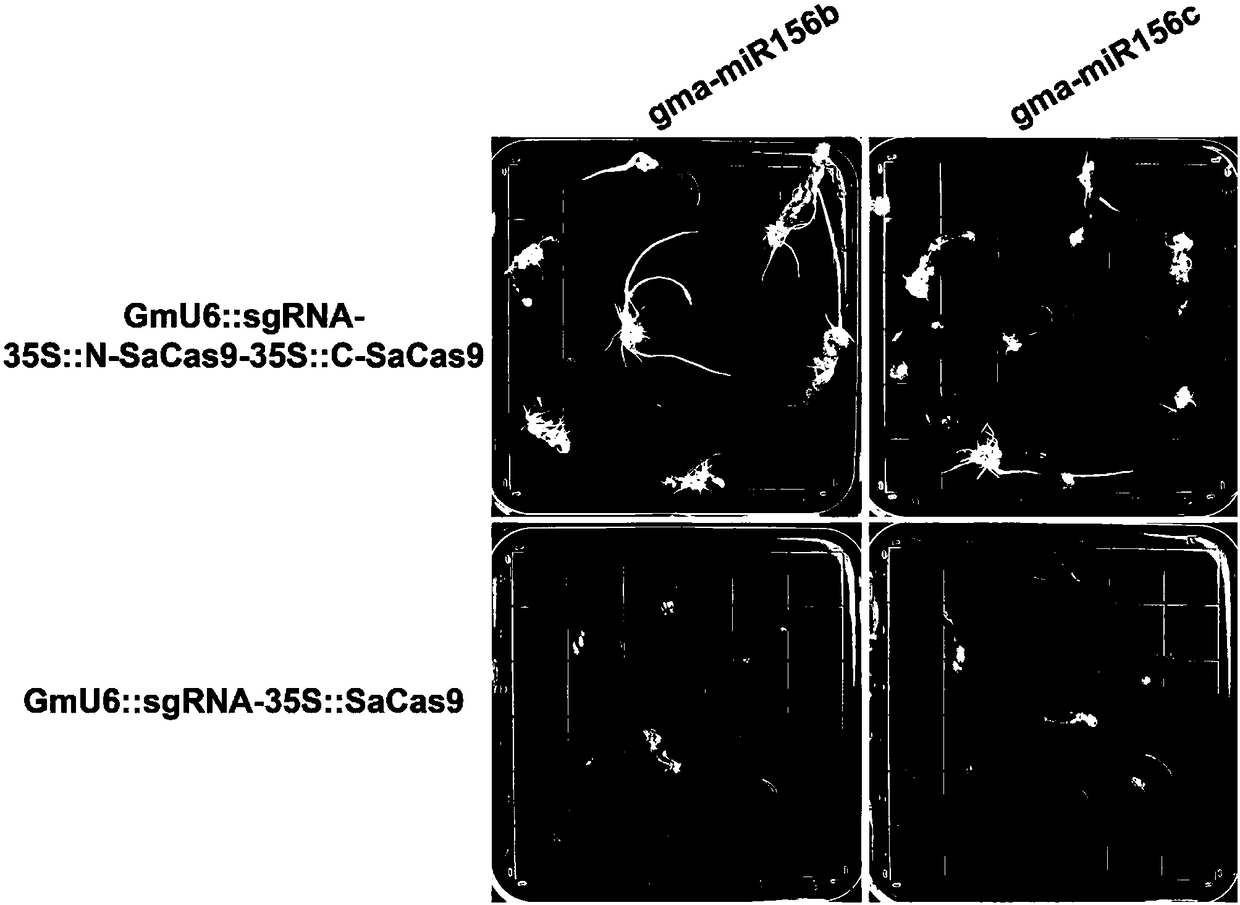
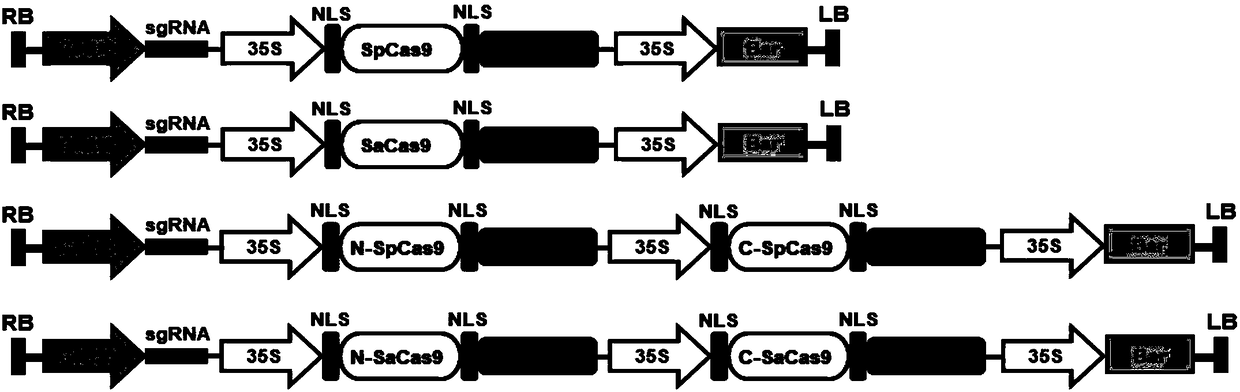
Construction method and application of high-efficiency soybean CRISPR/Cas9 system
A construction method and high-efficiency technology, applied in the field of genetic engineering, can solve problems such as editing efficiency needs to be further optimized, stable genetic transformation system to verify editing efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The sources of reagents and materials used in this embodiment:
[0034] 1) The pCAMBIA1300 vector is owned by the experiment;
[0035] 2) Primer synthesis and sequencing are entrusted to Beijing Qingke Xinye Biotechnology Co., Ltd.;
[0036] 3) The soybean variety is YC03-3;
[0037] 4) The reagents used to construct the vector are Vazyme ClonExpress Seamless Cloning Kit, NEB T4ligase, TIANAprep Mini Plasmid Kit II Plasmid Extraction Kit, TaKaRa MiniBest DNA FragmentPurification Kit, NEB BsaI enzyme, etc.
[0038] 1. Construction of Soybean CRISPR / Cas9 Editing Vector
[0039] 1. Pick a single colony of the pCAMBIA1300 plasmid, inoculate it in 50ml of liquid LB medium containing kana antibiotics, place it on a shaker at 37°C, and culture it at 220rpm / r overnight;
[0040] 2. Collect the bacteria by centrifugation, and extract the plasmid with TIANAprep Mini Plasmid Kit II plasmid extraction kit;
[0041]3. Select a suitable enzyme cutting site, digest the pCAMBIA1300...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com