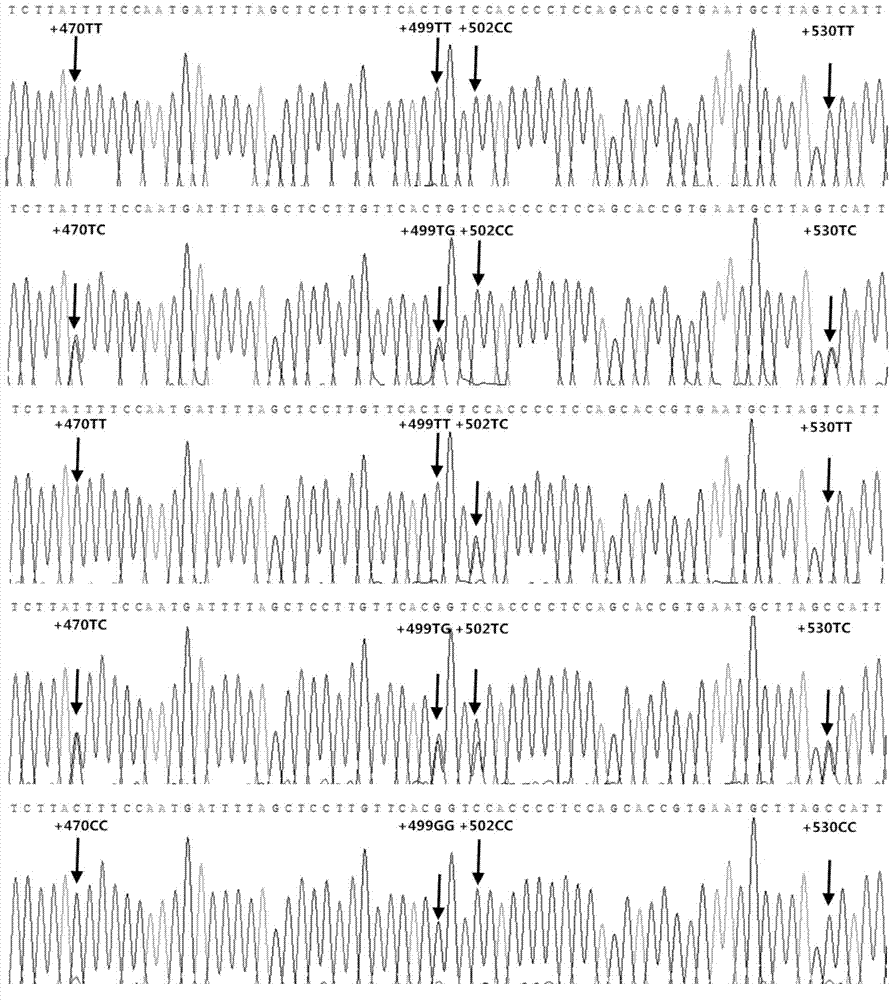
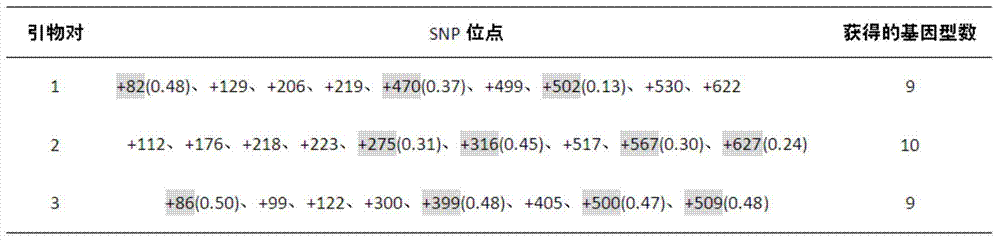
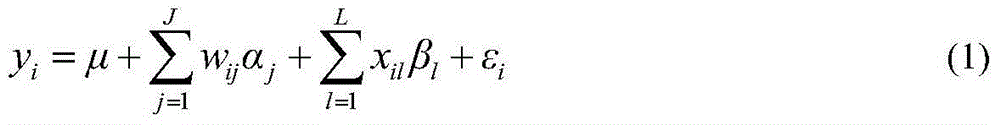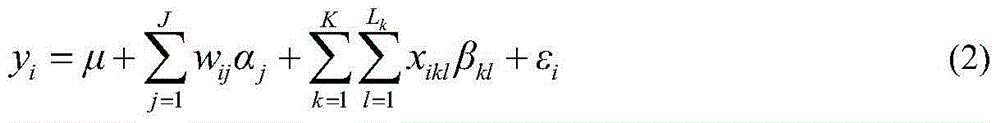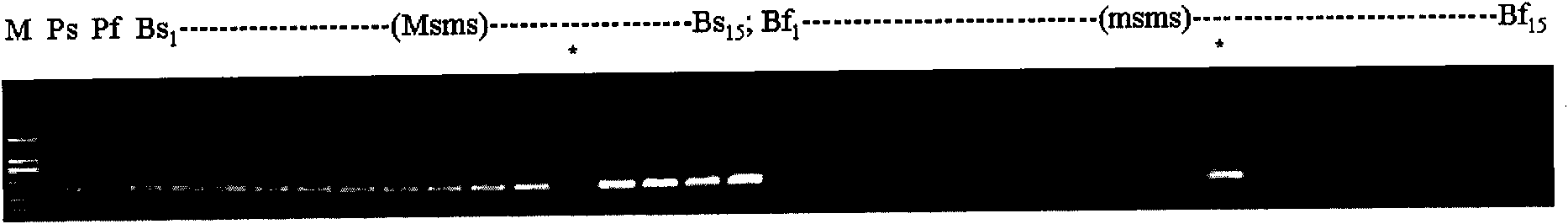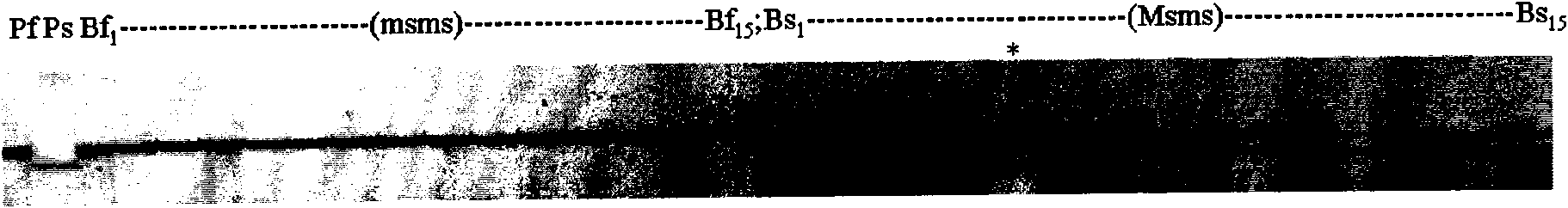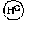Patents
Literature
50 results about "Multiple alleles" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Multiple alleles. Alleles are the pairs of genes occupying a specific spot called locus on a chromosome. Typically, there are only two alleles for a gene in a diploid organism. When there is a gene existing in more than two allelic forms this condition is referred to as multiple allelism.
Mutagenesis methods
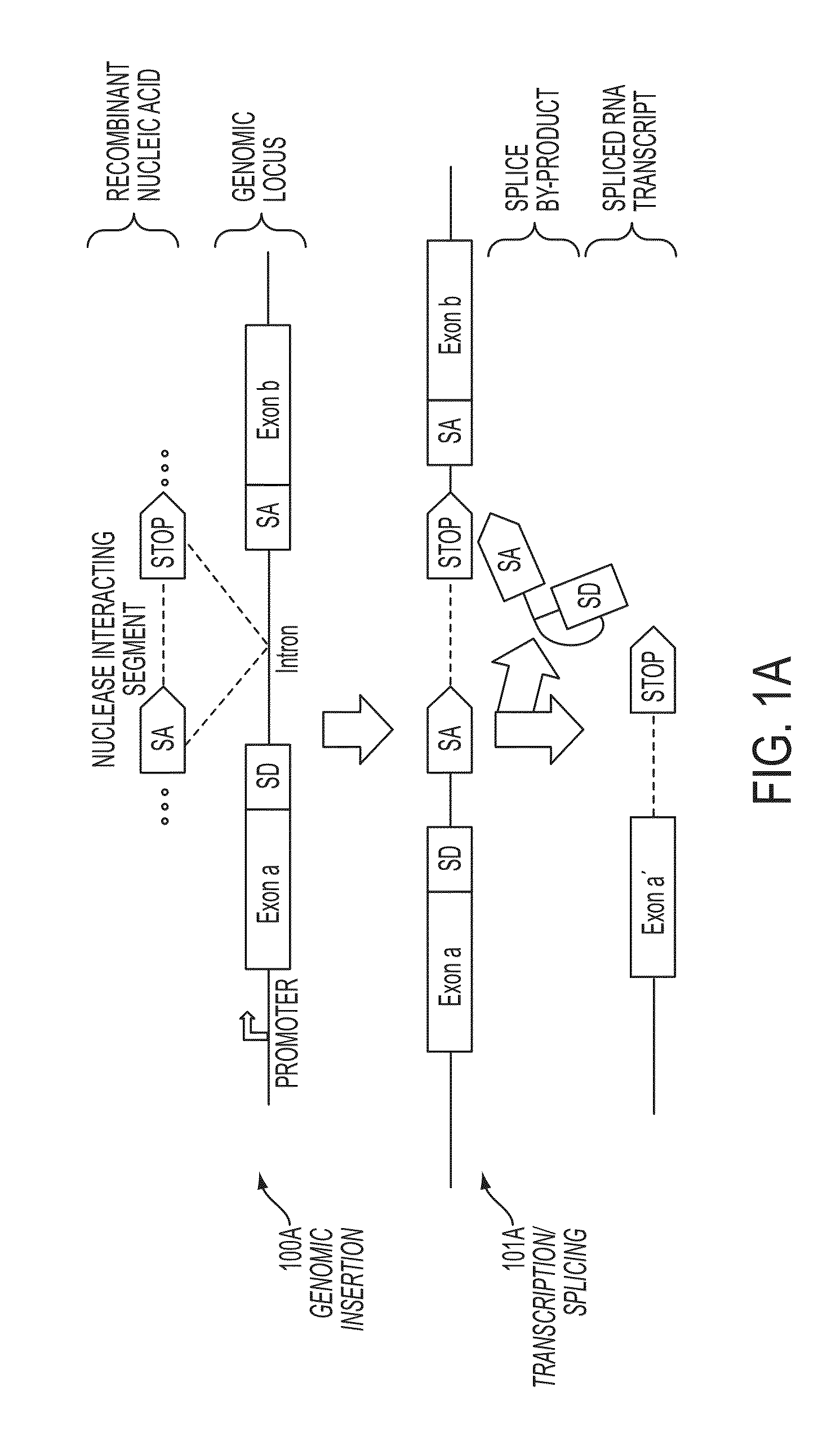
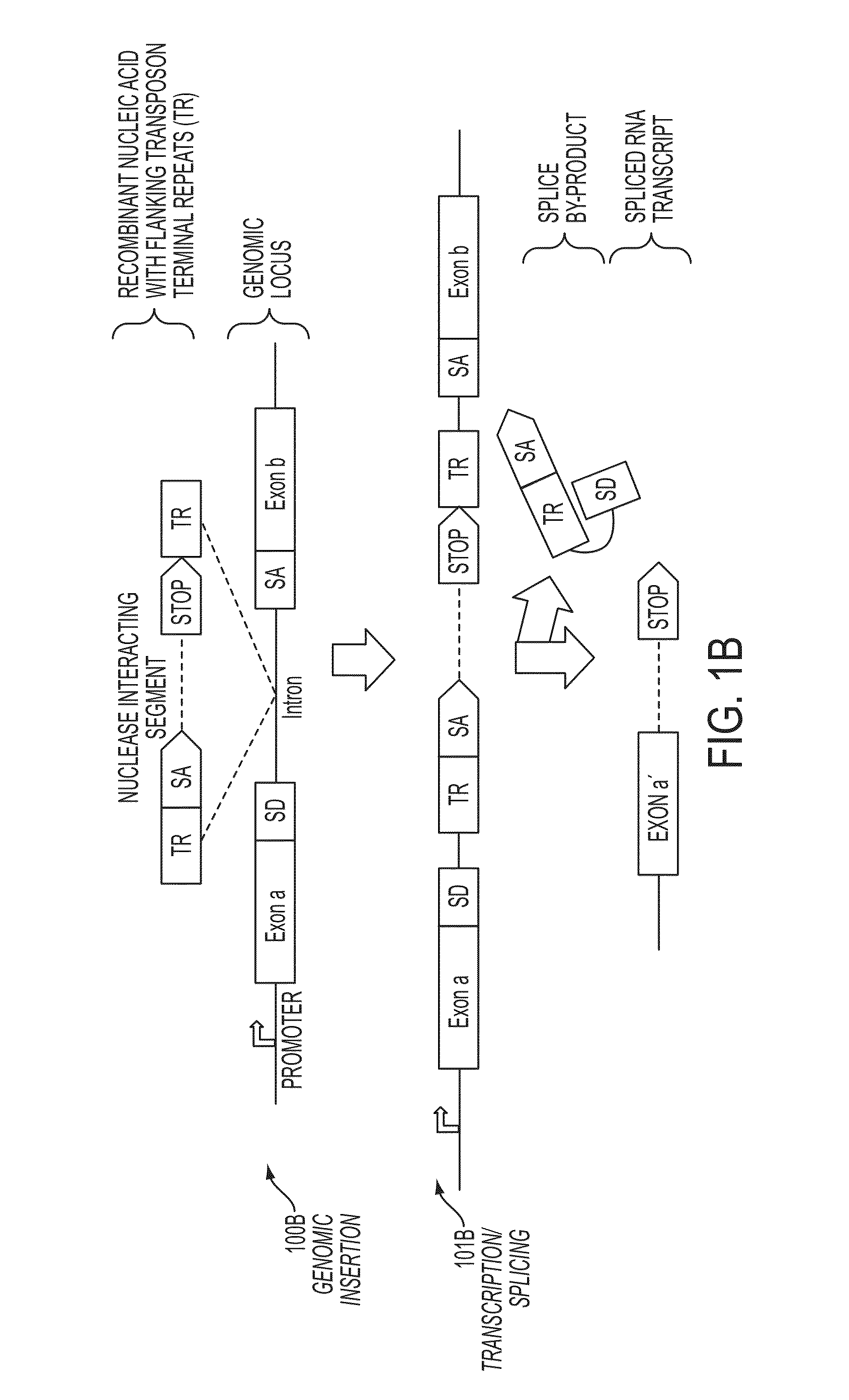
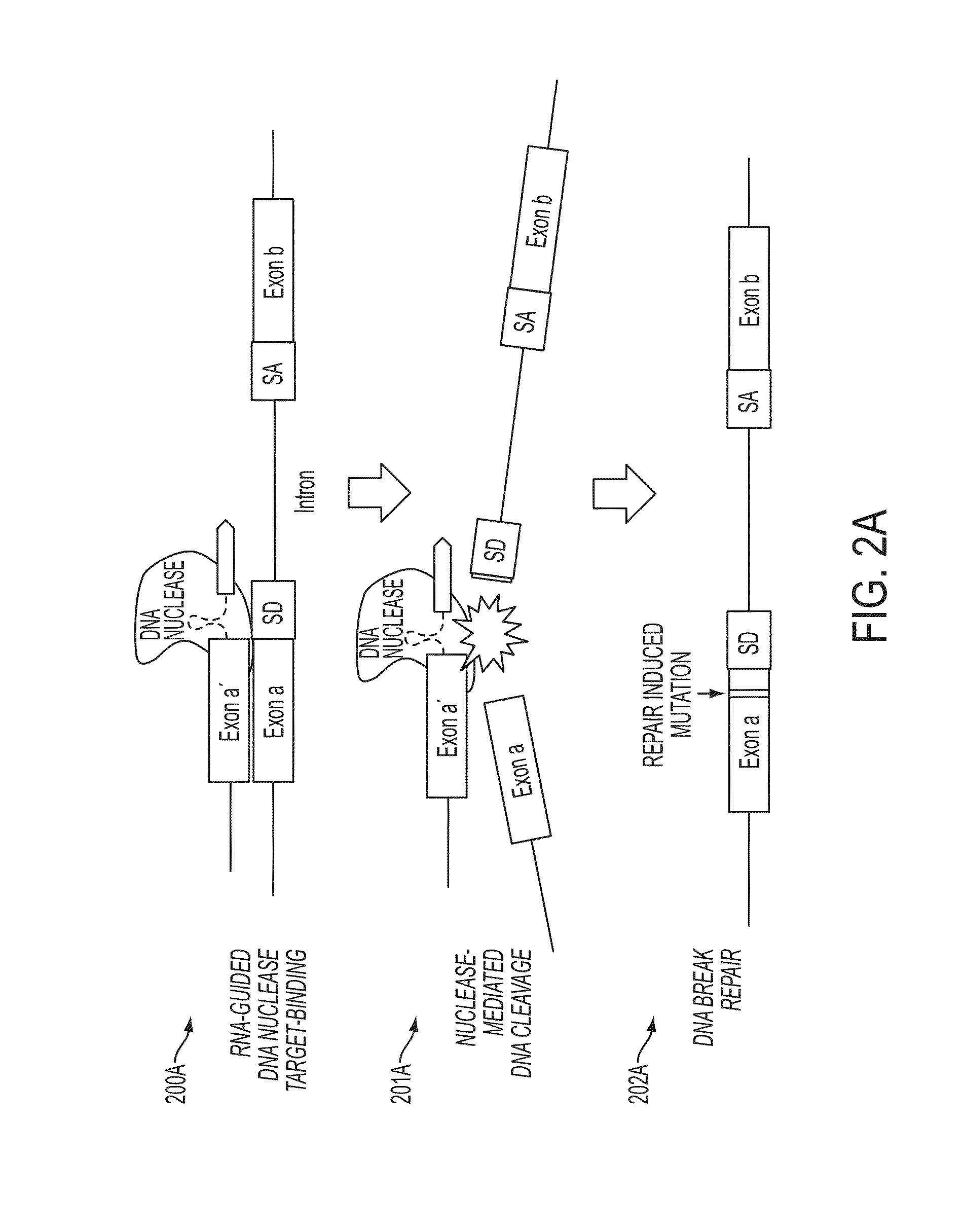
In some embodiments, aspects of the disclosure provide methods and compositions that are useful for modifying (e.g., mutating) one or more alleles of a genomic locus within a cell. In some embodiments, methods and compositions described herein involve producing a chimeric spliced RNA molecule that includes a transcribed exon spliced to a nuclease interacting RNA segment. In some embodiments, the chimeric spliced RNA guides a DNA modifying enzyme (e.g., a nuclease) to a genomic locus in a cell resulting in modification of the locus.
Owner:LAM THERAPEUTICS
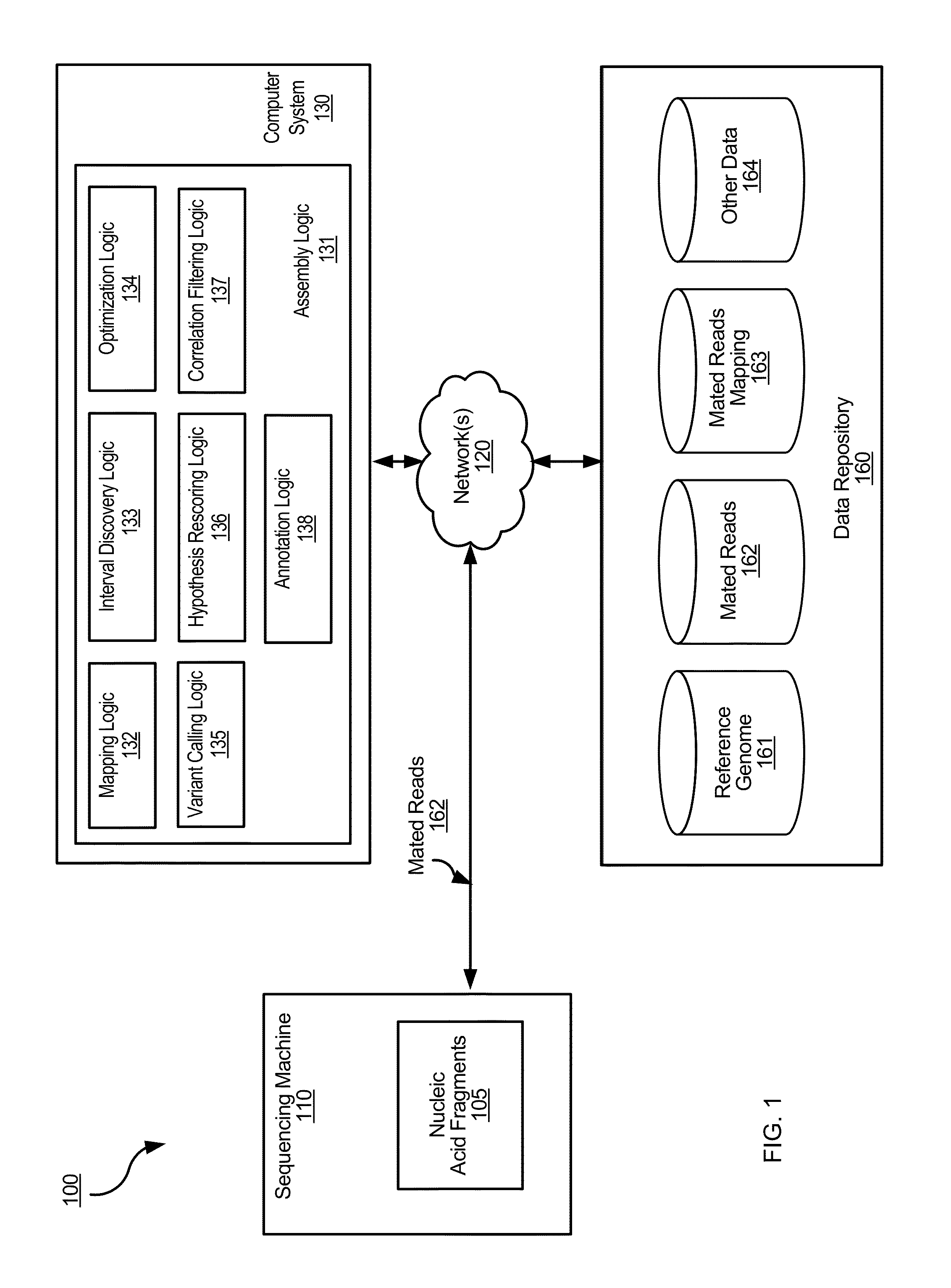
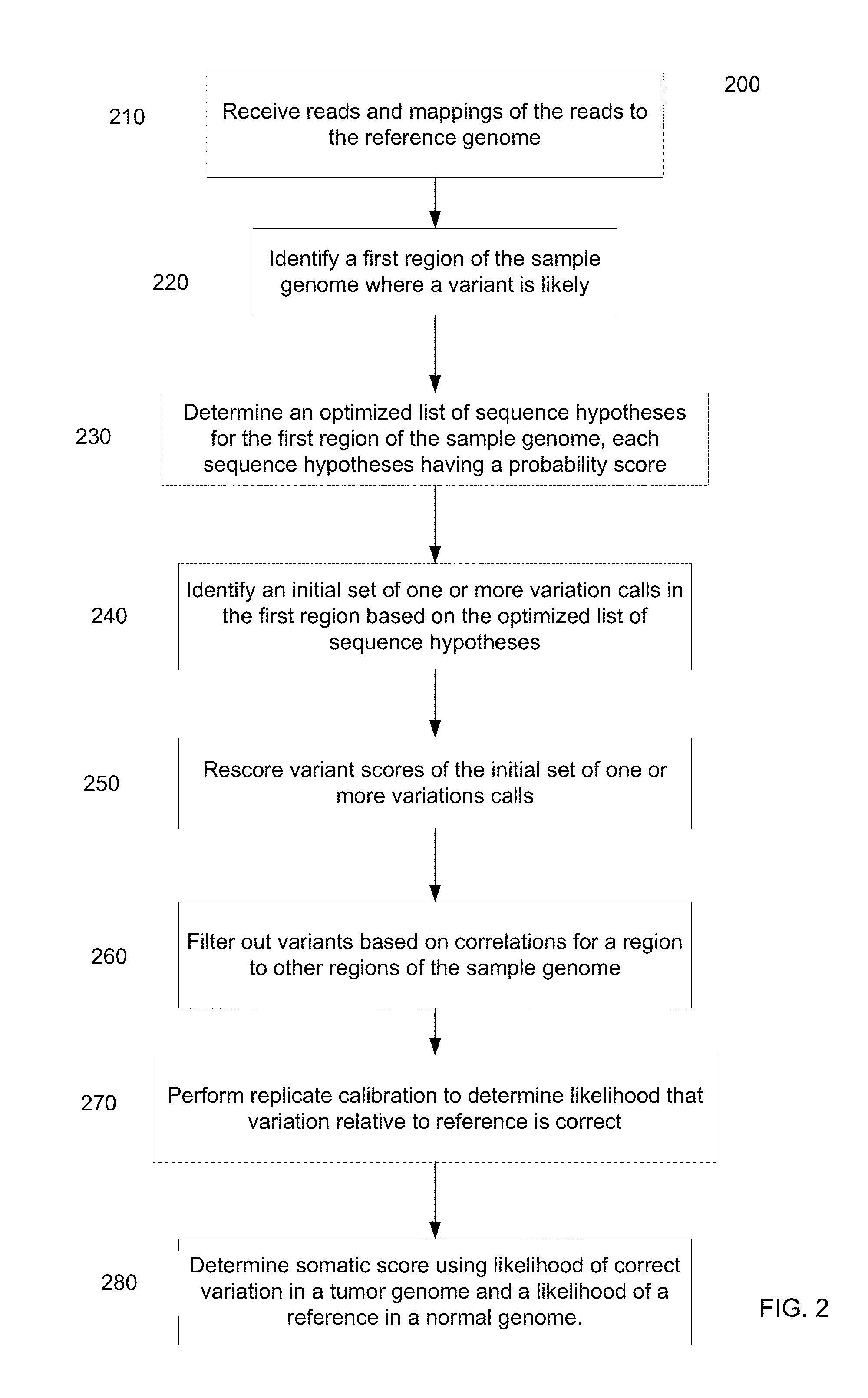
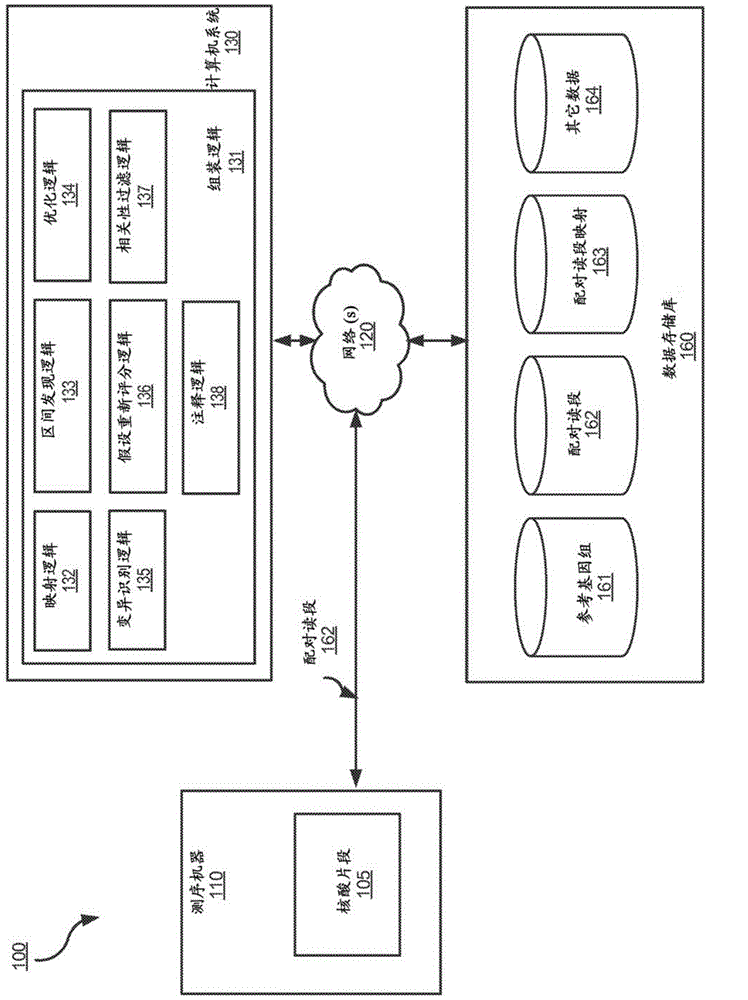
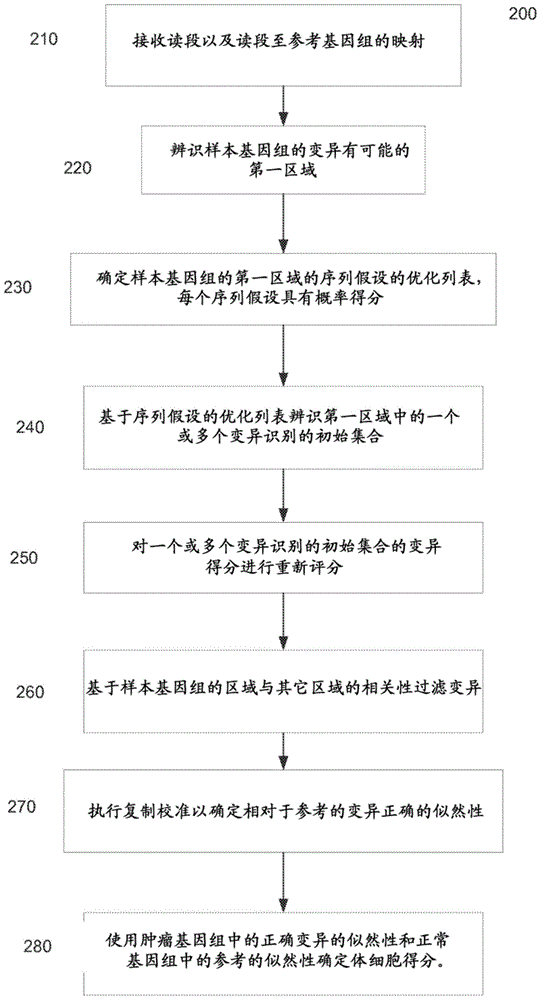
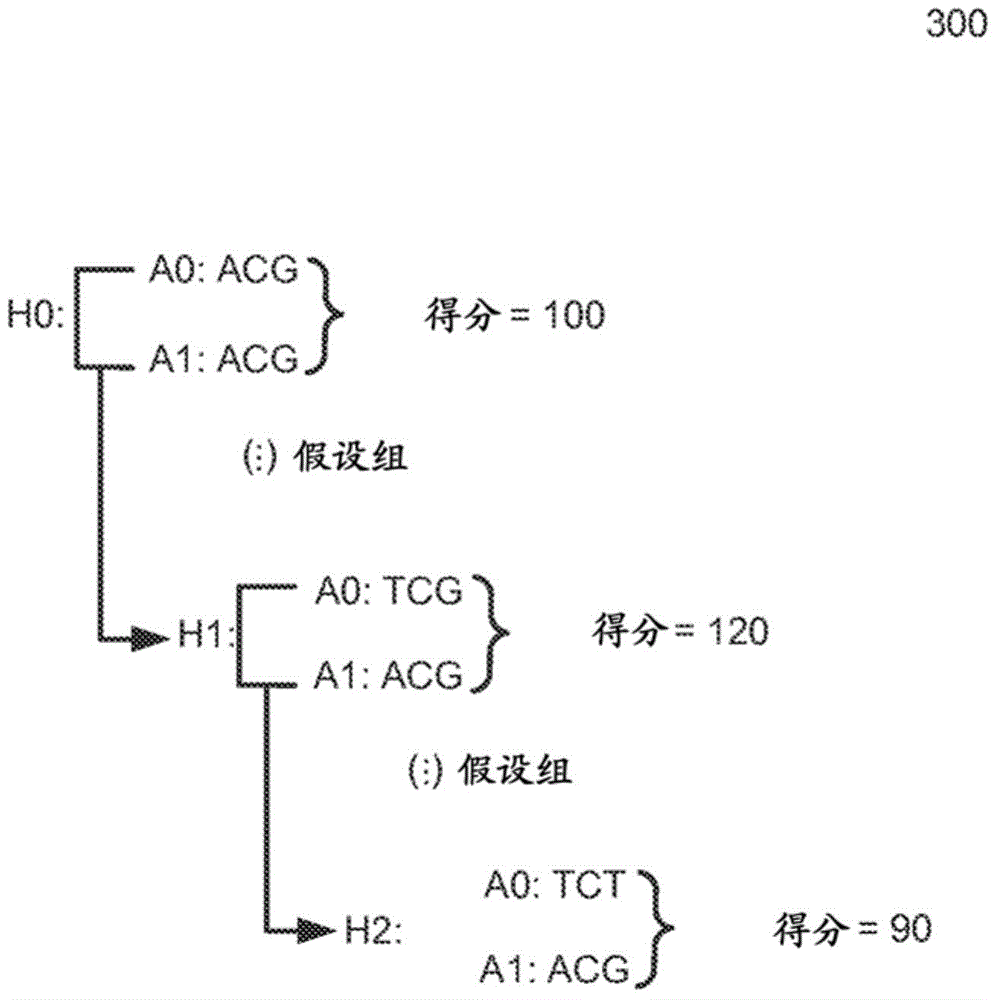
Determining variants in genome of a heterogeneous sample
InactiveUS20130110407A1Accurately determineBiostatisticsBiological testingSequence hypothesisHypothesis
After DNA fragments are sequenced and mapped to a reference, various hypotheses for the sequences in a variant region can be scored to find which sequence hypotheses are more likely. A hypothesis can include a specific variable fraction for the plurality of alleles that comprise the sequence hypothesis in the region. A likelihood of each hypothesis can be determined using a probability that accounts for the fraction of the alleles specified in the respective sequence hypothesis. Thus, other hypotheses besides standard homozygous and equal heterozygous (i.e., one chromosome with A and one with B in a cell) can be explored by explicitly including the variable fractions of the alleles as a parameter in the optimization. Also, a variant score can be determined for a variant relative to a reference. The variant score can be used to determine a variant calibrated score indicating a likelihood that the variant call is correct.
Owner:COMPLETE GENOMICS INC
Method for long range allele-specific PCR
InactiveUS20070184457A1Enhanced hybridizationSugar derivativesMicrobiological testing/measurementPolymerase LA-DNA
The present invention is directed to a method and kit for determining the molecular haplotype of a gene in a diploid DNA sample. The method discriminates between two haplotypes on the basis of a difference of one or more nucleotides using allele-specific PCR amplification in combination with long range PCR, using a DNA polymerase enzyme having 3′→5′ exonuclease activity, using annealing temperature conditions sufficiently greater than the predicted annealing temperature (Tm) to effect selective hybridization and extension of an allele-specific extension primer to the target allele relative to the variant allele. The present invention is particularly useful, for example, to determine the haplotype of a gene having multi-allelic genetic loci separated by a distance in which accurate PCR amplification of a single fragment containing the multiple genetic loci cannot be performed without using a DNA polymerase enzyme having 3′→5′ exonuclease activity, generally about 5 kilobases or more.
Owner:UNIV OF UTAH RES FOUND
Determining variants in a genome of a heterogeneous sample
InactiveCN104160391AMicrobiological testing/measurementBiostatisticsSequence hypothesisDNA fragmentation
After DNA fragments are sequenced and mapped to a reference, various hypotheses for the sequences in a variant region can be scored to find which sequence hypotheses are more likely. A hypothesis can include a specific variable fraction for the plurality of alleles that comprise the sequence hypothesis in the region. A likelihood of each hypothesis can be determined using a probability that accounts for the fraction of the alleles specified in the respective sequence hypothesis. Thus, other hypotheses besides standard homozygous and equal heterozygous (i.e., one chromosome with A and one with B in a cell) can be explored by explicitly including the variable fractions of the alleles as a parameter in the optimization. Also, a variant score can be determined for a variant relative to a reference. The variant score can be used to determine a variant calibrated score indicating a likelihood that the variant call is correct.
Owner:COMPLETE GENOMICS INC
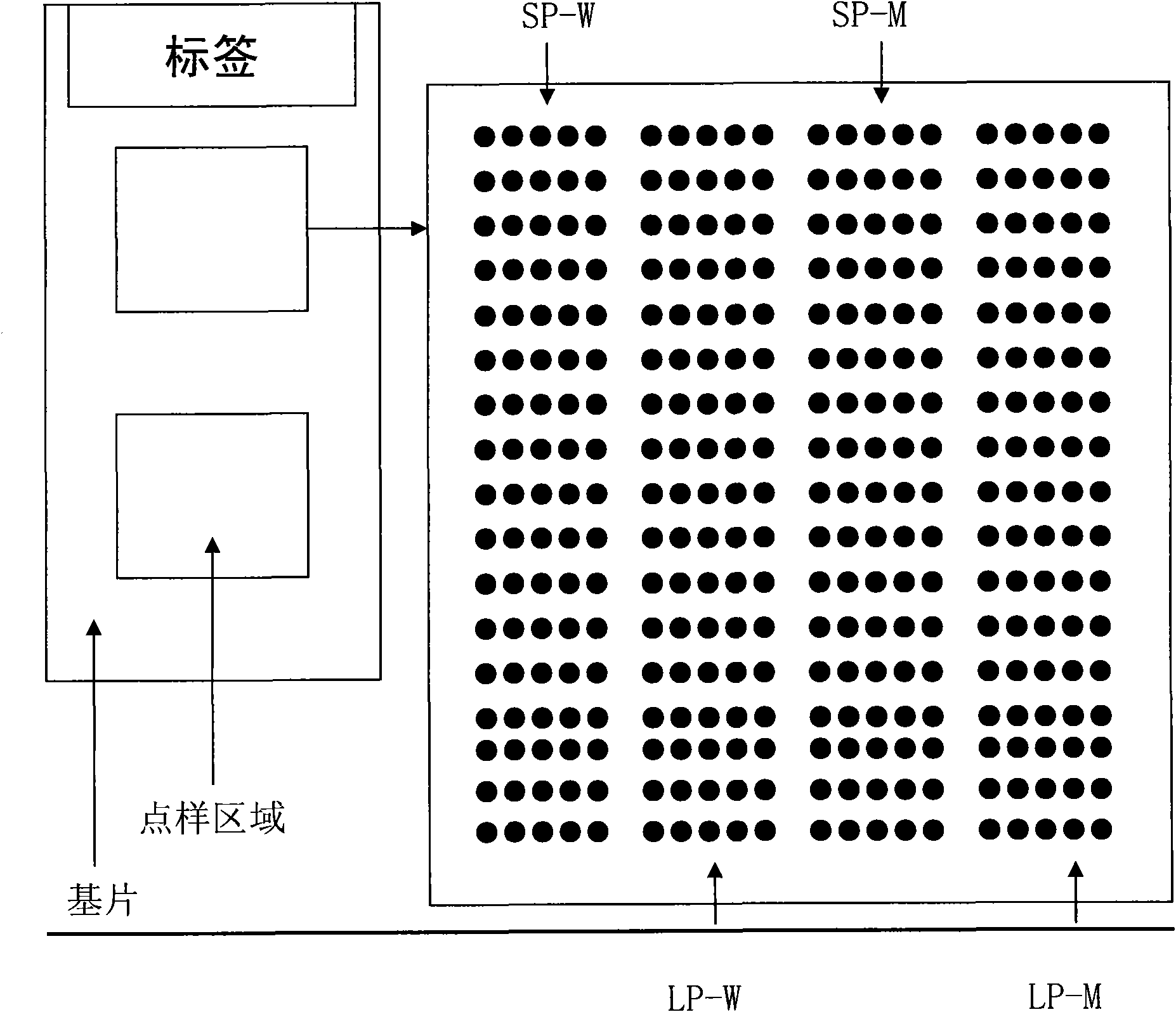
Double-probe gene mutation detecting method based on allele special amplification as well as special chip and kit thereof
ActiveCN101619352AEasy to detectAvoid the hassle of markingMicrobiological testing/measurementGenotypeOligonucleotide
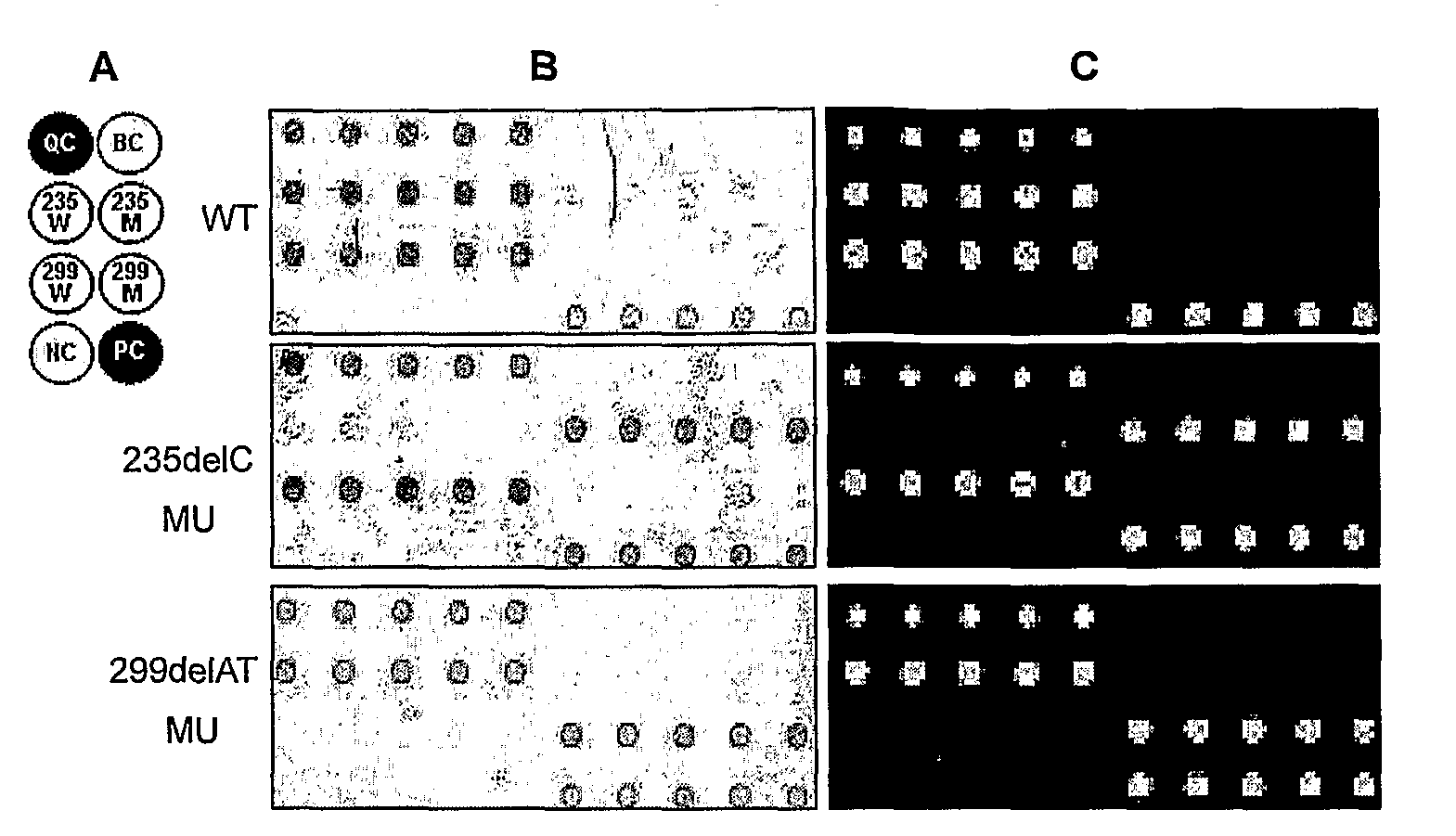
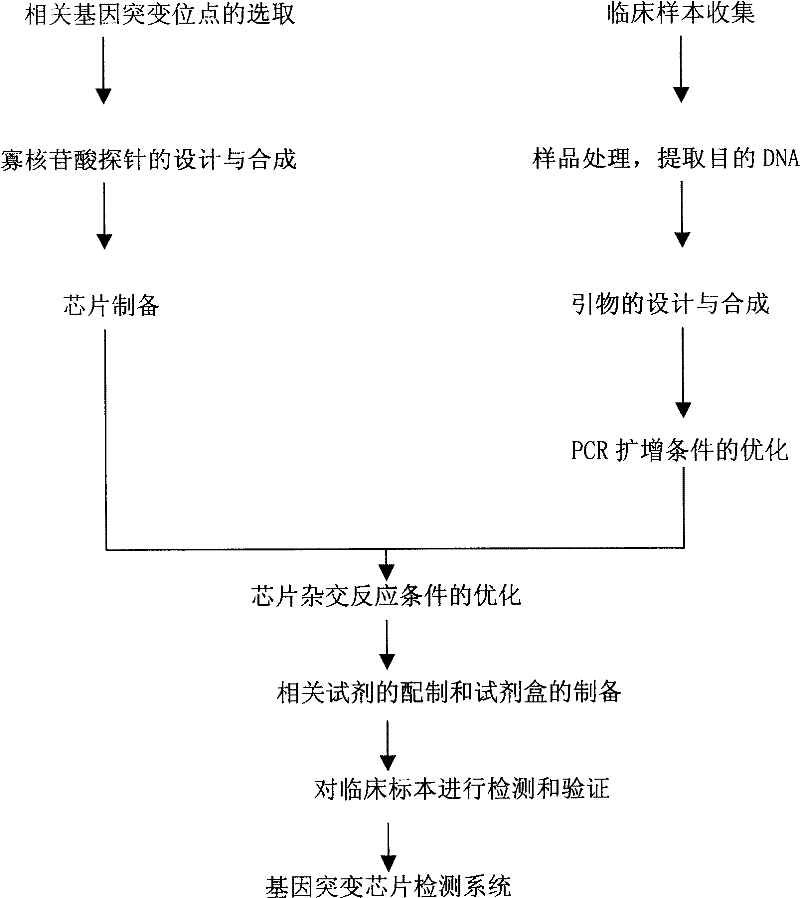
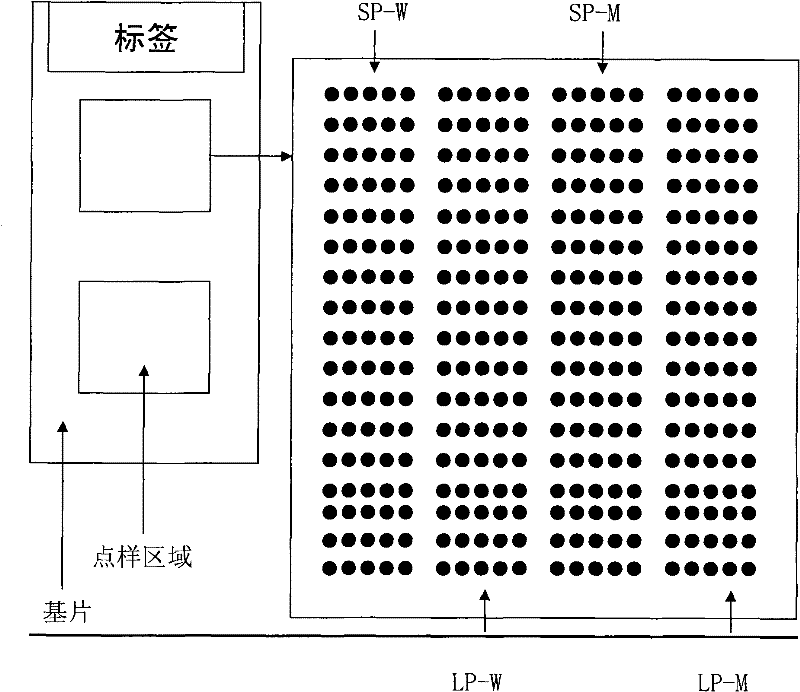
The invention relates to a method for identifying gene mutation types in the field of gene analysis as well as a special chip and a kit thereof. The gene mutation detecting method comprises the following steps: taking a genome to be detected from a human tissue as a template, carrying out multiple allele special PCR amplification by a primer group that is designed aiming at special mutant sites and DNA polymerase without 3'-5' end exonclease activity, then hybridizing the obtained PCR product and an oligonucleotide probe (allele special probe) on the gene chip, and confirming mutation types of all gene sites according to the hybridizing result. The allele special probe is designed aiming at special gene types of gene mutant sites to be detected. The invention can detect gene mutations in comprehensive, systemic and high-flux ways and has light environmental pollution as well as simple and rapid operation compared with PCR-RFLP and a sequencing method.
Owner:CENT SOUTH UNIV
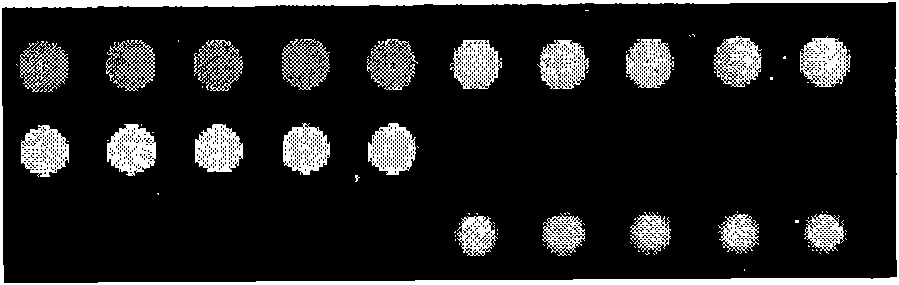
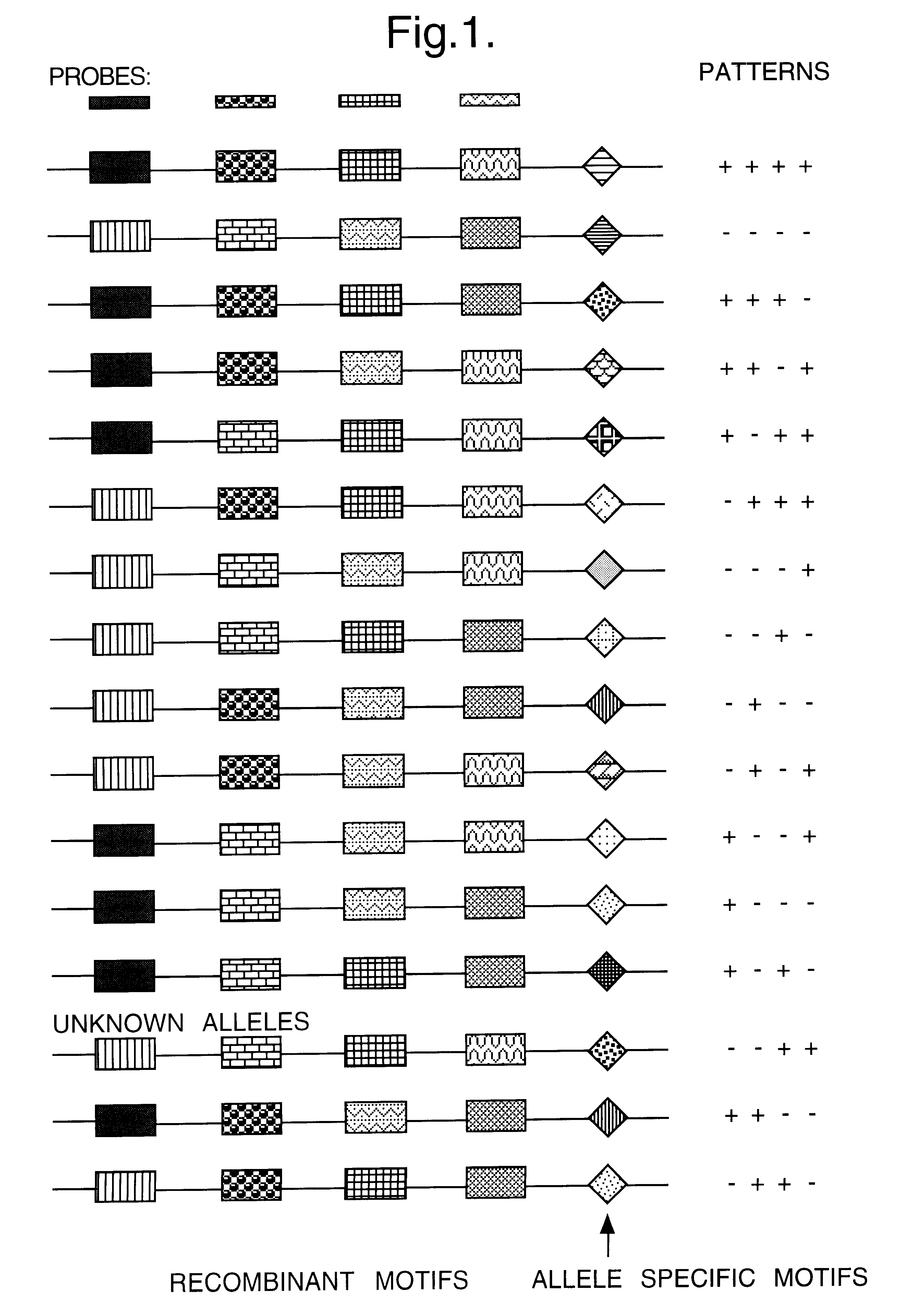
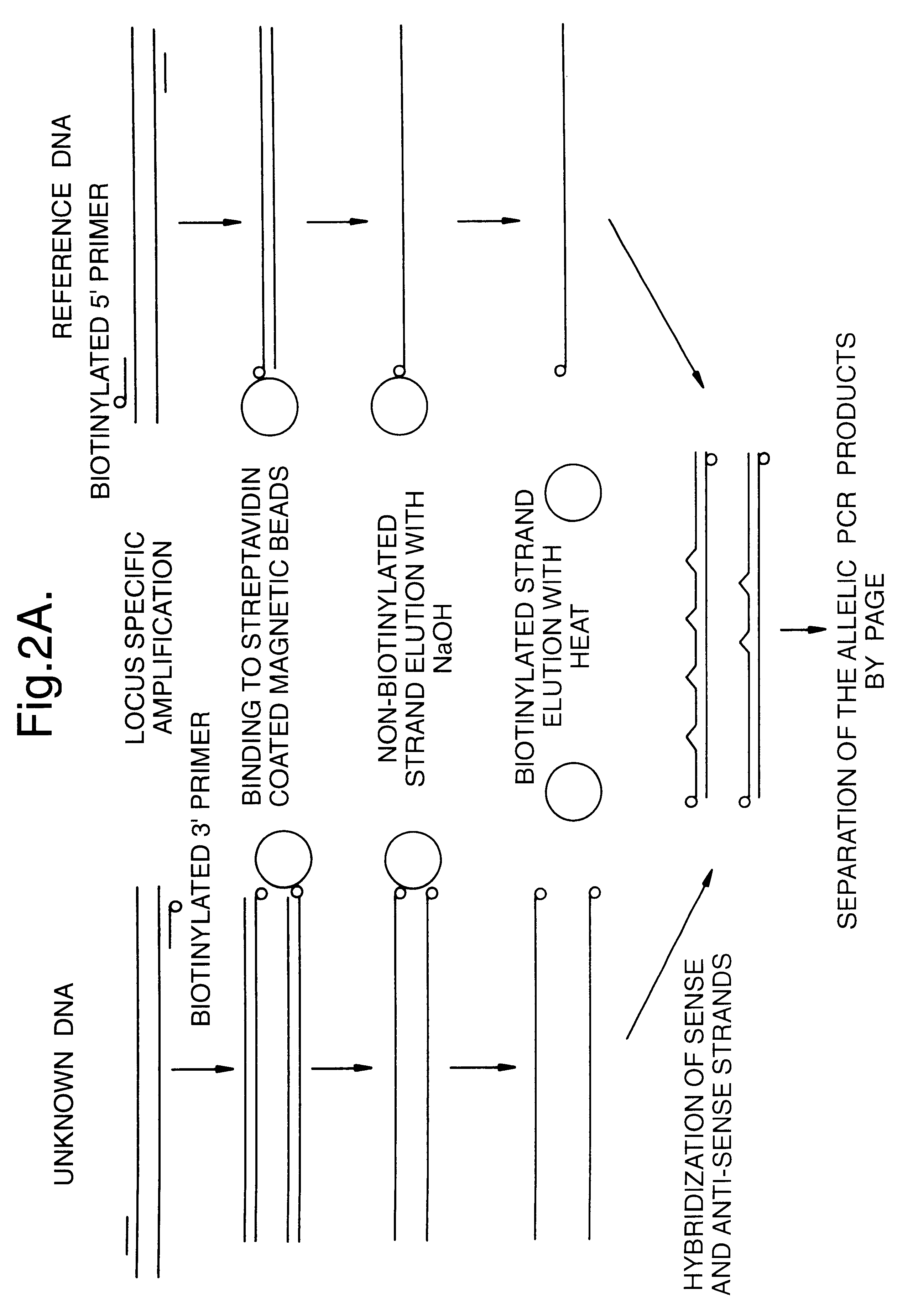
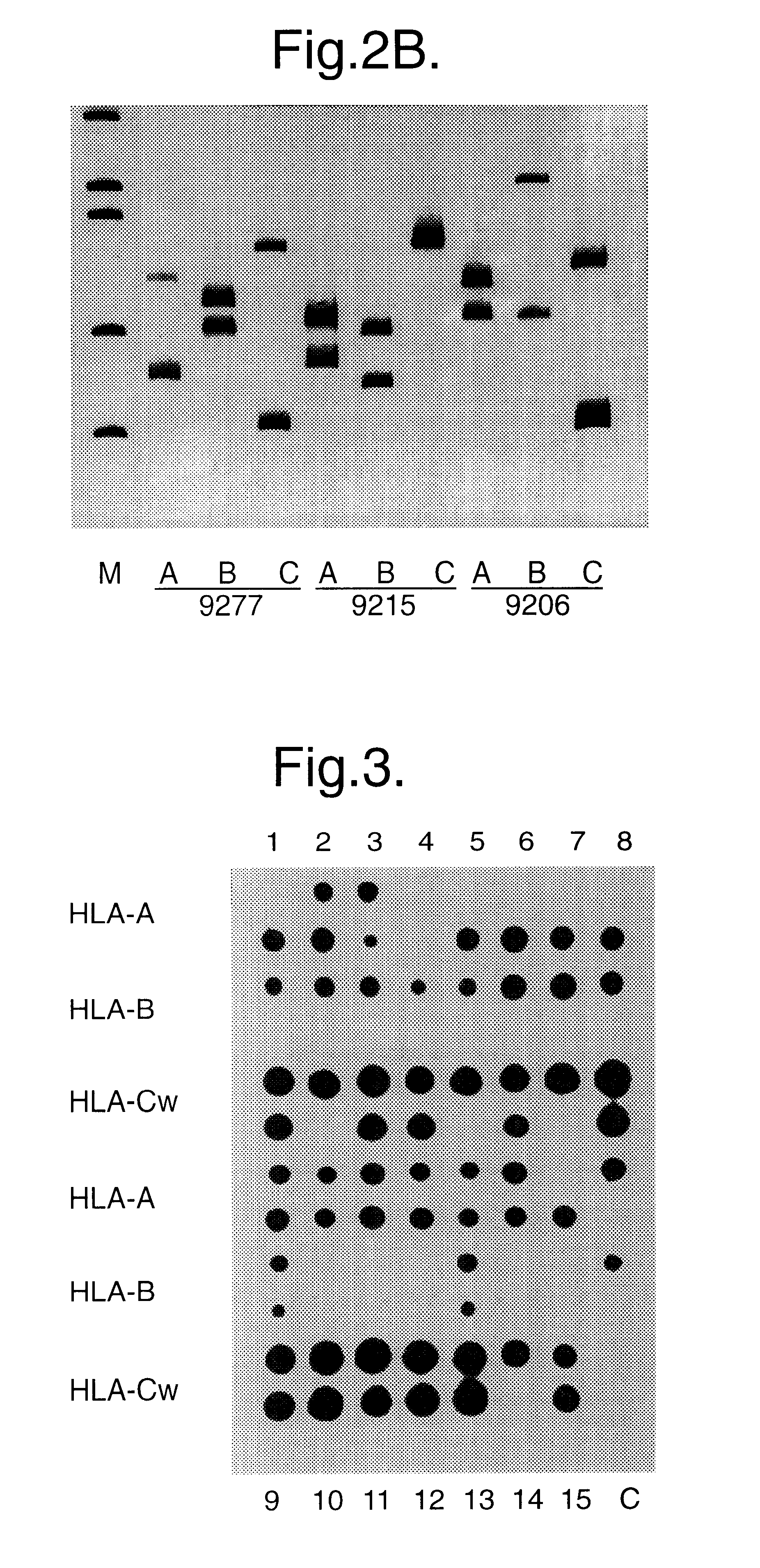
Method for identifying an unknown allele
The invention provides a method for identifying an unknown allele of a polyallelic gene, which method comprises (i) contacting the unknown allele with a panel of probes, each of which recognises a sequence motif that is present in some alleles of the polyallelic gene but not in others; (ii) observing which probes recognise the unknown allele so as to obtain a fingerprint of the unknown allele; and (iii) comparing the fingerprint with fingerprints of known alleles. The use of a panel of probes which each recognises a different motif allows identification of which motifs are present in the unknown allele. The alleles of the polyallelic gene each have a unique combination of motifs and so identification of this combination (or "fingerprint") leads to identification of the unknown allele.
Owner:ANTHONY NOLAN BONE MARROW TRUST
Methods for identification of alleles
ActiveUS20100041563A1Low homologyMicrobiological testing/measurementLibrary screeningPolymerase LGenomic DNA
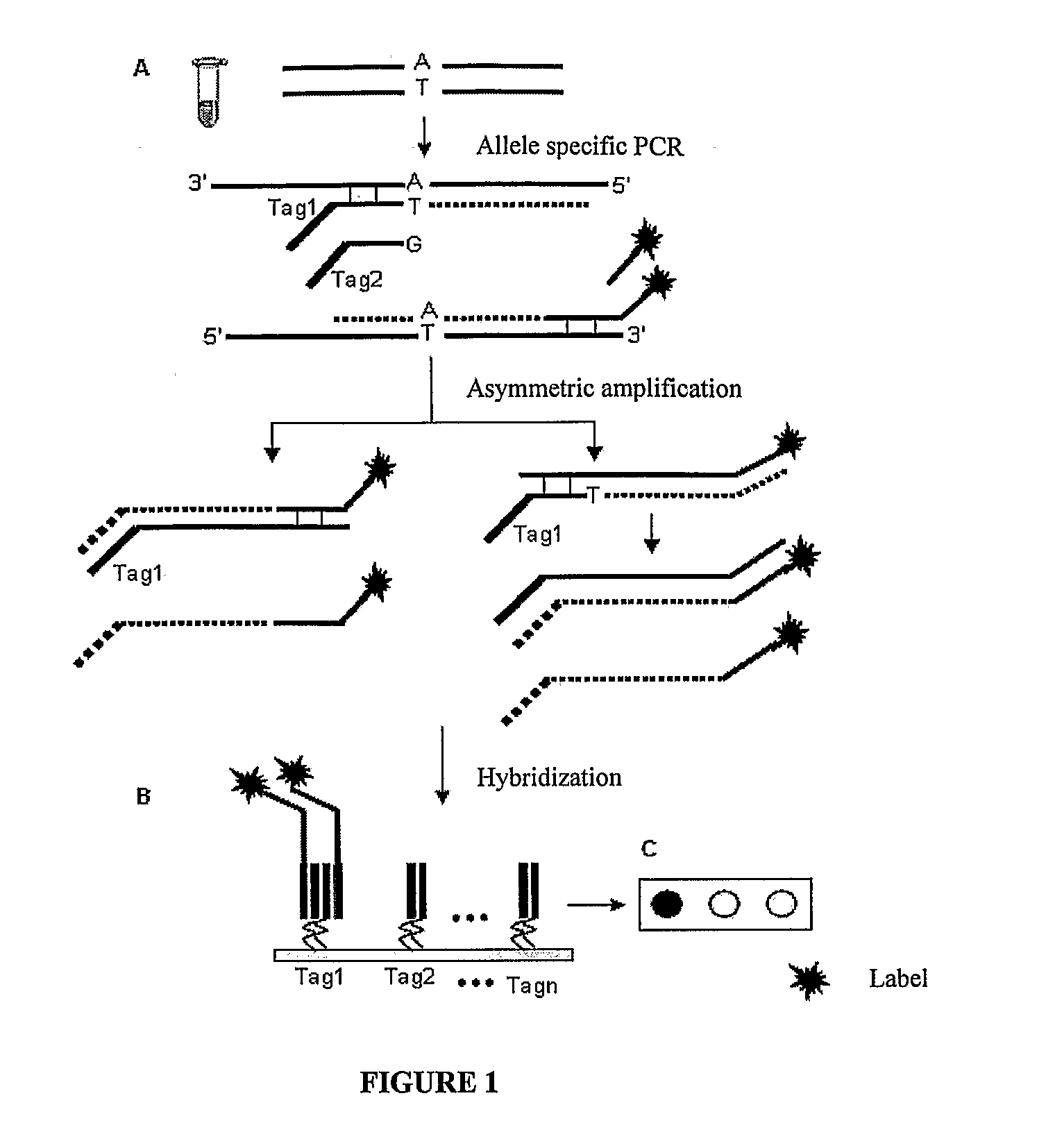
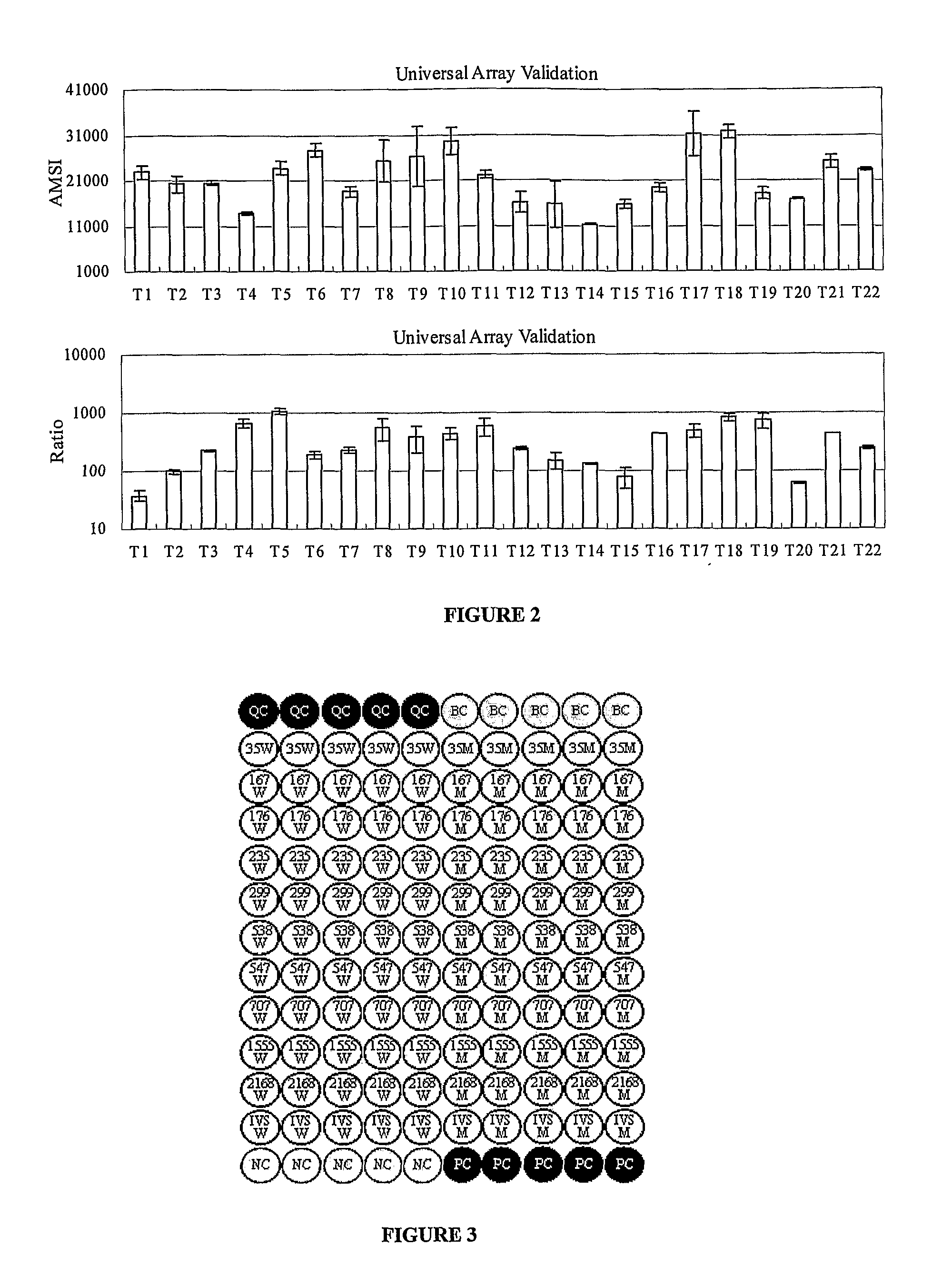
The invention provides a method for identification of alleles. In this method, genomic DNA is used as target. Multiple allele-specific PCR amplification are carried out with a group of primers comprising one or more allele-specific primers for a target gene, a universal primer, and a common primer; and a DNA polymerase without 5′ to 3′ exonuclease activity. The PCR products are hybridized with tag probes immobilized on a DNA chip. Results are determined based on the signal intensity and the position of the probe immobilized on the array. Each allele-specific primer comprises a unique tag sequence at the 5′ end. Each tag probe immobilized on the DNA chip comprises a sequence identical to its corresponding tag sequence; and each tag probe hybridizes only with the complementary sequence in the PCR amplification product.
Owner:CAPITALBIO CORP +1
Restrictive two-stage genome-wide association study (GWAS) method based on SNPLDB mark
InactiveCN104651517AShorten the attenuation distanceMultiple allelic variationMicrobiological testing/measurementProteomicsMulti siteRegression analysis
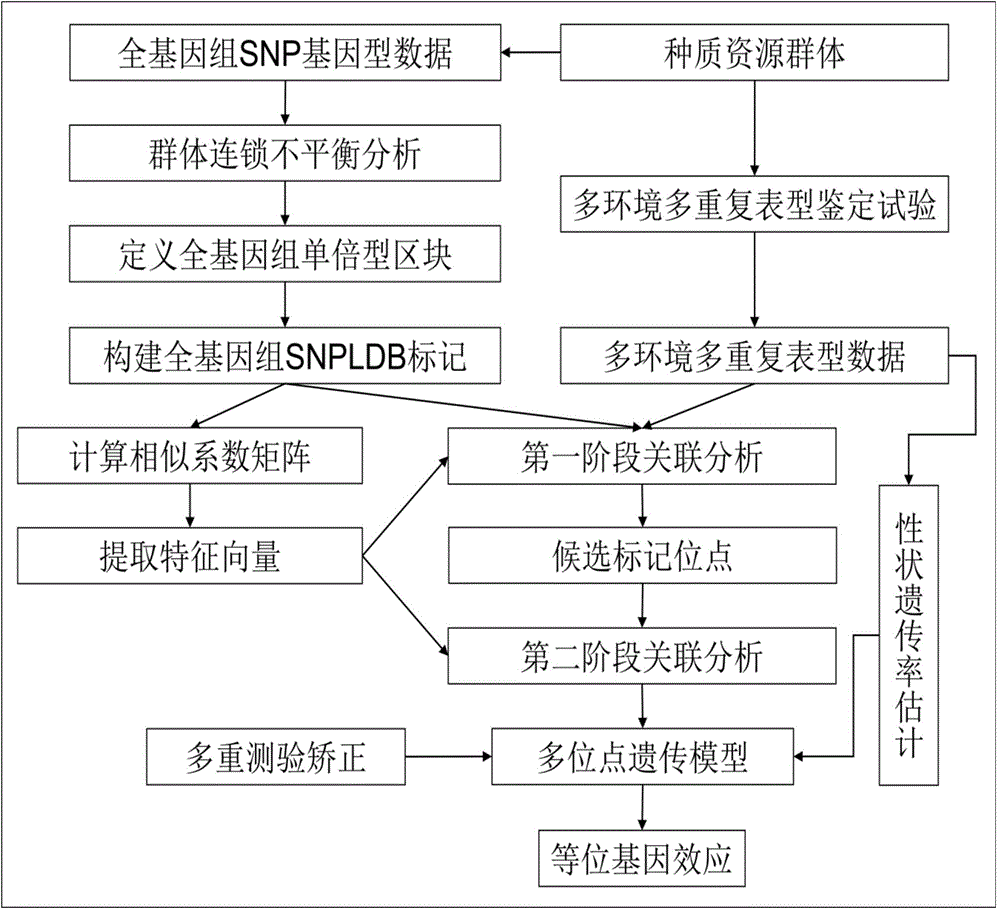
The invention discloses a restrictive two-stage genome-wide association study (GWAS) method based on an SNPLDB mark, and aims at solving the problems that a traditional method cannot be used for estimating multiple allele information, and is high in false positive rate and low in efficiency of detection on inbreeding crops. By combination of the SNPLDB mark constructed on the basis of a haplotype block, correction of deviation of an inbreeding population relation analysis model and a two-stage relation analysis strategy under a multi-site model, the GWAS method suitable for conventional breeding of inbreeding crops is built; the SNPLDB mark is applied to GWAS, so that a method is provided for multiple allele estimation; candidate sites are screened on the basis of a single-site model in the first stage, and are further screened on the basis of a progressive regression analysis method in the multi-site model in the second stage, so as to balance the problems of missing of heritability and over-high heritability estimation. Therefore, the interpretation ratio of a final genetic model is controlled at trait heritability. The positioning accuracy and efficiency are improved by a feature vector and an appropriate significant level of a similarity coefficient matrix estimated by the SNPLDB mark by GWAS.
Owner:NANJING AGRICULTURAL UNIVERSITY
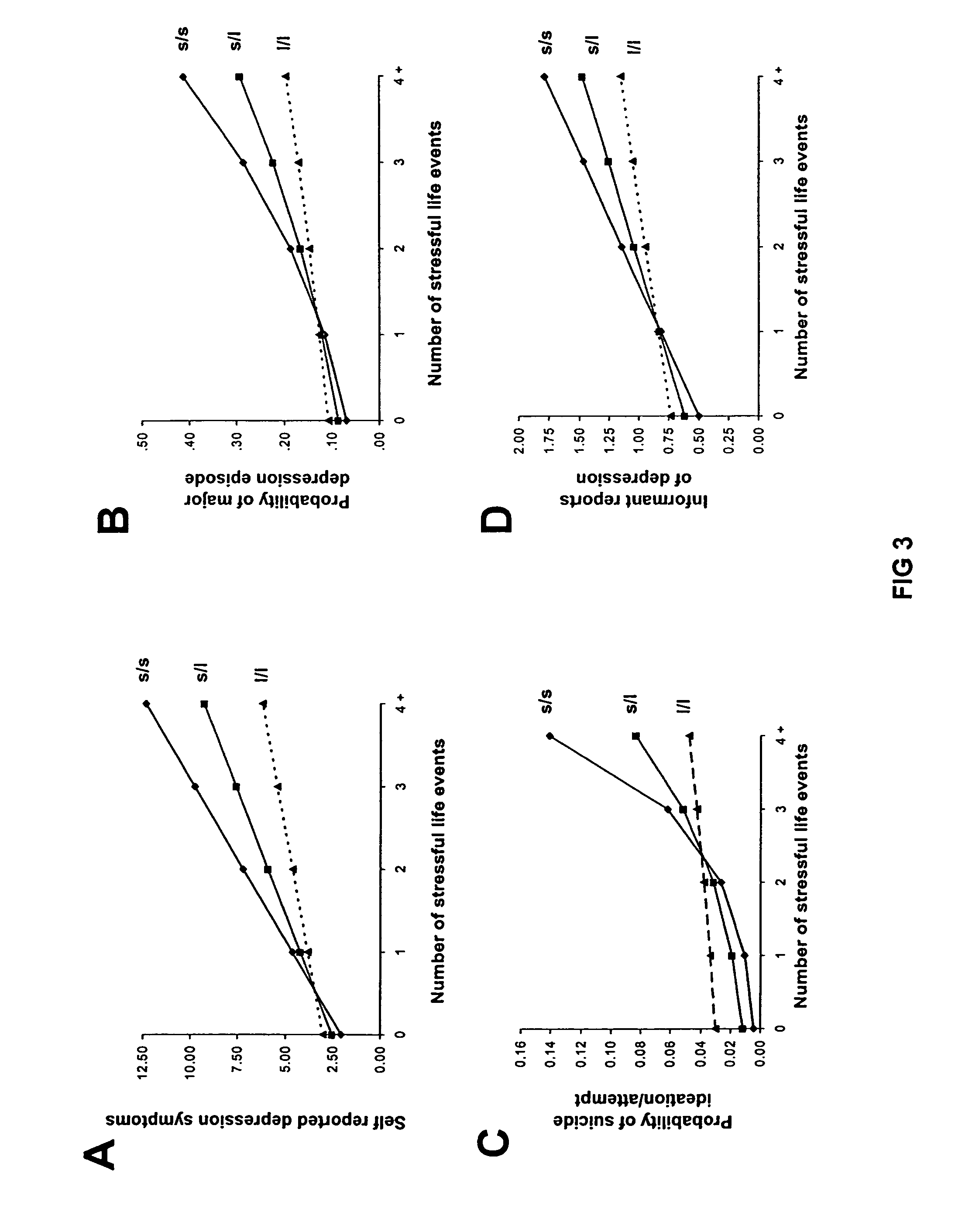
Method for assessing predisposition to depression
The present invention relates to diagnostic methods for assessing predisposition of a subject to a mental disorder phenotype having an association with an at-risk allele of a brain-functional gene having a plurality of alleles, the association being conditioned by a pathogenic environmental risk factor status condition. Additionally, the invention relates to methods for discovering a conditional association between a mental disorder phenotype and an at-risk allele of a brain-functional gene having a plurality of alleles, the association being conditioned by a pathogenic environmental risk factor status condition.
Owner:WISCONSIN ALUMNI RES FOUND
Preparation method and use of simple sequence repeat (SSR) marker for screening miscanthus by magnetic bead enrichment process
InactiveCN102080130AEfficient enrichmentCo-dominance is goodMicrobiological testing/measurementMagnetic beadGenetic diversity
The invention discloses a preparation method and use of a simple sequence repeat (SSR) marker for screening miscanthus by a magnetic bead enrichment process, which comprises: 1) extracting genomic DNA of miscanthus floridulus which belongs to miscanthus; 2) incising the extracted total DNA by using restriction incision enzyme to obtain a genomic DNA; 3) intercrossing a probe and a target fragment; 4) enriching the hybrid DNA molecules having an SSR sequence by using magnetic beads; 5) amplifying and purifying the DNA fragment having the SSR sequence; 6) cloning and sequencing the DNA fragment; and 7) designing an SSR primer according to the flanking sequence of SSR, and designing a specific primer by using primer design software for amplifying the satellite fragment at the locus. The method is easy and convenient for operation; meanwhile, the SSR marker has the characteristics of high codominance, high polymorphism, multiple allele property and the like and can be used for analyzing the genetic diversity and genetic relationship of miscanthus and used in construction of genetic map of miscanthus, calibration of a target gene and drawing of a fingerprint.
Owner:湖北光芒能源植物有限公司
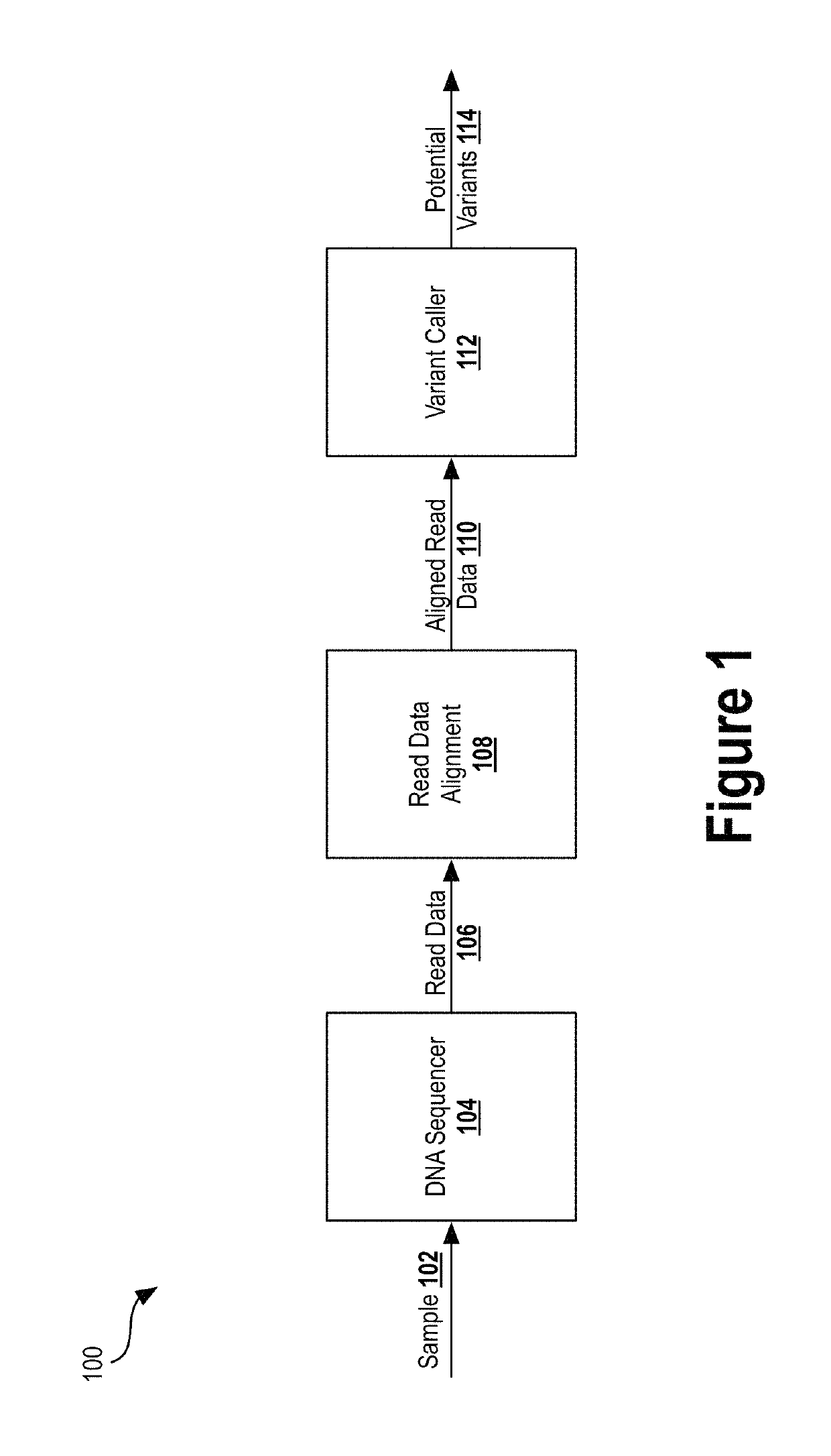
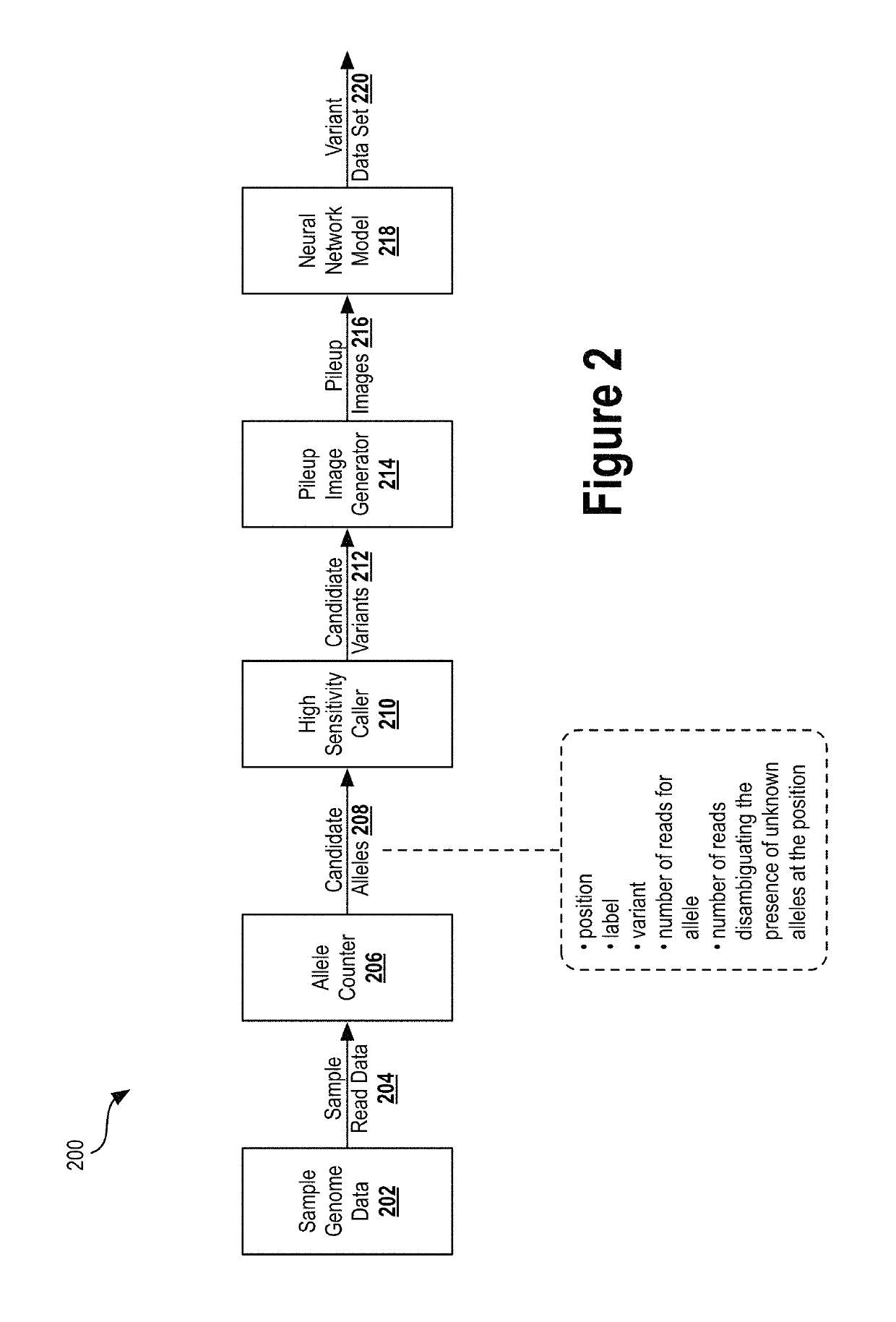
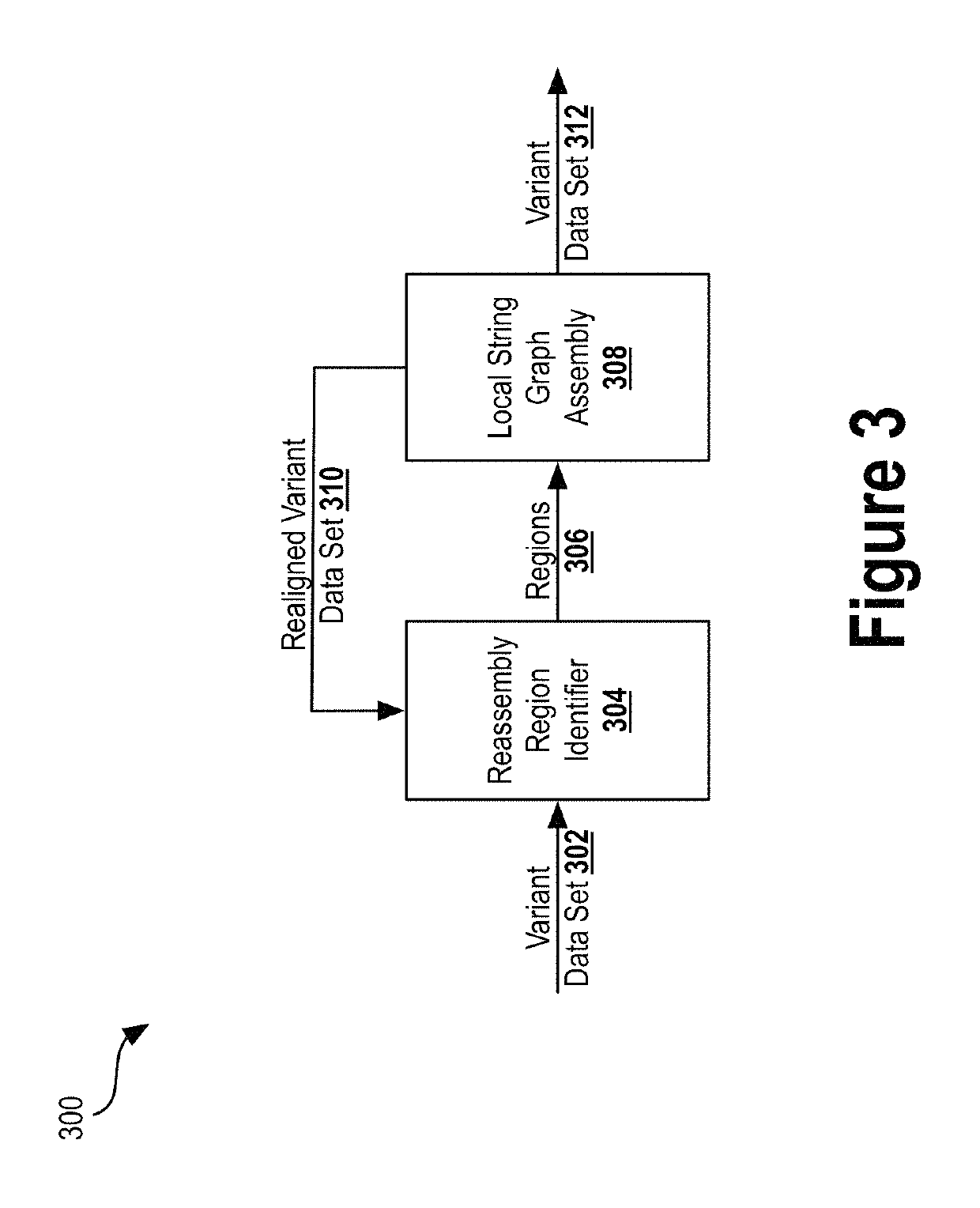
Deep learning analysis pipeline for next generation sequencing
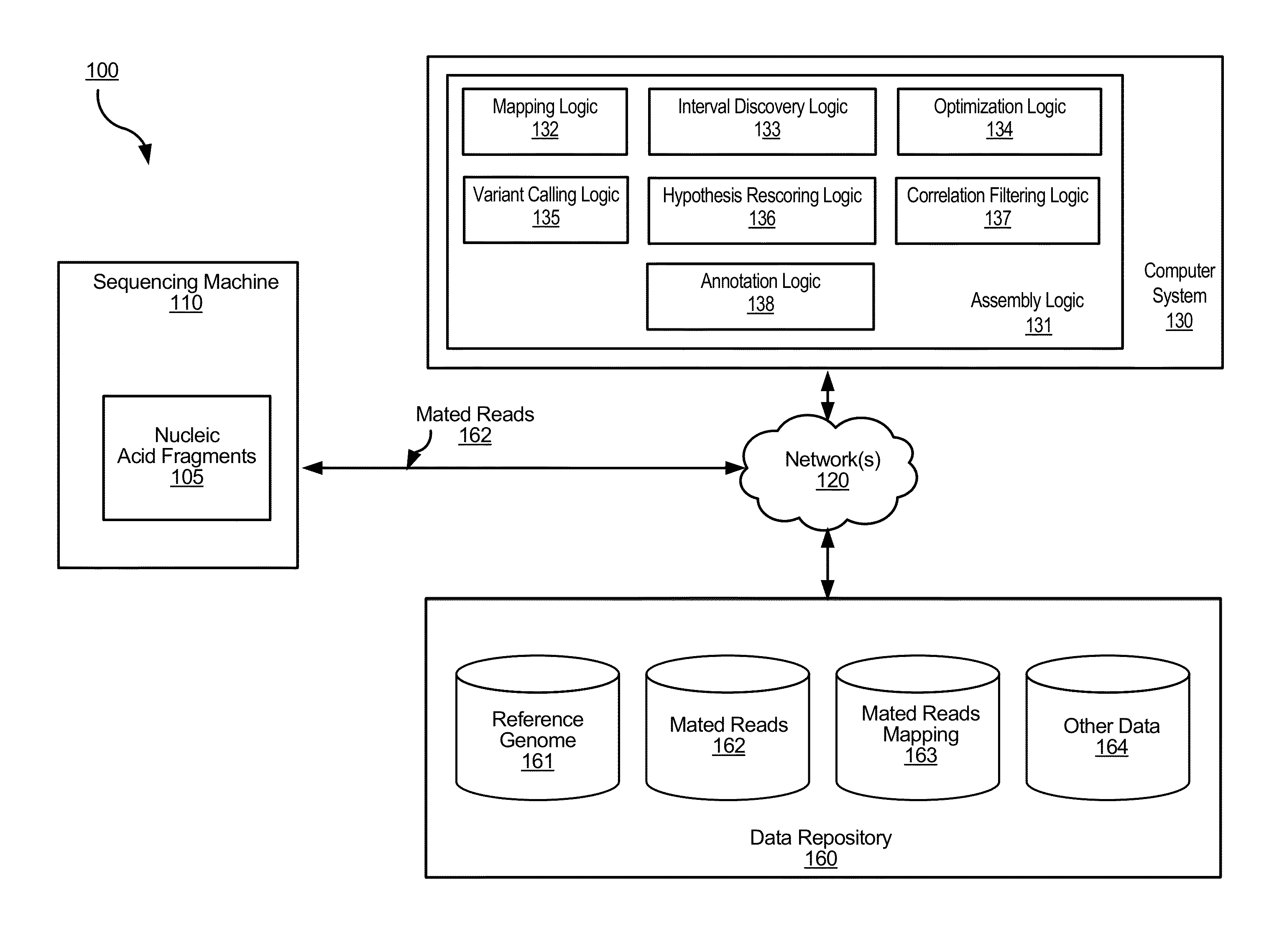
A method for variant calling in a next generation sequencing analysis pipeline involves obtaining a plurality of sequence reads that each include a nucleotide aligned at a nucleotide position within a sample genome. The method also involves obtaining a plurality of alleles associated with the nucleotide position. The method further involves determining that a particular allele of the plurality of alleles matches one or more sequence reads of the plurality of sequence reads, wherein the particular allele is located at the nucleotide position. Additionally, the method involves generating an image based on information associated with the plurality of sequence reads. Further, the method involves determining, by providing the generated image to a trained neural network, a likelihood that the sample genome contains the particular allele. The method may also involves providing an output signal indicative of the determined likelihood.
Owner:VERILY LIFE SCI LLC
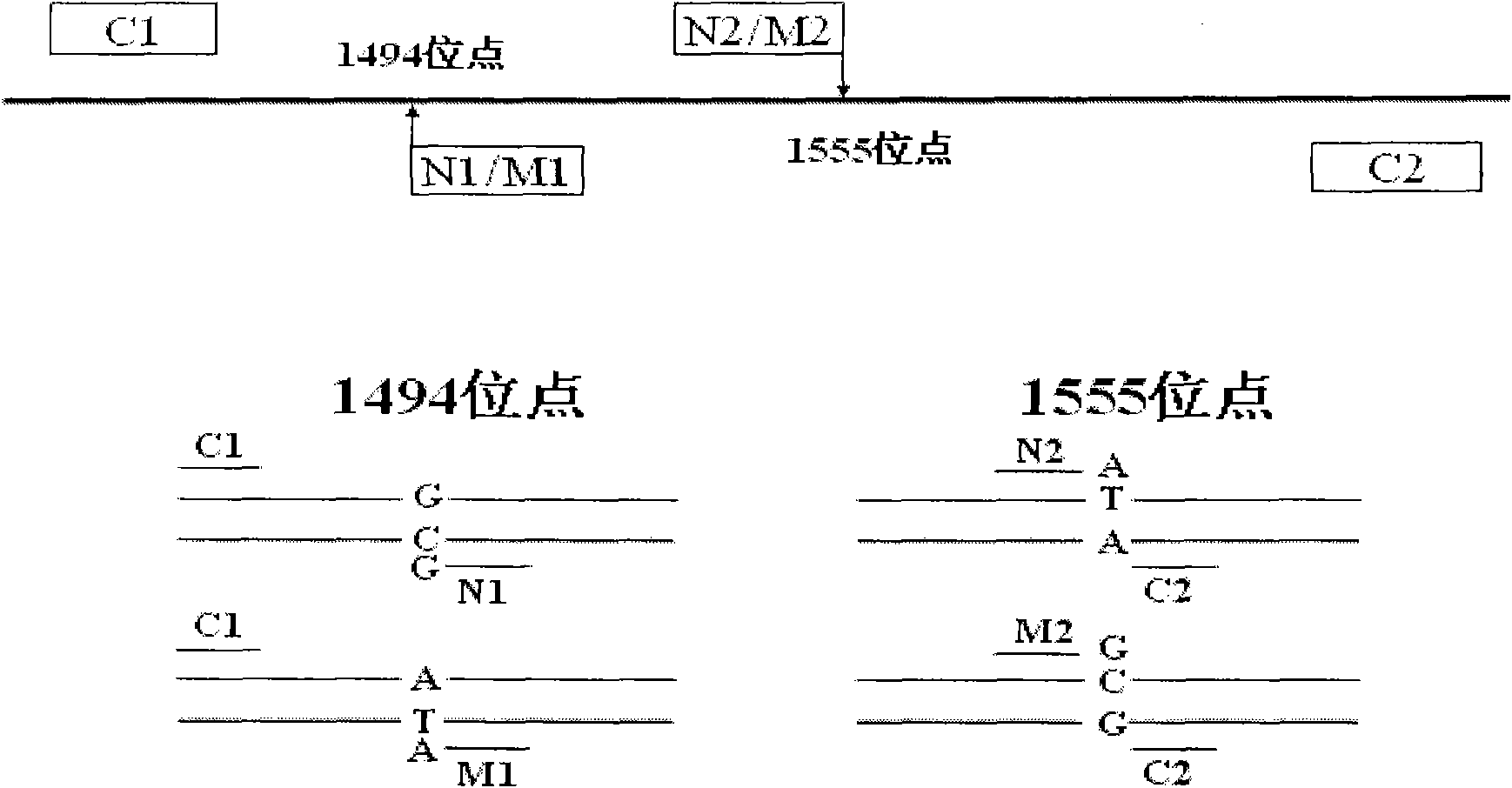
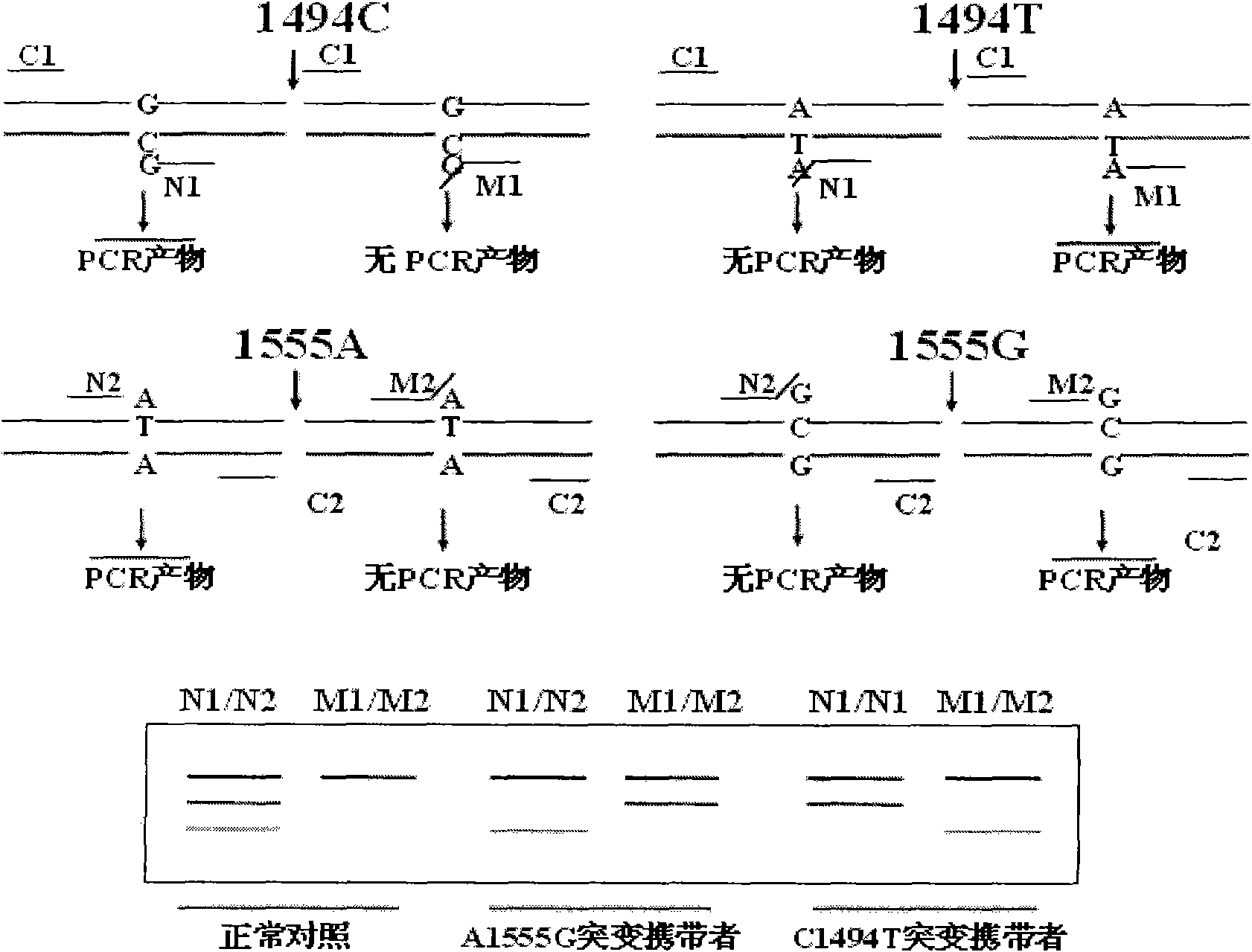
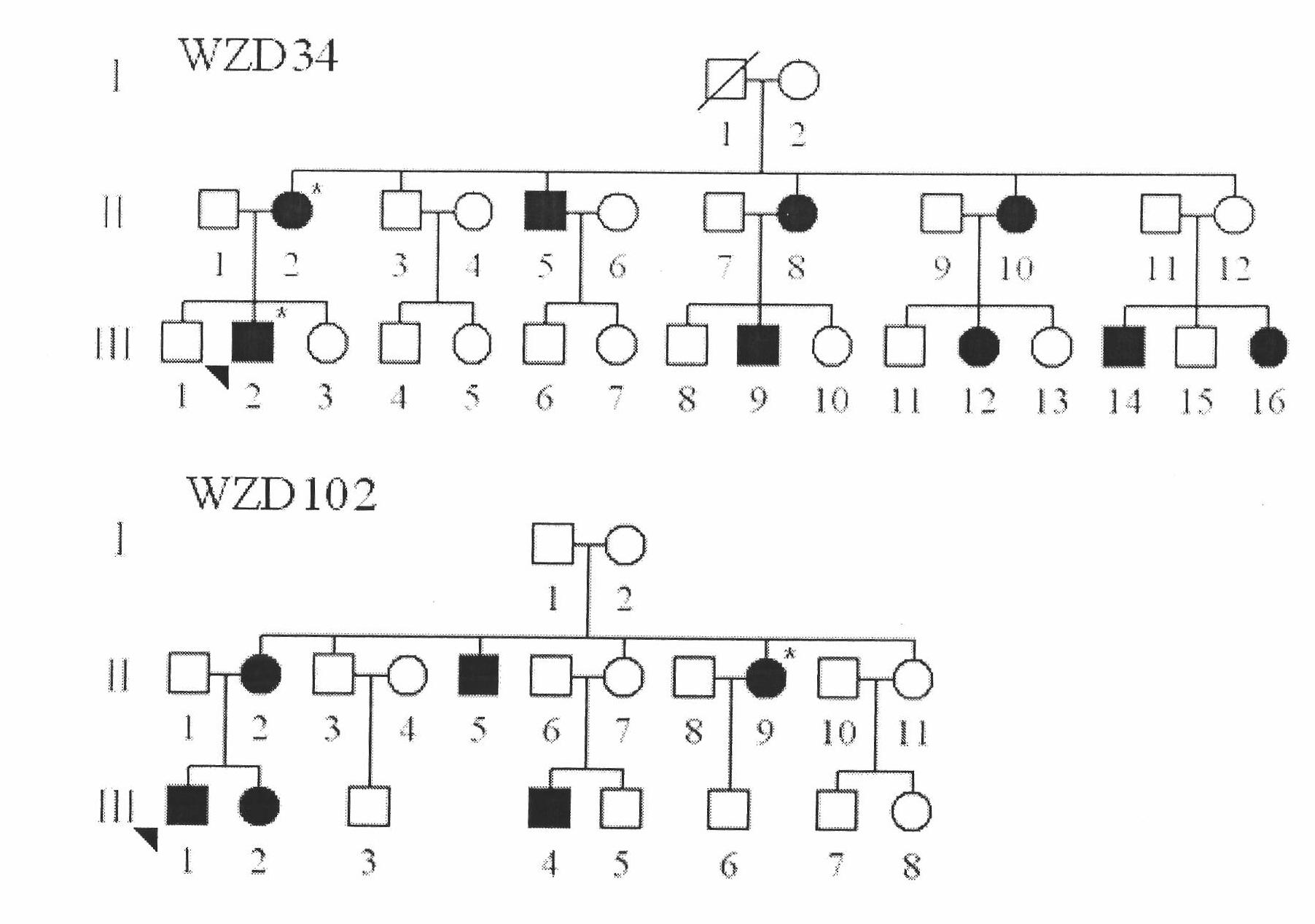
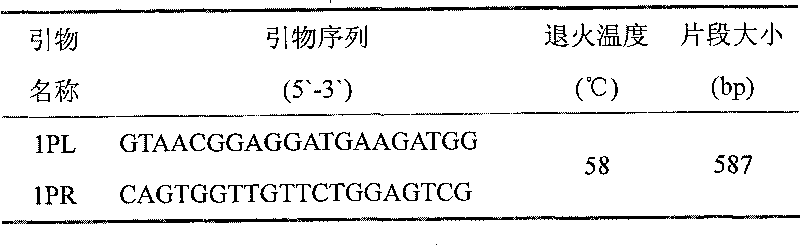
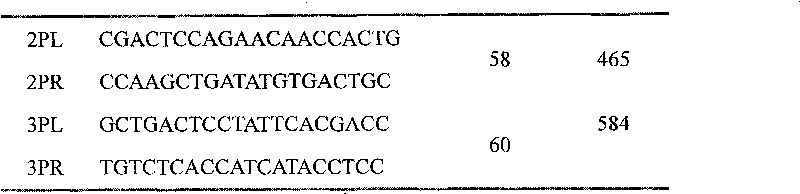
Kit for simultaneously detecting mutations in mitochondria DNA A1555G and C1494T and using method thereof
InactiveCN101768637ARelieve painSolve the problem of extracting DNAMicrobiological testing/measurementPositive controlGenomic DNA
The invention provides a kit for simultaneously detecting mutations in mitochondria DNA A1555G and C1494T related to maternally inherited drug-induced deafness and a using method thereof. The kit comprises a reagent for extracting sample genomic DNA, a PCR amplification reactive reagent, a primer mixed liquor, a positive control template and a negative control template. The using method of the kit mainly comprises the following steps: using blood, hair with follicle, oral mucosa doctor blade, saliva, and the like, as a sample; adopting a proteinase K digestion pyrolysis method to extract genomic DNA; and then simultaneously detecting mutations in A1555G and C1494T by multiple allele specific PCR. The kit is used for detecting the mutations in mitochondria DNA A1555G and C1494T related to maternally inherited drug-induced deafness and is more rapid, economical and simpler than the single detection for mutation in A1555G or C1494T, and the kit has low requirements for equipment and environment and is conducive to promotion and application.
Owner:WENZHOU MEDICAL UNIV
SNP molecular marker of swine and primer thereof
The invention screens a PSMB6 gene through swine breeds and combination widely used in commercial swine breeding, and searches out an SNP molecular marker for tracing DNA of a pork product; and a molecular marker of a (C / G) multiple allele SNP is in the 428th locus in the gene. The invention also provides a group of primers for identifying the SPN molecular marker of a swine PSMB6 gene sequence and a method thereof. The analysis of the distribution condition of the SNP locus allele can be used for tracing the DNA of the pork product.
Owner:SHANGHAI ACAD OF AGRI SCI
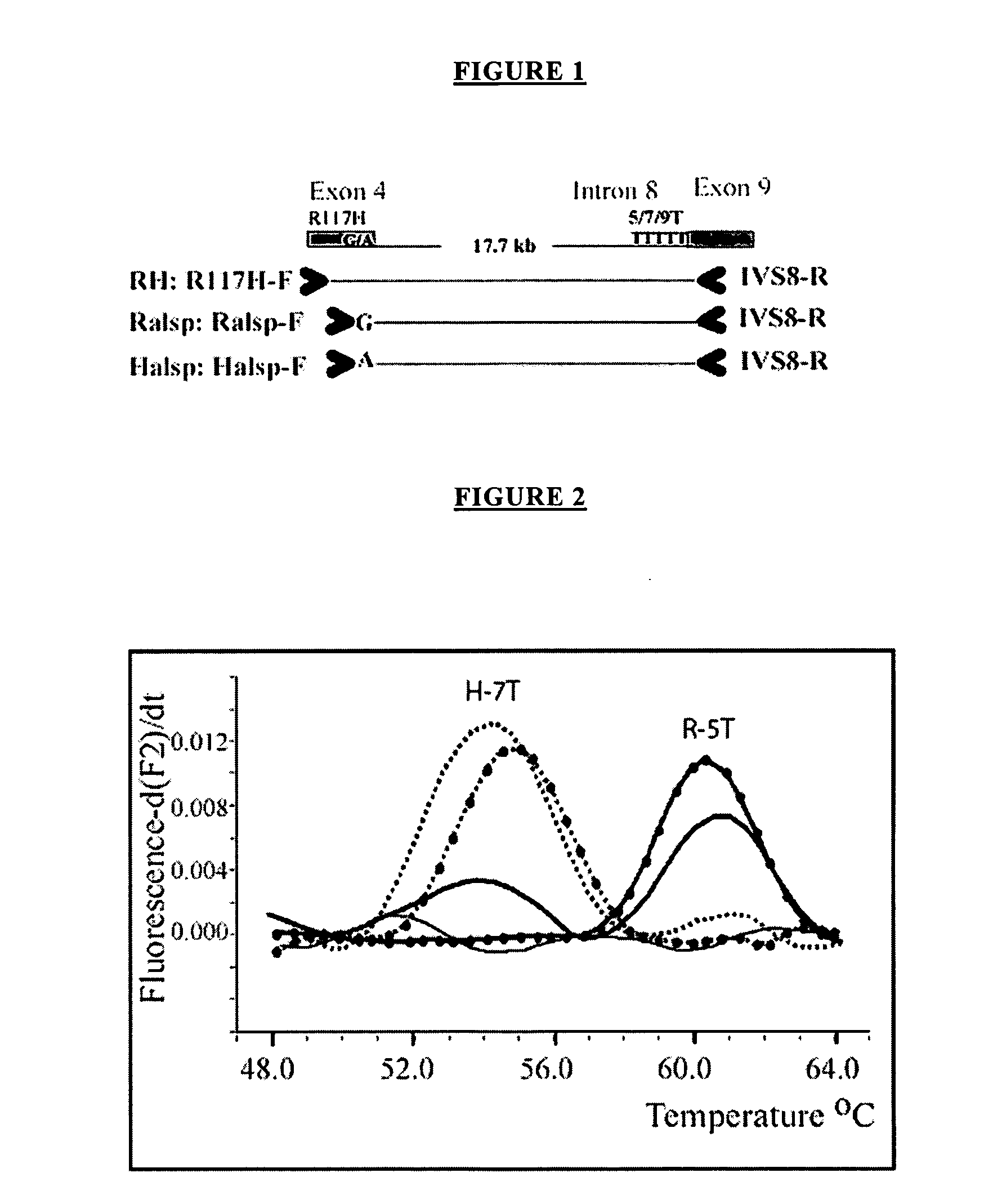
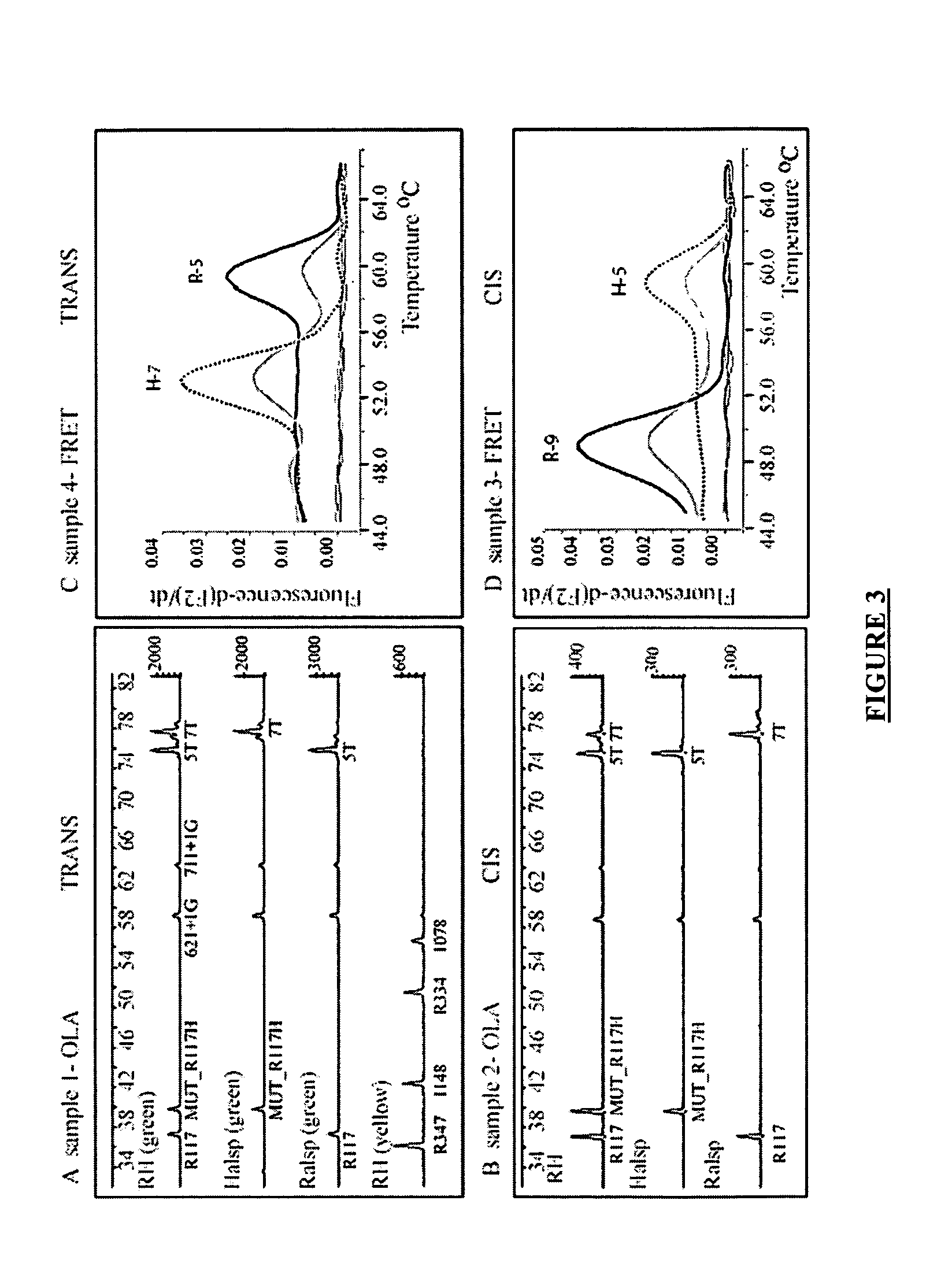
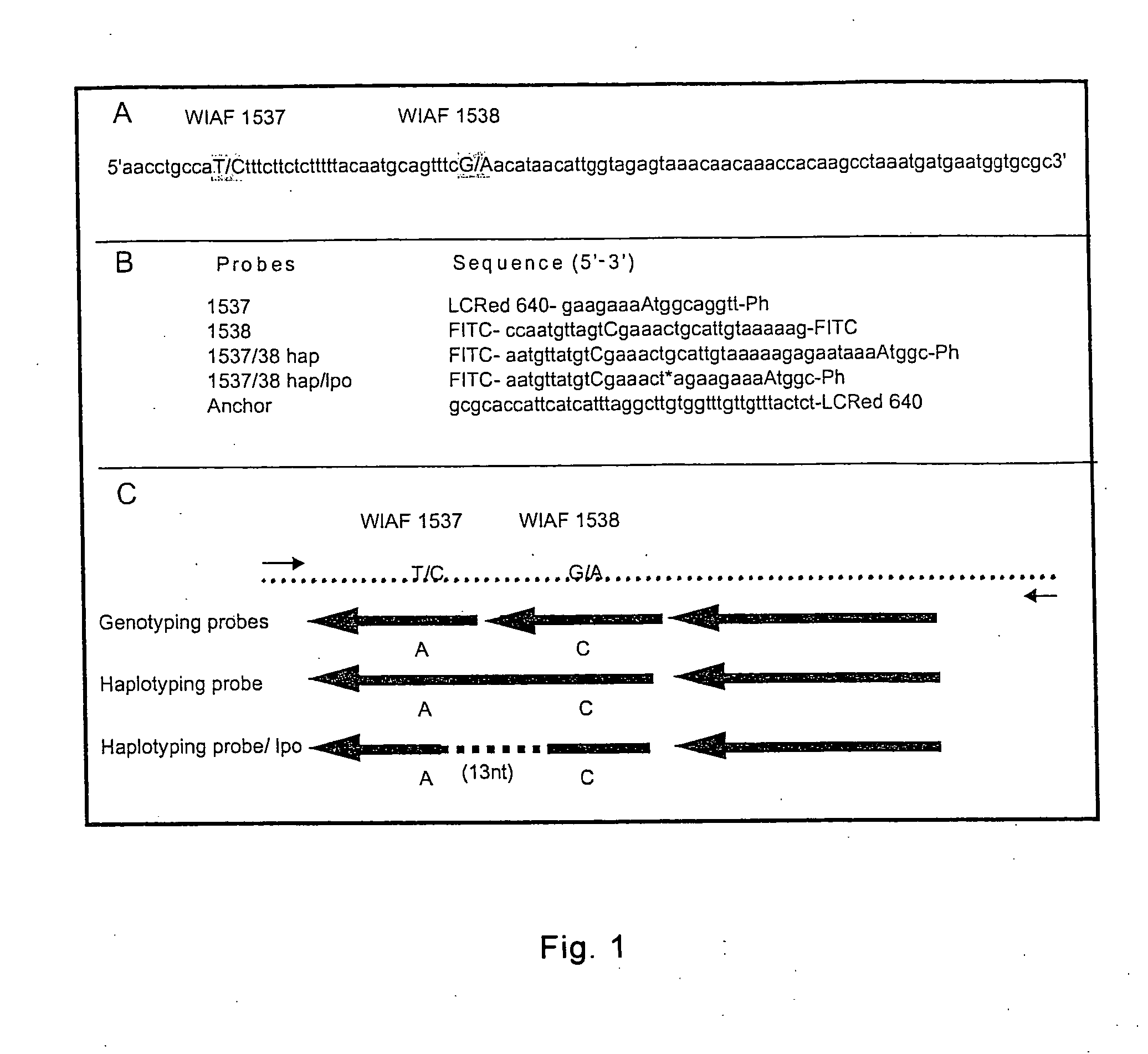
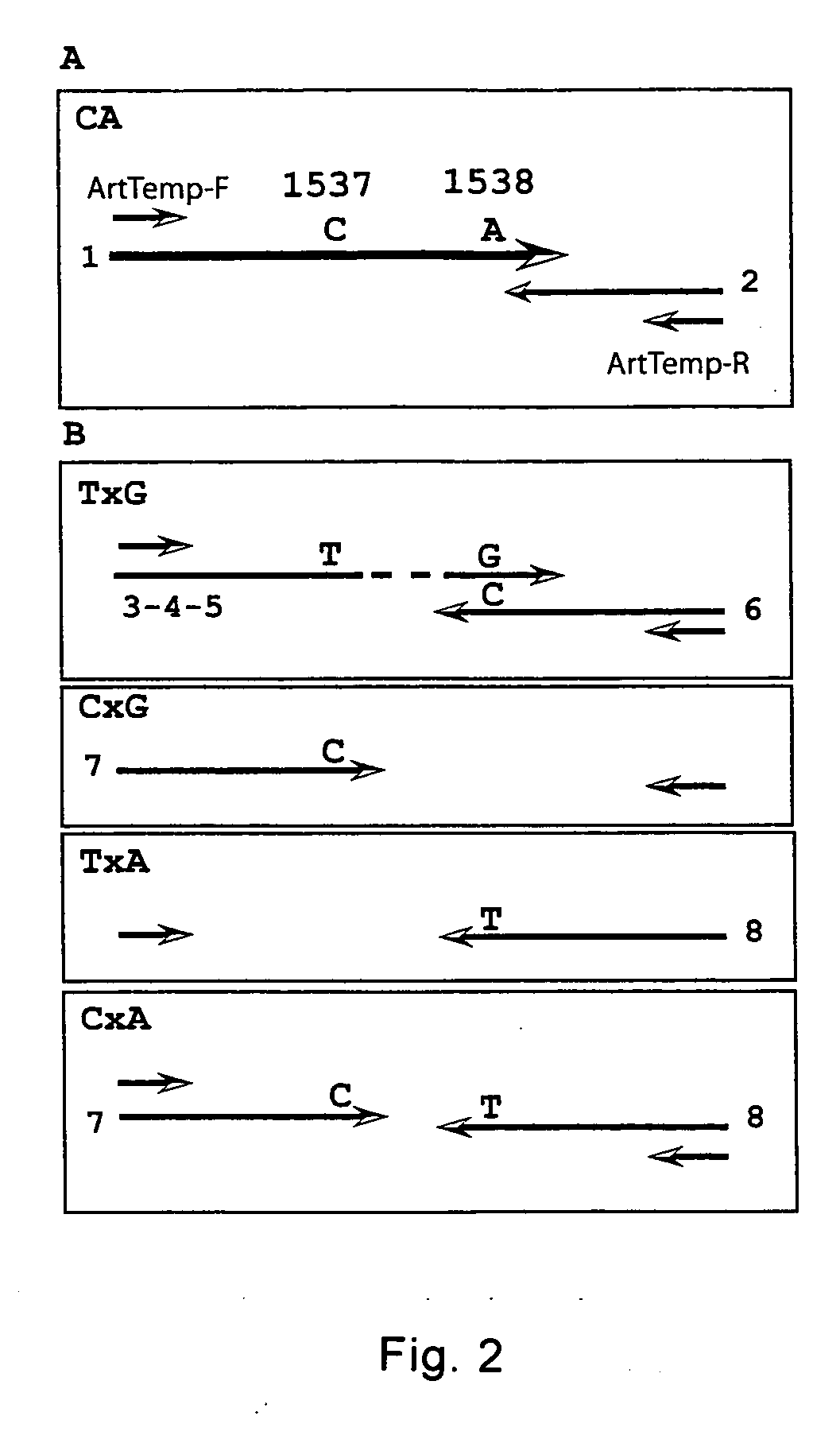
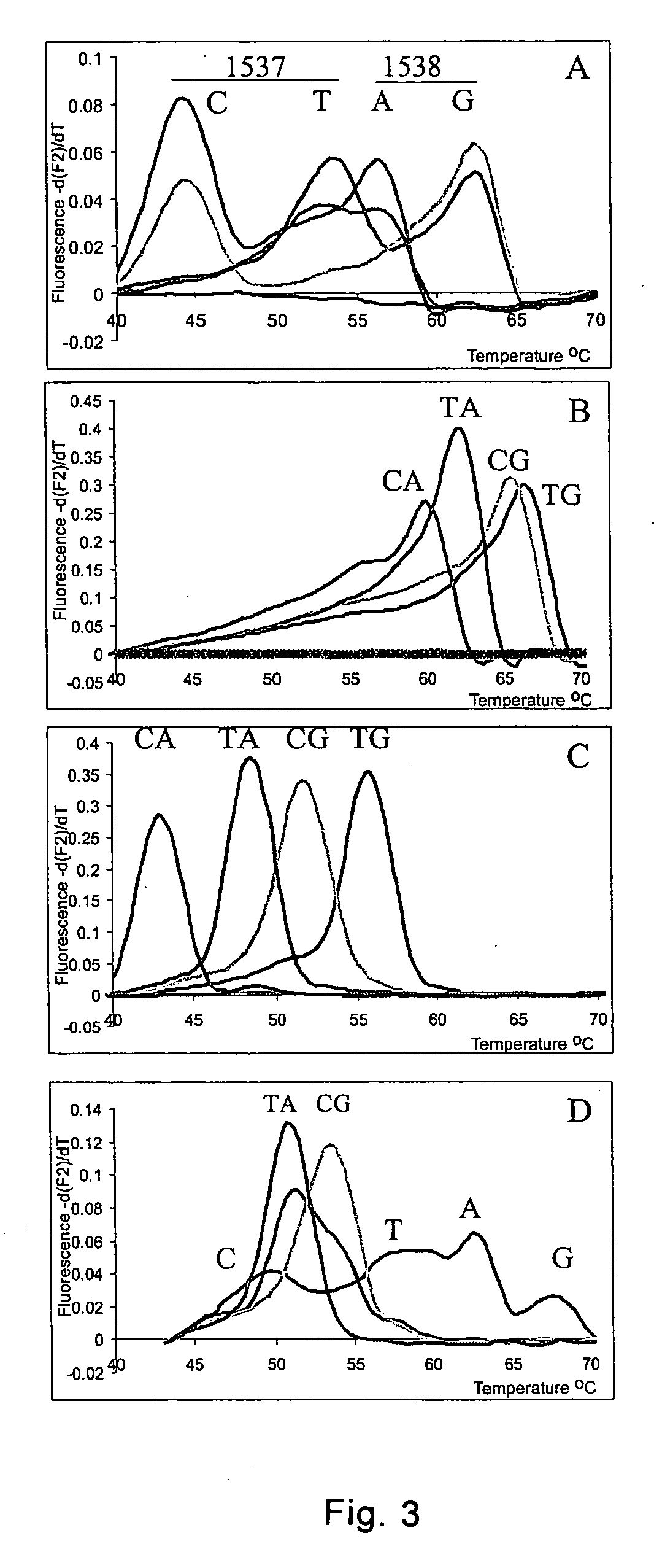
Method for haplotyping and genotyping by melting curve analysis of hybridization probes
InactiveUS20060183136A1Facilitate haplotypingFacilitate genotypingSugar derivativesMicrobiological testing/measurementHybridization probeNucleic Acid Probes
The present invention is directed to nucleic acid probes, complexes and methods of using such probes and complexes for molecular haplotyping and genotyping of mutations, using melting curve analysis of nucleic acid probes to discriminate between and determine the identity of multiple alleles at one or more loci.
Owner:UNIV OF UTAH RES FOUND
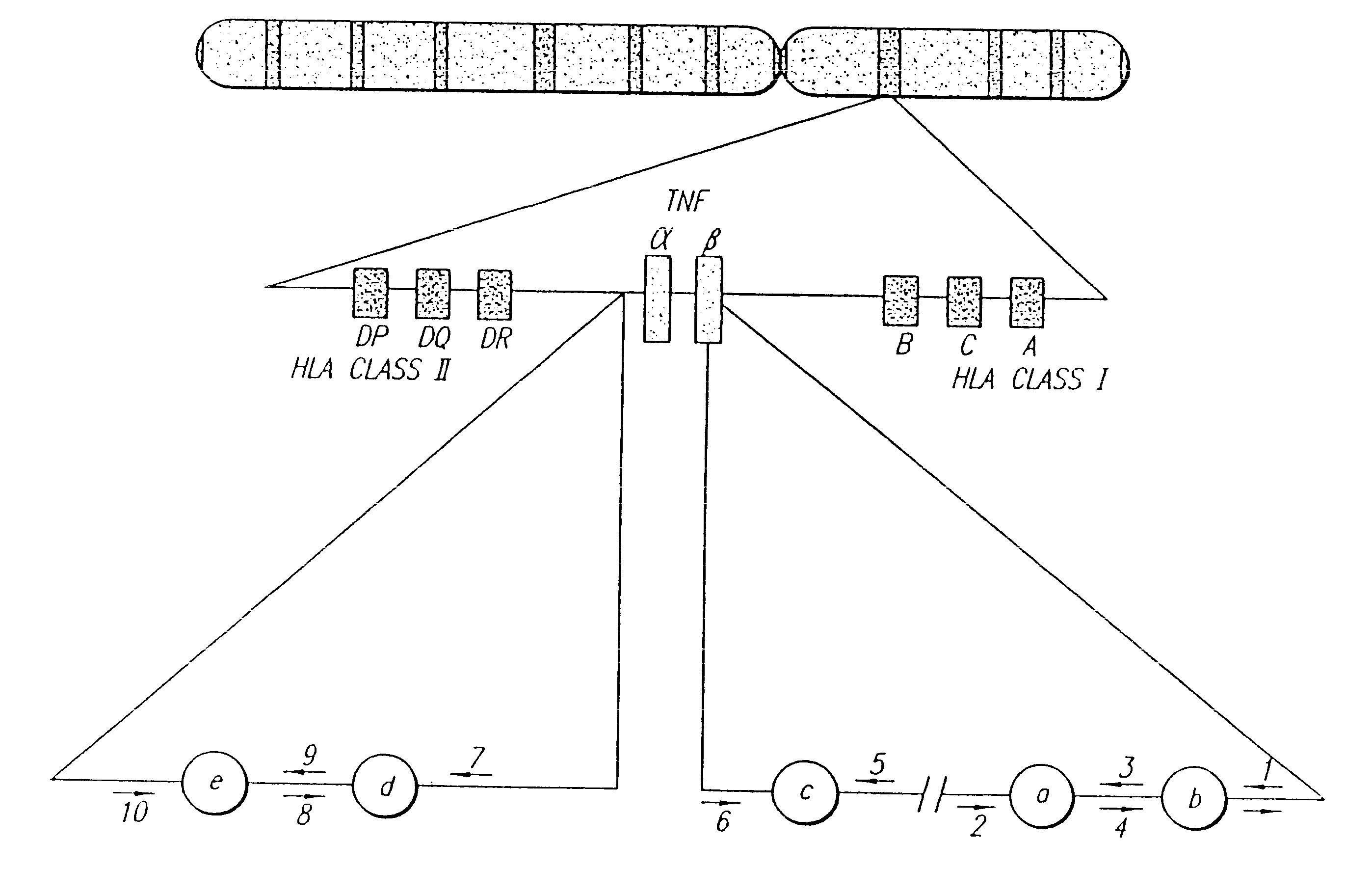
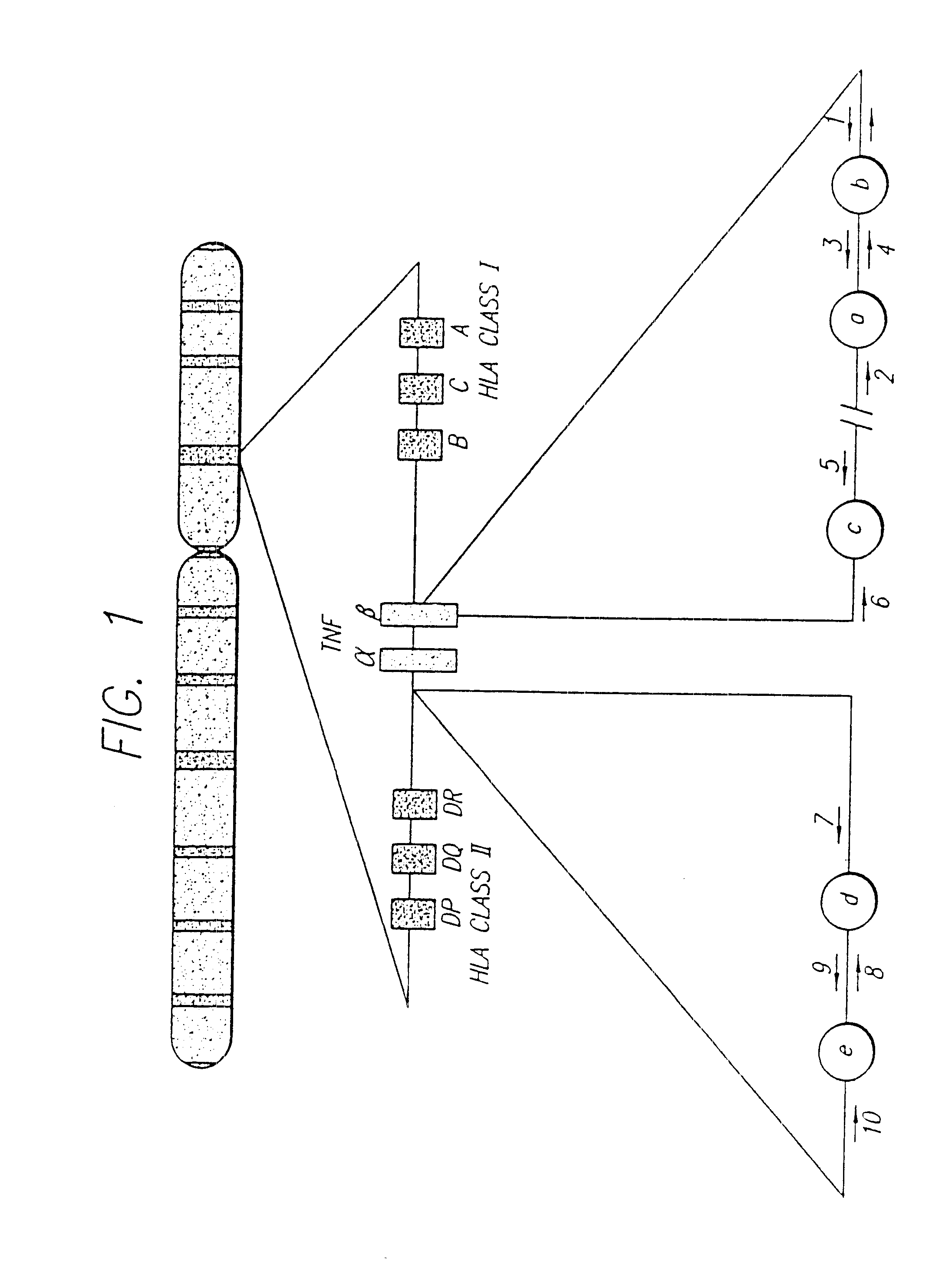
Methods of screening for Crohn's disease using TNF microsatellite alleles
InactiveUS6534263B1Reliable and goodSugar derivativesMicrobiological testing/measurementCrohn's diseaseTNF Microsatellite
A novel association between certain tumor necrosis factor microsatellite alleles and Crohn's disease has been discovered. In accordance with the present invention, there is provided methods for screening for Crohn's disease comprising detecting the presence or absence of nucleic acid of a subject encoding TNF microsatellite alleles associated with Crohn's disease, wherein the presence of nucleic acid encoding three or more of the alleles is indicative of Crohn's disease. Kits useful for screening for Crohn's disease are also provided which comprise nucleic acid encoding TNF microsatellite alleles associated with Crohn's disease.
Owner:CEDARS SINAI MEDICAL CENT
Evolutionary models of multiple sequence alignments to predict offspring fitness prior to conception
A system, device and method for receiving multiple aligned genetic sequences obtained from genetic samples of multiple organisms of one or more different species. A measure of evolutionary variation may be computed for one or more alleles at each of one or more aligned genetic loci. The aligned genetic loci in the multiple organisms may be derived from one or more common ancestral genetic loci or may be otherwise related. The measure of evolutionary variation may be a function of variation in alleles at corresponding aligned genetic loci in the multiple aligned genetic sequences. One or more likelihoods may be computed that an allele mutation at each of the one or more genetic loci in a simulated virtual progeny will be deleterious based on the measure of evolutionary variation of alleles at the corresponding aligned genetic loci for the multiple organisms.
Owner:ANCESTRY COM DNA
SCAR marks of genetic sterile multiple allele Ms of celery cabbage and application thereof
InactiveCN101575603AImprove breeding efficiencyShorten the breeding processMicrobiological testing/measurementDNA/RNA fragmentationNucleotide sequencingRepeatability
The invention provides two SCAR marks including an SCR01-378 and an SCR02-208 which are positioned at two sides of a genetic sterile multiple allele Ms of a celery cabbage and are in tight linage with the genetic sterile multiple allele Ms. The genetic distances of the two marks and the MS gene are 1.2 centimeters and 2.5 centimeters respectively. According to the nucleotide sequences of the two marks, two pairs of specific amplification primers are designed respectively and are used for molecular-assisted selection of the genetic sterile multiple allele of the celery cabbage. The two marks are used simultaneously in the process of the molecular-assisted selection, and the selection accuracy is over 97.5 percent. The two marks have the advantages of good repeatability, high reliability, low detection cost, time conservation and labor conservation.
Owner:SHENYANG AGRI UNIV
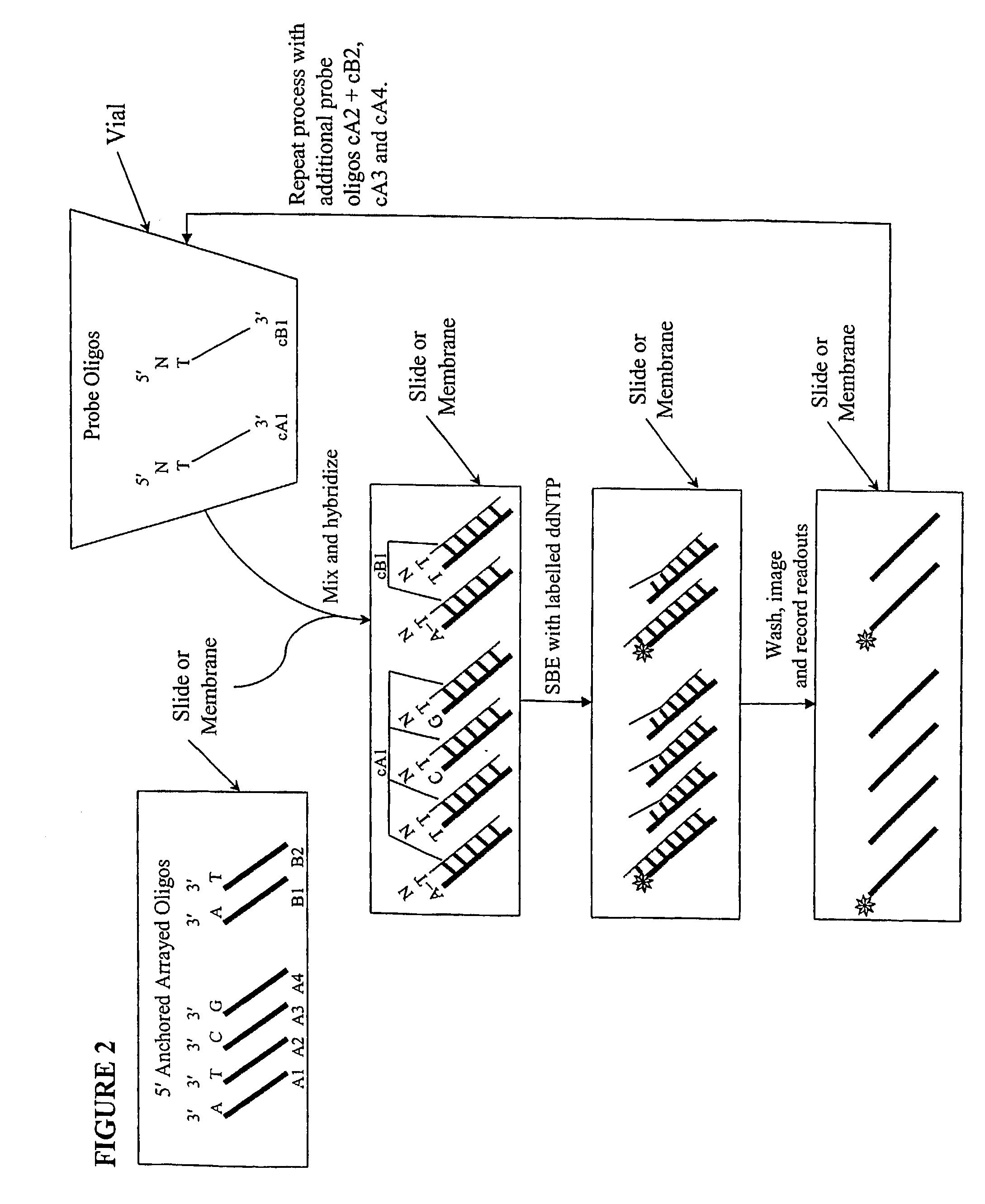
Quality control methods for arrayed oligonucleotides
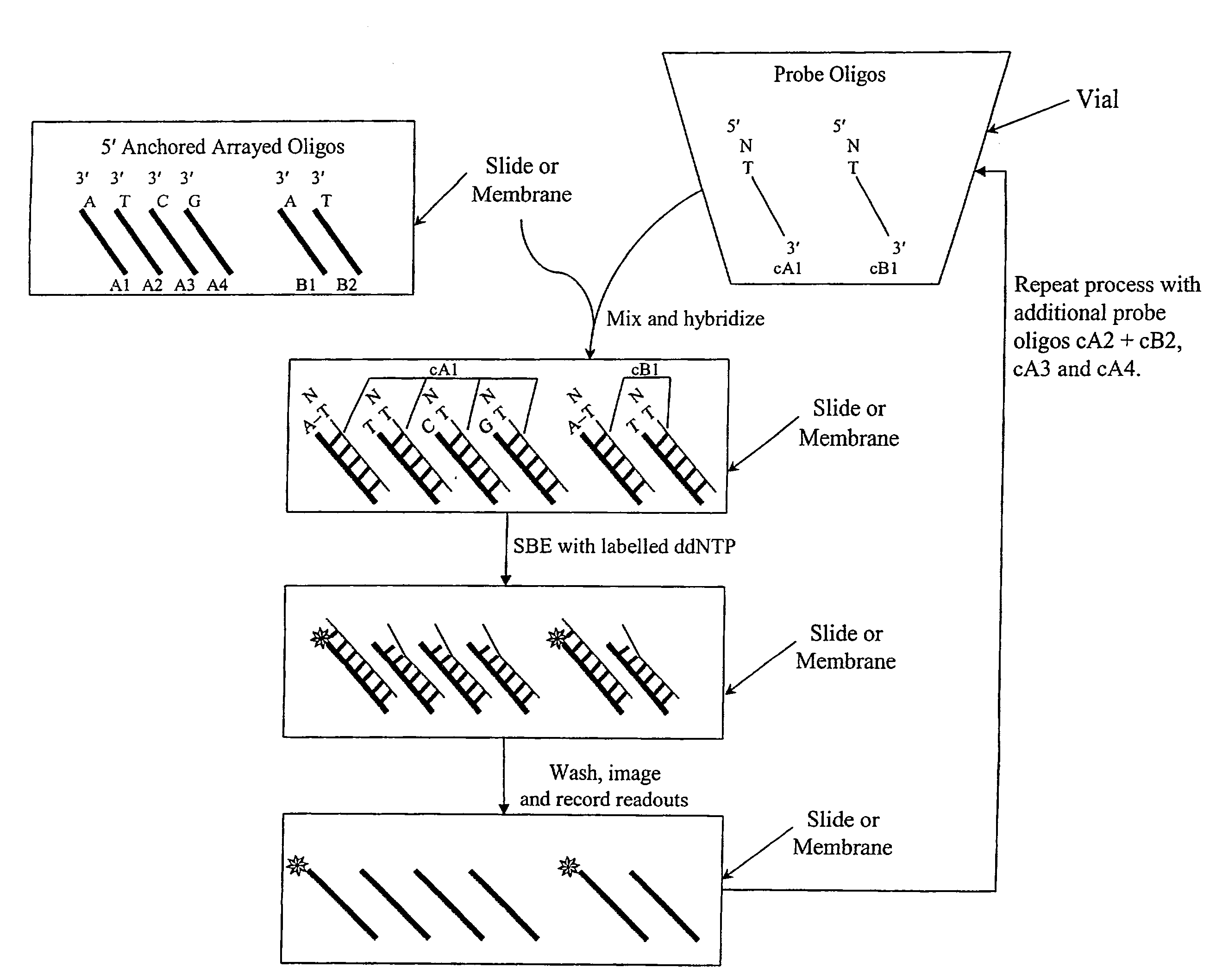
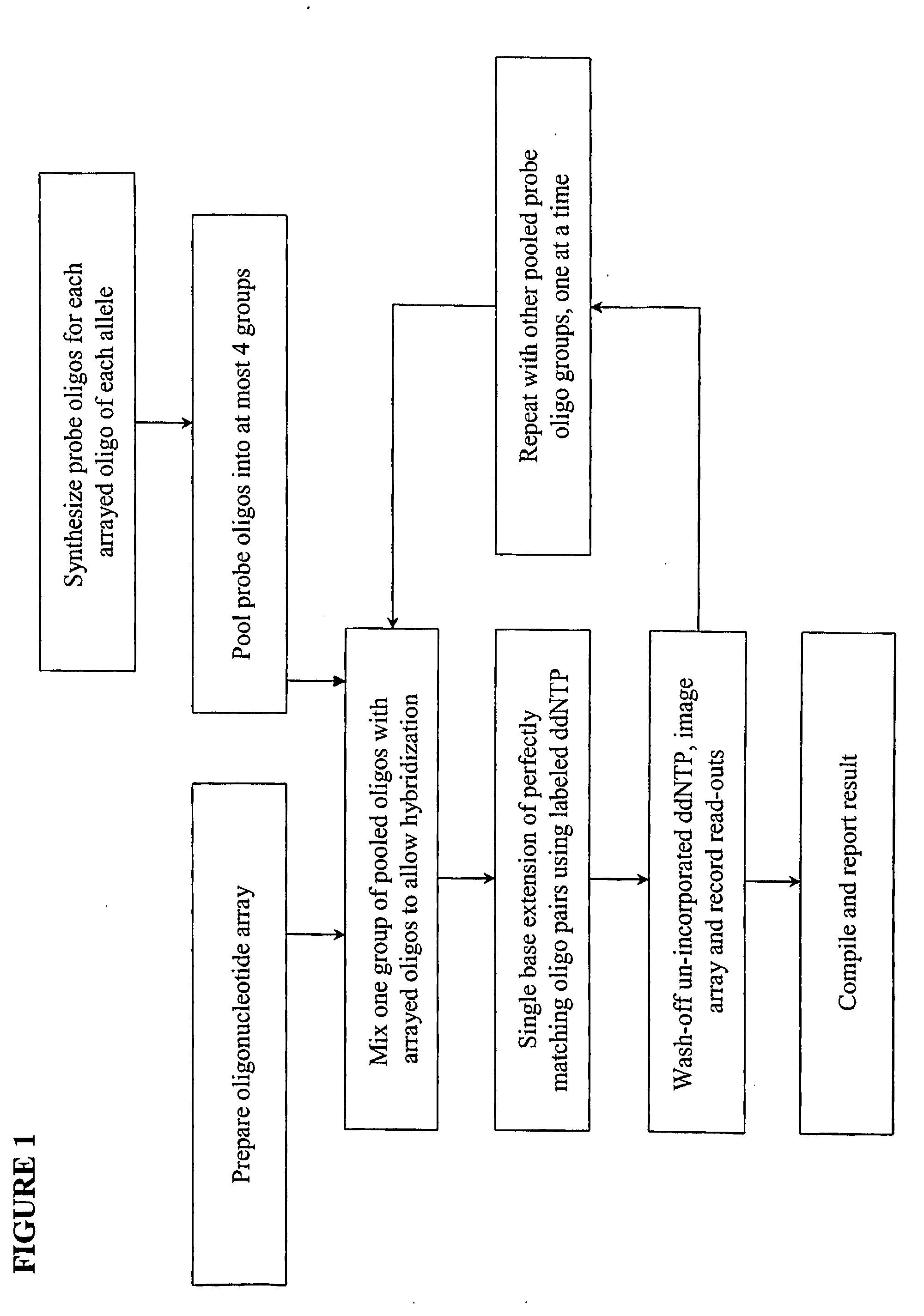
InactiveUS20090143234A1Quick and accurate verificationBioreactor/fermenter combinationsBiological substance pretreatmentsDideoxynucleotide TriphosphatesQuality control
We disclose quality controls methods that allow quick and accurate verification of a test oligonucleotide deposited on a solid support. It is especially useful for the verification of oligonucleotides representing alleles of a multi-allelic locus. It employs single base extension, with labeled dideoxynucleotides, to locate and verify the identity of test oligonucleotides. This approach involves synthesizing a complement probe oligonucleotide for each oligonucleotide being tested. Probe oligonucleotides are optionally grouped. They are then hybridized to test oligonucleotides, and the hybridized pair is subject to single base extension and detection. It requires the presence of one unique base, either in the last two bases at the free hanging end of the test oligonucleotide—as opposed to the end anchored to the solid support surface, or in the last two bases at one end of the probe oligonucleotide.
Owner:GE HEALTHCARE BIO SCI CORP
Composite amplification system for detecting 129 molecular genetic markers of four types based on NGS technology
The invention belongs to the field of innovative research and application of a new forensic medicine technology, and particularly relates to a composite amplification system for detecting multiple molecular genetic markers based on a next-generation sequencing platform. The method comprises the following steps: screening 48 AISNPs for different intercontinental and domestic group forensic medicine progenitor inference and analysis on the basis of an HGDP-CPEH database; 18 multi-allele InDel sites with high polymorphism distribution in domestic populations are screened on the basis of a thousand-person genome plan; 27 micro-haplotypes with high application value in domestic populations are screened based on previous research reports; 34 Y-SNP sites and 2 Y-InDel sites are selected for male individual haplogroup distribution research; and based on a next-generation sequencing platform, synchronous amplification detection analysis of the multiple genetic markers is realized by adopting a multiple PCR method. The system can be used for simultaneously realizing forensic individual identification, genetic relationship identification, mixed sample resolution, forensic initial ancestor analysis and male individual haplogroup distribution research aiming at a field physical evidence inspection material, and can provide more comprehensive and valuable biological information for forensic application research.
Owner:朱波峰 +3
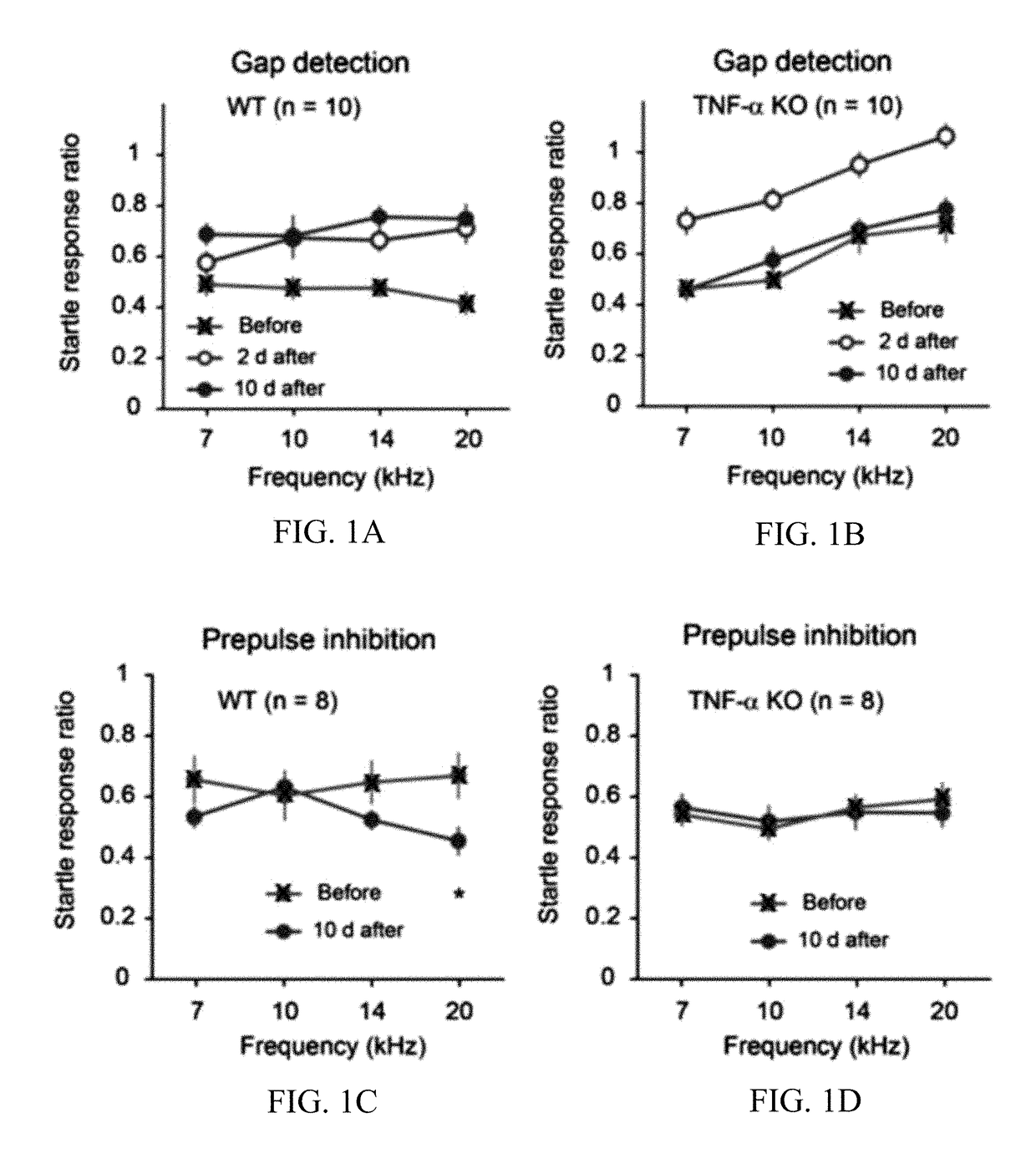
Methods of treating hearing disorders
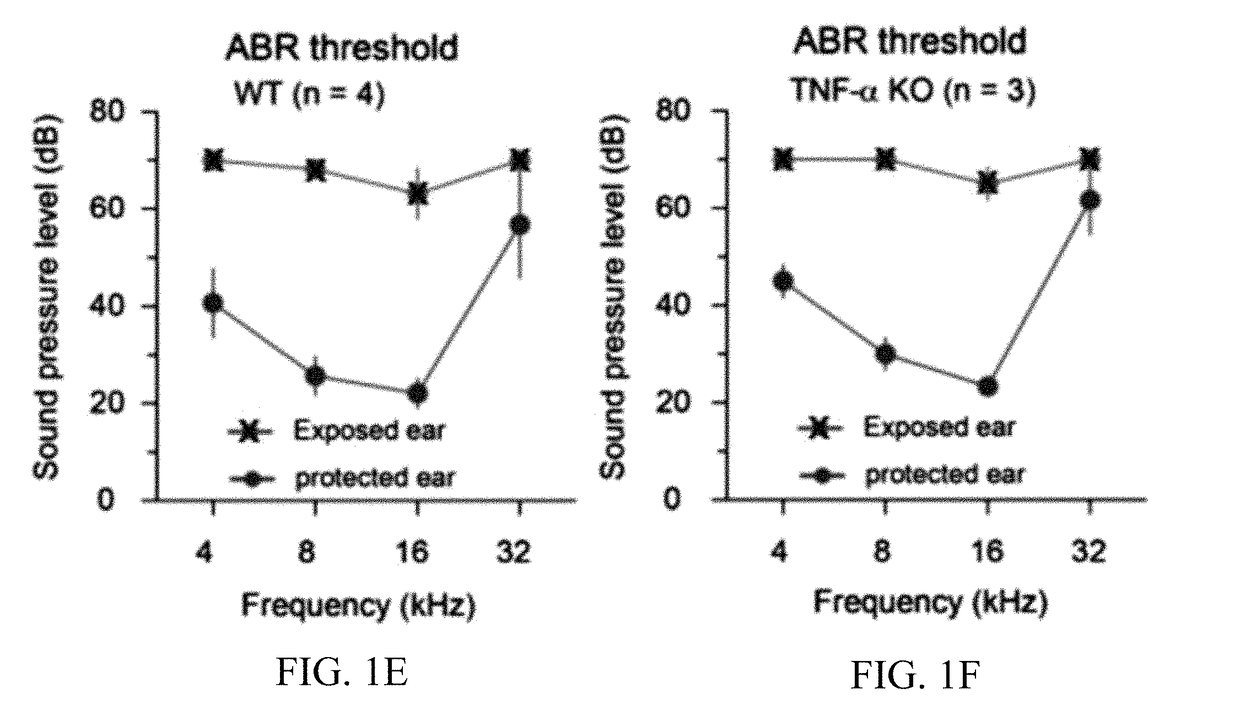
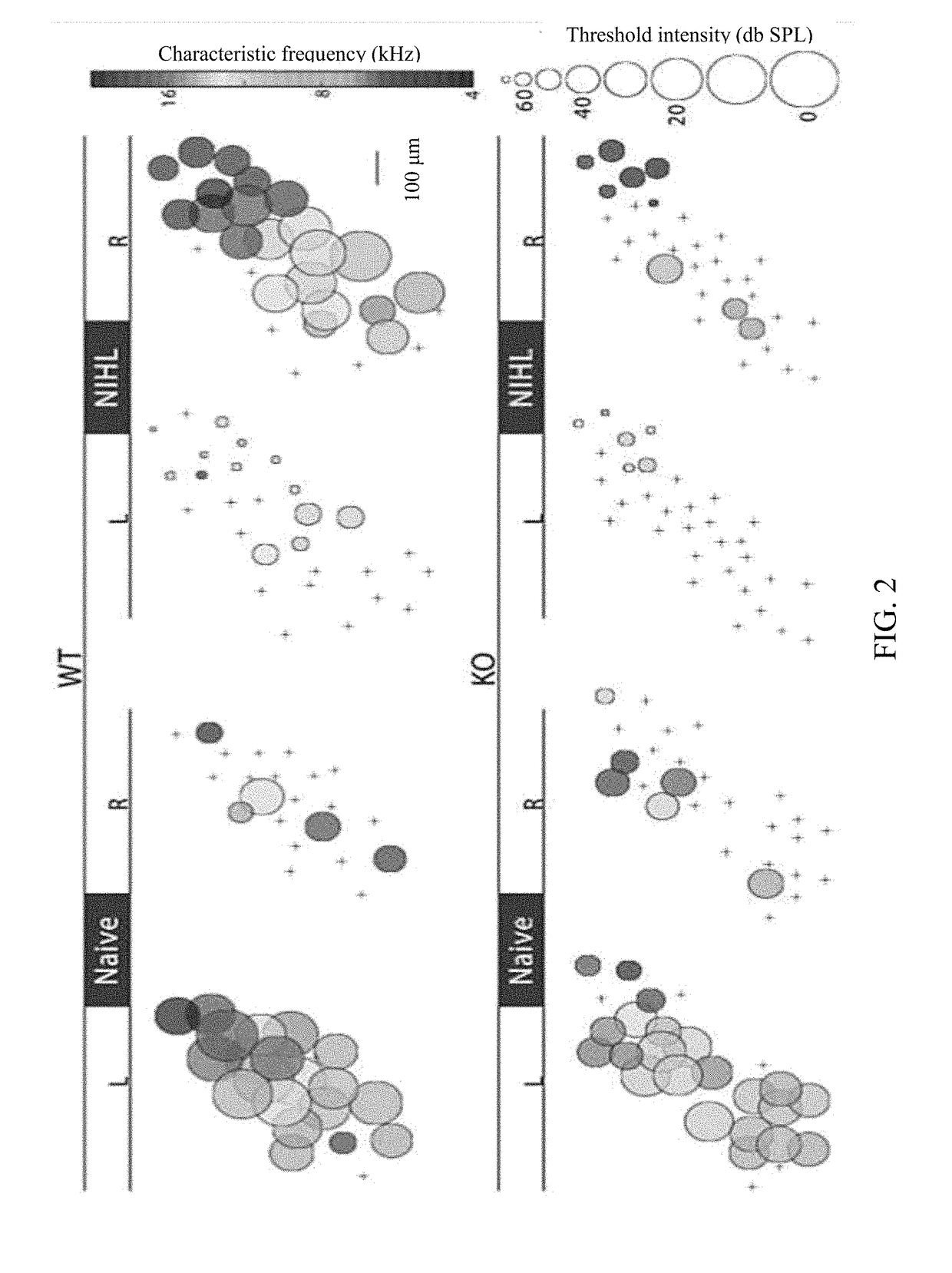
InactiveUS20180021315A1Reduce inhibitionIncrease changeOrganic active ingredientsPeptide/protein ingredientsSignaling Pathway GeneTumor necrosis factor alpha
The present disclosure provides methods for treating a hearing disorder associated with maladaptive neuroplasticity, reduction of inhibition, shift of excitation-to-inhibition balance, changes in central gain, and / or changes in neural sensitivity in a subject by inhibiting the function and / or production of tumor necrosis factor alpha (TNF-α) in the subject. The present disclosure provides methods of administering to the subject a TNF-α inhibitory agent in an amount effective to treat the subject for a hearing disorder associated with maladaptive neuroplasticity, reduction of inhibition, shift of excitation-to-inhibition balance, changes in central gain, and / or changes in neural sensitivity. TNF-α inhibitory agents of the subject disclosure include agents that inhibit the function TNF-α, inhibit the production of TNF-α, inhibit TNF-α signaling, inhibit TNF-α expression, or inhibit TNF-α signaling pathway genes in the subject. The present disclosure also provides methods for treating a hearing disorder associated with maladaptive neuroplasticity, reduction of inhibition, shift of excitation-to-inhibition balance, changes in central gain, and / or changes in neural sensitivity in a subject by disrupting one or more alleles of a TNF-α signaling pathway gene in a cell of the subject.
Owner:WAYNE STATE UNIV
Method of crossing flower color genotypes
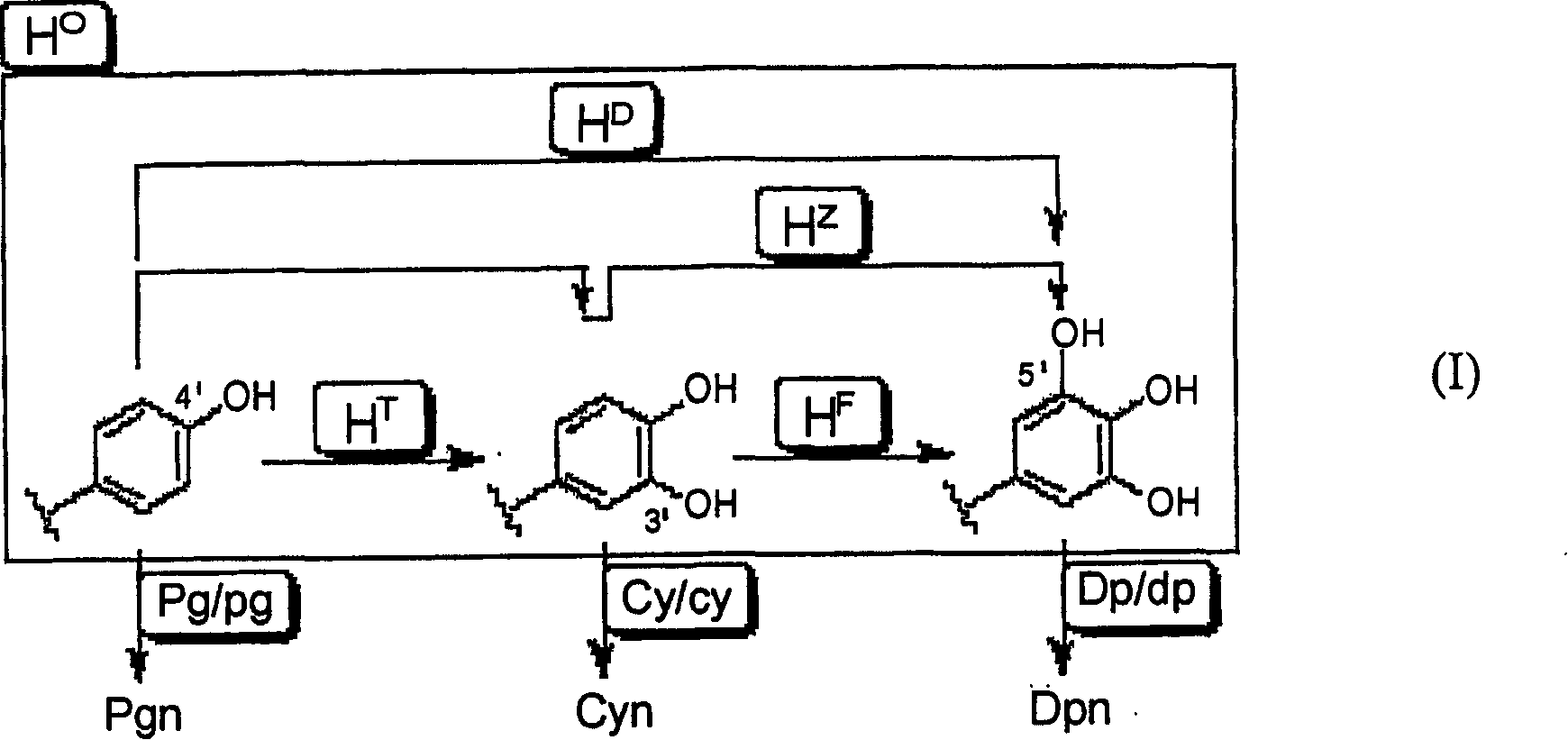
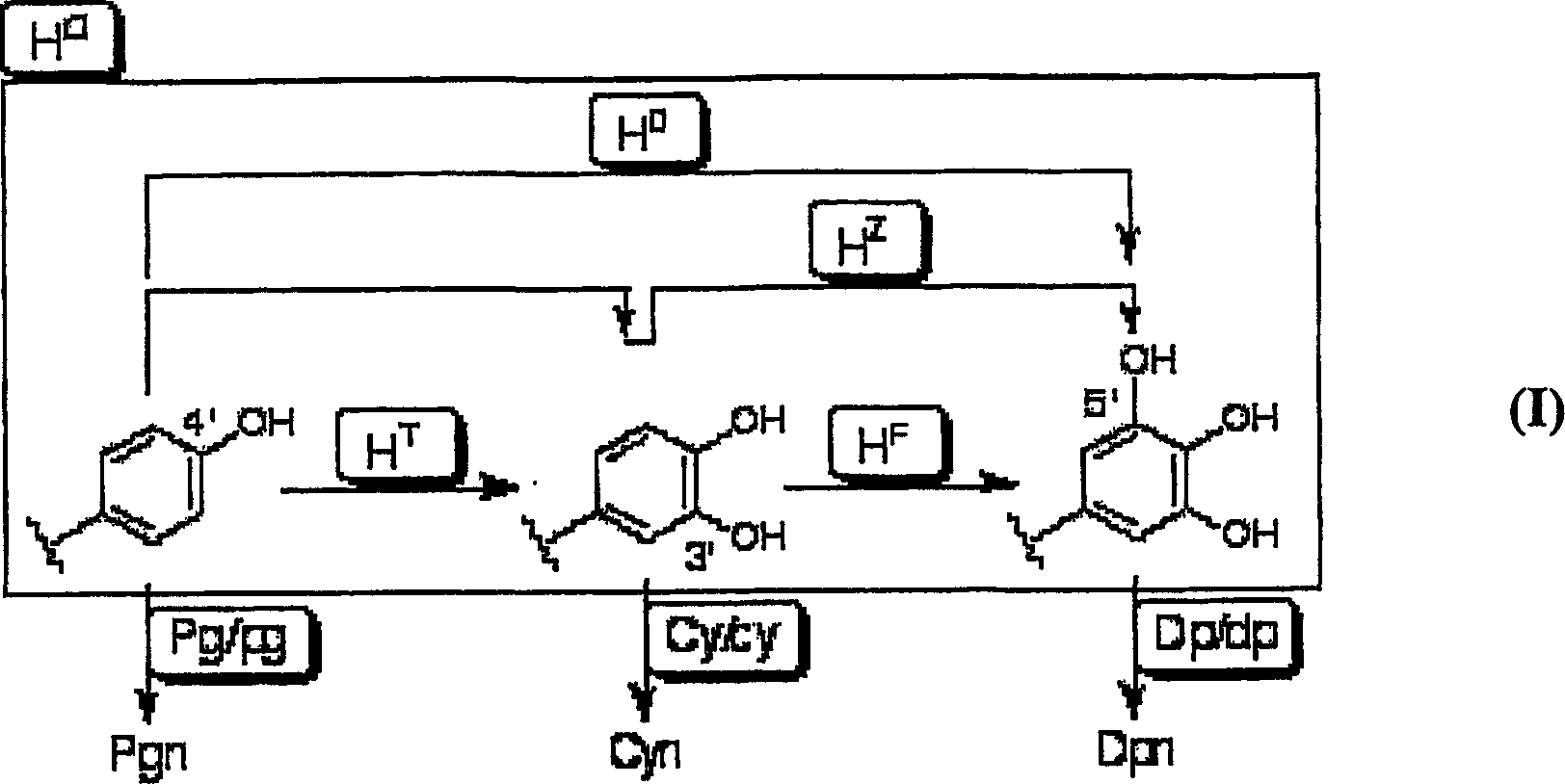
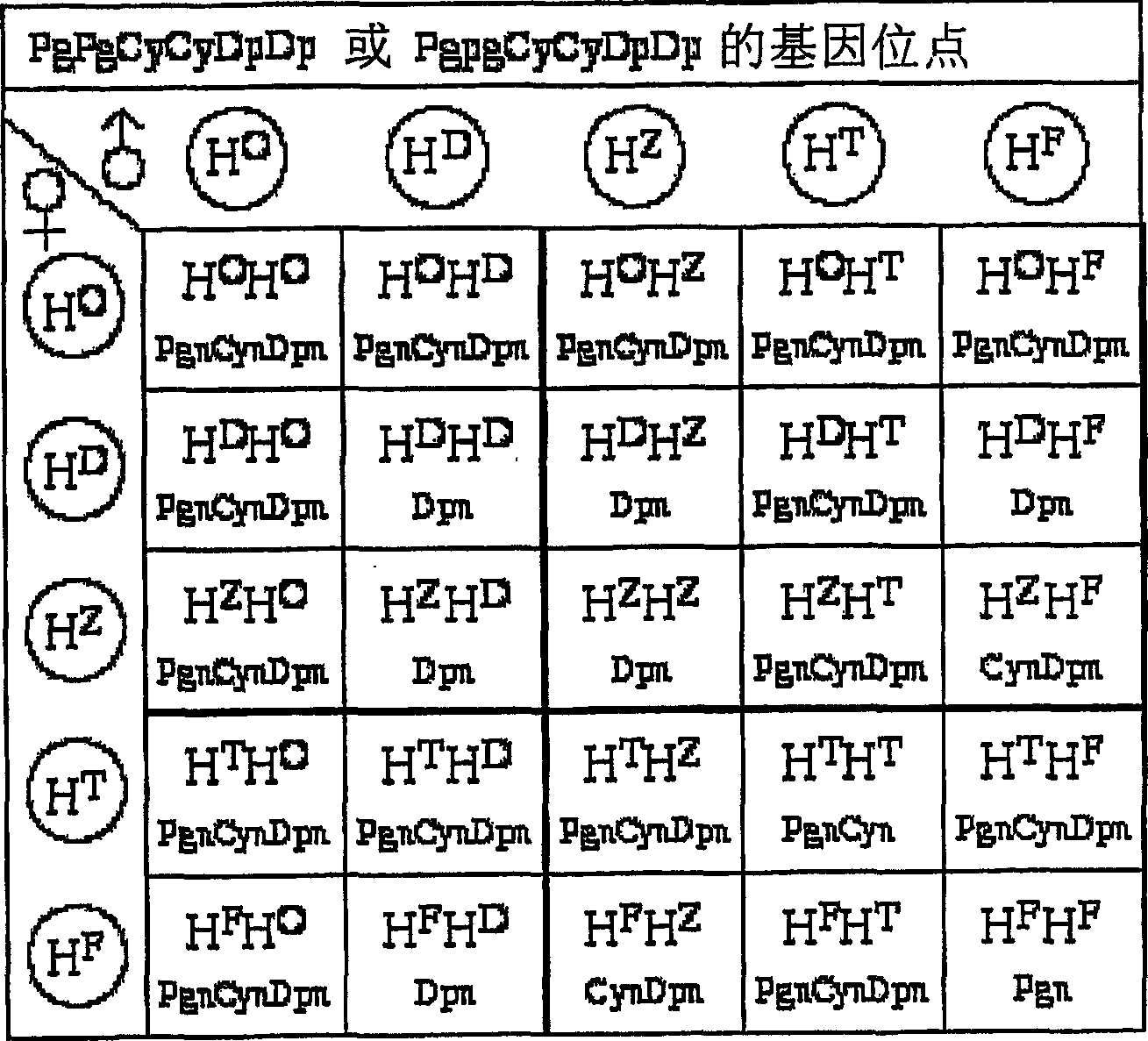
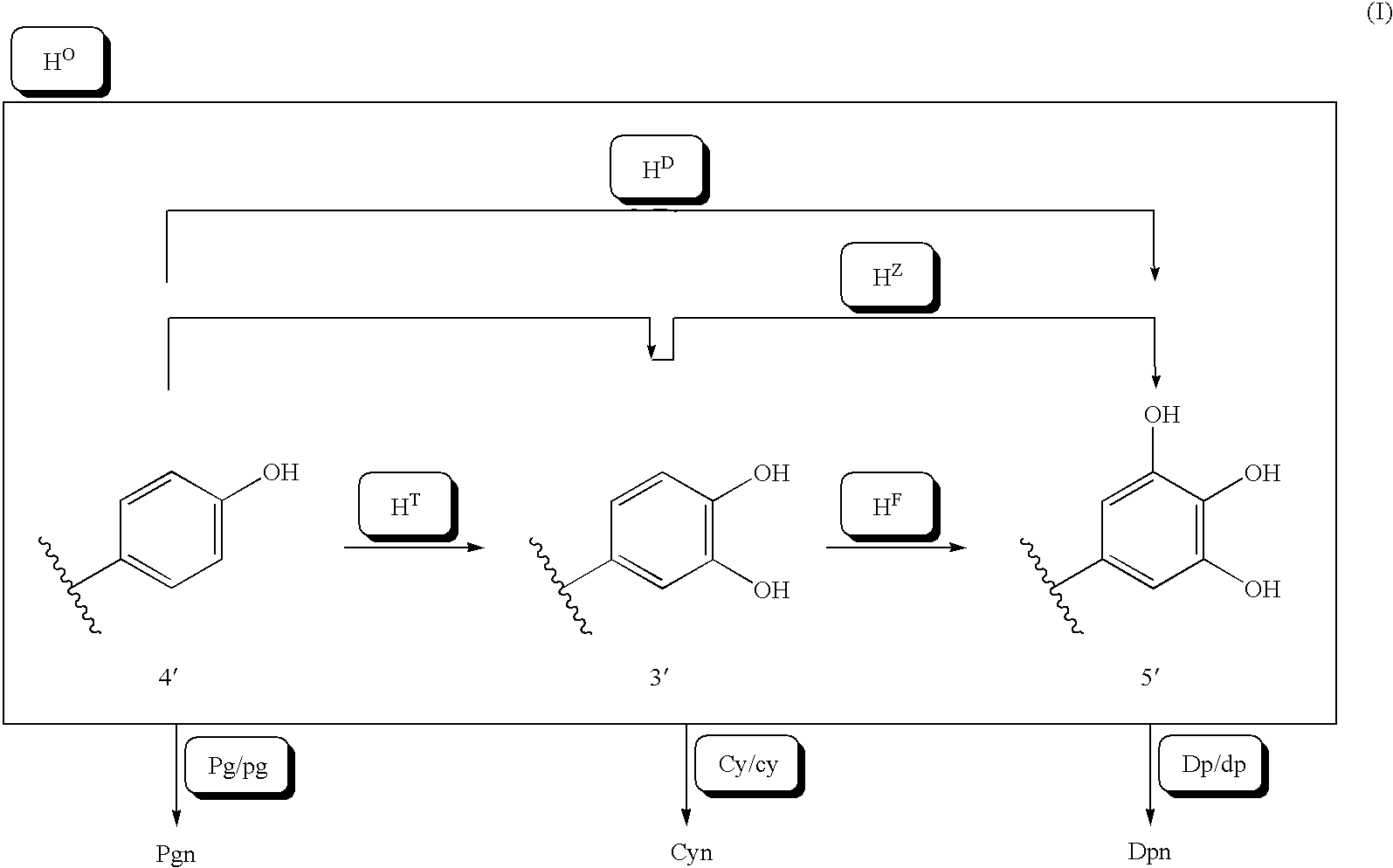
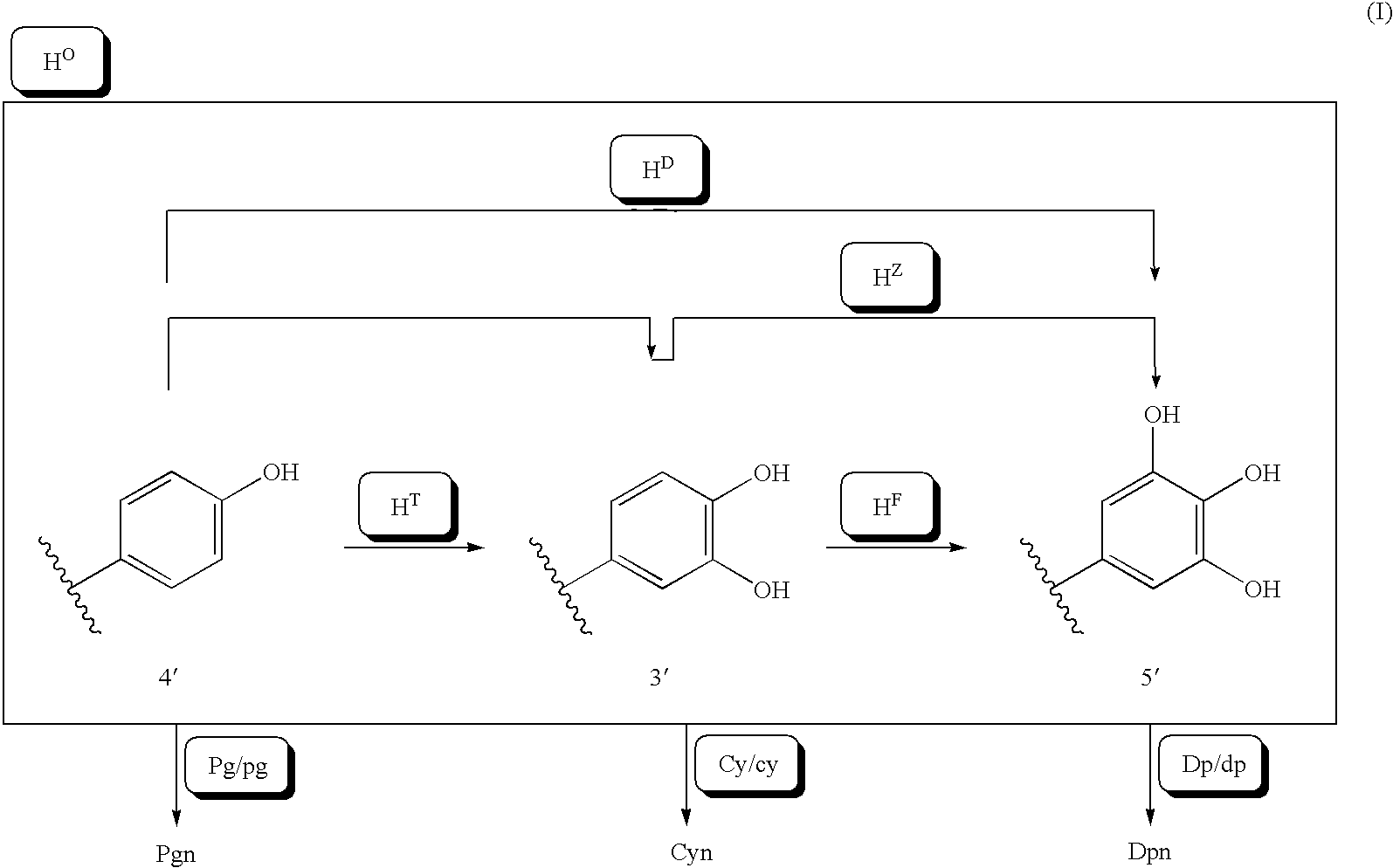
The purpose of the present invention is to clarify the genetic characteristics of anthocyanin biosynthesis and the relationship between flower color inheritance and pigment genotype, so as to provide a hybridization method for flower color genotypes that can create new flower colors in practice. The present invention has discovered a new rule: the flower color genotype is related to the biosynthesis of flavonoids shown in the pathway of formula (I), and flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′, 5′- The genetic characteristics of hydroxylase (F3', 5'H) are controlled by 5 multiple alleles. The result makes it possible to provide a method for creating new flower colors using genotypes D / d·E / e·HXHX·Pg / pg·Cy / cy·Dp / dp by which flower colors can be freely created according to pigment genotypes Without resorting to genetic recombination or mutations caused by radiation exposure, etc.
Owner:KAGOSHIMA TLO
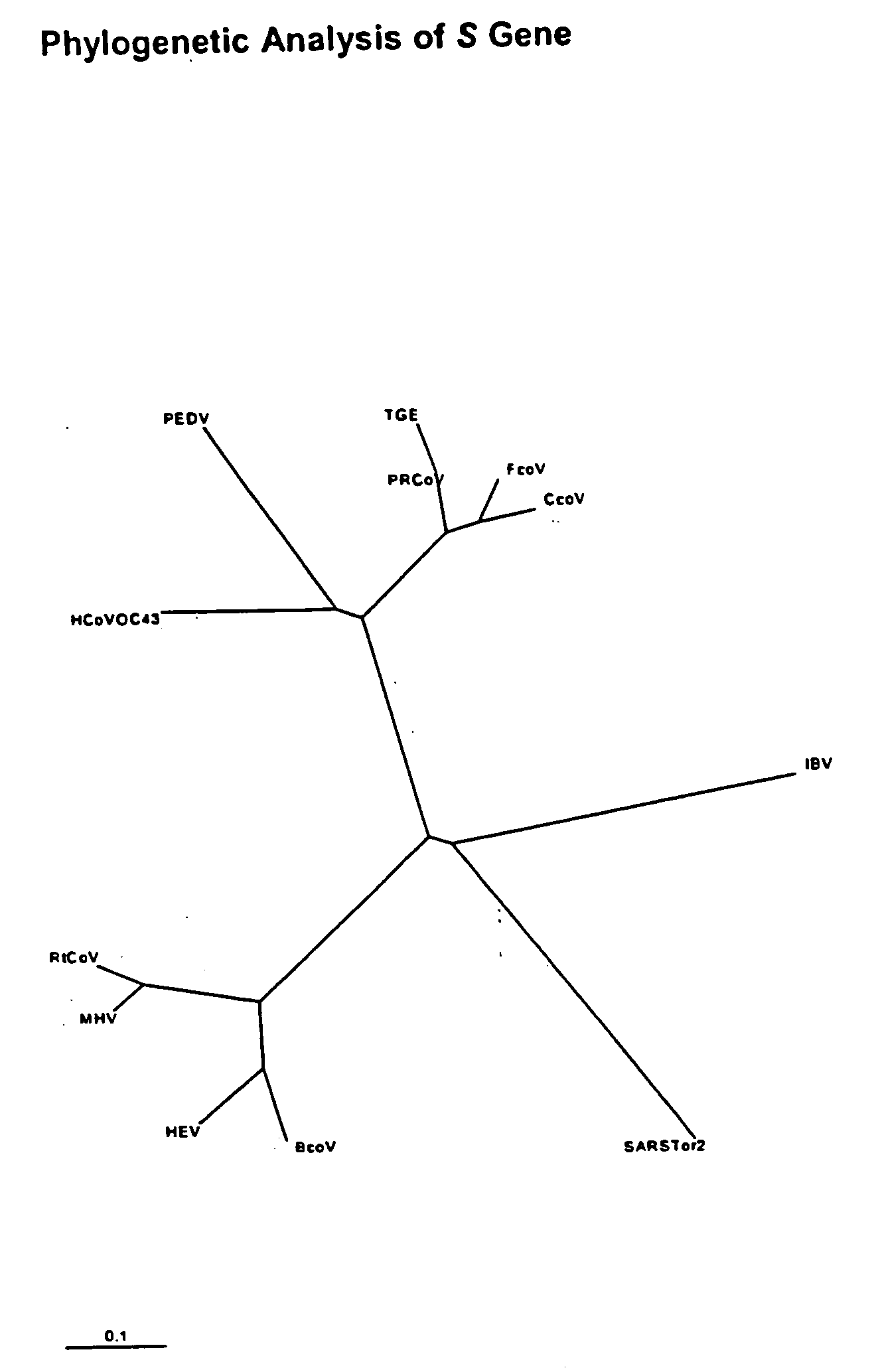
Multi-allelic molecular detection of SARS-associated coronavirus
InactiveUS20060003340A1Raise the possibilityMinimize the possibilityMicrobiological testing/measurementViruses/bacteriophagesQuantitative determinationSARS-associated coronavirus
The subject invention relates to a multiple-allelic RT-real-time polymerase chain reaction (PCR) assay for coronaviruses including the SARS virus. Multiple target sequences within the SARS-CoV, S, E, M and N genes are identified. The use of the four different targets enhances the likelihood that the fundamental genetic drift of the virus will not lead to a false negative result. Multiplex assays format for the assay are envisioned. Thus, the present invention allows for early diagnosis of a SARS infection. The assay would be useful in the context of monitoring treatment regimens, screening potential anti SARS agents, and similar applications requiring qualitative and quantitative determinations.
Owner:BIRCH BIOMEDICAL RES
Method of crossing flower color genotypes
InactiveUS20070011776A1Clarify the pigment genotypeGood colorOther foreign material introduction processesBiological testingGenotypePigment
It is intended to clarify heredity of the biosynthesis of flower pigments and the relationship between flower color heredities and pigment genotypes, thereby providing a method of crossing flower color genotypes which is practically available in creating a novel flower color. There is found out a novel rule that flower color genotypes relate to the biosynthesis of flavonoids represented by the pathway (I) and the heredities of flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′,5′H) are controlled by 5 multiple alleles. As a result, it becomes possible to provide a method of producing a novel flower color with the use of genotypes D / d.E / e.H<X>H<X>.Pg / pg.Cy / cy.Dp / dp by which flower colors can be freely created based on flower pigment genotypes without resort to gene recombination or mutation caused by exposure to radiation, etc.
Owner:KAGOSHIMA TLO
Double-probe gene mutation detecting method based on allele special amplification as well as special chip and kit thereof
ActiveCN101619352BLittle pollutionEasy to operateMicrobiological testing/measurementGenes mutationOligonucleotide
Owner:CENT SOUTH UNIV
Screening method and application of multiple-allele PCR (Polymerase Chain Reaction) amplified fragment
InactiveCN103966349AEasy to identifyImprove efficiencyMicrobiological testing/measurementGeneticsScreening method
The invention discloses a screening method and application of a multiple-allele PCR (Polymerase Chain Reaction) amplified fragment. The screening method comprises the steps of finding the published chromosome related data through NCBI (National Center for Biotechnology Information), and determining SNP (Single Nucleotide Polymorphism) aggregation suspected to exist; then, designing a primer for an SNP aggregation region suspected to exist, and carrying out PCR (Polymerase Chain Reaction); sequencing to detect whether a plurality of SNP sites really exist on a PCR amplified fragment; further carrying out correlation analysis on the SNP sites to determine whether the amplified fragment has multiple alleles so as to screen the multiple-allele PCR amplified fragment for animal individual identification, meat product tracing technology research and industrialization and the like.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
General codominance molecular marker for resisting nilaparvata lugens Stal BPH9 multiple alleles of rice, and detection method and application of general codominance molecular marker
ActiveCN109554494AEfficient and accurate screeningEliminate the effects of false negativesMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyBrown planthopper
The invention relates to a general codominance molecular marker for resisting nilaparvata lugens Stal BPH9 multiple alleles of rice, and a detection method and application of the general codominance molecular marker. The general molecular marker for resisting nilaparvata lugens Stal multiple alleles BPH1 / 10 / 18 / 21, BPH2 / 26, BPH7 and BPH9 of the rice, provided by the invention, is located at (22874098bp)th-(22874099bp)th position of a 12th chromosome of the rice, and the polymorphism is 5'-TG-3' / 5'-CC-3' / 5'-CA-3'. The general codominance molecular marker for resisting nilaparvata lugens Stal multiple alleles of rice provided by the invention can be generally used for identification of homozygous genotypes or heterozygous genotypes of the multiple alleles BPH1 / 10 / 18 / 21, BPH2 / 26, BPH7 and BPH9, and efficient accurate identification of BPH1 / 10 / 18 / 21, BPH2 / 26, BPH7 and BPH9 genotypes of nilaparvata lugens Stal resisting rice recourse is realized.
Owner:YUAN LONGPING HIGH TECH AGRI CO LTD +2
Engineering of plants to exhibit self-compatibility
InactiveUS20110265202A1Improve featuresPreventing self-pollinationOther foreign material introduction processesFermentationBiotechnologyTransmembrane protein
Owner:THE UNIV OF BIRMINGHAM
Assay For Bipolar Affective Disorder
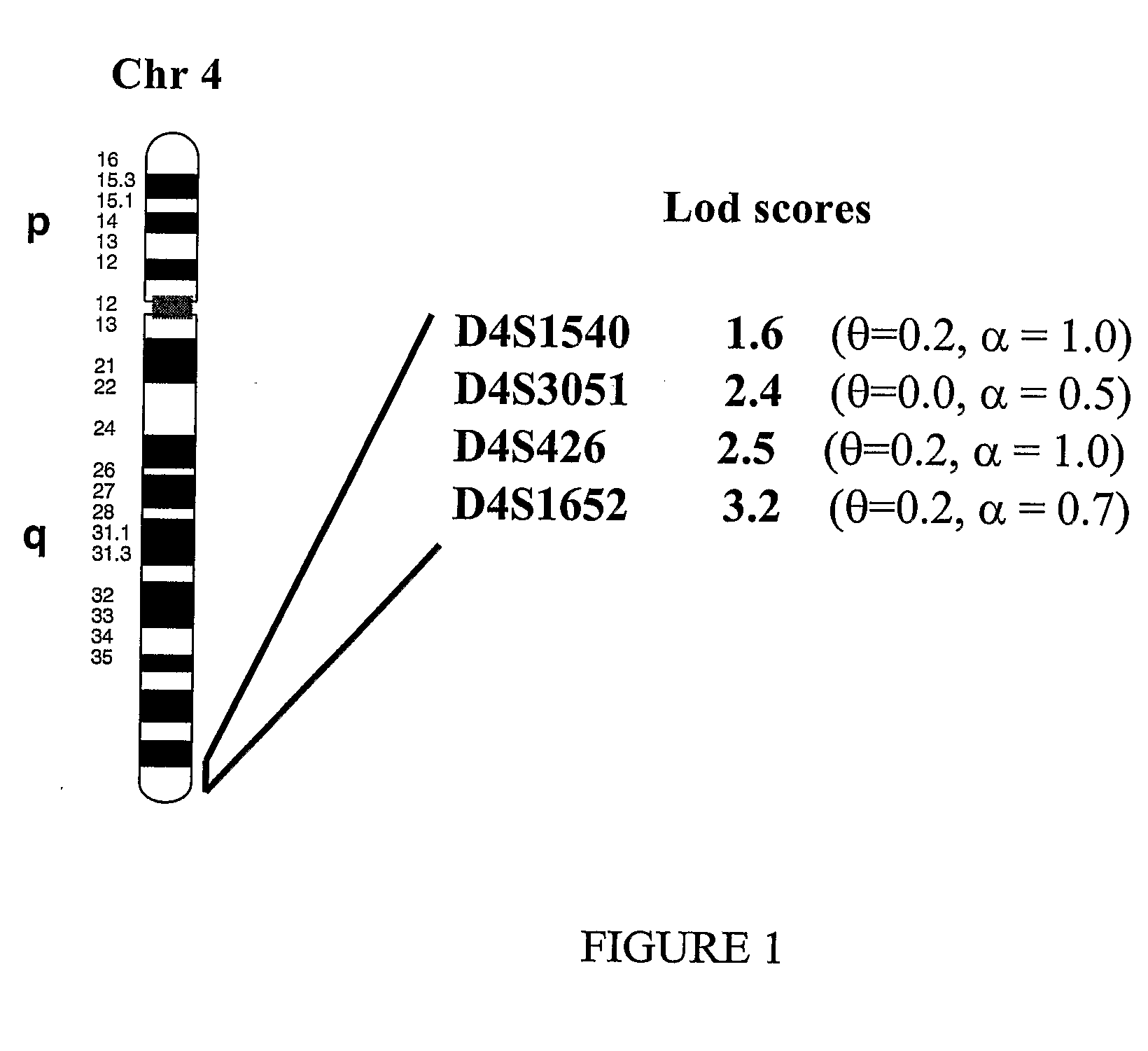
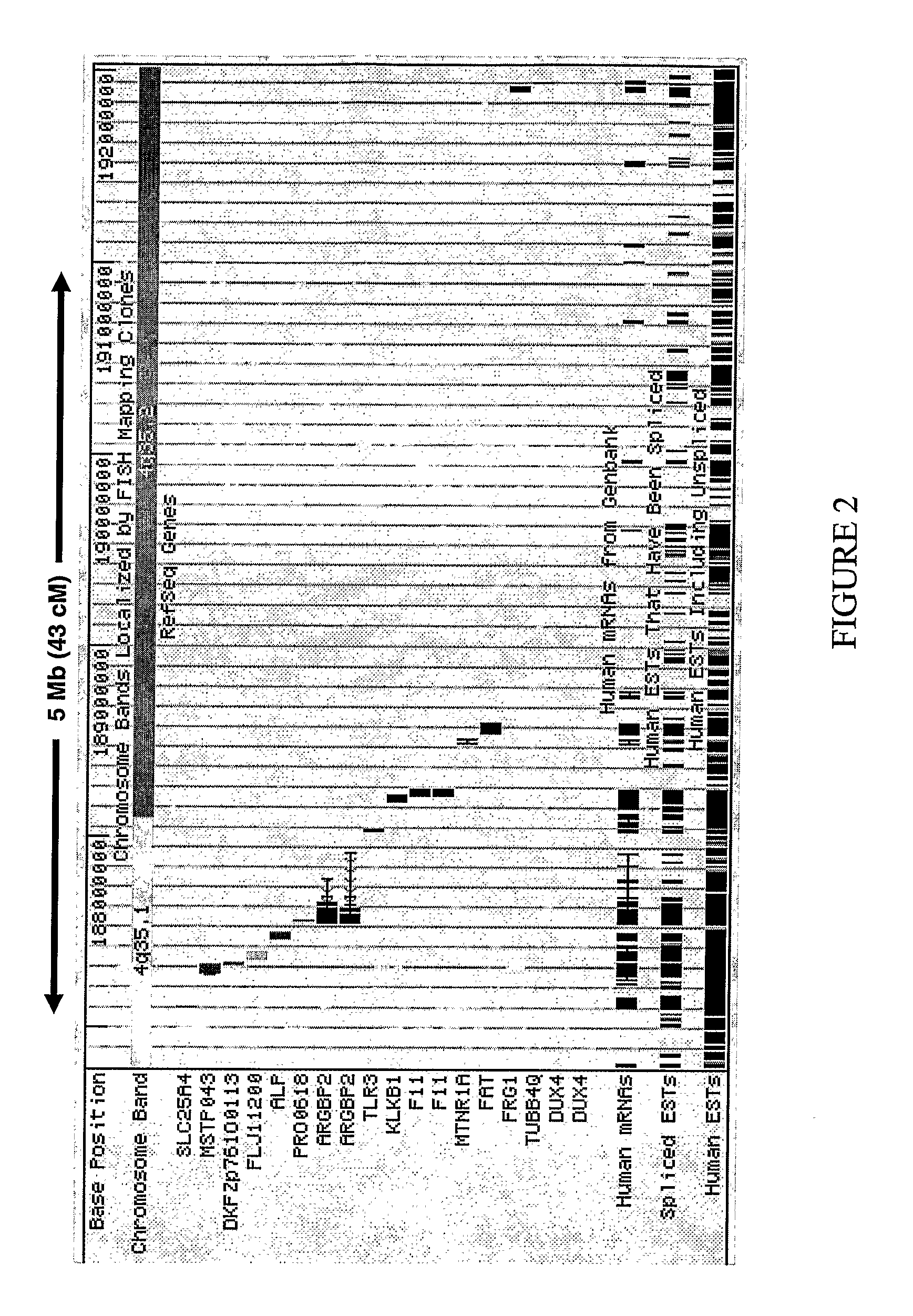
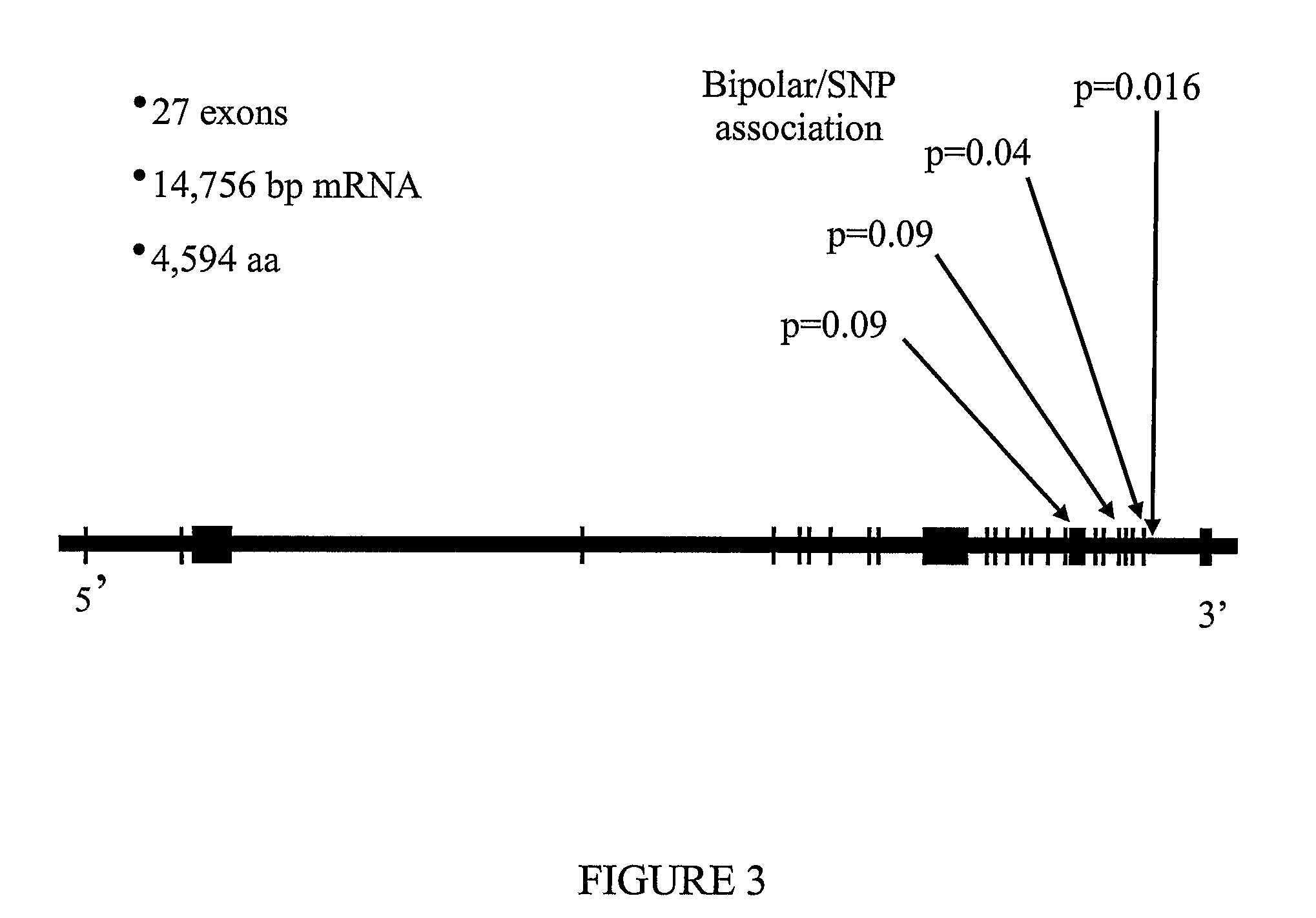
The present invention provides a method of diagnosing bipolar affective disorder (BAD) and for determining a predisposition of a subject to bipolar affective disorder. In particular, the methods of the present invention comprise detecting a marker that comprises one or more polymorphisms at position 4q35.2 of the human genome and / or one or more allelic variants of the FAT gene associated with BAD or linked thereto. The present invention also relates to identifying new markers that are diagnostic of bipolar affective disorder. Furthermore, the present invention relates to methods of identifying and producing candidate compounds for the treatment of bipolar affective disorder.
Owner:GARVAN INST OF MEDICAL RES
Method for producing a generation of hybridization seeds of Chinese cabbage by using multiple alleles gene male sterility line
InactiveCN101138316AReduce manufacturing costHigh purityPlant genotype modificationBrassicaAgricultural science
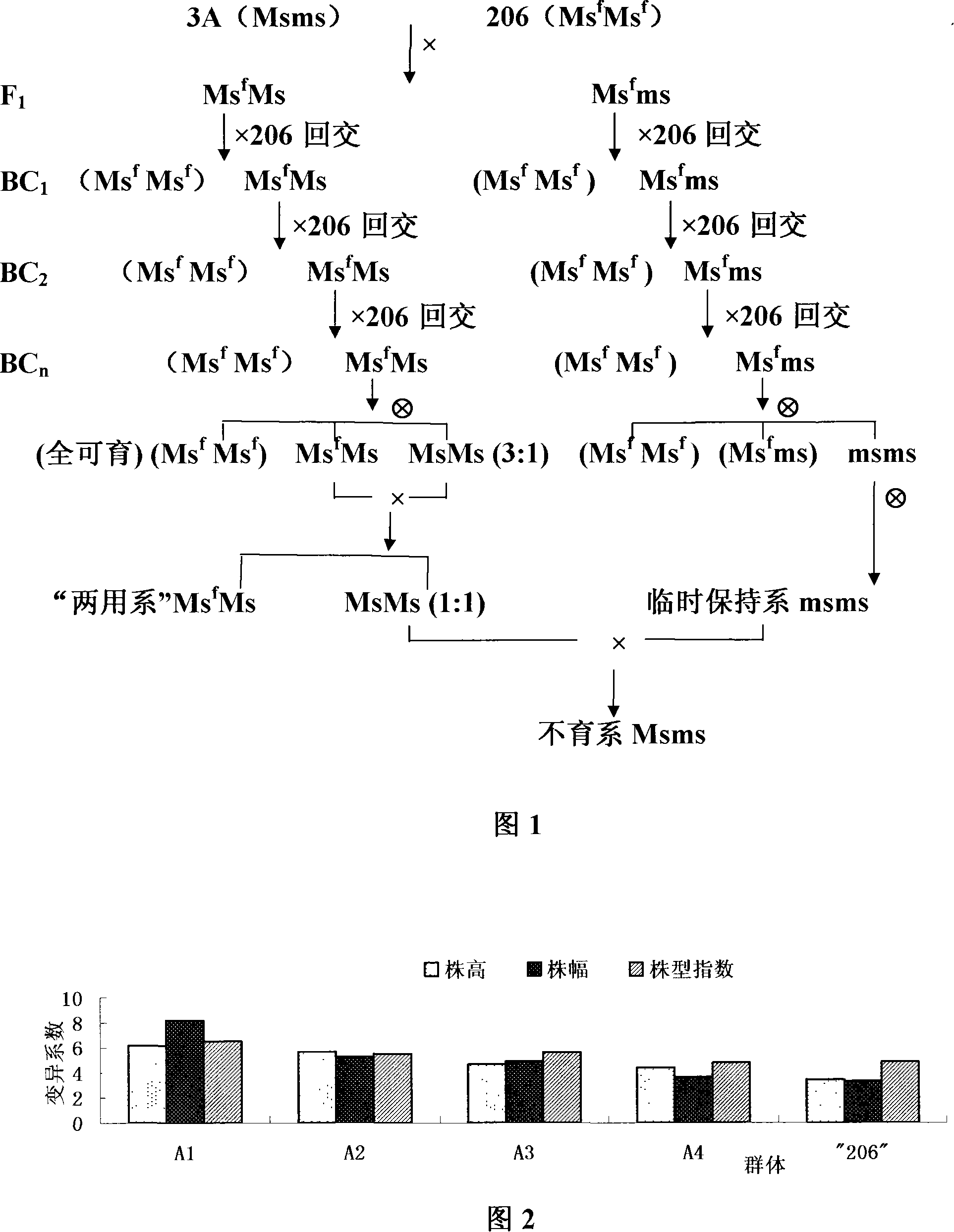
A method for producing cabbage F1 hybrid seeds with multiple allele male sterile line relates to the breeding and the manufacturing technology of cabbage multiple allele male sterile line (Brassica campstris L.ssp.pekinensis); the method belongs to the field of utilizing the plant hybrid seeds advantages. The present invention provides a method for producing cabbage F1 hybrid seeds with multiple allele male sterile line. The present invention comprises the following steps that the breeding of the maternal plant cabbage multiple allele male sterile line: the cabbage sterile line is taken as the sterile source and the cabbage fertile line is taken as the breeding material; through the approaches of hybrid, backcrossing and inbreeding, the multiple allele male sterile line is obtained; the breeding of paternal inbred line: the paternal plants are high-generation inbred lines after inbreeding and breeding of a plurality of years of the hybrids of cabbage; F1 hybrid seeds breeding: the cabbage multiple allele male sterile line is hybridized with the paternal inbred lines to get the F1 hybrid seeds.
Owner:SHENYANG AGRI UNIV
Multi-allelic molecular detection of sars-associated coronavirus
InactiveUS20070042351A1Raise the possibilityMinimize the possibilityMicrobiological testing/measurementFermentationAssayQuantitative determination
The subject invention relates to a multiple-allelic RT-real-time polymerase chain reaction (PCR) assay for coronaviruses including the SARS virus. Multiple target sequences within the SARS-CoV, S, E, M and N genes are identified. The use of the four different targets enhances the likelihood that the fundamental genetic drift of the virus will not lead to a false negative result. Multiplex assays format for the assay are envisioned. Thus, the present invention allows for early diagnosis of a SARS infection. The assay would be useful in the context of monitoring treatment regimens, screening potential anti SARS agents, and similar applications requiring qualitative and quantitative determinations.
Owner:BIRCH BIOMEDICAL RES
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com