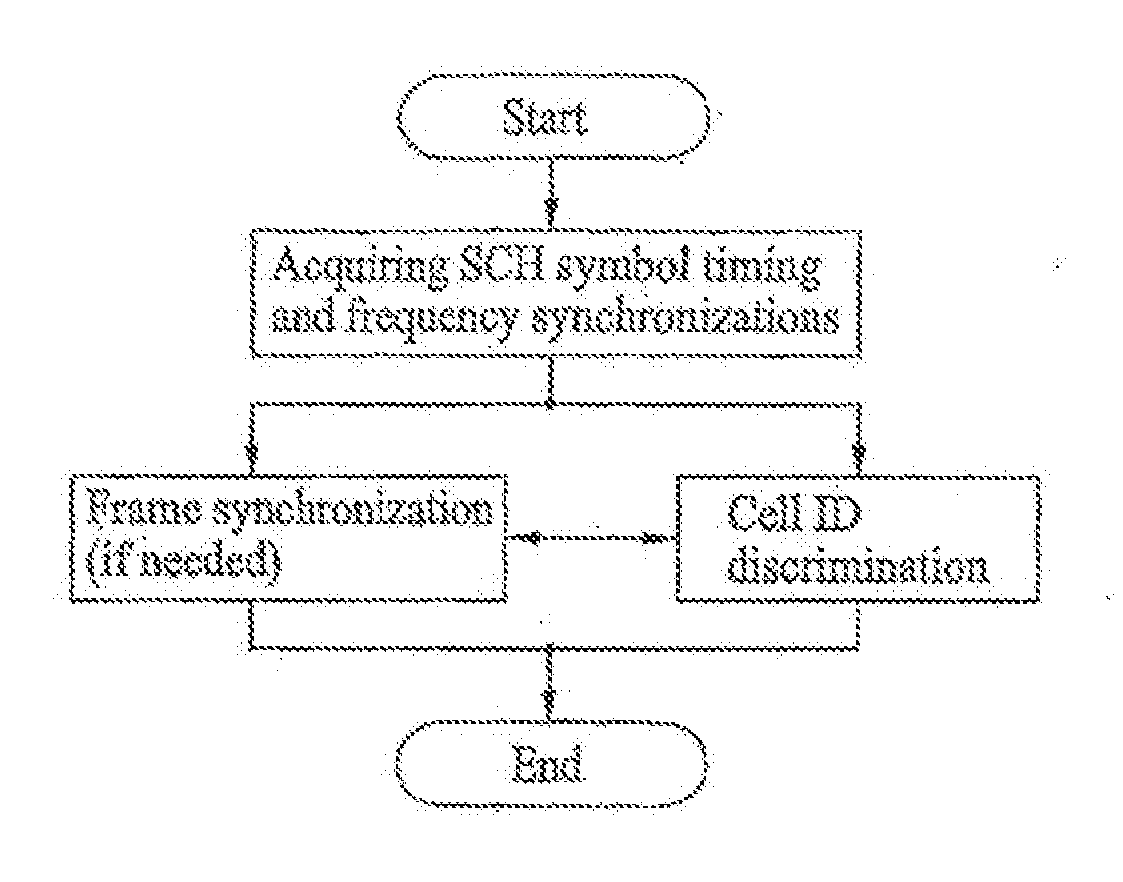
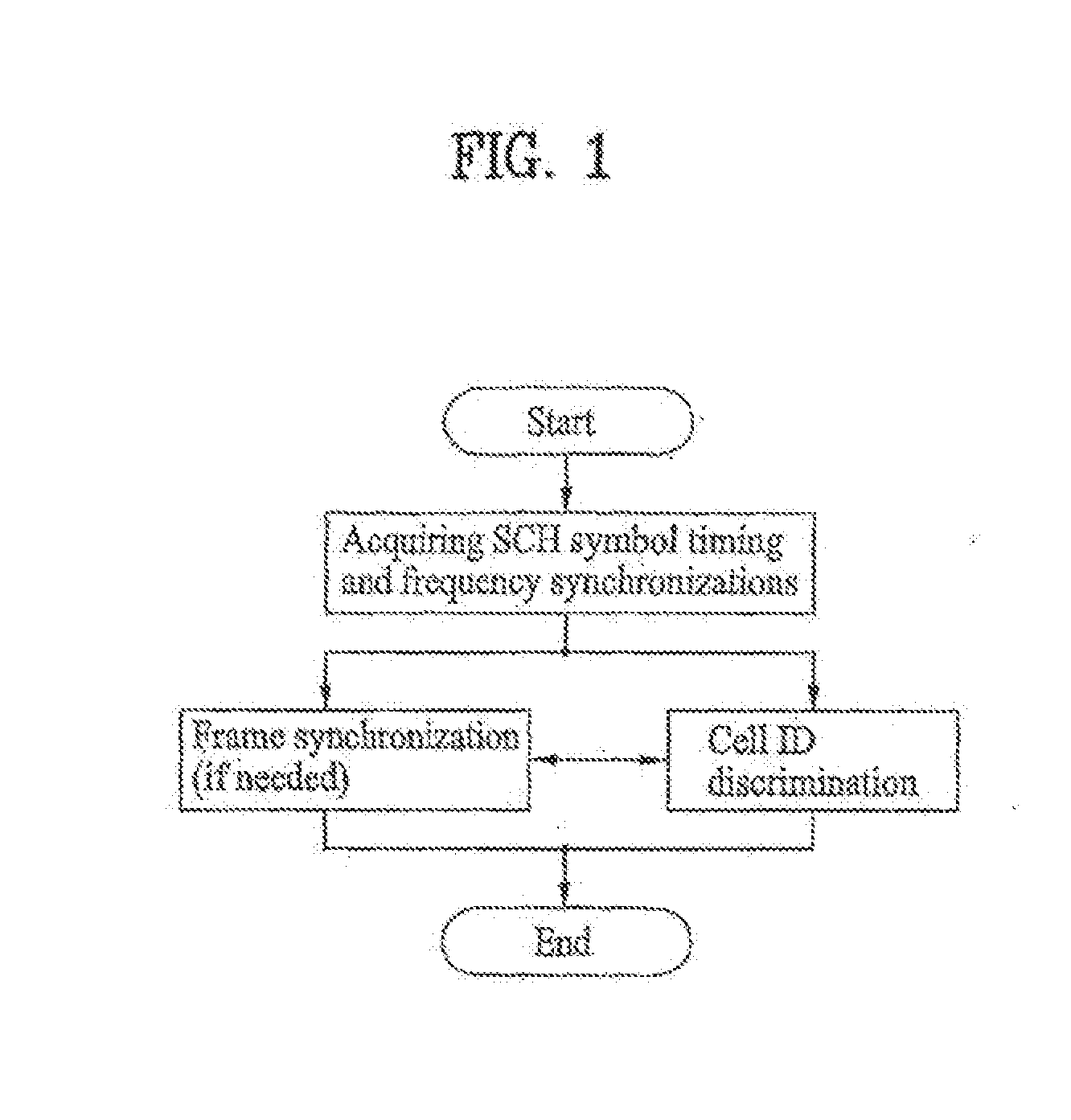
Method of generating code sequence and method of transmitting signal using the same
ActiveUS20090268602A1Synchronisation arrangementTransmission path divisionTime domainComputer science
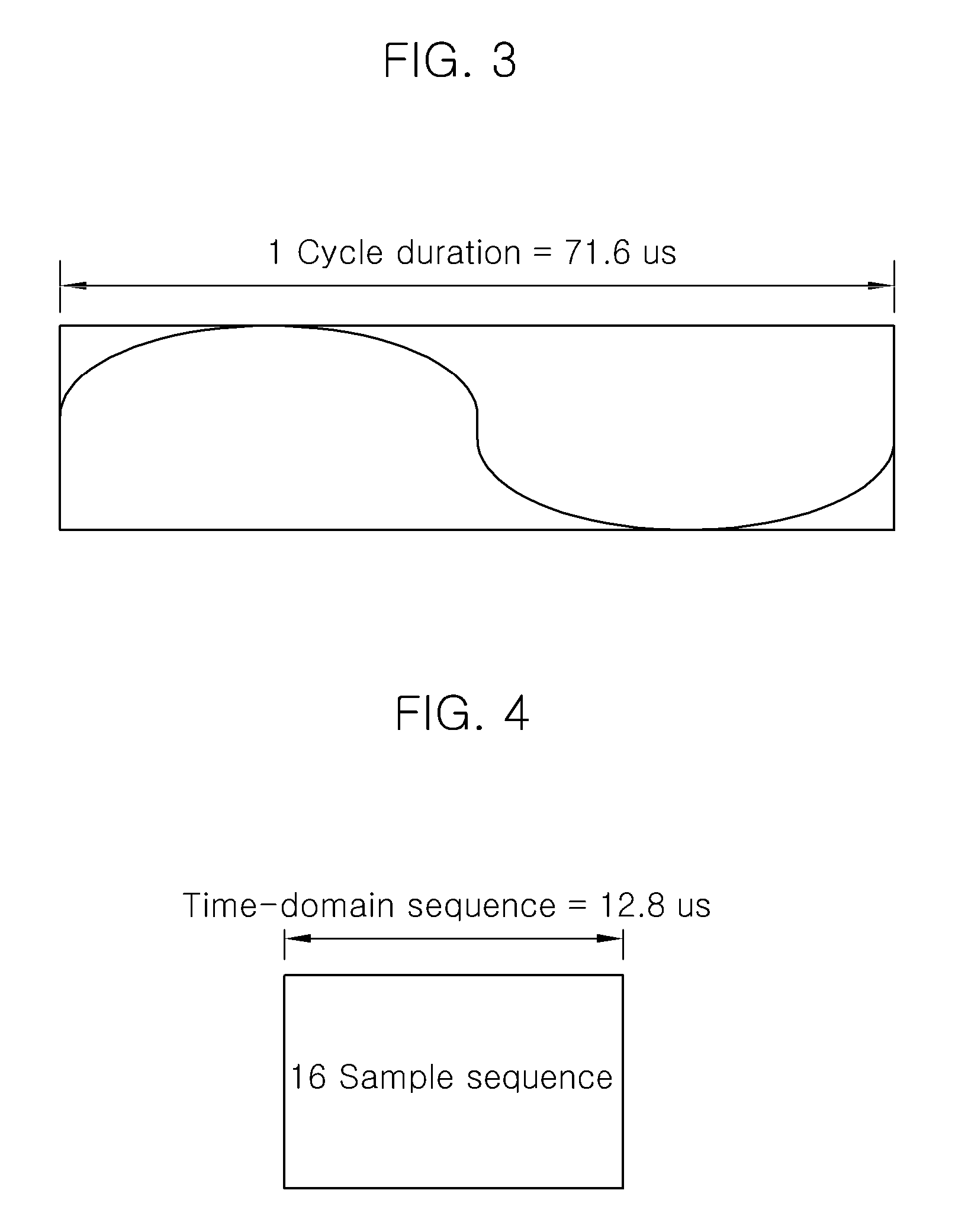
A method of generating a code sequence and method of adding additional information using the same are disclosed, by which a code sequence usable for a channel for synchronization is generated and by which a synchronization channel is established using the generated sequence. The present invention, in which the additional information is added to a cell common sequence for time synchronization and frequency synchronization, includes the steps of generating the sequence repeated in time domain as many as a specific count, masking the sequence using a code corresponding to the additional information to be added, and transmitting a signal including the masked sequence to a receiving end.
Owner:LG ELECTRONICS INC
Method for developing mung bean simple sequence repeat (SSR) primer based on transcriptome sequencing
InactiveCN103642912AImprove design success rateSimple designMicrobiological testing/measurementDNA/RNA fragmentationSequence databaseTranscriptome Sequencing
The invention provides a method for developing a mung bean simple sequence repeat (SSR) primer based on transcriptome sequencing. The method comprises the following steps: obtaining a set of mung bean genome-wide transcription, and forming a sequence database; splicing sequencing sequences into a transcriptome by Trinity; taking the longest transcript in each gene as Unigene; carrying out bioinformatics analysis of a Unigene sequence; carrying out SSR detection on the Unigene by adopting MISA1.0; carrying out SSR primer design by using a Primer 3, and carrying out SSR primer polymorphism identification. 13134 pairs of SSR primers are successfully designed by application of the method; 50 pairs of primers are randomly selected to verify 8 parts of mung bean deoxyribonucleic acids (DNAs) from different countries, wherein 32 pairs of polymorphic primers are formed in all; the mung bean materials with different geographical origins can be distinguished by using the 32 pairs of SSR primers. The method disclosed by the invention is convenient, fast and accurate, and low in cost, and a new thought is provided for development of the mung bean SSR primer.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
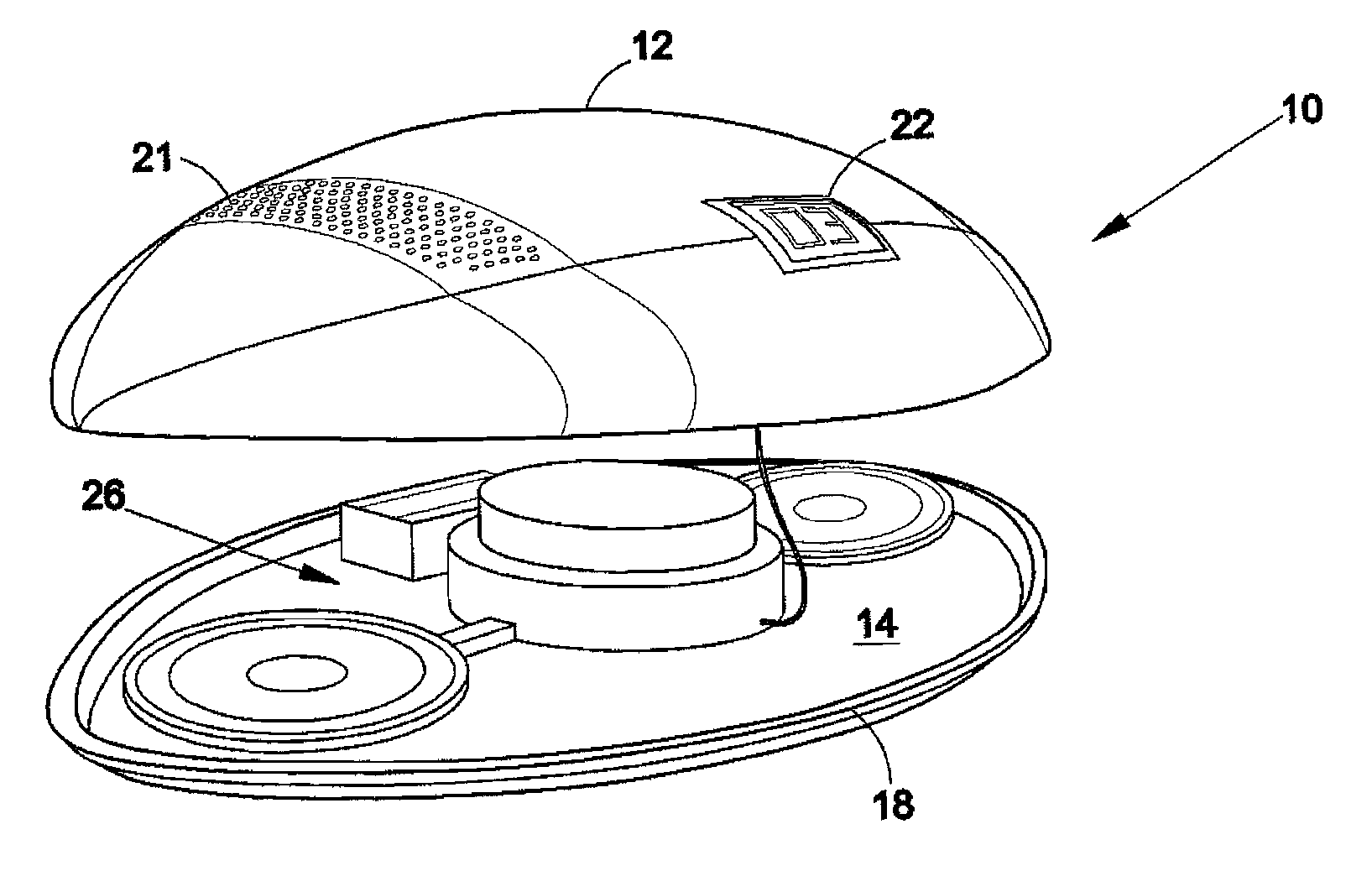
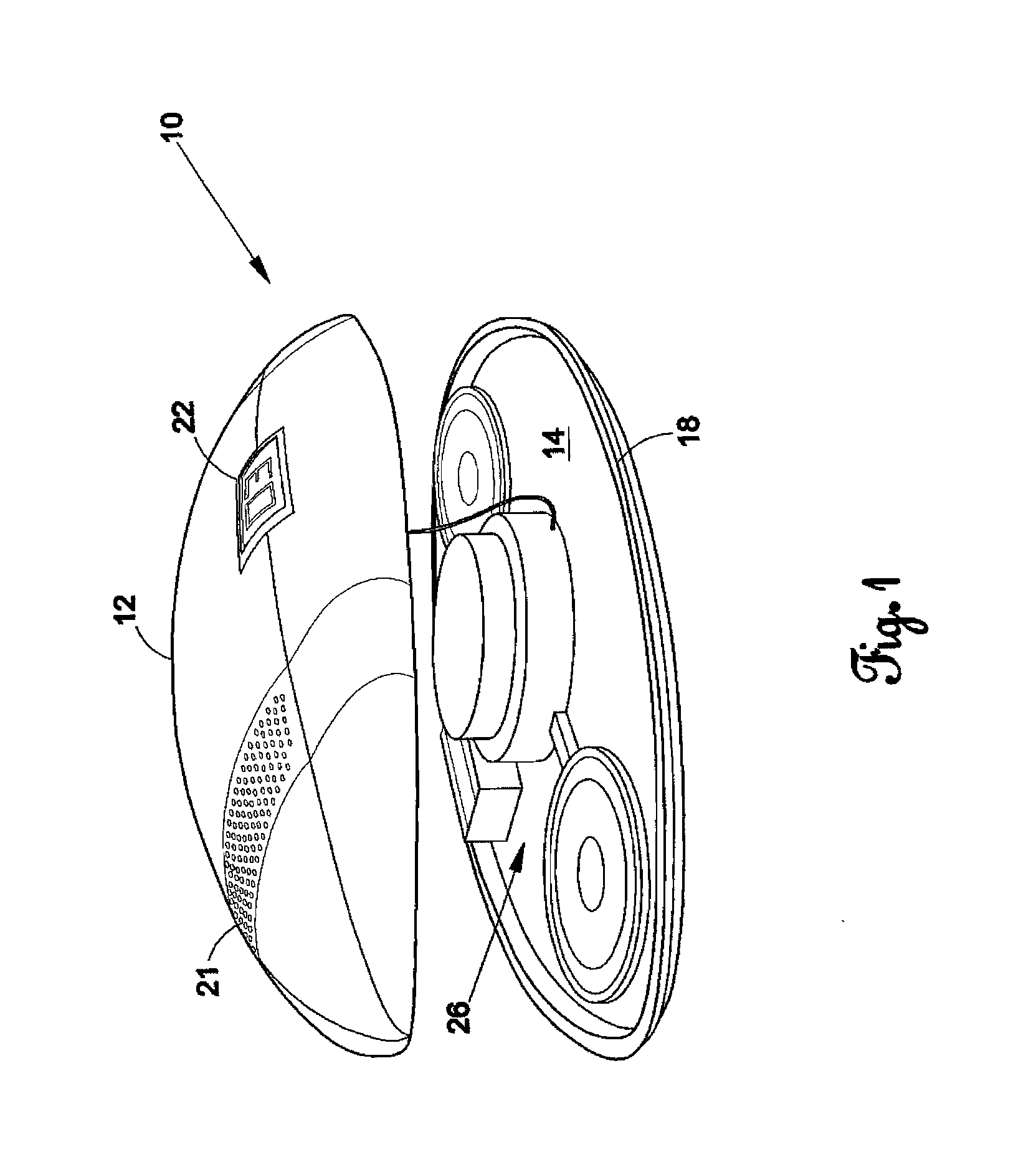
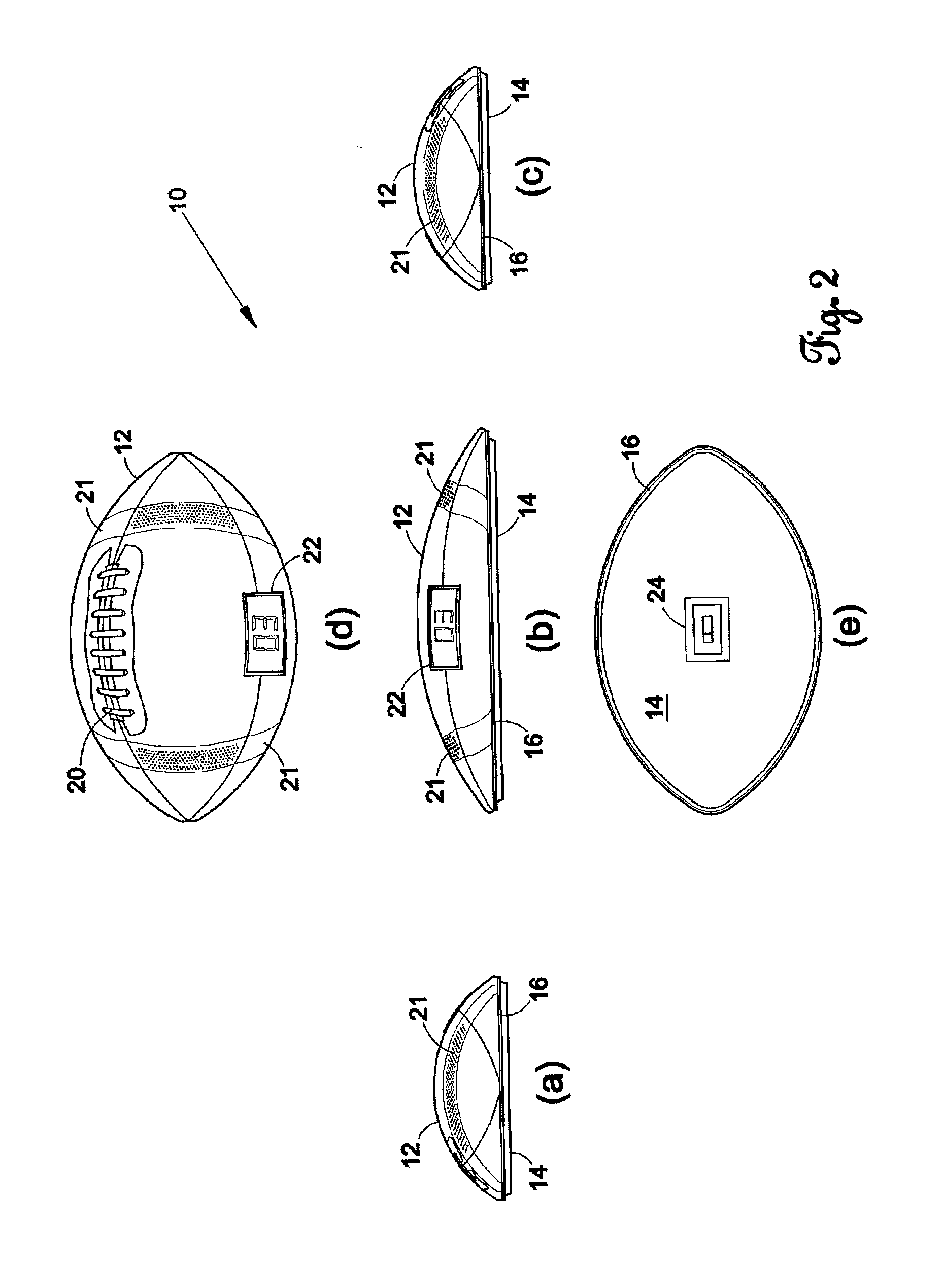
Football counting device
InactiveUS20120157246A1Hollow inflatable ballsHollow non-inflatable ballsMicrocomputerTelecommunications
The present invention is a football counting device that utilizes a microcomputer so that a proper count can be given for backyard or sandlot football games as to when the defense can rush the passer. A push-button switch will start the sequence and the microcomputer (1) controls an audio sound that counts “1 Mississippi”, “2 Mississippi” and (2) gives a visual display as to when the quarterback can be rushed. The length of time delay and the voice utilized can be selected by the players. The sequence repeats for each new down.
Owner:GLOVER ROBERT MICHAEL
Method for generating an intraoral volume image
ActiveUS20120307965A1Improve artEasy to adjustMaterial analysis using wave/particle radiationRadiation/particle handlingX-rayAngular orientation
A method for obtaining an intraoral x-ray image determines an initial spatial position and angular orientation of an x-ray source relative to a detector. A first x-ray image is obtained with the x-ray source at the initial spatial position and angular orientation and stored, associating the initial spatial position and angular orientation to the first x-ray image. A sequence repeats that calculates a next spatial position and angular orientation for the x-ray source, provides positional adjustment information between the x-ray source and detector for obtaining a next x-ray image, measures and records the actual spatial position and angular orientation of the x-ray source relative to the detector and obtains and stores the next x-ray image at the measured spatial position and angular orientation, and forms a composite image using image data from the first and from the one or more next x-ray images.
Owner:CARESTREAM DENTAL TECH TOPCO LTD
Method for generating an intraoral volume image
ActiveUS8670521B2Easy to adjustImage data processing detailsRadiation beam directing meansSoft x rayX-ray
A method for obtaining an intraoral x-ray image determines an initial spatial position and angular orientation of an x-ray source relative to a detector. A first x-ray image is obtained with the x-ray source at the initial spatial position and angular orientation and stored, associating the initial spatial position and angular orientation to the first x-ray image. A sequence repeats that calculates a next spatial position and angular orientation for the x-ray source, provides positional adjustment information between the x-ray source and detector for obtaining a next x-ray image, measures and records the actual spatial position and angular orientation of the x-ray source relative to the detector and obtains and stores the next x-ray image at the measured spatial position and angular orientation, and forms a composite image using image data from the first and from the one or more next x-ray images.
Owner:CARESTREAM DENTAL TECH TOPCO LTD
Method of generating code sequence and method of transmitting signal using the same
ActiveUS20110188465A1Eliminate the problemSynchronisation arrangementModulated-carrier systemsTime domainComputer science
A method of generating a code sequence and method of adding additional information using the same are disclosed, by which a code sequence usable for a channel for synchronization is generated and by which a synchronization channel is established using the generated sequence. The present invention, in which the additional information is added to a cell common sequence for time synchronization and frequency synchronization, includes the steps of generating the sequence repeated in time domain as many as a specific count, masking the sequence using a code corresponding to the additional information to be added, and transmitting a signal including the masked sequence to a receiving end.
Owner:LG ELECTRONICS INC
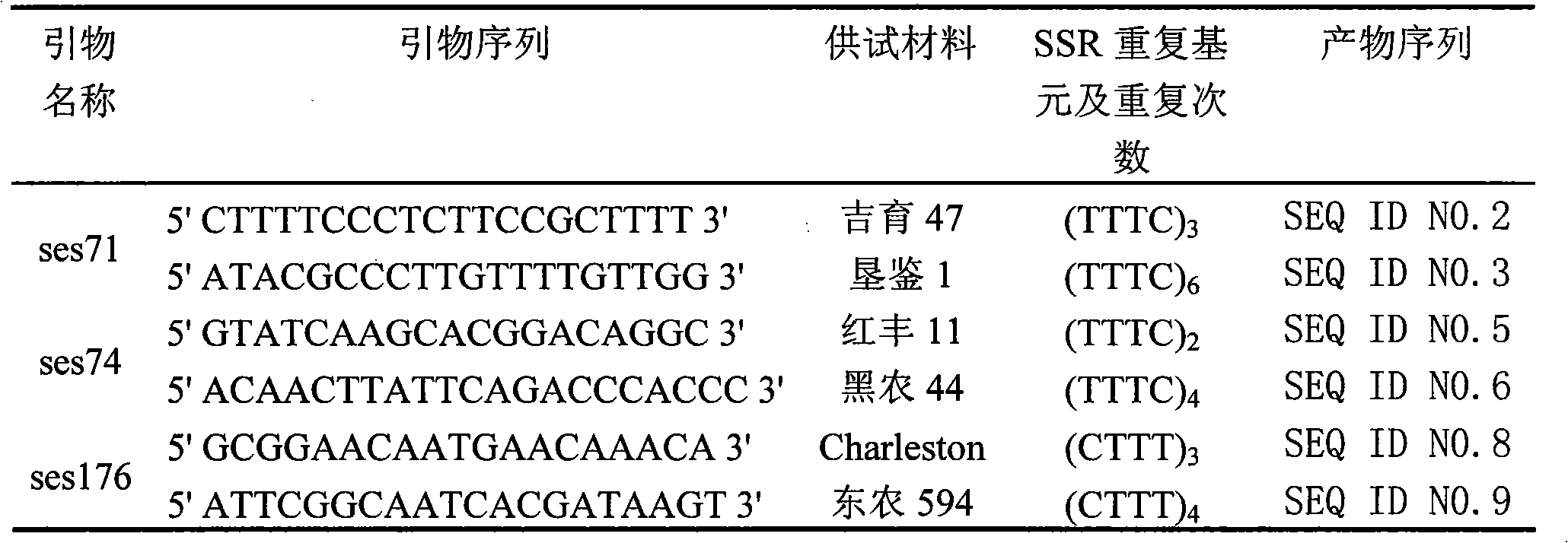
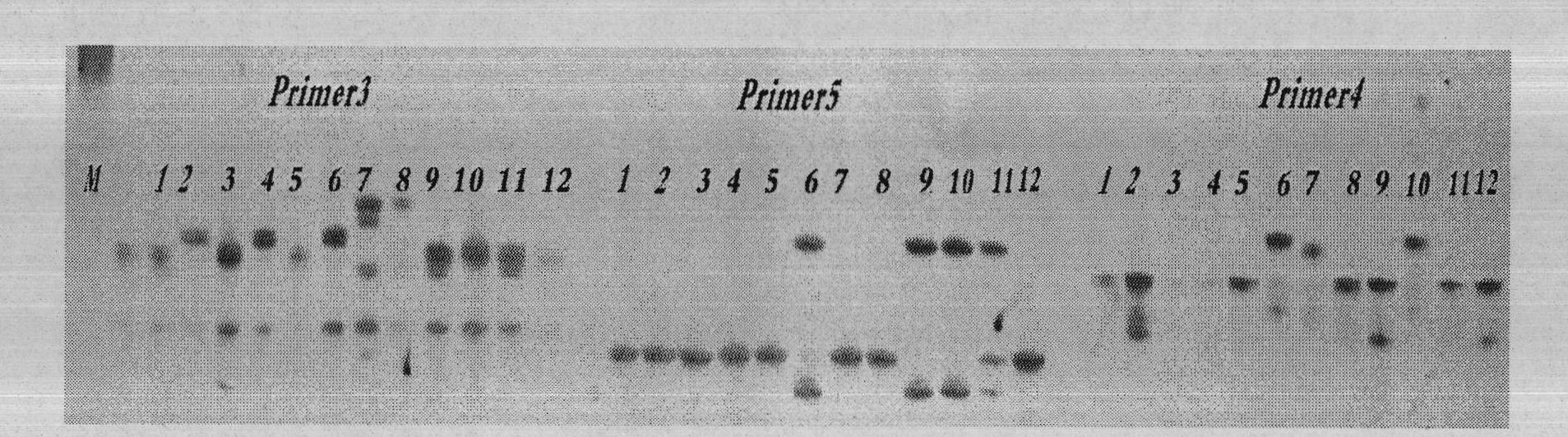
SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and application of SSR core primer group
ActiveCN103060318AImproved geneticsEvenly distributedMicrobiological testing/measurementDNA preparationAgricultural scienceGenetic diversity
The invention discloses an SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and an application of the SSR core primer group, and belongs to the technical field of molecular biology. The core primer group comprises 30 pairs of primers, wherein nucleotide sequences are represented by SEQ ID NO. 1-60. The primer has advantages of clear electrophoretic band and rich polymorphism, and is uniformly distributed and stable in amplification. The invention also provides the application of the SSR core primer group in identifying the genetic diversity and variety of the foxtail millet. The primer group can be used for precisely and quickly identifying the variety of the foxtail millet and precisely reflecting a genetic relationship among the varieties of the foxtail millet.
Owner:CROP RES INST SHANDONG ACAD OF AGRI SCI
Major QTL (Quantitative Trait Locus) of cotton high-strength fiber and molecular marker and application thereof
ActiveCN103255139AHigh strengthImprove fiber quality levelsMicrobiological testing/measurementVector-based foreign material introductionFiberAgricultural science
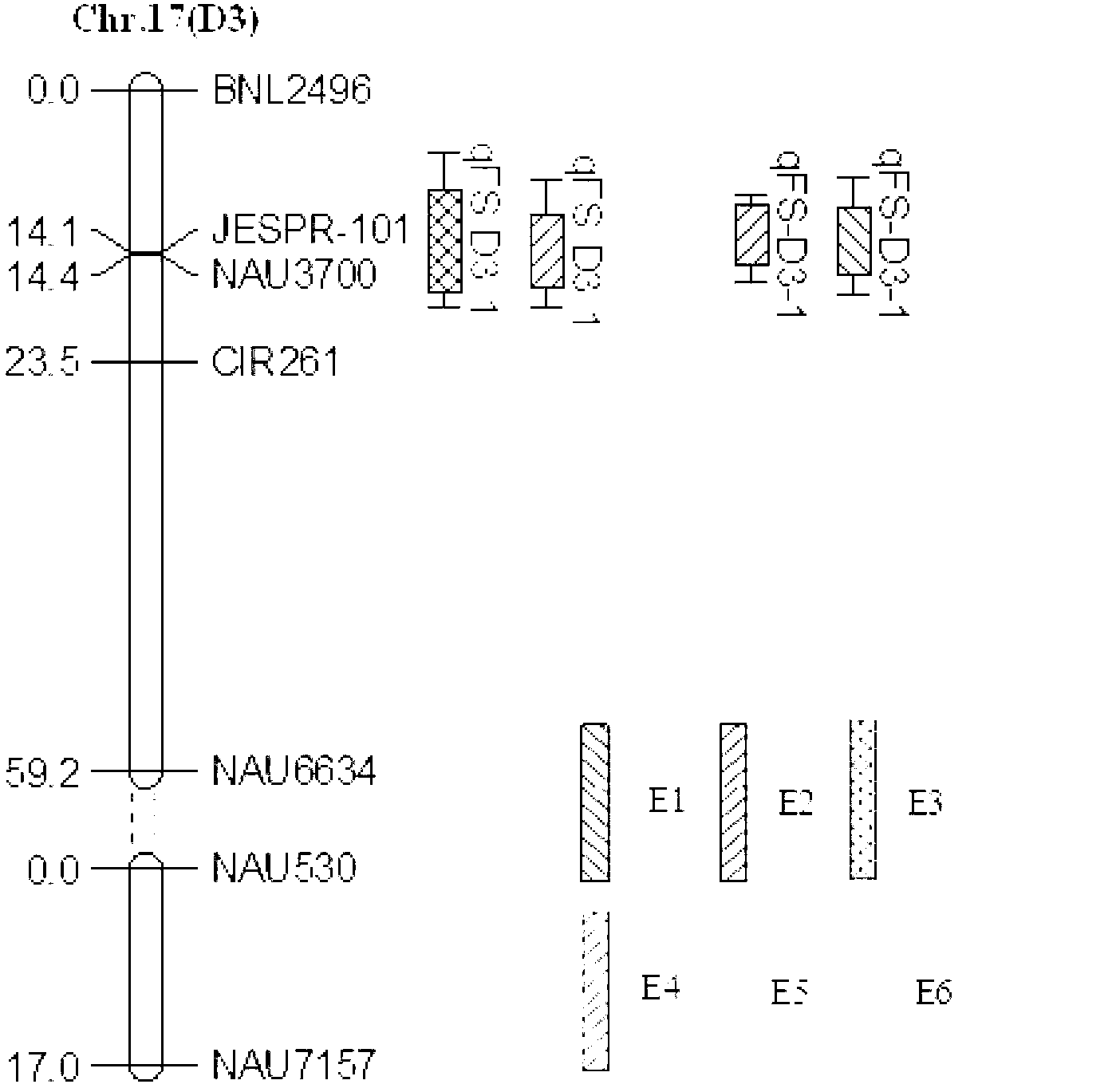
The invention discloses a major QTL (Quantitative Trait Locus) of a cotton high-strength fiber and a molecular marker and application thereof. The major QTL of the cotton high-strength fiber is linked with two SSR (Simple Sequence Repeat) markers, is positioned through markers NAU / SSR / FS1195 and JESPR / SSR / FS2127 and is 0.01-2.39 centimeters away from a marker NAU3700; and the major QTL locus can explain 4.51%-17.55% of phenotype variation and achieves the LOD (Limit Of Detection) value between 3.23 and 7.09. The major QTL disclosed by the invention overcomes the defects of high cost, high difficulty and slow progress of quality breeding in the prior art, greatly increases the selection efficiency of the high-strength fiber by utilizing the major QTL locus of a cotton high-strength fiber gene and the molecular marker thereof, can be used for exploring high-quality fiber gene resources, provides the gene resources and a marker auxiliary selection technology for the quality breeding of cotton and can be applied to the production and quality detection of high-strength fiber cotton varieties.
Owner:NANJING AGRICULTURAL UNIVERSITY
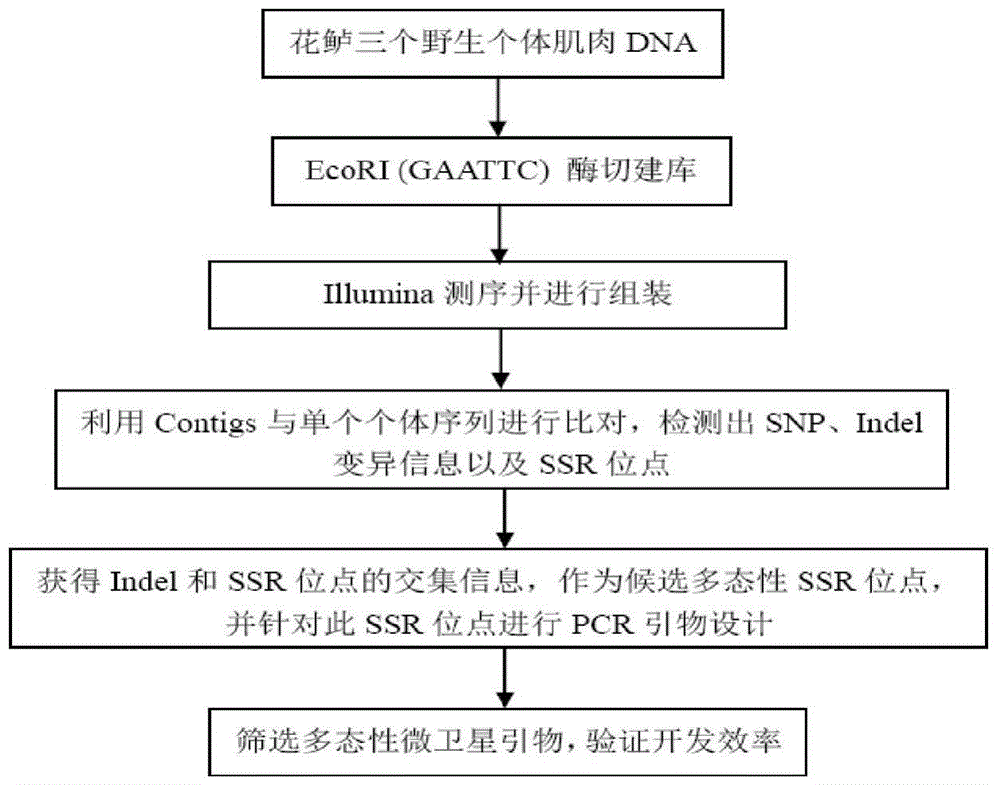
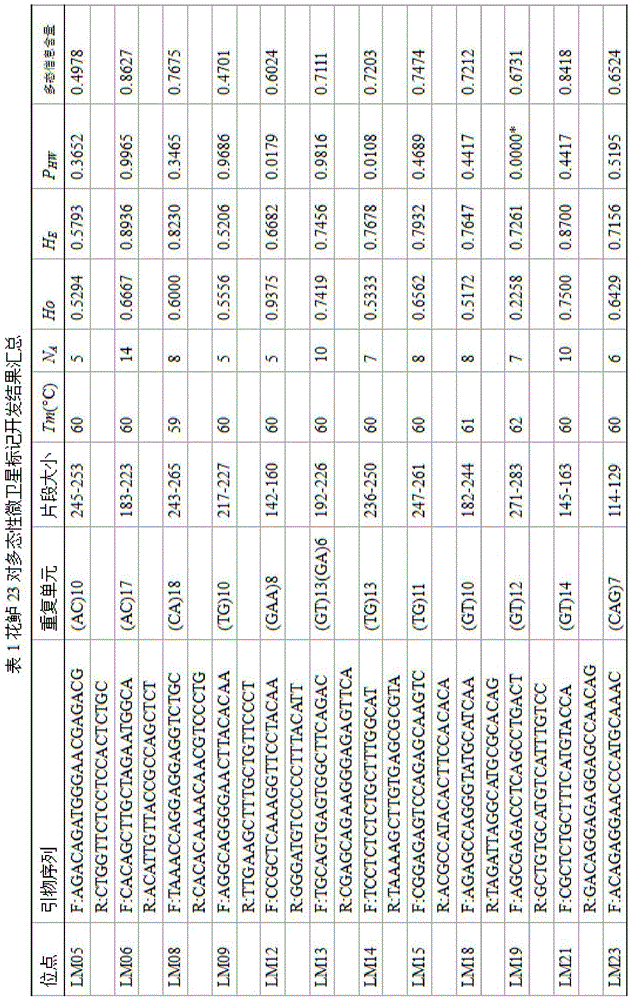
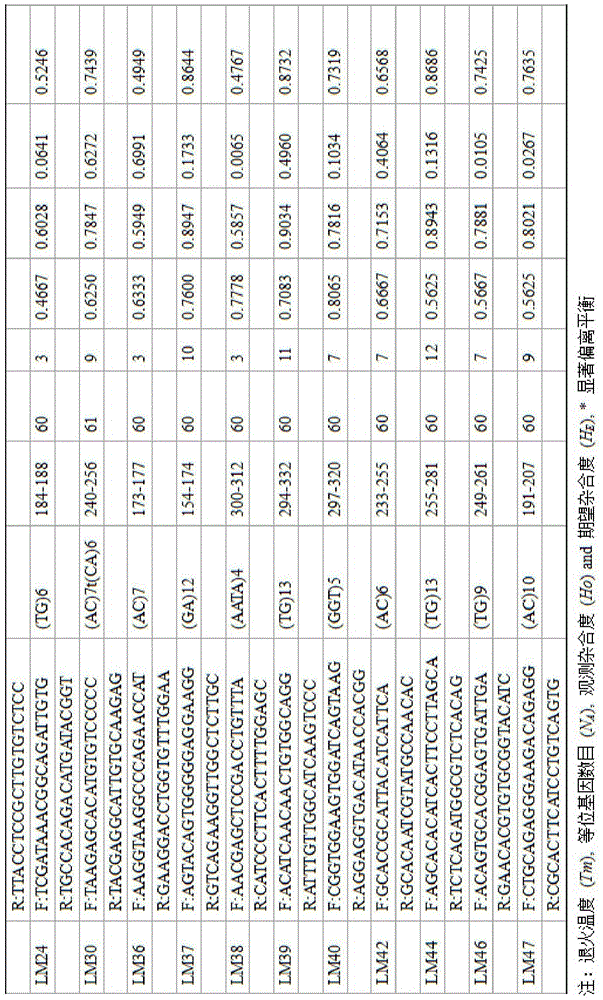
Method for massively and efficiently developing molecular markers on basis of Indel and SSR (simple sequence repeat) site techniques
ActiveCN105695572APromote rapid developmentShorten the timeMicrobiological testing/measurementDNA preparationContigAgricultural science
The invention discloses a method for massively and efficiently developing molecular markers on the basis of Indel and SSR (simple sequence repeat) site techniques. The method comprises the following steps: (1) selecting at least 3 samples to be developed, and respectively extracting DNAs (deoxyribonucleic acids) of the samples to be developed; (2) carrying out enzyme digestion on the DNA samples of the samples to be developed, establishing a sequencing library, and carrying out sequencing; (3) mixing the genomes of all the samples to be developed, and assembling to obtain Contigs; (4) comparing the Contigs with the sequences of the sample individuals to be developed, and acquiring SSR sites with Indel inside according to the Indel and SSR site information as a candidate polymorphism SSR site; (5) designing primers according to the obtained candidate polymorphism SSR site, carrying out PCR (polymerase chain reaction) amplification and sequencing, and selecting a ribbon-shaped stable clear strip as molecular marker primers to be verified; and (6) carrying out PCR amplification on different samples to be developed by using the obtained molecular marker primers, and selecting the molecular markers with diversity, thereby obtaining the molecular markers. The method enhances the molecular marker development efficiency; and the developed SSR molecular markers can be efficiently applied to research in the aspects of genetics, multiplication release evaluation and the like.
Owner:SOUTH CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Oil-tea SSR molecular marker
ActiveCN103667275AGood repeatabilityImprove reliabilityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCamellia oleifera
The invention discloses an oil-tea SSR molecular marker which comprises acquisition and pre-treatment of an oil-tea DNA sequence, retrieval of SSR loci in the DNA sequence, design and synthesis of an SSR primer, establishment of an SSR marker system, and screening of a polymorphic SSR marker. Oil-tea genomic sequences obtained by 454 sequencing and eligible sequences containing simple sequence repeat units in an EST sequence are retrieved and screened to obtain twelve polymorphic SSR markers: CO_gSSR11, CO_gSSR12, CO_gSSR14, CO_gSSR16, CO_gSSR19, CO_gSSR20, CO_eSSR01, CO_eSSR04, CO_eSSR16, CO_eSSR40, CO_eSSR44 and CO_eSSR48. The oil-tea SSR molecular marker disclosed by the invention is suitable for genetic evaluation, germplasm identification and molecular marker assisted breeding research of oil-tea germplasm resources, has the advantages of good repeatability and high reliability, and is an effective molecular marker.
Owner:JIANGXI ACAD OF FORESTRY
Method for obtaining EST-SSR mark
InactiveCN101619357AImprove development efficiencyReduce workloadMicrobiological testing/measurementFermentationContigConserved sequence
The invention discloses a method for obtaining an EST-SSR mark, comprising the following steps: (1) obtaining an EST sequence containing simple repeat sequence in genome; (2) in the EST sequence which contains the simple repeat sequence and is obtained in the step (1), classifying the EST sequences with the same simple sequence repeat unit into a same type; (3) performing sequence splicing on the EST sequences of the same type obtained in the step (2) to obtain an overlapping group with variable numbers of simple sequence repeat units, an overlapping group without variable numbers of simple sequence repeat units and an EST sequence without overlapping groups; (4) designing primers according to a side-vane conserved sequence of simple repeat sequence in the overlapping group with available numbers of simple sequence repeat units in the step (3), and detecting the polymorphism of the primers to obtain polymorphic primers, i.e. EST-SSR mark. Compared with the conventional method, the invention increases the development efficiency by 2-4 times, and reduces the work capacity and expenditure, thereby shortening the development time, reducing the development cost and simultaneously reducing the possibility of missing the locus of polymorphism SSR.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
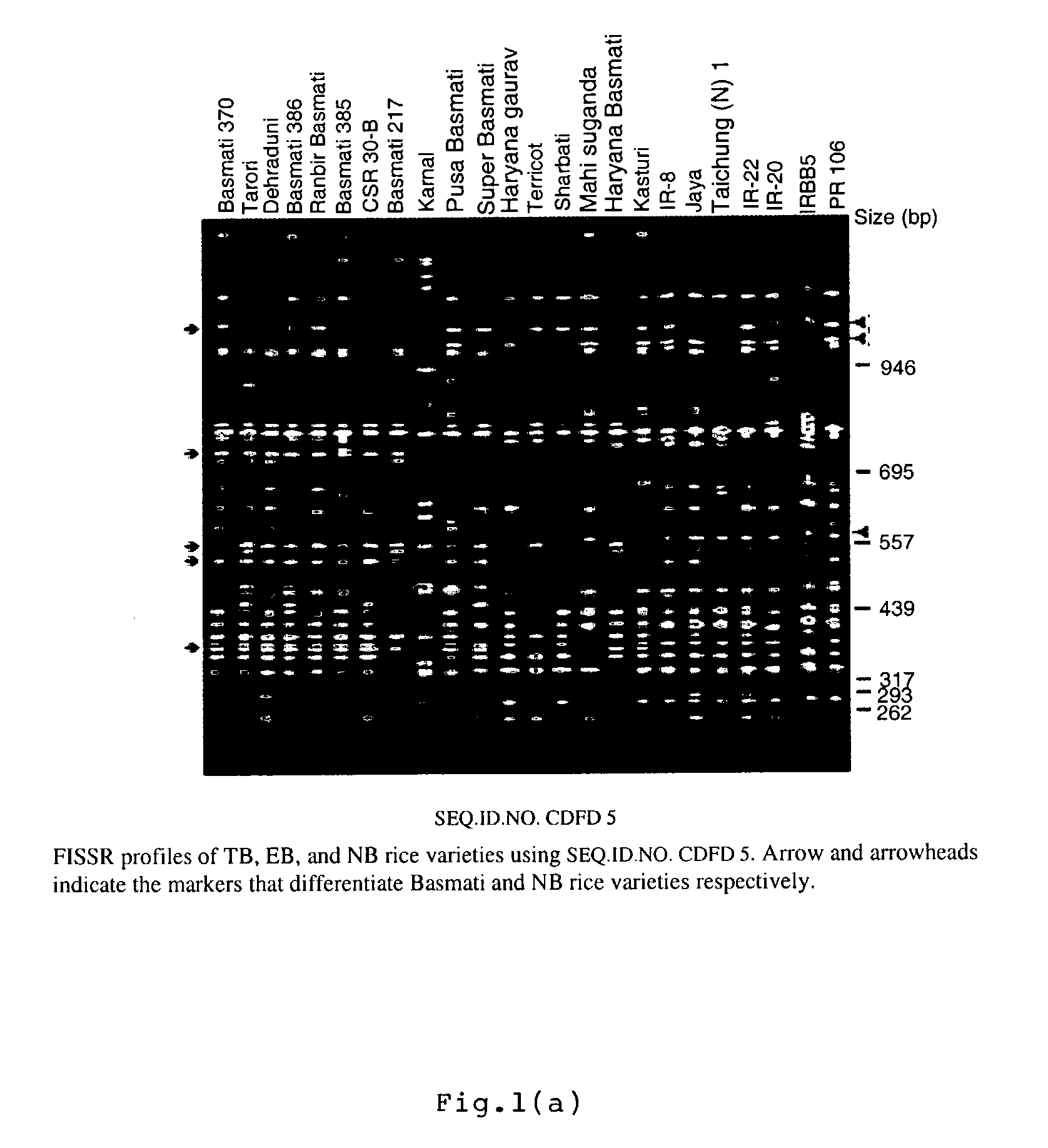
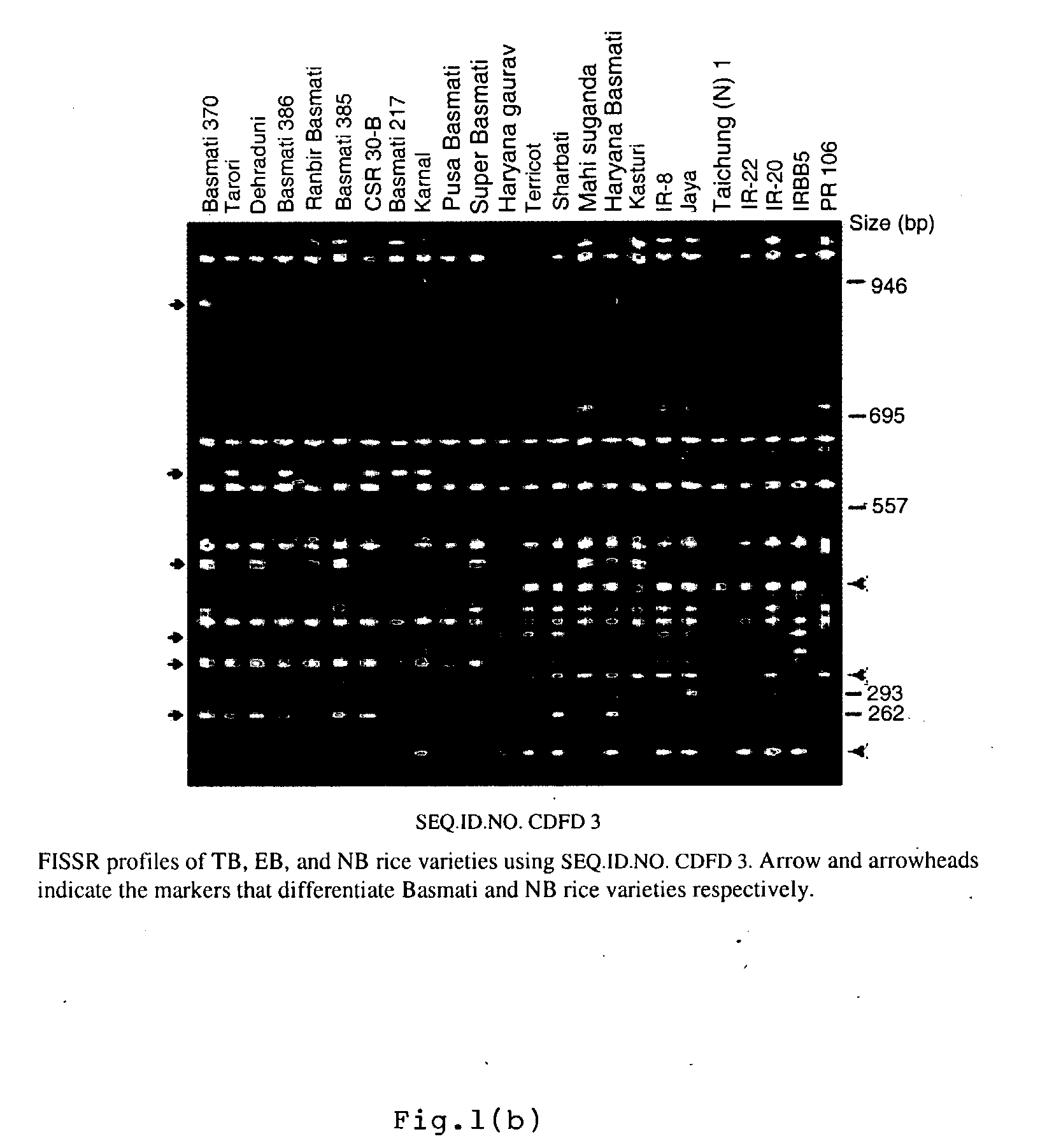
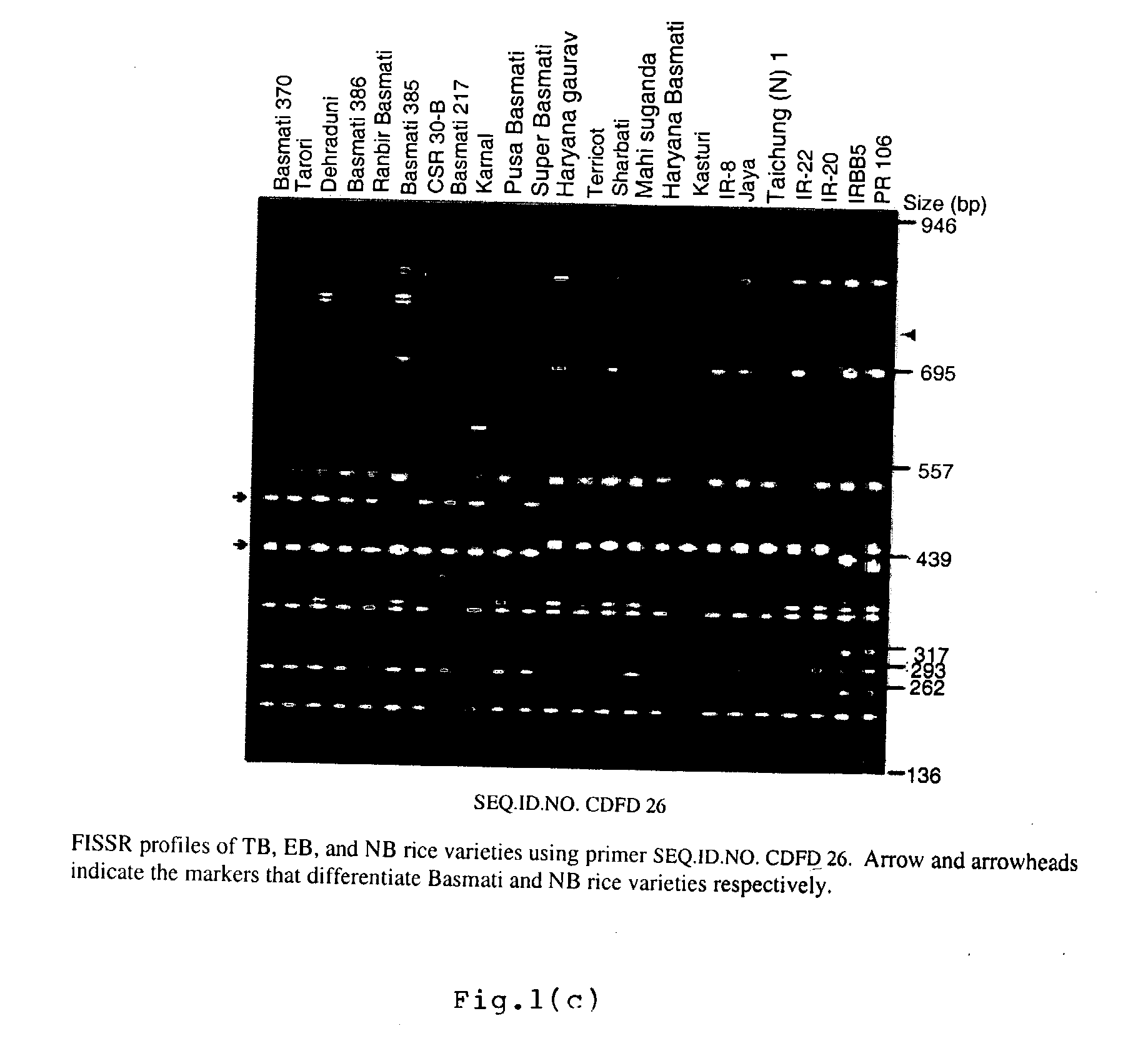
Novel FISSR-PCR primers and methods of identifying genotyping diverse genomes of plant and animal systems including rice varieties, a kit thereof
The present invention relates to set of inter-simple sequence repeats (ISSR)-PCR primers of SEQ ID Nos. 1 to 37 for genotyping eukaryotes and a method of genotyping diverse genomes of plant and animal systems using FISSR-PCR primers and SSR markers; more particularly, a FISSR and SSR method of distinguishing Basmati rice varieties from Non-Basmati (NB) rice varieties, and Traditional Basmati (TB) rice varieties from Evolved Basmati (NB) rice varieties, and also, a method for determining adulteration of Basmati rice with other rice varieties and a kit thereof.
Owner:CENTRE FOR DNA FINGERPRINTING AND DIAGNOSTICS
Method for identifying purity of tobacco variety Zhongyan 90 by using specific molecular marker method
InactiveCN102965443AIntuitive and reliable test resultsEasy to operateMicrobiological testing/measurementPhases of clinical researchBiology
The invention relates to a method for identifying the purity of a tobacco variety Zhongyan 90 by using a specific molecular marker method, wherein the purity of the flue-cured tobacco variety Zhongyan 90 is identified by using an SSR (Simple Sequence Repeats) molecular marker. According to the invention, based on a large amount of experiments, a specific primers of a characteristic strip of Zhongyan 90 are obtained through amplification, the specific primer sof Zhongyan 90 are used to carry out purity detection, detection results are reliable and visual, and compared with the conventional detection method, the method disclosed by the invention is simple to operate, high in detection efficiency and lower in cost. With adoption of the method, the detection period is short, that is, the purity of a seed can be detected in a seedling stage by using the method, in addition, standards are unified and are not influenced by human factors and environment, and results are accurate and reliable.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
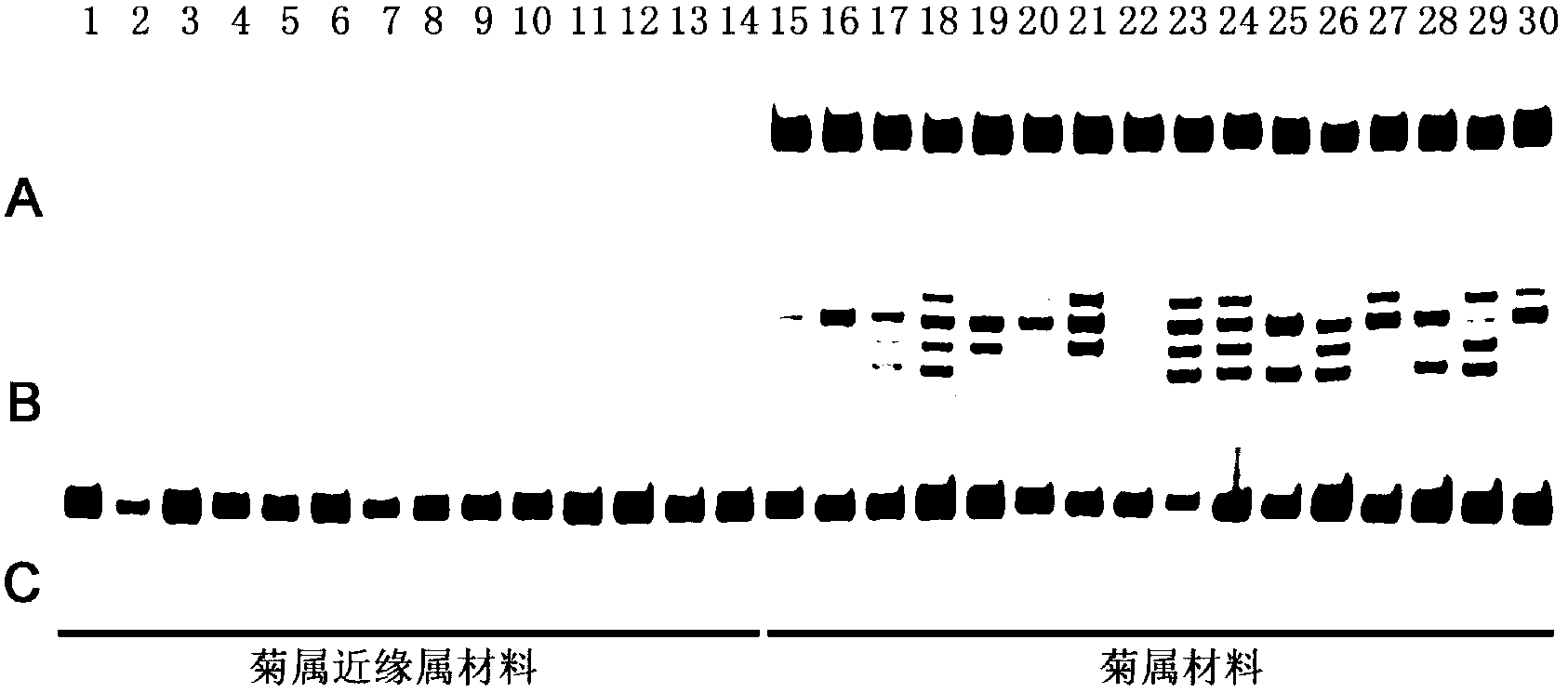
Method for developing dendranthema SSR (Simple Sequence Repeat) primer based on transcriptome sequencing
ActiveCN103233075AAdd raw dataOvercoming access difficultiesMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceLymphatic Spread
The invention belongs to biotechnology field, and relates to a method for developing a dendranthema SSR (Simple Sequence Repeat) primer based on transcriptome sequencing. A lot of sequence information is processed in batch for searching an SSR sequence and designing an SSR labeled primer based on the transcriptome sequencing by utilizing EST-SSR (Expressed Sequence Tags-Simple Sequence Repeat) interspecific metastasis and using a Perl (Practical Extraction and Reporting Language) programming language method in combination, so that the defects of low efficiency, long time consumption, high cost and the like of the SSR development are conquered. Different dendranthema materials are selected for verifying the designed SSR primer, and the primer is a successful SSR primer if detected out by any strip. By adopting the method, 1788 pairs of SSR primers are successively designed, so that a novel method and thinking are provided for the development of the dendranthema SSR primer to further achieve molecular marker assistant selection breeding and comparative genomics research.
Owner:NANJING AGRICULTURAL UNIVERSITY
Lysinecorn SSR (simple sequence repeat) molecular marker auxiliary selecting and breeding method
InactiveCN102154449AShort breeding cycleLow costMicrobiological testing/measurementAgricultural scienceMarker-assisted selection
The invention provides a lysinecorn SSR (simple sequence repeat) molecular marker auxiliary selecting and breeding method. By combining a backcross transforming method with a molecular marking method, the method successfully constructs the lysinecorn isogenic gene systems QZ58, Q478 and QC72. After the SSR molecular marker auxiliary selecting and breeding method is used the mono-clones which contain the O2 gene can be detected by SSR molecular marker in the O2 gene after one generation is backcrossed, and the second generation can be backcrossed without selfing, so that the breeding is short in period, low in cost, and high in efficiency. Aiming at different breeding target requirements, the method is used for performing the gene type detection to the backcross offspring seedling leaves; and the method is not influenced by the environment conditions and combines with south propagating and generation adding, so that the target genes are transferred into a good material within a shorter time.
Owner:HENAN ACAD OF AGRI SCI
Inter-simple sequence repeat ISSR-SCAR marker specific to E-group chromosomes of agropyron elongatum
The invention discloses an inter-simple sequence repeat ISSR-SCAR marker specific to E-group chromosomes of agropyron elongatum. The invention provides a kit for detecting the E-group chromosomes of agropyron elongatum. The kit comprises the following primer pair: one primer in the primer pair is nucleotide shown in a sequence 3 of a sequence table, and the other primer in the primer pair is nucleotide shown in a sequence 4 of the sequence table. The E-group chromosomes of thinopyrum elongatum are the basic chromosome group forming the polyploid species of elytrigia and carry the genes beneficial to the genetic breeding of wheat, the detection of exogenous E chromatins in wheat is the key point for identifying recombination lines and introgression lines of the wheat, namely the agropyron elongatum, the work of screening and identifying a large number of filial generations is quite complicated, and tests prove that the ISSR-SCAR marker developed by the invention can lay a foundation for quickly and accurately identifying the E-group chromatins or chromosomes of exogenous agropyron elongatum under the genetic background of wheat.
Owner:HARBIN NORMAL UNIVERSITY
Seedless litchi graft stock selecting technology
InactiveCN1887048AGrafting affinity is goodImprove survival rateHorticultureRootstockGenetic diversity
The present invention discloses the seedless litchi graft stock selecting technology. The technological process of selecting seedless litchi graft stock includes the following steps: 1. separating the simple sequence repeating (SSR) sequence through a combined selective amplifying microsatellite (SAM) and database searching method; 2. designing SSR primer with the flanking sequence of SSR sequence; 3. screening out the useful primer to obtain litchi specificity SSR marker; 4. applying the litchi specificity SSR marker in the genetic diversity analysis of partial litchi germplasm; 5. data analysis; and 6. selecting the litchi germplasm with near affinity as stock for seedless litchi grafting propagation based on the analysis result. The said process results in high survival rate and can obtain high quality seedless litchi variety.
Owner:INST OF TROPICAL BIOSCI & BIOTECH CHINESE ACADEMY OF TROPICAL AGRI SCI +1
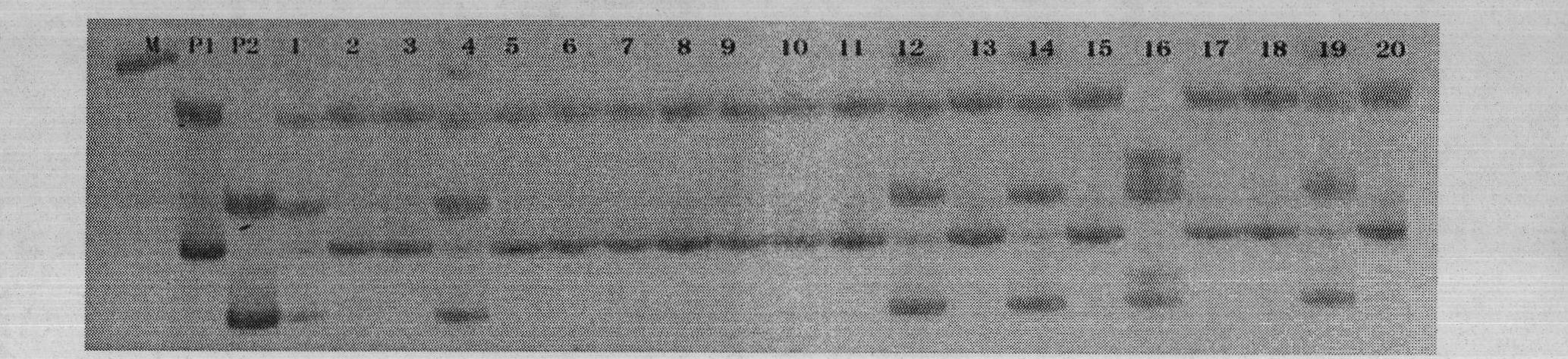
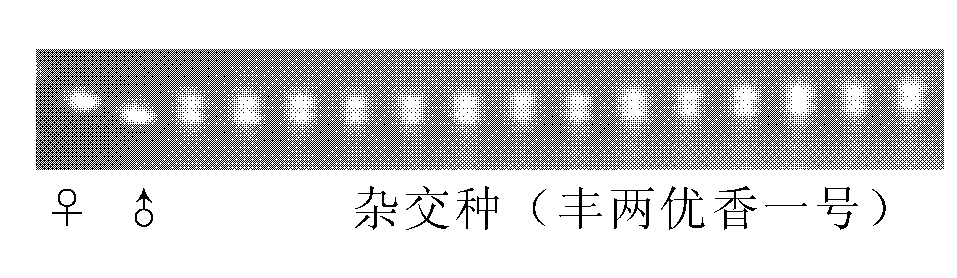
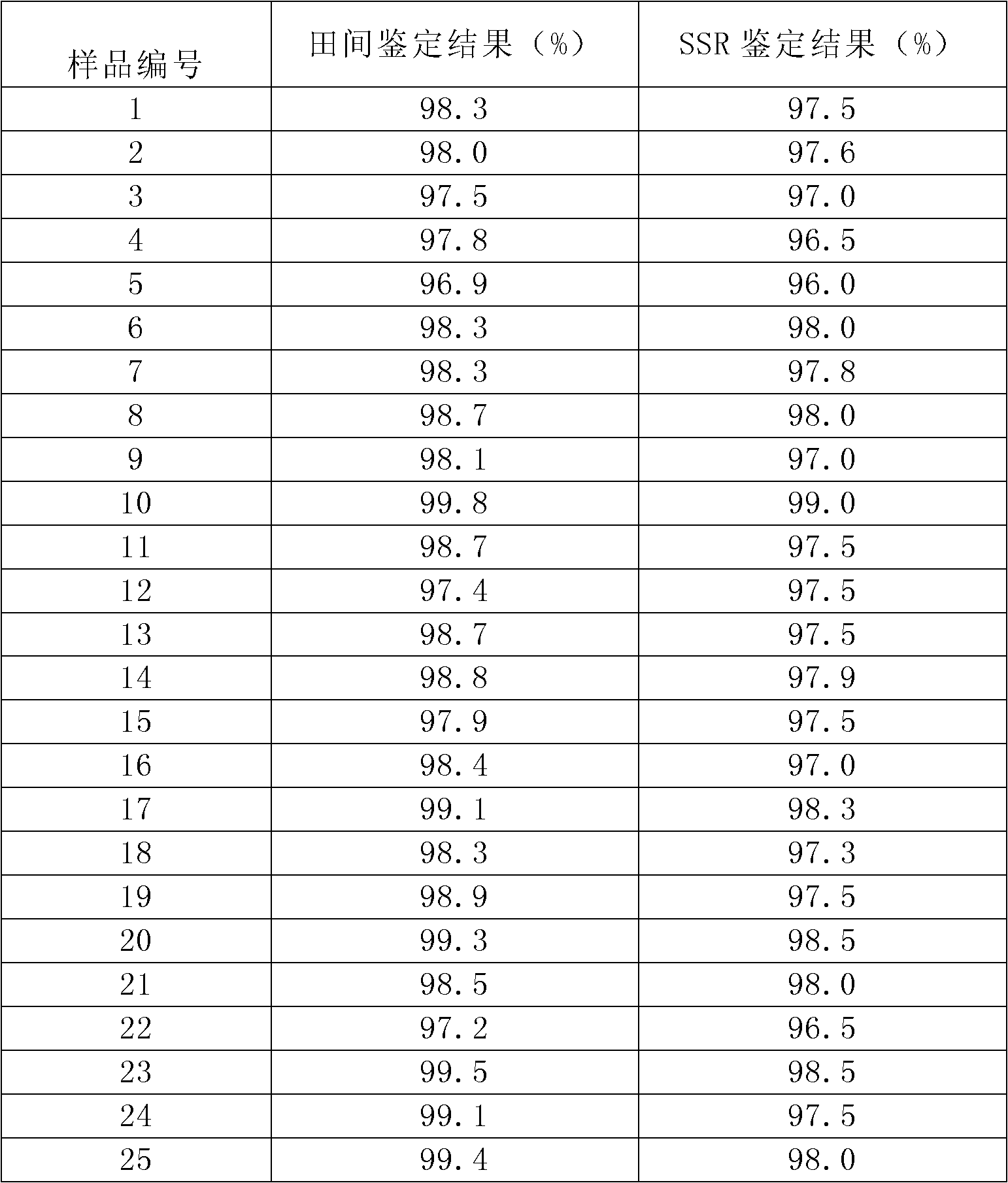
Method for inspecting purity of two-line hybrid rice seeds by utilizing SSR (Simple Sequence Repeat) method
InactiveCN102220428AAccurate purityHigh purityMicrobiological testing/measurementAgricultural scienceAgarose electrophoresis
The invention discloses a method for inspecting purity of two-line hybrid rice seeds by utilizing an SSR (Simple Sequence Repeat) method, in particular a method for inspecting the purity of Fengliangyou No. 1 rice or Fengliangyouxiang No. 1 rice seeds by utilizing SSR molecular markers. The method comprises the following steps of: performing amplification and agarose electrophoretic separation on genome DNA (Deoxyribonucleic Acid) of the two-line hybrid rice varieties Fengliangyou No. 1 and parents thereof or Fengliangyouxiang No. 1 and parents thereof respectively by utilizing primers P1 and P2 to find out a characteristic spectrum band of difference between a first-filial generation and the parents; inspecting the samples of Fengliangyou No. 1 or the Fengliangyouxiang No. 1 by calculating the purity of the inspected seeds; and comparing and contrasting the inspected purity with purity tested by field planting to confirm that the results of the two are highly consistent. The method is suitable for inspecting the authenticity and variety purity of the Fengliangyou No. 1 or Fengliangyouxiang No. 1 two-line hybrid rice and parents thereof.
Owner:HEFEI FENGLE SEED +1
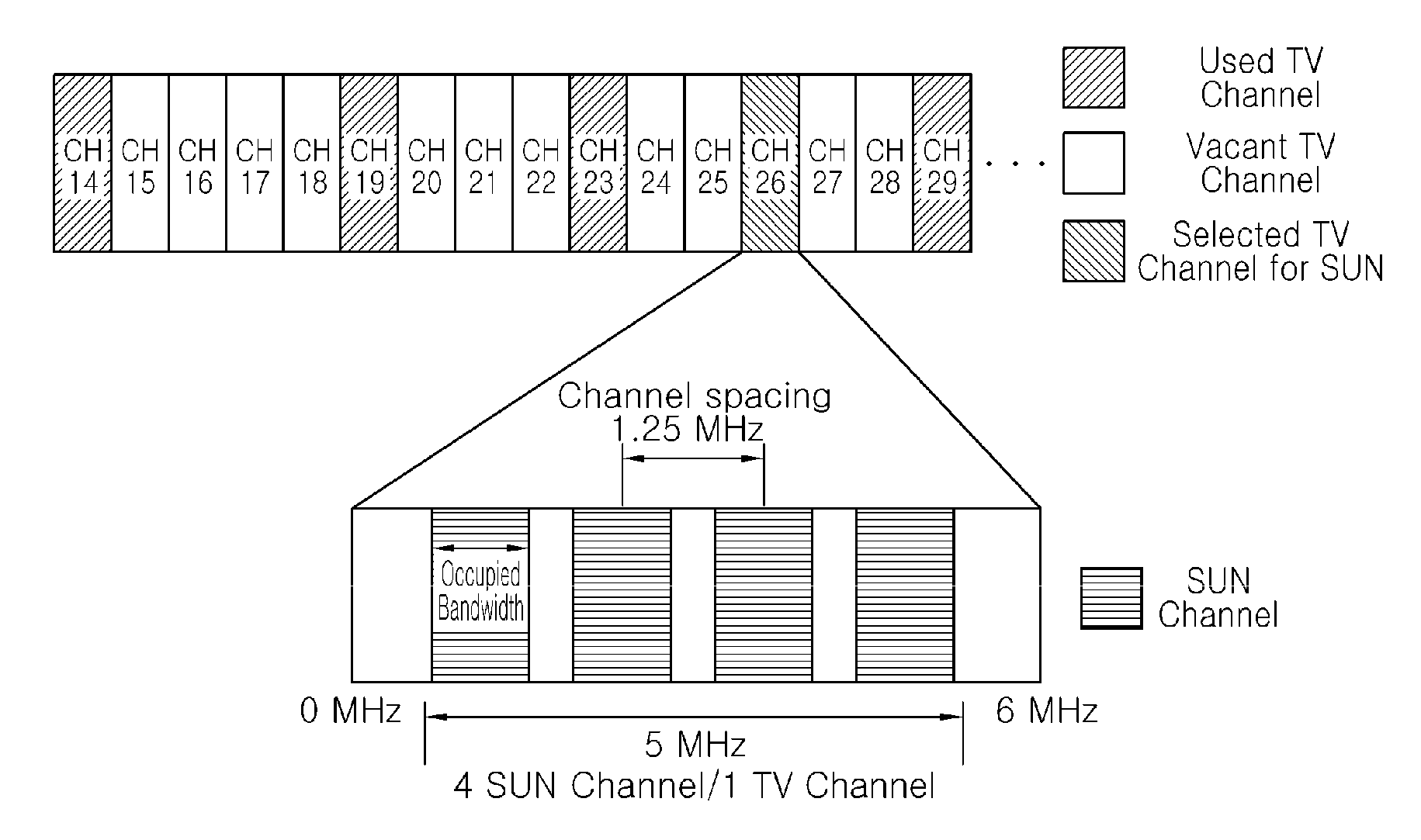
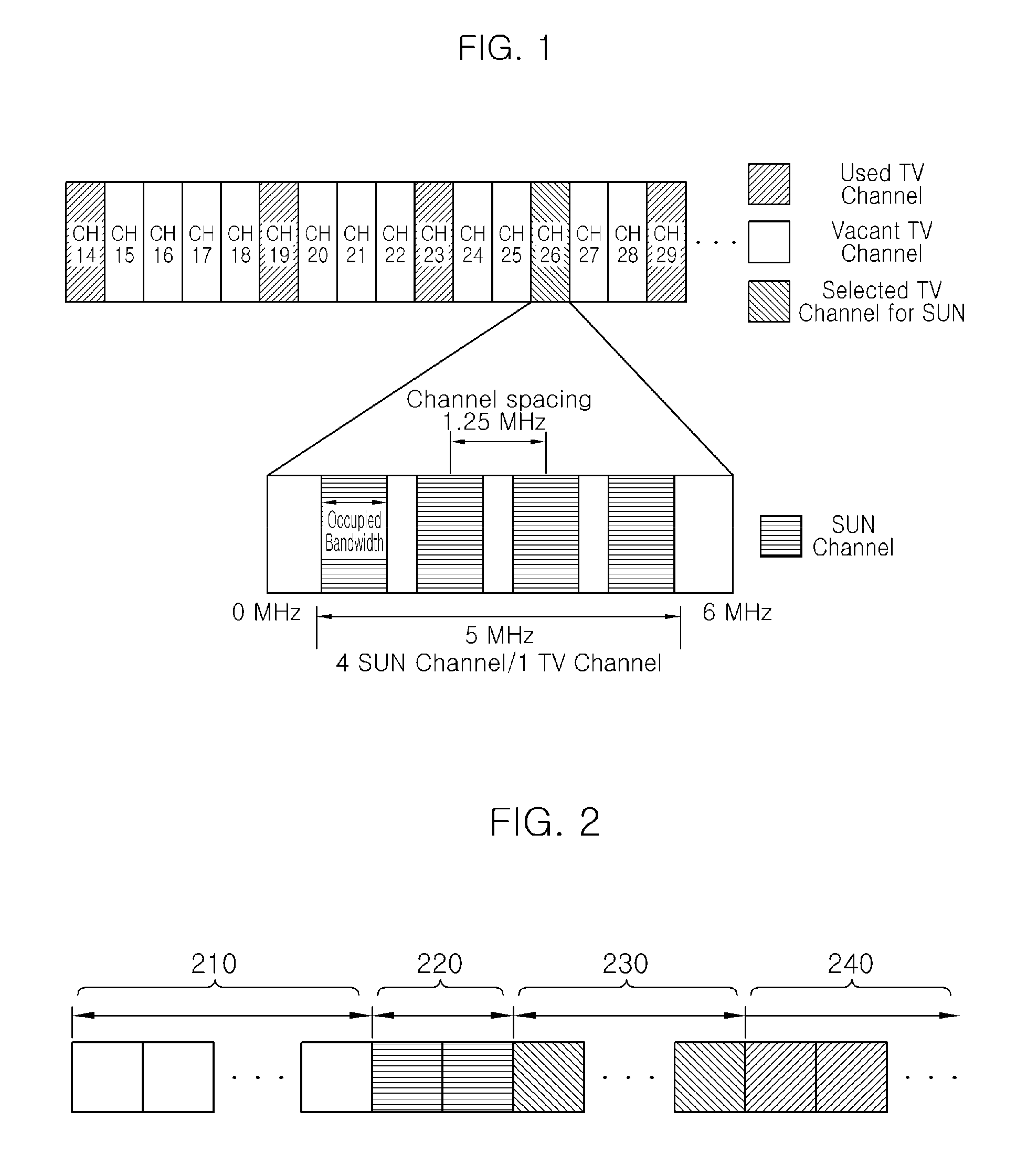
Method of communicating for smart utility network using TV white space and apparatus for the same
InactiveUS20110002416A1Satisfies requirementOvercome fadingSecret communicationPhase-modulated carrier systemsTime domainCyclic prefix
There are disclosed a method of communicating for a smart utility network using a TV white space and an apparatus for the same. The method of communicating for a smart utility network using a TV white space according to the present invention includes: generating a time domain sequence repeated every predetermined number of samples; generating an OFDM symbol having a cyclic prefix length corresponding to an FFT size divided by a natural number of 2 or more and including samples of a number corresponding to the sum of the FFT size and the cyclic prefix length; and generating an SUN packet to be transmitted through a TV channel band selected in the TV white space by using the time domain sequence and the OFDM symbol. Accordingly, it is possible to satisfy all requirements required by the IEEE 802.15.4g SUN standardization group.
Owner:ELECTRONICS & TELECOMM RES INST
Method for identifying barley varieties by SSR (simple sequence repeat) primers and applications of method
ActiveCN102787172ANot easy to influenceImprove test efficiencyMicrobiological testing/measurementBiotechnologyHordeum vulgare
The invention relates to a method for identifying barley varieties by SSR (simple sequence repeat) primers. The method comprises steps as follows: extracting DNA of samples to be tested; using 14 pairs of basic core SSR primers and 14 pairs of extended core SSR primers to perform PCR (polymerase chain reaction) amplification; performing gel electrophoresis, modified polyacrylamide gel electrophoresis and silver staining on the PCR amplified product; analyzing an SSR band, wherein the samples to be tested are at least two barley varieties; when the number of different sites of the varieties is larger than or equal to 3, the varieties are judged as different varieties; when the number of the different sites of the varieties is 1 or 2, the varieties are judged as similar varieties; and when the number of the different sites of the varieties is 0, the varieties are judged as suspected identical varieties. The method provided by the invention can be used for identifying the barley varieties to be tested and the known barley varieties, or establishing a barley variety database; the method has advantages of high efficiency, accuracy, low cost and convenient operation; and with the adoption of the method, the authenticity of the barley varieties is effectively monitored, crop varieties are protected, and fake varieties are prevented from entering the market.
Owner:JIANGSU ACAD OF AGRI SCI
Method for breeding high-yield high-oil peanut species
InactiveCN102067816ADeep leaf colorBig fruitPlant genotype modificationAngiosperms/flowering plantsHeterosisOperability
The invention discloses a method for breeding high-yield high-oil peanut species. In the method, a genetic relationship of the selected high-yield high-oil peanut species is found out by an SSR (Simple Sequence Repeat) molecule marker technology; the species with distant hybrid advantages and larger selection difference is used as a parent for preparing cross combination; the variation frequency is expanded by radiation effect, and the selecting range of a high-yield high-oil gene is expanded; the genetic linkage of an adverse gene is broken down by applying hybridization, and the recombination, polymerization and complementation of high yield, high oil and resistance are realized; an individual plant is directionally selected by using ideal characters as a target in stable generation; and the individual plant is inbred for one generation, a plant system with complete and ideal characters is further selected, and the high-yield high-oil plant system is obtained. The invention has the characteristics of rapidly breeding the high-yield high-oil peanut new species and having simple method and high operability.
Owner:山东鲁花农业科技推广有限公司
Method for identifying pennisetum forage grass by utilizing ISSR (Inter Simple Sequence Repeat) molecular marker technology
InactiveCN102586449AReduce demandImprove the problem of filling the good with the bad, mixing the good and the bad, and confusing the real with the fakeMicrobiological testing/measurementAnimal ForagingAgricultural science
The invention belongs to the field of forage grass germplasm resource identification, in particular to a method for identifying pennisetum forage grass by utilizing an ISSR (Inter Simple Sequence Repeat) molecular marker technology, wherein primers used by ISSR-PCR (Polymerase Chain Reaction) are UBC815 and UBC835, and the nucleotide sequences of the primers are respectively UBC815-CTCTCTCTCTCTCTCTG and UBC835-AGAGAGAGAGAGAGYC. The method fills the blank of distinguishing pennisetum forage grass categories (varieties) by applying the ISSR molecular marker method, and an electrophoresis ideograph finally drawn by the method for distinguishing seven pennisetum forage grass materials for tests can be used as a basis for distinguishing and identifying the seven materials. The method gets rid of unreliable factors of traditional morphology identification and can quickly and accurately distinguish the seven pennisetum forage grass materials, and an identification result can be used as a reliable basis for approving categories, varieties and strains so as to guarantee the great popularization of germplasm on production.
Owner:CHINA AGRI UNIV
Method for identifying commercial cigarette by applying SSR (Simple Sequence Repeat) molecular marker technique
InactiveCN103451296AThe identification result is accurate and reliableHigh sensitivityMicrobiological testing/measurementBiotechnologyNicotiana tabacum
The invention discloses a method for identifying a commercial cigarette by applying an SSR (Simple Sequence Repeat) molecular marker technique, belonging to the field of molecular biology. The method comprises the steps of carrying out genetic map analysis on a tobacco in single variety by applying the SSR technique, screening out SSR sites with moderate aberration rate and variety specificity, extracting total DNA (deoxyribonucleic acid) of commercial cigarette shreds, amplifying by using screened specific primers, comparing genetic variation type with the type of the single variety, distinguishing the variety of tobacco leaves contained in the commercial cigarette shreds according to the variety specificity sites, and contrasting the using condition of the variety of the tobacco leaves in the corresponding cigarette formula to conveniently identify the authenticity of the cigarette. Meanwhile, the method is also suitable for the identification of tobacco leaves in single variety, tobacco shred groups and tobacco shred samples in mixed varieties.
Owner:HONGYUN HONGHE TOBACCO (GRP) CO LTD
Method for identifying authenticity of sweet potato variety by using inter-simple sequence repeat (ISSR) molecular marker method
ActiveCN103484555AEasy to identifyRapid identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
The invention discloses a method for identifying authenticity of sweet potato variety by using an inter-simple sequence repeat (ISSR) molecular marker method. A disclosed primer group used for identifying the authenticity of the sweet potato variety in an assisted mode consists of a primer HBAAS-001, a primer HBAAS-815 and a primer HBAAS-825, wherein the nucleotide sequence of the primer HBAAS-001 is shown as SEQ ID No:1, the nucleotide sequence of the primer HBAAS-815 is shown as SEQ ID No:2, and the nucleotide sequence of the primer HBAAS-825 is shown as SEQ ID No:3. An ISSR fingerprint spectrum of a single primer of the sweet potato variety is successfully constructed according to an ISSR molecular marker, so that the method for identifying the authenticity of the sweet potato variety is simple, convenient, rapid, high in repeatability and high in stability, and a solid foundation is laid for allowing an ISSR molecular marker technology to be widely applied to identifying the authenticity of the sweet potato variety.
Owner:INST OF FOOD CROPS HUBEI ACAD OF AGRI SCI
Molecular marking method for major quantitative trait loci(QTL) for rice grain length
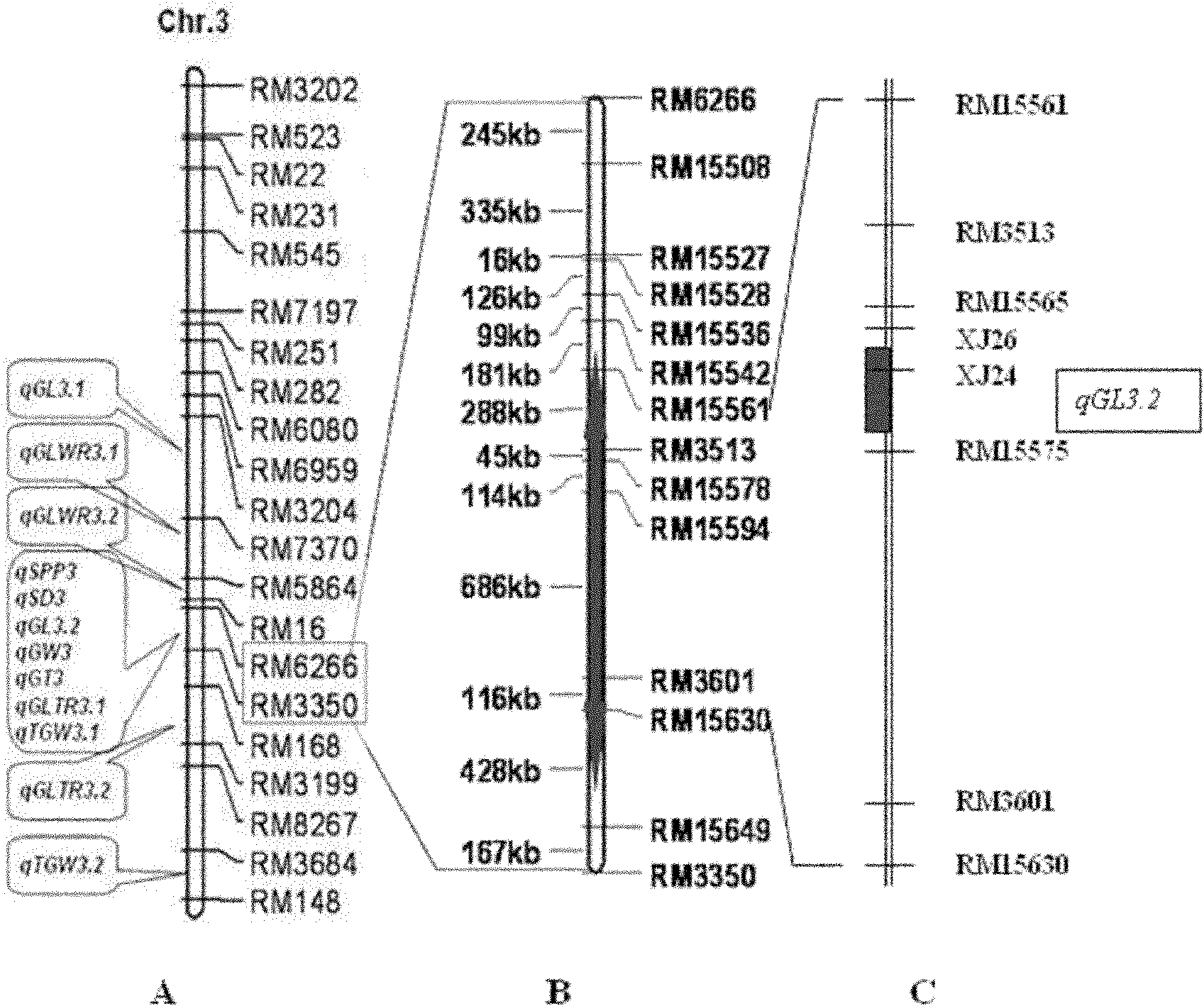
InactiveCN102154471AClear locationFine locationMicrobiological testing/measurementGermplasmRice grain
The invention belongs to the field of genetic breeding science and discloses a molecular marking method for a major quantitative trait loci (QTL) for rice grain length. In the method, Indel and simple sequence repeats (SSR) are used to mark one or more pairs of primers of XJ24 and XJ26 and RM15575 respectively to perform the polymerase chain reaction (PCR) amplification of rice genome DNA, PCR amplification products undergo an electrophoretic test on 8 percent non-denaturing polyacrylamide gel, and if DNA fragments in corresponding size are generated through amplification, qGL 3.2 synergisticallelic mutant genes exist. When the large-grain germplasm of rice and breeding populations constructed by various parents of rice are detected by the grain length QTL qGL3.2 coseparated and closely interlocked molecular marking method, the major gene qGL 3.2 for controlling grain length and weight can be quickly imported, the selecting efficiency of the major gene qGL 3.2 is improved, and the method contributes to appearance improvement and high-yield breeding of rice.
Owner:NANJING AGRICULTURAL UNIVERSITY
ISSR-SCAR (inter simple sequence repeat-sequence characterized amplified region) marker capable of identifying Wujiang brassica chinensis and identification method of marker
ActiveCN104894124AEasy to operateHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMolecular identification
The invention discloses an ISSR-SCAR (inter simple sequence repeat-sequence characterized amplified region) marker capable of identifying Wujiang brassica chinensis and an identification method of the marker. The method includes: performing total DNA (deoxyribose nucleic acid) extraction, PCR (polymerase chain reaction) amplification, primer selection and ISSR-SCAR detection on Wujiang brassica chinensis and other 10 brassica chinensis varieties, and finally determining an ISSR-SCAR molecular marker and a molecular identification method of the Wujiang brassica chinensis. A primer shown in the sequence such as SEQ ID NO:1 is used for amplification to obtain a specific fragment shown in the sequence such as SEQ ID NO:2. According to the sequence, a pair of primers in the sequences such as SEQ ID NO:3 and SEQ ID NO:4 is designed. The marker is simple to operate, stable and reliable, is the first molecular identification marker applied to the variety of Wujiang brassica chinensis, can be used for quickly identifying the variety and has significance of protecting germplasm resources of geographical indication products.
Owner:苏州市种子管理站
SSR (Simples sequence repeats) and InDel (insertion/deletion) molecular marker primer linked with brassica campestris orange head gene Br-or, and application thereof
InactiveCN103103184AEasy to detectStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationBrassicaAgricultural science
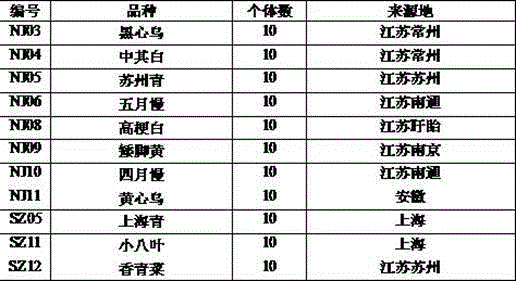
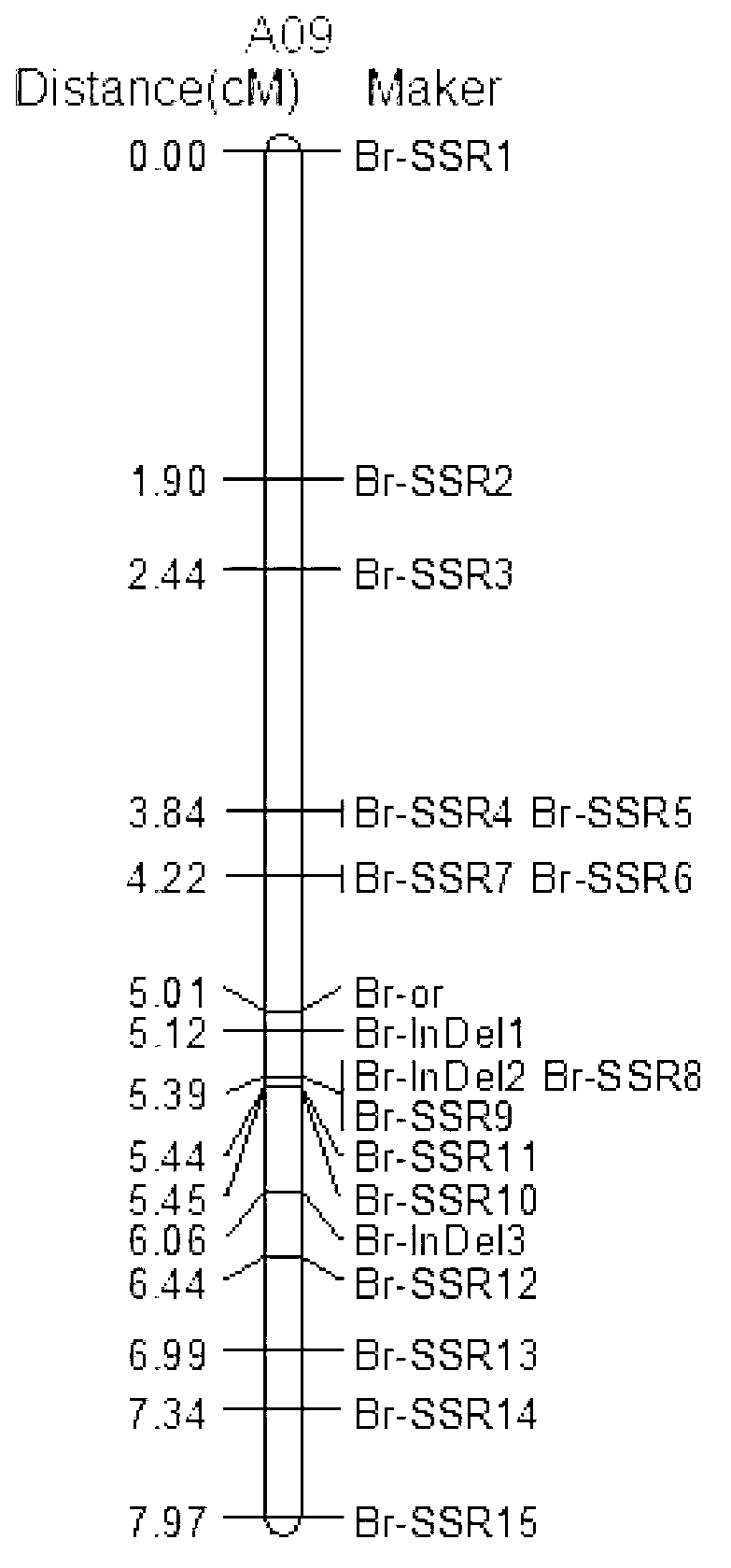
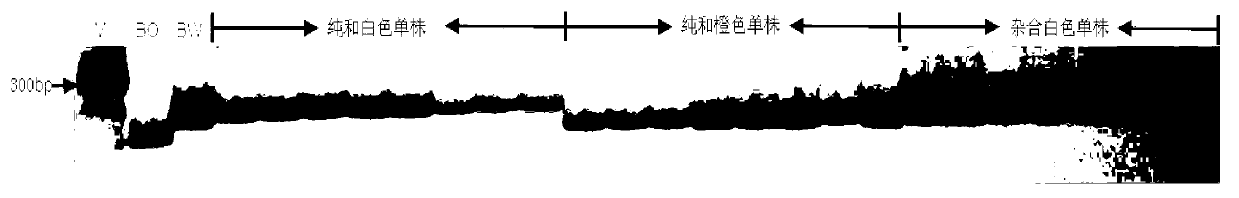
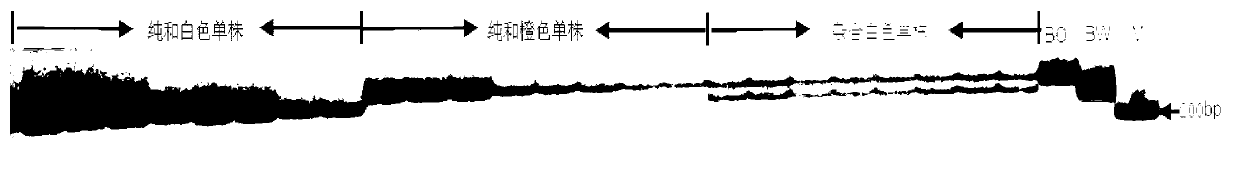
The invention discloses an SSR (simples sequence repeats) and InDel (insertion / deletion) molecular marker primer linked with brassica campestris orange head gene Br-or and an application of the molecular marker primer. The marker primer is obtained by the steps of: based on the genome DNA (deoxyribonucleic acid) of orange brassica campestris and normal white brassica campestris and F2S4 separation group as a template, carrying out polymorphism primer screening to orange head and white head single strain DNA mixed tank in F2S4 by using 480 pairs of SSR and InDel primer pairs, then analyzing single strain of F2S4 group, and screening the molecular marker linked with the brassica campestris orange head gene Br-or to finally obtain fifteen Br-SSR and three Br-InDel molecular markers linked with the Br-or gene, wherein a molecular genetic map of the brassica campestris orange head gene Br-or is created on the ninth link group (A09). According to the link analysis, the genetic distance of two markers most tightly linked with both sides of the Br-or gene are 0.11cM and 0.79cM; and the molecular marker primer has the advantages of being convenient for detection, stable in amplification, and high in repeatability and accuracy, and thus a basis is provided to the molecule assistant selective breeding of brassica campestris orange head character and the breeding process is accelerated.
Owner:NORTHWEST A & F UNIV
High sensitive and jettisonable multicomponent chemiluminescent imaging immunosensor
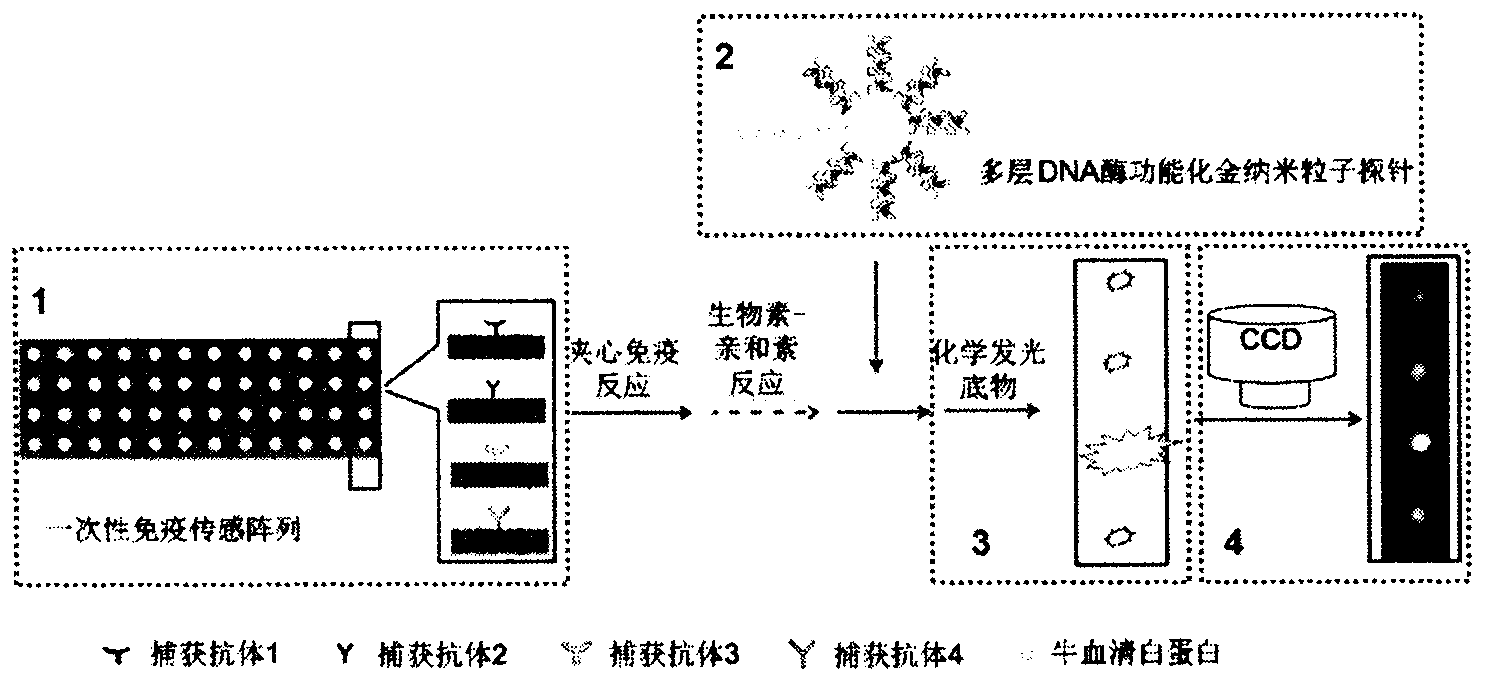
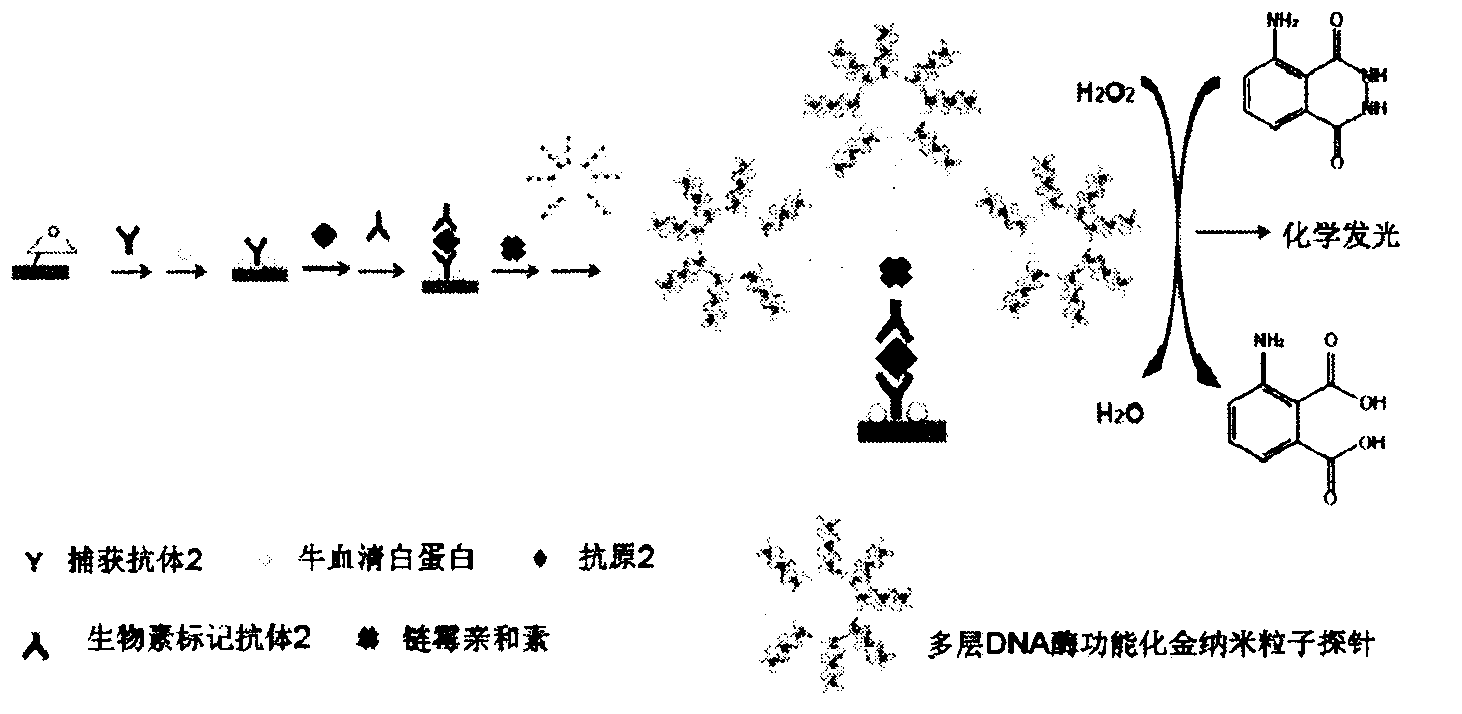
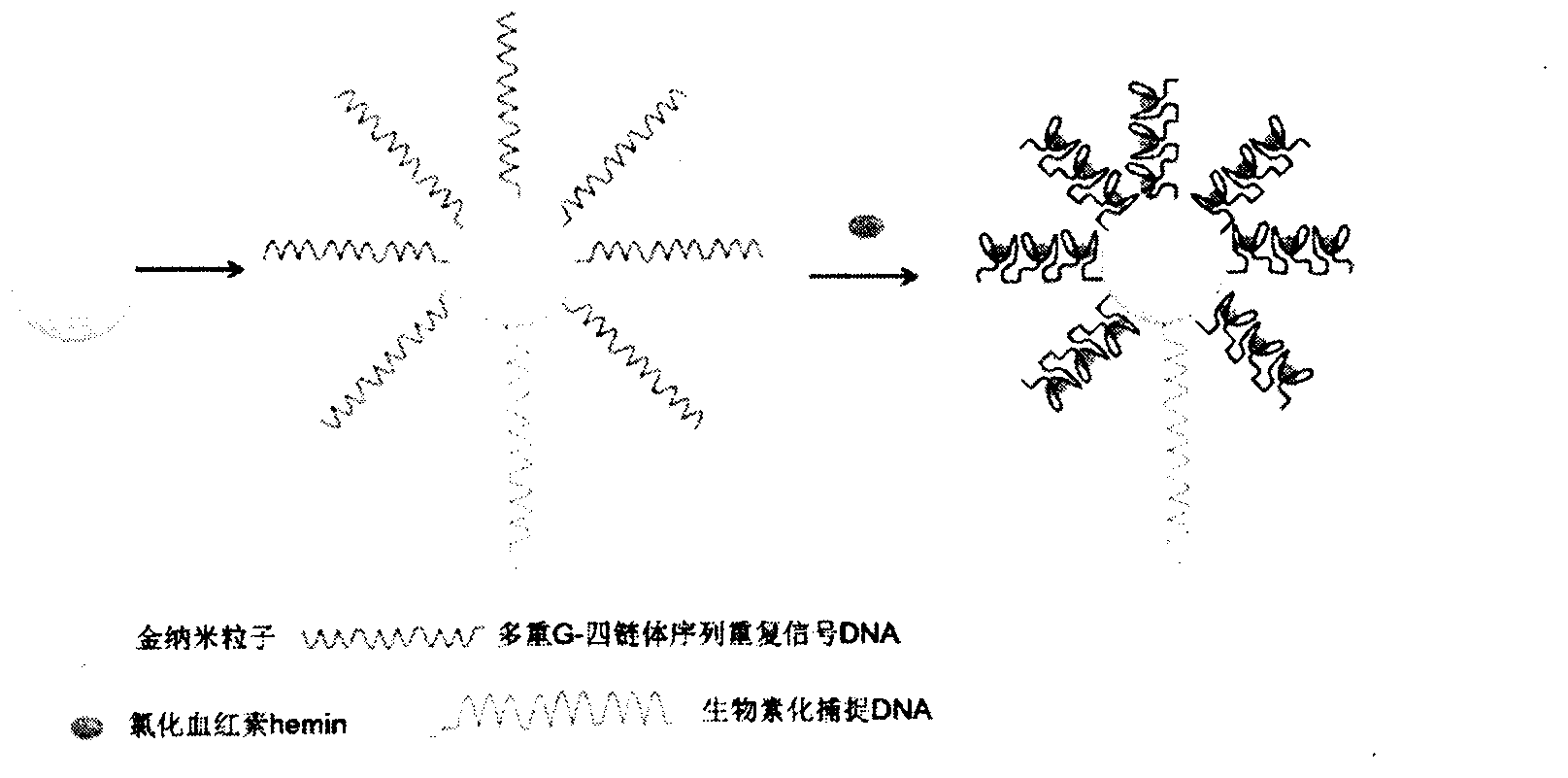
InactiveCN103575896AAchieving Simultaneous High Sensitivity Image ImmunoassaysEasy to makeMaterial analysis by optical meansBiological testingSensor arrayPeroxidase
The invention relates to a high sensitive and jettisonable multicomponent chemiluminescent imaging immunosensor. The immunosensor is prepared by: constructing a 4*12 array on a silanized glass slide by a screen printing technique, and coating the array points with different capture antibodies to construct a jettisonable multicomponent immune sensor array; immobilizing biotinylated capture DNA and multiple G-quadruplex sequence repeat signal DNA on gold nanoparticle surfaces simultaneously, combining G-quadruplex signal DNA with heme to form DNA enzyme so as to prepare a multilayer DNA enzyme functionalized gold nanoparticle probe; and based on sandwich immunoassay, forming a sandwich immune complex on the sensor array, carrying out biotin-avidin reaction, labeling different immune complexes with the multilayer DNA enzyme functionalized gold nanoparticle probe; and making use of the peroxidase characteristics of DNA enzyme to catalyze reaction between chemiluminescent substrate H2O2-luminol to obtain a sensitive chemiluminescent signal, thus realizing high sensitive image immunoassay of a variety of protein. The immunosensor has the advantages of simple design, low cost, high sensitivity, high throughput and good repeatability, etc., and has certain clinical application value.
Owner:JIANGSU CANCER HOSPITAL +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com