Promoter WY195 and application thereof
A WY195, promoter technology, applied in the field of genetic engineering, can solve the problem of molecular biology research lag and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1: PCR amplification of WY195 promoter fragment
[0037]Use the fungal genome DNA extraction kit (OMEGA, D3390-01) to extract the genome DNA of powdery mildew fungi, according to the sequence of WY195 promoter, design a pair of specific amplification primers (upstream primer WY195F, plus restriction enzyme cutting site HindⅢ and protection bases, downstream primer WY195R, plus restriction site BamHI and protection bases). Using the above-mentioned extracted Genomic DNA of Powdery mildew Hevea as a template, high-fidelity Ex Taq polymerase (TRANSGEN, `AP122) was used for PCR amplification. As shown in Table 1.
[0038] Table 1 PCR system for gene promoter amplification
[0039]
[0040] The PCR amplification program was as follows: pre-denaturation at 94°C for 5 min, then denaturation at 94°C for 60 s, annealing at 55°C for 50 s, extension at 72°C for 60 s, 35 reaction cycles, and finally extension at 72°C for 5 min.
[0041] Among them, the upstream pri...
Embodiment 2
[0044] Embodiment 2: Construction of pGEM-T easy-WY195 recombinant vector
[0045] The PCR amplification product obtained above was transformed into Escherichia coli by T / A cloning (pGEM-T easy plasmid, PROMEGA, A1360), and positive clones were picked and sequenced, which proved to be accurate.
[0046] Among them, the connection conditions of T / A clone are as follows:
[0047] T / A connection system: 10ul
[0048] pGEM-T EasyVector (PROMEGA, A137A): 1ul
[0049] 2×Rapid ligation Buffer: 5ul
[0050] PCR amplification product (recovered insert): 2ul
[0051] T4DNAligase: 1ul
[0052] wxya 2 O: 1ul
[0053] First place at room temperature for 1 hour, then ligate overnight at 4°C to obtain the pGEM-T easy-WY195 recombinant vector. The product after the above connection was transformed into Escherichia coli as follows:
[0054] Take out 100 μl of DH5α (Transgene, CD201) competent cells prepared according to the calcium chloride method shown in "Molecular Cloning Experiment...
Embodiment 3
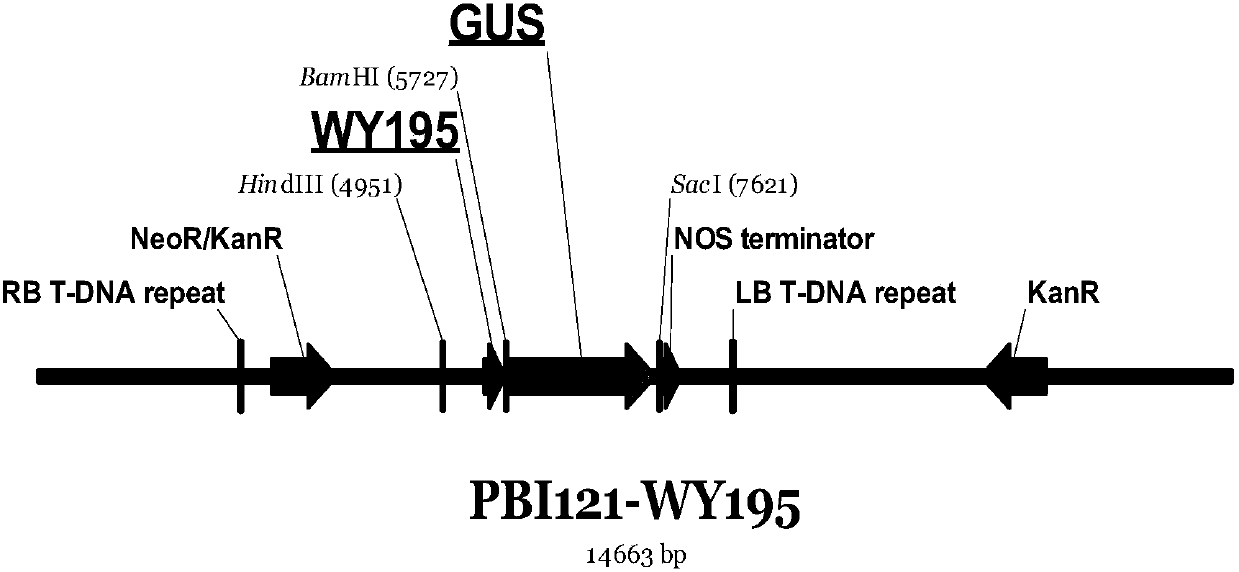
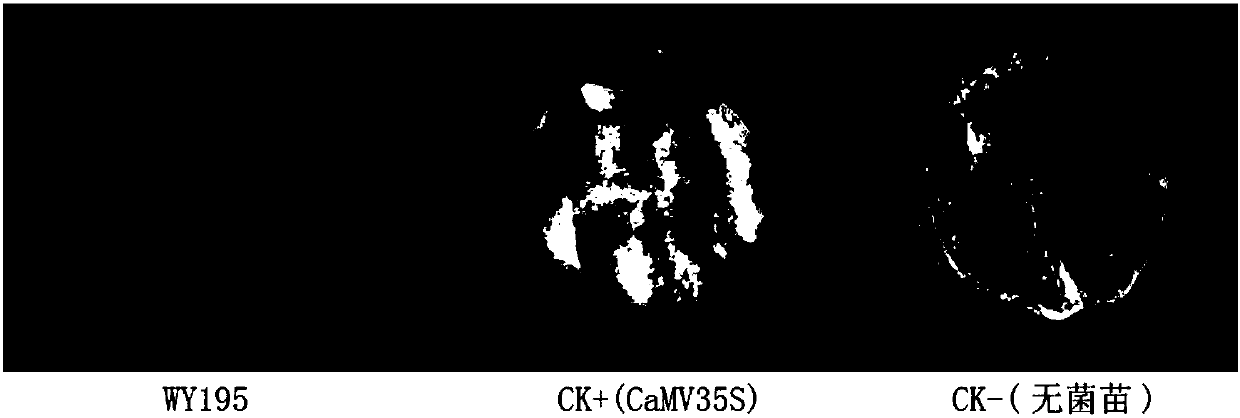
[0056] Embodiment 3: Construction of PBI121-WY195 recombinant vector
[0057] Pick a single colony of the DH5α-WY195 strain obtained from the above construction and shake it overnight at 220 rpm at 37°C. Extract the plasmid with OMEGA plasmid mini-extraction kit (D6943-01), and then use Hind Ⅲ (NEB, R0104S) and BamHI (NEB, R0136V) restriction endonuclease was used for double digestion, and the digested product was recovered with the OMEGA recovery kit (D2500-01) to recover the WY195 promoter fragment.
[0058] The recovered product obtained above was transformed into Escherichia coli by T / A cloning (PBI121 plasmid, TIANNZ, 60908-750y), and positive clones were picked and sequenced, which proved to be accurate.
[0059] Among them, the connection conditions of T / A clone are as follows:
[0060] T / A connection system: 10ul
[0061] PBI121Vector: 1ul
[0062] 10×T4 DNA Ligase Buffer: 1ul
[0063] Recovered product (WY195 promoter fragment): 6ul
[0064] T4DNALigase (TaKaRa, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com