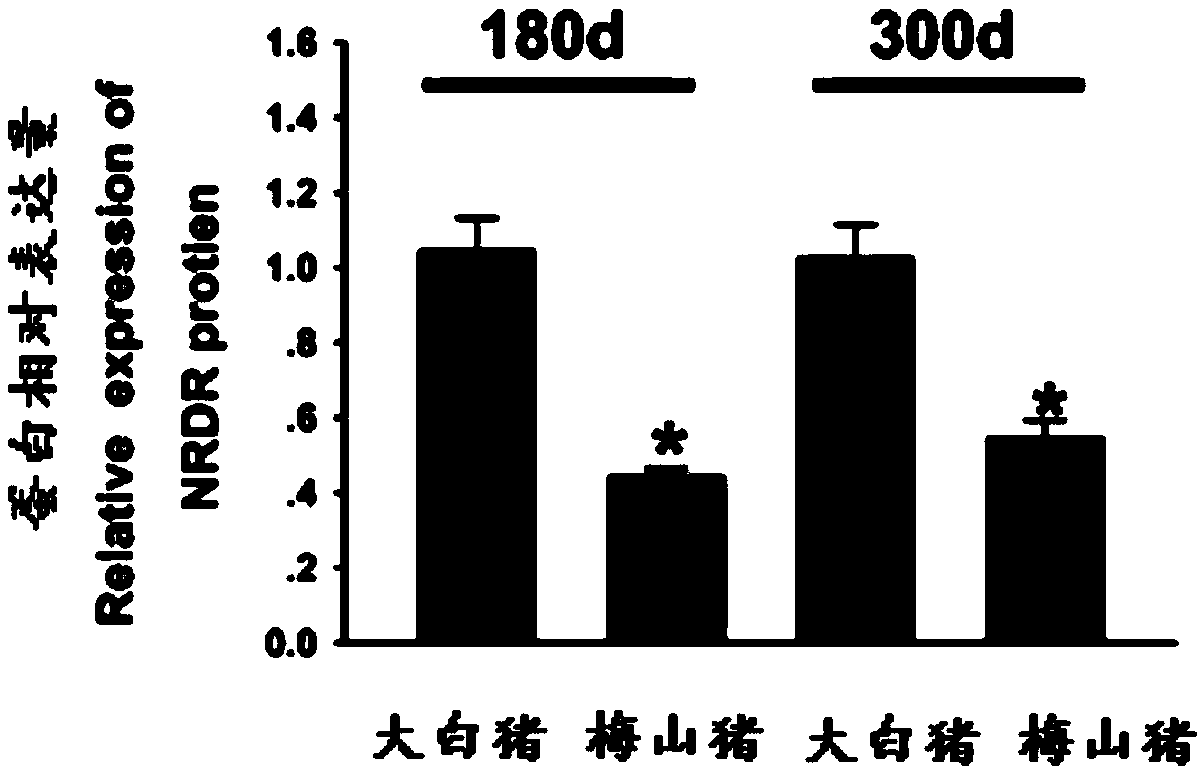
Patents
Literature
349results about How to "Valid identification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Nanoparticle tracer-based electrochemical DNA sensor for detection of pathogens-amplification by a universal nano-tracer (AUNT)
InactiveUS20110171749A1Rapid and sensitive detectionRapid and sensitive and valid identificationMaterial nanotechnologyNanomedicineSalmonella entericaEscherichia coli
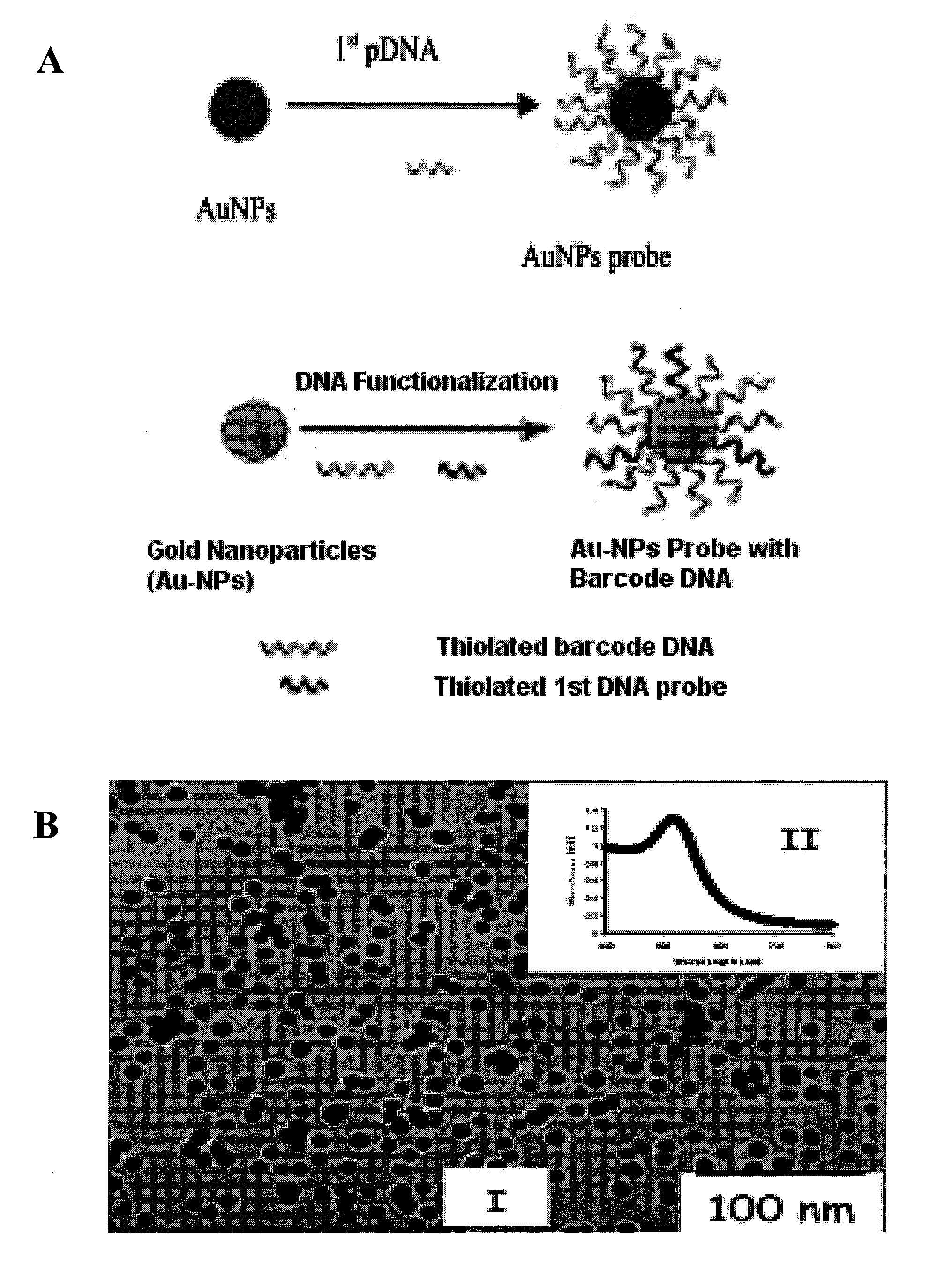
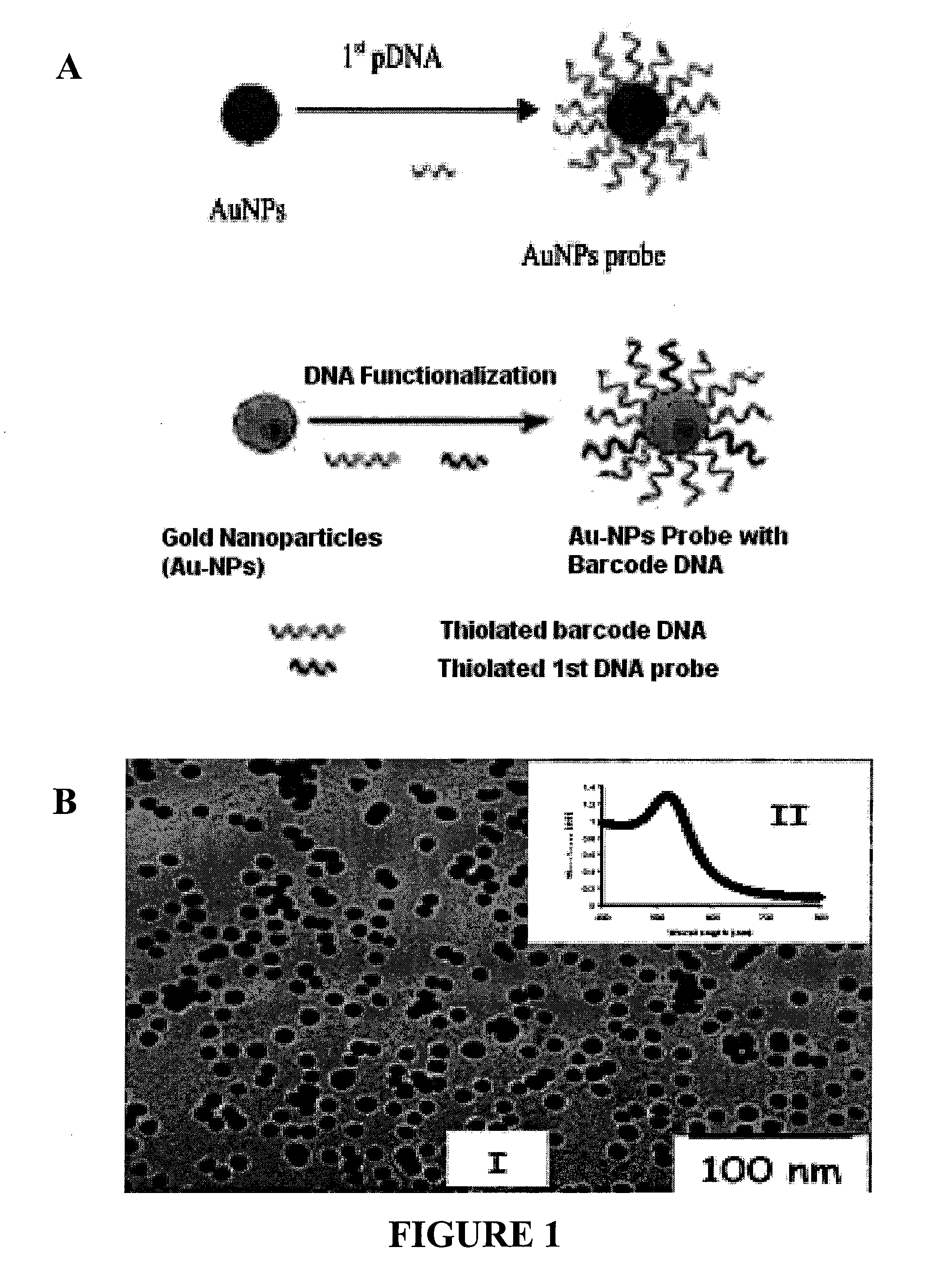
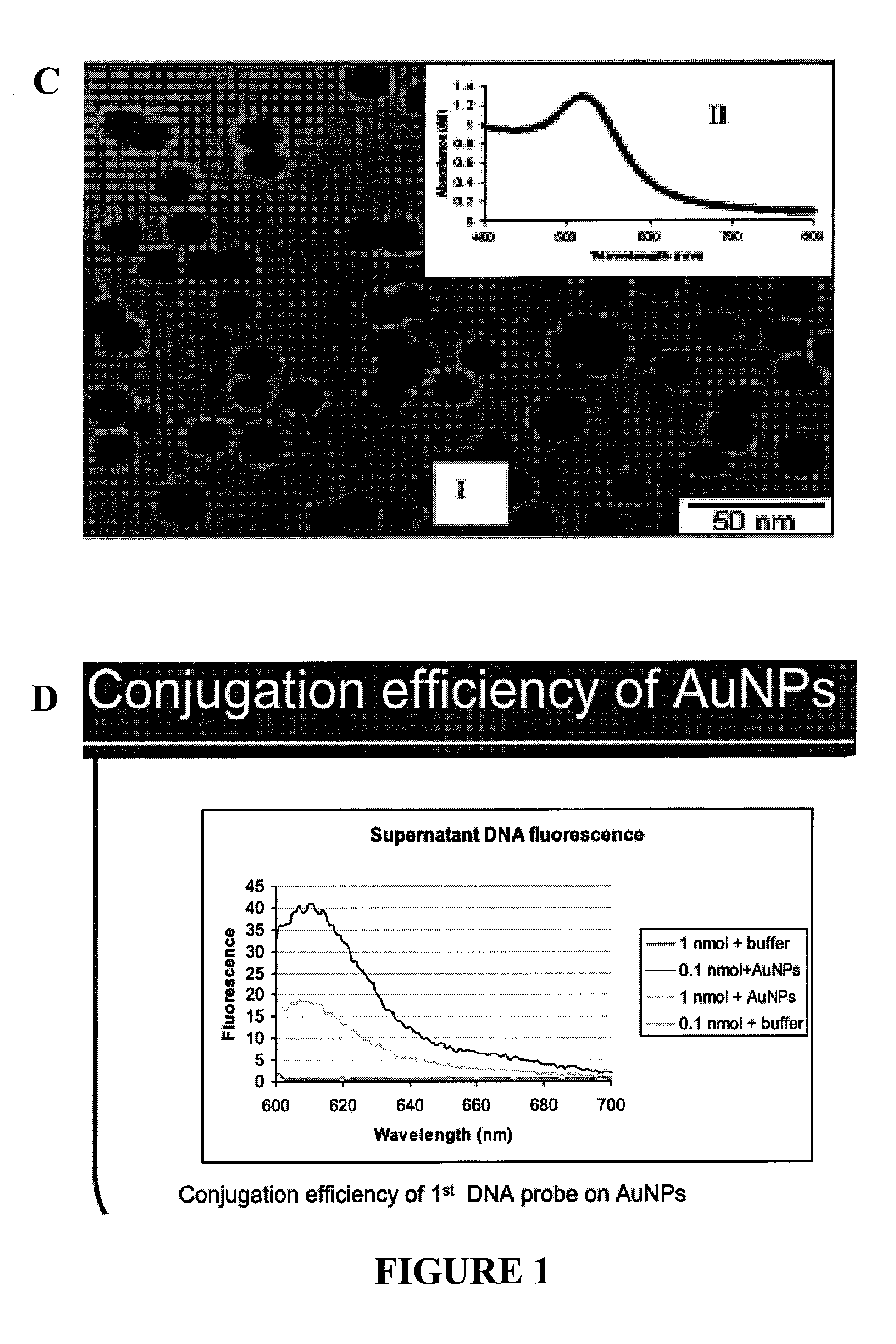
The present invention relates to methods and compositions for identifying a pathogen. The inventions provide an antibody-based biosensor probe comprising (AUNT) in combination with a polymer-coated magnetic nanoparticle. In particular, a nanoparticle-based biosensor was developed for detection of Escherichia coli O157:H7 bacterium in food products. Further described are biosensors for detecting pathogens at low concentrations in samples. Even further, a gold nanoparticle-based electrochemical biosensor detection and amplification method for identifying the insertion element gene of Salmonella enterica Serovar Enteritidis is described. The present invention provides compositions and methods for providing a handheld potentiostat system for detecting pathogens outside of the laboratory. The AUNT biosensor system has applications detecting pathogens in food, water, beverages, clinical samples, and environmental samples.
Owner:BOARD OF TRUSTEES OPERATING MICHIGAN STATE UNIV
Earthquake physical model and preparation method and application thereof
ActiveCN102443245AValid identificationExperimental investment is smallEducational modelsSeismic signal processingEpoxyKinematics
The invention provides an earthquake physical model and a preparation method and application thereof. The materials of the earthquake physical model comprise main materials, auxiliary materials and aids, wherein the main materials are epoxy resin and rubber, and the aids are curing agent, cross-linking agent, diluent, defoaming agent and the like. A novel earthquake physical model material is synthesized by special preparing equipment and processes and used for building an earthquake physical model according to a similarity principle to simulate a real geological structure; and the earthquake physical model can be used for research on seismic wave field and researches for other purposes, particularly used for researching kinematics and kinematic characteristics of seismic waves propagating in complicated areas; moreover the earthquake physical model can provide objective bases for verification of new methods and new theories in oil-gas exploration and development.
Owner:CHINA PETROLEUM & CHEM CORP +1
Cas9/RNA system and application thereof
InactiveCN107304435AEasy to synthesizeSpeed up breedingVectorsHydrolasesGenetic engineeringDouble stranded
The invention relates to genetic engineering transformation of nannochloropsis oculata and particularly relates to a method for carrying out genetic transformation by using Cas9 / CRISPR at a fixed point. A compound is formed by overexpressed Cas9 protein in the nannochloropsis oculata and gRNA, the Cas9 protein is used for carrying out site-specific cleavage on a genome double-stranded DNA under the guidance of the gRNA to cause gene mutation, and mutation is effectively screened by combining a sequencing technique. Fixed-point genetic engineering transformation is carried out on the nannochloropsis oculata by using a CRISPR / Cas technology, so that the algae species improvement efficiency of the nannochloropsis oculata can be significantly improved.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
Combined test method for combining software reliability tests with hardware reliability tests
ActiveCN103914353ASimulate the realReflect actual usageSoftware testing/debuggingCombined testPresent method
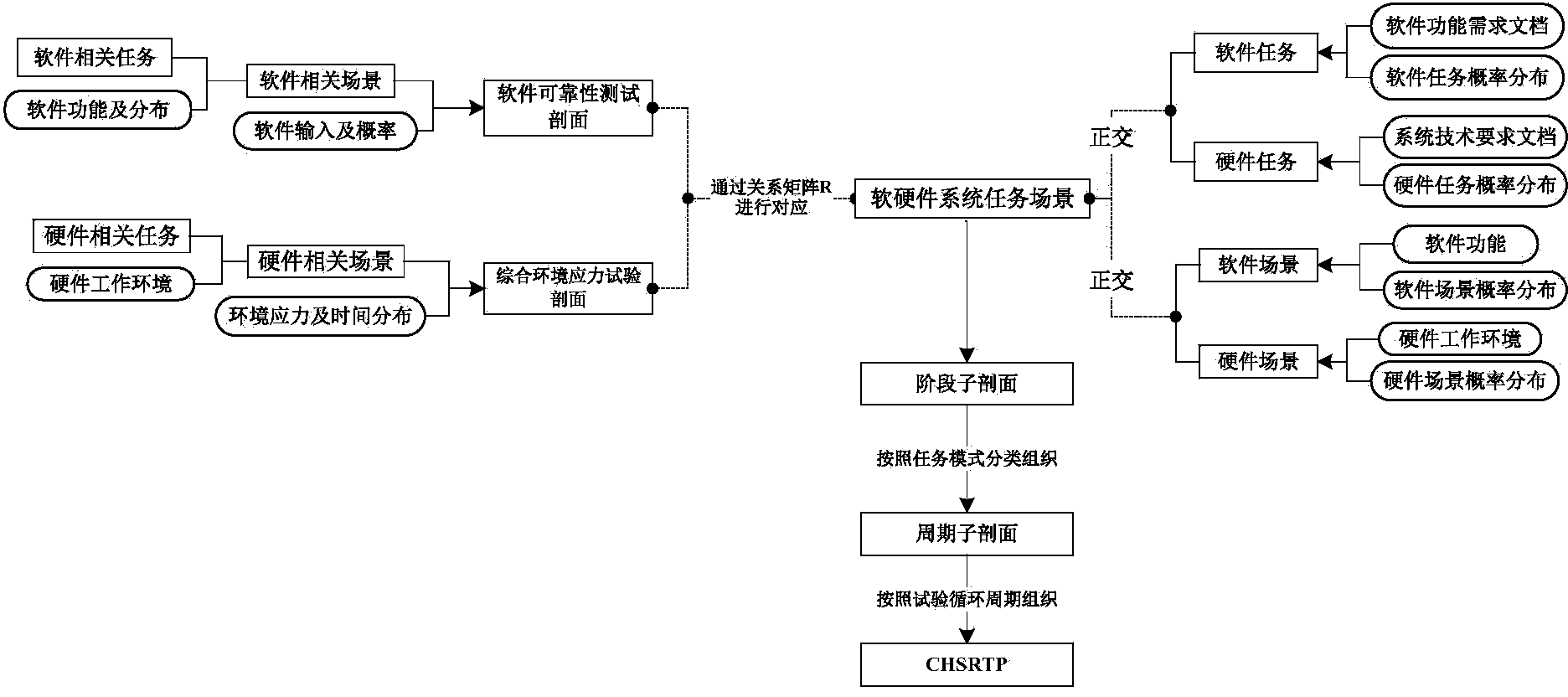
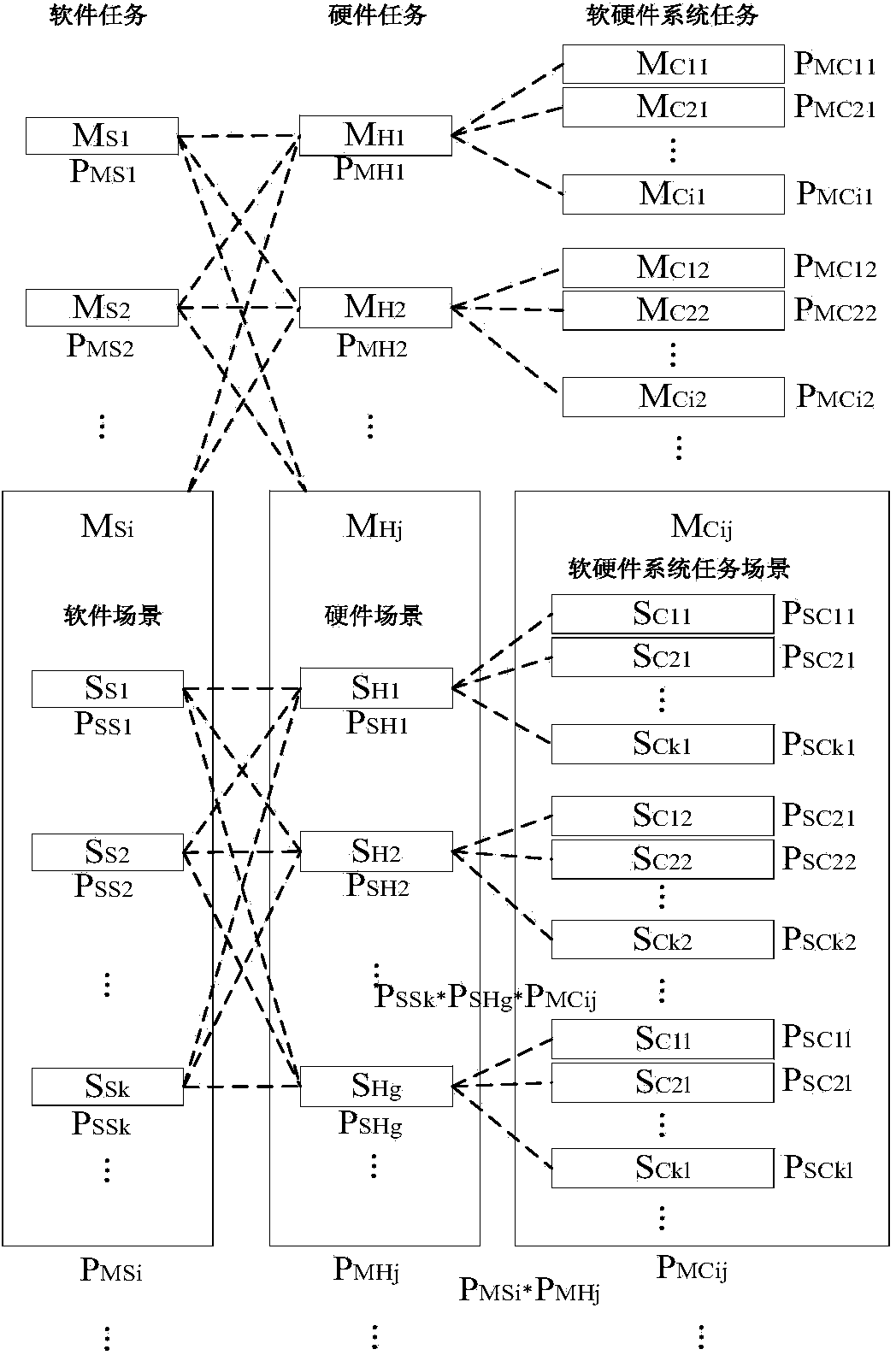
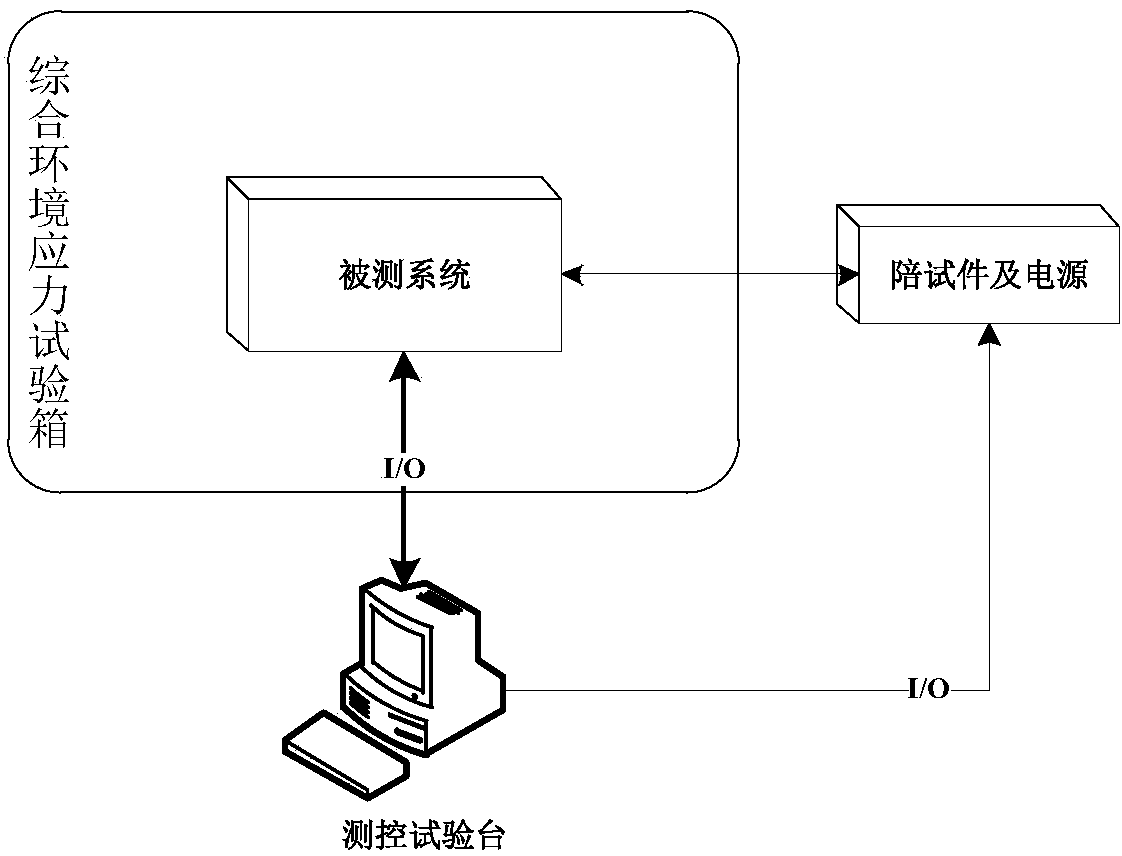
The invention discloses a combined test method for combining software reliability tests with hardware reliability tests. The combined test method is used for testing the reliability of software and hardware systems. The combined test method includes analyzing the software and hardware systems and constructing hardware reliability test profiles and software reliability test profiles; enabling software and hardware scenarios to be orthogonal to one another to acquire system task scenarios and determining execution probabilities of the system task scenarios; creating relation matrixes for determining whether the software reliability test profiles and hardware scenario profiles are associated with one another or not; combining corresponding hardware environmental stress test profiles with the associated software reliability test profiles to generate stage child profiles for each system task scenario; organizing the stage child profiles of the same system task to form a periodic child profile, organizing the periodic child profiles according to test cycle periods to generate final combined test profiles; generating test cases according to the combined test profiles and performing the tests. The combined test method has the advantages that the reliability of the systems can be accurately evaluated by the aid of test results generated by the method, and system failure which cannot be discovered in traditional reliability tests can be discovered by the aid of the combined test method.
Owner:BEIHANG UNIV
Classification test method and classification test system for edible oil and swill-cooked dirty oil
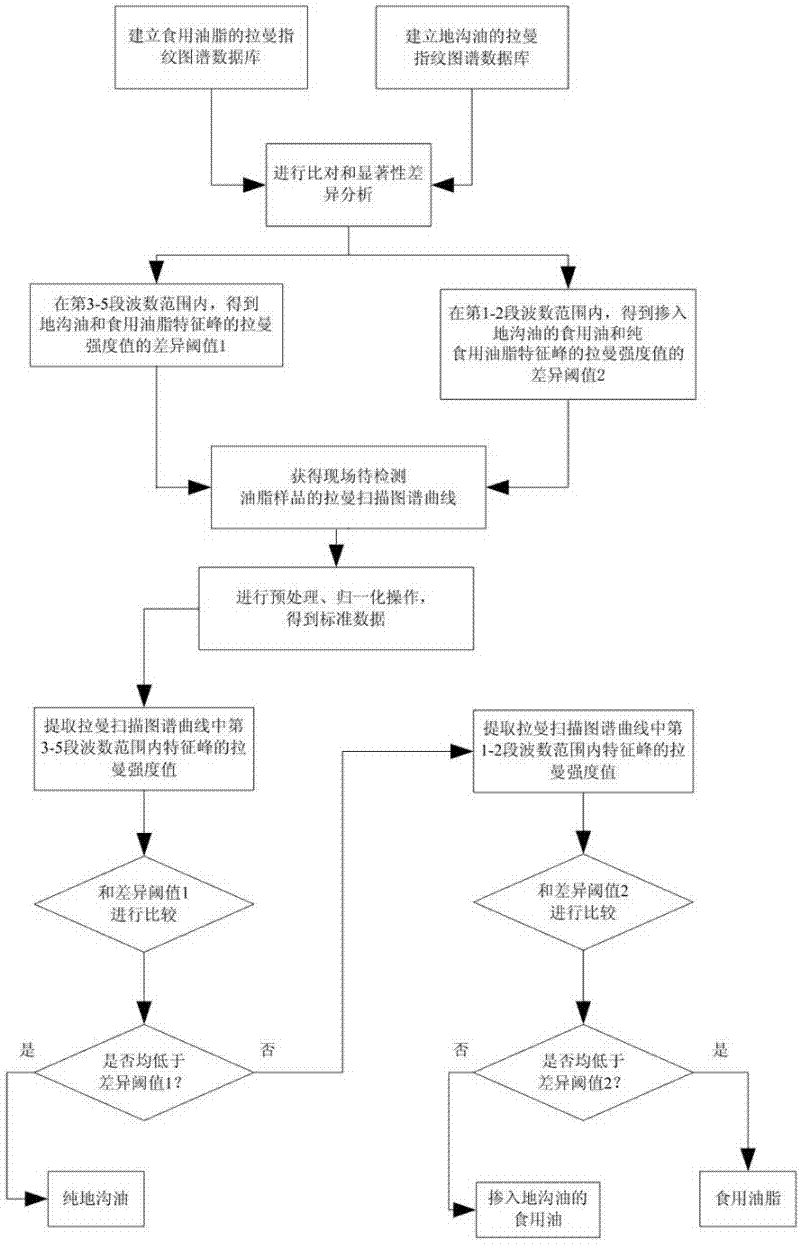
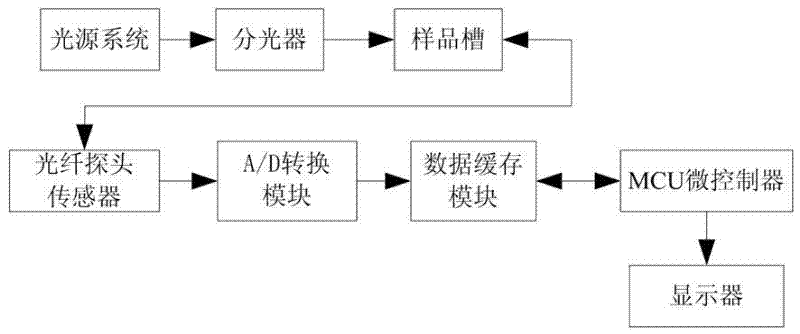
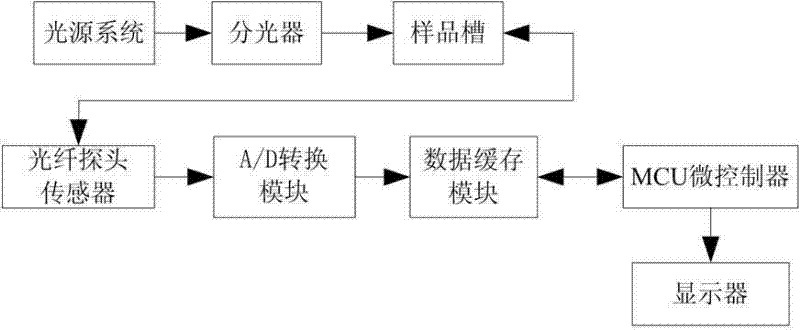
The invention discloses a classification test method and a classification test system for edible oil and swill-cooked dirty oil. The classification test method comprises the following steps that: Raman fingerprint databases for edible oil and swill-cooked dirty oil are first created, and the notable difference between the maximum Raman intensity values of the characteristic peaks of edible oil and swill-cooked dirty oil within a five-band wave number range and the threshold of the difference are then obtained by way of comparison; and in a practical test, the variety of a tested samples obtained by extracting the maximum Raman intensity value of the characteristic peak of the tested sample within the five-band wave number range and comparing the maximum Raman intensity value with the difference threshold. The classification test system comprises a light source system, an optical splitter, a sample box and a measurement control unit. The invention has the advantages of flexibility and high specificity, and can effectively test whether a tested sample is pure swill-cooked dirty oil, pure edible oil meeting the active national standard or edible oil mixed with swill-cooked dirty oil or not; and meanwhile, the operation is simple and convenient, the testing time is short, and swill-cooked dirty oil can be tested on the spot.
Owner:邹玉峰
Primer system for PCR (polymerase chain reaction) identification for deer, pig, cow, sheep, horse, donkey, rabbit and chicken
ActiveCN102876805AValid identificationEfficient identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceWhole blood product
The invention discloses a primer system for PCR (polymerase chain reaction) identification for a deer, a pig, a cow, a sheep, a horse, a donkey, a rabbit and a chicken, and can detect eight animal species in one time so as to achieve a purpose that common animal species can be almost covered. The primer of the primer system only carries out specific amplification on a respective species target segment without reacting with other species, and therefore the primer can be suitable for multiple PCR reaction characterized in that at least two primer pairs are induced in one-time PCR reaction so as to effectively shorten experiment time. In addition, when the PCR is carried out, an optimized specific PCR reaction system and reaction program can be used, a detection flow is simplified due to the adoption of a uniform PCR detection method, a quick and efficient identification mode with the high specificity is established so as to effectively identify whether the deer blood product is true and false and identify the adulteration mode, and a modern molecular biology detection means is provided for deer blood quality control. In addition, the primer system can be cooperated with relevant reagent to be prepared into a kit, thereby being convenient to use. Meanwhile, possibility is provided for the industrial production and application, and the primer system has an excellent application prospect.
Owner:苏州红冠庄国药股份有限公司
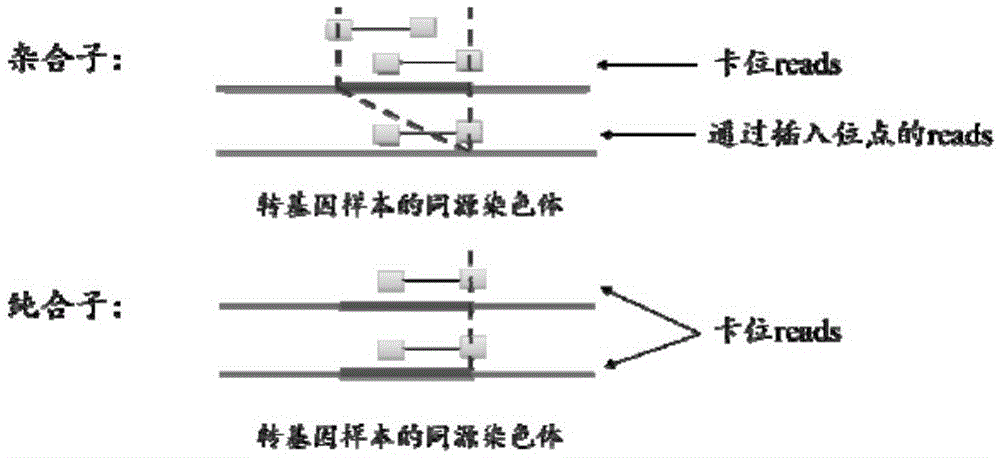
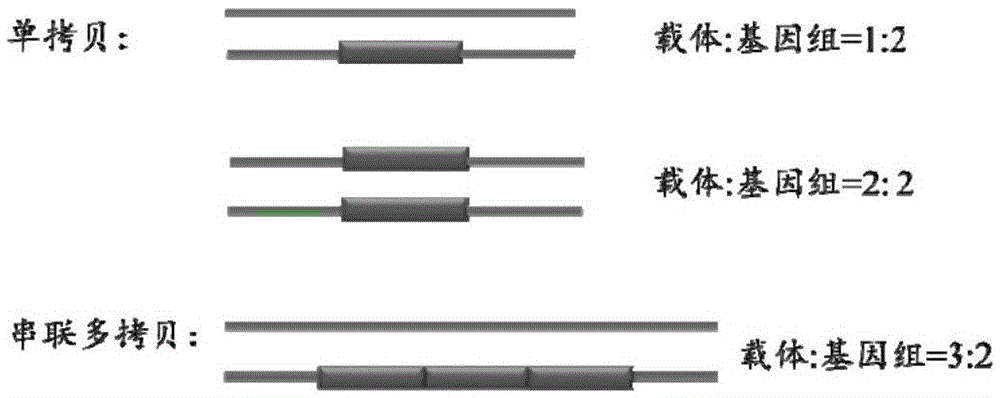
Method for identifying transgenic events through whole genome sequencing data
ActiveCN105631242AAccurate analysisValid identificationProteomicsGenomicsBiological bodyWhole genome sequencing
The invention provides a method for identifying transgenic events through whole genome sequencing data. The method comprises the steps that transgenic events are identified through a bioinformatics means, and the position, direction, copying number and flanking sequence information of foreign fragments inserted into genomes and the homozygous and heterozygous state of transgenic samples can be rapidly and effectively identified. Compared with traditional Genome walking, real-time quantitative PCR and other experiments methods, the method has the advantages that time is short, repeatability is high, the result is stable, and explaining is easier. The method is the most comprehensive in existing transgenic event detection methods, can accurately analyze influences of transgenic events on the living body within a short time, carries out safety evaluation on transgenic animals and plants, and pushes the transgenic technology to be applied in agriculture.
Owner:CHINA AGRI UNIV
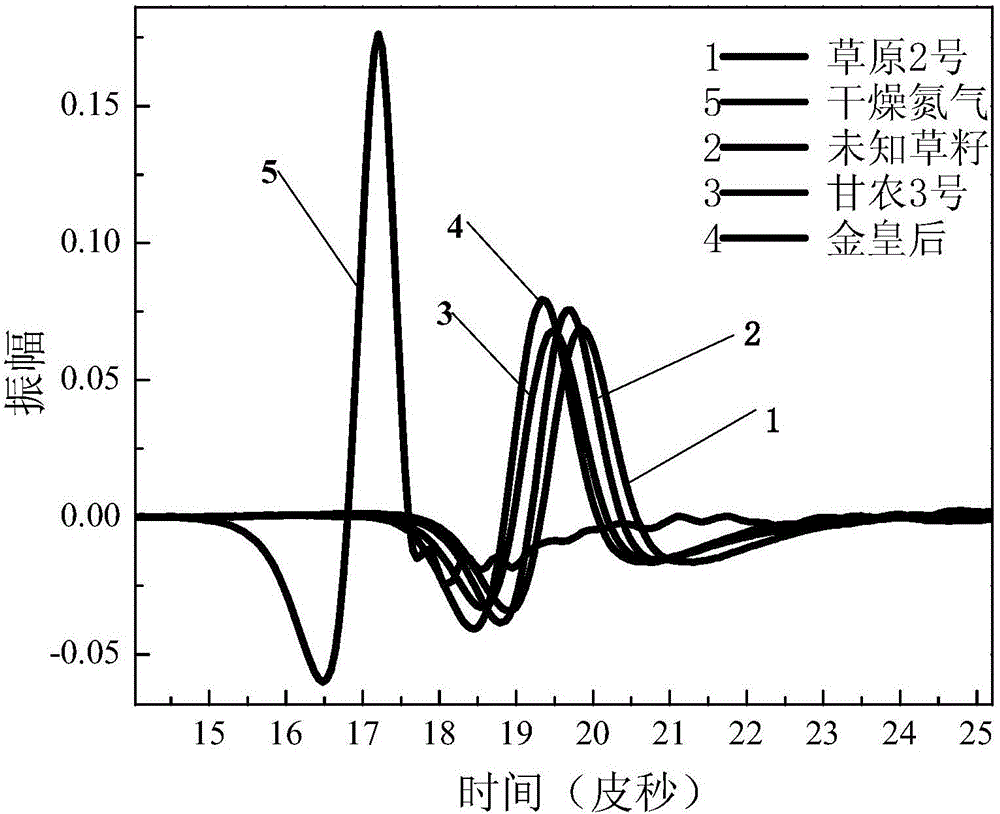
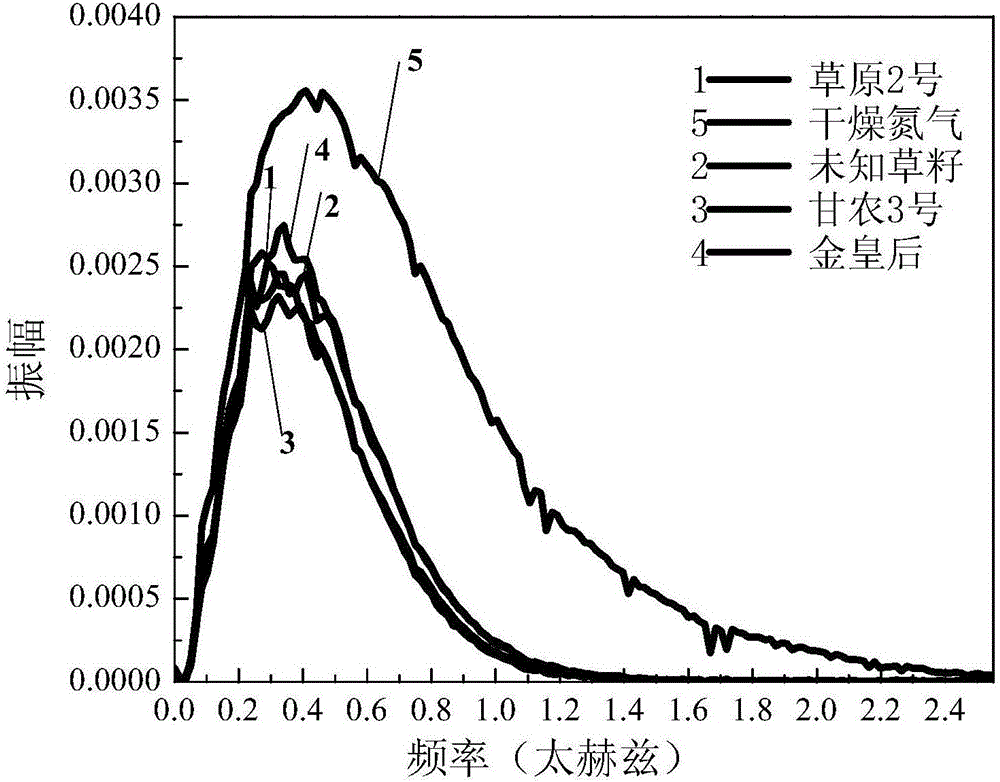
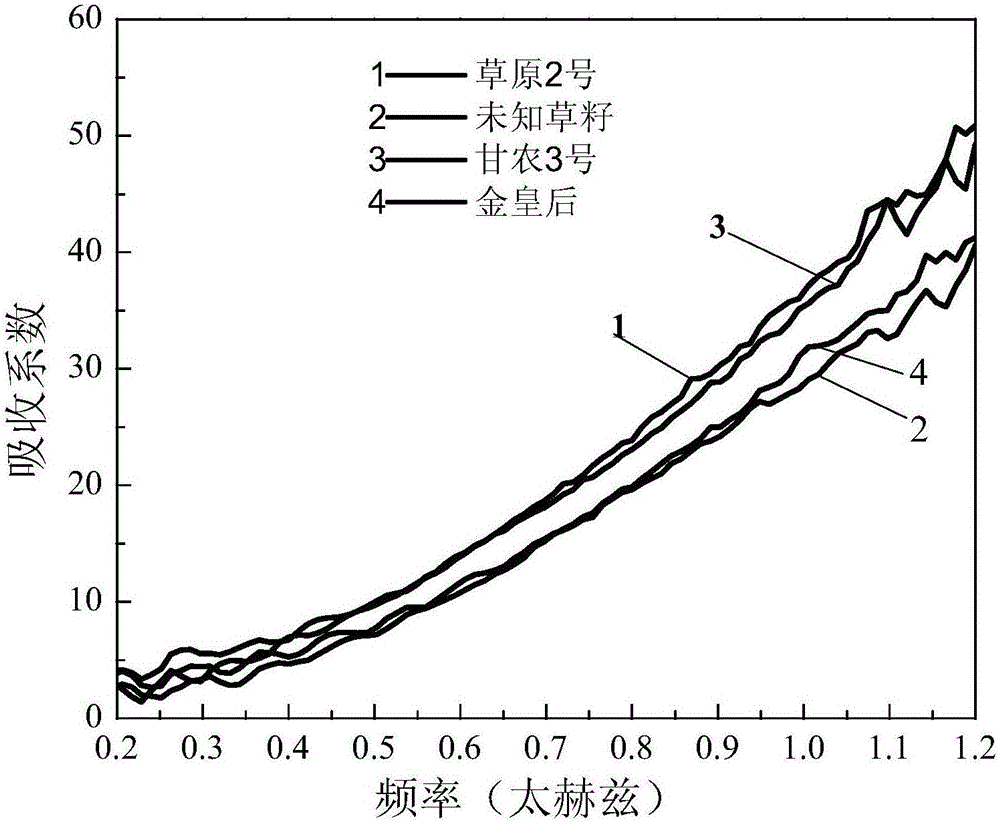
Dynamic multipoint grass variety identification and authentication method based on terahertz time domain spectroscopy
ActiveCN106248610AAvoid breakingValid identificationColor/spectral properties measurementsInvestigation of vegetal materialData treatmentSeed sample
The invention provides a dynamic multipoint grass variety identification and authentication method based on terahertz time domain spectroscopy. The method comprises the following steps: preparing a standard grass seed sample; respectively detecting the standard grass seed sample and dry nitrogen by using a terahertz time domain spectroscopy device to respectively obtain a terahertz pulse time domain waveform sum of the standard grass seed sample and the dry nitrogen, taking the terahertz pulse time domain waveform of the dry nitrogen as a reference signal, and taking the terahertz pulse time domain waveform of the standard grass seed sample as a sample signal; carrying out data processing on the terahertz pulse time domain waveform of the standard grass seed sample and the terahertz pulse time domain waveform of the dry nitrogen to construct a standard fingerprint spectrum library; and analyzing, calculating and establishing an identification database of unknown grass varieties according to optical parameters of the standard fingerprint spectrum library in combination with a mathematical statistical method, thereby carrying out qualitative analysis and recognition on unknown grass samples. By adoption of the method, the grass varieties can be effectively recognized and detected, the database can also be established for the unknown grass samples, and the unknown grass samples can be quickly distinguished and identified in batches.
Owner:CHINA UNIV OF PETROLEUM (BEIJING) +1
Pelteobagrus fulvidraco microsatellite family identification method
InactiveCN104357553ASave time and costConservation of water bodies and human managementMicrobiological testing/measurementPopulation geneticsGenomic DNA
The invention discloses a pelteobagrus fulvidraco microsatellite family identification method which comprises steps as follows: pelteobagrus fulvidraco parent and offspring genomic DNA extraction, polymorphic microsatellite marker screening and primer synthesis, microsatellite locus genotyping and family identification and the like. Six pairs of microsatellite primers are adopted and have nucleotide sequences shown from SEQ ID NO. 1 to SEQ ID NO.12 respectively. Fluorescently-labeled microsatellite markers are firstly utilized on pelteobagrus fulvidraco, a paternity test platform is established, the accurate rate of identification of a pelteobagrus fulvidraco microsatellite family is up to 98.9%, different families of pelteobagrus fulvidraco can be rapidly and effectively identified, and evidence is provided for breeding and breeding matching of pelteobagrus fulvidraco. The pelteobagrus fulvidraco microsatellite family identification method selects more microsatellite locus alleles with high polymorphism, can be used for population genetics evaluation, family tree authentication and paternity test of pelteobagrus fulvidraco and further can be used for molecular marker auxiliary family management and molecular marker auxiliary parent selection.
Owner:HUAZHONG AGRICULTURAL UNIVERSITY
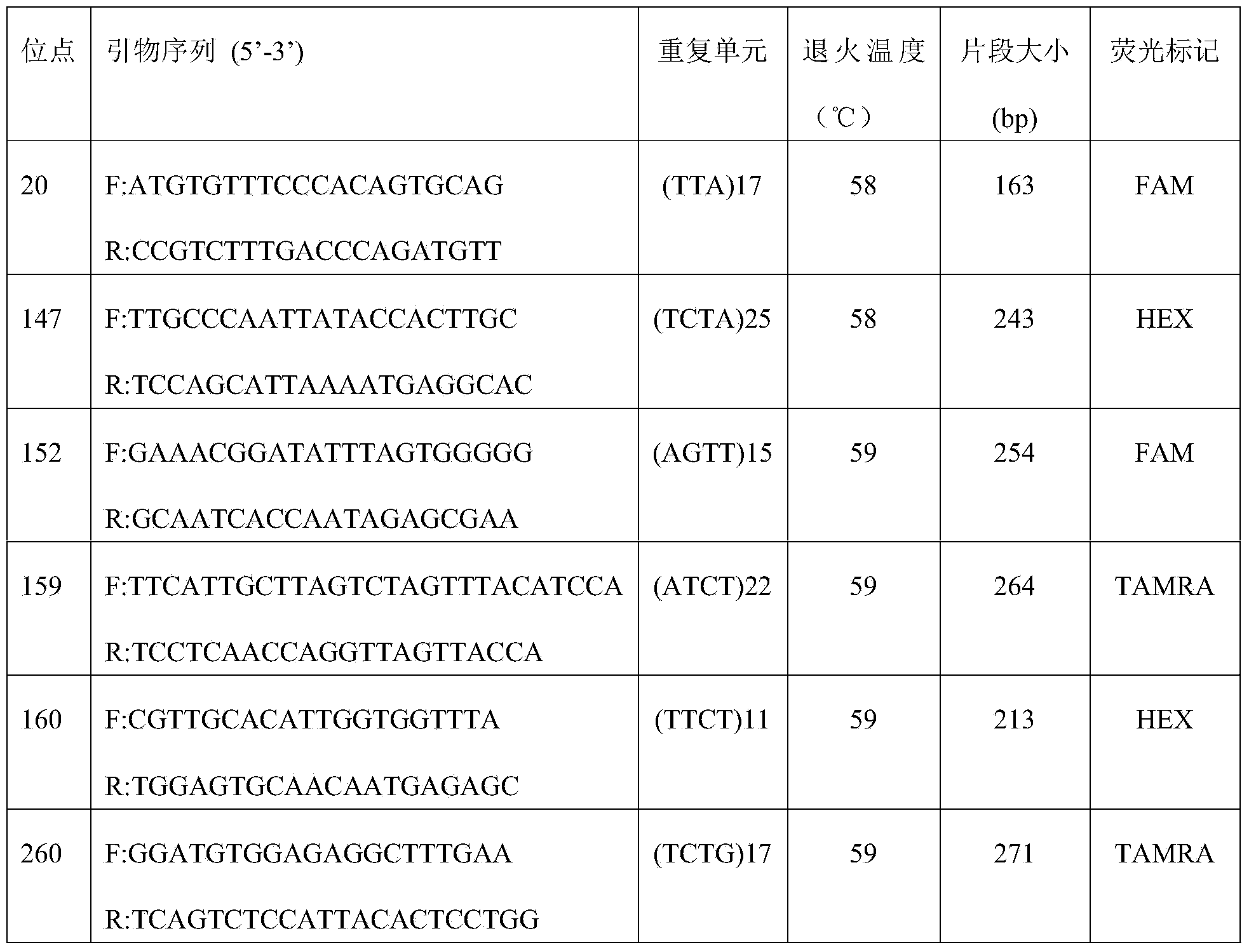
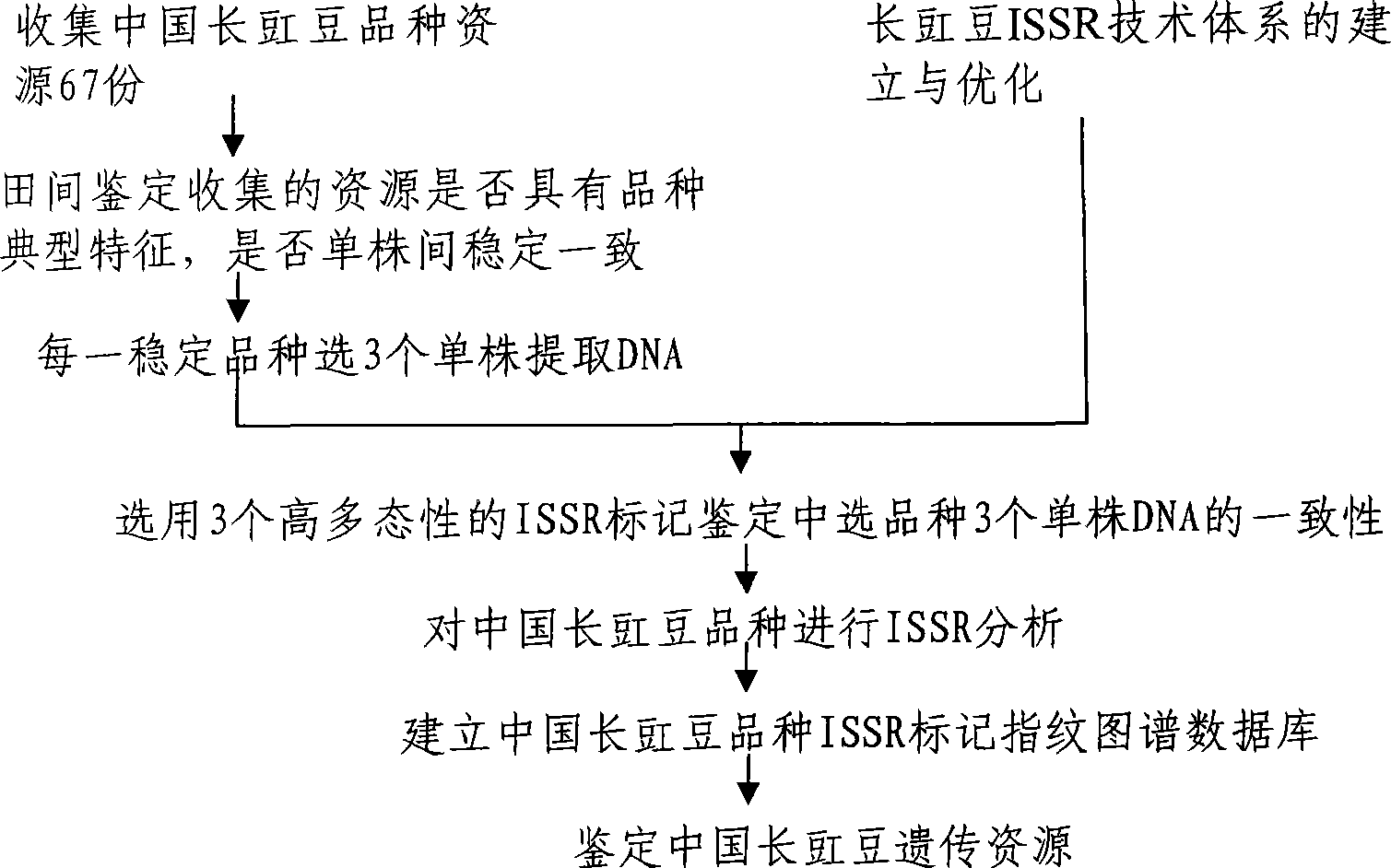
Method for constructing China asparagus bean genetic resource database based on ISSR molecular marker and uses thereof
InactiveCN101436229AValid identificationRapid identificationMicrobiological testing/measurementSpecial data processing applicationsDependabilityBiology
The invention discloses an ISSR molecular marking based method for constructing a Chinese asparagus bean generic resource database and application thereof. The invention also utilizes the data base to identify the trueness of a variety; according to field forms, the consistency and typicality among individual plants of the variety are identified, separated and atypical materials are given up, and DNA of the individual plant of the consistent and typical variety is picked up; by utilizing three ISSR markings with high polymorphism, the consistency of DNA level among the individual plants is primarily selected, and the variety and individual plant constructing a fingerprint are finally determined; and by utilizing the finally selected variety and individual plant, the ISSR analysis of the Chinese asparagus bean is made to construct a fingerprint database. The invention overcomes the defects existing in the construction of the prior generic resource database, such as separation among different individual plants of the variety and impurity on the DNA level, the reliability and accuracy of the Chinese asparagus bean ISSR fingerprint data base constructed by the method are greatly improved, and the planting varieties of the Chinese asparagus bean can be effectively and conveniently identified and distinguished.
Owner:JIANGHAN UNIVERSITY
Identification method of coping digital image
InactiveCN102521614AValid identificationFree from attackCharacter and pattern recognitionGraphicsPattern recognition
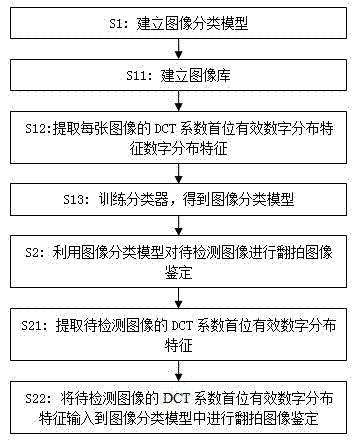
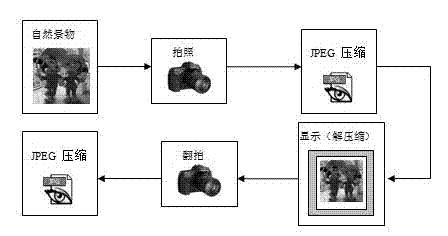
The invention belongs to the multimedia information safety field and particularly relates to an identification method of a coping digital image. The method comprises the following steps that: (1), an image classification model is established; more particularly, an image library is established and a copying image and a natural image are stored in the image library; discrete cosine transform (DCT) coefficient matrixes of all images in the image library are extracted and DCT coefficient first effective digit distribution characteristics of all the images are extracted; and a classifier is trained to obtain an image classification model; and (2), the image classification model is utilized to treat a detected image to carry out coping image identification. According to the invention, on the basis of a DCT coefficient first effective digit distribution change during a secondary image obtaining process, statistics of the DCT coefficient first effective digit distribution characteristics is carried out and the characteristics are input into the classifier to carry out study, so that the image classification model is obtained; and the classification model is utilized to analyze a digital image, so that it can be effectively identified whether the digital image is a copying image or not.
Owner:SUN YAT SEN UNIV
Enriching and analyzing method of circulating tumor cells of colorectal cancer
InactiveCN106282116AImprove capture efficiencyImprove the detection rate of CTCMicrobiological testing/measurementTumor/cancer cellsWilms' tumorMessenger ribonucleic acid
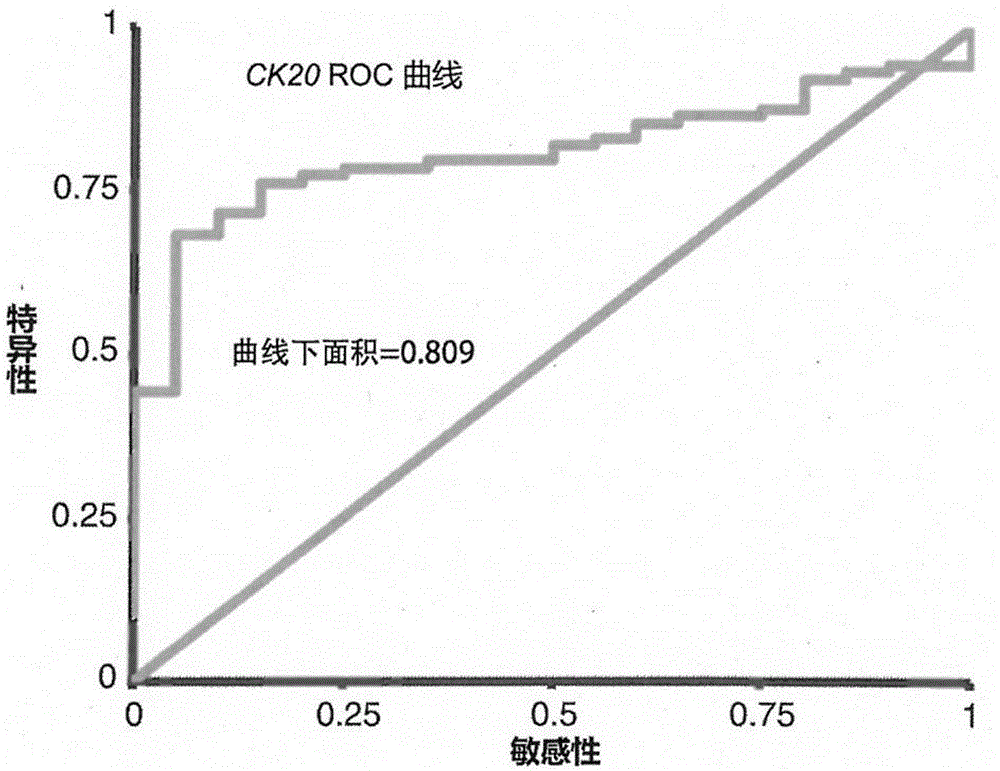
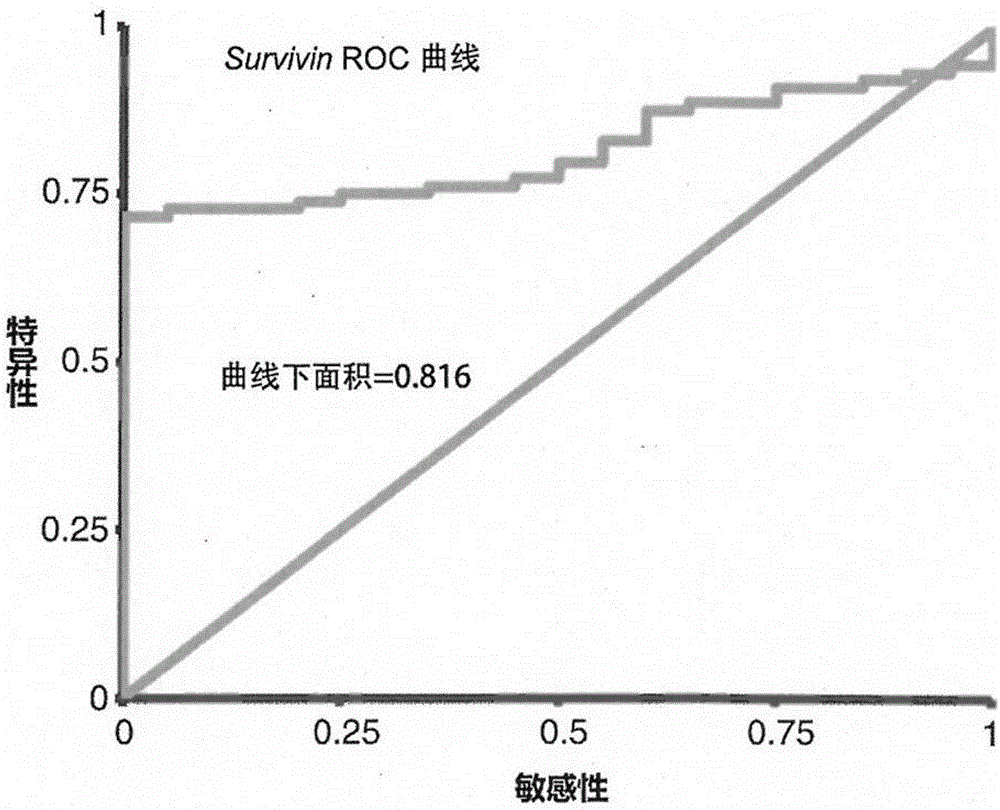
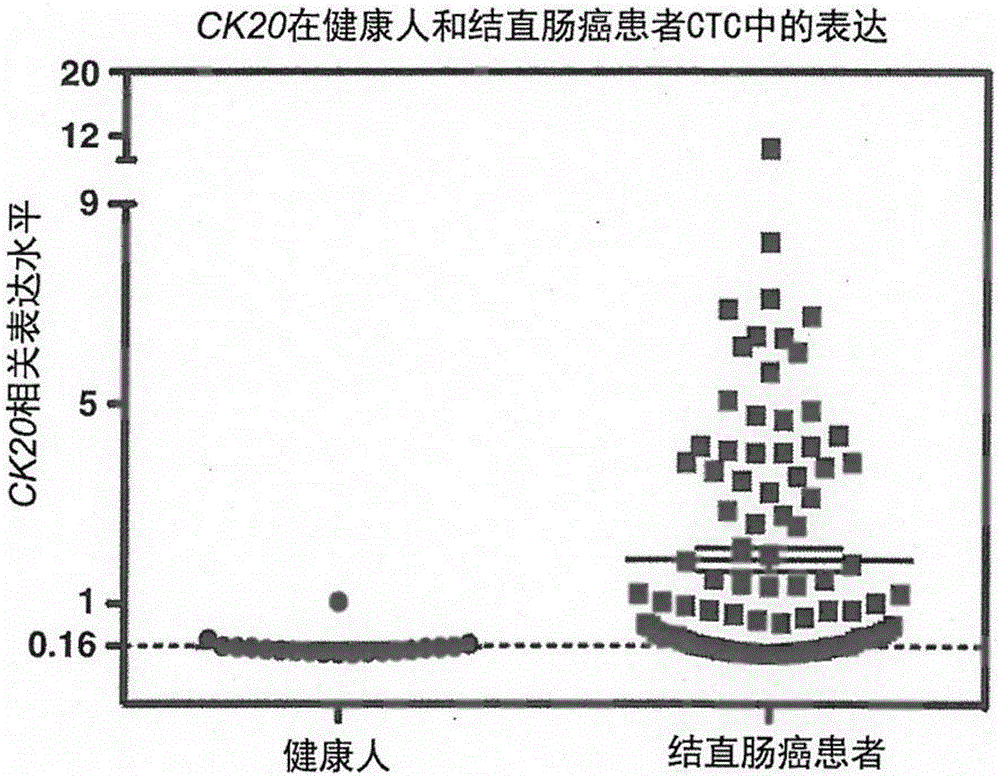
The invention provides an enriching and analyzing method of circulating tumor cells of colorectal cancer. The enriching and analyzing method is characterized by comprising: acquiring a human body blood sample and separating single nuclear cells in the human body blood sample; carrying out two times of enriching on the single nuclear cells by adopting two types of manners of backward enriching the single nuclear cells through CD45 magnetic beads and forward enriching the single nuclear cells through EpCAM magnetic beads, so as to obtain cell suspension; after cracking the cells, extracting mRNA (messenger Ribonucleic Acid); detecting expression levels of CK20 and Survivin genes of the circulating tumor cells of the colorectal cancer through an RT-qPCR detection method; after comparing the expression levels of the two genes with an existing expression level database of the CK20 and Survivin genes of normal people and colorectal cancer patients, analyzing to know whether the circulating tumor cells of the colorectal cancer exist or not and an expression degree. The enriching and analyzing method is a method for detecting the circulating tumor cells in blood of the colorectal cancer patients and has the advantages of high sensitivity and specificity and low cost.
Owner:BEIJING KEXUN BIOTECH CO LTD
Method for identifying pennisetum forage grass by utilizing ISSR (Inter Simple Sequence Repeat) molecular marker technology
InactiveCN102586449AReduce demandImprove the problem of filling the good with the bad, mixing the good and the bad, and confusing the real with the fakeMicrobiological testing/measurementAnimal ForagingAgricultural science
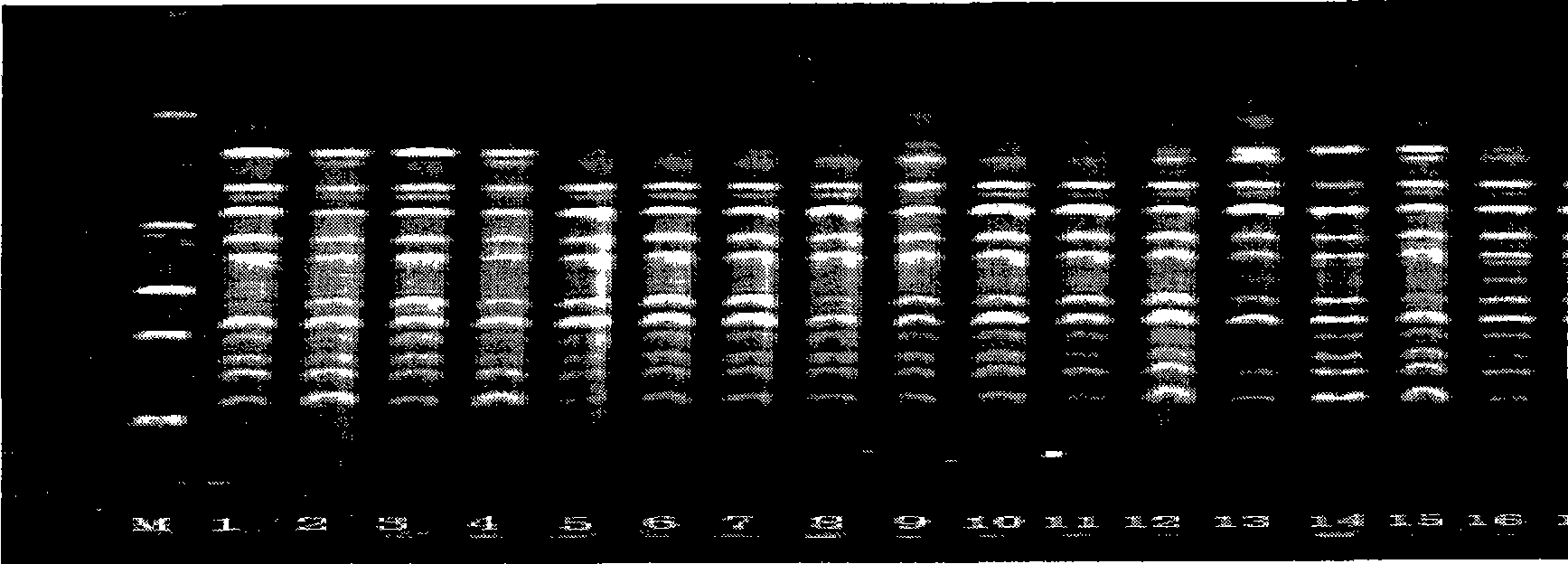
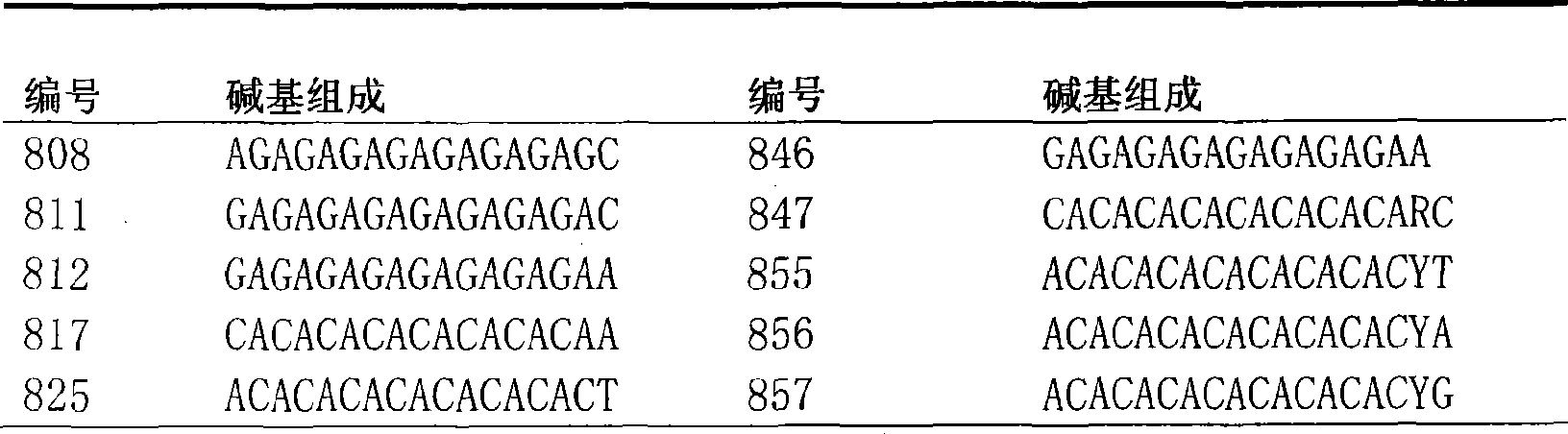
The invention belongs to the field of forage grass germplasm resource identification, in particular to a method for identifying pennisetum forage grass by utilizing an ISSR (Inter Simple Sequence Repeat) molecular marker technology, wherein primers used by ISSR-PCR (Polymerase Chain Reaction) are UBC815 and UBC835, and the nucleotide sequences of the primers are respectively UBC815-CTCTCTCTCTCTCTCTG and UBC835-AGAGAGAGAGAGAGYC. The method fills the blank of distinguishing pennisetum forage grass categories (varieties) by applying the ISSR molecular marker method, and an electrophoresis ideograph finally drawn by the method for distinguishing seven pennisetum forage grass materials for tests can be used as a basis for distinguishing and identifying the seven materials. The method gets rid of unreliable factors of traditional morphology identification and can quickly and accurately distinguish the seven pennisetum forage grass materials, and an identification result can be used as a reliable basis for approving categories, varieties and strains so as to guarantee the great popularization of germplasm on production.
Owner:CHINA AGRI UNIV
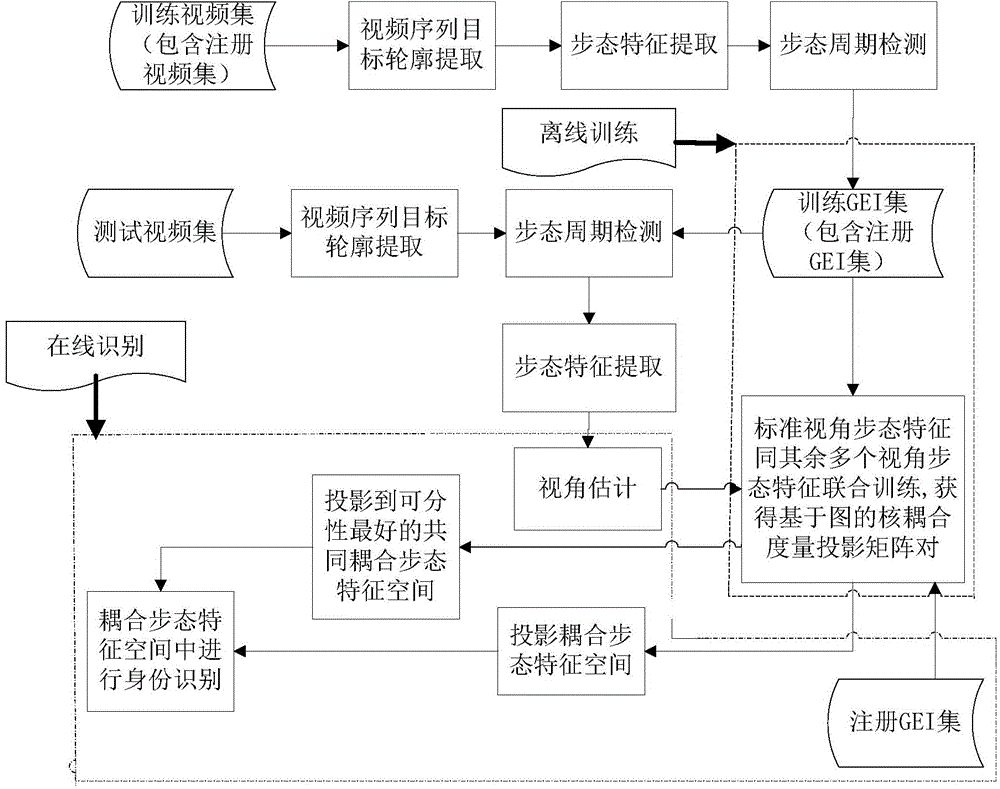
Multi-angle gait recognizing method based on semi-supervised coupling measurement of picture
ActiveCN104463099AAddress high storage requirementsValid identificationCharacter and pattern recognitionHat matrixCoupling
The invention belongs to the field of pattern recognizing, and particularly relates to a multi-angle gait recognizing method based on semi-supervised coupling measurement of a picture. The multi-angle gait recognizing method comprises the steps that a target outline sequence is obtained from video streaming through a codebook detecting method; the overall feature of a gait is extracted from a cycle through a gait energy picture; an off-line training stage of a multi-angle gait recognizing system is established, and a semi-supervised coupling projection matrix pair based on the picture is obtained through training; target outline extracting is carried out on a test video, the gait cycle is detected for the size-normalized outline sequence, the gait energy picture feature of the single cycle is generated, and the selected semi-supervised coupling projection matrix pair based on the picture is estimated through the viewing angle. The multi-angle gait recognizing method solves the problem that according to a traditional gait recognizing method, gait features of all viewing angles need to be stored, and the storage volume is high; the identity recognizing for gaits of walking of any angle is valid.
Owner:HARBIN ENG UNIV
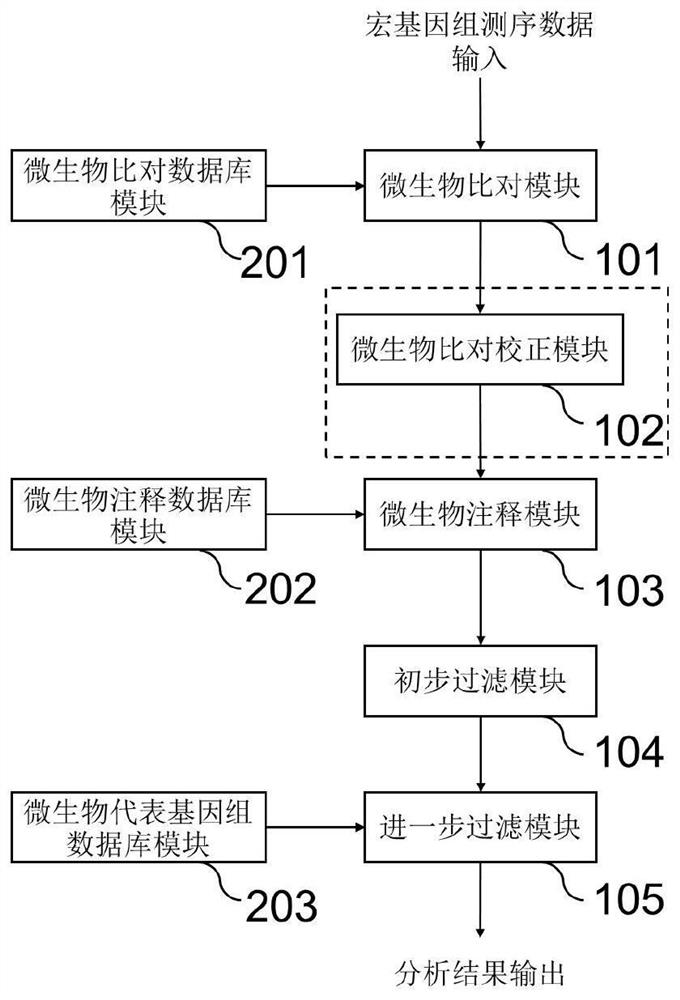
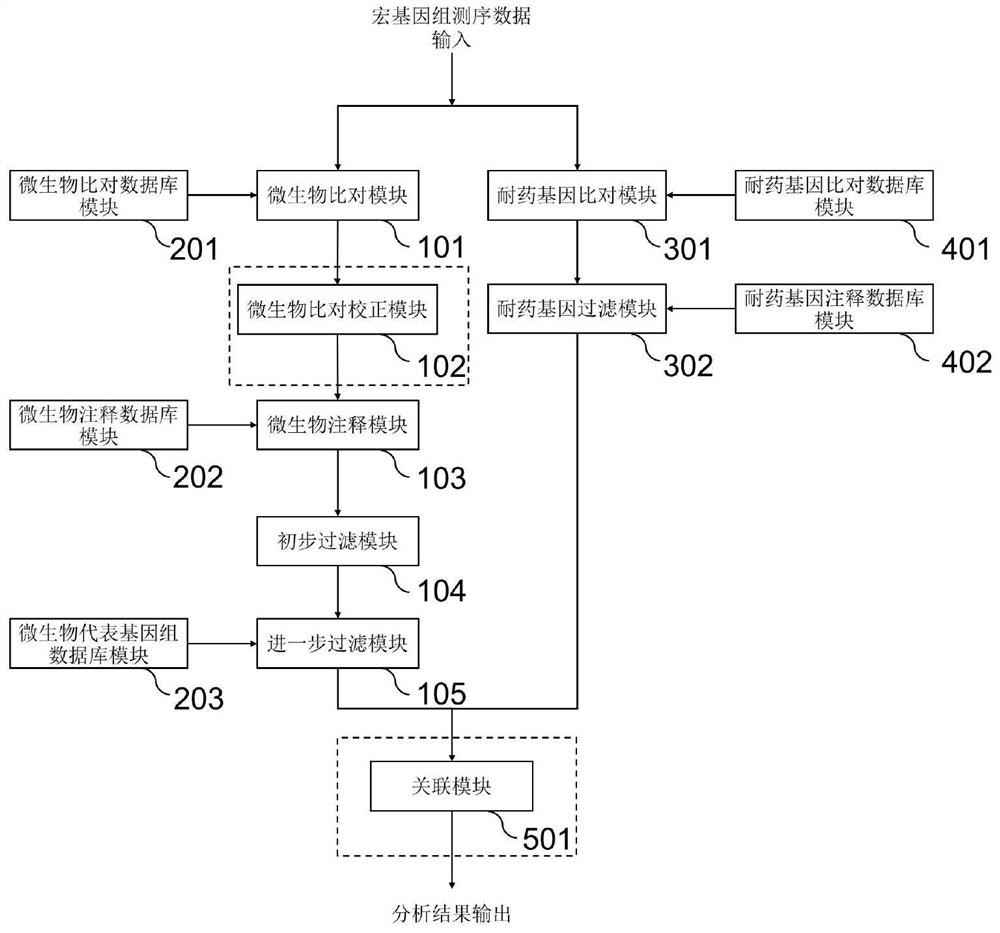
Method and system for detecting microorganisms and drug-resistant genes in sample
ActiveCN112530519AAccurate identificationAccurate detectionProteomicsGenomicsGenomic sequencingPathogenic microorganism
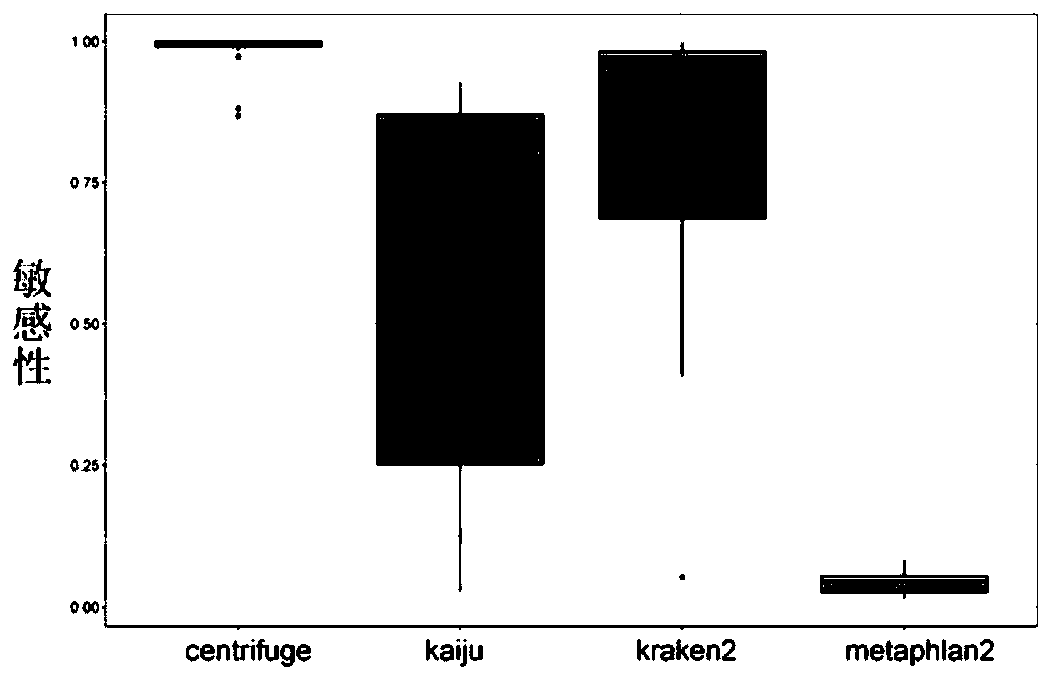
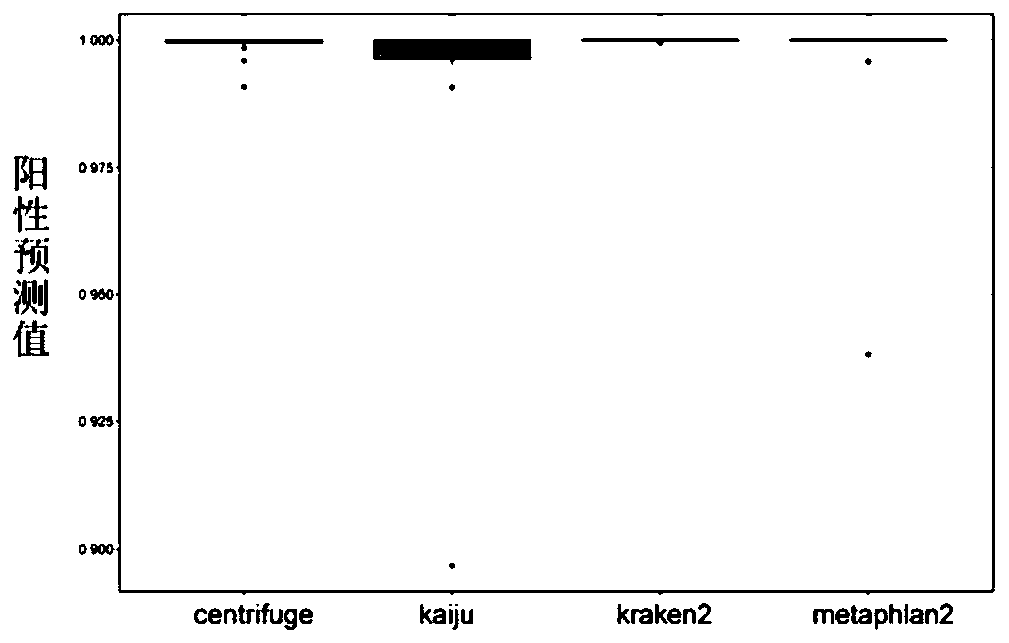
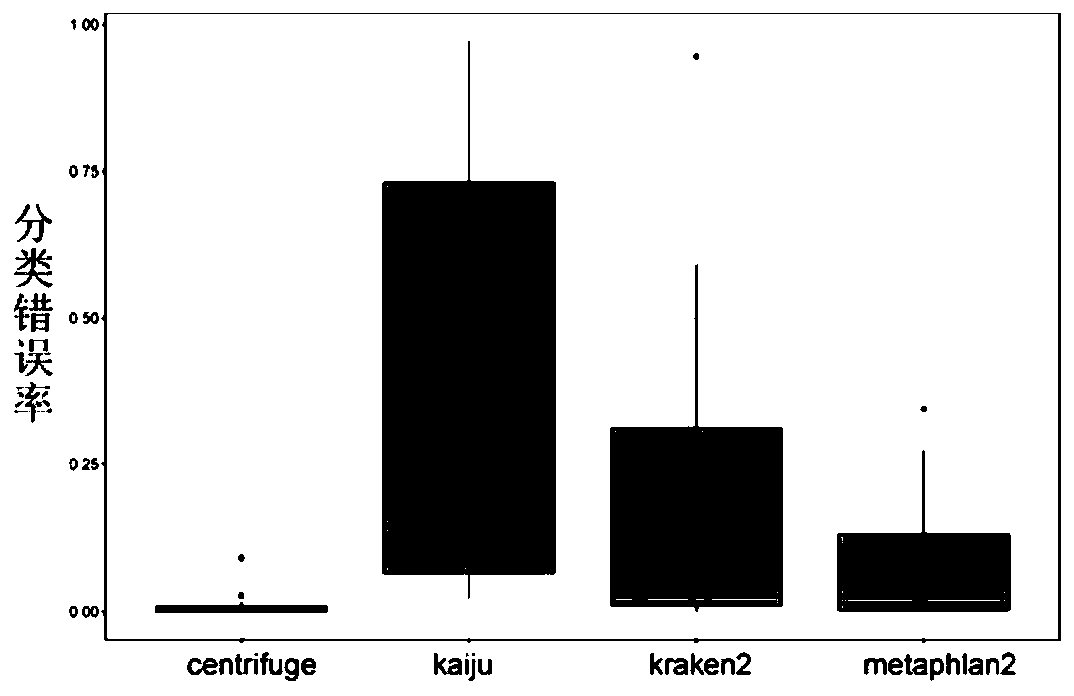
The invention discloses a method for detecting microorganisms in a sample, which belongs to the technical field of metagenome analysis and comprises the following steps: S1, acquiring metagenome sequencing data of the sample; and S2, performing species analysis on the metagenome sequencing data. The method further comprises a step of carrying out drug resistance gene analysis on the metagene sequencing data of the sample. The invention also discloses a system for detecting microorganisms and drug-resistant genes in a sample. On the basis of a comprehensive and accurate database and an intelligent analysis and screening algorithm, pathogenic microorganisms and drug-resistant genes are effectively identified by utilizing a metagenome sequencing method, false positive is effectively reduced,bacteria suspected to correspond to the detected drug-resistant genes can be prompted, and better technical support is provided for accurate diagnosis and treatment of infection; in addition, according to the method and the system, potential new pathogenic microorganisms can be accurately analyzed, and technical support is provided for early warning of new infectious diseases.
Owner:广东美格基因科技有限公司
Target bar code gene of bar codes of plants in18 species in melilotus miller and preparation method thereof
ActiveCN105603095AImprove accuracyAccurate classification and storageMicrobiological testing/measurementDNA/RNA fragmentationDNA barcodingGermplasm
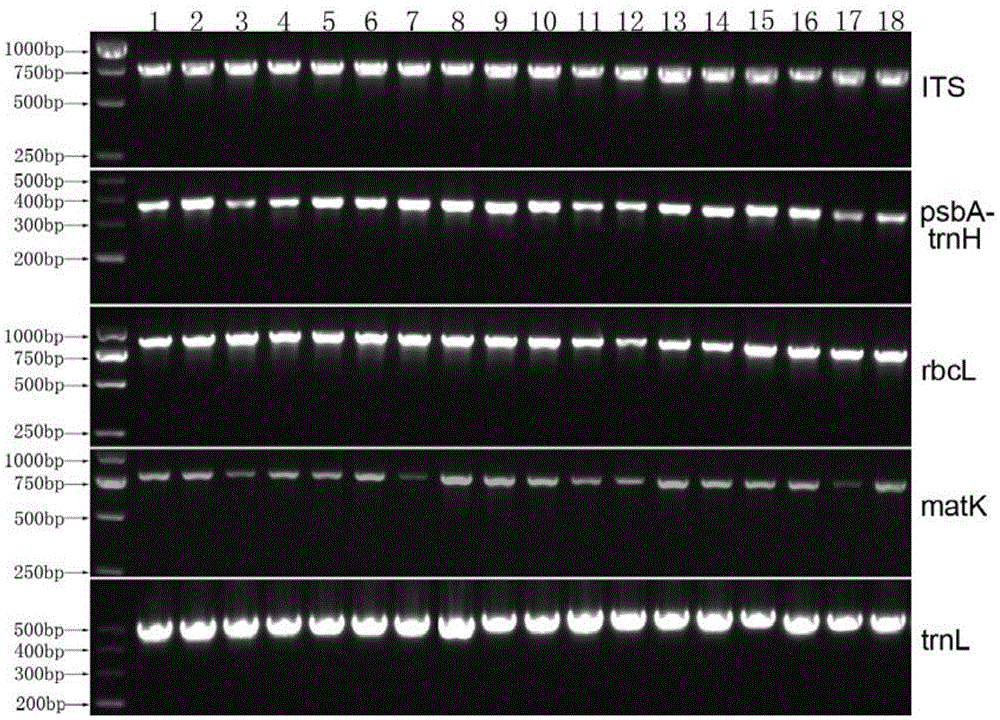
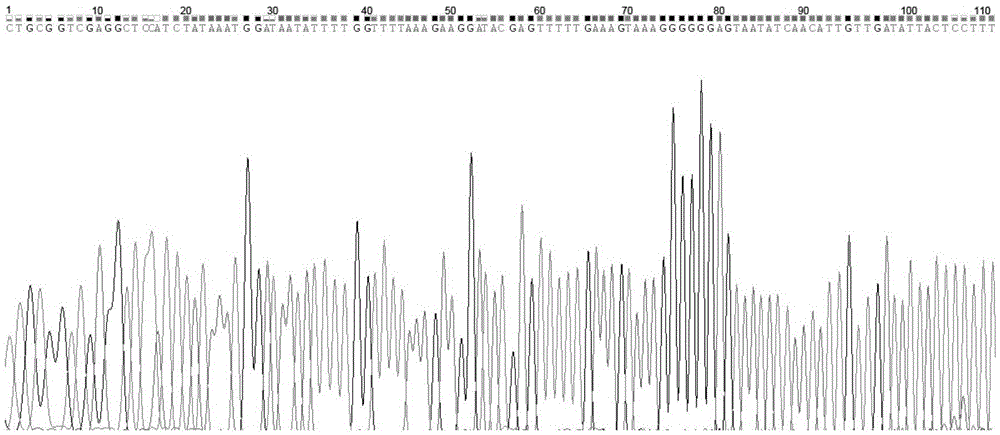
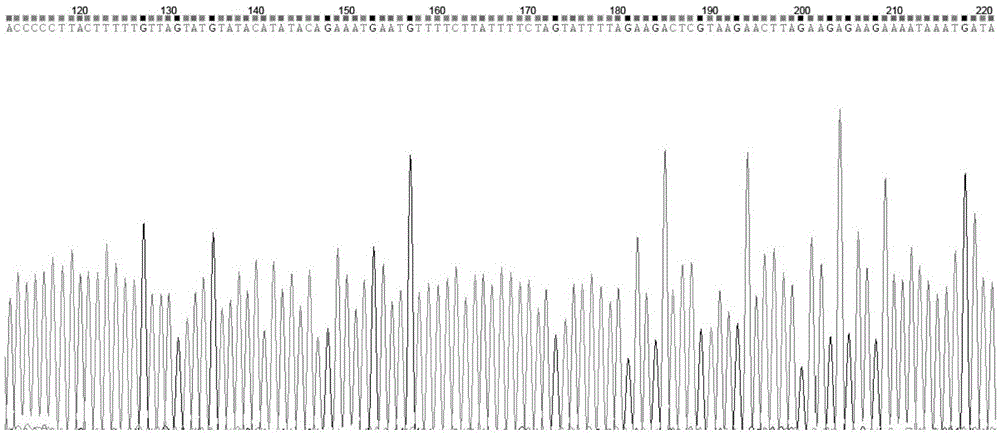
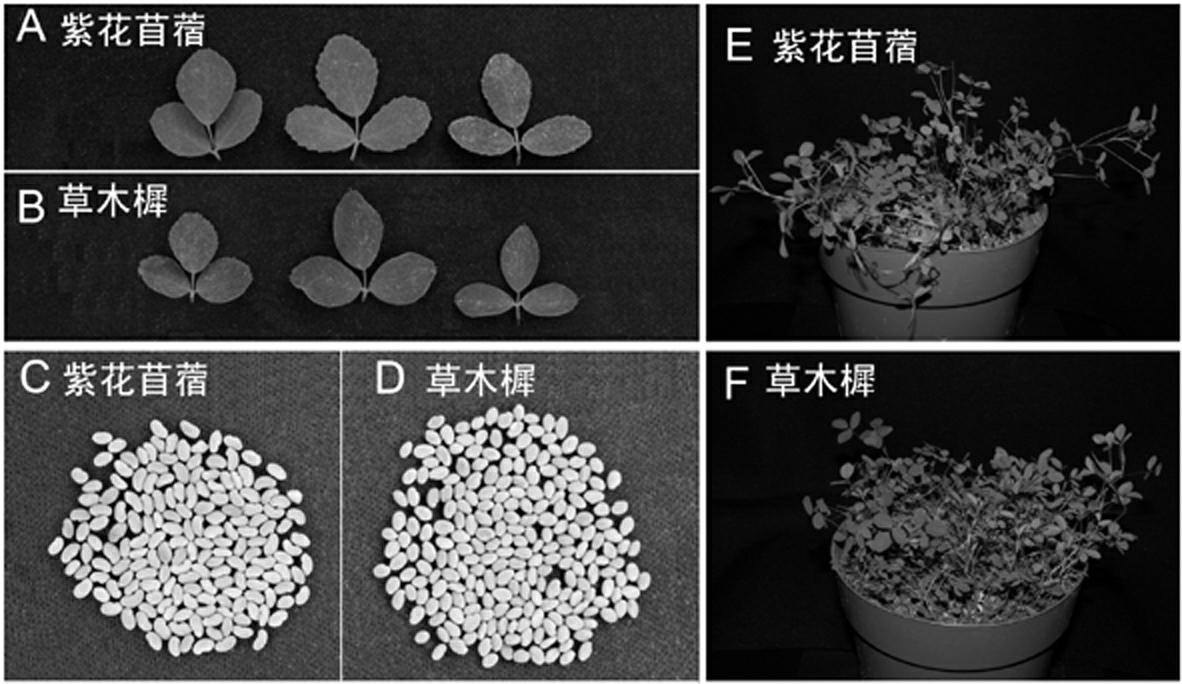
The invention relates to a technology for identifying DNA bar codes of plants in 18 species in melilotus miller and an application method thereof. Five genes of ITS, psbA-trnH, rbcL, matK and trnL are selected and integrated to serve as a target DNA bar code gene for identifying 18 species in melilotus miller. Based on data organization, data editing and analysis methods of ContigExpress, a multi-sequence alignment method of ClustalW, a Neighbor-Joining algorithm based on a minimum evolution principle and a no of difference method, a clustering chart is built, and a basic using method of the technology for identifying the DNA bar codes of plants in 18 species in melilotus miller is formed, and a standard classification bar code gene and a corresponding sequence are determined. By means of the technology, the fast, efficient and high-accuracy method for identifying the DNA bar codes of different species in melilotus miller is built, and the method has great significance in differentiation of various species in melilotus miller and utilization of germplasm resources.
Owner:LANZHOU UNIVERSITY
Kit for identifying authenticity of medicago sativa seed and detection method thereof
InactiveCN102643896ARapid identificationEfficient and accurate identificationMicrobiological testing/measurementDNA/RNA fragmentationMelilotus albusPastoralism
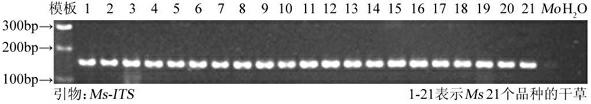
The invention mainly relates to a method for identifying authenticity of a medicago sativa seed and special primers thereof, in particular to a gene detection kit of medicago sativa and melilotus albus and a detection method thereof. The kit for identifying the authenticity of the medicago sativa seed is mainly characterized in that: the total volume of a medicago sativa PCR (Polymerase Chain Reaction) detection system is 20 mul; and the kit comprises 2*PCR Master, the special primer MsITS-F (5'-3'): TTTTTGAACGCAAGTTGCGC and the special primer MsITS-R (5'-3'): CAACCAACCACGAGACAGA and ddH2O. Detection result shows that the technology can be used for identifying the authenticity of the medicago sativa seed, has the characteristics of high sensitivity, high accuracy, high repeatability, quickness, convenience and the like, has broad application prospect in medicago sativa industrial promotion of China, can guarantee that the production of livestock husbandry of China is served by the medicago sativa well and lays a solid foundation for guaranteeing grain safety of China.
Owner:LANZHOU UNIVERSITY
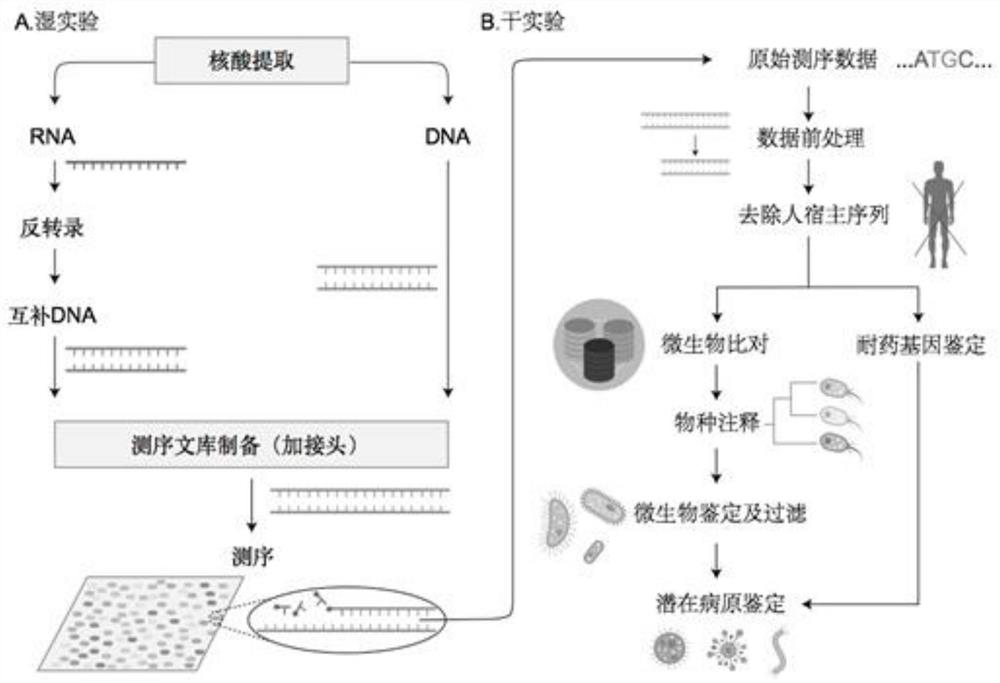
Method for detecting and identifying pathogens of children infectious diseases based on metagenomic sequencing
InactiveCN111394486AIncrease success rateImprove effectivenessMicrobiological testing/measurementSequence analysisGenomic sequencingInfective disorder
The invention relates to a method for detecting and identifying pathogens of children infectious diseases based on metagenomic sequencing. The method comprises the following specific steps: extractingtrace sample DNA after infection of a child, and carrying out concentration determination and quality control on the DNA; establishing a sequencing library of the sample DNA, and then carrying out high-throughput sequencing and screening; and comparing the treated sample sequence with a reference database to obtain a sequence with a comparison ratio of 70% or above and no multiple comparison, andthen carrying out screening of pathogenic microorganisms to complete the detection and identification method. According to the method disclosed by the invention, a DNA extraction method and a database establishment method are optimized aiming at trace samples of children, so that the success rate of database establishment of the trace samples and the effective data volume of sequencing are improved. A pathogenic microorganism screening and identifying strategy is established, laboratory and reagent pollution is effectively eliminated, and identification of pathogenic microorganisms can be effectively completed.
Owner:CHILDRENS HOSPITAL OF FUDAN UNIV
Multiple-PCR primer system for quickly testing animal origin ingredients of pigs, sheep and cows and testing method
ActiveCN105177150AShorten detection timeShorten experiment timeMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyAnimal science
The invention provides a multiple-PCR primer system for quickly testing animal origin ingredients of pigs, sheep and cows and a testing method. The testing method comprises the following steps that the total DNA of a to-be-tested sample is taken as a template, the multiple-PCR primer system composed of a first primer pair, a second primer pair and a third primer pair is utilized to perform a multiple-PCR amplification experiment, and after reacting is completed, a result is judged according to agarose gel electrophoresis; the total DNA template of the sample adopts one or multiple of a pork product, a mutton product and a beef product. According to the multiple-PCR testing method, the authenticity of the pork product, the mutton product and the beef product and whether adulteration exists or not can be quickly and highly specifically tested, the testing efficiency is greatly improved, the time is saved, the testing cost is reduced, and the testing method is particularly suitable for quick anti-counterfeiting authentication of the pork product, the mutton product and the beef product. In addition, the multiple-PCR primer system cooperates with relevant reagents to prepare a kit, use is convenient, and meanwhile possibility of industrial production and application is supplied.
Owner:SHANGHAI ACAD OF AGRI SCI
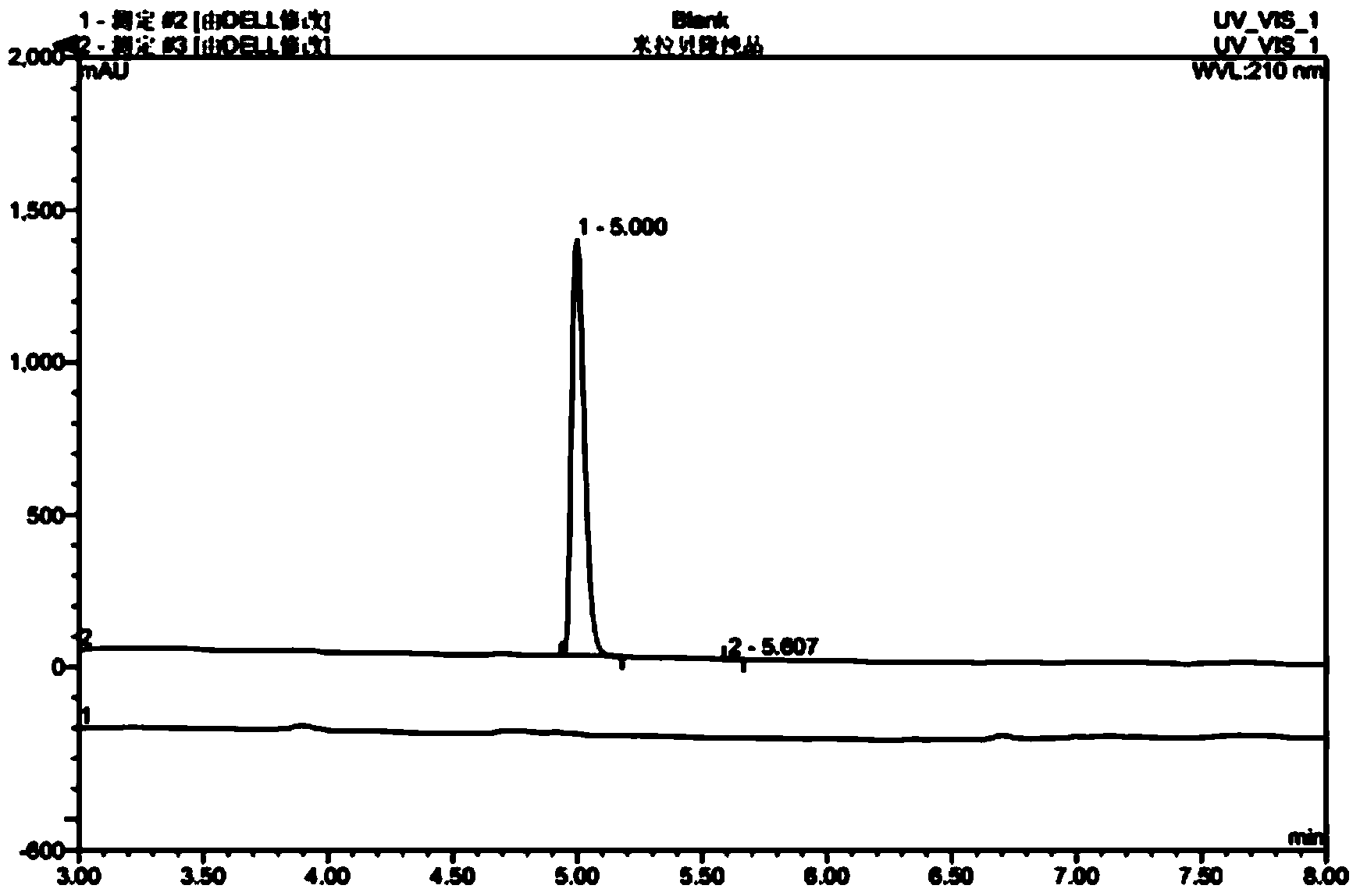
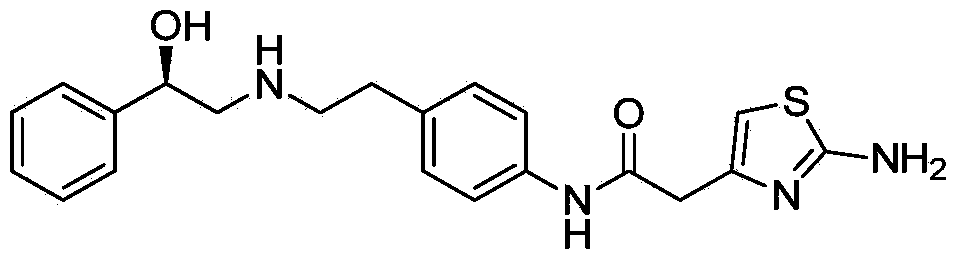
Mirabegron related substance or salt thereof, and preparation method and use thereof
The invention discloses a mirabegron related substance as shown in a formula I or a salt thereof, and a preparation method and a use thereof. The preparation method of the mirabegron related substance or the hydrochloride thereof disclosed by the invention comprises the following step: in a solvent, carrying out a condensation reaction as shown in specification between a compound as shown in a formula 6 and a compound as shown in a formula 7 or an active ester thereof under the action of a condensating agent to obtain the mirabegron related substance as shown in the formula I; or in the solvent, carrying out the condensation reaction between the hydrochloride of the compound as shown in the formula 6 and the compound as shown in the formula 7 or the active ester thereof under the action of the condensating agent to obtain the hydrochloride of the mirabegron related substance as shown in the formula I. The mirabegron related substance disclosed by the invention is capable of effectively identifying impurities generated in the synthesis of the mirabegron so as to control the medicine quality of the mirabegron.
Owner:CHINA NAT MEDICINES GUORUI PHARMA +1
Methods and means for nucleic acid sequencing
InactiveCN101460633AQuick analysisRapid identificationBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid sequencingNucleic acid sequence
The present invention provides a nucleic acid sequencing method. The method comprises enriching a nucleic acid sample for target nucleic acids, where the nucleic acid sample is enriched through at least a first round of hybridization selection and amplification, and a second round of hybridization selection and amplification. The enriched nucleic acids are in a form convenient for sequencing with the Cantaloupe sequencing technology, which employs shotgun sequencing by hybridization (SBH) of immobilized rolling circle amplicons.
Owner:GENIZON BIOSCI
Molecular identification method of Yunnan manyleaf Paris rhizome
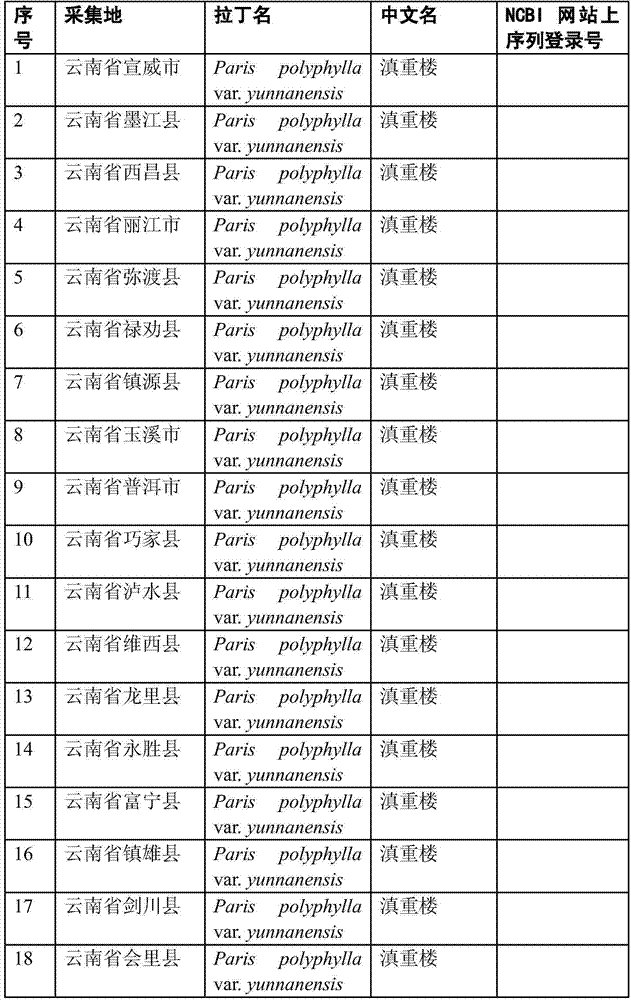
InactiveCN103484558AEnsure authenticityValid identificationMicrobiological testing/measurementMolecular identificationBarcode
The invention discloses a molecular identification method of Yunnan manyleaf Paris rhizome, belonging to the technical field of molecular markers. The method comprises the steps of performing PCR (polymerase chain reaction) on a sample to be identified to obtain an amplified product, sequencing the amplified product, then comparing with a barcode of the Yunnan manyleaf Paris rhizome, and judging that the sample to be identified is the Yunnan manyleaf Paris rhizome when the similarity between the sequence of the amplified product and a base sequence shown in the barcode of the Yunnan manyleaf Paris rhizome is more than 98.2%, the bases at sites 1-4 are CTGA, the base at the site 19 is deleted, the bases at the site 77 are all C, the bases at the site 138 are all C, the base at the site 792 is G, the bases at the sites 1110-1113 are GGCG, the base at the site 1116 is G and the bases at the sites 1119-1120 are CC. By the method disclosed by the invention, the technical problem that sensory evaluation or physical and chemical analysis and other methods are difficult to identify the authenticity of the Yunnan manyleaf Paris rhizome and different Paris plants under Paris can be overcome and the plants of the Yunnan manyleaf Paris rhizome can be effectively, simply, conveniently and quickly identified.
Owner:YUNNAN AGRICULTURAL UNIVERSITY
SNP molecular marker related to quantity of multiparity piglets of pigs, identification and application thereof
InactiveCN108893544AHigh yieldImprove stabilityMicrobiological testing/measurementBiological material analysisReference genesAllele
The invention discloses an SNP site related to the quantity of multiparity piglets of pigs. The SNP site is located on Chr7:75245594 of a pig reference genome Sscrofa11.1; the two equipotential genesof the SNP site are C and T; three genotypes corresponding to the SNP site are respectively CC, TT and TC; and compared with the pigs with genotypes of TT and TC, the pigs with genotypes of CC have higher quantity of multiparity piglets. The confirmation for the correlation between the SNP site and the quantity of multiparity piglets can effectively be used for identifying or assisting in identifying the quantity of multiparity piglets and can quicken the breeding process of the high-yield piglets.
Owner:FOSHAN UNIVERSITY
Multiplex PCR (polymerase chain reaction) primer system for synchronously detecting five animal-derived ingredients and detection method
PendingCN106148559AShorten detection timeShorten experiment timeMicrobiological testing/measurementDNA/RNA fragmentationMultiplex pcrsBiology
The invention provides a multiplex PCR (polymerase chain reaction) primer system for synchronously detecting five animal-derived ingredients and a detection method. Total DNA (deoxyribonucleic acid) of a sample to be detected is used as a template; a multiplex PCR primer system formed by a primer pair I, a primer pair II, a primer pair III, a primer pair IV and a primer pair V is used for multiplex PCR amplification tests; after the reaction is completed, the results are judged according to agarose gel electrophoresis, wherein the sample total DNA template is one or several of materials in cow, pig, sheep, chicken and duck meat products. When the multiplex PCR primer system provided by the invention is used for detecting the authenticity of the cow, pig, sheep, chicken and duck meat products and the adulteration, the detection speed is high; the specificity is high; the sensitivity is high; the detection efficiency can be greatly improved; the time is saved; in addition, the detection cost is reduced; the multiplex PCR primer system is particularly applicable to the fast anti-counterfeiting identification of cow, pig, sheep, chicken and duck meat products. In addition, the multiplex PCR primer system is matched with relevant reagents and can be made into a kit; the use is convenient; meanwhile, possibility is provided for industrial production and application.
Owner:SHANGHAI ACAD OF AGRI SCI +1
Method for molecular identification of Qinling Mountain illegal trade animal species and primer thereof
InactiveCN105441537AValid identificationCompensate for shape defectsMicrobiological testing/measurementDNA/RNA fragmentationSerowMolecular identification
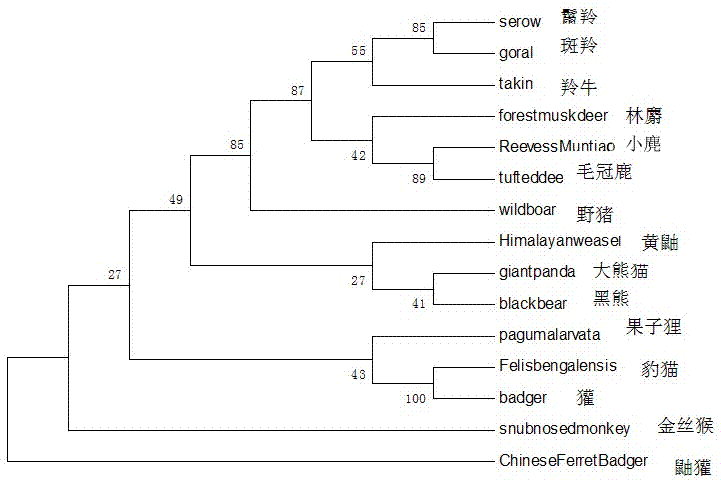
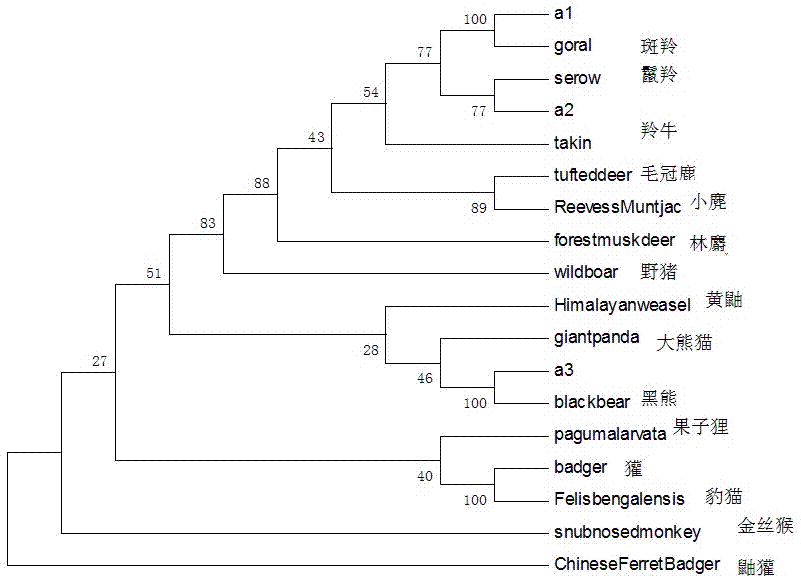
The invention relates to a method for molecular identification of Qinling Mountain illegal trade animal species and a primer thereof. At present, the conventional method that one primer is for one specie is adopted for the molecular identification of animal species, reaction conditions, reaction times and annealing temperatures of primers are all different, time and experimental materials are wasted, and the case investigating speed is influenced. According to the invention, DNA of a sample is extracted, then PCR amplification is carried out by utilizing a designed primer, PCR products are sequenced and are compared with the obtained sequences of 15 Qinling Mountain illegal trade animals, and the sample with the nearest clustering analysis relationship is namely from the Qinling Mountain illegal trade animals. The method and primer which are provided by the invention can be applied to the molecular identification of 15 Qinling Mountain illegal trade animal species in total such as takin, forest musk deer, goral, serow, elaphodus cephalophus, muntiacus reevesi, black bear, paguma larvata, leopard cat, wild boar, giant panda, snub-nosed monkey, yellow weasel, ferretbadger and badger, identification can be completed by virtue of only one primer, one PCR system and one experiment, the detection flow is simplified, the experimental period is effectively shortened, and the experimental cost is reduced.
Owner:SHAANXI INST OF ZOOLOGY NORTHWEST INSTOF ENDANGERED ZOOLOGICAL SPECIES
Mixed gas identification method based on convolutional neural network
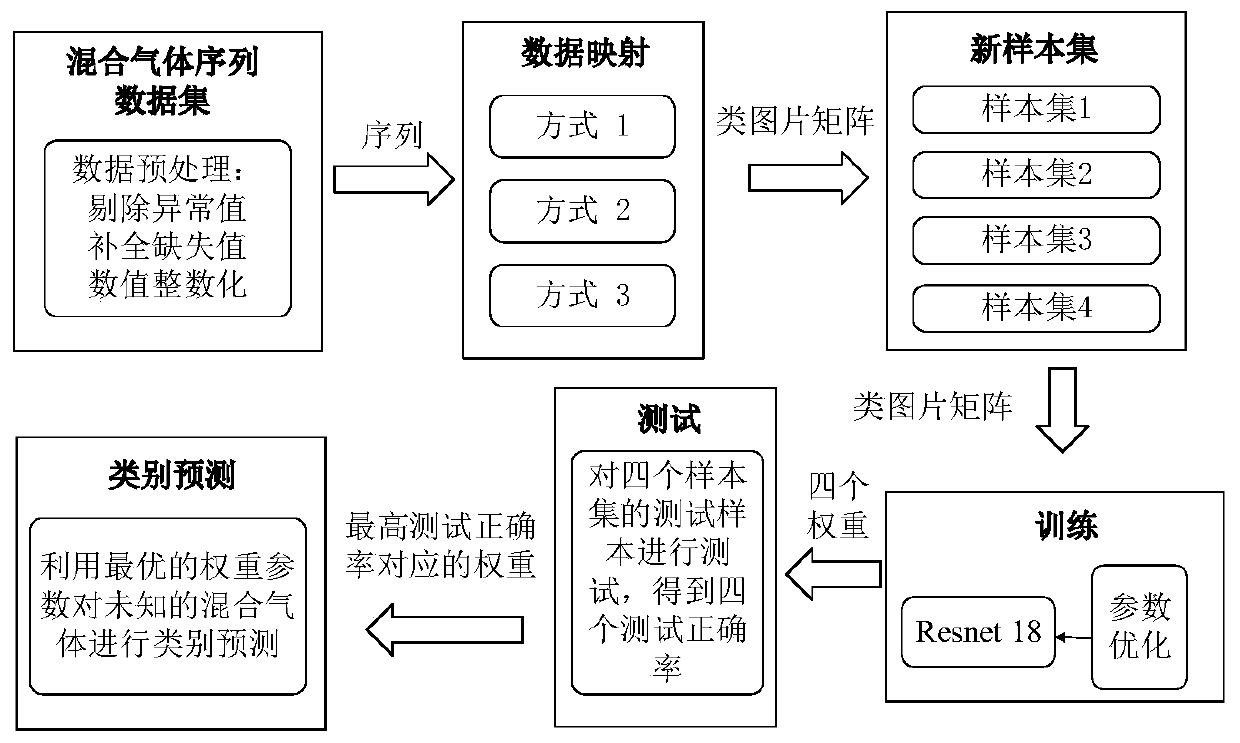
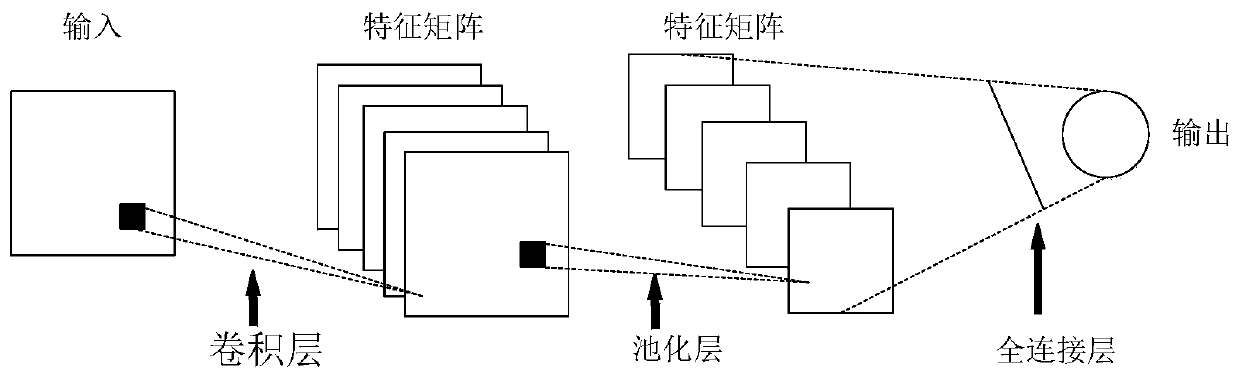
ActiveCN110309867AAccelerateImprove accuracyCharacter and pattern recognitionNeural architecturesConvolutionTime sequence
The invention discloses a mixed gas recognition method based on a convolutional neural network, and the method comprises the steps of mapping the original gas time sequence data obtained through a sensor into a similar image matrix in different modes, and carrying out the feature extraction and classification through a convolutional neural network model CNN, thereby achieving the classification ofthe mixed gas. The method is based on the classification advantages of the convolutional neural network, is applied to the classification field of the mixed gas of the time sequence, extracts more comprehensive features of the matrix data by using the convolution operation of the CNN, is high in speed, and also can obtain the higher accuracy. According to the method, the problem that in an existing mixed gas classification technology, due to the limitation of the input data, the VGG, Google-Net and other CNN networks for the image classification cannot be directly applied to classify the mixed gas data, can be solved.
Owner:BEIJING TECHNOLOGY AND BUSINESS UNIVERSITY +1
Method for identifying waste oil by using Caenorhabditis elegans dormancy as biological marker
InactiveCN102586387AValid identificationLow requirements for experimental conditionsMicrobiological testing/measurementEscherichia coliDistilled water
The invention relates to a method for identifying waste oil by using Caenorhabditis elegans dormancy as a biological marker, and relates to a waste oil identifying method. The method of the invention solves the problems of low sensitivity, poor accuracy, complex operation and high cost existed in the current waste oil identification method. The method comprises the following steps: 1, uniformly mixing NaCl, agar powder, peptone, a CaCl2 aqueous solution, a MgSO4 aqueous solution, a phosphate buffer and distilled water to obtain a nutrient solution, then sterilizing, and drying to obtain a medium; and 2) coating escherichia coli OP50 bacterial liquid in a prepared medium in the step 1), culturing for certain time, and culturing Caenorhabditis elegans larva for 46 hours. The method has the following advantages that 1, effective identification of high refining waste oil with low specific component trace can be realized; and 2, the operation is convenient, the cost is low, the method is easy to train and grasp. The method of the invention is mainly used for identifying the waste oil.
Owner:NORTHEAST INST OF GEOGRAPHY & AGRIECOLOGY C A S
Co-segregation SSR (Simple Sequence Repeat) codominant molecular marker of onion male sterility gene Ms site and application thereof
InactiveCN102586239AValid identificationOvercome deficienciesMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCo segregation
The invention discloses a co-segregation SSR (Simple Sequence Repeat) codominant molecular marker WH-SSR-1 of an onion male sterility gene Ms site. Nucleotide sequences of primers used for developing the marker are shown as SEQ ID NO. 3 and SEQ ID NO. 4, and nucleotide sequences of primers used for detecting an onion sample by applying the marker are shown as SEQ ID NO. 1 and SEQ ID NO. 2. The co-segregation SSR codominant molecular marker of the onion male sterility gene Ms site and the primers can be used for identifying the genotype of the onion male sterility gene Ms site by the followingsteps of: extracting leaf total DNA of the onion sample to be detected, amplifying with the primers through PCR, and detecting the size of the amplified product. By adopting the molecular marker provided by the invention, defects of the traditional molecular marker are overcome, a fussy screening process in the traditional method is avoided, and 4-6 years for crossing and test crossing as well asa great deal of manpower and material resource consumption in the process can be saved, therefore, the operation process is simpler and more efficient.
Owner:VEGETABLE RES INST OF SHANDONG ACADEMY OF AGRI SCI
PCR identification method of Chinese medicament turtle-derived component
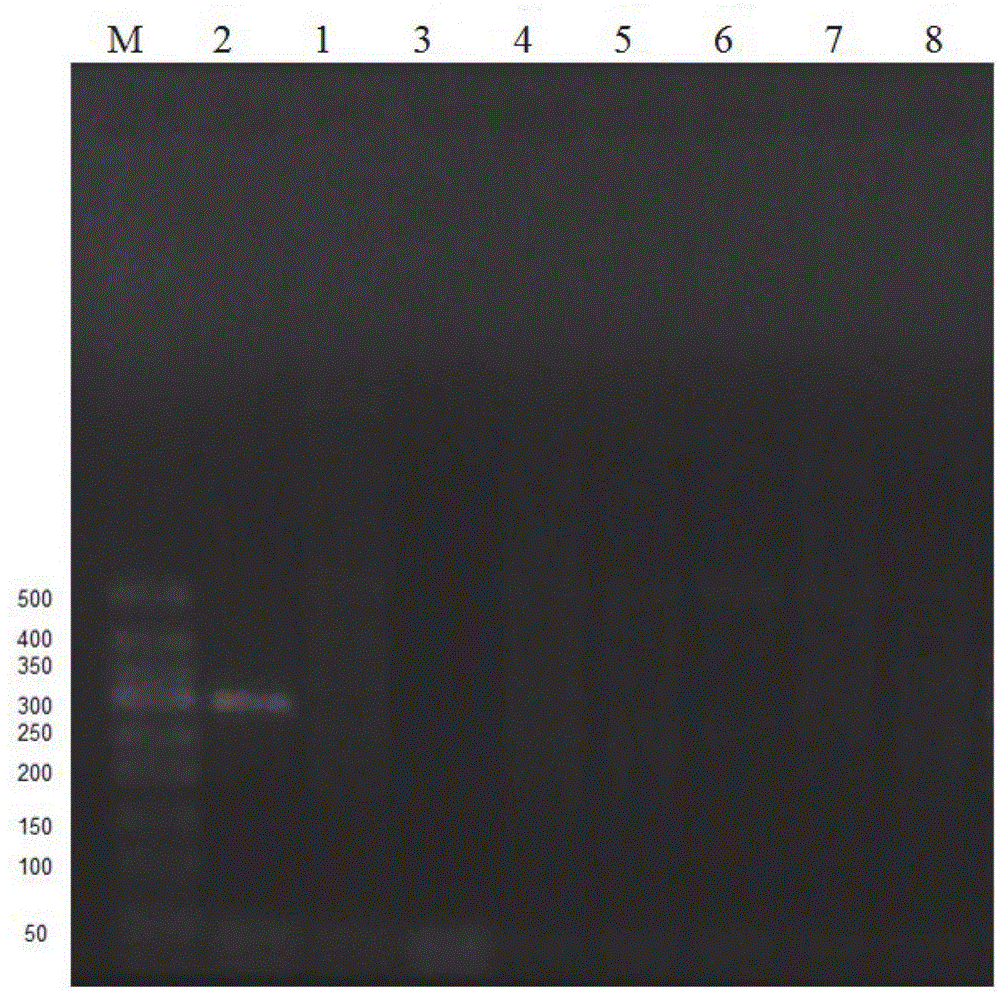
InactiveCN101838684ASensitive and fast identificationValid identificationMicrobiological testing/measurementElectrophoresisDna amplification
The invention provides a PCR identification method of a Chinese medicament turtle-derived component, which comprises the following steps: a, extracting DNA in a Chinese medicament; b, performing the PCR amplification of the DNA by using a specific primer pair designed according to the SINE sequence of a Cryptodira genome to obtain a amplified product; and c, subjecting the amplified product to electrophoresis, and judging if the DNA has an amplified positive amplified band according to the result of the electrophoresis, wherein if the DNA has the amplified positive amplified band, the Chinese medicament is derived from Cryptodira animals and has a turtle-derived component, and if the DNA has no amplified positive amplified band, the Chinese medicament is not derived from the Cryptodira animals and does not contain any turtle-derived component. The method of the invention is simple, flexible ad quick in operation and low in cost, can effectively identify turtle-derived products, particularly deeply processed tortoise-shell glue which has a very small amount of DNA and extremely short fragments due to deep processing. The method is suitable to be promoted and used in a large scale.
Owner:EAST CHINA UNIV OF SCI & TECH +1
PCR-RFLP method for rapidly identifying radix tetrastigme and various counterfeits and adulterants of radix tetrastigme
ActiveCN104611424ARapid identificationImprove accuracyMicrobiological testing/measurementMedicinal herbsEnzyme digestion
The invention belongs to the field of molecular markers and discloses a PCR-RFLP method for rapidly identifying radix tetrastigme and various counterfeits and adulterants of the radix tetrastigme. The method comprises the following steps: 1, extracting the NDA of the medicinal material (namely the radix tetrastigme); 2, carrying out PCR amplification by forward and reverse primers of internal transcribed spacer ITS2 sequences of a pair of amplified ribosomal DNAs; 3, digesting the PCR product by restriction enzyme NCO I; 4, carrying out agarose gel electrophoresis analysis. After the enzyme digestion of amplified products of the DNA of the radix tetrastigme, two DNA fragments with sizes about 335 bp and 200 bp are generated, and the various counterfeits and adulterants are not identified by the NCO I enzyme. By utilizing the differences between the DNA sequences of radix tetrastigme and the counterfeits and adulterants of the radix tetrastigme, a quick, convenient and reliable PCR-RFLP identification method is established and used for identifying whether counterfeits and adulterants are mixed in the radix tetrastigme or not, so that the technical problem that the truth and false cannot be identified by sensory or physical and chemical analysis methods is solved, and the medication safety of the radix tetrastigme is ensured.
Owner:ZHEJIANG PHARMA COLLEGE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com