Construction method of high-throughput sequencing library suitable for single-stranded DNA
A technology of sequencing library and construction method, applied in the field of high-throughput sequencing library construction, can solve problems such as inability to use single-stranded DNA, and achieve the effects of improving molecular utilization, avoiding molecular loss, and improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1. High-throughput sequencing library construction method suitable for single-stranded DNA
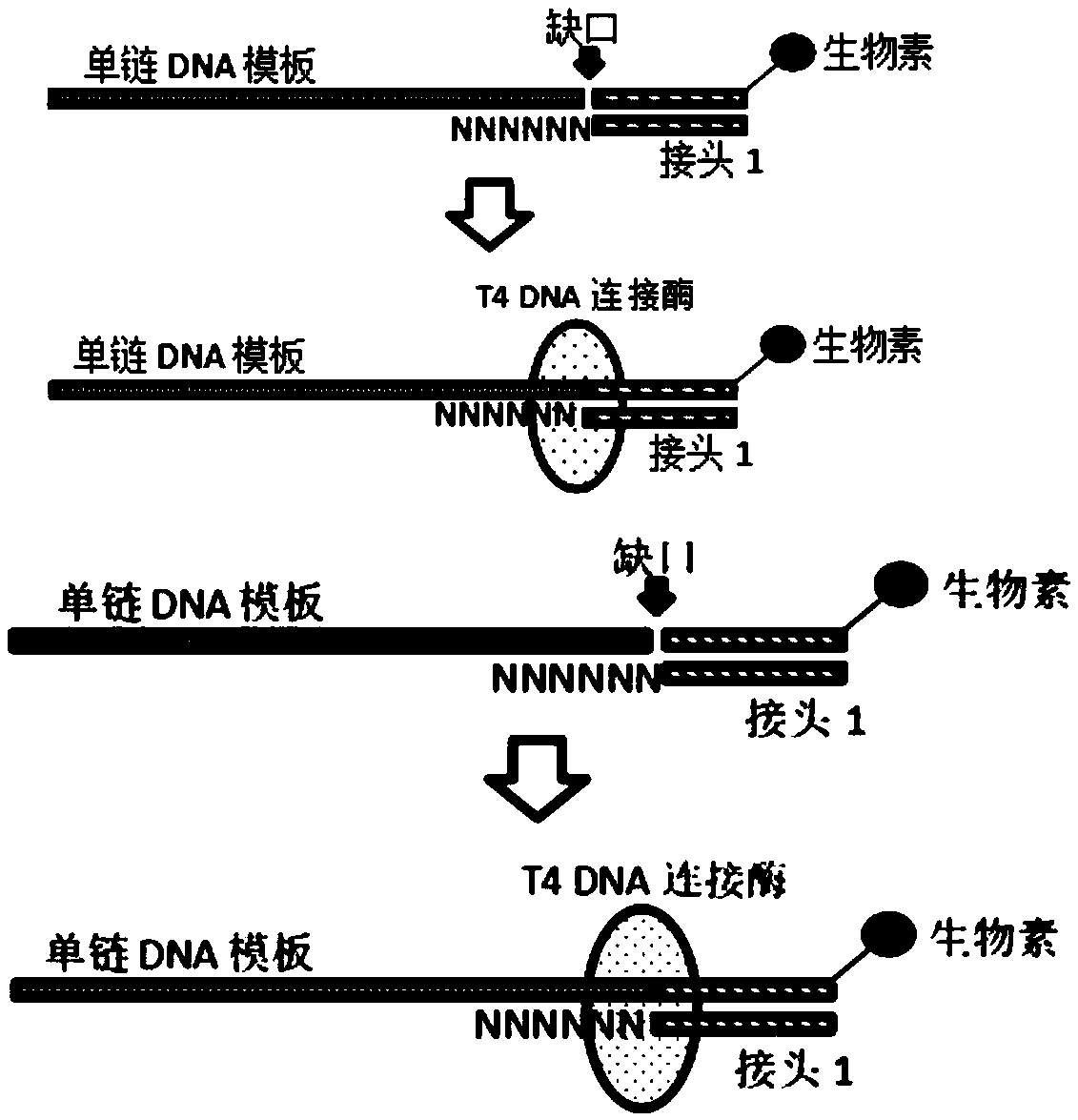
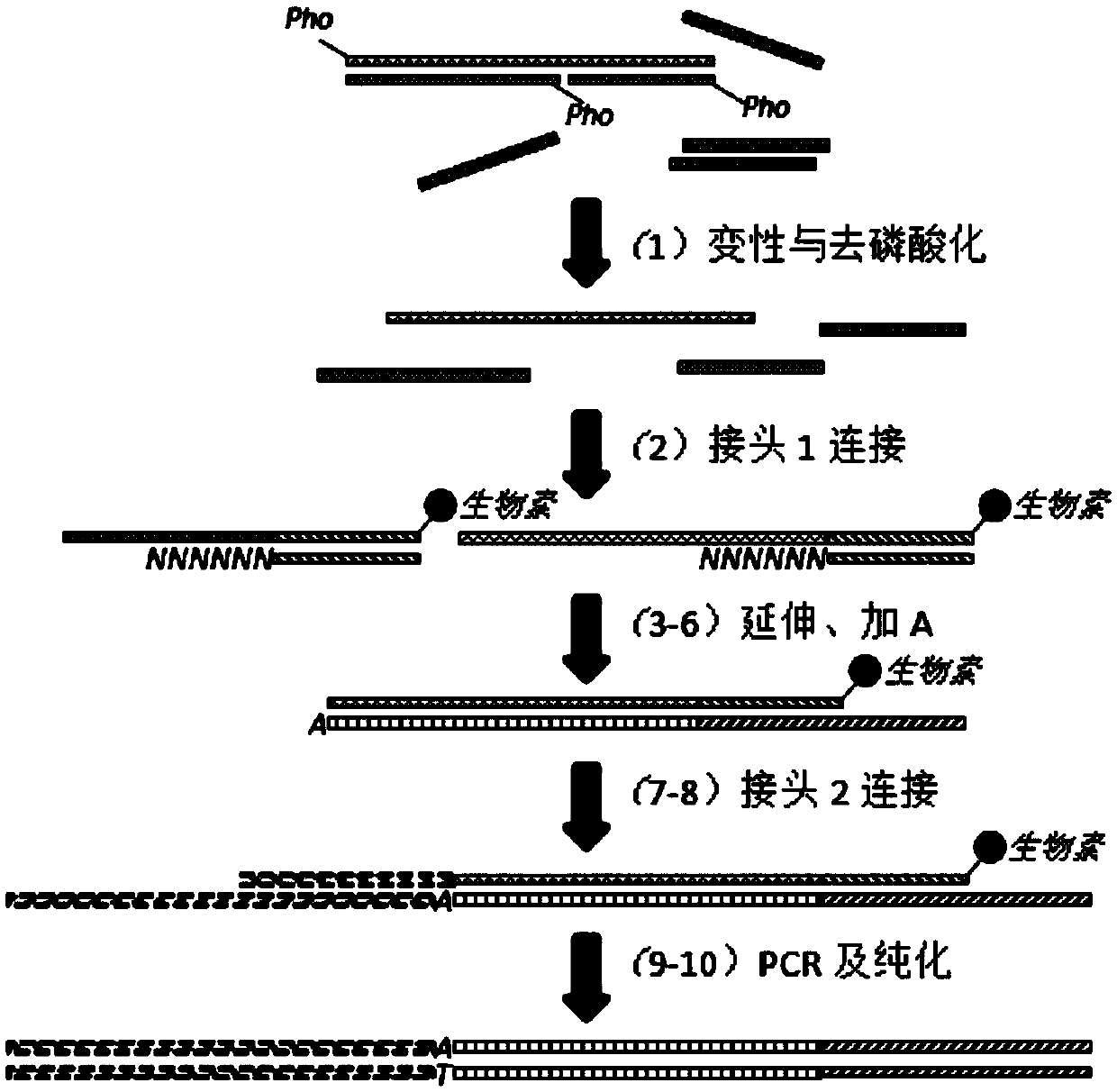
[0053] The present invention is a method for constructing a high-throughput sequencing library. Using this method, single-stranded DNA can be used as a template for sequencing library construction. It is applicable to but not limited to sequencing platforms such as BGISEQ-500, MGISEQ, and Illumina. Applicable sample types include but are not limited to. Limited to fragmented DNA, cfDNA, and bisulfite-treated DNA, it can realize whole-genome sequencing, whole-genome methylation sequencing, and capture sequencing.
[0054] The high-throughput sequencing library construction method suitable for single-stranded DNA provided in this example, its technical core lies in a special linker (linker 1) with 6N (N is A or T or C or G) to make single-stranded DNA It is possible to connect the template with the adapter. The double-stranded linker with 6N can be randomly combined to th...
Embodiment 2
[0148] Example 2. Practical application of the method for constructing a high-throughput sequencing library suitable for single-stranded DNA of the present invention
[0149] cfDNA whole genome methylation sequencing (WGBS) was performed according to the technical process introduced in Example 1.
[0150] Preparation of library template: use MagPure Circulating DNA Maxi Kit to extract healthy human cfDNA, take 20ng of extracted cfDNA, and use EZ DNA Methylation-Gold TM Kit performs bisulfite conversion.
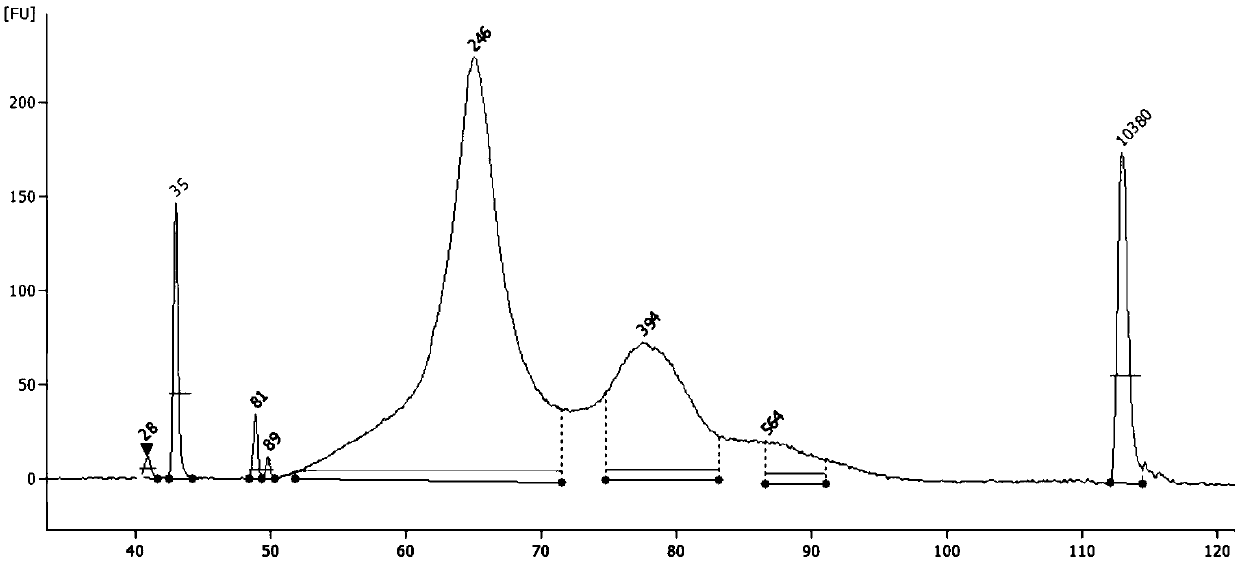
[0151] According to the relevant steps of Example 1, the WGBS library was constructed. After PCR purification, the library was quantified using Qubit. The total amount of the library was 378ng, and the library was 2100 such as image 3 shown. After the library construction is completed, the BGISEQ-500 platform is used for PE50 sequencing, and the expected WGBS sequencing data volume is 20G.
[0152] WGBS sequencing data analysis: BGISEQ-500 was used for sequencing, and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com