Nanoreporters and methods of manufacturing and use thereof
a reporter and molecule technology, applied in the field of compositions and methods for detection and quantification of individual target molecules, can solve the problems of insufficient supply of biological samples, method still requires significant amounts of biological samples, and the kinetics of hybridization on the surface of a microarray are less efficient than hybridization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
6. EXAMPLE 1
Nanoreporter Manufacturing and Protocol
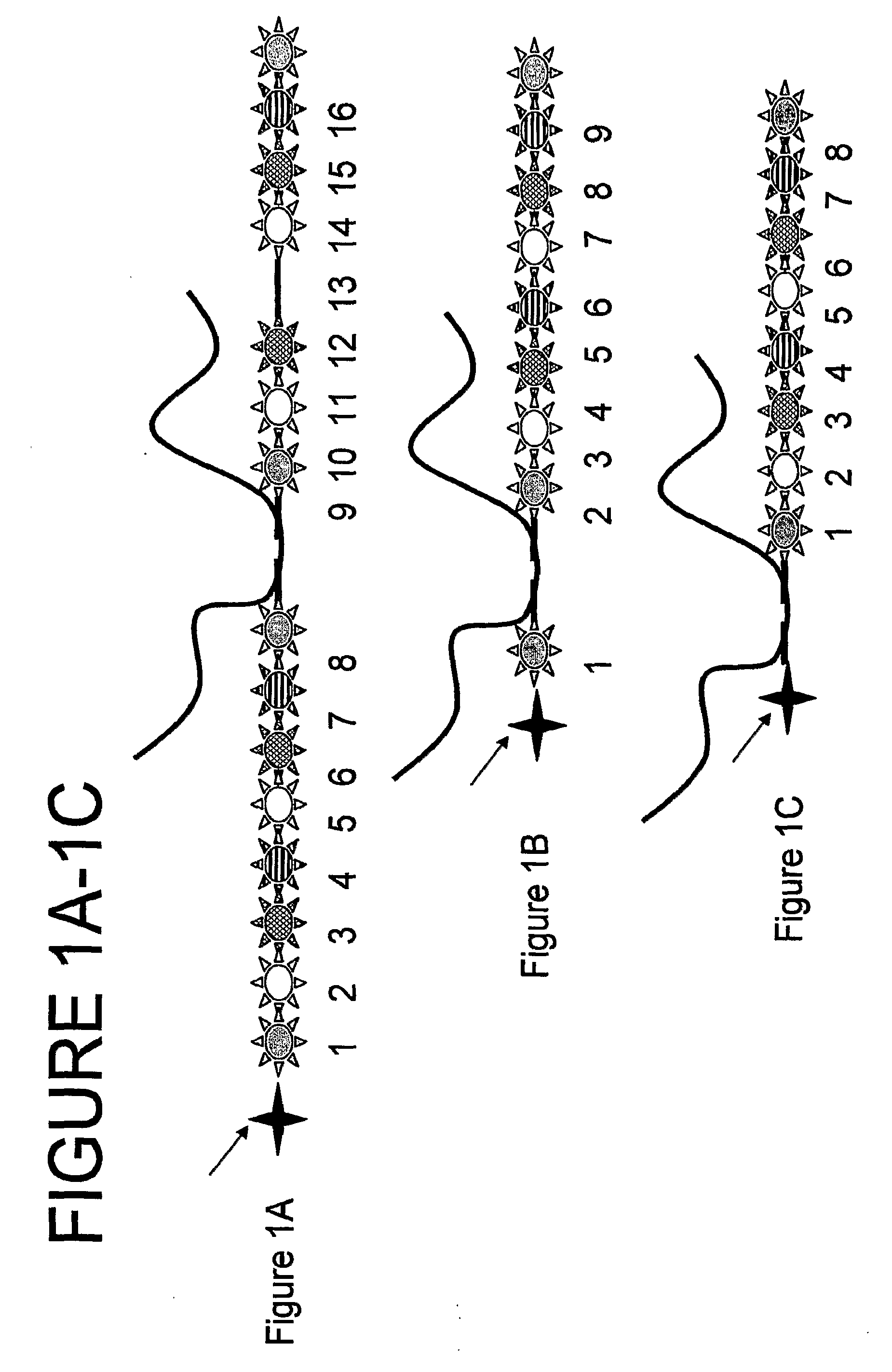
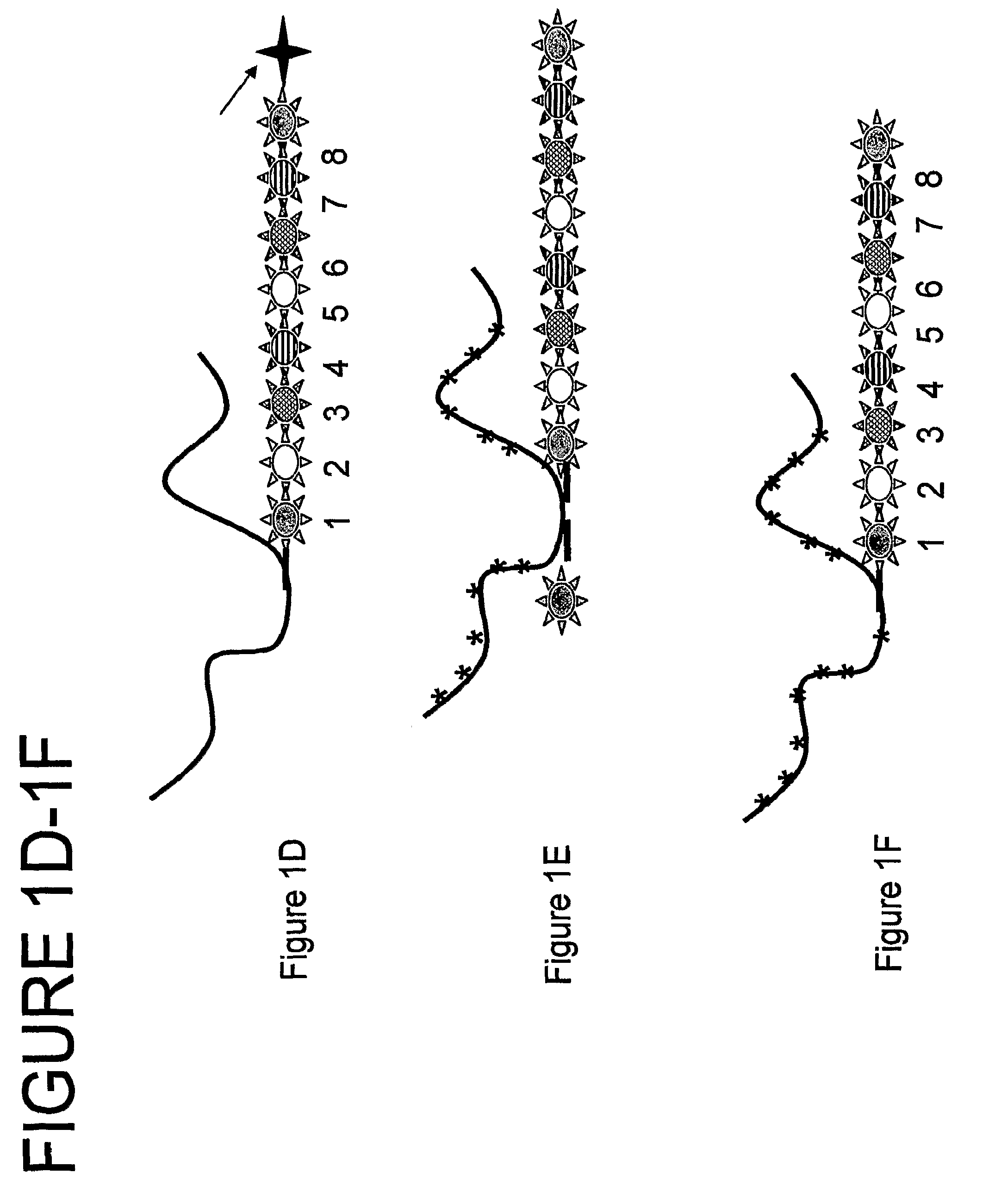
[0395]Herein is a step-by-step example of a method construction of a nanoreporter from various components.
[0396]It can be appreciated that various components can be constructed or added either at the same time, before or after other components. For example, annealing patch units or flaps to a scaffold can be done simultaneously or one after the other.
[0397]In this example the starting material is a circular M13mp18 viral vector. Using a single linear strand M13mp18, patch units are annealed to it to form a double stranded scaffold. Next, flaps are added then a target-specific sequence is ligated. Meanwhile purification steps aid to filter out excess, unattached patch units and flaps. Construction of labeled nucleic acids (patches and / or flaps and / or other labeled oligonucleotides) that bind the nanoreporter are also described.
[0398]Upon attachment (e.g., via hybridization) of a target molecule, the nanoreporter is attached to a surf...
example 2
7. EXAMPLE 2
Patch / Flap Nanoreporter Manufacturing Protocol
[0474]This example demonstrates another way of making a nanoreporter which consists of a single stranded linear M13mp18 viral DNA, oligonucleotide patch units and long flaps.
[0475]Nanoreporter label units were successfully generated using methods substantially as described in this example.
[0476]Pre-phosphorylated patch units and flaps are added together with the M13mp18 DNA vector and ligated together. After the ligation of the flaps to the patch units which are ligated to the M13mp18 DNA, the BamH1 enzyme is introduced to linearize the vector.
[0477]Prepare a batch of nanoreporters starting with 5 μg of M13mp18 as a scaffold. The hybridization may be scaled up accordingly to the desired amount. This process will take about 1-2 days to complete.
[0478]Materials:
QtyItemVendor20250 μg / μl M13mp18 viral ssDNANew EnglandBiolabs27μl0.74 pmol / μl Oligonucleotide PatchIDTUnit Mix8μlLong Flap Oligonucleotide A 100 pmol / μlIDT8μlLong Flap ...
example 3
8. EXAMPLE 3
Protocol for Production of RNA Nanoreporters
[0498]Nanoreporters were generated and successfully employed to detect target molecules using methods substantially as described in this example. An example of target detection using such this method is shown in FIG. 6.
[0499]8.1 Scaffold Production
[0500]Single-stranded circular M13mp18 DNA (USB Corporation) is annealed to a 10-fold molar excess of an oligonucleotide complementary to the Bam HI recognition site (Barn Cutter oligo) and cut with Bam HI restriction enzyme to yield a linear single-stranded DNA backbone. An oligonucleotide complementary to the Barn Cutter oligonucleotide (anti-Bam oligonucleotide) is subsequently added in 50-fold excess to the Barn Cutter oligonucleotide to sequester free Barn Cutter oligonucleotide and thus prevent recircularization of the M13 during later steps.
[0501]The linear M13 molecule serves as a scaffold onto which RNA patches, or RNA segments, with incorporated fluorophores can be annealed....
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com