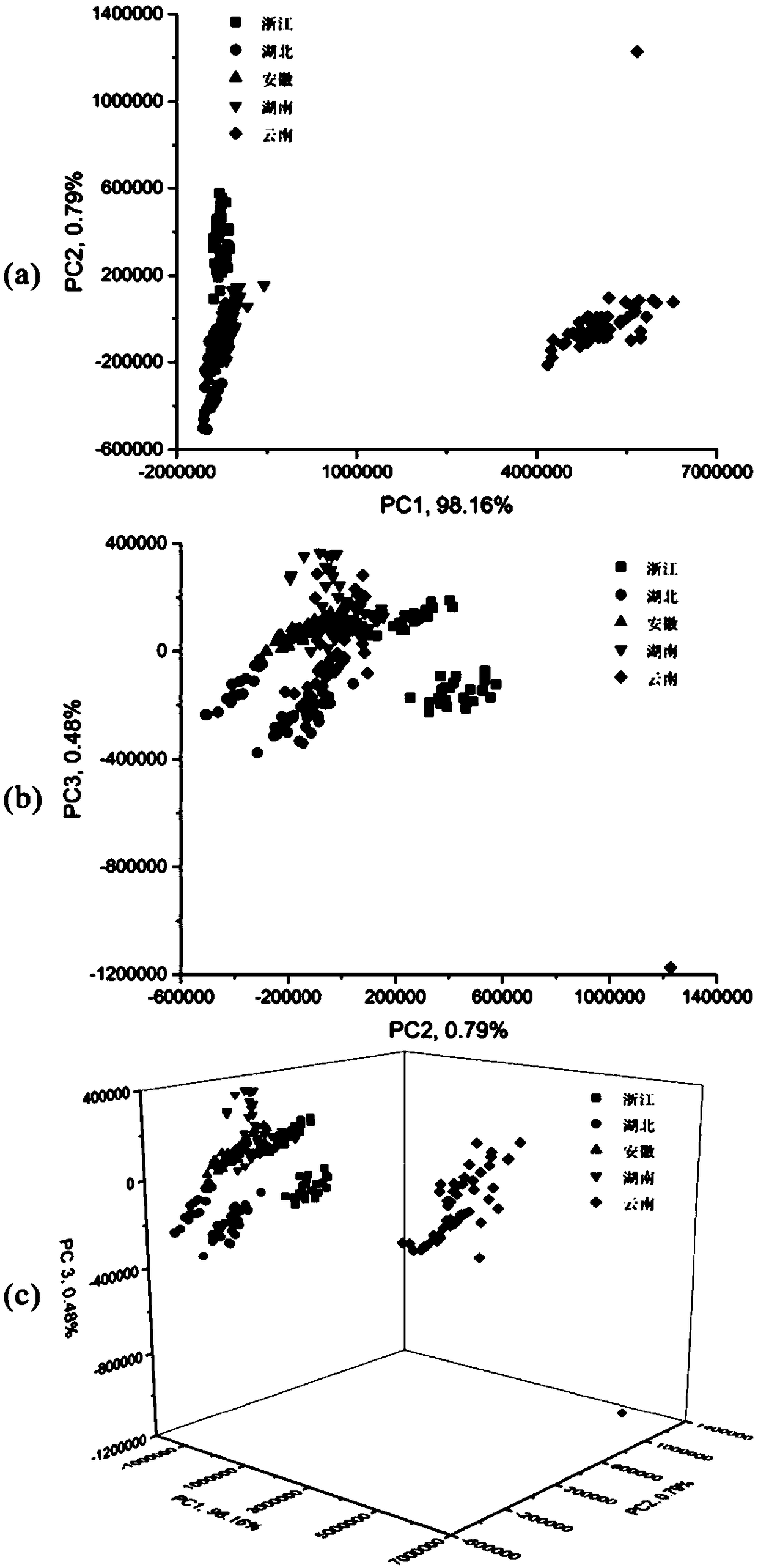
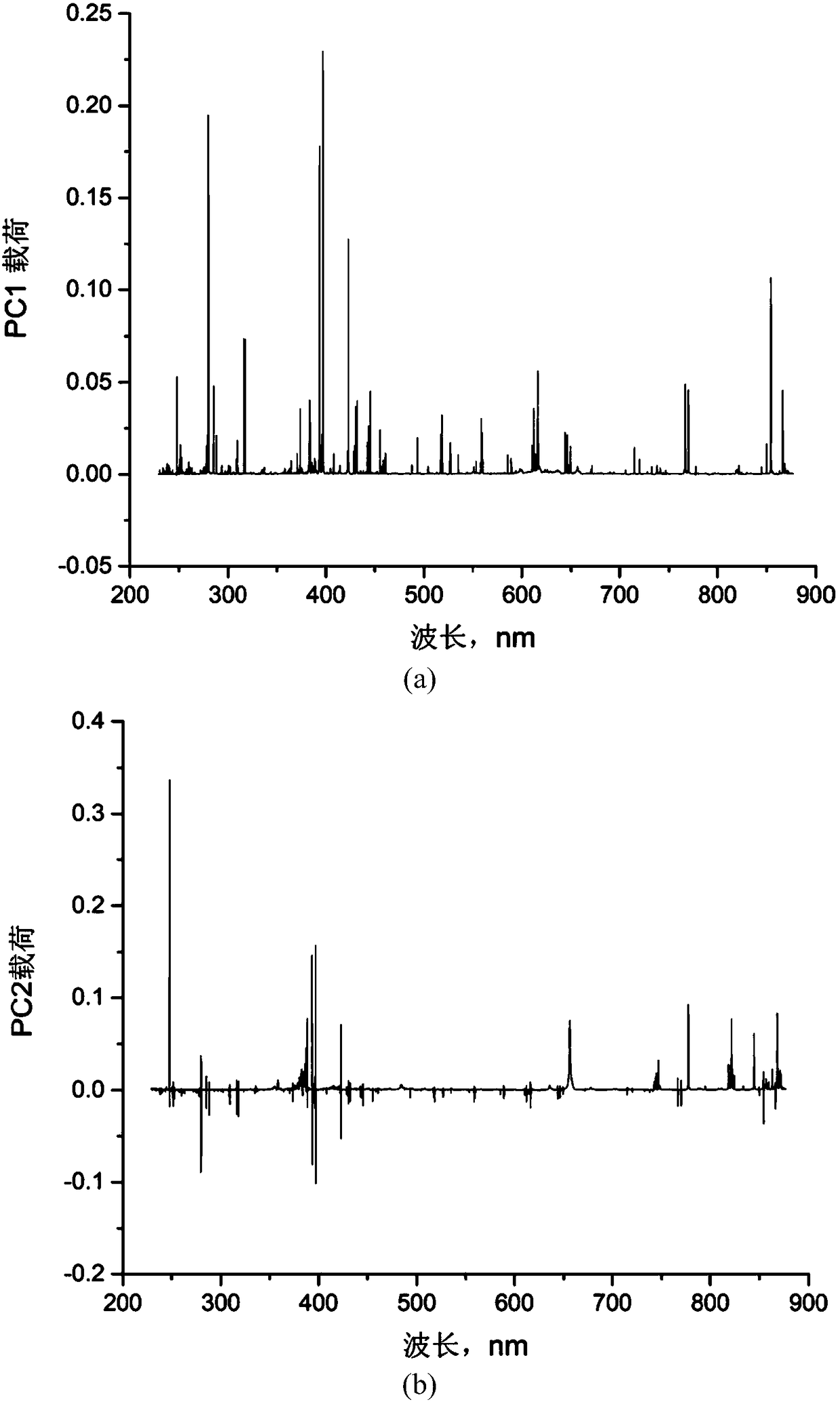
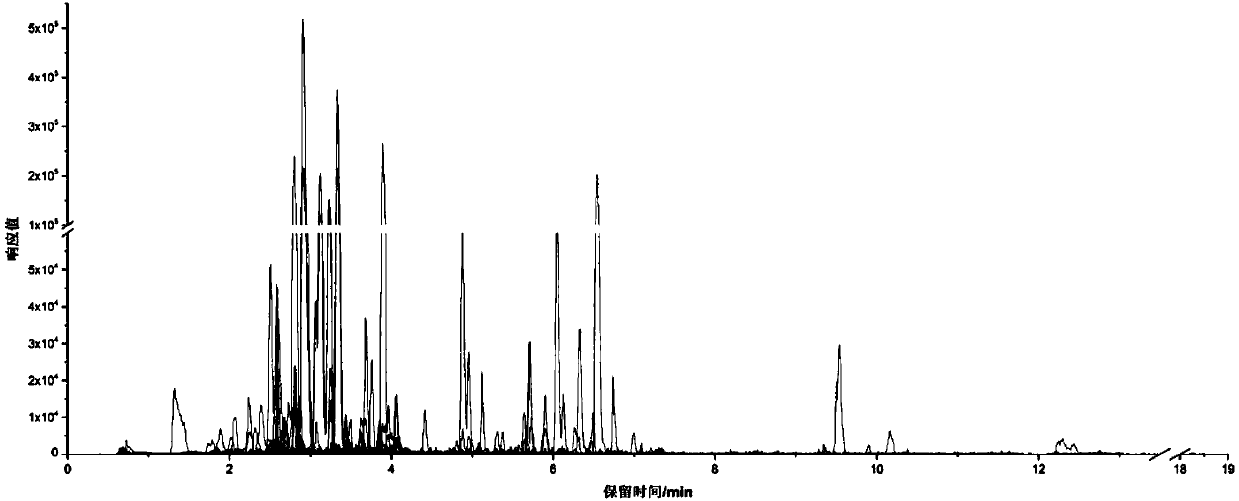
Patents
Literature
52results about How to "Rapid identification method" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
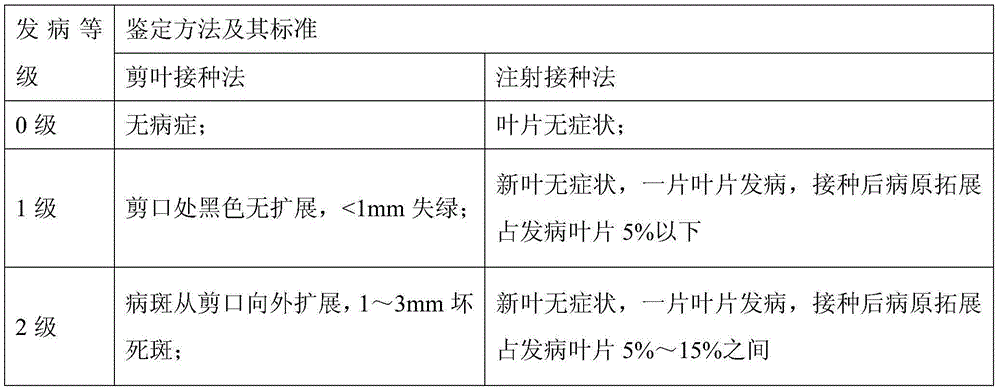
Sugarcane top rot resistance identification method
InactiveCN104450862AIntuitive resistance levelIntuitive performance resistance levelMicrobiological testing/measurementHorticulture methodsSporeDisease
The invention discloses a sugarcane top rot resistance identification method. The method comprises the steps of injecting a well-prepared sugarcane top rot pathogenic bacterium spore suspension onto a growth point of a sugarcane seedling, setting blank sterile water as a reference, culturing the sugarcane seedling for 10 to 15 days after the sugarcane seedling is inoculated, investigating the pathogenic bacterium breeding situation, and classifying the resistance of the sugarcane variety to the sugarcane top rot into an immunity grade, a disease-resisting grade, a medium-resisting grade and an infectious grade according to the disease degree and the disease situation. The sugarcane seedling top rot is identified by adopting an injection inoculation way, the method is closer to the real situation that the sugarcane seedling in the field is infected with the top rot, and the resistance of the sugarcane seedlings of various varieties to the sugarcane top rot can be directly reflected.
Owner:GUANGXI UNIV
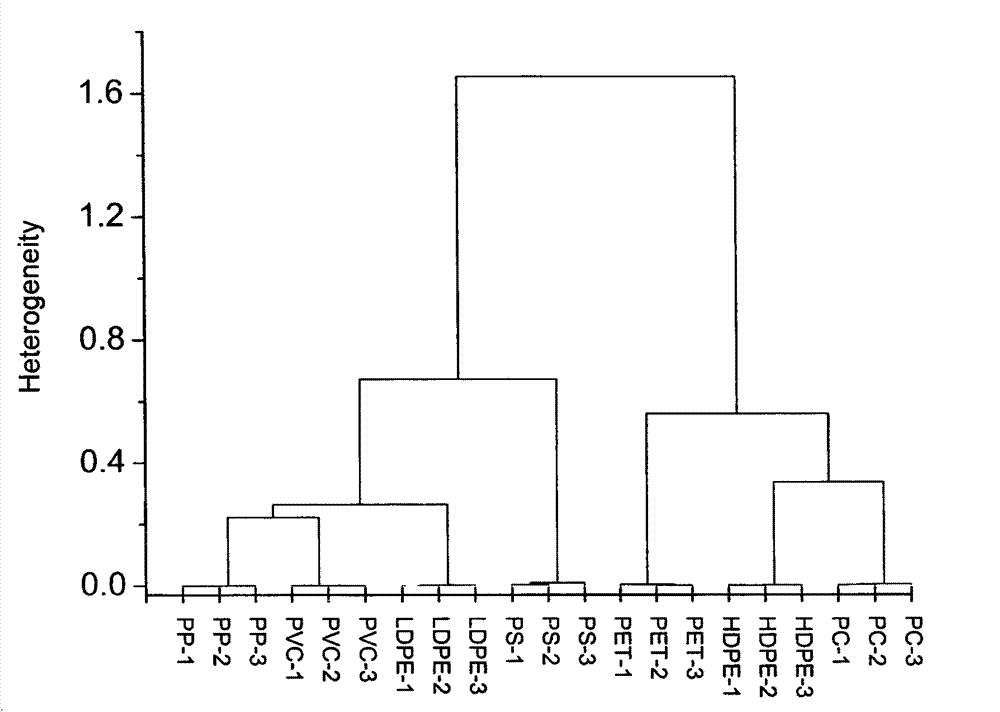
Fast and intelligent waste plastic sorting method
InactiveCN103499552AHigh speedIntelligent classification methodMaterial analysis by optical meansDimensionality reductionPrincipal component analysis
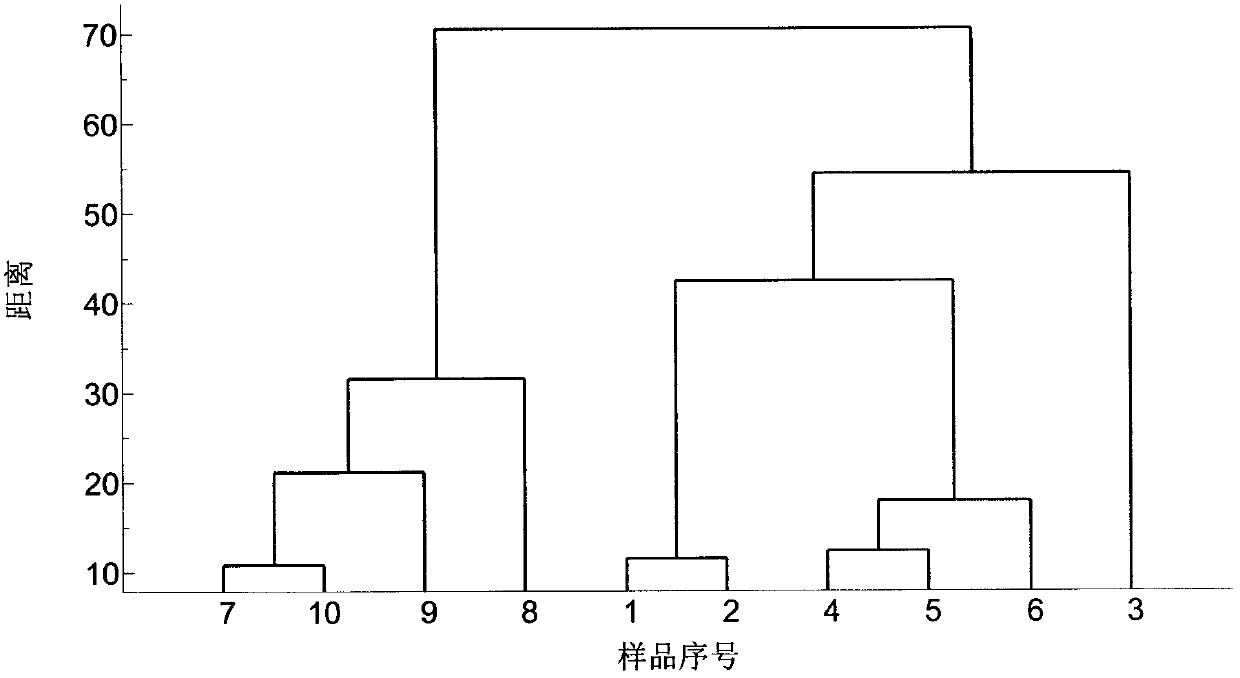
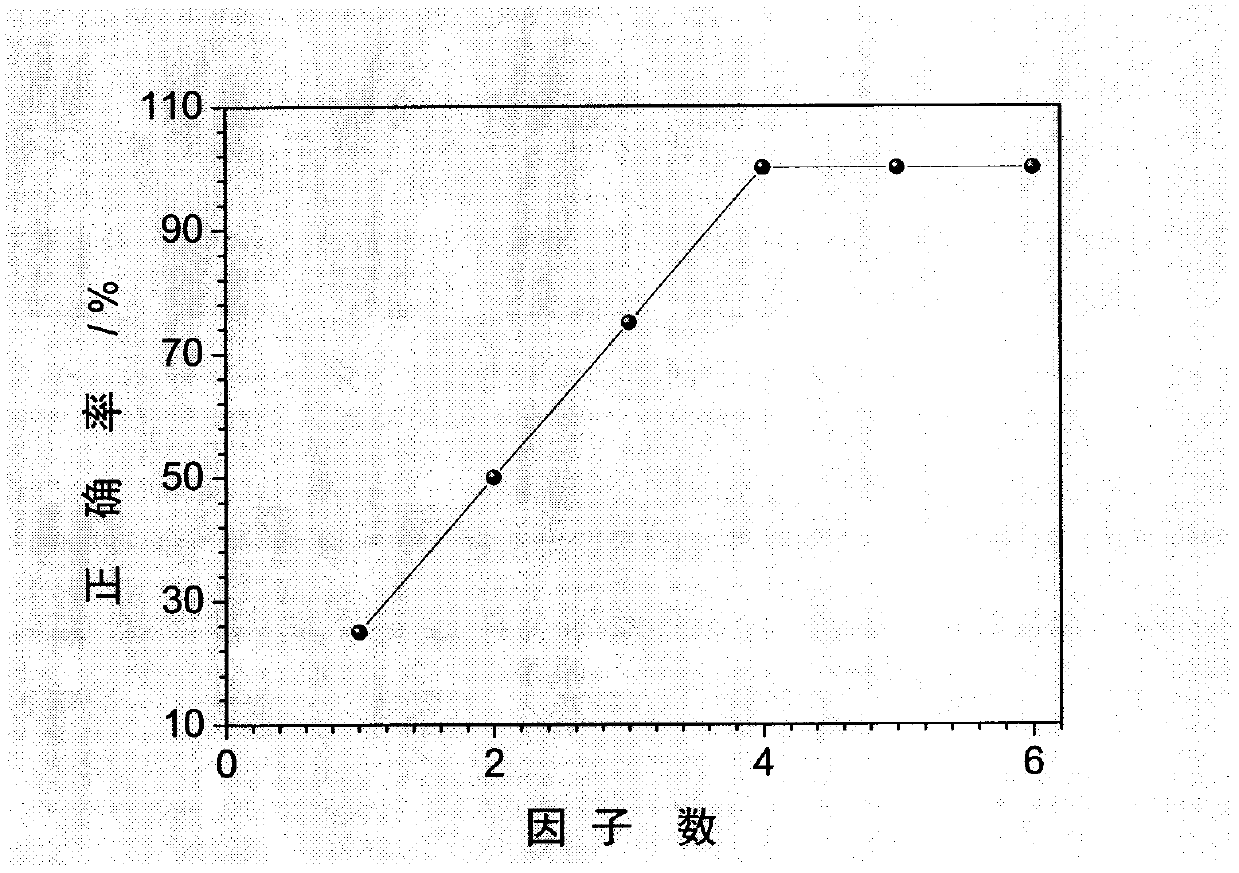
The invention relates to a fast and intelligent waste plastic sorting method. The method comprises the steps of scanning the infrared spectrum of a sample with an attenuated total reflection fourier transform infrared spectrograph to obtain an infrared spectrum data matrix, pre-processing the data matrix including standardization and principal component analysis and dimensionality reduction, and carrying out cluster analysis on data with dimensionality reduced by means of the hierarchical clustering method. The result shows that the sample testing method is free of sample preprocessing and high in sample testing speed, and the accuracy of sorting of waste plastic can reach 100% by means of the principal component analysis - hierarchical clustering identification method. Therefore, fast and intelligent waste plastic sorting is achieved by means of attenuated total reflection fourier transform infrared spectrum and chemical mode combined identification.
Owner:TIANJIN POLYTECHNIC UNIV
Wild ginseng and cultivated ginseng multiple polymerase chain reaction (PCR) test kit and identification method
InactiveCN102191308ASpecificity Accurate Simultaneous DistinctionThe identification method is simpleMicrobiological testing/measurementRibonucleosideOligonucleotide primers
The invention discloses a wild ginseng and cultivated ginseng multiple PCR test kit, which is characterized by containing: buffer solution, 12.5mM of deoxy-ribonucleoside triphosphate (dNTP), 0.1mM of one of a primer 1 and a primer 2, Taq DNA polymerase, a sample DNA to be tested and a double-distilled water identification reaction system; or the buffer solution, 12.5 mM of dNTP, 0.1 mM of one of the primer 1 and the primer 2, Taq DNA polymerase, wild ginseng and cultivated ginseng DNA 1:1 mixture and a double-distilled water positive reference reaction system; or the buffer solution, 12.5 mM of dNTP, 0.1 mM of one of the primer 1 and the primer 2, Taq DNA polymerase, araliaceae congeneric DNA 1:1 mixture, and a double-distilled water negative reference reaction system. The detection and identification method comprises the steps of designing two pairs of specific oligonucleotide primers of wild ginseng and cultivated ginseng mitochondrion DNAs, designing two pairs of specific oligonucleotide primers of synthetic wild ginseng and cultivated ginseng mitochondrion DNAs, determining a reaction process, determining result and the like. The method can accurately determine the specificity of both the wild ginseng and the cultivated ginseng, and the detection result is reliable.
Owner:BEIHUA UNIV
Method for establishing number of Fusarium sp. copies in rhizosphere soil in growth period of transgenic rice by fluorescence real-time quantitative PCR (polymerase chain reaction)
InactiveCN102517385AImprove linearityHigh amplification efficiencyMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceGenetically modified wheat
The invention relates to a method for establishing number of Fusarium sp. copies in rhizosphere soil in growth period of transgenic rice by fluorescence real-time quantitative PCR (polymerase chain reaction), which comprises the following steps: establishing five soil sampling points on a diagonal line and marking and locating, and collecting rhizosphere soil samples at the above soil sampling points before seeding and at the growing period; extracting total DNA from the soil samples collected at the soil sampling points, dissolving the total DNA of the samples in double distilled water, and carrying out PCR amplification of the sample DNA at each period with a Fusarium-specific universal primer pair SEQ ID No.2 and SEQ ID No.3 to obtain a specific fragment of size 418bp (base pairs); directly measuring the number of copies of the corresponding Fusarium sp. with a fluorescence quantitative PCR instrument according to the variation of fluorescence signals and a standard curve; and summarizing the number of Fusarium sp. copies in the soil samples at different periods to obtain the number of Fusarium sp. copies in rhizosphere soil in the growth period of transgenic rice.
Owner:JIANGSU ACAD OF AGRI SCI
Method for detecting quantity of pseudomonas fluorescens in rhizospheric soil during growth period of transgenic wheat by virtue of fluorescent quantitative PCR (Polymerase Chain Reaction)
ActiveCN103397099AConsistent repetitive responseEasy to distinguishMicrobiological testing/measurementFluorescence/phosphorescencePseudomonas fluorescensPseudomonas
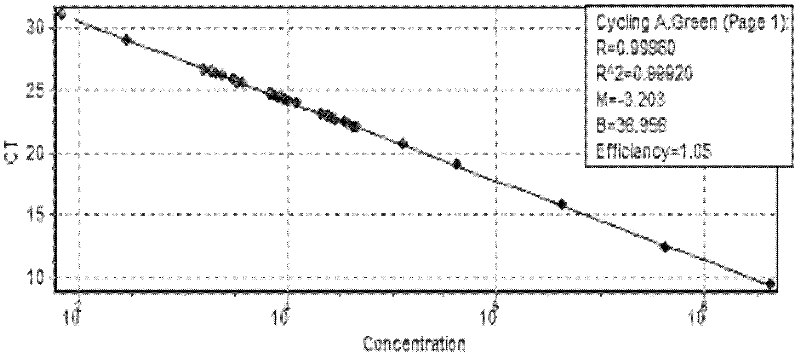
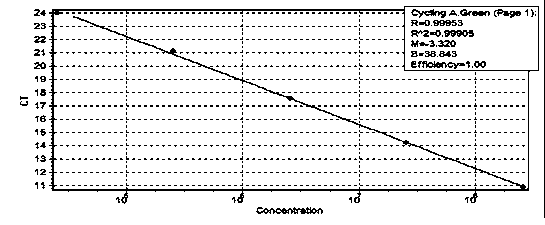
The influence of transgenic crops on rhizospheric microorganisms in soil is an important aspect in environmental biosafety research. The invention establishes a detection system of Real-Time QPCR (Real-Time Fluorescent Quantitative Polymerase Chain Reaction)) by carrying out absolute quantification on the quantity of pseudomonas fluorescens in rhizospheric soil during the growth period of transgenic disease-resistant wheat. By virtue of application of the reaction system, the absolute quantification is carried out on the copy number of the pseudomonas fluorescens in the rhizospheric soil during the whole growth period of the transgenic wheat. A result shows that the designed primer has better specificity; the intervals of cycle thresholds (Ct values) of plasmids with various gradient standards in an amplification curve of Real-Time QPCR are uniform, the peak value of a melting curve is more obvious, and for the standard curve, the correlation coefficient R2 is equal to 0.99905, the slope is minus 3.203, and the amplification efficiency E is equal to 100%. In the seeding stage, seedling establishment stage, watery stage and mature stage of the transgenic wheat, the quantity of the pseudomonas fluorescens in the rhizospheric soil takes on a trend of gradually rising.
Owner:JIANGSU ACAD OF AGRI SCI
Application of cabbage type rape floral leaf mutant
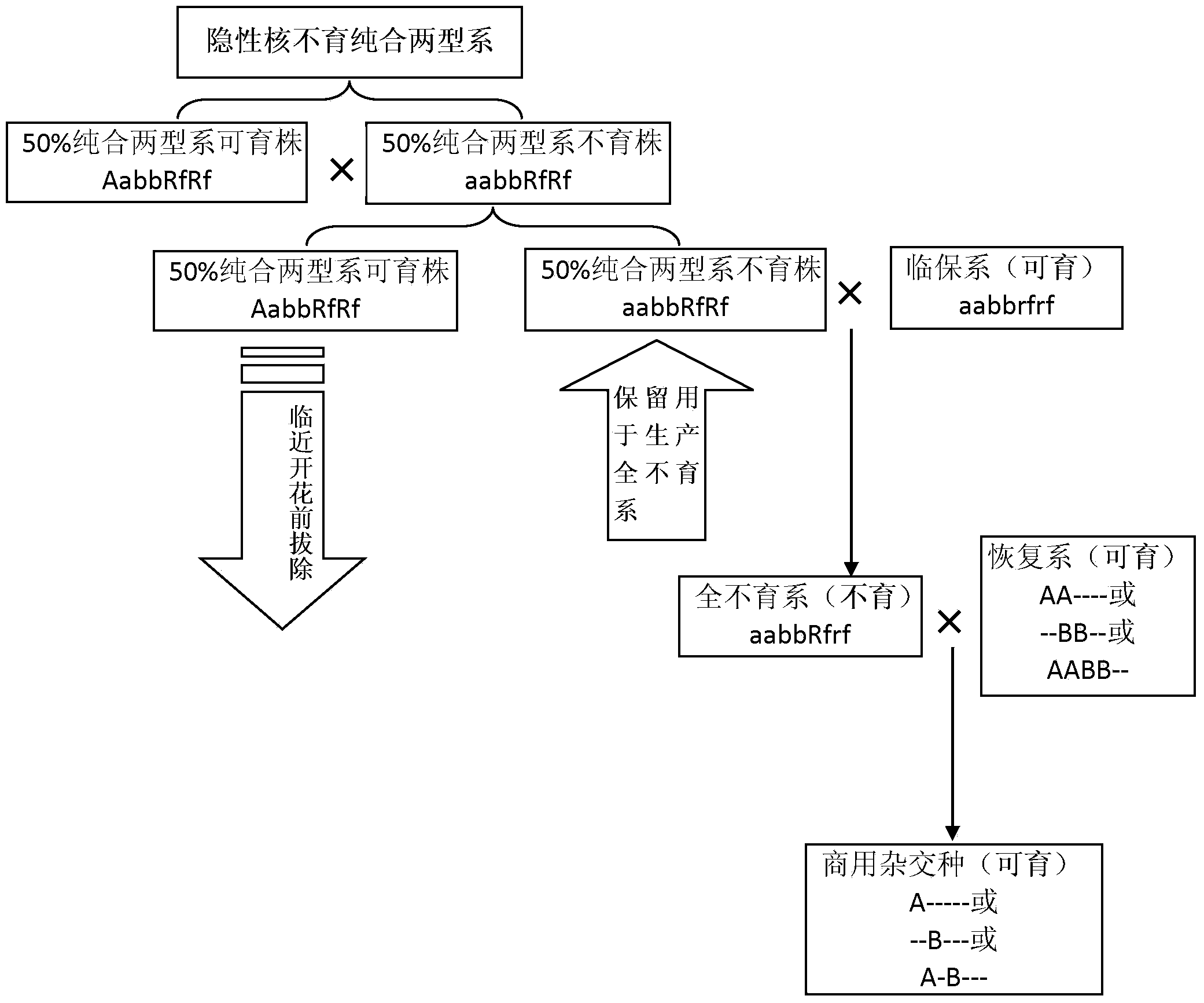
ActiveCN103931486AEfficient identificationEfficient removal of impuritiesPlant genotype modificationBiotechnologyHybrid seed
The invention relates to an application of a cabbage type rape floral leaf mutant. The cabbage type rape floral leaf mutant is applied by the following steps: hybridizing a sterile plant in a cabbage type rape recessive genic male sterile homozygous two-type line 20118AB serving as a female parent with a cabbage type rape floral leaf mutant serving as a male parent, bagging and performing inbreeding to obtain a floral leaf sterile plant; hybridizing the floral leaf sterile plant with a fertile plant in 20118AB to breed a new homozygous two-type line, namely, a floral leaf recessive genic male sterile homozygous two-type line; hybridizing the floral leaf sterile plant with a temporary maintaining line to obtain a new temporary maintaining line, namely, a floral leaf temporary maintaining line; and hybridizing the cabbage type rape floral leaf recessive genic male sterile homozygous two-type line with the floral leaf temporary maintaining line to produce a cabbage type rape floral leaf complete sterile line. Purity identification and impurity removal at the seedling stage of the complete sterile line are realized by using the character of a floral leaf, the purity of the complete sterile line is increased, the genic male sterile seed production procedure is simplified, and the labor amount of field hybrid seed production is reduced.
Owner:SHANGHAI ACAD OF AGRI SCI +1
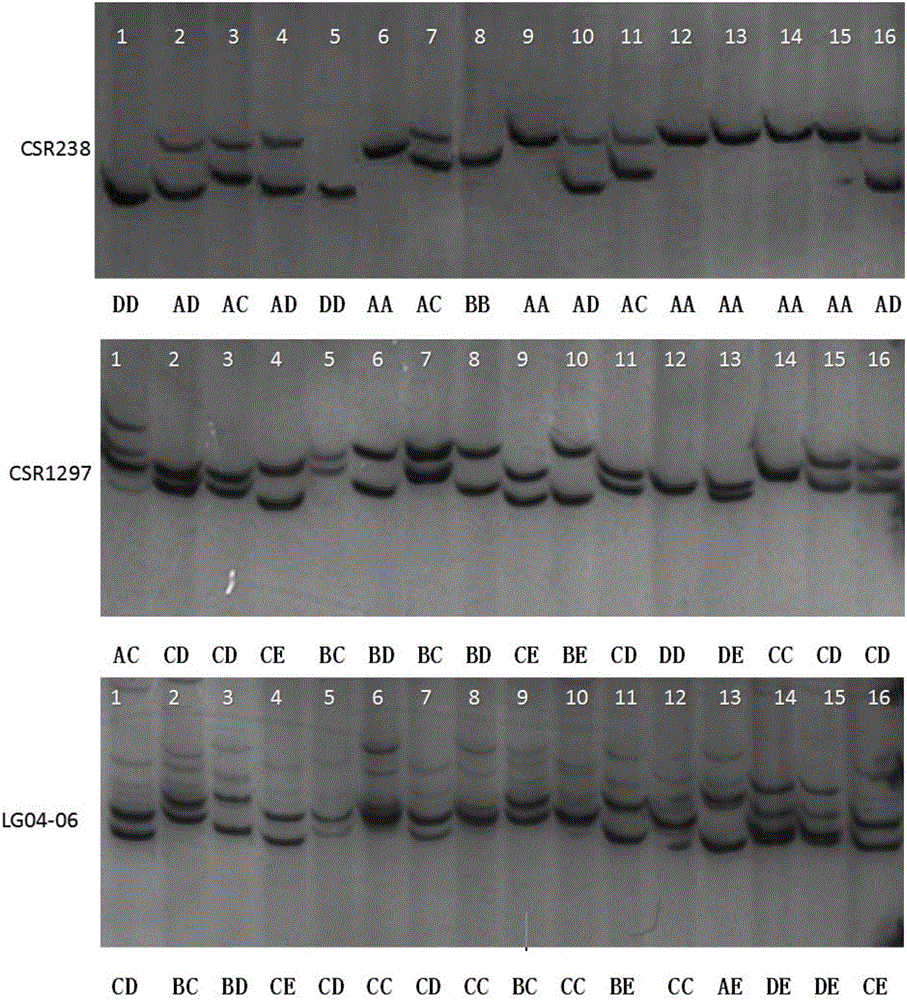
Molecular marker combination for rapidly identifying different albino tea tree varieties, method and application
ActiveCN106480224ASolve mixed problemsEasy to confirmMicrobiological testing/measurementDNA/RNA fragmentationTea leafMolecular marker
The invention discloses a molecular marker combination for rapidly identifying different albino tea tree varieties. The molecular marker combination adopts CSR238, CSR 1297 and LG04-06 primer combinations; the invention further discloses a method for rapidly identifying different albino tea tree varieties by using the molecular marker combination and a kit containing the molecular marker. The molecular marker combination can distinguish and identify the albino tea tree varieties on the market, and the identifying method is simple and fast, and can solve the problem of confused names of albino tea tree varieties on the market for a long time; the invention further discloses a finger-print of the albino tea tree varieties; tea selling enterprises can apply the two-dimensional code albino tea tree varieties generated by the invention to mark the product formed by processing different albino tea tree varieties, thus consumers can more conveniently learn about the product information.
Owner:TEA RES INST CHINESE ACAD OF AGRI SCI
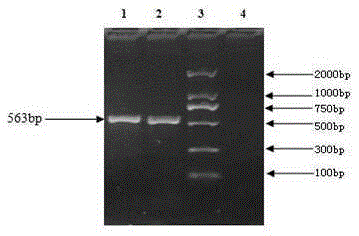
Deer fetus DNA identification kit and method
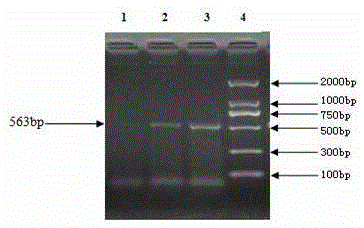
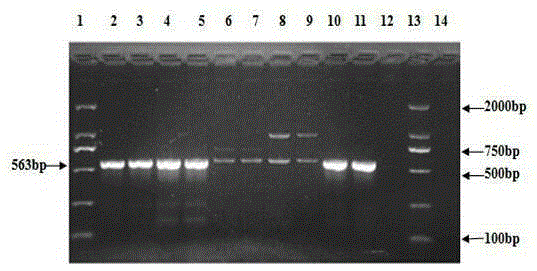
InactiveCN105238856AThe identification method is simpleRapid identification methodMicrobiological testing/measurementBiotechnologyDNA extraction
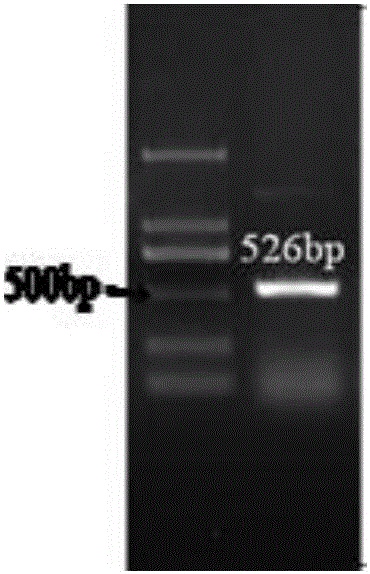
The invention relates to the technical field of traditional Chinese medicine detection, in particular to a deer fetus DNA identification kit and method. The deer fetus DNA identification kit comprises a mitochondria DNA extraction system, a PCR (polymerase chain reaction) system and a result observation system. The deer fetus DNA identification method includes the steps of detection sample pretreatment, mitochondria DNA extraction, PCR primer designing and synthesis, building of the PCR system and result judgment, wherein judgment standards includes that deer fetuses are judged to be qualified if 563bp strips appear and judged to be pseudo if only multiple strips or no strips appear. The PCR identification method has the advantages of simplicity, quickness and reliable detection result, and specificity of qualified and pseudo deer fetuses can be accurately differentiated and identified at the same time.
Owner:BEIHUA UNIV
Method for identifying cordyceps sinensis phorozoon by using immunohistochemistry
ActiveCN101900724AStrong specificitySpecificPreparing sample for investigationImmunoglobulins against fungi/algae/lichensSerum igeCordyceps militaris
The invention relates to a method for identifying phorozoon of cordyceps sinensis by using immunohistochemistry. The method comprises the following steps of: preparing and staining hypha antibodies, and comparing staining results; directly observing a staining picture, wherein positive staining (stained with color) means cordyceps sinensis, and negative staining (not stained with color) means not phorozoon of the cordyceps sinensis. The method creatively adopts hyphae of cordyceps militaris with highest cordyceps sinensis homology to absorb polyvalent serum and has the characteristic of high specificity. The identification method has the advantages of no need of instrument, simpleness, convenience, easy popularization, short staining time, higher speedability than that of the conventional identification method and the molecular biology method, and low cost and can be used for identifying the phorozoon of the cordyceps sinensis hyphae.
Owner:HEBEI UNIV OF ENG +2
Method for fast identifying frankliniella occidentalis
InactiveCN101805782AGood specificityProtect the environmentMicrobiological testing/measurementEcological environmentBiology
The invention discloses a method for fast identifying frankliniella occidentalis. The method is based on the identification results of forms of six kinds of adult thrips such as frankliniella occidentalis and the like for respectively carrying out PCR amplification by using two pairs of frankliniella occidentalis specific primers F.OL1 / F.OR and F.OL2 / F.OR to establish the method for fast identifying the frankliniella occidentalis. The invention has the advantages of high speed, accuracy and good specificity, provides an effective identification measure for the quarantine pest of the frankliniella occidentalis, and has important meaning for protecting the ecological environment.
Owner:CHECKOUT & QUARANTINE TECH CENT YUNNAN ENTRY &EXIT CHECKOUT & QUARANTINE BUR
American ginseng DNA detection reagent box and identification method
InactiveCN105039584AThe identification method is simpleRapid identification methodMicrobiological testing/measurementAMERICAN GINSENG ROOTBioinformatics
The invention relates to the technical field of traditional Chinese medicine, in particular to an American ginseng DNA detection reagent box and an identification method. The American ginseng DNA detection reagent box comprises five parts, namely a sample pretreatment solution, a genome DNA extracting system, a PCR reaction system, a restriction enzyme reaction system and an observation result system. The identification method comprises six steps, namely detection sample pretreatment, genome DNA extraction, PCR primer design and synthesis, PCR reaction establishment, restriction enzyme reaction and result judgment. According to judgment standards, a 100 bp band and a 50 bp band occur simultaneously, the American ginseng is a salable product, and if only one band occurs or no band occurs, the American ginseng is a fake product. The PCR-AFLP identification method has the advantages of being simple, convenient, fast, reliable in detection result and the like, and the specificity of the American ginseng and the fake product can be identified accurately at the same time.
Owner:BEIHUA UNIV
DNA barcode based premier for identifying dinodon rufozonatum, PCR-RFLP method, and kit
InactiveCN104164493ASave operating timeGuarantee the quality of medicinal materialsMicrobiological testing/measurementDNA/RNA fragmentationDNA barcodingBarcode
The invention belongs to the technical field of traditional Chinese medicine species identification, and provides a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for identifying dinodon rufozonatum. The sequence of the premier is DK1-CO1:5'-CAA CTA ACC ACA AAG ACA TCG G-3', DK1-CO2:5'-CTT CTG GCT GGC CGA AAA ATC A-3'; and the characteristic DNA base sequence of dinodon rufozonatum is 5'-GTGCAC-3', which can be recognized and cut by restriction endonuclease APaL I. The identification comprises the following steps: extracting the DNA of a sample; using premiers DK1-CO1 and DK1-CO2 to carry out PCR amplification; using restriction endonuclease APaL I to digest the PCR products; carrying out agarose gel electrophoresis to obtain the analysis result; and comparing the analysis result with the PCR-RFLP characteristic result of dinodon rufozonatum to judge whether the provided sample is dinodon rufozonatum or not. The invention also provides a kit for the identification method. The identification is accurate, rapid, and reliable, and is capable of effectively identifying dinodon rufozonatum from other snakes.
Owner:SOUTHERN MEDICAL UNIVERSITY
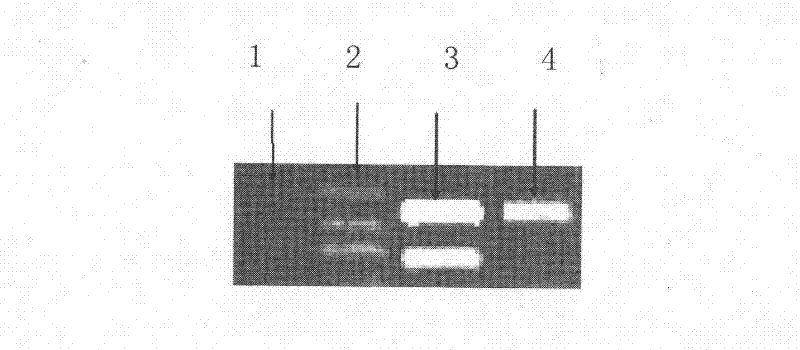
Method for identifying seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin
InactiveCN103293213AHigh priceIdentification method is simpleMaterial analysis by electric/magnetic meansBiotechnologyDismutase
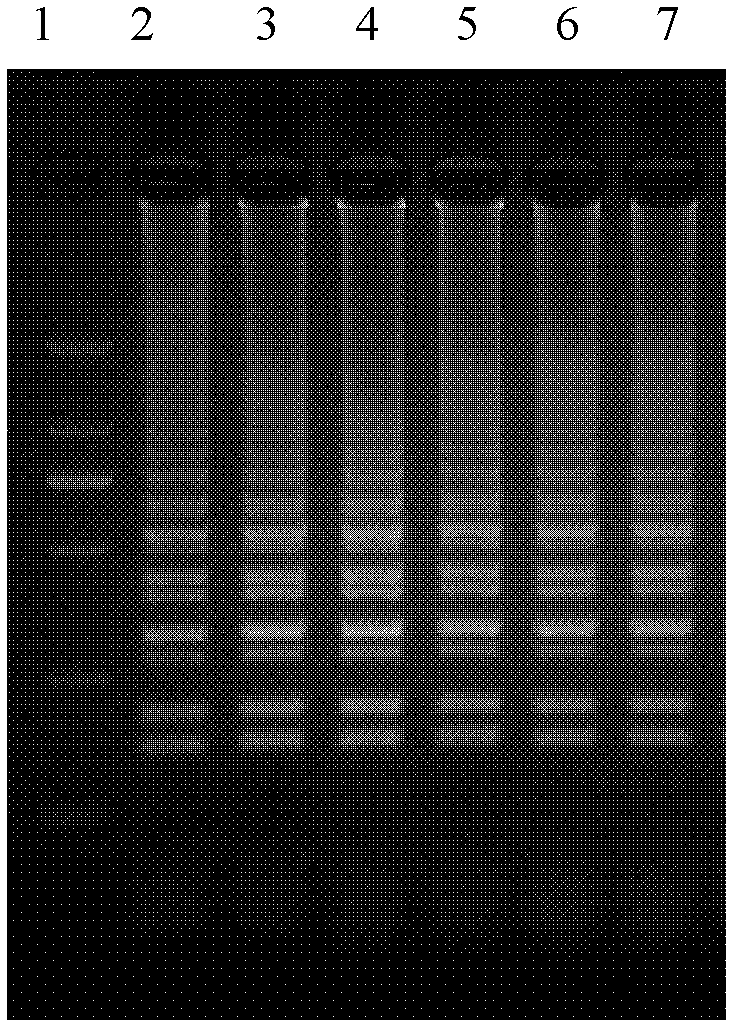
The invention discloses a method for identifying the seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin, which comprises the following steps of: preparation of enzyme liquid, preparation of a gel plate, sample application and electrophoresis, SOD (superoxide dismutase) isozyme dyeing process, variety conformation and the like. The method mainly makes use of the physiological and biochemical difference between the seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin, and takes the SOD isozyme zymogram as an index for identifying the seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin. If the SOD isozyme zymogram of the seed to be detected is the same as the two lanes on the left side of a standard zymogram, the seed is the glycyrrhiza uralensis seed; if the SOD isozyme zymogram of the seed is the same as the two lanes on the right side of the standard zymogram, the seed is the glycyrrhiza inflata batalin seed. The identifying method disclosed by the invention has the advantage of simple, quick and accurate identification.
Owner:TIANJIN NORMAL UNIVERSITY
Method for identifying lamb meat and adult mutton
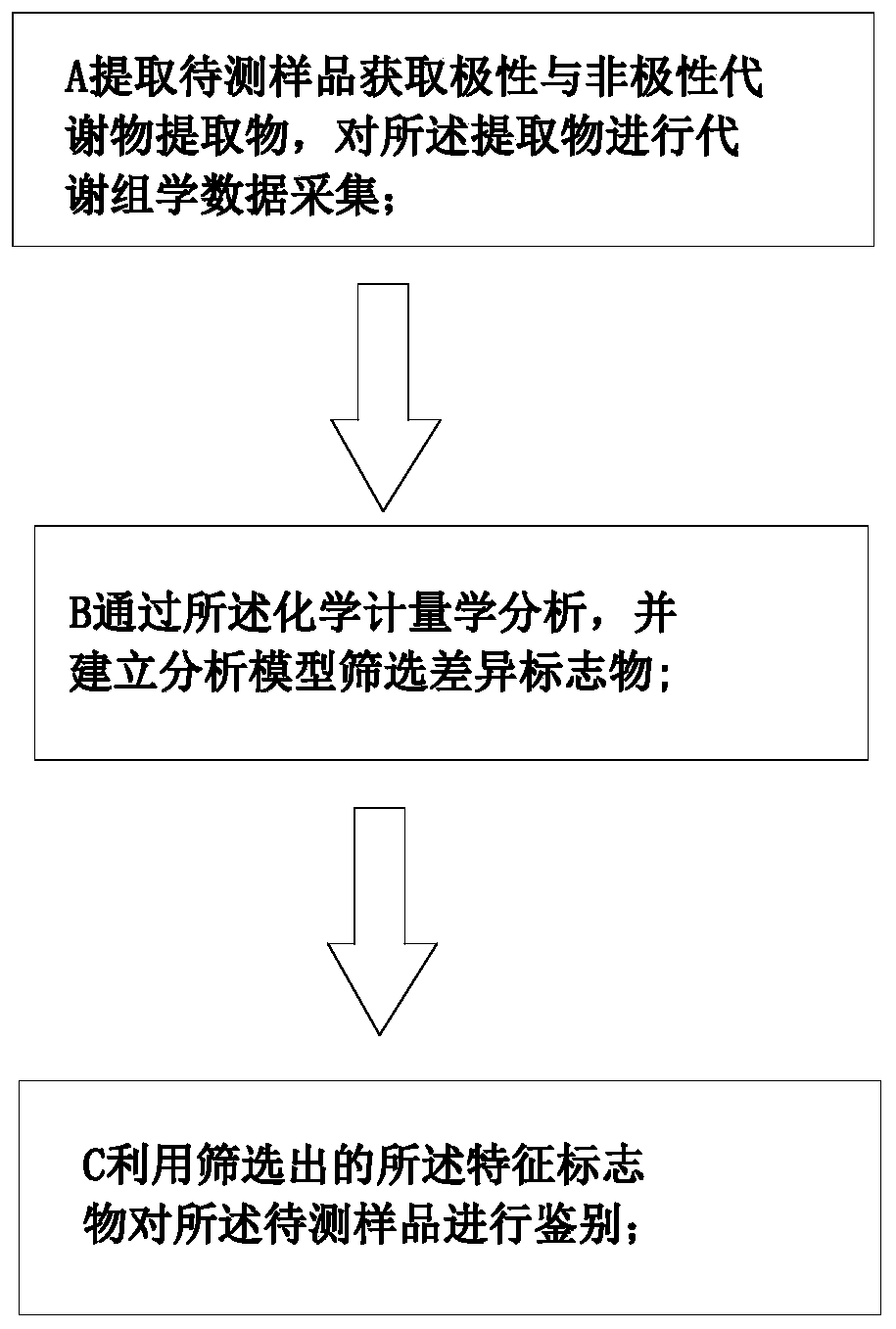
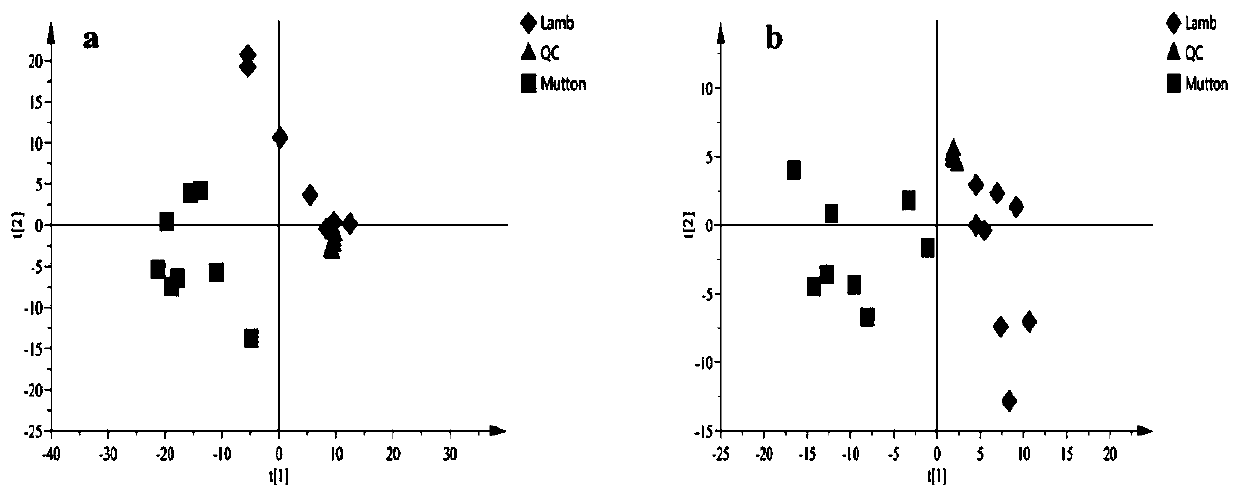
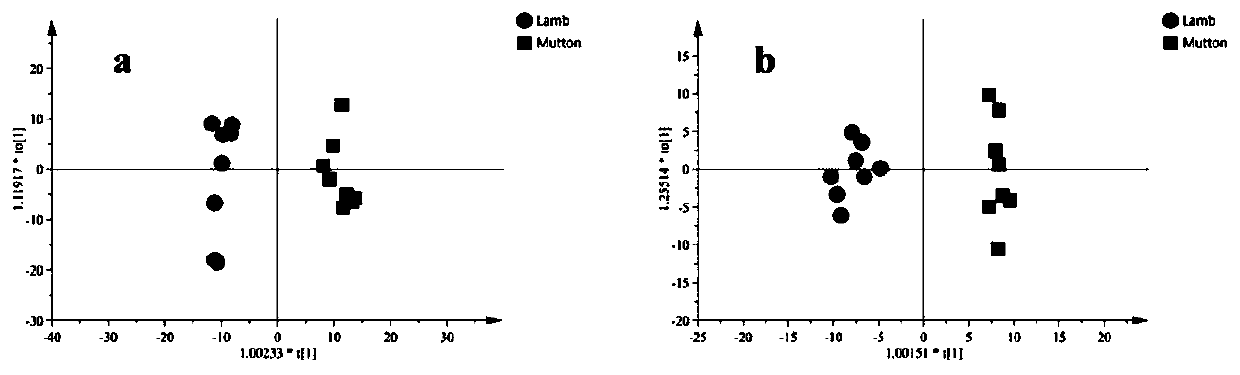
ActiveCN111413436ARapid identification methodSimple methodComponent separationAnimal scienceMetabolomics data
The invention discloses a method for identifying lamb meat and adult mutton, which comprises the following steps of extracting metabonomics data of a sample to be detected, and carrying out chemometrics analysis on the detection data to extract characteristic information, analyzing through the chemometrics, establishing an analysis model to screen difference markers, and using the screened characteristic marker to identify the sample to be detected. A reliable and rapid identification method is provided for distinguishing the lamb meat from the adult mutton, the method is convenient, sensitiveand accurate, and the rapid identification on the mutton can be achieved by screening the difference marker through the analysis model.
Owner:INST OF QUALITY STANDARD & TESTING TECH FOR AGRO PROD OF CAAS
Real time-loop-mediated isothermal amplification (RT-LAMP) detection primers for performing differential diagnosis on canine distemper virus wild strains and vaccine strains and application of primers
InactiveCN102586485AGood repeatabilityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationLoop-mediated isothermal amplificationWild strain
The invention discloses a set of real time-loop-mediated isothermal amplification (RT-LAMP) primers for differentiating canine distemper virus (CDV) wild strains from vaccine strains and application of the primers. The RT-LAMP primers comprise a pair of outer primers and a pair of inner primers, wherein the sequences of the outer primers are shown as SEQ ID NO:1 and SEQ ID NO:2 respectively; and the sequences of the inner primers are shown as SEQ ID NO:3 and SEQ ID NO:4 respectively. When the primers are used for detecting various genotypes of CDV wild strains according to an RT-LAMP detection method, the detection results are positive; and when the primers are used for detecting canine distemper vaccine strains, canine parvoviruses and other common canine viruses according to the RT-LAMPdetection method, the detection results are negative. Due to application of the primer sequence by the RT-LAMP detection method, the CDV wild strains and the CDV vaccine strains can be differentiatedaccurately; and the invention has the advantages of high specificity, high sensitivity, high repeatability and the like.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
Quick and accurate identification method of baijiu scent types
InactiveCN107796783AImprove qualityFlavor fastMaterial analysis by optical meansPattern recognitionOptimal modeling
The invention relates to a quick and accurate identification method of baijiu scent types, comprising the specific steps of using a near-infrared spectrometer to scan near-infrared spectrum of a sample to obtain near-infrared spectral data; pretreating a data matrix; using the two chemical pattern recognition methods of HCA (hierarchical cluster analysis) and PLS-DA (partial least squares discriminant analysis) to model the data, determining an optimal modeling method, and using the optimal modeling method to predict unknown scent types. The pattern recognition method of partial least squaresdiscriminant analysis is used herein to identify baijiu scent types and has high correct rate, high speed and good effect. The quick and accurate identification method of baijiu scent types is applicable to identification of baijiu with different scent types.
Owner:TIANJIN POLYTECHNIC UNIV
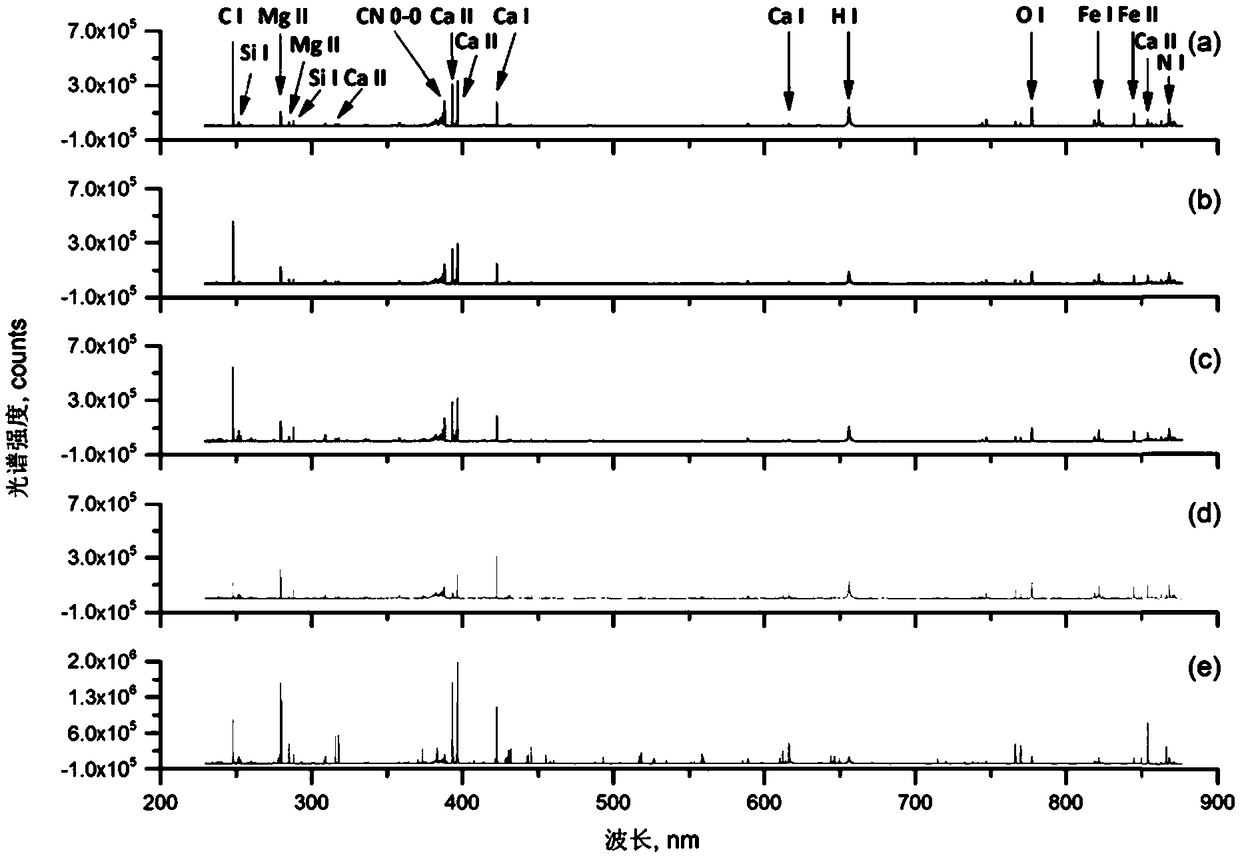
Identification method of radix puerariae powder of different production areas based on laser-induced-breakdown spectroscopy
InactiveCN109115748ARapid identification methodQuick checkAnalysis by thermal excitationMedicinePrincipal component analysis
The invention discloses an identification method of radix puerariae powder of different production areas based on a laser-induced-breakdown spectroscopy and belongs to the technical field of laser spectroscopy. The identification method comprises the following steps: (1) selecting pure radix puerariae powder samples of different production areas and adding classification tags of different production areas on the radix puerariae powder samples; (2) drying and grinding all the radix puerariae powder samples to prepare tablets; (3) collecting spectrum lines of the radix puerariae powder samples in the tablets by using the laser-induced-breakdown spectroscopy; (4) carrying out main component analysis of spectroscopic data of the radix puerariae powder samples and selecting characteristic wavelengths by using a main component analysis method; (5) dividing the radix puerariae powder samples of different production areas into modeling sets and prediction sets according to a uniform ratio; (6)taking the characteristic wavelengths as input, taking production area classification tags as output, respectively building discriminatory analysis models and comparing accuracy rate of the modelingsets and the prediction sets of different discriminatory analysis models to obtain an optimal discriminatory analysis model; and (7) identifying production areas of radix puerariae powder to be detected by using the optimal discriminatory analysis model.
Owner:ZHEJIANG FORESTRY UNIVERSITY
DNA barcode based premier for identifying garter snake, PCR-RFLP method, and kit
InactiveCN104164494AGuarantee the quality of medicinal materialsRapid identification methodMicrobiological testing/measurementDNA/RNA fragmentationDNA barcodingBarcode
The invention belongs to the technical field of traditional Chinese medicine species identification, and provides a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for identifying garter snake. The sequence of the provided PCR amplification primer is DK1-CO1:5'-CAA CTA ACC ACA AAG ACA TCG G-3', DK1-CO2:5'-CTT CTG GGT GGC CGA AAATC A-3'; the characteristic DNA base sequences of garter snake are 5'-ACTAGT-3', which is positioned at the 121st position to 126th position of the COI sequence, and 5'-GGAAACC-3', which is positioned at the 353rd position to the 359th position of the COI sequence; the restriction endonuclease Spe I can identify and cut the characteristic sequence 5'-GGAAACC-3', the restriction endonuclease BstE II cannot identify the characteristic sequence 5'-GGAAACC-3', and the two restriction endonucleases can only cut garter snake double-enzyme into two segments. The identification process comprises the following steps: extracting the DNA of a sample, using primers DK1-CO1 and DK1-CO2 to carry out PCR amplification, purifying the PCR products, using restriction endonucleases Spe I and BstE II to digest the PCR products; carrying out agarose gel electrophoresis to obtain the analysis result, comparing the analysis result with the PCR-RFLP characteristic result of garter snake to determine whether the provided sample is garter snake or not. The invention also provides a kit for the identification method. The identification method is accurate, rapid, and reliable, and can be widely applied to identification of traditional Chinese herbals.
Owner:晁志
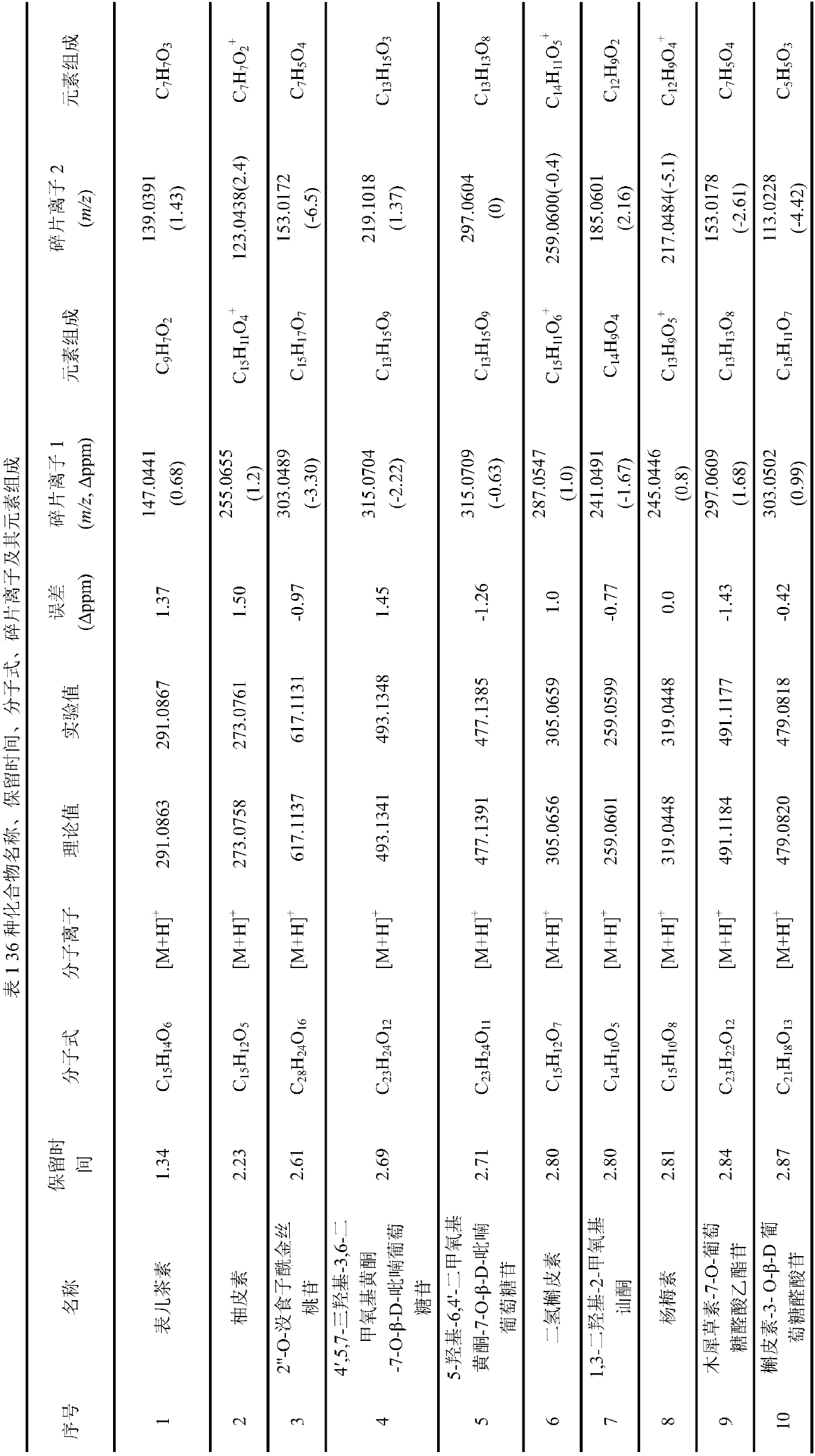
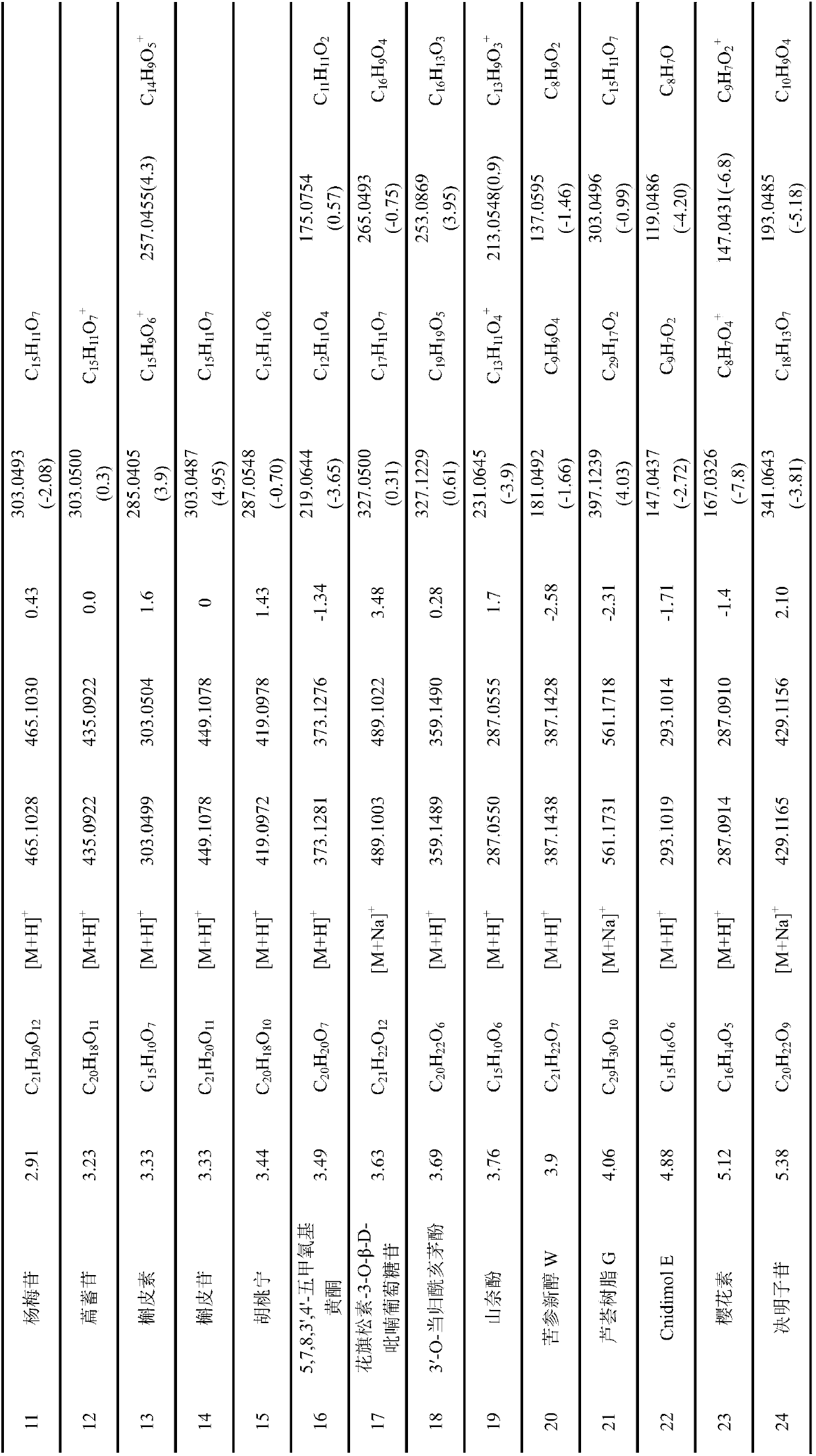
Method for analyzing flavonoid chemical components in walnut flowers
InactiveCN107607654ARapid identification methodEfficient identification methodComponent separationRefluxWalnut Nut
The invention discloses a method for analyzing flavonoid chemical components in walnut flowers. The method comprises the following steps: extracting the medicine walnut flowers by using an ethanol heating reflux method, concentrating, redissolving, carrying out ultrasonic treatment, and filtering to obtain a test solution; detecting the test solution by using an ultrahigh performance liquid chromatography-quadrupole-time of flight mass spectrometry so as to obtain secondary mass spectrometry information of compounds, and analyzing the information about the chemical components in the detectionresult. The method provided by the invention can be used for rapidly and efficiently analyzing the flavonoid compounds in the walnut flowers, thus providing a basis for the research and rapid and accurate identification of effective substances of the walnut flowers and being beneficial to the development and utilization of the walnut flowers.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE +1
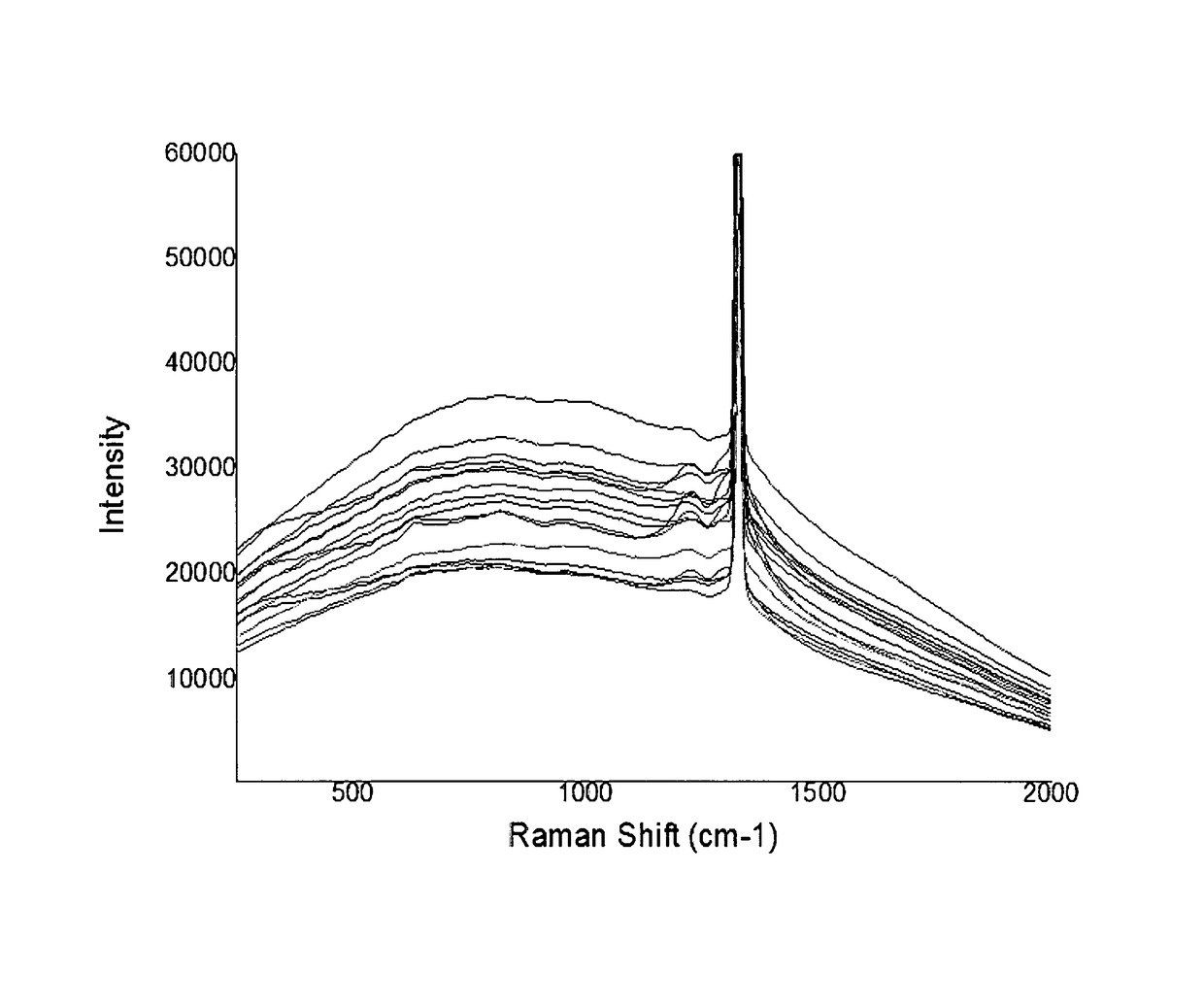
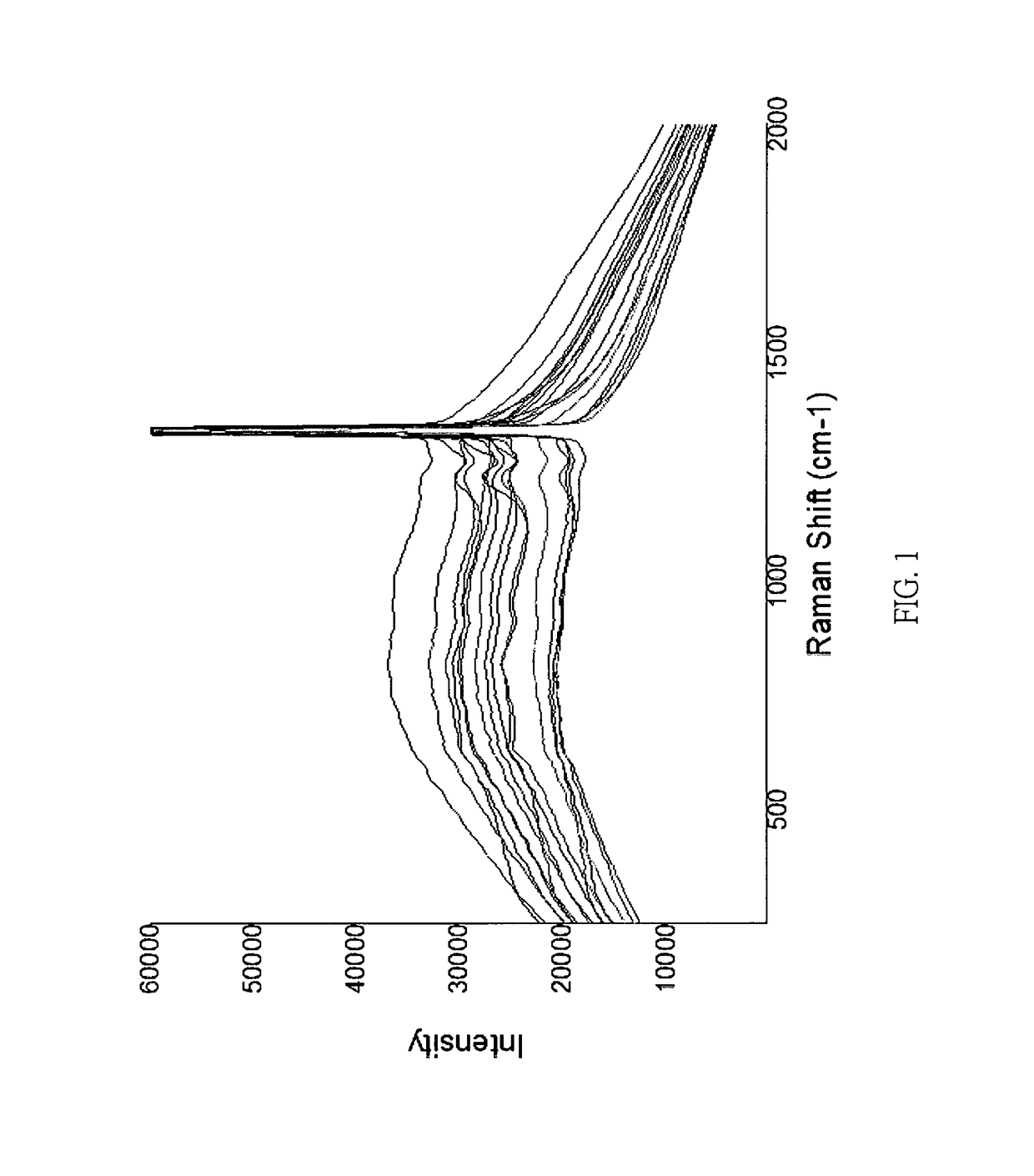
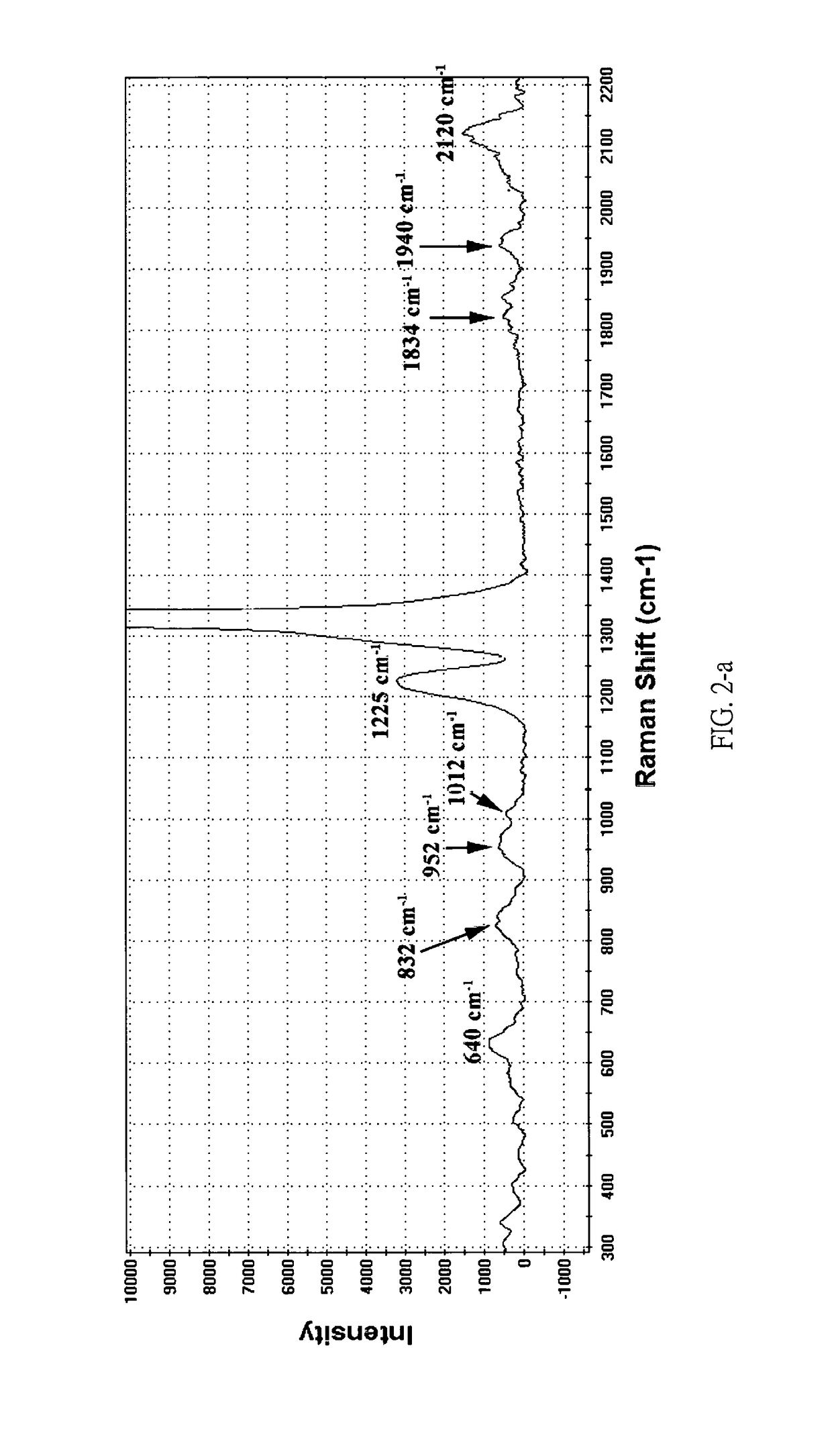
Rapid identification method of Argyle diamond's origin by charactristic Raman spectra
InactiveUS9625394B1Fast and easy differentiationRapid identification methodRaman scatteringInvestigating jewelsRapid identificationRefractive index
The origin of a gemstone often governs its value. Traditional jewelry appraisals attempted the recognition of the origin of a gemstone based on criteria of its physical properties include, but not limited to color, refractive indices, and microstructure. However, these criteria, in addition to its inclusions, are generally failed to resolve the locality and origin of a gemstone without doubt. Through careful examination and compilation of Raman spectroscopic data, Argyle pink diamonds can be classified as two types according to their characteristic Raman spectra. The method of the present invention provides sound basis for the rapid determination of the authenticity of the Argyle pink diamonds by Raman spectroscopy.
Owner:PAN DONG SHYOGN
Method for establishing number of Fusarium sp. copies in rhizosphere soil in growth period of transgenic rice by fluorescence real-time quantitative PCR (polymerase chain reaction)
InactiveCN102517385BImprove linearityHigh amplification efficiencyMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceDistilled water
The invention relates to a method for establishing number of Fusarium sp. copies in rhizosphere soil in growth period of transgenic rice by fluorescence real-time quantitative PCR (polymerase chain reaction), which comprises the following steps: establishing five soil sampling points on a diagonal line and marking and locating, and collecting rhizosphere soil samples at the above soil sampling points before seeding and at the growing period; extracting total DNA from the soil samples collected at the soil sampling points, dissolving the total DNA of the samples in double distilled water, and carrying out PCR amplification of the sample DNA at each period with a Fusarium-specific universal primer pair SEQ ID No.2 and SEQ ID No.3 to obtain a specific fragment of size 418bp (base pairs); directly measuring the number of copies of the corresponding Fusarium sp. with a fluorescence quantitative PCR instrument according to the variation of fluorescence signals and a standard curve; and summarizing the number of Fusarium sp. copies in the soil samples at different periods to obtain the number of Fusarium sp. copies in rhizosphere soil in the growth period of transgenic rice.
Owner:JIANGSU ACAD OF AGRI SCI
Multiplex PCR detection kit and identification method for mouse meat, goat meat and mutton
InactiveCN109988849ASpecificity Accurate Simultaneous DistinctionThe identification method is simpleMicrobiological testing/measurementPositive controlOligonucleotide Primer
The invention relates to the technical field of food detection, in particular to a multiplex PCR detection kit for mouse meat, goat meat and mutton, which is characterized by comprising an animal tissue mitochondrial DNA extraction system and three multiplex PCR reaction systems (a positive control reaction system, a negative control reaction system and an identification reaction system). The total PCR reaction system is 50 microliters, containing 5 microliters of reaction buffer solution, 2-5 microliters of dNTP, 23 microliters of MgCl, 1-5 microliters of Taq DNA polymerase, 0.5-2 microlitersof each primer, 1-3 microliters of DNA of a sample to be detected, and the balance being double distilled water. A multiplex PCR identification method for the mouse meat, the goat meat and the muttoncomprises the steps of designing three specific oligonucleotide primers by utilizing species specificity of cytochrome b, determining a reaction procedure, judging results and the like; the multiplexPCR identification method can simultaneously distinguish the specificity of the mouse meat, the goat meat and the mutton; and the multiplex PCR identification method for the mouse meat, the goat meatand the mutton has the advantages of simplicity, convenience, rapidness, reliable detection result and the like.
Owner:BEIHUA UNIV
PCR identifying method for nitrite bacteria
InactiveCN106755555AHigh sensitivityImprove accuracyMicrobiological testing/measurementBacteroidesElectrophoresis
The invention belongs to the technical field of bioengineering and provides a PCR identifying method for nitrite bacteria. A special feature gene-ammonia monooxygenase gene of nitrite bacteria is used as a template to design and synthesize a pair of specific primers; a nitrite bacteria genome DNA is used as a template for PCR amplification; the PCR amplification condition is optimized, and then the PCR amplification product is subjected to agarose level electrophoresis; and whether the bacterial strain belongs to nitrite bacteria or the nitrite bacteria is contained in the mixed flora can be judged according to the existence and size of the strip in a track lane. According to the method, the specific primer designed by taking the ammonia monooxygenase gene as a template is the PCR amplification primer; an experimental result is high in specificity; the experimental operation is simple; the result reliability is high; the identifying period of the nitrite bacteria is shortened in the manner of optimizing the PCR amplification condition; the amplification time is above 26% shorter than the conventional PCR amplification time; and a quick and effective identifying method is supplied for identifying the nitrite bacteria population.
Owner:CHAMBROAD CHEM IND RES INST CO LTD
Rapid identification method for fingerprint
ActiveUS10503951B2Rapid identification methodPrint image acquisitionTransmissionPattern recognitionComputer vision
A rapid identification method for fingerprint first provides a fingerprint identification apparatus having a fingerprint sensing area and divides the fingerprint sensing area into fingerprint sensing sub-regions. In a registration stage, the method performs fingerprint sensing for the entire fingerprint sensing area to obtain fingerprint image for a whole fingerprint sensing area, fingerprint minutiae and relevant locations for the fingerprint minutiae and then pre-stores those data. In an identification stage, the method performs fingerprint sensing on a part of the fingerprint sensing sub-regions for a user to be identified and detects fingerprint minutiae and relevant locations for the fingerprint minutiae in the part of the fingerprint sensing sub-regions. The method compares the fingerprint minutiae and relevant locations detected in the identification stage with respect to the corresponding fingerprint minutiae and relevant locations in the registration stage in order to determine whether the user can be granted with access right.
Owner:SUPERC TOUCH CORP
Multiple PCR detection kit for kangaroos, guinea pigs and sewer rats and method
PendingCN111118176AThe identification method is simpleReliable test resultsMicrobiological testing/measurementCytochrome bAnimal science
The invention relates to the technical field of food detection and discloses a multiple PCR detection kit for kangaroos, guinea pigs and sewer rats. The multiple PCR detection kit is characterized bycomprising an animal tissue mitochondria DNA extraction system and an authenticating reaction system (being similar to a positive control solution reaction system and a negative control solution reaction system). An authenticating reaction total system is 50 microliters and contains 5 microliters of a reaction buffer solution, 2-5 microliters of dNTP, 23 microliters of MgCl, 1-5 microliters of TaqDNA polymerase, 2.25 microliters of a kangaroo primer, 3 microliters of a guinea pig primer, 2 microliters of a sewer rat primer, 2-6 microliters of DNA of a to-be-detected sample and the balance ofdouble distilled water. A multiple PCR authentication method for kangaroo meat, guinea pig meat and sewer rat meat comprises the steps of designing three specific oligonucleotide primers by virtue ofspecies specificity of cytochrome b, determining reaction procedures, determining a result, and the like; and the specificities of three rat meat can be simultaneously distinguished; and the method has the advantages of rapidness, reliable detection result and the like.
Owner:BEIHUA UNIV
Molecular biology method of early embryo sex determination of transgenic dairy cattle
InactiveCN102477459AIncrease production capacityRapid identification methodMicrobiological testing/measurementEmbryoBiology
The invention belongs to the technical field of animal reproduction, and in particular relates to the field of early embryo sex determination of transgenic dairy cattle. The invention discloses a molecular biology method of early embryo sex determination of transgenic dairy cattle, which comprises the following steps of: incising 4-6 cells from an embryo to be detected by applying a micromanipulation technology, washing the incised cells by using sterile ultra-pure water and putting the cells in a PCR (Polymerase Chain Reaction) tube; adding a proper amount of the sterile ultra-pure water, boiling and placing in trash ice; cooling, centrifuging at a low temperature, and taking a supernatant liquor to perform PCR reaction; detecting PCR amplification products by using agarose gel electrophoresis so as to obtain an electrophoretogram; observing the electrophoretogram, determining the embryo as a bull when a 131bp stripe and a 250bp stripe are simultaneously existent in the electrophoretogram, and determining the embryo as a cow when only a 250bp stripe is existent in the electrophoretogram.
Owner:TIANJIN MENGDE GROUP
Seedling stage identification method of almond fragrance black tea resources
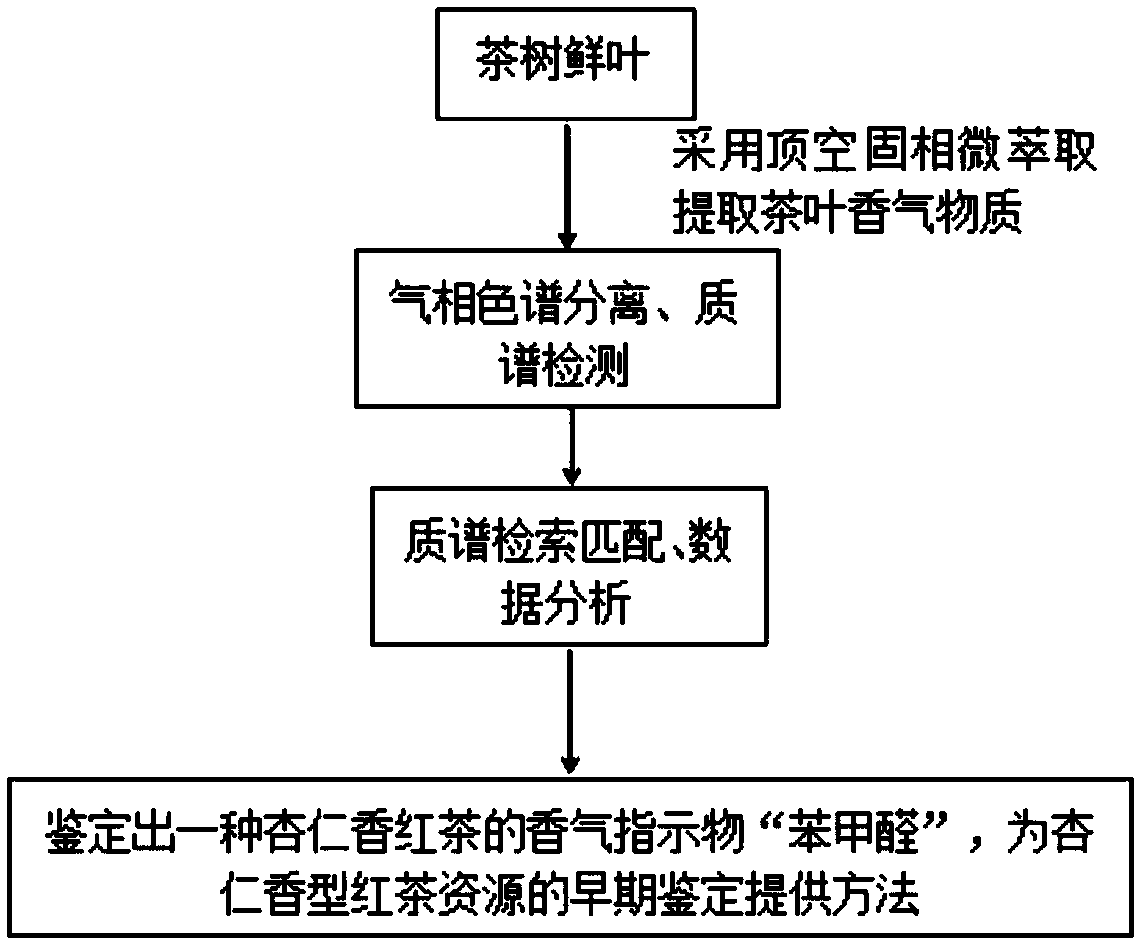
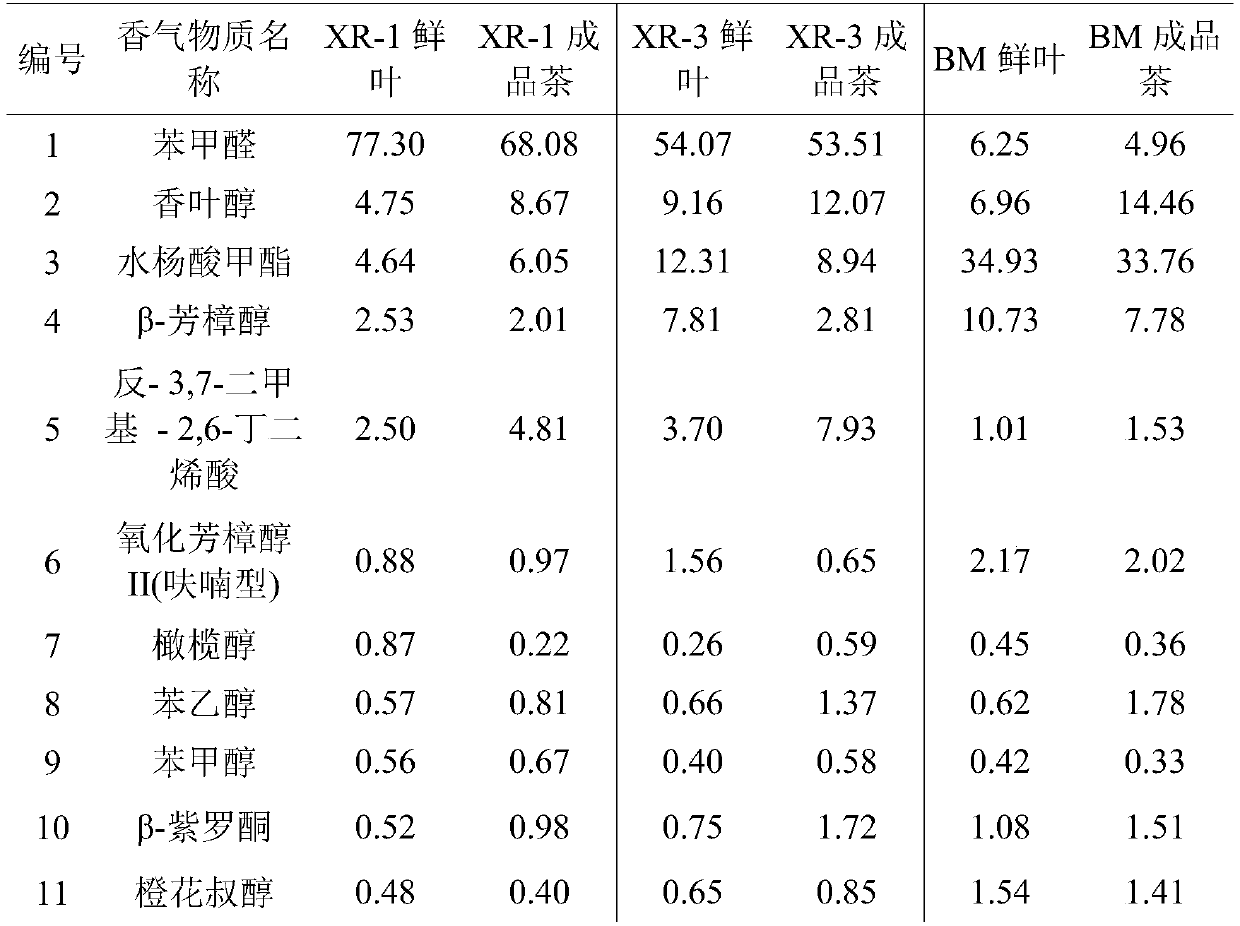
InactiveCN109632999AEasy extractionRapid identification methodComponent separationBenzaldehydeDesorption
The invention relates to a seedling stage identification method of almond fragrance black tea resources. The method comprises the following steps: (1) extracting aroma substances in fresh tea leaves by using a headspace solid-phase microextraction method: after freeze-drying and crushing a fresh leaf sample, adding water of 90-100 DEG C, soaking at 40-70 DEG C, and inserting an extraction head into a space above the liquid level for aroma adsorption; (2) after the adsorption is finished, inserting an extraction head into a gas chromatography sample inlet for desorption, carrying out gas chromatography separation, carrying out mass spectrometry detection, and analyzing data; and (3) determining the relative percentage content of benzaldehyde in the aroma substances. According to the method,any fresh tea leaves can be detected, whether the black tea product has the aroma characteristics of almond aroma or not can be judged without tea trial-production property study, and a rapid and accurate identification method is provided for seedling stage identification of almond aroma black tea resources.
Owner:TEA RES INST GUANGDONG ACAD OF AGRI SCI
Method for detecting quantity of pseudomonas fluorescent in rhizospheric soil during growth period of transgenic wheat by virtue of fluorescent quantitative PCR (Polymerase Chain Reaction)
ActiveCN103397099BConsistent repetitive responseEasy to distinguishMicrobiological testing/measurementFluorescence/phosphorescencePeak valuePseudomonas
The influence of transgenic crops on rhizospheric microorganisms in soil is an important aspect in environmental biosafety research. The invention establishes a detection system of Real-Time QPCR (Real-Time Fluorescent Quantitative Polymerase Chain Reaction)) by carrying out absolute quantification on the quantity of pseudomonas fluorescens in rhizospheric soil during the growth period of transgenic disease-resistant wheat. By virtue of application of the reaction system, the absolute quantification is carried out on the copy number of the pseudomonas fluorescens in the rhizospheric soil during the whole growth period of the transgenic wheat. A result shows that the designed primer has better specificity; the intervals of cycle thresholds (Ct values) of plasmids with various gradient standards in an amplification curve of Real-Time QPCR are uniform, the peak value of a melting curve is more obvious, and for the standard curve, the correlation coefficient R2 is equal to 0.99905, the slope is minus 3.203, and the amplification efficiency E is equal to 100%. In the seeding stage, seedling establishment stage, watery stage and mature stage of the transgenic wheat, the quantity of the pseudomonas fluorescens in the rhizospheric soil takes on a trend of gradually rising.
Owner:JIANGSU ACAD OF AGRI SCI
Molecular beacon probe for rapid detection of rifampin-resistant mycobacterium tuberculosis and detection method thereof
InactiveCN109880923AIdentification method is simpleRapid identification methodMicrobiological testing/measurementDNA/RNA fragmentationTrue positive rateBiology
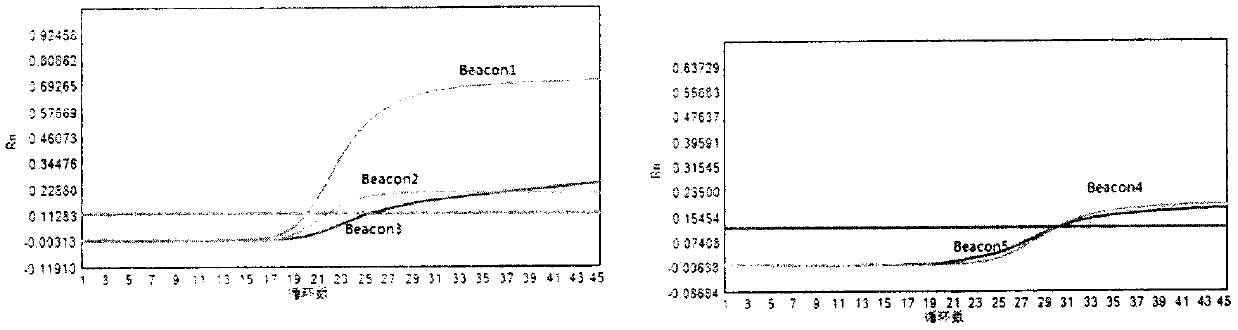
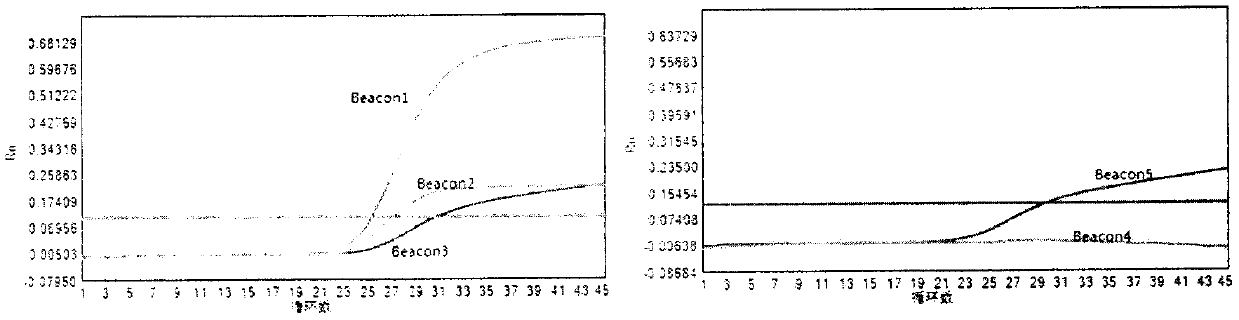
The invention discloses a molecular beacon probe for rapid detection of rifampin-resistant mycobacterium tuberculosis and a detection method thereof. The molecular beacon probe comprises Beacon 1, Beacon 2, Beacon 3, Beacon 4 and Beacon 5. All 81 sites of a rifampin-resistant rpoB gene are covered by the five probes, so that the rifampin-resistant mycobacterium tuberculosis can be detected. The molecular beacon probe has high detection sensitivity and specificity, and can rapidly and specifically detect the rifampicin-resistant mycobacterium tuberculosis. The invention also discloses a detection method of the molecular beacon probe for rapid detection of the rifampin-resistant mycobacterium tuberculosis.
Owner:北京岱美仪器有限公司
Method for rapidly identifying specificity of transgenic herbicide-resistant soybean ZH10-6 transformant
PendingCN112029892AEfficient identification methodRapid identification methodMicrobiological testing/measurementDNA/RNA fragmentationMultiplexPcr method
The invention discloses a method for quickly identifying the specificity of a transgenic herbicide-resistant soybean ZH10-6 transformant based on a multiplex PCR method. The method comprises the following steps of designing and synthesizing a specific primer by using G2-epsps and GAT exogenous insertion gene sequences of the transgenic herbicide-resistant soybean ZH10-6, and constructing a transformant specificity identification multiplex PCR reaction system through primer concentration optimization combination. Four pairs of transformant specific sequences are designed aiming at a unique expression mode (two repeated G2-epsps genes and one GAT gene are inserted to form two expression cassettes and four boundaries) of an exogenous insertion sequence of the transgenic herbicide-resistant soybean ZH10-6, and a soybean endogenous Lectin gene is introduced to construct a quintuple PCR reaction system. And an economic, efficient, accurate and reliable high-throughput technical means is provided for specific identification of the transgenic transformant based on the exogenous target gene.
Owner:天津市农业科学院 +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com