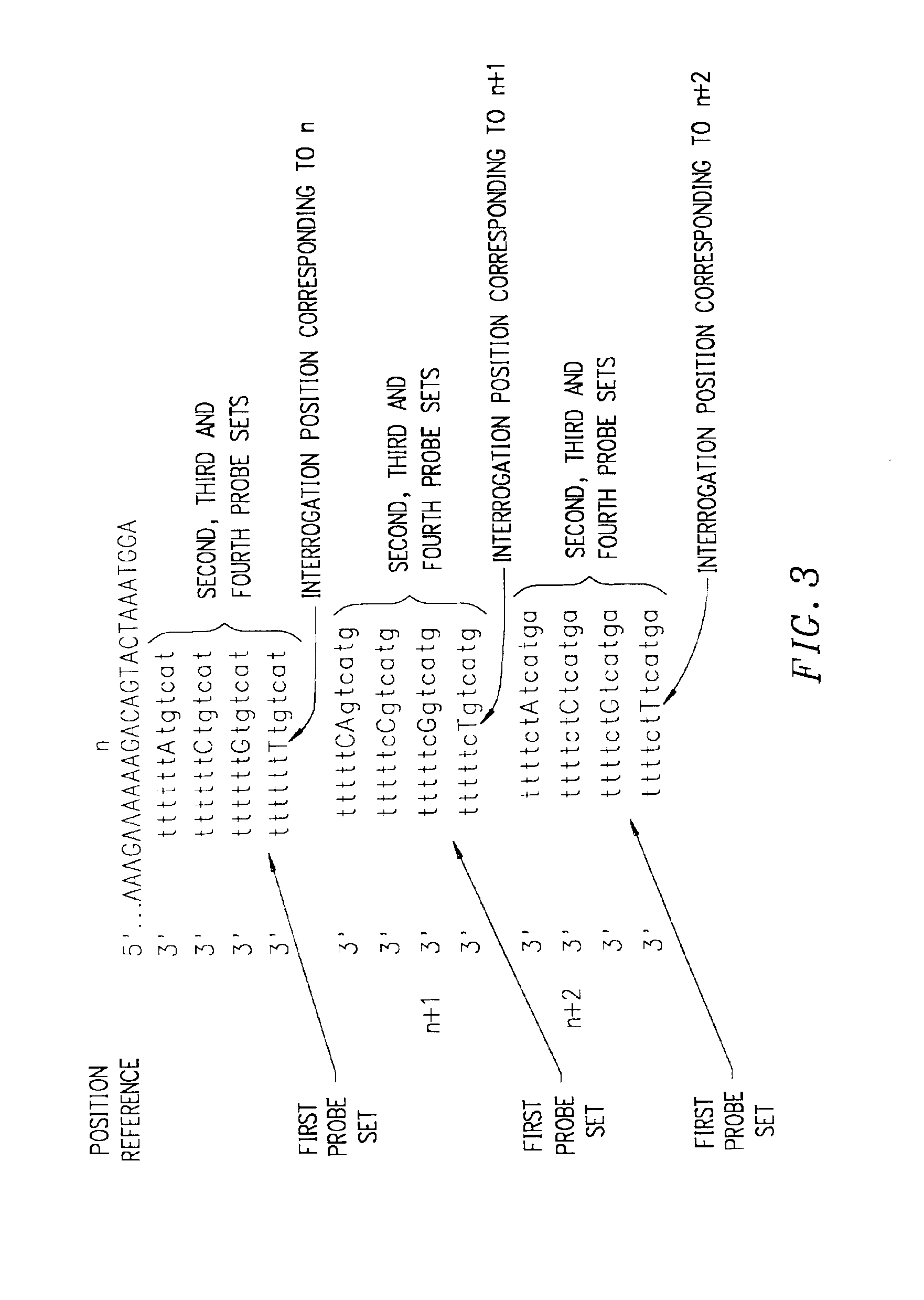
Patents
Literature
556 results about "Species identification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Identification of Species. Species identification is generally based at least initially on basic phenotypic characteristics such as size, shape and colouration. Related species generally have common characteristics allowing convenient groupings on various levels from kingdom to genus.
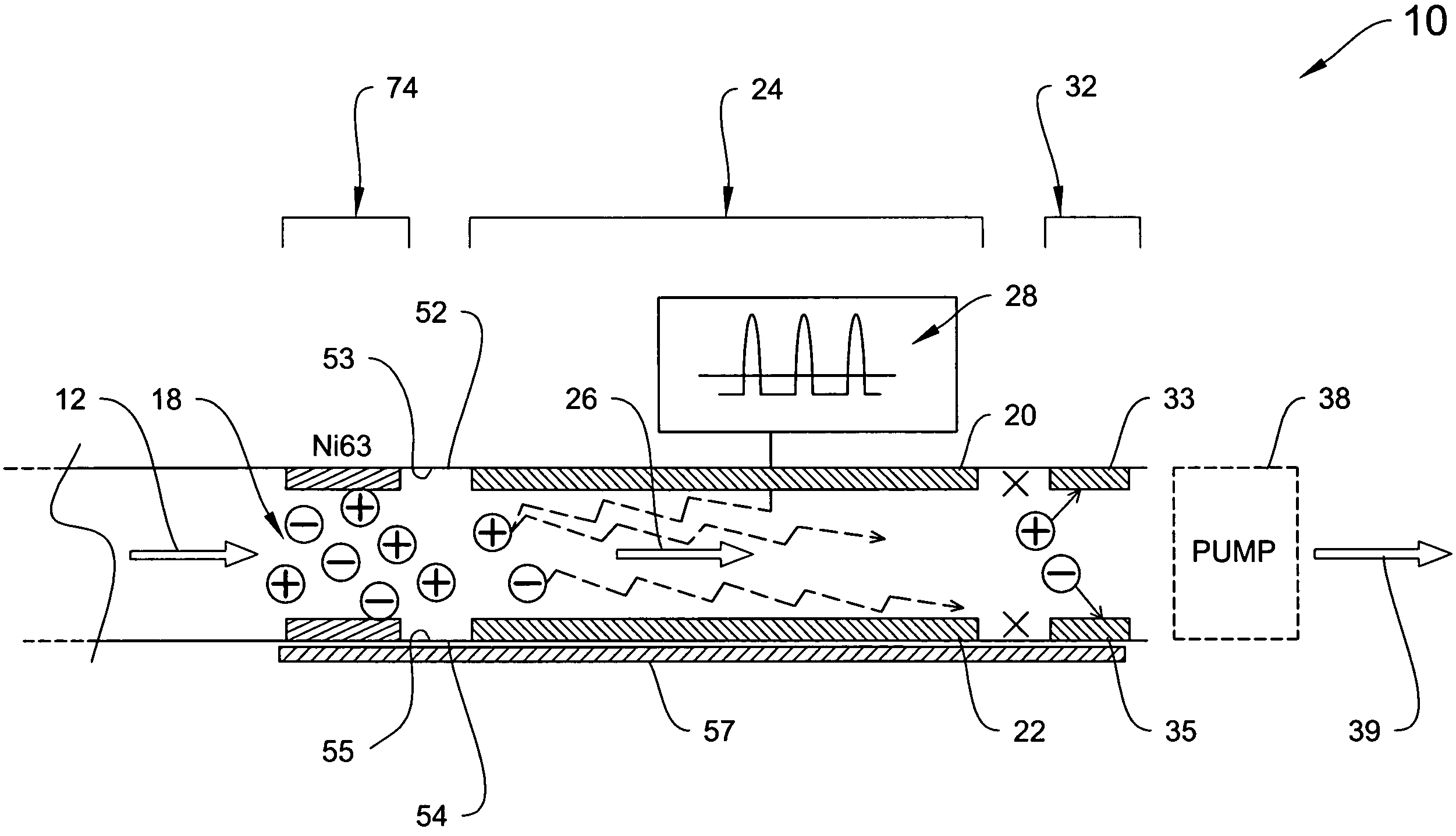
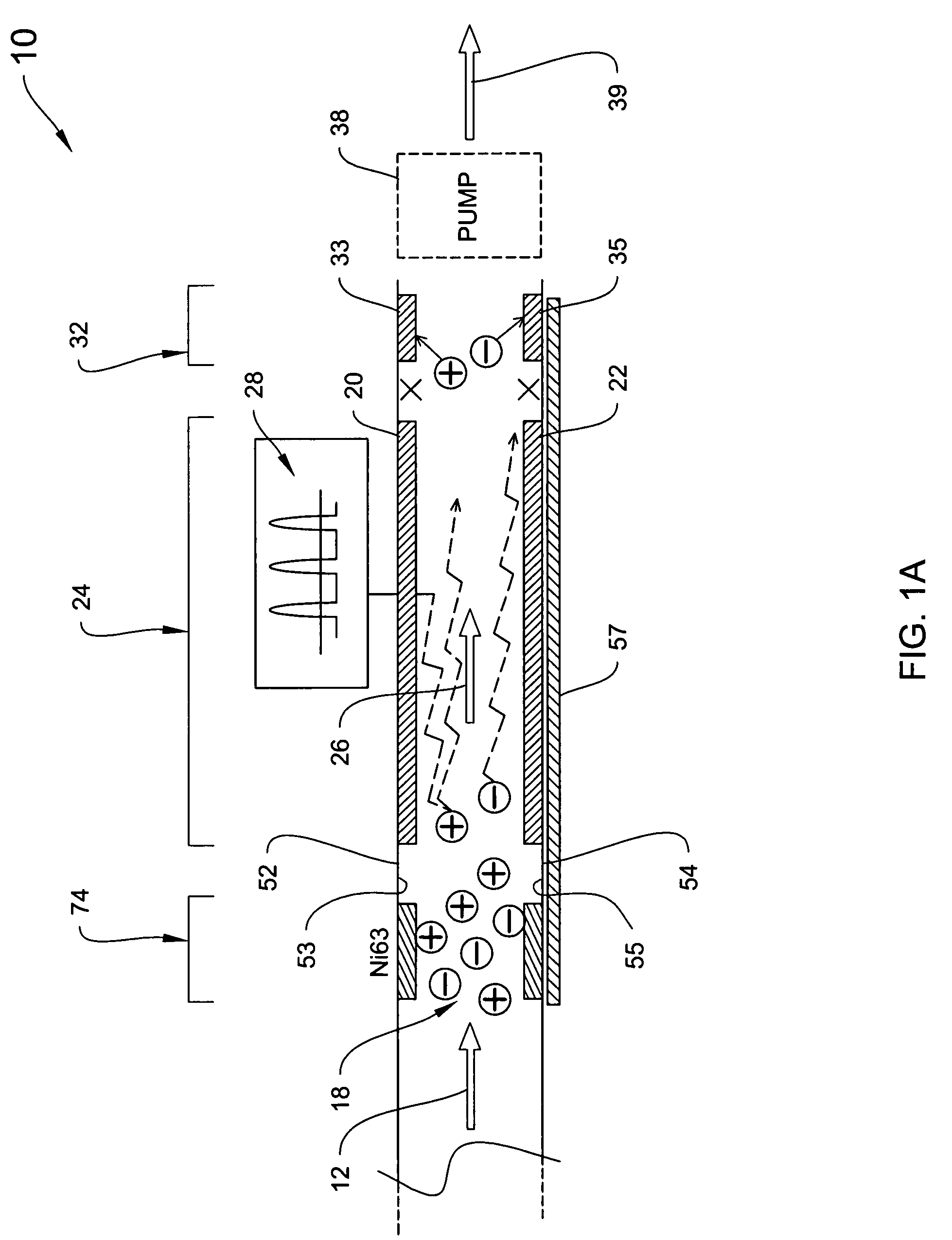
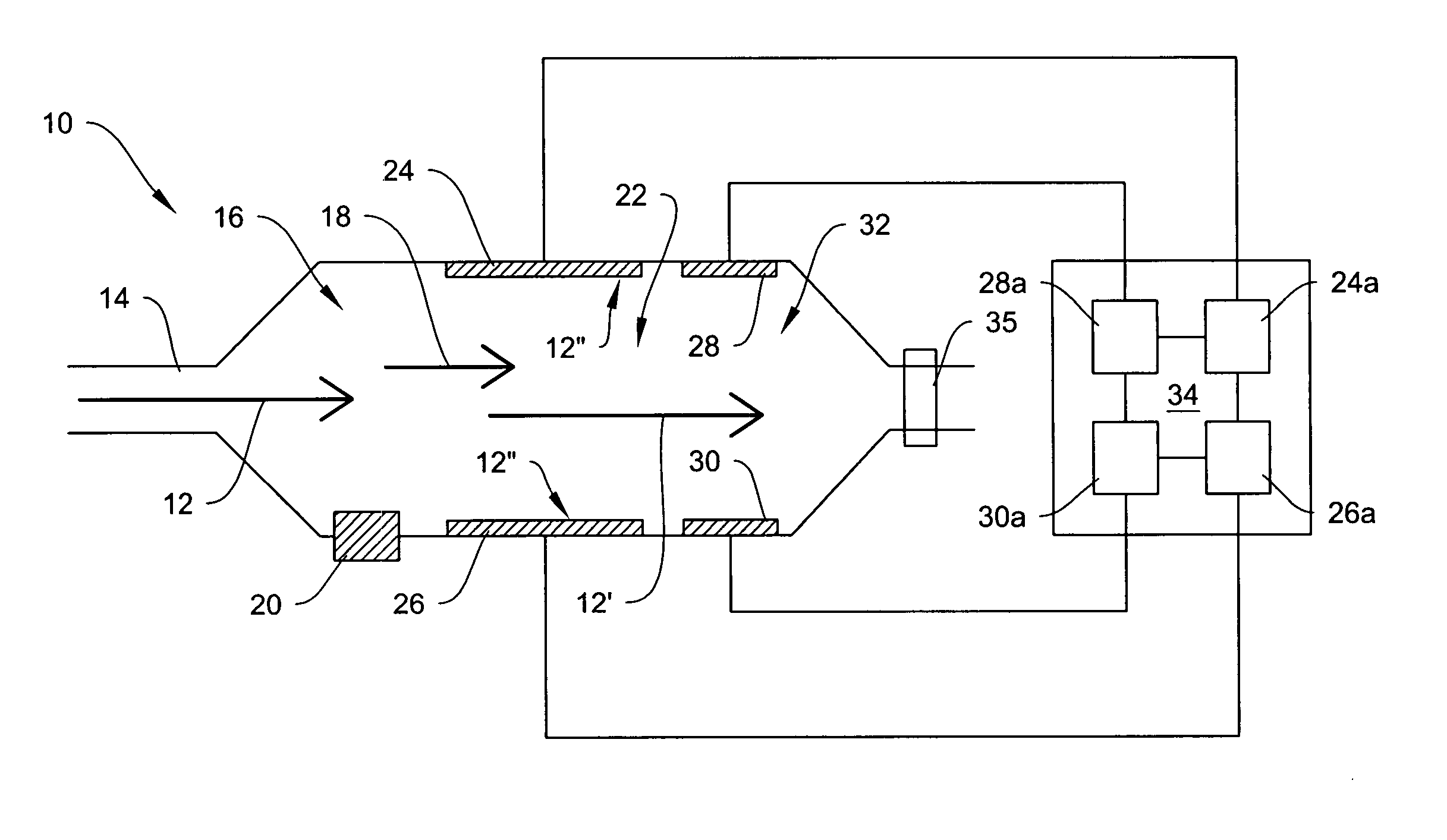
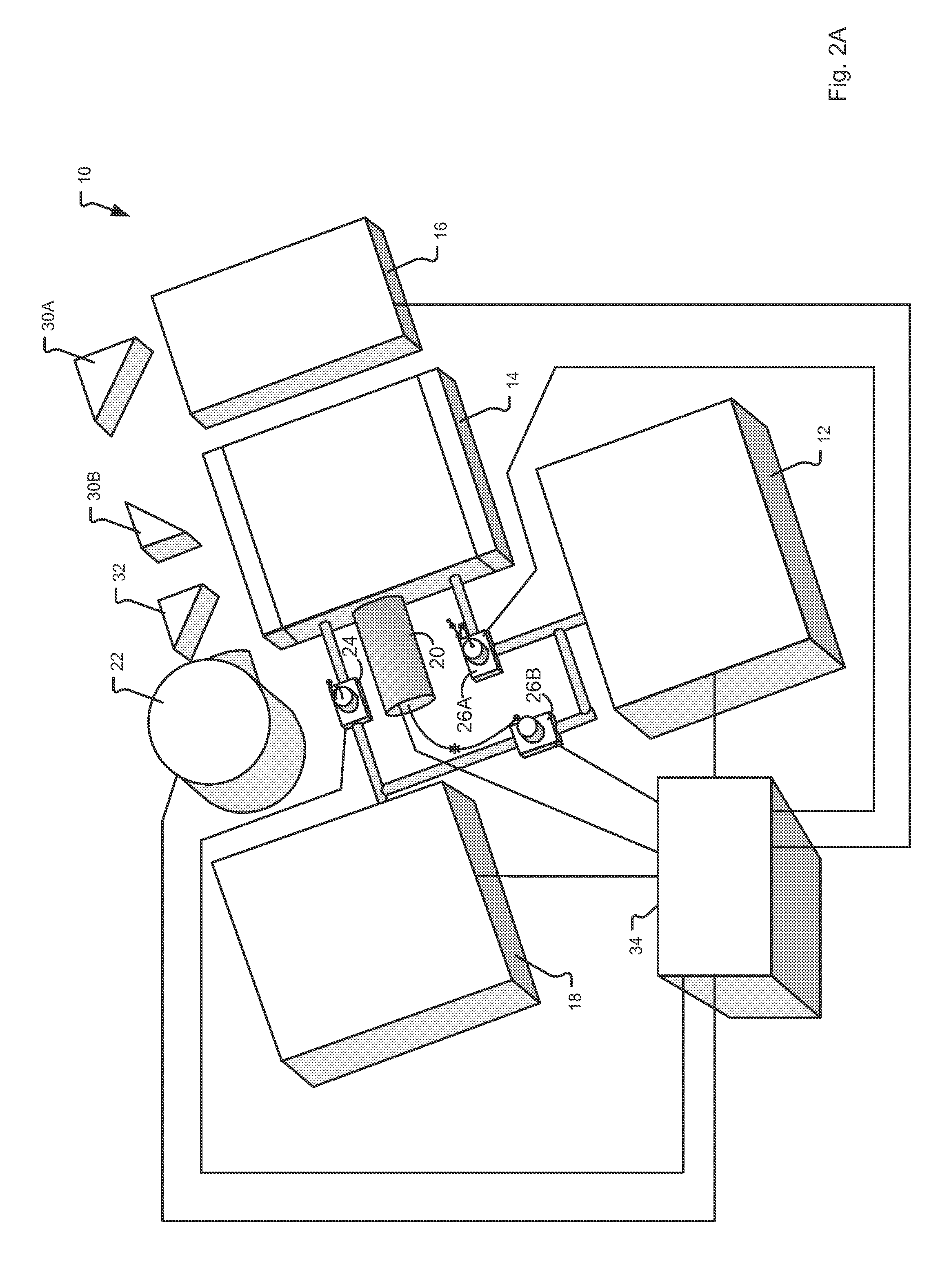
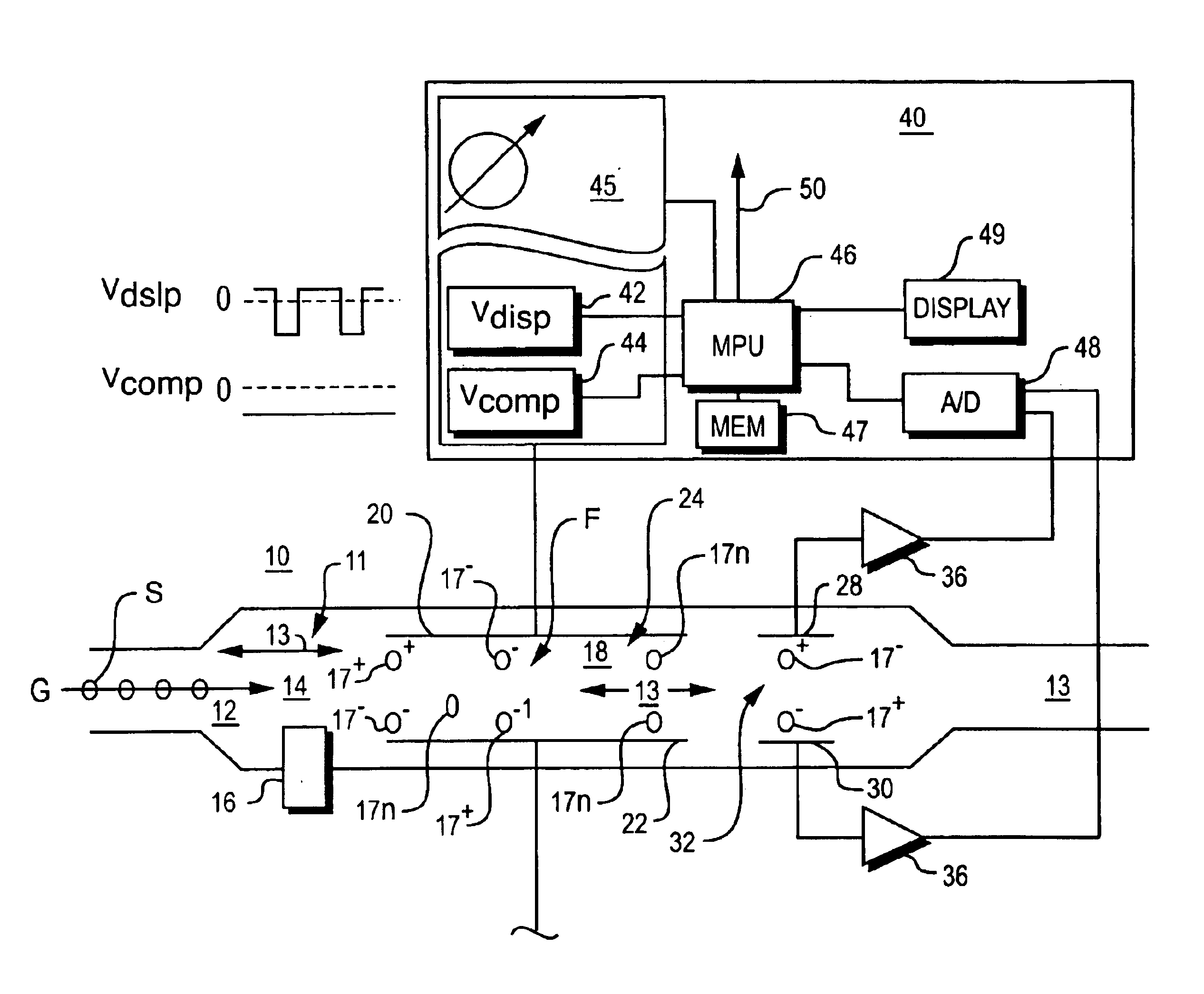
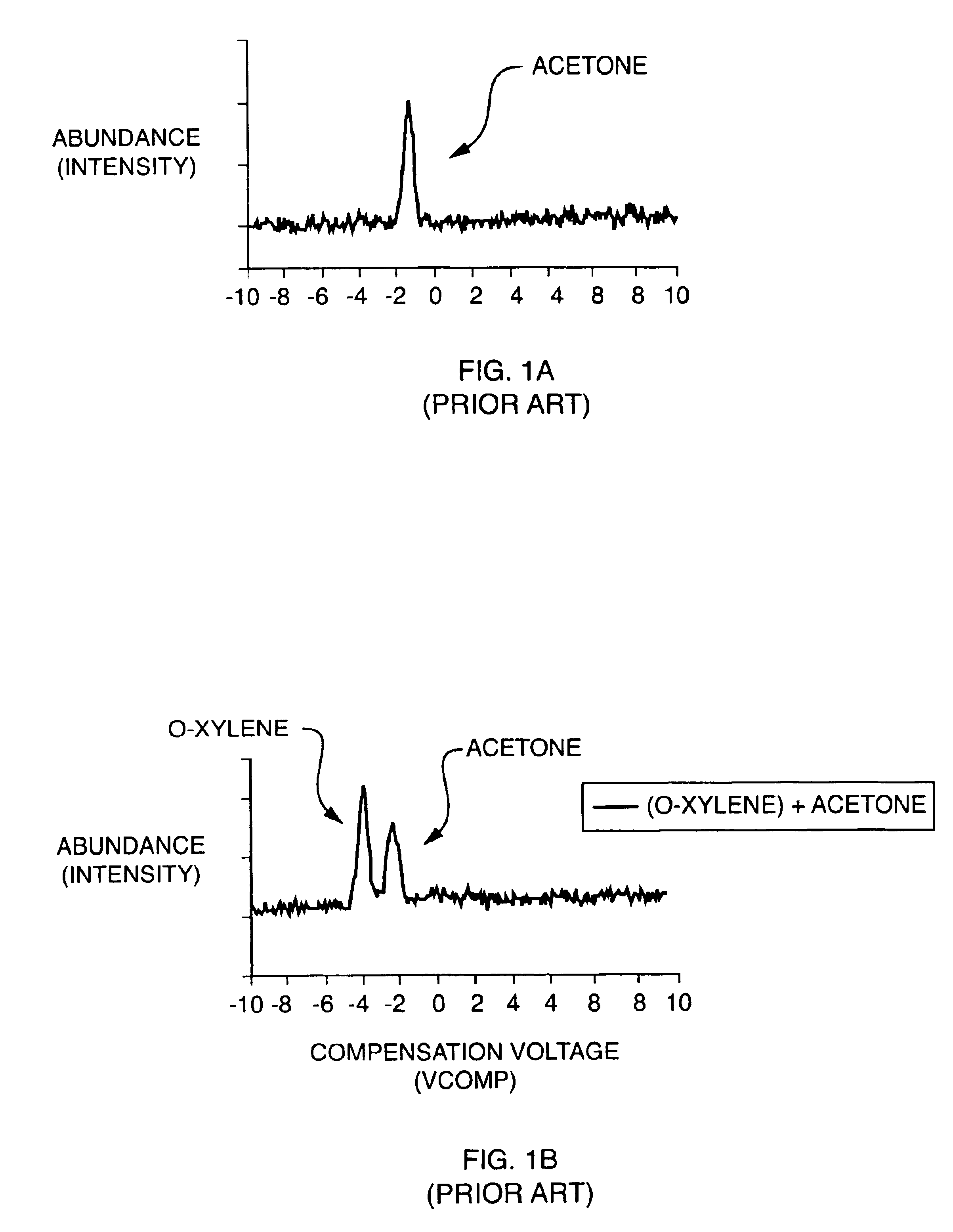
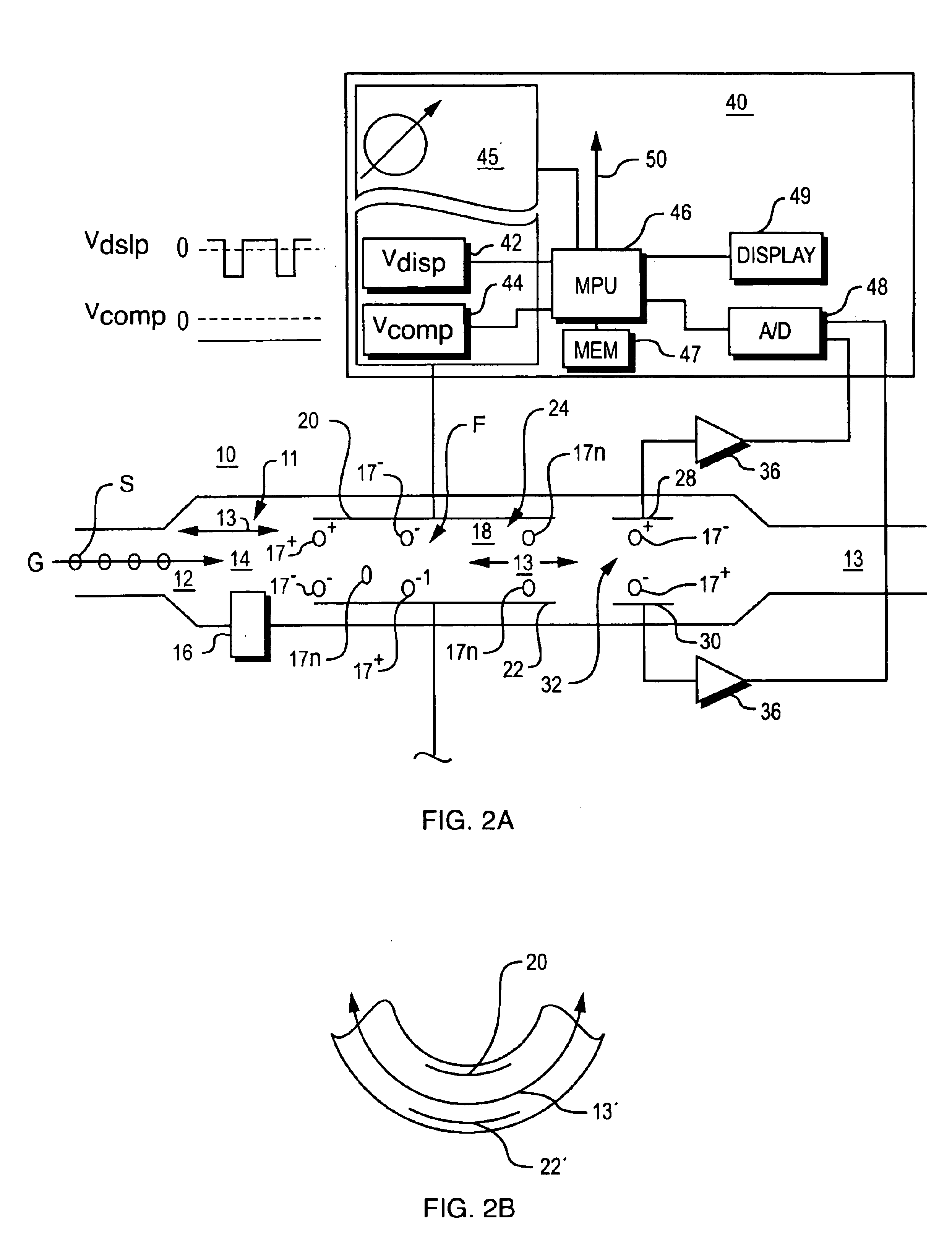
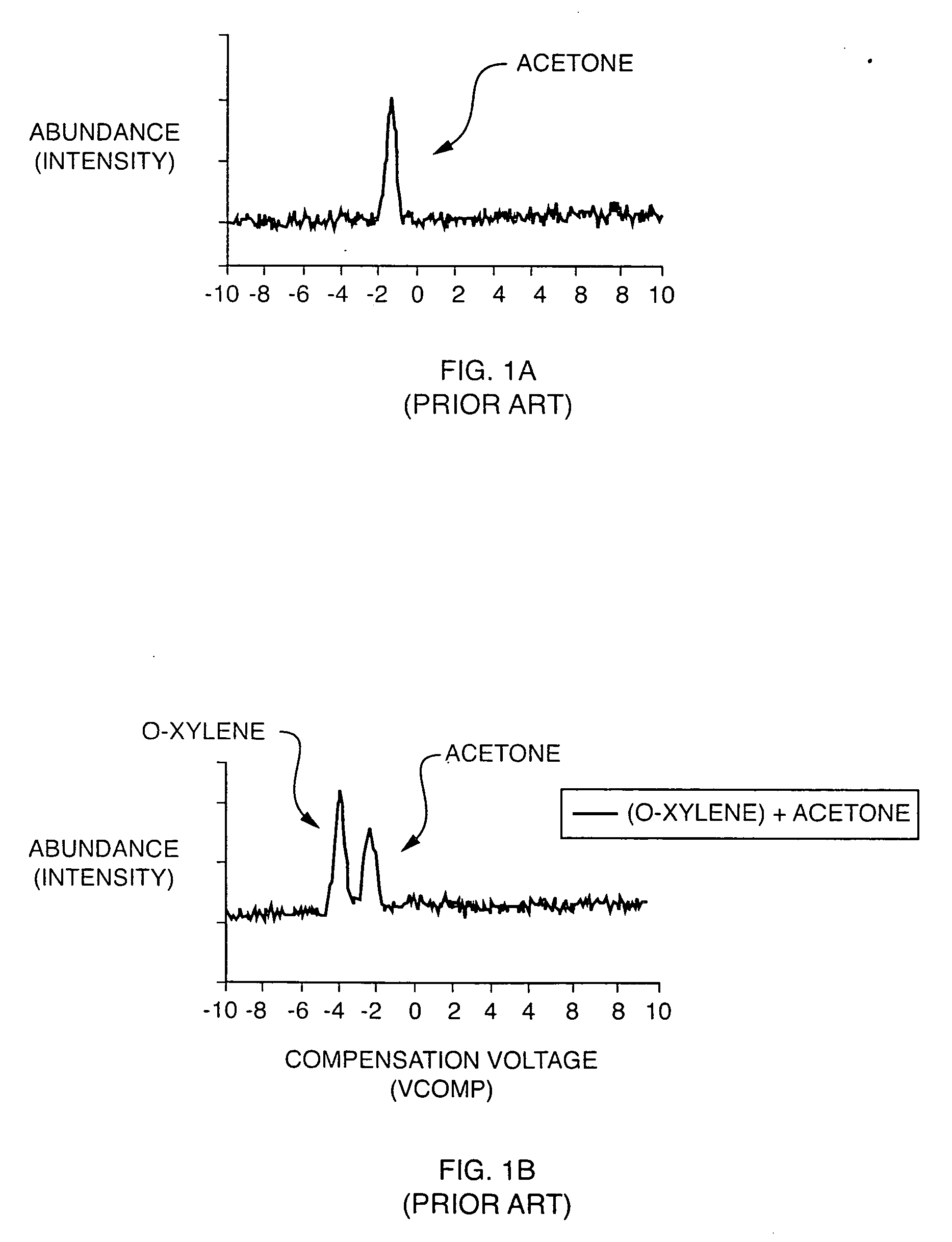
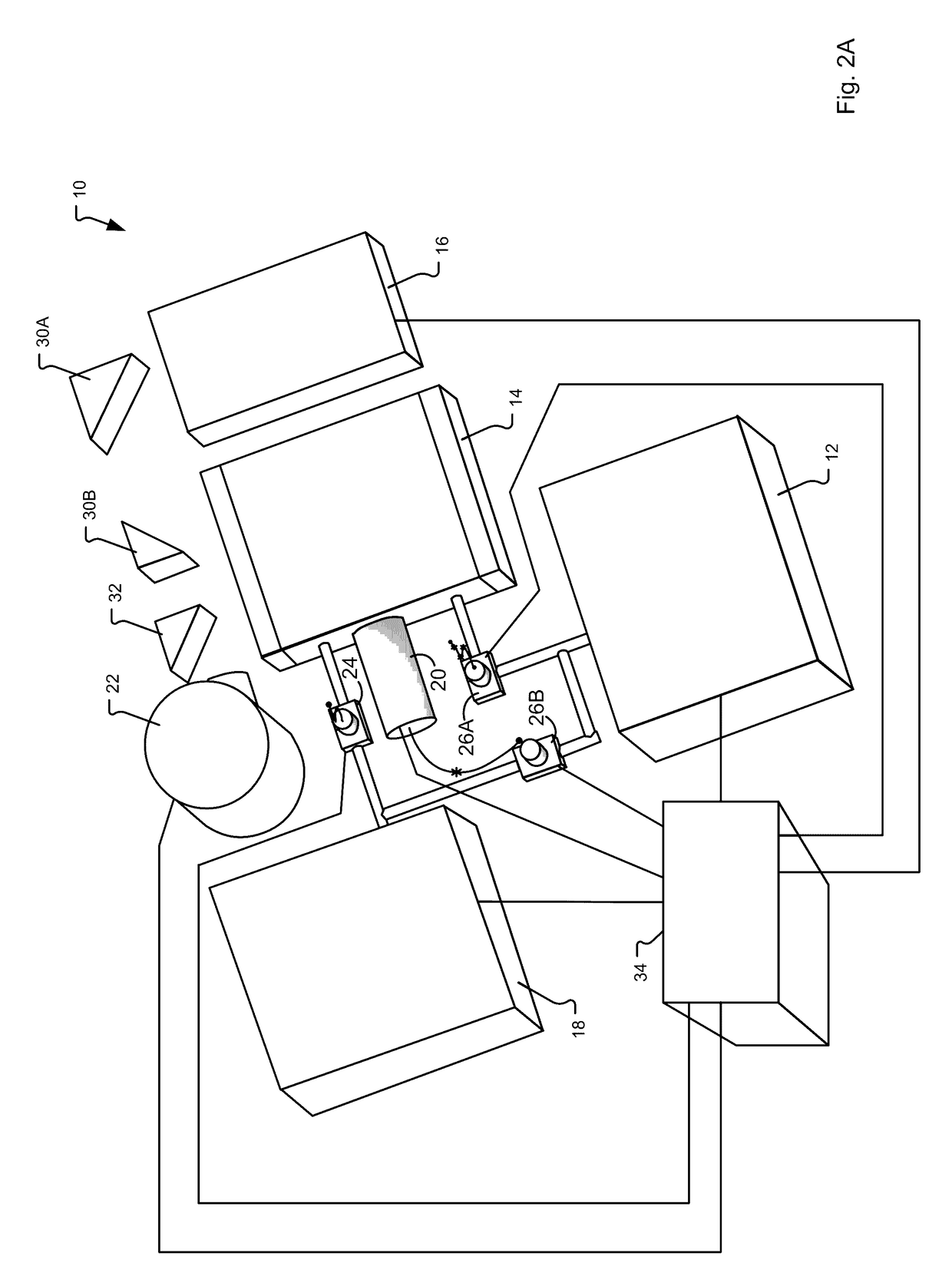
Explosives detection using differential ion mobility spectrometry
InactiveUS20050133716A1Easy to separateHigh resolutionTime-of-flight spectrometersFuel testingOptical spectrometerPhysical chemistry
System for control of ion species behavior in a time-varying filter field of an ion mobility-based spectrometer to improve species identification for explosives detection.
Owner:DH TECH DEVMENT PTE
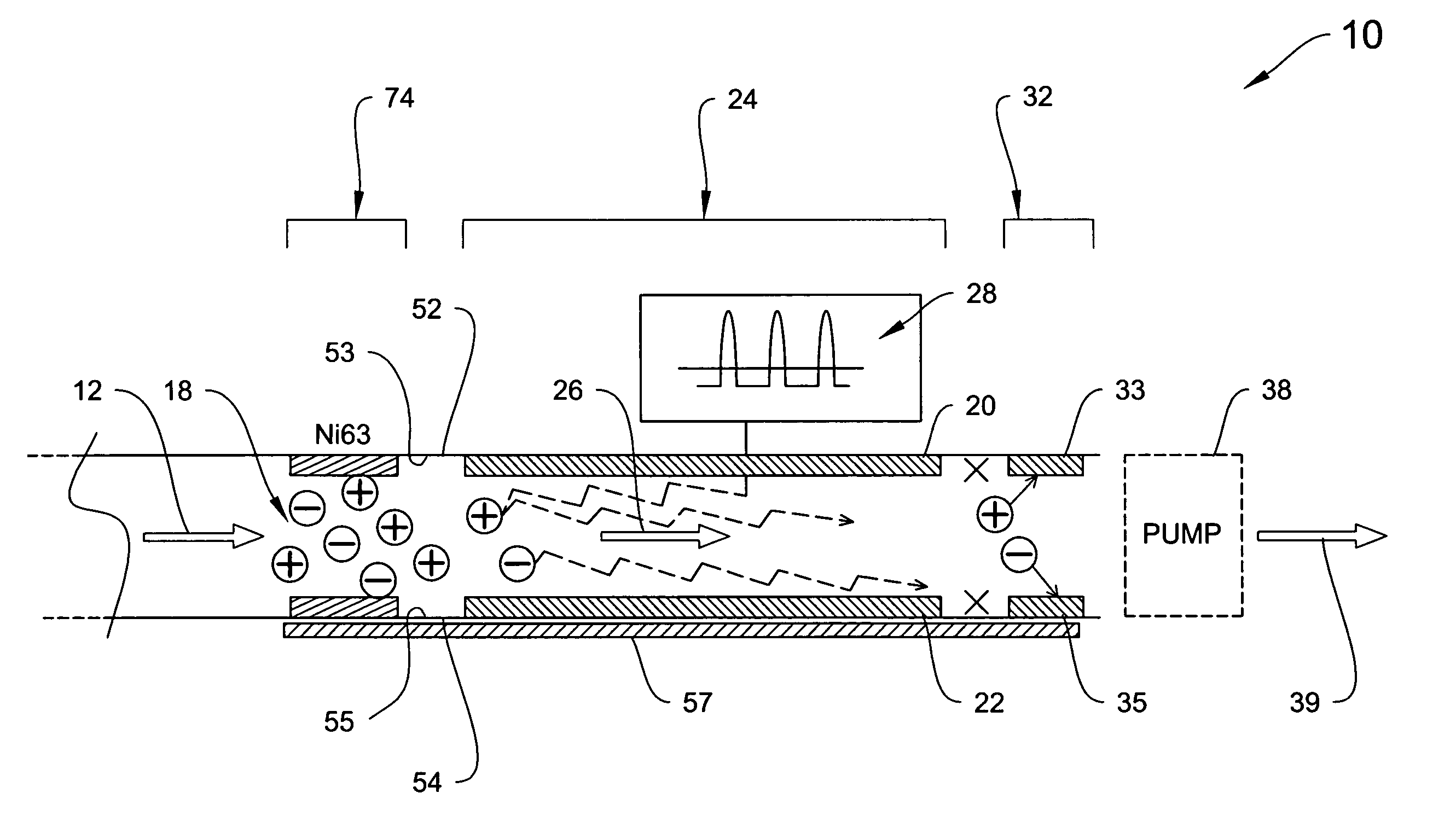
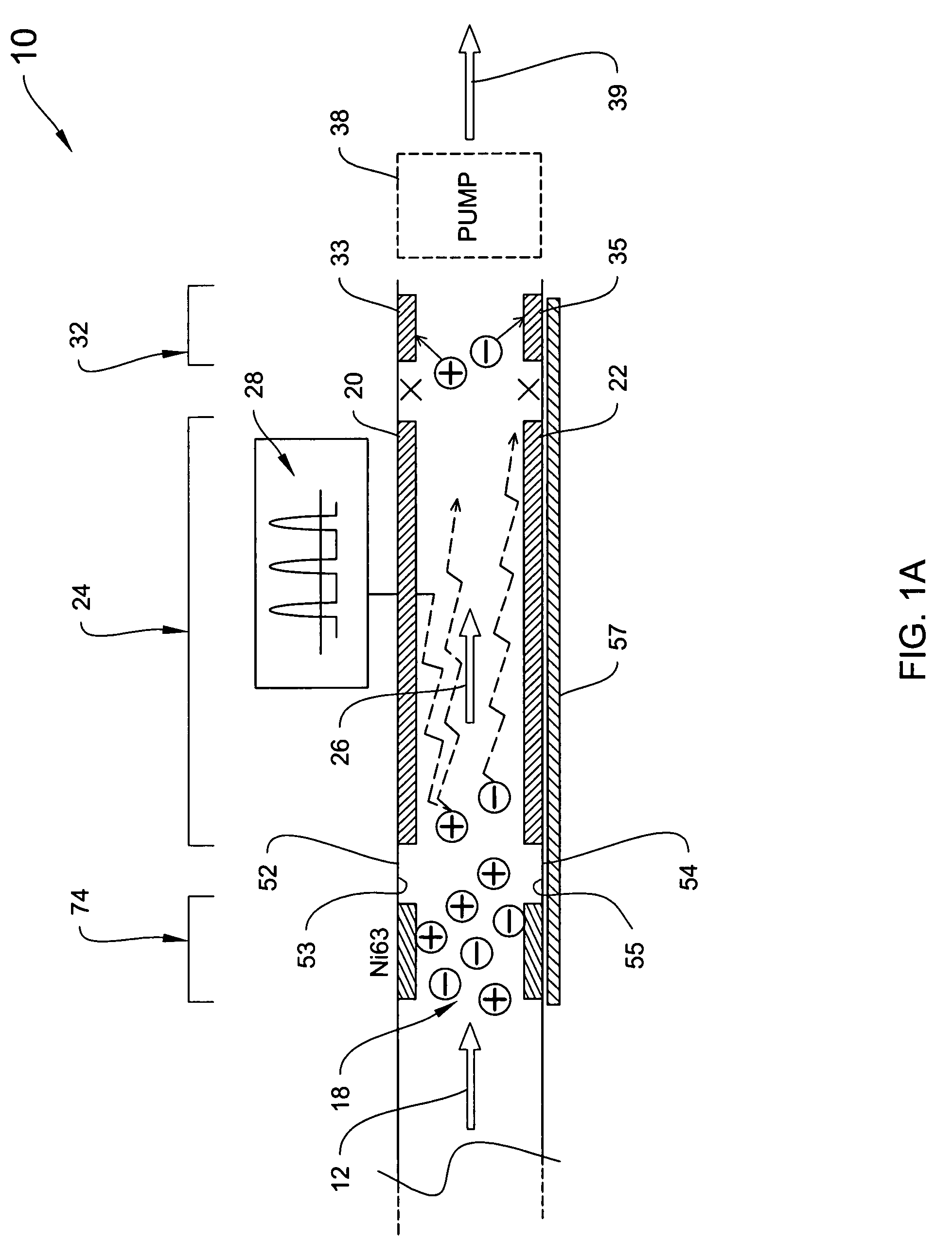
System for trajectory-based ion species identification
InactiveUS20050029449A1High sensitivityReduced false positiveStability-of-path spectrometersMaterial analysis by electric/magnetic meansChemical compoundPhysical chemistry
Method and apparatus for field ion mobility spectrometry for identification of chemical compounds in a sample by trajectory of sample ions in a transverse electric field.
Owner:DH TECH DEVMENT PTE
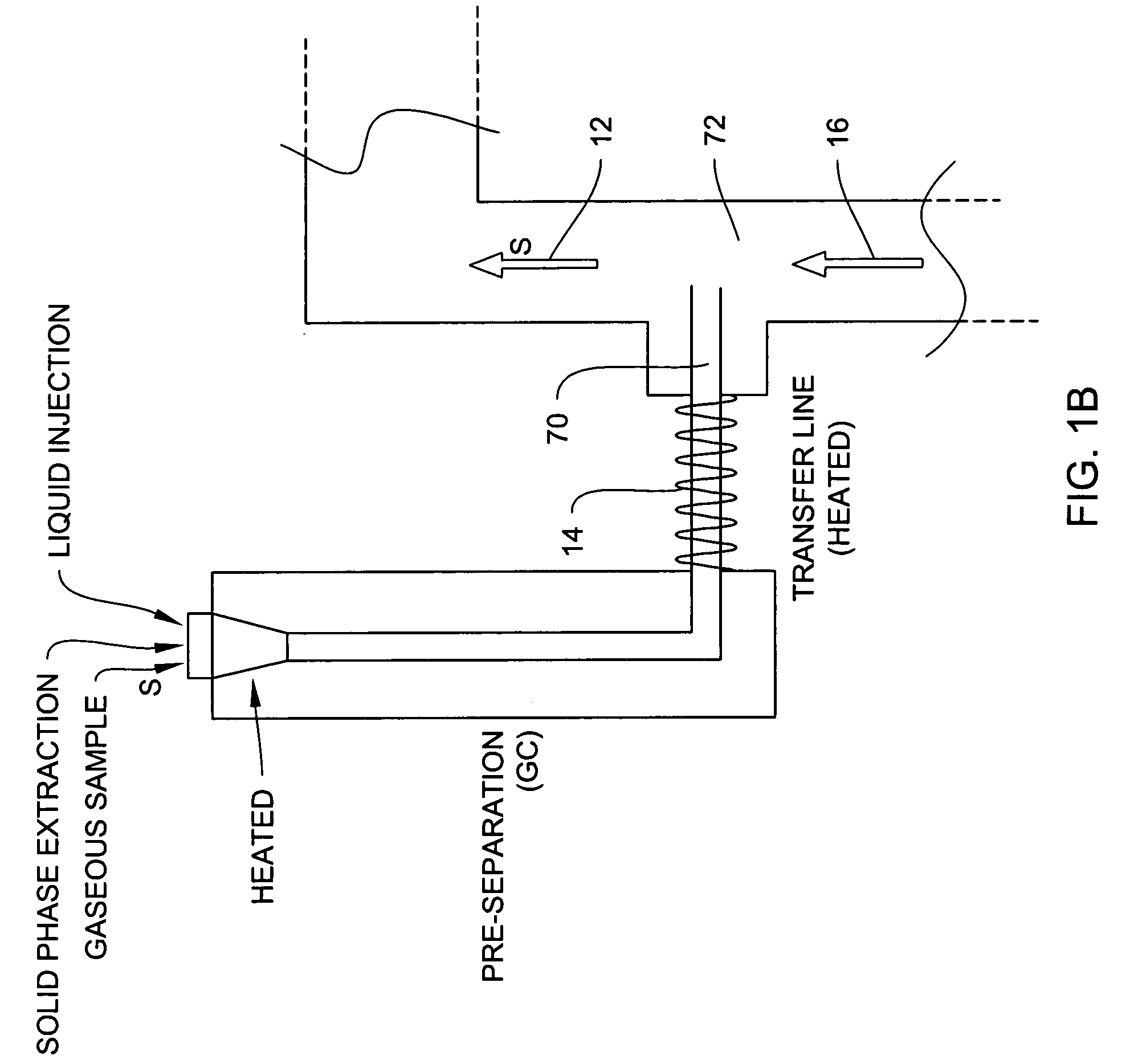
Process and system for rapid sample analysis
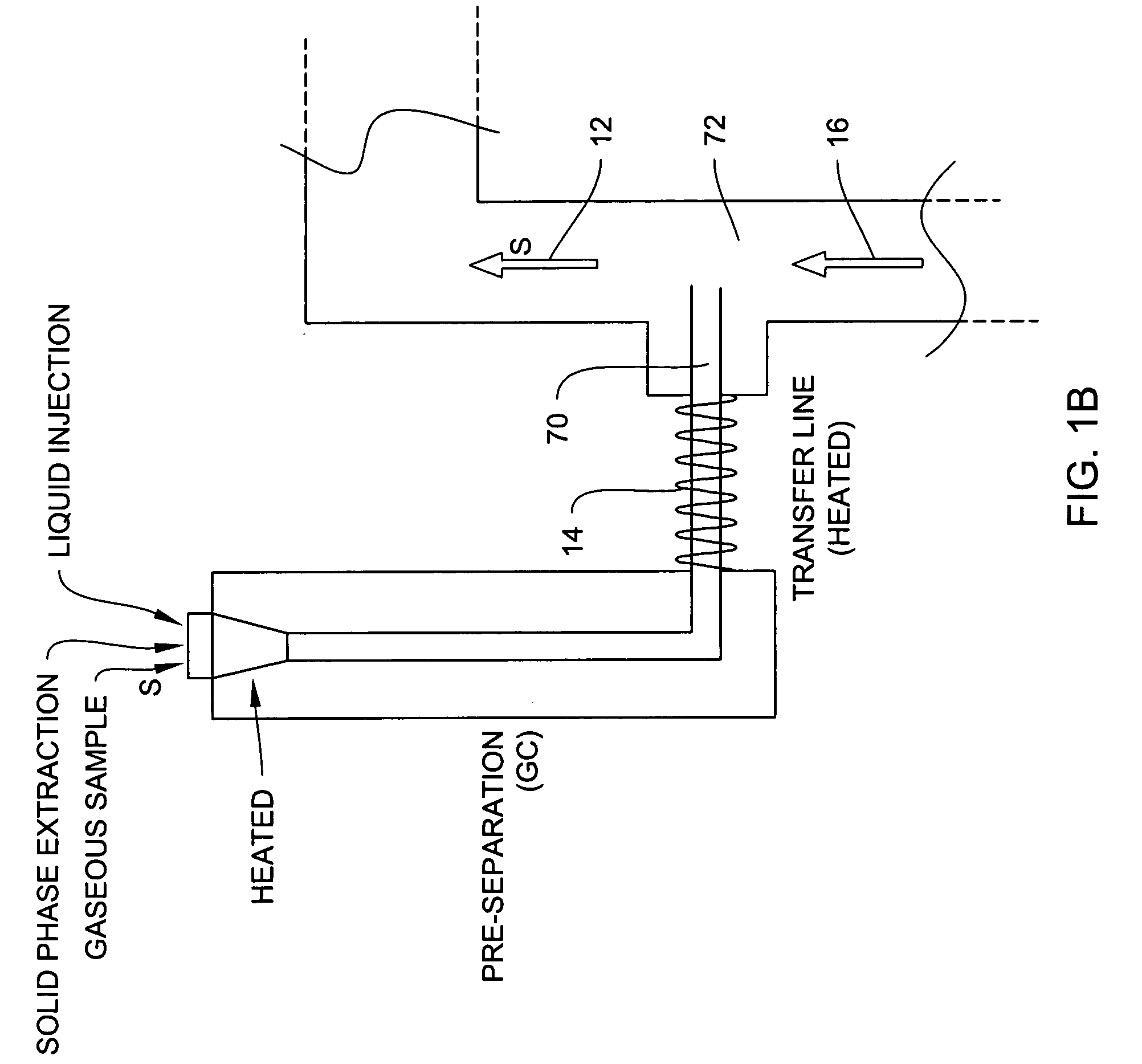
ActiveUS20150260695A1Low MDLsLow detection limitRadiation pyrometryComponent separationDistillationElectromagnetic radiation
Components resolved in time by a separator accumulate in a sample cell and are analyzed by electromagnetic radiation-based spectroscopic techniques. The sample cell can be configured for multiple path absorption and can be heated. The separator can be a gas chromatograph or another suitable device, for example a distillation-based separator. The method and system described herein can include other mechanical elements, controls, procedures for handling background and sample data, protocols for species identification and / or quantification, automation, computer interfaces, algorithms, software or other features.
Owner:MLS ACQ INC
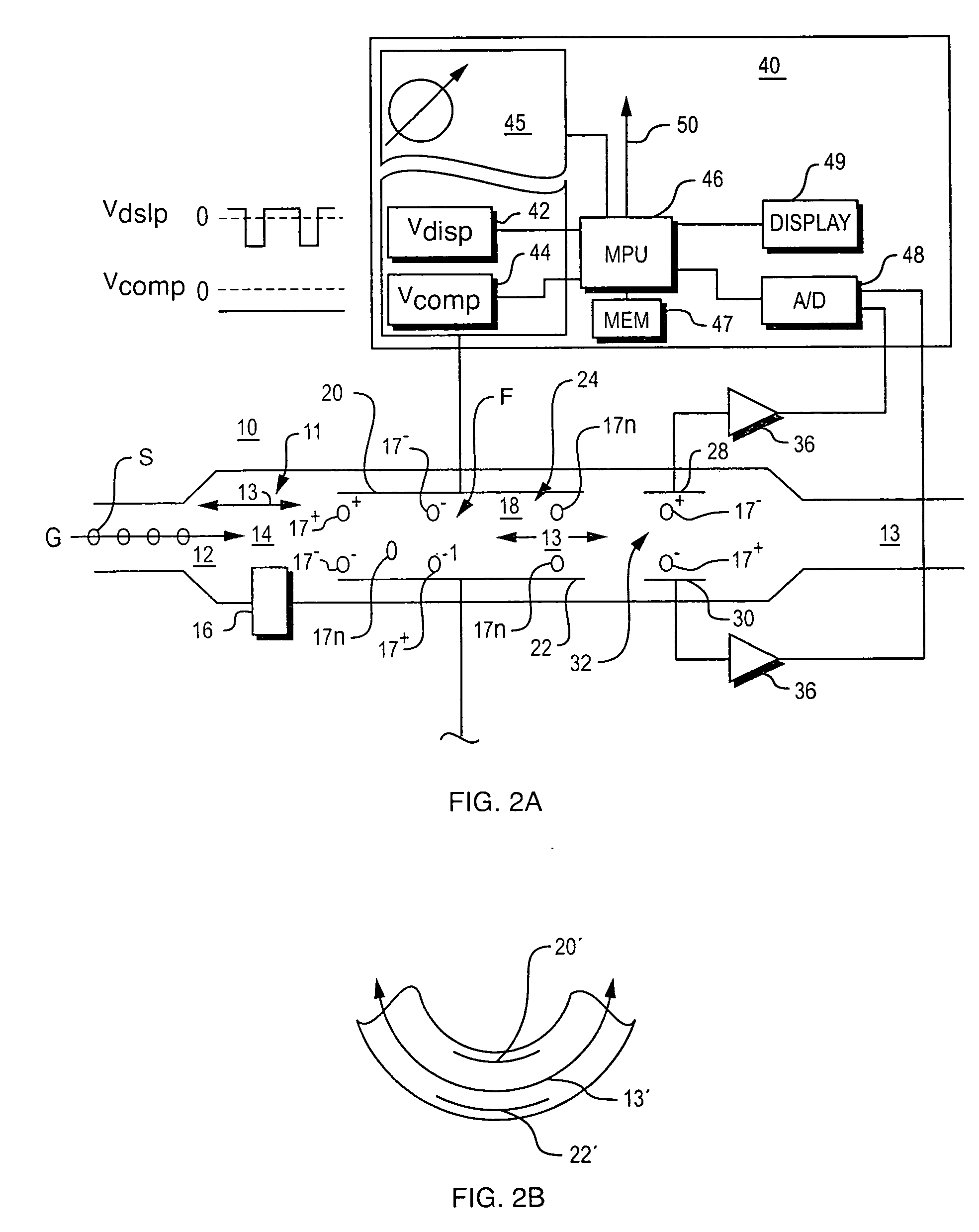
Method and apparatus for control of mobility-based ion species identification
InactiveUS7005632B2Improve system performanceGuaranteed accuracyTime-of-flight spectrometersFuel testingBiologySpectrometer
Owner:DH TECH DEVMENT PTE
Building method for rhubarb medicinal material trueness/falseness and species prediction model
The invention provides a building method for a rhubarb medicinal material trueness / falseness and species prediction model. According to the model, category identification of quality rhubarb, fake rhubarb and three rhubarb species is realized on the basis of a gas chromatography-mass spectrum technology in combination with partial least square discriminant analysis. The building method comprises the following steps: (1) collecting samples; (2) preparing the samples; (3) acquiring GC-MS data; (4) performing data processing and multivariate analysis, wherein cluster analysis and principal component analysis are performed by comparing the differences of chromatographic behaviors of rhubarb of different sources; (5) building the prediction model; and (6) verifying the model. Two built category prediction models are proved to have 100 percent identification accuracy and prediction accuracy. Through adoption of the method, trueness / falseness and species identification of rhubarb are realized by modeling, so that a reliable conclusion and high integrity are achieved; the defects of an existing method are overcome; and a more feasible and reliable new thought is provided for the trueness / falseness and species identification of rhubarb medicinal materials and decoction pieces.
Owner:青岛市食品药品检验研究院
Fluorescently-labeled gene multiplex amplification method for identifying sources of goat, sheep, pig and duck meat simultaneously
ActiveCN103397101AAvoid cross interferenceHigh detection throughputMicrobiological testing/measurementDNA/RNA fragmentationCytosineFluorescence
The invention provides a fluorescently-labeled gene multiplex amplification technology for detecting sources of goat, sheep, pig and duck meat simultaneously. A method comprises the steps that mitochondria 16S rRNA (ribosomal Robonucleic Acid) and Cyt (Cytosine) b genes of a goat, sheep, a pig and a duck serve as target genes, and are subjected to PCR (Polymerase Chain Reaction) multiplex amplification by two pairs of fluorescently-labeled primers; PCR amplification products are detected by a 3730xl full automatic gene analysis meter; and species identification is performed according to 16S rRNA gene amplified fragment length polymorphism. The method fills in a gap of a detection method of the four kinds of meat.
Owner:VEGETABLE RES INST OF SHANDONG ACADEMY OF AGRI SCI
Analysis method and system of metagenome data
ActiveCN108334750AImprove accuracyReduce processingSequence analysisSpecial data processing applicationsEndocarditisResistant genes
The invention relates to an analysis method and a system of metagenome data. According to the invention, a preliminary species identification result of a sample is obtained on the basis of a k-Mer algorithm, a part or all of supporting sequences are extracted on the basis of the preliminary species identification result, and the preliminary species identification result is verified by using a blast algorithm to judge whether the preliminary species identification result is a reported detected species or not. The method and system disclosed by the invention can lower false positivity, quickly and accurately obtain the reported detected species of the sample in a short time, and are compatible with various mainstream sequencing platforms, thereby being suitable for second-generation sequencing technologies and third sequencing technologies; the method and system of the invention can also accurately identify drug-resistant genes and drug-resistant mutation sites of the sample and map thedrug-resistant genes and the drug-resistant mutation sites of the sample to the reported detected species. Furthermore, the system disclosed by the invention can be used for identifying pathogenic microorganisms, especially endocarditis pathogens to overcome the defect that the endocarditis pathogens are difficultly cultured.
Owner:SIMCERE DIAGNOSTICS CO LTD +2
Method and apparatus for control of mobility-based ion species identification
InactiveUS20050156107A1Improve system performanceGuaranteed accuracyTime-of-flight spectrometersFuel testingBiologySpectrometer
System for control of ion species behavior in a time-varying filter field of an ion mobility-based spectrometer to improve species identification, based on control of electrical and environmental aspects of sample analysis.
Owner:DH TECH DEVMENT PTE
Explosives detection using differential ion mobility spectrometry
InactiveUS7129482B2Easy to separateHigh resolutionFuel testingMaterial analysis by electric/magnetic meansIon-mobility spectrometrySpectrometer
System for control of ion species behavior in a time-varying filter field of an ion mobility-based spectrometer to improve species identification for explosives detection.
Owner:DH TECH DEVMENT PTE
Anti-pollution portable gene detection method and device
InactiveCN105199940AQuickly and reliably determineEasy to operateBioreactor/fermenter combinationsBiological substance pretreatmentsAnimal husbandryGene
The invention relates to an anti-pollution portable gene detection method and device. The operation procedure comprises the steps that a PCR pipe containing amplification products is put into a device to conduct sealing, and by means of a series of simple manual operations, a detection result visible by eyes is obtained; target nucleic acid amplification products can be detected rapidly, nucleic acid amplification products can be prevented from being polluted, and a false positive result is avoided. Application can be achieved in the aspects of species identification on the gene level in the fields such as pathogen detection on clinical infectious diseases, food pathogenic microbe detection, agriculture, industry, customs and animal husbandry.
Owner:GUANGZHOU HEAS BIOTECH CO LTD
Carrot beta(1,2)xylosetransferase gene full sequence and plasmid construction of CRISPR (clustered regularly interspaced short palindromic repeats)/CAS9 for dicotyledon transfection
The invention belongs to the technical field of biology and particularly relates to a carrot beta(1,2)xylosetransferase gene full sequence and plasmid construction of CRISPR (clustered regularly interspaced short palindromic repeats) / CAS9 for dicotyledon transfection. The carrot beta(1,2)xylosetransferase gene full sequence is discovered and determined for the first time, two effective gRNAs are designed aiming at the sequence to realize synthesis of corresponding CRISPR / CAS9 plasmids, and thus knockout of beta(1,2)xylosetransferase in suspension carrot cell genomes can be realized. In addition, two STR loci are found in the beta(1,2)xylosetransferase gene sequence, which is remarkably significant to species identification and evolution of carrots.
Owner:SHANGHAI JIAO TONG UNIV
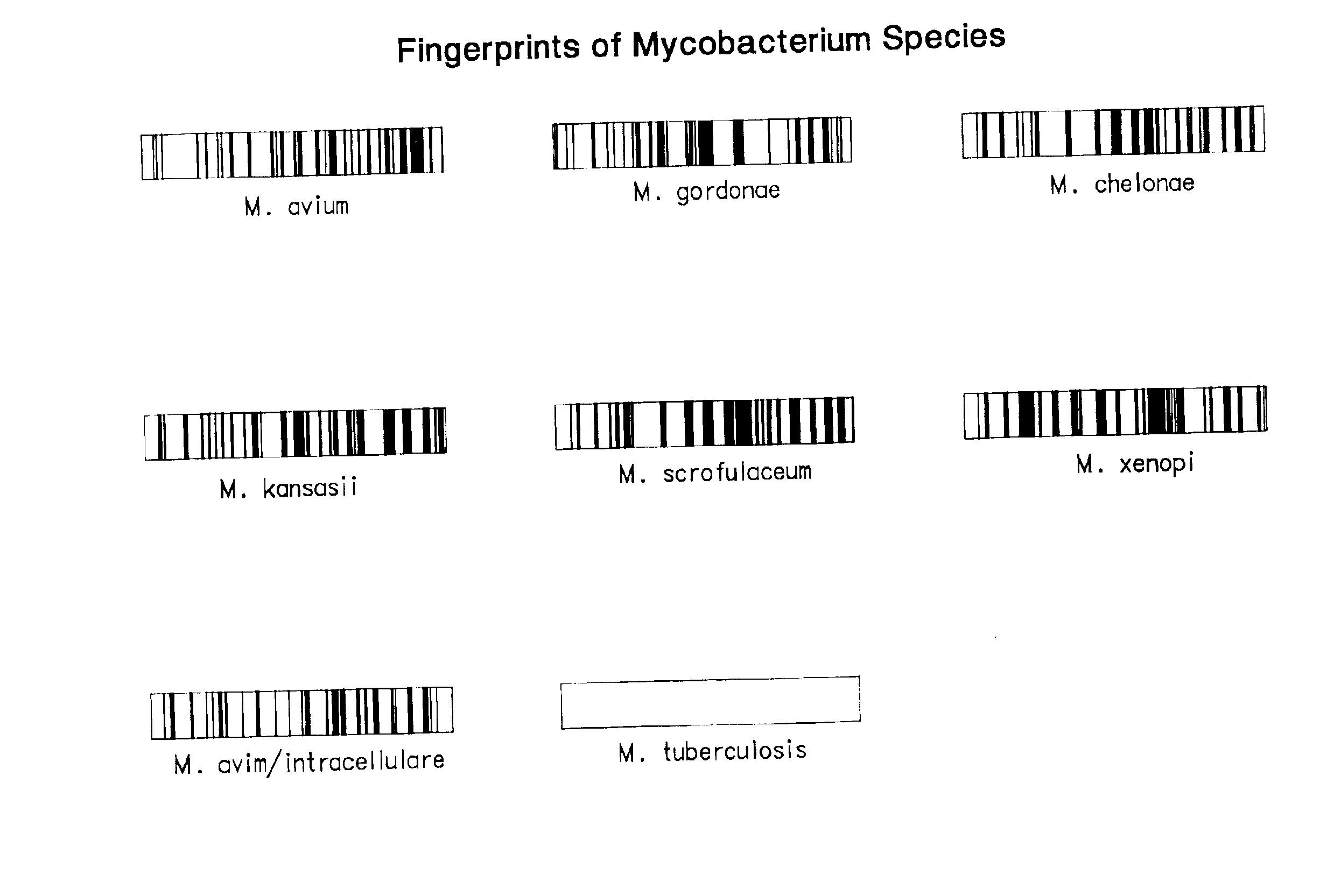
Chip-based species identification and phenotypic characterization of microorganisms
This invention provides oligonucleotide based arrays and methods for speciating and phenotyping organisms, for example, using oligonucleotide sequences based on the Mycobacterium tubercluosis rpoB gene. The groups or species to which an organism belongs may be determined by comparing hybridization patterns of target nucleic acid from the organism to hybridization patterns in a database.
Owner:AFFYMETRIX INC
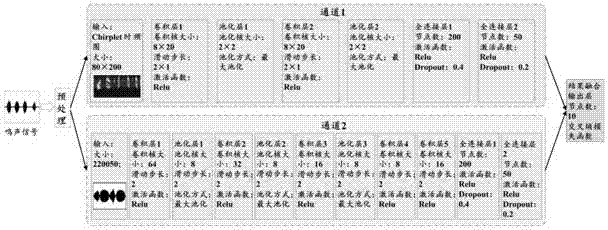
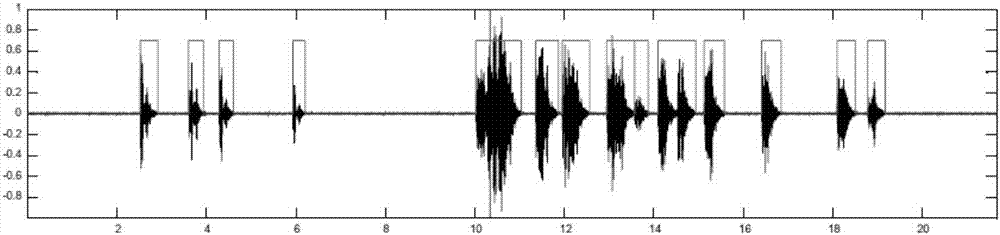
Bird species identification method based on dual-channel neural network
ActiveCN107393542AImprove efficiencyImprove accuracySpeech analysisNeural architecturesTime domainNerve network
The invention discloses a bird species identification method based on a dual-channel neural network. The bird species identification method comprises the steps that bird tweet signals of the known species are acquired and the preprocessed bird tweet signals are obtained through filtering, pre-emphasis and segmentation processing; signal sonagrams are generated based on linear frequency modulation wavelet transform; the bird tweet signals meeting the preset frame length range are cut to act as bird tweet time domain signals; the constructed preliminary identification model is trained with the signal sonograms acting as the input signals of a first channel, the bird tweet time domain signals acting as the input signals of a second channel and the corresponding bird species of the bird tweet signals acting as the identification result so as to obtain a bird species identification model; and the signals obtained by performing the same processing on the bird tweet signals to be identified are substituted in the bird species identification model to be identified so as to obtain the identification result. According to the bird species identification method based on the dual-channel neural network, the time domain characteristics and the time frequency characteristics of the tweet signals are fully utilized so that the efficiency and the accuracy of bird species identification can be enhanced.
Owner:BEIJING FORESTRY UNIVERSITY
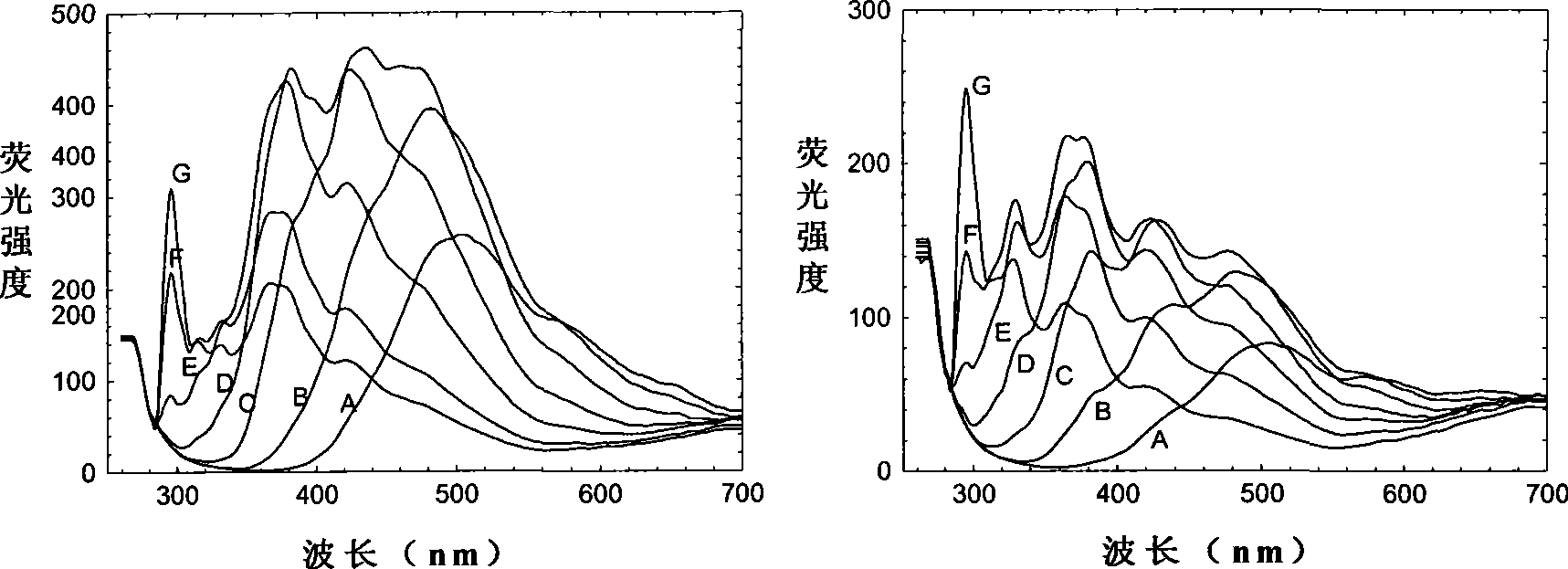
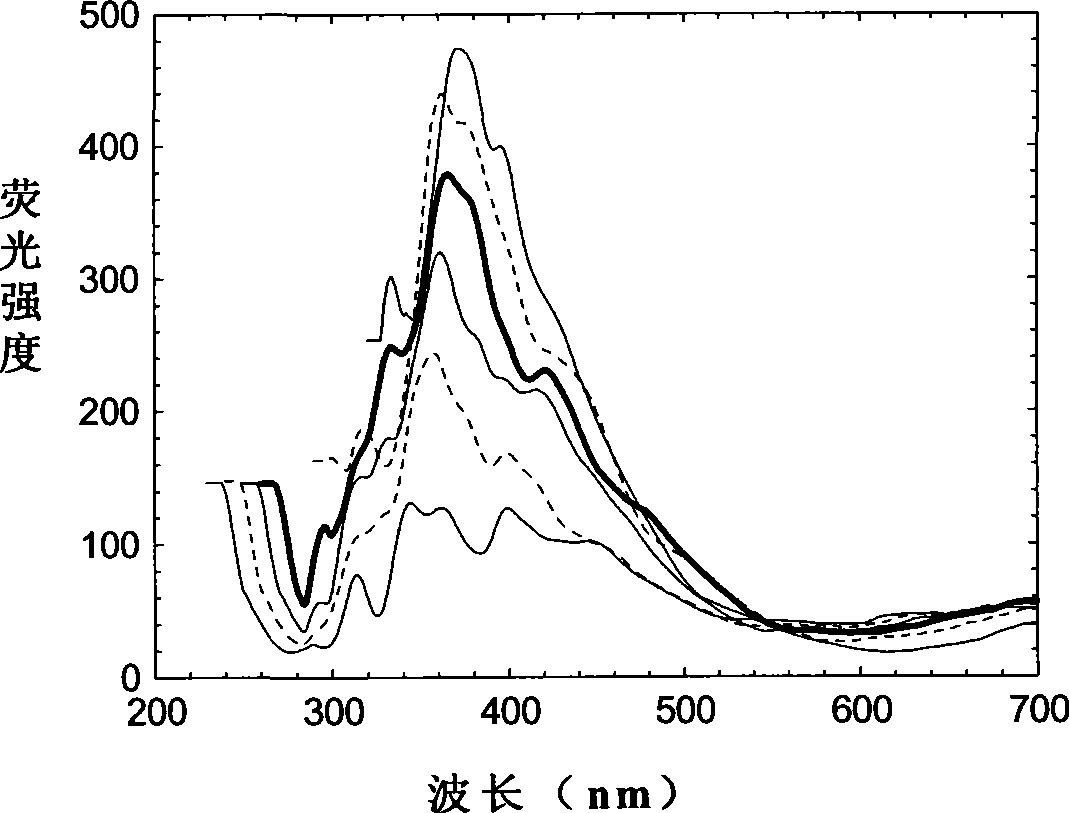
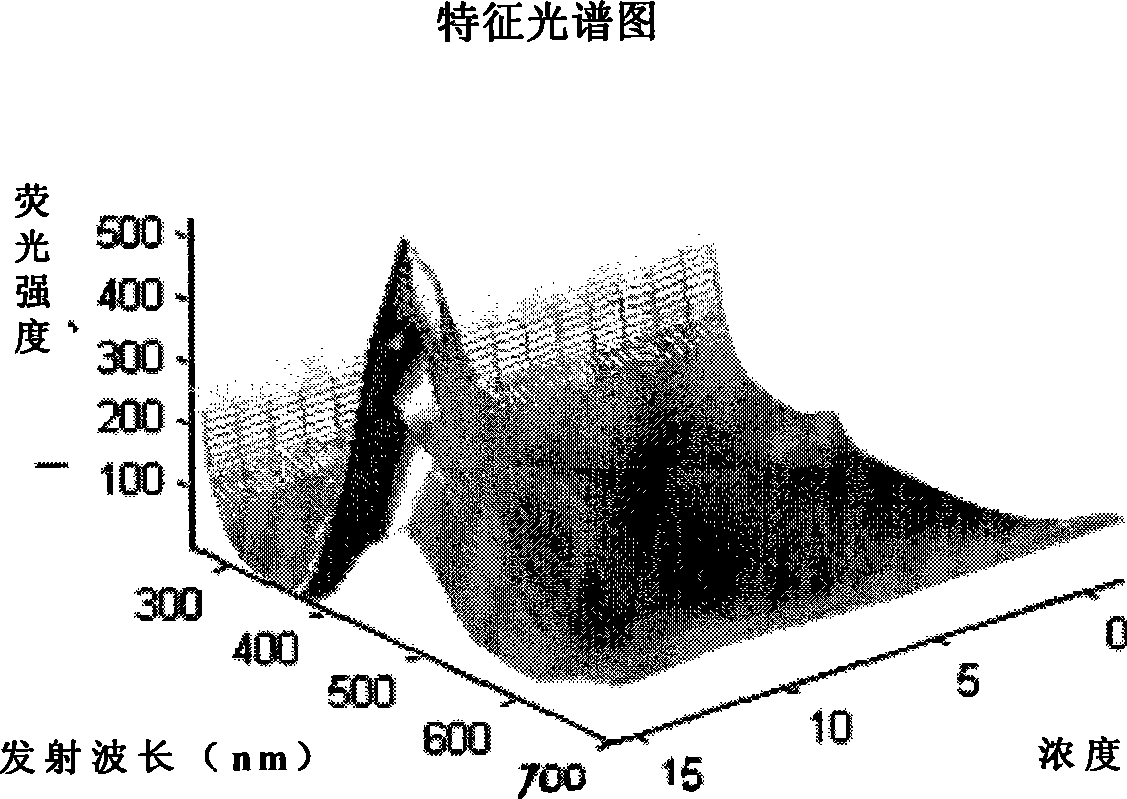
Oil species identification method by sea oil spill concentration auxiliary auxiliary parameter fluorescence spectrum
InactiveCN101458213AMaterial testing goodsFluorescence/phosphorescenceSpectral databaseFluorescence spectrometry
The invention discloses an oil type identification method. The oil spill fluorescence spectra of serial concentrations in a certain concentration range are taken as fingerprint identification indexes to identify oil types, and can be considered as a traditional fluorescence spectrum plus a dimensionality that is the concentration so as to obtain more fluorescence signals of an oil spill sample, thus improving capability of identifying the oil types. The identification method comprises the following steps: half-diluting the normal oil spill sample with a certain fixed original concentration to prepare extraction liquids of the serial concentrations, and measuring the fluorescence spectra of the extraction liquids; selecting a 3D fluorescence spectrum or a synchronous fluorescence spectrum according to requirements for detection precision and data processing speed, thus establishing a concentration parameter supplemental characteristic spectrum database of the normal oil spill samples of different oil sources sequentially. While identifying the oil type of a certain oil spill sample, the oil spill sample is extracted and diluted in the same way, and the corresponding characteristic spectrum is obtained and identified by being compared with the sample characteristic spectrum in the database, thus the optimal matched sample is the oil type of the oil spill sample.
Owner:WEIFANG UNIVERSITY
Process and system for rapid sample analysis
ActiveUS9606088B2Effectively integrating their spectral signatureEasy to measureComponent separationRaman scatteringDistillationElectromagnetic radiation
Components resolved in time by a separator accumulate in a sample cell and are analyzed by electromagnetic radiation-based spectroscopic techniques. The sample cell can be configured for multiple path absorption and can be heated. The separator can be a gas chromatograph or another suitable device, for example a distillation-based separator. The method and system described herein can include other mechanical elements, controls, procedures for handling background and sample data, protocols for species identification and / or quantification, automation, computer interfaces, algorithms, software or other features.
Owner:MLS ACQ INC D B A MAX ANALYTICAL TECH
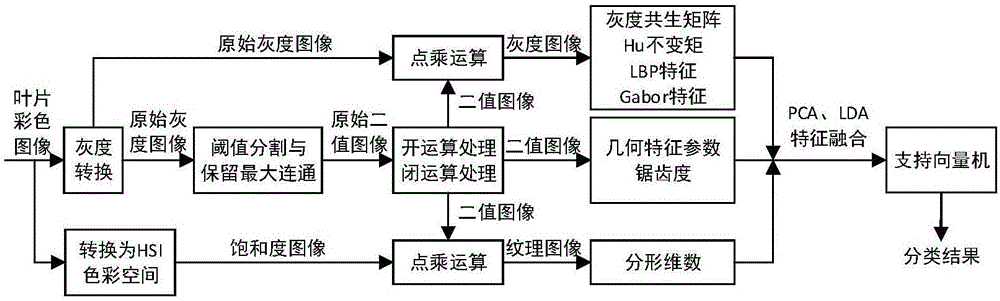
Leaf image multi-feature integrated plant species identification method and device
InactiveCN106295661AImprove accuracyImprove efficiencyCharacter and pattern recognitionImaging processingPrincipal component analysis
The invention relates to a leaf image multi-feature integrated plant species identification method and device. The method comprises the steps of obtaining plant leaf sample images of known species; obtaining binary images, leaf gray images and leaf texture images according to the leaf sample images; extracting leaf shape features and texture factures according to the leaf gray images, the binary images and the texture images; carrying out feature integration on the extracted leaf shape features and the texture factures through a main component analysis and linear judgment analysis combined method and carrying out dimension reduction on feature integration results, thereby obtaining training sample feature data; and training a support vector machine classifier according to the training sample feature data and corresponding plant species, thereby carrying out species identification on the leaf images of to-be-tested plants according to training results. According to the method and the device, image processing and machine learning principles are combined for plant species identification, and the plant classification and identification accuracy and efficiency are improved.
Owner:BEIJING FORESTRY UNIVERSITY
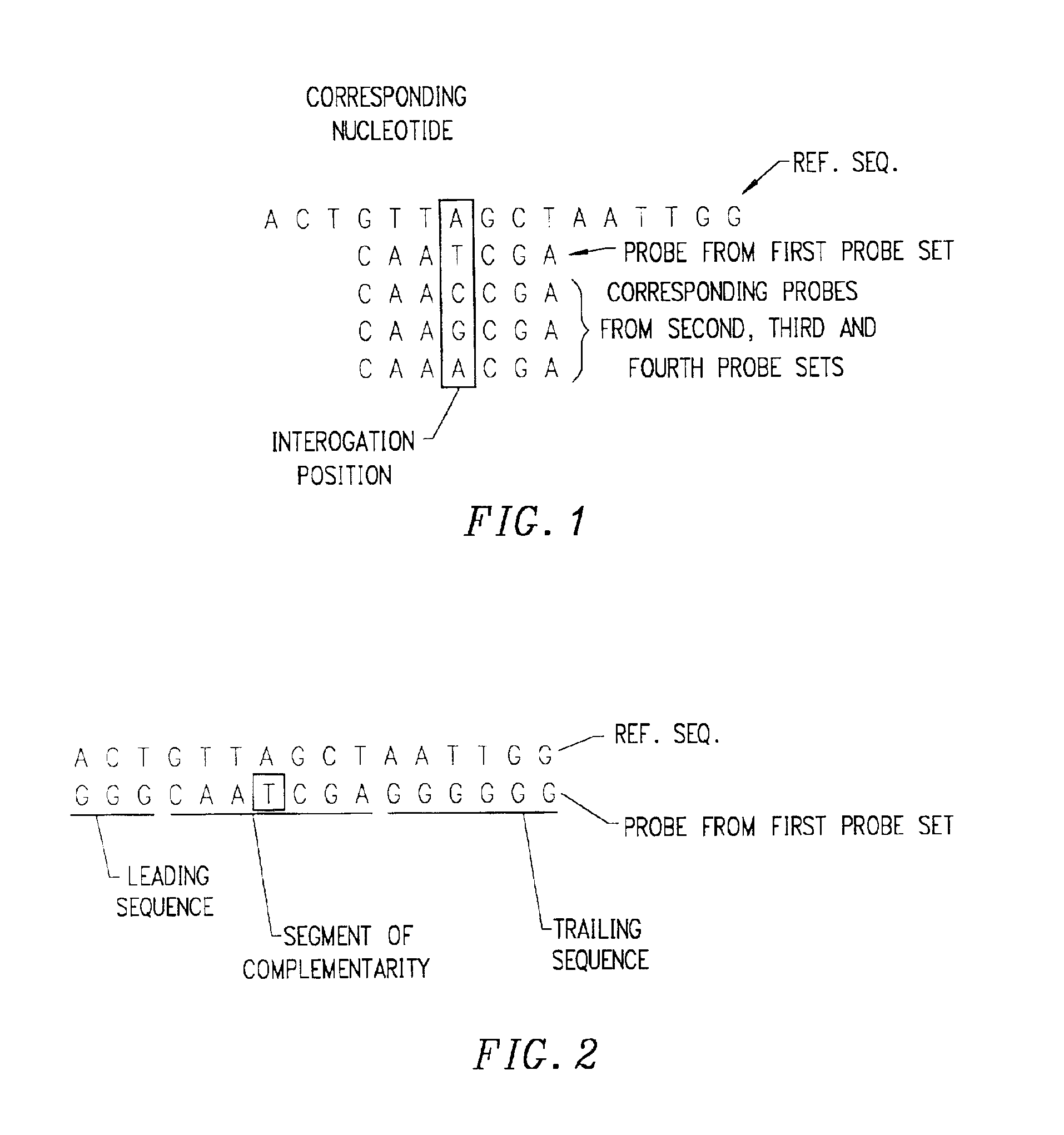
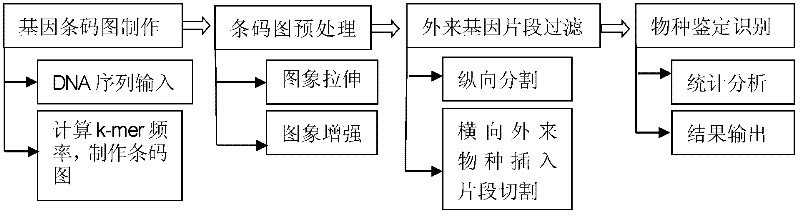
Biological species identification method based on genetic barcode
InactiveCN102332064AGenomic signature recognition at a glanceHigh precisionSpecial data processing applicationsNucleotideBiological species
The invention discloses a biological species identification method based on a gene barcode, which comprises the following steps of: 1. the production of a gene barcode image and a gene barcode image database: DNA (deoxyribonucleic acid) nucleotide sequences of 617 prokaryotes are downloaded from a website http: / / www. ncbi. nlm. nih. gov / , and the gene barcode image of species to be identified is produced according to the prior art; 2. the pre-processing of the gene barcode image: the gray scale of [0, L] of the gene barcode image is stretched to [0,255] by gray scale stretching, and the contrast of the gene barcode image is enhanced by gray scale enhancement; 3. the retrieval of foreign gene fragments of the gene barcode image: the longitudinal division of the gene barcode image is carried out, and the horizontal foreign gene fragments are searched; 4. species identification: the similarity quantity between the two species is determined, namely, the spatial distance between the two species is determined, and the species identification and result output are carried out according to the similarity quantity.
Owner:JILIN UNIV
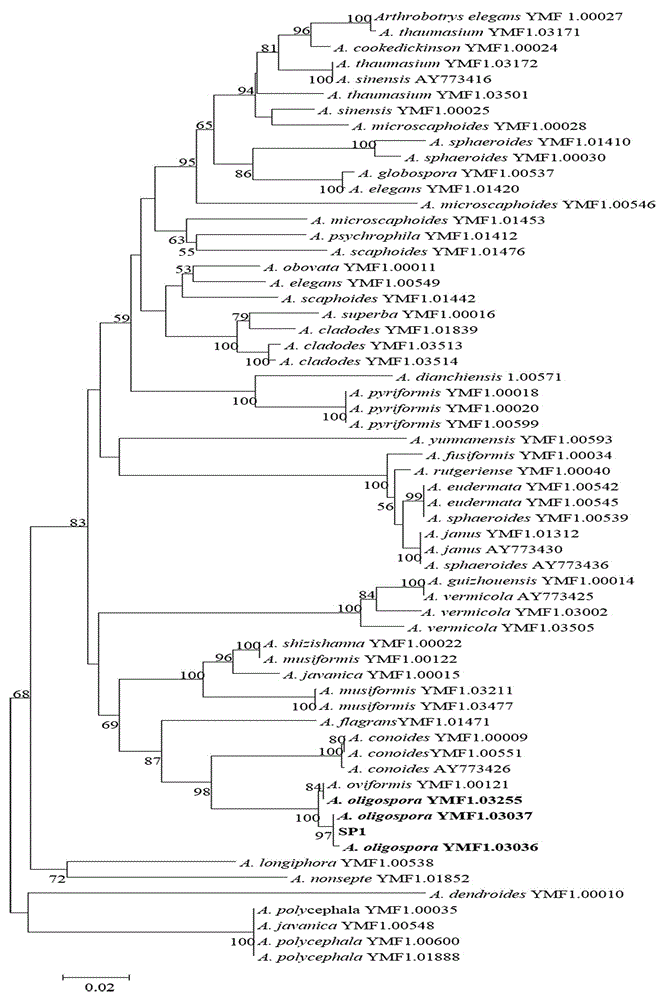
Method for identifying predator nematophagous hyphomycete arthrobotrys through DNA bar codes
InactiveCN105063761AEasy to expandEasy to compareLibrary creationSpecial data processing applicationsDNA barcodingGermplasm
The invention provides a method for identifying predator nematophagous hyphomycete arthrobotrys through DNA bar codes and belongs to the field of fungus species identification. According to the method, RPB2 genes serve as target DNA bar code genes for identifying the predator nematophagous hyphomycete arthrobotrys, combined arthrobotrys sample experiment data and target fungus data merged strategy is adopted to establish a high-cavity database, meanwhile, RPB2 genes to be identified are compared with a gene library, an identifying rule is established based on a system generation tree method of genetic distance method and clustering analysis of the Kimura-2-parameter probability to identify species. The method has the advantages that the RPB2 genes serve as the DNA bar codes most suitable for identifying the nematophagous hyphomycete arthrobotrys, and the method is universal and easy to amplify and compare. The identifying efficiency and the reliability and accuracy of the identifying method are greatly superior to those of a conventional DNA bar code method, and the method makes up for the blank of the nematophagous hyphomycete arthrobotrys DNA bar code molecular markers and the method provides a useful research tool for researches on germplasm resource excavation, biocontrol application and genetic diversity of nematophagous hyphomycete.
Owner:YUNNAN UNIV
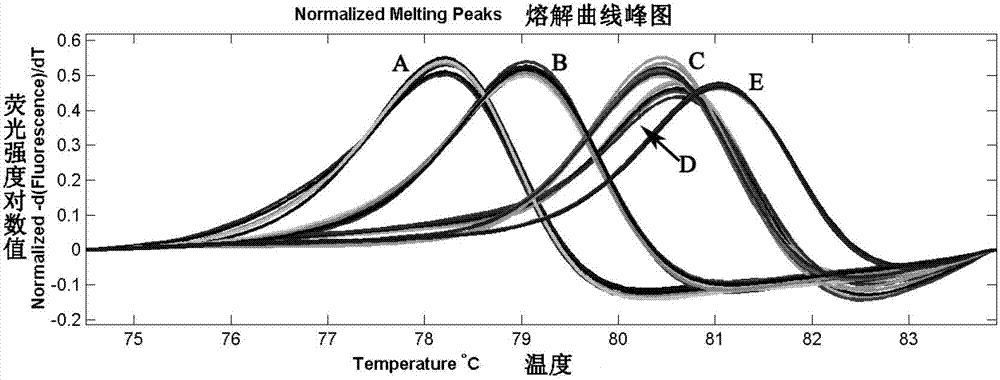
Method for identifying related species of oyster
ActiveCN102758009ASimple methodNo sequencing requiredMicrobiological testing/measurementOysterGenome
The invention relates to a related specie identification technology, in particular to a method for identifying related species of an oyster. The method specifically comprises the following steps of: by using an oyster genome DNA (Deoxyribonucleic Acid) as a template, designing a primer according to a tag sequence of the oyster specie genome; carrying out PCR (Polymerase Chain Reaction) amplification on the primer; and distinguishing oyster species by using Tm value difference in a high-resolution melting curve of an amplification product. Through the adoption of the method provided by the invention, the species can be identified rapidly, economically, simply and conveniently; and compared with a tag sequence sequencing result, an identifying result of the method provided by the invention is 100% in accuracy.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
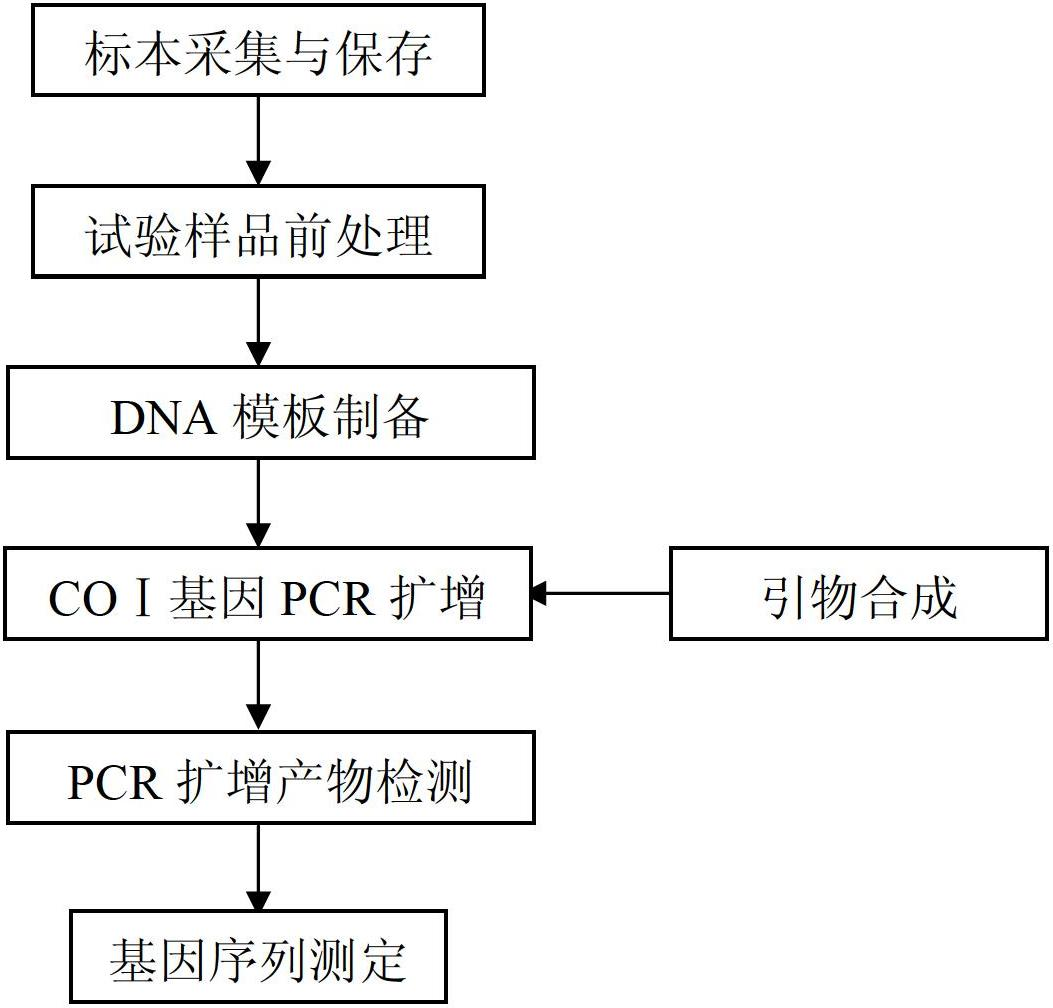
A DNA bar code standard gene sequence for Taiwan Lasiohelea
ActiveCN102690829AFacilitate the realization of molecular identificationShorten identification timeMicrobiological testing/measurementFermentationRapid identificationLasiohelea
The invention discloses a DNA bar code standard gene sequence for Taiwan Lasiohelea, which is characterized in that a bar code standard detection gene is COI gene, possessing gene sequence SEQ ID NO.1. Manual proofreading and sequence assembly are carried out to the sequencing result and Blast similarity search is carried out in NCBI to ensure that the obtained sequence is the target sequence. Species identification of Taiwan Lasiohelea is realized through morphological identification, thereby result reliability is guaranteed. The DNA bar code standard gene sequence for Taiwan Lasiohelea provided in the invention is in favor of realizing rapid identification of Taiwan Lasiohelea, and reduces identification time.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Molecular identification method for lasiohelea taiwana
ActiveCN103805609AAccurate identificationShorten identification timeMicrobiological testing/measurementFermentationBiotechnologyMolecular identification
The invention discloses a molecular identification method for lasiohelea taiwana. The method is characterized in that two different genes, namely a COI gene and an ITS2 gene are adopted to be combined, and the lasiohelea taiwana is subjected to species identification by comparison of the similarity of the gene sequences. By adopting the molecular identification method for lasiohelea taiwana provided by the invention, accurate and rapid identification of the lasiohelea taiwana is facilitated. A basis is provided for molecular identification of the lasiohelea taiwana. The new variety can play a certain role in transmission of plant pollen and environment harmless reaction.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Online plant species identification method based on geometric invariant shape features
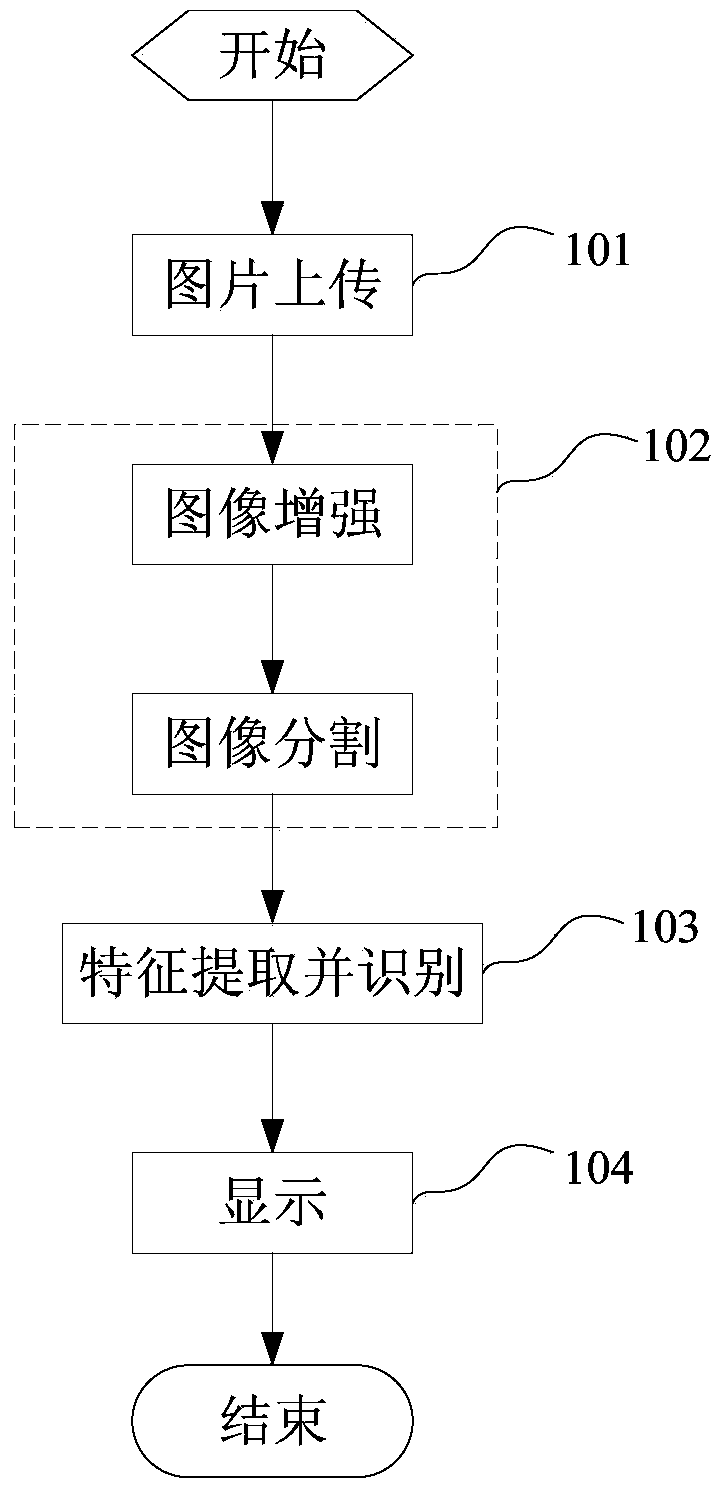
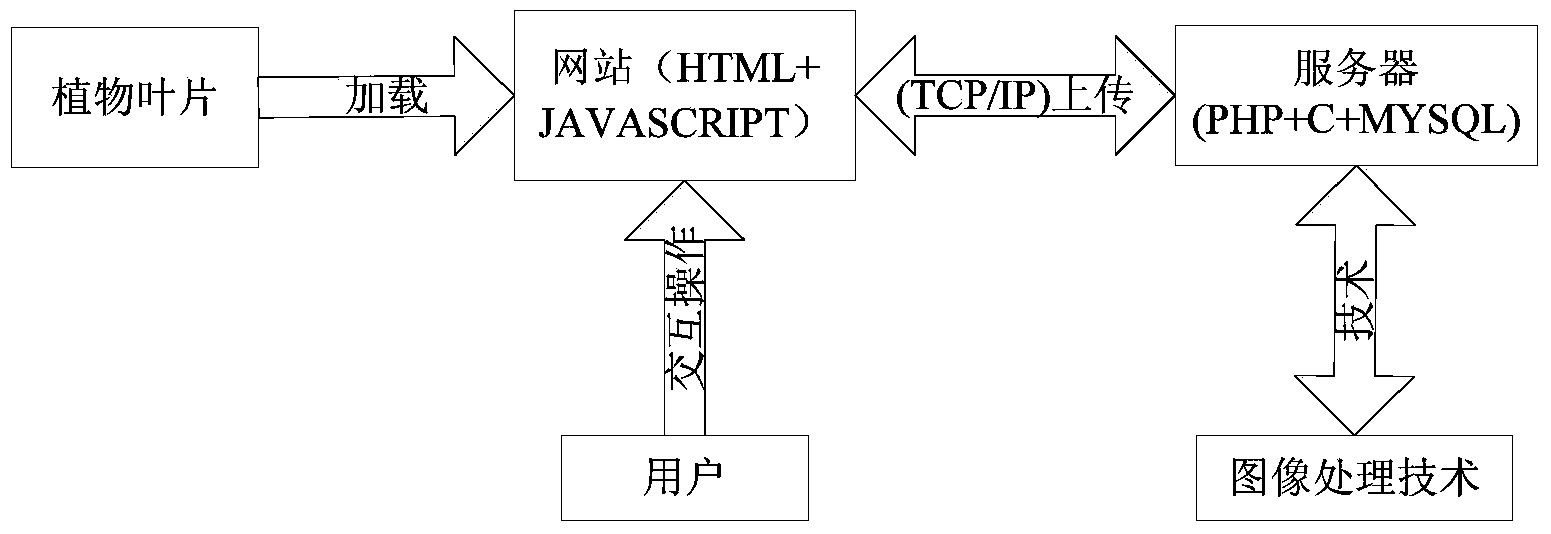
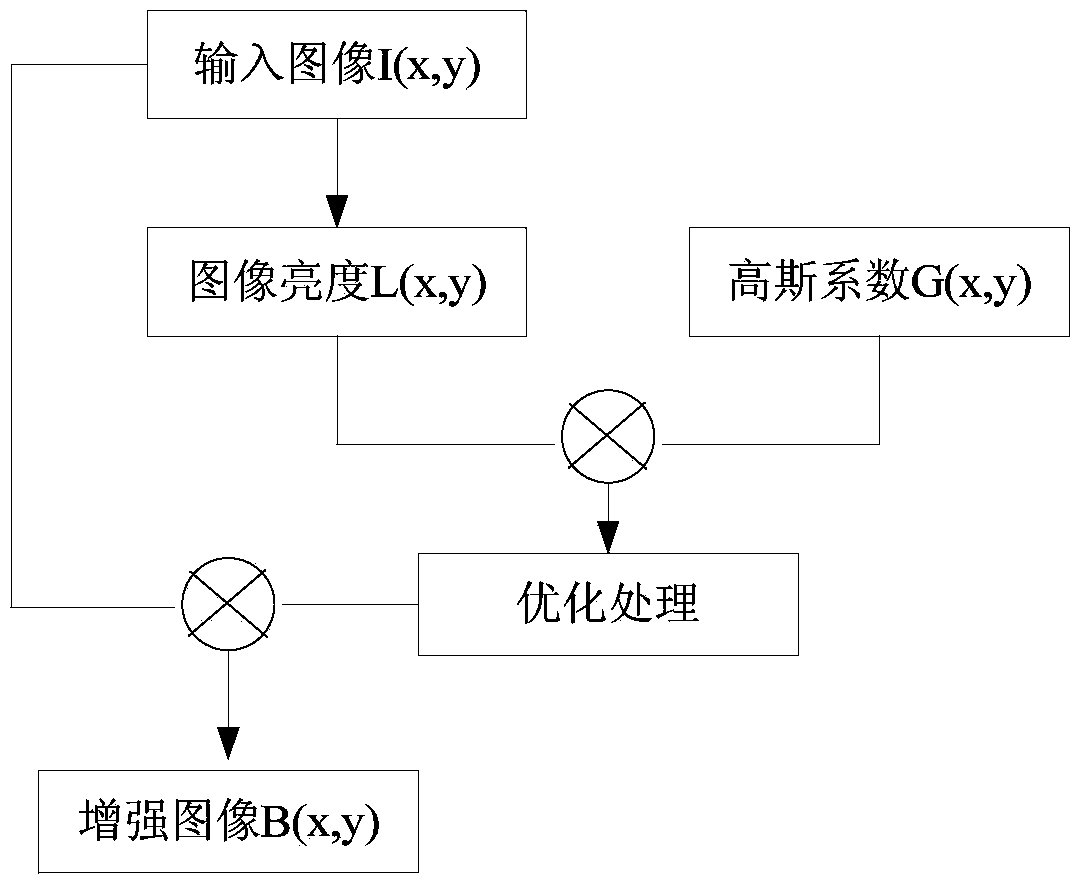
InactiveCN104361342AEasy pretreatmentRemove complex operationsCharacter and pattern recognitionFeature vectorHypersphere
The invention relates to an online plant species identification method based on geometric invariant shape features. The method includes 1, allowing a browser to acquire leaf image and transmit to a server after pre-processing; 2, allowing the server to calculate the leaf shape features according to pre-processed leaf images, establishing feature vectors, storing a trained moving center hypersphere classifier in the server, allowing the moving center hypersphere classifier to perform species identification according to the feature vectors, outputting the plant species code, querying a leaf information database according to the plant species code, and feeding the final identification result back to the browser; 3, allowing the browser to display the final identification result including the leaf, flower, fruit and seed images of the identified species and the scientific instruction information of the species. Compared with the prior art, the method has the advantages of fine real-time performance, convenience and high accuracy.
Owner:TONGJI UNIV
Food-borne pathogenic bacterium detection system and method based on high spectrum
ActiveCN103398994AResolve resolutionSolve efficiency problemsRaman scatteringColor/spectral properties measurementsBiological cellFood borne
The invention discloses a food-borne pathogenic bacterium detection system based on a high spectrum. The food-borne pathogenic bacterium detection system comprises a surface detection line, a sorting line, a qualified product conveying belt and a point detection line, which are connected with a control system respectively. The invention further discloses a detection method of the food-borne pathogenic bacterium detection system based on the high spectrum. The surface detection line based on a near-infrared high-spectrum system is used for rapidly positioning food-borne pathogenic bacteria; the sorting line is used for conveying pork polluted by the food-borne pathogenic bacteria to the point detection line; Raman chemical imaging is used for realizing species identification of the food-borne pathogenic bacteria. According to the food-borne pathogenic bacterium detection system disclosed by the invention, the problems that the resolution ratio of detecting a microorganism cell structure by the near-infrared high-spectrum system is limited and the large-area scanning and imaging efficiency of the Raman chemical imaging is low are solved; the food-borne pathogenic bacterium detection system based on the high spectrum has the advantages of convenience for utilization and obvious detection efficiency.
Owner:SOUTH CHINA UNIV OF TECH
Amplimer of large yellow croaker mitochondrion complete genome sequence and application thereof
ActiveCN102776183AEasy splicingIncrease success rateMicrobiological testing/measurementDNA preparationGerm plasmGene orders
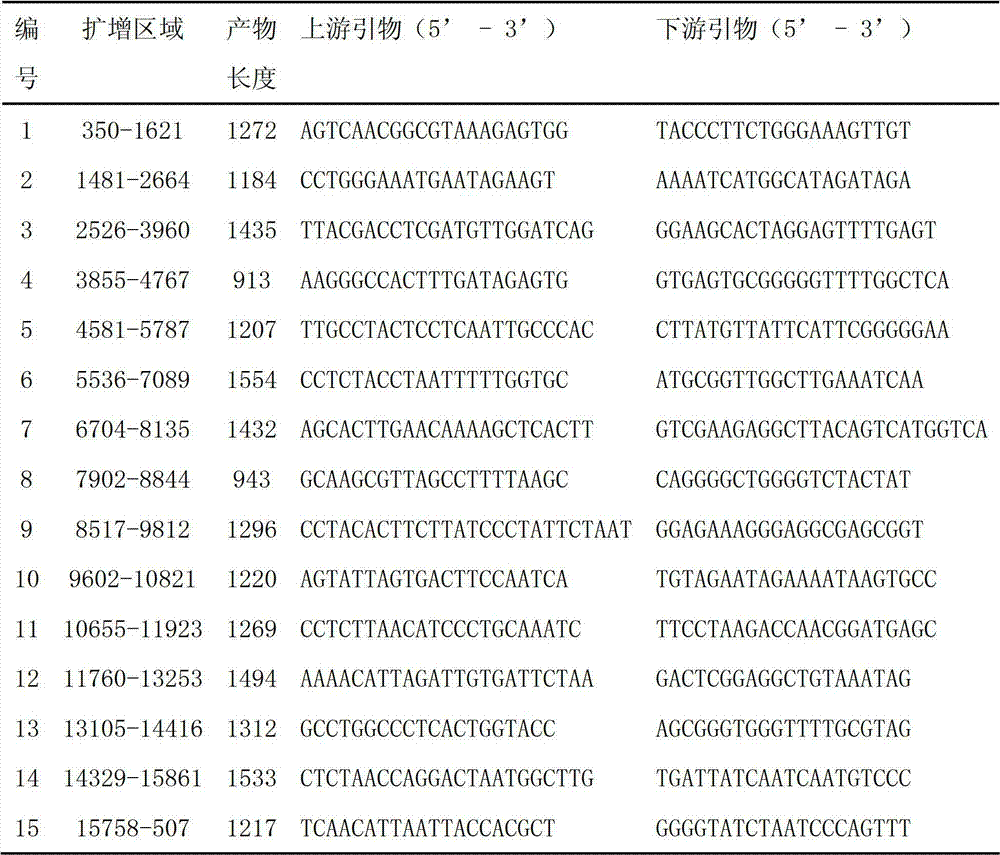
The invention discloses an amplimer of a large yellow croaker mitochondrion complete genome sequence and an application thereof. The 15 pairs of amplimer are applied, a gene order is respectively shown as SEQ ID No: 1-30, the amplimer is further used for obtaining a Tai Qu family large yellow croaker mitochondrion complete genome sequence and a Min-Yue East family large yellow croaker mitochondrion complete genome sequence which are respectively shown as SEQ ID No: 31 and SEQ ID No: 32, and an amplimer of large yellow croakers in a high variation region is obtained. The amplimer and the application have the advantages that the Tai Qu family large yellow croaker and Min-Yue East family large yellow croaker mitochondrion complete genome sequences can be quickly and efficiently obtained respectively through the 15 pairs of amplimers; and the amplimer of the large yellow croakers in the high variation region provides support for the research of species identification, geographical population identification, germ plasm resources and genetic diversity of the large yellow croakers, and further provide the basis for developing an identification technology of Tai Qu family large yellow croakers and Min-Yue East family large yellow croakers.
Owner:NINGBO UNIV
Probe identifying three aedes simultaneously on the basis of gene chip and kit
InactiveCN105039563AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationAedes albopictusAedes taeniorhynchus
The invention belongs to the field of biological detection and identification, and relates to a probe identifying three aedes of an aedes albopictus, an aedes aegypti and an aedes togoi on the basis of a gene chip, and a kit. The invention discloses the probe identifying the three aedes simultaneously on the basis of the gene chip, and the probe comprises the following three probes: the aedes albopictus: CTTTAACACTGCTGCTTTCTAGTTC; the aedes aegypti: GTGCTGAACTTAGCCACCCTGGT; the aedes togoi: AACTCTCCTGCTTTCAAGTAGT. According to gene sequences of the different aedes, conservative positions of the gene sequences are selected and a specific oligonucleotide detection probe is designed; mosquito species identification is carried out by utilizing a biological method; the problems that the number of identification experts is limited, the identification period is long, and the identification efficiency is low and the like in a traditional media mosquito species identification method are overcome; the quick and accurate identification on the three aedes of the aedes albopictus, the aedes aegypti and the aedes togo is realized. According to an experimental detection, the gene chip and the kit provided by the invention are high in specificity, high in sensitivity, and can be well used for media biological identification work of a port.
Owner:珠海国际旅行卫生保健中心
Misgurnus anguillicaudatus and paramisgurnus dabryanus specie identification primer and identification method
InactiveCN104498604ASolve misjudgmentMicrobiological testing/measurementDNA/RNA fragmentationSpecies identificationPhases of clinical research
The invention provides a molecular biology-based misgurnus anguillicaudatus and paramisgurnus dabryanus specie identification primer and an identification method. The misgurnus anguillicaudatus and paramisgurnus dabryanus specie identification primer is characterized by designing a specific primer sequence: upstream primer sequence being 5'-CCGCCCAGGAGTATTTTATG-3'(shown by SEQ ID NO.1); downstream primer sequence being 5'-TGGCATTTCATCAGGGGTG-3'(shown by SEQ ID NO.2). The identification method comprises the following steps: extracting DNA of misgurnus anguillicaudatus or paramisgurnus dabryanus samples by adopting a conventional method; performing PCR amplification; after the PCR amplification is completed, taking a proper amount of amplified products for performing gel electrophoresis detection to determine the size of amplified fragments, and identifying. The identification method is accurate and high-efficiency. The problems that the traditional morphological distinguishing method is difficult to operate and low in resolution success rate can be solved. Especially, the misgurnus anguillicaudatus and paramisgurnus dabryanu at the young and juvenile stages can be difficulty identified from appearance, and the invention provides a convenient and accurate identification method.
Owner:中国水产科学研究院北戴河中心实验站
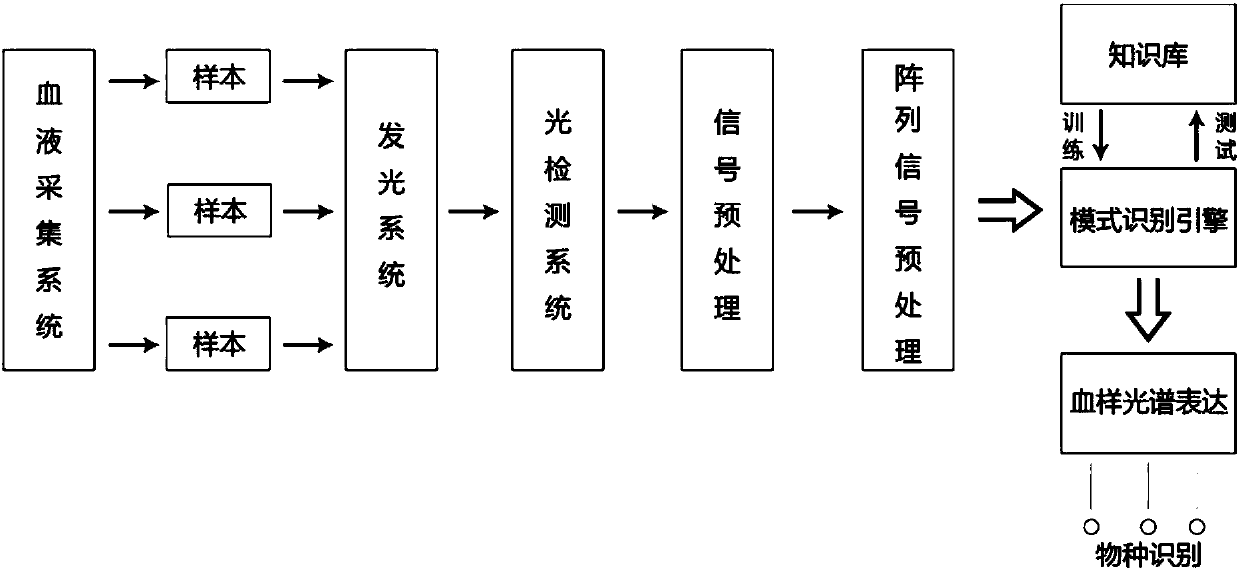
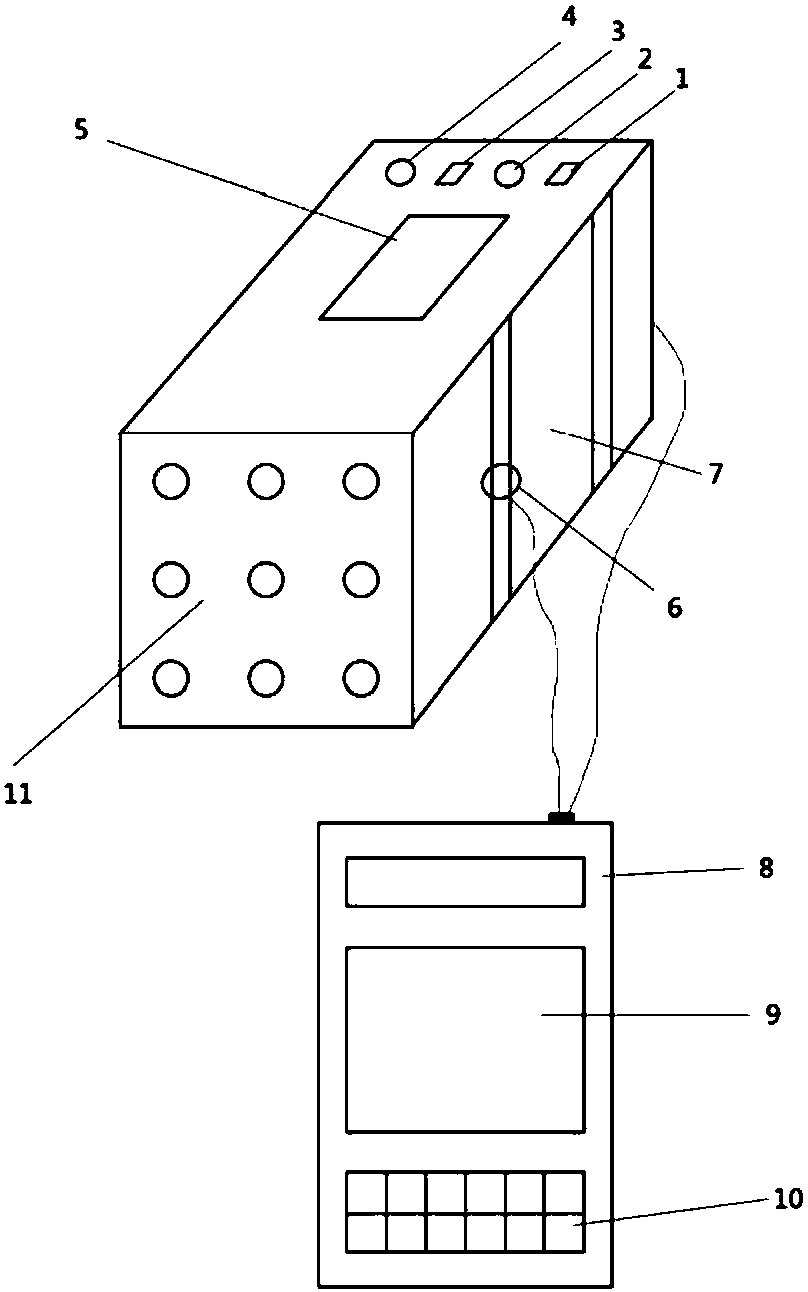
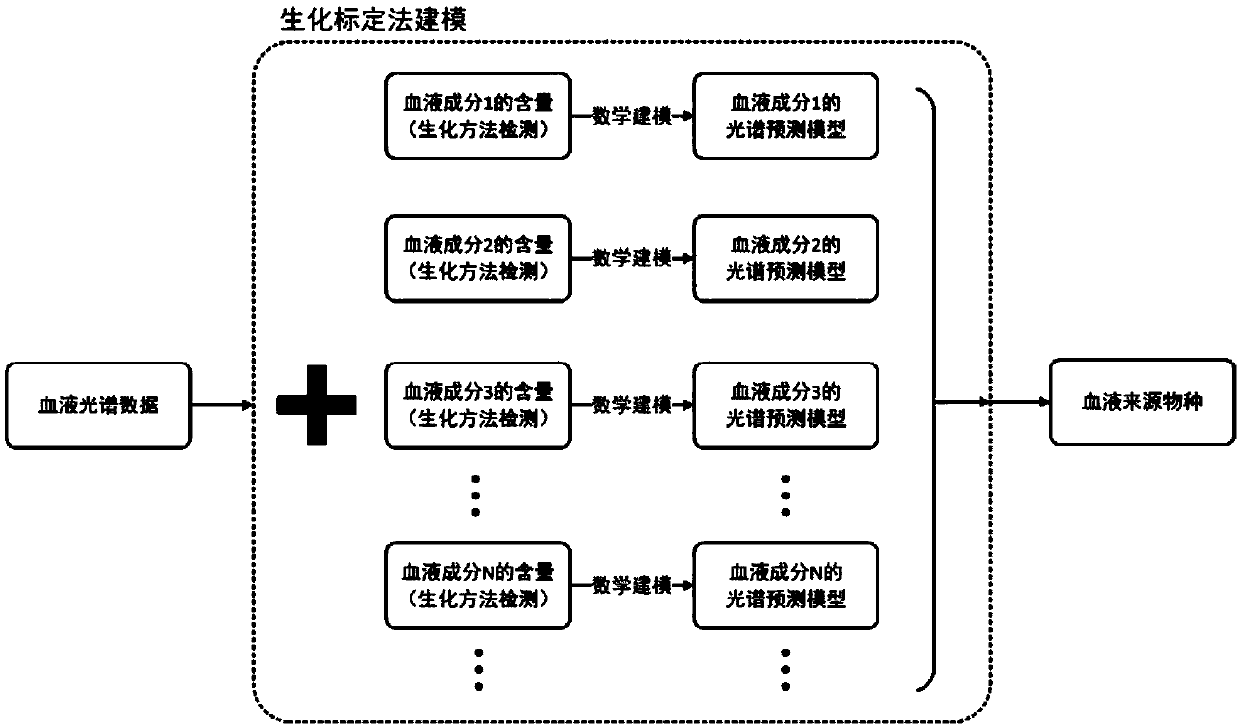
Spectrum-based blood species identification instrument and identification method
ActiveCN107045637AScattering properties measurementsColor/spectral properties measurementsData setData mining
The present invention relates to a method for identifying a species from which blood is derived based on the spectrum data of the blood. The method can be used for fast identifying a species from which a blood sample is derived. According to the implementation steps of the method, a sample data set is divided into a plurality of data sets with relative independence; an identification model is trained for each data set; the capacities of the identification models of the data sets to predict other data sets are compared, so that samples contained in each data set are dynamically adjusted; by means of a circulative iterative process, feature distribution in the spectrum data of the blood sample is de-centralized, so that the wide-area distribution of spectrum feature information is ensured; and the capacity of an overall model to express the spectrum feature information of the blood sample based on the integration of the prediction capacities of the plurality of predictive identification models, and therefore, the accuracy of the prediction of the overall model can be improved.
Owner:INST OF BIOMEDICAL ENG CHINESE ACAD OF MEDICAL SCI
Method for screening nitrification bacteria
InactiveCN101434905AGrowth inhibitionEasy to handleMicroorganismsMicrobiological testing/measurementActivated sludgeIndustrial waste water
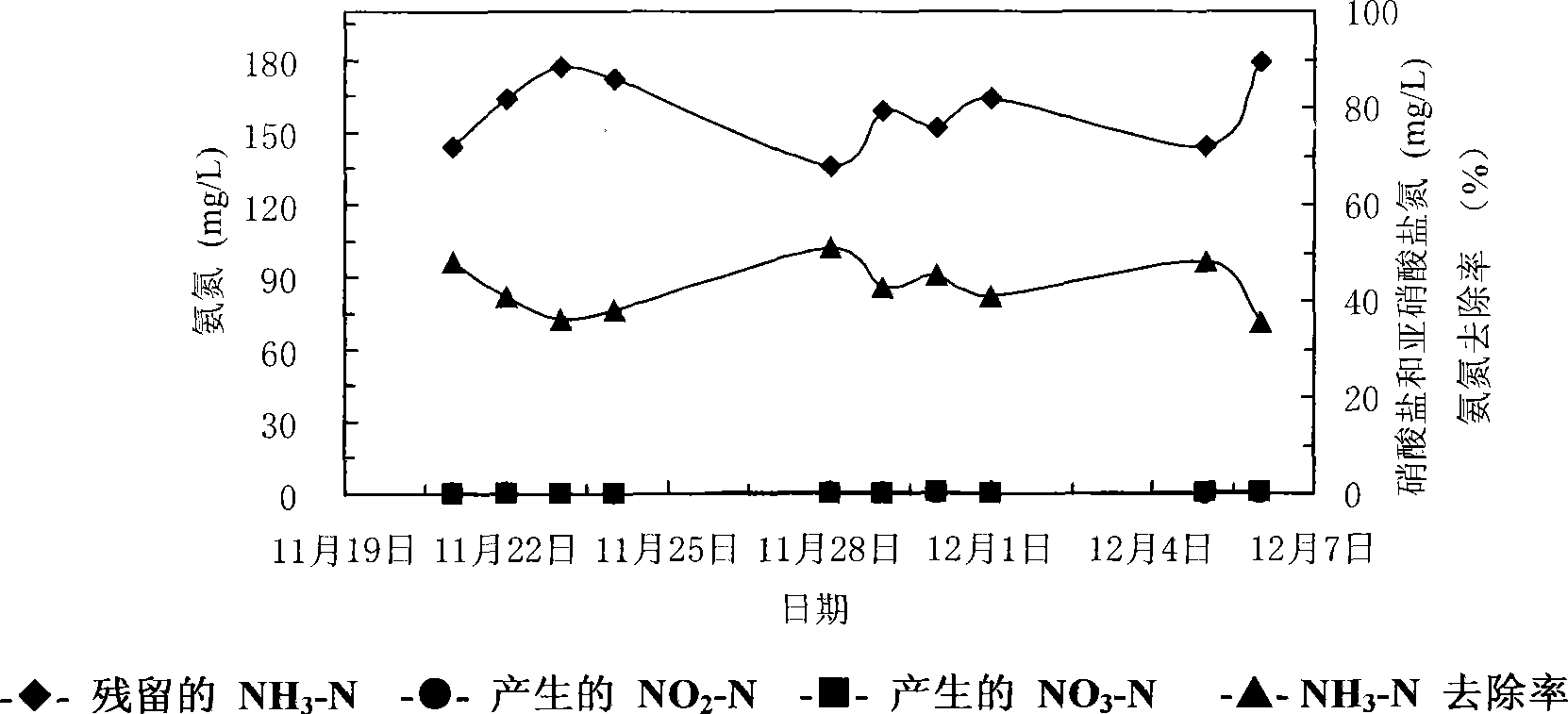
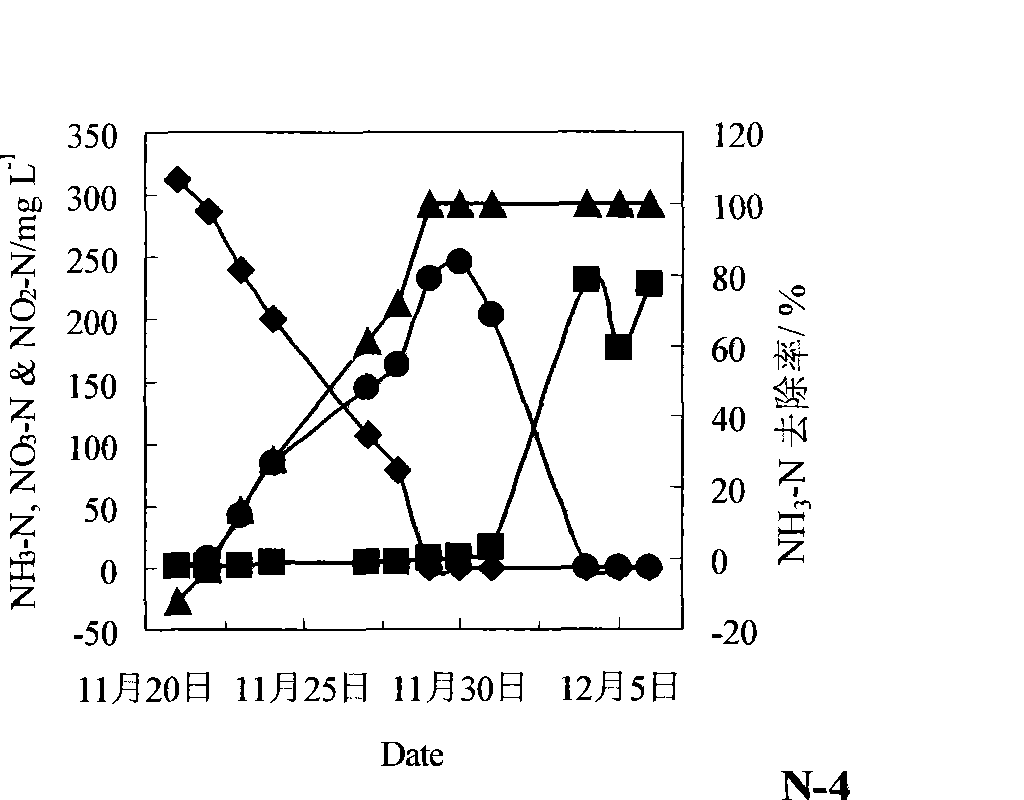
The invention relates to a screening method of heterotrophic nitrifier. The screening method comprises the following steps: (1) nitrifier-enriched activated sludge which can tolerate high ammonia nitrogen loads is cultured by a method of gradually improving the ammonia nitrogen loads; (2) the heterotrophic nitrifier is separated and purified from the nitrifier-enriched activated sludge; (3) the nitrification of the heterotrophic nitrifier which is separated and purified is inspected; and (4) the species identification of the heterotrophic nitrifier which is separated and purified is carried out. The heterotrophic nitrifier which is screened by utilizing the method has rapid growth rate and can not only carry out synchronous nitrification and denitrification to high-COD and high ammonia nitrogen industrial waste water, but also can carry out high-efficient nitrification to the low-COD and high ammonia nitrogen waste water.
Owner:CHINA PETROLEUM & CHEM CORP +1
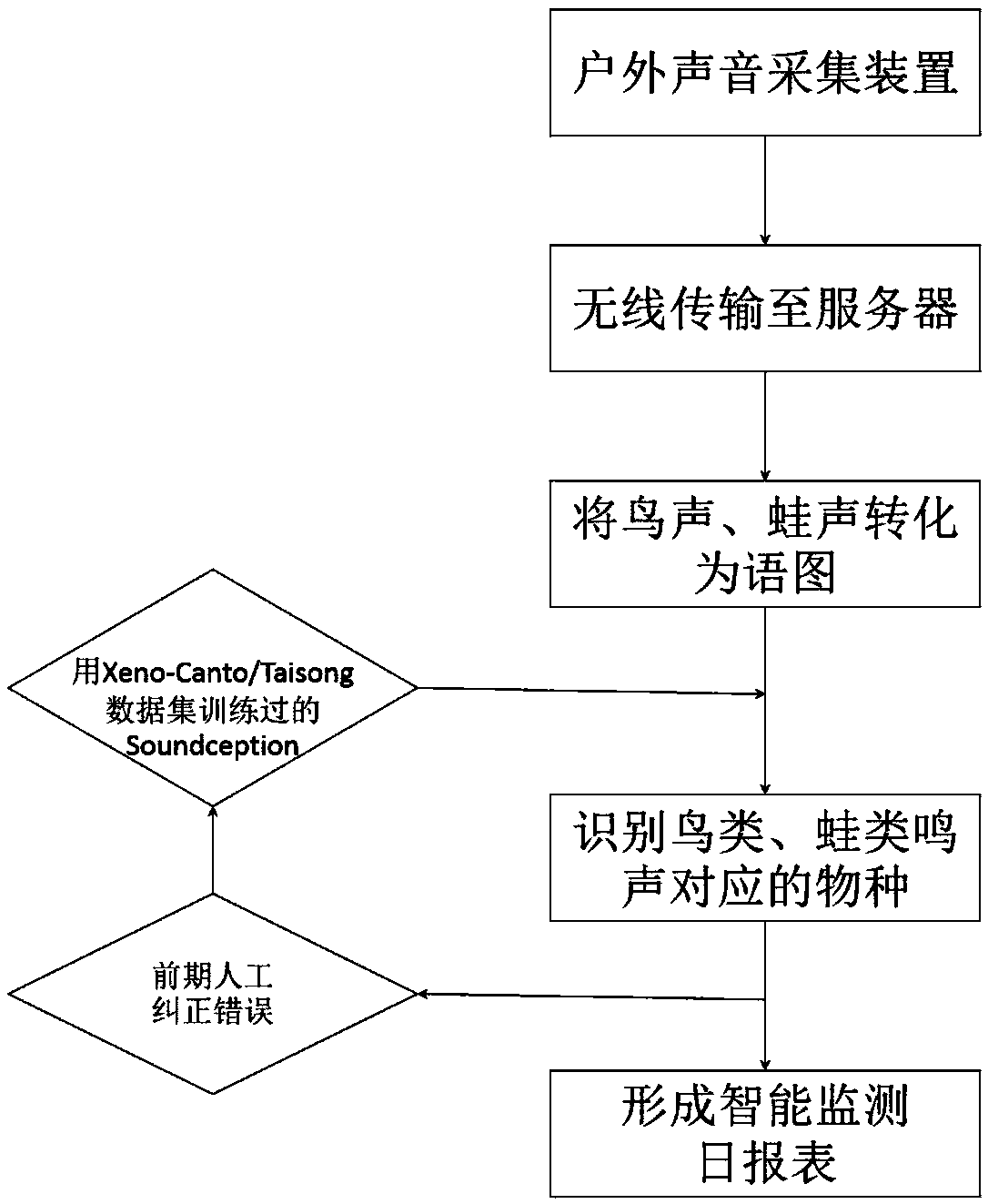
Bird and frog intelligent monitoring method based on deep learning
InactiveCN108922548ASolve the large investment of manpower and material resourcesSolve the requestSpeech analysisStatistical analysisHuman power
The invention discloses a bird and frog intelligent monitoring method based on deep learning, which is used for remote wireless ecological monitoring. The method comprises: arranging an outdoor soundacquisition device in a monitoring area, wherein the outdoor sound acquisition device is triggered by a sound triggering module to acquire a bird sound and a frog sound; allowing a wireless communication module to wirelessly transmit an audio file of the bird sound the frog sound to a computer server; allowing the computer server to convert the audio file of the bird sound and the frog sound intoa visual sonogram and performing pre-processing; allowing the computer server to perform image recognition and statistical analysis on the sonogram; and generating a daily monitoring form. The bird and frog intelligent monitoring method compensates for the problems that in manual ecological monitoring, the human and material cost is high, the monitoring time and frequency are limited, the data processing efficiency is low, and requirements of species for professional skills of a recognition person are high, and can be used for long-term ecological monitoring of natural protection sites.
Owner:深圳园林股份有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com