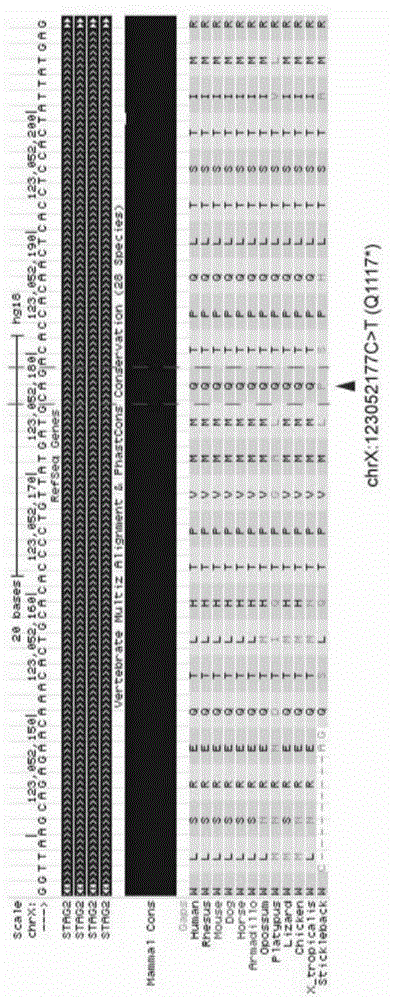
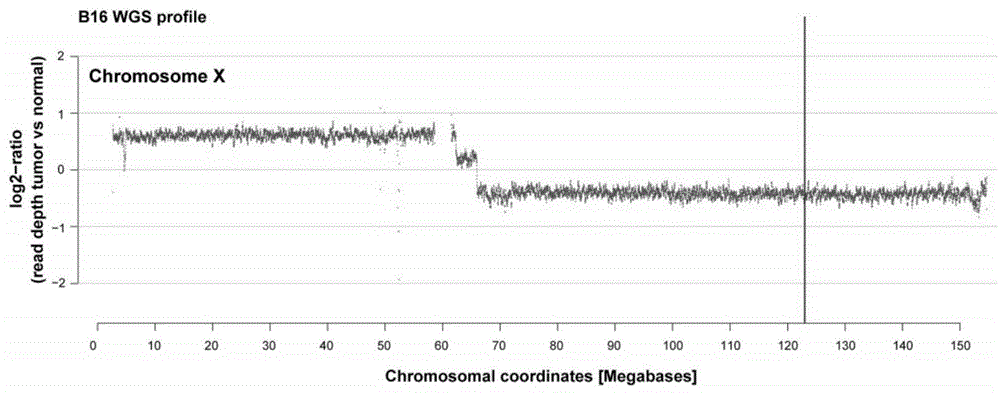
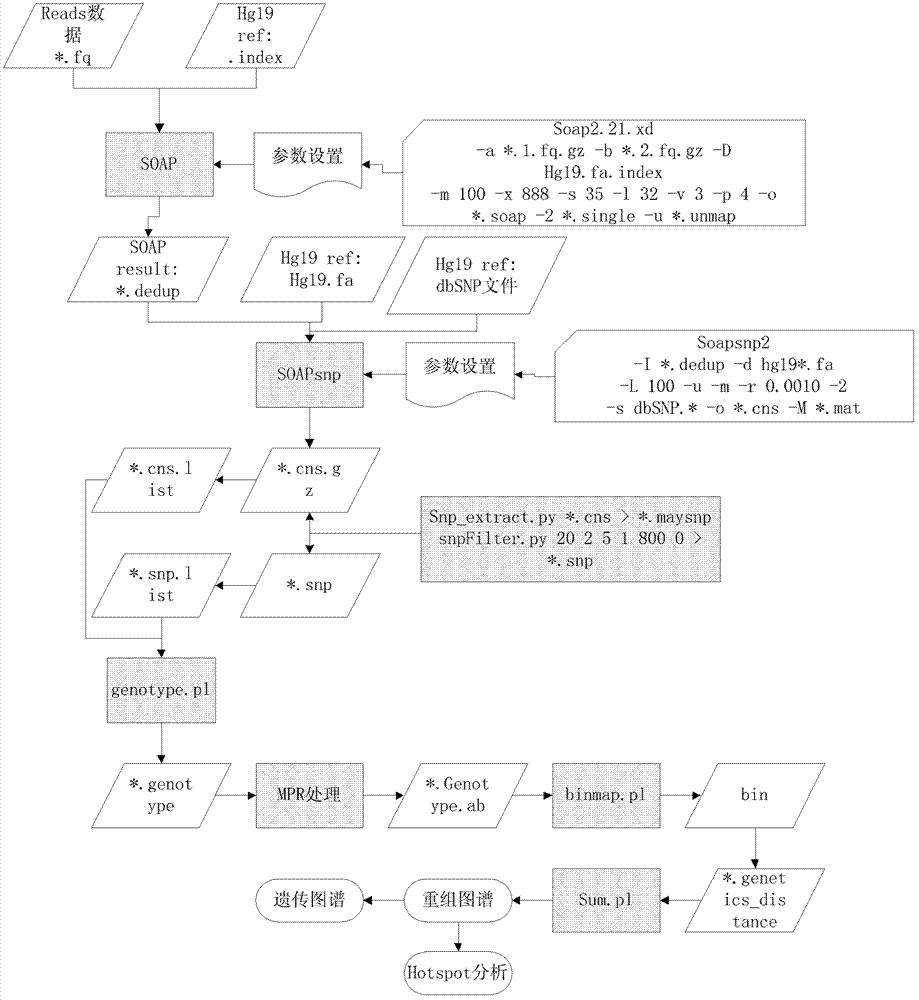
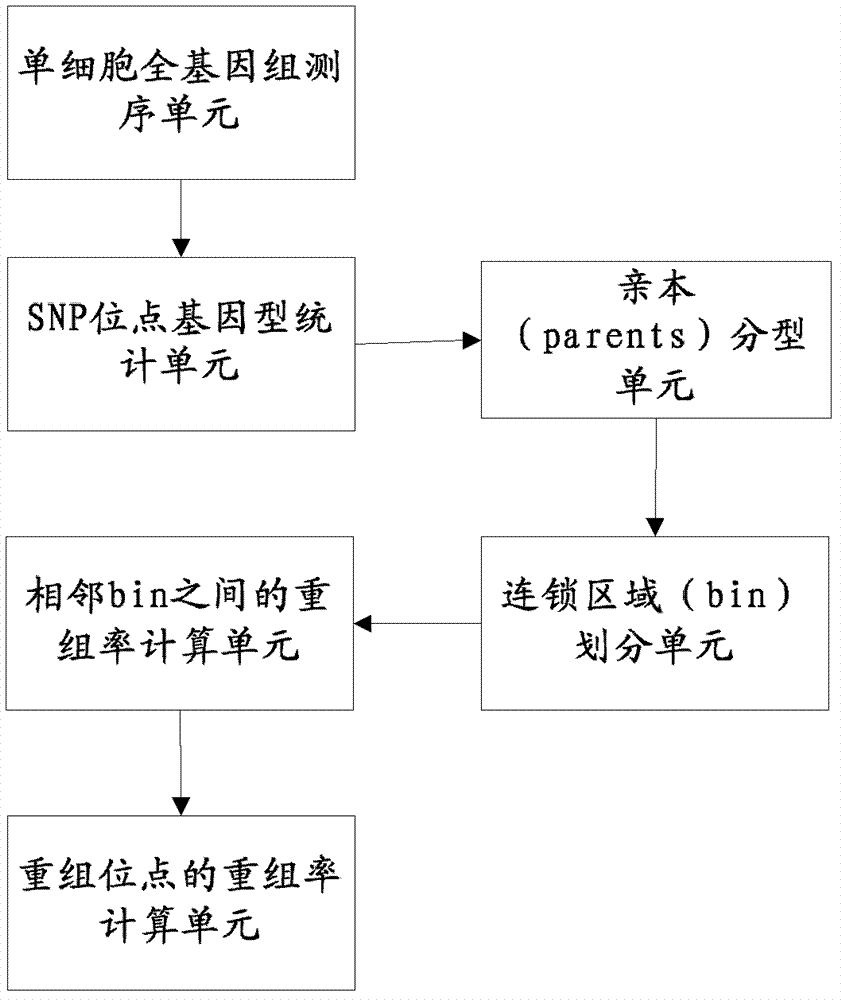
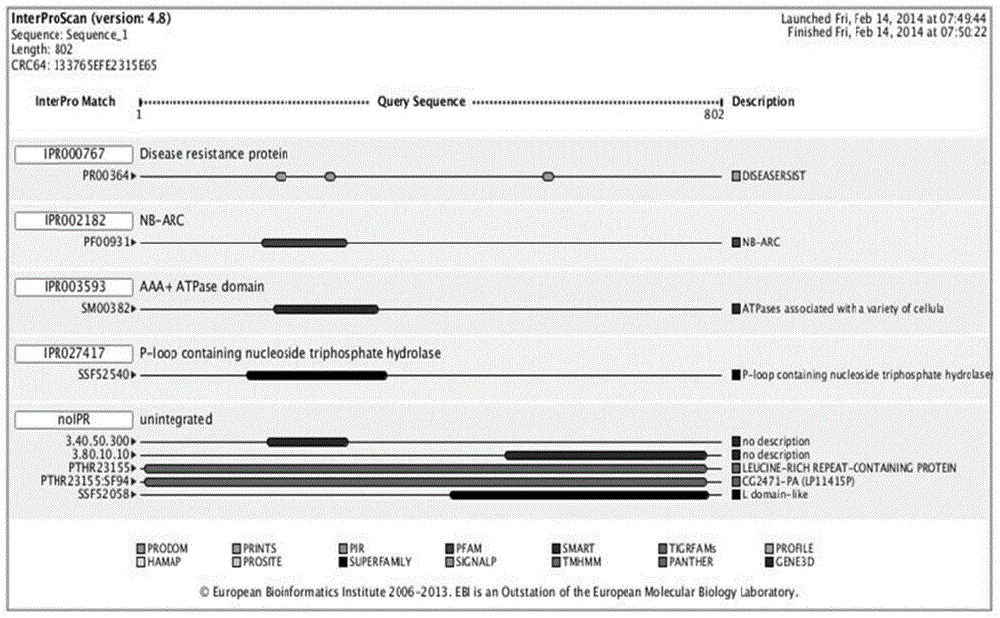
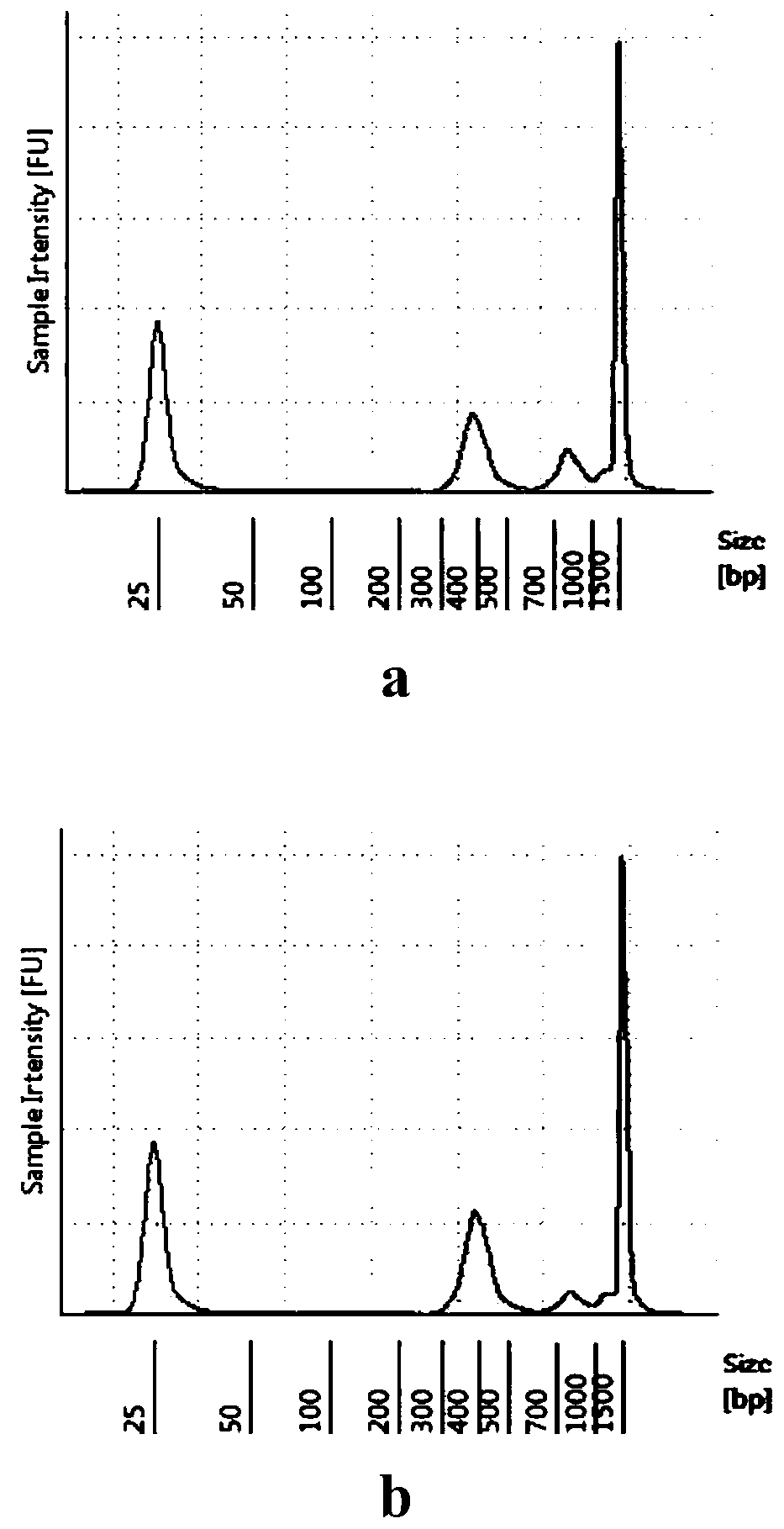
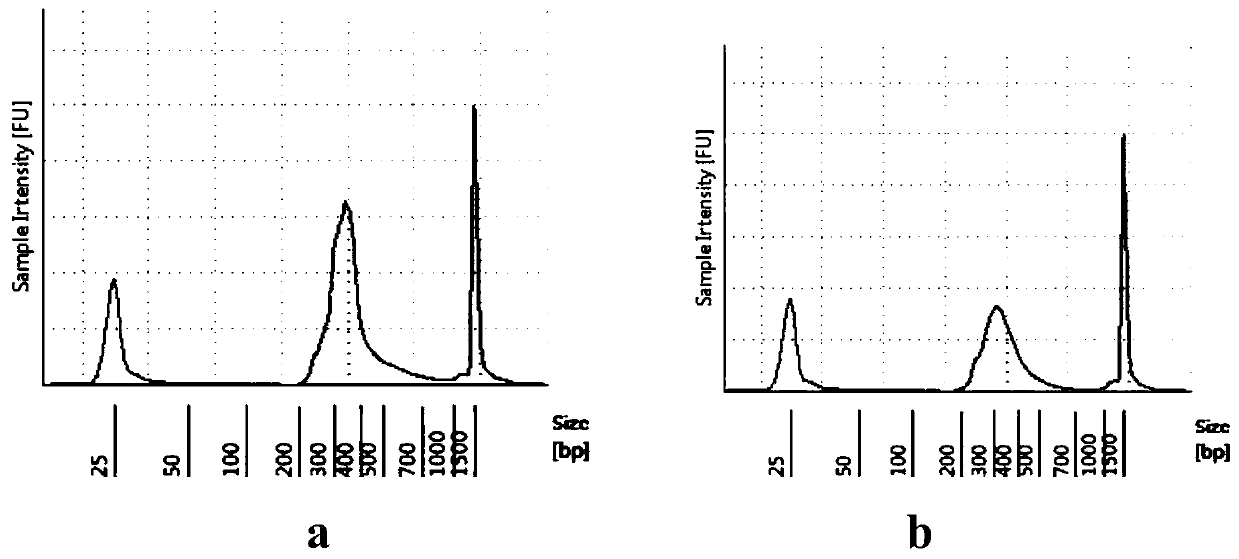
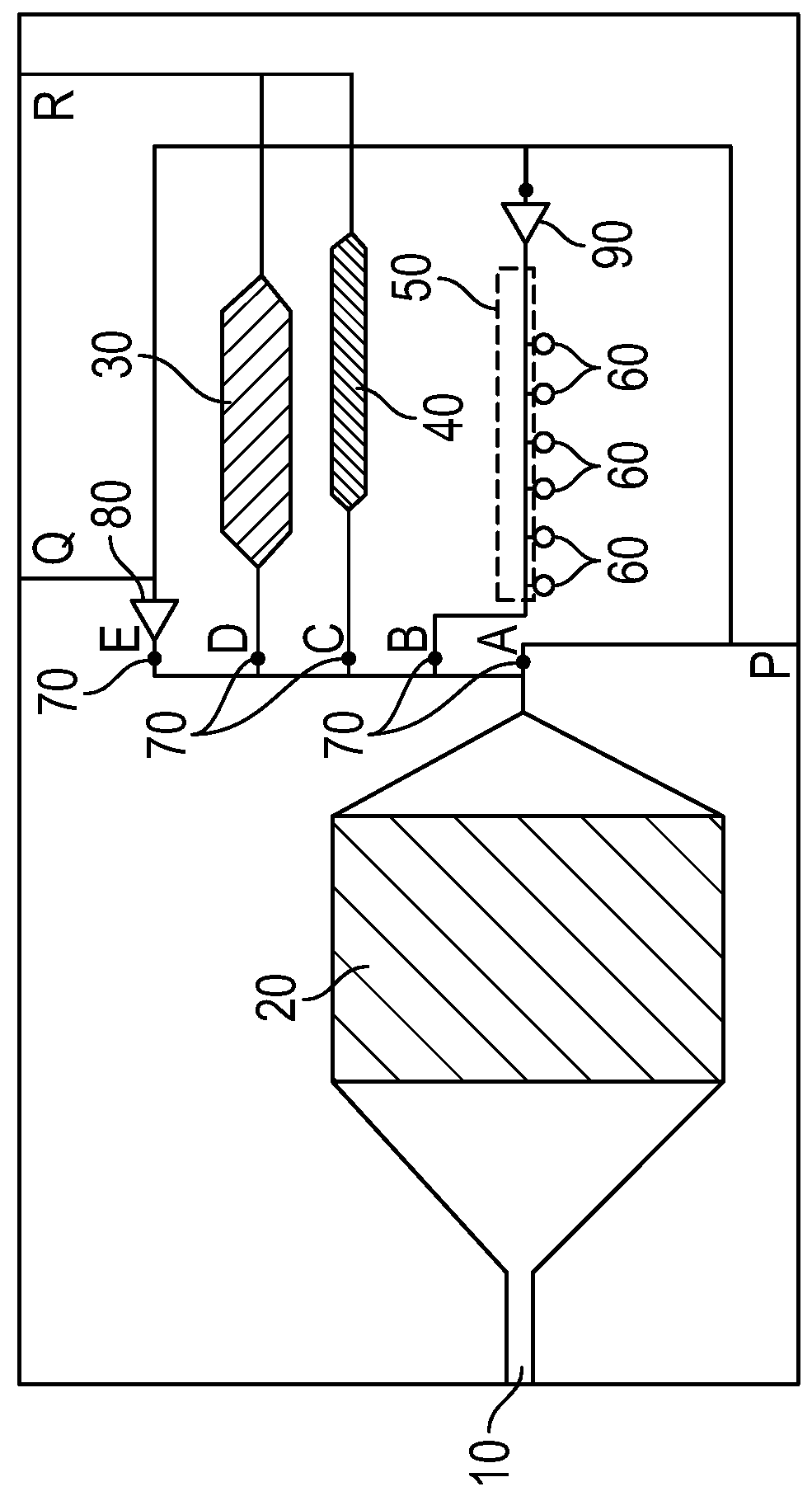
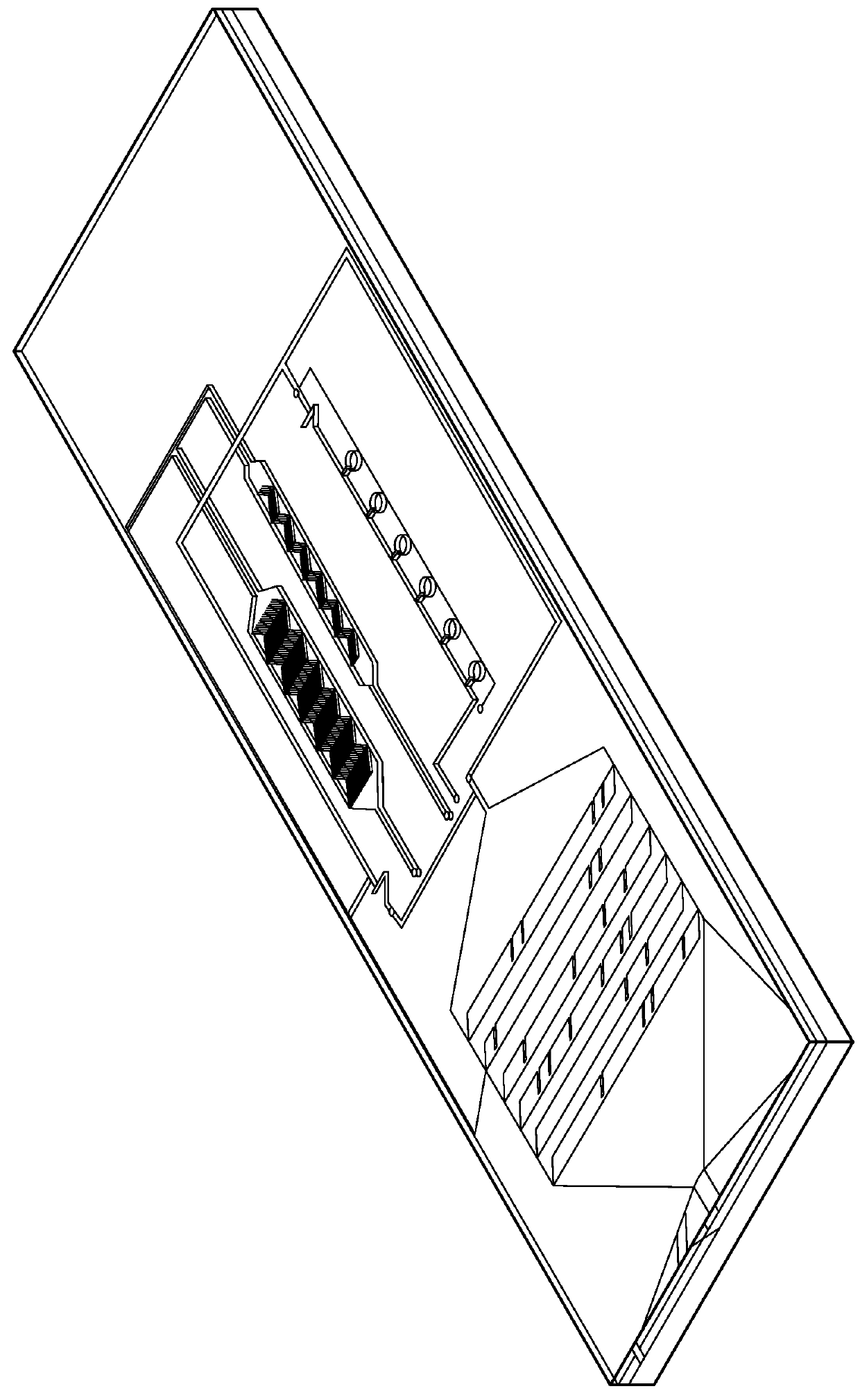
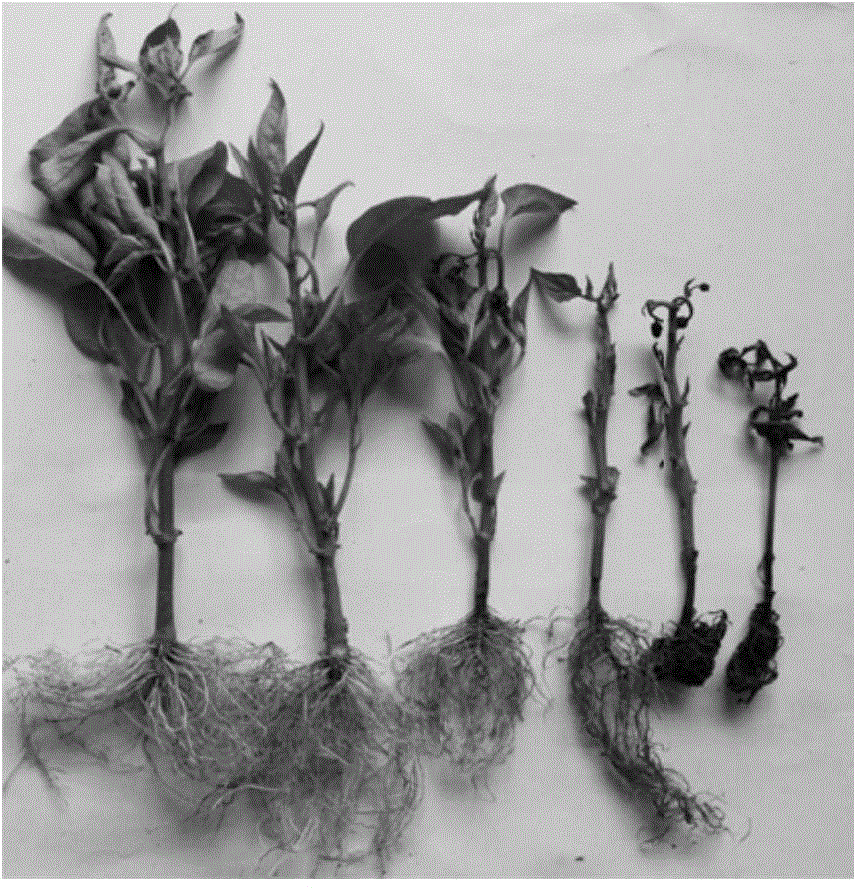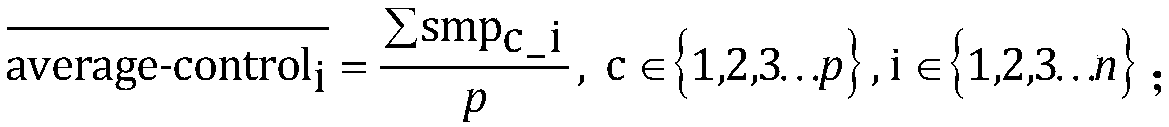Patents
Literature
340 results about "Whole genome sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Whole genome sequencing (also known as WGS, full genome sequencing, complete genome sequencing, or entire genome sequencing) is ostensibly the process of determining the complete DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast. In practice, genome sequences that are nearly complete are also called whole genome sequences.
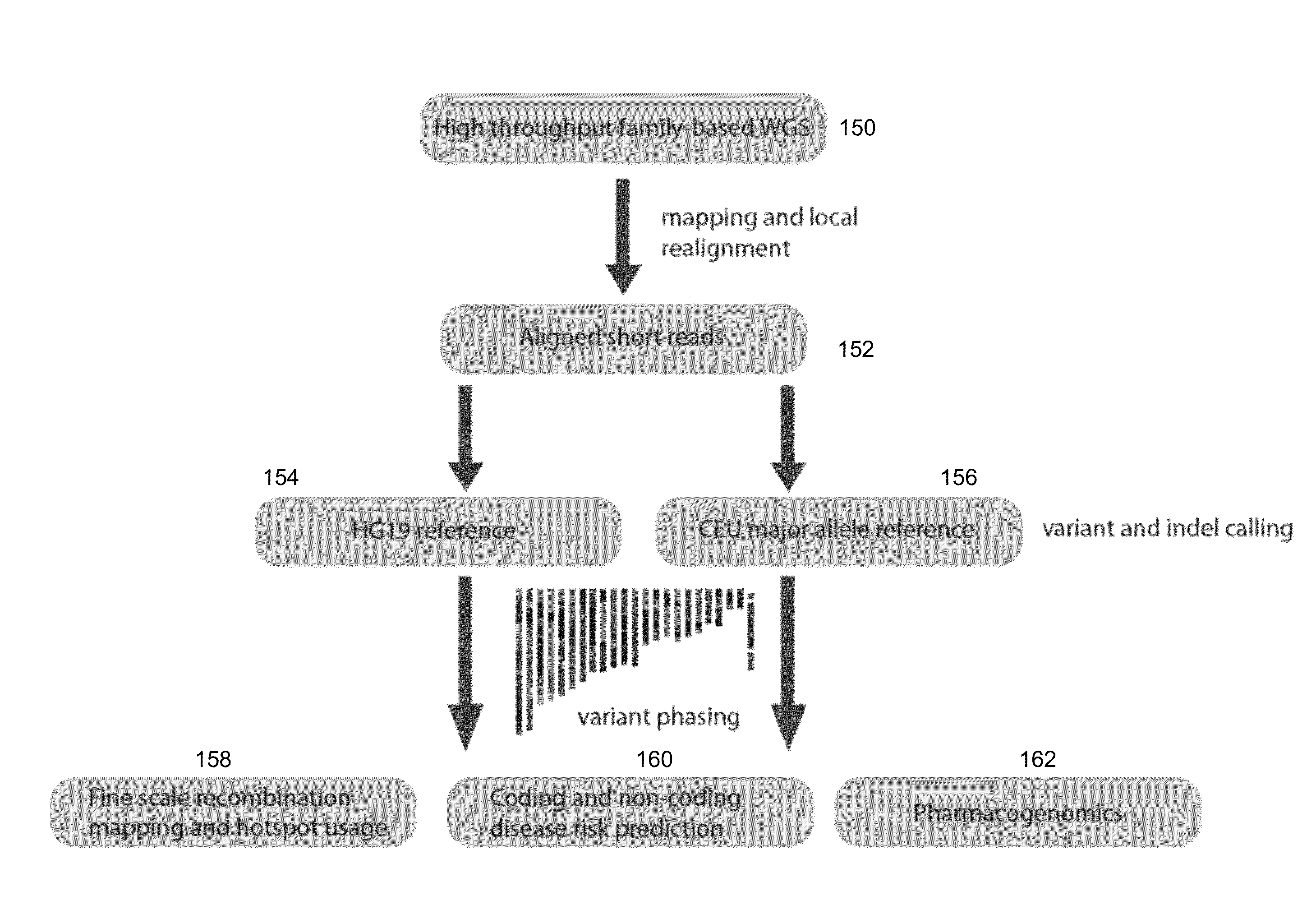
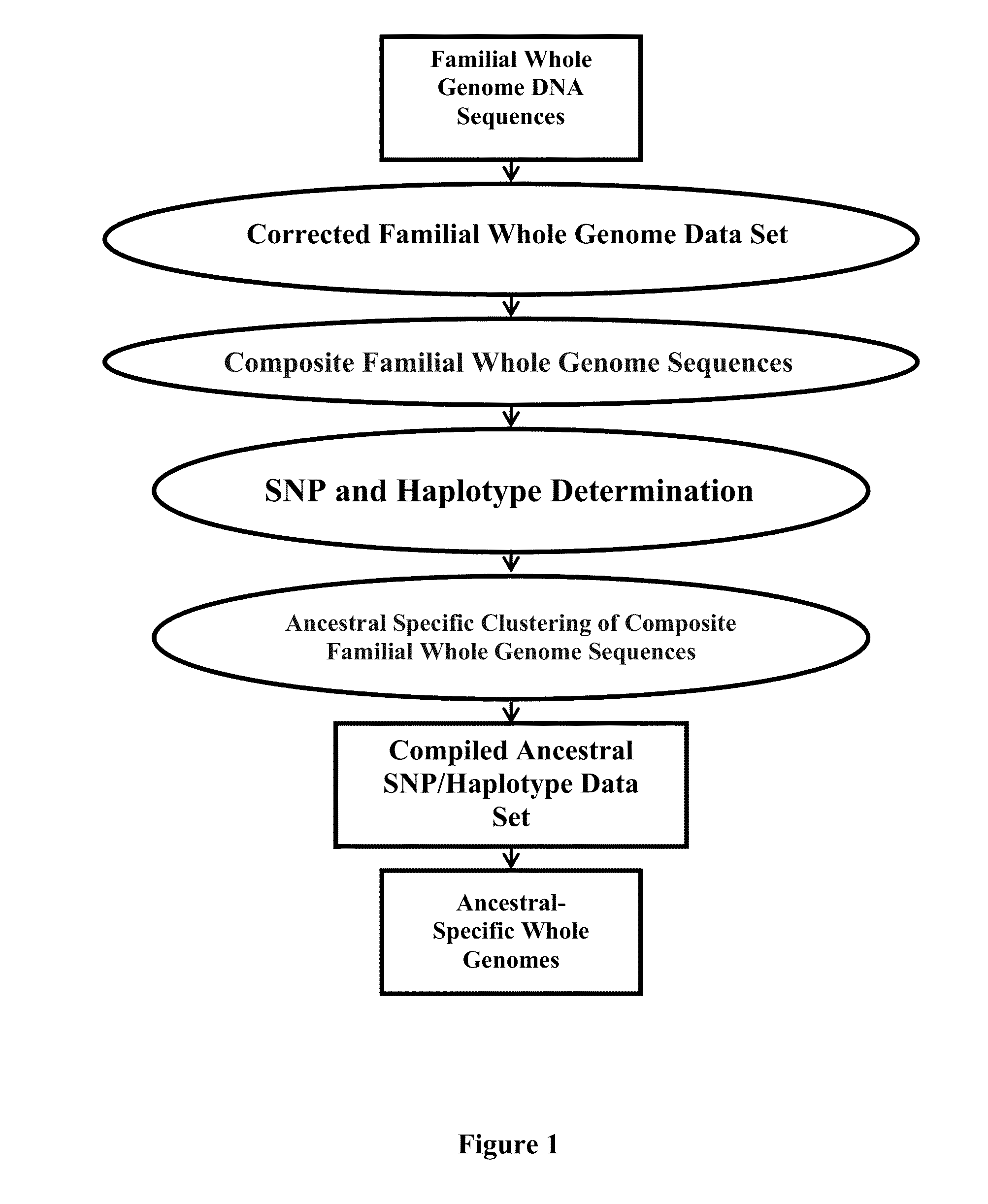
Phased Whole Genome Genetic Risk In A Family Quartet
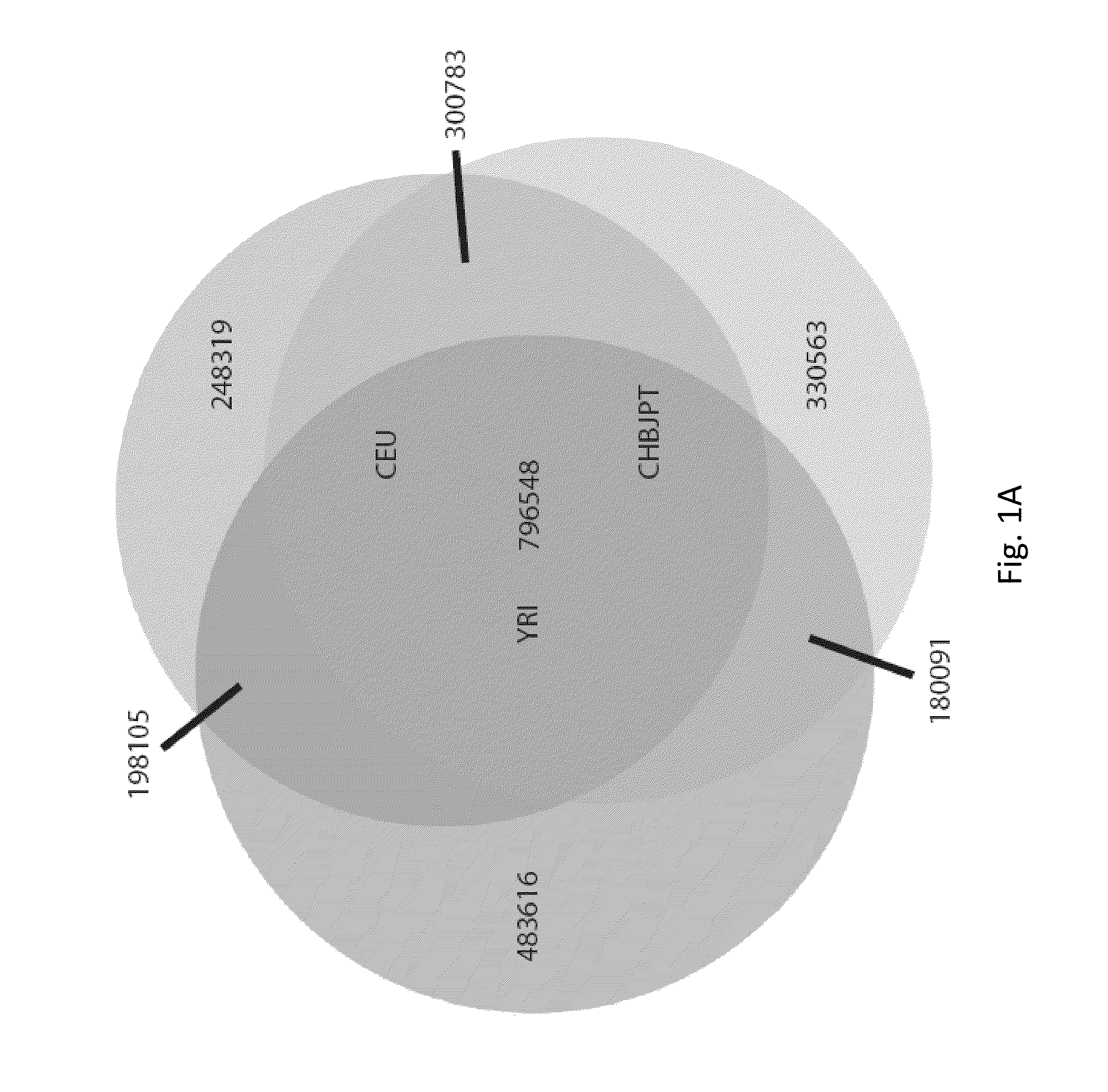
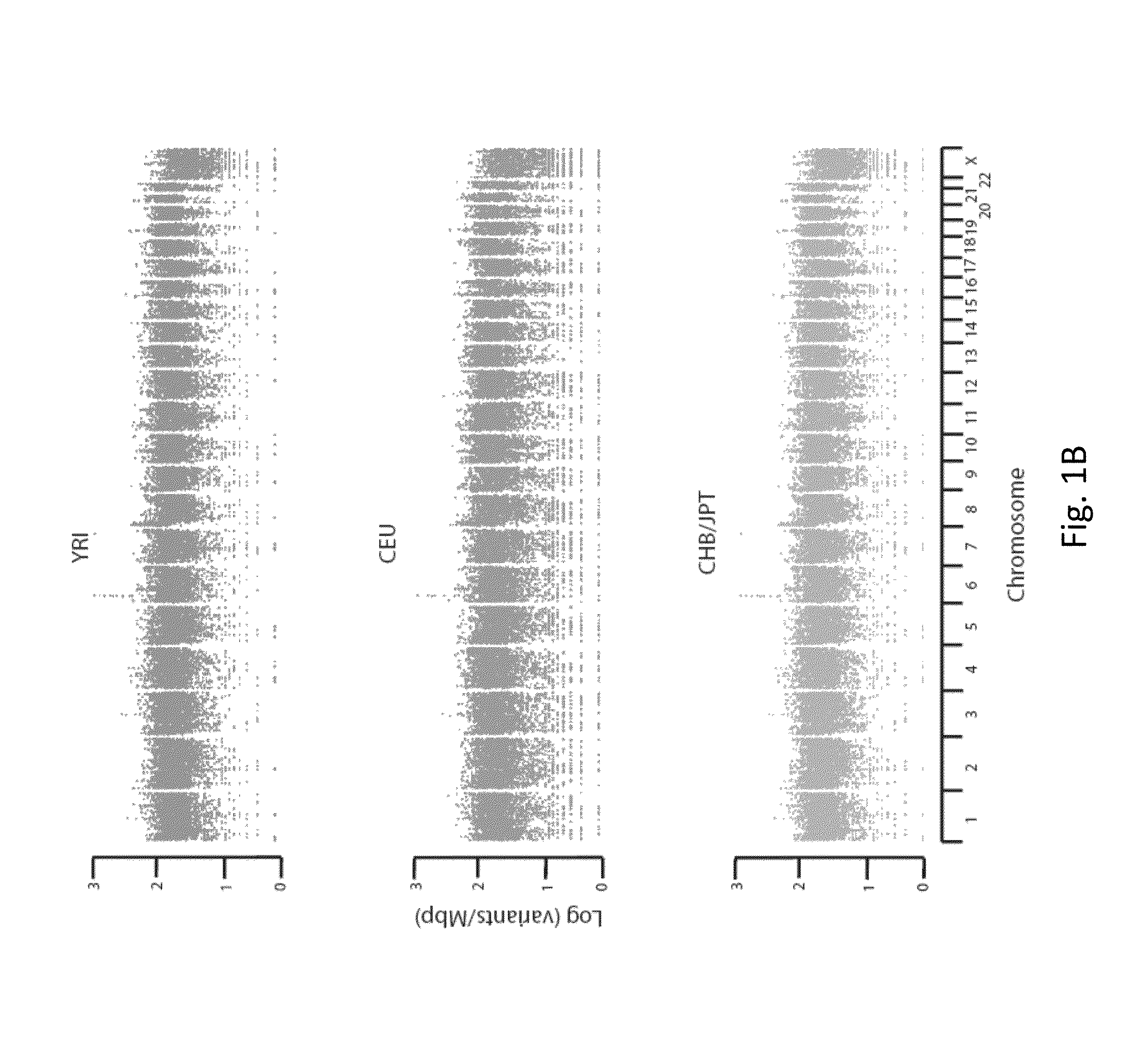
In an embodiment of the present invention, three novel human reference genome sequences were developed based on the most common population-specific DNA sequence (“major allele”). Methods were developed for their integration into interpretation pipelines for highthroughput whole genome sequencing.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
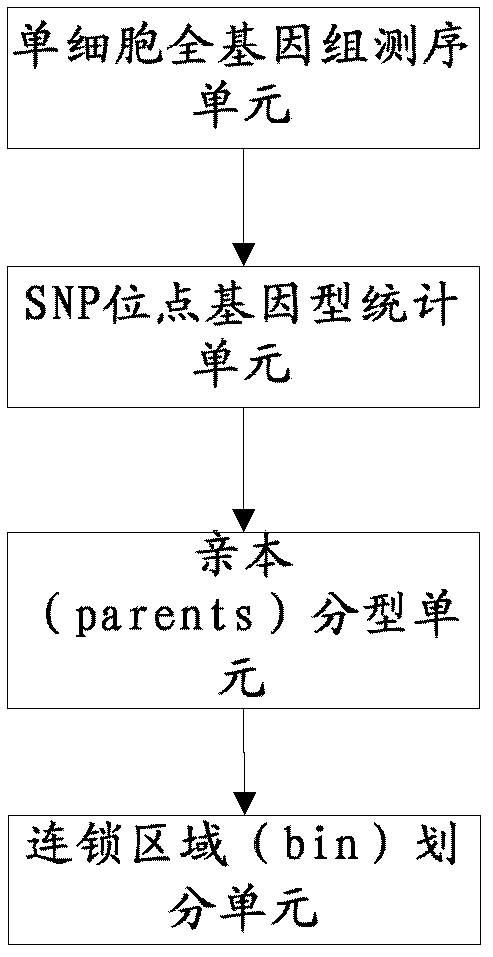
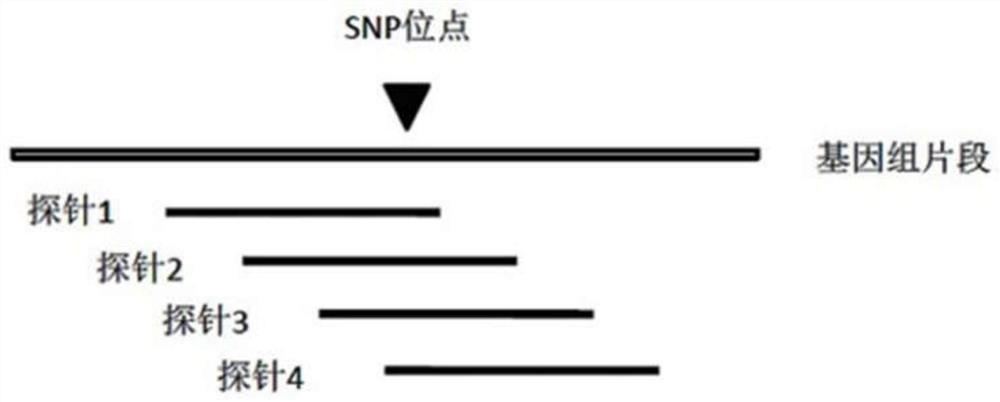
Genetic map construction method and device, haplotype analytical method and device
ActiveCN102952855AHigh resolutionUnderstand the genetic laws of reproductionBioreactor/fermenter combinationsBiological substance pretreatmentsGenomic sequencingTyping Classification
The invention relates to a genetic map construction and haplotype analytical method based on single-cell whole genome sequencing, and a device used for the method. The genetic map construction method comprises the following steps: comparing a whole genome sequence of a single cell of a species with a reference sequence to obtain a statistic result of a single nucleotide polymorphic site genotype; performing parent type classification of the genotype result to obtain a most appropriate male parent / female parent (a / b) result, so as to obtain a minimum recombination frequency of all the cells in the statistic result; according to the type classification result, dividing a linkage area (bin); calculating a change ratio of a / b to obtain recombination rates between all adjacent bins; and finally obtaining the genetic map according to the recombination rate of each recombinant site. The method of the invention can obtain a genetic map with high resolution by using a single cell, can not only applicable to human, but also applicable to some higher organisms with limited fertility, especially to endangered species.
Owner:TIANJIN MEDICAL LAB BGI
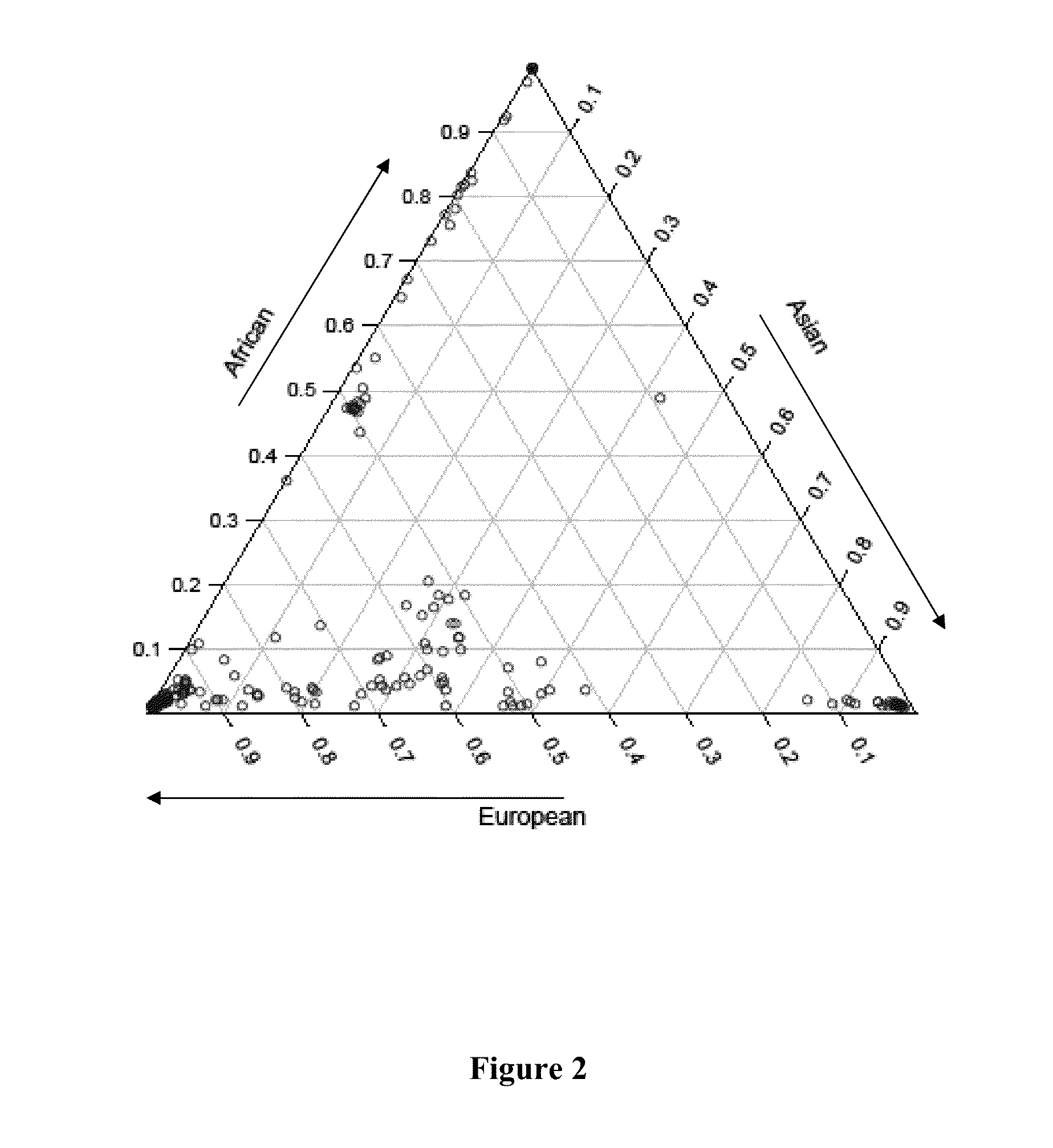
Ancestral-Specific Reference Genomes And Uses Thereof
ActiveUS20140067280A1Improve accuracyImprove targetingProteomicsGenomicsWhole genome sequencingMarker Discovery
Ancestry has a significant impact on the major and minor alleles found in each nucleotide position within the genome. Due to mechanisms of inheritance, ancestral-specific information contained within the genome is conserved within members of an ancestry. For this reason, individuals within a specific ancestry are more likely to share alleles in their genomes with other members of the same ancestry. Functionally, the combination of alleles at all positions within a group of individuals defines that group as having a common ancestry. Moreover, the aggregation of differences between alleles at all positions distinguishes one ancestry from another. The genomic similarities and differences between ancestries provides a mechanism to generate reference genomes that are specific for each ancestry. Reference genomes that are specific to an ancestry can be used to increase the accuracy of whole genome sequencing, DNA-based diagnostics and therapeutic marker discovery and in a variety of real-world DNA-based applications.
Owner:INOVA HEALTH SYST
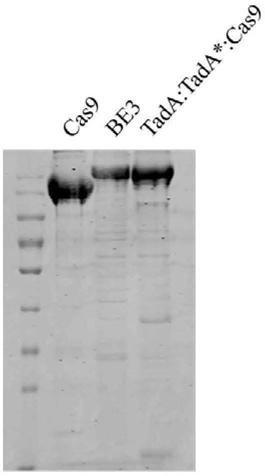
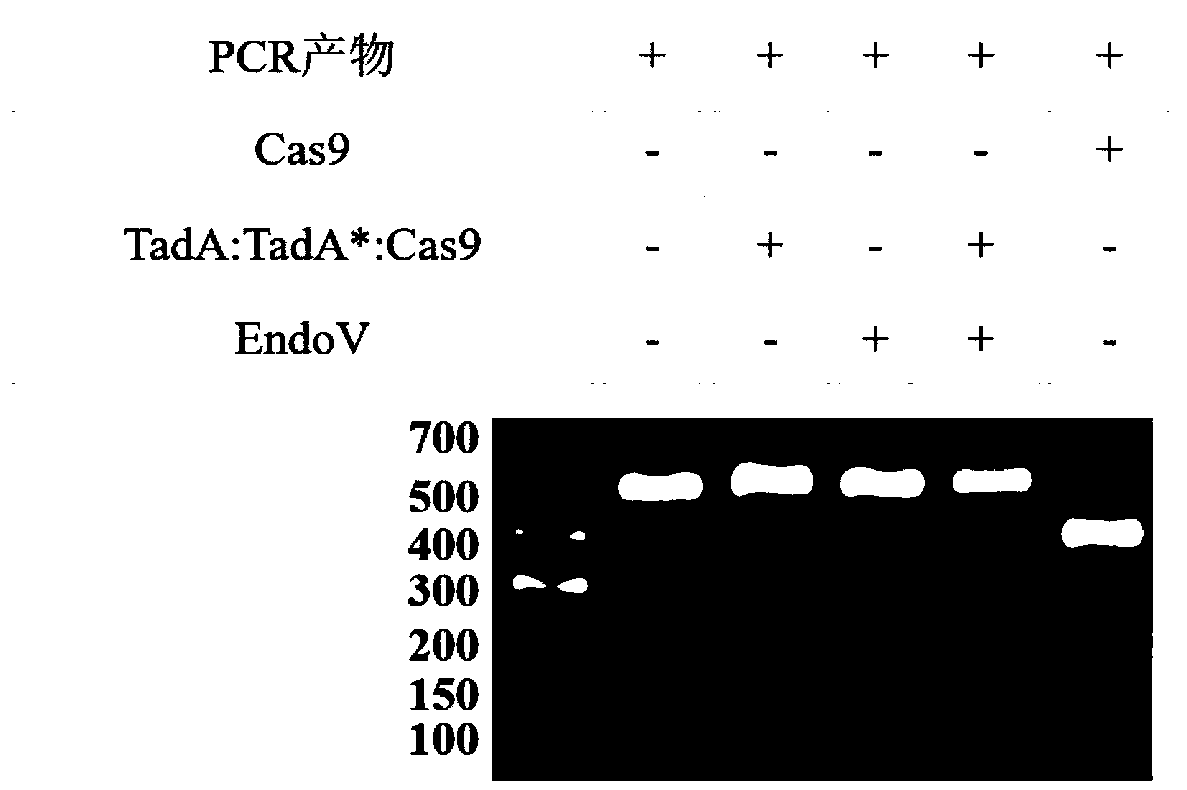
Method for detecting off-target effect of adenine base editor system based on whole genome sequencing, and its application in gene editing
PendingCN109295186AAccelerate cultivationMicrobiological testing/measurementAgainst vector-borne diseasesGenome editingWhole genome sequencing
The invention provides a method for detecting the off-target effect of an adenine base editor system based on whole genome sequencing, and its application in gene editing, The adenine base editor system comprises a TadA:TadA*:Cas9 fusion gene and gRNA. The system can catalyze the efficient replacement of adenine (A) at a target site with guanine (G), and has a broad application prospect in human disease gene editing therapy and disease model construction. The first method for detecting the off-target effect of the ABE system in a whole genome range, called EndoV-seq method for short, is developed in the invention. The EndoV-seq method provided by the invention has a broad application prospect in the field of gene editing, especially gene editing therapy.
Owner:SUN YAT SEN UNIV
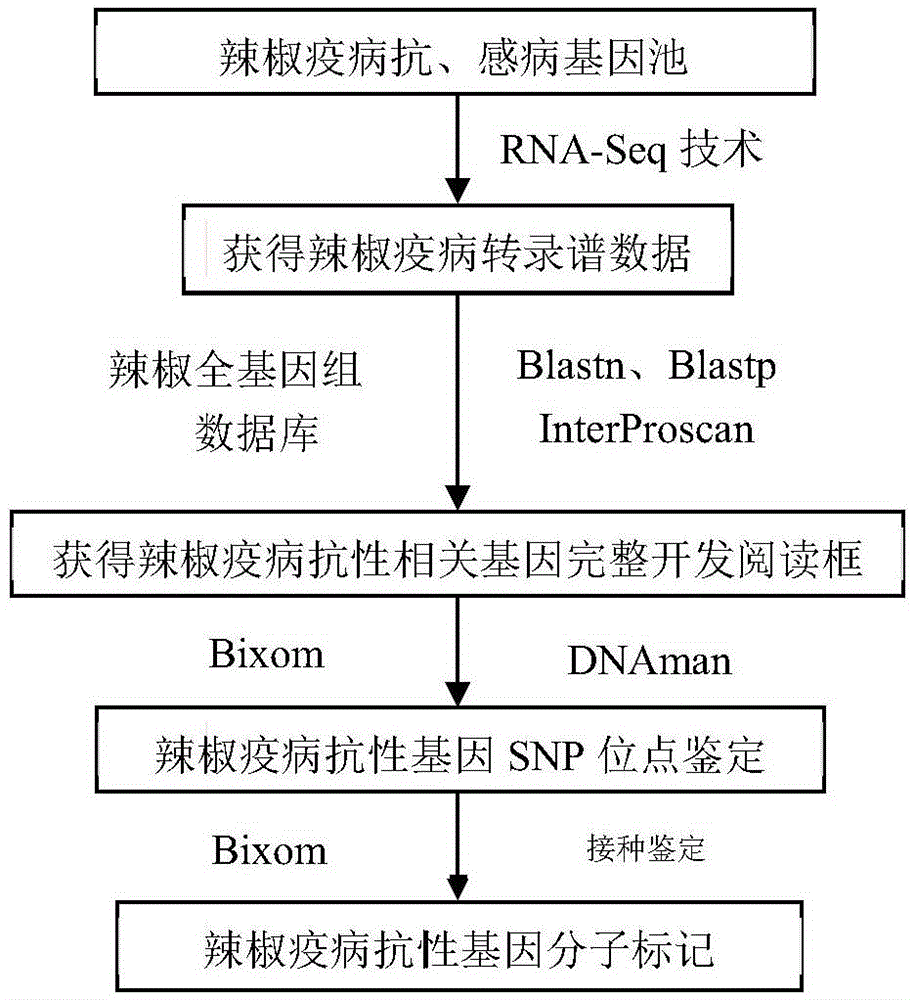
Method for obtaining capsicum phytophthora resistance candidate gene and molecular marker, and application
InactiveCN104560973AAccurate identificationLarge amount of data informationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyData information
The invention relates to a method for obtaining a capsicum phytophthora resistance candidate gene and a molecular marker, and application. The method is used for obtaining the capsicum phytophthora resistance candidate gene by utilizing capsicum phytophthora transcriptome and whole-genome sequencing data information, differentially-expressed gene identification, bioinformatics analysis, molecular marker development and phytophthora inoculation identification and belongs to the technical field of capsicum biology. The method comprises the following steps: sequencing a phytophthora resistant and susceptible gene pool transcriptome obtained after phytophthora inoculation of an F2 population constructed by capsicum highly-resistant and highly-susceptible phytophthora materials, performing expression analysis and functional annotation on differential genes, extracting DNAs (Desoxvribose Nucleic Acid) of a capsicum phytophthora highly-resistant and highly-susceptible phytophthora material genome, performing primer design and PCR (Polymerase Chain Reaction) amplification, performing sequence difference analysis and SNP site identification, performing SNP specific primer design and validity verification, and performing other steps to efficiently obtain the capsicum phytophthora resistance candidate gene and the molecular marker. According to the method, the capsicum phytophthora resistance candidate gene can be accurately identified, and the effective molecular marker can be developed.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
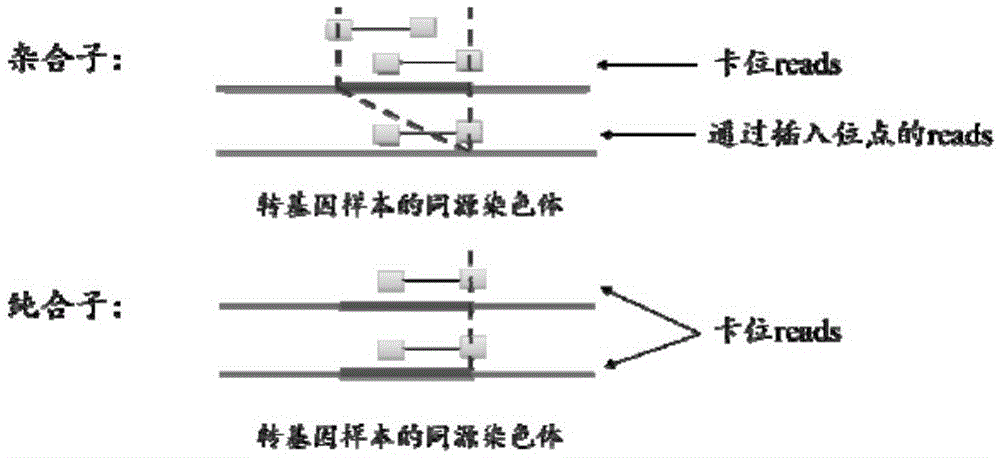
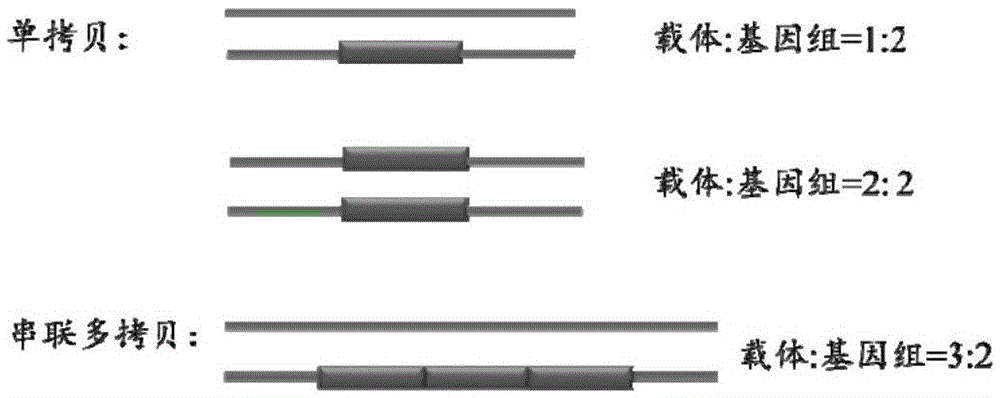
Method for identifying transgenic events through whole genome sequencing data
ActiveCN105631242AAccurate analysisValid identificationProteomicsGenomicsBiological bodyWhole genome sequencing
The invention provides a method for identifying transgenic events through whole genome sequencing data. The method comprises the steps that transgenic events are identified through a bioinformatics means, and the position, direction, copying number and flanking sequence information of foreign fragments inserted into genomes and the homozygous and heterozygous state of transgenic samples can be rapidly and effectively identified. Compared with traditional Genome walking, real-time quantitative PCR and other experiments methods, the method has the advantages that time is short, repeatability is high, the result is stable, and explaining is easier. The method is the most comprehensive in existing transgenic event detection methods, can accurately analyze influences of transgenic events on the living body within a short time, carries out safety evaluation on transgenic animals and plants, and pushes the transgenic technology to be applied in agriculture.
Owner:CHINA AGRI UNIV
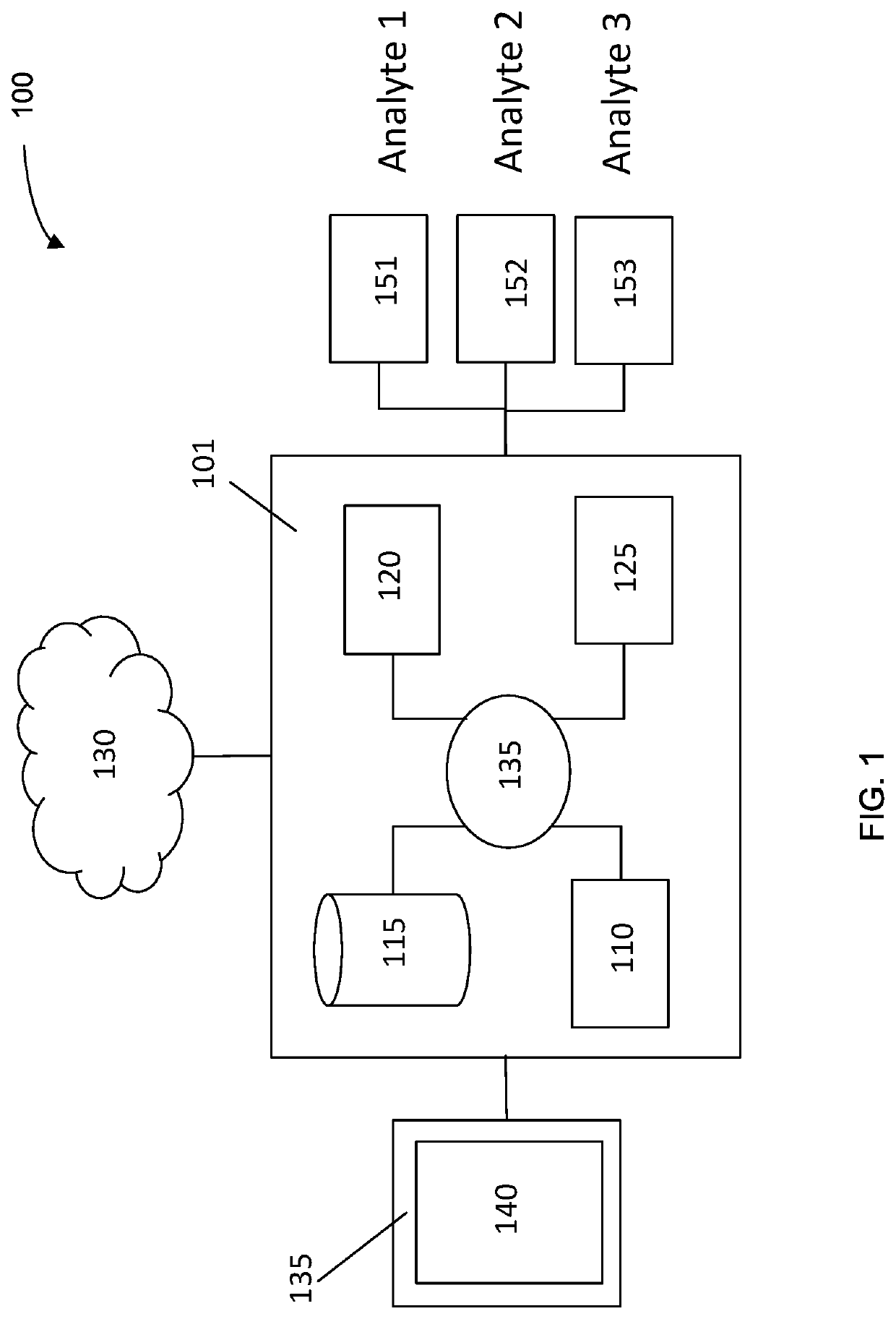
Machine learning implementation for multi-analyte assay development and testing
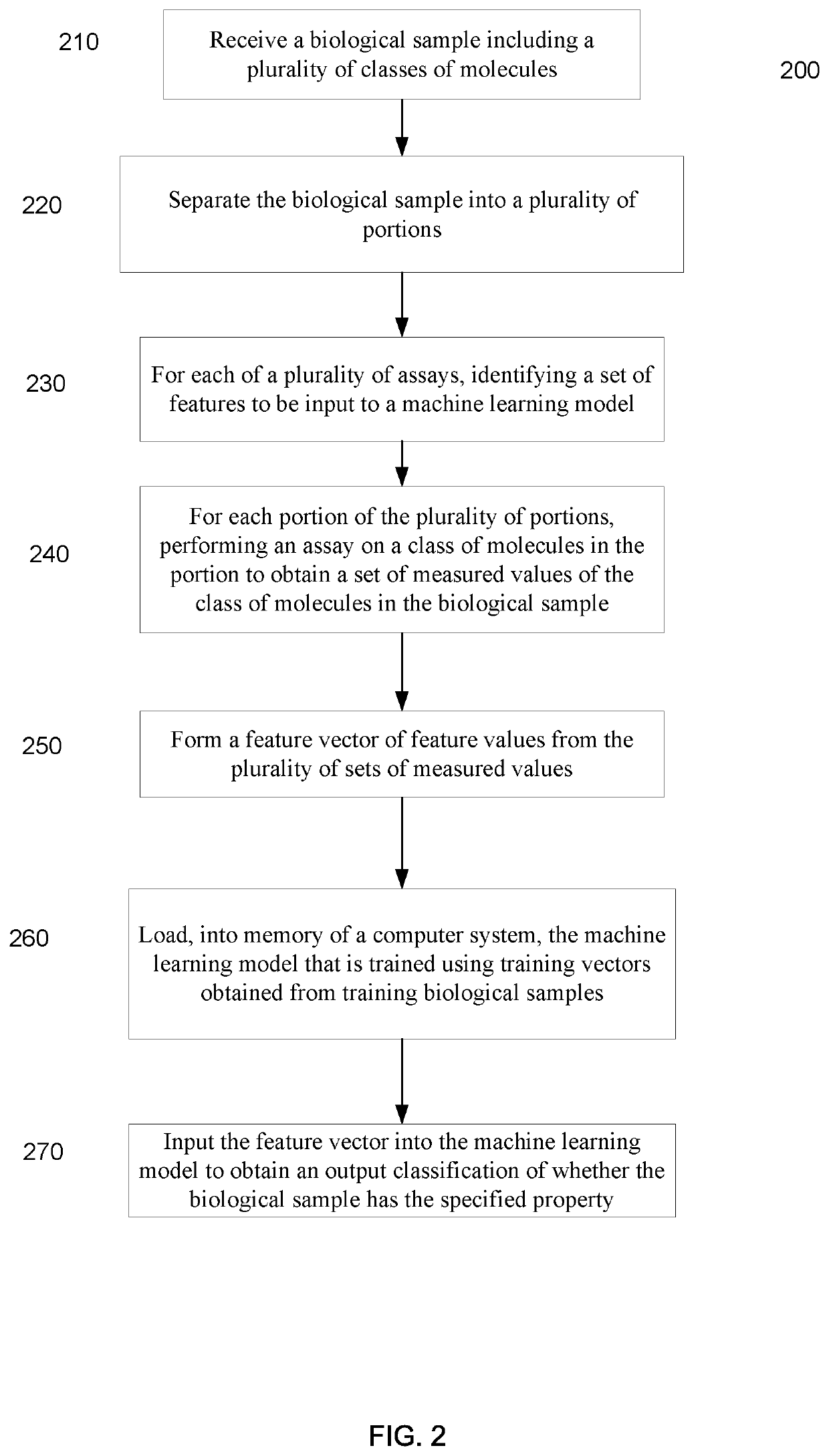
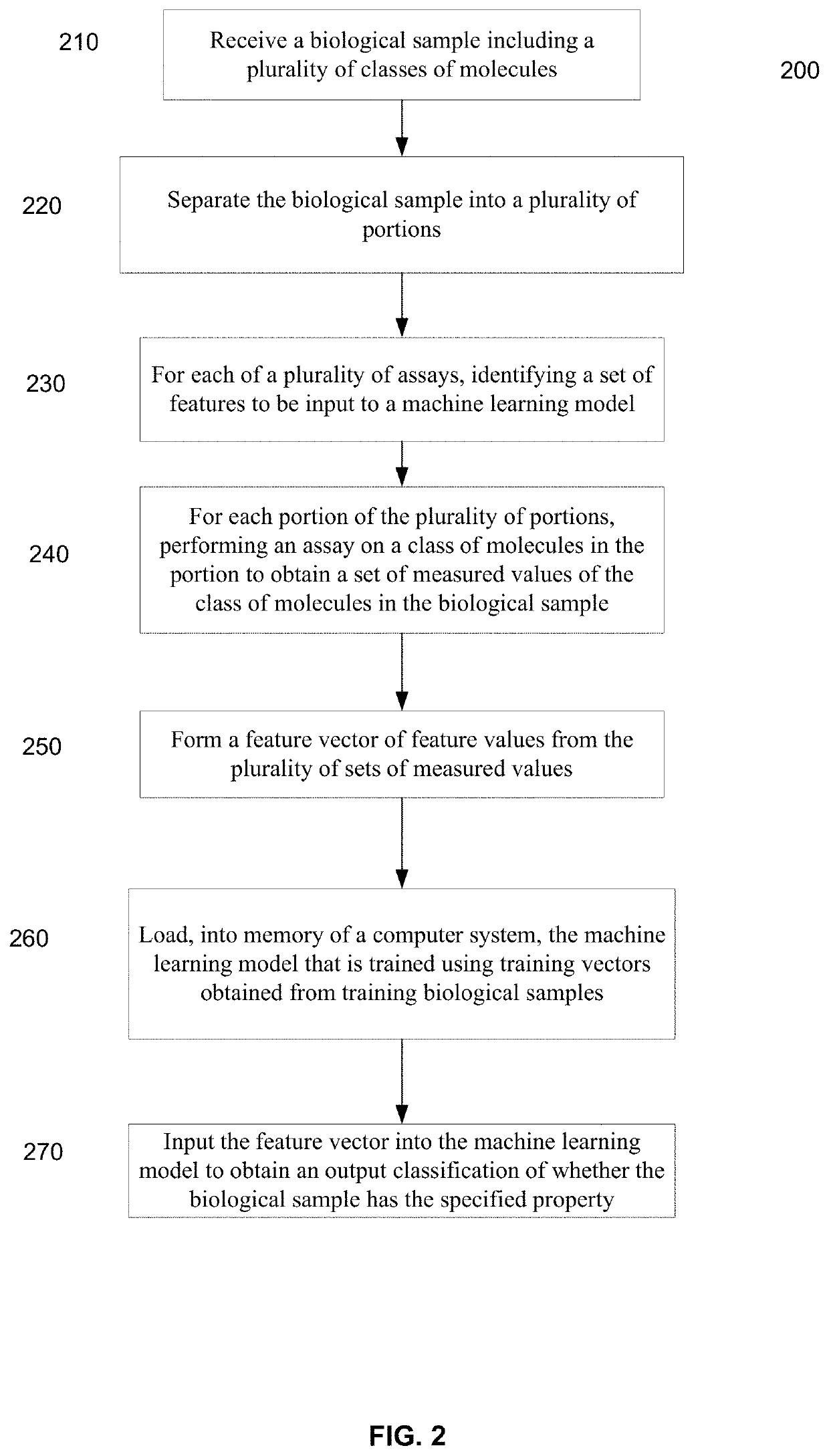
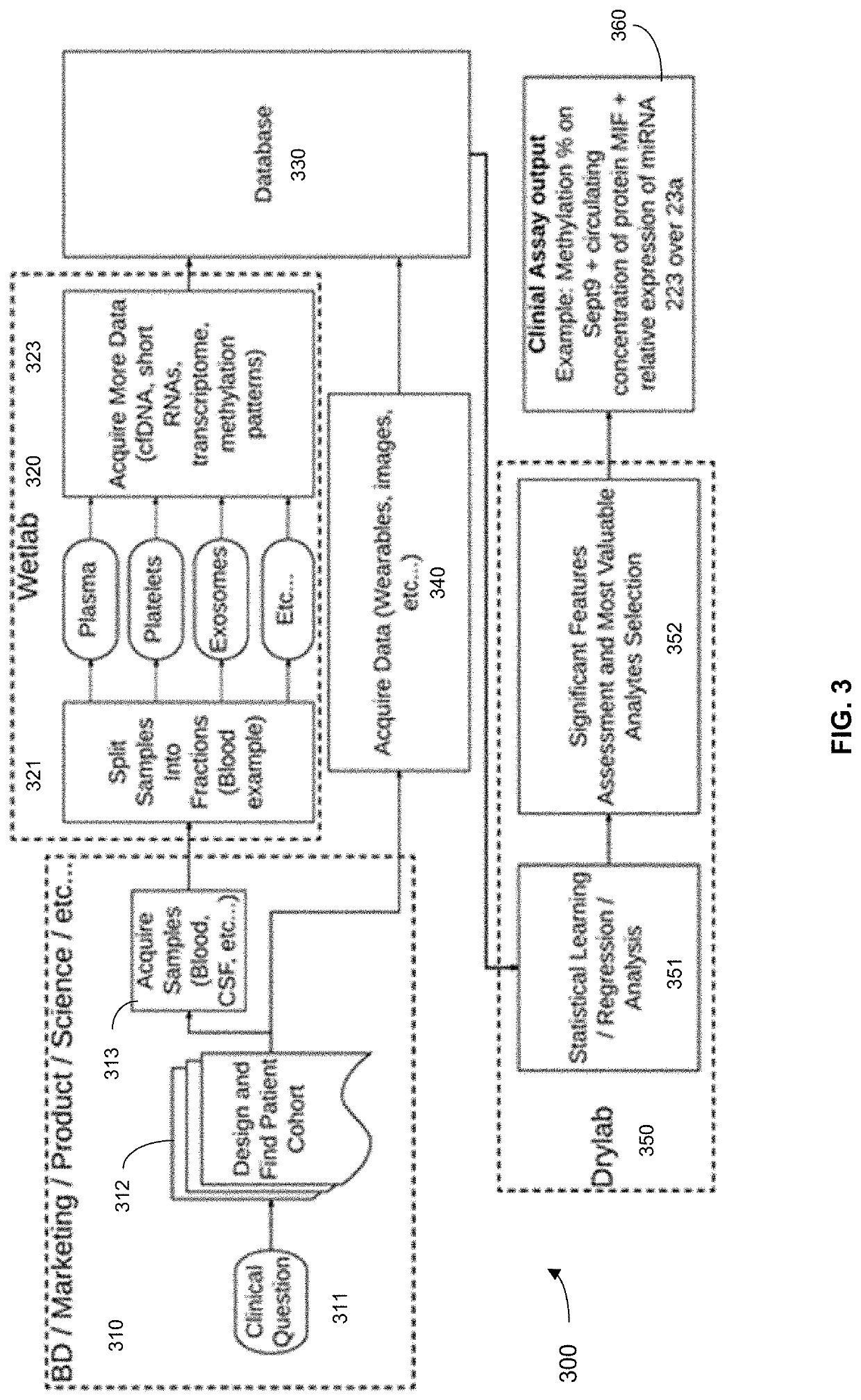
Systems and methods that analyze blood-based cancer diagnostic tests using multiple classes of molecules are described. The system uses machine learning (ML) to analyze multiple analytes, for example cell-free DNA, cell-free microRNA, and circulating proteins, from a biological sample. The system can use multiple assays, e.g., whole-genome sequencing, whole-genome bisulfite sequencing or EM-seq, small-RNA sequencing, and quantitative immunoassay. This can increase the sensitivity and specificity of diagnostics by exploiting independent information between signals. During operation, the system receives a biological sample, and separates a plurality of molecule classes from the sample. For a plurality of assays, the system identifies feature sets to input to a machine learning model. The system performs an assay on each molecule class and forms a feature vector from the measured values. The system inputs the feature vector into the machine learning model and obtains an output classification of whether the sample has a specified property.
Owner:FREENOME HLDG INC
Long insert-based whole genome sequencing
ActiveUS20150126379A1Microbiological testing/measurementLibrary member identificationGenomic sequencingWhole genome sequencing
The present invention is directed to a method of detecting a genomic rearrangement in a nucleic acid sample with Long Insert Whole Genome Sequencing (LI-WGS). The method may include obtaining a nucleic acid sample and then fragmenting the nucleic acid sample (e.g., via sonication). In particular, the fragmenting may result in the production of a plurality of inserts. Thereafter, the method comprises purifying the plurality of inserts using magnetic beads and then amplifying the purified plurality of inserts. In addition, the method further comprises sequencing the purified and amplified plurality of inserts. In some aspects, the plurality of inserts have a length of between about 800 and about 1,100 base pairs.
Owner:TRANSLATIONAL GENOMICS RESEARCH INSTITUTE
Molecular biology method for identifying purity of tobacco varieties
InactiveCN105734141ANot affectedPreventing and ensuring the safety of production seedsMicrobiological testing/measurementBiotechnologyWhole genome sequencing
The invention relates to a molecular biology method for identifying purity of tobacco varieties. The method comprises the following main steps: respectively extracting total genome DNA of a control group and a to-be-detected tobacco variety, performing whole genome sequencing of proper depth by adopting the latest sequencing technology, performing sequence splicing, assembling and whole genome sequence comparison on the control group and the to-be-detected tobacco variety based on a tobacco genome reference sequence by utilizing bioinformatics means, counting base differences of the two groups, and calculating the purity percentage of the to-be-detected tobacco variety relative to the control tobacco variety. The method disclosed by the invention is not influenced by environmental conditions and seasons, is accurate and reliable in result, can accurately identify the purity of the tobacco varieties from a single base variation level of the smallest hereditary unit, can be used for parent purification and authenticity identification of the tobacco varieties and has great significances for guaranteeing safety of tobacco production varieties, maintaining economic benefits of tobacco growers and effectively solving intellectual property disputes of the tobacco varieties.
Owner:HUBEI TOBACCO SCI RES INST
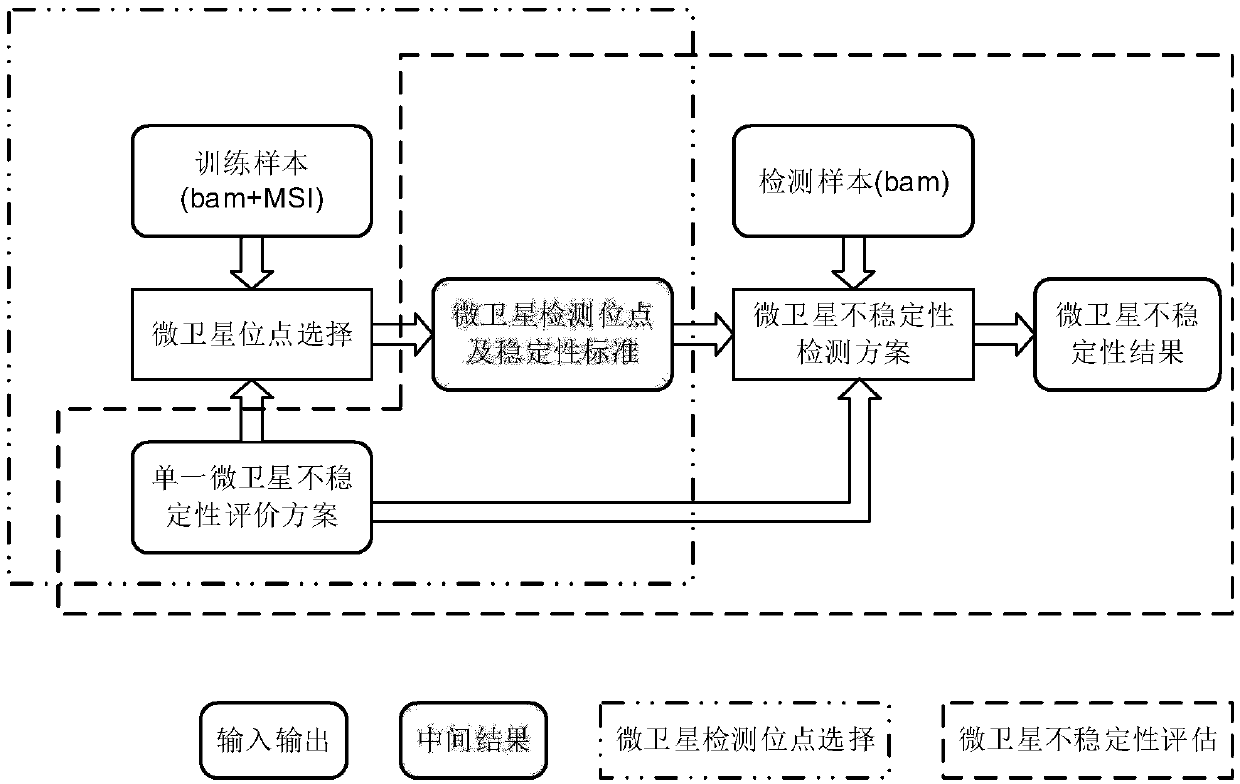
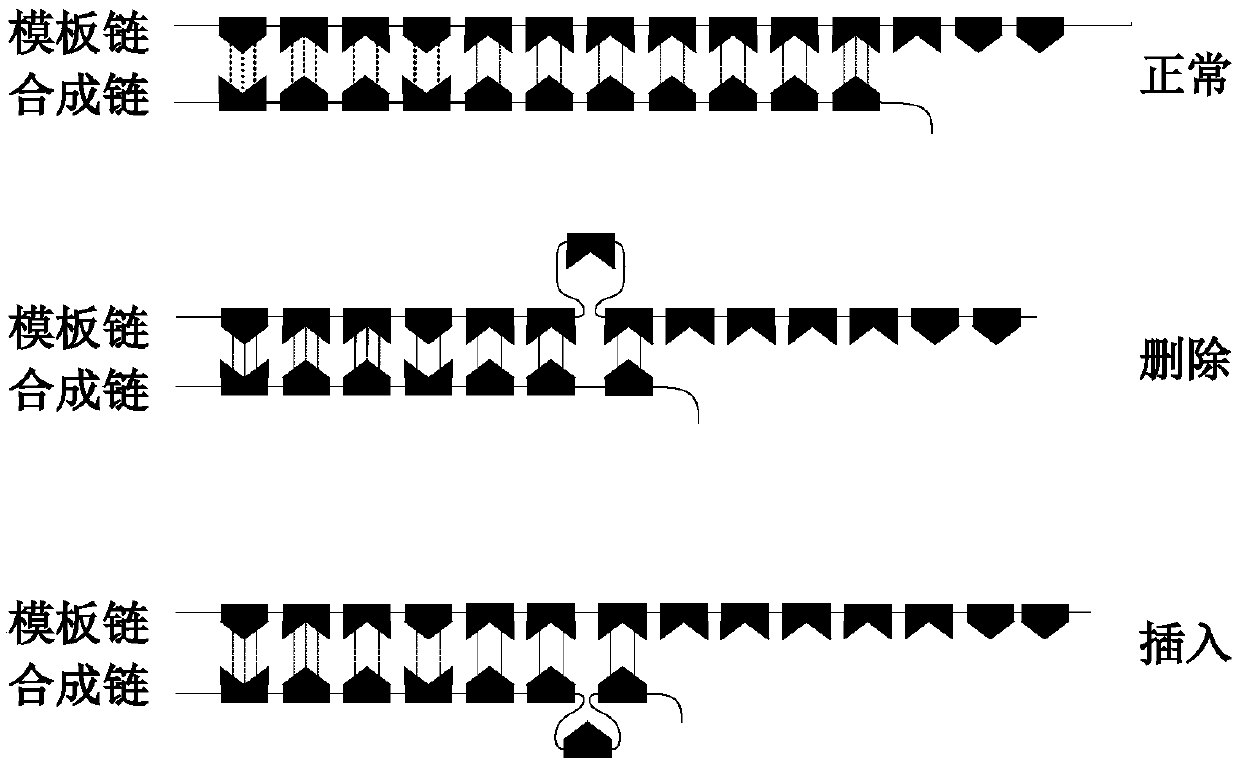
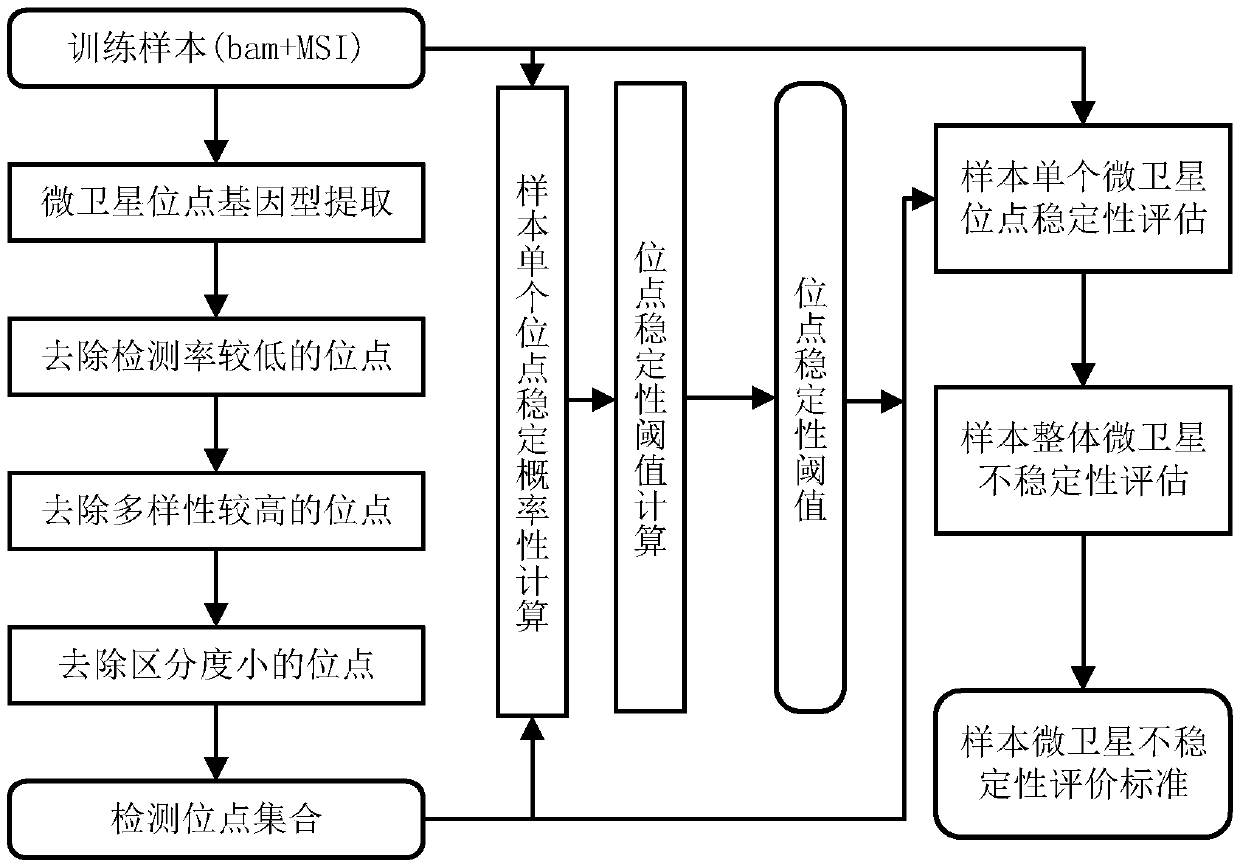
Microsatellite instability detecting system and method based on genome sequencing
ActiveCN109637590AReduce limitationsRelieve painSequence analysisInstrumentsGenomic sequencingWhole genome sequencing
The invention discloses a microsatellite instability detecting system and method based on genome sequencing. The method comprises the steps that microsatellite detecting site selection is conducted, wherein an effective detecting site is selected according to sequencing data of a certain tumor sample, the threshold value of the instability of a single microsatellite corresponding to the effectivedetecting site and the evaluation standard of the instability of the microsatellite of a certain tumor sample are computed; microsatellite instability detection is conducted on a detected sample according to the threshold value of the instability of the single microsatellite corresponding to the effective detecting site and the evaluation standard of the instability of the microsatellite of the certain tumor sample. The system and the method have the advantages that the system and the method do not rely on a control sample, the pain on a detected person caused by sampling can be reduced, all genetic information of the detected person is included in the control sample, the probability of privacy leakage of the detected person can be reduced by not using the control sample, and the detectioncost can be reduced by not detecting the control sample; the operation is convenient, the cost is low, and the confidence level is high.
Owner:XI AN JIAOTONG UNIV
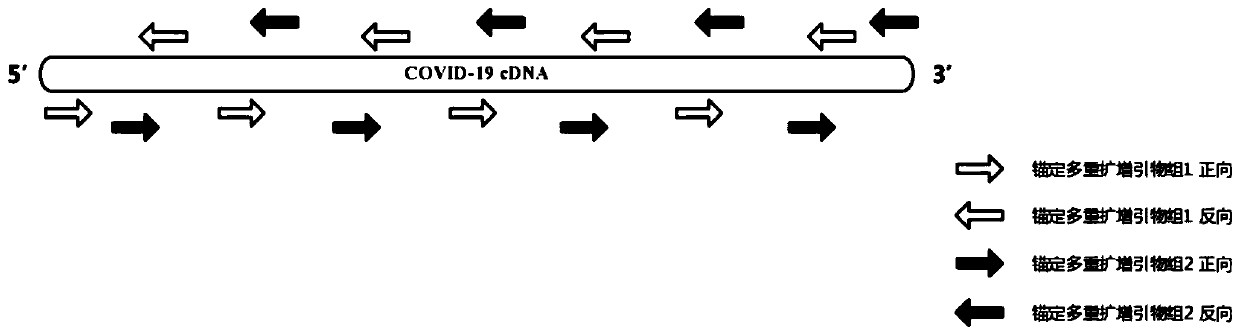
Construction method of novel coronavirus whole-genome high-throughput sequencing library, and kit for library construction
ActiveCN111334868AEfficient targeted enrichmentLow targetingMicrobiological testing/measurementMicroorganism based processesMultiplexGenomic sequencing
The invention provides a construction method of a novel coronavirus whole-genome high-throughput sequencing library, and a kit for library construction. The method comprises the following steps: 1) carrying out reverse transcription on virus RNA; (2) carrying out a first round of PCR reaction by utilizing multiple amplification primers of an anchoring part Illumina linker sequence; and 3) carryingout a second round of PCR reaction by using a labeled Illumina library amplification primer, and purifying the amplification product to obtain the high-throughput sequencing library. The anchoring multiplex amplification primer combination provided by the invention is utilized, so the efficient targeted enrichment can be performed on the genome of the novel coronavirus COVID-19, the influence that an existing method is low in targeting, low in experimental timeliness and prone to being brought into host background pollution is overcome, COVID-19 virus whole genome sequencing can be completedwithin a short time with the small sequencing data volume and cost, so differential diagnosis and virus mutation identification of the COVID-19 virus are achieved.
Owner:福州福瑞医学检验实验室有限公司 +1
Rare cell isolation device and method of use thereof
ActiveUS20180106805A1Faster cell captureQuick snapComponent separationMicrobiological testing/measurementPopulation sampleLymphatic Spread
This invention concerns patentable devices configured to capture cancer stem cells and cell clusters. After capturing such cells and / or clusters, the cancerous cells are subjected to whole genome sequencing. The resulting genomic sequence information is then compared to that for “normal” or non-diseased tissue (obtained, for example, from either the same patient, or a population sample, etc.) in order to identify the specific genetic mutation(s) present in the CSCs. Further analysis then correlates the genetic mutations with cell growth signaling pathways typically found with tumor metastases. Armed with this information, an oncologist can then develop a specifically targeted therapy that utilizes approved drugs or drug candidates undergoing clinical testing to address the identified driver mutations and thus effect a “targeted” therapy tailored to the particular patient's disease.
Owner:TUMORGEN INC
Single exon copy number variation predicting method based on target area sequencing
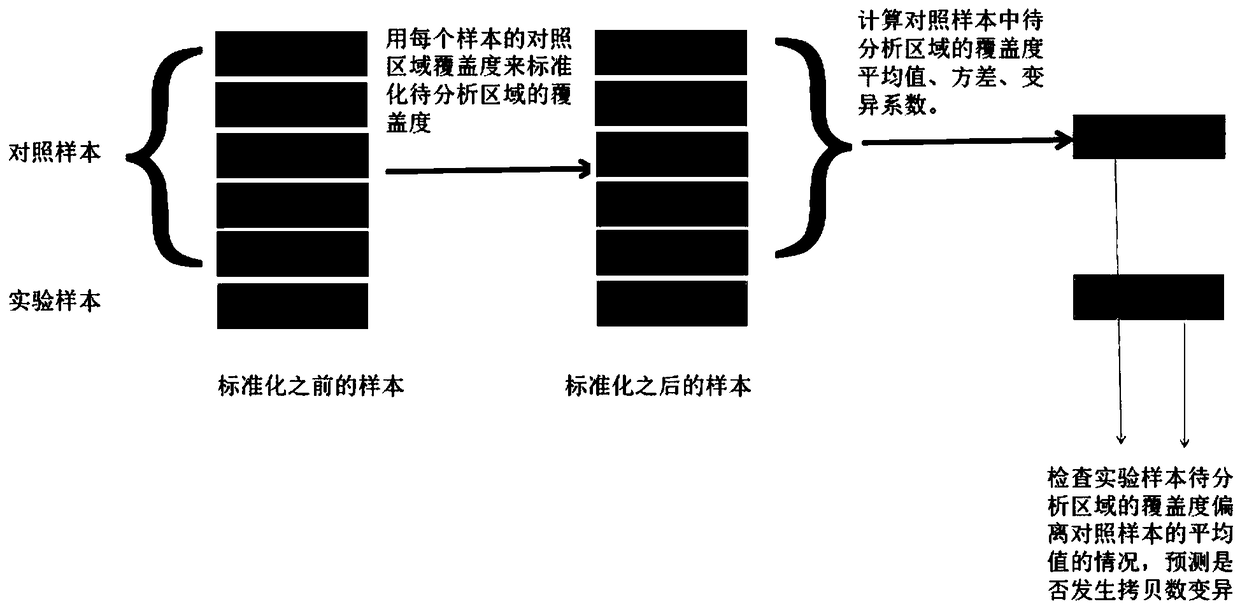
ActiveCN108920899ALow costReduce Analysis ComplexitySpecial data processing applicationsWhole genome sequencingExon
The invention relates to a single exon copy number variation predicting method based on target area sequencing. The method includes: processing sequencing data; predicting copy number variation, to bemore specific, statistically counting the total number of sequencing sequences and basic groups covering a target area, determining control exon areas, standardizing the coverage of the to-be-analyzed exon areas of each control sample and each experiment sample, calculating the average value, standard deviation and variation coefficient of the standardized coverage of the to-be-analyzed exon areas in the control samples, and predicting the copy number variation of to-be-analyzed exon areas. The method has the advantages that whole genome sequencing is not needed, the copy number variation ofexon level is analyzed by directly using the coverage information of the exon level, and the method is simple in analysis and does not need complex GC correction and modeling.
Owner:杭州迈迪科生物科技有限公司
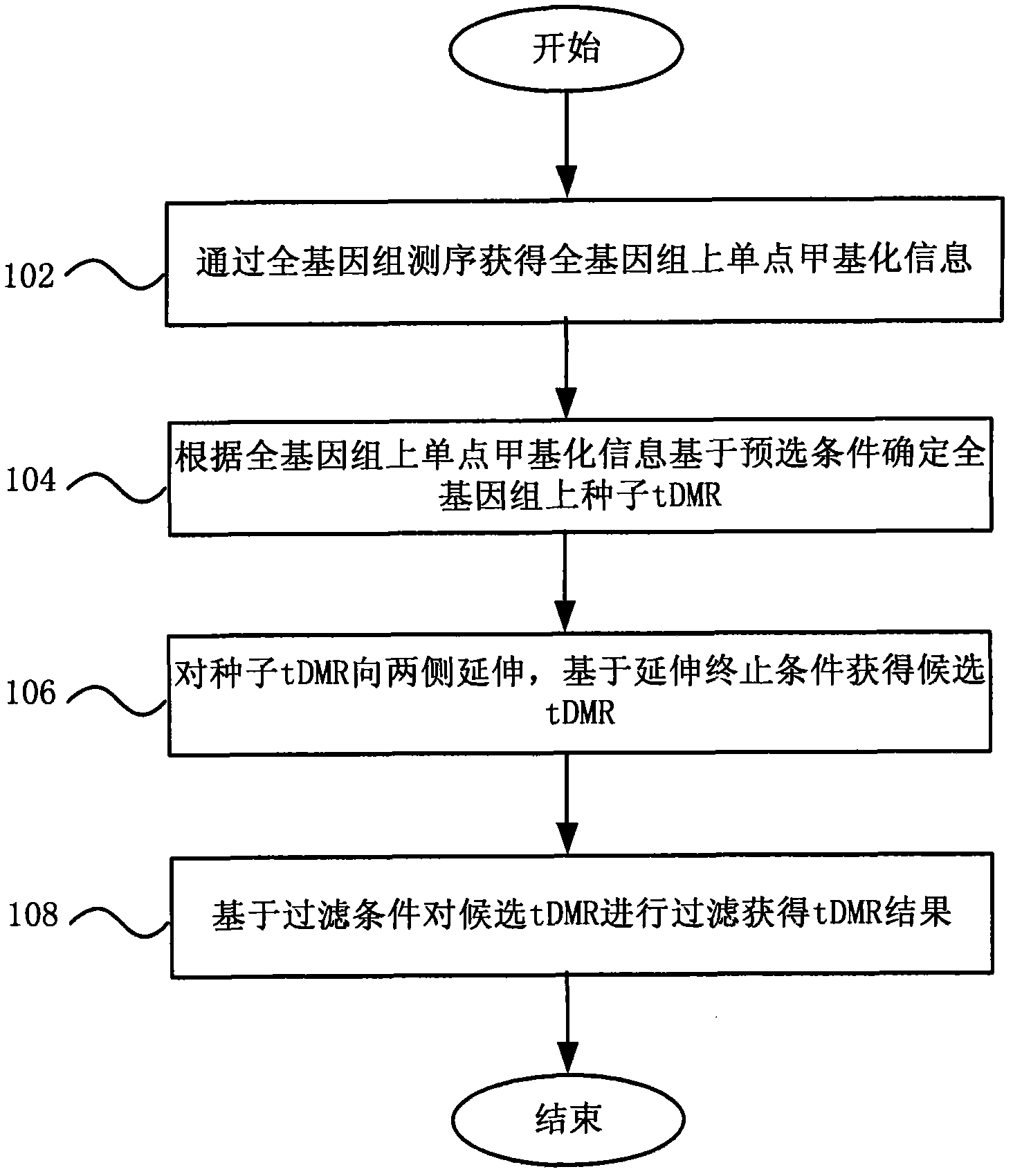
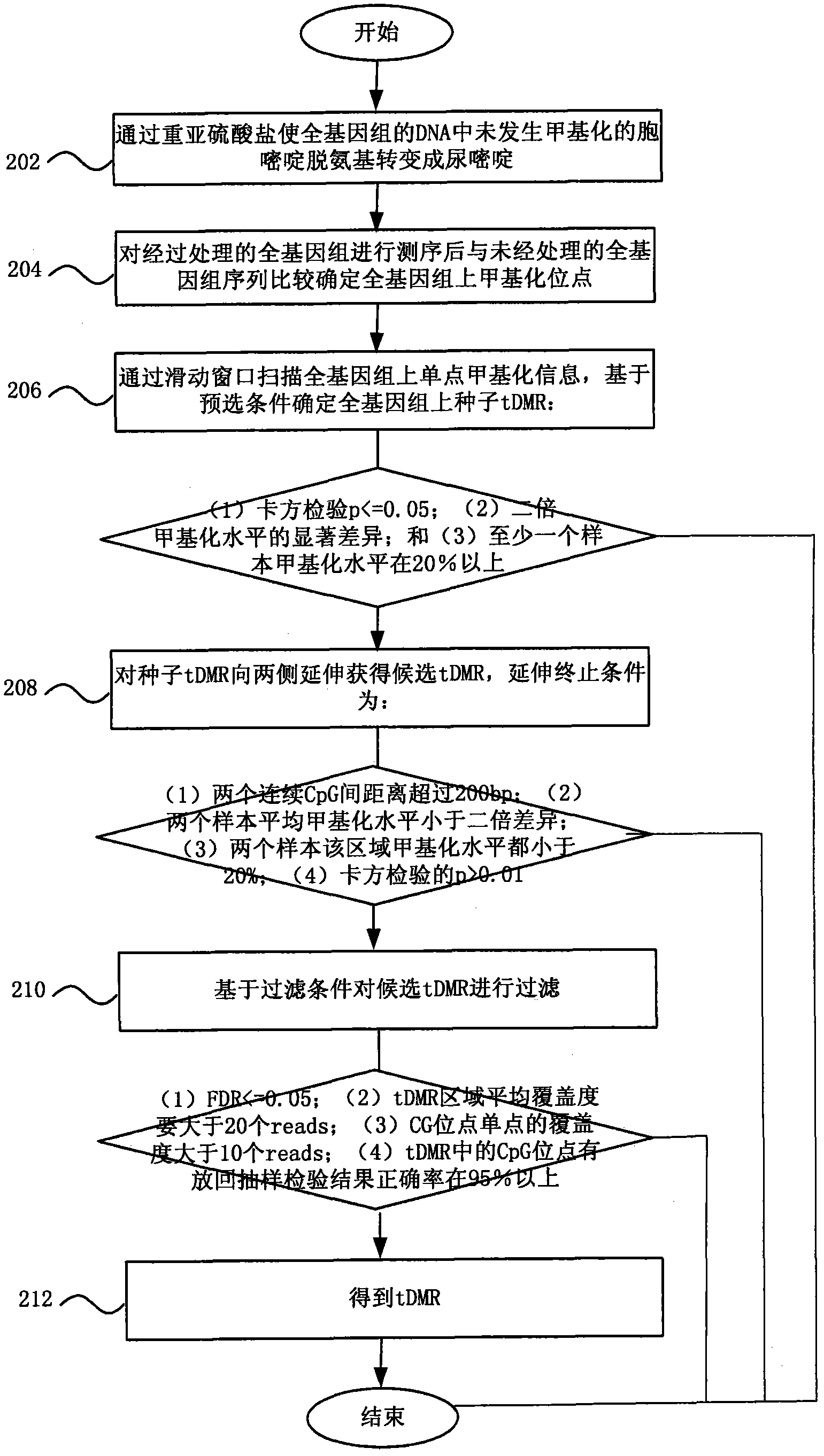
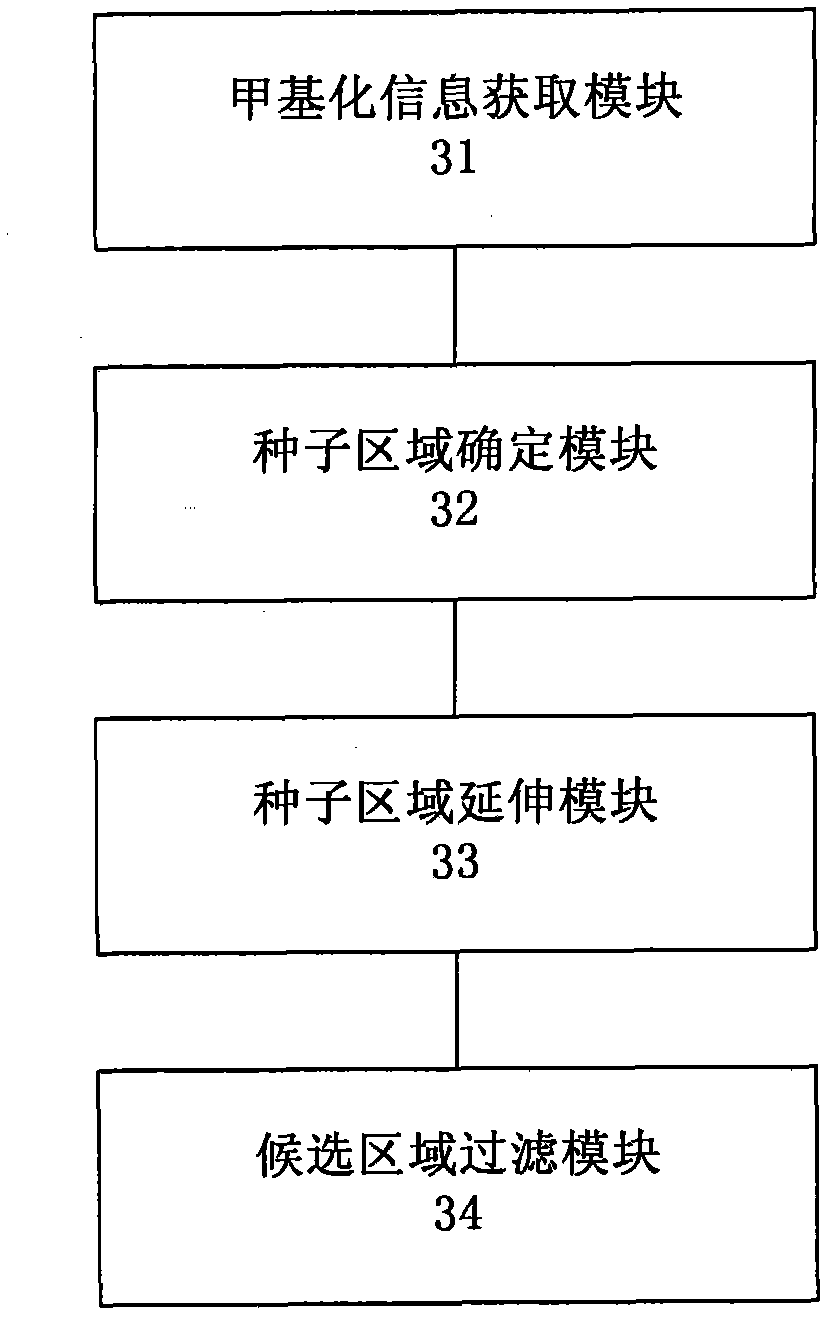
Method and system for detecting tissue-specific differentially methylated region (tDMR)
The invention discloses a method and a system for detecting a tissue-specific differentially methylated region (tDMR). The method comprises the following steps: obtaining single-point methylation information on a complete genome by genome-wide sequencing; determining a seed tDMR on the complete genome under a pre-selection condition according to the single-point methylation information on the complete genome; extending the seed tDMR along two sides, and obtaining candidate tDMRs based on an extension terminal condition; and filtering the candidate tDMRs based on a filtering condition to obtain the tDMR result. By means of the method and the system provided by the invention, the methylation state of each site on the complete genome can be defined so as to determine the tDMR within the range of the complete genome, thus greatly improving the detection efficiency and lowering the cost. Subsequent validation shows that the found tDMR has high accuracy rate up to 85%; and validation in a plurality of species (not only limited to mammals) shows that the method has higher accuracy and higher sensitivity compared with other methods such as vardhman Rakyan and the like.
Owner:BGI TECH SOLUTIONS
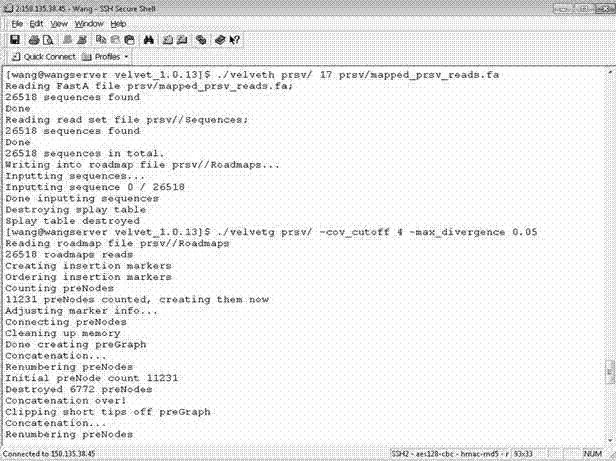
Method for obtaining genome sequence of papaya ringspot virus through high-throughput small RNA sequencing
The invention belongs to the fields of bioinformatics and genetic engineering. A papaya ringspot virus Hainan isolate PRSV-HN-y1 has genome sequences of SEQ ID No. 1 and SEQ ID No. 2 as shown in the specification. The invention further discloses a cloning method for obtaining the genome sequence of the papaya ringspot virus Hainan isolate PRSV-HN-y1 through a high-throughput small RNA sequencing. Research results firstly confirm that the high-throughput small RNA sequencing can be used for pathogen identification, detection and virus whole genome sequencing of samples infected by suspected papaya virus.
Owner:INST OF TROPICAL BIOSCI & BIOTECH CHINESE ACADEMY OF TROPICAL AGRI SCI
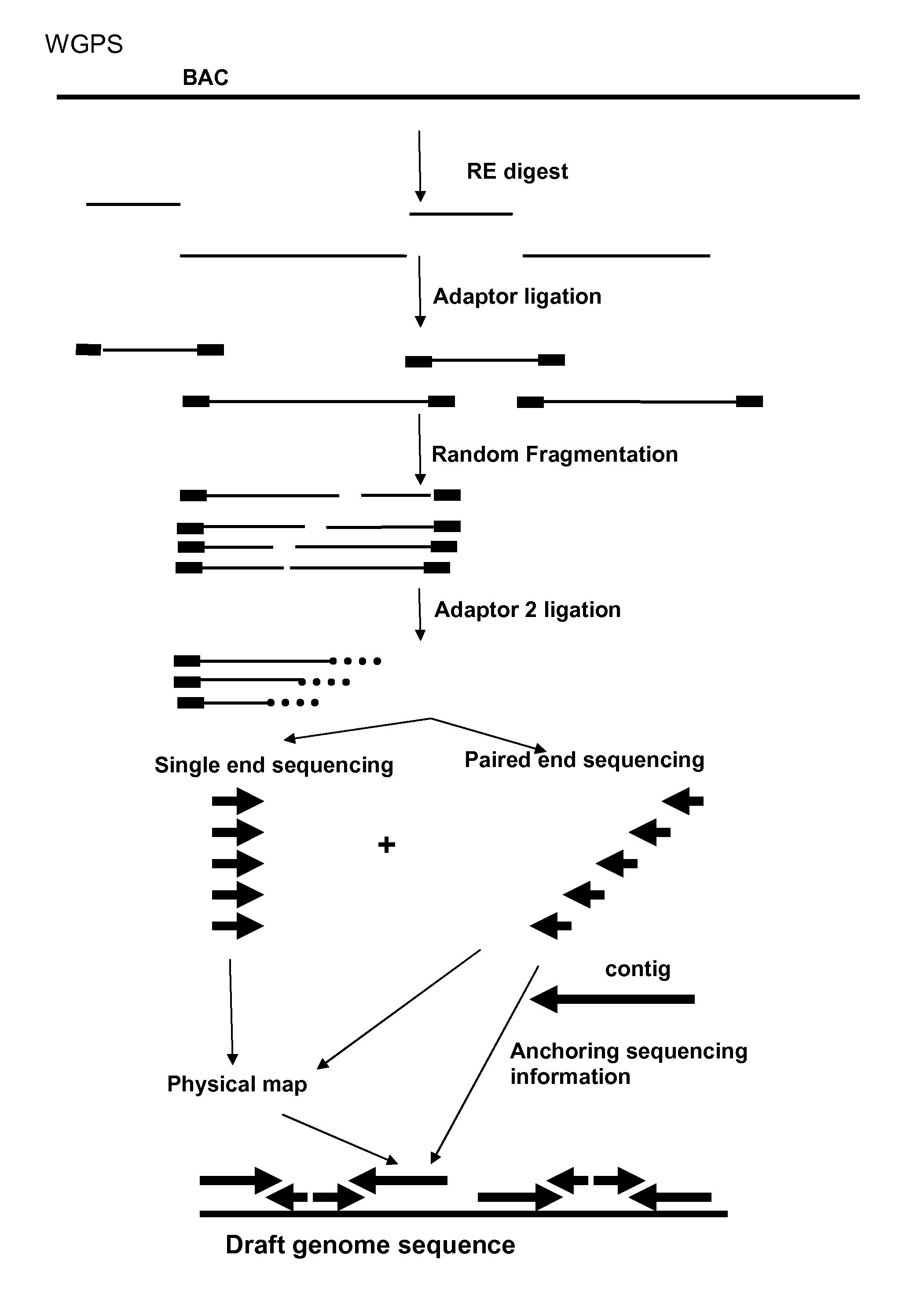
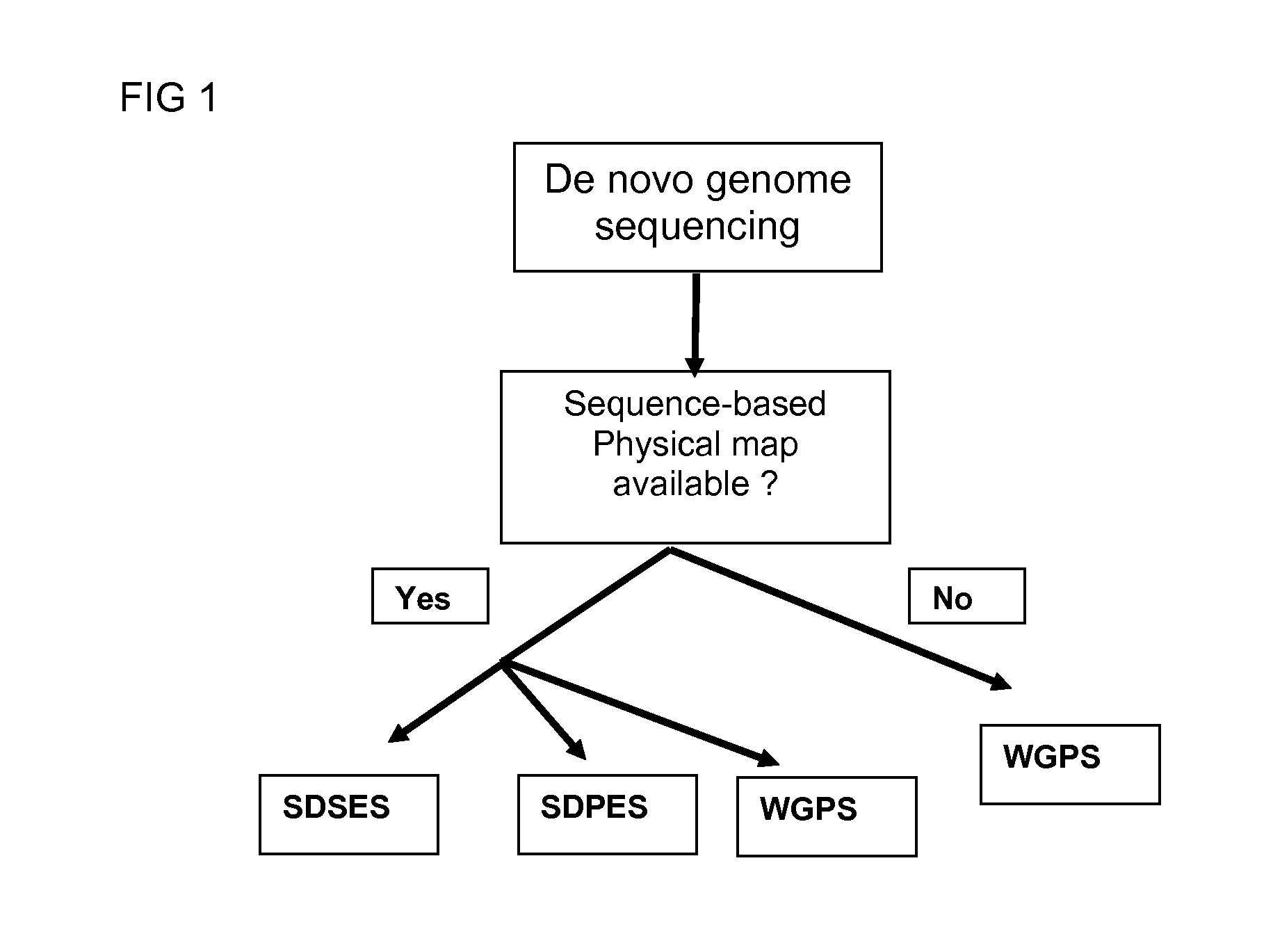
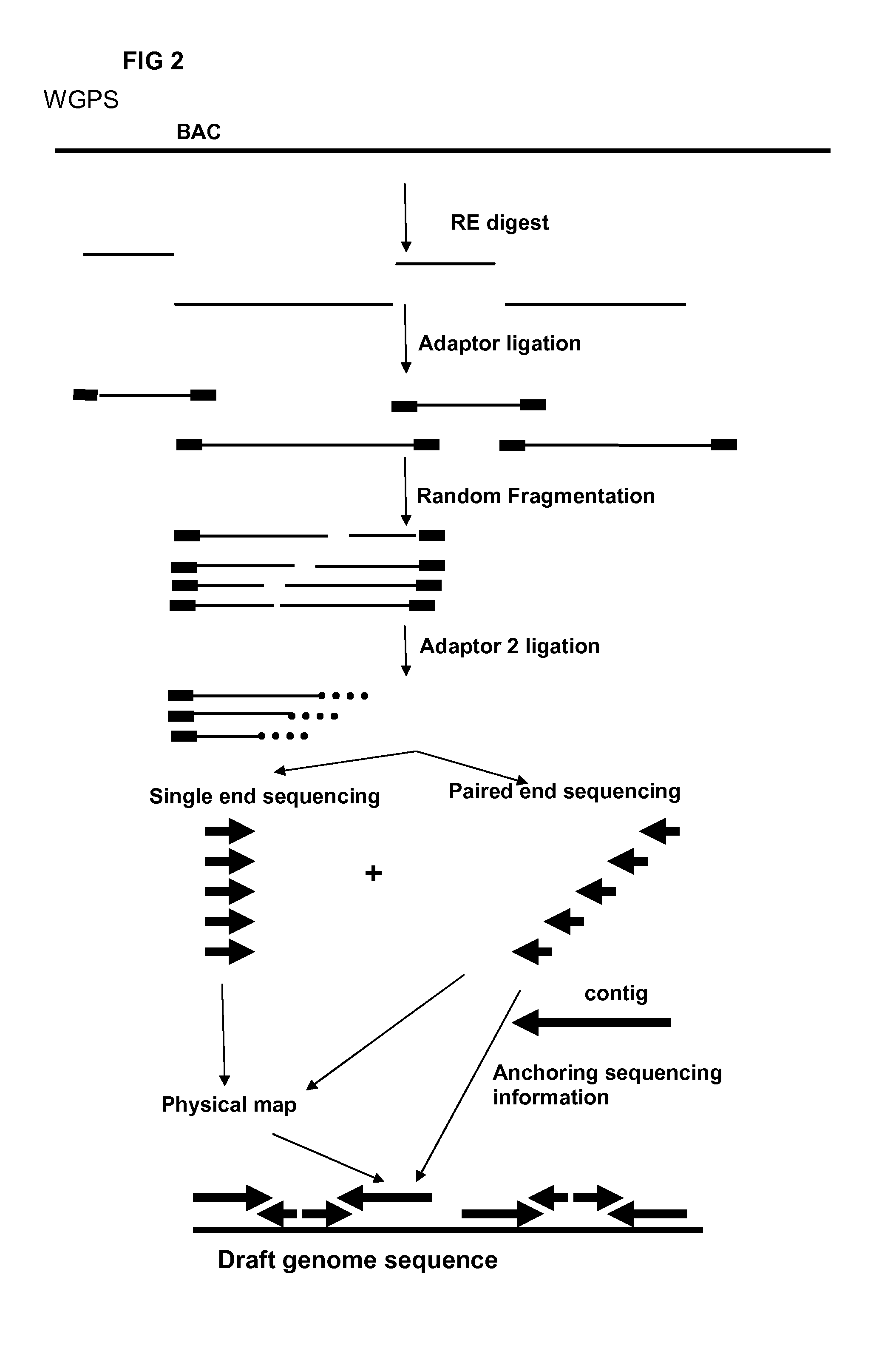
Restriction enzyme based whole genome sequencing
InactiveUS20120245037A1Maximize possibilityMicrobiological testing/measurementLibrary member identificationWhole genome sequencingPhysical Maps
Method for de novo whole genome sequencing based on a (sequence-based) physical map of a DNA sample clone bank based on end-sequencing tagged adapter-ligated restriction fragments, in combination with sequencing adapter-ligated restriction fragments of the DNA sample wherein the recognition sequence of the restriction enzyme used in the generation of the physical map is identical to at least part of the recognition sequence of the restriction enzyme used in the generation of the DNA sample.
Owner:KEYGENE NV
Whole genome DNA (Deoxyribonucleic Acid) extraction kit for blood and method thereof
ActiveCN104017800AReduce the chance of secondary pollutionImprove accuracyDNA preparationLiquid layerLithium chloride
The invention relates to a kit of extracting a whole genome DNA (Deoxyribonucleic Acid) from blood and a using method thereof. The kit is characterized by comprising a red blood cell lysate, a white blood cell scrubbing solution, digestive juice, proteinase K, a purifying liquid, gDNA salting out liquid, a gDNA scrubbing solution, a gDNA eluant and the like. The using method of the whole genome DNA extraction kit for blood is characterized by comprising the following steps: washing the red blood cell split to obtain the white blood cell; splitting the white blood cell by the digestive juice containing the proteinase K; and further purifying by an improved lithium chloride purifying liquid, salting out the liquid layer, and carrying out chromatography to obtain the high purity whole genome DNA. When the kit provided by the invention is used to extract the whole genome DNA in blood, plasma and serum in blood are not separated in advance but fresh or frozen anti-freezing whole blood is taken, wherein the lowest blood volume required reaches 20 microliters or blood cakes are required. According t the kit provided by the invention, the whole genome DNA with high purity can be fully unlinked and the PCR (Polymerase Chain Reaction) amplification is efficiently carried out, so that the kit is used for scientific research or clinical diagnostic analysis such as PCR amplification, gene expression, gene sequencing, whole genome sequencing, exome sequencing, gene mutation and single nucleotide polymorphism.
Owner:ZICHENG RUISHENGHUI BEIJING BIOTECH DEV CO LTD
Machine learning implementation for multi-analyte assay development and testing
Systems and methods that analyze blood-based cancer diagnostic tests using multiple classes of molecules are described. The system uses machine learning (ML) to analyze multiple analytes, for example cell-free DNA, cell-free microRNA, and circulating proteins, from a biological sample. The system can use multiple assays, e.g., whole-genome sequencing, whole-genome bisulfite sequencing or EM-seq, small-RNA sequencing, and quantitative immunoassay. This can increase the sensitivity and specificity of diagnostics by exploiting independent information between signals. During operation, the system receives a biological sample, and separates a plurality of molecule classes from the sample. For a plurality of assays, the system identifies feature sets to input to a machine learning model. The system performs an assay on each molecule class and forms a feature vector from the measured values. The system inputs the feature vector into the machine learning model and obtains an output classification of whether the sample has a specified property.
Owner:FREENOME HLDG INC
Mitochondrial DNA copy index variability detecting device
ActiveCN104694384AImprove accuracyImprove reliabilityBioreactor/fermenter combinationsBiological substance pretreatmentsInformation analysisPlasma samples
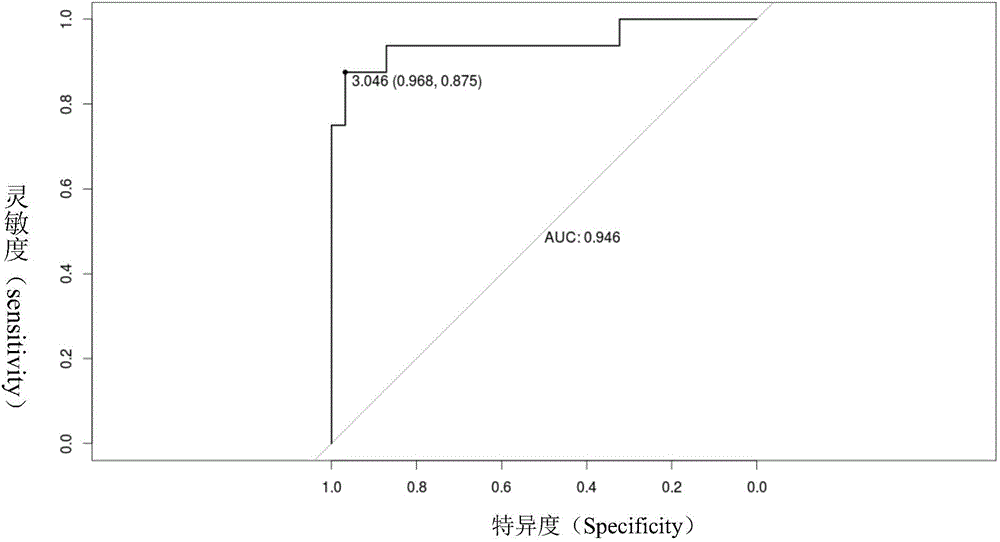
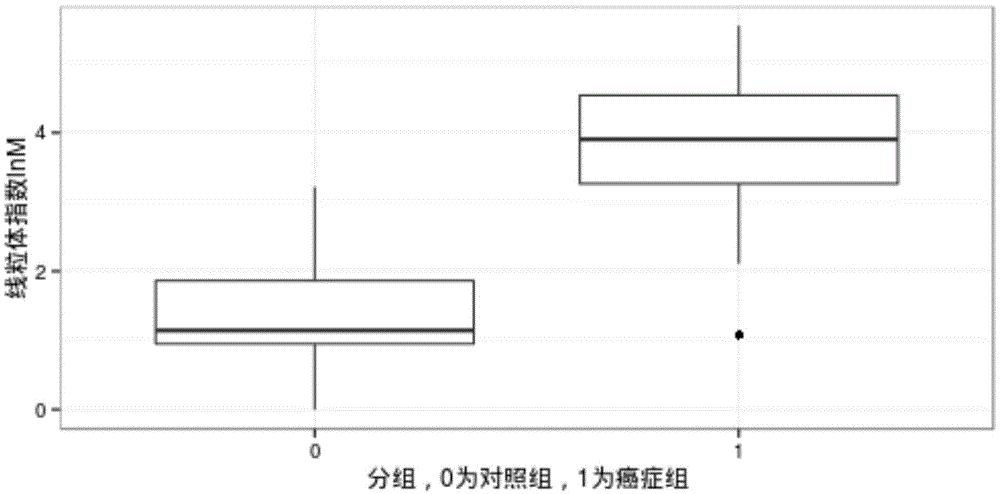
The invention relates to the technical field of medical detection, and discloses a mitochondrial DNA copy number variability detecting device. The device comprises a sequencing unit, a computing unit and a comparative judgment unit. The sequencing unit carries out whole genome sequencing on free DNA in a plasma sample of a person to be detected. The computing unit compares a sequencing result of the sample to be detected with a human reference genome and computes the mitochondrial DNA copy index of the sample to be detected. The comparative judgment unit compares the mitochondrial DNA copy index of the sample to be detected with a preset diagnosis boundary value, judges whether mitochondrial DNA copy number variability exists on the sample to be detected or not and can further judge whether the person to be detected has the cancered risk or not. According to the detecting device, mitochondrial DNA copy number abnormal conditions are recognized by combining the molecular biology sequencing technology and the biological information analysis technology and carrying out whole genome sequencing on the free DNA in blood, so that whether the person to be detected is cancered or not is judged, and early-stage noninvasive cancer screening is achieved.
Owner:SHANGHAI MAJORBIO BIO PHARM TECH
Gene scar for characterizing HRD homologous recombination repair defects and identification method
ActiveCN111883211AImprove robustnessImprove inspection accuracyMicrobiological testing/measurementSequence analysisAllele ImbalanceOriginal data
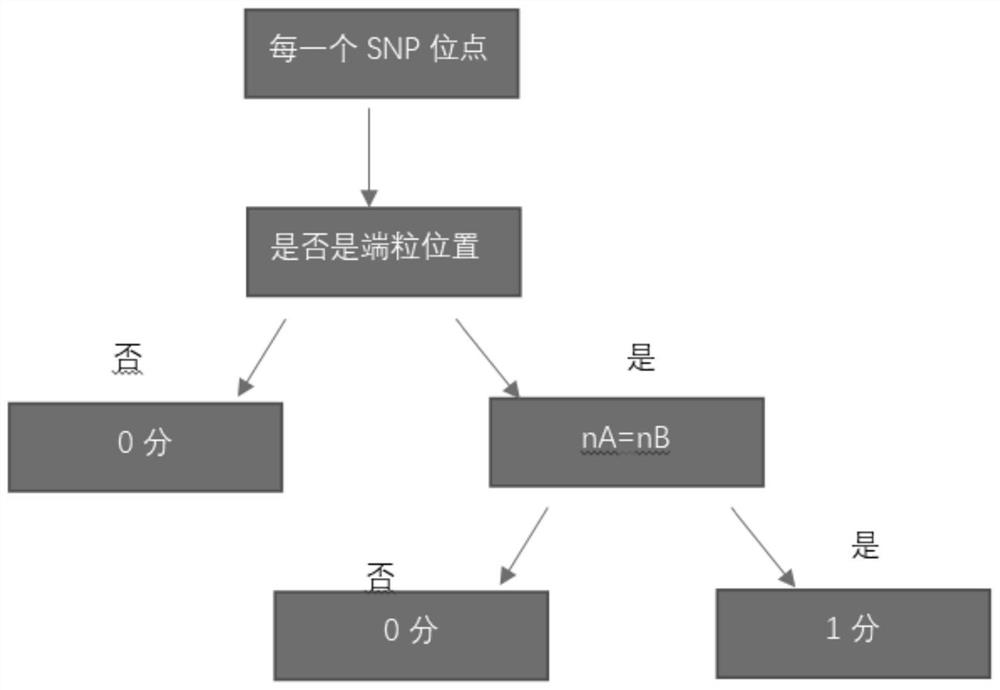
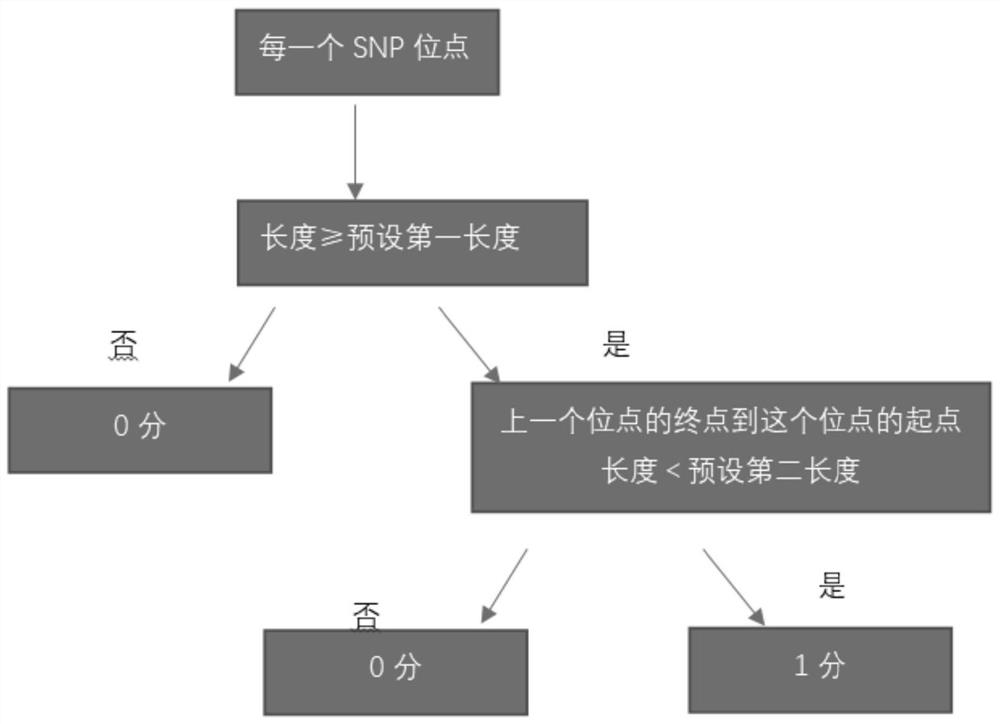
The invention relates to a gene scar for characterizing HRD homologous recombination repair defects and an identification method. The identification method comprises the following steps of step 1, performing whole genome sequencing to obtain gene original data; step 2, establishing a chromosome information matrix X (X1, X2, X3... Xn); step 3, performing data quality control; step 4, performing sequence comparison; step 5, deleting repeated reads; step 6, extracting an LRP file through signals copied by two genomes. step 7, comparing the number of high-quality non-reference bases with the number of the high-quality bases to extract a BAF file; step 8, generating fragmented LRP and BAF files for preprocessing to obtain an A allele copy number nA and a B allele copy number nB; step 9, obtaining a telomere allele imbalance TAI score through the nA and the nB; step 10, obtaining a large-scale transition LST score according to the length of each site; step 11, obtaining a heterozygosity deletion LOH score through judgment of the nA and the nB of each SNP site; and step 12, performing statistical calculation to obtain an HRD score and preforming judgment.
Owner:张哲 +1
Method for constructing environmental microbial genome draft
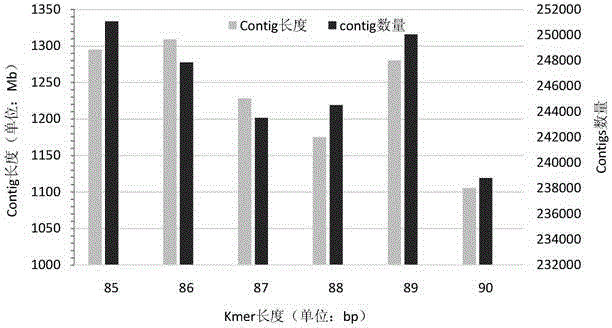
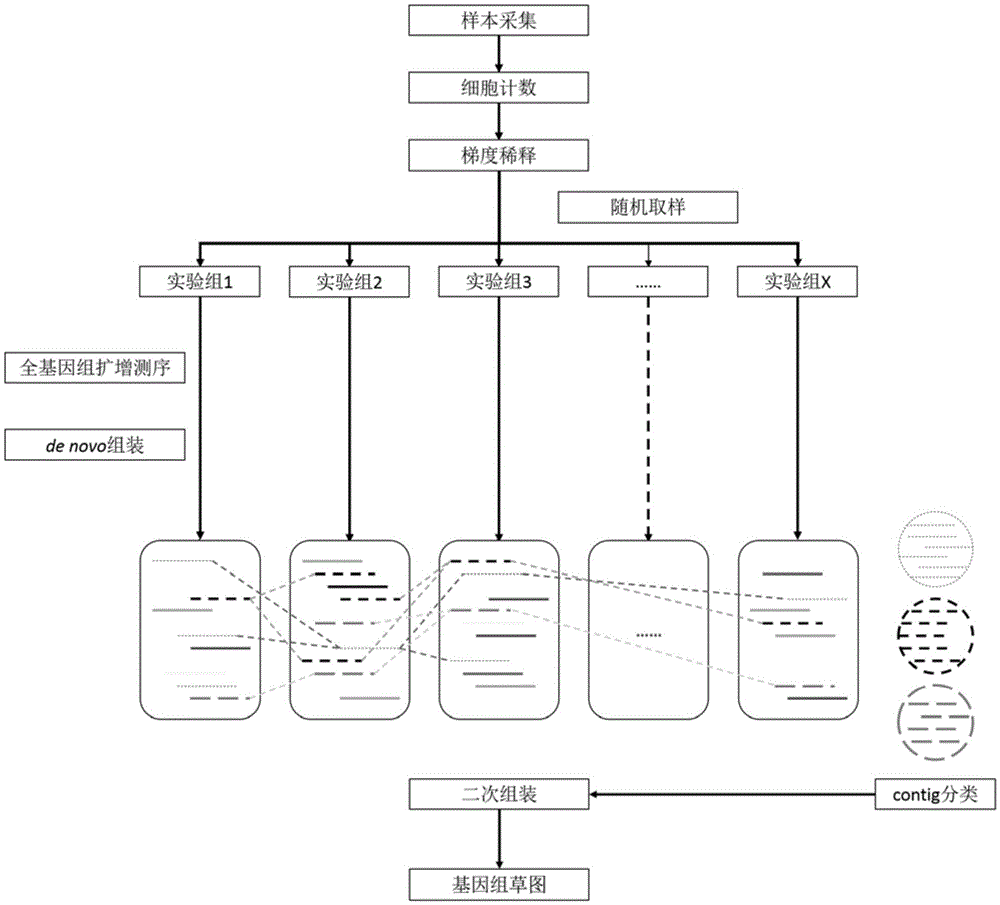
ActiveCN105420375AExcellent analysis resultsMicrobiological testing/measurementBiostatisticsMicrobial GenomesMicroorganism
The invention discloses a method for constructing an environmental microbial genome draft based on micro-cell whole genome sequencing. The method comprises the steps of conducting nucleic acid isolation and amplification on multiple environmental microorganism micro metagenome samples, constructing a high-pass sequencing library, conducting filtration, de novo assembly and open reading frame prediction on sequencing data, conducting sequence classification with the algorithm based on a Hidden Markov model, and obtaining the microbial genome draft. According to the method, the number of times of parallel tests is adjusted according to the complexity of an environment sample microflora, so that the optimal analysis result is obtained. The method can be applied to research of microflorae of various environments such as soil, air, water and human bodies.
Owner:PEKING UNIV
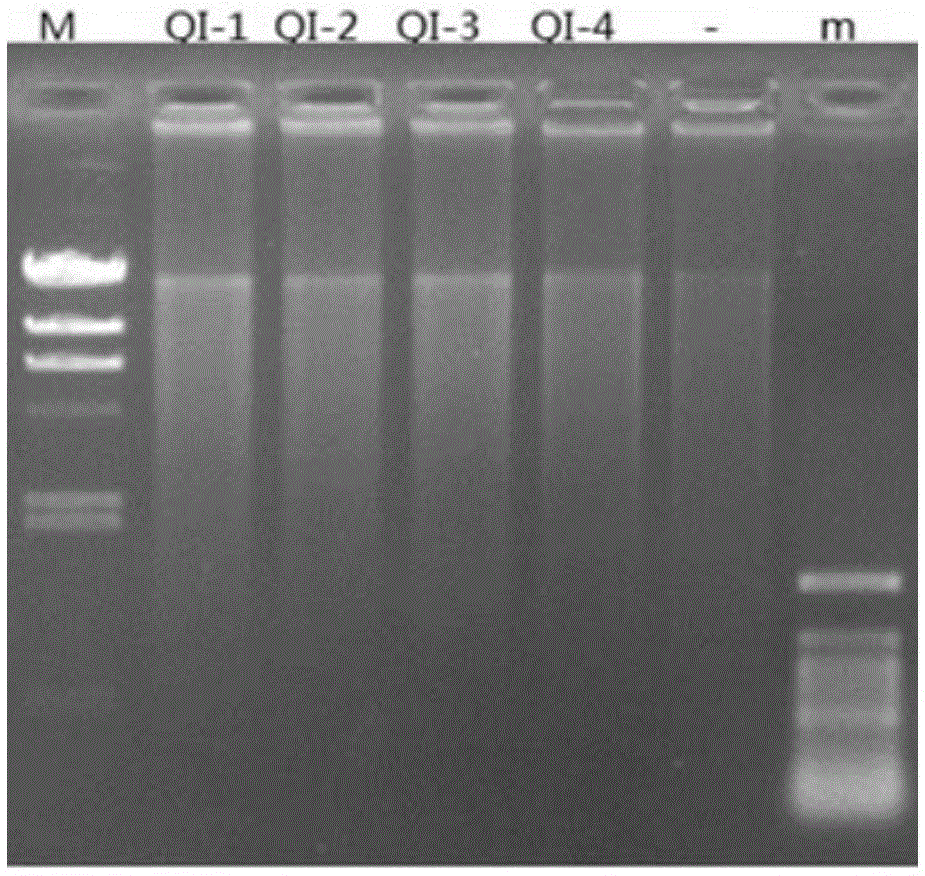
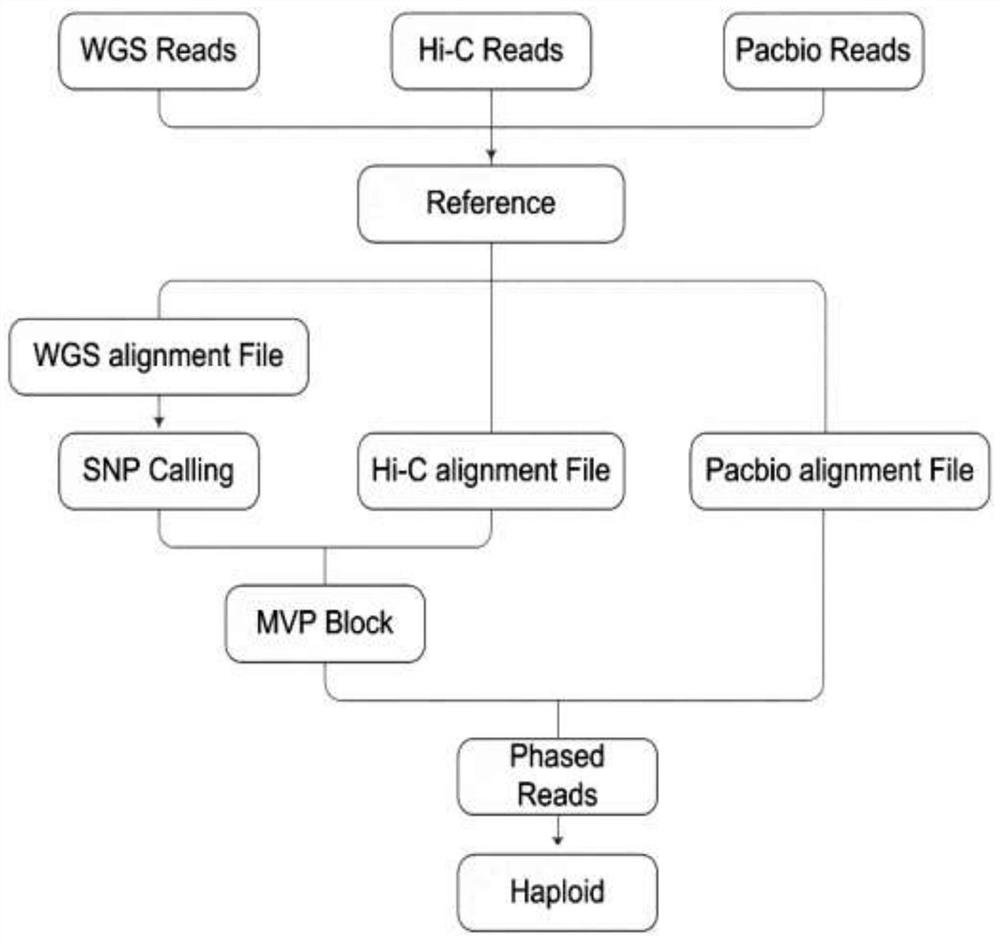
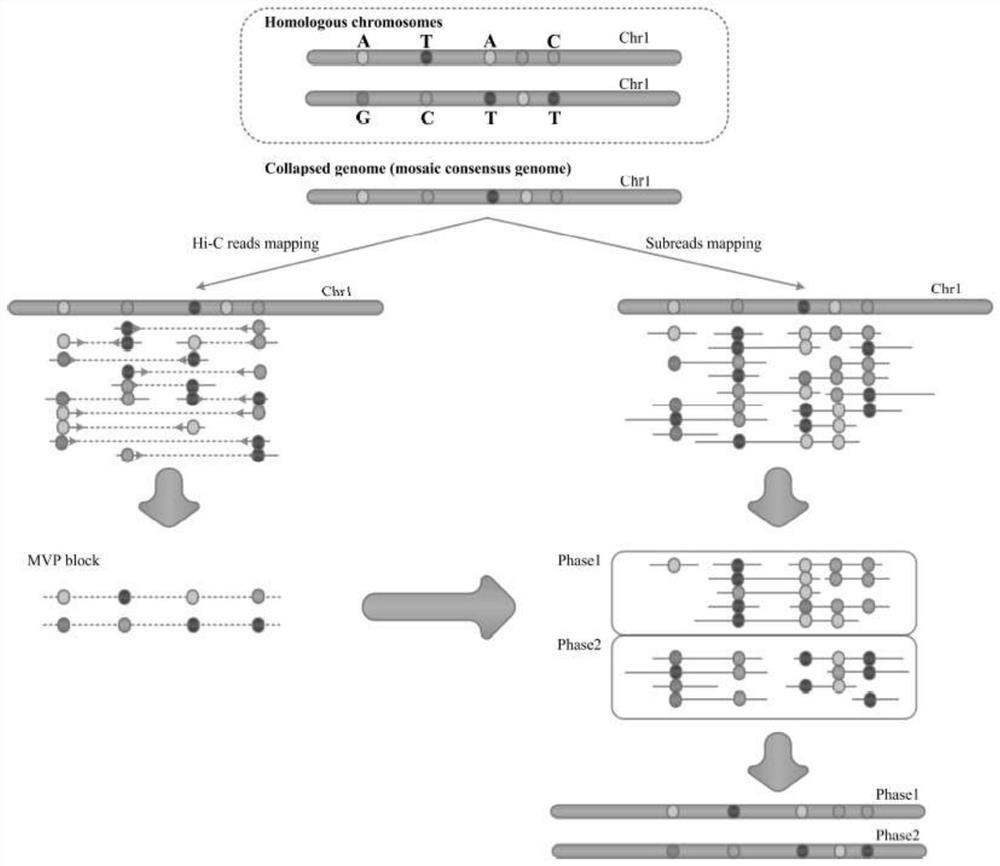
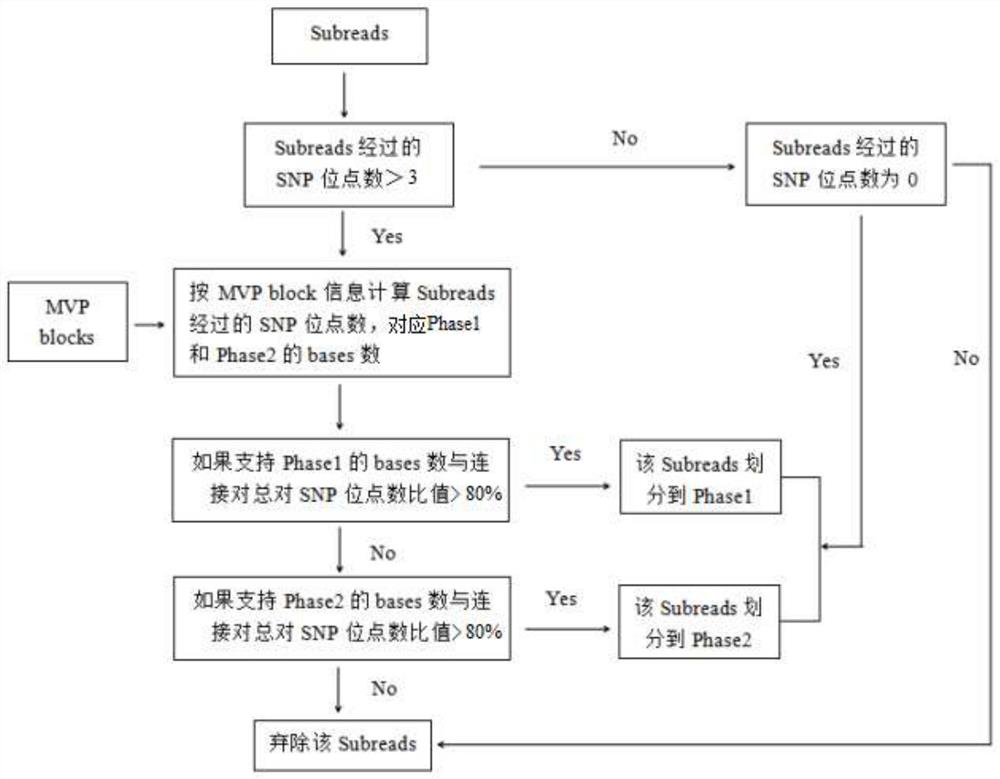
Whole genome typing method based on Pacio subreads and Hi-C reads
PendingCN111816248AGuaranteed accuracyReduced risk of errorProteomicsGenomicsRestriction Enzyme Cut SiteWhole genome sequencing
The invention relates to a whole genome typing method based on Pacbio subreads and Hi-C reads. The whole genome typing method comprises the following steps: 1) preparing a reference genome; 2) comparing second-generation sequencing data with the reference genome, and detecting all the SNP sites of each chromosome; 3) comparing Hi-C library building sequencing data with a reference genome, and building a linkage SNP group by adopting a combination of HapCUT2 with the SNP sites; 4) grouping the Pacbio subreads based on the MVP Block, and then respectively assembling the Pacbio subreads to finally obtain the sequence of each chromatid; and 5) performing whole genome sequencing on the parent genome, comparing a sequencing result to a chromatid sequence separated in the previous step, and dividing the chromatids into two groups according to a comparison result to correspond to male and female parent genomes. According to the method, the defect that contigs with too few restriction enzyme cutting sites cannot be assembled in the process of Hi-C data assembling is avoided, a linked SNP group is constructed from the aspect of whole genome, and then the Pacbio long reads are combined, so the error risk of typing is greatly reduced.
Owner:WUHAN FRASERGEN CO LTD
Method and system for determining individual chromosome structure abnormity
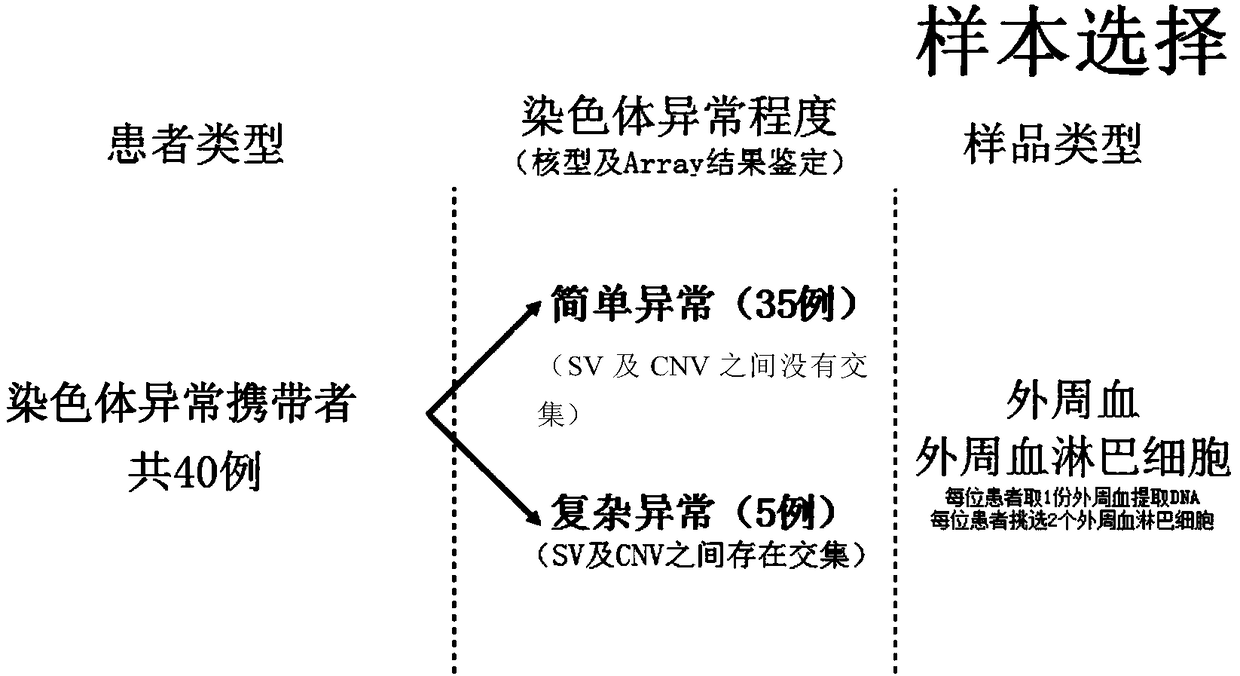
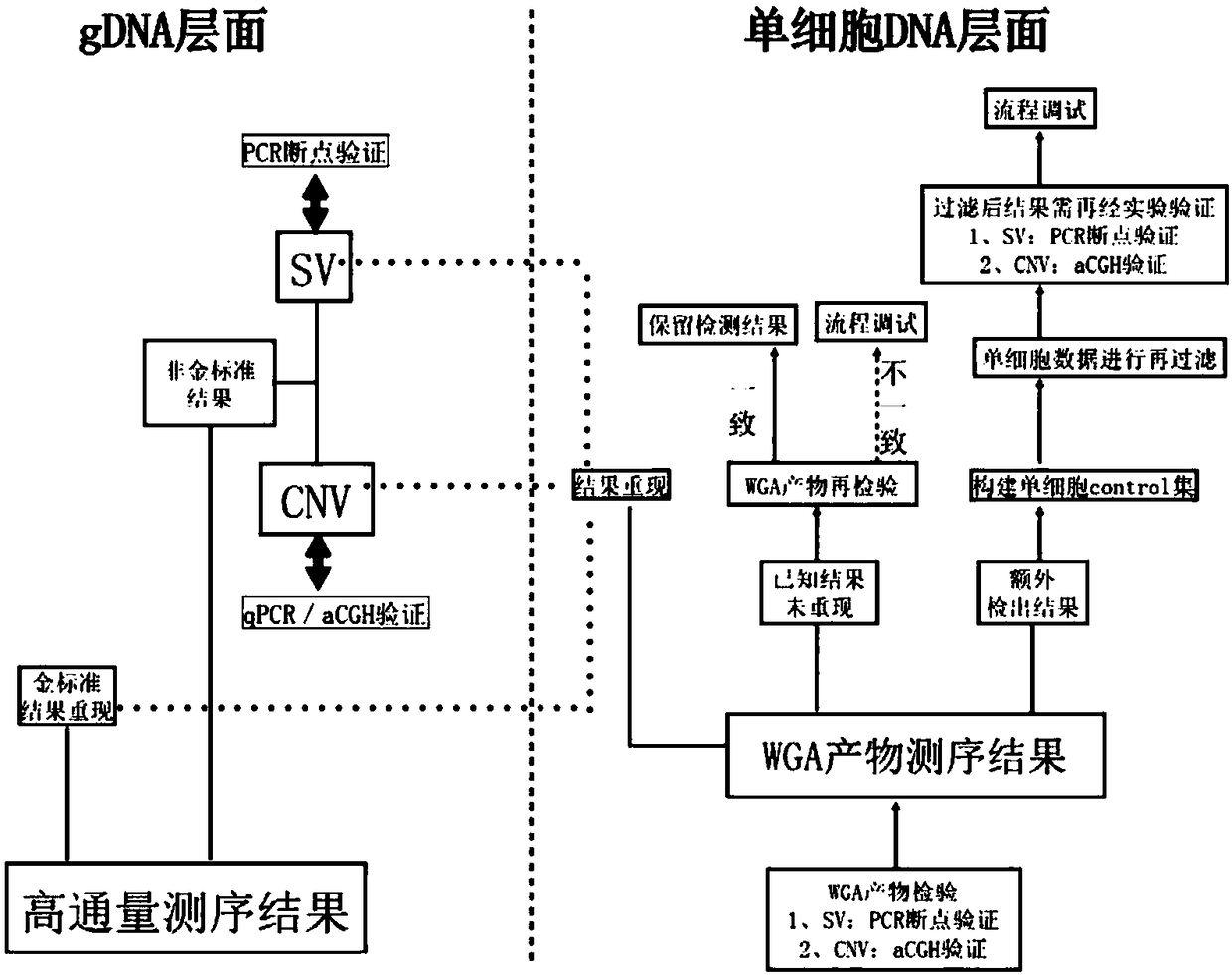
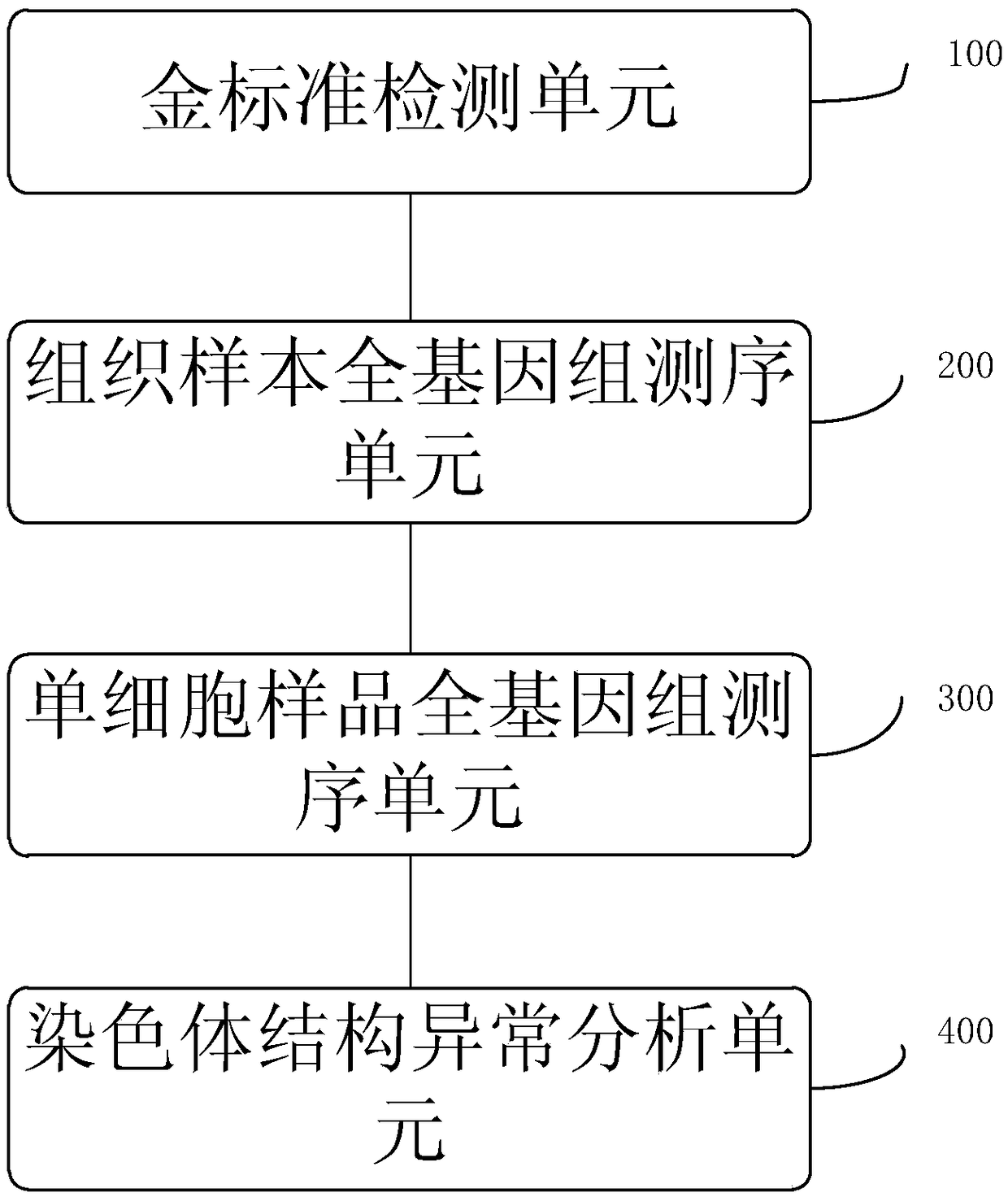
PendingCN109280702AImprove the detection rateMicrobiological testing/measurementWhole genome sequencingTissue sample
The invention provides a method for determining an individual chromosome structure abnormity. The method includes the following steps: (1) determining first candidate chromosome structure abnormity types of an individual by using at least one of karyotyping and microarray chip analysis to obtain a first candidate chromosome abnormality type set; (2) performing whole genome sequencing on tissue samples of the individual and preforming first data analysis on obtained sequencing results to obtain a second candidate chromosome abnormality type set; (3) performing whole genome sequencing on singlecell samples of the individual and preforming second data analysis on obtained sequencing results to obtain a third candidate chromosome abnormality type set; and (4) determining a final chromosome structure abnormity type of the individual based on the first candidate chromosome abnormality type set, the second candidate chromosome abnormality type set and the third candidate chromosome abnormality type set.
Owner:SHENZHEN HUADA GENE INST
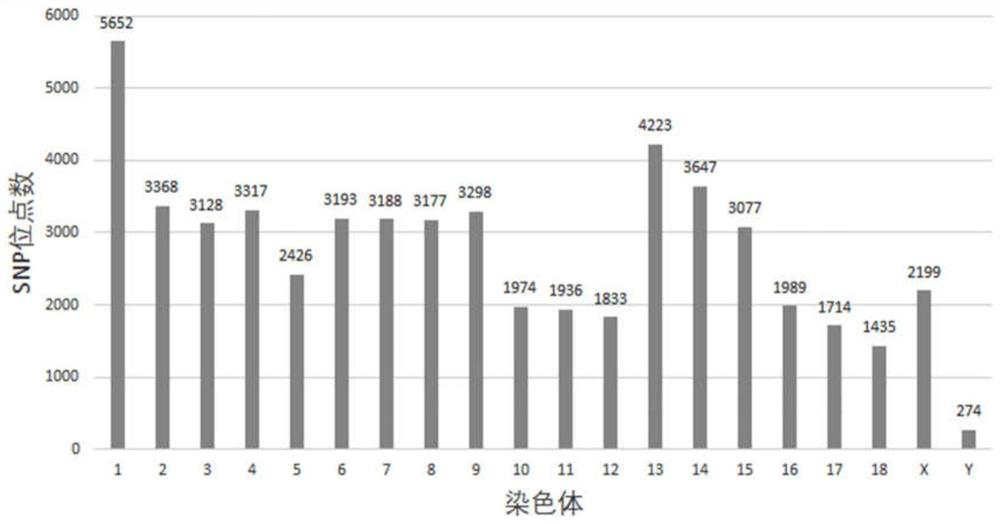
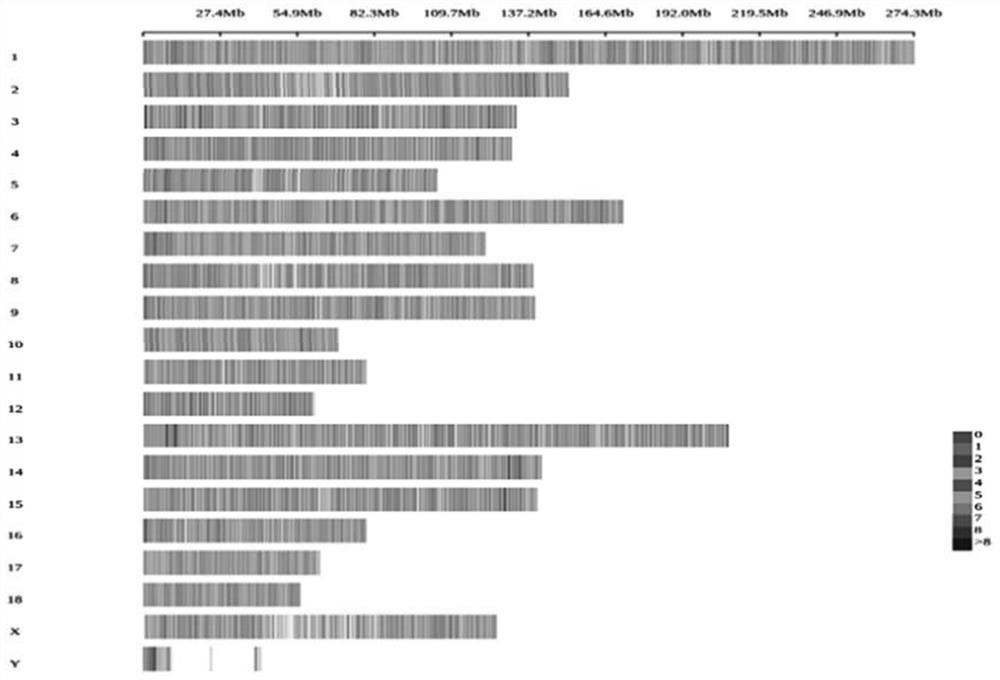
Golden snub-nosed monkey inheritance monitoring and breeding management method based on SNP site
InactiveCN107779499AHigh mutation rateWidely distributedMicrobiological testing/measurementSequence analysisWhole genome sequencingGenetic diversity
The invention discloses a golden snub-nosed monkey individual inheritance diversity monitoring method. The method includes steps of (1), collecting a golden snub-nosed monkey blood sample, and extracting gene group DNA of the blood sample; (2), performing gene group DNA sequencing by means of RAD-seq; (3), analyzing to obtain the SNP site; (4), performing genetic variation monitoring of population; (5), selecting the SNP site, and building a dynamic family relationship of a breeding group on the basis of the ties of consanguinity. By means of the acquired SNP site, the golden snub-nosed monkeyindividual inheritance diversity monitoring method can simply, exactly and comprehensively acquire the inheritance information and ties of consanguinity of the golden snub-nosed monkey individual, and is good for monitoring the inheritance diversity of the golden snub-nosed monkey breeding group for a long time and building up a dynamic breeding family pedigree. Compared with the existing inheritance marking method, the SNP site is high in mutation rate, large in site amount, and wide in distribution; more comprehensive inheritance information can be acquired, the result is more reliable; compared with a full gene group sequencing, the SNP site is simple, and data is easy to analyze.
Owner:INST OF FOREST ECOLOGY ENVIRONMENT & PROTECTION CHINESE ACAD OF FORESTRY
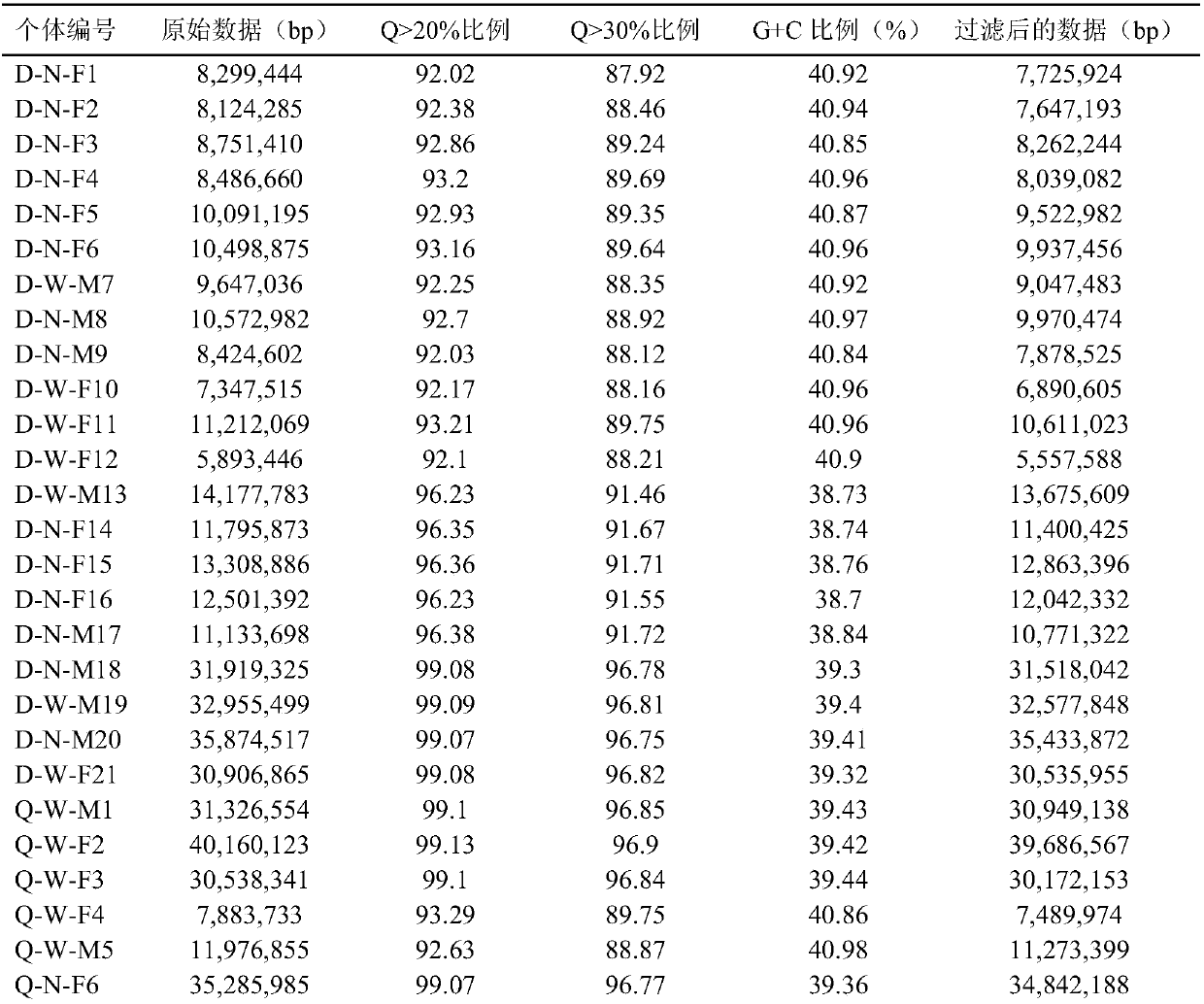
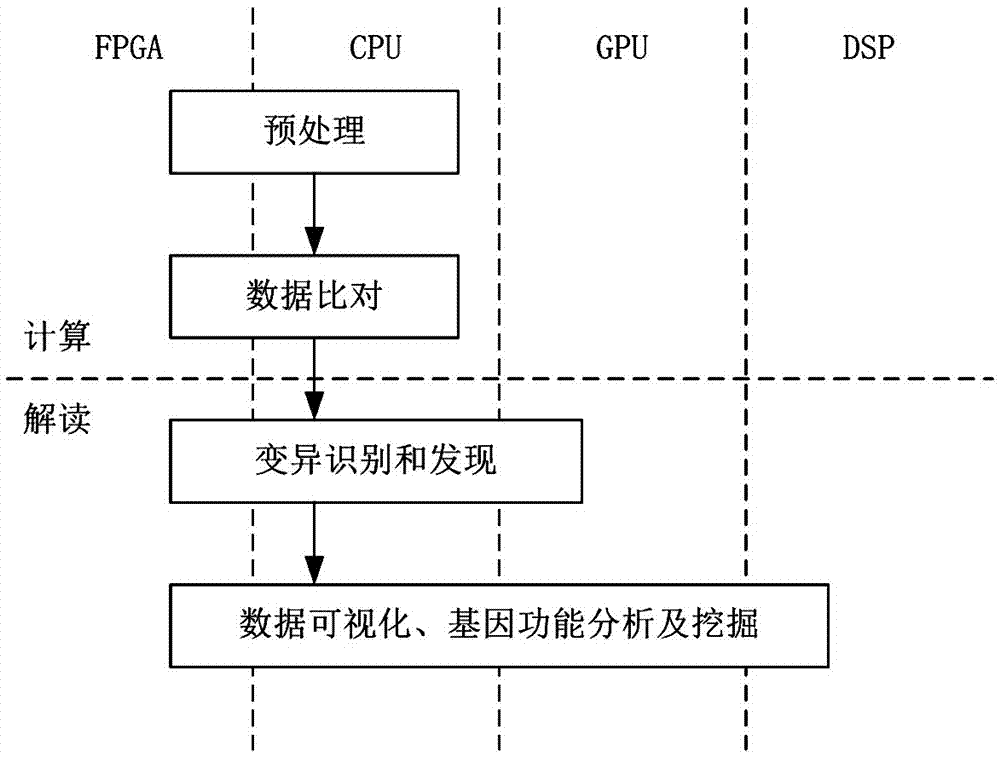
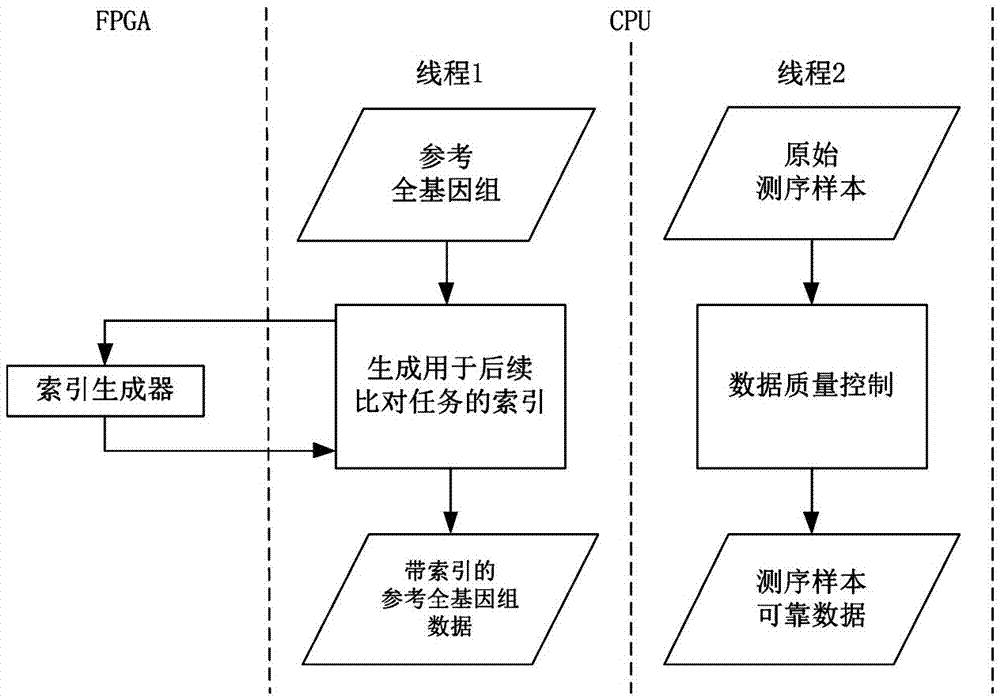
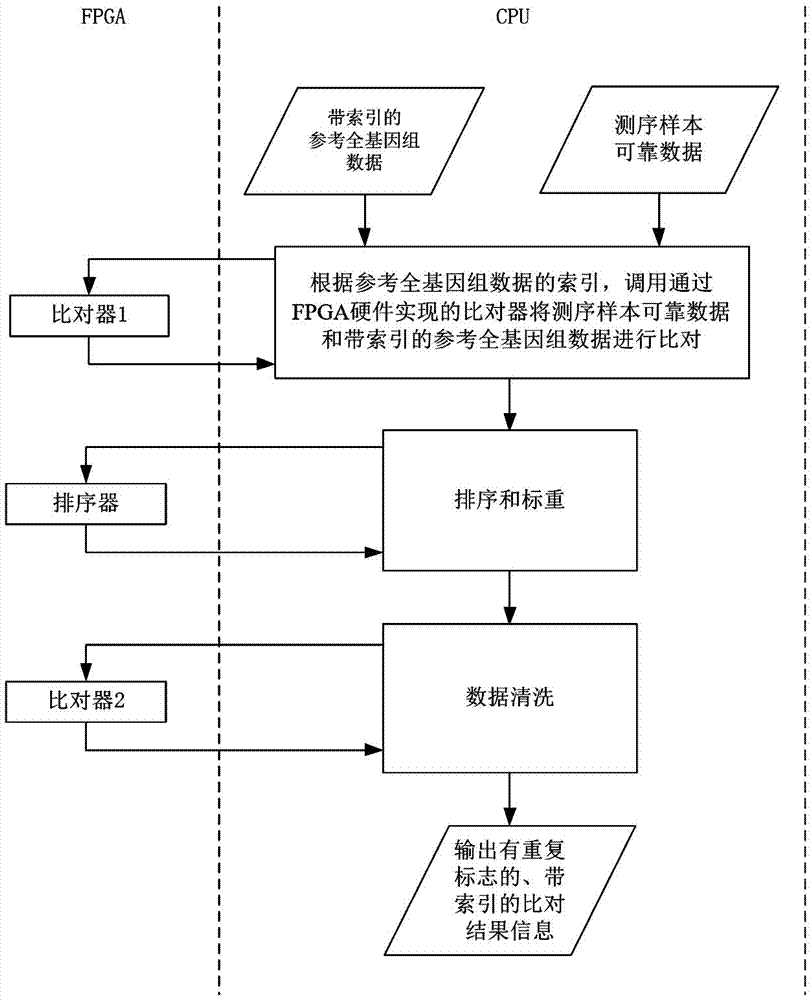
Whole genome sequencing data calculation interpretation method
InactiveCN107194204AImprove real-time performanceAdd depthBiostatisticsSequence analysisWhole genome sequencingGenomic data
The invention discloses a whole genome sequencing data calculation interpretation method, which comprises the following implementation steps that: inputting reference whole genome data used for whole genome sequencing and organic sequencing sample data, and carrying out preprocessing; calling an FPGA (Field Programmable Gate Array) by a CPU (Central Processing Unit) to be accelerated to compare sequencing sample reliable data with the reference whole genome data with an index to obtain a comparison result with a repeated sign and the index; calling the FPGA and a GPU (Graphics Processing Unit) by the CPU to be accelerated to carry out genome reassembling on the sequencing sample reliable data, and carrying out variant identification recognition on the comparison result with the repeated label and the index; and calling the GPU and a DSP (Digital Signal Processor) by the CPU to be accelerated to carry out visual processing, and calling a deep learning model realized by hardware on the FPGA by the CPU to carry out the analysis and the mining of the whole genome and a variant function on the basis of a visual processing result. By use of the method, the GPU, DSP and FPGA processors can be comprehensively utilized for acceleration, and the method has the advantages of being quick, real-time, accurate in penetration, popular and easy in understanding and varied in forms.
Owner:GENETALKS BIO TECH CHANGSHA CO LTD
Pig 50K liquid phase chip based on targeted capture sequencing, and application thereof
ActiveCN112921076AAvoid uniformityGuaranteed specific capture rateNucleotide librariesMicrobiological testing/measurementPig farmsNucleotide
The invention relates to a pig 50K liquid phase chip based on targeted capture sequencing, and application thereof. The pig 50K liquid phase chip is composed of pig 50K probe mixed liquor and a hybridization capture reagent which are independently packaged; pig 50K probes are DNA double-stranded probes and are nucleotide sequences designed and synthesized according to screened SNP sites; and the SNP sites are screened by comparing the sequencing results of the whole genome of the pig to a pig reference genome. The pig 50K liquid phase chip prepared by the invention is mixed with a pig DNA high-throughput sequencing library; DNA fragments containing target sites in the pig DNA high-throughput sequencing library are captured for amplification and purification; and, after a product is subjected to high-throughput sequencing, a sequencing result is replied to a pig reference genome for comparison, so that the genotyping of the genome of a to-be-detected pig is obtained. When the 50K liquid phase chip prepared by the invention is used for genotyping pigs; and the problems of high cost, inflexibility and incapability of large-scale use in pig farms in China in the prior art can be solved.
Owner:CHINA AGRI UNIV
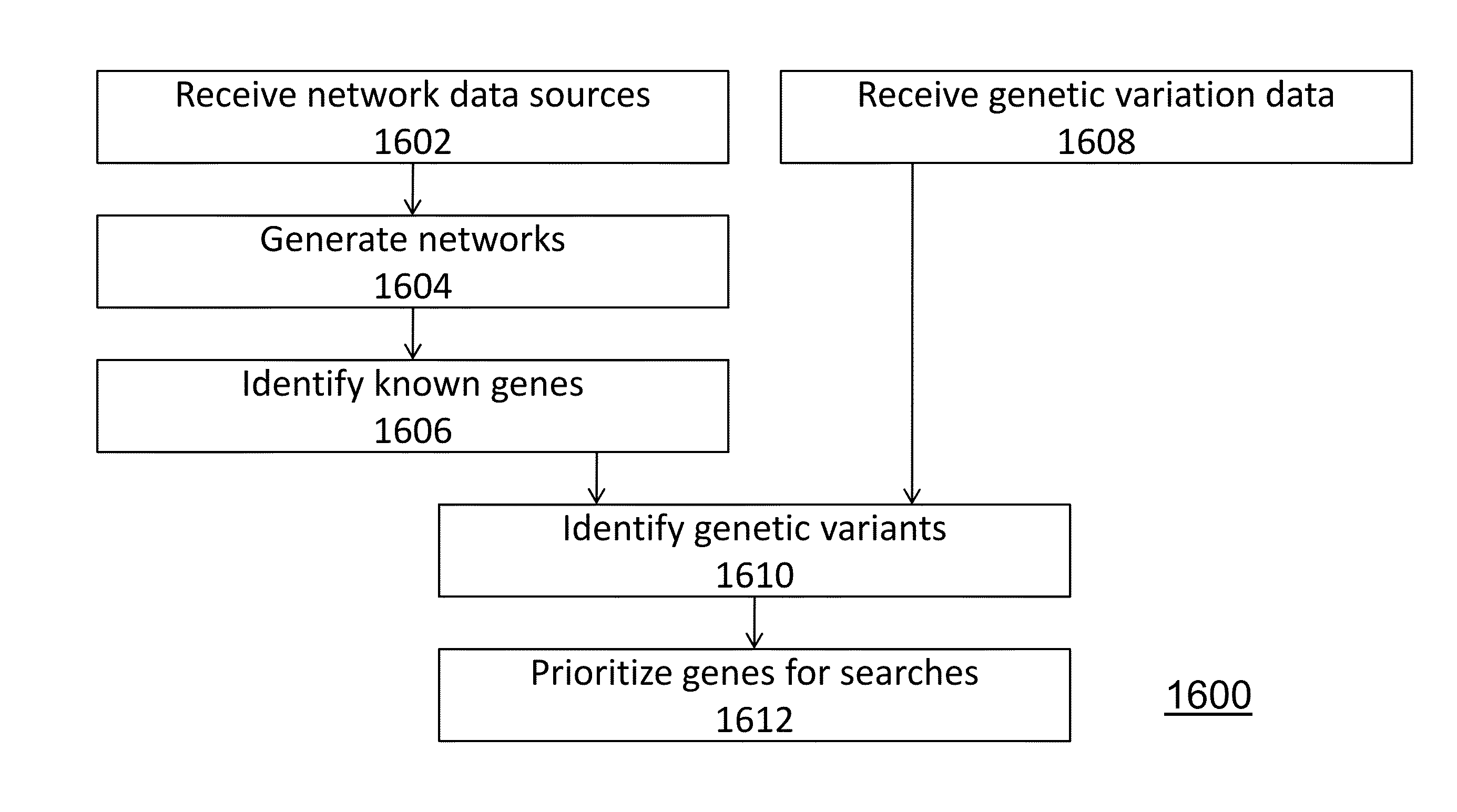
Method and system for network modeling to enlarge the search space of candidate genes for diseases
ActiveUS20130090908A1Analogue computers for chemical processesSystems biologyGenomic sequencingCandidate Gene Association Study
With the advent of low cost, high-throughput whole genome sequencing (“next generation sequencing”), tools are available to assay human genetic variation contributing to inherited disease syndromes. A method is disclosed for prioritization of genetic variants, and identification of disease genes, using network modeling of gene associations.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Cotton pachytene chromosome fluorescence in-situ hybridization method
InactiveCN1995396AEnhance functional geneImprove detection efficiencyMicrobiological testing/measurementFluorescence/phosphorescenceGenomic sequencingAgricultural science
The invention discloses a fluorescent home position hybrid technique, which is characterized by the following: adopting chromosome in the pachytene as target DNA to proceed fluorescent home position hybrid; drawing high-density physical atlas; fitting for separating cloning order gene; measuring sequence of full genome of cotton; improving the locating measuring efficiency of functional gene and molecular mark for cotton effectively; adopting chromosome in the pachytene with two homologous chromosomes to avoid the identifying defect; improving distinguishing efficiency greatly.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Biological marker relevant to vascular malformation and relevant detection reagent kit
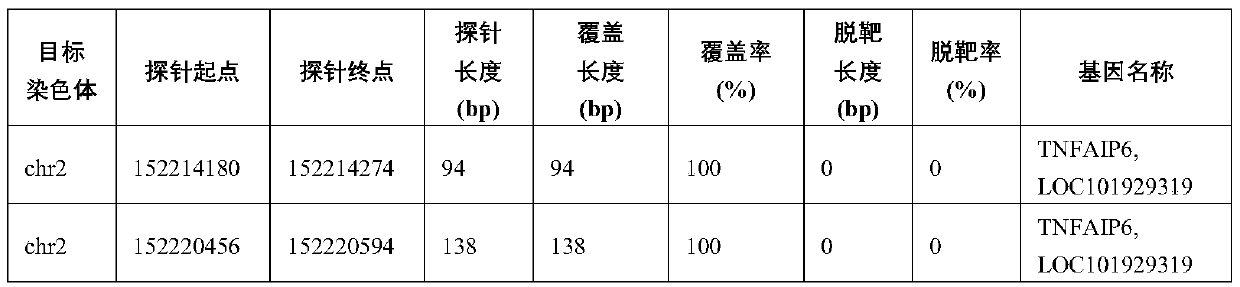
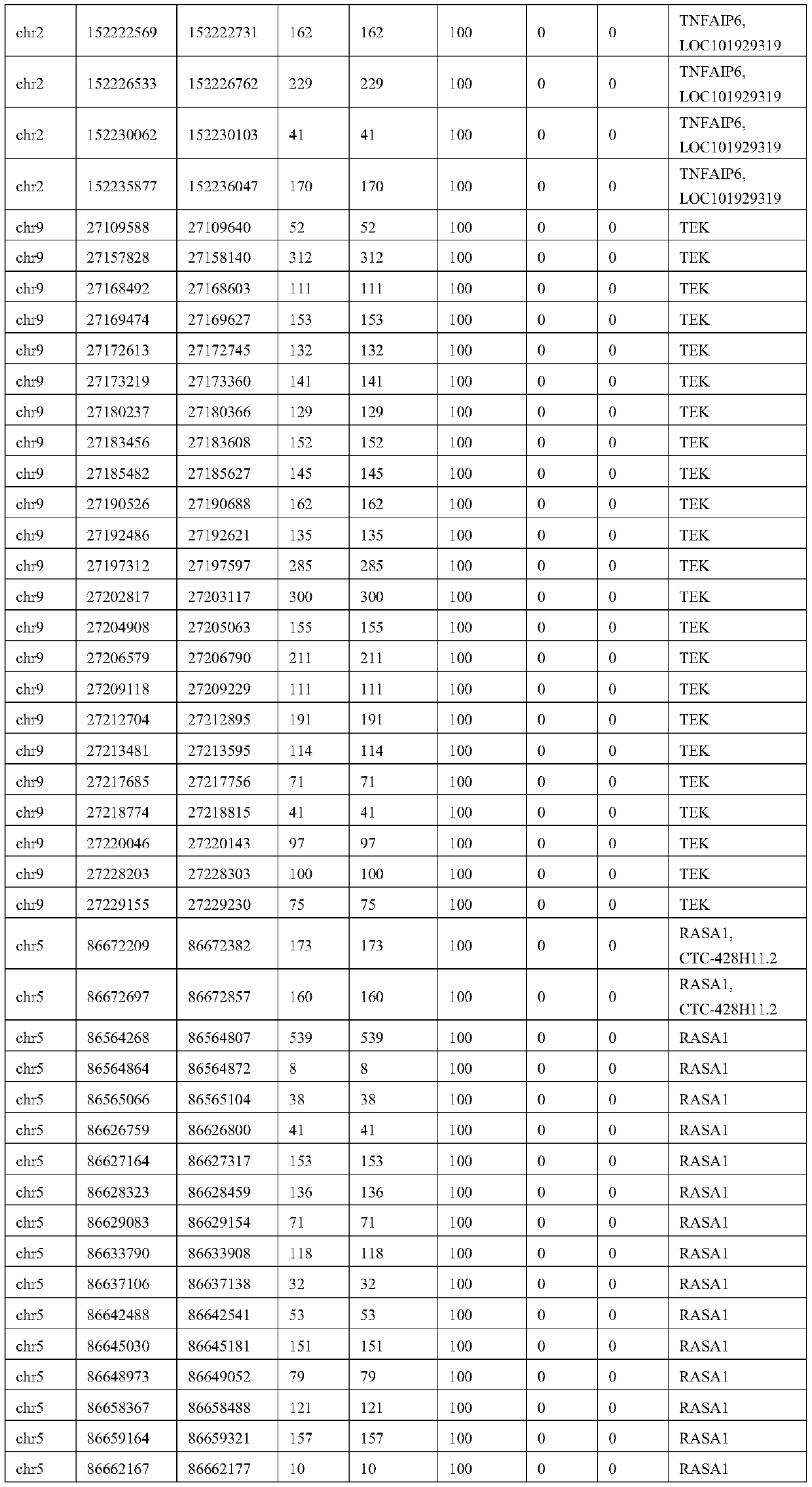
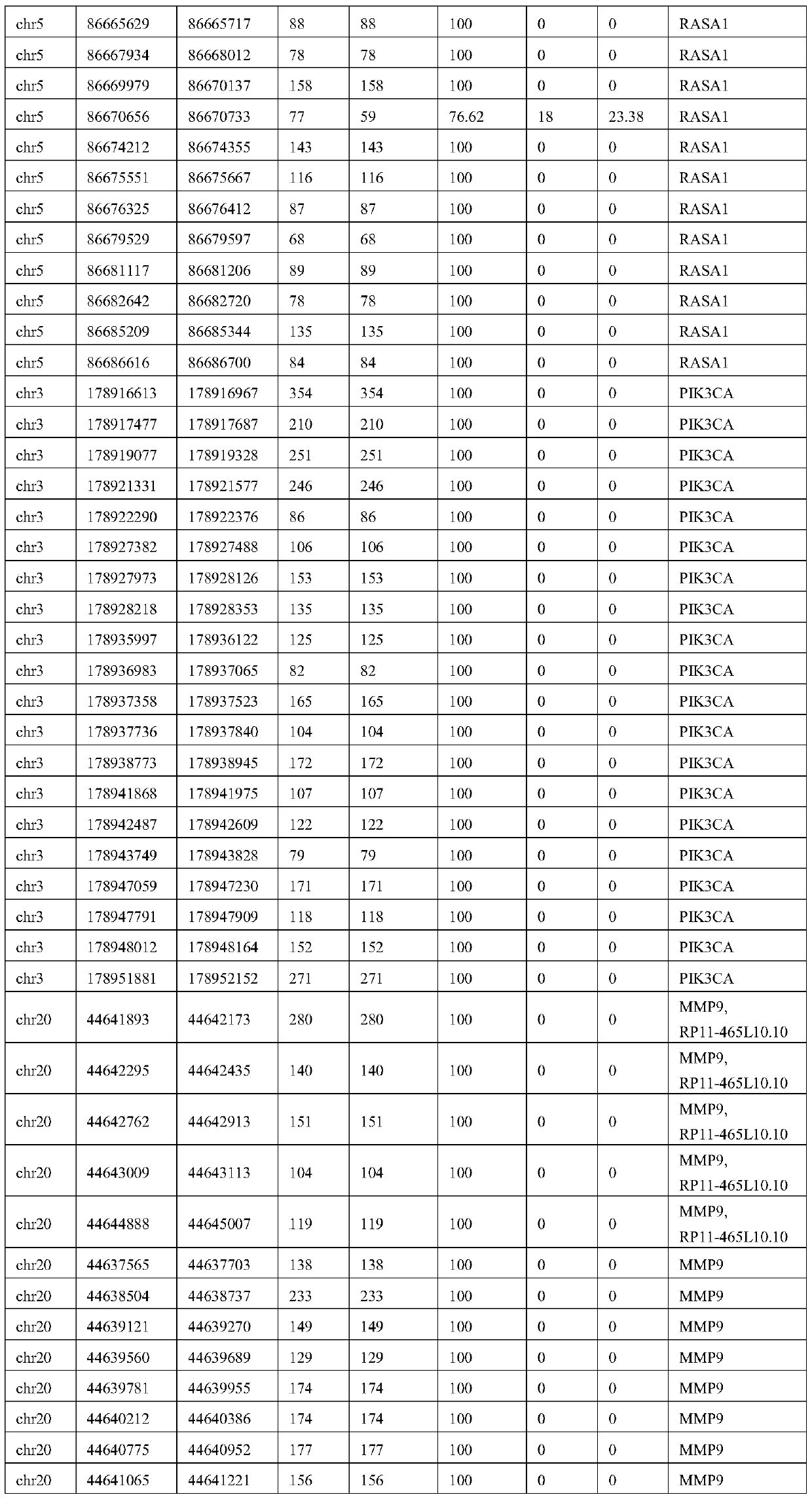
ActiveCN109852689AMicrobiological testing/measurementDNA/RNA fragmentationVascular diseaseWhole genome sequencing
The invention relates to the technical field of biology, in particular to a set of biological markers and a relevant detection reagent kit. The invention provides the use of a combination of a substance for detecting Q346P mutation in a TEK gene or an active fragment thereof, a substance for detecting Q279R / R574P mutation in a MMP9 gene or an active fragment thereof, a substance for detecting Y1213Y mutation in a FLT1 gene or an active fragment thereof, and a substance for detecting X278X mutation in a TNFAIP6 gene or an active fragment thereof in a prepared reagent kit. The reagent kit is used for diagnosing vascular diseases and / or assessing the risk of the vascular diseases. The biological marker provided by the invention can avoid complete genome sequencing, so that needed sequencing data number can be greatly reduced, and the biological marker has excellent detection sensibility and specificity, and is low in cost, high in depth and accurate in detection on relevant mutation.
Owner:SHANGHAI NINTH PEOPLES HOSPITAL SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com