Fast CRISPR-Cas9 working efficiency testing system and application thereof
A technology for testing system and work efficiency, applied in genetic engineering, virus/phage, microbial assay/inspection, etc., can solve the problems of gene editing efficiency and inability to accurately test the pros and cons of sgRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Application of the CRISPR / Cas9 work efficiency test system based on the pBGN plasmid containing the BSD-fsEGFP fusion gene (screening sgRNA targeting the mouse FBXW7 gene)
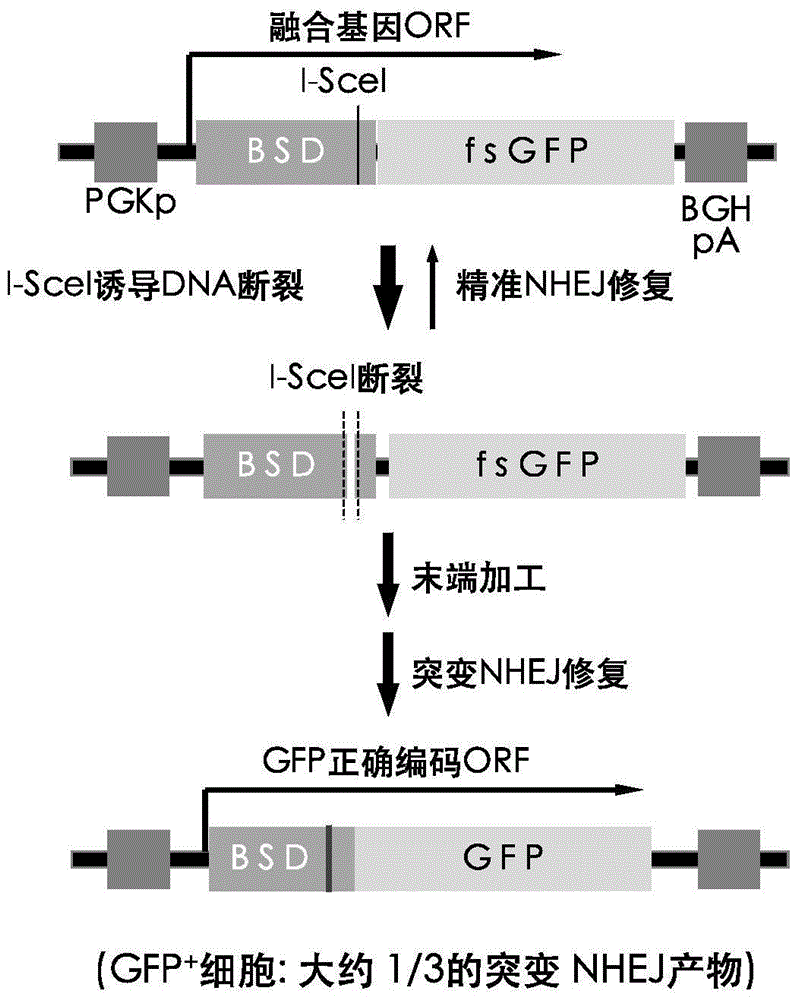
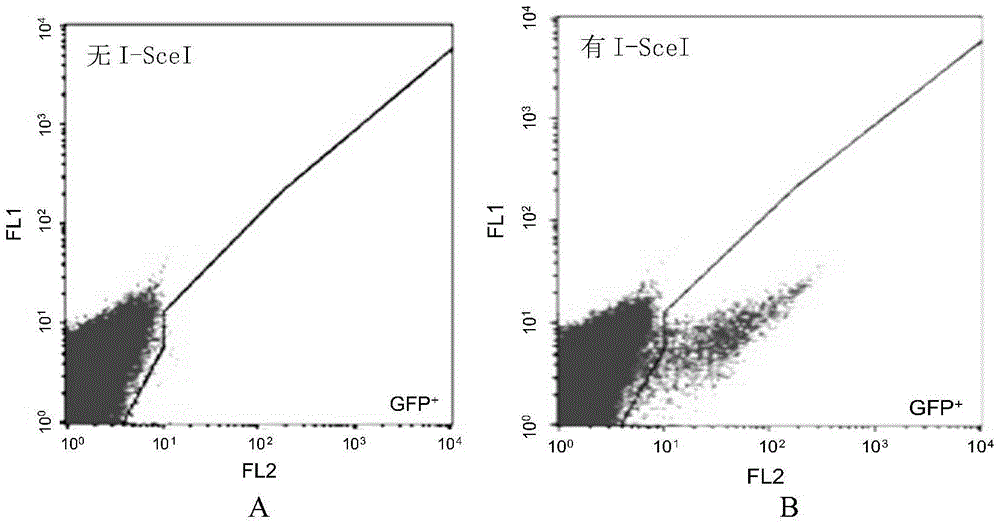
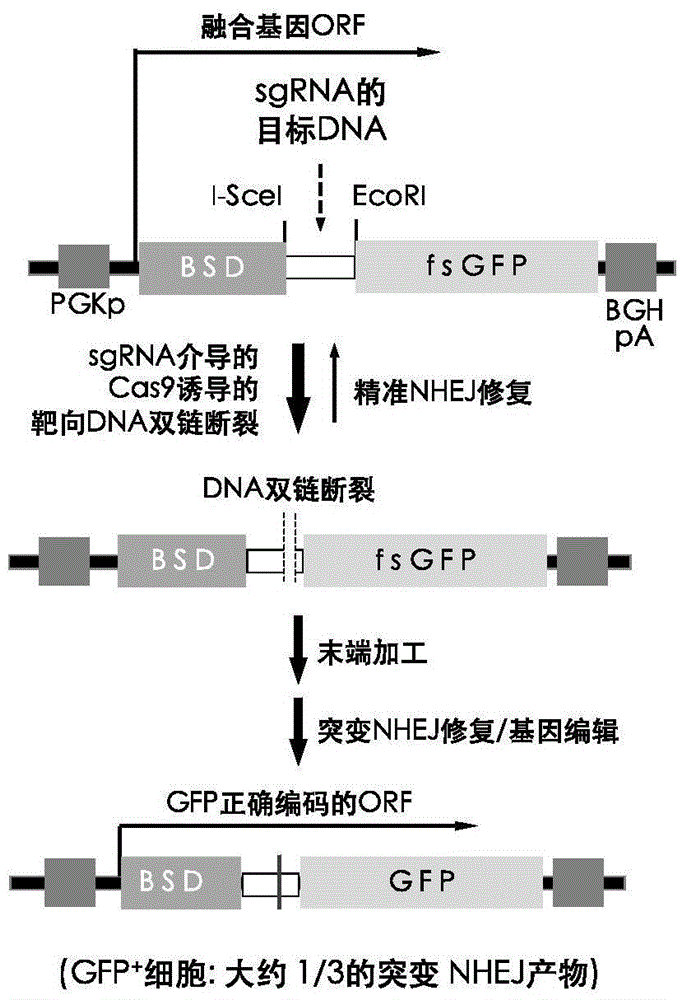
[0056] (1) BSD-fsEGFP fusion gene: use conventional PCR to amplify the known BSD gene, 5'-PCR primers with HindIII sites, and 3'-PCR primers to introduce I-SceI and EcoRI sites. The PCR product (BSD) is inserted into the HindIII and EcoRI sites between the CMV driver and the EGFP coding region in the EGFP plasmid (the EGFP nucleotide sequence is shown in SEQ ID NO.1, and the amino acid sequence is shown in SEQ ID NO.2), generating Plasmid pBGN containing BSD-fsEGFP fusion gene ( image 3 ), the nucleotide sequence of the BSD-fsEGFP fusion gene is shown in SEQ ID NO.3, and the amino acid sequence is shown in SEQ ID NO.4. The fusion gene is driven by CMV driver or PGK driver, but EGFP is inactive due to frameshift, so it is called fsEGFP.
[0057] 5'-PCR primers are
[0058] CTCAAGCTTAAC...
Embodiment 2
[0072] Example 2: Application of the CRISPR / Cas9 work efficiency test system based on the pBLuc plasmid containing the BSD-fsFLuc fusion gene (screening the sgRNA targeting the mouse MDC1 gene)
[0073] (1) Utilize conventional PCR amplification system and condition, PCR amplifies FLuc gene (nucleotide sequence is shown in SEQIDNO.5, aminoacid sequence is shown in SEQIDNO.6), 5'-PCR primer band KpnI site, 3 '-primer with NotI site. The PCR product (FLuc) replaced the EGFP part between KpnI and NotI in the plasmid pBGN (containing BSD-fsEGFP) to generate the BSD-fsFLuc plasmid pBLuc ( Figure 4 ), FLuc is inactive due to frameshift, so it is called fsFLuc. The nucleotide sequence of the BSD-fsFLuc fusion gene is shown in SEQ ID NO.7, and the amino acid sequence is shown in SEQ ID NO.8.
[0074] 5'-PCR primers are
[0075] GACGGTACCGCGGGCCCGGGATCCATCGCCACCATGGAAGATGCCAAAAAC,
[0076] The 3'-PCR primer was AGTCGCGGCCGCTTTACACGGCGATCTTGCCGC.
[0077] (2) Similar to Example 1,...
Embodiment 3
[0093] Example 3: Screening of sgRNA targeting human hypoxanthine phosphoribosyltransferase HPRT and application of human HPRT gene editing (knockout)
[0094] (1) The construction of pBGN is the same as in Example 1.
[0095] (2) Utilize the CRISPR / Cas9 gene editing test system of the present invention to screen the best sgRNA for gene editing (knockout) of human cell hypoxanthine phosphoribosyltransferase HPRT. Embodiment is with embodiment 1. First, seven different sgRNAs were designed for the human HPRT gene using www.crispr.mit.edu website.
[0096] Six human HPRTsgRNA expression sequences and targeting sequences, the underlined part indicates the PAM motif:
[0097] sgRNA0 expression sequence 5'to3'GTGCTTTGATGTAATCCAGC
[0098] sgRNA0 target sequence 5'to3'GTGCTTTGATGTAATCCAGC AGG
[0099] sgRNA1 expression sequence 5'to3'TAAATTCTTTGCTGACCTGC
[0100] sgRNA1 target sequence 5'to3'TAAATTCTTTGCTGACCTGC TGG
[0101] sgRNA2 expression sequence 5'to3'TGTAGCCCCTCTGTG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com