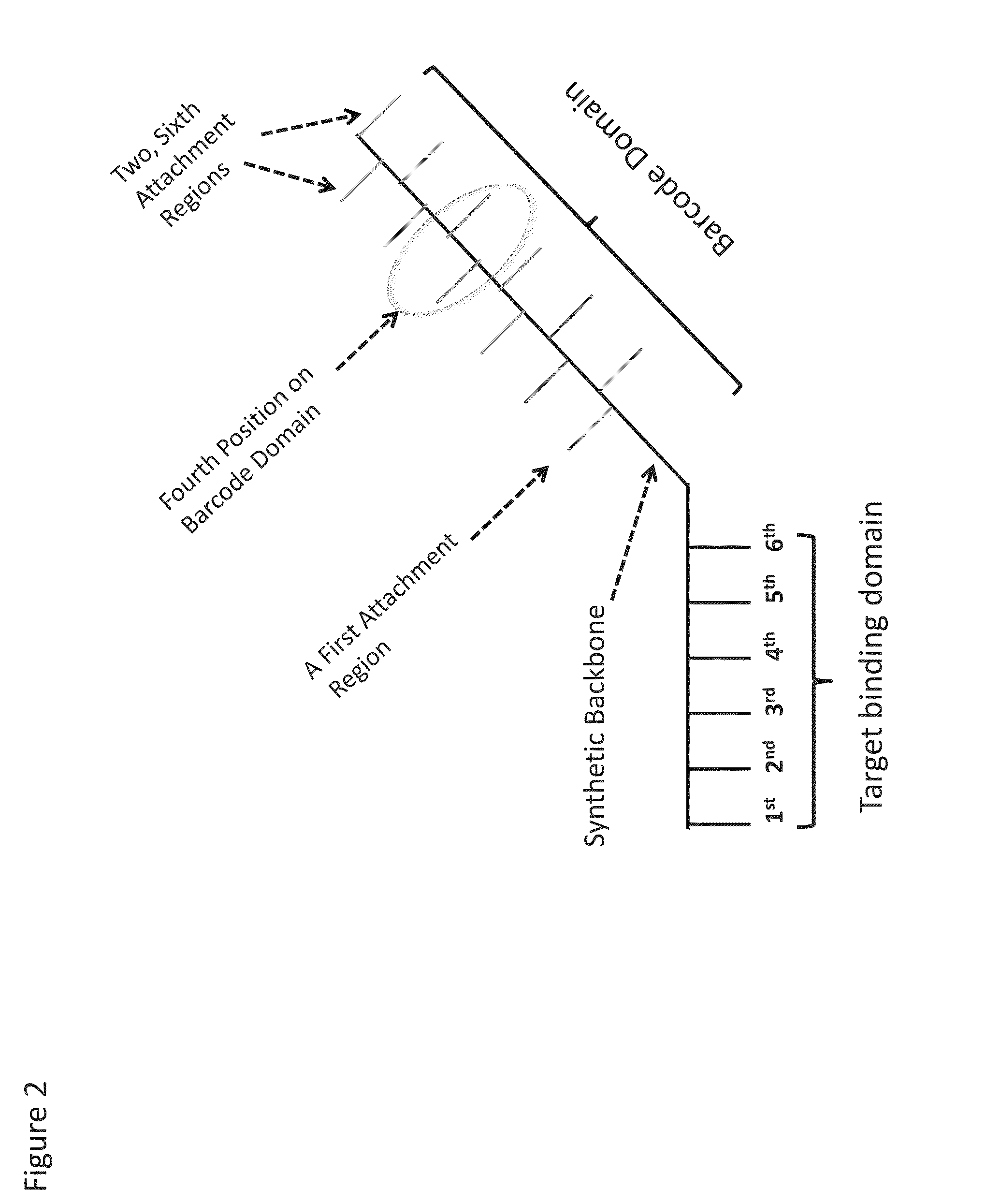
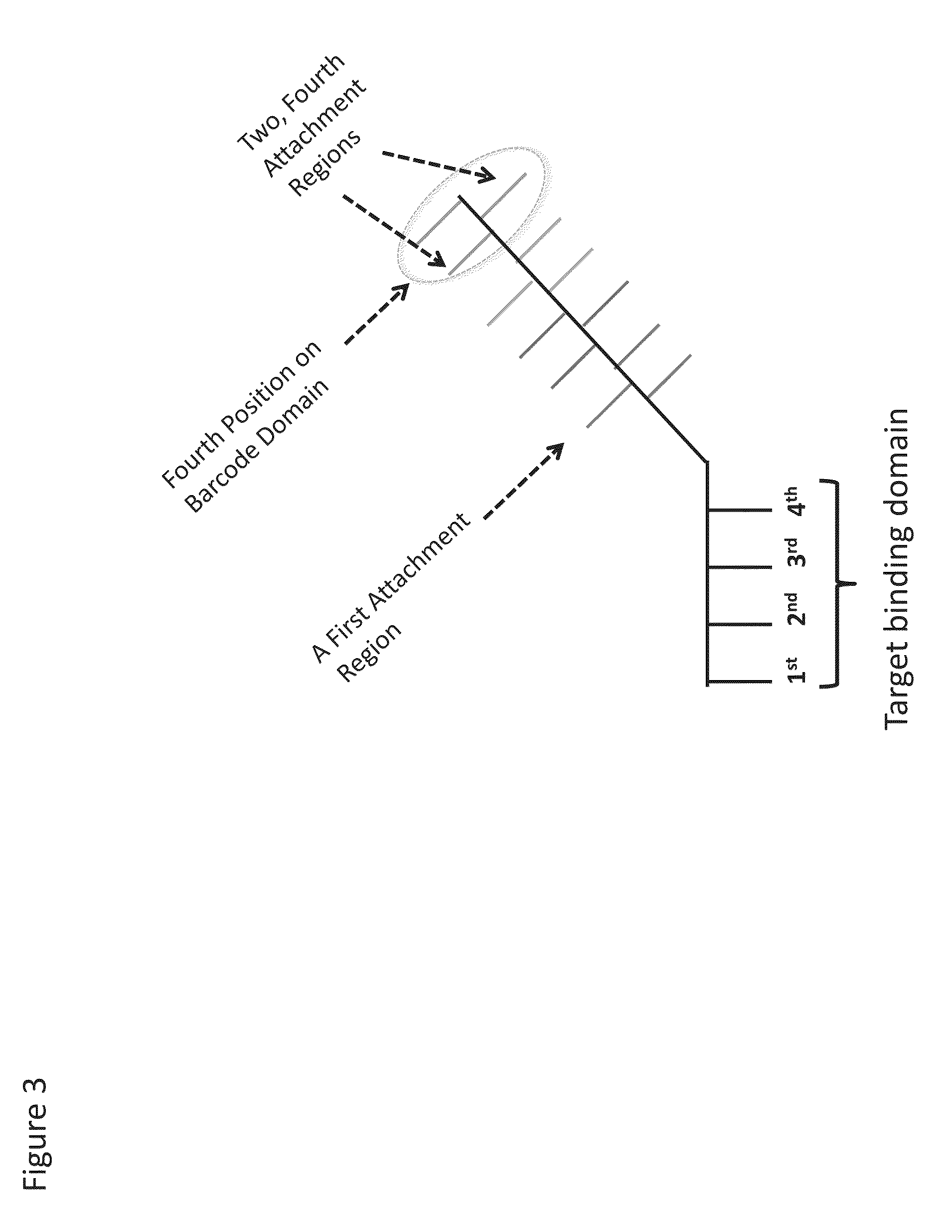
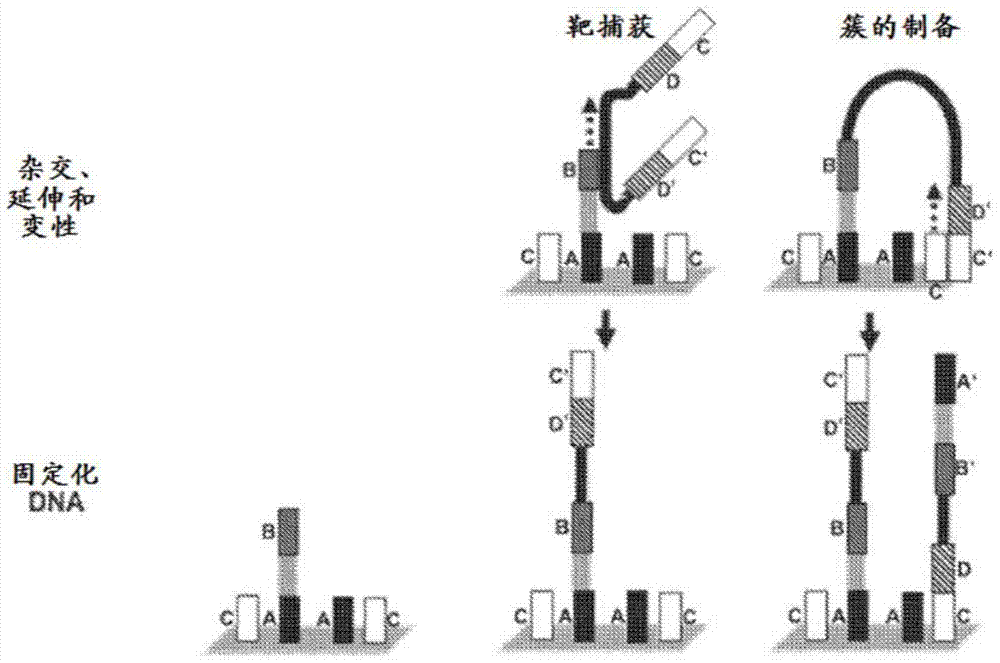
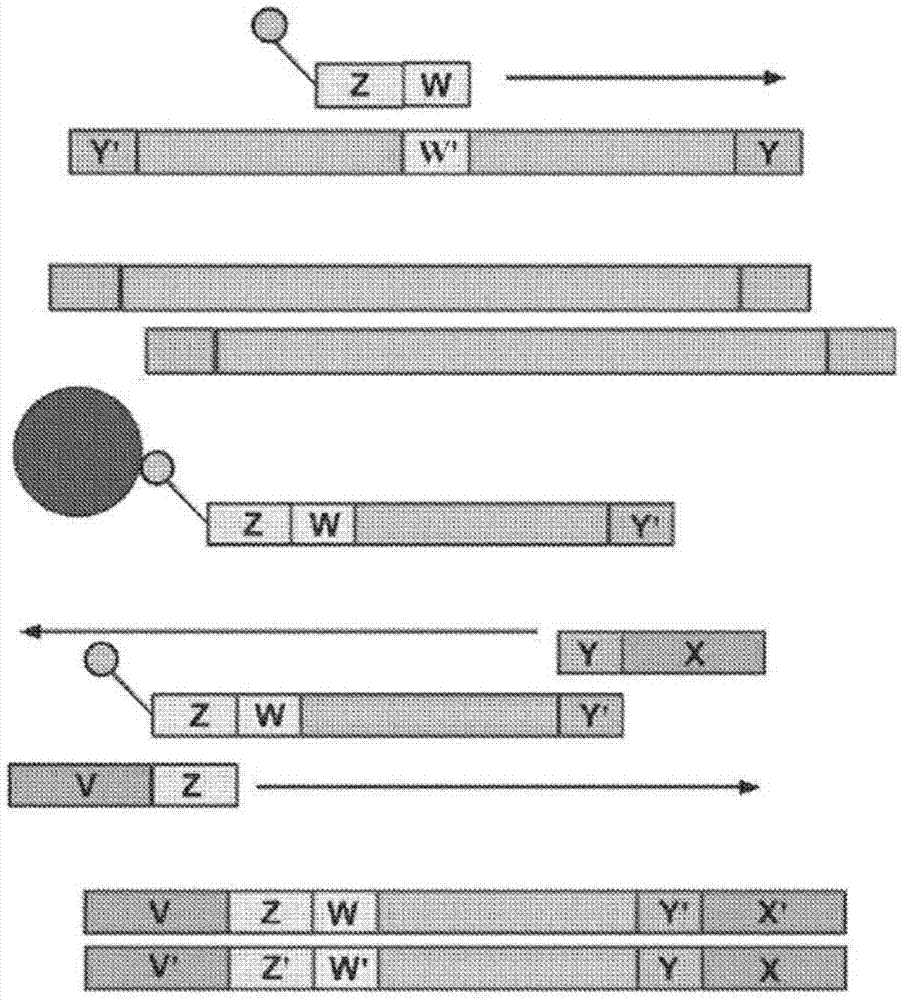
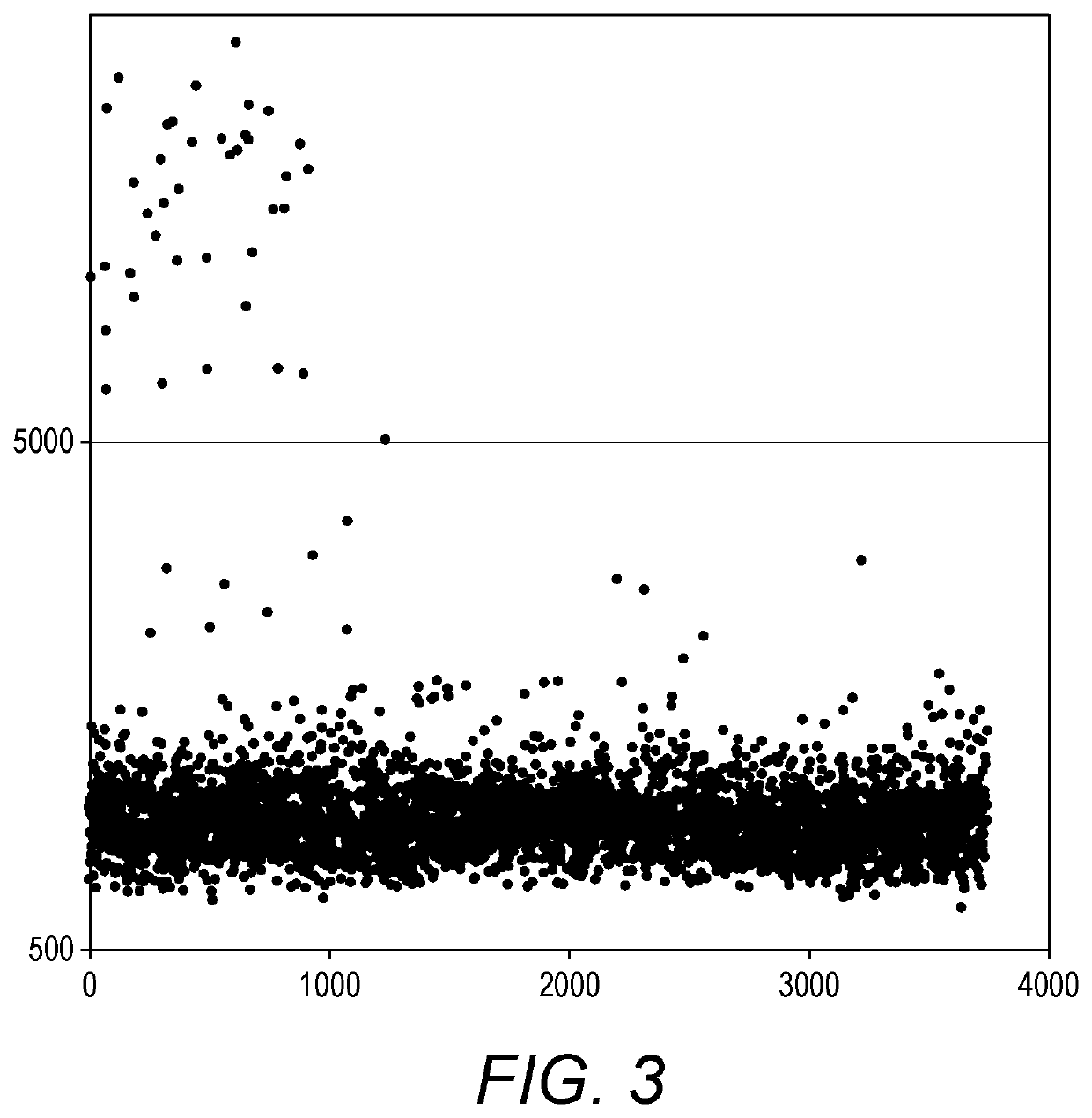
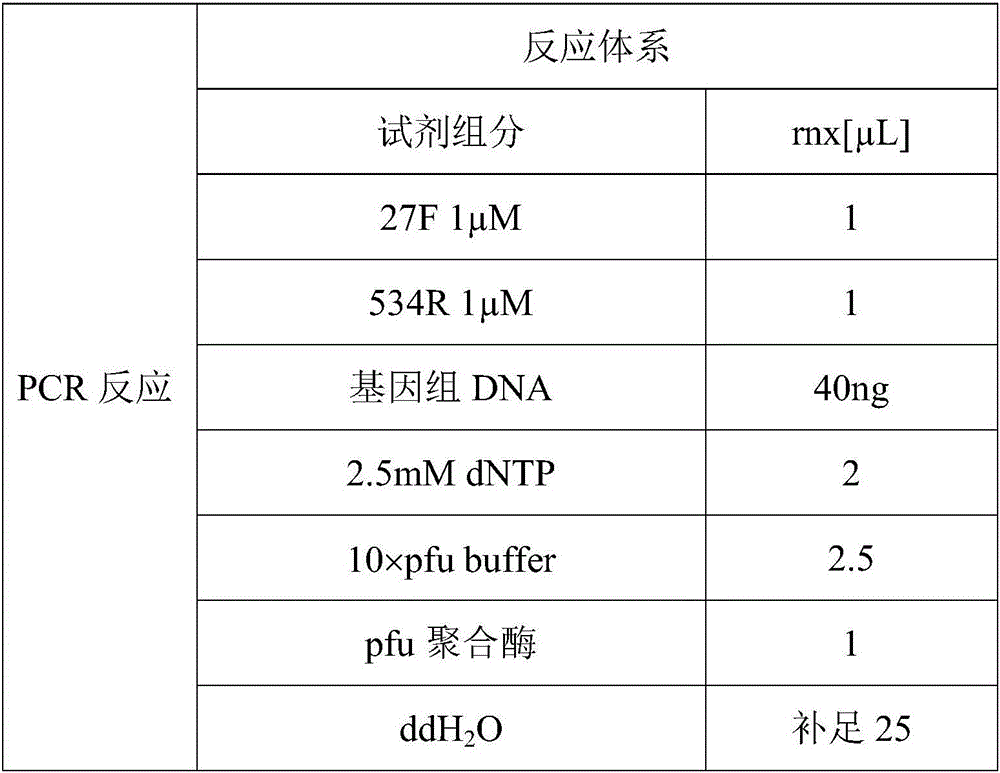
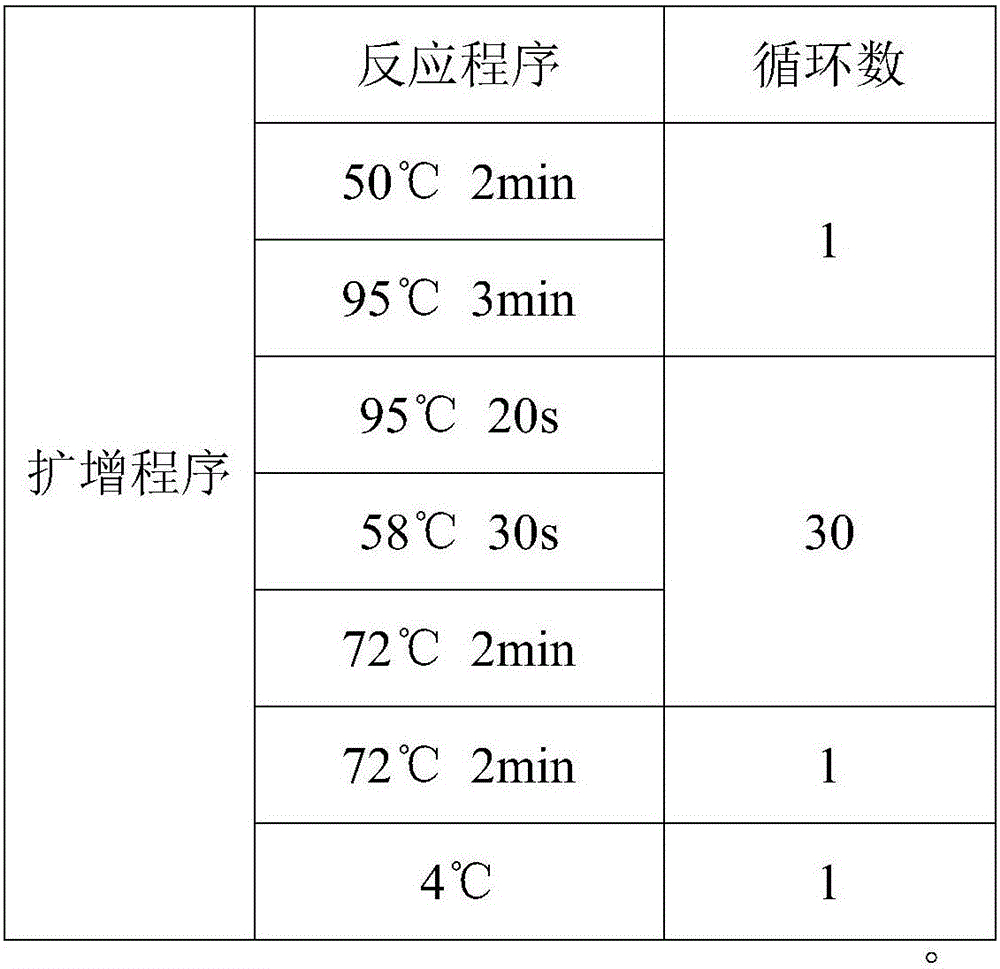
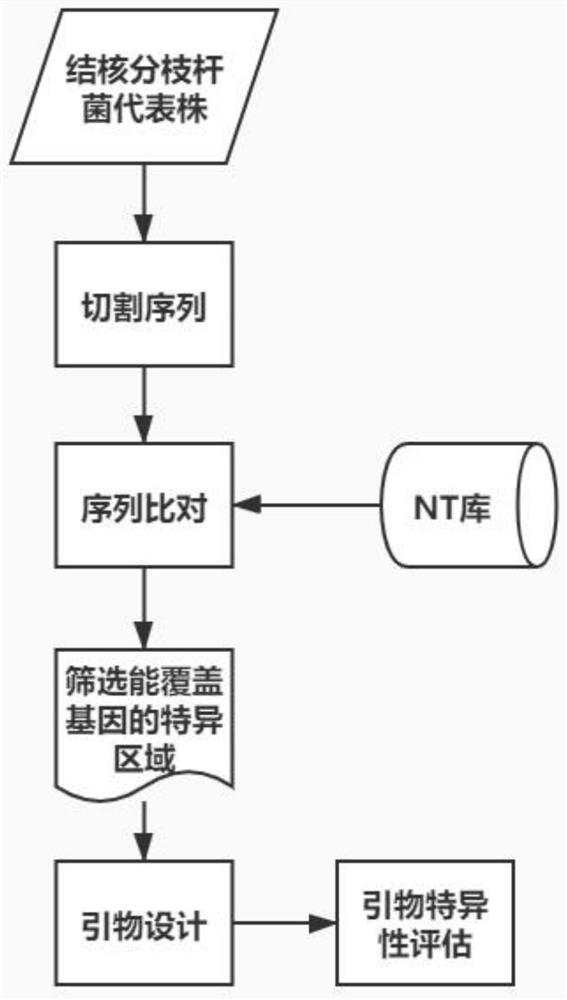
Patents
Literature
91 results about "Amplification/Sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
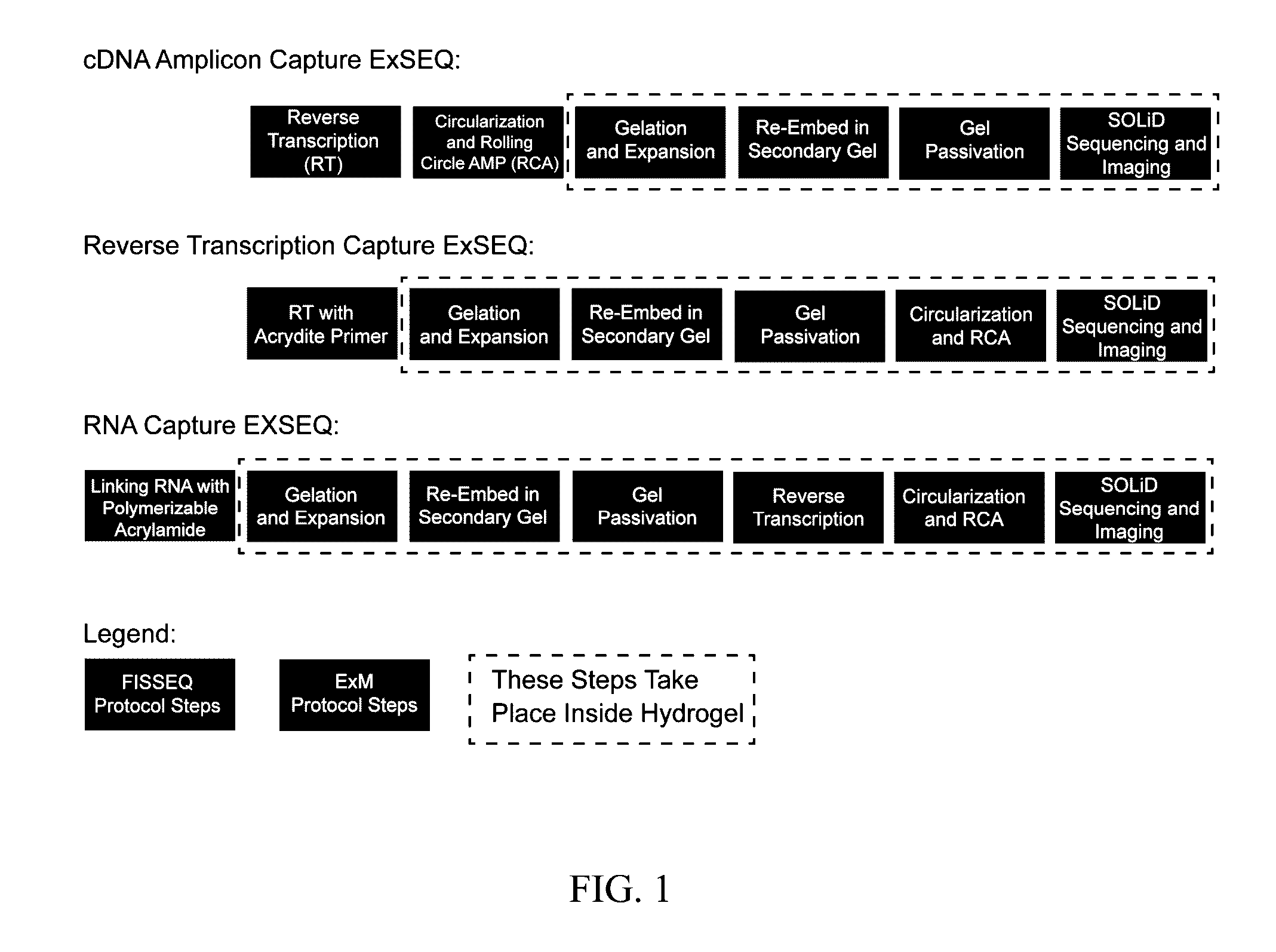
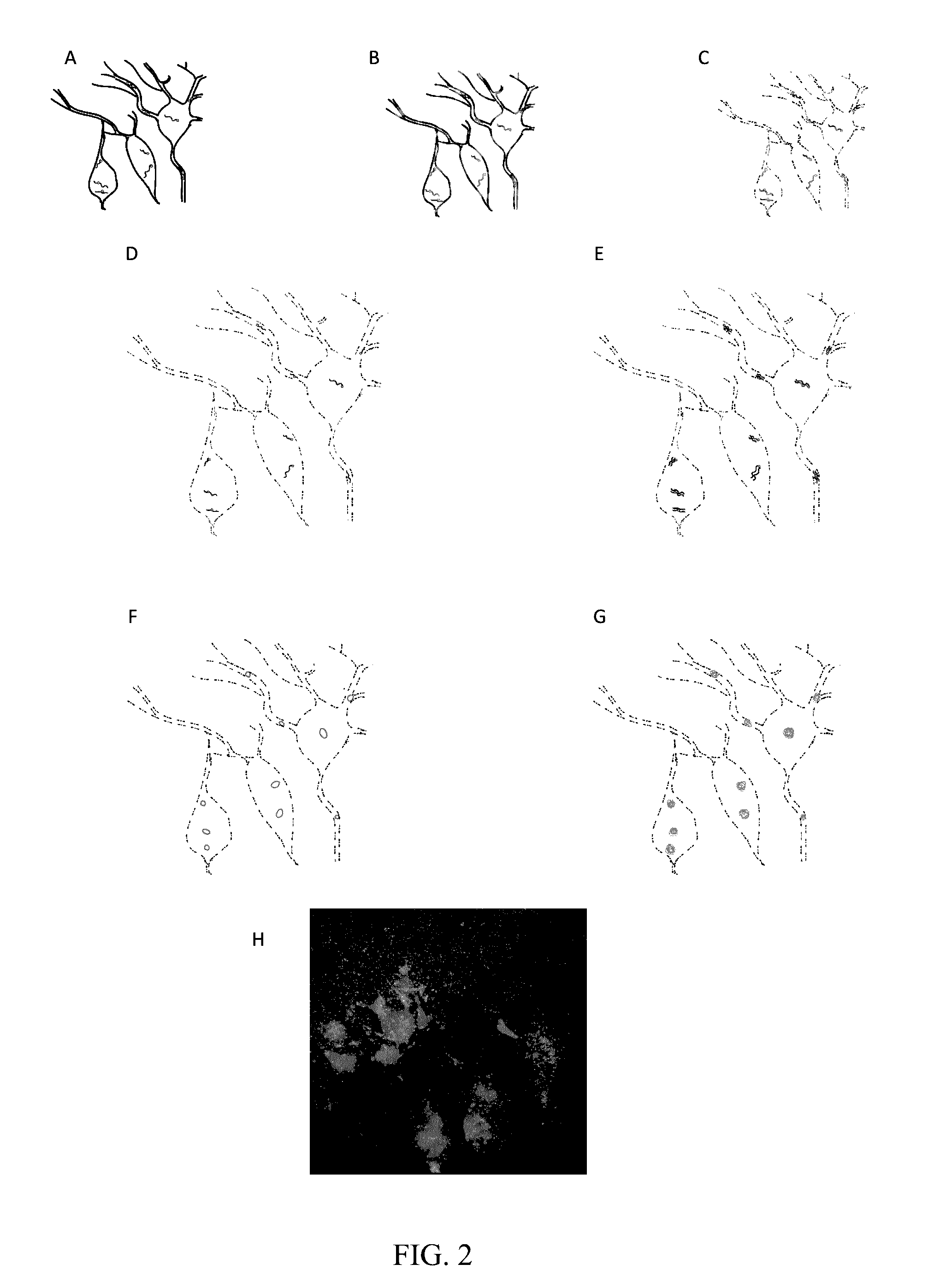
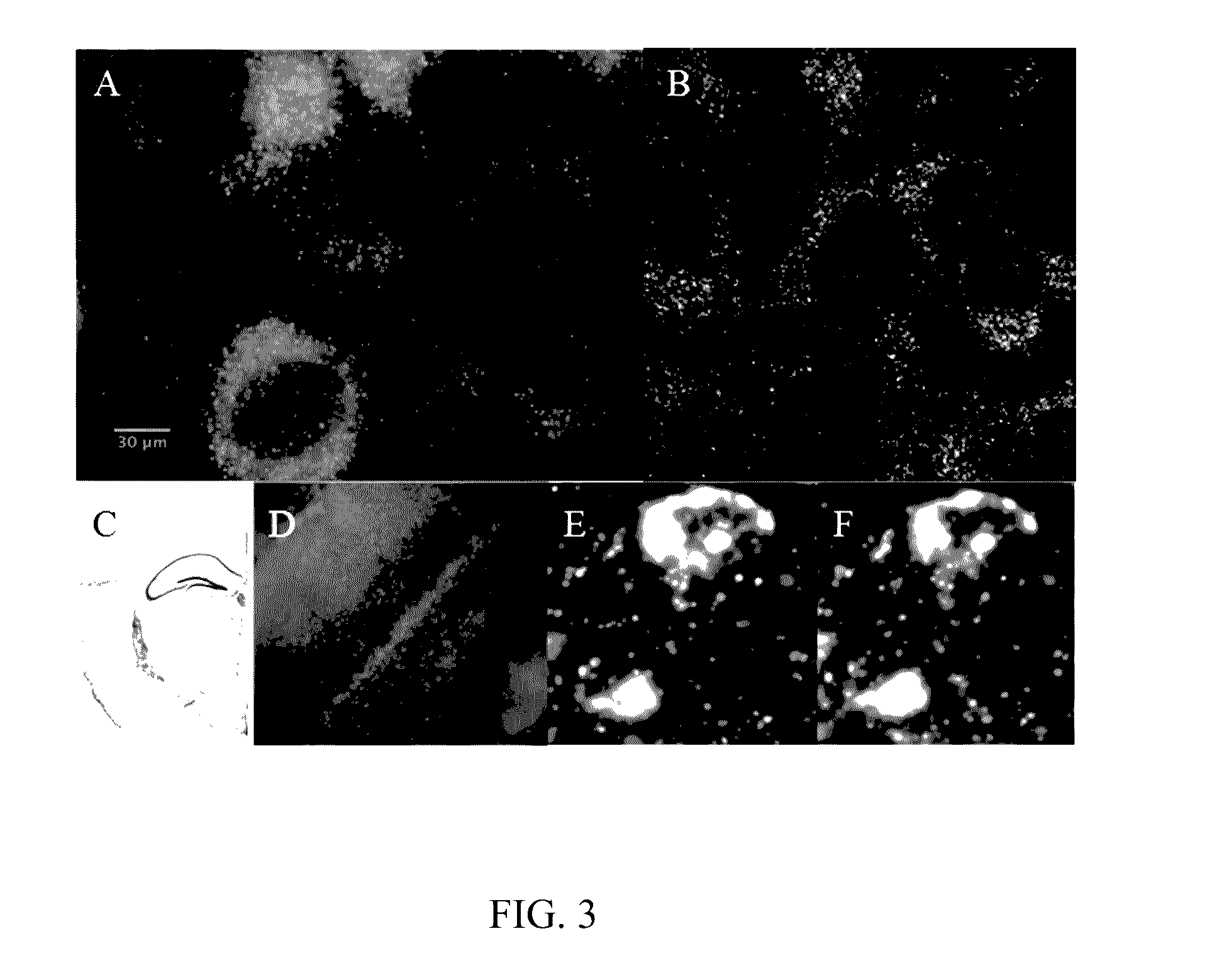
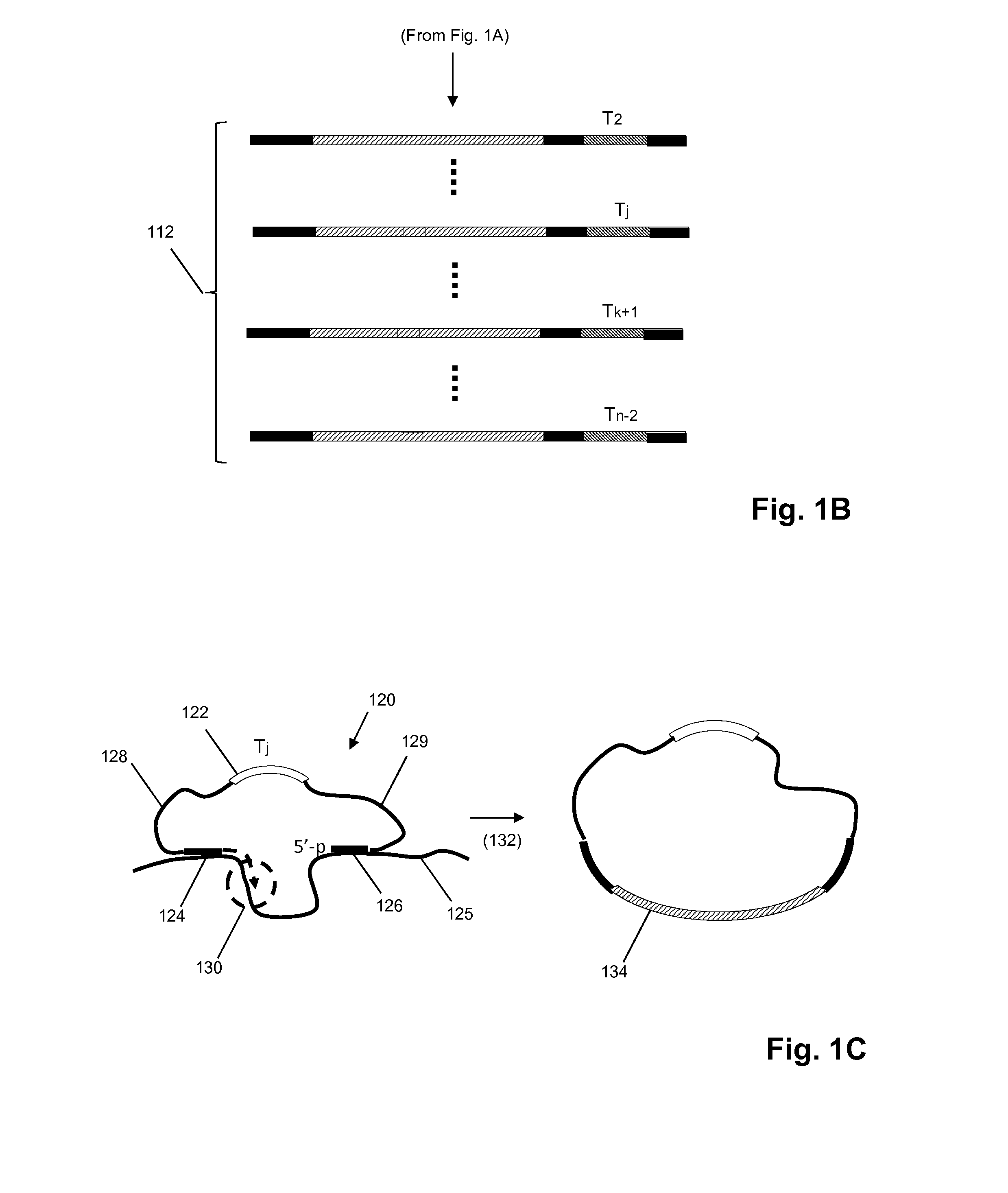
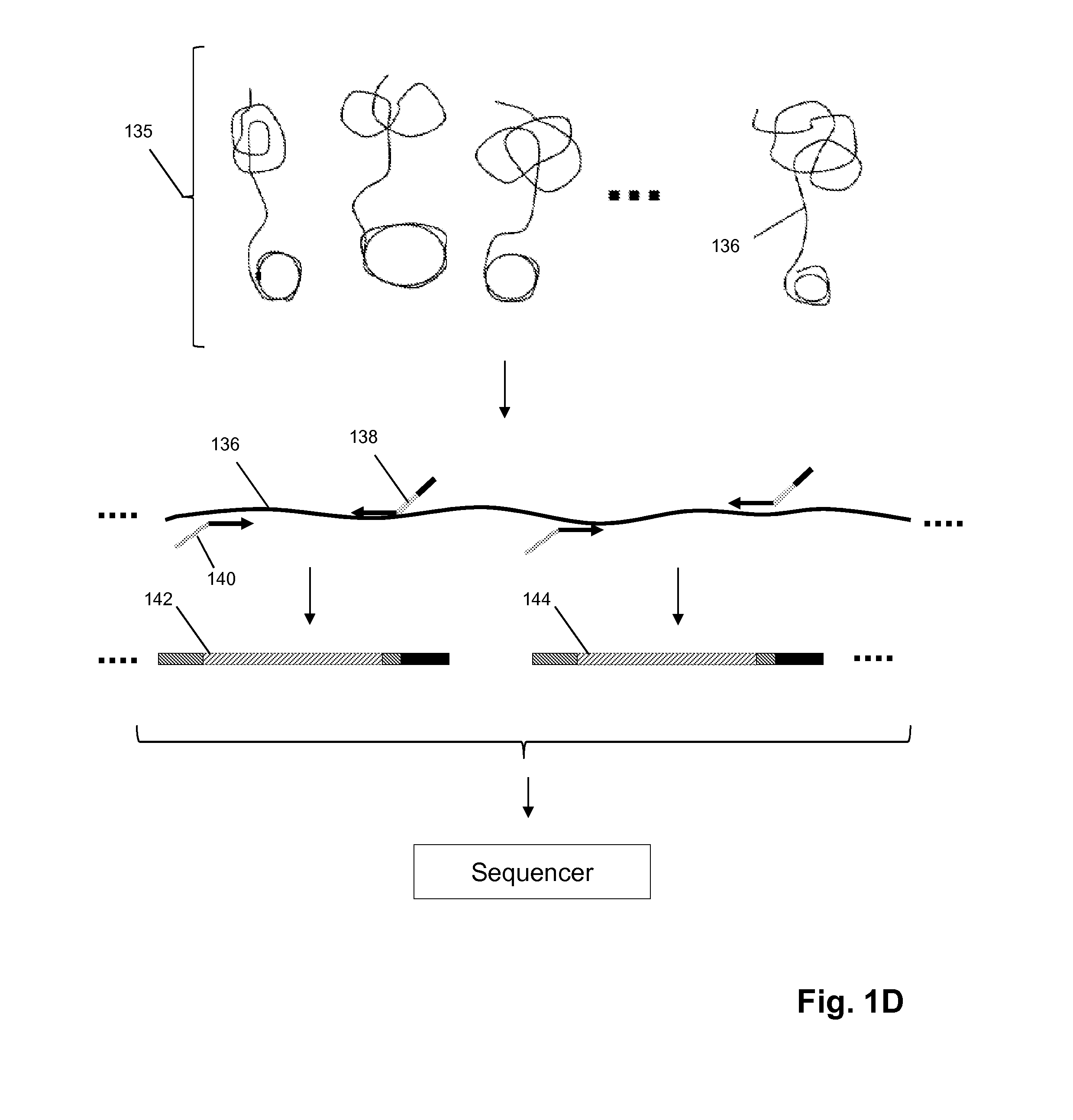
In situ nucleic acid sequencing of expanded biological samples
ActiveUS20160304952A1Microbiological testing/measurementLaboratory apparatusFluorescent in situ sequencingNucleic acid sequencing
The invention provides in situ nucleic acid sequencing to be conducted in biological specimens that have been physically expanded. The invention leverages the techniques for expansion microscopy (ExM) to provide new methods for in situ sequencing of nucleic acids as well as new methods for fluorescent in situ sequencing (FISSEQ) in a new process referred to herein as “expansion sequencing” (ExSEQ).
Owner:MASSACHUSETTS INST OF TECH +1
Enzyme- and amplification-free sequencing
InactiveUS20200010891A1Improve rendering capabilitiesReduce error rateMicrobiological testing/measurementNucleic acid sequencingEnzyme
The present invention relates to sequencing probes, methods, kits, and apparatuses that provide enzyme-free, amplification-free, and library-free nucleic acid sequencing that has long-read-lengths and with low error rate.
Owner:NANOSTRING TECH INC
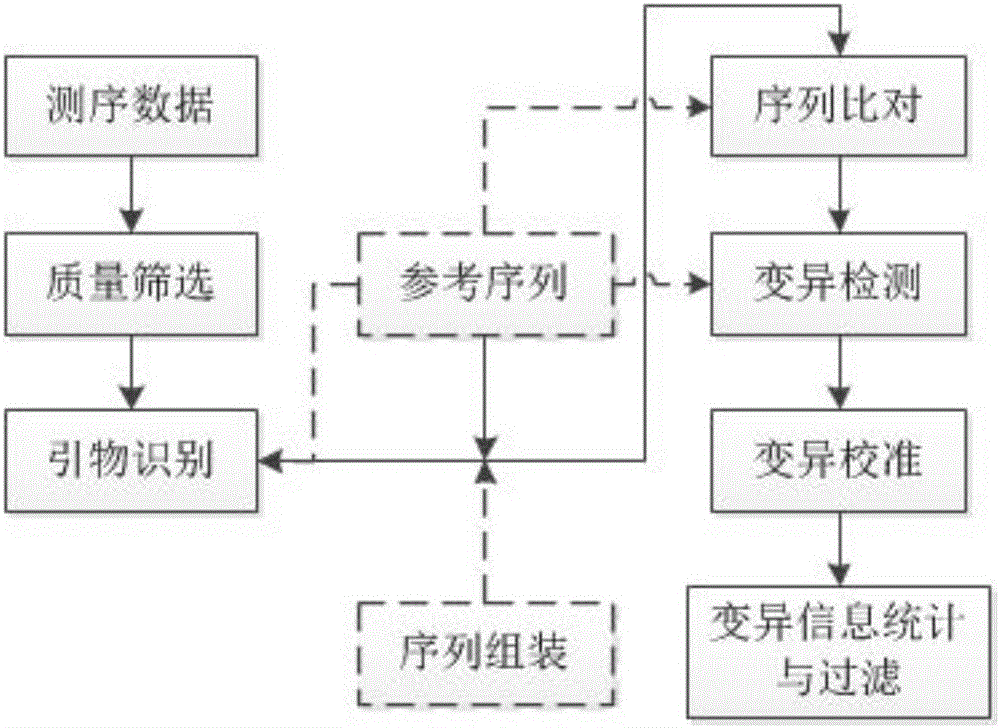
Method for detecting mutation information in multiplex amplification sequencing product of genome
ActiveCN106202991AEfficient identificationQuick identificationHybridisationSpecial data processing applicationsReference genesNatural abundance
The invention discloses a method for detecting mutation information in a multiplex amplification sequencing product of a genome. The method comprises steps as follows: sequencing data are subjected to quality assessment and preprocessing; a recognizable sequencing sequence is selected for sequence assembling; the recognizable sequencing sequence or a sequence obtained through assembling is compared with a reference gene sequence, and preliminary variation information is obtained; fine calibration of sequence variation is performed according to different types of conditions; a calibrated sequencing fragment is obtained; the homozygosis or heterozygosis state of a target fragment is obtained according to the type of the sequencing fragment with the highest abundance; finally, the mutation information in the multiplex amplification sequencing product of the genome is obtained. By means of the method, the amplification product can be rapidly, efficiently and accurately recognized, and the calculation resources are saved; the sequence assembling process is compatible, and the problem of reduction of the quality value of basic groups produced in the sequencing process can be effectively solved; the homozygosis / heterozygosis state of variation information can be more effectively and stably judged, and random errors introduced in the PCR (polymerase chain reaction) process and the sequencing process are eliminated.
Owner:AMOY DIAGNOSTICS CO LTD +1
High sensitivity mutation detection using sequence tags
InactiveUS20160115532A1Improve accuracyHigh sensitivityMicrobiological testing/measurementNucleotideMutation detection
The invention is directed to methods for increasing the sensitivity of high throughput sequencing, particularly for distinguishing true rare mutations from amplification, sequencing and other sample processing errors that occur in sequencing techniques. In one aspect, methods of the invention includes steps of (a) preparing templates from nucleic acids in a sample; (b) labeling by sampling the templates to form tag-template conjugates, wherein substantially every template of a tag-template conjugate has a unique sequence tag; (c) linearly amplifying the tag-template conjugates; (d) generating a plurality of sequence reads from the linearly amplified tag-template conjugates; and (e) determining a nucleotide sequence of each of the nucleic acids based on the frequencies, or numbers, of each type of nucleotide at each nucleotide position of each plurality of sequence reads having identical sequence tags.
Owner:ADAPTIVE BIOTECH
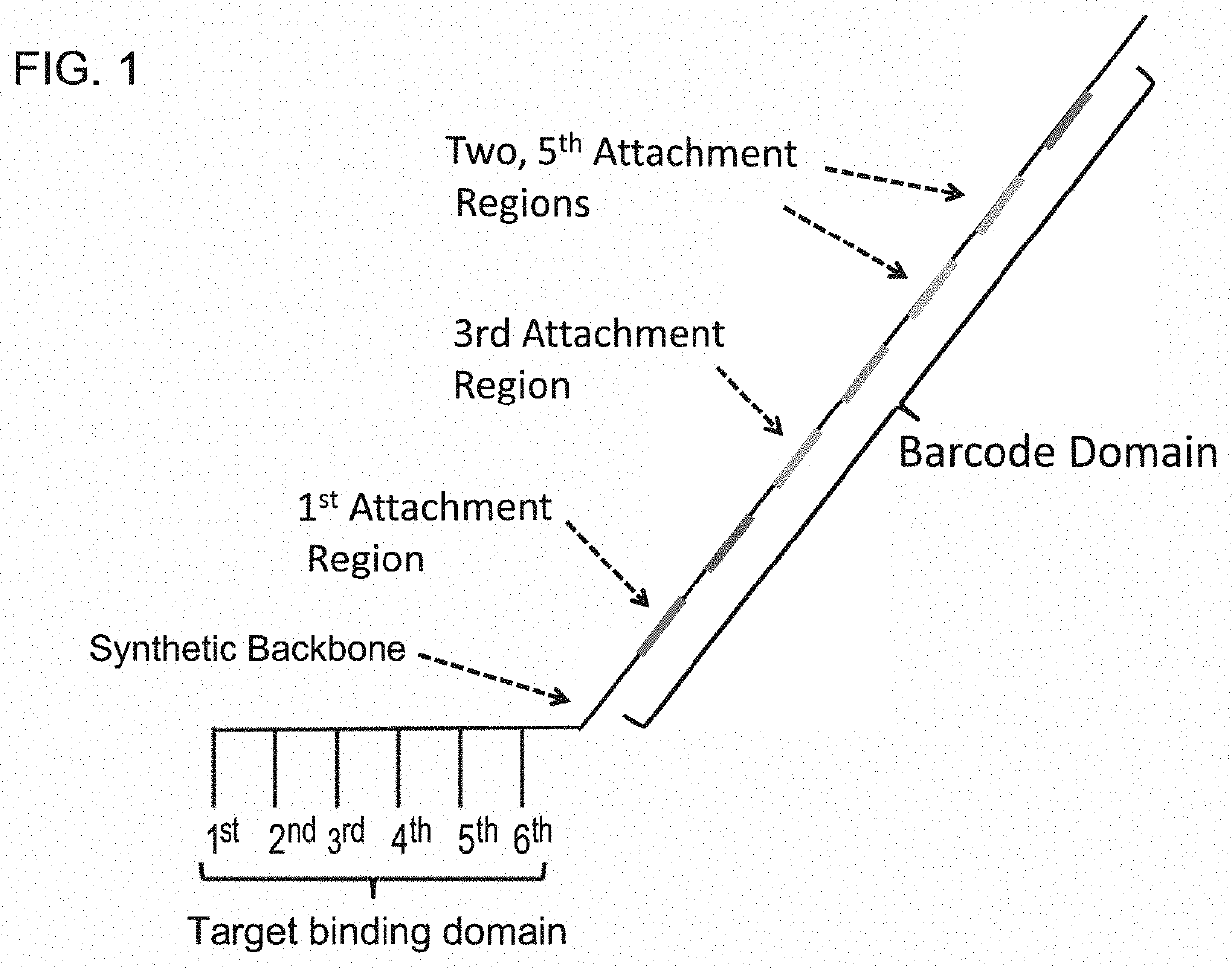
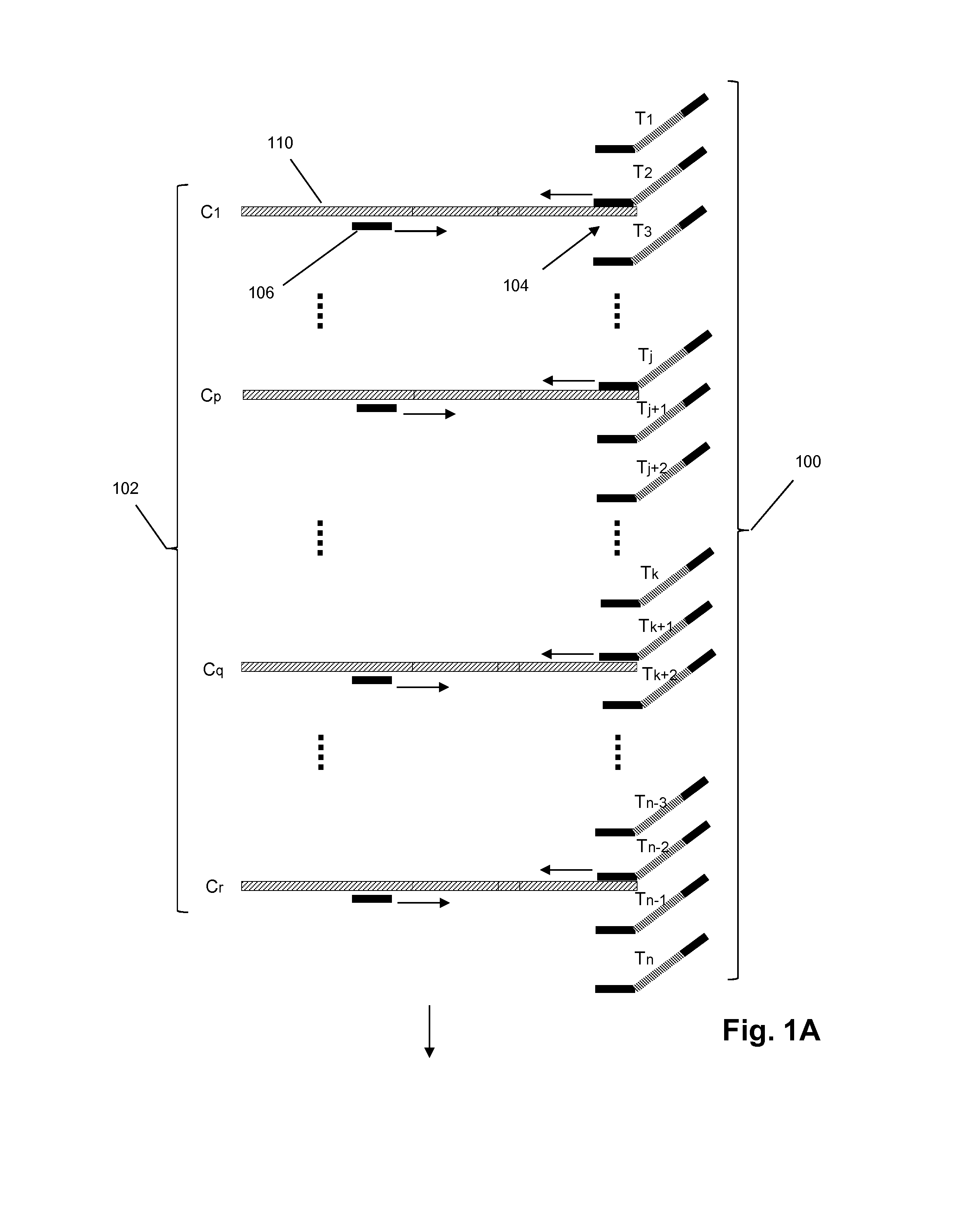
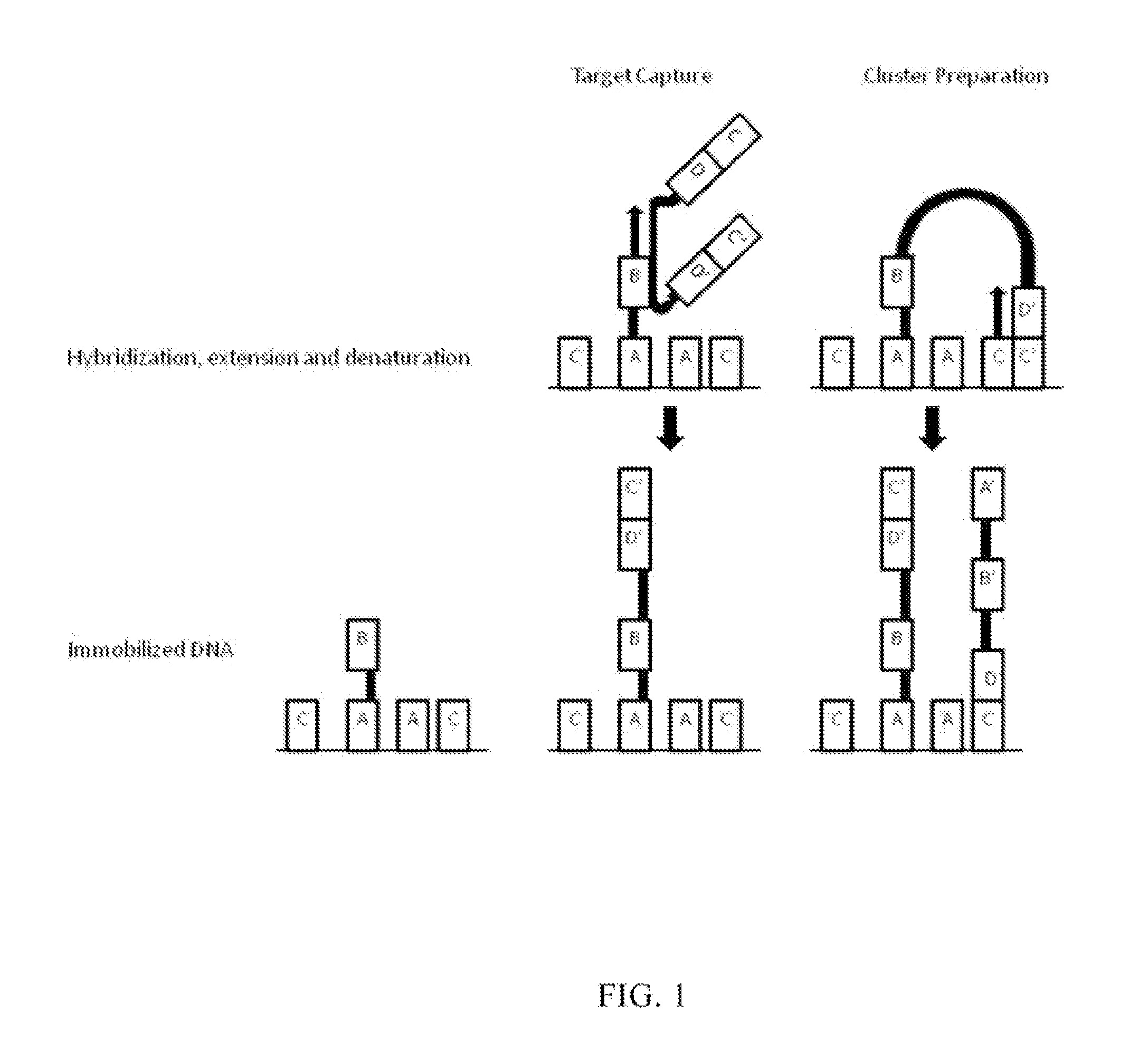
Methods and compositions for high-throughput sequencing
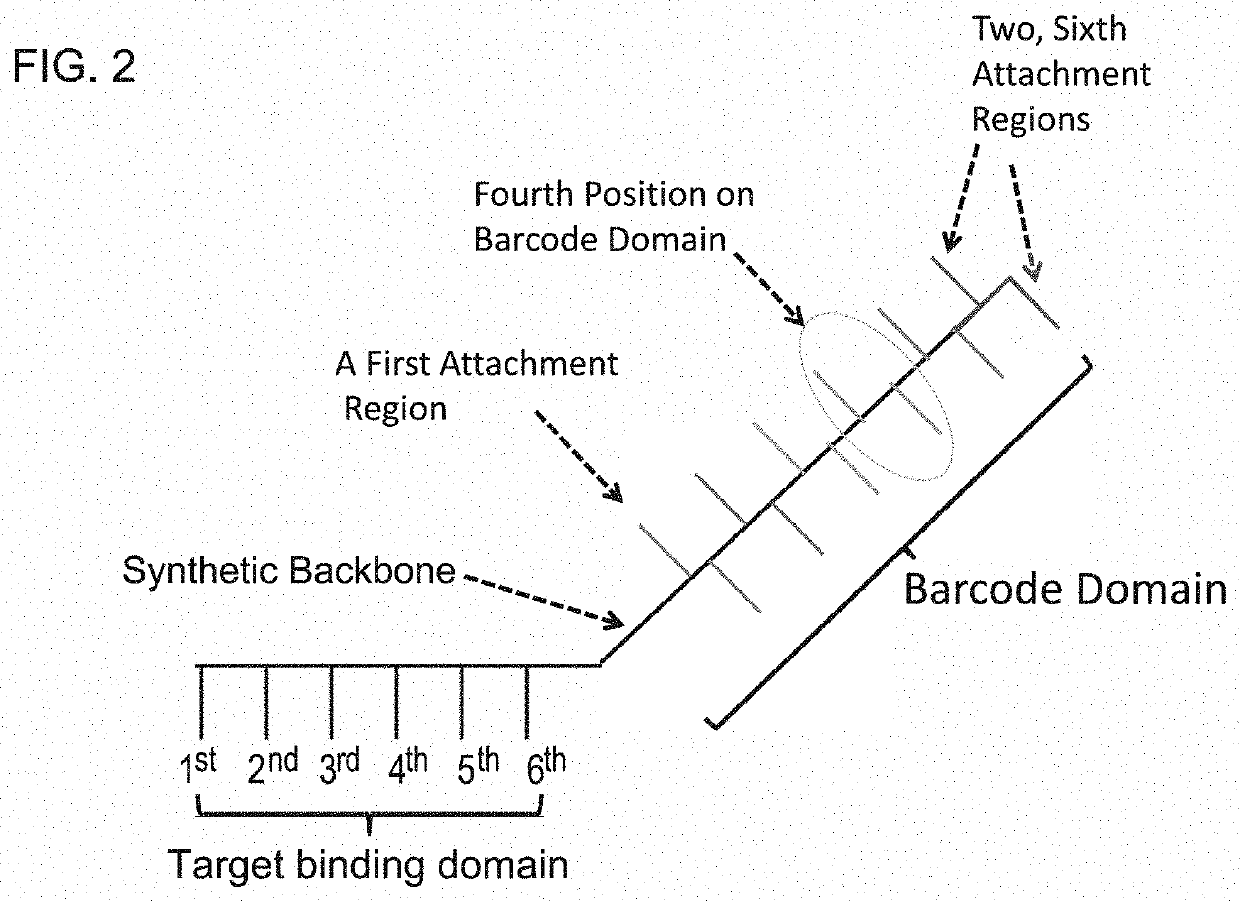
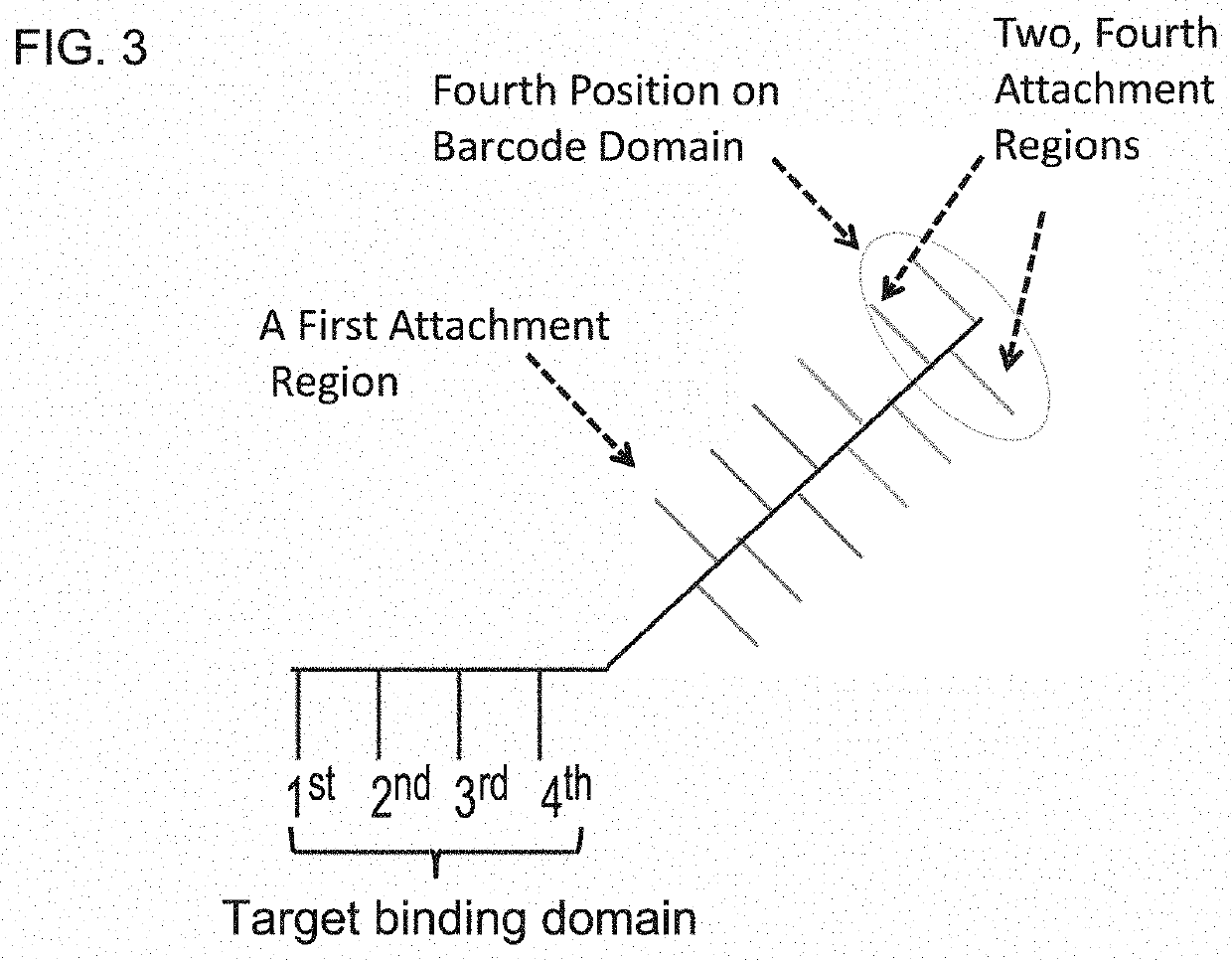
InactiveUS20140024541A1Microbiological testing/measurementLibrary screeningBarcodeHigh throughput sequence
The invention provides methods, apparatuses, and compositions for high-throughput amplification sequencing of specific target sequences in one or more samples. In some aspects, barcode-tagged polynucleotides are sequenced simultaneously and sample sources are identified on the basis of barcode sequences. In some aspects, sequencing data are used to determine one or more genotypes at one or more loci comprising a causal genetic variant.
Owner:COUNSYL INC
Apparatus and methods for high-throughput sequencing
InactiveUS20140024536A1Microbiological testing/measurementLibrary member identificationBarcodeGenotype
The invention provides methods, apparatuses, and compositions for high-throughput amplification sequencing of specific target sequences in one or more samples. In some aspects, barcode-tagged polynucleotides are sequenced simultaneously and sample sources are identified on the basis of barcode sequences. In some aspects, sequencing data are used to determine one or more genotypes at one or more loci comprising a causal genetic variant.
Owner:COUNSYL INC
Enzyme- and amplification-free sequencing
InactiveUS20160194701A1Reduce error rateRapid sample-to-answer capabilitySugar derivativesNucleotide librariesNucleic acid sequencingEnzyme
The present invention relates to sequencing probes, methods, kits, and apparatuses that provide enzyme-free, amplification-free, and library-free nucleic acid sequencing that has long-read-lengths and with low error rate.
Owner:NANOSTRING TECH INC
System and methods for detecting genetic variation
The invention provides methods, apparatuses, and compositions for high- throughput amplification sequencing of specific target sequences in one or more samples. In some aspects, barcode-tagged polynucleotides are sequenced simultaneously and sample sources are identified on the basis of barcode sequences. In some aspects, sequencing data are used to determine one or more genotypes at one or more loci comprising a causal genetic variant. In some aspects, systems and methods of detecting genetic variation are provided.
Owner:COUNSYL INC
Methods and compositions for enrichment of target polynucleotides
The invention provides methods, apparatuses, and compositions for high-throughput amplification sequencing of specific target sequences in one or more samples. In some aspects, barcode-tagged polynucleotides are sequenced simultaneously and sample sources are identified on the basis of barcode sequences. In some aspects, sequencing data are used to determine one or more genotypes at one or more loci comprising a causal genetic variant.
Owner:MYRIAD WOMENS HEALTH INC
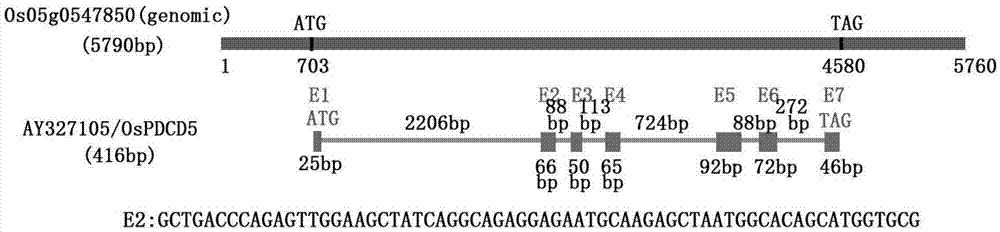
Method for fixed point knock-out of second exon of rice OsPDCD5 gene by using CRISPR/Cas9 system
ActiveCN106939316AIncrease productionEfficient breedingPlant peptidesVector-based foreign material introductionDouble strandedAmplification/Sequencing
The invention discloses a method for fixed point knock-out of the second exon of rice OsPDCD5 gene by using a CRISPR / Cas9 system. The method comprises the following steps: selecting a target fragment in the second exon region of the rice OsPDCD5 gene, constructing a plant CRISPR / Cas9 recombinant vector, introducing the plant CRISPR / Cas9 recombinant vector to rice calluses through an Agrobacterium infection technology, carrying out regeneration to form seedlings, shearing double strands of the second exon of the OsPDCD5 gene under the combined action of guide RNA and Cas9 nuclease, and realizing random insertion or random deletion of the target gene fragment through the self DNA repairing function of cells. Nine strains with the target fragment having point mutation are obtained through a PCR amplification sequencing technology, and pass phenotype identification, so the purpose of the increase of the plant height and the yield of rice is realized. The method provides an efficient breeding way for cultivating high-yield and high-quality rice varieties.
Owner:FUDAN UNIV
System and Methods for Detecting Genetic Variation
The invention provides methods, apparatuses, and compositions for high-throughput amplification sequencing of specific target sequences in one or more samples. In some aspects, barcode-tagged polynucleotides are sequenced simultaneously and sample sources are identified on the basis of barcode sequences. In some aspects, sequencing data are used to determine one or more genotypes at one or more loci comprising a causal genetic variant. In some aspects, systems and methods of detecting genetic variation are provided.
Owner:MYRIAD WOMENS HEALTH INC
HCM (Hypertrophic Cardiomyopathy) genotyping method and kit
The invention provides a technical scheme to provide an HCM (Hypertrophic Cardiomyopathy) genotyping method and a kit. The method comprises the following steps of: a, carrying out a gene mutation test on genes MYH7 (Myosin Heavy Chain 7), MYBPC3 (Myosin Binding Protein C 3) and TNNT2 (Troponin 2) in a sample DNA (Deoxyribonucleic Acid), wherein the gene mutation test comprises PCR (Polymerase Chain Reaction) amplification sequencing of MYH7, MYBPC3 and TNNT2 gene exons; and b, comparing sequences obtained by amplification in the step a with a standard sequence of a database to determine a genotyping result. The invention has the beneficial effects of: (1) assisting the early diagnosis: finding out asymptomatic generation sufferers and non-generation sufferers in family members; and (2) assisting to guide the selective birth so as to eliminate the spread of the HCM in the family.
Owner:泰普生物科学(中国)有限公司
Reagent kit for extracting DNA in Chinese alligator chorion film and use method thereof
InactiveCN101368179AHigh purityIncrease concentrationMicrobiological testing/measurementDNA preparationHigh concentrationChinese alligator
The invention discloses a kit for extracting DNA in the egg shell membrane of Yangtze alligator and a use method thereof, and the kit comprises the following reagents: a reagent A, a reagent B, a reagent C, a reagent D, a reagent E and a reagent F. The template DNA which is extracted by the kit has good purity, and the kit can be used for gene amplification sequencing and typing. In the amplification of the 12S rRNA gene, the mitochondrial D-loop region and the Cyt b gene, all the extracted shell membrane DNA templates can be successfully amplified to obtain the fragment products with expected identical sizes, high concentration and high specificity. Microsatellite DNA marker specificity primers are utilized to obtain typing data through the PCR amplification to be used for the identification in disputed paternity of Yangtze alligator. In addition, the method also can be applied to the extraction of other ovipara such as the turtles, the birds and the like. The obtained template DNA can be widely applied to the studies of the ovipara on the fields such as population genetic analysis, molecular evolution, evolution of relationship among individuals and the like.
Owner:ANHUI NORMAL UNIV
Novel broad-spectrum rice blast resistance allele pi21t and resistance application thereof
InactiveCN103409433ARapid determination of allelic typePlant peptidesGenetic engineeringBiotechnologyAllele
The invention belongs to the field of molecular genetics and relates to a novel broad-spectrum rice blast resistance allele pi21t and resistance application thereof. According to a cloned sequential design primer of the broad-spectrum rice blast resistance recessive allele pi21t, PCR amplification is performed on the wide compatibility variety 02428 which is used in high frequency in breeding, and sequencing comparative analysis is performed, so that the novel broad-spectrum rice blast resistance allele pi21t is identified in combination with a field rice blast inoculation identification result of the variety. The material has high resistance for popular microspecies of 6 rice blast groups in the Yangtze-Huaihe river basin, and provides new gene resources for breeding for rice blast resistance.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
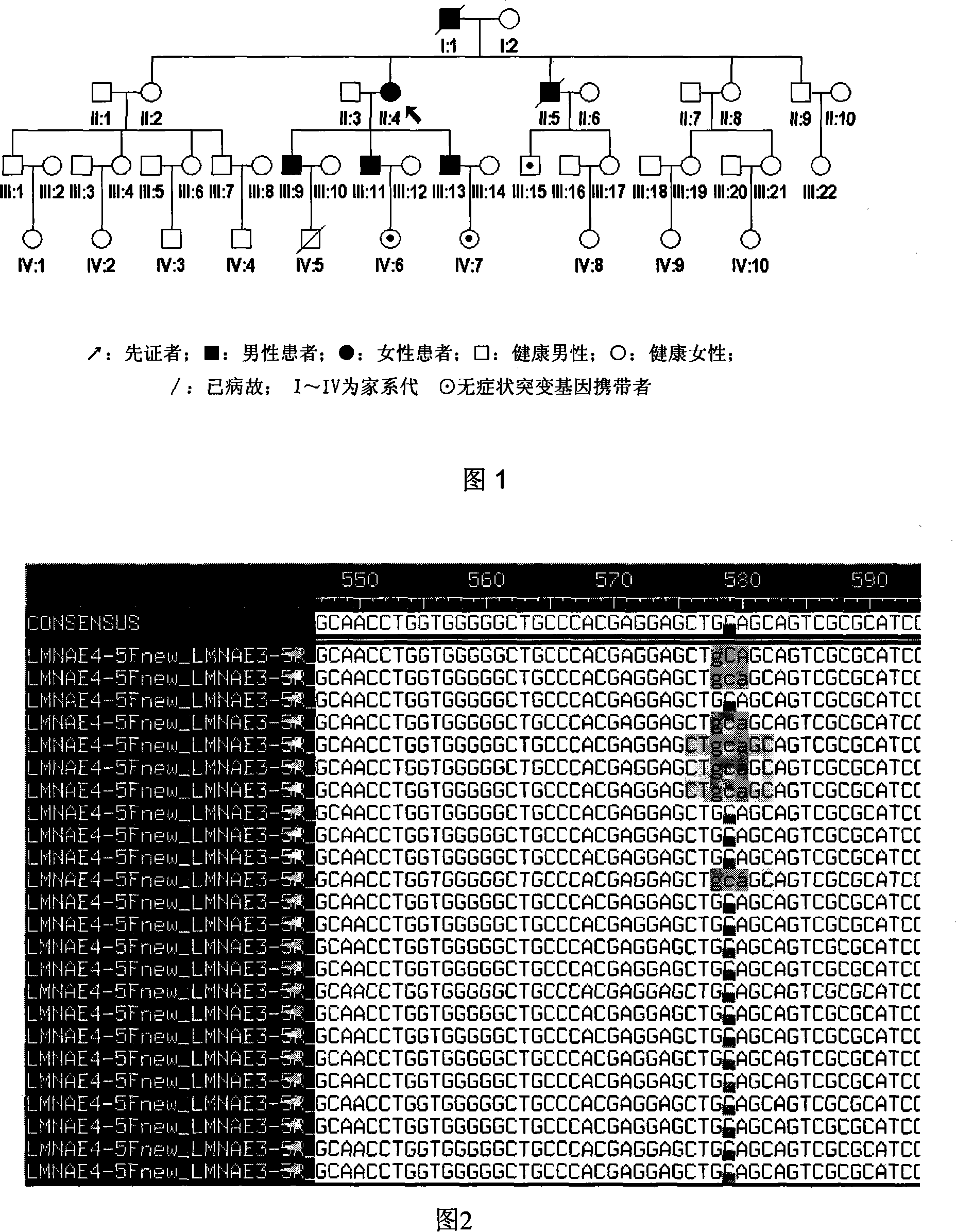
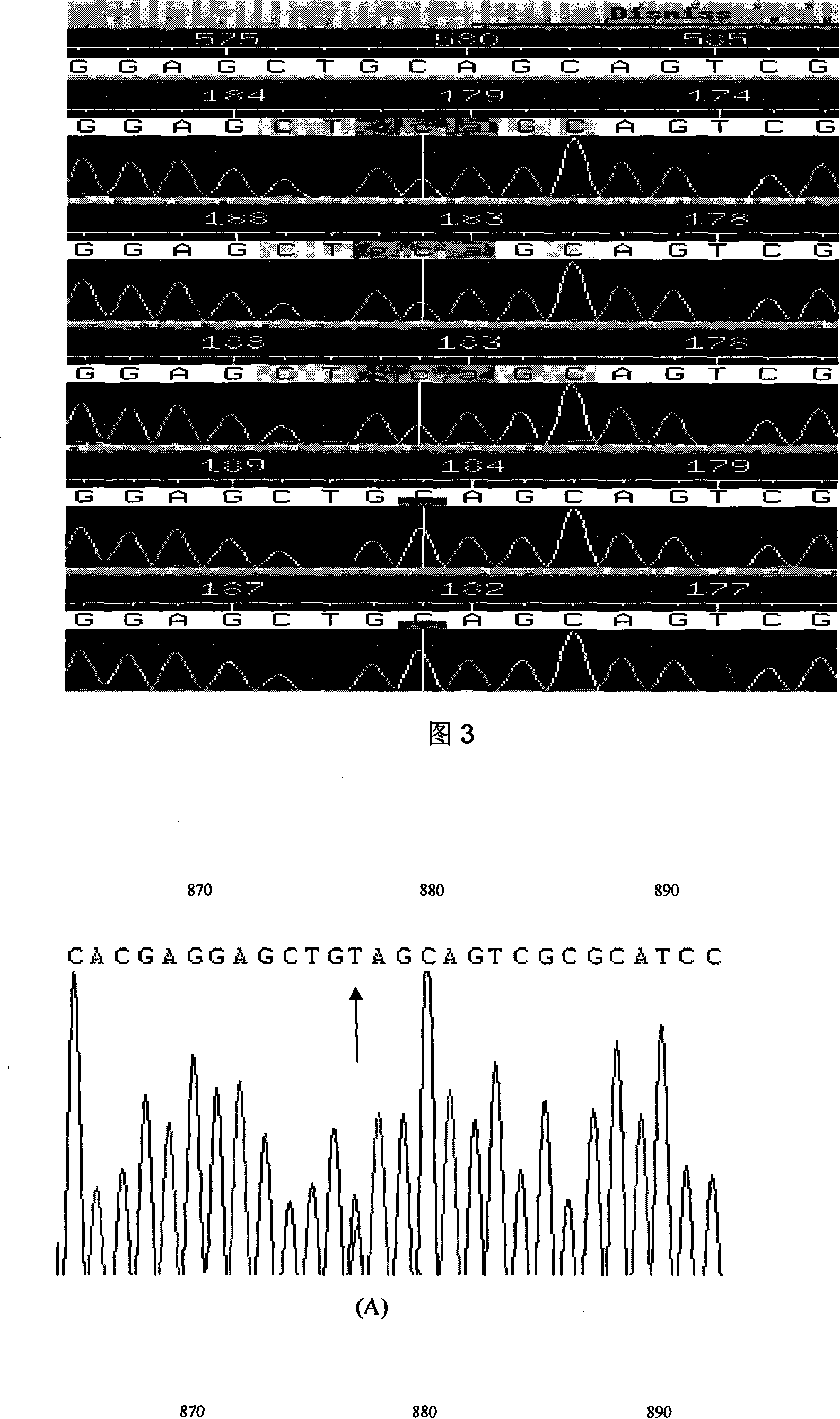
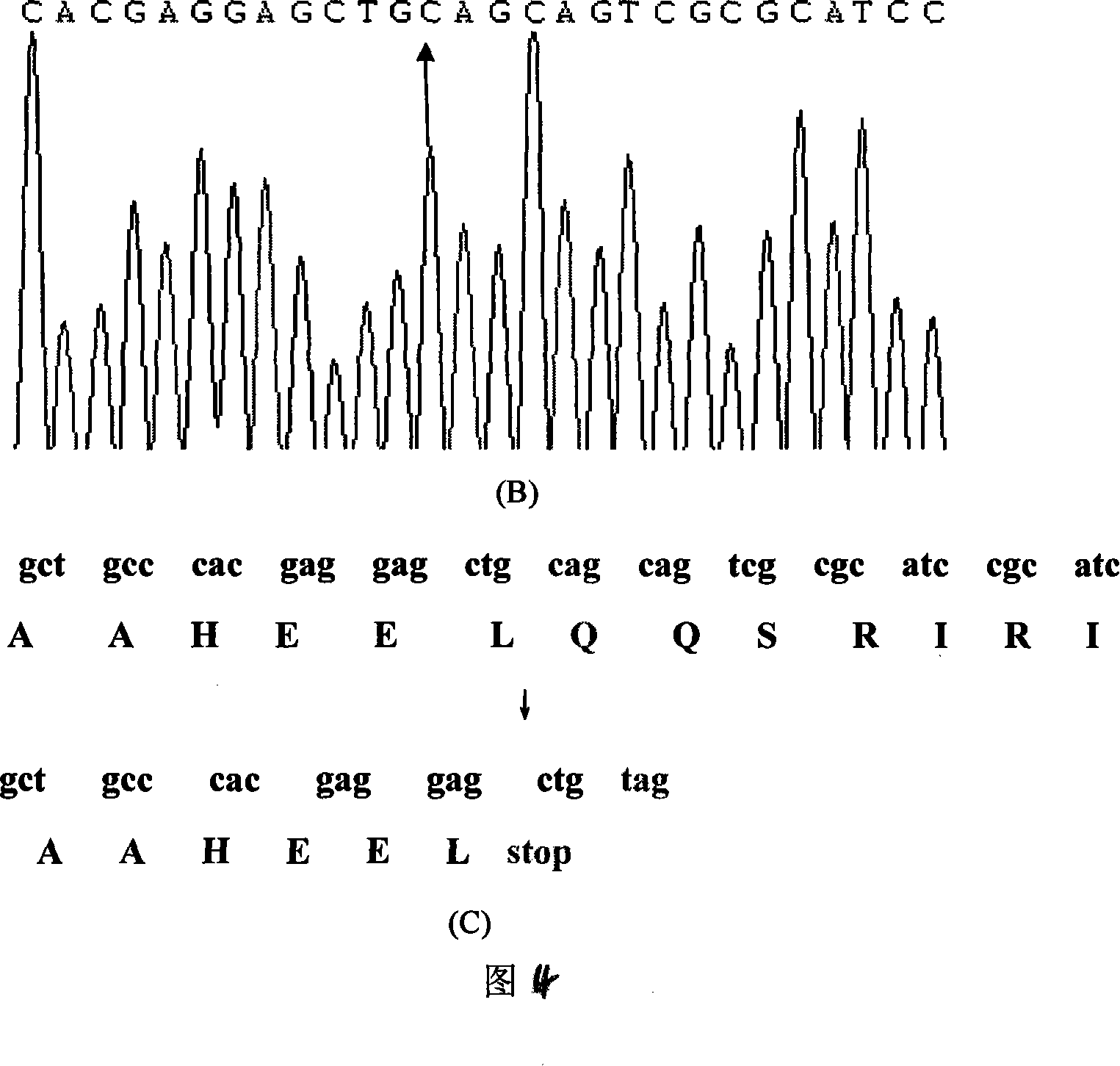
Expanding cardiomyopathy LMNA gene mutation and detecting method thereof
InactiveCN101134960AMicrobiological testing/measurementGenetic engineeringMolecular geneticsConducting system
The present invention relates to LMNA gene mutation of dilated cardiomyopathy and its detection method. The LMNA gene mutation is the C-->T heterozygous mutation of the No. 877 place base in the 5th exon of LMNA gene, and causes the coded amino acid in No. 293 place to change from glutamine (Q) to termination codon (X). Its detection method includes the following steps: 1. extracting peripheral blood DNA; 2. in vitro PCR amplification of LMNA gene exon; and 3. DNA sequencing analysis. The discovery of the LMNA gene mutation can define the molecular genetic mechanism of dilated cardiomyopathy combined with conducting system abnormality further.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY +1
System and methods for detecting genetic variation
The invention provides methods, apparatuses, and compositions for high- throughput amplification sequencing of specific target sequences in one or more samples. In some aspects, barcode-tagged polynucleotides are sequenced simultaneously and sample sources are identified on the basis of barcode sequences. In some aspects, sequencing data are used to determine one or more genotypes at one or more loci comprising a causal genetic variant. In some aspects, systems and methods of detecting genetic variation are provided.
Owner:COUNSYL INC
HDAC8 gene knockout BHK-21 cell line as well as construction method and application thereof
ActiveCN112980878ANo significant effect on growth rateHigh titerSsRNA viruses positive-senseHydrolasesVaccine ProductionCell growth rate
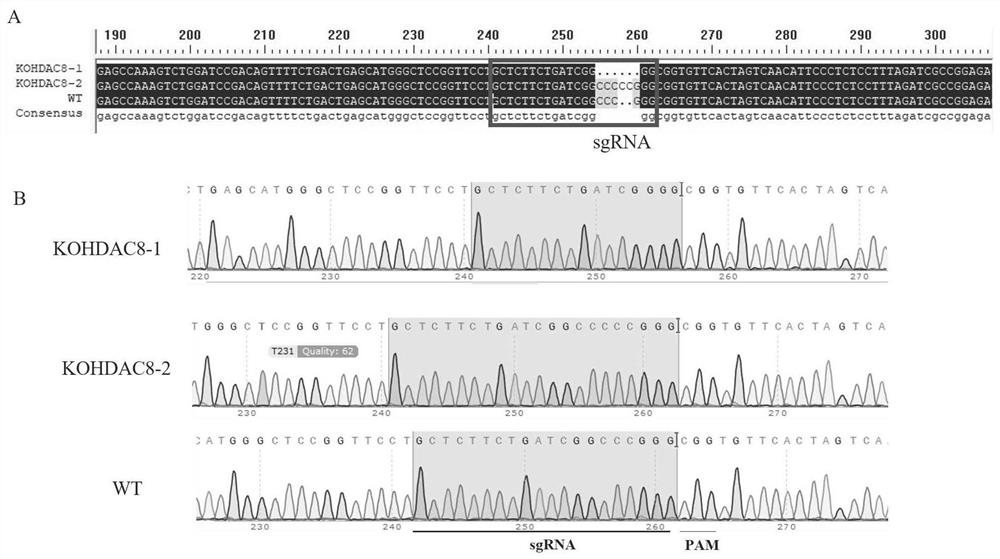
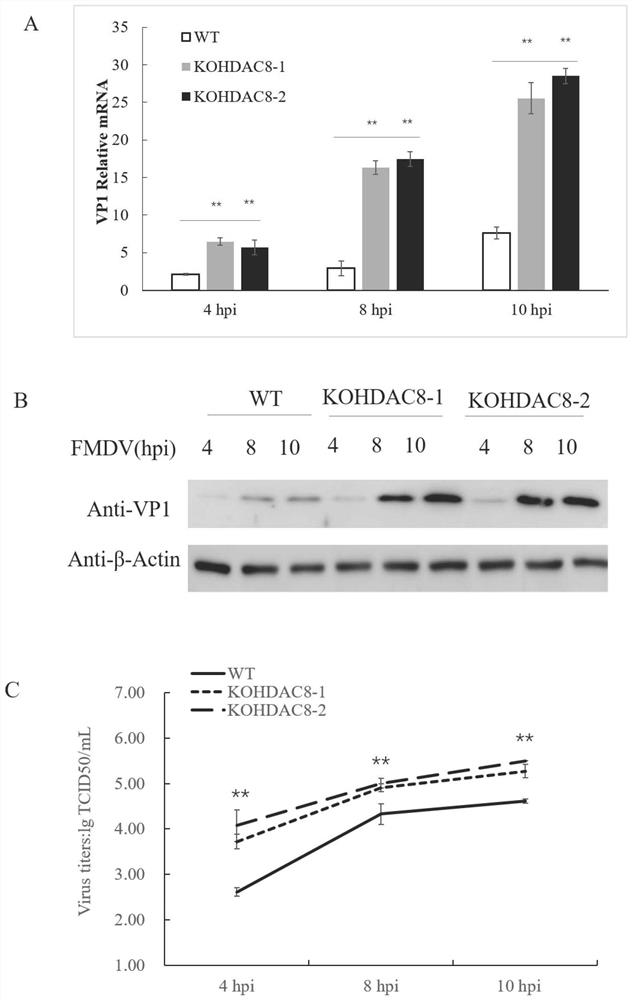
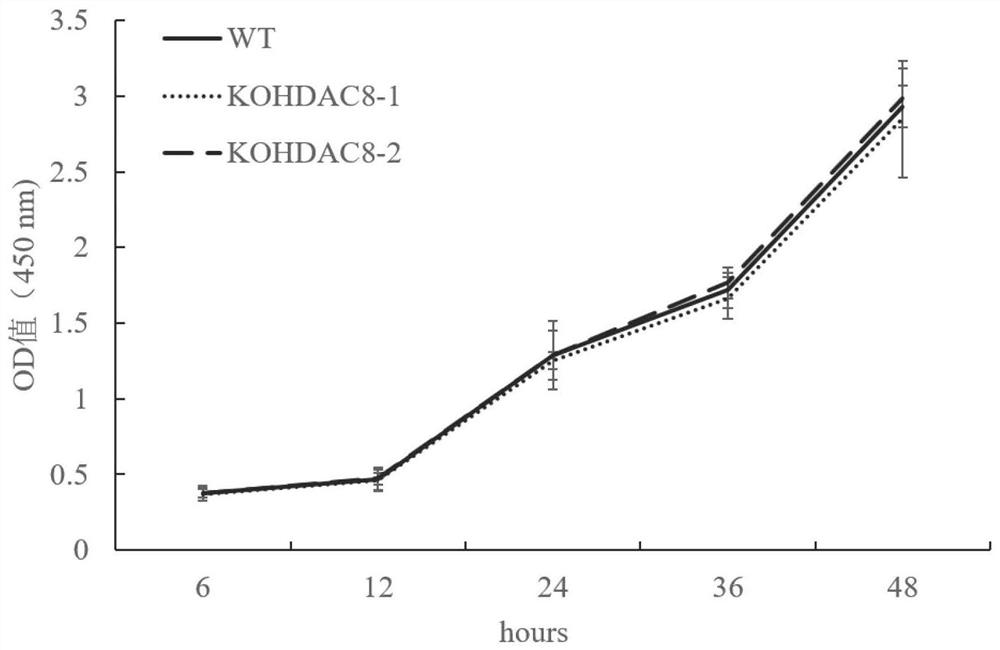
The invention discloses a construction method of an HDAC8 gene knockout BHK-21 cell line, which comprises the following steps: carrying out gene knockout on HDAC8 in a cell line BHK-21 for foot-and-mouth disease vaccine production by utilizing a CRISPR / Cas9 technology, transfecting BHK-21 cells by utilizing CRISPR plasmids for knocking out the HDAC8, and performing separation to obtain a plurality of cell clones by utilizing antibiotic screening in combination with gradient dilution and a cloning ring method. After genome DNA is extracted, PCR amplification and sequencing are carried out, and cell clones of which two HDAC8 genes are subjected to homozygous frameshift mutation are successfully identified. In the HDAC8 knockout BHK-21 cell line, the replication rate of foot-and-mouth disease virus is obviously accelerated, the final virus titer is obviously improved, and the HDAC8 knockout has no obvious influence on the cell growth rate, which shows that the HDAC8 knockout BHK-21 cell line has the prospect of being used for foot-and-mouth disease vaccine production. A foundation is laid for further knocking out HDAC8 from suspension culture type BHK-21 cells and directly applying HDAC8 to production of foot-and-mouth disease vaccines.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
Mitochondria whole genome sequencing method based on high-throughput sequencing
ActiveCN108949942AModerate lengthImprove the detection rateMicrobiological testing/measurementGenomic sequencingDetection rate
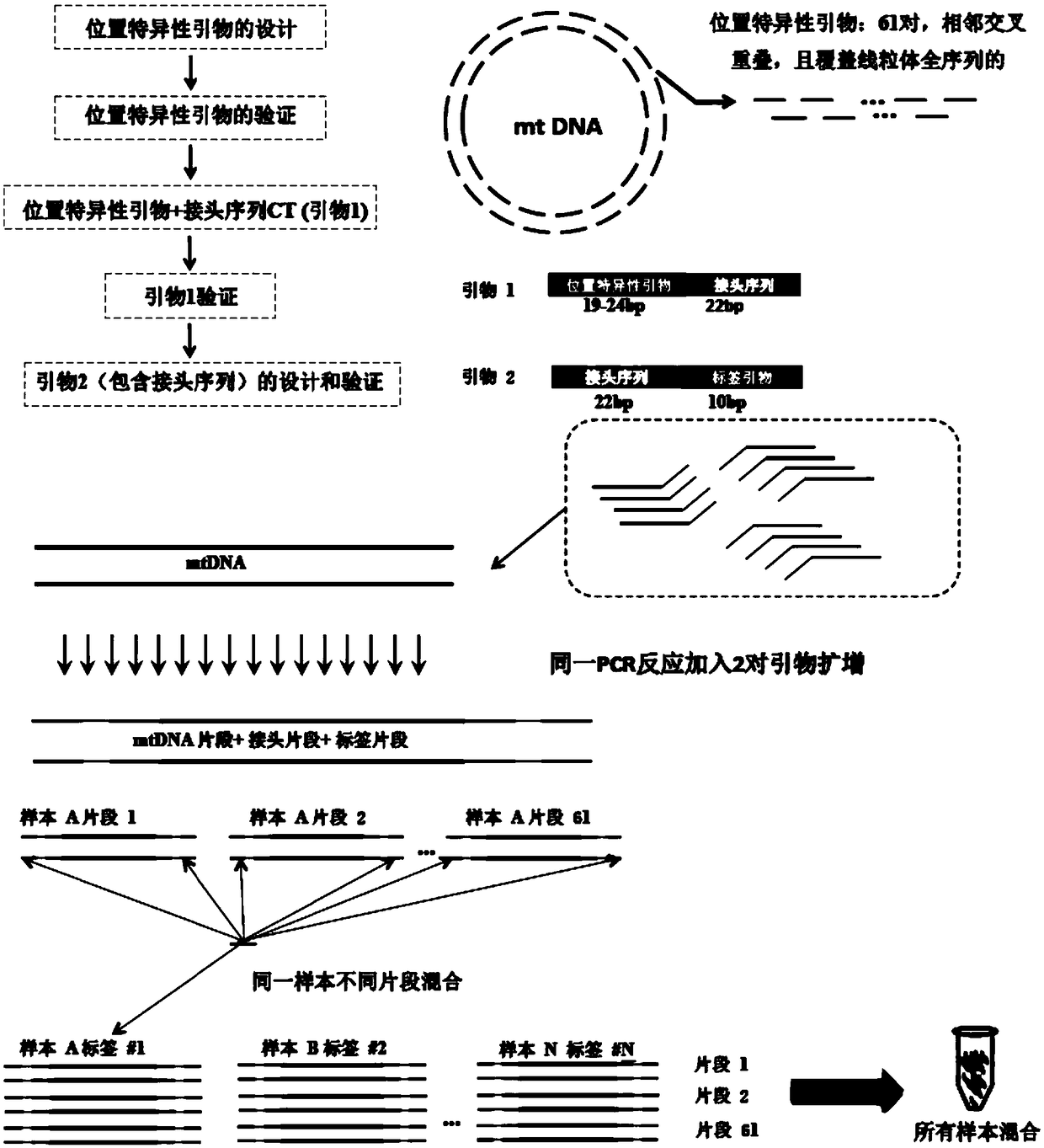
The invention discloses a simple, economical and accurate mitochondria whole genome sequencing barcode amplification sequencing method based on second generation sequencing. Two groups of matched primers are designed, wherein the first group of primers is composed of 61 pairs of target fragment primer sequences and primer joint sequences which are overlaid and cover the mitochondria whole genome.The second group of primers is composed of positive and negative label primers which comprise the joint sequences and are matched two by two for marking samples of different sources. By matching the two pairs of primers in use, two pairs of the primers are added in a primary PCR reaction for amplification to obtain amplicons which have marks, are moderate in length and comprise mitochondria wholegenome. The amplicons are mixed and used for establishing a library and sequencing directly to achieve multi-sample mixed sequencing. Sequencing data identifies mark sequences at the head and tail ends at the same time by means of applying a bioinformatics assembling technology to different sequences of different sources to obtain all mitochondria genome sequence information of all samples. By employing the method, the sequencing experiment steps are simplified, the sequencing cost is lowered greatly, and the detection rate of low-frequency mutation is reduced, thereby providing probability for human mitochondria whole genome associated researches.
Owner:ZHEJIANG UNIV +1
Primer group and method for rapidly identifying influenza A virus subtypes through combination of DNA barcodes and second-generation high-throughput sequencing
InactiveCN105255864AQuick judgmentRapid Influenza Typing DiagnosticsMicrobiological testing/measurementMicroorganism based processesPolymerase chain reactionAmplification/Sequencing
The invention discloses a primer group and a method for rapidly identifying influenza A virus subtypes through combination of DNA barcodes and second-generation high-throughput sequencing. The method comprises steps as follows: (1), genome extraction and inverse transcription; (2), primer design and PCR (polymerase chain reaction) amplification; (3), Ion Torrent sequencing; (4), bioinformatic analysis; and (5), phylogenetic analysis and genetic distance calculation. According to the primer group and the method, sequences of influenza A virus materials of 16 kinds of different areas and subtypes are analyzed, and the DNA barcodes which can be used for distinguishing the subtypes are found out; with the adoption of the technical idea of the DNA barcodes and a traditional RT-PCR (reverse transcription-polymerase chain reaction) detection method, whether the influenza virus exists can be rapidly judged with preference of a touch-down program, the subtype and a pathotype of the influenza A virus can be measured simultaneously, and one simpler and more rapid influenza typical diagnosis program is provided.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
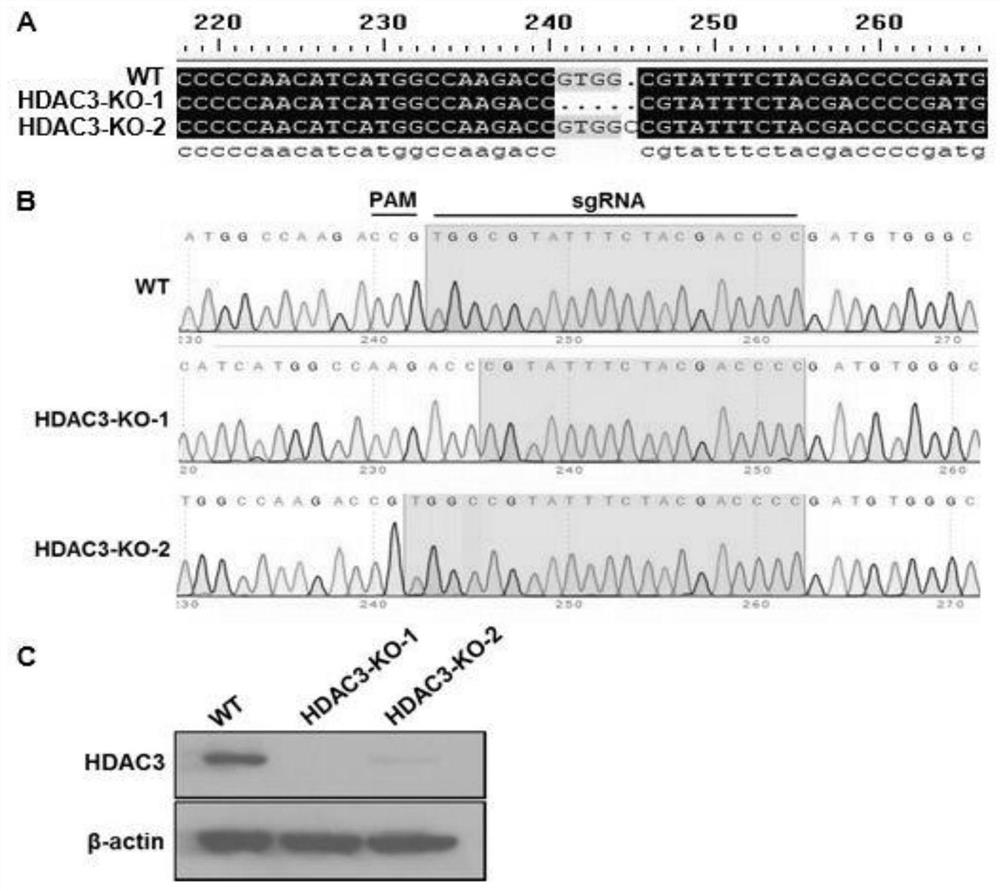
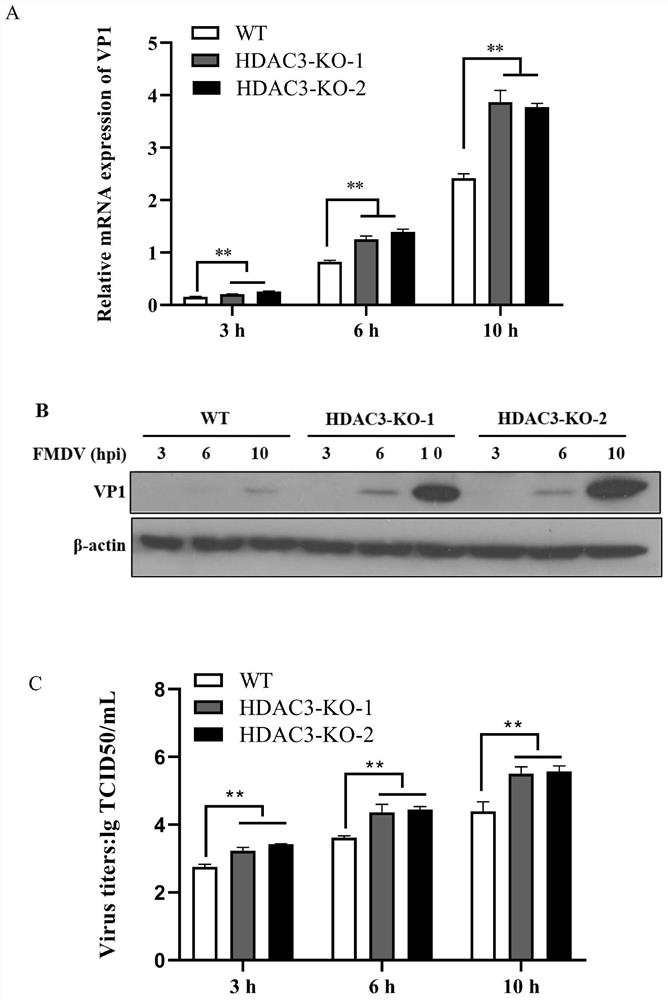
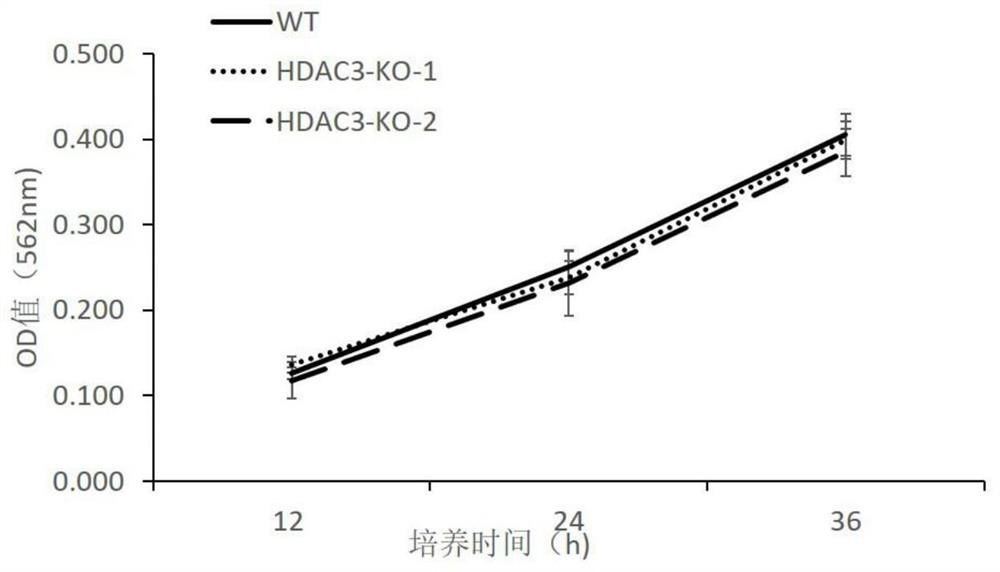
HDAC3 gene knockout BHK-21 cell line as well as construction method and application thereof
ActiveCN112852745ANo significant effect on growth rateHigh titerHydrolasesMicrobiological testing/measurementVaccine ProductionCell growth rate
The invention discloses a construction method of an HDAC3 gene knockout BHK-21 cell line, which comprises the following steps: carrying out gene knockout on HDAC3 in a cell line BHK-21 for foot-and-mouth disease vaccine production by utilizing a CRISPR / Cas9 technology, transfecting BHK-21 cells by utilizing CRISPR plasmids for knocking out the HDAC3, and separating the cells to obtain a plurality of cell clones by utilizing antibiotic screening in combination with gradient dilution and a cloning ring method; extracting the genome DNA, performing PCR amplification and sequencing, and successfully identifying the cell clones of which two HDAC3 genes are subjected to homozygous frameshift mutation. In the HDAC3 knockout BHK-21 cell line, the replication rate of foot-and-mouth disease virus is obviously accelerated, the final virus titer is obviously improved, and the HDAC3 knockout has no obvious influence on the cell growth rate, so that the HDAC3 knockout BHK-21 cell line has the prospect of being used for foot-and-mouth disease vaccine production. Therefore, the foundation is laid for further knocking out HDAC3 from suspension culture type BHK-21 cells and directly applying HDAC3 to foot-and-mouth disease vaccine production.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
Spider mitochondria COIII gene complete sequence amplification primer and identification method thereof
ActiveCN104450699AAmplification efficient and specificAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationGenetic resourcesAmplification/Sequencing
The invention discloses a spider mitochondria COIII gene complete sequence amplification primer and a method for rapidly identifying species of spiders by utilizing complete sequence sequencing information. The method comprises the following steps: extracting total DNA of spider genomes, designing the COIII gene complete sequence amplification primer, performing PCR amplification, sequencing and rapidly identifying the species of spiders based on the gene complete sequence. With the adoption of the universal amplification primer disclosed by the invention, the complete sequence of the multiple mitochondria COIII genes of spiders can be rapidly and efficiently amplified, a powerful tool is provided for researches such as rapid identification of the species of spiders, population diversity analysis and genetic resource conservation and utilization, and a foundation is laid for spider mitochondria whole genome sequencing and analysis and research on different taxonomic category system evolutions.
Owner:CHINA JILIANG UNIV
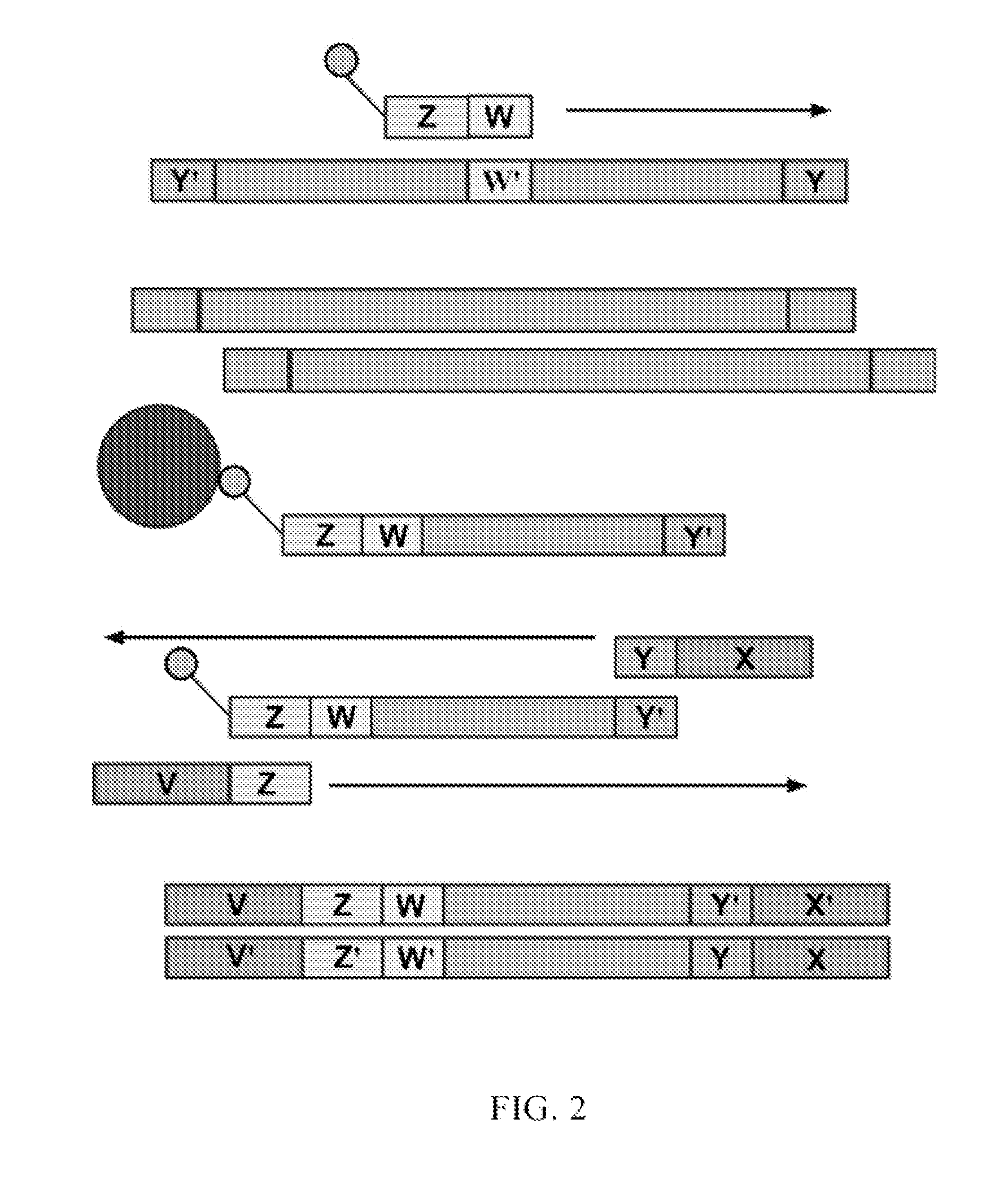
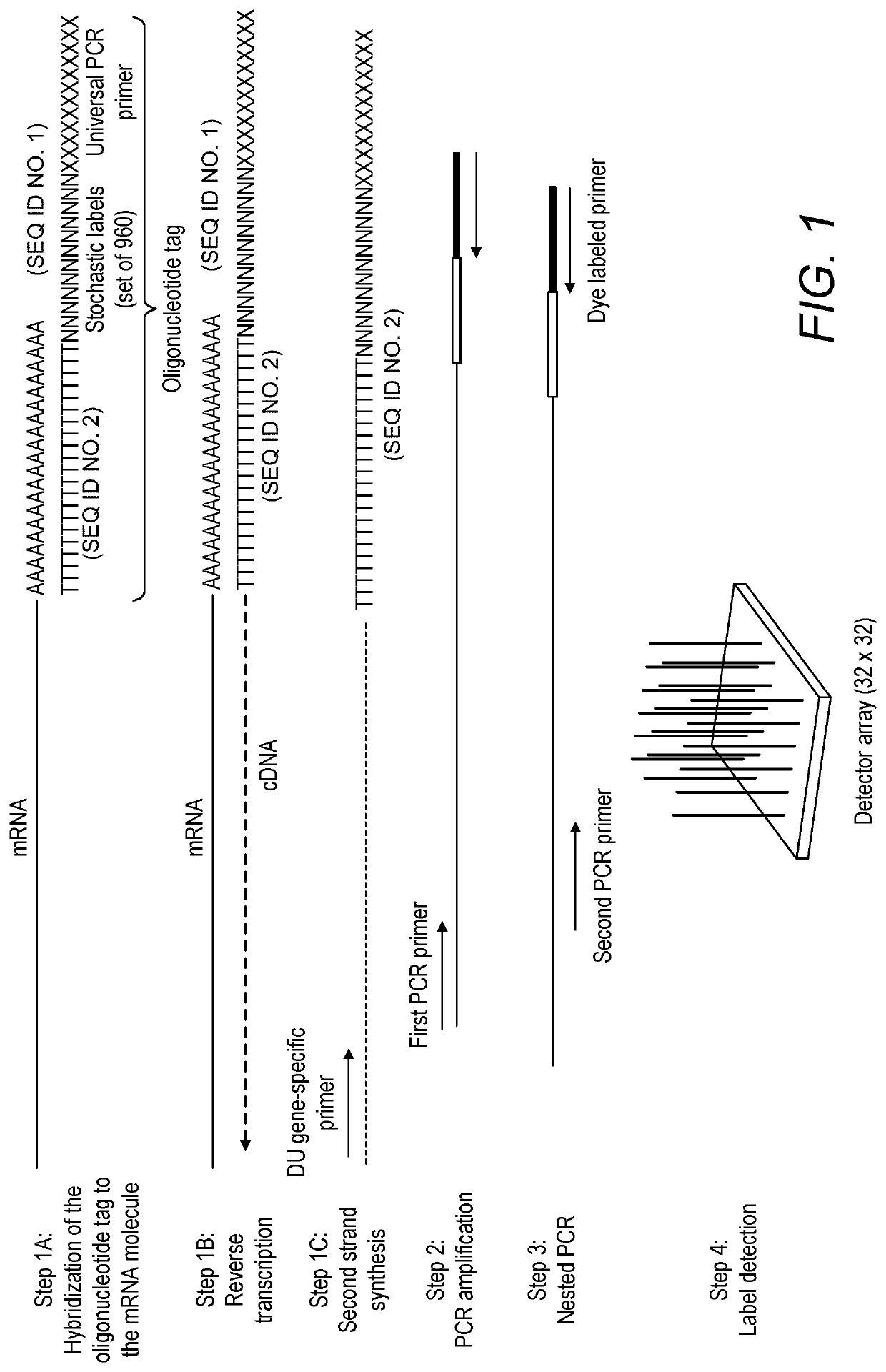
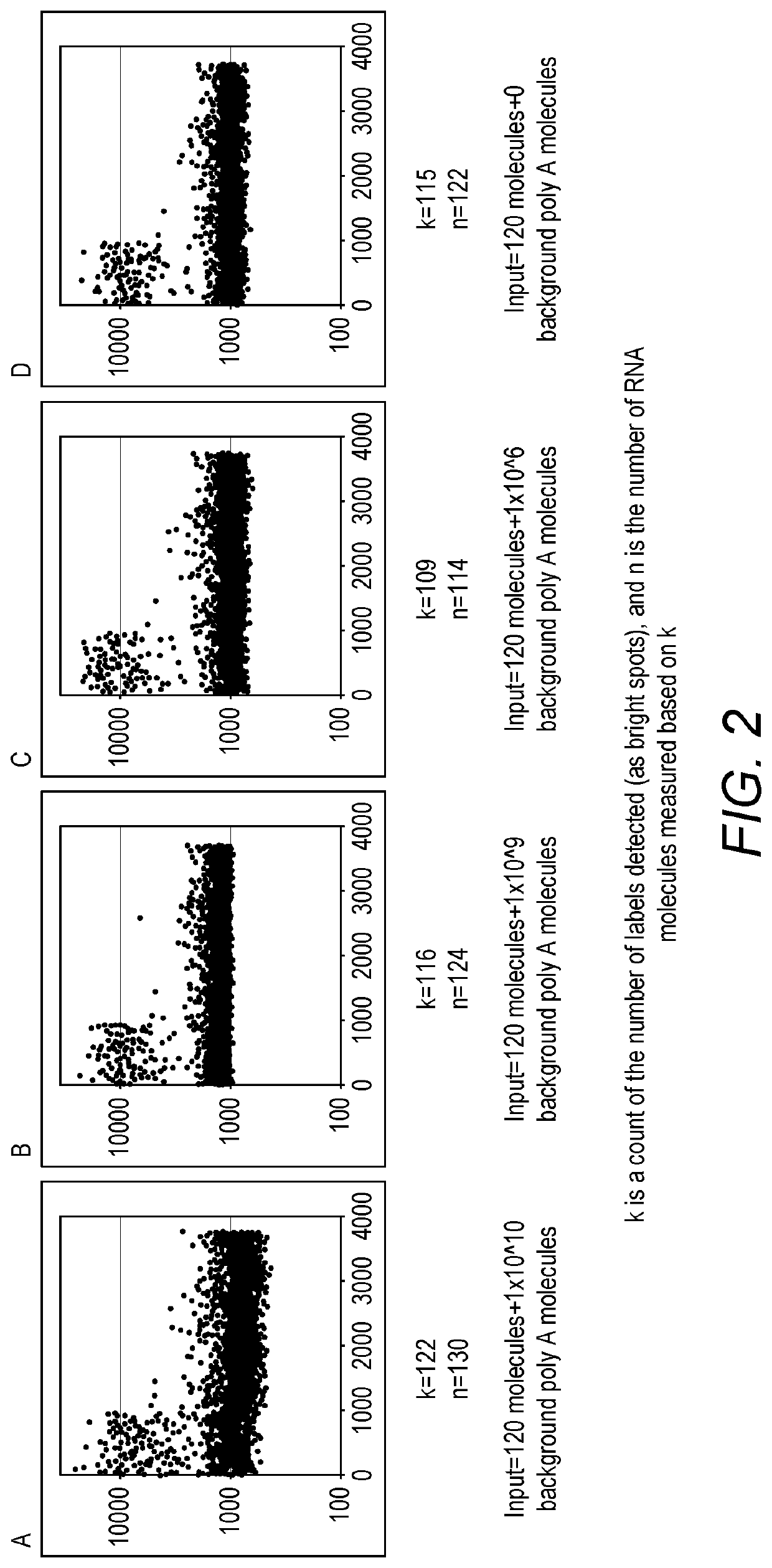
Compositions and kits for molecular counting
Methods, kits and systems are disclosed for analyzing one or more molecules in a sample. Analyzing the one or more molecules may comprise quantitation of the one or more molecules. Individual molecules may quantitated by PCR, arrays, beads, emulsions, droplets, or sequencing. Quantitation of individual molecules may further comprise stochastic labeling of the one or more molecules with a plurality of oligonucleotide tags to produce one or more stochastically labeled molecules. The methods may further comprise amplifying, sequencing, detecting, and / or quantifying the stochastically labeled molecules. The molecules may be DNA, RNA and / or proteins.
Owner:BECTON DICKINSON & CO
Method for detecting probiotics in milk powder by Illumina Miseq sequencing platform
InactiveCN106701912AAvoid untestabilityCredibility of sequencing resultsMicrobiological testing/measurementBiotechnologyCentrifugation
The invention relates to the technical field of milk powder detection, and particularly relates to a method for detecting probiotics in milk powder by an Illumina Miseq sequencing platform. The method comprises the following steps of: (1) extraction of DNA (deoxyribonucleic acid) of a milk powder genome, i.e., after brewing the milk powder, carrying out primary centrifugation to take supernatant, then carrying out secondary centrifugation to take precipitates, adding buffer solution to oscillate, and then after adding lysozyme to perform a reaction, carrying out extraction of the DNA of the genome; (2) PCR (Polymerase Chain Reaction) amplification, i.e., aiming at v1 to v3 regions of 16s rDNA of a bacterium, selecting a specific primer to carry out PCR amplification; (3) Illumina Miseq sequencing, i.e., carrying out Illumina Miseq sequencing on a PCR product. The Illumina Miseq sequencing method effectively avoids the defects of low flux, complex operation, low accuracy and the like by analysis, not only can simultaneous sequencing on a plurality of variable regions of a plurality of samples be implemented, but also both a sequencing speed and a sequencing flux are further, and a detection result is high in credibility.
Owner:PLANTS & ANIMALS & FOOD TESTING QUARANTINE TECH CENT SHANGHAI ENTRY EXIT INSPECTION & QUARANTINE BUREAU +1
Mycobacterium tuberculosis multi-line drug resistance gene identification method and device
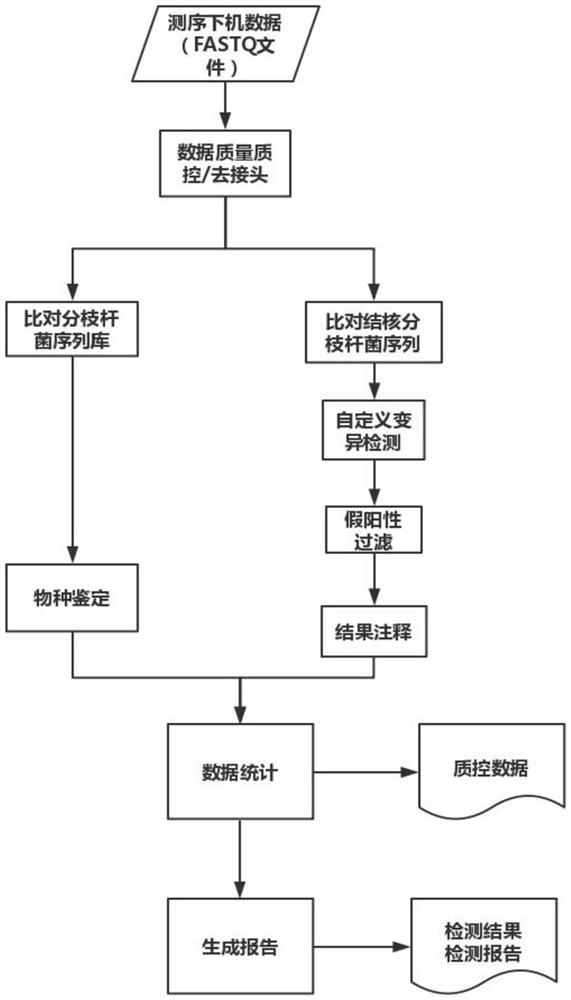
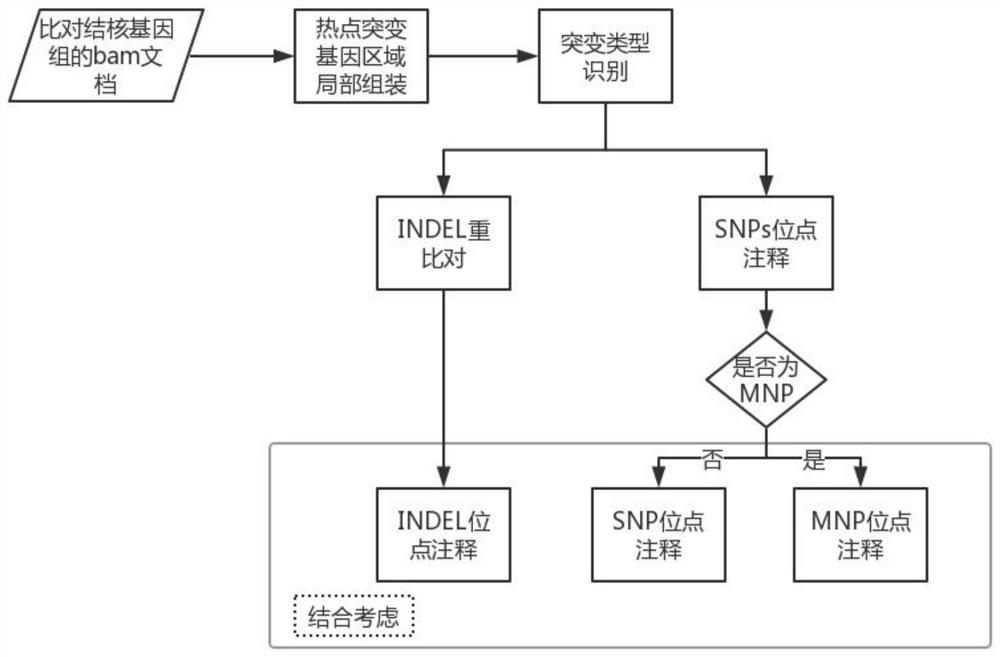
PendingCN113621716AImprove detection accuracyHelp identifyMicrobiological testing/measurementProteomicsGenomic sequencingGenetic Databases
The invention relates to a mycobacterium tuberculosis multi-line drug resistance gene identification method and device. The method comprises the following steps: acquiring a whole genome sequence obtained by metagenome sequencing of a sample or a sequence of a mycobacterium tuberculosis drug-resistant target region obtained on the basis of targeted amplification sequencing, and carrying out data quality control; comparing the sequence subjected to the data quality control to a mycobacterium tuberculosis genome, sequencing and screening out a comparison result meeting the quality requirement; carrying out local assembly and variation detection on the sequences of the gene regions where the variation sites of which the comparison positions are located in the mycobacterium tuberculosis drug resistance gene database are located; annotating a variation detection result to the mycobacterium tuberculosis drug resistance gene database according to the positions and mutation types of variation sites on the genome; and outputting a detection result of each drug resistance related mutation sites in the mycobacterium tuberculosis drug resistance gene database based on an annotation result. According to the invention, multiple drugs and corresponding drug resistance sites can be covered in one detection, and the detection precision is high.
Owner:深圳华大因源医药科技有限公司 +2
Screening method of gene editing site homozygote without transgenosis
InactiveCN112609019ADetection speedThe throughput of the library building process is lowMicrobiological testing/measurementBiotechnologyGenome editing
The invention relates to the technical field of gene editing, and particularly discloses a screening method of a gene editing site homozygote without transgenosis. Different from the existing screening method, the screening method disclosed by the invention has the advantages that high-throughput screening is carried out on T1-generation individuals subjected to gene editing of a target gene by utilizing a KASP marker primer designed for a transgenic vector, individuals containing transgenic components are removed, then amplification sequencing of a gene editing target section is carried out on non-transgenic individuals, and individuals homozygous at a target gene editing site are screened. The method can be applied to high-throughput identification and screening of various animal and plant gene editing offspring populations, and particularly to large-population editing offsprings, for example, tens of thousands to hundreds of thousands of gene editing individuals need to be identified and screened when a gene editing mutant library is constructed, and the screening efficiency can be remarkably improved by applying the technical scheme provided by the invention.
Owner:CHINA TOBACCO HUNAN INDAL CORP +1
Polyene macrolides compound, preparation and application thereof
InactiveCN101709058AEnhanced inhibitory effectHas antitumor activityAntibacterial agentsOrganic active ingredientsMetaboliteGene cluster
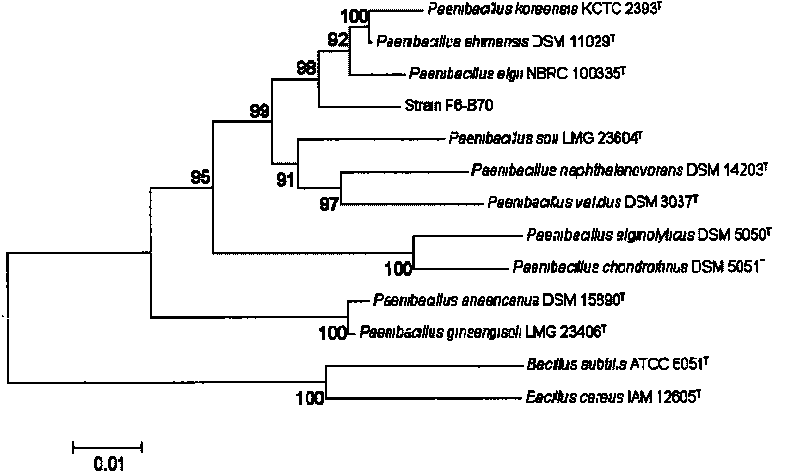
The invention provides a new compound having medicinal value, a preparation method thereof and a new bacterial strain which can generate the compound. The compound with formula (I) is chosen and prepared by rapidly choosing bacillus from soil sample with specific primer, using 16Ss rRNA gene to preliminarily identify and separate bacterial strain, amplifying, sequencing and comparing partial sequence of NRPSs / PKSs gene cluster after choosing potential new resources, analyzing the diversity of metabolite thereof, acquiring bacterial strain F6-B70 which contains new NRPSs / PKSs gene cluster and has new potential after choosing a large amount, fermenting in a fermentation medium, extracting active substance from bacterial cell and fermenting liquid, and then separating and purifying by using multiple purification techniques. The compound is a polyene macrolides compound which has an obvious function of restraining drug-resisting staphylococcus and an activity of resisting tumor.
Owner:ZHEJIANG UNIV +1
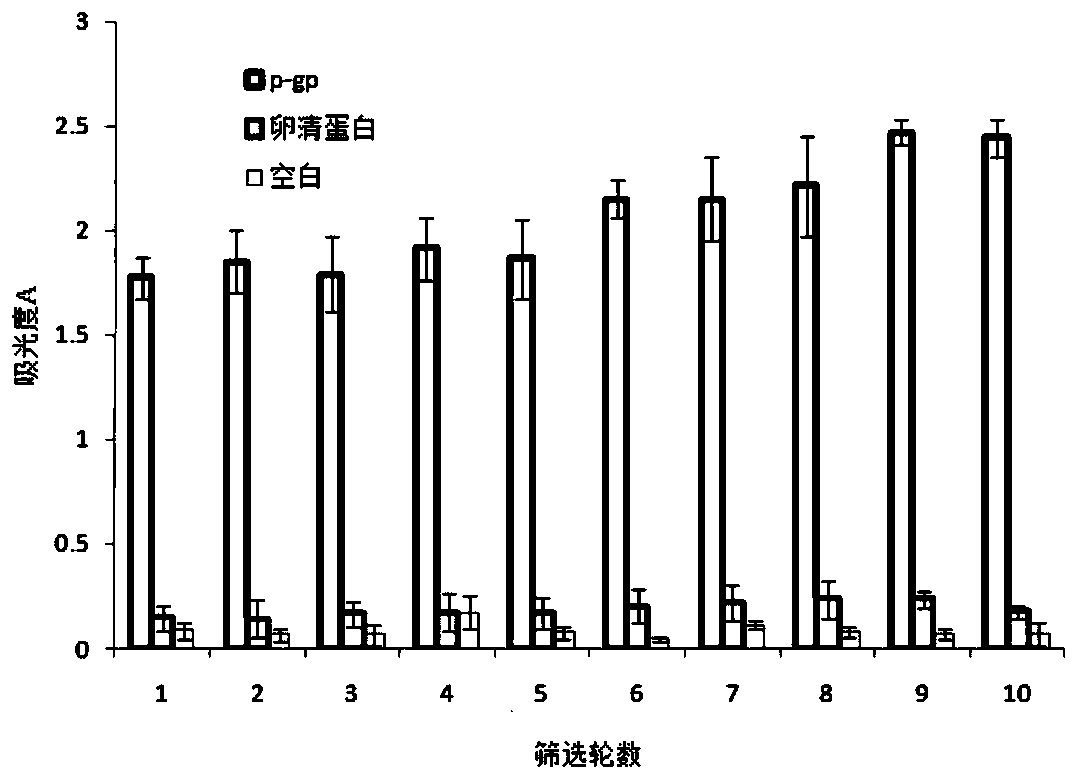
Nucleic acid aptamer specifically binding to P-glycoprotein, and preparation method and application of nucleic acid aptamer
ActiveCN111304208AInhibitory functionStrong specificityBiological testingDNA preparationAptamerChemical synthesis
The invention relates to a nucleic acid aptamer specifically binding to P-glycoprotein (P-gp), and a preparation method and application of the nucleic acid aptamer, and belongs to the technical fieldof biology. The nucleotide sequence of the nucleic acid aptamer is selected from SEQ ID No.1 to SEQ ID No.14 in sequence tables; the nucleic acid aptamer is prepared by screening, amplifying and sequencing an original random oligonucleotide library through an SELEX technology in combination with surface modified magnetic beads; the nucleic acid aptamer is small in molecular weight, can be chemically synthesized, and is low in cost; the affinity and the specificity are high; marking is facilitated; and the repeatability and the stability are high, and storage is easy. The nucleic acid aptamer can be made into a fluorescent molecular probe, and fluorescent visualization and quantification of P-gp on in-vitro tissues and cells of an organism are realized; and the nucleic acid aptamer can be specifically bound with P-gp, inhibits the function of P-gp, and becomes a potential nucleic acid type inhibitor of the function of P-gp.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
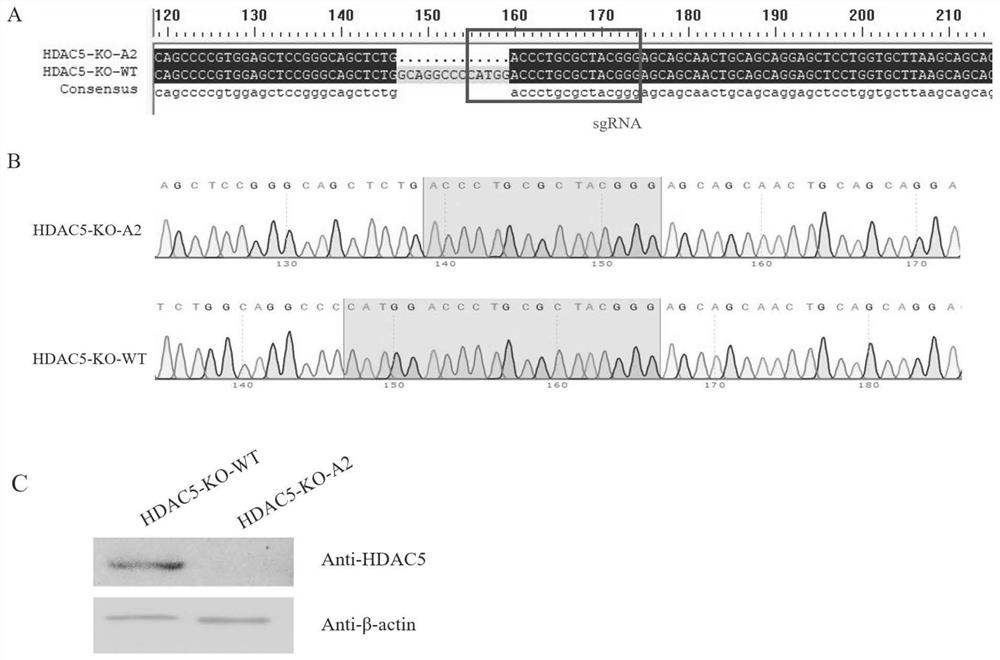
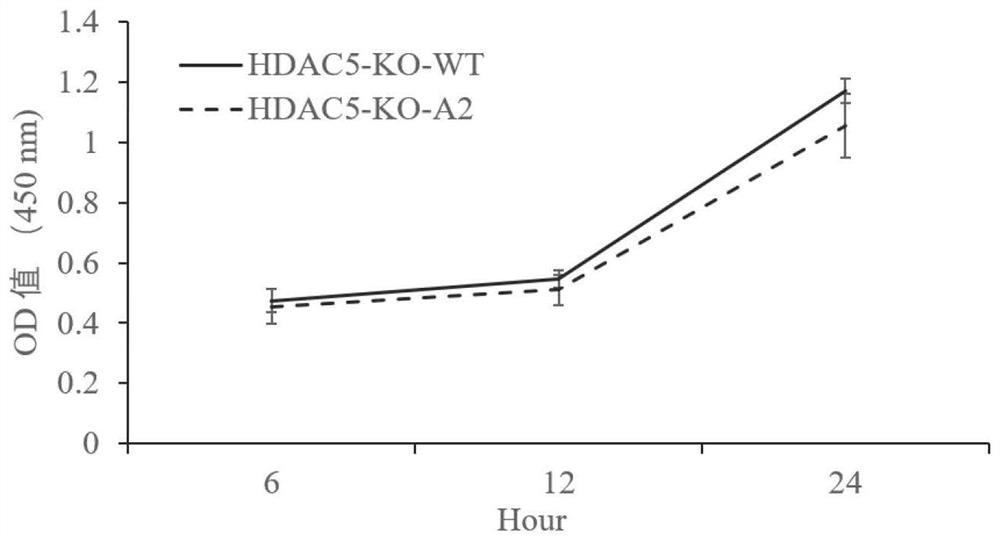
HDAC5 gene knockout BHK-21 cell line as well as construction method and application thereof
ActiveCN112852874ANo significant effect on growth rateHigh titerSsRNA viruses positive-senseHydrolasesVaccine ProductionCell growth rate
The invention discloses a construction method of an HDAC5 gene knockout BHK-21 cell line, which comprises the following steps: carrying out gene knockout on HDAC5 in a cell line BHK-21 for foot-and-mouth disease vaccine production by utilizing a CRISPR / Cas9 technology, transfecting BHK-21 cells by utilizing CRISPR plasmids for knocking out the HDAC5, and separating the cells to obtain a plurality of cell clones by utilizing antibiotic screening in combination with gradient dilution and a cloning ring method; extracting the genome DNA, performing PCR amplification and sequencing and successfully identifying the cell cloning of which one HDAC5 gene is subjected to homozygous frameshift mutation, wherein the HDAC5-KO-A2 has deletion of 13 basic groups at a Cas9 predetermined cutting position. In the HDAC5 knockout BHK-21 cell line, the replication rate of foot-and-mouth disease virus is obviously accelerated, the final virus titer is obviously improved, and the HDAC5 knockout has no obvious influence on the cell growth rate, so that the HDAC5 knockout BHK-21 cell line has the prospect of being used for foot-and-mouth disease vaccine production. Therefore, the foundation is laid for further knocking out HDAC5 from suspension culture type BHK-21 cells and directly applying HDAC5 to production of foot-and-mouth disease vaccines.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
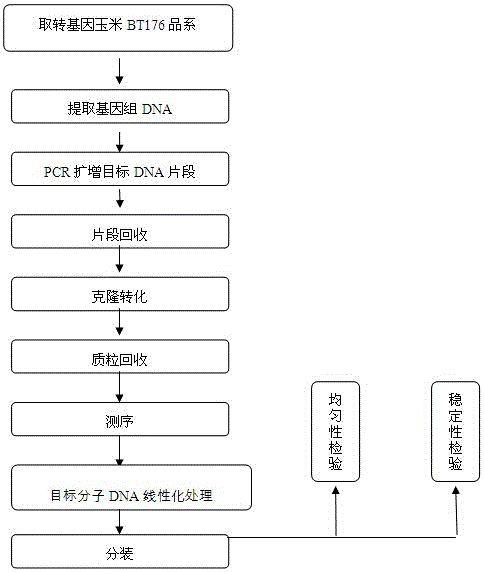
Transgenic maize BT176 nucleic acid standard sample and preparation method thereof
InactiveCN105695595ANo longer biologically activeBiologically activeMicrobiological testing/measurementVector-based foreign material introductionBiotechnologyPositive control
The invention belongs to the field of biotechnology, in particular to a standard sample of transgenic corn BT176 nucleic acid and a preparation method thereof. Take the mature transgenic corn BT176 strain as raw material, extract the total DNA, perform specific PCR amplification and sequencing, and prepare the plasmid. The transgenic corn BT176 nucleic acid standard sample contains the specific gene of the transgenic corn BT176 strain and the endogenous zein Gene. The nucleic acid standard sample prepared by the method of the present invention no longer has biological activity. In the process of construction, amplification and analysis and identification, the laboratory environment is monitored and analyzed, and there is no problem of biological contamination and infection, and the positive control of molecular biology is solved. The source is problematic, and the sample is stable and free of pollution. The preparation of the standard sample of the present invention is completed, which is helpful for comprehensive and in-depth research on the preparation technology and stability assurance technology of the molecular DNA standard sample for the detection of genetically modified components, and actively develops the development of the standard sample for the detection of genetically modified products in my country to fill the gap in the measurement field. has important practical significance.
Owner:曹际娟 +3
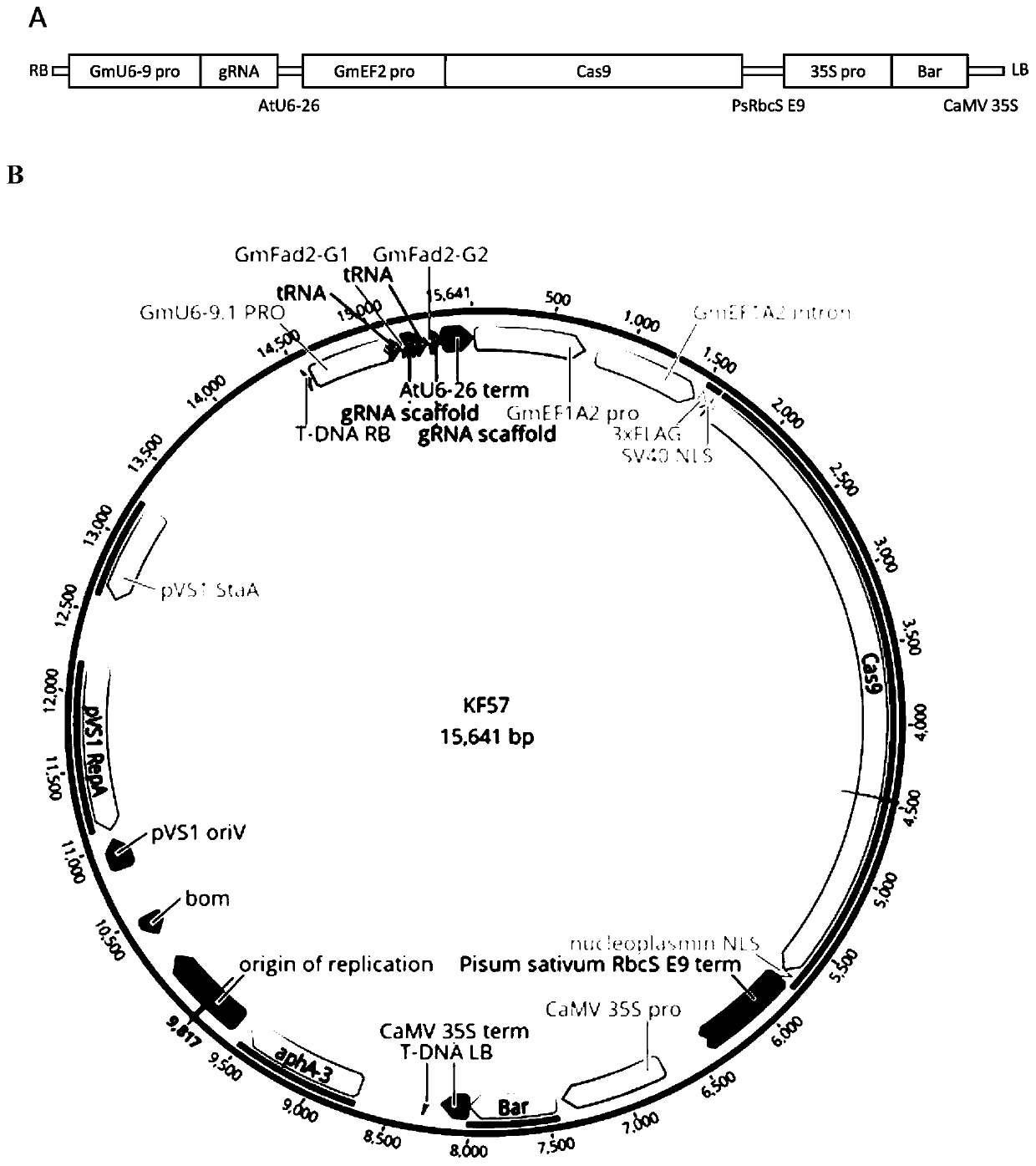
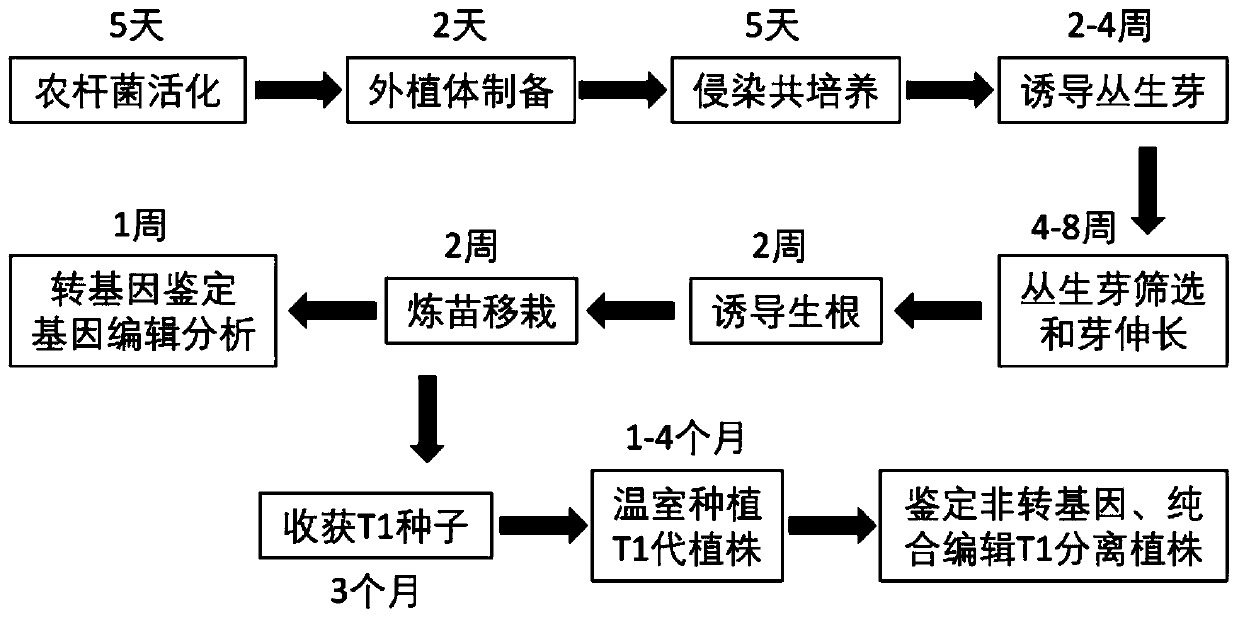
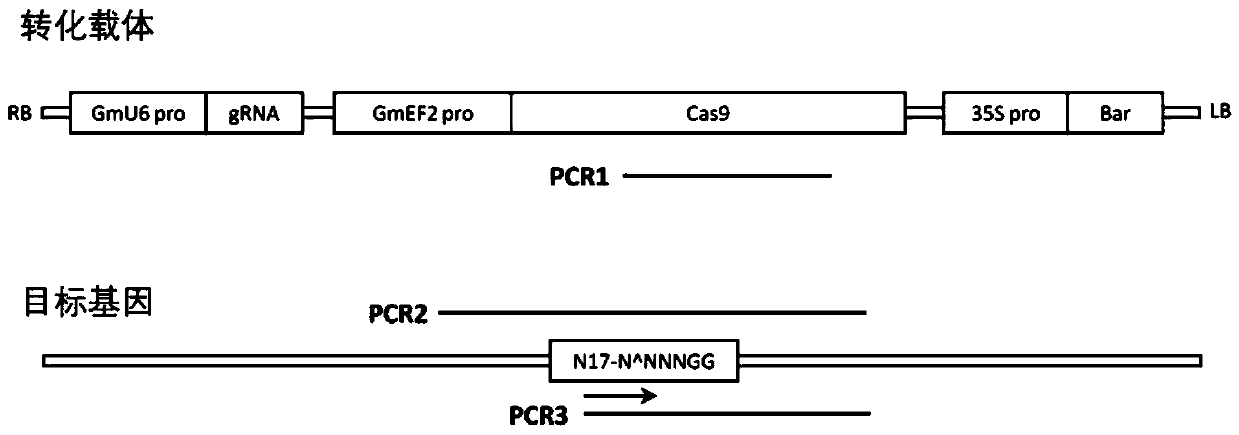
Genetic transformation, gene editing and analysis methods of main soybean varieties
PendingCN111073904ARapid genetic transformationEfficient genetic transformationPlant peptidesVector-based foreign material introductionVector (molecular biology)Promoter
The invention belongs to the field of crops, and particularly relates to genetic transformation, gene editing and analysis methods of main soybean varieties. The invention discloses a gene editing andtransforming vector which is based on a CRISPR / Cas9 system and takes an agrobacterium transforming vector pCambia3301 as a skeleton, and a promoter is designed for improving expression of Cas9 and gRNA. According to the invention, rapid and effective genetic transformation of the main soybean varieties is realized for the first time, and a simple and rapid PCR amplification sequencing analysis method for identifying the gene editing effect is developed. Through genetic separation and screening by molecular biology analysis technology, gene editing offspring plants which do not carry exogenousgenes of the soybean varieties for production are successfully obtained, and editing sites are stably inherited.
Owner:北大荒垦丰种业股份有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com