Carrot beta(1,2)xylosetransferase gene full sequence and plasmid construction of CRISPR (clustered regularly interspaced short palindromic repeats)/CAS9 for dicotyledon transfection
A carrot and gene technology, applied in the field of CRISPR/CAS9 plasmid construction, can solve the problems of missing nucleotides, insertion of break sites, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
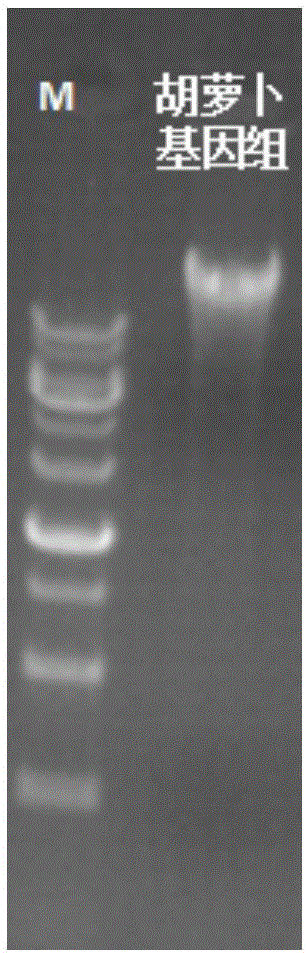
[0064] Example 1 Extraction of Carrot Genomic DNA
[0065] 1. Experimental method
[0066] This step uses the Plant Genomic DNA Extraction Kit from Tiangen Biochemical Technology Co., Ltd.
[0067]The specific operation steps include:
[0068] Take about 100 mg of fresh carrot cell mass, add liquid nitrogen and grind thoroughly. Quickly transfer the ground powder to a centrifuge tube pre-filled with 700 μL of 65°C preheating buffer GP1 (before the experiment, mercaptoethanol needs to be added to the preheated GP1, and it is prepared and used now to make the final concentration 0.1%). After quickly inverting and mixing, place the centrifuge tube in a 65°C water bath for 20 minutes, and invert the centrifuge tube several times every 5 minutes during the water bath to mix the samples. Add 700 μL of chloroform, mix thoroughly, and centrifuge at 12,000 rpm for 5 min. Transfer the upper aqueous phase obtained in the previous step into a new centrifuge tube, add 700 μL buffer GP2...
Embodiment 2
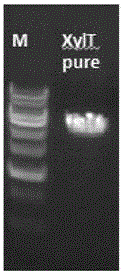
[0071] Example 2 Obtaining XylT Fragment Using Nested PCR Technology
[0072] 1. Experimental method
[0073] (1) PCR reaction:
[0074] Using the carrot genomic DNA obtained in Example 1 as a template, use gene homology to obtain 3 exon sequences from the carrot database, design two pairs of primers on the outside and inside according to this sequence, and use the outside pair of primers to carry out the first PCR reaction . Then use the 10-fold dilution of the first PCR product as a template, and use the inner pair of primers for the second PCR reaction. The PCR primer sequences are shown in Table 1, and the reaction components are shown in Table 2. The cycle parameters of the first PCR reaction were 98° C. for 1 min, 95° C. for 0.5 min, 55° C. for 0.5 min, 68° C. for 10 min, 35 cycles, and 68° C. for 10 min. The cycle parameters of the second PCR reaction were 98° C. for 1 min, 95° C. for 0.5 min, 55° C. for 0.5 min, 68° C. for 6 min, 35 cycles, and 68° C. for 10 min. ...
Embodiment 3
[0095] Example 3 PCR product sequencing after XylT purification
[0096] 1. Experimental method
[0097] The PCR product purified in Example 2 was sent to Shanghai Huajin Biotechnology Co., Ltd. for sequencing. The outer two pairs of PCR primers were used as the first pair of sequencing primers, and the sequencing was carried out in the middle at the same time. According to the sequencing results, two pairs of primers were designed to continue the sequencing. The sequences of the sequencing primers are listed in Table 3.
[0098] Table 3 Sequencing primer sequences
[0099] Sequencing primer name Primer sequence (5'-3') serial number W1F GCTTATTTTGGTAATGGCTTTACTCGAC SEQ ID NO.5 W1R CAAGAATAGGATGGTACTCTAATC SEQ ID NO.6 W2F AATCCTTTTCTGGAGTTGAG SEQ ID NO.7 W2R GGCTGTTGCCACAGATCATG SEQ ID NO.8 W3F CGTATTACTATGAGGCGAGG SEQ ID NO.9 W3R ATGCAGCATACCAATCTGTAACTG SEQ ID NO.10 W4F GATTTCTTGATGGCATGGTCAAG SE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com