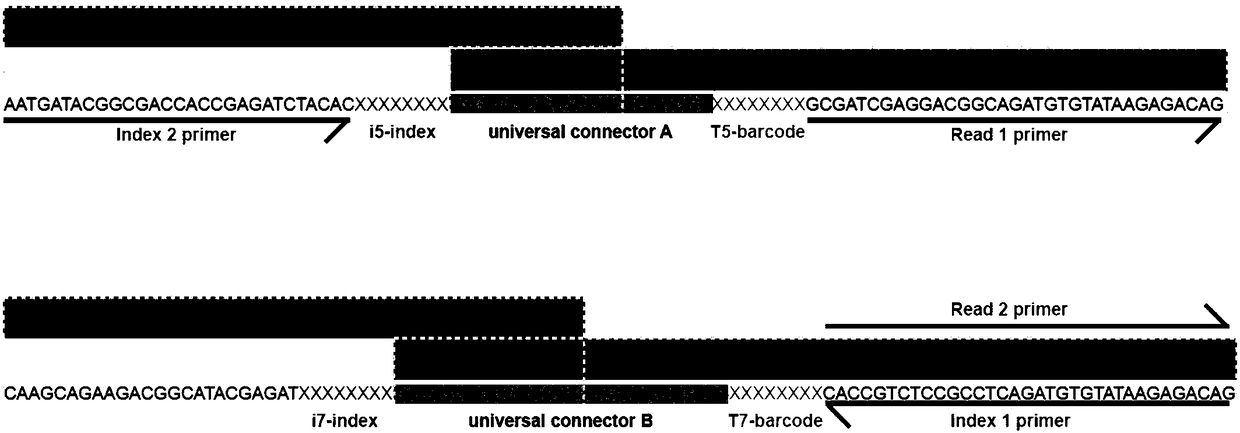
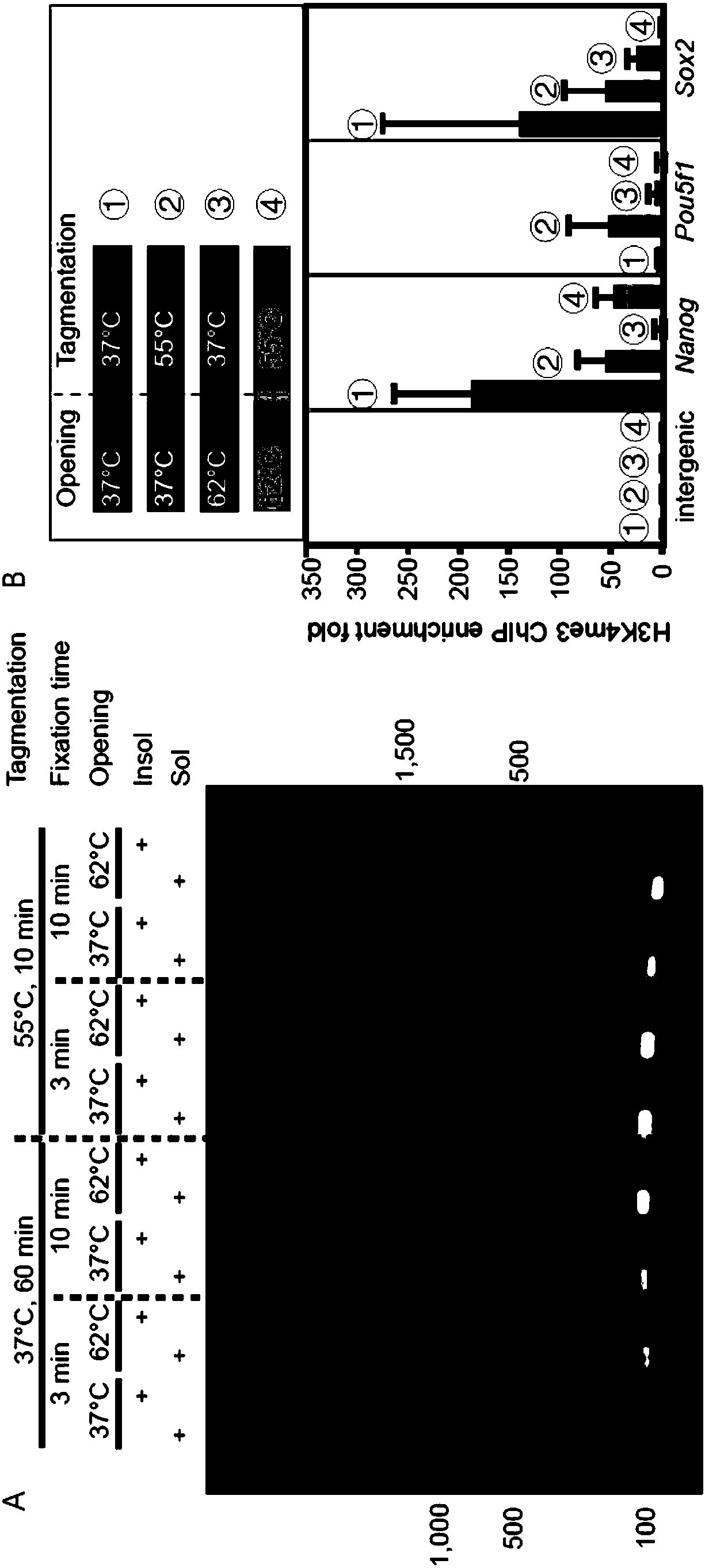
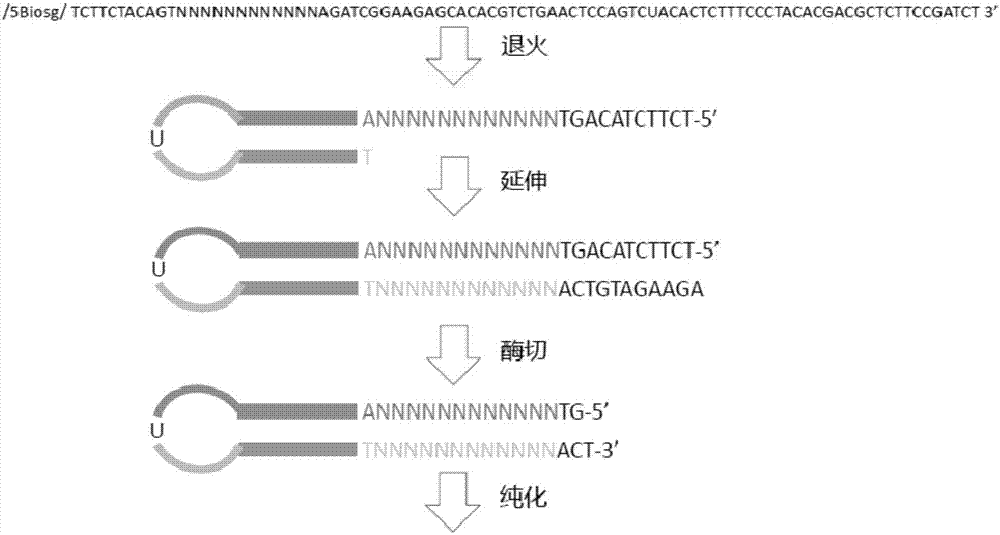
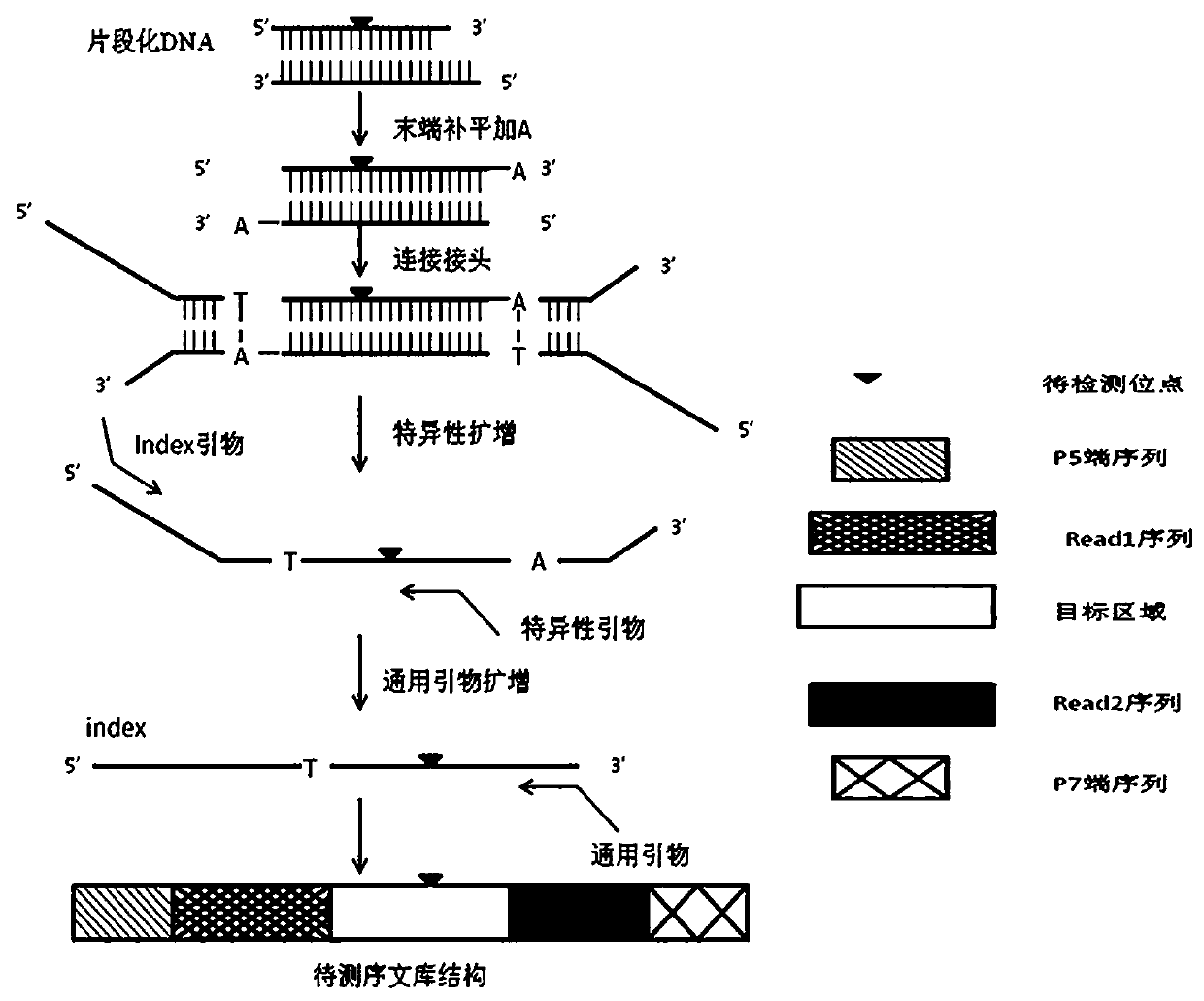
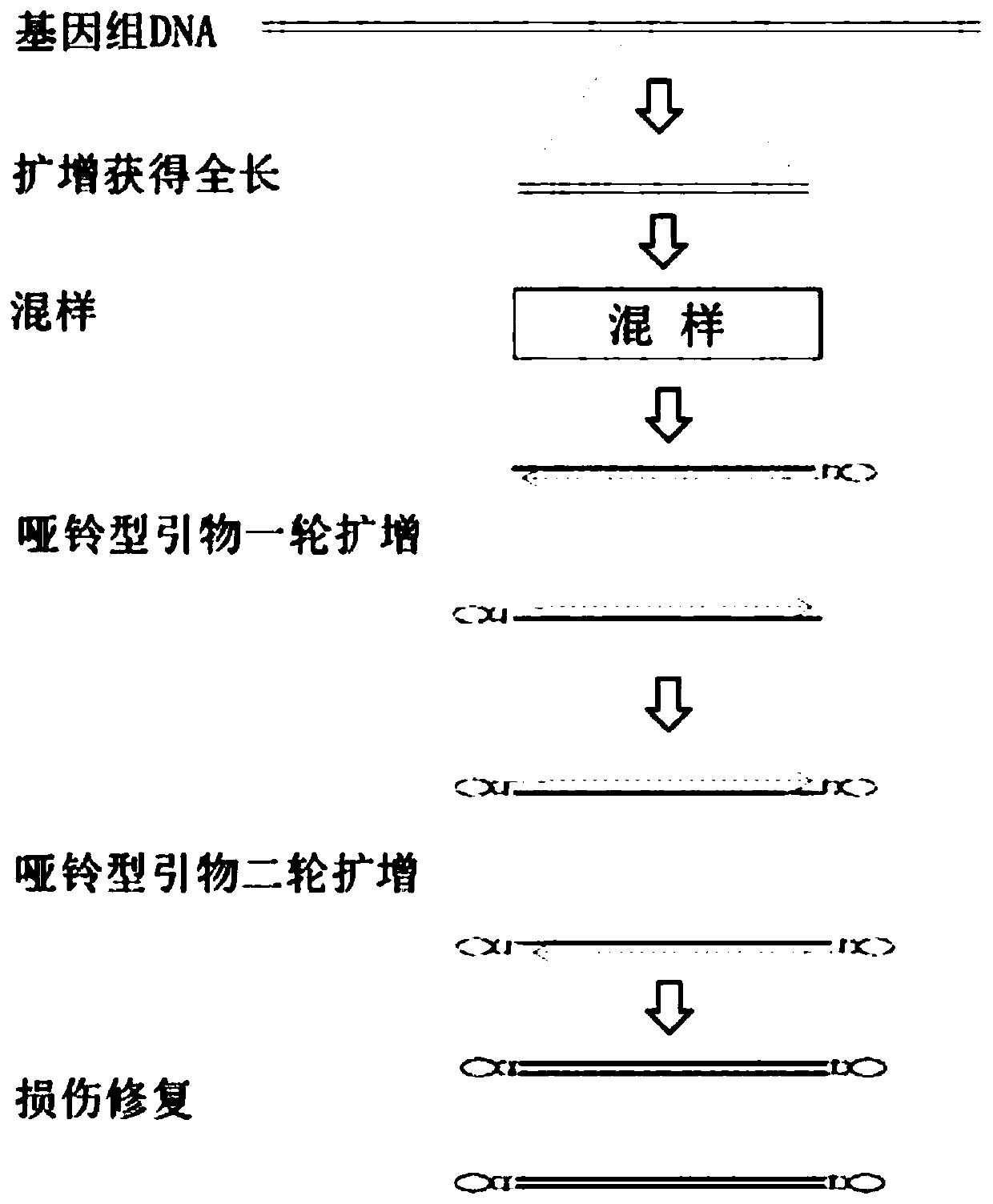
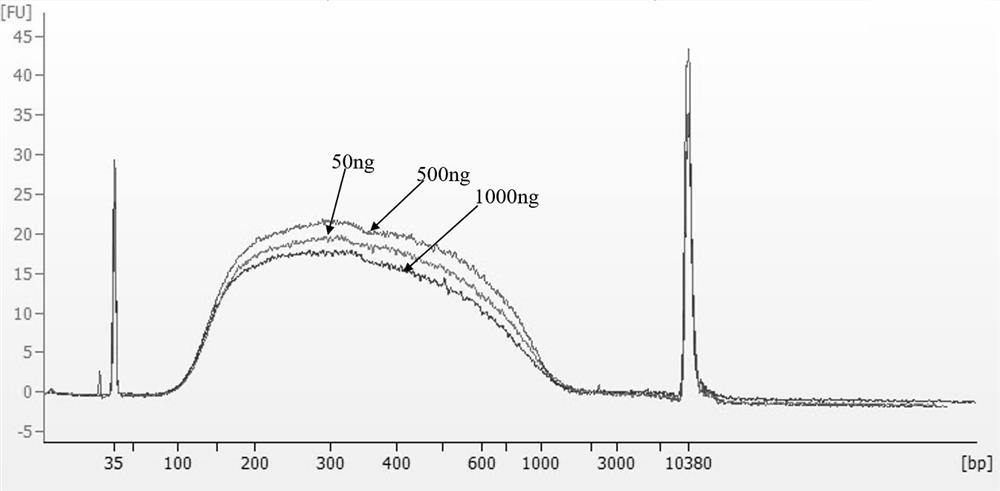
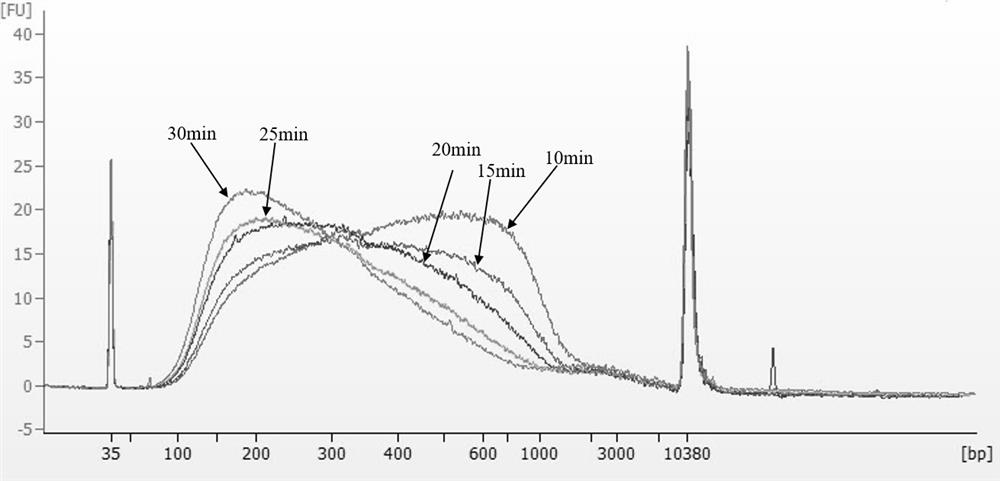
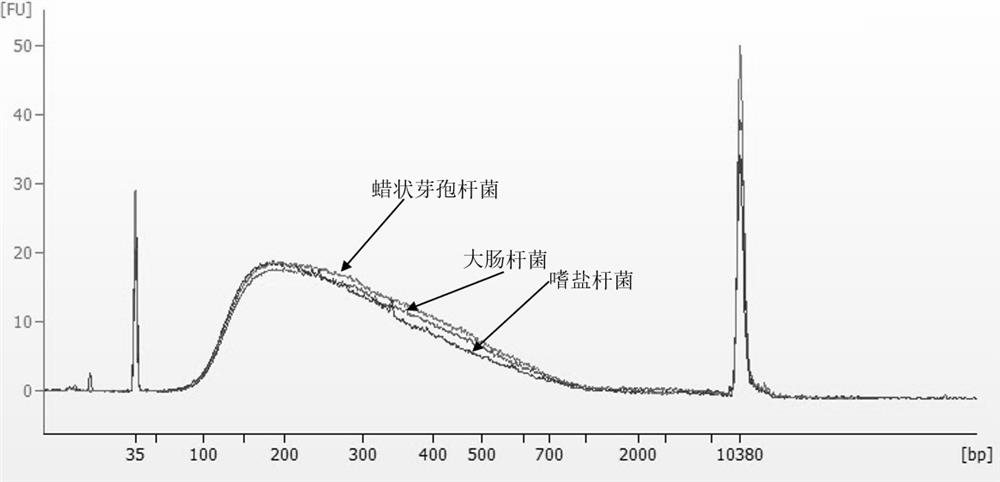
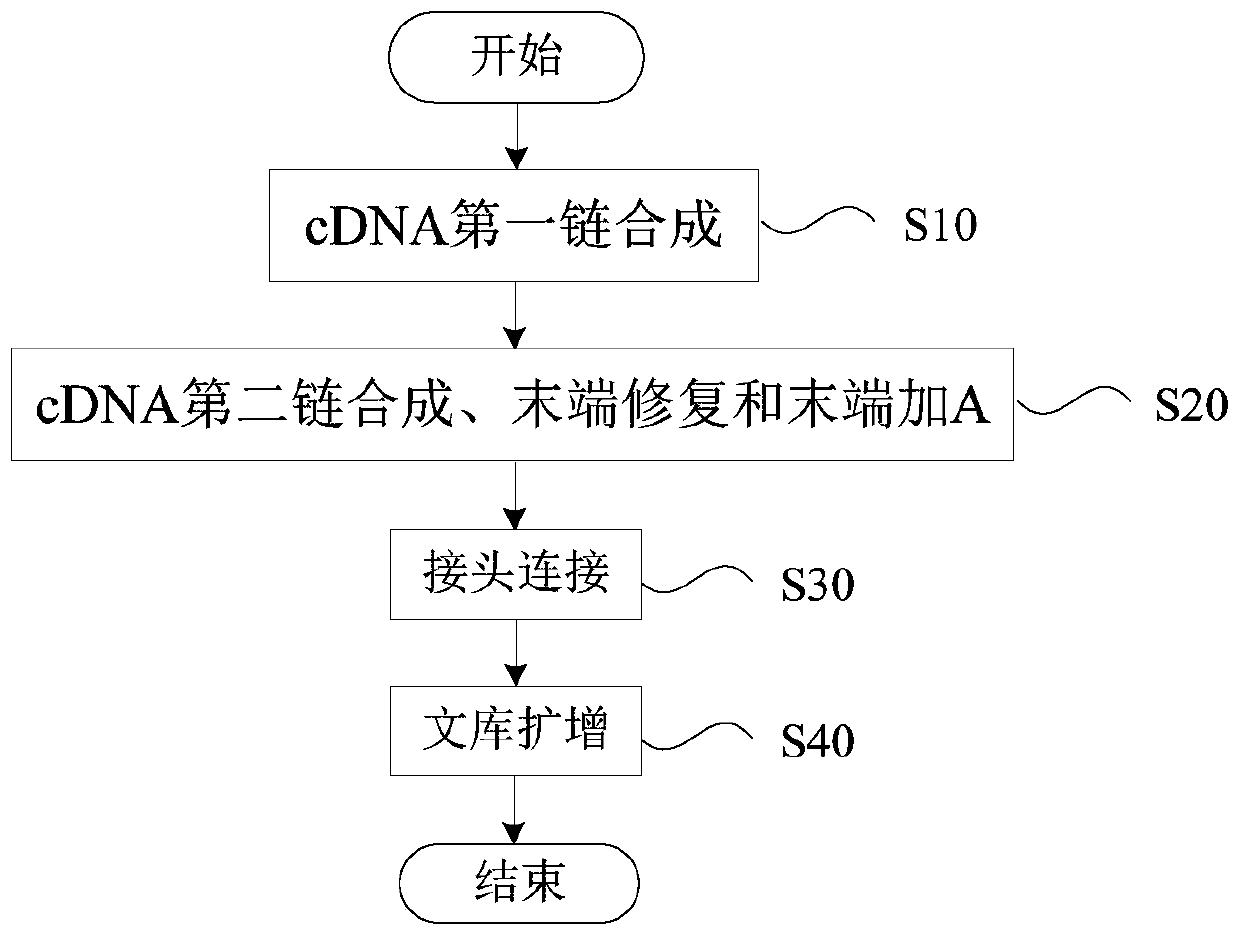
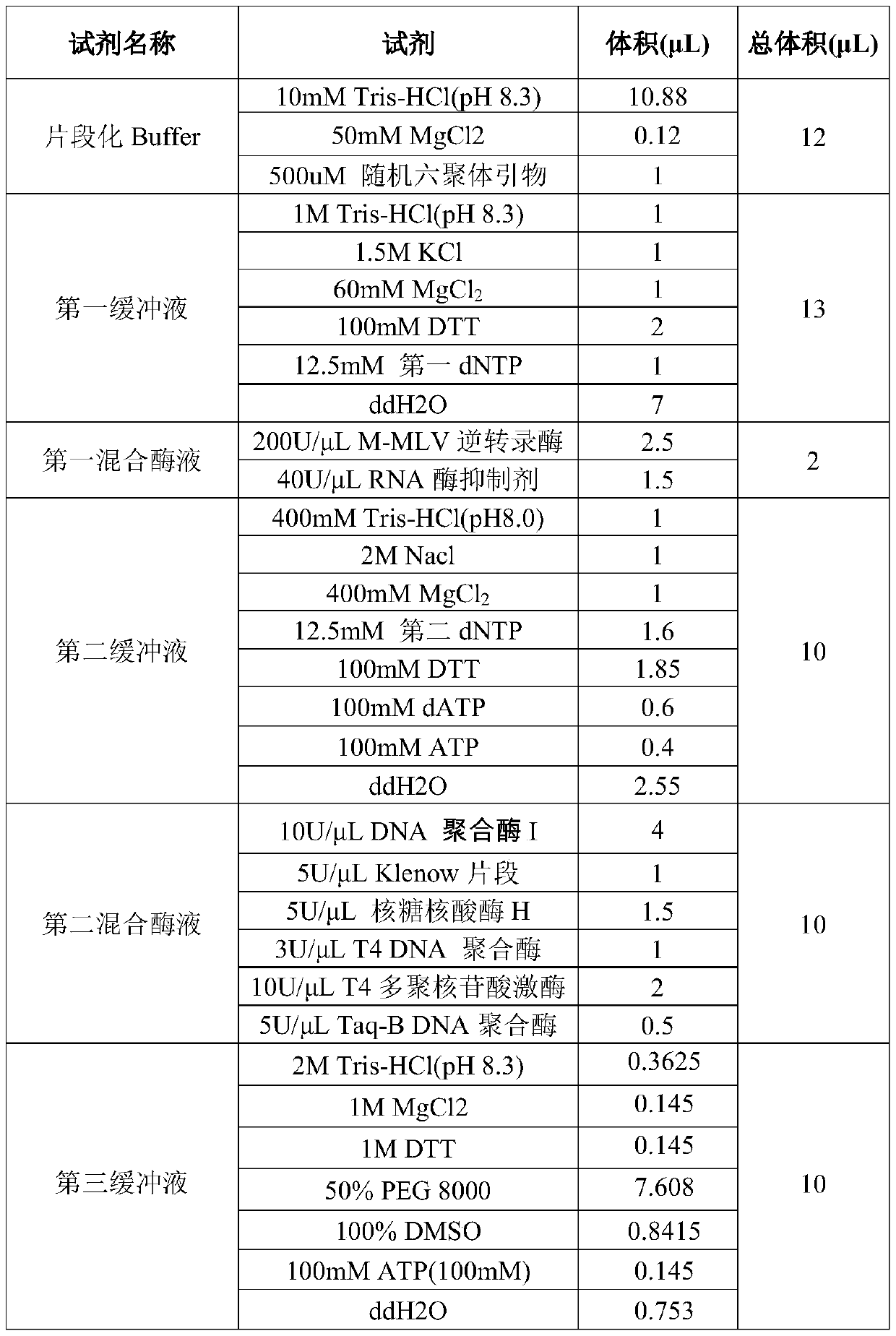
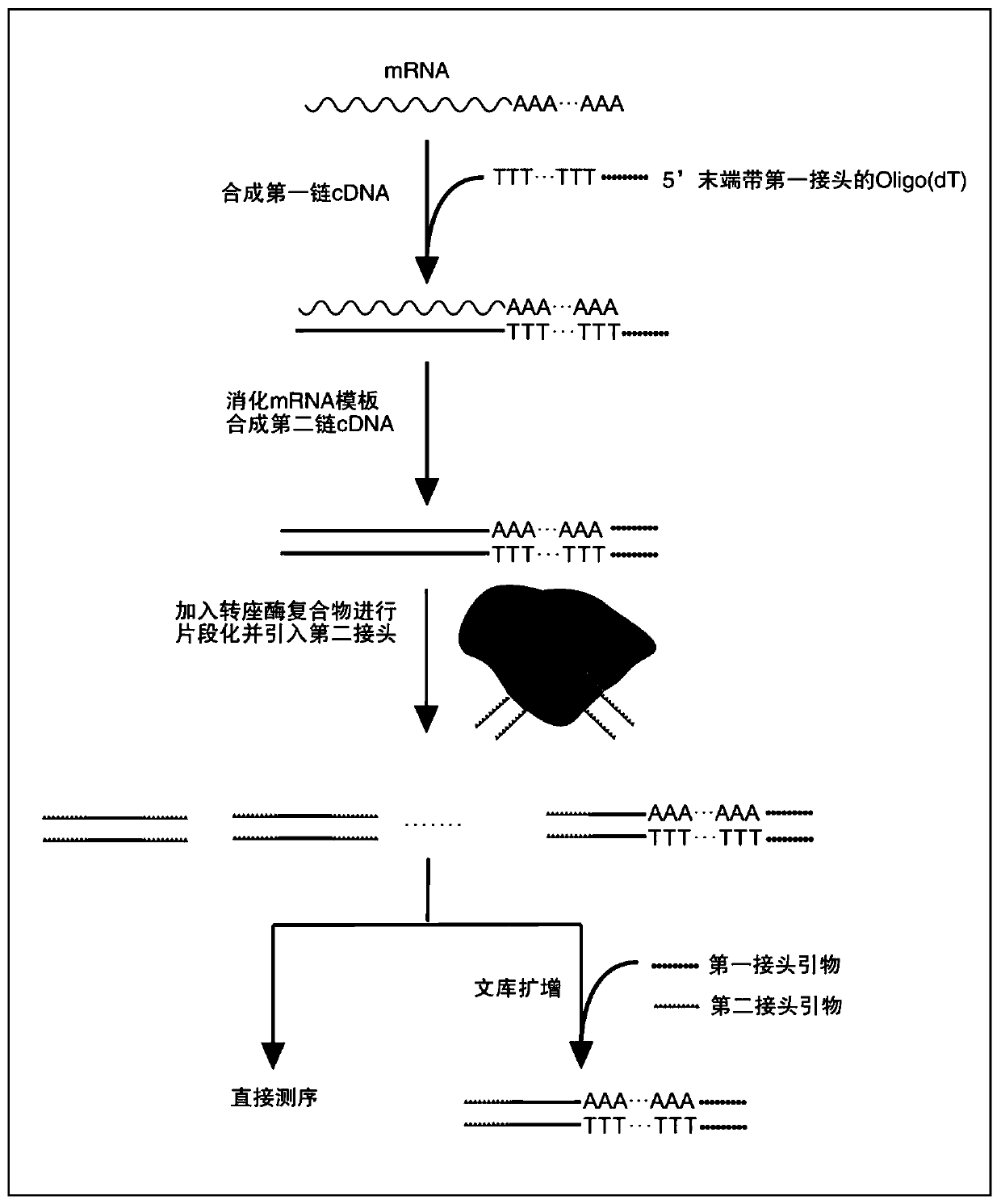
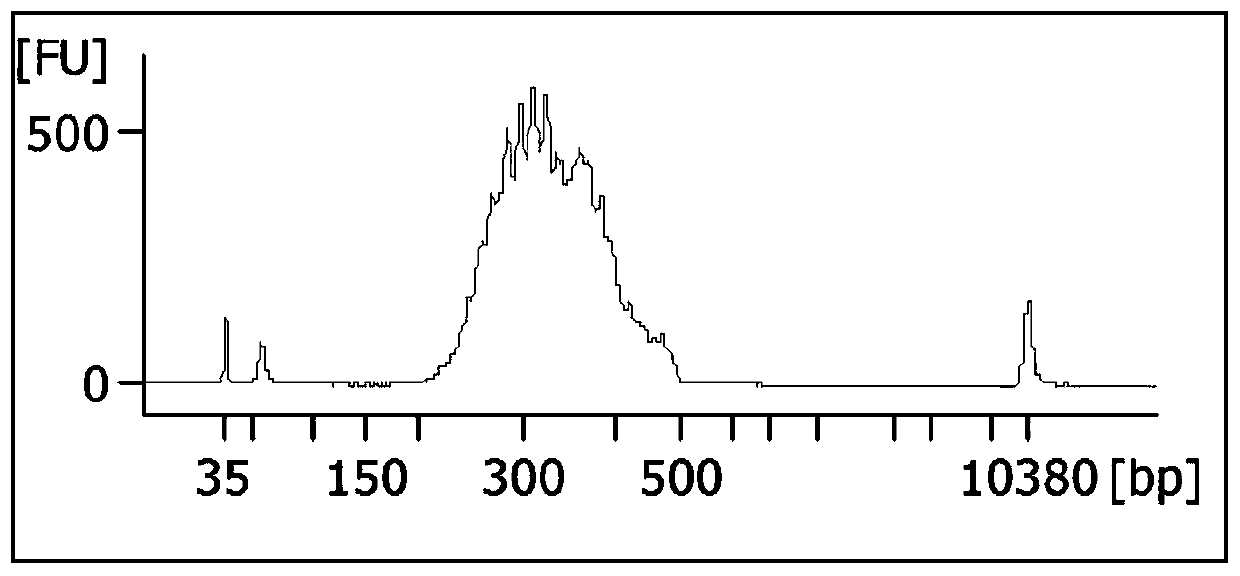
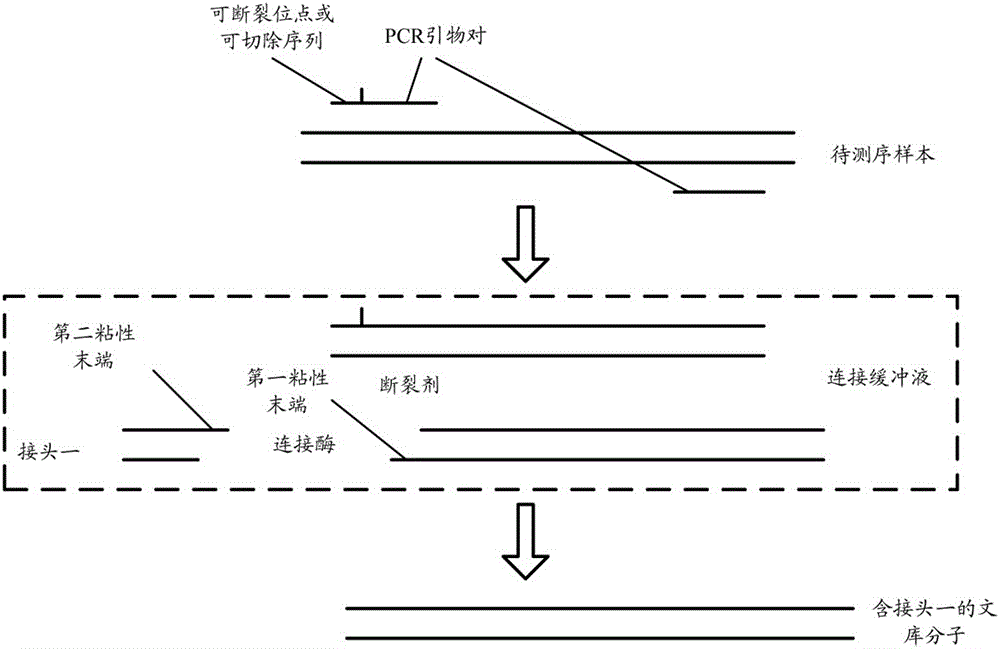
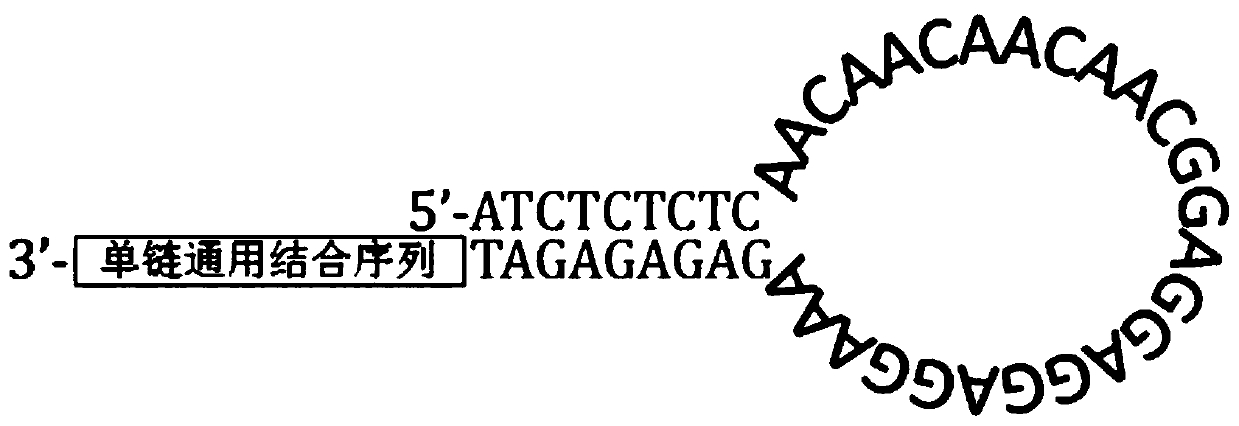Patents
Literature
60results about How to "Improve the efficiency of database construction" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
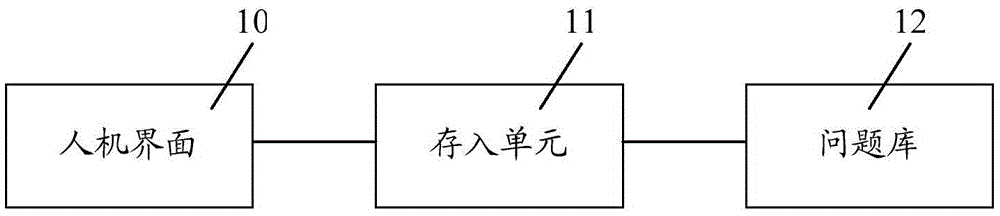
Intelligent question answering knowledge base establishment method, establishment device and establishment system
ActiveCN105608218AImprove build efficiencyImprove accuracySpecial data processing applicationsMachine learningA domain
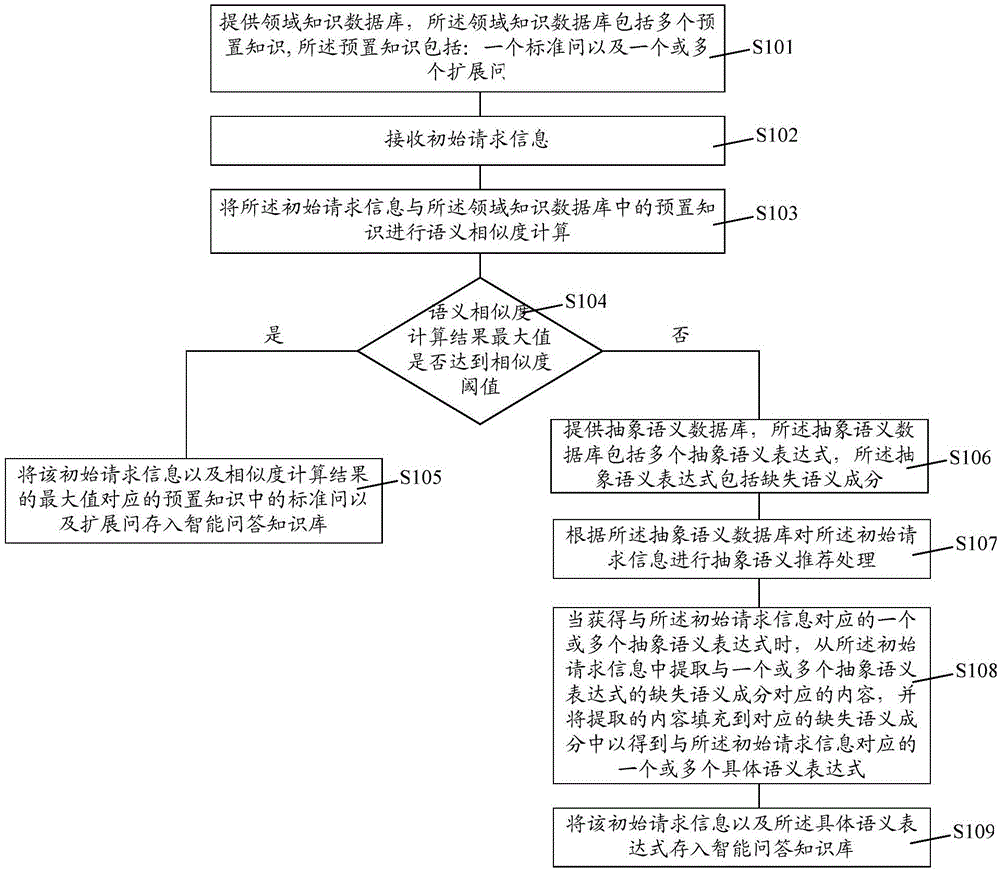
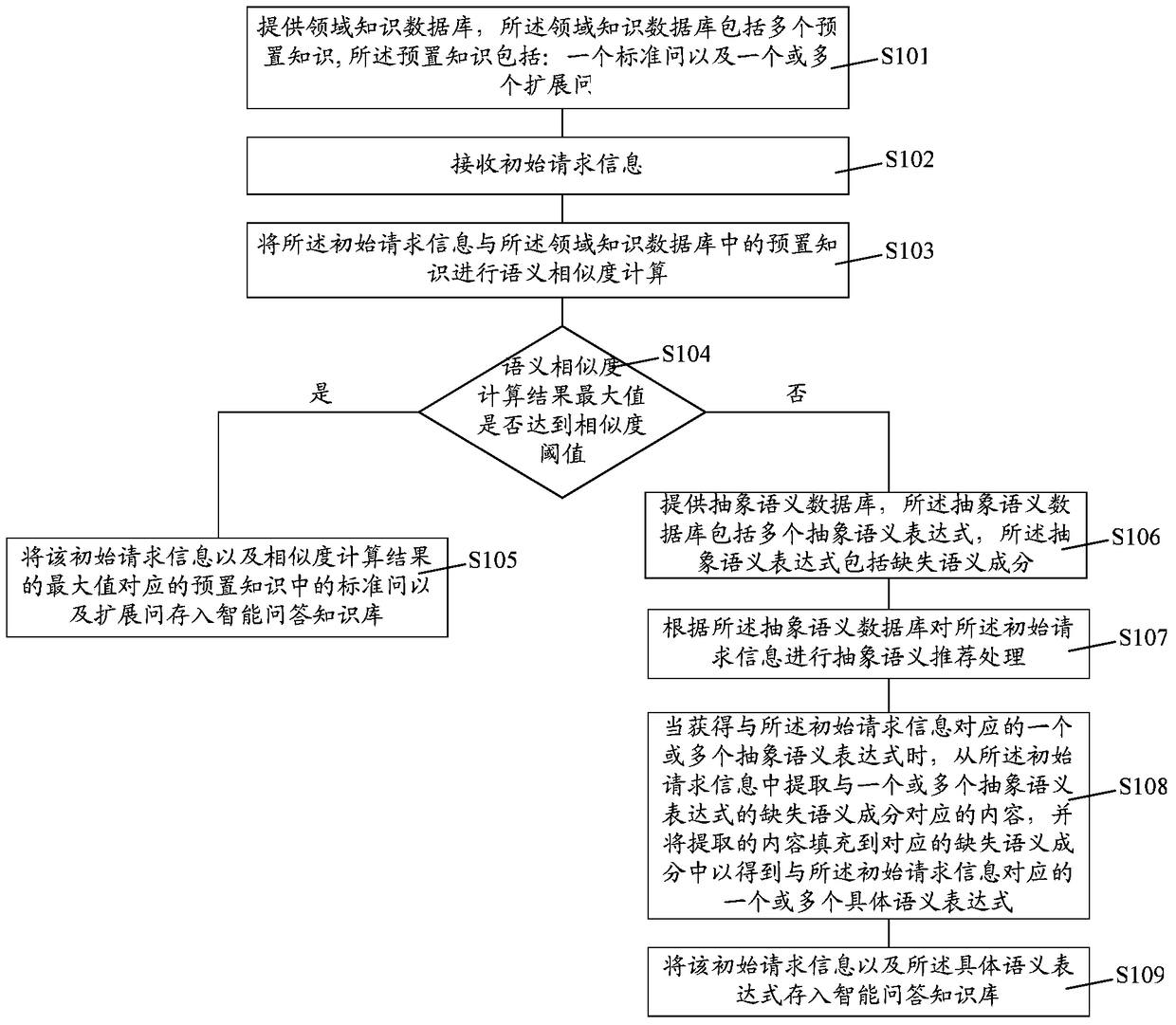
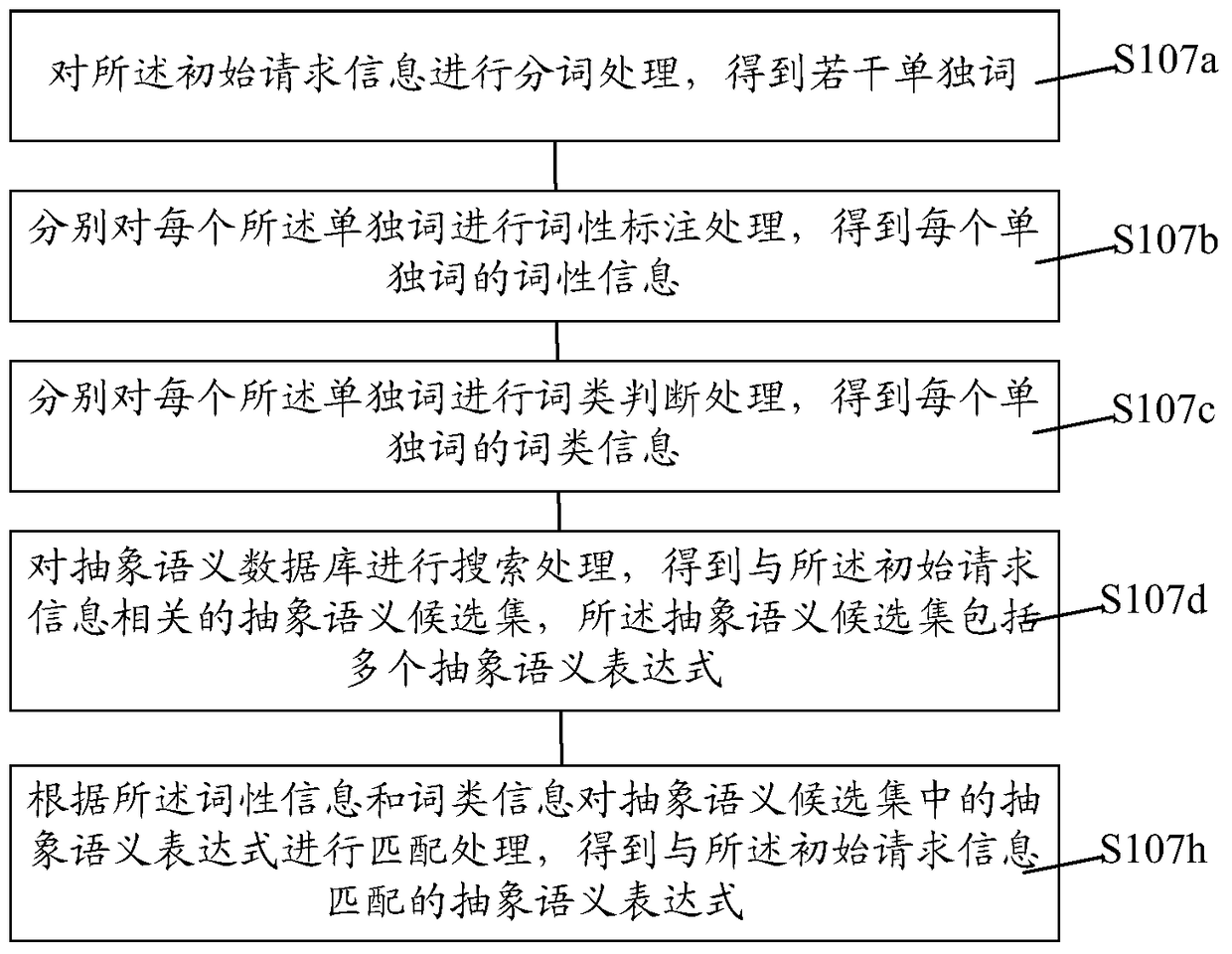
The invention relates to an intelligent question answering knowledge base establishment method, establishment device and establishment system. The establishment method comprises the following steps: providing a domain knowledge data base, wherein the domain knowledge data base comprises a plurality of pieces of preset knowledge; receiving initial request information; performing semantic similarity calculation on the initial request information and the preset knowledge in the domain knowledge data base, and judging whether the maximum value of the semantic similarity calculation results is larger than a similarity threshold or not; if the maximum value of the semantic similarity calculation results is larger than the similarity threshold, storing a standard question and an extended question in the preset knowledge corresponding to the initial request information and the maximum value of the semantic similarity calculation results in the intelligent question answering knowledge base; if the maximum value of the semantic similarity calculation results is smaller than the similarity threshold, carrying out the abstract semantic recommendation process so as to acquire one or more specific semantic expressions corresponding to the initial request information, and storing the initial request information and the specific semantic expressions in the intelligent question answering knowledge base. The establishment method provided by the invention can improve the establishment efficiency of the intelligent question answering knowledge base.
Owner:SHANGHAI XIAOI ROBOT TECH CO LTD
Method for screening drug target genes based on CRISPR/Cas9 high-throughput technology
InactiveCN106399377ADetermining infection efficiencyDetermine the MOI valueMicrobiological testing/measurementFermentationInfected cellCancer cell
The invention relates to a method for screening drug target genes based on a CRISPR / Cas9 high-throughput technology. The method comprises the steps of firstly establishing a sgRNA library; secondly packaging the sgRNA library by using slow viruses, and collecting the viruses; thirdly screening the sgRNA library in a cancer cell line; fourthly extracting cells obtained by screening and genome DNA of the cells before screening; and finally enriching sgRNA in genome DNA. Compared with the prior art, the method has the advantages that a CRISPR / Cas cell screening process is improved, the virus infection efficiency is determined with a simple and convenient method by utilizing puromycin resistance of infected cells, and MOI values of the viruses are determined; more importantly, a virus packaging method is greatly optimized, so that the virus packaging efficiency is improved to be more than 5 times that of a conventional method, and large-scale drug target screening cost can be greatly reduced; and the method is used for promoting industrialization of cancer drug target screening.
Owner:TONGJI UNIV
Fusion protein used for single-cell ChIP-seq library preparation and application thereof
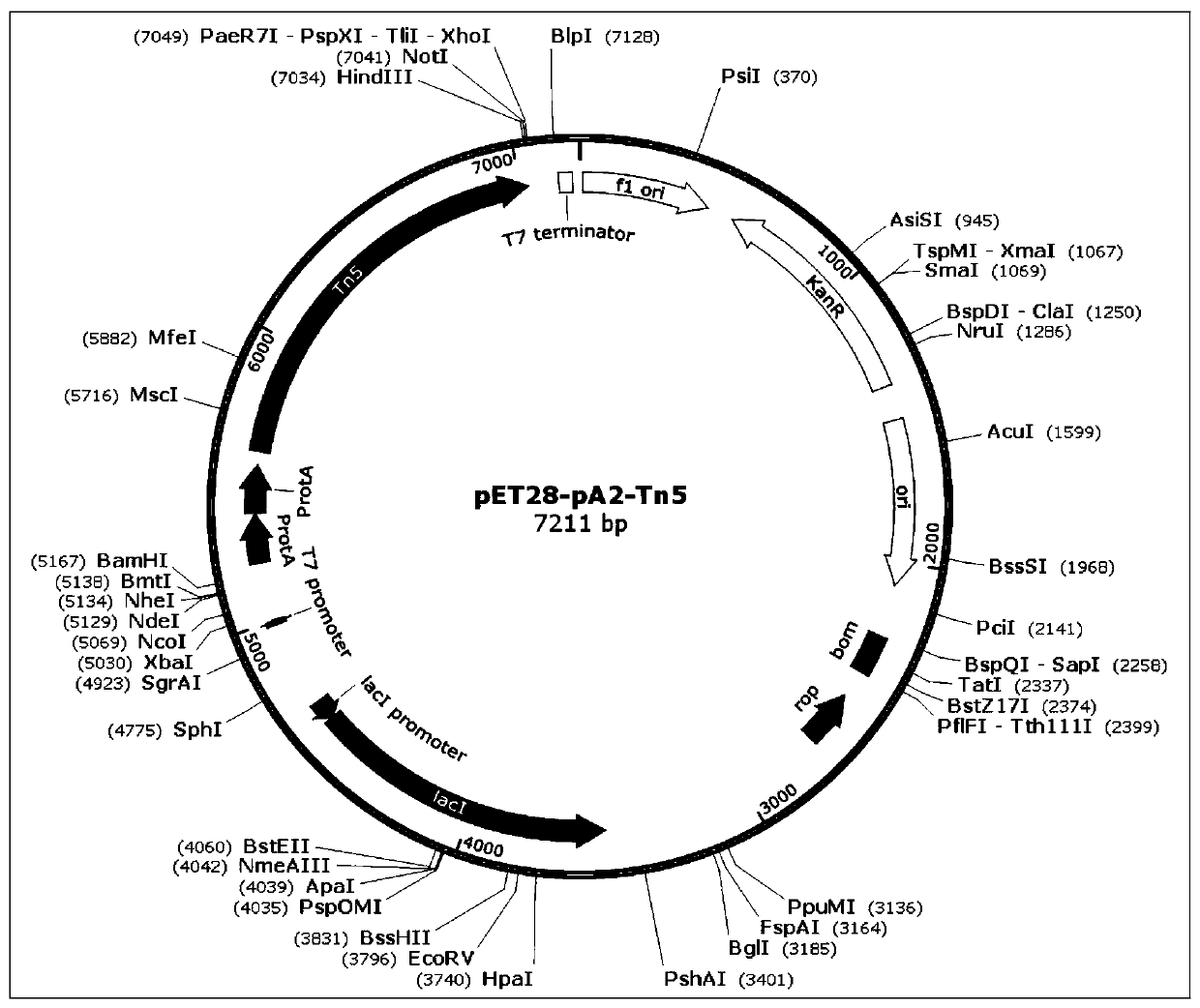
ActiveCN110372799AImprove accuracySimplify the experimental processPolypeptide with affinity tagMicrobiological testing/measurementTn5 transposaseFc binding
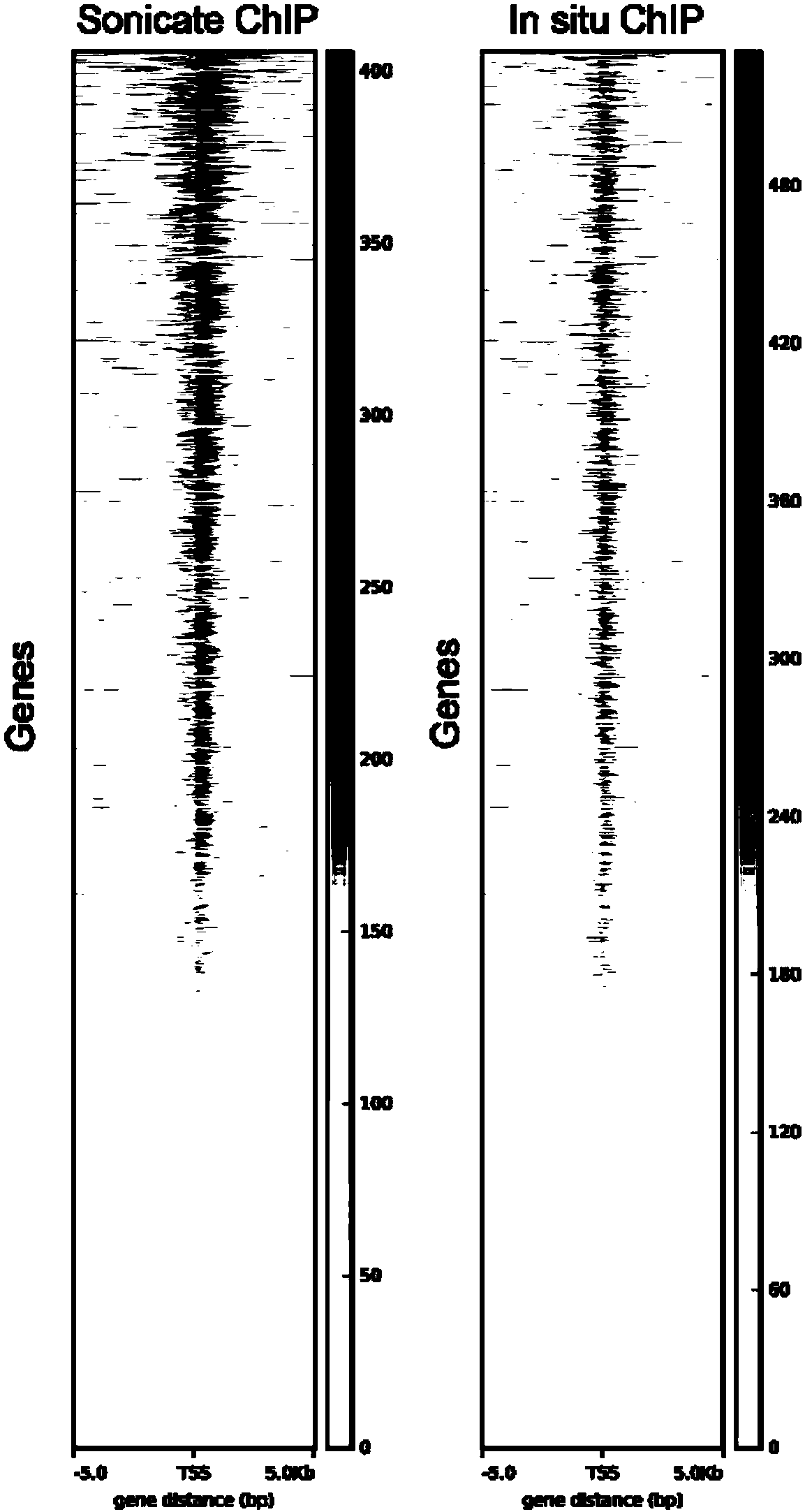
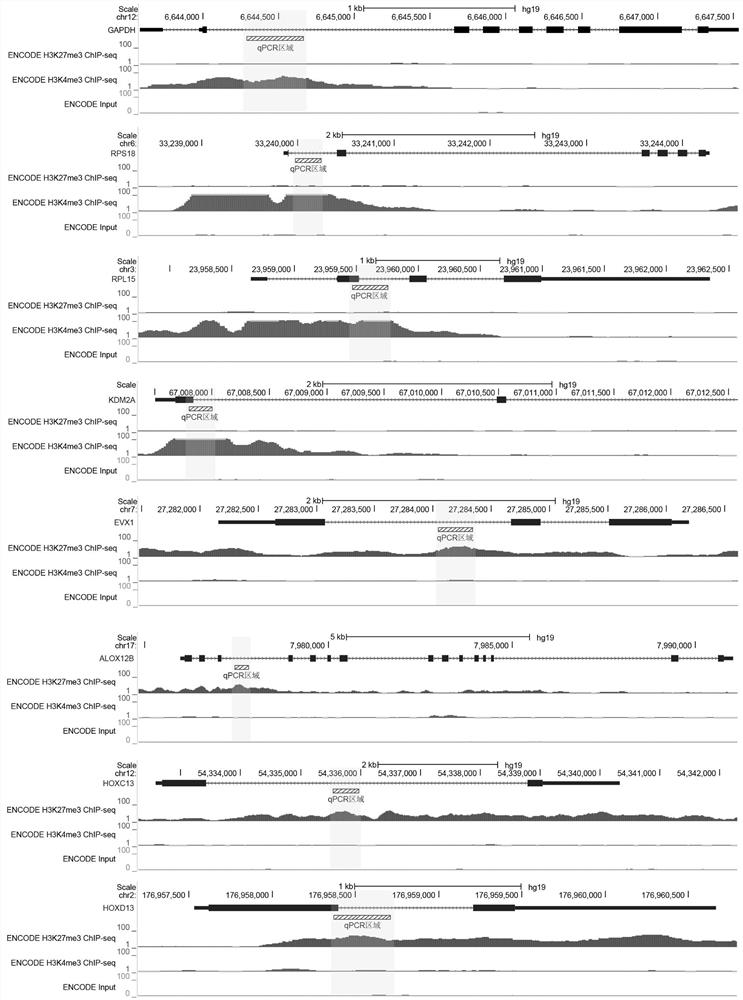
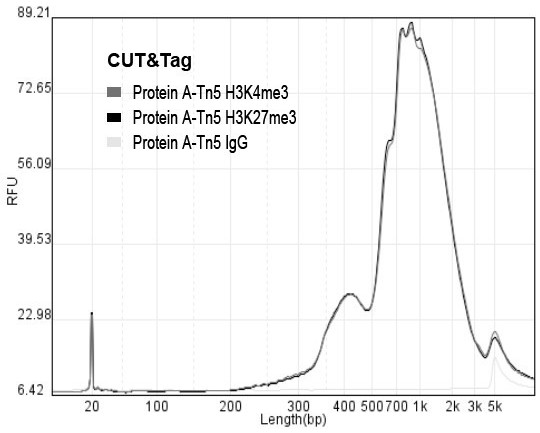
The invention relates to a fusion protein used for single-cell ChIP-seq library preparation and application thereof. The fusion protein comprises Tn5 transposase and an Fc binding protein; a kit comprises the fusion protein and other auxiliary detection reagents; a method uses the fusion protein or the kit for ChIP-seq detection. The fusion protein, the kit and the method can improve the library building efficiency and lower the library background in the ChIP-seq detection process, so that the accuracy of the ChIP-seq detection method can be improved, and the experimental flow of ChIP-seq canbe simplified.
Owner:PEKING UNIV
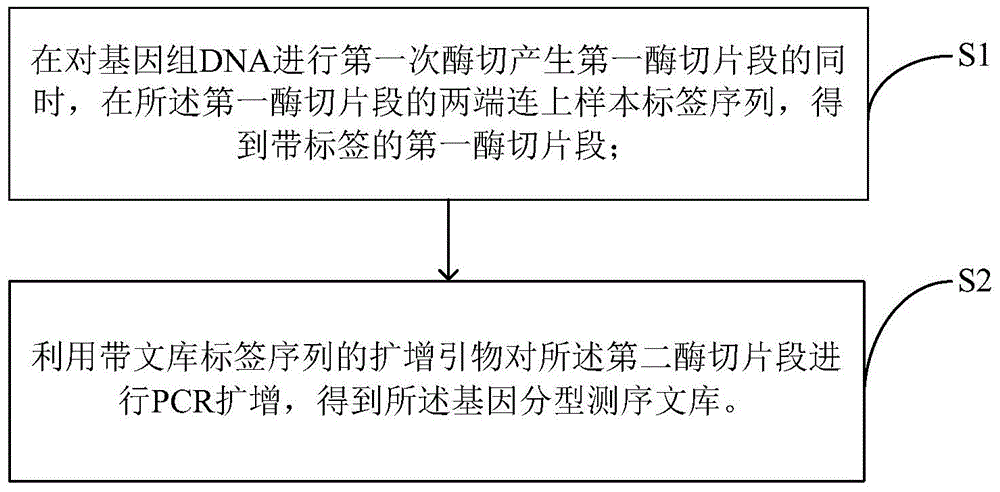
Construction method and sequencing method of genetic typing sequencing library
ActiveCN104561294AIncrease the lengthIncrease the effective amountMicrobiological testing/measurementLibrary creationEnzyme digestionInvalid Data
The invention discloses a construction method and a sequencing method of a genetic typing sequencing library. The construction method comprises the following steps: S1, bonding sample label sequences at the two ends of a first enzyme digestion fragment while a gene group DNA is subjected to enzyme digestion for the first time to produce the first enzyme digestion fragment so as to obtain a first enzyme digestion fragment with a label; S2, performing PCR amplification on the first enzyme digestion fragment by utilizing an amplification primer with a library label sequence to obtain the gene typing sequencing library. According to the construction method, the sample label sequences are directly bonded at the two ends of the first enzyme digestion fragment while the gene group DNA is subjected to enzyme digestion for the first time to produce the first enzyme digestion fragment, so that the constructed library can directly get into the next step of reading a target enzyme digestion fragment sequence after the sample label information is read during sequencing by a computer, and the reading length of the target enzyme digestion fragment sequence is increased, the invalid data volume is reduced, and the effective quantity of sequencing data is increased.
Owner:BEIJING NOVOGENE TECH CO LTD
Construction method of high-throughput sequencing library suitable for single-stranded DNA
PendingCN110904512AIncrease profitAvoid lostMicrobiological testing/measurementLibrary creationSingle strandHigh throughput sequence
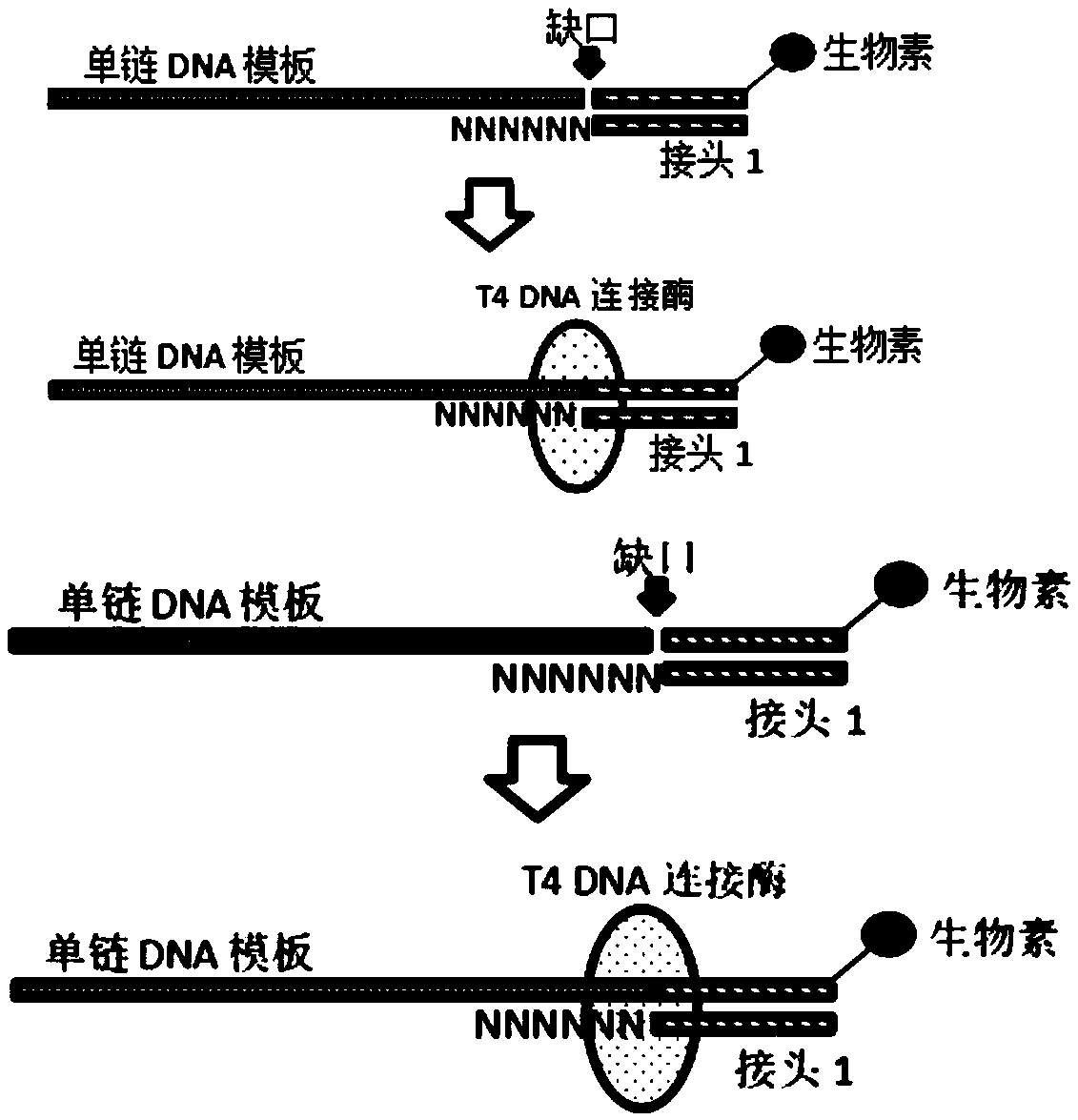
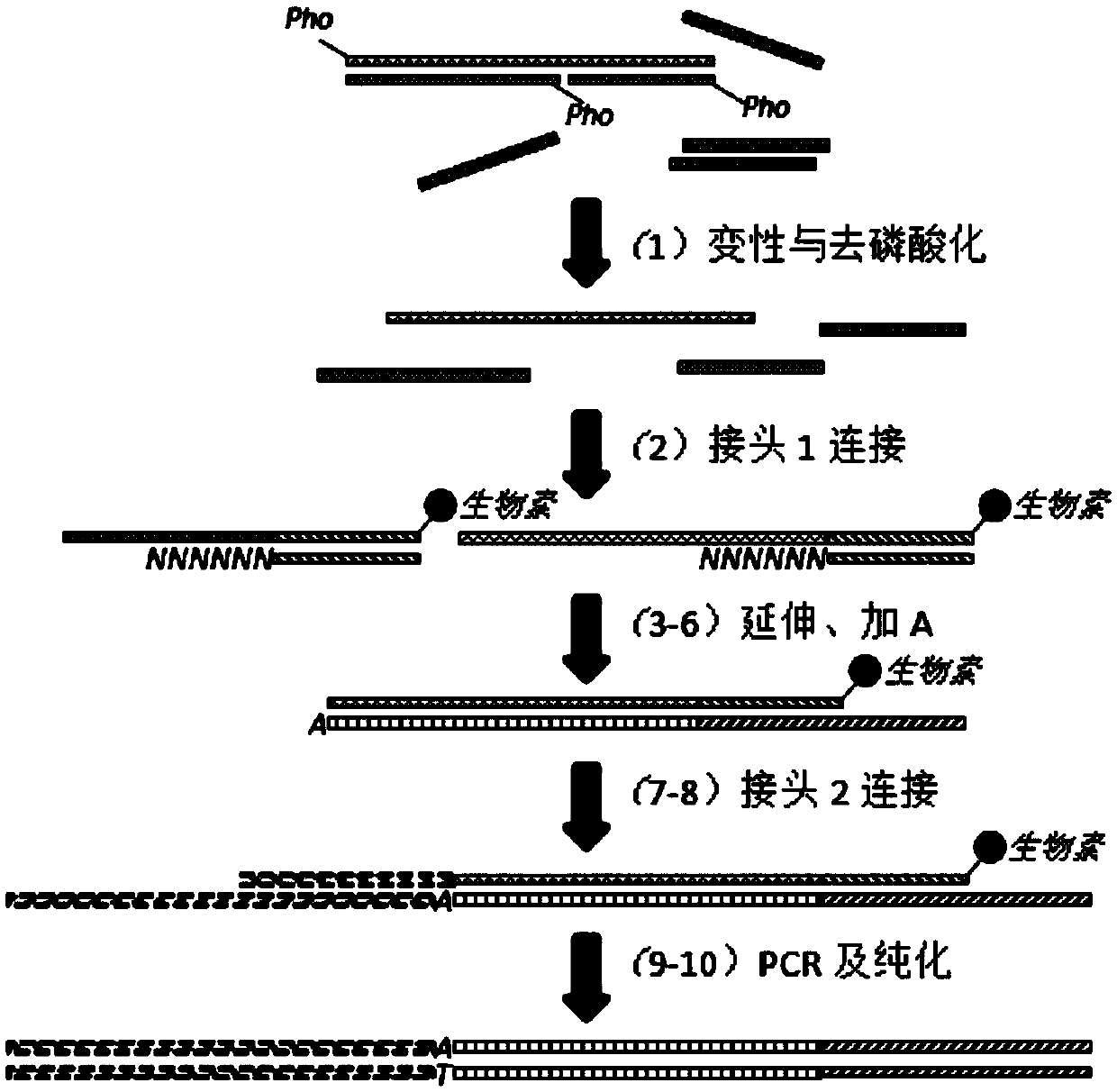
The invention discloses a construction method of a high-throughput sequencing library suitable for single-stranded DNA. The method comprises the following steps: linking a single-stranded template anda double-stranded linker 1 with degenerate bases (a special linker with degenerate bases enables the linking of the single-stranded template and the double-stranded linker to be possible, the double-stranded linker can be randomly combined to the single-stranded template to make a linker sequence be fully close to the template, and then the linking is completed by using T4 DNA ligase); extendingby using an extension primer to obtain a double-stranded product; linking the double-stranded product to a linker 2; and carrying out PCR amplification by taking the obtained linking product as a template to obtain the required library. According to the invention, the method can be used for constructing a high-throughput sequencing library, can further be used for constructing a single-stranded DNA library, can improve the utilization rate of single-stranded DNA in the construction of a high-throughput sequencing library, further achieves the purpose of low-initial-quantity library construction, and has key effect in early screening and prognosis monitoring application of tumors in the future.
Owner:ENVELOPE HEALTH BIOTECHNOLOGY CO LTD
Method for constructing high-flux and low-cost Fosmid library, label and label joint used in method
ActiveCN102409043AEfficient use ofImprove efficiencyNucleotide librariesLibrary creationHigh fluxLibrary preparation
Based on a test platform of the Illumina Company at present and aiming at the characteristic of a small section of a Fosmid library, a plurality of libraries are mixed together to form one library; furthermore, a purifying step is omitted, so a new Fosmid library preparation flow is established. The Fosmid library is constructed successfully; the Fosmid library is successfully used for sequencing; therefore, the library construction time is saved and the sequencing cost is reduced.
Owner:WUXI QINGLAN BIOLOGICAL SCI & TECH
Fusion protein, kit and CHIP-seq detection method
ActiveCN108285494AImprove binding efficiencyImprove cutting efficiencyHydrolasesAntibody mimetics/scaffoldsTn5 transposaseFc binding
The invention relates to fusion protein, a kit and a CHIP-seq detection method. The fusion protein comprises Tn5 transposase and Fc binding protein, the kit comprises the fusion protein and other auxiliary detection reagents, and the method is the CHIP-seq detection method by using the fusion protein or the kit. According to the fusion protein, the kit and the method, the library establishment efficiency can be improved in the CHIP-seq detection process, the library background is reduced, the accuracy of the CHIP-seq detection method is improved accordingly, and the experimental process of CHIP-seq is simplified.
Owner:PEKING UNIV
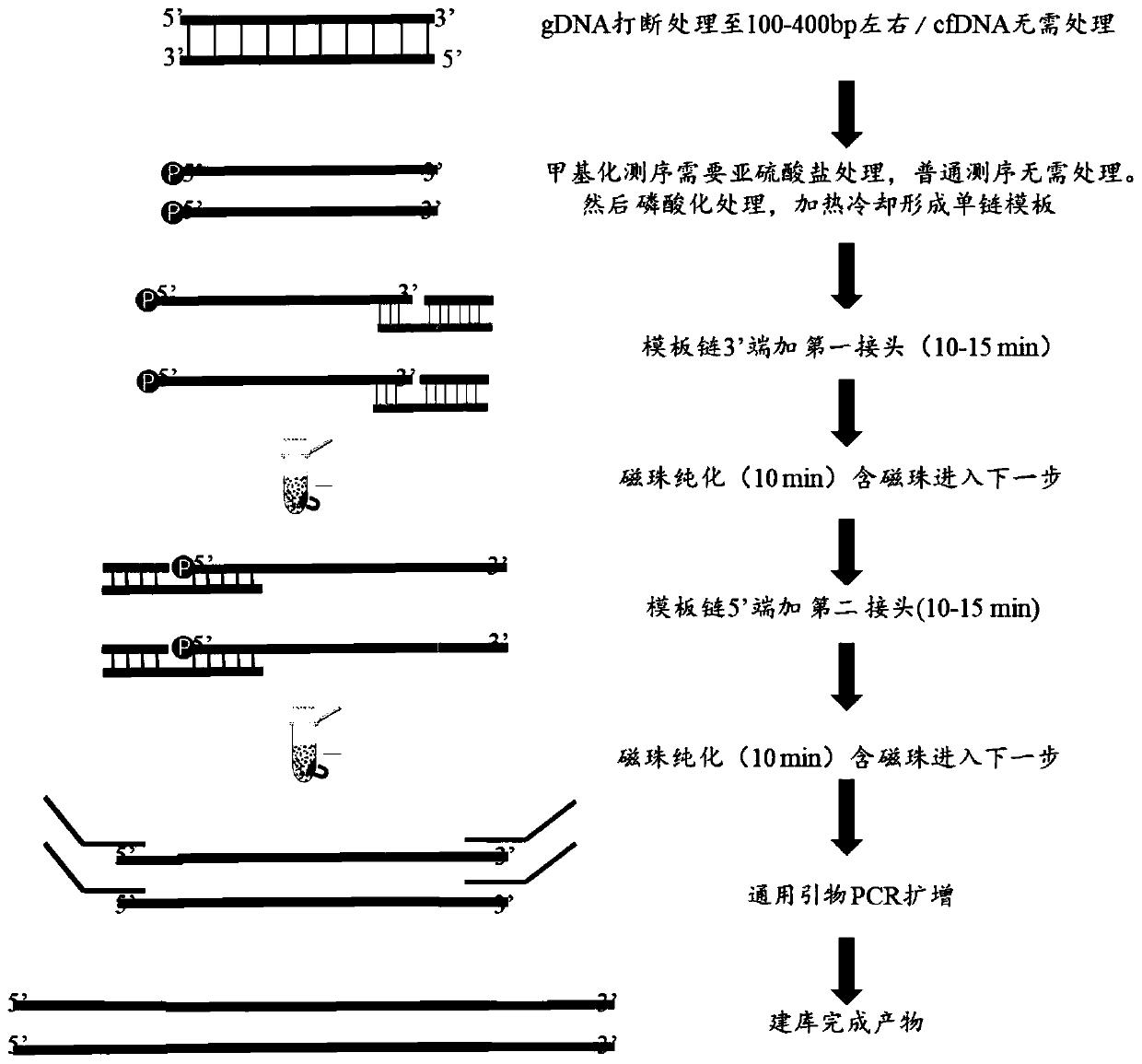
Rapid single-stranded library building method
The invention belongs to the technical field of gene sequencing libraries, and particularly relates to a rapid single-stranded library building method which comprises the following steps: providing sample DNA, and performing phosphorylation treatment on the sample DNA to obtain a single-stranded DNA template; connecting the 3' end of the single-stranded DNA template with a first joint, and then performing purification with a first magnetic bead to obtain a first connection product containing the first magnetic bead; connecting the 5' end of the first connection product with a second joint, andperforming purification with a second magnetic bead to obtain a second connection product containing the first magnetic bead and the second magnetic bead; performing PCR amplification on the second connection product by using universal primers to obtain a library; wherein the first joint and the second joint are double-stranded joints. According to the rapid single-stranded DNA library building method, the library building efficiency is improved, the library building time is saved, and library building of the single-stranded DNA can be achieved through the method so as to meet the requirements of a second-generation sequencing platform and even a third-generation sequencing platform.
Owner:深圳易倍科华生物科技有限公司
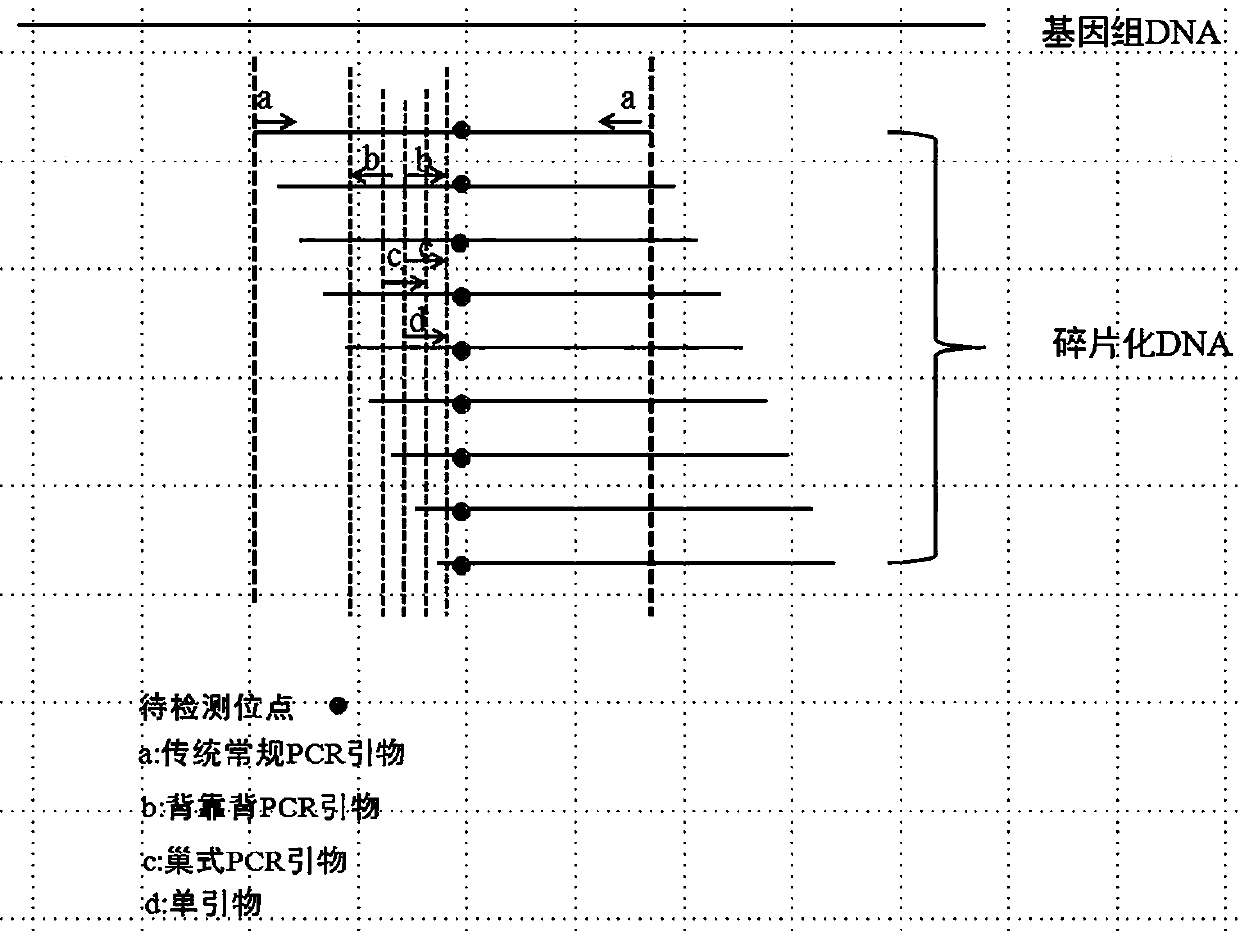
Detection method of low requency mutated DNA fragment and library building method
ActiveCN108192955AEasy to useImprove detection limitMicrobiological testing/measurementLibrary creationMutation detectionEngineering
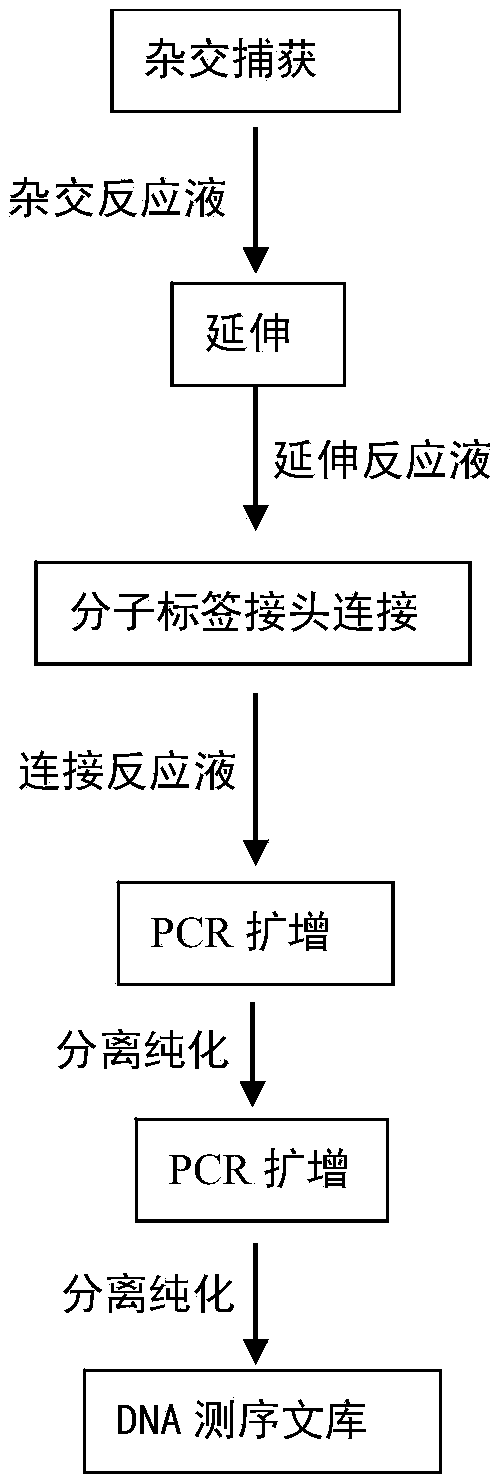
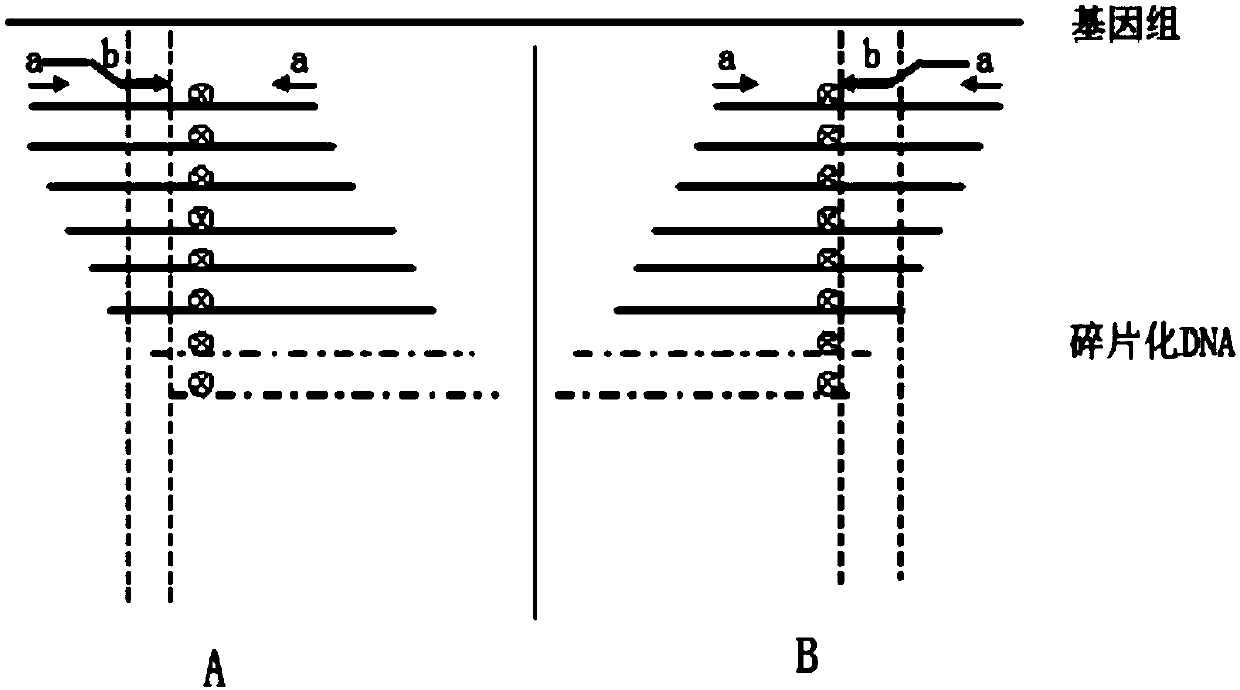
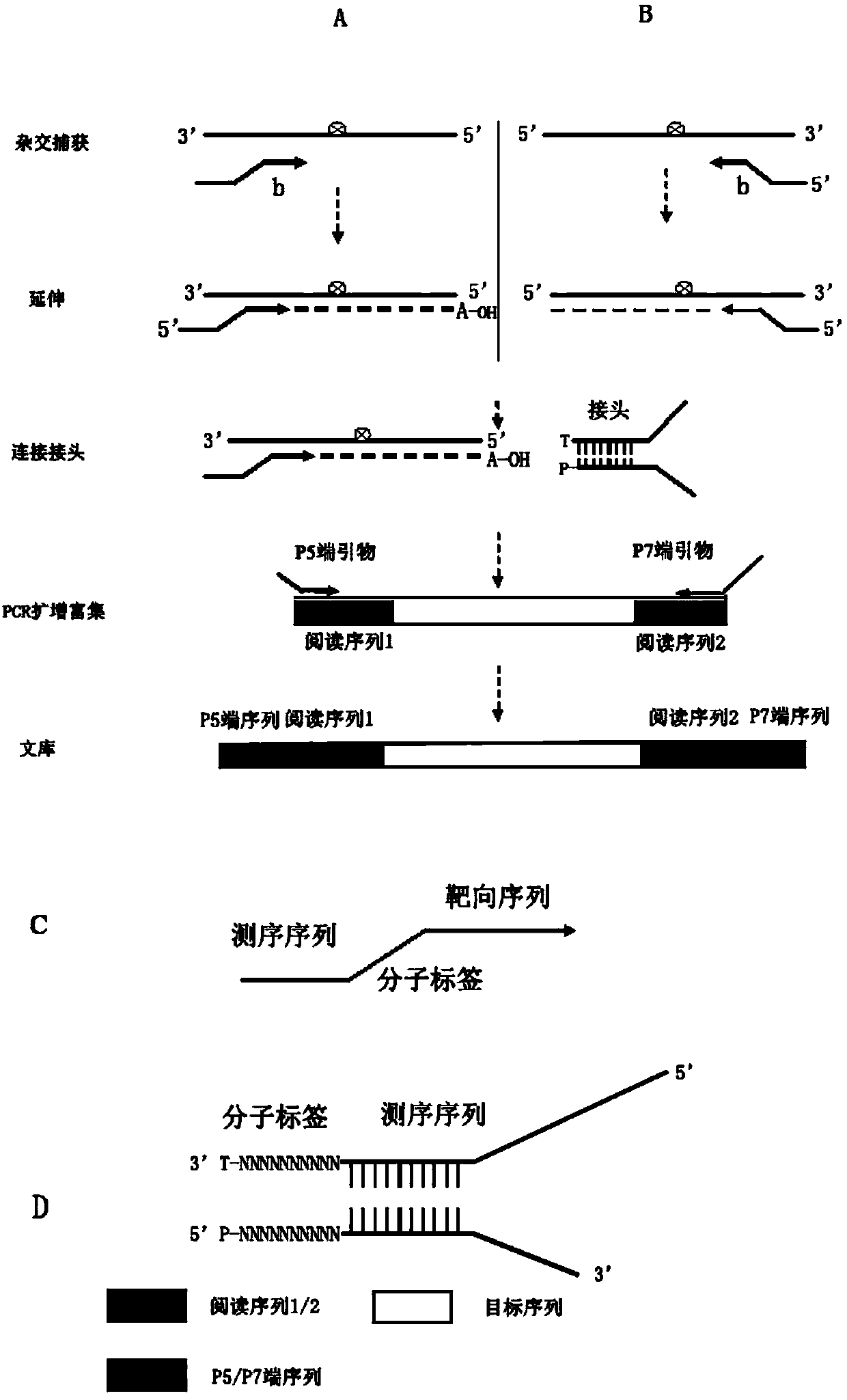
The invention belongs to the technical field of gene engineering, and particularly relates to a detection method of a low frequency mutated DNA fragment and a library building method. The technical problems that in the existing detection technique, the DNA fragment capturing efficiency is low, wrong mutation is introduced in the library building process, the detection limit is low, and the sensitivity is low are solved by adopting the hybrid capturing and molecular tagging technique and meanwhile combining with a special library building mode, the library specificity is improved, the false positive rate is lowered, and the template utilization rate is improved.
Owner:HUNAN YEARTH BIOTECHNOLOGICAL CO LTD
Rapid library building method for identifying target protein chromatin binding map
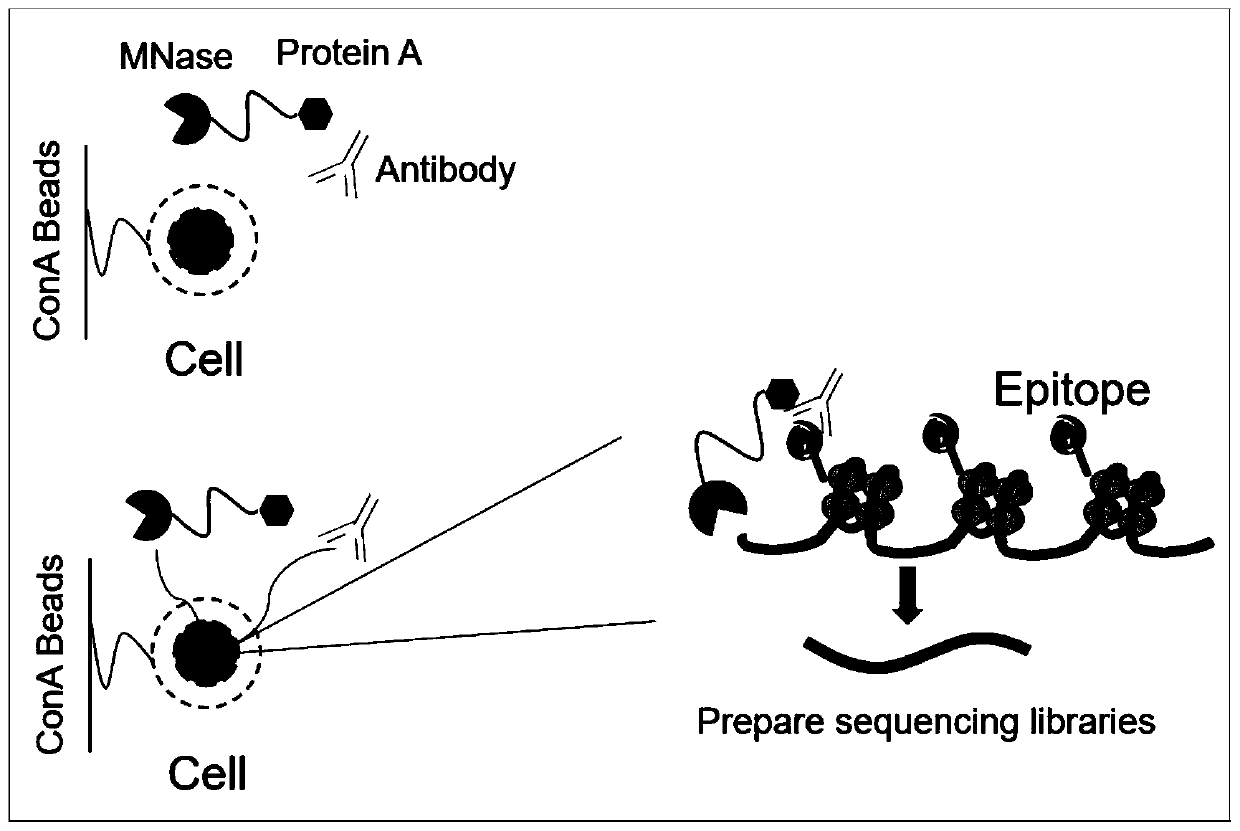
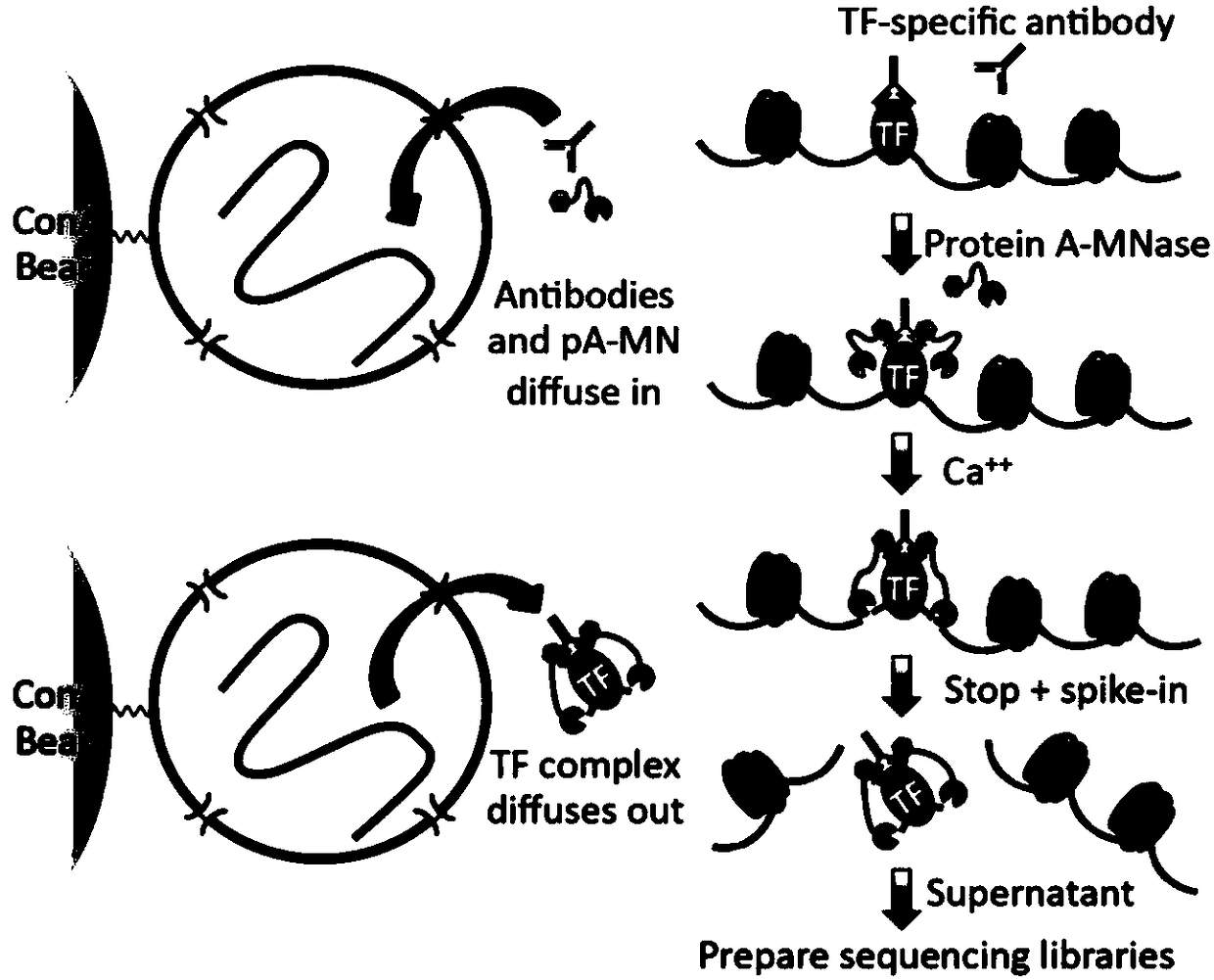
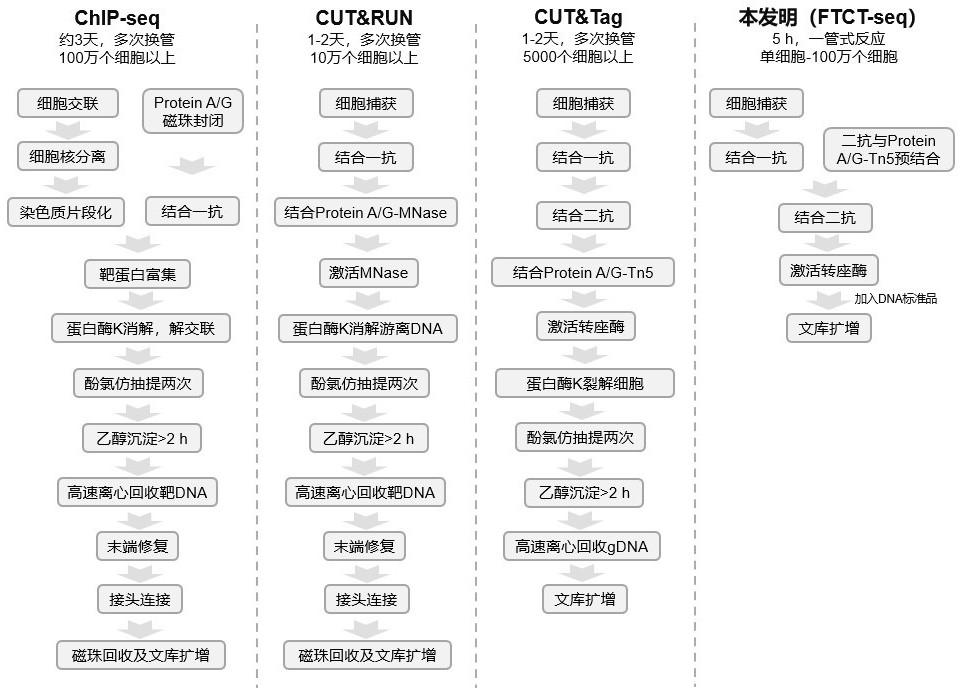
ActiveCN112553695ALower requirementReduce non-specificityLibrary creationProtein targetMagnetic bead
The invention discloses a rapid library building method for identifying a target protein chromatin binding map, which comprises the following steps: collecting sample cells, adding activated magneticbeads into the cells, incubating, and incubating the magnetic beads with a primary antibody; pre-binding a secondary antibody with recombinant fusion transposase; incubating the magnetic beads with the pre-combined secondary antibody and recombinant fusion transposase, adding a transposase activator and a cell perforating agent into the magnetic beads, and incubating; and terminating the transposase reaction, and carrying out library amplification. According to the method, the process of a traditional method is remarkably simplified, the library building time is shortened, the library buildingefficiency is improved, the loss is small, and the requirement for library building materials is lower. Meanwhile, three DNA standard substances with different concentrations are doped into the library building process, so that the effect of accurately quantifying the DNA copy number in the library can be achieved. The method is very suitable for researching a small amount of precious samples orsamples with different treatment states.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Method for standardized management of component packaging
ActiveCN110598263AEasy to operateImprove the efficiency of database constructionSpecial data processing applicationsWorking environmentSoftware engineering
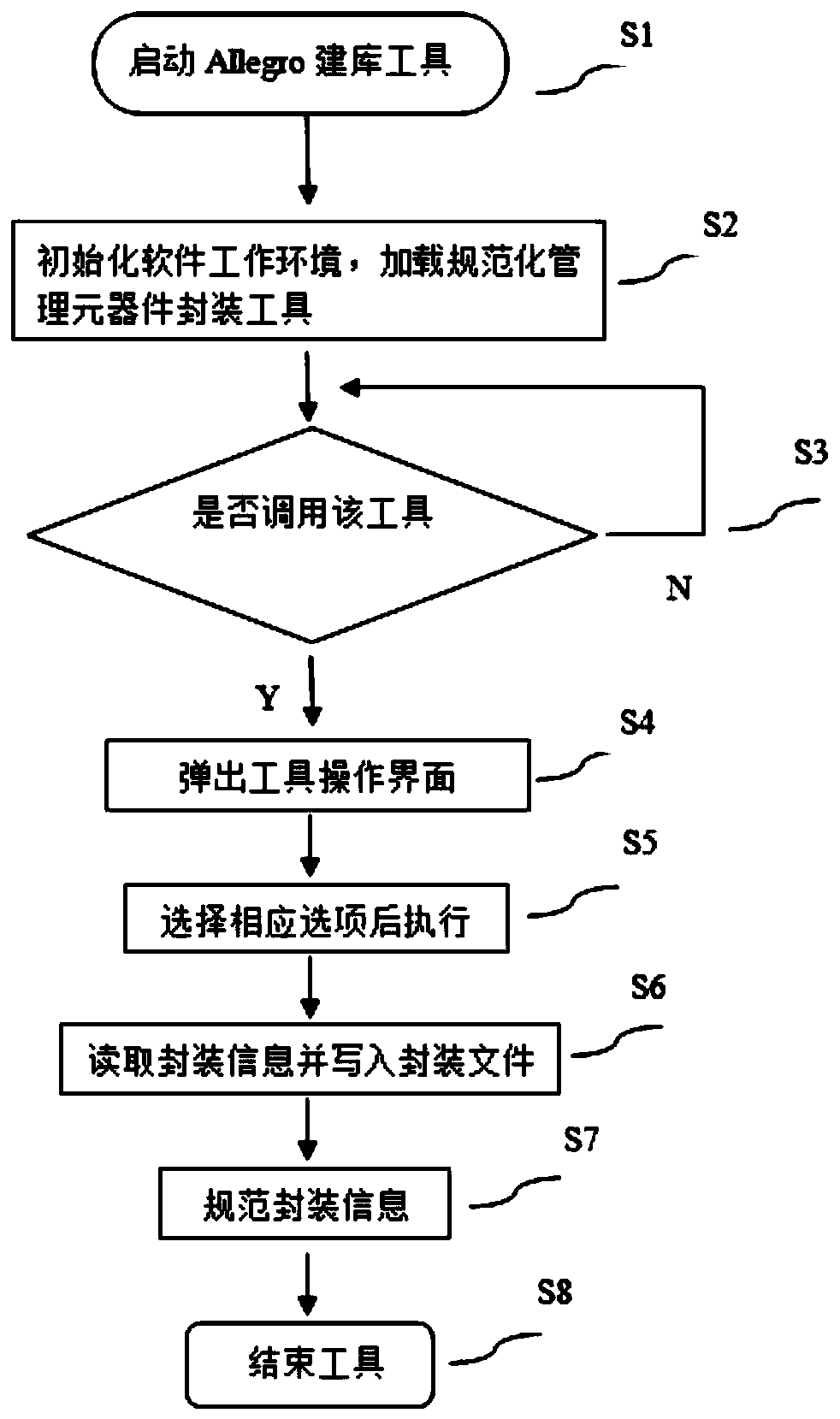
The invention discloses a method for standardized management of component packaging. The method comprises the following steps: S1, starting an Allegro library building tool; S2, initializing a software working environment, and configuring related parameters; S3, judging whether a packaging tool is called or not; S4, if the packaging tool is called, popping up a tool operation interface; S5, setting a packaging rule, selecting a corresponding option and then executing; S6, reading packaging information and writing the packaging information into a packaging file, acquiring the packaging information, and extracting and storing information required by specifications in a list by traversing packaged bonding pads, pins, symbols and packaging name information; and S7, standardizing the packaginginformation, and outputting a standardized packaging library in a PDF format. Due to the method, operation is simplified, and enterprises are facilitated to promote standardized management of component packaging; packaging information does not need to be queried manually, and library building efficiency is improved; packaging information does not need to be written manually, and library building efficiency is improved, and writing errors are reduced; and unified management and unified modification are facilitated.
Owner:SICHUAN JIUZHOU ELECTRIC GROUP
Construction method of cell Hi-C sequencing library
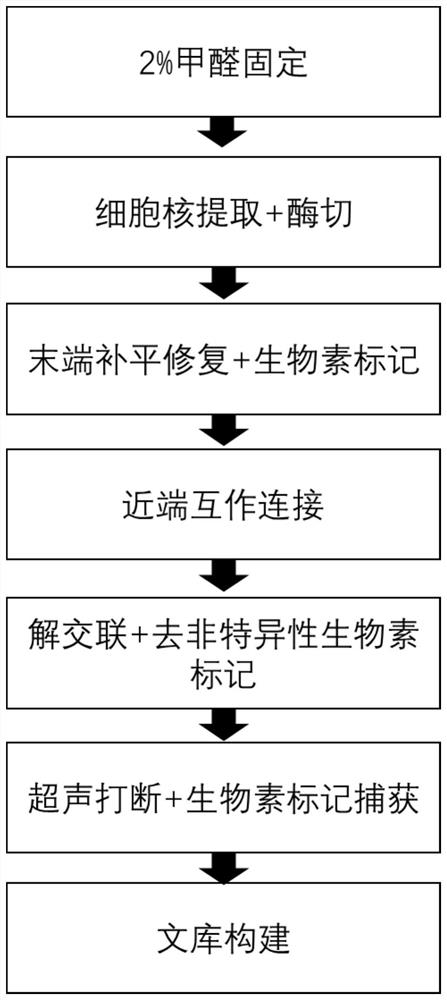
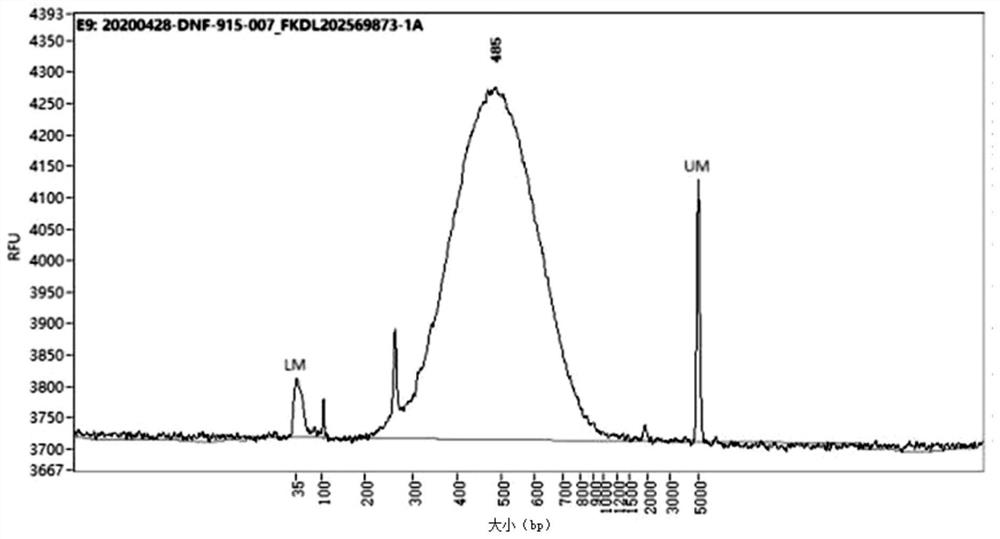
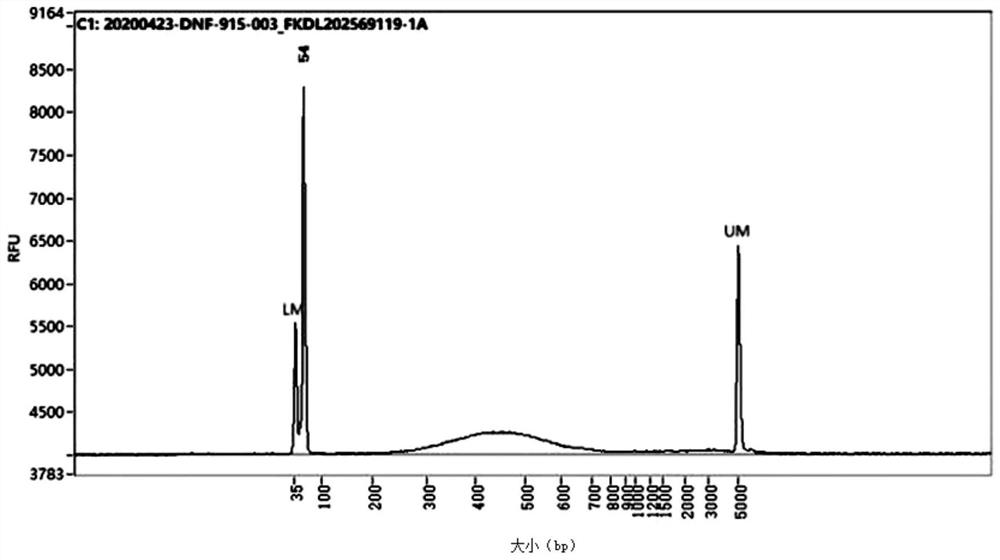
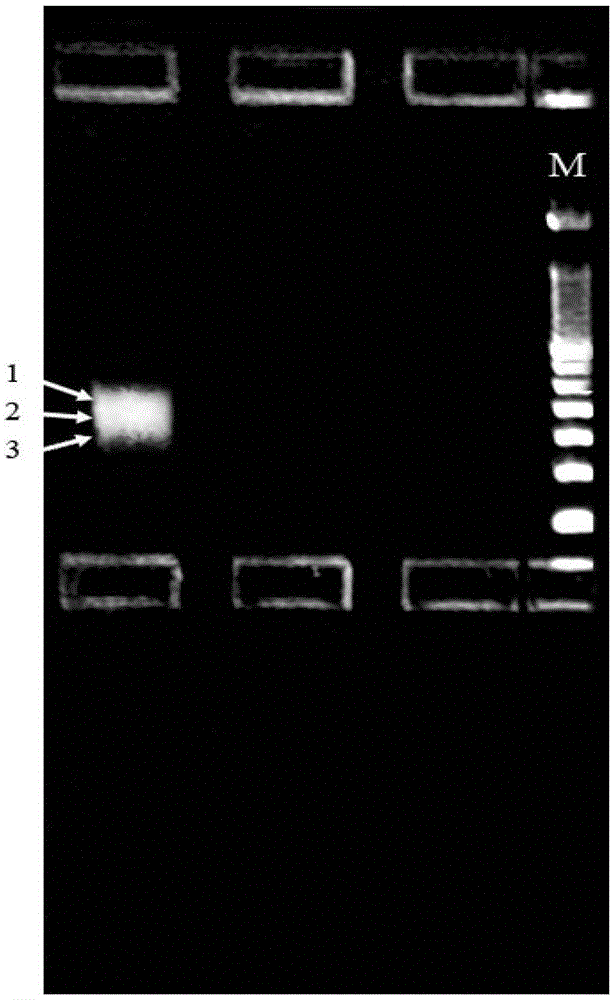
The invention provides a construction method of a cell Hi-C sequencing library. The construction method comprises the following steps: carrying out formaldehyde cross-linking immobilization on 10<5>-order cells by adopting a cross-linking system of which the volume is less than or equal to 800 muL and the final concentration of formaldehyde is 1.5-2.2%, and then treating the cross-linked cells byadopting an optimized cracking system and temperature to obtain a cell nucleus suspension; carrying out Hi-C library construction on the cell nucleus to obtain an Hi-C sequencing library. The cell membrane lysis solution comprises the following components with final concentrations: Tris-HCl with the pH value of 7.8 to 8.2 and the concentration of 9 to 11mM, NaCl with the concentration of 9 to 11mM, CA-630 with the concentration of 0.1 to 0.3 percent and a protease inhibitor with the concentration of 1*. A small-volume system fixes the initial quantity of low cells, optimizes a cell membrane lysis system and reaction conditions, ensures the integrity of cell nucleuses, and improves the quantity and purity of DNA of the cell nucleuses, thereby improving the library building efficiency (improving the proportion of effective data).
Owner:天津诺禾致源生物信息科技有限公司
Asymmetric high-throughput sequencing linkers capable of effectively improving library construction efficiency, and application of linkers
InactiveCN105154444AInefficient connectionReduce difficultyMicrobiological testing/measurementLibrary creationBinding siteDNA
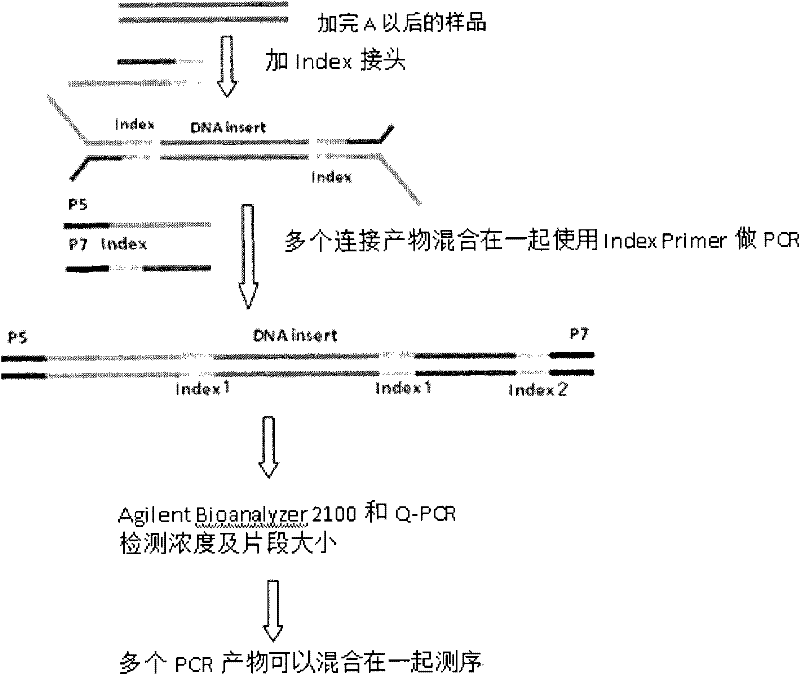
The invention relates to the field of high-throughput sequencing, and provides asymmetric high-throughput sequencing linkers for construction of an SOLiD sequencing system library, and a library construction method which is suitable for an SOLiD sequencing system and adopts the asymmetric high-throughput sequencing linker. The asymmetric high-throughput sequencing linkers are prepared by separately selecting one DNA single stranded from a P1 linker sequence and a P2 linker sequence which are inherent in the SOLiD sequencing system, and modifying and annealing the selected linkers. Asymmetric areas of the modified linkers contain binding sites of library amplification primers. The asymmetric high-throughput sequencing linkers provided by the invention are applied to the construction of the SOLiD sequencing system library, a phenomenon that two ends of one sequencing fragment are connected to the same linker during library construction in the prior art is avoided, and the library construction efficiency is effectively improved.
Owner:南京普东兴生物科技有限公司
Trace cell ChIP (Chromatin Immunoprecipitation) method
ActiveCN108315387AHigh catchImprove the efficiency of database constructionMicrobiological testing/measurementProtein targetEnzyme digestion
The invention relates to a trace cell ChIP (Chromatin Immunoprecipitation) method. The method comprises the following steps: 1) dividing a cell sample to be detected into n groups, wherein n is a natural number which is not equal to 0; 2) carrying out crosslinking fixation on the nth group sample; 3) treating the nth group sample by utilizing a cell membrane perforating agent; 4) carrying out enzyme digestion by utilizing Tn5 transposase and breaking a chromatin segment of the nth group sample, and connecting a barcode sequence and a primer sequence to two ends of a production segment DNA (Deoxyribonucleic Acid), wherein when the n is greater than or equal to 2, the barcode sequence and / or the primer sequence used in each group of step 4) are / is different; 5) when the n is greater than orequal to 2, combining samples of all groups; 6) enriching a DNA segment combined with target protein and taking a primer as an index for creating a database; sequencing and analyzing. When the methodis used for sequencing, the database creating efficiency is high; the capturing of transcription factor sites of an extremely trace amount of cells can be realized; relatively good efficiency is ensured and the stability and the accuracy are relatively good.
Owner:PEKING UNIV
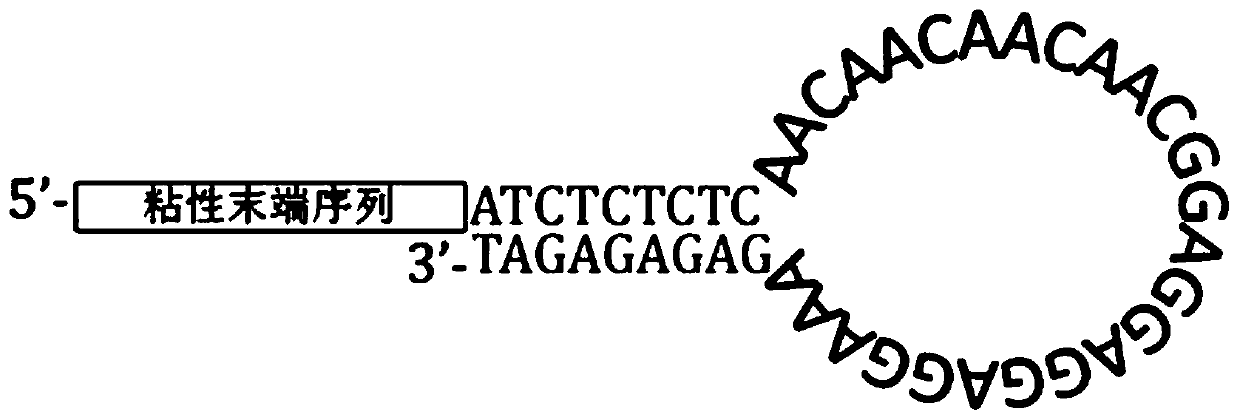
Molecular tag connector as well as preparation method and application thereof
PendingCN107988320AIncrease varietyLower ego connectionMicrobiological testing/measurementDNA/RNA fragmentationDNAGene mutation
The invention discloses a molecular tag connector as well as a preparation method and application thereof. According to the molecular tag connector, a nucleic acid molecule (molecular tag connector) (also named as nucleic acid molecule A) is firstly protected, which is as shown in sequence 5 or sequence 6 or sequence 7 or sequence 8 of a sequence list. The invention also protects a nucleic acid molecule mixture (also named as nucleic acid molecule A mixture) which consists of a nucleic acid molecule shown by sequence 5 of the sequence list, the nucleic acid molecule shown by sequence 6 of thesequence list, the nucleic acid molecule shown by sequence 7 of the sequence list and the nucleic acid molecule shown by sequence 8 of the sequence list. The molecular tag connector has great application and popularization value for detecting ultralow-frequency gene mutation. The mutation of the ultralow-frequency gene is generally ctDNA in total DNA of peripheral blood, so that by utilizing the molecular tag connector, the diagnosis can be more effectively assisted or the drug application can be more effectively instructed.
Owner:SHANGHAI ORIGIMED CO LTD
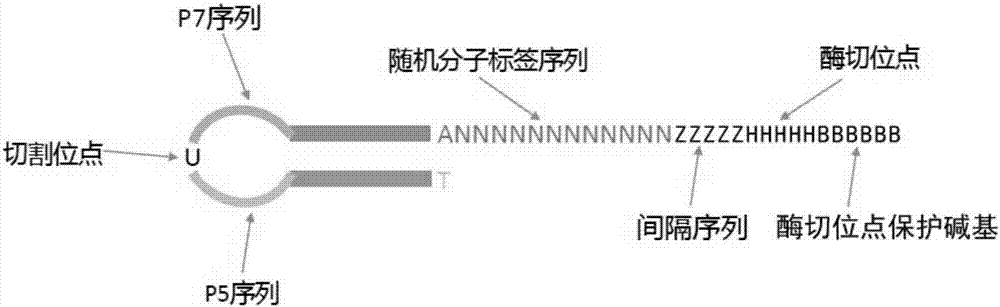
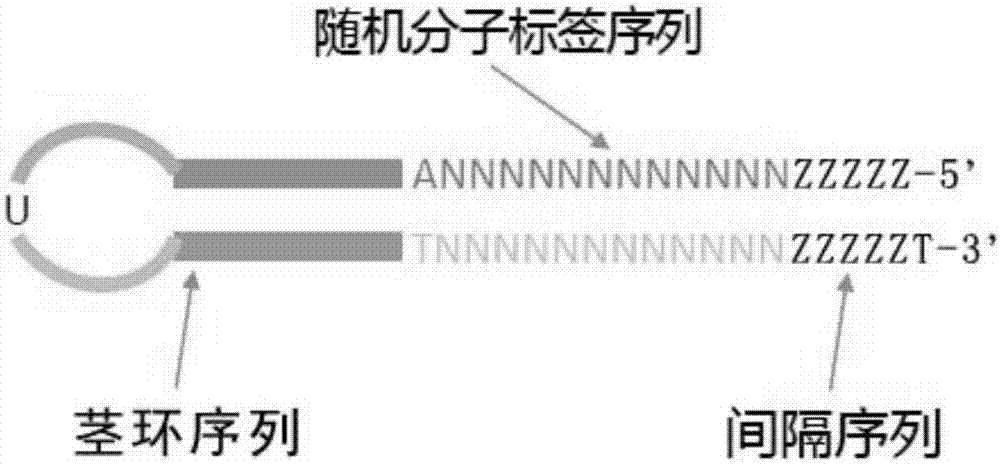
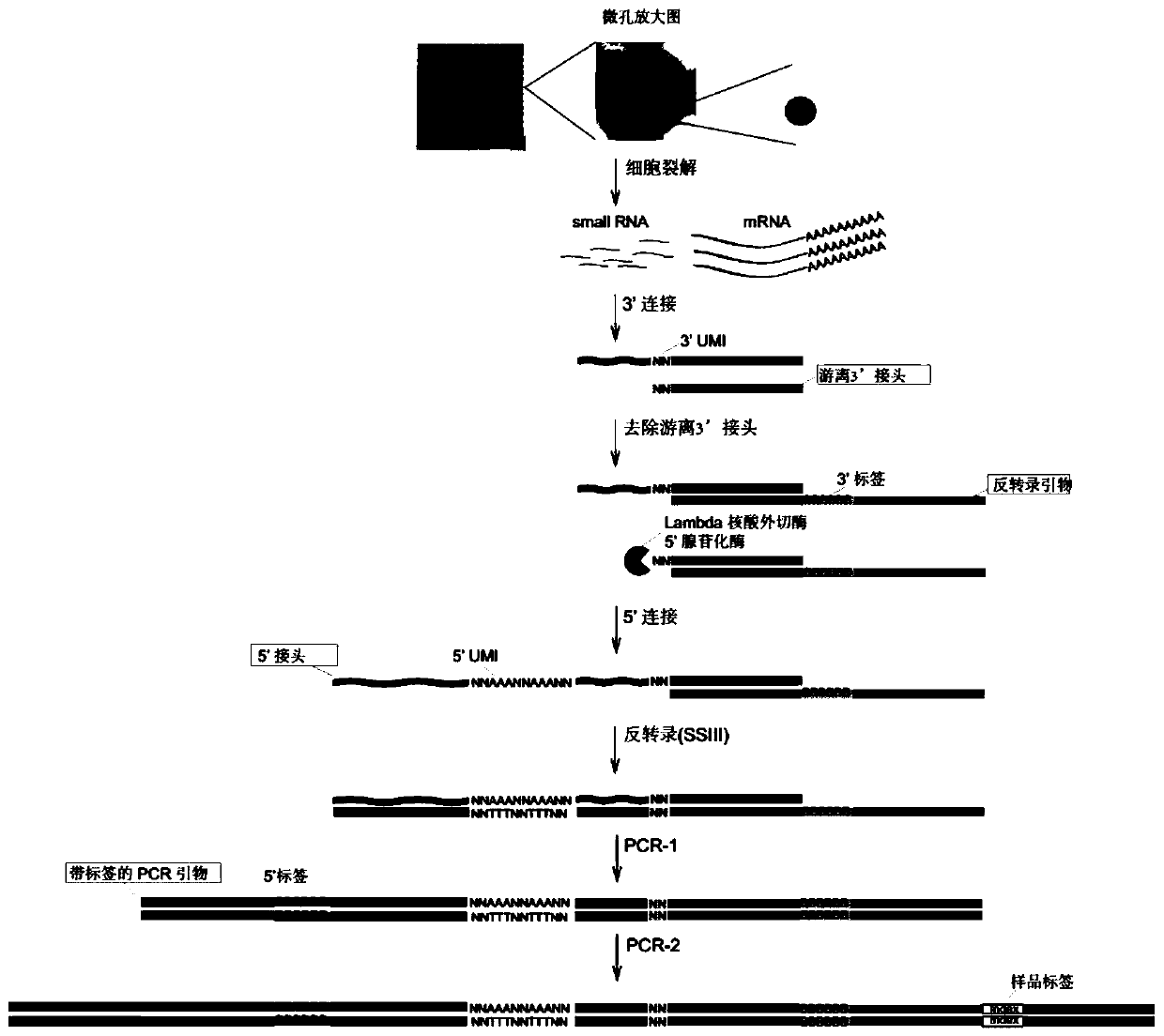
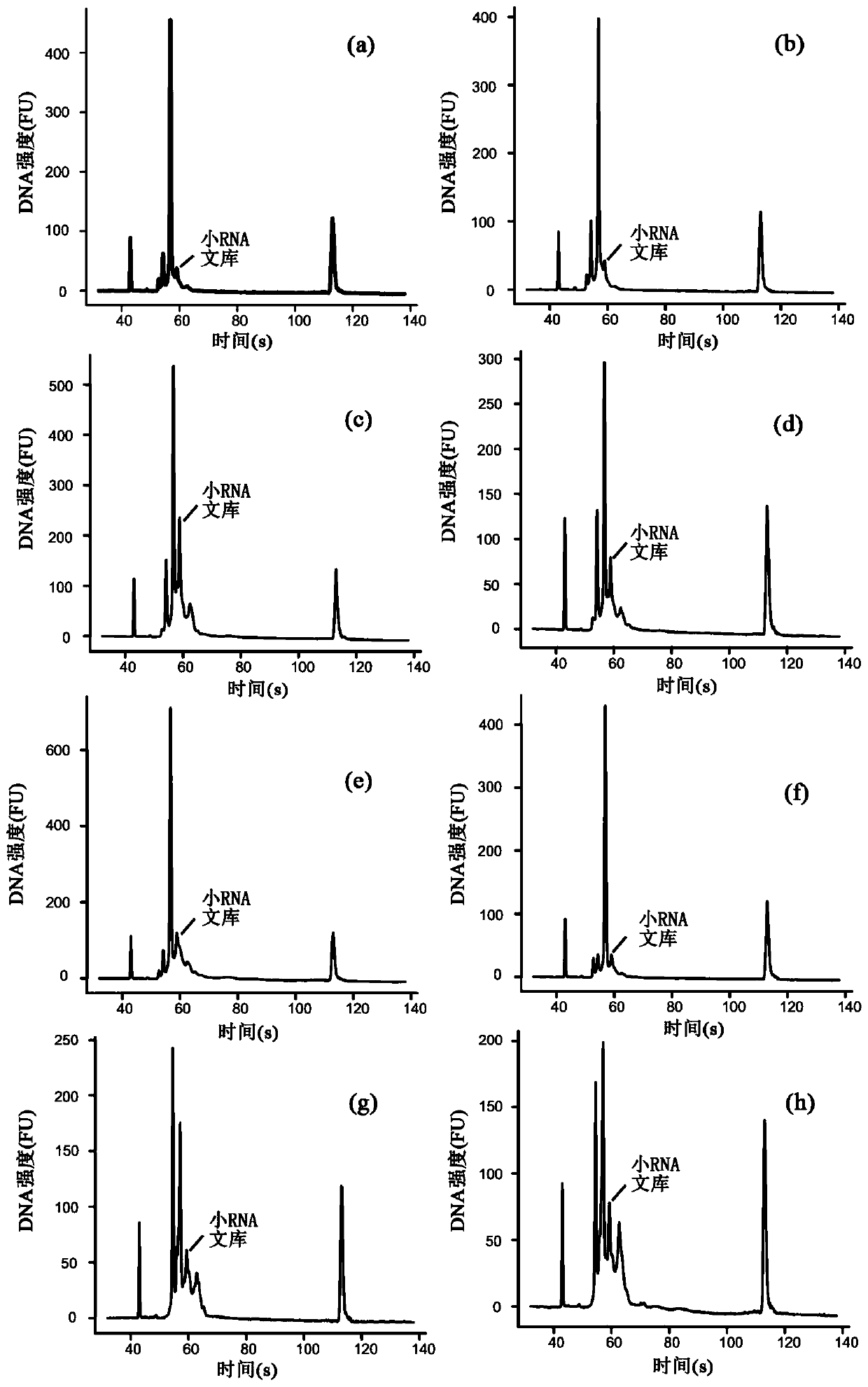
Method for establishing high-throughput single-cell small RNA (ribonucleic acid) library
ActiveCN110747514AImprove accuracyHigh sensitivityNucleotide librariesLibrary creationLysisNucleotide
The invention provides a method for establishing a high-throughput single-cell small RNA (ribonucleic acid) library. The method comprises the following steps: preparing a single-cell suspension, adding the single-cell suspension to micropores of a chip of a single-cell operating system, and selecting micropores of single living cells for experiment; performing a cell lysis reaction, a 3'-end connection reaction, a free junction removal reaction, a 5'-end connection reaction, a reverse transcription reaction and two PCR (polymerase chain reaction) reactions successively; and performing productpurification and fragment screening recycling, so as to obtain a single-cell small RNA library, wherein the nucleotide sequence of 3'-junction used in the 3'-end connection reaction is shown in SEQ IDNO: 1 in the description; and the nucleotide sequence of 5'-junction used in the 5'-end connection reaction is shown in SEQ ID NO: 2 in the description. The method provided by the invention has the advantages of high accuracy, high sensitivity and good repeatability.
Owner:NAT INST OF BIOLOGICAL SCI BEIJING
Rapid full-length amplicon library building method suitable for PacBio platform, universal primer and sequencing method
PendingCN111455036ASimplify the build processImprove the efficiency of database constructionMicrobiological testing/measurementLibrary creationEnzyme digestionMolecular biology
The invention relates to a rapid full-length amplicon library building method suitable for a PacBio platform, a universal primer and a full-length amplicon sequencing method based on the library building method. According to the library building method, aiming at the characteristic that full-length amplicon sequencing needs PCR, in combination with a dumbbell type library joint structure of the PacBio sequencing platform, the universal primer with a special structure aiming at a full-length amplicon is designed, and a PCR product with a specific sticky end is obtained through an amplificationreaction, and then sequencing joints with same sticky ends are connected to finish library construction of the PacBio sequencing platform. According to the method, an original PacBio standard libraryconstruction process is optimized to be only one-step amplification and enzyme digestion connection, so that the library construction process of the full-length amplicon is greatly simplified, the working efficiency is improved, and the sequencing cost is reduced.
Owner:WUHAN FRASERGEN CO LTD
Bladder cancer mutation gene specific primer, kit and library construction method
ActiveCN110468211AReduce lossIncrease profitMicrobiological testing/measurementLibrary creationBladder cancerWilms' tumor
The invention discloses a bladder cancer mutation gene specific primer, kit and library construction method. A mode of unilateral enrichment-single molecule specific primer (single primer) is used foramplifying sub enrichment technologies for the first time; the application scope of the primer is expanded, the use amount of an effective template is increased, especially the detection limit for detecting low-frequency mutation samples is greatly improved; and secondly, a single molecule specific primer is composed of specific sequences and sequencing sequences, the specificity and the primer use ratio of multiple PCR can be effectively provided. The bladder cancer mutation gene specific primer, kit and library construction method applies the technology to the field of bladder cancer tumormutation gene detection, prepares corresponding reagents, can detect the mutation condition of tumor genes related bladder cancer efficiently and sensitively, and provides guidance for clinical application.
Owner:HUNAN YEARTH BIOTECHNOLOGICAL CO LTD
Rapid full-length amplicon library construction method, primers and sequencing method applicable to PacBio platform
InactiveCN111455023ASimplify the build processImprove work efficiencyNucleotide librariesMicrobiological testing/measurementGeneticsEngineering
The invention relates to a rapid full-length amplicon library construction method and primers applicable to PacBio platform and a full-length amplicon sequencing method based on the library construction method. According to the characteristic that PCR is required for full-length amplicon sequencing, an amplicon primer with a general binding sequence and a dumbbell-shaped primer matched with the general binding sequence are designed in combination with a dumbbell-shaped library joint structure of the PacBio sequencing platform, the rapid full-length amplicon library construction method applicable to the PacBio platform is provided based on the two primers with special structures, through library construction by the method, addition of sequencing joints can be realized by increasing two cycles of annular primer amplification only after conventional amplification, and meanwhile, a PCR product becomes a standard library applicable to the PacBio sequencing platform under the action of damage repair enzyme. The method optimizes an original PacBio standard library construction process into only one-step amplification and repair, the library construction process of a full-length amplicon is greatly simplified, and the sequencing cost is significantly reduced.
Owner:WUHAN FRASERGEN CO LTD
Single-stranded rapid library building method and library building instrument
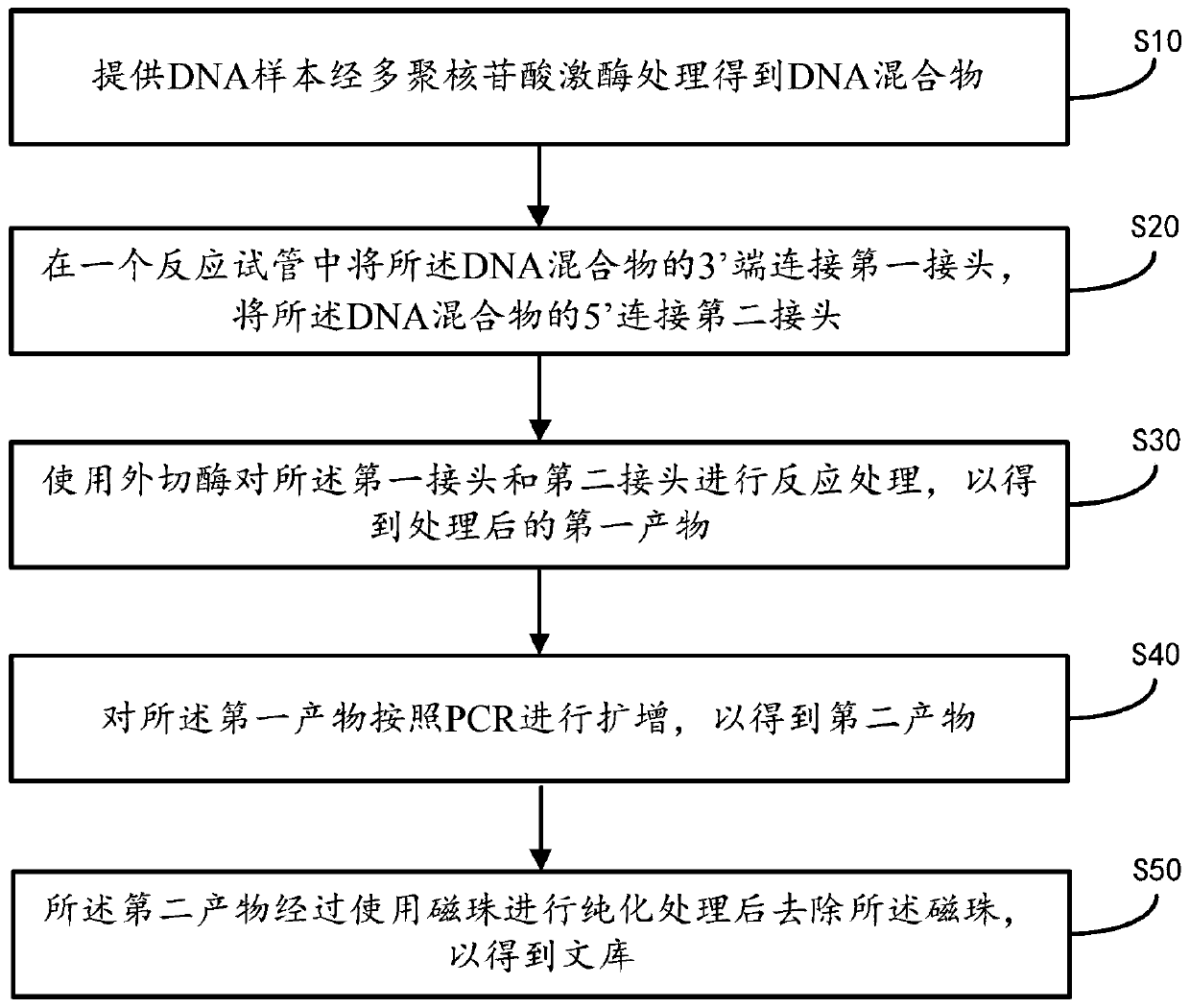
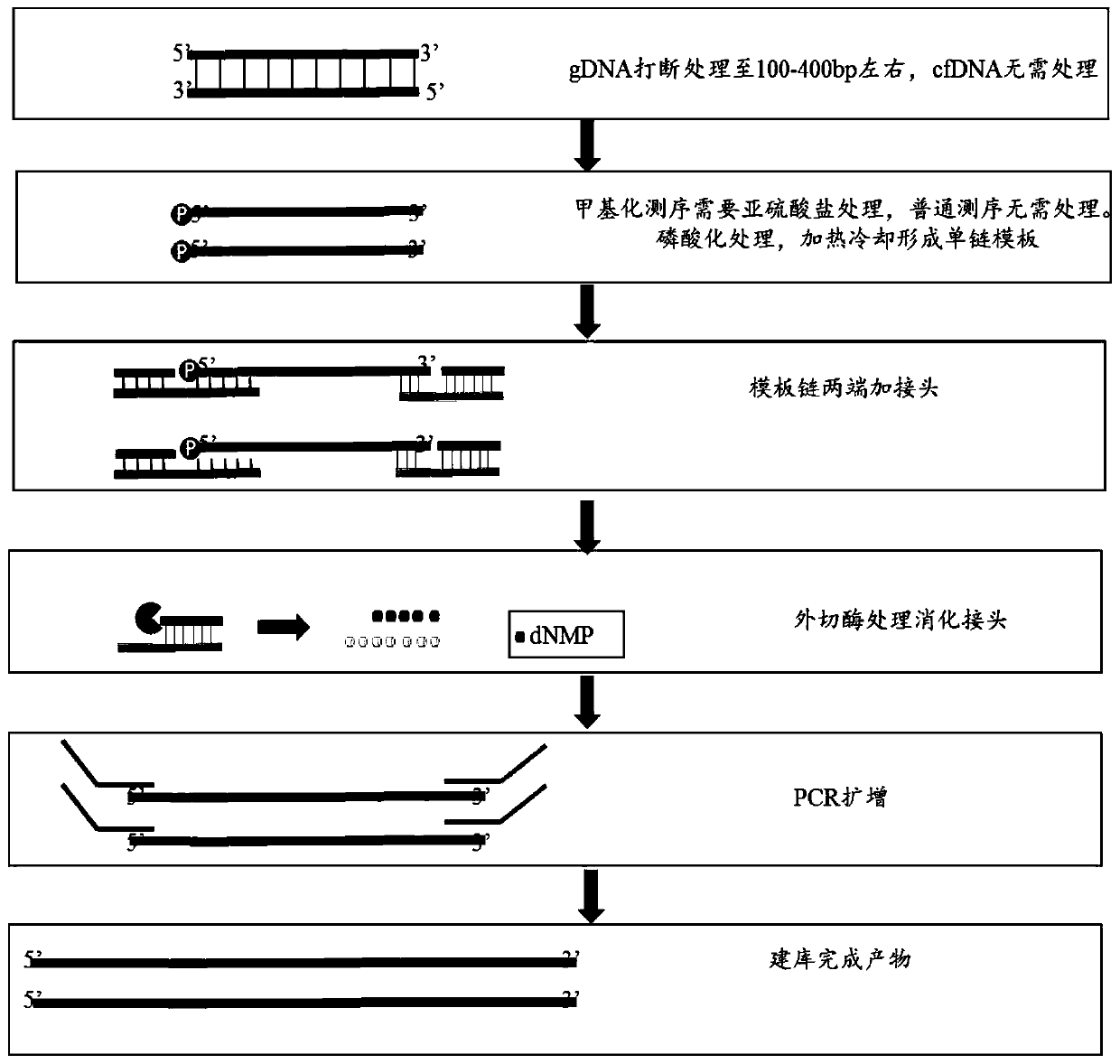
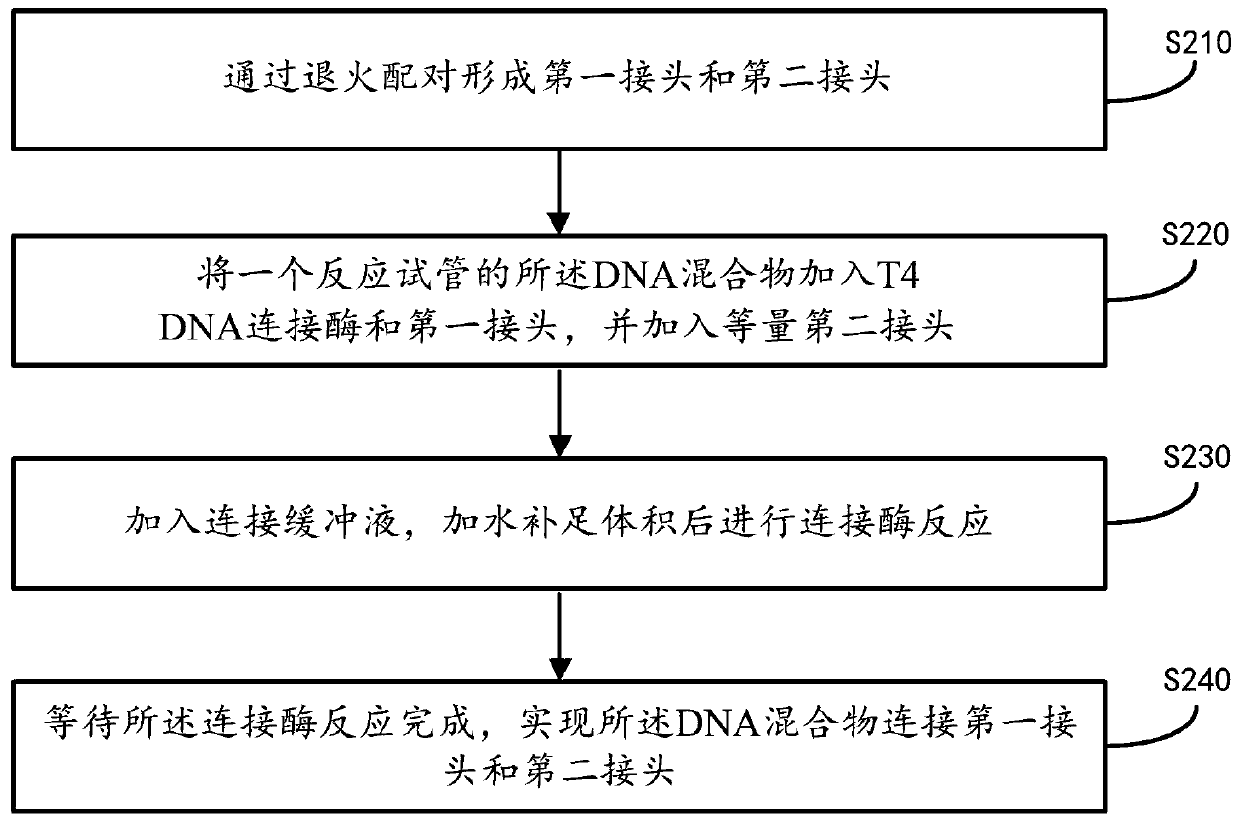
ActiveCN111455469AReduce formationShort reaction timeLibrary creationLibraries apparatusMagnetic beadEngineering
The invention provides a single-stranded rapid library building method. The method comprises the following steps: providing a DNA sample, and treating the DNA sample with polynucleotide kinase to obtain a DNA mixture; connecting the 3' end of the DNA mixture with a first linker in a reaction test tube, and connecting the 5' end of the DNA mixture with a second linker; carrying out reaction treatment on the first linker and the second linker by using excision enzyme to obtain a treated first product; amplifying the first product according to PCR to obtain a second product; and purifying the second product by using magnetic beads, and removing the magnetic beads to obtain a library. In addition, the invention further provides a library building instrument applying the method. According to the single-stranded rapid library building method provided by the invention, library building reaction and amplification reaction are completed in the same reaction test tube, and reaction time is saved; and connection of the linkers at the two ends is completed through a one-time connection reaction, so operation steps of single-stranded library building are reduced, operation difficulty is reduced, library building time is shortened, and library building efficiency is further improved.
Owner:深圳易倍科华生物科技有限公司
Establishment method, establishment device and establishment system of intelligent question answering knowledge base
ActiveCN105608218BImprove build efficiencyImprove accuracySpecial data processing applicationsTheoretical computer scienceQuestions and answers
The invention relates to an intelligent question answering knowledge base establishment method, establishment device and establishment system. The establishment method comprises the following steps: providing a domain knowledge data base, wherein the domain knowledge data base comprises a plurality of pieces of preset knowledge; receiving initial request information; performing semantic similarity calculation on the initial request information and the preset knowledge in the domain knowledge data base, and judging whether the maximum value of the semantic similarity calculation results is larger than a similarity threshold or not; if the maximum value of the semantic similarity calculation results is larger than the similarity threshold, storing a standard question and an extended question in the preset knowledge corresponding to the initial request information and the maximum value of the semantic similarity calculation results in the intelligent question answering knowledge base; if the maximum value of the semantic similarity calculation results is smaller than the similarity threshold, carrying out the abstract semantic recommendation process so as to acquire one or more specific semantic expressions corresponding to the initial request information, and storing the initial request information and the specific semantic expressions in the intelligent question answering knowledge base. The establishment method provided by the invention can improve the establishment efficiency of the intelligent question answering knowledge base.
Owner:SHANGHAI XIAOI ROBOT TECH CO LTD
Multi-amplification database building method for trace DNA and special kit
ActiveCN108929901AGood uniformity of amplificationEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationMutation detectionCommercial kit
The invention discloses a multi-amplification database building method for trace DNA and a special kit. The multi-amplification database building method can realize multiple amplification sequencing with trace DNA or even single-cell-level DNA as a template, and the whole reaction process can be realized in a single tube. Experiments prove that the database building method achieves 207-fold one-tube amplifier amplification at the single cell level, has the fidelity higher than that of conventional commercial kits, not only has the advantages of convenient operation, low cost, high sequencing quality and high sensitivity, but also can meet the sequencing requirements of two sequencing platforms, and is suitable for high-throughput and low-frequency mutation detection fields such as the ctDNA mutation detection field and the low-initial-quantity DNA embryonic mutation detection field such as single cell mutation detection and screening.
Owner:CAPITALBIO CORP
Kit for DNA library building and application of kit
ActiveCN108660135AReduce usageReduce lossesMicrobiological testing/measurementLibrary creationBlood plasmaKlenow fragment
The invention discloses a kit for DNA library building and an application of the kit, and in particular relates to a kit for peripheral blood cfDNA library building and an application of the kit. According to the invention, the kit for DNA library building is firstly provided, wherein the kit comprises a component A and a component B; the component A consists of the following ingredients at a ratio as follows: 1-3U of T4 PNK to 1-4.5U of Klenow Fragment to 0.25-0.5U of LA Taq DNA Polymerase to 0.3-0.9U of T4 DNA Polymerase to 0.5-2.5 nmol of dNTP to 0.5-2.5nmol of dATP; and the component B consists of the following ingredients at a ratio as follows: 2.5-8nmol of ATP to 26.5 132U of T4 DNA Ligase to 0.213 0.534[mu]L of PEG 8000. According to the kit and the application method provided by the invention, DNA loss in a library building process can be reduced to the greatest extent, the usage amount of plasma can be reduced, library building cost can be reduced, library building time can beshortened, operations can be simplified and more stable and reliable results can be obtained. The kit provided by the invention is significant for improving library building efficiency and reducing labor cost.
Owner:TIANJIN MEDICAL LAB BGI +2
DNase I mutants as well as coding nucleotide sequences and application thereof
ActiveCN113215132AEfficient and stable interruptionThe quality of library construction and sequencing is excellentHydrolasesGenetic engineeringNucleotideWild type
According to the invention, wild type DNase I related sites are mutated and screened to obtain three mutants with good effects, wherein two mutants are in double-site mutation, and mutation sites are respectively A249T, R42H and A249T, F110V; one mutant is in three-site mutation, and the mutation sites are A249T, R42H and F110V. The DNase I mutant with the three-site mutation has the best effect, and the DNase I mutant has an amino acid sequence shown in SEQ ID No.1 and a coding nucleotide sequence shown in SEQ ID No.2. The invention also discloses application of the DNase I mutants in construction of a high-throughput sequencing library. The DNase I mutants disclosed by the invention can efficiently and stably break genome DNA, fragmented products of the DNase I mutants can be applied to construction of a high-throughput sequencing library, and compared with wild type DNase I, library construction and sequencing quality is more excellent, and GC preference is lower. Low-cost and high-efficiency library building can be realized, and sequencing quality is excellent.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
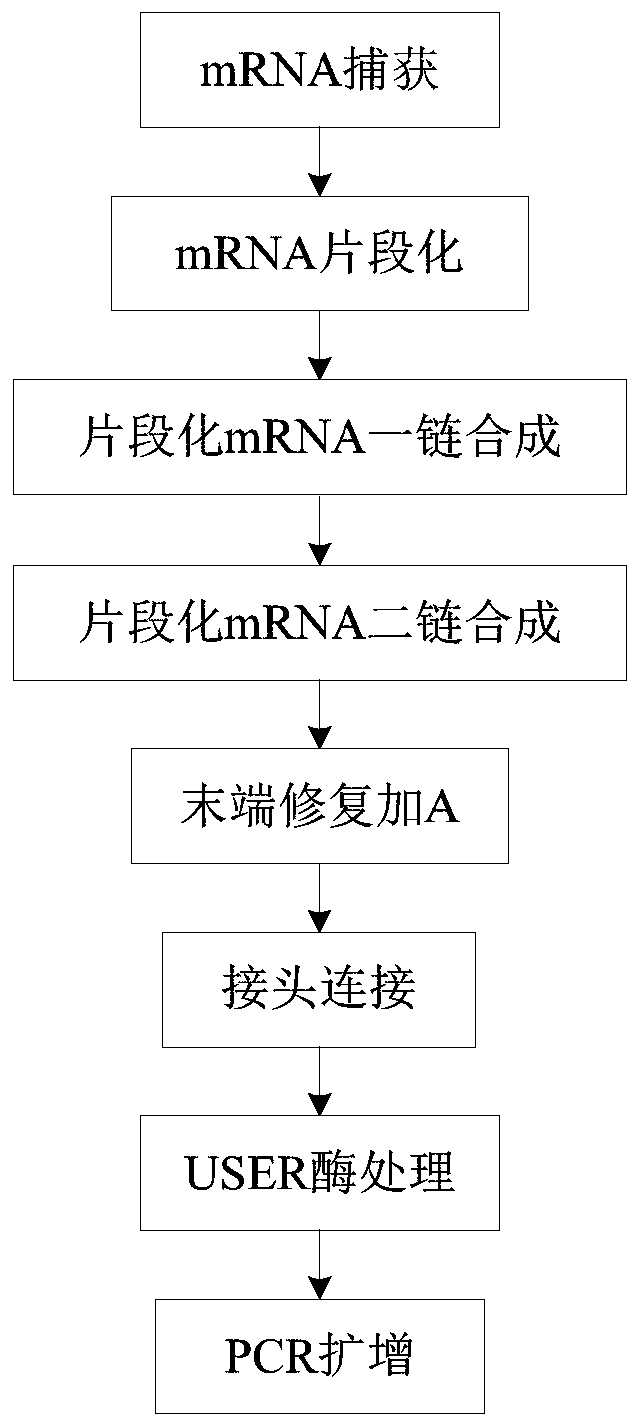
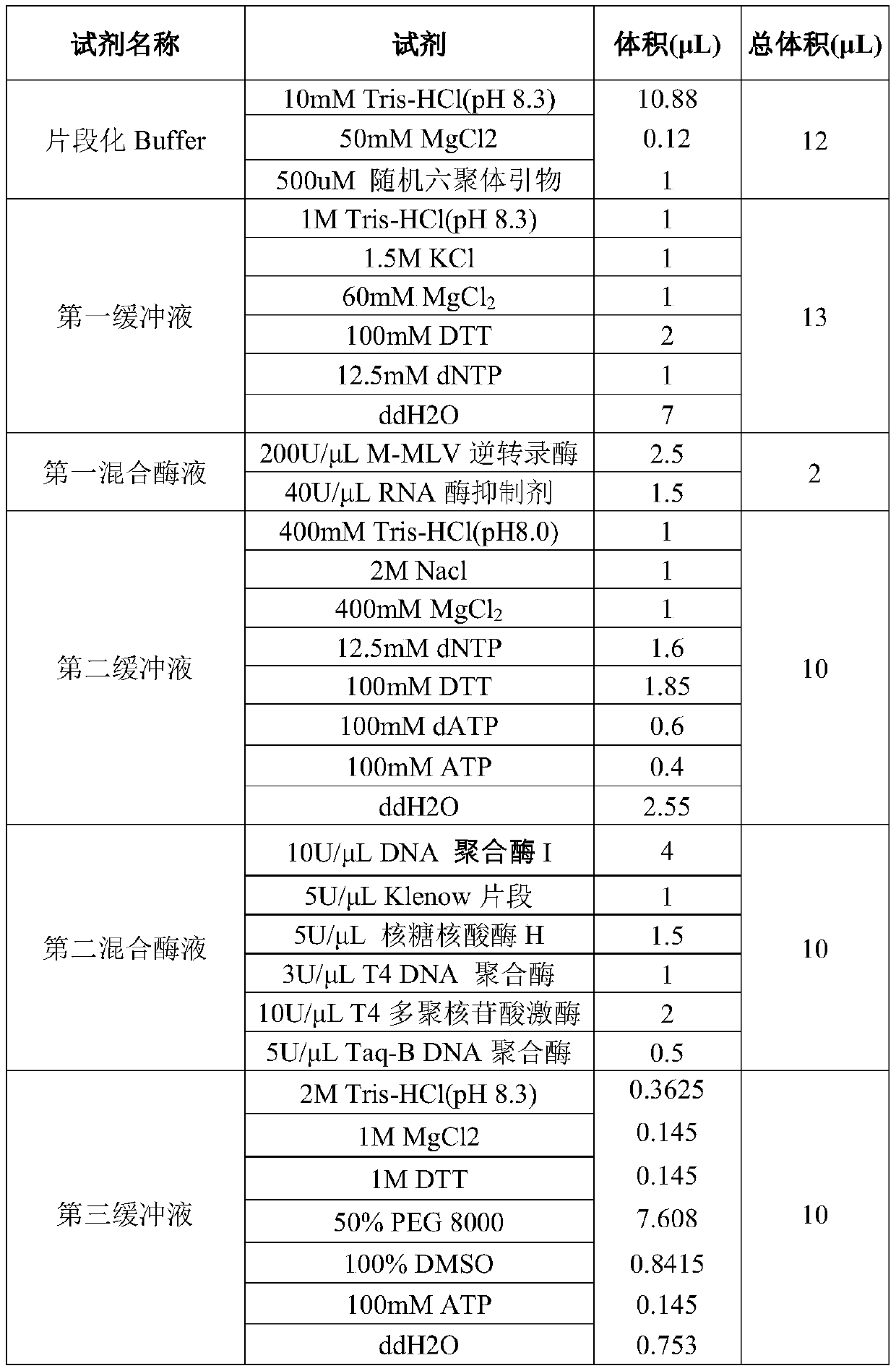
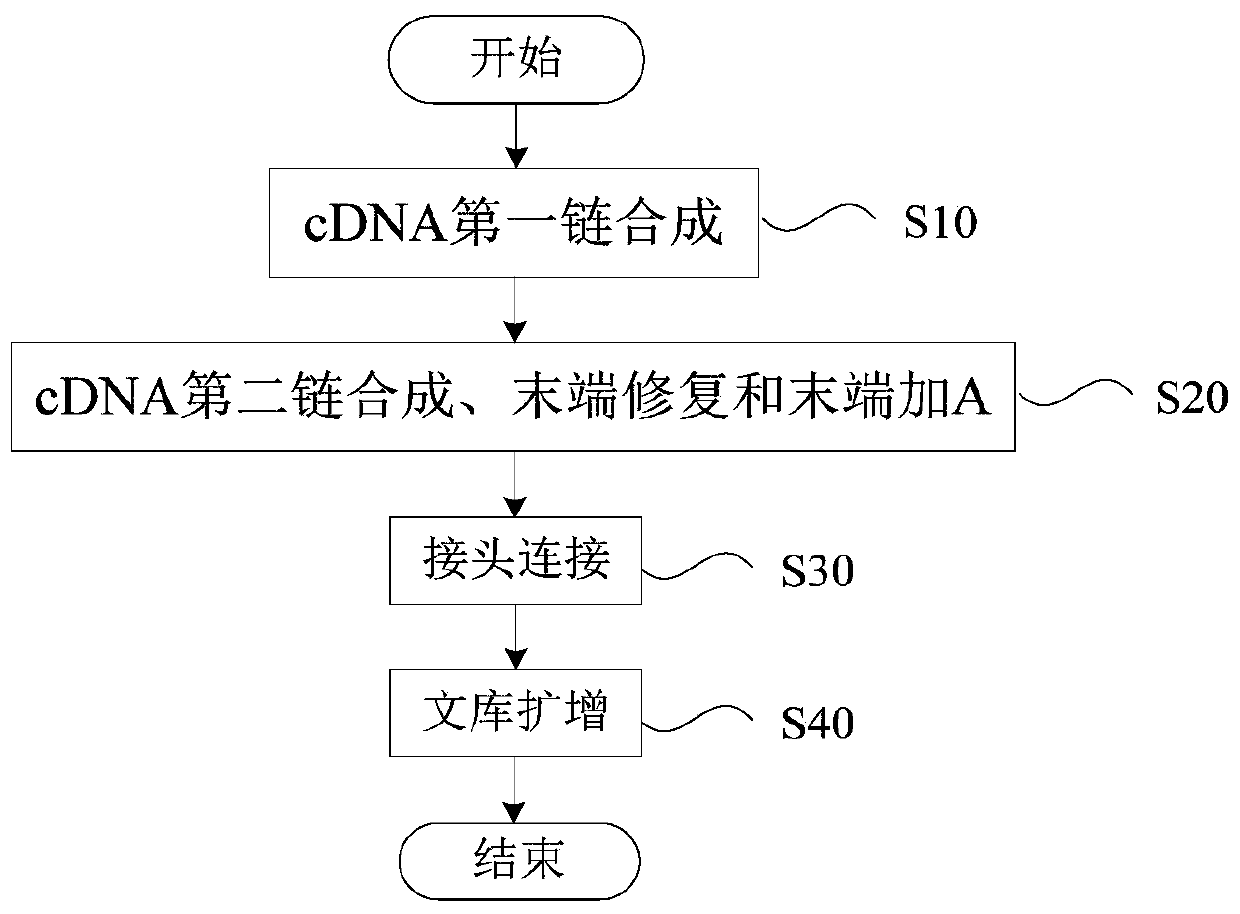
MRNA library construction method and kit
ActiveCN110846383AImprove the efficiency of database constructionMicrobiological testing/measurementLibrary creationPhysical chemistryProcess engineering
The invention provides an mRNA library construction method. The mRNA library construction method includes mixing an mRNA fragment with a first mixed enzyme solution and a first buffer solution to obtain a first reaction system, and subjecting the first reaction system to first heat treatment to obtain a first reaction solution; mixing the first reaction solution with a second mixed enzyme solutionand a second buffer solution to obtain a second reaction system, and subjecting the second reaction system to second heat treatment to obtain a second reaction solution; mixing the second reaction solution with T4DNA ligase, a third buffer solution and a Y-shaped joint to obtain a third reaction system, subjecting the third reaction system to third heat treatment, and after the third heat treatment, performing purification to obtain a third reaction solution; adding a pre-mixed solution and a amplification primer into the third reaction solution to obtain a fourth reaction system, subjectingthe fourth reaction system to fourth heat treatment, and after the fourth heat treatment, performing purification to obtain an mRNA library. The mRNA library construction method and a kit, which are beneficial to improvement of the library construction efficiency, can be provided.
Owner:江西海普洛斯医学检验实验室有限公司
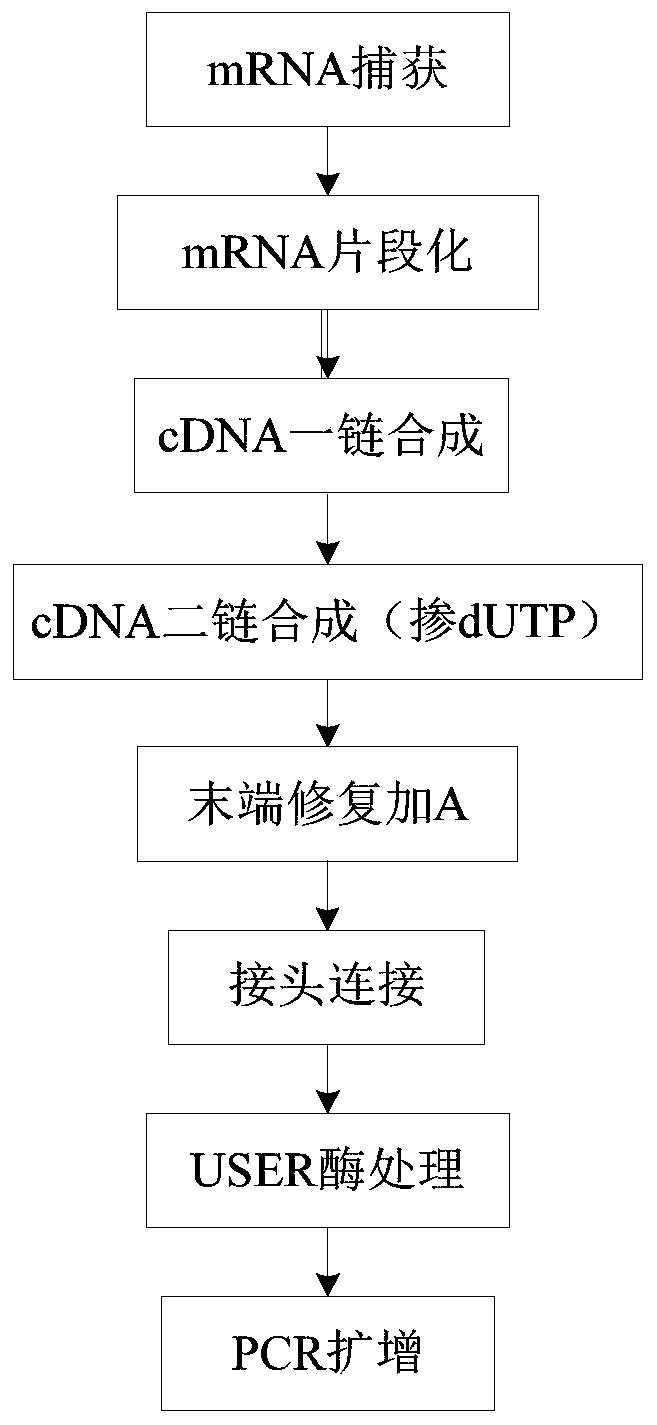
Method and kit for constructing mRNA chain specific library
The invention provides a method for constructing an mRNA chain specific library, and the method comprises the following steps: mixing an mRNA fragment, actinomycin D, a first mixed enzyme solution anda first buffer solution to form a first reaction system, and carrying out first heat treatment on the first reaction system to obtain a first reaction solution; mixing the first reaction solution with a second mixed enzyme solution and a second buffer solution to form a second reaction system, and carrying out second heat treatment on the second reaction system to obtain a second reaction solution; mixing the second reaction solution with T4DNA ligase, a third buffer solution and a Y-shaped linker to form a third reaction system, carrying out third heat treatment on the third reaction system,and purifying after the third heat treatment to obtain a third reaction solution; and adding a premixed solution, an amplification primer and UDG enzyme into the third reaction solution, mixing to form a fourth reaction system, carrying out fourth heat treatment on the fourth reaction system, and purifying after the fourth heat treatment to obtain the mRNA chain specific library. According to thepresent invention, the method and the kit for mRNA chain specific library construction can be provided, wherein the mRNA chain specific library construction method and the kit can easily improve thelibrary construction efficiency.
Owner:江西海普洛斯医学检验实验室有限公司
Method for rapid construction of RNA 3' terminal gene expression library
InactiveCN110257479AClear technical routeImprove the efficiency of database constructionMicrobiological testing/measurementLibrary creationDouble strandedTransposase
The invention discloses a method for rapid construction of a RNA 3' terminal gene expression library. The method includes: subjecting an mRNA sample to reverse transcription by adopting an Oligo (dT) reverse transcription primer with a first joint to obtain a first chain cDNA with the first joint; synthesizing a second chain cDNA by taking the first chain cDNA as a template to obtain a double-stranded cDNA with the first joint; processing the double-stranded cDNA by adopting a transposase compound with a second joint, and inserting the second joint during fragmentation to obtain a double-stranded cDNA fragment with the first joint and / or the second joint, wherein sequences of the first joint and the second joint are not the same. By adoption of the method, construction of a transcriptome library aiming at eukaryotic RNA 3' terminals can be realized effectively.
Owner:BEIJING TRANSGEN BIOTECH CO LTD
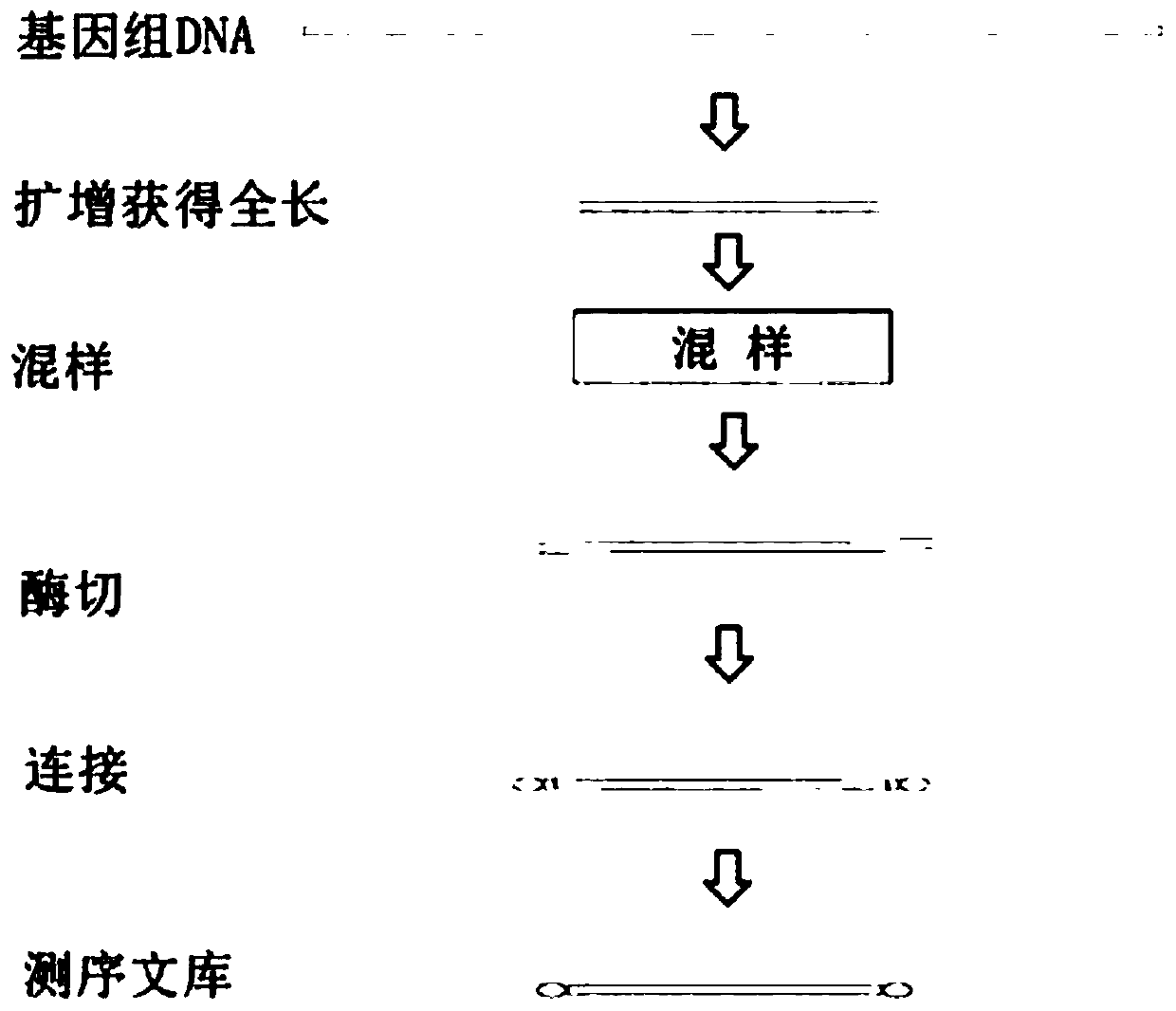
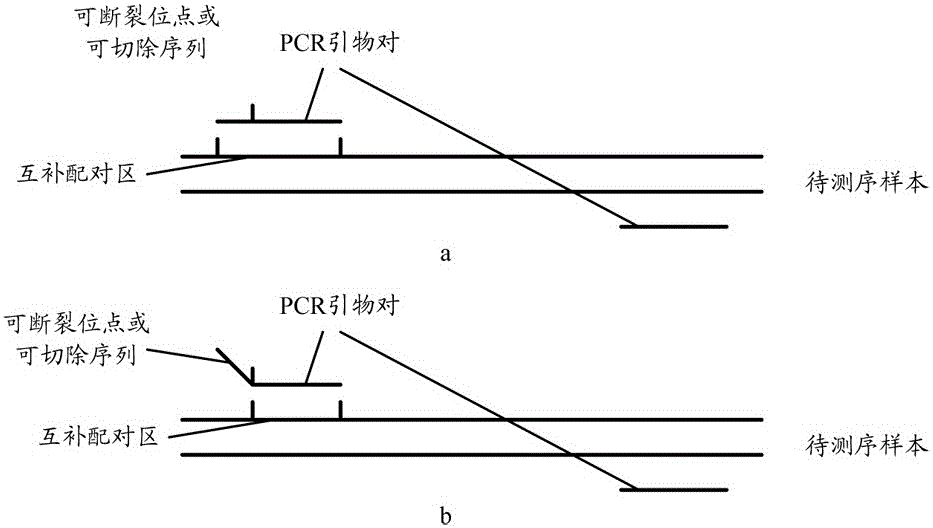
A method of building a database
ActiveCN104480534BSimplify the experimental stepsImprove the efficiency of database constructionMicrobiological testing/measurementLibrary creationComplementary pairGenotyping
Owner:SHENGZHEN CHINA GENE TECH COMPANY
A method for constructing a high-throughput sequencing library
ActiveCN104947197BLarge capacityIncrease success rateNucleotide librariesMicrobiological testing/measurementSmall fragmentHigh throughput sequence
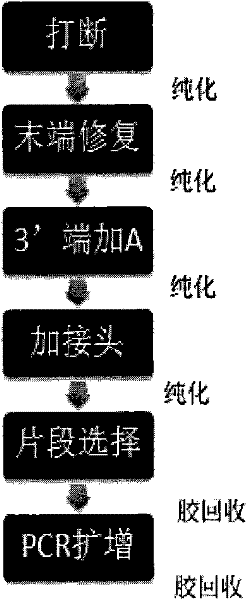
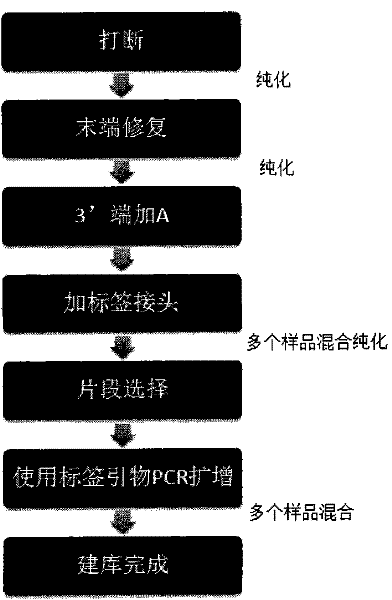
The invention belongs to the fields of genetic engineering and molecular biology, and in particular relates to a method for constructing a high-throughput sequencing library. The steps include (1) DNA fragmentation; (2) purification; (3) linker connection; (4) amplification; (5) small fragment removal; (6) library fixation and other steps. The joint connection efficiency of the present invention is high, and by using the method of the present invention, the sequencing plate can be used simply and efficiently, the loss of samples and reagents is reduced, and a high-throughput sequencing library can be constructed.
Owner:BEIJING ZHONGKEZIXIN TECH
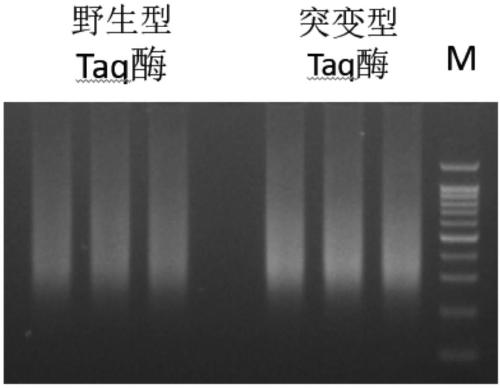
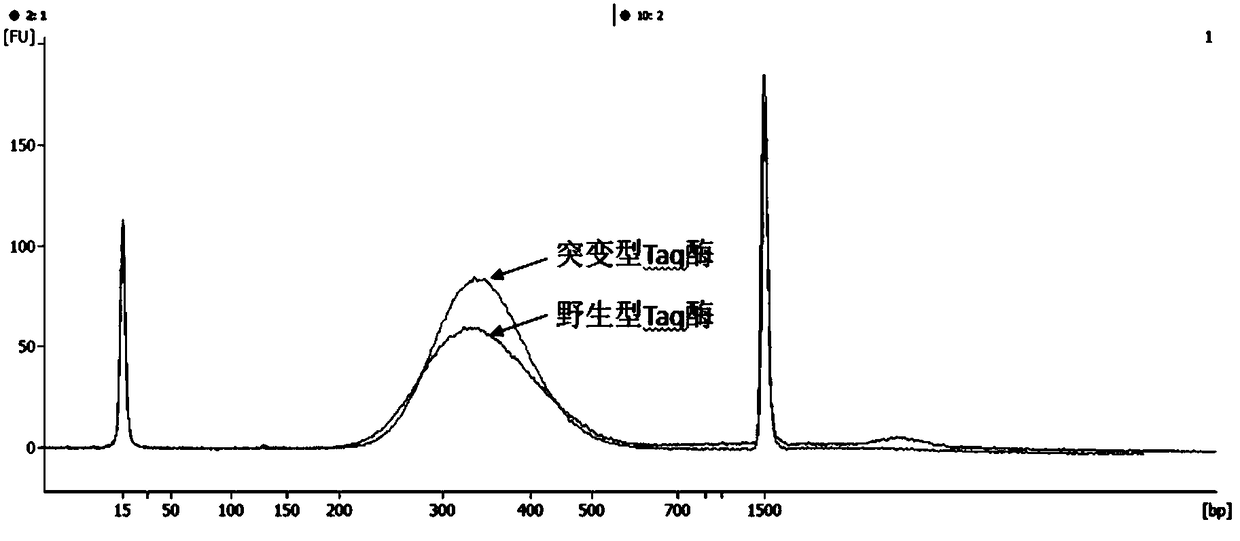
A mutant taq enzyme capable of improving the efficiency of adding a and its preparation method and application
ActiveCN106754812BAdd A to improve efficiencyImprove the efficiency of database constructionMicrobiological testing/measurementTransferasesArginineValine
The invention discloses mutant type Taq enzyme capable of improving the A adding efficiency as well as a preparation method and application of the mutant type Taq enzyme. The preparation method comprises the following specific steps: directionally mutating wild type Taq enzyme, and mutating 315th glutamic acid into lysine; mutating 388th glutamic acid into valine, mutating 507th glutamic acid into the lysine, mutating 578th aspartic acid into glycine, mutating 608th alanine into the valine, and mutating 747th methionine into arginine to obtain the mutant type Taq enzyme with the amino acid sequence shown as SEQ NO.1. The A adding efficiency of the mutant type Taq enzyme is improved, and the database building efficiency is improved; the mutant type Taq enzyme can be applied to a template with lower initial amount, and the completeness and the cover degree of building the database are improved.
Owner:VAZYME BIOTECH NANJING
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com