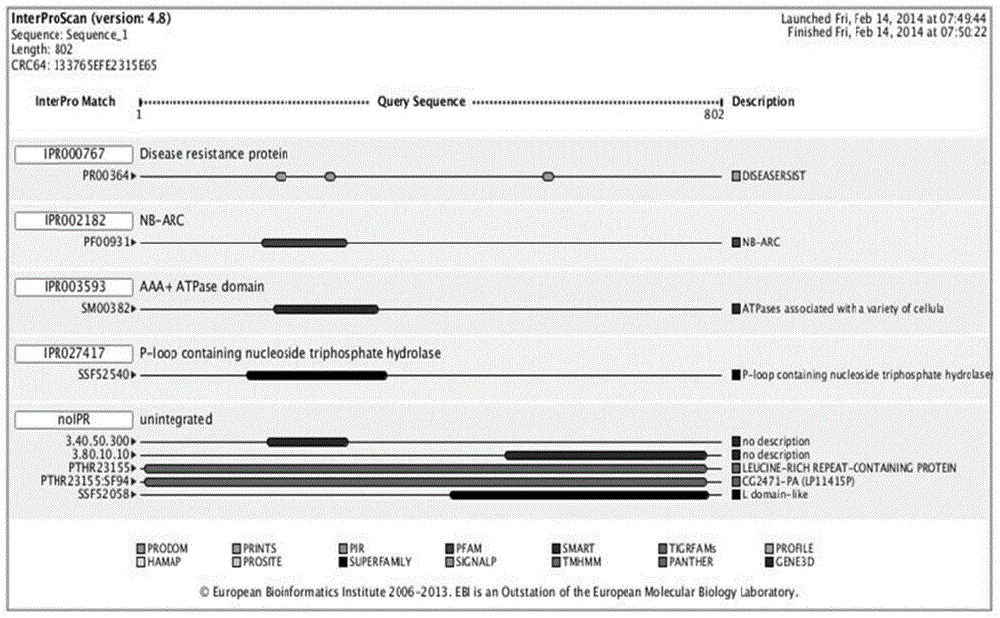
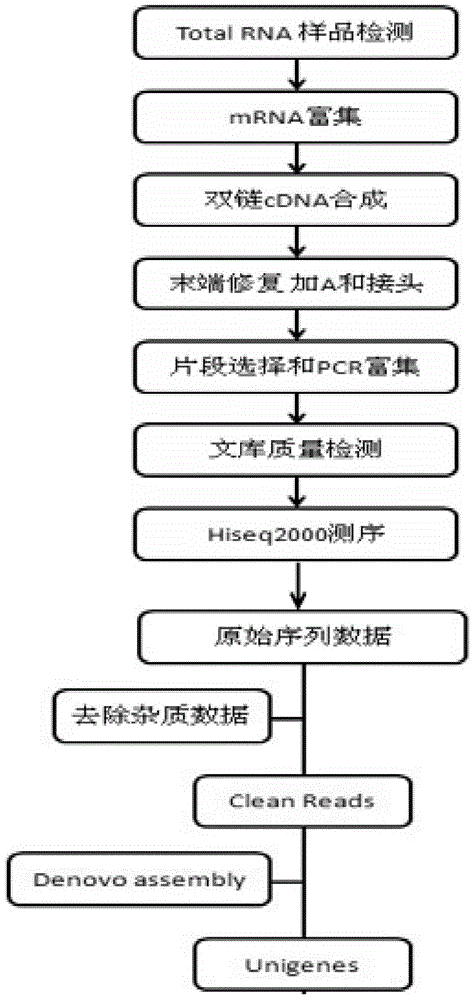
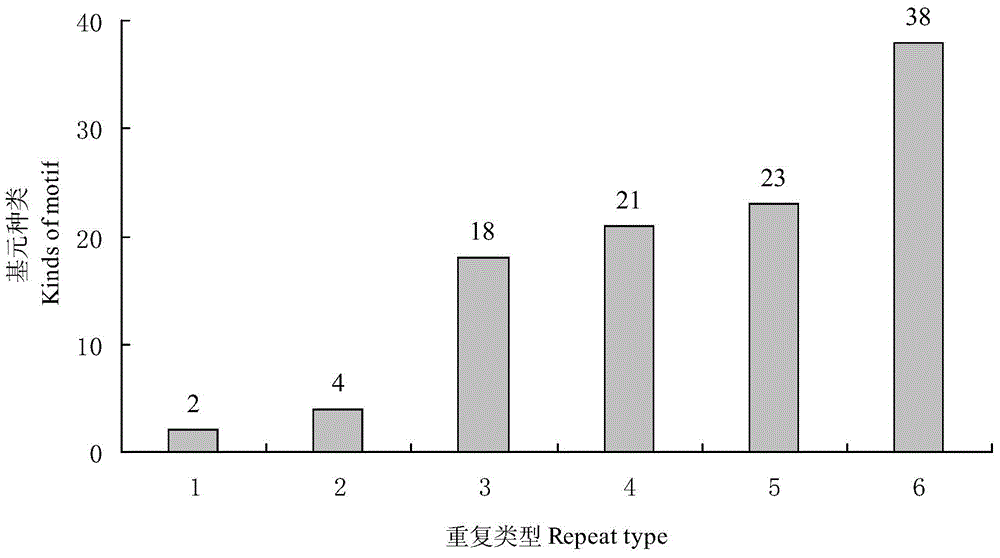
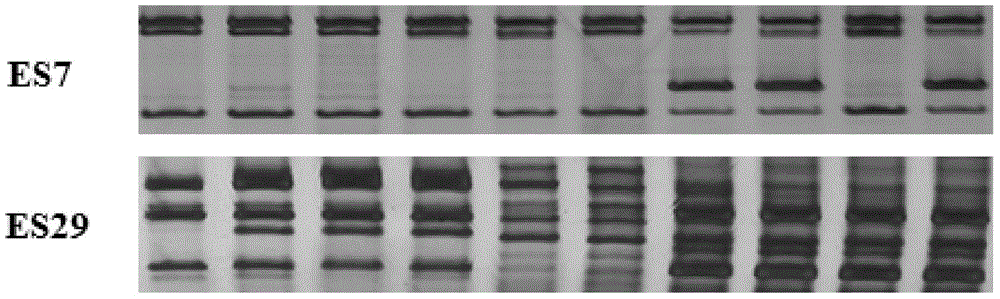
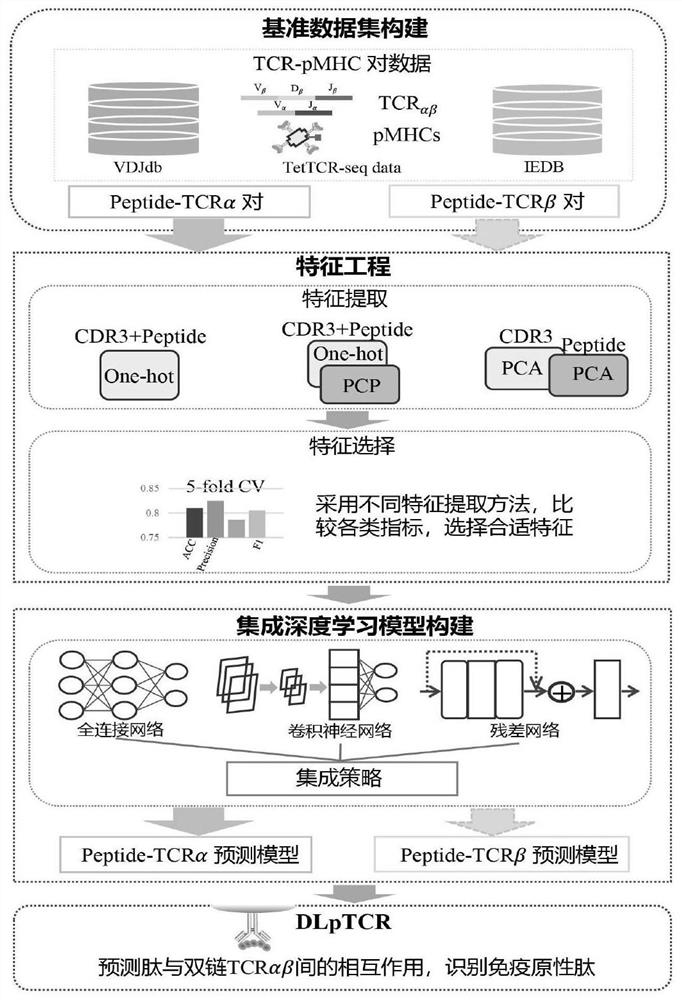
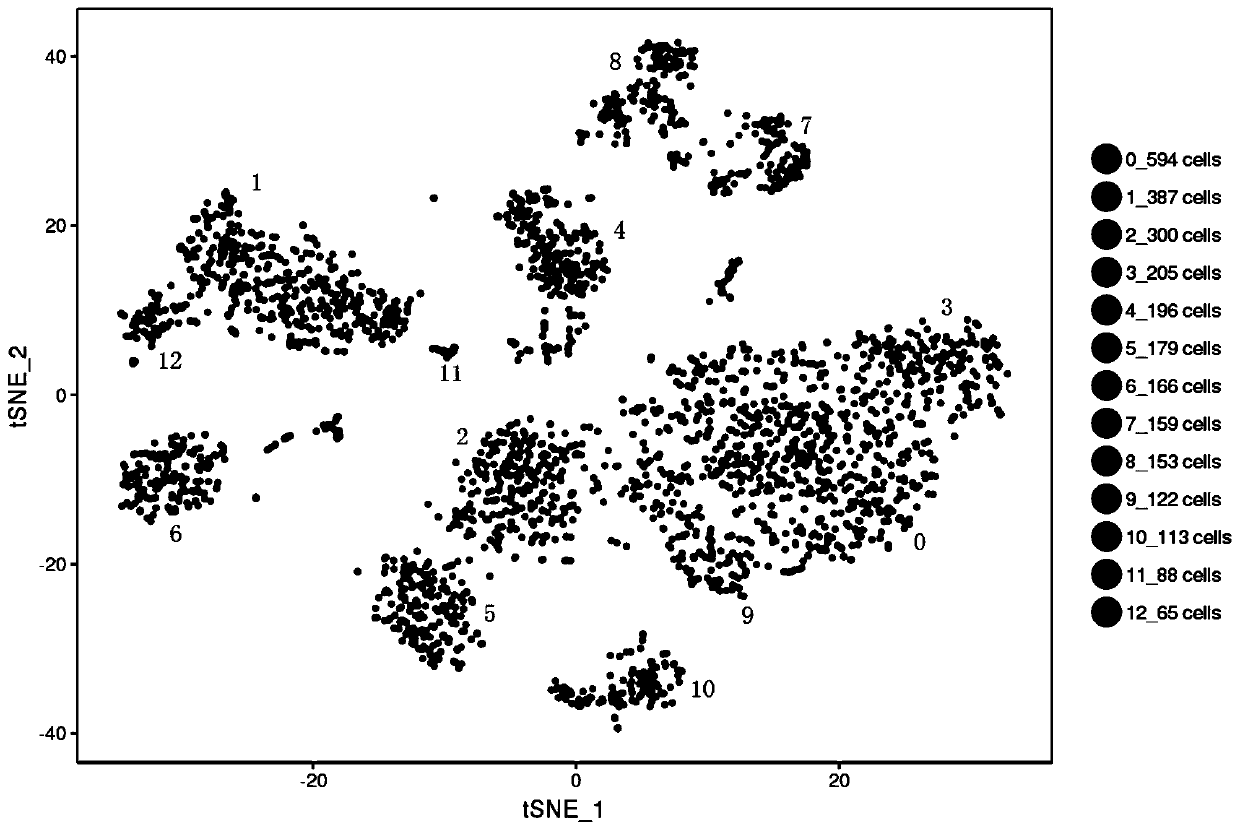
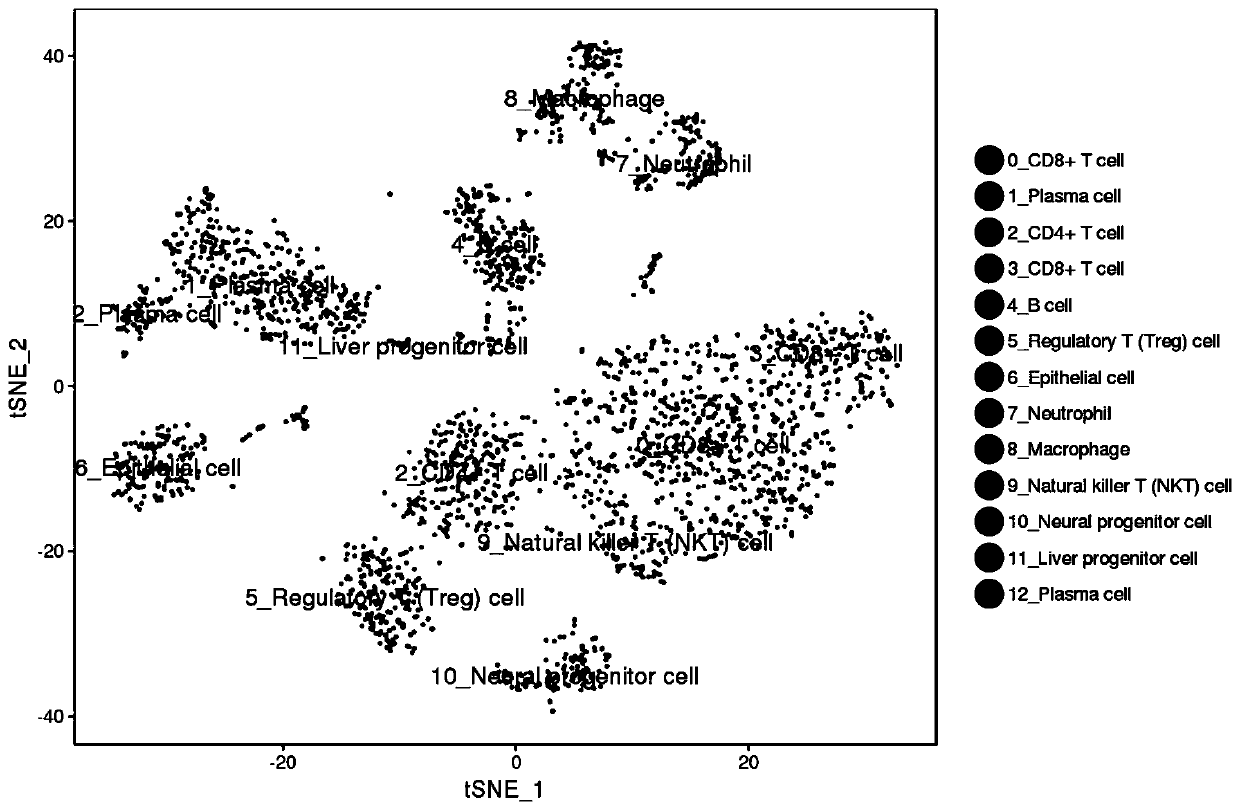
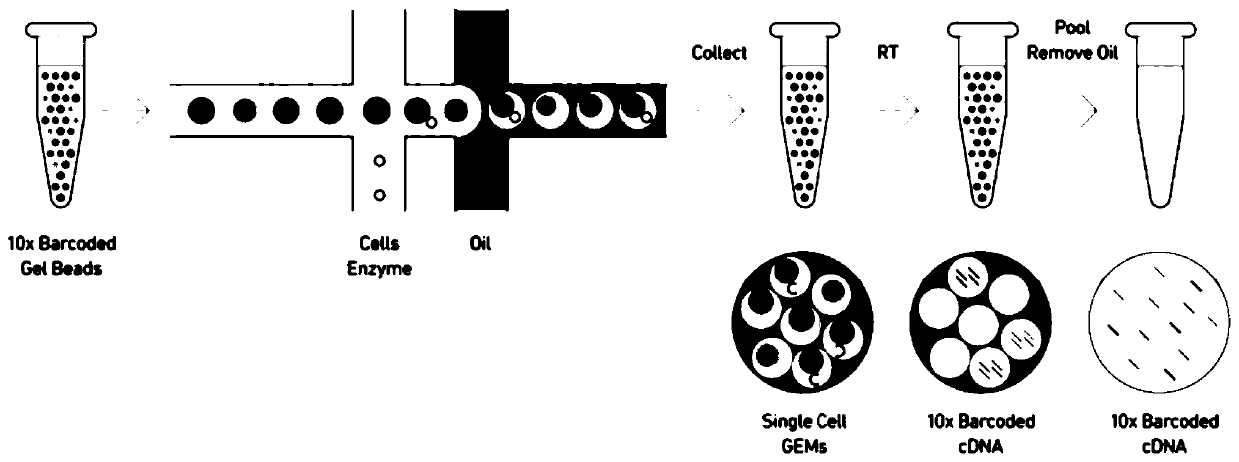
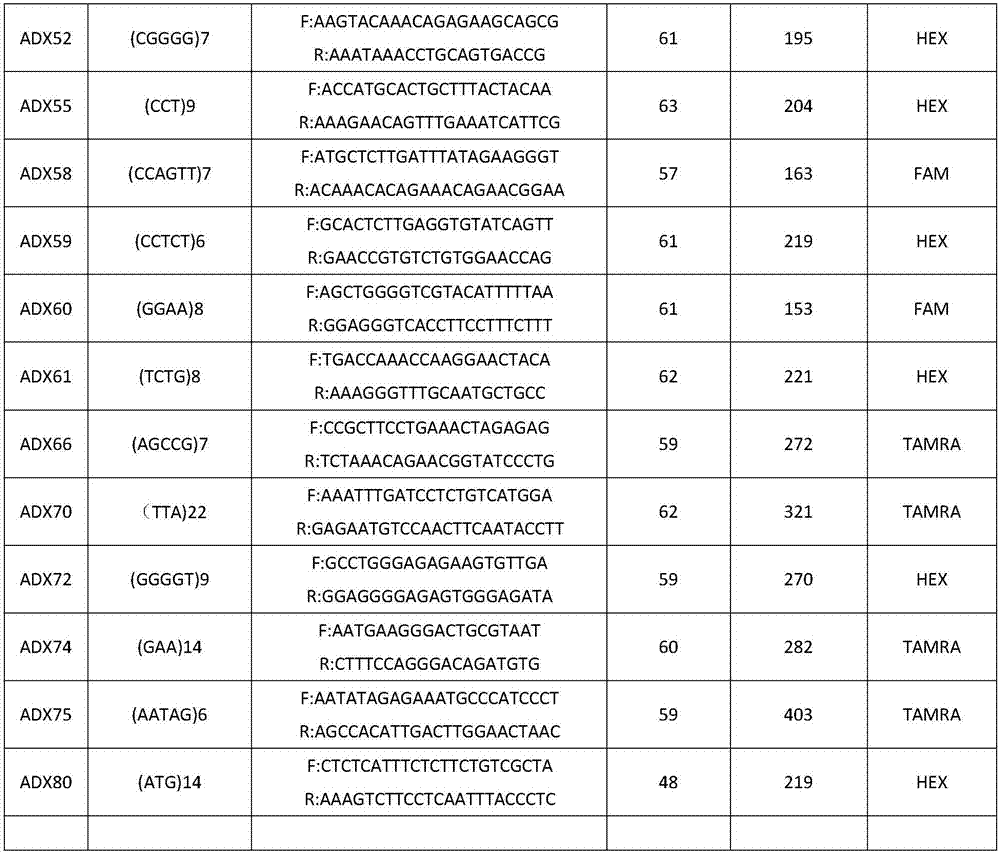
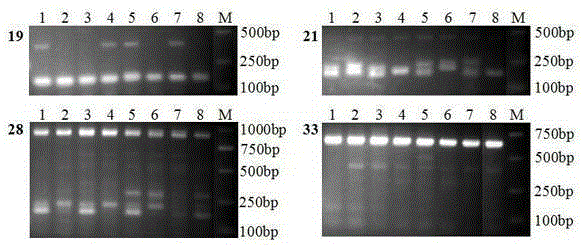
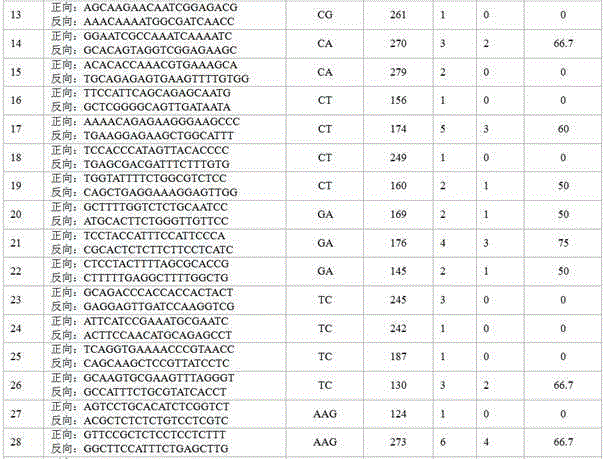
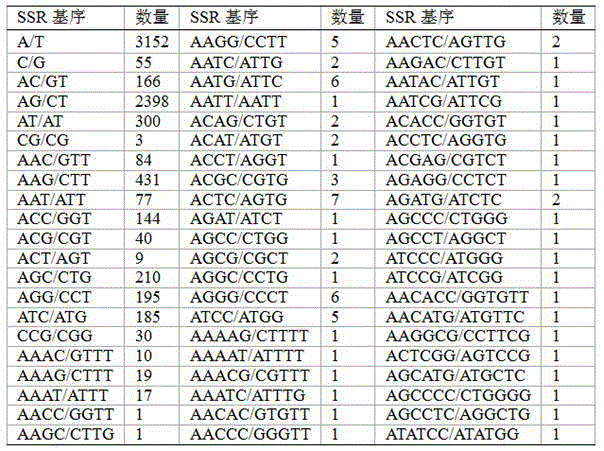
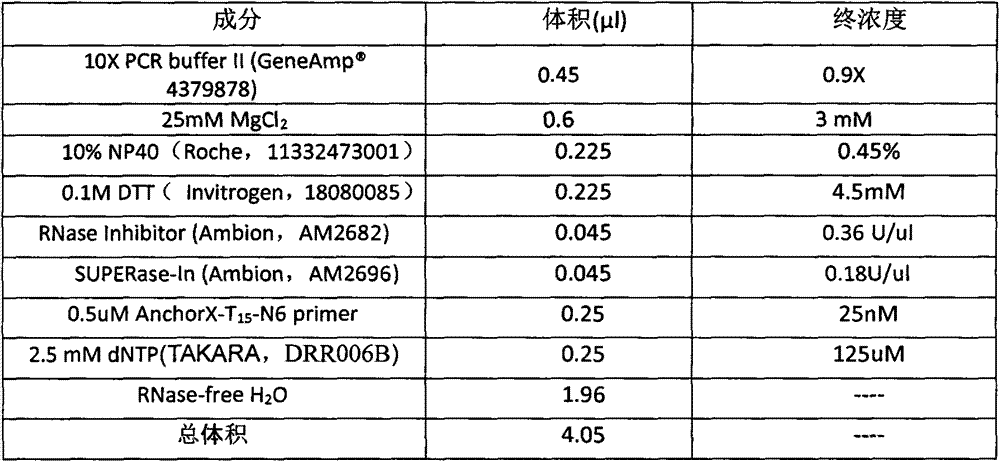
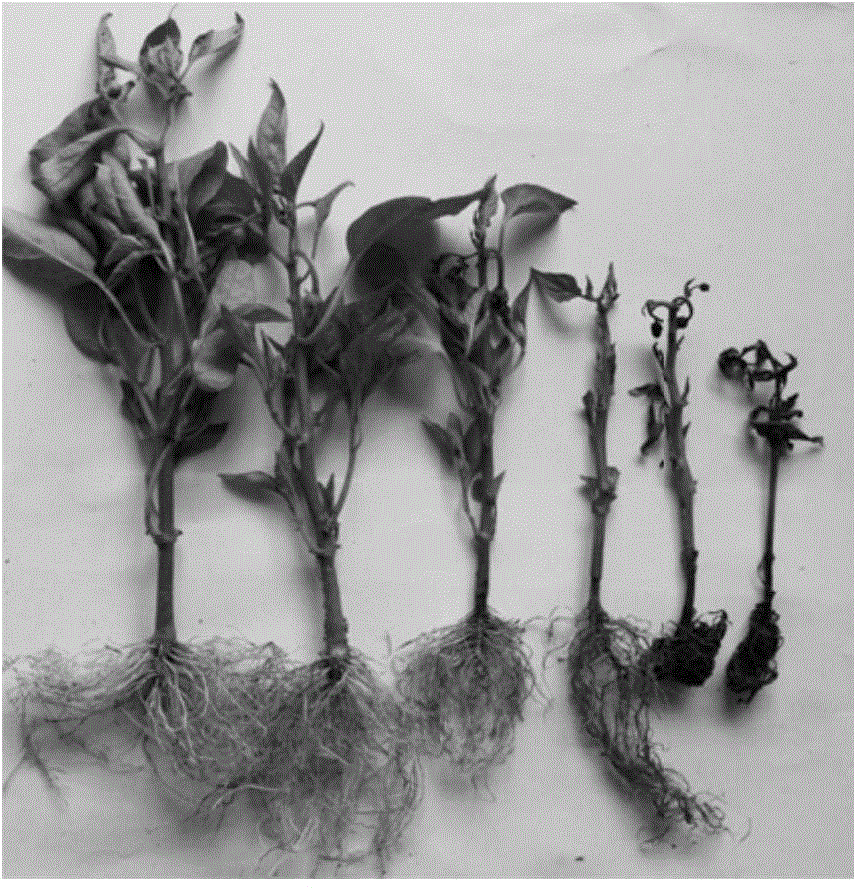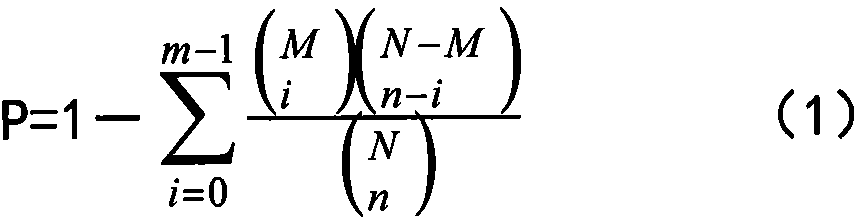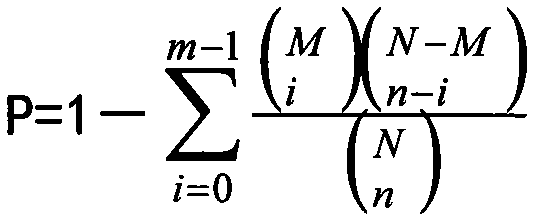Patents
Literature
477 results about "Transcriptome Sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
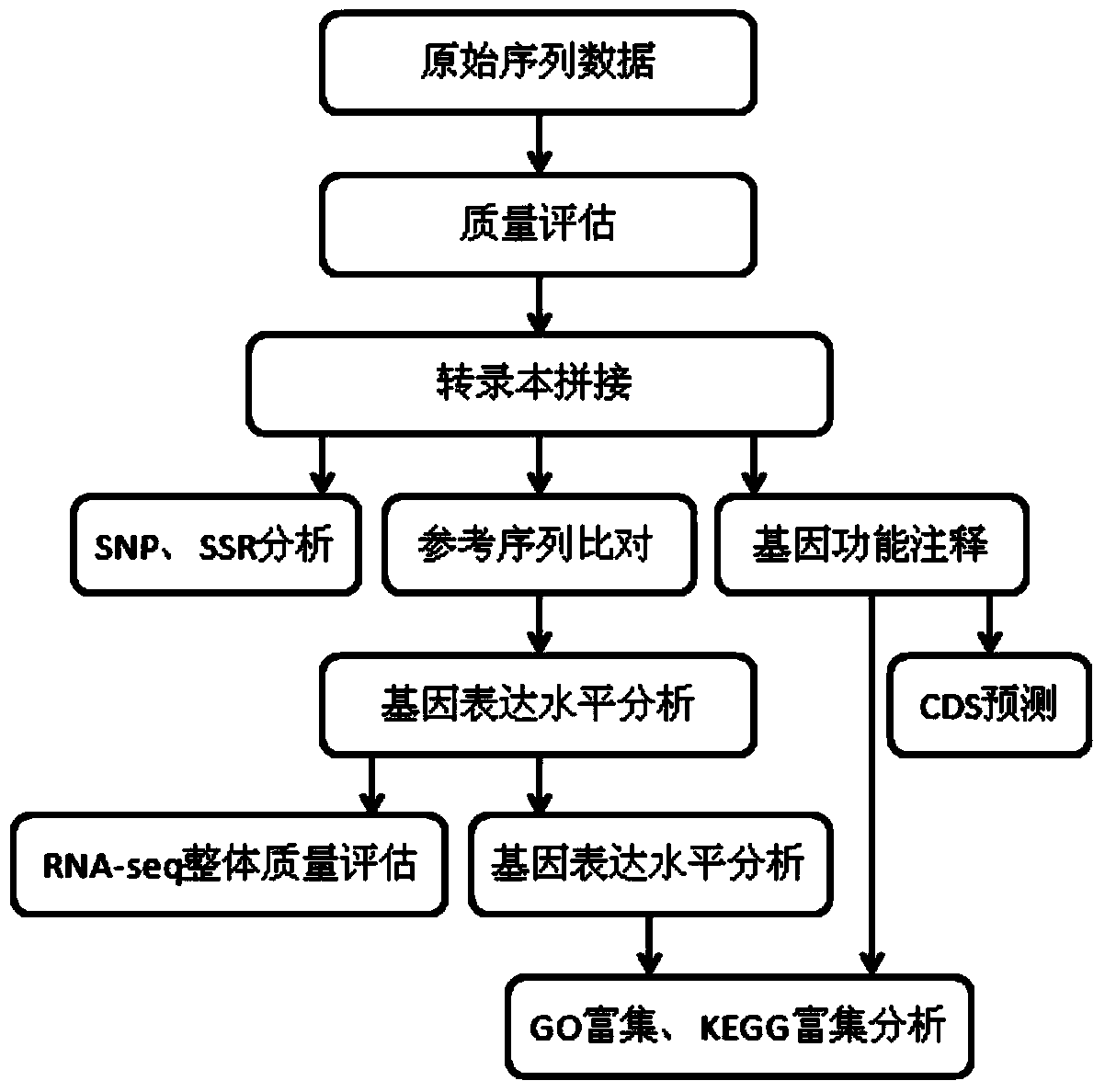
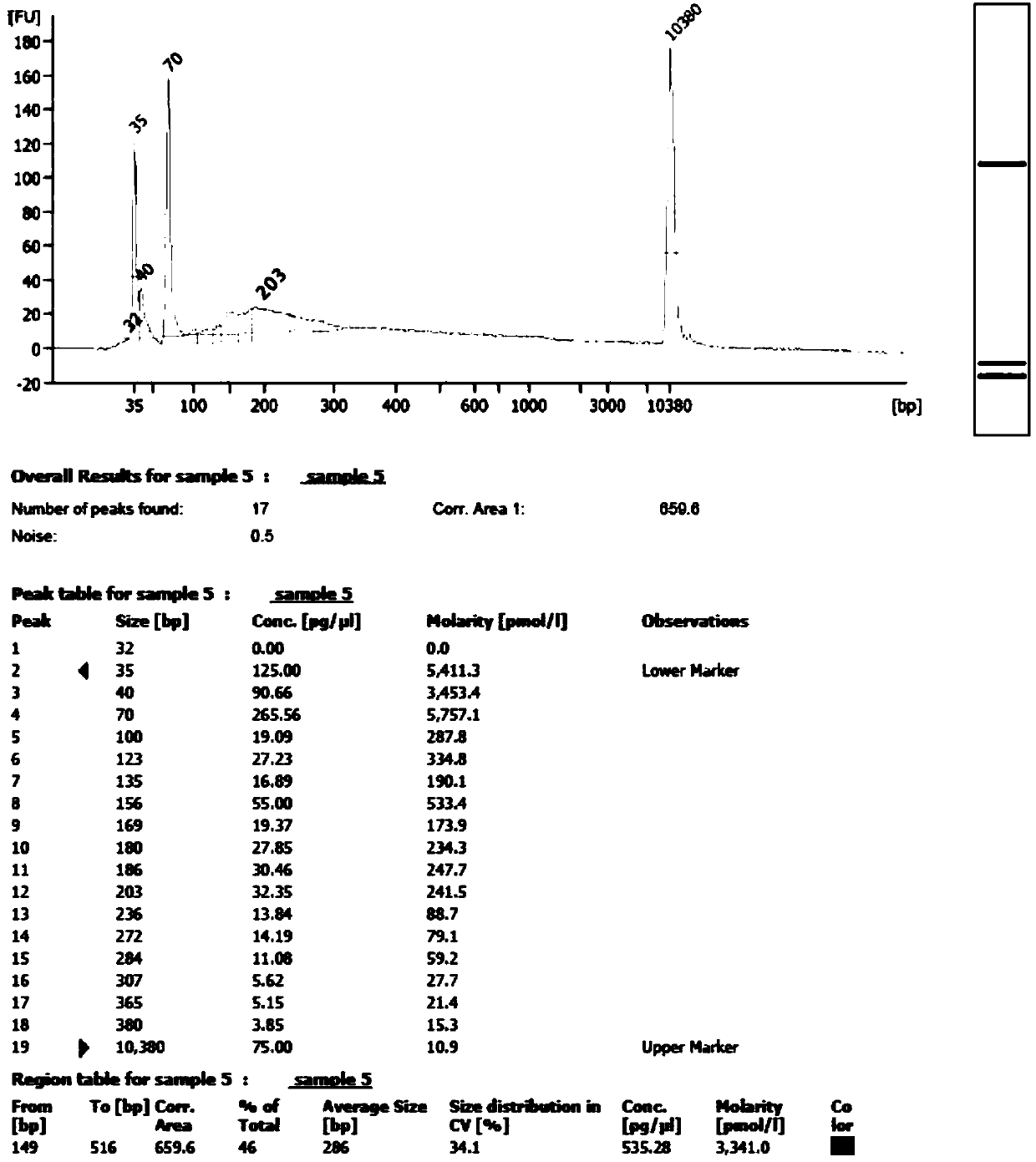
Transcriptome sequencing is used to reveal the presence, quantity and structure of RNA in a biological sample under specific conditions.
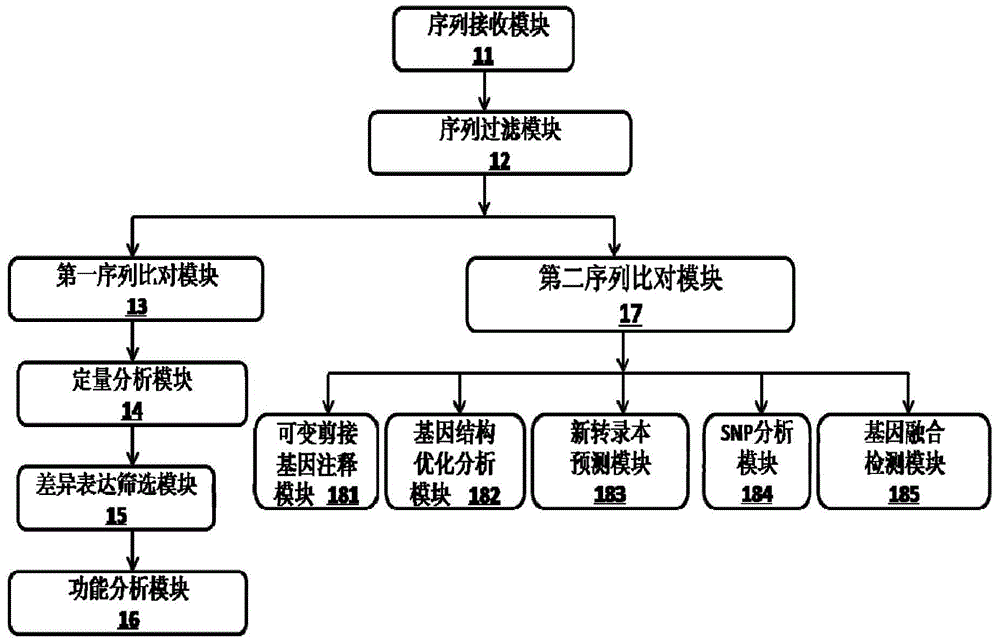
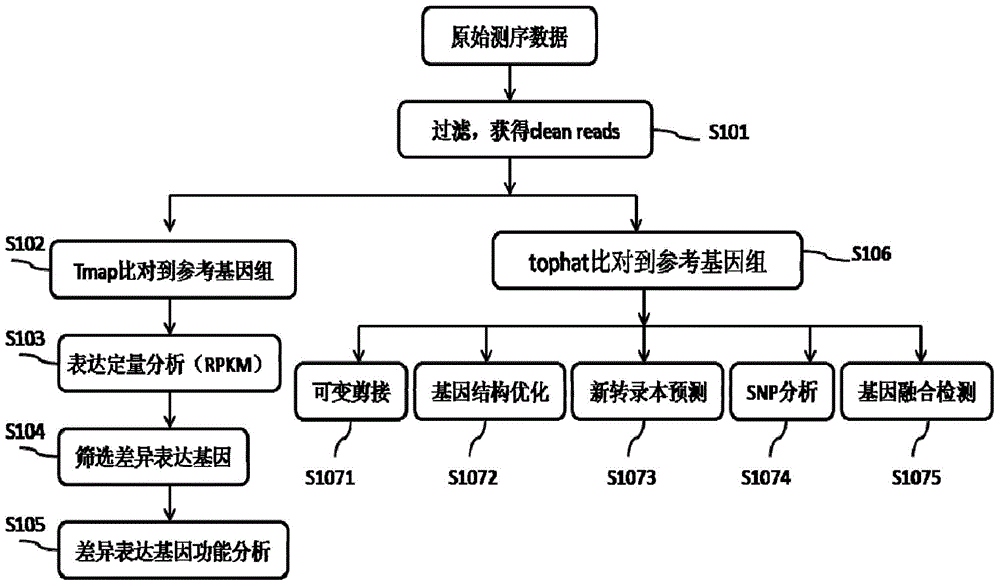
Proton-based transcriptome sequencing data comparison and analysis method and system
InactiveCN104657628AImprove accuracyImprove reliabilitySpecial data processing applicationsDifferentially expressed genesSingle-nucleotide polymorphism
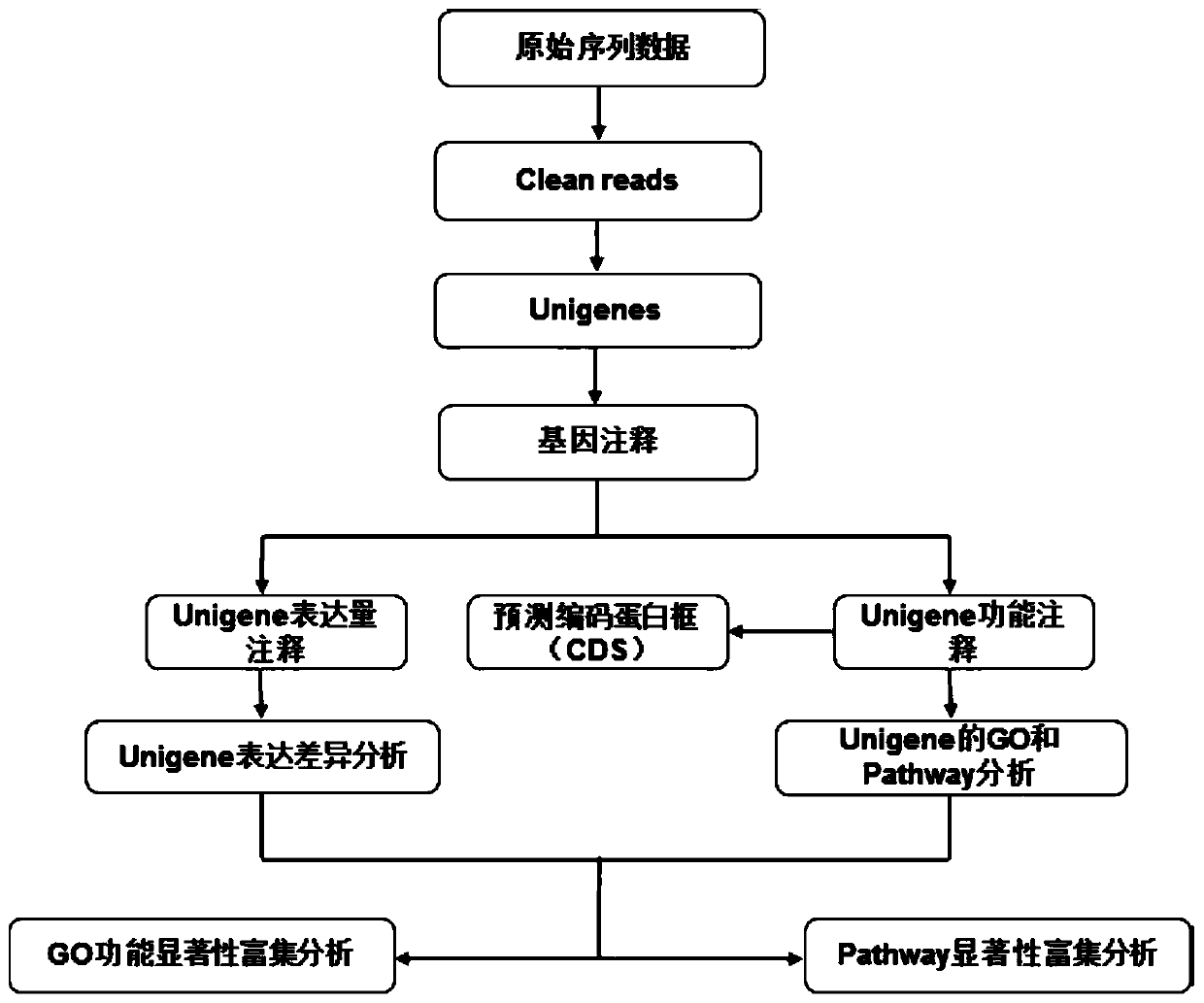
The invention provides a Proton-based transcriptome sequencing data comparison and analysis method and system. The method comprises the following steps: acquiring original sequencing data of at least two transcriptomes of a certain species by virtue of a Proton sequencing platform; filtering unqualified data to obtain clean reads; performing first-step analysis and second-step analysis, wherein the first-step analysis comprises the steps of comparing the clean reads with a reference genome of the species respectively, performing transcript quantitative analysis, screening significantly differently expressed genes and performing significantly differently expressed gene function analysis; the second-step analysis comprises the steps of comparing the clean reads to the reference genome of the species respectively, performing alternative splicing analysis, performing gene structure optimization analysis, performing new transcript prediction, performing SNP (Single Nucleotide Polymorphism) analysis and performing gene fusion detection. According to the method and the system, the transcriptome sequencing data comparison and analysis accuracy and reliability can be improved.
Owner:BGI TECH SOLUTIONS
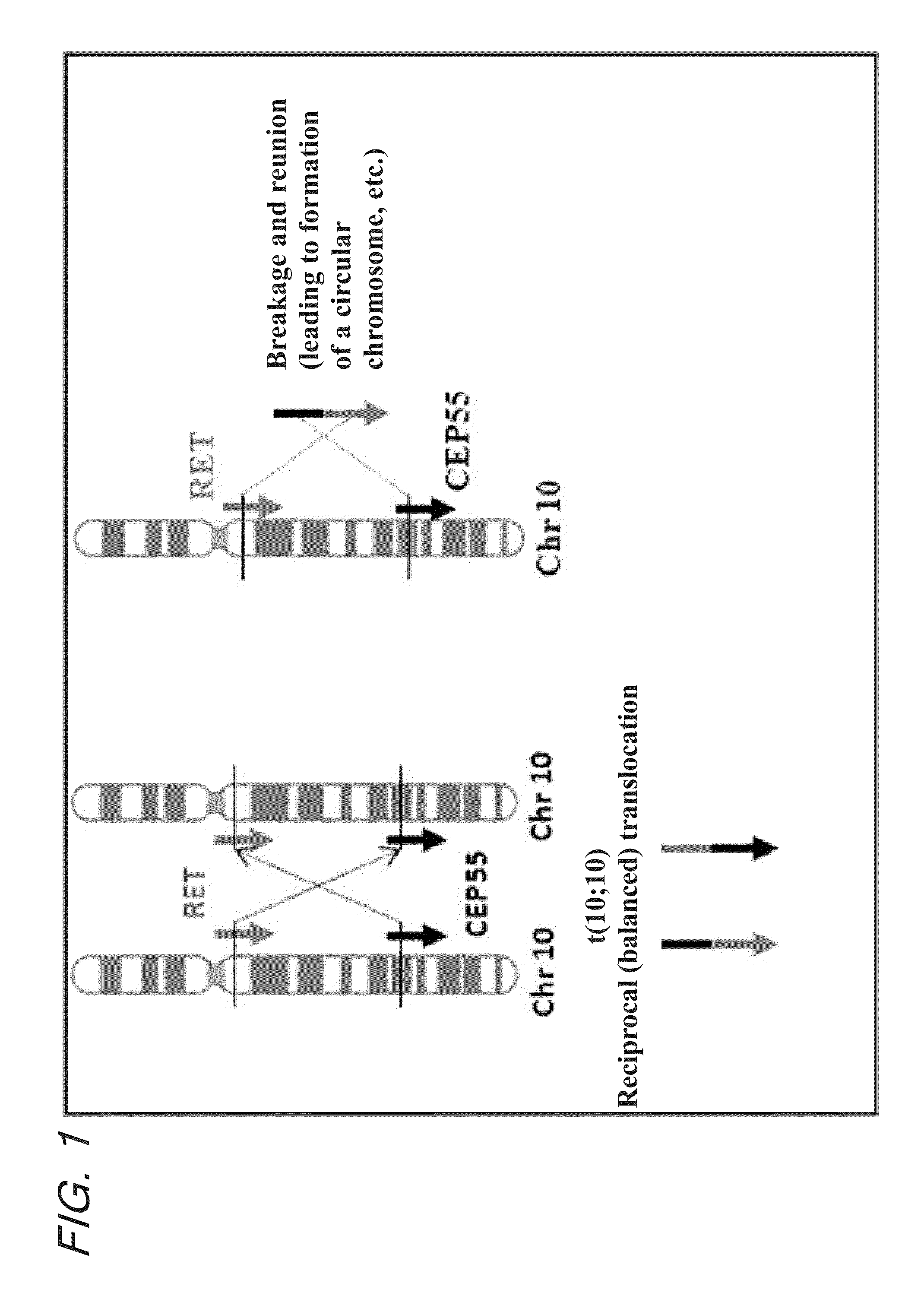
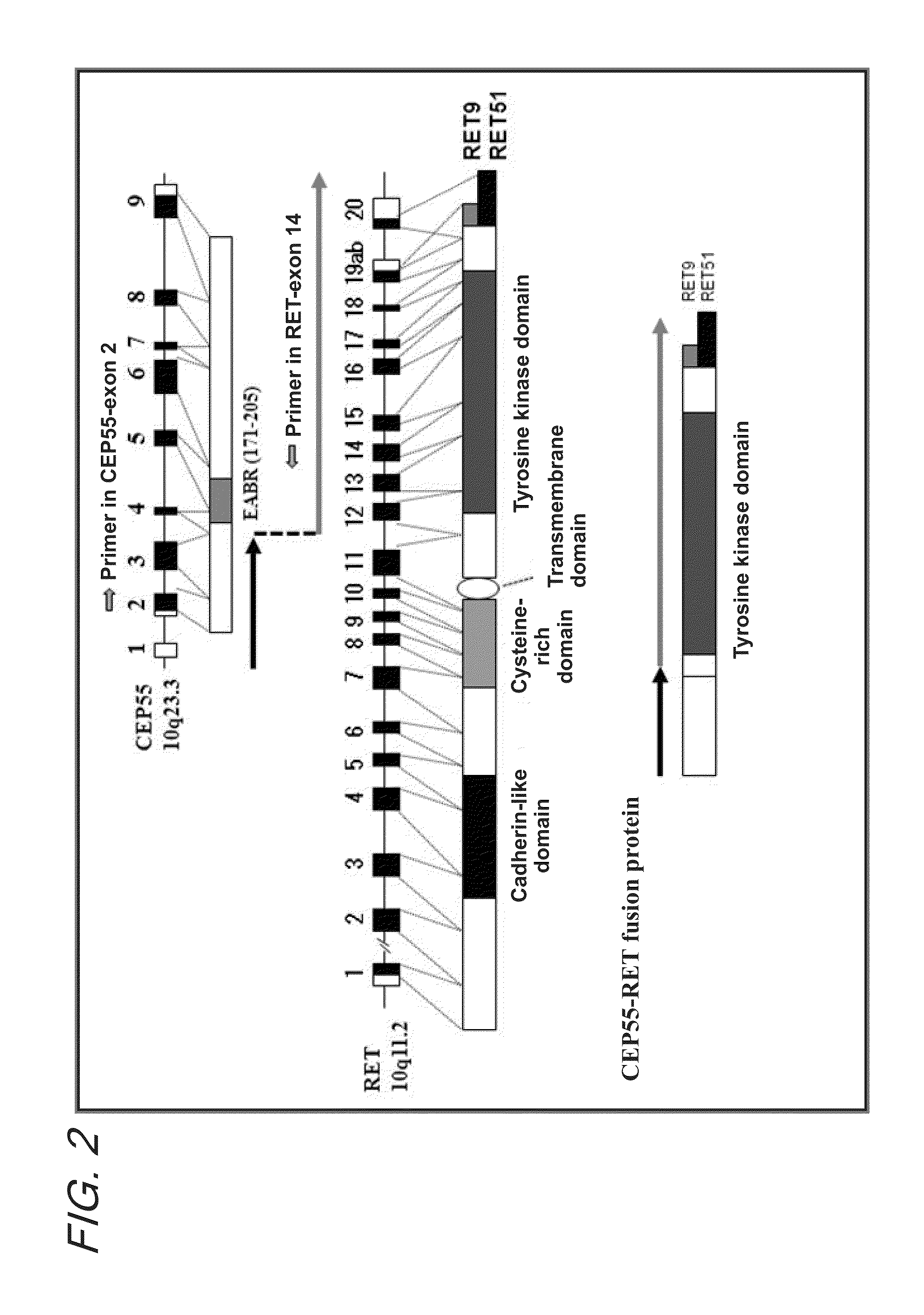
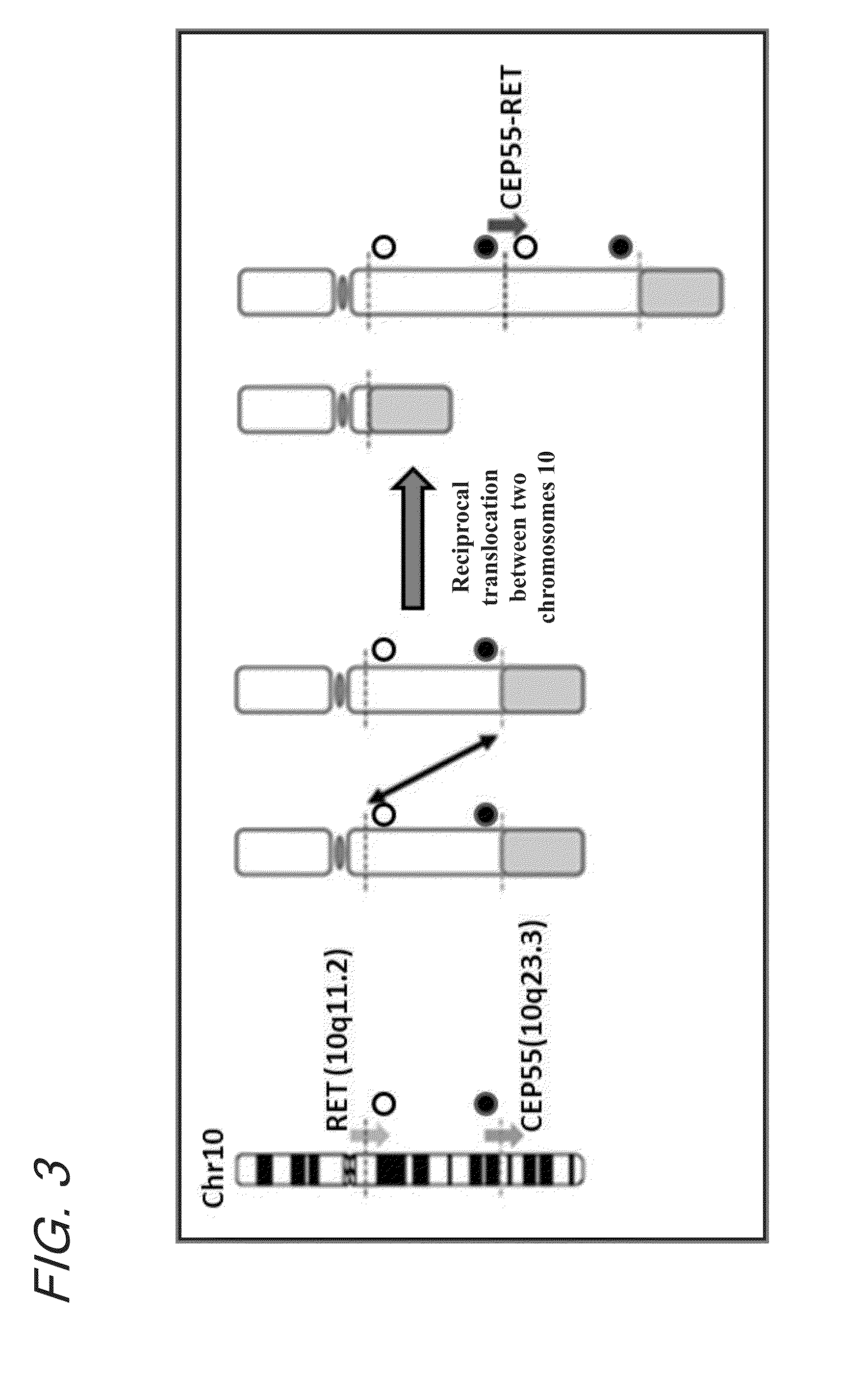
Fusion gene of cep55 gene and ret gene
InactiveUS20150177246A1Efficient detectionEffective treatment of cancerBiocideAntibody mimetics/scaffoldsRet geneTyrosine-kinase inhibitor
In order to identify genes that can serve as indicators for predicting the effectiveness of drug treatments in cancers and provide novel methods for predicting the effectiveness of treatments with drugs targeting said genes, transcriptome sequencing was performed of diffuse-type gastric cancer. As a result, in-frame fusion transcripts between the CEP55 gene and the RET gene were identified. It was also found that said gene fusions induce activation of RET protein, thereby causing canceration of cells. Further, it was demonstrated that the RET protein activation and canceration caused by said gene fusion can be suppressed by using a RET tyrosine kinase inhibitor, and that treatments with a RET tyrosine kinase inhibitor are effective in patients with detection of said gene fusion.
Owner:NAT CANCER CENT +1
Method for developing mung bean simple sequence repeat (SSR) primer based on transcriptome sequencing
InactiveCN103642912AImprove design success rateSimple designMicrobiological testing/measurementDNA/RNA fragmentationSequence databaseTranscriptome Sequencing
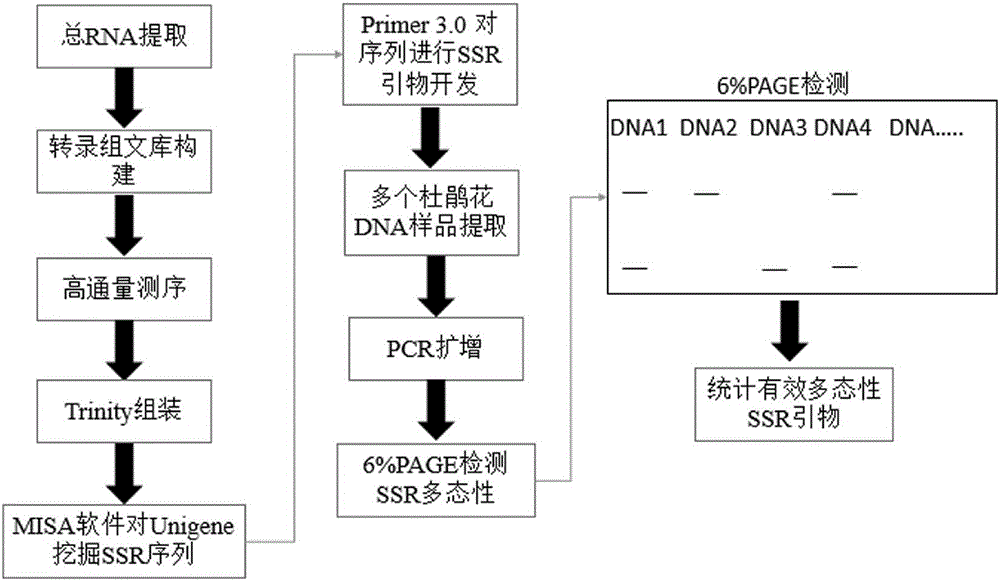
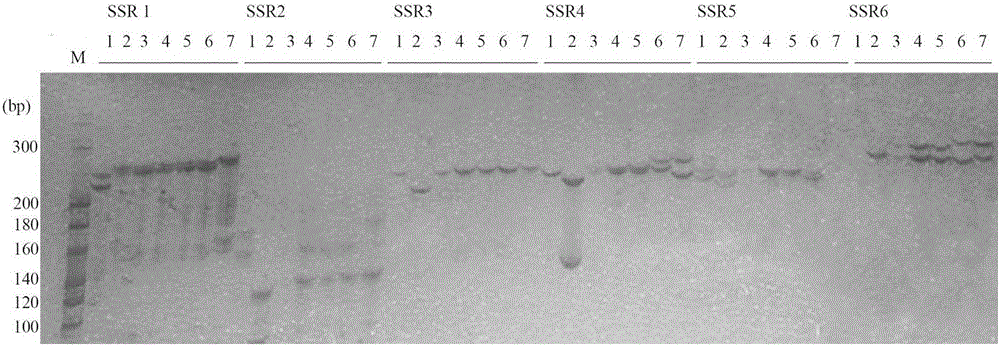
The invention provides a method for developing a mung bean simple sequence repeat (SSR) primer based on transcriptome sequencing. The method comprises the following steps: obtaining a set of mung bean genome-wide transcription, and forming a sequence database; splicing sequencing sequences into a transcriptome by Trinity; taking the longest transcript in each gene as Unigene; carrying out bioinformatics analysis of a Unigene sequence; carrying out SSR detection on the Unigene by adopting MISA1.0; carrying out SSR primer design by using a Primer 3, and carrying out SSR primer polymorphism identification. 13134 pairs of SSR primers are successfully designed by application of the method; 50 pairs of primers are randomly selected to verify 8 parts of mung bean deoxyribonucleic acids (DNAs) from different countries, wherein 32 pairs of polymorphic primers are formed in all; the mung bean materials with different geographical origins can be distinguished by using the 32 pairs of SSR primers. The method disclosed by the invention is convenient, fast and accurate, and low in cost, and a new thought is provided for development of the mung bean SSR primer.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Method and kit for preparing a target RNA depleted sample
InactiveUS20150275267A1Simple methodDepleting unwanted target RNAPeptide librariesMicrobiological testing/measurementTotal rnaNon-coding RNA
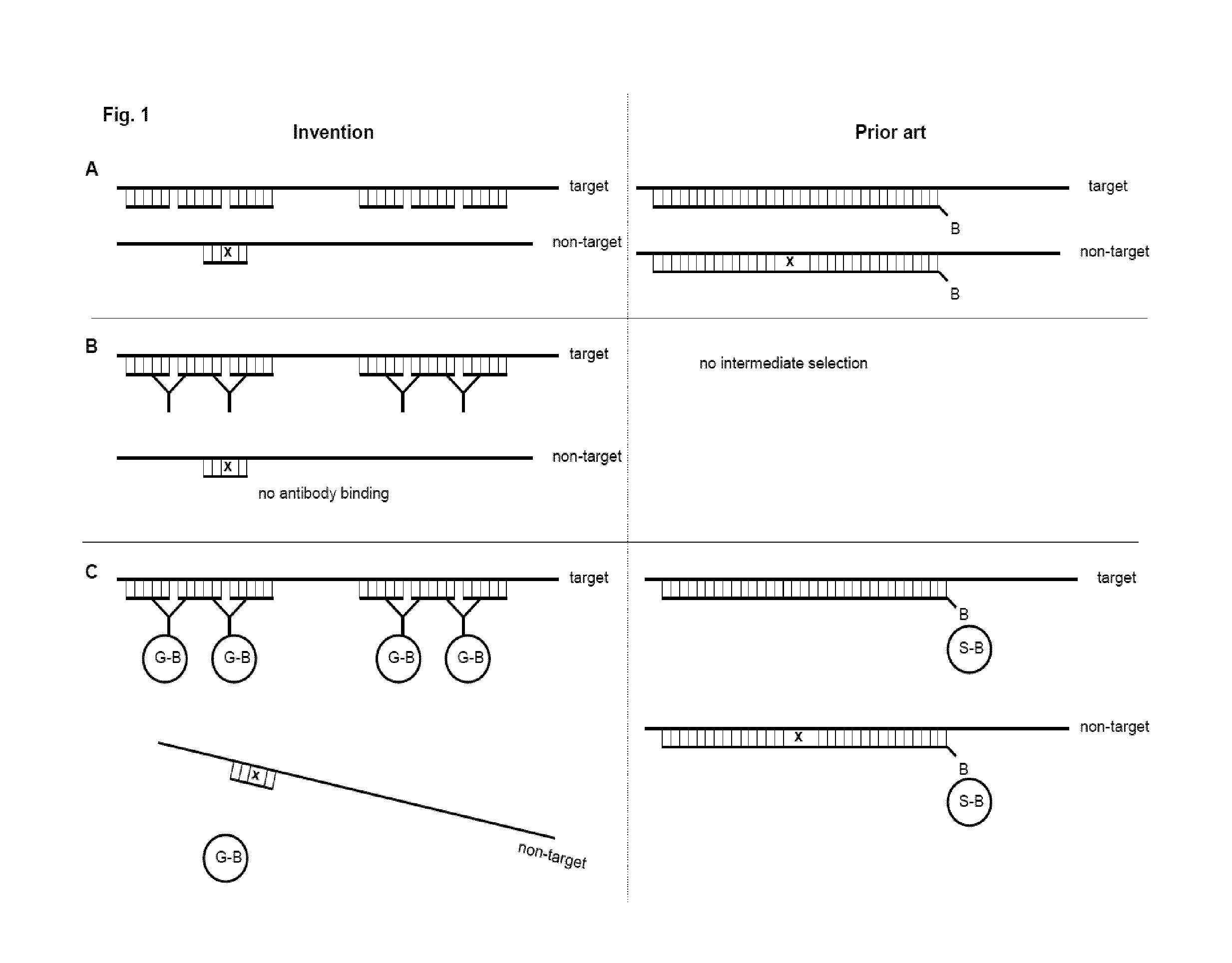
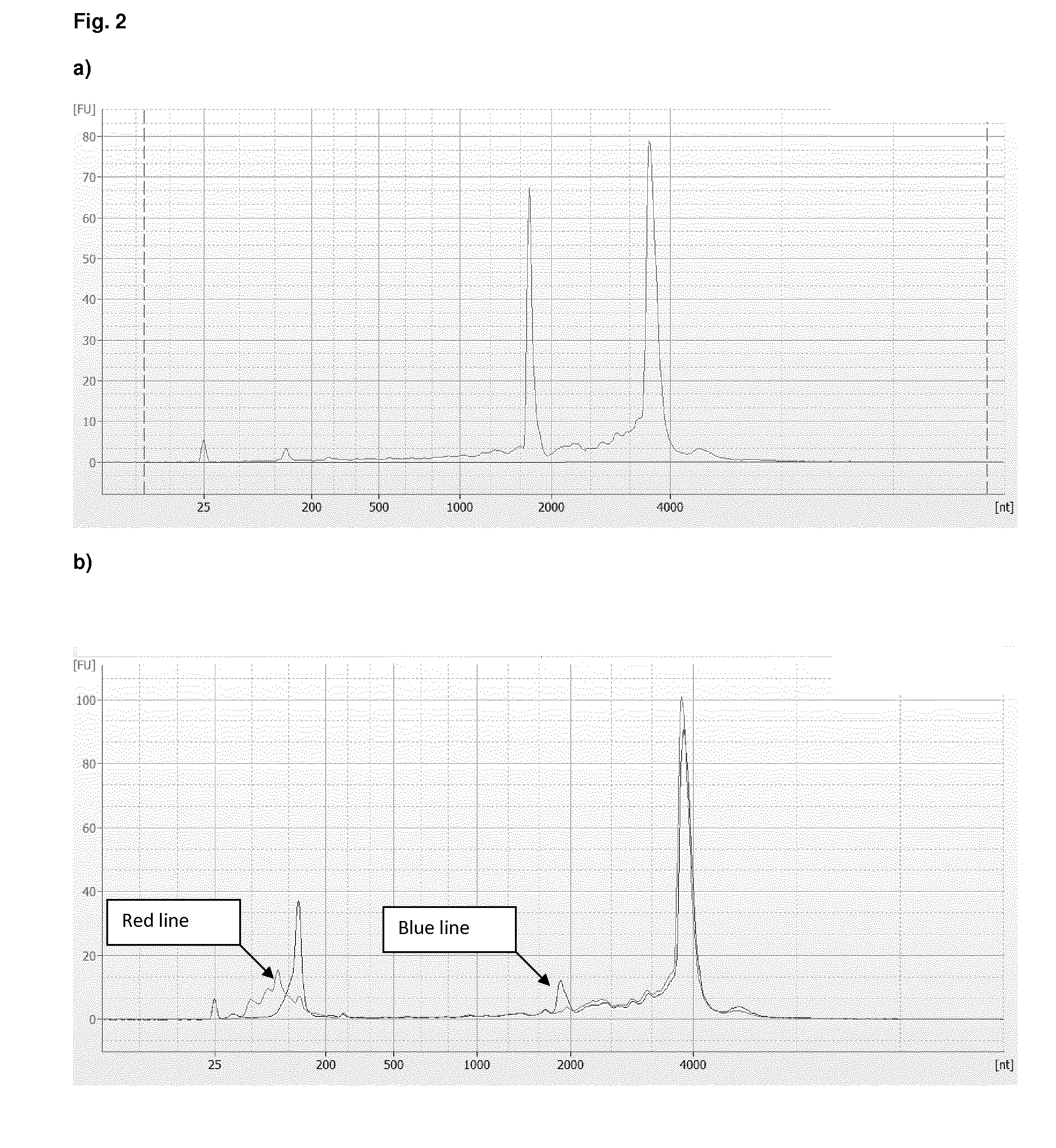
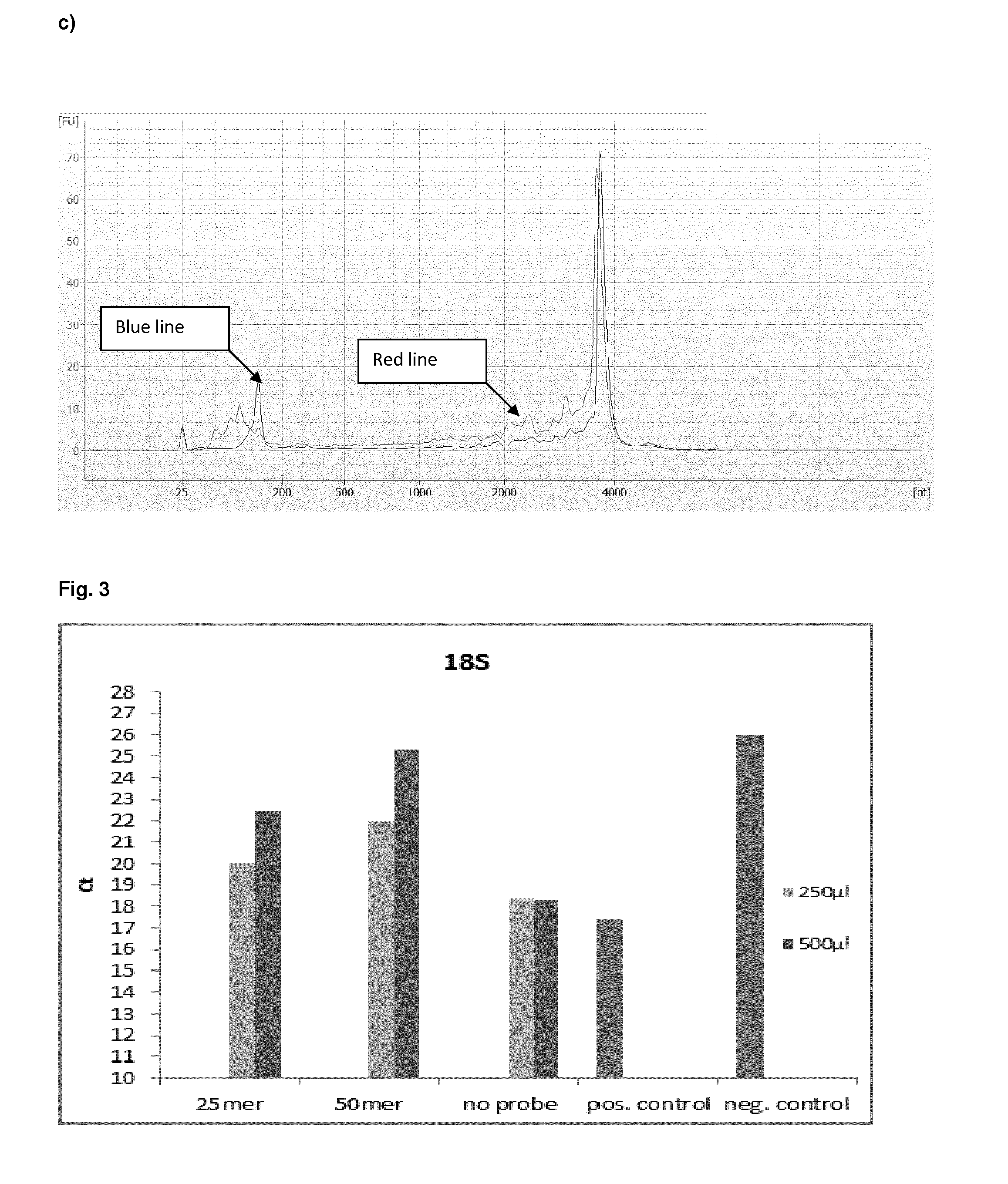
The present invention provides a method of preparing a target RNA depleted composition from an initial RNA containing composition, comprising a) contacting the initial RNA containing composition with one or more groups of probe molecules, wherein a group of probe molecules has the following characteristics: i) the group comprises two or more different probe molecules having a length of 100 nt or less; ii) the probe molecules comprised in said group are complementary to a target region of a target RNA; iii) when hybridized to said target region, the two or more different probe molecules are located adjacent to each other in the formed double-stranded hybrid; and generating a double-stranded hybrid between the target RNA and the probe molecules; b) capturing the double-stranded hybrid by using a binding agent which binds the double-stranded hybrid, thereby forming a hybrid / binding agent complex; c) separating the hybrid / binding agent complexes from the composition, thereby providing a target RNA depleted composition. By combining hybrid capturing with a unique probe design, an improved depletion method is provided which effectively and specifically removes unwanted target RNA such as ribosomal RNA (rRNA) from total RNA, while ensuring recovery of mRNA and noncoding RNA from various species, including human, mouse, and rat. By improving the ratio of useful data, decreasing bias, and preserving non-coding RNA species, the method provides high-quality RNA that is especially suited for next-generation sequencing (NGS) applications. By integrating said depletion method in common sequencing applications, in particular NGS applications such as transcriptome sequencing, improved methods for sequencing RNA molecules are provided.
Owner:QIAGEN GMBH
SSR molecular marker primer based on transcriptome data of azalea as well as screening method and application of SSR molecular marker primer
InactiveCN105969767AAvoid it happening againEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenetic diversity
The invention discloses an SSR molecular marker primer based on transcriptome data development and specific application of the SSR molecular marker primer, belonging to the technical field of biology. The primer is obtained based on development of a transcriptome sequence. By virtue of primer designs of carrying out batch screening on lots of sequencing data based on transcriptome sequencing to obtain an SSR sequence and carrying out SSR marking, the obstacles that the SSR molecular markers of the azalea are few, the development efficiency is low, and the like are overcome. The validity of the SSR primer is verified by virtue of different varieties of azaleas, and foundation is laid for the research of genetic diversity of the azaleas, the constitution of genetic linkage maps, the assistant breeding of the molecular marker, and the like.
Owner:HUANGGANG NORMAL UNIV
Method for obtaining capsicum phytophthora resistance candidate gene and molecular marker, and application
InactiveCN104560973AAccurate identificationLarge amount of data informationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyData information
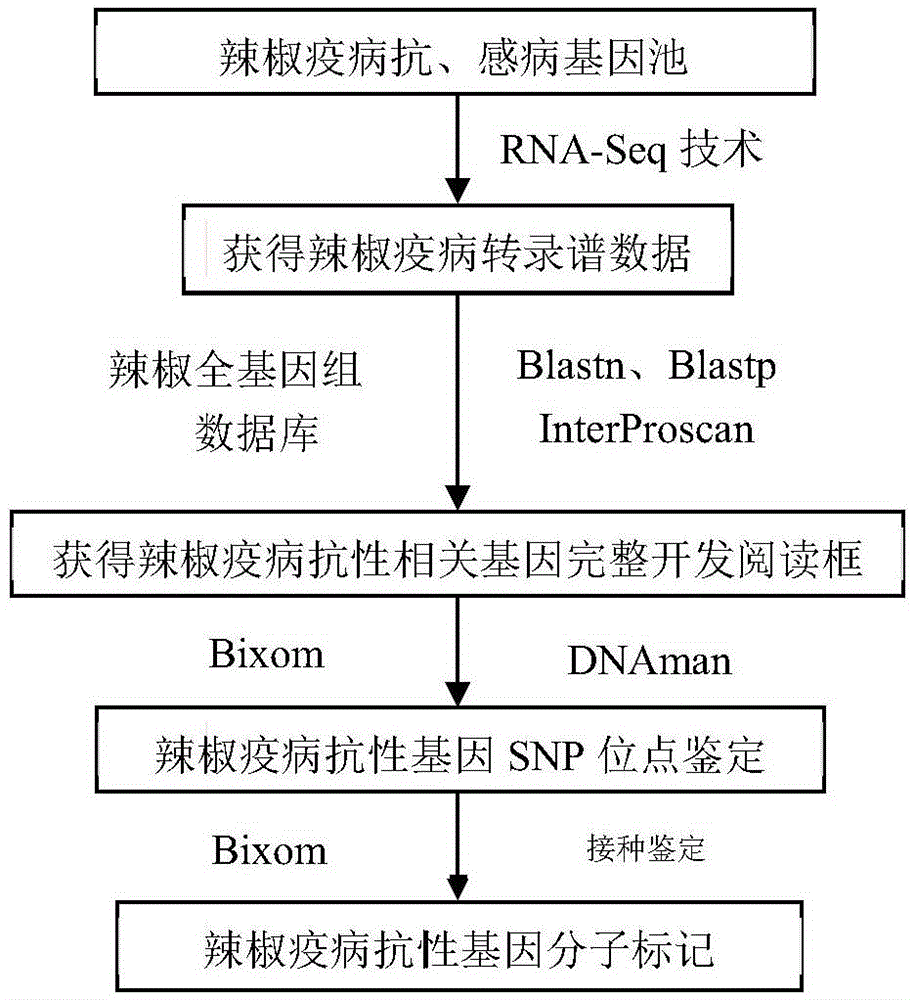
The invention relates to a method for obtaining a capsicum phytophthora resistance candidate gene and a molecular marker, and application. The method is used for obtaining the capsicum phytophthora resistance candidate gene by utilizing capsicum phytophthora transcriptome and whole-genome sequencing data information, differentially-expressed gene identification, bioinformatics analysis, molecular marker development and phytophthora inoculation identification and belongs to the technical field of capsicum biology. The method comprises the following steps: sequencing a phytophthora resistant and susceptible gene pool transcriptome obtained after phytophthora inoculation of an F2 population constructed by capsicum highly-resistant and highly-susceptible phytophthora materials, performing expression analysis and functional annotation on differential genes, extracting DNAs (Desoxvribose Nucleic Acid) of a capsicum phytophthora highly-resistant and highly-susceptible phytophthora material genome, performing primer design and PCR (Polymerase Chain Reaction) amplification, performing sequence difference analysis and SNP site identification, performing SNP specific primer design and validity verification, and performing other steps to efficiently obtain the capsicum phytophthora resistance candidate gene and the molecular marker. According to the method, the capsicum phytophthora resistance candidate gene can be accurately identified, and the effective molecular marker can be developed.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Method for sequencing and development of Asplenium nidus L. EST-SSR primers based on transcriptome
InactiveCN105274198AAdd raw dataOvercoming access difficultiesMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionBatch processing
The invention belongs to the molecular biology technology field, and concretely relates to a method for sequencing and development of Asplenium nidus L. EST-SSR primers based on transcriptome. The method comprises the following steps: firstly, a transcriptome library is constructed; secondly, transcriptome data is obtained, namely, sequencing data is subjected to splicing by utilization of a Trinity software and sequences are spliced into a complete transcriptome; thirdly, SSR site search is carried out, namely, combined with a Perl programming language method, a lot of sequence information is subjected to batch processing, and SSR site search is carried out; fourthly, batch design of EST-SSR primers is carried out. Different fiddlehead materials are selected to verify the designed SSR primers, if strips are detected, successful SSR primers are obtained. Through the method, 4063 pairs of SSR primers are designed, and a new method and thought are provided for development of Asplenium nidus L. EST-SSR primers and further for achieving molecule label auxiliary selection breeding.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Method for developing dendranthema SSR (Simple Sequence Repeat) primer based on transcriptome sequencing
ActiveCN103233075AAdd raw dataOvercoming access difficultiesMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceLymphatic Spread
The invention belongs to biotechnology field, and relates to a method for developing a dendranthema SSR (Simple Sequence Repeat) primer based on transcriptome sequencing. A lot of sequence information is processed in batch for searching an SSR sequence and designing an SSR labeled primer based on the transcriptome sequencing by utilizing EST-SSR (Expressed Sequence Tags-Simple Sequence Repeat) interspecific metastasis and using a Perl (Practical Extraction and Reporting Language) programming language method in combination, so that the defects of low efficiency, long time consumption, high cost and the like of the SSR development are conquered. Different dendranthema materials are selected for verifying the designed SSR primer, and the primer is a successful SSR primer if detected out by any strip. By adopting the method, 1788 pairs of SSR primers are successively designed, so that a novel method and thinking are provided for the development of the dendranthema SSR primer to further achieve molecular marker assistant selection breeding and comparative genomics research.
Owner:NANJING AGRICULTURAL UNIVERSITY
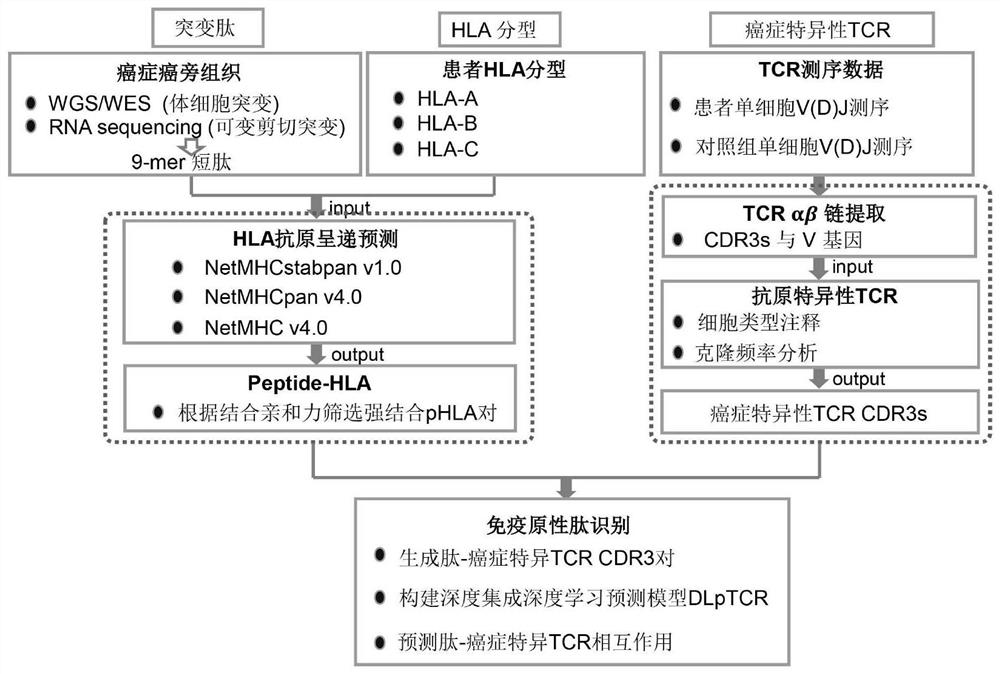
Tumor neoantigen screening method fused with single cell TCR sequencing data
ActiveCN113160887AOvercoming the high rate of misselection and omissionOvercoming problems such as insufficient immunogenicityMicrobiological testing/measurementProteomicsSingle cell transcriptomeCD8
The invention discloses a tumor neoantigen screening method fused with single cell TCR sequencing data. The tumor neoantigen screening method comprises the following steps: based on whole exon sequencing data and transcriptome sequencing data, carrying out quality control, comparison and other steps through software to obtain a neomutant peptide library; predicting an HLA-I type typing by using HLA typing prediction software; in combination with single-cell TCR sequencing and single-cell transcriptome sequencing, finding cancer-specific CD8 + T cell receptors through cell type annotation and clone frequency analysis; meanwhile, based on integrated deep learning, the short peptide immunogenicity is identified through a peptide-TCR interaction prediction model, a tumor neoantigen screening method fused with single cell TCR sequencing data is provided, and the problems that a traditional tumor antigen screening method is high in neoantigen misselection and missed selection rate, insufficient in immunogenicity and the like are solved.
Owner:HARBIN INST OF TECH
Method for building high-flux single-cell full-length transcriptome sequencing library and application of method
ActiveCN109811045AGuaranteed accuracyIntegrity guaranteedMicrobiological testing/measurementLibrary creationTn5 transposaseLysis
The application discloses a method for building a high-flux single-cell full-length transcriptome sequencing library and application of the method. The method comprises the following steps: recognizing single cells by adopting a customized micro-hole chip and an ICELL8 platform, and performing cell lysis, mRNA reverse transcription, cDNA pre-amplification and Tn5 transposase library building on the single cells, wherein a product can be directly applied to sequencing; in a process of mRNA reverse transcription or Tn5 transposase library building, introducing a dual-terminal Barcode sequence with a 5' terminal and a 3' terminal in order to obtain a single cell full-length transcriptome. By adopting the method, the single cell recognition flux is high, and the product can be directly appliedto sequencing. When the method is applied to single-cell sequencing, only two days are required for efficiently obtaining thousands of single-cell full-length transcriptome libraries at one time, sothat the labor cost and time cost are lowered greatly; moreover, the reagent cost is lowered greatly through a trace reaction system, and a basis is laid for large-scale single-cell full-length transcript sequencing.
Owner:MGI TECH CO LTD
Method for detecting variable spliceosome in third generation full-length transcriptome
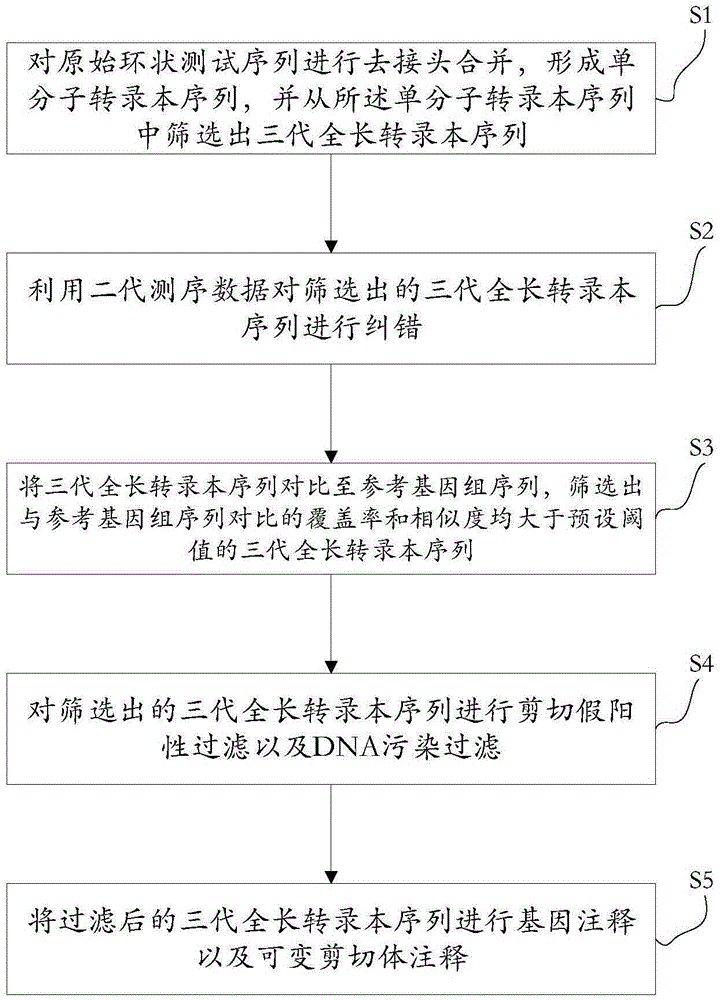
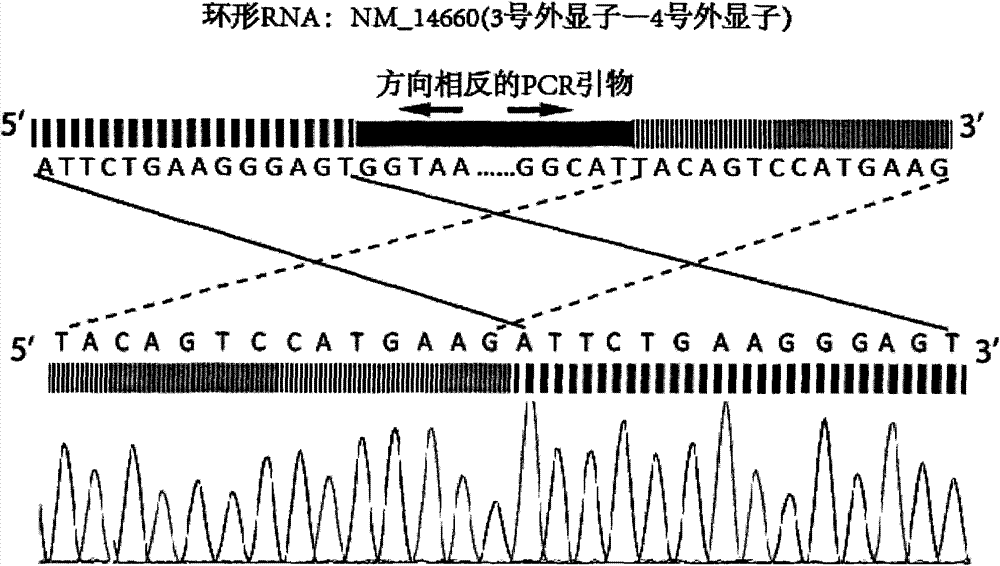
ActiveCN105389481AEfficient access to shear structuresPerfect commentSequence analysisSpecial data processing applicationsReference genome sequenceGene model
The invention discloses a method for detecting a variable spliceosome in a third generation full-length transcriptome. The method comprises the following steps: merging original annular test sequences with joints removed to form a monomolecular transcript sequence, and screening a third generation full-length transcript sequence; comparing the third generation full-length transcript sequence with a reference genome sequence, and screening a third generation full-length transcript sequence having coverage and similarity with the reference genome sequence larger than preset thresholds; carrying out splicing false positive filtration and DNA contamination filtration on the screened third generation full-length transcript sequence; and carrying out gene annotation and variable spliceosome annotation on the filtered third generation full-length transcript sequence. An overlong read length of a third generation sequencing technology mentioned in the method disclosed by the invention is large enough to cover most RNA, the third generation full-length transcript sequence can be obtained by SMRT sequencing transcriptomes without being assembled, and a splicing structure of a gene can be effectively obtained by third generation transcriptome sequencing, and more perfect gene model annotation can be constructed.
Owner:嘉兴菲沙基因信息有限公司
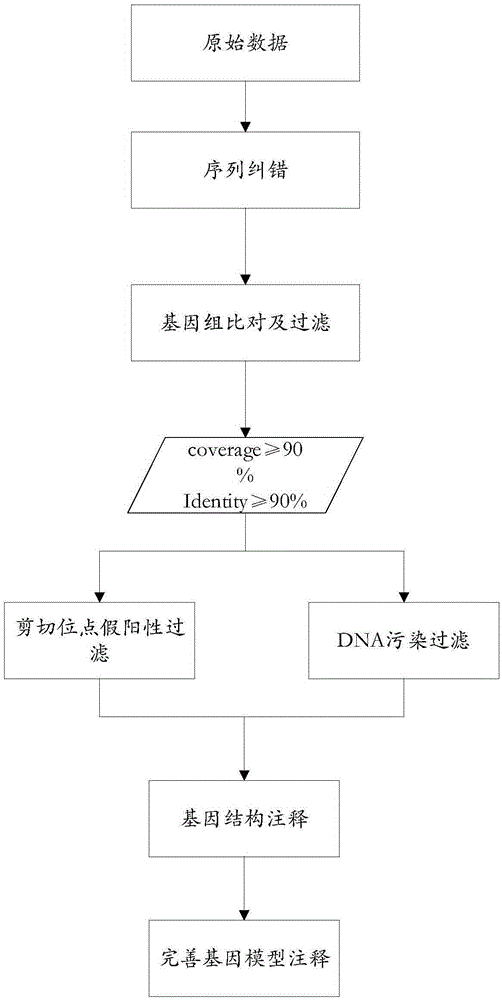
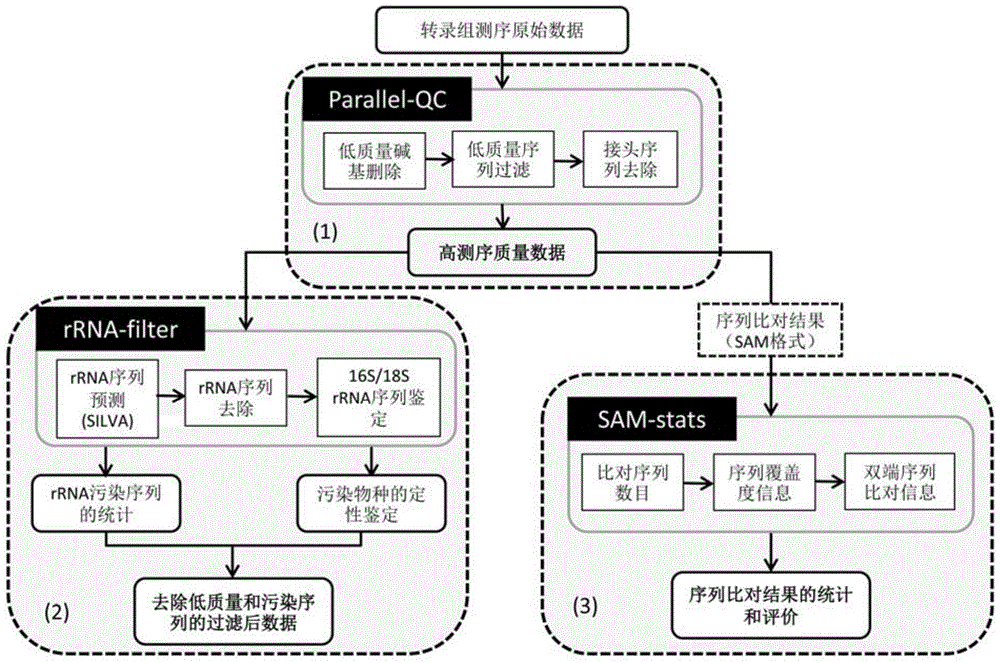
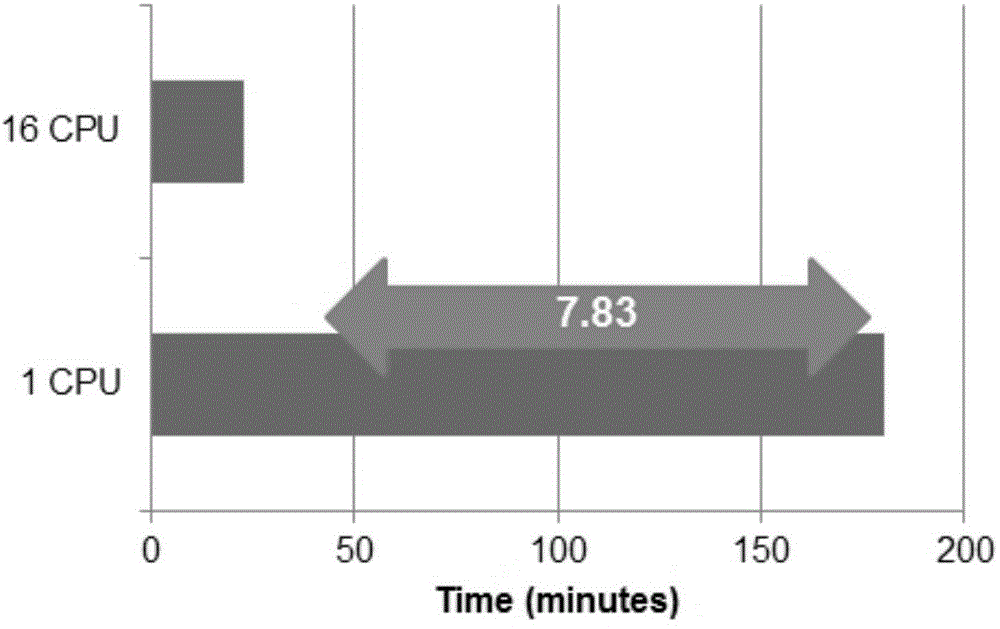
High-flux transcriptome sequencing data quality control method based on multi-core CPU (Central Processing Unit) hardware
ActiveCN105095686AEfficient Data Quality ControlOvercoming Computational Efficiency BottlenecksSpecial data processing applicationsHigh fluxQuality control
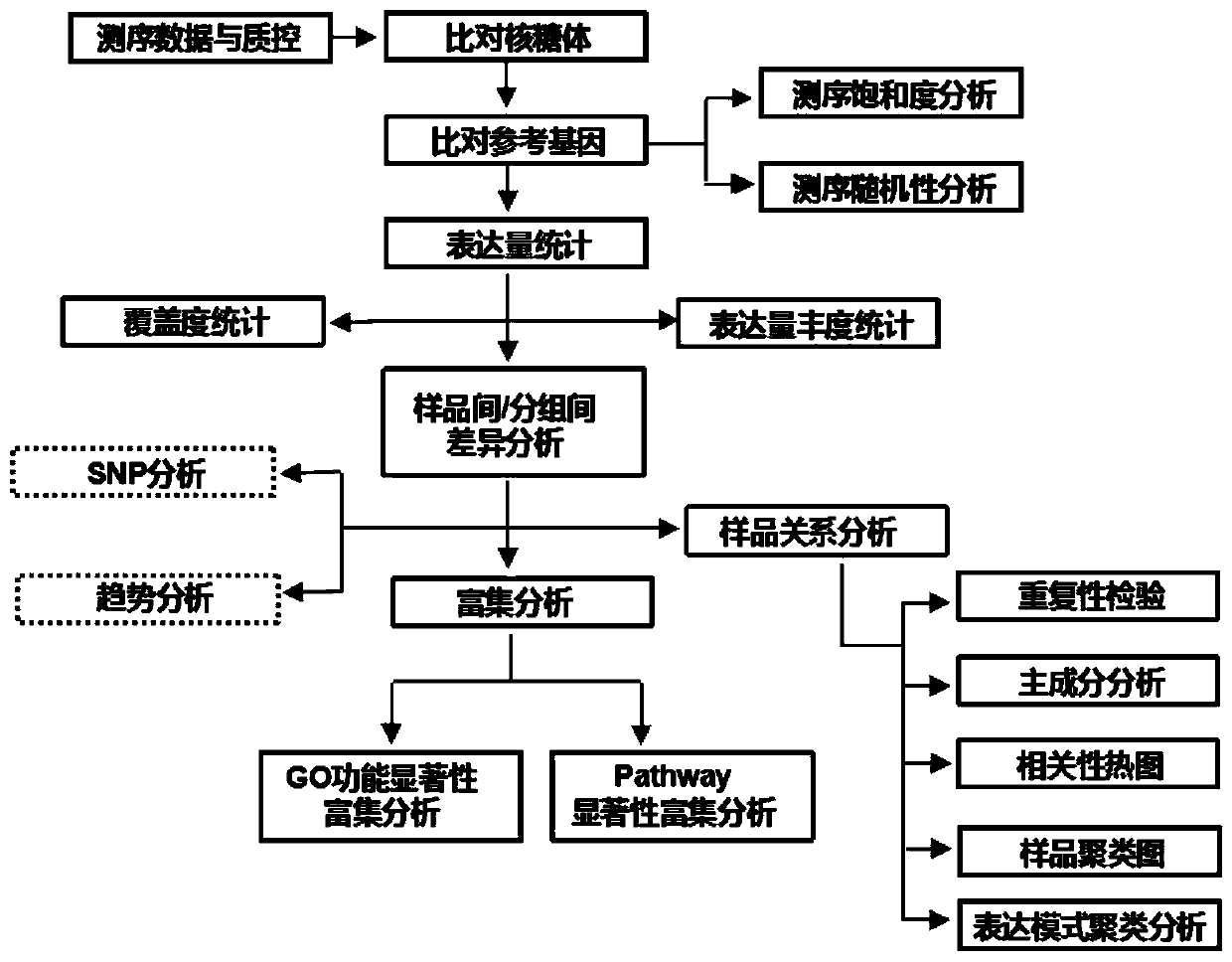
The present invention provides a high-flux transcriptome sequencing data quality control method based on multi-core CPU hardware. The method comprises: performing parallel processing on high-flux transcriptome sequencing data by using a multi-core CPU, so as to obtain data without low sequencing quality sequences; performing prediction and removal on rRNA sequences in the data without the low sequencing quality sequences by using the multi-core CPU, and performing qualitative identification on polluted sequences; and performing statistics and evaluation on a sequence comparison result. According to the high-flux transcriptome sequencing data quality control method based on the multi-core CPU hardware, provided by the present invention, based on a multi-core CPU computer, a computing efficiency bottleneck based on a single-core CPU hardware computer is overcome, so that high-flux transcriptome data quality control efficiency is increased by over 7 times; and by applying the high-flux transcriptome sequencing data quality control method, the accuracy and speed of the high-flux transcriptome data quality control are significantly improved, and rapid development of relevant researches of transcriptome sequencing is widely facilitated.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
Method and kit for screening target region of methylation PCR detection and application of target region
ActiveCN108410980AComprehensive forecastReliable predictionMicrobiological testing/measurementDNA/RNA fragmentationTrue positive rateTranscriptome Sequencing
The invention discloses a method and a kit for screening a target region of methylation PCR detection and an application of the target region. The method comprises the following steps: (1) acquiring ato-be-analyzed tumor methylation chip and corresponding transcriptome sequencing data from a database; (2) calculating methylation degree values of a normal group and a cancer group, and screening methylation sites, which keep obvious differences, in the various groups; (3) in combination with analysis on a transcriptome sequencing expression profile, calculating related coefficients, and screening negatively related sites; and (4) associating the methylation candidate sites which are acquired in the step (3) with related disclosed documents, screening sites which are supported and reported by many documents, keep a high inter-group methylation difference group and are in expression amount negative relation, and on the basis of a regression algorithm, acquiring a site set, namely the target region, achieving the optimal sensitivity and specificity. According to the method, through comprehensive analysis of the database, transcriptome sequencing and the documents and in combination with multiple data filtering and the regression algorithm, the target region of methylation PCR detection can be obtained sensitively and specifically.
Owner:ENVELOPE HEALTH BIOTECHNOLOGY CO LTD
Method for annotating cell identities based on single cell transcriptome clustering results
The invention discloses a method for annotating cell identities based on single cell transcriptome clustering results. The method comprises the following steps: S1, providing survival cells, establishing a library by using 10X genetics, and sequencing to obtain transcriptome sequencing data; S2, filtering the sequencing data obtained in the step S1, then carrying out initial analysis by using thesoftware cellranger, and outputting original data; S3, analyzing the original data output in the step S2; S4, performing cell identity annotation, including the steps: S41, classifying the tag genes of the Cell Marker database according to cell types; S42, sorting the tag genes screened out by the FindAllMarkers function according to a P value; S43, taking an intersection of the tag gene of each type of cells in the Cell Marker database and a cell subset tag gene screened by the FindAllMarkers function, and scoring according to the intersection gene; and S44, sorting according to the score ofthe gene intersection, and annotating the cell type corresponding to the highest score as the cell identity of the current subgroup. By adopting the method for annotating cell identities based on single cell transcriptome clustering results disclosed by the invention, the cells can be accurately and quickly classified and subjected to identity annotation.
Owner:广州序科码医学检验实验室有限公司
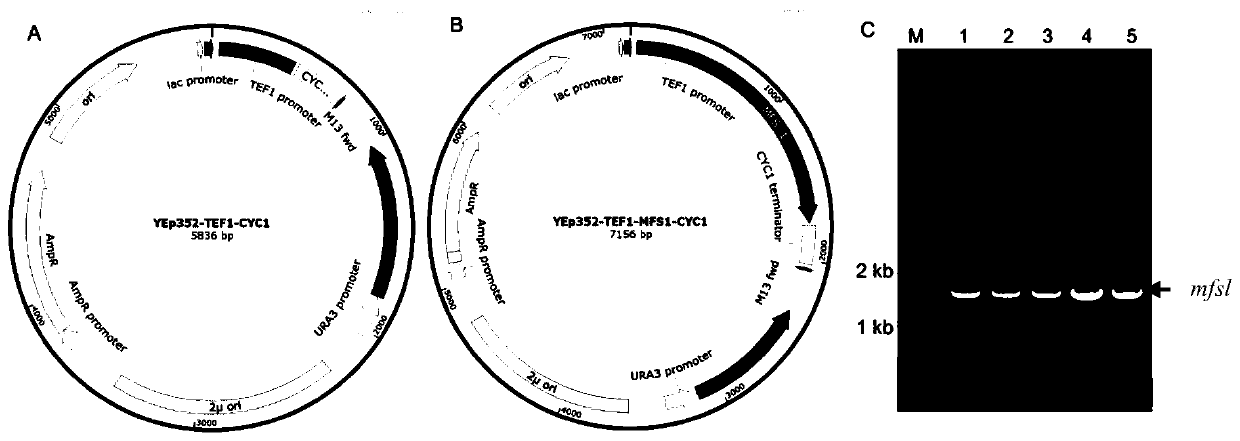
Myrothecium roridum A553 trichothecene-resistant self-protection gene mfs1, and application thereof
ActiveCN110904125AIncreased heterologous expression levelsIncreased resistance to trichothecenesDepsipeptidesFermentationCDNA libraryHeterologous
The invention discloses a myrothecium roridum A553 trichothecene-resistant self-protection gene mfs1 and an application thereof. The nucleotide sequence of the myrothecium roridum A553 trichothecene-resistant self-protection gene mfs1 is as shown in SEQ ID NO. 1. According to the invention, a transcriptome sequencing result is used to predict a gene sequence, a cDNA library is used as a template,and amplification is carried out to obtain the myrothecium roridum A553 trichothecene-resistant self-protection gene mfs1. The myrothecium roridum A553 trichothecene-resistant self-protection gene mfs1 can efficiently assist saccharomyces cerevisiae in resisting trichothecene toxins, and lay a molecular biological foundation for improving the trichothecene-resistant capacity of saccharomyces cerevisiae in the later period, improving the heterologous expression level of trichothecene toxins and obtaining novel trichothecene toxins.
Owner:东岱(济南)智能技术有限公司
Lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, method and application
InactiveCN107345256ALow costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSequence database
The invention provides a lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, a method and application. The method comprises the steps of obtaining a set of all root, stem and leaf transcripts of two lathyrus quinquenervius varieties of different flower colors at a seedling stage to form an original sequence database; removing low-quality sequencing data with a joint and forming a high-quality filtered sequence database; splicing high-quality filtered sequences, reducing the transcripts and carrying out BLAST comparison on the transcripts and a reference protein database NR, reserving the optimal comparison result and determining the maximal sequence as a Unigene representative sequence; carrying out SSR locus scanning in Unigene by using MISA 1.0; and carrying out SSR primer design by adopting primer 3.0, screening SSR primers and carrying out fingerprint map construction on 43 parts of lathyrus quinquenervius materials. The lathyrus quinquenervius EST-SSR primer group is convenient, fast, accurate, low in cost, and a new thought is provided for the development of lathyrus quinquenervius EST-SSR primers and germplasm resources identification.
Owner:山西省农业科学院农作物品种资源研究所 +1
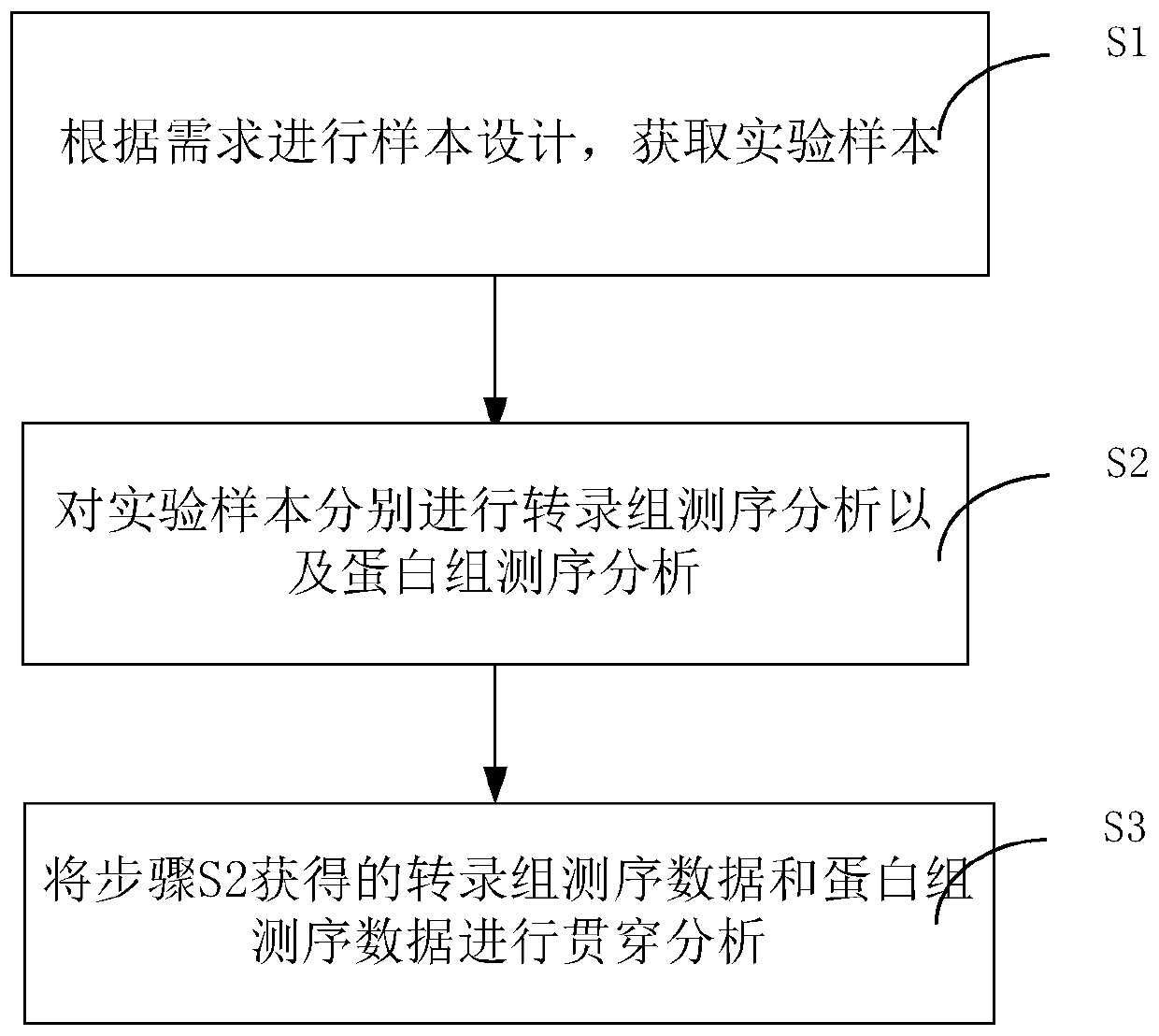
Penetration analysis method for transcriptome sequencing and proteomics sequencing data, and system
PendingCN109949864ALarge amount of informationHigh selectivityProteomicsGenomicsBiological bodySequence analysis
The invention discloses a penetration analysis method for transcriptome sequencing and proteomics sequencing data, and a system. The method comprises the following steps of S1, performing sample designing according to a requirement, and acquiring a test sample; S2, performing transcriptome sequencing analysis and proteomics sequencing analysis on the test sample; and S3, performing penetration analysis on the transcriptome sequencing data and the proteomics sequencing data in the step S2. The penetration analysis method can realize penetration analysis technology which is suitable for transcriptome sequencing and proteomics sequencing data of any organism.
Owner:GUANGZHOU GENE DENOVO BIOTECH
Method for developing SSR (Simple Sequence Repeat) primers of sisal hemp based on transcriptome sequencing
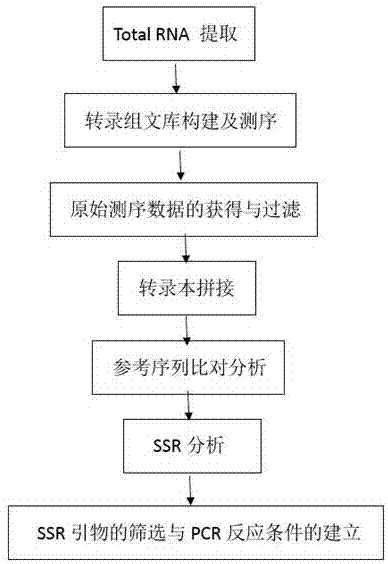
ActiveCN107201408AImprove development efficiencyFill the scarce gapsMicrobiological testing/measurementHybridisationAgricultural scienceTotal rna
The invention discloses a method for developing SSR (Simple Sequence Repeat) primers of sisal hemp based on transcriptome sequencing and belongs to the technical field of molecular biology. A preparation method of the SSR primers comprises the following five steps: extracting Total RNA (Ribonucleic Acid) and constructing a transcriptome library; carrying out transcriptome sequencing and sequence quality analysis; carrying out transcript assembly and SSR labeling analysis; designing the SSR primers; carrying out PCR (Polymerase Chain Reaction) amplification on the primers and electrophoresis analysis on amplified products. According to the method disclosed by the invention, a transcriptome sequence of the sisal hemp at a certain specific period is obtained by using a transcriptome sequencing method; then the SSR labeling analysis is carried out by bioinformatics analysis software, and further the SSR labeled primers of the sisal hemp are developed; the labeling and developing efficiency is high, and the blank of scarce SSR labels of the sisal hemp at present is filled up.
Owner:SOUTH SUBTROPICAL CROPS RES INST CHINESE ACAD OF TROPICAL AGRI SCI
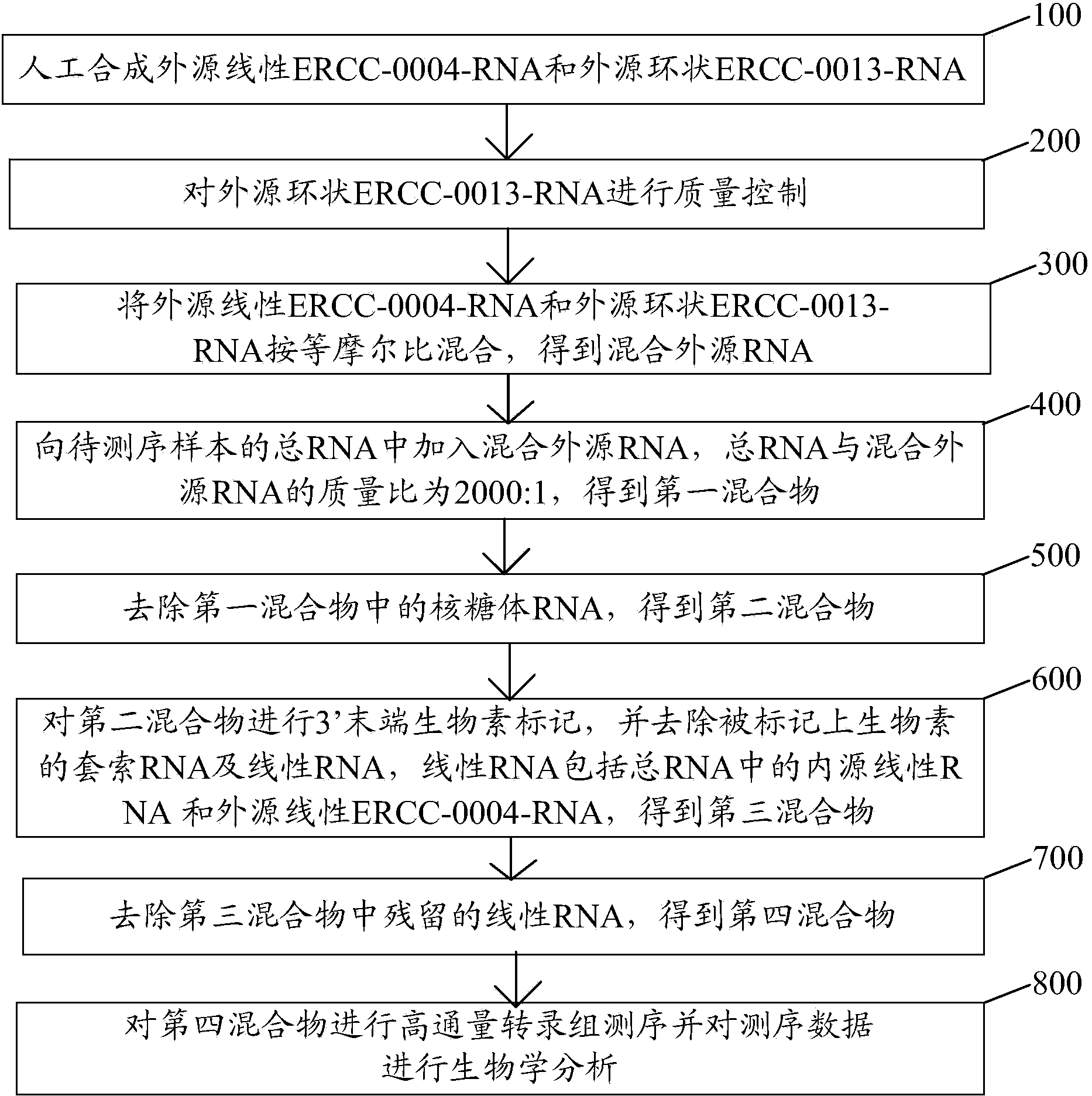
Method for sequencing high-flux circular RNA (ribonucleic acid)
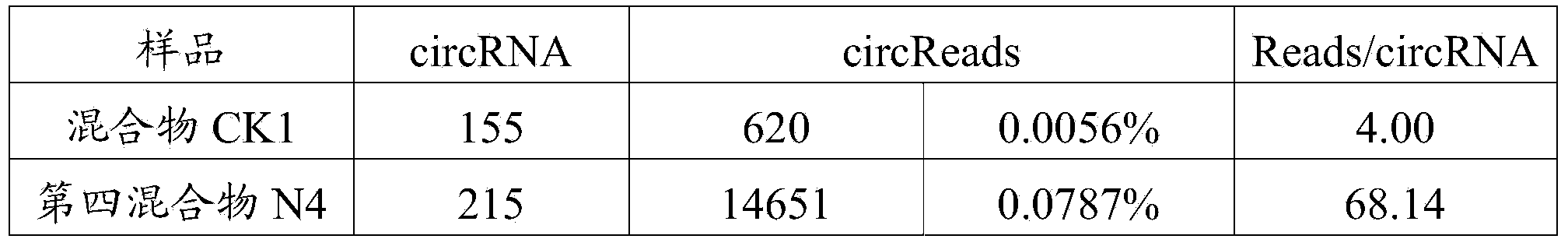
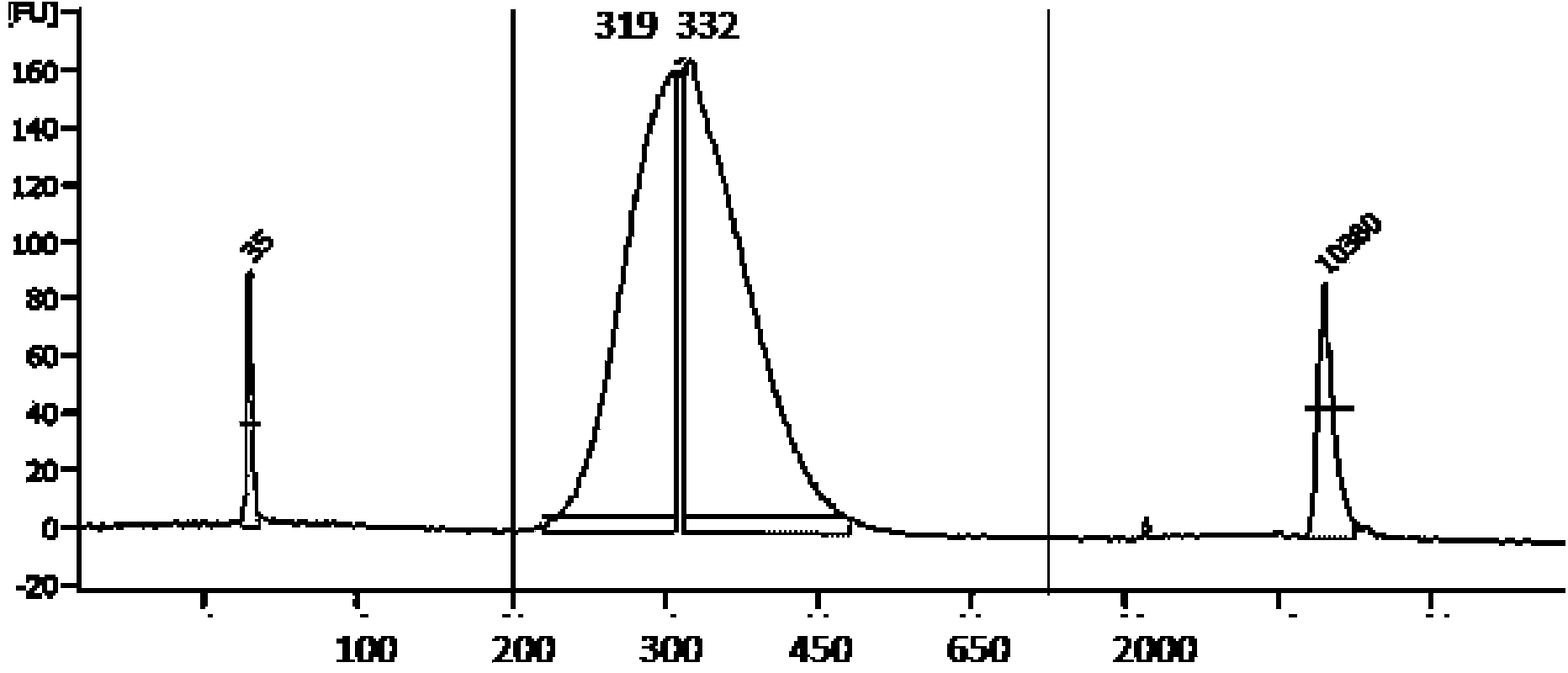
ActiveCN104388548APromote enrichmentReduce data volumeMicrobiological testing/measurementHigh fluxTotal rna
The invention discloses a method for sequencing high-flux circular RNA (ribonucleic acid), belonging to the field of molecular biology. The method comprises the following steps: artificially synthesizing exogenous linear ERCC-0004-RNA and exogenous circular ERCC-0013-RNA; performing quality control on the exogenous circular ERCC-0013-RNA; mixing the exogenous linear ERCC-0004-RNA and the exogenous circular ERCC-0013-RNA at an equal mole ratio to obtain a mixed exogenous RNA; adding the mixed exogenous RNA into total RNA of a to-be-sequenced sample to obtain a first mixture; removing ribosome RNA in the first mixture to obtain a second mixture; performing 3' tail end biotin marking on the second mixture, and removing lariat RNA and linear RNA marked with biotin to obtain a third mixture; removing linear RNA in the third mixture to obtain a fourth mixture; and performing standard high-flux transcriptome sequencing on the fourth mixture and performing biological analysis on sequencing data. By adopting the method disclosed by the invention, the expression of genes in an organism under a specific condition can be accurately reflected, and the cost is reduced.
Owner:JIANGHAN UNIVERSITY
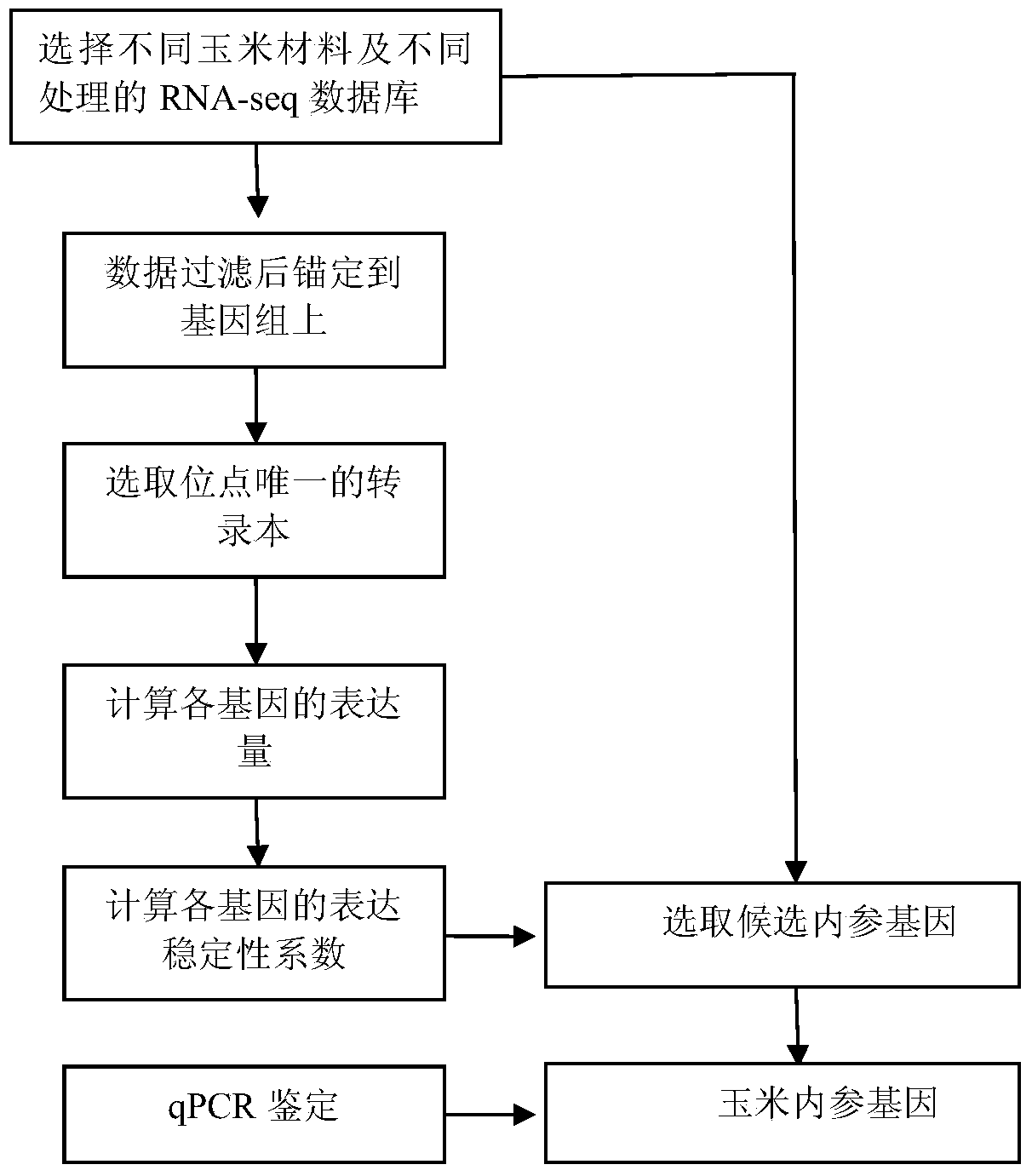
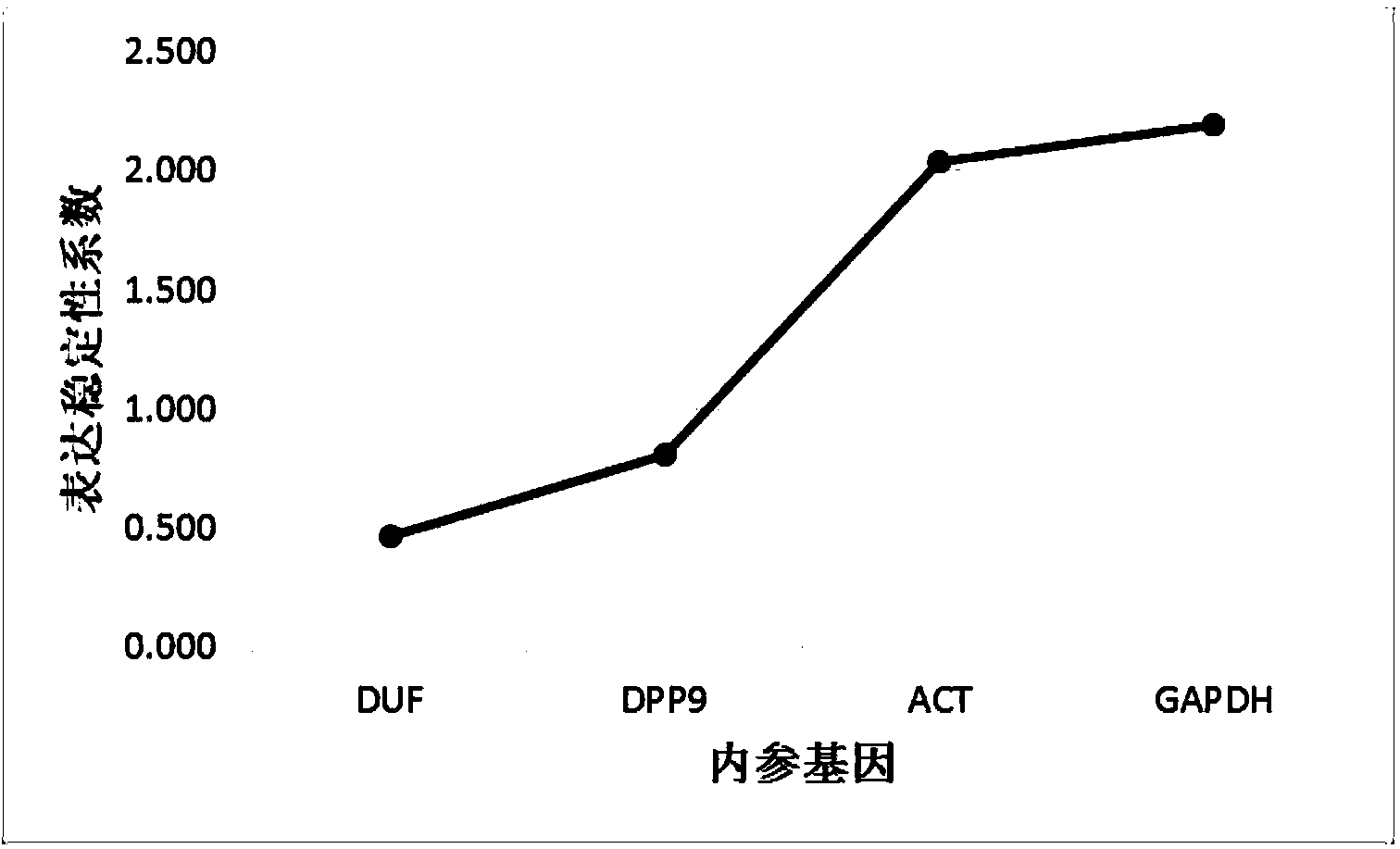
Method for high-throughput screening of maize reference gene
InactiveCN104131107AImprove selection efficiencyReduce experimental workloadMicrobiological testing/measurementReference genesHigh-Throughput Screening Methods
The invention discloses a method for high-throughput screening of a maize reference gene. The method comprises the following steps: by using a high-throughput transcriptome sequencing (RNA-seq) database which is sourced from different maize inbred lines with high diversity, different tissues and different developmental periods and is constructed after different stress treatment, analyzing the expression conditions of a whole transcriptome massively; screening a gene of which the expression is the most constant in the different developmental periods and under different conditions as the reference gene, and providing an accurate comparison for the study of transcriptome expression. The method disclosed by the invention is used for analyzing all the expressed genes of the whole transcriptome by using data of the whole transcriptome, is used for systematically screening the maize reference gene, and is more accurate and reliable compared with a conventional screening method.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Microsatellite marker primers for identification of Acipenser dabryanus family and identification method thereof
InactiveCN106939348APromote rapid developmentPrevent population quality declineMicrobiological testing/measurementDNA/RNA fragmentationDNA paternity testingMicrosatellite
The invention relates to microsatellite marker primers for identification of an Acipenser dabryanus family. Through a transcriptome sequencing technology, Unigenes of the screened Acipenser dabryanus are screened, microsatellite loci in the Unigenes sequence are detected, according to flanking sequences at two ends of each one of the microsatellite molecular marker loci, specific primers of the molecular marker are labeled, the specific primers are subjected to PCR amplification, the stability and polymorphism of the amplification result are detected, and 18 effective microsatellite markers are acquired. Through the microsatellite marker primers, according to the polymorphism of the microsatellite marker primers, a parentage identification platform of Acipenser dabryanus is built and the basis is provided for Acipenser dabryanus selection and breeding matching.
Owner:FISHERIES INST SICHUAN ACADEMY OF AGRI SCI
Plum SSR labeled primer pair exploited on basis of transcriptome sequence, and application thereof
ActiveCN105238781AOvercoming complexityOvercome workloadMicrobiological testing/measurementDNA/RNA fragmentationGeneticsGermplasm
The invention belongs to the fields of molecular marker technology exploitation and application, and concretely provides a plum SSR labeled primer pair exploited on the basis of a transcriptome sequence, and an application thereof in plum germplasm resource researches. The primer pair is exploited on the basis of the transcriptome sequence, batch treatment of a large amount of sequence information is carried out on the basis of transcriptome sequencing, and large batch exploitation of SSR labels can be realized through SSR site searching and SSR primer designing, so the exploitation efficiency is greatly improved, and the problems of complex steps, large workload and high exploitation cost of traditional SSR label exploitation methods are solved. The validity of SSR primers is verified through SSR primer screening and diversity analysis. Foundation is laid for plum SSR labeling primer exploitation and researches of the plum germplasm resource diversity, the kind identification and the genetic relationship by using the SSR molecular labels.
Owner:POMOLOGY RES INST FUJIAN ACAD OF AGRI SCI
Preparation method and application for Miscanthus Genic-SSR mark
InactiveCN102424826ASimple methodImprove efficiencyMicrobiological testing/measurementDNA preparationCritical energyTotal rna
The invention discloses a preparation method and application for a Miscanthus Genic-SSR mark. The method comprises the following steps: A, acquiring Miscanthus transcriptomic sequence, that is, selecting young and tender Miscanthus leaves, extracting total RNA respectively so as to obtain transcriptome Reads, removing the pollution of carrier RNA sequence and carrying out splicing so as to obtainthe Miscanthus transcriptomic sequence; B, identifying SSR sites in Miscanthus ESR sequence, that is, searching SSR sites in obtained non-redundant Unigene-EST sequence by using SSR retrieval software, with repeats being 2 to 6 bases, so as to obtain a series of Unigene sequences containing information about the SSR sites; C, designing Miscanthus Genic-SSR primers, that is, using primer design software to design primers according to the obtained Unigene sequences containing the SSR sites so as to obtain a series of the Miscanthus Genic-SSR primers. According to the invention, the method is simple, has high efficiency and is simple to operate; a huge amount of marks are obtained; EST sequence obtained through transcriptomic sequencing lays a foundation for genetic research, molecular improvement and the like of the critical energy plant Chinese silvergrass.
Owner:湖北光芒能源植物有限公司
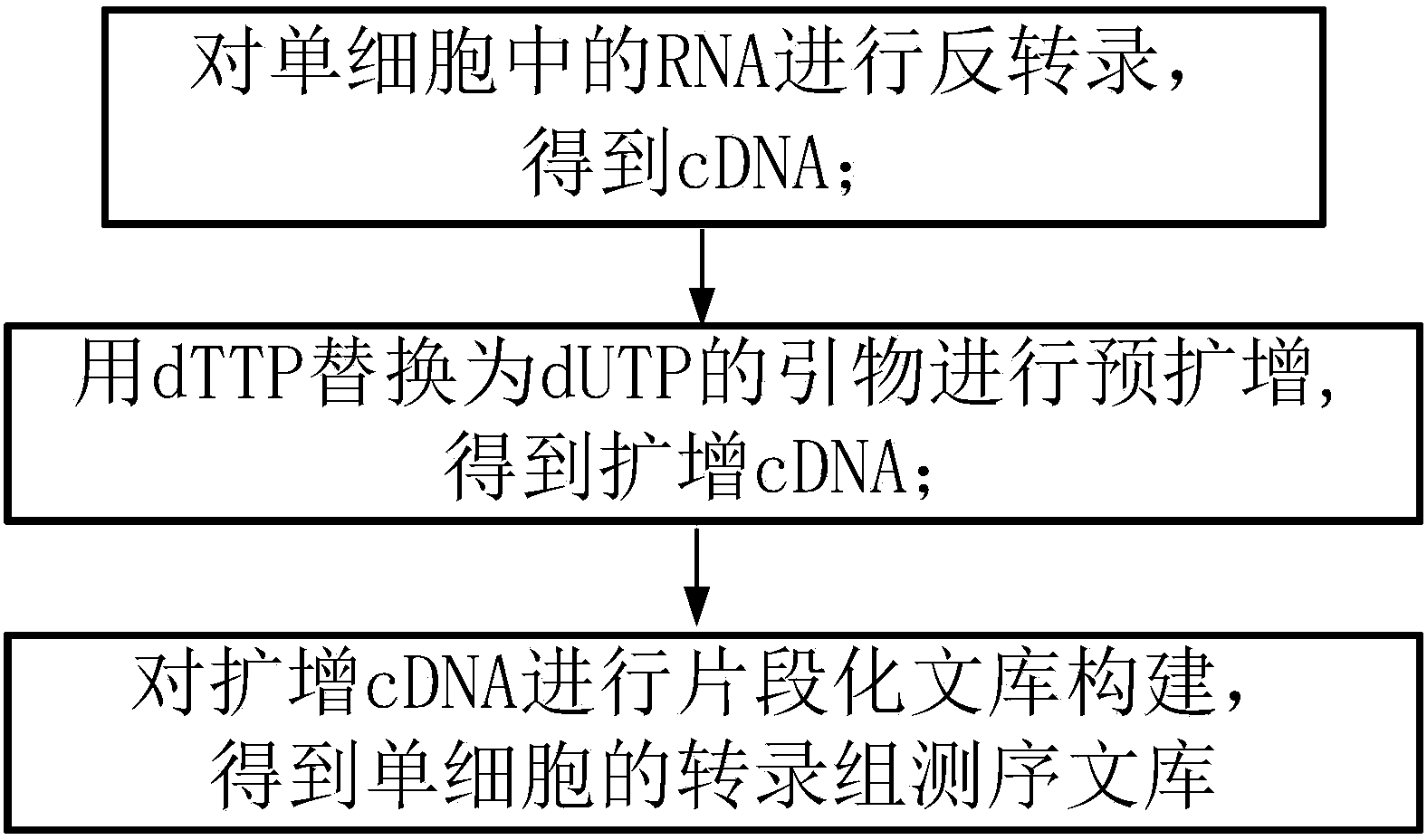
Method for establishing single cell transcriptome sequencing library and application of method
InactiveCN104389026AReduced amplificationIncrease the amount of effective dataMicrobiological testing/measurementLibrary creationSingle cell transcriptomeTranscriptome Sequencing
The invention discloses a method for establishing a single cell transcriptome sequencing library and application of the method. The method comprises the following steps: performing reverse transcription on RNA in a single cell, thus obtaining cDNA; performing pre-amplification on cDNA by using an amplification primer, thus obtaining amplified cDNA; and performing fragmentation library construction on the amplified cDNA, thus obtaining a transcriptome sequencing library of the single cell, wherein dTTP in the amplification primer is substituted by dUTP. According to the method disclosed by the invention, as the dTTP in the pre-amplification primer is substituted by dUTP, fragments containing the pre-amplification primer in a jointed fragment can be interrupted in an enzymic digestion step after the step of joint connection, and are removed in later high-temperature pre-denaturation and denaturation steps; furthermore, the amplification of fragments with the pre-amplification primer can be reduced, the ratio that the proportion of data with pre-amplification primer pollution in the obtained sequencing data is greatly reduced, and the effective data amount of the obtained data is greatly increased.
Owner:BEIJING NOVOGENE TECH CO LTD
DNA molecular marker method for identification of alien invasive species Spartina alterniflora population
InactiveCN103146816AGenetic stabilityIncrease success rateMicrobiological testing/measurementInvasive species monitoringA-DNAGenetic stability
The invention relates to a DNA molecular marker method for identification of the alien invasive species Spartina alterniflora population. The method includes: conducting high-throughput transcriptome sequencing and screening to obtain a target gene, carrying out a PCR reaction, subjecting the PCR product to sequencing so as to obtain a plurality of single nucleotide polymorphism (SNP) sites, then letting all effective SNP sites compose a haplotype for molecular markers, thus achieving genetic typing. According to the method provided in the invention, rich polymorphism information can be obtained in one experiment. SNP markers have genetic stability, can be used for samples of different sources, and are suitable for high-throughput automated analysis.
Owner:NANJING UNIV
Transcriptome sequencing method
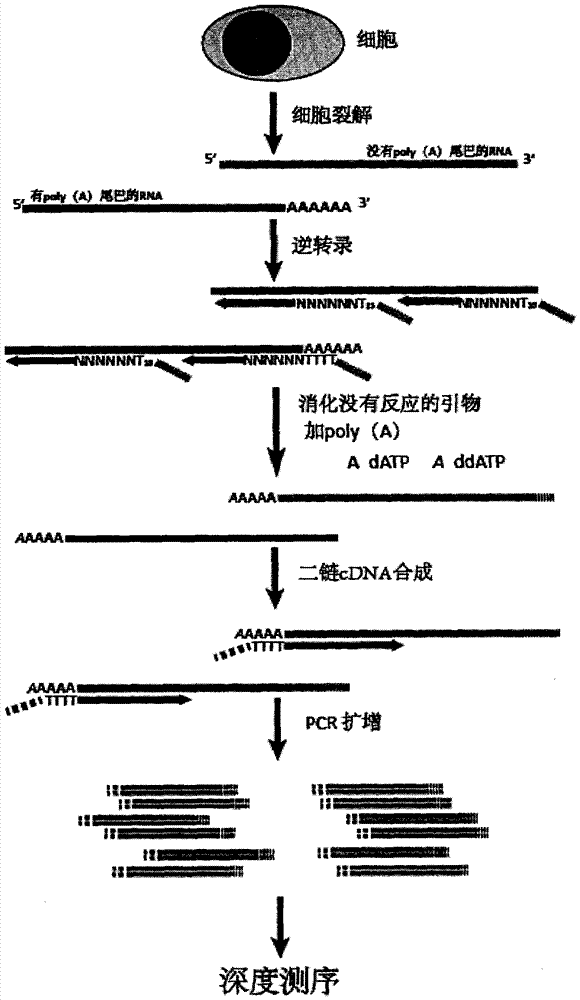
InactiveCN104711340AHigh technical repeatabilityRich researchMicrobiological testing/measurementEthylene HomopolymersTranscriptome Sequencing
The present invention provides a transcriptome sequencing method, wherein primers used during a reverse transcription process from RNA to cDNA comprise a random sequence and a specific sequence, the random sequence is positioned at the 3' terminal of the specific sequence, bases from 0 to 5 exist between the random sequence and the specific sequence, the random sequence comprises 4-12 random bases, the specific sequence is a homopolymer of 5-30 bases, and the base of the specific sequence is any one selected from A, G, C and T. According to the present invention, with the design on the reverse transcription primers, the RNA having the poly (A) tail can be sequenced while the RNA with no poly (A) tail can be sequenced, the rRNA removal step is not required during the cell transcriptome sequencing, and the method is especially suitable for the sequencing on the transcriptome having the low initial RNA amount and the low cell number.
Owner:PEKING UNIV
Method for cell quality control through unicellular transcriptome sequencing
ActiveCN106701995AControl UniformityControl homogeneityMicrobiological testing/measurementBiostatisticsSingle cell transcriptomePrincipal component analysis
The invention provides a method for cell quality control through unicellular transcriptome sequencing. The method comprises the following steps of: performing unicellular separation so as to obtain a total unicellular number; performing transcriptome amplification on each unicell; confirming that amplification can be implemented but the unicellular number is not sufficient for analysis on sequencing results; performing sequencing and analysis, including performing main component analysis and cluster analysis on the sequencing results, comparing with a standard unicellular database, and calculating the unicellular number off from total tendency; screening, including screening based on unicellular partitioning and screening based on expression gene types, wherein the step of screening based on unicellular partitioning comprises a step of screening cells of which the activity levels are greater than 75%, and the step of screening based on the expression gene types comprises a step of classifying expression genes into a good gene group, a neutral gene group and a malignant gene group according to gene functions. By adopting the method provided by the invention, the uniformity or the homogeneity of cells can be effectively controlled.
Owner:GENEIS TECH BEIJING CO LTD
Method for screening and annotating of longissimus dorsi differential expression genes of pigs of different varieties
InactiveCN104636638ADeepen understandingSpecial data processing applicationsLongissimus dorsiLongissimus
The invention discloses a method for screening and annotating of longissimus dorsi differential expression genes of pigs of different varieties. The method comprises the following steps that 1, sampling and sequencing are conducted; 2, original data are processed; 3, differential genes are screened; 4, function significance enrichment analysis is conducted; 5, Pathway significance enrichment analysis is conducted. According to the method, a high throughout transcriptome sequencing technology is adopted, the gene expression profiles of longissimus dorsi tissue of the pigs of different varieties are established, the differential expression genes are obtained, the differential expression genes are selected through GO analysis and pathway analysis, the key gene of controlling the meat quality trait is determined, control over muscle growth and development and fat metabolism can be better understood, and the molecular biology evidence is provided for the following research of the molecular mechanism forming the difference of the meat quality and the growth trait of domestic and oversea varieties.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Wheat BSR-Seq gene locating method
ActiveCN106202995ALow costHigh precisionSequence analysisSpecial data processing applicationsReference genome sequenceAllele frequency
The invention discloses a wheat BSR-Seq gene locating method. The wheat BSR-Seq gene locating method comprises the following steps of: constructing and sequencing mixing pools, performing quality variation mining, screening a transcript tightly linked to a target gene, developing and locating a molecular marker, etc. The next-generation transcriptome sequencing technology (transcriptome sequencing, RNA-Seq) and a mixing pool technology (Bulked Segregant Analysis BSA) are combined; a wheat sequencing draft sequence is used as a reference sequence at first; then, a lot of high-quality SNP heritable variations on the transcript is mined at high throughput by adopting the next-generation sequencing technology; the allele frequency is precisely calculated in combination with the mixing pool technology, so that the transcript tightly linked to the target gene possibly can be rapidly screened out; false positive is precisely checked and controlled through Fish; the method is independent of a reference genomic sequence, low in cost, rapid and high in precision; the wheat gene locating efficiency and precision are increased; the wheat polymorphic molecular marker developing cost is reduced; therefore, the wheat gene fine locating working time is reduced to several months from several years; the locating precision is reduced to a fraction or 0 cM from several cM; and the fine locating cost is reduced to several thousands from several ten thousands.
Owner:北京麦美瑞生物科技有限公司
Space transcriptome detection chip and method
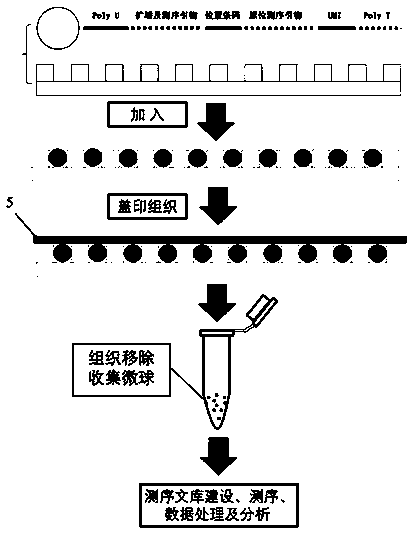
InactiveCN109762728AHigh resolutionNo damageBioreactor/fermenter combinationsBiological substance pretreatmentsCell specificMicrosphere
The invention provides a space transcriptome sequencing chip and method. The chip and the method are used for determining one or more single cell sequences in different space positions in tissues. Thechip comprises a micropore plate and coding sequencing microspheres, wherein the micropore plate is provided with the coding sequencing microspheres. The method comprises the steps of coupling with the microspheres using a plurality of random codes, wherein each random code comprises a molecular marker and a position marker barcode; the molecular marker can detect sequence information of single cells of the tissues, and the position markers are used for recording space position information of the single cells of the tissues. Based on the detection chip and the detection method, the single-cell specific transcriptome information of the tissues can be analyzed and detected, and the spatial positions of thousands of single cells in the tissues can be simultaneously detected.
Owner:SOUTHEAST UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com