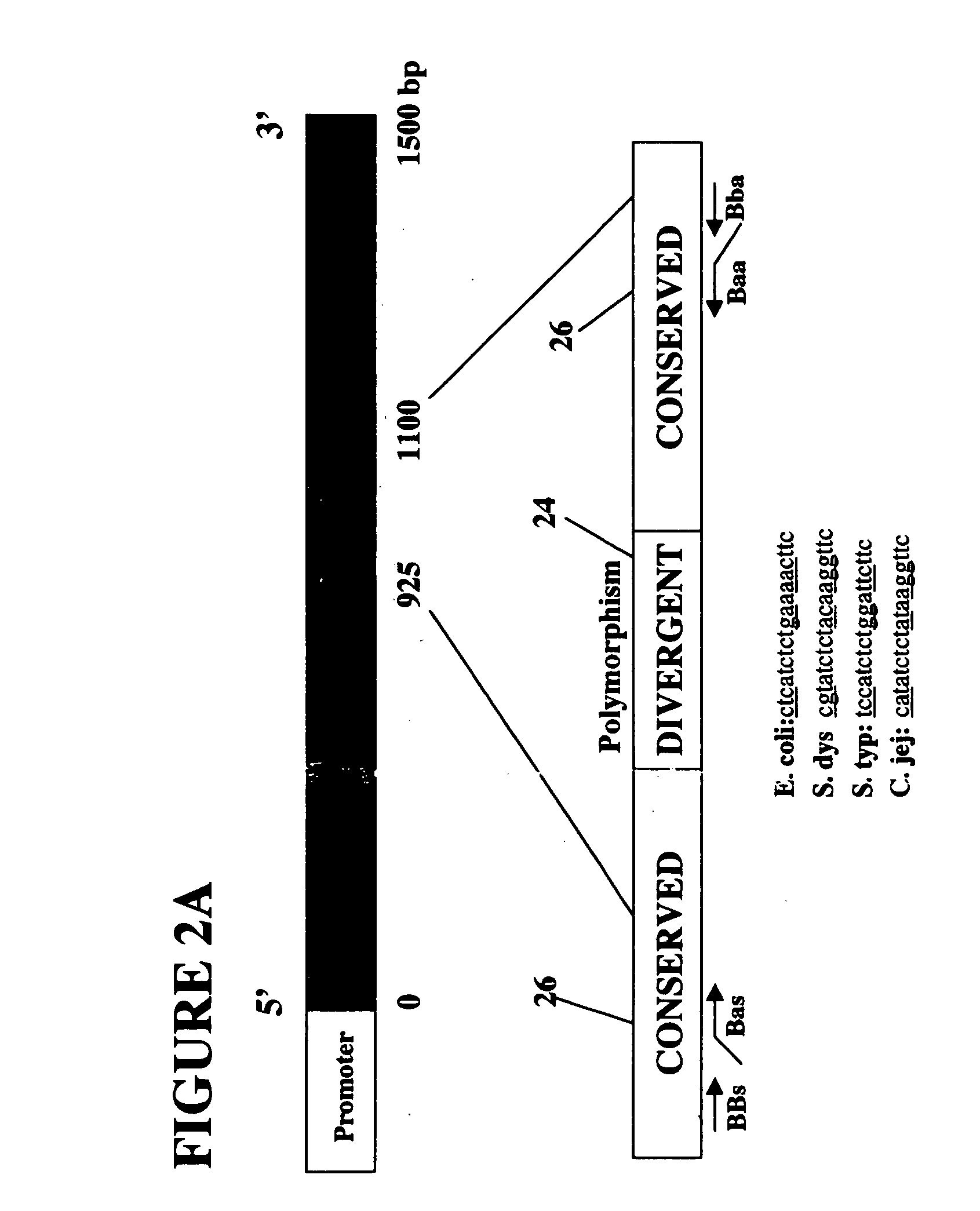
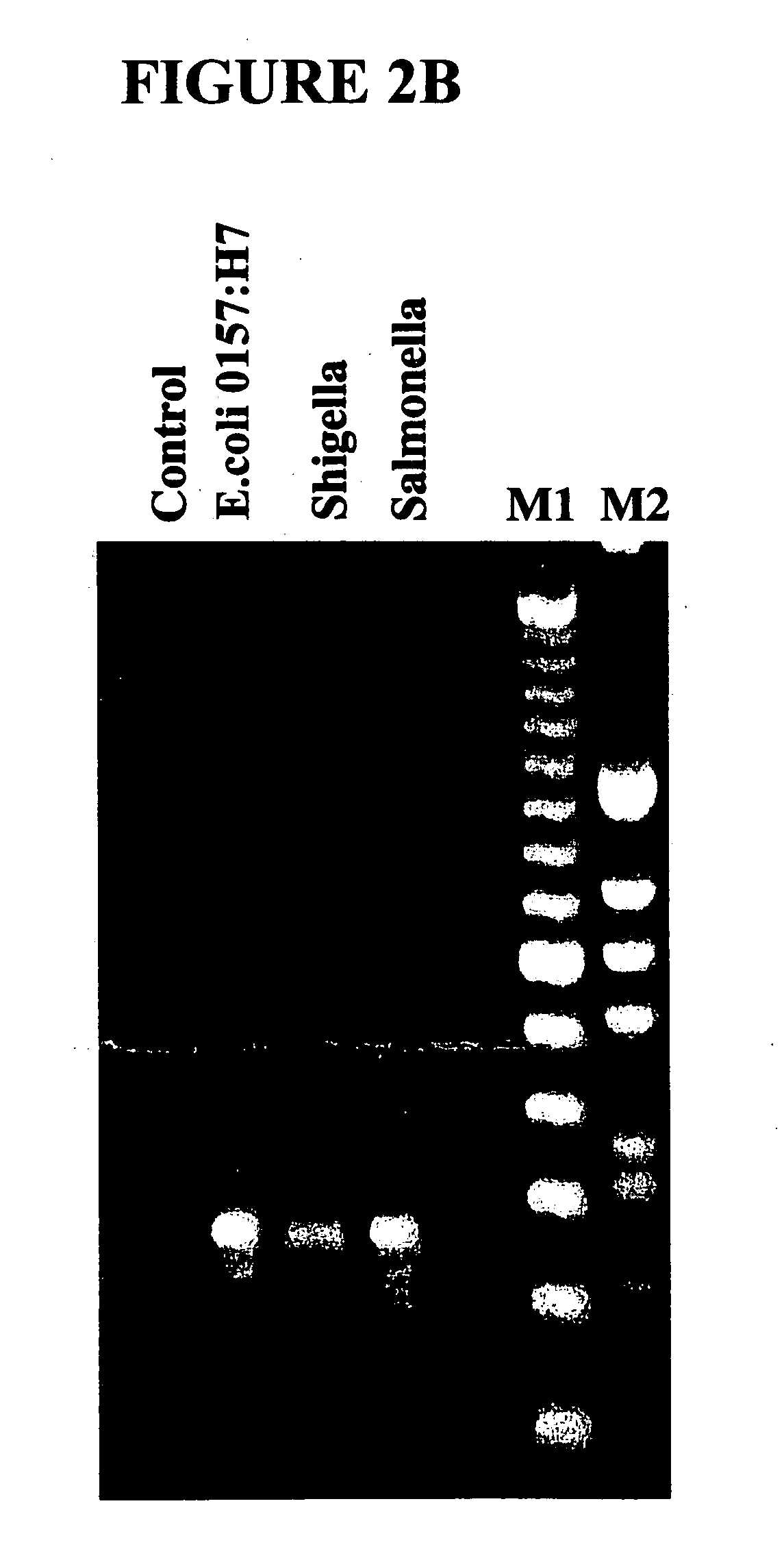
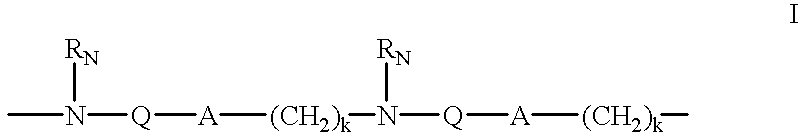
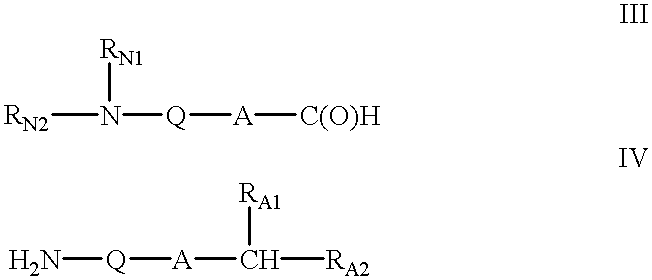
Patents
Literature
51results about How to "Enhanced hybridization" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
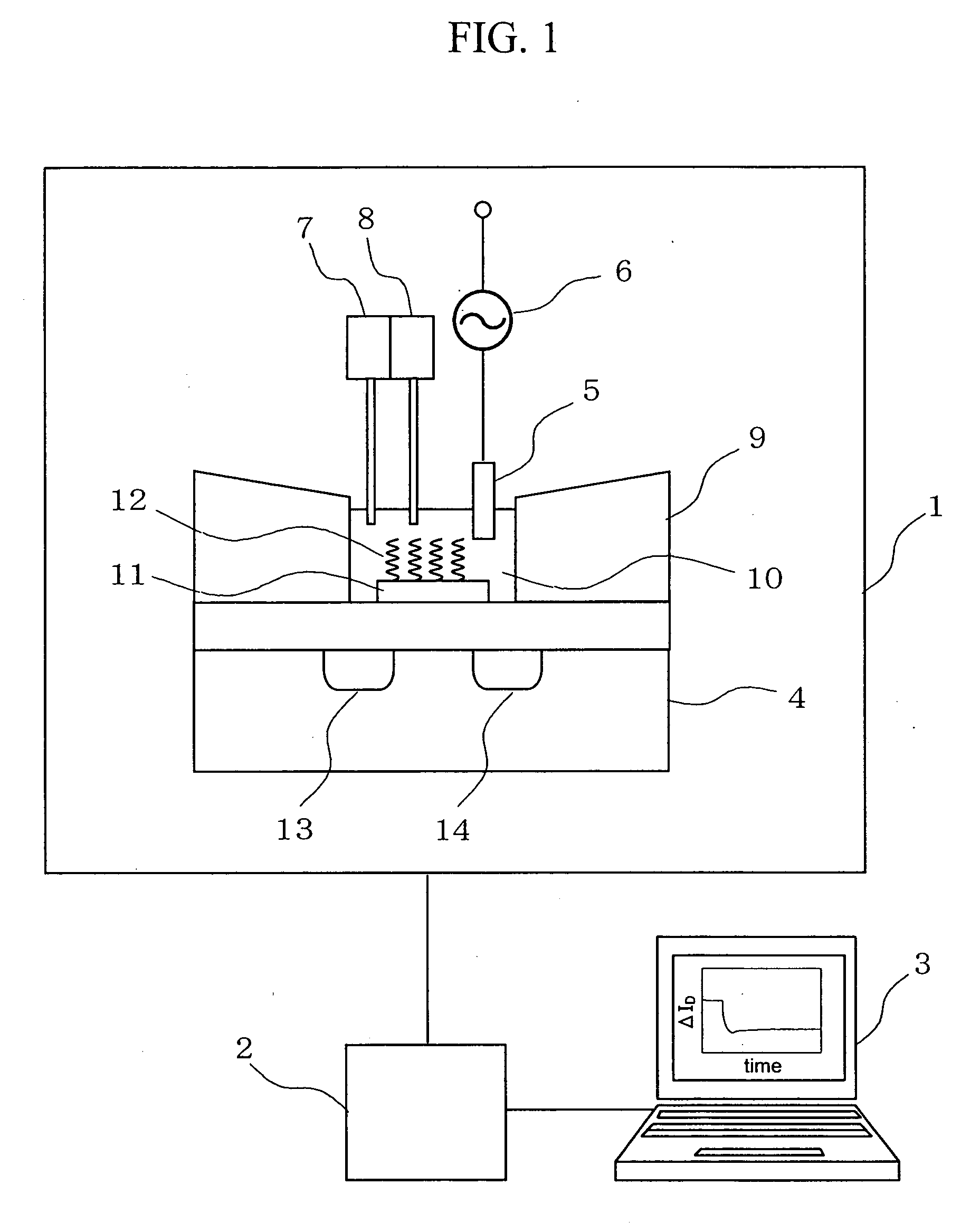
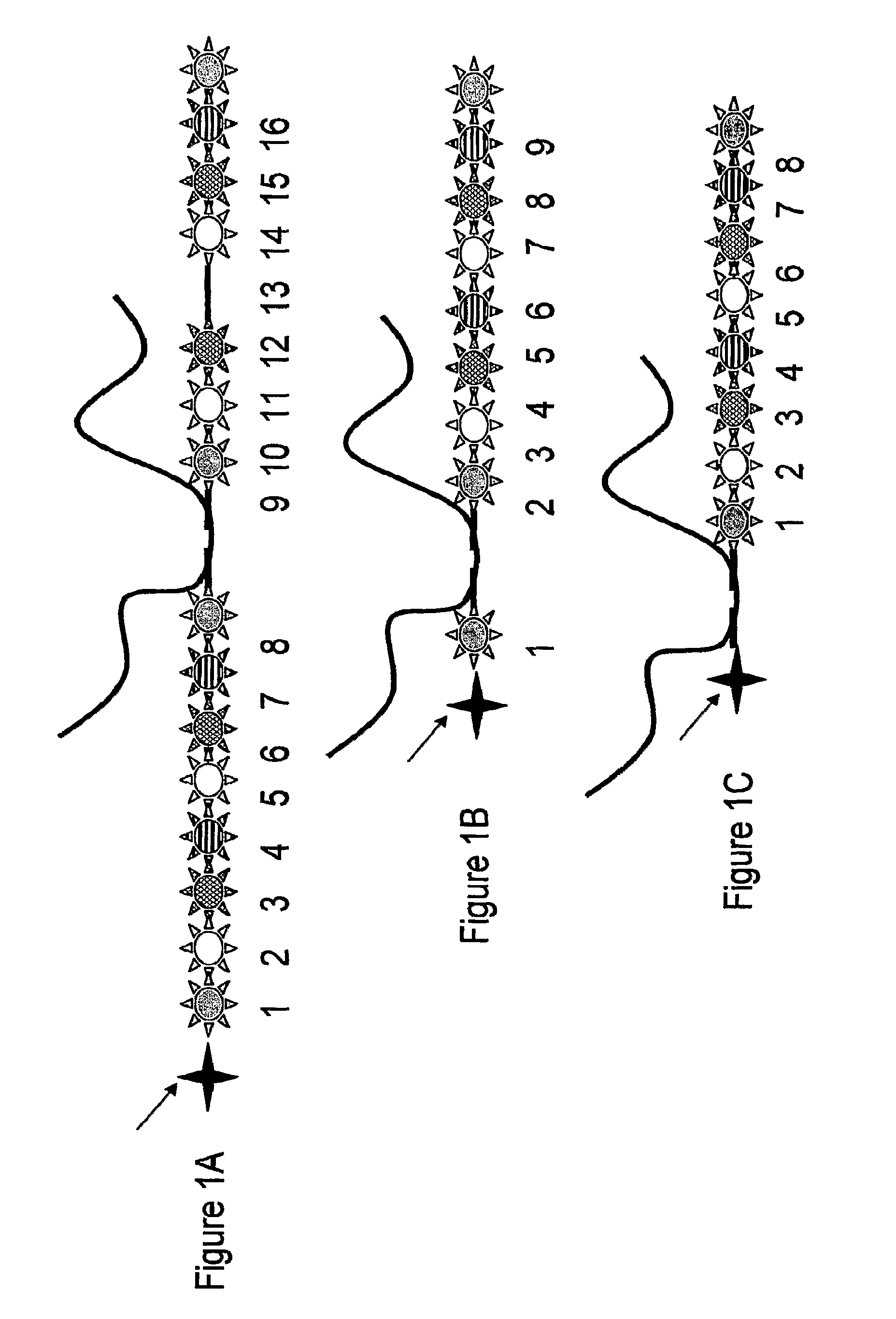
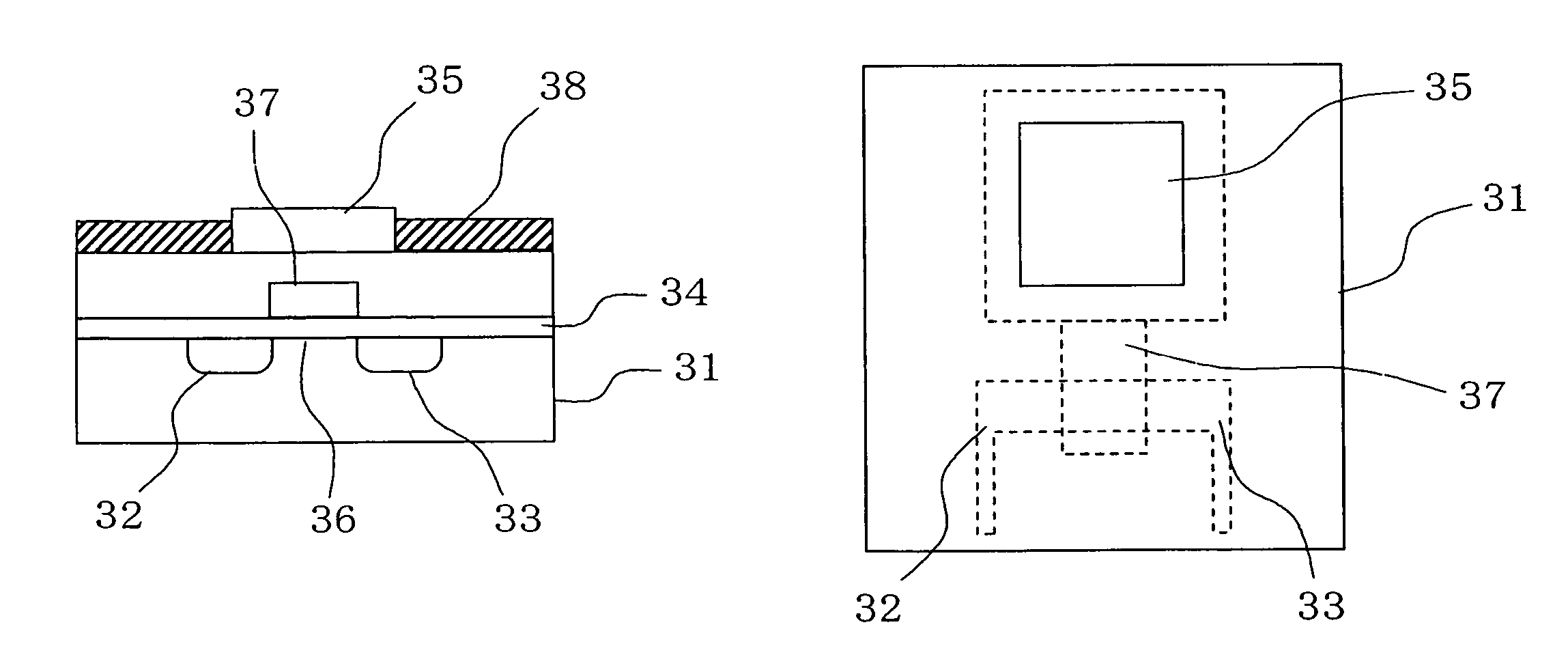
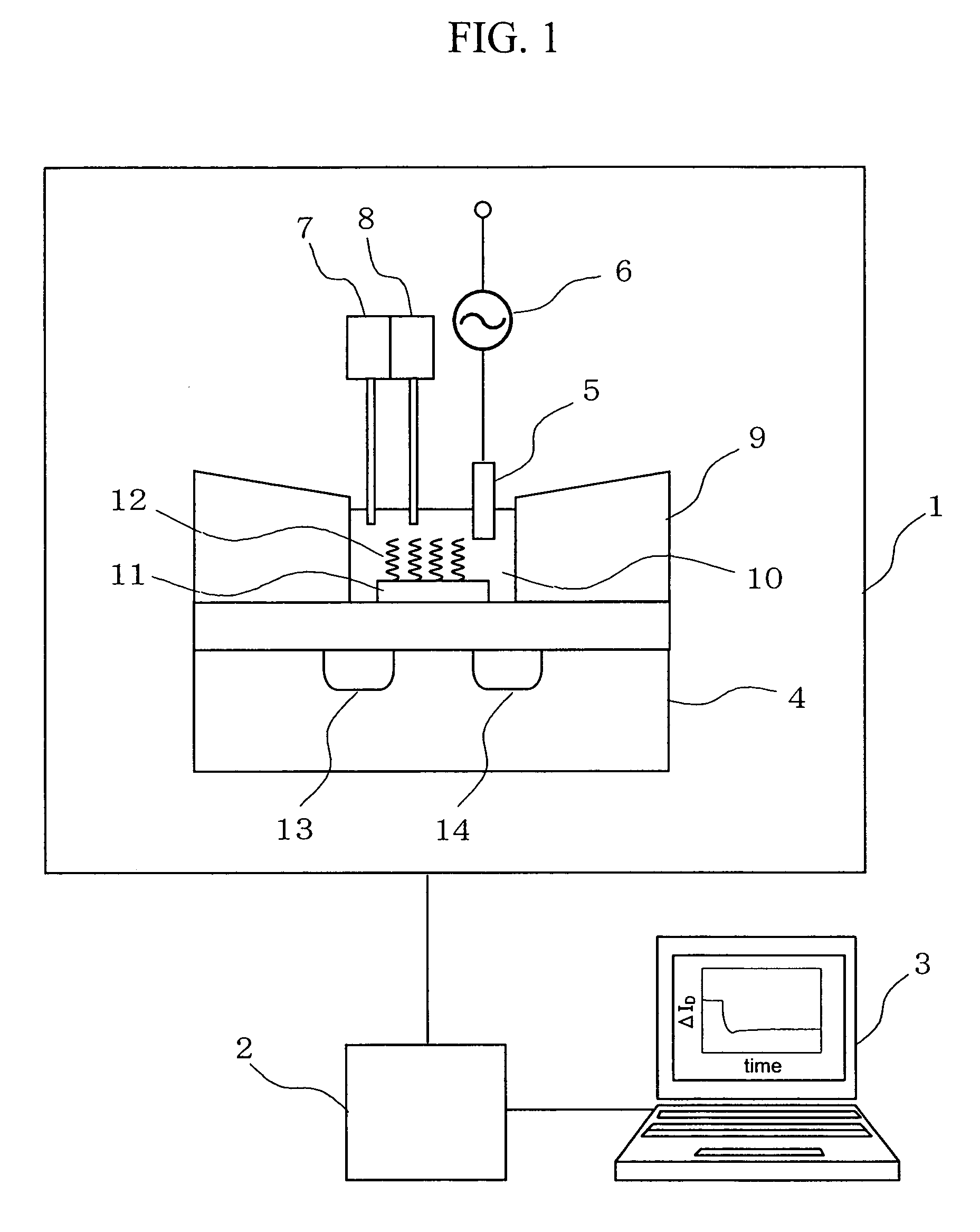
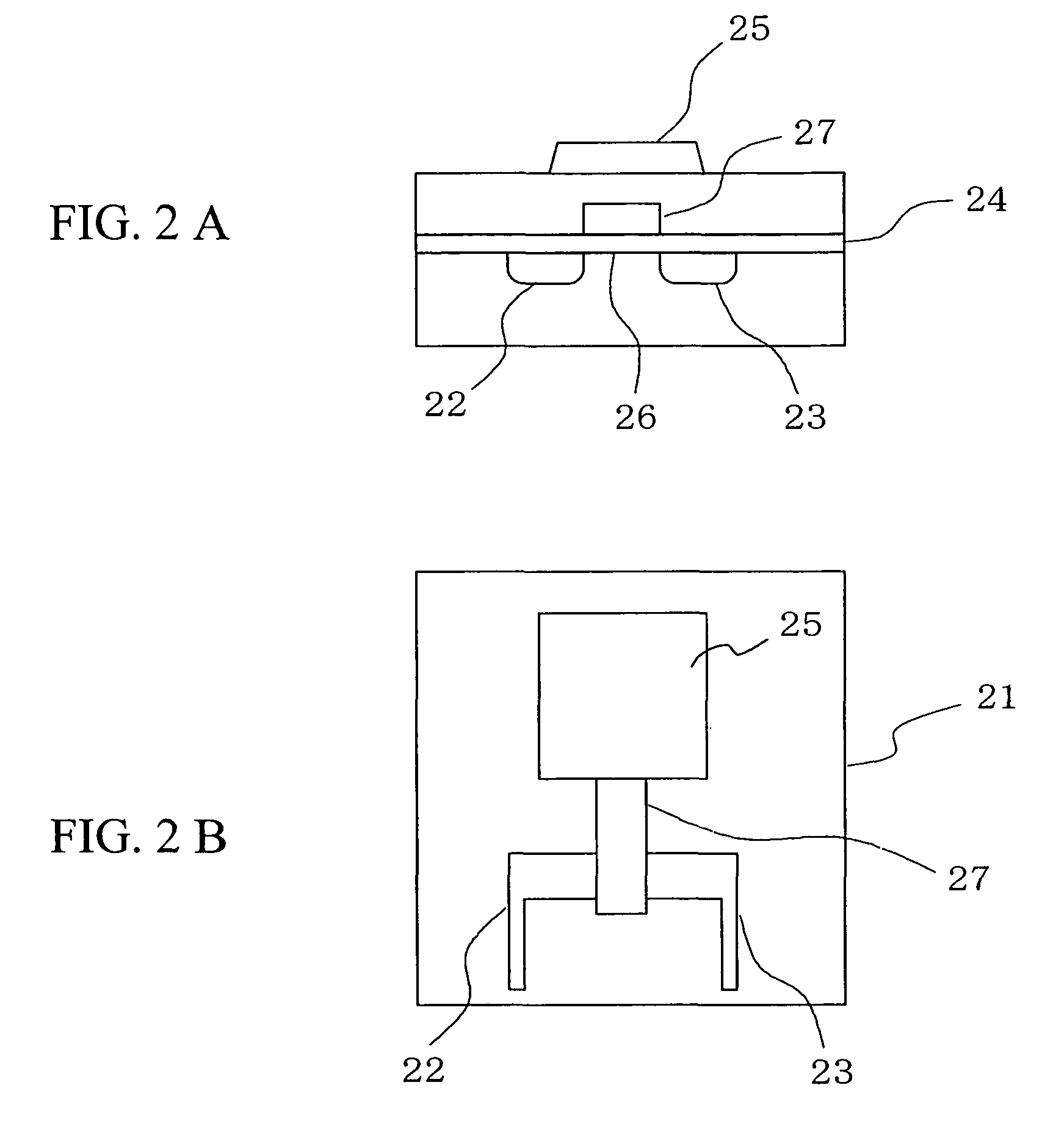
Deoxyribonucleic acid measuring apparatus and method of measuring deoxyribonucleic acid
InactiveUS20070059741A1Accelerate reaction cycleReduce timeMaterial nanotechnologyBioreactor/fermenter combinationsForeign matterPhosphate
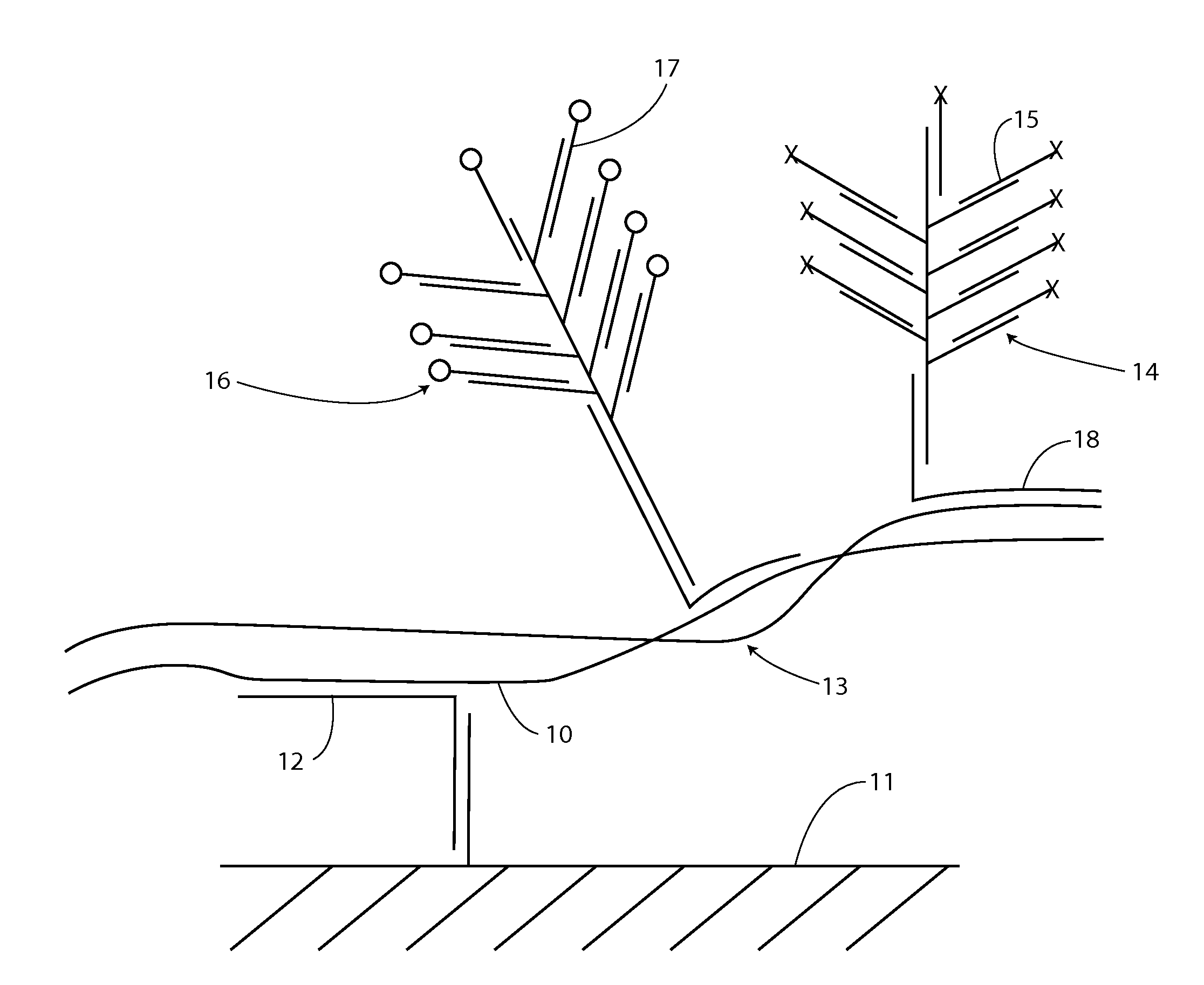
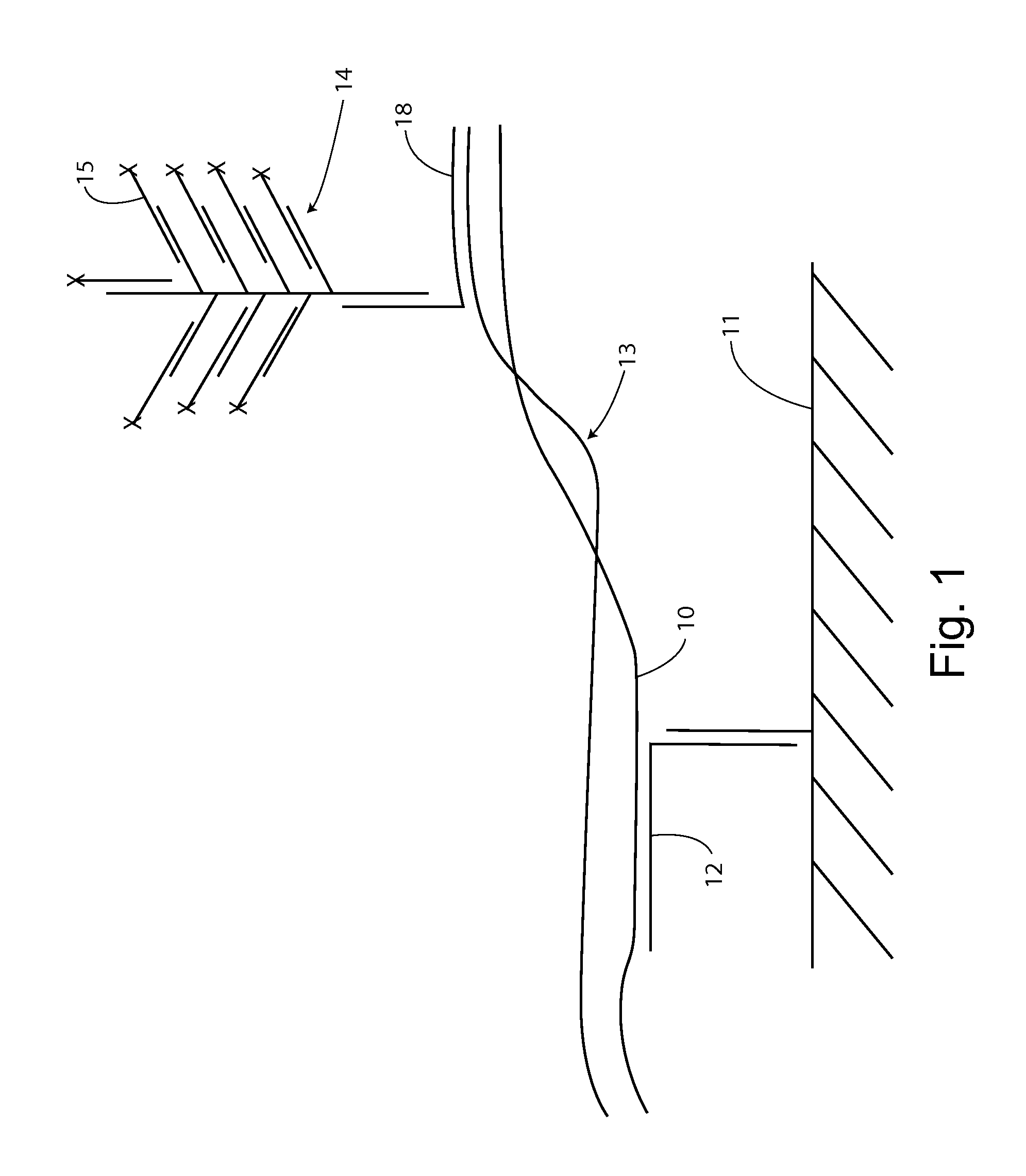
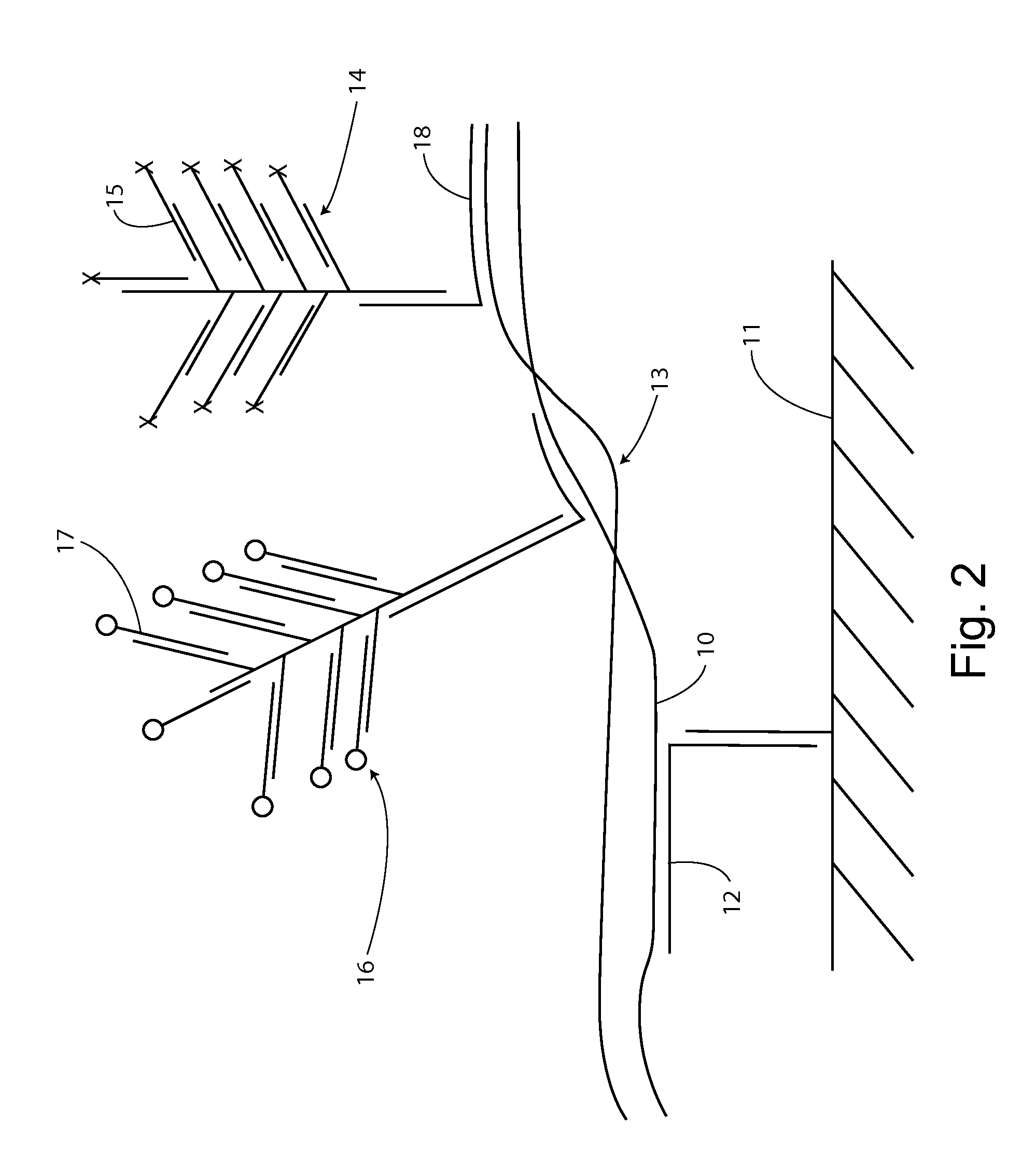
With an insulated gate field effect transistor in which deoxyribonucleic acid (DNA) probes are immobilized on a gold electrode, extension reaction on the gold electrode is performed with DNA polymerase to directly measure an increased amount of a phosphate group caused by the extension reaction, that is, negative charge, by means of a current change between a source and a drain of the insulated gate field effect transistor. Thus, presence / absence of hybridization of target DNAs with the DNA probes, and presence / absence of the extension reaction are detected. Optimum immobilization density of the DNA probes on the gold electrode is set at 4×1012 molecules / cm2. To reduce surface potential fluctuation caused by external variation (influences of foreign substances), which is a problem when using the gold electrode in a solution, a high-frequency voltage equal to or above 1 kHz is applied between the gold electrode and a reference electrode by a power source.
Owner:HITACHI LTD
Nanoreporters and methods of manufacturing and use thereof
InactiveUS20100015607A1Capable of detectingEnhanced hybridizationSugar derivativesMicrobiological testing/measurementBiochemistryCell biology
The present invention relates to compositions and methods for detection and quantification of individual target molecules in biomolecular samples. In particular, the invention relates to coded, labeled probes that are capable of binding to and identifying target molecules based on the probes' label codes. Methods of making and using such probes are also provided. The probes can be used in diagnostic, prognostic, quality control and screening applications.
Owner:INSTITUTE FOR SYSTEMS BIOLOGY +2
Detection of nucleic acids
ActiveUS9273349B2Enhanced hybridizationConfidenceSugar derivativesMicrobiological testing/measurementTissue sampleSingle strand
Owner:AFFYMETRIX INC
Compositions, methods, and kits for fabricating coded molecular tags
InactiveUS20100105886A1Minimise measureImprove method performanceSugar derivativesMicrobiological testing/measurementBiotechnologyAnalyte
Owner:LIFE TECH CORP
Methods and compositions for directed microwave chemistry
InactiveUS20040209303A1Enhanced hybridizationHeat is availableImmobilised enzymesMicrobiological testing/measurementPolymerase chain reactionChemical agents
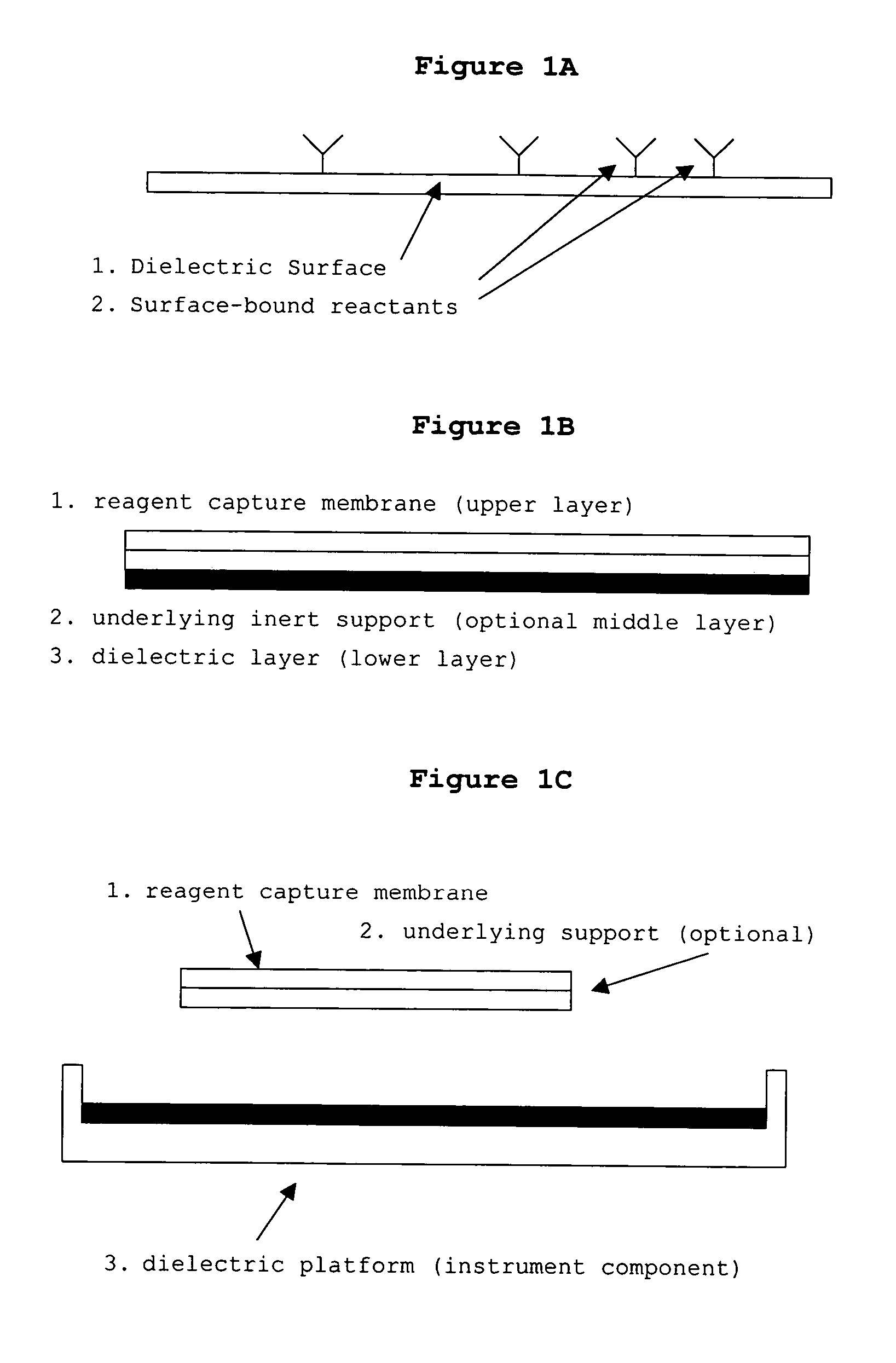
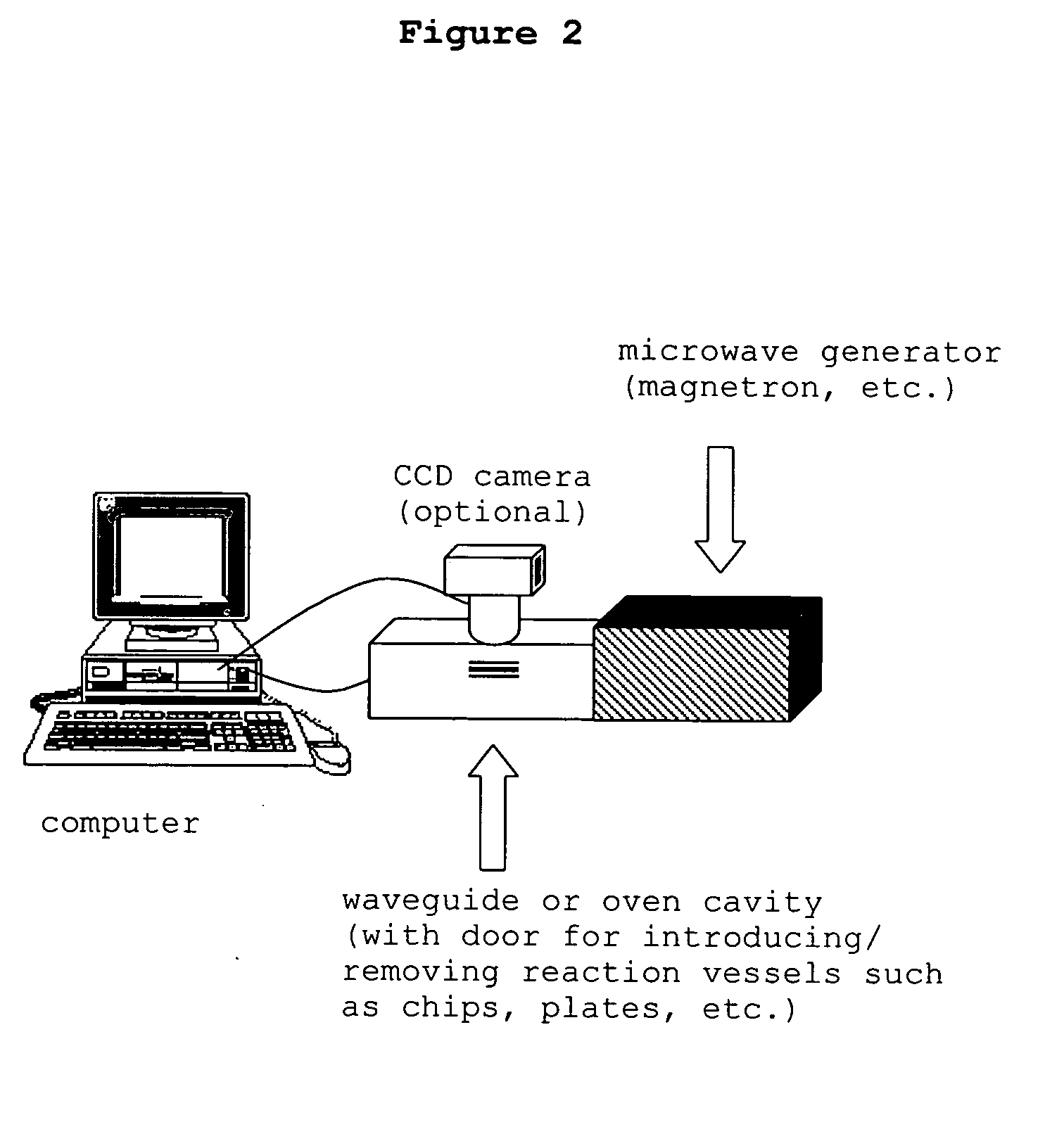
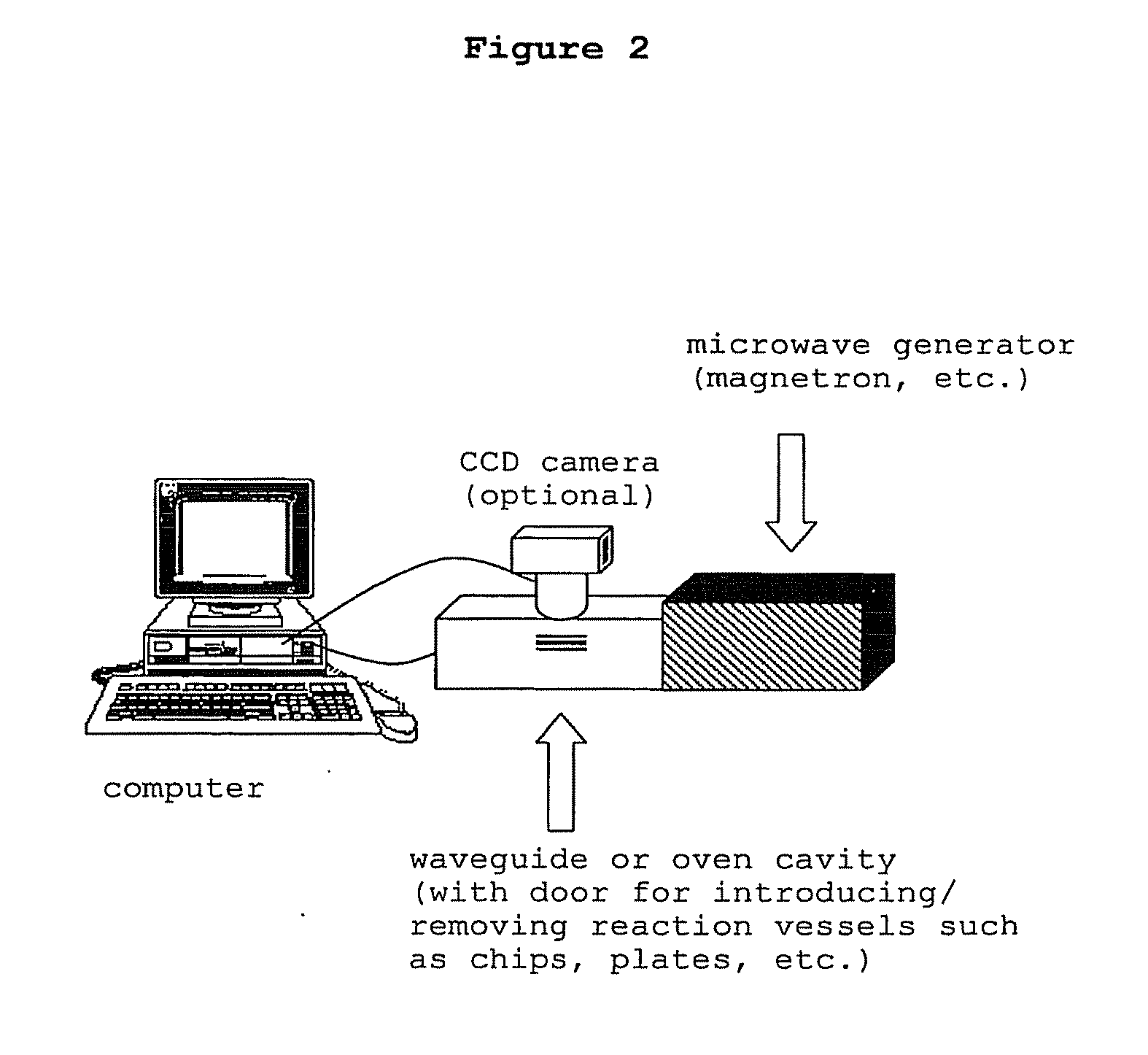
The present invention concerns a novel means by which chemical preparations can be made. Reactions can be accelerated on special cartridges using microwave energy. The chips contain materials that efficiently absorb microwave energy causing chemical reaction rate increases. The invention is important in many chemical transformations including those used in protein chemistry, in nucleic acid chemistry, in analytical chemistry, and in the polymerase chain reaction.
Owner:MIRARI BIOSCI
Nanoreporters and methods of manufacturing and use thereof
ActiveUS9371563B2Capable of detectingEnhanced hybridizationBioreactor/fermenter combinationsBiological substance pretreatmentsQuality controlBiology
The present invention relates to compositions and methods for detection and quantification of individual target molecules in biomolecular samples. In particular, the invention relates to coded, labeled probes that are capable of binding to and identifying target molecules based on the probes' label codes. Methods of making and using such probes are also provided. The probes can be used in diagnostic, prognostic, quality control and screening applications.
Owner:INSTITUTE FOR SYSTEMS BIOLOGY +1
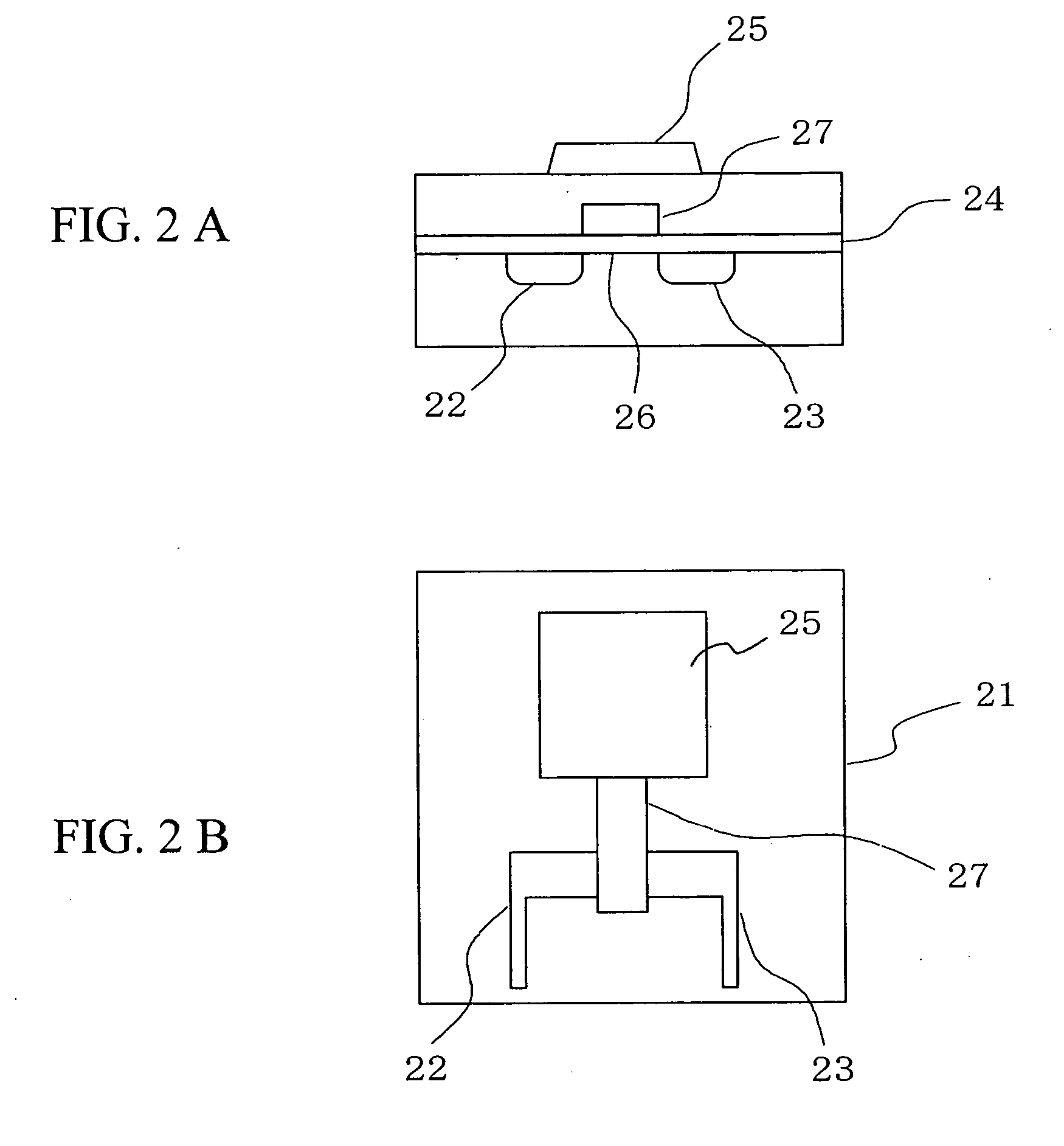
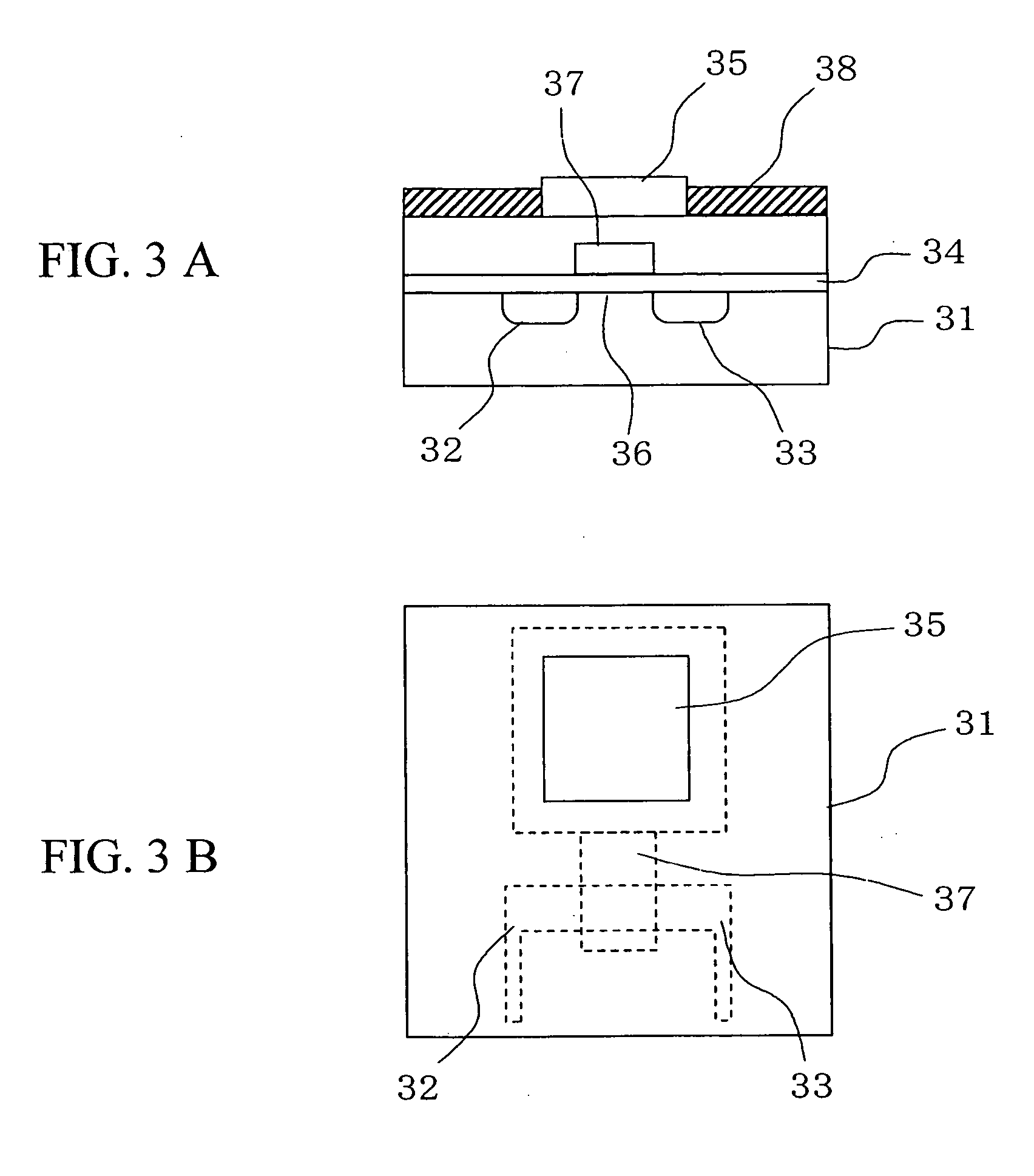
Deoxyribonucleic acid measuring apparatus and method of measuring deoxyribonucleic acid
InactiveUS7838226B2Optimize immobilization densityEfficient executionBioreactor/fermenter combinationsMaterial nanotechnologyDeoxyribonucleic acid probeField effect
With an insulated gate field effect transistor in which deoxyribonucleic acid (DNA) probes are immobilized on a gold electrode, extension reaction on the gold electrode is performed with DNA polymerase to directly measure an increased amount of a phosphate group caused by the extension reaction, that is, negative charge, by means of a current change between a source and a drain of the insulated gate field effect transistor. Thus, presence / absence of hybridization of target DNAs with the DNA probes, and presence / absence of the extension reaction are detected. Optimum immobilization density of the DNA probes on the gold electrode is set at 4×1012 molecules / cm2. To reduce surface potential fluctuation caused by external variation (influences of foreign substances), which is a problem when using the gold electrode in a solution, a high-frequency voltage equal to or above 1 kHz is applied between the gold electrode and a reference electrode by a power source.
Owner:HITACHI LTD
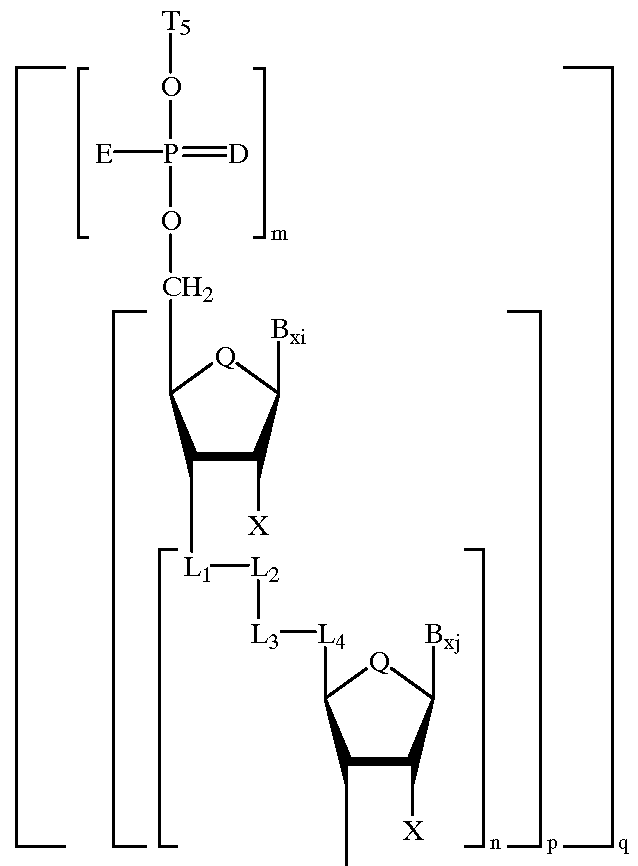
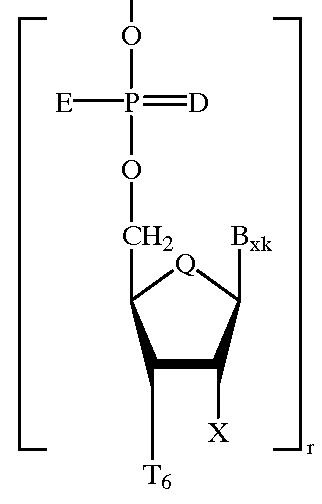
Heteroatomic oligonucleoside linkages
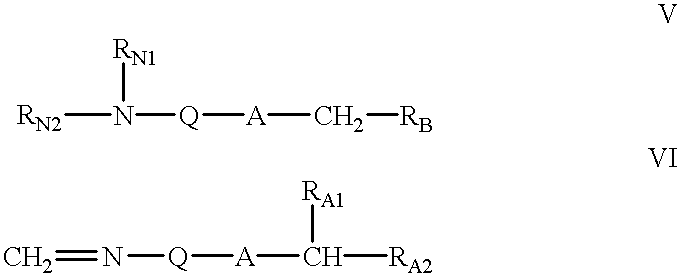
InactiveUS6087482AEnhanced cellular uptakeImprove propertiesGroup 4/14 element organic compoundsSugar derivativesRna expressionOligonucleotide
PCT No. PCT / US94 / 03536 Sec. 371 Date Sep. 18, 1995 Sec. 102(e) Date Sep. 18, 1995 PCT Filed Mar. 30, 1994 PCT Pub. No. WO94 / 22886 PCT Pub. Date Oct. 13, 1994Oligonucleotide-mimicking macromolecules that have improved nuclease resistance are provided. Replacement of the normal phosphorodiester inter-sugar linkages found in natural oligonucleotides with four atom linking groups provide unique compounds that are useful in regulating RNA expression and in therapeutics. Methods of synthesis and use also are disclosed.
Owner:IONIS PHARMA INC
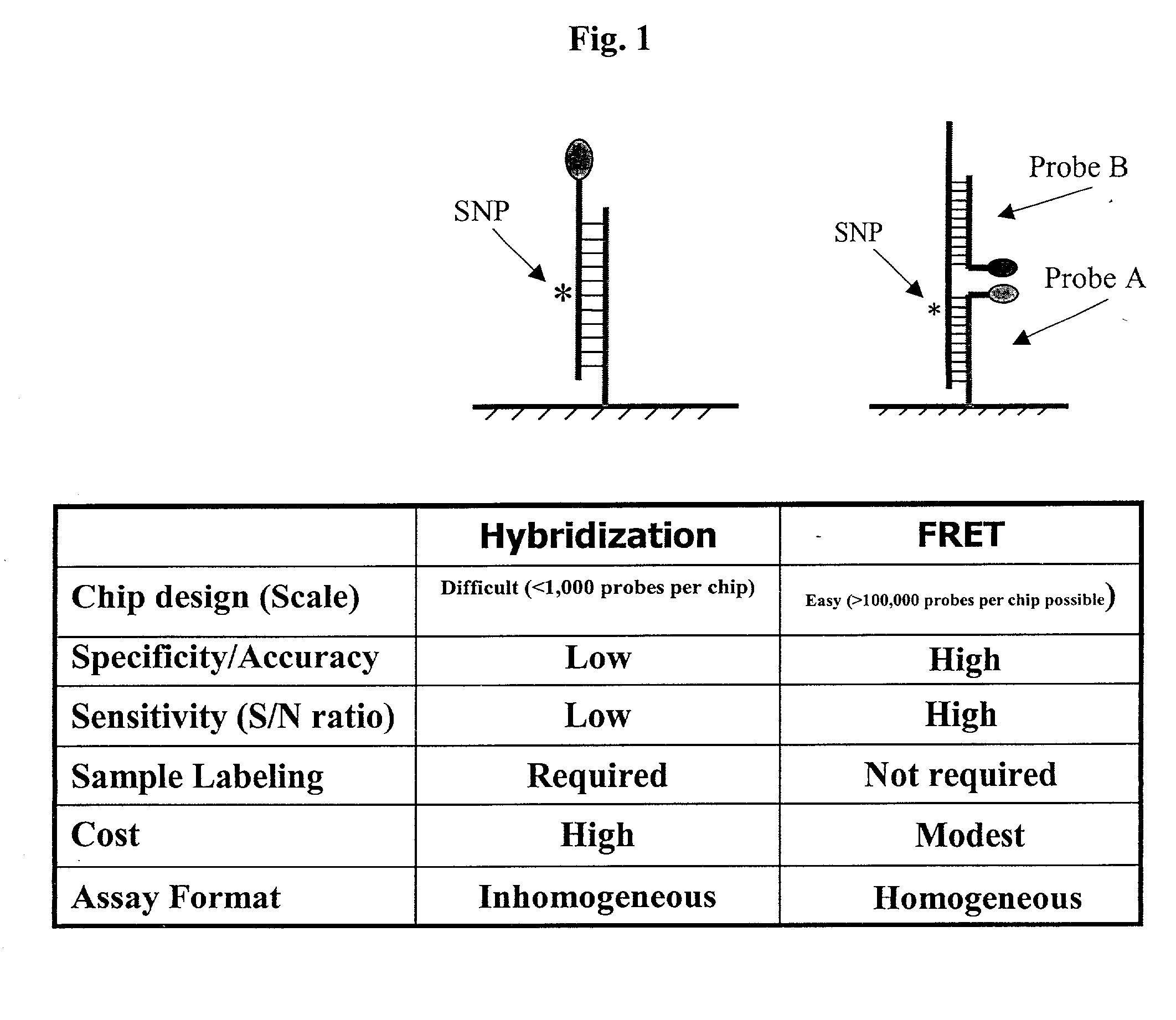
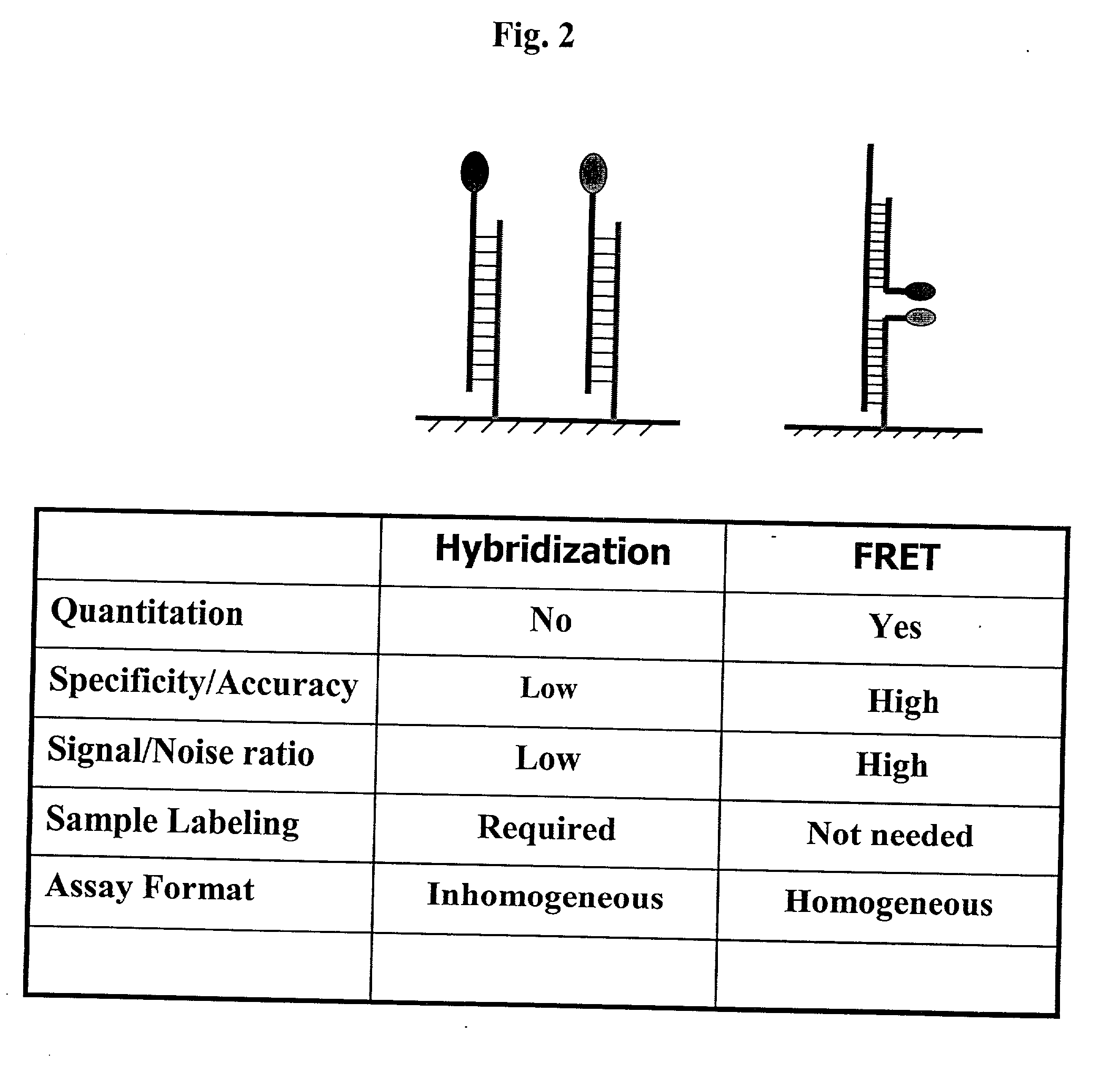
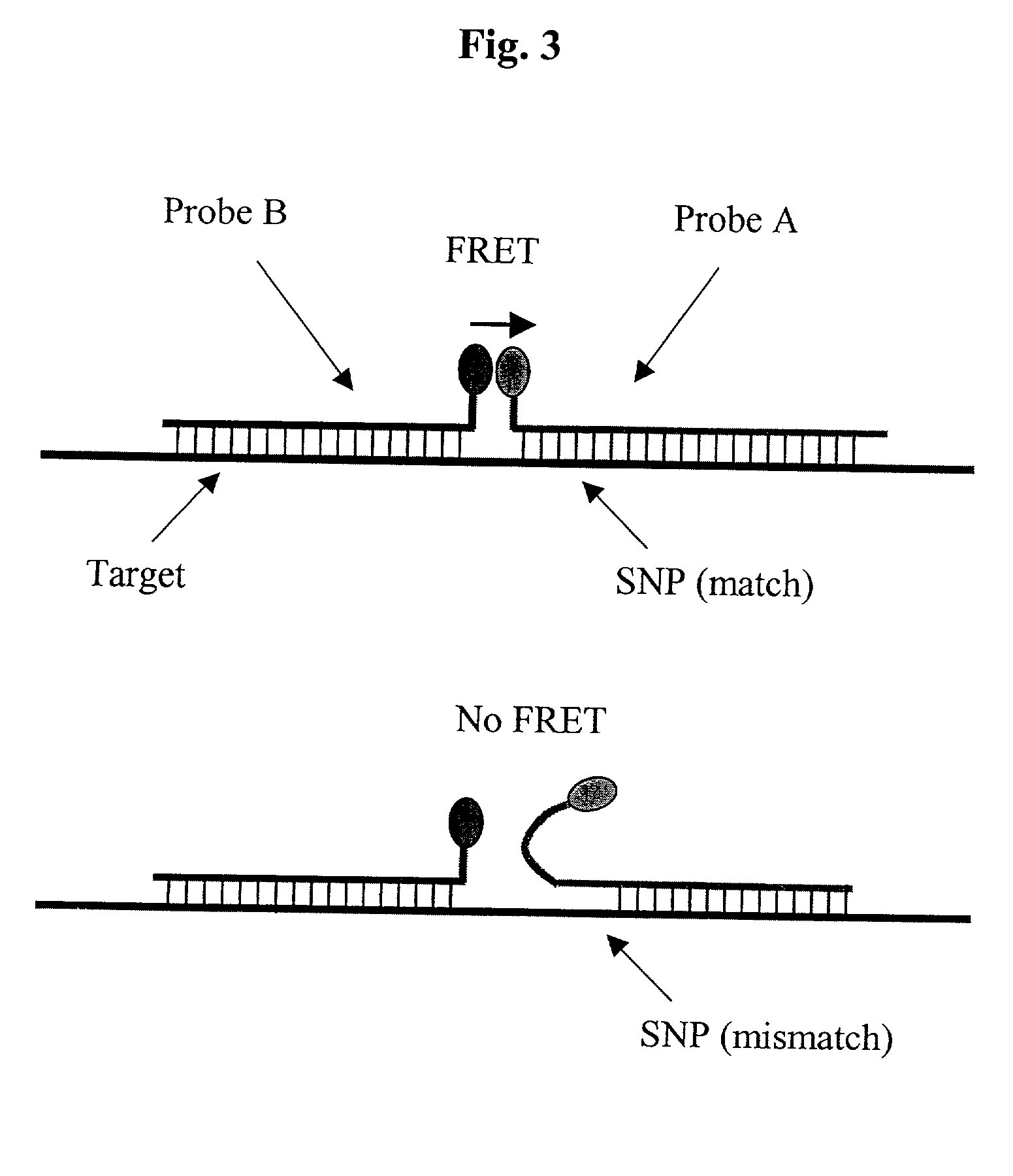
Pairs of nucleic acid probes with interactive signaling moieties and nucleic acid probes with enhanced hybridization efficiency and specificity
InactiveUS20020177157A1Improve efficiencyEnhanced hybridizationSugar derivativesMicrobiological testing/measurementNucleotideGenotype Analysis
The invention provides methods and kits for detecting and / or quantifying nucleic acid sequences of interest by using pairs of probes containing donor-acceptor moieties that when hybridized on a target polynucleotide, with one of the probes being hybridized to a sequence of interest in the target polynucleotide, places the donor-acceptor moieties in sufficiently close proximity such that a detectable signal is generated. Methods of the invention are particularly useful for genotyping analysis and gene expression profiling. Methods of the invention are easily adaptable to arrays and automation. The invention also provides probes with enhanced hybridization efficiency and / or specificity comprising a probe portion having a spacer element and / or a minor groove binder molecule, and methods and kits for using these probes.
Owner:GENOSPECTRA
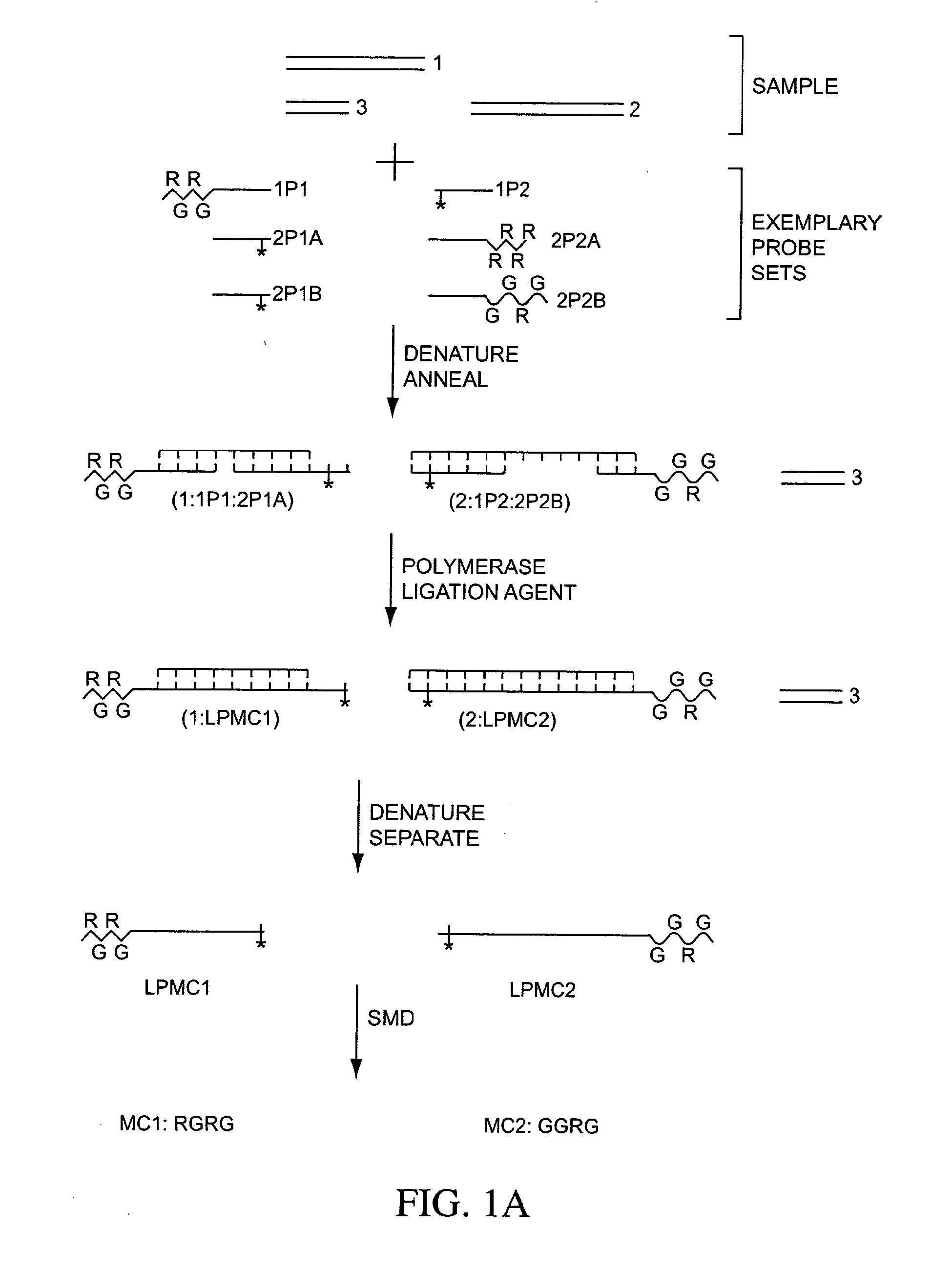
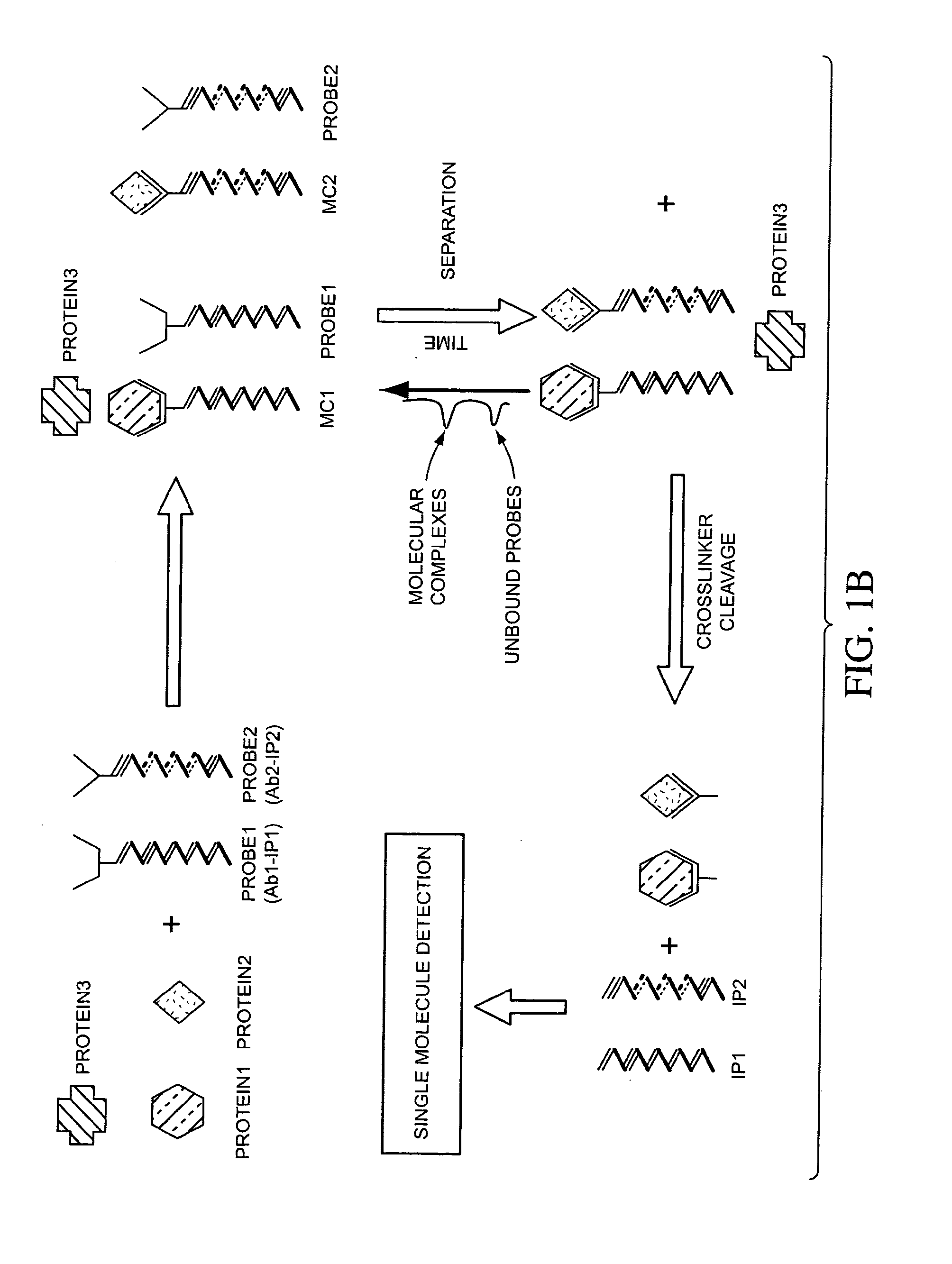
Multiplex detection compositions, methods, and kits
ActiveUS7198900B2Minimise measureImprove method performanceSugar derivativesMicrobiological testing/measurementMultiplexBio molecules
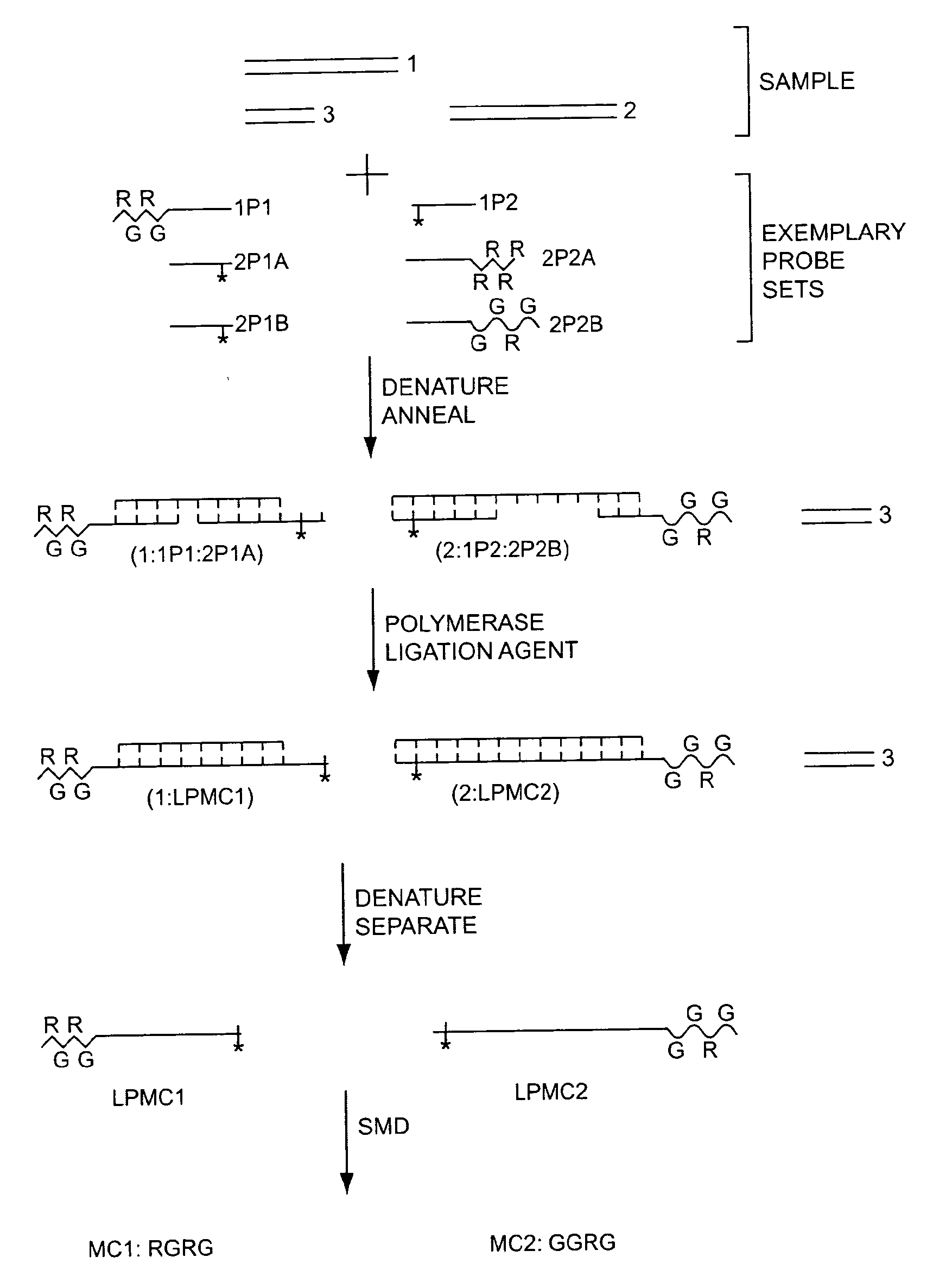
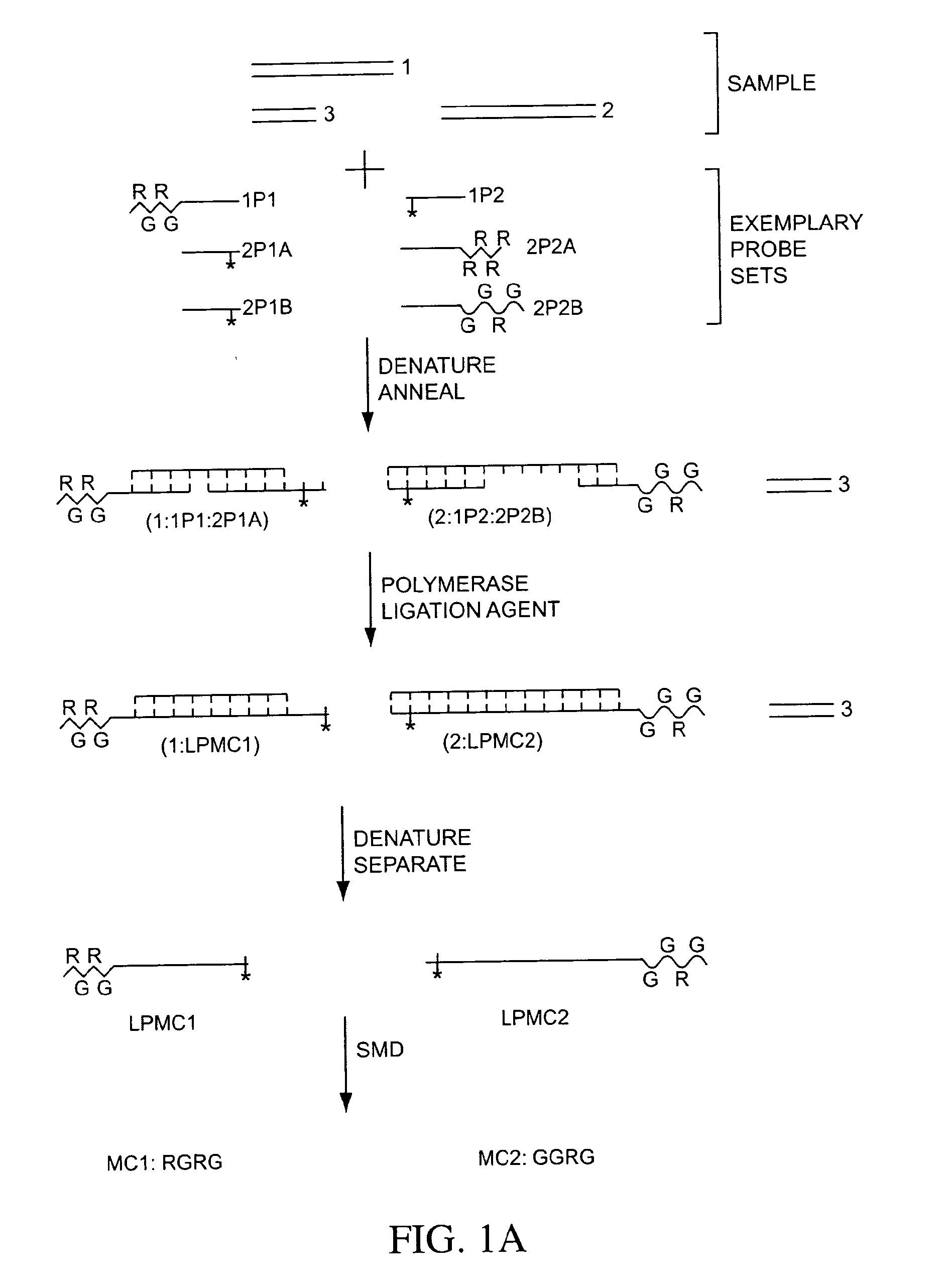
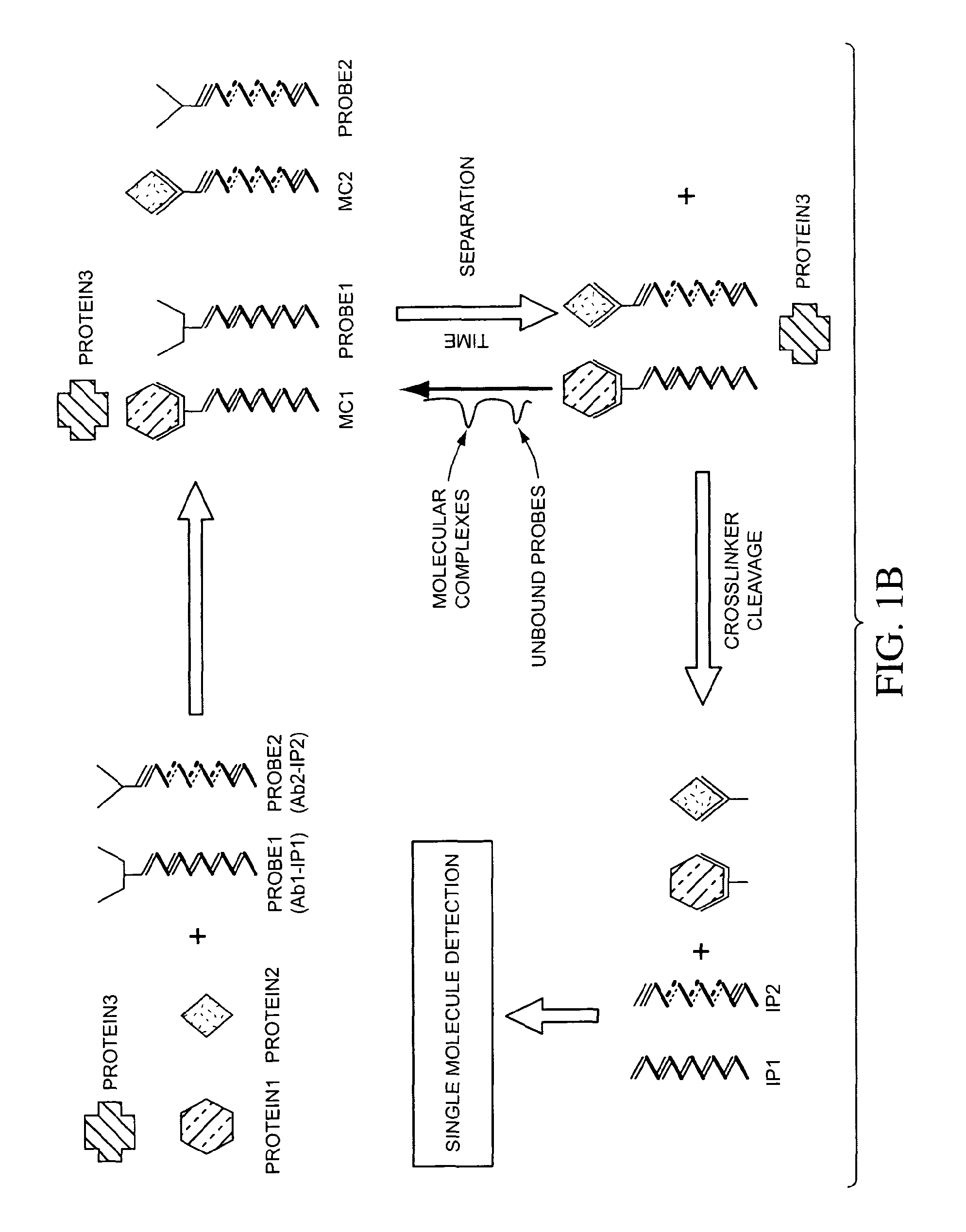
The present invention generally relates to the detection of analytes, particularly biomolecules in samples. The invention also relates to compositions, methods, and kits for detecting the presence of analytes, typically in multiplex detection formats. The invention also relates to methods for determining the presence of at least one analyte in a sample, the methods employing employ single molecule detection techniques to individually detect at least one molecular complex or at least part of a molecular complex.
Owner:APPL BIOSYSTEMS INC
Method for long range allele-specific PCR
InactiveUS20070184457A1Enhanced hybridizationSugar derivativesMicrobiological testing/measurementPolymerase LA-DNA
The present invention is directed to a method and kit for determining the molecular haplotype of a gene in a diploid DNA sample. The method discriminates between two haplotypes on the basis of a difference of one or more nucleotides using allele-specific PCR amplification in combination with long range PCR, using a DNA polymerase enzyme having 3′→5′ exonuclease activity, using annealing temperature conditions sufficiently greater than the predicted annealing temperature (Tm) to effect selective hybridization and extension of an allele-specific extension primer to the target allele relative to the variant allele. The present invention is particularly useful, for example, to determine the haplotype of a gene having multi-allelic genetic loci separated by a distance in which accurate PCR amplification of a single fragment containing the multiple genetic loci cannot be performed without using a DNA polymerase enzyme having 3′→5′ exonuclease activity, generally about 5 kilobases or more.
Owner:UNIV OF UTAH RES FOUND
Nanoreporters And Methods Of Manufacturing And Use Thereof
ActiveUS20130230851A1Capable of detectingEnhanced hybridizationSugar derivativesPeptide/protein ingredientsQuality controlBiology
The present invention relates to compositions and methods for detection and quantification of individual target molecules in biomolecular samples. In particular, the invention relates to coded, labeled probes that are capable of binding to and identifying target molecules based on the probes' label codes. Methods of making and using such probes are also provided. The probes can be used in diagnostic, prognostic, quality control and screening applications.
Owner:NANOSTRING TECH INC +1
Carbohydrate arrays
InactiveUS20070269818A1Increase hybridization stringencyEconomical and facileMicrobiological testing/measurementBiological material analysisHigh densityBiology
Methods of detecting a carbohydrate binding compound in a sample by providing a high density oligonucleotide array including a plurality of probe sequences, hybridizing a plurality of carbohydrates that include an oligonucleotide that is complementary to a probe sequence to the high density oligonucleotide array, hybridizing a sample including a plurality of carbohydrate binding compounds to the high density oligonucleotide array, and detecting hybridization of at least one carbohydrate binding compound to at least one carbohydrate to determine the presence of the carbohydrate binding compound in a sample are provided.
Owner:AFFYMETRIX INC
Novel hybridization probes and uses thereof
ActiveUS20160083783A1Improve hybridizationLow-cost and rapid and highly quantitative RNA profilingBioreactor/fermenter combinationsBiological substance pretreatmentsPathogenNucleic acid structure
The present invention relates to novel hybridization probes useful for rapid hybridization to realize a practicable and affordable pathogen diagnostic based on RNA signature detection, technology and hardware. In particular, the present invention relates to a nucleic acid structure which may comprise nucleic acid tiles, wherein each nucleic acid tile comprises nucleic acid oligomers, wherein each nucleic acid oligomer hybridizes to each other thereby forming a nucleic acid tile.
Owner:THE BROAD INST INC +1
Microarray system
ActiveUS20090280997A1High capacityPrevents total solubilizationPeptide librariesSequential/parallel process reactionsRNABiotechnology
This invention embodies a method that can be used to increase the porosity of the gel substrate during the fabrication of microchips. This increase in gel porosity will allow larger molecules such as non-fragmented RNAs and DNA fragments greater then 100 base pairs in length, and proteins to be immobilized and introduced within the gel microchips.
Owner:AKONNI BIOSYSTEMS INC
Method of binding a compound to a surface
InactiveUS20050238685A1Low mass production costIncrease storage spaceCell receptors/surface-antigens/surface-determinantsPeptide/protein ingredientsCompound (substance)Hydrophobin
The present invention relates to a method of binding a compound to at least a part of a surface of an object, the method comprising the step of adsorbing a hydrophobin-like substance to the surface. The invention provides a method of providing a surface of an object with a reactive compound comprising the steps of coating at least a part of the surface of the object with a coating of a hydrophobin-like substance and contacting the compound with the coated hydrophobin-like substance to form a non-covalent bond between the hydrophobin-like substance and the compound.
Owner:HEKTOR HARM JAN +5
Detection of Nucleic Acids
ActiveUS20140274756A1Improve abilitiesHigh sensitivityNucleotide librariesMicrobiological testing/measurementTissue sampleSingle strand
This invention provides compositions, methods, and systems for characterizing, resolving, and quantitating single stranded and double stranded DNA and RNA in-situ. Paired sense and anti-sense probes can signal the presence of double stranded nucleic acids. DNA and RNA can be distinguished in cell and tissue samples by hybridizing with probe sets adapted to highlight differences in these targets in-situ.
Owner:AFFYMETRIX INC
Selection Methods of Self-Pollination and Normal Cross Pollination in Poplation, Variety of Crops
InactiveUS20080098492A1Laborious and time-consumingHybridization is not heavyPlant genotype modificationFemale groupSelf-pollination
The invention relates to the field of crop selection and crop breeding, particularly to selection methods for breeding colony varieties of crops involving self-pollination and normal cross-pollination. Selection methods include crossing male parents with female parents to obtain crop populations, wherein the female parents are individual plants in a segregation population or self-crossed descendants of early segregation generations, said segregation population obtained by hybridizing pairs of parental plants with different desired characteristics to produce population F1, and then hybridizing pairs of F1 one more time. The male parents are homozygous breeding lines, varieties, F1, heterozygous plants in the segregation generations or individual plants produced in the same manner as for the female group. Crop populations and colony varieties of crops display characteristics of consistency, stability, specificity, disease resistance, high yield, and high adaptability while permitting farmers to reserve seeds properly and companies to exploit pedigreed seed on a large scale.
Owner:RICE RES INST GUANGDONG ACADEMY OF AGRI SCI +1
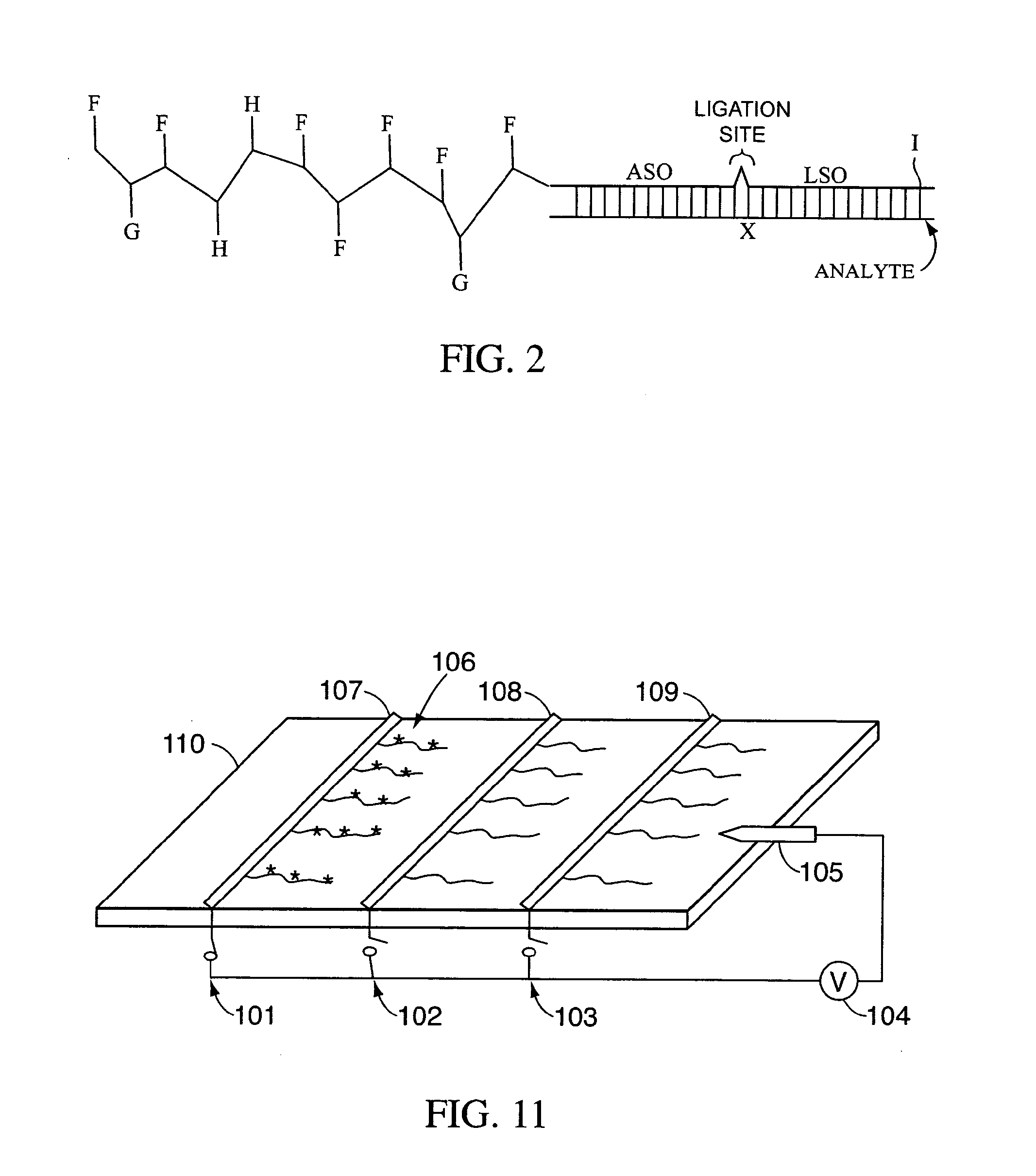
Amplification and separation of nucleic acid sequences using strand displacement amplification and bioelectronic microchip technology
InactiveUS20060110754A1Avoid uncertaintyHigh magnificationBioreactor/fermenter combinationsBiological substance pretreatmentsFluorescenceCombined use
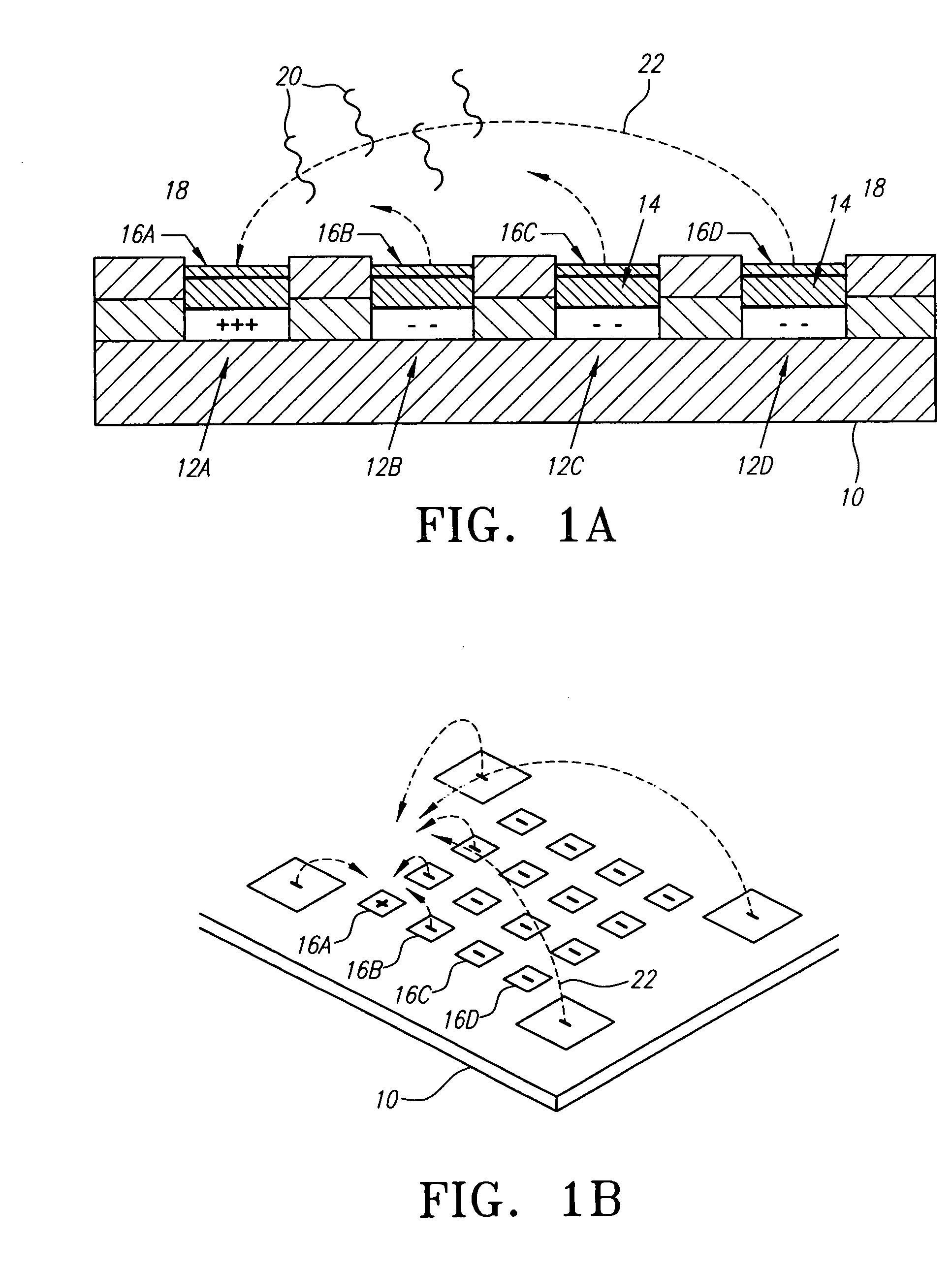
Described and disclosed are devices, methods, and compositions of matter for the multiplex amplification and analysis of nucleic acid sequences in a sample using novel strand displacement amplification technologies in combination with bioelectronic microchip technology. Specifically, a nucleic acid in a sample is amplified to form amplicons, the amplicons are addressed to specified electronically addressable capture sites of the bioelectronic microchip, the addressed amplicons are captured and labeled, and then the capture sites are analyzed for the presence of label. Samples may be amplified using strand displacement amplification. The invention is also amenable to other amplification methodologies well known by those skilled in the art. The capture and label steps may be by a method of universal capture with sequence specific reporter, or by a method of sequence specific capture with universal reporter. The label may be detected by fluorescence, chemiluminescence, elecrochemiluminescence, or any other technique as are well known by those skilled in the art. This invention further allows for analyzing multiple nucleic acid targets on a single diagnostic platform wherein the nucleic acids may be amplified while either in direct contact with microchip components or in solution above the microchip array.
Owner:NANOGEN INC
Oligonucleotide mimics having nitrogen-containing linkages
InactiveUS6271357B1Sequencing is facilitatedImprove propertiesGroup 4/14 element organic compoundsSugar derivativesNitrogenOligonucleotide
Owner:IONIS PHARMA INC
Use of Nucleic Acid Probes to Detect Nucleotide Sequences of Interest in a Sample
InactiveUS20090203017A1Reduce signalingHigh detection sensitivityMicrobiological testing/measurementNucleic Acid ProbesNucleic acid hybridisation
The invention relates to methods for the determination and detection of nucleic acids sequences in a sample. The nucleic acid may be RNA or DNA or both. The invention also relates to methods for the determination of the presence and species of various microorganisms in a sample. We have also identified a set of oligonucleotide nucleic acid sequences within the rRNAs of Gram-negative organisms that facilitates both the broad identification of Gram-negative organisms as a class when used as a pool, or in combination, for example in a hybridization assay. This set of oligonucleotides may detect sequences that are indicative of the presence of organisms of the broad class of Gram-negative organisms while exhibiting little or no false identification of Gram-positive organisms, and fungi, or other microorganisms. The assay includes concurrent incubation with at least one nucleotide sequence of interest, at least one nucleic acid probe, a fluorosurfactant, and a nuclease. The assay may further be employed to detect the presence of bacteria, fungi, or other microorganisms by use of additional specific probes, or to detect and / or identify target nucleic acid sequences in a sample. Further, the invention also relates to methods of reducing non-specific binding and facilitating complex formation in a binding assay. The binding assay may be, but is not limited to, a nucleic acid hybridization assay or an immunoassay. The invention also relates to methods of detection that employ at least one target of interest, which may be a nucleotide sequence, at least one probe, which may be a nucleic acid probe and a nuclease.
Owner:BIOVENTURES INC
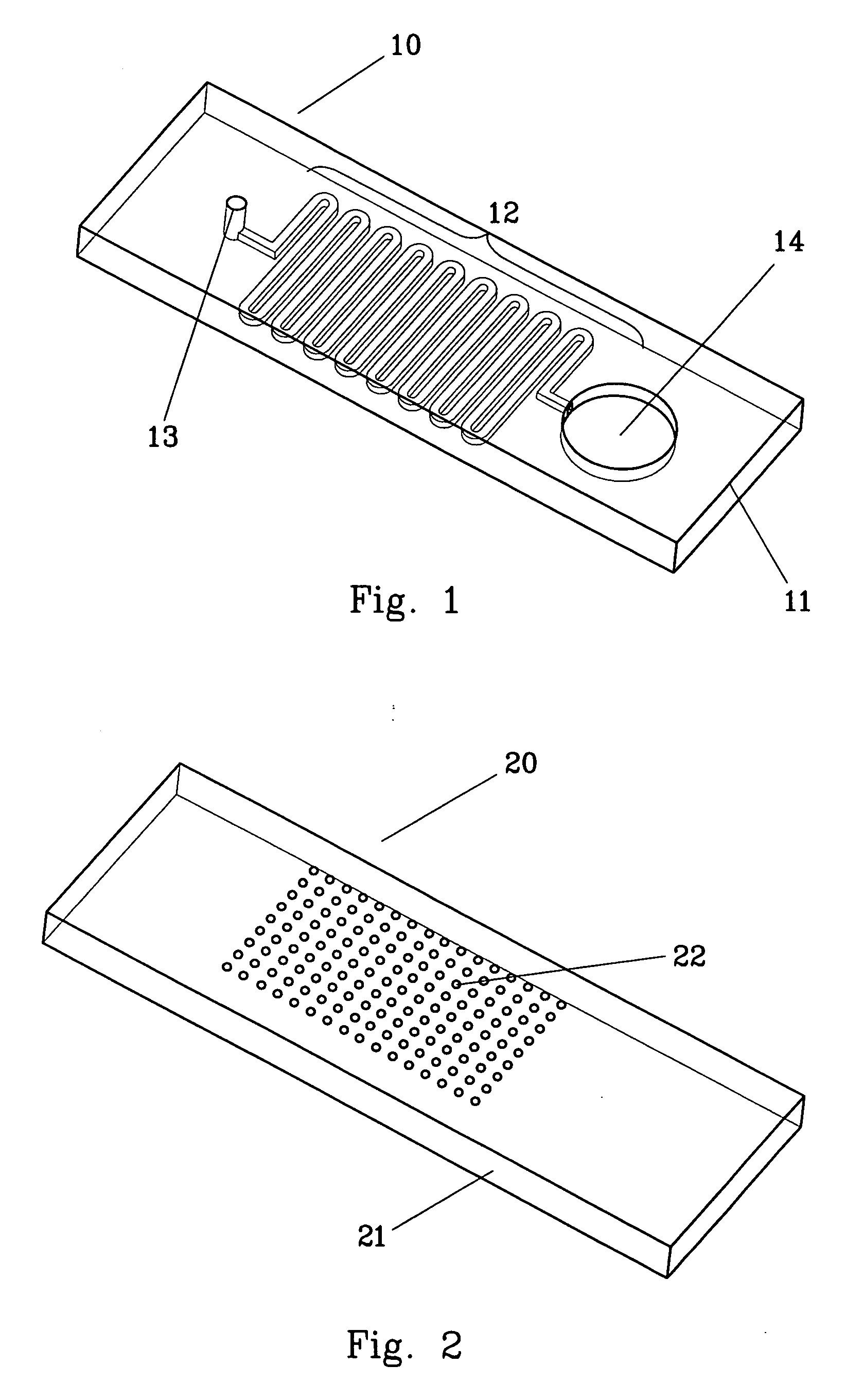
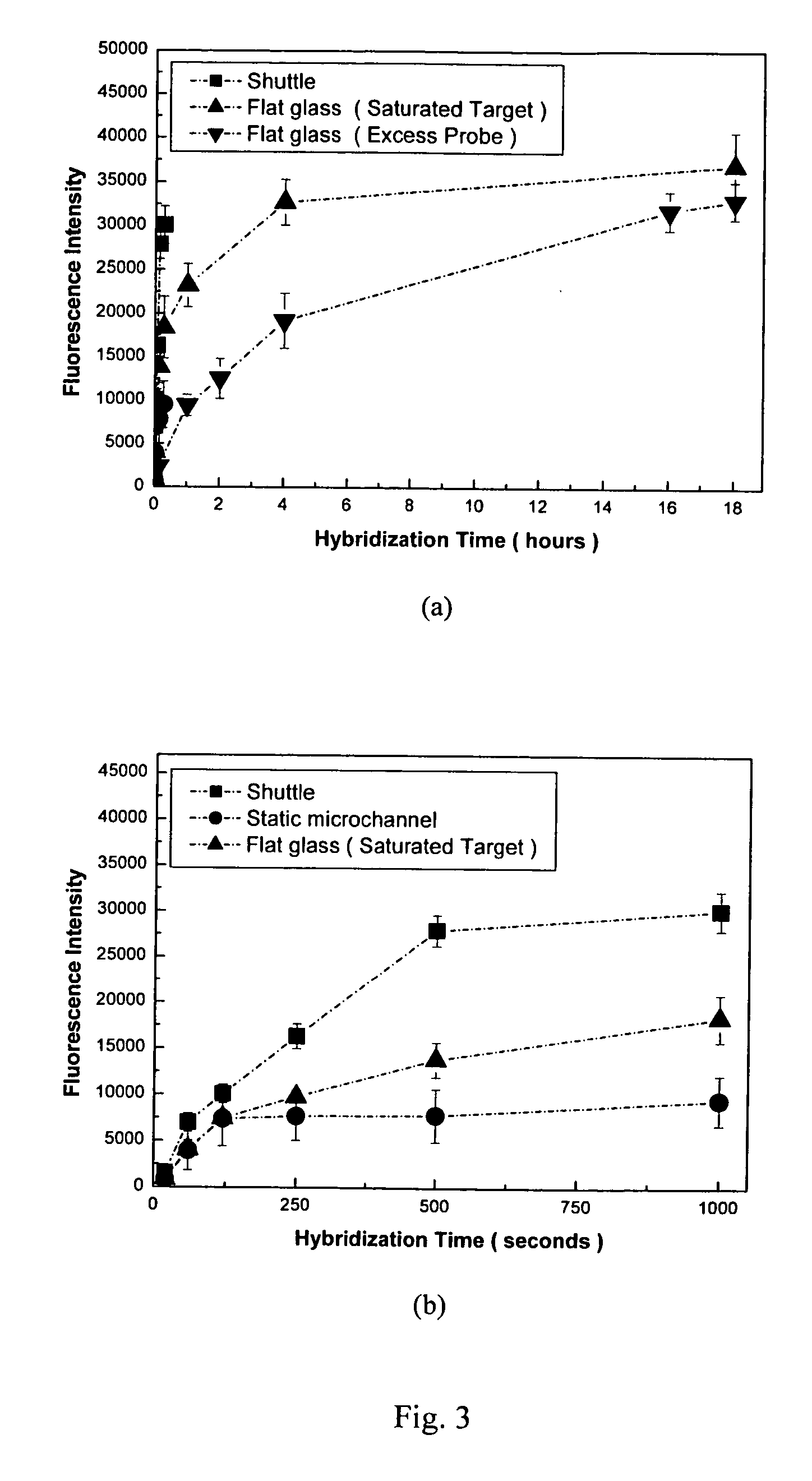
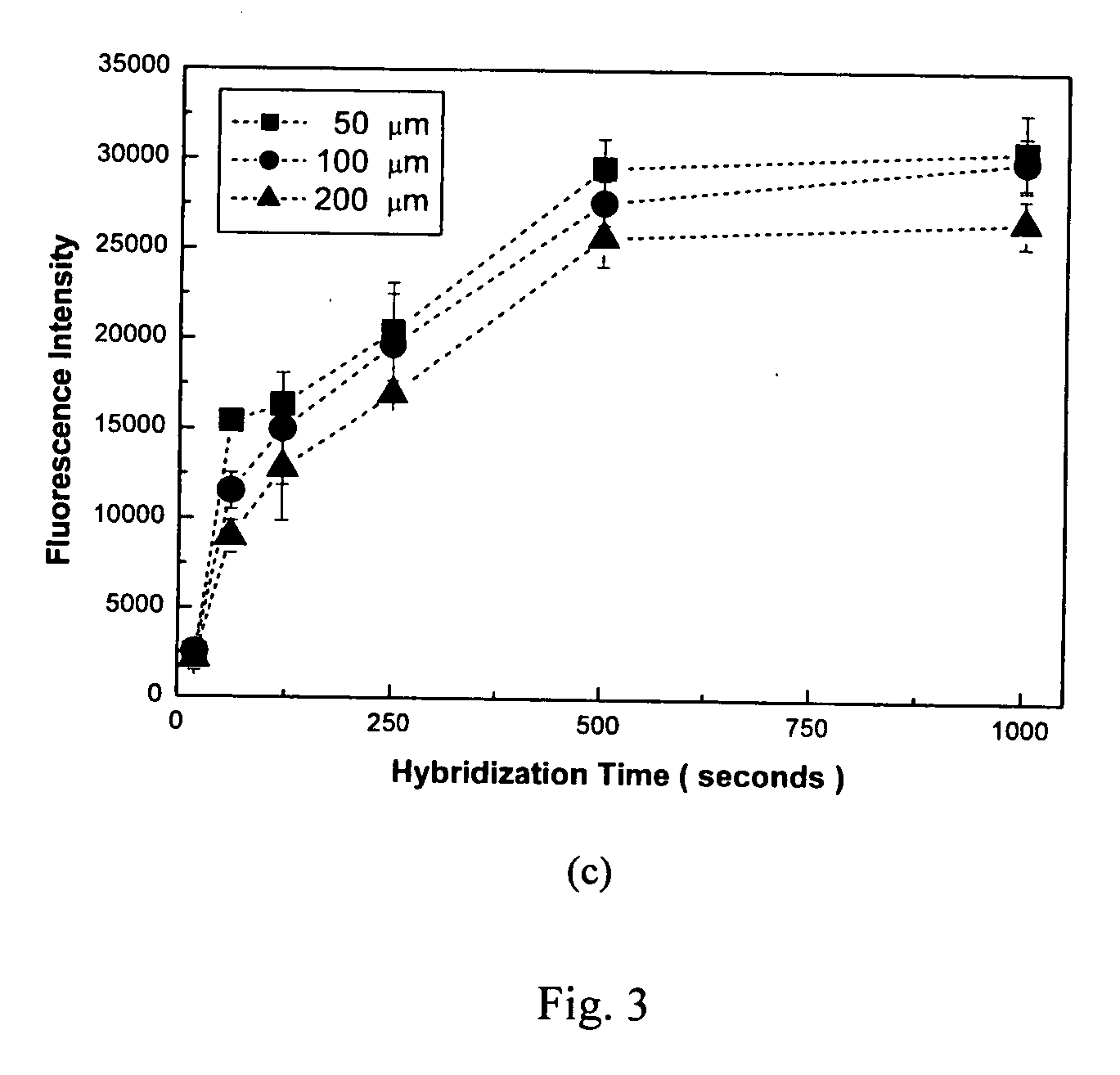
Microarray biochemical reaction device
InactiveUS20070087353A1Delayed reaction timeSample and reagent consumption for DNA hybridization may be reducedBioreactor/fermenter combinationsBiological substance pretreatmentsChaotic mixingDNA
The microarray biochemical reaction device of this invention integrates microfluidic trenches with a microarray to form a serpentine microchannel passing through all DNA probes provided in the microarray. A sample solution is introduced into the microchannel and scrambled into discrete plugs to induce droplet mixing. The discrete plugs are then shuttled through the entire microchannel (shuttle hybridization), sweeping over DNA probes to perform hybridization. Using chaotic mixing of droplets, the hybridization efficiency is enhanced and reaction time for hybridization is shortened. During shuttling, the plugs are thoroughly mixed by the natural re-circulating flows. Method for the preparation of the microarray biochemical reaction device is also disclosed.
Owner:ACAD SINIC
Oligonucleotide probes and uses thereof
InactiveUS20100086916A1Enhances probe hybridizationUseful in detectionSugar derivativesMicrobiological testing/measurementNucleic acid detectionHigh sensitive
The present invention provides a dually labeled oligonucleotide probe, methods of preparing and using the same. The subject probes are particularly useful for high-sensitive nucleic acid detection via hybridization assays including but not limited to template-directed polymerization reactions.
Owner:ALLELOGIC BIOSCIENCES CORP
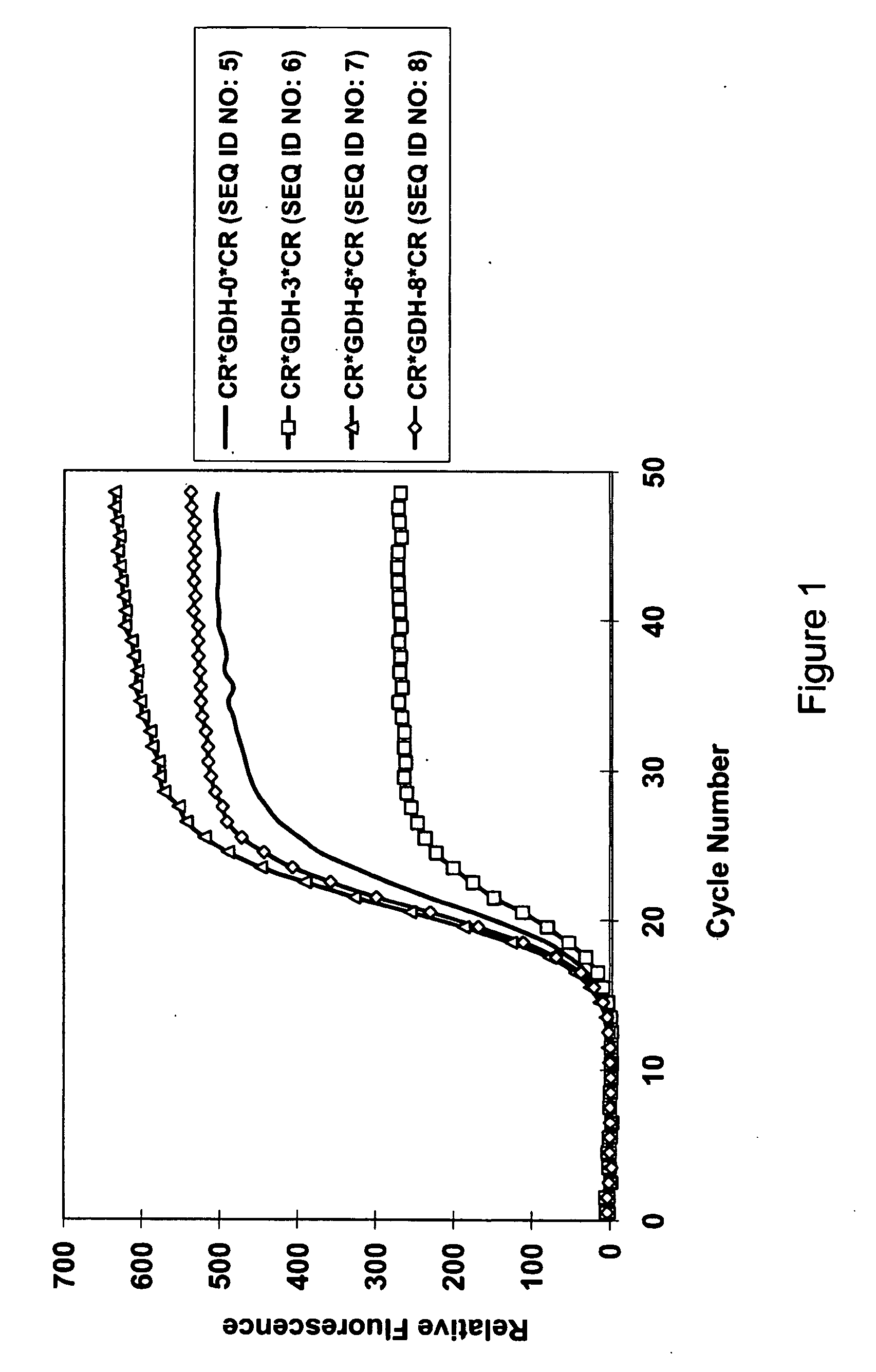
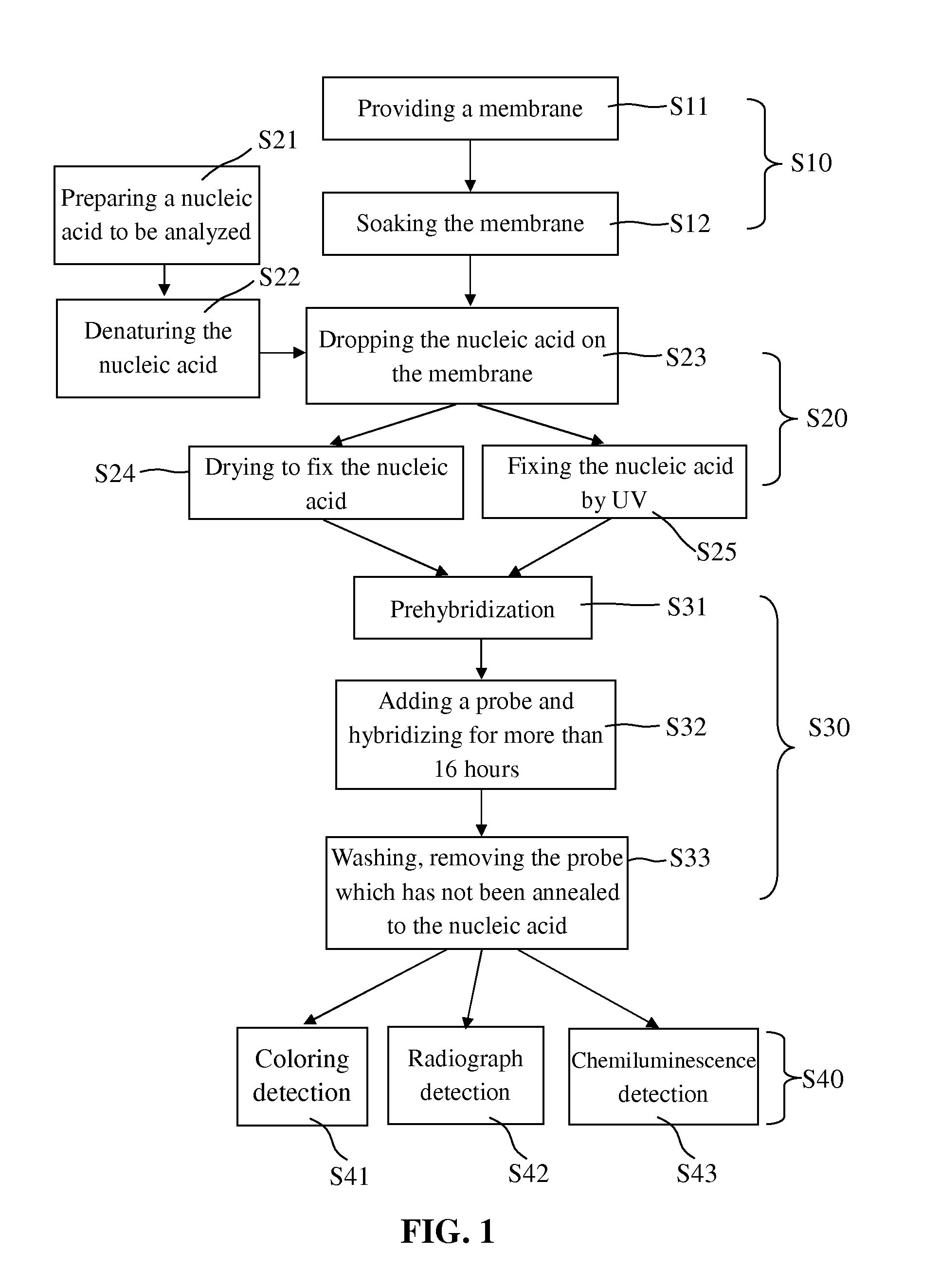
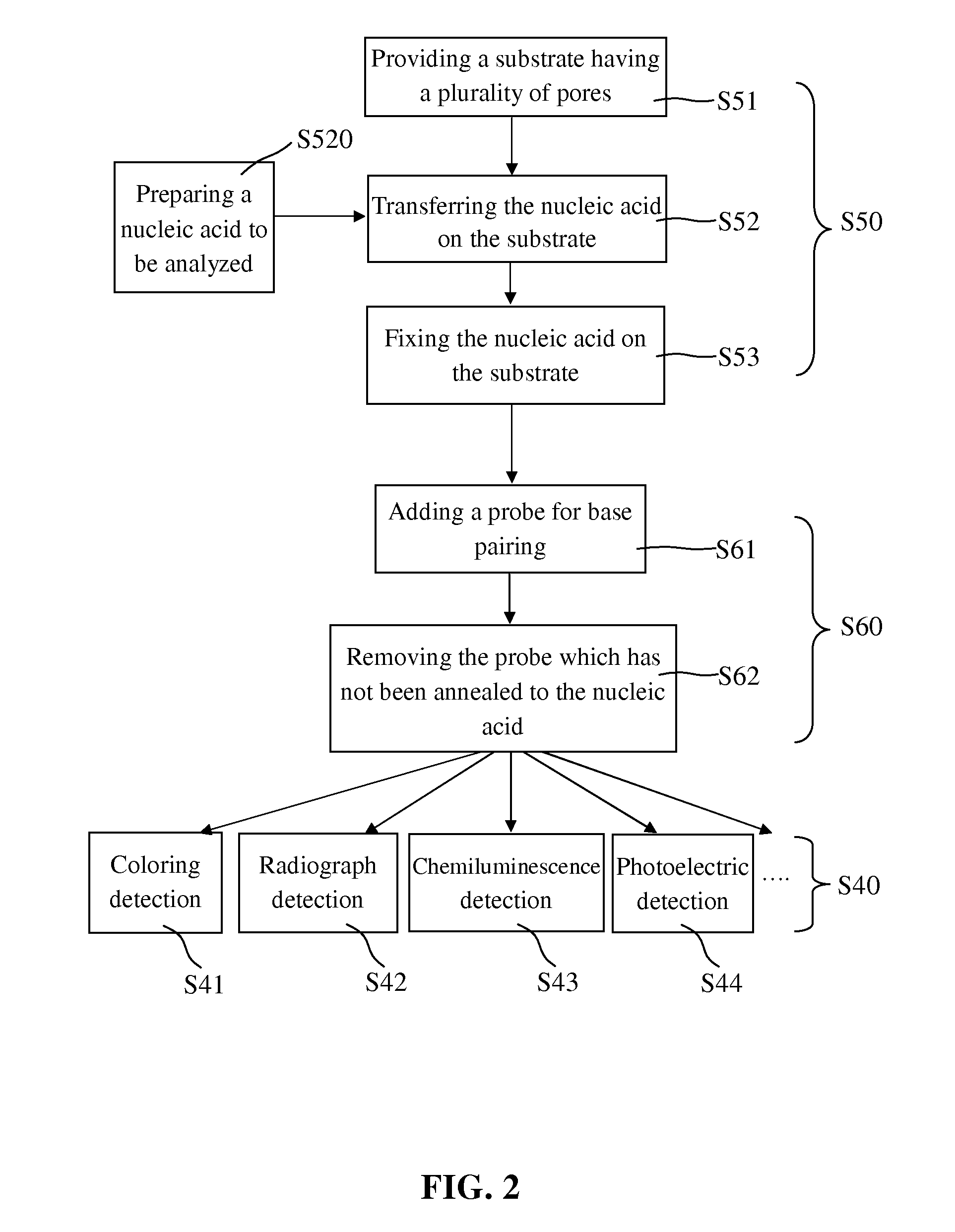
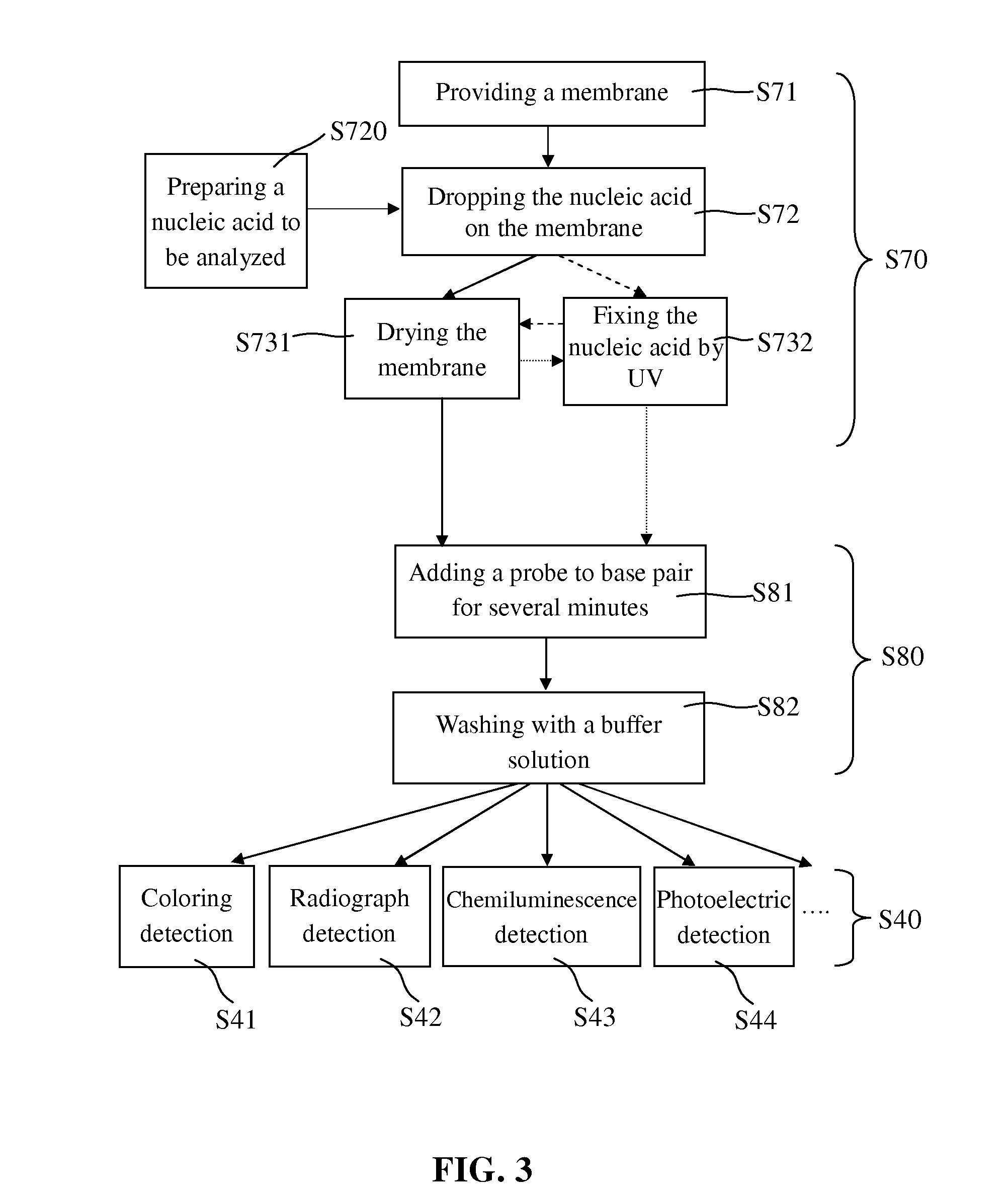
Blotting Method for Rapidly Analyzing Nucleic Acid
ActiveUS20080076125A1Simple procedureShorten the timeBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic Acid ProbesNucleic acid hybridisation
The present invention relates to a blotting method for rapidly analyzing nucleic acid comprising the steps of transferring a nucleic acid to be analyzed to the substrate and fixing the nucleic acid to be analyzed absorbed on the substrate; directing adding a nucleic acid probe to hybridize in a short time, without blocking the areas where the nucleic acid to be analyzed has not been fixed; removing the nucleic acid probe which has not been annealed to the nucleic acid to be analyzed by washing; and finally detecting the hybridization signal. According to the present invention, since the prehybridization is not needed and the hybridization and washing time is shortened, the time for the nucleic acid hybridization is dramatically shortened. Therefore, the whole blotting procedures for rapidly analyzing nucleic acid may be finished quickly.
Owner:CHANG CHUNG CHENG +2
Oligonucleotide probes and uses thereof
InactiveUS8354523B2Enhanced hybridizationSugar derivativesMicrobiological testing/measurementNucleic acid detectionHigh sensitive
The present invention provides a dually labeled oligonucleotide probe, methods of preparing and using the same. The subject probes are particularly useful for high-sensitive nucleic acid detection via hybridization assays including but not limited to template-directed polymerization reactions.
Owner:ALLELOGIC BIOSCIENCES CORP
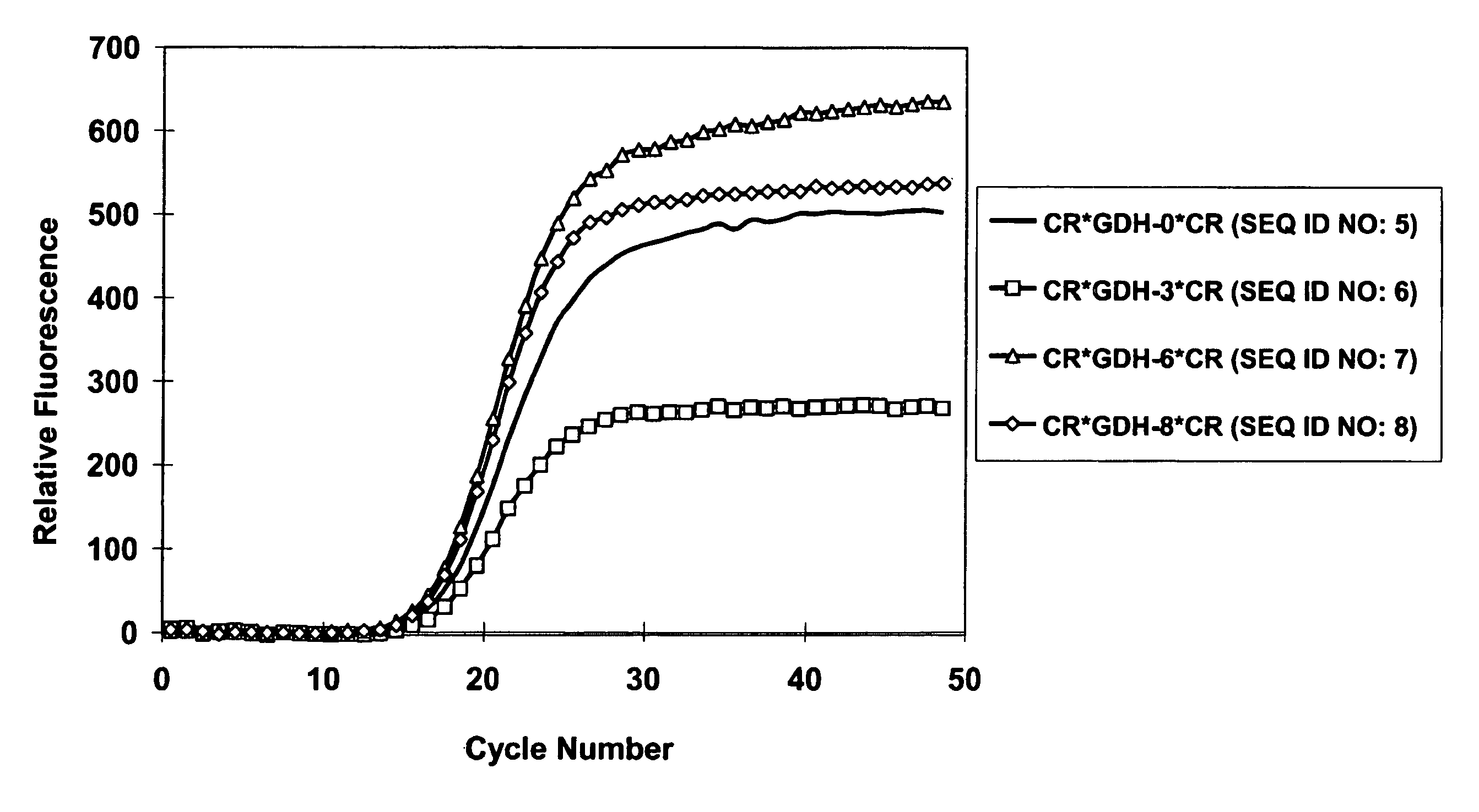
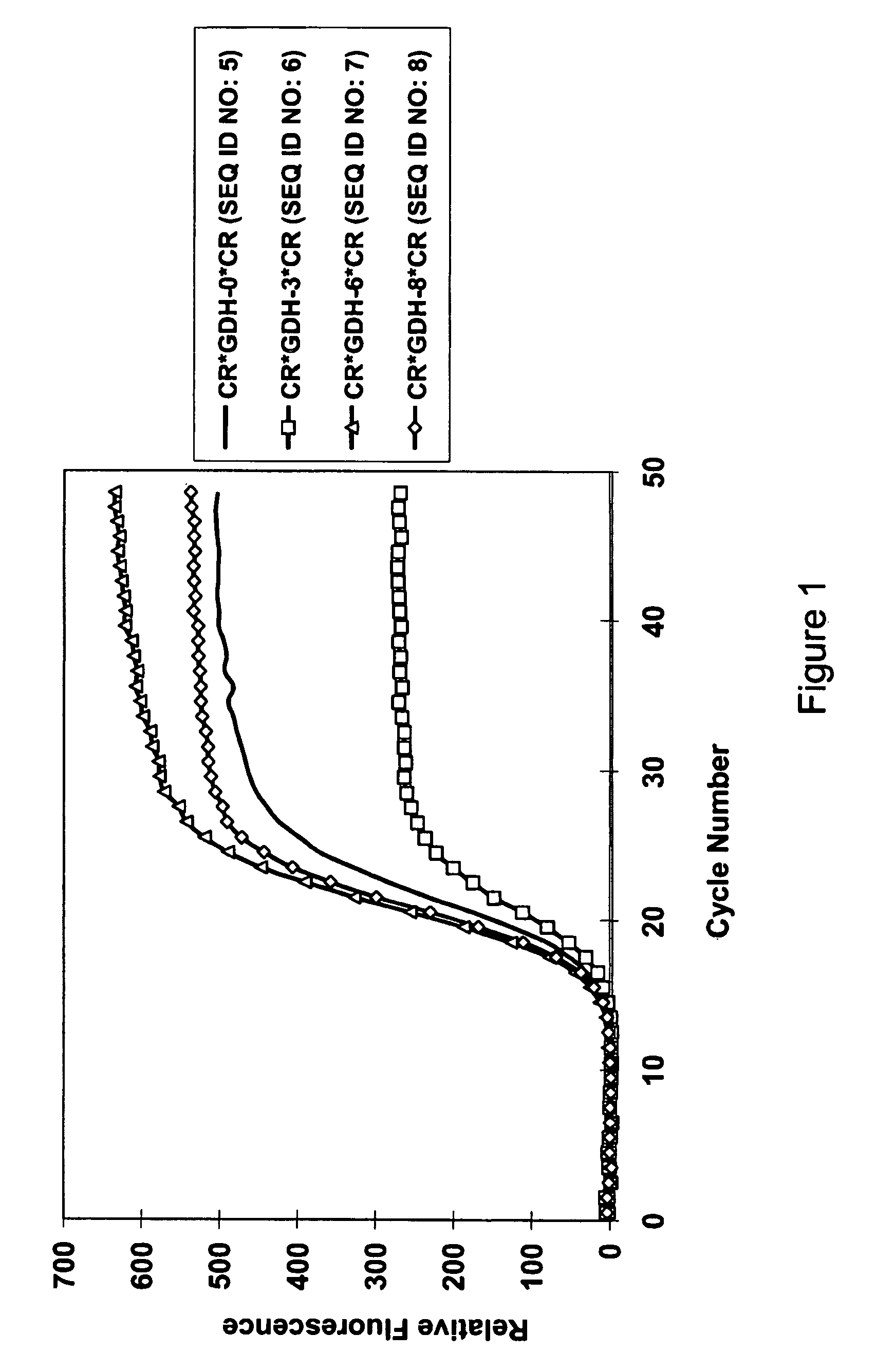
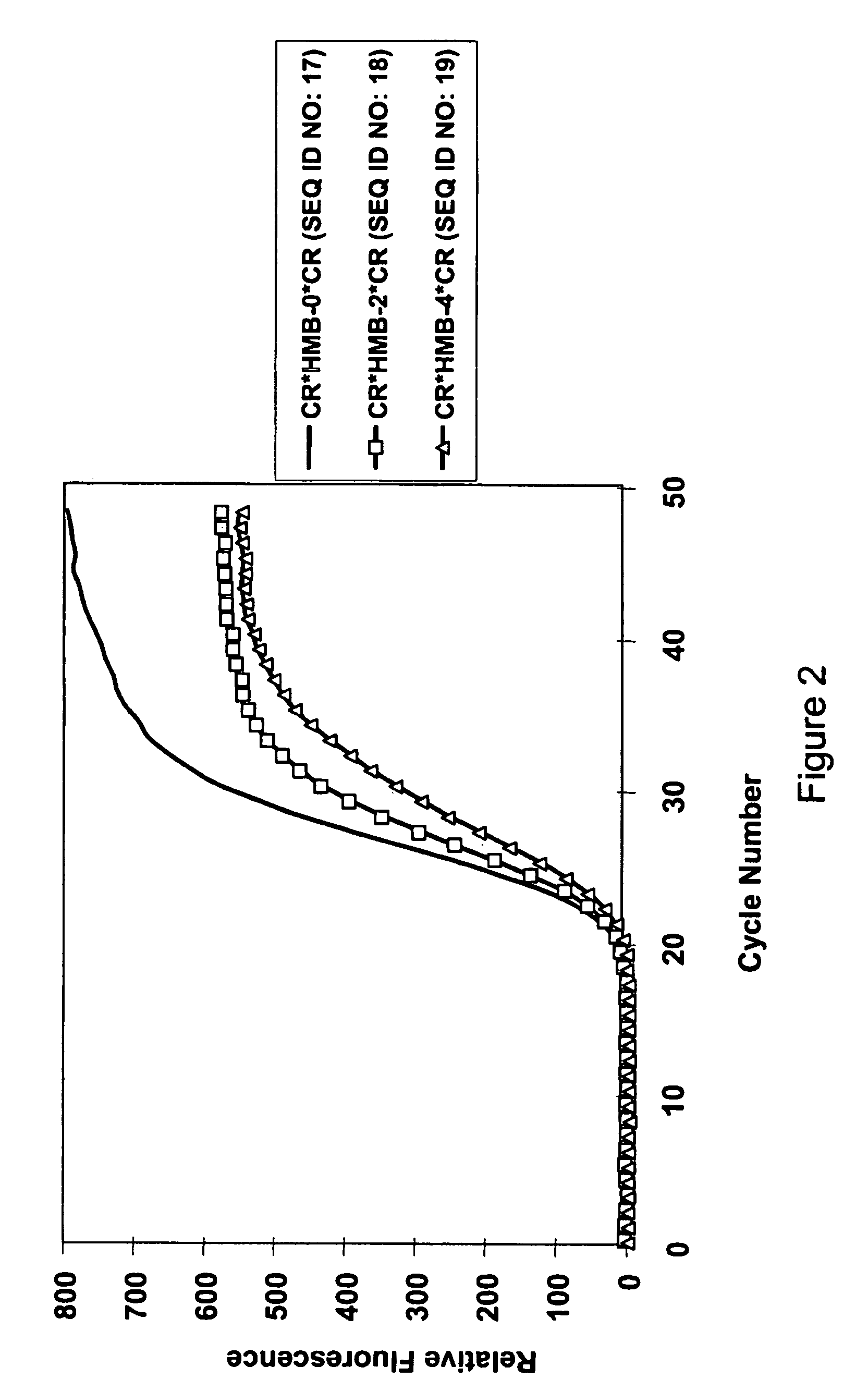
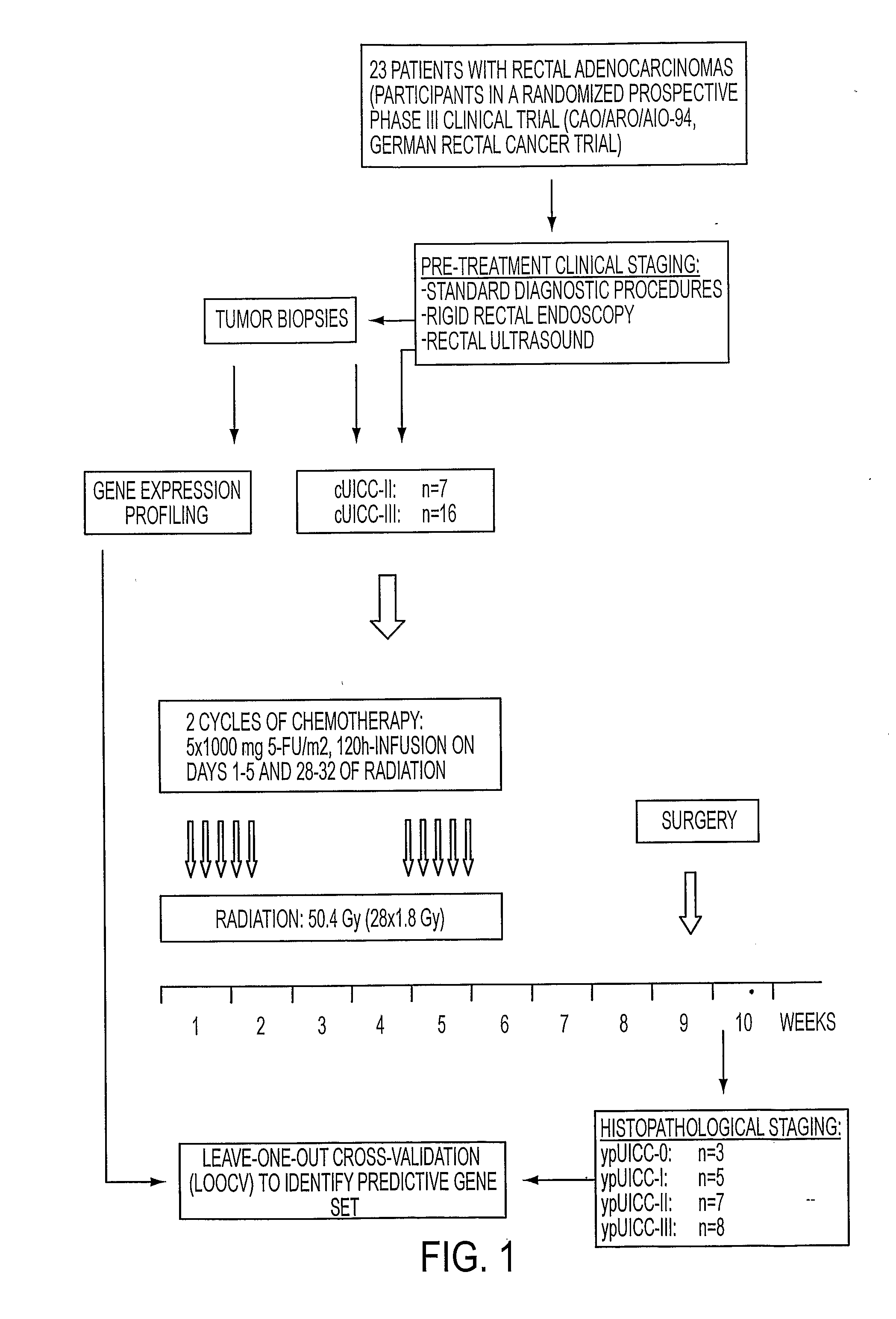
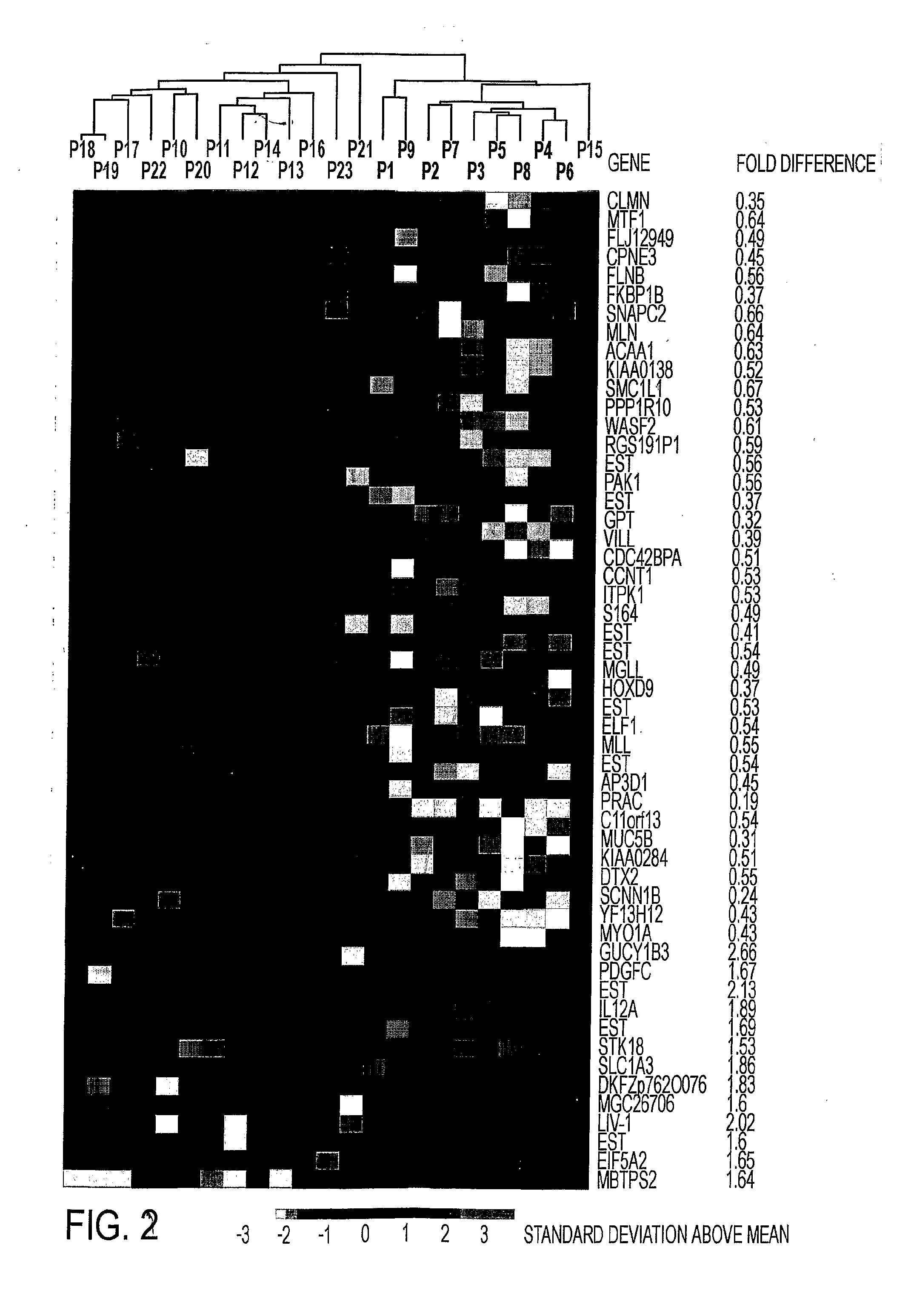
Composition for Detecting the Response of Rectal Adenocarcinomas to Radiochemotherapy
ActiveUS20080020373A1Enhanced hybridizationReduced structureSugar derivativesMicrobiological testing/measurementNon respondersGene
A cDNA array (9984 genes) was used for expression profiling in rectal adenocarcinoma. The expression data were correlated to responsiveness to chemotherapy followed by radiotherapy. A set of 54 genes was found that were differentially expressed in responders vs. non-responders. The genes may be used as prognostic markers for determining whether a rectal adenocarcinoma is responsive to radiochemotherapy.
Owner:UNITED STATES OF AMERICA
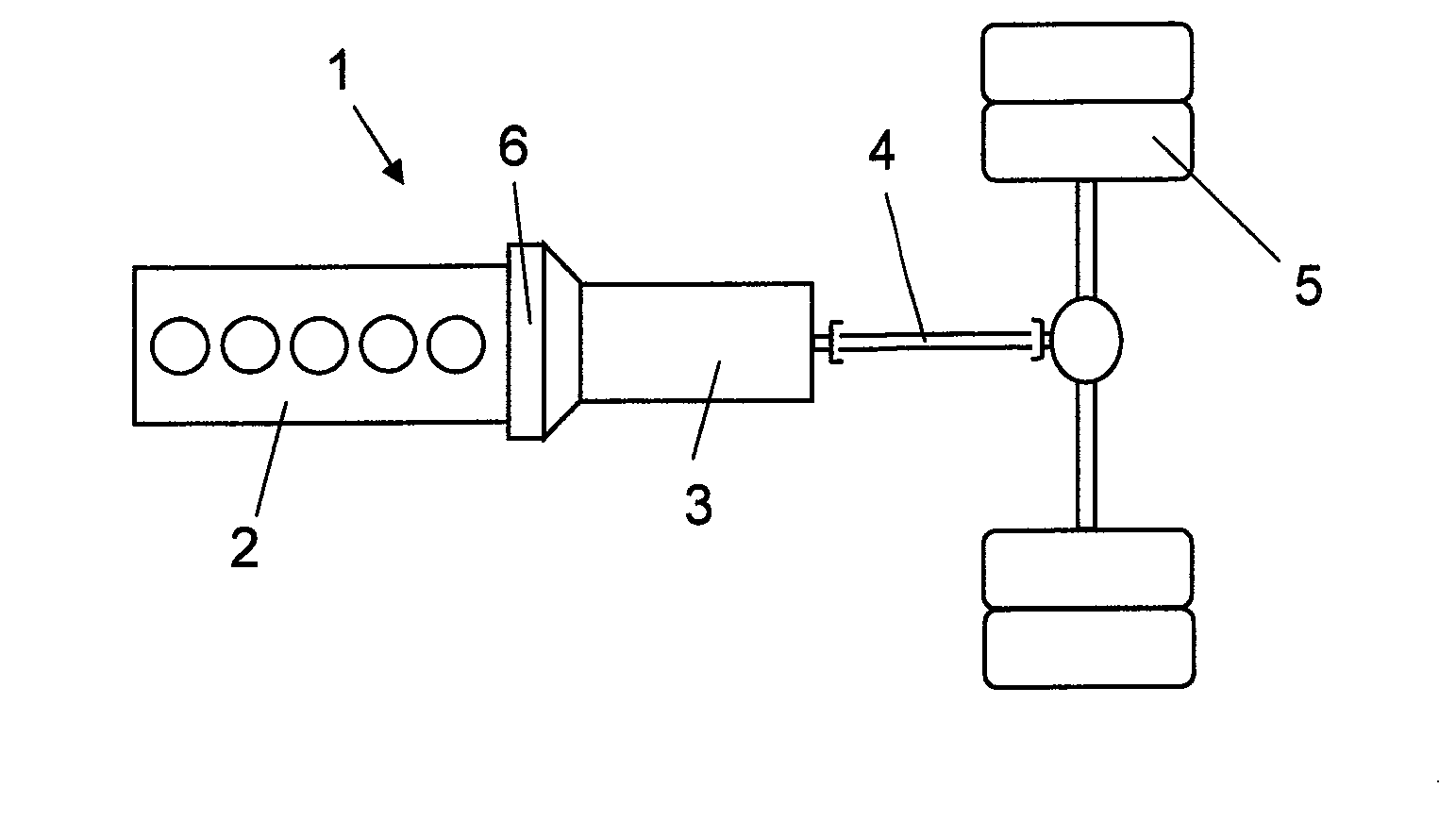
Method for accelerating a hybrid vehicle
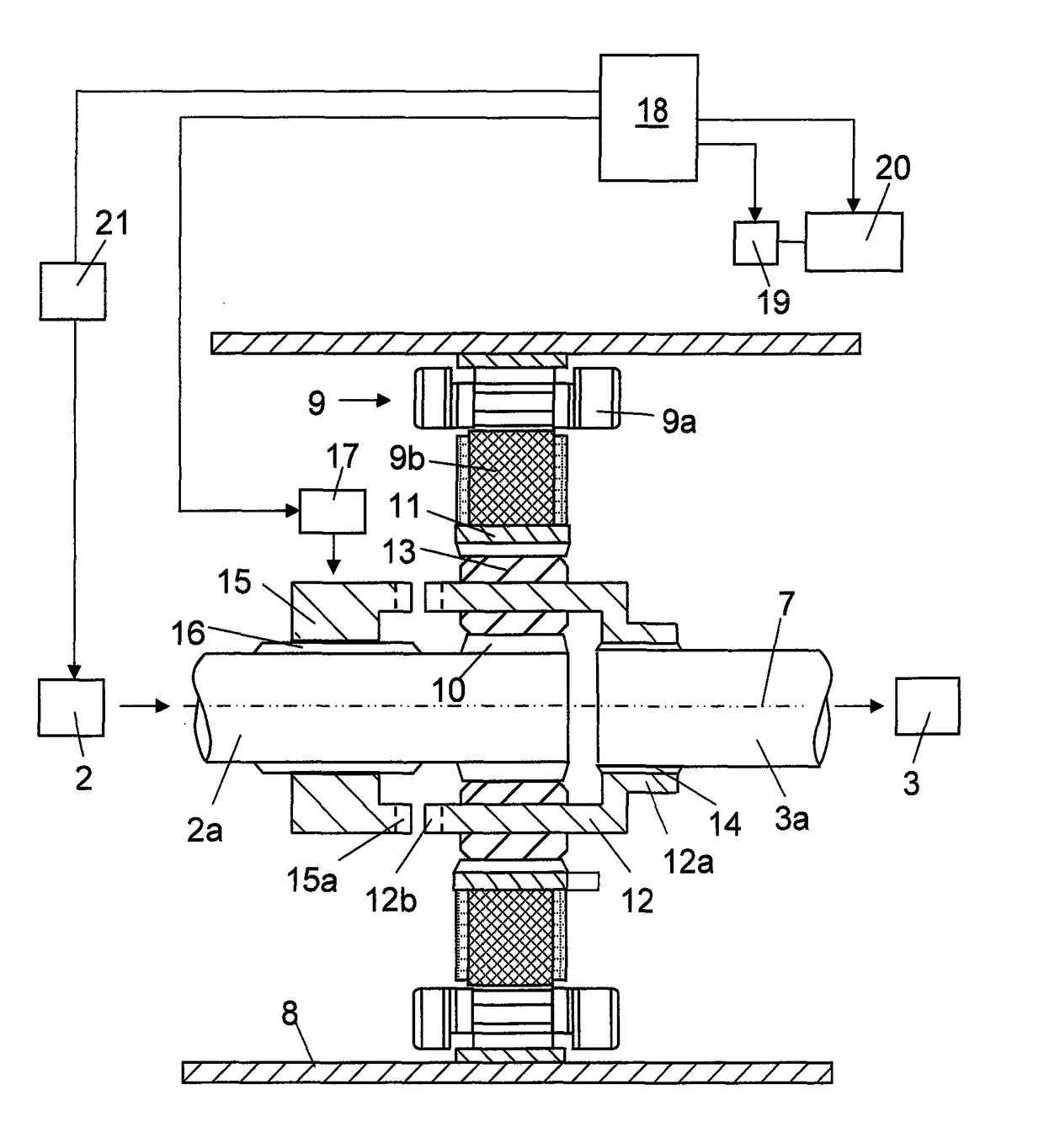
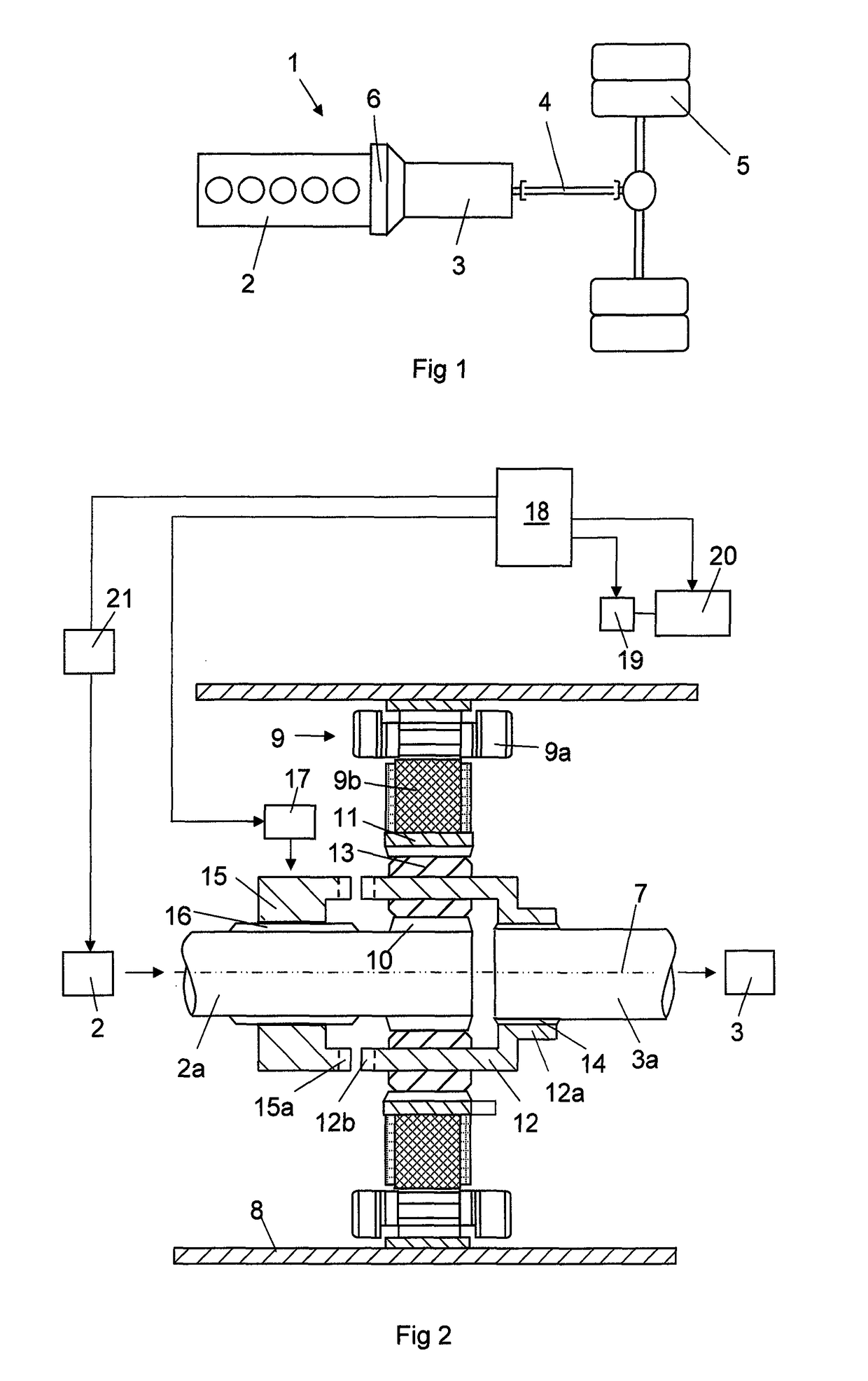
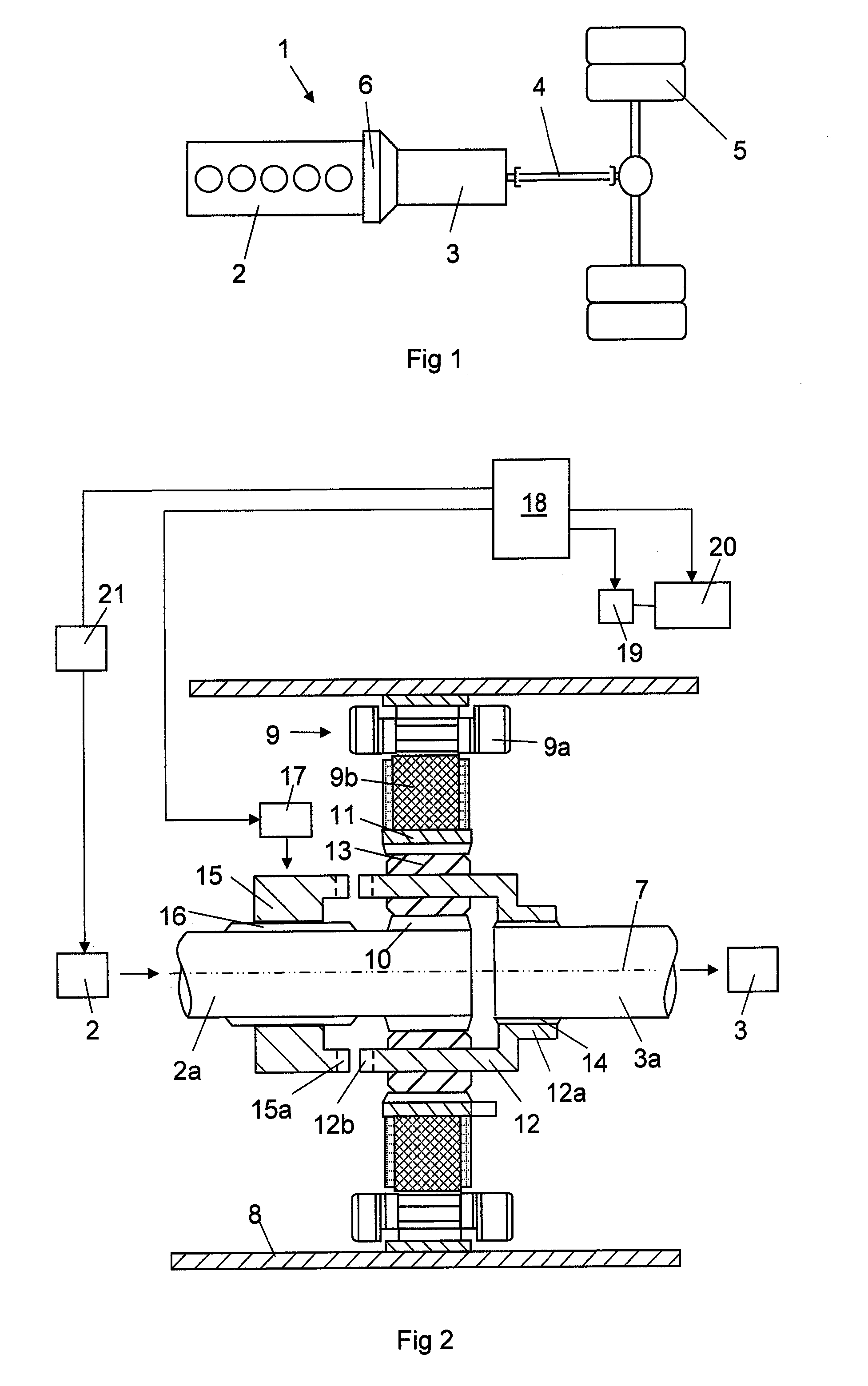
ActiveUS10046756B2Enhanced hybridizationHigher possible brake torqueGas pressure propulsion mountingVehicle sub-unit featuresCombustionElectric machine
A method for accelerating a vehicle driving forward, in which the vehicle has a propulsion system including a combustion engine with an output shaft (2a), a gearbox (3) with an input shaft (3a), an electric machine (9) comprising a stator and a rotor, and a planetary gear comprising a sun gear (10), a ring gear (11) and a planet wheel carrier (12). When accelerating the vehicle the torque of the electric machine is controlled and the rotational speed of the combustion engine is controlled until the components of the planetary gear have the same rotational speed and may be interlocked.
Owner:SCANIA CV AB
Compositions, methods, and kits for fabricating coded molecular tags
InactiveUS20100112572A1Minimise measureImprove method performanceMicrobiological testing/measurementMaterial analysisBiotechnologyAnalyte
Owner:LIFE TECH CORP
Method for gearchange in a hybrid vehicle
ActiveUS20150203098A1Shorter torque interruptionHigh moment of inertiaHybrid vehiclesElectric propulsion mountingGear wheelElectric machinery
A method for gearchange in a gearbox of a vehicle driving, said vehicle having a propulsion system comprising a combustion engine with an output shaft (2a), a gearbox (3) with an input shaft (3a), an electric machine (9) comprising a stator and a rotor, and a planetary gear comprising a sun gear (10), a ring gear (11) and a planet wheel carrier (12). The gearchange is carried out with the components of the planetary gear allowed to rotate with different rotational speeds and is finished by interlocking them. The combustion engine is controlled with respect to rotational speed during the gearchange towards the rotational speed required for the interlocking.
Owner:SCANIA CV AB
Methods and compositions for directed microwave chemistry
InactiveUS20120165209A1Enhanced hybridizationDissociation—can be acceleratedImmobilised enzymesMicrobiological testing/measurementNucleic acid chemistryChemical reaction
The present invention concerns a novel means by which chemical preparations can be made. Reactions can be accelerated on special cartridges using microwave energy. The chips contain materials that efficiently absorb microwave energy causing chemical reaction rate increases. The invention is important in many chemical transformations including those used in protein chemistry, in nucleic acid chemistry, in analytical chemistry, and in the polymerase chain reaction.
Owner:MIRARI BIOSCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com