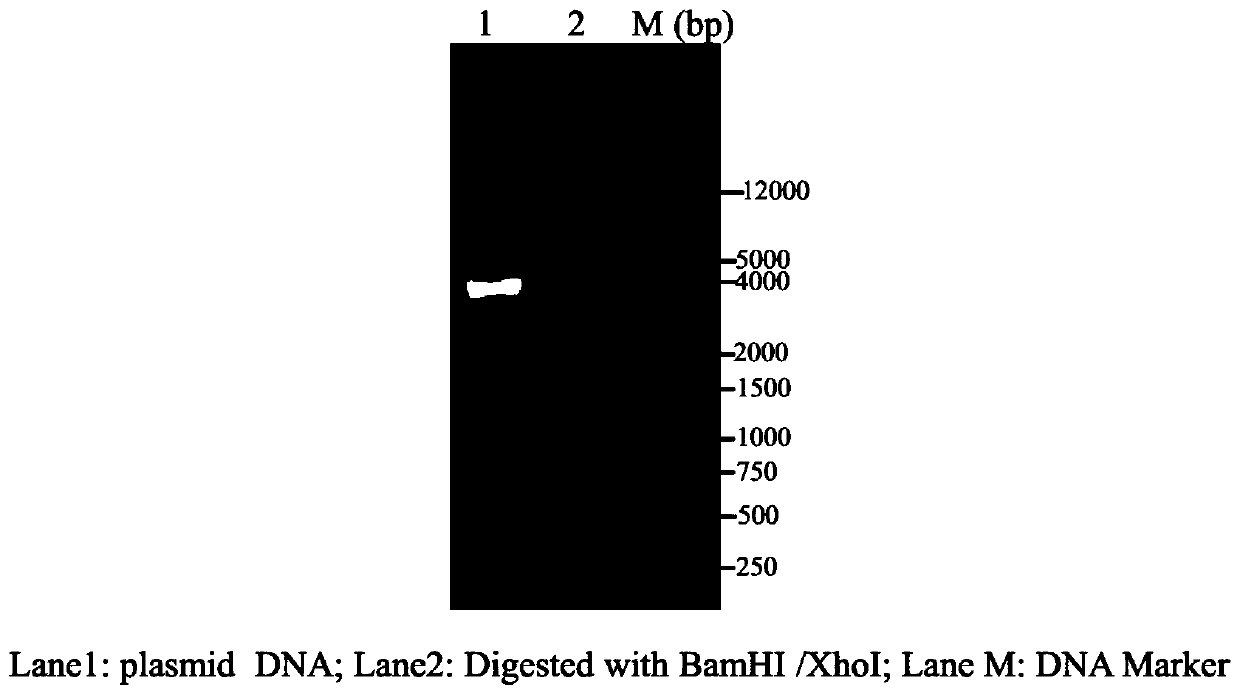
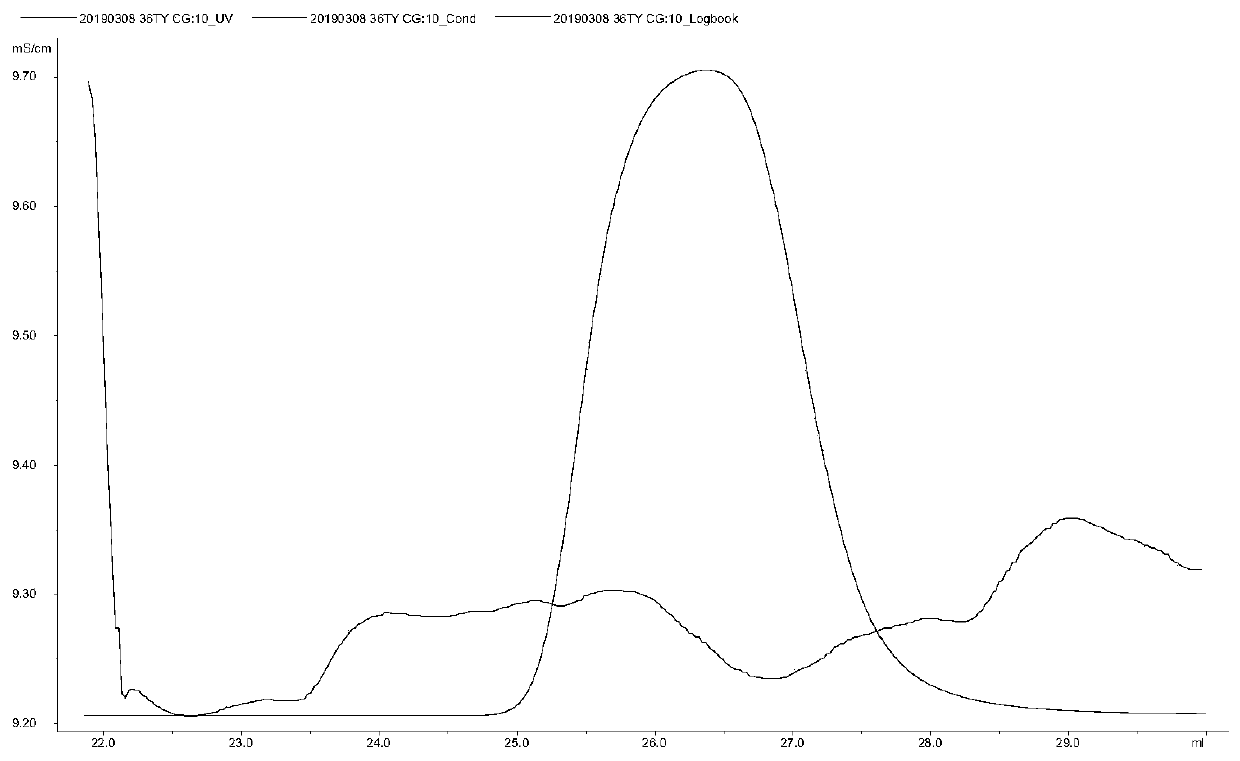
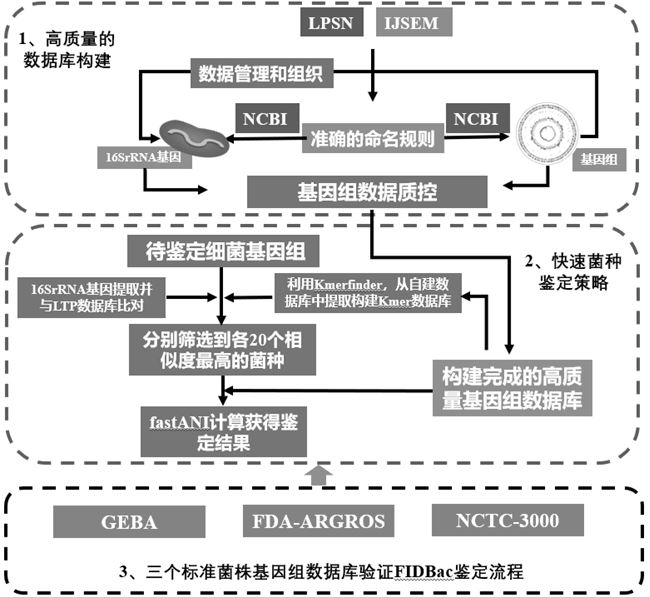
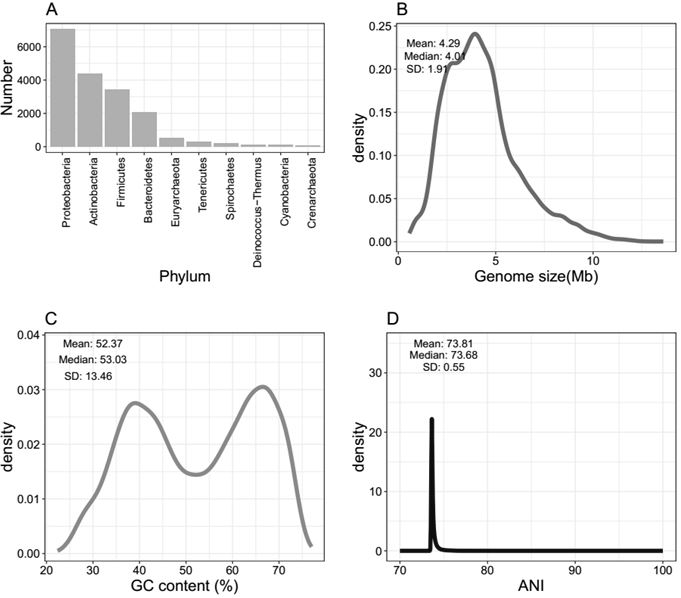
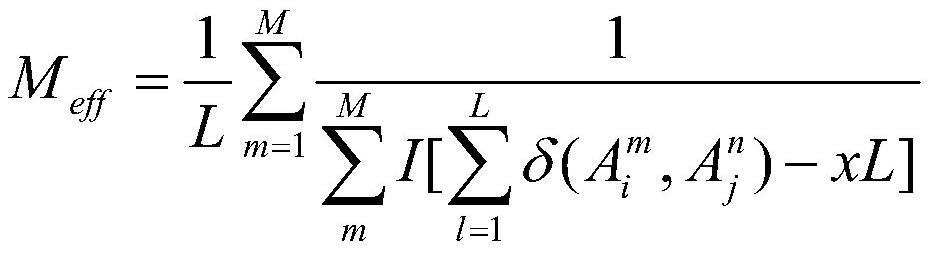Patents
Literature
40 results about "Genomic databases" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
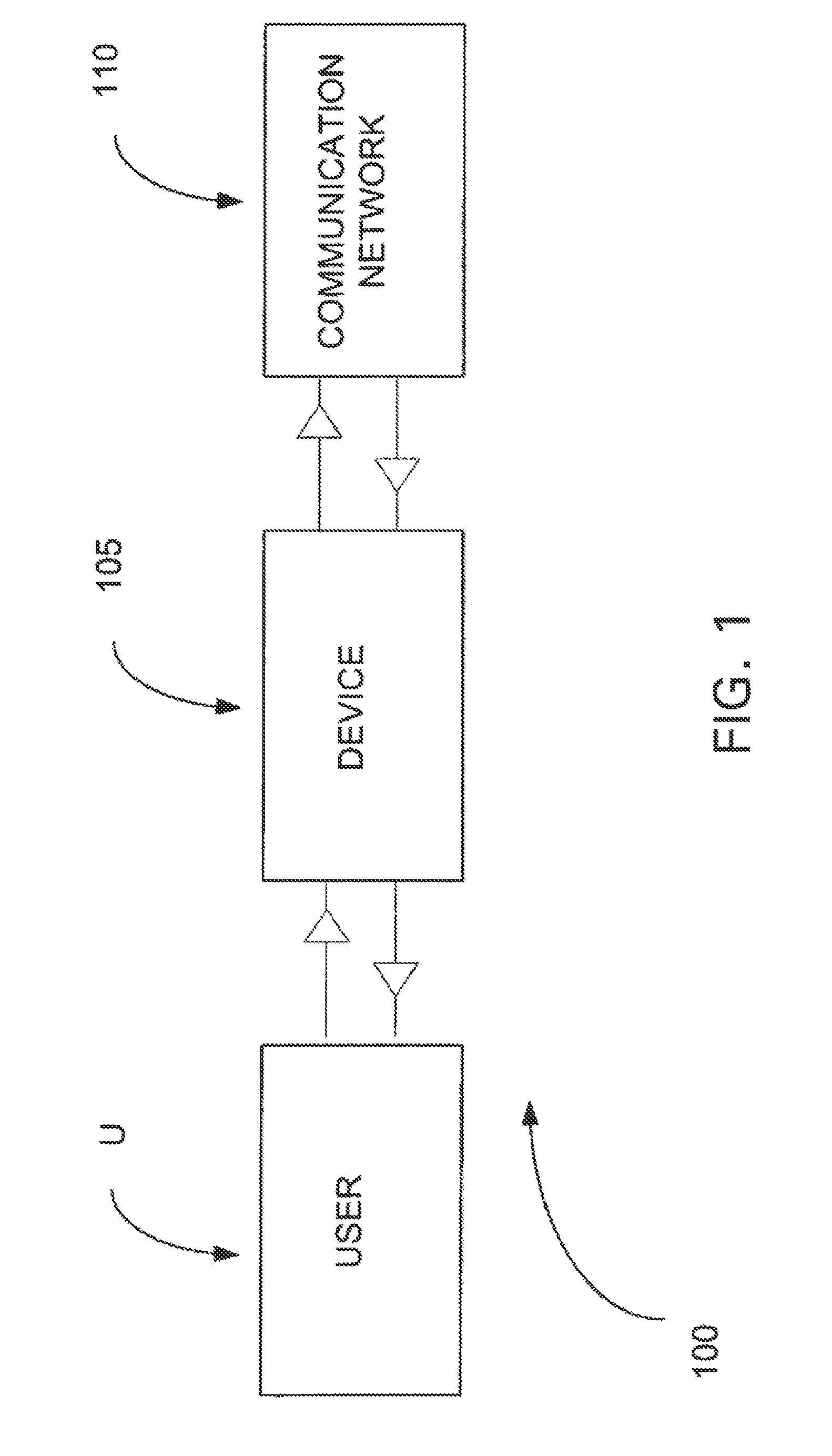
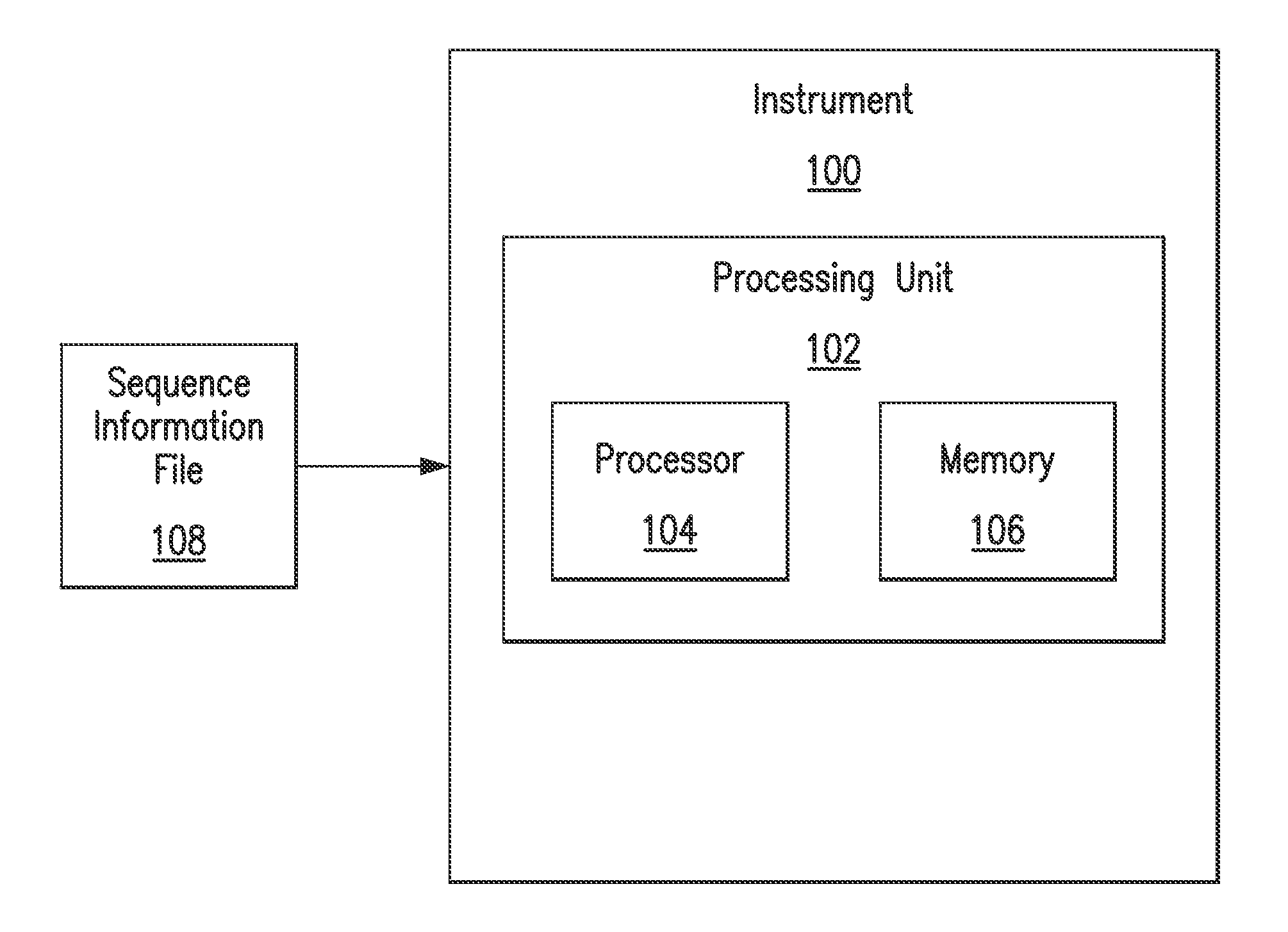
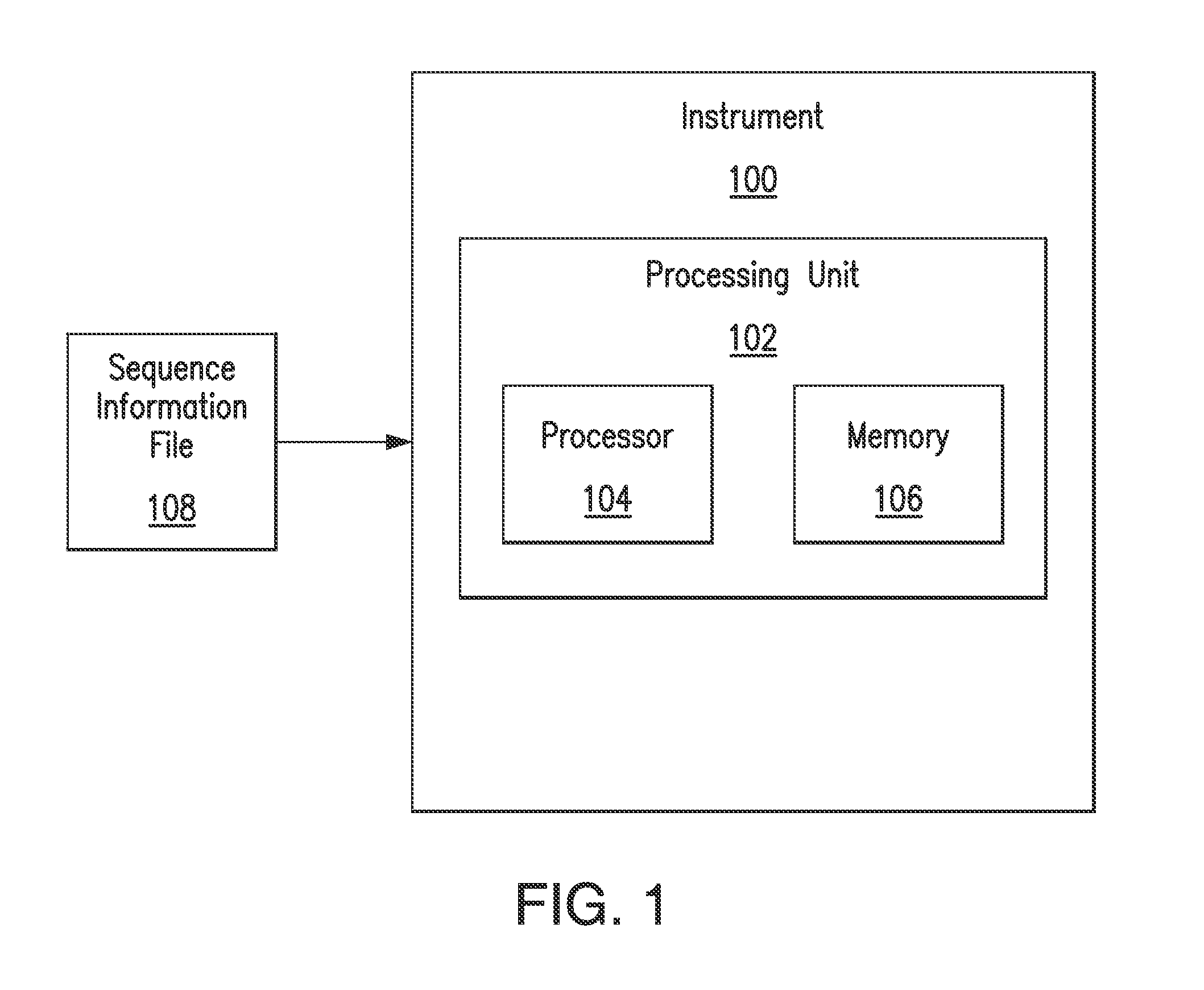
A genome database is a cross-referenced collection of information about one or more organisms, so one scientist can look at all the available genetic information to help him or her in research. Genomes are highly complex and contain billions of bases in the sequence of information.
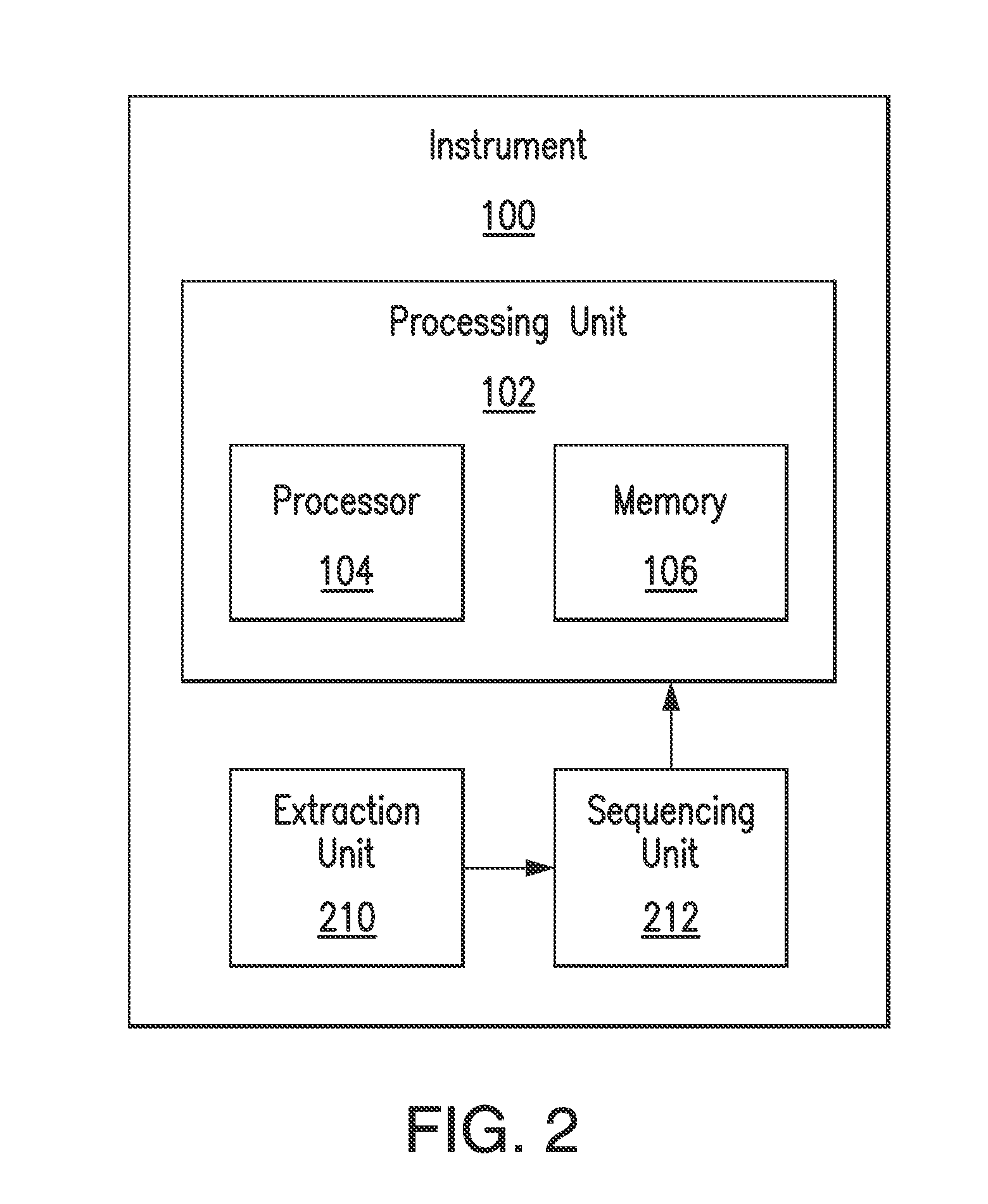
Direct identification and measurement of relative populations of microorganisms with direct DNA sequencing and probabilistic methods
ActiveUS8478544B2Quick identificationMicrobiological testing/measurementBiostatisticsGenomic SegmentProbabilistic method
Owner:COSMOSID INC
Direct identification and measurement of relative populations of microorganisms with direct DNA sequencing and probabilistic methods
ActiveUS20120004111A1Quick identificationMicrobiological testing/measurementLibrary screeningGenomic SegmentProbabilistic method
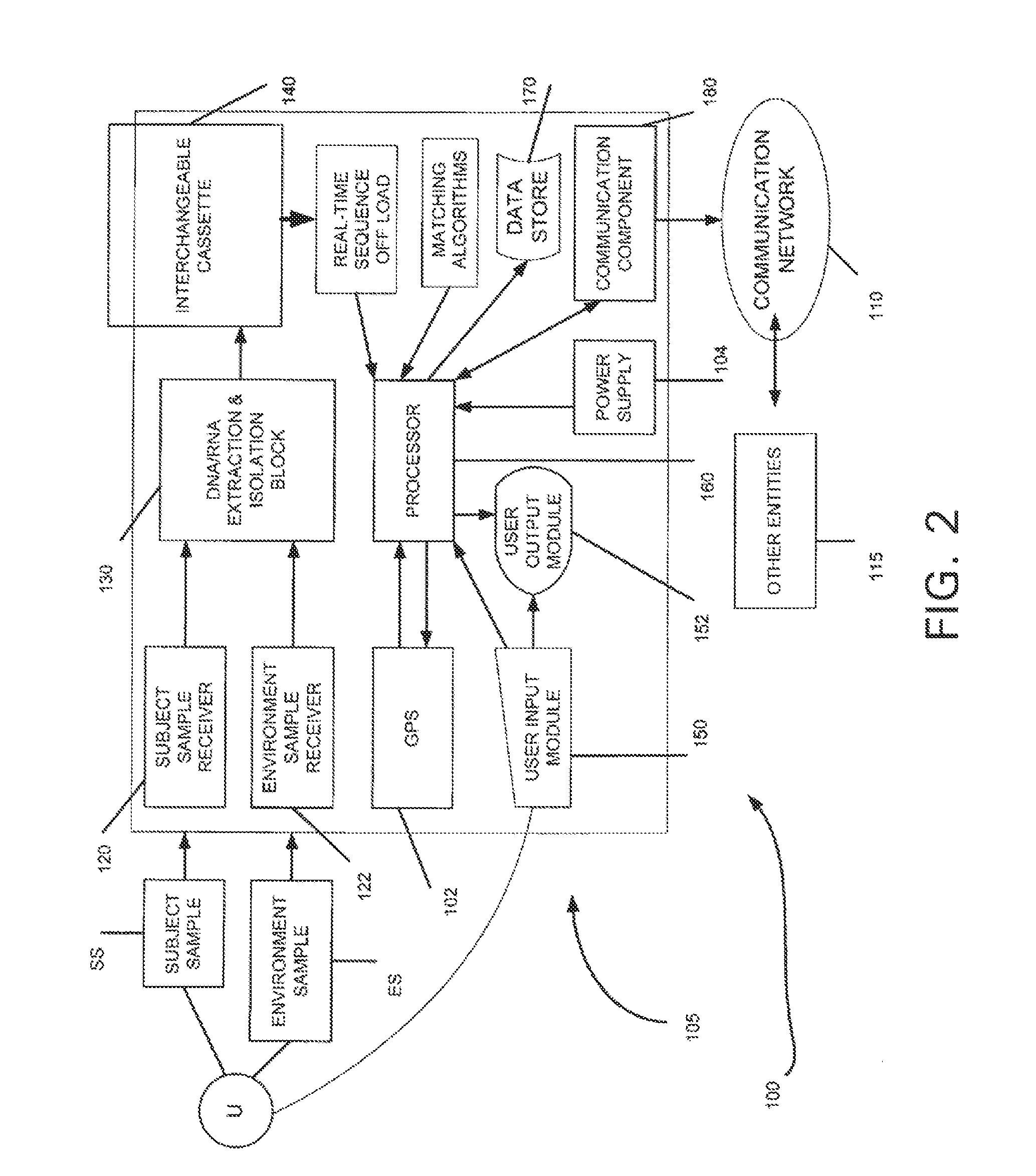
The present invention relates to systems and methods capable of characterizing populations of organisms within a sample. The characterization may utilize probabilistic matching of short strings of sequencing information to identify genomes from a reference genomic database to which the short strings belong. The characterization may include identification of the microbial community of the sample to the species and / or sub-species and / or strain level with their relative concentrations or abundance. In addition, the system and methods may enable rapid identification of organisms including both pathogens and commensals in clinical samples, and the identification may be achieved by a comparison of many (e.g., hundreds to millions) metagenomic fragments, which have been captured from a sample and sequenced, to many (e.g., millions or billions) of archived sequence information of genomes (i.e., reference genomic databases).
Owner:COSMOSID INC
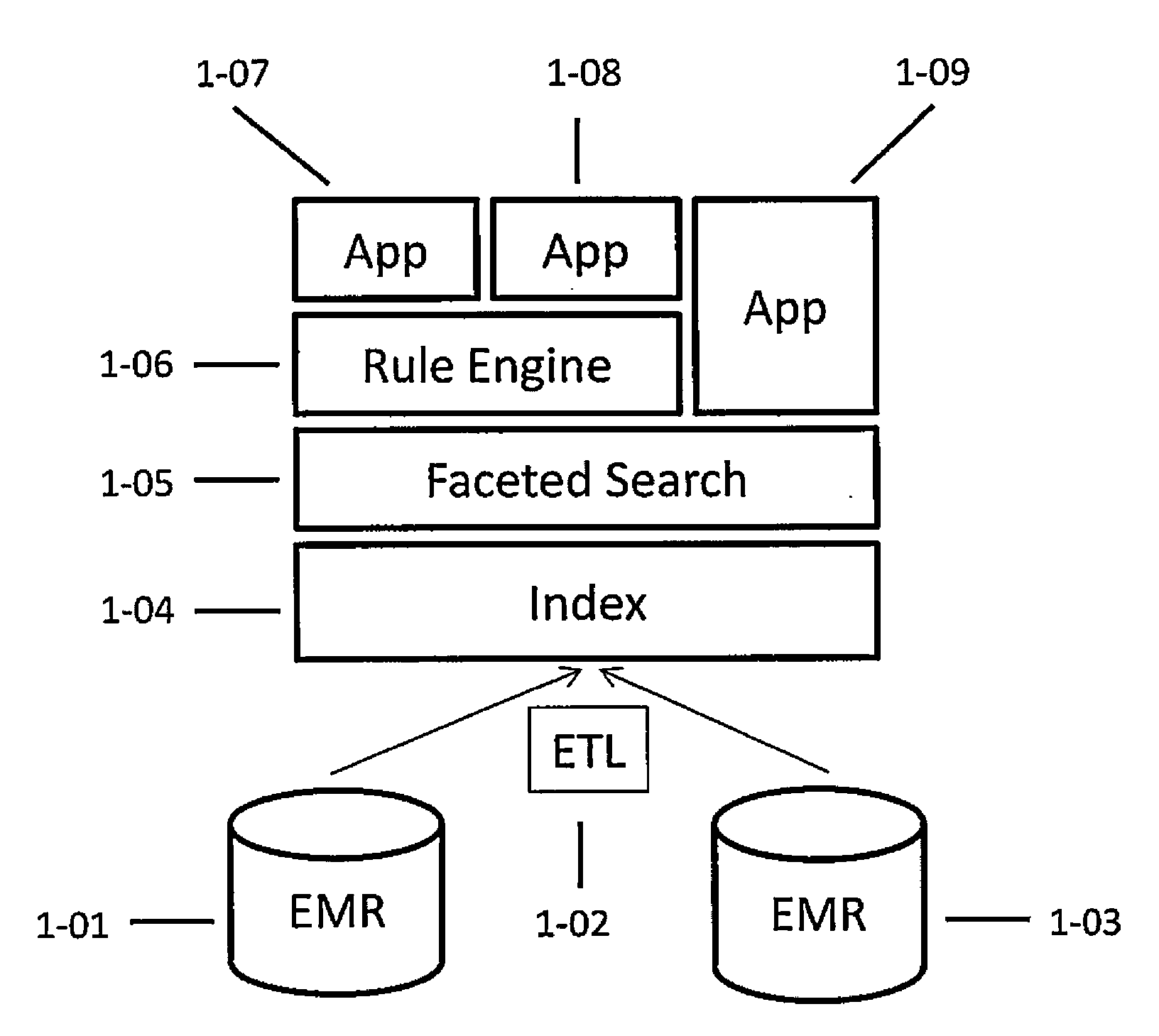
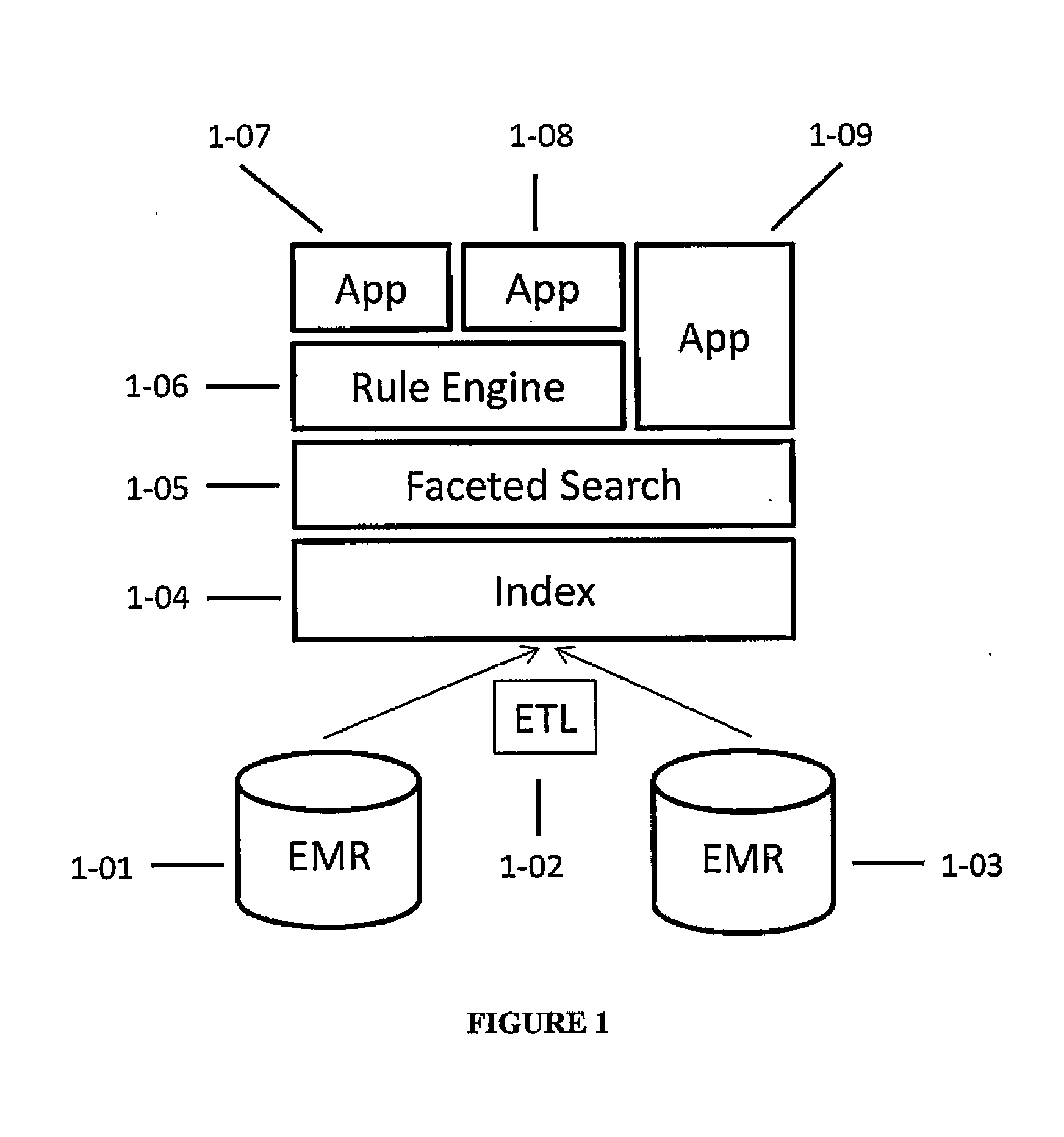
Systems and methods for searching genomic databases
InactiveUS20140046926A1Digital data processing detailsComputer-assisted medical data acquisitionClinical valueData source
The invention described herein solves the challenges encountered in searching for clinical and genomic information from multiple data sources. Systems, methods, and devices of the invention allow a user to search a number of dissimilar information sources simultaneously, and view, process, and perform correlations on the information. The invention uses faceted search to process clinical values, genomic data, subject characteristics, and population characteristics, thereby providing a user with an array of information useful for monitoring or improving the state of health of a subject or a subject population. The invention allows a user to evaluate clinical and research information in a subject-centric way, and analyze information at either the individual or the population level.
Owner:MYCARE
Pathogenic microorganism genome database and establishment method thereof
ActiveCN110473594AReduce data volumeAvoid false positivesBioinformaticsInstrumentsPathogenic microorganismHemidesmosome assembly
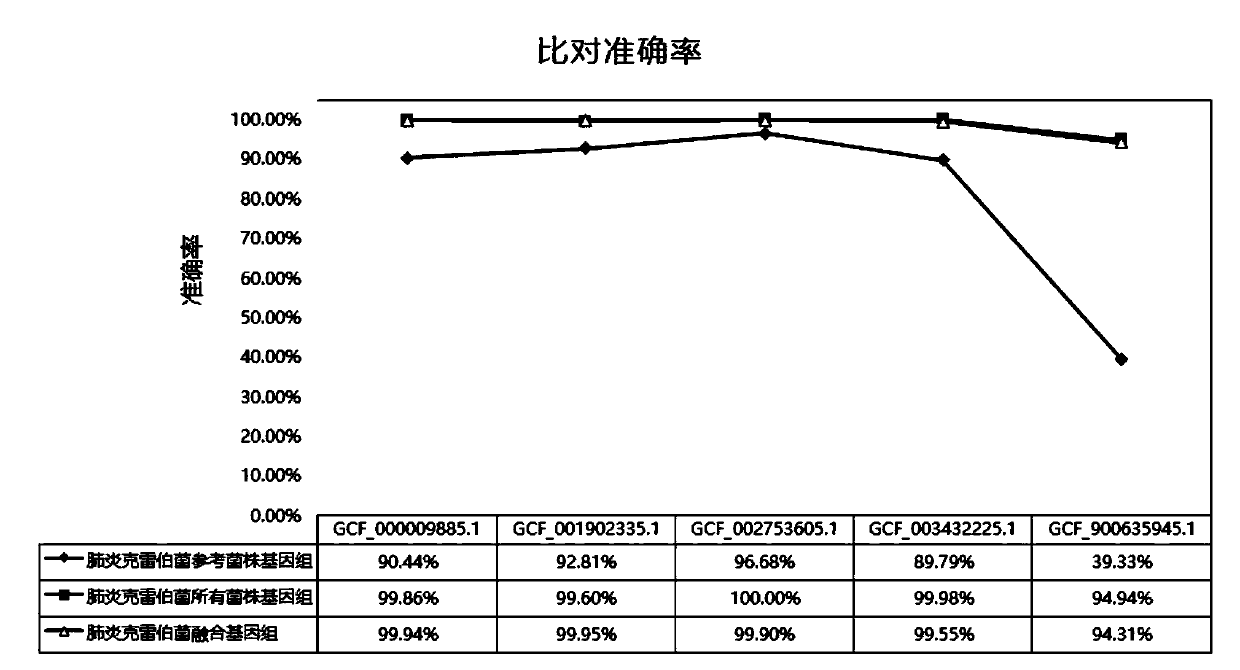
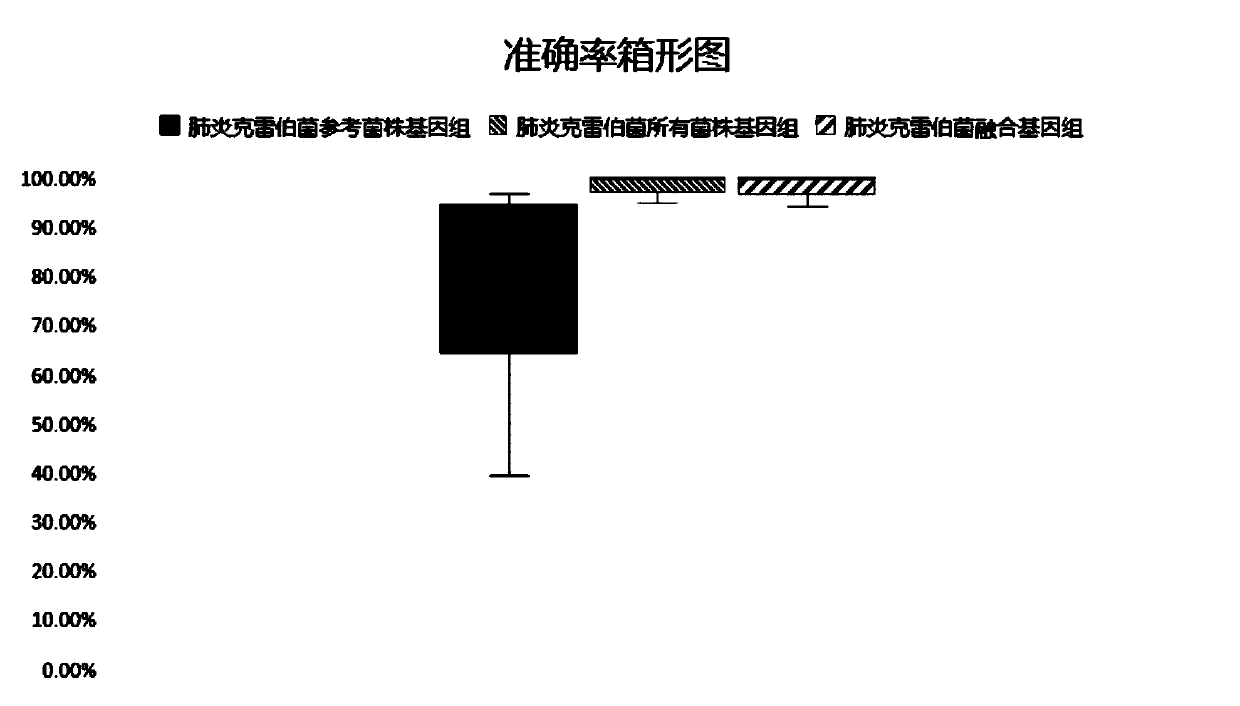
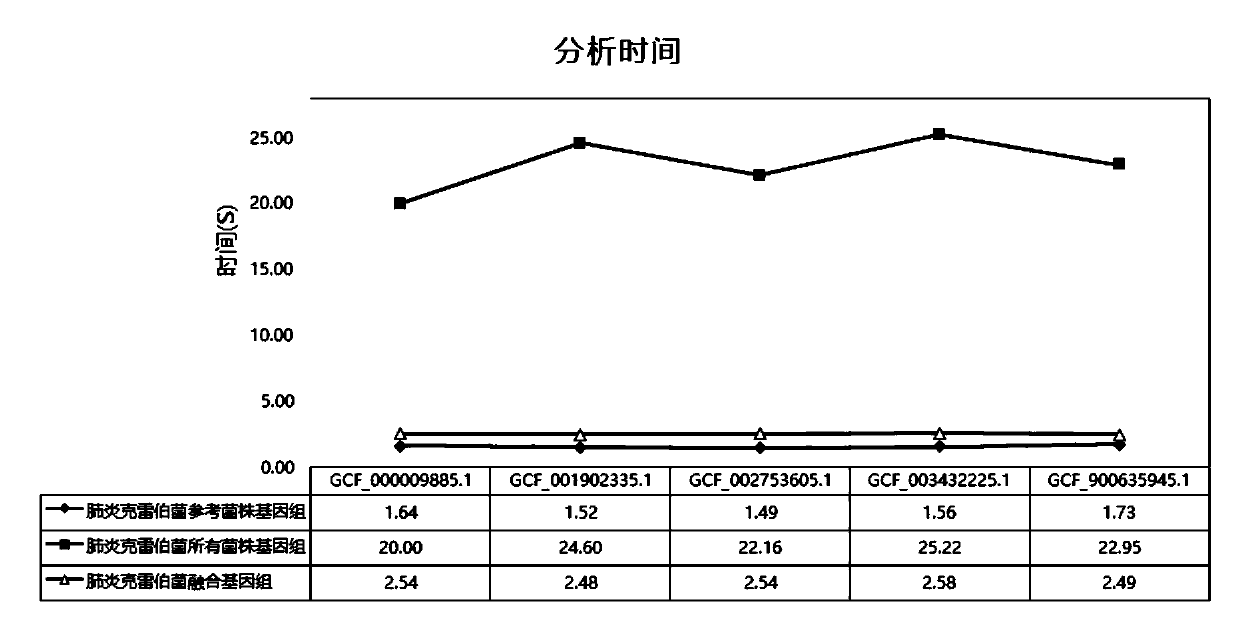
The invention relates to a pathogenic microorganism genome database and an establishment method thereof, and belongs to the technical field of meta-genomes. The method comprises the following steps ofdata acquisition, wherein pathogenic microorganism genome data is obtained; strain genome screening, wherein species strain genomes are selected according to a predetermined screening rule; plasmid sequence removal, wherein plasmid sequences existing in the strain genomes obtained in the last step are removed; filtration, wherein according to a predetermined filtering rule, strains with incorrectlabeling information, incomplete chromosome assembly and incorrect classification are removed to obtain a reference strain genome of the species; fusion genome construction, wherein the reference strain genome is interrupted, redundancy is removed, reassembly is performed, and the sequences are reassembled to obtain a fusion genome of the species; database assembly, wherein the above steps are repeated to obtain the fusion genome of the predetermined species, summary is performed, and the pathogenic microorganism genome database is obtained. The genome database has the advantages of not onlyhaving a high precision rate, but also having short analysis time and reducing the cost.
Owner:GZ VISION GENE TECH CO LTD +4
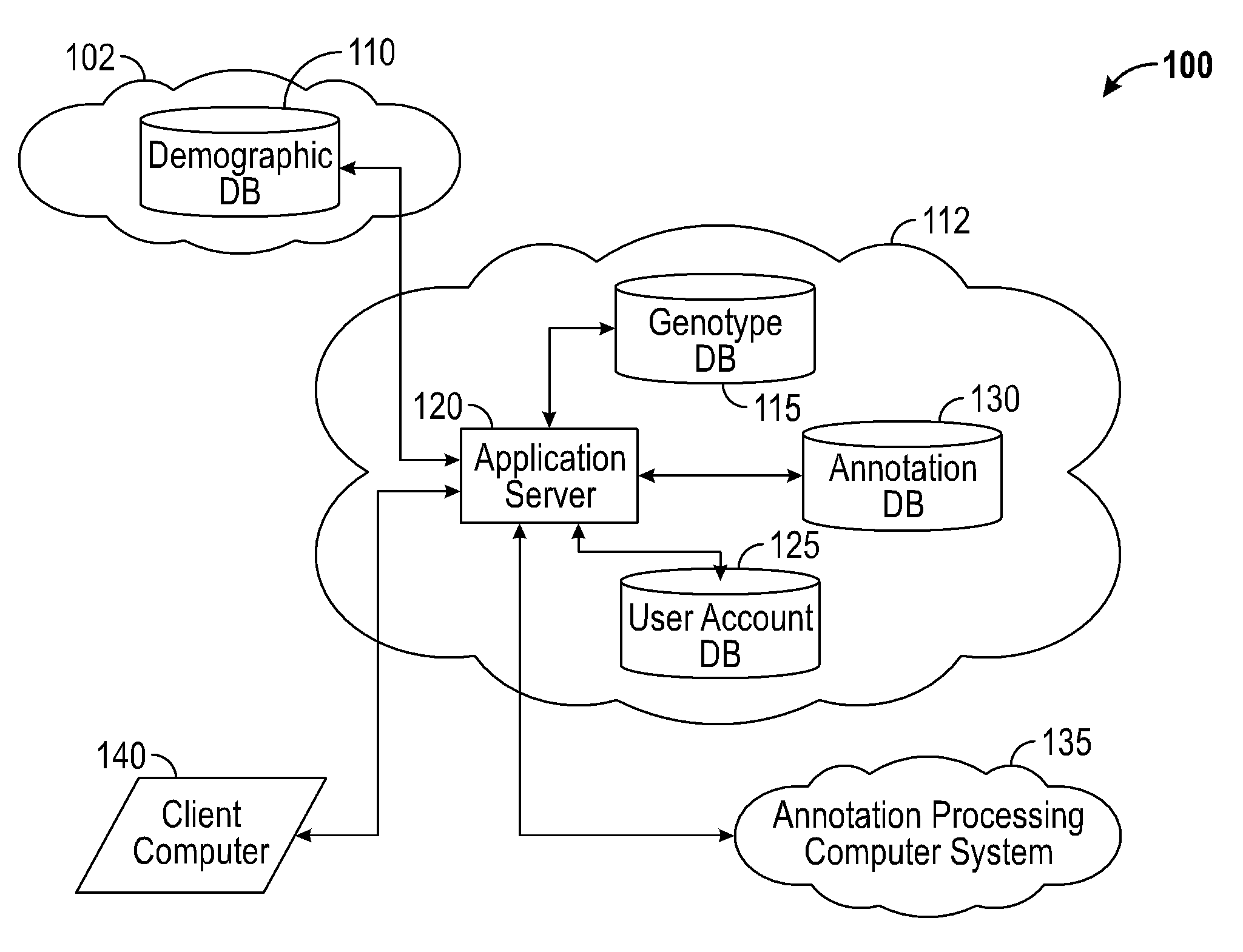
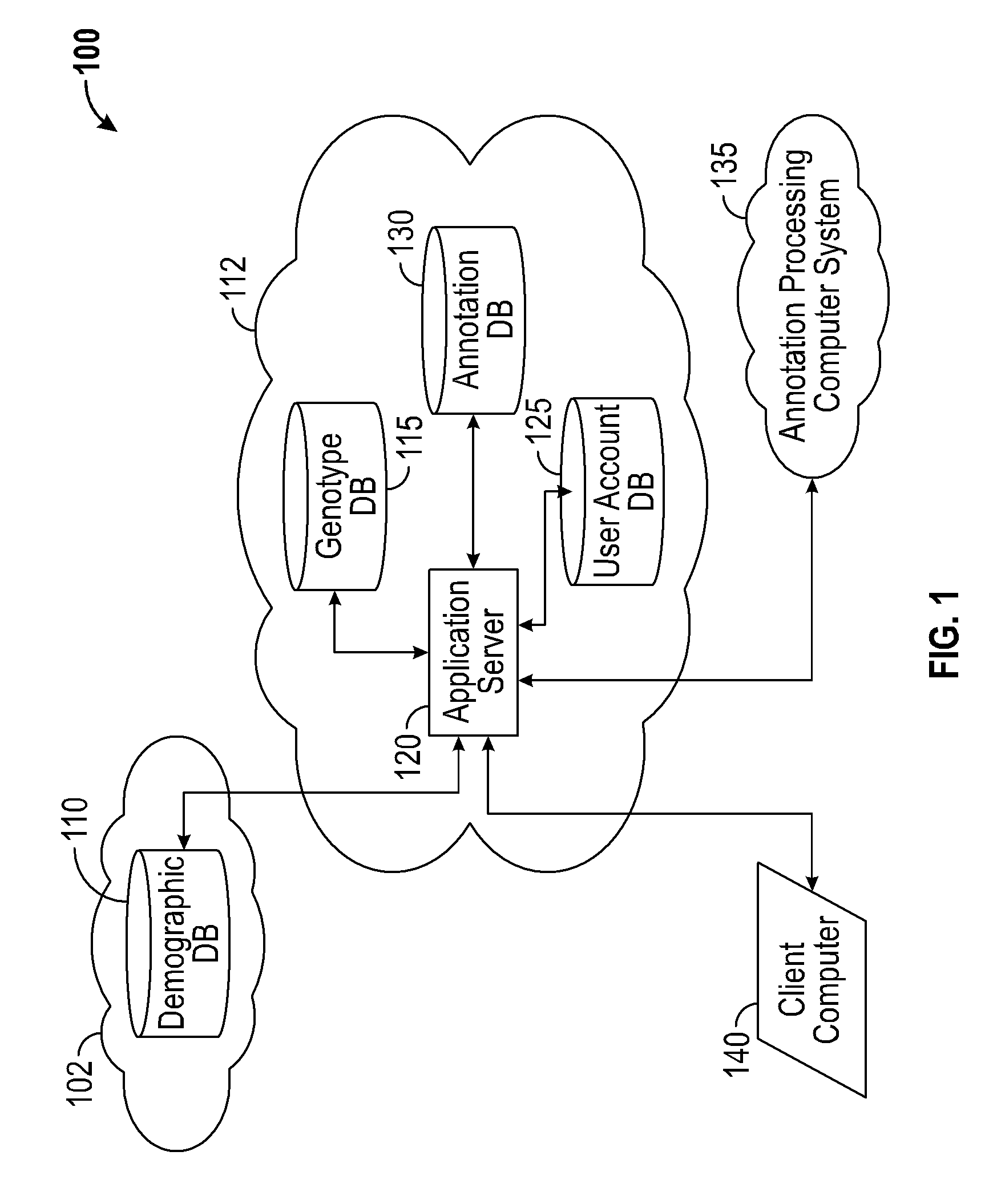
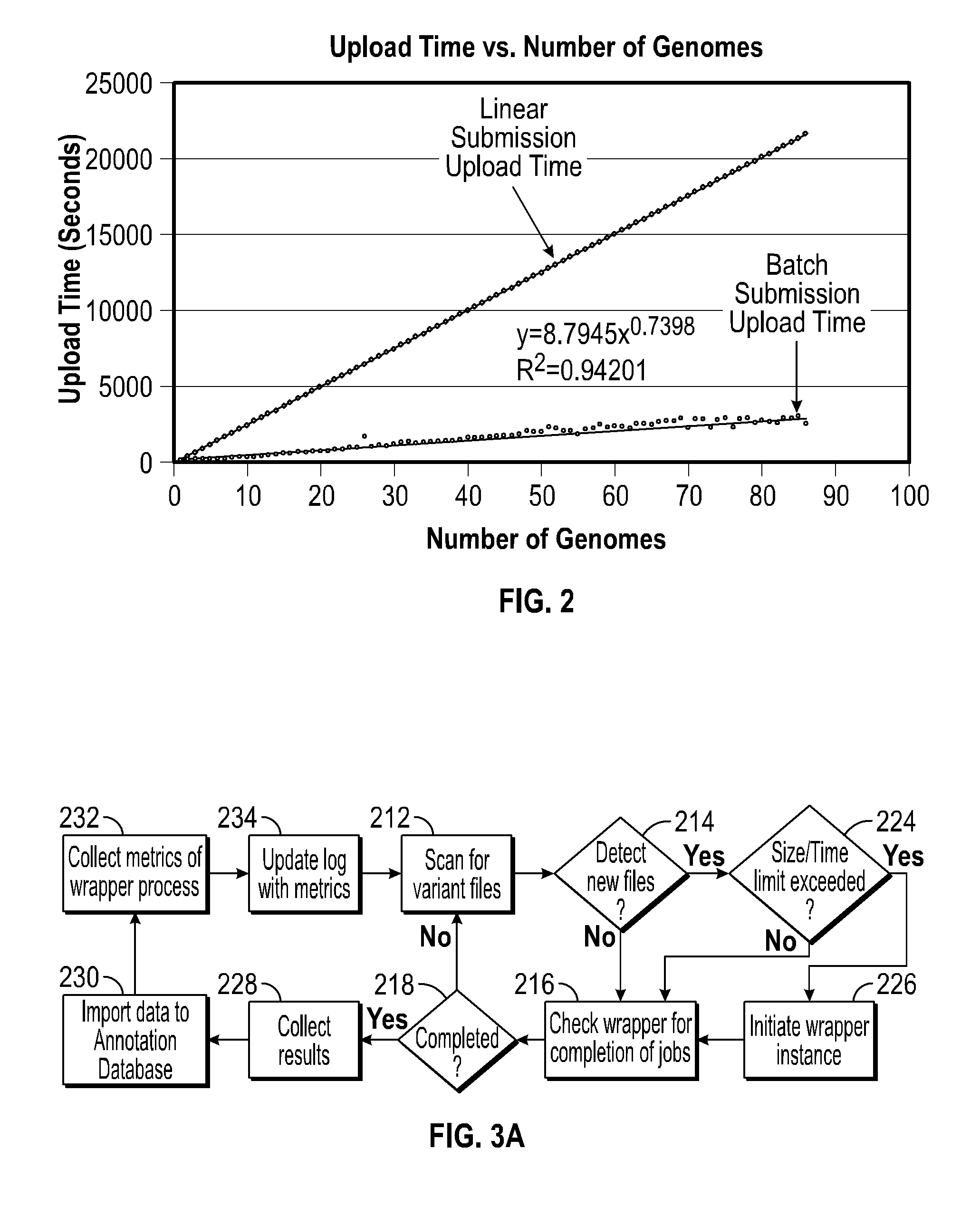
Systems and methods for genomic variant annotation
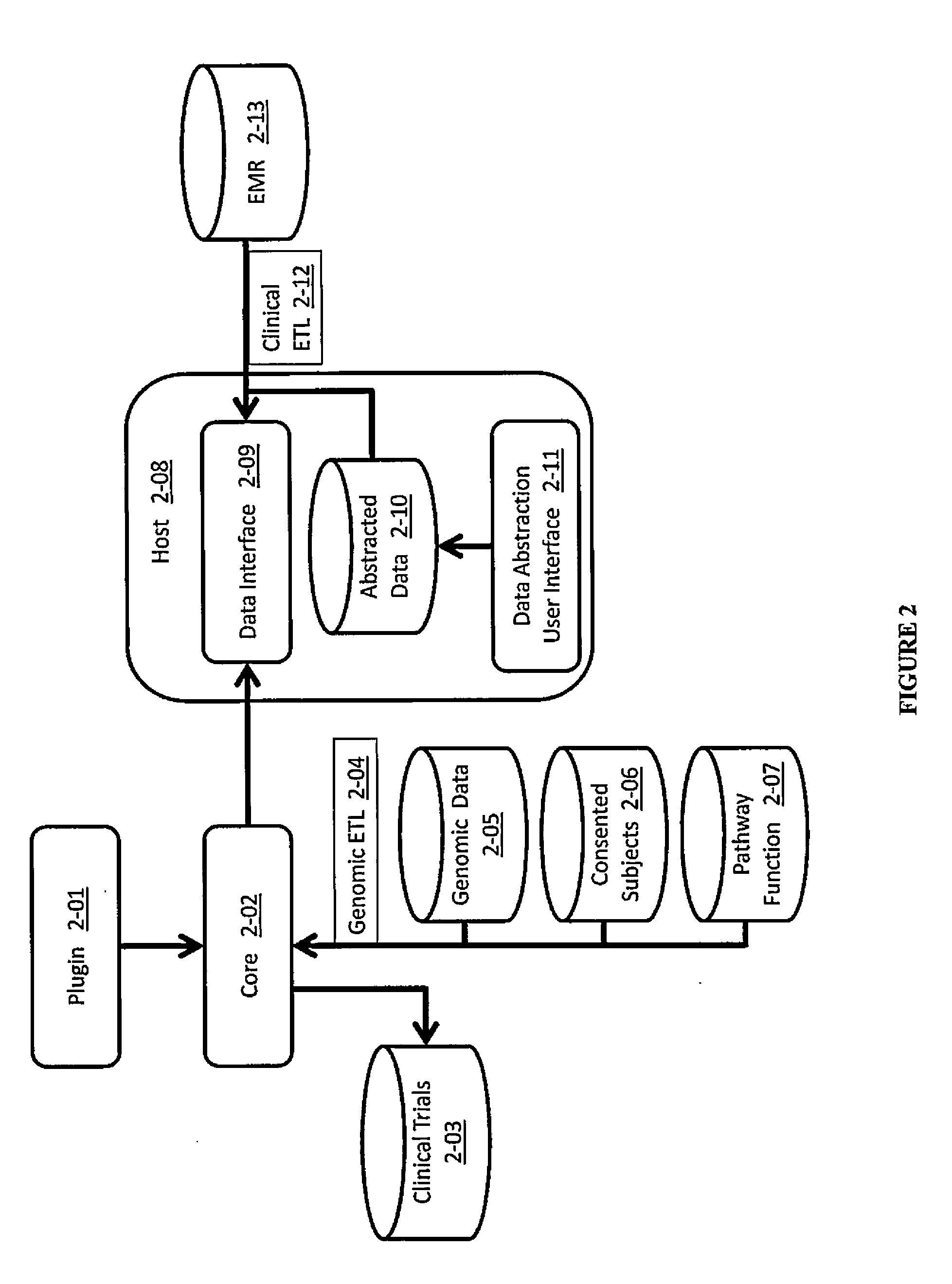
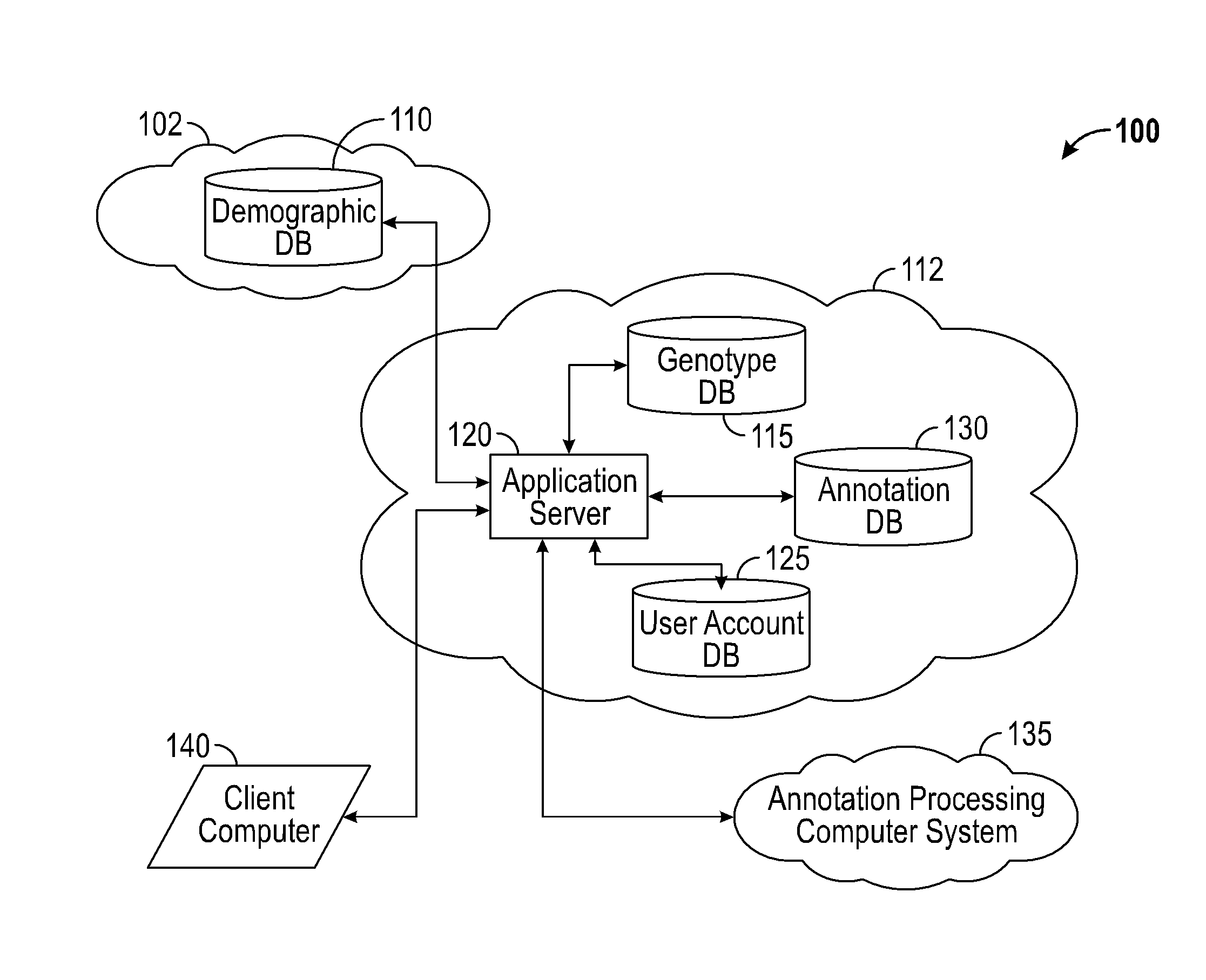
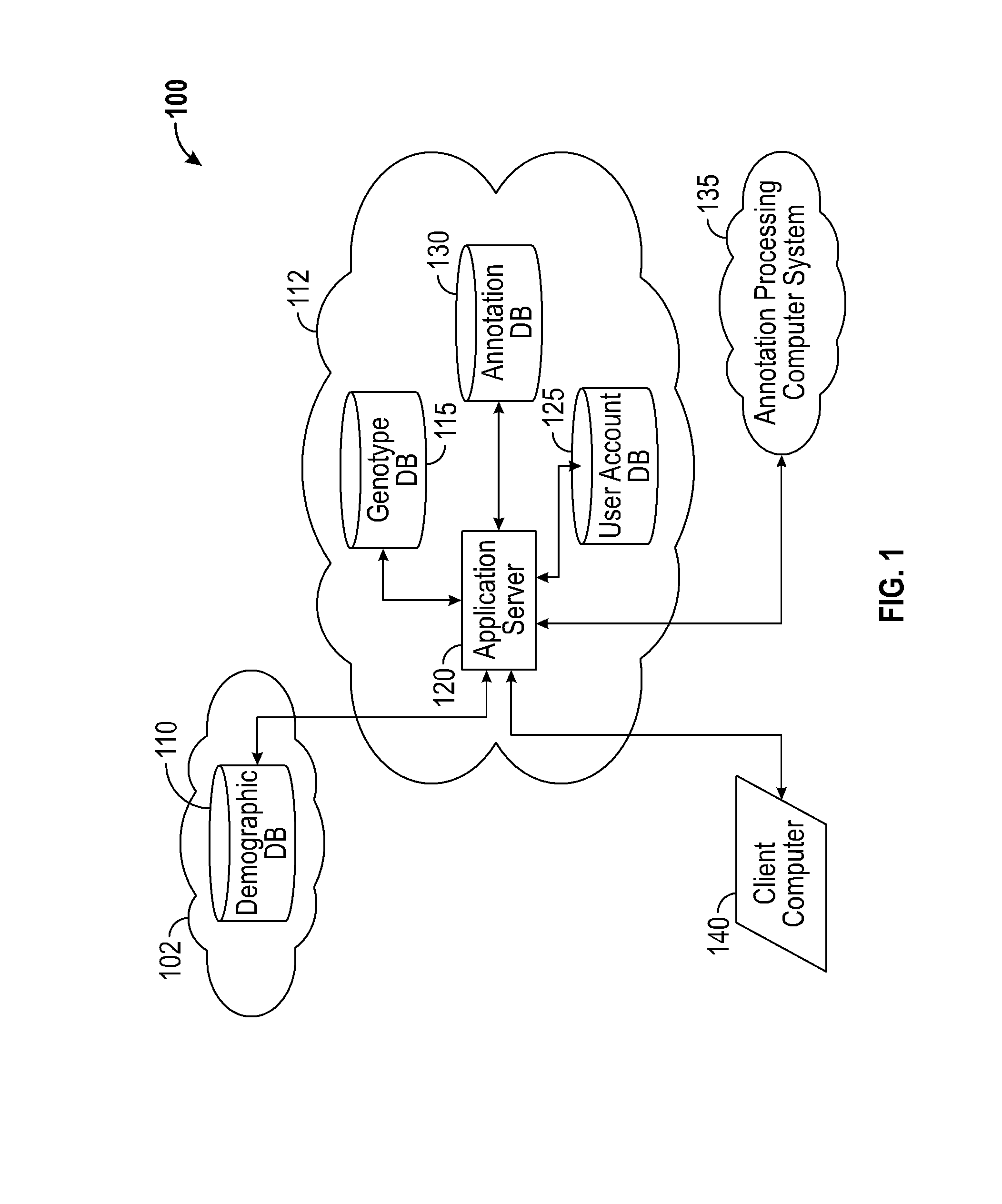
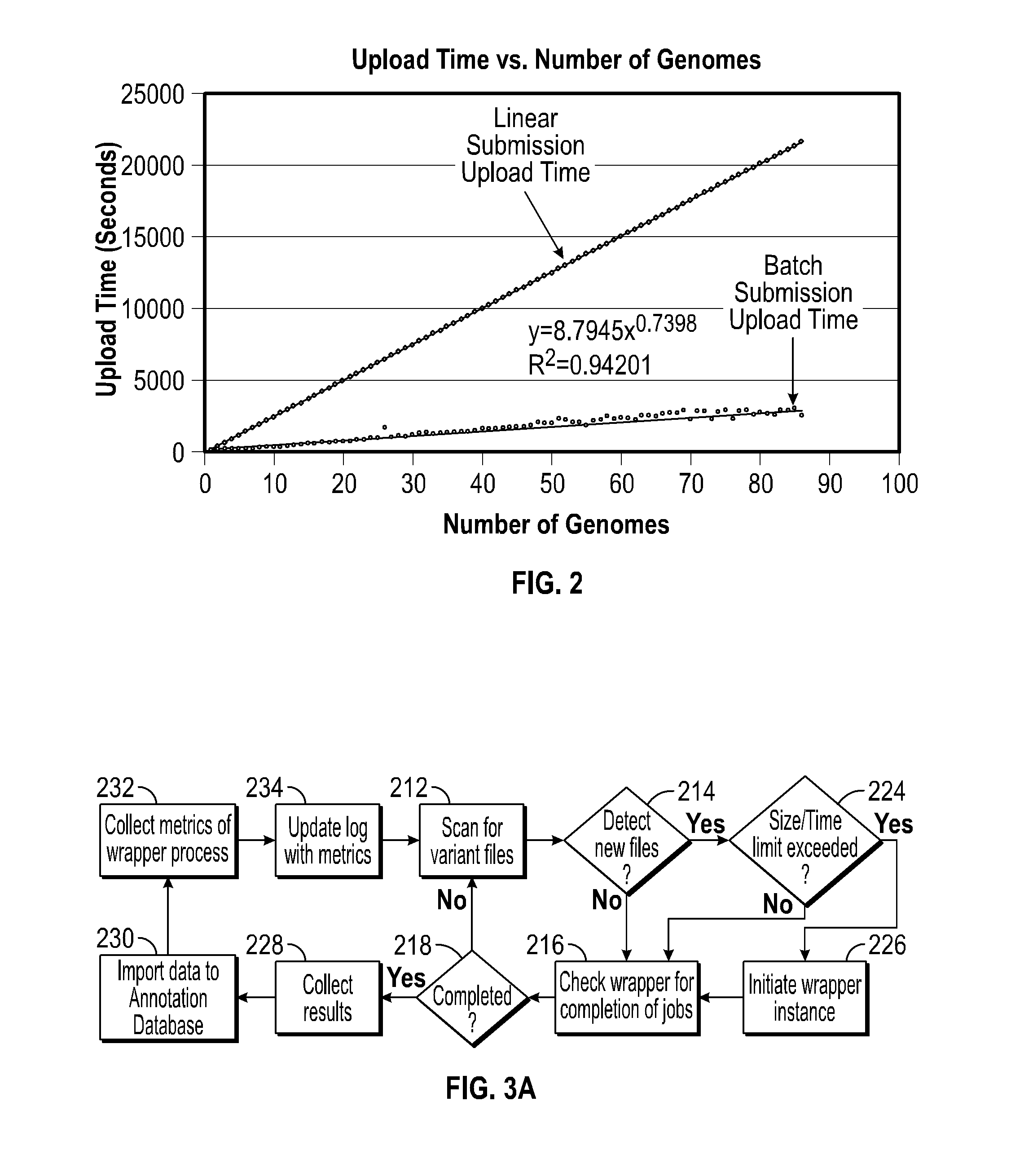
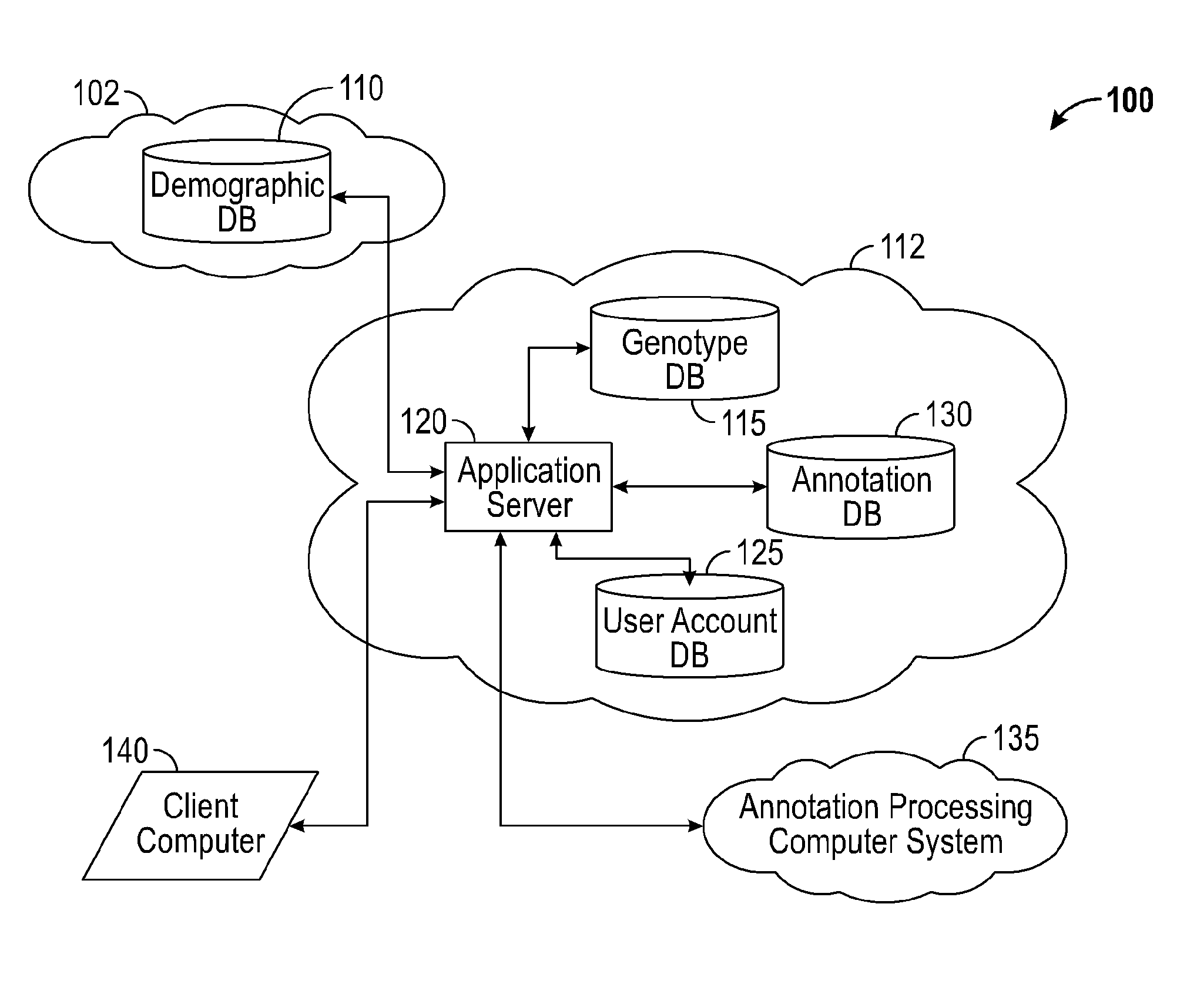
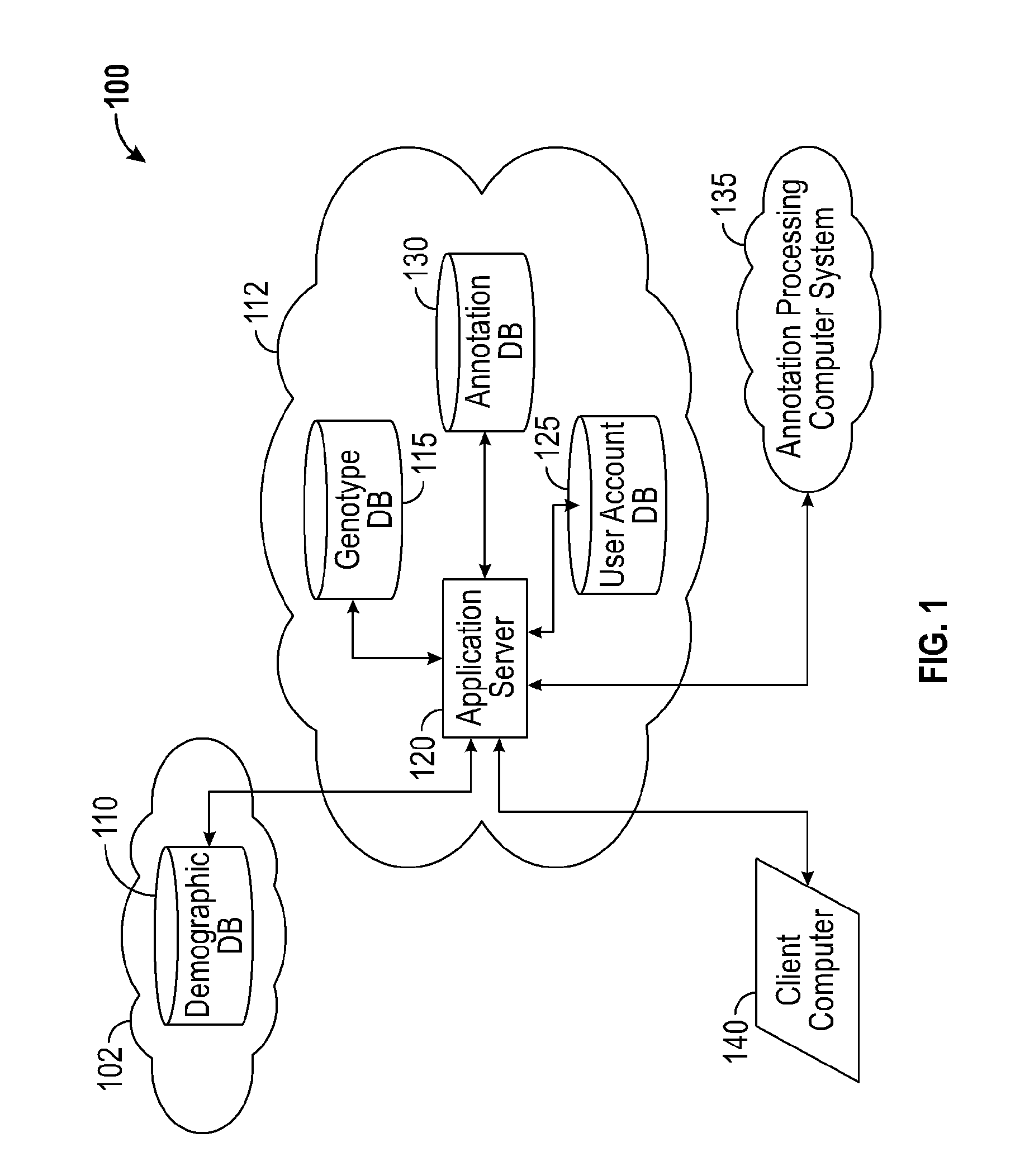
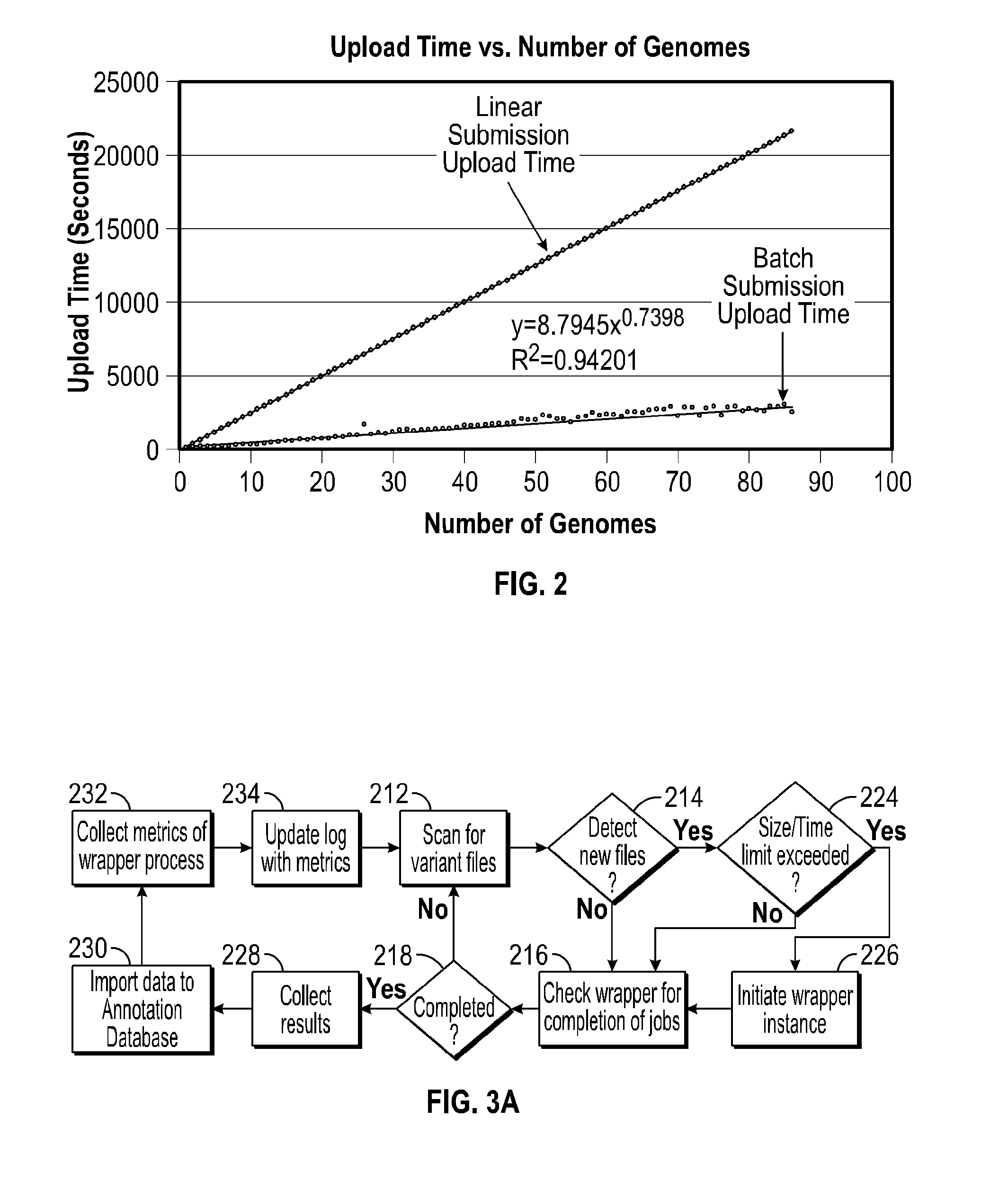
A system for annotating genomic variant files includes an application server, an annotation database, a genomic database, and an annotation processing computer system. The genomic database may be graph-oriented. The annotation processing computer system processes can process variant files in batch modes and includes annotation modules designed to improve the speed of the annotation process. The batch modes may include batch transmission, and / or batch annotation.
Owner:CYPHER GENOMICS
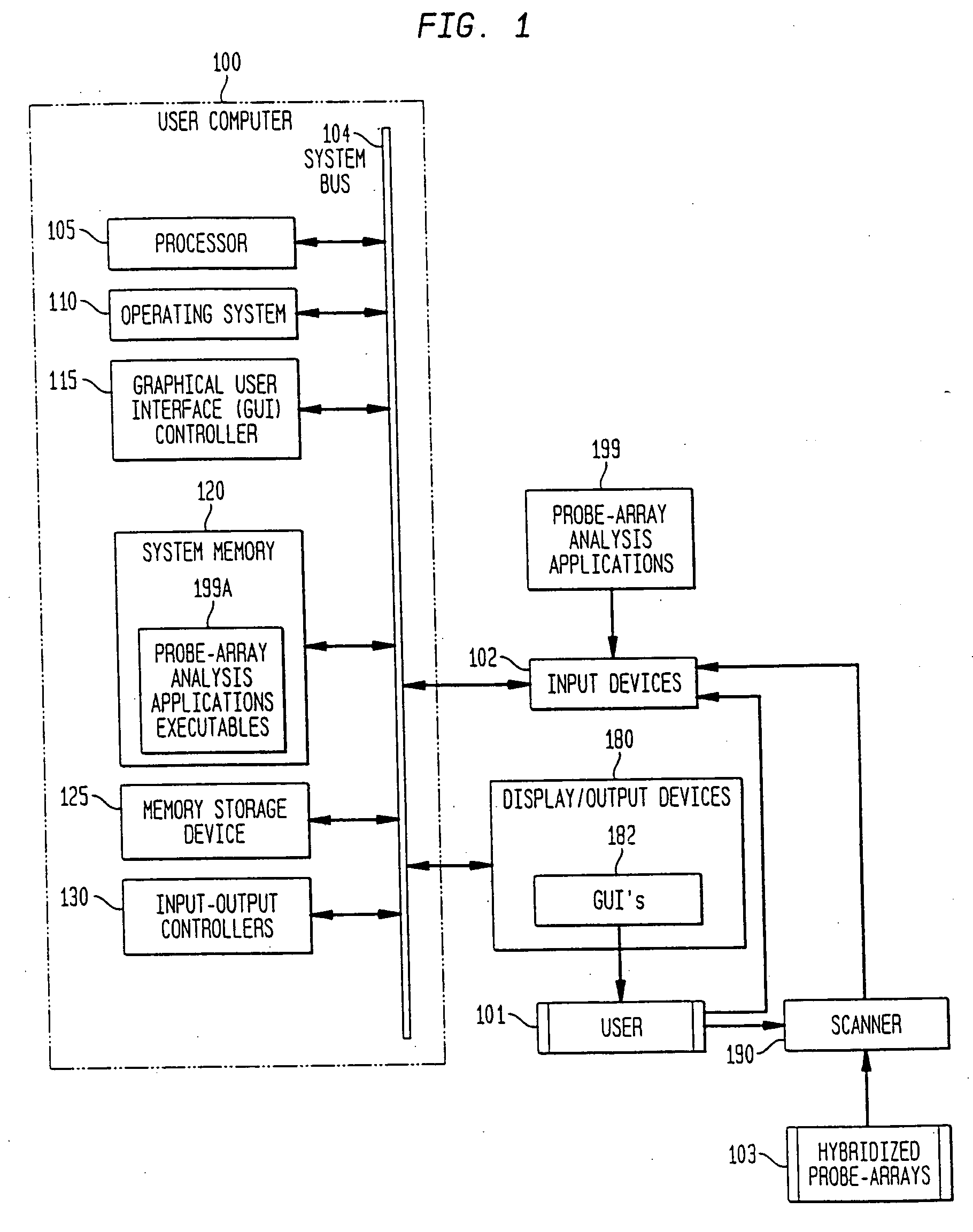
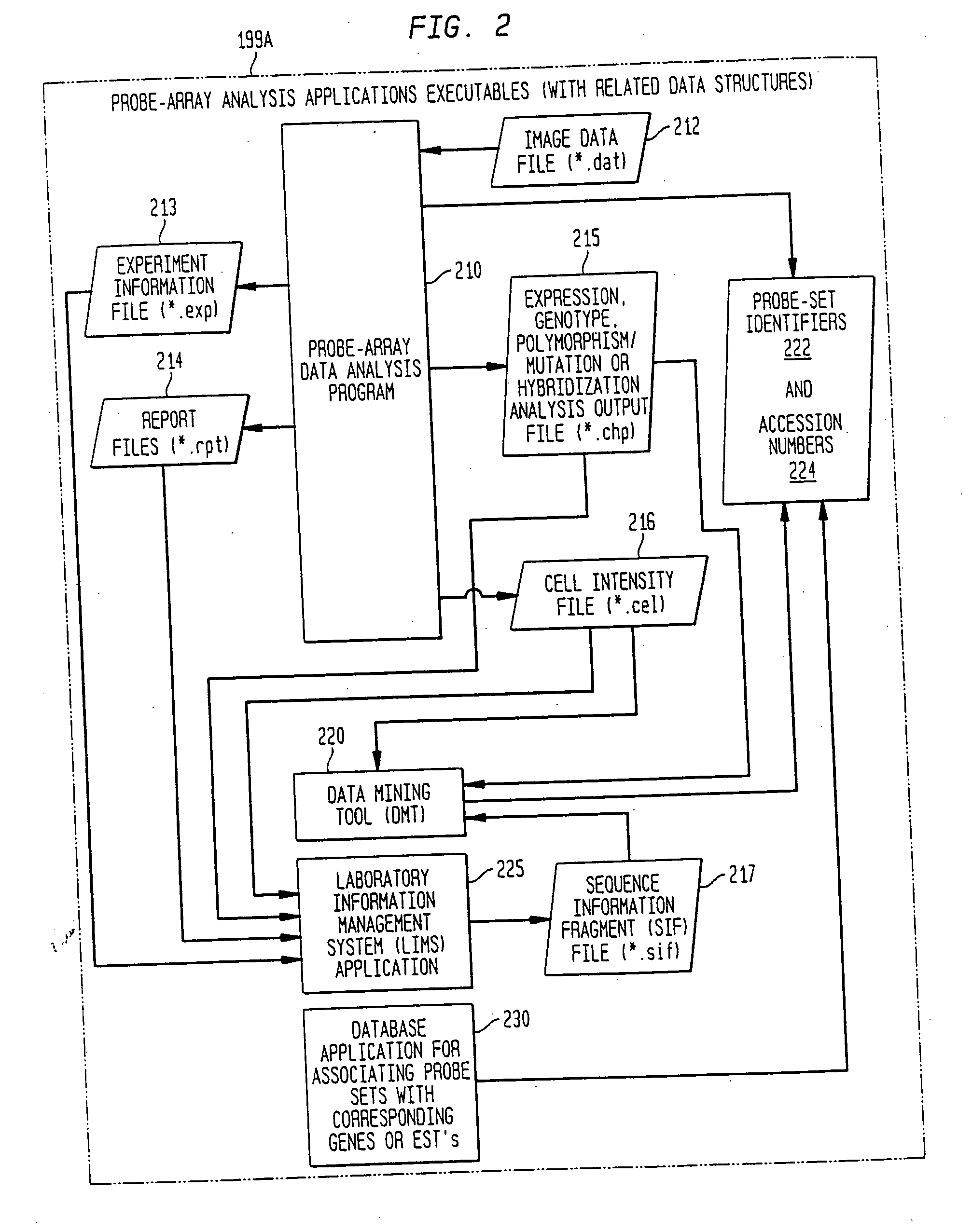
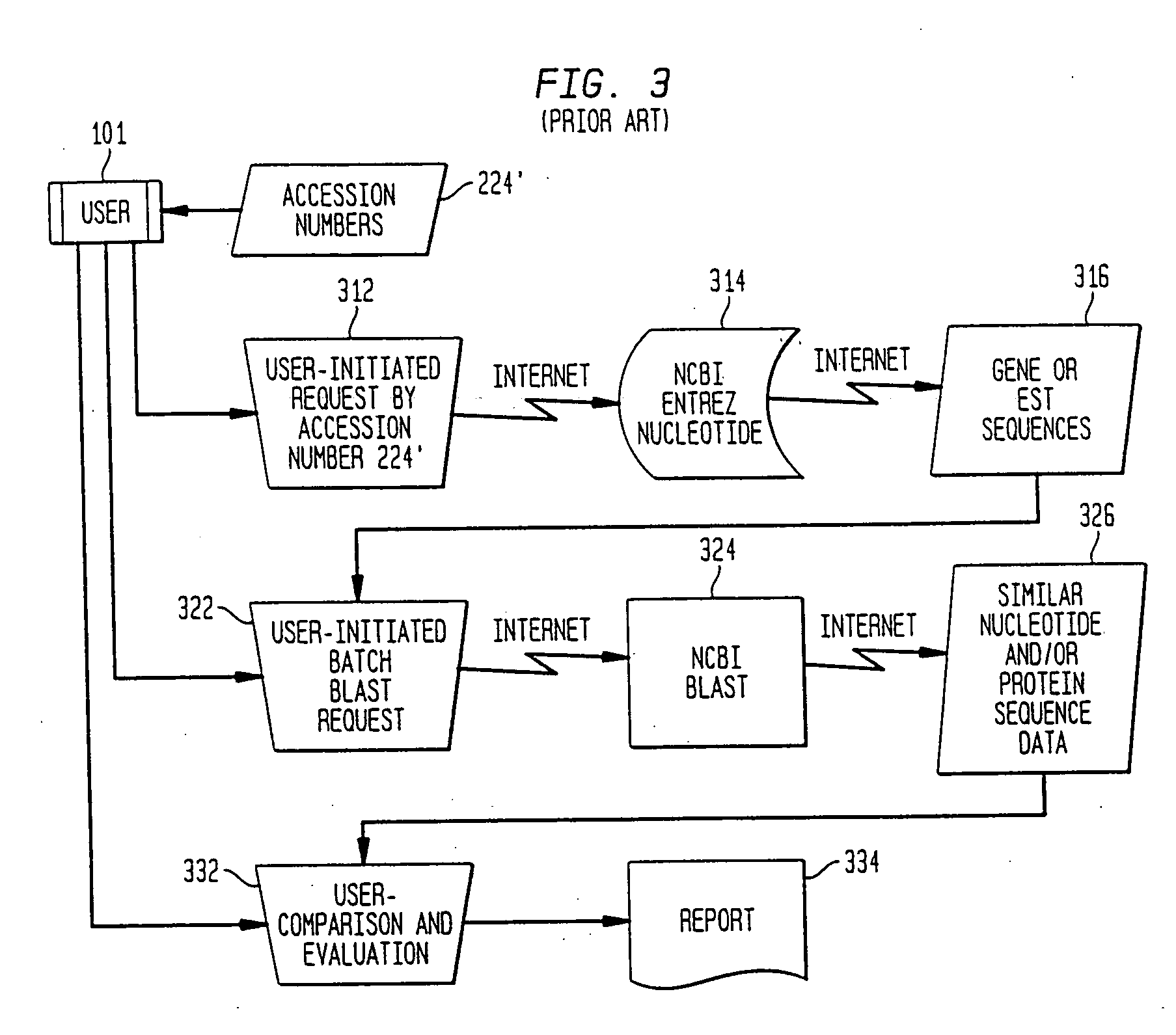
Method, system, and computer software for providing a genomic web portal
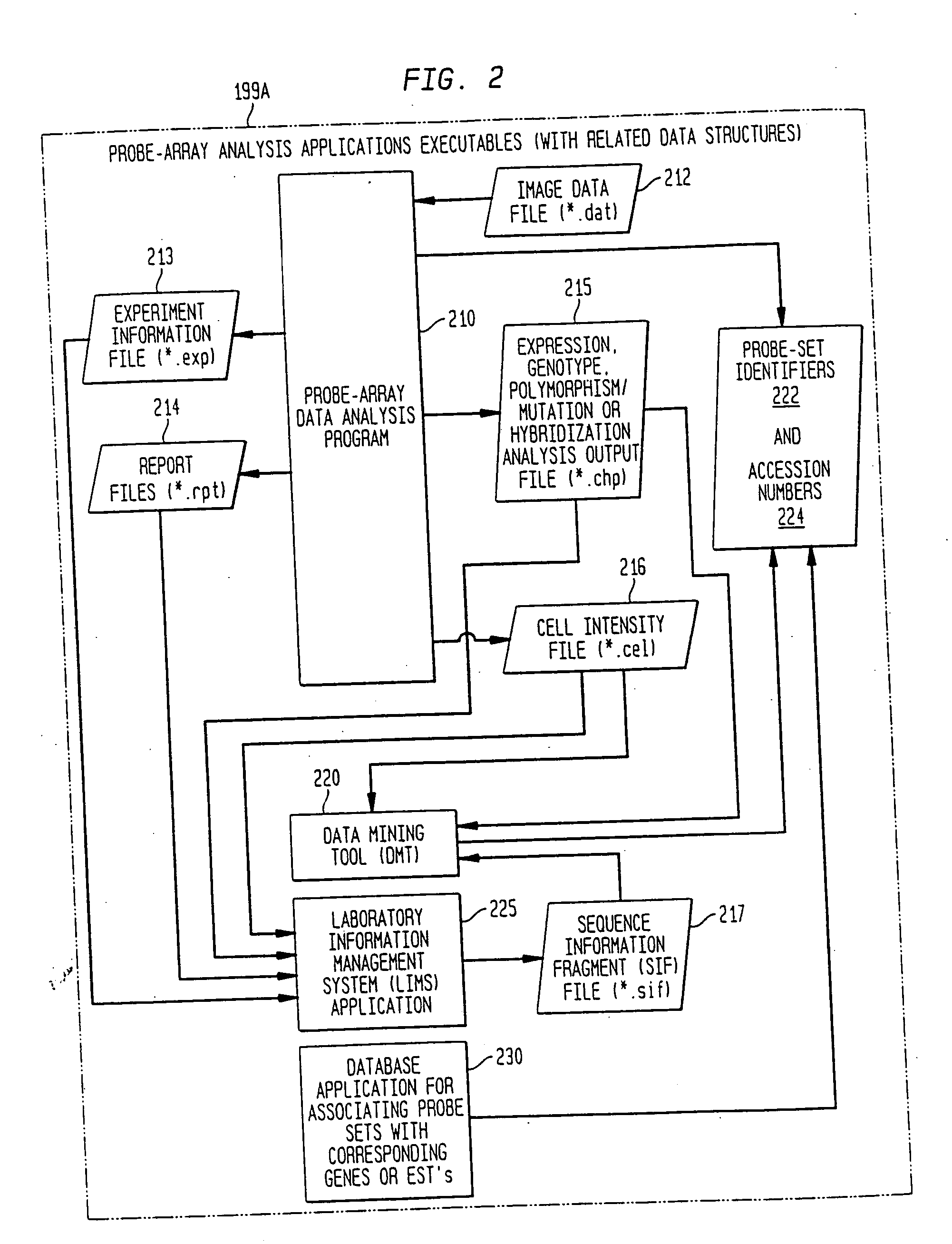
InactiveUS20050009078A1Significant delayMicrobiological testing/measurementBiological testingGenomic databasesComputer software
Systems, methods, and computer program products are described that process inquiries or orders regarding purchase of biological devices, substances, or related reagents. In some implementations, a user selects probe-set identifiers that identify microarray probe sets capable of enabling detection of biological molecules. Corresponding genes or EST's are identified and are correlated with related product data, which is provided to the user. Further, the user may select products for purchase based on the product data. If so, the user's account may be adjusted based on the purchase order. In the same or other implementations, a local genomic database is periodically updated. In response to a user selection of probe-set identifiers, data related to corresponding genes or EST's is provided to the user from the local genomic database.
Owner:AFFYMETRIX INC
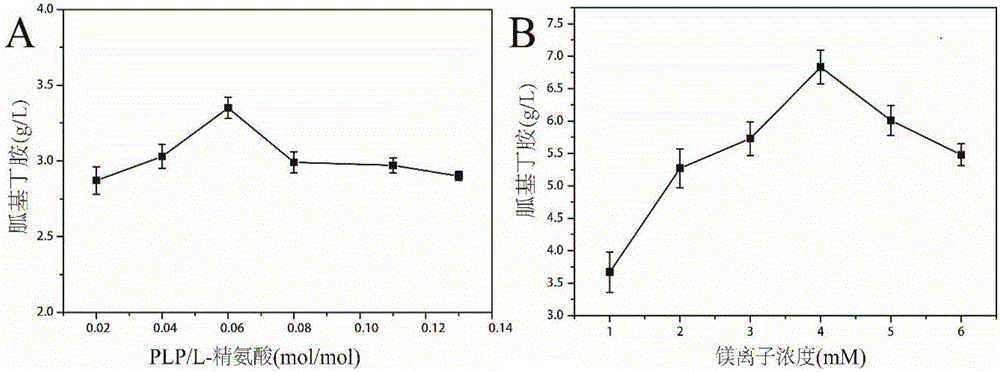
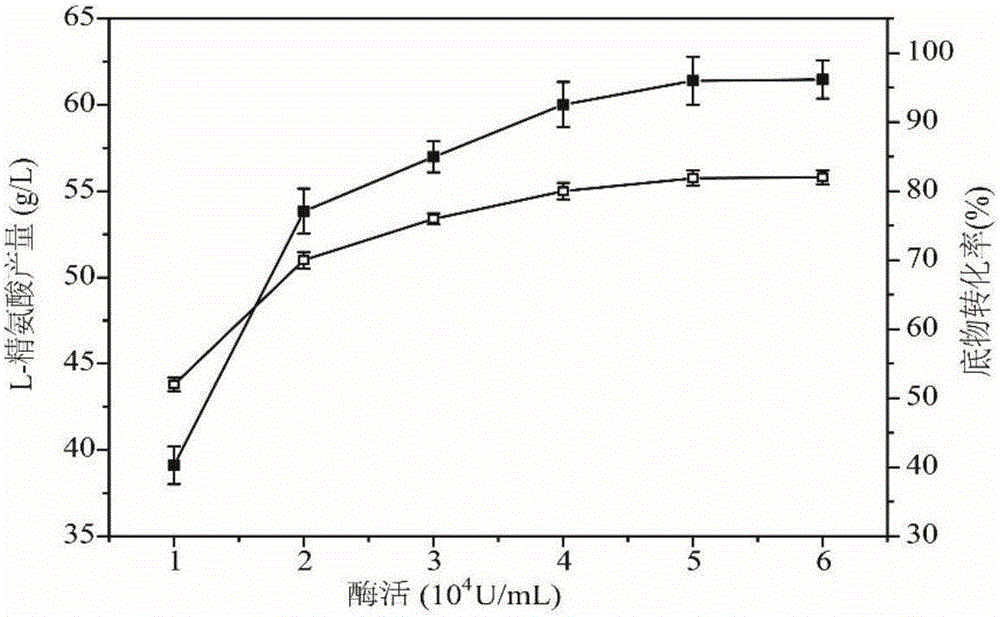
Arginine decarboxylase and application thereof
InactiveCN105861529AIncrease vitalityMeet the needs of industrial scale productionFermentationGenetic engineeringEscherichia coliArginine
The invention discloses arginine decarboxylase and application thereof and belongs to the technical field of biological engineering. Shewanella putrefaciens is cloned by adopting genome database mining and homologous sequence alignment and other virtual screening means and combining with a molecular biological means to obtain an arginine decarboxylase gene. The molecular biological means, process optimization and the like are adopted, the enzyme production capacity of arginine decarboxylase production strains is improved, and a platform for efficiently producing gamatine through biological catalysis is established by optimizing a catalytic system. Crude enzyme liquid obtained through fermentation is purified and then is converted, a reaction system is clear in composition, and extraction and purification of follow-up products are promoted. Under the condition of 37 DEG C, the decarboxylase has high activity, meanwhile is beneficial to the growth of escherichia coli of host cells, and at the temperature, the reaction speed is improved greatly, a conversion period should be 24 hours, the gamatine yield can be up to 61-71 g / L, and the conversion rate can be up to 68-82%.
Owner:JIANGNAN UNIV
Systems and methods for genomic variant annotation
InactiveUS20160048633A1Digital data information retrievalDigital data processing detailsGraphicsApplication server
A system for annotating genomic variant files includes an application server, an annotation database, a genomic database, and an annotation processing computer system. The genomic database may be graph-oriented. The annotation processing computer system processes can process variant files in batch modes and includes annotation modules designed to improve the speed of the annotation process. The batch modes may include batch transmission, and / or batch annotation.
Owner:CYPHER GENOMICS
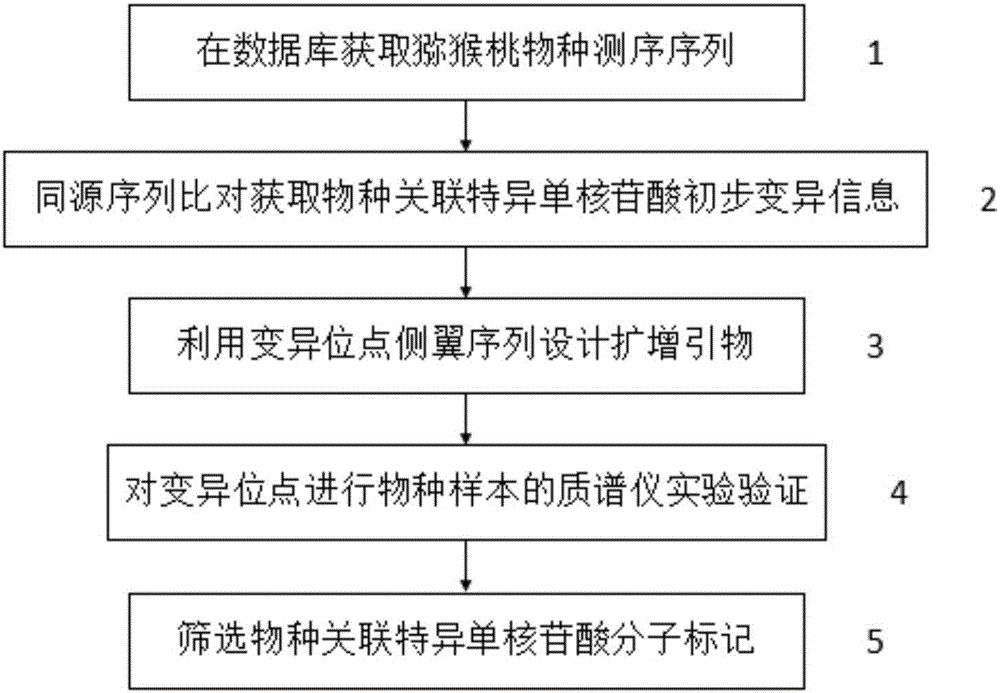
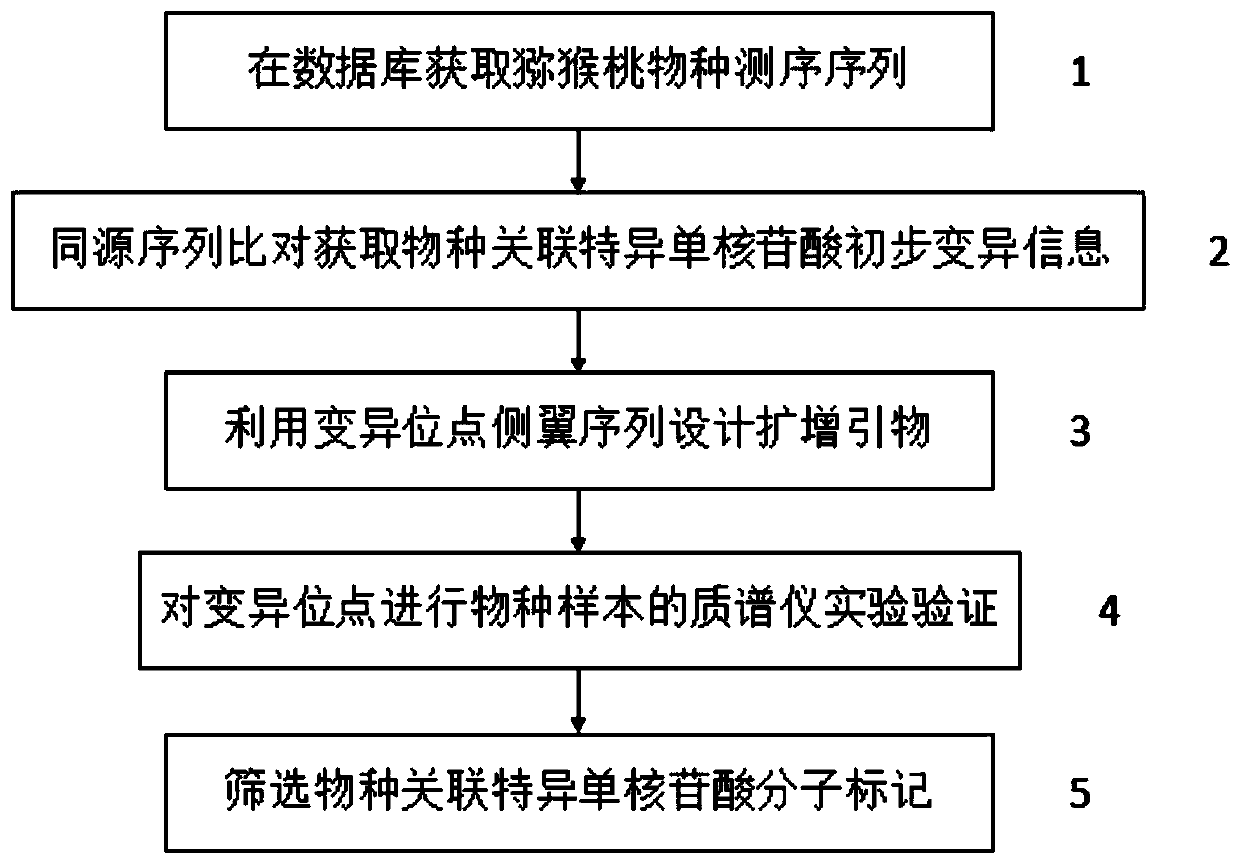
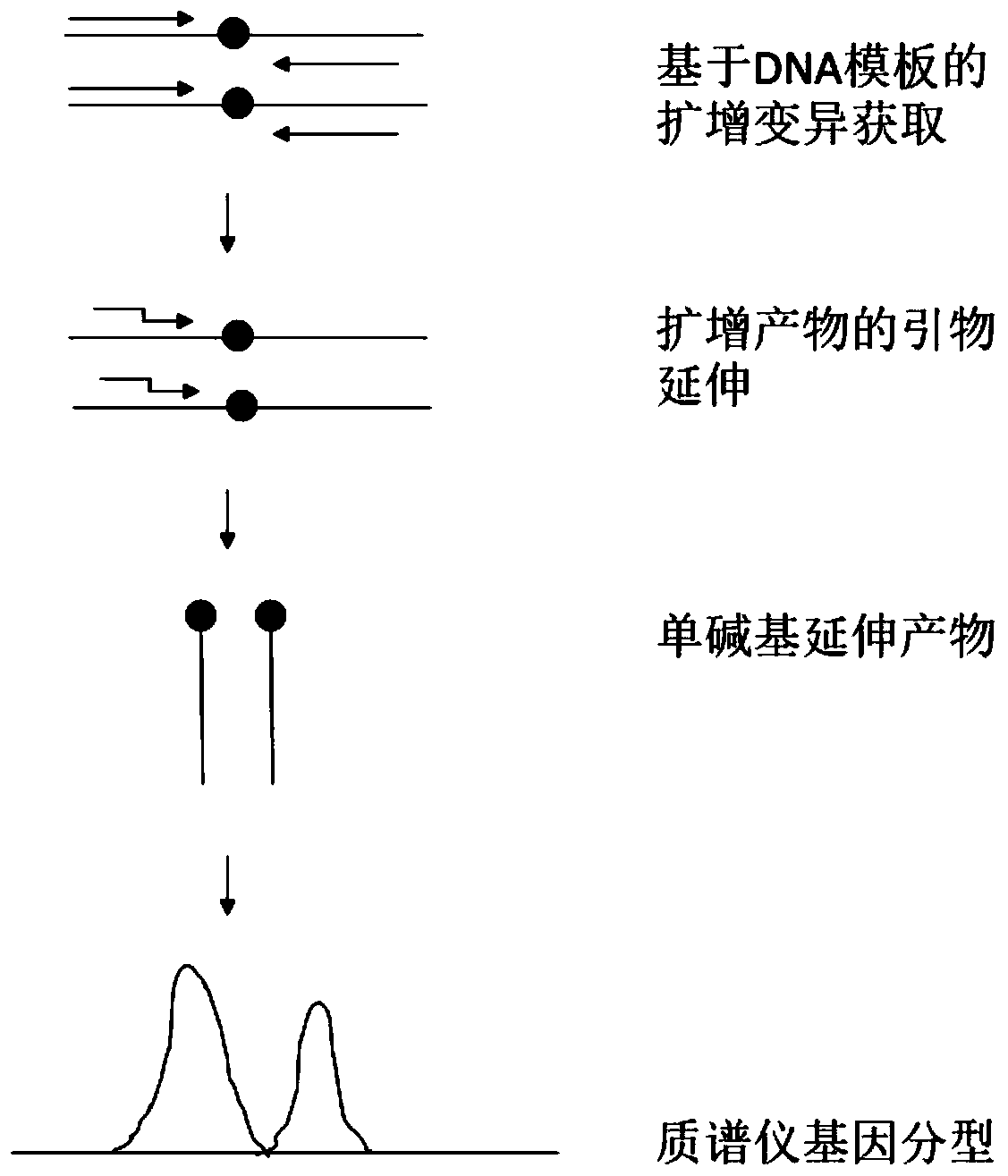
Kiwi fruit species association specific mononucleotide molecular markers and detection primer set and application thereof
ActiveCN106367496AThe identification result is accurateFacilitate the determination of parental sourceMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
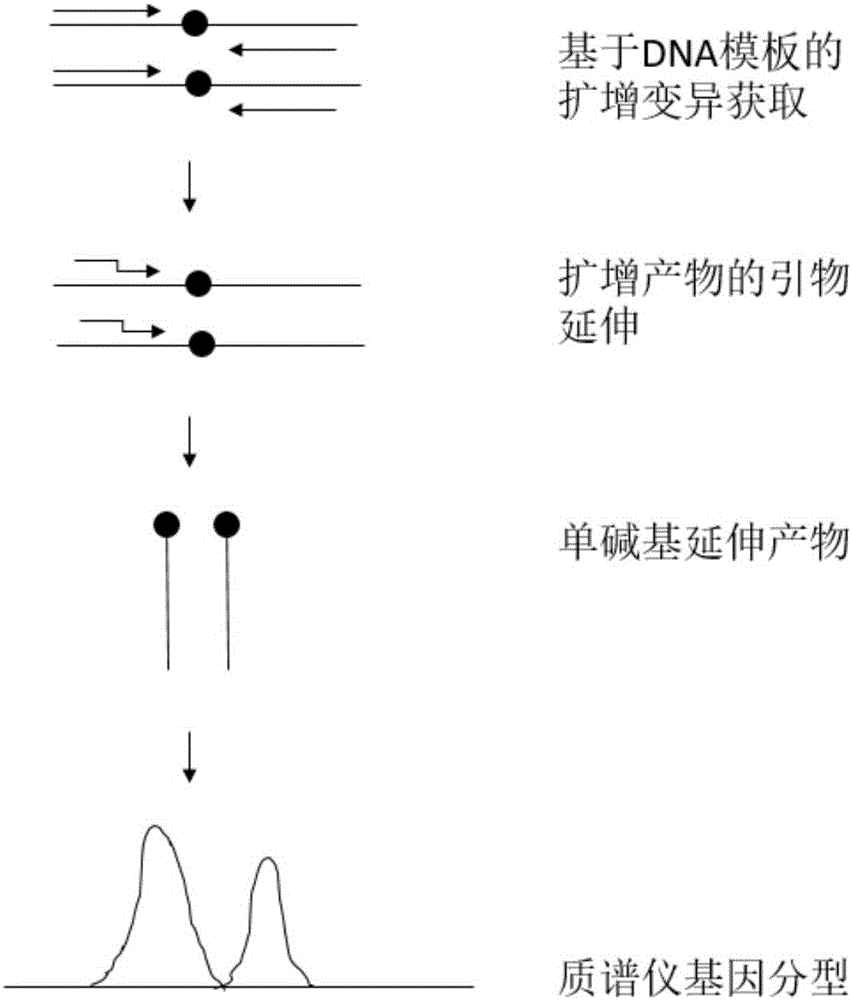
The invention discloses kiwi fruit species association specific mononucleotide molecular markers and a detection primer set and application thereof. By means of sequencing sequences of different kiwi fruit species existing in a kiwi fruit genome database, high-diversity mononucleotide variation regions are looked for in a sequence alignment reference genome mode, a mononucleotide molecular marker primarily selected set is formed, on the basis, a novel mass spectrometer gene typing platform is used for carrying out experimental verification and genotype acquisition on mononucleotide variation sites, species association specific mononucleotide markers are screened through association analysis with the species, finally, a set of molecular markers for kiwi fruit hybrid resource parent species source identification are obtained, and the technical scheme is formed. Identification of kiwi fruit hybrid resource parents can promote understanding of formation of hybrid resource characters and biological characters and accelerate genetic evaluation and development and utilization of hybrid resources, and great theoretical academic significance and high economic application value are achieved.
Owner:SOUTH CHINA BOTANICAL GARDEN CHINESE ACADEMY OF SCI
Use of Fibroblast Growth Factor Fragments
InactiveUS20090093398A1Provide goodEasy to understandSenses disorderPeptide/protein ingredientsWhole OrganismMulti organ
A discovery process beginning with an in vivo screening of proteins, peptides, natural products, classical medicinal compound or other substances. The administration of compounds to the animal can be either direct or indirect, such as by the administration and expression of cDNA-containing plasmids. Since the discovery process of the invention is based on a non-preconceived hypothesis and whole organism multi-organ analysis, a compound can be selected for testing in the absence of any biological selection criteria. The resulting organism-wide pattern of the gene expression changes in the transcriptome provides an overview of the activities at the molecular and organism-wide levels. The discovery process of the invention then integrates in vivo profiling and internal and external genomic databases to elucidate the function of unknown proteins, typically within few months. The invention further relates to medical uses of fibroblast growth factor 23 (FGF-23), FGF-23 fragments, FGF-23 C-terminal polypeptides, FGF-23 homologs and / or FGF-23 variants.
Owner:BOLLEKENS JACQUES +6
Characterization of biological material in a sample or isolate using unassembled sequence information, probabilistic methods and trait-specific database catalogs
The present invention relates to systems and methods for the characterization of biological material within a sample or isolate. The characterization may utilize probabilistic methods that compare sequencing information from fragment reads to sequencing information of reference genomic databases and / or trait-specific database catalogs. The characterization may be of the identities and / or relative concentrations or abundance of one or more organisms contained in the sample or isolate. The identification of the organisms may be to the species and / or sub-species and / or strain level with their relative concentrations or abundance. The characterization may additionally or alternatively be of one or more traits (i.e., characteristics) of the biological material contained in the sample or isolate. The characterization of the one or more traits may be with the relative abundance of the traits.
Owner:COSMOSID INC
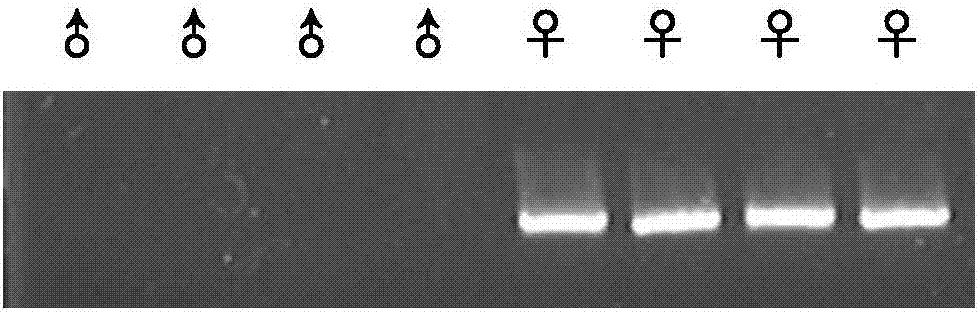
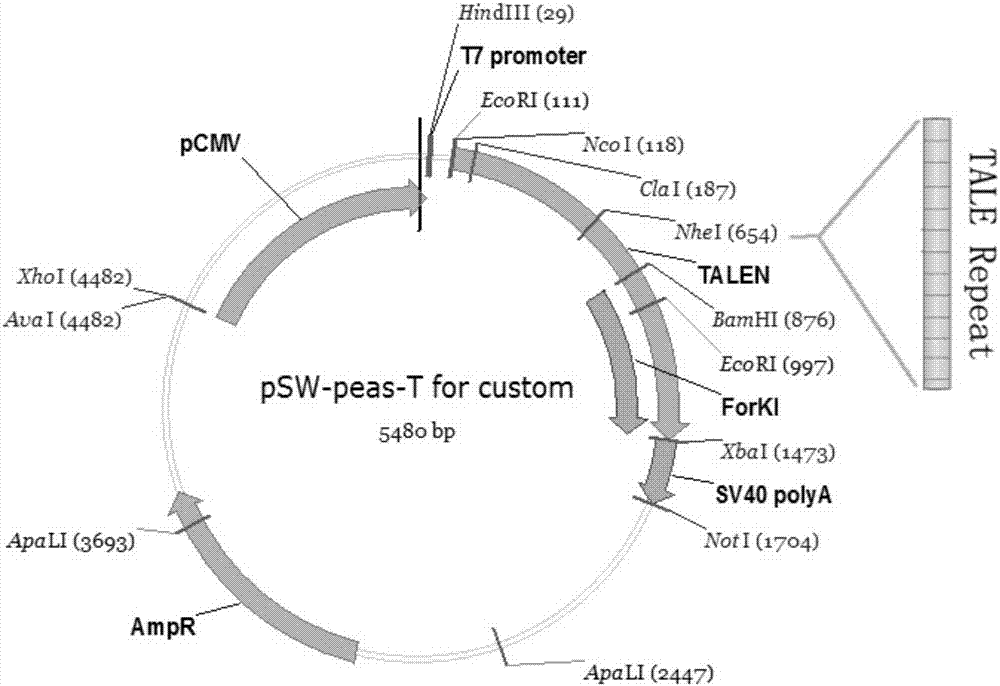
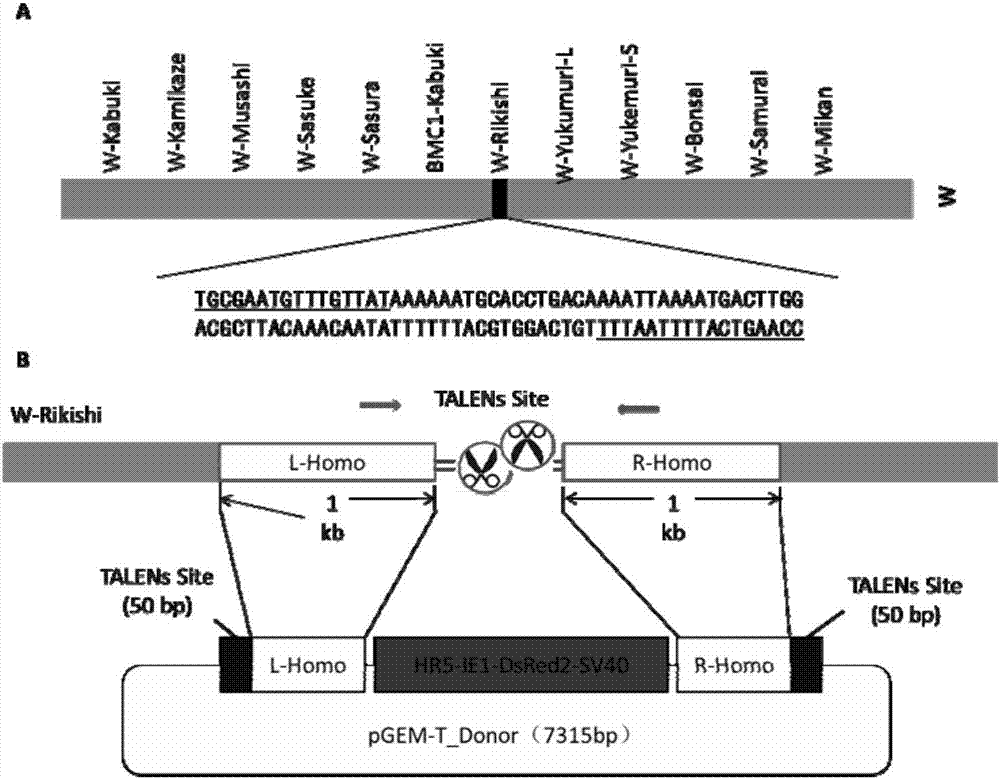
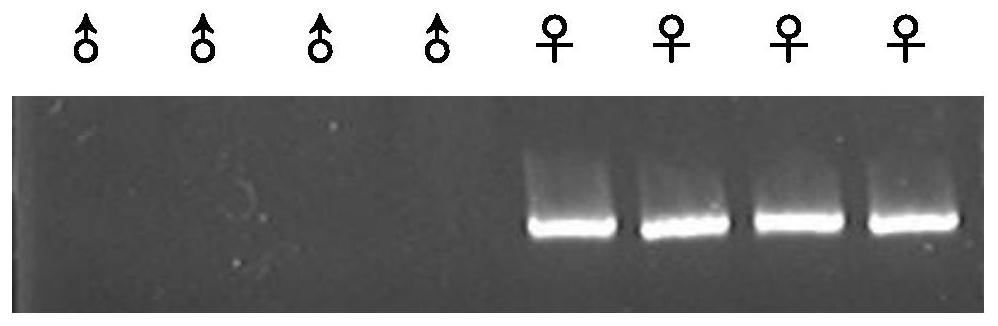
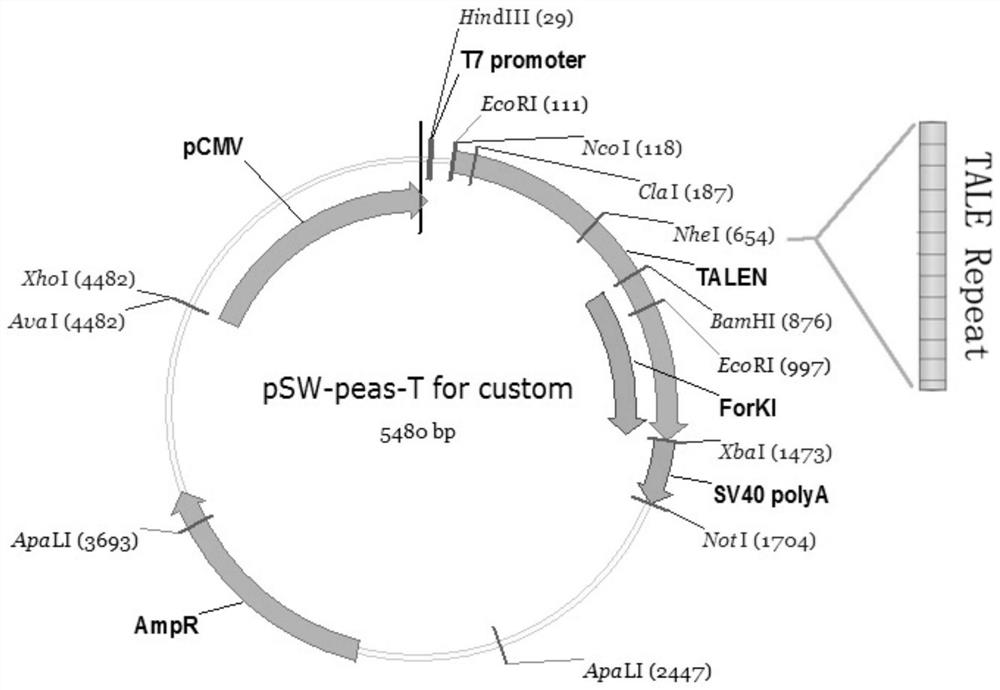
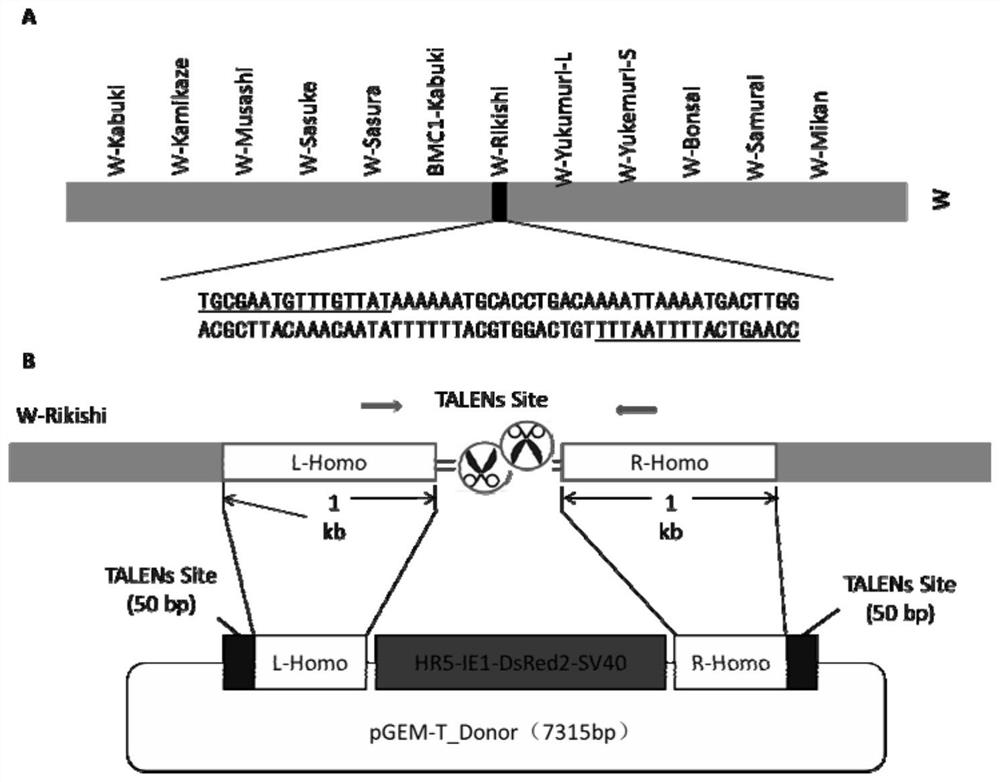
Method for inserting exogenous genes into W chromosomes of bombyx mori in fixed-point mode
ActiveCN107384967AAccurate resolutionGuaranteed hybridization rateHydrolasesStable introduction of DNAFluorescent proteinMrna expression
The invention belongs to the field of biotechnology (genome editing), and relates to a method for inserting exogenous genes into W chromosomes of bombyx mori in a fixed-point mode through mediating of transcriptional activator-like nuclease, and application of the method to bombyx mori sex discrimination. The method for inserting exogenous genes into W chromosomes of bombyx mori in the fixed-point mode specifically comprises the steps that W chromosome molecular markers in a bombyx mori genome database are analyzed, a TALENs targeting site is screened, TALENs mRNA expression plasmid is designed and obtained by synthesis, homologous recombinant carrier plasmid for expressing exogenous (reporter) genes (such as red fluorescent protein) is constructed, the microinjection technique is used for injecting the TALENs mRNA expression plasmid and the homologous recombinant (donor) carrier plasmid into eggs of the bombyx mori Nistara strain, the exogenous genes are inserted into the W chromosomes of bombyx mori in the fixed-point mode, the exogenous genes are only expressed in female individuals, and thus the purpose of rapid and efficient sex discrimination is achieved.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
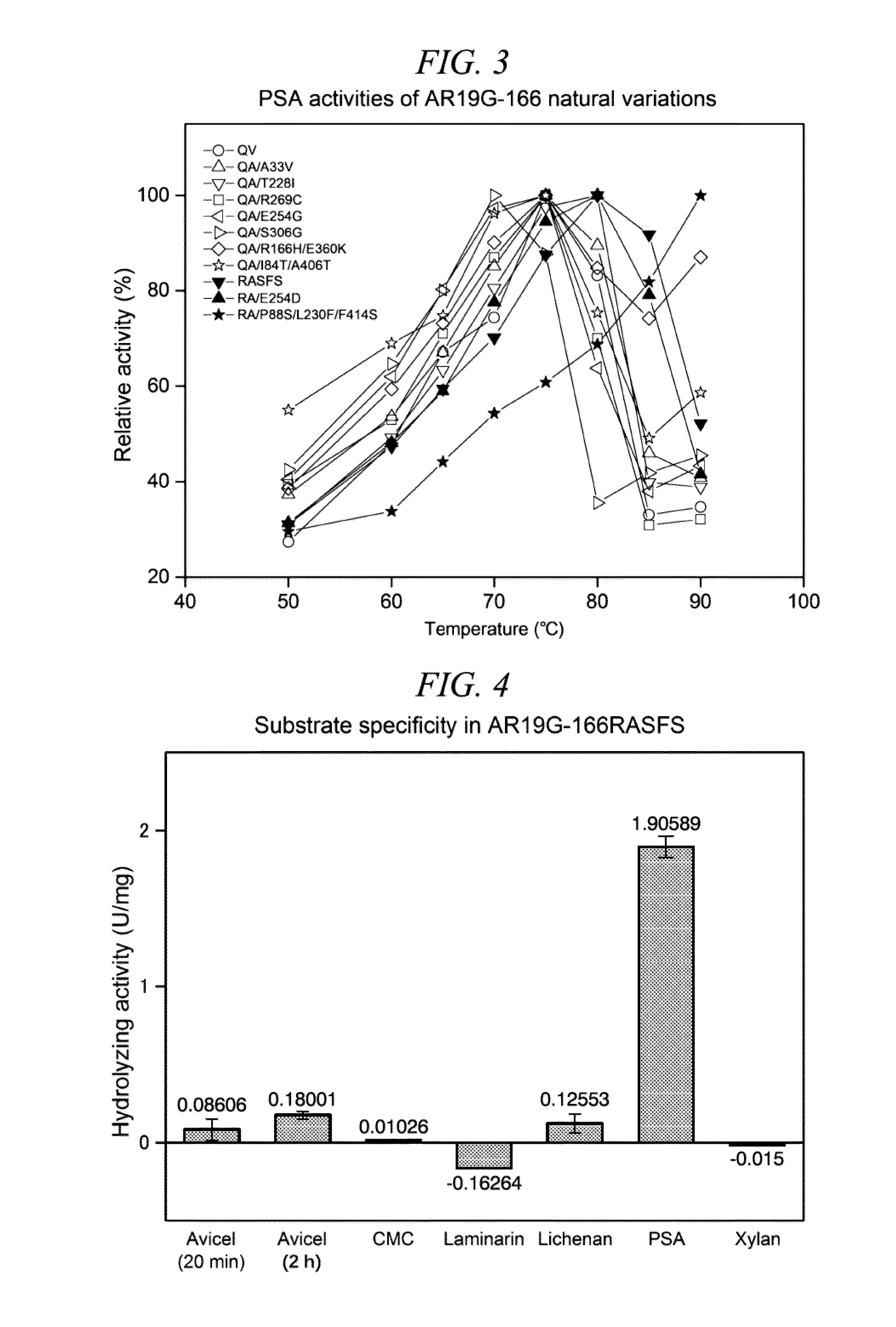
Method for obtaining natural variant of enzyme and super thermostable cellobiohydrolase
InactiveUS20150259659A1Efficiently obtainedIncrease enzyme activityMicroorganismsLibrary screeningGenome databaseEnzyme
A method for selectively obtaining a natural variant of an enzyme having activity includes (1) a step of detecting an ORF sequence of a protein having enzyme activity from a genome database including base sequences of metagenomic DNA of environmental microbiota; (2) a step of obtaining at least one PCR clone including the ORF sequence having a full length, a partial sequence of the ORF sequence, or a base sequence encoding an amino acid sequence which is formed by deletion, substitution, or addition of at least one amino acid residue in an amino acid sequence encoded by the ORF sequence, by performing PCR cloning on at least one metagenomic DNA of the environmental microbiota by using a primer designed based on the ORF sequence; (3) a step of determining a base sequence and an amino acid sequence which is encoded by the base sequence for each PCR clone obtained in the step (2); and (4) a step of selecting a natural variant of an enzyme having activity by measuring enzyme activity of proteins encoded by each PCR clone obtained in the step (2).
Owner:HONDA MOTOR CO LTD
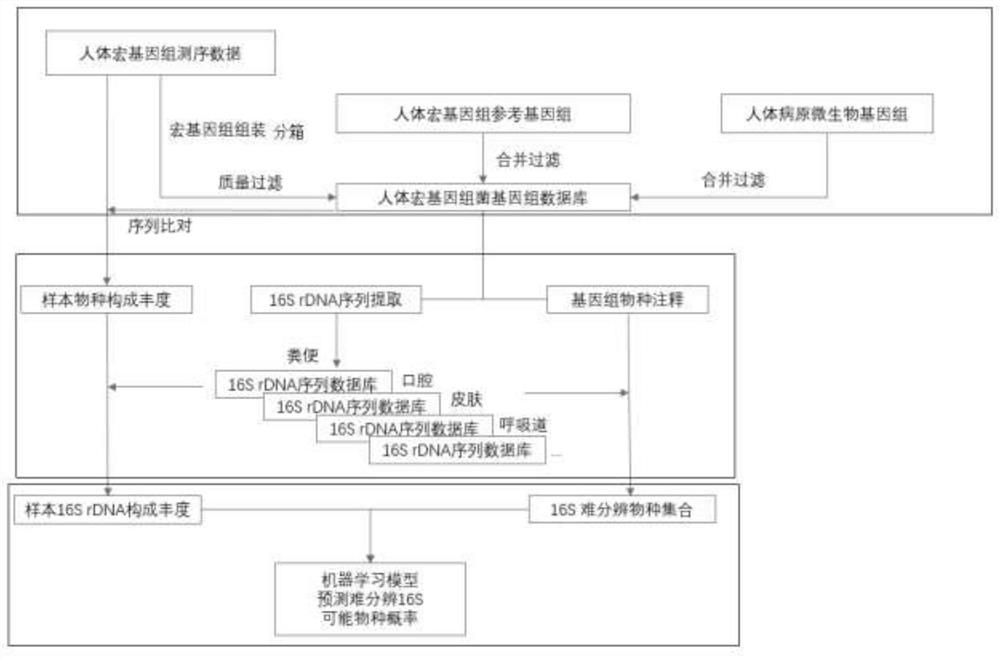
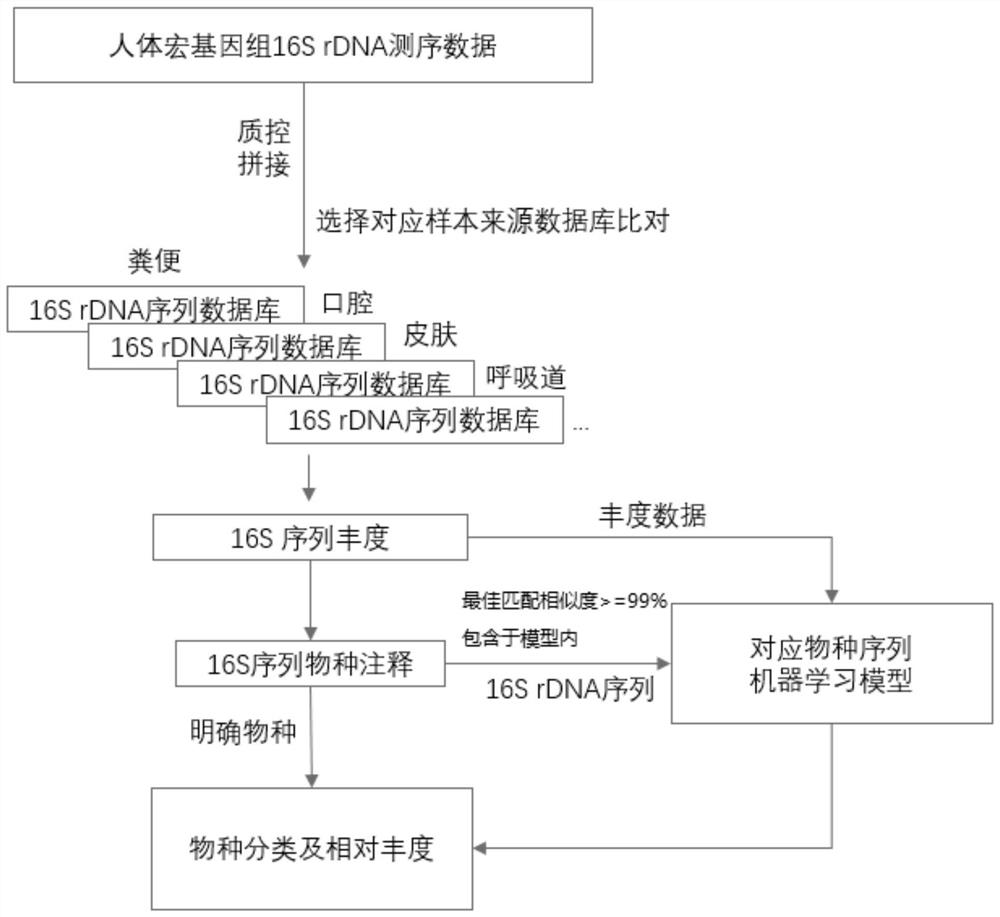
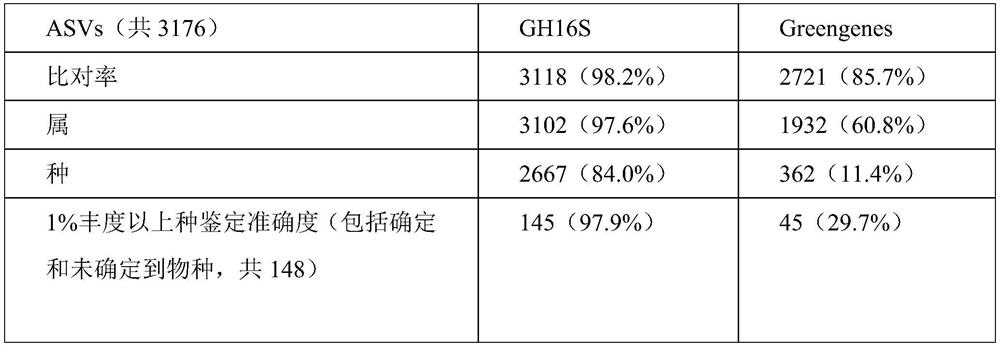
Optimization method for accurate species identification of human flora through high-throughput 16S rDNA sequencing
PendingCN111816258AReduce similar sequence interferenceImprove comparison efficiencyBiostatisticsSequence analysisMicroorganismGenomic databases
The invention discloses an optimization method for accurate species identification of human flora through high-throughput 16S rDNA sequencing. The optimization method comprises the following steps: 1)establishing a human metagenome bacterium genome database based on human microorganism samples; 2) extracting full-length 16S rDNA sequences of the human metagenomic bacterium genome database, and establishing 16S rDNA species annotation databases of different human body parts; and 3) establishing a sample-strain 16S sequence abundance correlation network, and training a machine learning model byusing the data. The invention also provides a method for performing species identification and abundance calculation on the high-throughput sequencing 16S rDNA sequences by using the constructed database and model. According to the invention, the species classification resolution and accuracy of 16S rDNA can be greatly improved, so an analyst can find more accurate and clear strains according toresults.
Owner:HANGZHOU GUHE INFORMATION TECH CO LTD
Methods and systems for the optimization of a biosynthetic pathway
PendingUS20210256394A1Improving phenotypic performanceImprove toleranceMathematical modelsMachine learningProtein targetGenomic databases
The present disclosure provides methods and systems for identifying variants of a given target protein or target gene that perform the same function and / or improve the phenotypic performance of a host cell transformed with such a variant. To enhance the diversity of identified candidate sequences, the methods may implement the use of a metagenomic database and / or machine learning methods. The methods and systems may be implemented in optimizing a biosynthetic pathway, e.g., to improve the production of a target molecule of interest.
Owner:ZYMERGEN INC
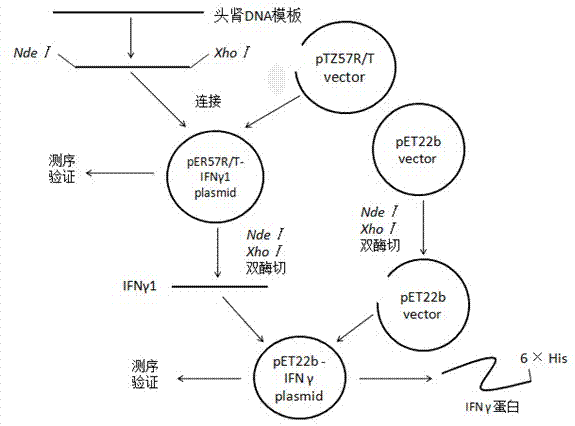
Epinephelus coioides interferon IFNgamma1 and preparation method and application thereof
ActiveCN103243106AEnriching System Signaling Pathway TheoryMicroorganism based processesAntibody medical ingredientsNucleotideEpinephelus bruneus
The invention relates to an epinephelus coioides interferon IFNgamma1 gene, a protein coded by the gene and an application of the protein in preparing a novel immunopotentiator. According to the invention, an opened reading frame of the IFNgamma1 gene is cloned from head-kidney RNAs (Ribonucleic Acid) of epinephelus coioides through a method of screening a genome database and designing specific primer amplification. The nucleotide sequence of the gene is shown as SEQIDNO:2. The amino acid sequence of the protein coded is shown as SEQIDNO:1. Through initial verification, the epinephelus coioides interferon IFNgamma1 protein can induce associated expression of immunogenes and can be developed to natural immunopotentiators or immunologic adjuvants for fishes.
Owner:SUN YAT SEN UNIV
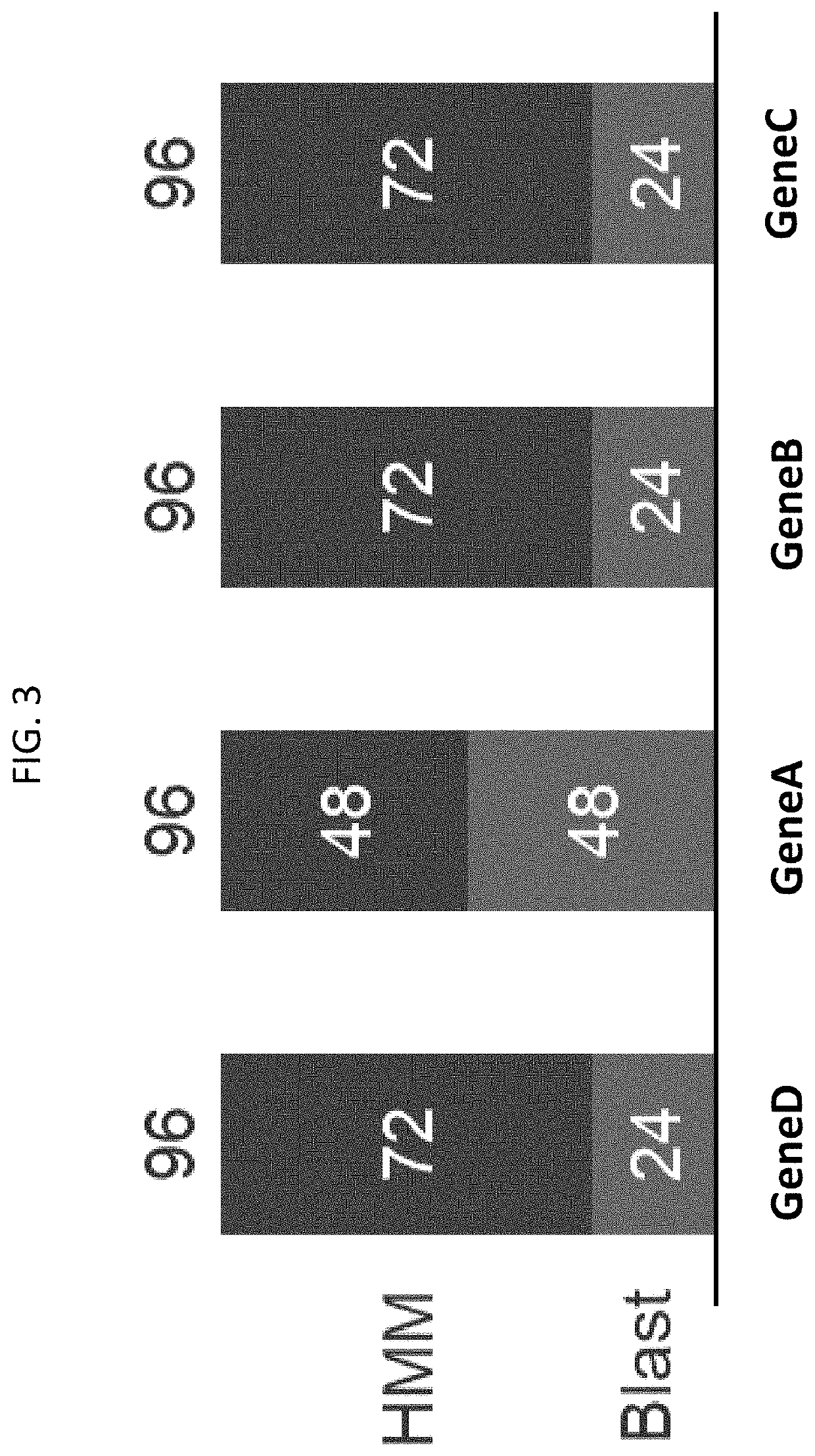
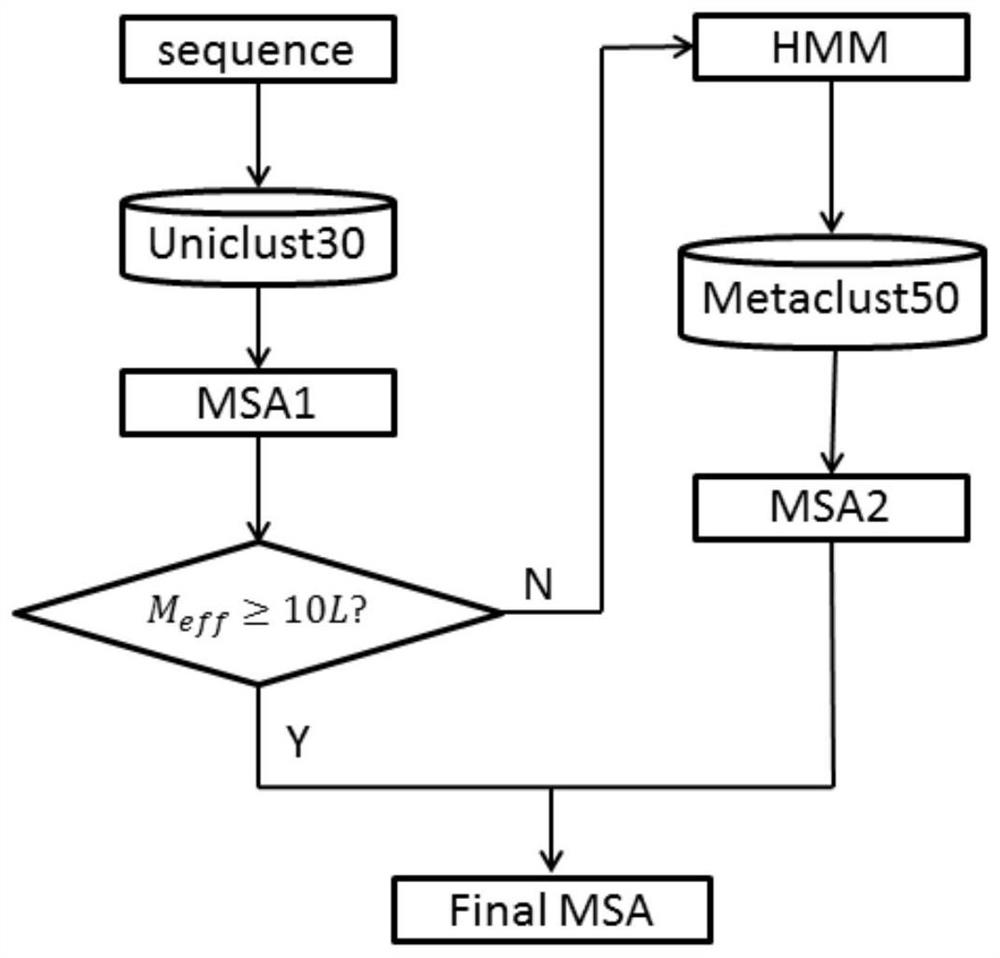
Macrogenome-based method for multiple-sequence alignment of proteins
InactiveCN113257337AEasy extractionIncrease the number of effective sequencesBiostatisticsInstrumentsProtein targetAlgorithm
The invention relates to a macrogenome-based method for multiple-sequence alignment of proteins, which comprises the following steps: performing initial search on a UniClust30 database by using HHblits according to a sequence of a target protein, filtering out a sequence with residue gap exceeding 50% the target sequence length in searched MSA, and iterating the search process for three times; filtering obtained multi-sequence comparison files by using hhfilter to generate a sequence with a gap proportion exceeding 25% in the MSA, so as to obtain MSA1; calculating the valid sequence number Meff of MSA1, if Meff is larger than or equal to 10 L, ending the search, and taking the MSA1 as an output result of multi-sequence alignment; otherwise, constructing a hidden Markov model HMM of the MSA1, searching a Metaclust50 metagenome database to obtain multiple-sequence alignment MSA2, and combining the MSA1 with the MSA2 to obtain final MSA. The invention not only enhances the number and quality of the searched homologous sequences, but also improves the calculation efficiency.
Owner:ZHEJIANG UNIV OF TECH
Systems and methods for genomic variant annotation
A system for annotating genomic variant files includes an application server, an annotation database, a genomic database, and an annotation processing computer system. The genomic database may be graph-oriented. The annotation processing computer system processes can process variant files in batch modes and includes annotation modules designed to improve the speed of the annotation process. The batch modes may include batch transmission, and / or batch annotation.
Owner:CYPHER GENOMICS
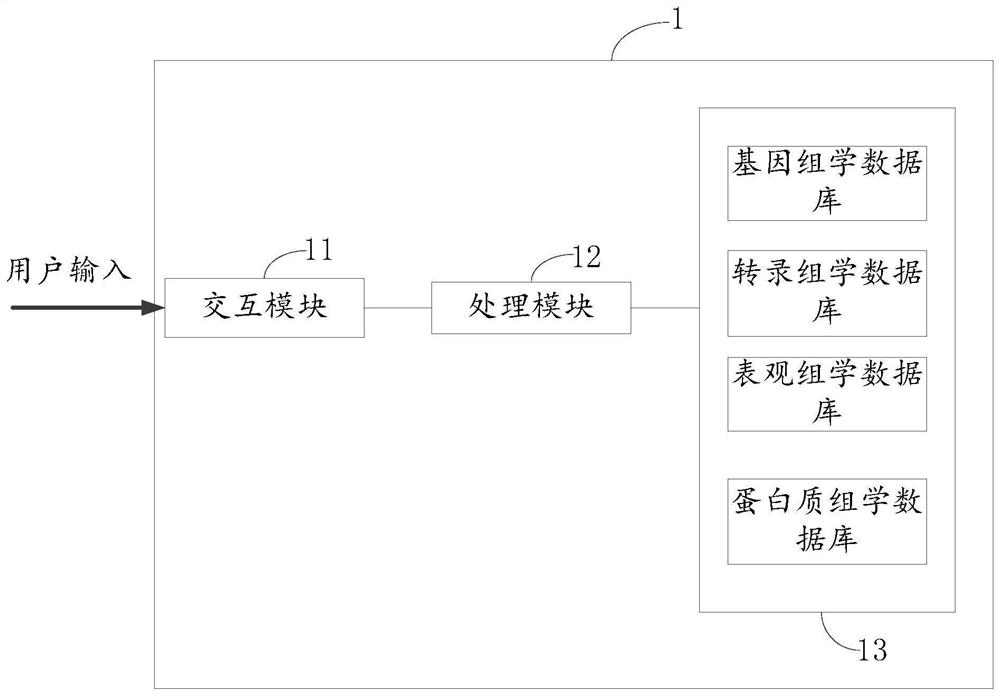
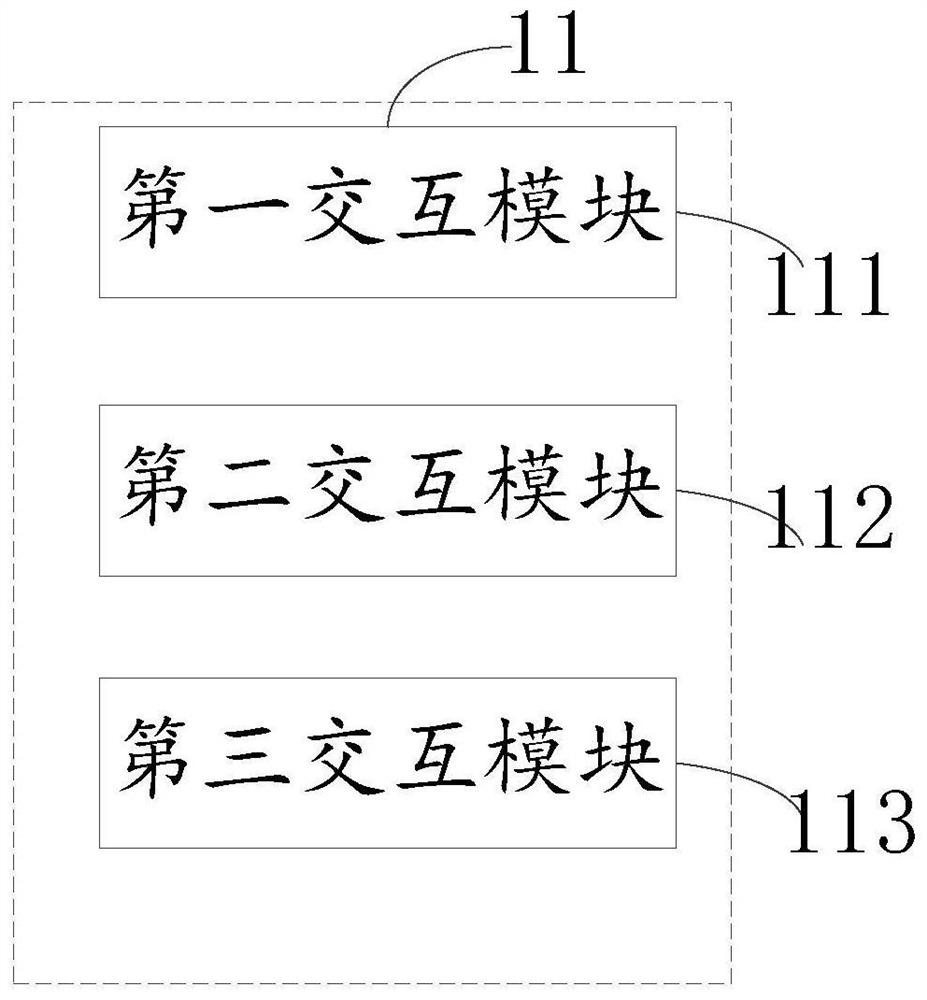
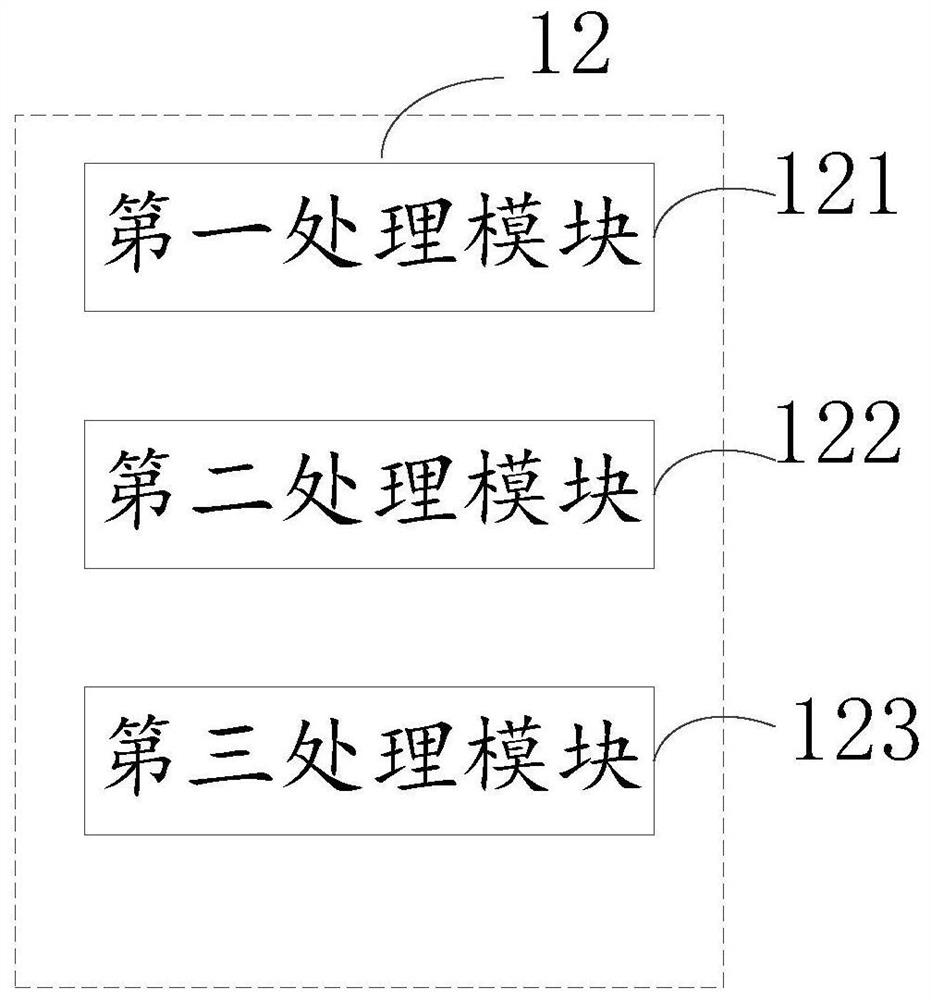
Multi-omics data analysis system and data conversion method thereof
PendingCN113377765AShow flexibleFlexible Analysis ResultsDatabase management systemsVisual data miningGenomicsData transformation
The invention relates to a multi-omics data analysis system and a data conversion method thereof. The analysis system comprises an interaction module, a processing module and a plurality of omics databases, the interaction module is used for providing an interactive form for a user so as to collect data analysis demand information of the user on one or more omics databases; the processing module is used for extracting or converting associated data matched with the analysis demand information from one or more omics databases according to the data analysis demand information and returning the associated data to the interaction module; and the plurality of omics databases includes a genomics database, a transcriptomics database, an epimics database, and a proteomics database. Different data analysis requirements of the user for one or more omics are collected in a mode of interacting with the user, and different analysis and mining of multiple omics data are realized by utilizing a mutual mapping relation of different omics data.
Owner:BGI TECH SOLUTIONS
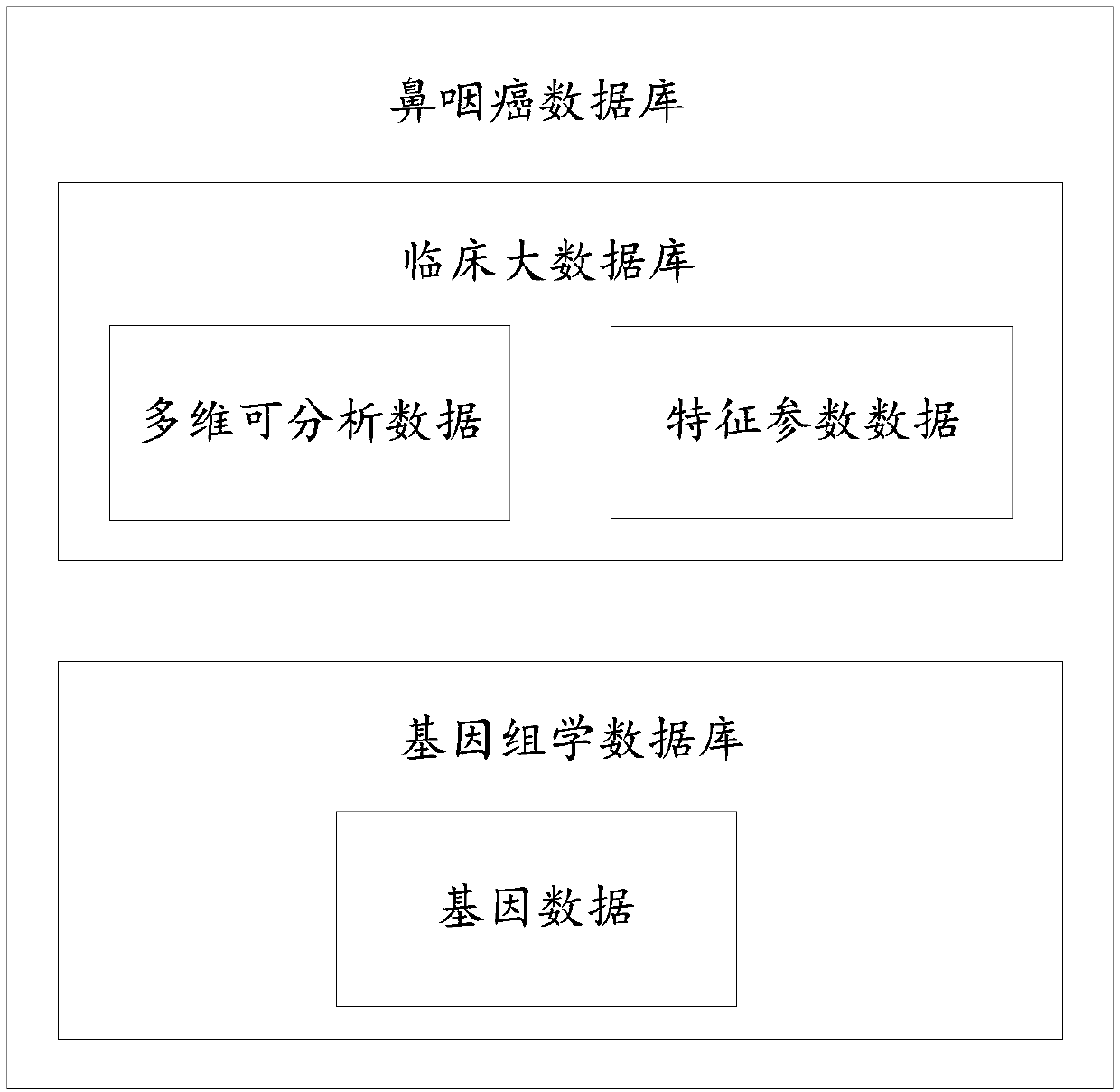
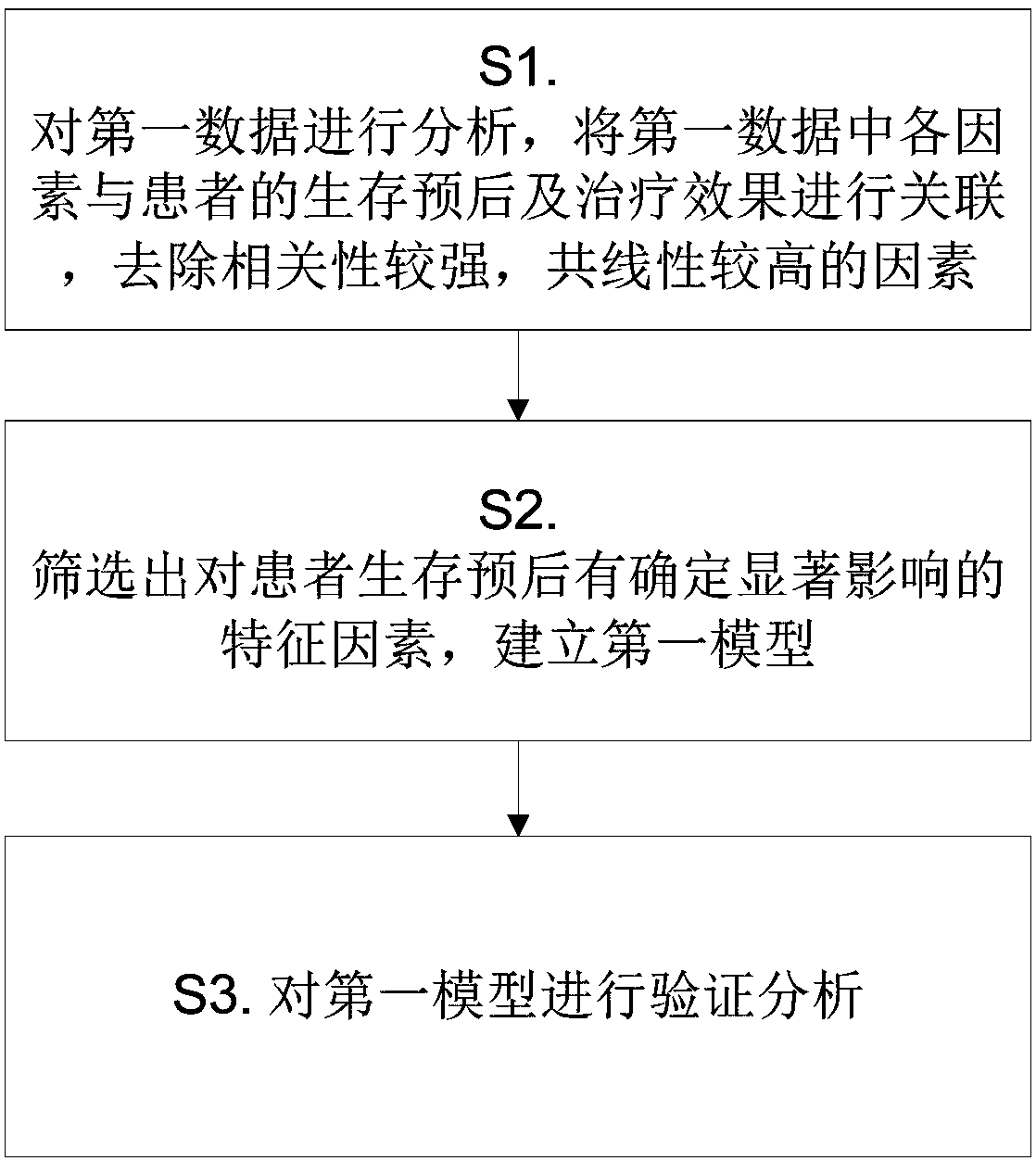
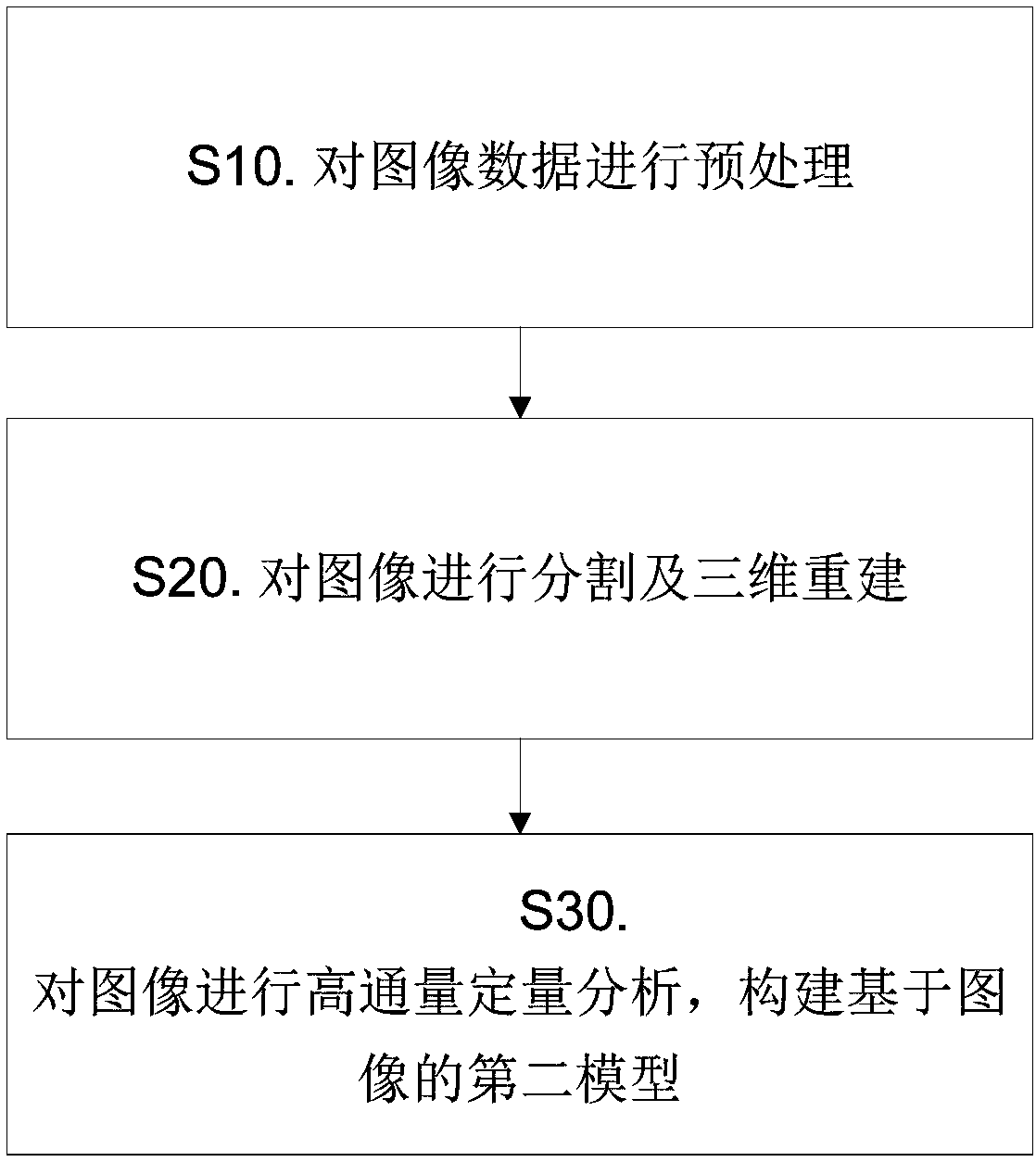
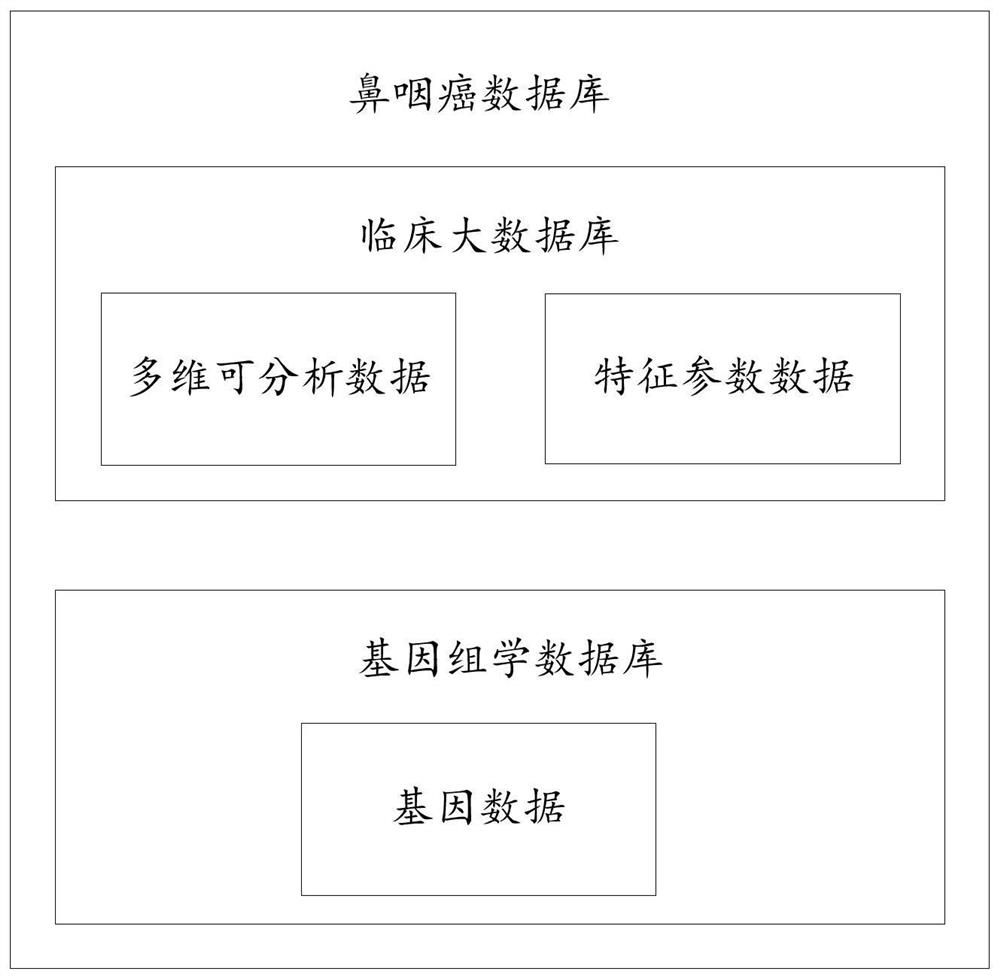
Nasopharyngeal carcinoma database and comprehensive diagnosis and treatment decision method based on same
The invention provides a nasopharyngeal carcinoma database which comprises a clinical large database and a genomics database. The clinical large database records first data formed by clinical alphanumeric information and second data extracted and recognized according to clinical image information. The first data, the second data and / or genomics data in the nasopharyngeal carcinoma database are accompanied by the time attributes thereof. The data in the nasopharyngeal carcinoma database are used to support a diagnosis and treatment decision server to carry out diagnosis and treatment decision based on a first model, a second model, a third model, a fourth model and / or a fifth model. Multi-omics data are input into the fourth model and the fifth model. Single-omics data are input into the first model, the second model and the third model. The nasopharyngeal carcinoma database provided by the invention provides a user with accurate intelligent diagnosis decision services based on artificial intelligence, and has a broad application prospect.
Owner:SUN YAT SEN UNIV
A method for site-directed insertion of exogenous gene into w chromosome of silkworm
ActiveCN107384967BAccurate resolutionGuaranteed hybridization rateHydrolasesStable introduction of DNAGenomic databasesFluorescent protein
The invention belongs to the field of biotechnology (genome editing), and relates to a method for inserting exogenous genes into W chromosomes of bombyx mori in a fixed-point mode through mediating of transcriptional activator-like nuclease, and application of the method to bombyx mori sex discrimination. The method for inserting exogenous genes into W chromosomes of bombyx mori in the fixed-point mode specifically comprises the steps that W chromosome molecular markers in a bombyx mori genome database are analyzed, a TALENs targeting site is screened, TALENs mRNA expression plasmid is designed and obtained by synthesis, homologous recombinant carrier plasmid for expressing exogenous (reporter) genes (such as red fluorescent protein) is constructed, the microinjection technique is used for injecting the TALENs mRNA expression plasmid and the homologous recombinant (donor) carrier plasmid into eggs of the bombyx mori Nistara strain, the exogenous genes are inserted into the W chromosomes of bombyx mori in the fixed-point mode, the exogenous genes are only expressed in female individuals, and thus the purpose of rapid and efficient sex discrimination is achieved.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Method, system, and computer software for providing a genomic web portal
InactiveUS20050004768A1Significant delayBiological testingHybridisationGenomic databasesComputer software
Systems, methods, and computer program products are described that process inquiries or orders regarding purchase of biological devices, substances, or related reagents. In some implementations, a user selects probe-set identifiers that identify microarray probe sets capable of enabling detection of biological molecules. Corresponding genes or EST's are identified and are correlated with related product data, which is provided to the user. Further, the user may select products for purchase based on the product data. If so, the user's account may be adjusted based on the purchase order. In the same or other implementations, a local genomic database is periodically updated. In response to a user selection of probe-set identifiers, data related to corresponding genes or EST's is provided to the user from the local genomic database.
Owner:AFFYMETRIX INC
Sorghum 14-3-3 protein GF14a gene and recombinant vector and expression method thereof
The invention discloses a sorghum 14-3-3 protein GF14a gene with a nucleotide sequence shown in SEQ ID NO.1. The invention also discloses a protein encoded by the sorghum 14-3-3 protein GF14a gene, with an amino acid sequence shown in SEQ ID NO.2. The present invention also discloses a recombinant vector comprising the above sorghum 14-3-3 protein GF14a gene and expression method of the above sorghum 14-3-3 protein GF14a gene. By the protein sequence of the corn GF14a gene, the homologous gene GF14a of sorghum is obtained from a sorghum genomic database, the gene is 780 bp in length, a prokaryotic expression vector is constructed according to the gene and is purified by a protein purification system, and the result thereof lays the foundation for further research on the crystal structure and biological characteristics of the protein, to further provide a basis for improving the stress resistance of sorghum by genetic editing methods.
Owner:GUIZHOU UNIV
Method for Compiling A Genomic Database for A Complex Disease And Method for Using The Compiled Database to Identify Genetic Patterns in The Complex Disease to Establish Diagnostic Biomarkers
PendingUS20200176086A1Low costShorten the timePhysical therapies and activitiesDrug and medicationsGeneticsGenomic databases
The invention provides a method for compiling a genomic database for a complex disease, such as Myalgic Encephalomyelitis / Chronic Fatigue Syndrome (ME / CFS), and a method for using the database for identifying genetic patterns that can potentially categorize, diagnose, and / or predict therapeutics for the complex disease.
Owner:NOVA SOUTHEASTERN UNIVERSITY
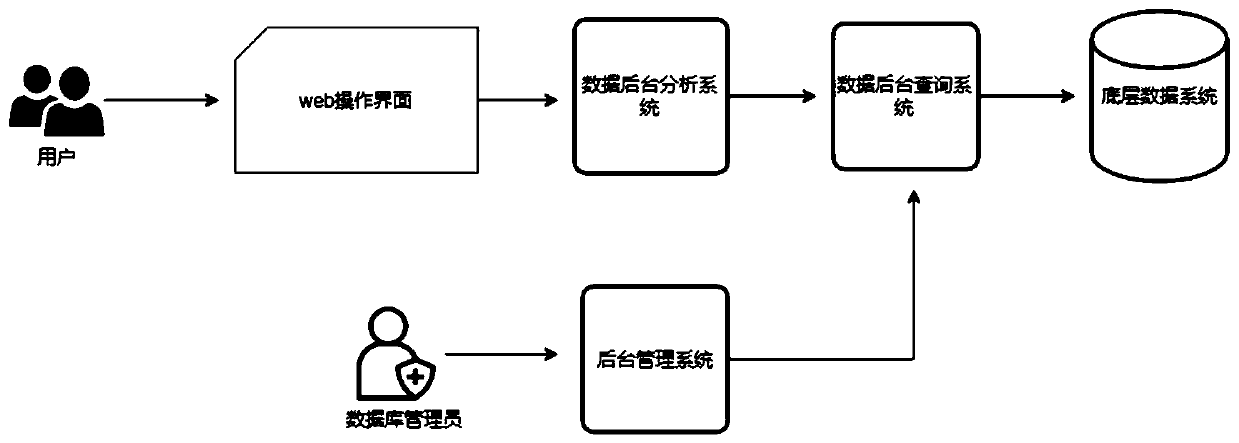
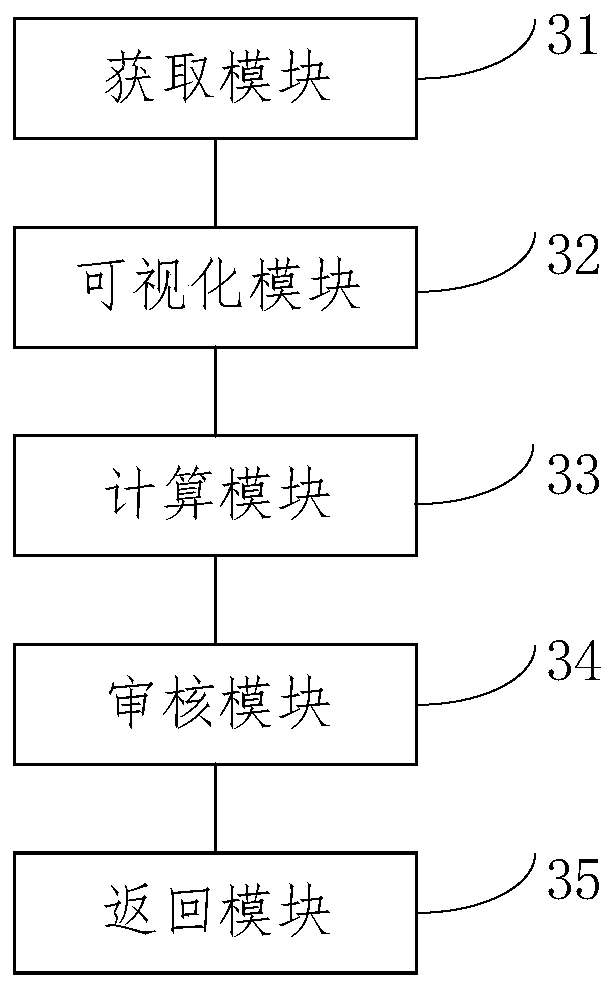
Query method and system for species genomics database
PendingCN111161804ASecurity Data Audit VerificationBioinformaticsInstrumentsGenomicsGenomic databases
Embodiments of the invention provide a query method and system for a species genomics database. The method comprises the steps of acquiring a query instruction input by a user; performing visual operation on species underlying data in the species genomics database based on the query instruction to generate a visual operation instruction; adopting a preset analysis tool to analyze and calculate thevisual operation instruction to obtain a data analysis result; performing permission auditing on the data analysis result based on a species data auditing mechanism, and feeding back a login auditingresult to the user; and based on the login auditing result, extracting a visual return result from the species genomics database for operation management by the user. According to the embodiments ofthe invention, the method capable of realizing efficient data analysis, safe data auditing and verification and standard data management is provided for the establishment of a species genome library by adopting a method of standardizing management and analysis of data via a plurality of subsystems.
Owner:BIOMARKER TECH
Bacterial identification and typing analysis genomic database and identification and typing analysis methods
ActiveCN112863606BImprove accuracyRapid identificationMicrobiological testing/measurementBioinformaticsBacteria identificationGenomic databases
Owner:杭州微数生物科技有限公司
A nasopharyngeal carcinoma database and a comprehensive diagnosis and treatment decision method based on the database
The invention provides a nasopharyngeal cancer database, including a large clinical database and a genomics database; the first data formed by clinical alphanumeric information and the second data extracted and identified according to clinical image information are recorded in the large clinical database. data; the first data, second data and / or genomics data in the nasopharyngeal cancer database are all accompanied by their time attributes; the data in the nasopharyngeal cancer database are used to support the diagnosis and treatment decision server to carry out based on the first model, the second model, the third model, the fourth model and / or the fifth model for diagnosis and treatment decisions, the fourth model and the fifth model are all input with multi-omics data, and the first model, the second model and the third model Both single-omics data were used as input. The invention can provide users with accurate intelligent diagnosis and decision-making services based on artificial intelligence, and has broad application prospects.
Owner:SUN YAT SEN UNIV
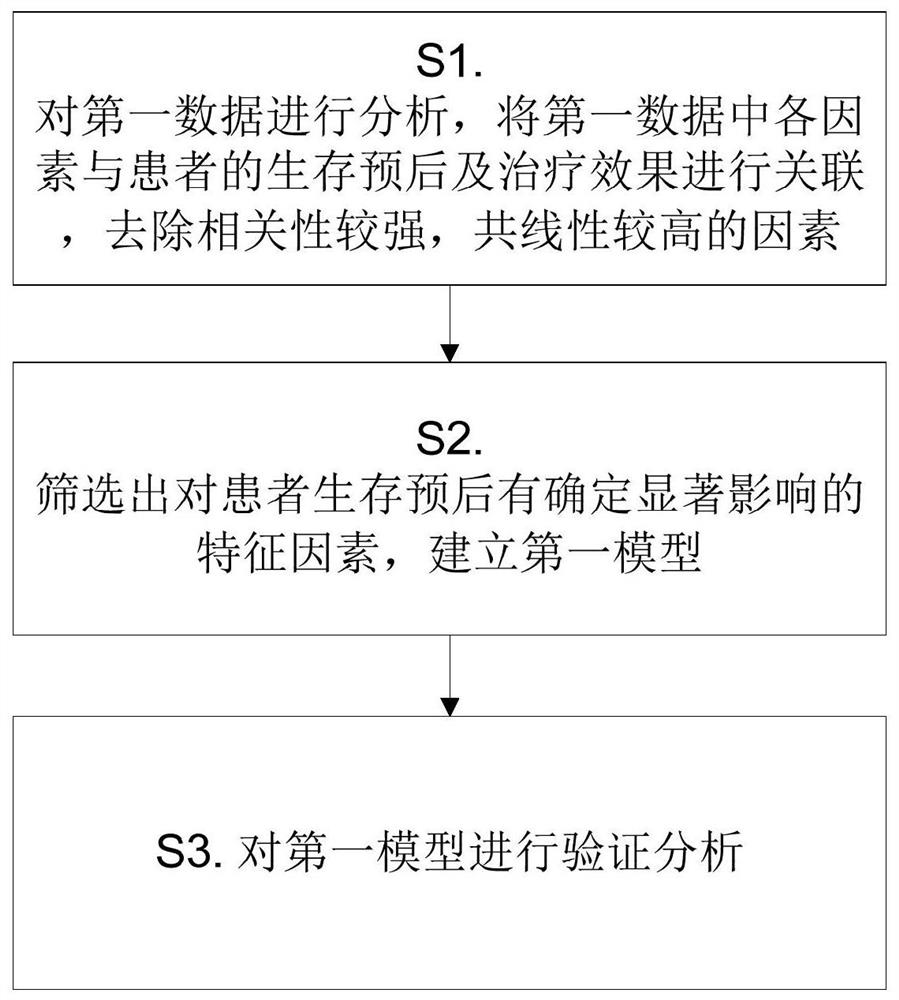
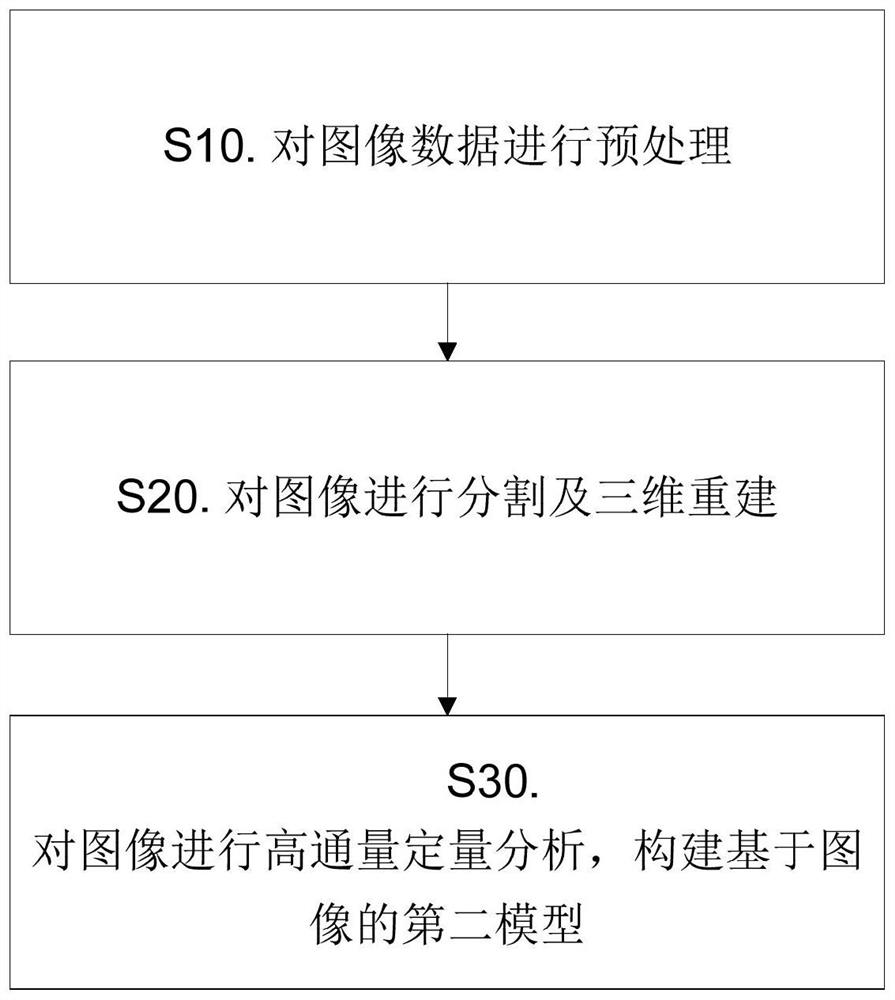
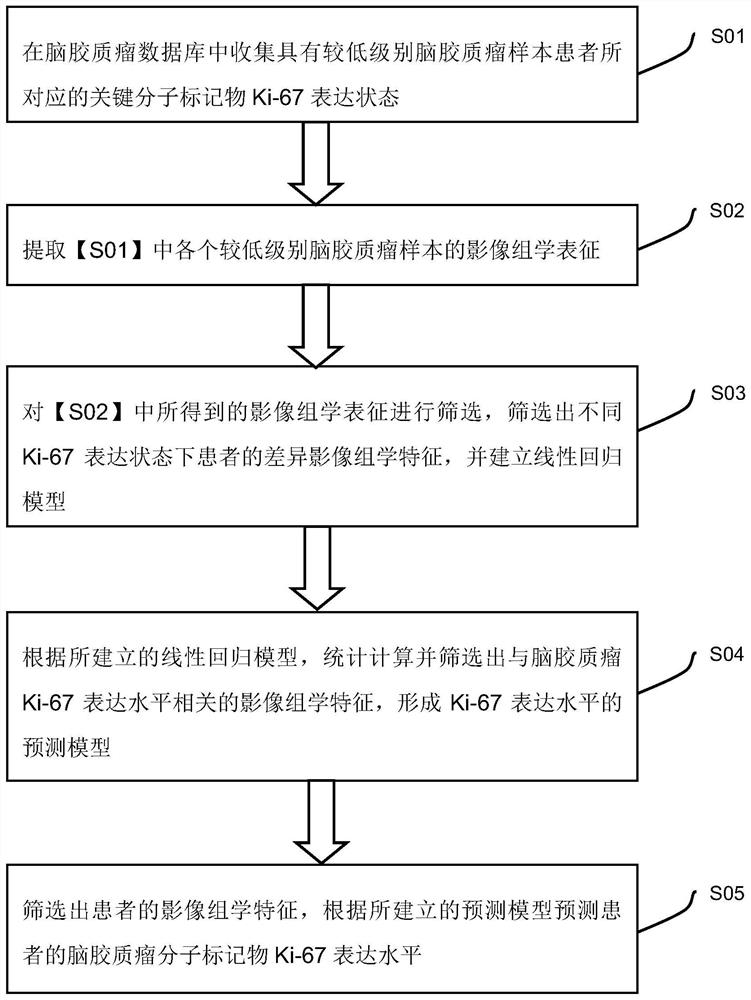
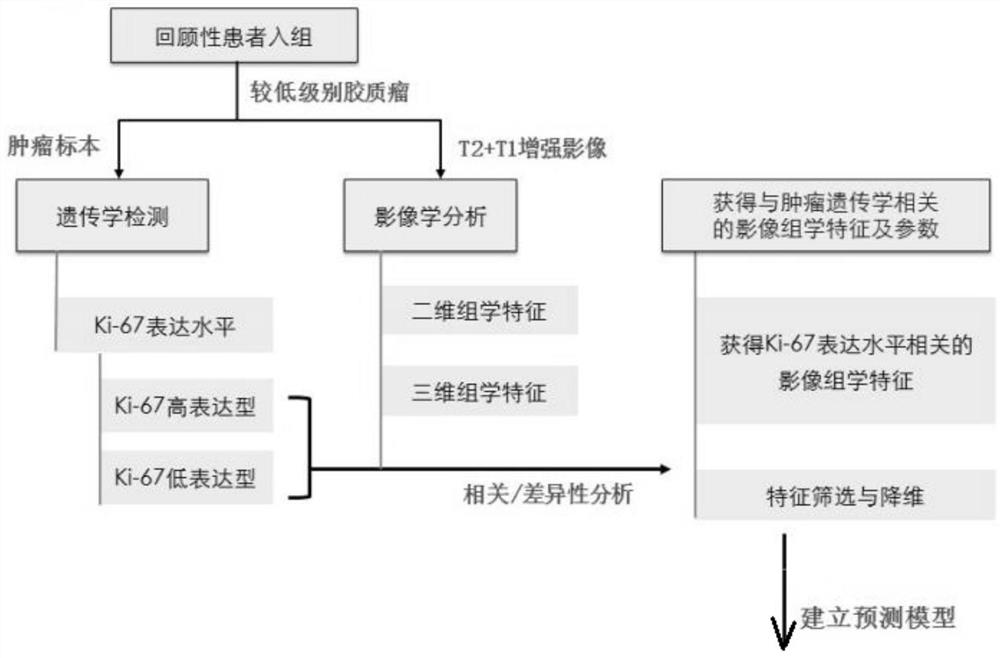
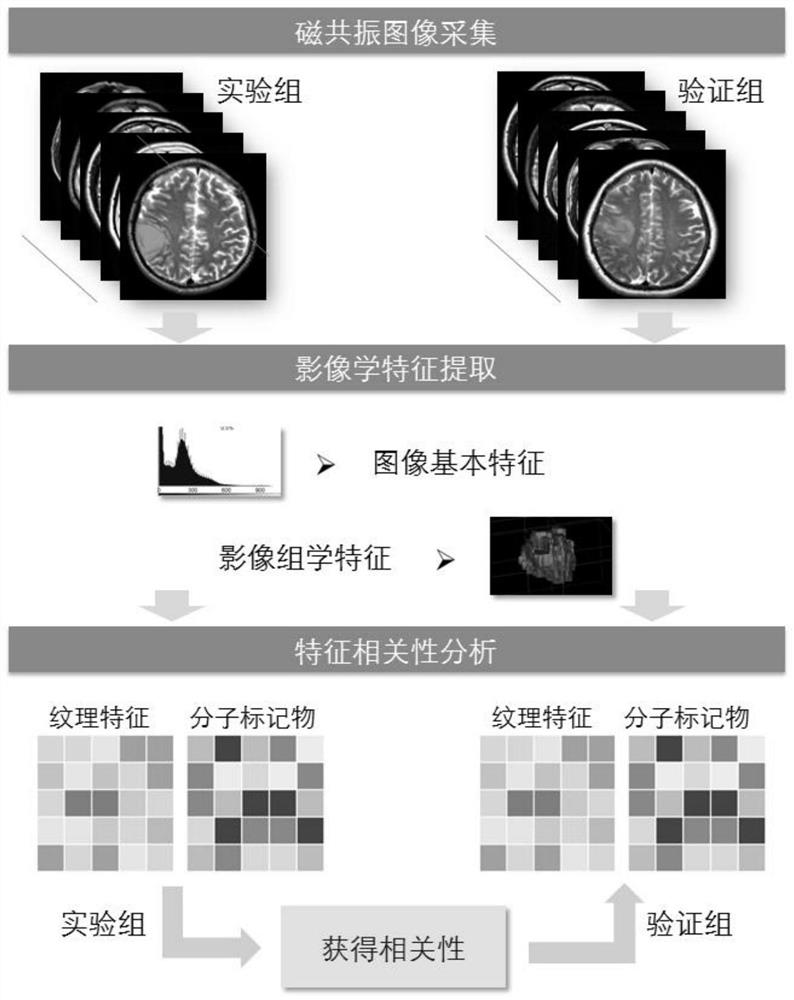
A radiomics prediction method for the expression level of ki-67 in glioma
ActiveCN108376565BAccurate analysisAccurate predictionImage enhancementImage analysisGenomicsGenomic databases
The invention discloses a radiomics prediction method for the expression level of glioma Ki-67. The key molecular marker Ki-67 corresponding to patients with low-grade glioma samples is collected in the glioma database. Expression status; extract the radiomics characterization of each lower-grade glioma sample; screen out the differential radiomics characteristics of patients with different Ki-67 expression status, and establish a linear regression model; according to the established linear regression model, statistical Calculate and screen out the radiomics features related to the expression level of Ki-67 in gliomas to form a prediction model; screen out the radiomics features of patients, and predict the molecular marker Ki-67 of glioma in patients according to the prediction model The expression level. Based on the existing glioma imaging and genomics database, the present invention adopts the method of linear regression model, and obtains the imaging omics characterization that can reflect the genetic characteristics of the tumor through rapid and accurate analysis of the established prediction model, and more accurately predicts Ki‑67 expression levels in glioma patients.
Owner:北京北卓医疗科技发展有限责任公司
A set of kiwifruit species-associated specific single nucleotide molecular markers and its detection primer set and application
ActiveCN106367496BThe identification result is accurateFacilitate the determination of parental sourceMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
The invention discloses kiwi fruit species association specific mononucleotide molecular markers and a detection primer set and application thereof. By means of sequencing sequences of different kiwi fruit species existing in a kiwi fruit genome database, high-diversity mononucleotide variation regions are looked for in a sequence alignment reference genome mode, a mononucleotide molecular marker primarily selected set is formed, on the basis, a novel mass spectrometer gene typing platform is used for carrying out experimental verification and genotype acquisition on mononucleotide variation sites, species association specific mononucleotide markers are screened through association analysis with the species, finally, a set of molecular markers for kiwi fruit hybrid resource parent species source identification are obtained, and the technical scheme is formed. Identification of kiwi fruit hybrid resource parents can promote understanding of formation of hybrid resource characters and biological characters and accelerate genetic evaluation and development and utilization of hybrid resources, and great theoretical academic significance and high economic application value are achieved.
Owner:SOUTH CHINA BOTANICAL GARDEN CHINESE ACADEMY OF SCI
Polymer encapsulated aluminum particulates
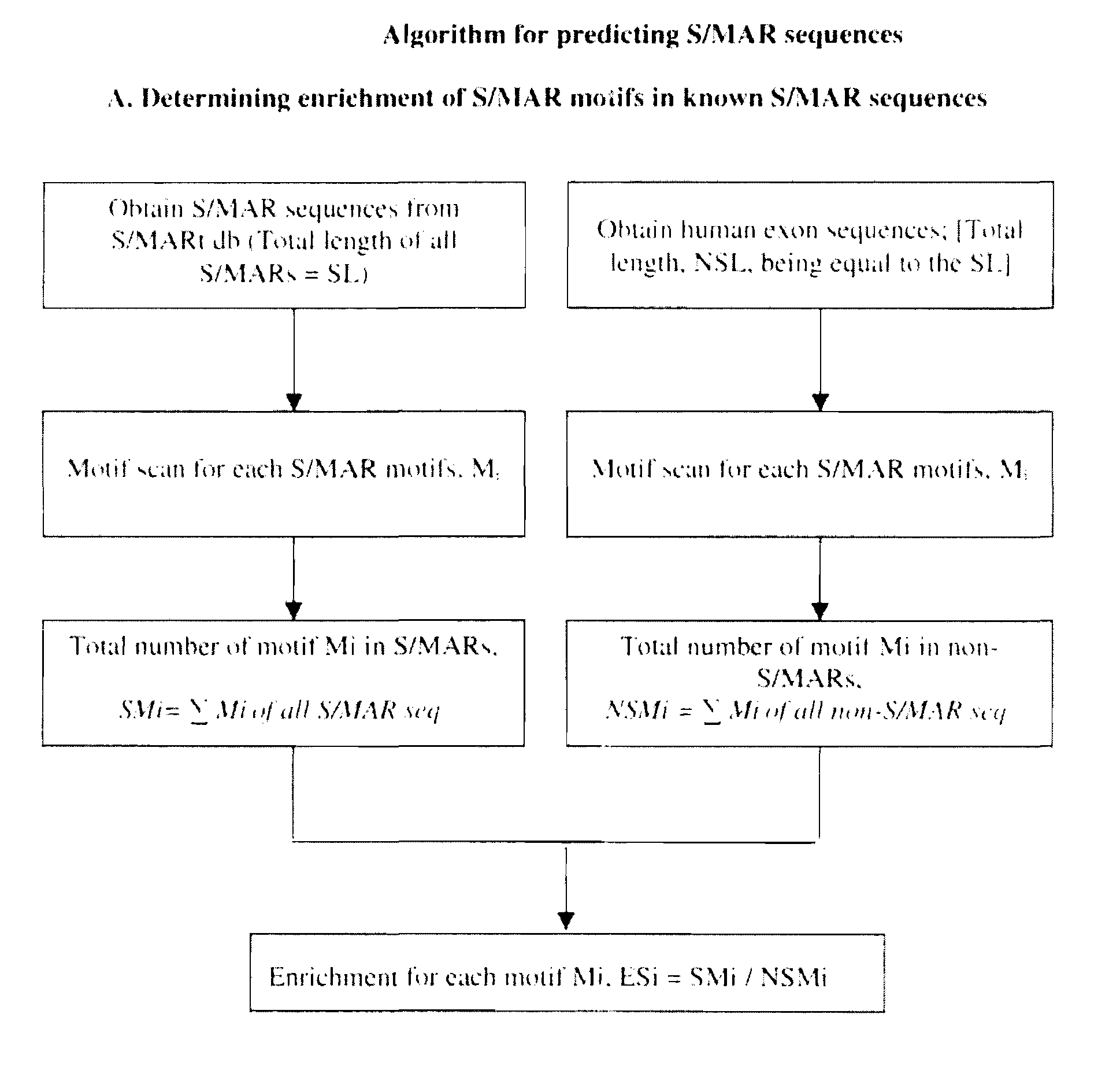
InactiveUS20110190482A1High protein yieldIncrease gene expressionSugar derivativesLibrary member identificationParticulatesMedicine
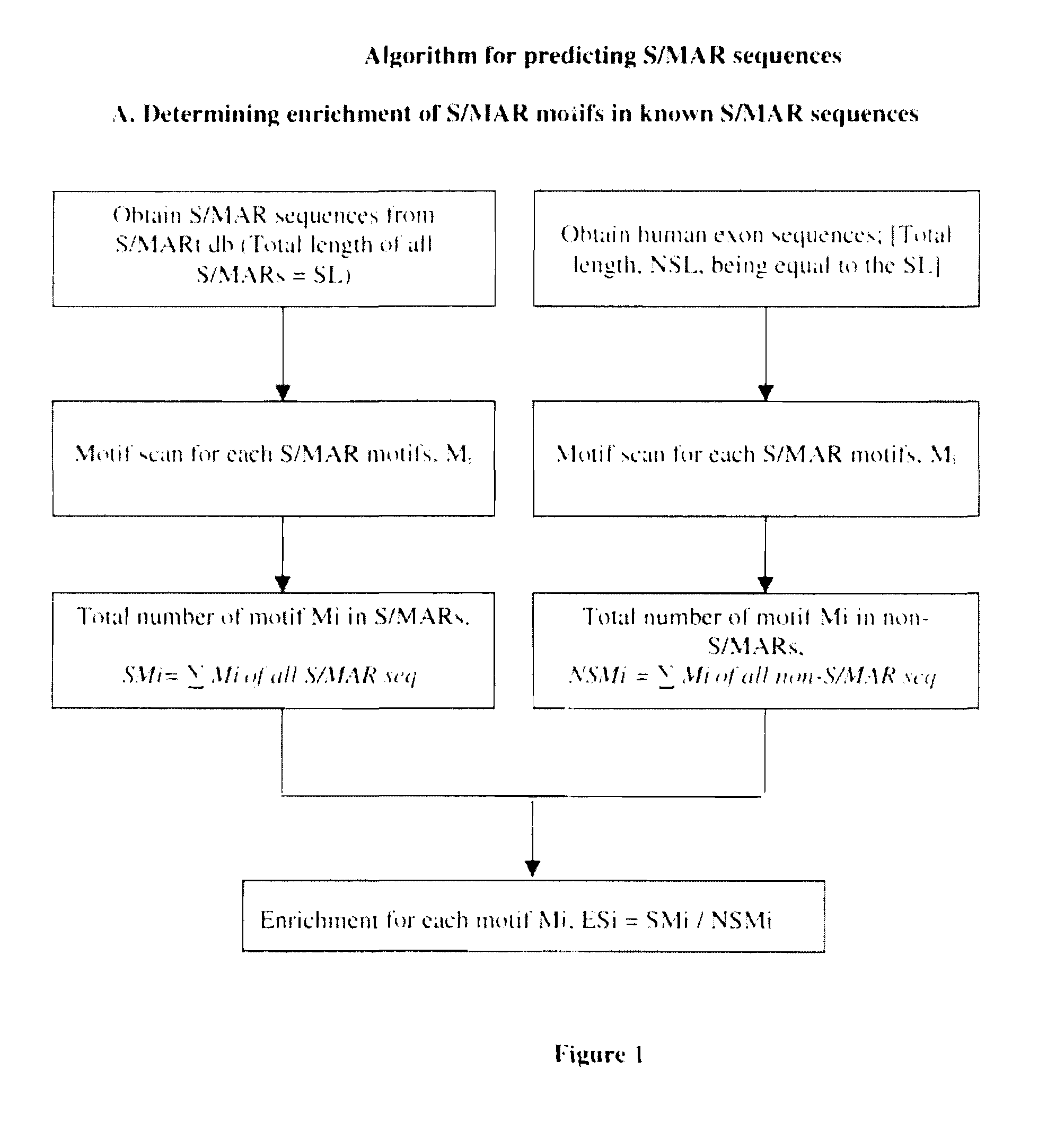
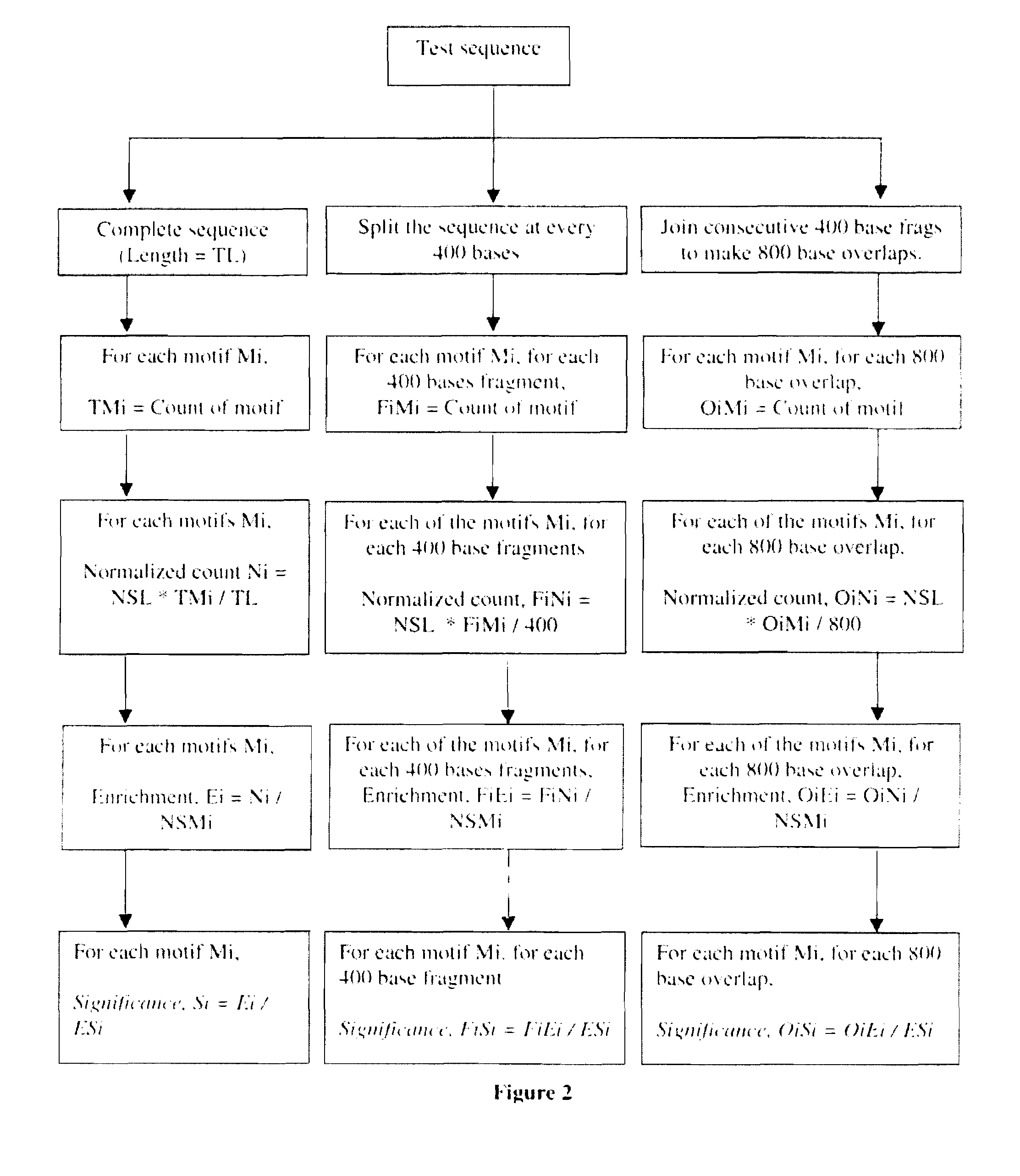
The present invention relates to use of novel bioinformatics approach for predicting and identifying Scaffold / Matrix attachment regions (S / MARs) from different genomic database.
Owner:AVESTHAGEN LIMITED DISCOVERER
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com