DNA library construction method for high-throughput sequencing
A technology for sequencing library and construction method, which is applied in the field of high-throughput sequencing DNA library construction, and can solve the problems of wasting time, heavy workload, cumbersome steps, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Example 1: Forensic kinship identification
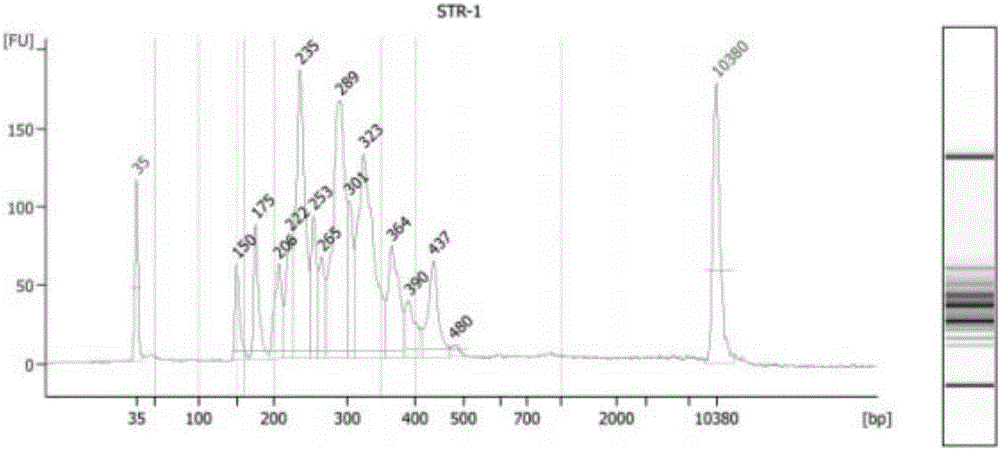
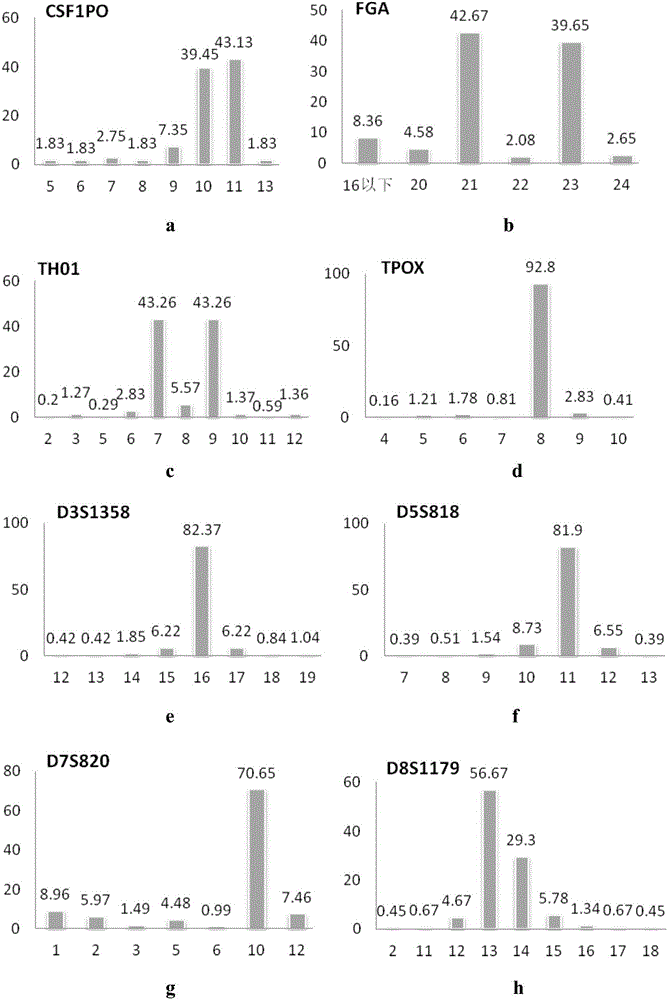
[0088] The purpose of this example is to detect the number of repeats of the repeat sequence in the STR, wherein the NGS sequencing protocol is used.
[0089] 1. Collection and processing of oral epithelial cells
[0090] (1) Collect human oral epithelial cells with a buccal swab collection tube.
[0091] (2) Oral epithelial cell DNA was extracted with buccal swab genome extraction kit (DP322).
[0092] (3) OD260nm and OD280nm were measured using a Nanodrop 2000 spectrophotometer to confirm that the purity was high, and the concentration was measured.
[0093] 2. Design of primers for multiplex PCR reaction
[0094] (1) The primers are designed as fusion primers, including the specific primer sequence of the target fragment and the sequencing linker sequence, which contains the index sequence.
[0095] (2) The fusion primer structure is:
[0096] Upstream primer: 5'-CCTCTCTATGGGGCAGTCGGTGAT-3'+ target fragment specific f...
Embodiment 2
[0127] Example 2: Detection of which cell line an unknown cell line belongs to
[0128] In this example, two unknown human cell lines were used, and the number of repetitions of the repeat sequence in the STR was detected to confirm which cell line the unknown cell line belonged to. Which uses the NGS sequencing protocol.
[0129] 1. DNA extraction from human cell lines
[0130] (1) Human cell line DNA was extracted using a peripheral blood DNA kit (Qiagen, Cat. No. 937236).
[0131] (2) OD260nm and OD280nm were measured using a Nanodrop 2000 spectrophotometer to confirm that the purity was high, and the concentration was measured. The final concentration was 51 ng / μl.
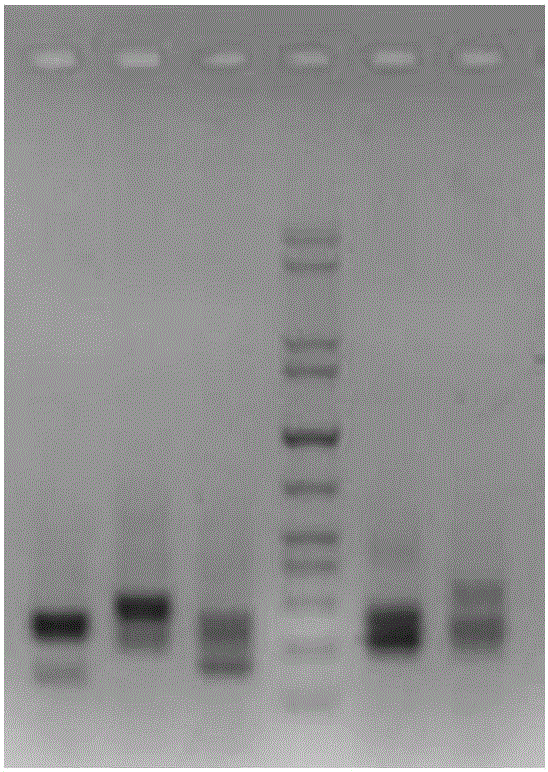
[0132] 2. Multiplex PCR reaction library construction and polyacrylamide PAGE gel nucleic acid electrophoresis
[0133] (1) The primers are designed as fusion primers, including the specific primer sequence of the target fragment and the sequencing linker sequence, which contains the index sequence.
[013...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com