Gene editing method based on streptomyces virginiae IBL14 gene cas7-5-3
A technology of gene editing and Streptomyces, applied in biochemical equipment and methods, viruses/bacteriophages, using vectors to introduce foreign genetic materials, etc., can solve problems that have not been discovered, and have not seen CRISPR-CasI type systems, etc., to achieve effective gene The effect of editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 (Three-plasmid one-step co-transformation method to knock out EC JM109 lacZ Gene)
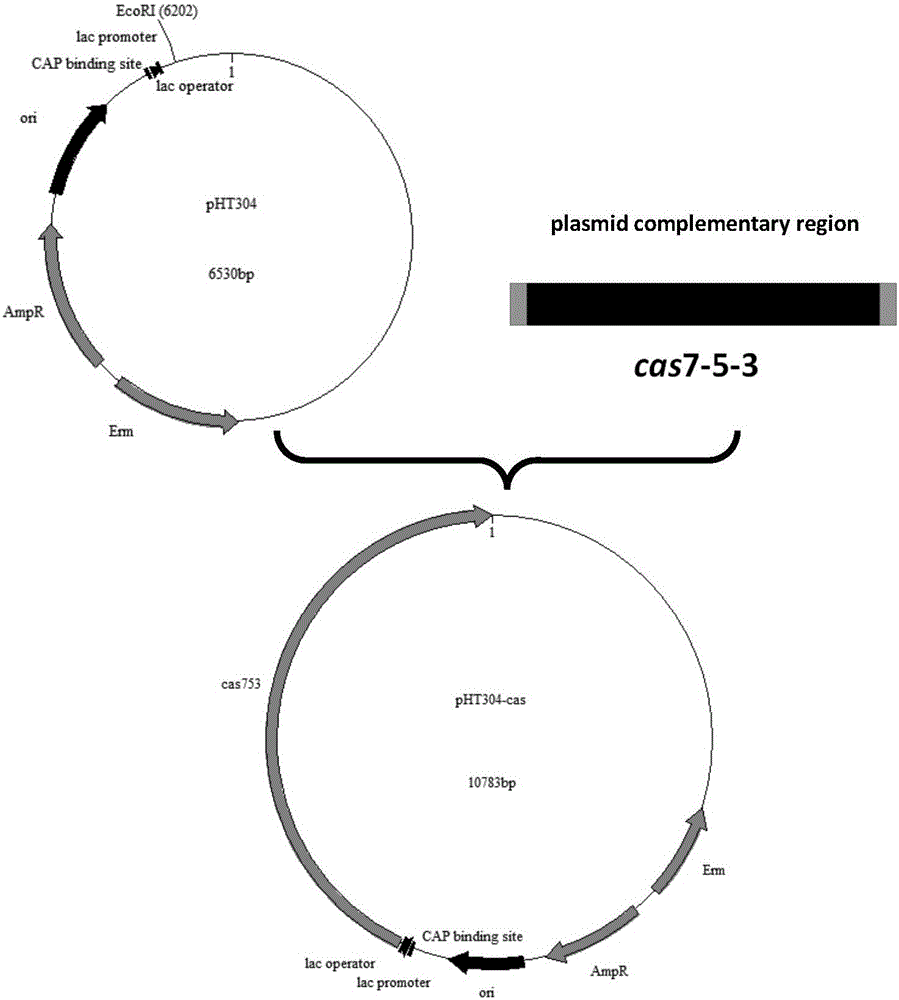
[0034] (1) Protein expression plasmid pHT304- cas 7-5-3 Construction
[0035] according to SV IBL14 whole-genome sequencing information and plasmid pHT-304 sequence information, designed with the complementary sequence of plasmid pHT-304 cas 7-5-3 Gene-specific primers cas -F and cas -R (Table 2). extract SV IBL14 genomic DNA was processed using TransStart FastPfu DNA Polymerase produced by Quanshijin Biotechnology Co., Ltd. cas Gene PCR amplification, reaction conditions: 95°C for 5 min, 94°C for 30 s, 55°C for 30 s, 72°C for 2 min, 2.5 U Pfu DNA Polymerase produced by Sangon Bioengineering (Shanghai) Co., Ltd. (50 μl reaction system ), 30 cycles at 72°C for 10 min. The PCR product was detected by 1% agarose electrophoresis, recovered by the kit, and purified cas full-length gene. in one step cas The full-length gene sequence was connected with the plasmid p...
Embodiment 2
[0052] Embodiment 2 (two-step co-transformation knockout of three plasmids EC JM109 lacZ Gene)
[0053] (1) Protein expression plasmid pHT304- cas 7-5-3 Construction
[0054] Same as embodiment 1 step (1)
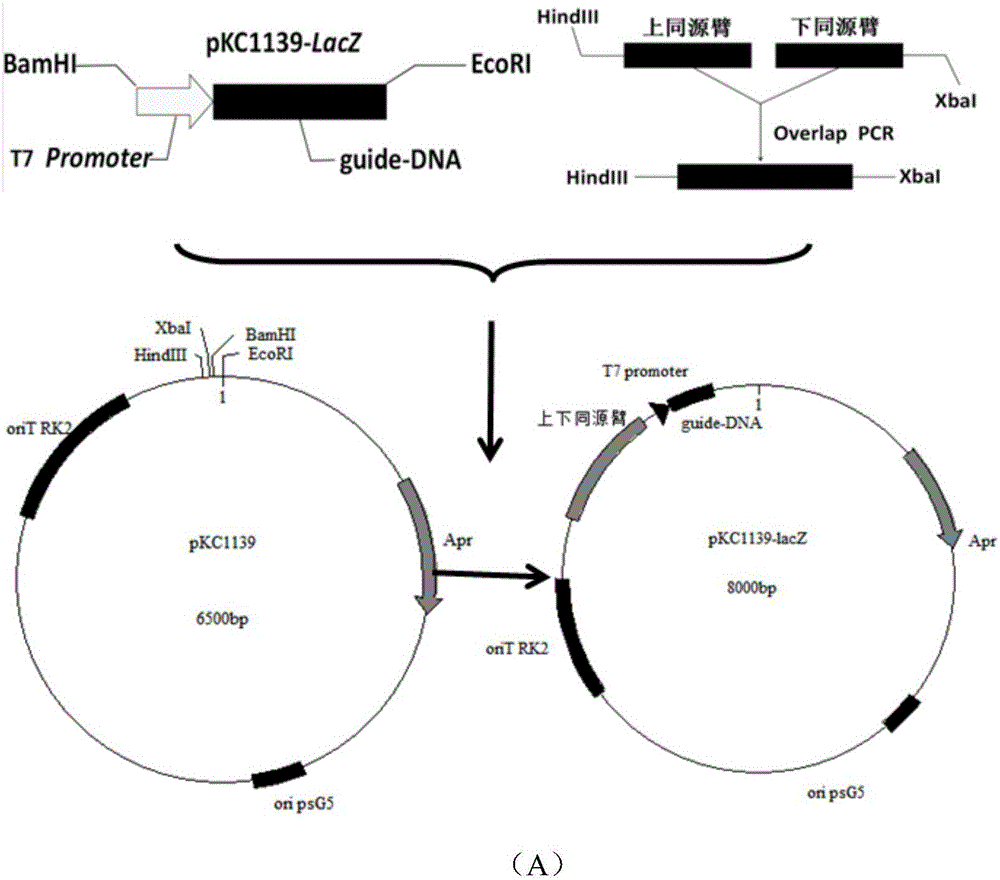
[0055] (2) Gene editing plasmid pKC1139- lacZ - Construction of t-DNA
[0056] Same as embodiment 1 step (2)
[0057] (3) Acquisition and inspection of recombinants
[0058] (A) Plasmid pHT304- cas 7-5-3 Transformation and EC JM109-pHT304- cas 7-5-3 Competent preparation
[0059] The plasmid pHT304- cas 7-5-3 Transforms to EC In the JM109 competent state, the transformant was screened by erythromycin resistance, and then the transformant was used to prepare a competent state to obtain a plasmid containing pHT304- cas 7-5-3 EC JM109-pHT304- cas Competence of 7-5-3 strain. The competent preparation method is the same as step (3A) in Example 1.
[0060] (B) Plasmids pKD46 and pKC1139- lacZ - Co-transformation of t / g-DNA
[0061] Plasmids pKD46 and...
Embodiment 3
[0064] Embodiment 3 (two plasmid one-step co-transformation method knockout EC JM109 lacZ Gene)
[0065] (1) Protein expression plasmid pHT304- cas 7-5-3 Construction
[0066] Same as embodiment 1 step (1)
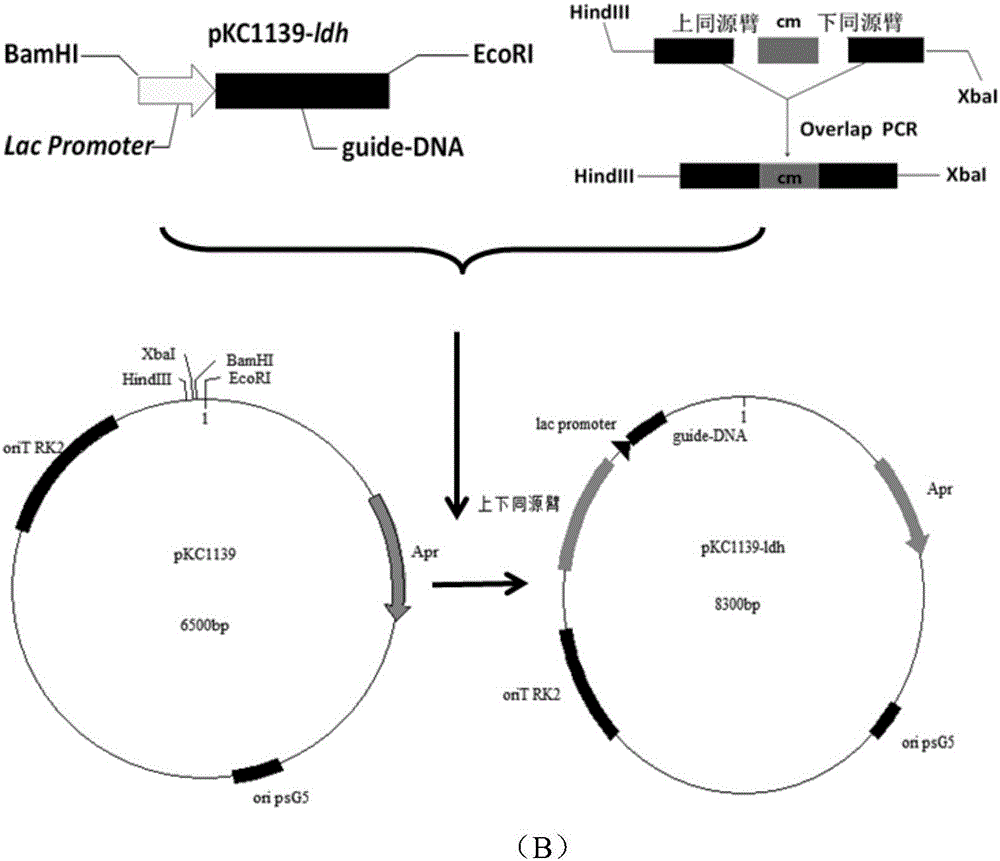
[0067] (2) Gene editing plasmid pKD46- lacZ - Construction of t / g-DNA
[0068] In addition to the homology arm and guide DNA- lacZ The target gene fragments are sequentially connected to the plasmid pKD46 to form the gene editing plasmid pKD46- lacZ Except for -t / g-DNA, other steps are the same as step (2) in Example 1.
[0069] (3) Acquisition and inspection of recombinants
[0070] Same as embodiment 1 step (3)
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com