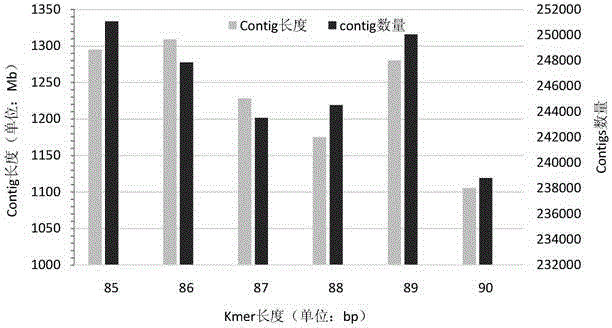
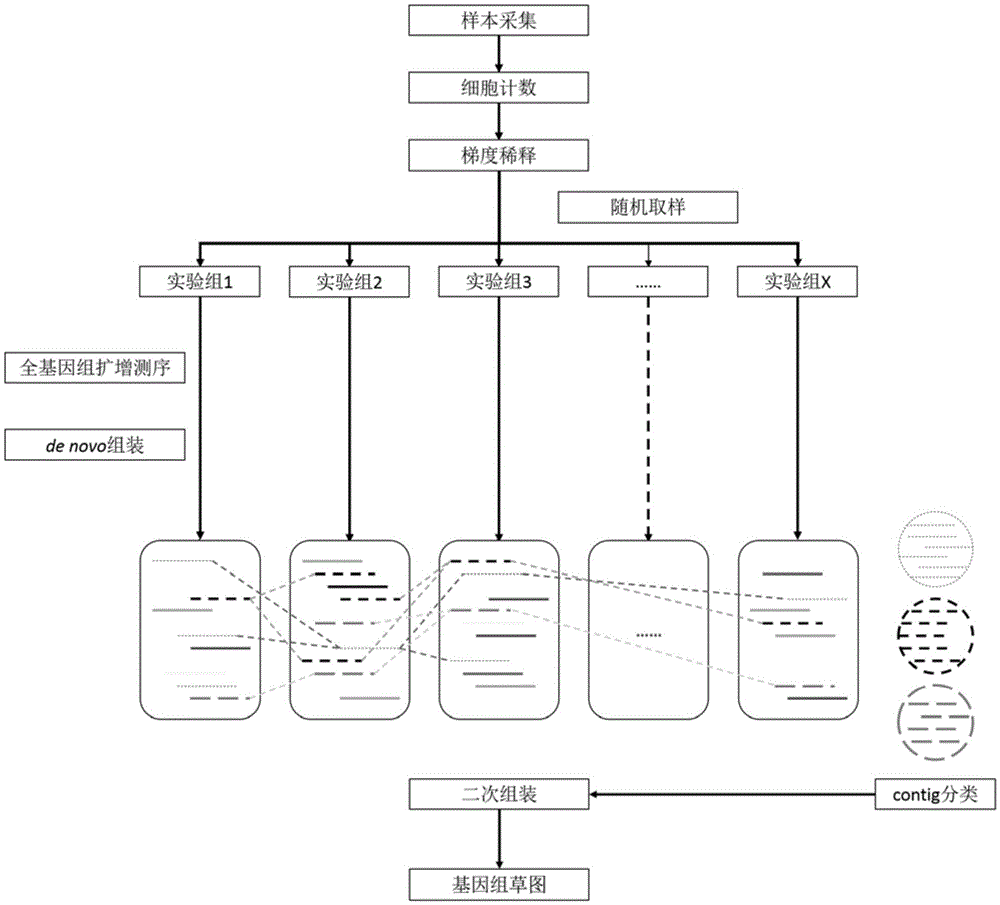
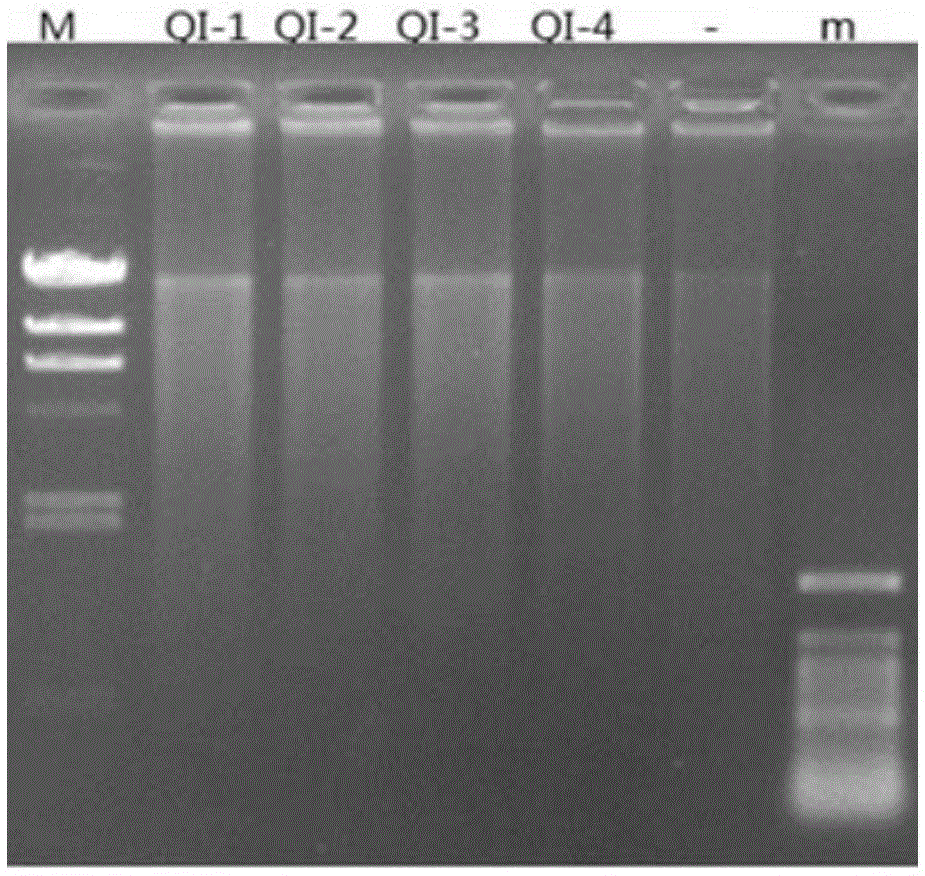
Method for constructing environmental microbial genome draft
An environmental microorganism and construction method technology, applied in the field of construction of environmental microorganism genome drafts, can solve problems such as inability to explain species and species, inability to directly analyze and assemble the genome sequence information of different individual microorganisms, incomplete microbial species genome information, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1. Construction of the Draft Human Gut Microbe Genome
[0071] 1. Stool sample processing
[0072] (1) Take 0.5g of feces, put it in a sterile 15mL centrifuge tube, add 5mL of PBS buffer, vortex shake and mix, centrifuge at 700g for 1min, and take the supernatant;
[0073] (2) Dilute the bacterial solution in a gradient, observe under a microscope, select an appropriate dilution factor, and use a hemocytometer for counting. Set α to 1 / 1000 to obtain all microorganisms with an abundance higher than 1 / 1000 in the stool sample For the draft genome, according to formula 1, determine that the final number of cells N used in a single reaction is 50, and the number of parallel experiments X is 100;
[0074] 2. Whole genome amplification sequencing
[0075] Whole genome amplification was performed according to the Qiagen REPLI-gSingleCellkit protocol.
[0076] (1) Cell lysis: Take 1 μL of diluted bacterial solution (50cells / μL) into a sterile 0.2mL PCR tube, add 3 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com