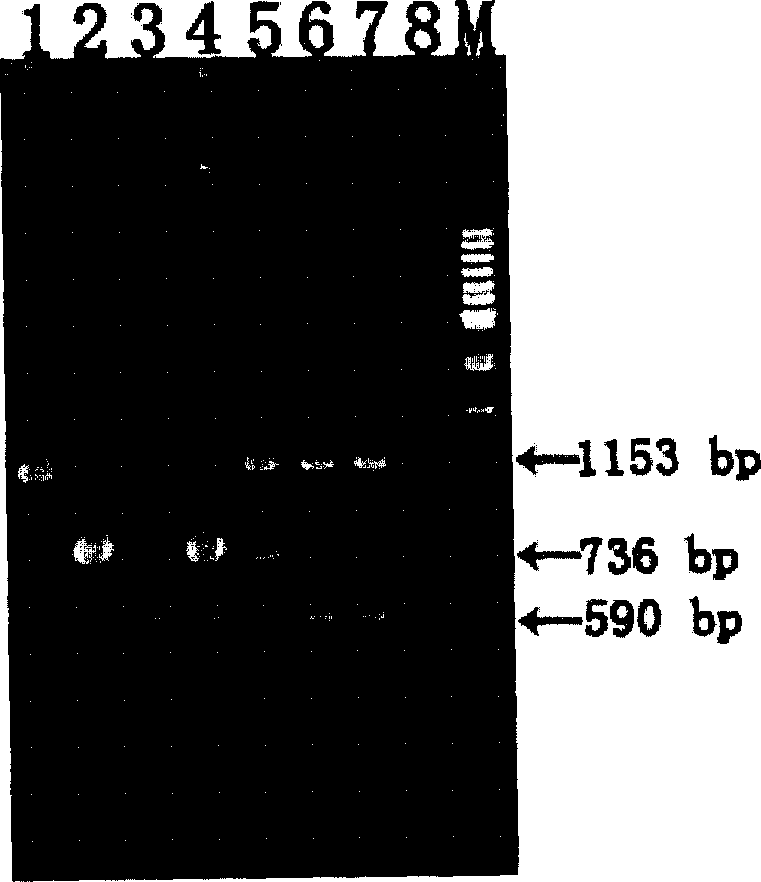
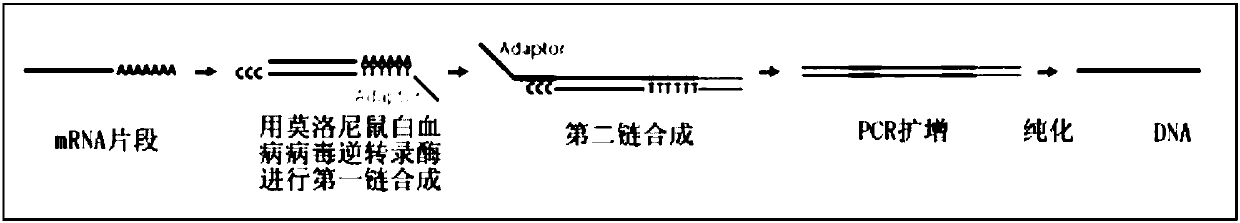
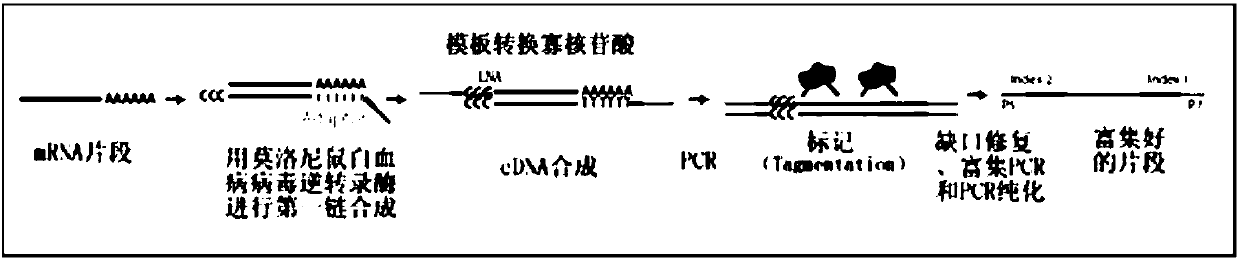
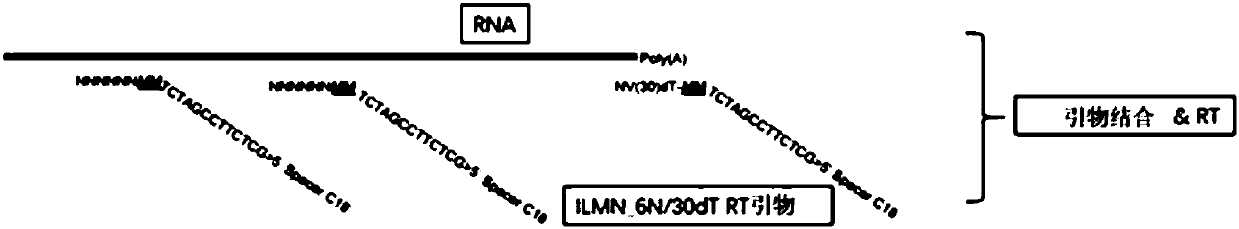
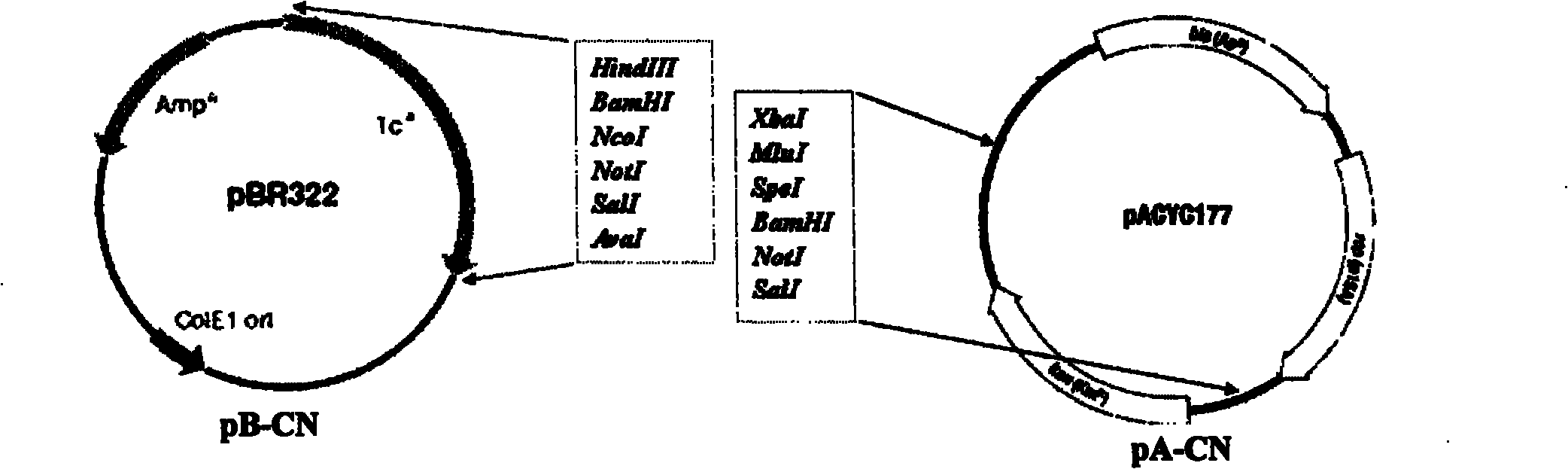
Patents
Literature
1567 results about "Total rna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Total RNA. Ribonucleic acid (RNA) is vital to the functioning of life. It is present in every cell in the body and has nearly limitless functions – from carrying genetic information to the production of deoxyribonucleic acid (DNA).
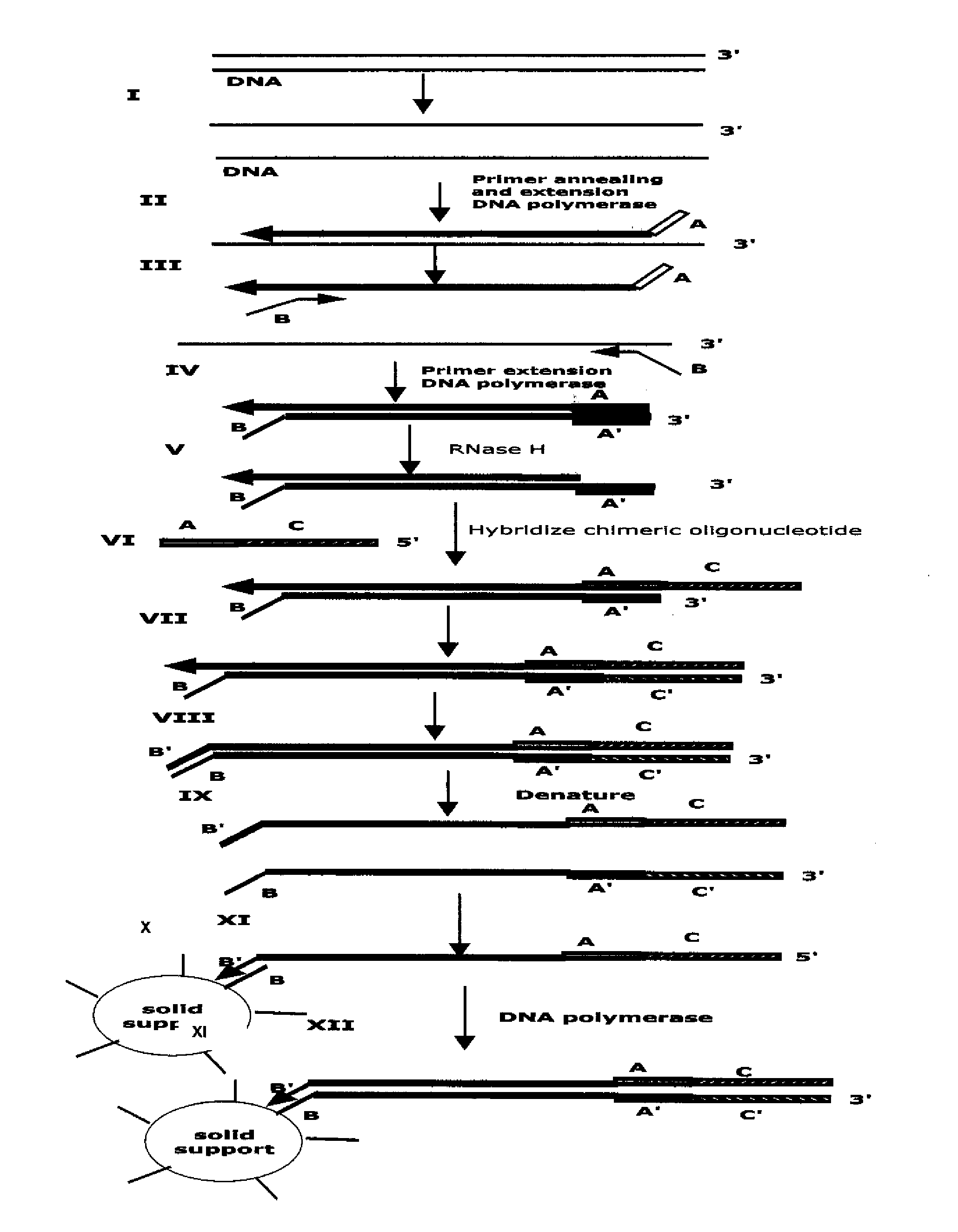
Method for Archiving and Clonal Expansion
InactiveUS20090203531A1Microbiological testing/measurementLibrary member identificationPresent methodTotal rna
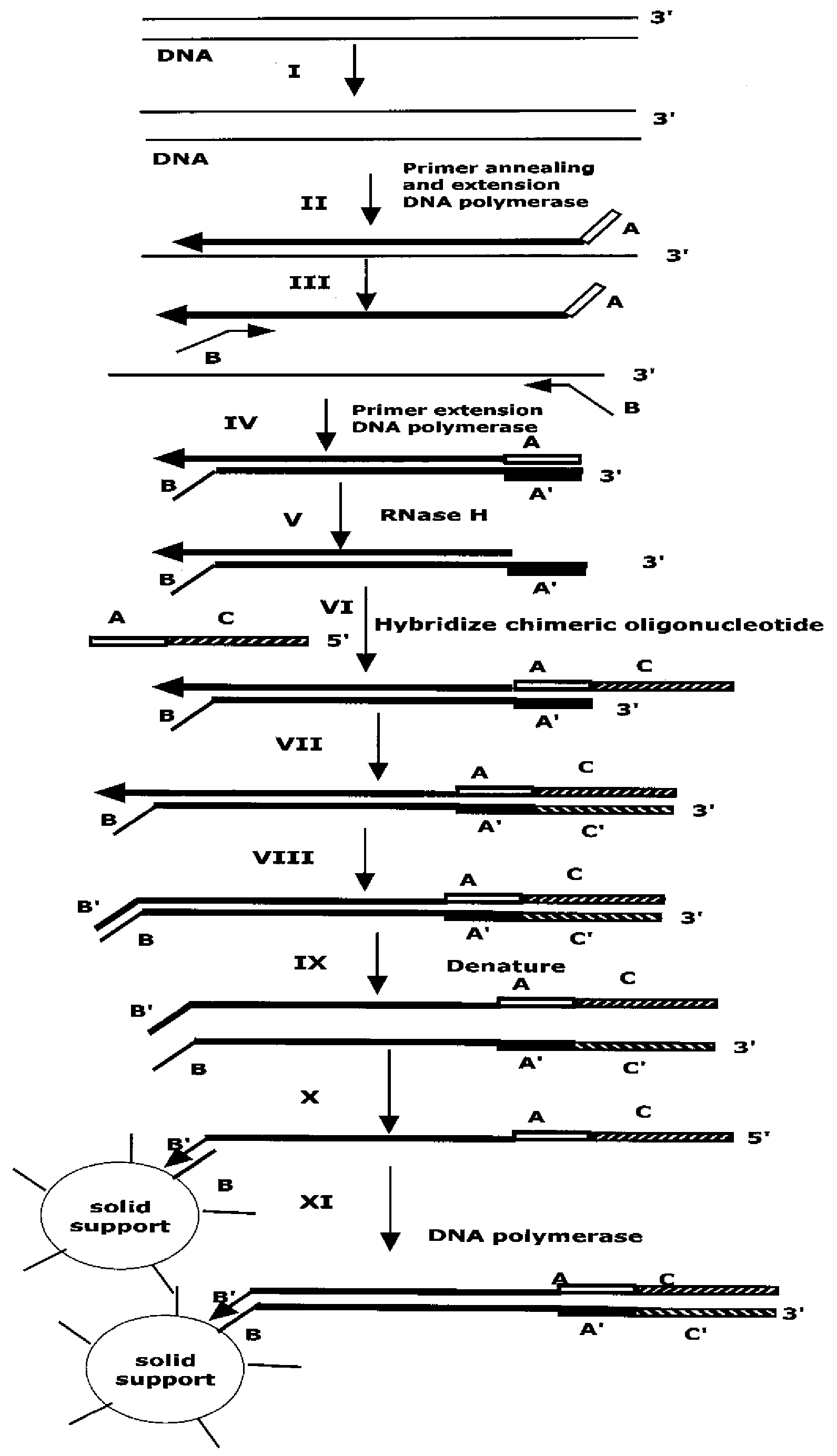
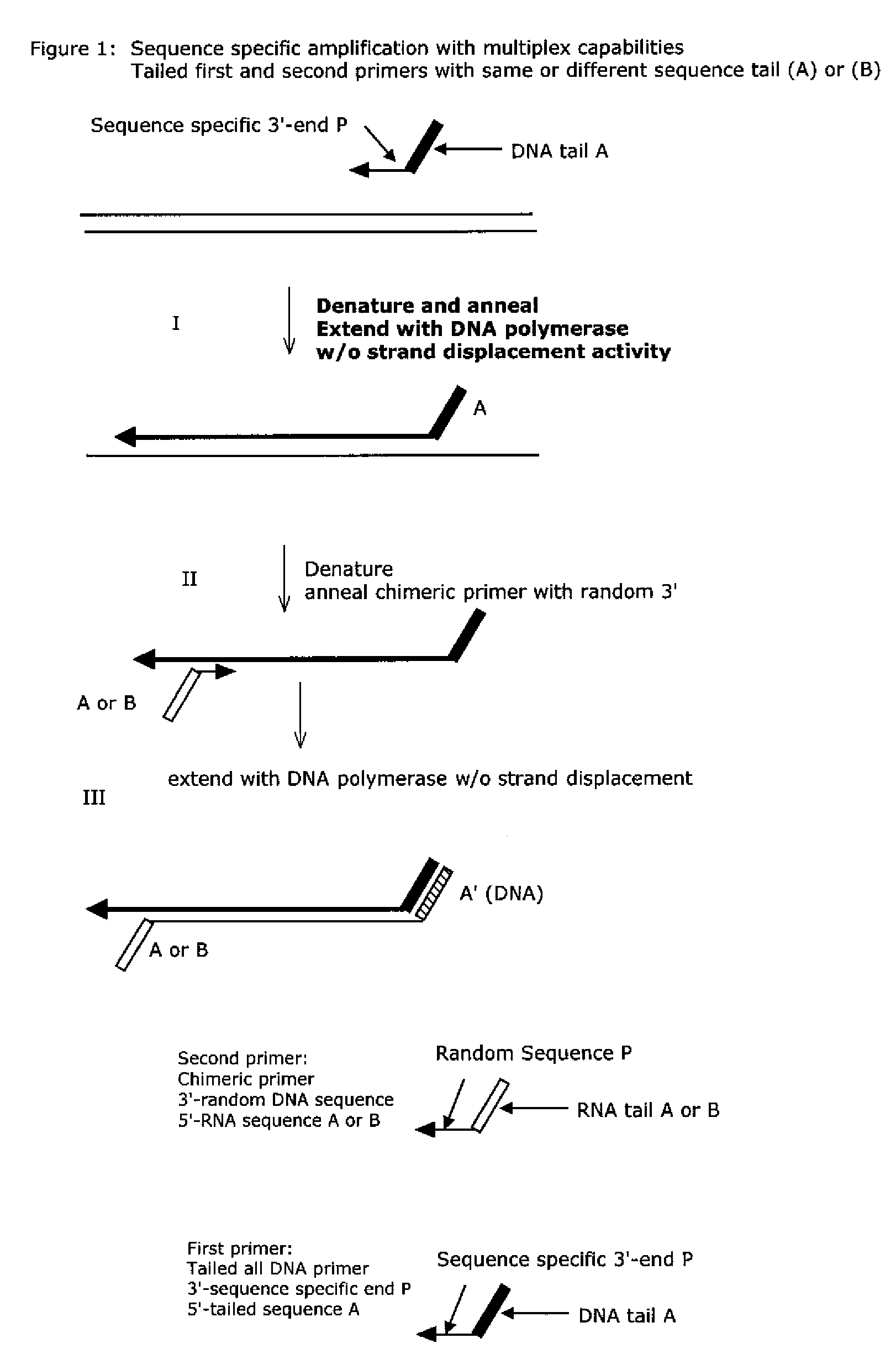
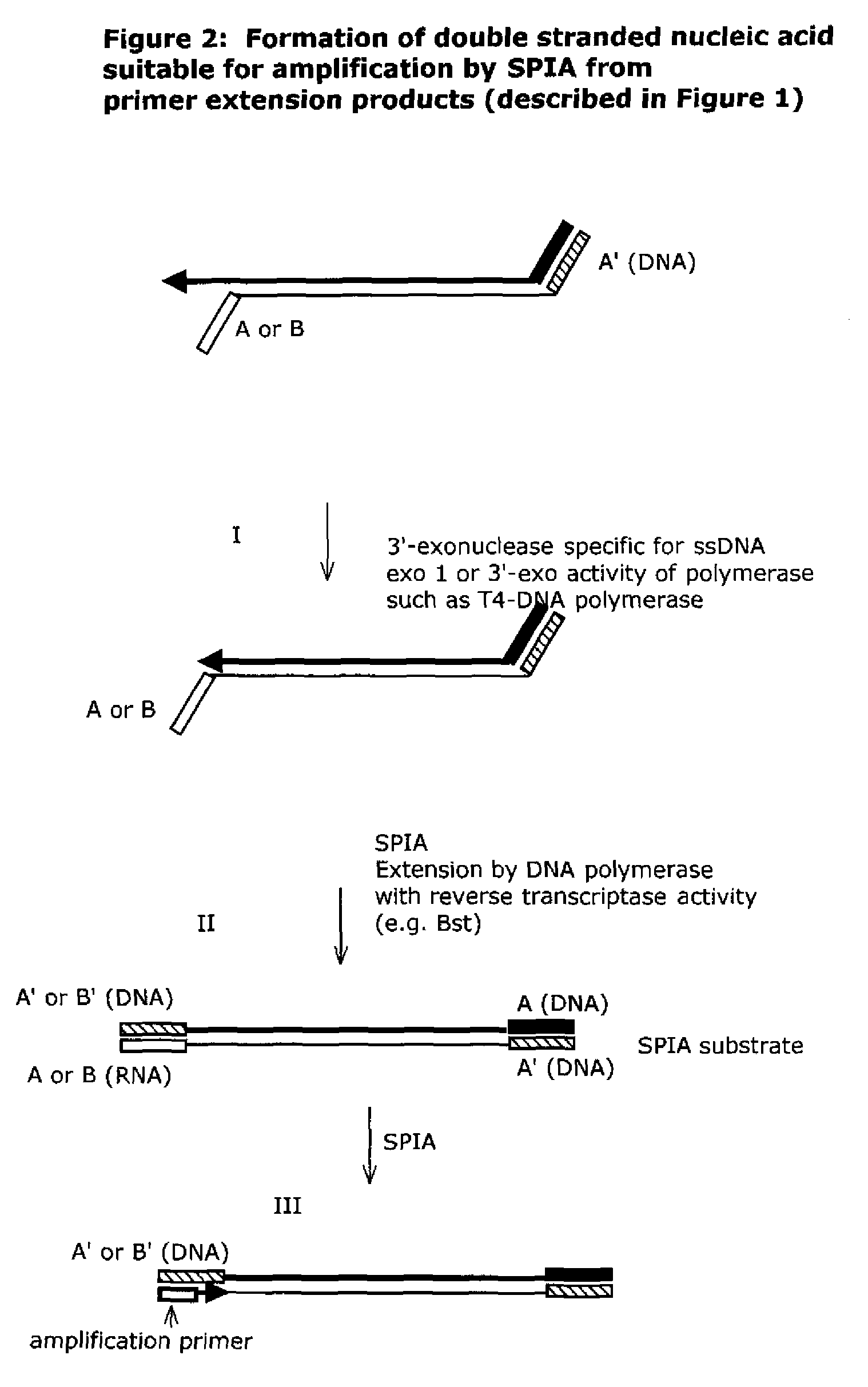
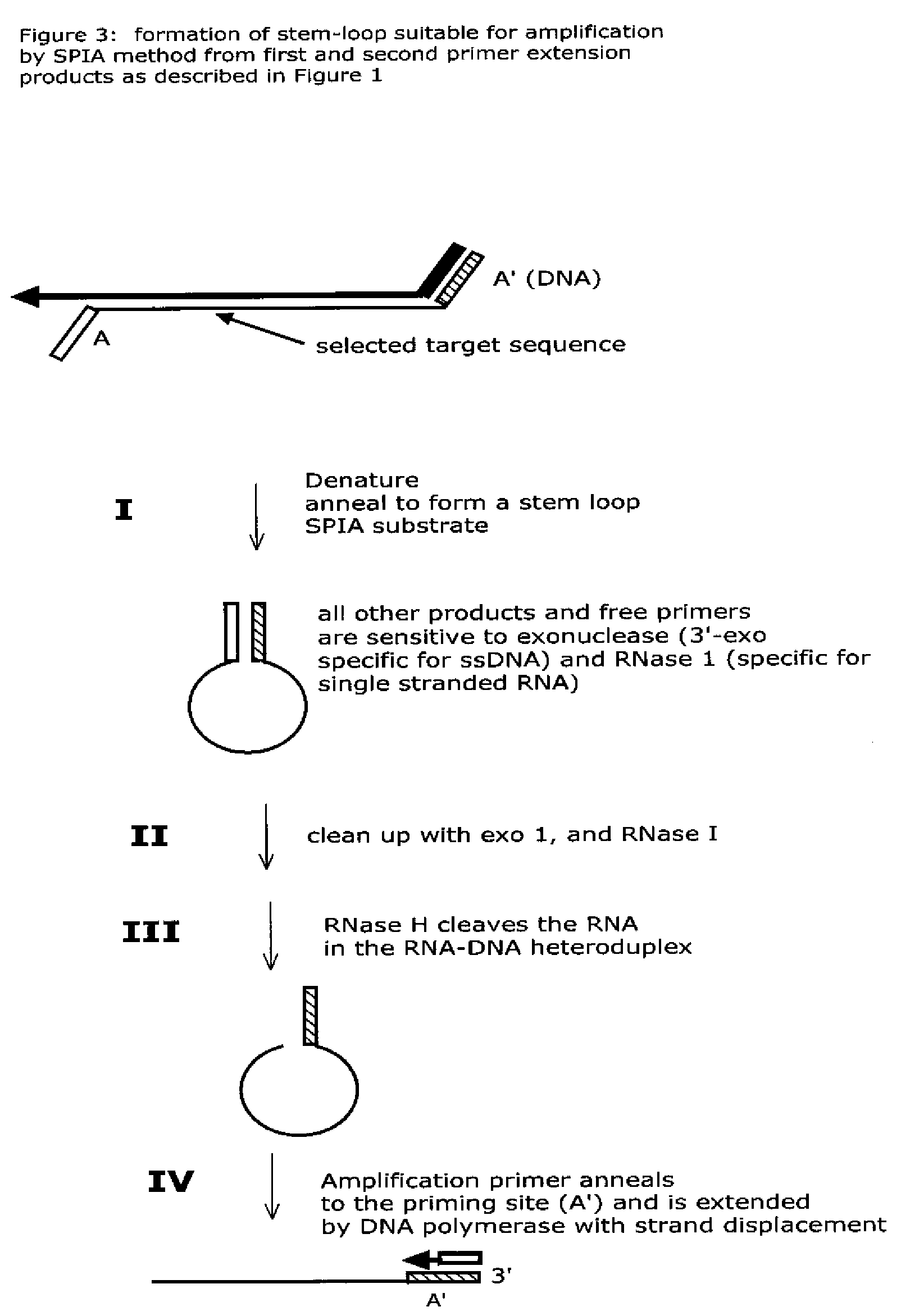
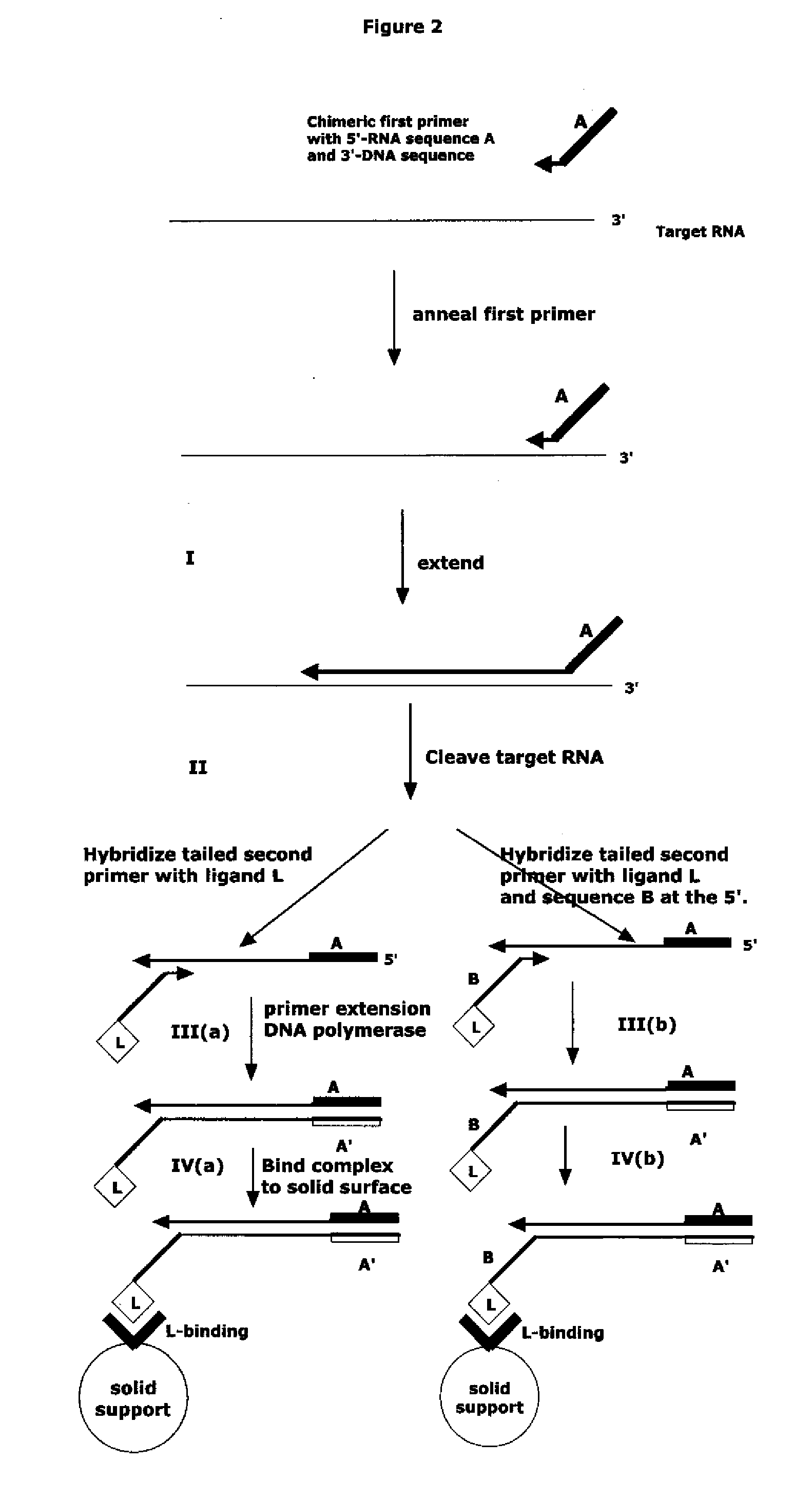
The present method provides methods, libraries, and kits related to the archiving and clonal expansion of sequences related to target polynucleotide sequences. The method allow for the attachment of polynucleotides with defined 3′ and or 5′ sequences to solid surfaces. The polynucleotides attached to the solid substrates can be stored or archived as libraries and can subsequently be retrieved for analysis, for example by clonal expansion. In some embodiments, nucleotides attached to solid surfaces can be used for sequencing of nucleotide sequences related to target RNA or target RNA. The methods are applicable to total RNA and / or total DNA analysis.
Owner:NUGEN TECH
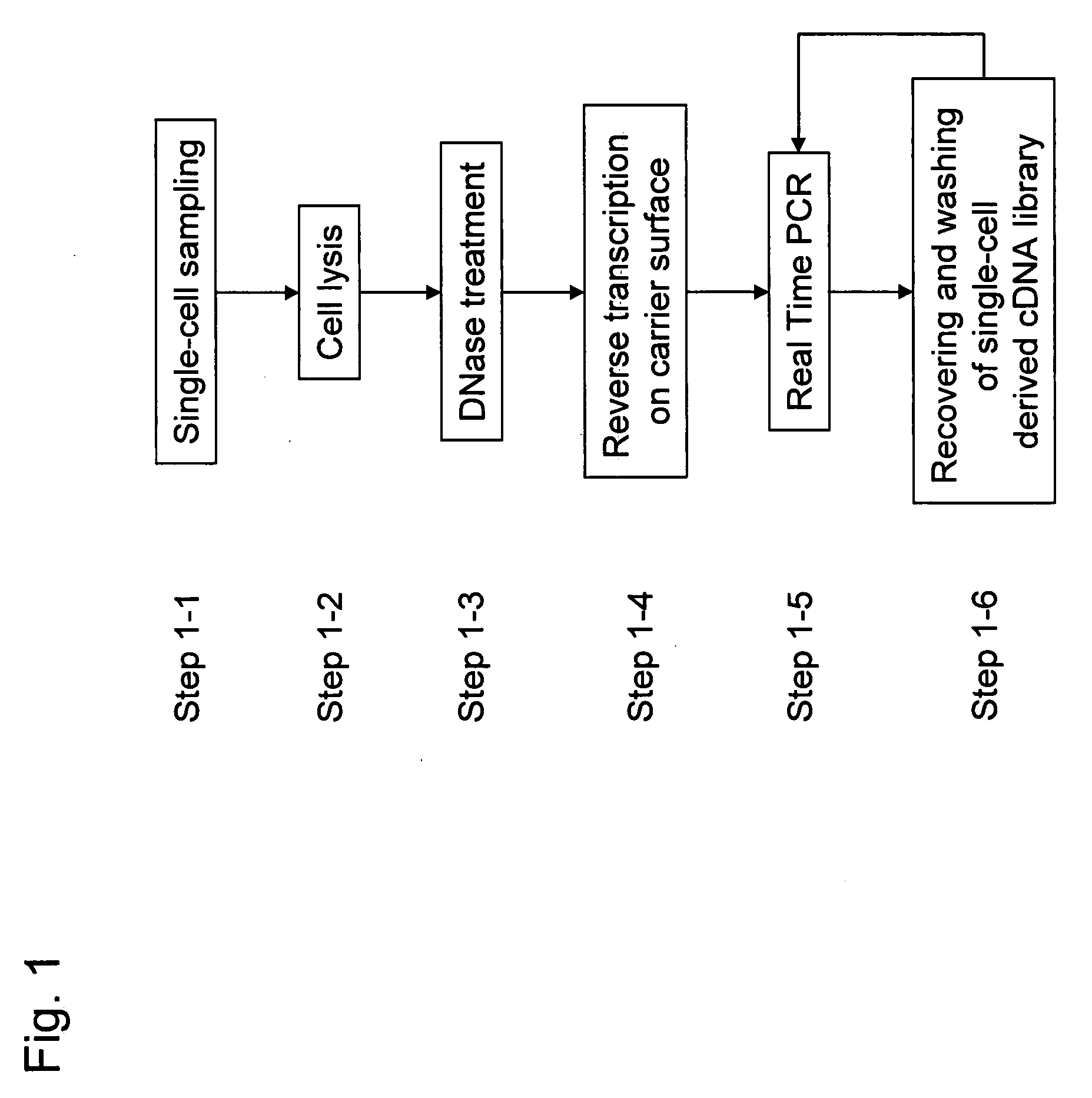
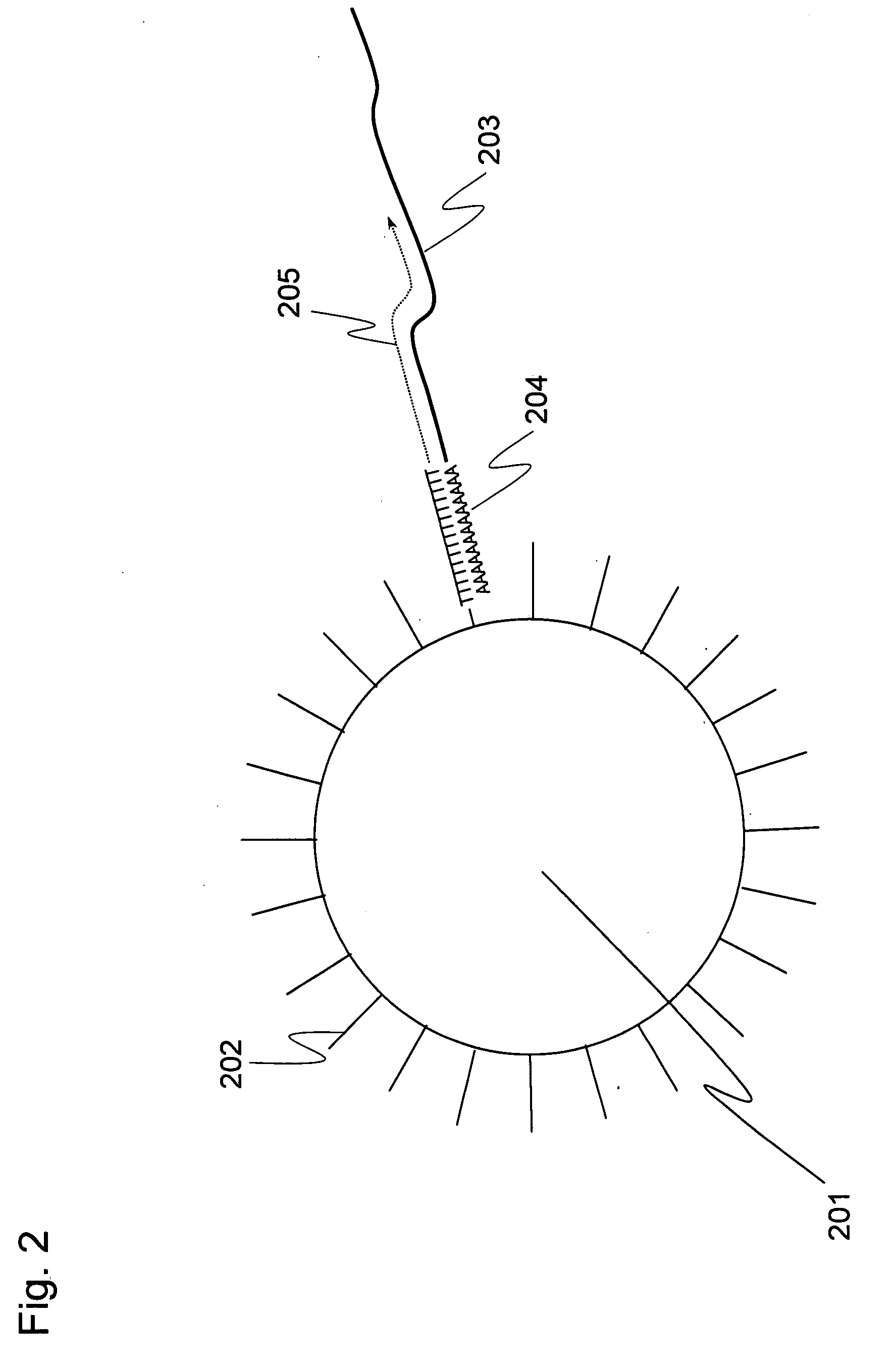
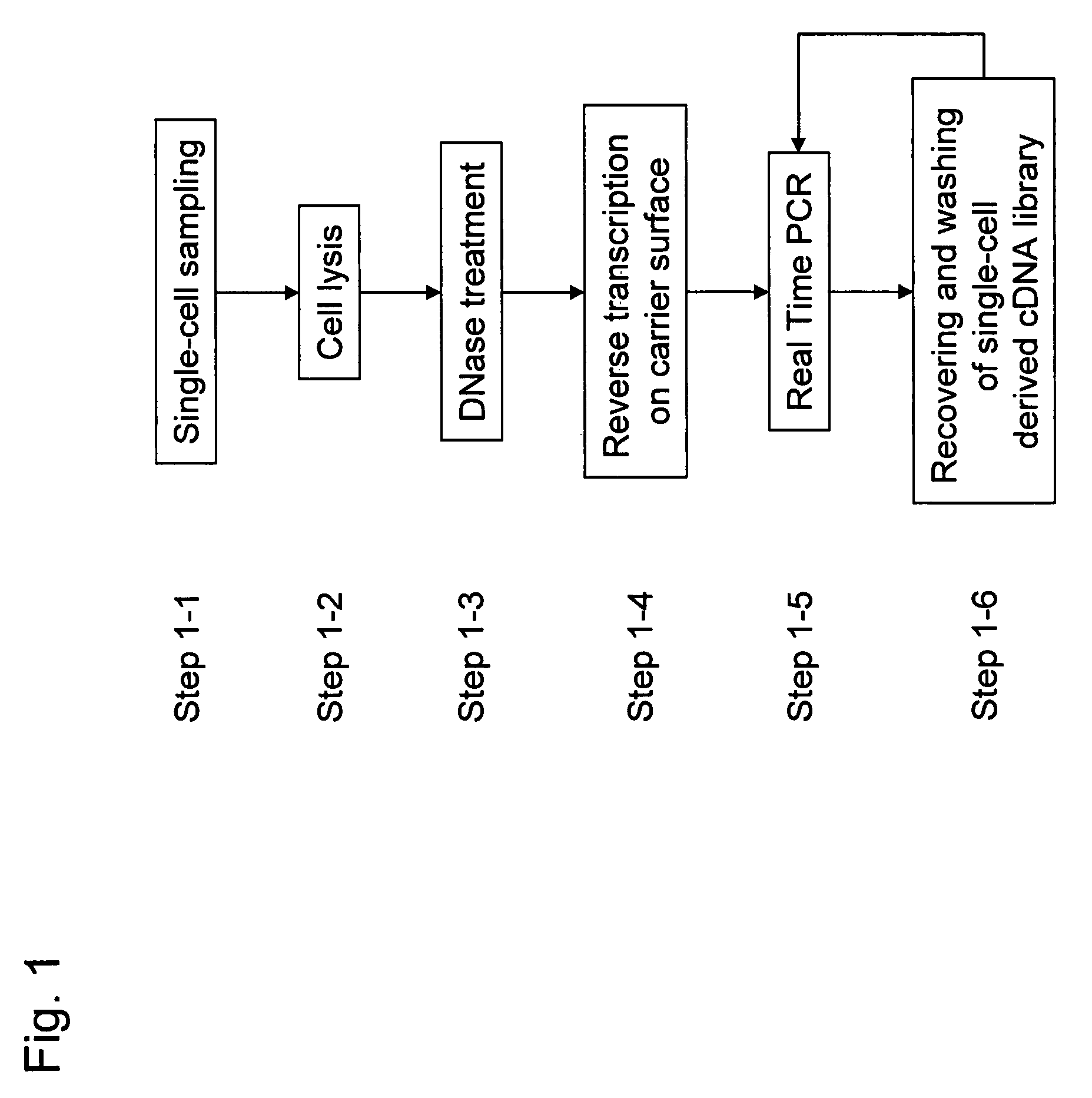
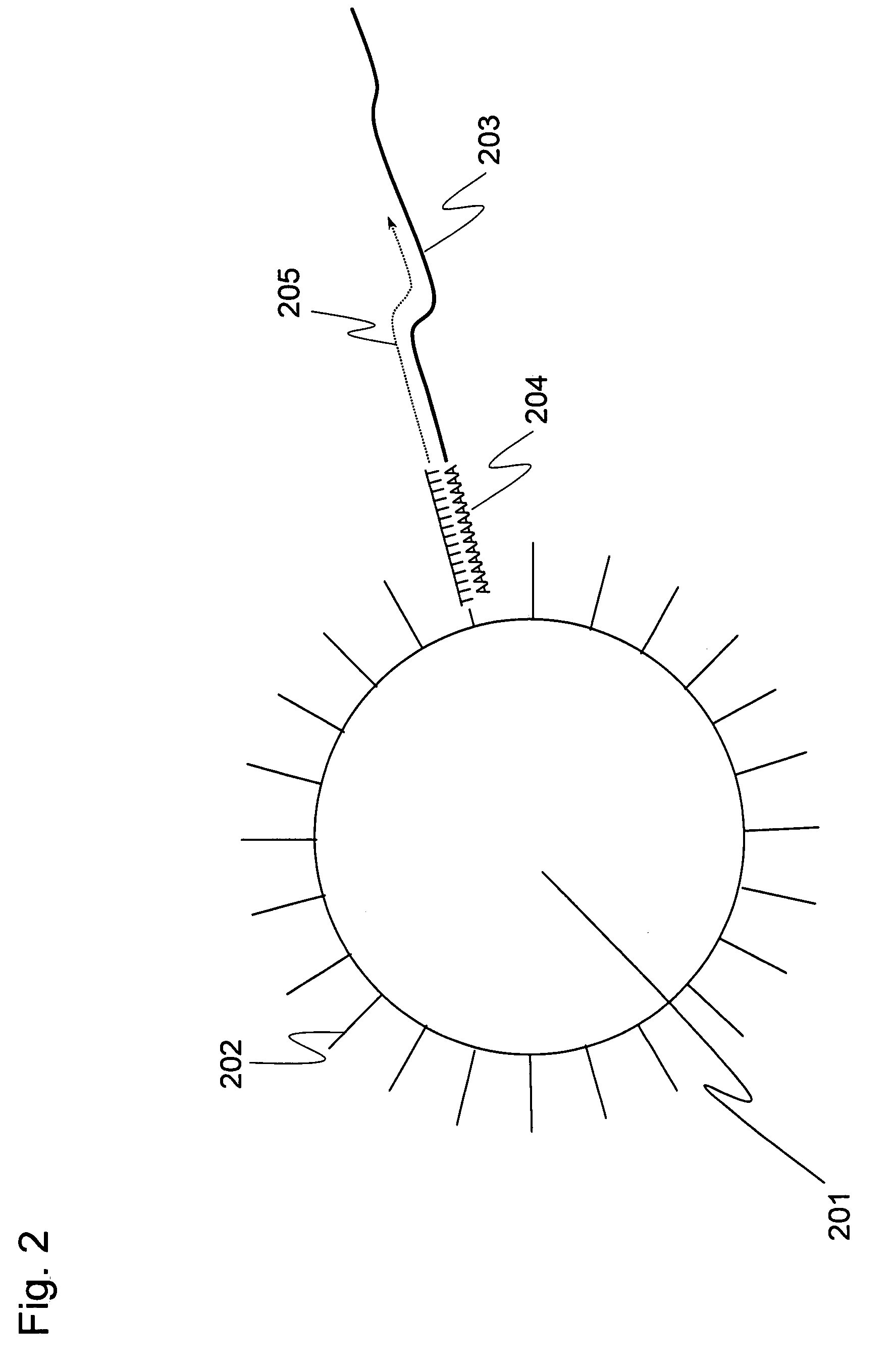
Methods for quantitative cDNA analysis in single-cell
ActiveUS20070281313A1High sensitivityPurification of smallMicrobiological testing/measurementDNA preparationCDNA libraryLysis
It is an object to provide a method of suitably analyzing the amount of gene expression of a single-cell.A method of detecting a nucleic acid comprisinga step of sampling a single-cell from a sample containing at least a single-cell,a cell lysis step of lysing cell membrane of the sampled single-cell and extracting nucleic acids from the cell,a DNase treatment step of degrading DNA of the extracted nucleic acids with DNase,a step of hybridizing mRNA of the total RNA contained in the single-cell with oligo (dT) fixed onto a carrier,a step of performing reverse transcription of the mRNA hybridized with the oligo (dT) to fix cDNA derived from the single-cell onto the carrier, thereby preparing a single-cell derived cDNA library fixed onto a carrier, anda step of amplifying cDNA fixed onto the carrier and simultaneously detecting an amplification amount of the cDNA.
Owner:HITACHI LTD
Methods for quantitative cDNA analysis in single-cell
ActiveUS8802367B2Purification of smallSmall recoverySugar derivativesMicrobiological testing/measurementCDNA libraryLysis
It is an object to provide a method of suitably analyzing the amount of gene expression of a single-cell.A method of detecting a nucleic acid comprisinga step of sampling a single-cell from a sample containing at least a single-cell,a cell lysis step of lysing cell membrane of the sampled single-cell and extracting nucleic acids from the cell,a DNase treatment step of degrading DNA of the extracted nucleic acids with DNase,a step of hybridizing mRNA of the total RNA contained in the single-cell with oligo (dT) fixed onto a carrier,a step of performing reverse transcription of the mRNA hybridized with the oligo (dT) to fix cDNA derived from the single-cell onto the carrier, thereby preparing a single-cell derived cDNA library fixed onto a carrier, anda step of amplifying cDNA fixed onto the carrier and simultaneously detecting an amplification amount of the cDNA.
Owner:HITACHI LTD
Compositions and methods for negative selection of non-desired nucleic acid sequences
ActiveUS20150299767A1Decrease abundance ratioReduce the ratioNucleotide librariesMicrobiological testing/measurementTotal rnaRibosomal RNA
The present invention provides methods, compositions and kits for the generation of next generation sequencing (NGS) libraries in which non-desired nucleic acid sequences have been depleted or substantially reduced. The methods, compositions and kits provided herein are useful, for example, for the production of libraries from total RNA with reduced ribosomal RNA and for the reduction of common mRNA species in expression profiling from mixed samples where the mRNAs of interest are present at low levels. The methods of the invention can be employed for the elimination of non-desired nucleic acid sequences in a sequence-specific manner, and consequently, for the enrichment of nucleic acid sequences of interest in a nucleic acid library.
Owner:NUGEN TECH
Methods for removing artifact from biological profiles
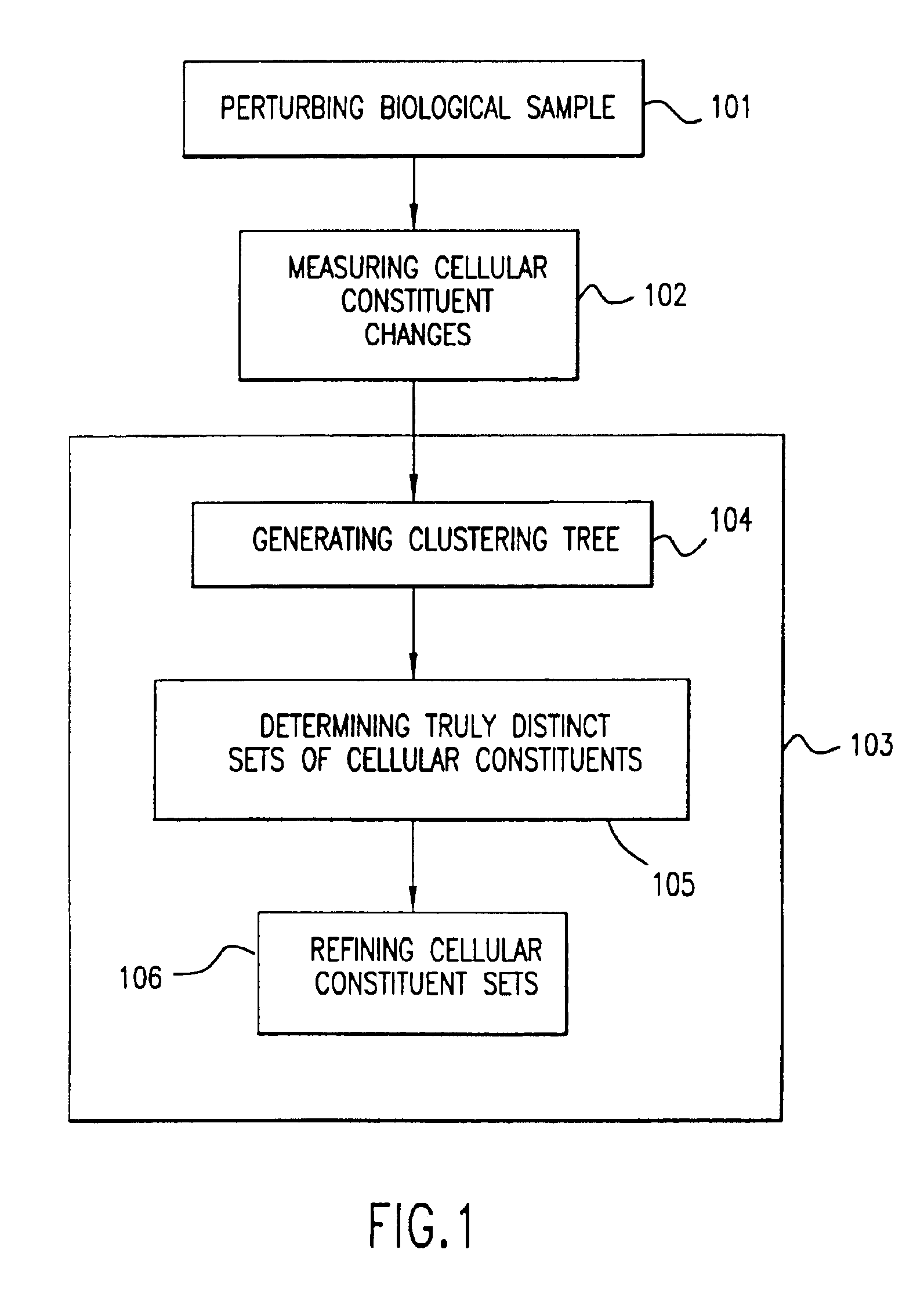
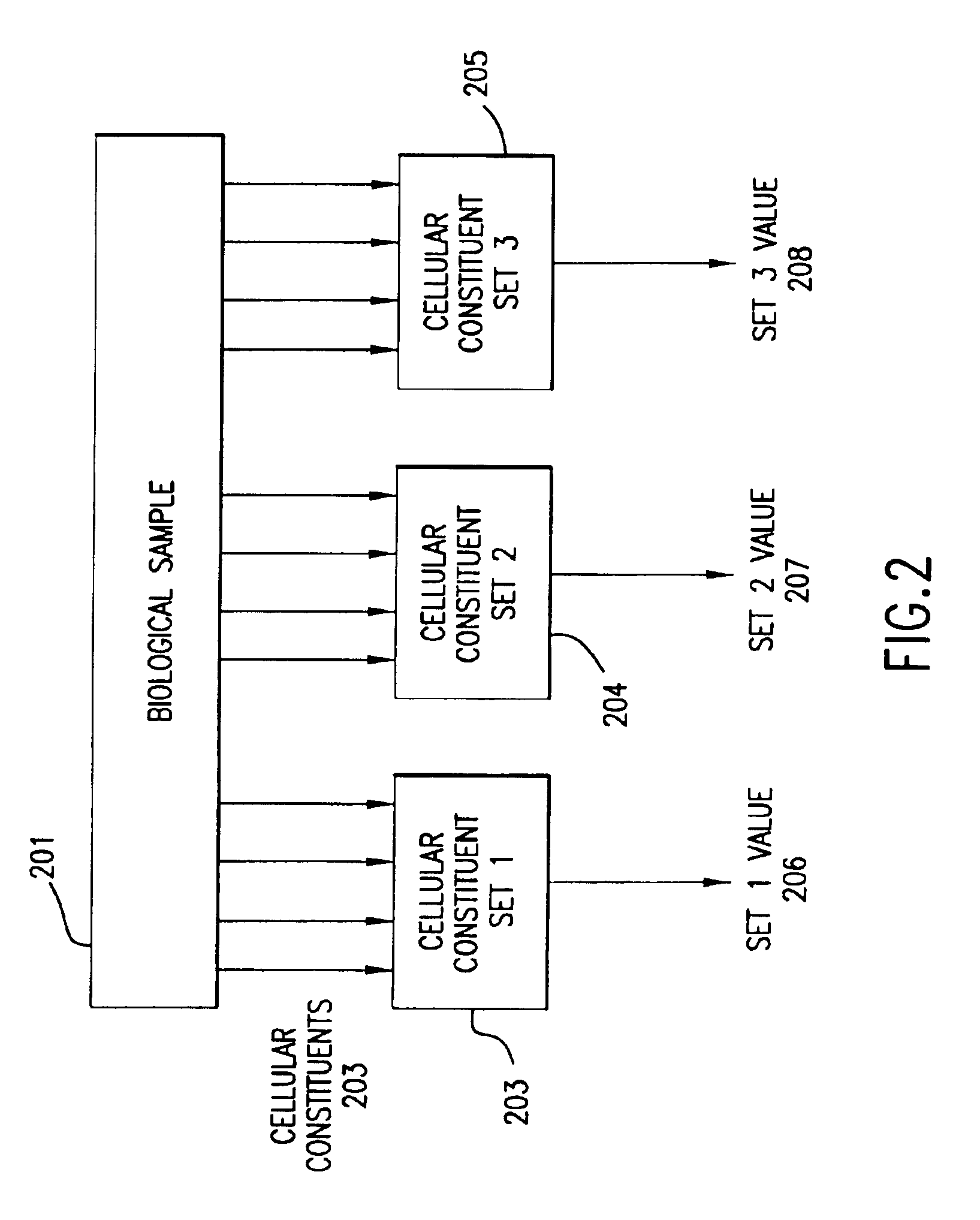
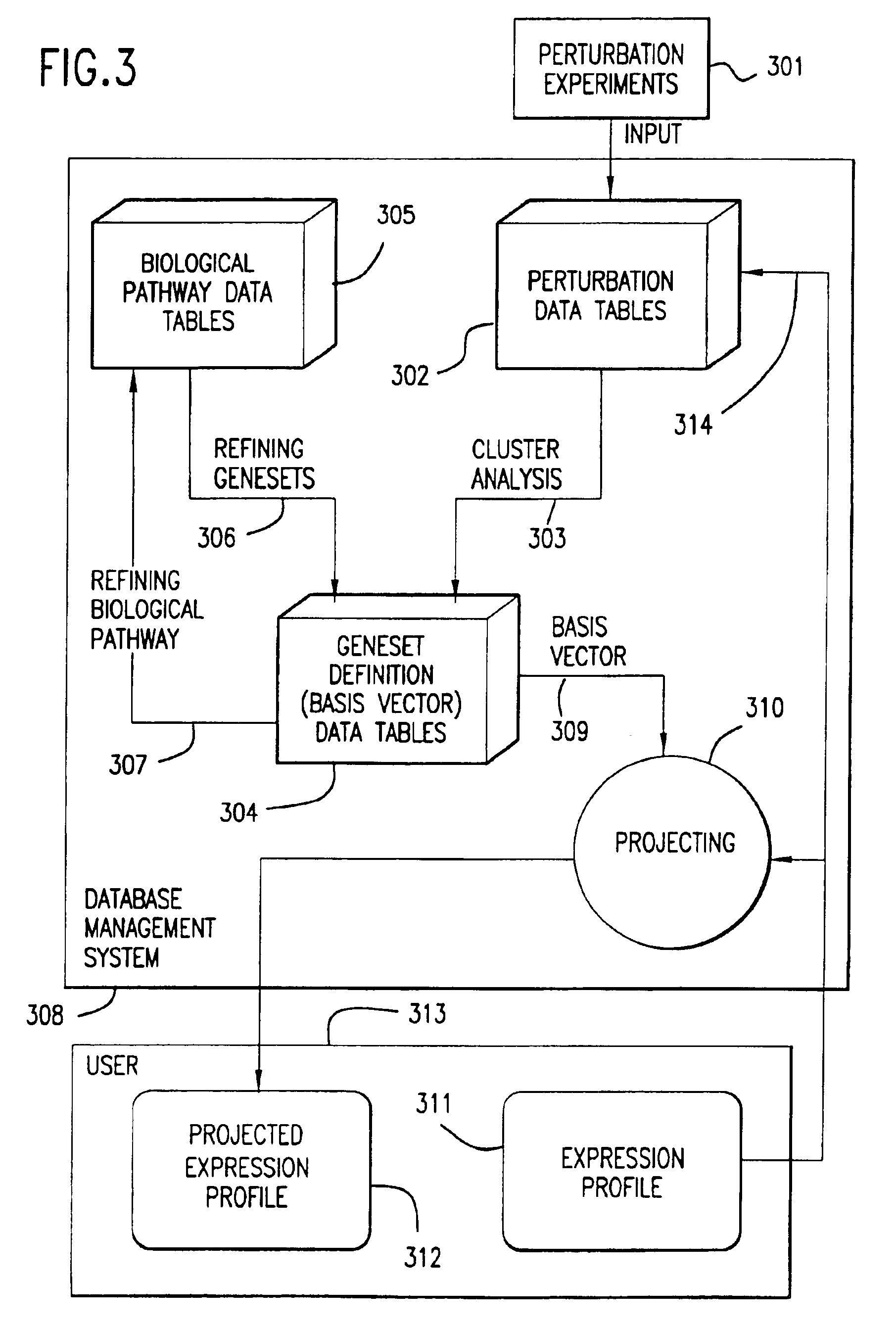
InactiveUS6950752B1Bioreactor/fermenter combinationsBiological substance pretreatmentsCellular componentTotal rna
The invention provides methods for removing unwanted response components (i.e., “artifacts”) from a measured biological profile comprising measurements of a plurality of cellular constituents of a cell or organism in response to a perturbation. The methods involve subtracting from the measured biological profile one or more artifact patterns, each of which comprises measurements of changes in cellular constituents as a result of deviation of one or more experimental variables, such as cell culture density and temperature, hybridization temperature, as well as concentrations of total RNA and / or hybridization reagents, from desired values.
Owner:ROSETTA INPHARMATICS LLC +1
Isothermal nucleic acid amplification methods and compositions
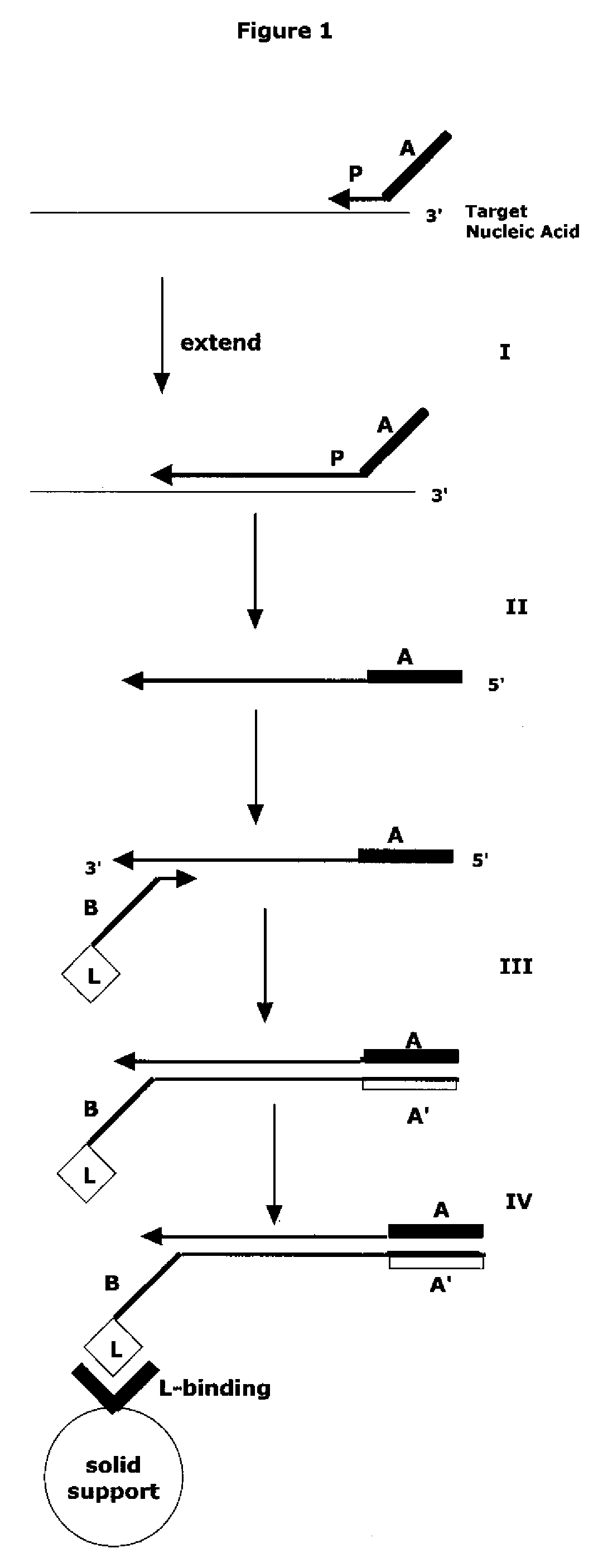
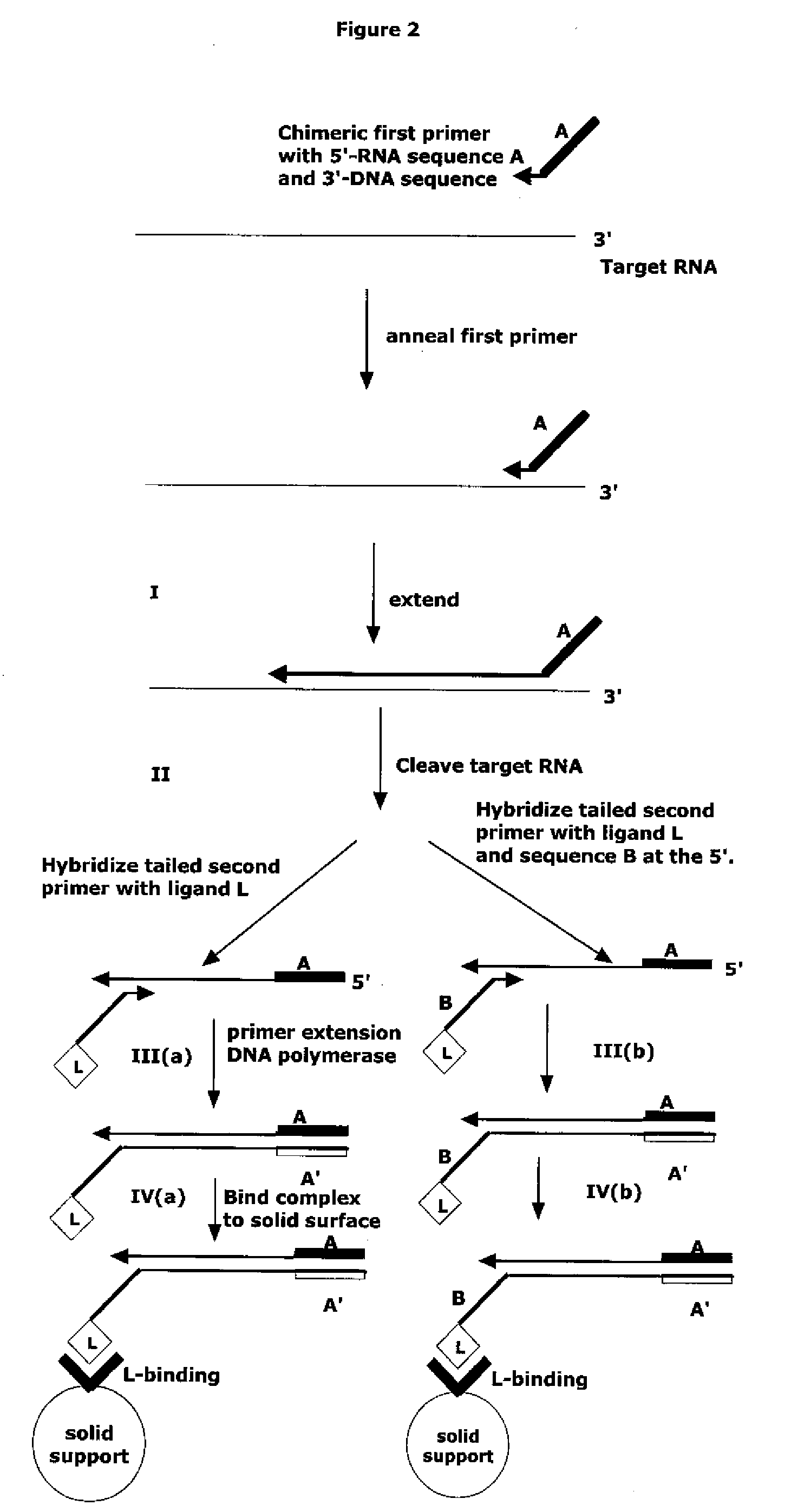
Methods and compositions are provided related to the amplification of target polynucleotide sequences as well as total RNA and total DNA amplification. In some embodiments, the methods and compositions also allow for the immobilization and capture of target polynucleotides with defined 3′ and or 5′ sequences to solid surfaces. The polynucleotides attached to the solid surfaces can be amplified or eluted for downstream processing. In some cases, nucleotides attached to solid surfaces can be used for high throughput sequencing of nucleotide sequences related to target DNA or target RNA.
Owner:NUGEN TECH
Method and kit for preparing a target RNA depleted sample
InactiveUS20150275267A1Simple methodDepleting unwanted target RNAPeptide librariesMicrobiological testing/measurementTotal rnaNon-coding RNA
The present invention provides a method of preparing a target RNA depleted composition from an initial RNA containing composition, comprising a) contacting the initial RNA containing composition with one or more groups of probe molecules, wherein a group of probe molecules has the following characteristics: i) the group comprises two or more different probe molecules having a length of 100 nt or less; ii) the probe molecules comprised in said group are complementary to a target region of a target RNA; iii) when hybridized to said target region, the two or more different probe molecules are located adjacent to each other in the formed double-stranded hybrid; and generating a double-stranded hybrid between the target RNA and the probe molecules; b) capturing the double-stranded hybrid by using a binding agent which binds the double-stranded hybrid, thereby forming a hybrid / binding agent complex; c) separating the hybrid / binding agent complexes from the composition, thereby providing a target RNA depleted composition. By combining hybrid capturing with a unique probe design, an improved depletion method is provided which effectively and specifically removes unwanted target RNA such as ribosomal RNA (rRNA) from total RNA, while ensuring recovery of mRNA and noncoding RNA from various species, including human, mouse, and rat. By improving the ratio of useful data, decreasing bias, and preserving non-coding RNA species, the method provides high-quality RNA that is especially suited for next-generation sequencing (NGS) applications. By integrating said depletion method in common sequencing applications, in particular NGS applications such as transcriptome sequencing, improved methods for sequencing RNA molecules are provided.
Owner:QIAGEN GMBH
Anti-tumor T cell as well as preparation method and anti-tumor drug thereof
InactiveCN104630145AMammal material medical ingredientsBlood/immune system cellsAbnormal tissue growthDendritic cell
The invention provides a method for preparing an anti-tumor T cell in vitro through DC (dendritic cell, namely antigen presenting cell), wherein the anti-tumor T cell is a tumor total RNA-loaded DC amplification and enrichment anti-tumor T cell. The invention also provides a preparation method of the anti-tumor T cell and an anti-tumor drug or inoculation vaccine taking the anti-tumor T cell as an active ingredient. According to the invention, a specific anti-tumor T cell is amplified in vitro by taking the tumor total RNA-loaded DC as a platform, a small amount of tumor total RNA is amplified to an enough amount in vitro so as to realize clinical application, and the in-vivo anti-tumor effect in an IC (intracranial) tumor-bearing model is evaluated, including how the mature state of DC and the cell factor combination adjust the differentiation of the anti-tumor T cell as well as the synergistic anti-tumor effect thereof in a preclinical experiment model.
Owner:深圳市中美康士生物科技有限公司
Plant flavonoid synthesis regulation gene and its application
Owner:WUHAN BOTANICAL GARDEN CHINESE ACAD OF SCI
Extraction method of soil microbe genome DNA and total RNA
The invention provides a method for effectively extracting crop rhizosphere soil microbe genome DNA and total RNA, which comprises the following steps: firstly, removing humus and other impurities which severely disturb nucleic acid extraction in soil samples by aluminum sulfate; further crushing soil microbe cells by glass beads; adding sodium dodecyl sulfonate (SDS) and LiCl solutions for cracking cells and dissociating nucleic acids; adding the mixture of phenol, chloroform and isoamylol for extracting; and then, depositing the extraction solution by isopropanol and sodium acetate to finally acquire the soil microbe metagenome DNA and total RNA. The method is suitable for extraction of the DNA and total RNA of soil in different regions. The invention establishes an extraction technology which has the advantages of simple and easy operation processes and higher purity of the prepared sample, and can provide an important basis for the research of soil metagenomics.
Owner:FUJIAN AGRI & FORESTRY UNIV
Method for rapidly identifying rice black-streaked dwarf virus and southern rice black-streaked dwarf virus
InactiveCN101691614ARapid identification methodThe identification method is simpleMicrobiological testing/measurementMicroorganism based processesRice black-streaked dwarf virusTotal rna
The invention provides a method for rapidly identifying the rice black-streaked dwarf virus and southern rice black-streaked dwarf virus, belonging to the field of plant protection. Three primers are designed according to the nucleotide sequence of the rice black-streaked dwarf virus and southern rice black-streaked dwarf virus, which are respectively [A] RB1_S9_F, [B] RB2_S9_F and [C] RB_S9_R, wherein the combination of A and C detects the rice black-streaked dwarf virus and the combination of B and C detects the southern rice black-streaked dwarf virus. After the total RNA is extracted fromthe samples, the two viruses of the rice black-streaked dwarf virus and the southern rice black-streaked dwarf virus can be detected by only one RT-PCR, thus realizing the identification of two viruses. Compared with the traditional method in which one RT-PCR can only detect one virus, the invention greatly reduces cost, reduces time, and is a detection method which is special, sensitive, economic and convenient.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Method of simultaneously extracting microorganism macrogenome DNA and total DNA from sea precipitate
A method for simultaneously extracting the macrogenomic DNA and total RNA of microbe from the sea sediment includes such steps as treating the sediment by modifying solution, adding sand, oscillating, adding proteinase and lysozyme, cracking adding SDS and PVPP solution, cracking further, adding extrant, extracting, depositing in isopropanol and purifying by nucleic acid adsortion resin.
Owner:THIRD INST OF OCEANOGRAPHY STATE OCEANIC ADMINISTATION
Detection and application of new molecular marker hsa-circ-0001017 of gastric cancer
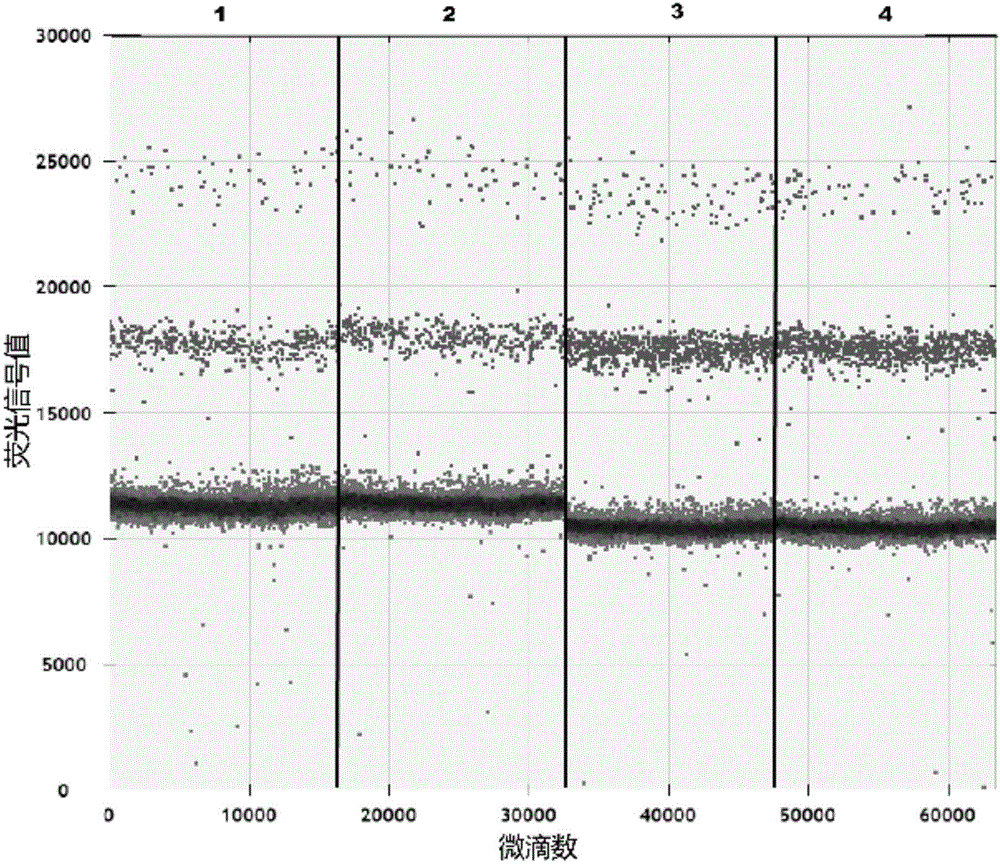
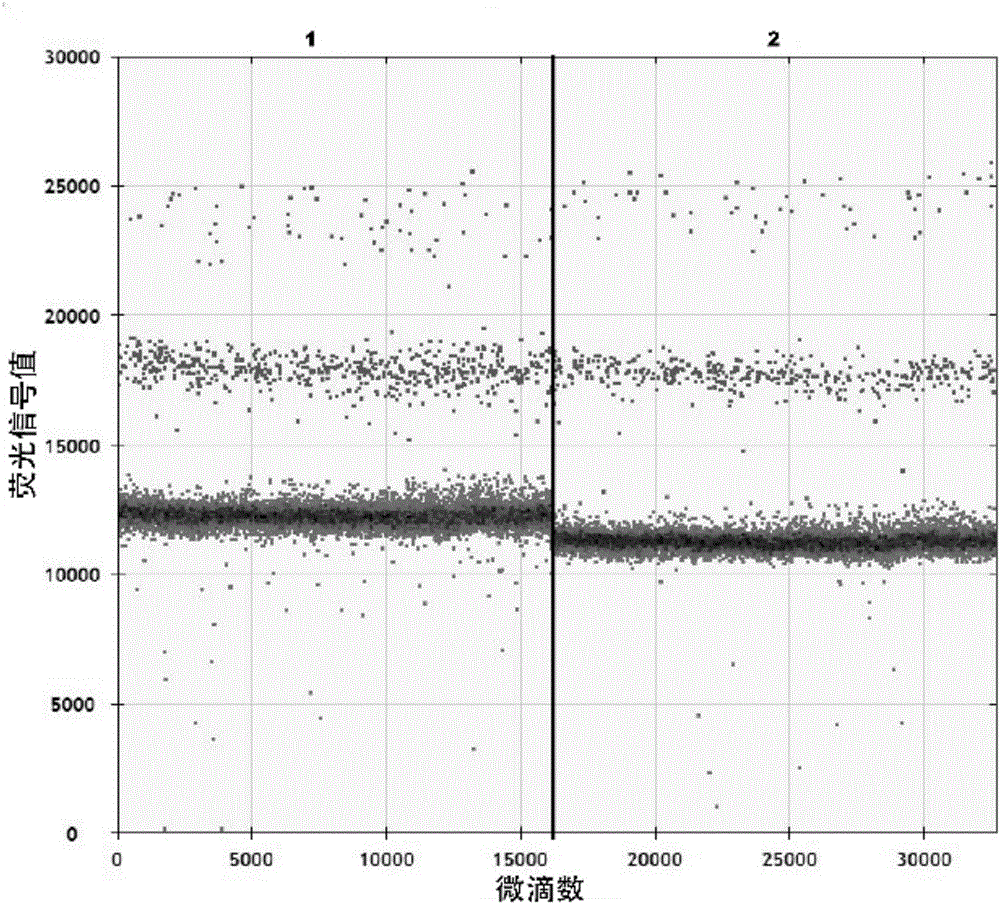
ActiveCN106591428AReduce yieldHigh yieldMicrobiological testing/measurementDNA/RNA fragmentationHousekeeping geneFluorescence
The invention relates to a cyclic RNA molecular marker for diagnosis of gastric cancer, the cyclic RNA molecular marker is characterized in that the cyclic RNA is hsa-circ-0001017, the invention also provides a method for detection of the cyclic RNA molecular marker in plasma, and the method comprises the following steps: (1) collecting blood, and extracting total RNA in the plasma; (2) performing reverse transcription of the total RNA into cDNA; (3) performing droplet digital PCR detection of a cDNA solution of the step (2) by use of specific amplification back-to-back primers and amplification upstream and downstream primers of housekeeping gene GAPDH, after the completion of the reaction, detecting fluorescence signal values of all droplets, setting a threshold, and determining whether the droplets include the cyclic RNA or the housekeeping gene GAPDH, wherein the droplets higher than the threshold are positive droplets, and the droplets below the threshold are negative droplets; and (4) counting the number of the positive droplets, and calculating the copy number of the hsa-circ-0001017 and the housekeeping gene GAPDH in the plasma for quantitative detection of the hsa-circ-0001017 and the housekeeping gene GAPDH in the plasma. Compared with the prior art, the advantages are that the hsa-circ-0001017 can be specifically expressed in plasma in patients with gastric cancer, and can be used as a new molecular marker for diagnosis of the gastric cancer.
Owner:NINGBO UNIV
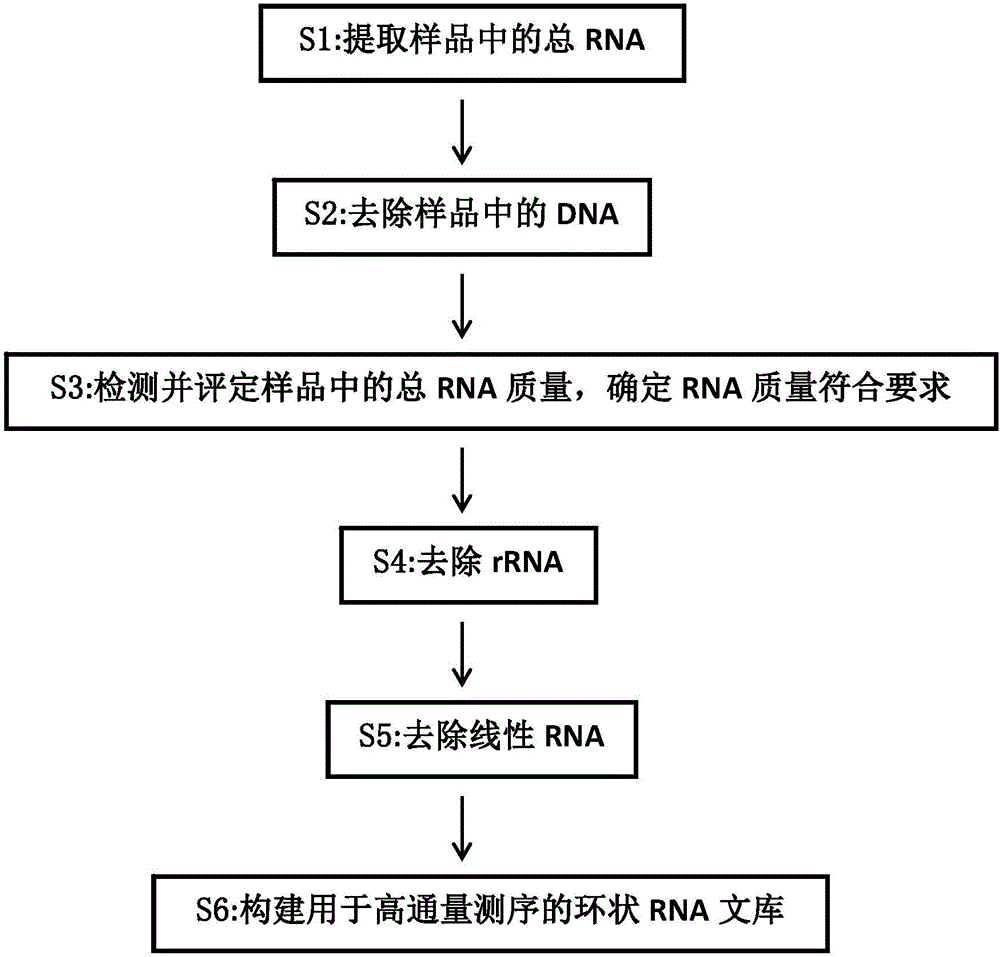
Establishing method of circular RNA high-throughput sequencing library and kit thereof
ActiveCN106801050AImprove detection efficiencyGood data repeatabilityMicrobiological testing/measurementLibrary creationTotal rnaRepeatability
The invention discloses an establishing method of a circular RNA high-throughput sequencing library and a kit thereof. The establishing method sequentially comprises the following steps: (S1) extracting total RNA from a sample; (S2) removing DNA in the sample; (S3) detecting and evaluating the quality of the total RNA in the sample, and determining that the RNA quality meets a requirement; (S4) removing rRNA; (S5) removing linear RNA; (S6) constructing the circular RNA library for high-throughput sequencing. The establishing method of the circular RNA high-throughput sequencing library, provided by the invention, is high in efficiency, stable, low in residual ratio of the rRNA, high in circular RNA detection efficiency, good in data reproducibility, and high in sequencing result verifying success rate, and is especially applicable to an FFPE tissue and other samples with poor RNA quality.
Owner:GUANGZHOU FOREVERGEN BIOTECH CO LTD +1
Kit for detecting 7 genetic markers of peripheral blood in early diagnosis of nasopharyngeal darcinoma
ActiveCN101956014AIncreased sensitivityImprove featuresMicrobiological testing/measurementFluorescence/phosphorescenceRNA extractionTotal rna
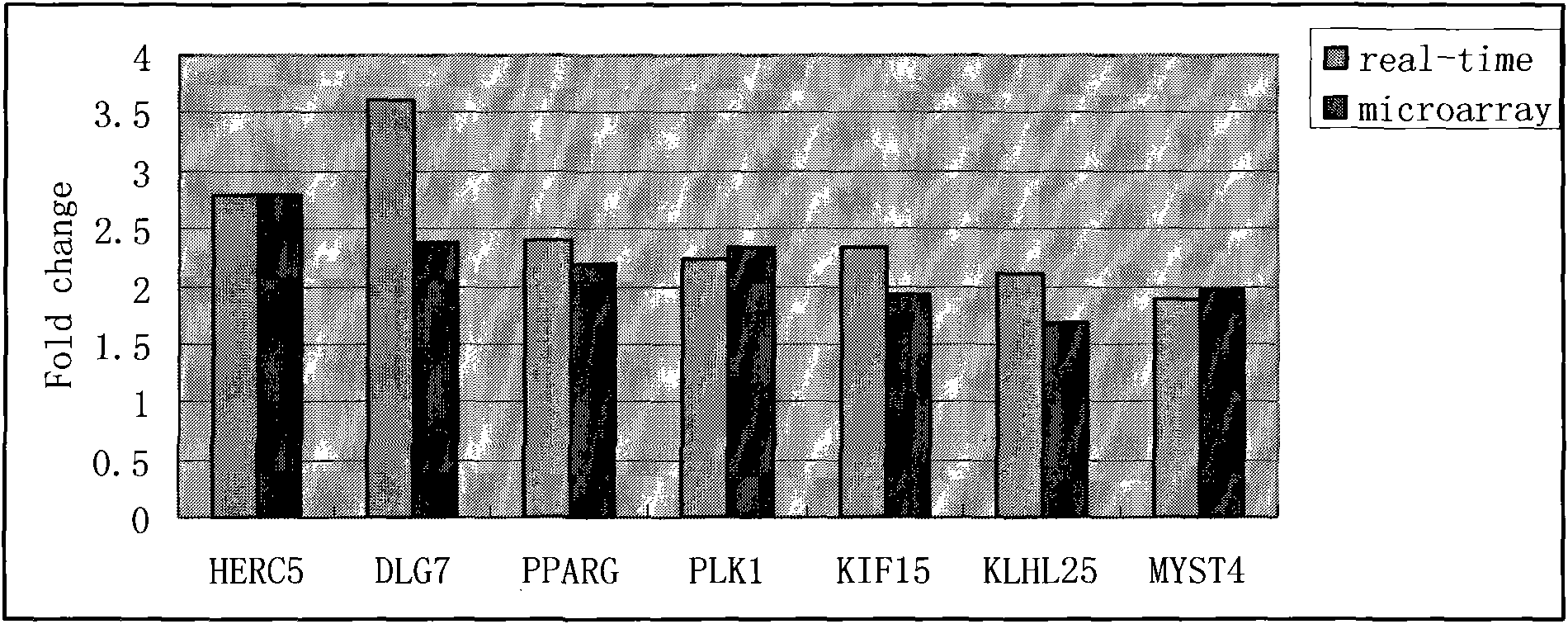
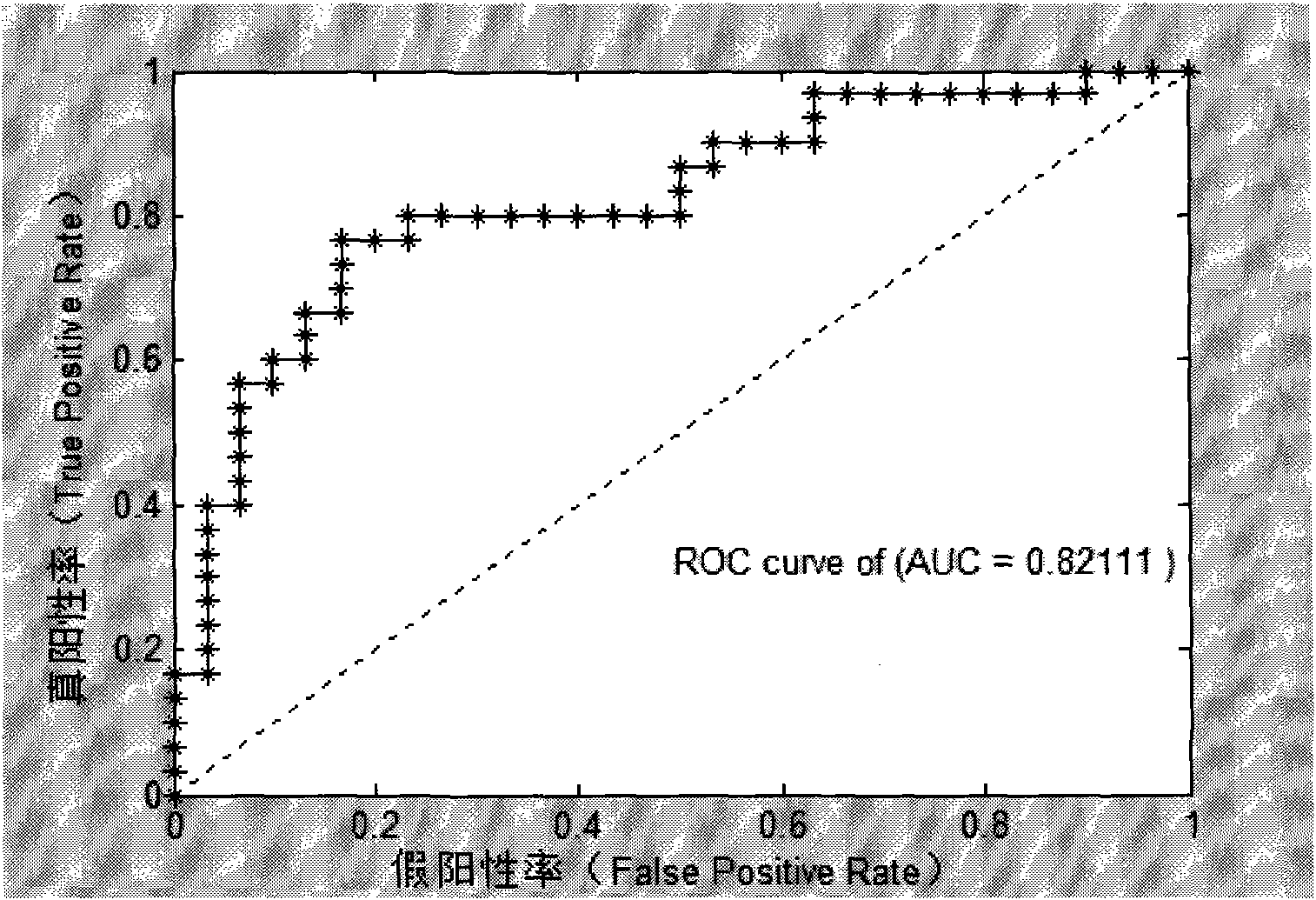
The invention provides a kit for detecting 7 genetic markers of peripheral blood in early diagnosis of nasopharyngeal darcinoma. The kit comprises a peripheral blood total RNA extracting reagent, RT-PCR reaction liquid, time fluorescence quantitative RT-PCR reaction liquid and an SVM nasopharyngeal darcinoma diagnosis model, wherein the time fluorescence quantitative RT-PCR reaction liquid comprises primers of which the sequences are shown as SEQ IDNO:1-16; the primer sequences are SYBR Green fluorescence quantitative RT-PCR detection primers of the 7 genetic markers and reference gene GAPD; and the 7 genetic markers are HERC5, DLG7, PPARG, PLK1, KIF15, KLHL25 and MYST4. The invention also provides a using method for the kit. The kit has the advantages of high sensitivity, specificity and convenience, and can be used as aided diagnosis and early diagnosis technology for the nasopharyngeal darcinoma.
Owner:SUN YAT SEN UNIV
Method for Archiving and Clonal Expansion
The present method provides methods, libraries, and kits related to the archiving and clonal expansion of sequences related to target polynucleotide sequences. The method allow for the attachment of polynucleotides with defined 3′ and or 5′ sequences to solid surfaces. The polynucleotides attached to the solid substrates can be stored or archived as libraries and can subsequently be retrieved for analysis, for example by clonal expansion. In some embodiments, nucleotides attached to solid surfaces can be used for sequencing of nucleotide sequences related to target RNA or target RNA. The methods are applicable to total RNA and / or total DNA analysis.
Owner:NUGEN TECH
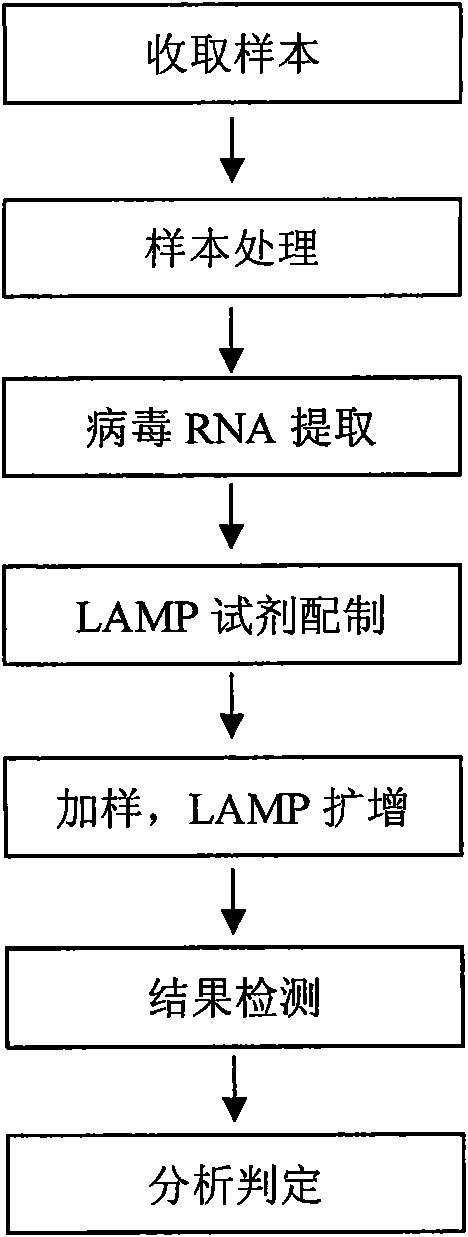
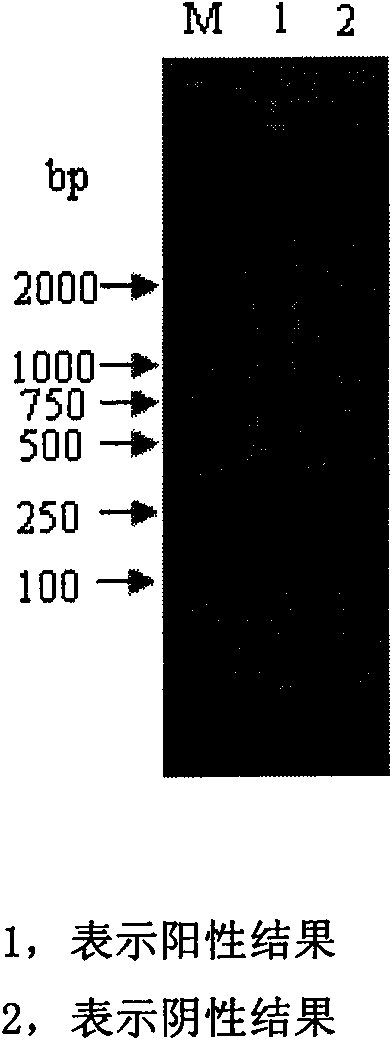
Method for detecting tobacco virus using reverse transcription loop-mediated isothermal amplification technique
InactiveCN102286637AImprove detection efficiencyReduce testing costsMicrobiological testing/measurementFluorescence/phosphorescenceNicotiana tabacumTobacco mosaic virus
The invention relates to a method for detecting tobacco viruses by a reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) technology, which comprises the steps of tobacco virus total ribonucleic acid (RNA) extraction, RT-LAMP reaction, electrophoresis detection and virus type determination, wherein tobacco viruses are selected from two or more than two kinds of viruses from cucumber mosaic viruses (CMV), potato viruses Y (PVY), tobacco etch viruses (TEV), tobacco mosaic viruses (TMV) and tobacco vein banding mosaic viruses (TVBMV). LAMP primers are designed according to a case protein gene conserved region of the tobacco viruses, and a fast, sensitive and high-specificity method is built for detecting various main tobacco viruses by the one-step RT-LAMP technology. A newmeasure is provided for the fast detection of the tobacco viruses.
Owner:SHAANXI TOBACCO RES INST +1
Method of synchronous detecting lily mottle virus, lily symptom less virus and cucumbus flomer leaf virus
InactiveCN1793858AReduce in quantityReduce interactionMaterial analysis by observing effect on chemical indicatorMicrobiological testing/measurementTotal rnaElectrophoresis
A method for synchronously detecting lily mottling virus, lily silent virus and cucumber mosaic virus includes carrying out amplification on three viruses of LMoV, LSV and CMV infected by lily in the same reaction tube and under the same heat circulation condition after detection processes of pick ¿C up total RNA, multiple RT ¿C PCR and electrophoresis; then carrying out separation and detection by means of gel electrophoresis.
Owner:YUNNAN AGRICULTURAL UNIVERSITY
Single-cell RNA (ribonucleic acid) reverse transcription and library construction method
ActiveCN108103055AEfficient and uniform transcriptAvoid pollutionMicrobiological testing/measurementLibrary creationCDNA libraryTotal rna
The invention belongs to the field of transcriptome analysis and relates to a quick single-cell RNA (ribonucleic acid) reverse transcription and library construction method. According to the method, 20-500ng high-quality full-length double-chain cDNA (complementary deoxyribonucleic acid) is amplified by taking 1-2000 cells or 10pg-20ng extracted eukaryote total RNA as an initial, and a high-quality cDNA library meeting downstream analysis requirements is obtained. The method effectively avoids 3' preference and genome DNA contamination in a cDNA synthesis process; an expression quantitation molecule label can assist in gene expression quantity calculation; and at the same time, the expression quantitation molecule label can also keep chain source information during complete amplification of RNA sequence information. The method can achieve a reverse transcription and amplification library construction success rate of above 95%; the cDNA library can be connected with an Illumina main stream sequencing platform; lower computer data (5M Reads) can detect gene expression of above 90%; the gene expression consistency exceeds 90%; amplification has no obvious bias; and a required sample input amount is smaller.
Owner:XUKANG MEDICAL SCI & TECH (SUZHOU) CO LTD
Method for constructing transcriptome library
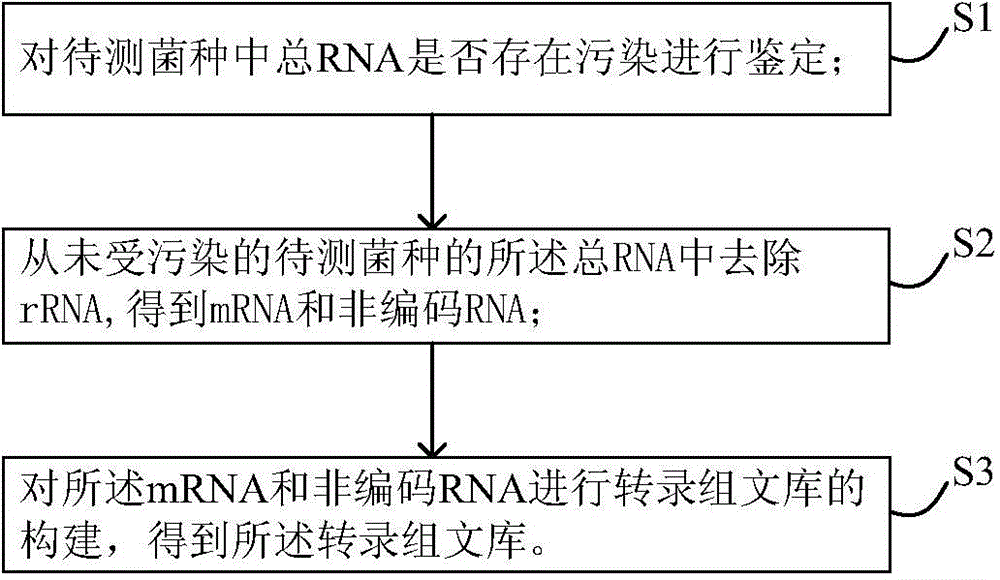
InactiveCN104630206AAvoid wasting human and financial resourcesReduce adverse effectsMicrobiological testing/measurementLibrary creationTotal rnaNon-coding RNA
The invention discloses a method for constructing a transcriptome library. The construction method comprises the following steps: S1, verifying whether total RNA in a strain to be measured is polluted; S2, removing rRNA from the total RNA of the unpolluted strain to be measured to obtain mRNA and non-coding RNA; and S3, using the mRNA and the non-coding RNA to construct the transcriptome library to obtain the transcriptome library. In the method, a step of controlling the quality of the total RNA is added to ensure that the RNA sample is not polluted, and a subsequent transcriptome library construction step is carried out to avoid the waste of manpower and financial resources caused by the polluted RNA and perfect the problem of low comparison rate of subsequent sequencing data. Few samples are needed in the verification step, the operation is simple, a result is obtained quickly and accurately, and the verification step has high throughput capacity, so that the verification step can be widely used in the transcriptome library construction step to verify the total RNA to the strain to be measured.
Owner:BEIJING NOVOGENE TECH CO LTD
Multiple RT-PCR (reverse transcription-polymerase chain reaction) detection method for SPVD (sweet potato virus disease)
ActiveCN102108419AEffectively distinguish strain typesTimely warning of the degree of dangerMicrobiological testing/measurementForward primerDisease
Owner:INST OF PLANT PROTECTION HENAN ACAD OF AGRI SCI
Extraction method of apostichopus japonicus body-wall total RNA
InactiveCN101864414AMeet Gene Expression AnalysisFulfil requirementsDNA preparationWater bathsTotal rna
The invention discloses a high-extraction-purity, good-integrity and high-yield extraction method of apostichopus japonicus body-wall total RNA (Ribonucleic Acid). The method comprises the following steps of: quickly freezing apostichopus japonicus body-wall tissue in liquid nitrogen; grinding and putting the frozen tissue in lysate to homogenate, centrifugate and take supernatant fluid; adding chloroform to the supernatant fluid and centrifugating to take the supernatant fluid to another centrifuge tube; then, adding a high-salt solution and isopropyl alcohol and filtering precipitation with ethanol; dissolving the precipitation with DEPC (diethypyrocarbonate) treated water to have constant volume; sequentially adding a DNA enzyme buffer solution without RNA enzymes, DNA enzymes without RNA enzymes and an RNA enzyme inhibitor to the dissolved solution to mix; carrying out a water bath at 37 DEG C to obtain DNA lysate; adding phenol and chloroform in a ratio of 5:1 to the DNA lysate to mix, centrifugate and take supernatant fluid; adding a glycogen solution, a potassium acetate solution and pre-cooling ethanol to the supernatant fluid; mixing and staying over night; centrifugating to discard the supernatant fluid; washing and dying the precipitation with ethanol; dissolving the solution with the DEPC treated water to have constant volume of 20 mu l; and storing at 80 DEG C below zero.
Owner:DALIAN OCEAN UNIV
H3 subtype flu quick-detecting type classifying method based on RT-LAMP technology
InactiveCN101608242AReduced Pollution ChancesShort amplification timeMicrobiological testing/measurementMicroorganism based processesWater bathsSequence analysis
The invention discloses a H3 subtype flu quick-detecting type classifying method based on an RT-LAMP technique. The method comprises the following steps: firstly, analyzing the HA gene sequence of the H3 subtype flu virus in a flu virus gene group sequence database, designing highly special and conserved RT-LAMP special primers and six primers corresponding to eight areas of target genes, wherein four primers are FIP, BIP, F3 and B3, and the other two primers are loop primers LP1 and LP2; secondly, treating a sample to be detected in advance, and extracting the total RNA of the sample to be used as a reaction template; thirdly, configuring an RT-LAMP reaction system; fourthly, mixing the RNA template into the RT-LAMP reaction system, arranging the RT-LAMP reaction system in a water bath, and carrying out RT-LAM enlargement at constant temperature; and fifthly, identifying the enlarged reaction product. The result shows that the reaction product contains H3 subtype flu virus when the reaction product is positive; and the reaction product is H3 subtype flu virus when the reaction product is negative.
Owner:TAIZHOU QINHELI BIO TECH
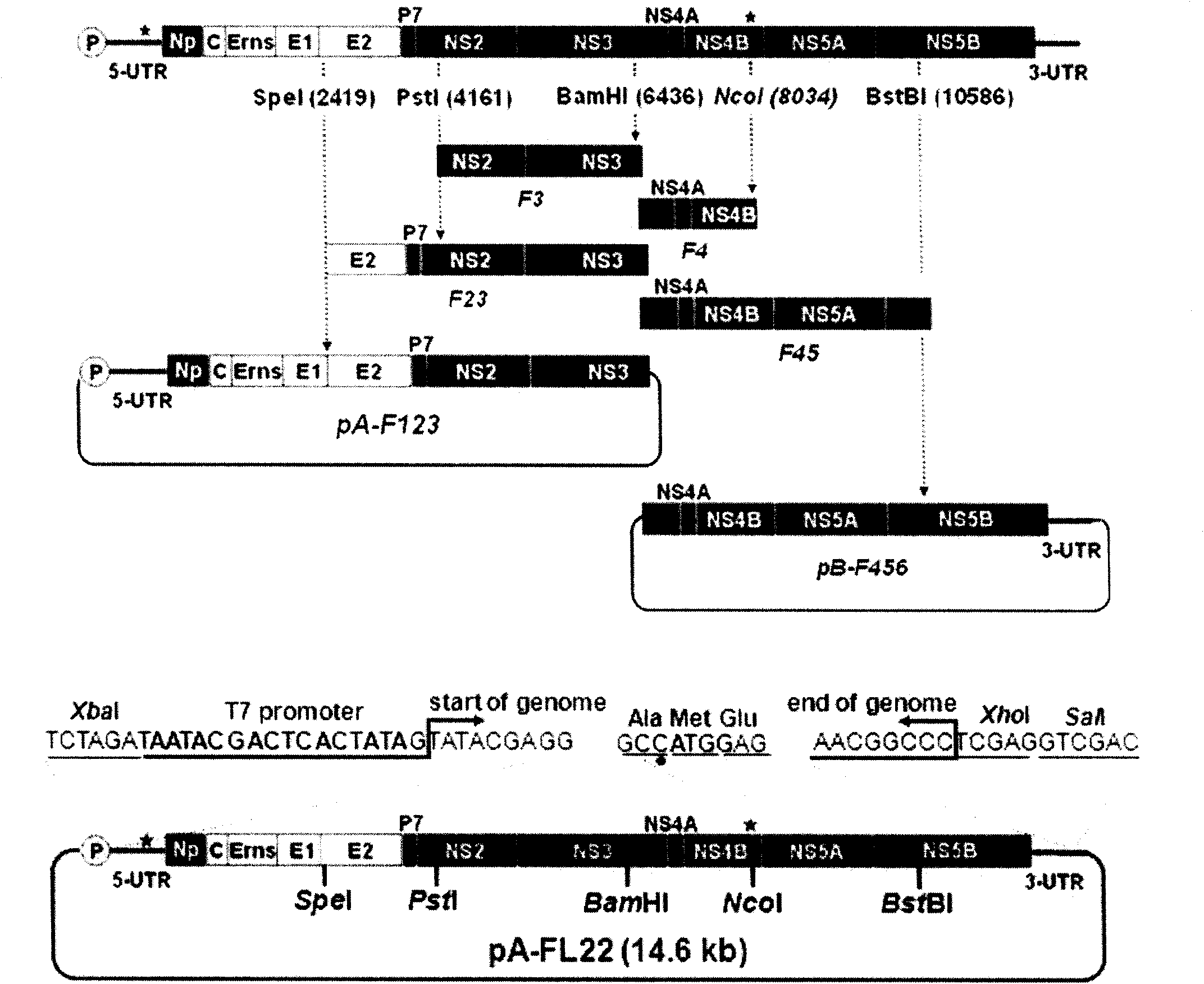
Method for constructing hog-cholera virus infectious cDNA carrier having molecule mark
ActiveCN101864445AFermentationVector-based foreign material introductionTotal rnaTechnological system
The invention relates to a virus antibody research and provides a method for constructing a hog-cholera virus infectious cDNA carrier having a molecule mark. The method comprises the following steps of: (1) diluting a hog-cholera live vaccine to 1 portion per milliliter as the experimental material, after extracting the head RNA, amplifying 6 corresponding cDNA fragments through RT-PCR section according to designed 6 pairs of primers, (2) leading in the corresponding molecule marks from F1 and F5 of cDNA fragments, (3) reforming plasmids required by overall length cDNA cloning, and (4) constructing the overall length cDNA carrier. A hog-cholera virus lapinized vaccine C stem infectious cDNA carrier pA-FL22 having a molecule mark can be acquired through the method of the invention. The permissive cells of RNA electro transferred hog-cholera virus acquired by the carrier can save the infectious progeny virus which can stably inherit the molecule mark led in the construction. By using the saving technique system of the infectious cDNA carrier and the progeny virus, the copy stem and the pathopoiesia reason of the virus can be deeply researched and the foundation for developing new marked vaccines can be settled.
Owner:ZHEJIANG UNIV
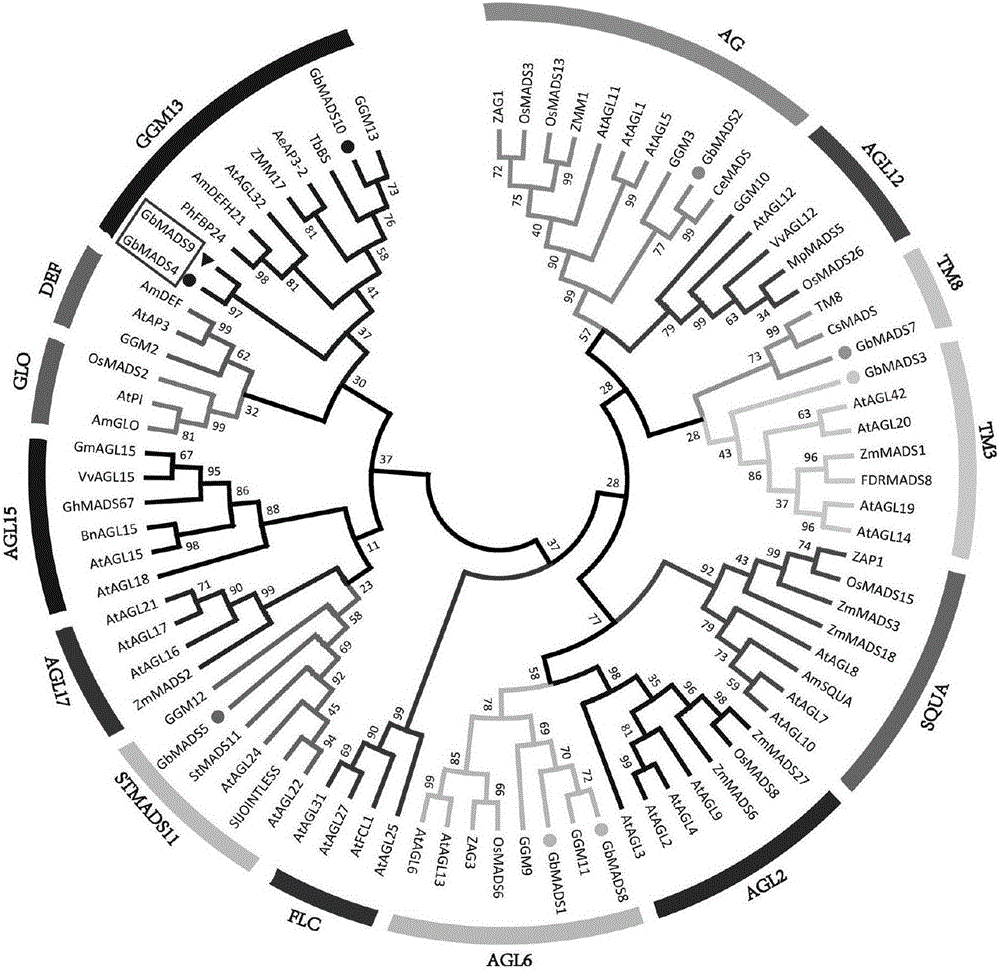
Gingko MADS-box transcription factor gene GbMADS9 for controlling blossoming of plants and encoding protein and application of Gingko MADS-box transcription factor gene GbMADS9
InactiveCN106591322AShortened childhoodImprove tolerancePlant peptidesGenetic engineeringTotal rnaPcr method
The invention provides Gingko MADS-box transcription factor gene GbMADS9 for controlling blossoming of plants; the gene is acquired by subjecting cDNA as a template to PCR (polymerase chain reaction) amplification, wherein the cDNA is acquired by reverse transcription of total RNA of gingko female flowers. The gene has the advantages that the gene GbMADS9 is capable of controlling blossoming of plants and shortening juvenile phase of plants, and under induction of GA3, ABA, salt stress, drought stress and cold damage stress, the overexpressed gene GbMADS9 is capable of enhancing plant tolerance to osmotic stress; the gene is applicable to controlling the blossoming phase of plants.
Owner:YANGTZE UNIVERSITY
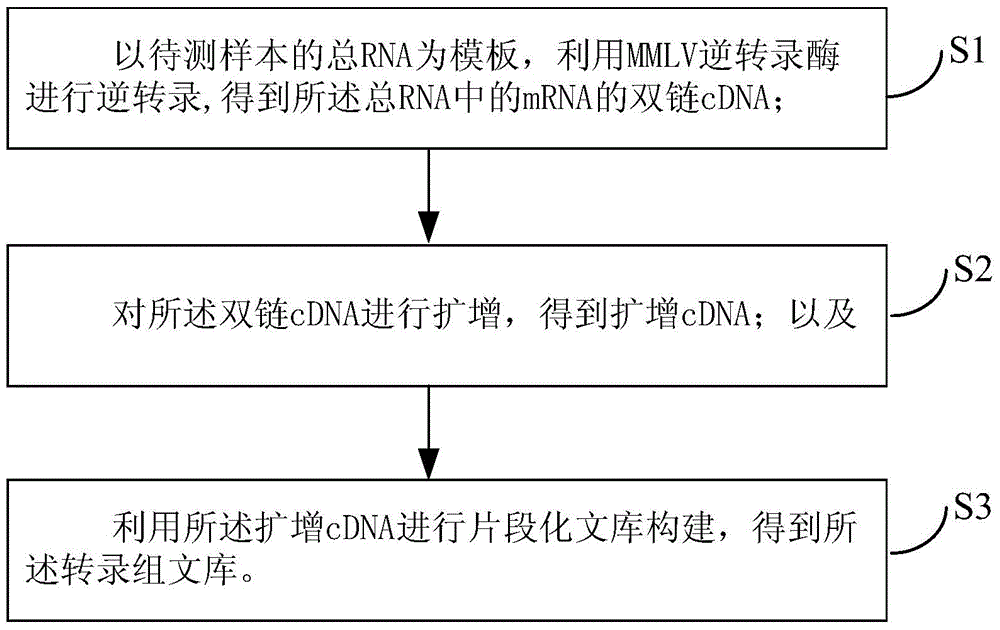
Transcriptome library and construction method thereof
InactiveCN105002569AThe build was successfully implementedImplement the buildNucleotide librariesLibrary creationTotal rnaReverse transcriptase
The invention provides a transcriptome library and a construction method thereof. The construction method comprises the following steps: total RNA of a sample to be detected is adopted as a template; reverse transcription is carried out with MMLV reverse transcriptase, such that double-stranded cDNA of the mRNA in the total RNA is obtained; the double-stranded cDNA is amplified, such that amplified cDNA is obtained; and the amplified cDNA is used in fragmented library construction, such that the transcriptome library is obtained. According to the invention, mRNA in the total RNA is reverse-transcribed into full-length fragment double-stranded cDNA with the MMLV reverse transcriptase, and the double-stranded cDNA is amplified, such that a DNA amount needed by library construction is reached, and subsequent segmented library construction can be realized smoothly. With the method, a complete full-length transcript can be enriched; cDNA can be amplified with high sensitivity and no preference; and the fullness of the transcripts of original mRNA can be maintained, such that the construction of trace sample transcriptome library can be realized.
Owner:BEIJING NOVOGENE TECH CO LTD
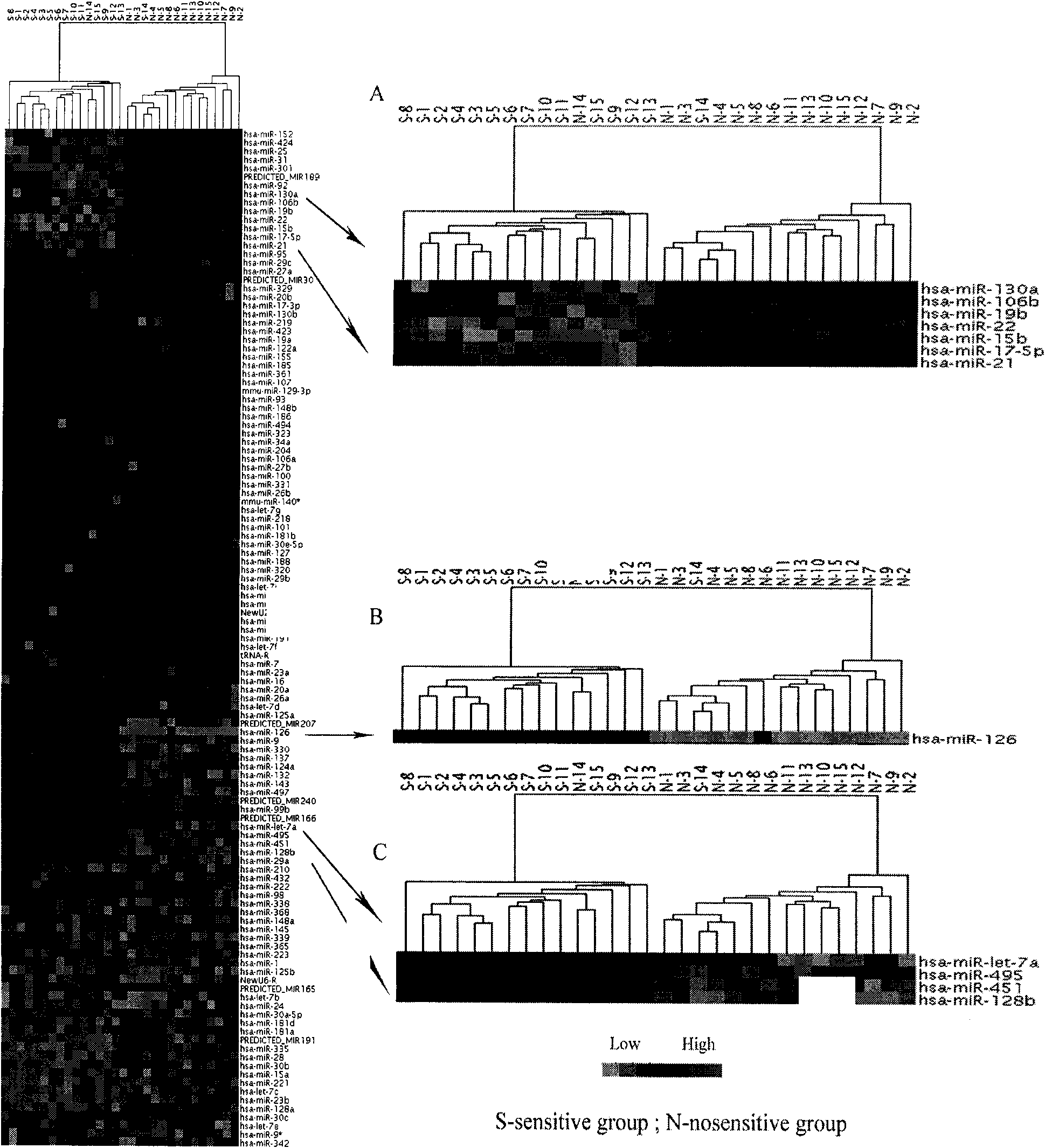
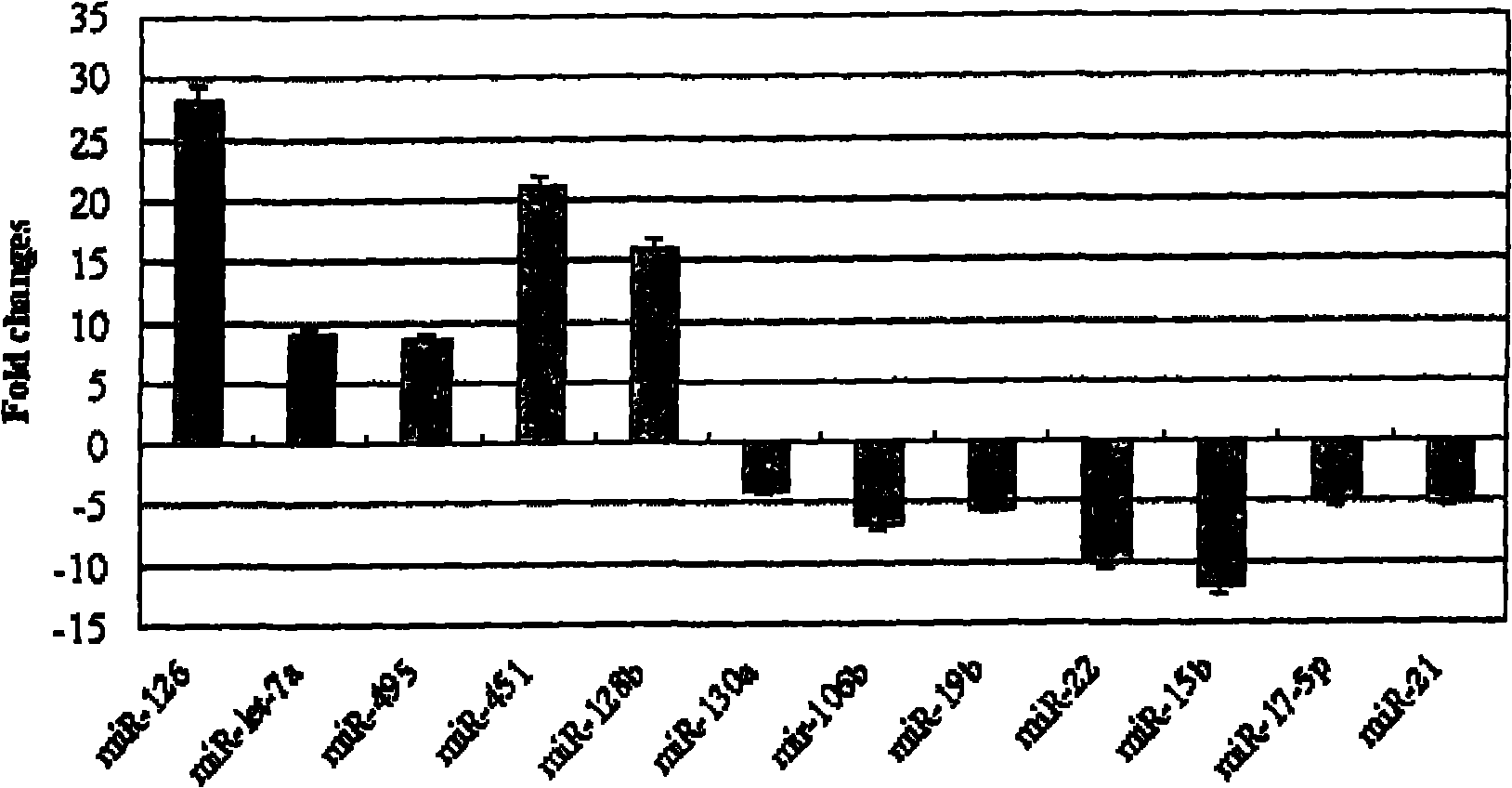
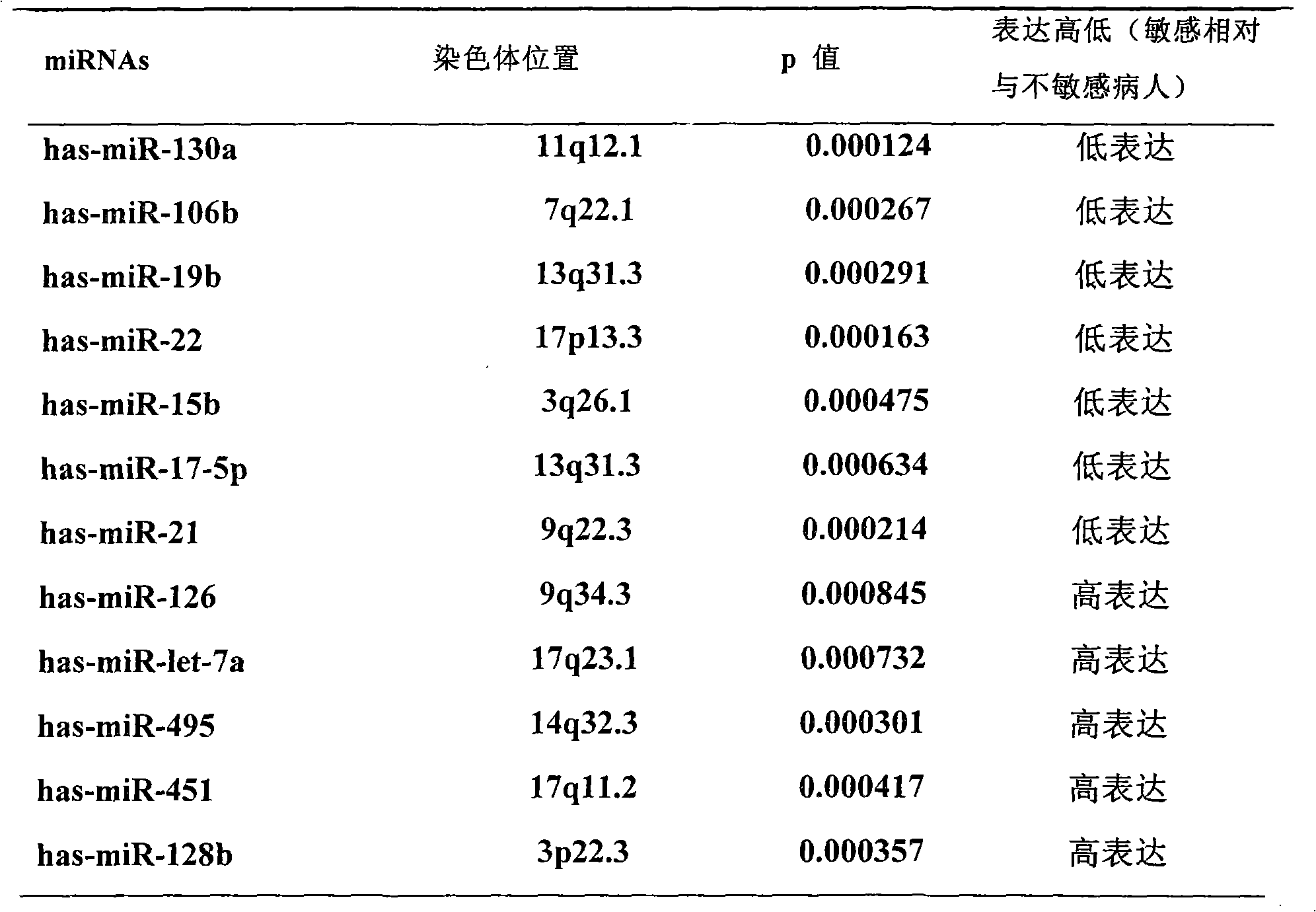
Application of miRNA (Micro-Ribonucleic Acid) expression profile in predicting sensibilities of lung cancer patients to radiotherapy
InactiveCN101824464AReduce overtreatmentLittle side effectsMicrobiological testing/measurementAbnormal tissue growthTotal rna
The invention discloses application of a miRNA (Micro-Ribonucleic Acid) expression profile in predicting the sensibilities of lung cancer patients to the radiotherapy. A preparation method for the invention comprises the following steps of: selecting lung cancer patients who are clinically unusually sensitive and highly tolerant to the radiotherapy, drawing 5ml of peripheral blood, and extracting total RNA by using a TRIZOL kit (Invitrogen); then hybridizing by using a MicroRNA expression profile chip; carrying out data processing, and analyzing the difference of MicroRNA expression profiles of radiotherapy sensitive and non-sensitive patients by using a cluster analysis method; and finally, checking a result by using a Real-time PCR (Polymerase Chain Reaction) method. The inventor finds that an expression profile consisting of 12 miRNA can regulate and control the expression of genes relevant to tumor radiosensitivity and be used for predicting the sensitivity of the lung cancer patients to the radiotherapy and providing an experimental foundation of an early period for the individualized radiotherapy of lung cancer.
Owner:INST OF RADIATION MEDICINE CHINESE ACADEMY OF MEDICAL SCI
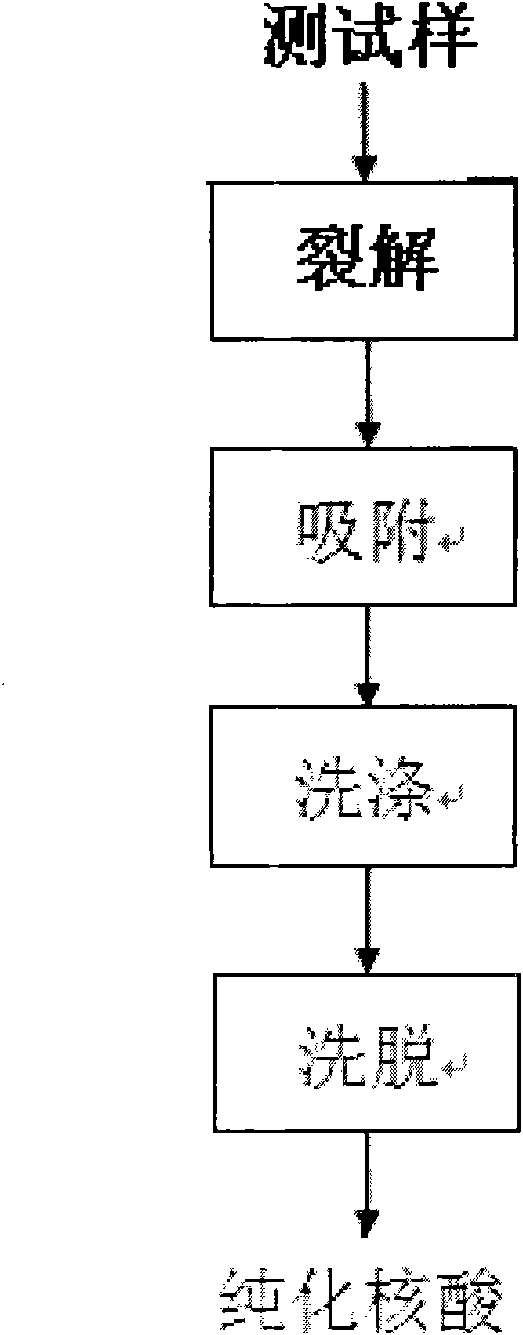
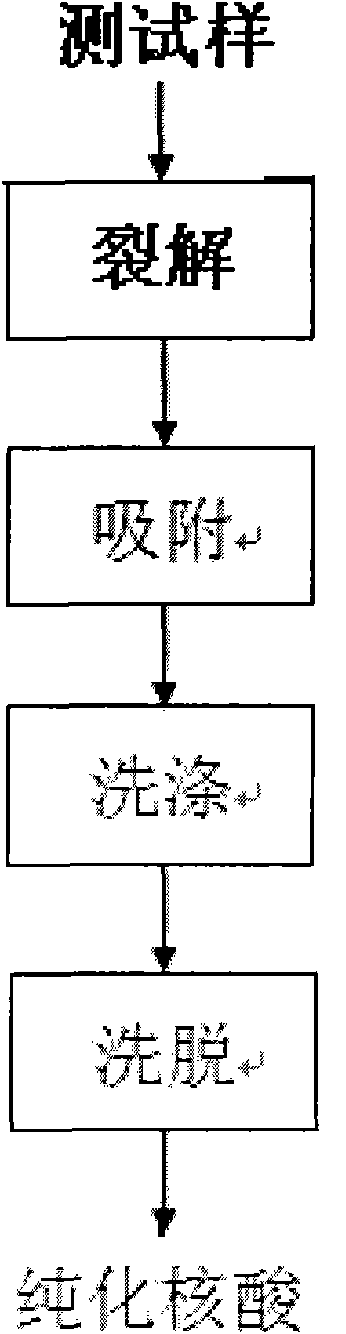
Simple nucleic acid purifying method
The invention discloses a simple nucleic acid simplifying method which comprises processes of cracking, adsorption promotion, washing and elution. The method is characterized by comprising preparation methods and use methods of a simple purifying column, cracking liquid, adsorption promoting liquid and washing liquid special for nucleic acid purification, wherein the simple purifying column is a 1.5 ml centrifuge tube with a hole on the bottom; a small mass of absorbent cotton is compacted on the tube bottom; 0.3-1.0 g of glass powder with diameter of 120-130 mum is added as a purifying column body; an uncovered 2.0 ml centrifuge tube is sleeved outside the column body to serve as a collection tube, therefore, the collection tube is easily manufactured by using a common EP (Eppendorf) tube, cotton and glass powder. With a wide application range, the method is directly applied to the extraction and purification of genome DNA, plasmid DNA and total RNA, as well as the recycling, centrifugation and purification of PCR (polymerase chain reaction) products; the operation steps are concise and easy to master, and the requirements on the experiment operation technique are not high; moreover, the method has the advantages of high purification purity, high integrity, short purification time and low experiment cost; and as toxic substances such as phenol and chloroform are not used in the purification process, the method is environmentally friendly and energy-saving.
Owner:GUANGXI UNIV
Method for detecting lncRNA (long noncoding RNA) in plasma
InactiveCN103966311AReduce degradationQuality improvementMicrobiological testing/measurementTotal rnaFluorescence
The invention discloses a method for detecting lncRNA in plasma. Through steps of TrizolLS pretreatment of theplasma, chloroform and isoamyl alcohol mixed liquid extraction, isopropanol precipitation, ethanol washing, RNA RT (reverse transcription), PCR (polymerase chain reaction) detection and the like, a TrizolLSreagentcan well remove impurities such as polysaccharide, mucoprotein and the like in blood, RNA degradation is less, the phenomenon thattrichloromethane is easy to emulsify under the TrizolLS strong acid condition is effectively eliminated by a chloroform and isoamyl alcohol mixed liquid, high-quality and high-content extraction of RNA in the plasma is realized, fluorescence quantitative RT-PCR detection is performed on the quality of lncRNA, and meanwhile, a reference Ct (cycle threshold) value of the total RNA (that is, cDNA) mass can be detected.
Owner:NINGBO UNIV
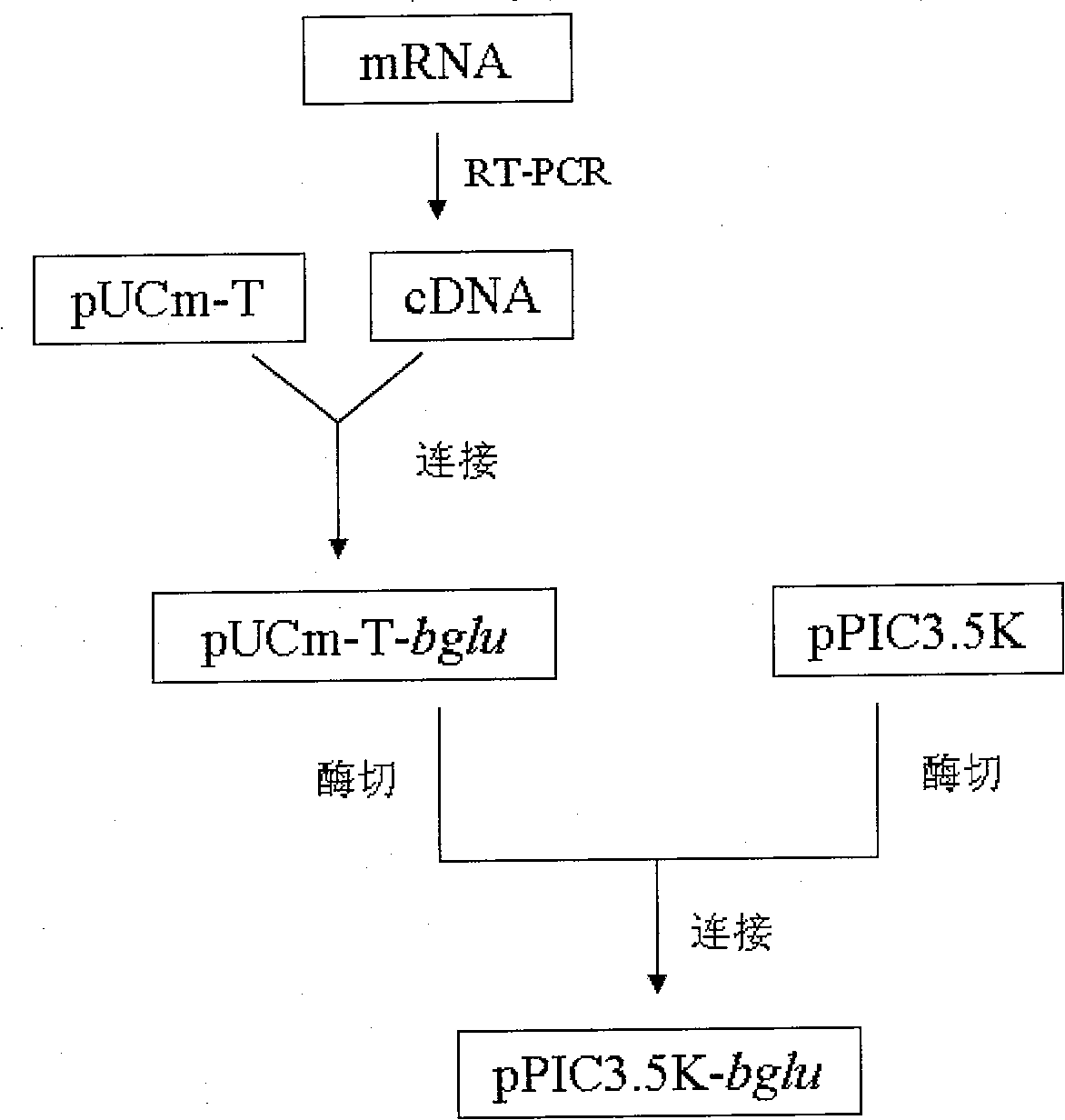
Preparation method for recombination strains for generating Beta glucosaccharase
ActiveCN101654680AHigh industrial application valueFungiMicroorganism based processesAlglucerasePichia pastoris
The invention discloses a preparation method for recombination strains for generating Beta glucosaccharase, comprising the steps of extracting gross RNA from strains which can provide Beta glucosaccharase gene, acquiring Beta glucosaccharase c DNA sequence by utilizing RT-PCR technique, attaching the c DNA sequence to a carrier to acquire a recombinant plasmid transferred host to form the recombination strains for generating Beta glucosaccharase. By adopting the preparation method of the invention, Pichia pastoris which can highly generate Beta glucosaccharase is prepared and is applied to generating Beta glucosaccharase, with high industry using value.
Owner:安徽丰原集团有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com