Patents
Literature
373 results about "Sequence comparison" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Comparative mapping and assembly of nucleic acid sequences
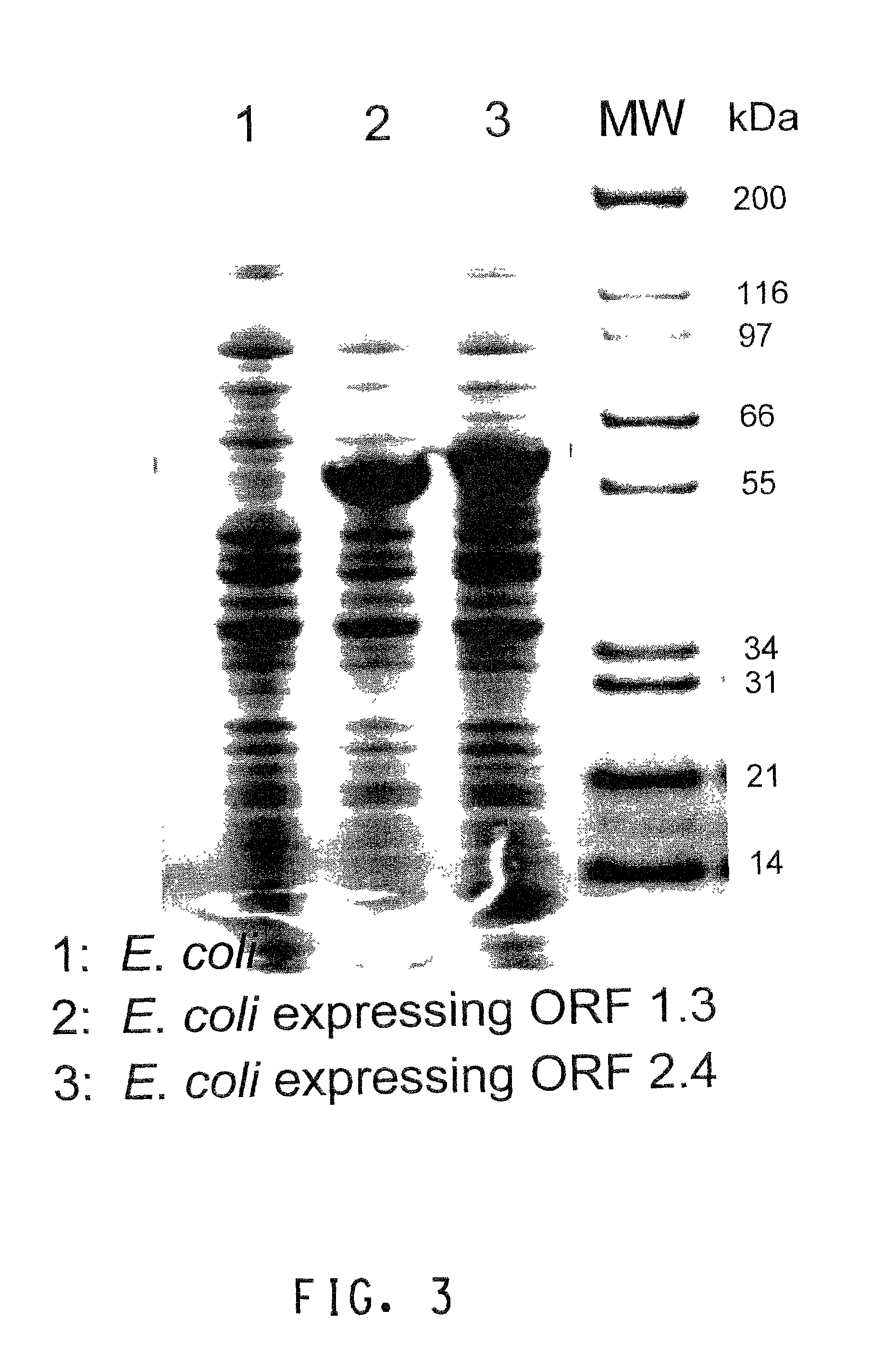
InactiveUS7809509B2Improve accuracyImprove completenessMicrobiological testing/measurementAnalogue computers for chemical processesNucleic acid sequencingBacterial artificial chromosome
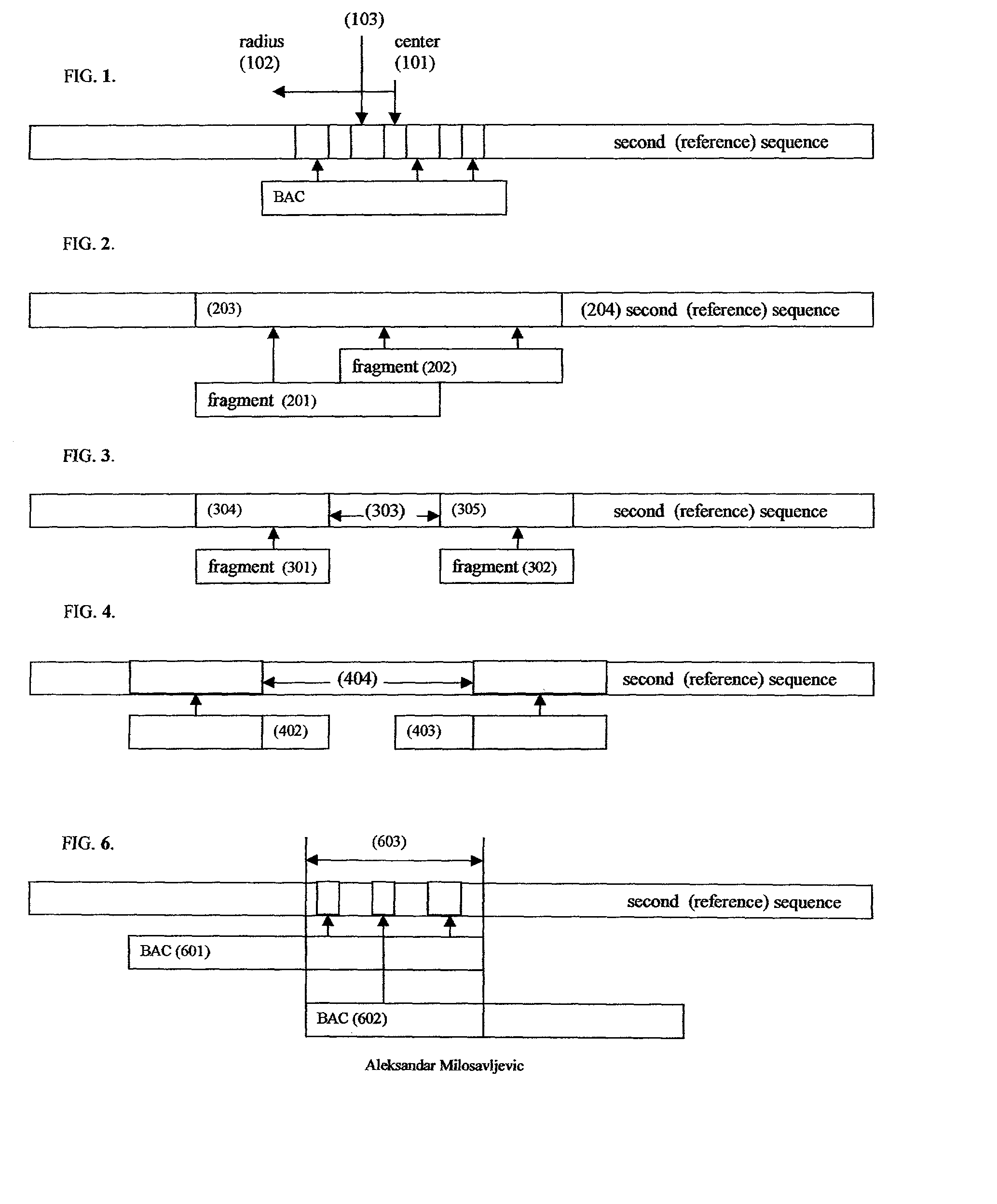
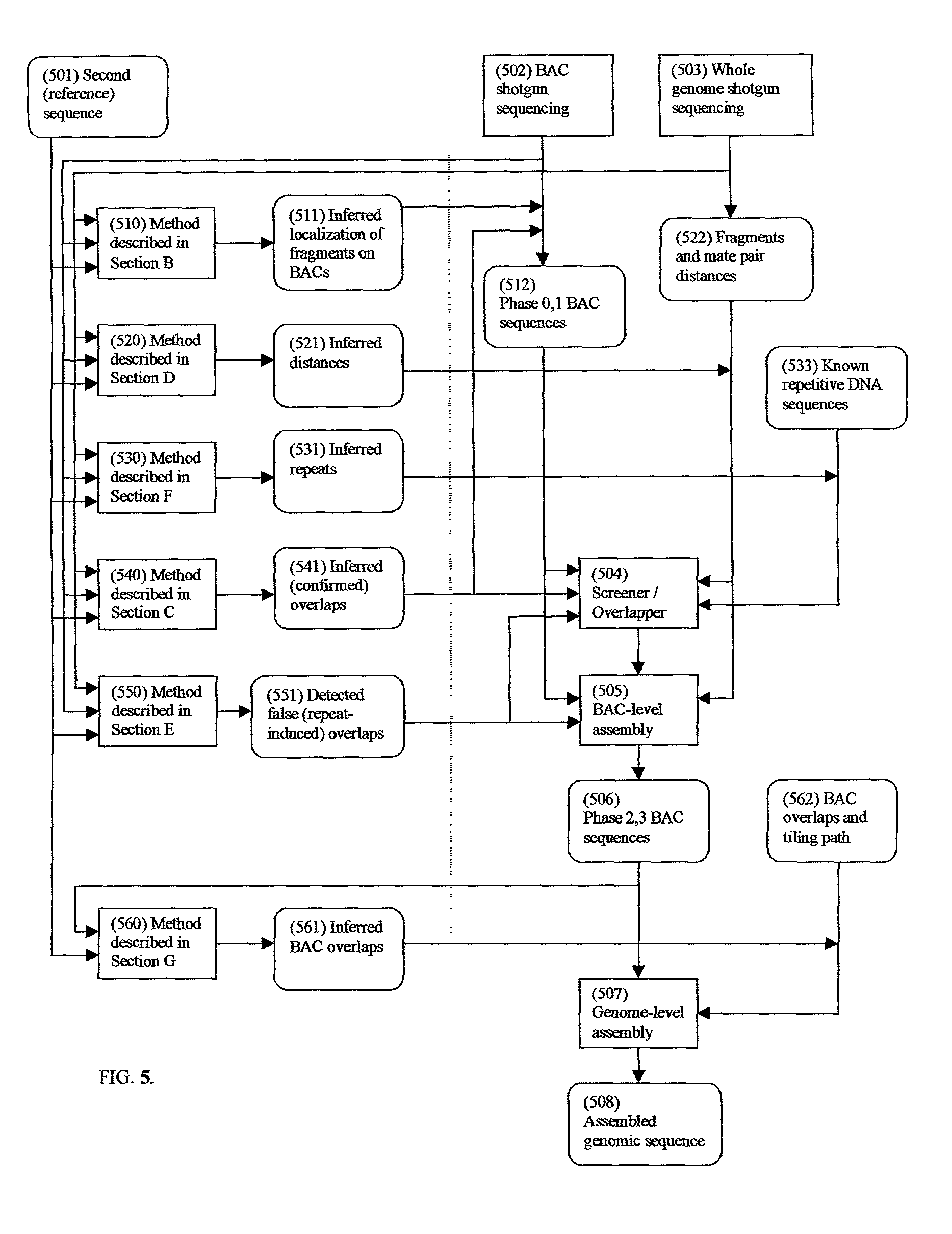
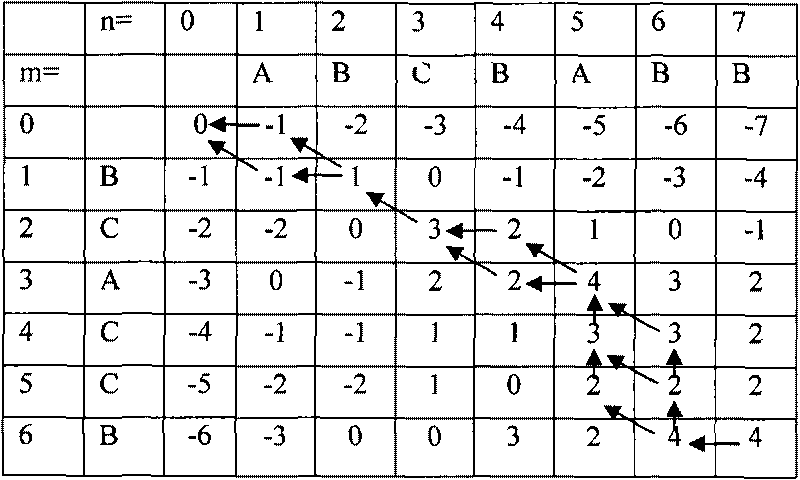
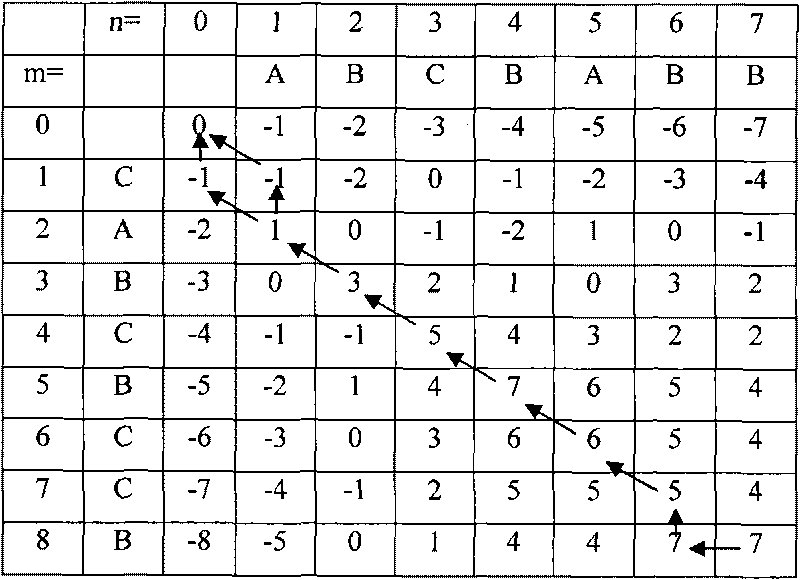
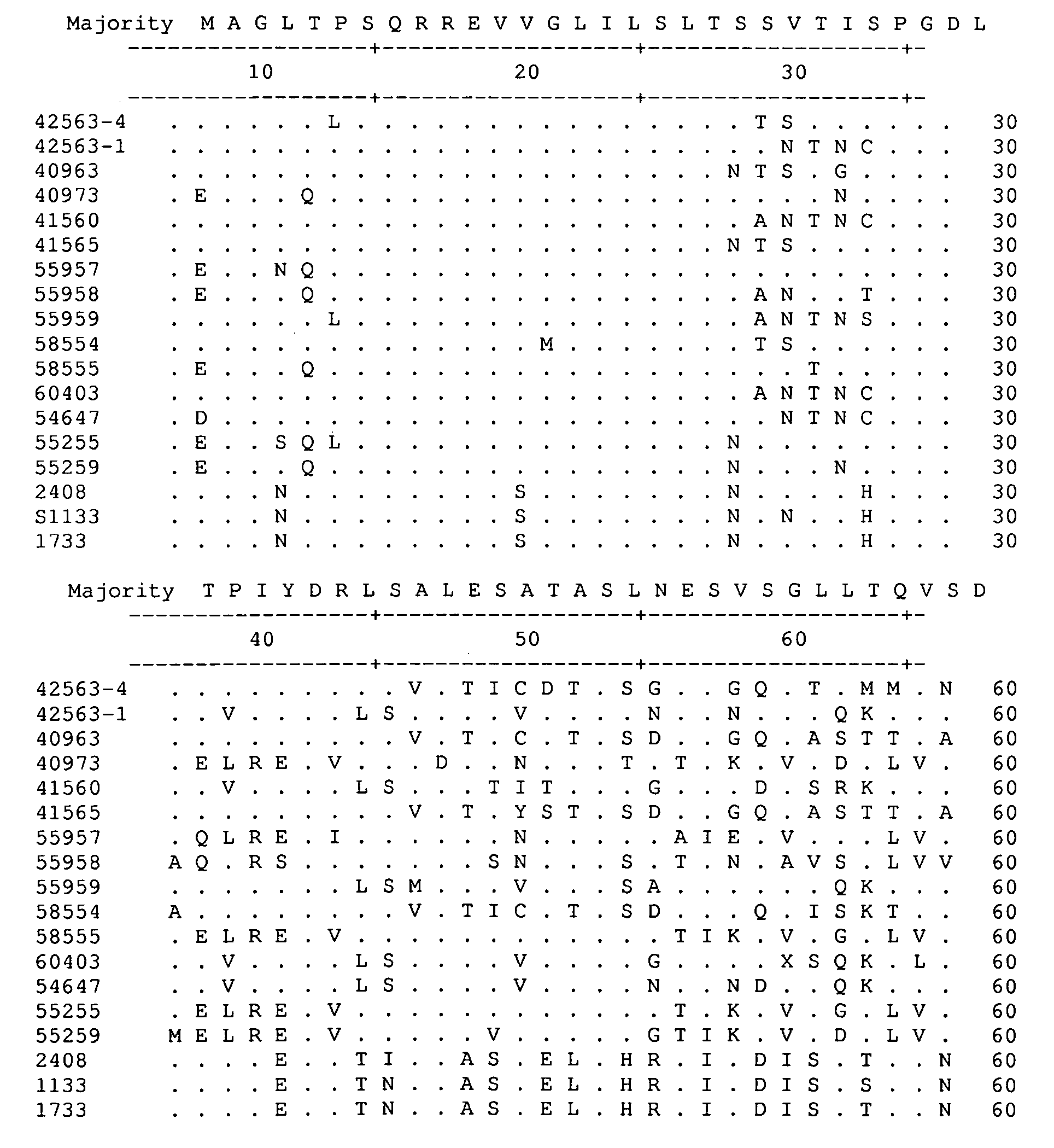
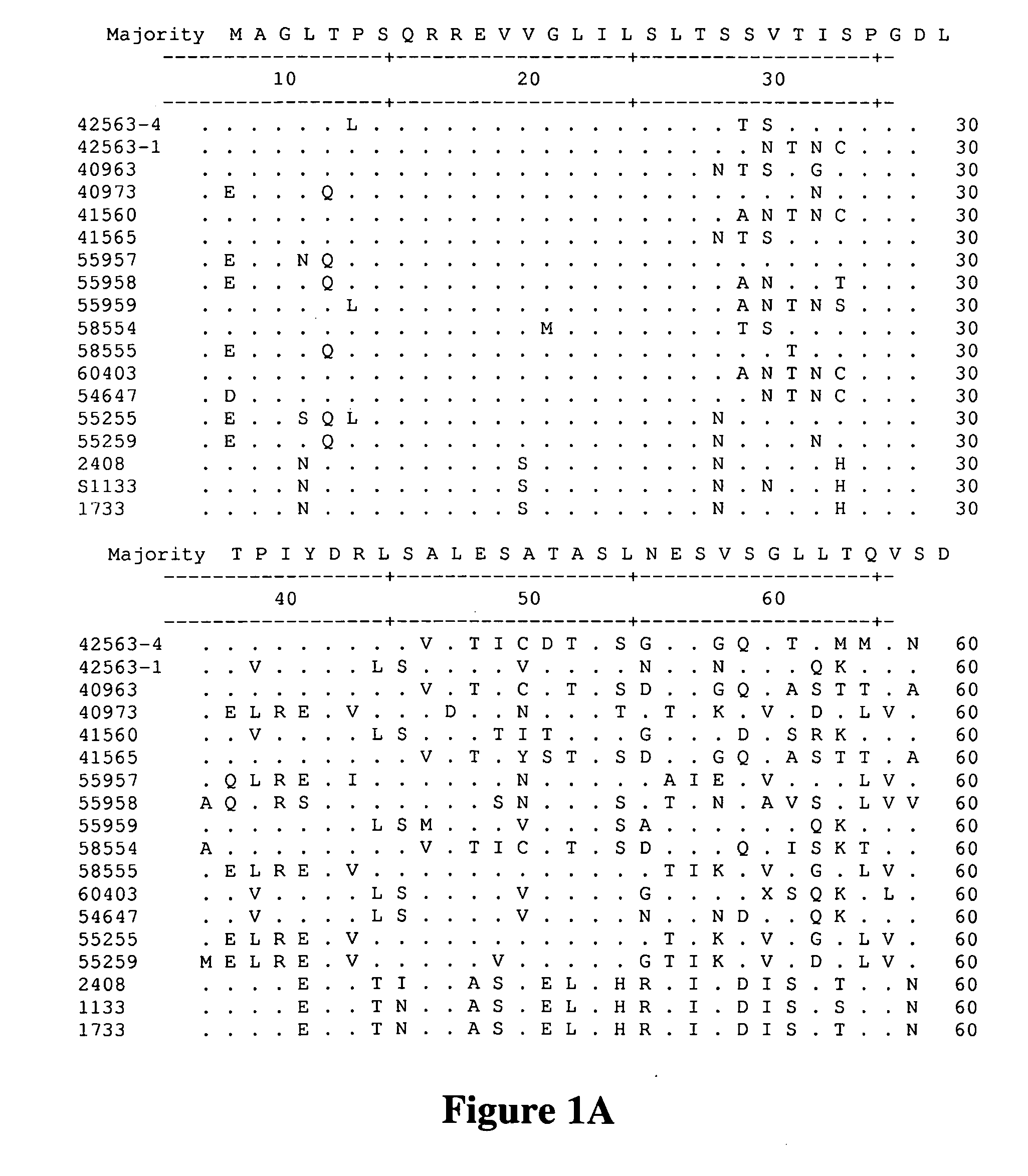
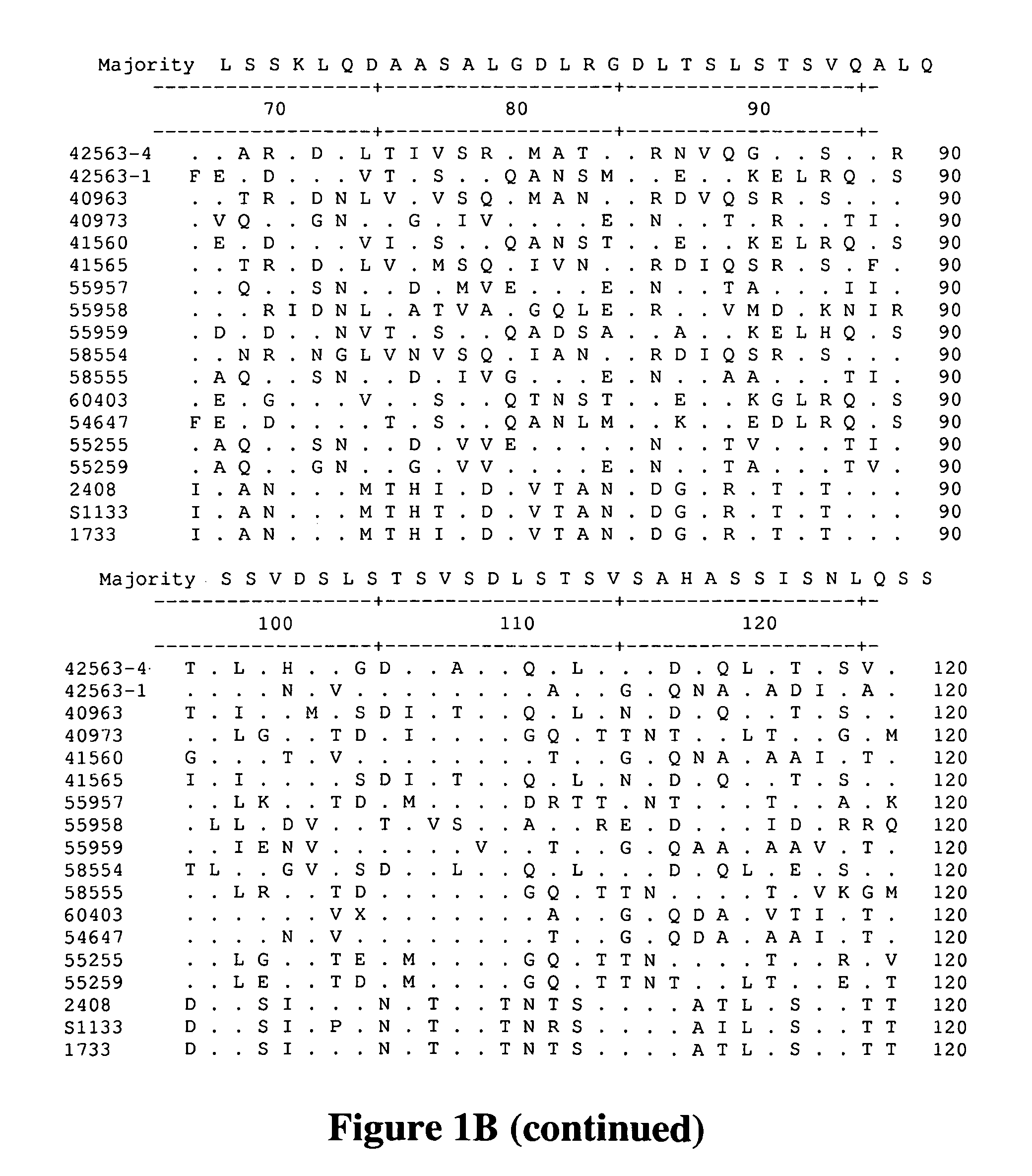
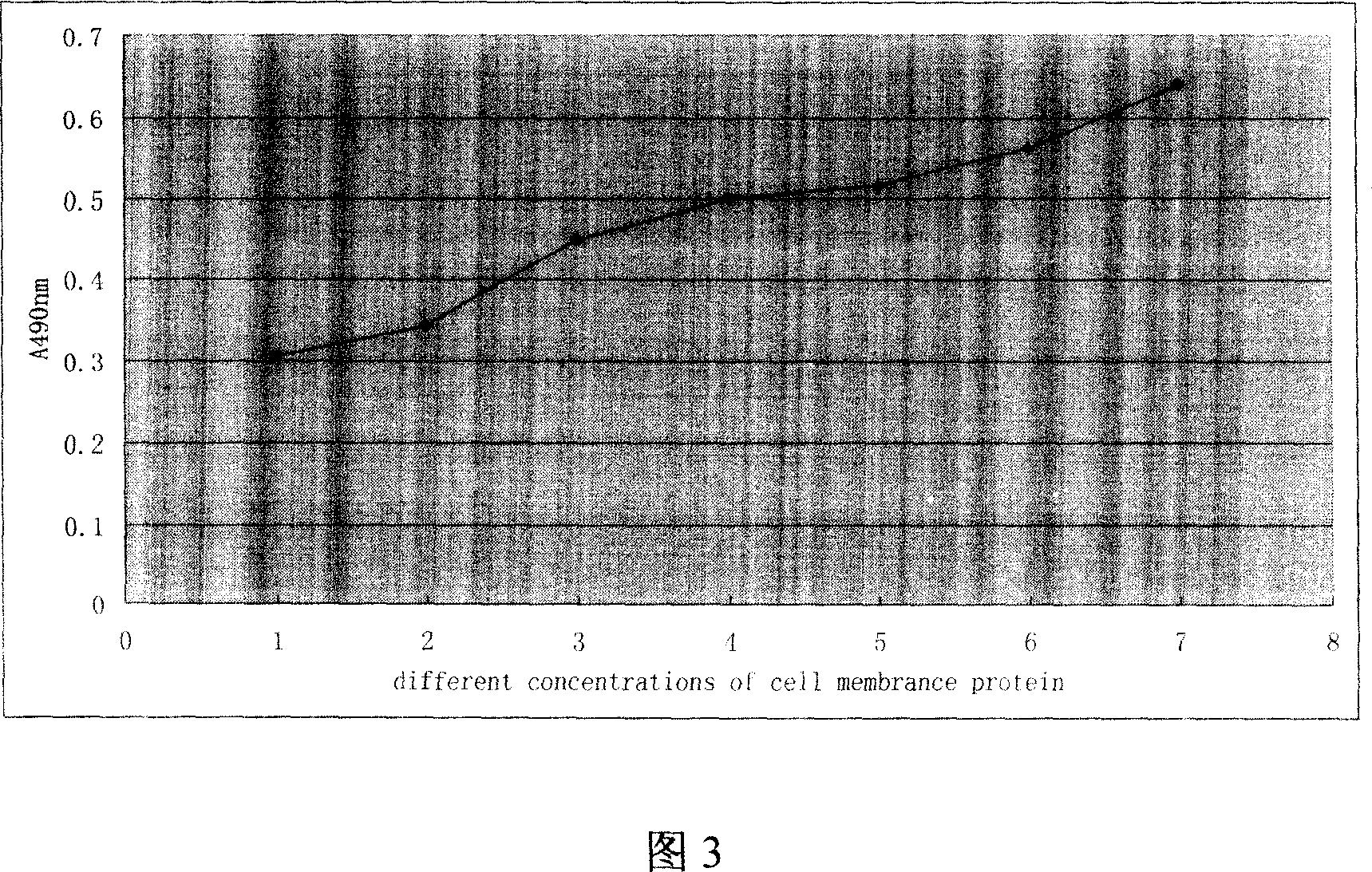
A method for assembling nucleic acid sequence fragments is disclosed. The fragments are assembled using information about their relative position inferred by comparison of the fragments against a known sequence of a related nucleic acid (FIG. 3). Additionally, the method localizes fragments to bacterial artificial chromosomes (FIG. 1) and determines relative position of bacterial artificial chromosomes using sequence comparison information (FIG. 6). The method utilizes the information about relative orientation, mutual distance, fragment localization to bacterial artificial chromosomes, and relative position of bacterial artificial chromosomes to constrain the assembly process (FIG. 5), thus resulting in a more accurate assembly requiring fewer sequencing reactions.
Owner:IP GENESIS
Method for probabilistic error-tolerant natural language understanding
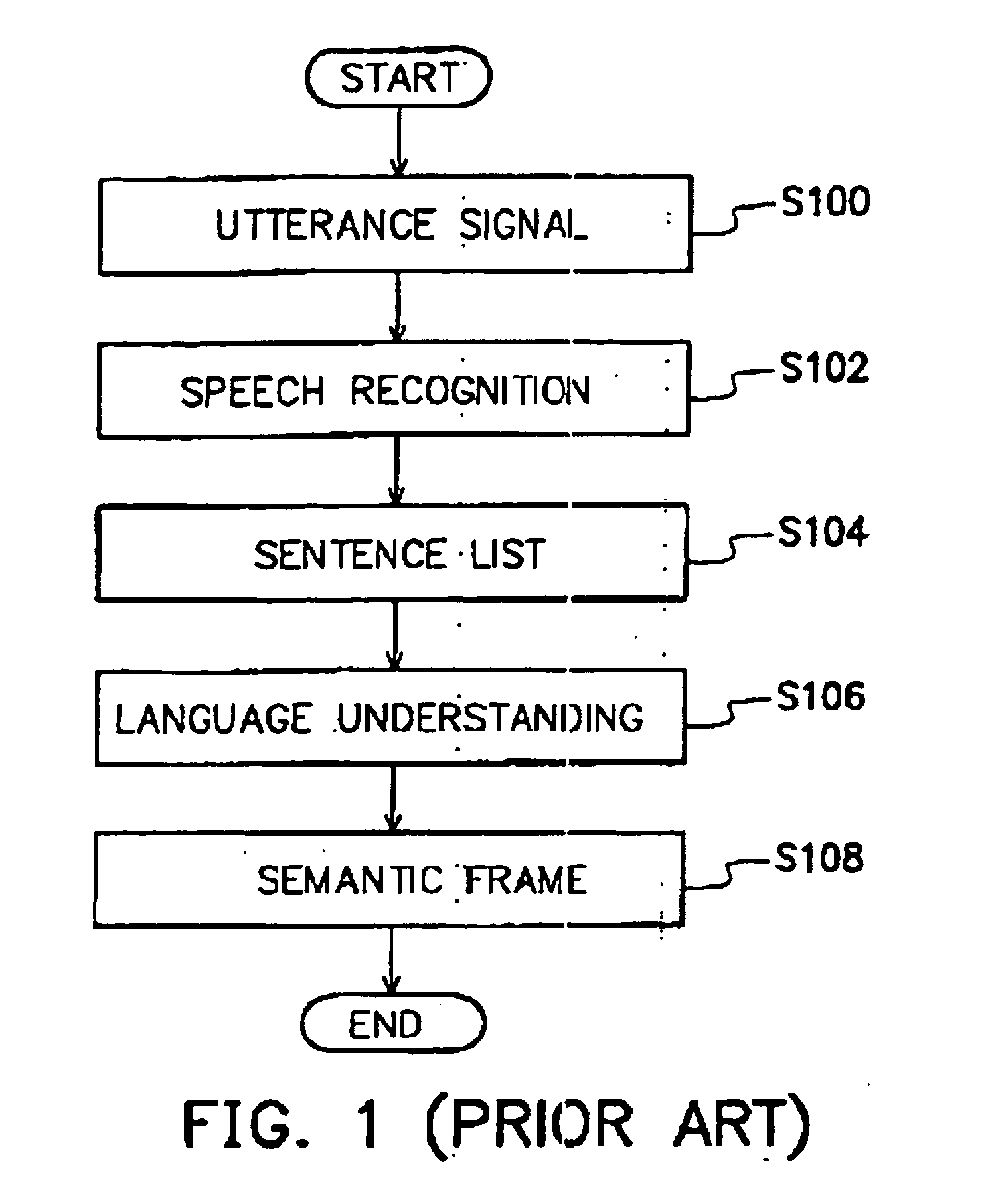
InactiveUS6920420B2Speech recognitionSpecial data processing applicationsLanguage understandingNatural language understanding
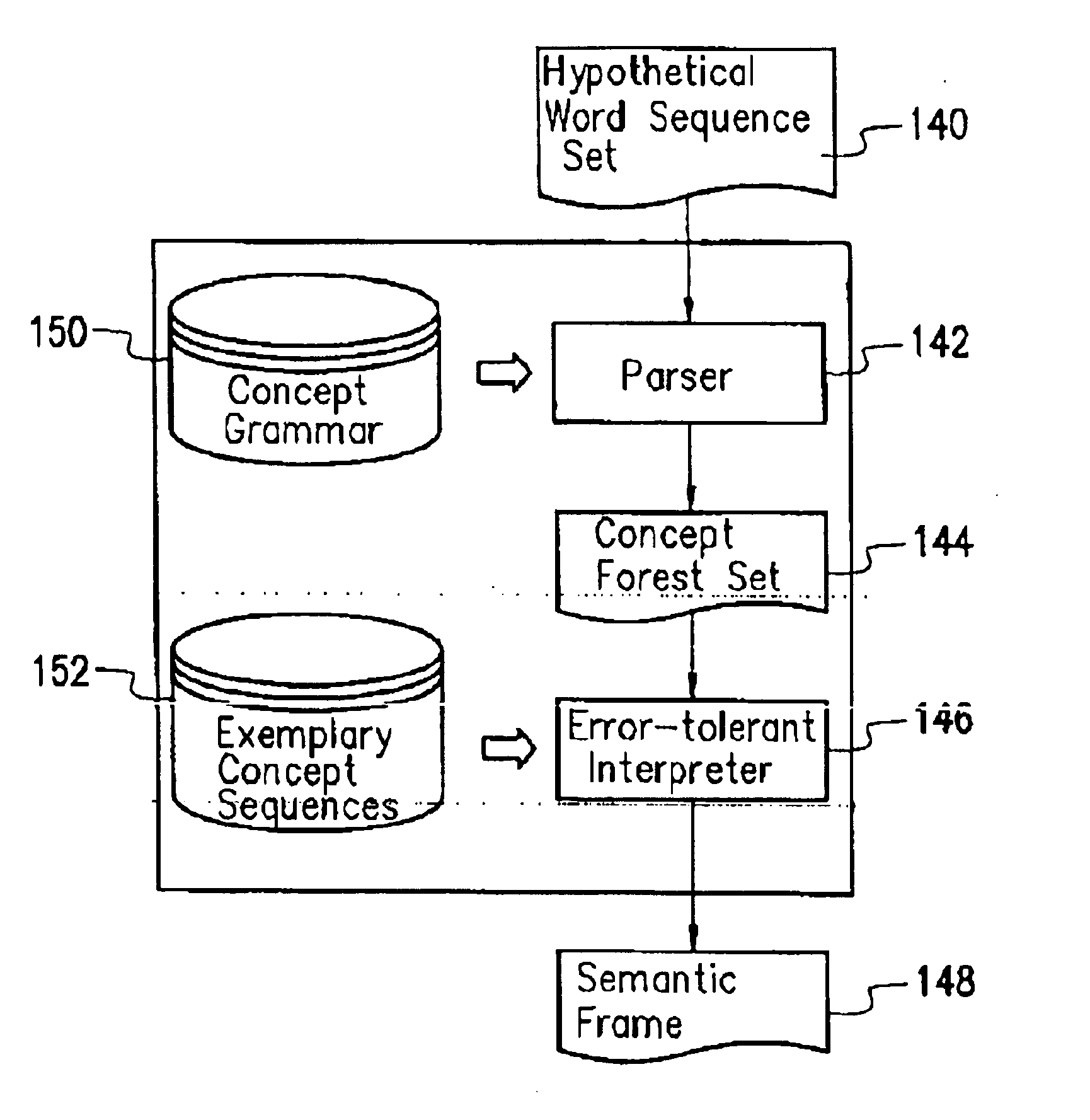
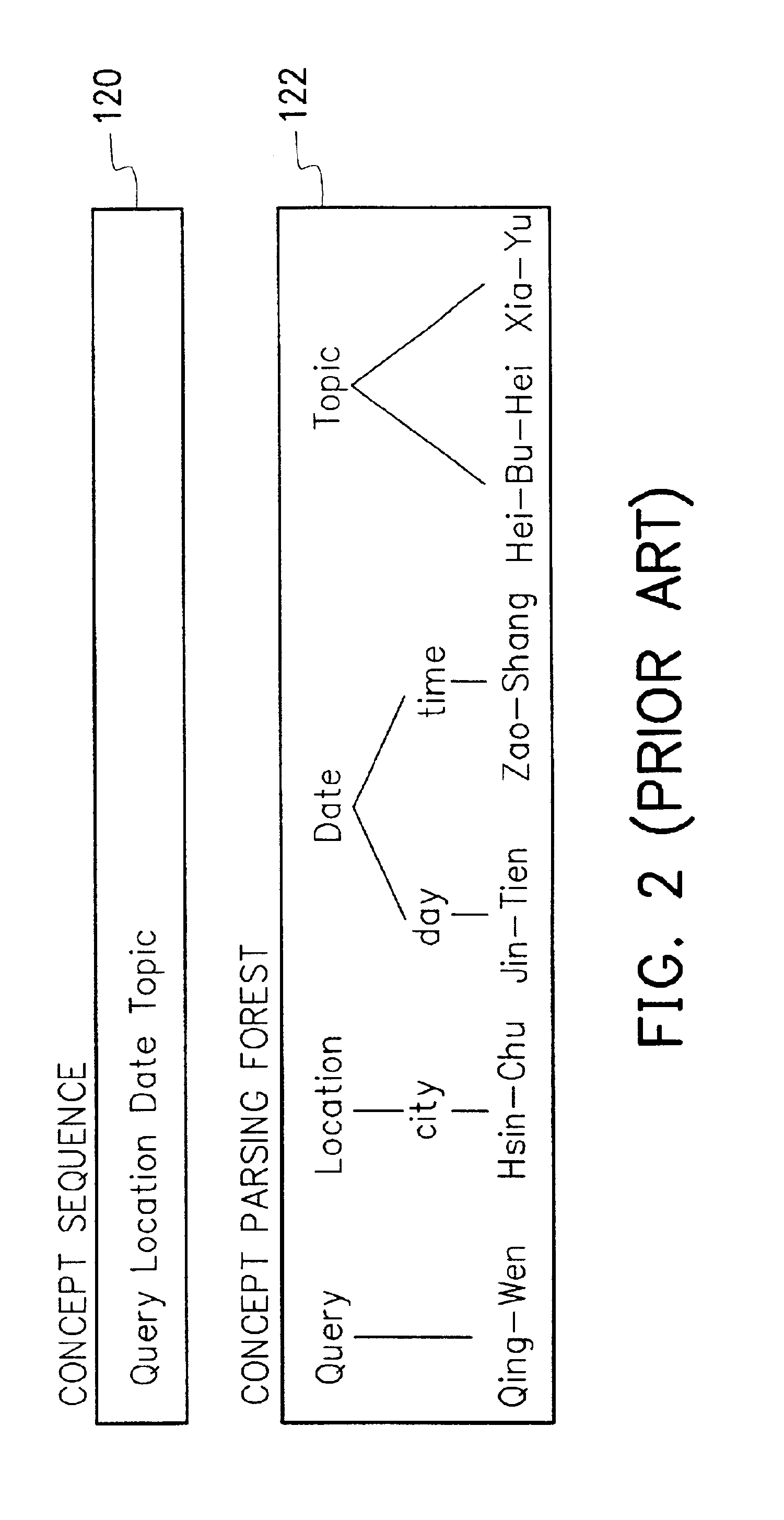
A method of probabilistic error-tolerant natural language understanding. The process of language understanding is divided into a concept parse and a concept sequence comparison steps. The concept parse uses a parse driven by a concept grammar to construct a concept parse forest set by parsing results of speech recognition. The concept sequence comparison uses an error-tolerant interpreter to compare the hypothetical concept sequences included by the concept parse forest set and the exemplary concept sequences included in the database of the system. A most possible concept sequence is found and converted into a semantic framed that expresses the intention of the user. The whole process is led by a probability oriented scoring function. When error occurs in the speech recognition and a correct concept sequence cannot be formed, the position of the error is determined and the error is recovered according to the scoring function to reduce the negative effect.
Owner:IND TECH RES INST
Processors for multi-dimensional sequence comparisons
InactiveUS20050228595A1Requirement to reduce computing speedEffect on its overall performanceDigital computer detailsBiological testingRecursive analysisHigh dimensional
Improved processors and processing methods are disclosed for high-speed computerized comparison analysis of multiple linear symbol or character sequences, such as biological nucleic acid sequences, protein sequences, or other long linear arrays of characters. These improved processors and processing methods, which are suitable for use with recursive analytical techniques such as the Smith-Waterman algorithm, and the like, are optimized for minimum gate count and maximum clock cycle computing efficiency. This is done by interleaving multiple linear sequence comparison operations per processor, which optimizes use of the processor's resources. In use, a plurality of such processors are embedded in high-density integrated circuit chips, and run synchronously to efficiently analyze long sequences. Such processor designs and methods exceed the performance of currently available designs, and facilitate lossless higher dimensional sequence comparison analysis between three or more linear sequences.
Owner:COOKE LAURENCE H +1
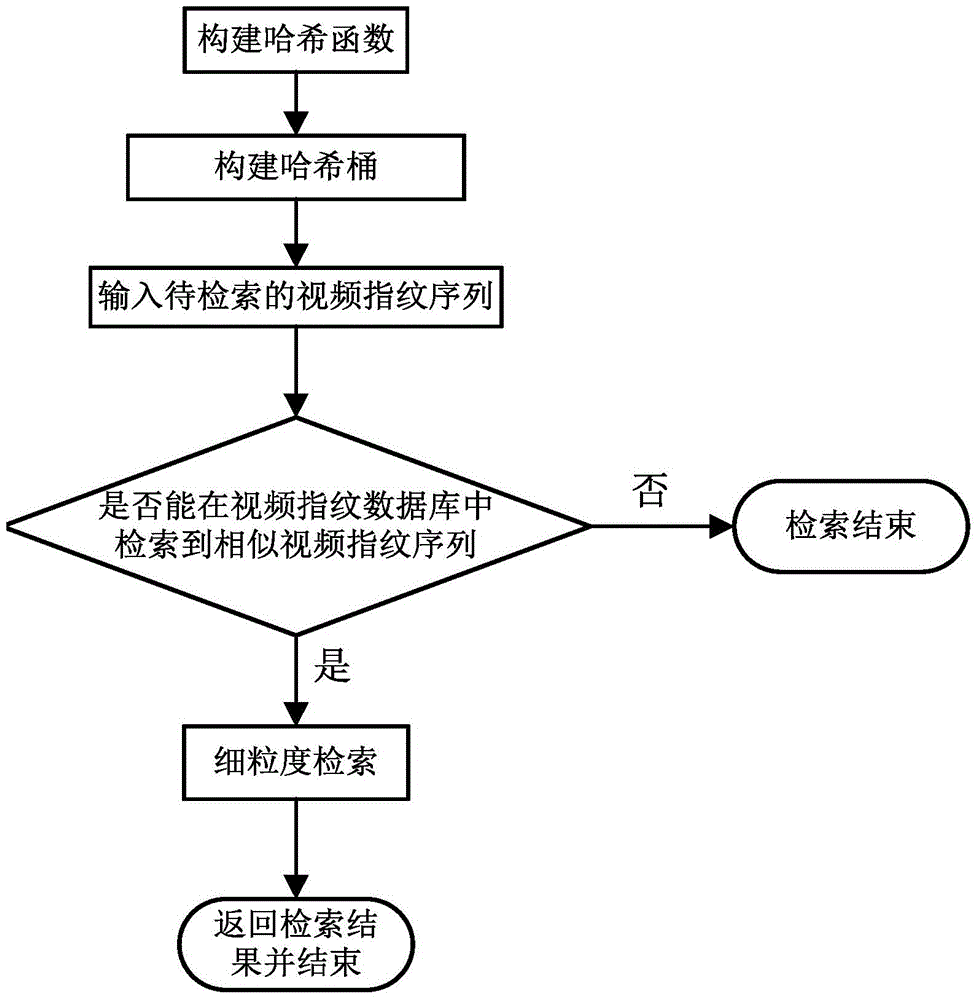
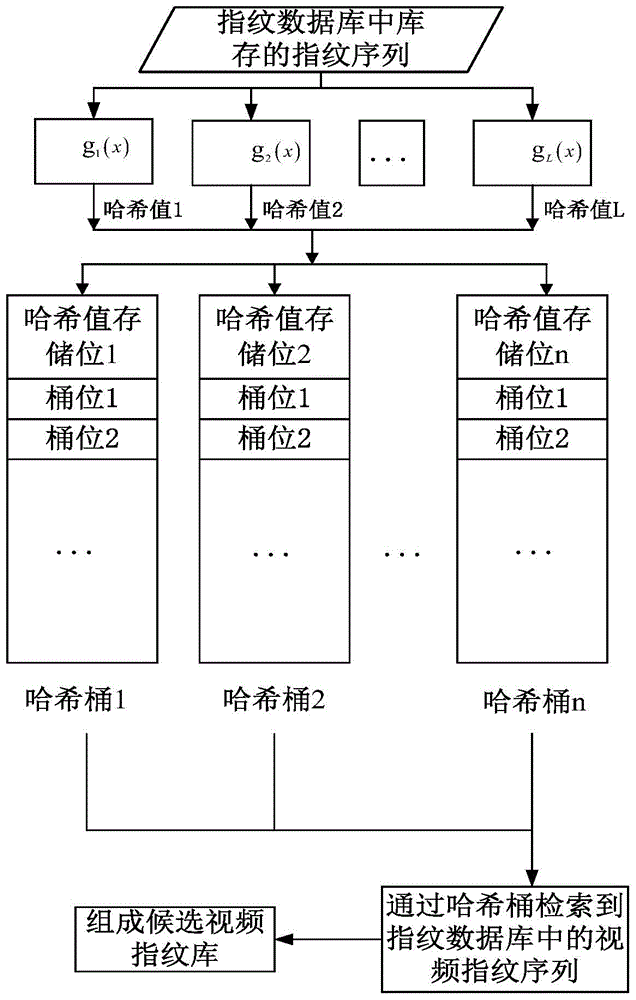
Video fingerprint retrieval method based on coarse and fine granularity
ActiveCN104142984AReduce time overheadGuaranteed accuracySpecial data processing applicationsPattern recognitionVideo retrieval
The invention discloses a video fingerprint retrieval method based on coarse and fine granularity. According to the method, a video fingerprint is subjected to retrieval comparison in different-layer and different granularity retrieving mode. A local sensitive Hash algorithm is used for carrying out Hash processing on all video fingerprints in a video fingerprint database and the video fingerprint to be retrieved, so that the video fingerprint most similar to the video fingerprint to be retrieved is found, and the coarse granularity finding is completed, and the preparation is made for effectively reducing the time expenditure of the video retrieval; during the fine granularity finding, through the fast video fingerprint matching mechanism improved based on a biological sequence comparison technology BLAST, whether the video fingerprint to be retrieved exists in the video fingerprint database or not is fast found, the video segment matching finding can also be carried out, and in addition, the affiliated complete videos of the video segments to be retrieved and the concrete occurring time positions of the video segments to be retrieved in the complete videos can be determined according to the features of the video fingerprint, so that the accuracy and the real-time performance of the video finding are ensured.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
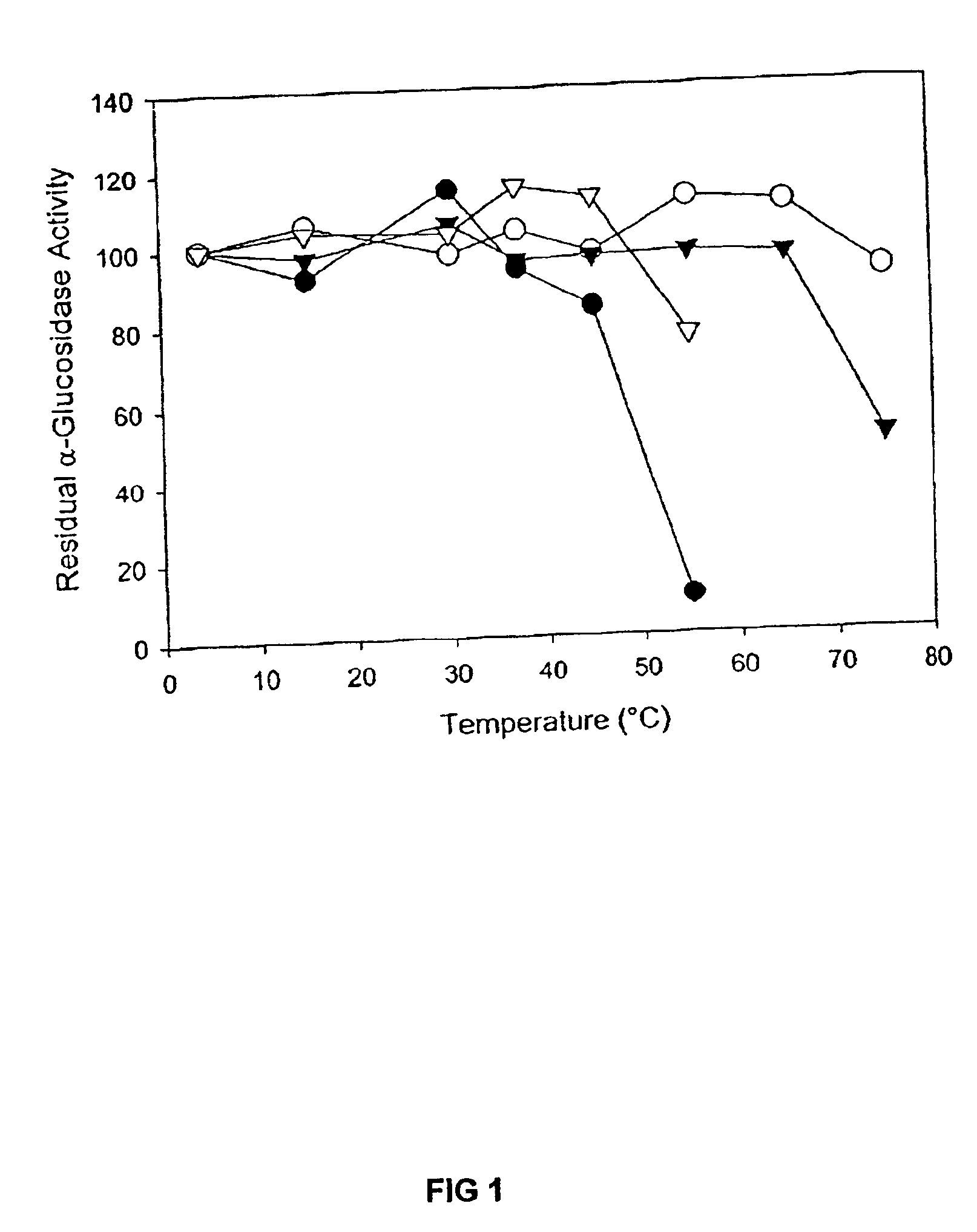
Modified barley α-glucosidase
Barley α-glucosidase is an important enzyme in the conversion of barley starch to fermentable sugars during the industrial production of ethanol, as in brewing and fuel ethanol production. The enzyme is, however, relatively thermolabile, a disadvantage for an enzyme useful in industrial processes which are preferably conducted at elevated temperatures. Site directed mutagenesis has been conducted to make mutant forms of barley α-glucosidase which have improved thermostability. The sites for this site-directed mutagenesis were selected by sequence comparisons with the sequences of other α-glucosidase proteins which are more thermostable. The recombinant mutant enzymes thus produced have been demonstrated to improve the thermostability of the enzyme.
Owner:WISCONSIN ALUMNI RES FOUND +1
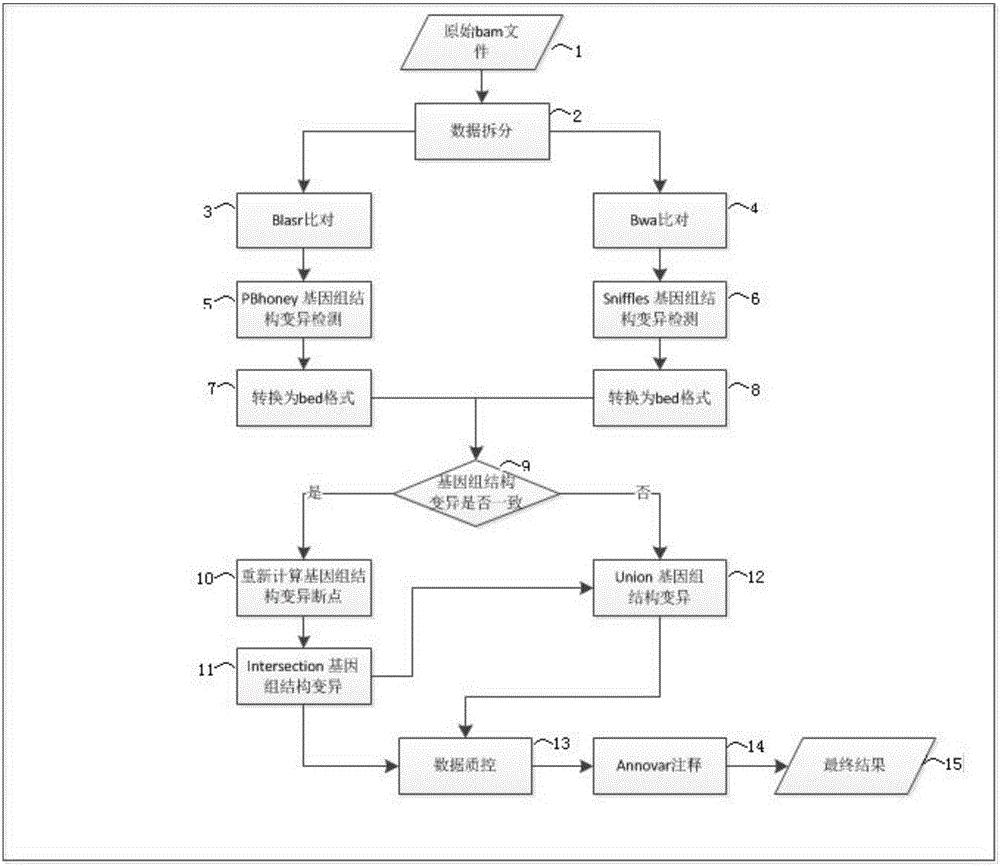
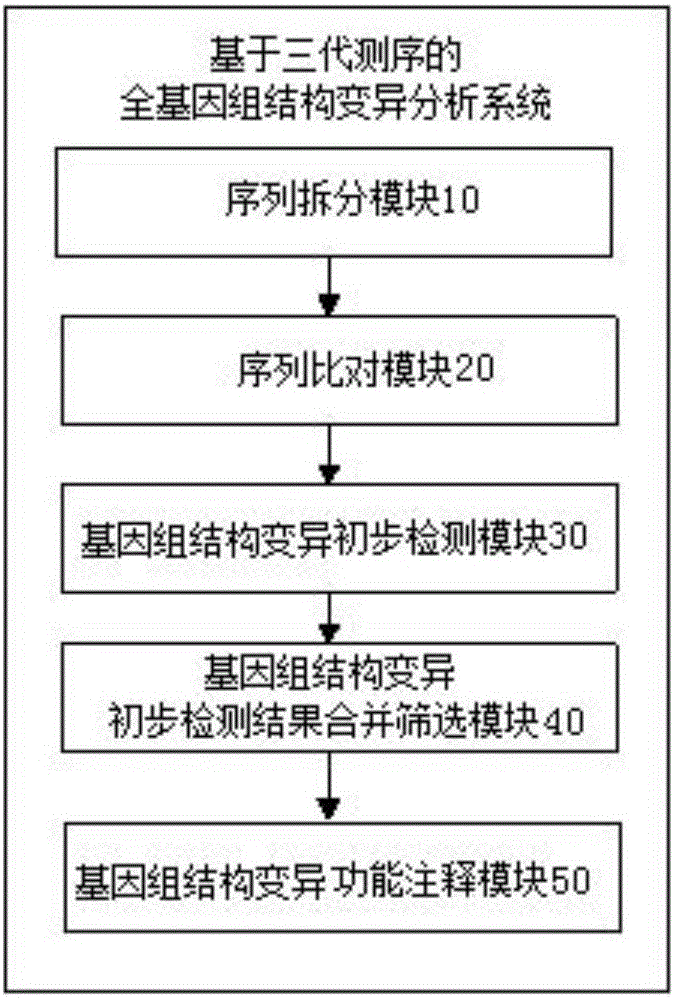
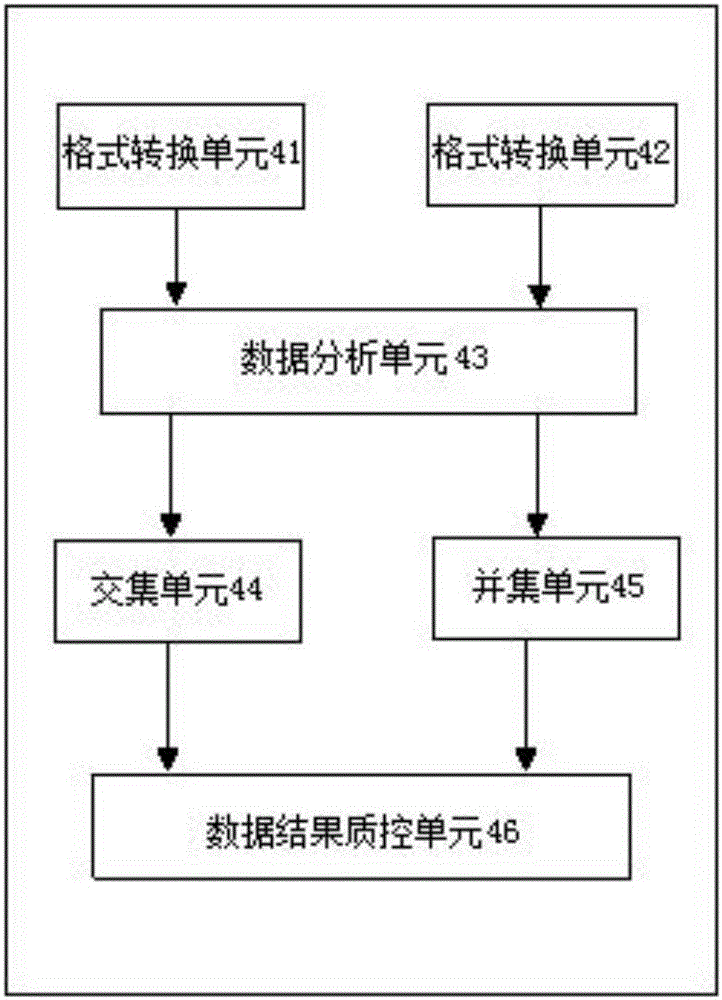
Three-generation sequencing-based whole genome structure variation analysis method and system
The invention discloses a three-generation sequencing-based whole genome structure variation analysis method and system. The method comprises the steps of 1) performing sequence splitting; 2) performing sequence comparison; 3) performing genome structure variation preliminary detection; 4) combining and screening genome structure variation preliminary detection results; and 5) performing genome structure variation function annotation. The system comprises a sequence splitting module, a sequence comparison module, a genome structure variation preliminary detection module, a genome structure variation preliminary detection result combining and screening module and a genome structure variation function annotation module. According to the method and the system, by integrating existing three-generation genome structure variation detection technologies PBhoney and Sniffles, the accuracy and sensitiveness of genome structure variation detection under low coverage degree can be effectively improved, and the reliability of the detection results is ensured while the detection cost is reduced.
Owner:BEIJING GRANDOMICS BIOTECH
Application of nucleotide sequence of rDNA (recombinant deoxyribonucleic acid) ITS (internal transcribed spacer)-D3 region in establishment of DNA (deoxyribonucleic acid) bar code identification system for medicinal plants
InactiveCN102191318AImprove versatilitySequence alignment is accurateMicrobiological testing/measurementSpecial data processing applicationsNucleotideDNA barcoding
The invention relates to a method for identifying medicinal plants by utilizing nucleotide sequences, in particular to an application of a nucleotide sequence of an rDNA (recombinant deoxyribonucleic acid) ITS (internal transcribed spacer)-D3 region in establishment of a DNA (deoxyribonucleic acid) bar code identification system for medicinal plants. The application comprises the following steps of: firstly, detecting the nucleotide sequences of the ITS-D3 regions of medicinal plants, and establishing a DNA bar code database; then, detecting the nucleotide sequence of the ITS-D3 region of a sample to be identified; constructing a clustering tree by the detected nucleotide sequence and the nucleotide sequences of the ITS-D3 regions in the DNA bar code database; determining the medicinal plant having the closest genetic relationship with the sample to be identified in the DNA bar code database according to the clustering tree, and comparing the difference between the nucleotide sequences of the ITS-D3 regions of the medicinal plant and the sample to be identified; and then, by taking the name of the species of the medicinal plant having the closest genetic relationship with the sample to be identified in the database as a reference and combining information such as morphological characteristics and the like, determining the name of the species of the sample to be identified. In the invention, the DNA bar codes have the characteristics of good primer universality, accurate sequence comparison and strong species distinguishing capability.
Owner:GUANGZHOU UNIVERSITY OF CHINESE MEDICINE +1
Protein secondary structure engineering prediction method based on large margin nearest central point
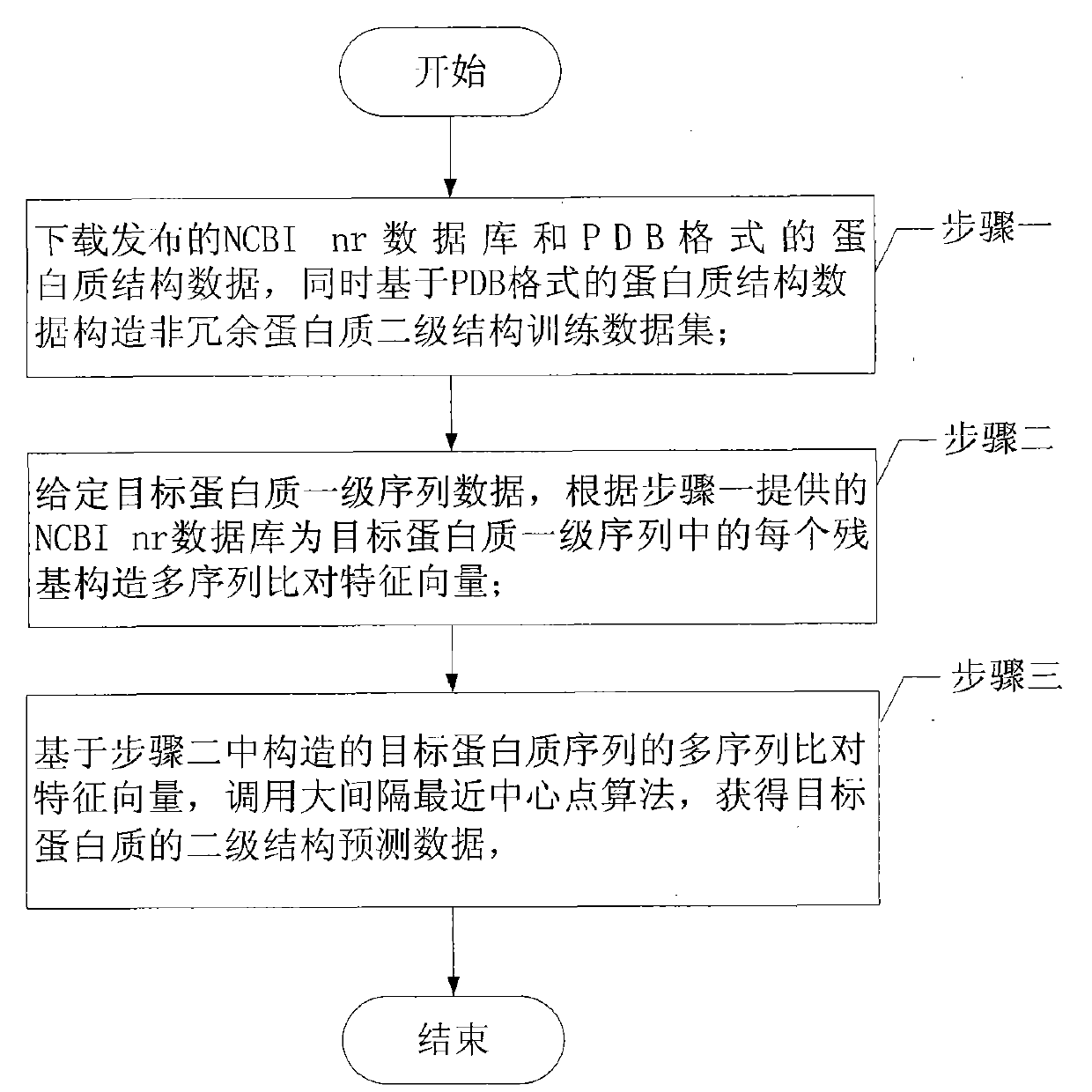
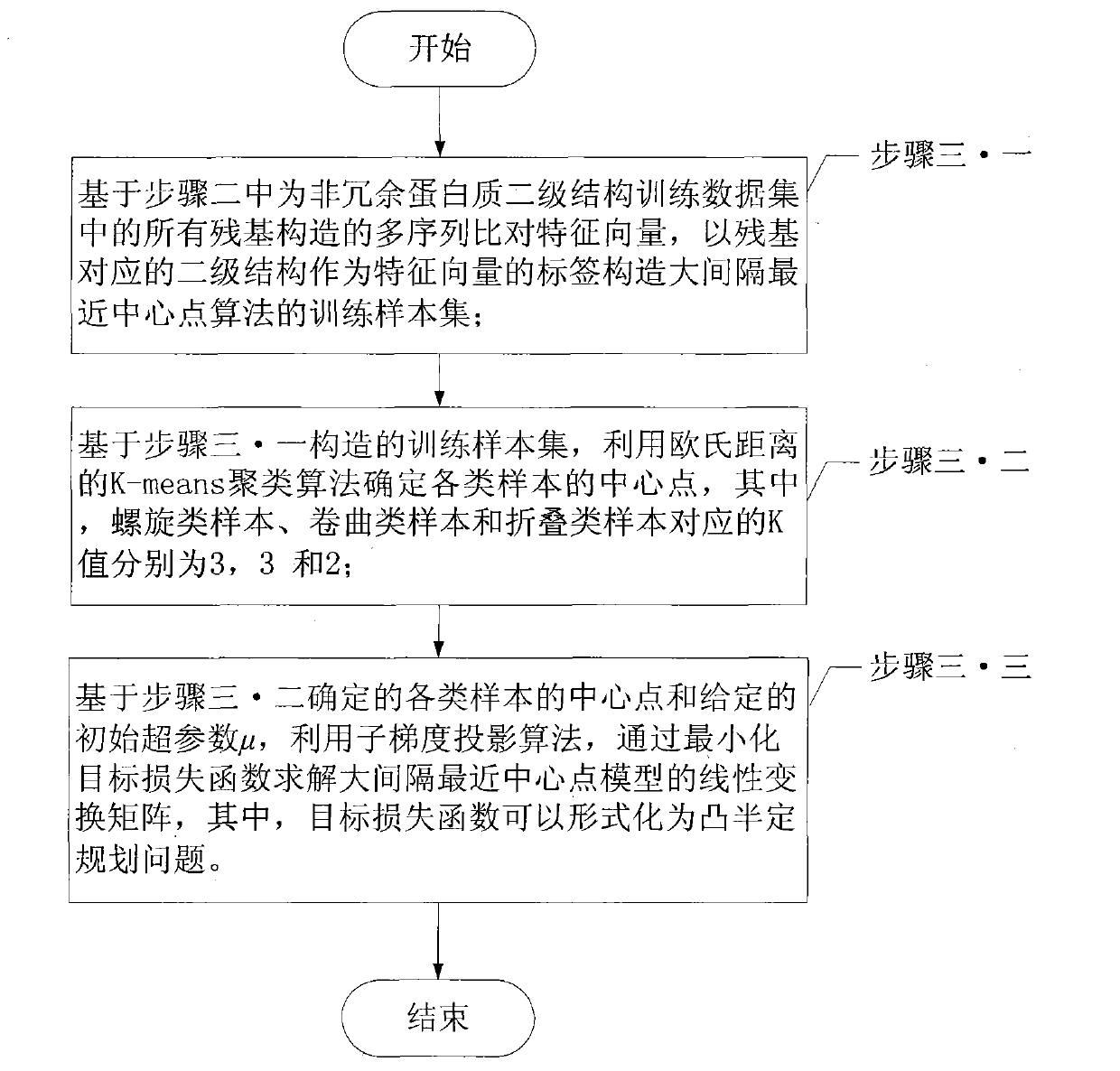
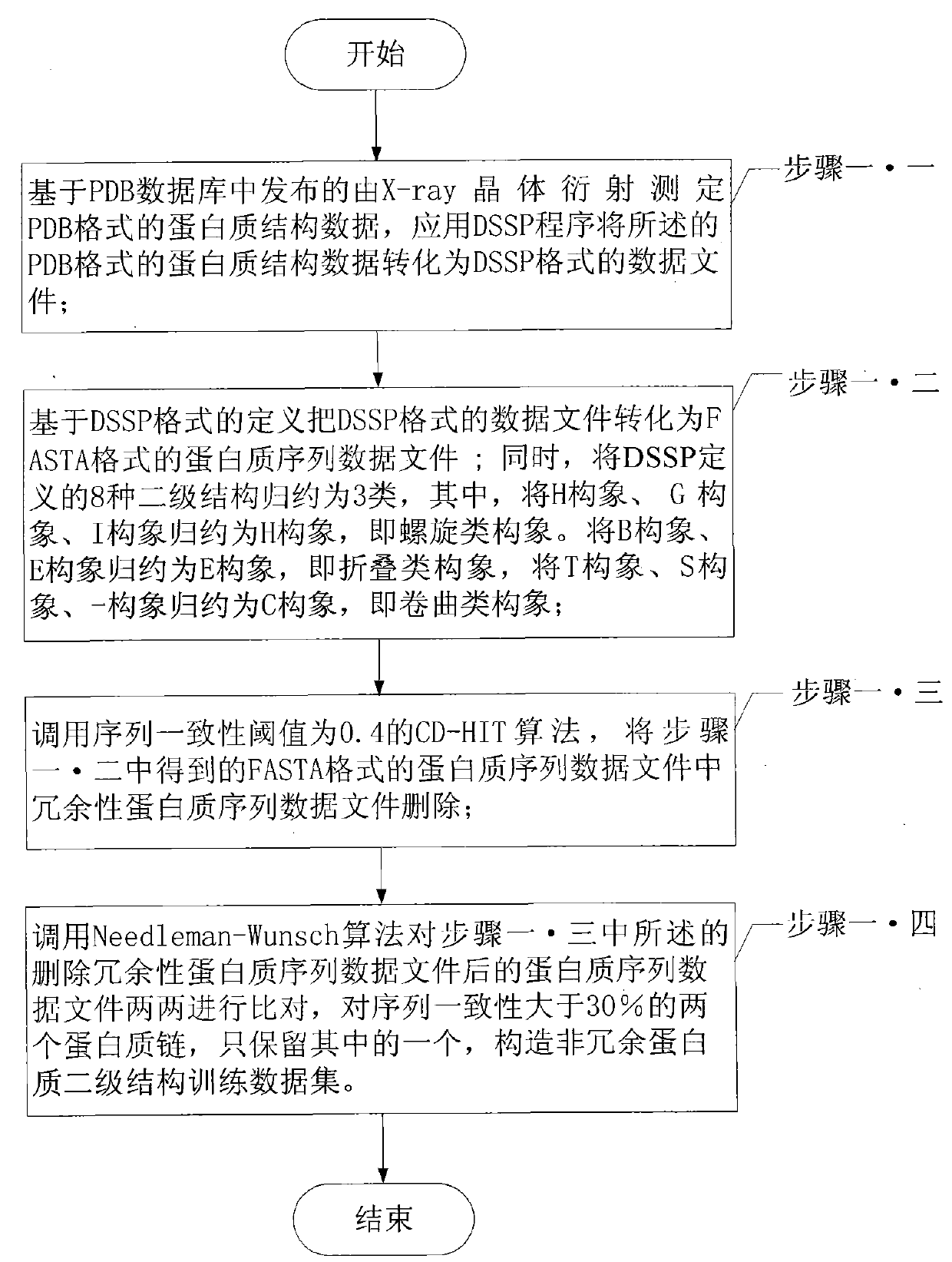
The invention relates to a protein secondary structure engineering prediction method based on large margin nearest central point, belonging to the protein secondary structure engineering prediction method field and solving the problems that the existing protein secondary structure prediction method has local minimum of data weight number and low prediction efficiency when adopting machine learning algorithm. The protein secondary prediction method of the invention includes that: firstly a non-redundant protein secondary structure training data set is constructed based on PDB database, then a multi-sequence comparison characteristic is constructed for a target protein chain based on NCBT nr database, and finally the large margin nearest central point algorithm is utilized to build a protein secondary structure prediction model. The large margin nearest central point algorithm utilizes Euclidean distance K-means clustering algorithm to determine the central point of each sample, and linear transformation of input space is learned by a minimization target loss function. The invention realizes fast, high-efficiency and high-precision protein secondary structure prediction and is applicable to protein secondary structure prediction.
Owner:HARBIN INST OF TECH
Method for quantitatively recovering carbonate rock stratum denudation quantity
ActiveCN105044797ARealize quantitative calculationAccurate calculationGeological measurementsGeomorphologyDecomposition
The invention discloses a method for quantitatively recovering carbonate rock stratum denudation quantity, comprising steps of performing system sampling on the field outcrop and corestone, performing analysis and measurement on the conodont to form a conodont sequence, forming a complete conodont sequence plate in a research area, performing conodont sequence comparison to determine a stratum deletion region and a stratum non-deletion region, sorting out single-well GR curves of the stratum deletion region and the stratum non-deletion region, performing Fourier changing on the GR curve which has gone through the non-deletion region decomposition to carry out time-frequency transformation so as to obtain the frequency of gyration of different levels, determining gyration of various levels to determine gyration thickness of the corresponding level, determining stratum gyration number of various levels, determining complete stratum depositing time, performing calculation on the -deletion region to obtain the gyration frequency, rotation time and rotation thickness of various levels, utilizing the ratio of the non-deletion complete depositing time to the deletion region alternative level gyration time to obtain an original stratum thickness and performing difference calculation on the original stratum thickness and the current residual thickness.
Owner:PETROCHINA CO LTD
Verification method of sea horses
InactiveCN104017899AExclude phenotypic plasticityRule out genetic variabilityMicrobiological testing/measurementSystematicsOrganism
The invention relates to a verification method of sea horses, belonging to the field of molecular biology. The invention aims to provide a verification method of sea horses. The method is characterized by comprising the following steps: a. extracting sea horse total DNA (deoxyribonucleic acid); b. PCR (deoxyribonucleic acid) amplification: carrying out PCR amplification on the sea horse total DNA to obtain a COI gene sequence; c. carrying out PCR product sequencing; and d. carrying out sequence comparison to determine the species of the sea horse. The method is free from the restriction of morphological characteristics, can identify the species of the sea horse under the condition of being incapable of utilizing the traditional morphological classification process due to lack of morphological characteristics, has the advantages of high accuracy and the like, provides scientific references for sea horse systematics and sea horse resource researches in China, and also provides reliable references for biological diversity of sea horse resources and conservation of resources.
Owner:CHENGDU UNIV OF TRADITIONAL CHINESE MEDICINE
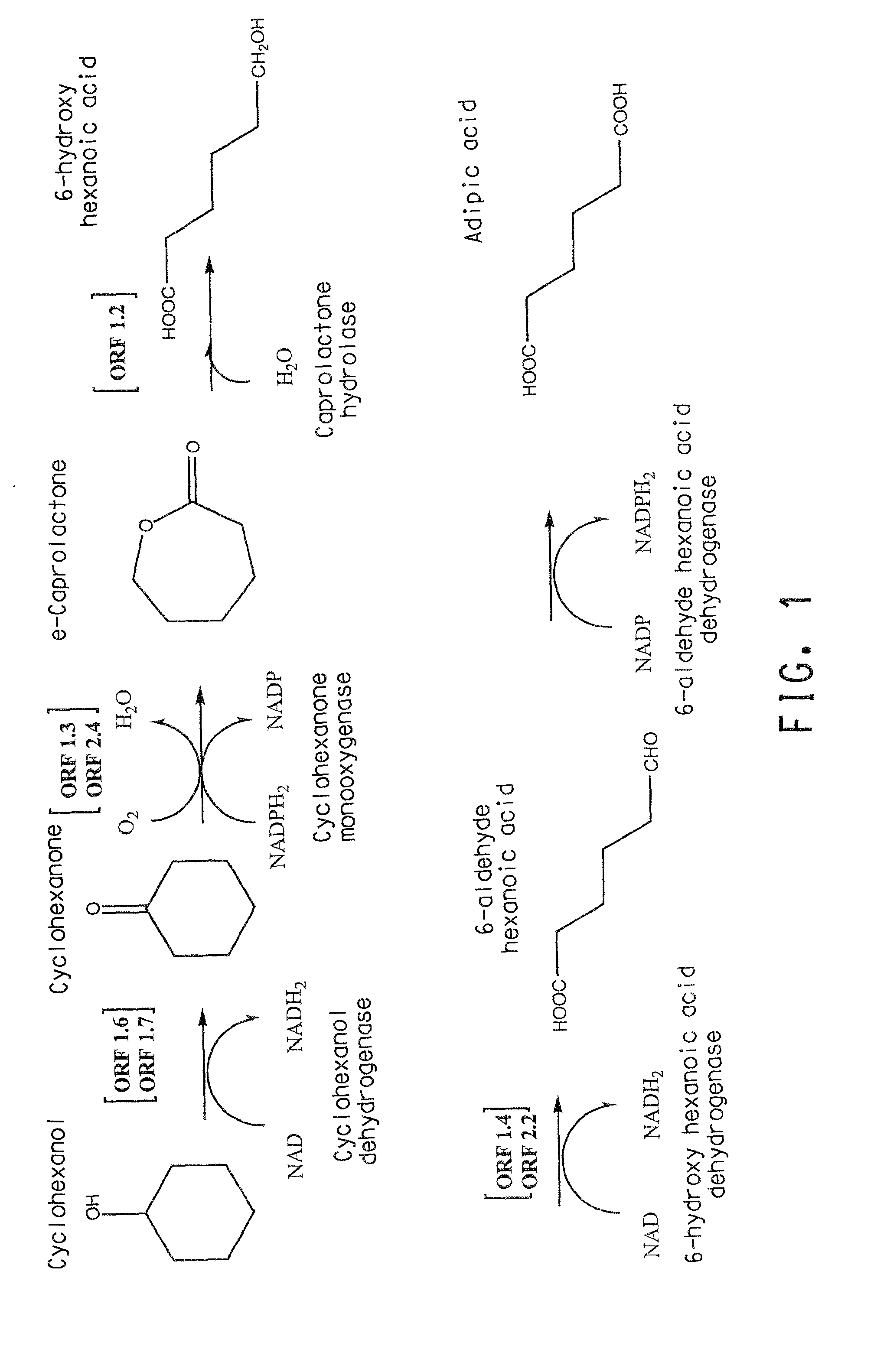
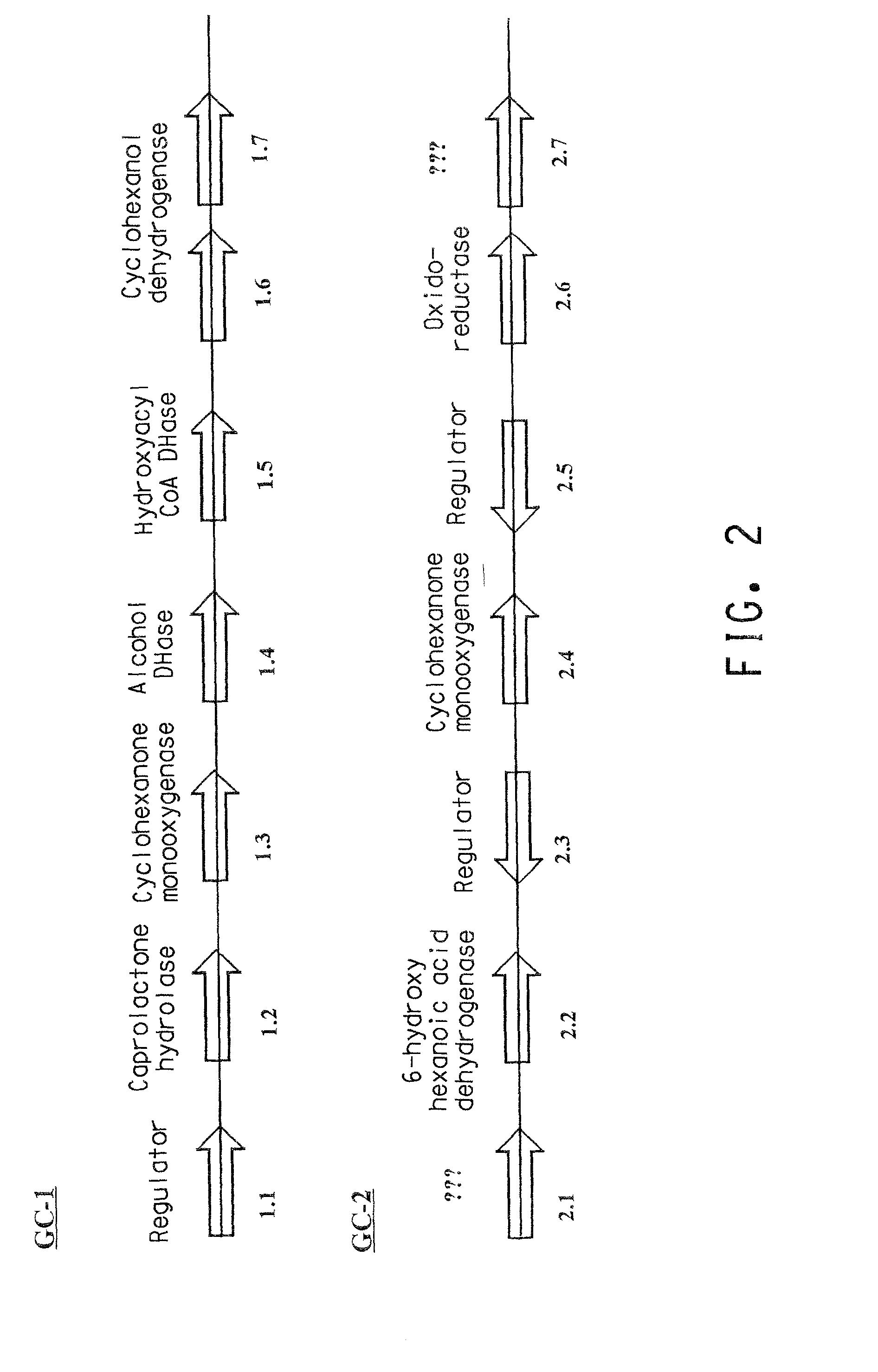
Genes and enzymes for the production of adipic acid intermediates
Two gene clusters have been isolated from an Brevibacterium sp HCU that encode the enzymes expected to convert cyclohexanol to adipic acid. Individual open reading frames (ORF's) on each gene cluster are useful for the production of intermediates in the adipic acid biosynthetic pathway or of related molecules. All the ORF's have been sequenced. Identification of gene function has been made on the basis of sequence comparison and biochemical analysis.
Owner:EI DU PONT DE NEMOURS & CO
Bacterial nucleic acid sequencing identification method based on DNA bar code
InactiveCN106701914AClear scopeClear lengthMicrobiological testing/measurementSequence analysisDNA barcoding
The invention discloses a bacterial nucleic acid sequencing identification method based on a DNA bar code. Common bacterial contaminants in a medicine production environment can be effectively and accurately identified. The bacterial nucleic acid sequencing identification method comprises the steps that 1, a monoclonal bacterial strain is obtained through culture; 2, a genome DNA of the monoclonal bacterial strain obtained in the step 1 is extracted; 3, PCR is performed by using a general nucleic acid sequencing primer to amplify a specific sequence of nucleic acids; 4, a PCR product is extracted and purified; 5, a nucleic acid sequence of the PCR product obtained in the step 4 is determined; 6, a sequence analysis software is applied to perform sequence splicing, positioning is performed by using a general nucleic acid sequencing primer pair, a primer region is removed, and a DNA sequence of a corresponding fragment is obtained; 7, the DNA sequence of the corresponding fragment in the step is imported into a DNA bar code standard sequence core database or a GenBank database for sequence comparison, and bacteria are identified.
Owner:SHANGHAI INST FOR FOOD & DRUG CONTROL
Molecular biology method for identifying purity of tobacco varieties
InactiveCN105734141ANot affectedPreventing and ensuring the safety of production seedsMicrobiological testing/measurementBiotechnologyWhole genome sequencing
The invention relates to a molecular biology method for identifying purity of tobacco varieties. The method comprises the following main steps: respectively extracting total genome DNA of a control group and a to-be-detected tobacco variety, performing whole genome sequencing of proper depth by adopting the latest sequencing technology, performing sequence splicing, assembling and whole genome sequence comparison on the control group and the to-be-detected tobacco variety based on a tobacco genome reference sequence by utilizing bioinformatics means, counting base differences of the two groups, and calculating the purity percentage of the to-be-detected tobacco variety relative to the control tobacco variety. The method disclosed by the invention is not influenced by environmental conditions and seasons, is accurate and reliable in result, can accurately identify the purity of the tobacco varieties from a single base variation level of the smallest hereditary unit, can be used for parent purification and authenticity identification of the tobacco varieties and has great significances for guaranteeing safety of tobacco production varieties, maintaining economic benefits of tobacco growers and effectively solving intellectual property disputes of the tobacco varieties.
Owner:HUBEI TOBACCO SCI RES INST
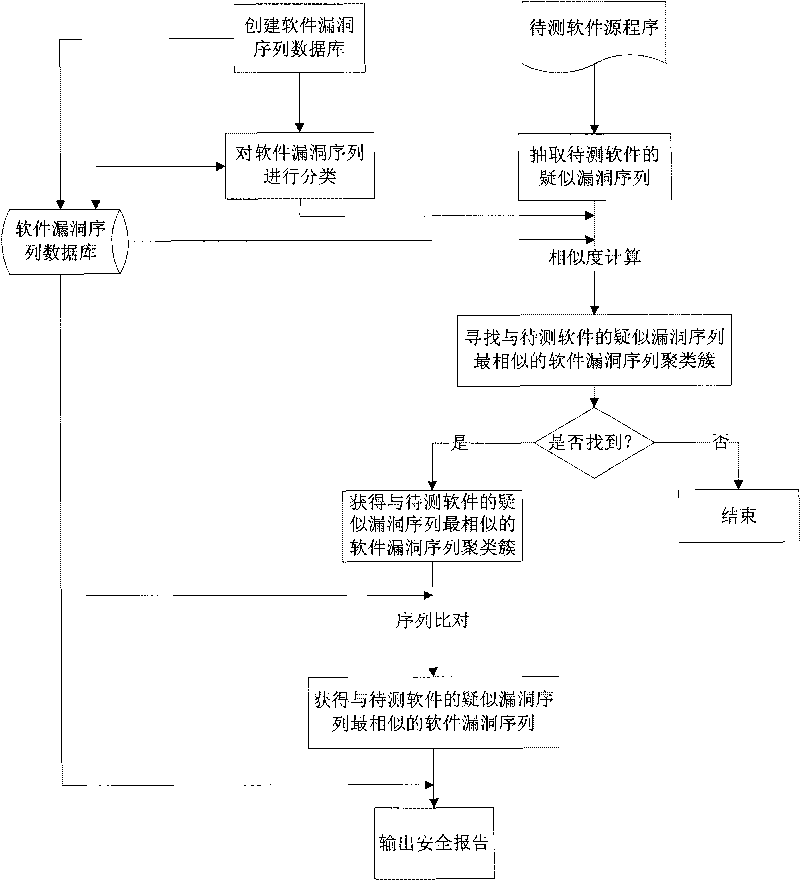
Method for analyzing characteristic of software vulnerability sequence based on cluster
InactiveCN101739337AImprove analysis efficiencyEasy to operateSoftware testing/debuggingSequence databaseSoftware engineering
The invention discloses a method for analyzing the characteristic of a software vulnerability sequence based on a cluster, which belongs to the technical field of information security. The method comprises the following steps: firstly, establishing a software vulnerability sequence database by using the conventional software vulnerability sequence; secondly, analyzing the software vulnerability sequence in the database by using clustering technology so as to generate a plurality of software vulnerability sequence clustering clusters; thirdly, finding the software vulnerability sequence clustering cluster which is most similar to a doubtful vulnerability sequence of software to be tested through similarity computation; fourthly, comparing the doubtful vulnerability sequence of the software to be tested with all software vulnerability sequences in the most similar software vulnerability sequence clustering cluster through a sequence comparison method, and further finding the software vulnerability sequence which is most similar to the doubtful vulnerability sequence of the software to be tested from the software vulnerability clustering cluster; and finally, outputting related vulnerability information, in the software vulnerability sequence database, corresponding to the most similar software vulnerability sequence as a security report. The method improves the analysis efficiency of the software vulnerability sequence.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
Reovirus compositions and methods of use
The present invention relates to novel strains of avian reovirus that were isolated from severe cases of Runting Stunting Syndrome in young broiler chickens in southeast United States. The invention is directed to avian reoviruses that impair digestion in poultry, diagnostic assays using nucleotide- or amino acid-specific components of such viruses, and to vaccines that protect chickens from disease caused by such viruses. Nucleotide sequences for the SI gene, encoding the sigma C minor outer capsid protein, were amplified, and the nucleotide and predicted amino acid sequences were compared with sequences from other recently isolated reovirus field isolates and vaccine strains. Antigenic and molecular characterization of the newly isolated reoviruses revealed a lack of homogeneity with current U.S. isolates, with less than 60% percent amino acid similarity across the sigma C protein. Sequence comparisons with previously reported malabsorption isolates from Europe and Asia revealed a higher amino acid similarity, approaching 80%.
Owner:UNIV OF GEORGIA RES FOUND INC
Rapid identification method for pathogenic fungi
The invention relates to a rapid identification method for pathogenic fungi, which specially comprises the following steps: (1) culturing fungi on a solid culture medium; (2) taking fresh fungi, and extracting the DNA of the fungi by using a microwave method; (3) amplifying DNA fragments between ribosomal spacers of the fungi by using ITS1 and ITS4 primers through PCR (polymerase chain reaction); (4) after recovering the PCR fragments, carrying out sequence determination on the fragments; and (5) carrying out comparison on the obtained sequences by using the Blast of NCBI. According to the invention, the identification of unknown pathogenic fungi can be performed within a shorter time and under relatively simple experimental conditions, thereby facilitating the rapid diagnosis for diseases occurring in fields so as to guide the prevention and control of diseases. According to the method, through the identification on 21 kinds of pathogenic fungi on different crops, and the coincidence rate is 100% through sequence comparison. The method is not only applicable to the identification on fresh mycelia, but also is applicable to the identification on conidium and sclerotium of pathogenic fungi, and has the characteristics of rapidness and high efficiency.
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
Primers for triple PCR of three types of sheep pathogenic mycoplasmas and detection method
InactiveCN106434919AOvercome limitationsQuick checkMicrobiological testing/measurementDNA/RNA fragmentationEpidemiologic surveyMycoplasma pneumonia
The invention provides primers for triple PCR of three types of sheep pathogenic mycoplasmas and a detection method. The three pairs of special primers MO, MCC and MCCP are synthesized respectively according to the designs of sheep mycoplasma pneumoniae, a goat mycoplasma mycoide subspecies and a goat mycoplasma pneumonia subspecies, and a triple PCR optimum reaction system and reaction conditions for the three types of sheep pathogenic mycoplasmas are provided. The simultaneous pathogenic detection of the sheep mycoplasma pneumoniae, the goat mycoplasma mycoide subspecies and the goat mycoplasma pneumonia subspecies can be rapidly carried out without cloning, sequencing and sequence comparison, and the detection method has the advantages of being rapid, accurate, strong in specificity, good in repeatability and the like, suitable for rapid detection of the sheep mycoplasma pneumoniae, the goat mycoplasma mycoide subspecies and the goat mycoplasma pneumonia subspecies and large-scale epidemiological investigation and has great economic and social benefits.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
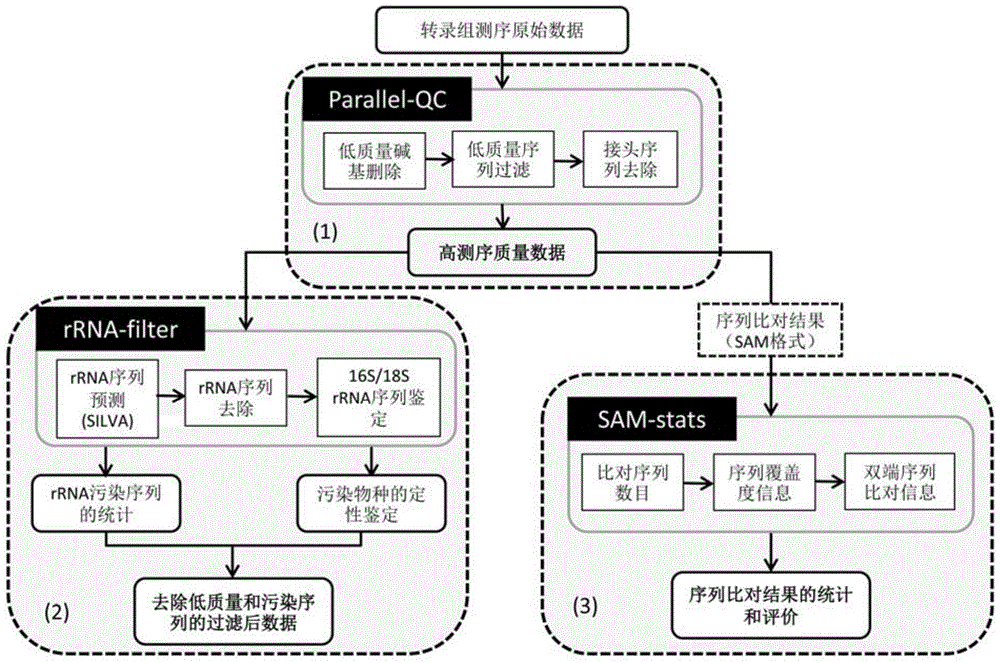
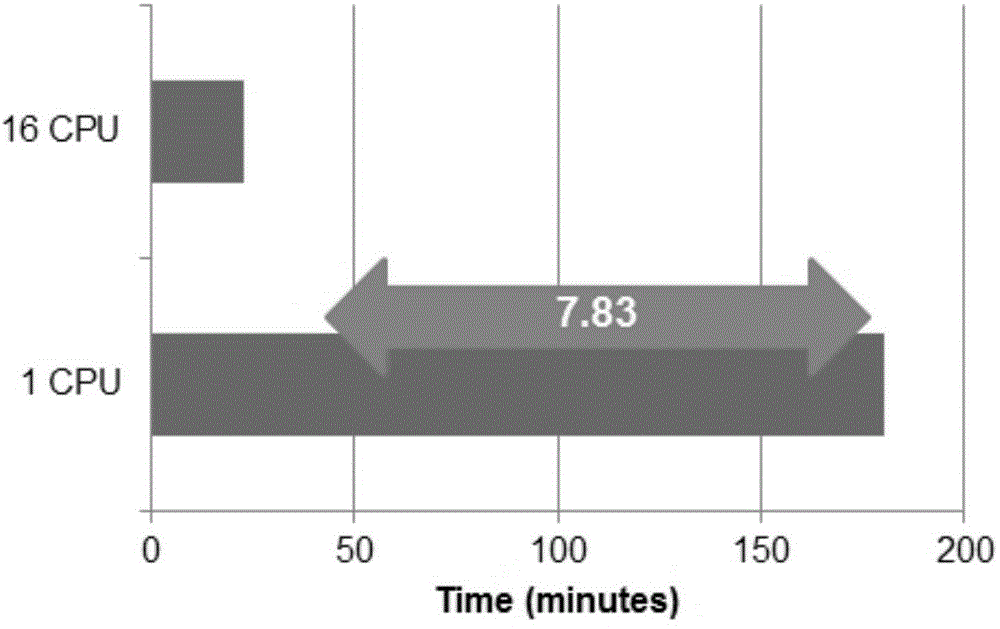
High-flux transcriptome sequencing data quality control method based on multi-core CPU (Central Processing Unit) hardware
ActiveCN105095686AEfficient Data Quality ControlOvercoming Computational Efficiency BottlenecksSpecial data processing applicationsHigh fluxQuality control
The present invention provides a high-flux transcriptome sequencing data quality control method based on multi-core CPU hardware. The method comprises: performing parallel processing on high-flux transcriptome sequencing data by using a multi-core CPU, so as to obtain data without low sequencing quality sequences; performing prediction and removal on rRNA sequences in the data without the low sequencing quality sequences by using the multi-core CPU, and performing qualitative identification on polluted sequences; and performing statistics and evaluation on a sequence comparison result. According to the high-flux transcriptome sequencing data quality control method based on the multi-core CPU hardware, provided by the present invention, based on a multi-core CPU computer, a computing efficiency bottleneck based on a single-core CPU hardware computer is overcome, so that high-flux transcriptome data quality control efficiency is increased by over 7 times; and by applying the high-flux transcriptome sequencing data quality control method, the accuracy and speed of the high-flux transcriptome data quality control are significantly improved, and rapid development of relevant researches of transcriptome sequencing is widely facilitated.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
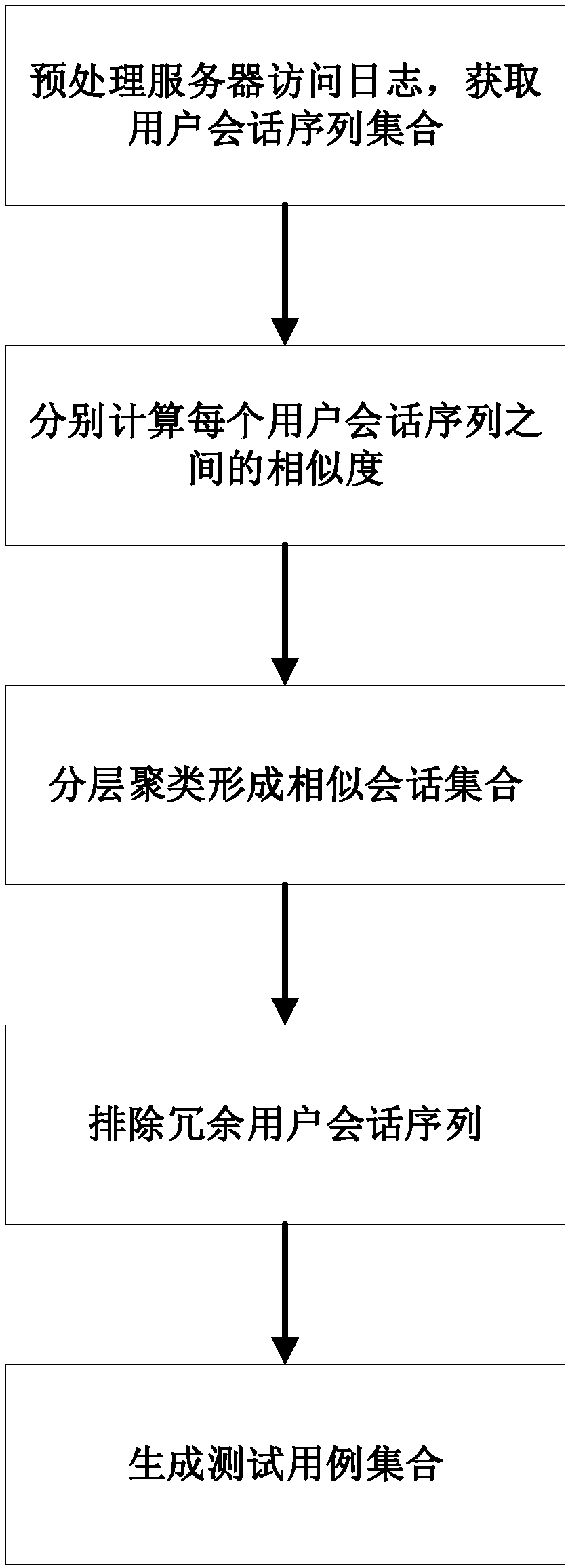
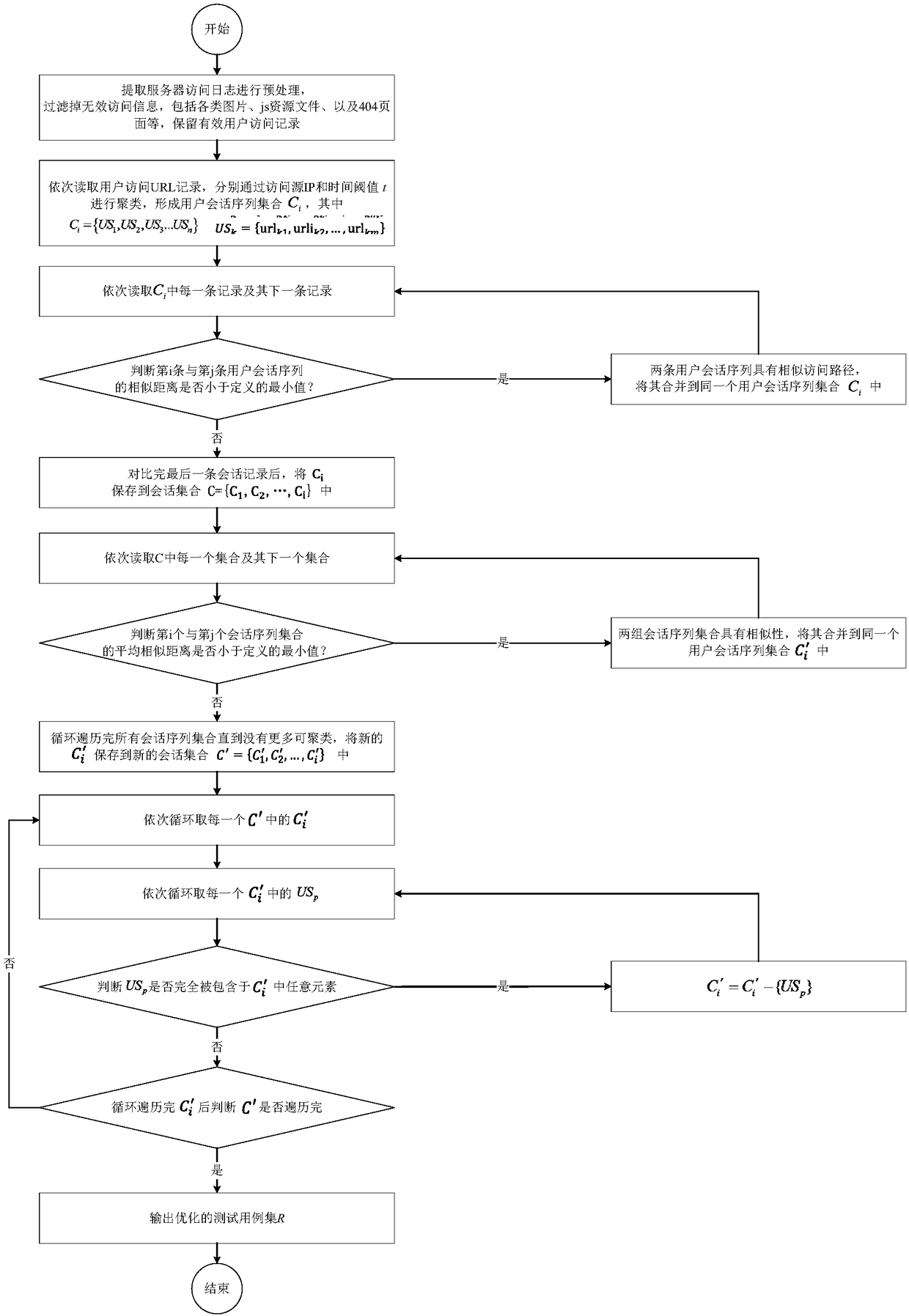
Test-case selection method based on user sessions and hierarchical clustering algorithm
InactiveCN108388508AGood choiceImprove production efficiencyCharacter and pattern recognitionSoftware testing/debuggingSequence alignment algorithmHierarchical cluster algorithm
The invention discloses a test-case selection method based on user sessions and a hierarchical clustering algorithm. The method includes the following steps: acquiring server access logs, and carryingout sorting according to time; carrying out preprocessing and clustering to form a user session sequence set; calculating similarity distances among all user session sequences through using an improved user-session-sequence comparison algorithm; employing the improved condensing hierarchical clustering algorithm to cluster the user session sequences, and outputting final clustering results of test cases; and optimizing selection of the test cases through deleting redundant test cases. According to the method of the invention, representative user operation sequences can be quickly mined from the large number of server access logs to use the same as test cases, automation of test-case generation and optimization of test-case selection are realized, and subsequent work of automated functiontests of a server, performance tests, user behavior analysis and the like is facilitated.
Owner:SOUTH CHINA UNIV OF TECH
Newcastle disease virus/avian influenza virus H9 subtype/infectious bronchitis virus triplex fluorescence quantification detection reagent and detection method
InactiveCN105671204AHigh sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesFluorescenceQuarantine
The invention relates to a newcastle disease virus / avian influenza virus H9 subtype / infectious bronchitis virus triplex fluorescence quantification detection reagent and a detection method and belongs to the technical field of animal quarantine. A newcastle disease virus M gene coding region specific sequence, an avian influenza virus H9 subtype H gene coding region specific sequence and a chicken infectious bronchitis virus M gene coding region specific sequence are selected as target regions, and on the basis of multi-sequence comparison, primer and probe design is conducted. The length of primers is about 20 basic groups, the GC content is 50-60%, a two-stage structure and repeatability do not exist in the primers, no complementary sequence exists between the primers or in the primers, and the melting temperature (Tm value) difference between the primers is smaller than 5 DEG C. In order to guarantee universal use of a newcastle disease virus probe, the length of the probe is only 13 basic groups, the probe is modified by LAN, and the Tm value of the probe is increased. The lengths of the other two virus probes are both about 25 basic groups, and the Tm values are about 5 DEG C higher than those of the primers.
Owner:山东省动物疫病预防与控制中心 +1
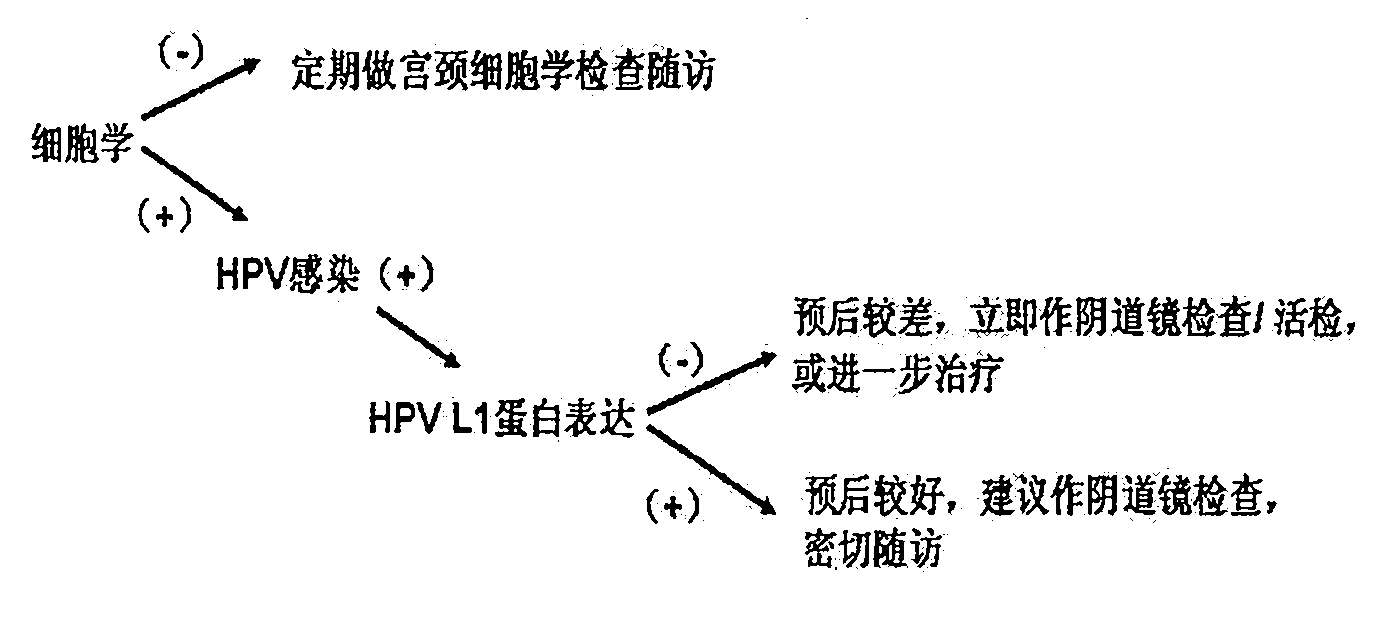
Human papilloma virus (HPV) capsid protein L1 polypeptide and preparation and application thereof
The invention relates to a human papilloma virus capsid protein L1 (HPVL1) peptide and preparation and application thereof. The HPVL1 peptide is a peptide segment of which the amino acid of the sequence is N end-Cys Thr Leu Thr Ala Asp Val Met listed Thr Tyr Ile His-C end, or is a peptide segment comprising the amino acid of the sequence and with a length less than 50 amino acids. In the invention, the peptide which is a very conservative peptide segment located on the surface of protein is sieved out by performing epitope forecasting and multi-sequencing comparison on the L1 proteins of 18 low-risk types and high-risk type HPV subtypes (6, 11, 16, 18, 52, 58 and the like); the antibody generated from induction can react with multiple types of HPVL1, and can be used for the detection of the multiple types of HPVL1, in particular to the application of precancerous lesions detection of human cervical carcinoma; and the antibody aiming at the peptide segment also can be used for purifying and preparing the multiple types of HPVL1. In addition, the invention also provides the applications of the peptide segment used as multiple types of HPV antiserum and detected matters of monoclonal antibody.
Owner:SICHUAN UNIV
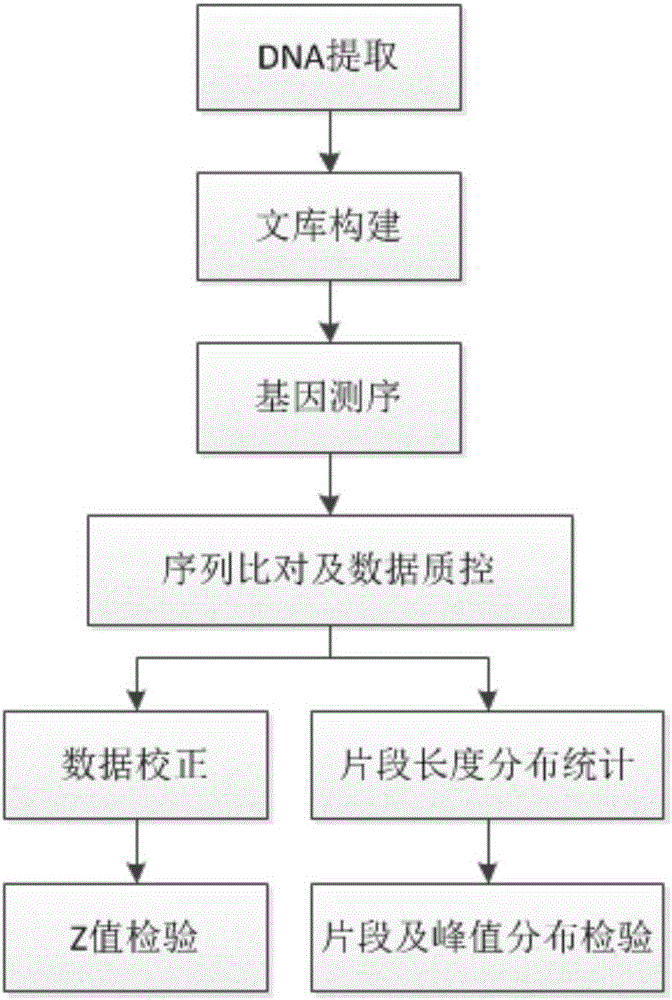
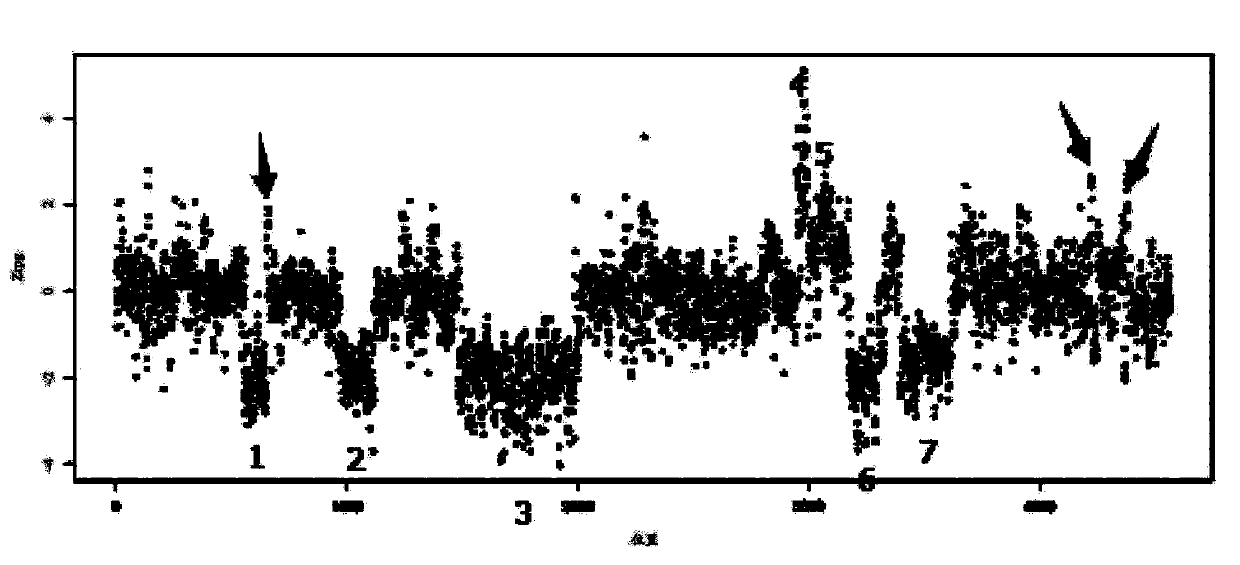
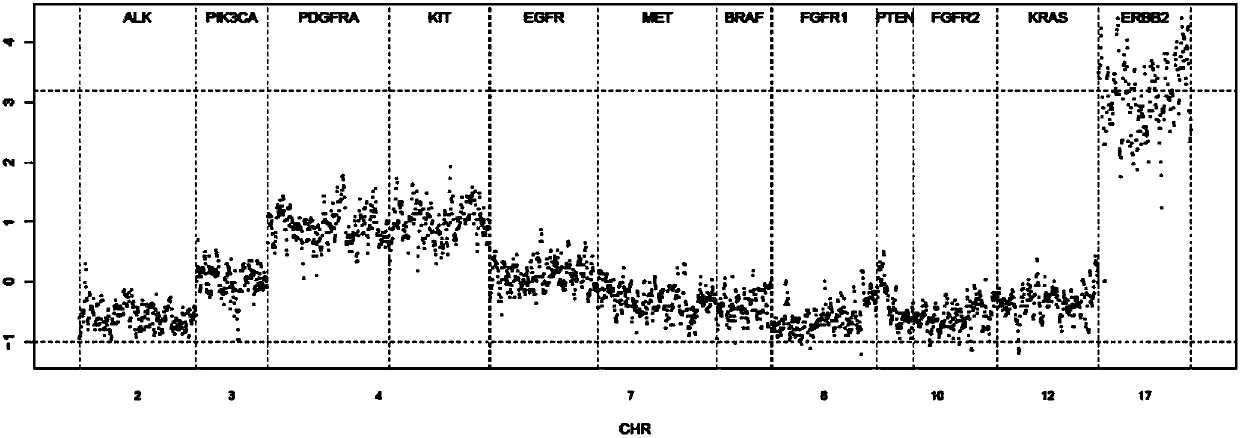
Tumor ctDNA information statistical method
ActiveCN106156543AEfficient StatisticsEasy to collect samplesBiostatisticsSequence analysisVenous bloodDNA extraction
The invention discloses a tumor ctDNA information statistical method. The tumor ctDNA information statistical method comprises the following steps: (1) DNA extraction and sequencing, (2) sequence comparison, (3) basic statistics, (4) sequencing batch correction, (5) sequence feature correction, (6) copy number variation detection, and (7) fragment difference statistics. The tumor ctDNA information statistical method takes ctDNA as a detection index, and detection can be carried out by only collecting a small amount of peripheral venous blood of a testee. By the tumor ctDNA information statistical method, it is convenient and simple to collect samples, and a detection range can be expanded to terminal patients who are not suitable for collection of samples of biopsy tissues.
Owner:AMOY DIAGNOSTICS CO LTD
Liver cancer cell specificity internalization short peptide and its in vitro screening and identification
InactiveCN101033251AStrong specificityGood effectLibrary screeningPeptidesSequence analysisWilms tumour
This invention relates to the special short peptide of hepatoma cell internalization and its screen and identification in vitro, belonging to the technology of tumor cell peptides screening and identification. It takes hepatoma cell BEL-7404 as the screening target cell, displays the random 12-peptide library for bacteria invader to conduct affinity panning. Through three times of screen, it randomly selects twenty negative colonies for amplification and sequence, to deduct the amino acid sequence of random peptide. Through cell ELISA, immunofluorescence, flow cytometry and other method, it further identifies the combination and internalization of bacteria invader clones and hepatoma cells. From sequencing and sequence analysis of the positive clones, four obtained different sequences have good combination with hepatoma cells, and the screened monoclone can internalize cells.
Owner:EAST CHINA NORMAL UNIVERSITY
Computer generation of concept sequence correction rules
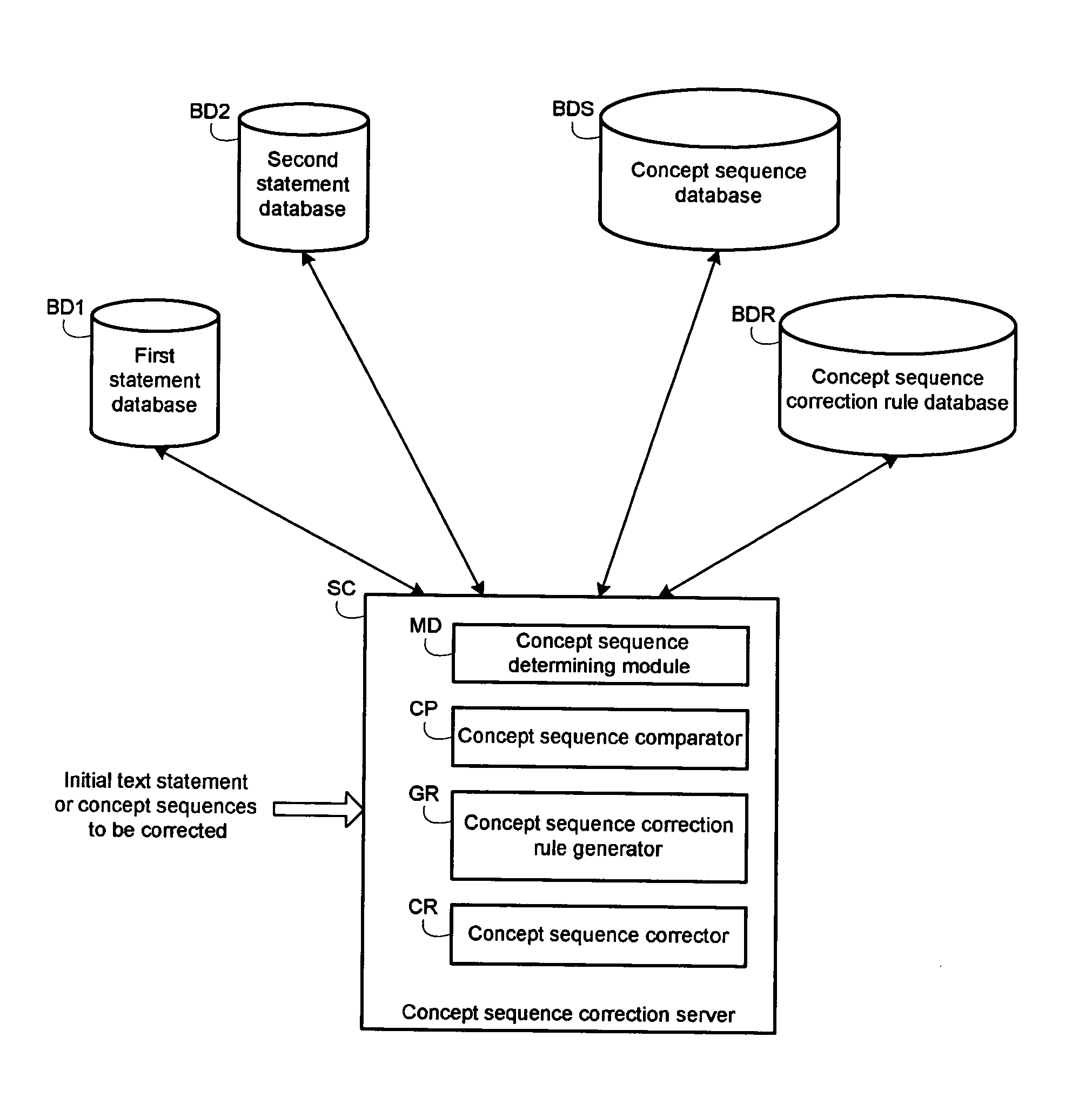
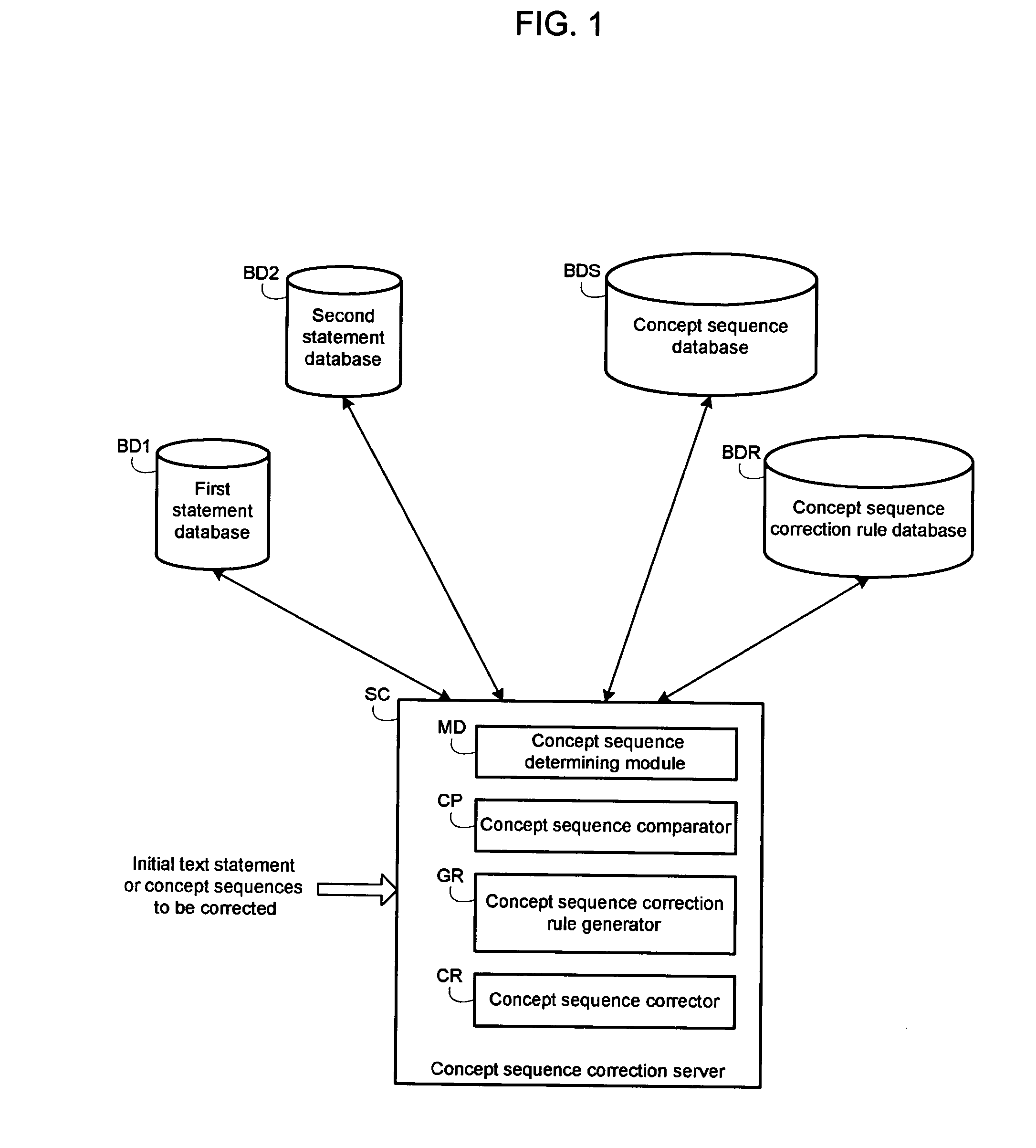
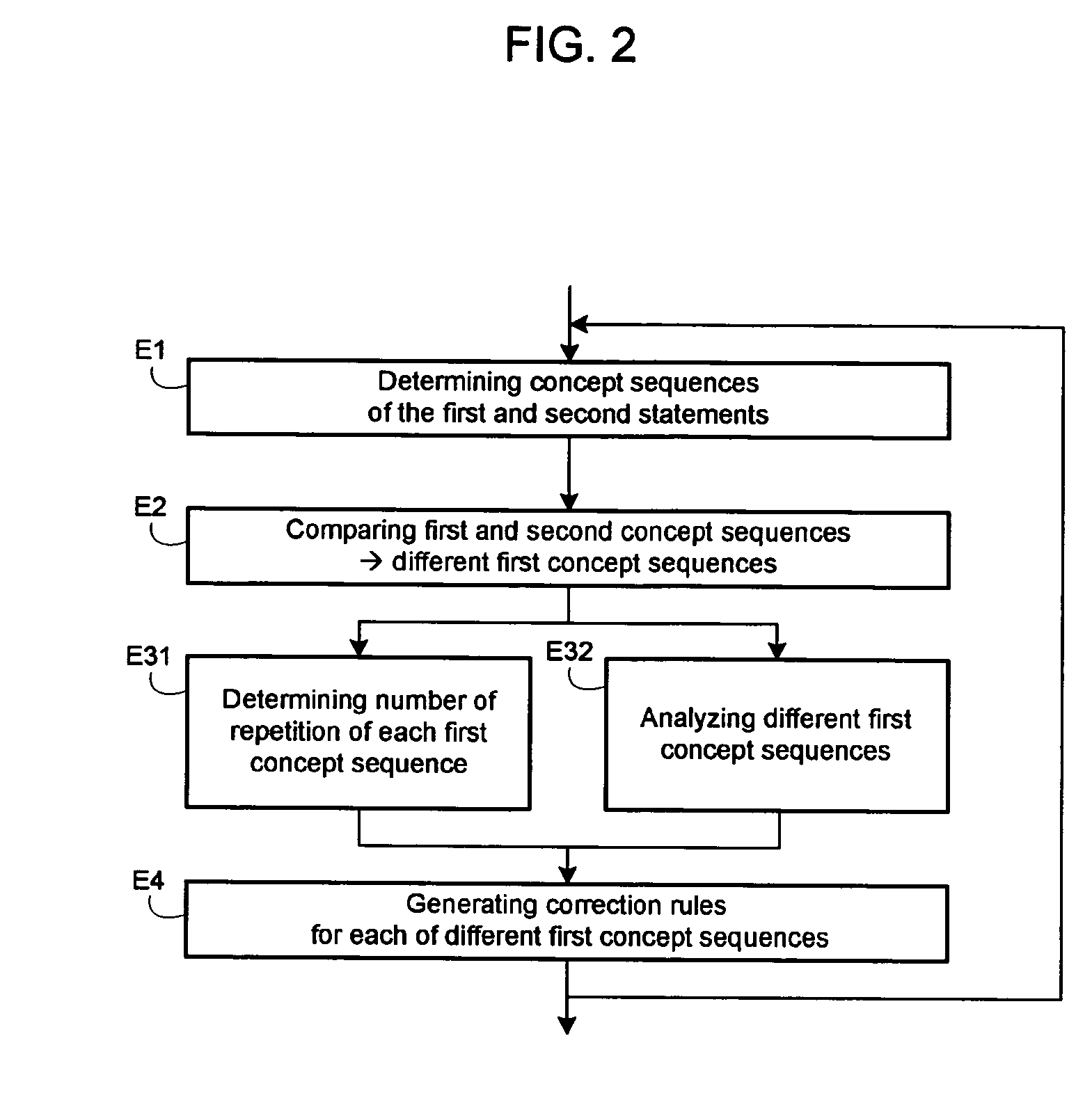
InactiveUS20060100854A1Semantic analysisSpecial data processing applicationsComparatorComputer generation
A system generates rules for the correction of a concept sequence, a concept sequence coming from a text statement. A module determines from first text statements, a set of first concept sequences liable to be corrected and from second text statements, a set of second concept sequences deemed to be valid. A comparator compares the two sets of concept sequences thereby selecting first concept sequences different from second concept sequences. A generator analyzes the selected first concept sequences and estimates at least one characteristic for each first concept sequence analyzed. The generator generates at least one concept sequence correction rule as a function of said at least one estimated characteristic.
Owner:FRANCE TELECOM SA
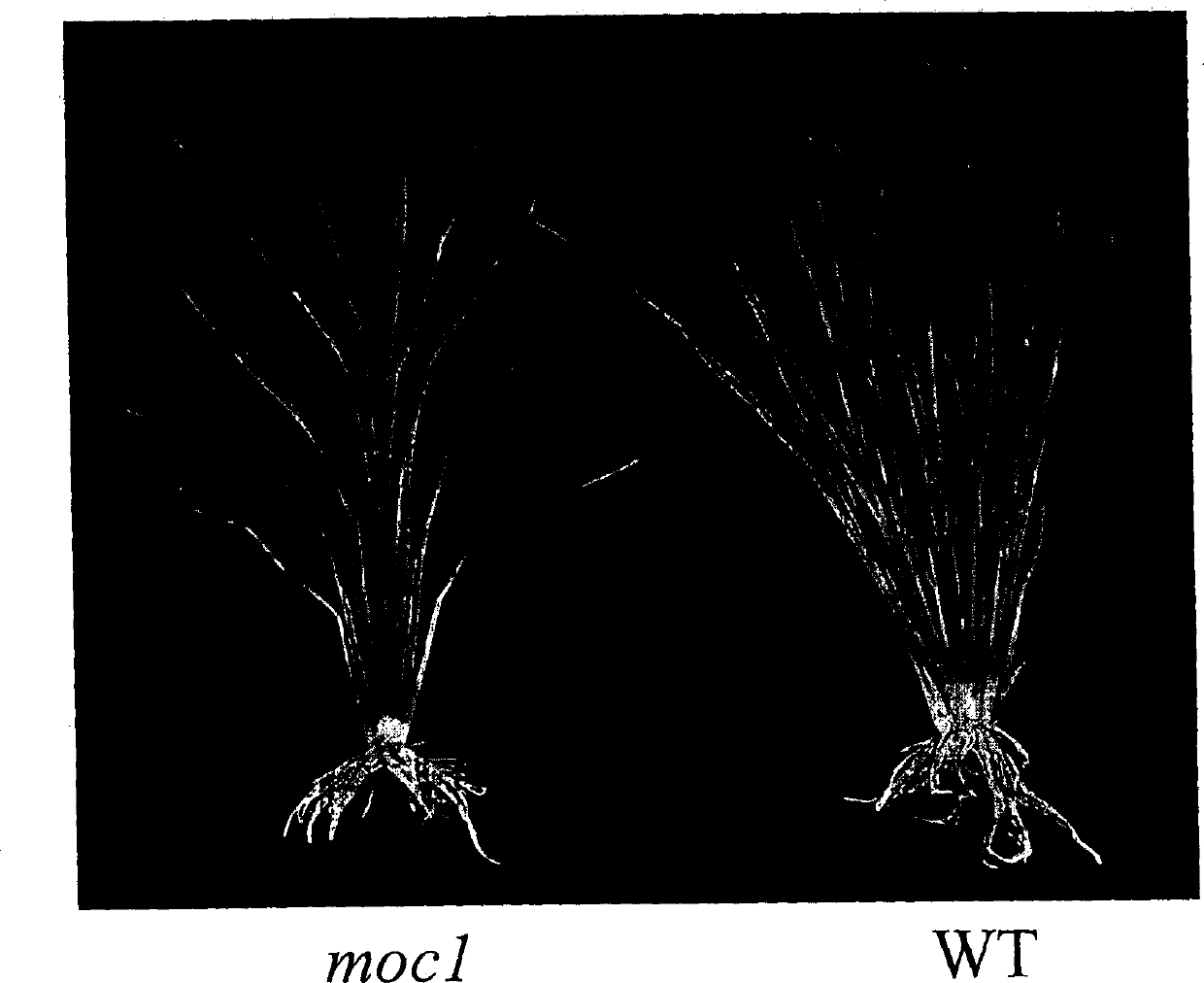
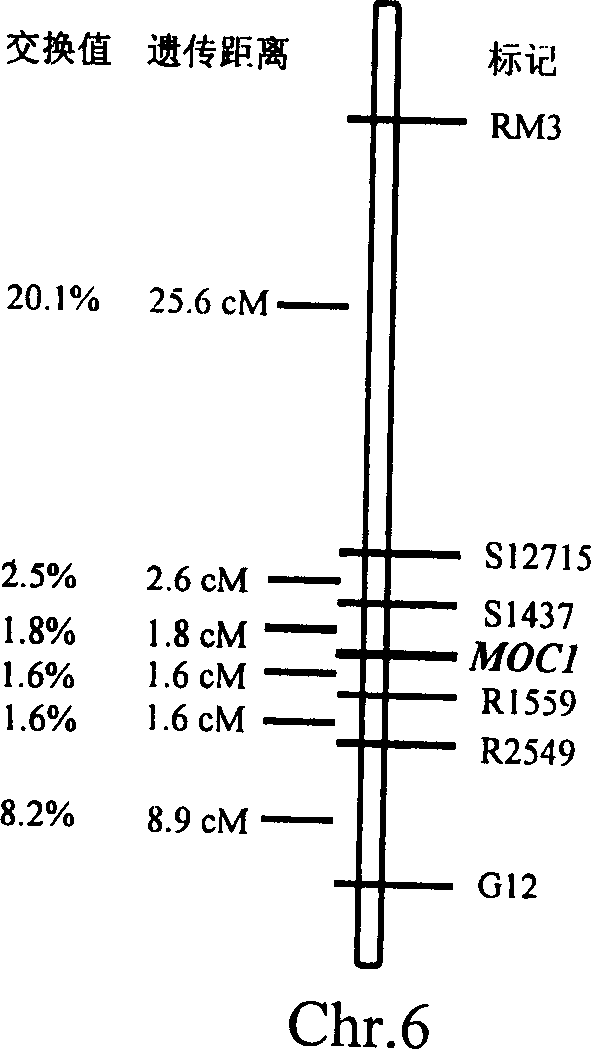
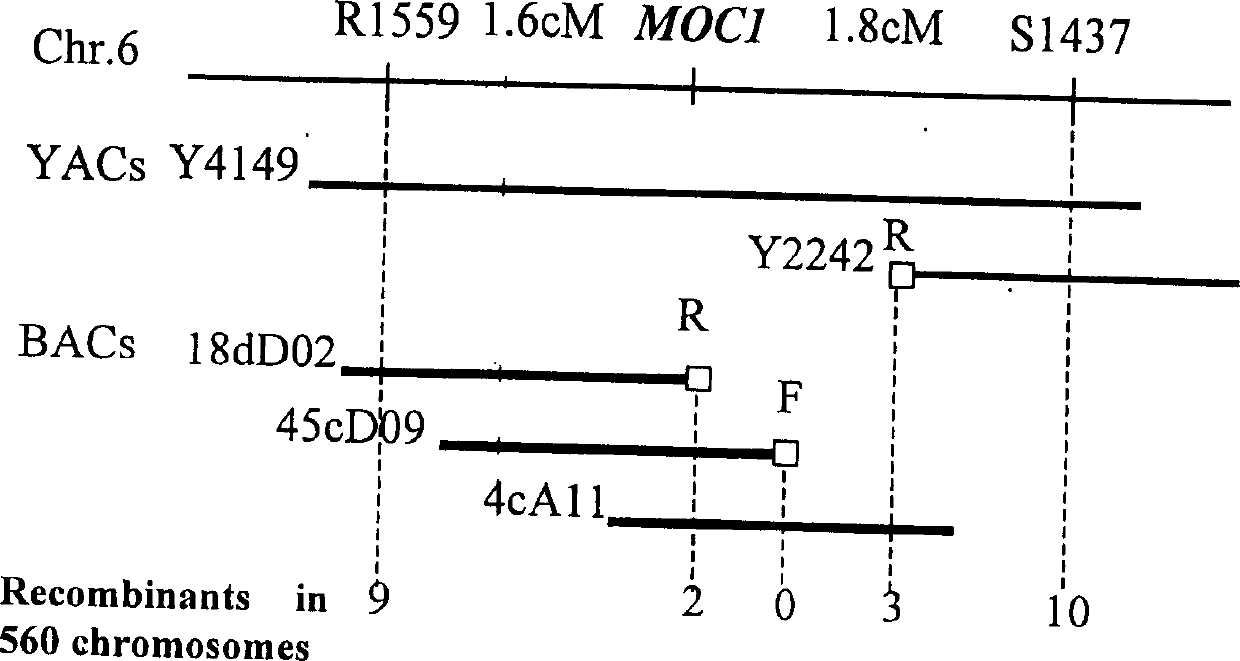
Rice tiller control gene MOC1 and its application
InactiveCN1477112AIncrease productionQuality improvementClimate change adaptationPlant peptidesGenetically modified riceAgricultural science
The present invention utilizes rice single tiller mutant Mono culm 1(moc1) to clone and identify the gene capable of controlling rice tiller, and its name is MOC1. The transgenic function complementary assay shows that MOCI is the gene for controlling rice tiller, and the amino acid sequence comparison analysis shows that said gene belongs to the GRAS transcription factor family. The gene can be used for regulating tiller character of rice.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
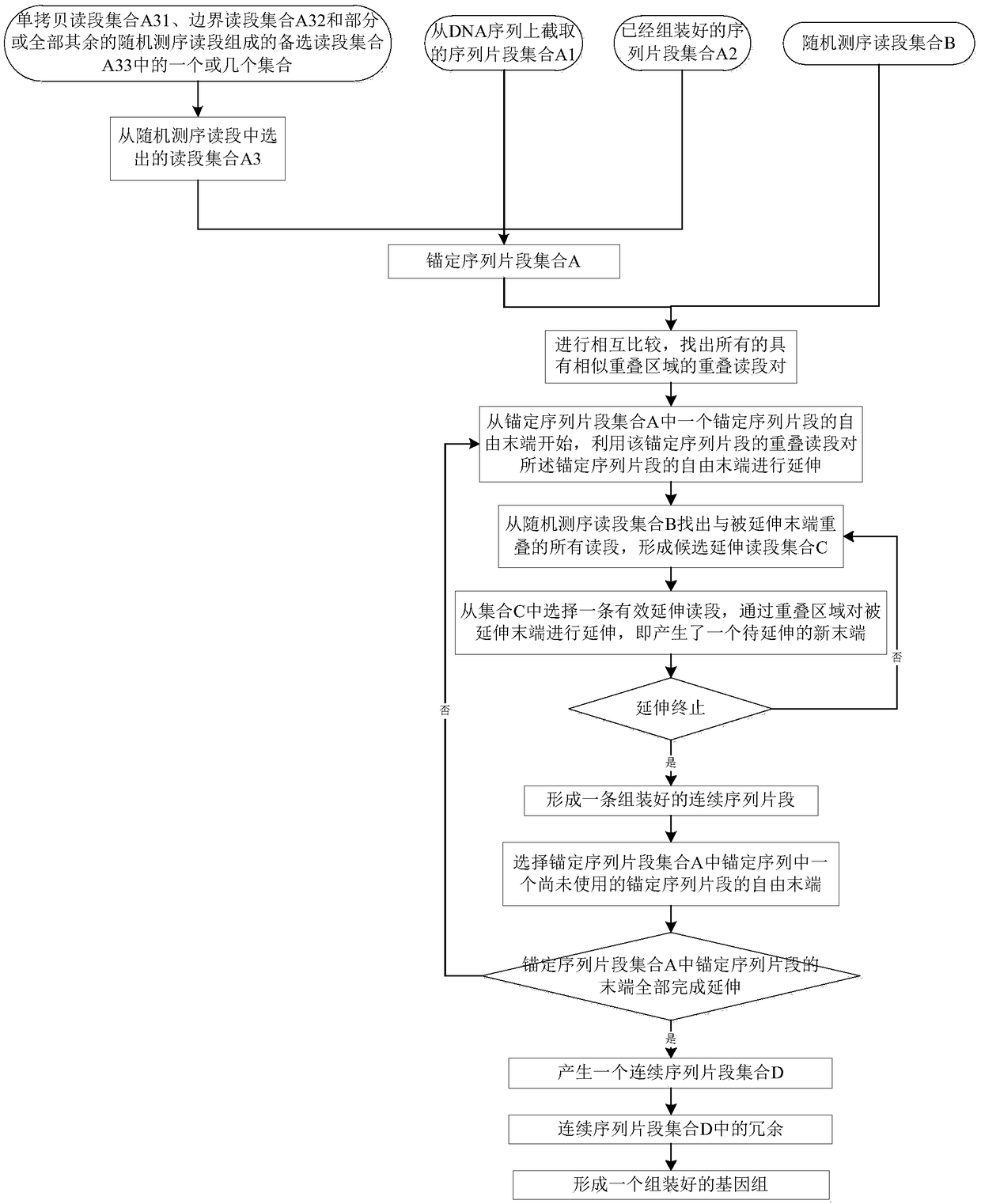
Genome assembling method
ActiveCN109234267AEfficient assemblySimplify the implementation process of assemblyDNA preparationComputational biologyChromosome
The invention discloses a genome assembling method, comprising the following four steps: sequence comparison, sequence extension, completion of extension and removal of redundancy. Genome-wide assembly is divided into two main steps: assembly of single copy sequence and assembly of remaining sequence, which simplifies the implementation process, makes the whole method fast, efficient and error-free, and greatly improves the continuity of the assembly sequence fragments and the assembly quality. By assembling the whole genome sequence with the method of the invention, the whole genome sequencecan be recovered quickly and efficiently, and the whole chromosome and the whole genome sequence can be recovered more easily. The genome assembling method of the invention can also be used for filling the sequence of the blank region in the genome sequence, in particular, the assembling effect can be greatly improved by combining the genome optical map information or the chromosome grouping sorting information. And for judging whether there is a connection between any two sequences or for estimating a distance between two adjacent sequences.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
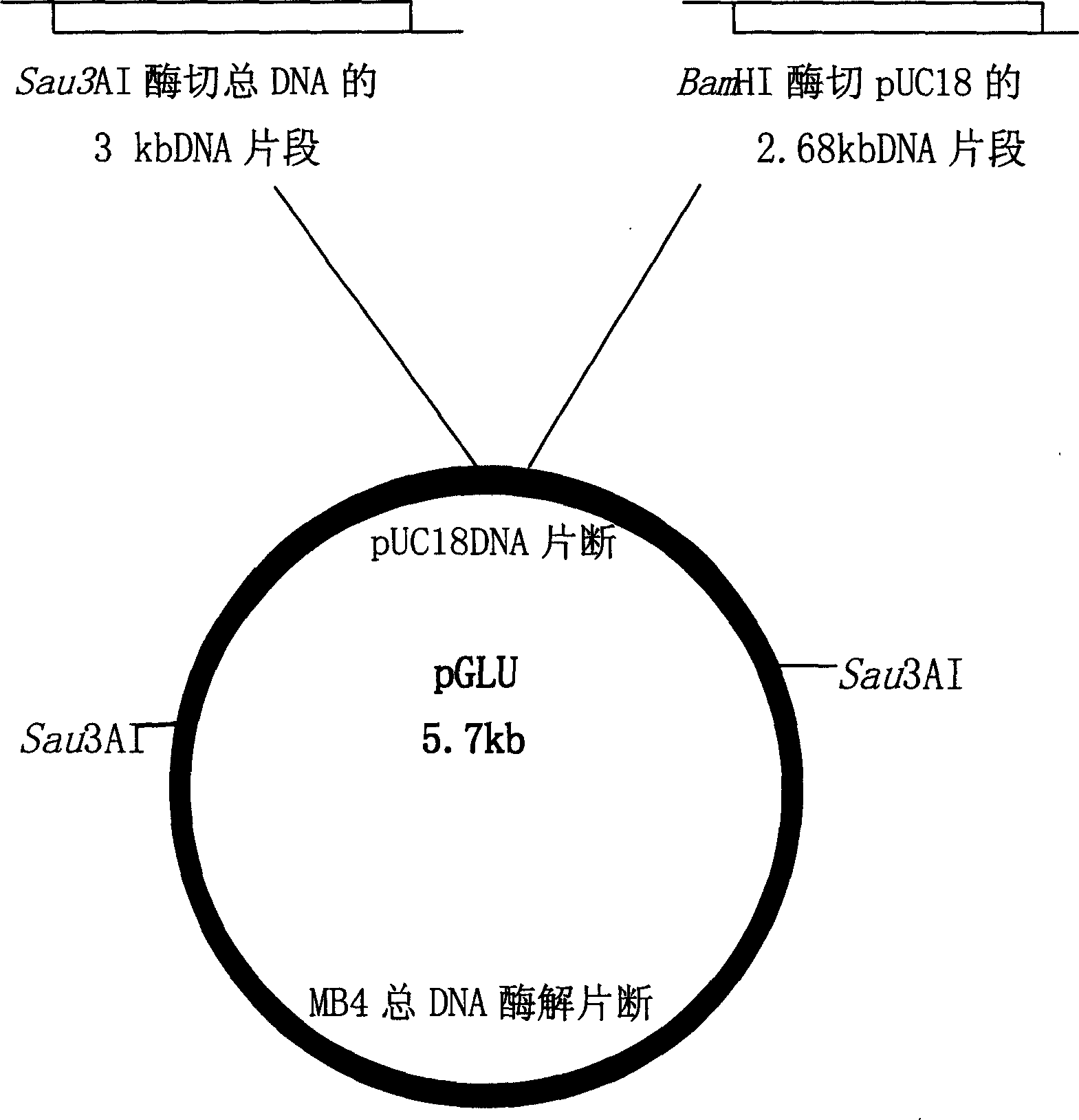
High temperature lipase, coding gene order and uses thereof
High temperature lipase gene is obtained from Thermoanaerobacter tengcongensis MB4 total DNA to constitute prokaryotic expression plasmid and transferred to colibacillus for expression. Amino acid sequence comparison shows that the lipase is one new kind of lipase.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
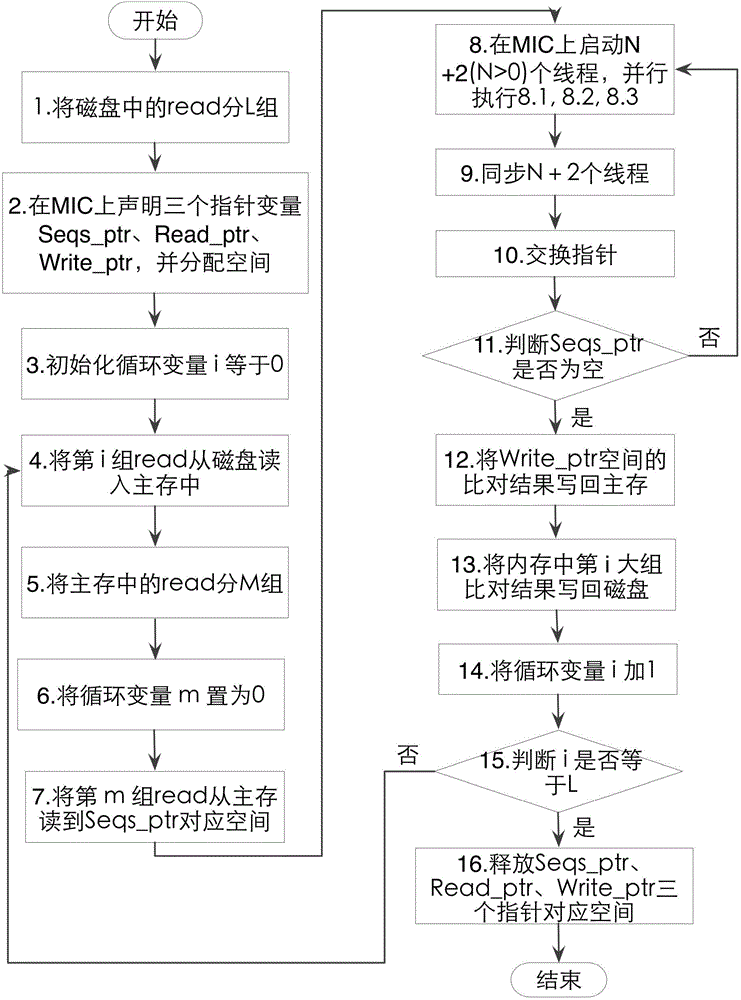
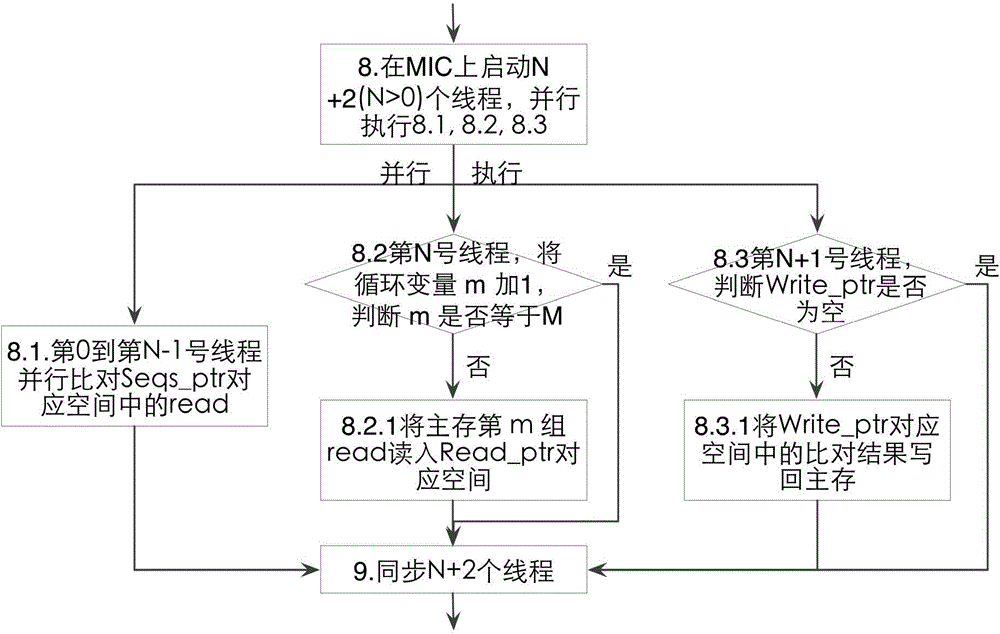
Three-level flow sequence comparison method based on many-core co-processor
ActiveCN104375807AFaster thanImprove comparison efficiencyConcurrent instruction executionThree levelMicrowave
The invention discloses a three-level flow sequence comparison method based on a many-core co-processor. The purpose of increasing the comparison speed of sequence comparison software is achieved. According to the technical scheme, sequence comparison is performed in a multi-threading manner by an MIC (microwave integrated circuit) many-core co-processor; three serial steps of reading sequences from a main memory to an MIC, comparing the sequences and writing a comparison result to the main memory in a sequence comparison process of the MIC are in a three-level flow mode, namely, sequences required for next comparison are read during sequence comparison, a previous comparison result is written into the main memory, and a reading and writing operation and a comparison operation are carried out simultaneously. By the three-level flow sequence comparison method based on the many-core co-processor, three main processes of sequence reading, sequence comparison and comparison result returning are carried out simultaneously, the comparison efficiency is improved, and the comparison time is shortened. Compared with a two-channel eight-core CPU (central processing unit), the three-level flow sequence comparison method has the advantages that the speed of a comparison process can be increased 2.3 times at least, a large amount of memory space is prevented from being copied, and the space-time efficiency of a procedure is improved.
Owner:NAT UNIV OF DEFENSE TECH
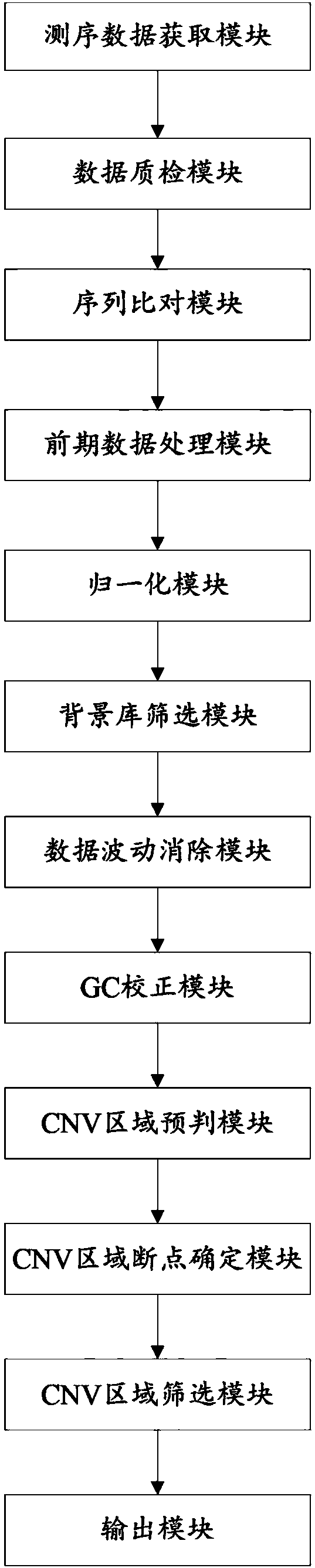
Copy number variation detection apparatus
The invention relates to a copy number variation detection apparatus, which is high in detection sensitivity. The copy number variation detection apparatus comprises a sequence comparison module, an early stage data processing module, a normalization module, a background library screening module, a data fluctuation elimination module, a GC correction module, a CNV region prejudgment module, a CNVregion breakpoint determination module, a CNV region screening module and an output module.
Owner:ZHEJIANG ANNOROAD BIO TECH CO LTD +2
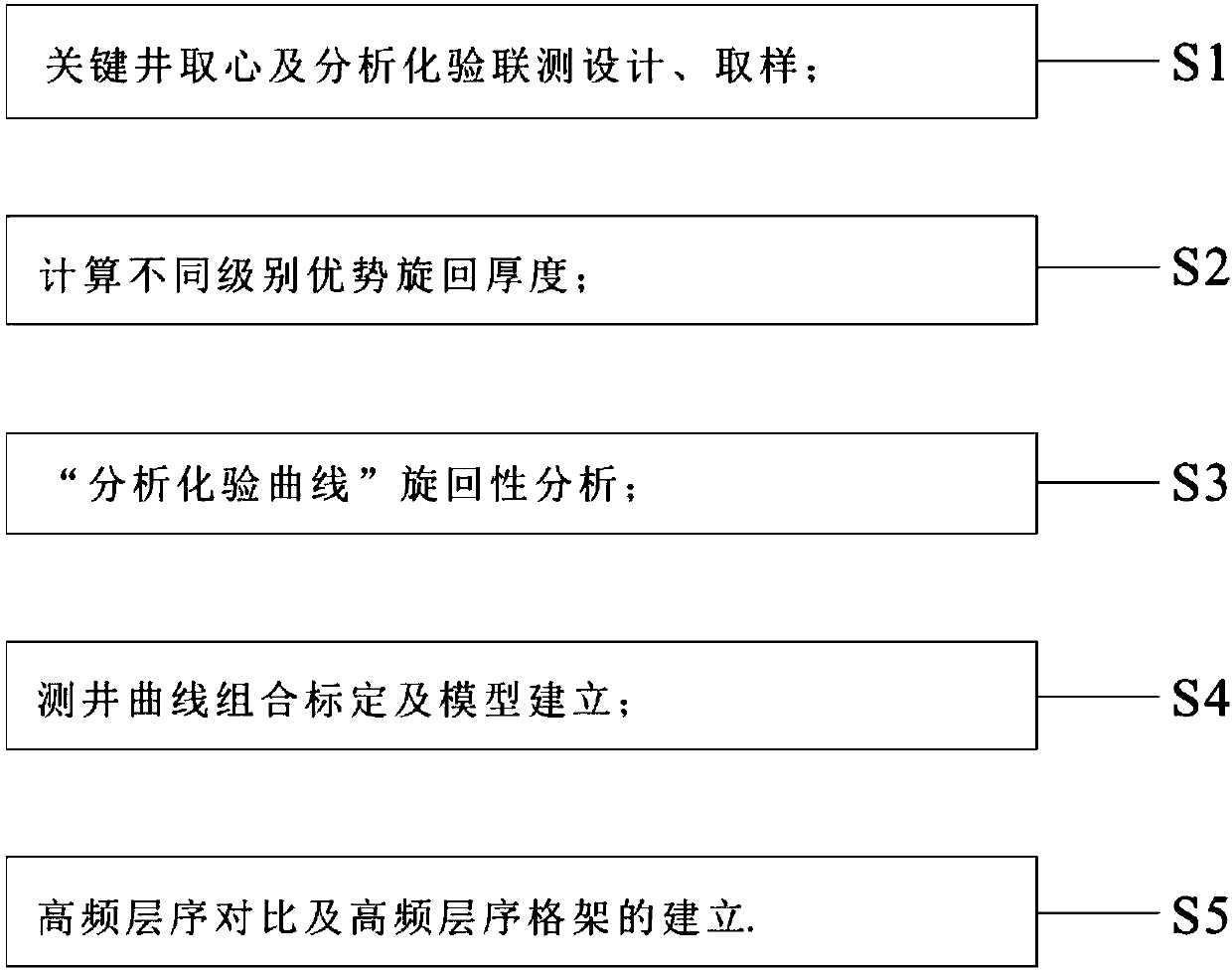
High-resolution sequence stratigraphic division and comparison method
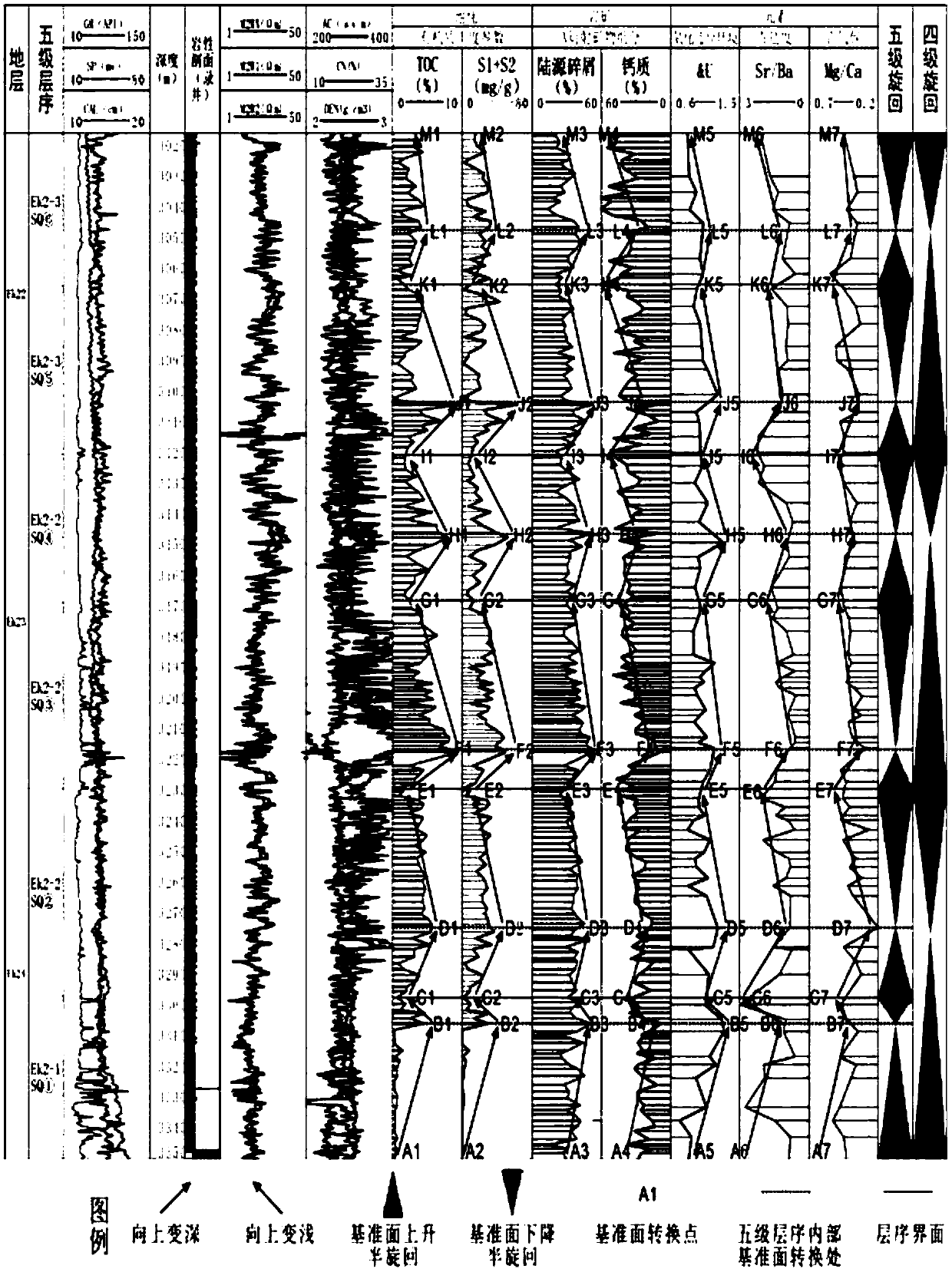
InactiveCN107688206AImprove research precisionMeet the needs of fine explorationGeological measurementsLaboratory testCoring
The invention discloses a high-resolution sequence stratigraphic division and comparison method. The high-resolution sequence stratigraphic division and comparison method comprises the following steps: S1, designing combine detection of key well coring and an analysis laboratory test, and sampling; S2, calculating superior cycle thicknesses with different grades; S3, carrying out analysis laboratory test curve type cyclic analysis; S4, carrying out logging curve combined calibration and model establishment; S5, carrying out high-frequency sequence comparison and establishing a high-frequency sequence framework. The high-resolution sequence stratigraphic division and comparison method disclosed by the invention has the beneficial effects that a practicable technical system is provided for exploration and development of oil gas of a fine-grain phase region, especially longitudinal and transverse spreading ranges of a sweet spot and a distribution rule thereof, preference of a well site and the like. The high-resolution sequence stratigraphic division and comparison method has high research precision of sequence identification and division and can be used for relatively accurately carrying out identification and division on five-grade, six-grade, even seven-grade sequences, so that requirements on current refined exploration are met; however, a method in the prior art only can berelatively accurately applied to division of low-frequency sequences of a conventional coarse grain phase belt.
Owner:DAGANG OIL FIELD OF CNPC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com