SSA (single-strand annealing) repair-based gene seamless editing method utilizing CRISPR/Cas9 technology
A genetic, seamless technology, applied in the field of seamless gene editing based on SSA repair using CRISPR/Cas9 technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
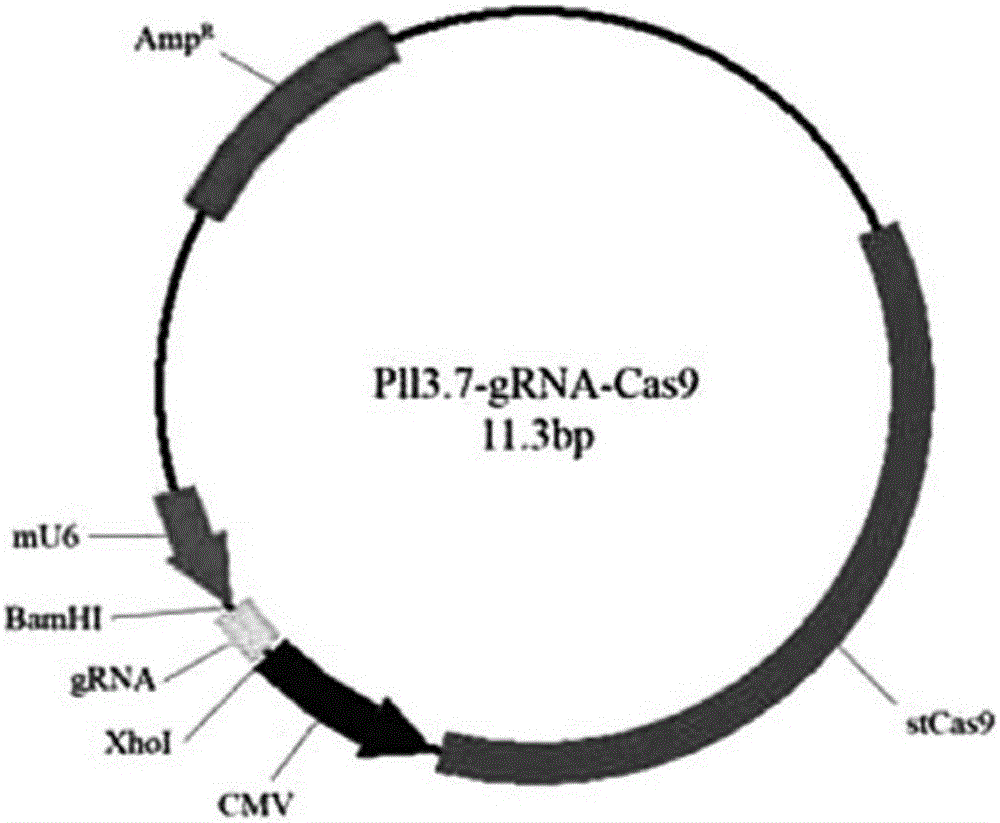
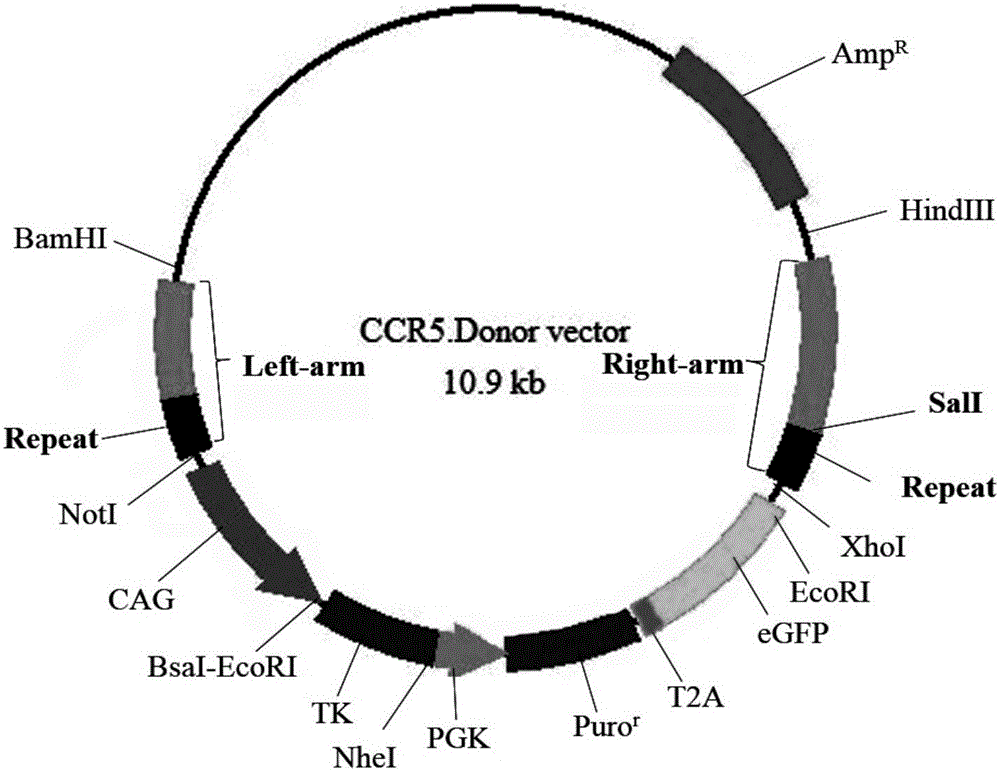
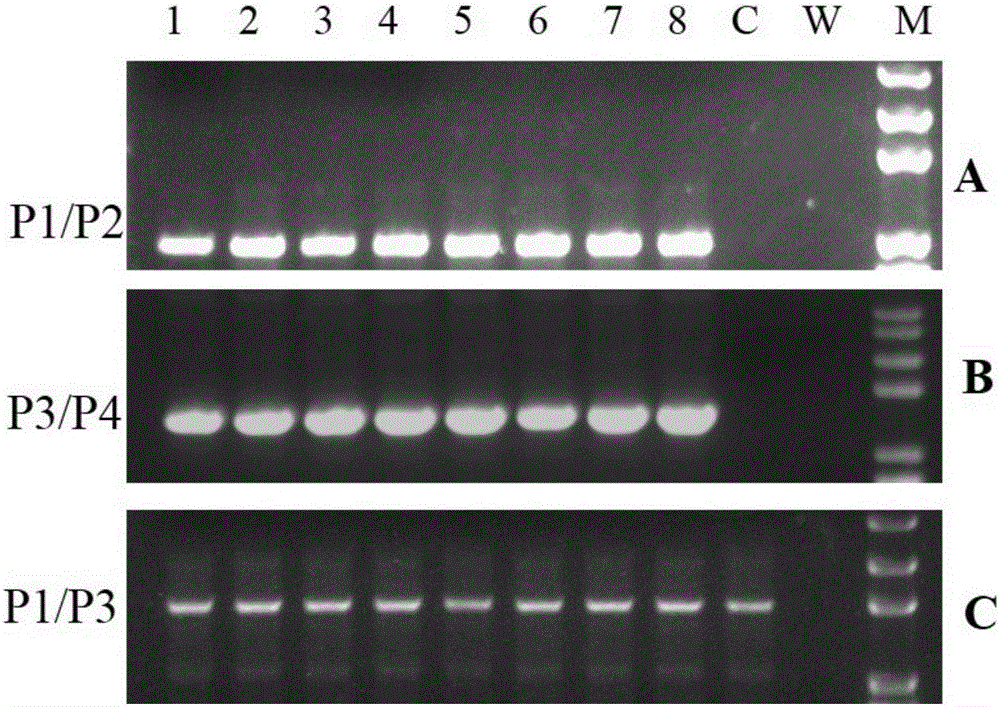
[0027] A method for seamless gene editing based on SSA repair using CRISPR / Cas9 technology. The seamless gene editing method first constructs the CCR5.CRISPR / Cas9 targeting the CCR5 gene and the eGFP.CRISPR / Cas9 expression vector targeting eGFP, and Construction of the donor CCR5-Donor vector with pXL-BACII-L-arm-CAG-TK-PGK-Puro R -T2A-eGFP-bGH pA-R-arm structure, where L-arm and R-arm are left and right homology arms, CAG-TK-PGK-Puro in the middle of L-arm and R-arm R -T2A-eGFP-bGH pA sequence is the structure of screening marker components; through two transfections, two CRISPR / Cas9 technologies are used to target the CCR5 gene and eGFP target sites and the intracellular HR and SSA repair mechanism, through the positive Negative screening for seamless editing.
[0028] Construction of CCR5.CRISPR / Cas9 targeting CCR5 gene and eGFP.CRISPR / Cas9 targeting eGFP expression vector:
[0029] Taking the construction of CRISPR / Cas9 expression vector as the prior art, the structure o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com