Multi-amplification database building method for trace DNA and special kit
A technology of multiple amplification and kits, applied in the biological field, can solve the problems of high cost, high error rate of indel detection, cumbersome steps, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
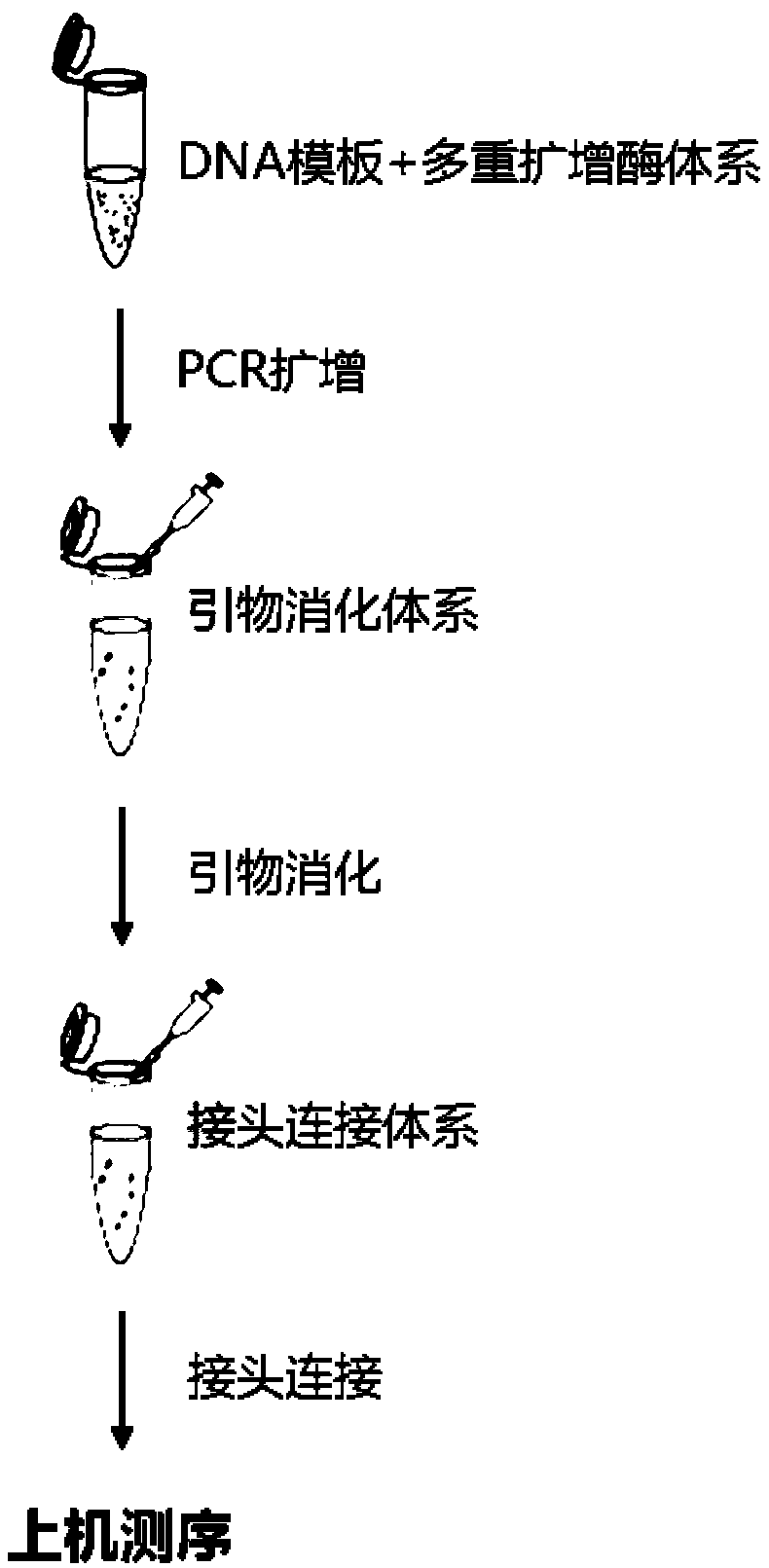
[0064] Example 1. The sequencing linker of the present invention and its application in building a trace DNA library.
[0065] 1. The sequencing adapter of the present invention
[0066] 1. Preparation of linker sequences
[0067] Prepare the linker sequence as follows:
[0068]adoligo1: T*A*A*TA*CACTCTTTCCCTACACGACGCTCTTCCG*A*T*C*T(sequence 1);
[0069] adoligo2: G*C*C*T*GTGACTGGAGTTCAGACGTGTGCTCTTCCG*A*T*C*T (sequence 2);
[0070] adlogo3: A*G*A*T*CGGAA*G*A*G*C (sequence 3);
[0071] Among them, * indicates thio modification.
[0072] 2. Preparation of Adaptor1 and Adaptor2 linkers
[0073] (1) Prepare adoligo1, adoligo2, and adoligo3 respectively into storage solutions with a final concentration of 500 μM.
[0074] (2) Mix adoligo1 and adoligo3 in equal molar numbers, configure the reaction system shown in Table 1, and react according to the procedure shown in Table 3 to form Adaptor1.
[0075] Mix adoligo2 and adoligo3 in equimolar amounts, configure the reaction sy...
Embodiment 2
[0155] Embodiment 2, the fidelity comparison between the amplification library construction system of the present invention and the Ampliseq kit
[0156] The cfDNA quality control products with a mutation frequency of 5%, 1% and 0.5% (the cfDNA quality control products with a mutation frequency of 5% and 1% were purchased from Horizon Discovery Company in the United States, and the cfDNA quality control products with a mutation frequency of 0.5% The cfDNA quality control products with a mutation frequency of 0% and 1% purchased from Horizon Discovery in the United States were mixed and prepared according to the mass ratio of 1:1) as the samples to be tested, and the amplification library construction system of the present invention and Ampliseq were used respectively. The kit was used for amplification and library construction, and the fidelity of the two methods was compared. Specific steps are as follows:
[0157] 1. Construction of library
[0158] (1) Amplification libra...
Embodiment 3
[0177] Example 3, Multiple amplification at the single cell level and method for building a library
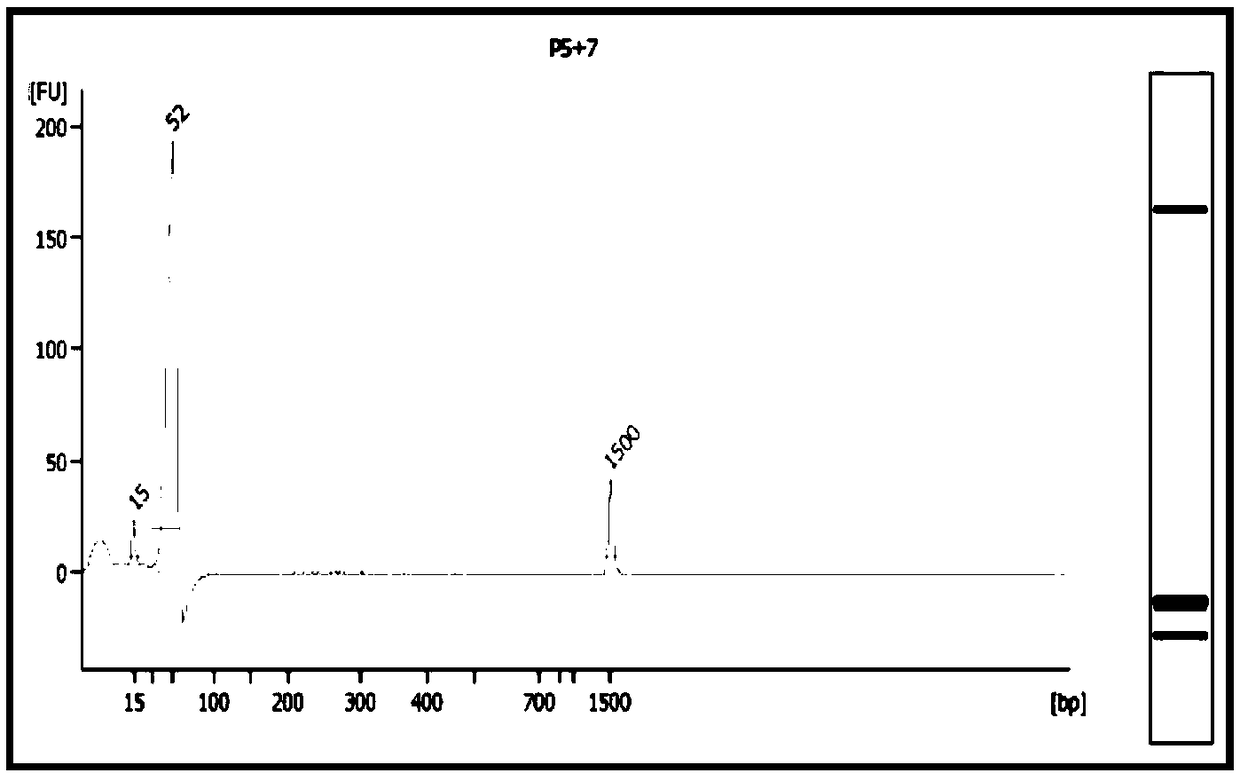
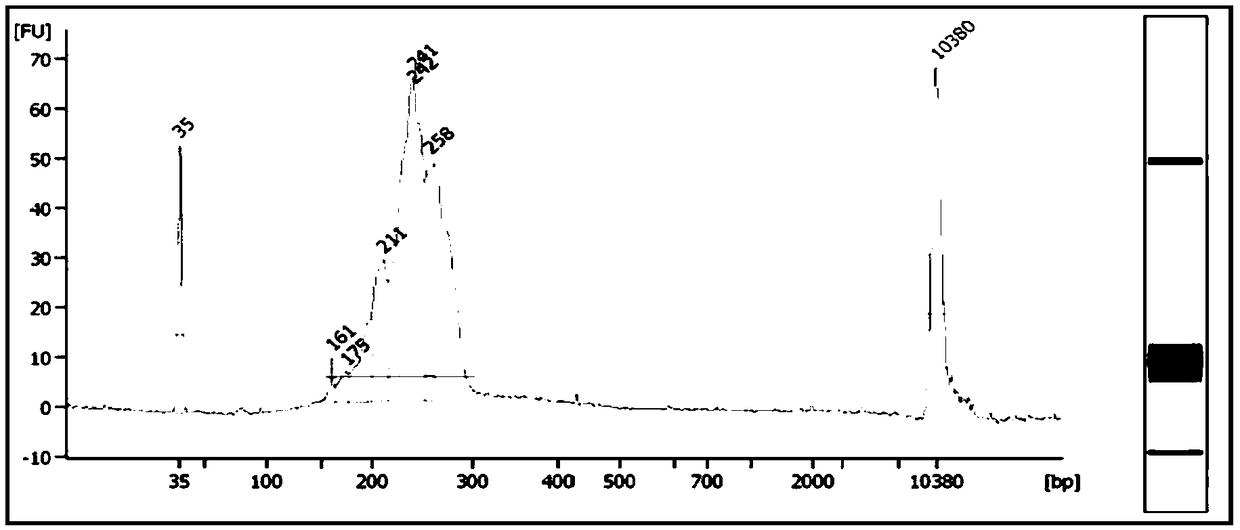
[0178] In this embodiment, 12 pg of gDNA at the single-cell level is used as a starting template to amplify and build a library (the preparation method of gDNA at the single-cell level is as follows: the fragmented human genome gDNA is fragmented with an ultrasonic breaker to obtain an average of 200 bp left and right fragmented human genome gDNA, which is then serially diluted to obtain gDNA at the single-cell level), and the amplification performance of the amplification library construction system of the present invention on picogram-level samples is tested. Specific steps are as follows:
[0179] 1. Multiple amplification of DNA at the single cell level
[0180] (1) Configure the amplification reaction system according to Table 4, blow and mix evenly, and centrifuge briefly to collect the liquid.
[0181] (2) Place it on the PCR instrument and run the program in Table 5....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com