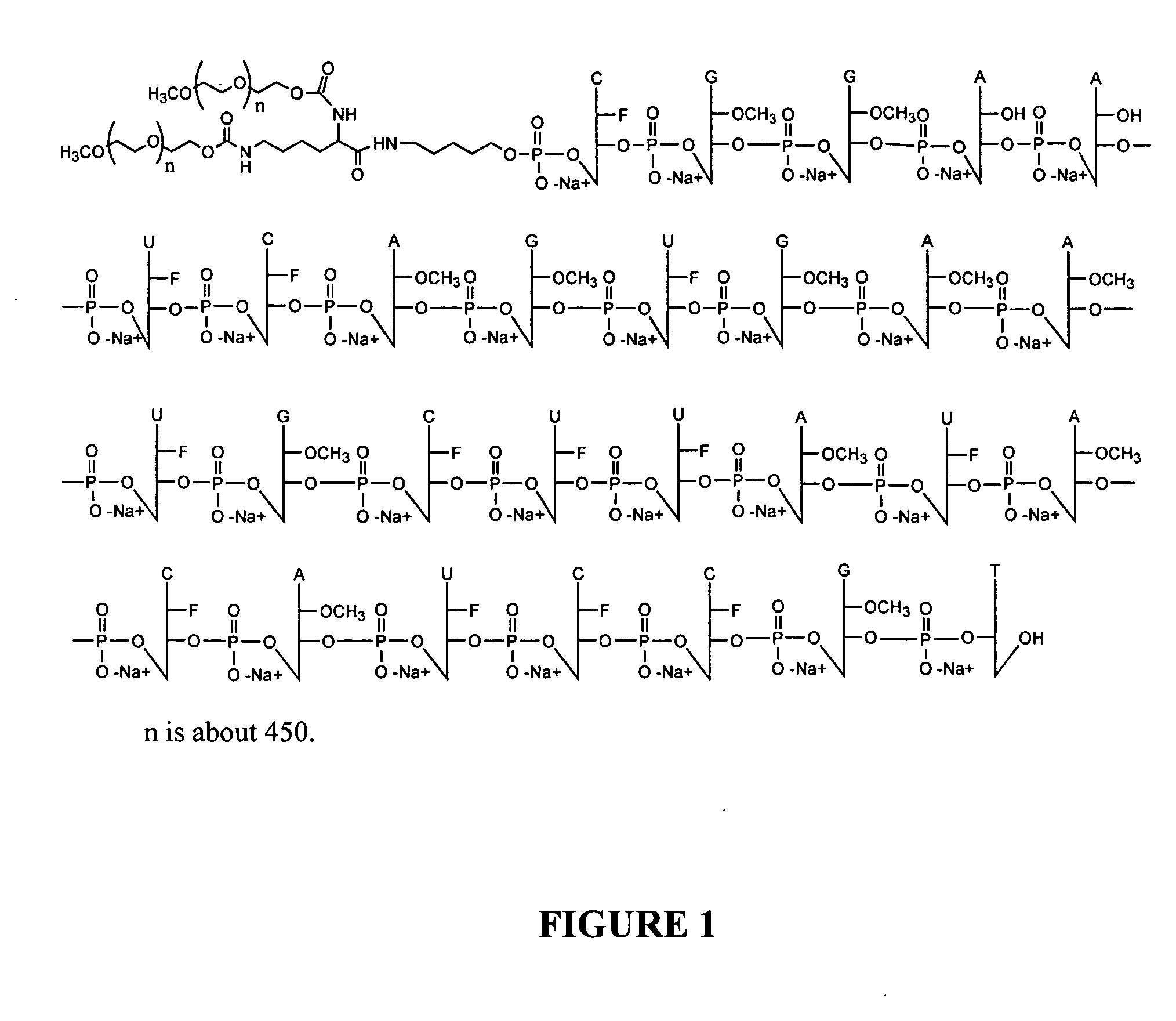
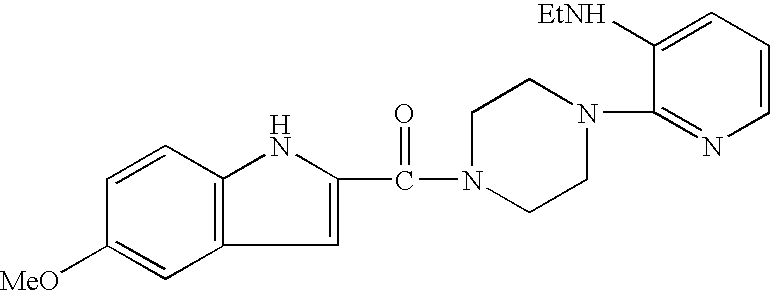
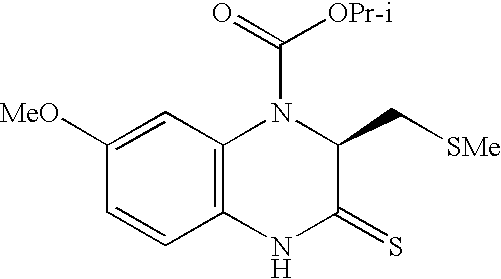
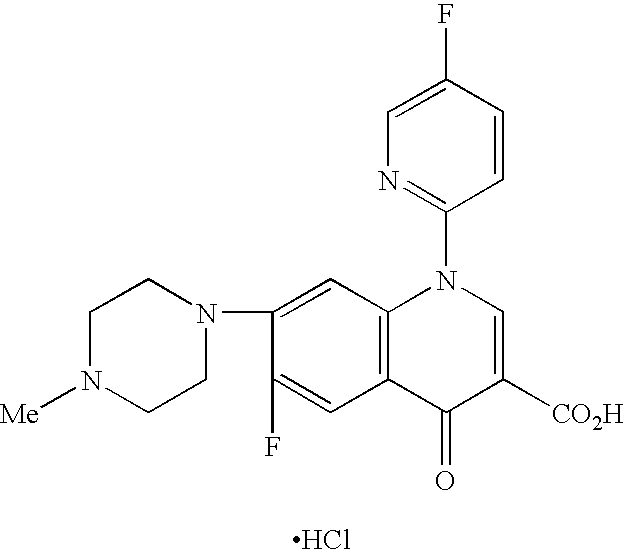
Patents
Literature
2715 results about "Aptamer" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
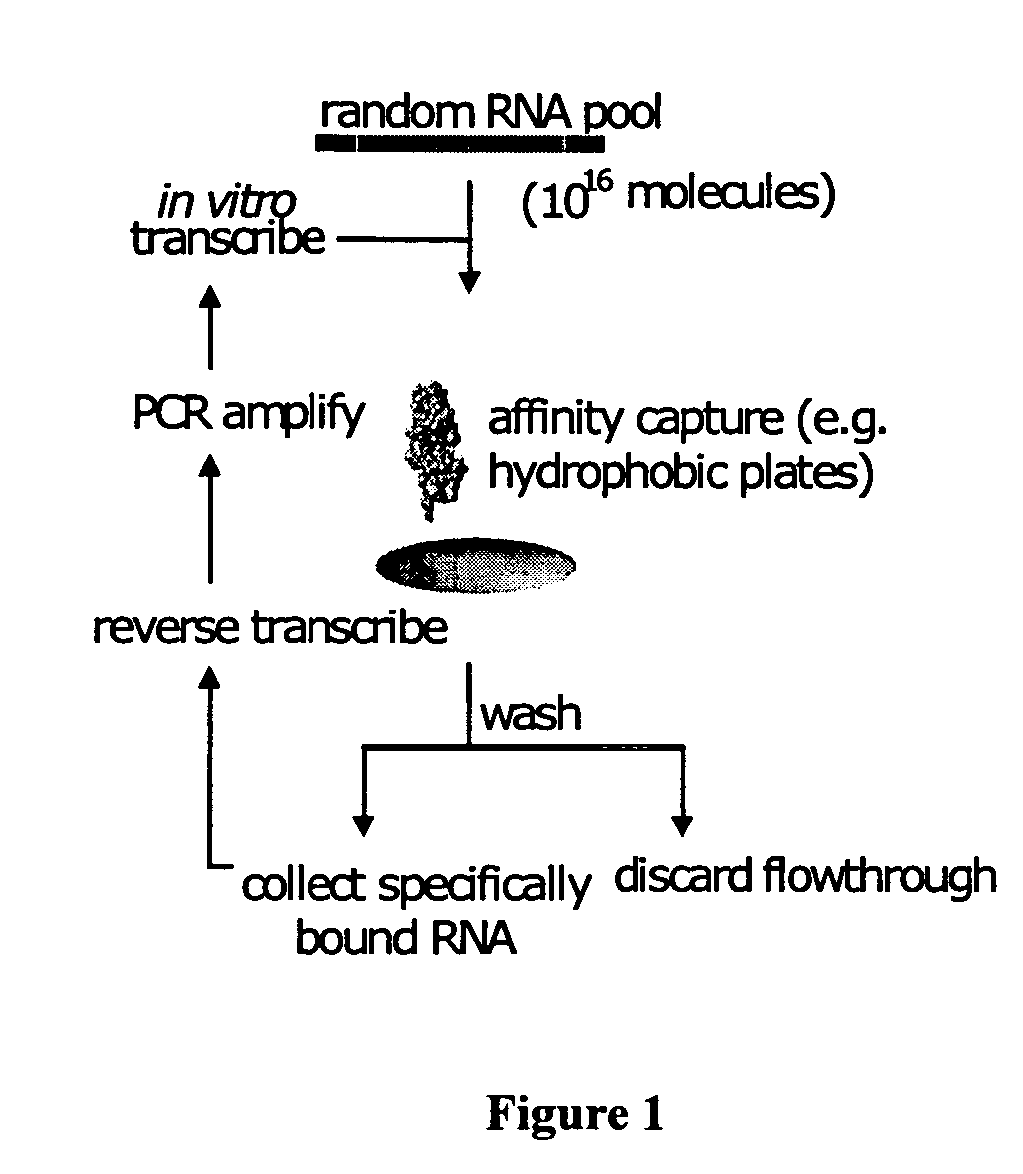
Aptamers (from the Latin aptus – fit, and Greek meros – part) are oligonucleotide or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist in riboswitches. Aptamers can be used for both basic research and clinical purposes as macromolecular drugs. Aptamers can be combined with ribozymes to self-cleave in the presence of their target molecule. These compound molecules have additional research, industrial and clinical applications.
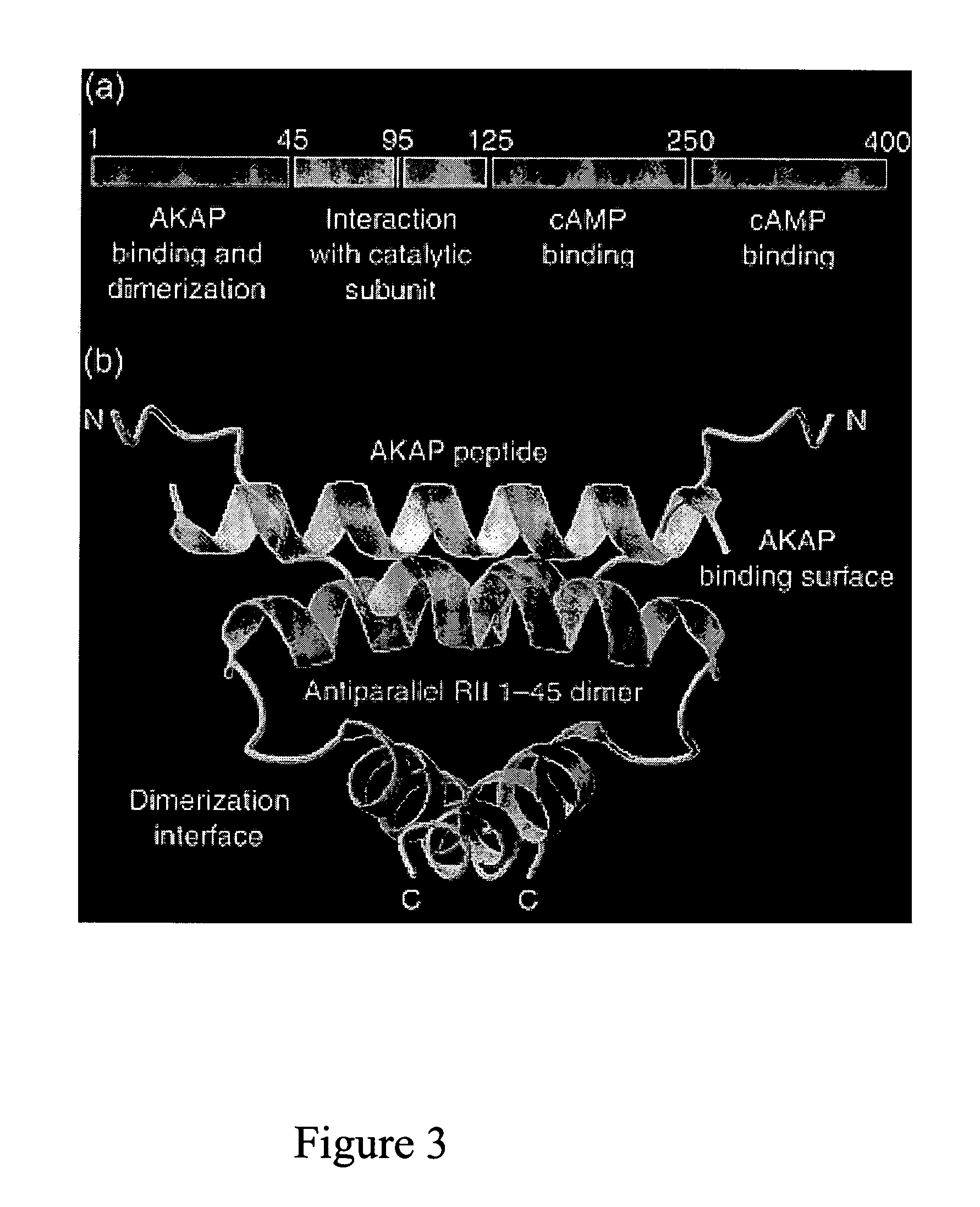
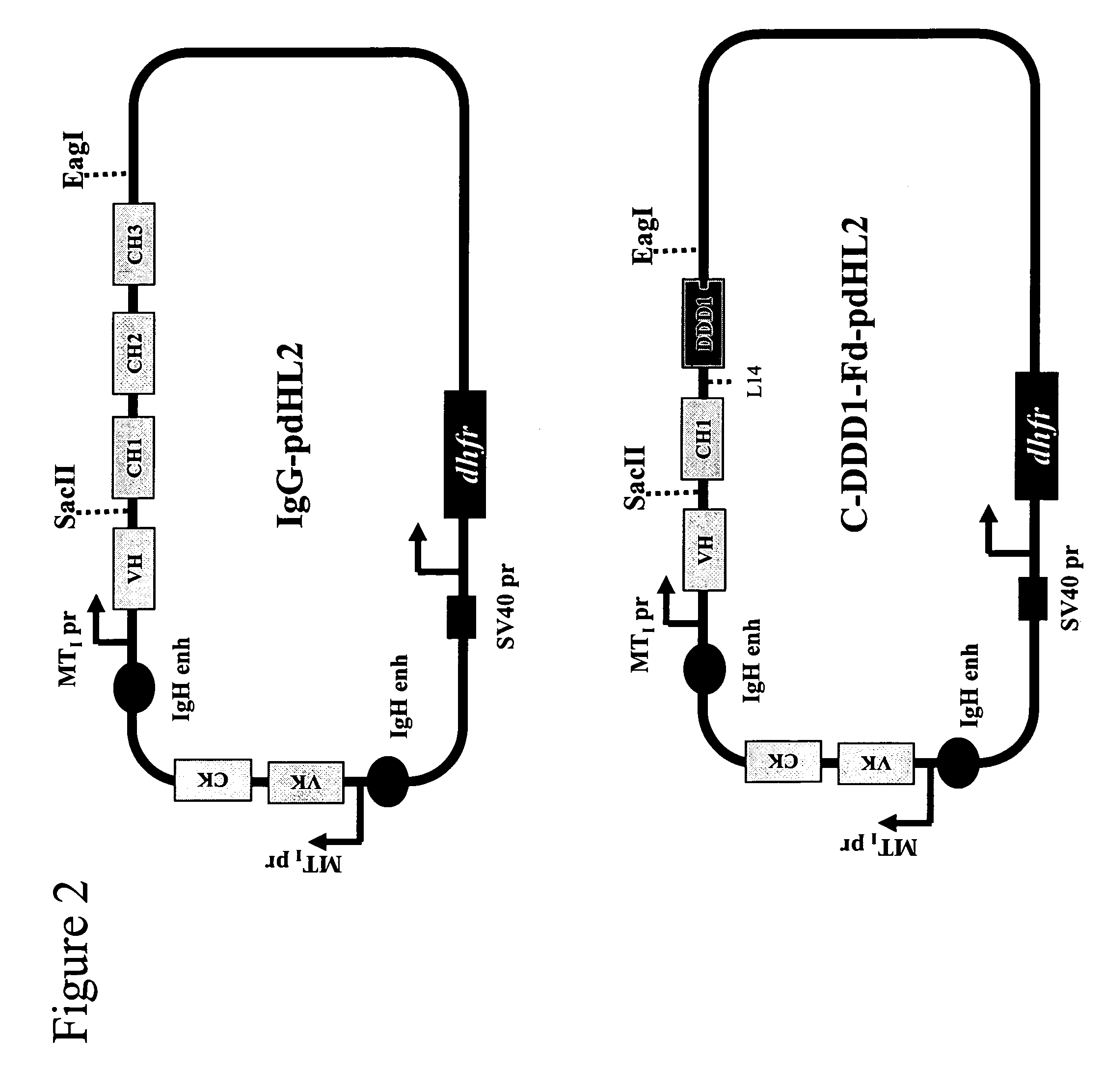
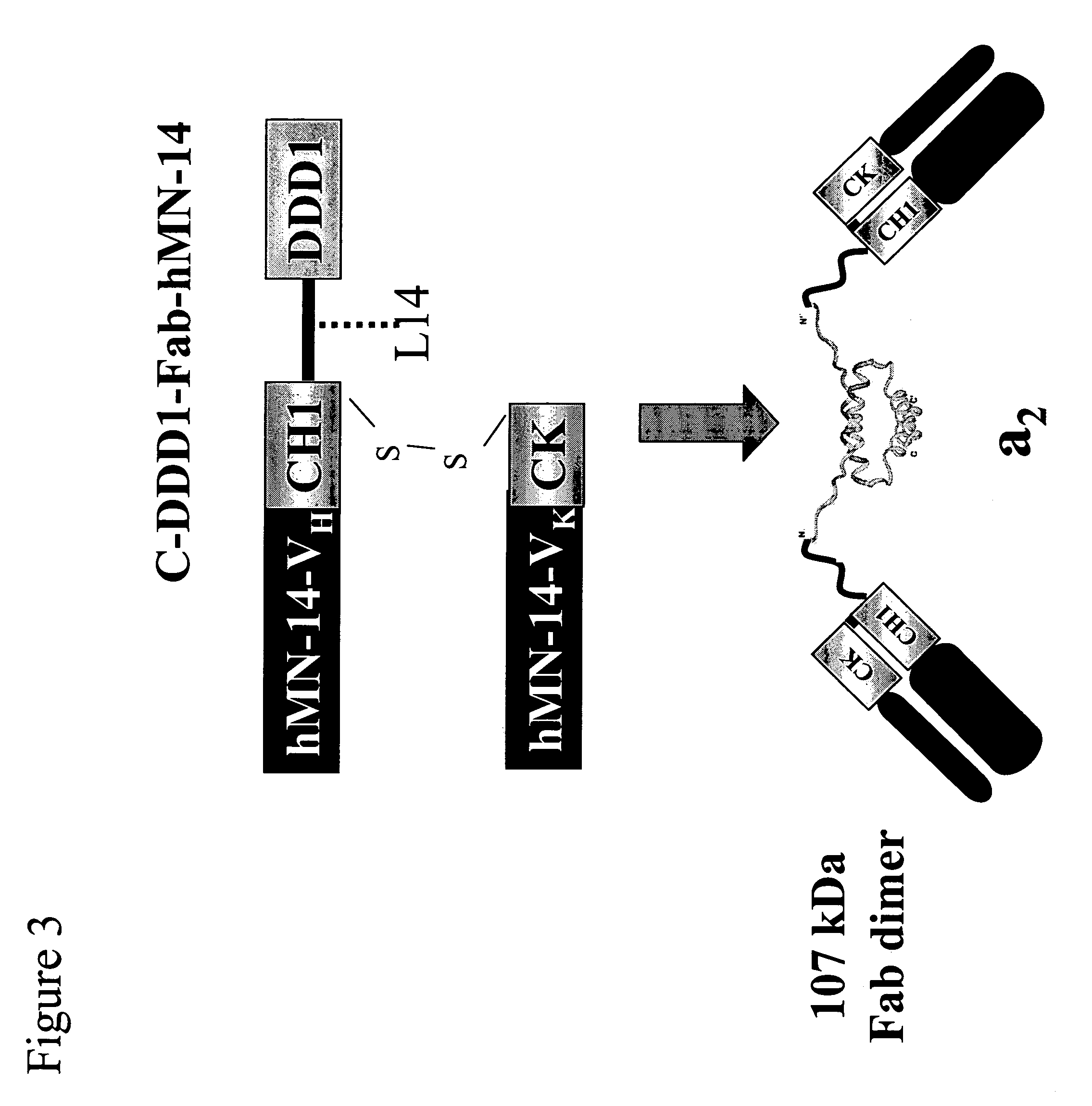
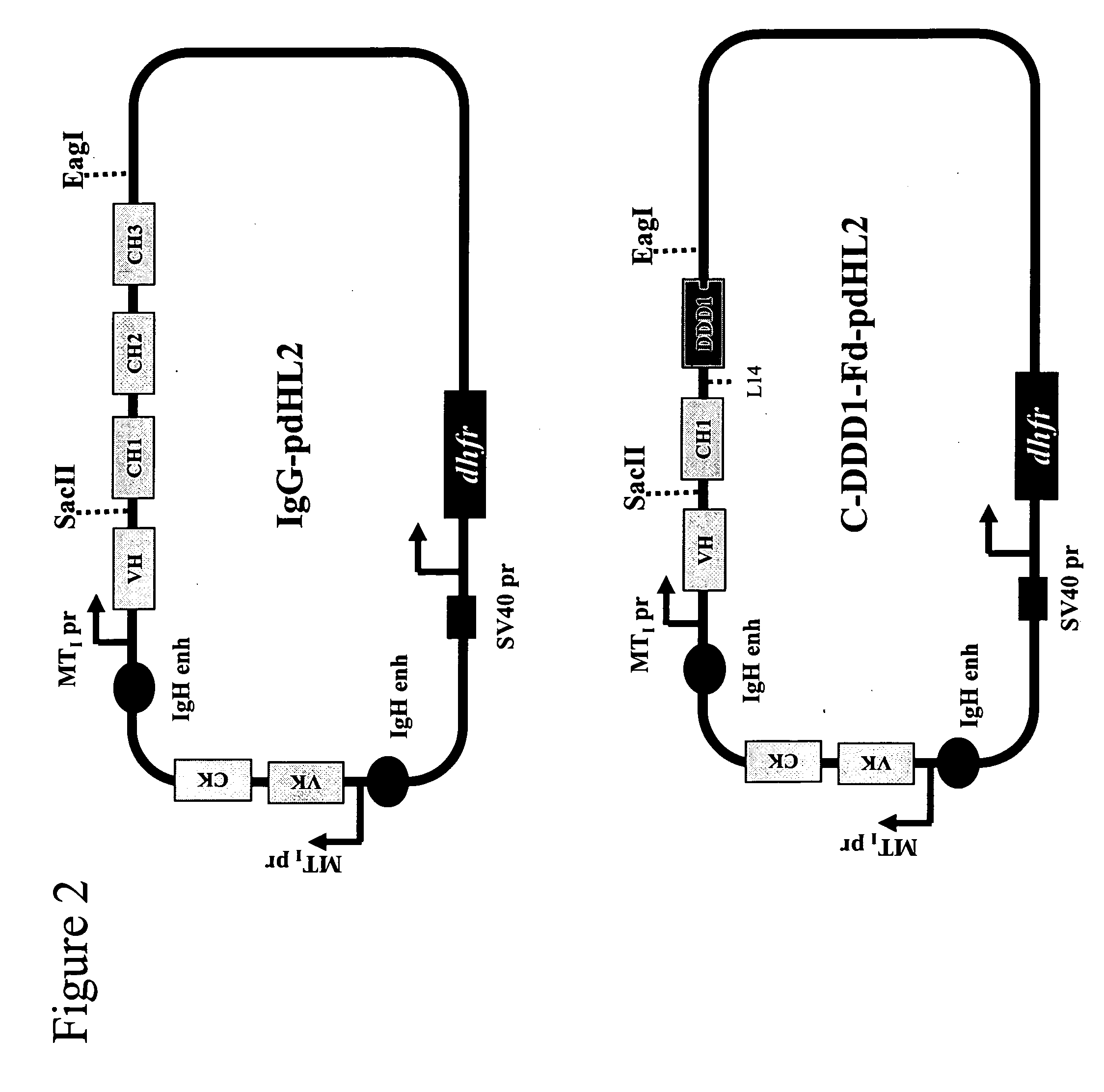
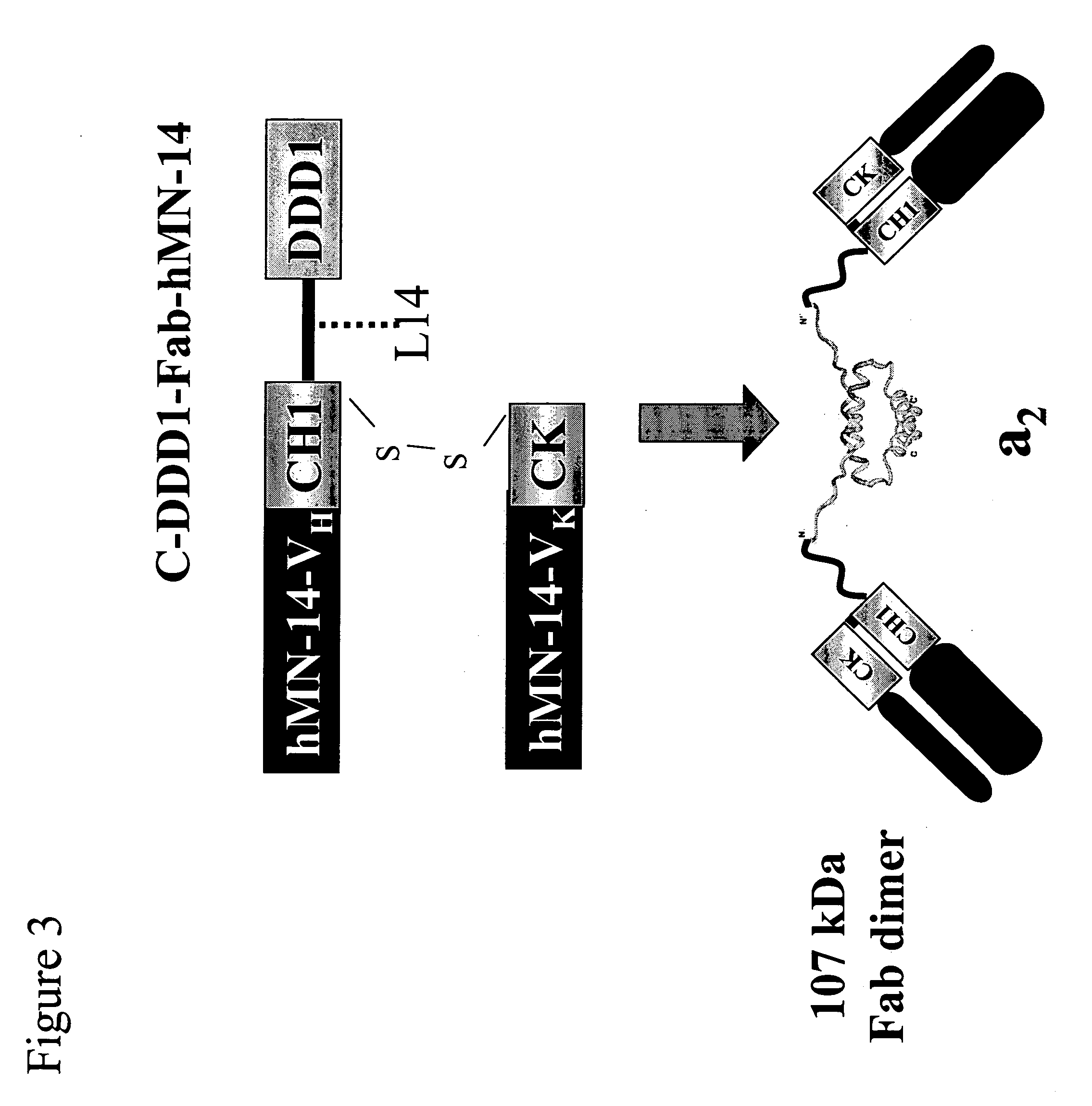
Stably tethered structures of defined compositions with multiple functions or binding specificities
Owner:IBC PHARMACEUTICALS INC
Methods for generating stably linked complexes composed of homodimers, homotetramers or dimers of dimers and uses
ActiveUS7550143B2Improve functionalityStrong specificityPeptide/protein ingredientsAntibody mimetics/scaffoldsHomotetramerAptamer
The present invention concerns methods and compositions for stably tethered structures of defined compositions, which may have multiple functionalities and / or binding specificities. Particular embodiments concern homodimers comprising monomers that contain a dimerization and docking domain attached to a precursor. The precursors may be virtually any molecule or structure, such as antibodies, antibody fragments, antibody analogs or mimetics, aptamers, binding peptides, fragments of binding proteins, known ligands for proteins or other molecules, enzymes, detectable labels or tags, therapeutic agents, toxins, pharmaceuticals, cytokines, interleukins, interferons, radioisotopes, proteins, peptides, peptide mimetics, polynucleotides, RNAi, oligosaccharides, natural or synthetic polymeric substances, nanoparticles, quantum dots, organic or inorganic compounds, etc. Other embodiments concern tetramers comprising a first and second homodimer, which may be identical or different. The disclosed methods and compositions provide a facile and general way to obtain homodimers, homotetramers and heterotetramers of virtually any functionality and / or binding specificity.
Owner:IBC PHARMACEUTICALS INC
Methods for generating stably linked complexes composed of homodimers, homotetramers or dimers of dimers and uses
ActiveUS20060228357A1Improve functionalityStrong specificityPeptide/protein ingredientsAntibody mimetics/scaffoldsChemistryToxin
The present invention concerns methods and compositions for stably tethered structures of defined compositions, which may have multiple functionalities and / or binding specificities. Particular embodiments concern homodimers comprising monomers that contain a dimerization and docking domain attached to a precursor. The precursors may be virtually any molecule or structure, such as antibodies, antibody fragments, antibody analogs or mimetics, aptamers, binding peptides, fragments of binding proteins, known ligands for proteins or other molecules, enzymes, detectable labels or tags, therapeutic agents, toxins, pharmaceuticals, cytokines, interleukins, interferons, radioisotopes, proteins, peptides, peptide mimetics, polynucleotides, RNAi, oligosaccharides, natural or synthetic polymeric substances, nanoparticles, quantum dots, organic or inorganic compounds, etc. Other embodiments concern tetramers comprising a first and second homodimer, which may be identical or different. The disclosed methods and compositions provide a facile and general way to obtain homodimers, homotetramers and heterotetramers of virtually any functionality and / or binding specificity.
Owner:IBC PHARMACEUTICALS INC
Inhibitor nucleic acids
InactiveUS20050256071A1Low melting pointQuality improvementOrganic active ingredientsNervous disorderAptamerHalf-life
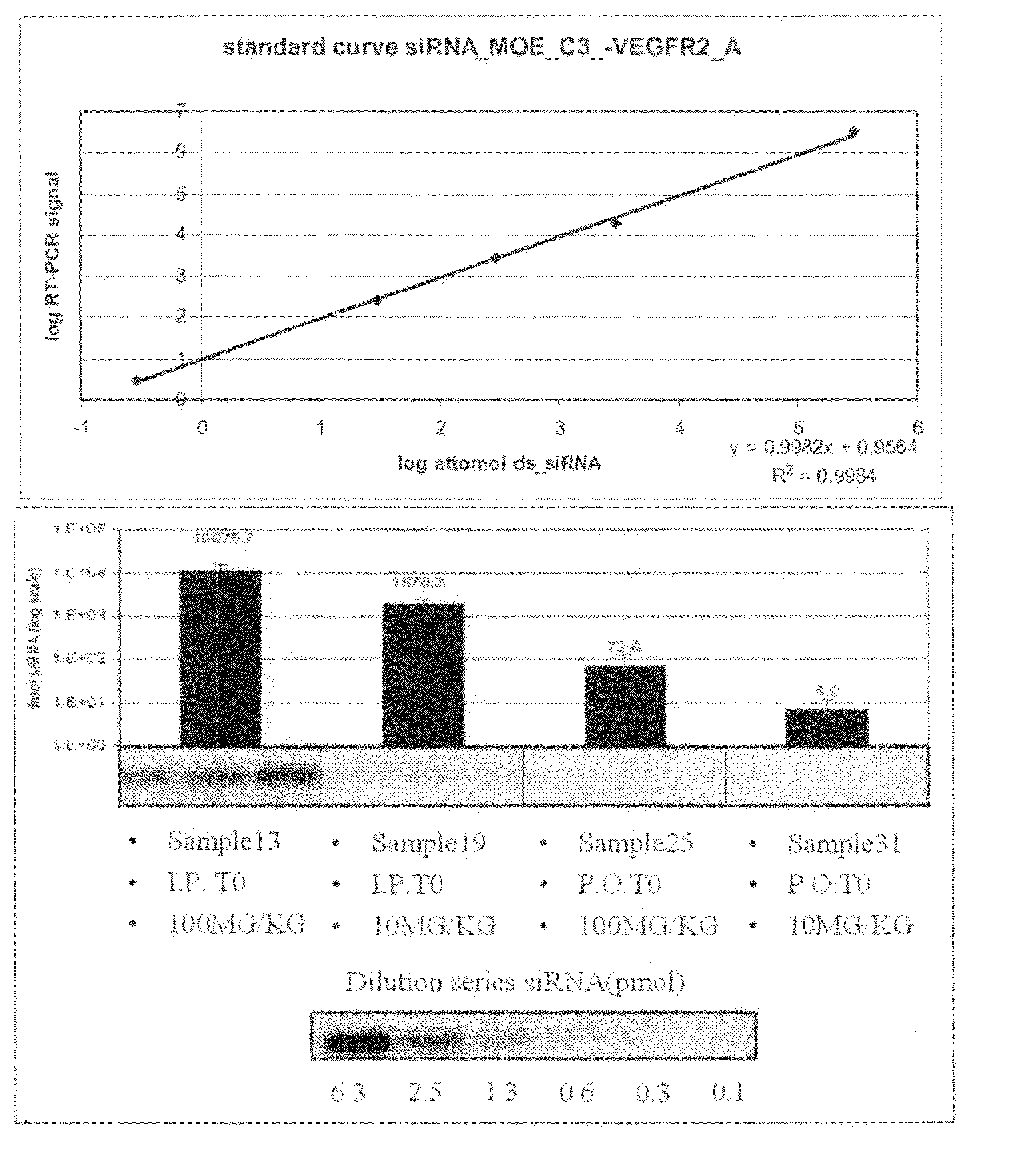
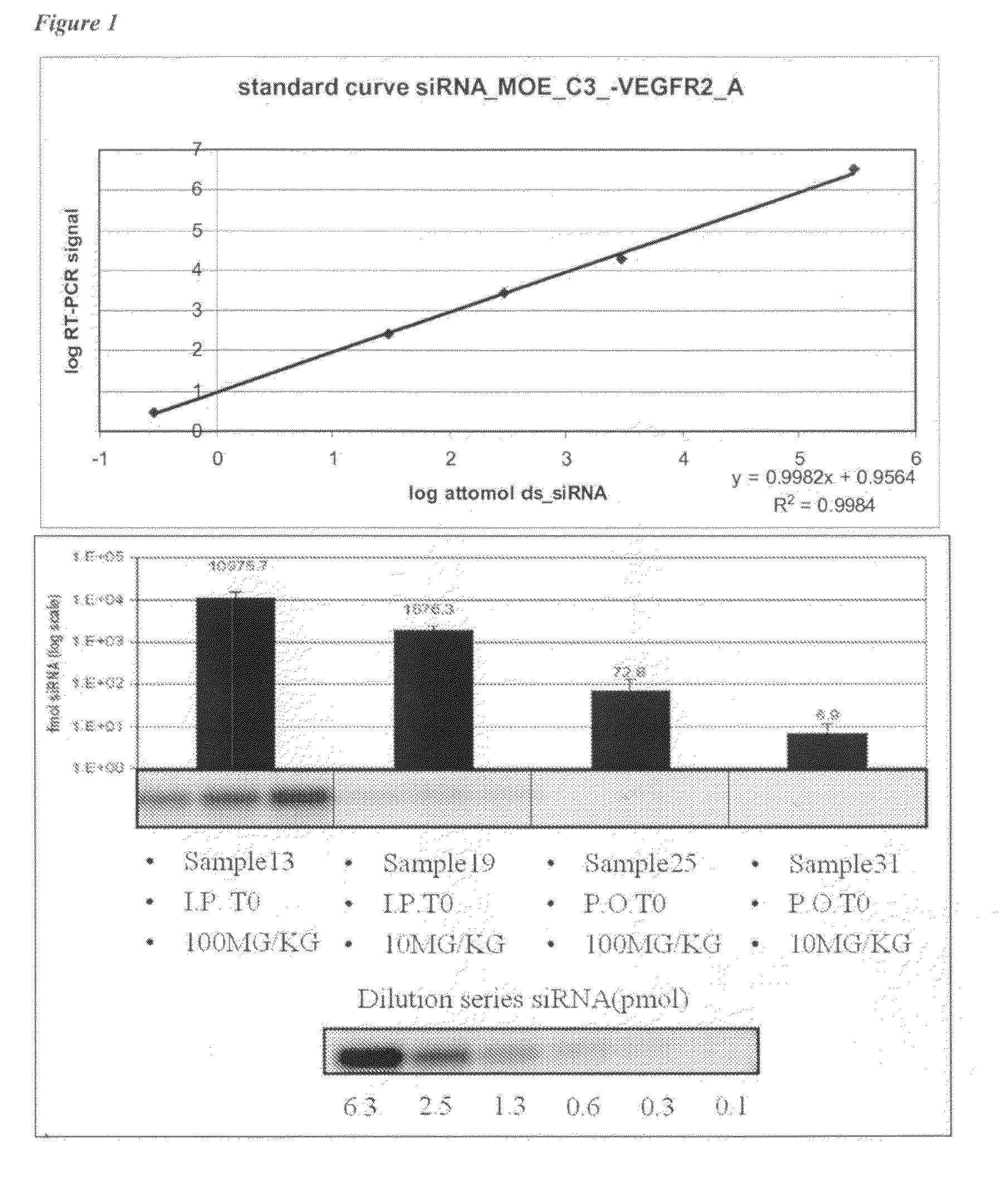
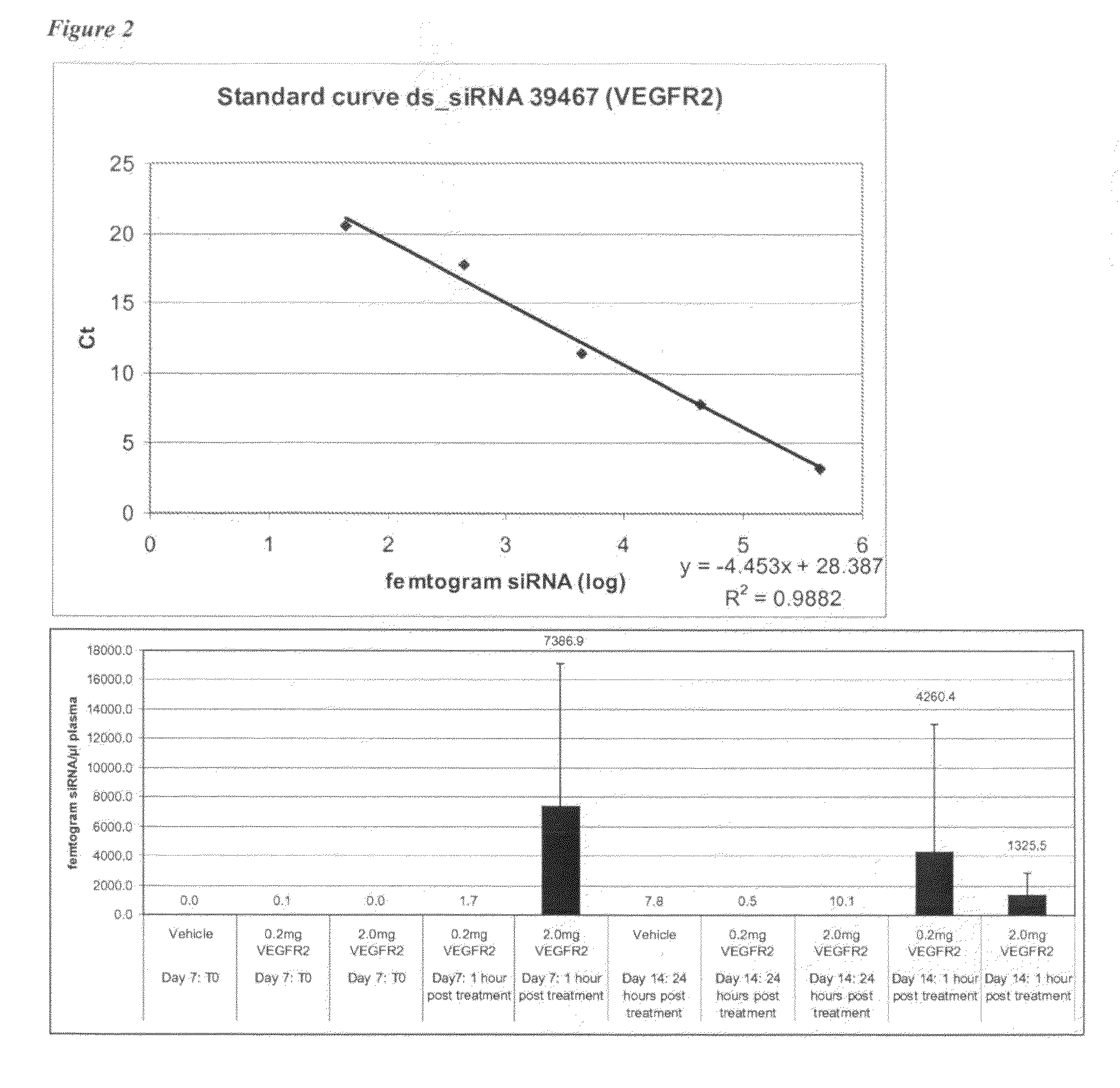
The present invention provides methods and compositions for attenuating expression of a target gene in vivo. In general, the method includes administering RNAi constructs (such as small-interfering RNAs (i.e., siRNAs) that are targeted to particular mRNA sequences, or nucleic acid material that can produce siRNAs in a cell), in an amount sufficient to attenuate expression of a target gene by an RNA interference mechanism. In particular, the RNAi constructs may include one or more modifications to improve serum stability, cellular uptake and / or to avoid non-specific effect. In certain embodiments, the RNAi constructs contain an aptamer portion. The aptamer may bind to human serum albumin to improve serum half life. The aptamer may also bind to a cell surface protein that improves uptake of the construct.
Owner:CALIFORNIA INST OF TECH
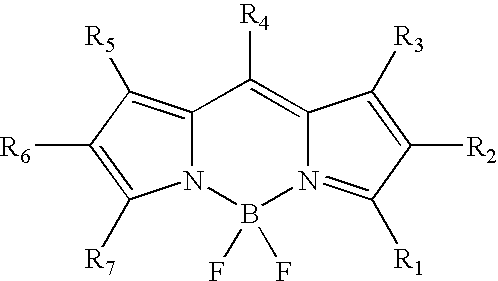
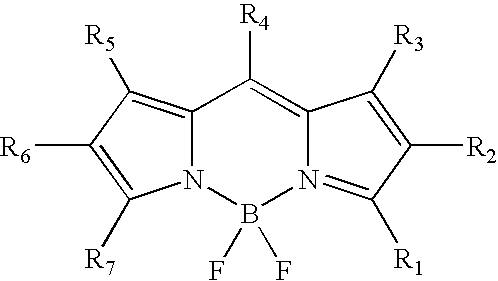
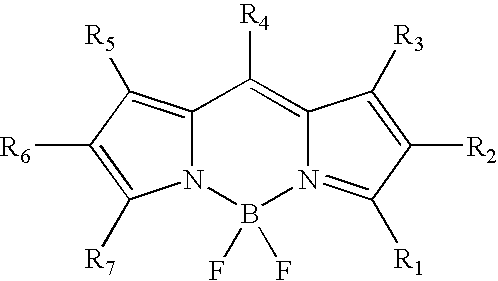
Labeling of immobilized proteins using dipyrrometheneboron difluoride dyes
InactiveUS6972326B2Hybrid immunoglobulinsChemiluminescene/bioluminescenceFluorescenceImmobilized protein
The invention describes methods for labeling or detecting of immobilized poly(amino acids), including peptides, polypeptides and proteins, on membranes and other solid supports, using fluorescent dipyrrometheneboron difluoride dyes. Such immobilized poly(amino acids) are labeled or detected on blots or on arrays of poly(amino acids), or are attached to immobilized aptamers.
Owner:MOLECULAR PROBES
Particulate labels
InactiveUS20120045748A1High sensitivitySolve the poor convenience of useMicrobiological testing/measurementParticulatesMolecular diagnostics
A methodology for bioassays and diagnostics in which a particulate label (ranging in size from nm-scale molecular assemblages to organisms on the scale of tens or hundreds of microns), such as, but not limited to, nanoparticles, bacteria, bacteriophage, Daphnia, and magnetic particles, serve carriers for analytes bound by molecular recognition elements such as antibodies, aptamers, etc. The described methodology is generally applicable to most pathogen assays and molecular diagnostics and also leads to enhanced sensitivity and convenience of use.
Owner:WILLSON RICHARD C +2
Aptamer-based methods for identifying cellular biomarkers
InactiveUS20090117549A1Easy to fixEasy to synthesizeElectrolysis componentsParticle separator tubesBiotin-streptavidin complexCancer cell
In this invention, a biomarker discovery method has been developed using specific biotin-labeled oligonucleotide ligands and magnetic streptavidin beads. In one embodiment, the oligonucleotide ligands are firstly generated by whole-cell based SELEX technique. Such ligands can recognize target cells with high affinity and specificity and can distinguish cells that are closely related to target cells even in patient samples. The targets of these oligonucleotide ligands are significant biomarkers for certain cells. These important biomarkers can be captured by forming complexes with biotin-labeled oligonucleotide ligands and collecting the complexes using magnetic streptavidin beads, whereupon the captured biomarkers are analyzed to identify the biomarkers. Analysis of biomarkers include HPLC-Mass Spectroscopy analysis, polyacrylamide gel electrophoresis, flow cytometry, and the like. The identified biomarkers can be used for pathological diagnosis and therapeutic applications. Using the disclosed methods, highly specific biomarkers of any kinds of cells, in particular cancer cells, can easily be identified without prior knowledge of the existence of such biomarkers.
Owner:TAN WEIHONG +1
Methods for Detecting Oligonucleotides
InactiveUS20110097716A1Quick checkRapid quantificationMicrobiological testing/measurementAptamerBioinformatics
The invention provides methods and compositions for detecting and / or quantifying nucleic acid oligonucleotides. These methods and compositions are useful for detecting and quantifying diagnostic and / or therapeutic synthetic target oligonucleotides, such as aptamers, RNAi, siRNA, antisense oligonucleotides or ribozymes in a biological sample.
Owner:NOVARTIS AG
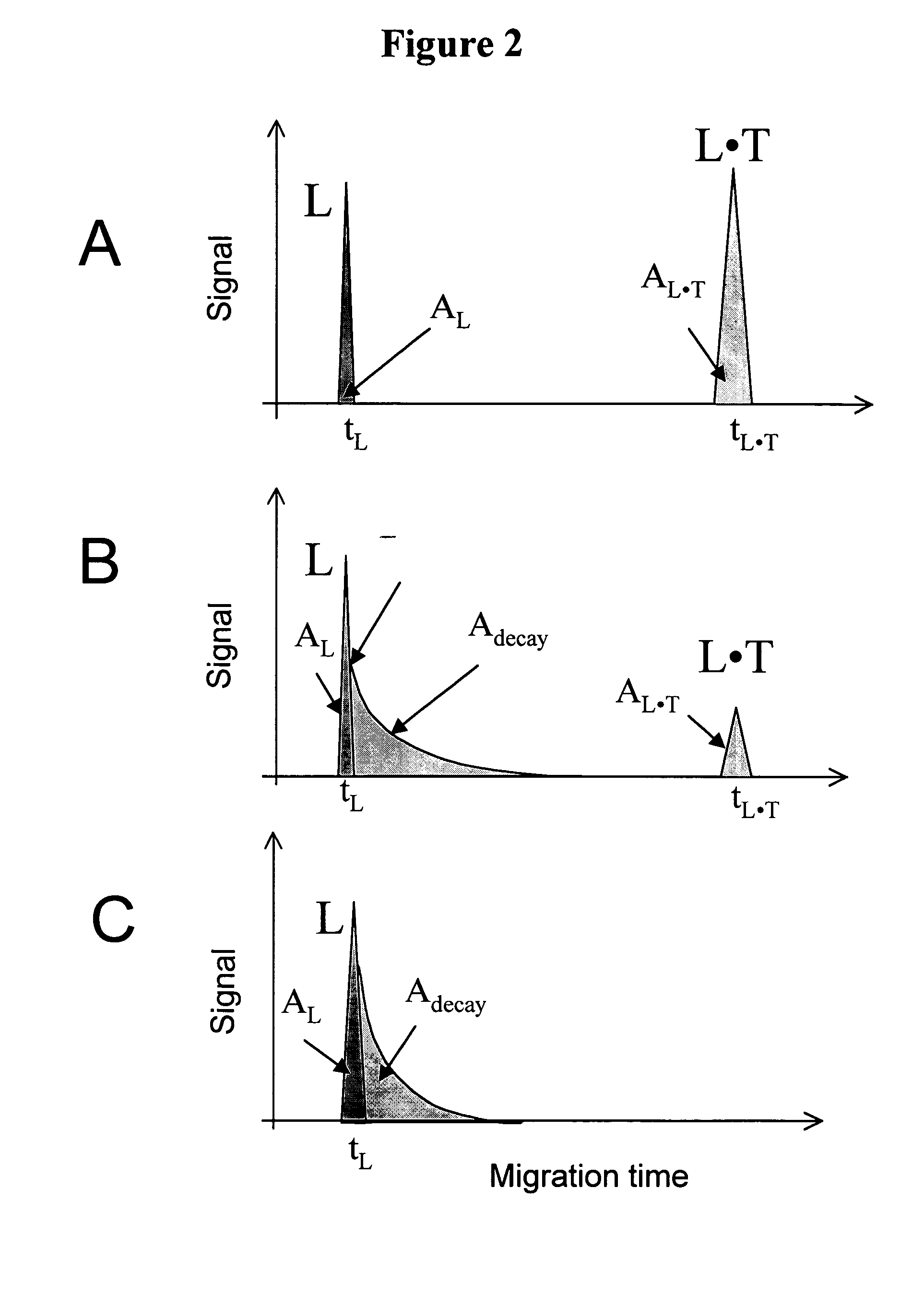
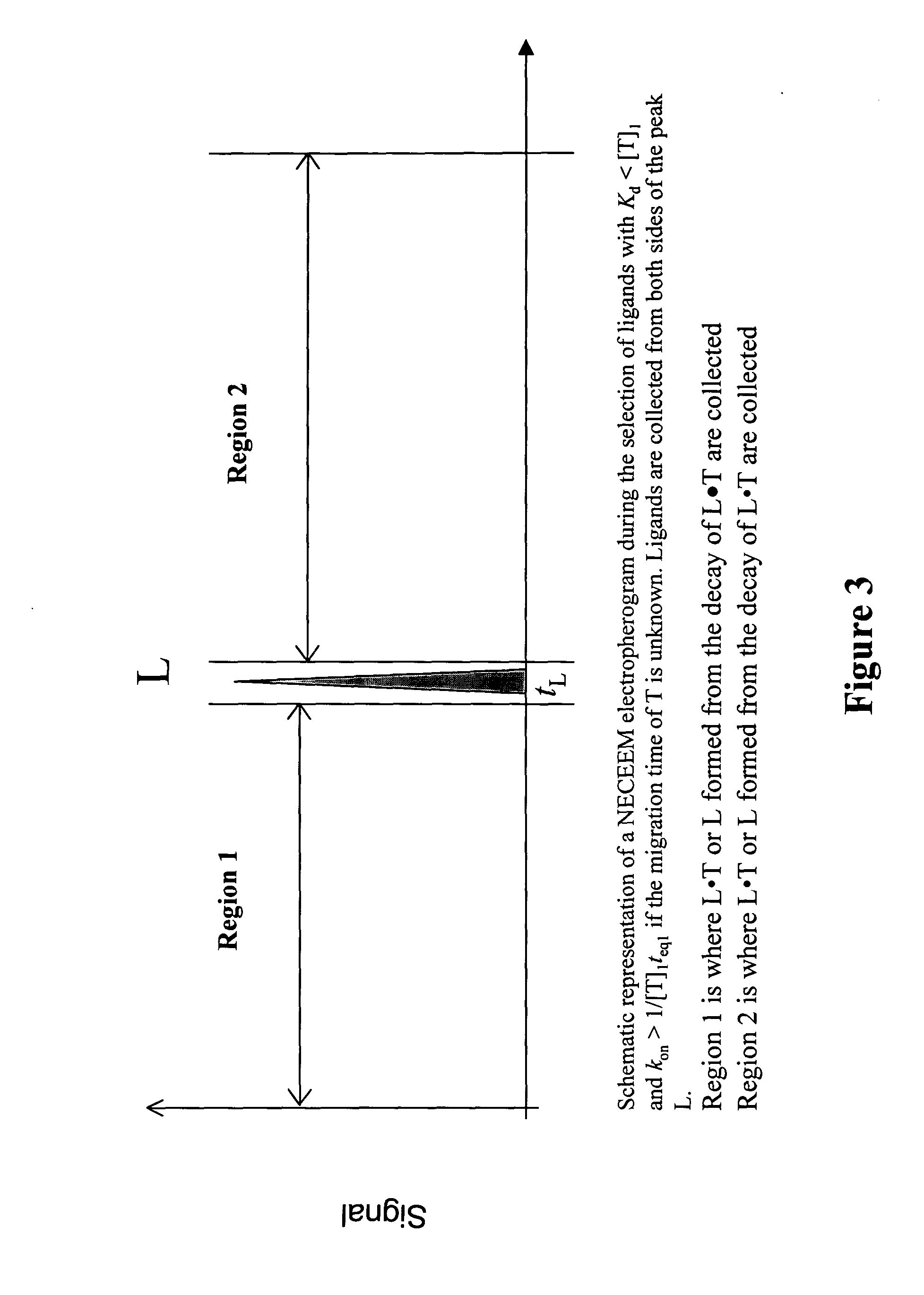
Non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM) - based methods for drug and diagnostic development
ActiveUS20050003362A1Enhances electrophoretic separationEasy to separateCompound screeningApoptosis detectionSolid substrateNucleotide Aptamers
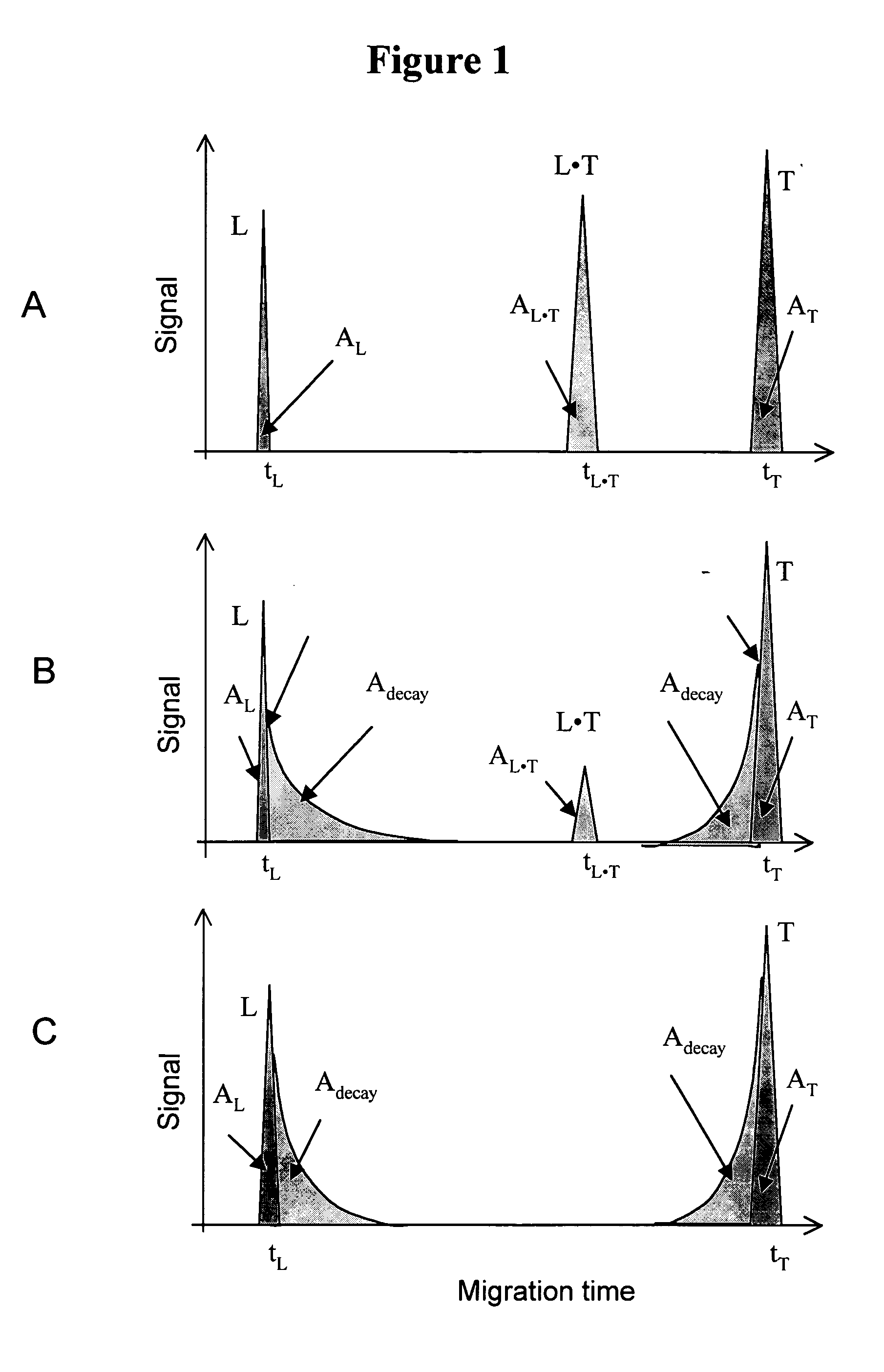
The invention discloses a Non-Equilibrium Capillary Electrophoresis of Equilibrium Mixtures (NECEEM) method and NECEEM-based practical applications. The NECEEM method is a homogeneous technique, which, in contrast to heterogeneous methods, does not require affixing molecules to a solid substrate. The method of the invention facilitates 3 practical applications. In the first application, the method allows the finding of kinetic and thermodynamic parameters of complex formation. It advantageously allows for revealing two parameters, the equilibrium dissociation constant, Kd, and the monomolecular rate constant of complex decay, koff, in a single experiment. In the second practical application, the method of this invention provides an approach for quantitative affinity analysis of target molecules. It advantageously allows for the use of affinity probes with relatively high values of koff. In the third practical application, the method of this invention presents a new and powerful approach to select target-binding molecules (ligands) from complex mixtures. Unique capabilities of the method in its third application include but not limited to: (a) the selection of ligands with pre-determined ranges of kinetic and thermodynamic parameters of target-ligand interactions, (b) the selection of ligands present in minute amounts in complex mixtures of biological or synthetic compounds such as combinatorial libraries of oligonucleotides, and (c) the selection of ligands for targets available in very low amounts. In particular, the method of this invention provides a novel approach for the selection of oligonucleotide aptamers. The NECEEM-based method can be used for discovery and characterization of drug candidates and the development of new diagnostic methods.
Owner:KRYLOV SERGEY
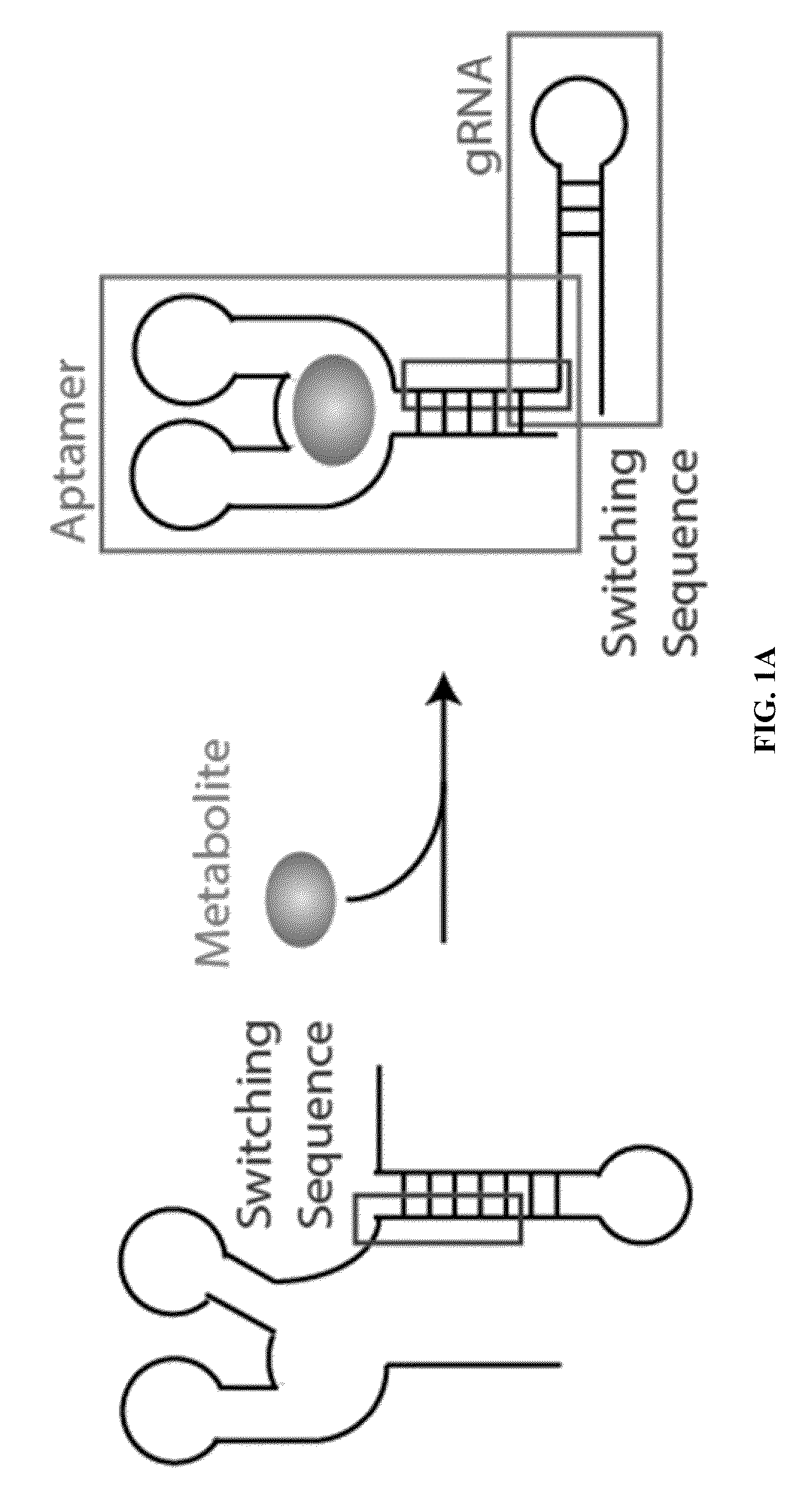
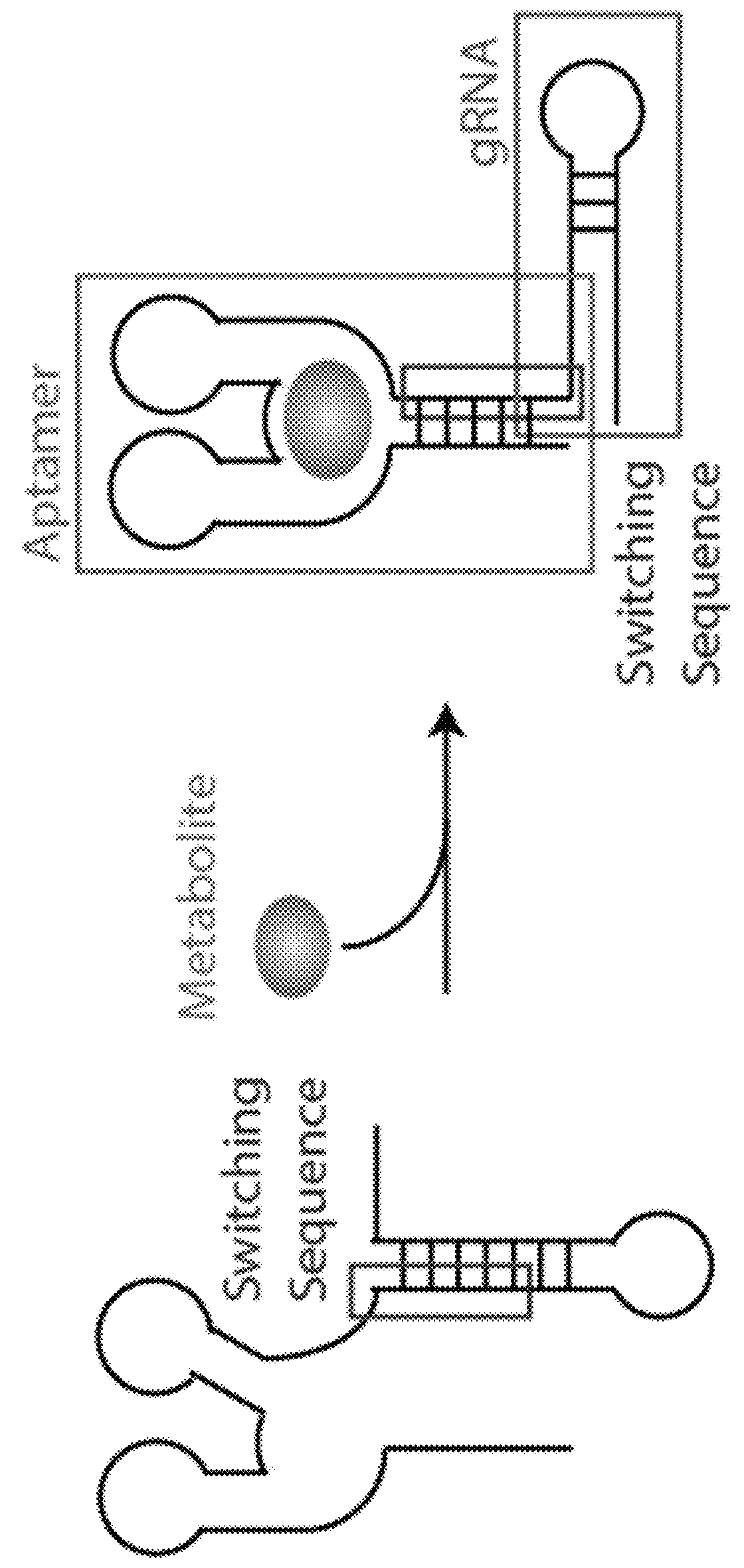
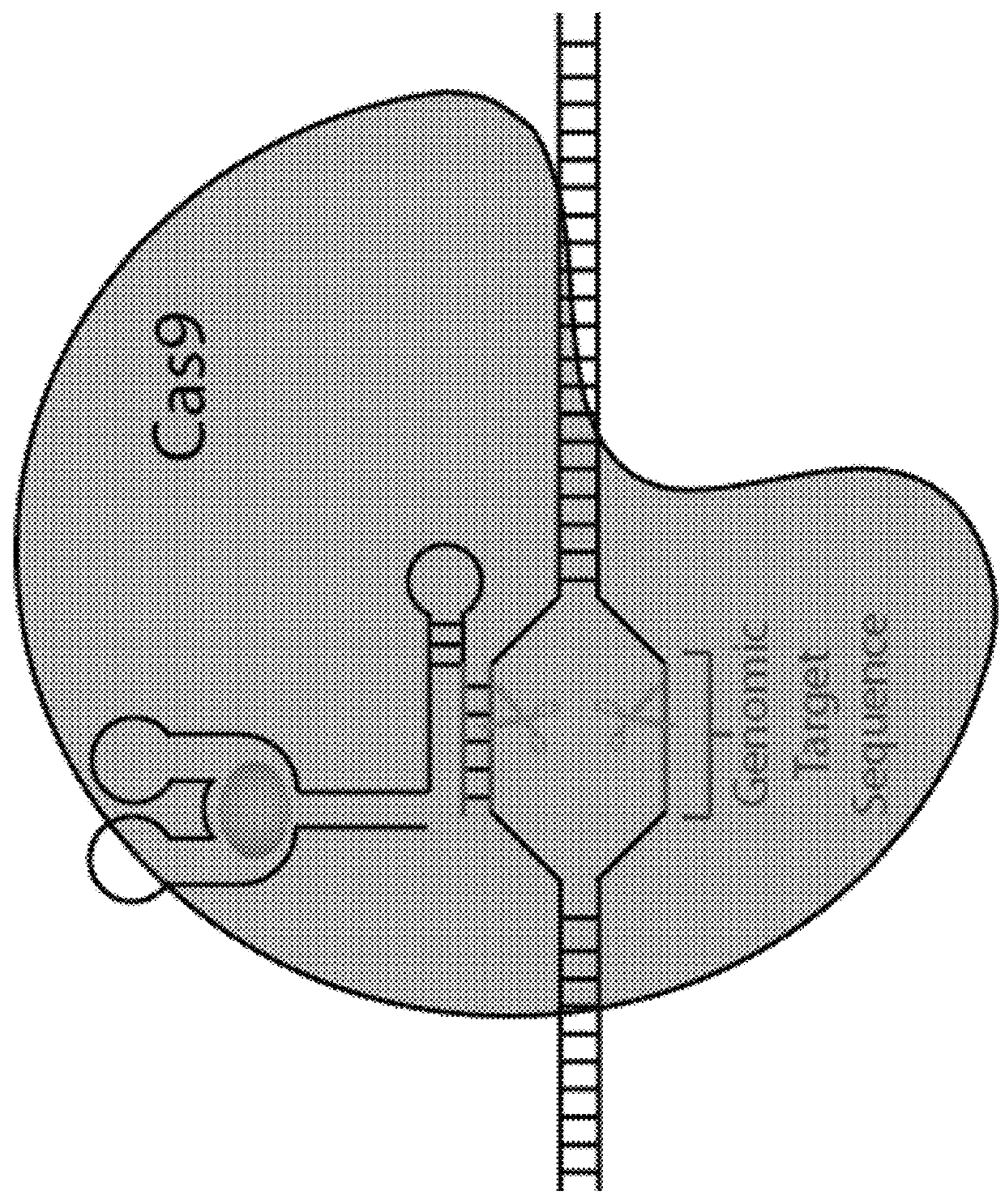
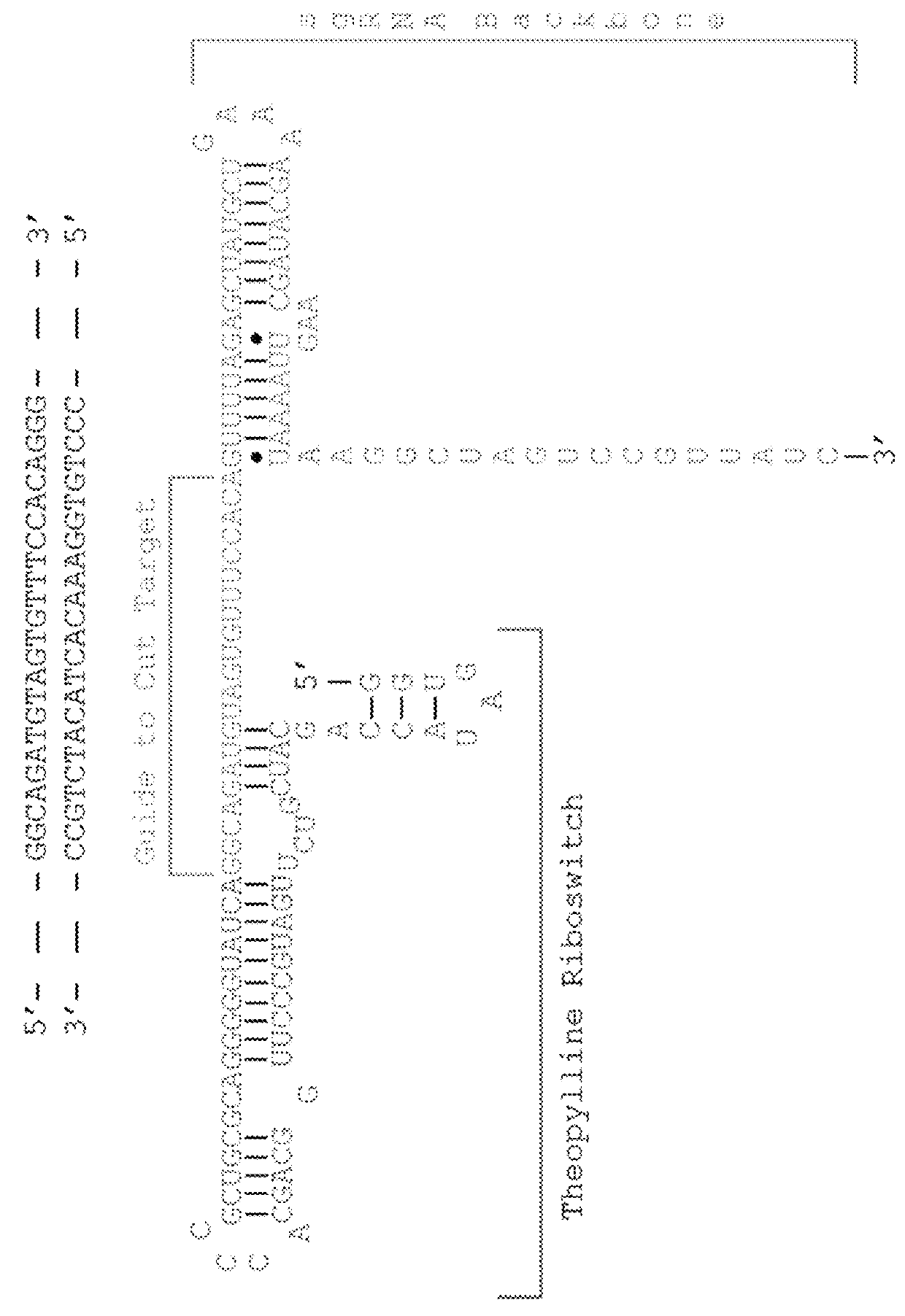
Switchable grnas comprising aptamers
ActiveUS20150071900A1Reduce the possibilityHydrolasesPeptide/protein ingredientsBiologyDNA Endonuclease
Some aspects of this disclosure provide compositions, methods, systems, and kits for controlling the activity and / or improving the specificity of RNA-programmable endonucleases, such as Cas9. For example, provided are guide RNAs (gRNAs) that are engineered to exist in an “on” or “off” state, which control the binding and hence cleavage activity of RNA-programmable endonucleases.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Switchable gRNAs comprising aptamers
Some aspects of this disclosure provide compositions, methods, systems, and kits for controlling the activity and / or improving the specificity of RNA-programmable endonucleases, such as Cas9. For example, provided are guide RNAs (gRNAs) that are engineered to exist in an “on” or “off” state, which control the binding and hence cleavage activity of RNA-programmable endonucleases.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Method for obtaining oligonucleotide aptamers and uses thereof
The present invention relates to a method for obtaining nucleic acid aptamers that bind to cancer cell-surface epitopes, to the aptamers generated using this method and their use for therapeutic, diagnostic and prognostic purposes.
Owner:CONSIGLIO NAT DELLE RICERCHE
Method for the detection and neutralization of bacteria
The present invention relates to the identification of bacteria present at the site of infection and the treatment of the infection using bacteriophage. In certain embodiments, the present invention provides methods and compositions for treating bacterial infections by identifying at least one bacteria species in the infection based on its interaction with bacteria-specific aptamers, selecting one or more bacteriophage that infect the identified bacteria species, and administering an effective amount of the bacteriophage to the subject to treat the infection.
Owner:KCI LICENSING INC
Stabilized aptamers to platelet derived growth factor and their use as oncology therapeutics
Materials and methods are provided for producing and using aptamers useful as oncology therapeutics capable of binding to PDGF, PDGF isoforms, PDGF receptor, VEGF, and VEGF receptor or any combination thereof with great affinity and specificity. The compositions of the present invention are particularly useful in solid tumor therapy and can be used alone or in combination with known cytotoxic agents for the treatment of solid tumors. Also disclosed are aptamers having one or more CpG motifs embedded therein or appended thereto.
Owner:ARCHEMIX CORP
Immunomodulatory compositions and uses therefor
The poxvirus proteins designated A41L and 130L bind to three receptor-like protein tyrosine phosphatases (RPTP), leukocyte common antigen related protein (LAR), RPTP-δ, and RPTP-σ, that are present on the cell surface of immune cells. When a host is infected with the poxvirus, binding of A41L to cell surface proteins on the host cells results in suppression of the immune response. The present invention provides agents such as antibodies, and antigen-binding fragments thereof, small molecules, aptamers, small interfering RNAs, and peptide-IgFc fusion polypeptides that interact with one or more of LAR, RPTP-δ, and RPTP-σ expressed by immune cells or interact with a polynucleotide encoding the RPTP. Also provided are RPTP Ig domain oligomers and Fc fusion polypeptides. Such agents are useful for treating an immunological disorder in a subject according to the methods described herein.
Owner:VIRAL LOGIC SYST TECH CORP
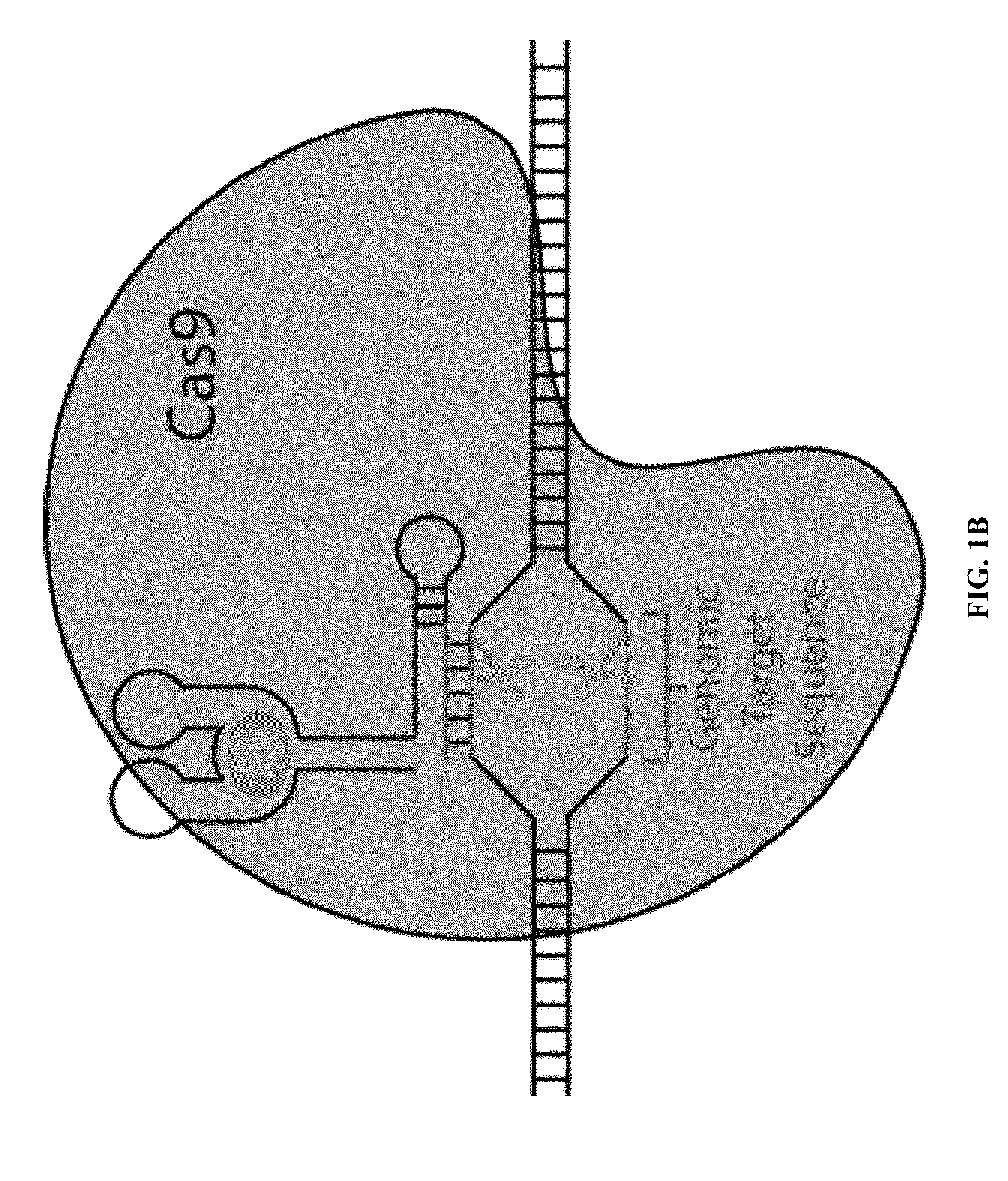
Gene control device and method based on CRISPR-Cas9
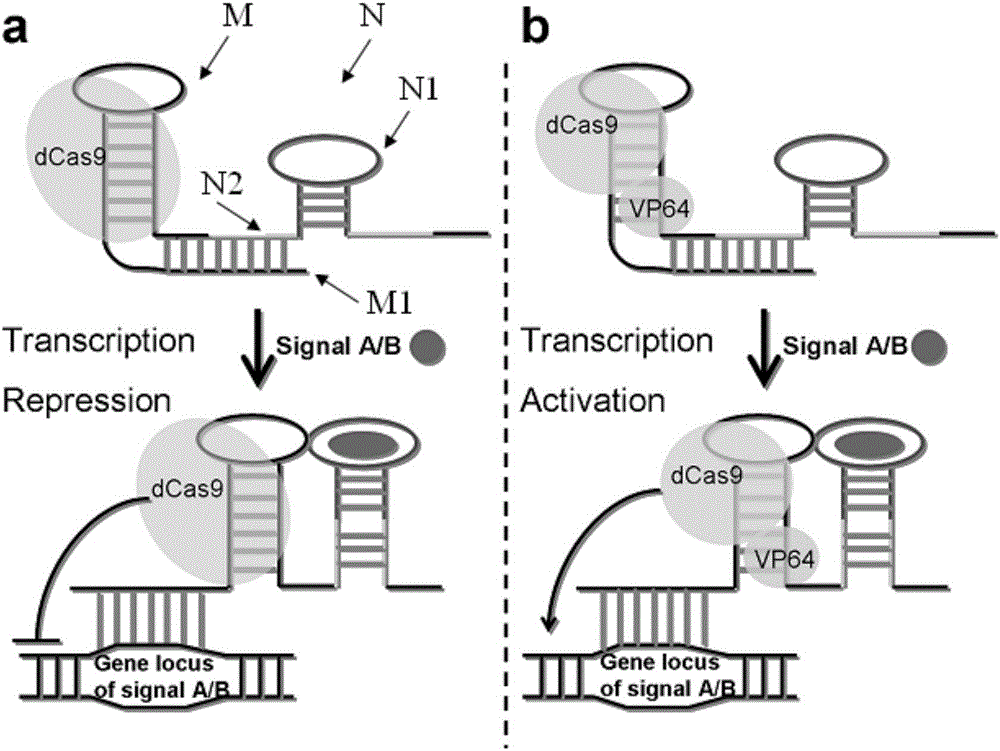
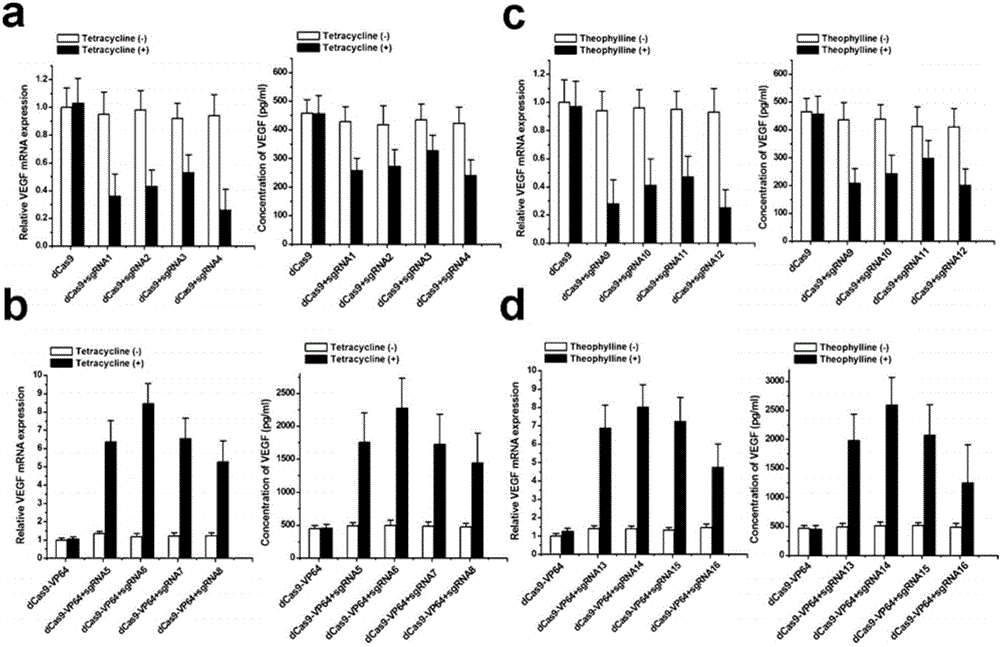
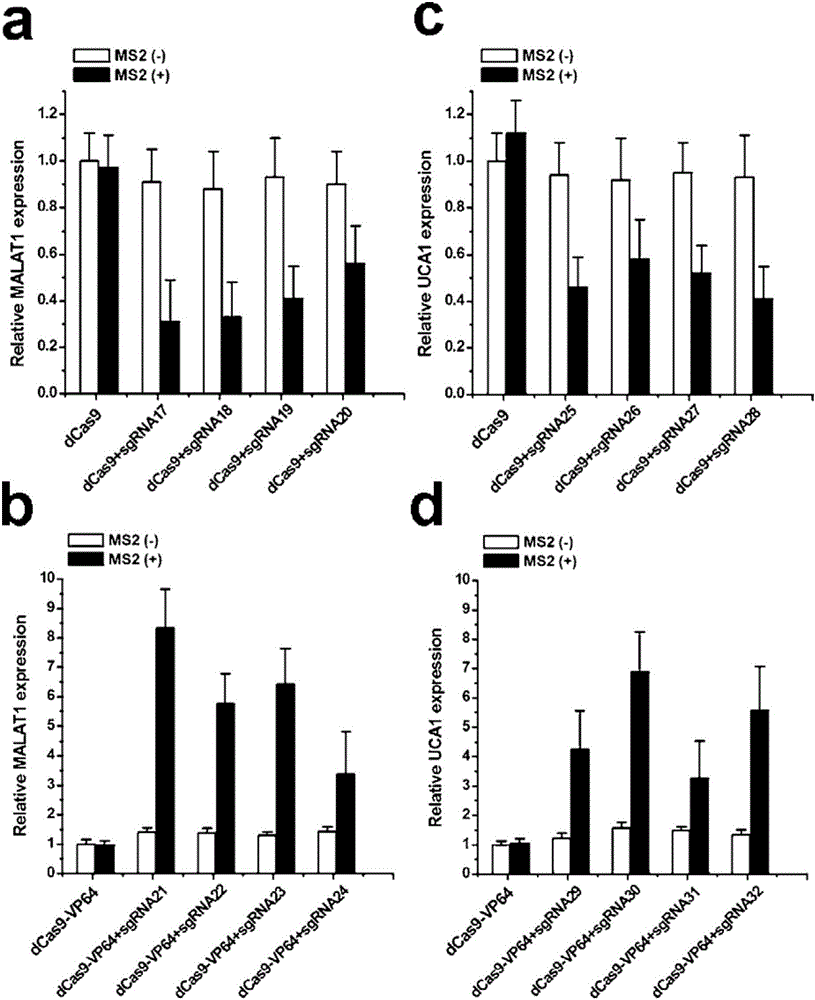
The invention discloses a gene control device and method based on CRISPR-Cas9. The device comprises a nucleotide sequence, wherein the nucleotide sequence is an sgRNA itself or can be transcribed into an sgRNA, the sgRNA structurally comprises a first part bonded with inactivated Cas9 protein and a second part connected to the 3'end of the first part, the 5'end of the first part is provided with an antisense sequence, and the second part is an aptamer sequence part which comprises a ligand binding part and an aptamer stem part; when no ligand exists, the aptamer sequence is pair-bonded with the aptamer stem part; when a ligand exists, the aptamer sequence is dissociated from the aptamer stem part so as to be pair-bonded with a target DNA. By integrating an RNA riboswitch bonded with a specific biological signal into the sgRNA, a bridge of cell signal intensity and gene expression intensity is established, any target gene can be activated or silenced on the basis that biological signal identification is achieved, and artificial connection is built between signal ligands and cytogene.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Gene knock-down by intracellular expression of aptamers
InactiveUS20050250106A1Increase over-expressionMaximally knock-down gene activitySplicing alterationSugar derivativesGene Knock-DownGene
Materials and methods are provided for target validation by gene knock-down with intracellularly expressed aptamers and siRNAs. The aptamers produced by the materials and methods of the invention are useful in target validation for therapeutics development.
Owner:ARCHEMIX CORP
Small molecule probe based on nano-gold and aptamer and preparation method of small molecule probe
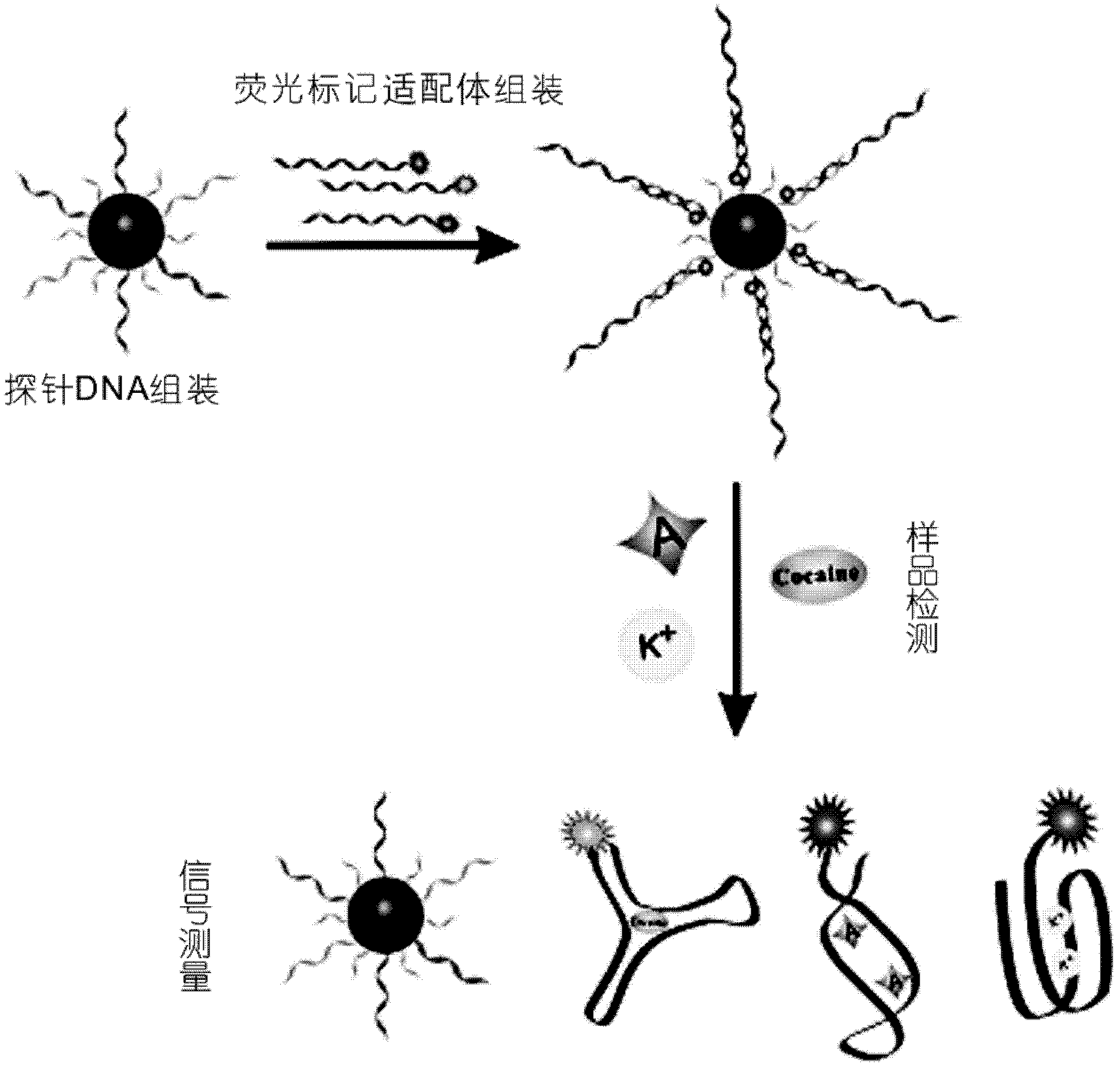
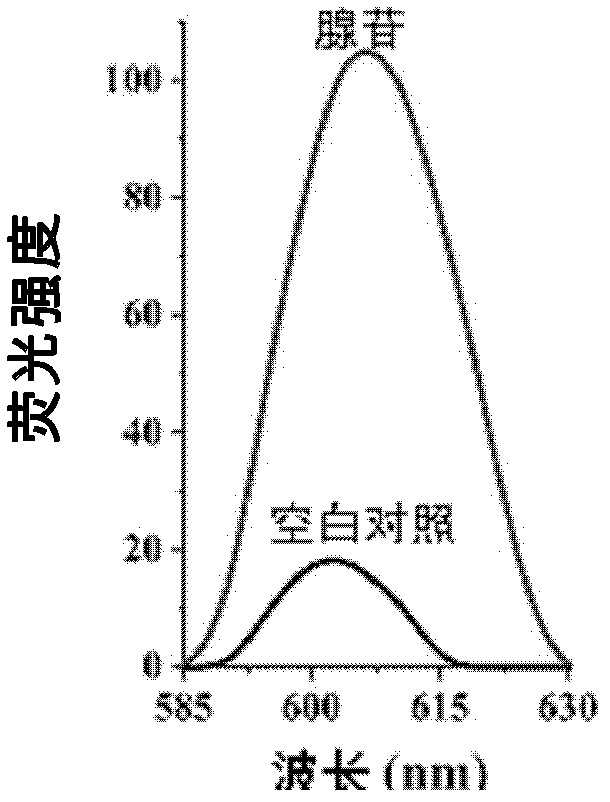
InactiveCN102676508AHigh detection sensitivityHigh quenching capacityMicrobiological testing/measurementFluorescence/phosphorescenceAptamerGold particles
The invention discloses a small molecule probe based on nano-gold and aptamer and a preparation method of the small molecule probe. The small molecule probe comprises nano-gold particles, more than one type of sulfydryl modifying deoxyribonucleic acid (DNA) fragment assembled on the nano-gold particles and more than one type of aptamer which is marked by fluorescent groups, and at least a part of fragments of the more than one type of the aptamer are complemented and hybridized with the more than one type of sulfydryl modifying DNA group. The small molecule probe is short in detecting time, simple in operation, high in specificity, good in repeatability, high in accuracy, and the false positive result is effectively reduced. By means of the aptamer technology, a high detection sensibility is achieved, and various molecules can be simultaneously detected.
Owner:华森新科(苏州)纳米技术有限公司
E-selectin antagonist compounds, compositions, and methods of use
ActiveUS9109002B2Enhancing hematopoietic stem cell survivalReduce the likelihood of occurrenceEsterified saccharide compoundsOrganic active ingredientsAptamerDisease
Methods and compositions using E-selectin antagonists are provided for the treatment and prevention of diseases and disorders treatable by inhibiting binding of E-selectin to an E-selectin ligand. Described herein are E-selectin antagonists including, for example, glycomimetic compounds, antibodies, aptamers and peptides that are useful in methods for treatment of cancers, and treatment and prevention of metastasis, inhibiting infiltration of the cancer cells into bone marrow, reducing or inhibiting adhesion of the cancer cells to endothelial cells including cells in bone marrow, and inhibiting thrombus formation.
Owner:GLYCOMIMETICS
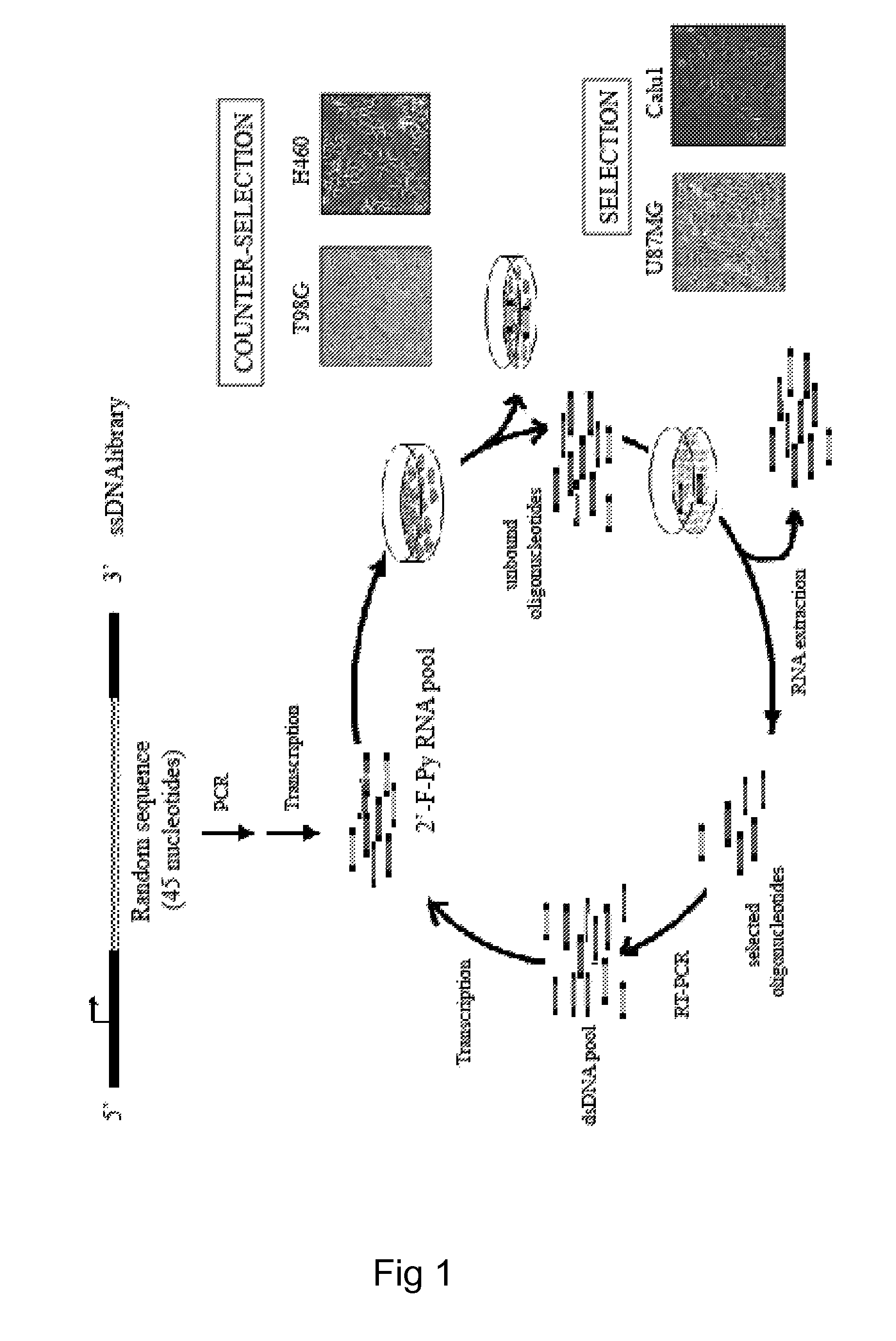
Focused Libraries, Functional Profiling, Laser Selex, and Deselex
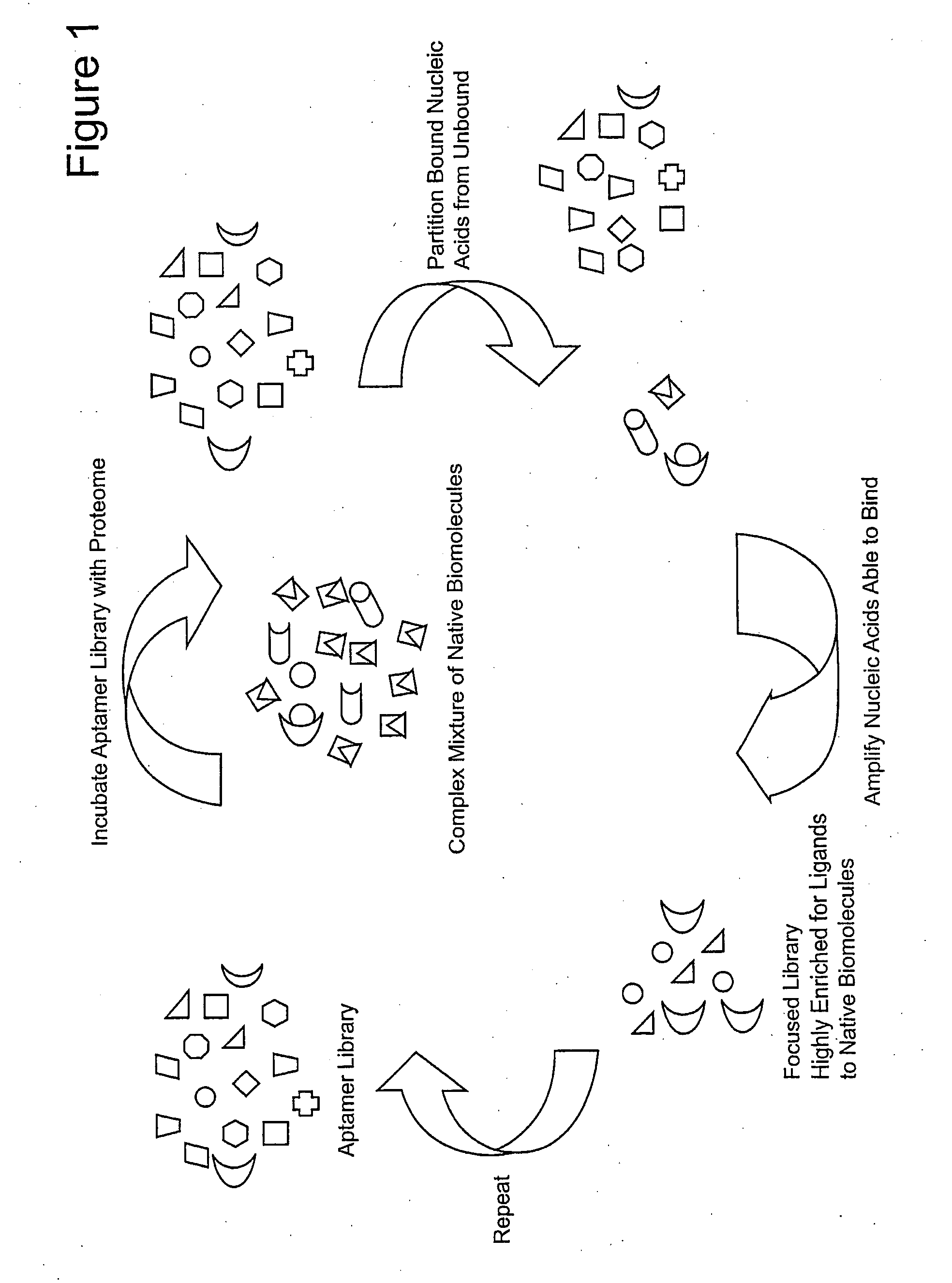
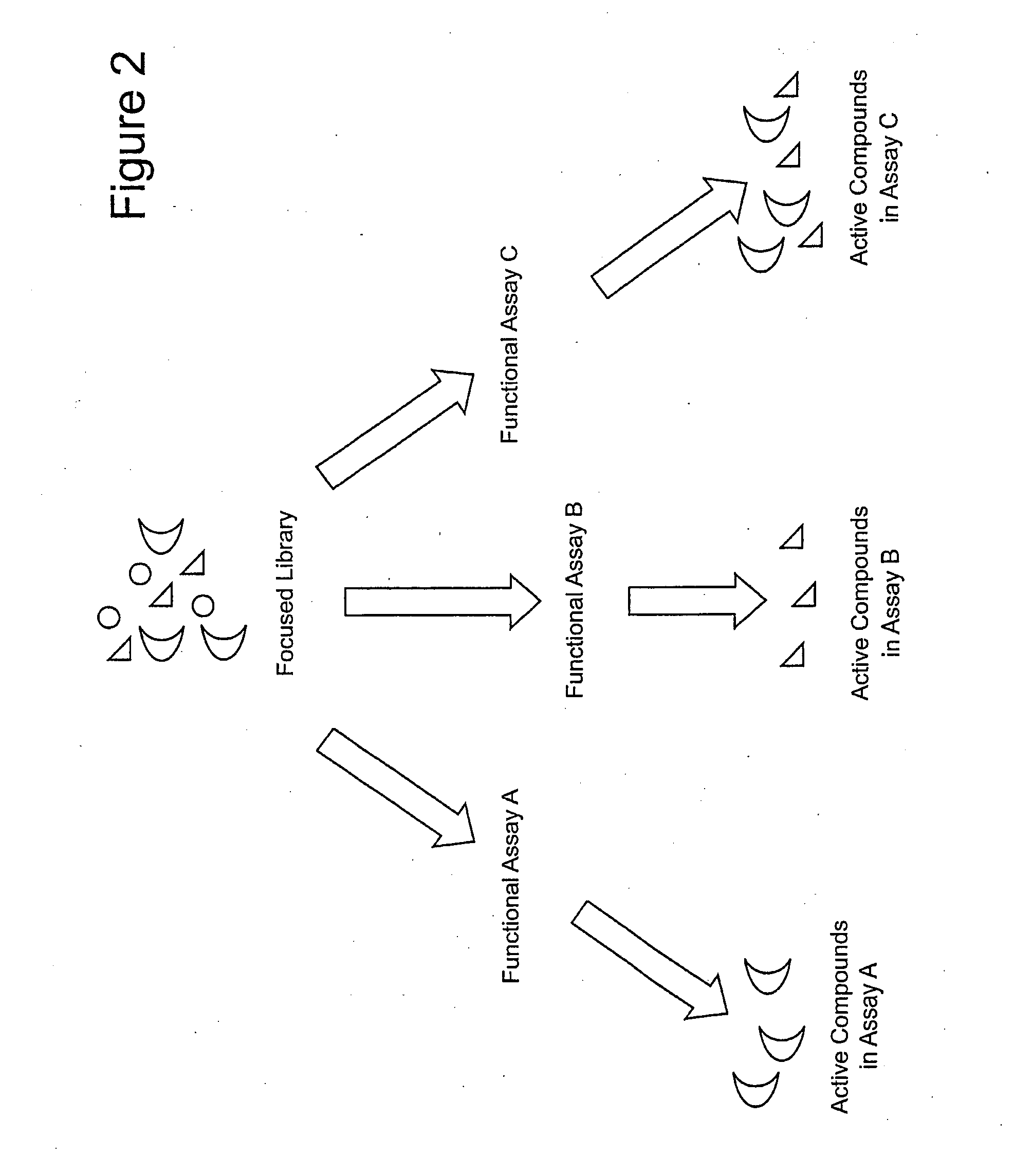
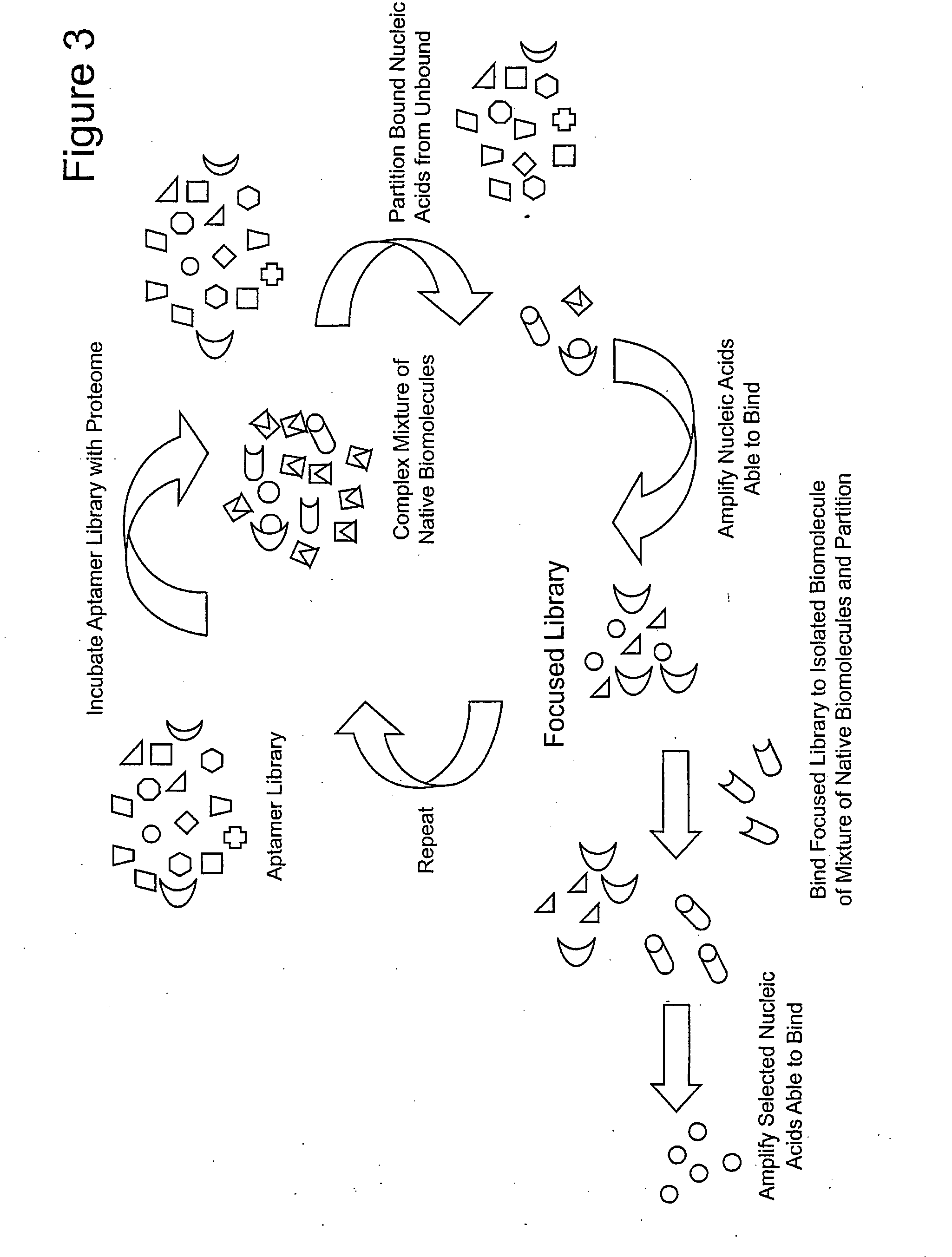
ActiveUS20090264508A1Reduce frequencyOrganic active ingredientsSugar derivativesFunctional profilingBiology
Focused aptamer libraries are constructed in accordance with a proteome (i.e., complex mixture of native biomolecules). The libraries may be screened to identify one or more candidate aptamers with desired biological activities other than specific binding to a target. Aptamers which are selected or derivatives thereof may be used for those specific activities in biological systems. Any combination of deconvoluting a focused library (functional profiling), increasing frequencies of particular aptamers in a focused library (Laser SELEX), and decreasing frequencies of particular aptamers in a focused library (DeSELEX) may be performed prior to assaying biological activity.
Owner:DUKE UNIV
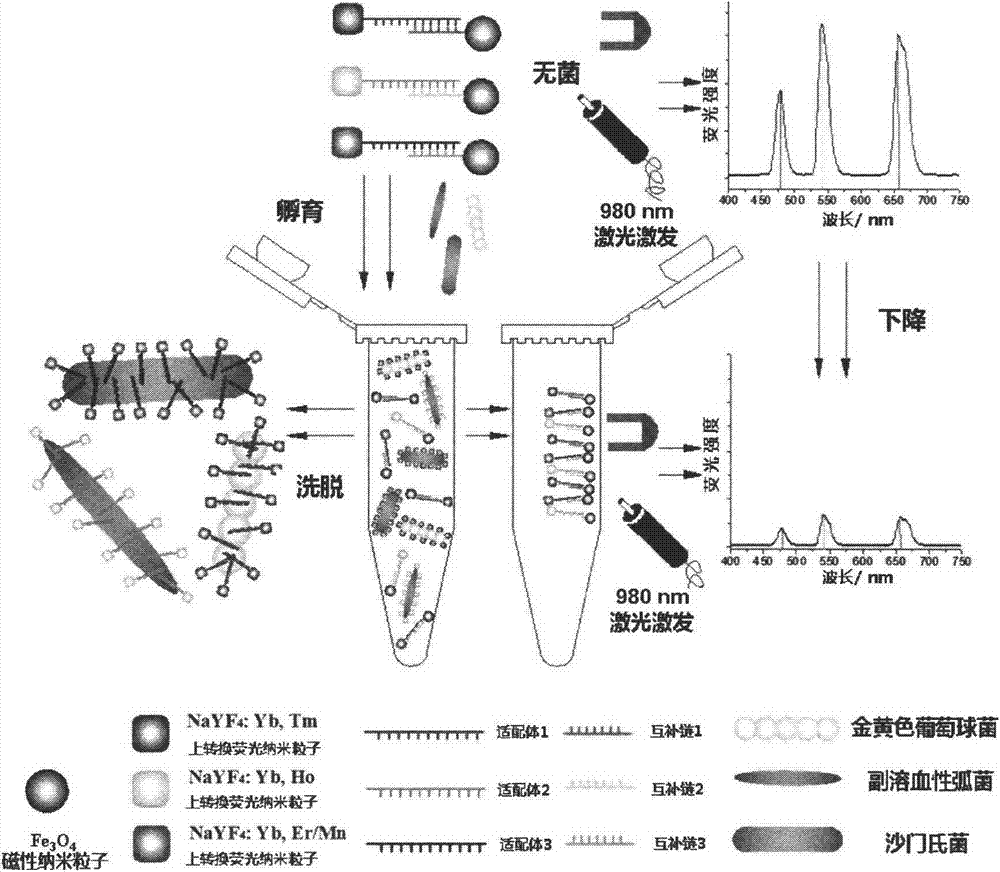
Method used for simultaneous detection of three food-borne pathogenic bacteria based on multicolor upconversion fluorescence labeling
ActiveCN103940792AImprove accuracy and stabilityHigh sensitivityFluorescence/phosphorescencePathogenic bacteriaFluorescence spectrometry
The invention provides a method used for simultaneous detection of three food-borne pathogenic bacteria based on multicolor upconversion fluorescence labeling. According to the method, three upconversion materials with differentiable fluorescence spectrums are used for forming multicolor upconversion fluorescent nanoprobes via respective connection with aptamers of staphylococcus aureus, vibrio parahaemolyticus, and salmonella, and complementary oligonucleotide single chains of the aptamers are connected with magnetic nanoparticles so as to form nano-composites. When bacteria to be tested are in a detection system, double chain unwinding is realized because of specific binding of the pathogenic bacteria with corresponding aptamers; it is possible to realize simultaneous quantitative determination of staphylococcus aureus, vibrio parahaemolyticus, and salmonella by monitoring upconversion fluorescence signal strength at 477nm, 550nm, and 660nm, detection linear range ranges from 50 to 1000000cfu / ml, and detection limits are 25cfu / ml, 10cfu / ml, and 15cfu / ml respectively. The method is used for detection of pathogenic bacteria, is high in sensitivity, is rapid and convenient, and can be used for detection of the three pathogenic bacteria in food such as milk and shrimp meat; and results are accurate and reliable.
Owner:JIANGNAN UNIV
Aptamer-targeted costimulatory ligand aptamer
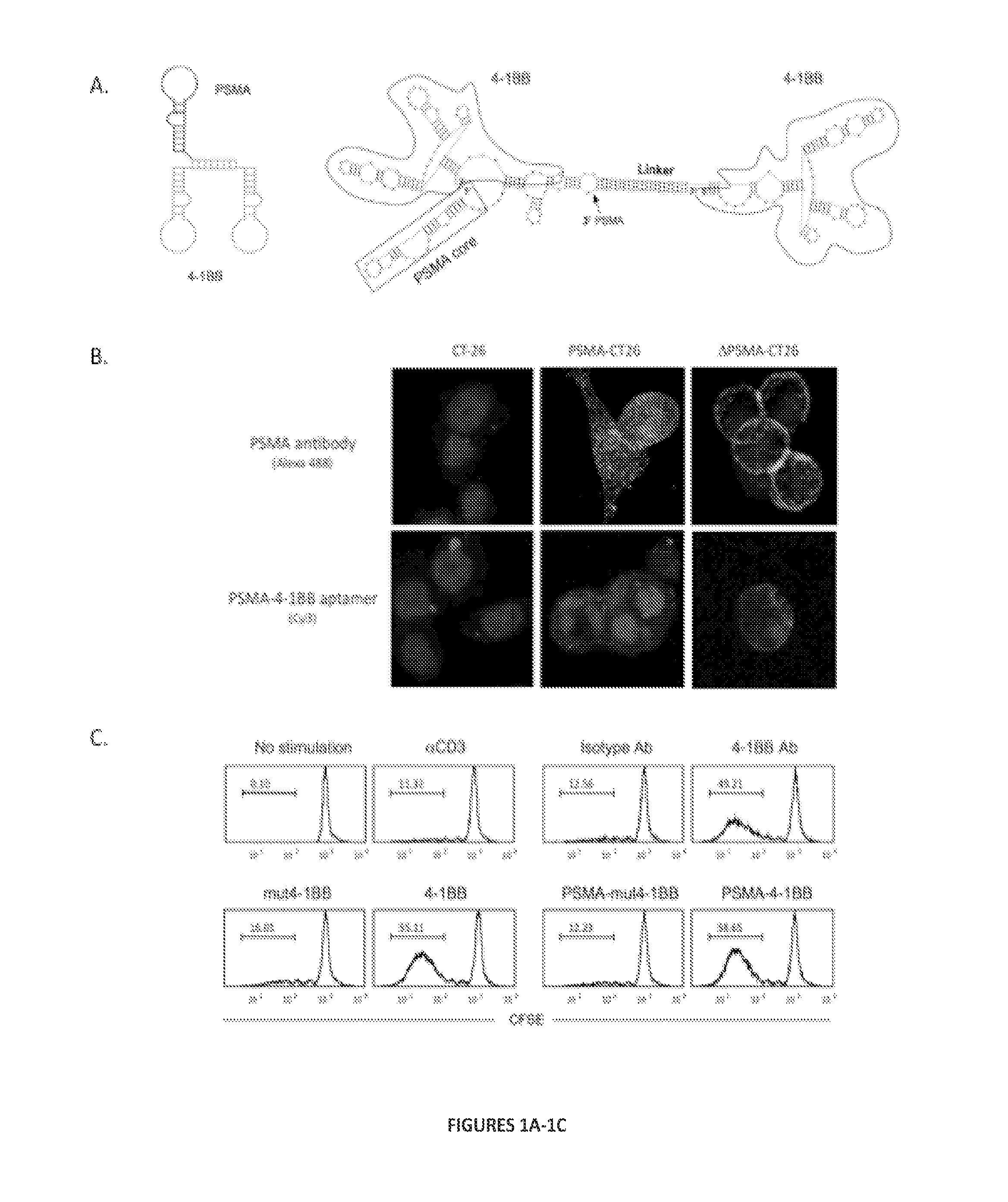
Compositions for inducing or enhancing immunogenicity of a tumor comprise bi- and multi-specific aptamers binding to a tumor cell and an immune cell. These compositions have broad applicability in the treatment of many diseases, including cancer.
Owner:UNIV OF MIAMI
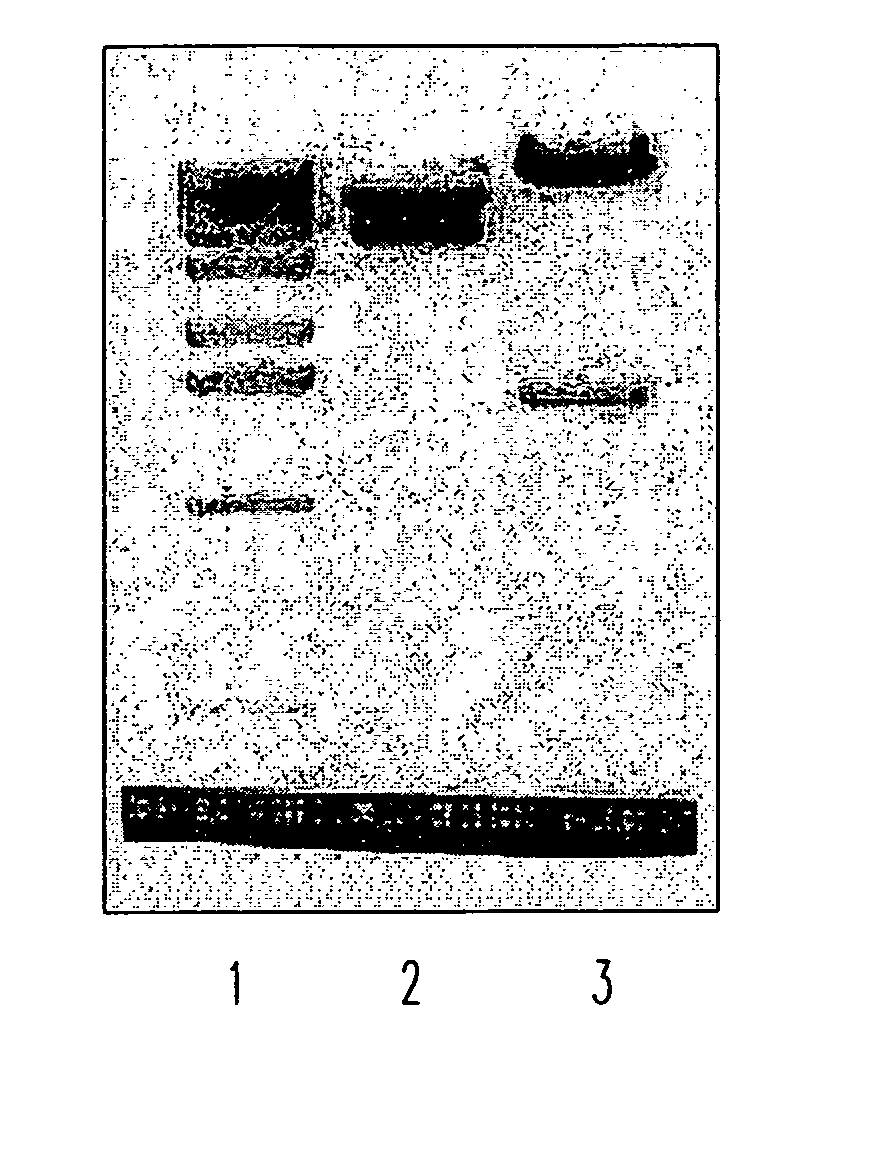
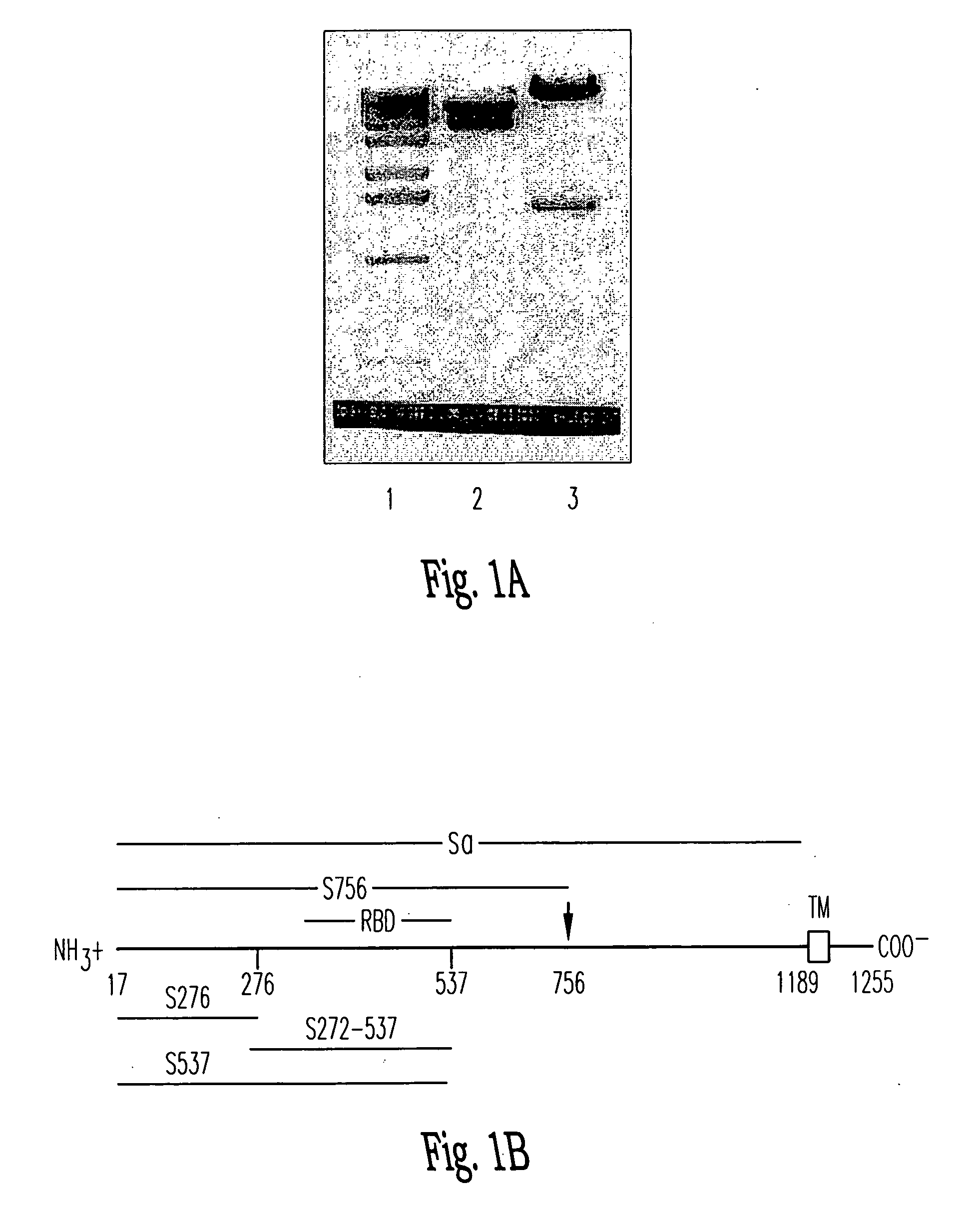
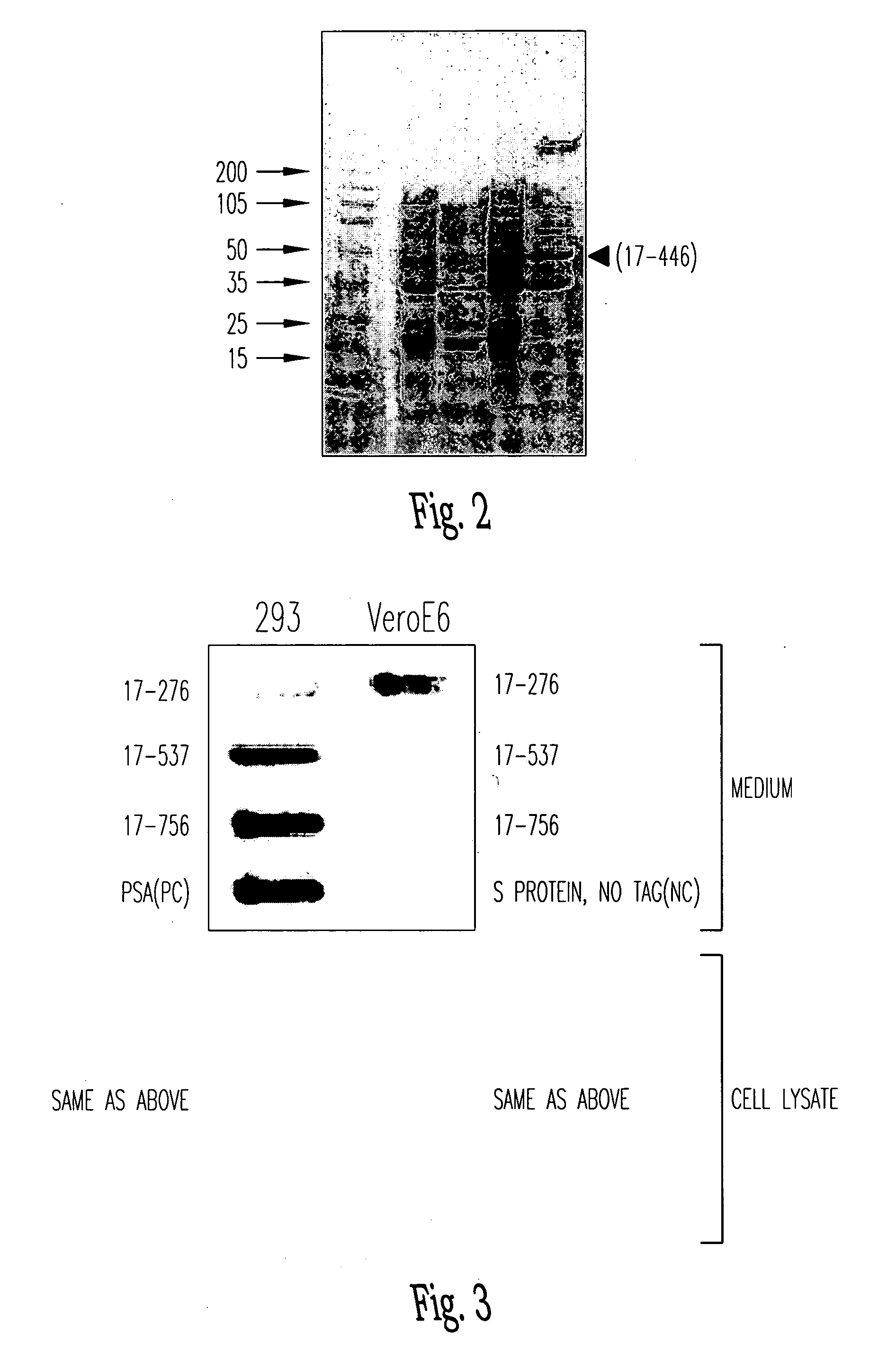
Soluble fragments of the SARS-CoV spike glycoprotein
InactiveUS20060240515A1Easy to produceImprove resistance to degradationSsRNA viruses positive-senseAntibody mimetics/scaffoldsAptamerVaccination
The invention relates to the spike protein from the virus (SARS-CoV) that is etiologically linked to severe acute respiratory syndrome (SARS); polypeptides and peptide fragments of the spike protein; nucleic acid segments and constructs that encode the spike protein, polypeptides and peptide fragments of the spike protein, and coupled proteins that include the spike protein or a portion thereof; peptidomimetics; vaccines; methods for vaccination and treatment of severe acute respiratory syndrome; antibodies; aptamers; and kits containing immunological compositions, or antibodies (or aptamers) that bind to the spike protein.
Owner:HEALTH & HUMAN SERVICES GOVERNMENT OF US SEC THE DEPT OF
Aptamer-based microfluidic chip capable of capturing cancer cells and preparation thereof as well as separation method of cancer cells
InactiveCN103087899AFast preparation methodAvidin immobilization method is simpleBioreactor/fermenter combinationsBiological substance pretreatmentsHigh cellAptamer
The invention discloses an aptamer-based microfluidic chip capable of capturing cancer cells, which is formed by reversibly sealing an upper-layer polymer chip and a lower-layer slide carrier, wherein a cell capturing channel is arranged between the upper-layer polymer chip and the lower-layer slide carrier; the bottom surface of the channel is rough; and avidin is fixed on the bottom surface. The preparation of the microfluidic chip comprises the steps of making a chip die by photoetching, and casting to obtain a polymer chip; processing the slide carrier with hydrofluoric acid and bonding with the polymer chip; and fixing avidin in the cell capturing channel to obtain a microfluidic chip. The microfluidic chip disclosed by the invention can be used for separating cancer cells; and the separation comprises the steps of adding biotin-modified aptamer into a sample, and incubating; and leading the mixed solution after the incubation into the microfluidic chip, wherein the target cancer cells in the mixed solution are combined with the avidin through the biotin to realize separation of the target cancer cells. The microfluidic chip disclosed by the invention has the advantages of high cell capturing efficiency, convenience in operation, strong generality, low cost and the like.
Owner:HUNAN UNIV
Circulating tumor cell detection chip and application thereof
InactiveCN105861297AReduce the probability of collisionImprove capture efficiencyBioreactor/fermenter combinationsBiological substance pretreatmentsAptamerBiology
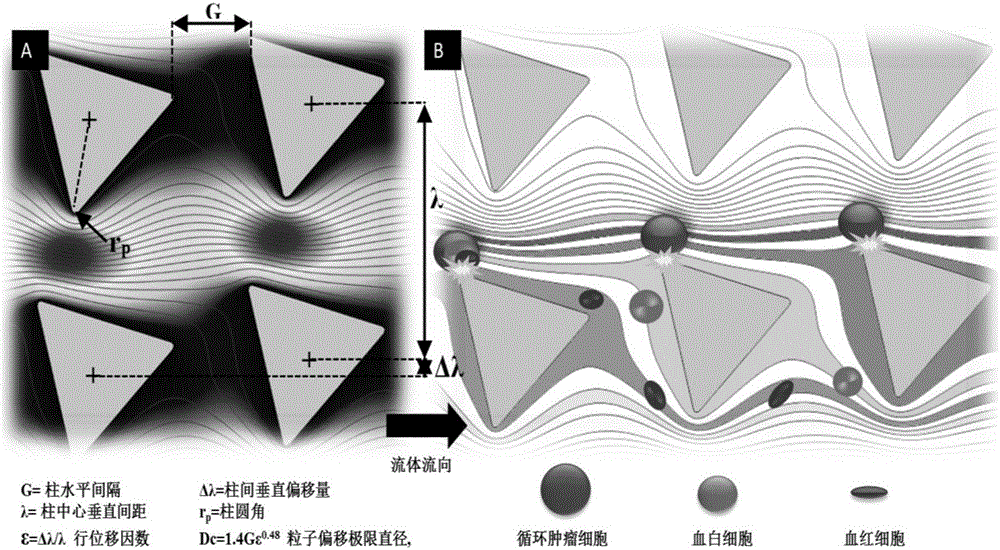
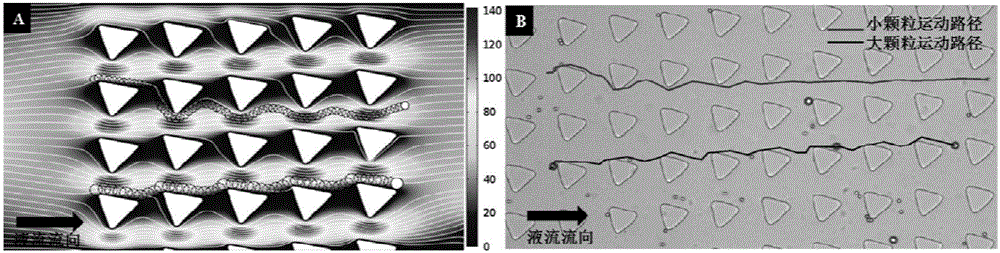
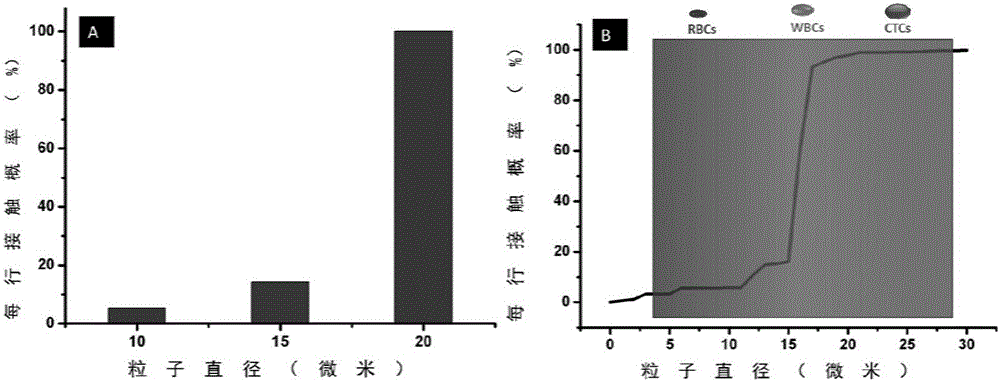
The invention relates to a circulating tumor cell detection chip and an application thereof. The above micro-fluidic chip is combined with deterministic lateral displacement and specific bio-affinity identification principles to realize synergistic high-efficiency and high-purity capturing and detection of circulating tumor cells in peripheral blood. A triangular microcolumn array modified with affinity identification molecules, such as a nucleic acid aptamer or antibody, and having a specific geometric arranging shape, is designed in the micro-chip. The rotating angle, the distance, the offset angle and other parameters of triangular arrays are regulated to control the flow behaviors of different dimensions going through micro-channels. The chip can realize three functions in order to improve the capture efficiency and the purity of the circulating tumor cells. A gas-driven sample introduction system is also designed in the invention, and can realize simultaneous control of a plurality of fluid paths through one gas pressure path.
Owner:WISDOM HEALTHY BIOTECH XIAMEN CO LTD
Aptamer capable of being in specific binding to TS/MDEP protein in gastric cancer cells
InactiveCN104818278ALow costSimple methodBiological testingDNA/RNA fragmentationAptamerProtein target
The invention discloses a targeted artificial TS / MDEP protein nucleic acid aptamer and a sequence thereof. A new combinatorial chemistry technology SELEX (Systematic Evolution of Ligands by Exponential Enrichment) is applied, the TS / MDEP protein serves as a target protein, and a DNA aptamer capable of being in specific binding to the TS / MDEP protein is screened from a single-chain RNA random library. The aptamer can form a special stem-loop structure in a random sequence area and can be in specific and high-affinity binding to the TS / MDEP protein or targeted binding to gastric cancer cells. The RNA aptamer provides a specific and efficient marker molecule for the gastric cancer diagnosis and treatment field, as well as a new choice for developing gastric cancer diagnosis reagents and gastric cancer treatment medicines.
Owner:刘红卫
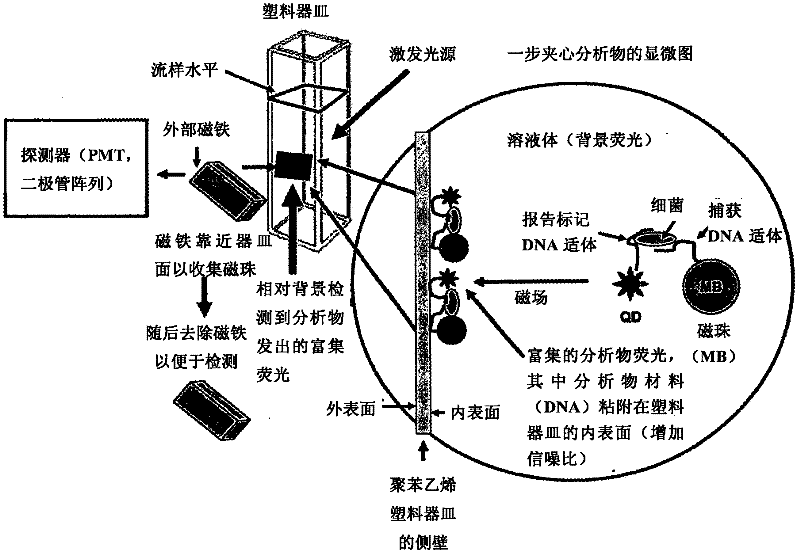
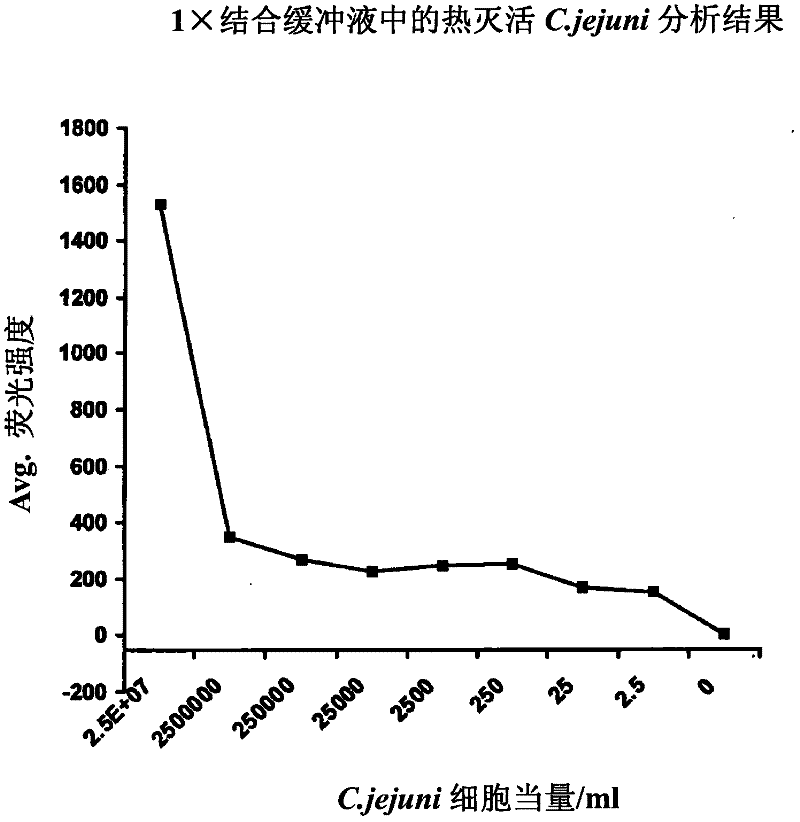
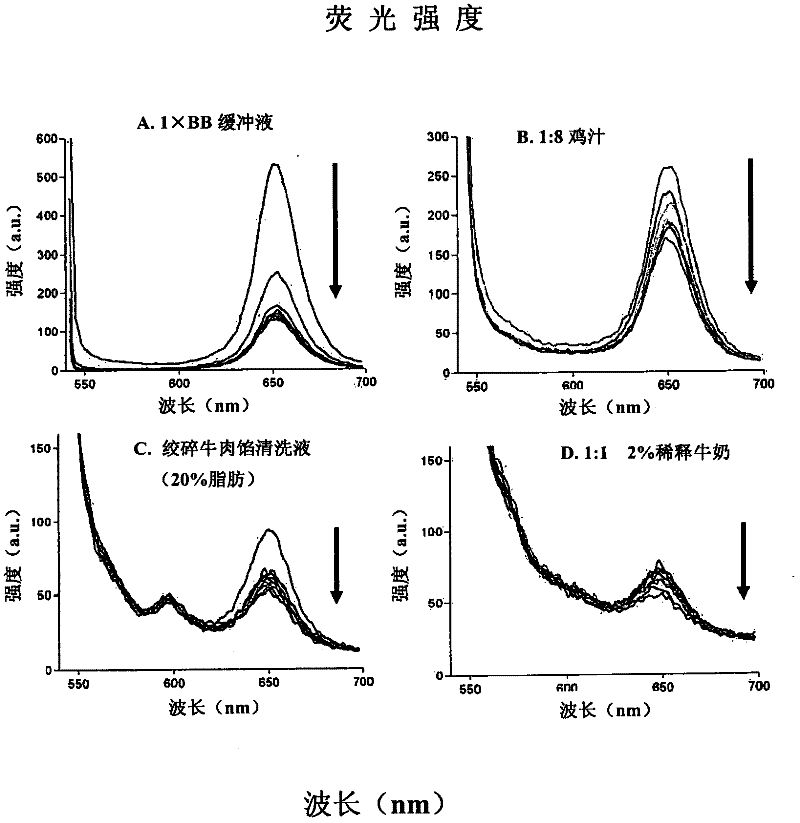
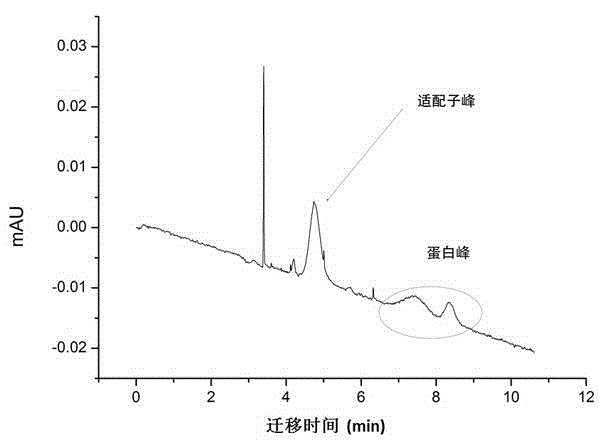
Methods of producing homogeneous plastic-adherent aptamer-magnetic bead-fluorophore and other sandwich assays
Methods are described for assembly of DNA aptamer-magnetic bead ('MB') conjugate plus aptamer-quantum dot ('QD') aptamer-fluorescent nanoparticle or other aptamer-fluorophore, aptamer-chemiluminescent reporter, aptamer-radioisotope or other aptamer-reporter conjugate sandwich assays that enable adherence to glass, polystyrene and other plastics. Adherence to glass or plastics enables detection of surface-concentrated partitioning of fluorescence versus background (bulk solution) fluorescence in one step (without a wash step) even when the external magnetic field for concentrating the assay is removed. This assay format enables rapid, one-step (homogeneous) assays for a variety of analytes without wash steps that do not sacrifice sensitivity.
Owner:OTC BIOTECH
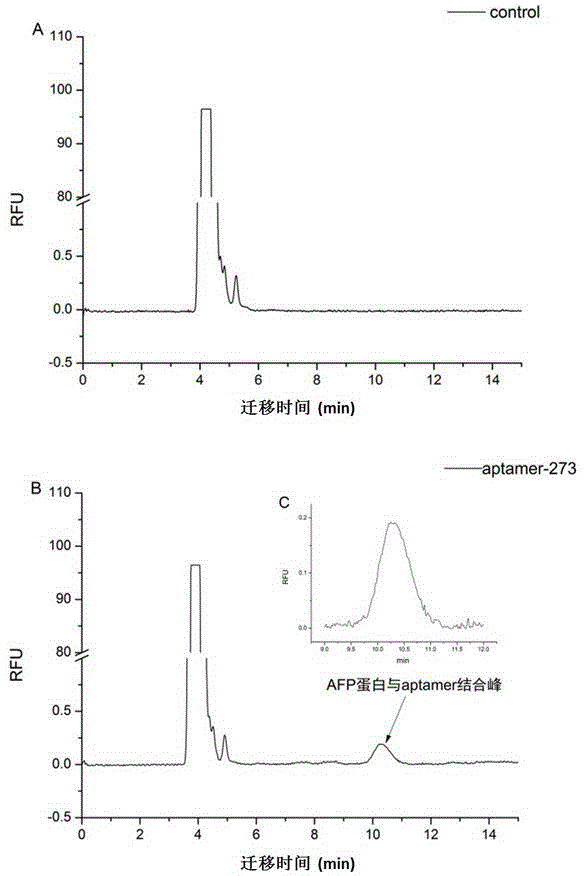
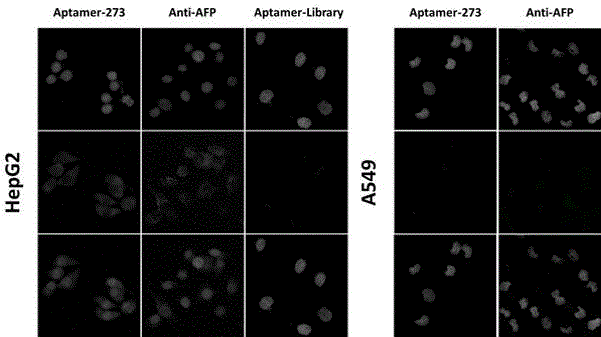
Method for screening aptamer specifically bound with alpha-fetoprotein
InactiveCN104131105AStrong interactionIncrease workloadMicrobiological testing/measurementDNA preparationAptamerFetuin b
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV +1
Enhanced biologically active conjugates
InactiveUS20050260651A1Improve propertiesImprove abilitiesOrganic active ingredientsSenses disorderAptamerComparable size
The invention provides compositions and methods for making and using sterically enhanced antagonist aptamer conjugates that include a nucleic acid sequence having a specific affinity for a target molecule and a soluble, high molecular weight steric group that augments or facilitates the inhibition of binding to, or interaction with, the target molecule binding partner by the target molecule when bound to the aptamer conjugate. The present invention also provides methods and formulations for ocular delivery of a biologically active molecule by attaching a charged moiety to the biologically active molecule and delivering the biologically active molecule by iontophoresis. Iontophoresis of a biologically active molecule that is conjugated to a high molecular weight neutral moiety, in enhanced by substituting the high molecular weight neutral moiety with a charged molecule of comparable size.
Owner:EYETECH
Identification and use of effectors and allosteric molecules for the alteration of gene expression
InactiveUS20050239061A1Eliminate the problemLimited utilityMicrobiological testing/measurementDrug compositionsCatalytic RNACatalytic function
The present invention relates to the construction of an allosteric control module in which a catalytic RNA forms a part of or is linked to an effector-binding RNA domain or aptamer. These constructs place the activity of the catalytic RNA under the control of the effector and require the presence of an appropriate effector for activation or inactivation. The present invention provides means to identify useful effector molecules as well as their use to evolve cognate aptamers. The invention involves both the evolution of RNA sequences which bind the effector and a selection proces in which the allosteric control modules are identified by their catalytic function in the presence and absence of the effector. The resulting regulatable catalytic RNAs may be used to alter the expression of a target RNA molecule in a controlled fashion.
Owner:AMGEN INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com