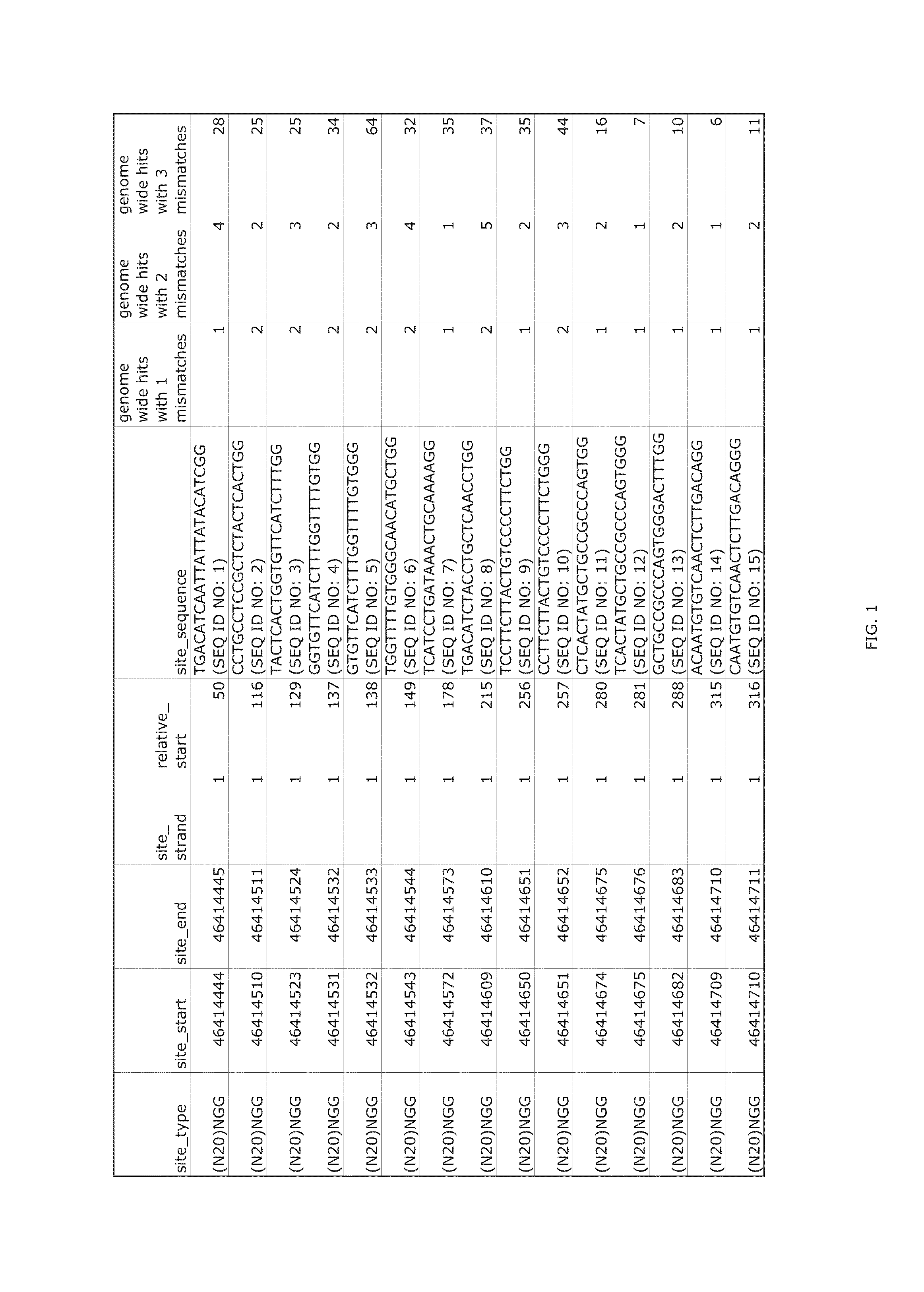
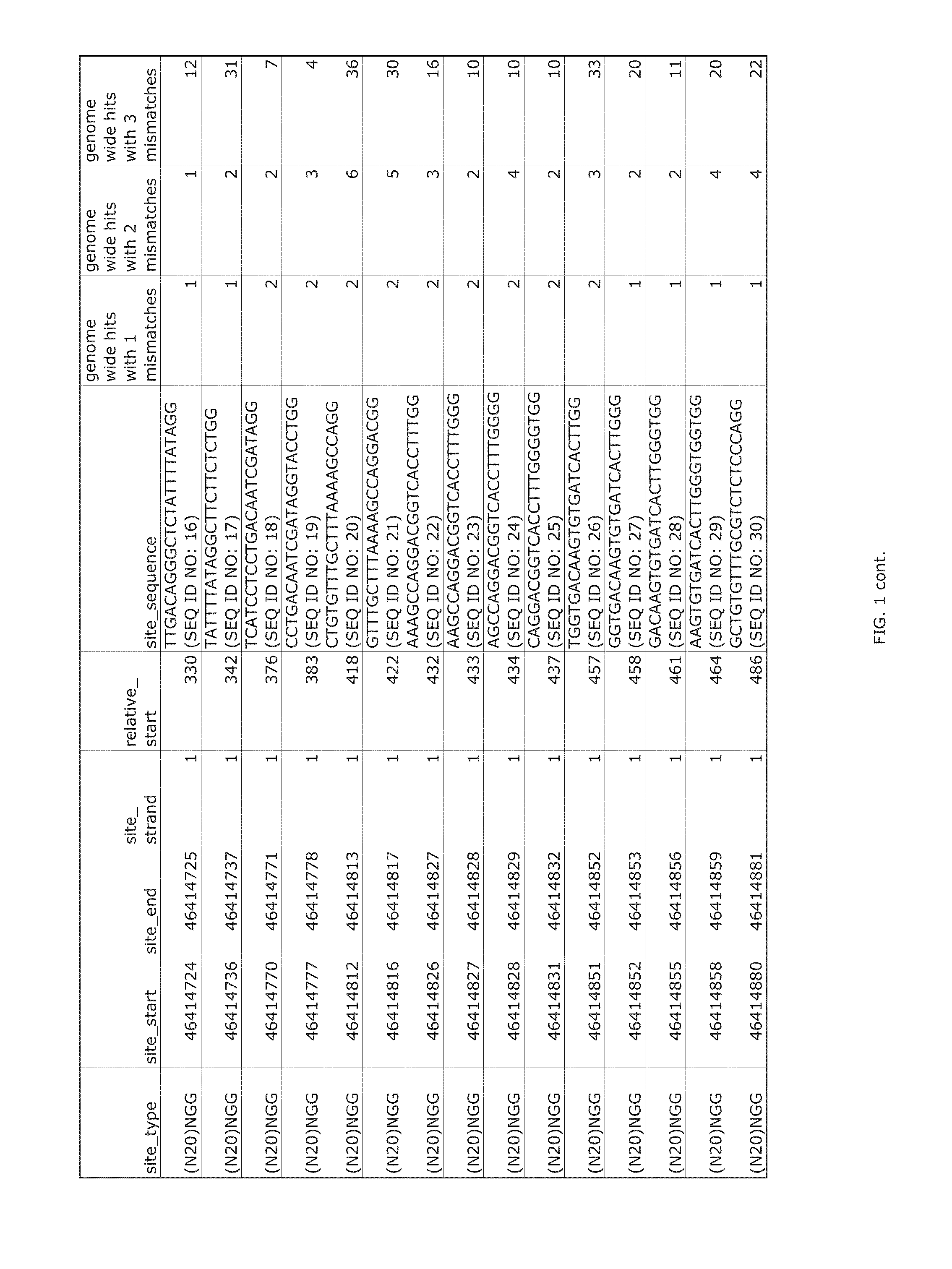
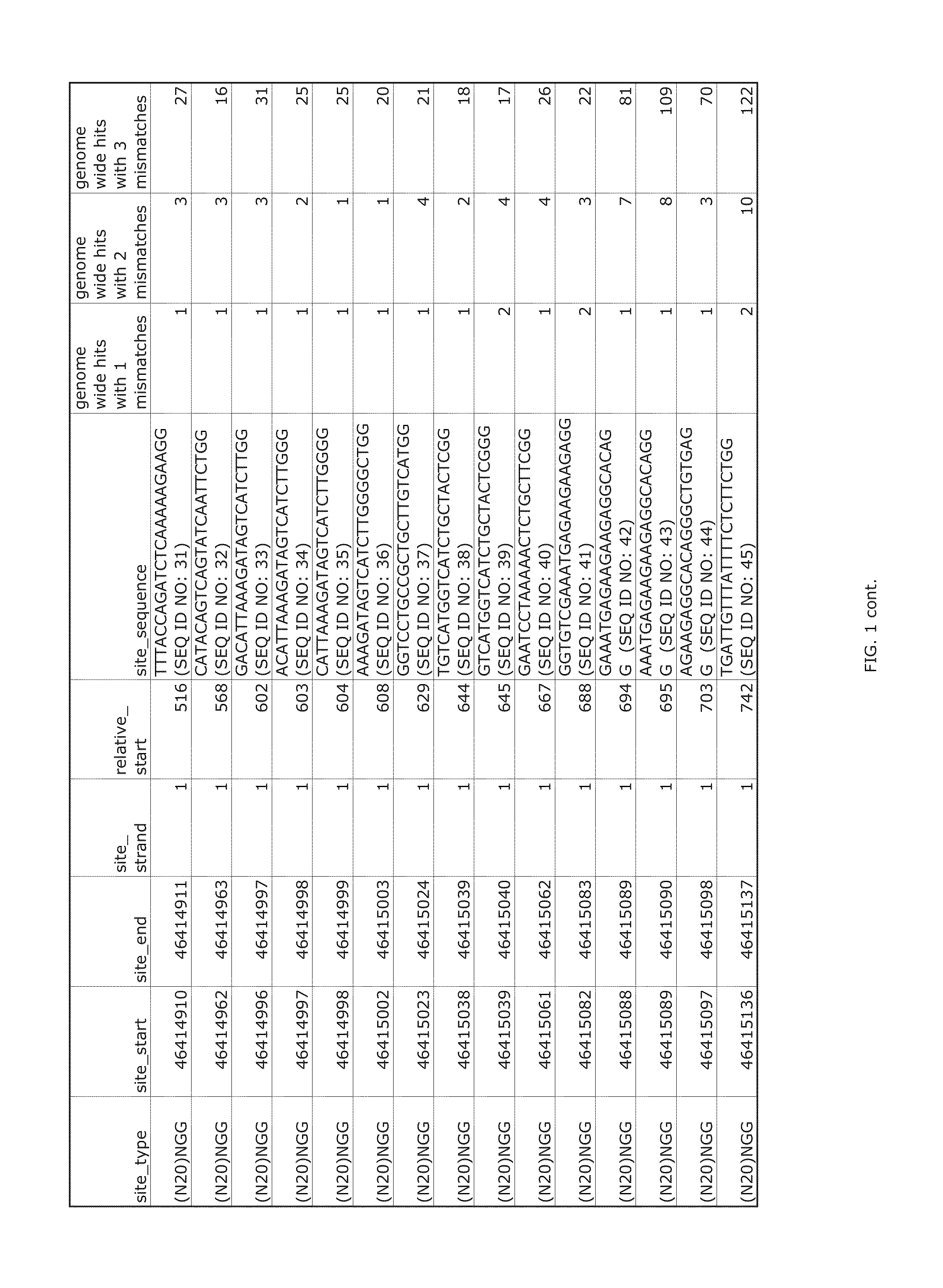
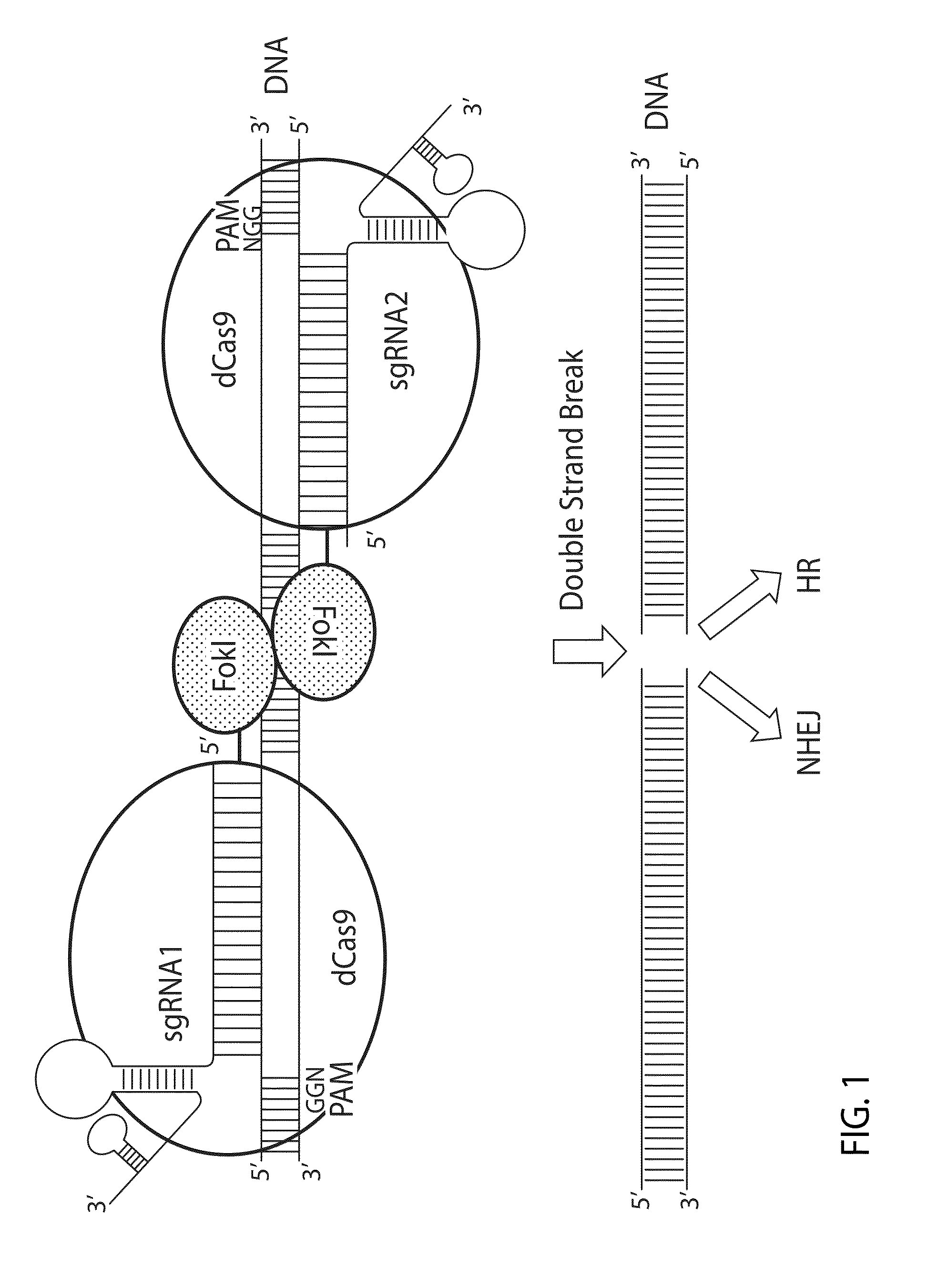
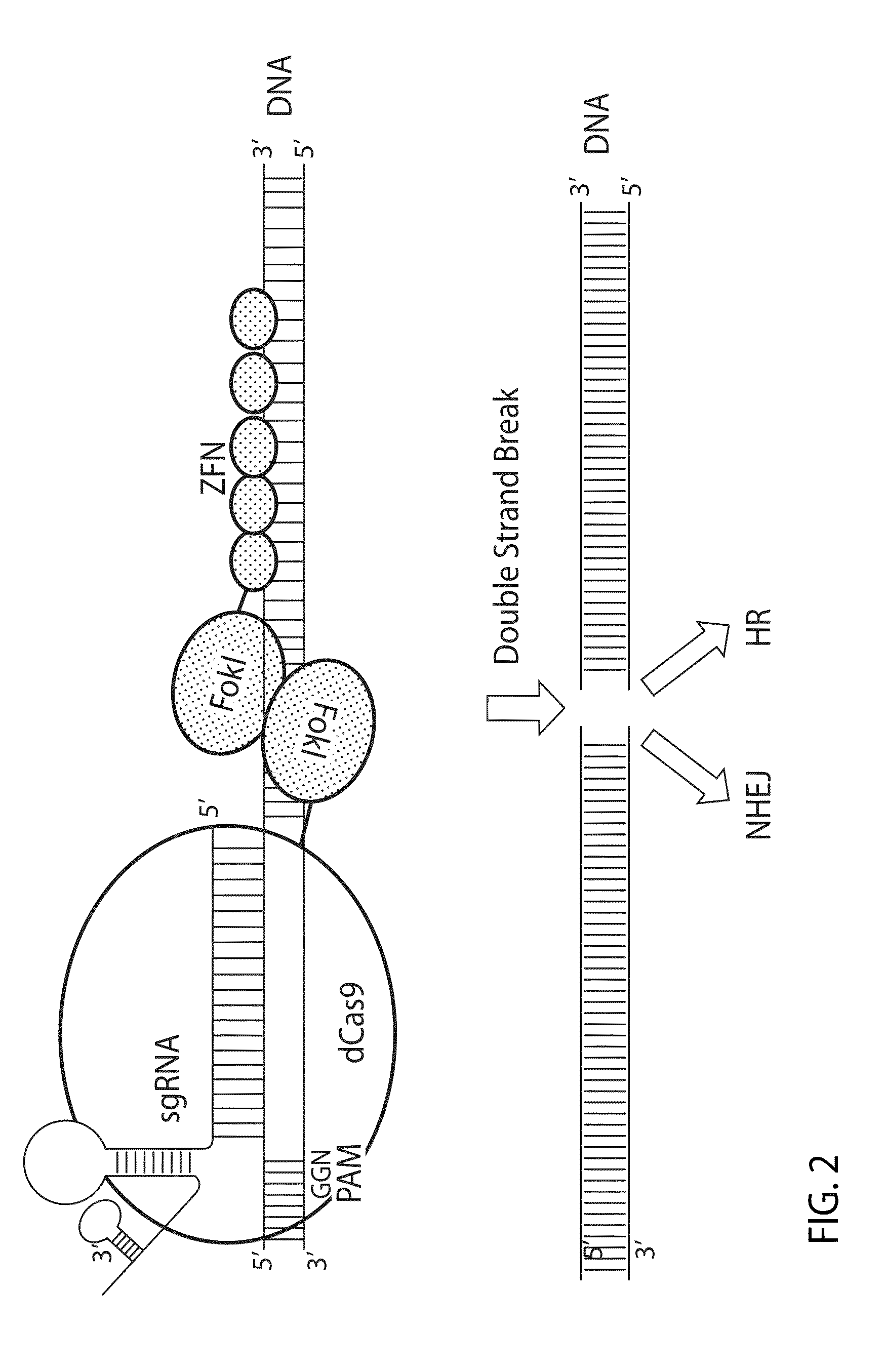
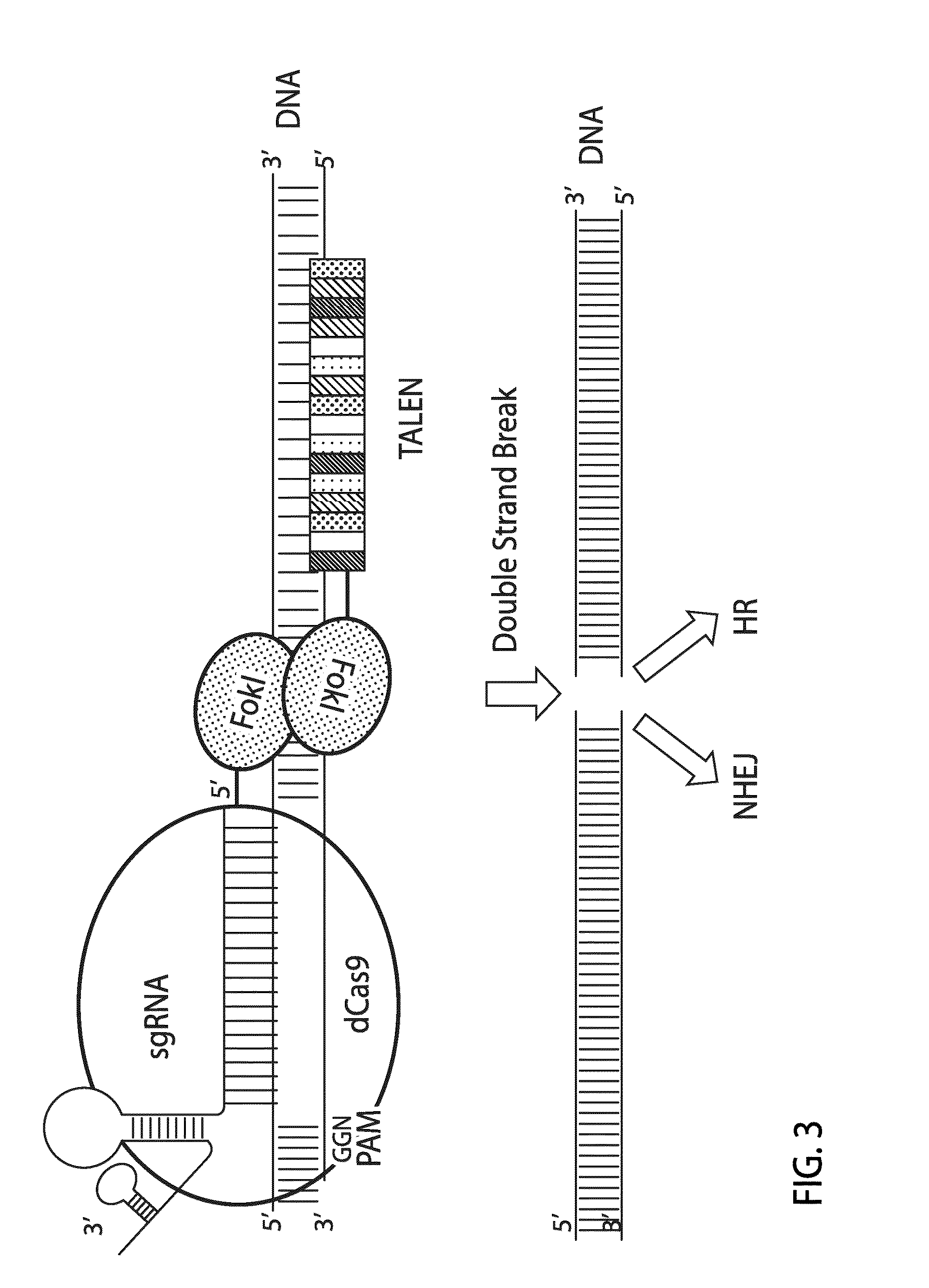
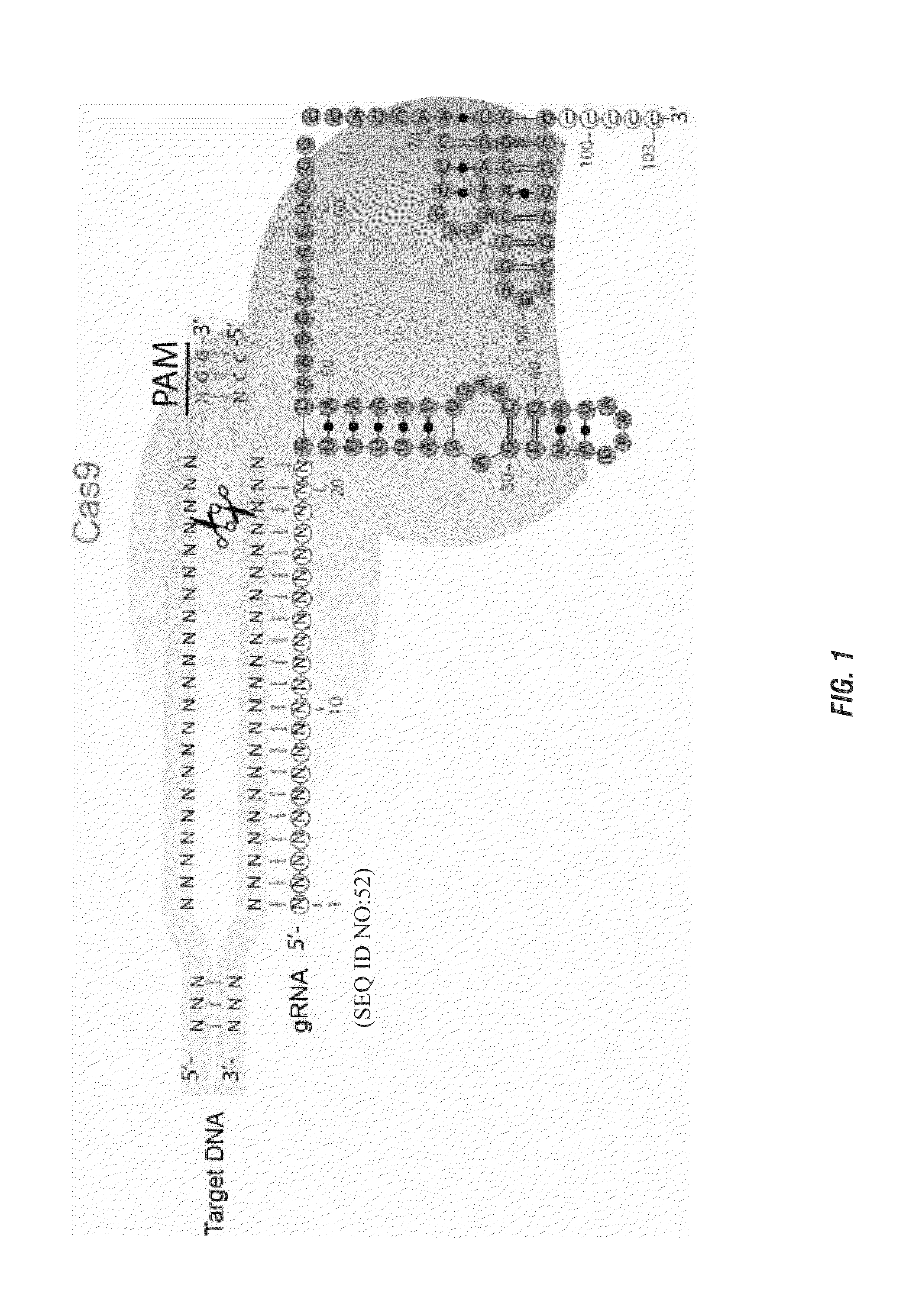
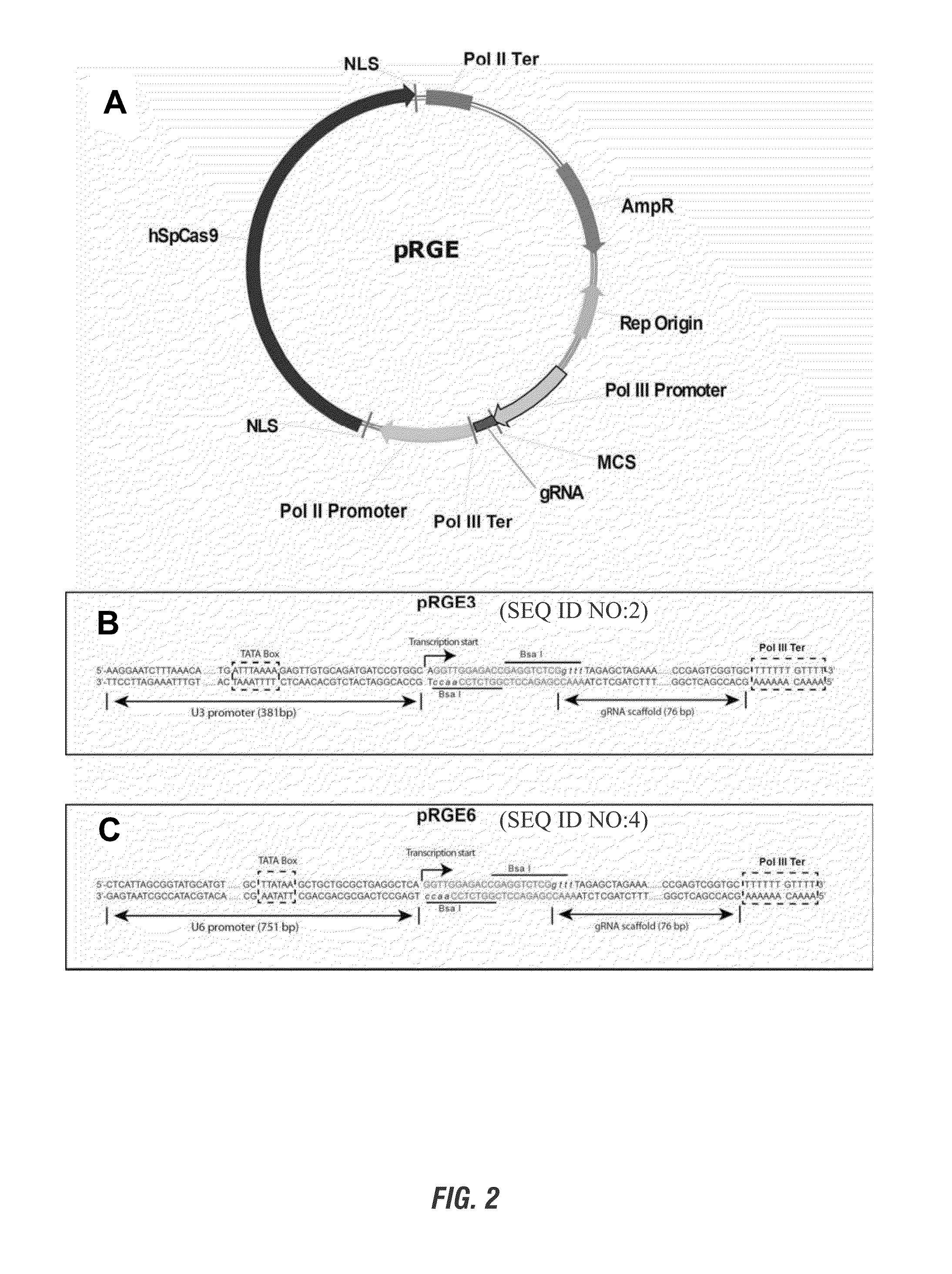
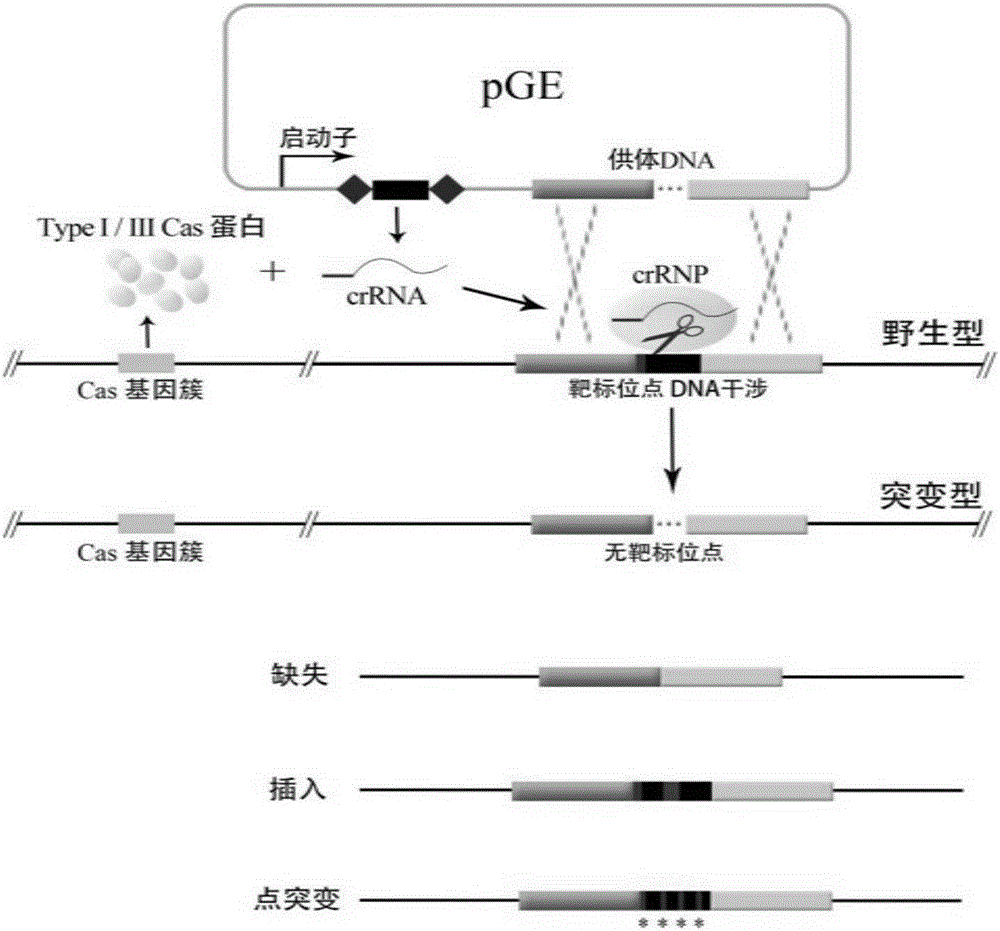
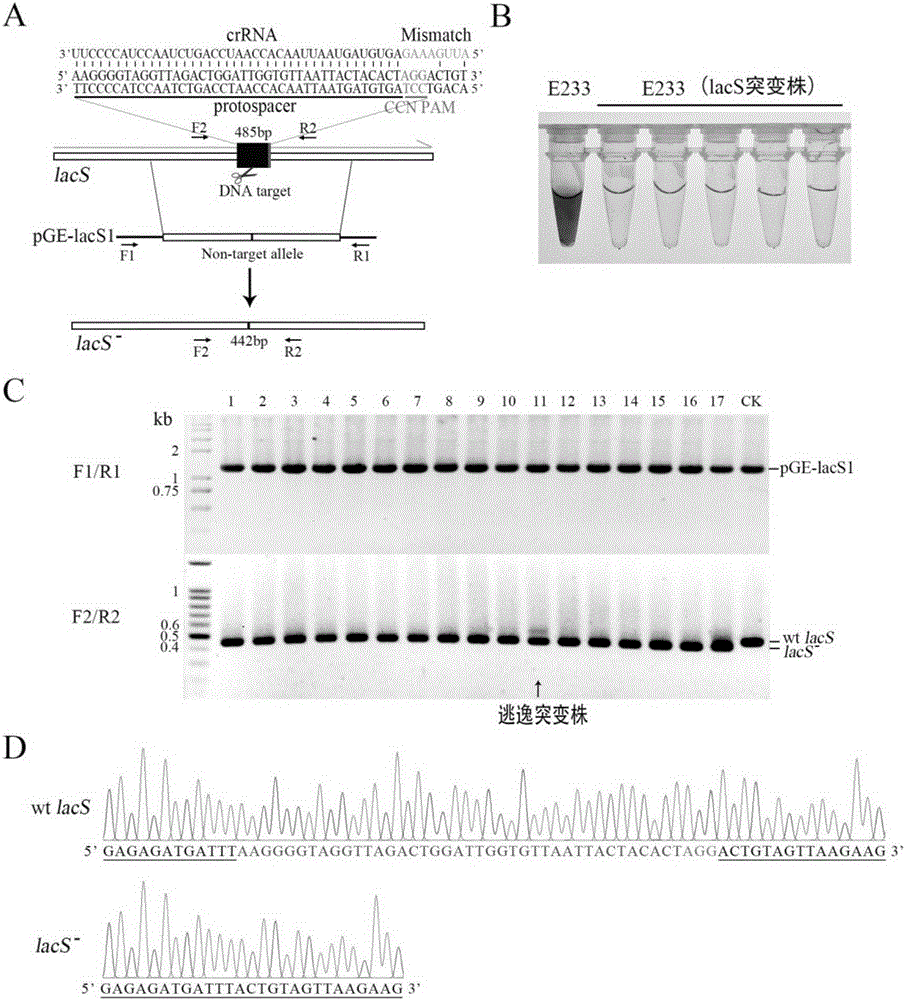
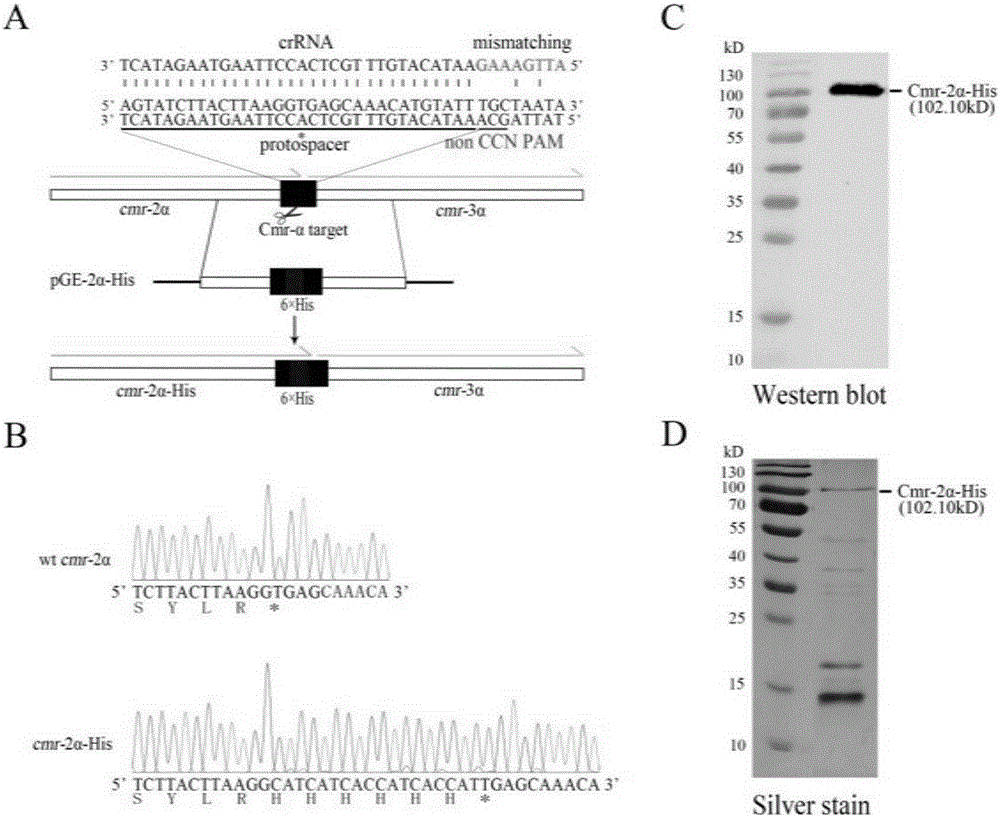
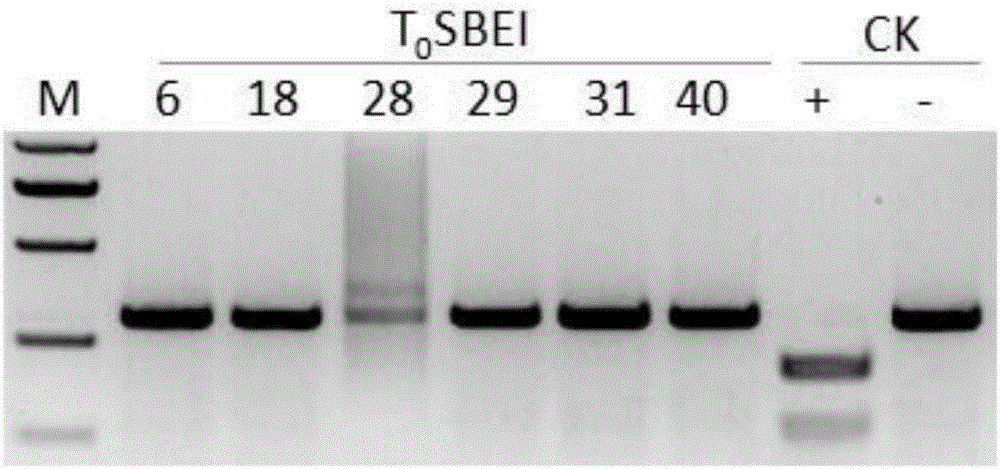
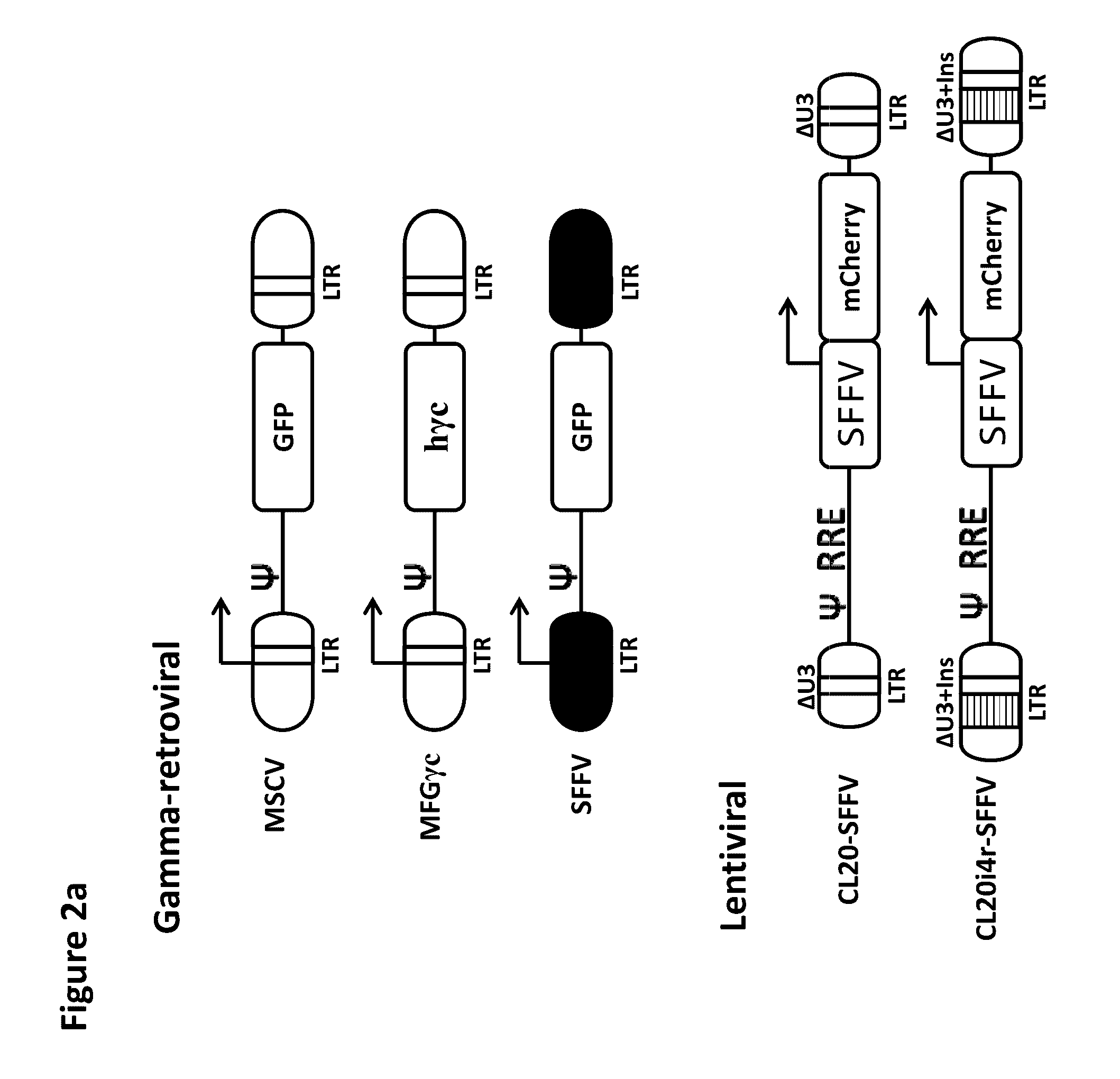
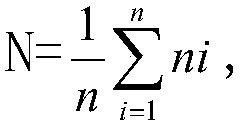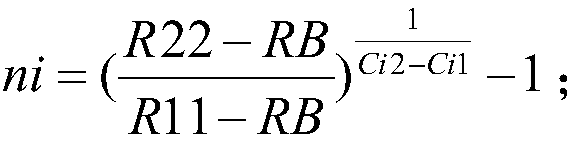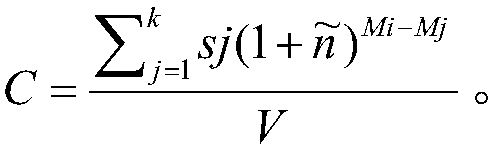Patents
Literature
963 results about "Genome editing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
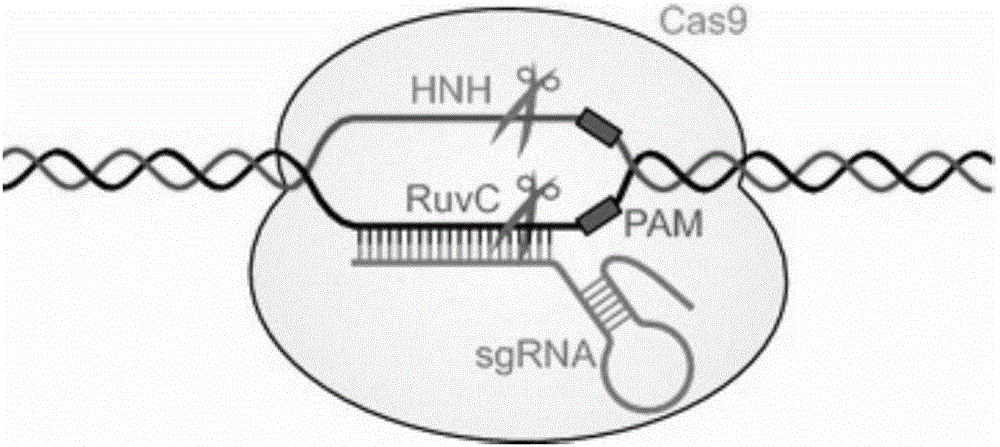
Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site specific locations.
THERAPEUTIC USES OF GENOME EDITING WITH CRISPR/Cas SYSTEMS
InactiveUS20150176013A1Effectively deletes target polynucleotide sequencesEfficiently deletingBiocideGenetic material ingredientsGenome editingBioinformatics
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +1
Crispr/cas system-based novel fusion protein and its applications in genome editing
An inactive CRISPR / Cas system-based fusion protein and its applications in gene editing are disclosed. More particularly, chimeric fusion proteins including an inCas fused to a DNA modifying enzyme and methods of using the chimeric fusion proteins in gene editing are disclosed. The methods can be used to induce double-strand breaks and single-strand nicks in target DNAs, to generate gene disruptions, deletions, point mutations, gene replacements, insertions, inversions and other modifications of a genomic DNA within cells and organisms.
Owner:SAGE LABS
THERAPEUTIC USES OF GENOME EDITING WITH CRISPR/Cas SYSTEMS
ActiveUS20150071889A1Effectively deletes target polynucleotide sequencesEfficiently deletingBiocideGenetic material ingredientsGenome editingBioinformatics
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +1
Gene targeting and genetic modification of plants via rna-guided genome editing
InactiveUS20150067922A1Improve stateFermentationVector-based foreign material introductionGenome editingGenetically modified crops
The present invention provides compositions and methods for specific gene targeting and precise editing of DNA sequences in plant genomes using the CRISPR (cluster regularly interspaced short palindromic repeats) associated nuclease. Non-transgenic, genetically modified crops can be produced using these compositions and methods.
Owner:PENN STATE RES FOUND
Detecting mutations for cancer screening and fetal analysis
ActiveUS20170073774A1Accurate detectionMicrobiological testing/measurementProteomicsCell freeEmbolization Therapy
Embodiments are related to the accurate detection of somatic mutations in the plasma (or other samples containing cell-free DNA) of cancer patients and for subjects being screened for cancer. The detection of these molecular markers would be useful for the screening, detection, monitoring, management, and prognostication of cancer patients. For example, a mutational load can be determined from the identified somatic mutations, and the mutational load can be used to screen for any or various types of cancers, where no prior knowledge about a tumor or possible cancer of the subject may be required. Embodiments can be useful for guiding the use of therapies (e.g. targeted therapy, immunotherapy, genome editing, surgery, chemotherapy, embolization therapy, anti-angiogenesis therapy) for cancers. Embodiments are also directed to identifying de novo mutations in a fetus by analyzing a maternal sample having cell-free DNA from the fetus.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
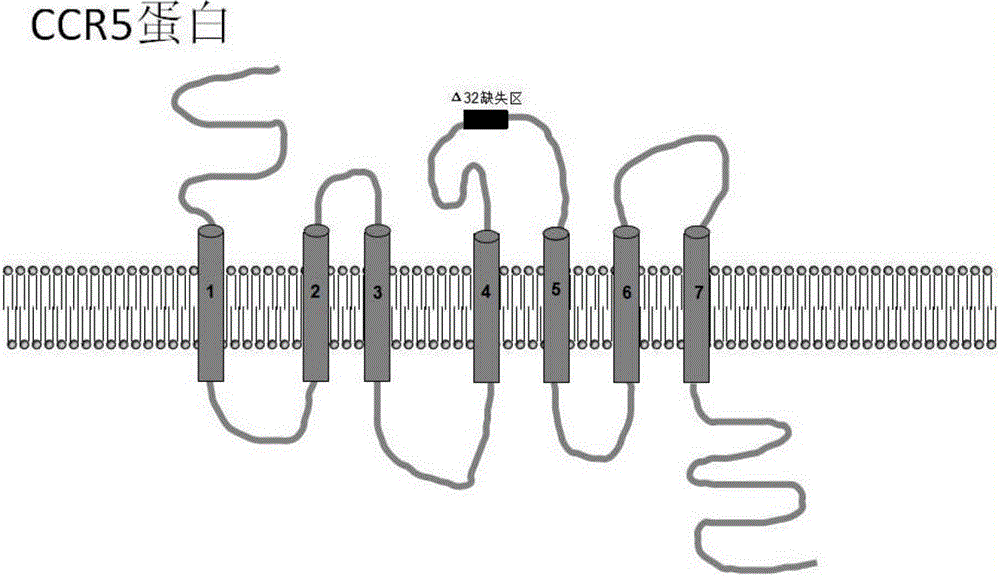
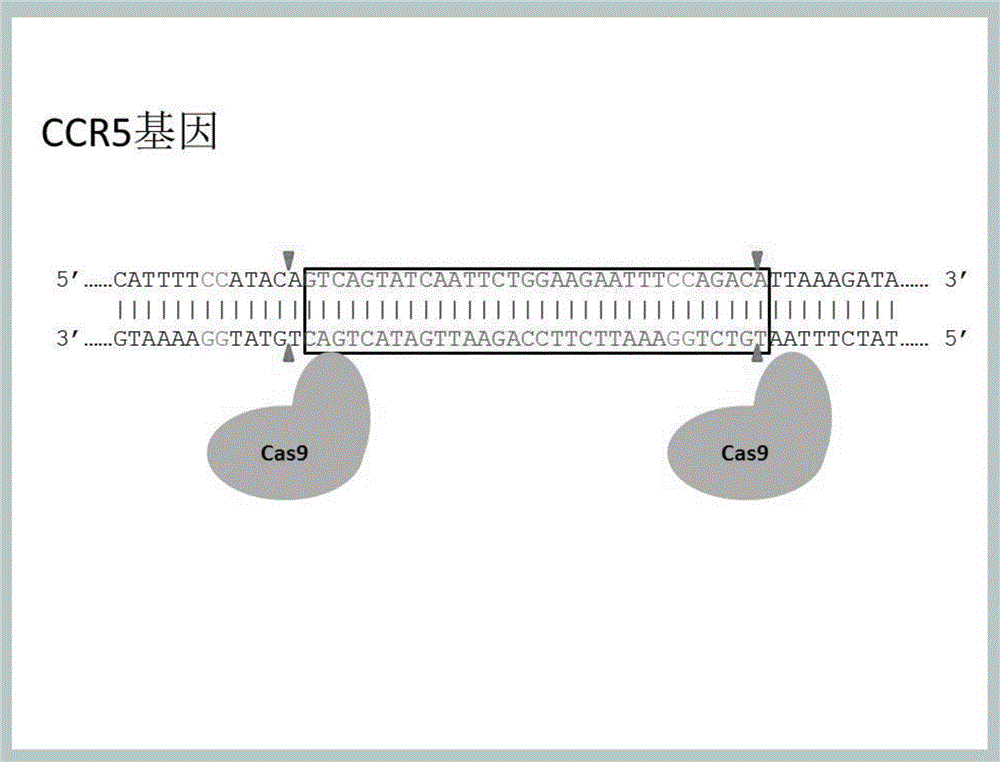
Method for inducing CCR5-delta32 deletion with genome editing technology CRISPR-Cas9
The invention relates to a method for successfully inducing cell chemokine receptor CCR5 genes to be mutated into CCR5-delta32 deletion-type genes with a new genome editing technology CRISPR-Cas9. CCR5 is an important receptor for human immunodeficiency viruses (HIV) to invade personal host cells. CCR5-delta32 deletion means deletion of 32 basic groups occurs in a CCR5 coding region, so that the sequence after the 185th amino acid is changed, and early termination occurs. CCR5-delta32 biallelic-gene homozygous deletion has natural resistance to HIV infection, and can not be infected by HIV. By means of the method, a slow virus packaging system and the CRISPR technology are used at the same time; as the slow virus infecting host range is wide, the method can be applied to cells such as bone marrow stem cells and CD4T cells, and the CCR5-delta32 deletion-type genes hopefully become medicine for treating acquired immune deficiency syndrome or other diseases.
Owner:NANKAI UNIV
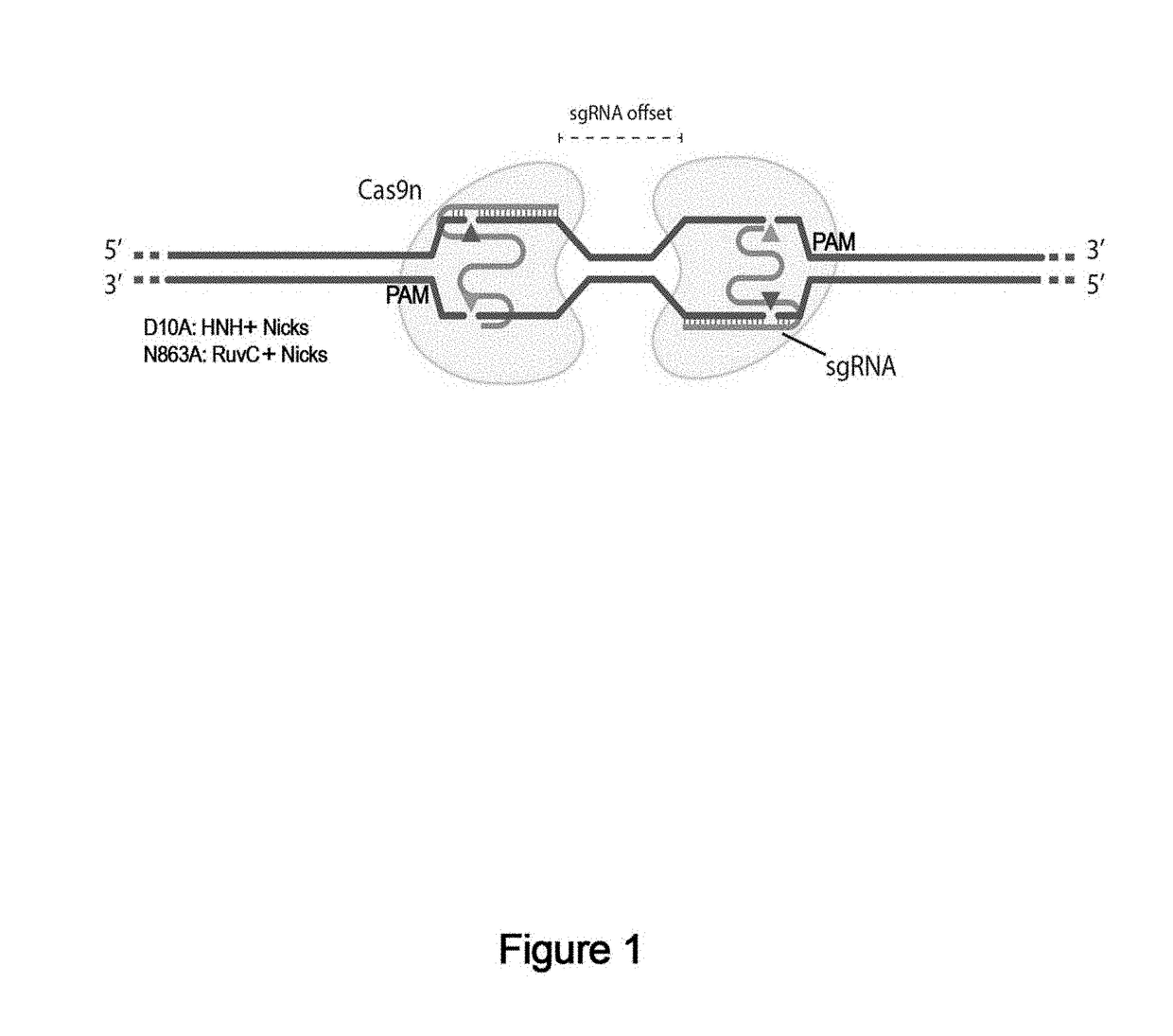
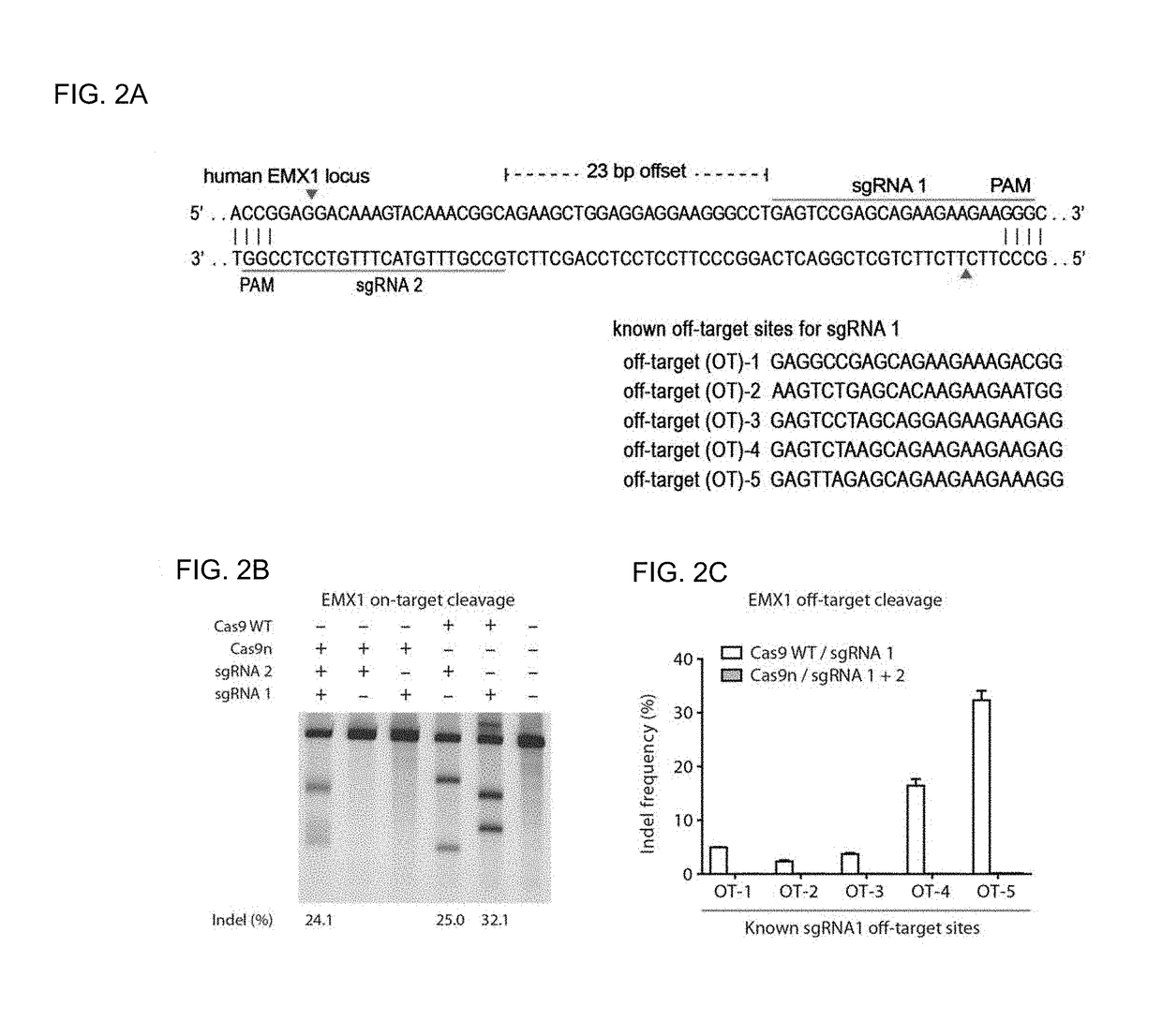
Genome editing using cas9 nickases
InactiveUS20170175144A1Low efficiencyStimulate HDRHydrolasesGenetic material ingredientsGenome editingVector system
The invention provides for delivery, engineering and optimization of systems, methods, and compositions for manipulation of sequences and / or activities of target sequences. Provided are vectors and vector systems, some of which encode one or more components of a CRISPR complex, as well as methods for the design and use of such vectors. Also provided are methods of directing CRISPR complex formation in prokaryotic and eukaryotic cells to ensure enhanced specificity for target recognition and avoidance of toxicity.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +2
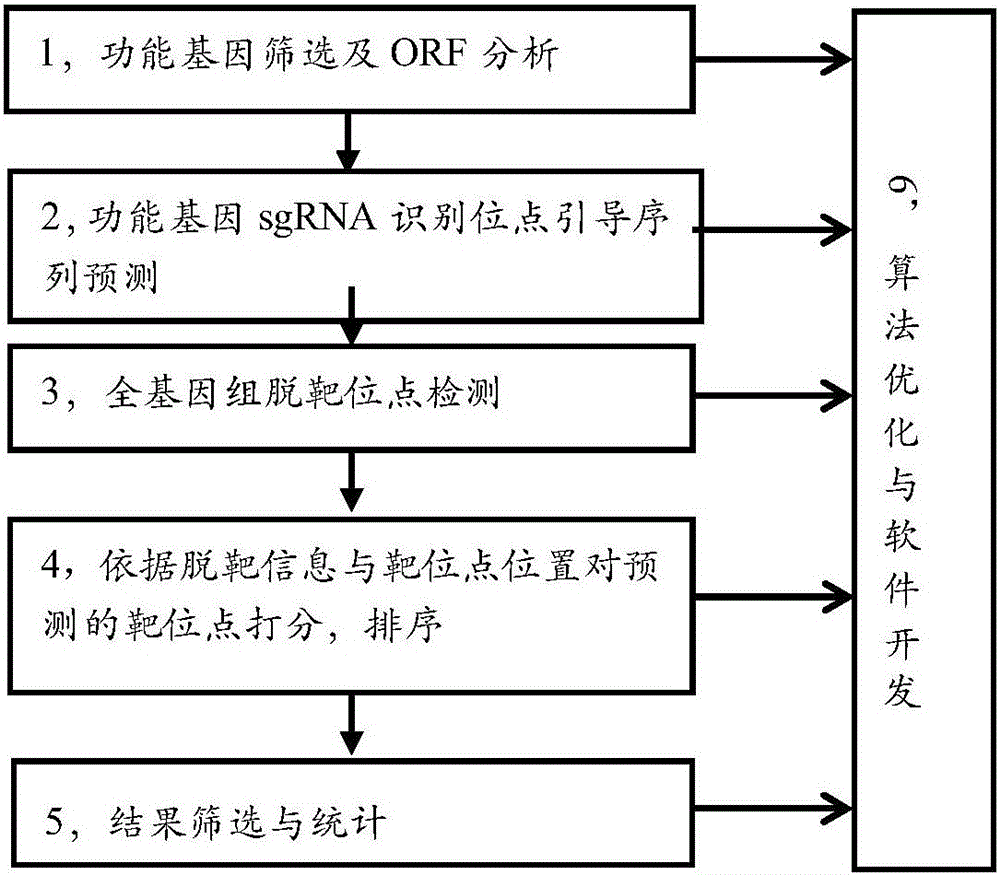
Efficient specific sgRNA recognition site guide sequence for pig gene editing and screening method thereof
ActiveCN105886616AMicrobiological testing/measurementDNA/RNA fragmentationGenome editingScreening method
The invention discloses an efficient specific sgRNA recognition site guide sequence for pig gene editing and a screening method thereof. The screening method includes: screening functional genes, performing ORF analysis, predicting a functional gene sgRNA recognition site guide sequence, detecting whole-genome off-target sites, grading predicted target sites according to off-target information and target site positions, sequencing, screening and statistically counting results, optimizing algorithms and developing software. The efficient specific sgRNA recognition site guide sequence and the screening method thereof have the advantages that the pig specific sgRNA recognition site guide sequence is obtained through strict screening and inspection and includes the sgRNA recognition site guide sequences, for CRISPR-Cas9 gene editing, of all pig protein encoding genes; the authenticating, grading and inspecting algorithms for specific sgRNA recognition and software corresponding to the algorithms and used for predicting and evaluating the pig functional gene sgRNA recognition site guide sequence are widely applicable to the sgRNA specific site prediction of non-model species with whole-genome sequences.
Owner:AGRO BIOLOGICAL GENE RES CENT GUANGDONG ACADEMY OF AGRI SCI
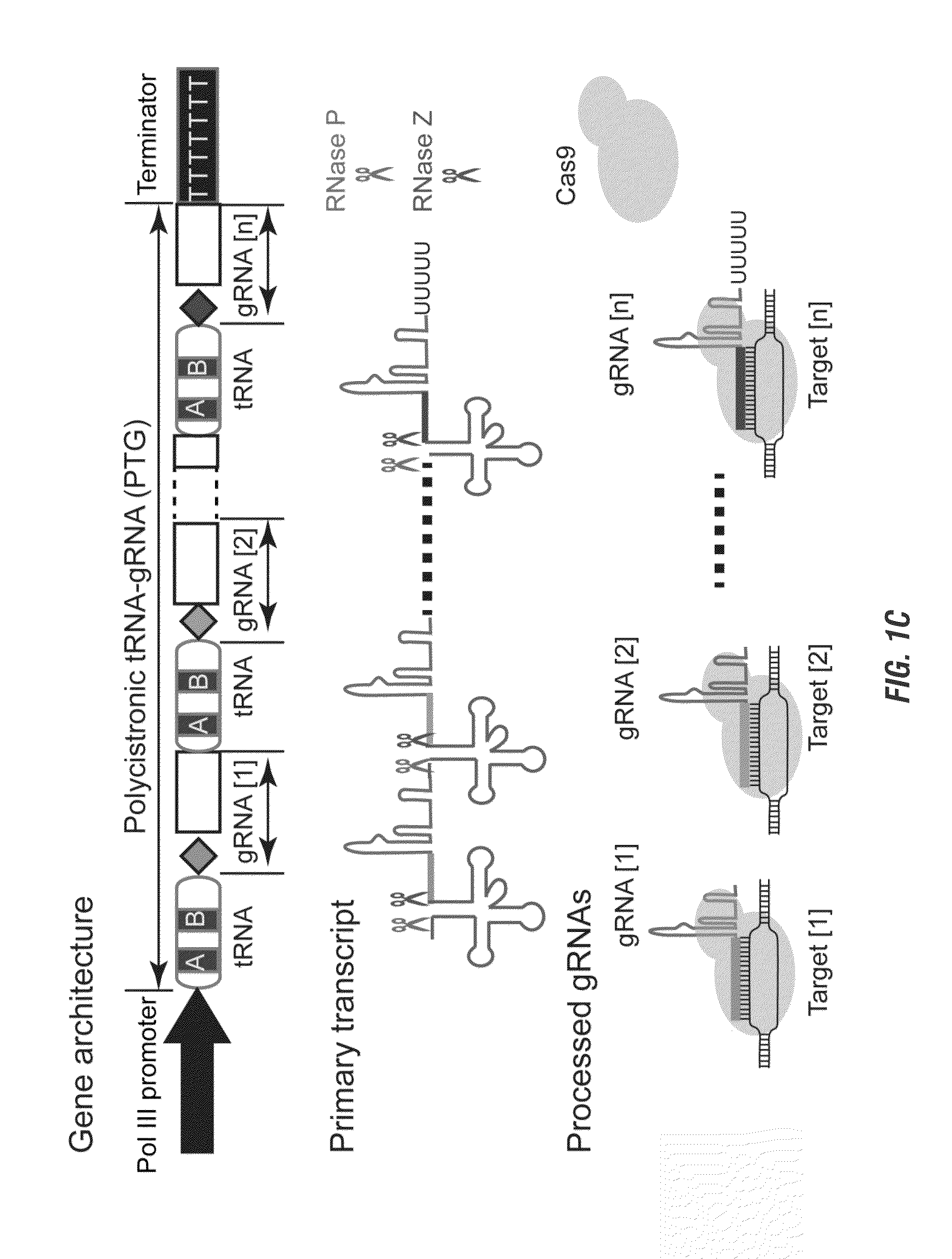
Methods and compositions for multiplex RNA guided genome editing and other RNA technologies
ActiveUS20160264981A1Facilitate RNA-guided multiplex genome editingVector-based foreign material introductionDNA/RNA fragmentationGenome editingGene silencing
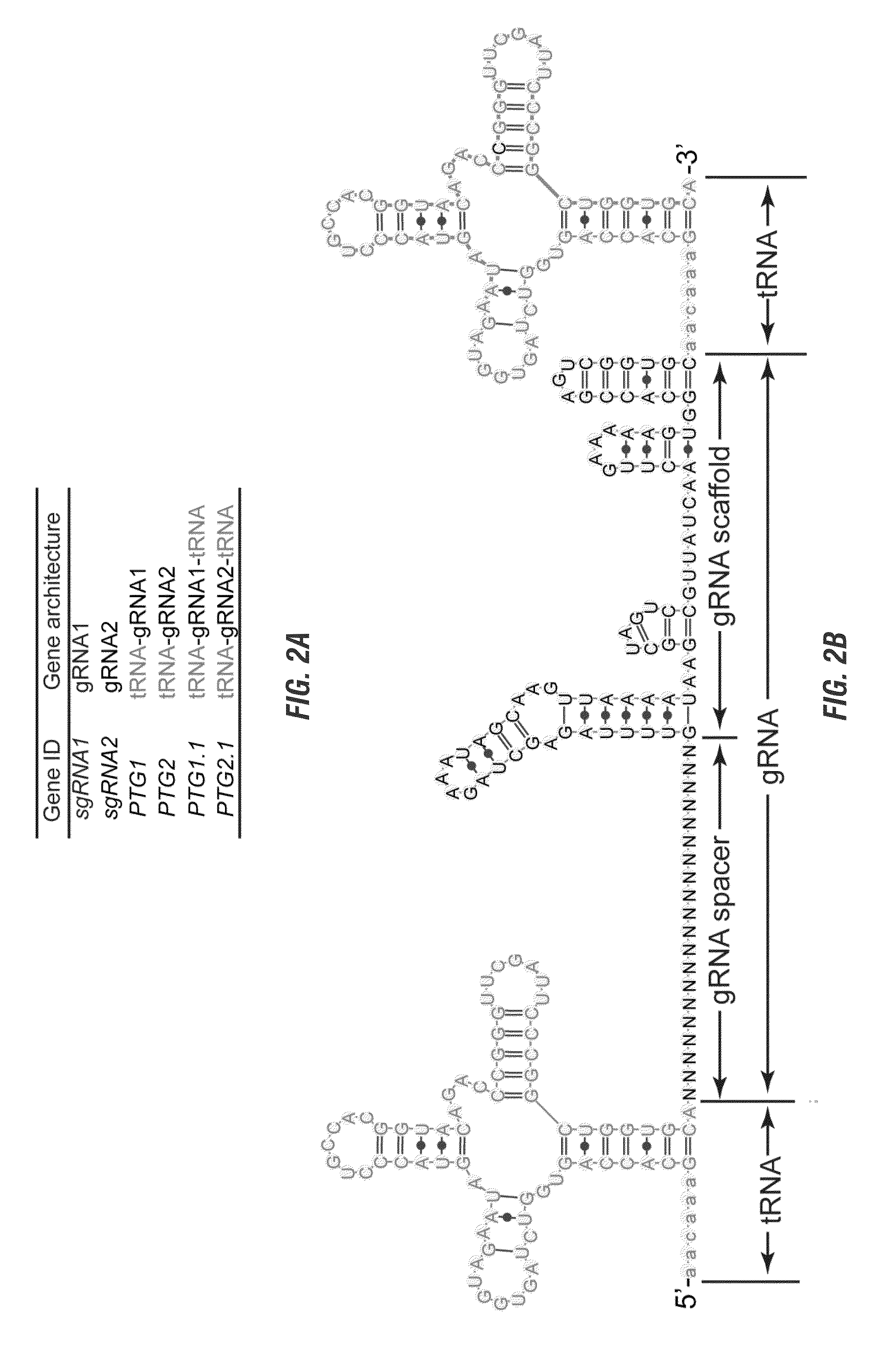
The invention includes materials and methods to generate numerous small RNAs from one polynucleotide construct (synthetic gene) to facilitate RNA-guided multiplex genome editing, modification, inhibition of expression and other RNA-based technologies. The synthetic gene / polynucleotide construct encodes polycistronic RNA components separated by tRNAs, and preferably also includes regulatory components such as a promoter or terminator to form an expression cassette. Once transcribed in a cell, the transcript is processed by the cell to multiple RNA molecules by the endogenous tRNA processing system. The system can be used for any RNA based gene manipulation method including RNA-mediated genome editing, artificial microRNA mediated gene silencing, small RNA mediated genetic manipulation, double-stranded RNA mediated gene silencing, antisense mechanisms and the like.
Owner:PENN STATE RES FOUND
CRISPR/Cas9 single transcription unit directionally modified backbone vector and application thereof
ActiveCN105132451AComplete efficientlyCo-transcriptionVector-based foreign material introductionGenome editingGene engineering
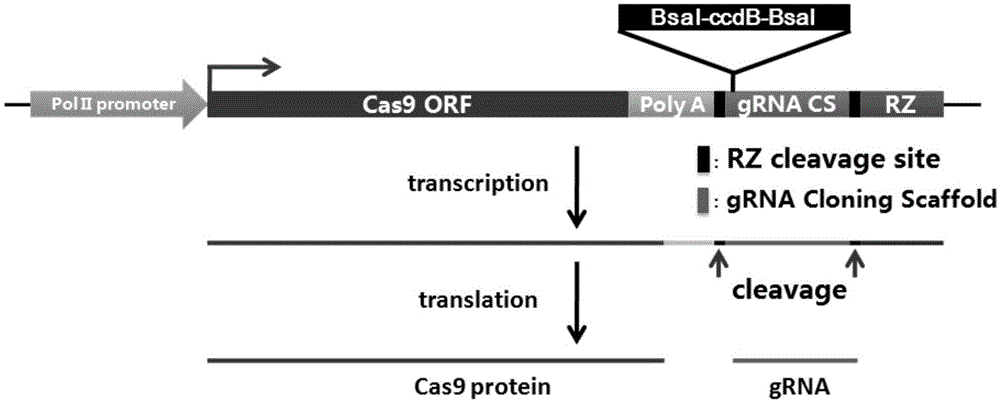
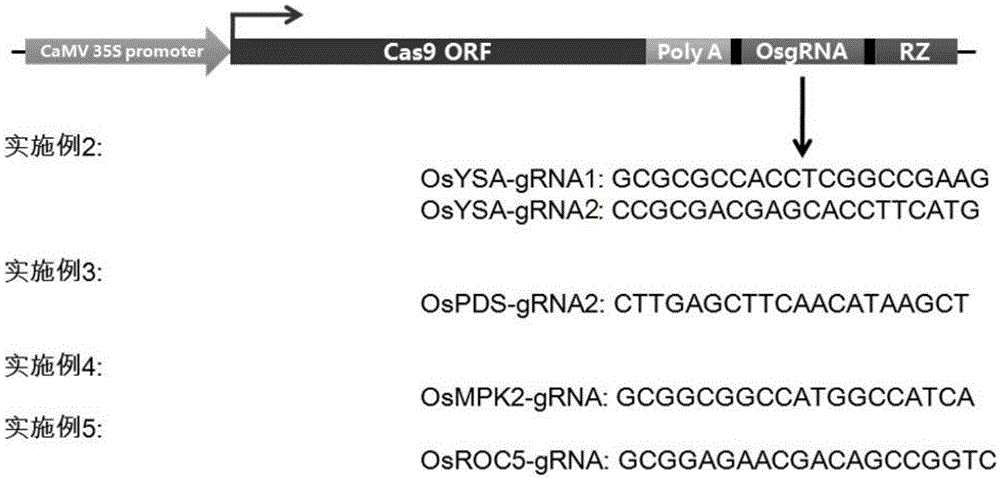
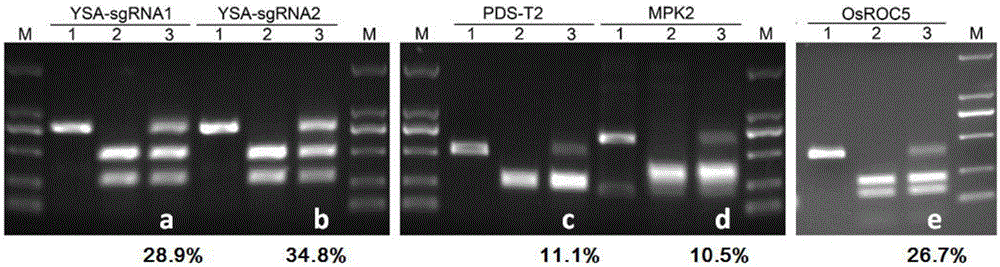
The invention belongs to the technical field of gene engineering, and concretely relates to a CRISPR / Cas9 single transcription unit directionally modified backbone vector and an application thereof. Technical problems to be solved in the invention comprise low species versatility and difficult realization of Cas9 protein expression and gRNA transcription cooperation of present CRISPR / Cas9 genome editing systems. A technical scheme in the invention is characterized in that a CRISPR / Cas9 single transcription unit backbone vector is constructed, and transcription of Cas9 and a guide RNA core unit are regulated by a promoter. The invention also provides a method for constructing a target site specifically modified Cas9-gRNA recombinant vector by adopting the CRISPR / Cas9 single transcription unit backbone vector. The efficient CRISPR / Cas9 single transcription unit backbone vector can effectively realize Pol II promoter driving-based Cas9 and gRNA unit cooperative transcription, and also can realize simple, fast and efficient genome directional heredity modification of various eukaryotes.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
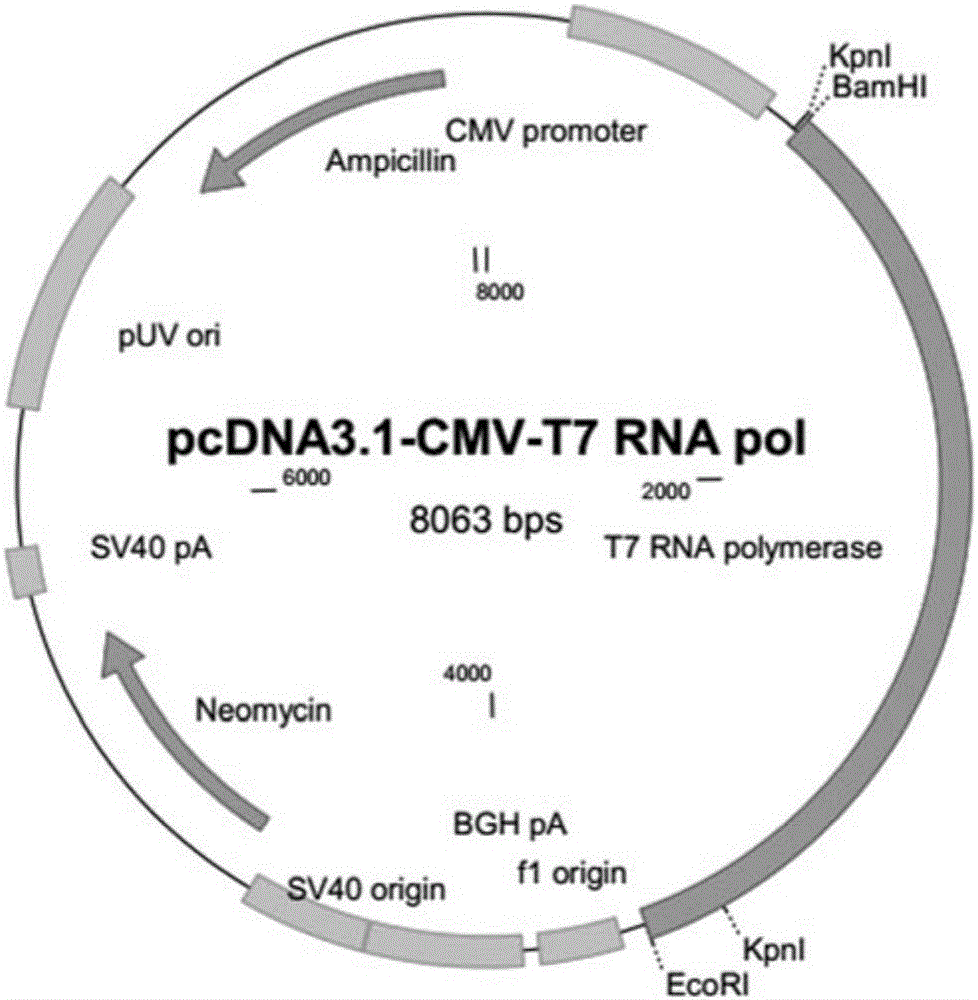
Establishment method of T7-RNA-polymerase-mediated CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 gene editing system
InactiveCN105861552ASimplify design stepsEasy to useVector-based foreign material introductionT7 RNA polymeraseGenome editing
The invention mainly belongs to the technical field of higher organism genome editing, and particularly relates to an establishment method of a T7-RNA-polymerase-mediated CRISPR (clustered regularly interspaced short palindromic repeats) / Cas9 gene editing system. The method is based on the highly specific recognition principle for the T7 RNA polymerase and T7 promoter, and comprises the following steps: establishing T7 promoter-sgRNA, transforming the T7 promoter-sgRNA, a Cas9 expression plasmid and a T7 RNA polymerase expression plasmid into cells, and catalyzing the T7 promoter to promote the transcription of sgRNA by using the T7 RNA polymerase so as to guide the Cas9 to perform cutting at the target, thereby completing the gene editing process. By using the T7 promoter to express the sgRNA, the method simplifies the design steps of the CRISPR / Cas9 system, so that more people can quickly and easily research the interested gene locus by using the CRISPR / Cas9 gene editing tool.
Owner:NORTHWEST A & F UNIV
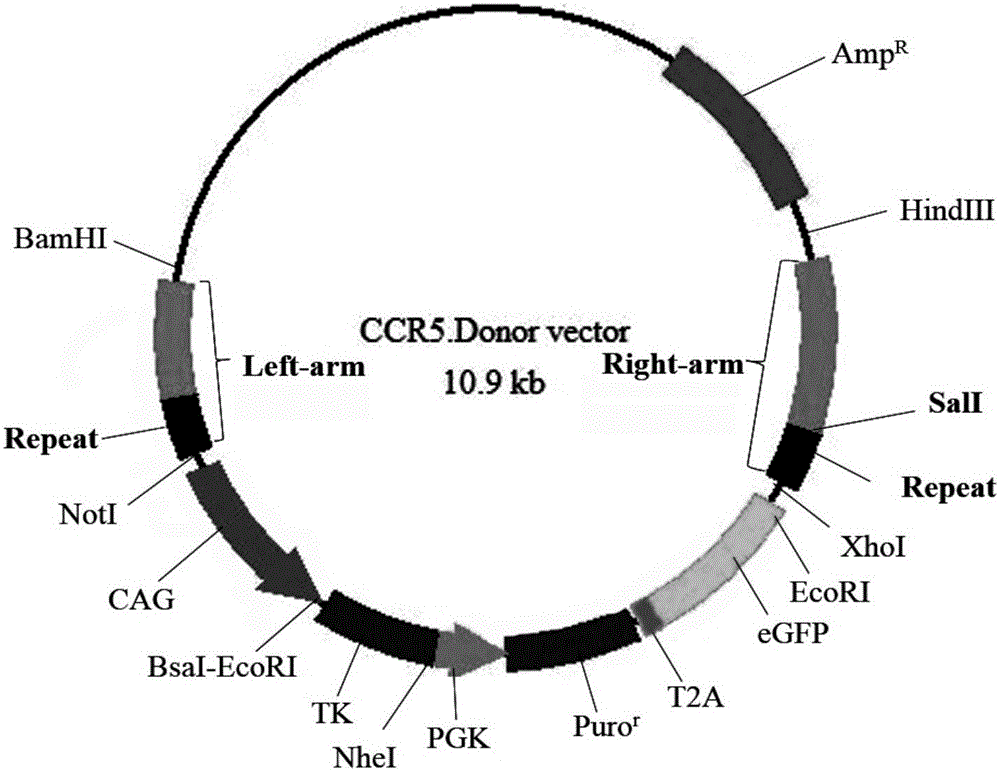
SSA (single-strand annealing) repair-based gene seamless editing method utilizing CRISPR/Cas9 technology
InactiveCN106011171ASeamless editingNucleic acid vectorVector-based foreign material introductionGenome editingPlasmid Vector
The invention mainly belongs to the technical field of gene editing and particularly relates to an SSA (single-strand annealing) repair-based gene seamless editing method utilizing a CRISPR / Cas9 technology. According to the gene seamless editing method, CRISPR / Cas9 expression vectors CCR5.CRISPR / Cas9 and eGFP.CRISPR / Cas9 targeting CCR5 and eGFP are constructed firstly, a donor CCR5-Donor vector is constructed, a CCR5-Donor plasmid vector adopts the pXL-BACII-L-arm-CAG-TK-PGK-Puro<R>-T2A-eGFP-bGH pA-R-arm structure, L-arm and R-arm are left and right homologous arms, and CAG-TK-PGK-Puro<R>-T2A-eGFP-bGH pA between L-arm and R-arm is a screening marking component; through two times of transfection, seamless editing is completed by using HR and SSA repair mechanisms in the CRISPR-Cas9 system and cells through positive and negative screening of puromycin and ganciclovir of the cells.
Owner:NORTHWEST A & F UNIV
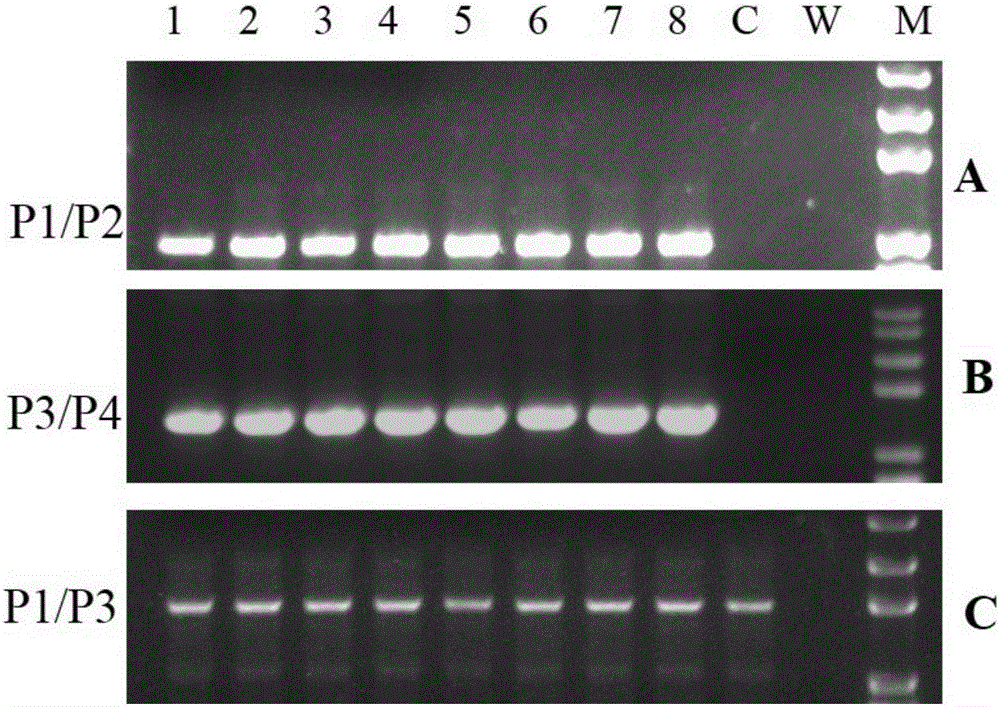
Constructs and methods for genome editing and genetic engineering of fungi and protists
Provided herein are constructs for genome editing or genetic engineering in fungi or protists, methods of using the constructs and media for use in selecting cells. The construct include a polynucleotide encoding a thymidine kinase operably connected to a promoter, suitably a constitutive promoter; a polynucleotide encoding an endonuclease operably connected to an inducible promoter; and a recognition site for the endonuclease. The constructs may also include selectable markers for use in selecting recombinations.
Owner:WISCONSIN ALUMNI RES FOUND
Method for acquiring gene editing sheep by RNA-mediated specific double-gene knockout and special sgRNA for method
ActiveCN105132427AStrong specificityPrecise targeted modificationArtificial cell constructsVertebrate cellsAnimal scienceGenome editing
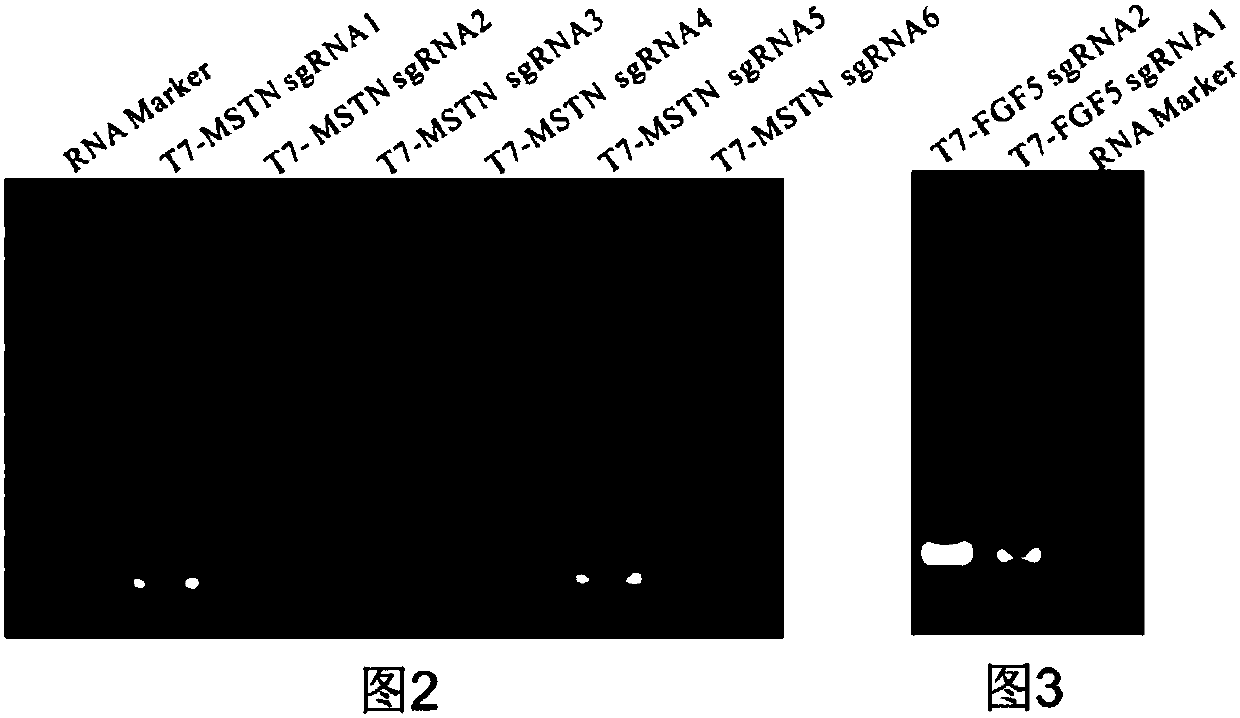
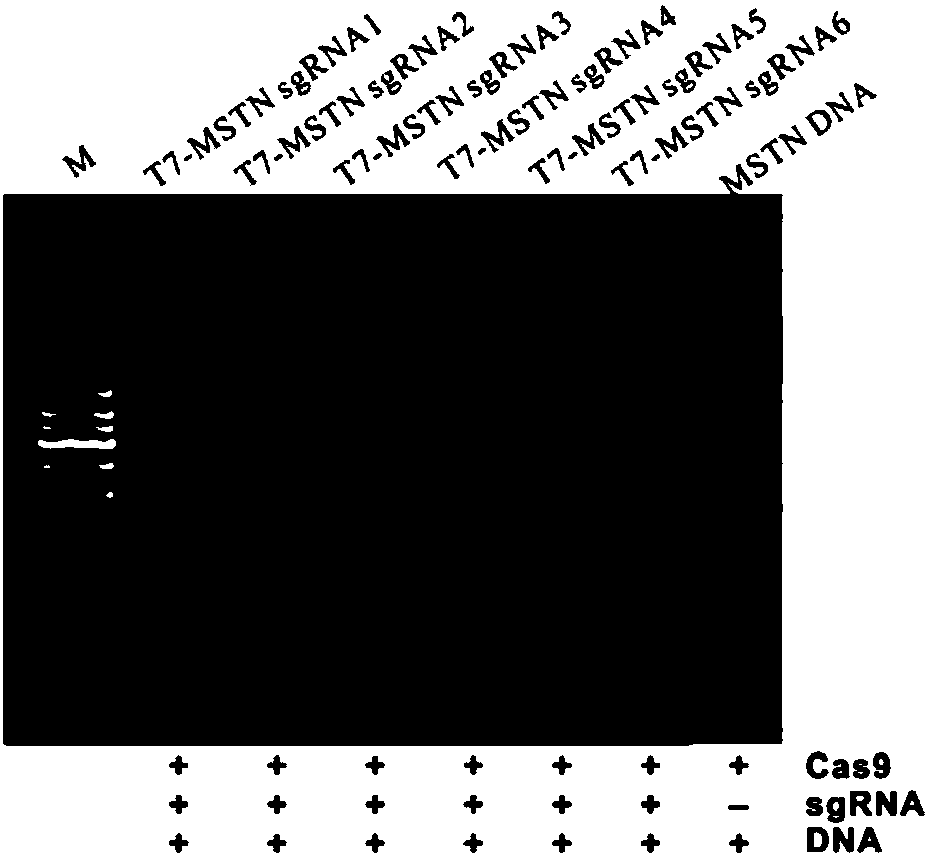
The invention discloses a method for acquiring gene editing sheep by RNA-mediated specific double-gene knockout and special sgRNA for the method. The sgRNA combination consists of sgRNAMSTN-1 and sgRNAFGF5-1, wherein sgRNAMSTN-1 is sgRNA which can realize specific targeted modification of sheep MSTN gene and is RNA shown in the second to 21st nucleotides in a sequence 6 or RNA with the second to 21st nucleotides of the sequence 6; sgRNAFGF5-1 is sgRNA which can realize specific targeted modification of sheep FGF5 gene and is RNA shown in the second to the 21st nucleotides in a sequence 8 or RNA with the second to 21st nucleotides of the sequence 8. According to the method for acquiring gene editing sheep by RNA-mediated specific double-gene knockout and special sgRNA for the method, the CRISPR / Cas9 genome editing technology and the micro-injection technology are combined, so that the sheep targeting efficiency is higher and more accurate, sheep double-gene knockout is realized for the first time in the generation, improvement on sheep meat production and wool production is greatly promoted, and a larger space and a more effective technical tool are provided for breeding of new sheep varieties.
Owner:新疆畜牧科学院生物技术研究所
Pluripotent stem cell-hereditary cardiomyopathy cardiac muscle cell and preparation method thereof
InactiveCN105039399AWide variety of sourcesLong-term in vitro cultureVector-based foreign material introductionForeign genetic material cellsDiseaseDisease phenotype
The invention belongs to the field of researching and application of biomedicine and particularly relates to human pluripotent stem cell-hereditary cardiomyopathy cardiac muscle cells and a preparation method thereof. The invention provides a human hereditary cardiomyopathy-pluripotent stem cell, which is constructed by means of TALEN or CRISPR / CAS9 genome editing technology. The human hereditary cardiomyopathy cardiac muscle cell can be combined with any scaffold materials to culture various in-vitro human hereditary cardiomyopathy cardiac muscle tissues. The human hereditary cardiomyopathy cardiac muscle cells in the invention have disease phenotypes and electrophysiology change being similar as the cardiac muscle cells of human hereditary cardiomyopathy patients. The human hereditary cardiomyopathy cardiac muscle cells are wide in sources and can be cultured in-vitro for a long time. The invention provides excellent tools and platforms for researching an effective new therapy means and researching an effective corresponding treatment medicine.
Owner:FUDAN UNIV
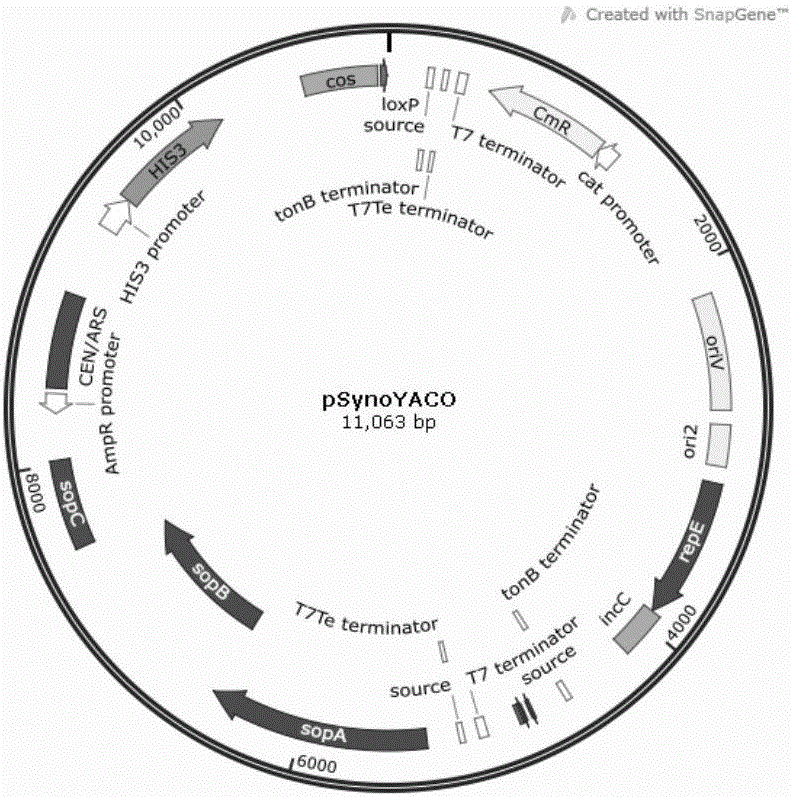
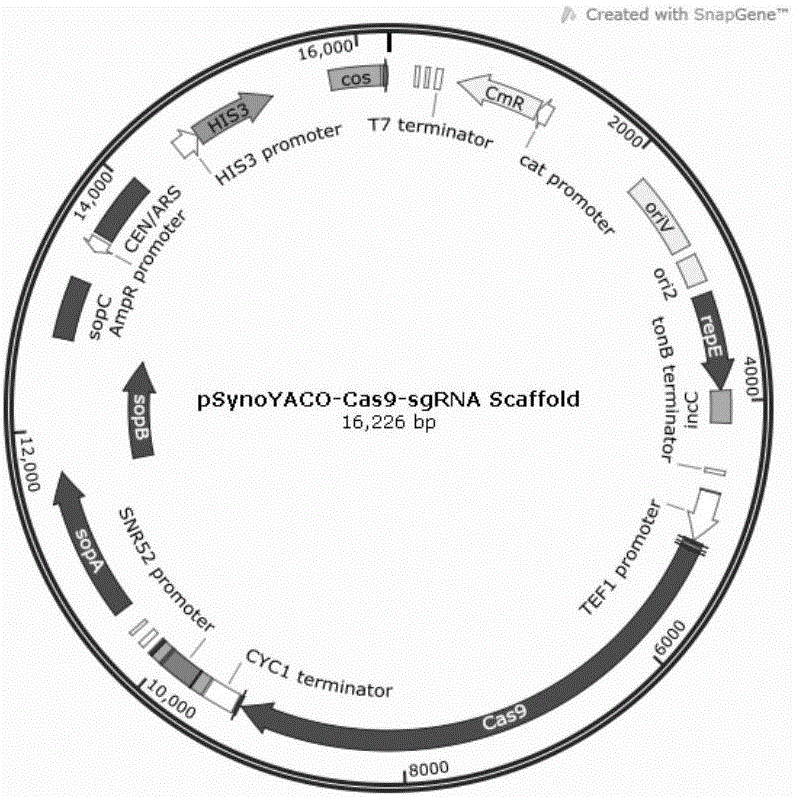
Saccharomyces cerevisiae genome editing carrier based on CRISPR-Cas9 system and application of carrier
InactiveCN106086061ASimple and fast operationStringent controlFungiMicroorganism based processesGenome editingCarrier system
The invention relates to a saccharomyces cerevisiae genome editing carrier based on a CRISPR-Cas9 system and application of the carrier. According to the saccharomyces cerevisiae genome editing carrier, a Cas9 protein expression cassette and an sgRNA scaffold expression cassette are integrated in the carrier, and a carrier-Cas9-sgRNA scaffold is obtained; any one of multiple sgRNA fragments which are designed and synthesized by taking ADE1 as a target site is integrated in the carrier-Cas9-sgRNA scaffold, and the saccharomyces cerevisiae genome editing carrier is obtained. According to the saccharomyces cerevisiae genome editing carrier based on the CRISPR-Cas9 system and the application of the carrier, a single plasmid and single copy carrier system is adopted, one plasmid is adopted to express Cas9 protein and sgRNA scaffold simultaneously, construction and conversion of a target gene editing system only need one step, and operation is easy and convenient.
Owner:SUZHOU HONGXUN BIOTECH CO LTD
Process of knocking out Wnt3a gene and verification method thereof
InactiveCN106434752AHigh targeting accuracyShort experiment cycleFermentationVector-based foreign material introductionValidation methodsWNT3A gene
The invention discloses a process of knocking out Wnt3a gene and a verification method thereof. The knockout and verification of Wnt3a gene are finished through the following steps: establishment of a Cas9 lentiviral vector for Wnt3a gene, culture and passage of HepG2 cell, lentivirus infection and screening of target cell, verification of gene knockout efficiency through a mispairing enzyme method, cell protein analysis and cell proliferation detection by a CCK-8 method. The invention has the following advantages: the Wnt3a gene is knocked out by establishing a Cas9 double-vector lentivirus system for the first time; Crispr / Cas9 is a technology for accurately editing specific site of the genome of any species, and the cell-level single gene or multiple genes can be knocked out by the technology; compared with other gene editing technologies, the method has the advantages that the targeting accuracy is higher; only if the RNA target sequence is completely matched with the genome sequence, can the Cas9 cut the DNA and realize simultaneous knockout of multiple sites of the target gene; and moreover, the experimental period of vector establishment is short, the time and the cost are remarkably saved, and species limit is avoided.
Owner:AFFILIATED HOSPITAL OF NANTONG UNIV
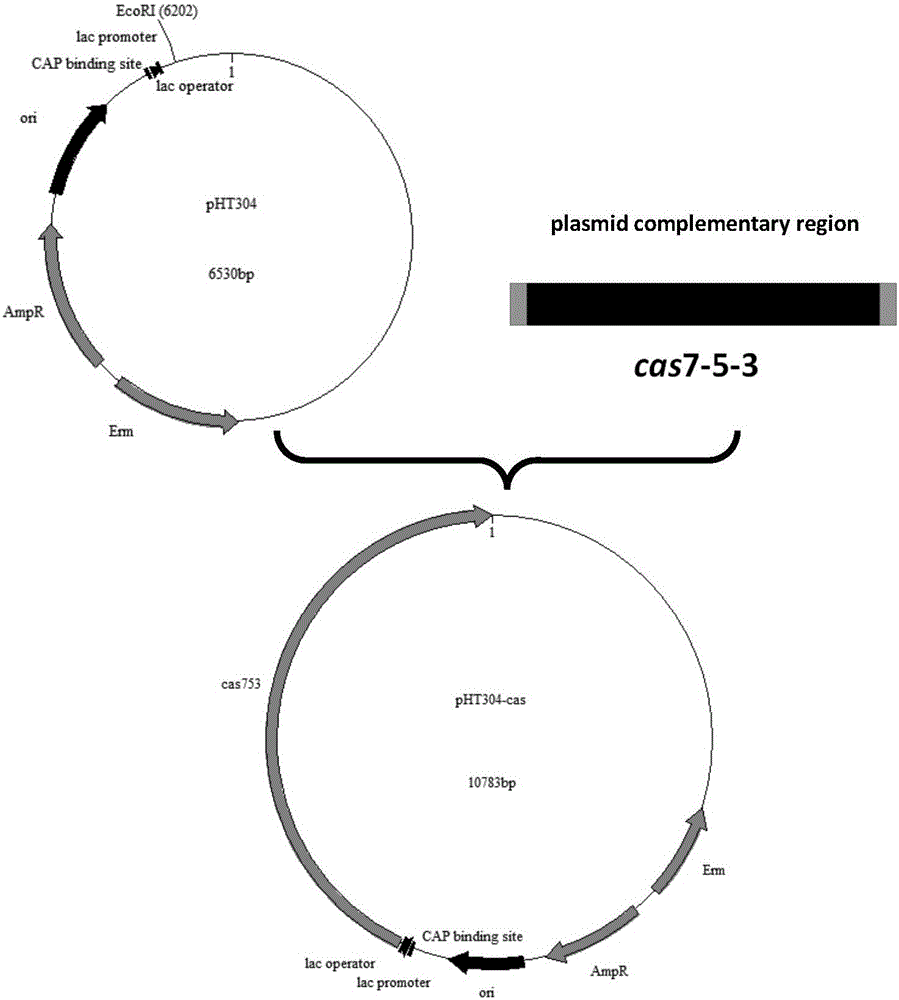
Gene editing method based on streptomyces virginiae IBL14 gene cas7-5-3
PendingCN106834323AGene editing achievedEasy gene editingNucleic acid vectorVector-based foreign material introductionEscherichia coliGenome editing
The invention discloses a gene editing method based on a streptomyces virginiae IBL14 gene cas7-5-3. The gene editing method has the advantages that the gene editing on prokaryotic genomes is realized for the first time by CRISPR-Cas-I system; a new supplementing and selection type is provided for the gene editing on the CRISR-Cas II type CAS; by using a system, the prokaryotic genomes, such as escherichia coli and bacillus subtilis, can be conveniently, quickly and effectively subject to gene editing; the optimized gene editing system may be applied to the gene editing of other organisms.
Owner:ANHUI UNIVERSITY
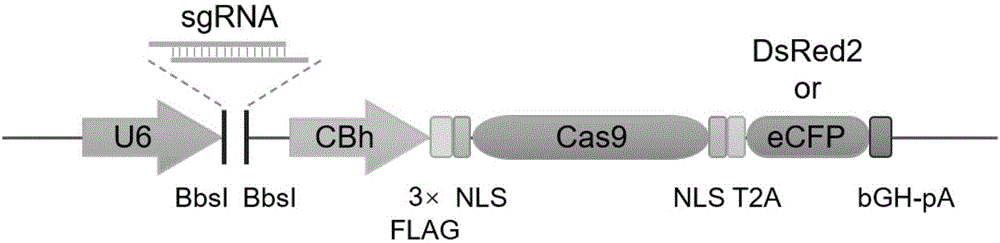
Cell line gene knock-out method for obtaining large fragment deletion through CRISPR/Cas9 system
ActiveCN107435051AEasy accessImprove knockout productivityVectorsStable introduction of DNAFluorescenceLarge fragment
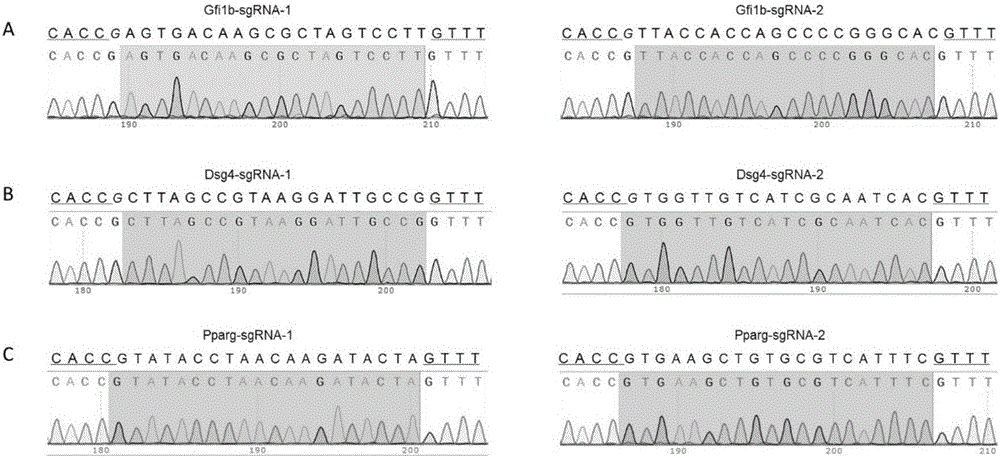
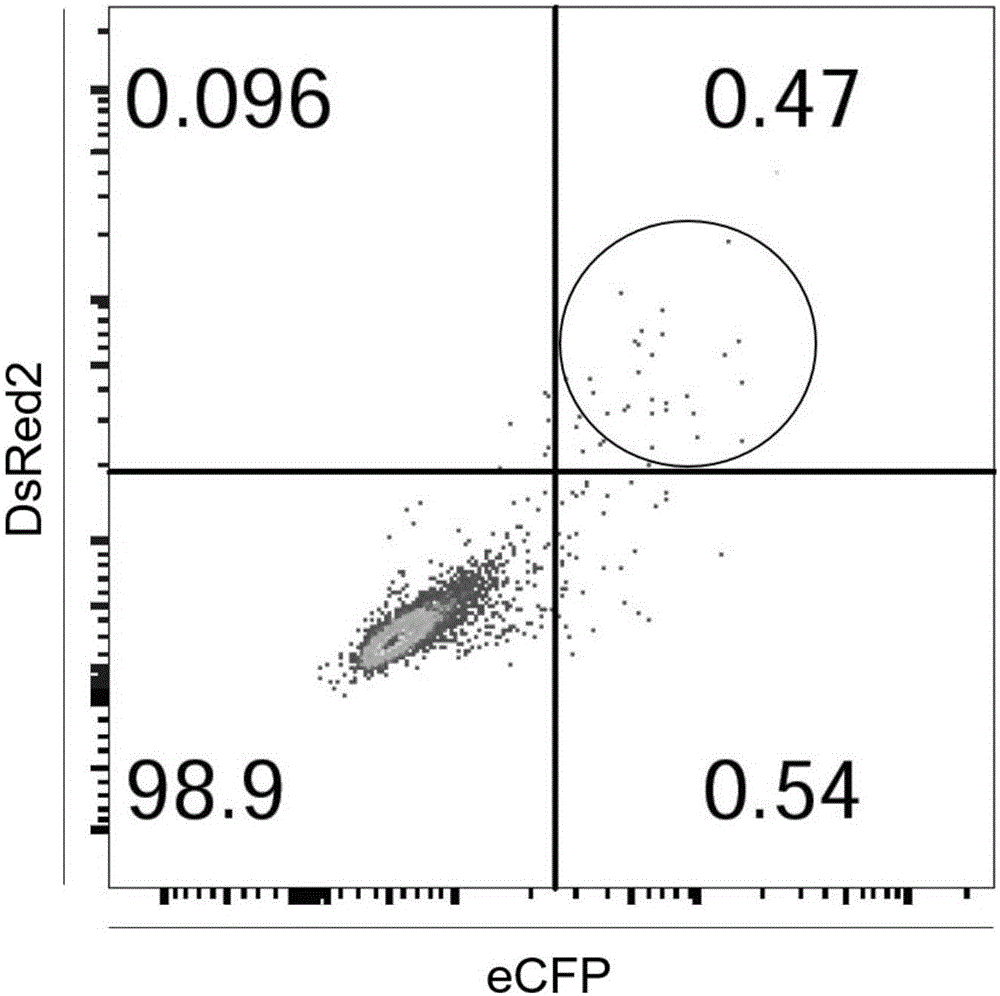
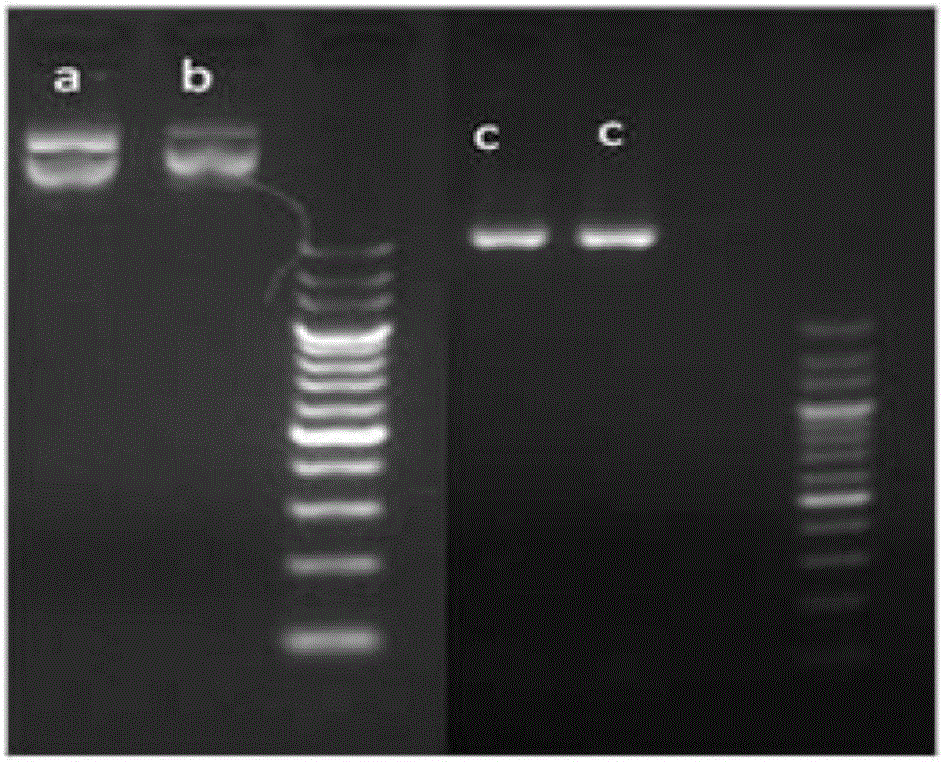
The invention relates to a cell line gene knock-out method for obtaining large fragment deletion through a CRISPR / Cas9 system, and belongs to the field of genetic engineering and genetic modification. A pX458 vector is modified, so that the pX458 vector is provided with DsRed2 and ECFP (Enhanced Cyan Fluorescence Protein); then, a plurality of specific sgRNA sites are designed by aiming at a target gene and are connected into the modified vector; after the cell line transfection, cell groups are sorted by a flow cytometry; single cells subjected to genome editing can be very fast obtained; then, a single-cell DNA (Deoxyribonucleic Acid) sequence is subjected to PCR (Polymerase Chain Reaction) amplification; and single cells with large fragment deletion can be picked out from the single cells through gel electrophoresis. The CRISPR / Cas9 system, the flow cytometry single-cell sorting and fluorescent protein screening on the expression vector are combined, so that the positive monoclonal cells with the large fragment deletion can be obtained in a short time; and the work efficiency of the cell line gene knock-out is greatly improved.
Owner:XINXIANG MEDICAL UNIV
Establishment method of caffeine synthetase CRISPR/Cas9 genome editing vector
InactiveCN105821075ANucleic acid vectorVector-based foreign material introductionEnzyme digestionGolden Gate Cloning
The invention relates to an establishment method of a caffeine synthetase CRISPR / Cas9 genome editing vector. The method comprises the following steps: designing two target sequences T1 and T2 on the basis of tea tree caffeine synthetase genes; respectively establishing sgRNA expression cassettes of the target sequences T1 and T2; and carrying out enzyme digestion-coupling reaction on a double-element expression vector pYLCRISPR / Cas9P35S-H, the T1sgRNA expression cassette and the T2sgRNA expression cassette so that the T1sgRNA expression cassette and the T2sgRNA expression cassette are connected and inserted into the double-element expression vector, thereby obtaining the CRISPR / Cas9 genome editing vector. By taking the tea tree caffeine synthetase as the example, conventional PCR (polymerase chain reaction), Overlapping PCR and Golden Gate Cloning techniques are combined to establish the CRISPR / Cas9 gene editing vector comprising the tea tree caffeine synthetase double targets, thereby laying a solid foundation for application of the CRISPR / Cas9 gene editing technique in tea trees, and providing technical supports for the establishment of CRISPR / Cas9 gene editing vectors of other plants.
Owner:HUNAN AGRICULTURAL UNIV
Method for editing prokaryotic genomes using endogenic CRISPR-Cas (CRISPR-associated) system
ActiveCN105331627AReduce workloadWide range of application hostsBacteriaVector-based foreign material introductionGenome editingWorkload
The invention relates to a method for editing prokaryotic genomes using an endogenic CRISPR-Cas (CRISPR-associated) system. An editor plasmid carrying both an artificial CRISPR cluster and a donor DNA is required to be established for the genome editing of a prokaryote containing the endogenic CRISPR-Cas system, after NDA interference is caused by the endogenic CRISPR-Cas to the genome, and the genome is edited through homologous recombination. The method has the advantages that a range of applicable hosts is wide, all bacteria and archaea containing the endogenic CRISPR-Cas being available for operation; the method is applicable to operations in various editing manners, such as deletion, insertion and point mutation; the editing efficiency is higher, screening positive rage is high, and the background is low; a flow is simple, a short period of time is required, and the workload of prokaryotic genome editing is greatly reduced.
Owner:HUAZHONG AGRI UNIV
Method of increasing content of resistant starch in rice through genome editing and sgRNA special for same
ActiveCN106086028AAddressing the lack of low-glycemic foodsAvoid problemsNucleic acid vectorPlant peptidesBiotechnologyAmylase
The invention discloses a method of increasing the content of resistant starch in rice through genome editing and sgRNA special for the same. The method provides a method of increasing the content of resistant starch and / or amylase in rice seeds. The method includes the following steps that expression of SBEIIb genes in the rice is inhibited; the SBEIIb genes are genes of encoding SBEIIb protein. The invention further provides a method of increasing the content of resistant starch in rice and includes the step of inhibiting the activity of the SBEIIb protein in the rice. By using the CRISPR / Cas9 technology, the rice SBEIIb genes are edited at fixed points, the rice SBEIIb genes are knocked off by causing frameshift mutation, and the new generation of new rice germplasm with the content of amylase and resistant starch obviously increased is obtained.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
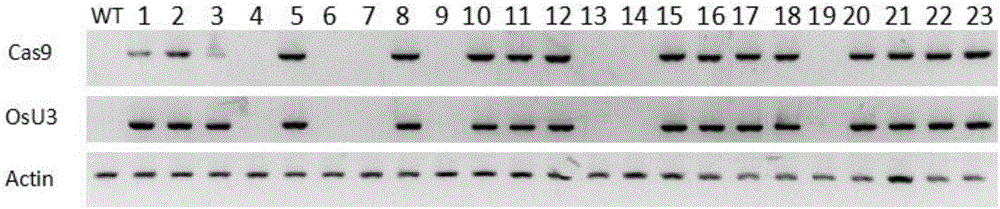
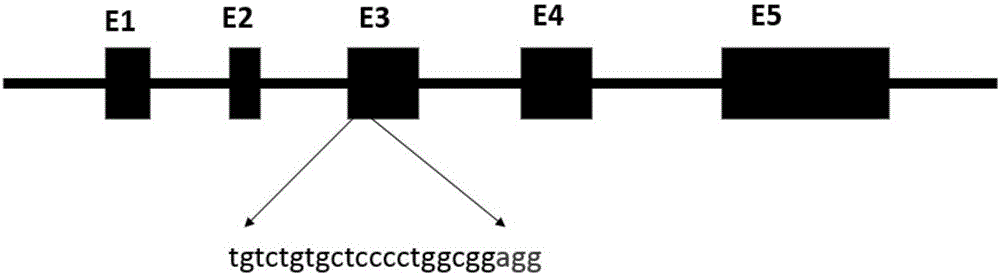
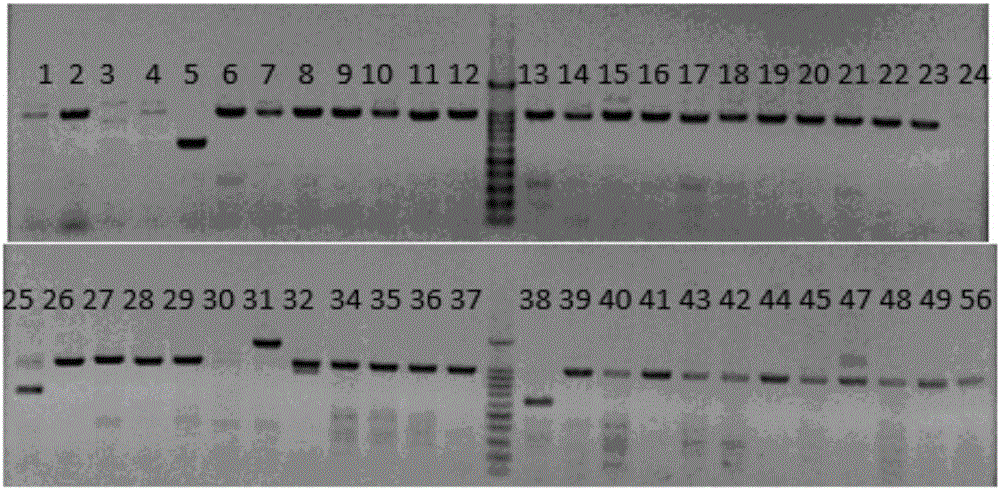
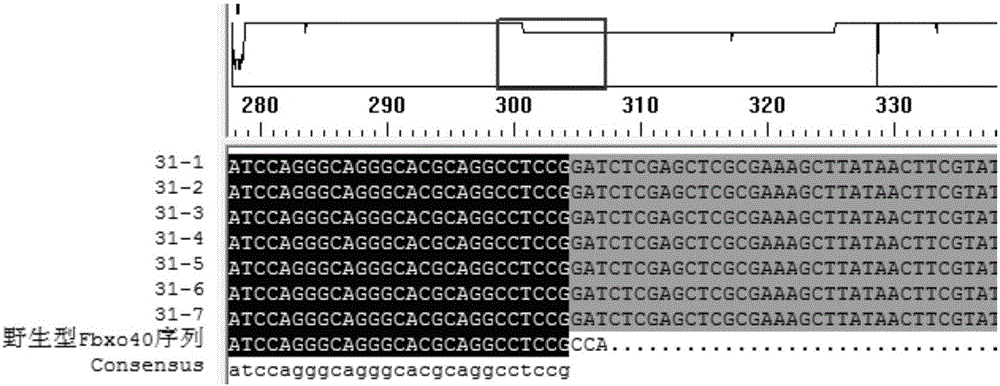
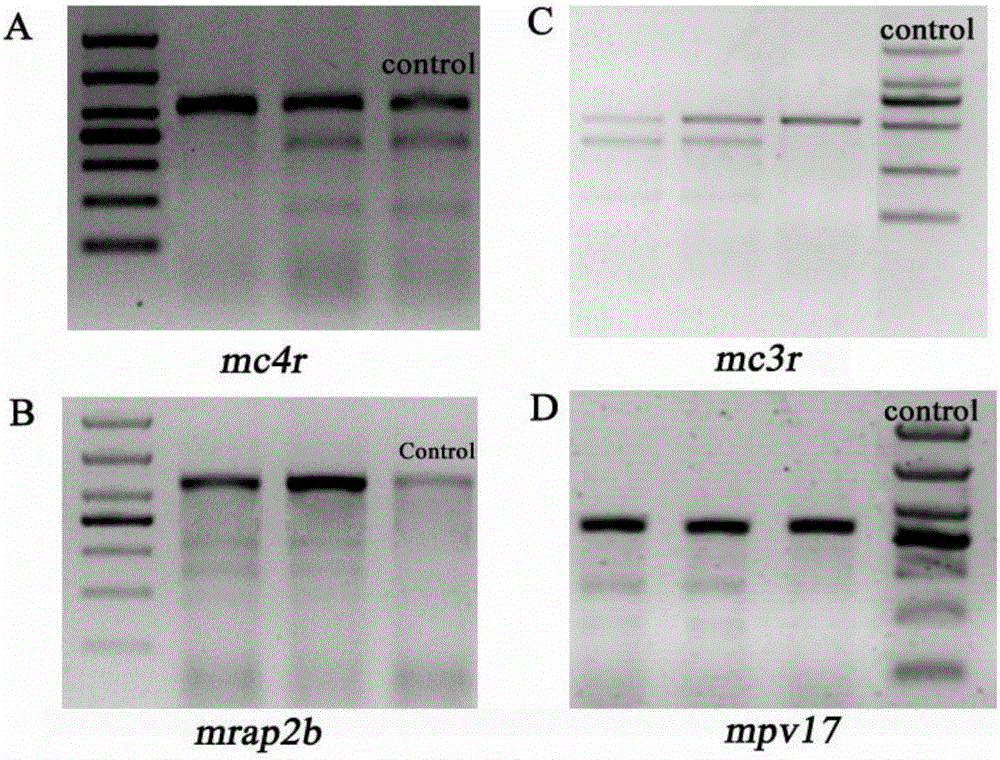
Production method for Fbxo40 gene knockout pigs
InactiveCN105821049AConvenient researchLow costNucleic acid vectorFermentationDiseaseAgricultural science
The invention provides a production method for Fbxo40 gene knockout pigs. The production method comprises the steps that a CRISPR-Cas9 targeting vector and a PGK-Neo resistant gene are jointly transfected into a porcine embryonic fibroblast to obtain a G418-resistant positive monoclonal cell, an Fbxo40 gene in the positive monoclonal cell generates insertion or deletion mutation, a reading frame generates frame shift and stops in advance, then the obtained cell is cloned to serve as a donor cell for nuclear transfer, and an oocyte serves as a receptor oocyte for nuclear transfer; cloned embryos are obtained through a somatic cell nuclear transfer technique, the high-quality cloned embryos are transferred into the fallopian tube of an oestrous sow, and the Fbxo40 gene knockout pigs are obtained through whole-period development. According to the production method, the Fbxo40 knockout pigs are efficiently obtained through a CRISPR-Cas9 gene editing technique at the low cost, and animal models are supplied to research of muscle development and diseases related to the muscles.
Owner:CHINA AGRI UNIV
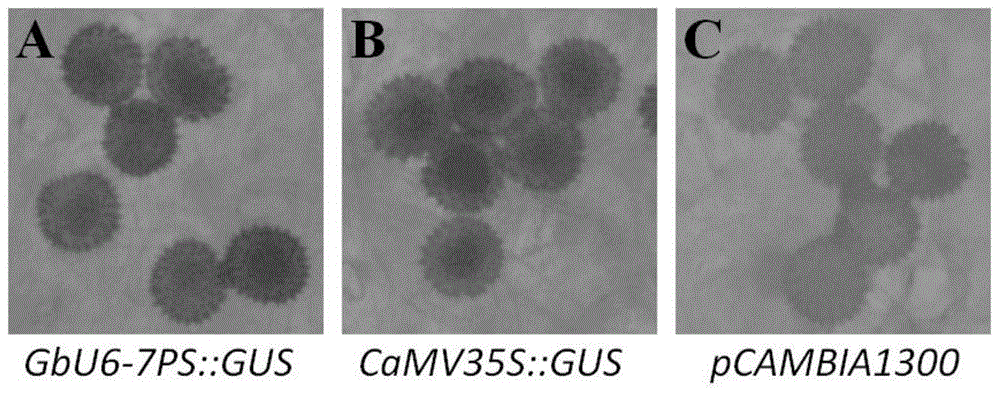
Cotton promoter GbU6-7PS and application
InactiveCN105779448AImprove editing efficiencyIncreased transcript levelsVector-based foreign material introductionDNA preparationBiotechnologyGenome editing
The invention provides a cotton promoter GbU6-7PS and application. The nucleotide sequence of the promoter is as shown in SEQ ID NO.1, and the nucleotide sequence of a complementary strand of the promoter is as shown in SEQ ID NO.2. The cotton promoter GbU6-7PS is obtained by conducting the first turn of PCR amplification so as to obtain a segment which covers full-length GbU6-7 promoter sequence, so that GbU6-7P is obtained; and then, conducting the second turn of PCR amplification, taking the GbU6-7P as a donor plasmid template and GbU6-5P::GUS as an acceptor plasmid template, and truncating and cloning the GbU6-7 promoter by virtue of Transfer PCR method, so that the cotton promoter GbU6-7PS is obtained. The cotton promoter GbU6-7PS disclosed by the invention is not only applicable to cotton, but also quite short in segment which is just 190bp long, conforming to the requirement of a CRISPR / Cas9 genome editing vector, the transcriptional level of sgRNA is significantly improved, and subsequently a genome editing efficiency may be increased.
Owner:XINJIANG AGRI UNIV
Fish breeding method for improving CRISPR-Cas9 gene editing and passage efficiency through fish roe preserving fluid
ActiveCN106191124AHigh target shooting efficiencyImproving gene editing passaging efficiencyMicroinjection basedAnimal husbandryFisheryMicroinjection
The invention belongs to the field of fish molecular breeding, and particularly relates to a fish breeding method for improving CRISPR-Cas9 gene editing and passage efficiency through fish roe preserving fluid. According to the method, the fish roe preserving fluid technology, the microinjection technology an the CRISPR-Cas9 gene editing technology are ingeniously combined, the targeting efficiency of the CRISPR-Cas9 gene editing technology and gene editing passage efficiency are greatly improved, the screening time needed by a gene editing method for fish breeding is obviously saved, and the method is of great significance in promoting rapid development of gene editing fish breeding.
Owner:CHONGQING INST OF GREEN & INTELLIGENT TECH CHINESE ACADEMY OF SCI
Cas9 plasmid, genome editing system and method of escherichia coli
ActiveUS20170226522A1Improve efficiencyImprove fidelityHydrolasesStable introduction of DNAEscherichia coliGenome editing
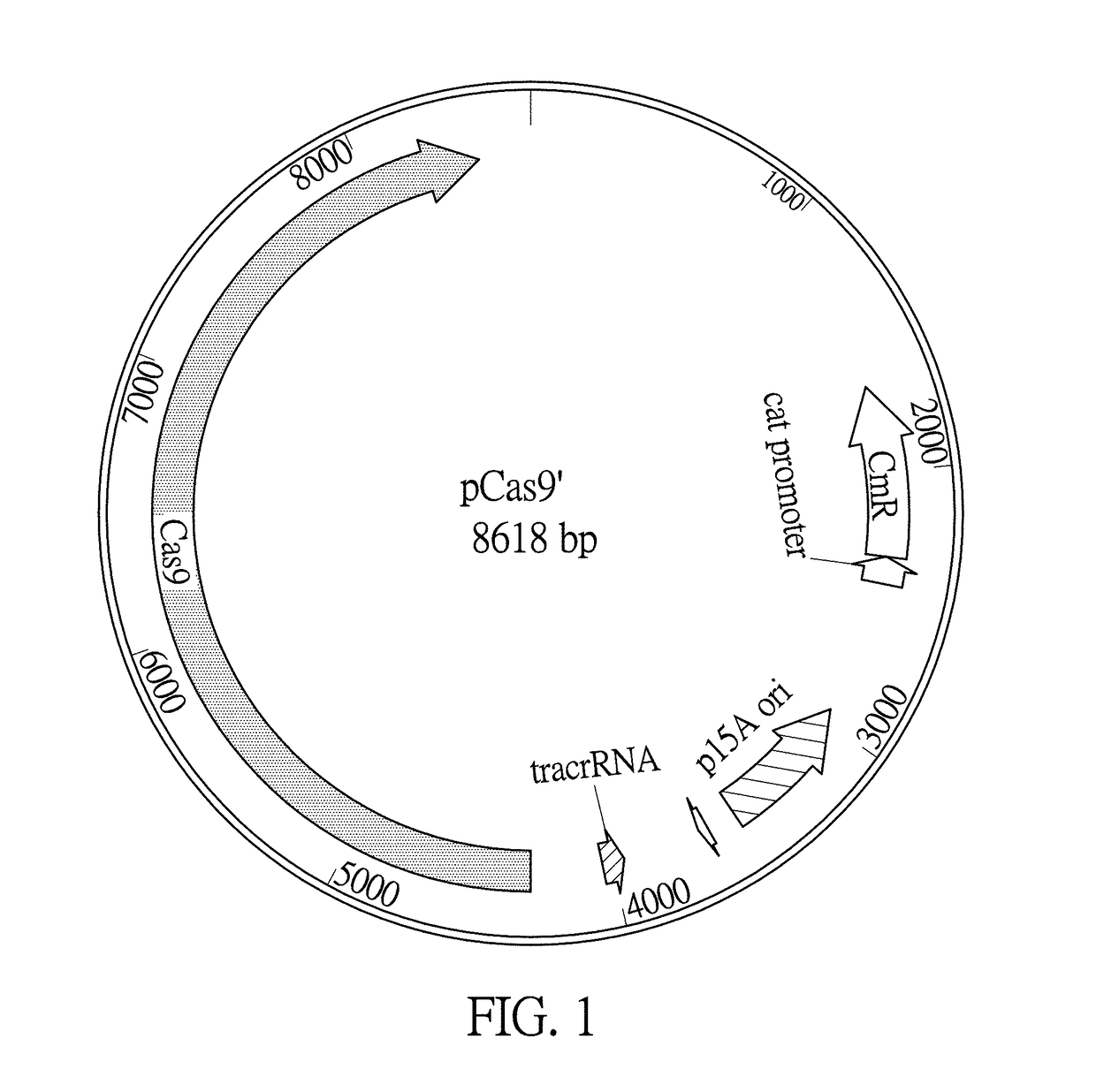
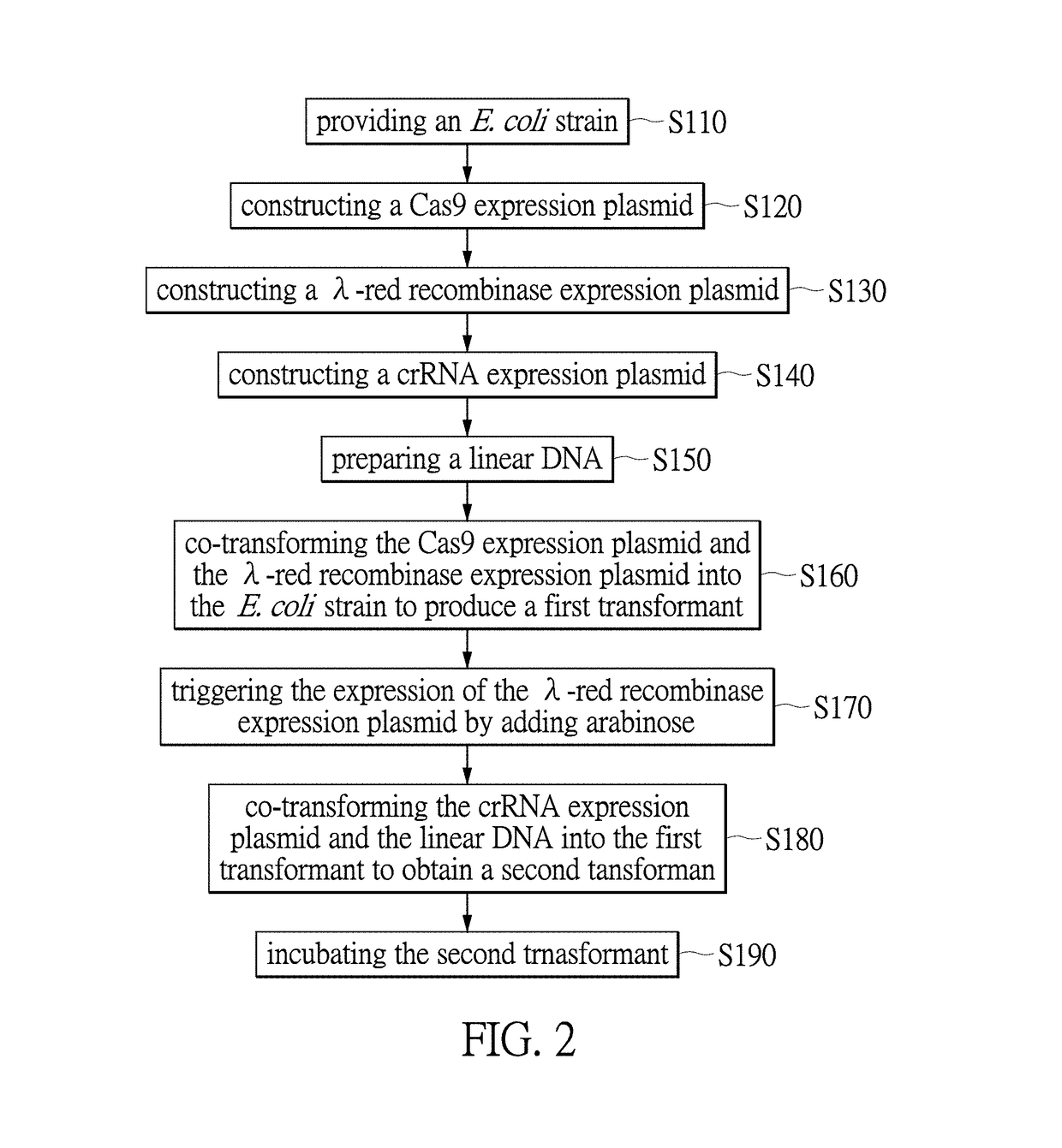
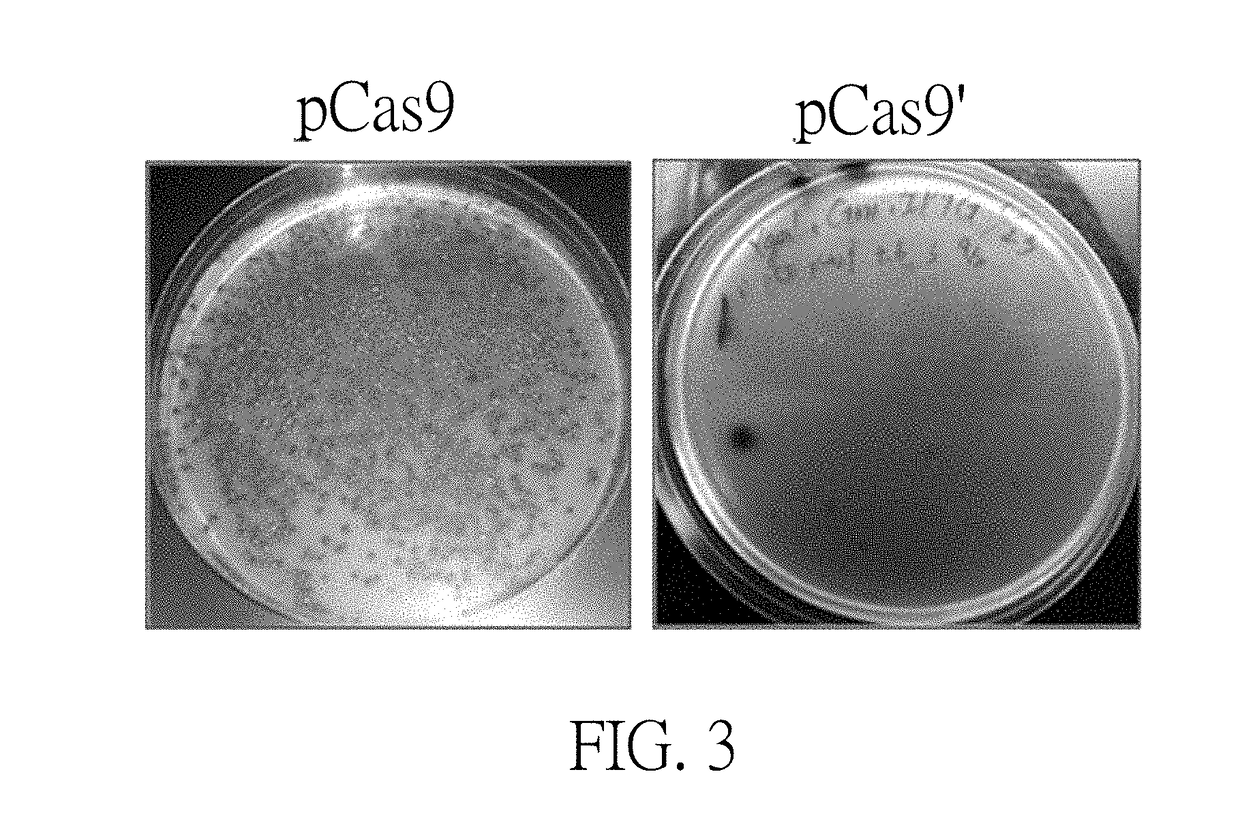
A Cas9 expression plasmid, a genome editing system and a genome editing method for Escherichia coli are provided. The Cas9 expression plasmid includes a tracrRNA sequence, a Cas9 gene and a chloramphenicol resistance gene (CmR). The Cas9 expression plasmid is applied to CRISPR / Cas-coulped λ-red recombineering system for editing genomes of E. coli with high efficiency.
Owner:NATIONAL TSING HUA UNIVERSITY
Perennial ryegrass breeding method based on CRISPR/Cas system
InactiveCN105219799AOther foreign material introduction processesVector-based foreign material introductionGenome editingMutant
The invention provides a perennial ryegrass breeding method based on a CRISPR / Cas system. A CRISPR / Cas genome-editing technique is applied to breed improvement of perennial ryegrass, gene site-directed knock-out is conducted on lignin synthesized key enzyme genes LpCCR and LpCOMT, homozygous and transformation carrier deleted mutants are selected out, the breeding material of the perennial ryegrass of which the lignin content is decreased is bred, and important significance for breeding research and breed improvement of pasture is achieved.
Owner:TIANJIN GENOVO BIOTECH CO LTD
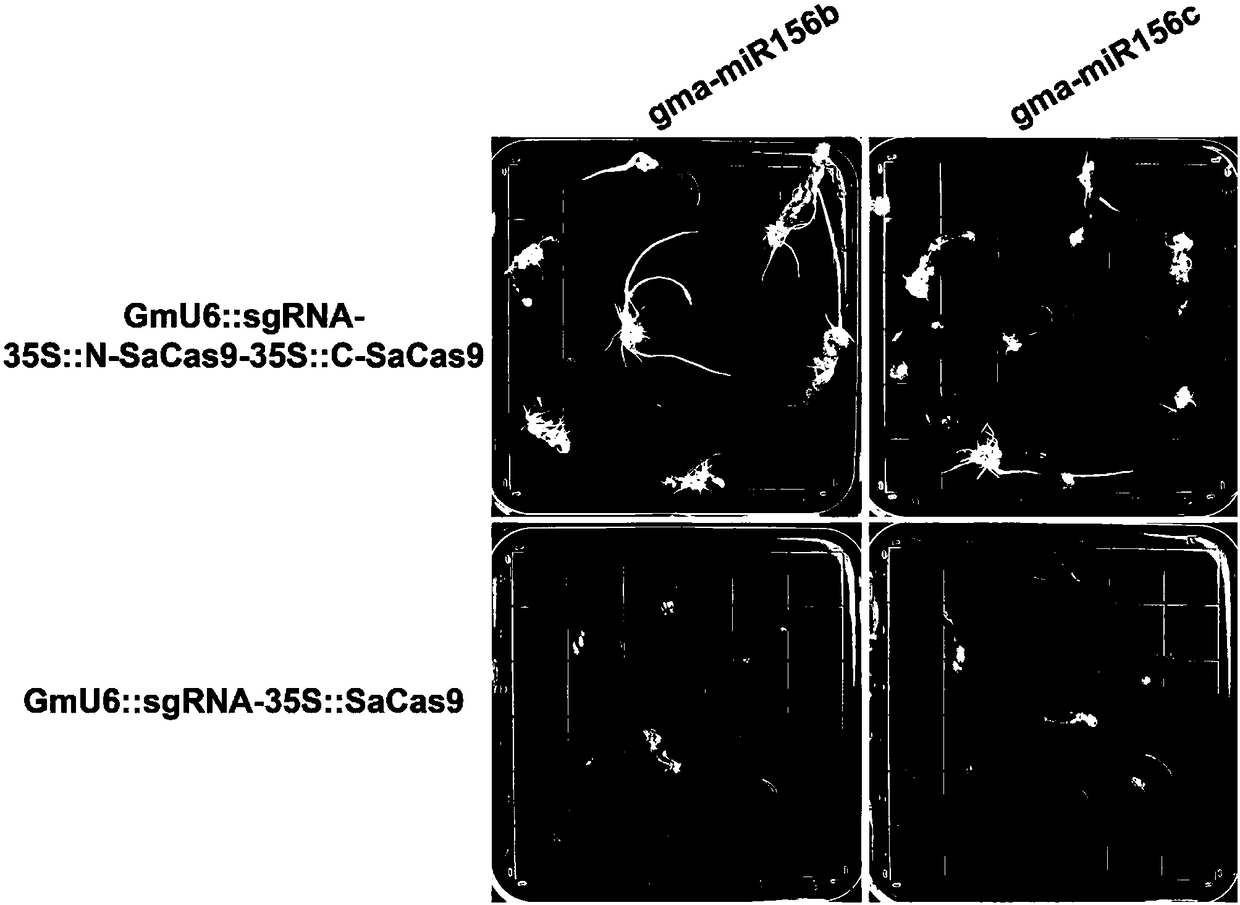
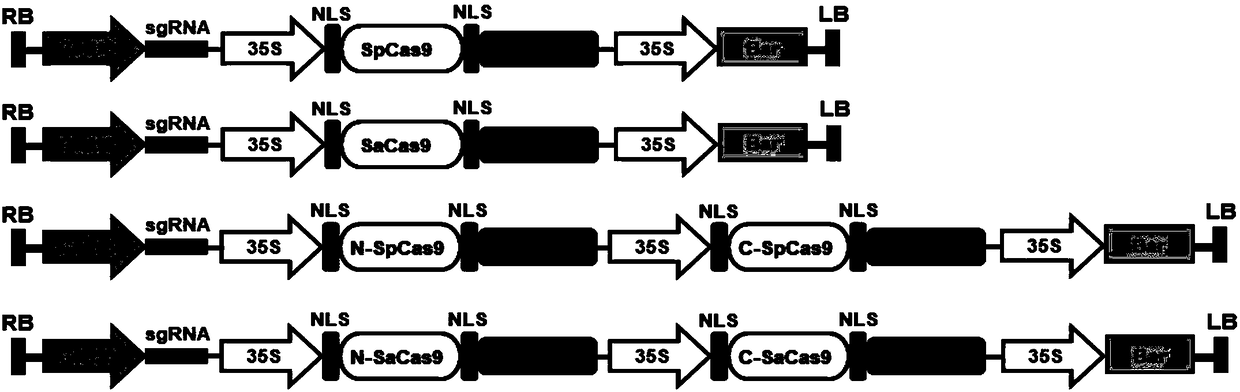
Construction method and application of high-efficiency soybean CRISPR/Cas9 system
InactiveCN108588128AImprove editing efficiencyImprove application breadthStable introduction of DNANucleic acid vectorAgricultural scienceGenome editing
The invention discloses a construction method and an application of the high-efficiency soybean CRISPR / Cas9 system. Staphylococus aureus Cas9 protein undergoes split expression; and in order to improve editing efficiency and broad spectrum, a soybean SaCas9 editing system is constructed. According to the construction method provided by the invention, a low-efficiency editing system is transformedinto the high-efficiency editing system, so that the application scope of editing soybean genome is widened, and meanwhile, a new thought is provided for solving a problem on crop genome editing efficiency in the future.
Owner:NANCHANG UNIV
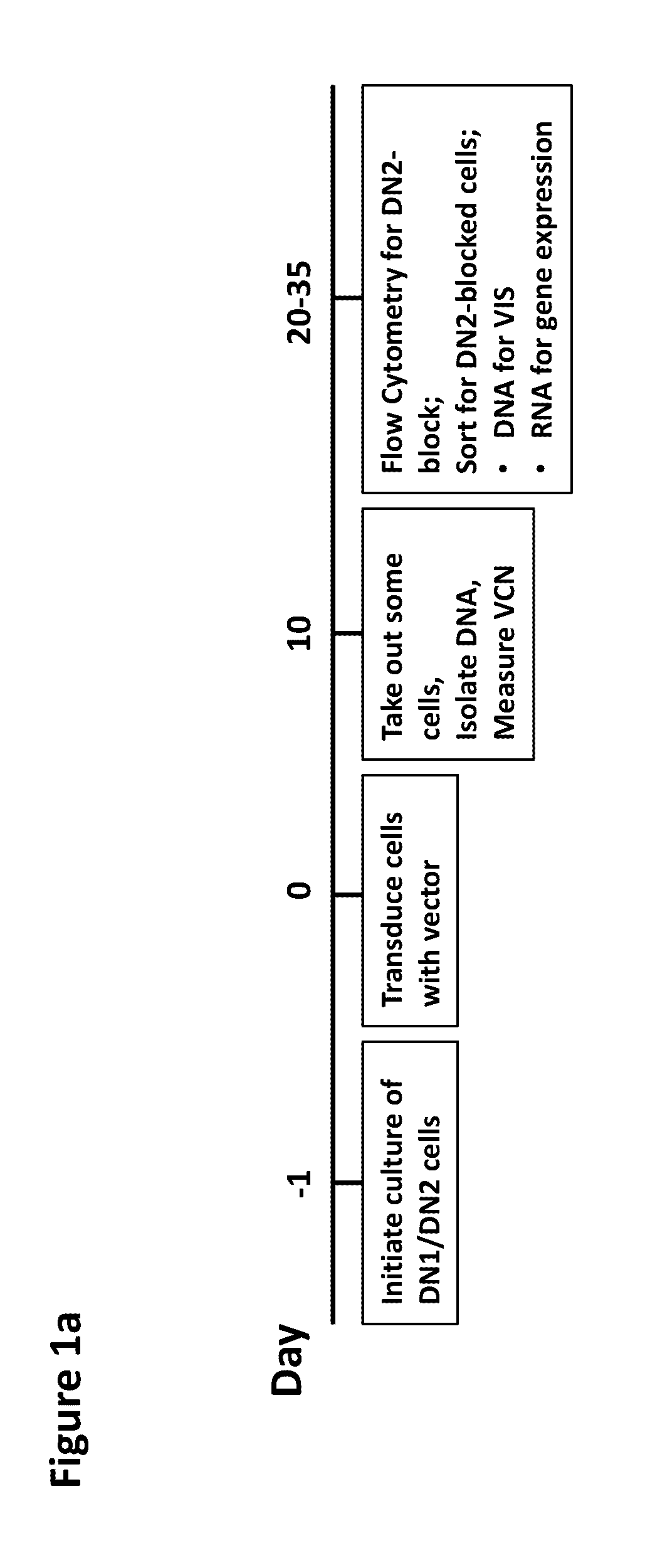
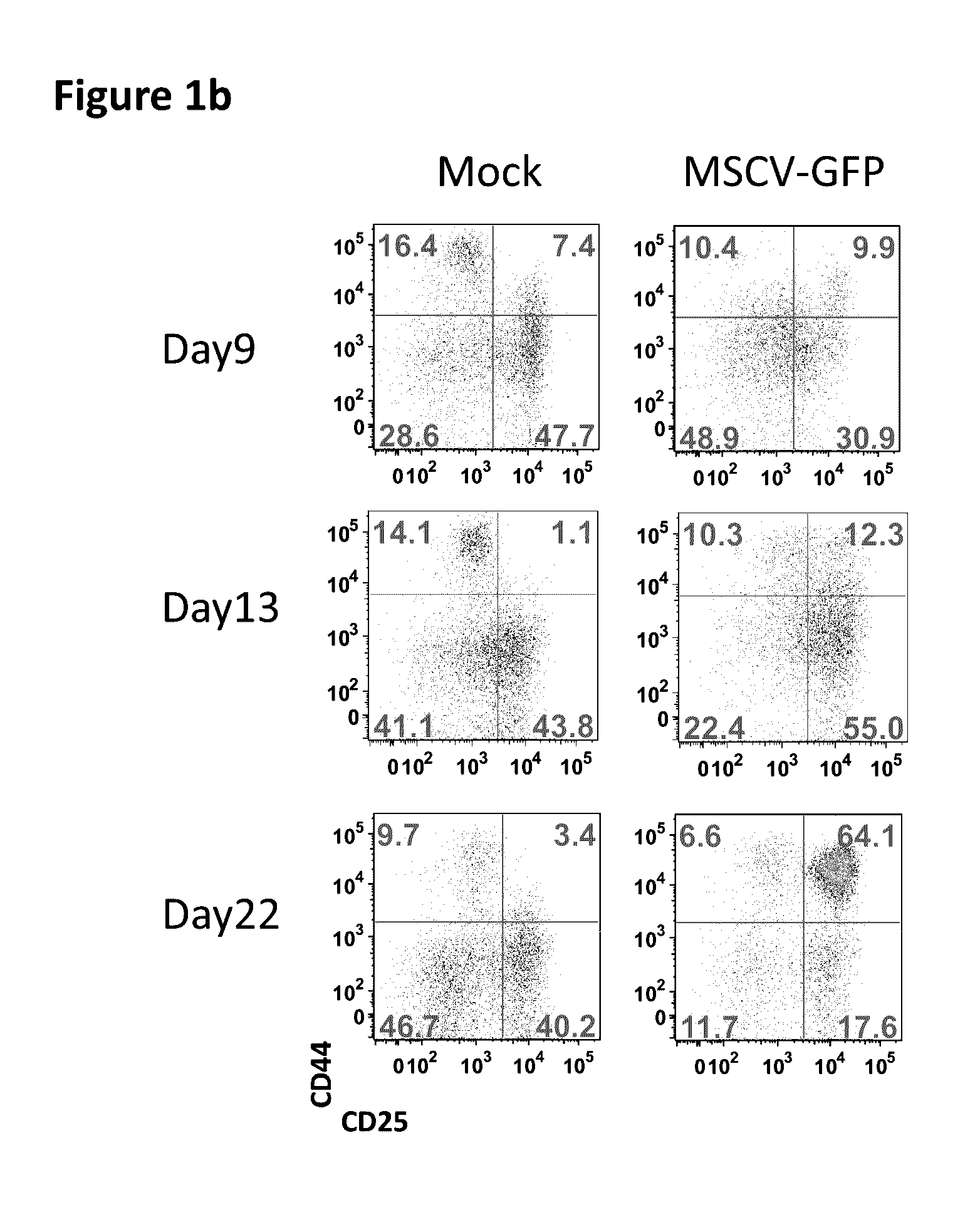
Assay for safety assessment of therapeutic genetic manipulations, gene therapy vectors and compounds
ActiveUS20160312304A1Microbiological testing/measurementGenome editingHematopoietic progenitor cells
The invention is directed to methods of assessing the safety of therapeutic compounds and therapeutic genetic manipulations, including integrating gene therapy vectors and genome editing. In particular, the invention provides a method, wherein the oncogenic potential of therapeutic compounds and therapeutic genetic manipulations, including integrating gene therapy vectors and genome editing, is determined by determining the percentage of differentiation blocked hematopoietic progenitor cells.
Owner:ST JUDE CHILDRENS RES HOSPITAL INC
Quantitative detection method for specific nucleic acid fragments based on CRISPR technology
ActiveCN108823291ASolve the problem of quantitative detectionQuantitative detection is fast and efficientMicrobiological testing/measurementGenome editingFluorescence
The invention discloses a quantitative detection method for specific nucleic acid fragments based on the CRISPR technology. The quantitative detection method comprises the following steps: acquiring atreated target nucleic acid sample, and adding the target nucleic acid sample and an aqueous-phase solution into a droplet generation device to generate reaction droplets, wherein the reaction droplets comprise at least one nucleic acid fragment to be tested; amplifying the nucleic acid fragment to be tested in the reaction droplets by using a recombinase polymerase amplification technique, and allowing target gene signals in the nucleic acid fragment to be tested to realize cascade amplification by using a gene editing technique; collecting a positive signal corresponding to a target gene, and converting the positive signal into image data; processing a fluorescence threshold in the image data by using the Poisson distribution principle so as to obtain the test corresponding to the number of the target gene, corresponding to the positive signal, in the nucleic acid fragment to be tested. Thus, rapid efficient quantitative detection of the concentration of the target gene is realizedbased on the CRISPR technology.
Owner:领航医学科技(深圳)有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com