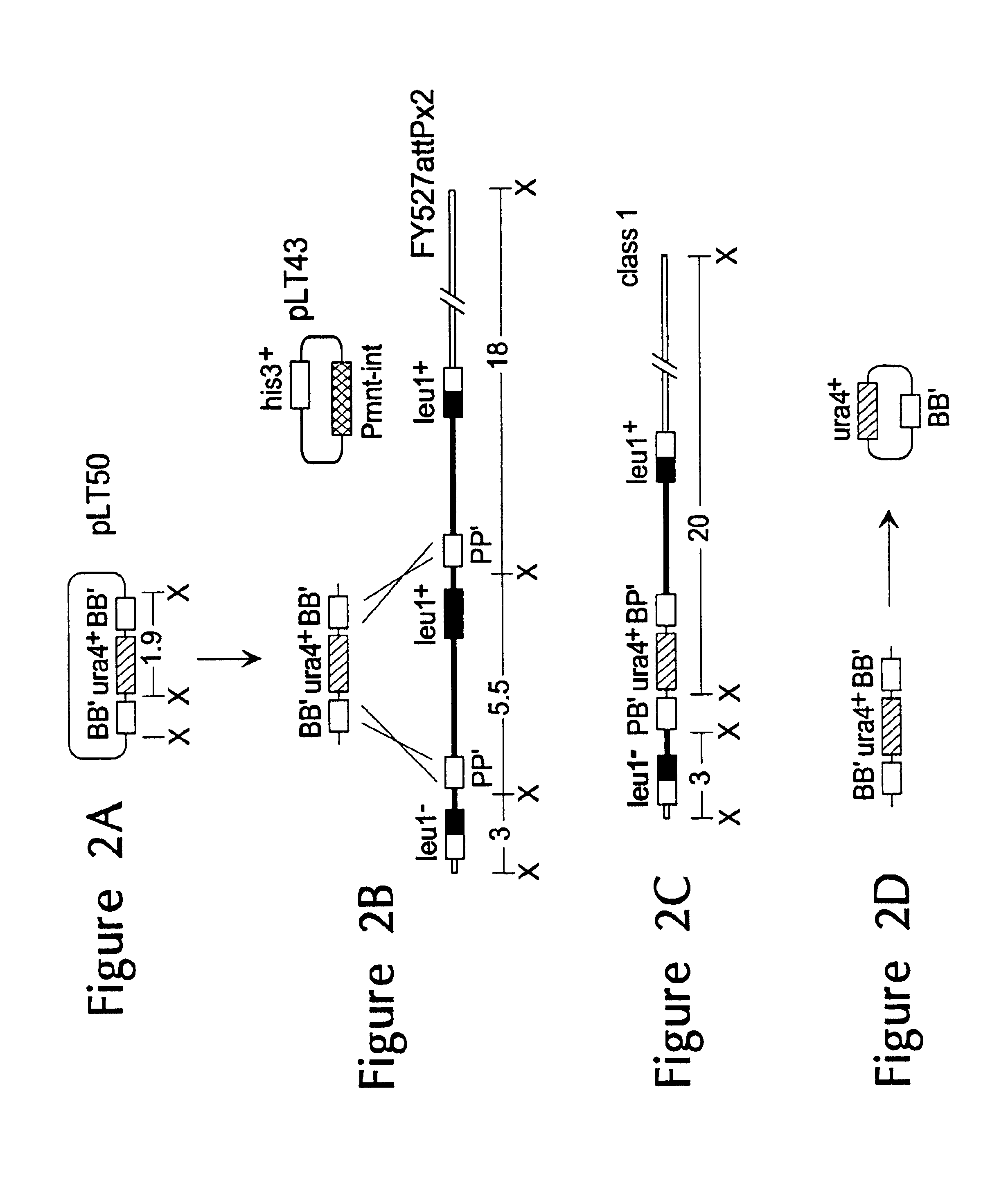
Patents
Literature
40 results about "Eukaryotic genome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Eukaryotic Genomes The genome sizes of eukaryotes are tremendously variable, even within a taxonomic group (so-called C-value paradox). Eukaryotic genomes are divided into multiple linear chromosomes; each chromosome contains a single linear duplex DNA molecule.
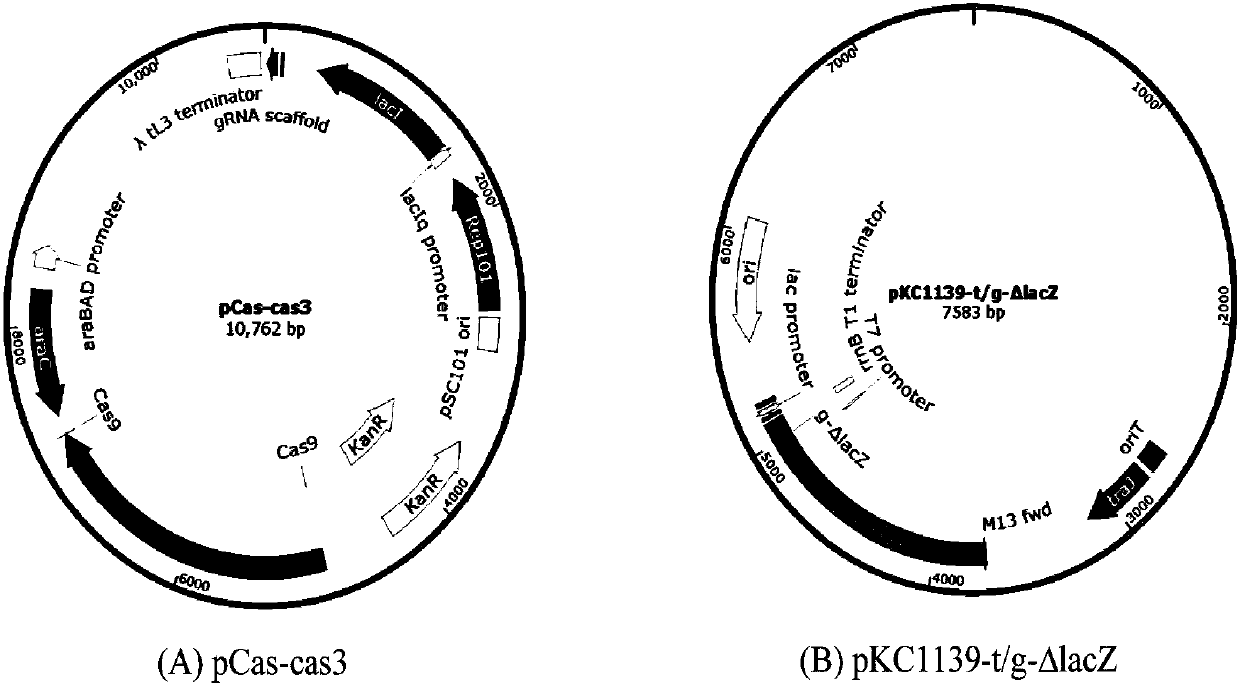
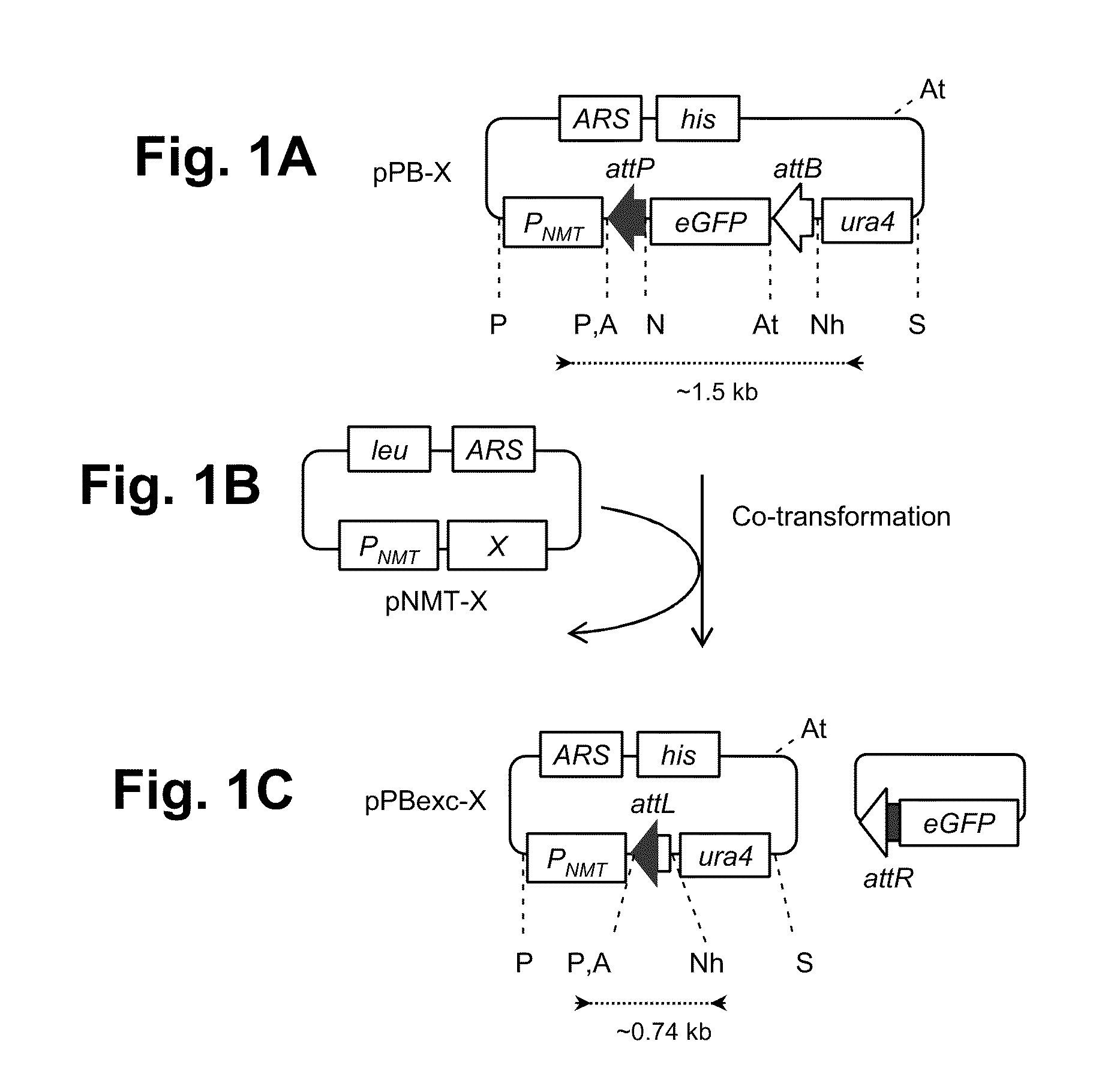
Gene editing method based on gene cas3 of I-B type CRISPR-Cas system
PendingCN107557373ASmall molecular weightGene editing is correctHydrolasesStable introduction of DNABiological cellEucoenogenes
The invention discloses a novel gene editing system which is established based on gene cas3 of an I-B type CRISPR-Cas system in a chromosome of Virginia streptomycete IBL14, the gene editing of an I type CRISPR-Cas system to a biological cell genome is realized for the first time, and new supplement and choice are provided for the gene editing system which is established by II type commercializedCas9. In the system, a target DNA can be specifically cut by the Cas3 through crRNA guide or t-DNA location. By adopting the system, error-free, simple and rapid gene editing can be performed on prokaryotic and eukaryotic genomes. The optimized gene editing system is expected to be superior to the commercialized gene editing system which is established based on Cas9 in multiple fields due to low molecular weight and ability to be guided by DNA.
Owner:ANHUI UNIVERSITY
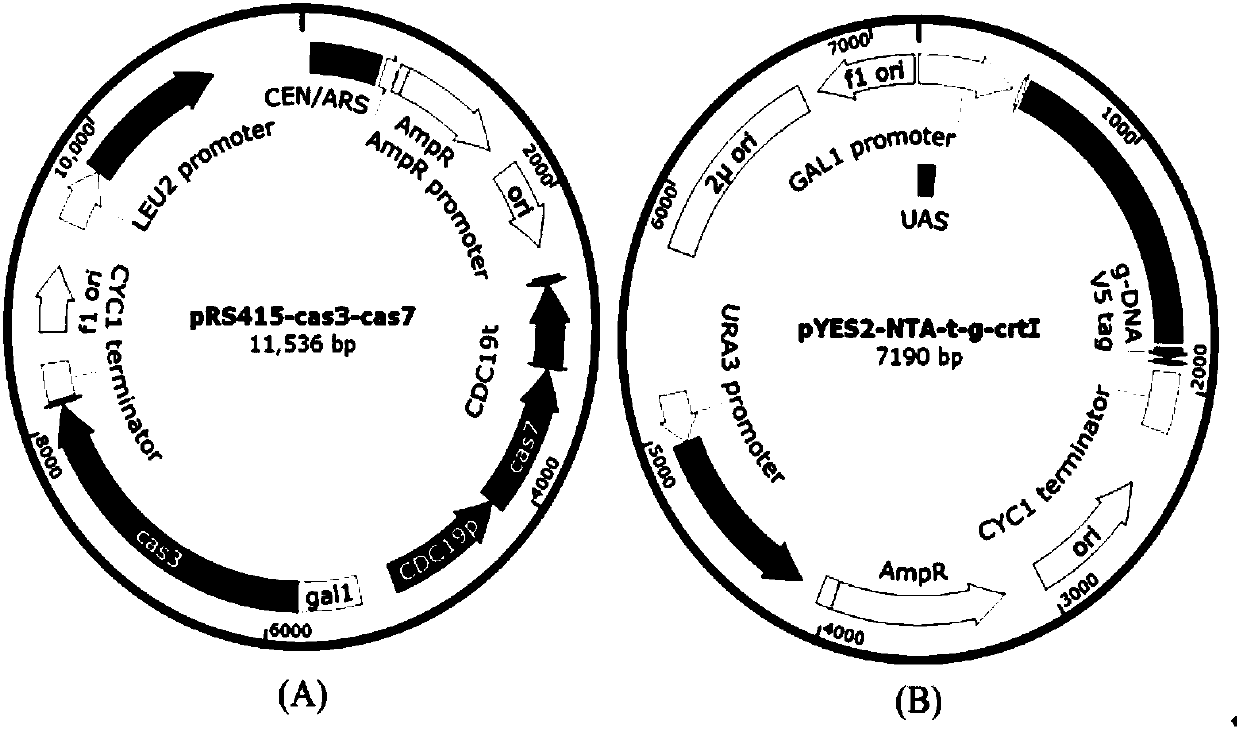
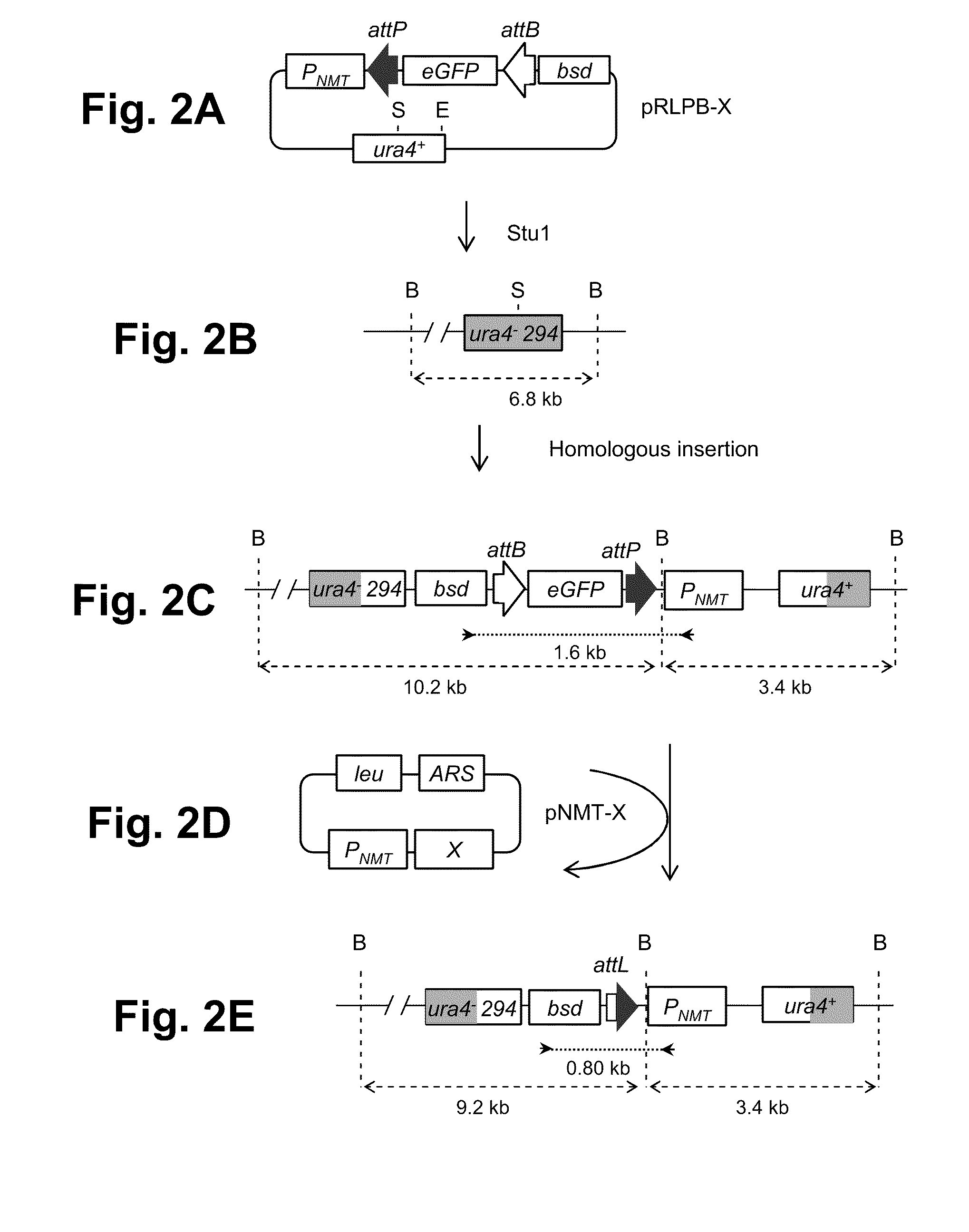
Eukaryotic gene editing method based on gene cas7-3 in I type CRISPR-Cas system
PendingCN107557378AEasy gene editingGene editing fastStable introduction of DNAVector-based foreign material introductionEukaryotic geneEucoenogenes
The invention discloses gene knockout and gene insertion on eukaryotic genomes carried out by two Cas proteins (Cas7 and Cas3) in a 1 class I type CRISPR-Cas system for the first time. The developmentof the method breaks the limitation of dependence on single-gene cas9 and cpf1, and provides a new perspective for performing gene editing on the eukaryotes by multi-gene. By applying the system, thegene editing can be conveniently, rapidly and effectively carried out on eukaryote brewer's yeast genomes. The optimized tool is expected to be widely used in the gene editing of other eukaryotes.
Owner:ANHUI UNIVERSITY
Diagnostic sequencing by a combination of specific cleavage and mass spectrometry
InactiveUS20060252061A1Rapid and reliableQuick fixMicrobiological testing/measurementDiseaseSomatic cell
The present invention is in the field of nucleic acid-based diagnostic assays. More particularly, it relates to methods useful for the "diagnostic sequencing" of regions of sample nucleic acids for which a prototypic or reference sequence is already available (also referred to as "re-sequencing"), or which may be determined using the methods described herein. This diagnostic technology is useful in areas that require such re-sequencing in a rapid and reliable way: (i) the identification of the various allelic sequences of a certain region / gene, (ii) the scoring of disease-associated mutations. (iii) the detection of somatic variations, (iv) studies in the field of molecular evolution, (v) the determination of the nucleic acid sequences of prokaryotic and eukaryotic genomes; (vi) identifying one or more nucleic acids in one or more biological samples; (vii) and determining the expression profile of genes in a biological sample and other areas.
Owner:AGENA BIOSCI
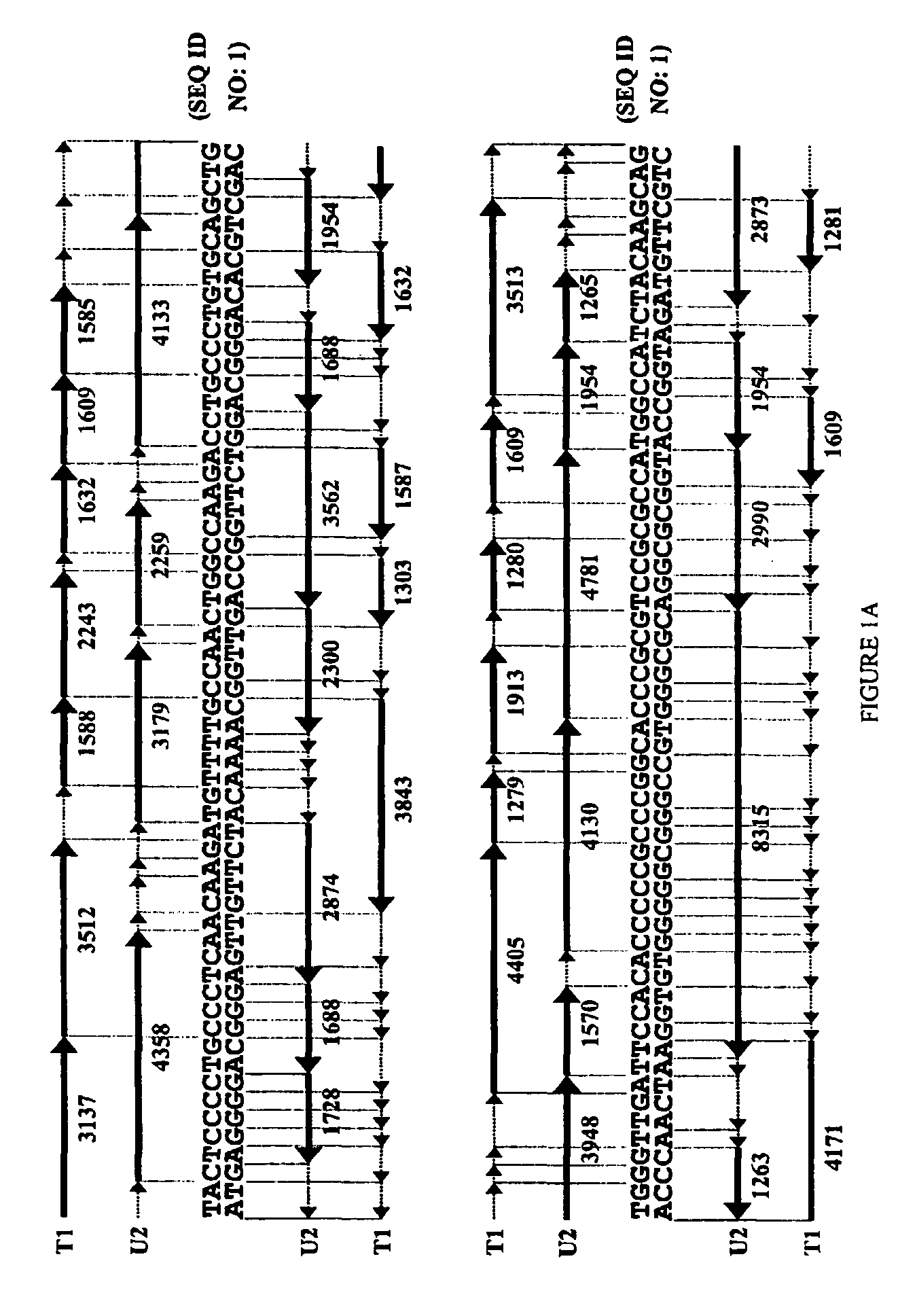
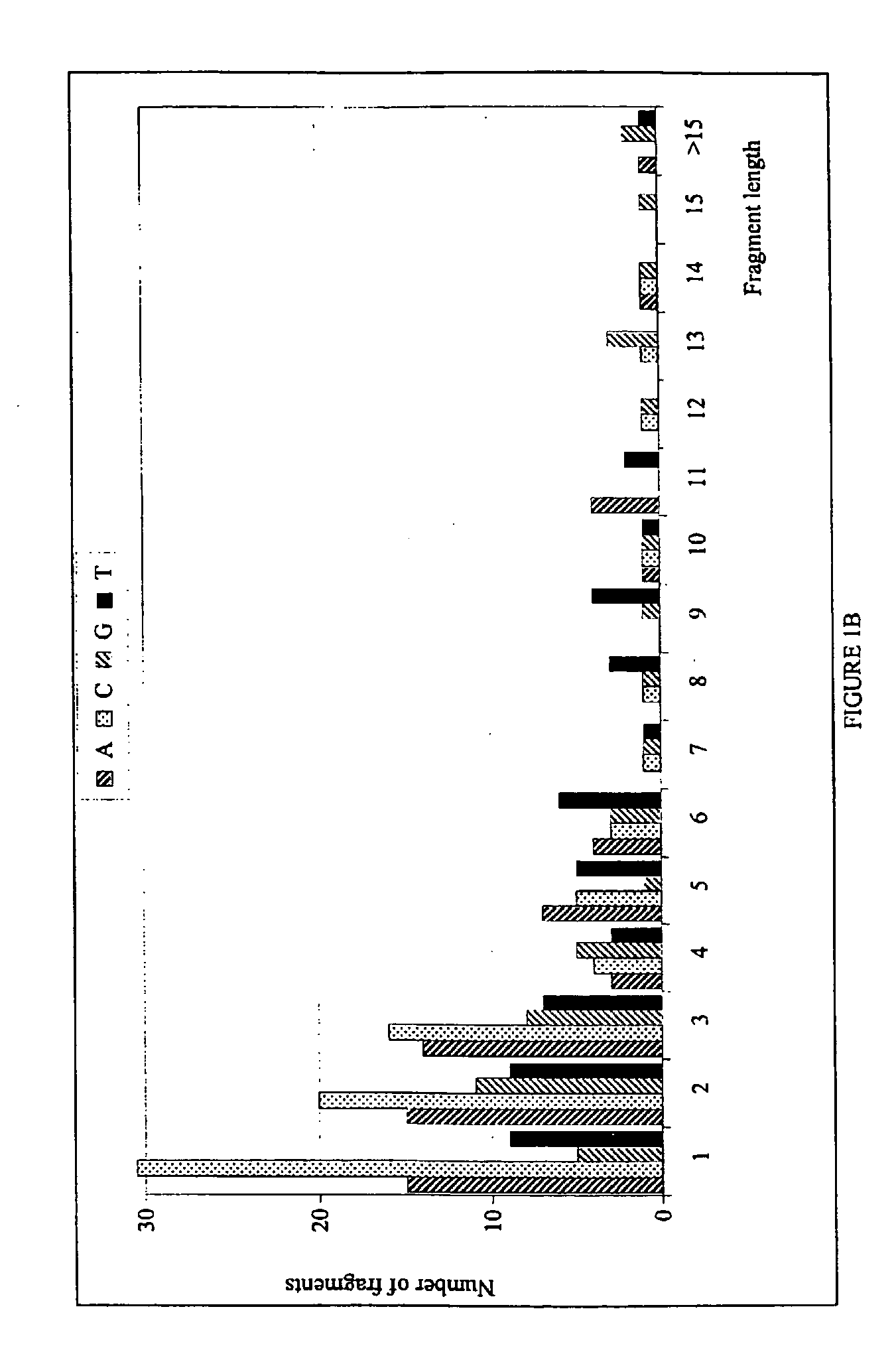
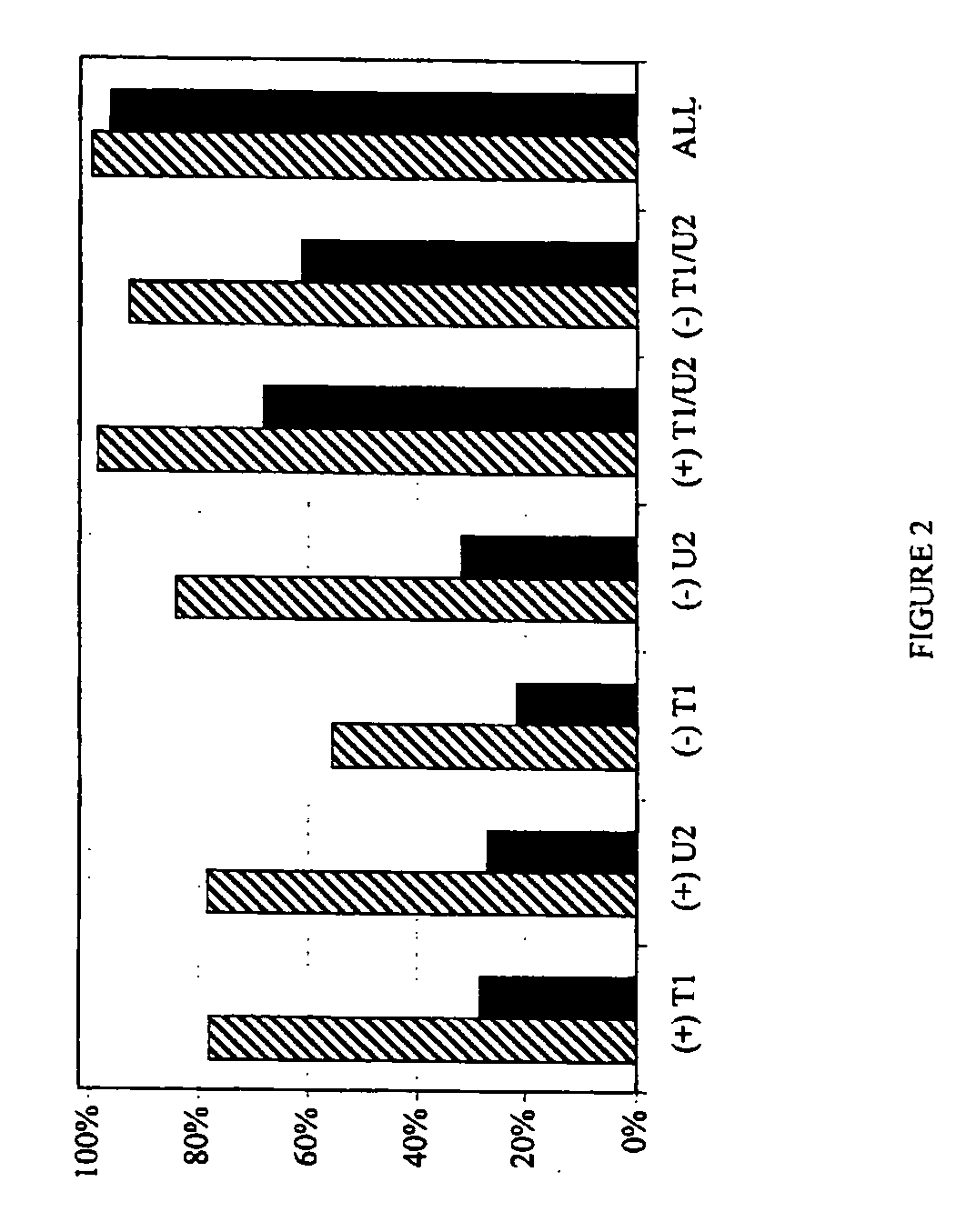
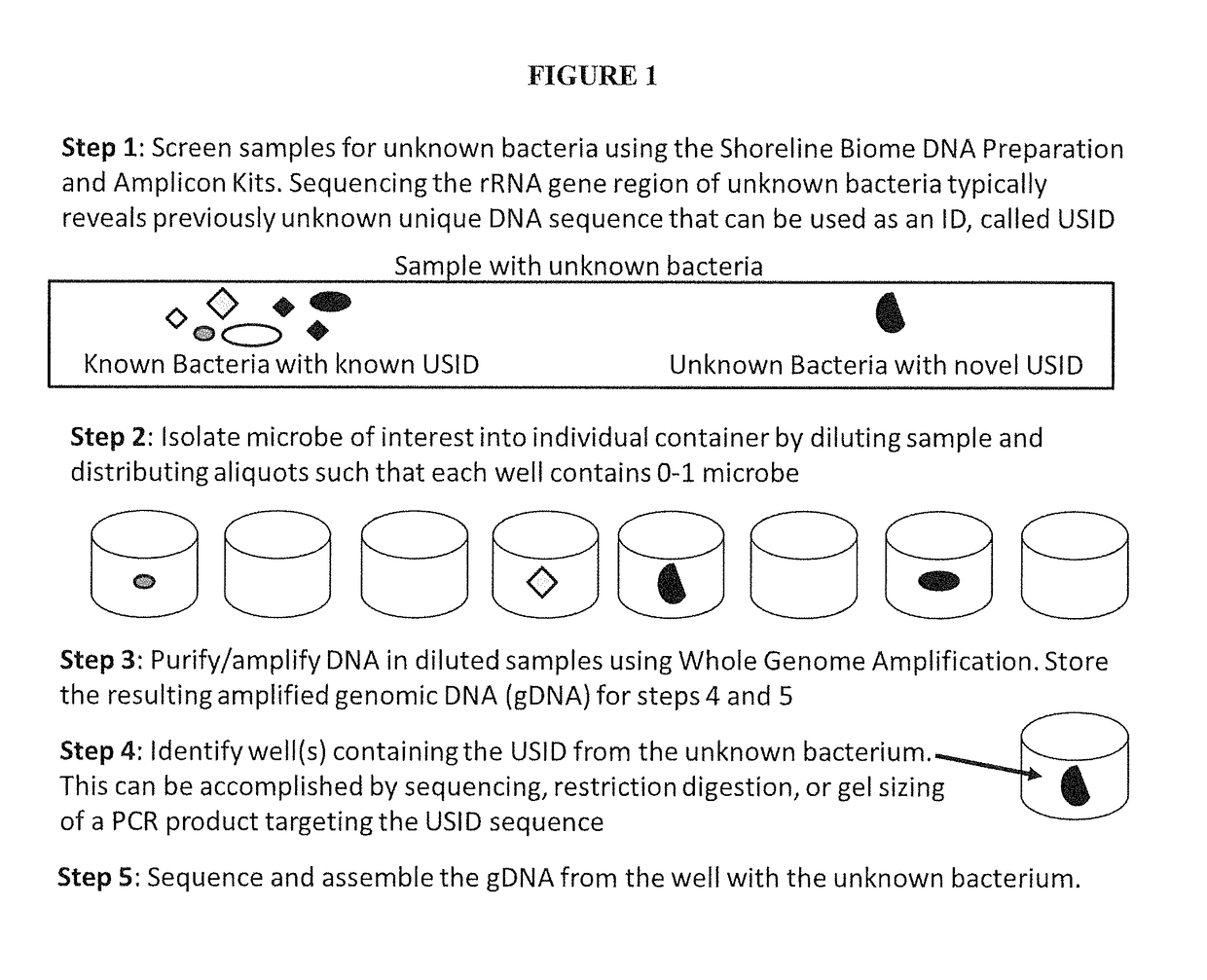
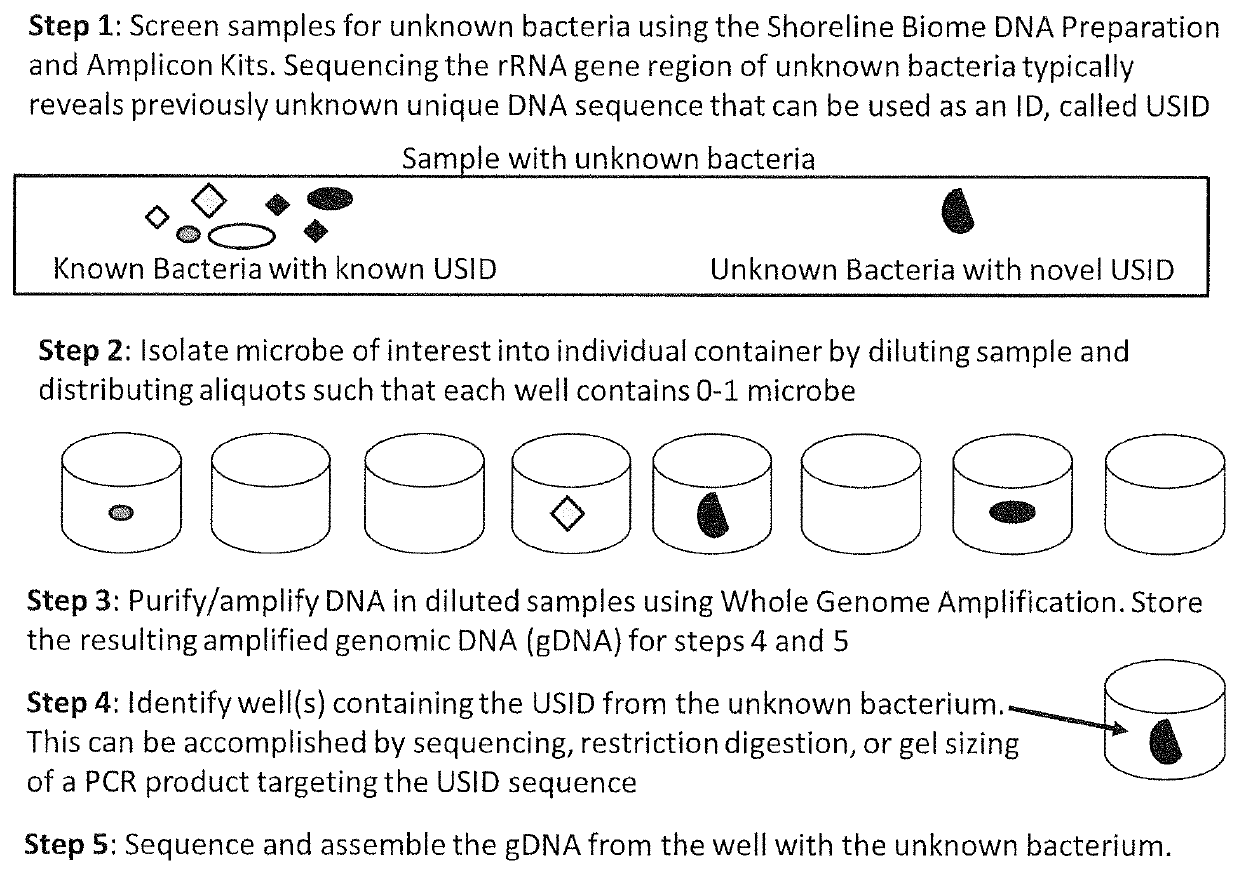
High Throughput Method for Identification and Sequencing of Unknown Microbial and Eukaryotic Genomes from Complex Mixtures
Disclosed are methods for screening biological samples for the presence unknown microbes, such as bacteria and archaea or unknown eukaryotes using rRNA gene sequences or other highly conserved genetic regions, across multiple biological samples using a unique sequence tag (barcode) corresponding to the sample. The screening process tracks the unknown microbe or eukaryote in a diluted sample where the DNA has been prepared using whole genome amplification. The whole genome of the unknown microbe or eukaryote is then sequenced and assembled.
Owner:SHORELINE BIOME LLC
Universal fingerprinting chips and uses thereof
InactiveUS20110105346A1Avoid formingNucleotide librariesMicrobiological testing/measurementNucleotideOrganism
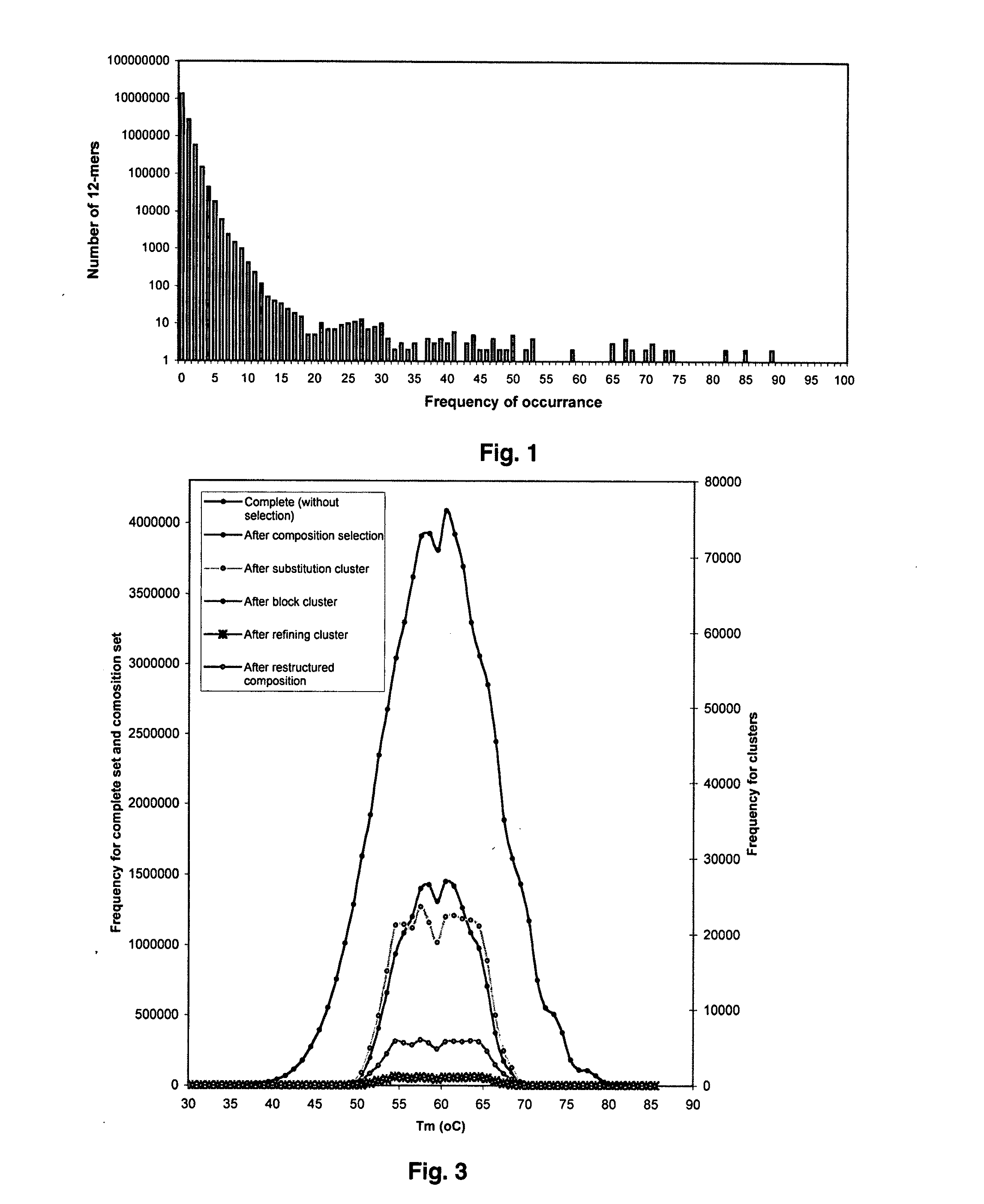
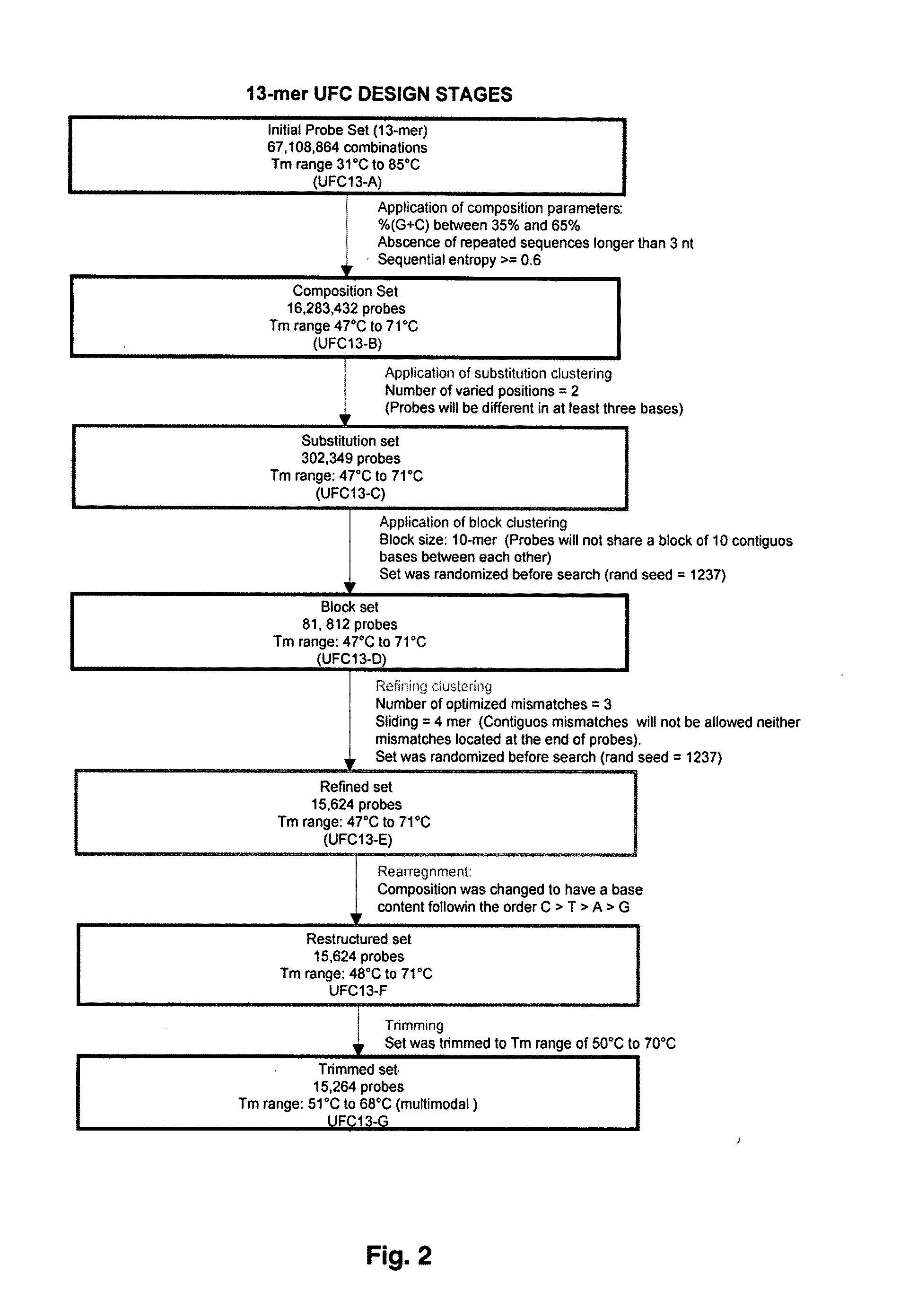
The present invention discloses a designing strategy for constructing a set of probes useful for analyzing all or most prokaryotic and eukaryotic genomes. A set of capture probes with optimal fingerprinting properties and highly representative of all possible sequences of an organism can be selected by six sequential steps. Fingerprinting potential of such probes is validated by phylogenetic analysis, which generates results that strongly correlate with phylogenetic trees produced by sequence alignment. The probes generated by the instant methods can be used for detecting an organism, for establishing phylogenetic relationships between different organisms, for detection of single nucleotide polymorphisms and a wide variety of other applications that require genetic analysis.
Owner:BEATTIE KENNETH L +4
Methods for the replacement, translocation and stacking of DNA in eukaryotic genomes
InactiveUS6936747B2Reduce in quantityImprove efficiencySugar derivativesStable introduction of DNAMammalPlant cell
The present invention includes compositions and methods for site-specific polynucleotide replacement in eukaryotic cells. These methods include single polynucleotide replacement as well as gene stacking methods. Preferred eukaryotic cells for use in the present invention are plant cells and mammalian cells.
Owner:US SEC AGRI
Use of argonaute endonucleases for eukaryotic genome engineering
InactiveUS20170367280A1Low costHigh inherent stabilityVector-based foreign material introductionPlant genotype modificationBiotechnologyEndonuclease
The present invention relates to the use of Argonaute systems in plants for genome engineering, and compositions used in such methods.
Owner:KWS SAAT SE
PCR determination method for Chinese medicine or Chinese medicinal crops derived from eukaryote
ActiveCN101265500ASensitive and fast identificationValid identificationMicrobiological testing/measurementProcessing typeEucoenogenes
The invention provides a PCR identification method derived from eucaryote for Chinese traditional medicine and traditional Chinese medicinal materials. PCR augmentation is processed on DNA samples to obtain augmentation production according to a species-specific primer, wherein, the species-specific primer is designed according to a SINE sequence of eukaryotic genomes. The DNA samples are extracted from the Chinese traditional medicine and traditional Chinese medicinal materials, in particular from the further processing type Chinese traditional medicine. Species categories of eukaryotic species in the further processing type Chinese traditional medicine are estimated by analyzing the augmentation production, and the truth of the identified samples is further estimated. The PCR identification method is suitable for Chinese traditional medicine and traditional Chinese medicinal materials, is particularly suitable for the processing type Chinese traditional medicine, especially for the Chinese traditional medicine which is with extremely little DNA content and extremely short segment due to the further processing. The identification method has the advantages of simple operation, rapidness and sensitivity, low cost and effective truth identification of the Chinese traditional medicine and the traditional Chinese medicinal materials.
Owner:EAST CHINA UNIV OF SCI & TECH +1
Diagnostic sequencing by a combination of specific cleavage and mass spectrometry
InactiveUS6994969B1Rapid and reliableQuick fixSugar derivativesMicrobiological testing/measurementDiseaseMass spectrometry imaging
The present invention is in the field of nucleic acid-based diagnostic assays. More particularly, it relates to methods useful for the “diagnostic sequencing” of regions of sample nucleic acids for which a prototypic or reference sequence is already available (also referred to as “re-sequencing”), or which may be determined using the methods described herein. This diagnostic technology is useful in areas that require such re-sequencing in a rapid and reliable way: (i) the identification of the various allelic sequences of a certain region / gene, (ii) the scoring of disease-associated mutations, (iii) the detection of somatic variations, (iv) studies in the field of molecular evolution, (v) the determination of the nucleic acid sequences of prokaryotic and eukaryotic genomes, (vi) identifying one or more nucleic acids in one or more biological samples', (vii) and determining the expression profile of genes in a biological sample and other areas.
Owner:AGENA BIOSCI
Site-specific recombination systems for use in eukaryotic cells
InactiveUS20060046294A1Reduce probabilityStable introduction of DNANucleic acid vectorSite-specific recombinationProtein translocation
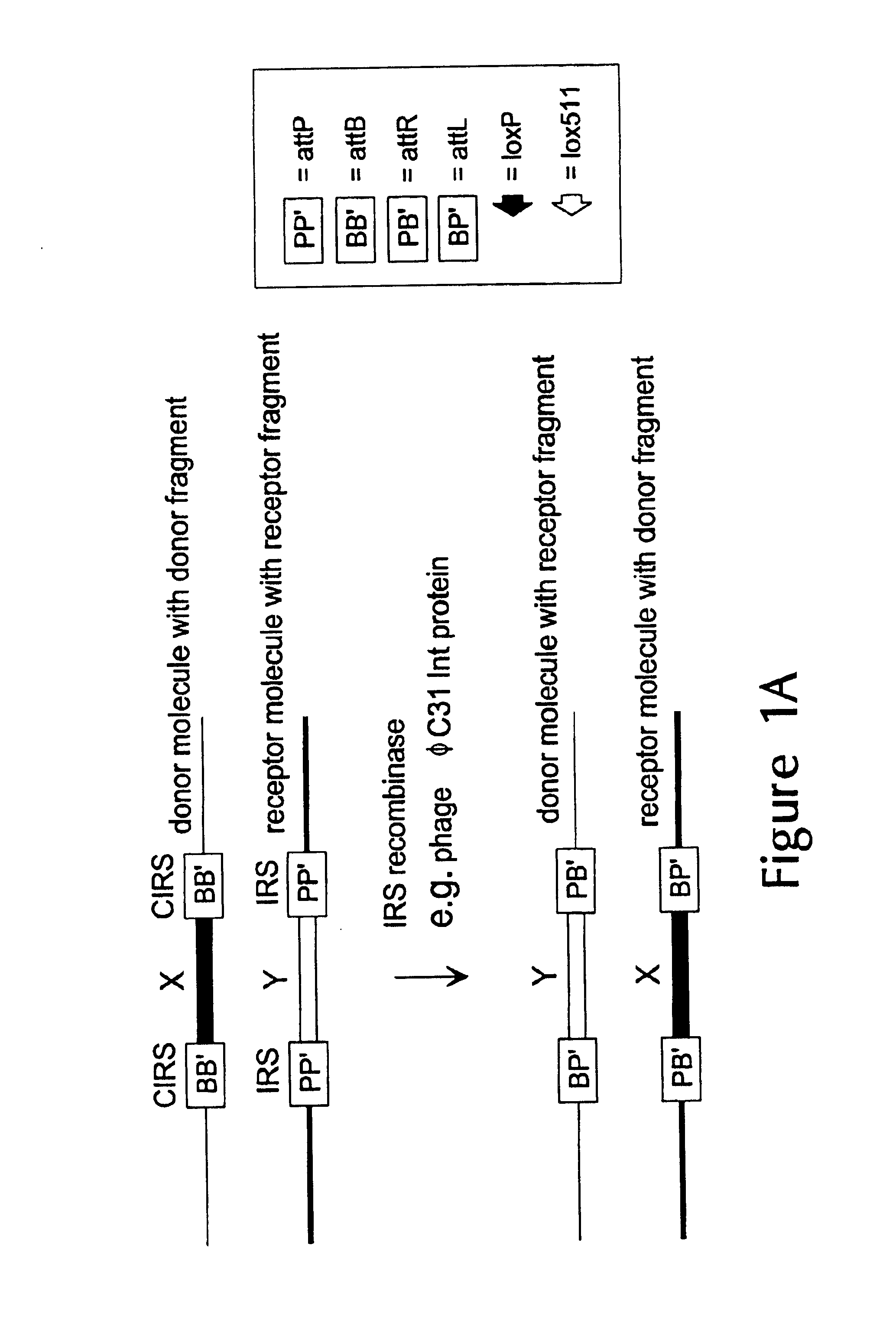
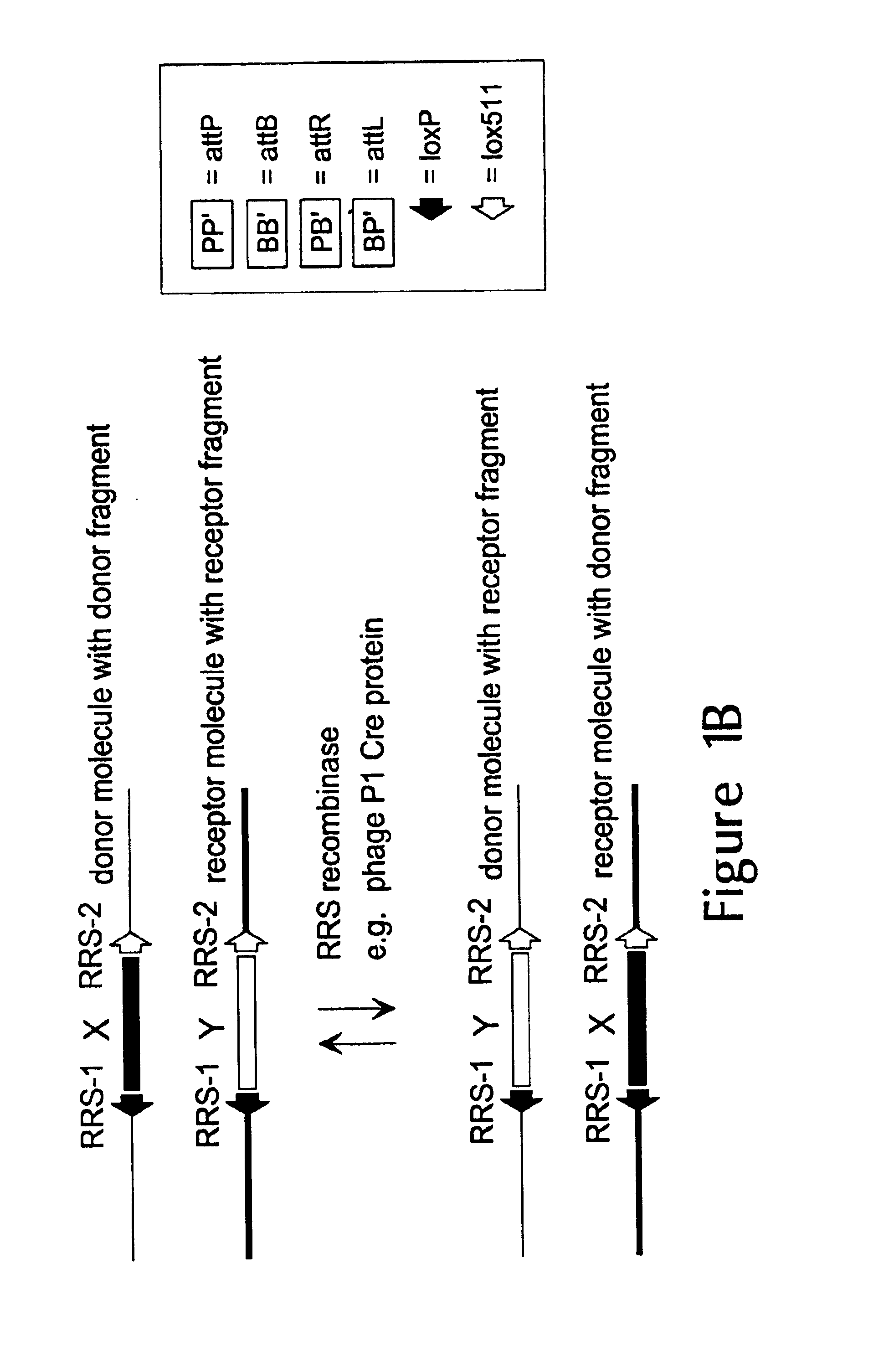
Prokaryotic recombination systems have been adapted to function in eukaryotes in order to achieve one or more of the following: DNA site specific excision, translocation, integration and inversion. These recombination systems are identified as seven members of the small serine resolvase subfamily: CinH, ParA, Tn1721, Tn5053, Tn21, Tn402, and Tn501 and three members of the large serine resolvase subfamily: Bxb1, U153, and TP901-1. These recombination systems represent new tools for the genetic manipulation of eukaryotic genomes.
Owner:AGRI THE UNITED STATES OF AMERICA REPRESENTED BY THE SEC OF
Method of constructing vectors for homologous recombination directed mutagenesis
InactiveUS6924146B1Easy constructionAvoid disadvantagesSugar derivativesGenetic material ingredientsVector systemOrganism
The present invention provides a novel vector system and thereby a novel method for the simplified construction of recombinant vectors for directed mutagenesis. Said vector system is used to modify the eukaryotic genome, particularly of embryonic stem cells, at precise and predefined loci by the means of homologous recombination. Furthermore, said system finds its usage in the generation of new strategies for gene therapy and in the generation of genetically modified higher eukaryotic organisms.
Owner:WATTLER SIGRID +1
Detection of epigenetic abnormalities and diagnostic method based thereon
The present invention provides a method of detecting an epigenetic abnormality associated with a disease. The method comprises identifying, within a eukaryotic genome, a locus having a hypomethylated sequence specific for the disease and an endogenous multi-copy DNA element. The method can also comprise separate steps of identifying a disease-specific hypomethylated sequence and identifying an endogenous multi-copy DNA element, where the steps may be performed in any order, so long as a locus is identified that has both a disease-specific hypomethylated sequence and an endogenous multi-copy DNA element. The disease-specific hypomethylated sequences detected in accordance with the present invention indicate putative regions of epigenetic dys-regulation and indicate aberrantly regulated nucleic acid sequences that may cause or predispose a patient to disease, such as, but not limited to, Huntingdon s disease, cancers, diabetes, schizophrenia, or bipolar disorder.
Owner:CENT FOR ADDICTION & MENTAL HEALTH
Process and apparatus for using the sets of pseudo random subsequences present in genomes for identification of species
InactiveUS20050255459A1Rapid increase of computational complexityEasy to analyzeMicrobiological testing/measurementLibrary member identificationHuman speciesGenomic DNA
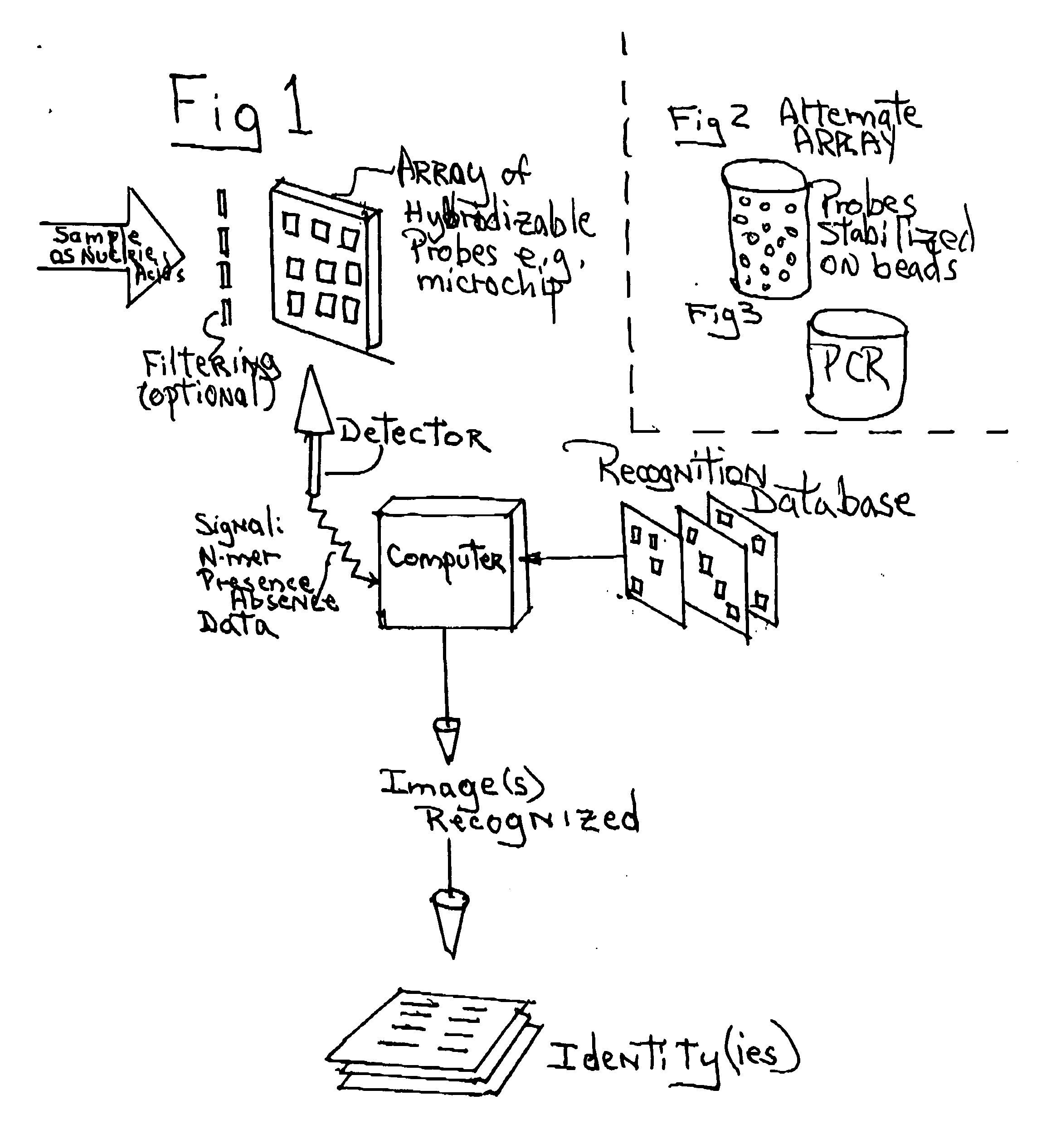
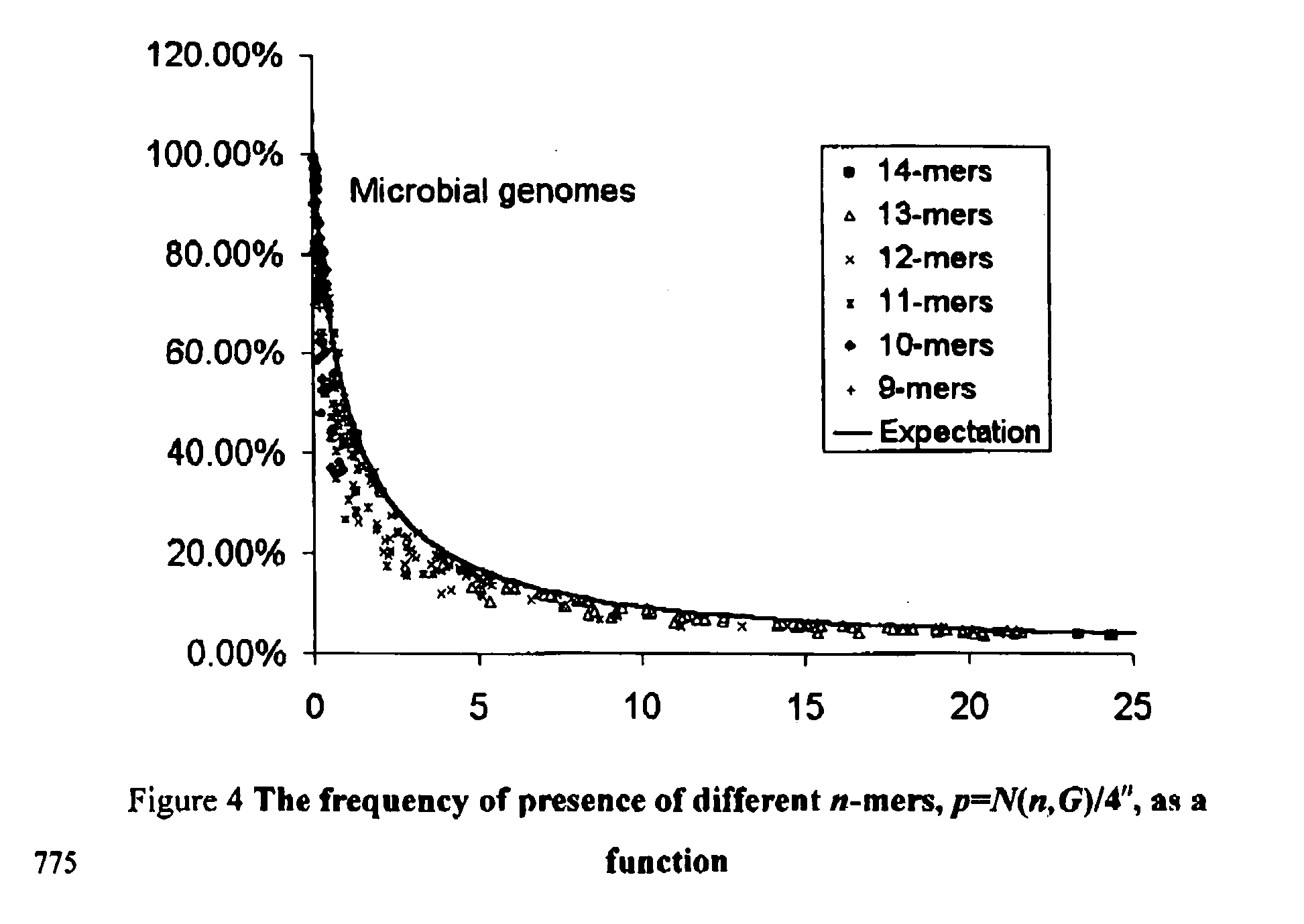
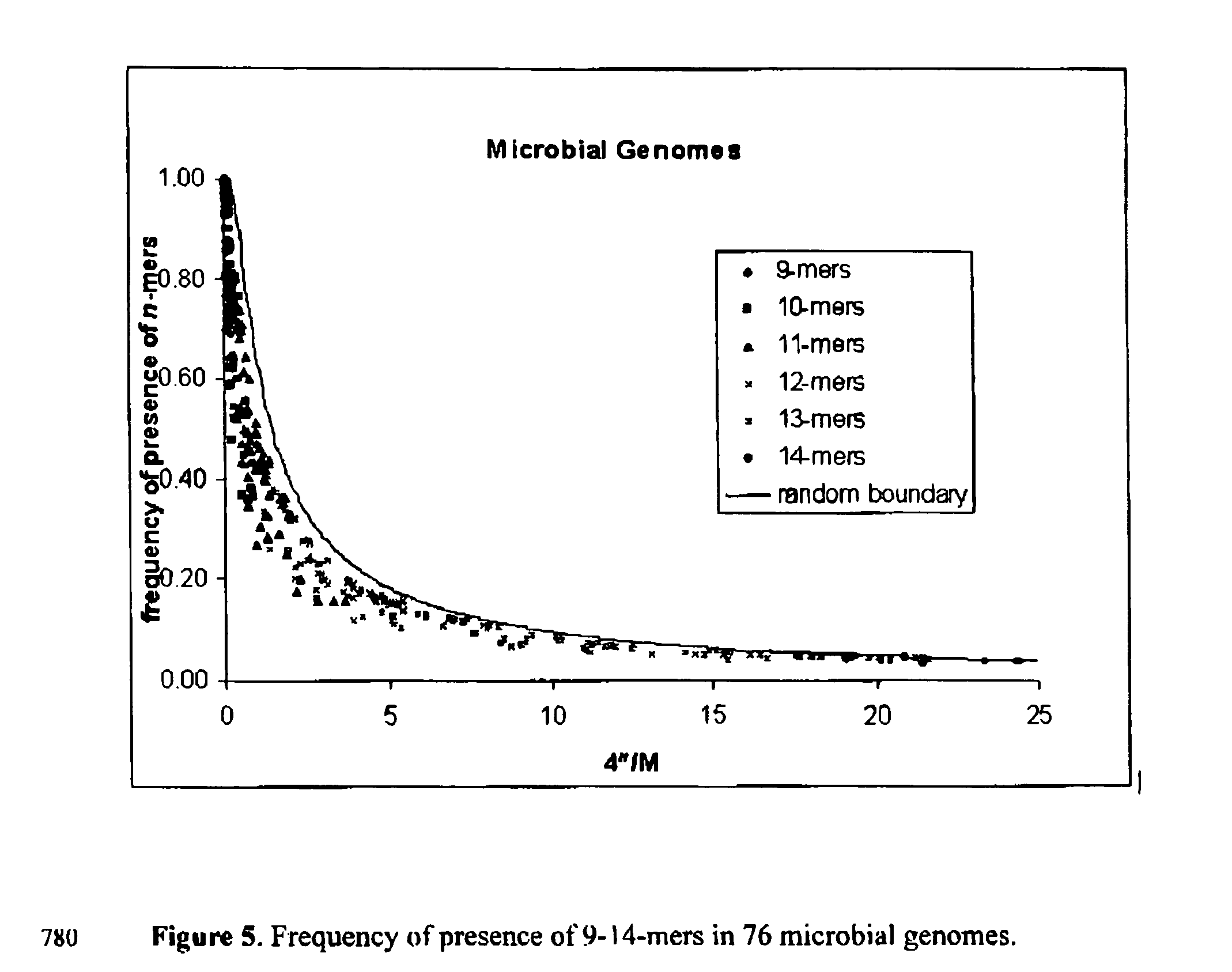
Our research conducted with the genome sequences of more than 250 species of organisms (including viral, microbial, and multi-cellular organisms, and human) results in the discovery that the occurrence of a particular subsequence (the so-called “motifs” or “n-mers,” (n being the length of the subsequences), which can be up to 25 and higher) in the genome of a particular species can be considered as a nearly random event; and that the occurrences of a particular subsequence in the genome sequences of different species can be considered as nearly independent events (with the exception of the cases where extremely closely related species are compared). The set of subsequences that occur in a particular species' genome can therefore be used as a genomic “fingerprint” of this species. This discovery leads to the concept of utilizing a set of pseudo-randomly designed subsequences for species identification or discrimination. These subsequences (probes, primers, motifs, n-mers) can be used with hybridization-based technologies (including, but not limited to, the microarray or PCR technologies) and any other technology allow to identity the fact of presence / absence of particular subsequence in genomic DNA for identification of species. The same approach can also be used to identify individuals of the same species (including the human species), to estimate the genome size of unknown organisms, and to estimate the total genome size in samples containing several viral, microbial, and eukaryotic genomes. The identification methods currently in use for these purposes require sequencing of the genomic sequences of the species or the individuals of interest. The introduction of the proposed computational method eradicates such requirement, and will tremendously reduce the expense of these tests.
Owner:FOFANOV YURIY +3
Identification and assignment of functionally corresponding regulatory sequences for orthologous loci in eukaryotic genomes
The present invention relates to a method and computer program product for identifying a regulatory sequence of a coding sequence within the genome of a eukaroytc organism.
Owner:GENOMATIX SOFTWARE
Methods for the replacement, translocation and stacking of DNA in eukaryotic genomes
InactiveUS7972857B2Reduce in quantityImprove efficiencyStable introduction of DNAFermentationPlant cellPolynucleotide
The present invention includes compositions and methods for site-specific polynucleotide replacement in eukaryotic cells. These methods include single polynucleotide replacement as well as gene stacking methods. Preferred eukaryotic cells for use in the present invention are plant cells and mammalian cells.
Owner:US SEC AGRI
Integrase fusion proteins and their use with integrating gene therapy
InactiveUS20090011509A1Avoid infectionPromote safe and targeted integrationHydrolasesPeptide/protein ingredientsIntegrasesTransgene
In a method of targeting intergration of a transgene comprising retrovirus-like DNA into a eukaryotic genome, the genome is cleared by an endonuclease and the transgene is introduced at the site of cleavage, wherein the endonuclease is specific to a site in an abundant rDNA locus and is fused to an integrase that mediates the introduction of the transgene. The fusion protein may be new.
Owner:ARK THERAPEUTICS
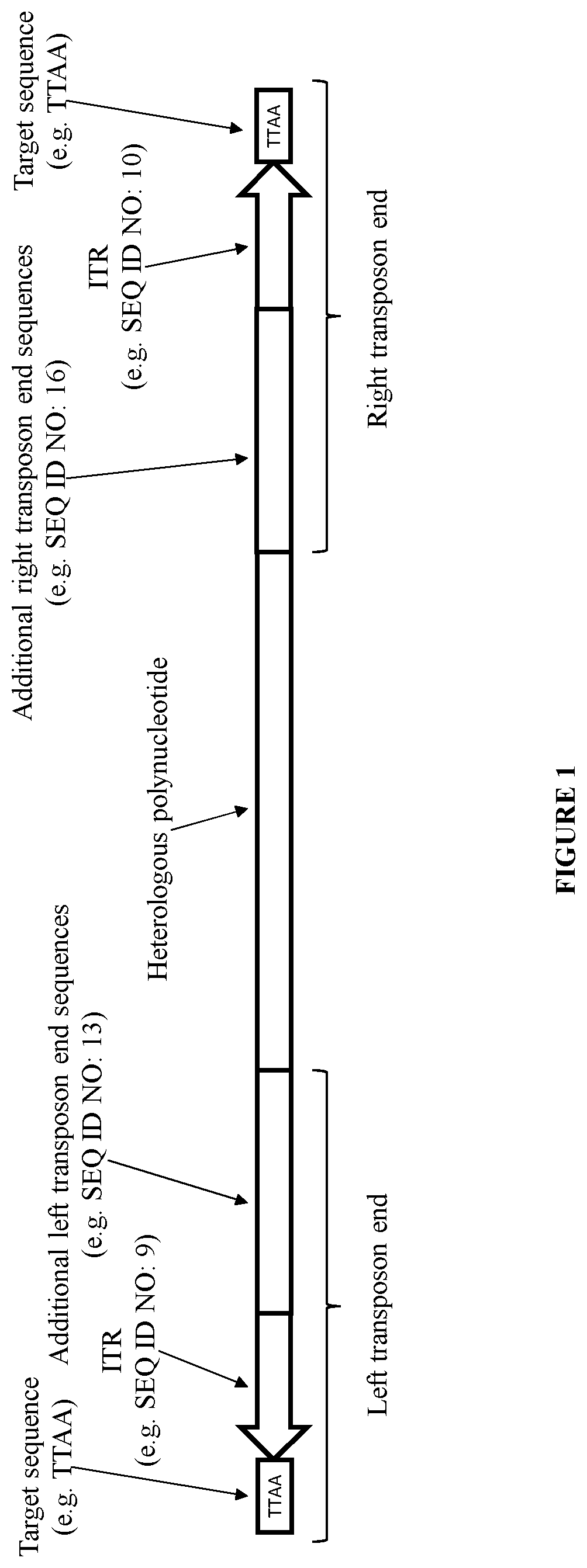
Transposition of nucleic acid constructs into eukaryotic genomes with a transposase from amyelois
ActiveUS20200318135A1Heterologous gene expression can be improvedPolypeptide with localisation/targeting motifPeptide/protein ingredientsHeterologousProcessed Genes
The present invention provides polynucleotide vectors for high expression of heterologous genes. Some vectors further comprise novel transposons and transposases that further improve expression. Further disclosed are vectors that can be used in a gene transfer system for stably introducing nucleic acids into the DNA of a cell. The gene transfer systems can be used in methods, for example, gene expression, bioprocessing, gene therapy, insertional mutagenesis, or gene discovery.
Owner:DNA TWOPOINTO
Regulatory Nucleic Acid Elements
InactiveUS20080124760A1Increase transcriptionHigh expressionAnimal cellsSugar derivativesBiotechnologyPromoter
The invention relates to DNA-sequences, especially transcription- or expression-enhancing elements (TE elements) and their use on an expression vector in conjunction with an enhancer, a promoter, a product gene and a selectable marker.The invention describes Sequence No. 1 and TE elements TE-01, -02, -03, -04, -06, -07, -08, -10, -11 or -12. Because of their small size, TE-06, TE-07 or TE-08 are particularly preferred. Sequence No. 1 originates from a sequence region located upstream from the coding region of the Ub / S27a gene from CHO cells.TE elements bring about an increase in the expression of the product gene, particularly when stably integrated in the eukaryotic genome, preferably the CHO-DG44 genome. Chromosomal positional effects are thereby overcome, shielded or cancelled out. In this way the proportion of high producers in a transfection mixture and also the absolute expression level are increased up to seven-fold.
Owner:BOEHRINGER INGELHEIM PHARM KG
DNA vectors, transposons and transposases for eukaryotic genome modification
ActiveUS10435696B2Polypeptide with localisation/targeting motifMicroorganismsHeterologousPolynucleotide
The present invention provides polynucleotide vectors for high expression of heterologous genes. Some vectors further comprise novel transposons and transposases that further improve expression. Further disclosed are vectors that can be used in a gene transfer system for stably introducing nucleic acids into the DNA of a cell. The gene transfer systems can be used in methods, for example, gene expression, bioprocessing, gene therapy, insertional mutagenesis, or gene discovery.
Owner:DNA2 0
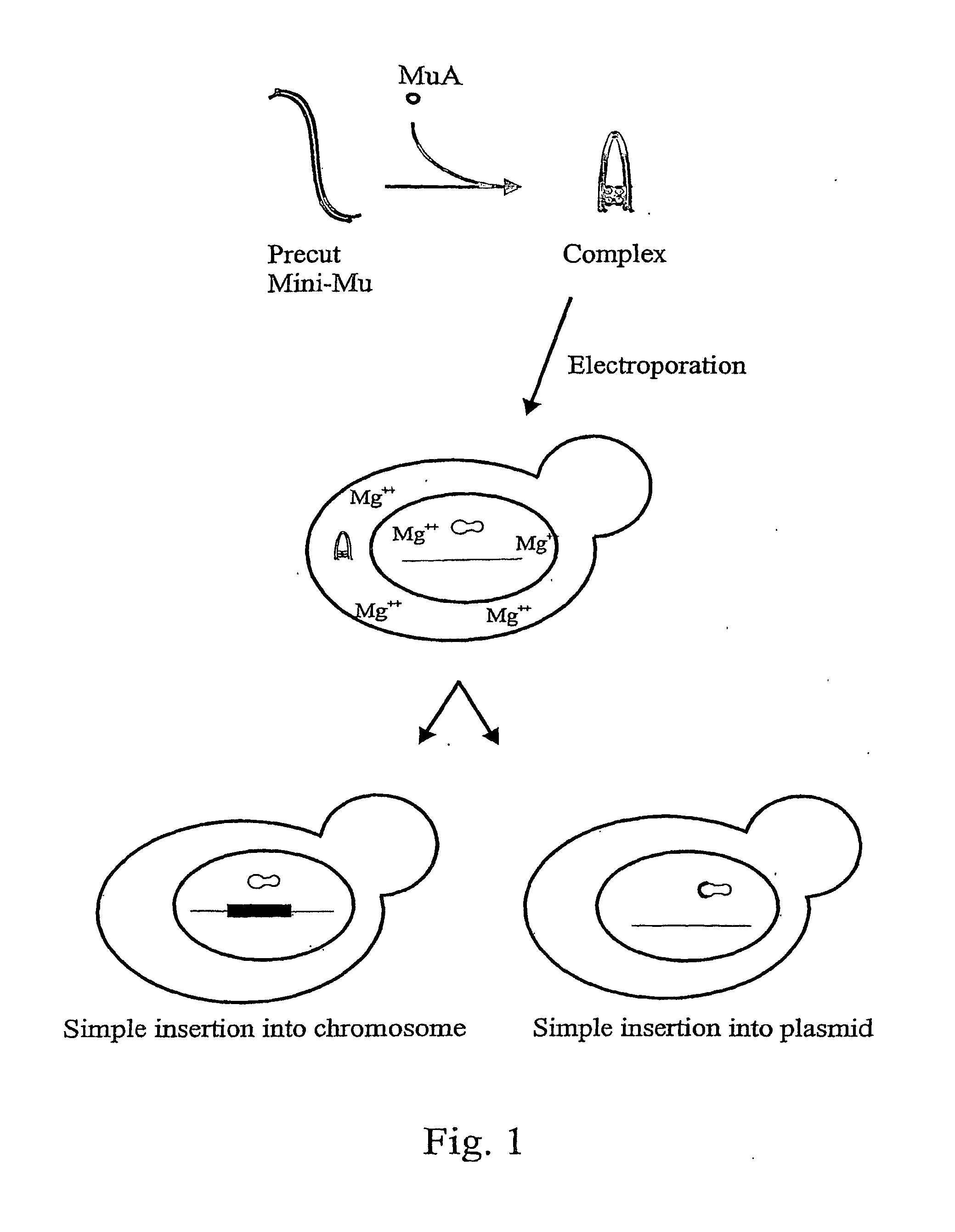
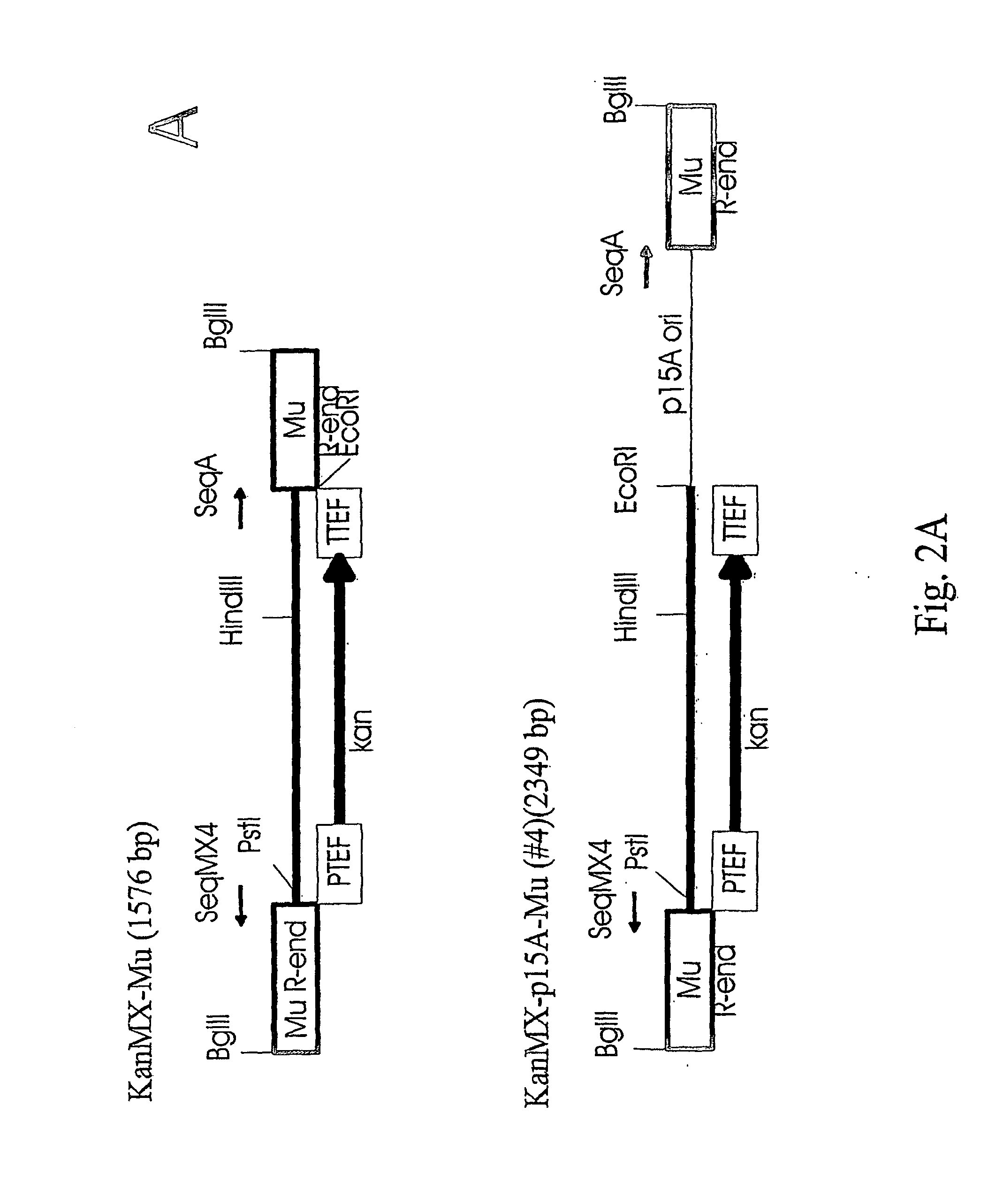
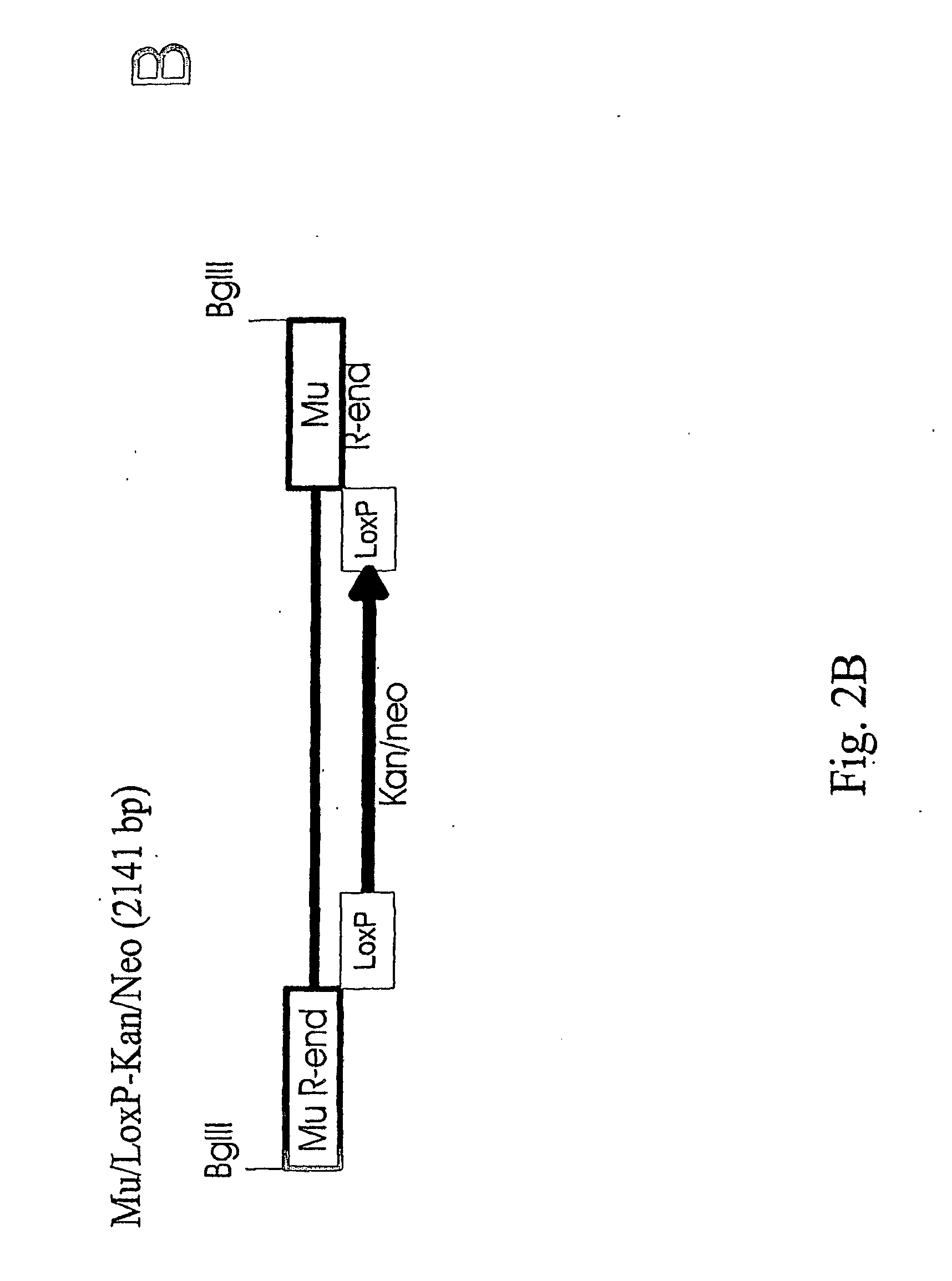
Method for delivering nucleic acid into eukaryotic genomes
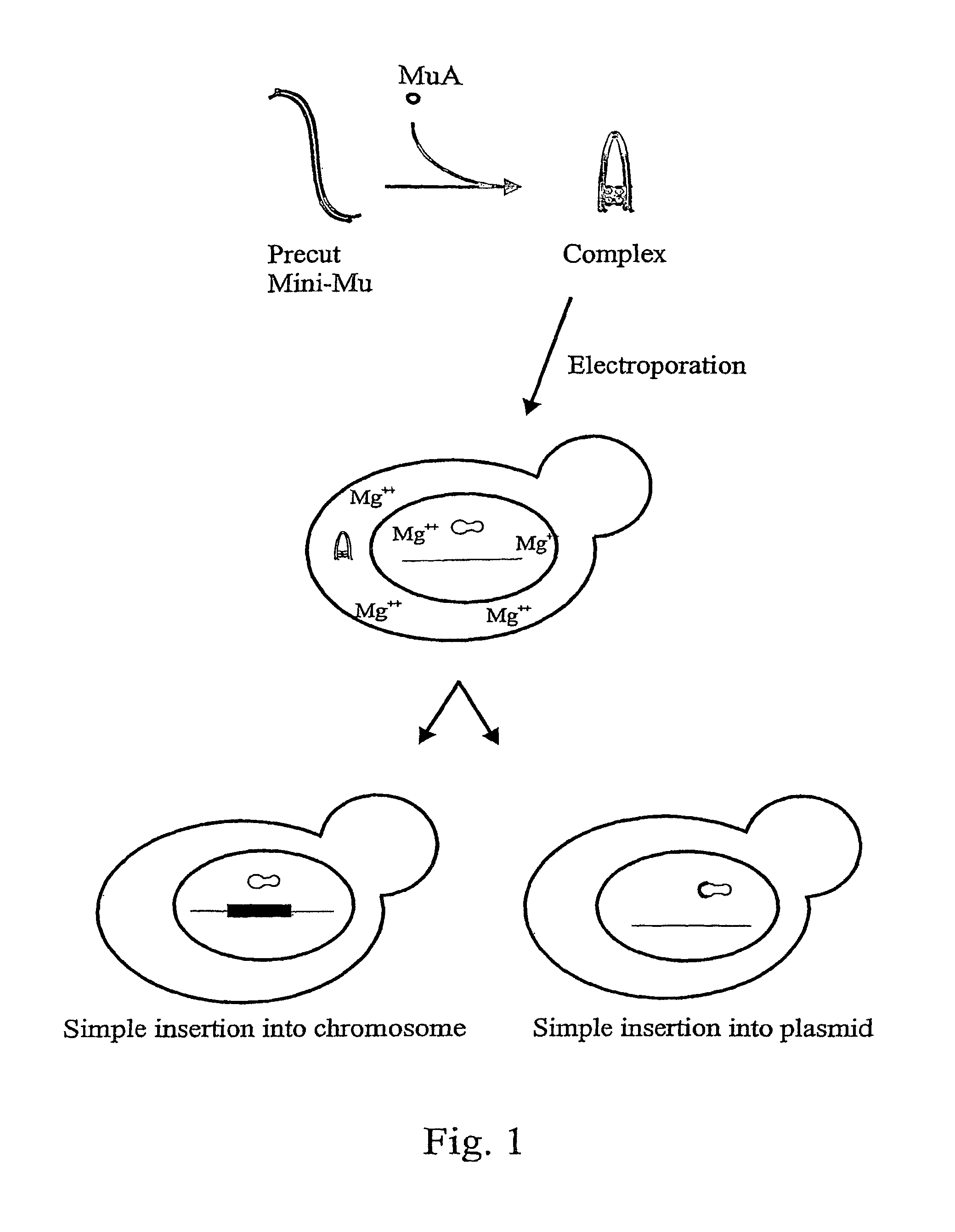
ActiveUS20120015831A1Improve applicabilitySugar derivativesHydrolasesBacteriophage MuGenetic engineering
The present invention relates to genetic engineering and especially to the use of DNA transposition complex of bacteriophage Mu. In particular, the invention provides a gene transfer system for eukaryotic cells, wherein in vitro assembled Mu transposition complexes are introduced into a target cell and subsequently transposition into a cellular nucleic acid occurs. The invention further provides a kit for producing insertional mutations into the genomes of eukaryotic cells. The kit can be used, e.g., to generate insertional mutant libraries.
Owner:FINNZYMES
Site-specific recombination systems for use in eukaryotic cells
InactiveUS20100068815A1Reduce probabilityStable introduction of DNANucleic acid vectorSite-specific recombinationGenetics manipulation
Prokaryotic recombination systems have been adapted to function in eukaryotes in order to achieve one or more of the following: DNA site specific excision, translocation, integration and inversion. These recombination systems are identified as seven members of the small serine resolvase subfamily: CinH, ParA, Tn1721, Tn5053, Tn21, Tn402, and Tn501 and three members of the large serine resolvase subfamily: Bxb1, U153, and TP901-1. These recombination systems represent new tools for the genetic manipulation of eukaryotic genomes.
Owner:UNITED STATES OF AMERICA
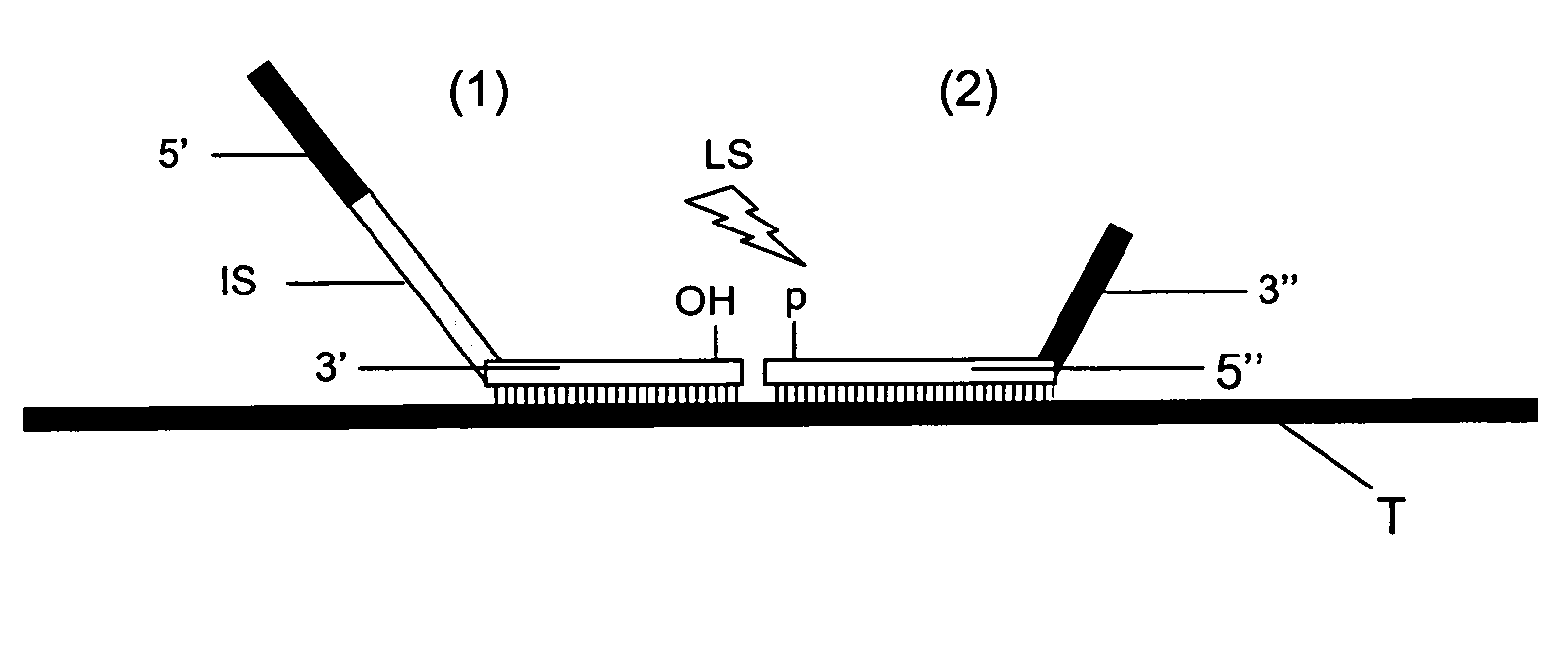
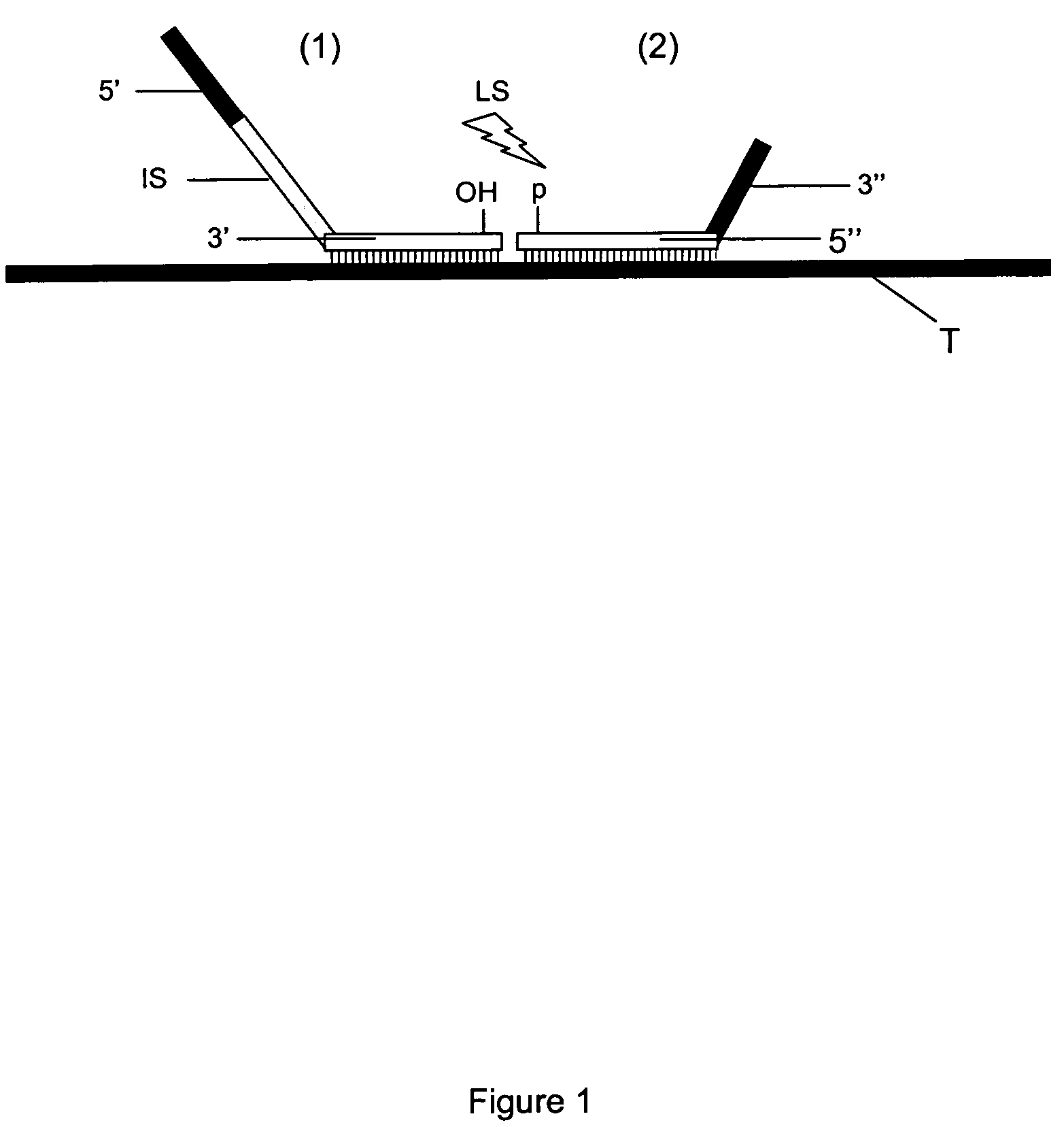
Method for Detection and Quantification of Target Nucleic Acids in a Sample
InactiveUS20090117552A1Minimised secondary structure formationLess-accurate detectionMicrobiological testing/measurementProtein nucleotide librariesPhosphoric acidNucleic acid sequencing
The present invention relates to methods for multiplex detection and quantification of target nucleic acid sequences in a sample comprising the steps of: (i) providing a solid support having immobilized thereon an array of detector oligonucleotides, wherein said array of detector oligonucleotides is designed by random selection of non-eukaryotic genomic sequences followed by random selection of oligonucleotide sequences and subsequent conversion of these oligonucleotide sequences such that these are composed of only three types of nucleotides; (ii) providing a sample having added thereto a fixed amount of control nucleic acid of known sequence; (iii) contacting said sample with at least two probes that hybridise to adjacent sites of a target sequence under conditions favouring hybridisation between the sample nucleic acids and the said at least two probes, wherein, a) a first probe is composed of a 5′ end sequence part for hybridisation to a PCR primer and a 3′ end sequence part for hybridisation to the target nucleic acid; and b) a second probe is composed of a 5′ end sequence part for hybridisation to the target nucleic acid, and a 3′ end sequence part for hybridisation with a PCR primer, and c) an intermediate sequence is present in between said 5′ and 3′ end sequence parts of said first or second probe; and d) said second probe is characterized by having 5′ phosphate group allowing ligation with a 3′ hydroxyl group at the said first probe forming a ligation-mediated probe; (iv) ligation of the said hybridised first and second probes to form ligation-mediated probes; (v) contacting a set of detectable labelled PCR primers with the ligation-mediated probes allowing amplification thereof; (vi) detection and quantification of sample nucleic acids via hybridisation of the said intermediate parts within the amplified ligation-mediated probes onto the array of detector oligonucleotides provided in The present invention also relates to the use of said methods as well as microarrays and kits for performing said methods.
Owner:PAMGENE
High-throughput and high-fidelity automatic synthetic method for long-chain template-free deoxyribonucleic acid (DNA)
The invention discloses a high-throughput and high-fidelity automatic synthetic method for a long-chain template-free deoxyribonucleic acid (DNA) and provides a method with an automatic liquid-removing workstation as a core. The method can finish automatic synthesis under extremely low human interference and even can automatically synthesize a DNA sequence of an eukaryotic gene group (10000,000 basepair). The lowest human interference is required by an automatic synthetic center and the whole synthesis process is performed under a sterile condition, so the pollution to a sample is reduced to the great extent. The automatic liquid-removing workstation can very precisely extract and release quantitative reagents, so errors caused by human operation are avoided, and the success rate is greatly improved. The automatic synthetic center is highly higher than human operation in operation speed and can work without stop at full speed, so the production efficiency is improved, simultaneously the labor and the time are reduced by reducing the errors, high-throughput and high-fidelity automatic synthesis on a large segment of DNA is realized, and the synthesis cost is greatly reduced.
Owner:陈怡露 +1
High throughput method for identification and sequencing of unknown microbial and eukaryotic genomes from complex mixtures
Disclosed are methods for screening biological samples for the presence unknown microbes, such as bacteria and archaea or unknown eukaryotes using rRNA gene sequences or other highly conserved genetic regions, across multiple biological samples using a unique sequence tag (barcode) corresponding to the sample. The screening process tracks the unknown microbe or eukaryote in a diluted sample where the DNA has been prepared using whole genome amplification. The whole genome of the unknown microbe or eukaryote is then sequenced and assembled.
Owner:SHORELINE BIOME LLC
DNA vectors, transposons and transposases for eukaryotic genome modification
ActiveUS10233454B2Stably introducingHeterologous gene expression can be improvedPolypeptide with localisation/targeting motifMicroorganismsHeterologousGene transfer
The present invention provides polynucleotide vectors for high expression of heterologous genes. Some vectors further comprise novel transposons and transposases that further improve expression. Further disclosed are vectors that can be used in a gene transfer system for stably introducing nucleic acids into the DNA of a cell. The gene transfer systems can be used in methods, for example, gene expression, bioprocessing, gene therapy, insertional mutagenesis, or gene discovery.
Owner:DNA TWOPOINTO
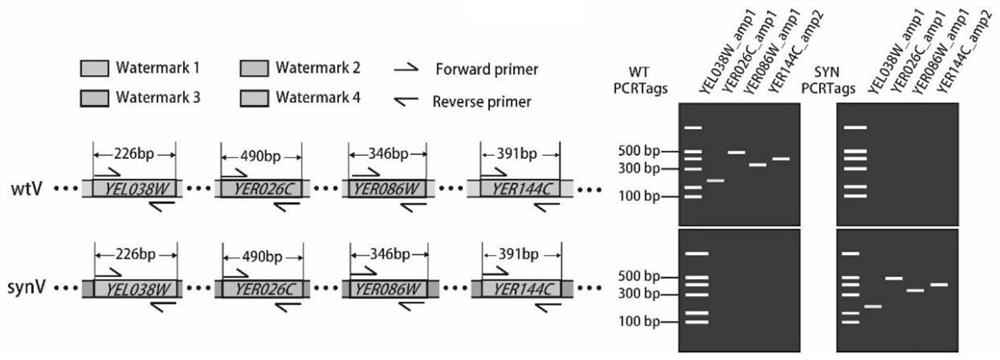
Detection method of yeast mitosis recombination hotspot
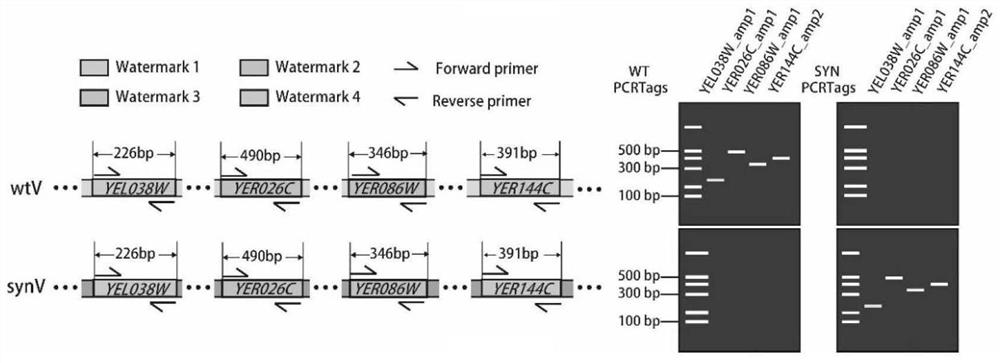
ActiveCN113549682AMicrobiological testing/measurementLibrary creationSynthetic biologyRapid identification
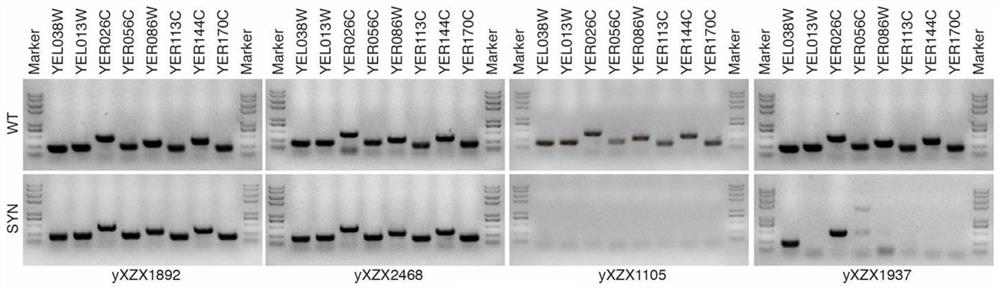
The invention relates to the technical field of synthetic biology, in particular to a detection method of yeast mitosis recombination hotspot. A PCRTag tag combination provided by the invention can be used for rapidly detecting eukaryotic genome mitosis recombination hotspots. By utilizing the tag combination, the mitosis recombination hotspot of the homologous chromosome can be quickly identified through a specific watermark tag, and a convenient method is provided for researching the eukaryotic mitosis recombination event.
Owner:TIANJIN UNIV
Method for delivering nucleic acid into eukaryotic genomes
ActiveUS8192934B2Improve applicabilityHydrolasesMicrobiological testing/measurementBacteriophage MuGenetic engineering
Owner:FINNZYMES
Method for introducing changes into a eukaryotic genome in vivo and a kit
InactiveUS20100279416A1Improve survivabilityGood model for in vivo experimentationGenetically modified cellsNucleic acid vectorA-DNAIn vivo
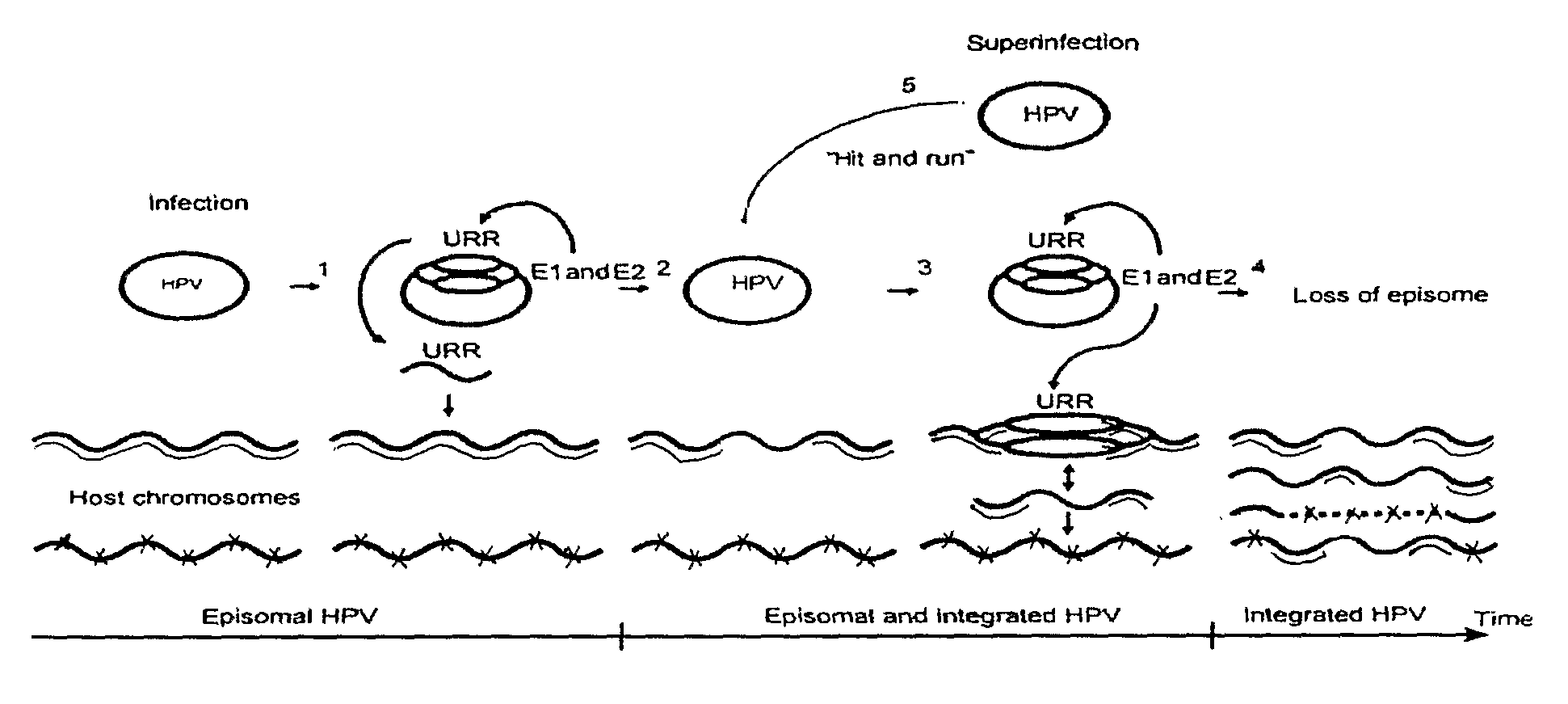
The invention relates to a method for introducing changes into a eukaryotic genome in vivo wherein the HPV genome, which comprises HPV replication origin sequence, is used together with HPV early proteins in order to achieve DNA replication in vivo. There is also disclosed a kit for in vivo amplification, excision, translocation and / or inversion of a DNA sequence, which comprises a vector carrying HPV genome or a part of HPV genome including HPV replication origin sequence, and expression vector or vectors encoding HPV early proteins.
Owner:UNIV OF TARTU
Regulatory nucleic acid elements
ActiveUS20100311121A1Increase transcriptionHigh expressionAnimal cellsSugar derivativesCoding regionTransfection
The invention relates to DNA-sequences, especially transcription- or expression-enhancing elements (TE elements) and their use on an expression vector in conjunction with an enhancer, a promoter, a product gene and a selectable marker.The invention describes Sequence No. 1 and TE elements TE-01, -02, -03, -04, -06, -07, -08, -10, -11 or -12. Because of their small size, TE-06, TE-07 or TE-08 are particularly preferred. Sequence No. 1 originates from a sequence region located upstream from the coding region of the Ub / S27a gene from CHO cells.TE elements bring about an increase in the expression of the product gene, particularly when stably integrated in the eukaryotic genome, preferably the CHO-DG44 genome. Chromosomal positional effects are thereby overcome, shielded or cancelled out. In this way the proportion of high producers in a transfection mixture and also the absolute expression level are increased up to seven-fold.
Owner:BOEHRINGER INGELHEIM PHARMA KG
Marker combination and application thereof in mitosis recombination hotspot detection
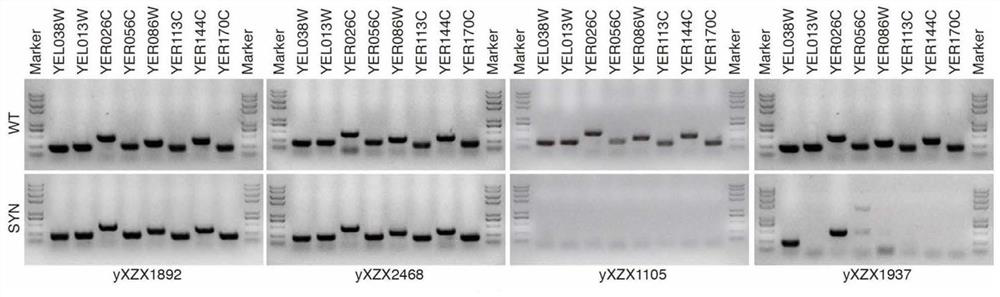
ActiveCN113528699AMicrobiological testing/measurementMicroorganism based processesSynthetic biologyRapid identification
The invention relates to the technical field of synthetic biology, in particular to a marker combination and application thereof in mitosis recombination hotspot detection. The PCRTag tag combination provided by the invention can be used for rapidly detecting eukaryotic genome mitosis recombination hotspots. By utilizing the tag combination, the mitosis recombination hot spot of the homologous chromosome can be quickly identified through the specific watermark tag, and a convenient method is provided for researching the eukaryotic mitosis recombination event.
Owner:TIANJIN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com