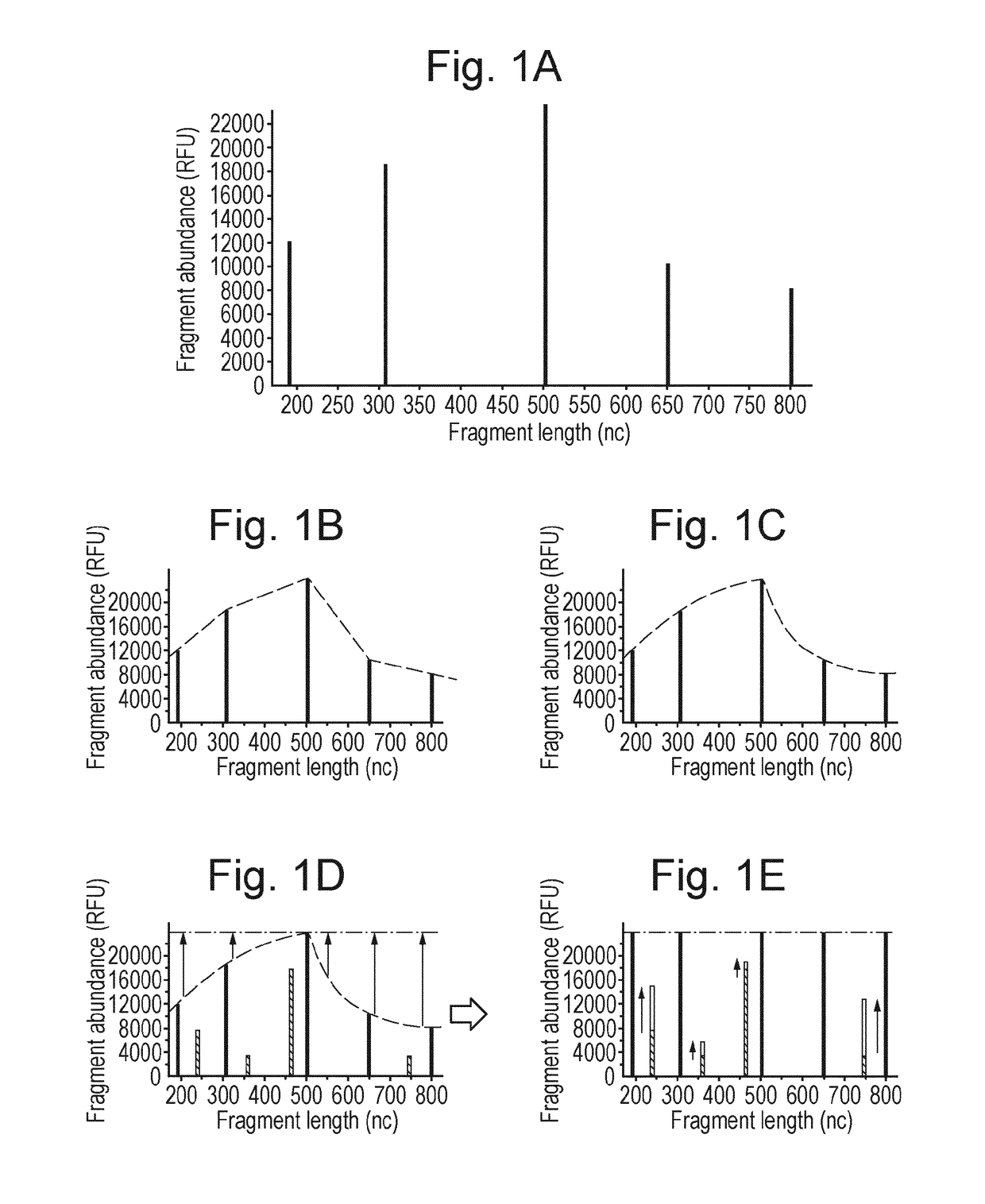
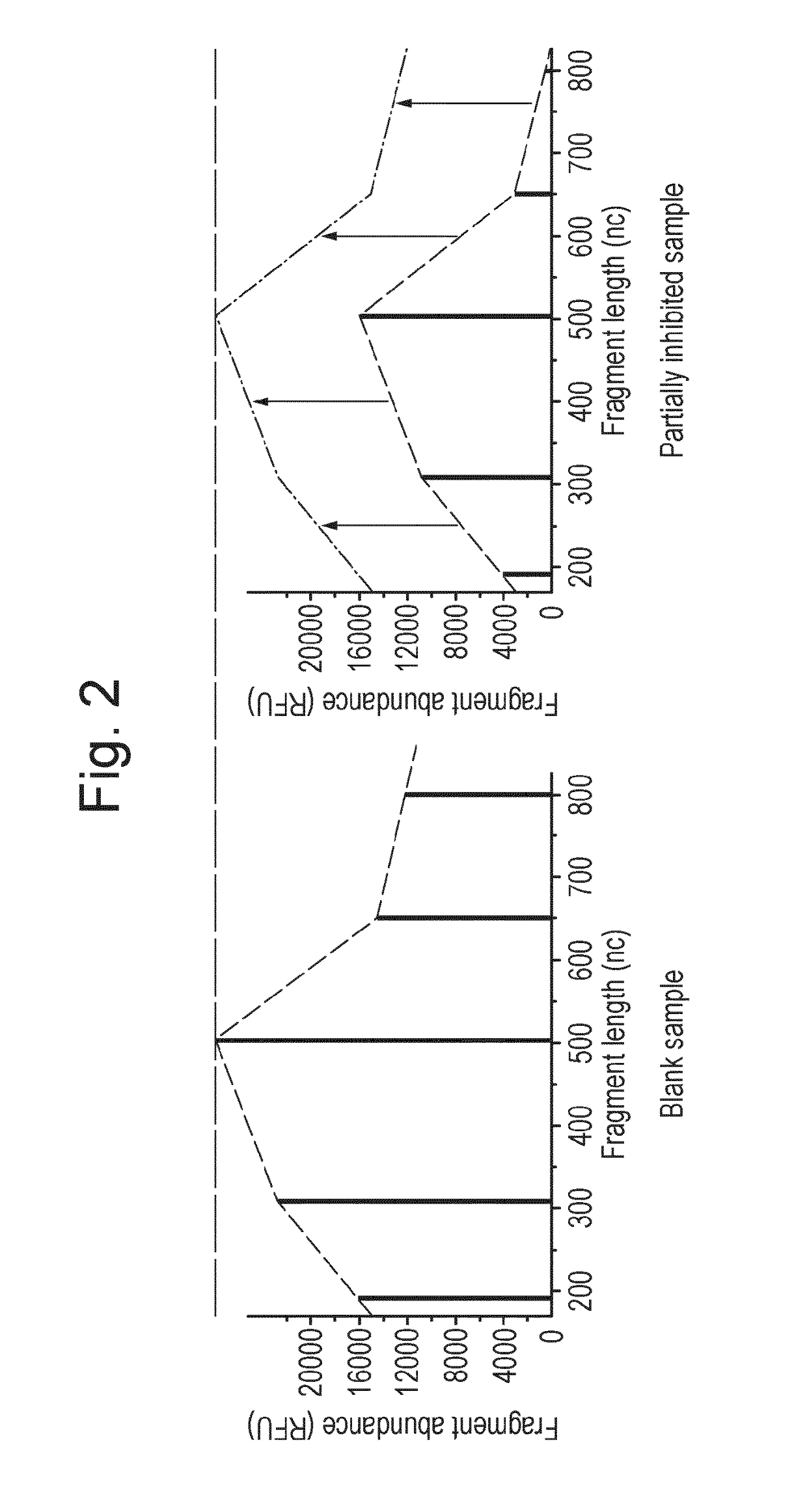
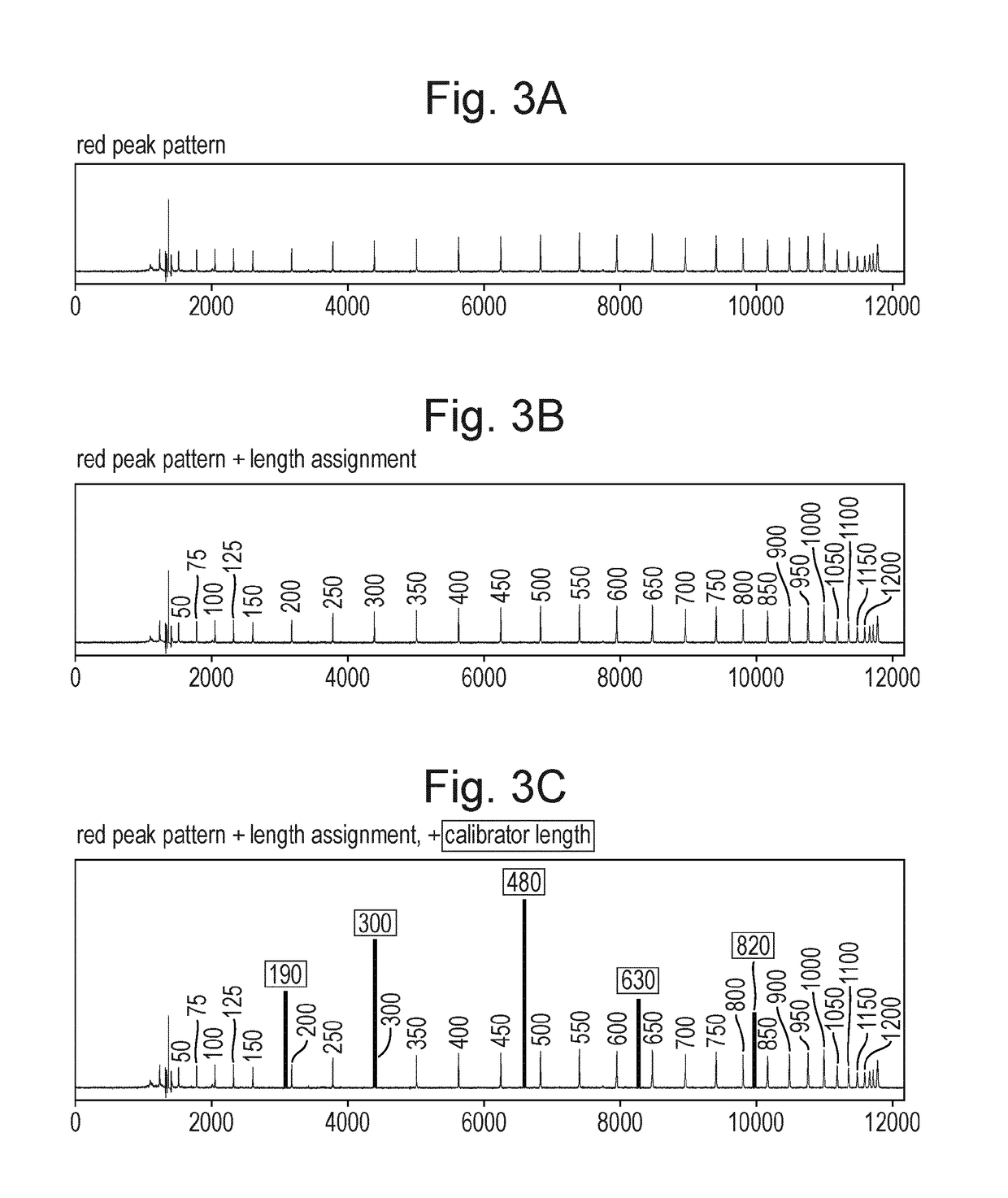
Patents
Literature
120 results about "Microbial genome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
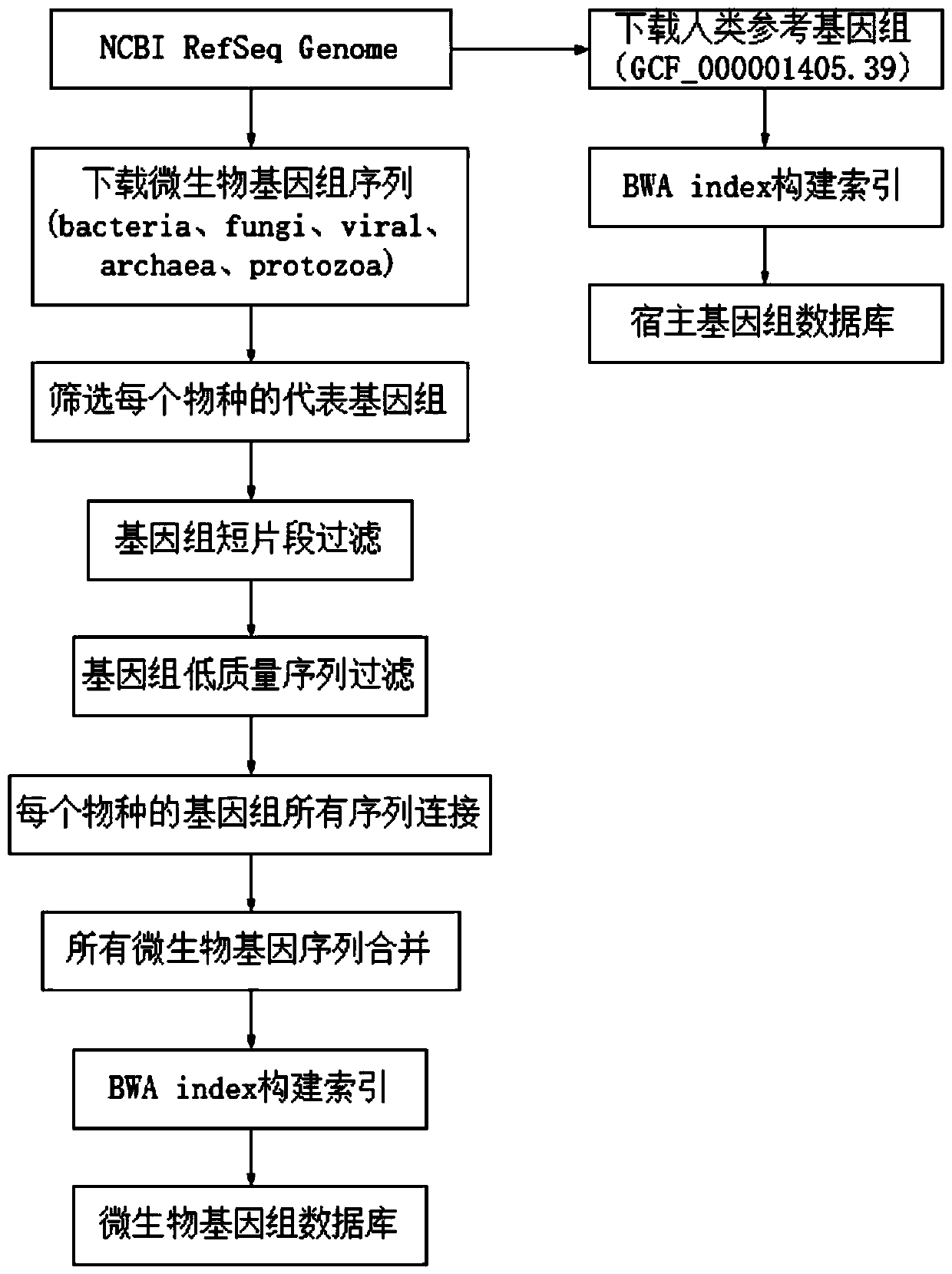
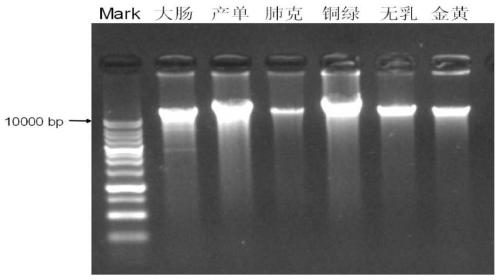
Pathogenic microorganism genome database and establishment method thereof
ActiveCN110473594AReduce data volumeAvoid false positivesBioinformaticsInstrumentsPathogenic microorganismHemidesmosome assembly
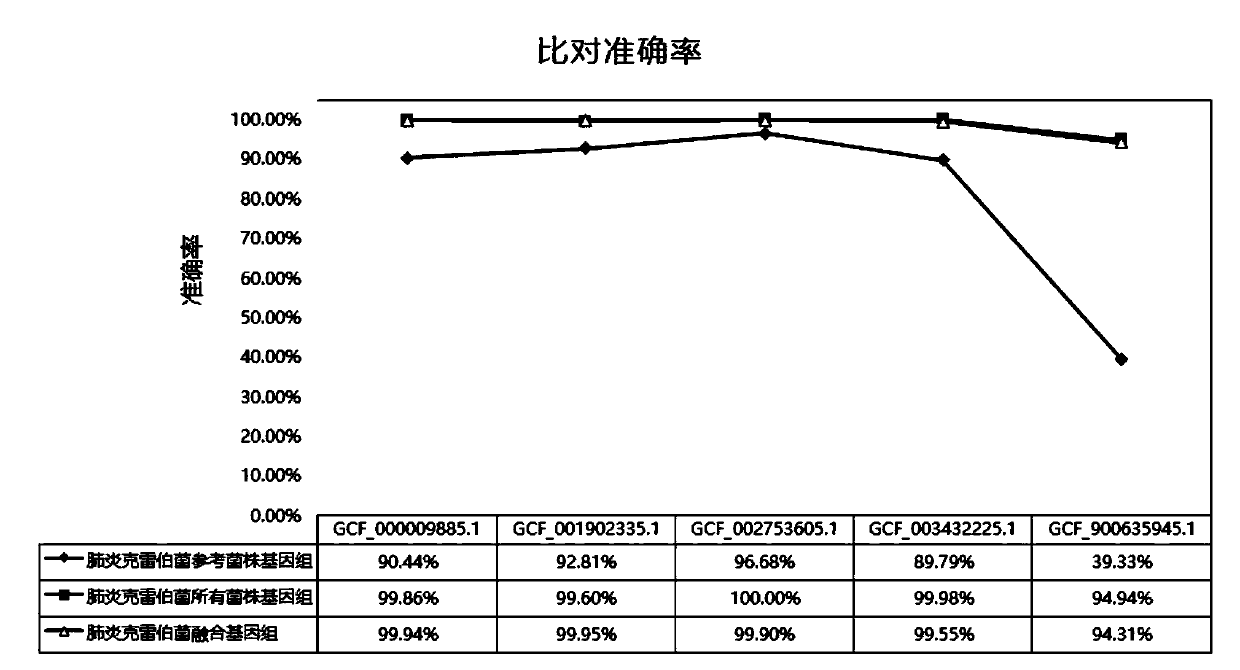
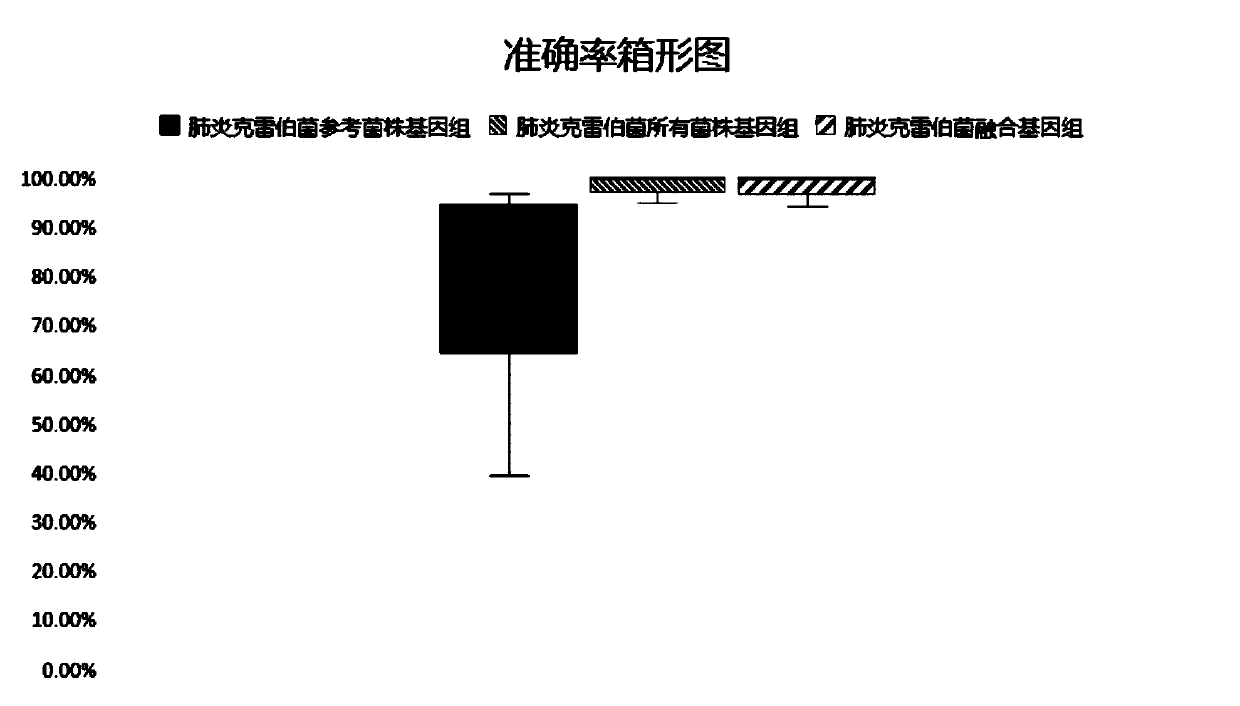
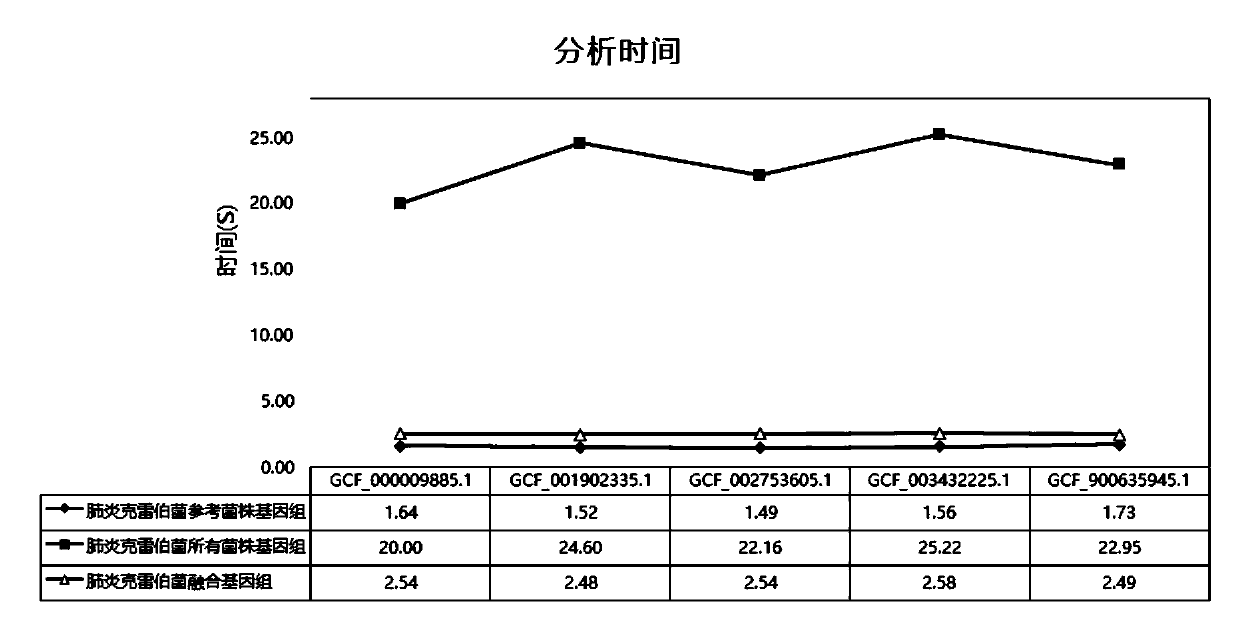
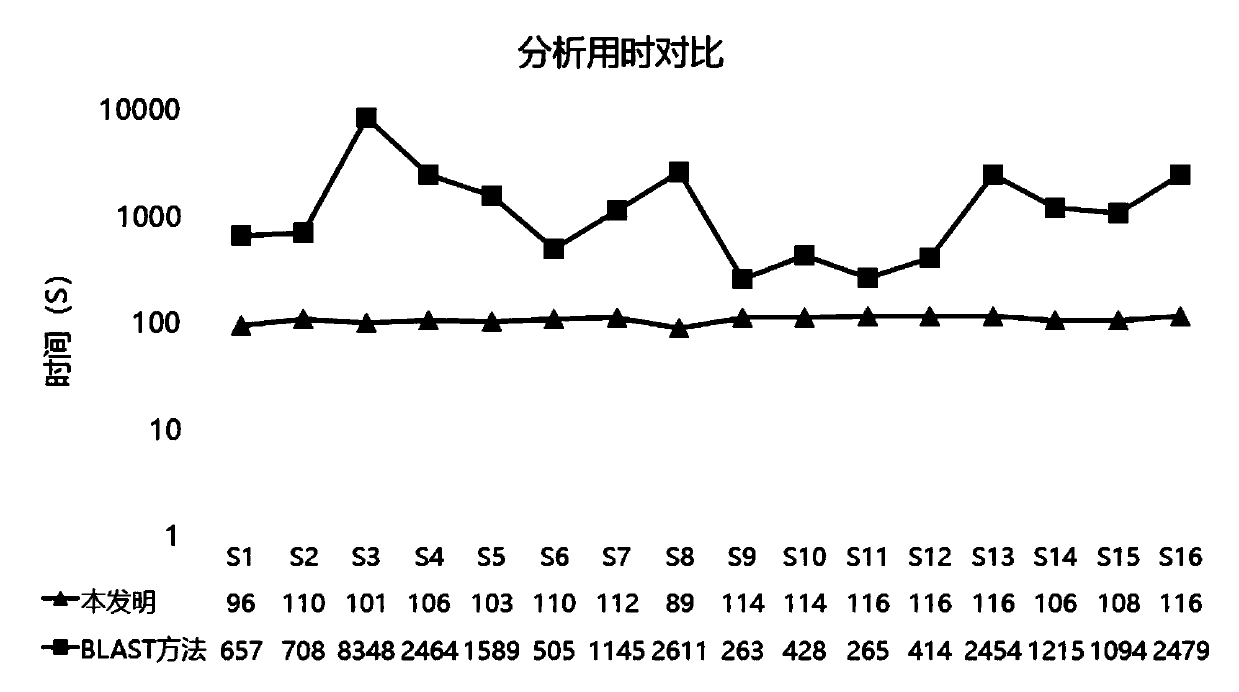
The invention relates to a pathogenic microorganism genome database and an establishment method thereof, and belongs to the technical field of meta-genomes. The method comprises the following steps ofdata acquisition, wherein pathogenic microorganism genome data is obtained; strain genome screening, wherein species strain genomes are selected according to a predetermined screening rule; plasmid sequence removal, wherein plasmid sequences existing in the strain genomes obtained in the last step are removed; filtration, wherein according to a predetermined filtering rule, strains with incorrectlabeling information, incomplete chromosome assembly and incorrect classification are removed to obtain a reference strain genome of the species; fusion genome construction, wherein the reference strain genome is interrupted, redundancy is removed, reassembly is performed, and the sequences are reassembled to obtain a fusion genome of the species; database assembly, wherein the above steps are repeated to obtain the fusion genome of the predetermined species, summary is performed, and the pathogenic microorganism genome database is obtained. The genome database has the advantages of not onlyhaving a high precision rate, but also having short analysis time and reducing the cost.
Owner:GZ VISION GENE TECH CO LTD +4
Extraction method of soil microbe genome DNA and total RNA
The invention provides a method for effectively extracting crop rhizosphere soil microbe genome DNA and total RNA, which comprises the following steps: firstly, removing humus and other impurities which severely disturb nucleic acid extraction in soil samples by aluminum sulfate; further crushing soil microbe cells by glass beads; adding sodium dodecyl sulfonate (SDS) and LiCl solutions for cracking cells and dissociating nucleic acids; adding the mixture of phenol, chloroform and isoamylol for extracting; and then, depositing the extraction solution by isopropanol and sodium acetate to finally acquire the soil microbe metagenome DNA and total RNA. The method is suitable for extraction of the DNA and total RNA of soil in different regions. The invention establishes an extraction technology which has the advantages of simple and easy operation processes and higher purity of the prepared sample, and can provide an important basis for the research of soil metagenomics.
Owner:FUJIAN AGRI & FORESTRY UNIV
Microbial population analysis
ActiveUS20170159108A1Increase diversityMicrobiological testing/measurementICT adaptationMicroorganismIts region
The present invention relates to a method of typing a microbiome for having a desirable or undesirable signature, comprising analyzing the composition of the population of microorganisms in said microbiome based on taxonomic variation in the DNA sequence of the microbial 16S-23S rRNA internal transcribed spacer (ITS) regions in the genomic DNA of said microorganisms, wherein the sequences of conserved DNA regions comprised in the 16S and 23S rRNA sequences flanking said ITS region in the genome of said microorganisms comprise primer binding sites for amplification of said ITS regions.
Owner:IS DIAGNOSTICS
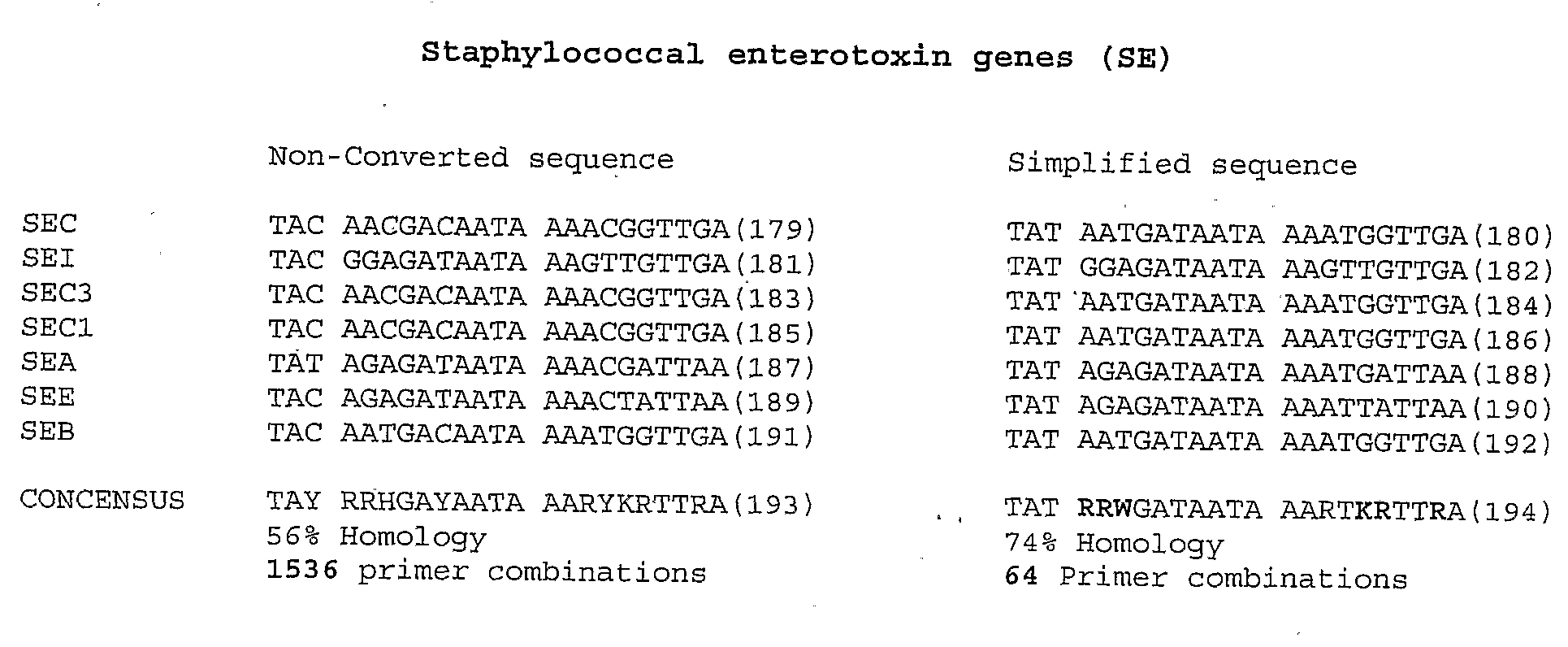
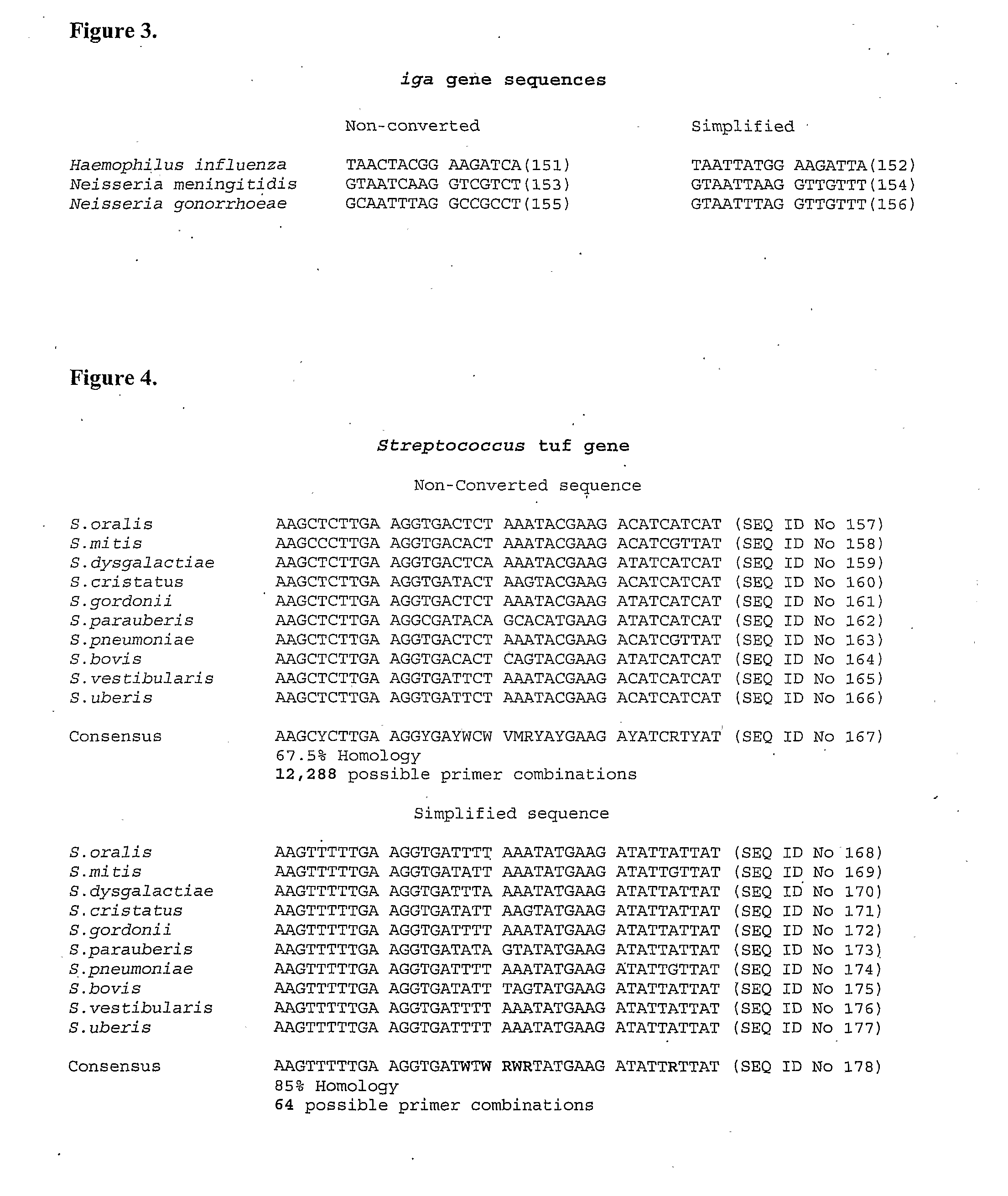
Methods for simplifying microbial nucleic acids by chemical modification of cytosines
ActiveUS20090042732A1High similaritySimplify the differenceSugar derivativesMicrobiological testing/measurementMicroorganismCytosine
A method for simplification of a microbial genome or microbial nucleic acid comprising treating microbial genome or nucleic acid with an agent that modifies cytosine to form derivative microbial nucleic acid and amplifying the derivative microbial nucleic acid to produce a simplified form of the microbial genome or nucleic acid.
Owner:HUMAN GENETIC SIGNATURES PTY LTD
Genetic assays
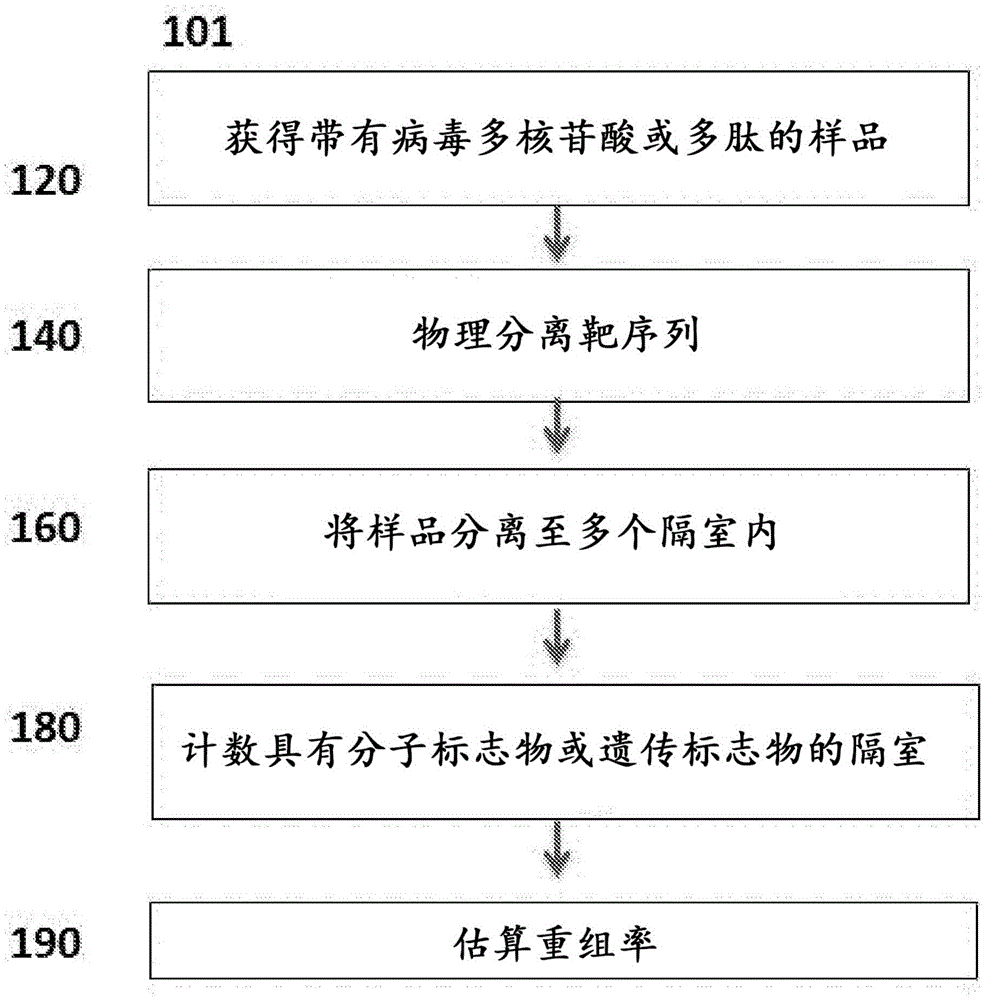
Provided herein are methods, compositions, systems, and kits for recombination assays, many of which involve amplification reactions such as PCR or droplet digital PCR. The methods, compositions, kits and systems are particularly useful for minimizing artifactual biases arising from recombination between viral or other genomes (e.g., any microbial genome) during the assay process. In some aspects, provided herein are methods of performing a recombination assay.
Owner:BIO RAD LAB INC
Compositions and methods for use thereof in modifying the genomes of microorganisms
InactiveUS7056728B2Degree of improvementInhibit transcriptionSugar derivativesBacteriaGram-positive bacteriumBacterial luciferase
The present invention relates to bacterial luciferase transposon cassettes suitable for conferring bioluminescence properties on a Gram-positive bacteria, Gram-negative bacteria, and other organisms of interest. The invention further includes cells transformed with vectors carrying the transposon cassettes, cells whose genomes have been modified by introduction of such cassettes, and methods of making and using such transposon cassettes, transposon cassette vectors, and cells containing the transposons.
Owner:XENOGEN CORP
Method for analyzing diversity and differences of intestinal and oral florae
ActiveCN107058490AAmplification Universal PrimerMicrobiological testing/measurementMicroorganismFlora
The invention provides a method for analyzing the diversity and the differences of intestinal and oral florae based on a new generation of high-throughput sequencing technology. The method comprises the following steps of (1) collecting a to-be-detected sample and carrying out microbial genome DNA extraction to obtain a genome DNA extracting solution; (2) detecting the genome DNA extracting solution to determine the concentration and the purity of a microbial genome DNA in the sample; (3) carrying out PCR amplification on the genome DNA extracting solution by using 16SrDNA-specific V3-V4 region primers and carrying out library construction; (4) sequencing by using an IlluminaMiSeq sequencing system; and (5) carrying out bioinformatics analysis by using FLASH, QIIME and usearch61. Compared with a traditional flora detection method, by using the new generation of high-throughput technology, compositions and functions of the florae can be analyzed more deeply in more detail.
Owner:山东探克生物科技股份有限公司
Kit for extracting fecal microbial genome by magnetic bead method and extraction method thereof
InactiveCN109266642AHigh yieldQuality improvementMicrobiological testing/measurementDNA preparationMagnetic beadGenomic DNA
The invention discloses a kit for extracting fecal microbial genome by magnetic bead method and an extraction method thereof, belongs to the field of biotechnology; the technical scheme adopted is; akit for extracting fecal microbial genome by magnetic bead method include lysis fluid, impurity removal reagent, genomic adsorbent and eluent, in buff a, B and C combine lysis microorganisms, the humic acid and other impurities are removed by the unique precipitating solution, then adsorbing genomic DNA with magnetic beads, further removing foreign proteins and salt ions with special buffer G andbuffer H, and finally eluting the purified genomic DNA; a method for extract genomic DNA of faeces microorganism includes such step as extracting genomic DNA of faeces microorganism with high efficiency and sufficient quantity, extracting genomic DNA of faeces microorganism with high quality, extracting genomic DNA of faeces microorganism with simple procedure, high speed, safety and no pollution,shortening extracting time, and can be used for manual operation and automatic platform, and is widely suitable for extracting animal faeces genomic DNA of faeces microorganism with high speed, efficiency and economy for customers, and has wide application prospect.
Owner:TIANGEN BIOTECH BEIJING
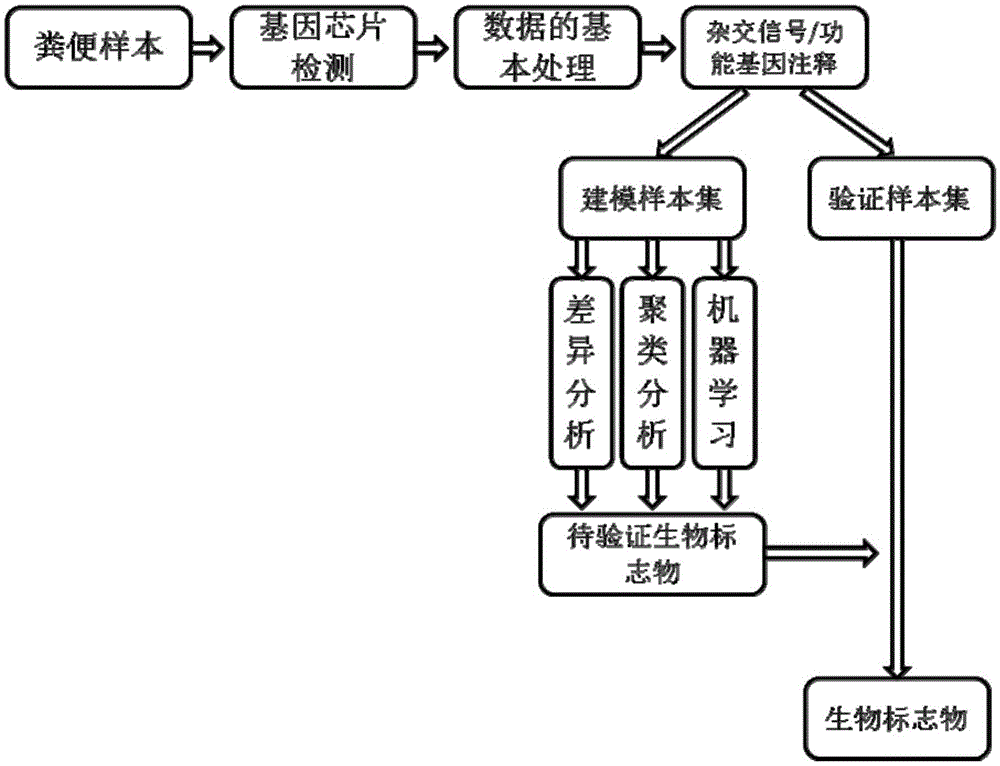
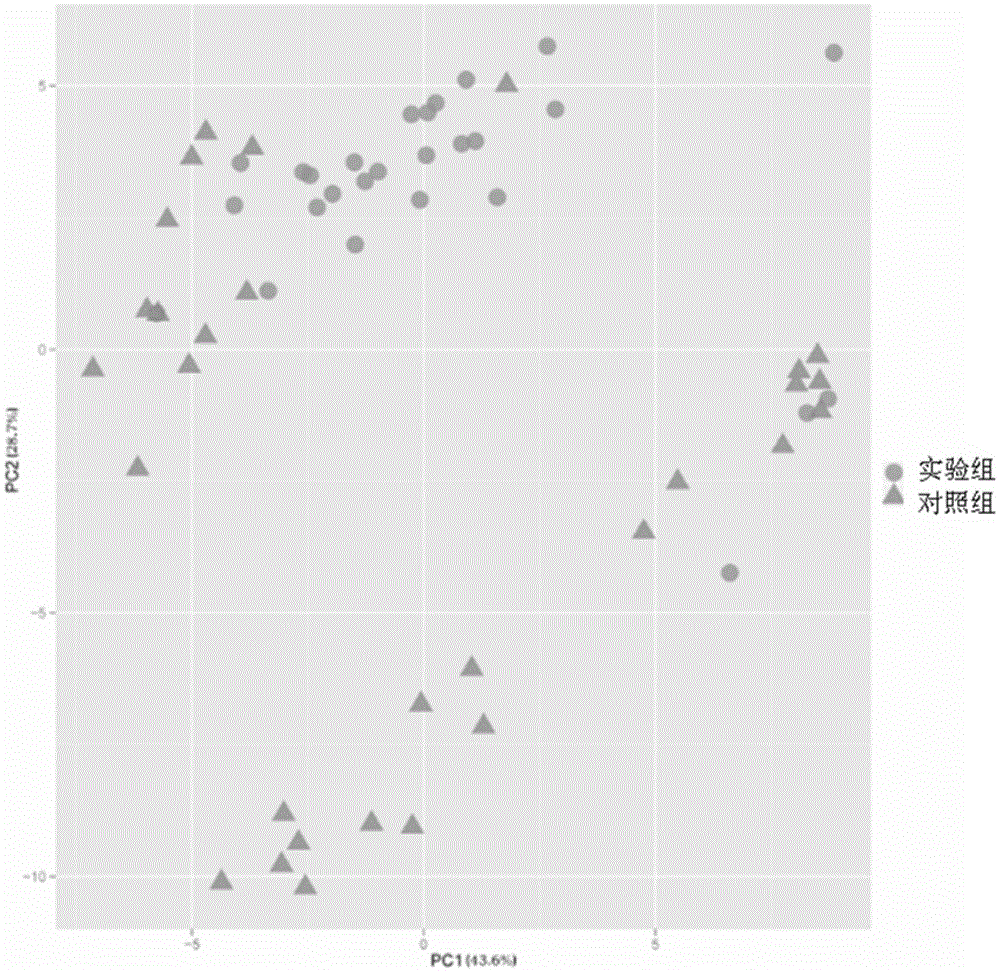
Mental disorder biomarker and application thereof
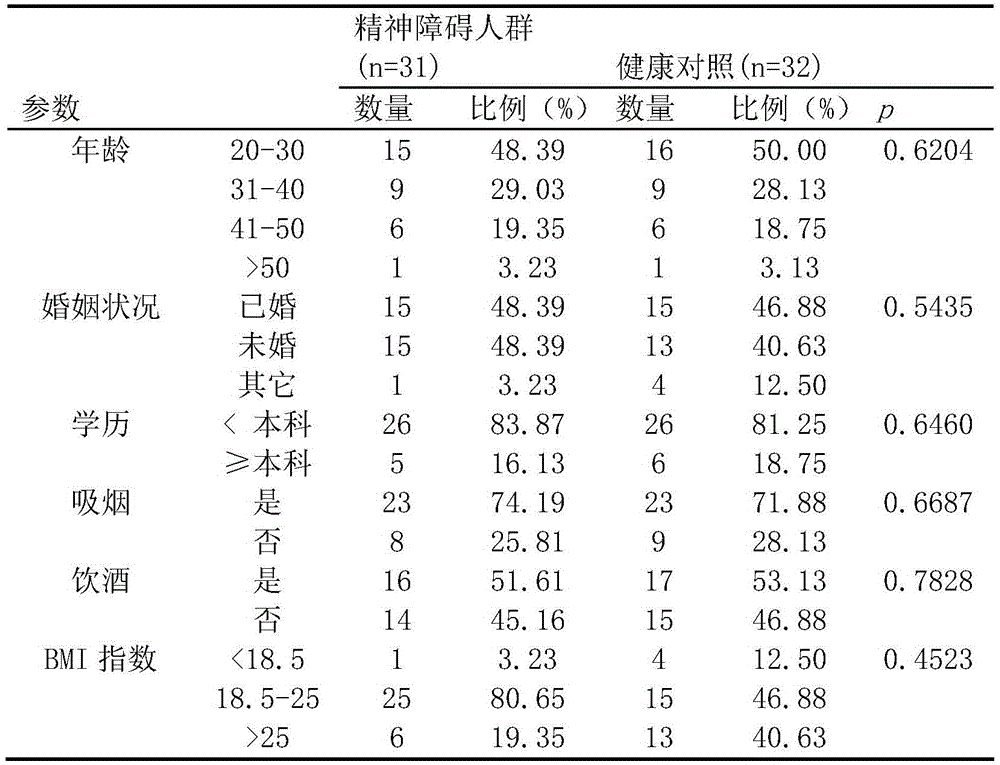
ActiveCN105543369AMonitoring the Effects of InterventionsMaster recoveryNervous disorderMicrobiological testing/measurementFecesGut flora
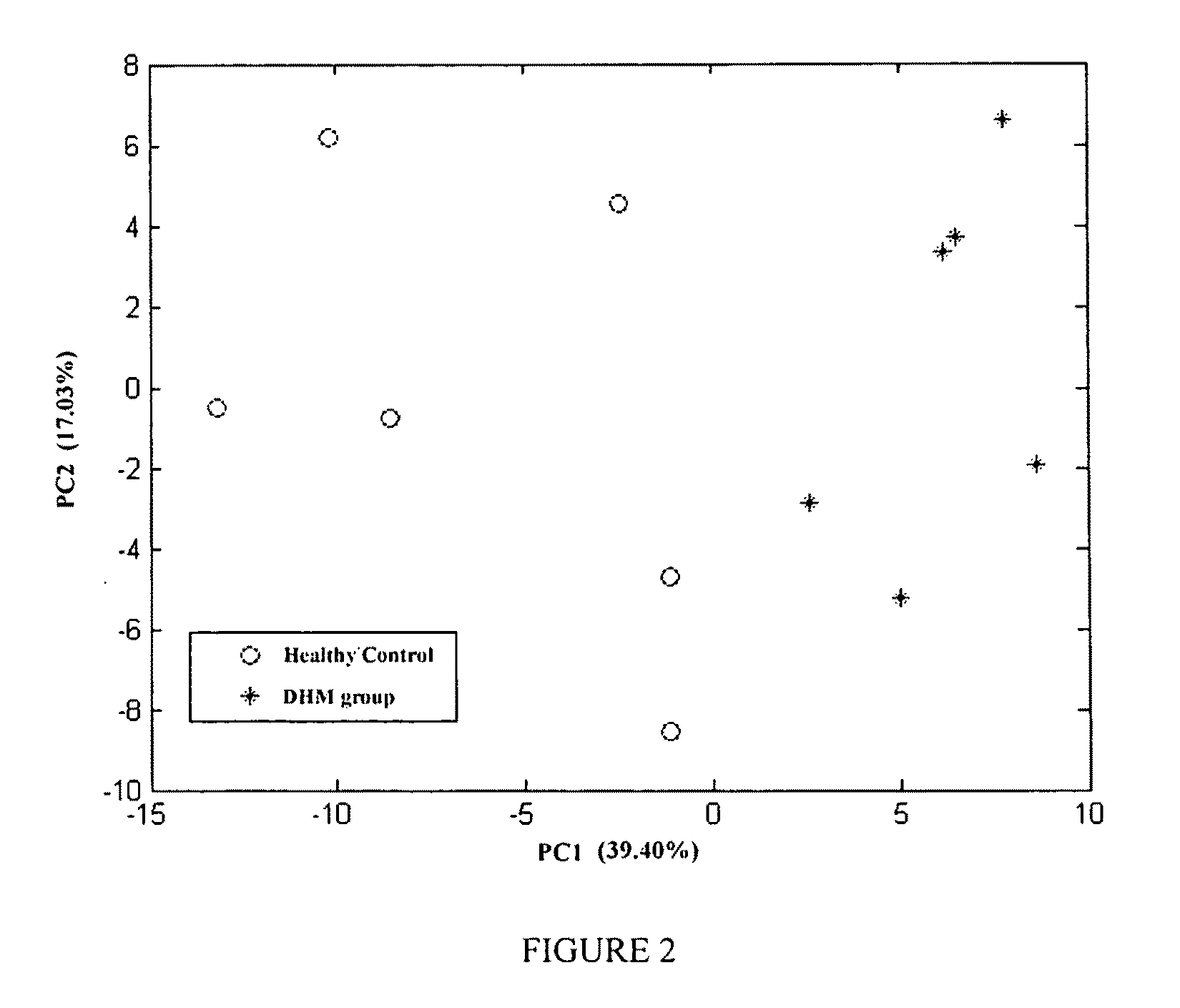
The invention discloses a mental disorder biomarker and application thereof. Microorganism gene chip hybridization research is conducted on intestinal flora microbial genomes in fecal samples of a mental disorder group and fecal samples of a healthy control group to describe characteristics of fecal microflorae and functional genes. By means of difference testing, discriminant analysis and machine learning, fifteen functional genes are recognized finally and can serve as biomarkers to highly accurately distinguish the mental disorder group. The biomarker is selected from at least ten genes in the fifteen functional genes. The fifteen functional genes have obvious differences in mental disorder people and the healthy people. The invention further discloses a drug which is capable of changing the number or expression of genes and is used for treating mental disorder, a method for producing or screening the drug for treating mental disorder and a kit for detecting mental disorder.
Owner:金锋 +1
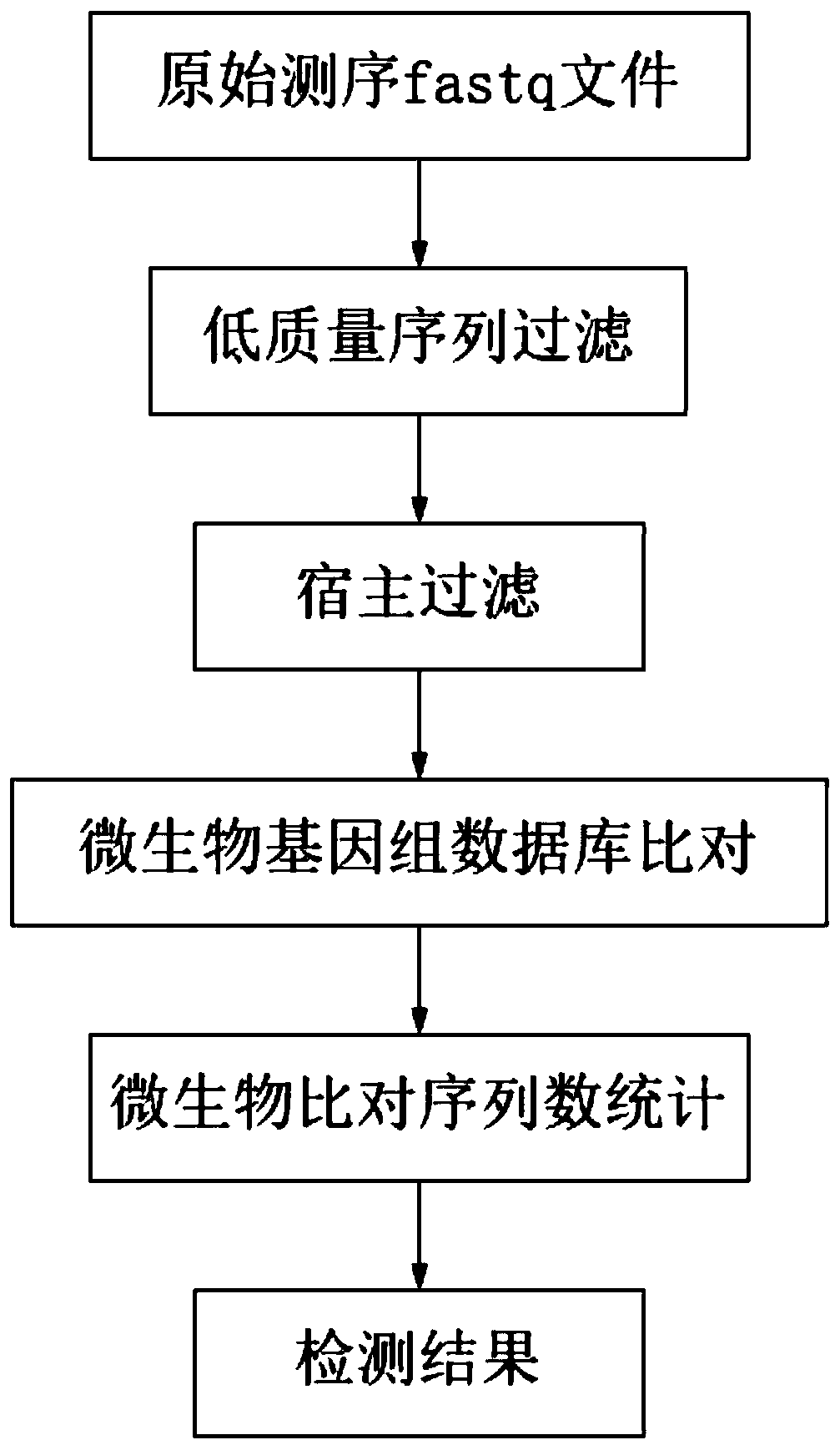
Method and device for microbiological analysis of host sample
ActiveCN111009286AFast and accurate analysisFast and accurate detectionProteomicsGenomicsMicrobial GenomesMicroorganism
The invention relates to the field of microbiological detection, in particular to a method and a device for microbiological analysis of a host sample. The method comprises the following steps: (1) performing first filtering processing on a sequencing data set from a host sample by adopting a host genome database so as to remove sequencing data which can be compared with the host genome database from the sequencing data set; (2) performing second filtering processing on the sequencing data set by adopting a homologous database so as to remove the sequencing data which can be compared with the homologous database from the sequencing data set; and (3) comparing the sequencing data set subjected to the first filtering treatment and the second filtering treatment with a microbial genome database so as to determine microbial sequencing data from the microorganisms in the sequencing data set. By applying the method and the device provided by the invention, rapid and accurate analysis and detection of microorganisms in a host can be realized.
Owner:深圳华大因源医药科技有限公司 +1
Pathogenic microorganism analysis and identification system and application thereof
ActiveCN111462821AImprove accuracyReduce mistakesSequence analysisInstrumentsMicrobial profilingOrganism
The invention relates to a pathogenic microorganism analysis and identification system and application thereof, and belongs to the technical field of gene detection and analysis. The pathogenic microorganism analysis and identification system comprises: a data acquisition module used for acquiring gene sequencing data obtained by high-throughput sequencing; a data filtering module used for performing low-quality sequence filtering and host sequence filtering in sequence; a data comparison module used for comparing sequences to a pathogenic microorganism genome database; a species comparison module used for counting to-be-analyzed sequences; a data analysis module used for calculating the similarity S and the average similarity value SMSi of each species on each sequence comparison in the common comparison sequence set; a species sequence module used for calculating the total comparison sequence number SNTi of the species; and a result output module used for carrying out bioinformaticsanalysis to obtain a pathogenic microorganism analysis and identification result. The pathogenic microorganism analysis and identification system has the advantages of short analysis time and high accuracy, and can accurately detect mixed infection and obtain specific pathogen information.
Owner:广州微远医疗器械有限公司 +3
Extraction method of oil reservoir microbial genome DNA
InactiveCN104630204AHigh purityReduce extraction costsDNA preparationMicrobial enhanced oil recoveryHelicase
The invention relates to the technical field of microbial enhanced oil recovery, and in particular relates to an extraction method of oil reservoir microbial genome DNA. The method comprises the following steps: removing interference of crude oil in samples with extraction by means of organic solvent extraction; releasing microbes on an oil-water interface; filtering water phase by using a hollow fiber membrane to enrich microbes; removing organic macromolecular components such as proteins by phenol and chloroform in cooperation with lysis cells such as lysozyme, helicase, protease and lauryl sodium sulfate according to a physical wall-breaking method under a high-salinity condition; subsequently, performing nucleic acid extraction and precipitation to obtain the oil reservoir microbial genome DNA. The method is quick and simple, and the whole extraction process can be completed in 10 hours; the extraction efficiency is high, and the purity of the obtained genome is high.
Owner:CHINA PETROLEUM & CHEM CORP +1
Kit for extracting microbial genome DNA from soil and its method
InactiveCN1936005AReduce lossesReduce physical health damageFermentationPlant genotype modificationImpurityMicroorganism
The invention relates to an agent box to distill microorganism gene group DNA from earth and the distilling method. The agent box includes agent A(immersion fluid), agent B(pretreatment fluid), agent C(cracking fluid), and agent D that is made up of RNA enzyme A, agent E Al2(SO4)3, agent F hexadecyl trimethyl amine, and agent G(DNA cleaning fluid). The distilling method includes the following steps: dipping the sample, taking pretreatment to the sample, cracking the microorganism, removing humus, taking DNA coupling, removing protein impurity, adhering DNA, washing DNA, and taking elution to DNA. The advantages of the invention are rapid, little DNA loss, little harmful to researchers, high yield, and low cost, etc.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Kit and method for extracting microbial genome DNA by magnetic bead method
InactiveCN110283816AAvoid interferenceProtection from cleavageMicrobiological testing/measurementDNA preparationMicroorganism preservationLithium chloride
The invention relates to a kit and method for extracting a microbial genome DNA by a magnetic bead method. The kit comprises a microbial preservation buffer solution, a bacteriolytic agent solution I, proteinase K, silica gel magnetic beads, isopropanol and an ethanol water solution, and further comprises a bacteriolytic agent II, a lysis adsorption solution and a reinforced scrubbing solution. The bacteriolytic agent II comprises MES, EDTA, Triton X-100, lysozyme and a lysozyme enhancer. The lysis adsorption solution comprises Tris-HCl, sodium chloride, EDTA, TrionX-100 and a nucleic acid separation and absorption promoter; and the reinforced scrubbing solution comprises Tris-HCl, lithium chloride and ethanol. By adopting the kit provided by the invention, cell walls of gram-positive bacteria and various fungi can be completely destroyed to form protoplasts, DNA of the protoplast cells can be rapidly released under the action of the lysis adsorption solution, the DNA is efficiently adsorbed on the silica gel magnetic beads, and after the DNA is washed by the scrubbing solution and eluted by an elution solution, microbial DNA with high concentration and purity can be obtained.
Owner:WUXI NO 2 PEOPLES HOSPITAL
High-throughput mulberry pathogenic bacteria identification and species classification method and application thereof
ActiveCN106636433AMicrobiological testing/measurementMicroorganism based processesDiseaseRibosomal DNA
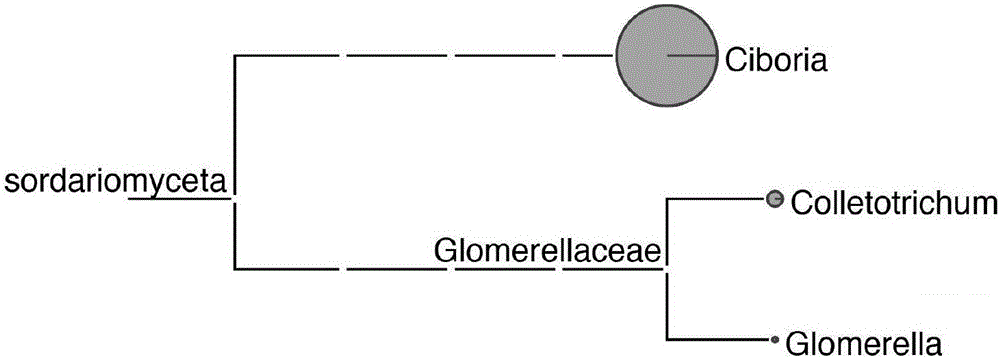
The invention discloses a high-throughput mulberry pathogenic bacteria identification and species classification method. The method comprises the following steps that diseased mulberries are collected; the total DNA of the diseased mulberries is extracted; an Illumina DNA library is created; Illumina high-throughput sequencing is carried out; a mulberry genome sequence in sequencing data is removed; microbial genome sequences are assembled; complete ribosomal DNA sequences are assembled; microbial ribosomal DNA sequences are screened and labeled; the ribosomal DNA sequences are comparatively analyzed to classify species, and thereby the mulberry pathogenic bacteria identification and species classification are fulfilled. A result shows that three species of fungi are identified in total when the method disclosed by the invention is applied to carry out the identification of pathogenic bacteria of popcorn disease and species classification, wherein the Ciboria pathogenic bacteria has the highest relative abundance, hereby the pathogenic Ciboria shiraiana is determined as Ciboria, and according to a comparison result, the pathogenic Ciboria shiraiana is determined as Ciboria carunculoides. Most of the species are phytopathogenic bacteria, and can lead to symptoms, such as mummification and swelling, appearing on fruits and seeds of plants, which are identical with the symptoms of the popcorn disease.
Owner:SOUTH CHINA AGRI UNIV
Soil microbe genome DNA extracting method
InactiveCN101372689AHigh purityReduce extraction costsFermentationPlant genotype modificationReproductionNucleic acid
The invention relates to the technical field of molecular biology, in particular to a method for extracting soil microbial genome DNA. The method comprises the following concrete steps: a soil sample and quartz sand are mixed and put into extracting buffer solution in a centrifugal tube, and the centrifugal tube is placed in a nucleic acid extraction apparatus for vibrating for 40-60 seconds, or on a turbine mixer for vibrating for 10-20 minutes; the mixture obtained is added with 50-100mul of lysozyme, treated by warm bath at the temperature of 30-40 DEG C for 30-60min, added with 75-125mul of SDS and mixed evenly, and further treated by the warm bath at the temperature of 50-60 DEG C for 30-60min, and supernatant is obtained after centrifugation; and the supernatant is further extracted and purified to obtain the soil microbial genome DNA. The soil microbial genome DNA extracted by the method has high purity, the OD260nm / OD280nm can reach over 1.6, the output can be up to 100ug / ml, and the whole process takes about 10 hours; the method can be directly used for PCR reproduction without purification kit for further purification in the whole process; and by taking the obtained DNA as a template, and the target product can be smoothly reproduced.
Owner:SHENYANG INST OF APPLIED ECOLOGY - CHINESE ACAD OF SCI
Methods for detecting therapeutic effects of anti-cancer drugs
precancerous lesions, and subject having precancerous lesions and being treated with an anti-cancer agent; (2) isolating total microbial genomic DNA from the fecal samples to provide total microbial genomic DNA; (3) comparing the total microbial genomic DNA using fingerprint spectrum analysis; (4) identifying key fingerprint bands correlated with the effect of the anti-cancer agent; (5) identifying key microorganisms associated with the key fingerprint bands; (6) designing microbial sequence-specific primers and probes; and (7) determining the quantitative differences of the key microorganisms in fecal samples to identify an indicator microorganism for monitoring the effect of the anti-cancer agent.
Owner:SHANGHAI JIAO TONG UNIV +1
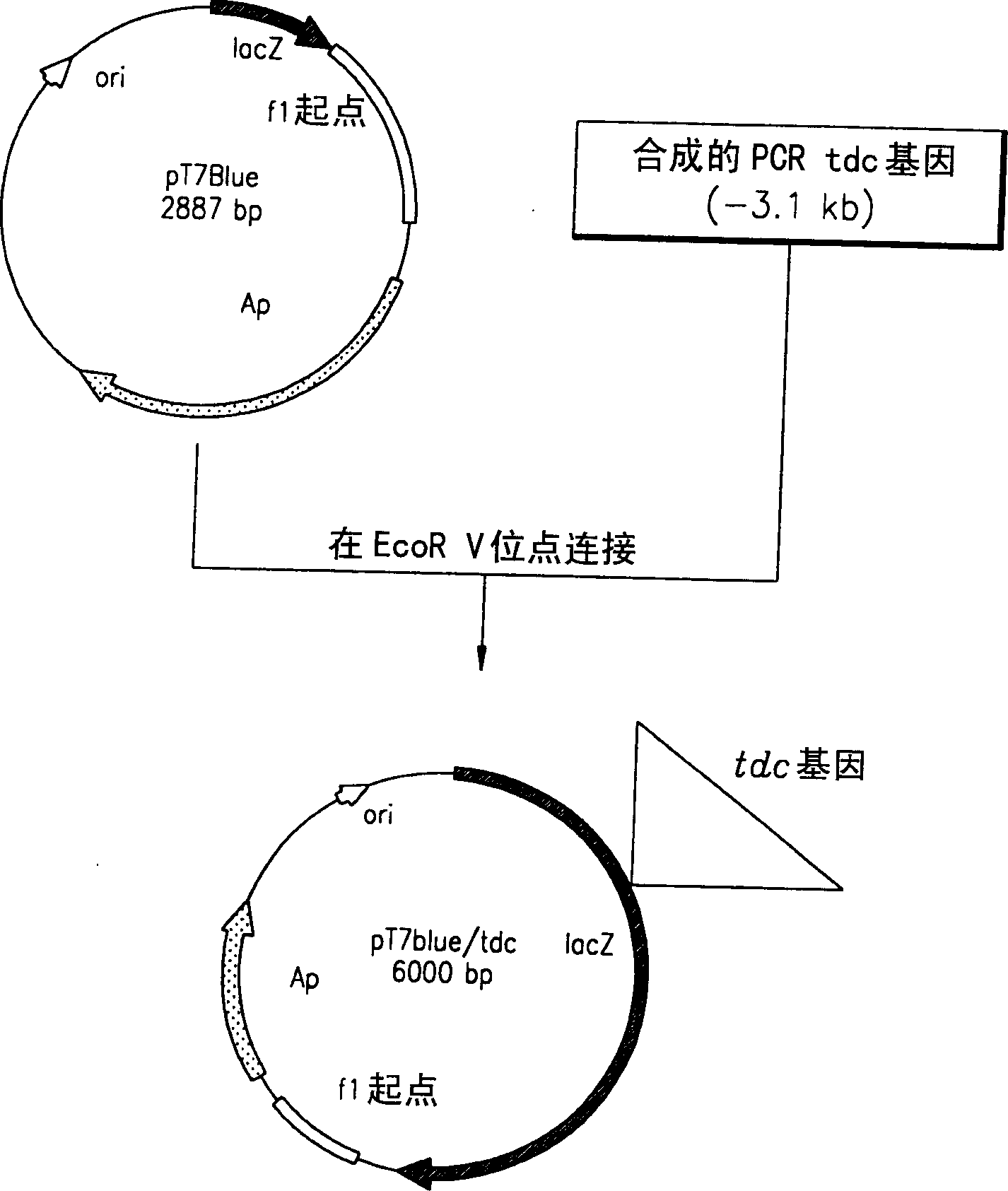
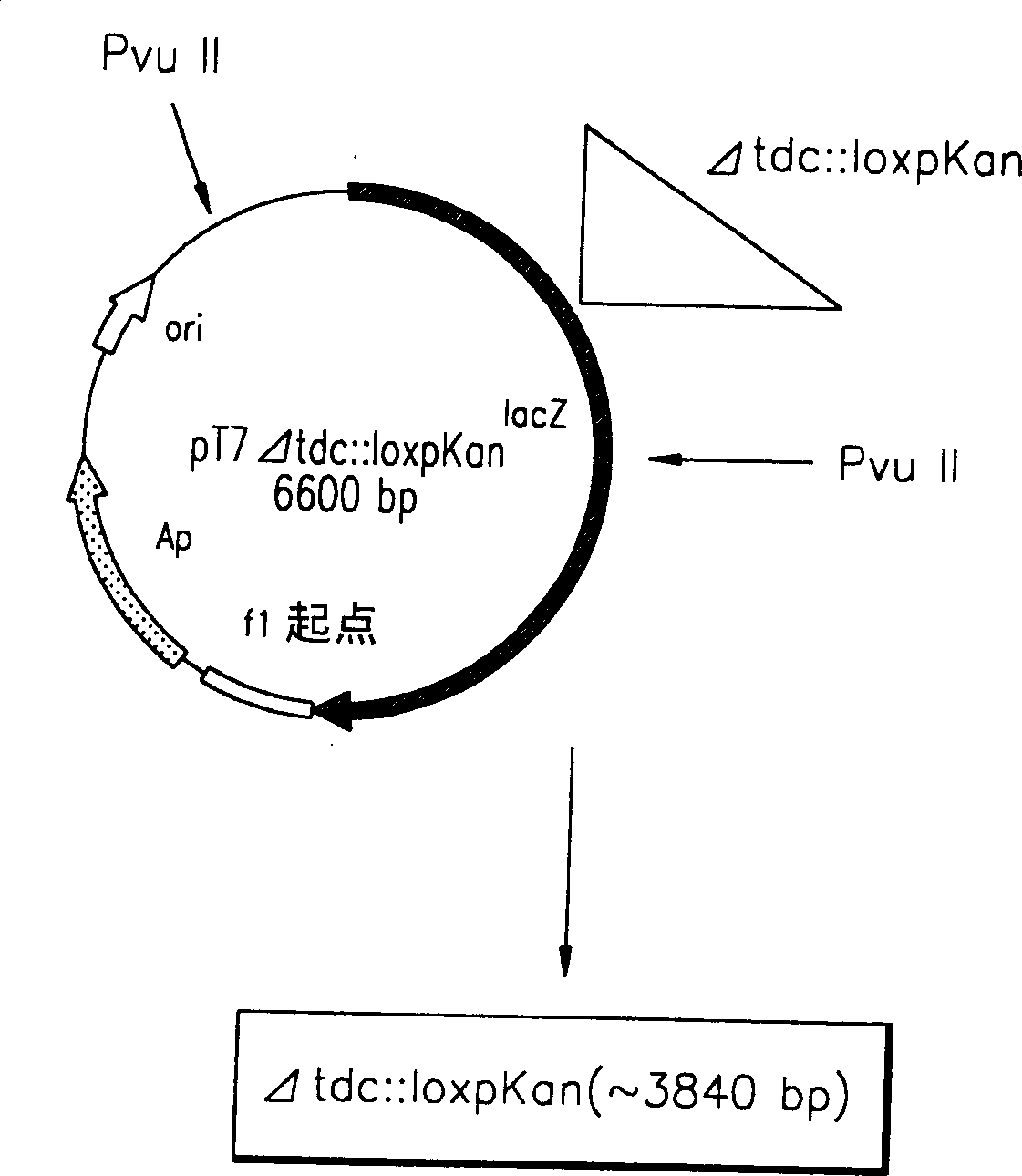
Process for prodn. of L-Thr
A method for producing L-threonine using a microorganism is provided. In the method, the threonine dehydratase (tdc) gene existing in the genomic DNA of the microorganism is partially deactivated using a recombination technique. For a microorganism strain with enhanced activity of threonine operon-containing enzymes and the phosphoenolpyruvate carboxylase (ppc) gene, the tdc gene engaged in one of the four threonine metabolic pathways is specifically deactivated, thereby markedly increasing the yield of L-threonine.
Owner:CJ CHEILJEDANG CORP
High-quality DNA (Deoxyribonucleic Acid) extraction method of spirit brewing microorganism
The invention relates to a high-quality DNA (Deoxyribonucleic Acid) extraction method of a spirit brewing microorganism. The high-quality DNA extraction method is characterized by comprising the following steps of: (1) performing simple pre-treatment on a sample to obtain thallus mixture; (2) adding DNA extract, various enzyme solution, SDS (Sodium Dodecyl Sulfonate), protease and the like into the mixture to fully release DNA in thallus; (3) adding phenol: chloroform: isoamyl alcohol and chloroform: isoamyl alcohol to extract miscellaneous protein so as to purify the DNA; and (4) settling the DNA by the isoamyl alcohol at a low temperature and washing with absolute ethyl alcohol to obtain high-quality microbial genome total DNA. The high-quality DNA extraction method has the advantages of capability of realizing that all used reagents are the conventional reagents, low cost, wide application range, easiness in operation, mild conditions, high purity of extracted DNA, capability of performing subsequent PCR (Polymerase Chain Reaction) and DGGE (Denaturing Gradient Gel Electrophoresis) analysis without purifying the genome DAN and the like.
Owner:SICHUAN UNIV
Method for the simultaneous monitoring of individual mutants in mixed populations
InactiveUS6528257B1Improved and efficientAccurately and quantitatively monitorMicrobiological testing/measurementOther foreign material introduction processesGenomic SegmentGenomic DNA
The present invention relates to an improved and efficient method for simultaneous monitoring of abundance of individual mutants of a microbe in mixed populations where insertion of a known transposon in the genome of a microbe such that each mutant caries a single transposon insertion, isolating the genomic DNA of the mixed population of mutants, fragmenting the same with a frequently cutting restriction enzyme, ligating a double standard adapter to the genomic DNA fragments, amplifying the DNA fragments adjoining transposon insertions thereby generating a set of DNA fragments corresponding only to mutated genes resolving the said amplified DNA fragments followed by comparing the intensity of DNA fragments obtained from population of mutants before selection with that obtained from the population subjected to selection and finally sequencing the DNA fragments that change in abundance to identify the mutated genes.
Owner:COUNCIL OF SCI & IND RES
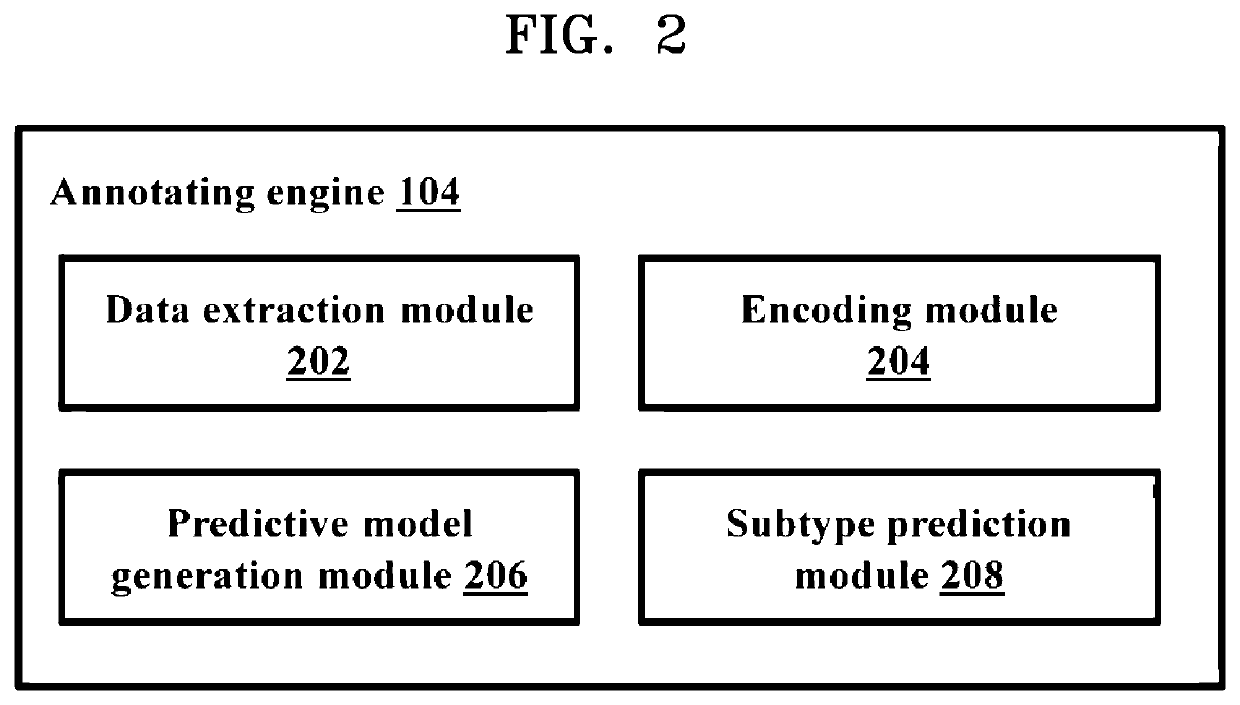
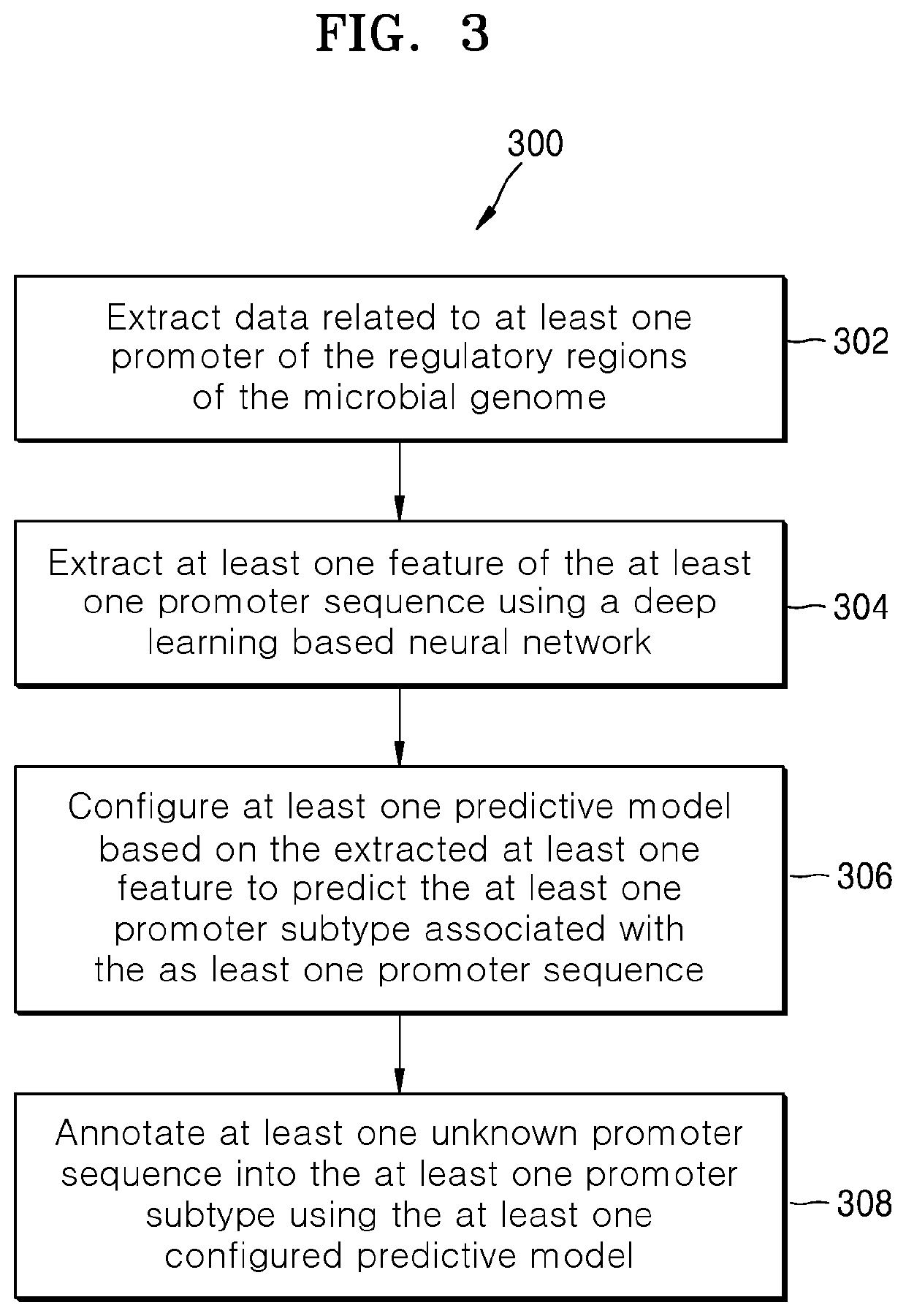
Methods and systems for annotating regulatory regions of a microbial genome
Methods and systems for annotating regulatory regions of a microbial genome. A method disclosed herein includes extracting data related to at least one promoter of the regulatory regions of the microbial genome, wherein the data includes at least one promoter sequence and data available for at least one promoter subtype. Based on extracted at least one feature of the at least one promoter sequence, the method further includes configuring at least one predictive model using the deep learning based neural network to predict the at least one promoter subtype associated with the at least one promoter sequence. The method further includes annotating at least one unknown promoter sequence into the at least one promoter subtype using the at least one configured predictive model.
Owner:SAMSUNG ELECTRONICS CO LTD
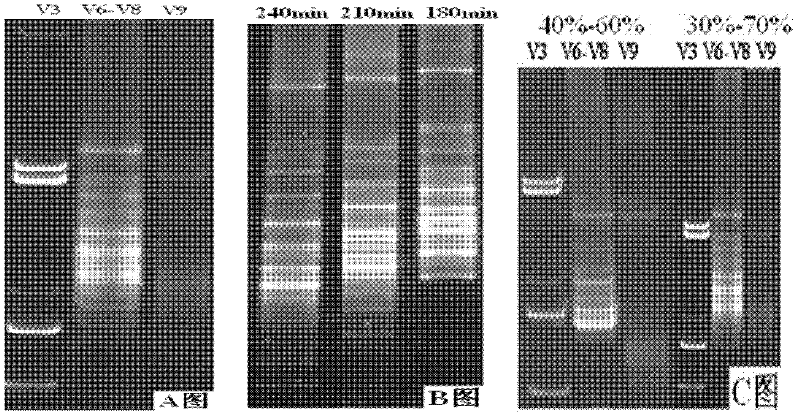
Biological active early warning marking method for chemical industry wastewater treatment
InactiveCN102408161ASolve problems that cannot be effectively and stably operatedGuarantee continuous and stable operationBiological water/sewage treatmentBiotechnologyActivated sludge
The invention relates to a biological active early warning marking method for chemical industry wastewater treatment, which specifically comprises the following steps of: extracting microorganism genome total DNA in active sludge of a sewage treatment system, and screening bacterial seeds by enrichment culture method and identifying; amplifying the microorganism genome total DNA by universal primers of V6-V8 areas of bacterial 16SrDNA; carrying out PCR-DGGE (Polymerase Chain Reaction Denaturing Gradient Gel Electrophoresis) analysis to the PCR product, and creating community succession mode in normal running system; finding bacterial strains closely related with COD (Chemical Oxygen Demand) change in the system, and using RAPD (Random Amplified Polymorphic DNA) technique to obtain specific molecular marker genes of pure culture strains; and directly judging if functional bacteria are existent for monitoring the sewage treatment system by PCR of specific molecular marker genes of functional bacteria. The invention solves the difficult problem that the current sewage treatment system cannot effectively and stably run, and the method is simple, convenient and fast to operate and can effectively monitor running situation of the sewage treatment system.
Owner:NANKAI UNIV
Process for extracting microbe genome DNA in complex solid-state environment
The invention provides a method for directly extracting microbial genome DNA in the complex solid environment. The method comprises specific steps of: firstly adding an Extraction Buffer in a sample, then crushing bacteria, next using CTAB to remove protein and organic macromolecular substances under high ionic concentration, and finally extracting DNA from a solution by the conventional method; and the method is fast and convenient, and the acquired DNA can be used for operating the follow-up PCR equimolecular experiment.
Owner:SHANGHAI UNIV
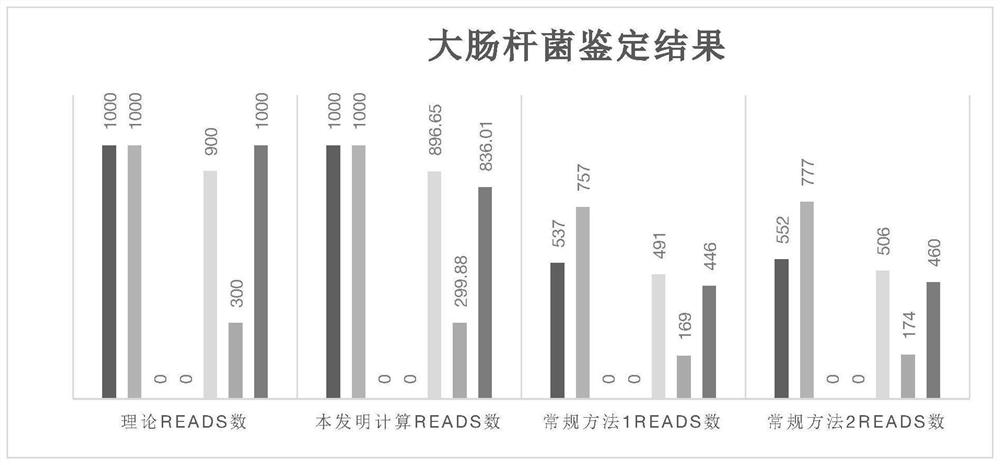
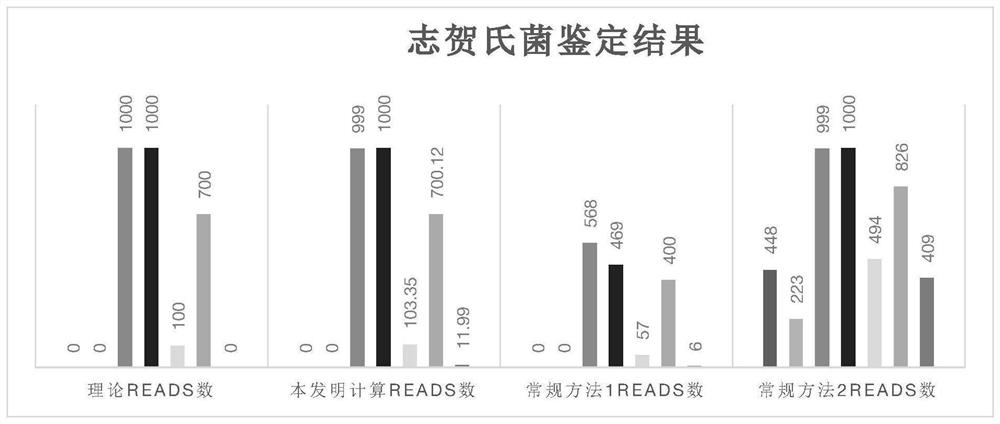
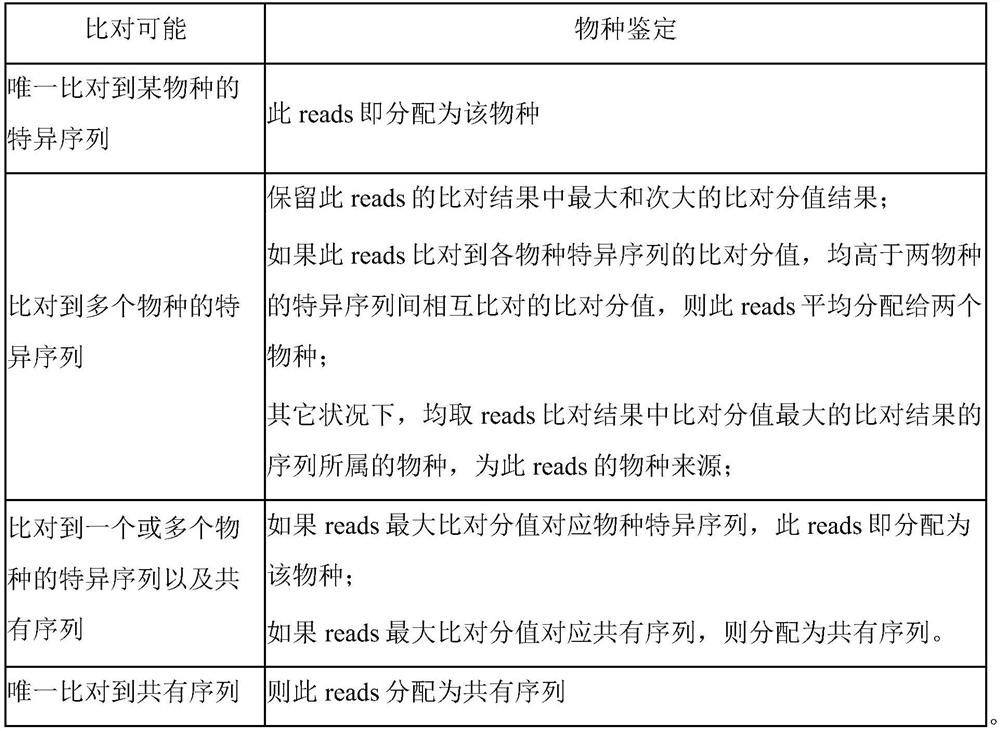
Microbial genome database construction method and application thereof
ActiveCN112992277ALow chance of false positivesImprove compatibilitySequence analysisInstrumentsMicrobial GenomesMicroorganism
The invention provides a microbial genome database construction method and application thereof. According to the invention, the construction method of the microbial genome database comprises the steps: constructing the database in a manner of labeling after genome breaking, labelling consensus sequences and specific sequences of multiple species, and constructing a comparison score matrix among the specific sequences of the species, so a sequence source is quickly and accurately obtained.
Owner:NANJING SIMCERE MEDICAL LAB CO LTD +2
Method for extracting microbial genome DNA from poplar wood
ActiveCN106754872AReduce degradationAvoid damageMicrobiological testing/measurementDNA preparationGenomic sequencingMicroorganism
The invention discloses a method for extracting microbial genome DNA from poplar wood. According to the method, a CTAB (cetyltrimethyl ammonium bromide) method and an SDS (sodium dodecyl sulfonate) method are combined, and technical parameters of various steps are optimized, so that the genome DNA extracted by virtue of the method has the advantages of being high in concentration and free from pollution; determined by a spectrophotometer, OD260 / OD230 and OD260 / OD280 values are close to standard values, completely satisfying requirements of 16S rDNA / ITS sequencing and metagenomics sequencing adopted in the later period; therefore, the method can be directly applied to a molecular operation.
Owner:HUAZHONG AGRI UNIV +1
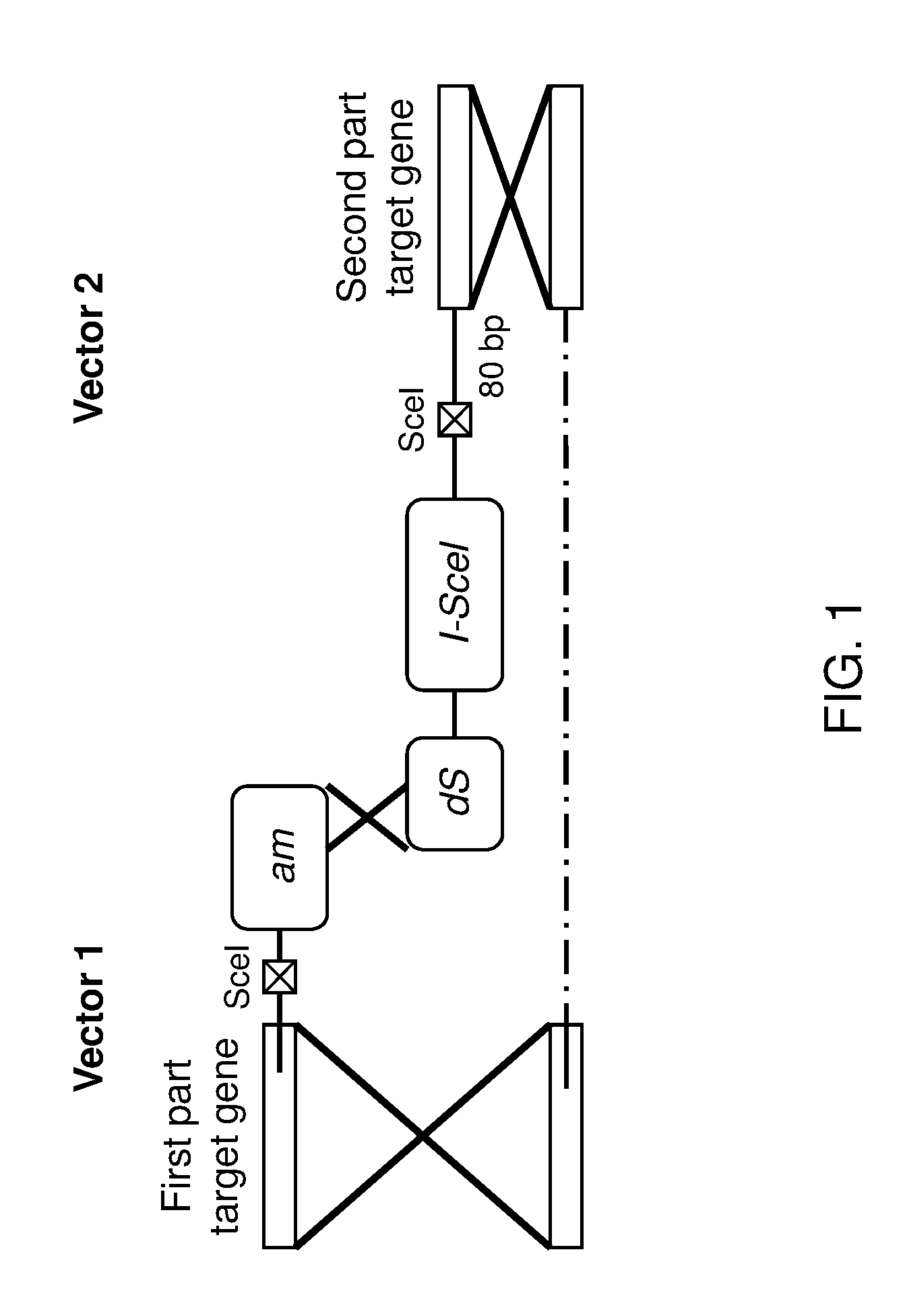
Novel genome alteration system for microorganisms
InactiveUS20160369300A1Stable introduction of DNAVector-based foreign material introductionSelectable markerBioinformatics
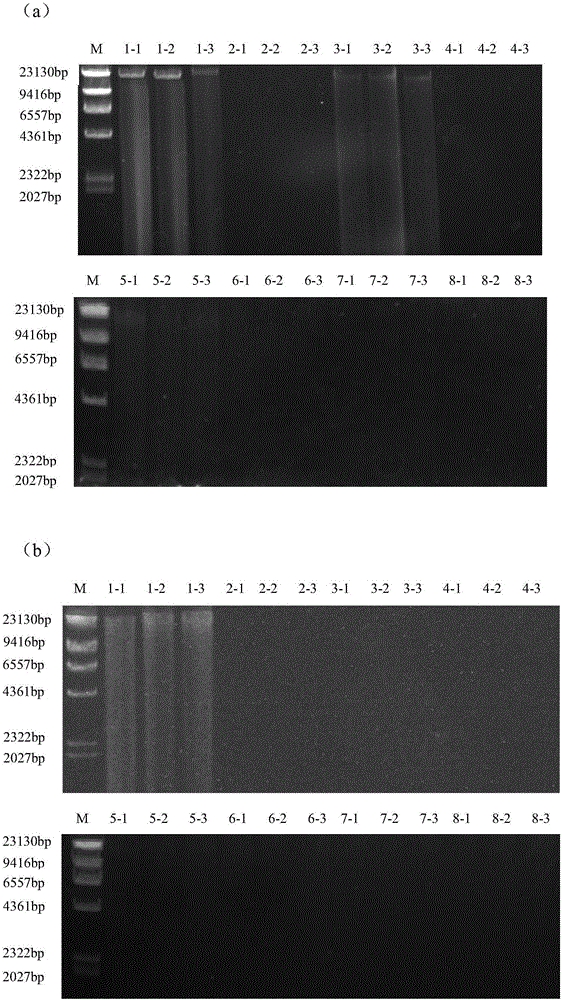
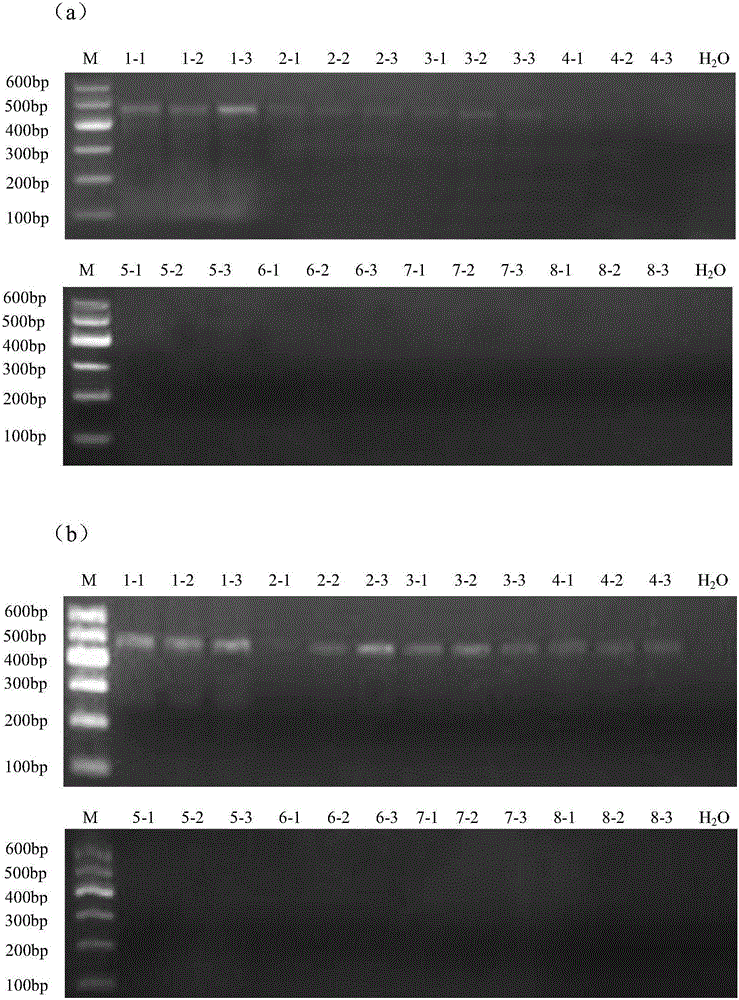
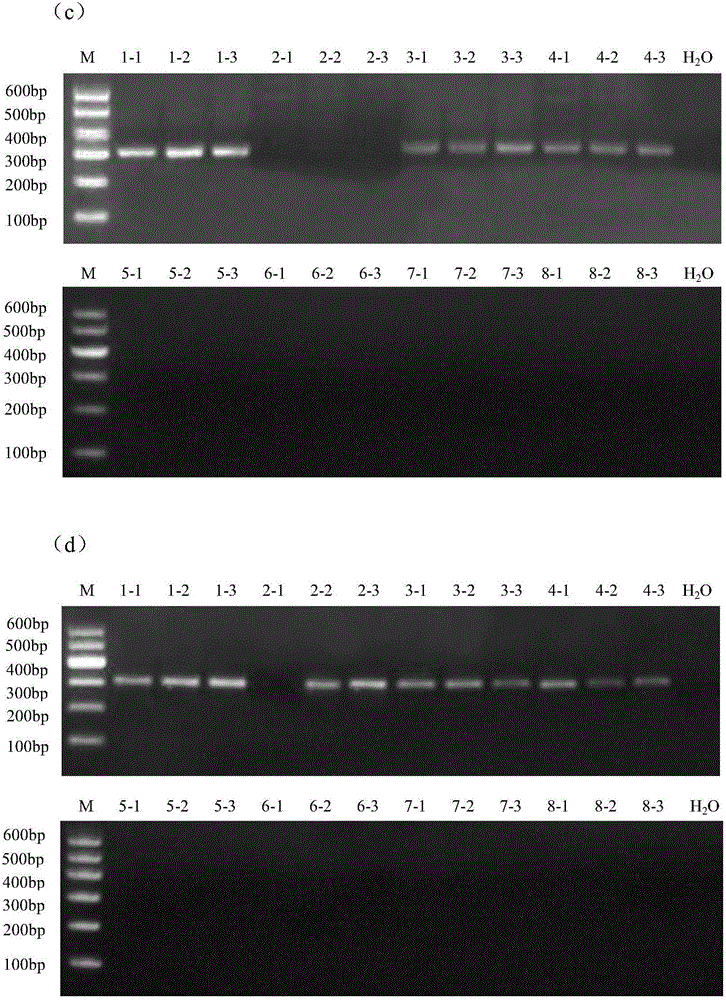
Novel genome alteration system for microorganisms The invention relates to a set of targeting constructs, comprising a first construct comprising a recognition site for an endonuclease, a first region of homology with a target gene of a microorganism, and a first part of a selection marker, and a second construct comprising a second part of the selection marker, a second region of homology with the target gene of the microorganism, and a copy of the endonuclease recognition site. The invention further relates to methods for altering a target gene in a microorganism, to methods for producing a microorganism, and to microorganisms that are produced by the methods of the invention.
Owner:HEINEKEN SUPPLY CHAIN BV
Method for simultaneously extracting microbial genome deoxyribonucleic acid (DNA) and total ribonucleic acid (RNA) in mining area environmental sample
The invention discloses a method for simultaneously extracting microbial genome deoxyribonucleic acid (DNA) and total ribonucleic acid (RNA) in a mining area environmental sample. The method comprises the following steps: S1, pretreatment of an environmental sample: collecting microorganisms in a liquid sample by a centrifuging method, or removing impurities out of a solid sample by a filtering method; S2, crushing cells: mixing the sample pretreated in the step S1 with quartz sand, adding liquid nitrogen, grinding for three times, adding PIPES extraction buffer of which the pH value is 7.0 and a sodium dodecyl sulfate solution, splitting cells at 65 DEG C for 1 hour; S3, purifying and participating nucleic acid: collecting supernate by the centrifuging method at the end of splitting of cells, adding an extraction agent to the supernate to centrifugally extract protein and lipids, participating the supernate by using isopropanol, and centrifuging so as to obtain the total ribonucleic acid; separating the total ribonucleic acid, so as to obtain the metagenome DNA and the total RNA. The method is low in cost and is capable of simultaneously extracting the metagenome DNA and the total RNA with high purity and good integrity from the mining area environmental sample.
Owner:CENT SOUTH UNIV
Extraction method of microbe DNA for high-throughput sequencing in sample
InactiveCN105176970AHigh yieldReduce the degree of degradationMicrobiological testing/measurementLibrary creationMicrobe DNAMicroorganism
The invention provides an extraction method of a microbe DNA for high-throughput sequencing in a sample and belongs to the field of a genome DNA extraction technology. According to the invention, DNA of a microbe in a sample is extracted by a SDS method; an inhibitor is removed after precipitation by the use of a sodium chloride solution and a aluminum ammonium sulfate solution, and then a supernatant is taken; and separation and purification are carried out by a silica gel film-column to obtain soil microbe genome DNA for high-throughput sequencing. The aluminum ammonium sulfate solution is prepared from 50-200 mM of Al2(SO4)3 and 50-200 mM of (NH4)2SO4 according to volume ratio of 1:1. The sample's microbe genome DNA extracted by the above method has high purity, good universality and fast extraction speed, can be directly used in a downstream high-throughput sequencing experiment and has a good application prospect.
Owner:BIOMARKER TECH
Methods for high-resolution microbiome analysis
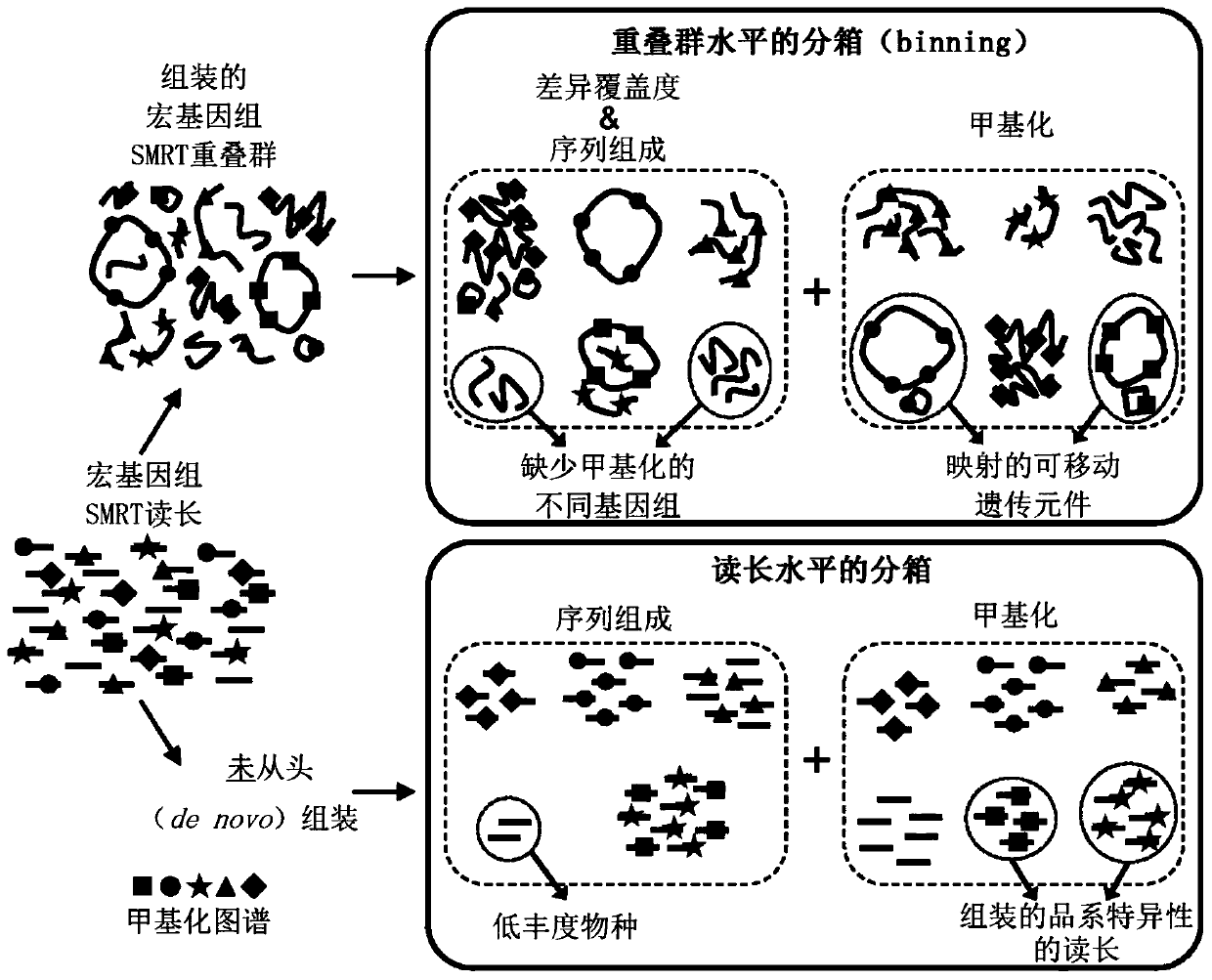
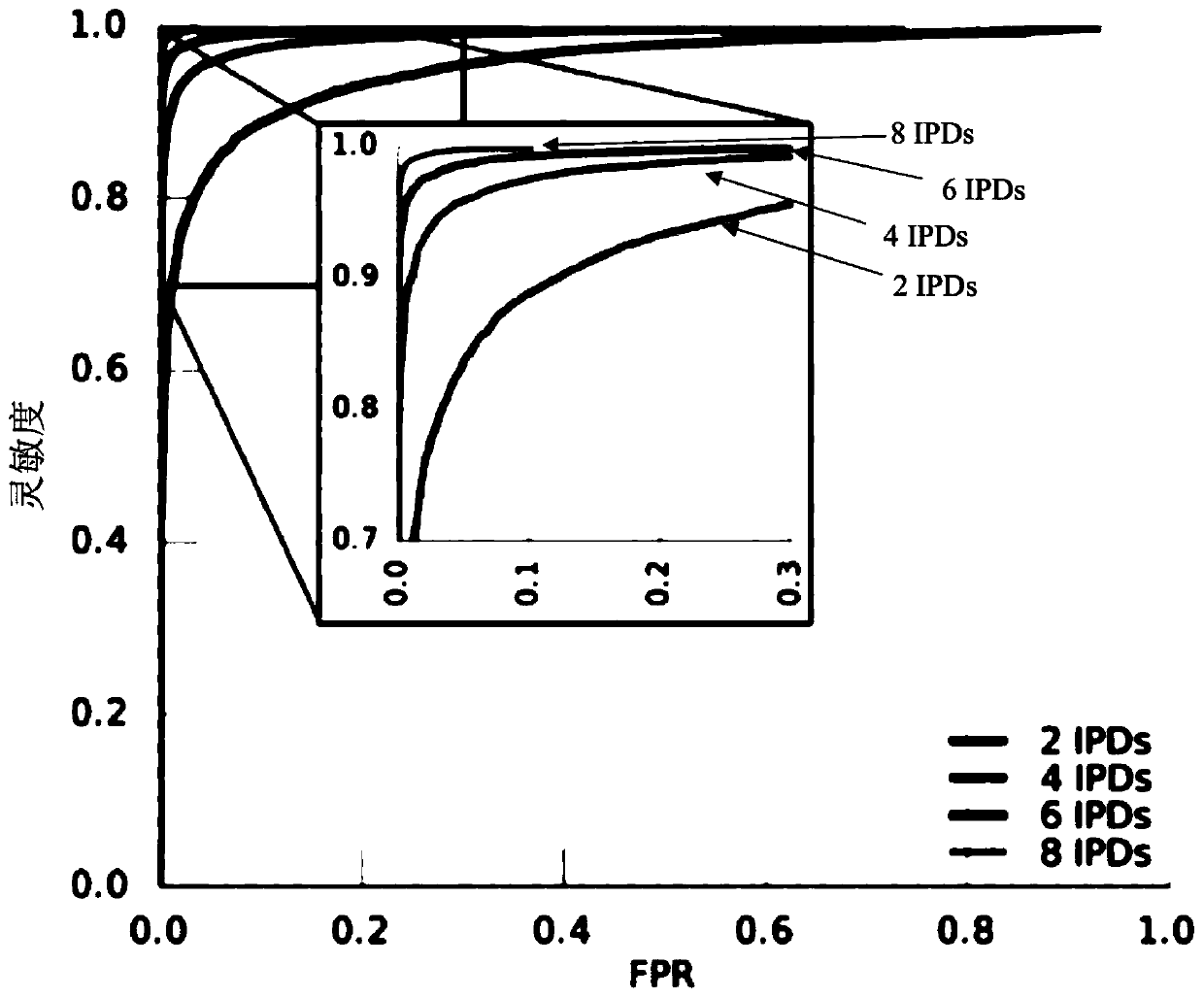
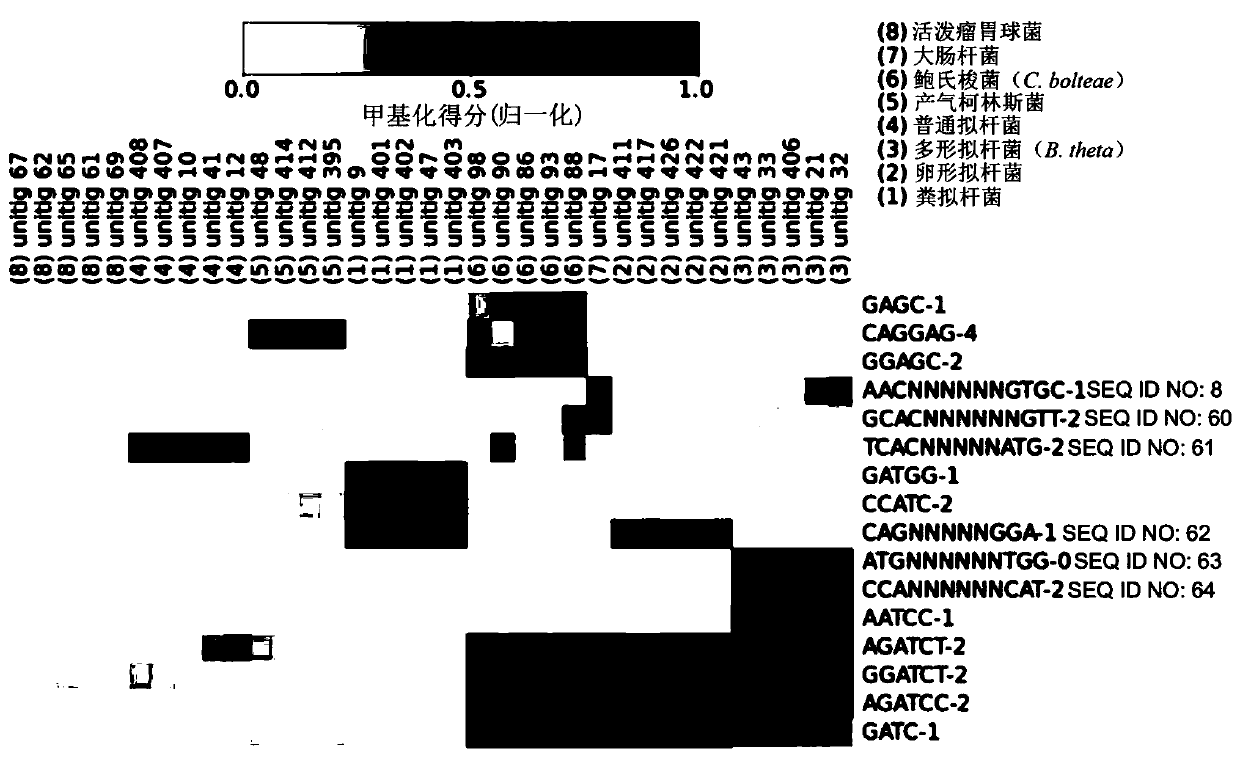
Methods are presented for binning metagenomic sequences that leverage long reads from a single-molecule long-read sequencing technology and utilize DNA methvlation signatures inferred from these readsto resolve individual reads and assembled contigs into species- and strain-level clusters. Methods for deconvolving prokaryotic organisms in a microbiome sample are presented. Methods for mapping mobile genetic elements to their host organisms in a microbiome sample are also presented.
Owner:MT SINAI SCHOOL OF MEDICINE
Method for obtaining high-quality fermented grain microorganism genome DNA (deoxyribonucleic acid) by pretreatment
InactiveCN102816757ADid not affect diversityThe method is simple and fastDNA preparationBiotechnologyMicroorganism
The invention relates to a method for obtaining a high-quality fermented grain microorganism genome DNA (deoxyribonucleic acid) by pretreatment and belongs to the technical field of molecular biology. The technical scheme of the method for obtaining the high-quality fermented grain microorganism genome DNA by pretreatment comprises the following steps of: pretreatment of a fermented grain sample, and extraction of a microorganism genome DNA. The microorganism genome DNA in fermented grains extracted by using the method has high purity and few impurities, and can be used for classification of fermented grain microorganisms, diversity research and the like.
Owner:LUZHOU PINCHUANG TECH CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com