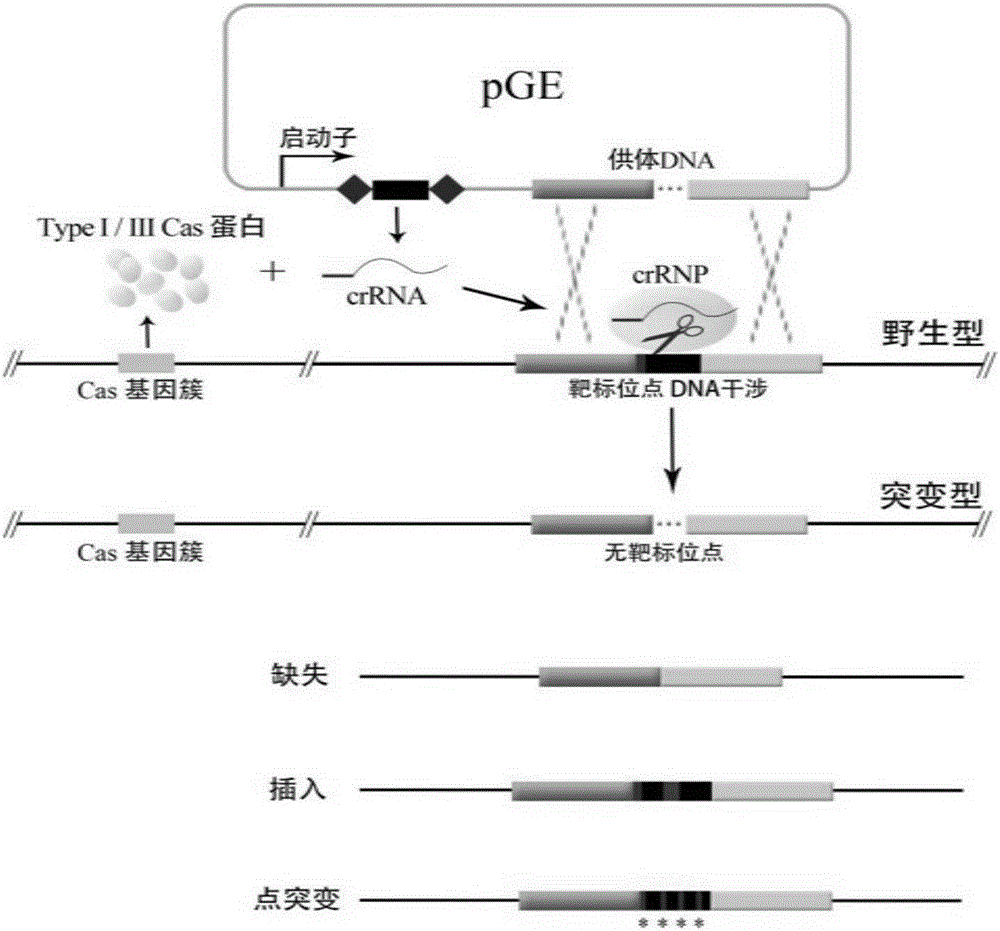
Method for editing prokaryotic genomes using endogenic CRISPR-Cas (CRISPR-associated) system
A genome editing and prokaryotic technology, applied in the field of prokaryotic genome editing, can solve the problems of limiting the application of CRISPR/Cas9 system, affecting the activity of Cas9 protein, affecting the specificity of gene editing, etc., achieving short time period, reducing workload and screening The effect of high positive rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] A method for editing a prokaryotic genome using an endogenous CRISPR-Cas system, comprising the following steps:
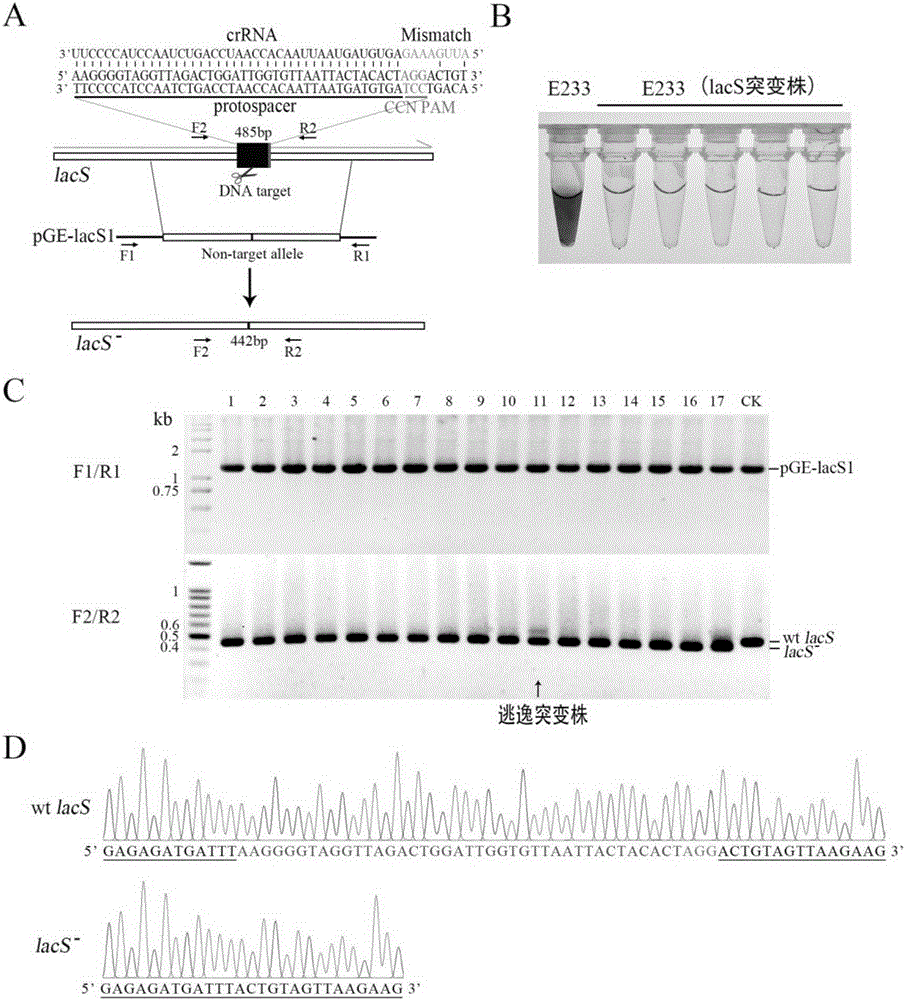
[0039] This example uses the endogenous (Type I-A CRISPR system and Type III-B CRISPR system) CRISPR system of S. islandicus REY15A to carry out precise deletion mutation of its lacS gene as an example to illustrate:
[0040] 1. Construction of Editing Plasmids
[0041] (1) A total of 40 bases from +933 to +972 were selected from the S. islandicus REY15Aβ-galactosidase gene lacS (SIRE_RS11295) as the protospacer, and its reverse complementary sequence was 5'-AGTGTAGTAATTAACACCAATCCAGTCTAACCTACCCTT-3', It is next to a CCT-PAM (Protospacer Adjacent Motif), so it can be targeted by the Type I-A CRISPR system; at the same time, because the 5' end sequence of crRNA is mismatched with the corresponding target site sequence, it can also be targeted by the Type III-B CRISPR system at the same time. Based on this protospacer, two primers (LacS-E-SpF / LacS-E-SpR) (Ta...
Embodiment 2
[0054] A method for editing a prokaryotic genome using an endogenous CRISPR-Cas system, comprising the following steps:
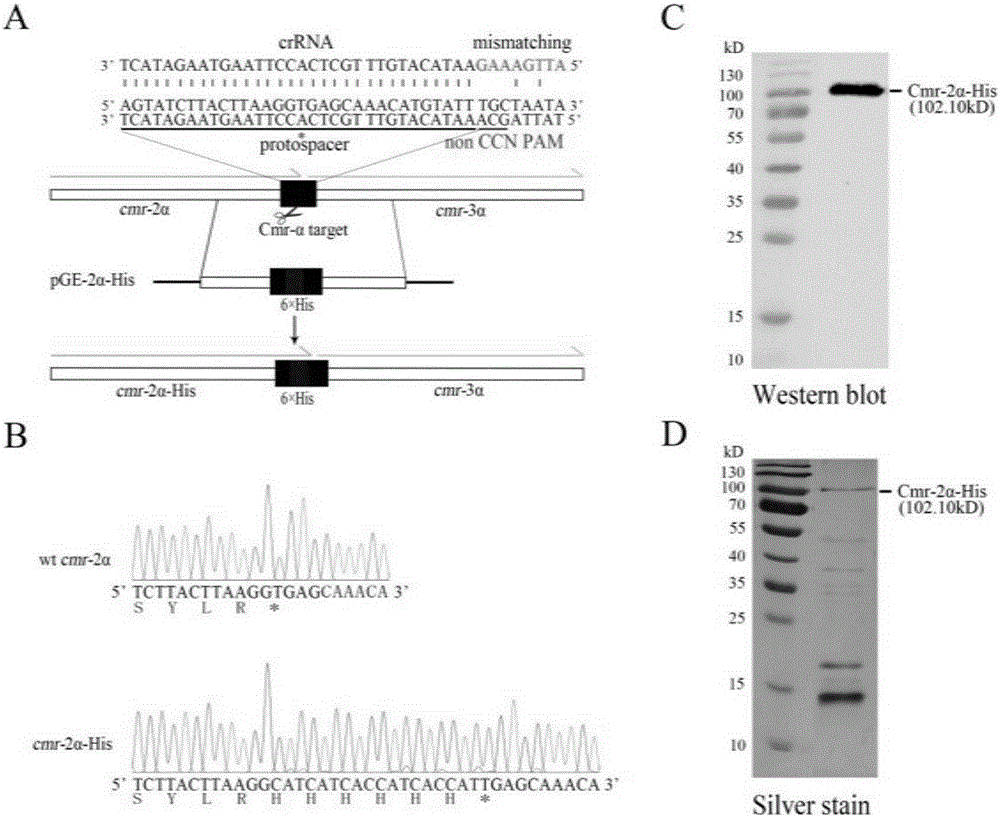
[0055] In the embodiment of the present invention, the C-terminus of the Cmr-2α protein of S. islandicus REY15A endogenous III-B type CRISPR system is used as an example to illustrate the 6×His tag insertion mutation.
[0056] 1. Construction of Editing Plasmids
[0057] (1) Select the last 25 bases of the cmr-2α gene (SIRE_RS04505) and the first 7 bases of the cmr-3α gene on the S. islandicus REY15A genome as a protospacer, and its reverse complement The sequence is 5'-AGTGTAGTAATTAACACCACAATCCAGTCTAACCTACCCCT-3', without CCN-PAM in front of it, so it only has the DNA interference activity of type III-B CRISPR. Based on this protospacer, two primers (2α-His-SpF / 2α-His-SpR) (Table 1) were designed, and the two primers were annealed to generate spacer fragments with sticky ends at both ends;
[0058] (2) The artificial CRISPR vector pSe-Rp is digested with...
Embodiment 3
[0070] A method for editing a prokaryotic genome using an endogenous CRISPR-Cas system, comprising the following steps:
[0071] This example uses the endogenous Type I-A CRISPR system of Sulfolobus Icelandicus REY15A to carry out multi-site mutation of its Cmr-2α protein HDdomain as an example to illustrate.
[0072] 1. Construction of Editing Plasmids
[0073] (1) On the S. islandicus REY15A genome, select 40 bases at the four conserved amino acids H / D / K / I of the N-terminal HDdomain of the cmr-2α gene as the protospacer, and its sequence is 5'-CGACCCTCCTTGGAAGGCATGGGTAATTACAAGGAATATT-3 ', preceded by CCA-PAM. Based on this protospacer, two primers (2α-HDmut-SpF / 2α-HDmut-SpR) were designed, and the two primers were annealed to generate a spacer fragment with sticky ends at both ends;
[0074] (2) The artificial CRISPR vector pSe-Rp is digested with BspMI, and the digested product is enzyme-ligated with the above spacer fragment to obtain an artificial CRISPR plasmid;
[0075...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com