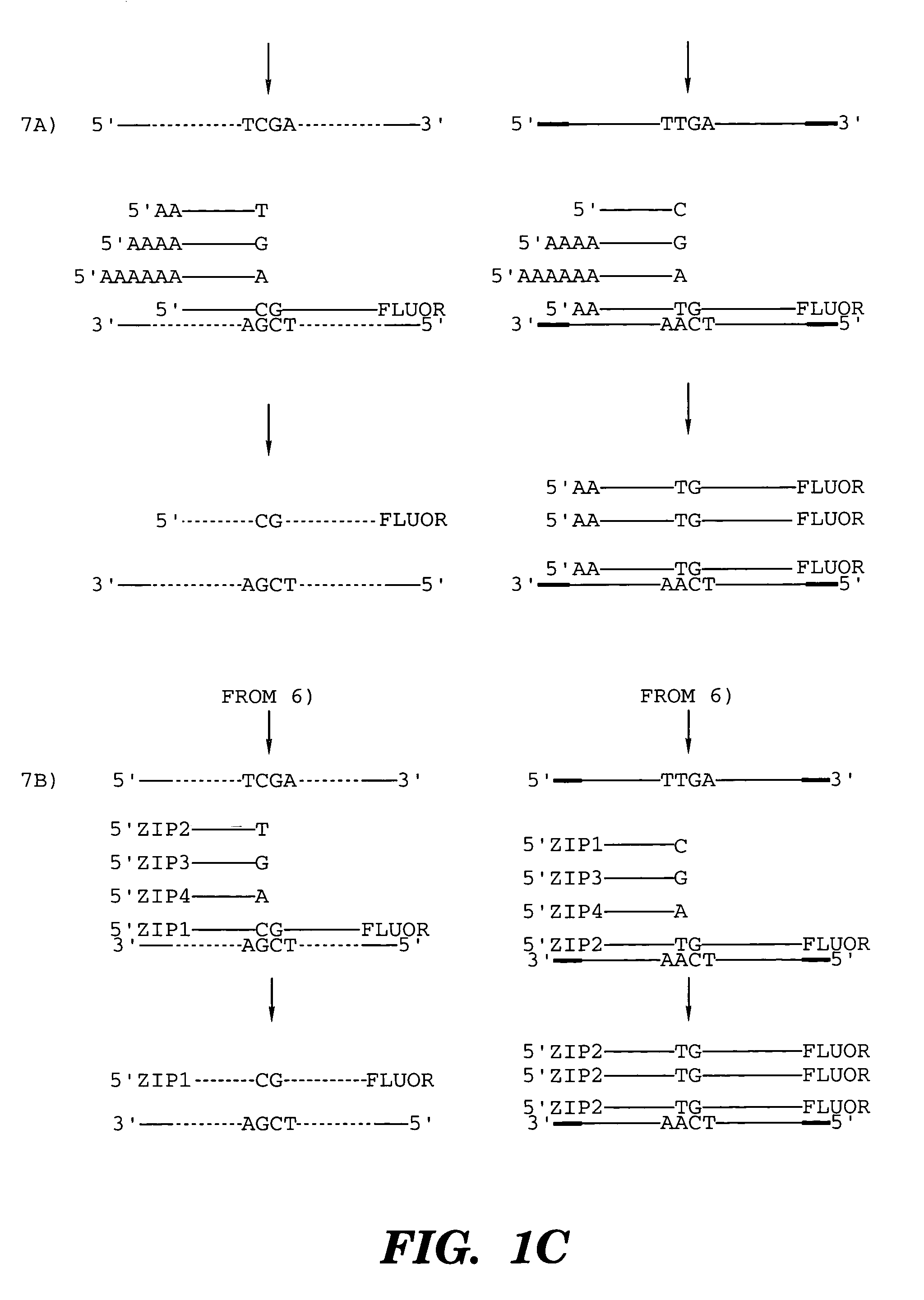
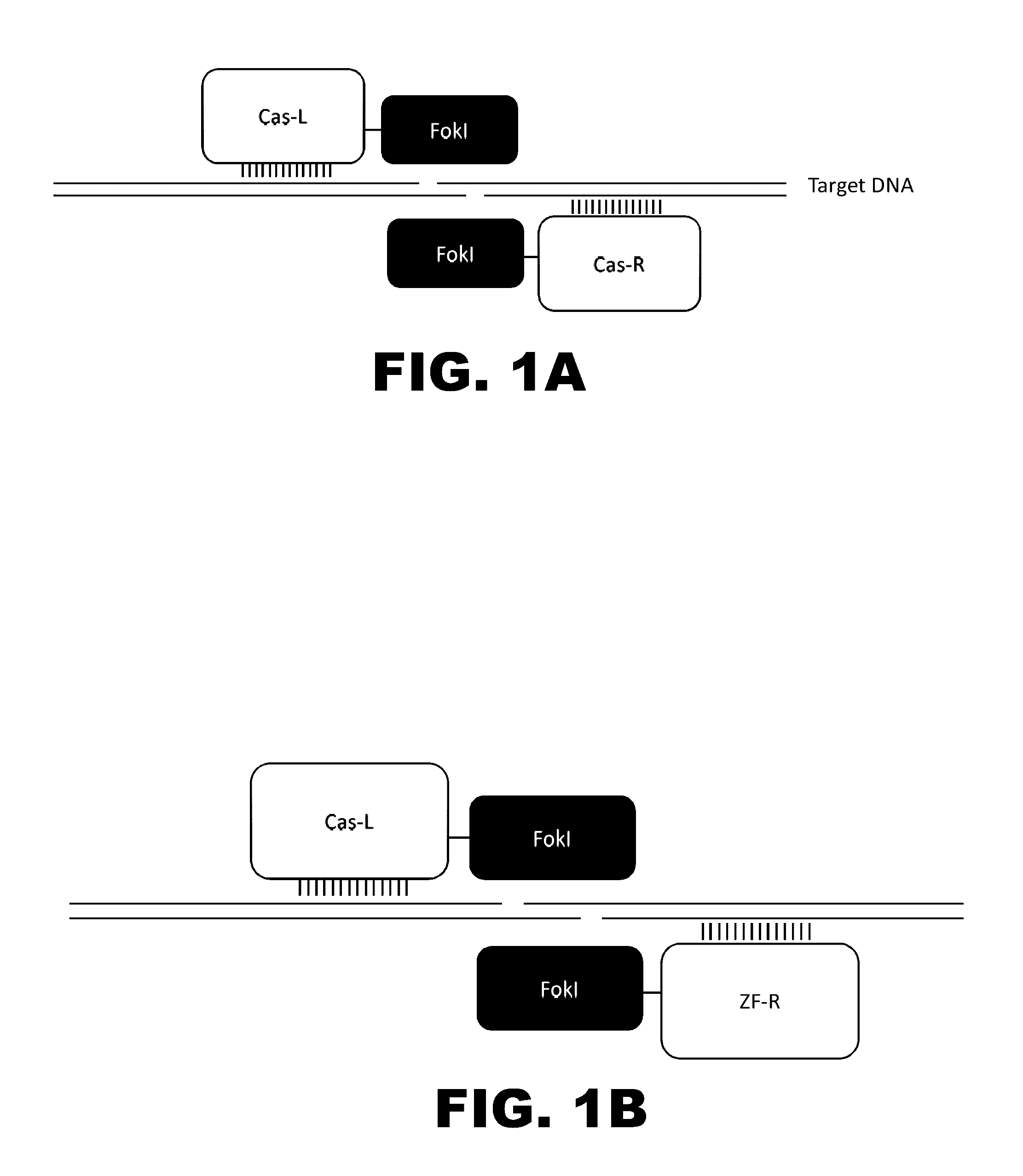
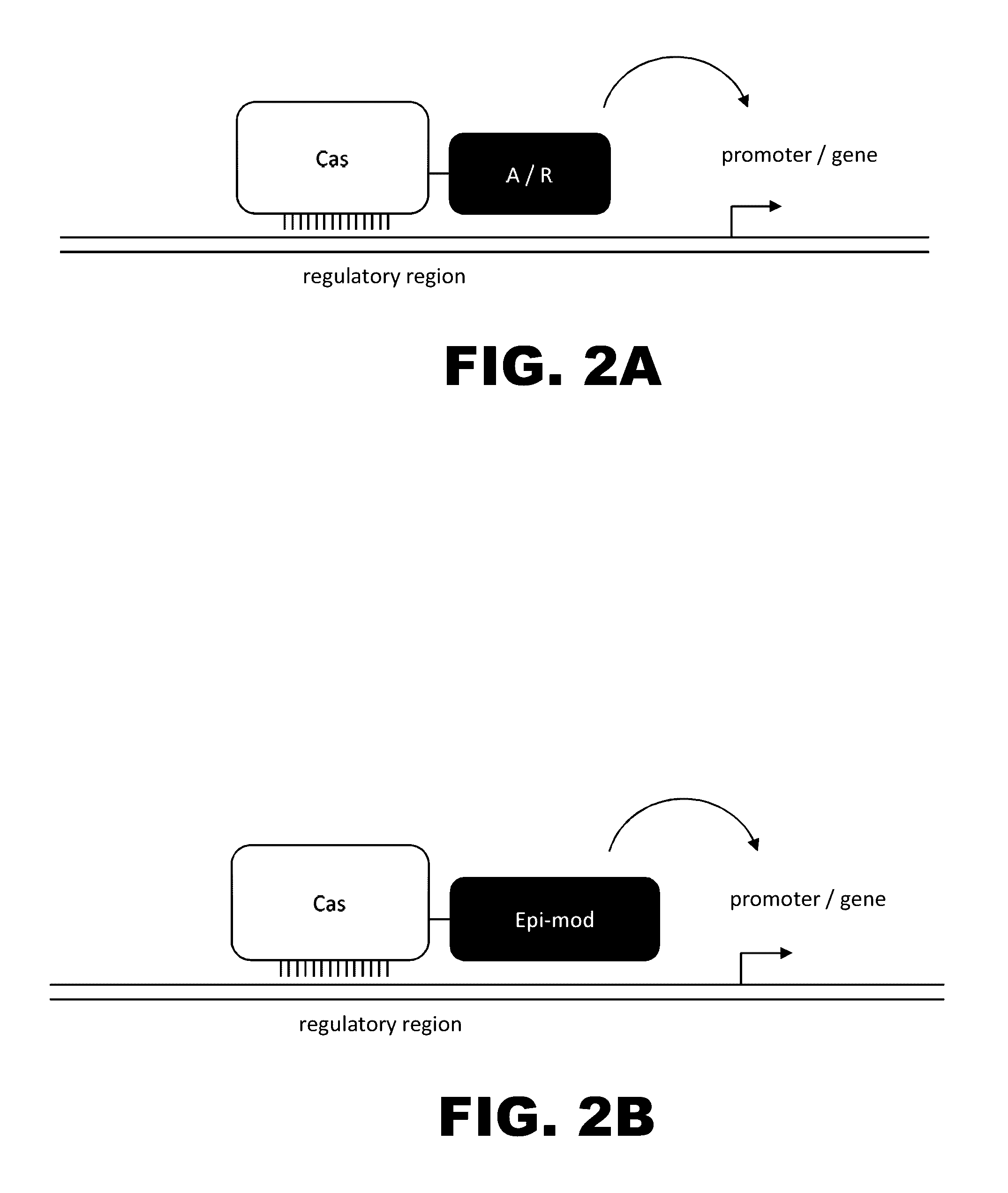
Patents
Literature
1126 results about "DNA Endonuclease" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Endonucleases play a role in DNA repair. AP endonuclease, specifically, catalyzes the incision of DNA exclusively at AP sites, and therefore prepares DNA for subsequent excision, repair synthesis and DNA ligation. For example, when depurination occurs, this lesion leaves a deoxyribose sugar with a missing base.
Modified nucleotide compounds
ActiveUS7495088B1Increased nuclease resistanceSugar derivativesMicrobiological testing/measurementBiologyModified nucleosides
Disclosed is a nuclease resistant nucleotide compound capable of hybridizing with a complementary RNA in a manner which inhibits the function thereof, which modified nucleotide compound includes at least one component selected from the group consisting of MN3M, B(N)xM and M(N)xB wherein N is a phosphodiester-linked modified 2′-deoxynucleoside moiety; M is a moiety that confers endonuclease resistance on said component and that contains at least one modified or unmodified nucleic acid base; B is a moiety that confers exonuclease resistance to the terminus to which it is attached; and x is an integer of at least 2.
Owner:ENZO BIOCHEM
PROKARYOTIC RNAi-LIKE SYSTEM AND METHODS OF USE
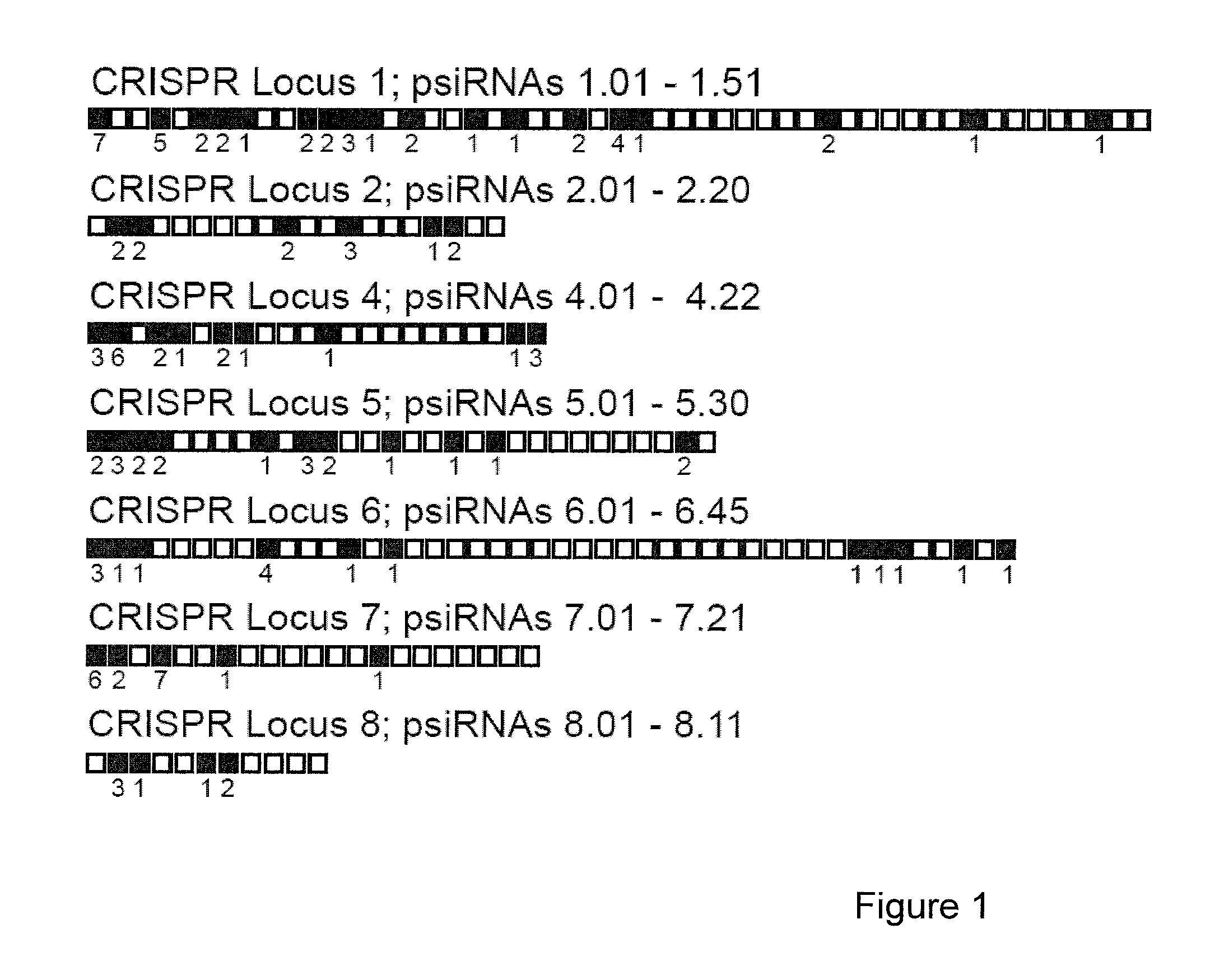
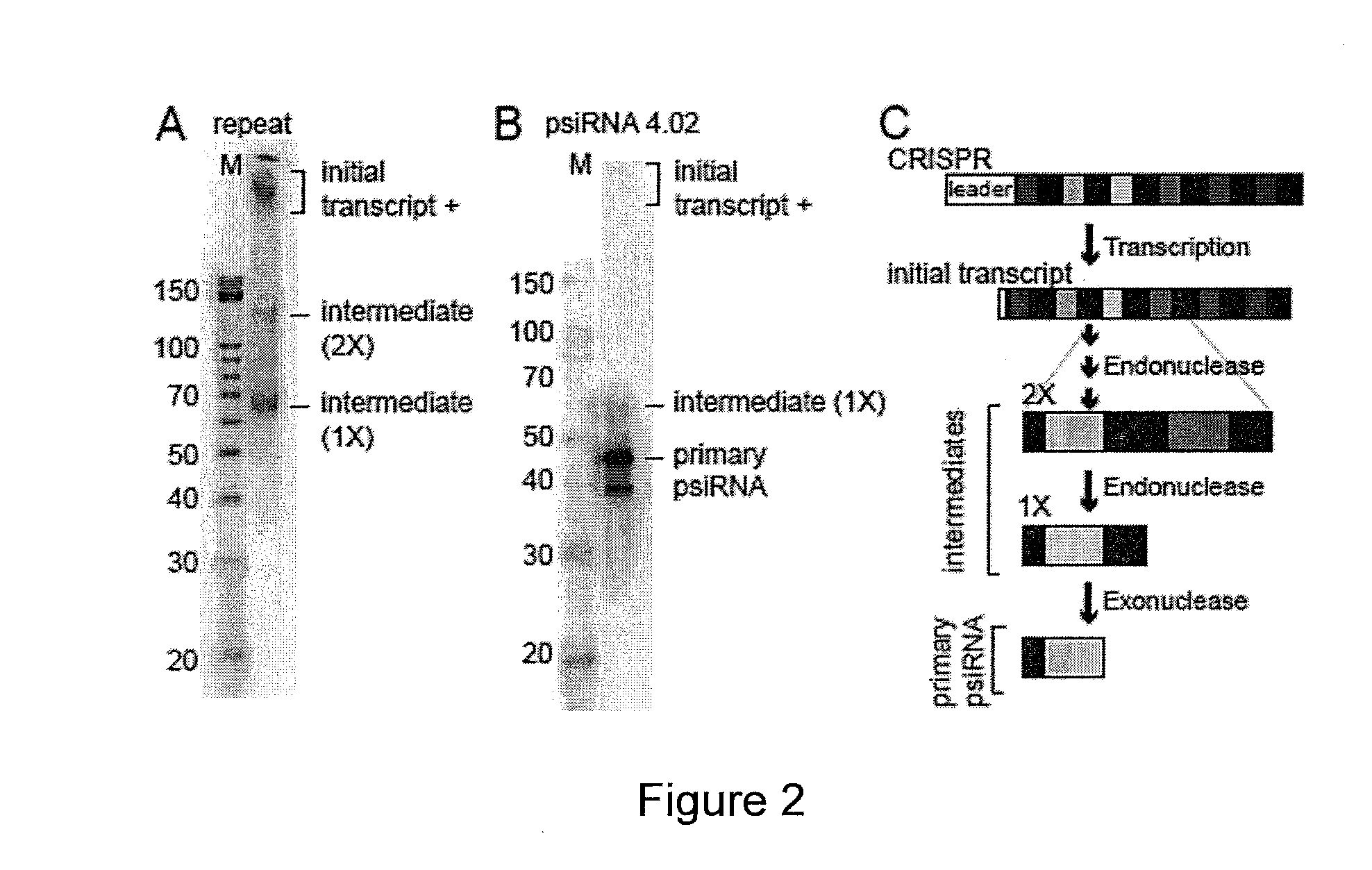
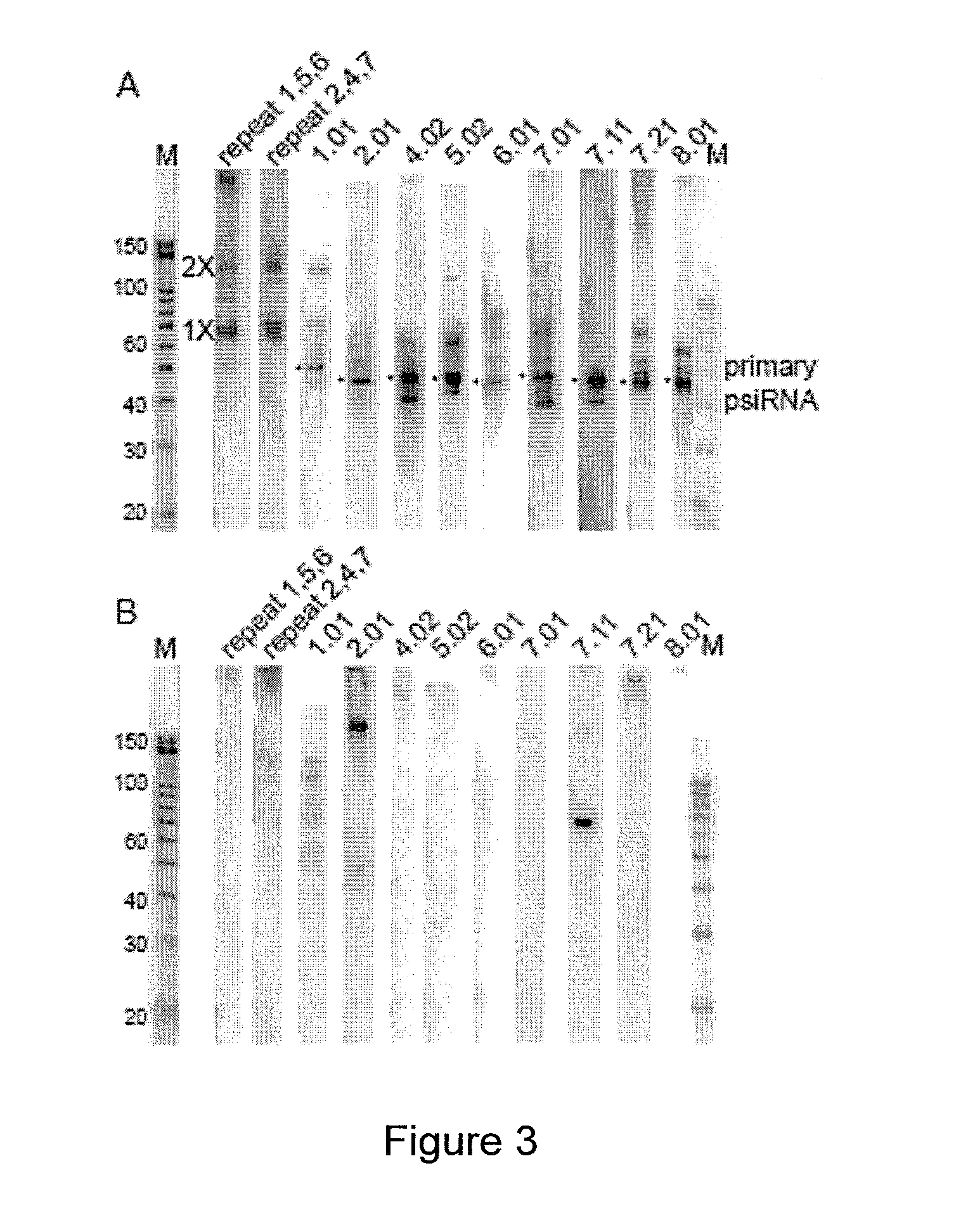
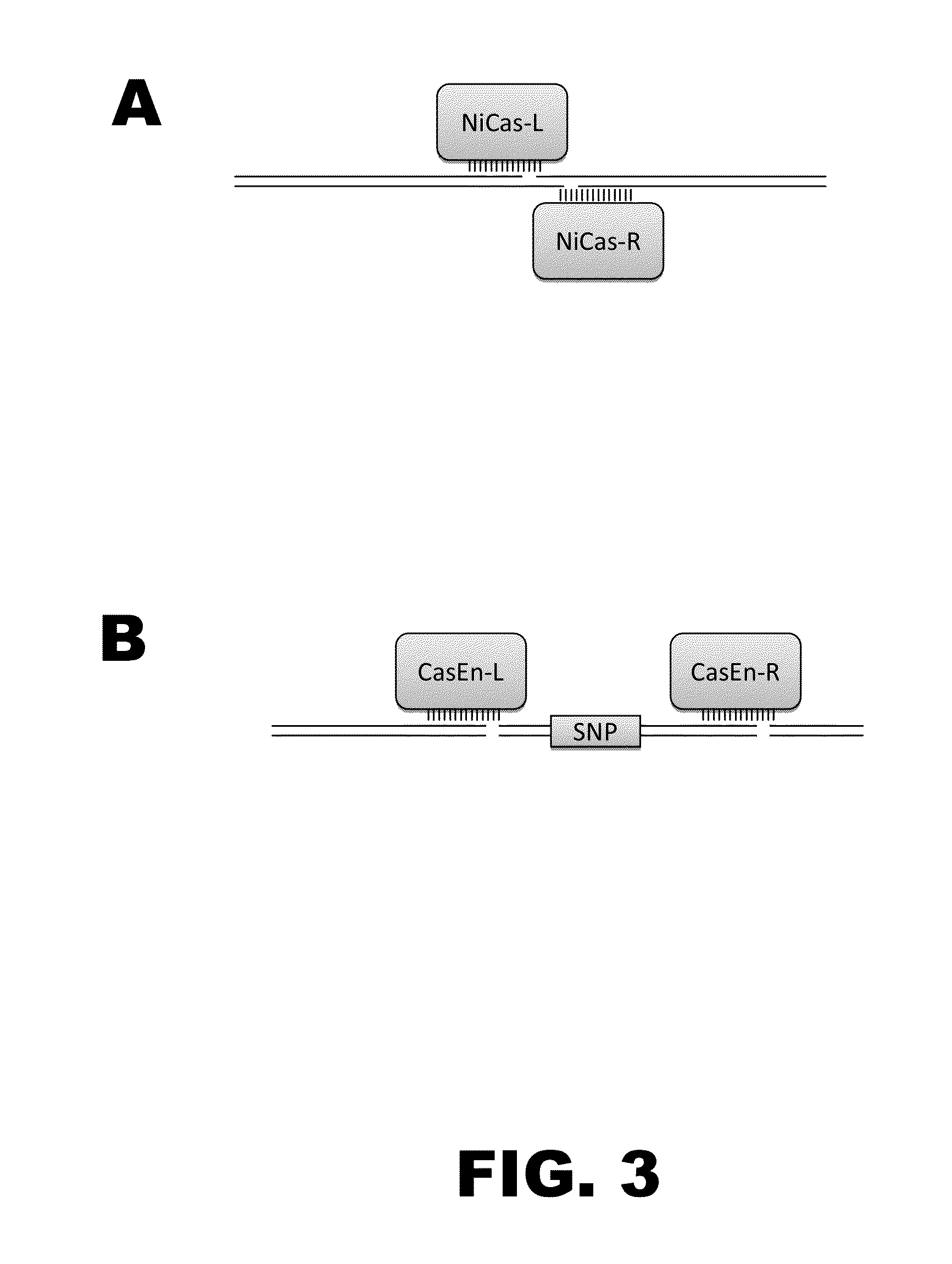
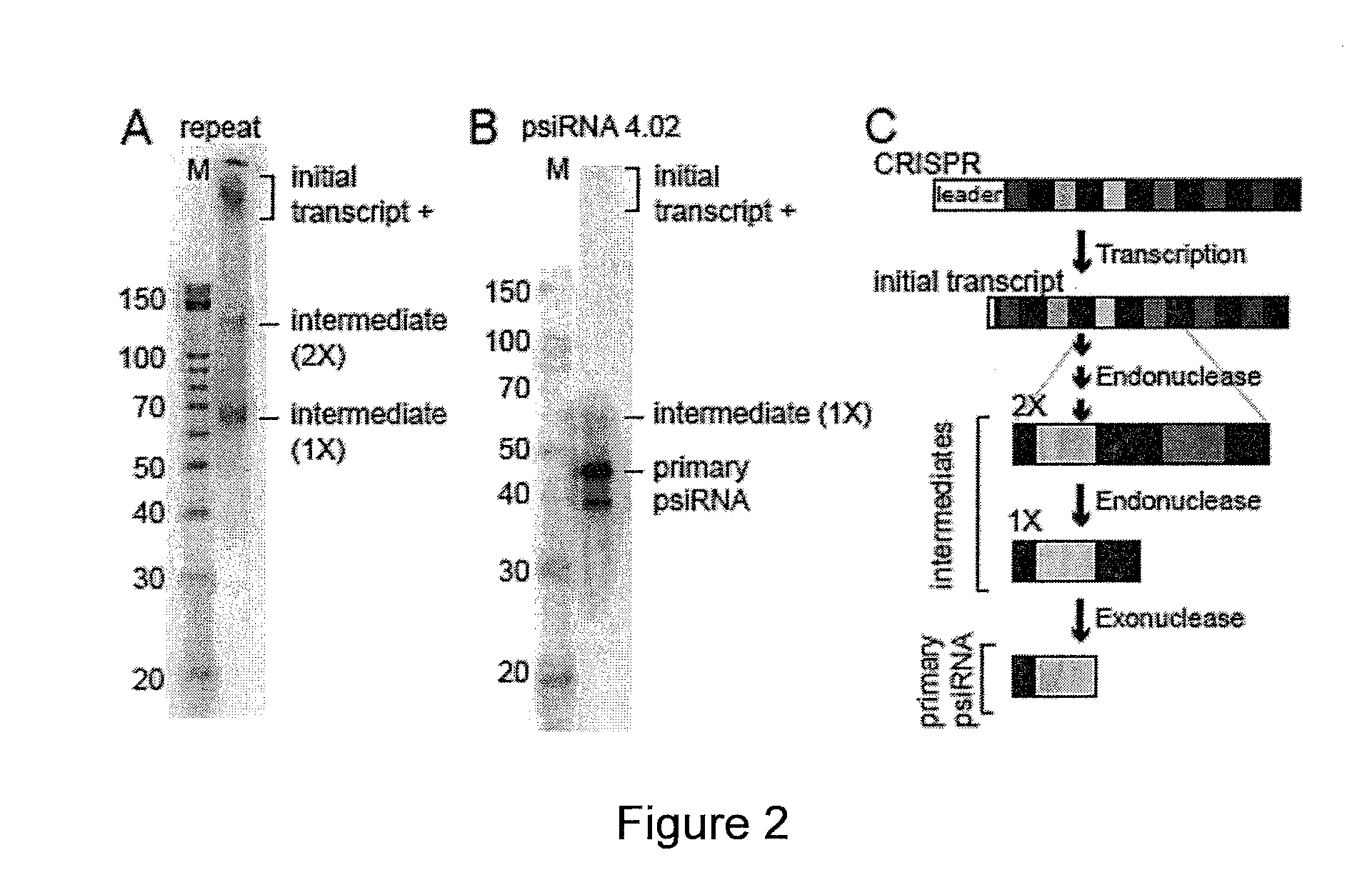
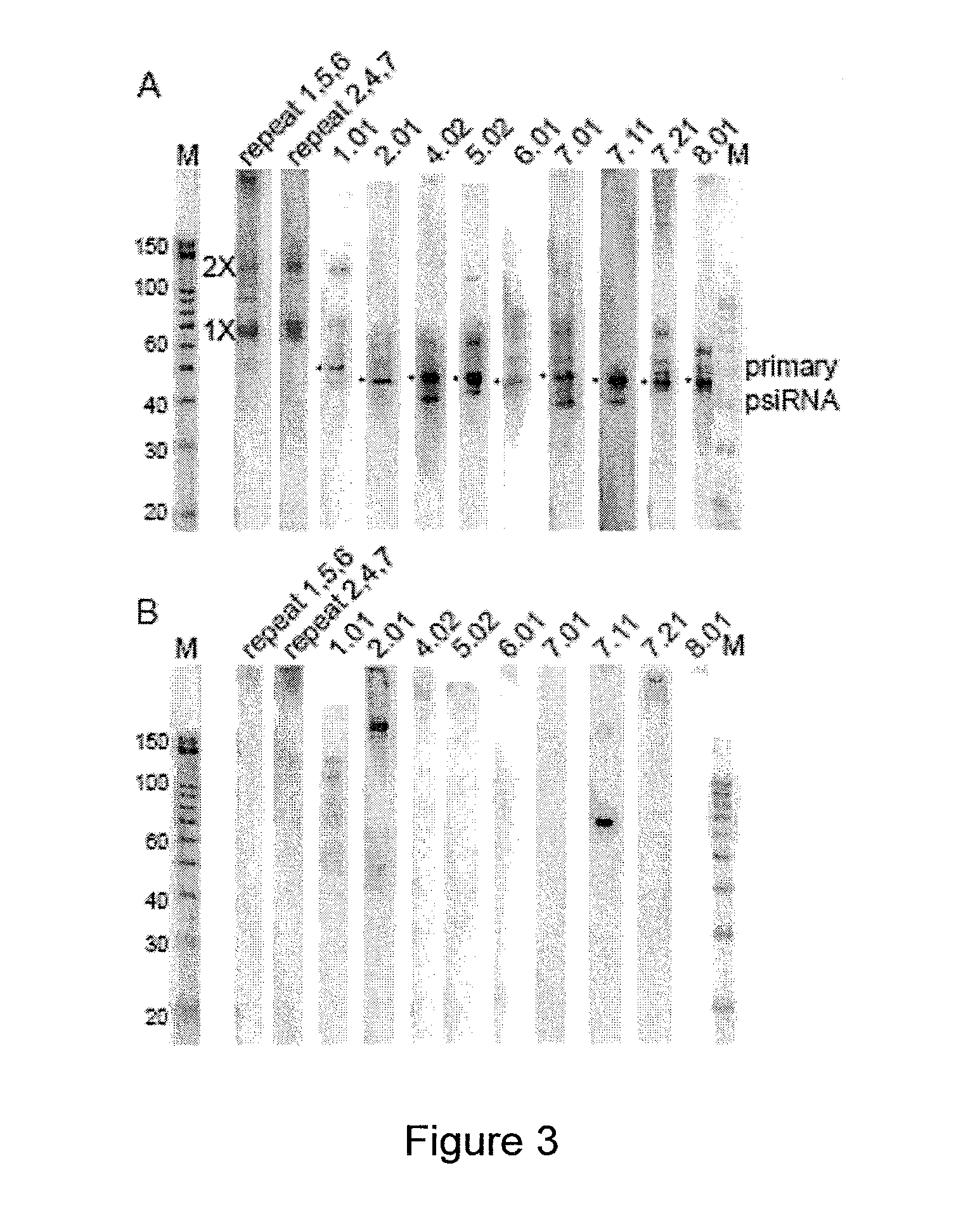
Provided herein are methods for inactivating a target polynucleotide. The methods use a psiRNA having a 5′ region and a 3′ region. The 5′ region includes, but is not limited to, 5 to 10 nucleotides chosen from a repeat from a CRISPR locus immediately upstream of a spacer. The 3′ region is substantially complementary to a portion of the target polynucleotide. The methods may be practiced in a prokaryotic microbe or in vitro. Also provided are polypeptides that have endonuclease activity in the presence of a psiRNA and a target polynucleotide, and methods for using the polypeptides.
Owner:UNIV OF GEORGIA RES FOUND INC
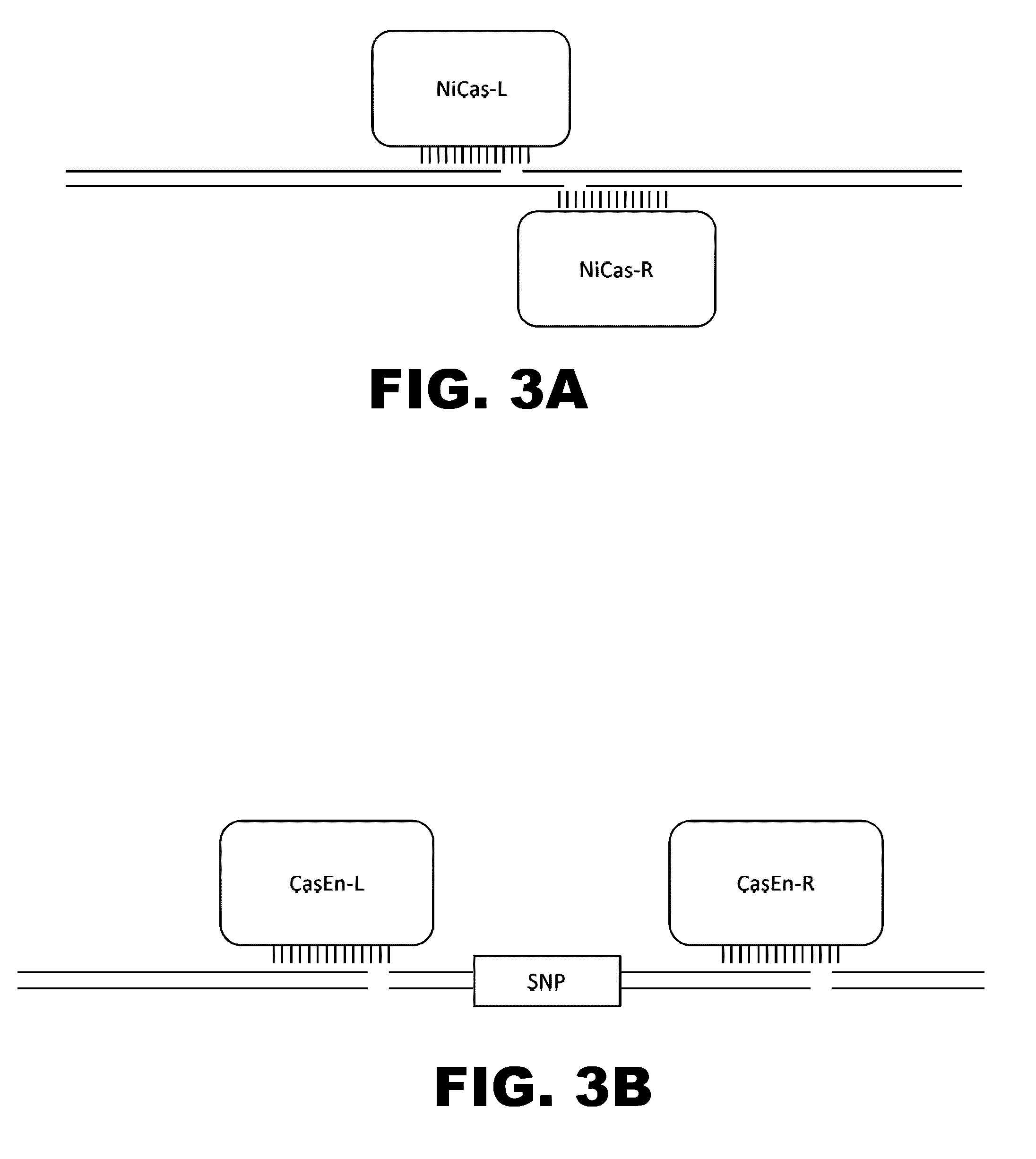
Crispr-based genome modification and regulation
InactiveUS20140273230A1Polypeptide with localisation/targeting motifHydrolasesGenomeDNA Endonuclease
Owner:SIGMA ALDRICH CO LLC
Prokaryotic RNAi-like system and methods of use
Owner:UNIV OF GEORGIA RES FOUND INC
Method and compositions for ordering restriction fragments
InactiveUS7598035B2High density physicalPrecise positioningSugar derivativesMicrobiological testing/measurementPhysical MapsNucleotide sequencing
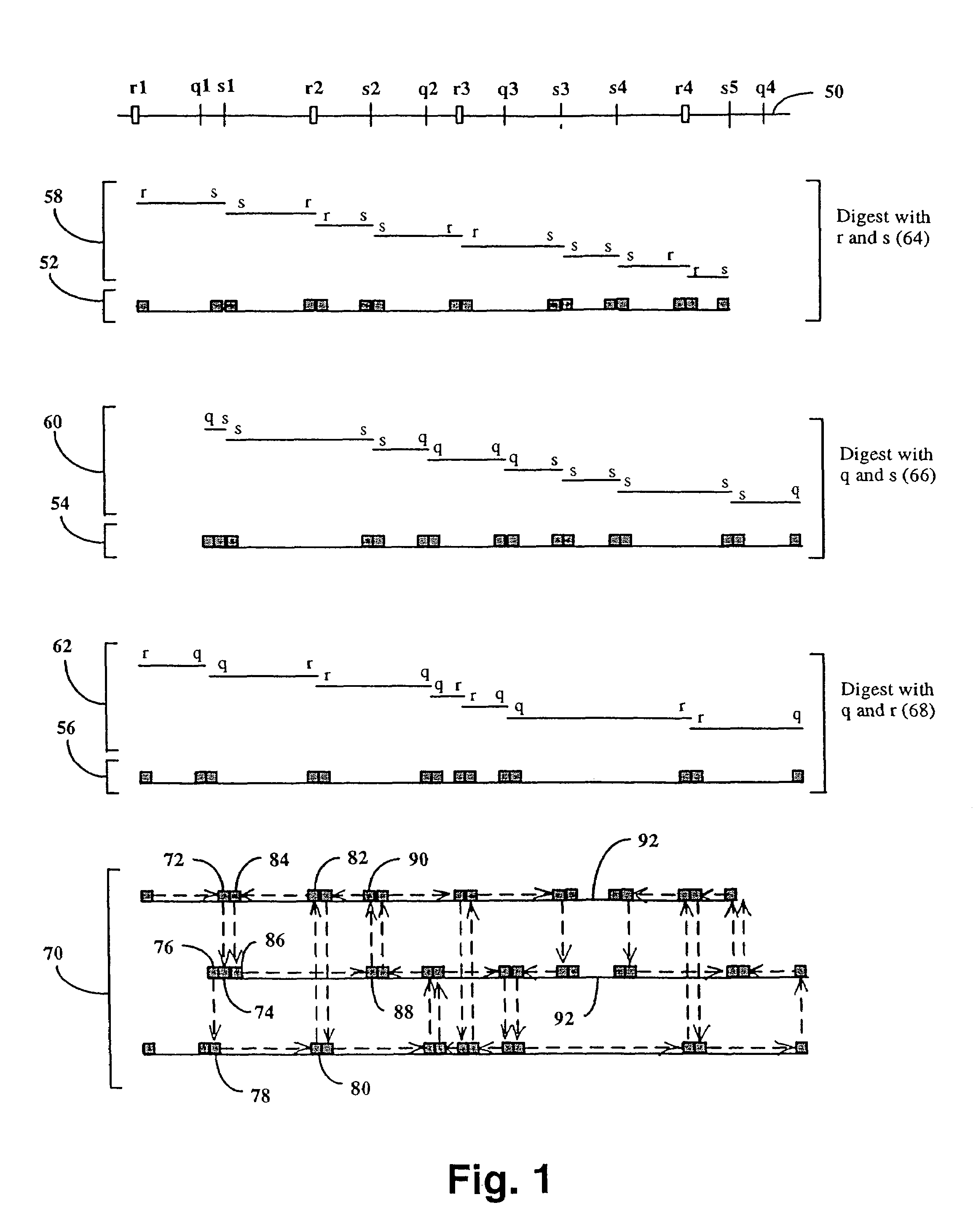
The invention provides a method for constructing a high resolution physical map of a polynucleotide. In accordance with the invention, nucleotide sequences are determined at the ends of restriction fragments produced by a plurality of digestions with a plurality of combinations of restriction endonucleases so that a pair of nucleotide sequences is obtained for each restriction fragment. A physical map of the polynucleotide is constructed by ordering the pairs of sequences by matching the identical sequences among the pairs.
Owner:ILLUMINA INC
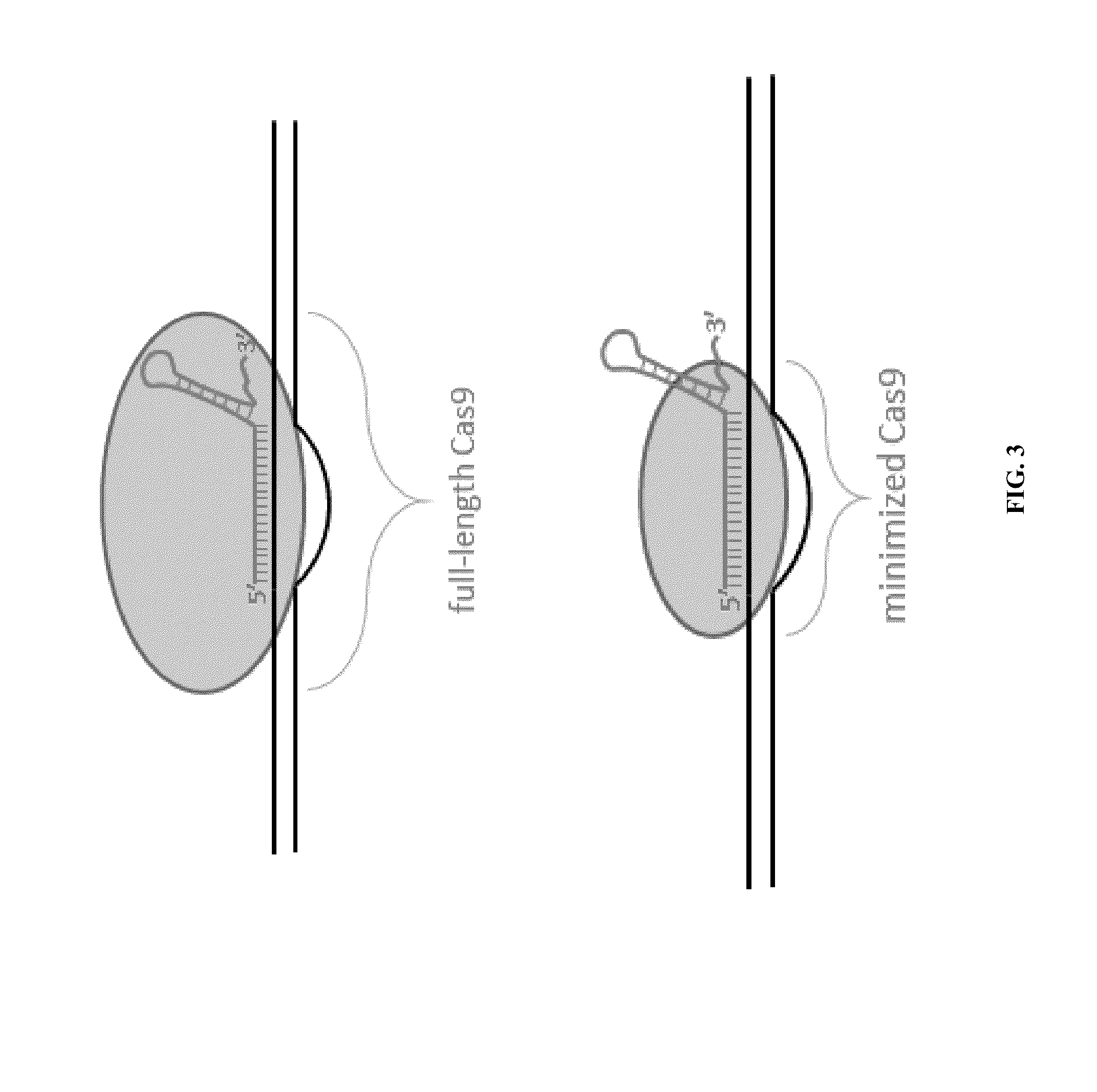
Cas9-recombinase fusion proteins and uses thereof
ActiveUS20150071898A1Strong specificityReduce the possibilityFusion with DNA-binding domainBacteriaSite-specific recombinationResearch setting
Some aspects of this disclosure provide compositions, methods, and kits for improving the specificity of RNA-programmable endonucleases, such as Cas9. Also provided are variants of Cas9, e.g., Cas9 dimers and fusion proteins, engineered to have improved specificity for cleaving nucleic acid targets. Also provided are compositions, methods, and kits for site-specific recombination, using Cas9 fusion proteins (e.g., nuclease-inactivated Cas9 fused to a recombinase catalytic domain). Such Cas9 variants are useful in clinical and research settings involving site-specific modification of DNA, for example, genomic modifications.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
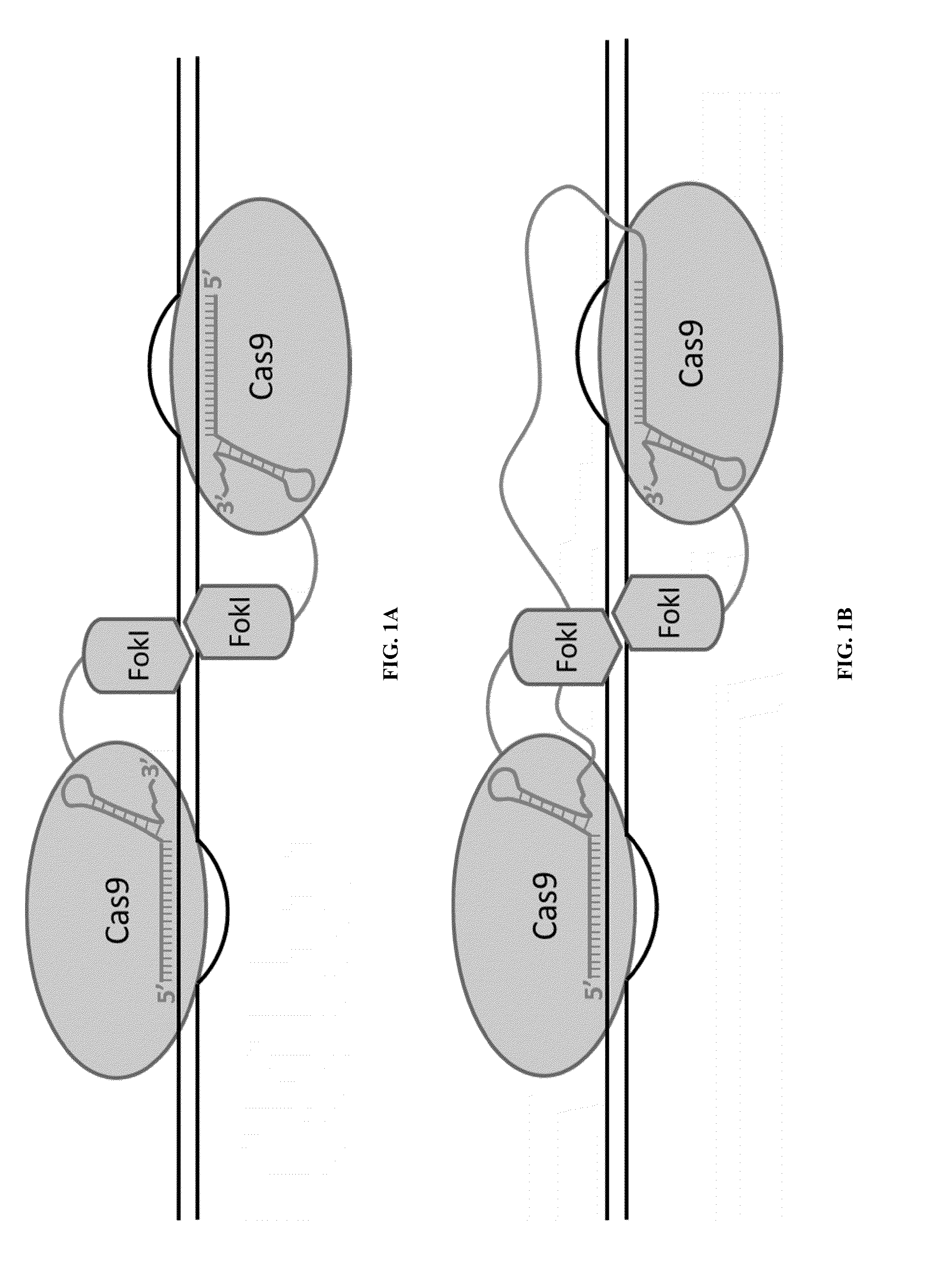
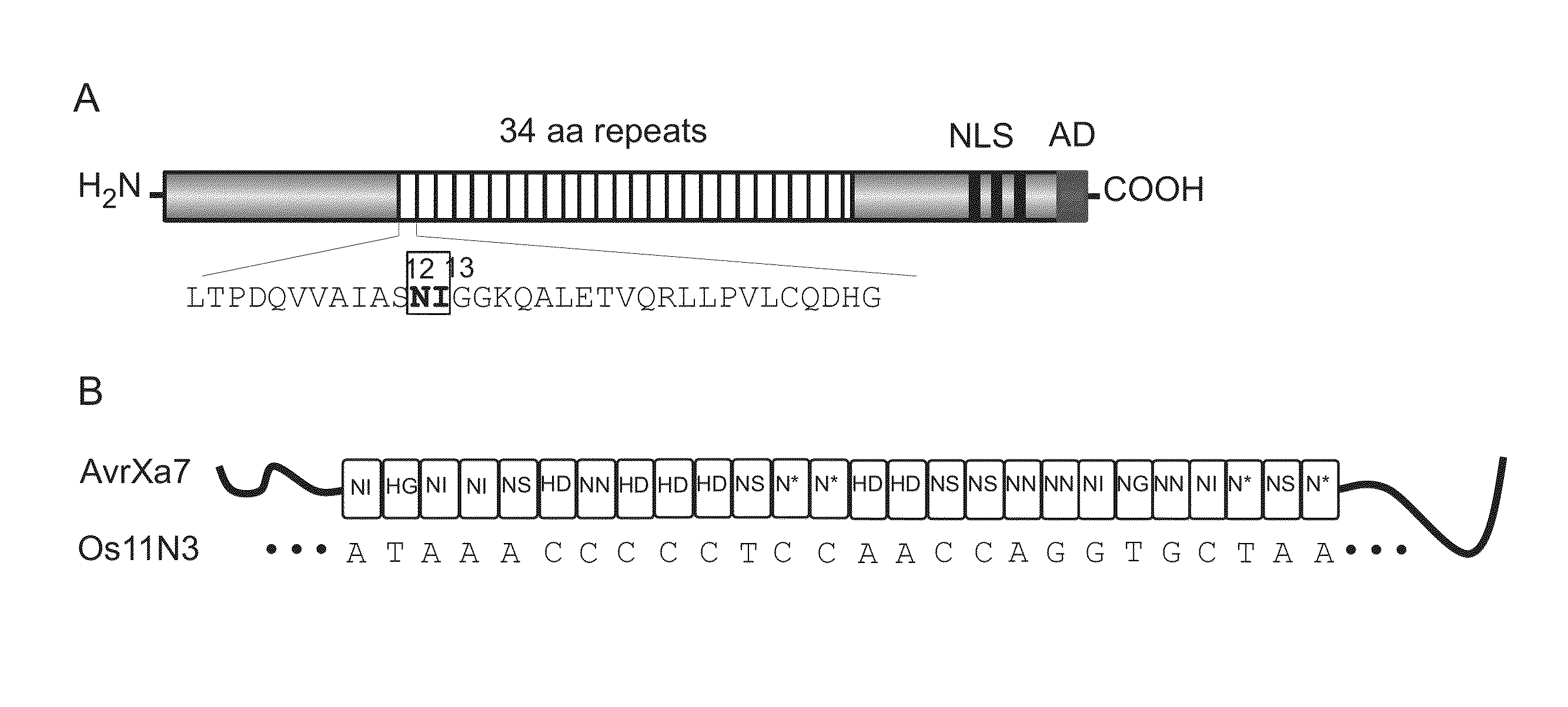
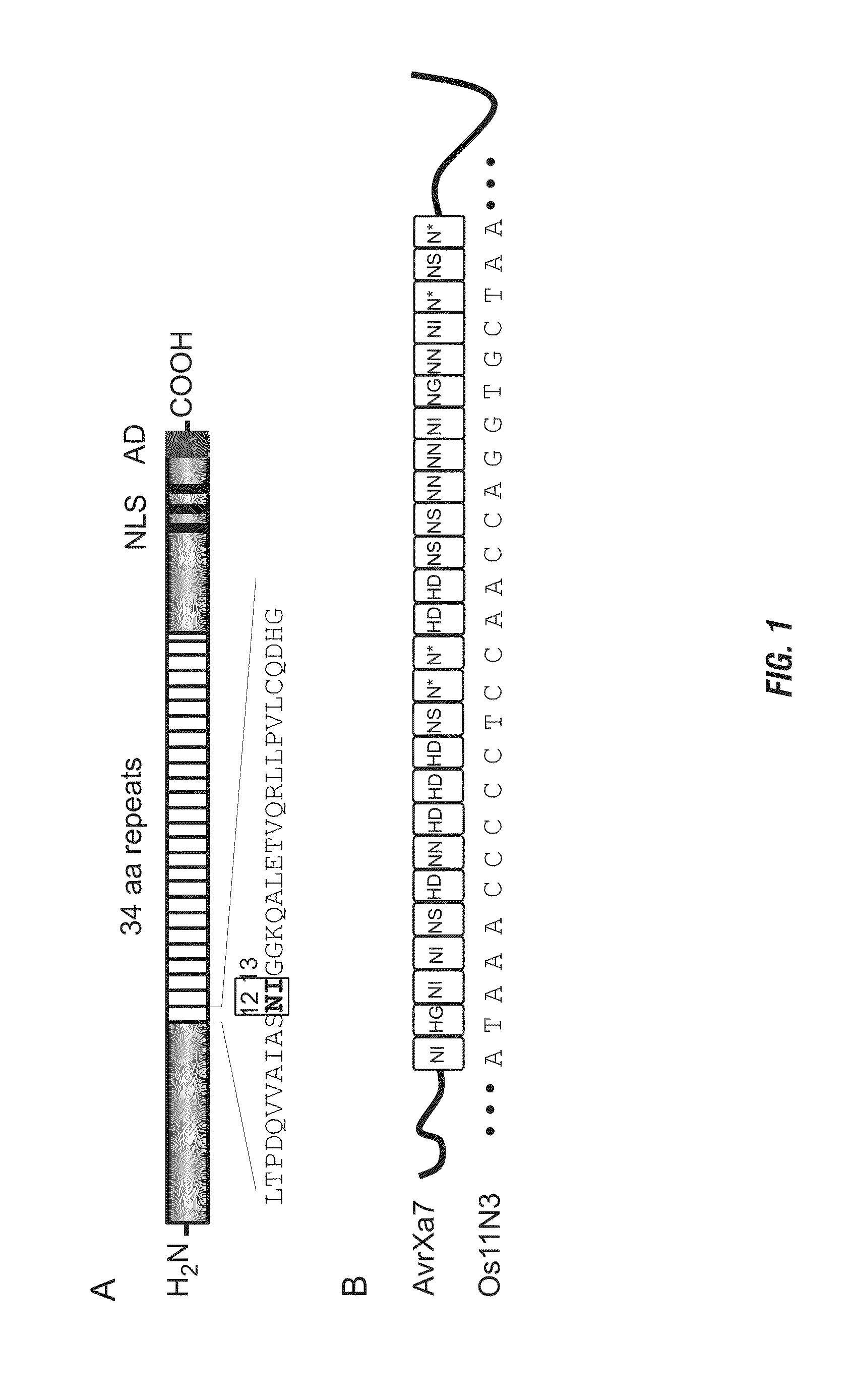
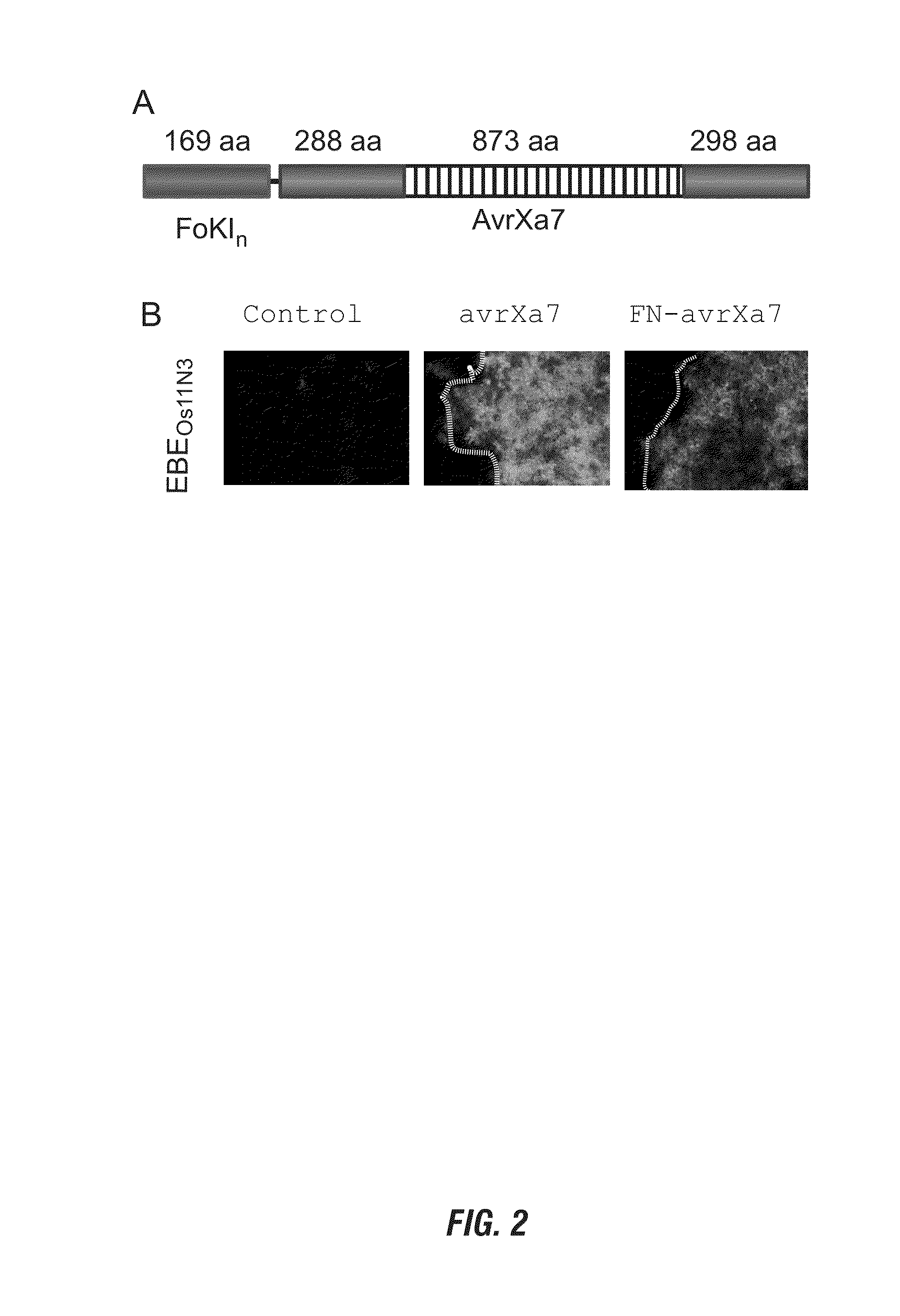
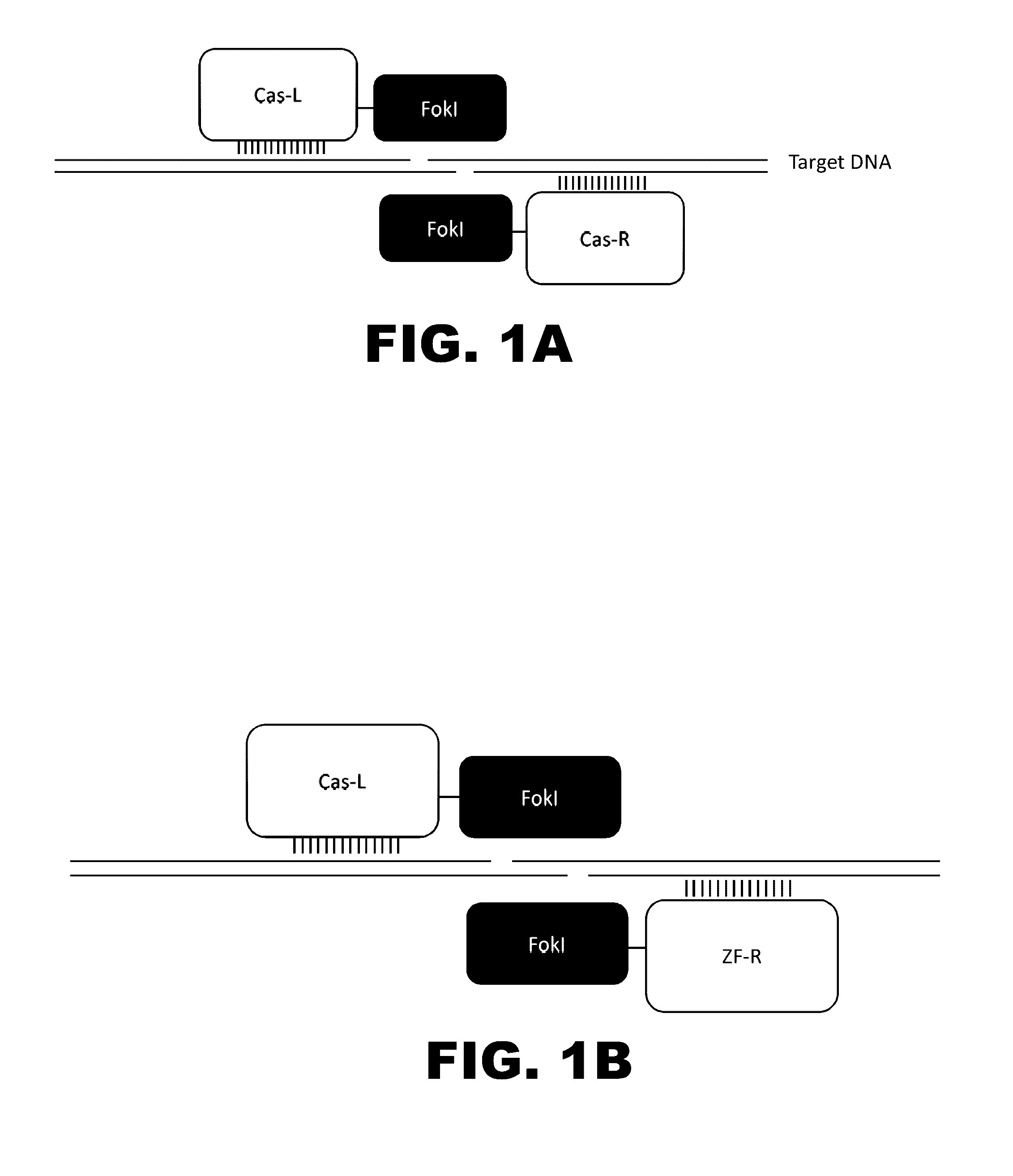
Nuclease activity of tal effector and foki fusion protein
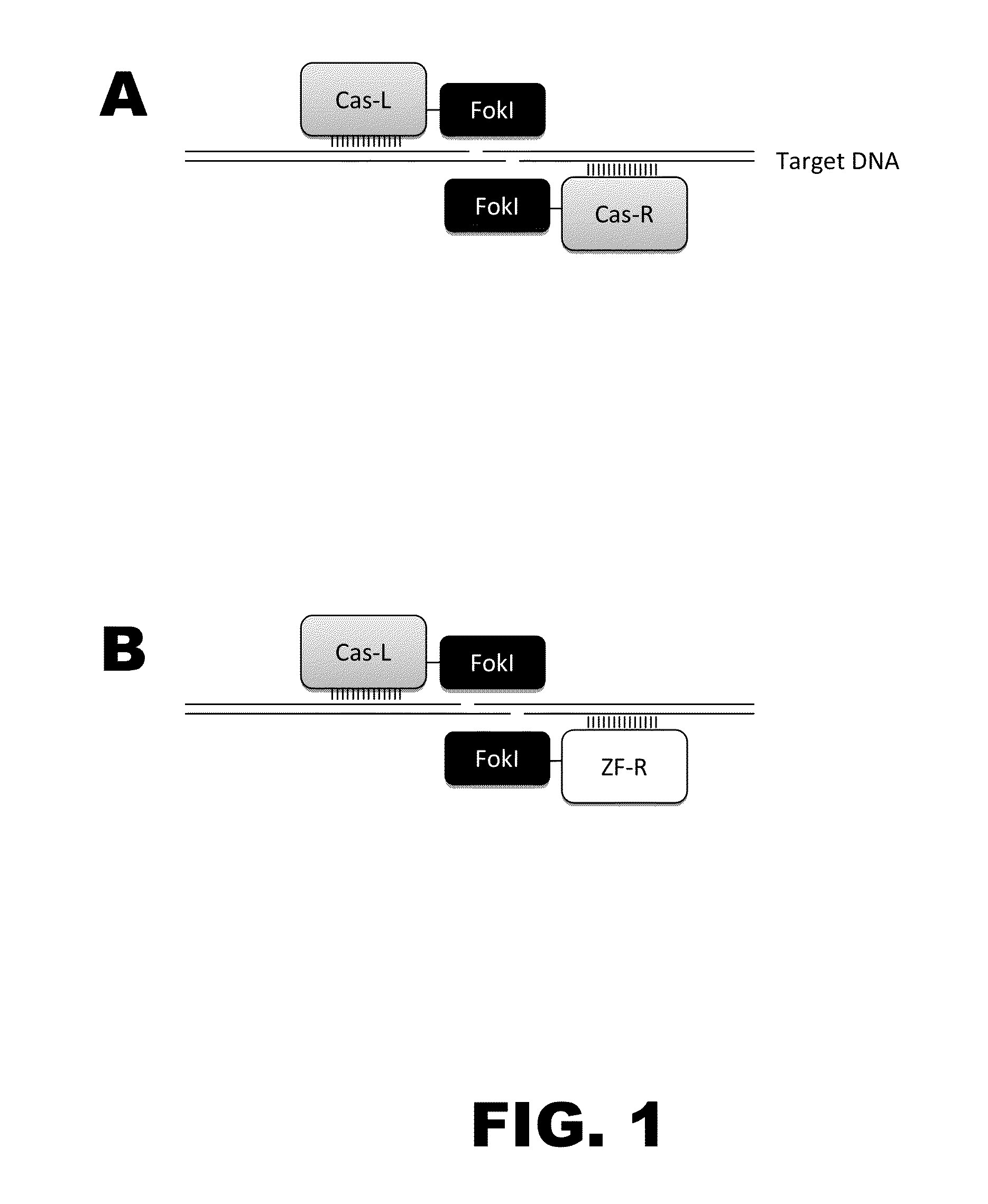
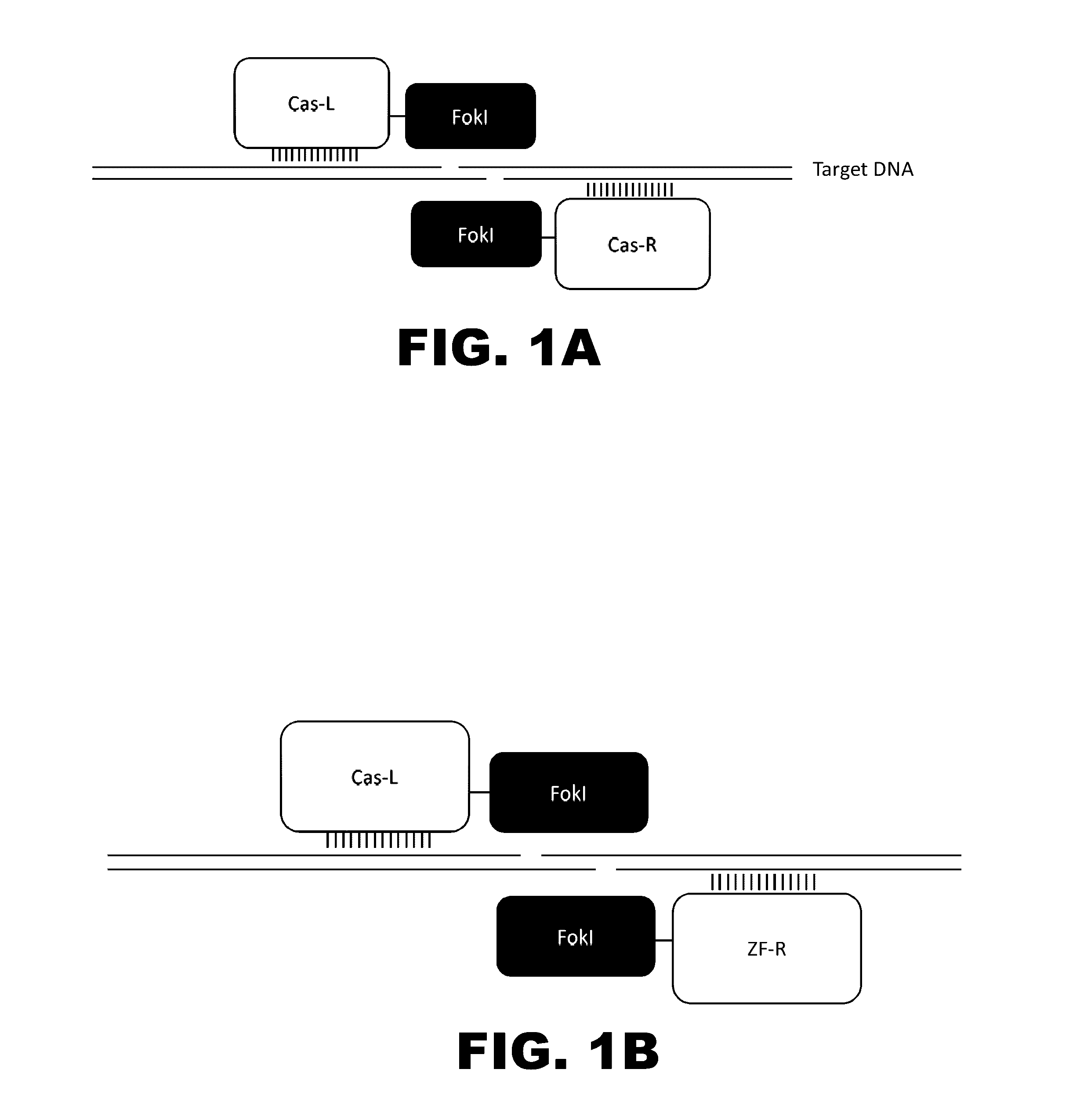
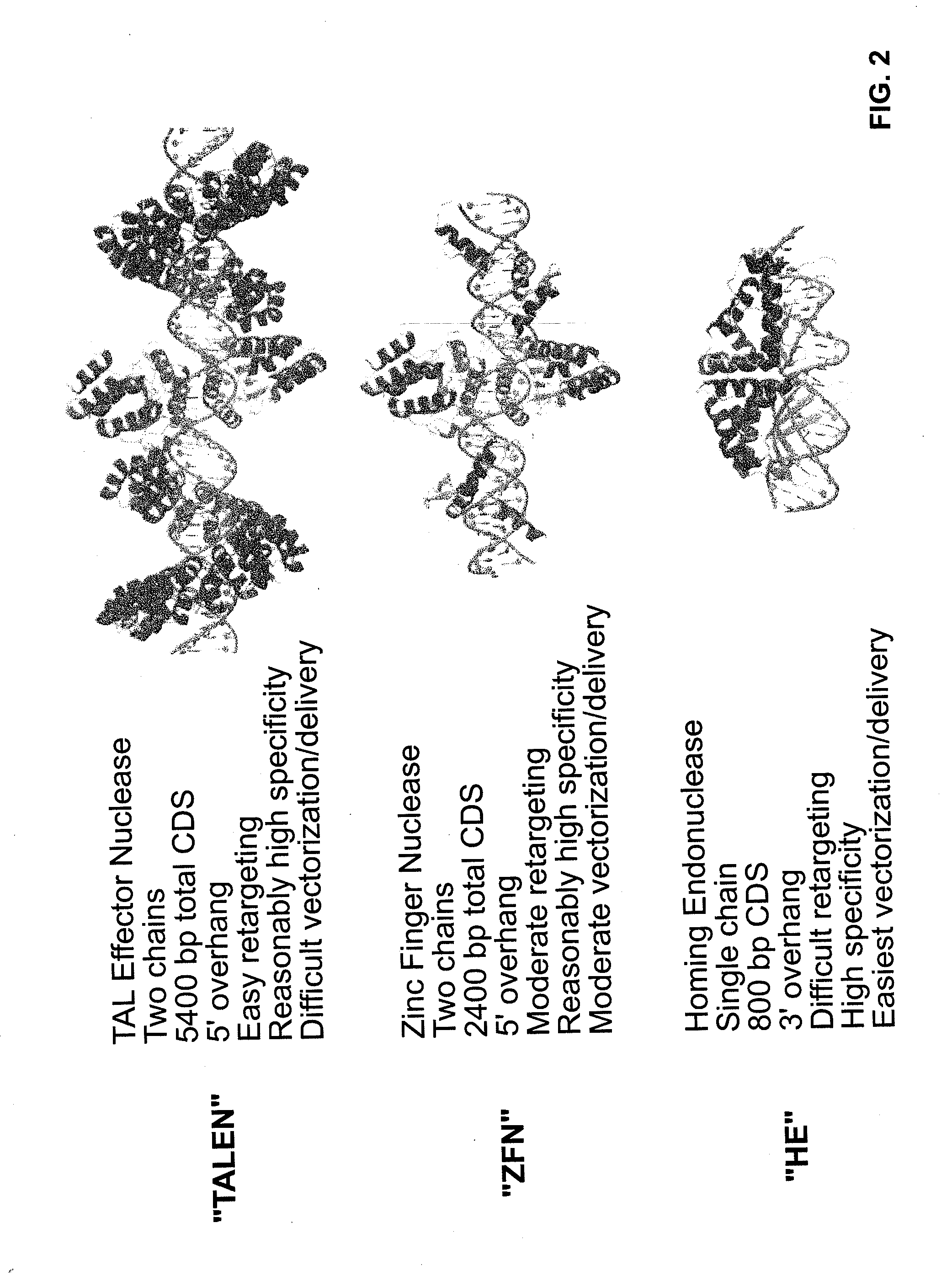
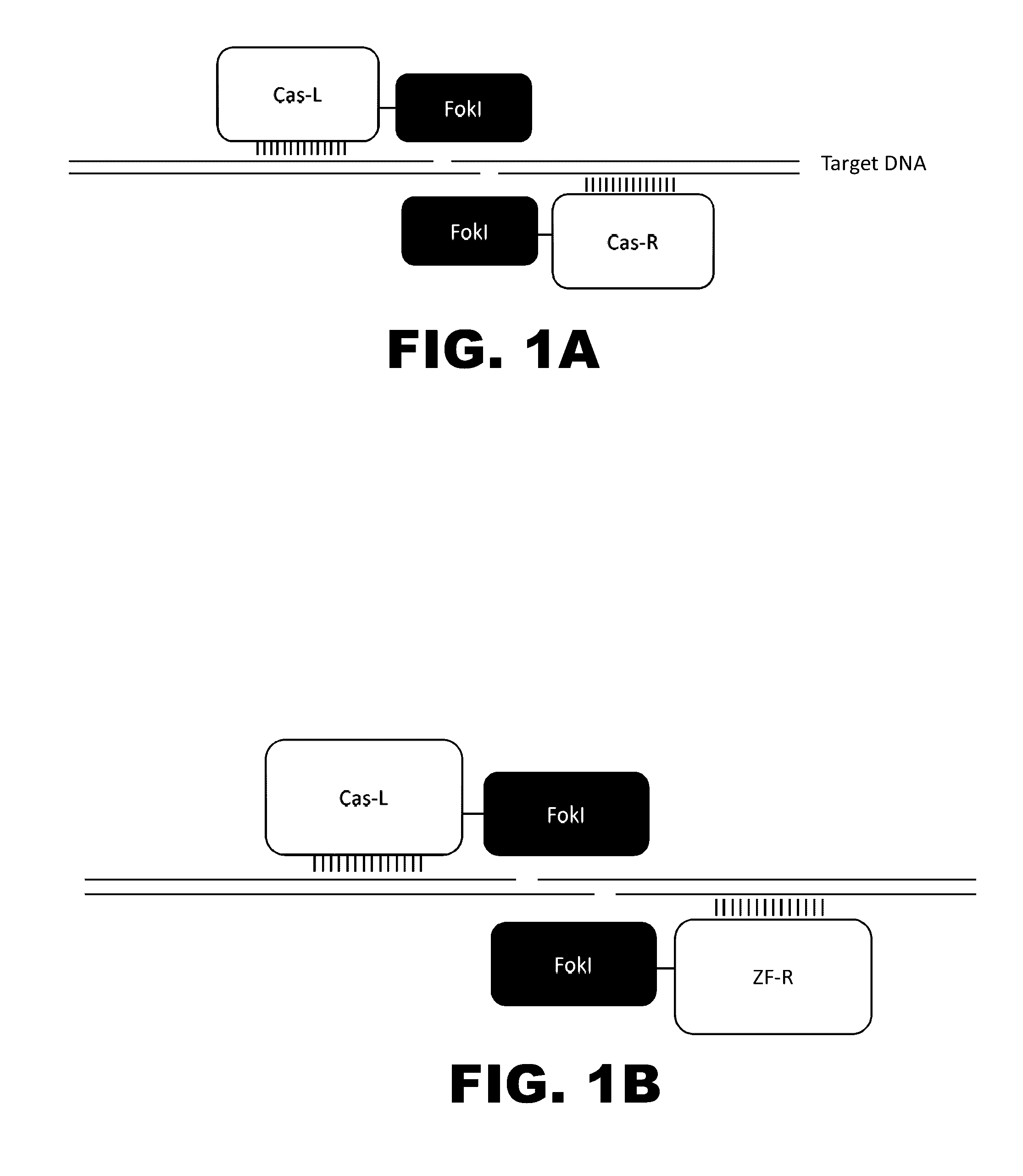
The present invention provides compositions and methods for targeted cleavage of cellular chromatin in a region of interest and / or homologous recombination at a predetermined site in cells. Compositions include fusion polypeptides comprising a TAL effector binding domain and a cleavage domain. The cleavage domain can be from any endonuclease. In certain embodiments, the endonuclease is a Type IIS restriction endonuclease. In further embodiments, the Type IIS restriction endonuclease is FokI.
Owner:IOWA STATE UNIV RES FOUND
Method for making linear, covalently closed DNA constructs
InactiveUS6451563B1Bulking digestionHigh processivitySugar derivativesHydrolasesDNA constructGenomic DNA
A process to obtain linear double-stranded covalently closed DNA "dumbbell" constructs from plasmids by restriction digest, subsequent ligation with hairpin oligodesoxyribonucleotides, optionally in the presence of restriction enzyme, and a final digestion with endo- and exonucleolytic enzymes that degrade all contaminating polymeric DNA molecules but the desired construct. The invention also provides a process to obtain said dumbbell constructs employing endonuclease class II enzymes. Furthermore, the invention provides a process to obtain linear, covalently closed DNA molecules, such as plasmids, free from contamination by genomic DNA, by submitting the DNA preparation to a facultative endonucleolytic degradation step and an obligatory exonucleolytic degradation step.
Owner:MOLOGEN AG +1
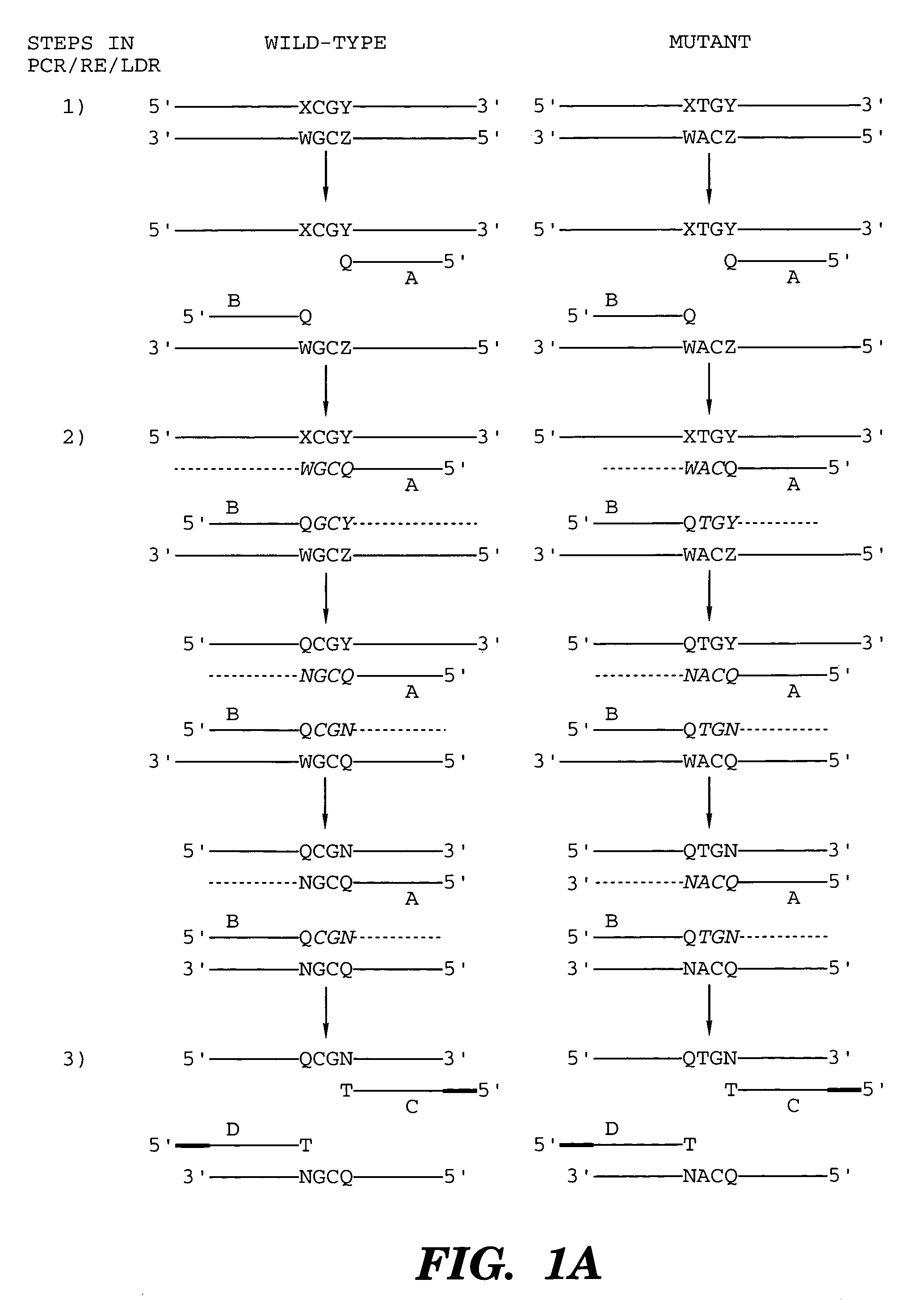
Coupled polymerase chain reaction-restriction-endonuclease digestion-ligase detection reaction process
InactiveUS7014994B1Sensitive highOptimizationSugar derivativesMicrobiological testing/measurementNucleotideWild type
The present invention provides a method for identifying one or more low abundance sequences differing by one or more single-base changes, insertions, or deletions, from a high abundance sequence in a plurality of target nucleotide sequences. The high abundance wild-type sequence is selectively removed using high fidelity polymerase chain reaction analog conversion, facilitated by optimal buffer conditions, to create a restriction endonuclease site in the high abundance wild-type gene, but not in the low abundance mutant gene. This allows for digestion of the high abundance DNA. Subsequently the low abundant mutant DNA is amplified and detected by the ligase detection reaction assay. The present invention also relates to a kit for carrying out this procedure.
Owner:LOUISIANA STATE UNIV +1
Promoter and plasmid system for genetic engineering
This invention provides a series of low-copy number plasmids comprising restriction endonuclease recognition sites useful for cloning at least three different genes or operons, each flanked by a terminator sequence, the plasmids containing variants of glucose isomerase promoters for varying levels of protein expression. The materials and methods are useful for genetic engineering in microorganisms, especially where multiple genetic insertions are sought.
Owner:EI DU PONT DE NEMOURS & CO
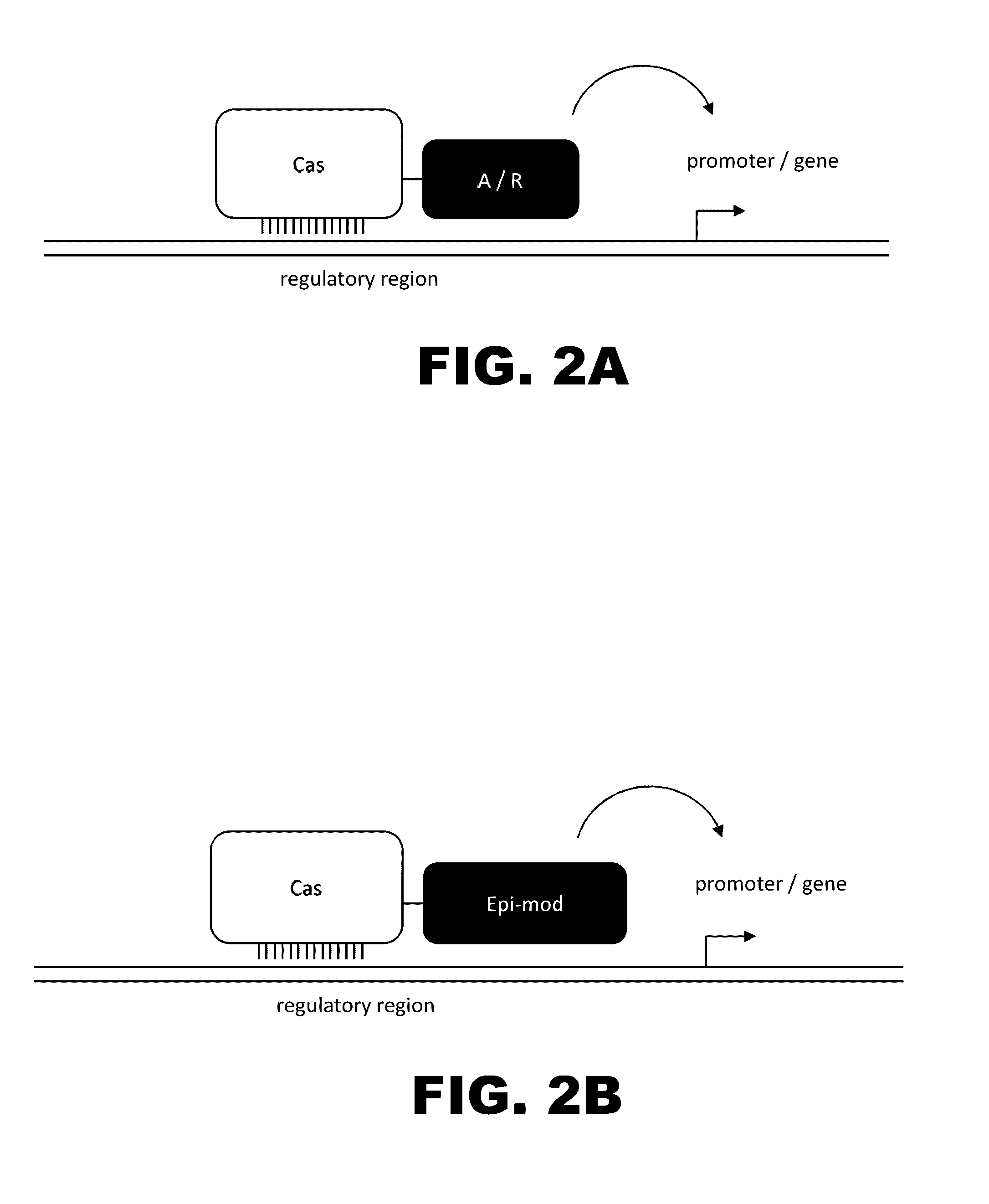
Crispr-based genome modification and regulation
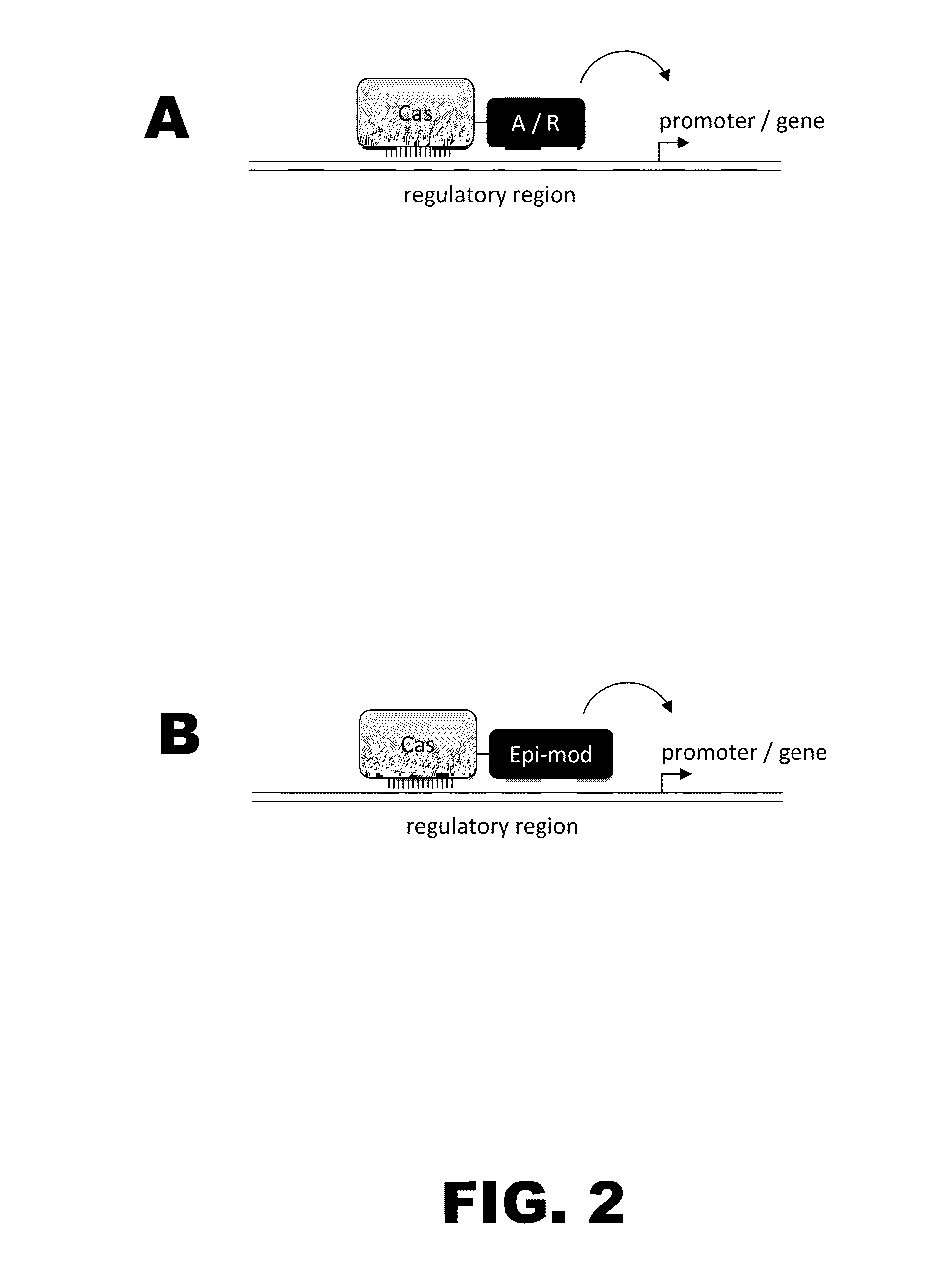
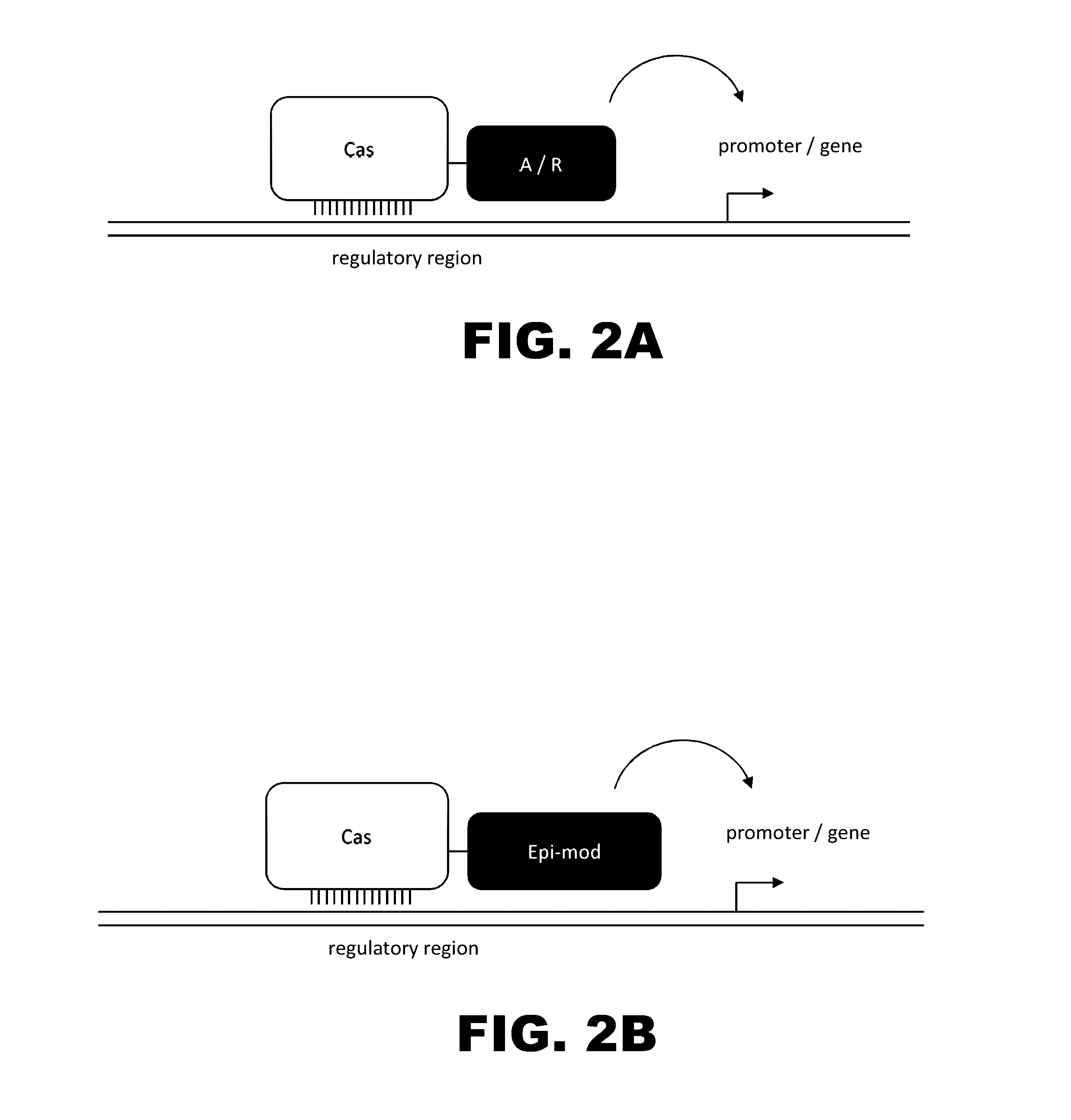
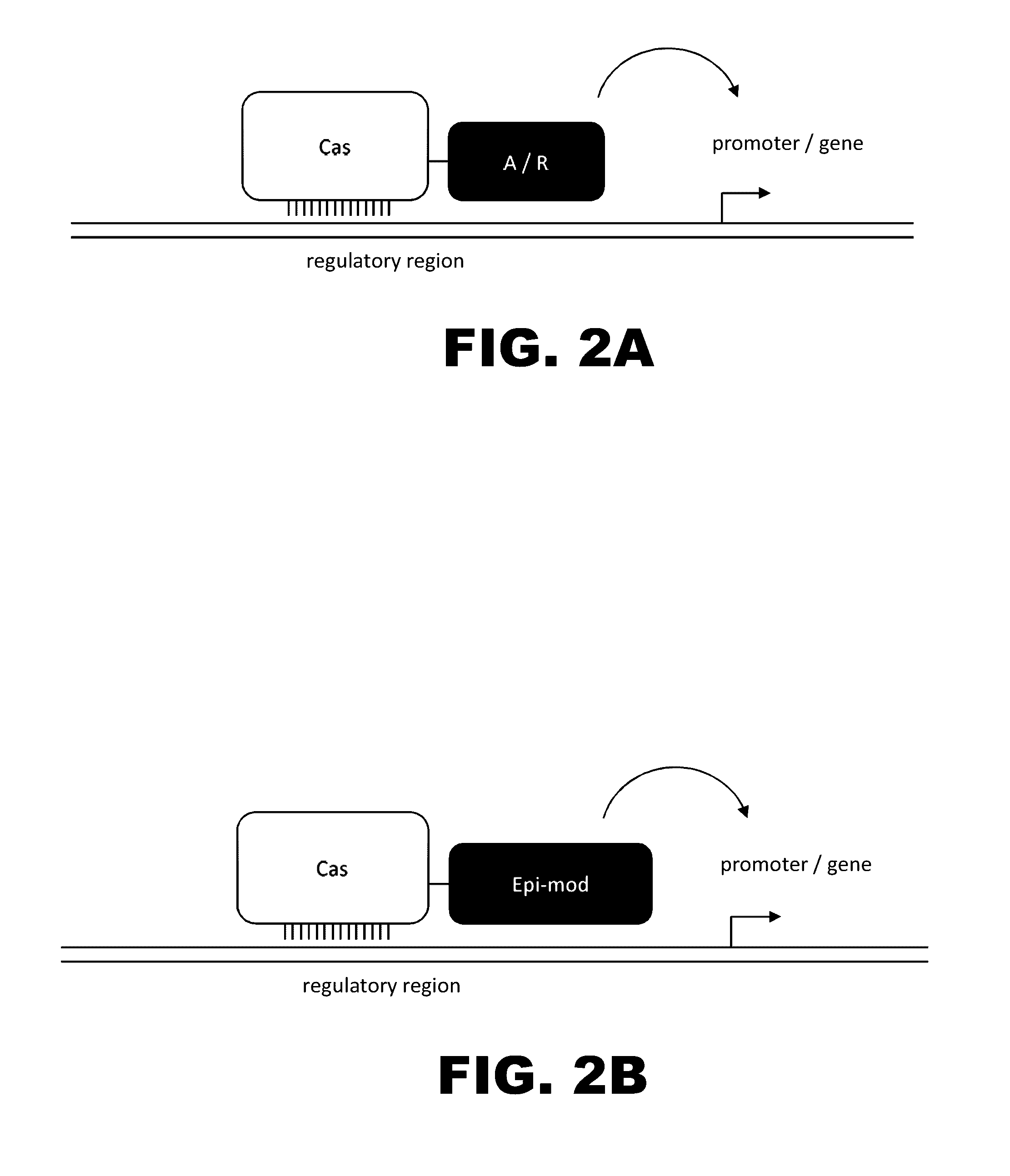
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
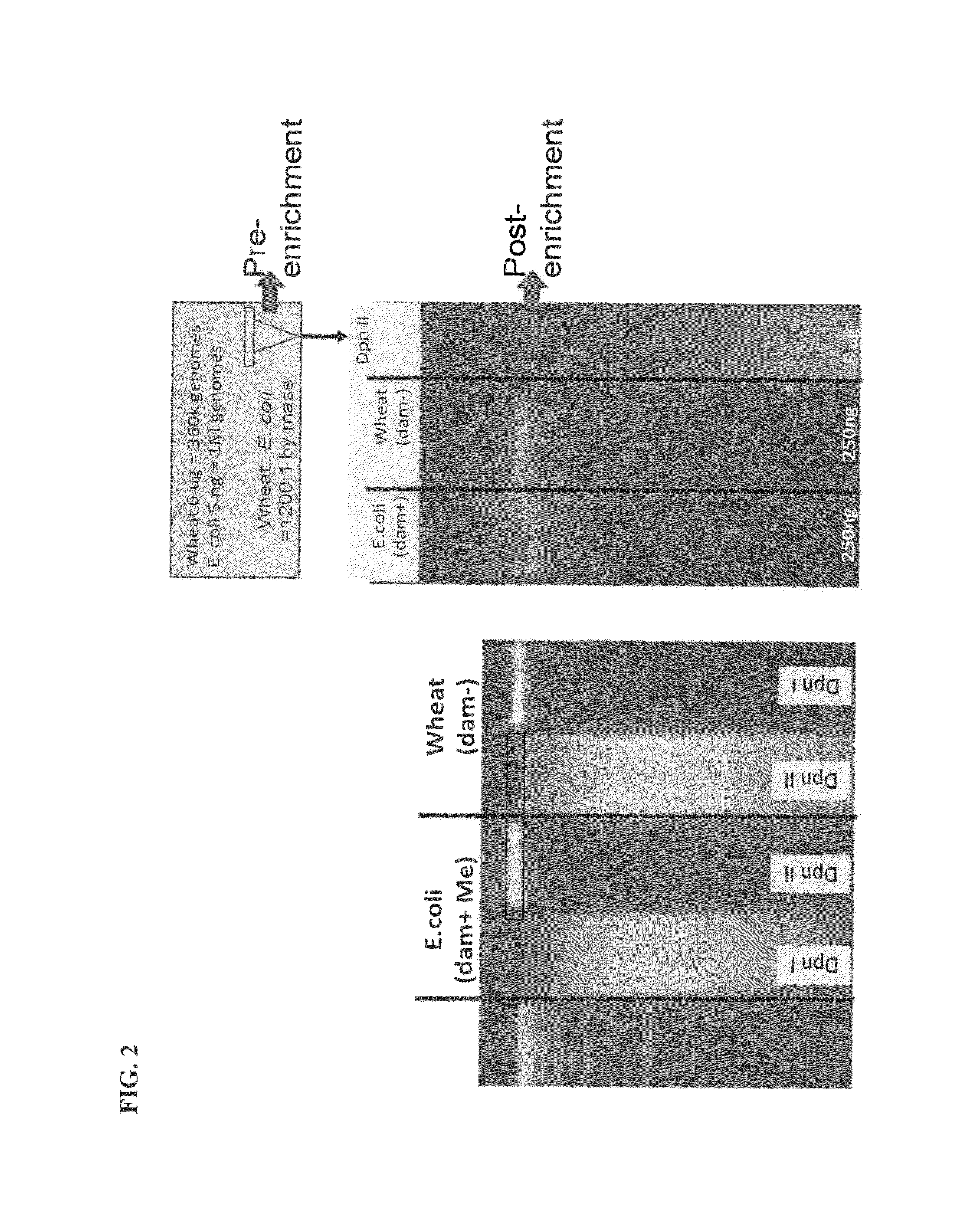
Methods and compositions for segregating target nucleic acid from mixed nucleic acid samples
InactiveUS8927218B2Rapid and efficient isolation and identificationMicrobiological testing/measurementDNA preparationDNA EndonucleaseMethylation
The invention provides methods, compositions and kits for segregating a target nucleic acid from a mixed nucleic acid sample. The methods, compositions and kits comprise a non-processive endonuclease (e.g., a restriction enzyme) or an antibody that binds the target nucleic acid (e.g., has methylation specificity). The mixed nucleic acid sample can comprise prokaryotic and eukaryotic nucleic acid and / or nucleic acid from more than one prokaryotic or eukaryotic organisms.
Owner:FLIR DETECTION
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
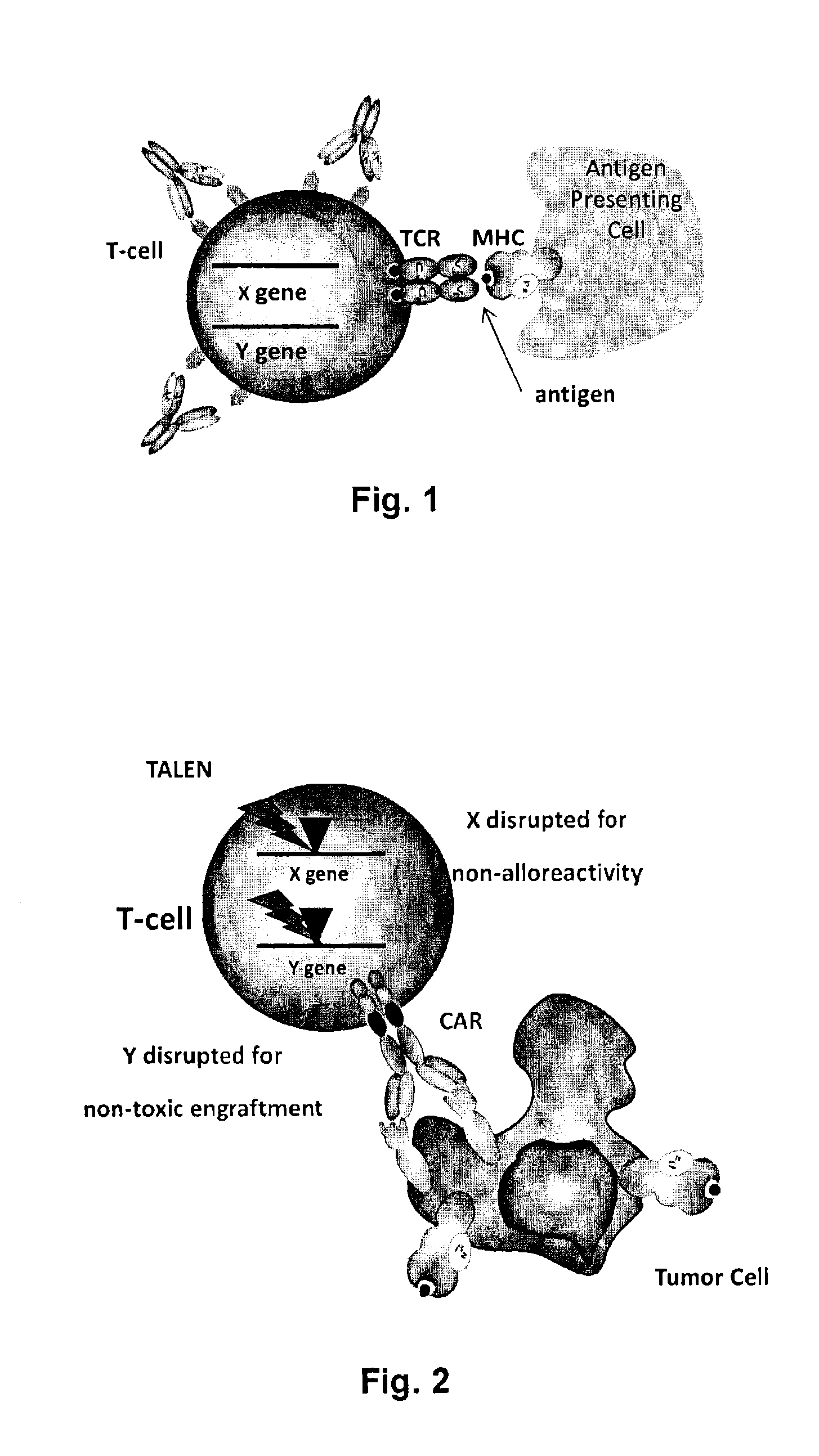
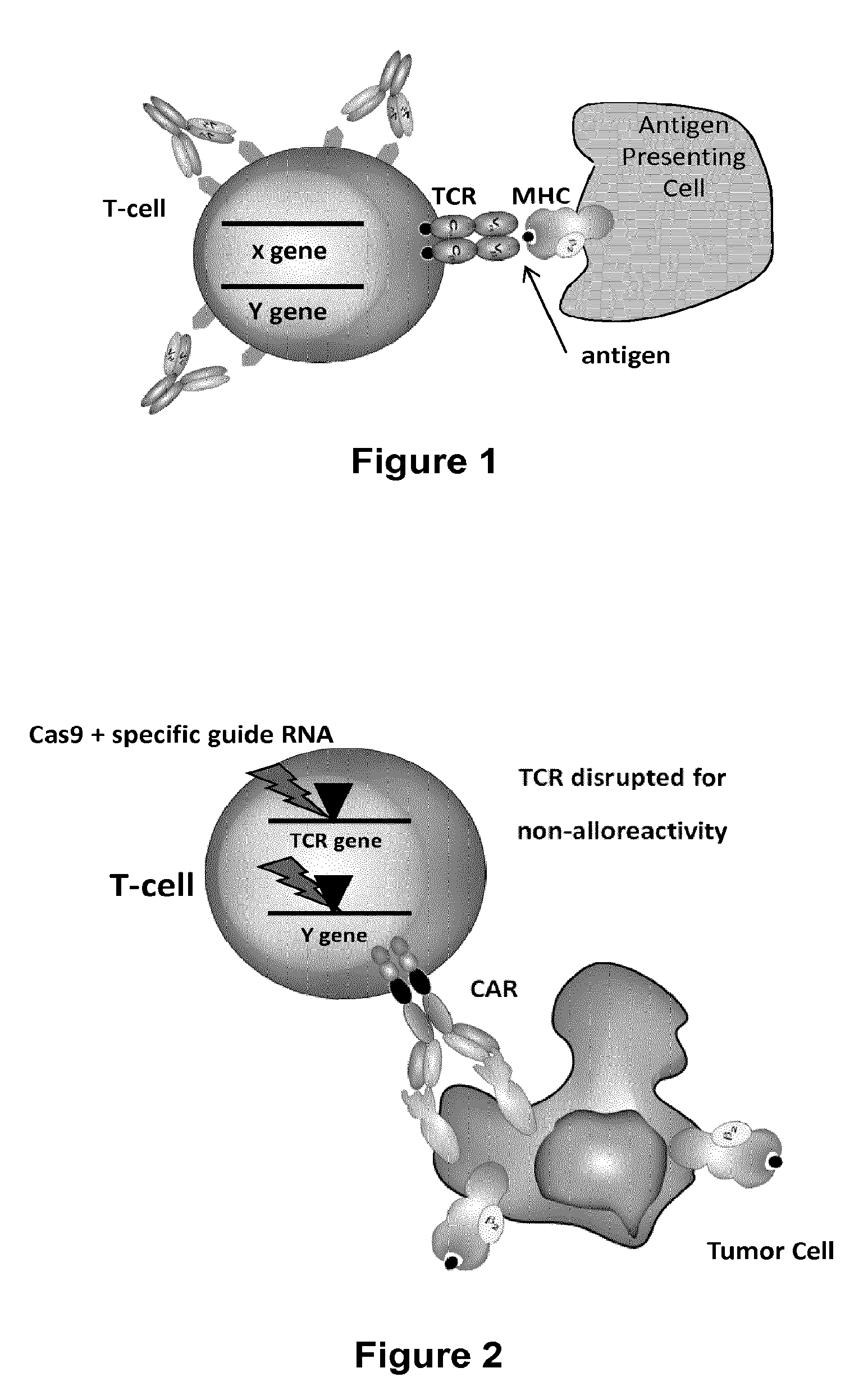
Methods for engineering highly active t cell for immunotheraphy
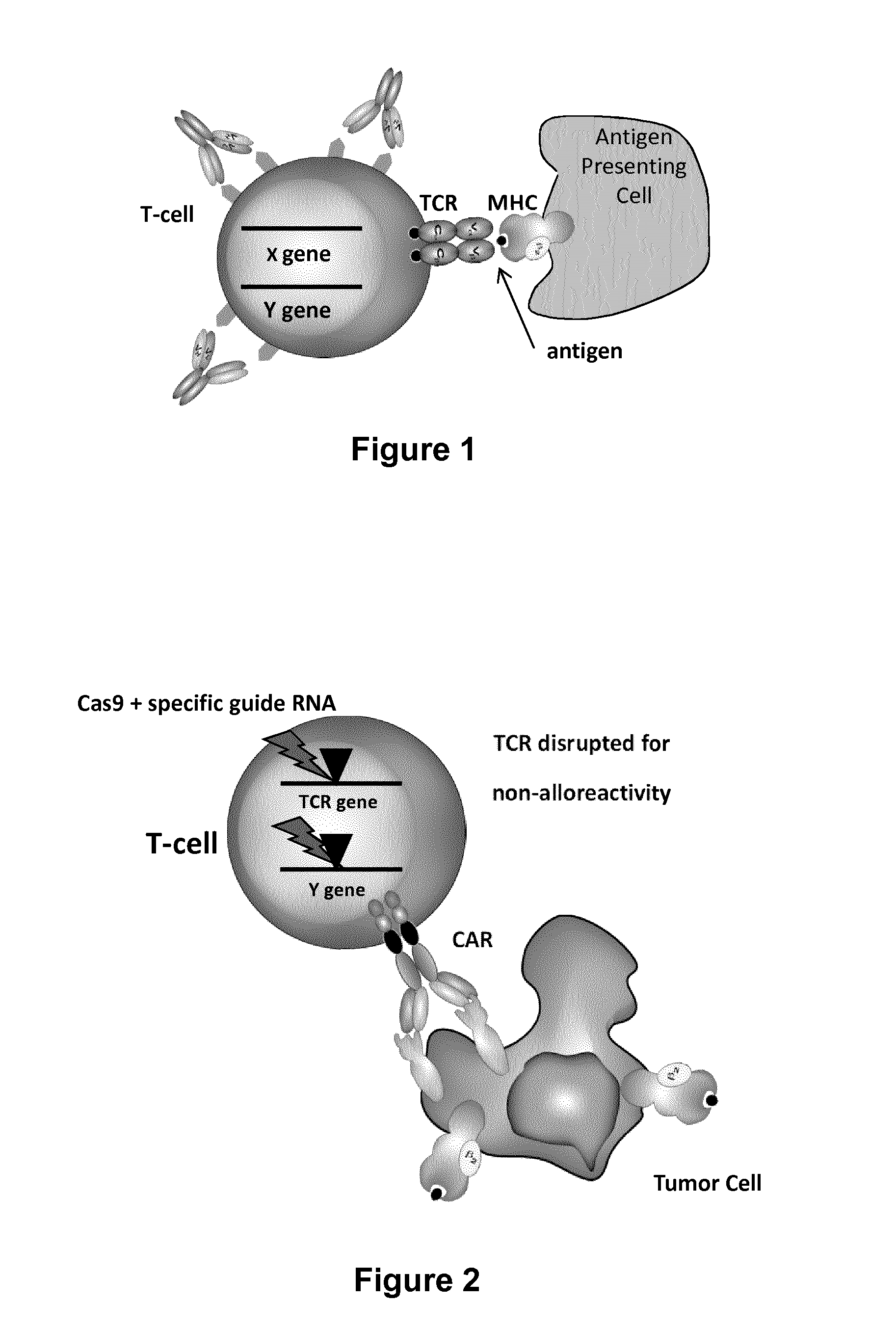
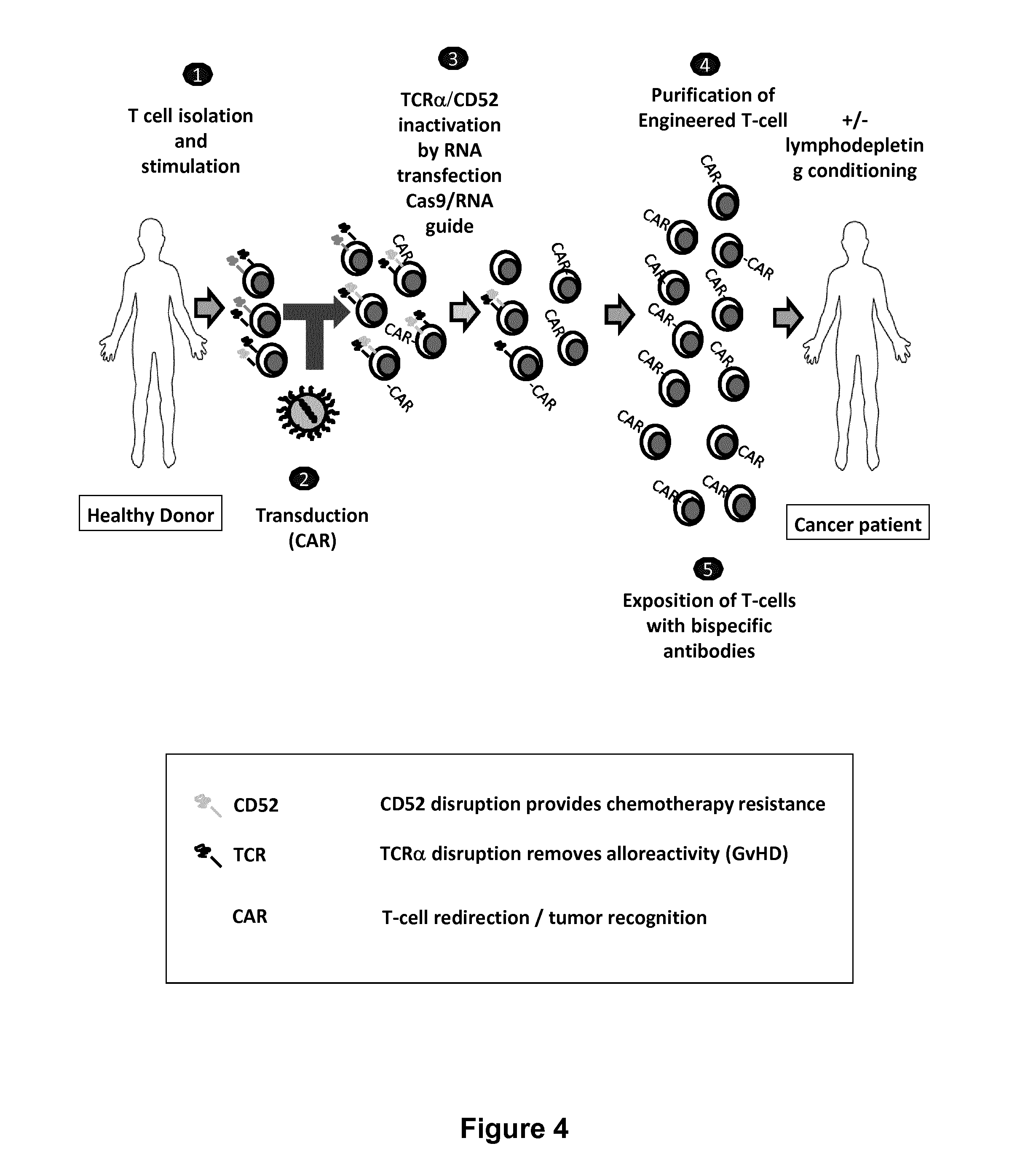
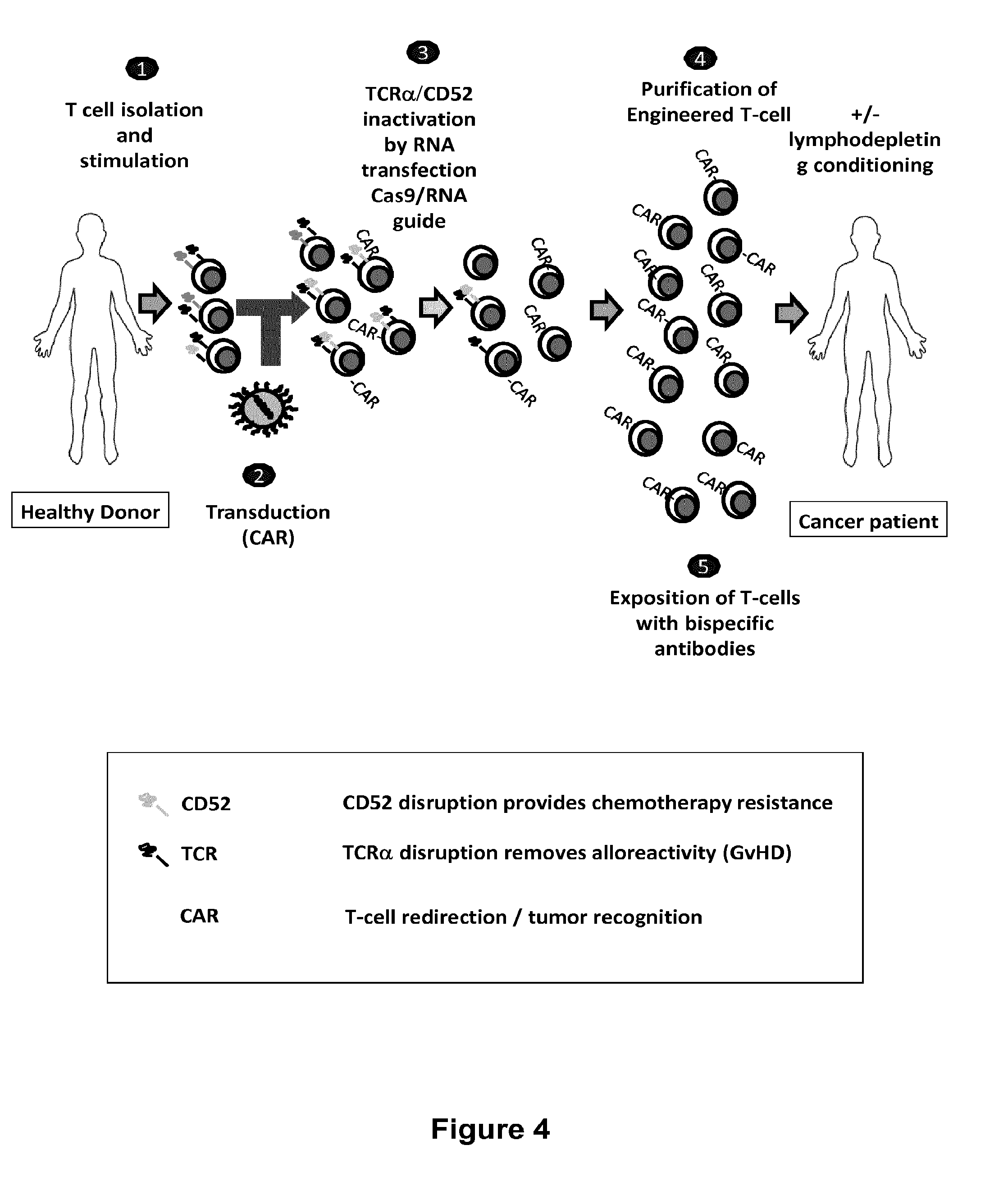
The present invention relates to methods for developing engineered T-cells for immunotherapy and more specifically to methods for modifying T-cells by inactivating at immune checkpoint genes, preferably at least two selected from different pathways, to increase T-cell immune activity. This method involves the use of specific rare cutting endonucleases, in particular TALE-nucleases (TAL effector endonuclease) and polynucleotides encoding such polypeptides, to precisely target a selection of key genes in T-cells, which are available from donors or from culture of primary cells. The invention opens the way to highly efficient adoptive immunotherapy strategies for treating cancer and viral infections.
Owner:CELLECTIS SA
Determination of methylated DNA
InactiveUS20070231800A1Microbiological testing/measurementFermentationNucleic Acid ProbesNucleic acid hybridisation
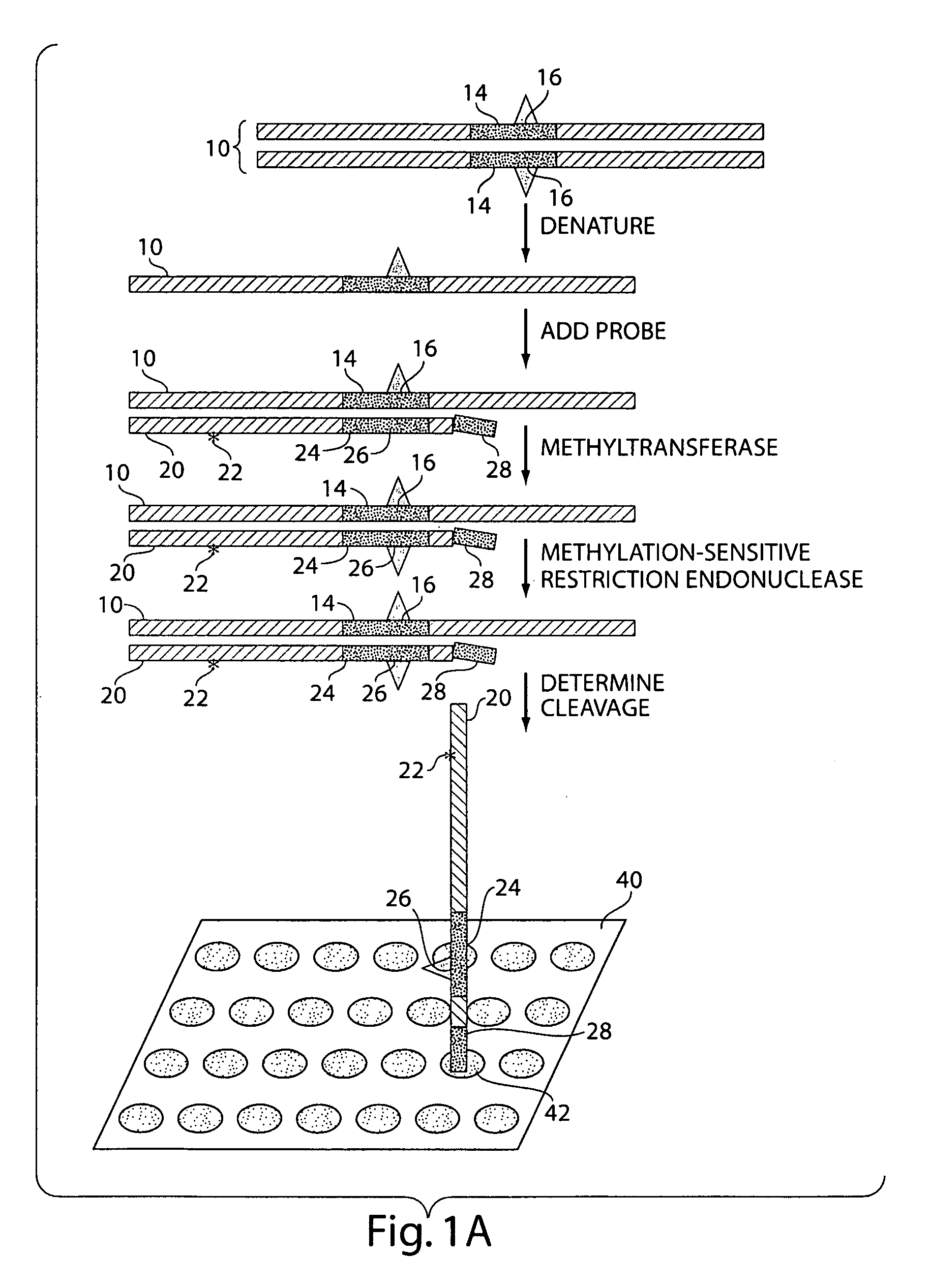
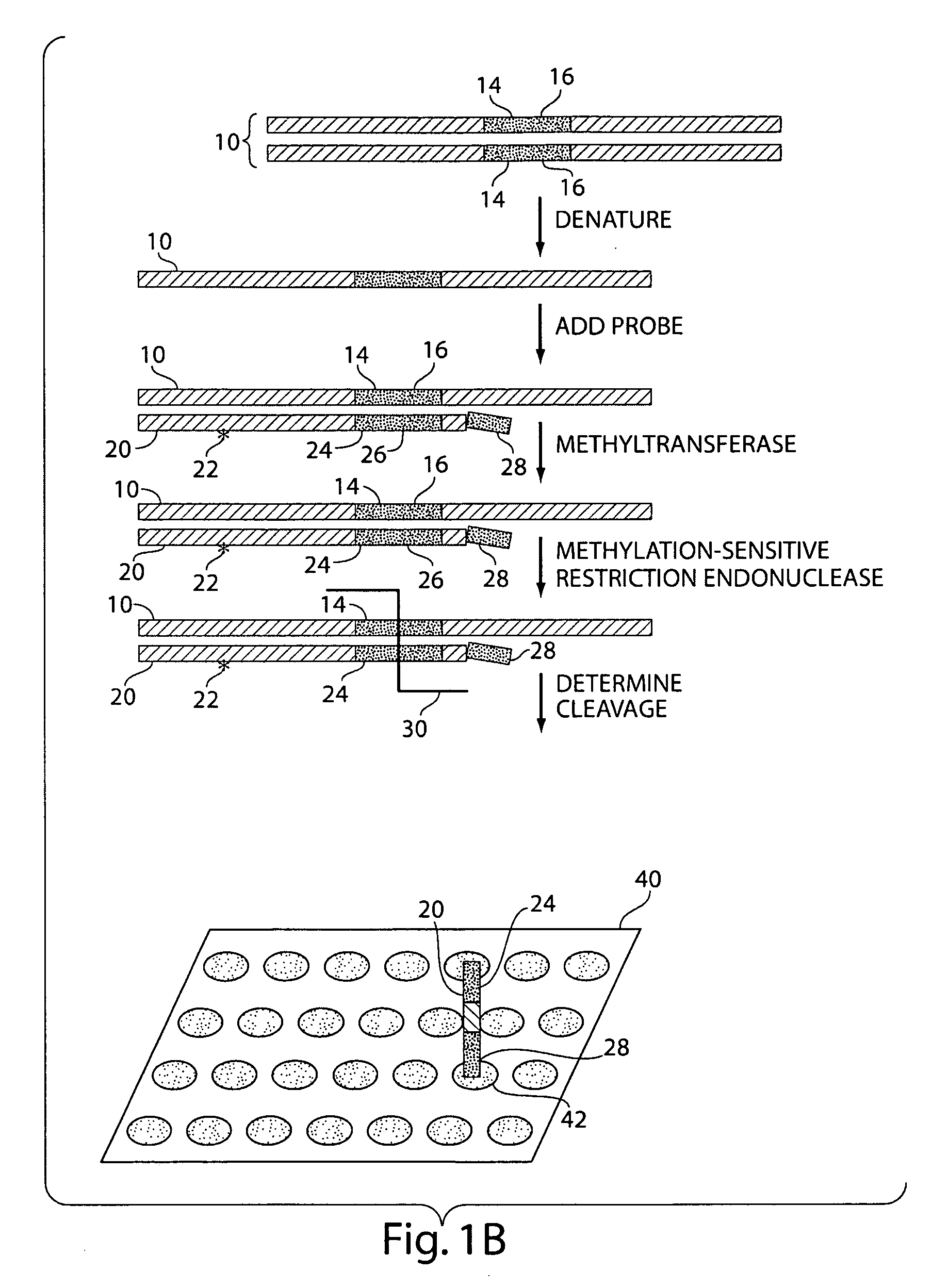
The present invention generally relates to the determination of the state of one or more locations within a nucleic acid and, in particular, to the determination of the methylation state of one or more methylation sites within a nucleic acid such as DNA. In one aspect of the invention, a nucleic acid, such as DNA, that is suspected of being methylated is exposed to a nucleic acid probe able to hybridize the nucleic acid at or near the methylation site. After hybridization, the nucleic acid-probe hybrid is exposed to a methylation-sensitive restriction endonuclease able to bind at or near the methylation site. The restriction endonuclease is not able to cleave the nucleic acid-probe hybrid if the DNA is methylated at the methylation site, but is able to cleave the nucleic acid-probe hybrid if the nucleic acid is not methylated at the methylation site. Determination of the cleavage state of the probe can thus be used to determine the state of the methylation site.
Owner:AGILENT TECH INC
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Compositions and methods for the treatment of hemoglobinopathies
InactiveUS20150166969A1Improve the level ofAvoid the insertion of vector sequencesFusion with DNA-binding domainSugar derivativesThalassemiaGlobin genes
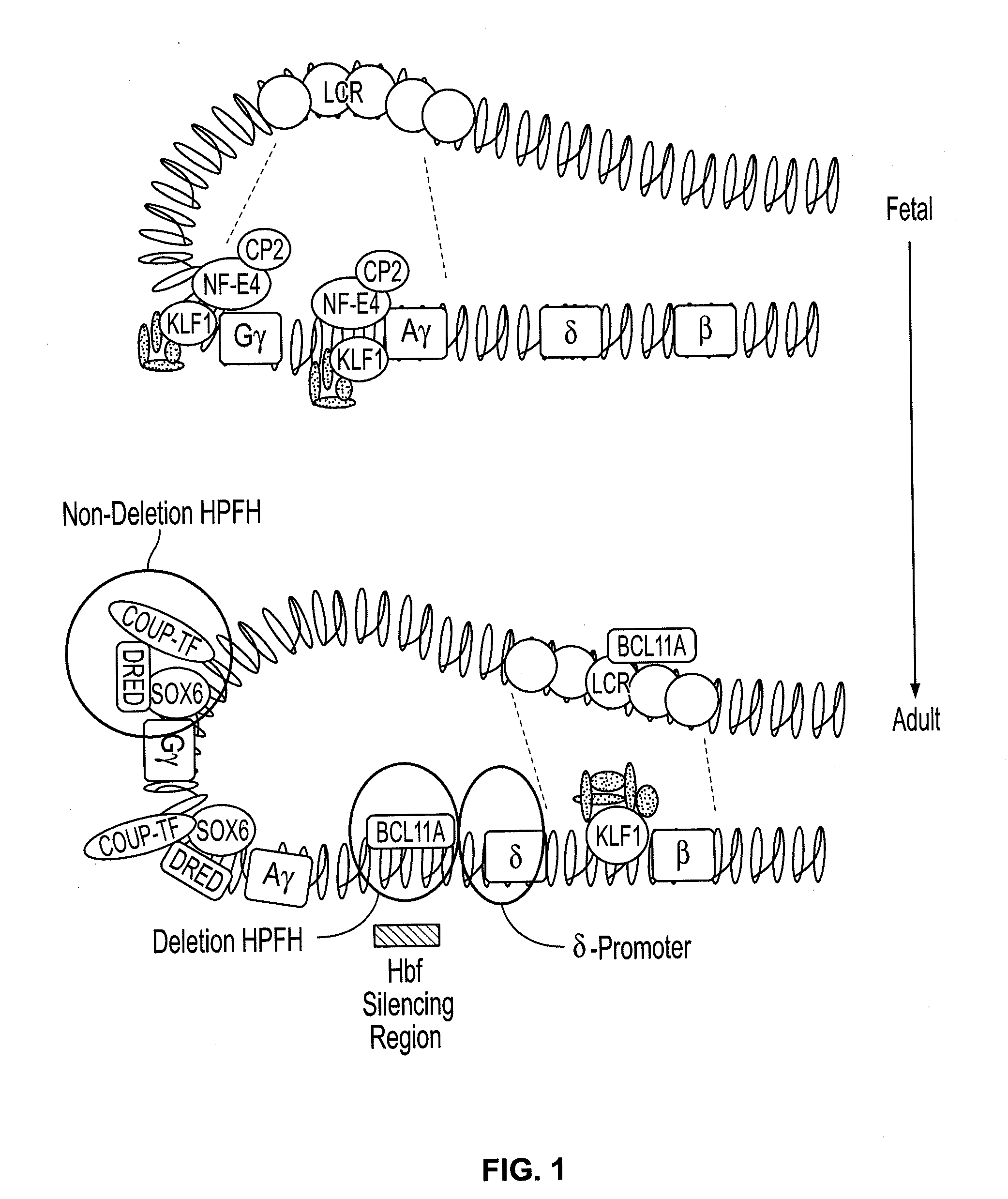
Provided are compositions and methods for the treatment of hemoglobinopathies such as thalassemias and sickle cell disease. Compositions and methods include one or more endonuclease(s) or endonuclease fusion protein(s), including one or more homing endonuclease(s) and / or homing endonuclease fusion protein(s) and / or CRISPR endonuclease(s) ad / or CRISPR endonuclease fusion protein(s): (a) to disrupt a Bcl11a coding region; (b) to disrupt a Bcl11a gene regulatory region; (c) to modify an adult human β-globin locus; (d) to disrupt a HbP silencing DNA regulatory element or pathway, such as a Bcl11a-regulated HbP silencing region; (e) to mutate one or more γ-globin gene promoter(s) to achieve increased expression of a γ-globin gene; (f) to mutate one or more δ-globin gene promoter(s) to achieve increased expression of a δ-globin gene; and / or (g) to correct one or more β-globin gene mutation(s).
Owner:NAT INST OF HEALTH DIRECTOR DEITR
FEN endonucleases
InactiveUS7122364B1Improve performanceA large amountHydrolasesMicrobiological testing/measurementPolymerase LNucleic acid sequencing
The present invention provides novel cleavage agents and polymerases for the cleavage and modification of nucleic acid. The cleavage agents and polymerases find use, for example, for the detection and characterization of nucleic acid sequences and variations in nucleic acid sequences. In some embodiments, the 5′ nuclease activity of a variety of enzymes is used to cleave a target-dependent cleavage structure, thereby indicating the presence of specific nucleic acid sequences or specific variations thereof.
Owner:GEN PROBE INC
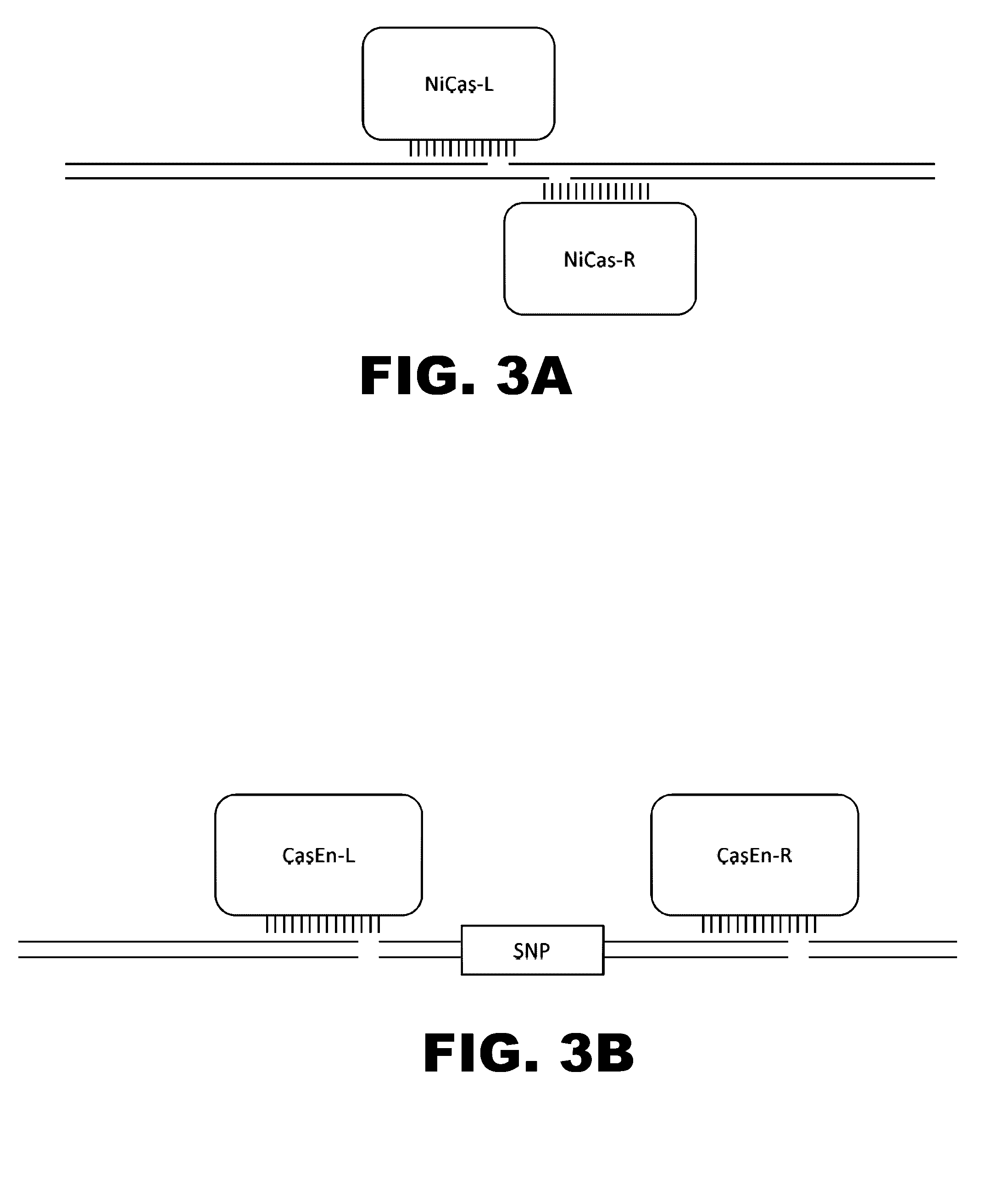
Fast CRISPR-Cas9 working efficiency testing system and application thereof
InactiveCN105647968ATest accurateTested and reliableMicrobiological testing/measurementPeptidesEnzyme digestionRapid testing
The invention discloses a fast CRISPR-Cas9 working efficiency testing system and application thereof. The testing system comprises a plasmid used for expressing sg RNA, a plasmid used for expressing Cas9 and a reporting system used for testing the CRISPR-Cas9 gene editing efficiency; the reporting system can splice the C-terminal of a nucleotide segment capable of coding effective protein and the N-terminal of a reporter gene and insert two restriction endonuclease enzyme digestion sites into the splicing position; before a special gene is edited (knocked out) through the CRISPR-Cas9 system, selection of a target sequence is vital, and the selection can influence the recognition efficiency of the sg RNA to target DNA, the binding efficiency of the sg RNA with the target DNA, the targeted cutting efficiency of the Cas9 and the NHEJ repairing efficiency. According to the system, the gene editing efficiency of different sgRNA-target DNA sequences can be quantitatively compared, the sgRNA with the best working effect can be determined within a short time, the actual knockout success rate is increased, therefore, the working cost can be lowered, the working efficiency can be improved, and the working progress can be promoted.
Owner:ZHEJIANG UNIV
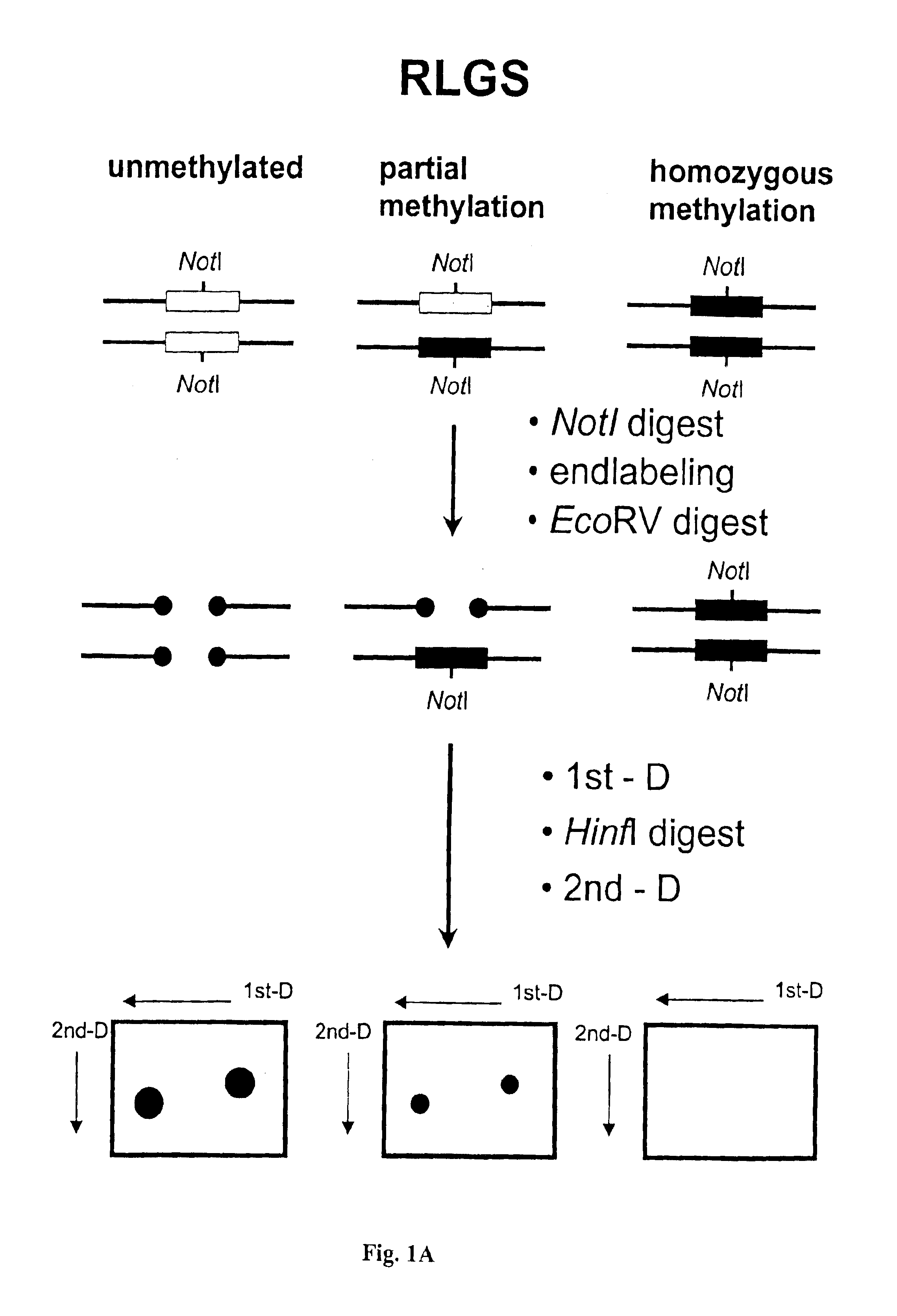
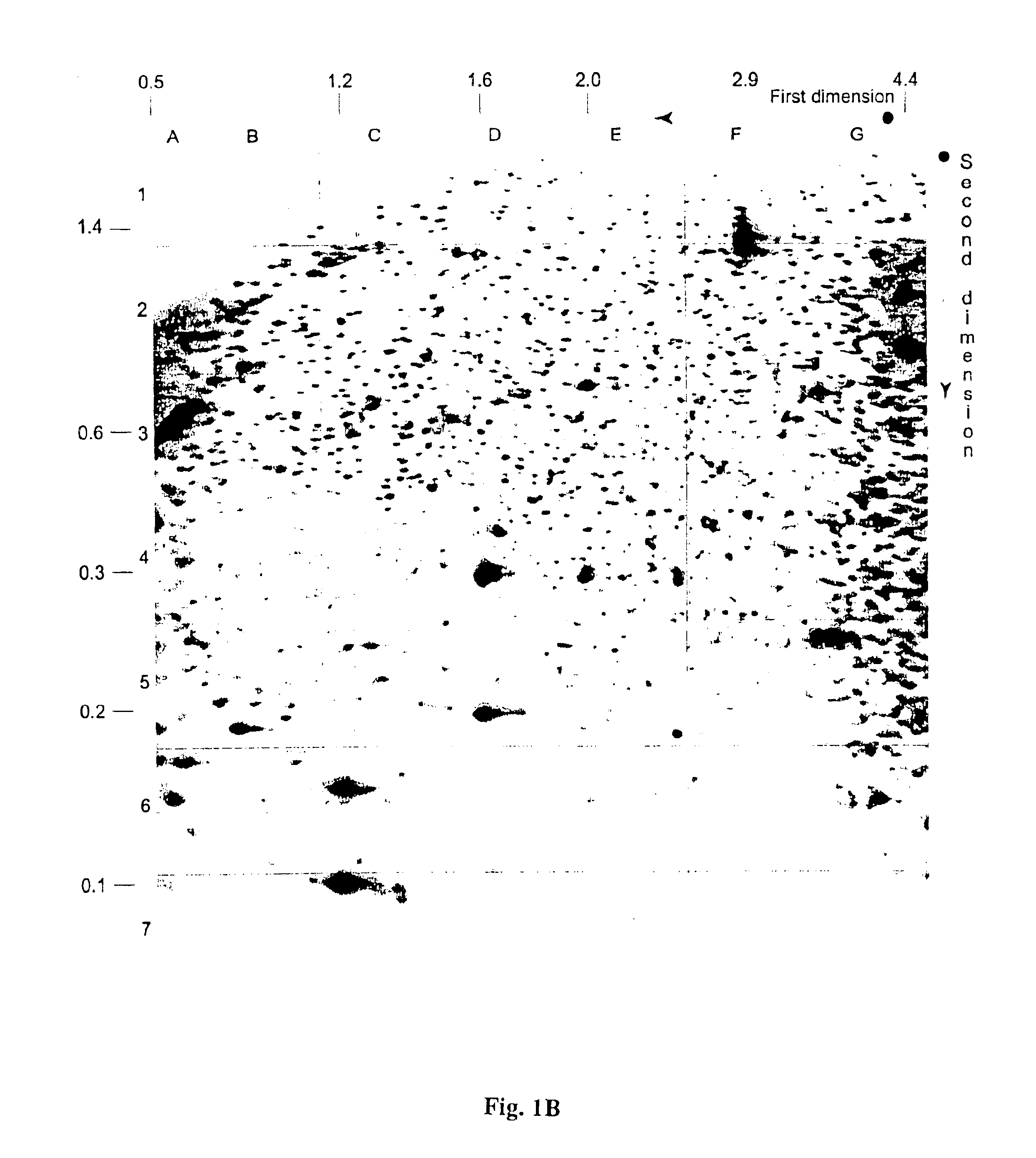
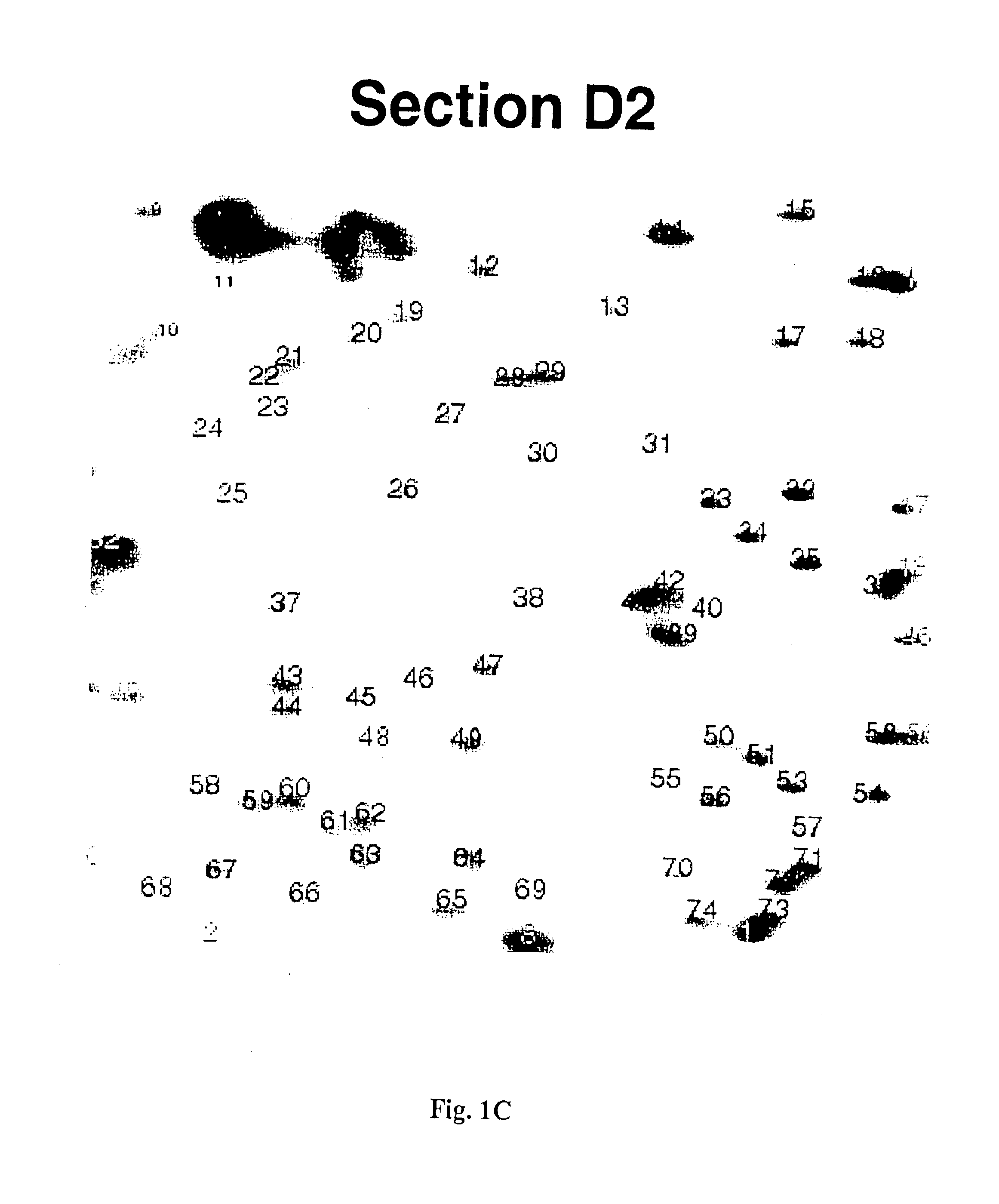
Detection of methylated CpG rich sequences diagnostic for malignant cells
InactiveUS6893820B1Microbiological testing/measurementMaterial analysis by electric/magnetic meansAbnormal tissue growthCancer cell
The present invention provides methods for determining the methylation status of CpG-containing dinucleotides on a genome-wide scale using infrequent cleaving, methylation sensitive restriction endonucleases and two-dimensional gel electrophoretic display of the resulting DNA fragments. Such methods can be used to diagnose cancer, classify tumors and provide prognoses for cancer patients. The present invention also provides isolated polynucleotides and oligonucleotides comprising CpG dinucleotides that are differentially methylated in malignant cells as compared to normal, non-malignant cells. Such polynucleotides and oligonucleotides are useful for diagnosis of cancer. The present invention also provides methods for identifying new DNA clones within a library that contain specific CpG dinucleotides that are differentially methylated in cancer cells as compared to normal cells.
Owner:THE OHIO STATE UNIV RES FOUND
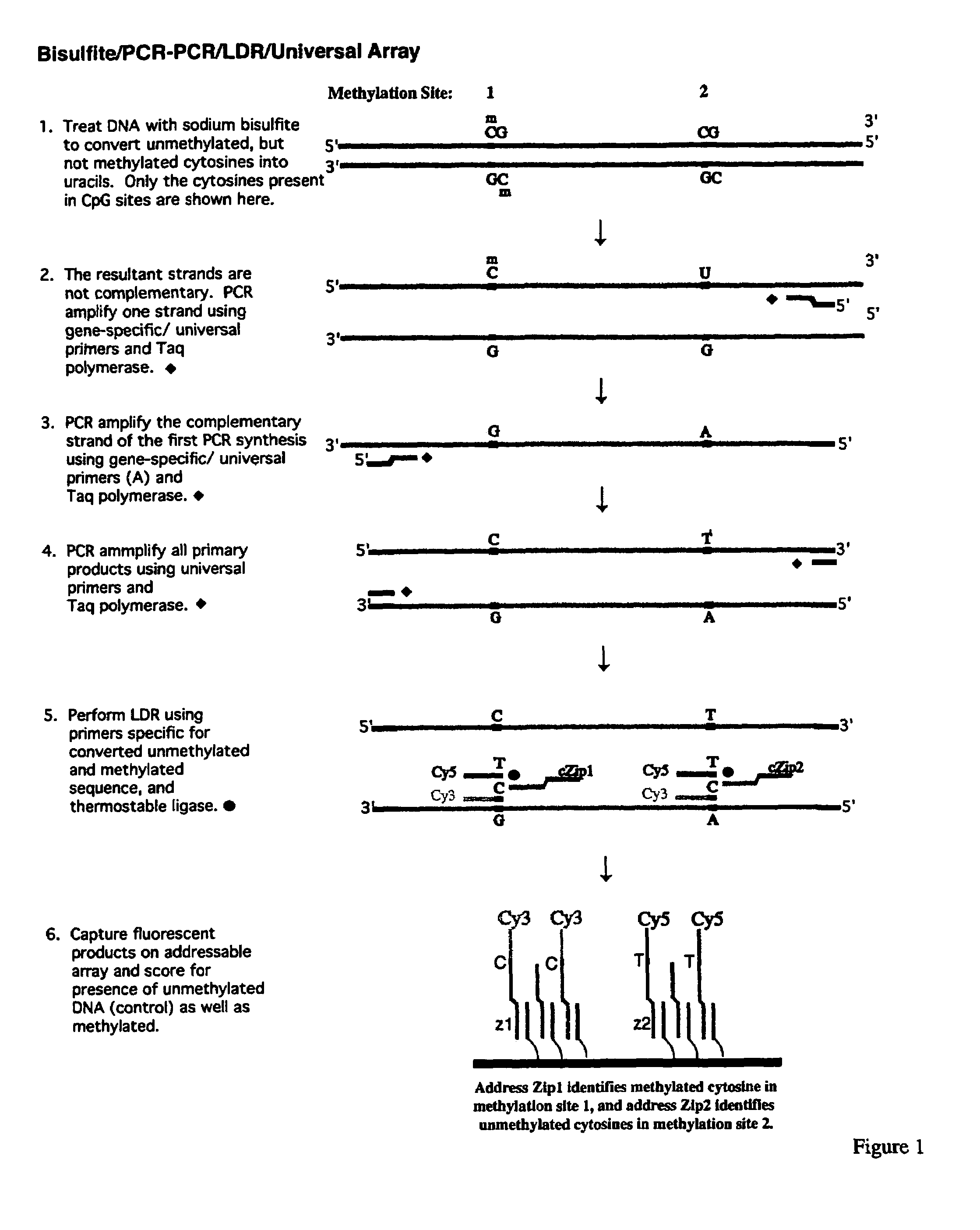
Method for detection of promoter methylation status
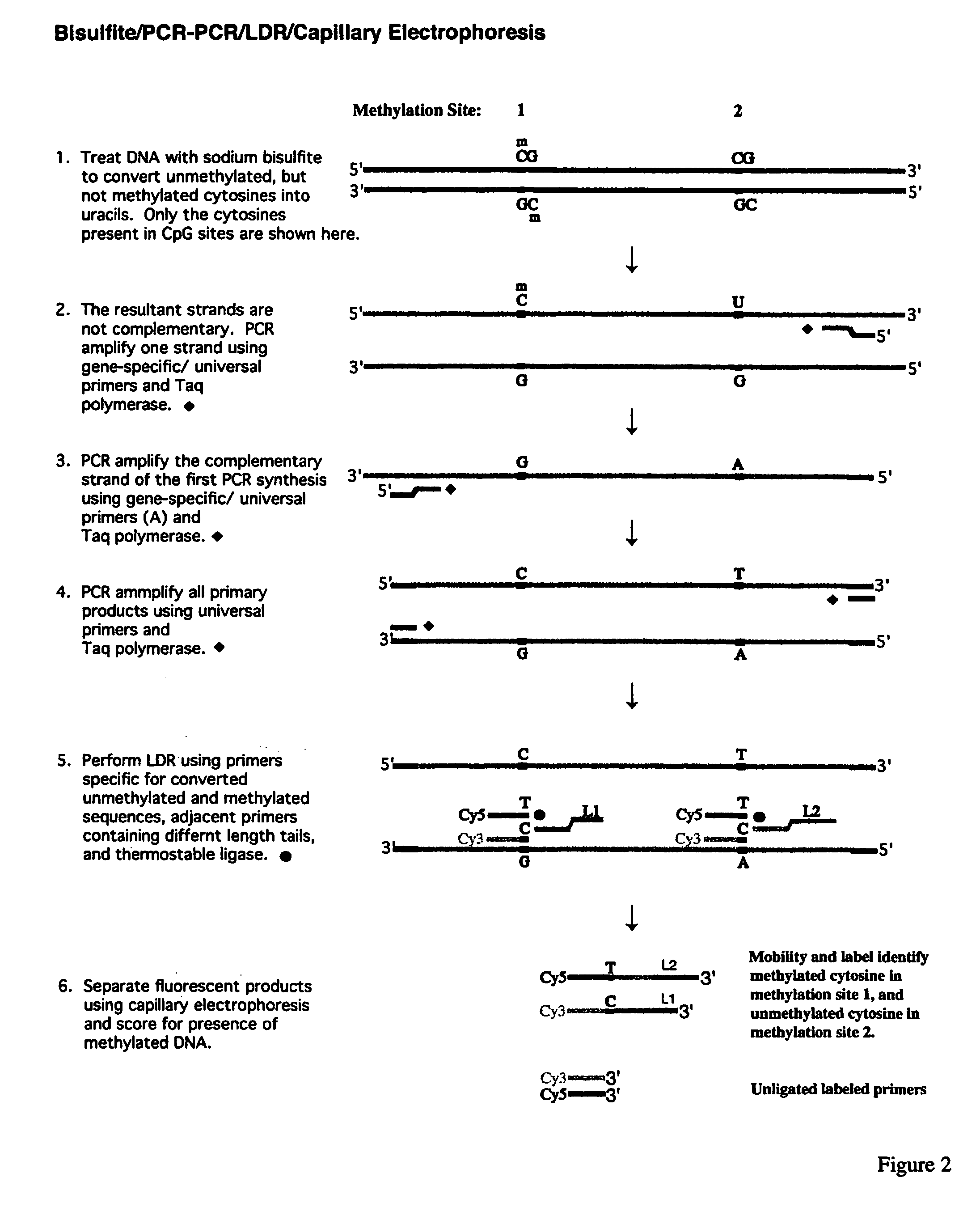
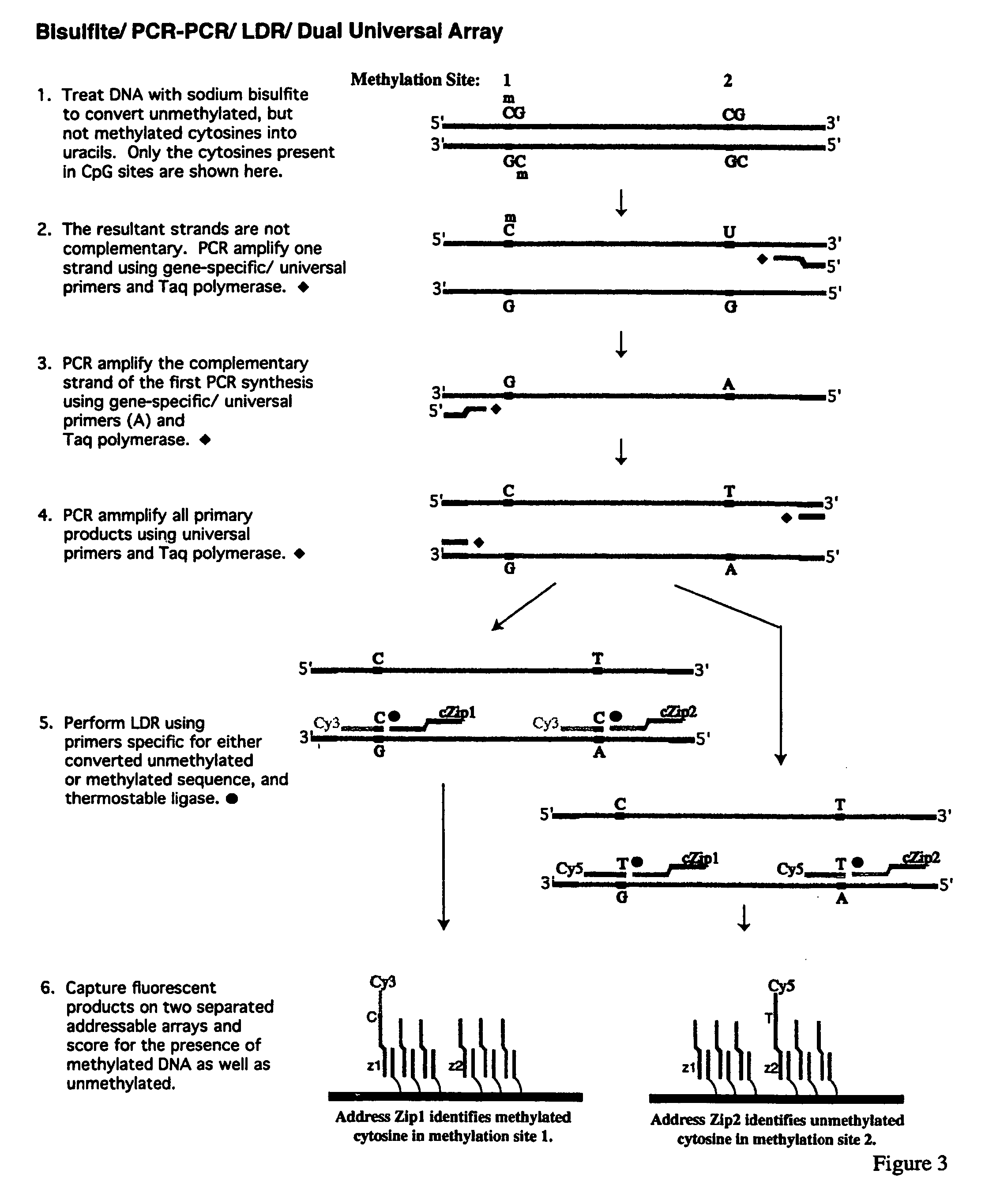
InactiveUS7358048B2Precise “ molecular signature ”Accurately methylation statusMicrobiological testing/measurementFermentationCapillary electrophoresisElectrophoresis
The present invention relates to the detection of promoter methylation status using a combination of either modification of methylated DNA or restriction endonuclease digestion, multiplex polymerase chain reaction, ligase detection reaction, and a universal array or capillary electrophoresis detection.
Owner:CORNELL RES FOUNDATION INC
Method and apparatus for identifying, classifying, or quantifying DNA sequences in a sample without sequencing
InactiveUS6418382B2Rapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsMicrobiological testing/measurementSample sequenceSingle sequence
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
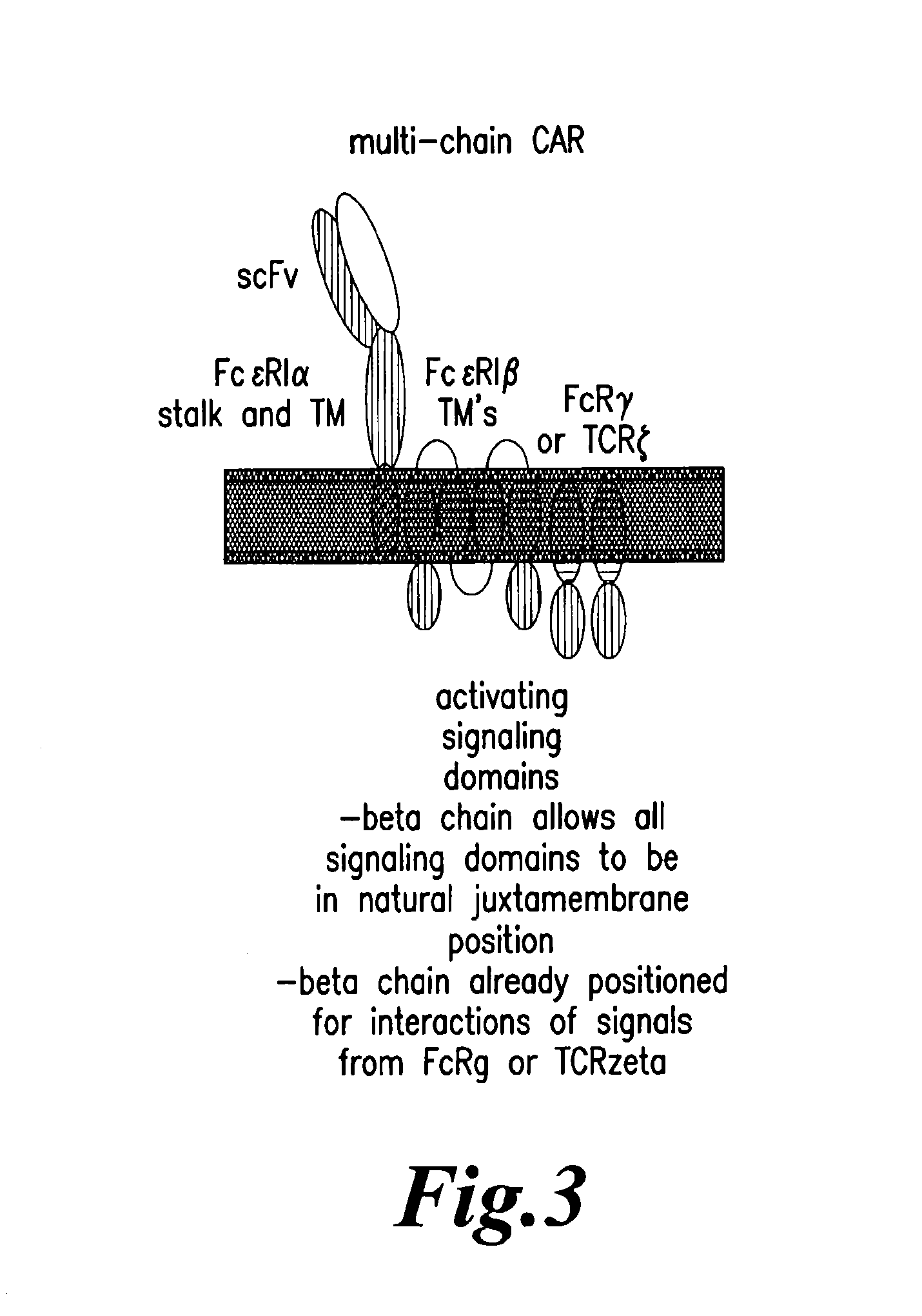
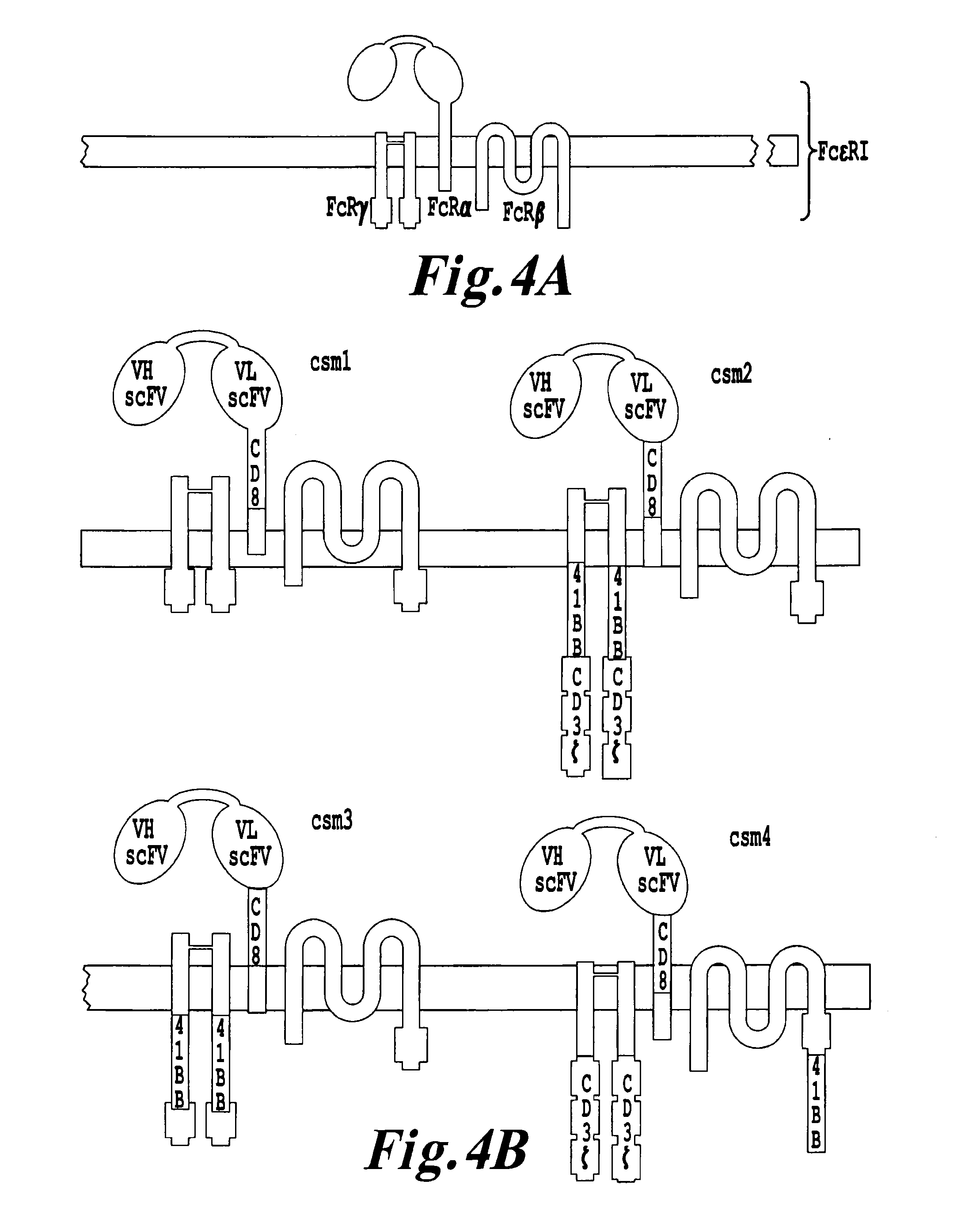
Methods for engineering t cells for immunotherapy by using rna-guided cas nuclease system
ActiveUS20160272999A1Accurate modificationHydrolasesGenetically modified cellsInfected cellAntigen receptors
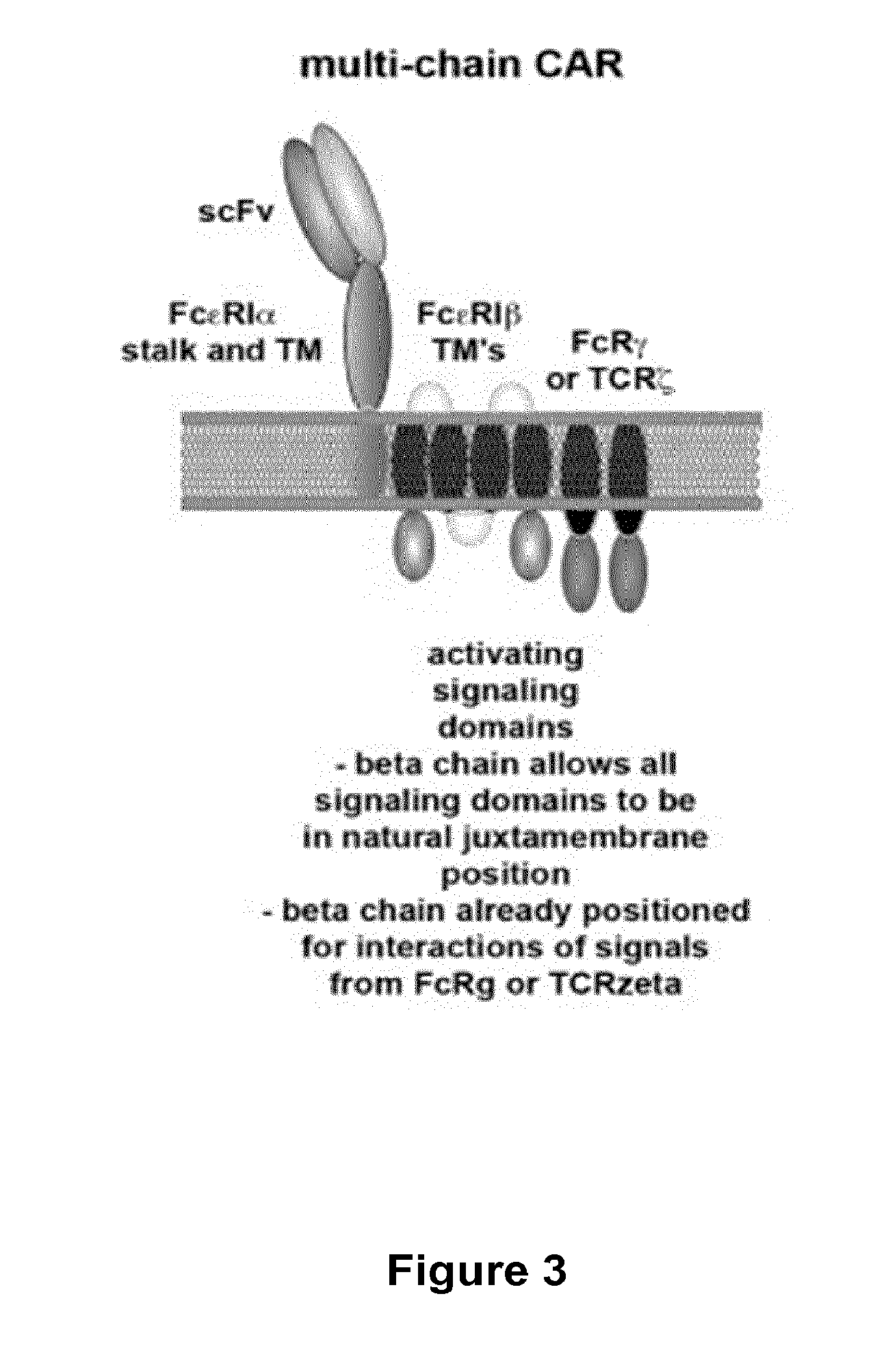
The present invention relates to methods of developing genetically engineered, preferably non-alloreactive T-cells for immunotherapy. This method involves the use of RNA-guided endonucleases, in particular Cas9 / CRISPR system, to specifically target a selection of key genes in T-cells. The engineered T-cells are also intended to express chimeric antigen receptors (CAR) to redirect their immune activity towards malignant or infected cells. The invention opens the way to standard and affordable adoptive immunotherapy strategies using T-Cells for treating cancer and viral infections.
Owner:CELLECTIS SA
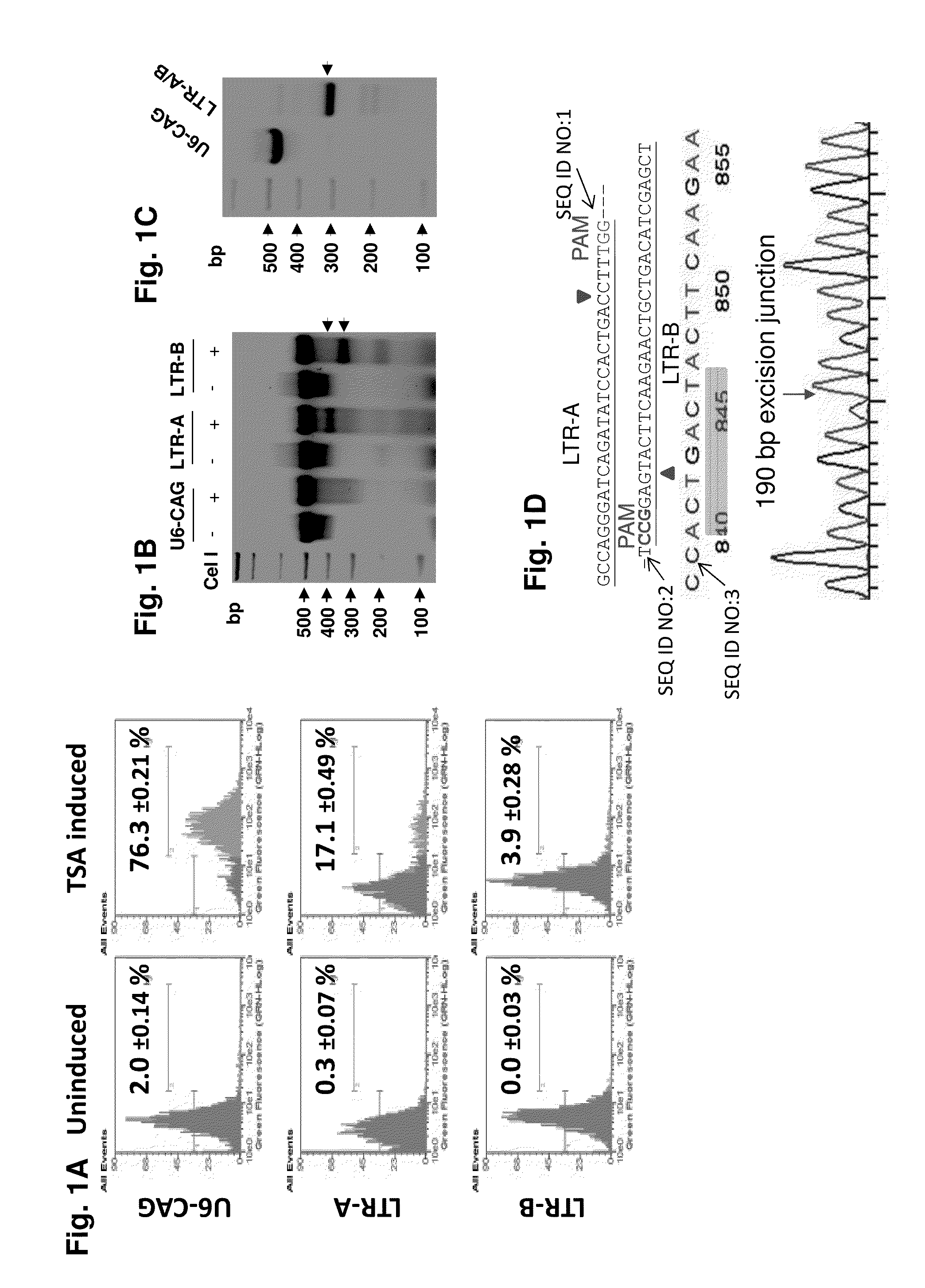
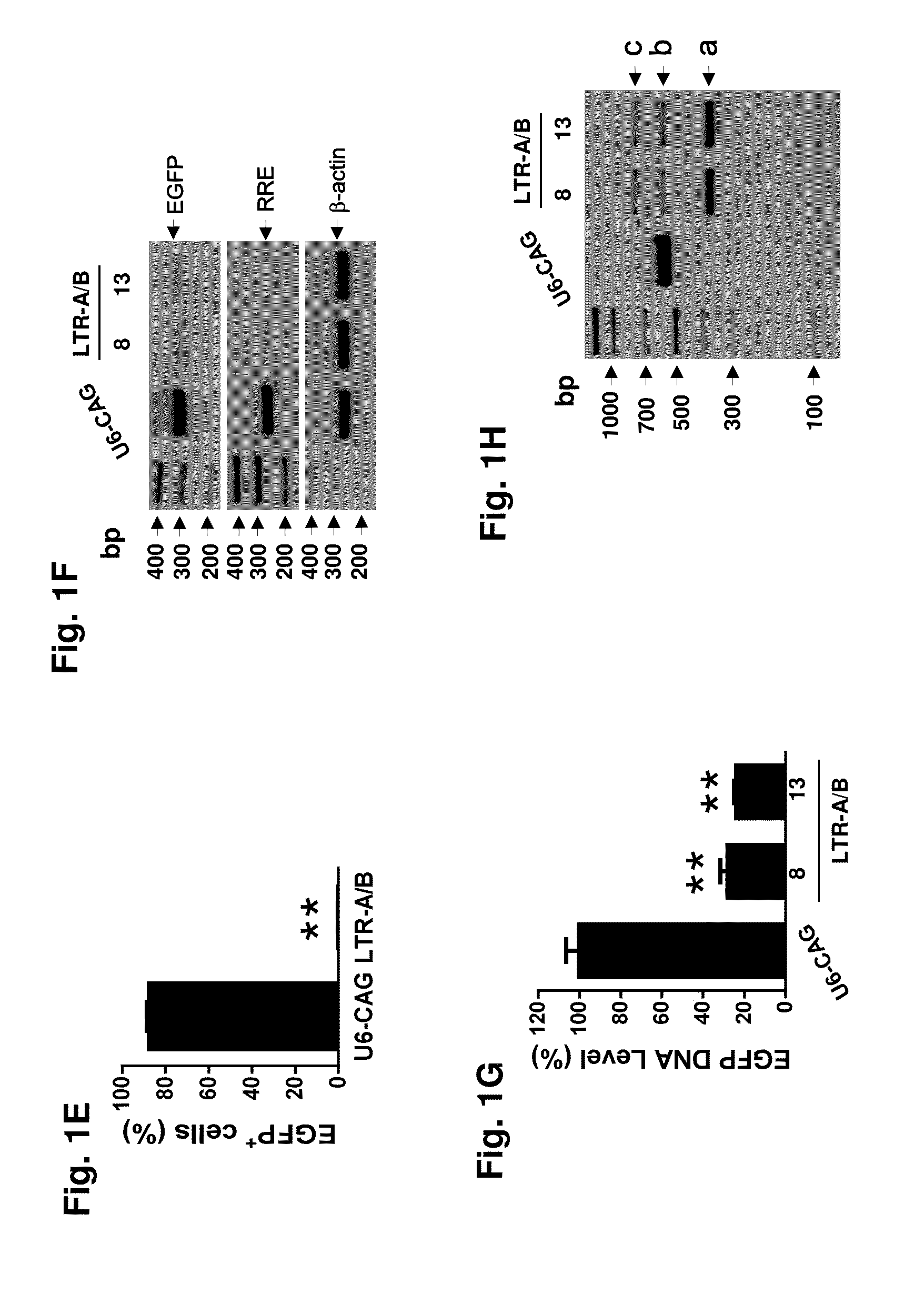
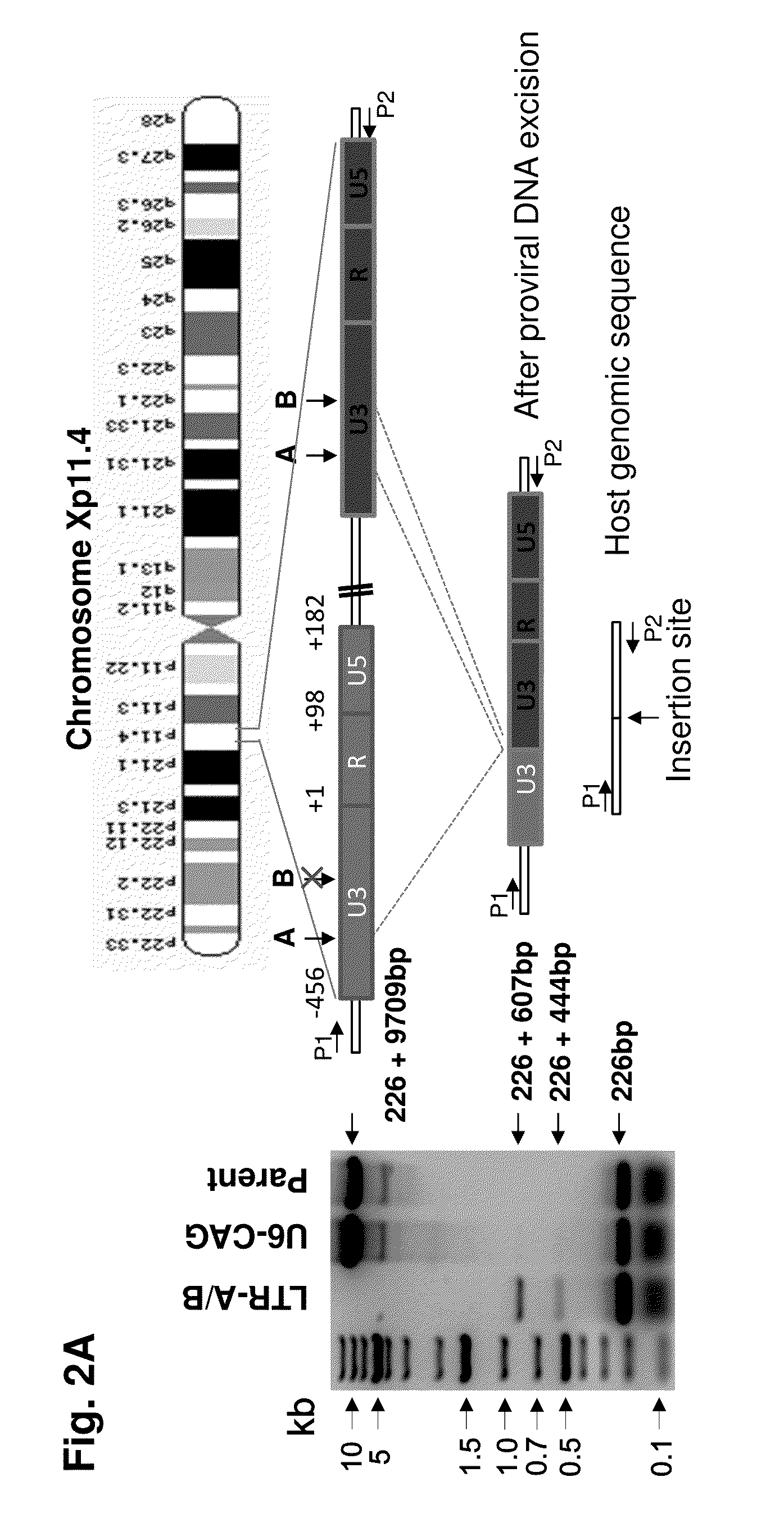
Methods and compositions for rna-guided treatment of HIV infection
A method of inactivating a proviral DNA integrated into the genome of a host cell latently infected with a retrovirus by treating the host cell with a composition comprising a Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated endonuclease, and two or more different guide RNAs (gRNAs), wherein each of the at least two gRNAs is complementary to a different target nucleic acid sequence in a long terminal repeat (LTR) in the proviral DNA, and inactivating the proviral DNA. A composition for use in inactivating a proviral DNA integrated into the genome of a host cell latently infected with a retrovirus including isolated nucleic acid sequences comprising a CRISPR-associated endonuclease and a guide RNA, wherein the guide RNA is complementary to a target sequence in a human immunodeficiency virus.
Owner:TEMPLE UNIVERSITY
Methods for engineering t cells for immunotherapy by using rna-guided cas nuclease system
ActiveUS20160184362A1Accurate modificationHydrolasesGenetically modified cellsInfected cellAntigen receptors
The present invention relates to methods of developing genetically engineered, preferably non-alloreactive T-cells for immunotherapy. This method involves the use of RNA-guided endonucleases, in particular Cas9 / CRISPR system, to specifically target a selection of key genes in T-cells. The engineered T-cells are also intended to express chimeric antigen receptors (CAR) to redirect their immune activity towards malignant or infected cells. The invention opens the way to standard and affordable adoptive immunotherapy strategies using T-Cells for treating cancer and viral infections.
Owner:CELLECTIS SA
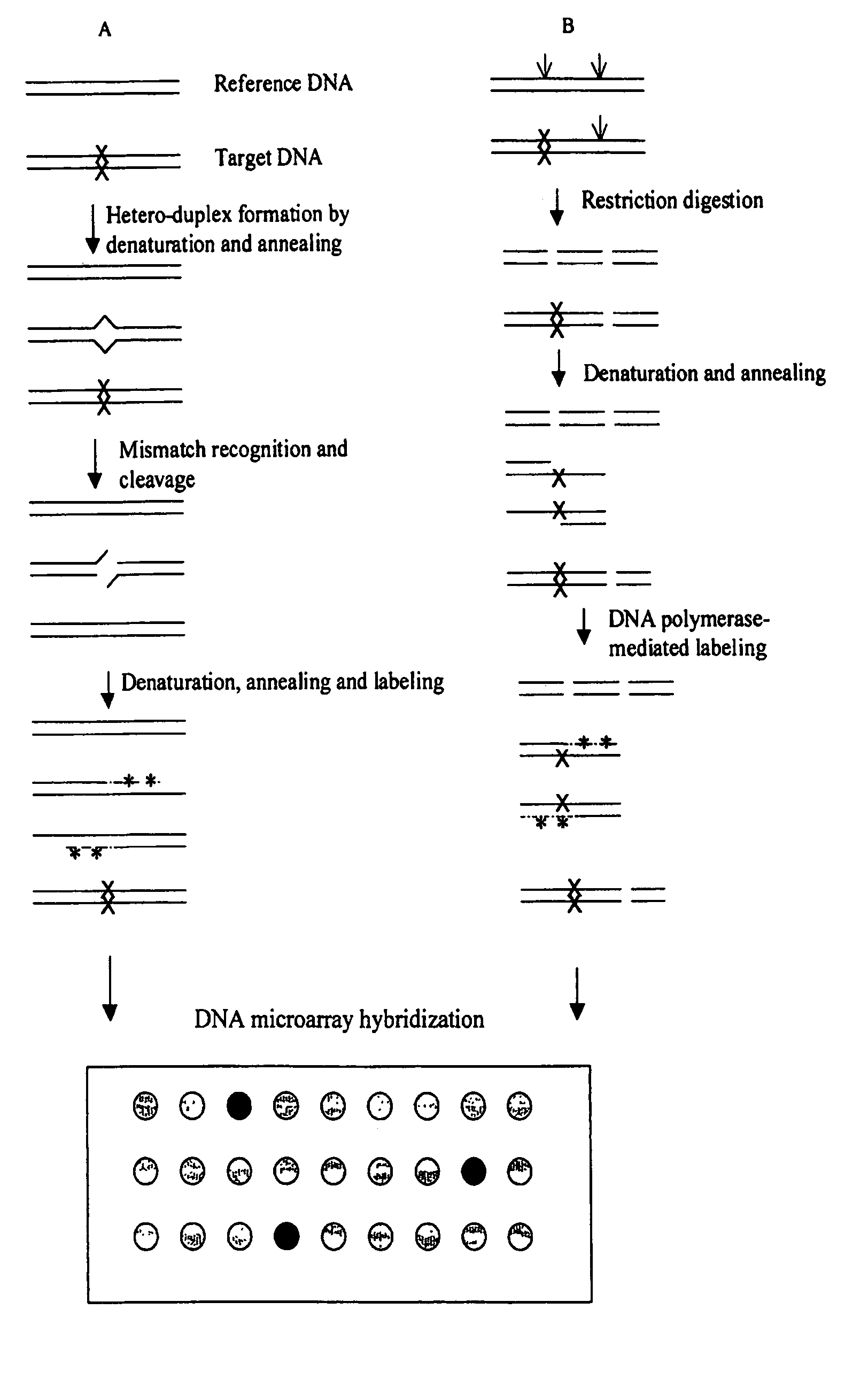
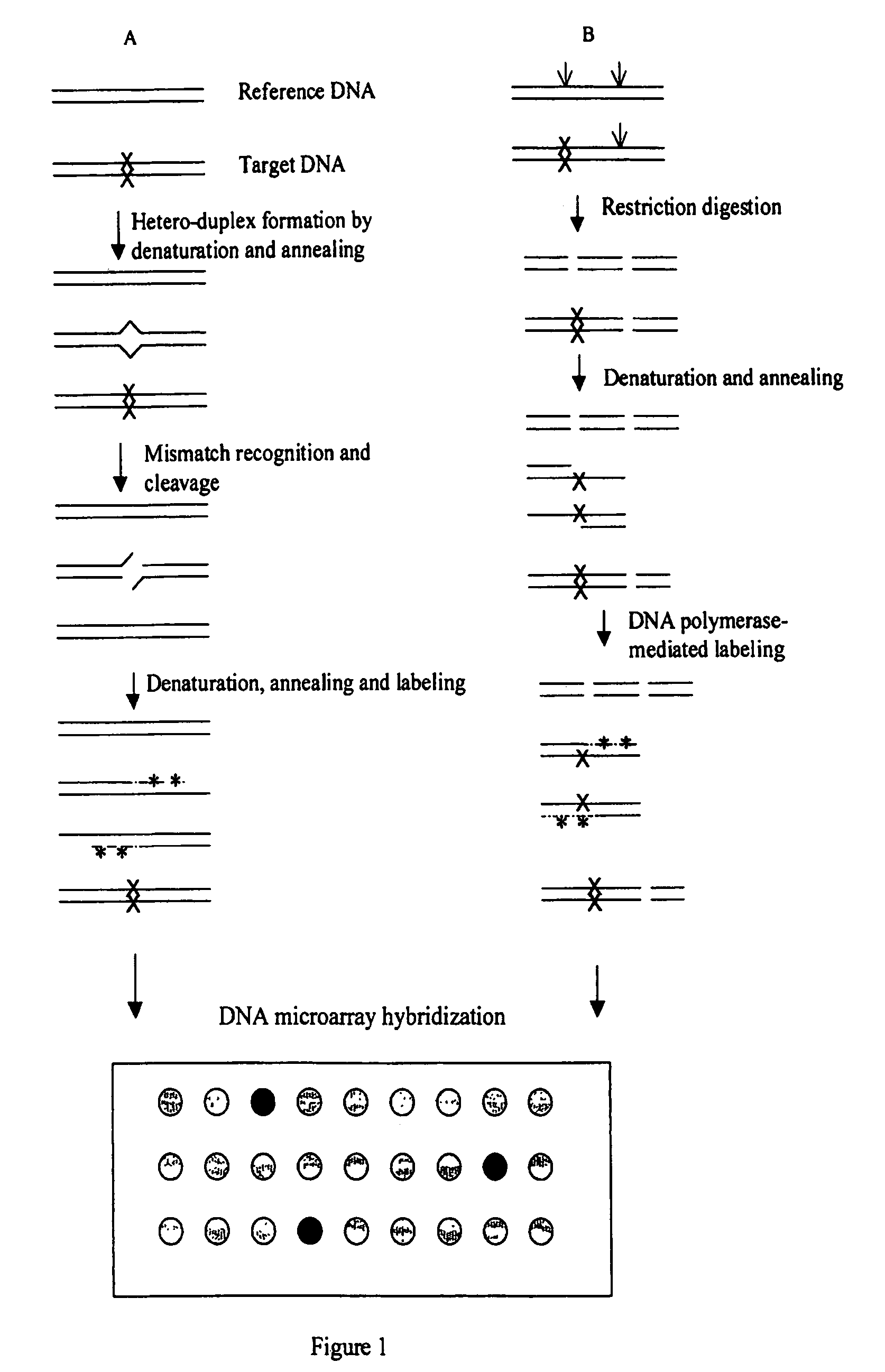
Methods for detecting and localizing DNA mutations by microarray
InactiveUS7378245B2Reduce impactMicrobiological testing/measurementFermentationDNA microarrayHeteroduplex
Owner:UNIVERSITY OF OREGON
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
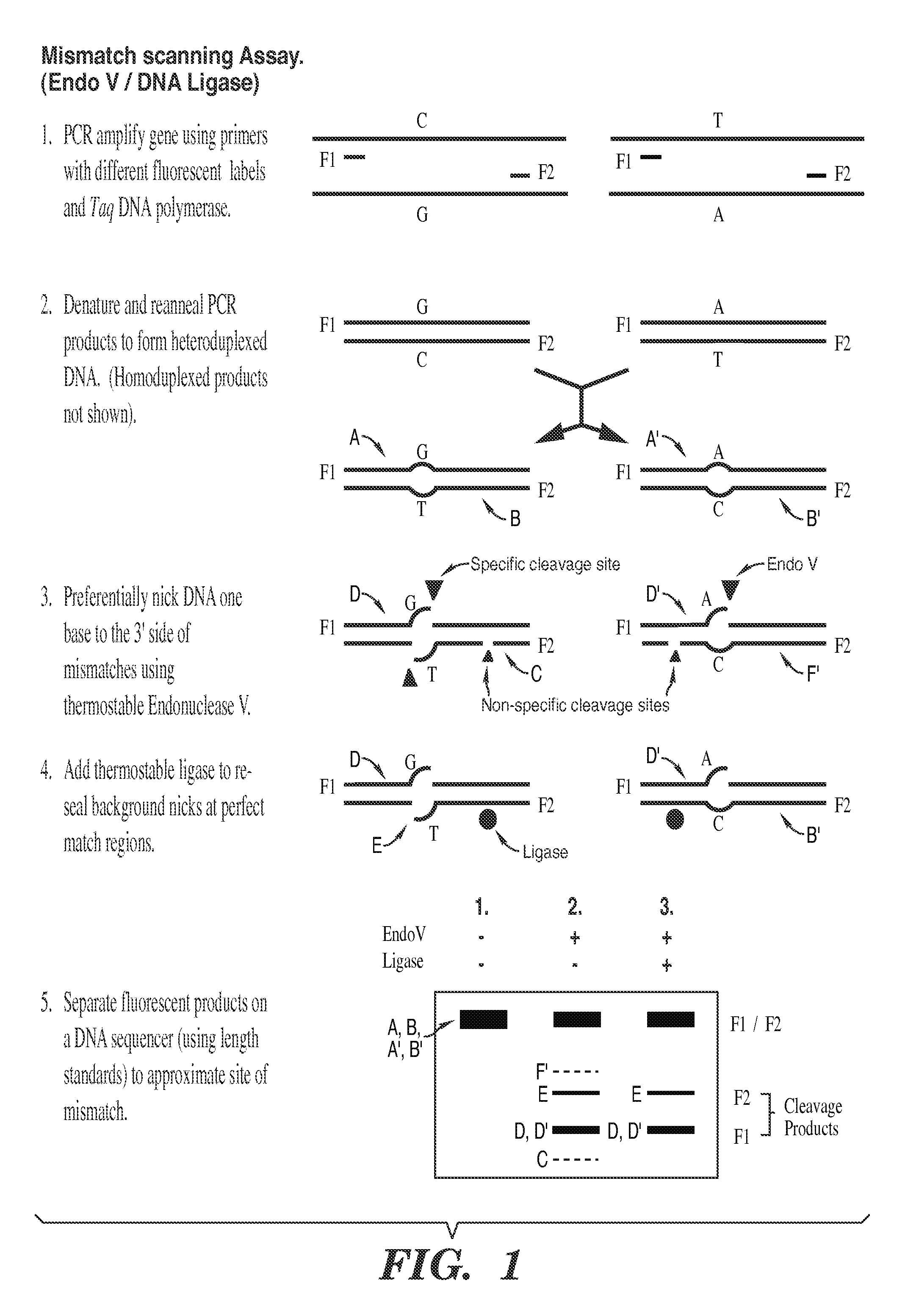
Detection of nucleic acid differences using combined endonuclease cleavage and ligation reactions
InactiveUS7807431B2Wide applicabilityImprove throughputSugar derivativesHydrolasesHeteroduplexSingle nucleotide mutation
The present invention is a method for detecting DNA sequence differences including single nucleotide mutations or polymorphisms, one or more nucleotide insertions, and one or more nucleotide deletions. Labeled heteroduplex PCR fragments containing base mismatches are prepared. Endonuclease cleaves the heteroduplex PCR fragments both at the position containing the variation (one or more mismatched bases) and to a lesser extent, at non-variant (perfectly matched) positions. Ligation of the cleavage products with a DNA ligase corrects non-variant cleavages and thus substantially reduces background. This is then followed by a detection step in which the reaction products are detected, and the position of the sequence variations are determined.
Owner:CORNELL RES FOUNDATION INC
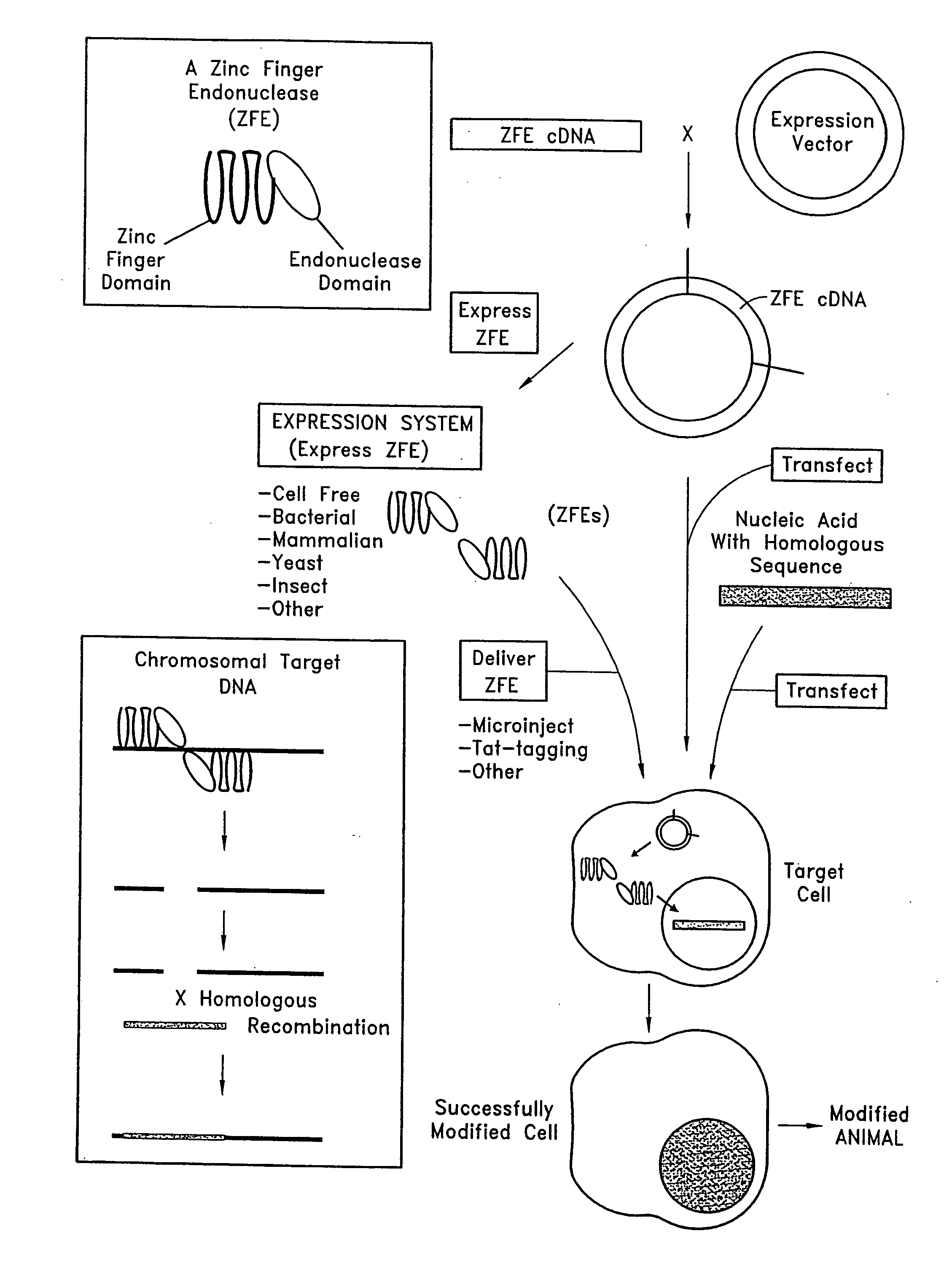
Methods and compositions for using zinc finger endonucleases to enhance homologous recombination
Owner:SANGAMO BIOSCIENCES INC
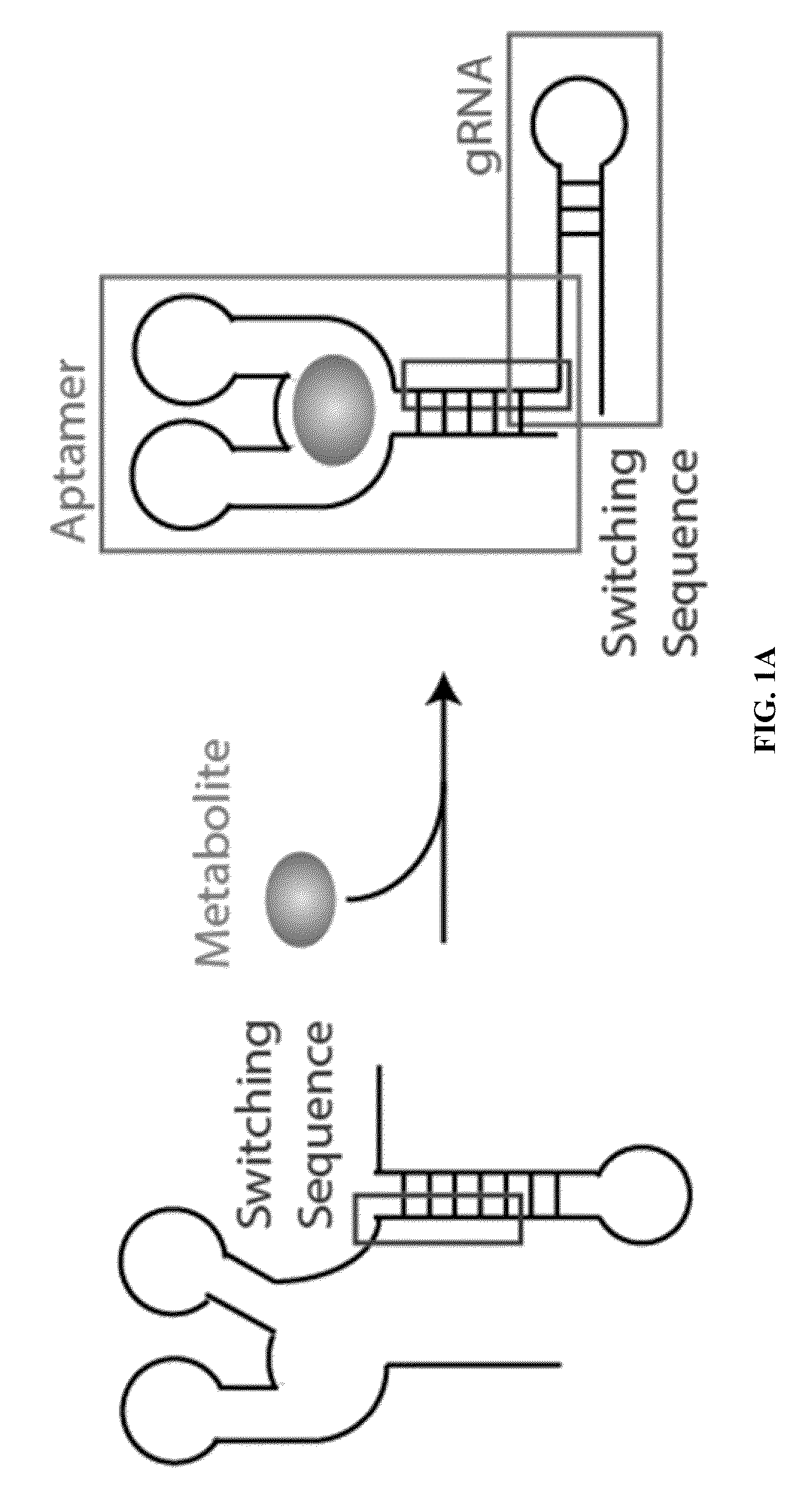
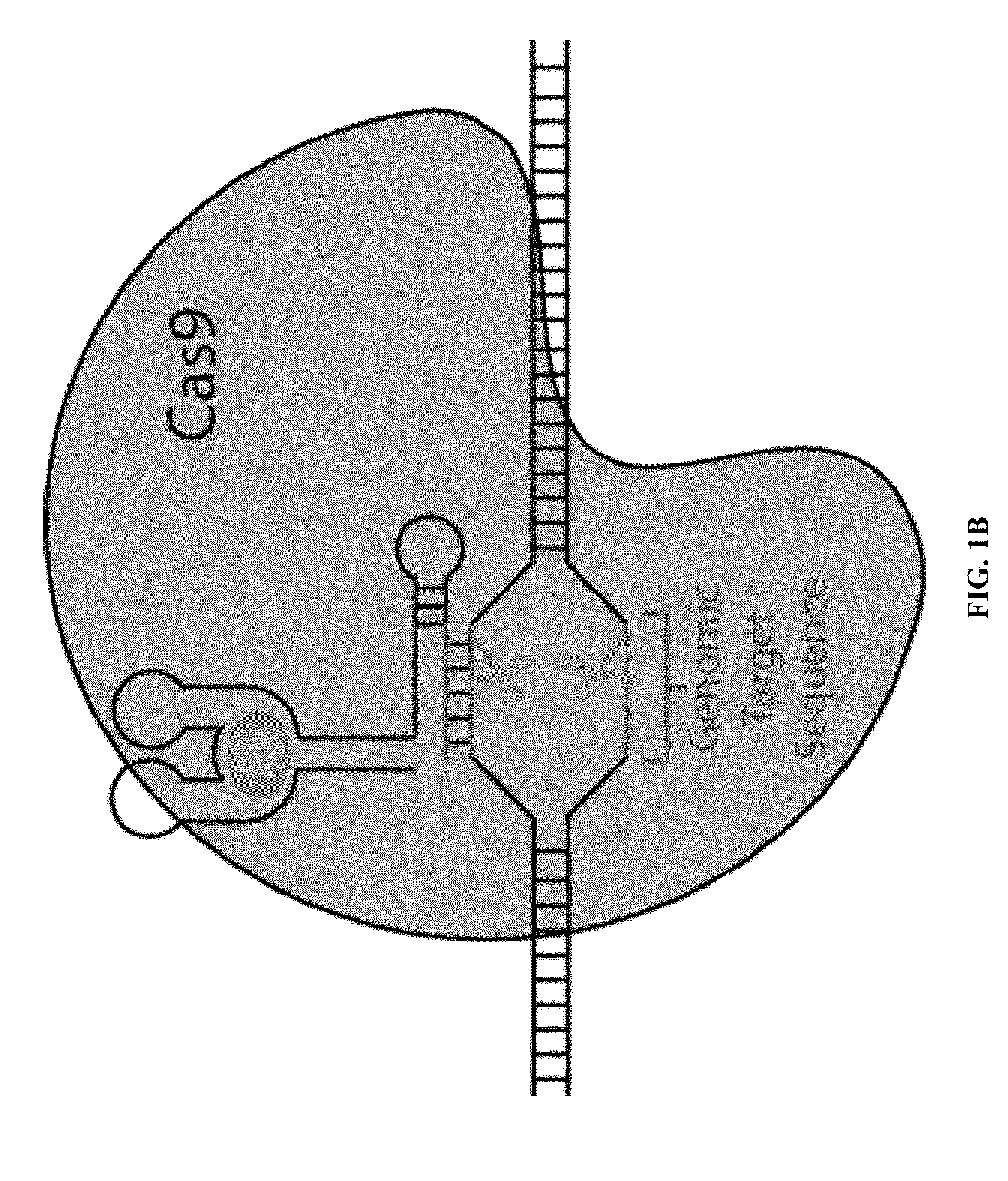
Switchable grnas comprising aptamers
ActiveUS20150071900A1Reduce the possibilityHydrolasesPeptide/protein ingredientsBiologyDNA Endonuclease
Some aspects of this disclosure provide compositions, methods, systems, and kits for controlling the activity and / or improving the specificity of RNA-programmable endonucleases, such as Cas9. For example, provided are guide RNAs (gRNAs) that are engineered to exist in an “on” or “off” state, which control the binding and hence cleavage activity of RNA-programmable endonucleases.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com