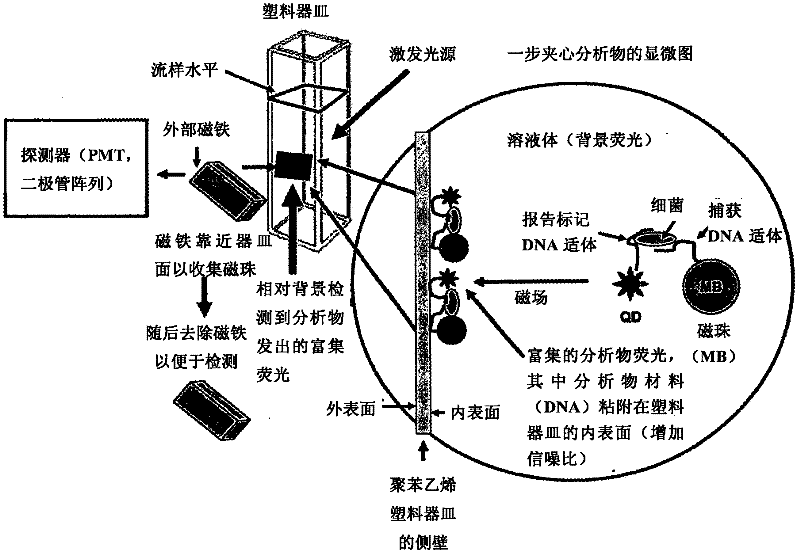
Methods of producing homogeneous plastic-adherent aptamer-magnetic bead-fluorophore and other sandwich assays
A sandwich analysis, fluorophore technology, applied in biochemical equipment and methods, microbial determination/inspection, biological testing, etc., can solve problems such as not very good effect, not very good adhesion effect, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
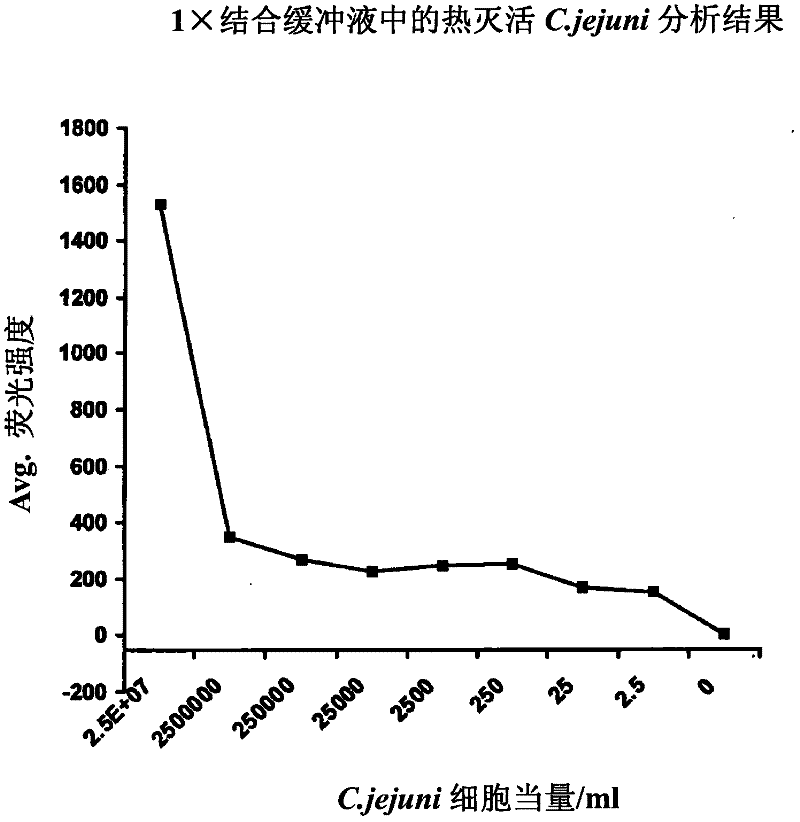
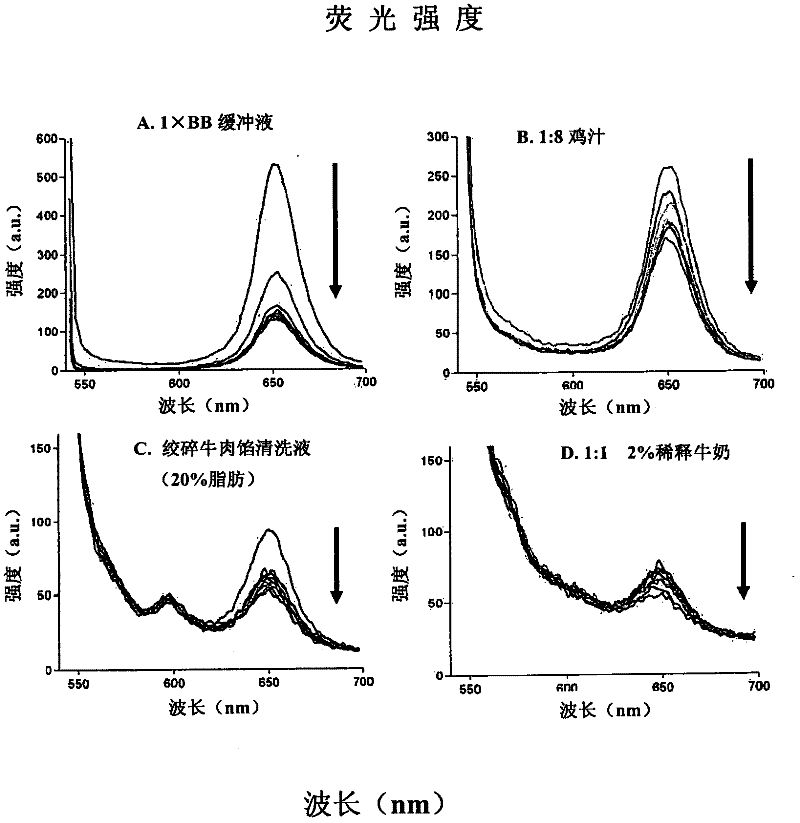
[0055] One-step wash-free ultrasensitive detection of Campylobacter jejuni (C. jejuni) in various food matrices
[0056] The present invention is used to detect such as Figure 2-4 Pure buffers and various food matrices shown contained as few as 2 live or dead cells of C. jejuni (a common foodborne pathogen). Among these analytes, there were two distinct C. jejuni sequences (represented by C2 and C3 or SEQ ID NO.2 and SEQ ID NO.3) whose 2'-amino groups were modified during solid-phase DNA synthesis , and then combined with 1,000 tosyl-M280 (diameter 2.8 μm) Dynal (Invitrogen) magnetic beads or 0.24 pL Q-dot 655ITK reagent (Invitrogen) for each test. The C2 aptamer (SEQ ID 2) is used for capture on the surface of tosyl-magnetic beads, and the C3 aptamer (SEQ ID NO.3) is passed through BS 3 (bis-suberate bifunctional linker from Pierce Chemical Co.) was used as a reporter reagent after conjugation with Q-dot 655ITK reagent. These reagents are purified, mixed, and lyophilized ...
Embodiment 2
[0058] One-step wash-free ultrasensitive detection of Escherichia coli O157:H7 and other toxin-producing (shiga or vero toxin) E. coli in various food matrices
[0059] By combining the aptamer with the outer sugar of O157 lipopolysaccharide (LPS) and the H7 flagellar antigen, the invention can be used to detect enterohemorrhagic E. coli O157:H7 in various foods or on the surface of foods. Aptamer sequences from SEQ ID NOs.7-20 can be used for capture (aptamer-magnetic bead conjugate) or reporter (aptamer-fluorophore conjugate) function, and for detecting E. coli O157:H7. Alternatively, outer membrane proteins (OMPs) shared by many E.coli can also be used for aptamer-magnetic bead capture (or identification) of E.coli bacterial cells, followed by LPS-specific aptamer-quantum dot The reporter reagent is used for specific identification of E.coli strains or serotypes. The aptamer SEQ ID NOs.279-322 can be used for the recognition and capture of E.coli OMP. In another example,...
Embodiment 3
[0061] One-step no-wash ultrasensitive detection of Listeria monocytogenes (Listeria monocytogens) in various food matrices
[0062] By combining with listerolysin (listerolysin, LO) surface protein, the present invention can be used to detect deadly bacteria L. monocytogens in various foods or on the surface of foods. Aptamer sequences from SEQ ID NOs.21-52 can be selected for capture (aptamer-magnetic bead conjugate) or reporter (aptamer-fluorophore conjugate) function, and for detection of LO and L. monocytogens.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com