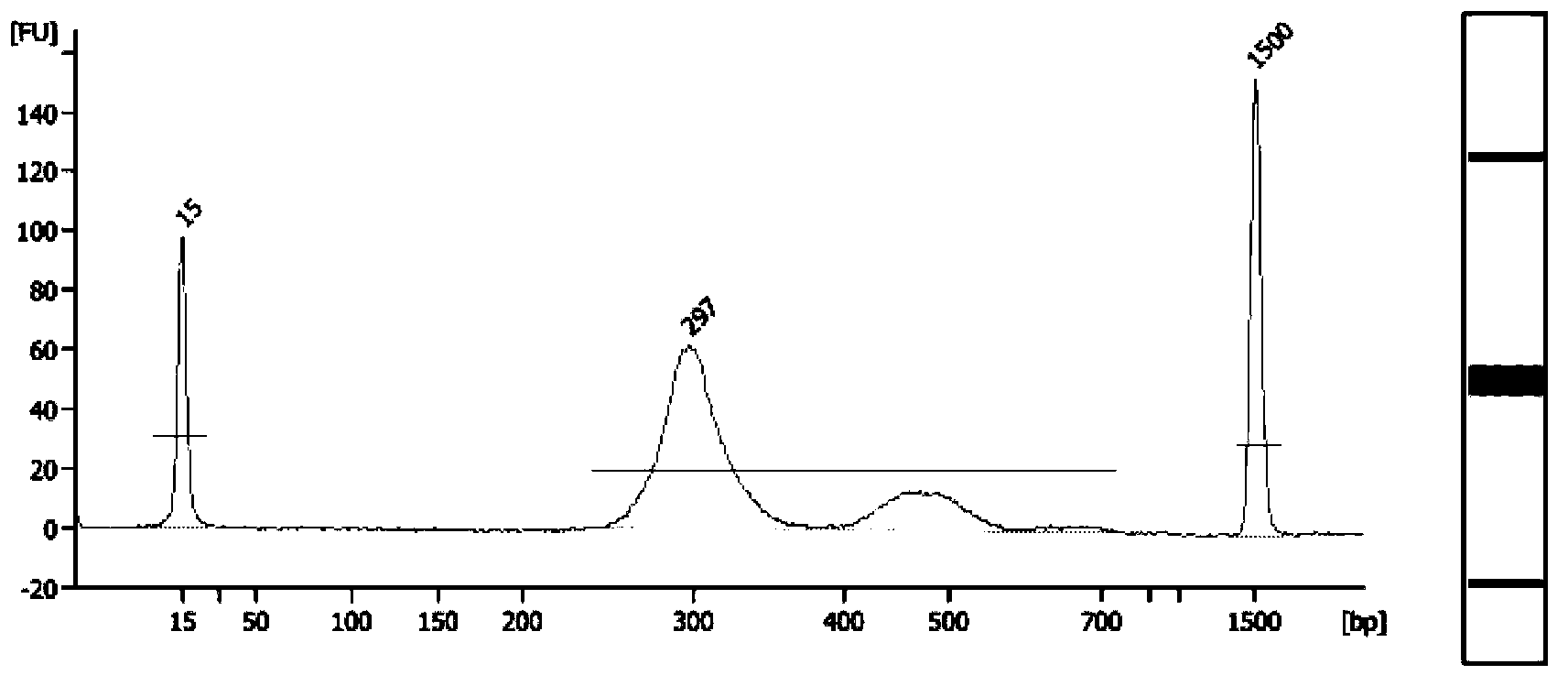
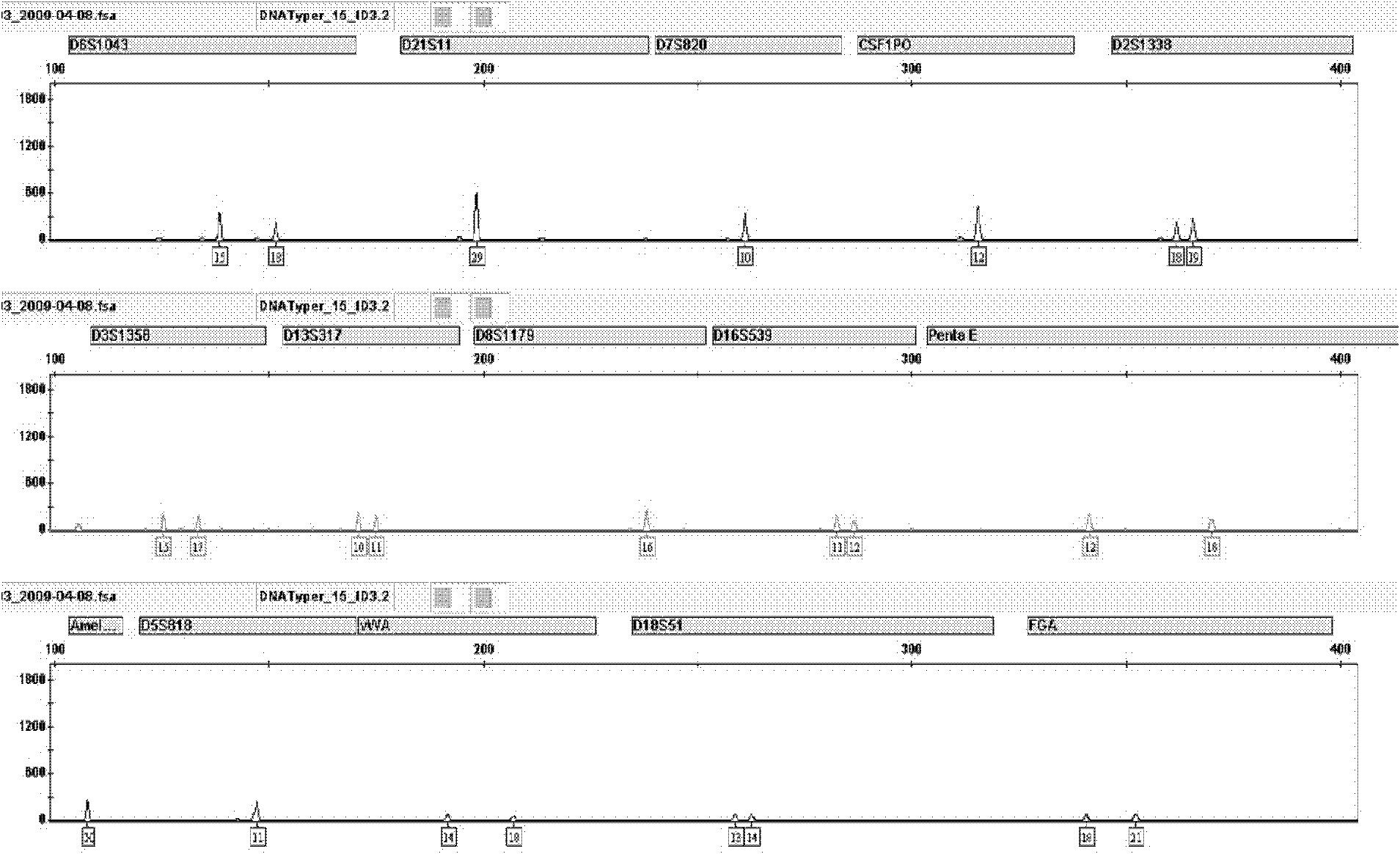
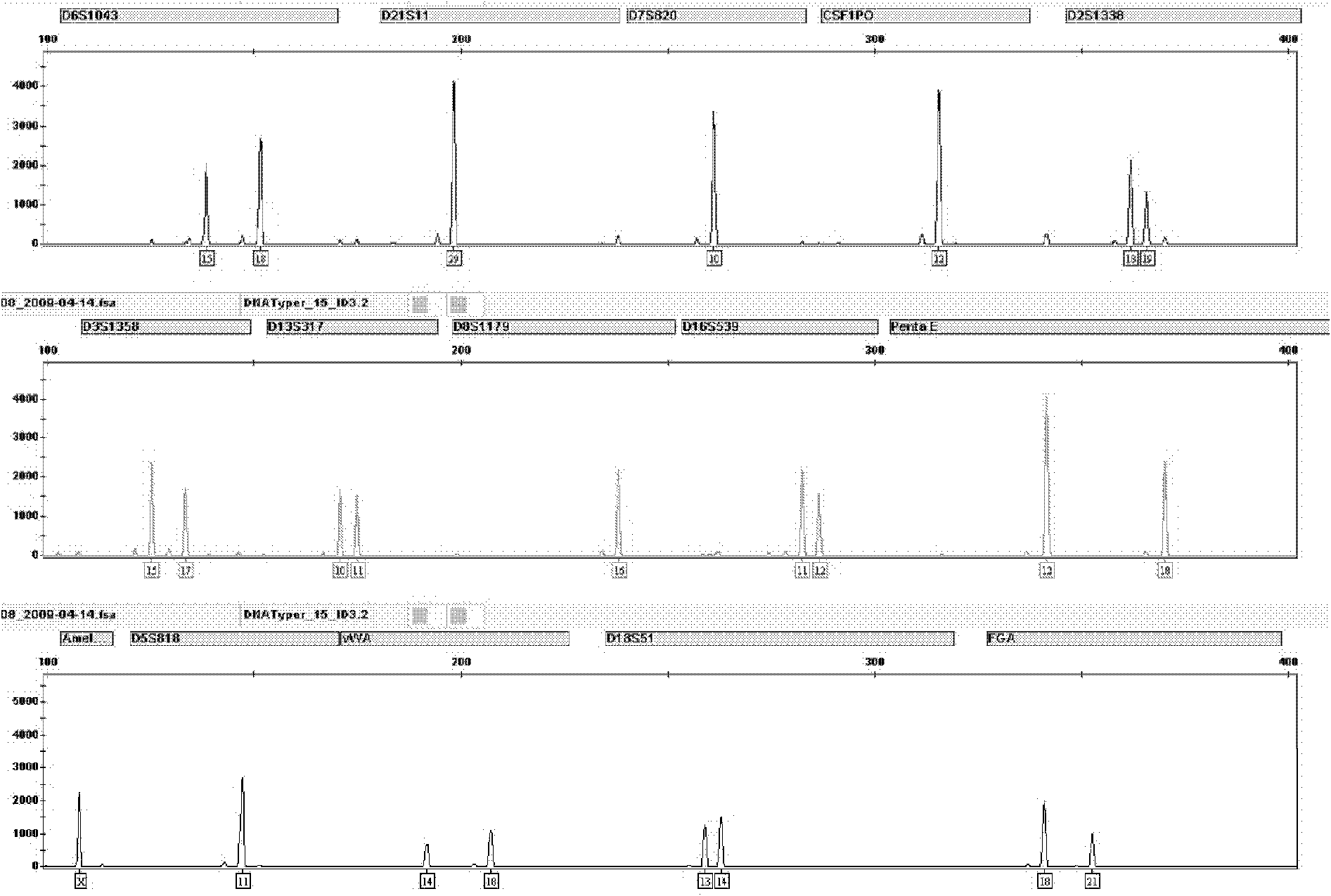
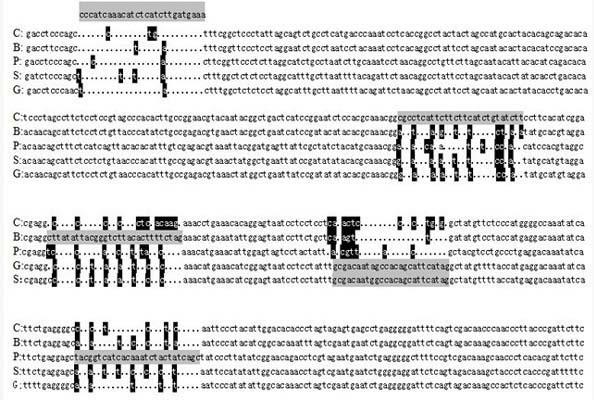
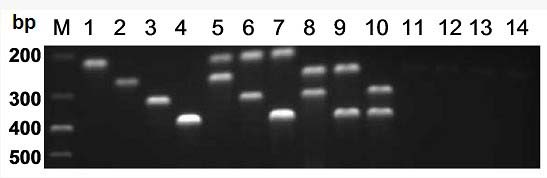
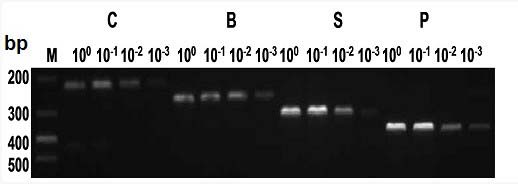
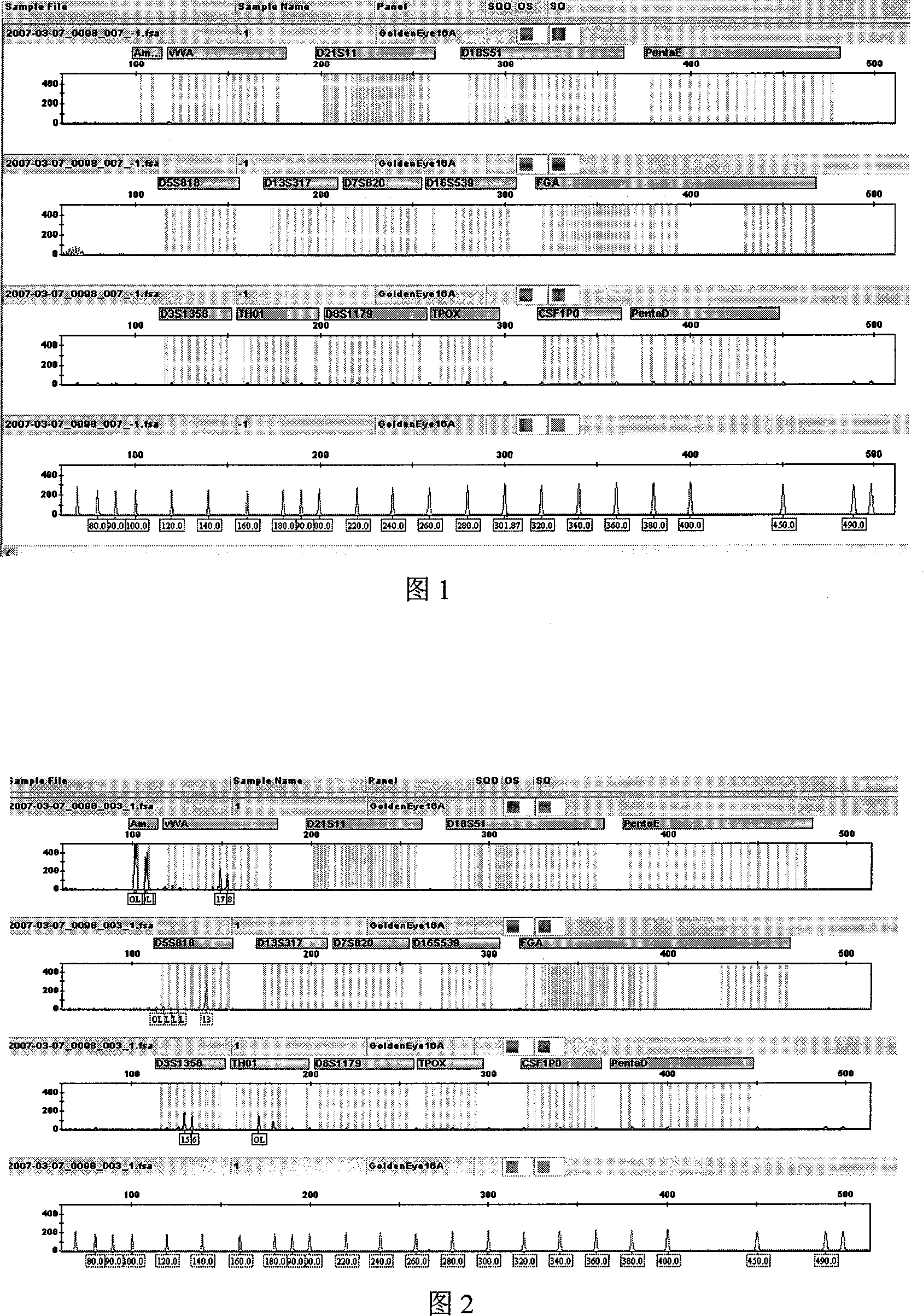
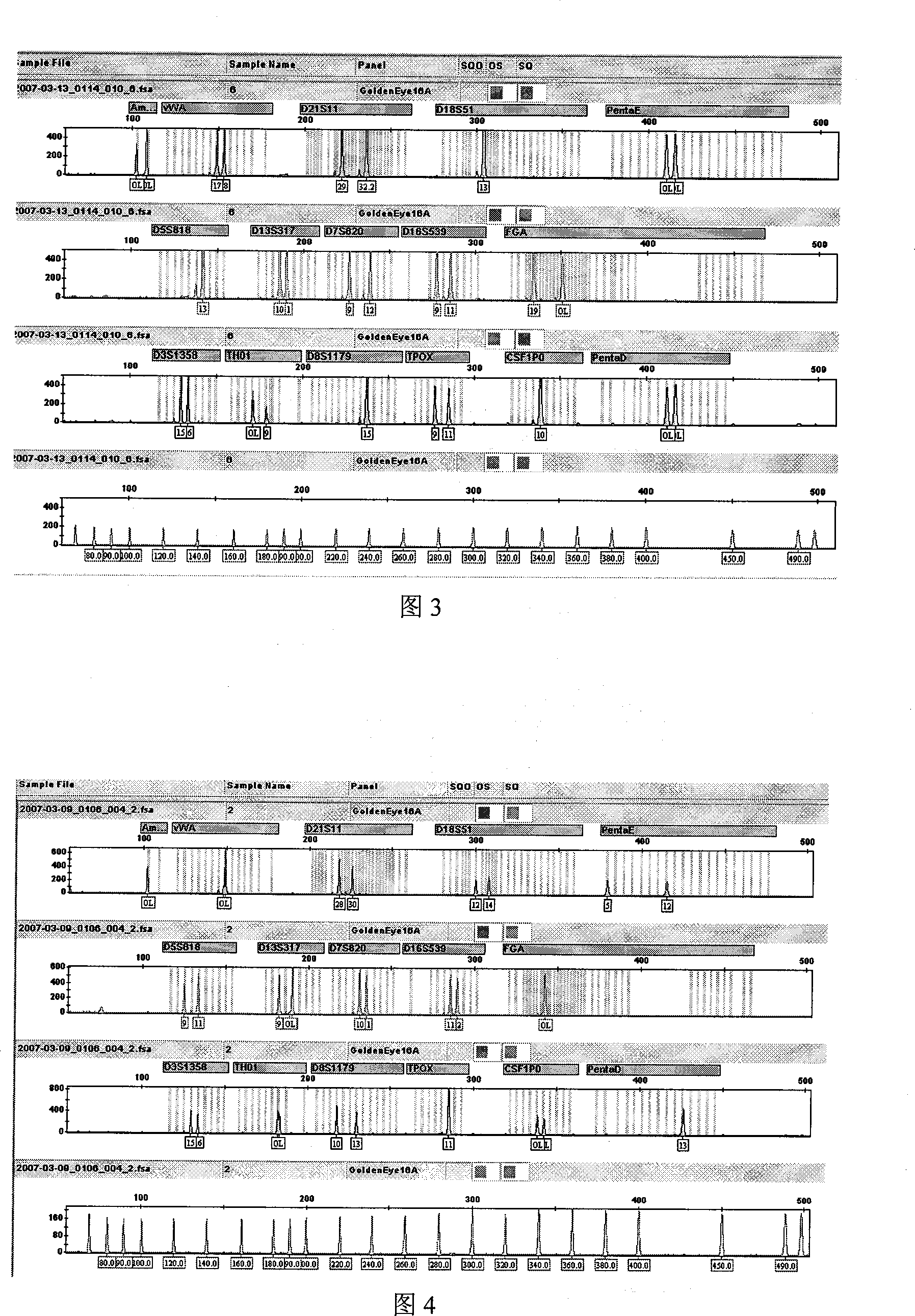
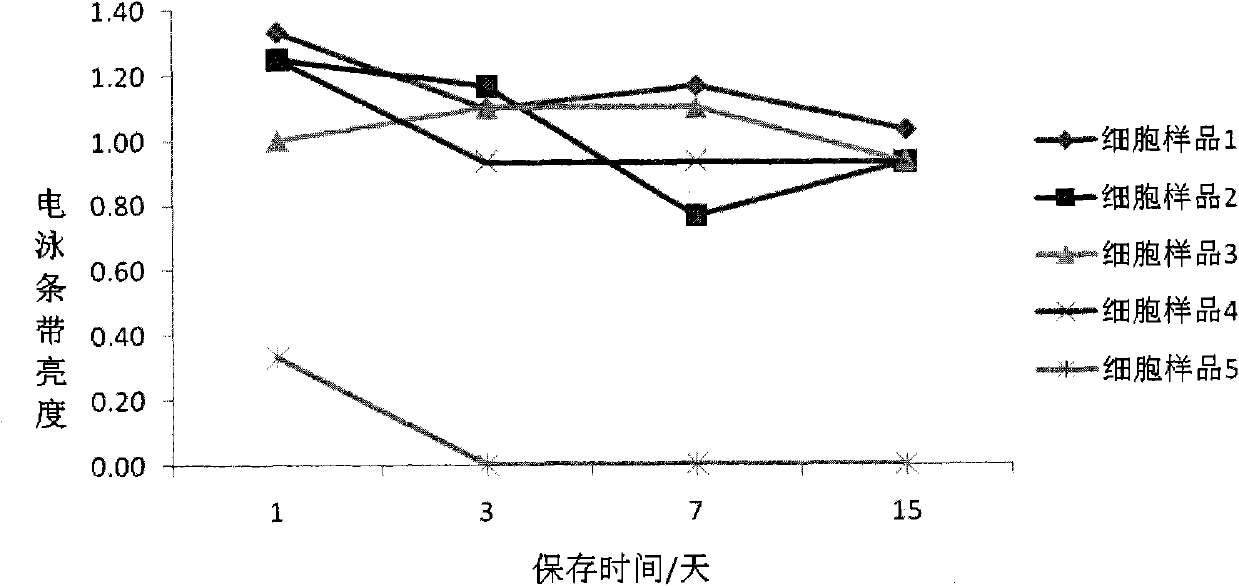
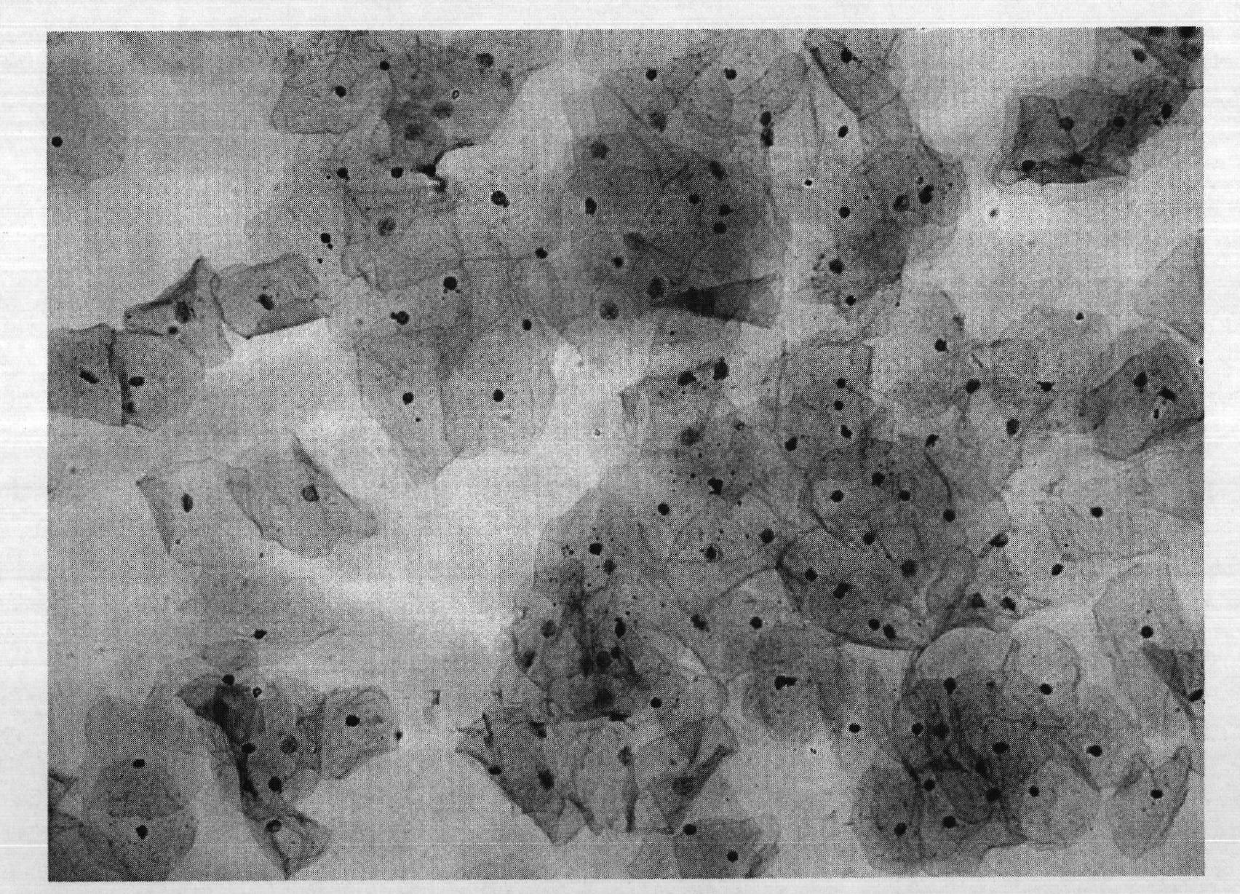
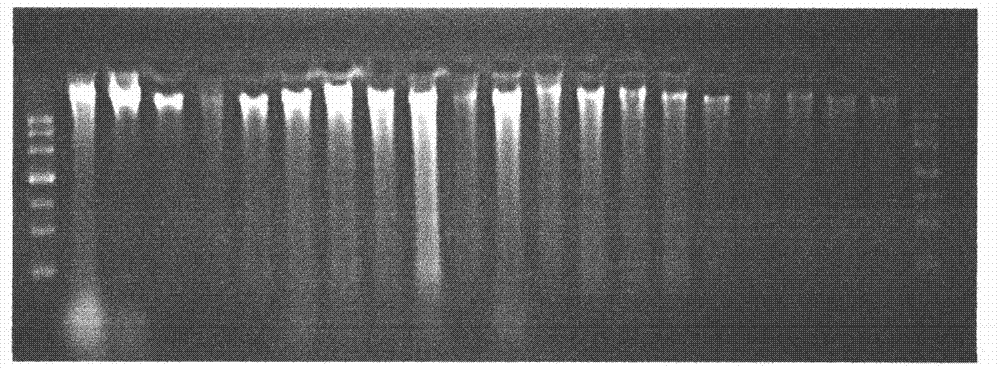
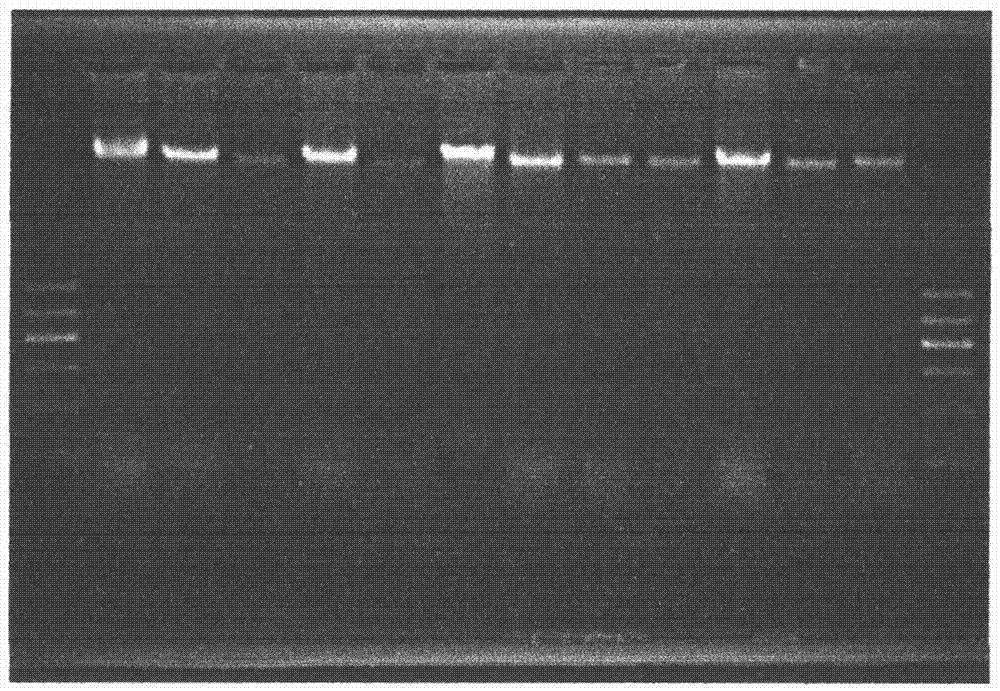
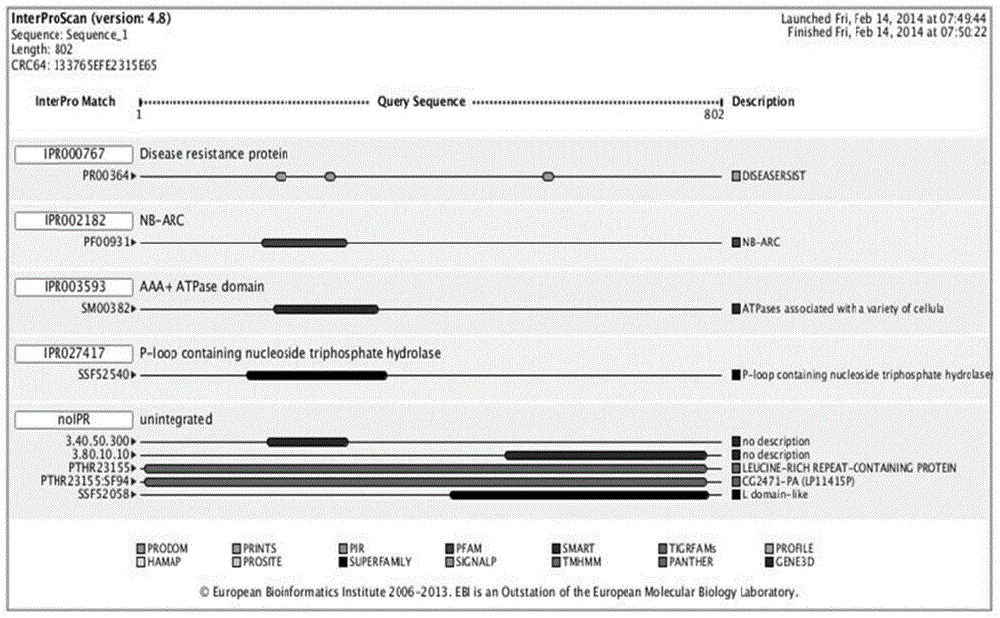
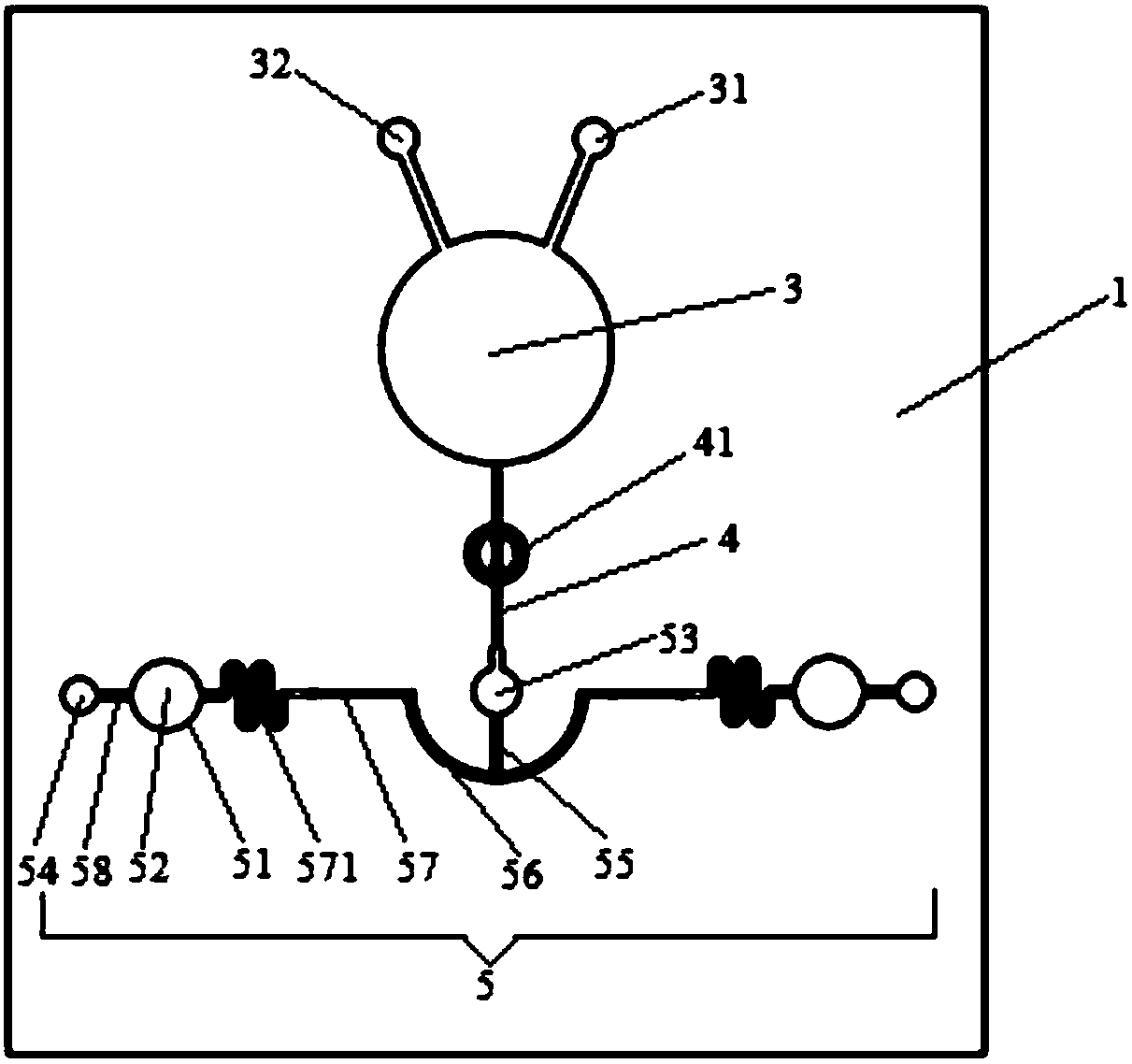
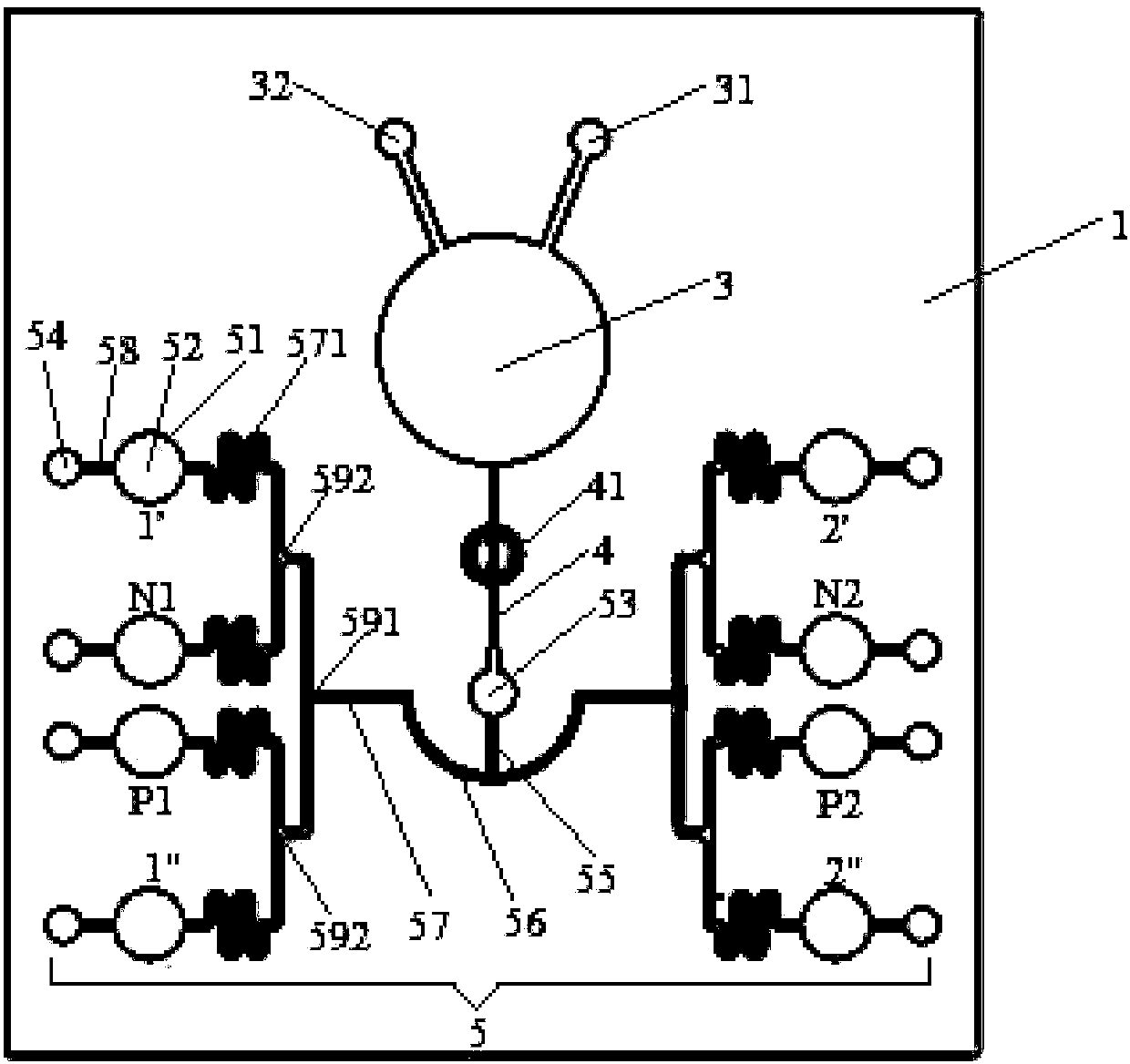
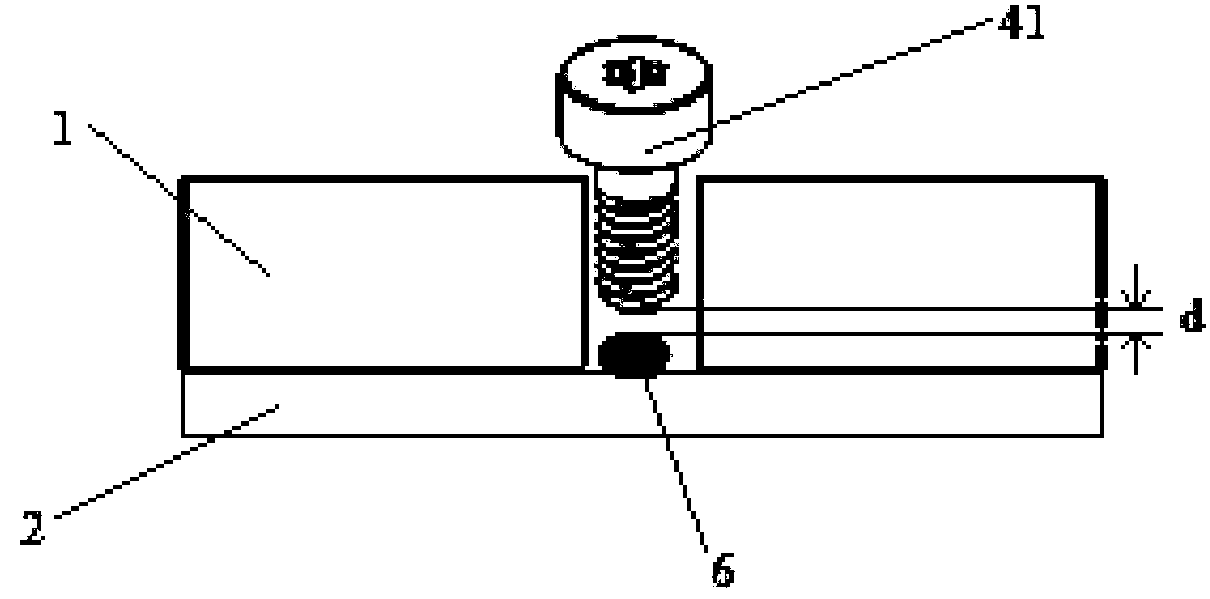
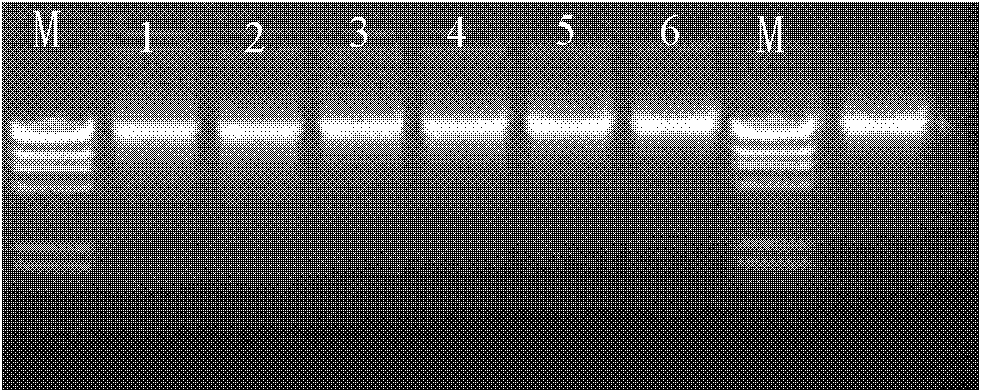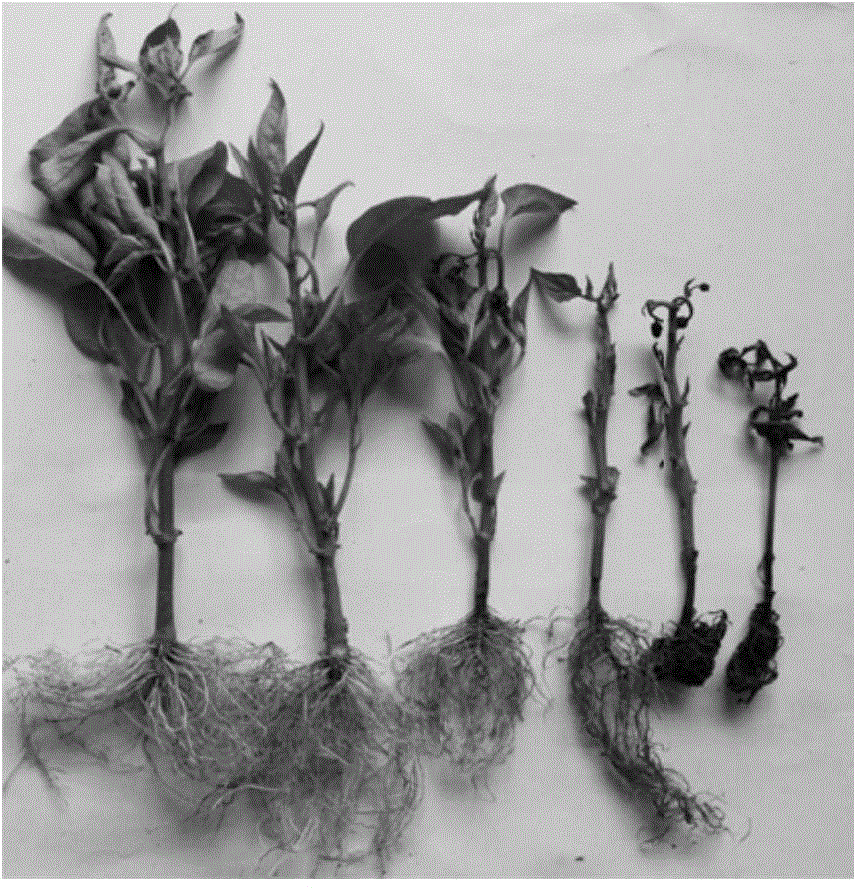Patents
Literature
2226 results about "DNA extraction" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA isolation is a process of purification of DNA from sample using a combination of physical and chemical methods. The first isolation of DNA was done in 1869 by Friedrich Miescher. Currently it is a routine procedure in molecular biology or forensic analyses. For the chemical method, there are many different kits used for extraction, and selecting the correct one will save time on kit optimization and extraction procedures. PCR sensitivity detection is considered to show the variation between the commercial kits.
Methods for the isolation of nucleic acids and for quantitative DNA extraction and detection for leukocyte evaluation in blood products
InactiveUS6958392B2Sugar derivativesMicrobiological testing/measurementWhole blood productWhite blood cell
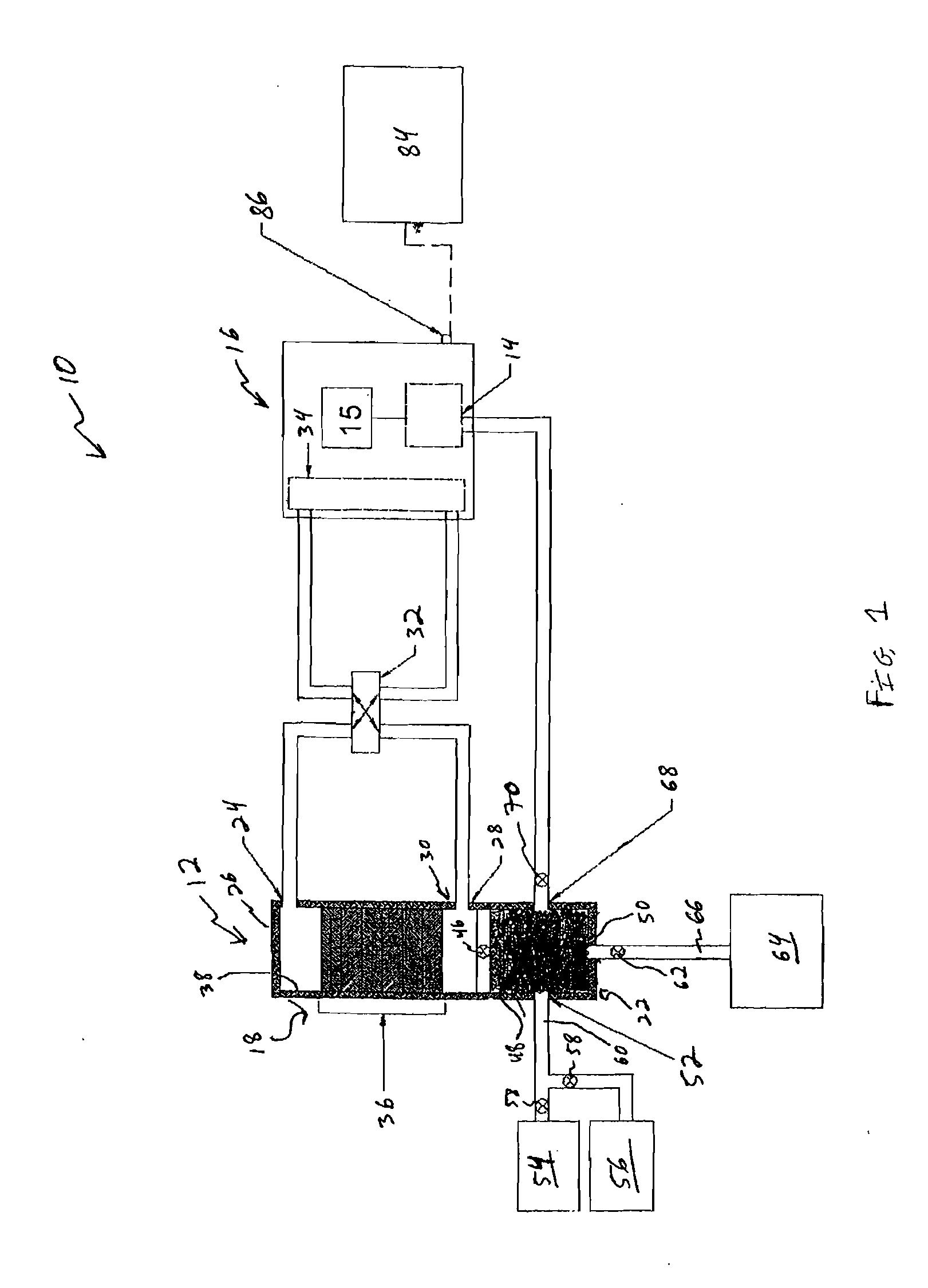
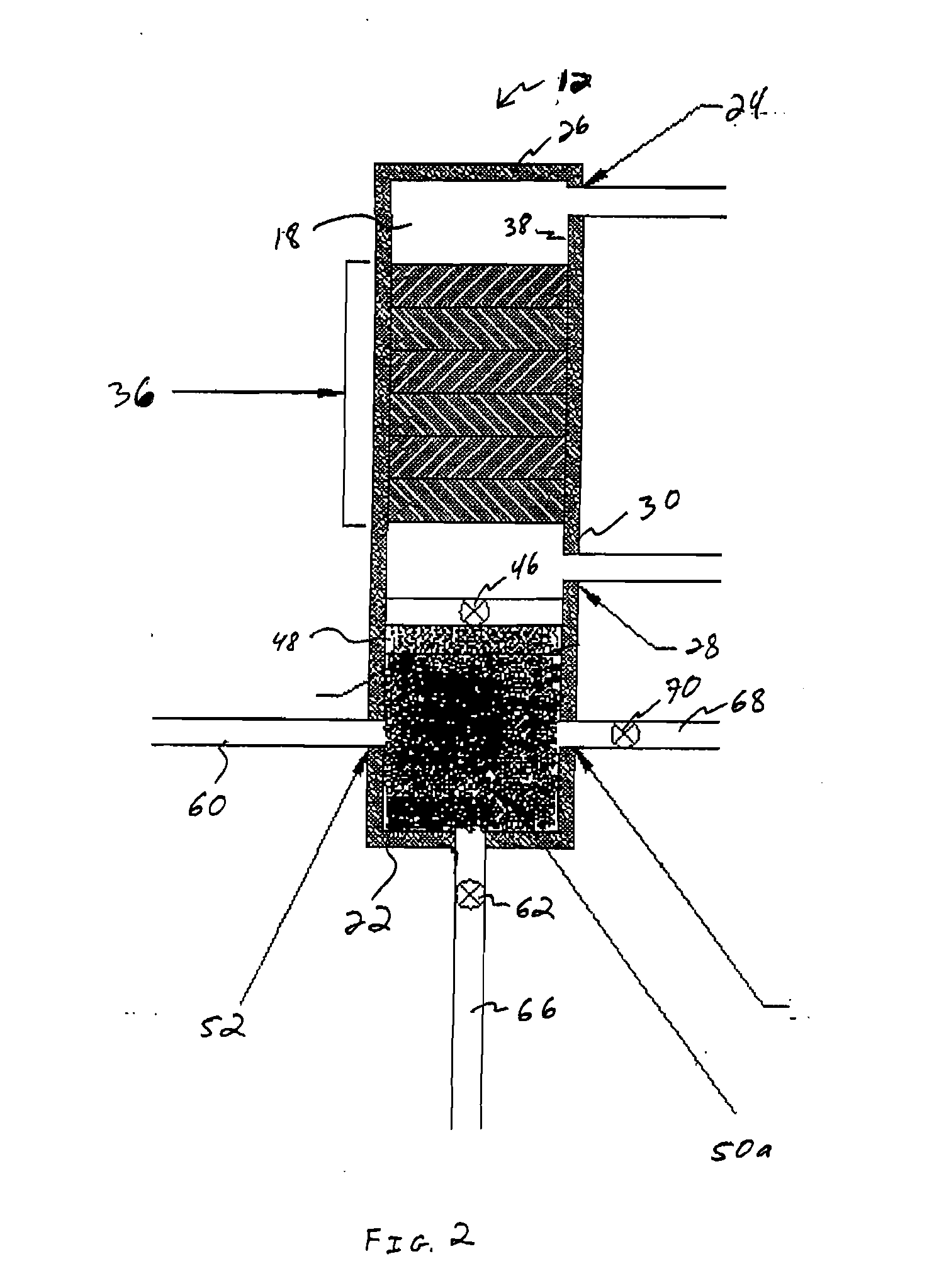
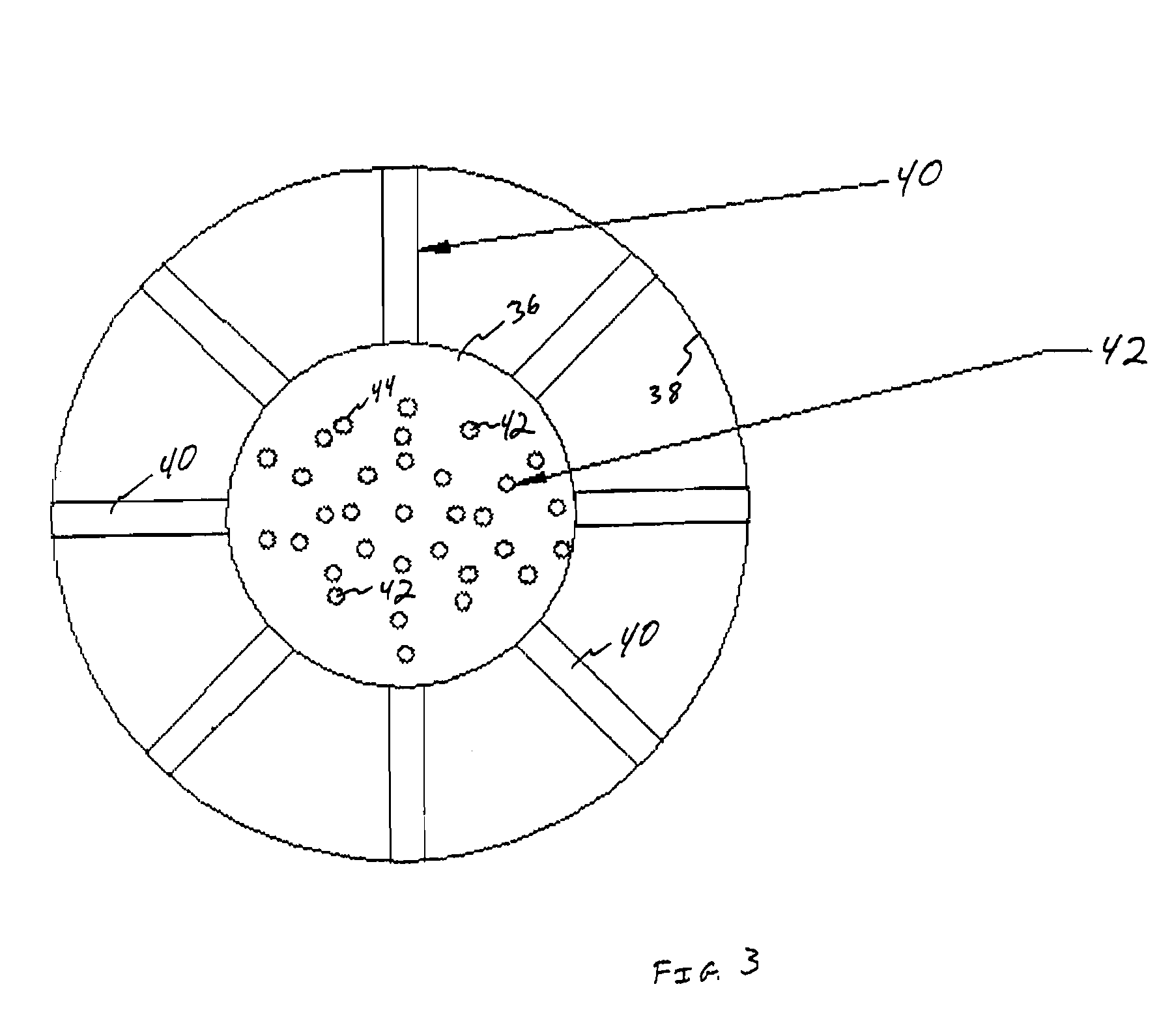
A method for isolating nucleic acid which comprises:(a) applying a sample comprising cells containing nucleic acid to a filter, whereby the cells are retained as a retentate and contaminants are removed;(b) lysing the retentate from step (a) while the retentate is retained by the filter to form a cell lysate containing the nucleic acid;(c) filtering the cell lysate with the filter to retain the nucleic acid and remove remaining cell lysate;(d) optionally washing the nucleic acid retained by the filter; and(e) eluting the nucleic acid, wherein the filter composition and dimensions are selected so that the filter is capable of retaining the cells and the nucleic acid.Additionally, there is provided a substrate for lysing cells and purifying nucleic acid having a matrix and a coating and an integrity maintainer for maintaining the purified nucleic acid. Also provided is a method of purifying nucleic acid by applying a nucleic acid sample to a substrate having an anionic detergent affixed to a matrix, the substrate physically capturing the nucleic acid, bonding the nucleic acid to a substrate and generating a signal when the nucleic acid bonds to the substrate indicating the presence of the nucleic acid. A kit for purifying nucleic acid containing a coated matrix and an integrity maintenance provider for preserving the matrix and purifying nucleic acid is also provided. Further, there is provided a method for quantifying DNA, such as double-stranded or genomic DNA, isolated from cells, such as leukocytes to determine the numbers of leukocytes in a sample of leukoreduced blood.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
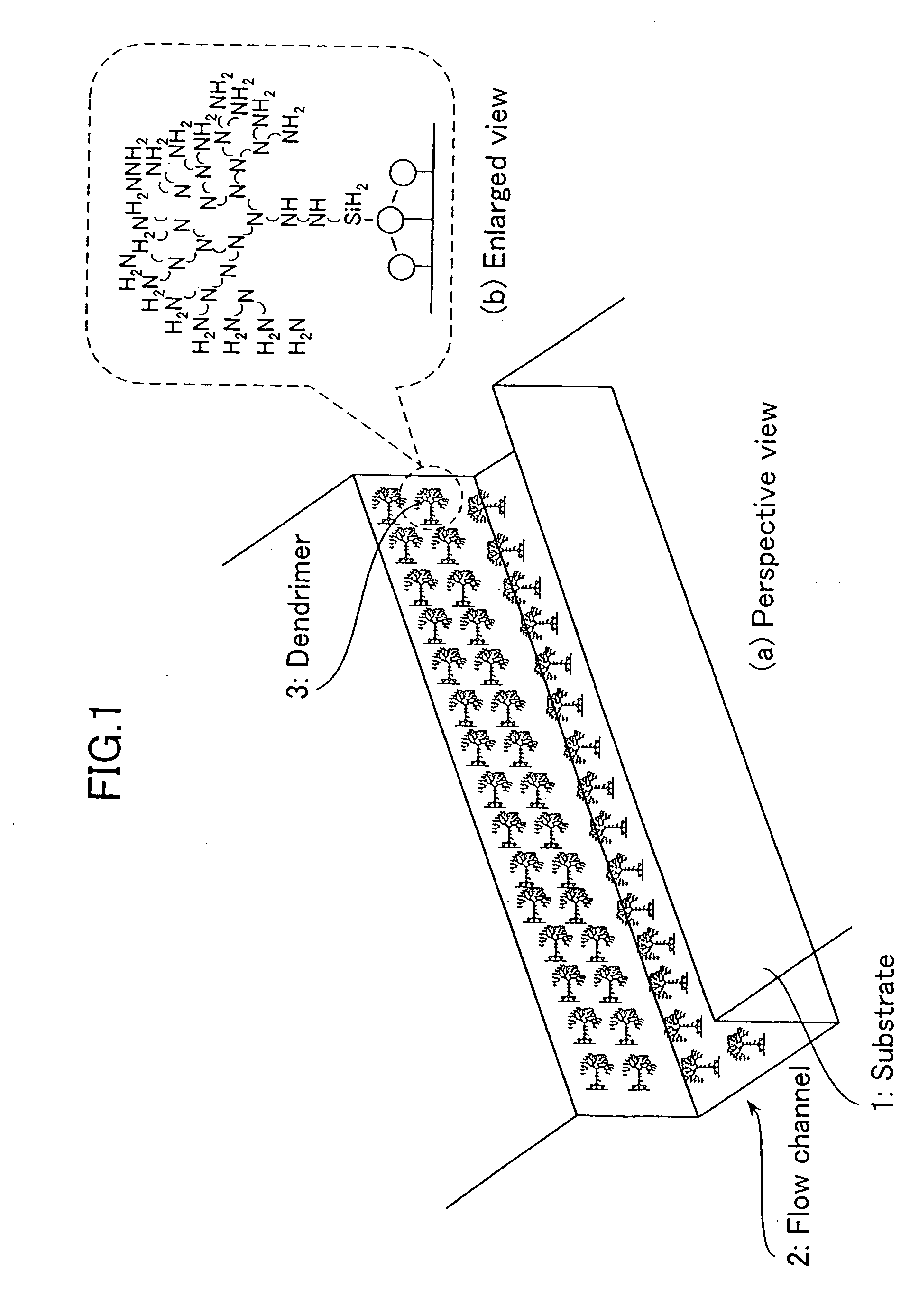
Dendrimer-based DNA extraction methods and biochips
InactiveUS20060269961A1MiniaturizationBioreactor/fermenter combinationsBiological substance pretreatmentsDendrimerBiopolymer
The present invention provides a dendrimer-based biochip, wherein a flow channel through which a solution containing biopolymer molecules is flowed is formed in the substrate of the biochip, a plurality of dendrimer molecules one end of each of which is bound to the walls of the flow channel are formed thereon, and probe biopolymer or antibody molecules are bound to the tips of the dendrimer molecules so that, if the probe biopolymer molecules are bound, then target biopolymer molecules can be captured by means of a complementary combination and, if the antibody molecules are bound, then protein can be extracted by means of antigen-antibody reaction, whereby biopolymers can be retrieved in a highly efficient manner.
Owner:YOKOGAWA ELECTRIC CORP +1
Multifunctional integrated microfluidic nucleic acid analysis chip and preparation and analysis method thereof
ActiveCN105349401AQuick buildEasy to operateBioreactor/fermenter combinationsBiological substance pretreatmentsInjection mouldingMultiple layer
The invention provides a multifunctional integrated microfluidic nucleic acid analysis chip. The chip is prepared by preparing a combination mould according to 3D printing or micromachining technology, generating a microfluidic substrate according to an injection moulding method, and completing bonding of multiple layers including a chip substrate, a liquid circuit layer, an elastic film, a gas circuit layer and the like by means of plasma cleaning to finally obtain the complete chip. The chip integrates various structures including reagent cavities, mixing cavities, a splitting cavity, a DNA (deoxyribonucleic acid) extraction cavity, amplification reaction cavities, a microvalve, a microchannel and the like and has functions of sample loading, reagent mixing, heating for splitting, DNA adsorption, cleaning, eluting, amplification reaction and the like. The multifunctional integrated microfluidic nucleic acid analysis chip has the advantages that totally-closed automatic operation of 'sample input-result output' is realized, complicated manual operations are avoided, and detection efficiency is improved.
Owner:合肥中科易康达生物医学有限公司
Rapid fluorescence PCR detection kit for ASFV (African swine fever virus)
PendingCN109593893ADetection fitReduce manual operationsMicrobiological testing/measurementDNA/RNA fragmentationSerum igeFluorescence
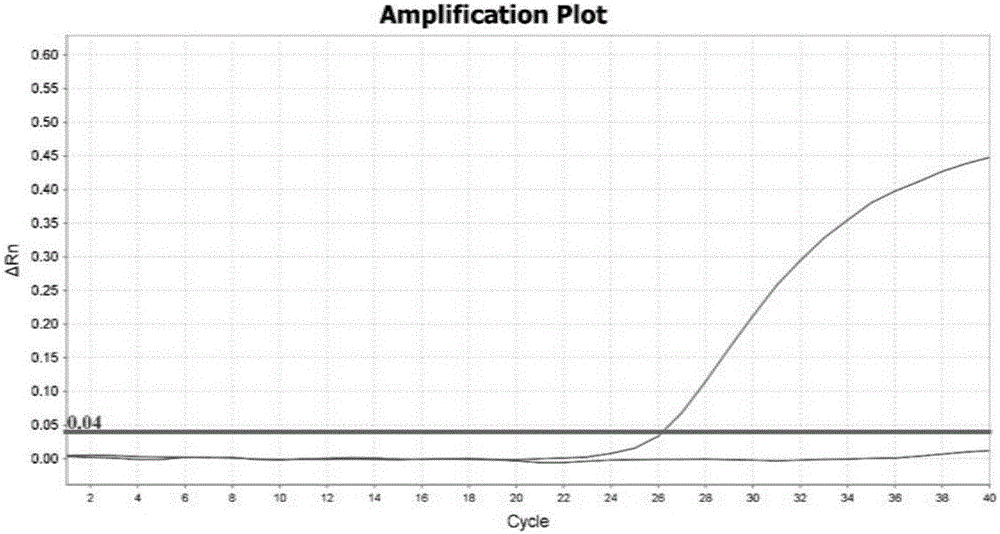
The invention discloses a rapid fluorescence PCR detection kit for ASFV (African swine fever virus). The kit comprises specific primers ASFV-F and ASFV-R as well as a TaqMan probe ASFV-P, the 5' end of the probe labels fluorescence dye as FAM and the 3' end labels a fluorescence quenching group as BHQ-1. The kit can be used for detecting the ASFV in nasal swabs, blood, serum, plasma and tissue ofswine to realize rapid detection of the ASFV. In the whole ASFV detection process, only 40 min is taken from DNA extraction to obtaining of detection results, so that the detection time is greatly shortened, and the detection efficiency is improved.
Owner:ZHENGZHOU ZHONGDAO BIOTECHNOLOGY CO LTD +2
Primer pair, probe and kit used for noninvasive polygene methylation combination detection for early stage colorectal cancer and applications thereof
ActiveCN108103195AEasy to sampleSampling non-invasiveMicrobiological testing/measurementDNA/RNA fragmentationDNA methylationFluorescence
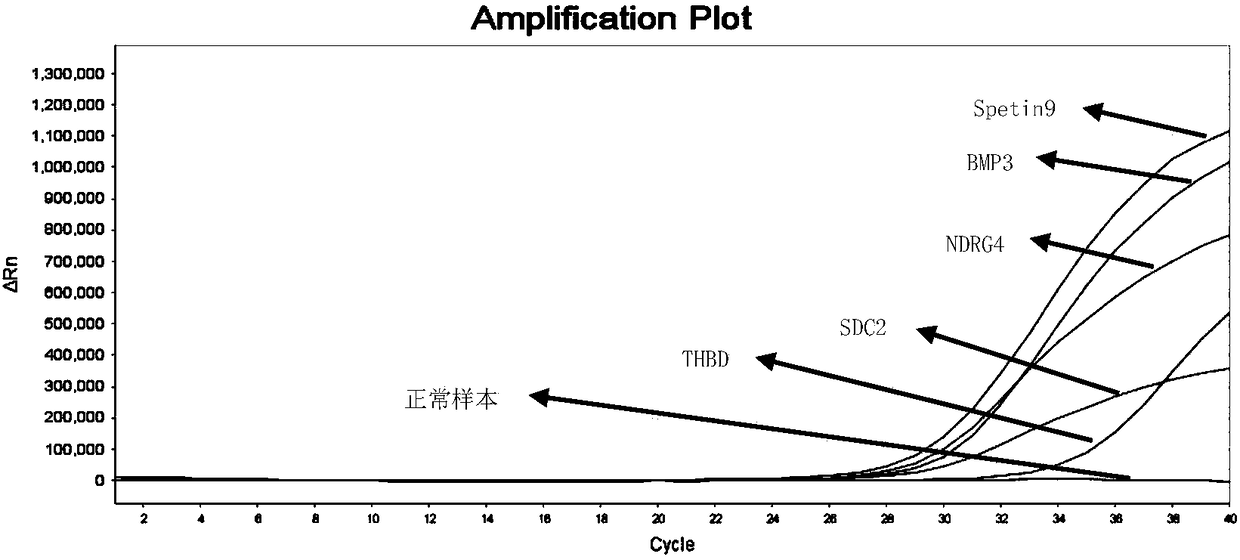
The invention relates to a primer pair and a probe used for noninvasive polygene methylation combination detection for early stage colorectal cancer, and includes the primer pair and the probe used for detecting methylation of genes Spetin9, NDRG4, BMP3, THBD and SDC2 and the primer pair and the probe for internal reference ACTB; the sequences of the primer pair and the probe are represented as the SEQ ID No.1 to the SEQ ID No.18. The invention also provides a kit containing the primer pair and the probe and applications thereof. The application method includes free DNA extraction from a plasma specimen, sulfite conversion, PCR amplification reaction, fluorescent signal detection and result determination. The kit and the method are suitable for methylation detection of the five genes Spetin9, THBD, SDC2, NDRG4 and BMP3 in human peripheral blood; compared with a conventional colorectal cancer diagnosis method, the application method fully utilizes the free DNA extraction from a plasma specimen, the DNA methylation and QPCR associated technologies, thus developing the kit having high sensitivity and specificity. The primer pair, probe and kit are used for performing early stage noninvasive screening to human colorectal cancer.
Owner:上海酷乐生物科技有限公司
Kit for separating genome DNA by using magnetic balls and application thereof
InactiveCN101792757AIncrease productionHigh purityMicrobiological testing/measurementDNA preparationMagnetic beadPhenol
The invention provides a kit for separating genome DNA by using magnetic balls and application thereof. The kit comprises magnetic balls, a magnetic frame, a genome DNA extraction reagent (lysing solution, binding solution, rinsing solution A, rinsing solution B and eluent) and specifications; the main steps of extracting the gene DNA comprise cell lysis, nucleic acid absorption, impurity removing and nucleic acid elution. The application of the kit for extracting the genome DNA does not need to use large-toxicity organic solvents of phenol, chloroform and the like, has good safety, and simple, fast and time-saving operation, and simultaneously can extract a plurality of samples. The kit can extract the genome DNA from materials of animals, plants, bacteria, fungus, blood, virus, animal source feed stuff, samples in the forensic medicine and the like, the DNA has high yield and purity, and the obtained genome DNA can be used for experiments such as PCR amplification, gene cloning, construction of genomic library, sequence measurement, molecular hybridization, molecular marking and the like. The kit can be stored at the temperature of 4 DEG C, also can be placed at room temperature, and is convenient to transport.
Owner:上海鼎国生物技术有限公司
Whole blood DNA (deoxyribonucleic acid) extraction kit based on paramagnetic particle method and application of extraction kit
The invention discloses a whole blood DNA (deoxyribonucleic acid) extraction kit based on a paramagnetic particle method and an application of the extraction kit. The kit comprises a cell lysis solution, a binding solution, a washing solution I, a washing solution II and a nucleic acid eluent. Compared with kits in the prior art, the kit has the advantages of simple and quick operation, use safety, high extraction efficiency, low manufacturing cost and the like; no toxic reagents or organic solvents are used when the kit is used, a blood sample is not required to be pre-treated, the extraction process can be finished manually and can also be automatically finished in a full-automatic nucleic acid extraction instrument, 1-16 animal tissue samples can be treated each time with a 96-hole reaction plate during automatic extraction, other operation is not required in the extraction process, time is saved for whole blood DNA extraction and detection, and the cost is reduced; DNA in the blood can be quickly extracted with the adoption of the kit, the concentration and the purity of extracted nucleic acid are high, the repeatability is good, and the kit can be used for scientific research and clinical detection.
Owner:NANJING INST OF ADVANCED LASER TECH
Microfluidic apparatus and method for DNA extraction, amplification and analysis
InactiveUS20110244467A1Reduce manufacturing costEasy to analyze and useBioreactor/fermenter combinationsBiological substance pretreatmentsGel basedA-DNA
An integrated gel based microfluidic sample processing device, suitable for forensic investigations at the scene of a crime, including a substrate having a plurality of micro-channels to form at least a DNA extraction chamber in fluidic cooperation with an amplification chamber which in turn is in fluidic cooperation with a separation and detection channel. The micro-channels containing a DNA extraction material and gel based reaction reagents necessary for processing the sample. The device further having electrical contacts for coupling to an external power source and capable of inducing electro-kinetic manipulation of the gel based reagents and DNA extracted from the sample throughout the device.
Owner:UNIVERSITY OF HULL
Listeria monocytogenes nucleic acid chromatography detection kit, its detection method and application thereof
InactiveCN102520172AGuaranteed SensitivityGuaranteed FeaturesMaterial analysisDNA extractionMicrobiology
The invention discloses a listeria monocytogenes nucleic acid chromatography detection kit, its detection method and an application thereof, and belongs to the field of molecular biology and immunology. According to the invention, with the combination of a polymerase chain reaction technology in molecular biology and an immunochromatography technology, rapid detection of listeria monocytogenes in food is accomplished. The kit provided by the invention includes three parts of DNA extraction, PCR amplification and strip detection. The combination of the molecular biology technology and the immunochromatography technology is accomplished by modification of primers and selection of corresponding coatings and markers in strip preparation so as to achieve specific amplification product detection of listeria monocytogenes in food. Compared with a traditional biochemistry method, the method provided by the invention is used to greatly save detection time and simplify operation steps, has characteristics of accuracy, rapidity, convenience and safety, and is suitable for detection of a large number of samples.
Owner:北京陆桥技术股份有限公司
Fluorescence quantitative PCR detection kit of hepatitis B virus and application thereof
ActiveCN101701267AStrong specificityHigh purityMicrobiological testing/measurementMicroorganism based processesPositive controlFluorescence
The invention discloses a fluorescence quantitative PCR detection kit of hepatitis B virus and an application thereof. The kit is composed of the following independent components: DNA extraction solution I, DNA extraction solution II, DNA extraction solution III, DNA extraction solution IV, positive control interior label, PCR reaction liquid, probe HBV-SP, enzyme mixed liquor containing heat resistant DNA polyase and uracil DNA glycosylase, quantitative hepatitis B virus reference material, hepatitis B virus positive control serum and hepatitis B virus negative control serum, wherein DNA extraction solution I contains 0.2-1.0% of lauryl sodium sulphate (mass / volume), 1.0-4.0% of Triton (volume / volume) and 0.2-1.0mol / L of guanidinium isothiocyanate; DNA extraction solution II contains 100-300mmol / L of 4-HEPES, 100-300mmol / L of sodium chloride with pH of 6.5+ / -0.2 and 100-400 mu g / ml of magnetic beads; DNA extraction solution III contains 0.1-1.0% of Triton (volume / volume) and 100-300mmol / L of sodium chloride; DNA extraction solution IV contains mineral oil. The fluorescence quantitative PCR detection kit of hepatitis B virus of the invention can be used for detecting the HBV-DNA concentration in samples of serum, blood plasma or latex and the like.
Owner:SANSURE BIOTECH
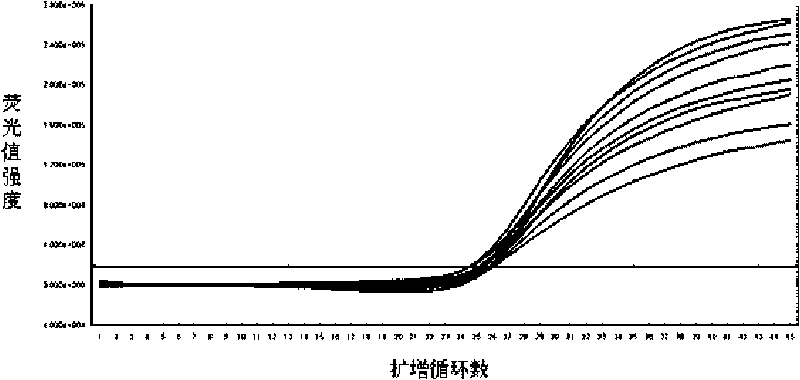
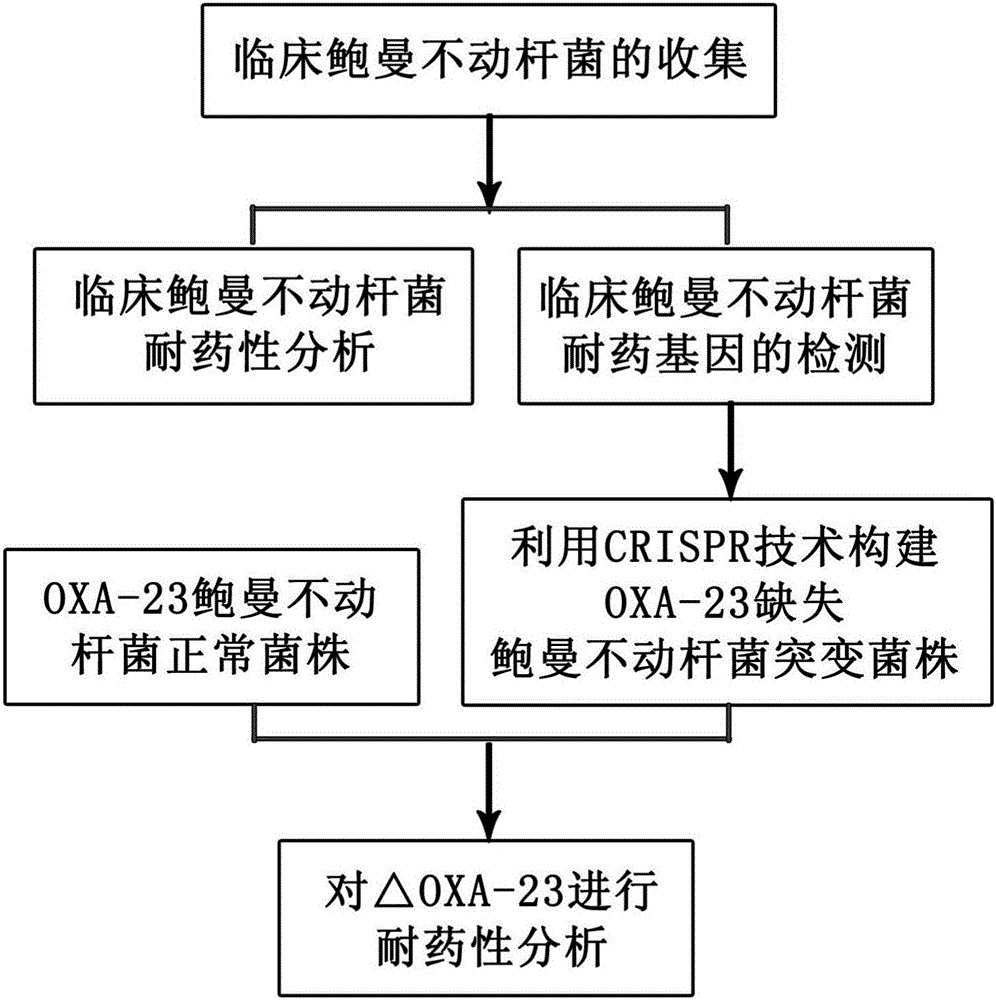
Method for deleting drug resistant genes of acinetobacter baumannii (AB) through CRISPR-Cas9
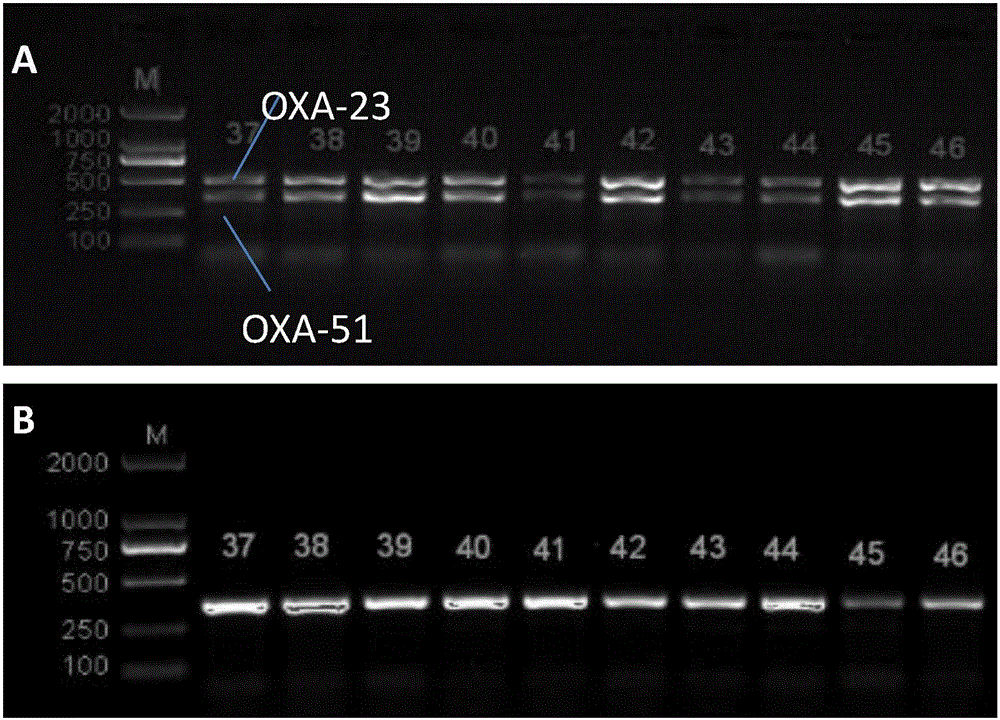
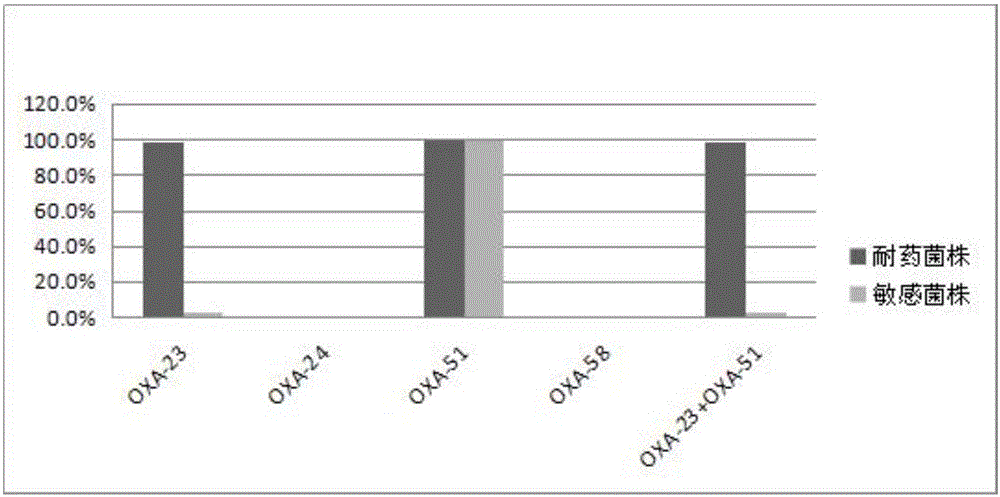
InactiveCN106544353ASensitivity reversalEasy to operateVectorsMicroorganism based processesBacteroidesMulti drug resistant
The invention discloses a method for deleting drug resistant genes of acinetobacter baumannii (AB) through CRISPR-Cas9. The method comprises the steps that firstly, AB is collected clinically, and drug resistance analysis and statistics are conducted after the AB is subjected to drug resistance measurement through a drug sensitive slip method; secondly, the multi-drug resistant AB is subjected to DNA extraction through a lysis-boiling method, and then amplification analysis is conducted by mean of well designed drug resistant gene primers of the AB; and thirdly, according to drug resistant gene detection in the former step, the AB containing an OXA-23 gene is selected, CRISPR / Cas9 and sgRNA plasmids are established and transferred into the AB containing the OXA-23 gene, an OXA-23 gene deletion AB mutant strain is established, and drug resistance analysis is conducted on the OXA-23 gene deletion AB mutant strain. The method is easy to operate and high in knocking-out efficiency, and a novel method and a novel concept are provided for preventing spreading of the drug resistant genes and treating drug resistant bacteria.
Owner:GENERAL HOSPITAL OF NINGXIA MEDICAL UNIV
Fluorescent quantitative PCR (Polymerase Chain Reaction) detection reagent, kit and detection method for African swine fever virus (ASFV)
ActiveCN103757134AImprove throughputNo biological safety hazardMicrobiological testing/measurementDimethyl pyrocarbonateBiology
The invention discloses a fluorescent quantitative PCR (Polymerase Chain Reaction) detection reagent, a kit and a detection method for an African swine fever virus (ASFV). The detection reagent is targeted to a conserved segment of an ASFV VP72 gene, and comprises a pair of specific primers, a specific probe and an internal labeling probe. The kit comprises a PCR mixed solution, Taq DNA polymerase, DEPC (diethylpyrocarbonate)-H2O, an ASFV positive quality product, an ASFV negative quality product, a quantification standard substance and a pseudovirus internal labeling solution, wherein the PCR mixed solution comprises a pair of specific primers, a specific probe and an internal labeling probe. The detection method comprises the steps of extraction of total DNAs, preparation of reaction components, making of a standard curve by diluting the standard substance, amplifying and result judgment. The reagent and the kit have the advantages of high specificity, high sensitivity, no pollution and high flux, and can be used for accurately detecting infection, inapparent infection or continuous carrying of a toxic host of low-content ASFV. The detection method is quick, specific and sensitive, and can be used for quantitatively detecting a large quantity of samples at the same time.
Owner:深圳澳东检验检测科技有限公司
Process for abstracting total DNA of swine waste sample
The invention relates to the microbiology and molecular biology technical field, in particular to a method for extracting pig manure sample total DNA. The method mainly comprises manure sample pretreatment and sample total DNA extraction. Moreover, the method comprises the following steps that: collected fresh manure sample or frozen manure sample is taken as a material; the pretreatment of the manure sample is carried out by means of a PBS buffer solution, a paraformaldehyde solution and anhydrous alcohol so as to obtain manure sample bacteria suspension; and a CTAB solution and beta-mercaptoethanol, etc. are used to carry out cell fragmentation, thereby releasing DNA. The method has the characteristics of simple operation, quickness, high efficiency and ideal repeatability and economical efficiency, etc.; moreover, the extracted DNA has high concentration, purity and yield, and can be directly used in subsequent molecular biology experimental researches such as PCR amplification without purification.
Owner:SICHUAN UNIV
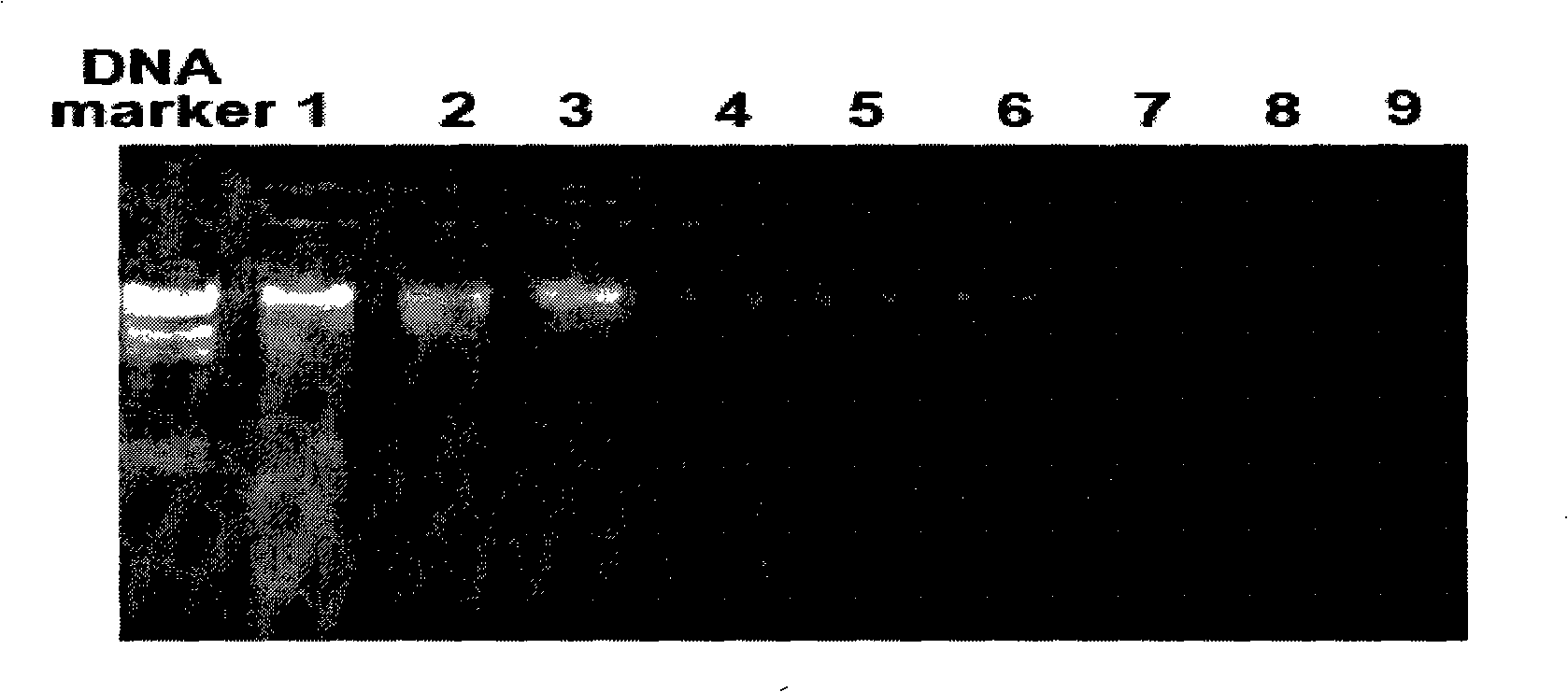
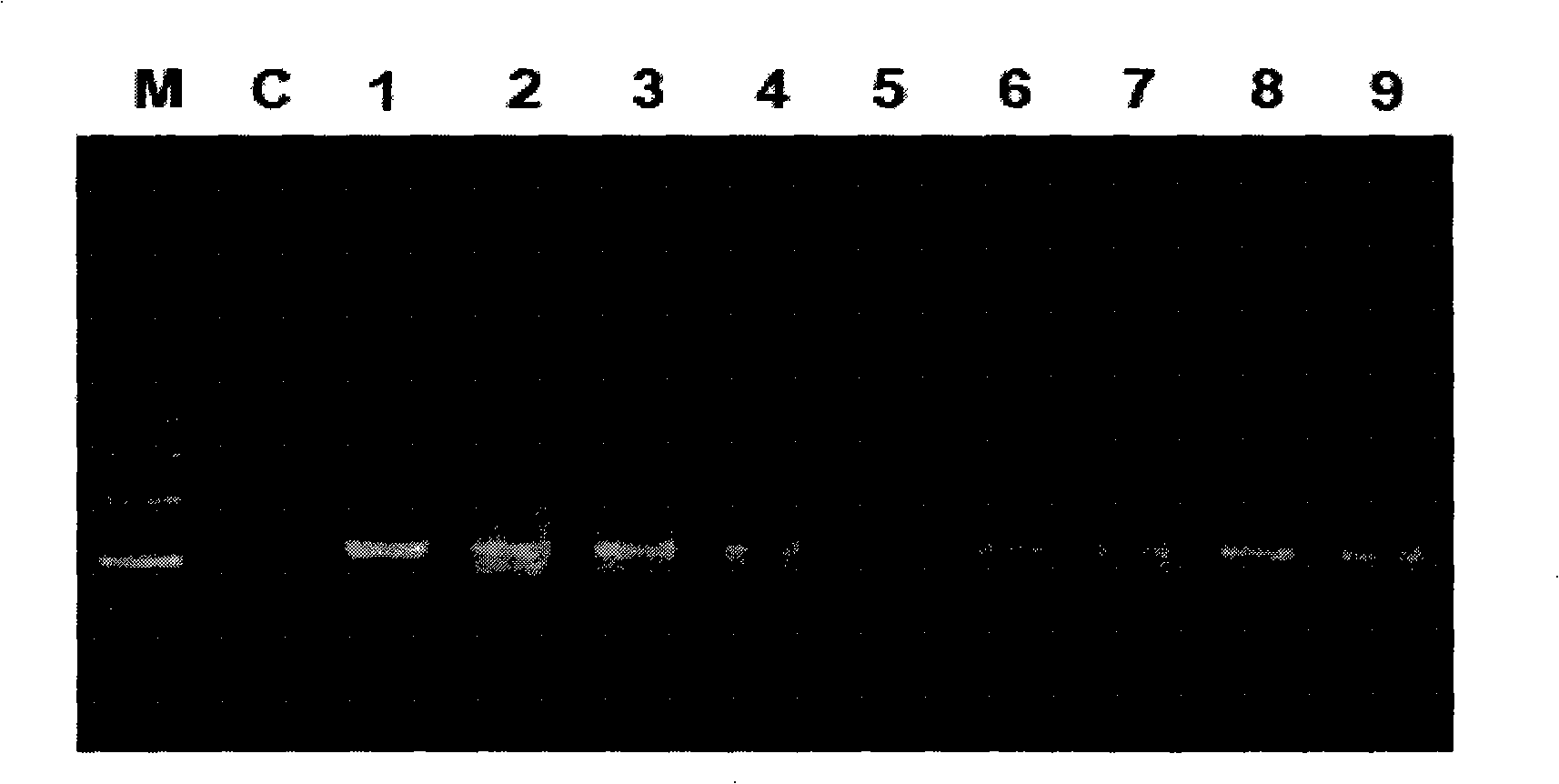
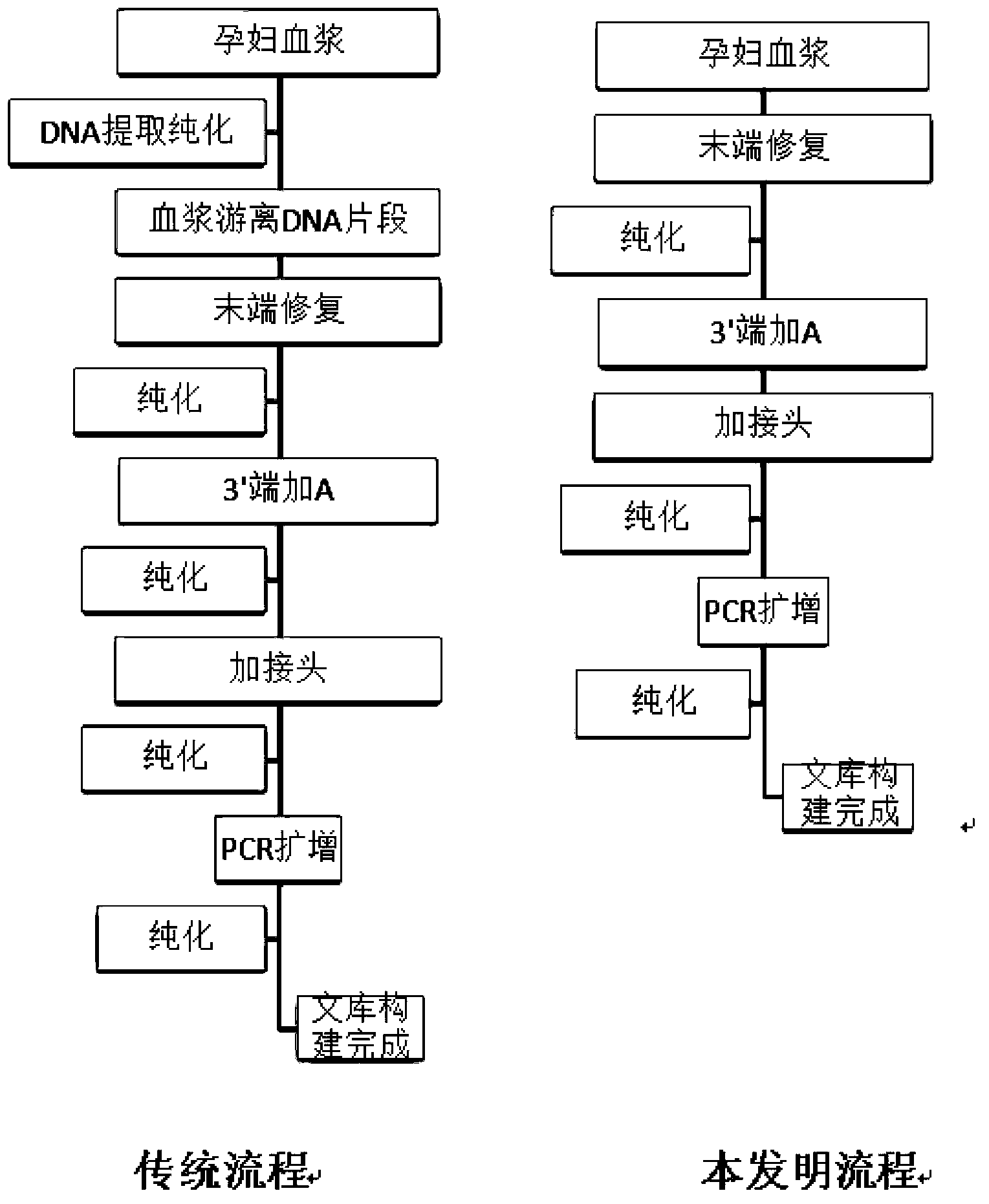
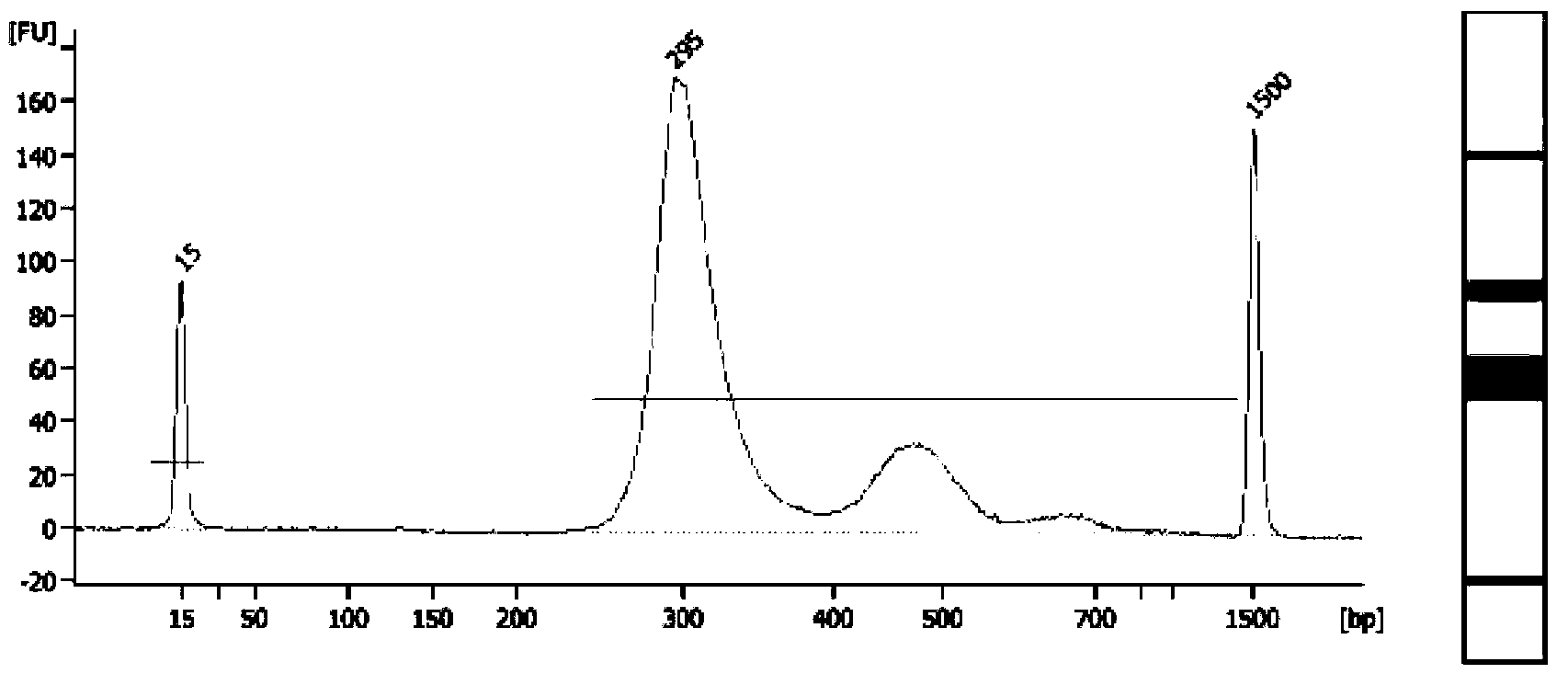
DNA library construction method for prenatal diagnosis
InactiveCN103361743AQuality improvementLow costMicrobiological testing/measurementLibrary creationCDNA libraryDNA extraction
The invention provides a DNA library construction method for prenatal diagnosis. According to the DNA library construction method for the prenatal diagnosis, pregnant woman blood plasma is directly used for library construction, and a DNA extraction step is omitted, so that use amount of the pregnant woman blood plasma can be reduced, at the same time, possibility of deviation of fetal DNA content can be reduced, and cost and time for the DNA library construction are greatly reduced. Another aspect of the invention relates to DNA libraries prepared by any of above preparation methods. The third aspect of the invention relates to a DNA sequencing method. The DNA library construction method has good application prospect.
Owner:ANNOROAD GENE TECH BEIJING
Method for extracting and purifying spittle DNA
InactiveCN102220310AMeet the extraction requirementsEasy to operateDNA preparationBiological cellLysis
The invention discloses a method for extracting and purifying spittle DNA, which comprises the following: 1) a step of pre-treatment, which is to contact the spittle with pretreatment lysis solution, remove solid matters adhered onto cells and obtain coarse lysis solution; 2) a step of nucleic acid absorption, which is to mix the coarse lysis solution obtained by the step 1) with lysis binding solution and magnetic bead suspension to form a solution system containing magnetic nano microsphere and DNA composite and is to collect the magnetic nano microsphere and DNA composite from the solutionsystem; and 3) a step of washing, which is to wash the magnetic nano microsphere and DNA composite obtained by the step 2) with washing liquid I and washing liquid II in turn, and dissolve out DNA byusing eluent. When the method provided by the invention is used, the DNA extraction efficiency may reach 90 percent, the extracted DNA can be used in short tandem repeat (STR) composite amplification, DNA sequencing, DNA quantification and other downstream analysis and operation.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Multiplex PCR method for distinguishing four meat components in food at the same time
InactiveCN102605090ARapid identificationAvoid cross interferenceMicrobiological testing/measurementForward primerGenomic DNA
The invention belongs to the technical field of food quality and safety detection and particularly relates to a method for rapidly detecting pork, beef, mutton and chicken components in food at the same time through animal genomic DNA (deoxyribonucleic acid) extraction, primer design and UP-M-PCR (universal primers-multiplex-PCR). The four meat components can be distinguished rapidly at the same time according to differential sites of mitochondrial cytochrome b of the animal by using the forward primer sharing and reverse primer specificity strategy and an M-PCR system including five primers. The result shows that the method has the advantages that the detection limit can be up to the pictogram level, the components of the reaction system are free from cross interference and the meat products on market can be detected, verified and distinguished accurately by sampling 80 meat products randomly according to the specificity of the four target meat components. In a word, the invention provides a specific, sensitive and practical multiplex PCR method for rapidly screening four common meat components in food at the same time.
Owner:NANJING INST OF PROD QUALITY INSPECTION
Blood DNA conserving card and method for making the same
ActiveCN101153263AAvoid degradationEasy accessBioreactor/fermenter combinationsBiological substance pretreatmentsBiotechnologyFiber
The invention relates to 'a blood DNA preserving card and production method' and belongs to the biology technical field. A blood DNA preserving card comprises fiber-shaped media with the ability of absorbing DNA, and is characterized in that: the salinity is absorbed in the media and contains cell lysing reagent, nuclease inhibitor, a substance allowing the pathogenic bacterium and the virus protein to be denaturalized as well as anti-oxidization inhibitor. These salinities can inhibit the activities of the viruses to prevent the infection, can prevent the nuclease from gradating DNA, so that the blood DNA can be preserved indoor for a long-term. The pure DNA can be obtained from the preserved DNA in the card of the invention by bleaching the salt ions and blood corpuscle leftovers on the card media with the water, thereby avoiding the extraction process, ensuring the operation is simpler, being convenient to be operated in batches and saving the expense of the DNA extraction agent.
Owner:DINGSHENG TECH BEIJING +1
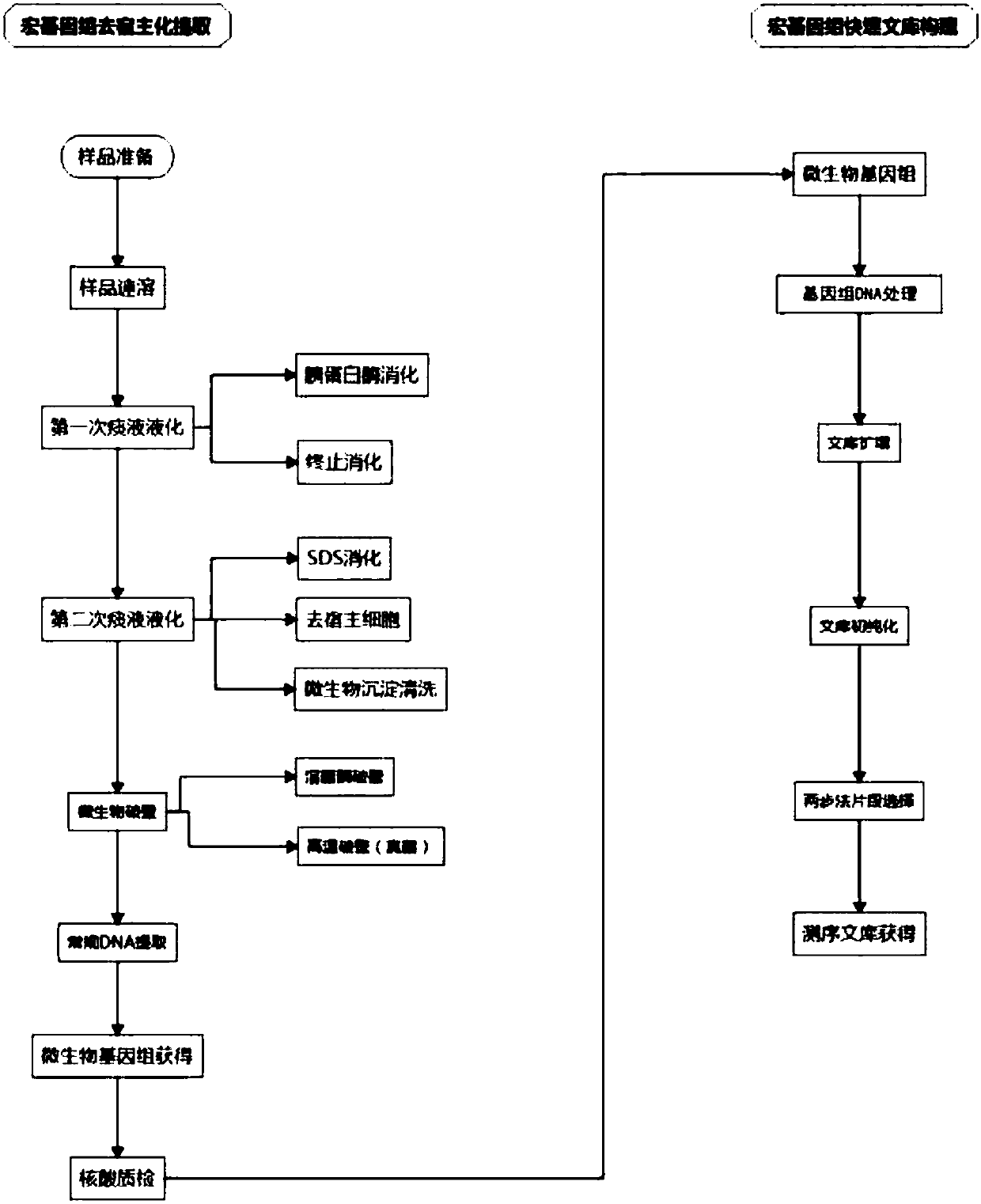
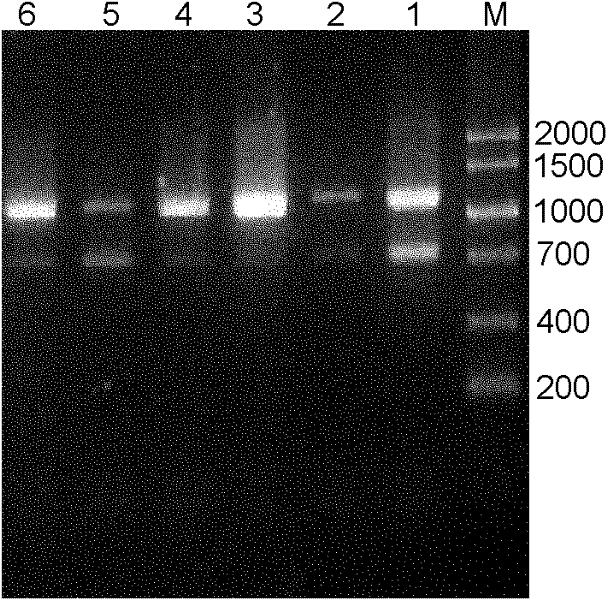
Host removal extraction and database building method for sputum microorganism metagenome
ActiveCN108048450AFully consider the physical characteristicsTaking biological differences into accountMicrobiological testing/measurementMicroorganism lysisCell wallDigestion
The invention belongs to the technical field of microorganism molecular biological detection, and specifically discloses a host removal extraction method for a sputum microorganism metagenome. The method comprises primary sputum liquefaction, secondary sputum liquefaction, microorganism wall breaking, DNA extraction and nucleic acid quality detection. The invention further discloses a database building method for a sputum microorganism metagenome, wherein the method comprises genome DNA extraction, genome DNA treatment, library amplification, library pre-purification, two-step fragment selection and computer detection. A mild digestion method combining trypsin with SDS is adopted to release a host genome to a solution, thereby obtaining microorganism with complete cell wall structure maintained and extracting nucleic acid from microorganisms with host removed. The provided extraction method and database building method are good in compatibility, and can be widely used to a mainstream sequencing platform. The methods are flexible and simple, efficient and fast, and cost-saving, and have bright popularization application prospects.
Owner:湖南赛哲智造科技有限公司
Extraction and purification method of total plant endophyte genome DNA for colony analysis
InactiveCN102174509AQuality improvementIncrease credibilityFermentationPlant genotype modificationPurification methodsPhosphate
The invention discloses an extraction and purification method of total plant endophyte genome DNA for colony analysis. The method comprises the following steps of: shearing and grinding the surface bacteria removed fresh plant tissues into paste, soaking the plant tissues into sterilized phosphate buffer solution, performing thermostatic shaking for 1 hour in a table concentrator to separate out microbial cells from the plant tissues, standing, transferring the suspension to a centrifuge tube, performing centrifugal collection on microbial bacteria, then adding lysing solution, shaking and uniformly mixing the solution to release DNA from the cells, repeatedly extracting protein by chloroform-isoamylol, performing centrifugal sedimentation with isopropanol and washing with 70 percent ice ethanol, performing purification by using a DNA specific centrifugal adsorption column to obtain total crude extracted genome DNA, and finally, performing polymerase chain reaction (PCR) amplificationby using special primers to obtain high-quality plant endophyte specific DNA fragments for subsequent analysis. The invention has the advantages that: the method is simple and effective, high in quality, low in pollution and the like, and provides high-quality guarantee for comprehensively and completely researching a plant endophyte colony structure.
Owner:HUNAN UNIV
Preparation of DNA-containing extract for PCR amplification
Environmental samples typically include impurities that interfere with PCR amplification and DNA quantitation. Samples of soil, river water, and aerosol were taken from the environment and added to an aqueous buffer (with or without detergent). Cells from the sample are lysed, releasing their DNA into the buffer. After removing insoluble cell components, the remaining soluble DNA-containing extract is treated with N-phenacylthiazolium bromide, which causes rapid precipitation of impurities. Centrifugation provides a supernatant that can be used or diluted for PCR amplification of DNA, or further purified. The method may provide a DNA-containing extract sufficiently pure for PCR amplification within 5–10 minutes.
Owner:LOS ALAMOS NATIONAL SECURITY
PCR-RFLP identification method for seven crassostrea oyster on south China coast
ActiveCN101130815AAccurate identificationImprove reliabilityMicrobiological testing/measurementGeneticsCrassostrea
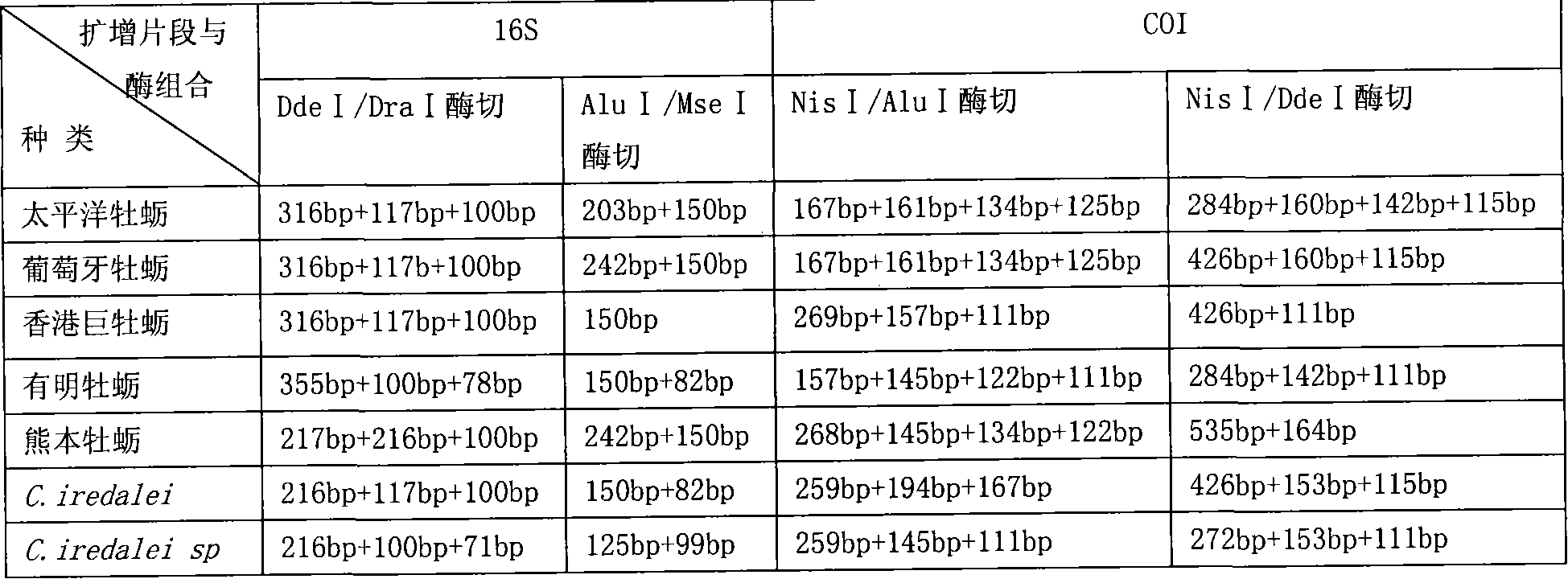
The present invention relates to a PCR-RFLP identification method for 7 kinds of Chinese south inshore oysters. Said identification method includes the following several main steps: making genome DNA extraction, PCR amplification of object fragment, selecting proper incision enzyme combination, utilizing selected incision enzyme to enzyme-cut the PCR product and utilizing agarose gel electrophoresis to make detection.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
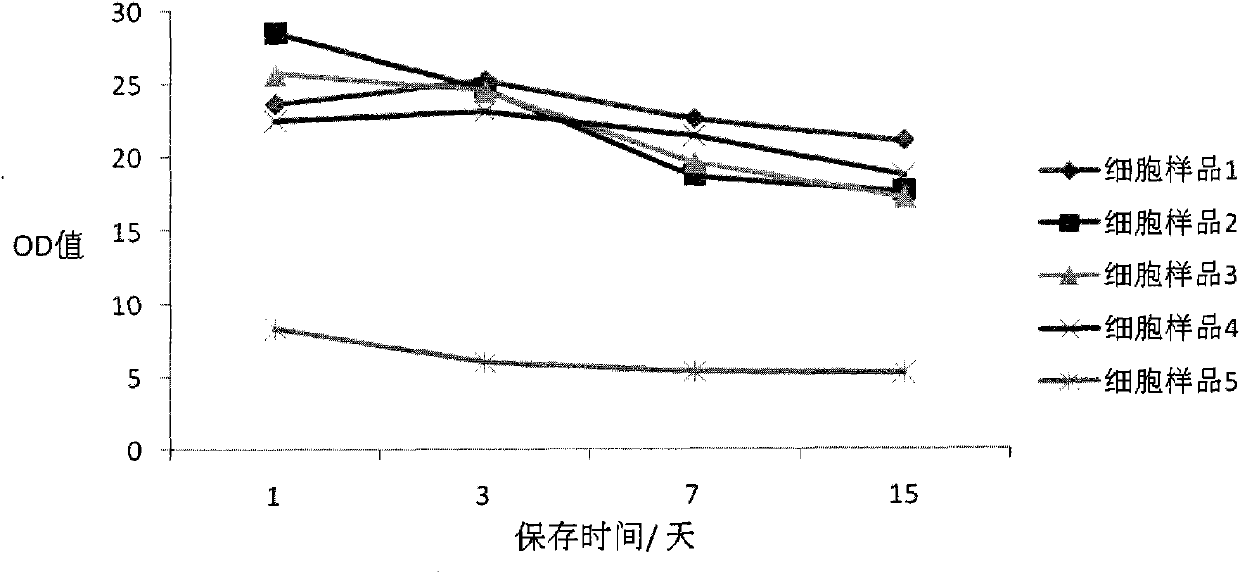
Cell preserving fluid and preparation method and use thereof
ActiveCN101999343AAvoid degradationConducive to follow-up detectionMicrobiological testing/measurementDead animal preservationDisulfide bondChemistry
The invention belongs to the field of cell biology, relating to a cell preserving fluid and a preparation method and use thereof. Concretely, the cell preserving fluid comprises a fixing agent, a fixing agent assistant, an anticoagulant, a cushion fluid and an ionic strength maintenance agent. The cell preserving fluid is characterized in that the content of the anticoagulant is 0.01% to 1.5%(w / w); the content of the ionic strength maintenance agent is 0.01% to 1%(w / w); and the cell preserving fluid does not contain a disulfide bond open reagent. The cell preserving fluid can effectively preserve cell DNA (deoxyribonucleic acid) and virus DNA infecting the cell, is convenient to collect the cast-off cells and extract DNA and can give attention to storage of the cell structure. The invention also relates to the preparation method and the use of the cell preserving fluid. The invention also relates to a kit and a method for detecting HPV (Human Papilloma Virus) or HPV DNA.
Owner:BGI GENOMICS CO LTD
DNA extraction method for polysaccharide-rich plant dried leaves
The present invention discloses a DNA extraction method for polysaccharide plant dried leaves. The DNA extraction method comprises dried leaf grinding, desaccharifying, cracking, extraction, precipitation, washing, detection and other steps, wherein desaccharifying is the key step, a pre-heated desaccharifying buffer solution is added before the nuclear membrane is not cracked and the genome DNA is not released, the leaf tissue solution is suspended under a 65 DEG C water bath condition so as to dissolve polysaccharides and other secondary metabolites in the buffer solution, and low speed centrifugation is adopted to remove the polysaccharides and other impurities, such that the final DNA obtained through precipitation has high concentration and high purity. The DNA extraction method is suitable for dried mature leaves of all cerasus plants and other polysaccharide-rich plants, wherein the concentration of the obtained DNA can be increased if the leaves are the young and tender leaves, and the more DNA with high quality can be obtained from the mature leaves.
Owner:SICHUAN AGRI UNIV
Portable microorganism detection unit
InactiveUS20100216225A1Easy to chopSmall apertureBioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismDNA extraction
The present invention is a portable device for detecting microorganisms in a food sample. The device includes a DNA extraction tube, a chip, and a portable control unit. The DNA extraction tube extracts DNA from the sample in an aqueous solution. The extraction tube includes successive chambers that are configured to mince, filter, and purify DNA in the form of an aqueous eluate. The chip includes reaction chambers disposed for a polymerase chain reaction to amplify a specific DNA fragment in the eluate for detection, and the portable control unit is configured to receive the chip and analyze the amplified DNA on the chip.
Owner:DEFENSE SYSTEMS
Kit for simultaneously detecting SLCO1B1, APOE and LDLR gene multisite mutation
The invention belongs to the technical field of gene mutation detection, and concretely discloses a kit for simultaneously detecting SLCO1B1, APOE and LDLR gene multisite mutation. Through meticulous design, multi-time verification, screening and optimization, specific primers and probes based on a Taqman allelic gene resolution analysis method are obtained; eight functional variation of the SLCO1B1, APOE and LDLR genes can be detected; the time from DNA (Deoxyribonucleic Acid) extraction to fluorescent PCR (Polymerase Chain Reaction) to result obtaining is less than four hours; and the manual operation time is less than two hours. The kit comprising the primer pairs and the probe pairs has the advantages that the time is saved; convenience is realized; the sensitivity is high; and both the positive conformity rate and the negative conformity rate of a sample are higher than 99 percent, and the like. A detection method provided by the invention is mainly used for the personalized medication auxiliary diagnosis of statins such as simvastatin, atorvastatin, fluvastatin and rosuvastatin.
Owner:钟诗龙
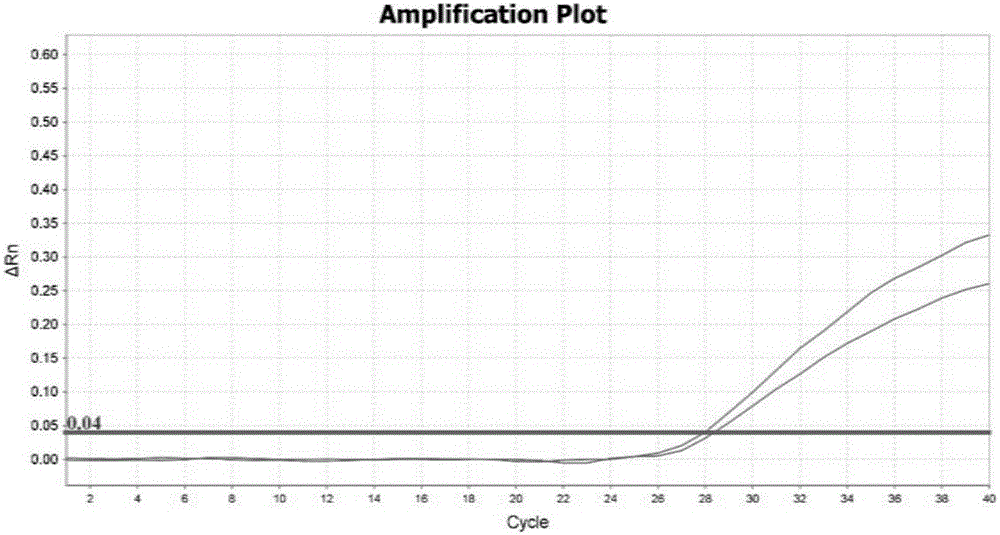
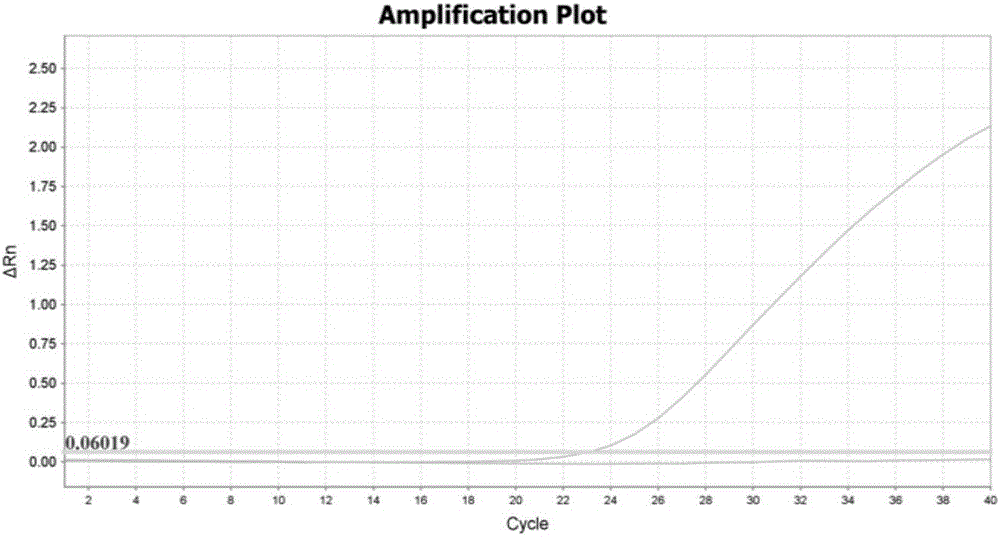
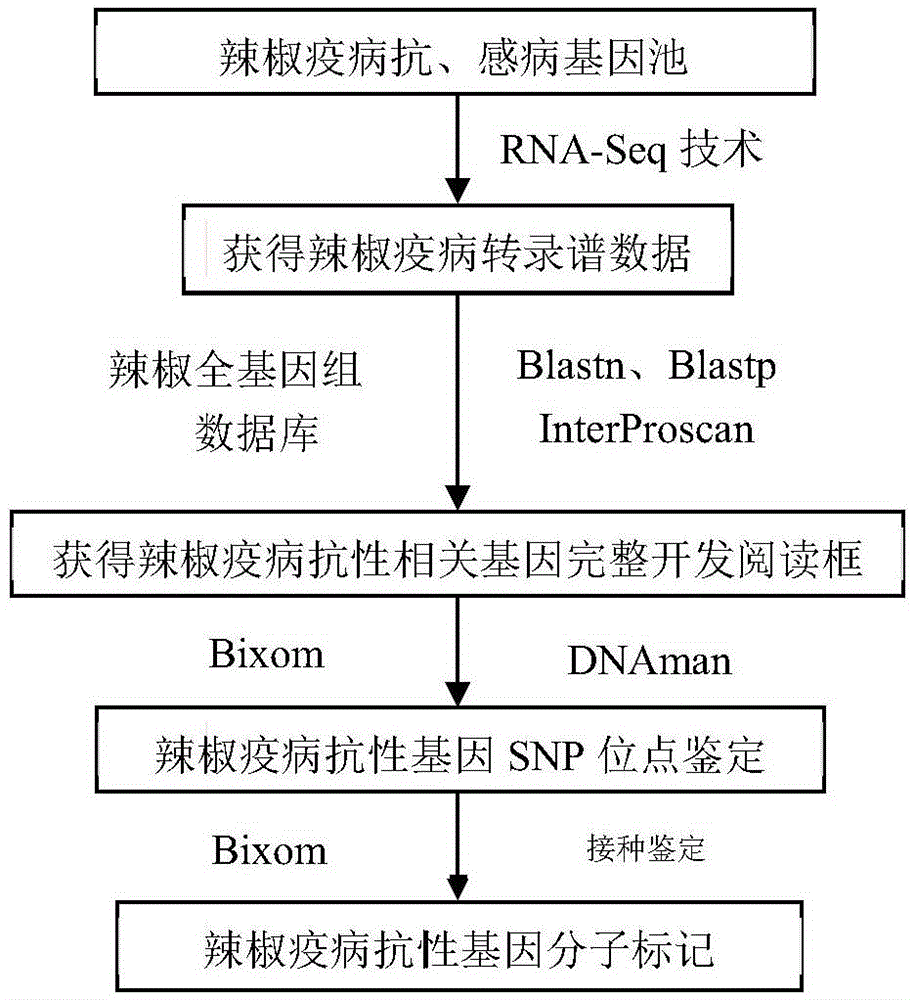
Method for obtaining capsicum phytophthora resistance candidate gene and molecular marker, and application
InactiveCN104560973AAccurate identificationLarge amount of data informationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyData information
The invention relates to a method for obtaining a capsicum phytophthora resistance candidate gene and a molecular marker, and application. The method is used for obtaining the capsicum phytophthora resistance candidate gene by utilizing capsicum phytophthora transcriptome and whole-genome sequencing data information, differentially-expressed gene identification, bioinformatics analysis, molecular marker development and phytophthora inoculation identification and belongs to the technical field of capsicum biology. The method comprises the following steps: sequencing a phytophthora resistant and susceptible gene pool transcriptome obtained after phytophthora inoculation of an F2 population constructed by capsicum highly-resistant and highly-susceptible phytophthora materials, performing expression analysis and functional annotation on differential genes, extracting DNAs (Desoxvribose Nucleic Acid) of a capsicum phytophthora highly-resistant and highly-susceptible phytophthora material genome, performing primer design and PCR (Polymerase Chain Reaction) amplification, performing sequence difference analysis and SNP site identification, performing SNP specific primer design and validity verification, and performing other steps to efficiently obtain the capsicum phytophthora resistance candidate gene and the molecular marker. According to the method, the capsicum phytophthora resistance candidate gene can be accurately identified, and the effective molecular marker can be developed.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Micro-fluidic chip for visual instant detection of pathogen nucleic acid as well as preparation method and detection method thereof
PendingCN107904161AQuick testSimultaneous real-time inspectionBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid detectionEngineering
The invention relates to a micro-fluidic chip for visual instant detection of pathogen nucleic acid. The micro-fluidic chip comprises an upper layer substrate and a lower layer substrate, wherein theupper layer substrate is provided with a micro-fluidic passage; the lower layer substrate is sealed with the upper layer substrate; the upper layer substrate is provided with a sample processing and DNA extraction region, a fluid control region and a visual detection region which are sequentially communicated; the visual detection region comprises a detection pool; the detection pool is provided with chromatography paper for covering specific primers or DNA; the fluid control region is provided with a screw valve used for performing fluid control. The invention also relates to a preparation method and a detection method of the micro-fluidic chip. The invention provides the micro-fluidic chip integrating sample treatment, pathogen nucleic acid extraction, amplification and visual detection;the simple, fast and visual beside pathogen nucleic acid detection can be realized; the detection method using the micro-fluidic chip has the characteristics of high speed, simplicity and no need ofspecial instruments; the micro-fluidic chip and the method can be applied to the instant inspection of various kinds of pathogen nucleic acids in respiratory tracts and genital tract samples.
Owner:RENJI HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Plant endophyte 16S rRNA gene amplification method and application
ActiveCN106282165ABig amount of dataThe test result is completeMicrobiological testing/measurementDNA preparationSequence analysisAmplicon sequencing
The invention discloses a plant endophyte 16S rRNA gene amplification method and application. The amplification method includes the following steps of plant pretreating; plant-sample total DNA extracting; sample 16S rRNA gene amplifying, wherein sample 16S rRNA gene amplifying comprises 16S rRNA gene total-length emulsion PCR amplifying and 16S rRNA gene hypervariable-region / conserved-region amplifying sub-amplifying; high-throughput sequencing and biological information analyzing based on a Illumina platform, wherein high-throughput sequencing and biological information analyzing based on the Illumina platform comprises plant endophyte 16S rRNA gene amplicon purifying and recycling, amplicon sequencing library establishing, Illumina HiSeq sequencing and sequencing data bioinformatics analysis. The gene amplification method is used for high-throughput-sequencing plant disease detection and phytophagous animal enteric microorganism detection. Sequencing analysis of plant endophytic bacteria is carried out with the high-throughput sequencing technology, the data size is larger, the detection result is more complete, pollution of plant hosts is reduced to the maximum degree, the result multiformity is higher, and the cost is low.
Owner:成都罗宁生物科技有限公司
Human feces DNA extraction method
ActiveCN102864138AQuality improvementHigh repeat stabilityDNA preparationTE bufferBovine serum albumin
The invention discloses a human feces DNA extraction method. The prior art is complicated and time-consuming; and the trace DNAs extracted by the previous sampling method is too little to perform molecular diagnosis. The extraction method uses selective sample collection PSC equipment for feces samples and adopts a low temperature preservation method. The extraction method comprises the following steps: picking a sample by using a sterilized gun head or sterilized blade and directly placing the picked sample on ice; and adding a feces lysis solution, fully cracking and evenly mixing, adding bovine serum albumin, centrifuging to remove the supernatant, adding protease K to perform a digestion reaction at 58 DEG C, then adding a protein precipitation solution to precipitate the protein, centrifuging to obtain the supernatant, adding a silica gel membrane binding solution to fully bind DNAs, centrifuging to obtain precipitates, adding a silica gel membrane washing solution to wash twice, centrifuging, and adding a TE (Tris + EDTA) buffer solution to finally obtain a preservation solution with DNAs. The human feces DNA extraction method has the advantages of high repeatability and stability, simpleness and short time.
Owner:ZHEJIANG CENTURY CONDOR MEDICAL SCI & TECHNOLOTY CORP
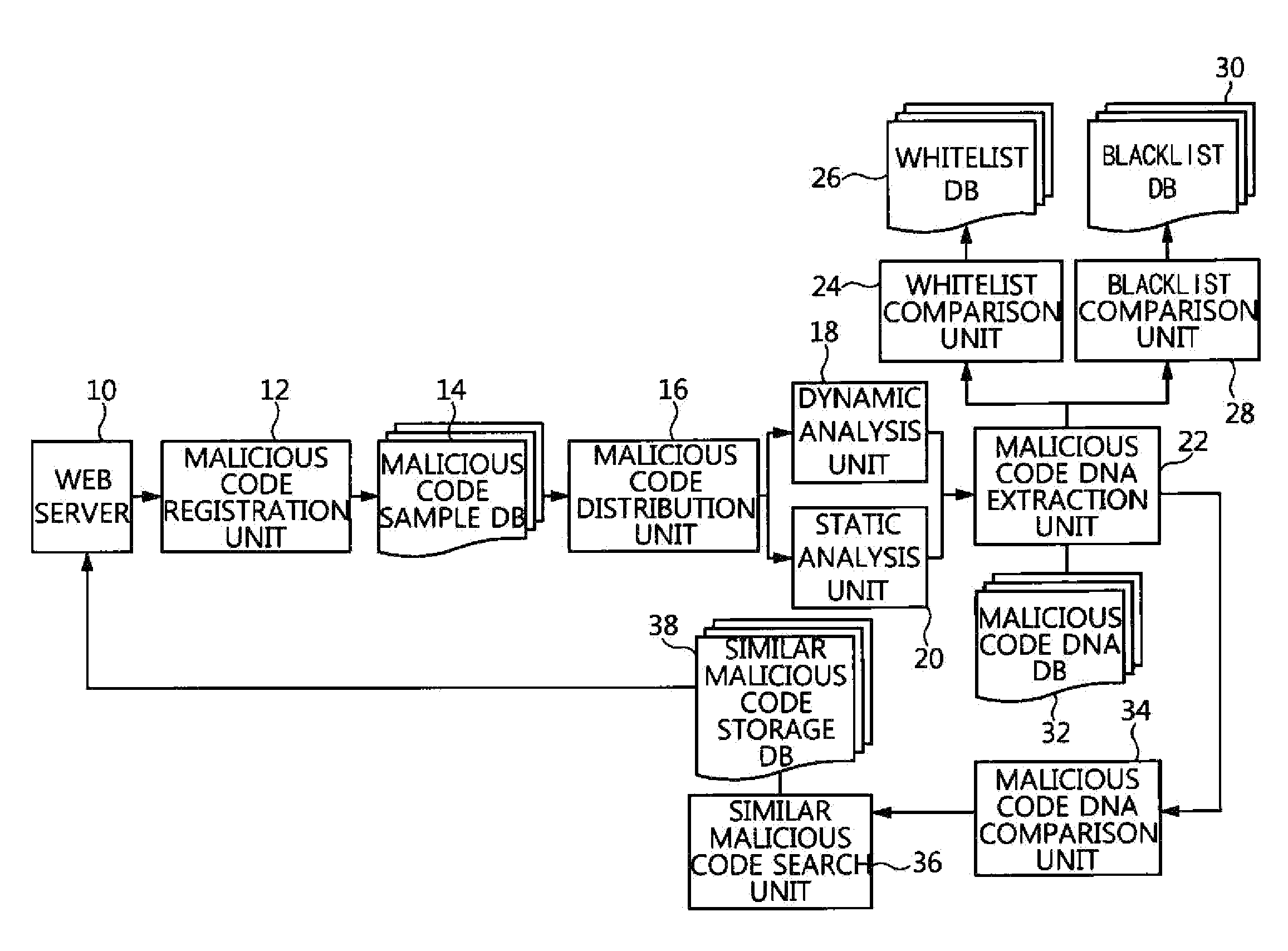
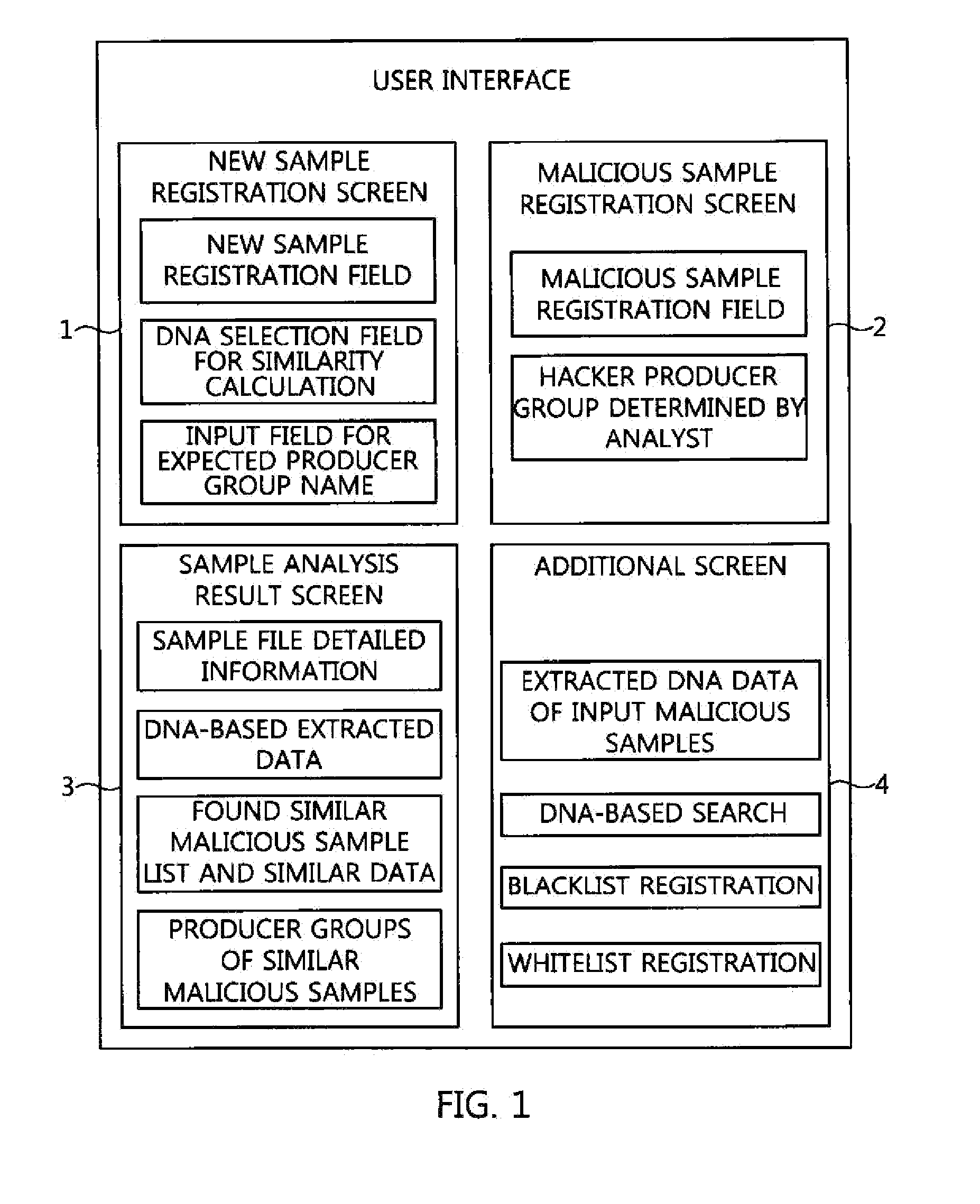
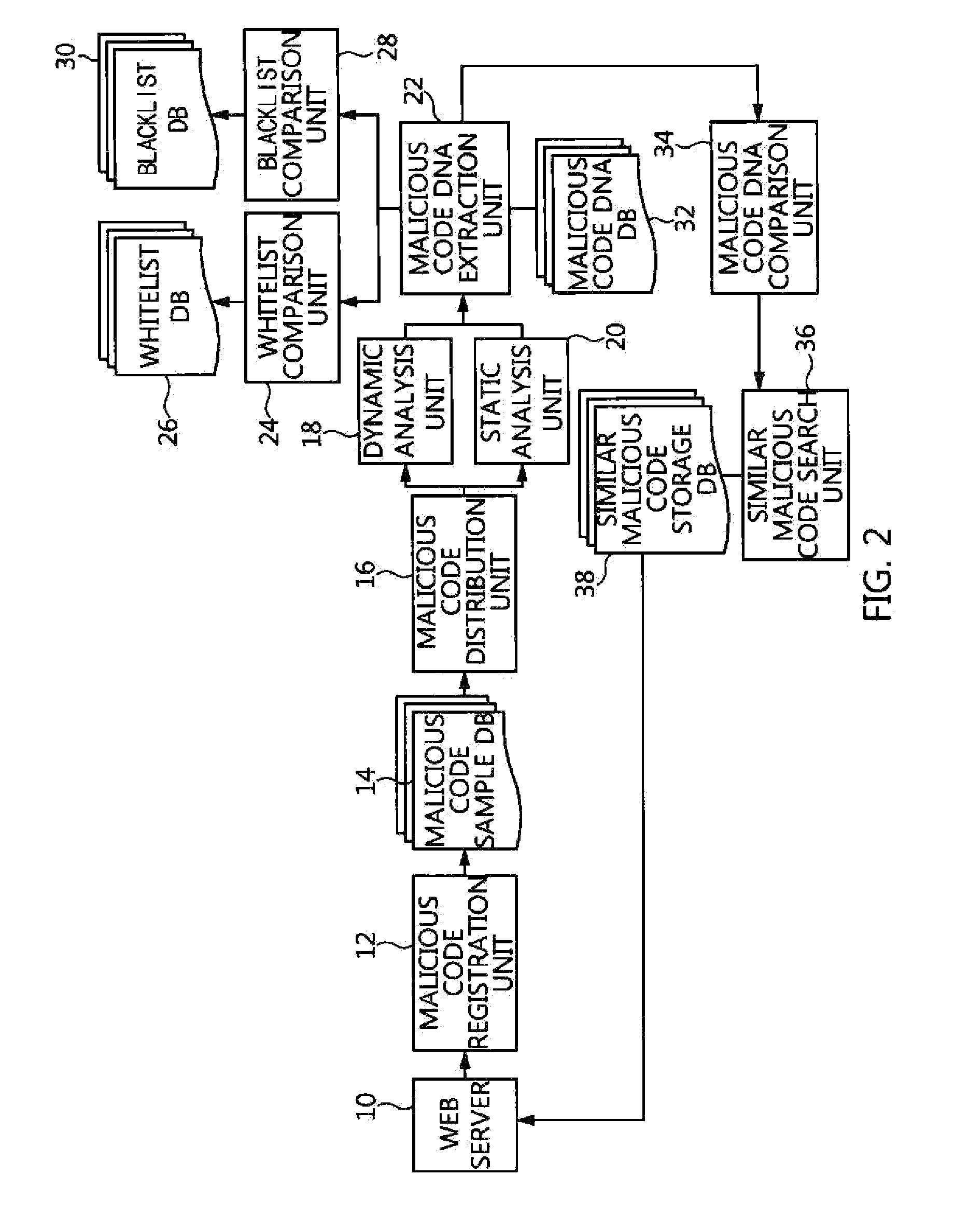
Apparatus and method for searching for similar malicious code based on malicious code feature information
ActiveUS20160072833A1High similarityDigital data information retrievalMemory loss protectionDNA extractionInformation searching
An apparatus and method for searching for similar malicious code based on malicious code feature information. The apparatus includes a malicious code registration unit for registering input new malicious code as a new malicious code sample, and extracting and registering detailed information of the new malicious code sample, a malicious code analysis unit for analyzing the detailed information of the new malicious code sample, a malicious code DNA extraction unit for extracting malicious code DNA information including malicious code feature information, a malicious code DNA comparison unit for comparing the extracted malicious code DNA information with malicious code DNA information of prestored malicious code samples, and calculating similarities therebetween, and a similar malicious code search unit for calculating, based on the calculated similarities, all similarities between the new malicious code sample and prestored malicious code samples, and extracting a specific number of malicious code samples.
Owner:ELECTRONICS & TELECOMM RES INST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com