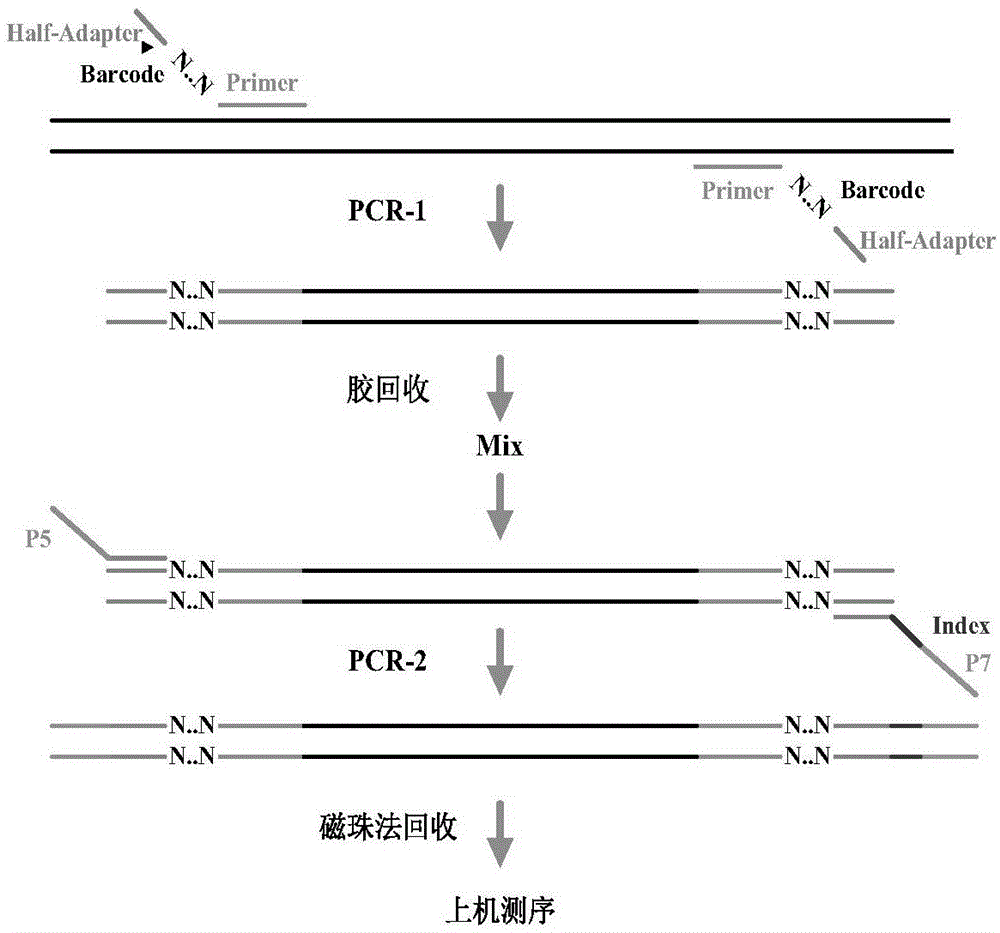
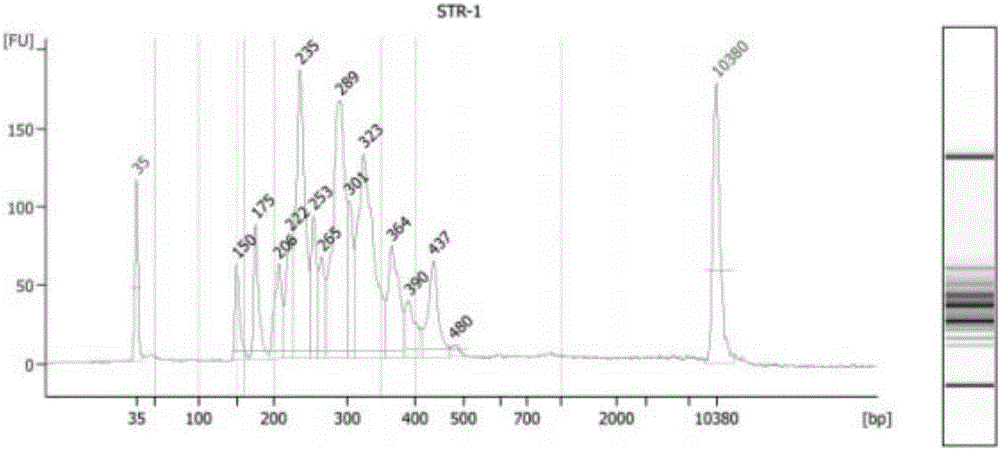
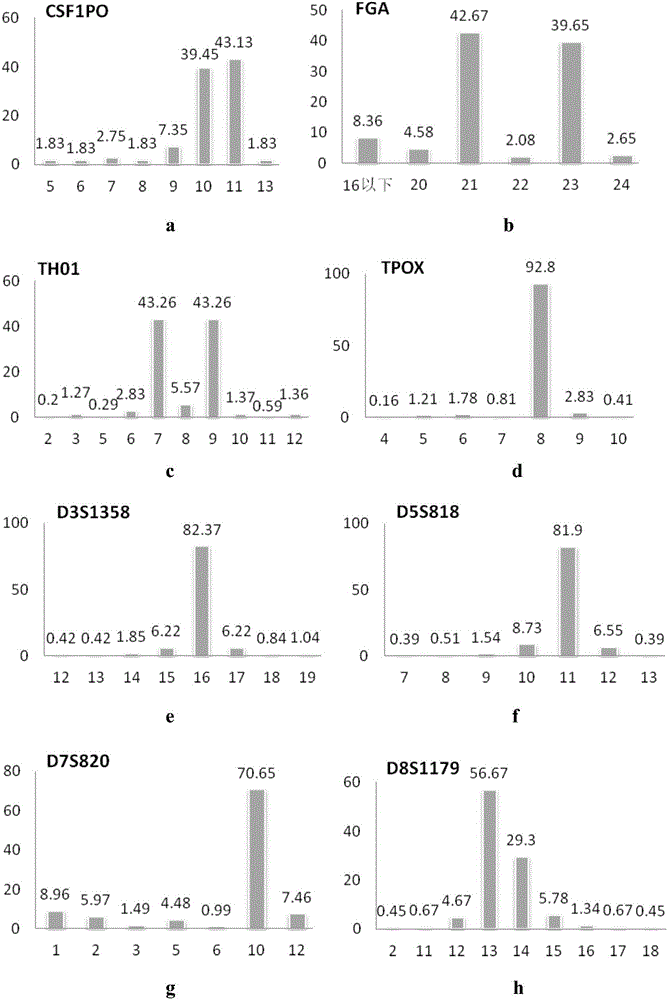
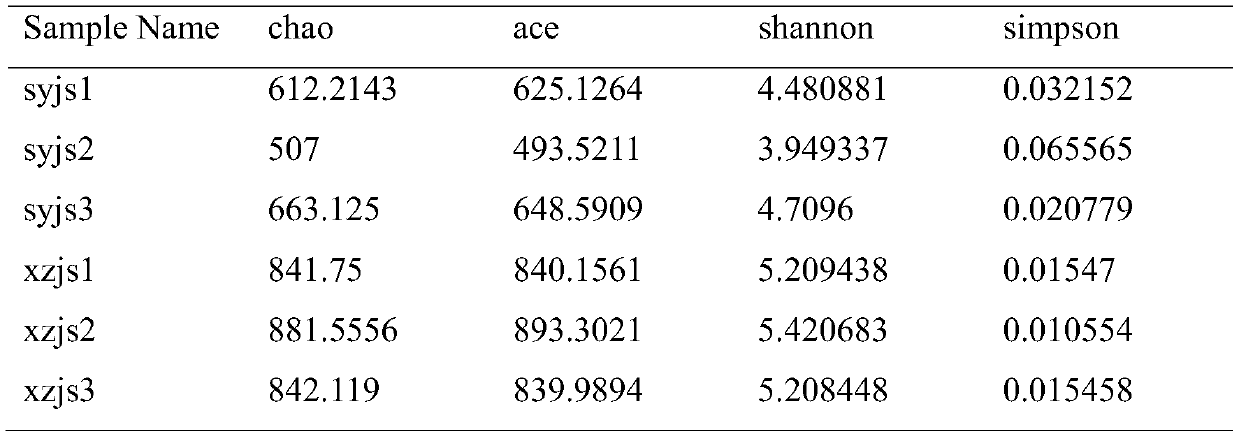
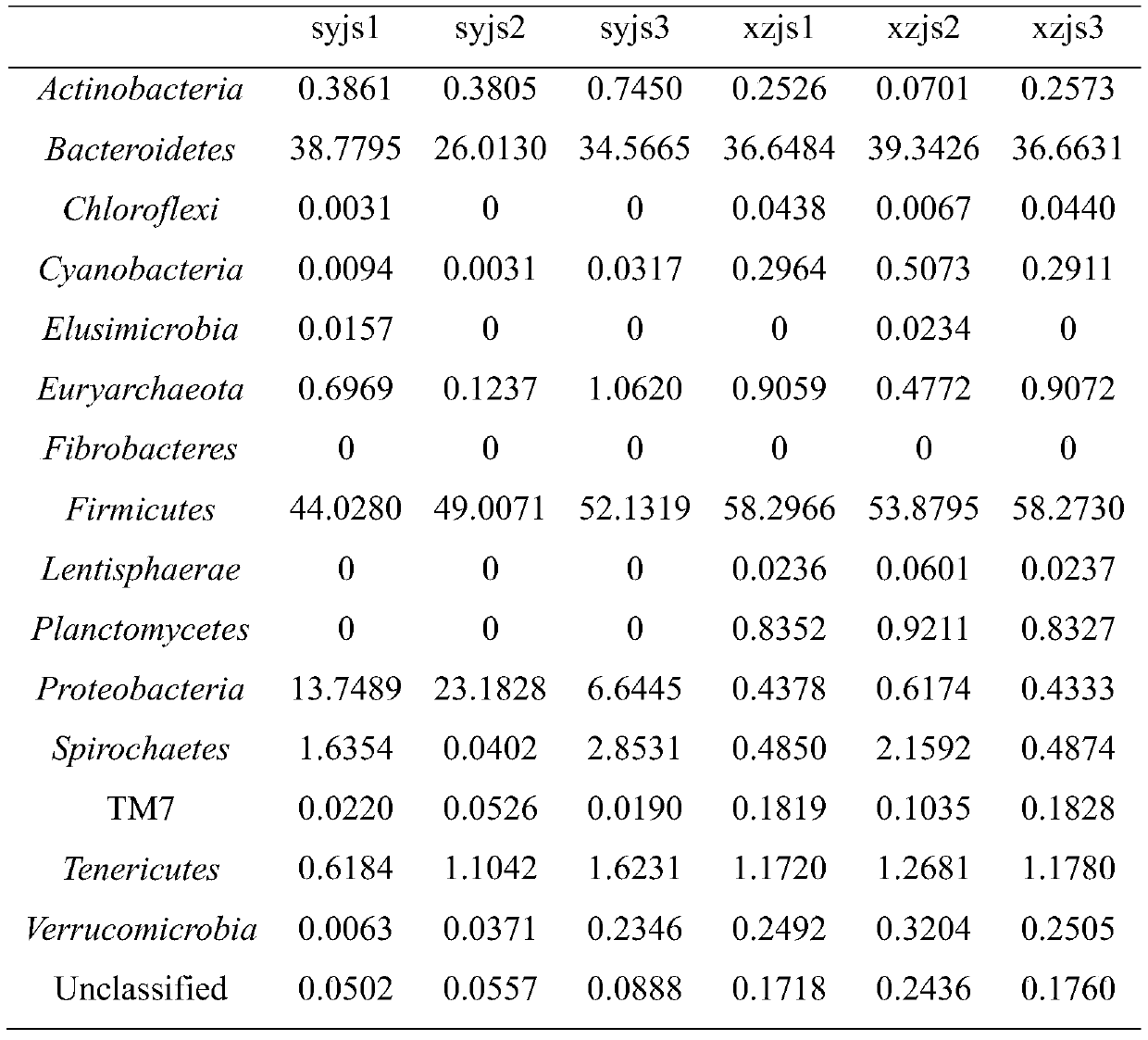
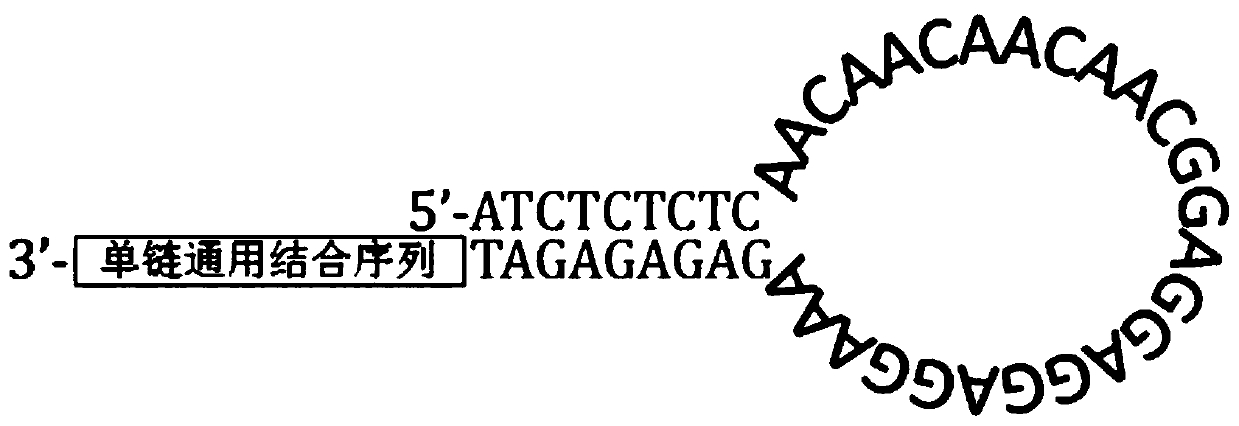Patents
Literature
63 results about "Amplicon sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
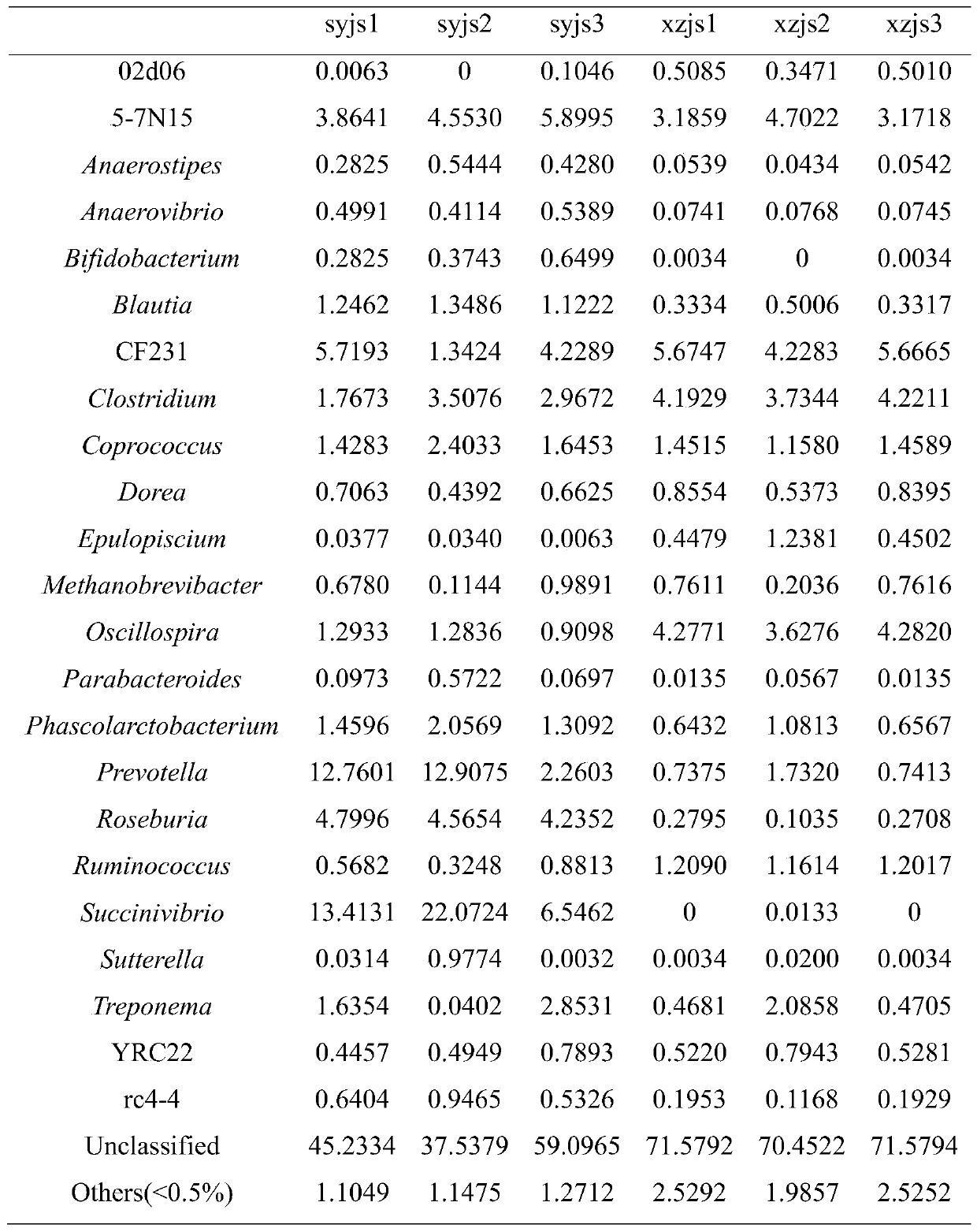
Amplicon sequencing is a highly targeted approach that enables researchers to analyze genetic variation in specific genomic regions.
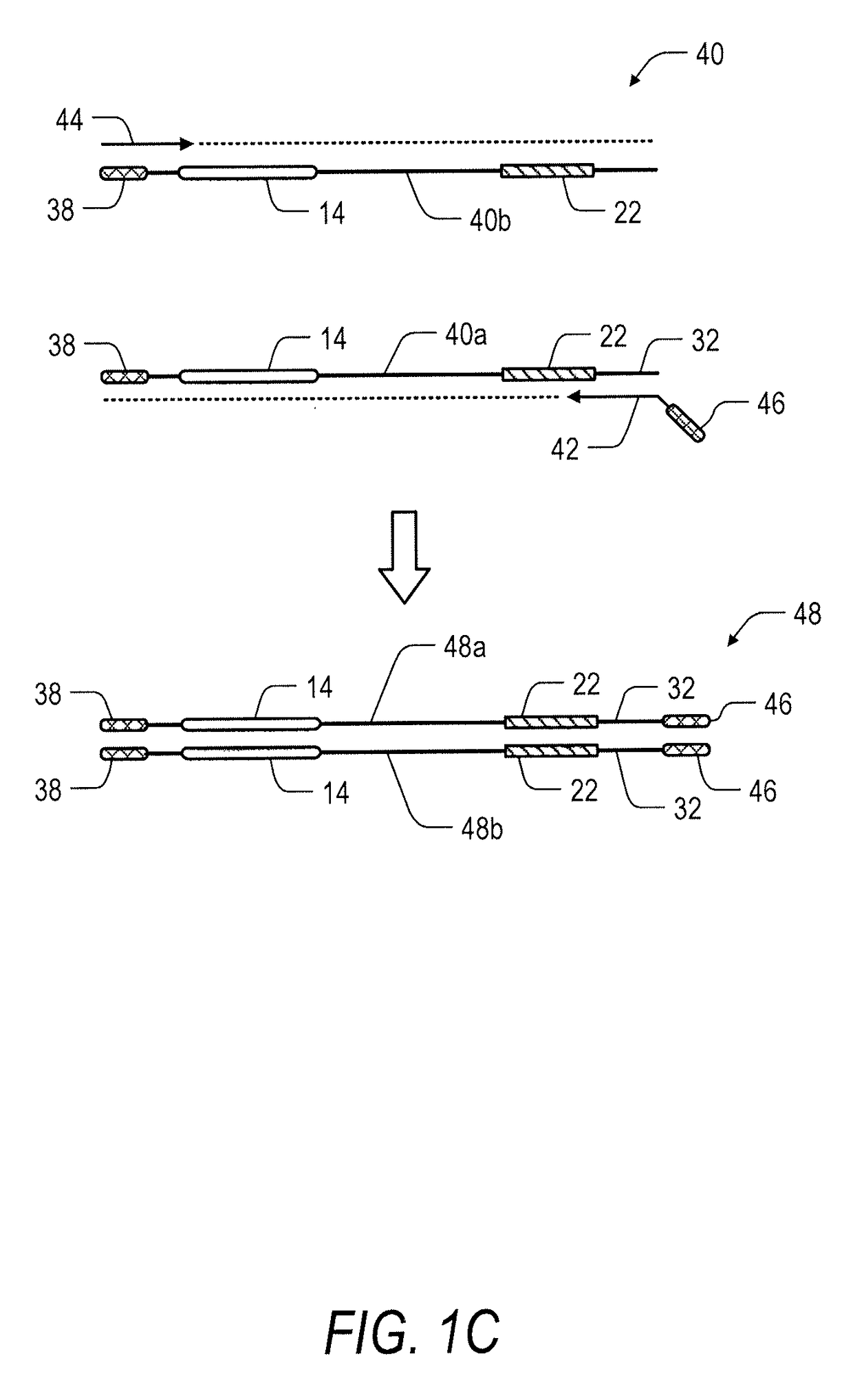
Primer applied to amplicon sequencing library construction and method for constructing amplicon sequencing library
InactiveCN104263726AQuality improvementImprove efficiencyLibrary creationDNA/RNA fragmentationForward primerOn board
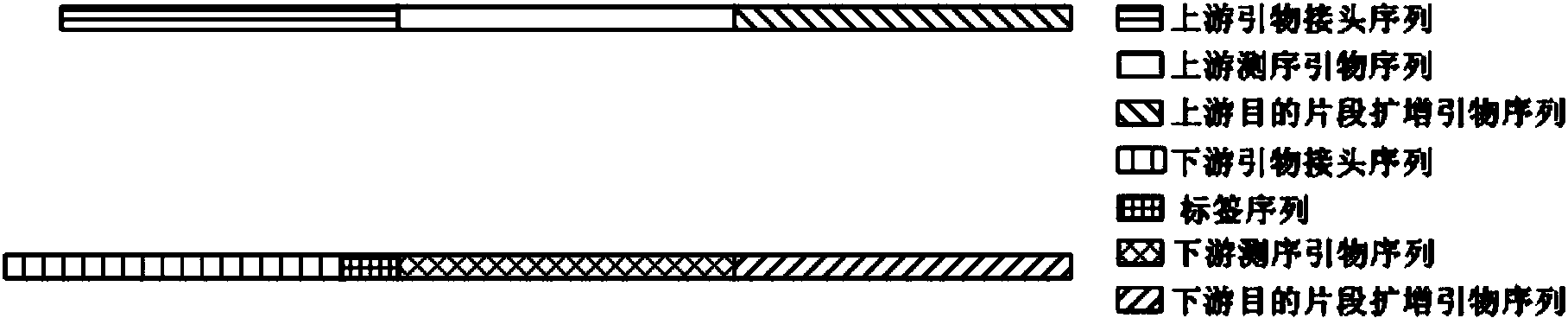
The invention discloses a primer applied to amplicon sequencing library construction and a method for constructing an amplicon sequencing library. The primer consists of a forward primer and a reverse primer, and the forward primer and the reverse primer comprise a joint sequence, a sequencing primer sequence and a target fragment amplification primer sequence from a sequence from the 5' end to the 3' end. According to the primer provided by the invention, the joint sequence, the sequencing primer sequence and the target fragment amplification primer sequence are integrated on a pair of primers, so that library construction can be finished in one step by utilizing the primer. The sequencing library quality and library construction efficiency are improved, the constructed amplicon sequencing library can perform high-throughput sequencing due to the joint sequence and the sequencing primer sequence used on a conventional sequencing platform by utilizing a common reagent for on-board sequencing, and the phenomena that an extra sequencing primer is provided and the mixed sequencing primers of the PHiX library are changed are avoided.
Owner:天津诺禾致源生物信息科技有限公司
Primer applicable to amplicon sequencing library construction, construction method, amplicon library and kit comprising amplicon library
InactiveCN104293783AImprove throughputImprove accuracyNucleotide librariesMicrobiological testing/measurementAmplicon sequencingComputational biology
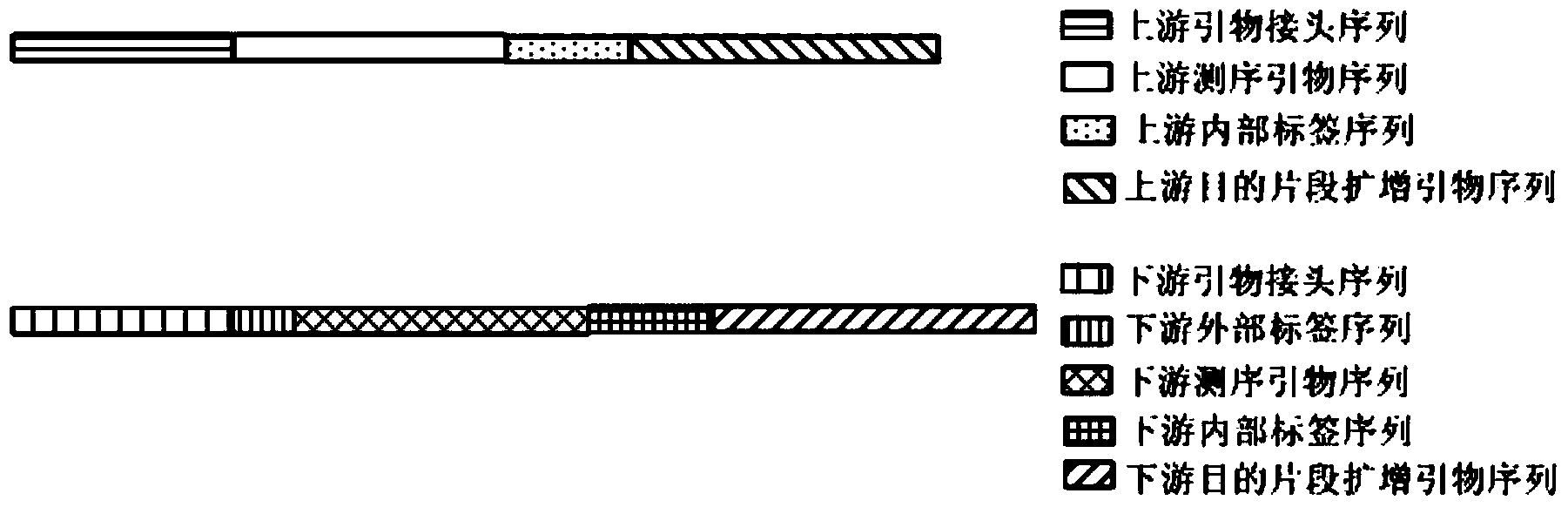
The invention discloses a primer applicable to amplicon sequencing library construction, a construction method, an amplicon library and a kit comprising the amplicon library. The primer comprises a target fragment amplification primer sequence, a sequencing primer sequence and an adaptor sequence, wherein at least one end of a target fragment amplification primer sequence, obtained from the target fragment amplification primer sequence, the sequencing primer sequence and the adaptor sequence by virtue of an amplification step, in the amplicon library is provided with an internal label sequence; the internal label sequence is formed by using 6-10 basic groups in different permutation and combination patterns. The primer enables the constructed amplicon library to have the internal label, and therefore, the number of the mixed samples of the library can be increased and the flux of sequencing can be increased, and besides, contaminated data generated in the library construction process can be differentiated and removed and the accuracy of late data analysis can be improved; as a result, the abundance ratio of different floras in an original sample can be reflected more really.
Owner:天津诺禾致源生物信息科技有限公司
Database-building method for amplicon sequencing
The invention relates to the technical field of high-flux sequencing and discloses a database-building method for amplicon sequencing. The database-building method comprises the following steps of 1, object region enrichment based on PCR amplification: designing primer sequences aiming at an object region of genome DNA of a sample to be detected, wherein ends 5' of a forward primer and a reverse primer in the primer sequences are provided with random base sequences and linking sequences, carrying out PCR amplification and recovering the PCR product, and 2, sequencing linker introduction based in PCR amplification: carrying out mixing on the PCR product obtained by the step 1, designing primer sequences comprising sequences complementary with the linking sequences obtained by the step 1, carrying out PCR amplification on the mixture, and recovering the PCR product so that a DNA database for amplicon sequencing is obtained. The database-building method reduces building time and cost of the DNA database for amplicon sequencing, reduces error of bioinformatics analysis of the sequencing result and improves accuracy and authenticity of the analysis result.
Owner:SHANGHAI MAJORBIO BIO PHARM TECH
Plant endophyte 16S rRNA gene amplification method and application
ActiveCN106282165ABig amount of dataThe test result is completeMicrobiological testing/measurementDNA preparationSequence analysisAmplicon sequencing
The invention discloses a plant endophyte 16S rRNA gene amplification method and application. The amplification method includes the following steps of plant pretreating; plant-sample total DNA extracting; sample 16S rRNA gene amplifying, wherein sample 16S rRNA gene amplifying comprises 16S rRNA gene total-length emulsion PCR amplifying and 16S rRNA gene hypervariable-region / conserved-region amplifying sub-amplifying; high-throughput sequencing and biological information analyzing based on a Illumina platform, wherein high-throughput sequencing and biological information analyzing based on the Illumina platform comprises plant endophyte 16S rRNA gene amplicon purifying and recycling, amplicon sequencing library establishing, Illumina HiSeq sequencing and sequencing data bioinformatics analysis. The gene amplification method is used for high-throughput-sequencing plant disease detection and phytophagous animal enteric microorganism detection. Sequencing analysis of plant endophytic bacteria is carried out with the high-throughput sequencing technology, the data size is larger, the detection result is more complete, pollution of plant hosts is reduced to the maximum degree, the result multiformity is higher, and the cost is low.
Owner:成都罗宁生物科技有限公司
Amplicon next-generation sequencing based small fragment insertion and deletion detection method and device
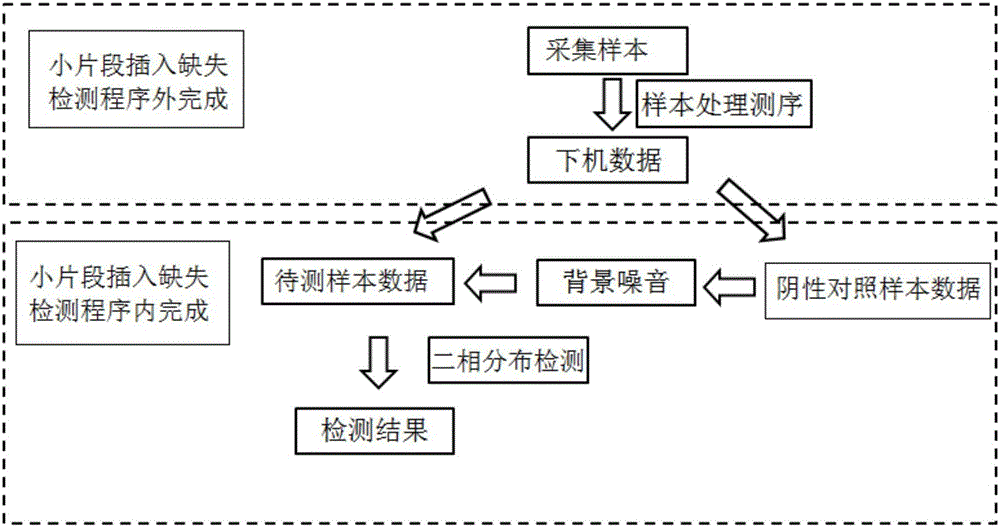
ActiveCN106355045ANo false positive detectionsHybridisationSpecial data processing applicationsReference genesSmall fragment
The invention discloses amplicon next-generation sequencing based small fragment insertion and deletion detection method and device. The amplicon sequencing based small fragment insertion and deletion detection method includes the steps of S1, respectively extracting DNA of a to-be-detected sample and a negative control sample, and amplifying a target region of a mutation related to the target trait by multiple amplicons; S2, carrying out the next-generation sequencing to obtain sequence of the target region; S3, comparing the sequence of the target region with a reference genome, wherein bases which are not matched with the reference genome are subjected to mispairing; S4, processing to obtain background noise of hotspot small fragment insertion and deletion related to the target trait according to the negative control sample, modeling the background noise by means of binomial distribution, distinguishing the hotspot short fragment insertion and deletion and the background noise, on each base, of the to-be-detected sample, and determining the mispairing bases as the small fragment insertion and deletion through positive detection if the proportion of the mispairing bases of the to-be-detected sample to the reference genotype is much different from that of the background noise. By the amplicon next-generation sequencing based small fragment insertion and deletion detection method and device, accuracy of small fragment insertion and deletion detection is improved.
Owner:天津诺禾致源生物信息科技有限公司
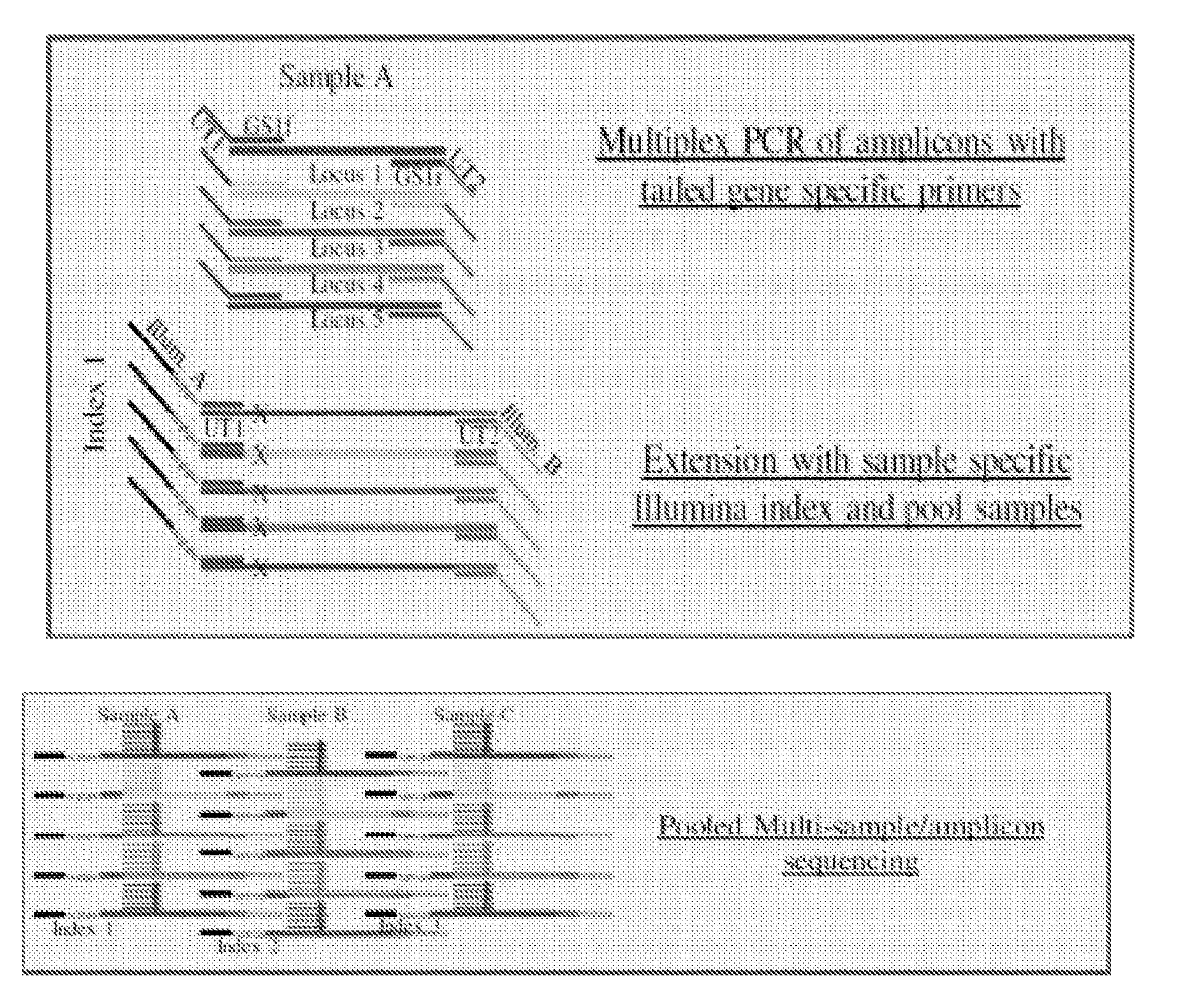
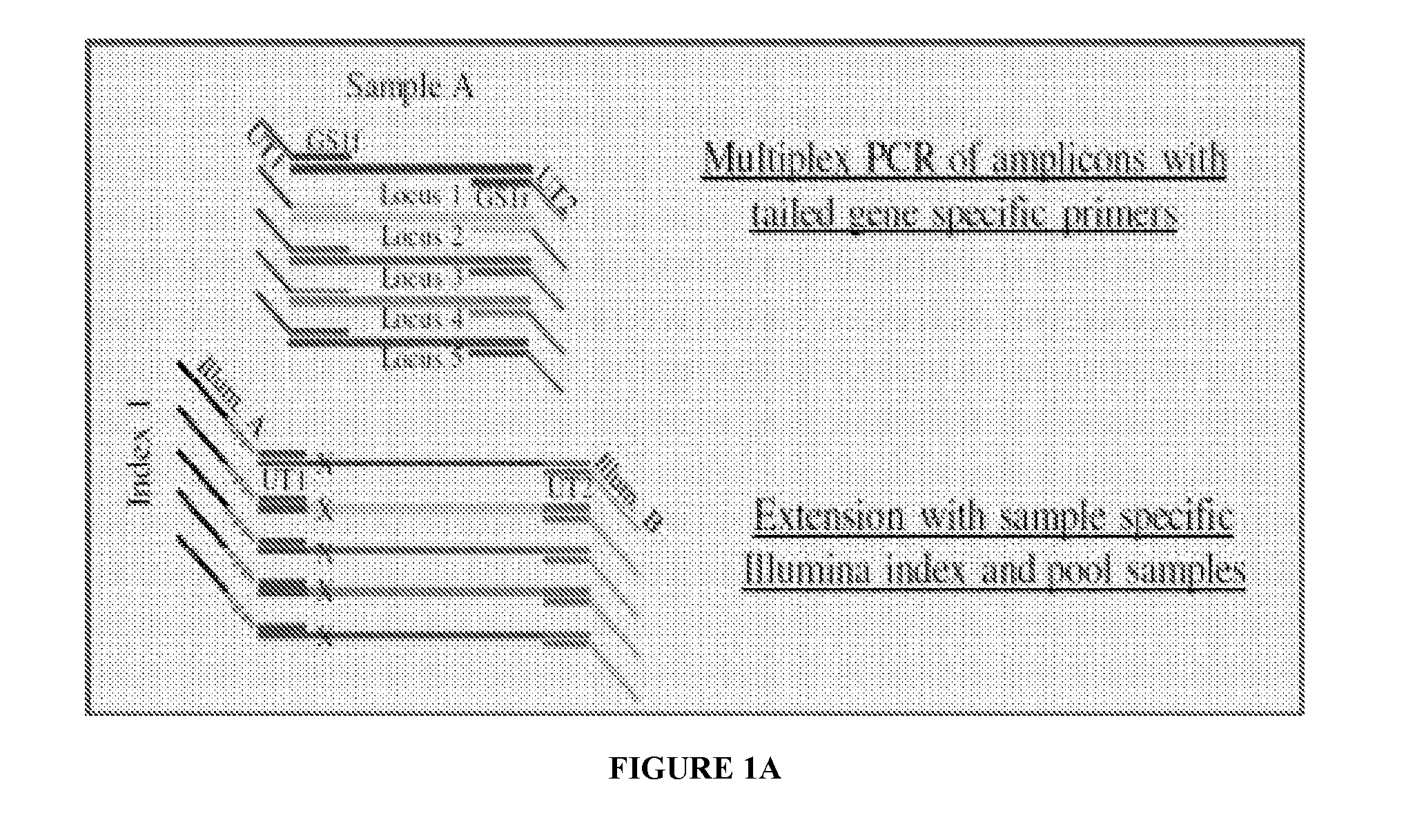
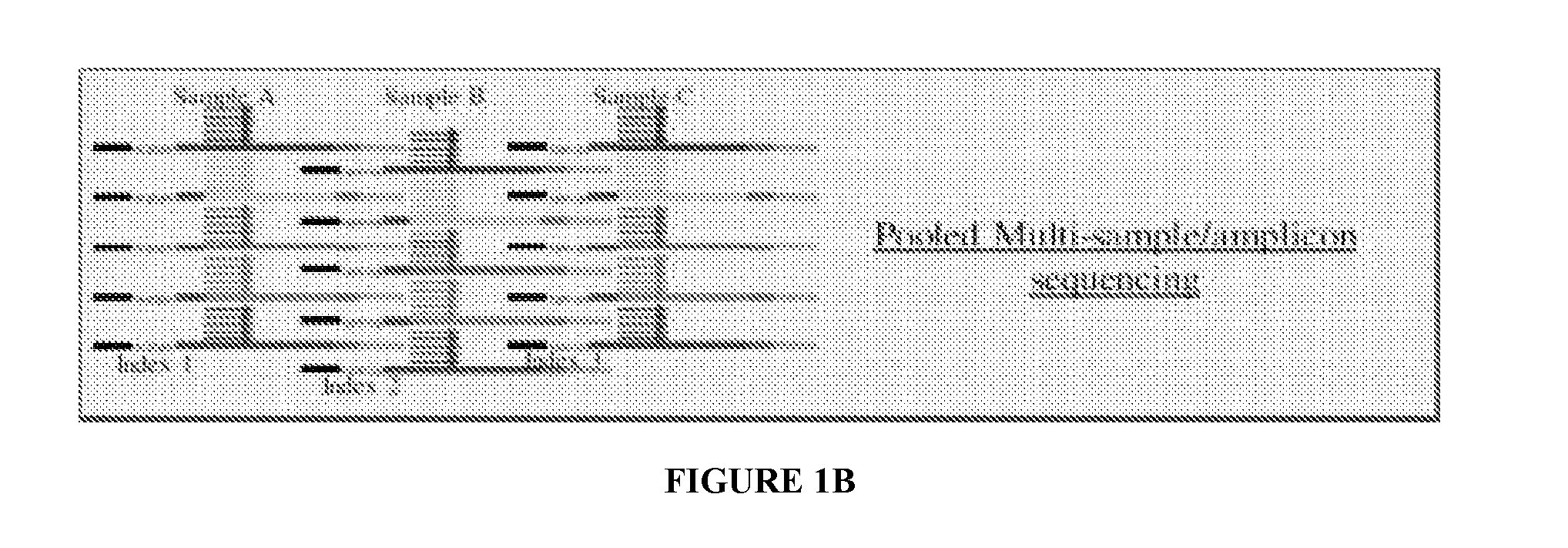
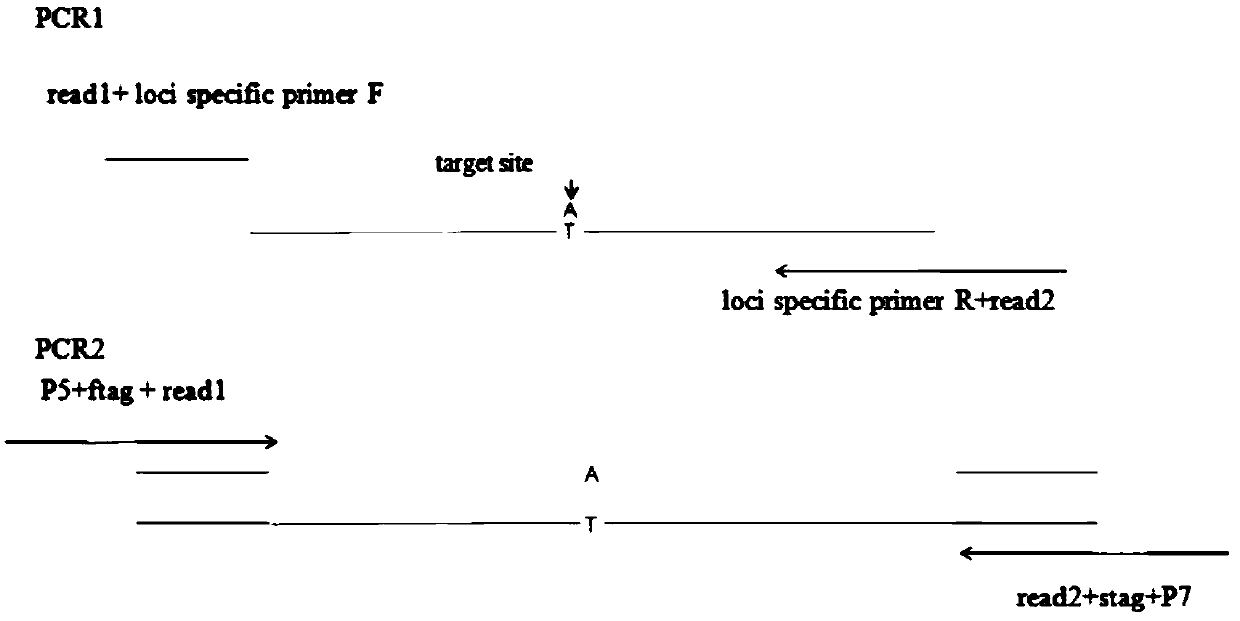
Systems and methods for universal tail-based indexing strategies for amplicon sequencing
Some embodiments of the invention include a method of preparing a sample for sequencing that includes receiving a sample and amplifying at least one marker within the sample. In some embodiments, amplification of the first marker may include mixing the sample with a first oligonucleotide that comprises a first universal tail sequence and a second oligonucleotide that comprises a second universal tail sequence. In some aspects of the invention, the first universal tail sequence and the second universal tail sequence are different sequences.
Owner:TRANSLATIONAL GENOMICS RESEARCH INSTITUTE +1
DNA library construction method for high-throughput sequencing
ActiveCN106350590AShorten the timeLow costMicrobiological testing/measurementLibrary creationAgricultural scienceTest sample
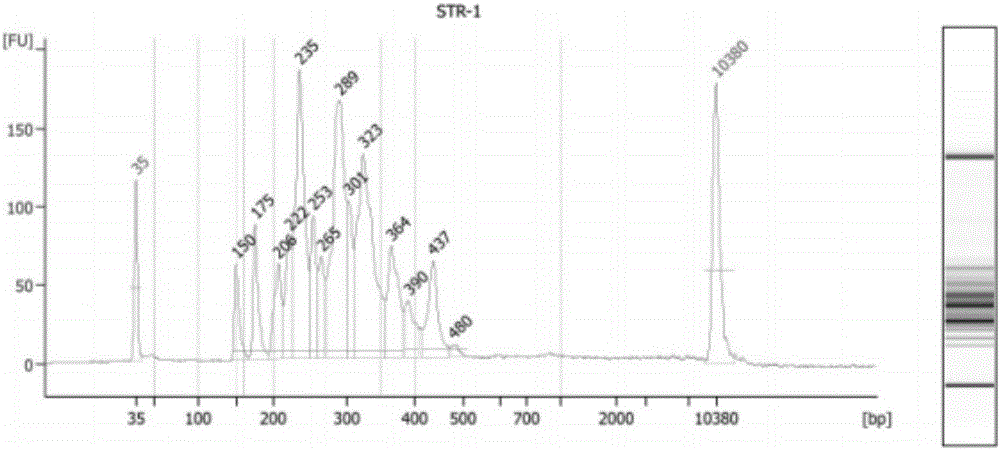
The invention discloses a DNA library construction method for high-throughput sequencing. The DNA library construction method includes the steps of carrying out multiple PCR (polymerase chain reaction) to a DNA template obtained from a to-be-tested sample through multiple pairs of fusion primers, then recycling PCR products to obtain a sequencing library. The multiple pairs of fusion primers respectively aim at different target fragments of the DNA template, and each fusion primer comprises specificity primer sequence aiming at the target fragment and linker sequence for sequencing. The multiple PCR is carried out on reaction conditions: reacting at 95 DEG C for 2 minutes; reacting within 38 cycles, wherein each cycle includes processes of preserving at the temperature of 95 DEG C for 30 seconds, cooling to the temperature from 76 DEG C to 55-58 DEG C slowly at the speed of 0.1 DEG C each second, holding the temperature for 20 seconds once cooling to the temperature of 58 DEG C, and then preserving at 72 DEG C for 30 seconds; after that, preserving at 72 DEG C for 2 minutes, and finally preserving at 4 DEG C. The invention further discloses application of the library construction method in testing STR locus and paternity identification on the basis of high-throughput sequencing.
Owner:承启医学(深圳)科技有限公司
Method for predicting health and immunity levels of introduced milk cows based on intestinal flora
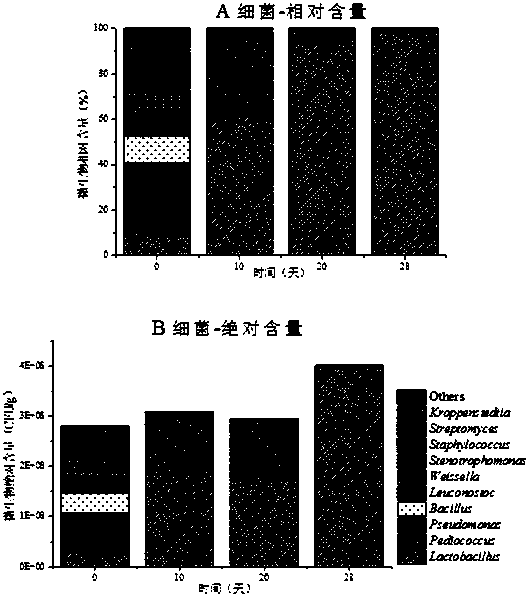
The invention relates to the technical field of biotechnology, in particular to a method for extracting and analyzing the DNA of introduced milk cow feces samples to predict the health and immunity levels of the introduced milk cows. The method comprises the processes such as sample selection, 16S amplicon sequencing, sequencing data analysis, intestinal flora diversity and abundance analysis, user report push and the like. According to the method, a monitoring index of intestinal flora diversity and abundance analysis is linked with the health and immunity levels of the introduced milk cows,after experiment studies and data analysis, fresh introduced (fewer than half a year) and early introduced (more than half a year) milk cow feces samples are collected, a hypervariable region is amplified, and the method for predicting the health and immunity levels of the introduced milk cows is realized. The samples are easy to collect, the analysis speed is rapid, the cost is low, and the assessment results are consistent with the existing studies, making the method for predicting the health and immunity levels of the introduced milk cows based on the intestinal flora possible.
Owner:DALIAN UNIV
Molecular marker detection method based on next-generation sequencing technology
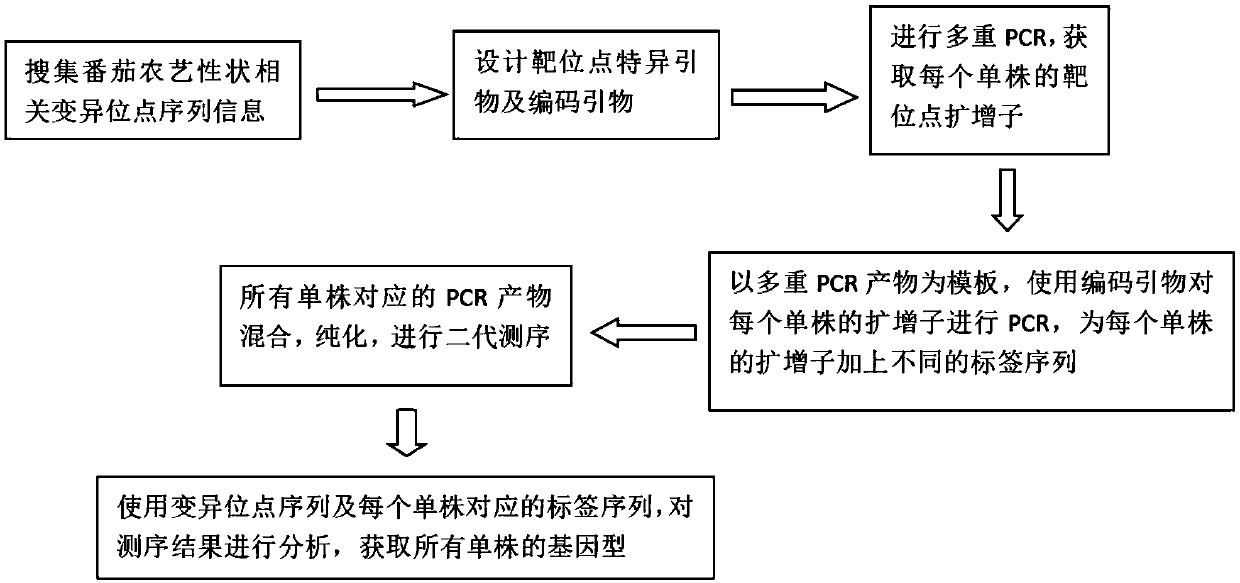
InactiveCN109022559AQuick checkAccurate detectionMicrobiological testing/measurementAgricultural scienceGenotype
The invention belongs to the technical field of molecular marker-assisted breeding, and discloses a molecular marker detection method based on a next-generation sequencing technology. Amplicons of allsingle plants at important agronomic trait-related loci are obtained at a time through multiple PCR by using designed specific primers of the multiple loci and a PCR reaction system and an analysis process according to the principle of amplicon sequencing, primers containing different tag sequences are used to carry out one-time amplification on the amplicons, the amplicons of all the plants areexpanded. With the different tag sequences, the amplicons undergo next-generation sequencing, and sequencing results are analyzed to obtain the genotypes of all the single plants at a time. A high-throughput detection system constructed in the invention is used to quickly and accurately detect the disease-resistant quality-related genes which are widely used in existing breeding practices.
Owner:HUAZHONG AGRI UNIV
Primer set for building 16S rRNA gene amplicon sequencing library and building method
ActiveCN107829146AIncrease diversityReduce wasteMicrobiological testing/measurementLibrary creationAmplicon sequencingBase sequence
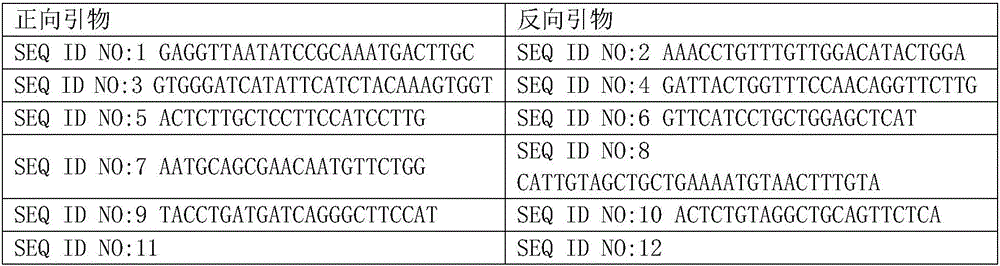
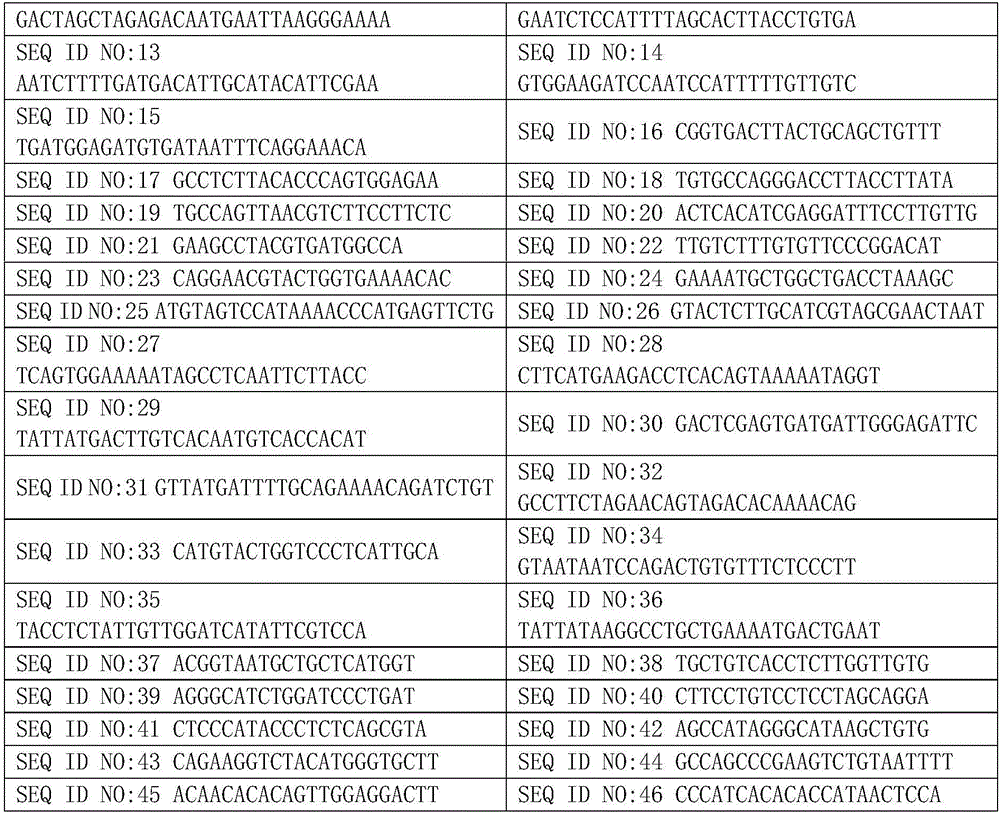
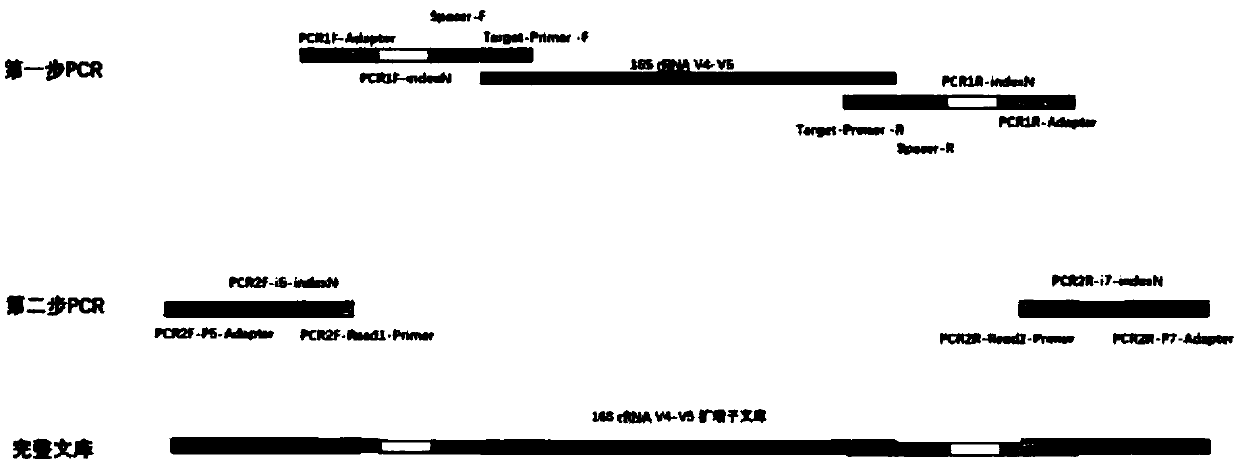
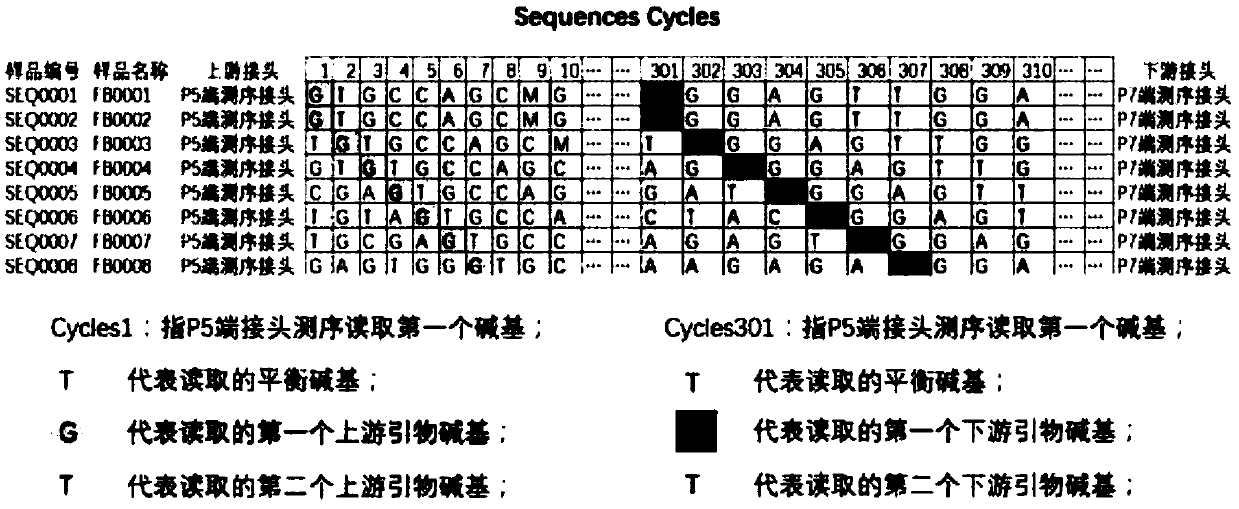
The invention discloses a quadruple-tag primer set for building a 16S rRNA gene amplicon sequencing library. The quadruple-tag primer set is composed of a first-step PCR double-tag primer pair and a second-step PCR double-tag primer pair, the sequence of an upstream primer of the first-step PCR double-tag primer pair is one of SEQ ID NO:1-8 while that of a downstream primer of the same is one of SEQ ID NO:9-20, and the sequence of an upstream primer of the second-step PCR double-tag primer pair is one of SEQ ID NO:21-44 while that of a downstream primer of the same is one of SEQ ID NO:45-68. Abalanced base sequence is added into the quadruple-tag primer, so that diversity of the library is improved, and sequencing quality is improved; during online sequencing, only a small amount of PhiXlibrary needs to be mixed, and staggered sequencing needs to be performed, so that waste of sequencing flux is reduced, and sequencing quality is ensured.
Owner:湖南赛哲智造科技有限公司
Sensitive detection of bacteria by improved nested polymerase chain reaction targeting the 16S ribosomal RNA gene and identification of bacterial species by amplicon sequencing
InactiveUS7309589B2Easy to detectRaise the possibilitySugar derivativesMicrobiological testing/measurementConserved sequenceGenetics
A method for identifying an RNA form of a bacteria, comprising reverse transcribing RNA material; conducting PCR using primers for a first highly conserved genetic sequence generic of the bacteria; conducting nested PCR using primers for a second highly conserved genetic sequence within the first genetic sequence of the bacteria; and identifying the bacteria based on unconserved amplified sequences linked to the conserved sequences.
Owner:VIRONIX
Absolute quantitative method for microflora based on multi-internal standard system
ActiveCN110305979AQuantitatively effectiveReduce mistakesMicrobiological testing/measurementSequence analysisMicroorganismInternal standard
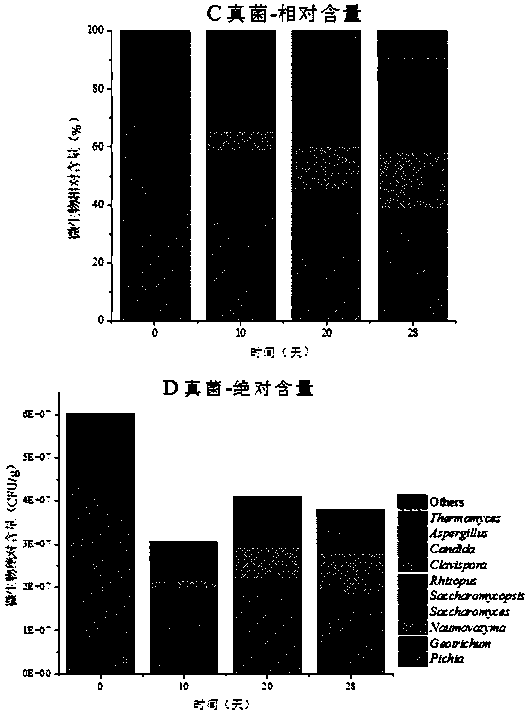
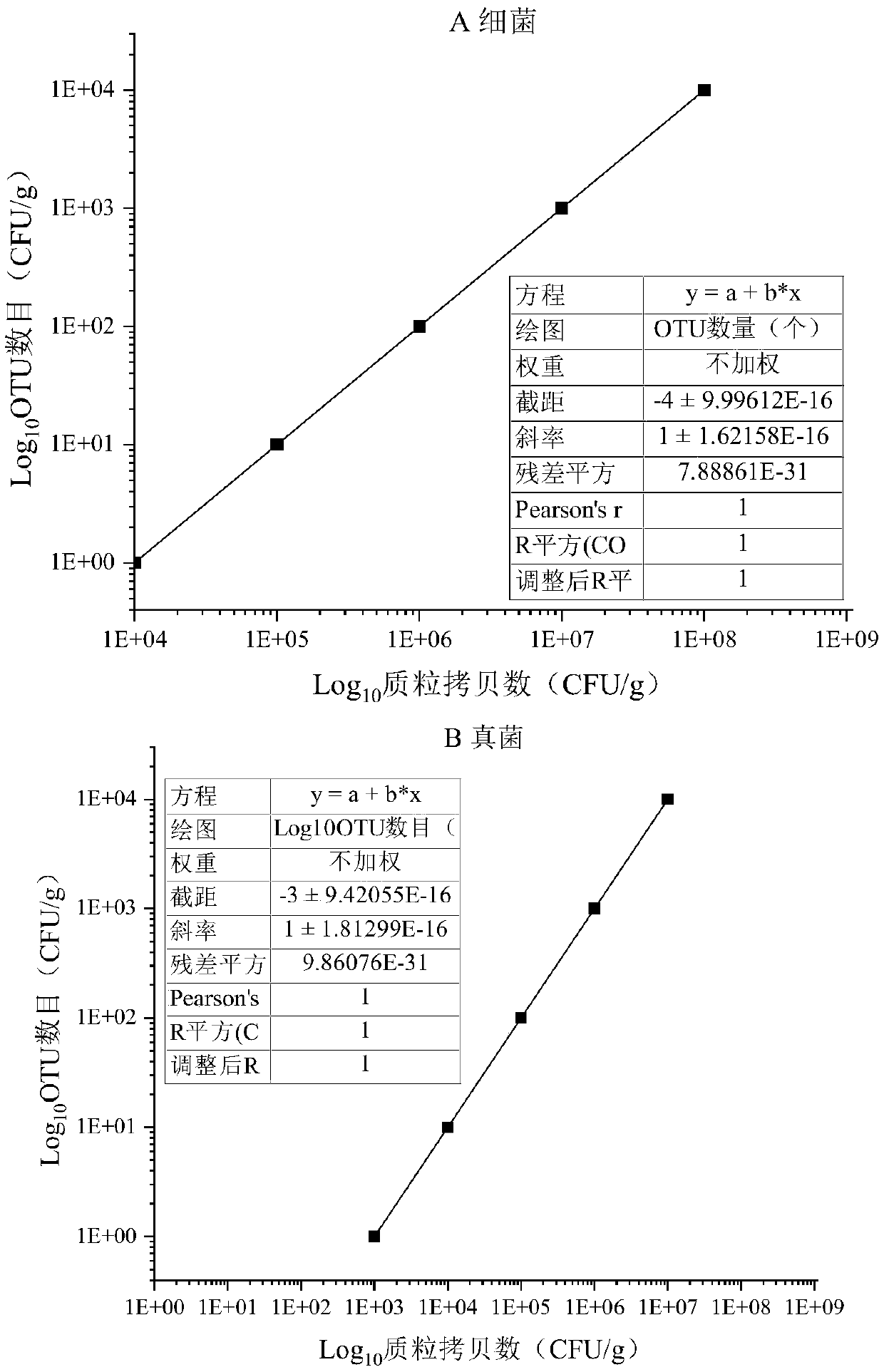
The invention discloses an absolute quantitative method for microflora based on a multi-internal standard system, which belongs to the field of biotechnology and omics analysis. The present inventionprovides a method for efficiently quantifying microbes during amplicon sequencing, the method is characterized in that the target plasmid with a corresponding gradient is added according to the copy number of the total amount of microbes in a sample, the steps of DNA separation, quality control, and sequencing are carried out to obtain an OTU classification unit, and the absolute number of corresponding microbes in the sample is calculated based on a proportional relationship between the amount of target substance added and the detection amount. The method can realize the absolute quantification of the sample to-be-tested, greatly improves the accuracy of the measurement result, and has the advantages of low cost and the like, and plays an important role in promoting the progress of a sequencing technology.
Owner:JIANGNAN UNIV
Library building kit for high flux detection of STR genetic markers
ActiveCN106399496AFast Downstream Forensic ApplicationsLow costMicrobiological testing/measurementLibrary creationHigh fluxA-DNA
The invention provides a library building kit for high flux detection of STR genetic markers. The kit comprises multiple pairs of fusion primers for amplifying target STR loci, each pair of fusion primer respectively aims at different STR loci on a DNA template, and each pair of fusion primer comprises a sequencing joint sequence and a specific primer sequence which aims at the STR locus from a 5'end to a 3'end in order. The kit can be used for realizing construction of a STR locus amplicon sequencing library by one-time amplification, the operation process of library building is simplified, time and cost for building the library are reduced, and sequencing quality is not influenced.
Owner:承启医学(深圳)科技有限公司
Construction method of amplicon library for detecting low-frequency mutation of target gene
PendingCN107604045AEasy to operateHigh standardNucleotide librariesMicrobiological testing/measurementA-DNAAmplicon sequencing
The invention discloses a construction method of an amplicon library for detecting low-frequency mutation of a target gene. The construction method of the amplicon library for detecting the low-frequency mutation of the target gene, provided by the invention, comprises the following steps: 1) designing and synthesizing a Barcode primer F1, an upstream primer F2, a downstream outer primer R1 and adownstream inner primer R2; 2) carrying out further PCR (Polymerase Chain Reaction) amplification on a cfDNA (cell-free Deoxyribonucleic Acid) of a sample to be detected by utilizing the Barcode primer F1, the upstream primer F2, the downstream outer primer R1 and the downstream inner primer R2, so as to obtain an amplified product, namely a DNA library for amplicon sequencing. By adopting the method disclosed by the invention, a tissue sample can be detected and different regions of free DNAs in samples including blood, urine, cerebrospinal fluid and the like can be rapidly, conveniently, sensitively and specifically subjected to target amplification and mutation which is as low as a 0.1 percent level is efficiently detected; the experiment operation is greatly simplified, the library loss and pollution are effectively avoided, the cost is remarkably reduced and the efficiency is improved.
Owner:GENETRON HEALTH (BEIJING) CO LTD +1
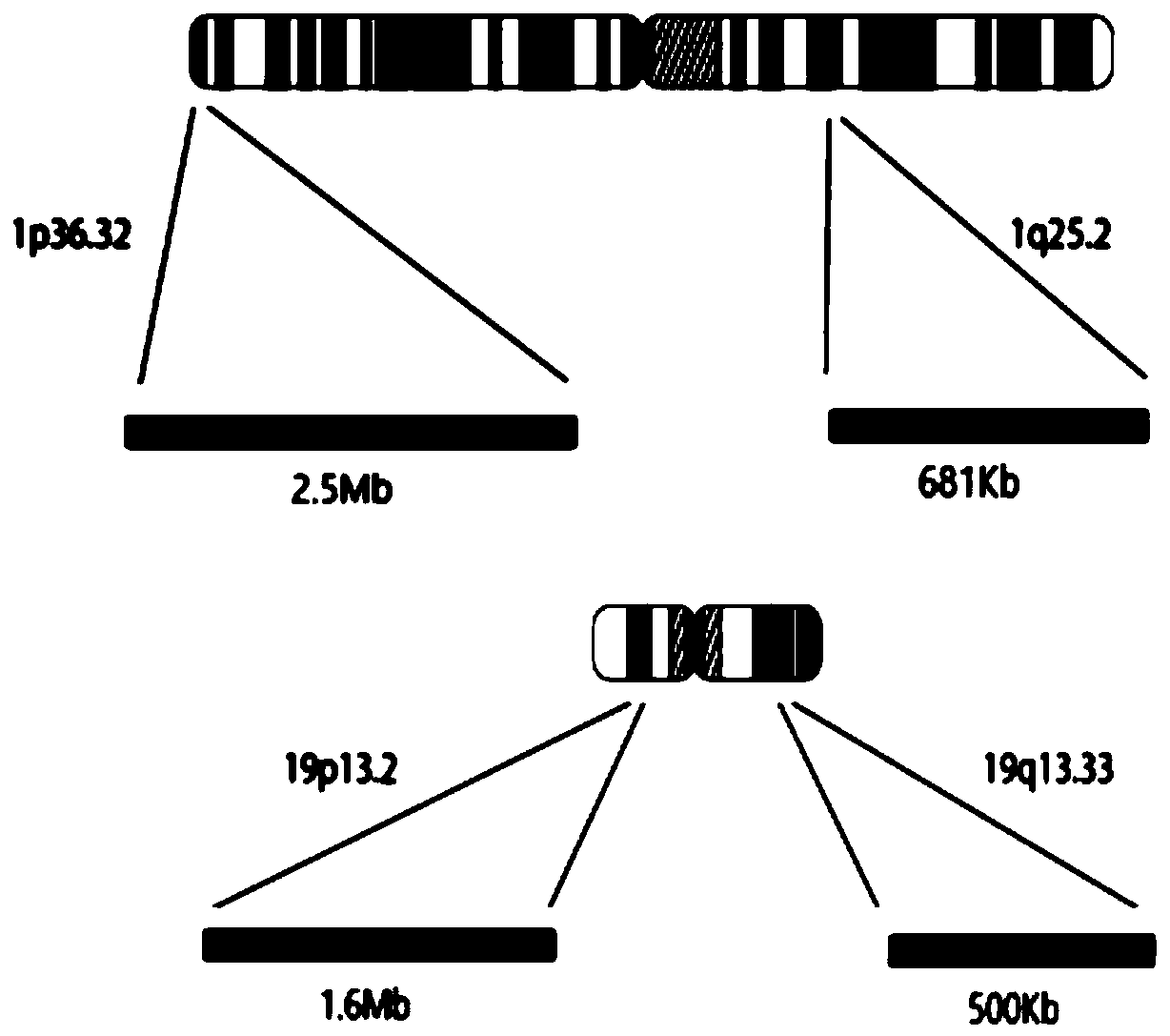
Kit and method for detecting chromosome heterozygosity loss on basis of amplicon sequencing
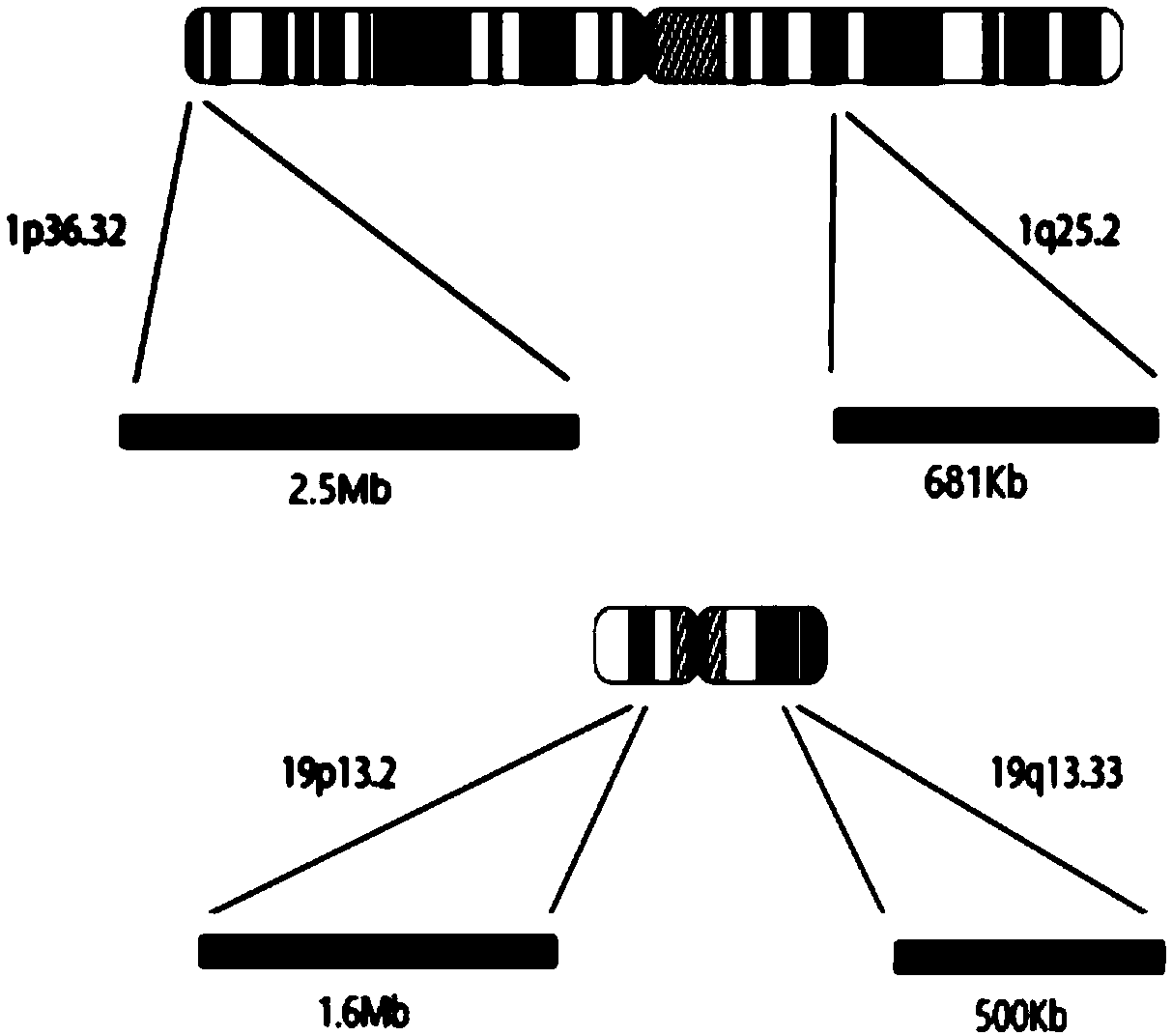
ActiveCN109504770AComprehensive, true and accurate responseReduce the amount of solutionMicrobiological testing/measurementDNA/RNA fragmentationNucleic acid detectionChromosome regions
The invention discloses a kit and method for detecting chromosome heterozygosity loss on the basis of amplicon sequencing, belongs to the field of molecular biology nucleic acid detection, and relatesto the kit and method for detecting chromosome heterozygosity loss by means of amplicon sequencing. The kit contains a mixture of 14 groups of primer pairs used for detecting a 1p36.32 region of a human chromosome 1, 6 groups of primer pairs used for detecting a 1q25.2 region of the human chromosome 1, 14 groups of primer pairs used for detecting a 19p13.2 region of a human chromosome 19 and 7 groups of primer pairs used for detecting a 19q13.33 region of the human chromosome 19, 20 kinds of tag primers used for distinguishing samples, a GC stabilizer, a PCR enzyme mixture, purifying magneticbeads, a quality control standard substance and ddH2O. Through two-time PCR amplification and two-time product purification and sequencing, through a ratio of READS(1p) / READS(1q) to READS(19q) / READS(19p), the heterozygosity loss state of 1p19q in the samples is analyzed. By adopting the method, the variation situation of the 1p19q can be reflected comprehensively, truly and accurately, and gene variation of the 96 samples can be subjected to high-throughput detection and analysis simultaneously, so that the detection cost is greatly reduced.
Owner:艾普拜生物科技(苏州)有限公司
Modeling method of non-rodent animal model for studying intestinal flora
InactiveCN107898796AShorten the timeReduce mortalityMicrobiological testing/measurementSaccharide peptide ingredientsKanamycinMortality rate
The invention discloses a modeling method of a non-rodent animal model for studying intestinal flora. A male rhesus monkey of 12-months old is taken as a laboratory animal and fed with mixed antibiotic for 21 days to clear symbiotic intestinal flora in the body. The antibiotic includes neomycin, streptomycin, ampicillin sodium, kanamycin, metronidazole and vancomycin; different antibiotic formulasare adopted for the first 10 days and later 11 days of feeding. The modeling result is verified by combining a Realtime-PCR method and an amplicon sequencing method. The method disclosed by the invention has the advantages of simplicity and easiness in implementation, short time, high repeatability, low death rate and stable model. The method has an important application value in the treatment offlora-related metabolic diseases, inflammatory bowel diseases and infectious diseases, evaluation of intestinal flora in the immune system, development of new medicines and the like.
Owner:INST OF MEDICAL BIOLOGY CHINESE ACAD OF MEDICAL SCI
System and method for transposase-mediated amplicon sequencing
InactiveUS20180016632A1Microbiological testing/measurementDNA preparationTarget enrichmentAmplicon sequencing
The present invention provides a method for targeted enrichment of nucleic acids including contacting a nucleic acid including at least one region of interest with a plurality of transposase complexes. Each of the transposase complexes includes at least a transposase and a first polynucleotide having a transposon end sequence and a first label sequence. The method further includes incubating the nucleic acid and the transposase complexes under conditions whereby the nucleic acid is fragmented into a plurality of nucleic acid fragments including first polynucleotide attached to each 5′ end of the nucleic acid fragments. The method further includes selectively amplifying the nucleic acid fragments, thereby enriching for a portion of the nucleic acid fragments including the at least one region of interest relative to a remaining portion of the nucleic acid fragments, and sequencing the enriched nucleic acid fragments.
Owner:KAPA BIOSYST
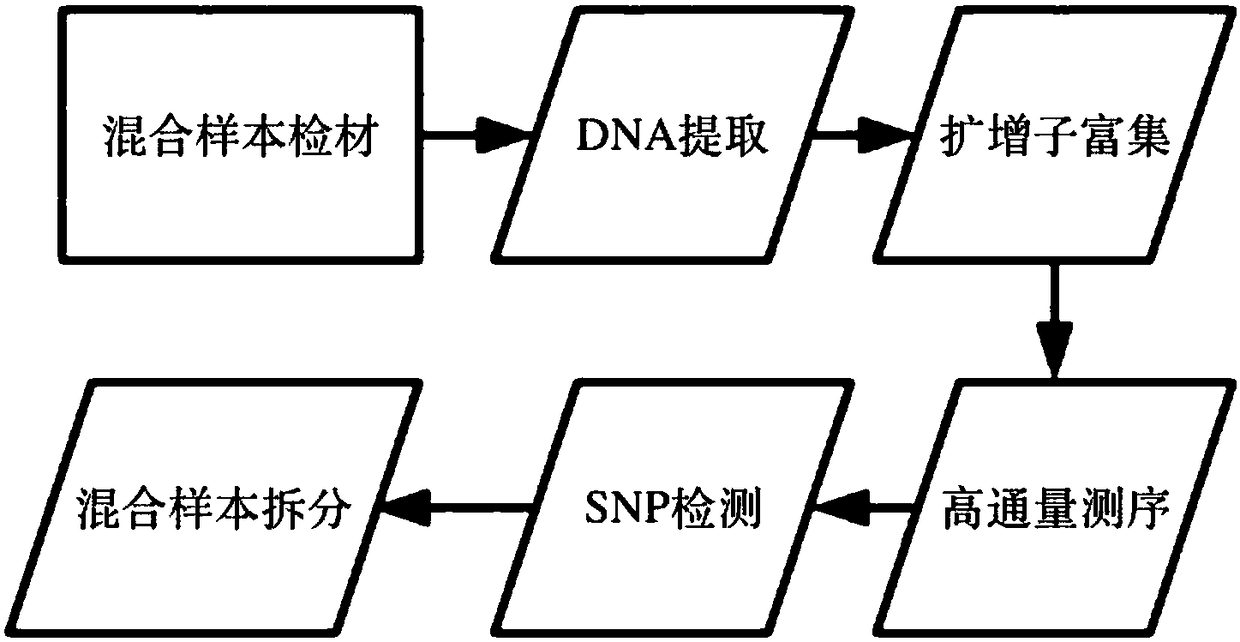
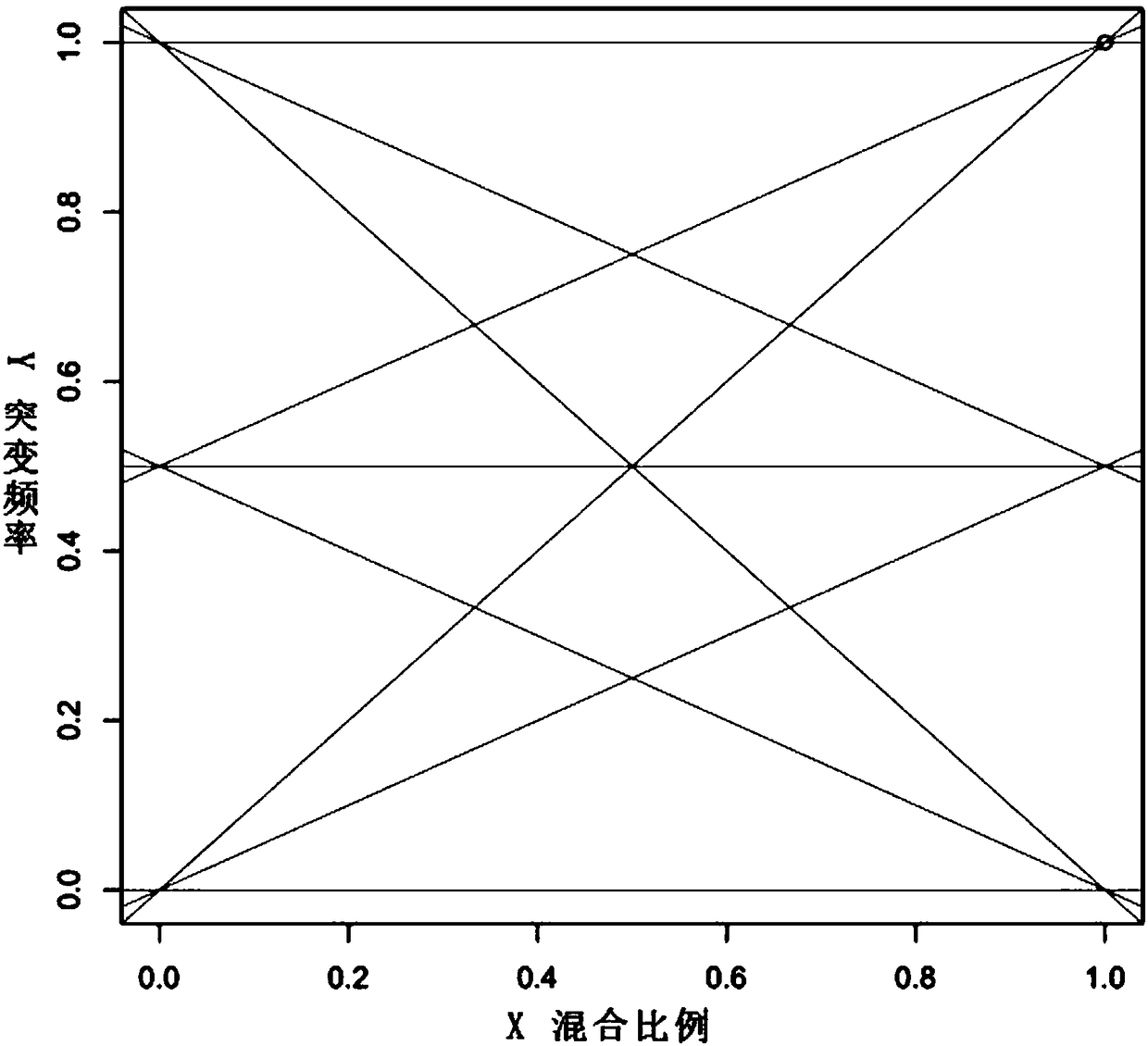
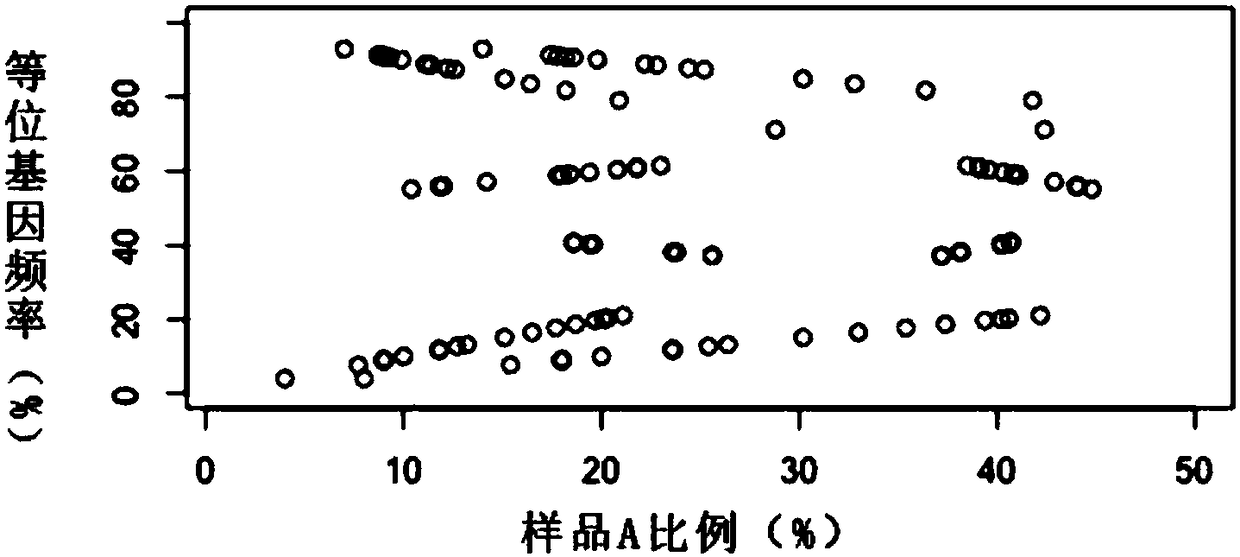
Method for separating mixed samples by SNP detection technology
ActiveCN109280696AReduce the starting amountHigh detection sensitivityMicrobiological testing/measurementNucleotideDNA fragmentation
The invention discloses a method for separating mixed samples by an SNP detection technology. The method uses a high-throughput gene sequencing platform (NGS) to detect multiple single nucleotide polymorphisms (SNPs) by amplicon sequencing, which can solve the problems that a mixed sample of two persons cannot be separated happened in criminal evidence identification, civil judicial identificationand clinic. The method includes the following technical points: 1) in order to ensure that amplification efficiency of amplicons is uniform and stable, the 219 amplicons are obtained to achieve simultaneous amplification in the same tube after strict condition design and screening; 2) in order to ensure successful amplification of a degraded sample (the length of a DNA fragment being 160 bp), theSNPs are selected as biomarkers, wherein the SNPs are located in the middle of the 140 bp amplicons; and 3) in order to distinguish whether the sample is a sample of one person or a mixed sample of two persons, a mixing proportion of the mixed sample and SNPs genotype sets of the mixed sample of two persons are further separated.
Owner:安塞斯(北京)生物技术有限公司 +1
Sensitive detection of bacteria by improved nested polymerase chain reaction targeting the 16S ribosomal RNA gene and identification of bacterial species by amplicon sequencing
InactiveUS20060057616A1Easy to detectRaise the possibilitySugar derivativesMicrobiological testing/measurementConserved sequenceAmplicon sequencing
A method for identifying an RNA form of a bacteria, comprising reverse transcribing RNA material; conducting PCR using primers for a first highly conserved genetic sequence generic of the bacteria; conducting nested PCR using primers for a second highly conserved genetic sequence within the first genetic sequence of the bacteria; and identifying the bacteria based on unconserved amplified sequences linked to the conserved sequences.
Owner:VIRONIX
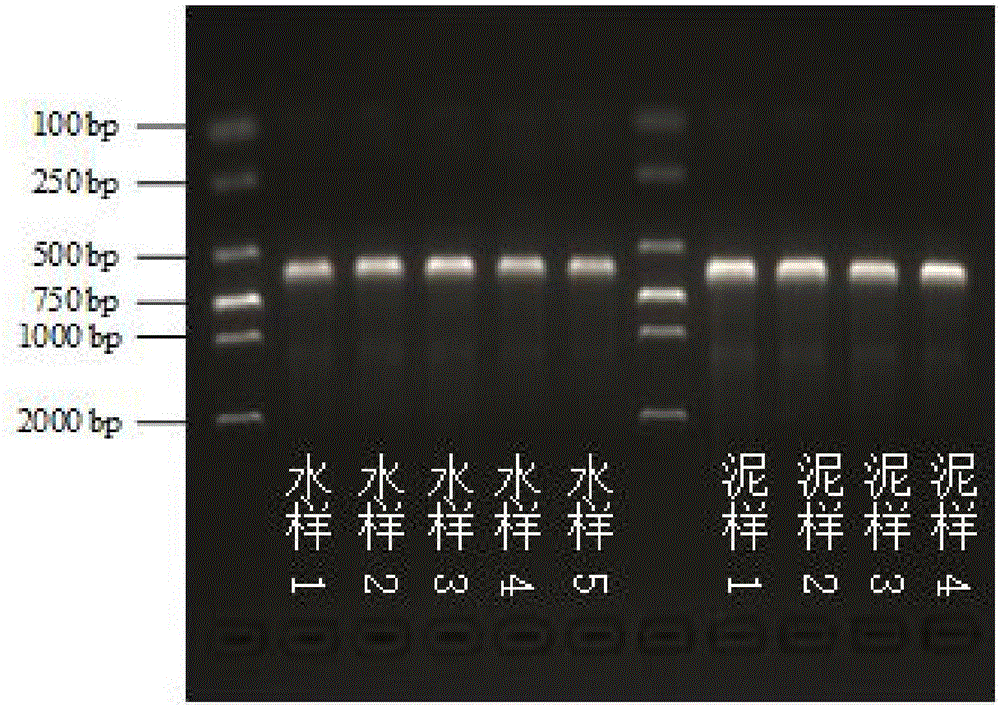
Fast construction primer and method for water/mud sample DNA amplicon sequencing library in sewage treatment system
InactiveCN106554958AQuality improvementImprove efficiencyMicrobiological testing/measurementLibrary creationStructure analysisDNA extraction
The invention discloses a fast construction primer and method for a water / mud sample DNA amplicon sequencing library in a sewage treatment system, and belongs to the field of high-throughput sequencing. The method comprises the steps of firstly, sample DNA extraction, secondly, target fragment amplification, thirdly, amplification product purification, fourthly, amplification product quality determination, fifthly, amplification product mixing, sixthly, mixed library quality determination and seventhly, mixed library preparing and sequencing. The primer comprises a connector sequence, an index sequence, a heterogeneity spacer sequence and a target fragment amplification sequence. The primer is used for further amplification, library construction can be fast completed, the sequencing cost is reduced, and a large amount of library construction time is saved. According to the fast construction primer and method, the problem about high diversity bacterial flora structure analysis in complex sewage / mud samples is solved, and the technical requirement that the fast construction primer and method are applied to water / mud sample bacterial flora deep analysis in the sewage treatment system based on the high-throughput DNA sequencing technology is met.
Owner:NANJING UNIV YIXING ENVIRONMENTAL PROTECTION RES INST +1
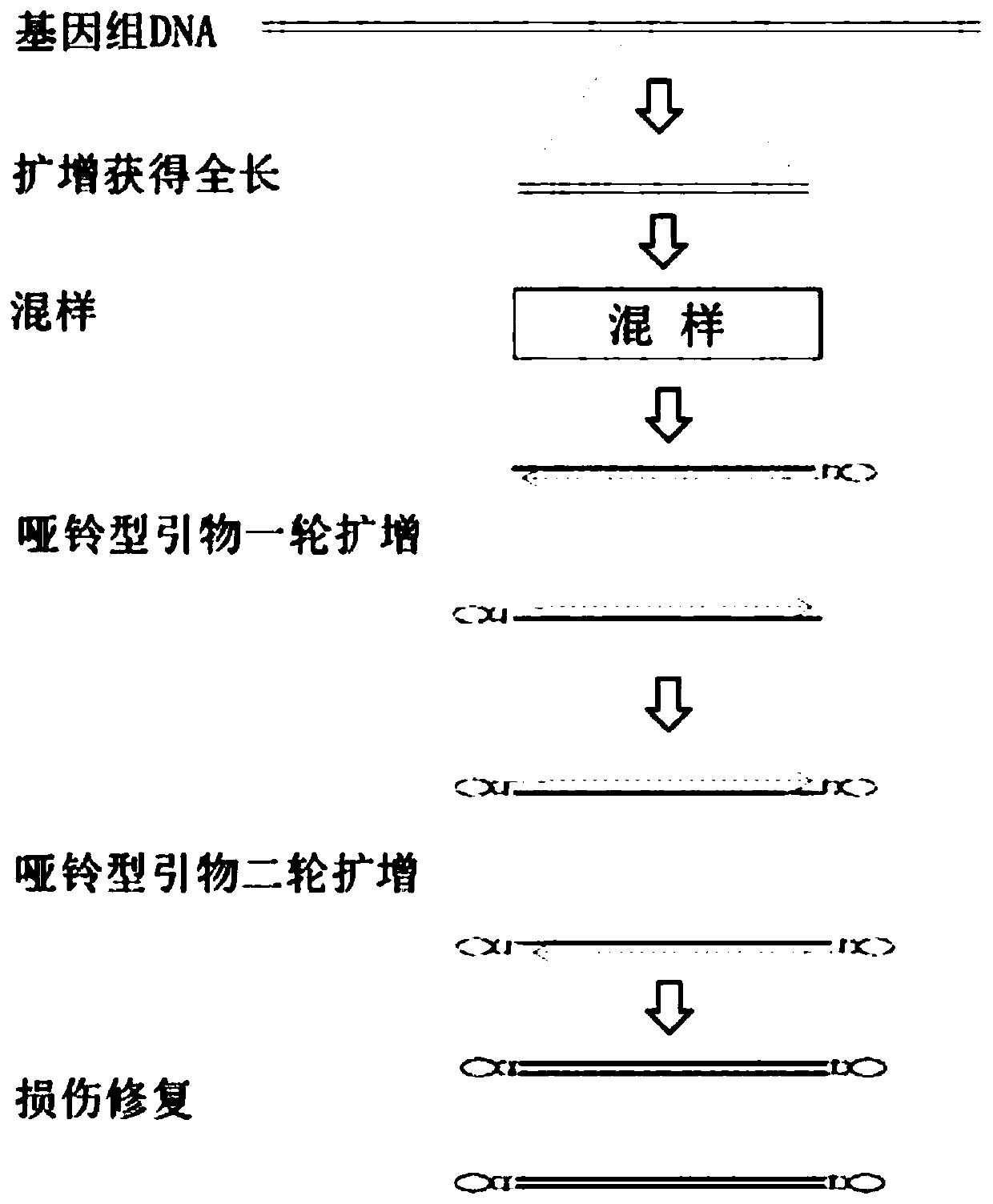
Rapid full-length amplicon library construction method, primers and sequencing method applicable to PacBio platform
InactiveCN111455023ASimplify the build processImprove work efficiencyNucleotide librariesMicrobiological testing/measurementGeneticsEngineering
The invention relates to a rapid full-length amplicon library construction method and primers applicable to PacBio platform and a full-length amplicon sequencing method based on the library construction method. According to the characteristic that PCR is required for full-length amplicon sequencing, an amplicon primer with a general binding sequence and a dumbbell-shaped primer matched with the general binding sequence are designed in combination with a dumbbell-shaped library joint structure of the PacBio sequencing platform, the rapid full-length amplicon library construction method applicable to the PacBio platform is provided based on the two primers with special structures, through library construction by the method, addition of sequencing joints can be realized by increasing two cycles of annular primer amplification only after conventional amplification, and meanwhile, a PCR product becomes a standard library applicable to the PacBio sequencing platform under the action of damage repair enzyme. The method optimizes an original PacBio standard library construction process into only one-step amplification and repair, the library construction process of a full-length amplicon is greatly simplified, and the sequencing cost is significantly reduced.
Owner:WUHAN FRASERGEN CO LTD
Universal primer for plant-derived component identification and application of universal primer
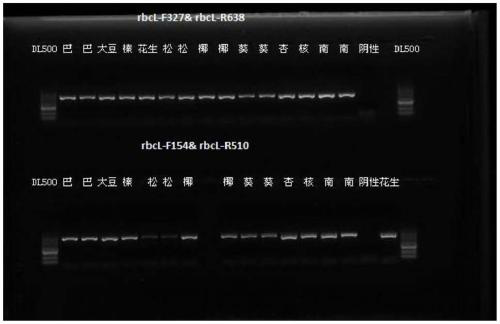
InactiveCN111455093ALow costReduce detection work intensityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologySequence analysis
The present invention discloses a universal primer for plant-derived component identification and an application of the universal primer. An upstream primer sequence of the universal primer is shown as SEQ No.1 and a downstream primer sequence of the universal primer is shown as SEQ No.2. An amplicon sequencing analysis method is used to identify at least 10 plant-derived components of walnuts, peanuts, soybeans, sesame seeds, hazelnuts, apricot kernels, coconuts, pine nuts, pumpkin seeds, Badam, etc. and can solve a problem that current PCR and ELISA detection methods can only conduct one-to-one targeted screening of the targeted components, only one experiment is needed to conduct non-targeted screening of the plant components of the product, intensity of detection work is reduced, costof synthezing primers and probes is saved, and in the case of saving time, labor and material costs, the universal primer can be flexibly used in a variety of food containing allergenic ingredients tomeet screening work of large quantities of plant component-containing samples, realizes development from target detection to target-free screening, improves a level of adulteration detection and provides an effective technical means for supervision.
Owner:NAT INST FOR FOOD & DRUG CONTROL
Sensitive detection of bacteria by improved nested polymerase chain reaction targeting the 16S ribosomal RNA gene and identification of bacterial species by amplicon sequencing
InactiveUS20080213773A1Easy to detectRaise the possibilityMicrobiological testing/measurementConserved sequenceAmplicon sequencing
A method for identifying an RNA form of a bacteria, comprising reverse transcribing RNA material; conducting PCR using primers for a first highly conserved genetic sequence generic of the bacteria; conducting nested PCR using primers for a second highly conserved genetic sequence within the first genetic sequence of the bacteria; and identifying the bacteria based on unconserved amplified sequences linked to the conserved sequences.
Owner:VIRONIX
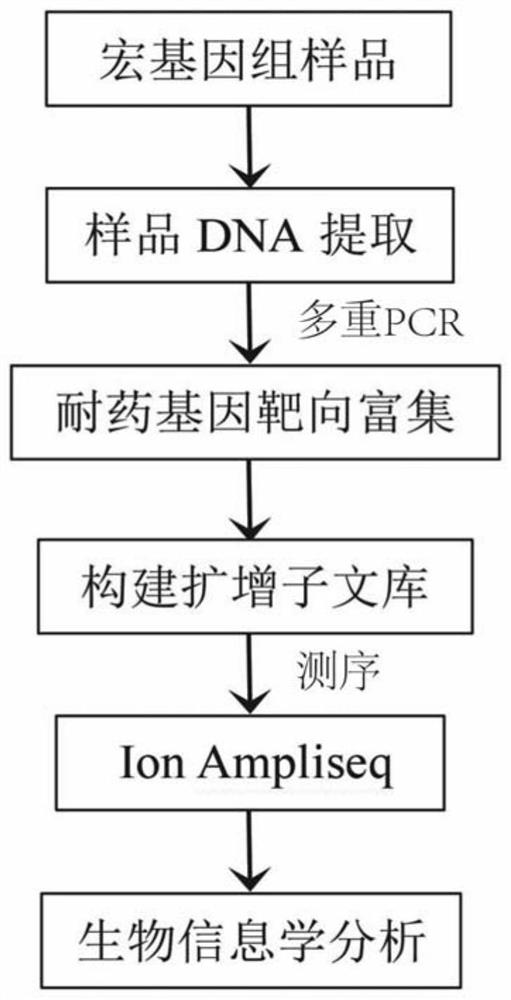
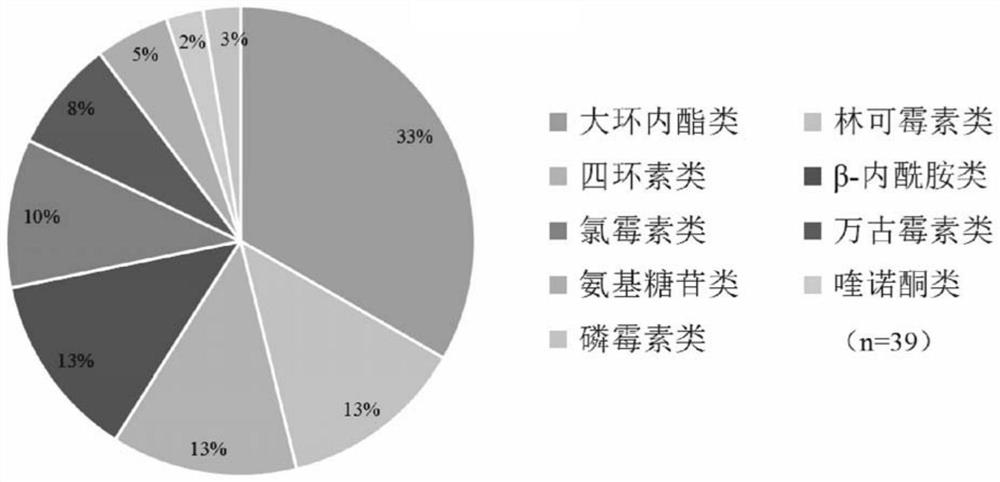
Primer group for gram-positive bacterium drug-resistant gene high-throughput amplicon sequencing and application
ActiveCN112852938AEfficient Targeted AmplificationSpecific target amplificationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyResistant genes
The invention discloses a primer group for gram-positive bacterium drug-resistant gene high-throughput amplicon sequencing and application of the primer group. The invention provides a complete set of primers for gram-positive bacterium drug-resistant gene high-throughput amplicon sequencing. The complete set of primers provided by the invention consists of primers as shown in sequences 1-78. The invention discloses an amplicon sequencing detection analysis method for 39 clinical common drug-resistant genes. The method has the technical advantages of rapidness, high efficiency, high targeting, strong specificity and the like, and is suitable for epidemic and difference analysis of various drug-resistant genes in clinic and livestock breeding industry.
Owner:CHINA AGRI UNIV
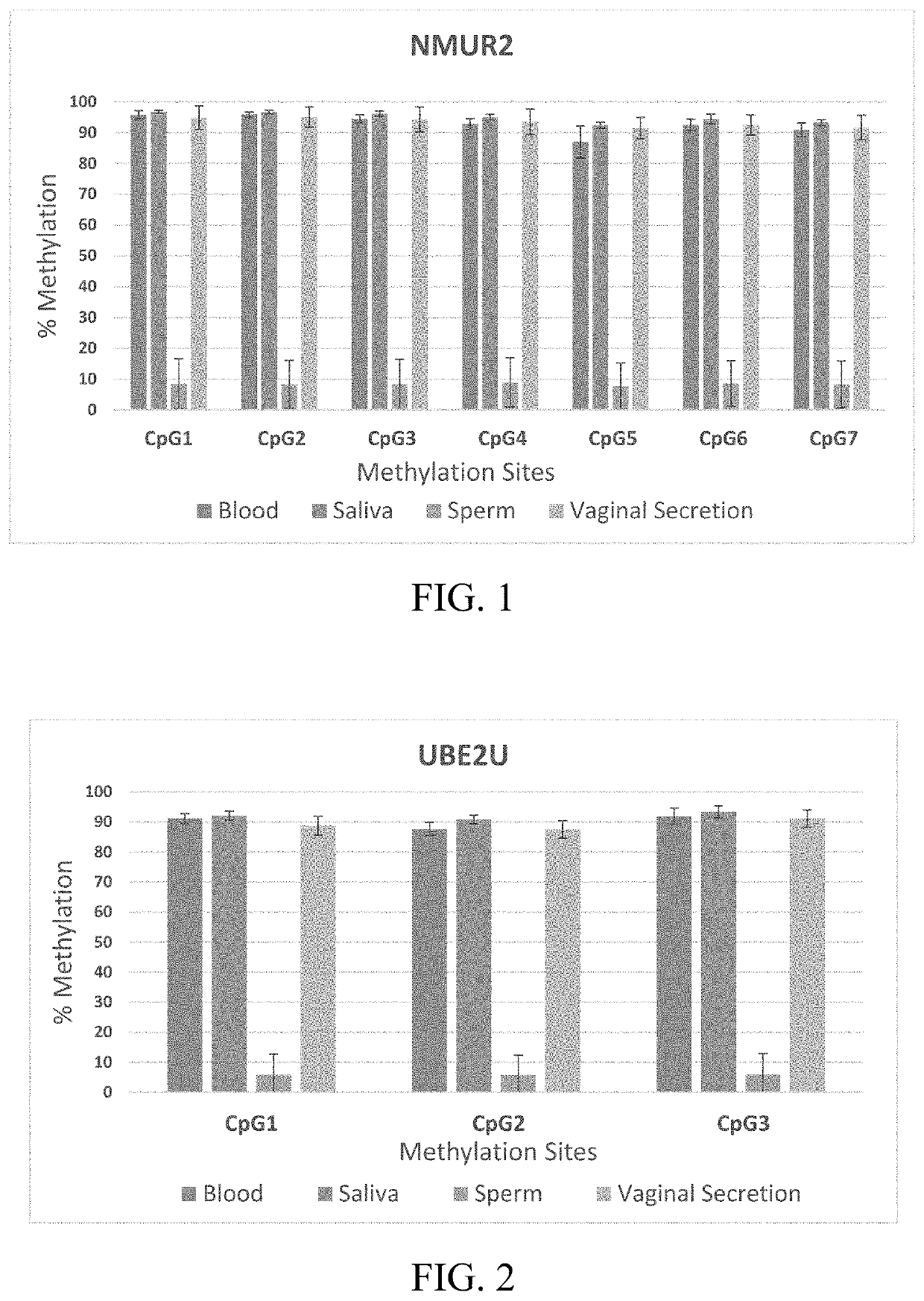
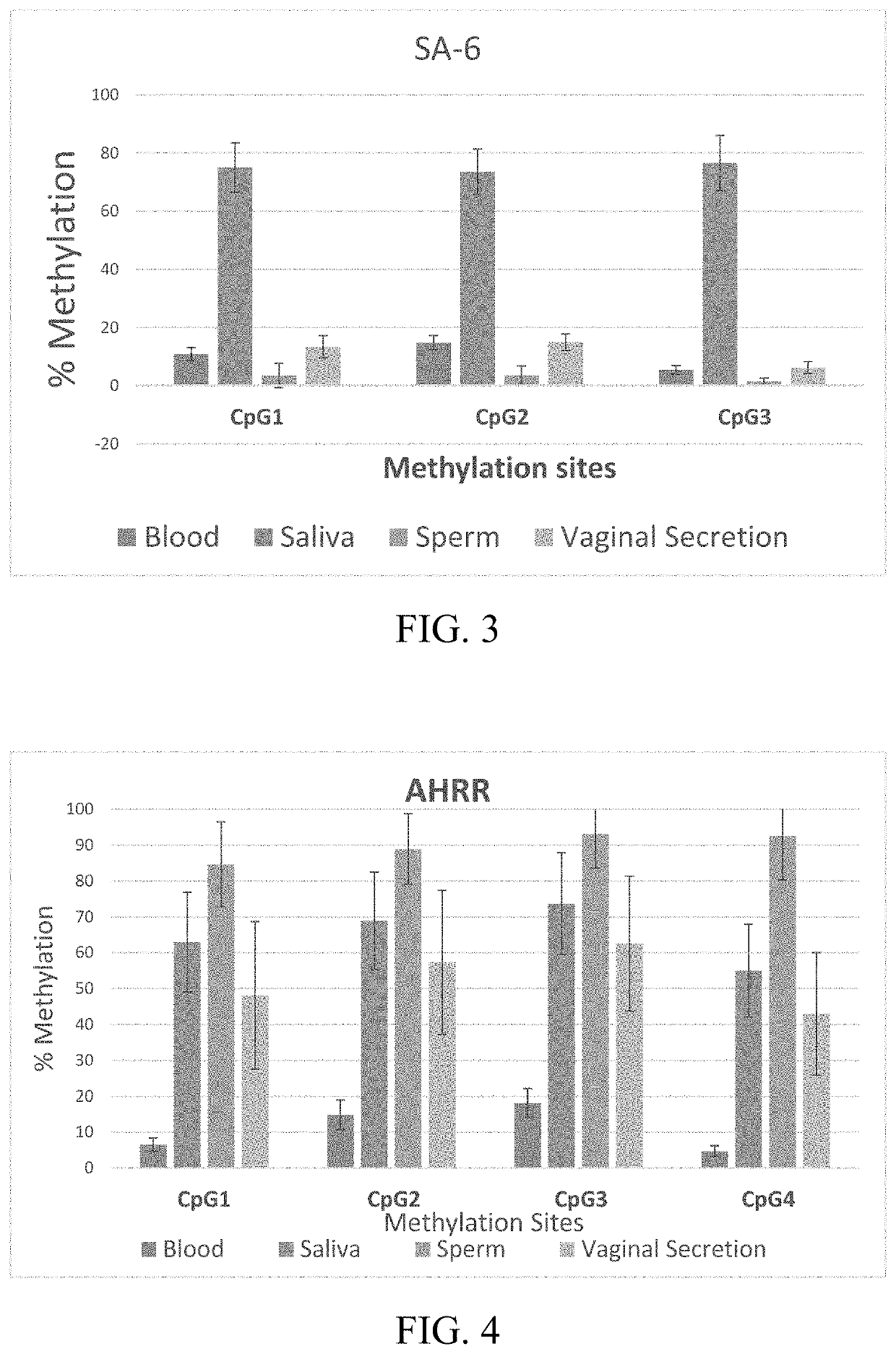
DNA methylation assays for body fluid identification
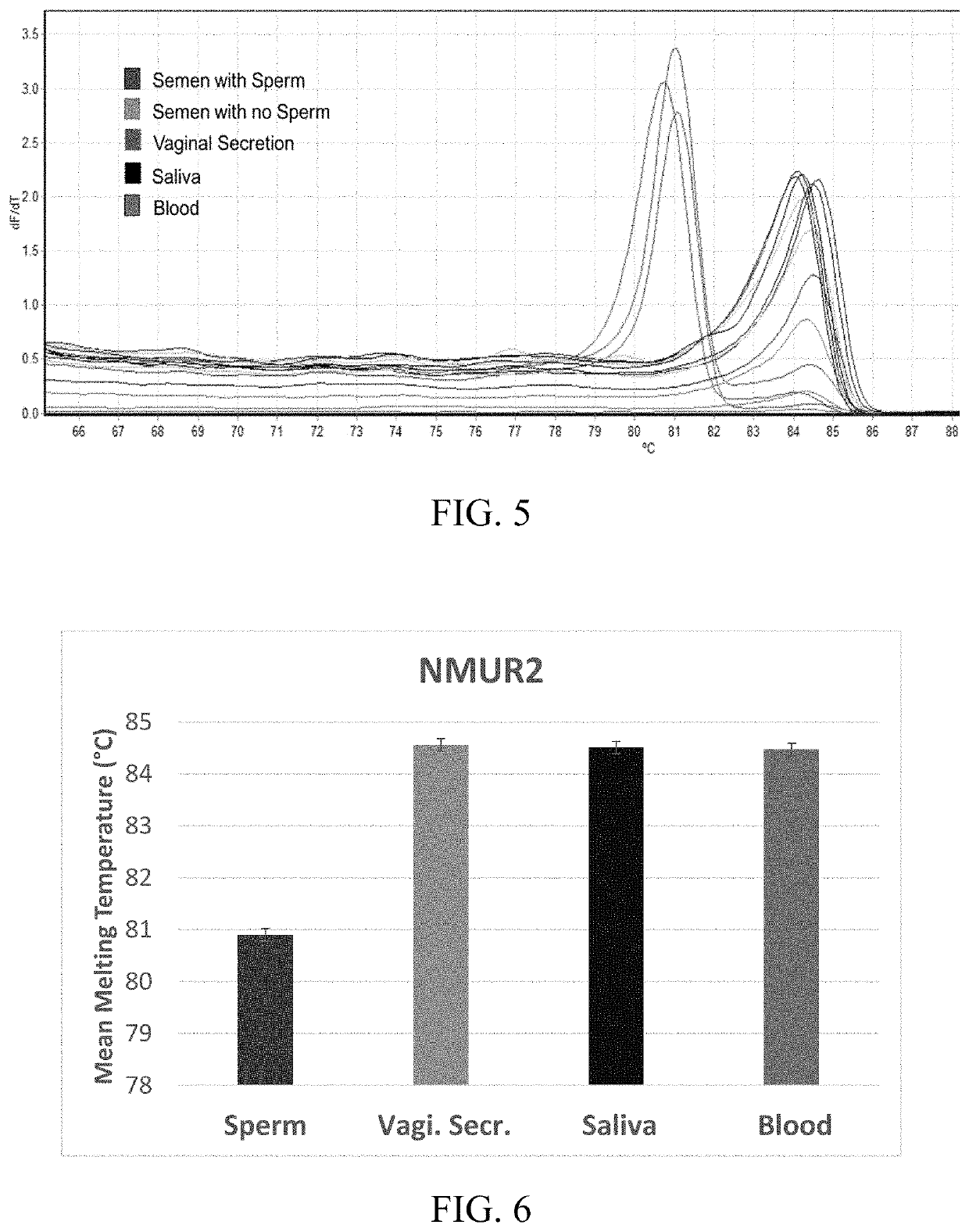
ActiveUS10704089B1Highly tissue specific and sensitive analysisMicrobiological testing/measurementDNA methylationSemen
The invention pertains to analyzing the DNA methylation levels at specific genetic loci. The DNA methylation levels at the specific genetic loci can be used to detect the presence in a sample of a specific body fluid, for example, semen containing sperm, saliva, or blood. Particularly, the DNA methylation levels at the genetic loci corresponding to SEQ ID NOs: 1, 7, 13, and 19 are used to detect sperms, saliva or blood cells. The DNA methylation levels at the specific loci can be determined by high-resolution melt (HRM) analysis or sequencing of the amplicons produced using specific primers designed to amplify the specific loci. Kits containing the primers and reagents for carrying out the methods disclosed herein are also provided.
Owner:FLORIDA INTERNATIONAL UNIVERSITY
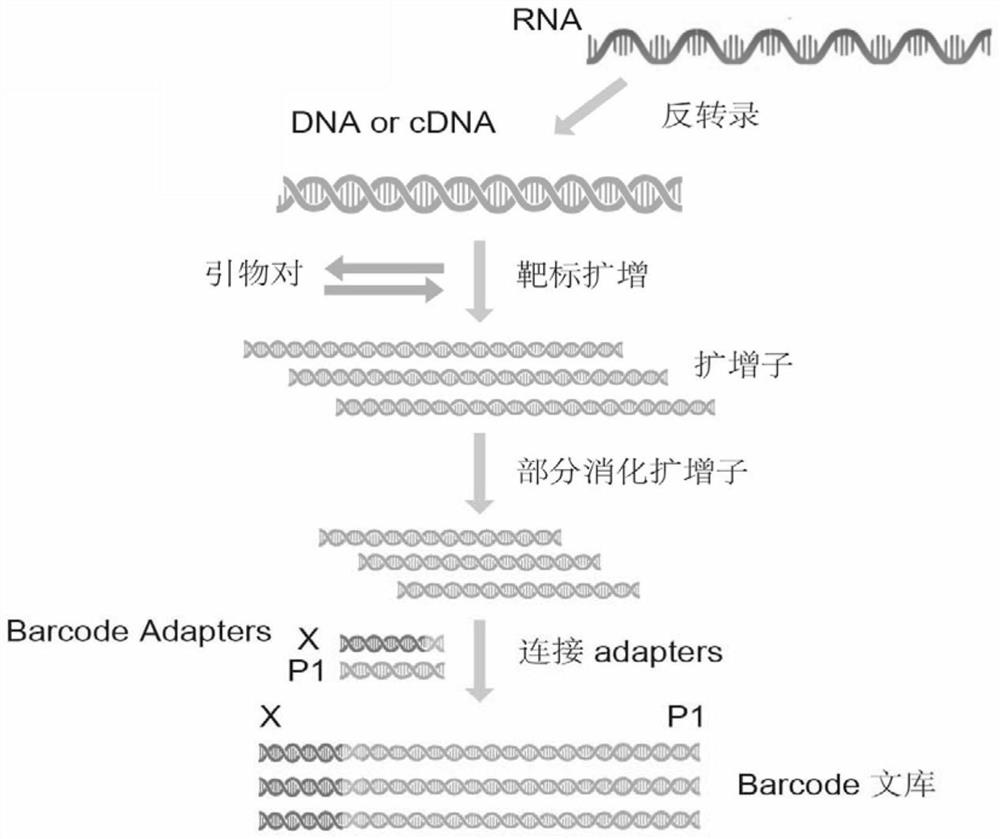
Primer for amplicon sequencing and two-step PCR database building method
ActiveCN110878334ALower Sequencing CostsImprove quality indicatorsMicrobiological testing/measurementLibrary creationMultiplexGenomic DNA
The invention provides a primer for amplicon sequencing and a two-step PCR database building method. The method comprises the following steps of: 1) extracting genomic DNA of a species sample to be tested; 2) respectively connecting universal primer sequences to 5' ends of forward and reverse primers of the multiplex PCR, and taking the genomic DNA as a template to carry out a first round of multiplex PCR to obtain a PCR product with universal primer sequences at both ends; 3) purifying the product of the first round of PCR; 4) respectively connecting primer sequences matched with the first round of PCR universal primer sequences at both ends of the sequencing label sequence to obtain a PCR product carrying the label sequence, and using the PCR product as a template to carry out a second round of PCR; and 5) purifying the PCR product, and sequencing on the machine after passing the quality inspection. The invention adopts the two-step PCR method to build a database, so that the amplicon sequencing cost is reduced, the detection cost is reduced by more than 50%, and the quality index of the amplicon is effectively improved. That is to say, the detection rate of the site and the datauniformity are improved, and the method has extremely high economic value.
Owner:BEIJING COMPASS BIOTECHNOLOGY CO LTD
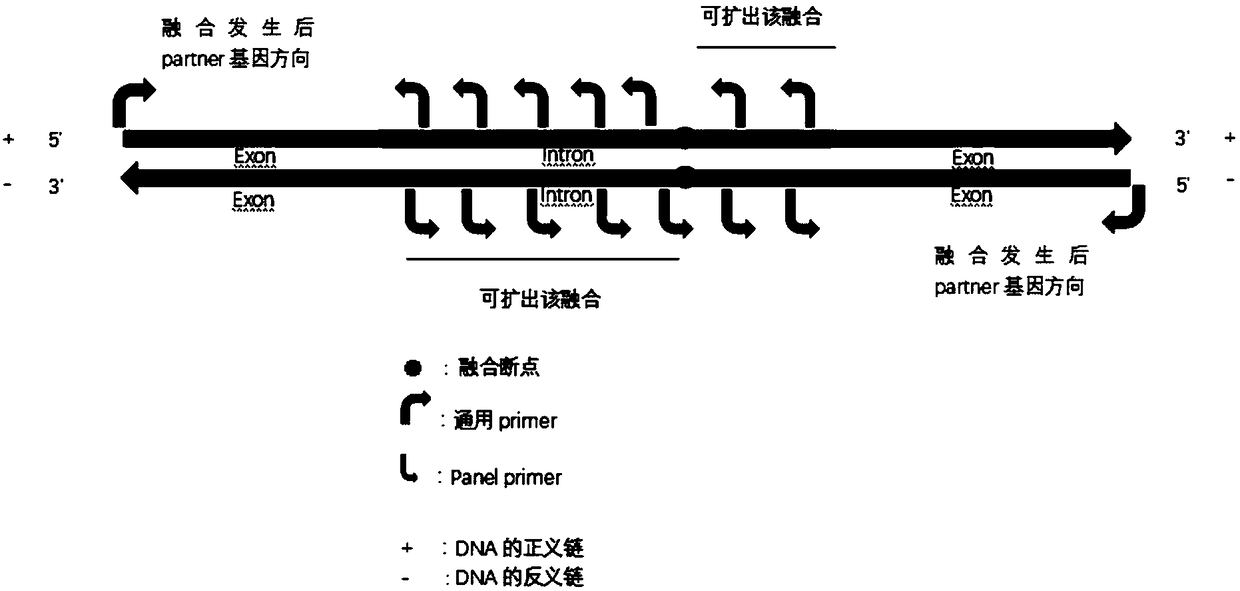
Method for detecting whether fusion mutation of target gene occurs, primer combination, kit and application thereof
ActiveCN109234357ATargeted Enrichment ImplementationOvercome the disadvantage of being unable to target and enrich unknown fusion genesMicrobiological testing/measurementLibrary creationRet geneFgfr3 gene
The invention relates to a method for detecting whether fusion mutation of target gene occurs, a primer combination, a kit and an application thereof. A plurality of primers are sequentially designedaccording to the possible gene fusion occurrence direction by the primer design method of the invention, forming a primer combination, wherein the primers of the primer combination extend along the direction in which gene fusion occurs, can carry out directional enrichment on the target gene in which unknown fusion occurs, overcomes the disadvantage that the existing amplicon sequencing method cannot enrich the unknown fusion gene targetedly, and is conducive to the discovery and detection of the unknown fusion type of the gene; At the same time, the interval between two adjacent primers is about 0 - 130 bp, which is good for highly fragmented samples (such as cfDNA, FFPE samples). The invention also designs primer combinations for detecting fusion mutations of lung cancer genes (includingALK gene, NTRK1 gene, RET gene, ROS1 gene, FGFR3 gene and NTRK3 gene), After multiple rounds of detection and optimization of the primer sequence, the homogeneity of the sequencing data was more than85%, and the capture efficiency was more than 92%. Finally, the effective detection of unknown fusion mutation of various lung cancer genes was successfully achieved.
Owner:SIMCERE DIAGNOSTICS CO LTD +1
A kit and method for detecting chromosome heterozygosity loss based on amplicon sequencing
ActiveCN109504770BComprehensive, true and accurate responseReduce the amount of solutionMicrobiological testing/measurementDNA/RNA fragmentationNucleic acid detectionAmplicon sequencing
The invention discloses a kit and method for detecting chromosome heterozygosity loss on the basis of amplicon sequencing, belongs to the field of molecular biology nucleic acid detection, and relatesto the kit and method for detecting chromosome heterozygosity loss by means of amplicon sequencing. The kit contains a mixture of 14 groups of primer pairs used for detecting a 1p36.32 region of a human chromosome 1, 6 groups of primer pairs used for detecting a 1q25.2 region of the human chromosome 1, 14 groups of primer pairs used for detecting a 19p13.2 region of a human chromosome 19 and 7 groups of primer pairs used for detecting a 19q13.33 region of the human chromosome 19, 20 kinds of tag primers used for distinguishing samples, a GC stabilizer, a PCR enzyme mixture, purifying magneticbeads, a quality control standard substance and ddH2O. Through two-time PCR amplification and two-time product purification and sequencing, through a ratio of READS(1p) / READS(1q) to READS(19q) / READS(19p), the heterozygosity loss state of 1p19q in the samples is analyzed. By adopting the method, the variation situation of the 1p19q can be reflected comprehensively, truly and accurately, and gene variation of the 96 samples can be subjected to high-throughput detection and analysis simultaneously, so that the detection cost is greatly reduced.
Owner:艾普拜生物科技(苏州)有限公司
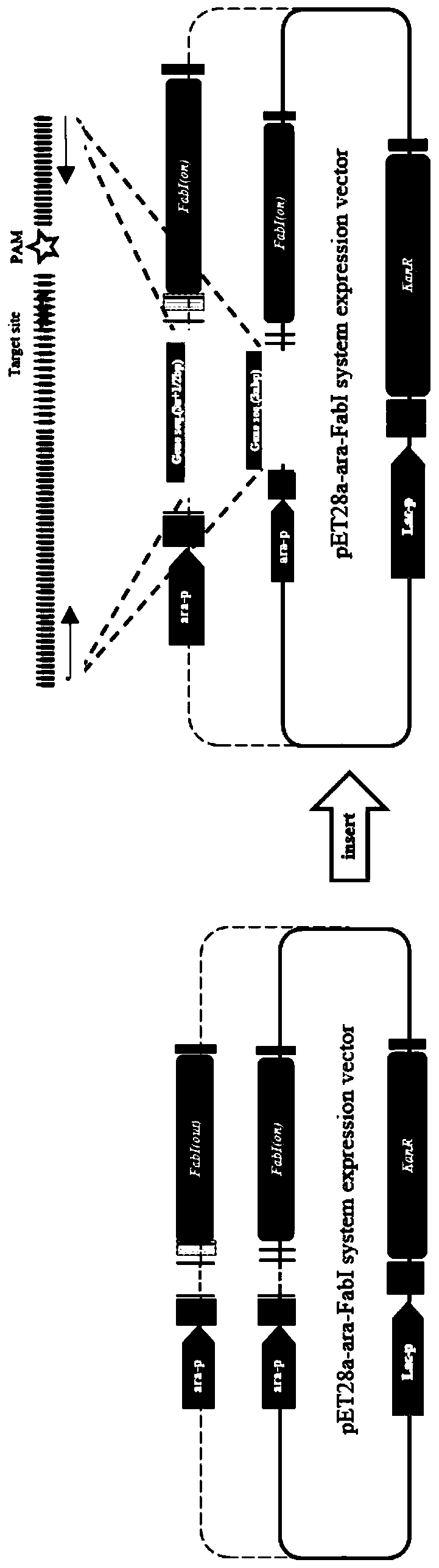
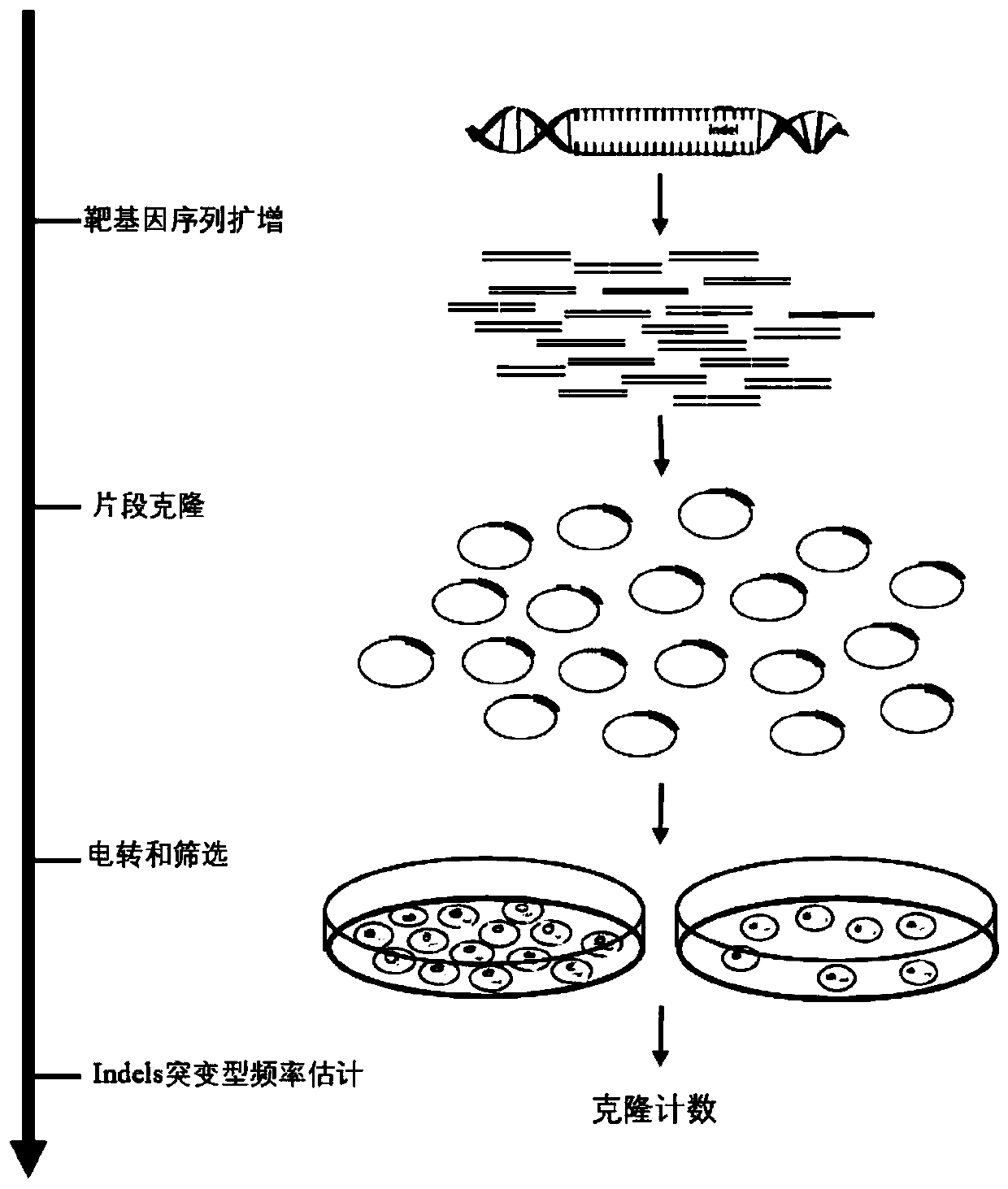
Construction and application of high-flux detection system for nuclease induced insertion and deletions (Indels)
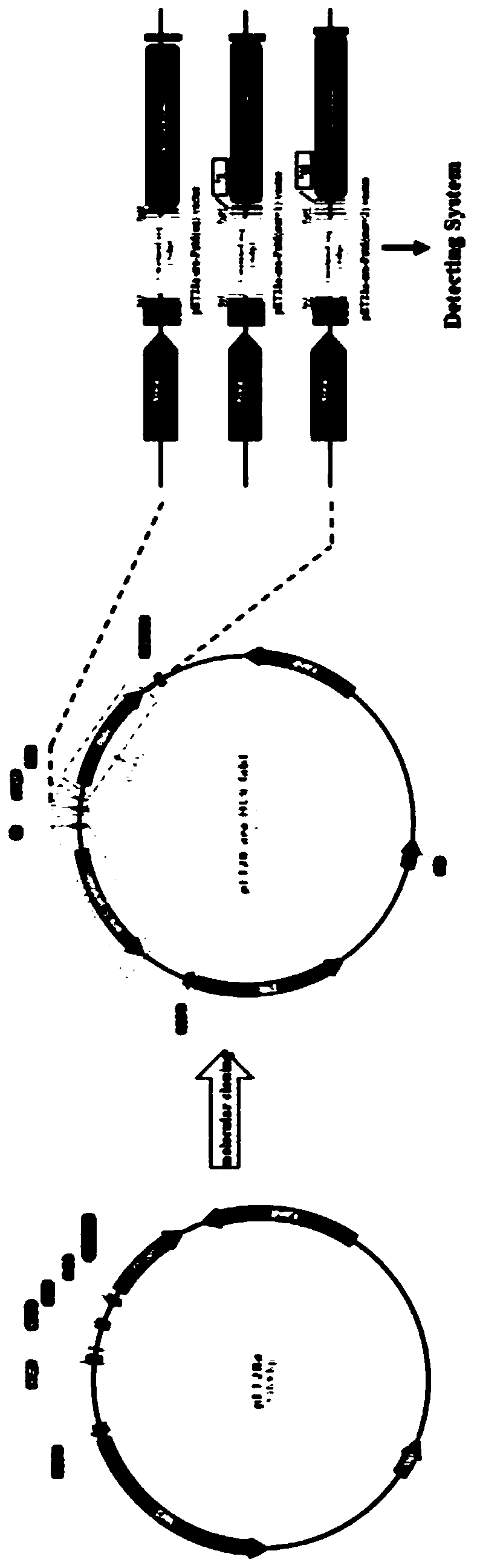
PendingCN110305888ARealize Quantitative RevealMicrobiological testing/measurementBiological material analysisOpen reading frameMutation frequency
The invention belongs to the field of gene engineering and particularly relates to construction and application of a mutation detection system for detecting artificial nuclease induced insertion and deletions (Indels). According to the adopted specific technical scheme, the ara-mFabI detection system is provided and comprises three detection carriers, and each carrier is separately responsible forquantitative and qualitative reveal of one type of insert fragment. The three carriers are distinguished in that the types of open reading frames (ORFs) of mFabI genes are different; and nine basic groups, ten basic groups and eleven basic groups are introduced through upstream primers of amplification primers of the mFabI genes correspondingly so as to change the ORFs of the mFabI genes. By using the detection system, quantitative reveal of the mutation frequency of all the types of nuclease induced Indels can be realized; and the mutation frequency of the Indels close to amplicon sequencingand type distribution can be realized.
Owner:NORTHWEST A & F UNIV
Method of correcting amplification deviation in amplicon sequencing
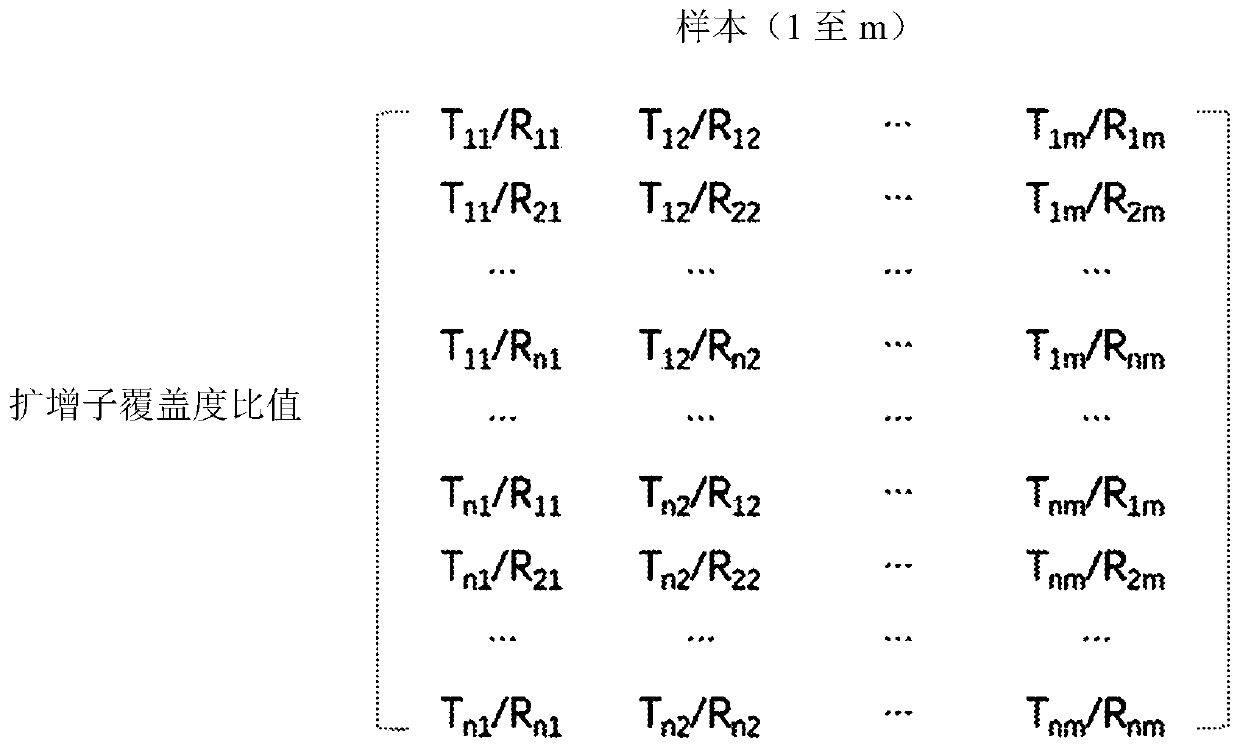
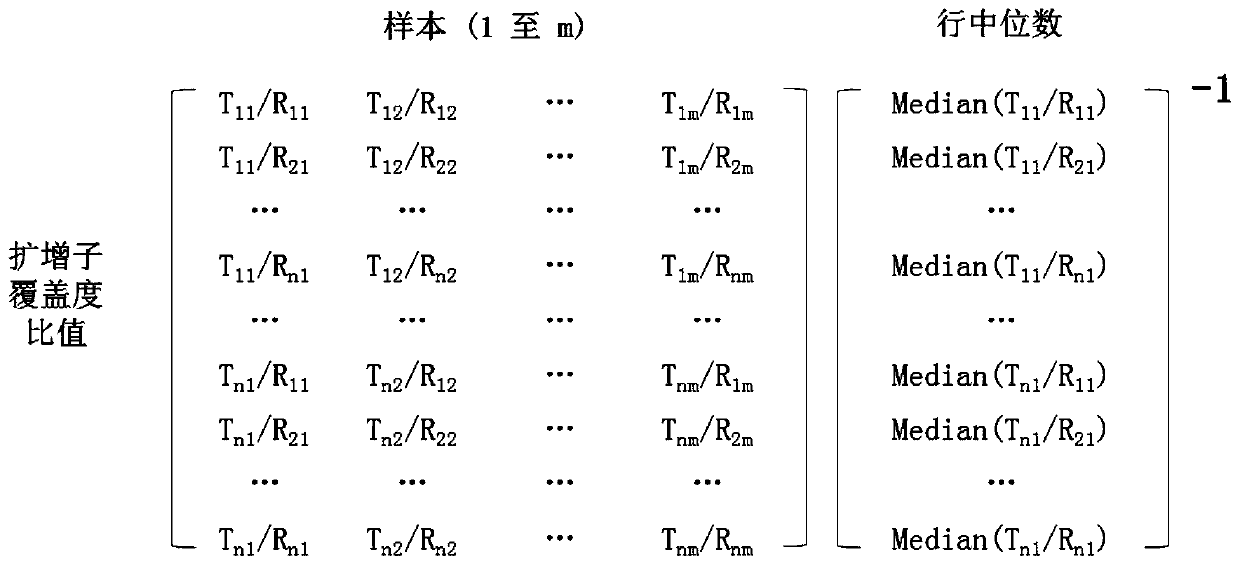
ActiveCN110741094AMicrobiological testing/measurementBiostatisticsAmplification biasAmplicon sequencing
A method to correct amplification deviation in amplicon sequencing is disclosed. Through steps of amplifying the target nucleic acid to obtain the amplicon coverage of the target nucleic acid, calculating the amplicon coverage ratio between the target nucleic acid in each test genomic region and the target nucleic acid in the reference genomic region, removing outliers, applying a formula to normalize the amplicon coverage ratio, calculating the differences between parameters of the test genomic region amplicon and the reference genomic region amplicon, and applying another formula to fit thedata, and other steps, a regression parameter obtained by fitting is used for correcting the amplification deviation to obtain a normalized amplicon coverage ratio after removing the amplification deviation, thereby eliminating the amplification deviation caused by experimental factors during the multiple PCR amplification process. Elimination of the amplification deviation facilitates accurate calculation of the copy number for a target genomic region, thus enabling detection of minor copy number variation using amplicon sequence data.
Owner:CELULA CHINA MED TECH CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com