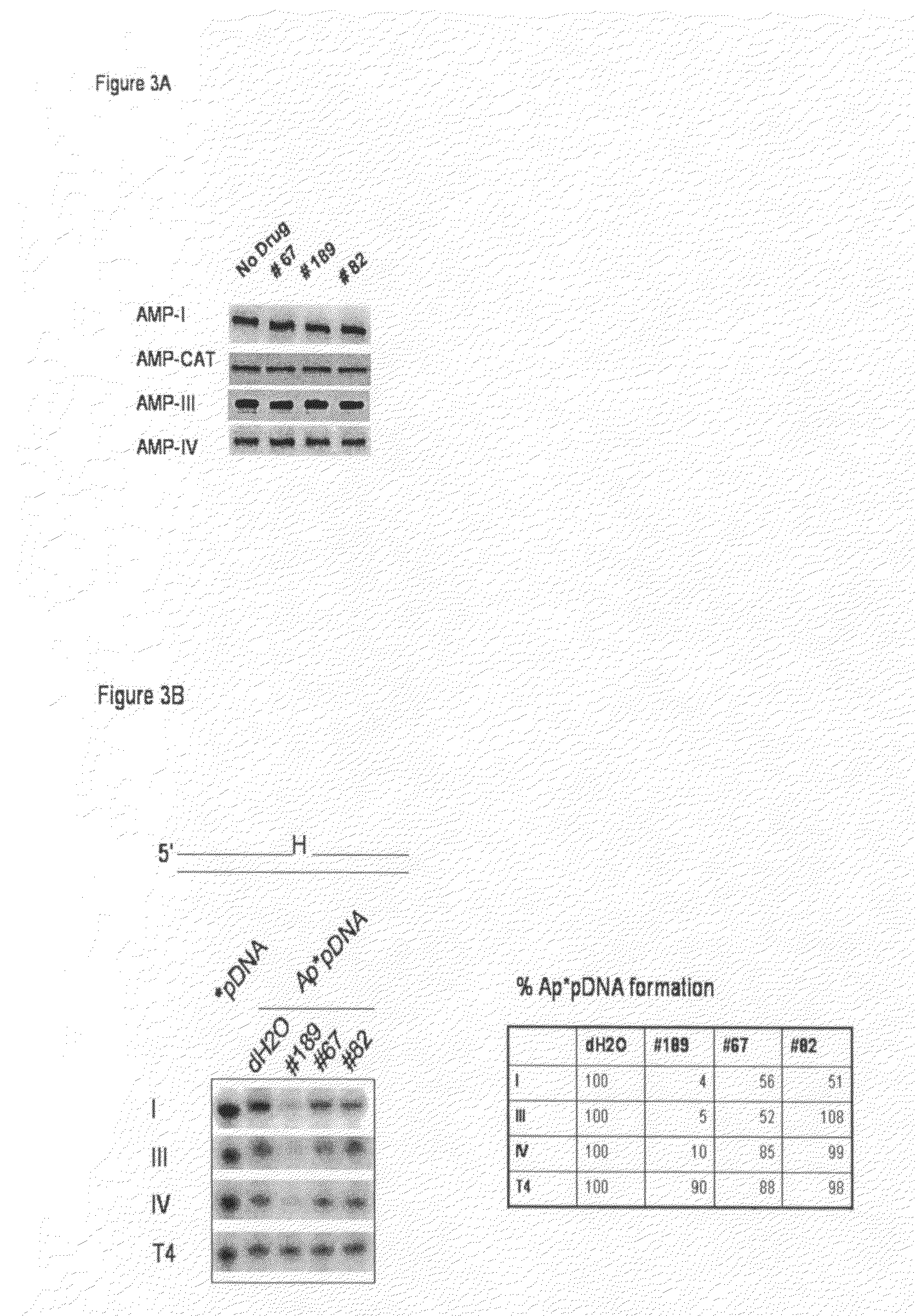
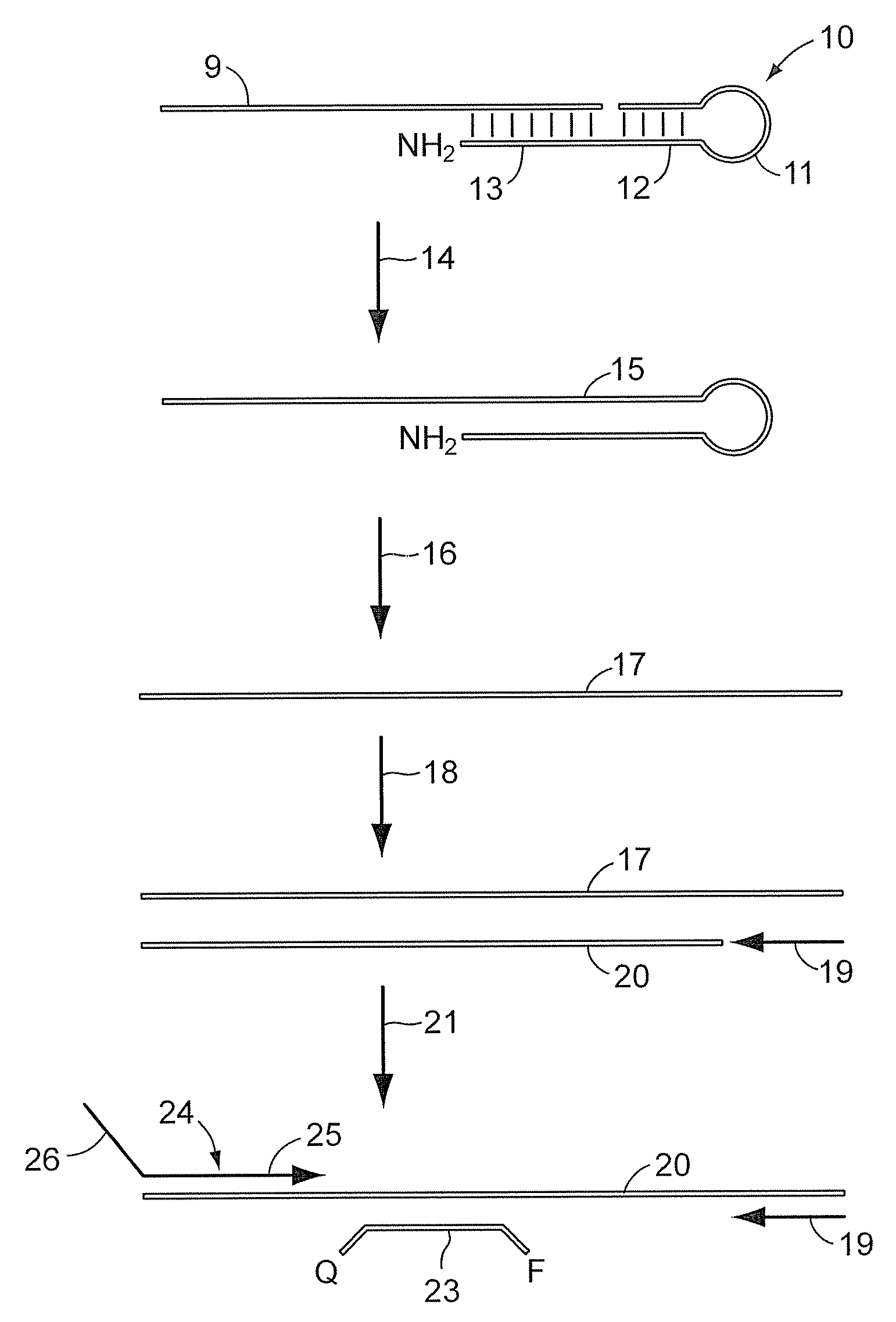
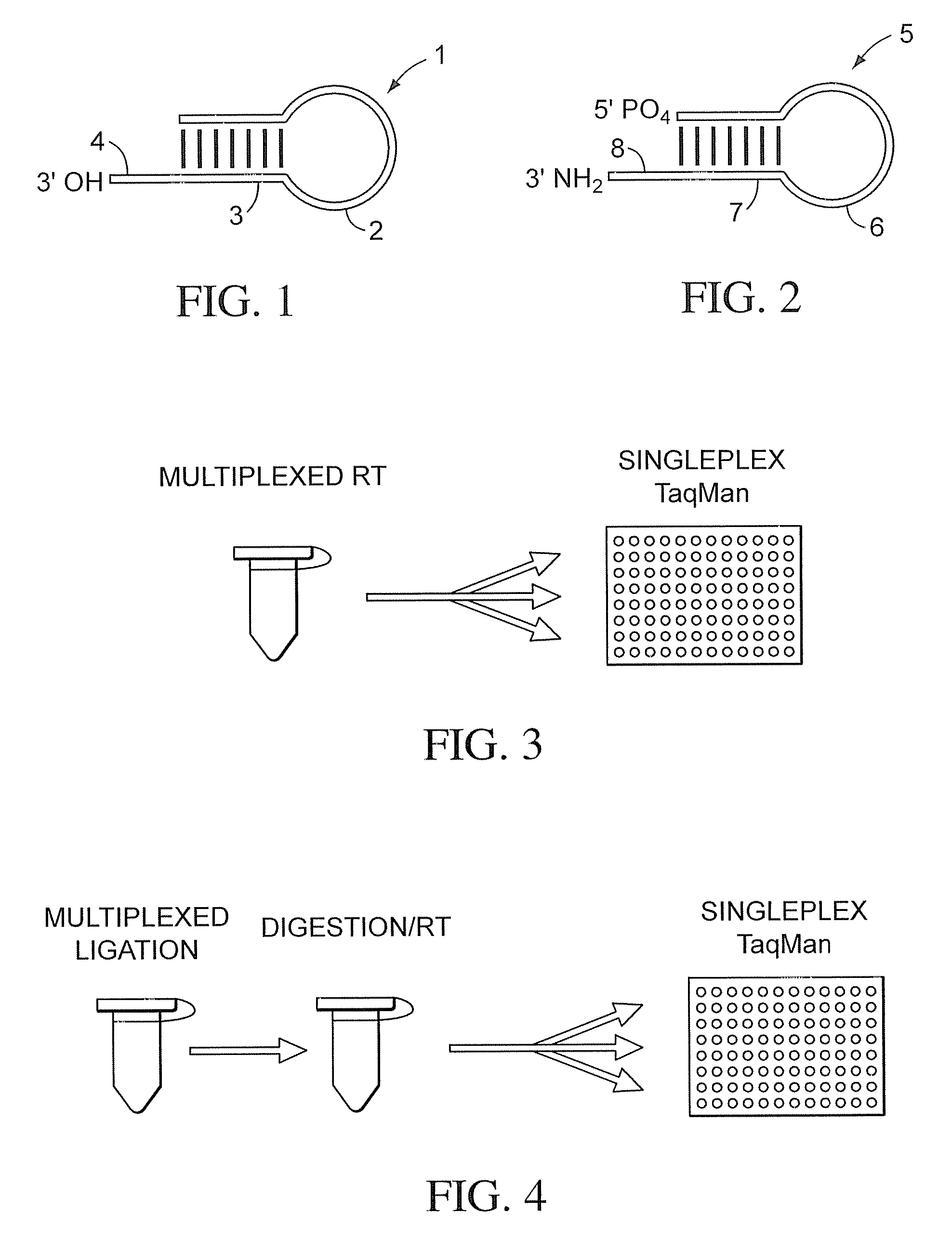
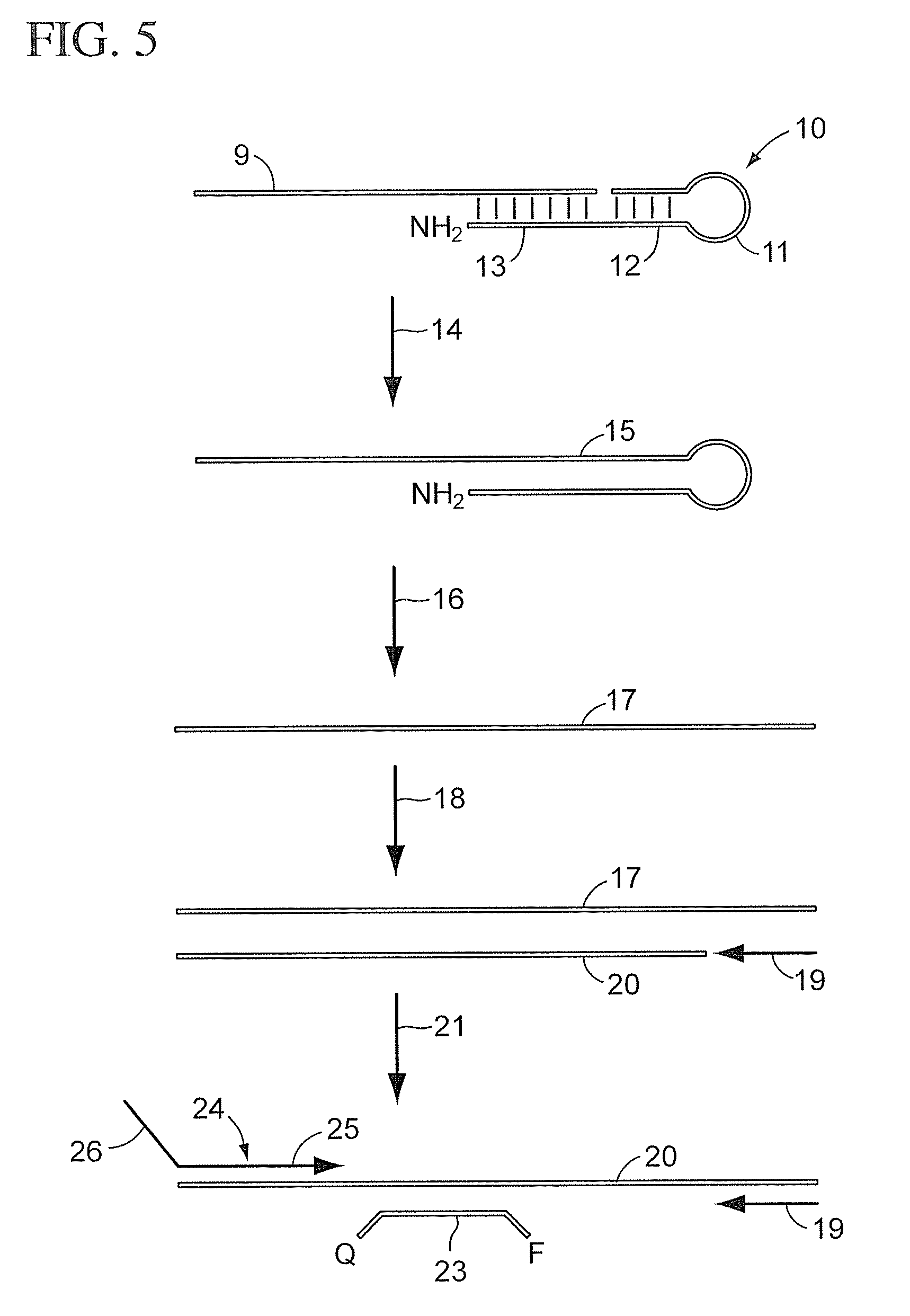
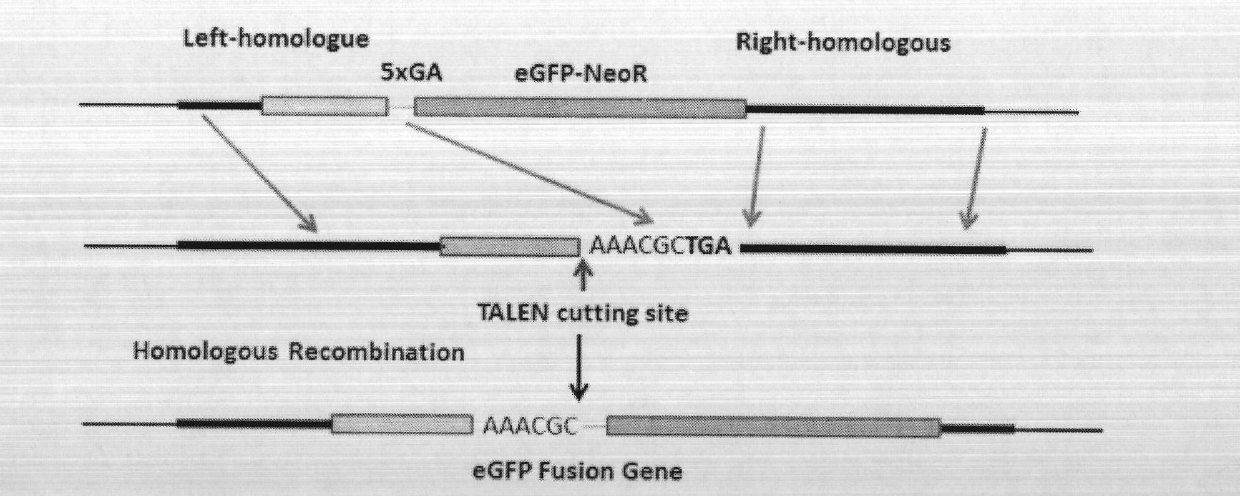
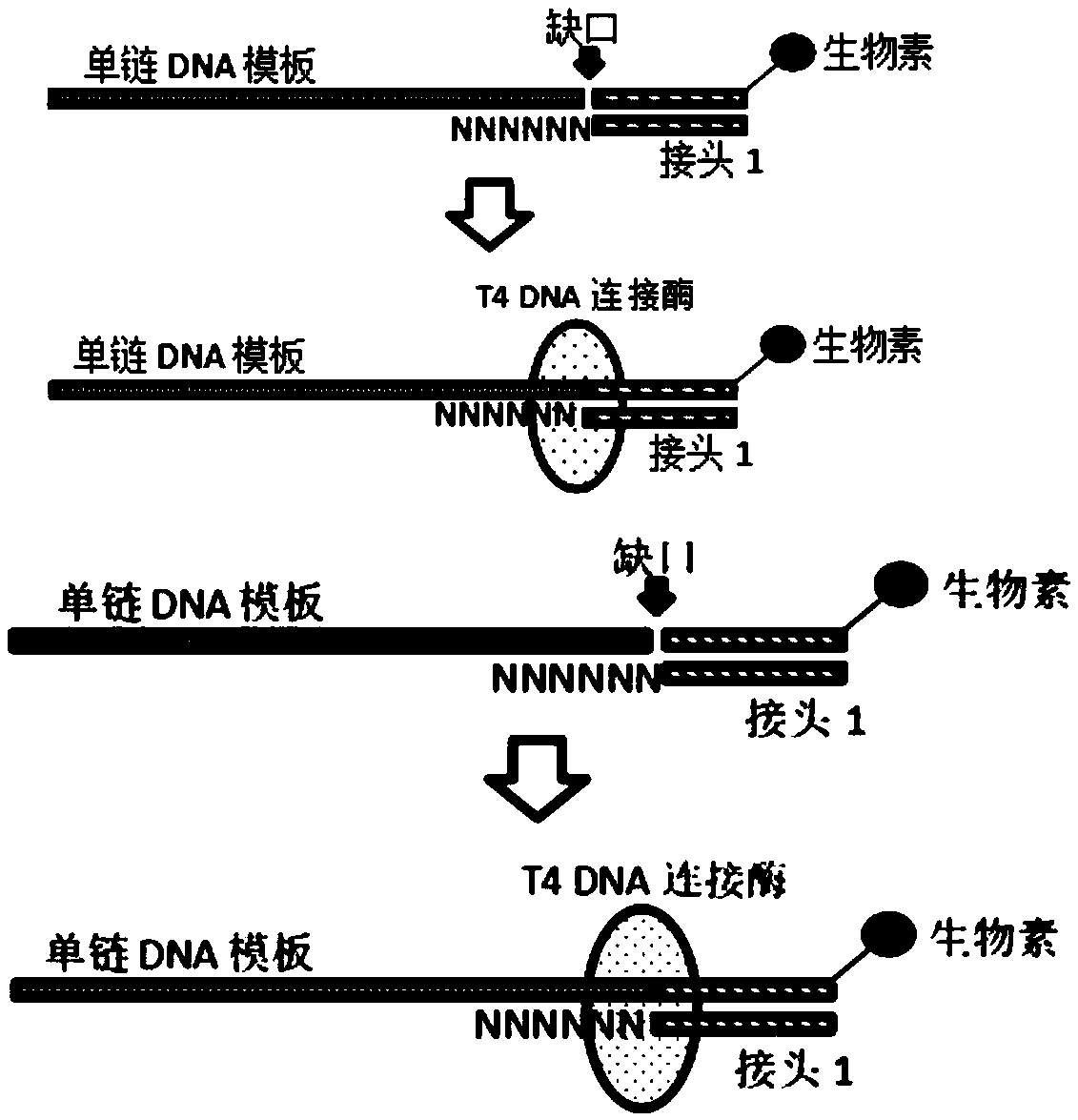
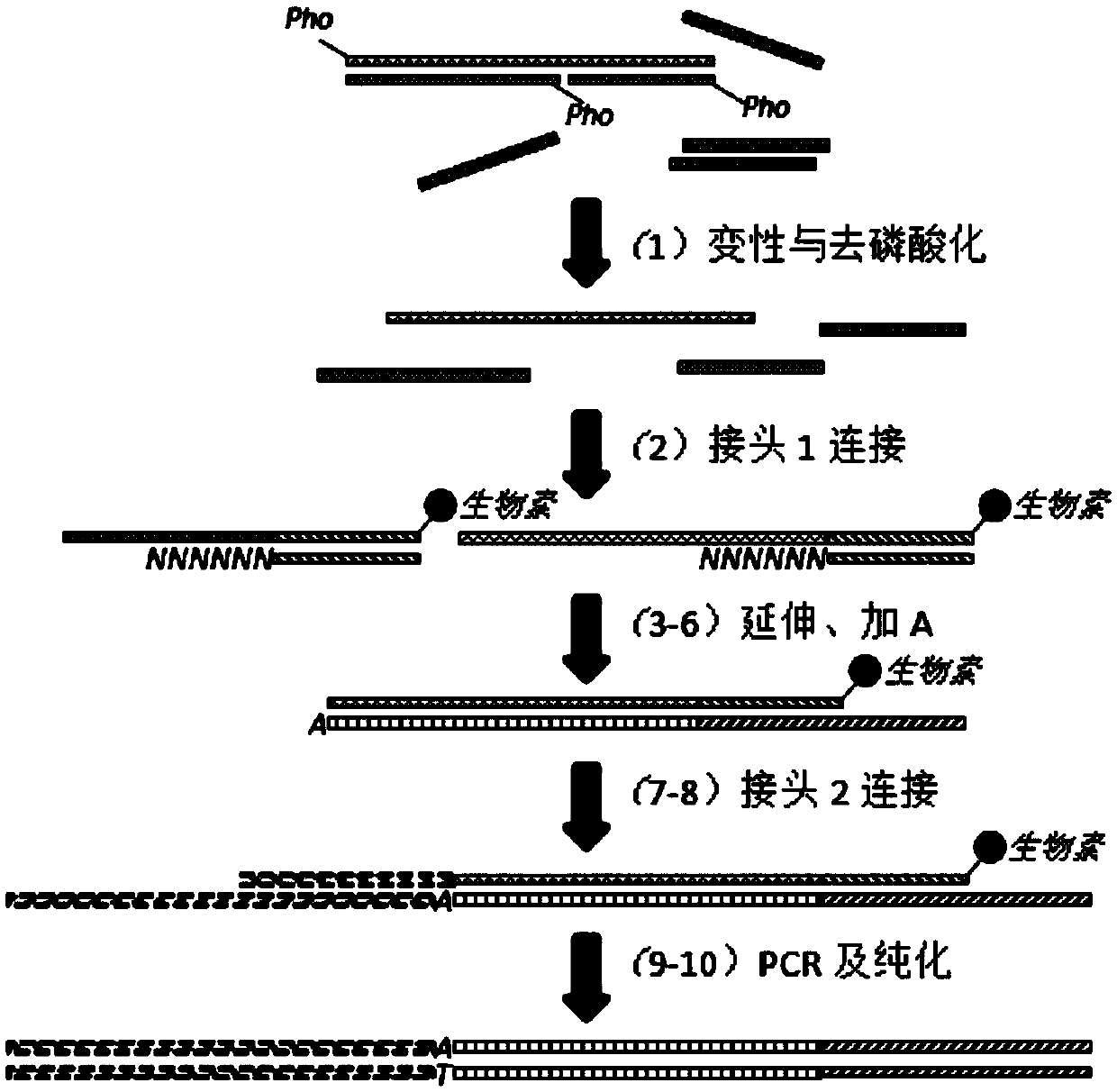
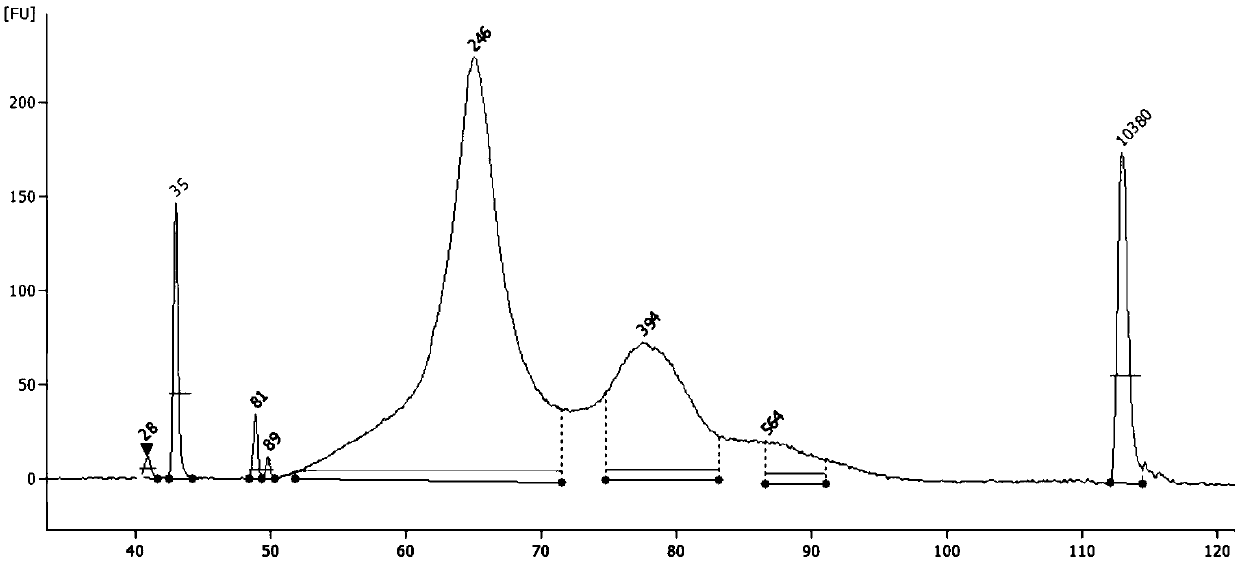
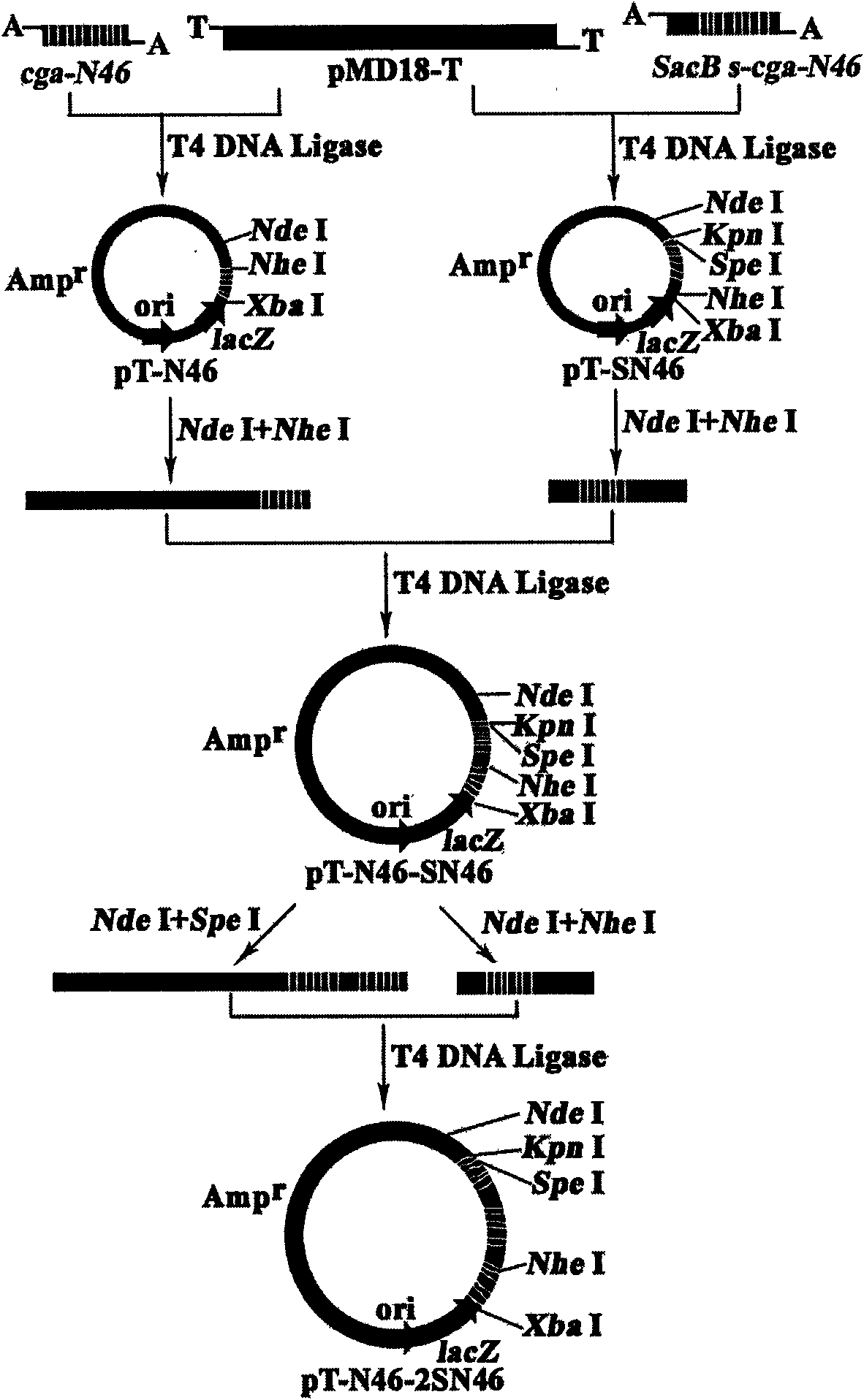
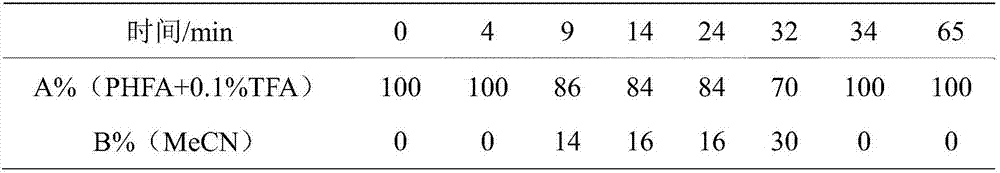
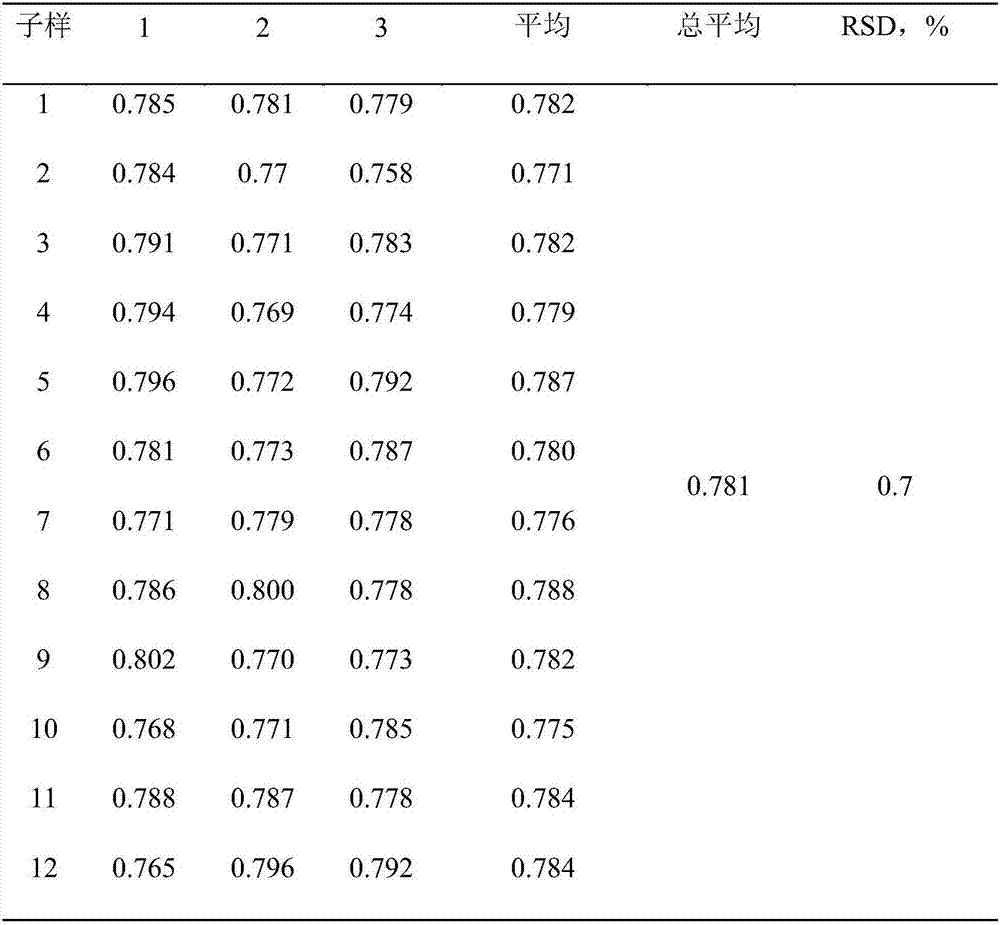
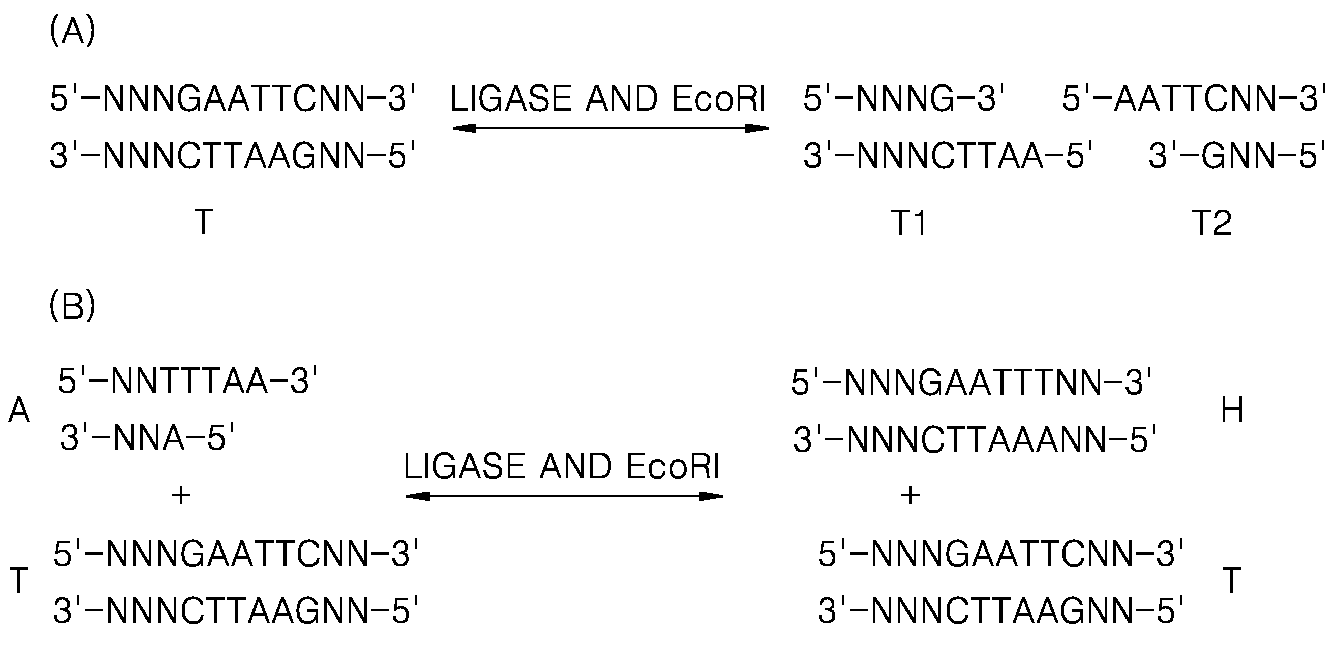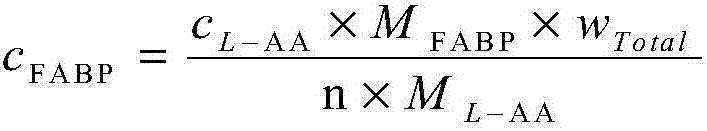Patents
Literature
131 results about "DNA ligase" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA ligase is a specific type of enzyme, a ligase, (EC 6.5.1.1) that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such as DNA ligase IV) may specifically repair double-strand breaks (i.e. a break in both complementary strands of DNA). Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.
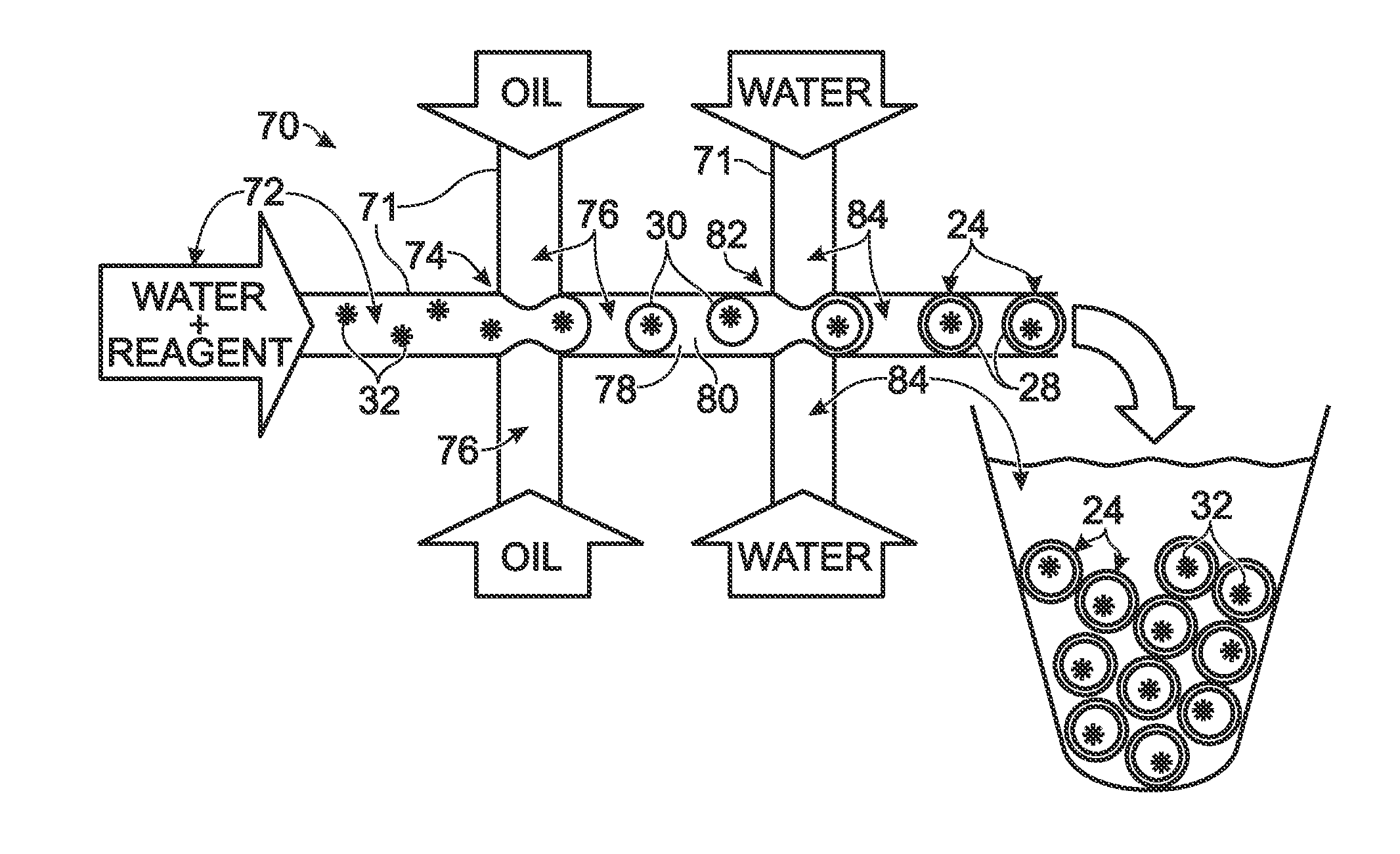
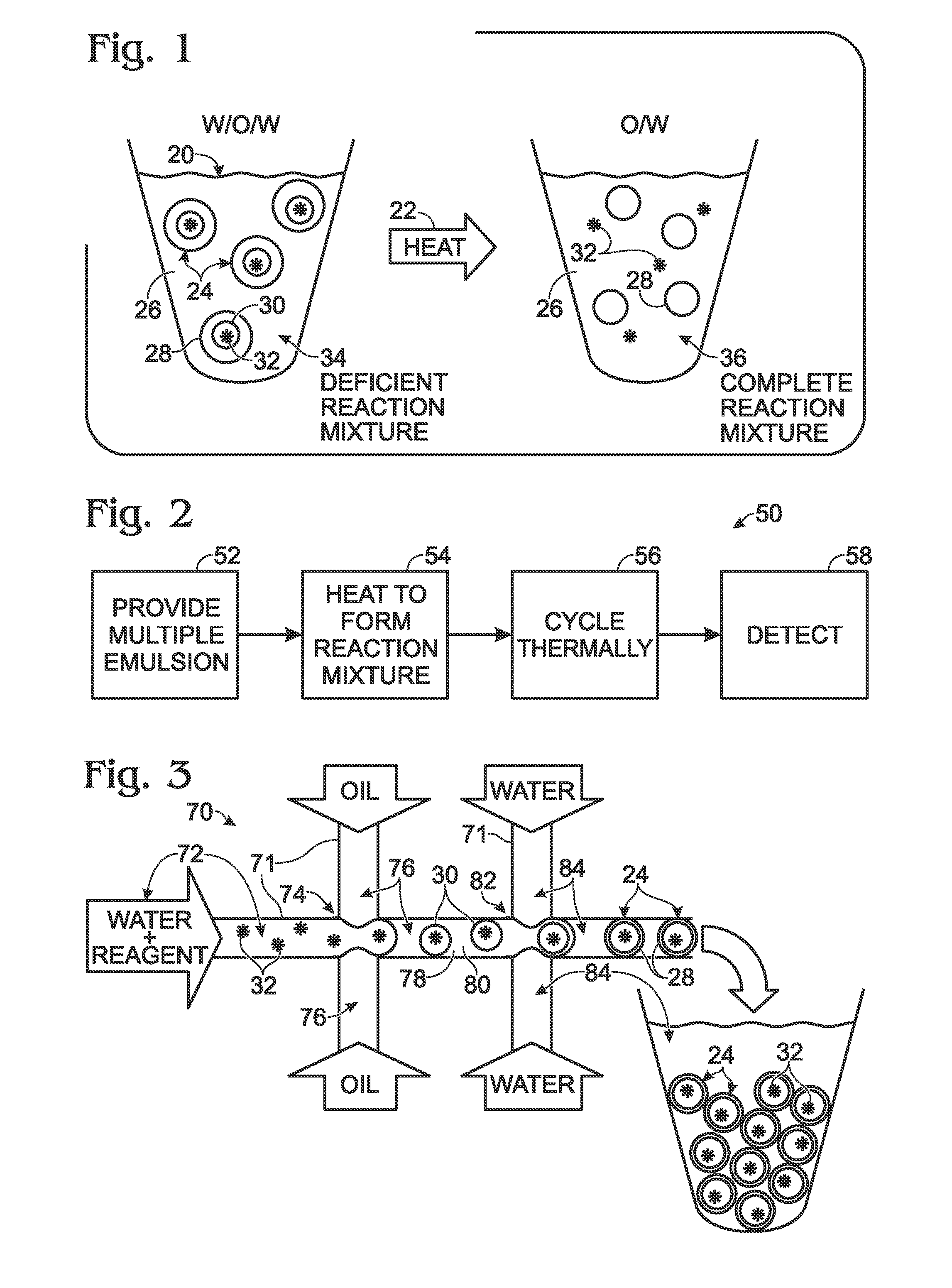
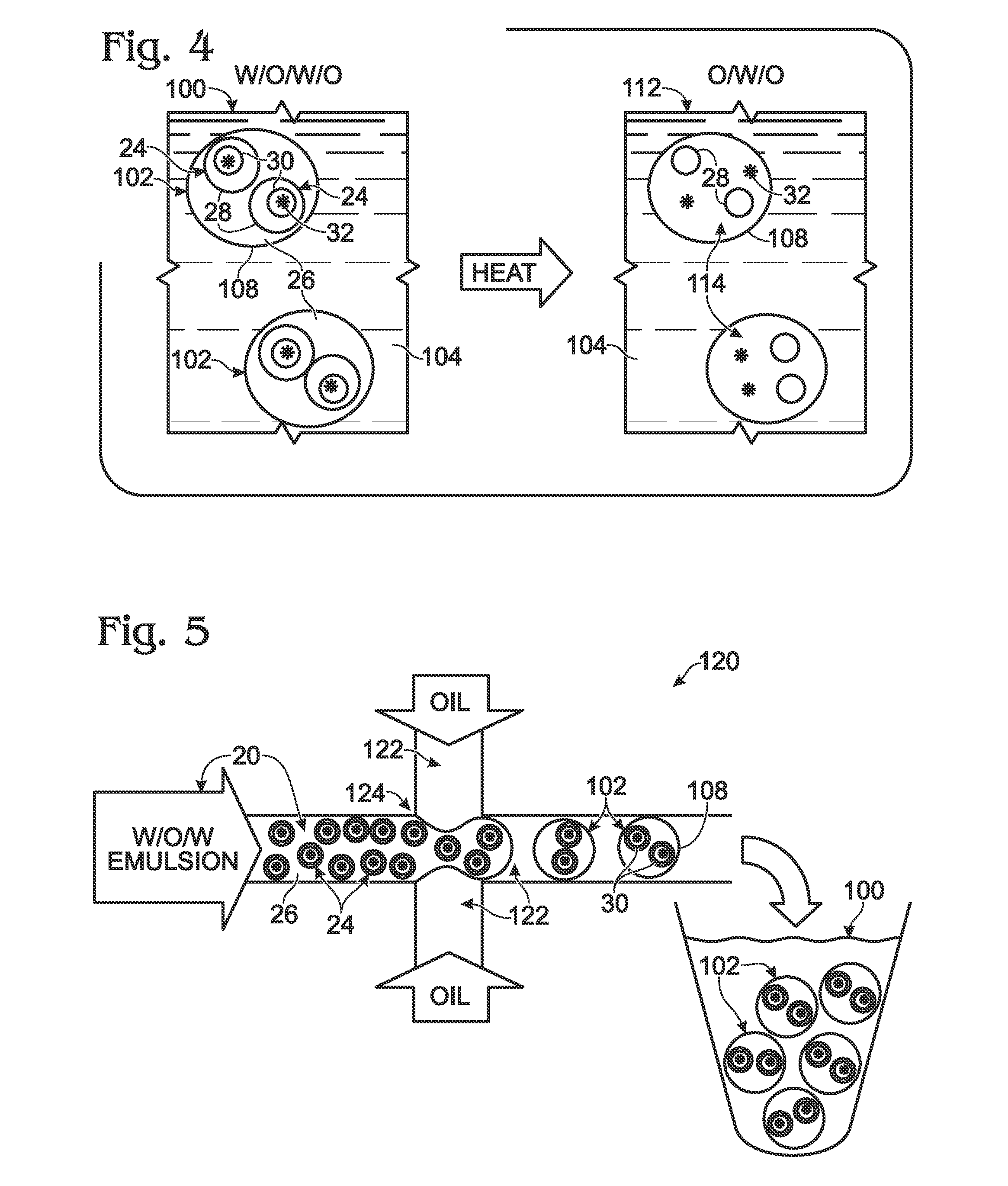
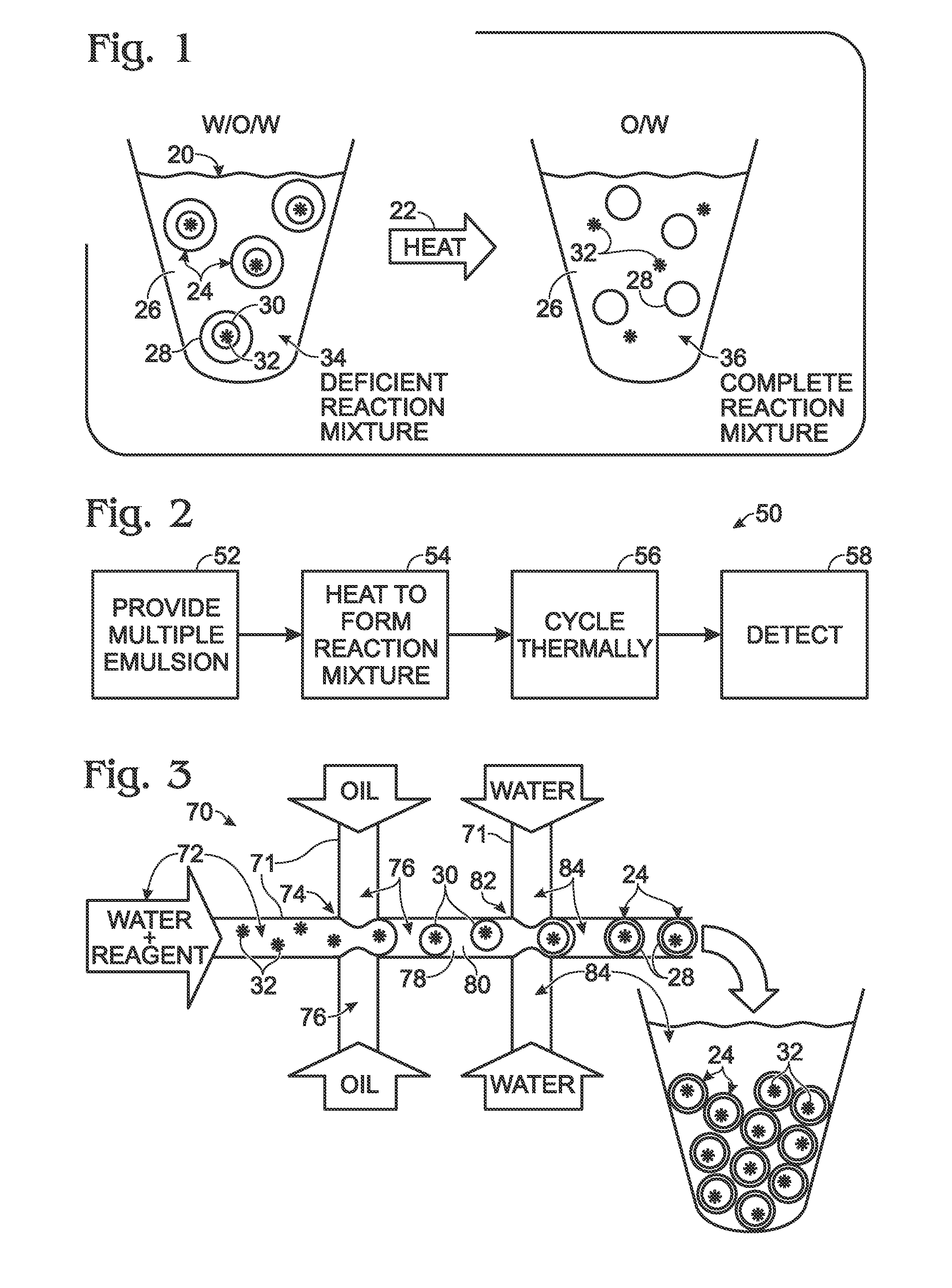
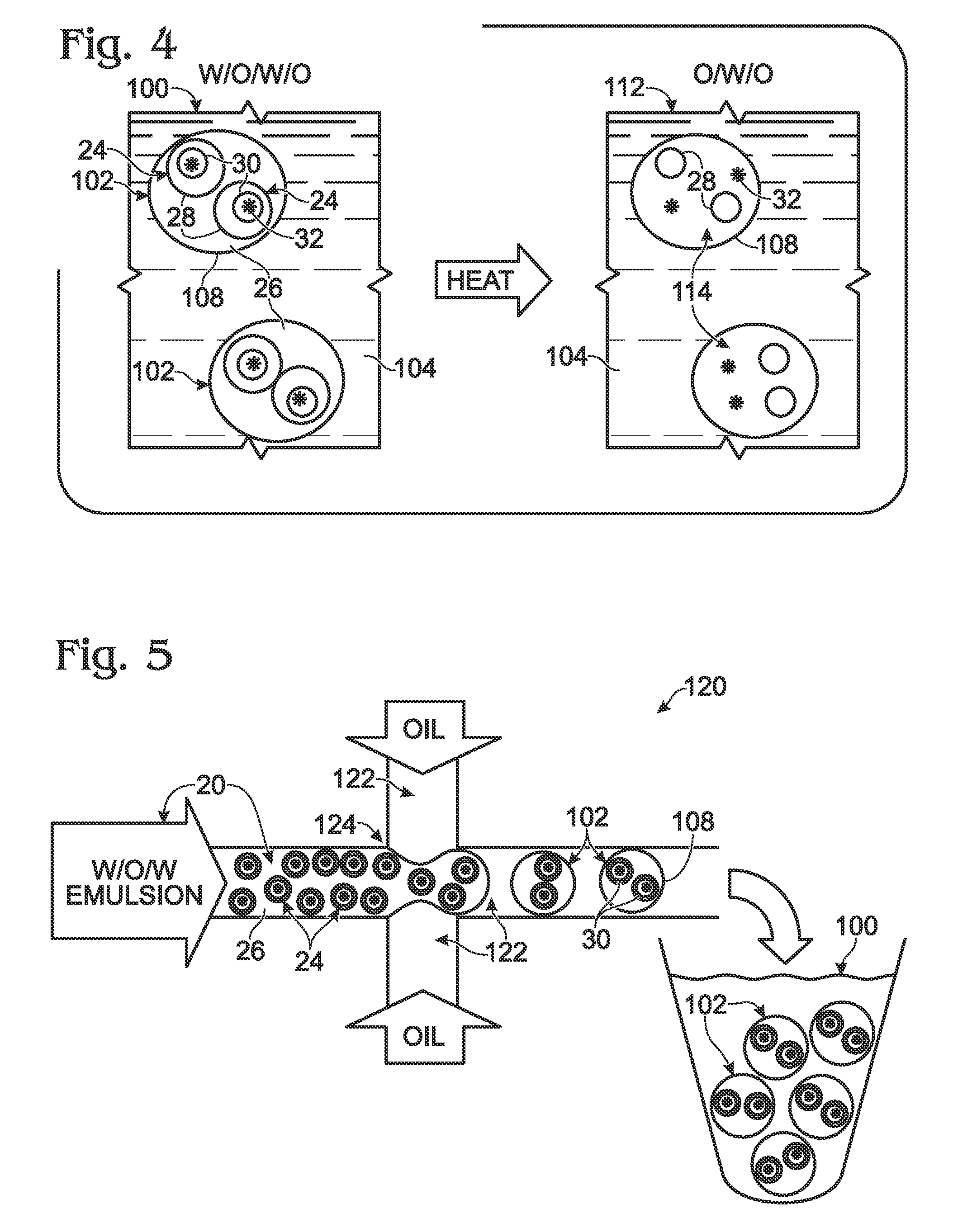
System for hot-start amplification via a multiple emulsion
System, including methods, apparatus, compositions, and kits, for making and using compound droplets of a multiple emulsion to supply an amplification reagent, such as a heat-stable DNA polymerase or DNA ligase, to an aqueous phase in which the compound droplets are disposed. The compound droplets may be induced to supply the amplification reagent by heating the multiple emulsion, to achieve hot-start amplification.
Owner:BIO RAD LAB INC
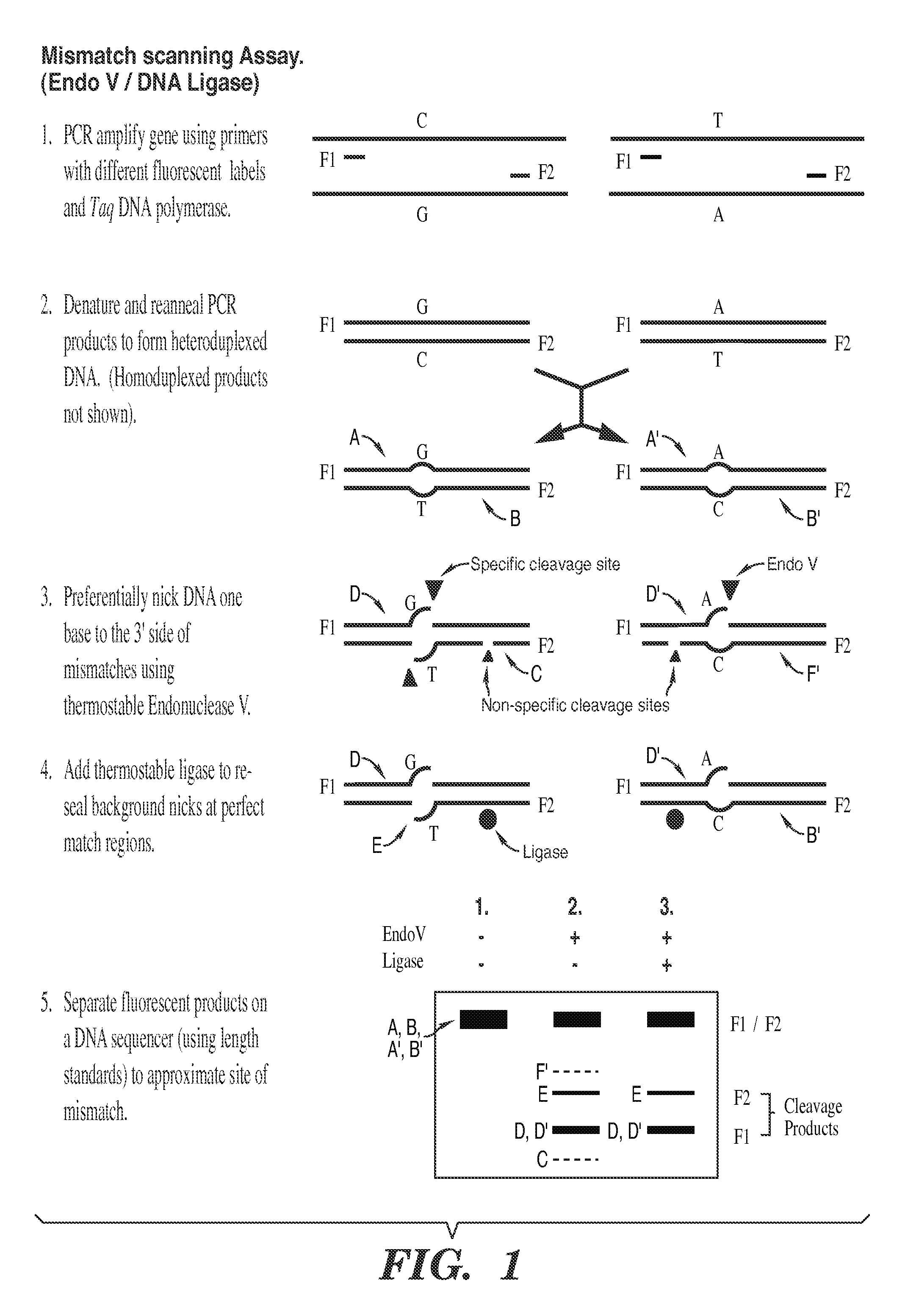
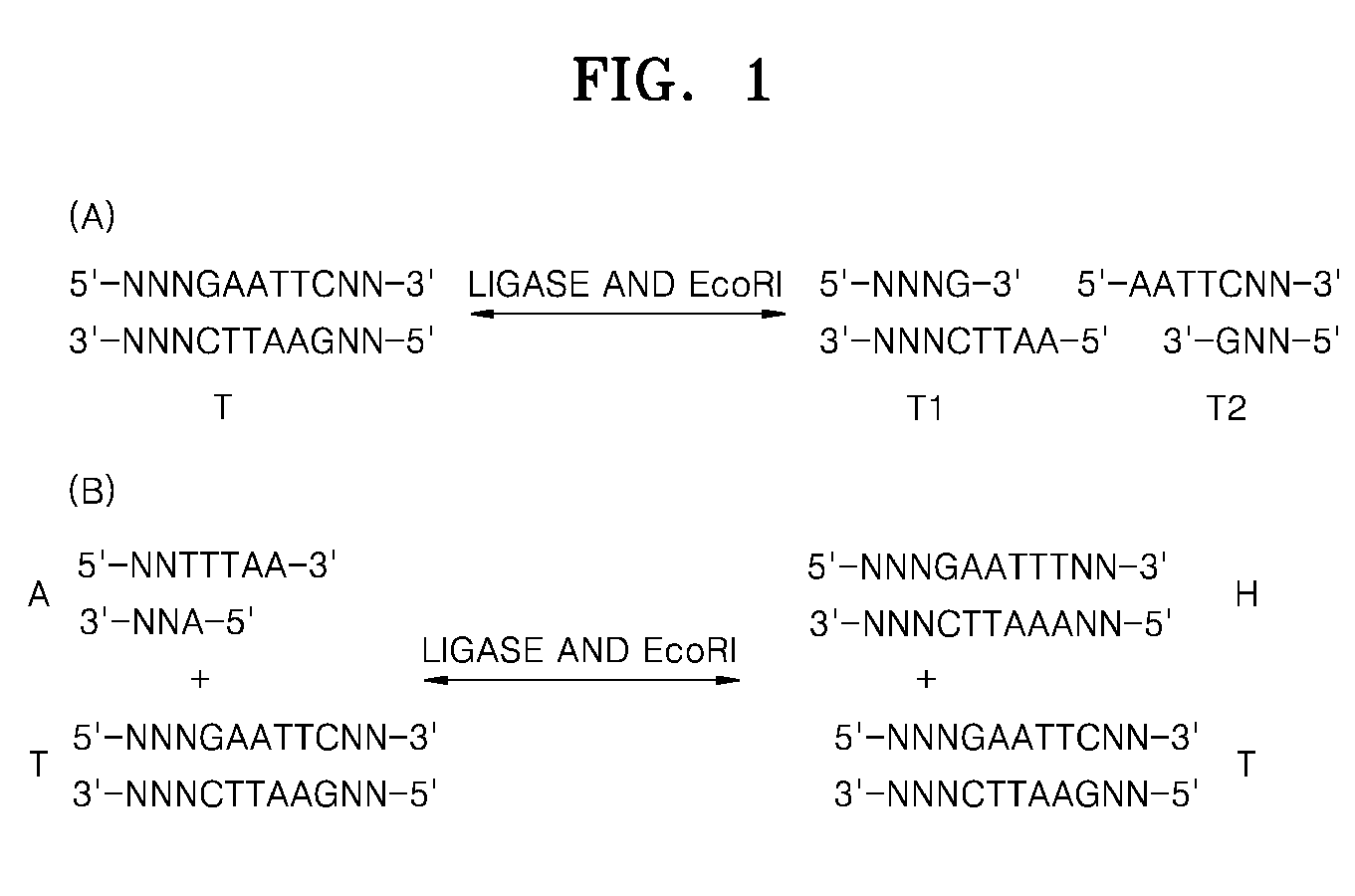
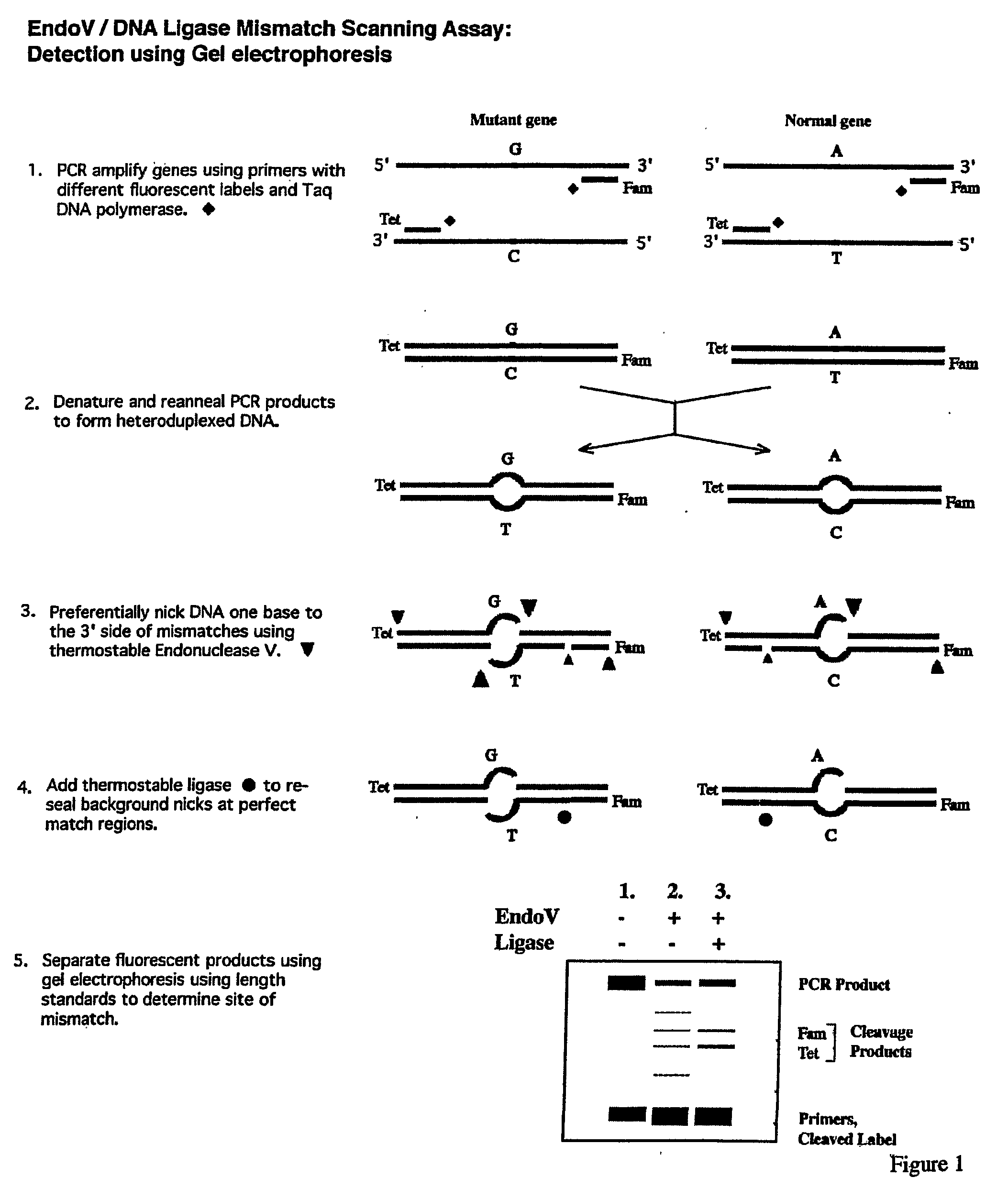
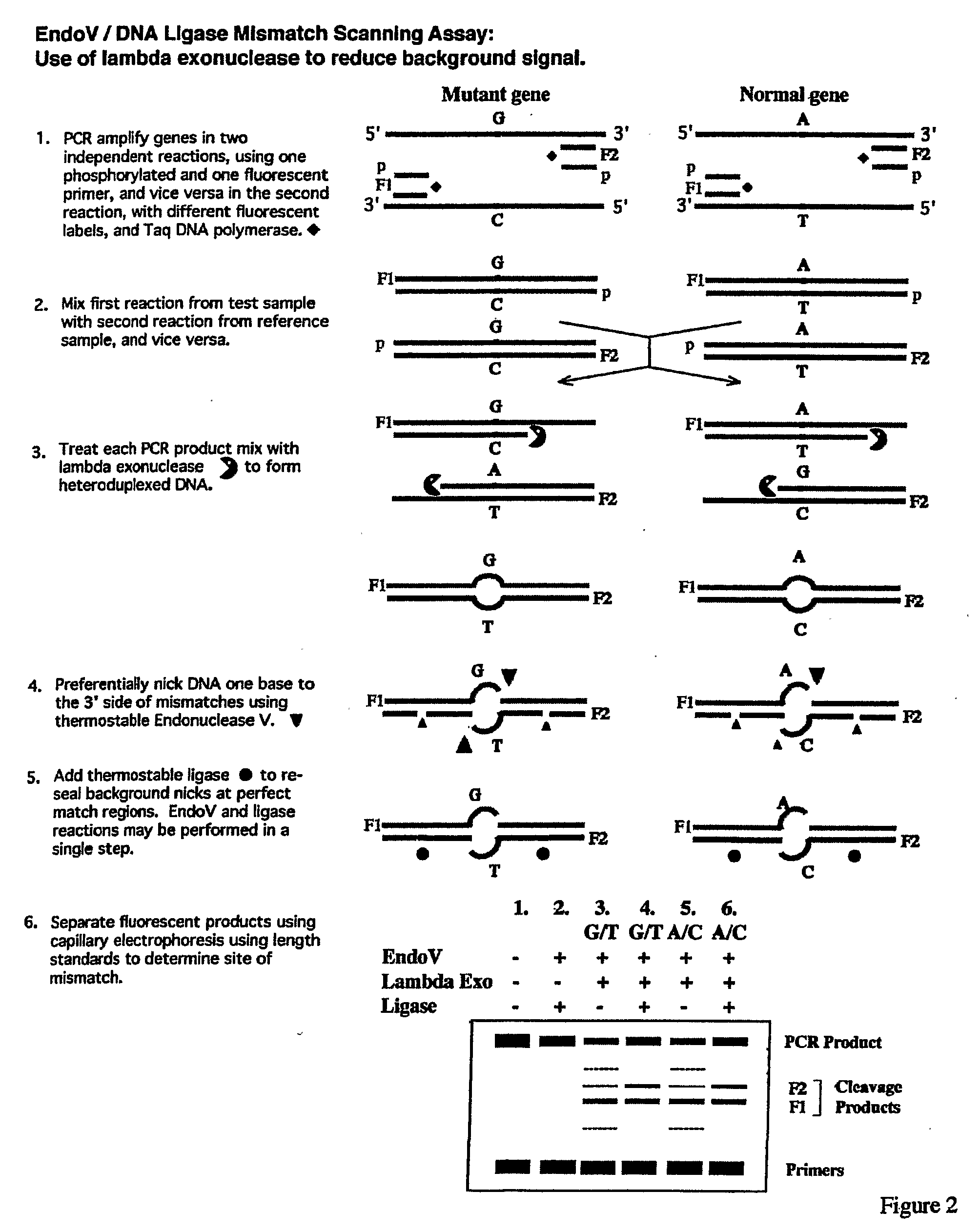
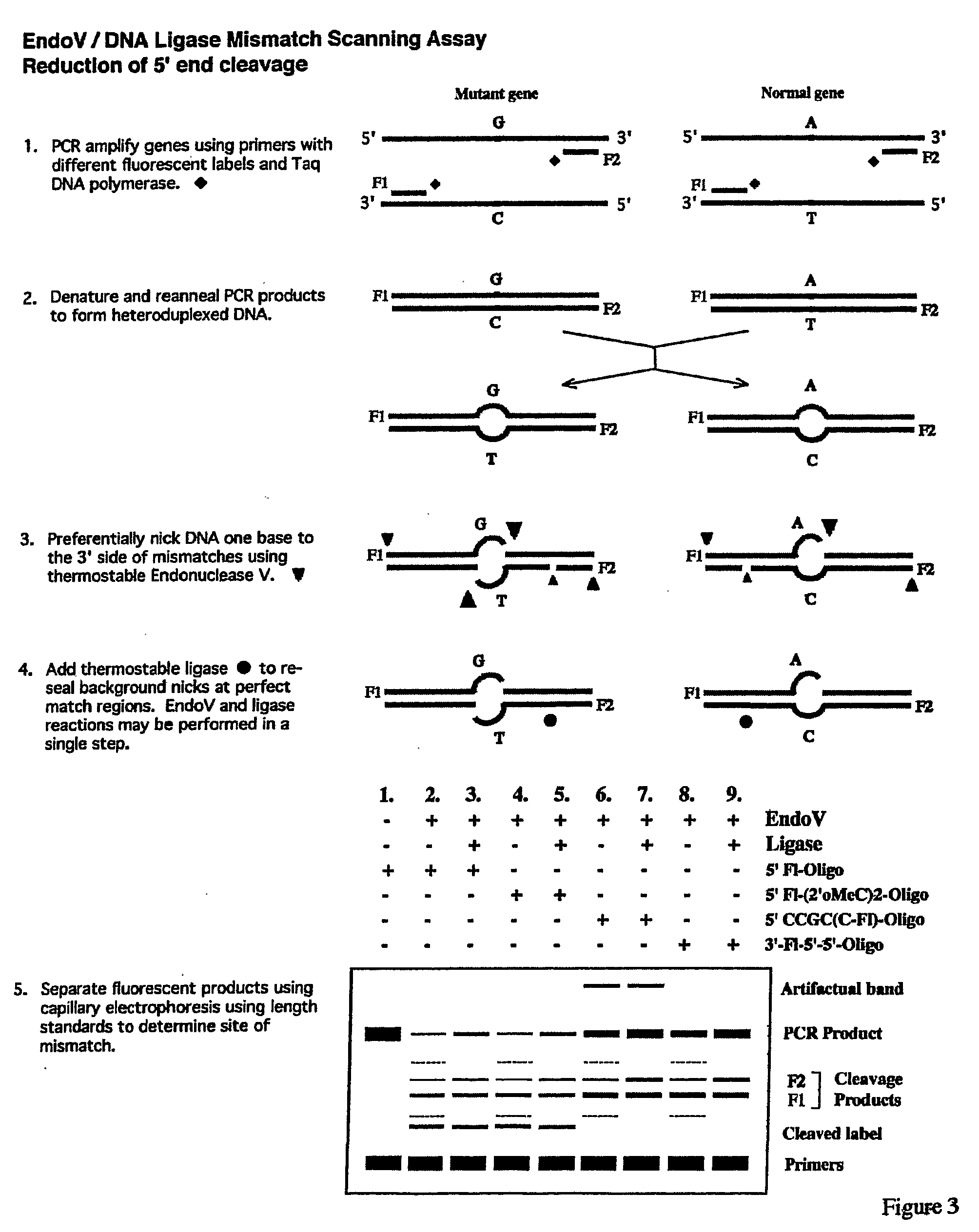
Detection of nucleic acid differences using combined endonuclease cleavage and ligation reactions
InactiveUS7807431B2Wide applicabilityImprove throughputSugar derivativesHydrolasesHeteroduplexSingle nucleotide mutation
The present invention is a method for detecting DNA sequence differences including single nucleotide mutations or polymorphisms, one or more nucleotide insertions, and one or more nucleotide deletions. Labeled heteroduplex PCR fragments containing base mismatches are prepared. Endonuclease cleaves the heteroduplex PCR fragments both at the position containing the variation (one or more mismatched bases) and to a lesser extent, at non-variant (perfectly matched) positions. Ligation of the cleavage products with a DNA ligase corrects non-variant cleavages and thus substantially reduces background. This is then followed by a detection step in which the reaction products are detected, and the position of the sequence variations are determined.
Owner:CORNELL RES FOUNDATION INC
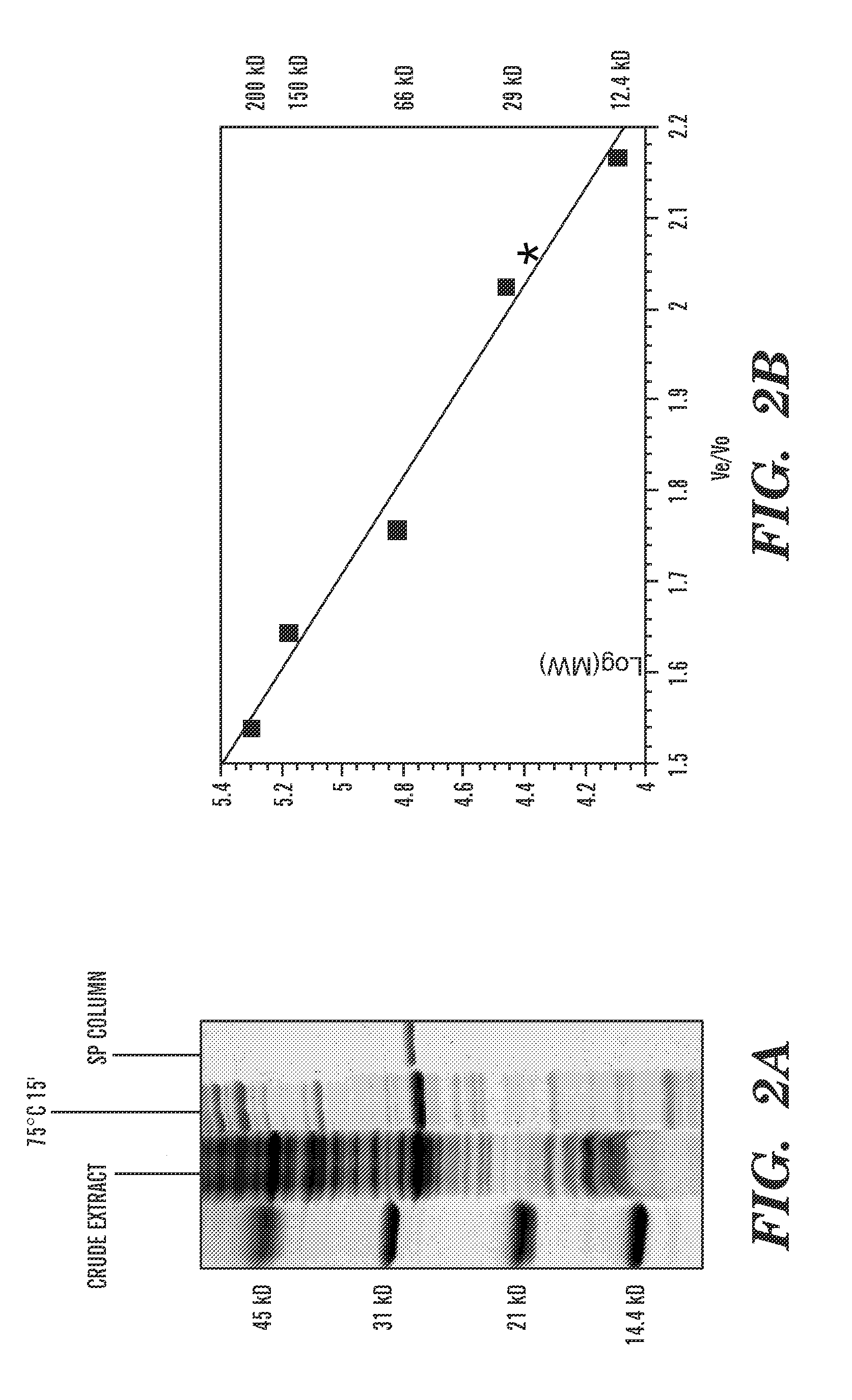
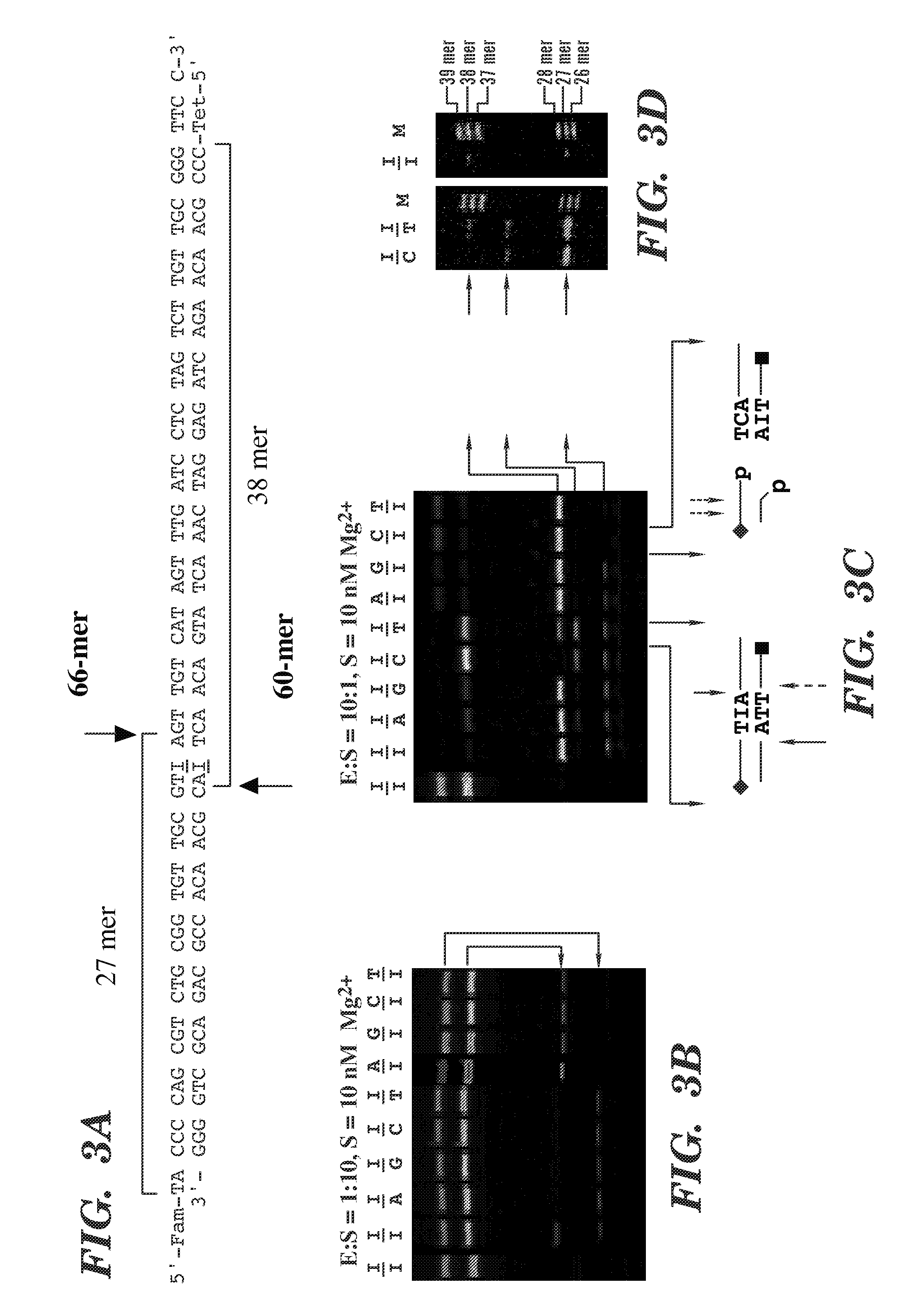
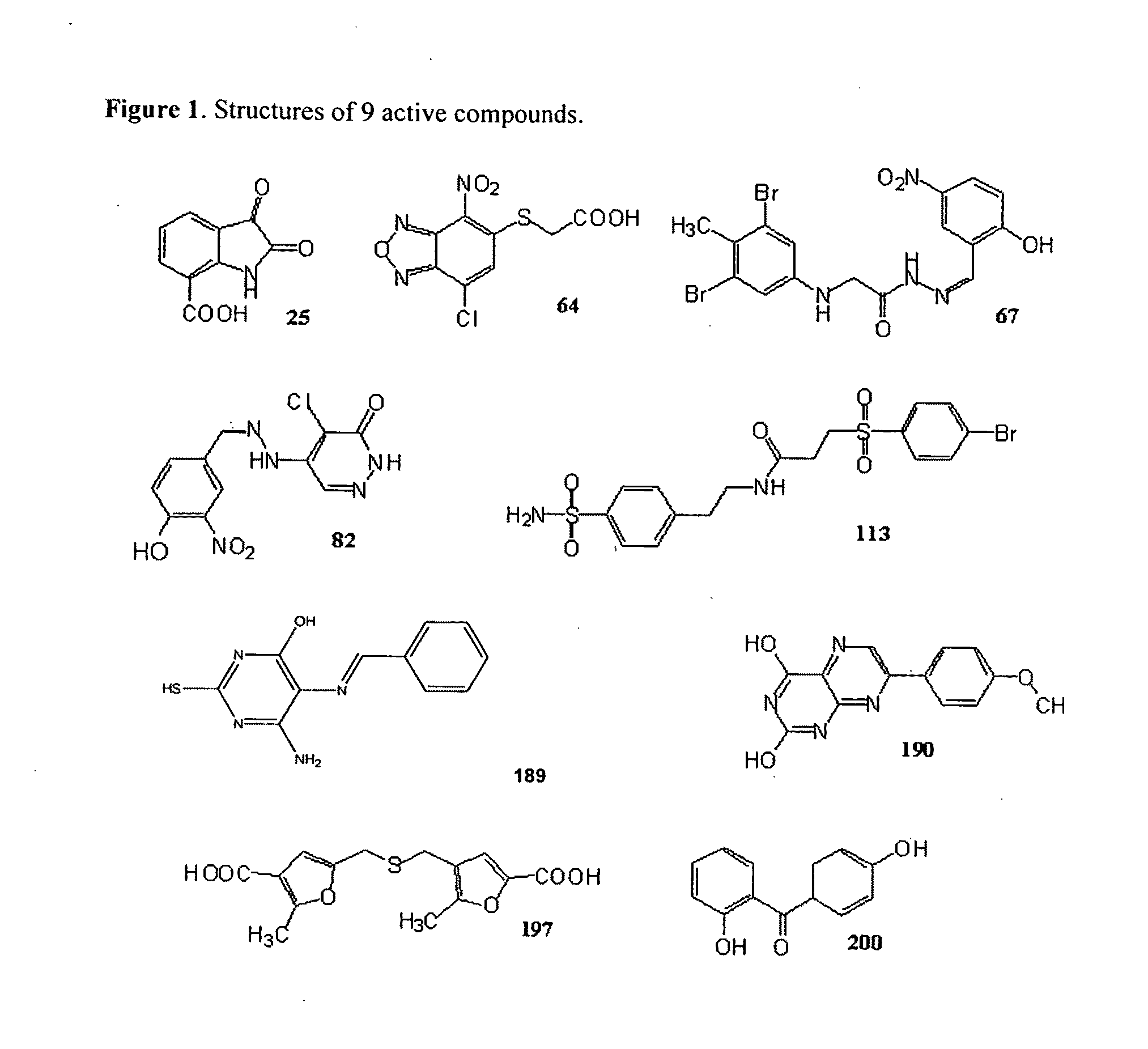
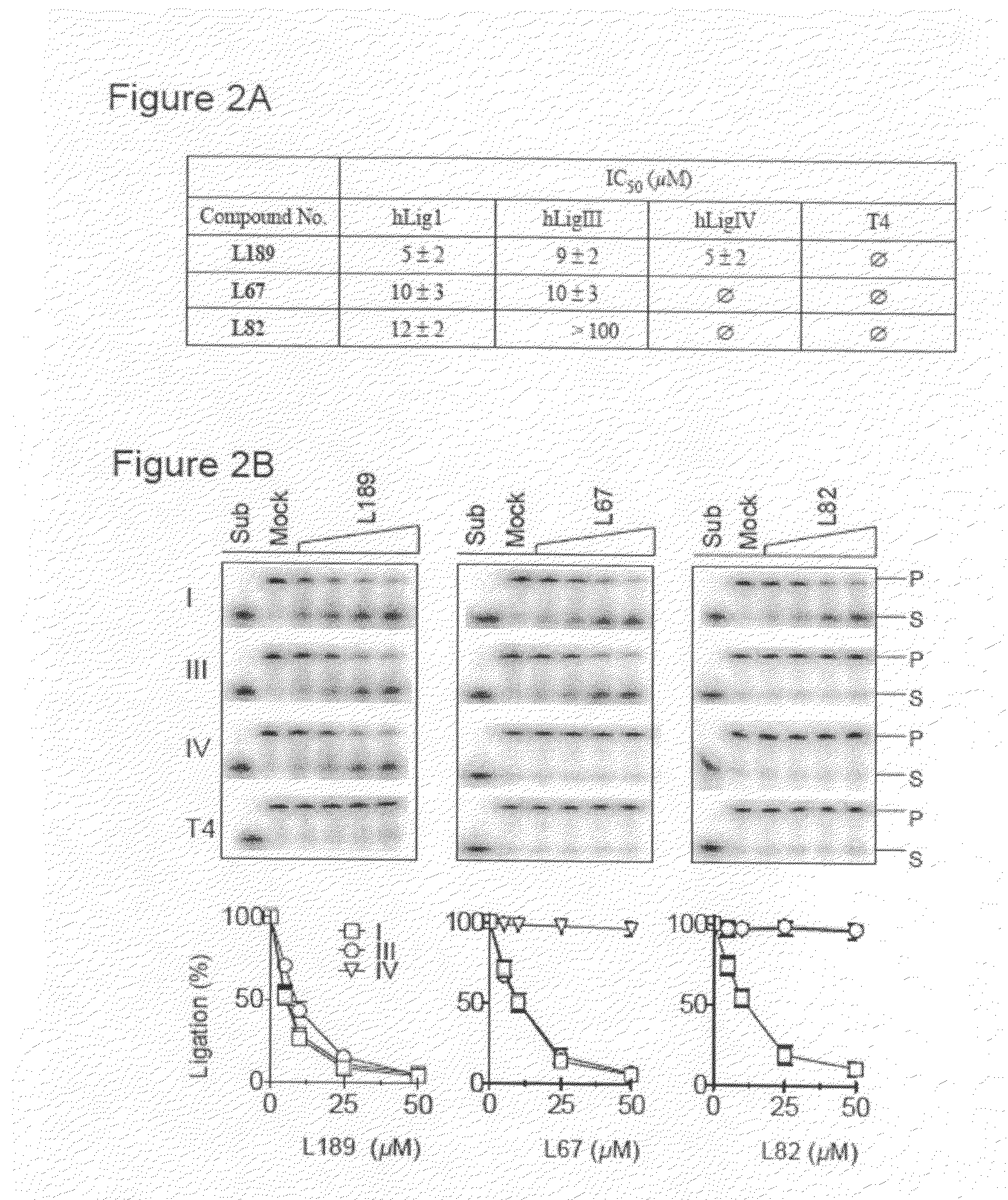
Compounds that inhibit human DNA ligases and methods of treating cancer
ActiveUS20100099683A1Strong cytotoxicityInhibit cell growthBiocideOrganic chemistryScreening methodBiochemistry
Methods for treating cancer using compounds that inhibit human DNA ligases. Methods for using compounds that inhibit human DNA ligases to provide insights into the reaction mechanisms of human DNA ligases, for example to identify the human DNA ligase involved in different DNA repair pathways. Screening methods for compounds that inhibit human DNA ligases.
Owner:UNIV OF MARYLAND
Method to Quantify siRNAs, miRNAs and Polymorphic miRNAs
The present teachings provide methods, compositions, and kits for quantifying target polynucleotides. In some embodiments, a reverse stem-loop ligation probe is ligated to the 3′ end of a target polynucleotide, using a ligase that can ligate the 3′ end of RNA to the 5′ end of DNA using a DNA template, such as T4 DNA ligase. Following digestion to form an elongated target polynucleotide with a liberated end, a reverse transcription reaction can be performed, followed by a PCR. In some embodiments, the methods of the present teachings can discriminate between polymorphic polynucleoitides that vary by as little as one nucleotide.
Owner:APPL BIOSYSTEMS INC
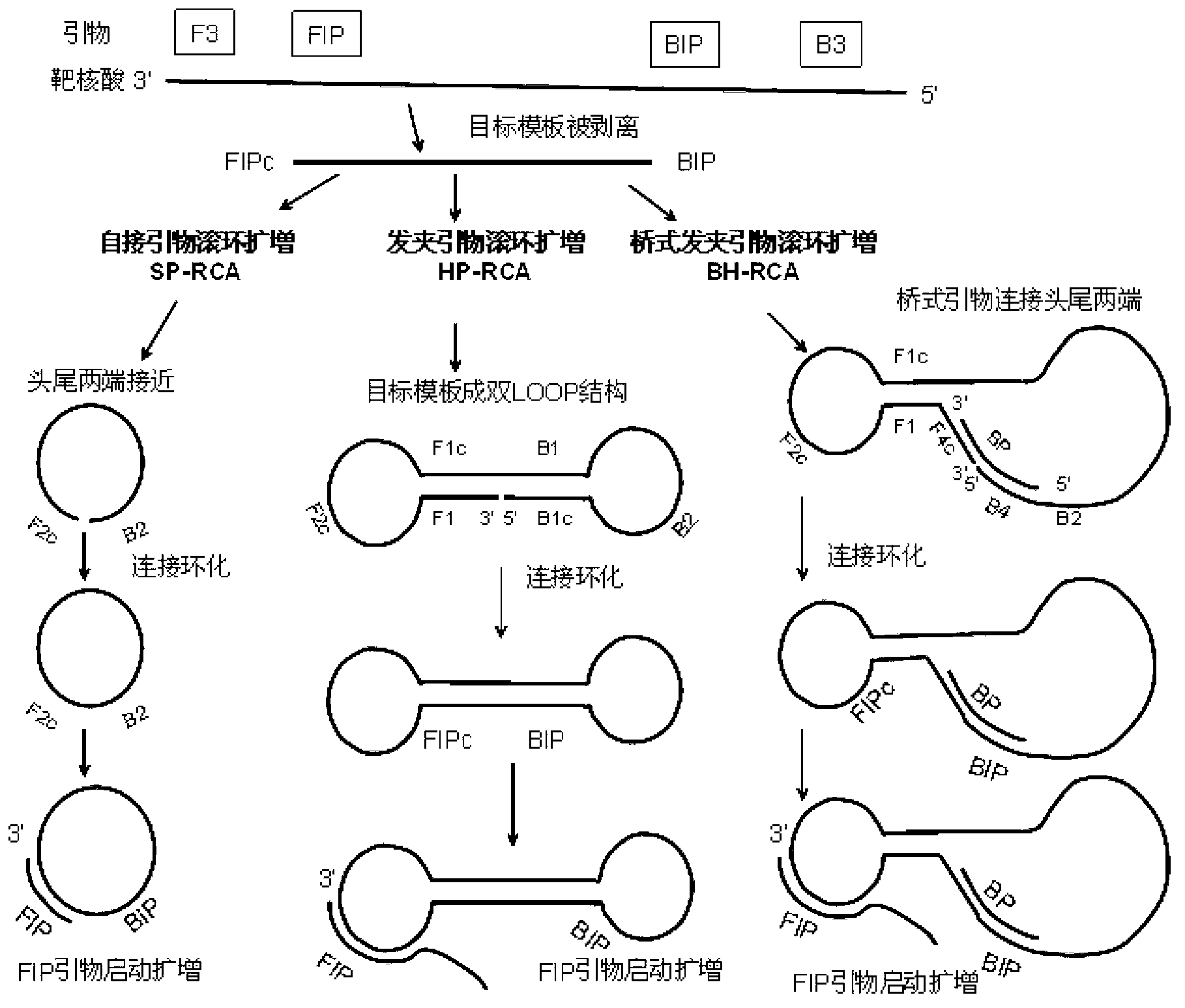
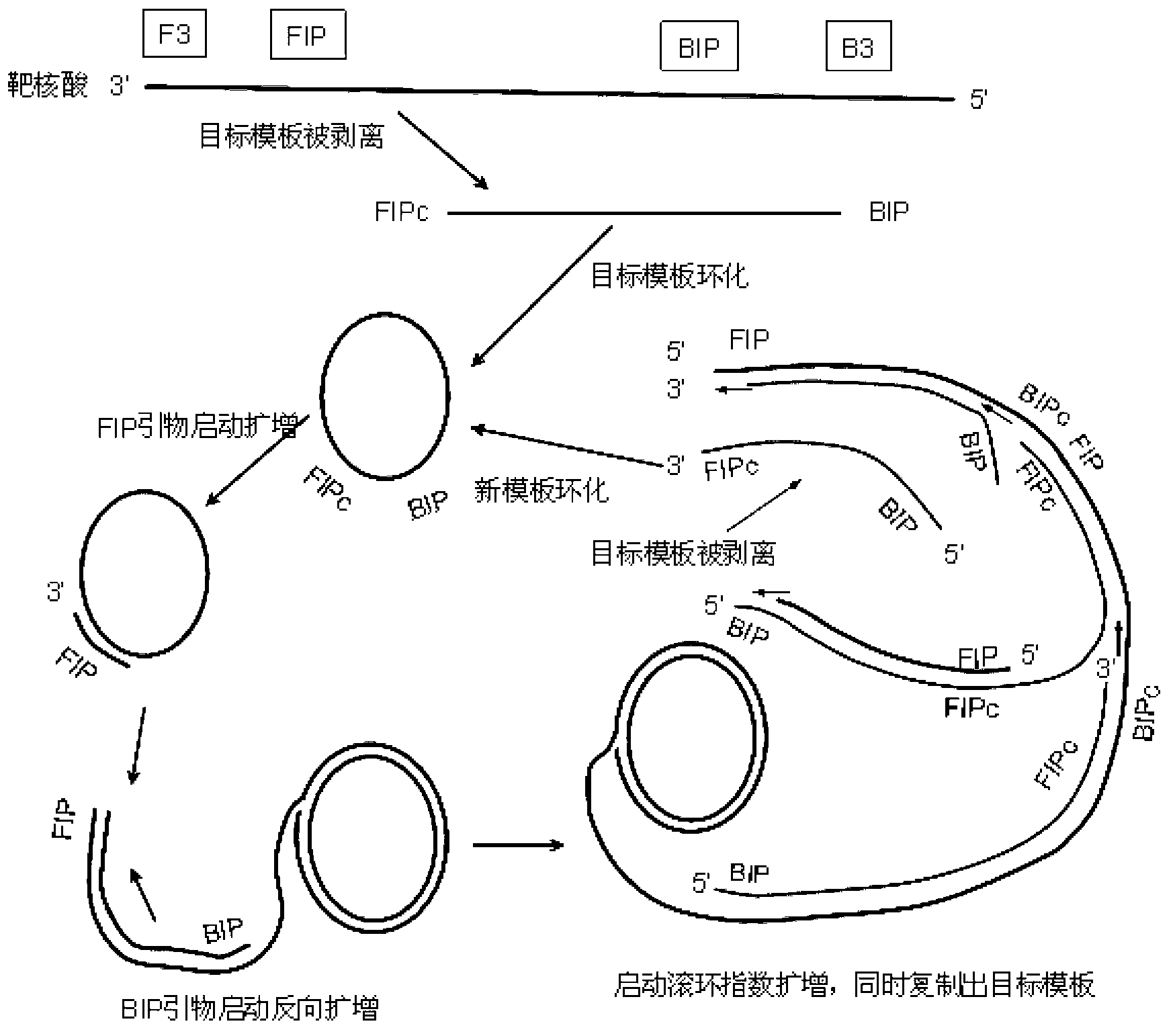
Primer-mediated cyclized constant-temperature nucleic acid rolling circle amplification method and kit
ActiveCN103255227AIncrease the lengthAvoid pollutionMicrobiological testing/measurementA-DNASingle strand dna
The invention discloses a primer-mediated cyclized constant-temperature nucleic acid rolling circle amplification method and a kit. The method comprises the following steps of: 1) designing an oligonucleotide sequence; 2) amplifying a single-stranded DNA (deoxyribonucleic acid) target template from target nucleic acid; 3) cyclizing the DNA target template through a DNA ligase; and 4) performing rolling circle amplification on the cyclized DNA target template. According to the method disclosed by the invention, a long padlock probe with high cost does not need to be synthesized, the length of the sequence of the target nucleic acid can be allowed to be longer, the rolling circle amplification can be performed on any sequence of the target nucleic acid, a solution can be provided for preventing a sample from being polluted by an amplified product, and a new era that gene detection enters applications in basic level units is opened.
Owner:QUICKING BIOTECH
Method of amplifying a target nucleic acid by rolling circle amplification
Provided is kit for and a method of amplifying a nucleic acid using rolling cyclic amplification (RCA), including amplifying a nucleic acid together with formation of a single-strand circular DNA template using RCA by reacting a reaction solution including: (a) two hairpin oligos, (b) a target nucleic acid, (c) a DNA ligase,(d) an endonuclease, (e) a DNA polymerase, and (f) a primer.
Owner:SAMSUNG ELECTRONICS CO LTD
CRISPR (clustered regularly interspaced short palindromic repeat) based DNA detecting and typing method and application thereof
ActiveCN107828874ASimple detectabilitySimplified typesMicrobiological testing/measurementNucleic acid detectionSpecific detection
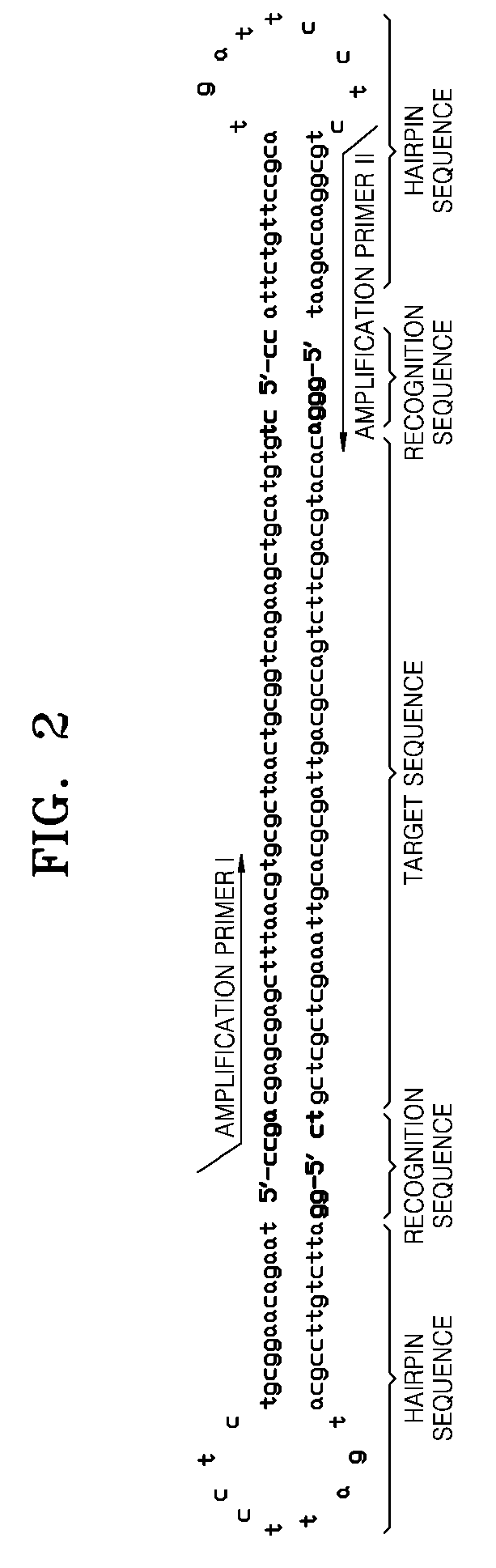
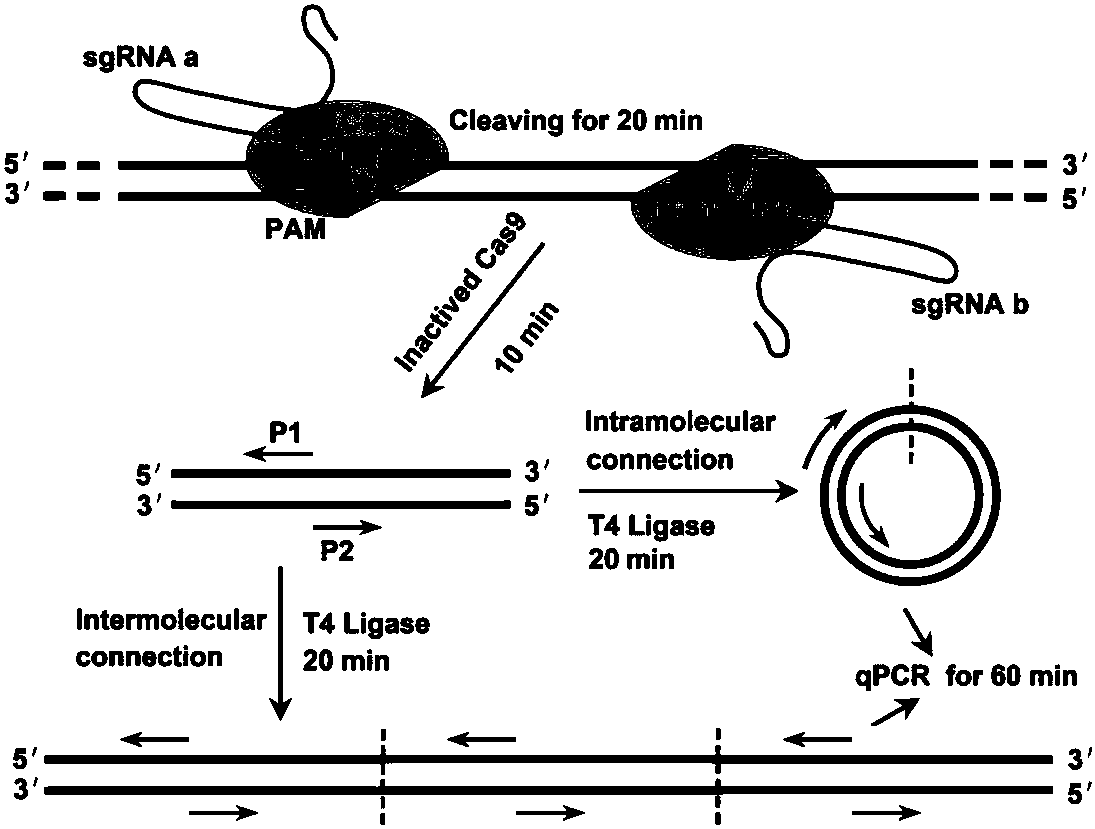
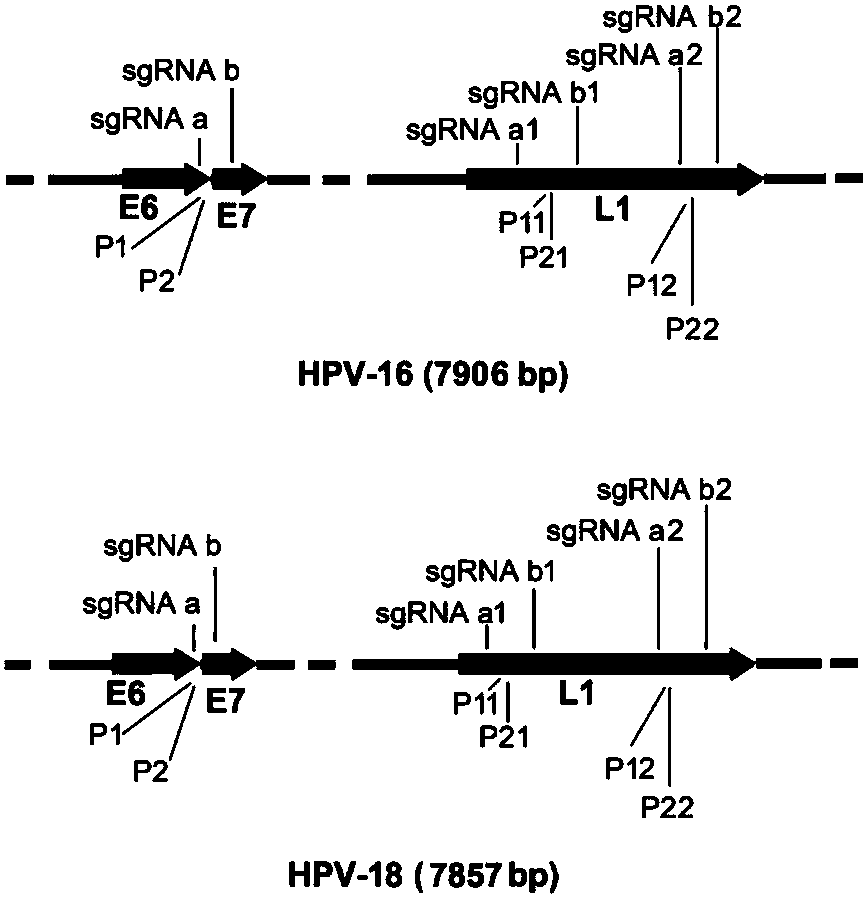
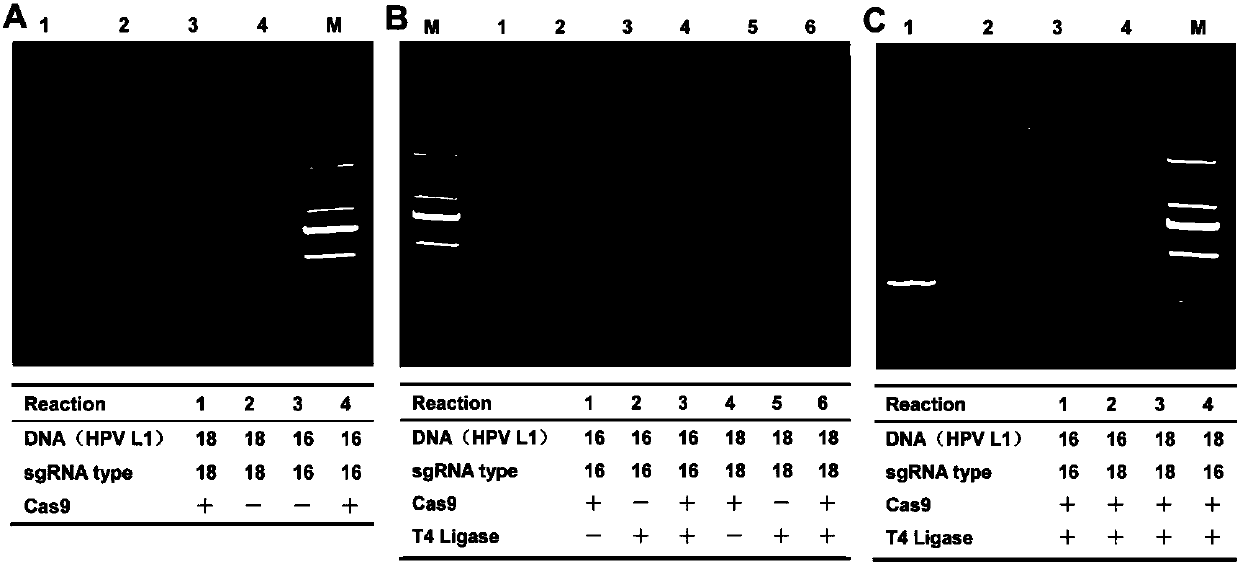
The invention discloses a CRISPR (clustered regularly interspaced short palindromic repeat) based DNA detecting and typing method and an application thereof. The method comprises following steps: (1),target DNA is subjected to PCR (polymerase chain reaction) amplification by a pair of universal primers; (2), the amplified target DNA is cut by Cas9 / sgRNA; (3), the cut target DNA is linked with DNAligase; (4), the linked target DNA is subjected to PCR amplification. According to the method, the target DNA can be subjected to specific detection and typing simply, rapidly and sensitively according to the specific identification cutting characteristics of the CRISPR technology on DNA, the method is one novel DNA detection method with high specificity and sensitivity, and the critical bottleneck problems about nucleic acid hybridization, specific PCR primer design and the like in current nucleic acid detection and typing fields are solved successfully. With the application of the method, L1 and E6 / E7 genes of HPV16 and HPV18 in human cervical cancer cells are detected successfully.
Owner:SOUTHEAST UNIV
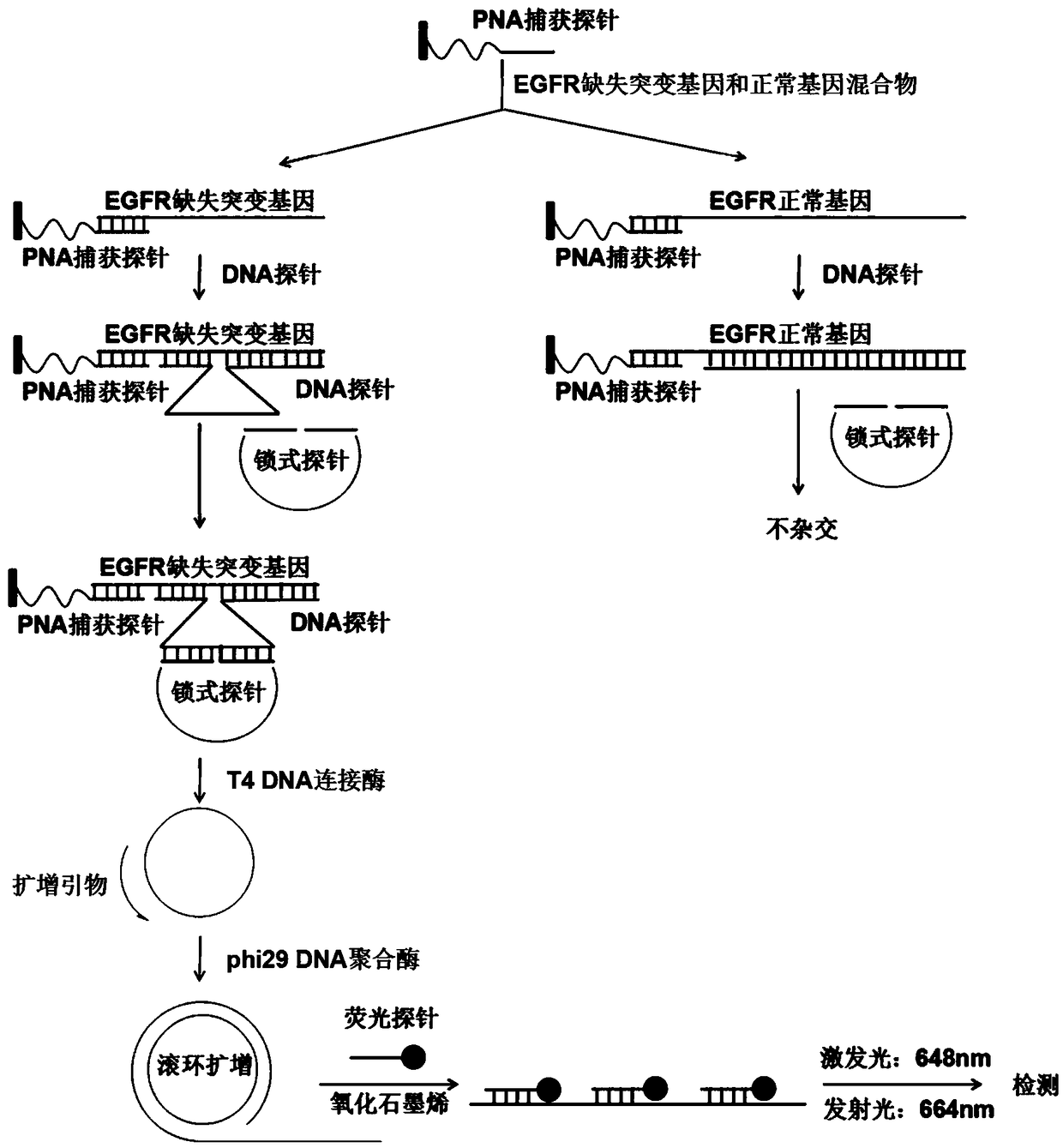
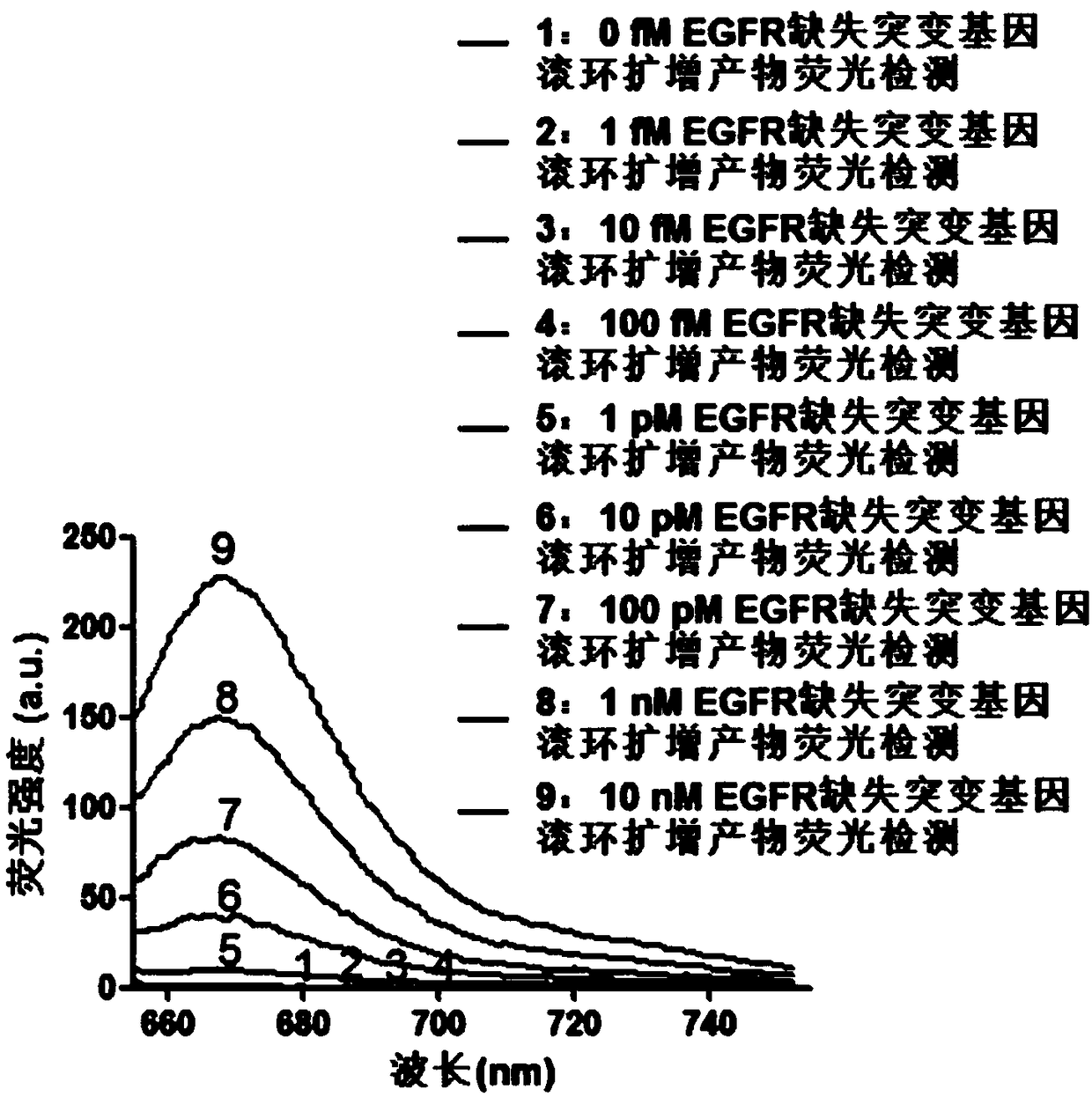
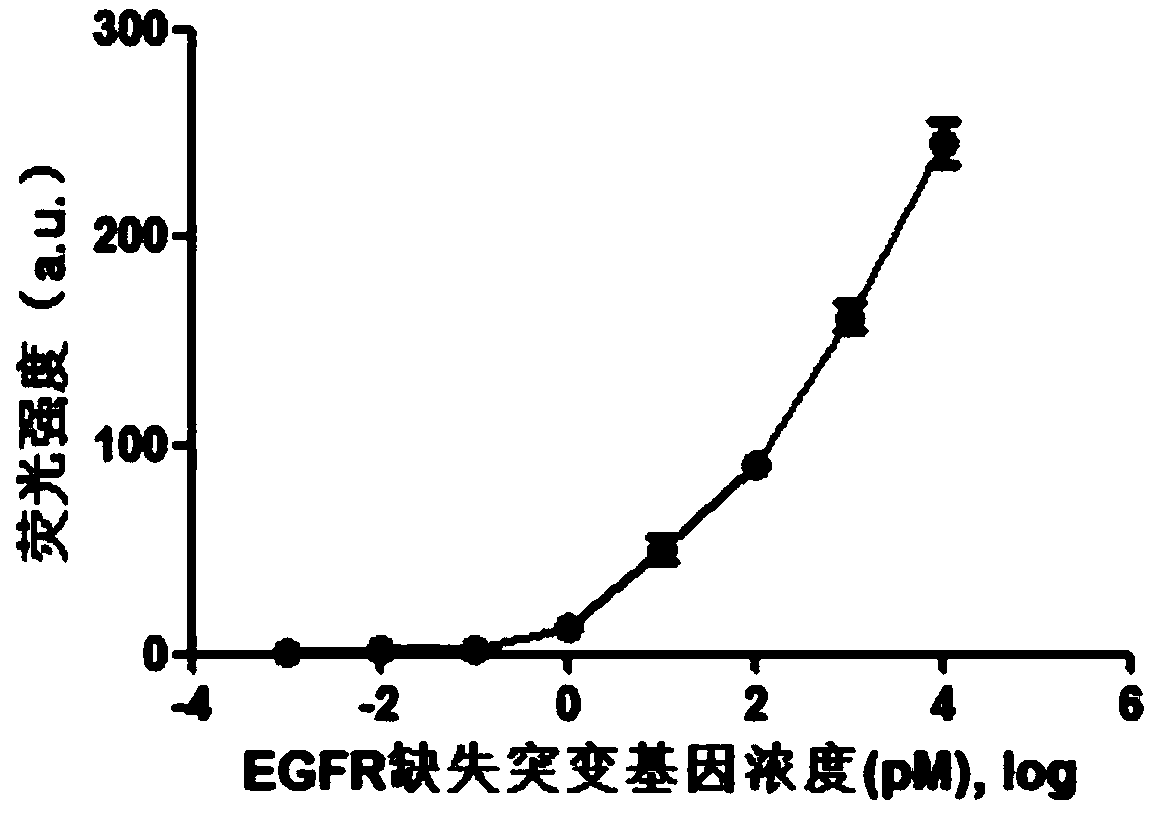
Fluorescence detection kit and fluorescence detection method for deletion mutation of gene
ActiveCN108949924AHigh sensitivityStrong specificityMicrobiological testing/measurementPolymerase LA-DNA
The invention provides a fluorescence detection kit for a deletion mutation gene. The kit includes: a PNA capture probe, a DNA probe, a padlock probe, a DNA polymerase, a DNA ligase, a rolling circleamplification primer, and a fluorescence probe. The invention also provides a method of using the kit to perform the fluorescence detection on the deletion mutation gene, wherein the method includes:1) immobilizing the PNA capture probe on the bottom of a pore plate, and hybridizing the probe with a target gene with a buffer solution; 2) hybridizing the target gene immobilized on the bottom of the pore plate with the DNA probe; 3) hybridizing a single chain section on the DNA probe, which is hybridized with the target gene, with the padlock probe, cyclizing the hybridized product with the DNAligase, and performing rolling circle amplification under effect of the rolling circle amplification primer and the DNA polymerase; 4) hybridizing the fluorescence probe with a rolling circle amplification product, quenching background fluorescence, and detecting the change on fluorescence intensity.
Owner:CIXI INST OF BIOMEDICAL ENG NINGBO INST OF MATERIALS TECH & ENG CHINESE ACAD OF SCI +1
Detection of nucleic acid differences using endonuclease cleavage/ligase resealing reactions and capillary electrophoresis or microarrays
InactiveUS20090123913A1Reduce noiseObstruction is producedMicrobiological testing/measurementNanoinformaticsHeteroduplexA-DNA
The present invention is directed to various methods for detecting DNA sequence differences, including single nucleotide mutations or polymorphisms, one or more nucleotide insertions, and one or more nucleotide deletions. Labeled heteroduplex PCR fragments containing base mismatches are prepared. Endonuclease cleaves the heteroduplex PCR fragments both at the position containing the variation (one or more mismatched bases) and, to a lesser extent, at non-variant (perfectly matched) positions. Ligation of the cleavage products with a DNA ligase corrects non-variant cleavages and thus substantially reduces background. This is then followed by a detection step in which the reaction products are detected, and the position of the sequence variations are determined.
Owner:CORNELL RES FOUNDATION INC
System for hot-start amplification via a multiple emulsion
Owner:BIO RAD LAB INC
Construction method of single-tube and high-flux sequencing library
The invention discloses a construction method of a single-tube and high-flux sequencing library. The method comprises the following steps: 1, respectively designing multiple pairs of basic amplimers for multiple target genes; 2, designing a pair of asymmetrically connected probes and a general primer not complementary to a mankind genome; 3, amplifying a template, the multiple pairs of basic amplimers, the asymmetric probes and the general primer in a PCR reaction system containing a DNA ligase and a DNA end modification enzyme to obtain an amplification product, wherein the amplification program sequentially comprises initial denaturation, first stage amplification and second stage amplification; and 4, purifying the amplification product to obtain the high-flux sequencing library. The construction method of the single-tube and high-flux sequencing library realizes single-tube rapid library completion of multiple target sequences can effectively solve the difficulty of polygene and multi-target detection of somatic cells of present clinic tumors, genetic diseases and other diseases on the basis of a small amount of clinic samples through combining a high-flux sequencing platform, and has low cost.
Owner:XIAMEN SPACEGEN BIOTECH CO LTD
Anti tumor translocation peptide of scorpion, preparation method and application
InactiveCN101003788AIncrease productionEffective anti-metastatic peptideBacteriaPeptide/protein ingredientsCDNA libraryEscherichia coli
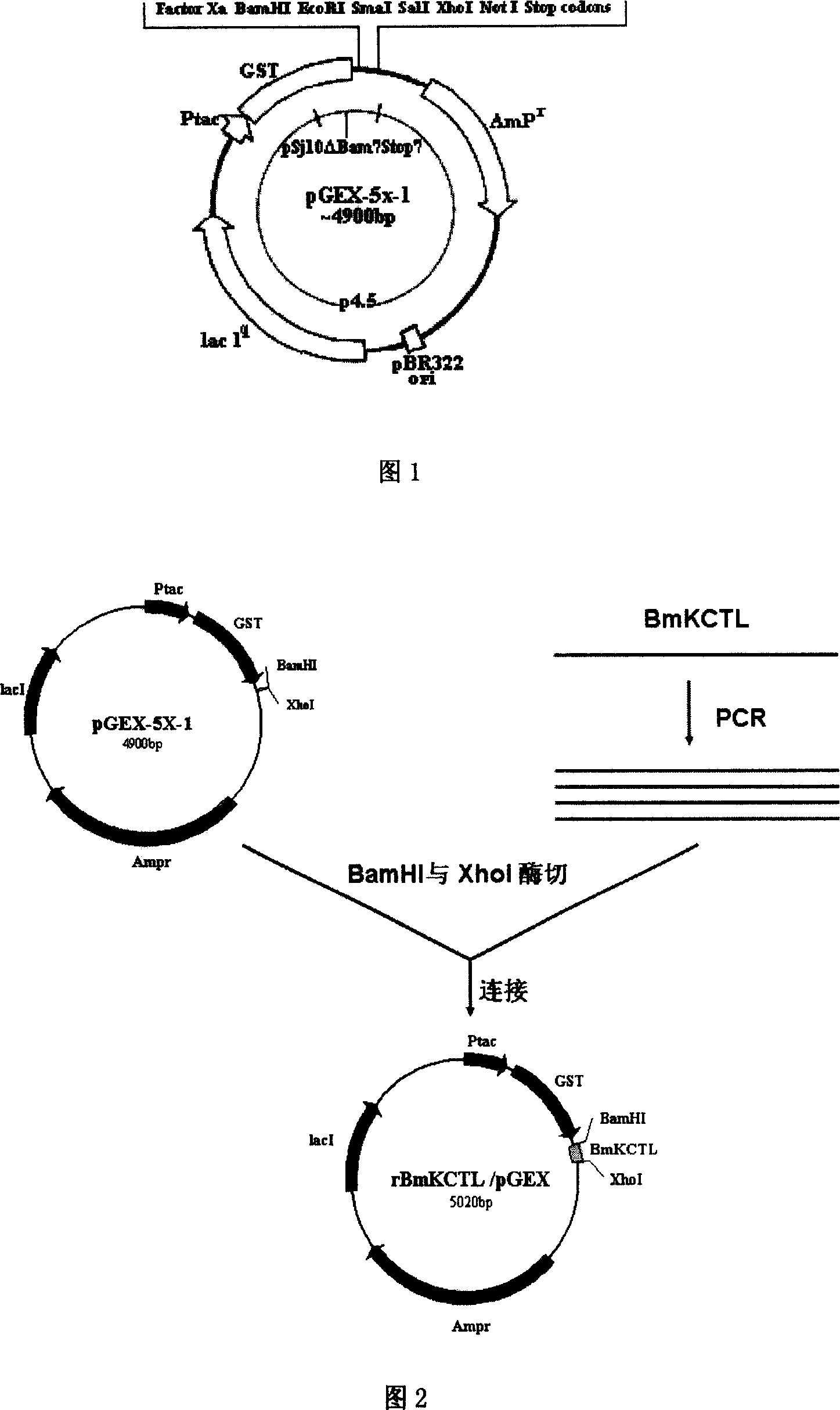
This invention discloses a method for preparing anti-tumor peptide of Buthus martensii, and its application. The method comprises: (1) screening cDNA sequence of anti-tumor peptide gene from cDNA library of Buthus martensii venom gland, designing primers, and performing PCR with the screened cDNA as a template to obtain anti-tumor peptide gene or gene fragment containing a digestion site of restriction endonuclease; (2) digesting the PCR product and expression plasmid; (3) ligating with T4 DNA ligase to obtain recombinant vector rBmKCTL / pGEX; (4) transferring into Eschericia coli BL21 to obtain engineering Eschericia coli BL21 / rBmKCTL / pGEX (CCTCC No. M206094). The method has such advantages as simple process, high safety, low cost, easy purification, and high product purity. The anti-tumor peptide can be used to manufacture drugs for preventing or treating tumor metastasis.
Owner:WUHAN UNIV
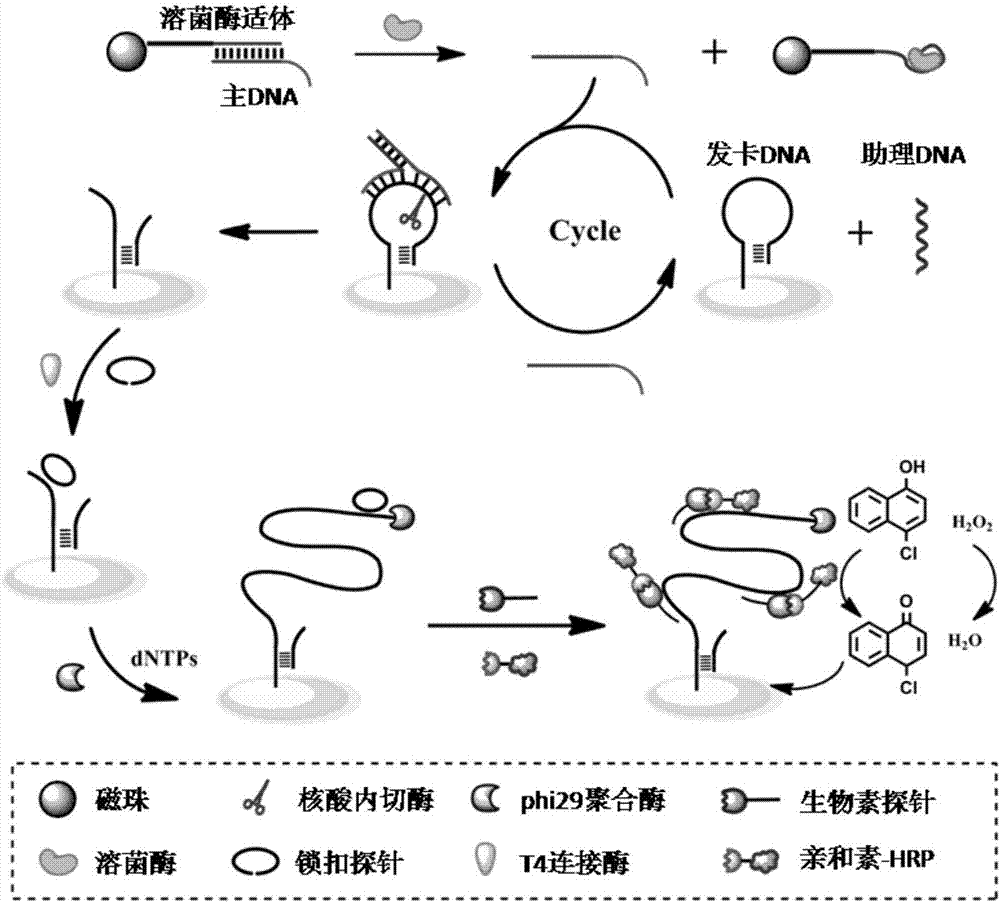
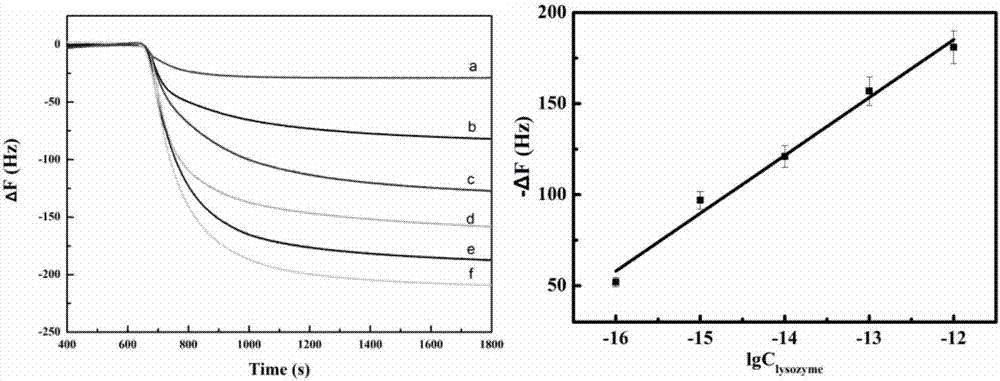
QCM detection method for detecting lysozyme based on multiple signal amplification technologies and application
InactiveCN107419005AEnhanced signalEasy to operateMicrobiological testing/measurementAptamerBiotin-streptavidin complex
The invention discloses a QCM detection method for detecting lysozyme based on multiple signal amplification technologies and application. The QCM detection method comprises the following steps that DNA hybridizes with lysozyme aptamer partially in a complementary mode, and through the specific binding reaction of the lysozyme and the lysozyme aptamer, the DNA is released; a Y-shaped structure is formed by complementary hybrid of the released DNA with hairpin DNA and assistant DNA modified on a gold leaf, and under the action of restriction enzyme, the hairpin DNA is cut and opened through specific identification sites; under the action of DNA ligase and DNA polymerase, using locking-ring-shaped DNA as a template chain, polymerization growing along the opened hairpin DNA is carried out, and a single chain with a large number of repeated sequences is formed; a signal probe marked with biotin hybridizes with the generated repeated sequences in a complementary mode, and after binding with streptavidin marked by HRP, hydrogen peroxide is catalyzed to oxidize 4-chloro naphthol, and precipitation reaction is generated; and accordingly the chip surface quality is increased, and the high-sensitivity detection of the QCM to the lysozyme is realized.
Owner:QINGDAO UNIV
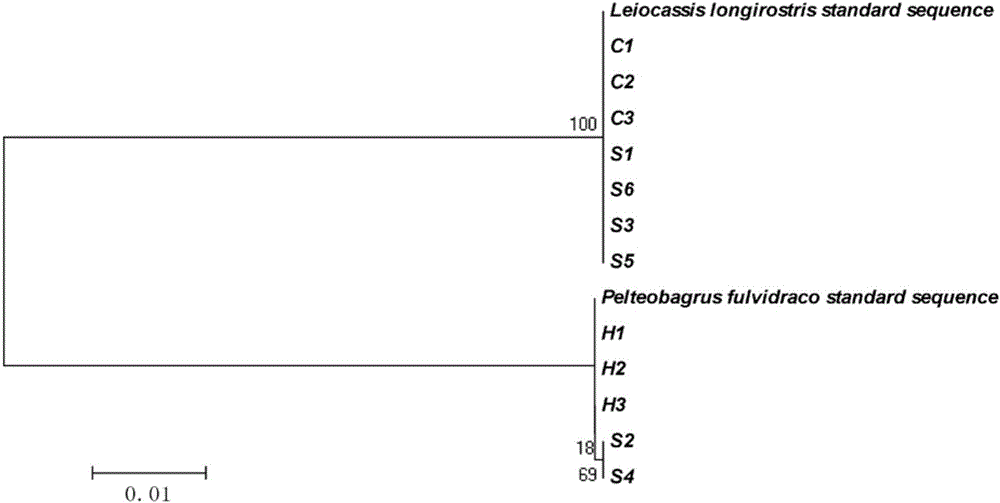
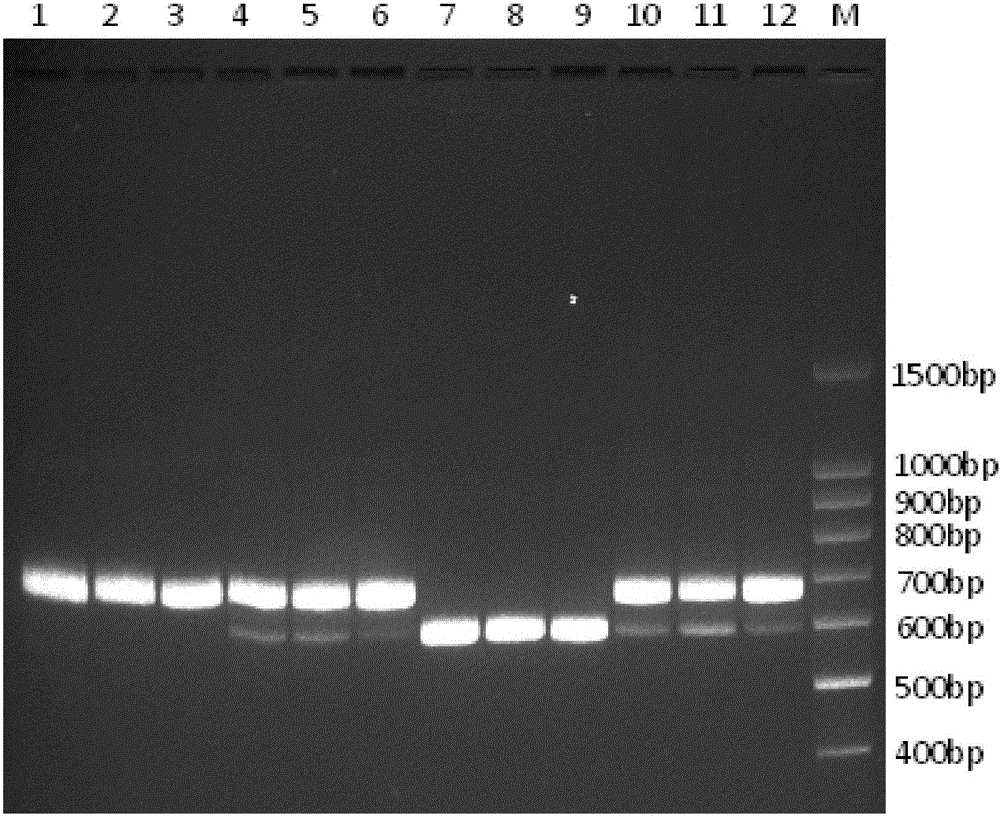
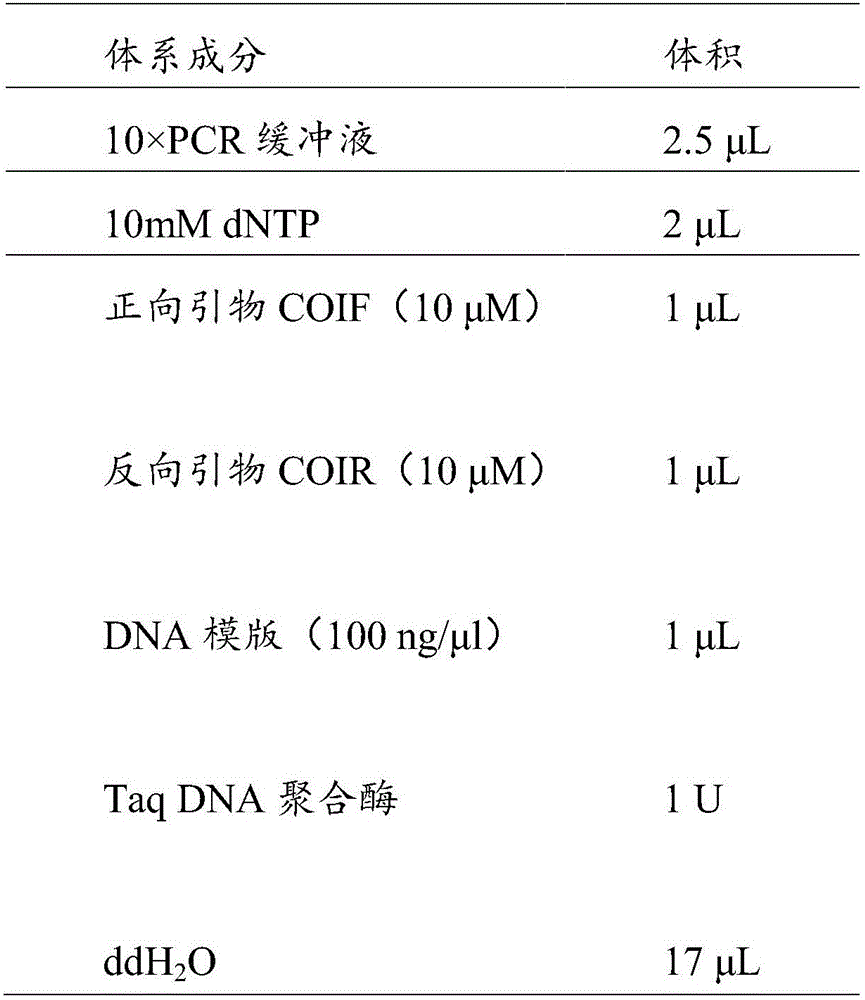
Primer group, kit and method for identifying tachysurus fulvidraco and leiocassis longirostris hybrid species
InactiveCN105695590AEfficient managementMonitor negative impactsMicrobiological testing/measurementDNA/RNA fragmentationAquaculture industryA-DNA
The invention provides a primer group for identifying a male parent and a female parent of tachysurus fulvidraco and leiocassis longirostris hybrid species. The primer group comprises a primer pair for amplifying COI genes and a primer pair for amplifying an ITS sequence, wherein the primer pair for amplifying COI genes consists of a primer COIF and a primer COIR; the primer pair for amplifying an ITS sequence consists of a primer ITSF and a primer ITSR. The invention also provides a kit which is for identifying the male parent and the female parent of tachysurus fulvidraco and leiocassis longirostris hybrid species and comprises the primer group. The kit also comprises any one or more of the following reagents: a buffer solution for PCR, dNTP, heat-resistant DNA polymerase, a cloning vector, a DNA ligase, and a buffer solution for ligation. The invention also provides a method for identifying the male parent and the female parent of tachysurus fulvidraco and leiocassis longirostris hybrid species. The primer group, the kit or the method can be used for accurately identifying the male parent and the female parent of the tachysurus fulvidraco and leiocassis longirostris hybrid species, thus effectively managing a breeding plan and monitoring negative influence thereof, and has great significance in sustainable development of aquaculture industry.
Owner:YANGTZE RIVER FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
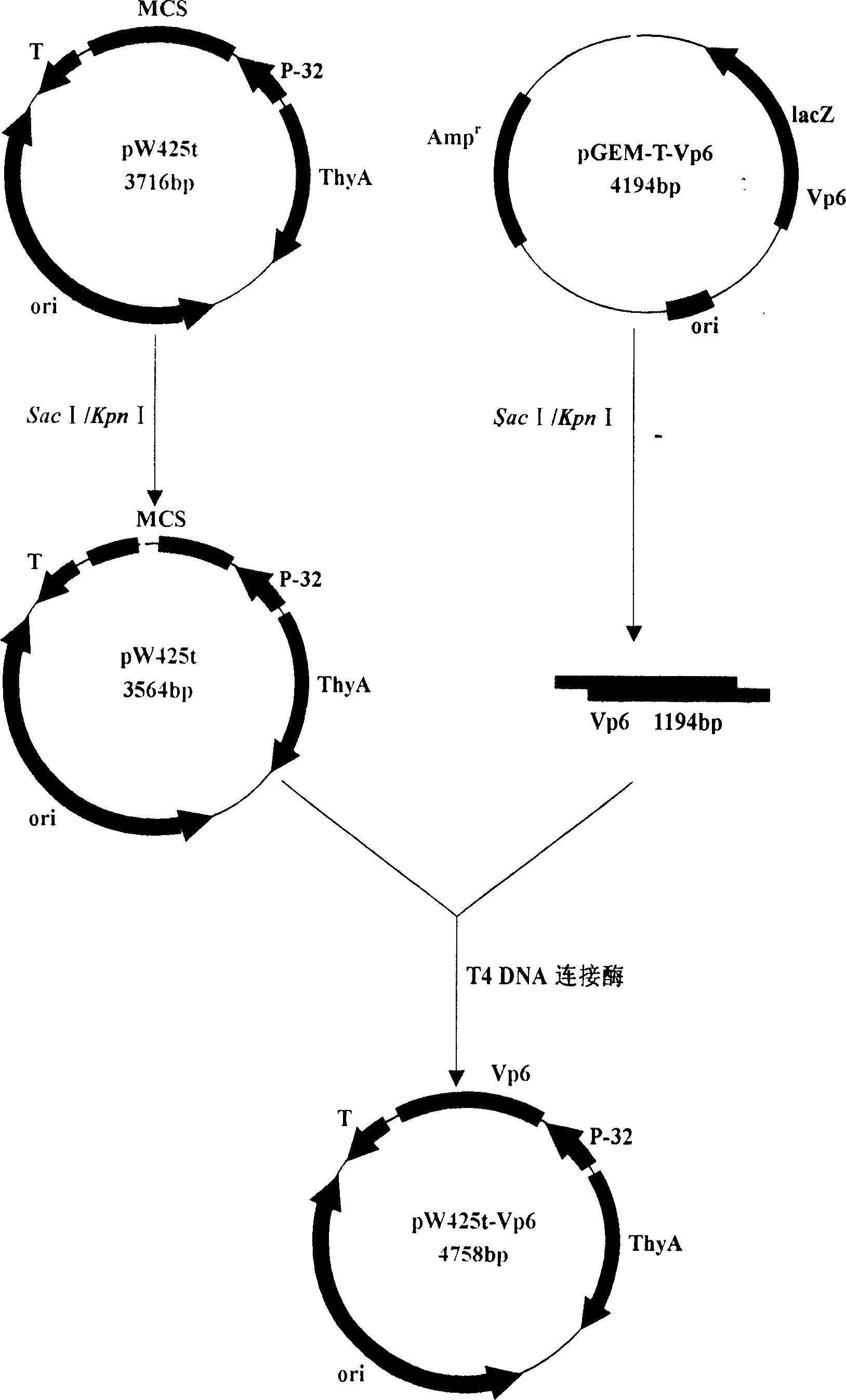
Recombination and expression for non-antibiotic expression vector of rotavirus Vp6 gene and lactic acid bacteria
InactiveCN1952156AFight reinfectionImprove immunityVirus peptidesFermentationEscherichia coliRotavirus RNA
The invention belongs to the field of biological gene, disclosing the recombination and expression of a rotavirus Vp6 gene and Bacterium acidi lactic nonresistance expression vector and it is characterized in: digesting the cloning vector and vector taking thyA gene as a selection pressure using Sac I and KpnI, purifying and recovering, linking the products with T4 DNA ligase, transforming to thyA gene-deficient E. coli competence E. coli X13, culturing in the medium, and screening to obtain prokaryotic recombinant expression plasmid through growing functional redeem, inducing positive bacteria for expression using threonine to obtain high expression of the recombinant plasmid. Molecular weight of fusion protein is about 44.88 KD. Beneficial effects are: VP6 protein is RV-group-specific antigen, locating at the virus endoconch, occupying 51%of the virus particles. As VP6 mainly stimulates organism to generate mucosal immune antibody sIgA, it plays a very important role in the mucosal immune, thus advanced development of rotavirus Vp6 mediated gene oral mucosal immune genetically engineered vaccine has important significance.
Owner:JILIN AGRICULTURAL UNIV +3
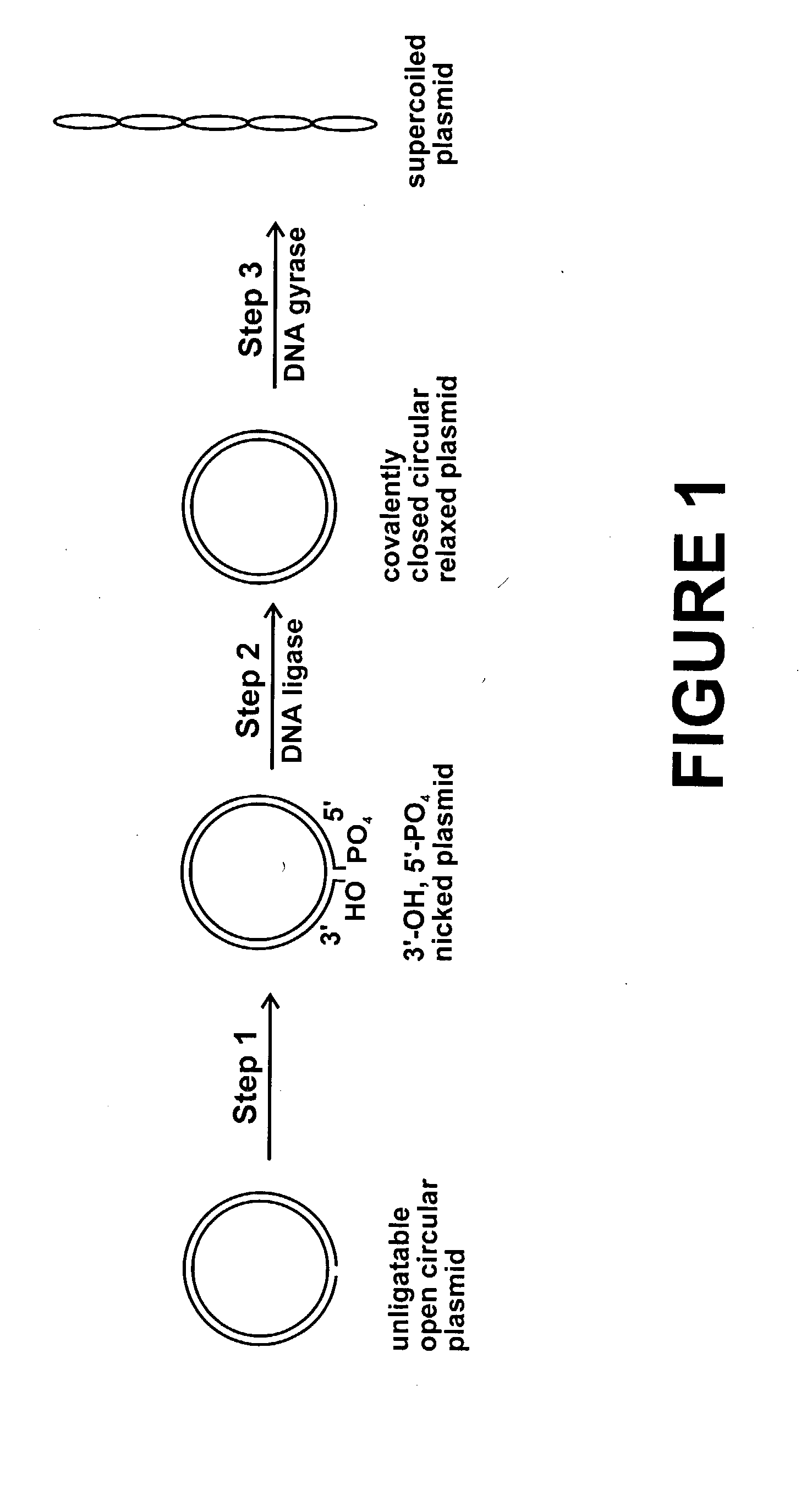
Method for plasmid preparation by conversion of open circular plasmid
InactiveUS20040191871A1Increase percentageFermentationVector-based foreign material introductionNucleotidePhosphoric acid
In accordance with the invention, there is provided a method for preparing plasmid from host cells which contain the plasmid, comprising the steps: (a) preparing a cleared lysate of the host cells, wherein the cleared lysate comprises unligatable open circular plasmid, wherein the open circular plasmid is not 3'-hydroxyl, 5-phosphate nicked plasmid; (b) incubating the unligatable open circular plasmid with one or more enzymes in the presence of their appropriate nucleotide cofactors, whereby the unligatable open circular plasmid is converted to 3'-hydroxyl, 5'-phosphate nicked plasmid; (c) incubating the 3'-hydroxyl, 5'-phosphate nicked plasmid with DNA ligase in the presence of DNA ligase nucleotide cofactor, whereby 3'-hydroxyl, 5'-phosphate nicked plasmid is converted to relaxed covalently closed circular plasmid; and (d) incubating the relaxed covalently closed circular plasmid with DNA gyrase in the presence of DNA gyrase nucleotide cofactor, whereby relaxed covalently closed circular plasmid is converted to negatively supercoiled plasmid. Preferably, the enzymatic steps (b), (c), and (d) are performed in a single step using an enzyme mixture comprising DNA polymerase, DNA ligase, and DNA gyrase. Preferably, the mixture further comprises a 3' terminus deblocking enzyme, such as exonuclease III or 3'-phosphatase. Preferably, the mixture further comprises one or more regenerating enzymes and a high energy phosphate donor, whereby the nucleotide by-products of the nucleotide cofactors generated by DNA ligase and DNA gyrase are converted to back to nucleotide cofactor. Preferably, the enzyme mixture further comprises one or more exonucleases, such as ATP dependent exonuclease, whereby linear chromosomal DNA is selectively degraded.
Owner:HYMAN EDWARD DAVID
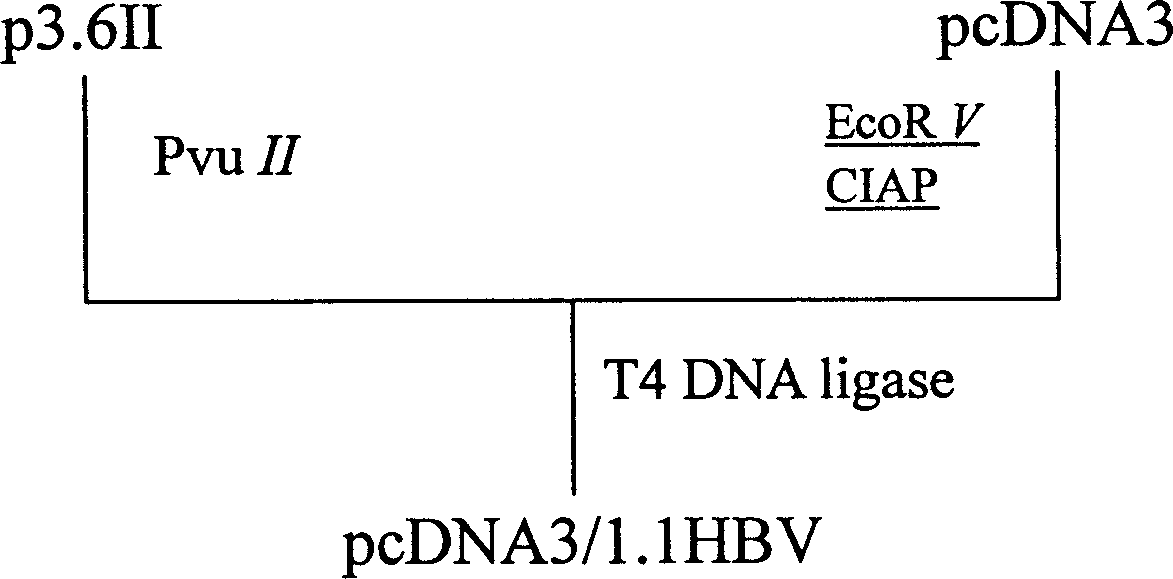
Production of mouse model of viral hepatitis B with high expression HBsAg
InactiveCN1679967AOvercome injectionOvercoming disadvantages of portal vein injection of naked DNAIn-vivo testing preparationsEscherichia coliPhosphorylation
A process for preparing VHB mouse model with high expression of HBsAg includes such steps as linearizing carrier pcDNA3 by EcoRV, dephosphorylating by alkaline phosphatase of ox's small intestine, recombining the clonal carrier p3.6II, enzyme severing by PvuII to obtain 1.1-time HBV gene fragment, linking it with linear pcDNA3, transforming colibacillus competent cell DH5 alpha, screening resistance, enzyme serving, sequencing to verify the recombinant plasmid, injecting it in the mouse body via tail vein, and verifying the model.
Owner:SHANDONG UNIV
Primers and probes for detecting genes associated with schizophrenia, bipolar affective disorders and major depression and kits and preparation methods thereof
InactiveCN102146479AMicrobiological testing/measurementDNA/RNA fragmentationMicrobiologySchizophrenia
The invention discloses a kit for detecting genes associated with schizophrenia, bipolar affective disorders and major depression, which belongs to the field of biotechnology, and consists of a polymerase chain reaction (PCR) reagent group and a ligase detection reaction (LDR) reagent group, wherein the PCR reagent group comprises buffer solution, deoxyribonucleotide triphosphate (dNTP) mixed solution, Taq DNA polymerase, pure water, an upstream primer represented by SEQ ID No.1 and a downstream primer represented by SEQ ID No.2; and the LDR reagent group comprises buffer solution, dNTP mixed solution Taq DNA polymerase, pure water, a probe represented by SEQ ID No.3, a probe represented by SEQ ID No.4 and a probe represented by SEQ ID No.5. In the invention, the operation is simple and convenient, the cost is low, and the kit is developed for bipolar affective disorders. The result is reliable, the stability is high and the sensitivity is high.
Owner:SHANGHAI JIAO TONG UNIV +1
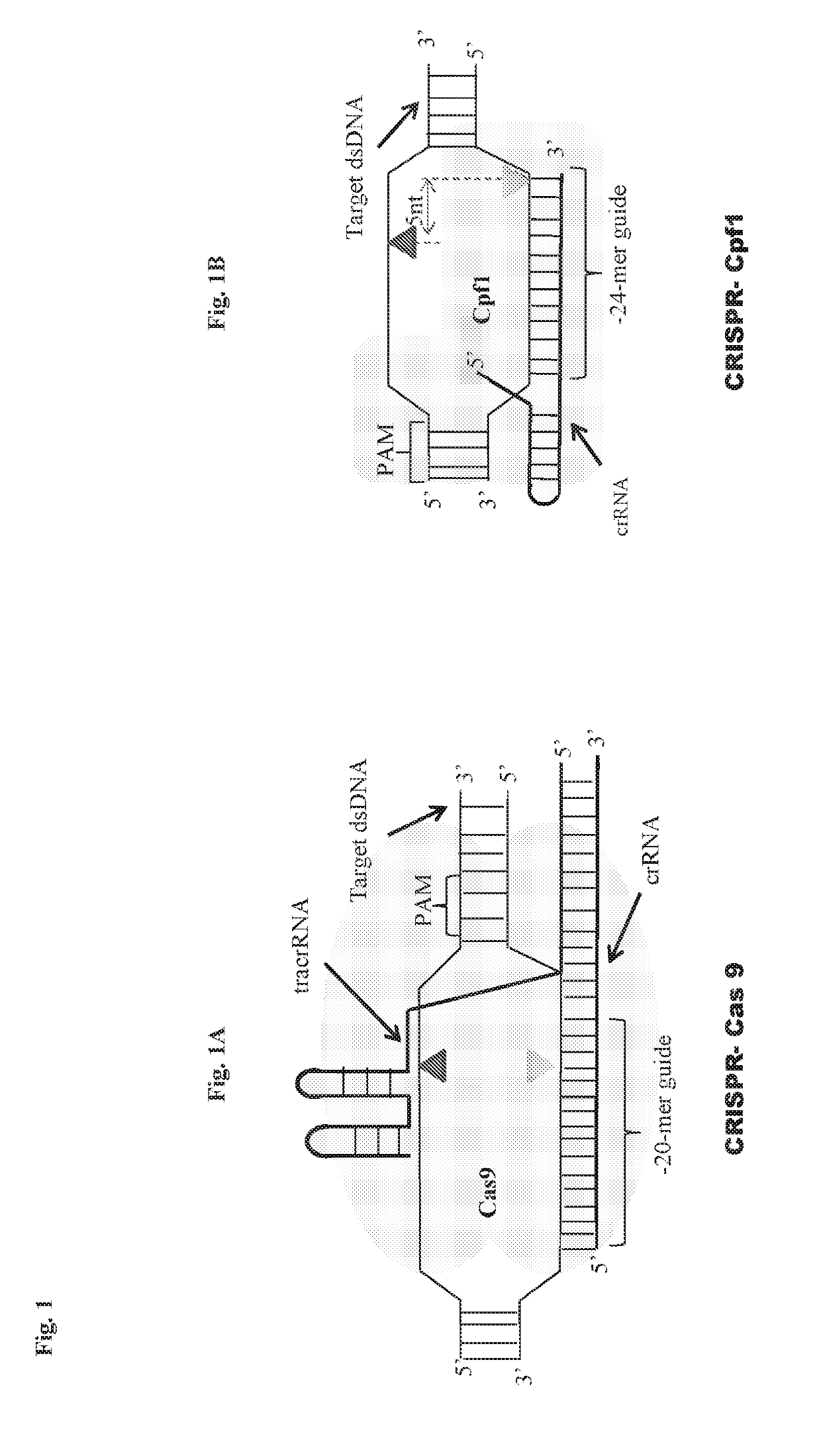
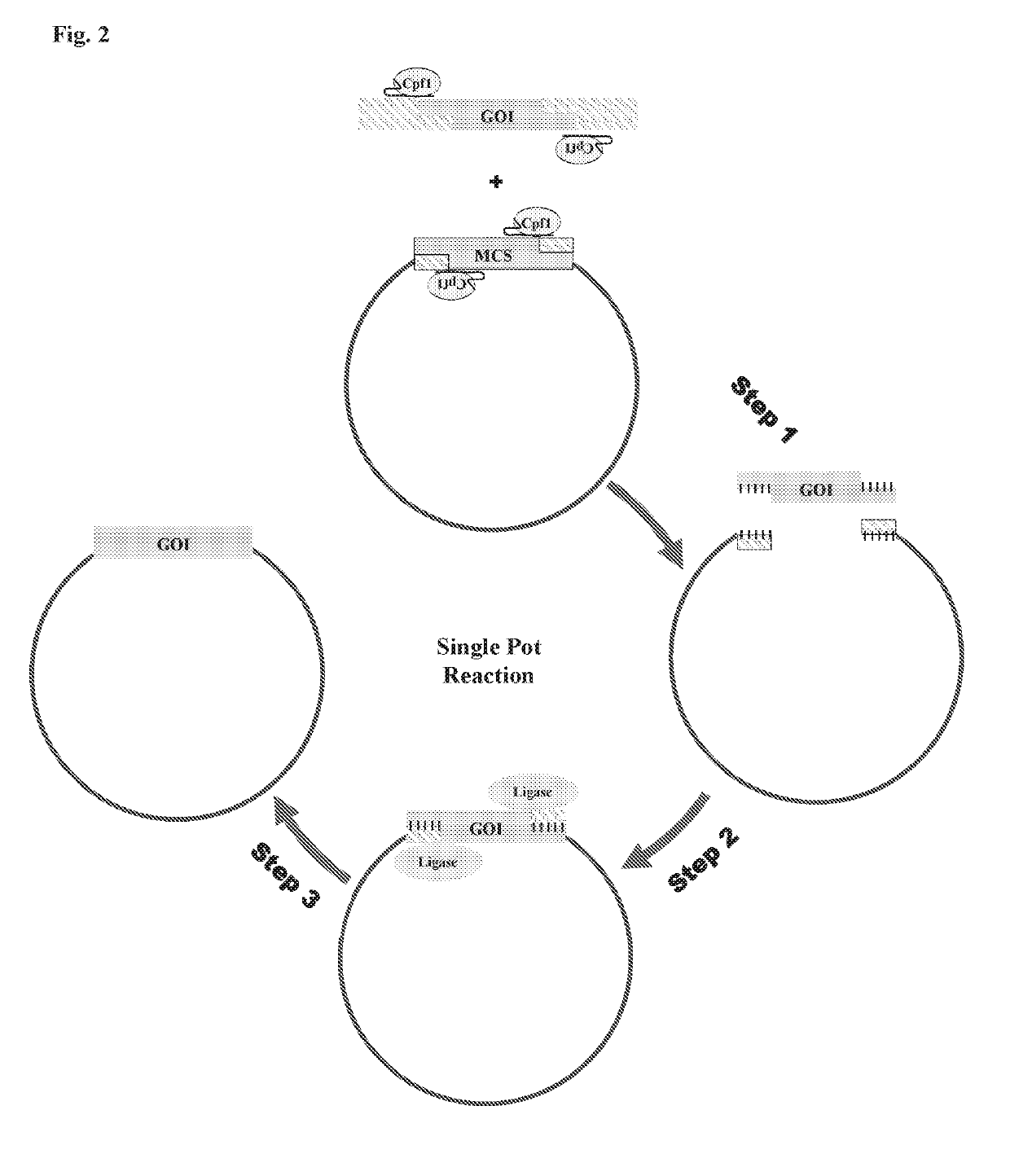
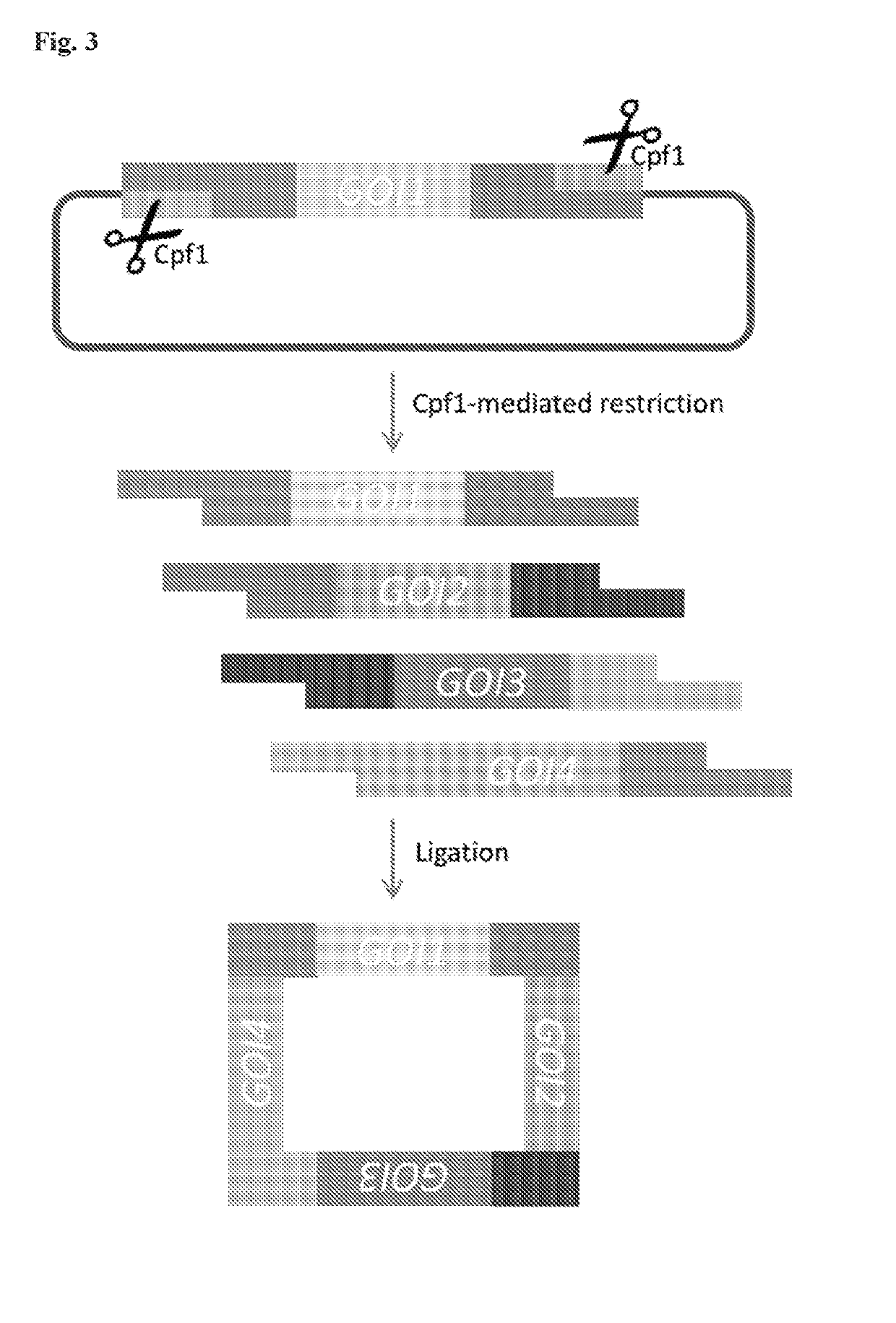
Scarless DNA assembly and genome editing using crispr/cpf1 and DNA ligase
InactiveUS20190330659A1Improve efficiencyMicrobiological testing/measurementStable introduction of DNAEpigenome editingNucleotide
The disclosure describes a scarless DNA assembly and genome editing methodology termed “CLIC” (CRISPR and Ligase Cloning), which utilizes a CRISPR / Cpf1 complex and DNA ligase to perform programmable gene editing and nucleotide assembly. The CLIC process is highly amenable to applications in vitro for the scarless assembly of a plurality of DNA parts simultaneously or in vivo for the site-specific insertion of one or more DNA molecules into the host genome.
Owner:ZYMERGEN INC
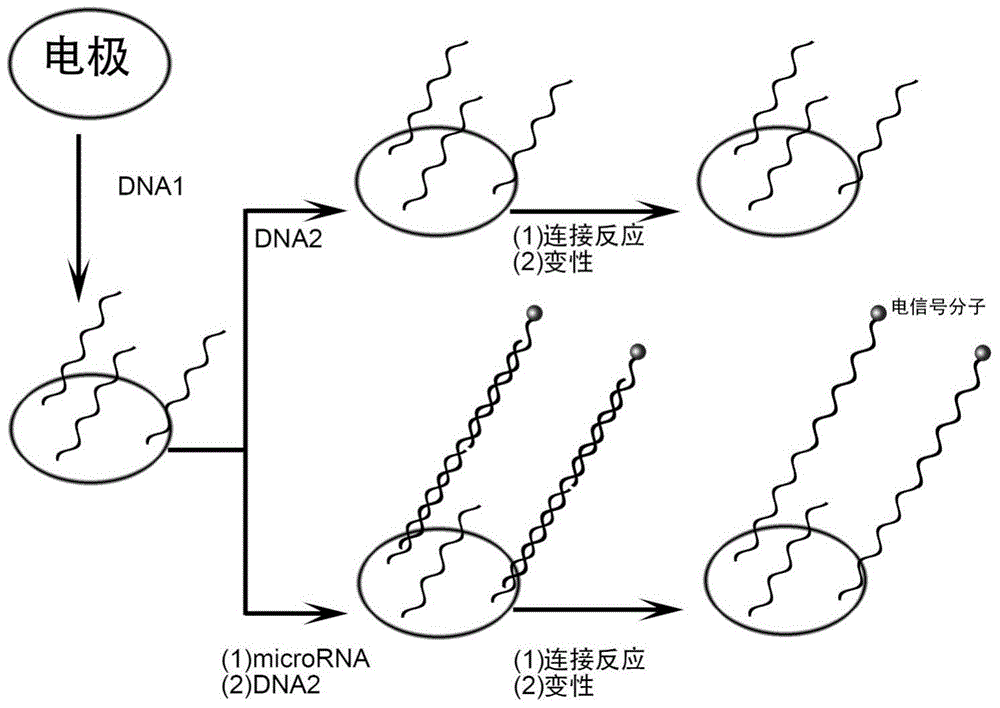
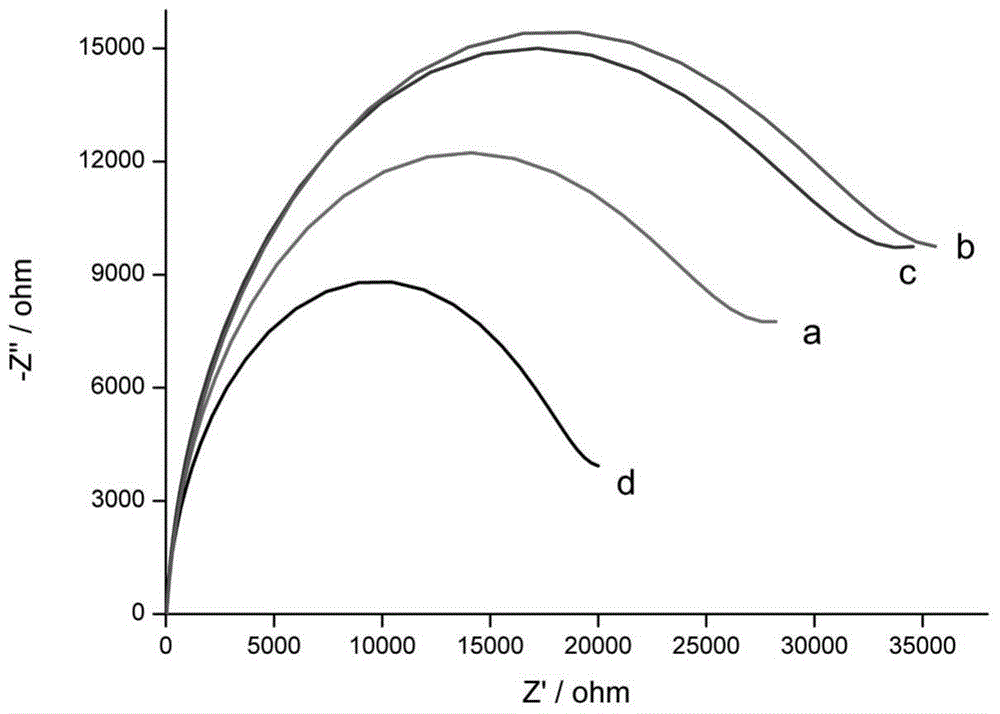
Method for detecting microRNA content of to-be-detected liquid
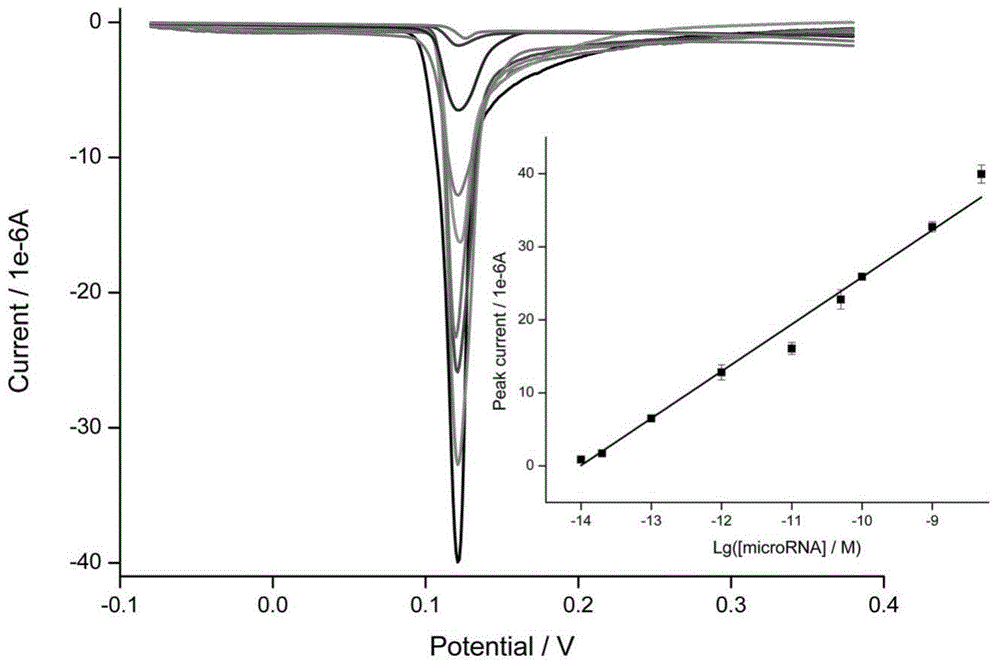
ActiveCN104561274AGuaranteed specificityHigh sensitivityMicrobiological testing/measurementSignalling moleculesComplementary pair
The invention discloses a method for detecting the microRNA content of to-be-detected liquid, which comprises the following steps that: 1, molecules of a first single-stranded DNA are modified on the surface of a working electrode of an electrochemical device; 2, a sample is added, wherein a target microRNA in the sample and the first single-stranded DNA are in partial complementary pairing so as to form a double-stranded structure; 3, a second single-stranded DNA marked with electrical single molecules is added, and due to the existence of partial complementarily-paired sequences between the second single-stranded DNA and the microRNA, a hybrid structure of DNA / microRNA / DNA is formed; 4, the first DNA and the second DNA are connected to be a single-stranded DNA by a DNA ligase, and by heated denaturation, the microRNA is removed from the surface of the electrode so as to effectively reduce background signals; and 5, by detecting electrical signal molecules at the tail end of the DNA, whether the sample contains the target microRNA and corresponding concentrations of the target microRNA can be judged.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
Kit for detecting hereditary deafness susceptibility gene mutation
ActiveCN104694629AHigh sensitivityImprove accuracyMicrobiological testing/measurementMultiplex ligation-dependent probe amplificationHigh-Throughput Screening Methods
The invention relates to a multiplex ligation-dependent probe amplification kit for detecting hereditary deafness susceptibility gene mutation. The multiplex ligation-dependent probe amplification kit comprises a detection probe for detecting mutation site specificity, a universal primer, a positive DNA template, a DNA ligase, a connection buffer solution, a hybridization solution and a PCR reaction reagent, wherein the detection probe is composed of a pair of short probe and long probe and corresponds to one mutation site. The method is high in sensitivity and is accurate in result, and compared with the traditional detection technology, the kit can be used for completing high-throughput screening of multiple key hereditary deafness gene mutation diagnosis at one time, and has the advantages of high economical efficiency, high efficiency and high accuracy.
Owner:江苏佰龄全基因生物医学技术有限公司
Fusion polypeptides and uses thereof
InactiveCN102597006ADoes not affect functionReserved functionFusion with RNA-binding domainFusion with DNA-binding domainBinding domainA-DNA
The invention relates to fusion polypeptides comprising a polynucleotide-binding domain, such as a DNA-binding domain, and a ligase domain, such as a DNA ligase domain, methods for the production of such fusion polypeptides, and uses of the fusion polypeptides, for example in a range of molecular biological techniques as well as applications in the diagnostics, protein production, pharmaceutical, nutraceutical and medical fields.
Owner:MASSEY UNIVERISTY
Solid-phase synthesis method of transcription activator-like effector
The invention relates to a solid-phase synthesis method of transcription activator-like effector (TALE). The method comprises the following steps of: immobilizing a segment of nucleic acid adaptation sequence (DNA (Deoxyribonucleic Acid) linker) to a solid-phase interface, and shearing with a restriction endonuclease to generate 3'-cohesive end; contacting a product with TALE linkage unit which is treated by the restriction endonuclease and is provided with 5'-cohesive end, connecting under the action of a DNA ligase, and eluting unconnected matrix; treating the product obtained in the previous step with restriction endonuclease to shear and generate 3'-cohesive end on DNA again, contacting a product with TALE linkage unit which is treated by the restriction endonuclease and is provided with 5'-cohesive end, connecting under the action of a DNA ligase, and eluting unconnected matrix; repeatedly operating the steps for at least one time; shearing the connected DNA molecule from the solid-phase interface with restriction endonuclease, and separating and purifying to remove the unconnected matrix; and expressing the DNA molecule into protein molecule, namely fusion protein (such as TALEN (TALE nuclease) or TALEA (TALE activator)) containing target TALE.
Owner:BEIJING VIEWSOLIDBIOTECH
Construction method of high-throughput sequencing library suitable for single-stranded DNA
PendingCN110904512AIncrease profitAvoid lostMicrobiological testing/measurementLibrary creationSingle strandHigh throughput sequence
The invention discloses a construction method of a high-throughput sequencing library suitable for single-stranded DNA. The method comprises the following steps: linking a single-stranded template anda double-stranded linker 1 with degenerate bases (a special linker with degenerate bases enables the linking of the single-stranded template and the double-stranded linker to be possible, the double-stranded linker can be randomly combined to the single-stranded template to make a linker sequence be fully close to the template, and then the linking is completed by using T4 DNA ligase); extendingby using an extension primer to obtain a double-stranded product; linking the double-stranded product to a linker 2; and carrying out PCR amplification by taking the obtained linking product as a template to obtain the required library. According to the invention, the method can be used for constructing a high-throughput sequencing library, can further be used for constructing a single-stranded DNA library, can improve the utilization rate of single-stranded DNA in the construction of a high-throughput sequencing library, further achieves the purpose of low-initial-quantity library construction, and has key effect in early screening and prognosis monitoring application of tumors in the future.
Owner:ENVELOPE HEALTH BIOTECHNOLOGY CO LTD
Methods, compositions and kits for a one-step DNA cloning system
ActiveUS9206433B2Increase productionImprove purification effectVector-based foreign material introductionA-DNARecognition sequence
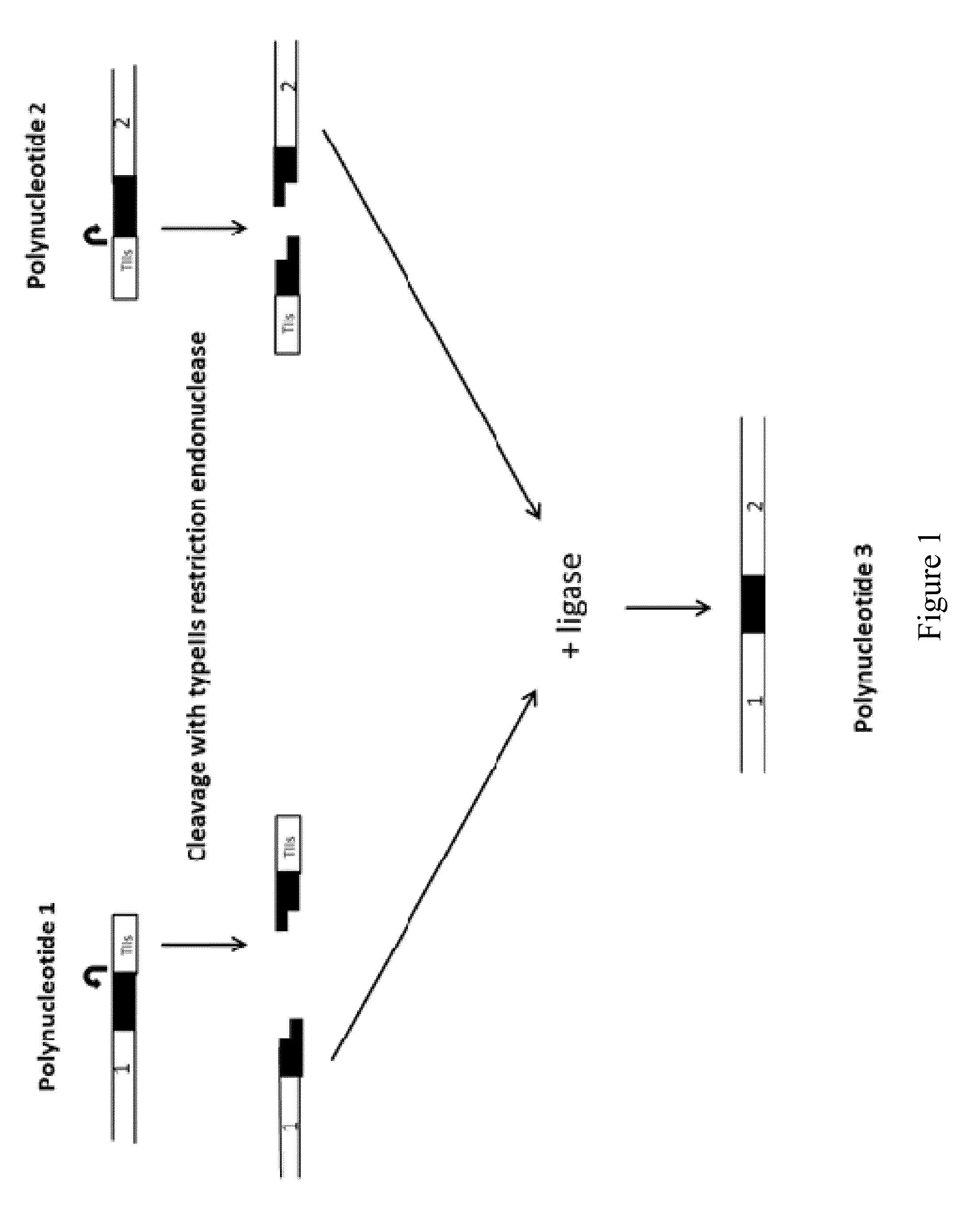
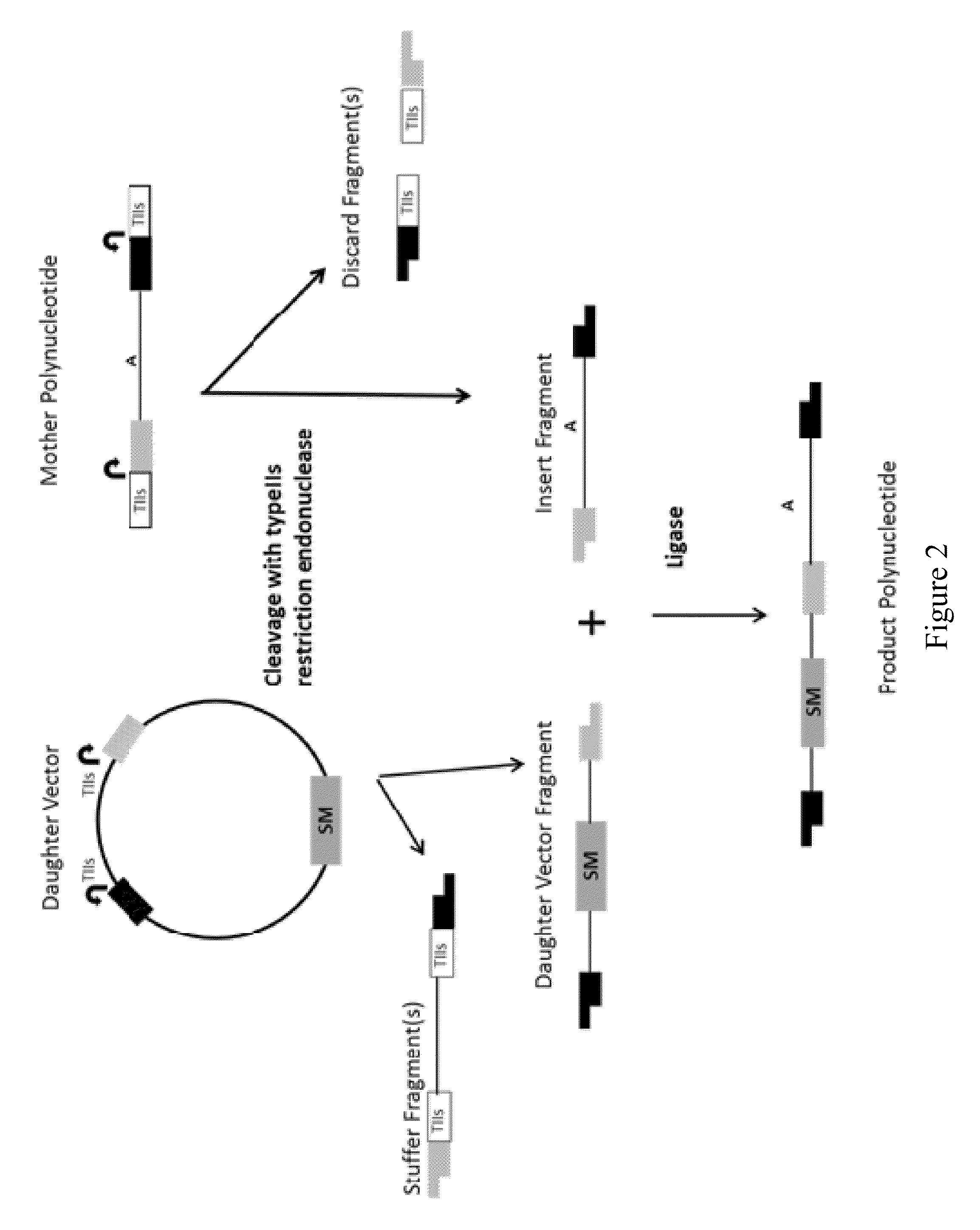
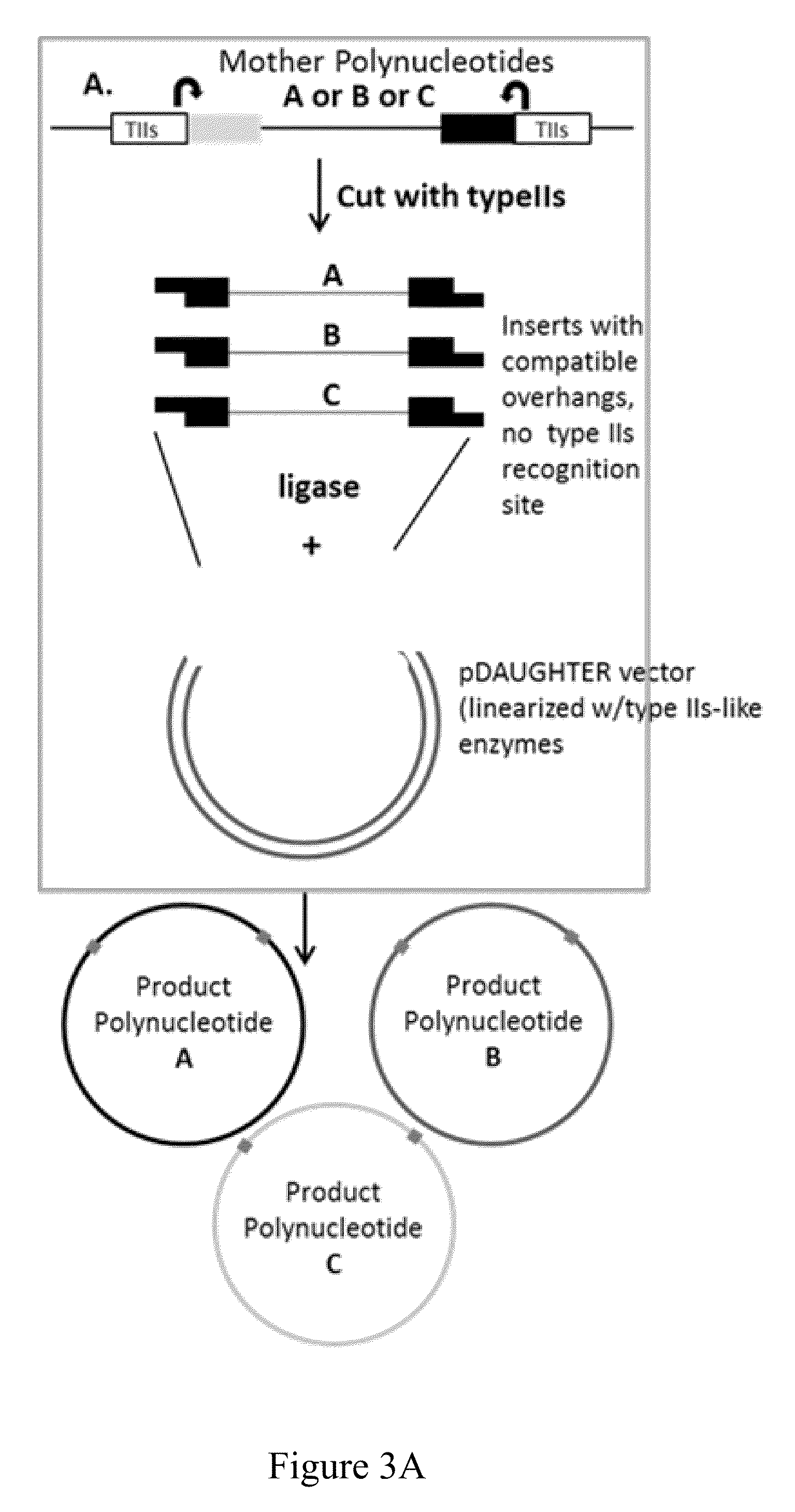
Methods and kits for joining two or more polynucleotides to form a product polynucleotide are provided. A mixture contains a first polynucleotide comprising a selectable marker. The mixture further contains a second polynucleotide comprising a first typeIIs recognition sequence and a second typeIIs recognition sequence. The second polynucleotide is other than the first polynucleotide. The mixture further contains a first typeIIs restriction endonuclease that cleaves the first typeIIs recognition sequence to produce a first end, a second typeIIs restriction endonuclease that cleaves the second typeIIs recognition sequence to produce a second end, and a DNA ligase. The first end is not compatible with the second end. The combined actions of the enzymes in the mixture join the first polynucleotide to the second polynucleotide forming a product polynucleotide, which is obtained by transforming the mixture into a host cell.
Owner:DNA TWOPOINTO
Method and kit for sensitively detecting human EGFR (epidermal growth factor receptor) gene mutation on basis of Sanger sequencing
ActiveCN105256021AGrowth inhibitionImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationMedicineEGFR Gene Mutation
The invention belongs to the field of gene mutation detection and particularly relates to a method and a kit for sensitively detecting human EGFR (epidermal growth factor receptor) gene mutation on the basis of Sanger sequencing. In order to overcome the defects of a conventional human EGFR gene mutation detection method and kit, the method and the kit for flexibly detecting the human EGFR gene mutation on the basis of the Sanger sequencing are provided; heatproof DNA ligase and a specific connecting primer system aiming at a mutation site are introduced into an EGFR gene segment amplification system, so that amplification of wild type EGFR gene segments in a sample is greatly inhibited, and a wild type sample can be directly determined on the basis of amplified segment bands; or the proportion of mutation segments in a final PCR (polymerase chain reaction) product of a sample with low mutation content is enabled to be higher than 50%, accordingly, the accurate identification of the Sanger sequencing on a heterozygous gene segment system is guaranteed; the method and the kit can detect the EGFR gene mutation with the content as low as 1% in the sample on the basis of the Sanger sequencing, the accuracy rate is very high, the detection cost is low, and new unknown mutation can be possibly detected.
Owner:FUJIAN MEDICAL UNIV
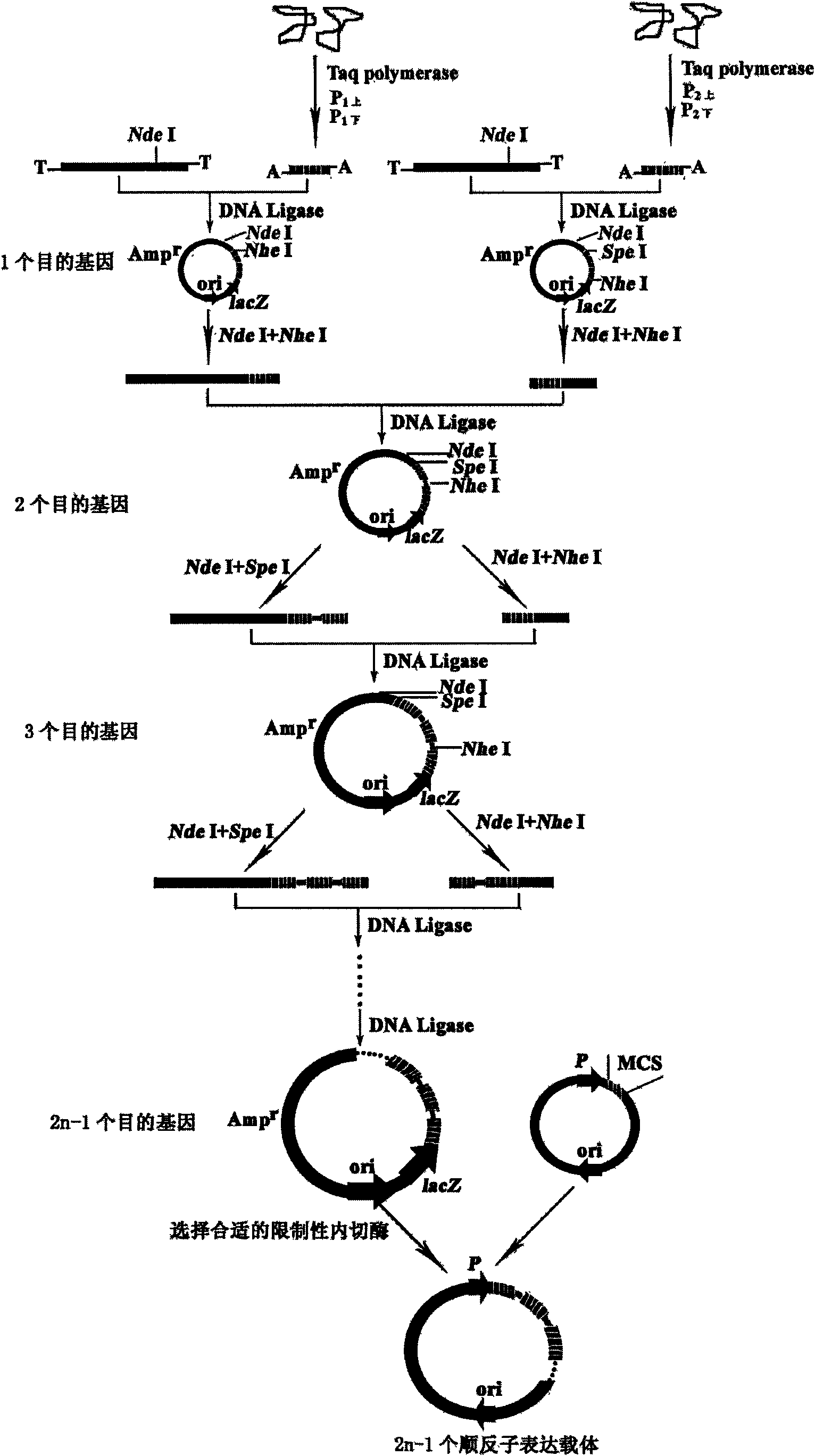
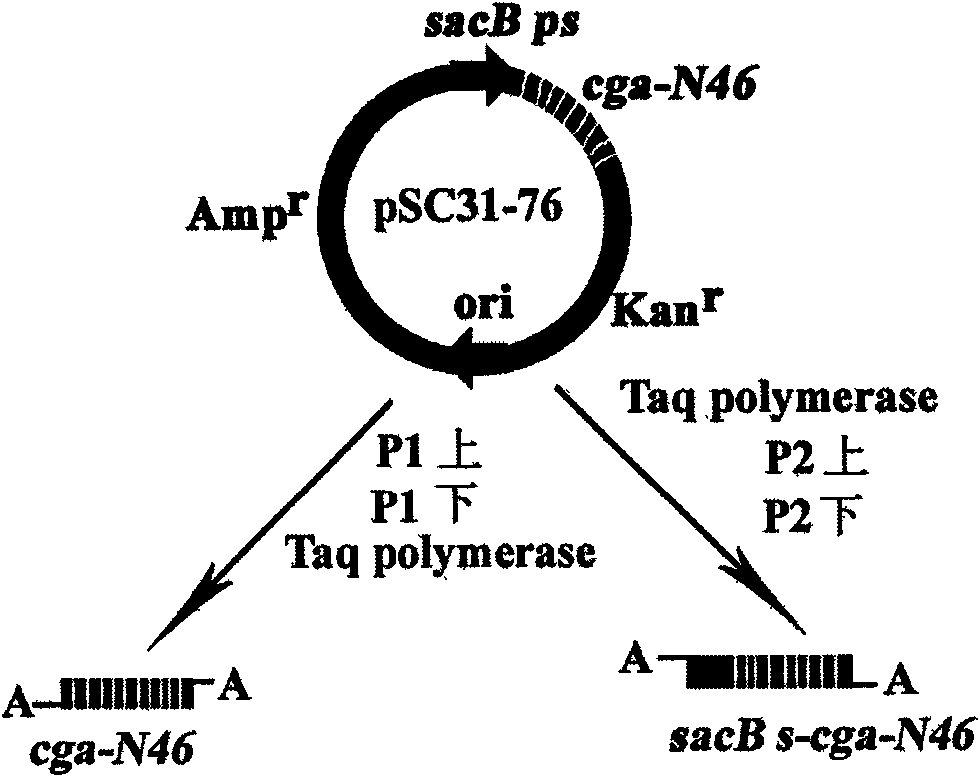
Construction method of polycistron expression vector
InactiveCN101608187AWide range of applicable genesReduce typesMicroorganism based processesVector-based foreign material introductionDNA ligaseRibosomal binding site
The invention relates to a construction method of a polycistron expression vector, which comprises the following steps: designing two pairs of primers by utilizing a commercial T-vector as an intermediate vector, introducing Nhel and Spel enzyme cutting sites and a ribosome bind site (RBS) in each primer, connecting an amplified product with the T-vector, cutting the obtained recombinant T-vector by using Nhel and Spel enzymes, and connecting an obtained DNA segment by using a DNA joining enzyme to enable an individual heterologous gene to increase progressively in the manner of 2n-1 (n is Gene number of the back recombinant T-vector), finally cutting the DNA segment containing a plurality of heterologous genes through a restriction enzyme corresponding to the enzyme cutting sites at two ends of the DNA segment, subcloning to the downstream of an expression vector promoter, and then obtaining the polycistron expression vector. The invention has the advantages of simple operation and wide application scope, and is applicable to all proteins, in particular to the expression of small peptides. The constructed expression vector can be directly converted into a procaryotic host cell and provide a technical platform for solving the bottleneck problem of low expression in the prokaryotic protein gene engineering expression.
Owner:HENAN UNIVERSITY OF TECHNOLOGY
Digital PCR-based protein active concentration determination method reference method
The invention discloses a digital PCR-based protein active concentration determination method reference method. The method comprises the following steps: (1) preparing the antibody of a target protein to be tested; (2) labeling the obtained antibody with biotin; (3) synthesizing three fragments of single-strand oligonucleotides; (4) respectively labeling the 5'-end and the 3'-end of two of the three oligonucleotide segments with avidin; (5) designing a Taqman probe and a corresponding primer; (6) labeling antibodies with the oligonucleotides by using the amplification affinity reaction between the biotin and the avidin; (7) diluting the target protein to be tested with a buffer solution until the concentration is 5000 molecules / [mu]L or less, adding the above obtained two antibodies labeled with the oligonucleotides, the third oligonucleotide fragment and a DNA ligase to form an antigen-antibody compound, carrying out a digital PCR amplification reaction, and recording the obtained copy number as c0; (8) carrying out the digital PCR amplification reaction, and marking the obtained copy number as c1; and (9) calculating the concentration of the target protein to be tested according to a formula of c0 - c1.
Owner:NAT INST OF METROLOGY CHINA
Method for plasmid preparation by conversion of open circular plasmid to supercoiled plasmid
InactiveUS20060057683A1Increase productionUniversal procedureFermentationVector-based foreign material introductionPhosphateGenetics
In one embodiment of the invention, a method is provided for preparing plasmid from host cells which contain the plasmid, comprising: (a) providing a plasmid solution comprised of unligatable open circular plasmid; (b) reacting the unligatable open circular plasmid with one or more enzymes and appropriate nucleotide cofactor(s), such that unligatable open circular plasmid is converted to 3′-hydroxyl, 5′-phosphate nicked plasmid; (c) reacting the 3′-hydroxyl, 5′-phosphate nicked plasmid with a DNA ligase and DNA ligase nucleotide cofactor, such that 3′-hydroxyl, 5′-phosphate nicked plasmid is converted to relaxed covalently closed circular plasmid; and (d) reacting the relaxed covalently closed circular plasmid with a DNA gyrase and DNA gyrase nucleotide cofactor, such that relaxed covalently closed circular plasmid is converted to negatively supercoiled plasmid. In other embodiments, DNA gyrase is replaced with reverse DNA gyrase or reaction (d) is not performed.
Owner:HYMAN EDWARD D
Method for marking non-heading cabbage molecule by utilizing SAMPL technique
The invention relates to a method for loose head cabbage molecular markers by utilizing the SAMPL technology comprising DNA extraction, DNA enzyme digestion-connection system optimization, SAMPL pre-amplification system optimization, SAMPL amplification system optimization, electrophoresis, silver staining color development and cluster analysis, wherein, a DNA sample is performed enzyme digestion through using EcoRI, MseI or PstI enzyme, and then corresponding joints of EcoRI,MseI or PstI are connected on the DNA sample through using T4-DAN ligase; the SAMPL pre-amplification is performed through using a 10mu L reaction system; a pre-amplification primer is corresponding to the chosen enzyme in the enzyme digestion; the SAMPL amplification is performed through using a 20mu L reaction system, one amplification primer is corresponding to the pre-amplification primer, and the other is an SAMPL primer. The method for loose head cabbage molecular markers by utilizing the SAMPL technology performs optimization and primer design for the complicated system of the SAMPL, constructs an SAMPL reaction system applicable to loose head cabbages and realizes loose head cabbage molecular markers.
Owner:SHANGHAI JIAO TONG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com