Gene editing method based on gene cas3 of I-B type CRISPR-Cas system
A gene editing and gene technology, applied in the field of gene editing in the field of biotechnology, can solve the problems of off-target, large molecular weight, etc., and achieve the effect of small molecular weight and rapid gene editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 Knockout of lacZ gene in EC JM109 (DE3)
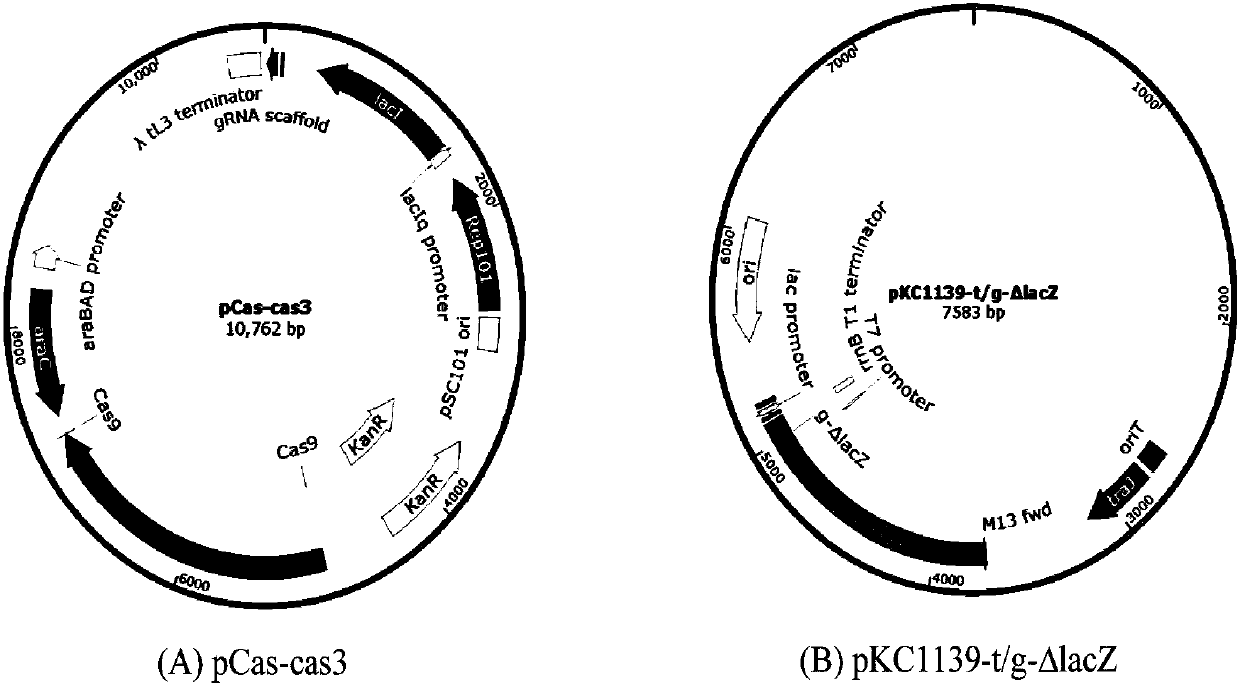
[0055] (1) Construction of Cas3 expression vector pCas-cas3
[0056]According to the DNA sequence information of the SV IBL14 gene cas-3 and the vector pCas, the specific forward primer pCas-cas3-F and the reverse primer pCas-cas3-R with the gene cas3 complementary to the vector pCas were designed. Extract SV IBL14 genomic DNA, and amplify the gene cas3 by conventional PCR. The PCR reaction system (50 μL) includes: 5 μL 10×Pfu buffer, 5 μL dNTPs (2.5 mM each), 1 μL 10 μM cas3-F, 1 μL 10 μM cas3-R, 5 μL DMSO, 0.5μL Pfu DNAPolymerase, 0.5μL SV IBL14 genomic DNA, 32μL sterile water (nuclease-free), reaction conditions: 95℃ for 5min, 94℃ for 60±30s, 55±3℃ for 30s, 72℃ for 90±30s, 2.5±0.5 U DNA polymerase (TransStartFastPfu DNA Polymerase, Quanshijin Biotechnology Co., Ltd.), 30 cycles, 72°C for 10min. The PCR product was detected by 1% agarose electrophoresis, recovered by the kit, and the purified full-length cas3 gene ...
Embodiment 2
[0079] Example 2 Knockout of ldh gene and insertion of chloramphenicol resistance gene cat in BS 168
[0080] (1) Construction of Cas3 expression vector pHT304-cas3
[0081] According to the DNA sequence information of SV IBL14 gene cas3 and carrier pHT304, design has the specific forward primer and reverse primer PHT304-cas3-F / R of the gene cas3 complementary to carrier pHT304, all the other steps are with embodiment 1 step (1 ).
[0082] (2) Construction of gene editing vector pKC1139-t / g-Δldh::cat
[0083] Design g-Δldh according to the BS 168 gene ldh sequence and design t-Δldh::cat inserted between the upper and lower homology arms of the ldh gene to express the chloramphenicol resistance enzyme gene cat sequence, and the remaining steps are the same as in Example 1 ( 2).
[0084] The synthetic t-Δldh::cat sequence is shown in Table 4, in which the single underline represents the promoter; the double underline metabolic terminator.
[0085] Table 4 t-Δldh::cat sequenc...
Embodiment 3
[0100] Example 3 Double plasmid knockout of SC BY4741crtE gene
[0101] (1) Construction of Cas3 expression vector pRS415-cas3
[0102] In addition to the DNA sequence information of the vector pRS415, design the specific forward primer and reverse primer pRS415-cas3-F / R with the gene cas3 complementary to the vector pRS415, and the upstream of the gene cas3 is the gal1 promoter, and the downstream end has a nucleus The signal sequence and the CYC1 terminator are located, and the rest of the steps are the same as step (1) in Example 1.
[0103] (2) Construction of gene editing vector pYES2-NTA-t / g-ΔcrtE
[0104] Except for designing g-ΔcrtE and designing and synthesizing t-ΔcrtE according to the SC BY4741crtE gene sequence, other steps are the same as step (2) of Example 1.
[0105] The synthesized t-ΔcrtE sequence is shown in Table 7.
[0106] Table 7 t-ΔcrtE sequence list
[0107]
[0108] The synthesized g-ΔcrtE sequence is shown in Table 8. Capital letters in the ta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com