Method for developing mung bean simple sequence repeat (SSR) primer based on transcriptome sequencing
A transcriptome sequencing and transcriptome technology, applied in the fields of molecular biology and bioinformatics, can solve the problem of a small number of mung bean SSR primers and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 RNA-seq analysis and design of SSR primers
[0043] 1. Acquisition of transcriptome data
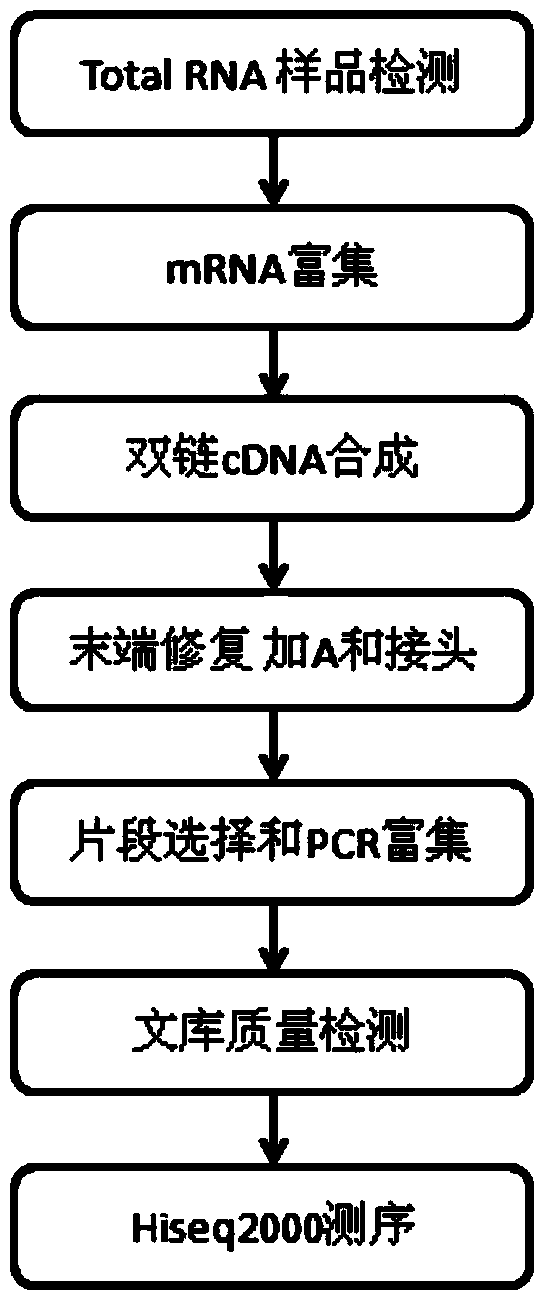
[0044] The total RNA of mung bean seedlings was extracted by Trizol reagent, and the mRNA was enriched by Oligo (dT) magnetic beads. Add fragmentation buffer to break mRNA into short fragments, use mRNA as a template, use six base random primers to synthesize the first cDNA strand, then add buffer, dNTPs, RNase H and DNA polymerase I to synthesize the second cDNA strand, in After purification with QiaQuick PCR kit and elution with EB buffer, end repair was performed, A was added and sequencing adapters were connected, and then agarose gel electrophoresis was used for fragment size selection, and finally PCR amplification was performed. The constructed sequencing library was used with IlluminaHiseq2000 Perform sequencing.
[0045] Reverse transcribe and synthesize double-stranded cDNA, purify cDNA, perform end repair, add A and connect sequencing adapters, then use agar...
Embodiment 2
[0069] Example 2 Discovery of mung bean high-throughput SSR sites
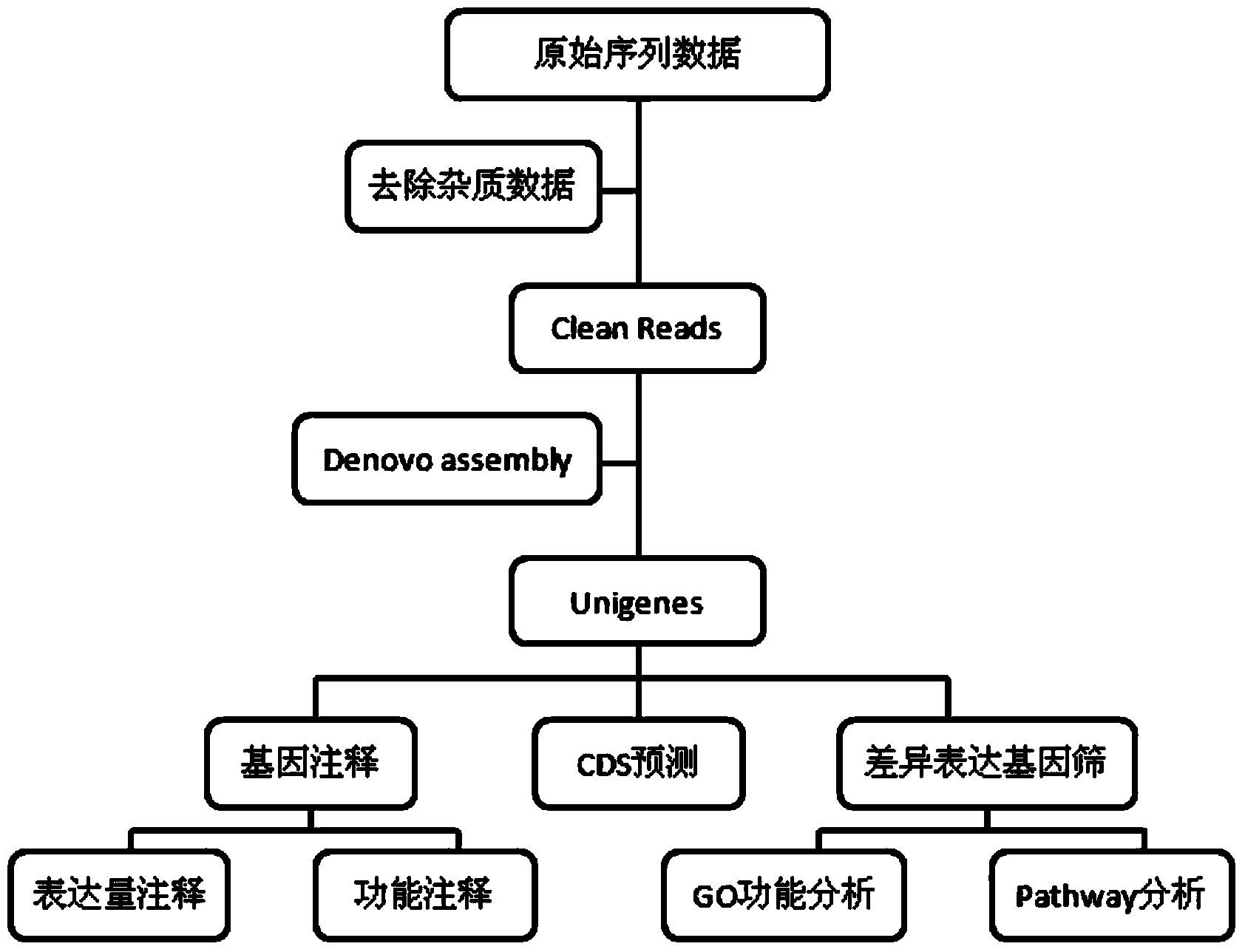
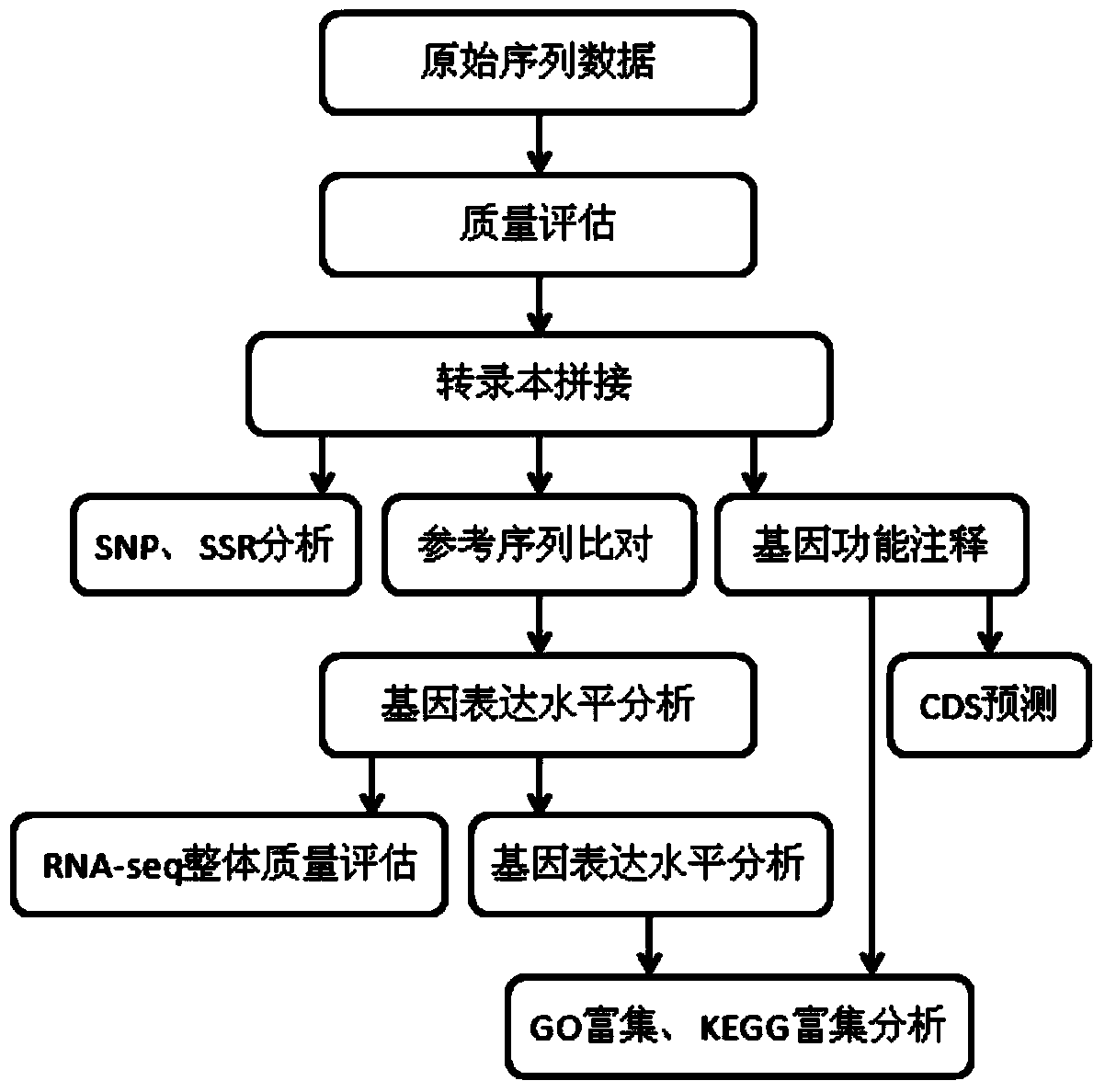
[0070]Using the above method, mung bean leaves were used as materials for high-throughput sequencing, and high-throughput SSR sites were excavated from mung bean transcriptome sequences using Perl language, and 83,542 transcriptome sequences and 48,693 unigenes were obtained (Table 1). The SSR density distribution has the highest frequency of single-base microsatellites, and the highest proportion is A / T, followed by tetranucleotides (Table 2, Figure 5 , Figure 6 ).
[0071] Table 1 Frequency distribution of splicing length
[0072]
[0073] Table 2 Repeated primitives
[0074]
[0075] A total of 13134 pairs of SSR primers were designed using primer3.0 batch design software. Randomly select 50 pairs of primers (see the nucleotide sequence list) from it, and use mung bean leaf DNA to carry out primer design success rate detection. The results show that a total of 46 pairs of SSR primers detect clea...
Embodiment 3
[0076] Example 3 Using SSR primers to identify polymorphisms in 8 mung bean DNAs from different countries
[0077] The DNA of 8 parts of mung bean was extracted, and its quality was detected by 0.8% agarose gel electrophoresis. The DNA concentration was diluted to 50 ng / μL and stored at -20°C for later use. The DNA of the materials used for primer development was used to identify the success rate of primer design by PCR. The PCR reaction system uses a 10 μL reaction system, which includes 40ng of genomic DNA, 1 × Taq enzyme buffer (10mmolL -1 Tris-HCl, pH8.8; 10mmol L -1 KCl; 10mmol L -1 (NH 4 ) 2 SO 4 ;1.5mmol L -1 MgCl 2 ; 0.1%Triton X-100), 1mmol L -1 dNTPs, upstream and downstream primers 0.25 μmol L -1 and 1U Taq DNA polymerase. The SSR reaction program was: pre-denaturation at 95°C for 5 min, deformation at 95°C for 30 s, annealing at 51-60°C for 45 s, extension at 72°C for 45 s, 32-35 cycles, and finally extension at 72°C for 5 min. After the reaction, add 2 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com