Marker combination and application thereof in mitosis recombination hotspot detection
A technology of mitosis and tag combination, applied in the field of synthetic biology, can solve the problems of insufficient research on yeast recombination hotspots and cold spots, screening of restricted recombination strains, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
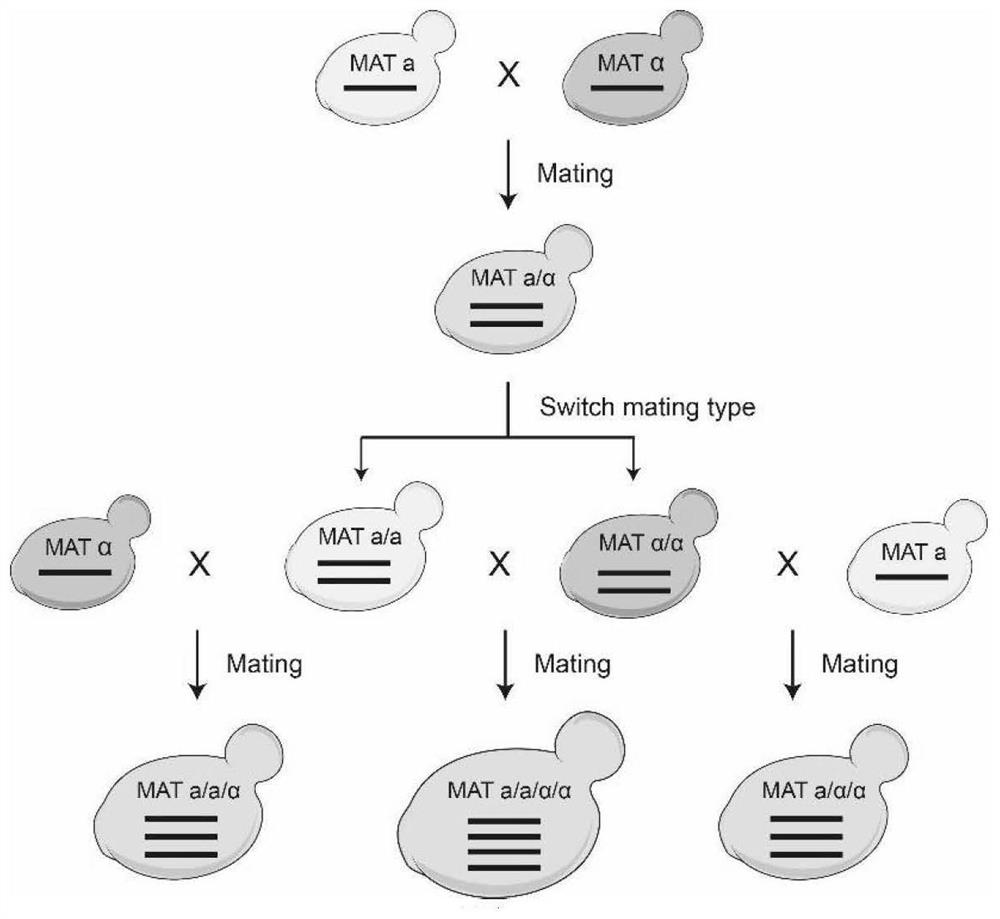
[0047] 1. Construction of rearrangement strain library
[0048] 1.1 Strains
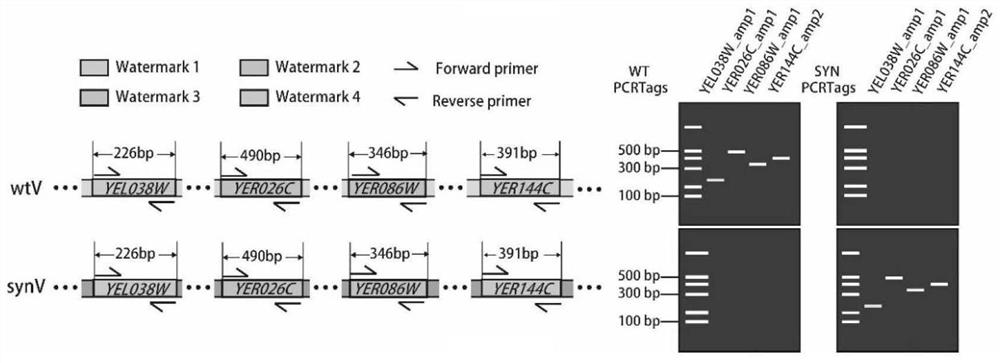
[0049] ① The diploid Saccharomyces cerevisiae strain yLHM120 was used as the starting strain, which contained a wild-type V chromosome (wtV) and a synthetic V chromosome (synV), and was from Tianjin University.
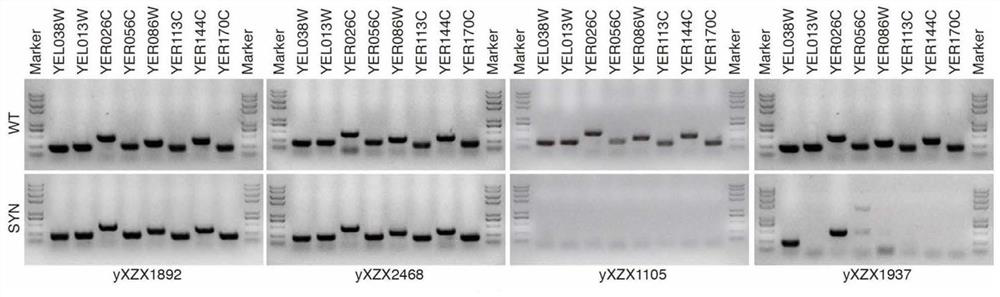
[0050] ②Triploid Saccharomyces cerevisiae strain yXZX1892 was used as the starting strain, which contained one wild-type V chromosome (wtV) and two synthetic V chromosomes (synV), and was from Tianjin University.
[0051] ③Tetraploid Saccharomyces cerevisiae strain yLHM086 was used as the starting strain, which contained two wild-type V chromosomes (wtV) and two synthetic V chromosomes (synV), and was from Tianjin University.
[0052] 1.2 Induced rearrangement:
[0053] A single colony carrying pCRE4 was picked from the SC-His plate, inoculated into 3 mL of SC-His medium, and cultured with shaking at 30°C for 24 hours.
[0054] Centrifuge at 5000 rpm for 2 min at room temperature, remove ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com