Double-probe gene mutation detecting method based on allele special amplification as well as special chip and kit thereof
An allele-specific and allele-based technology, applied in the field of gene analysis, can solve problems such as high cost, inability to be widely used, and difficult operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] [Example 1] Preparation of a dual-probe gene mutation chip detection system based on allele-specific amplification.
[0079] The design and synthesis of probes and primers, chip preparation, etc. in this example are all skills mastered by those skilled in the art. For detailed operation steps, please refer to "Molecular Cloning Test Guide" (([US] J. Sambrook Author, Science Press)
[0080] 1. Preparation process
[0081] See attached Figure 11 .
[0082] 2. The main raw and auxiliary materials and equipment required for preparation
[0083] Table 6
[0084] Reagents and equipment
place of origin
Reagent purity / concentration
Blank Ultraflat
Shanghai Baiao Technology Co., Ltd.
/
Aminosilane reagent
Acros (United States)
99wt.%
Acros (United States)
25wt.%
95% ethanol
Shanghai Chemical Plant
Analytical pure
Glacial acetic acid
Shanghai Chemical Plant
Analytica...
Embodiment 3
[0126] [Example 3] Comparison between the gene chip detection system of the present invention and the conventional SSOP gene chip detection system.
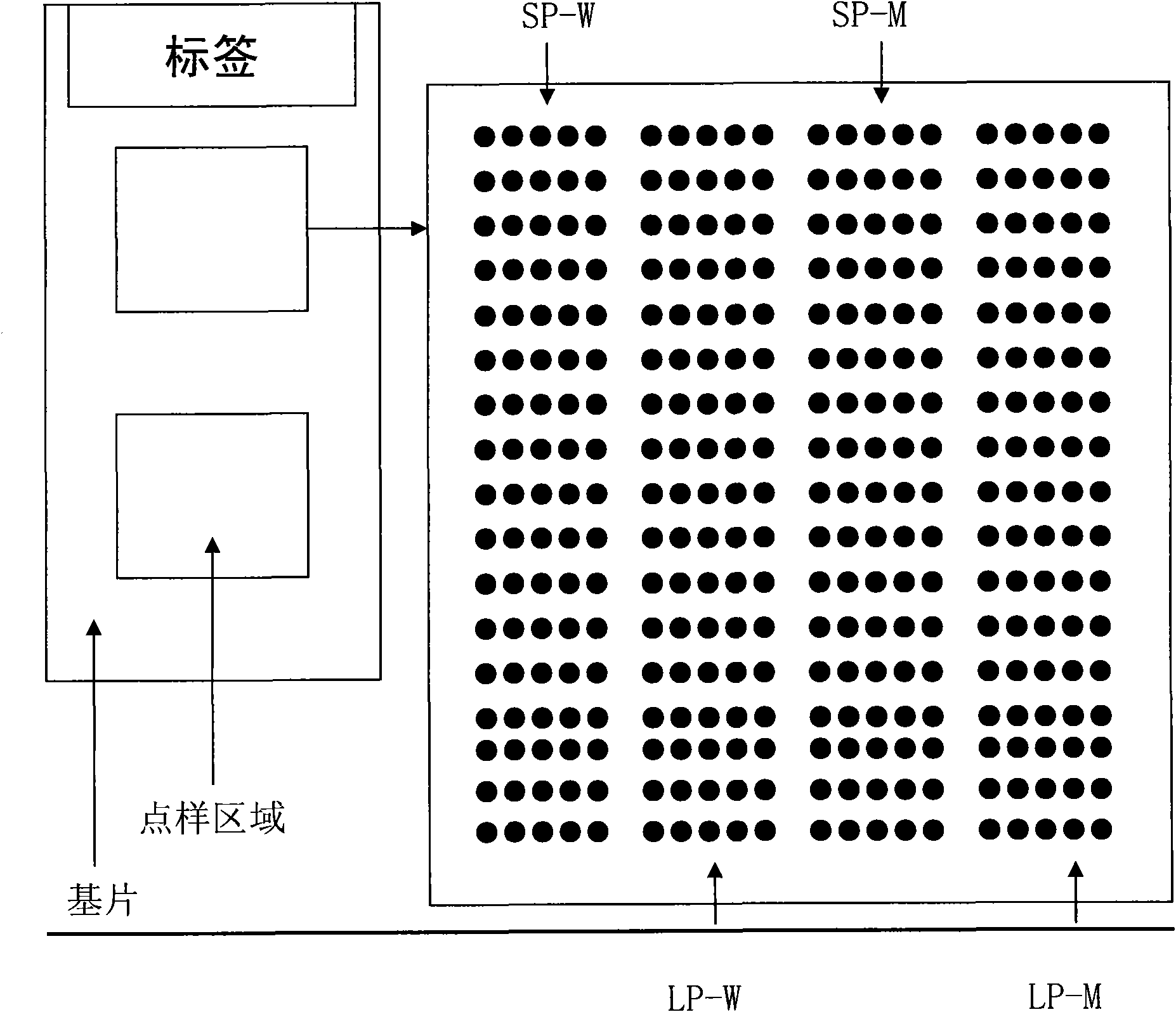
[0127] The method described in Example 1 is used to prepare a dual-probe gene chip detection system for detecting rs1057910 (CYP2C9*3), rs4986893 (CYP2C19*3) and rs1065852 (CYP2D6*10), and the probe arrangement array is
[0128] Rows
site
1-5 points
6-10 o'clock
11-15 o'clock
16-20 o'clock
1
rs1057910
SP-W
LP-W
SP-M
LP-M
2
rs3758581
SP-W
LP-W
SP-M
LP-M
3
rs17878459
SP-W
LP-W
SP-M
LP-M
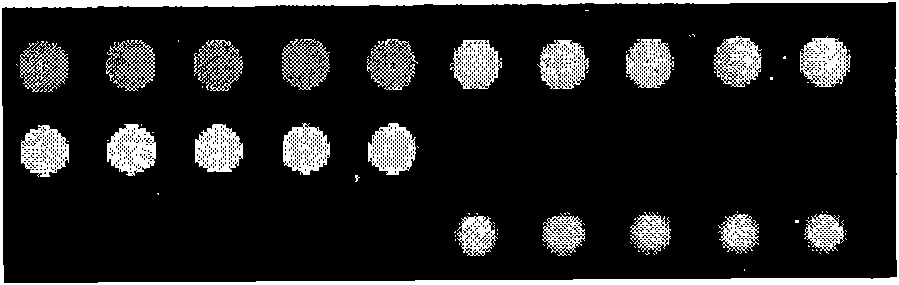
[0129] According to the method disclosed in CN 101054601A invention patent (an oligonucleotide probe and a gene chip for detecting mutation sites of cytochrome P450 enzymes), it was prepared for detecting rs1057910 (CYP2C9*3), rs4986893 (CYP2C19 *3) and rs1065852 (CYP2D6*10) conventional SSOP gene chip detection system,...
Embodiment 4
[0133] [Example 4] The influence of the introduction of artificial mismatched bases at the 3' end of the allele-specific inner primer on the detection results of the gene chip.
[0134] Select SEQ ID NO: 19 and SEQ ID NO: 20 as alternative sequences of primers in the wild-type and mutant alleles of rs2231142 (BCRP 421C>A), and prepare the primers for detecting the mutation according to the method described in Example 1. Chip A and Chip B.
[0135] The primer sequence (Inner P-W_A) and mutant allele inner primer sequence (Inner P-M_A) of chip A are respectively:
[0136] Inner P-W_A: GACGGTGAGAGAAAACTTAC (no base mismatch)
[0137] Inner P-M_A: AAGAGCTGCTGAGAACT T (no mismatched bases)
[0138] The primer sequence (Inner P-W_B) and mutant allele inner primer sequence (Inner P-M_B) of chip B are respectively:
[0139] Inner P-W_B: GACGGTGAGAGAAAACT A C (The penultimate position introduces a mismatched base)
[0140] Inner P-M_B: AAGAGCTGCTGAGAA T T (The penultimate po...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com