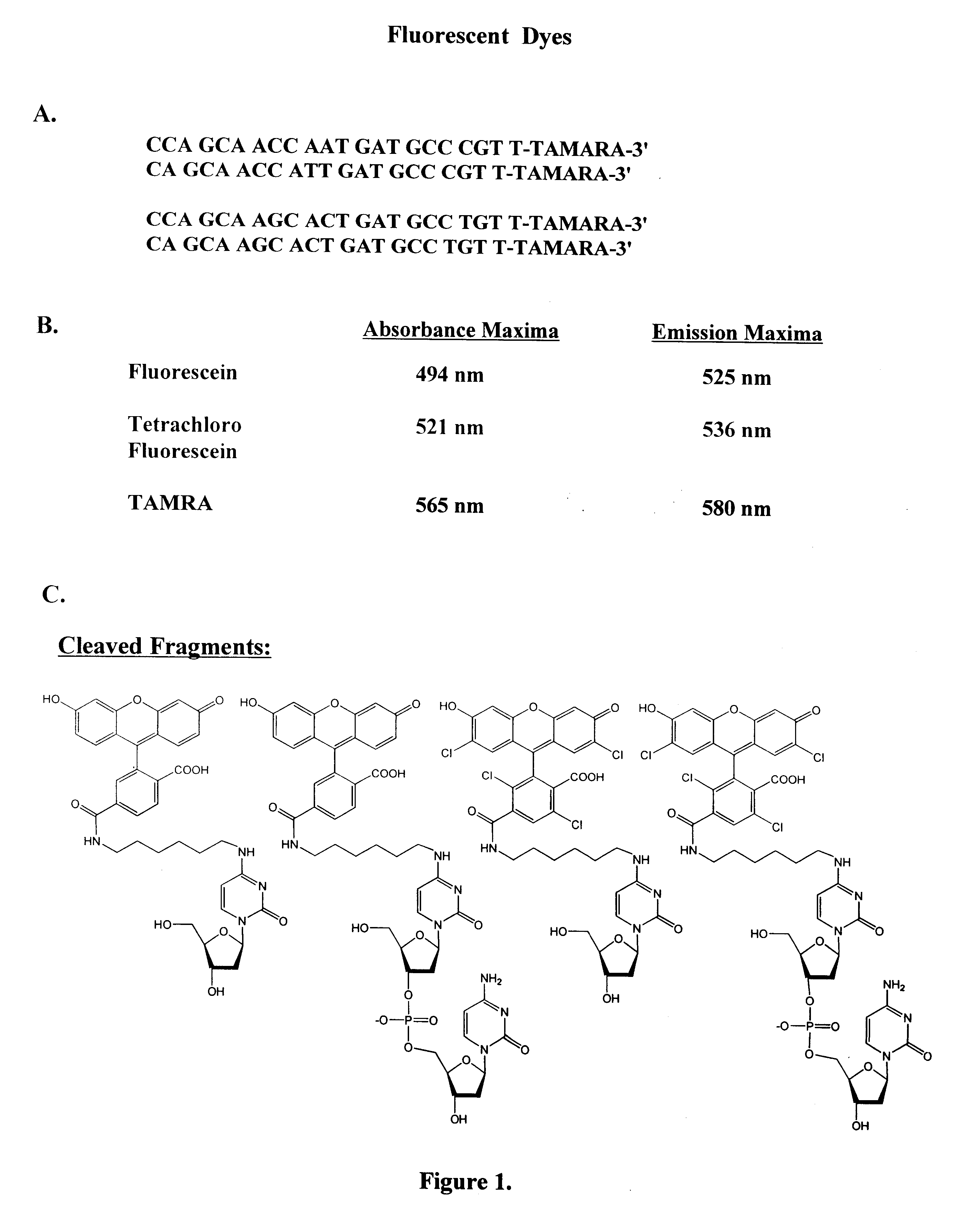
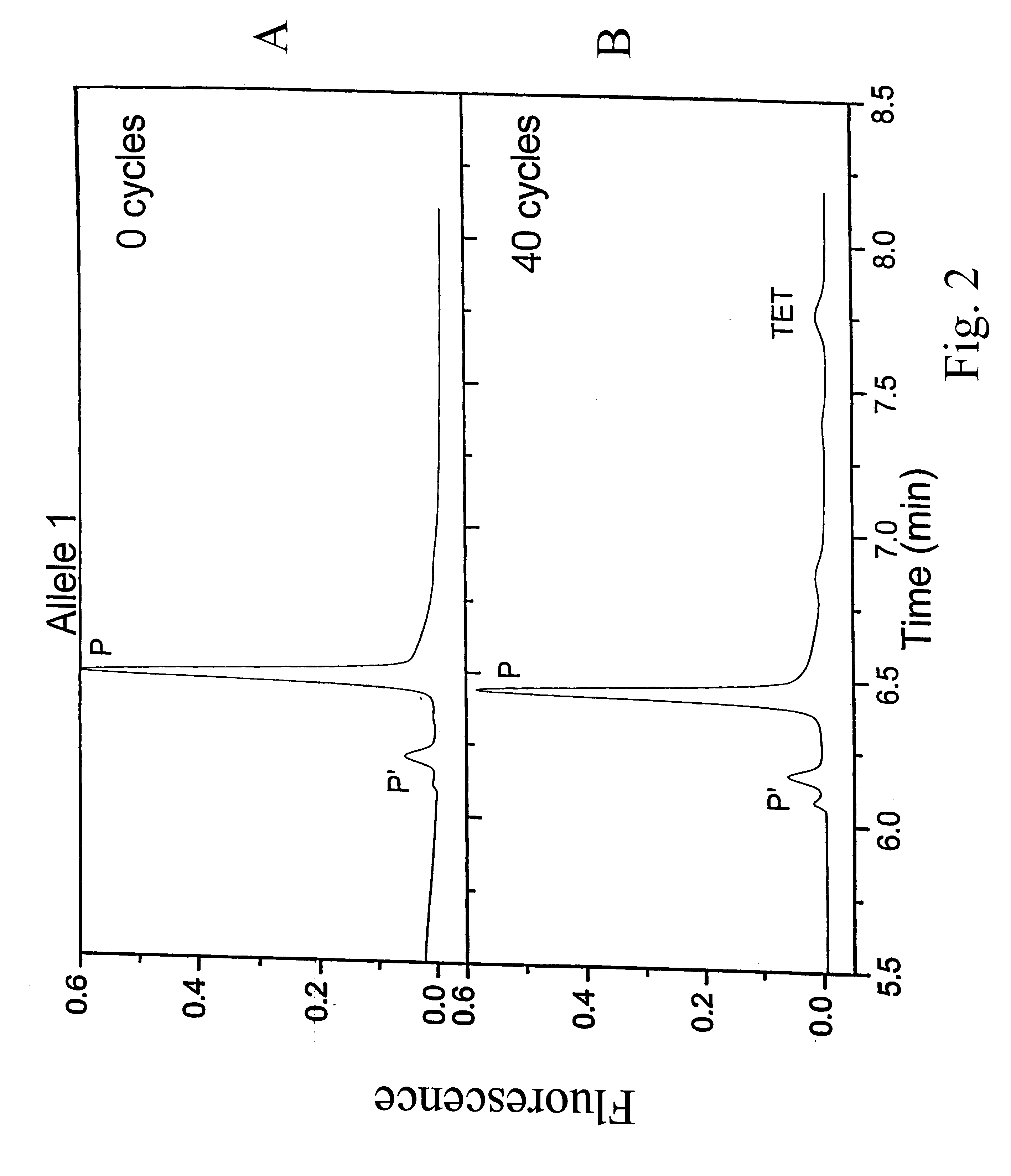
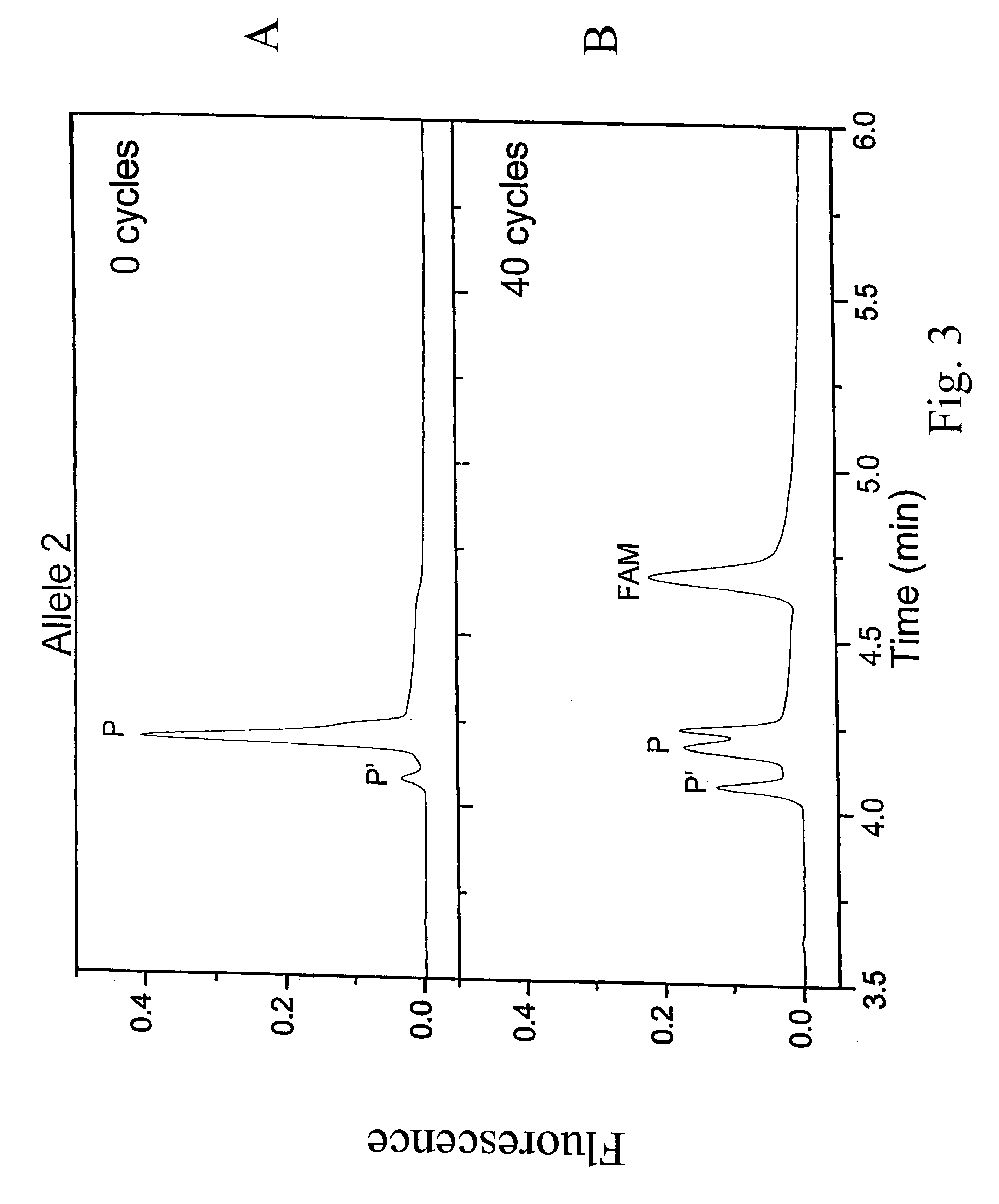
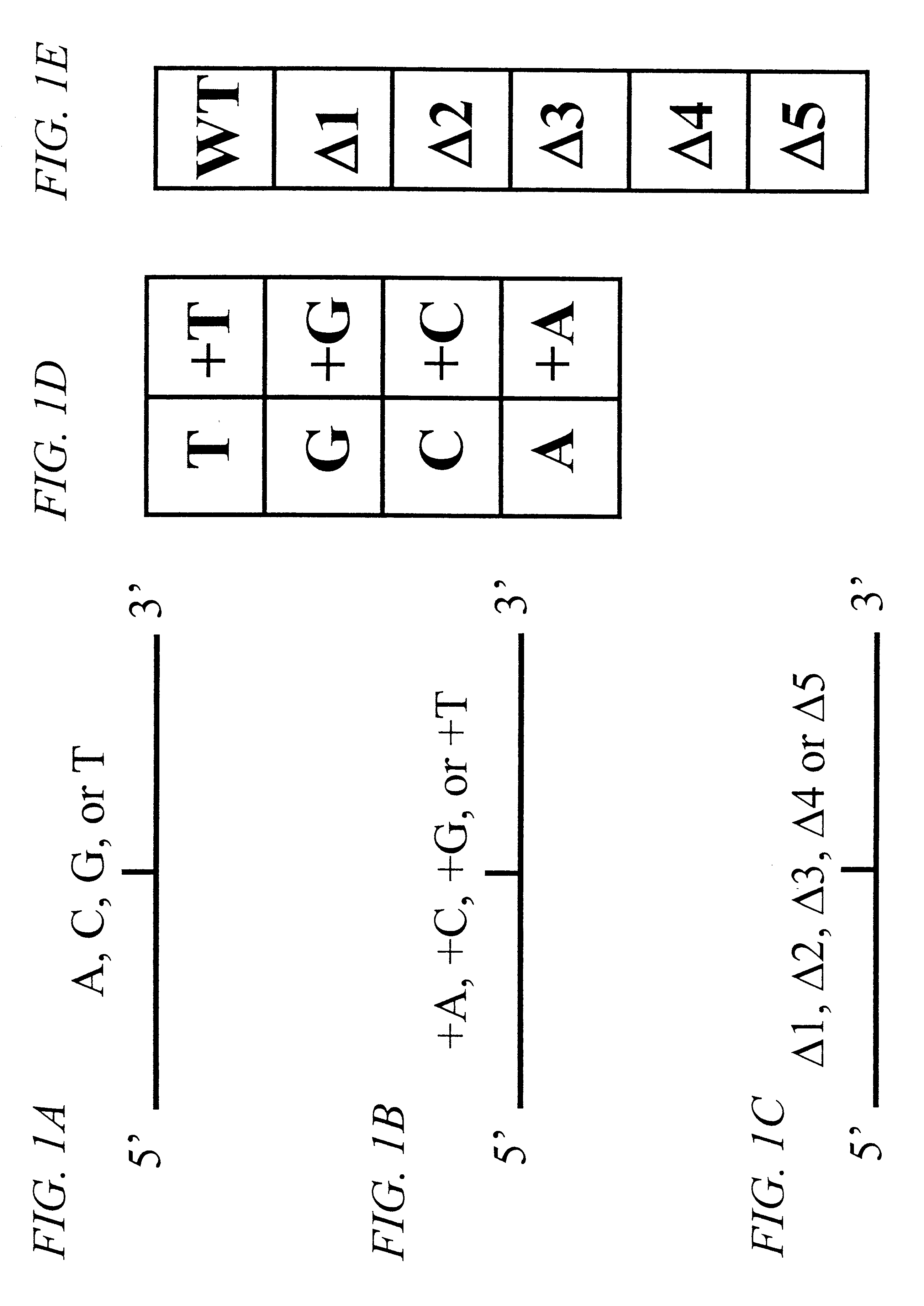
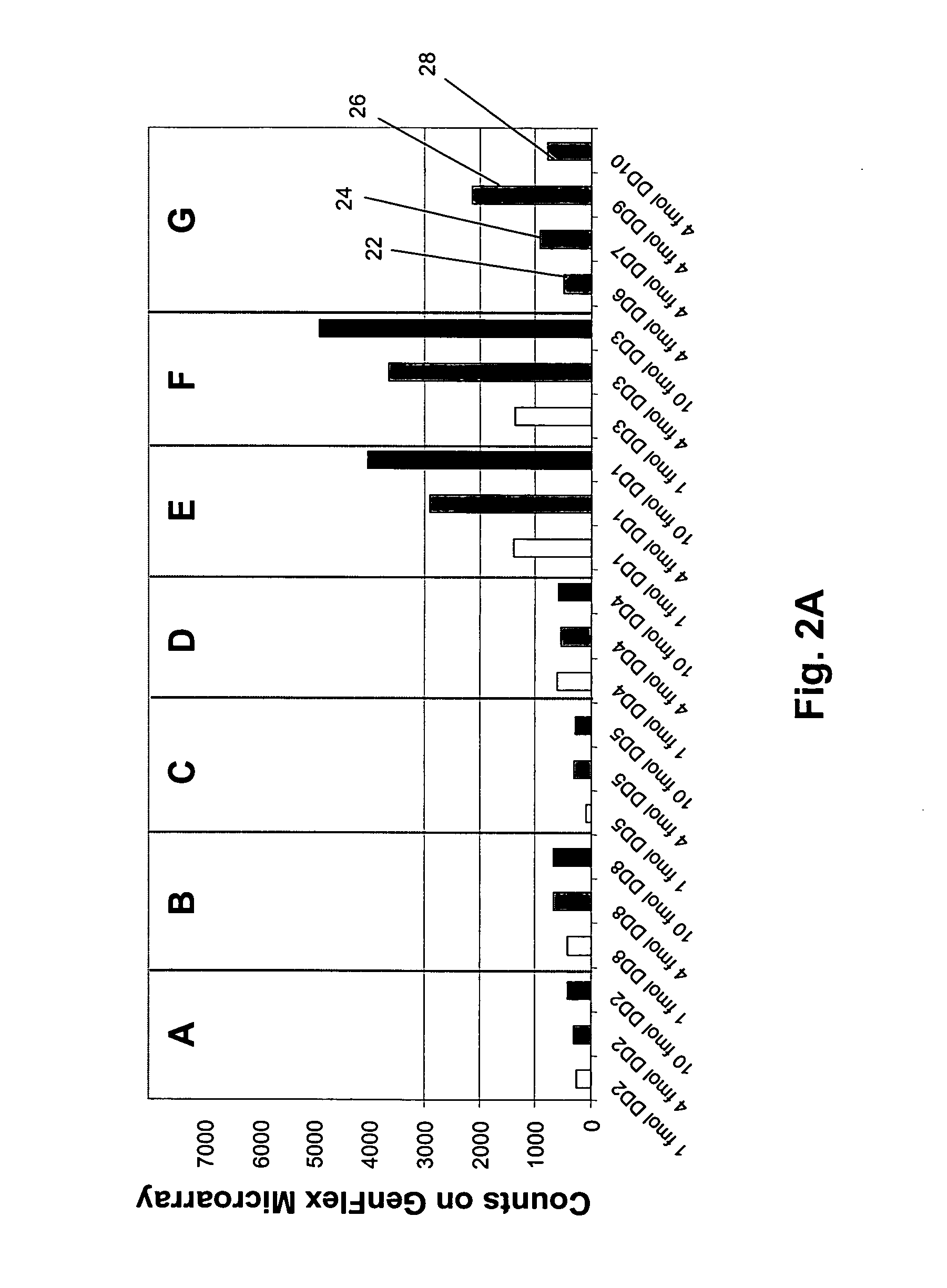
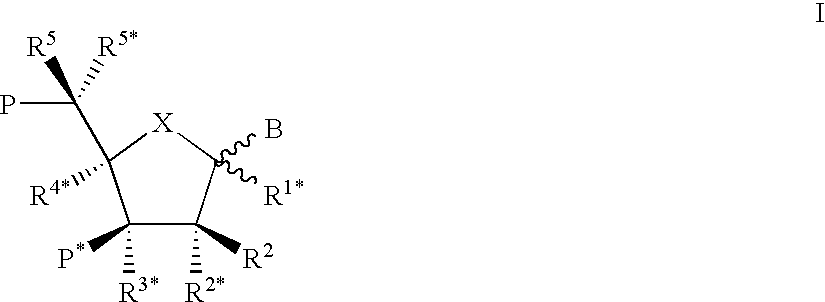Patents
Literature
4059 results about "Genomic clone" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
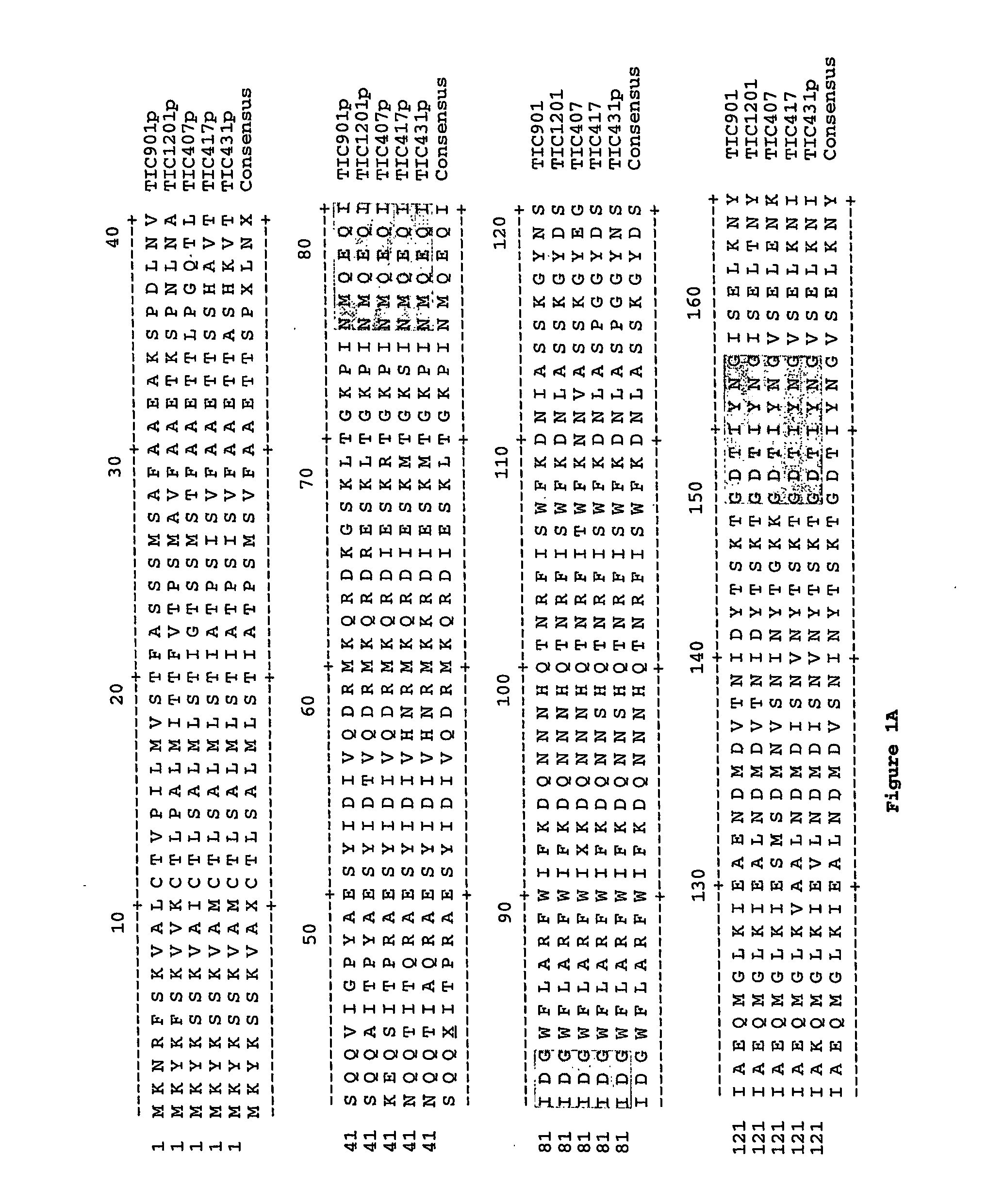
Genomic clone a cell with a vector containing a fragment of DNA from a different organism. A cell with a vector containing a fragment of DNA from a different organism. pertaining to a genome. see clone. the DNA sequences making up the genome of an individual. see gene bank.
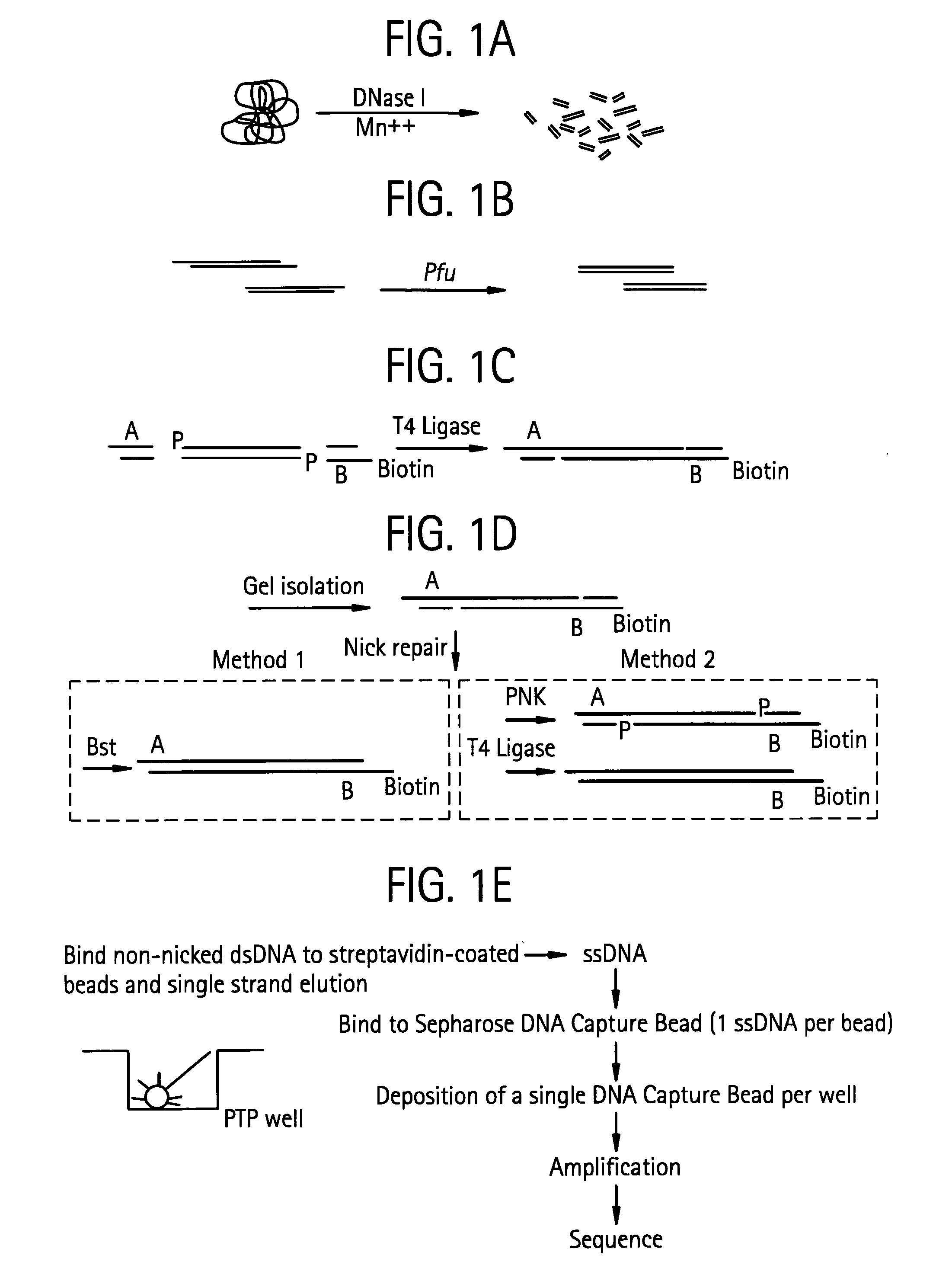
Methods of amplifying and sequencing nucleic acids
An apparatus and method for performing rapid DNA sequencing, such as genomic sequencing, is provided herein. The method includes the steps of preparing a sample DNA for genomic sequencing, amplifying the prepared DNA in a representative manner, and performing multiple sequencing reaction on the amplified DNA with only one primer hybridization step.
Owner:454 LIFE SCIENCES CORP
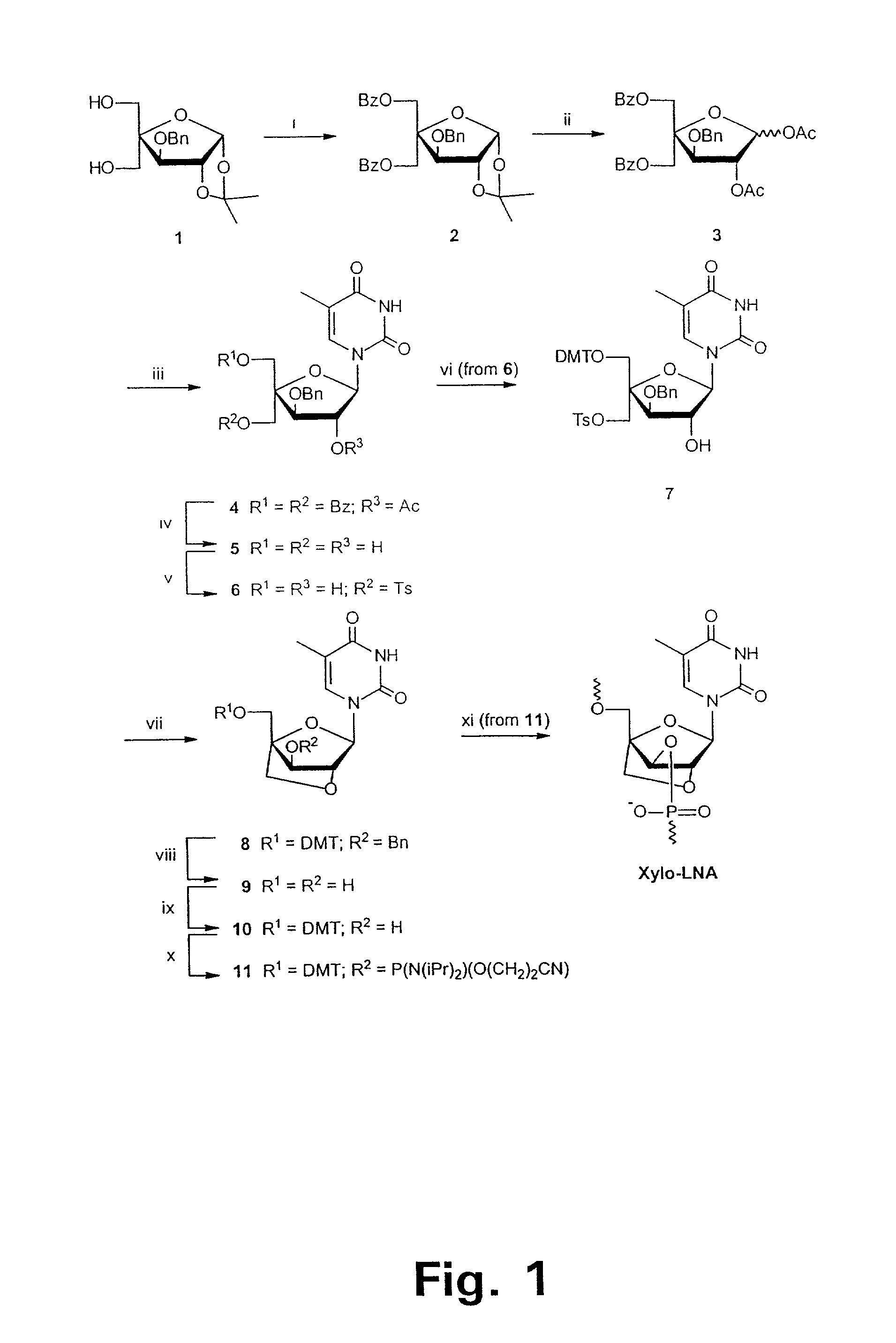
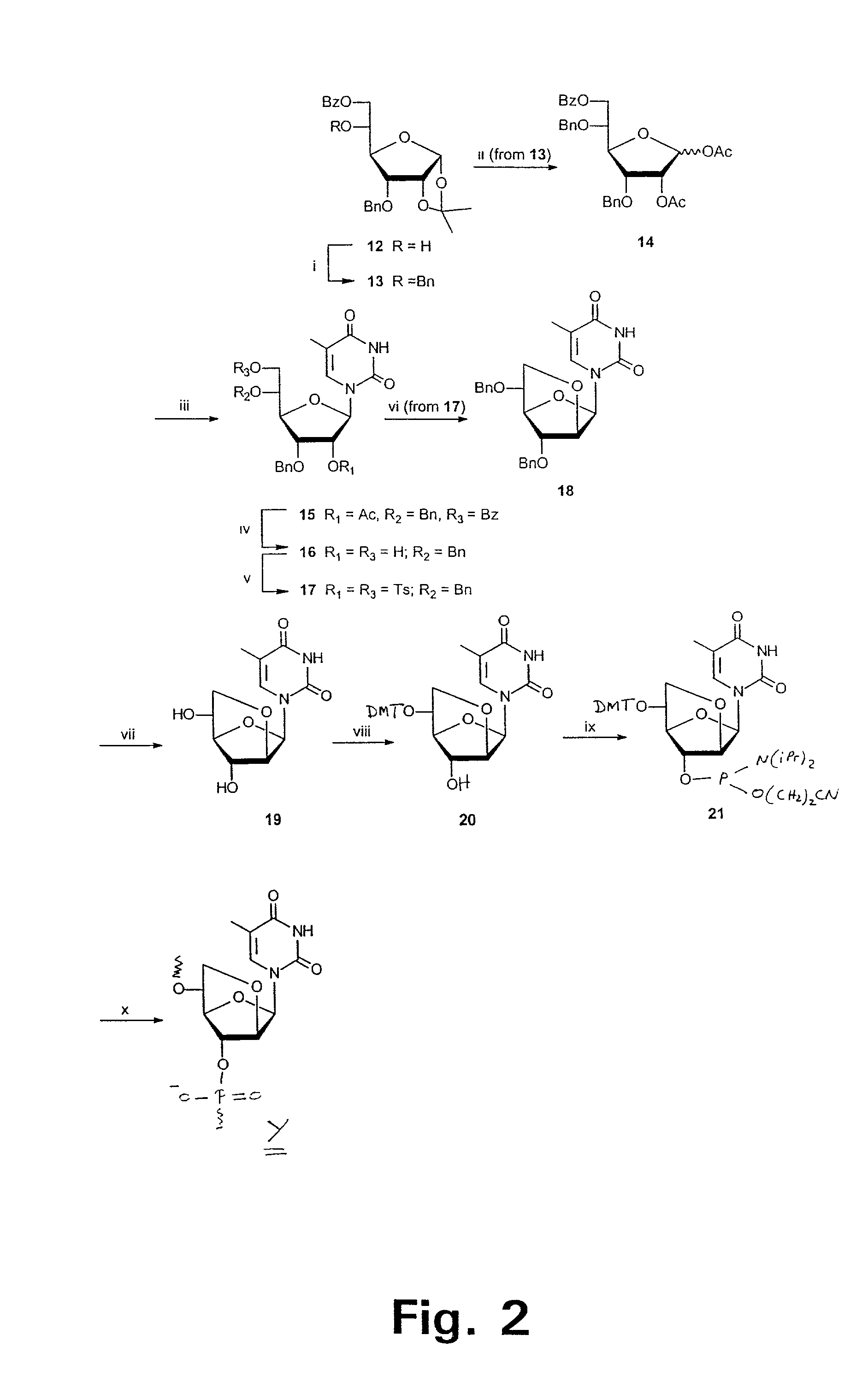
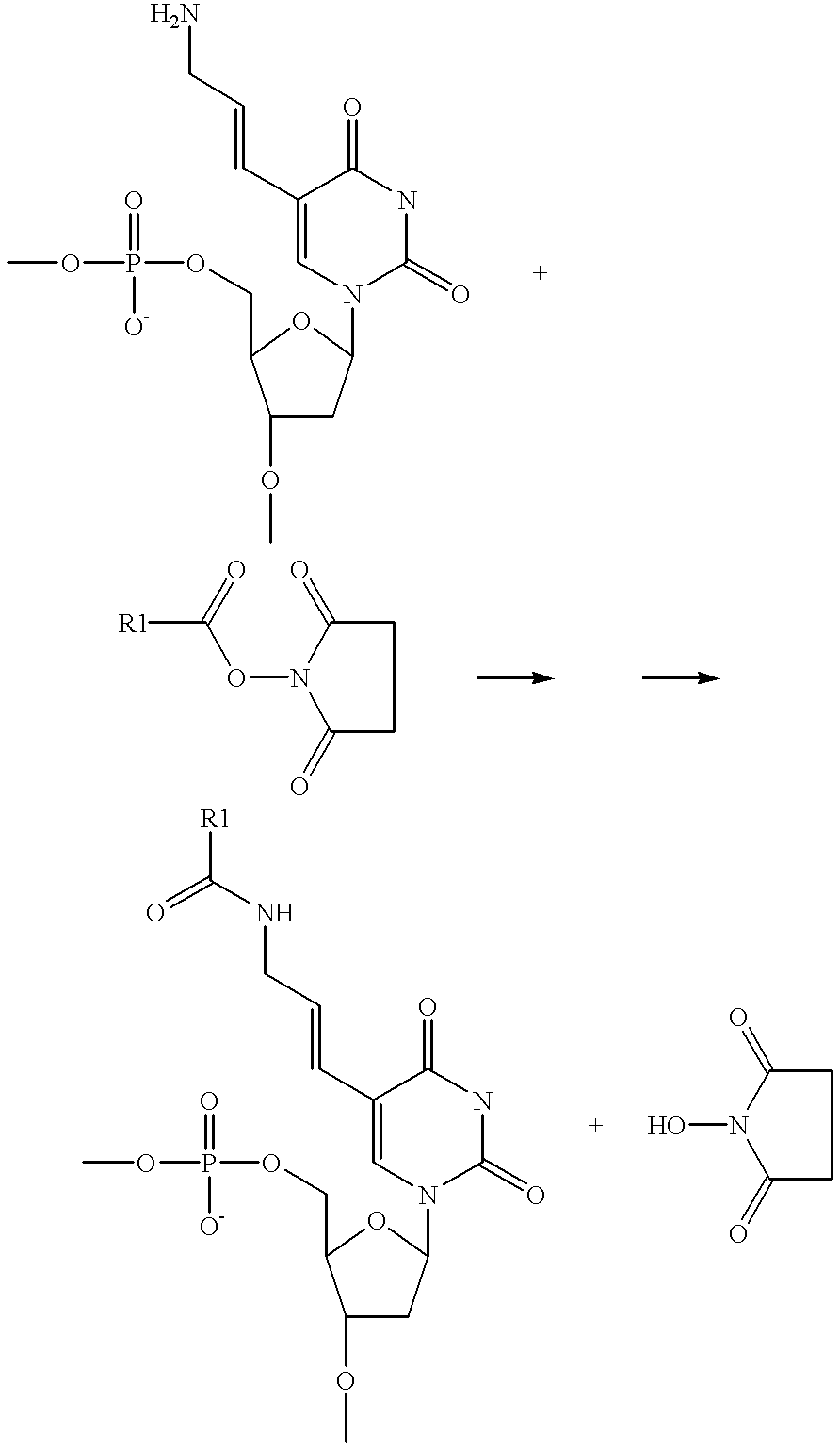
Xylo-LNA analogues
Based on the above and on the remarkable properties of the 2′-O,4′-C-methylene bridged LNA monomers it was decided to synthesise oligonucleotides comprising one or more 2′-O,4′-C-methylene-β-D-xylofuranosyl nucleotide monomer(s) as the first stereoisomer of LNA modified oligonucleotides. Modelling clearly indicated the xylo-LNA monomers to be locked in an N-type furanose conformation. Whereas the parent 2′-deoxy-β-D-xylofuranosyl nucleosides were shown to adopt mainly an N-type furanose conformation, the furanose ring of the 2′-deoxy-β-D-xylofuranosyl monomers present in xylo-DNA were shown by conformational analysis and computer modelling to prefer an S-type conformation thereby minimising steric repulsion between the nucleobase and the 3′-O-phopshate group (Seela, F.; Wömer, Rosemeyer, H. Helv. Chem. Acta 1994, 77, 883). As no report on the hybridisation properties and binding mode of xylo-configurated oligonucleotides in an RNA context was believed to exist, it was the aim to synthesise 2′-O,4′-C-methylene-β-D-xylofuranosyl nucleotide monomer and to study the thermal stability of oligonucleotides comprising this monomer. The results showed that fully modified or almost fully modified Xylo-LNA is useful for high-affinity targeting of complementary nucleic acids. When taking into consideration the inverted stereochemistry at C-3′ this is a surprising fact. It is likely that Xylo-LNA monomers, in a sequence context of Xylo-DNA monomers, should have an affinity-increasing effect.
Owner:QIAGEN GMBH
Modified nucleotide compounds
ActiveUS7495088B1Increased nuclease resistanceSugar derivativesMicrobiological testing/measurementBiologyModified nucleosides
Disclosed is a nuclease resistant nucleotide compound capable of hybridizing with a complementary RNA in a manner which inhibits the function thereof, which modified nucleotide compound includes at least one component selected from the group consisting of MN3M, B(N)xM and M(N)xB wherein N is a phosphodiester-linked modified 2′-deoxynucleoside moiety; M is a moiety that confers endonuclease resistance on said component and that contains at least one modified or unmodified nucleic acid base; B is a moiety that confers exonuclease resistance to the terminus to which it is attached; and x is an integer of at least 2.
Owner:ENZO BIOCHEM
Methods of labeling nucleic acids for use in array based hybridization assays
Methods and kits are provided for labeling nucleic acids, e.g. for use in array based hybridization assays. In the subject methods, target nucleic acid is generated from an initial nucleic acid source, e.g. mRNA, where the target nucleic acid is characterized by having at least one reactive functionality that is not a moiety found on naturally occurring nucleic acids. Functionalized label is then conjugated to the target nucleic acid, either before or after it has been hybridized to array of nucleic acids stably associated with the surface of a solid support. The subject methods find use in a variety of array based hybridization assays, including differential expression assays.
Owner:TAKARA BIO USA INC
Oligonucleotide arrays and their use for sorting, isolating, sequencing, and manipulating nucleic acids
InactiveUS6322971B1Increase in hybridization specificityBioreactor/fermenter combinationsMaterial nanotechnologyHybridization ArrayBioinformatics
Ligation methods for manipulating nucleic acid stands and oligonucleotides utilizing hybridization arrays of immobilized oligonucleotides. The oligonucleotide arrays may be plain or sectioned, comprehensive or non-comprehensive. The immobilized oligonucleotides may in some cases be binary oligonucleotides having constant as well as variable segments. Some embodiments include amplification of ligated products.
Owner:UNIV OF MEDICINE & DENTISTRY OF NEW JERSEY
Rolling circle replication of padlock probes
InactiveUS6558928B1Avoid splittingStrong specificitySugar derivativesMicrobiological testing/measurementNucleic acid hybridisationOligonucleotide
Rolling circle replication of a padlock primer is inhibited when it is hybridized to a target nucleic acid that is long or circular. The invention provides methods of addressing this problem including cutting the target nucleic acid near or preferably at the site which hybridizes with the padlock probe, whereby a 3'-end of the cut target nucleic acid acts as a primer for rolling circle replication of the padlock probe. Also included is a method of assaying for a polyepitopic target by the use of two affinity probes each carrying an oligonucleotide tag and of a padlock probe for rolling circle replication in association with the two affinity probes
Owner:SIGMA ALDRICH CO LLC
Method for producing complex DNA methylation fingerprints
InactiveUS6214556B1Accurately determineSugar derivativesMicrobiological testing/measurementFingerprintGenomic DNA
Method for characterizing, classifying and differentiating tissues and cell types, for predicting the behavior of tissues and groups of cells, and for identifying genes with changed expression. The method involves obtaining genomic DNA from a tissue sample, the genomic DNA subsequently being subjected to shearing, cleaved by means of a restriction endonuclease or not treated by either one of these methods. The base cytosine, but not 5-methylcytosine, from the thus-obtained genomic DNA is then converted into uracil by treatment with a bisulfite solution. Fractions of the thus-treated genomic DNA are then amplified using either very short or degenerated oligonucleotides or oligonuclcotides which are complementary to adaptor oligonucleotides that have been ligated to the ends of the cleaved DNA. The quantity of the remaining cytosines on the guanine-rich DNA strand and / or the quantity of guanines on the cytosine-rich DNA strand from the amplified fractions are then detected by hybridization or polymerase reaction, which quantities are such that the data generated thereby and automatically applied to a processing algorithm allow the drawing of conclusions concerning the phenotype of the sample material.
Owner:EPIGENOMICS AG
Integrated active flux microfluidic devices and methods
InactiveUS20040248167A1Increase speedImprove accuracyBioreactor/fermenter combinationsFlow mixersAntigenHybridization probe
The invention relates to a microfabricated device for the rapid detection of DNA, proteins or other molecules associated with a particular disease. The devices and methods of the invention can be used for the simultaneous diagnosis of multiple diseases by detecting molecules (e.g. amounts of molecules), such as polynucleotides (e.g., DNA) or proteins (e.g., antibodies), by measuring the signal of a detectable reporter associated with hybridized polynucleotides or antigen / antibody complex. In the microfabricated device according to the invention, detection of the presence of molecules (i.e., polynucleotides, proteins, or antigen / antibody complexes) are correlated to a hybridization signal from an optically-detectable (e.g. fluorescent) reporter associated with the bound molecules. These hybridization signals can be detected by any suitable means, for example optical, and can be stored for example in a computer as a representation of the presence of a particular gene. Hybridization probes can be immobilized on a substrate that forms part of or is exposed to a channel or channels of the device that form a closed loop, for circulation of sample to actively contact complementary probes. Universal chips according to the invention can be fabricated not only with DNA but also with other molecules such as RNA, proteins, peptide nucleic acid (PNA) and polyamide molecules.
Owner:CALIFORNIA INST OF TECH
Oligonucleotides for genotyping thymidylate synthase gene
ActiveUS20070031829A1Sensitive and convenient detectionEasy to aimSugar derivativesMicrobiological testing/measurementGeneticsGenomic DNA
Oligonucleotides for genotyping the thymidylate synthase gene are provided. The number of tandem repeats in the promoter region of the thymidylate synthase gene can be identified based on the hybridization of an oligonucleotide of the invention to the genomic DNA of a subject. Therefore, the genotype of the thymidylate synthase gene can be identified based on the number of tandem repeats. The genotype relates to the responsiveness of a subject towards an antitumor agent.
Owner:F HOFFMANN LA ROCHE & CO AG
Methods and compositions for isolating nucleic acid sequence variants
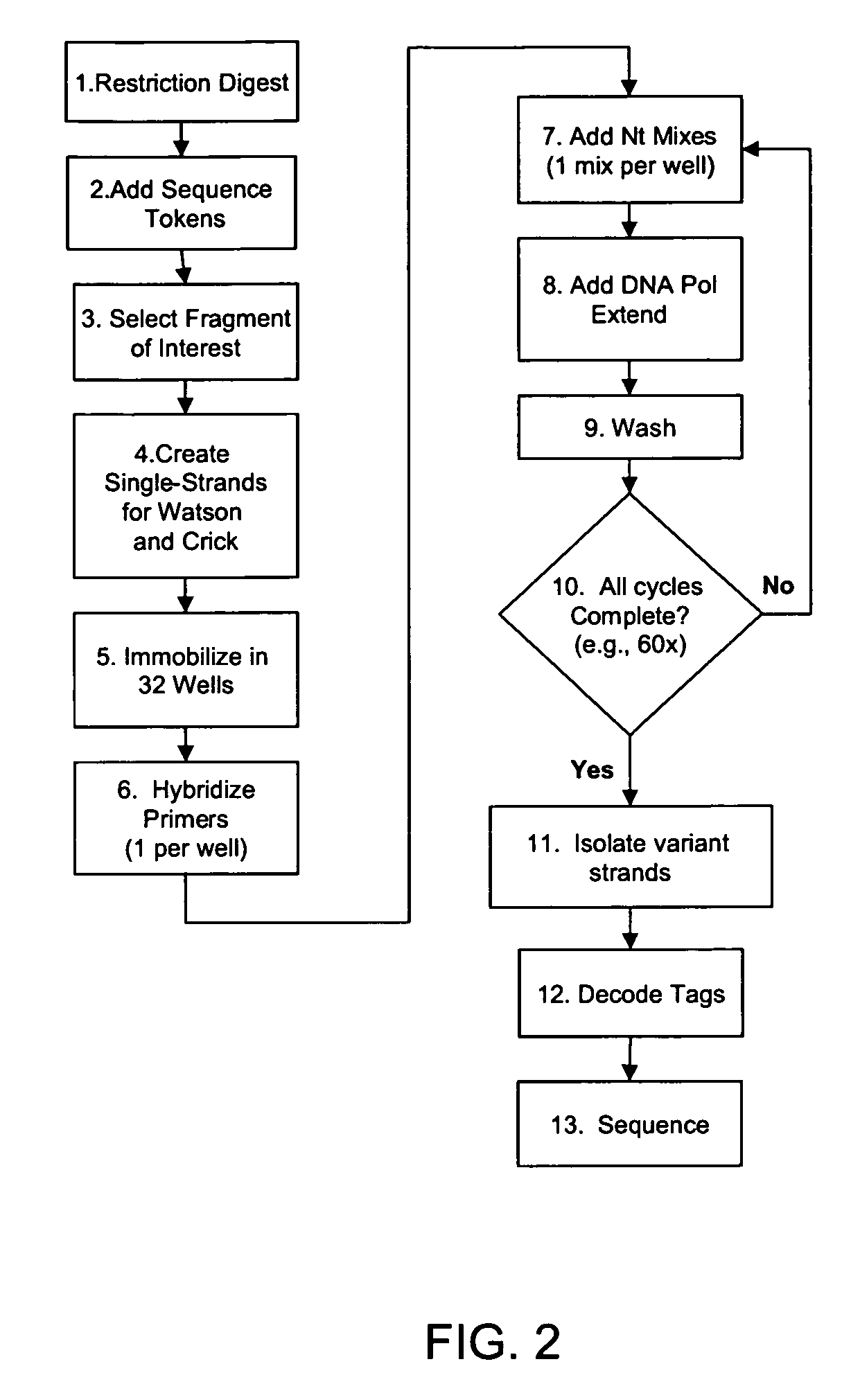
The invention is drawn to isolating sequence variants of a genetic locus of interest using a modified iterative primer extension method. The nucleic acids analyzed are generally single stranded and have a reference sequence which is used as a basis for performing iterative single nucleotide extension reactions from a hybridized polymerization primer. The iterative polymerization reactions are configured such that polymerization of the strand will continue if the sequence of the nucleic acid being analyzed matches the reference sequence, whereas polymerization will be terminated if the nucleic acid being analyzed does not match the reference sequence. Nucleic acid strands that have mutations can be isolated using a variety of methods and sequenced to determine the precise identity of the mutation / polymorphism. By performing the method on both strands of the nucleic acid being analyzed, virtually all possible mutations can be identified.
Owner:AGENCY FOR SCI TECH & RES
DNA amplification and subtraction techniques
InactiveUS6107023AImprove concentrationSugar derivativesMicrobiological testing/measurementHybrid speciesDna amplification
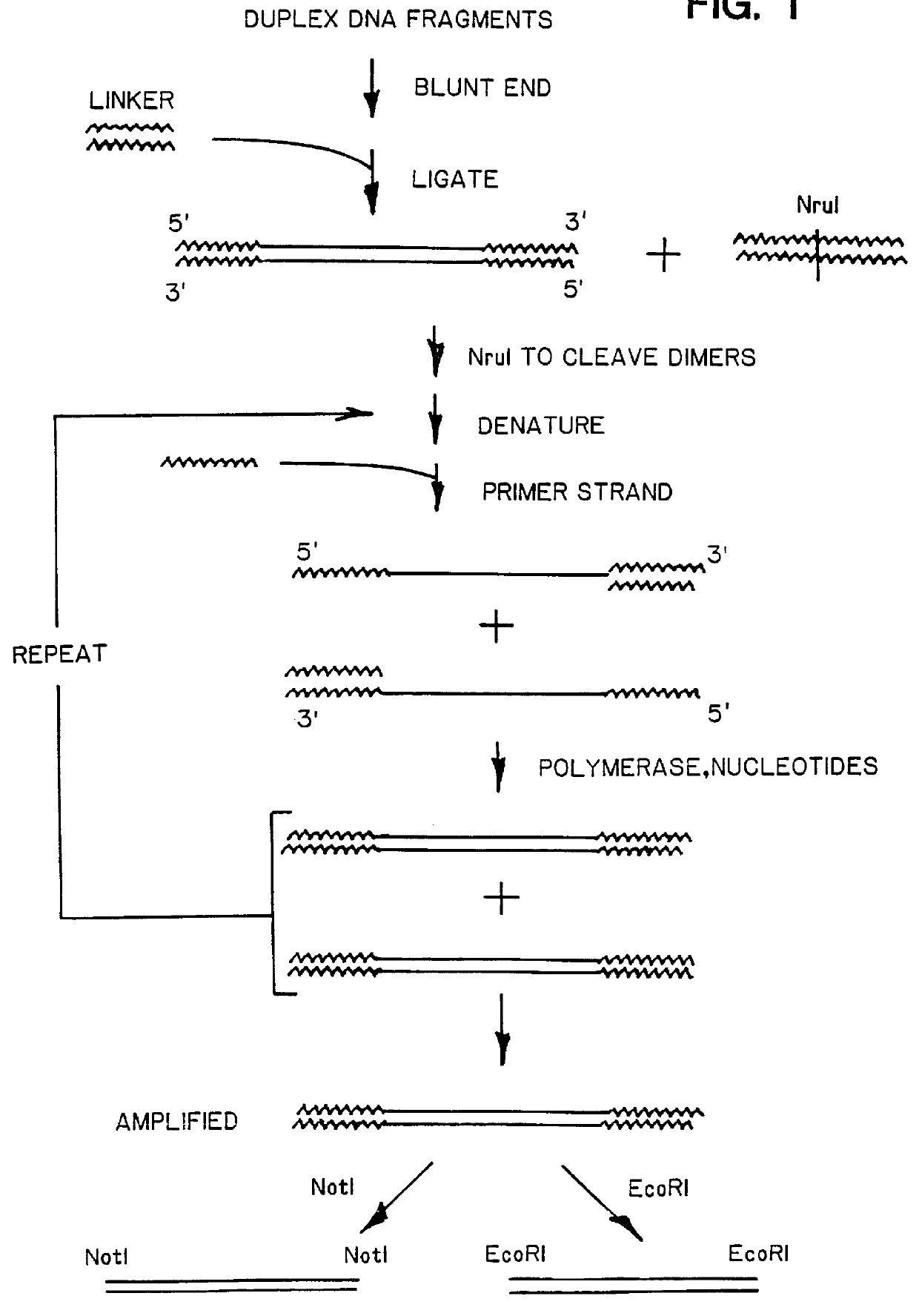
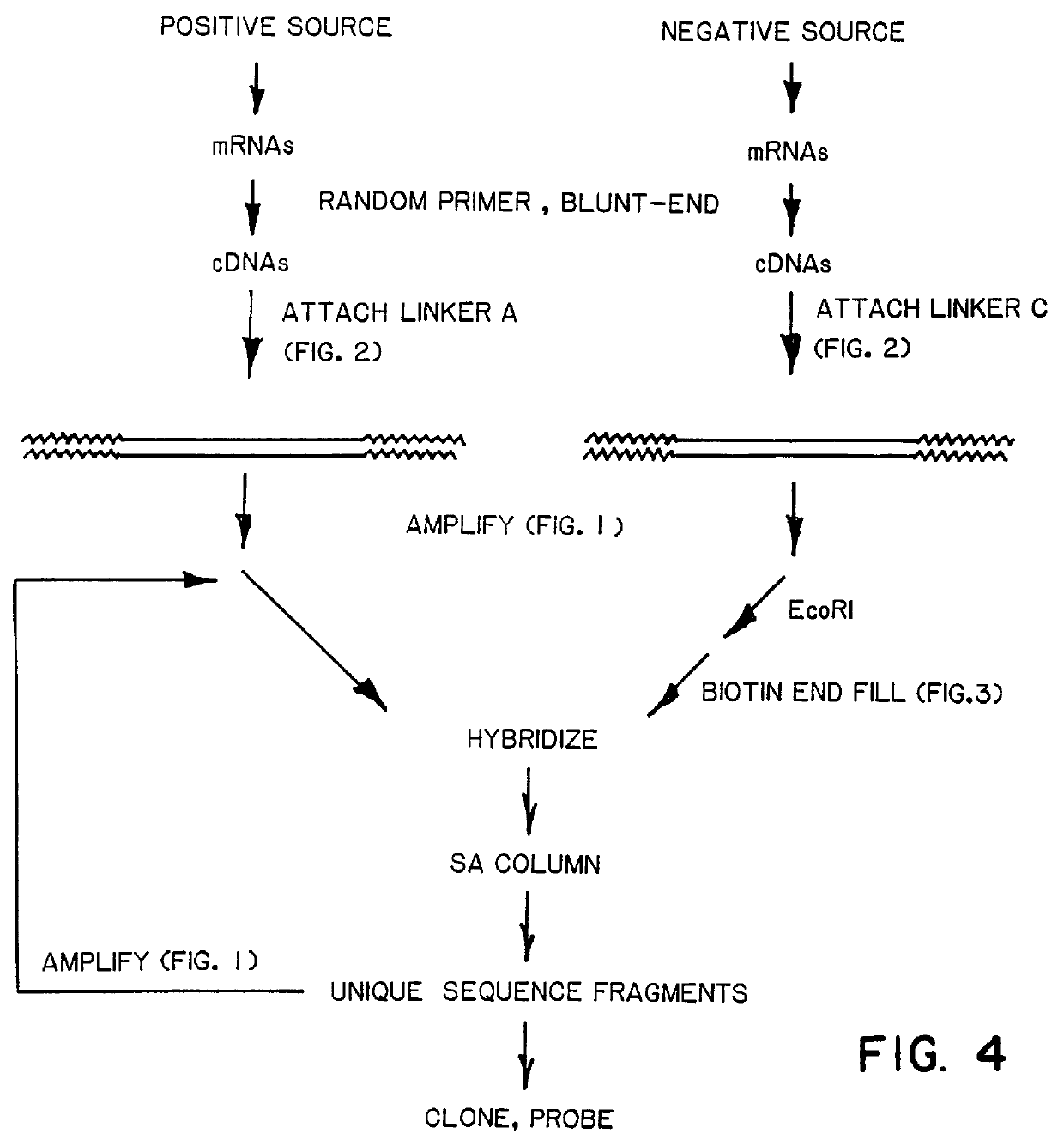
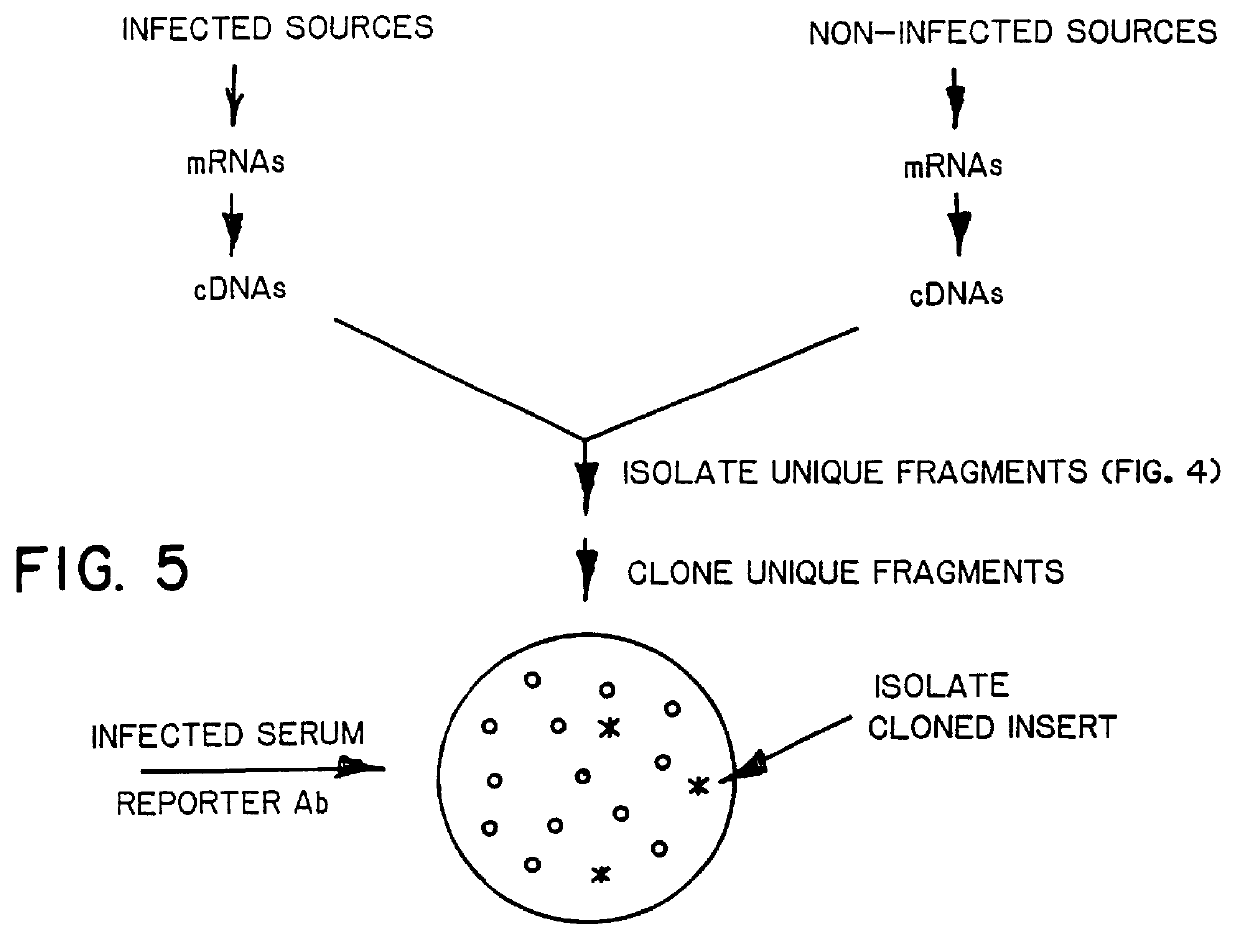
A method of isolating genomic or RNA-derived duplex fragments which are unique to one of two fragment mixtures. The fragments in positive-source and negative-source mixtures are separately equipped with end linkers, and each mixture is amplified by successive primed-strand replications, using a single primer which is homologous to the associated linker. The second-source linker is biotinylated, and the fragments in this mixture are hybridized in molar excess with the fragments in the positive-source mixture. DNA species which are not hybridized with the biotinylated species, i.e., species that are unique to the positive-source mixture, are isolated after removal of hybridized species by affinity chromatography. Also disclosed is a method of amplifying a mixture of DNA fragments by repeated linker / primer replication.
Owner:ILLUMINA INC +1
Spatially distinguished, multiplex nucleic acid analysis of biological specimens
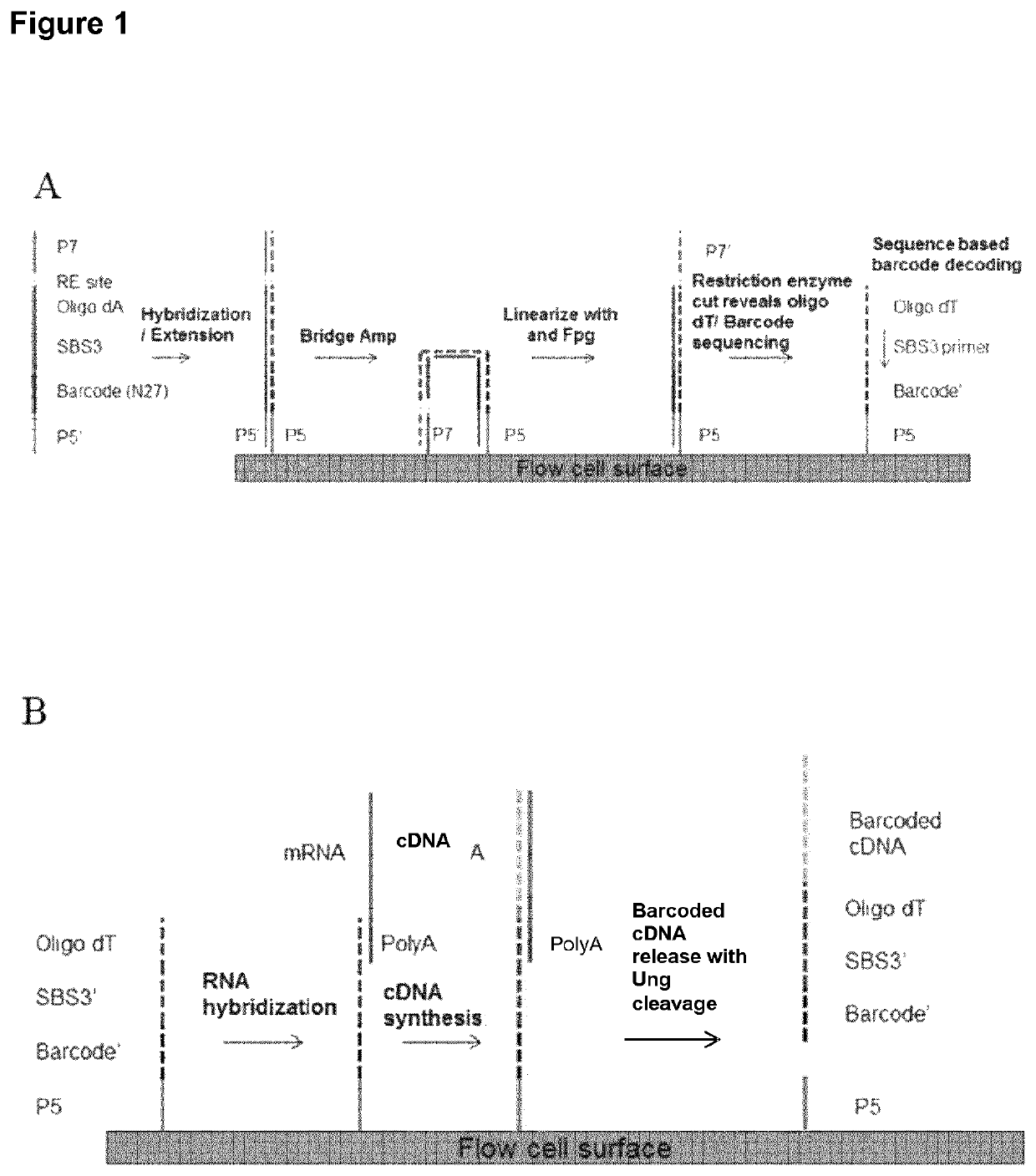
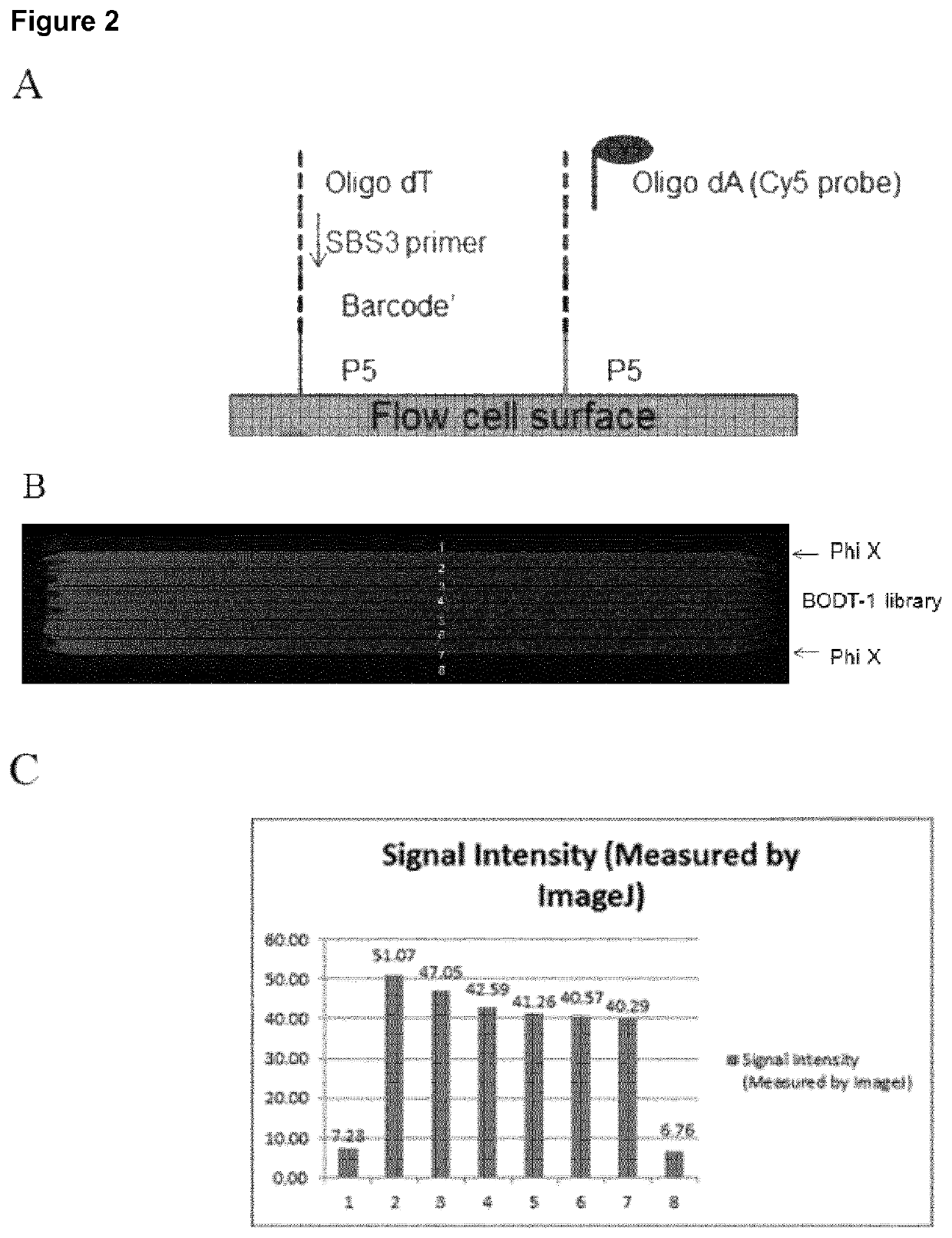
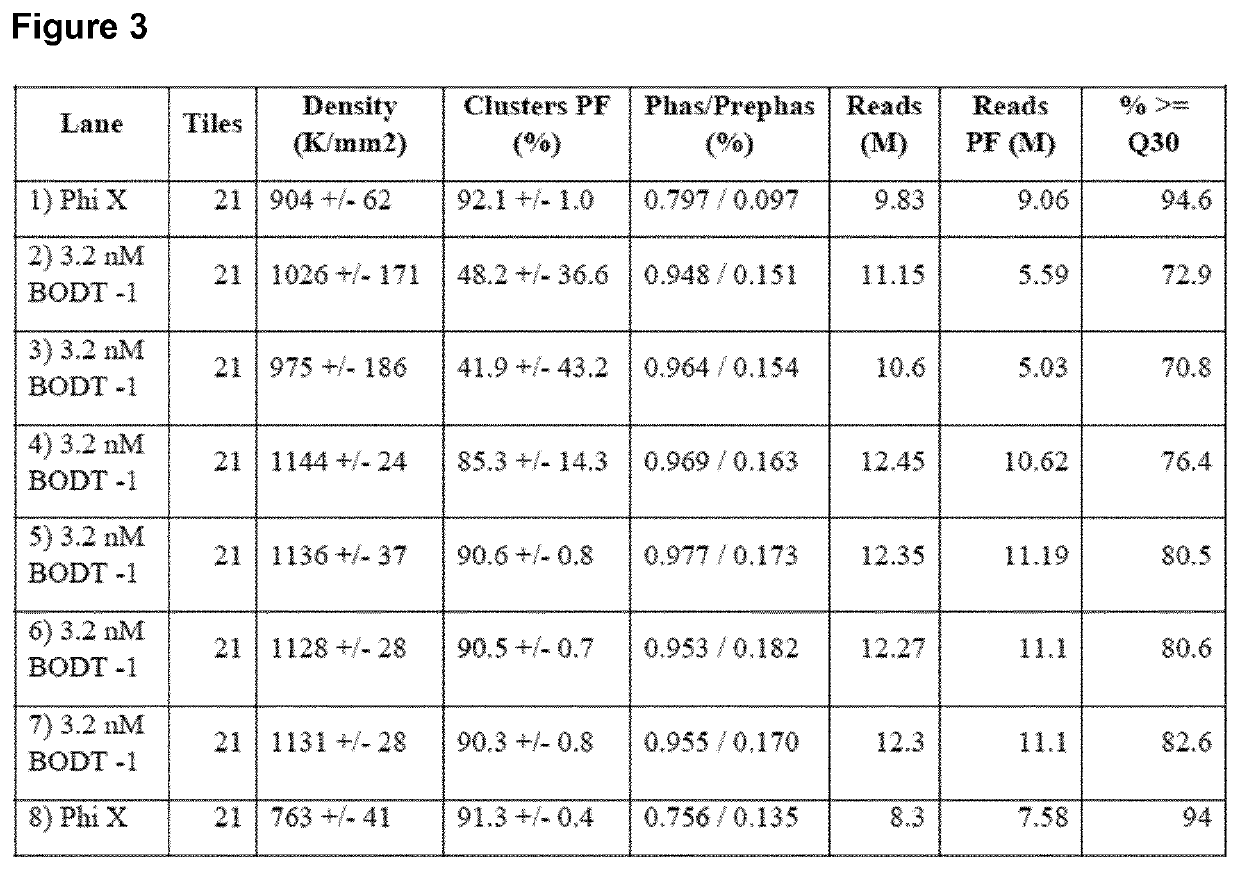
A method for spatially tagging nucleic acids of a biological specimen, including steps of (a) providing a solid support comprising different nucleic acid probes that are randomly located on the solid support, wherein the different nucleic acid probes each includes a barcode sequence that differs from the barcode sequence of other randomly located probes on the solid support; (b) performing a nucleic acid detection reaction on the solid support to locate the barcode sequences on the solid support; (c) contacting a biological specimen with the solid support that has the randomly located probes; (d) hybridizing the randomly located probes to target nucleic acids from portions of the biological specimen; and (e) modifying the randomly located probes that are hybridized to the target nucleic acids, thereby producing modified probes that include the barcode sequences and a target specific modification, thereby spatially tagging the nucleic acids of the biological specimen.
Owner:X GENOMICS SWEDEN AB +1
Insecticidal proteins secreted from bacillus thuringiensis and uses therefor
ActiveUS20060191034A1Improvement in insect resistance managementEffective controlBiocidePeptide/protein ingredientsNucleotideTransgene
The present invention relates to the isolation and characterization of nucleotide sequences encoding novel insecticidal proteins secreted into the extracellular space from Bacillus thuringiensis and related strains. The proteins are isolated from culture supernatants of Bacillus thuringiensis and related strains and display insecticidal activity against coleopteran insects including Colorado potato beetle (Lymantria dispar) and Southern Corn Rootworm (Diabrotica undecempunctata). Insecticidal proteins encoded by nucleotide sequences that hybridize to the isolated and characterized nucleotide sequences are disclosed. Also disclosed are methods of making and using transgenic cells and plants comprising the novel nucleotide sequence of the invention.
Owner:MONSANTO TECH LLC
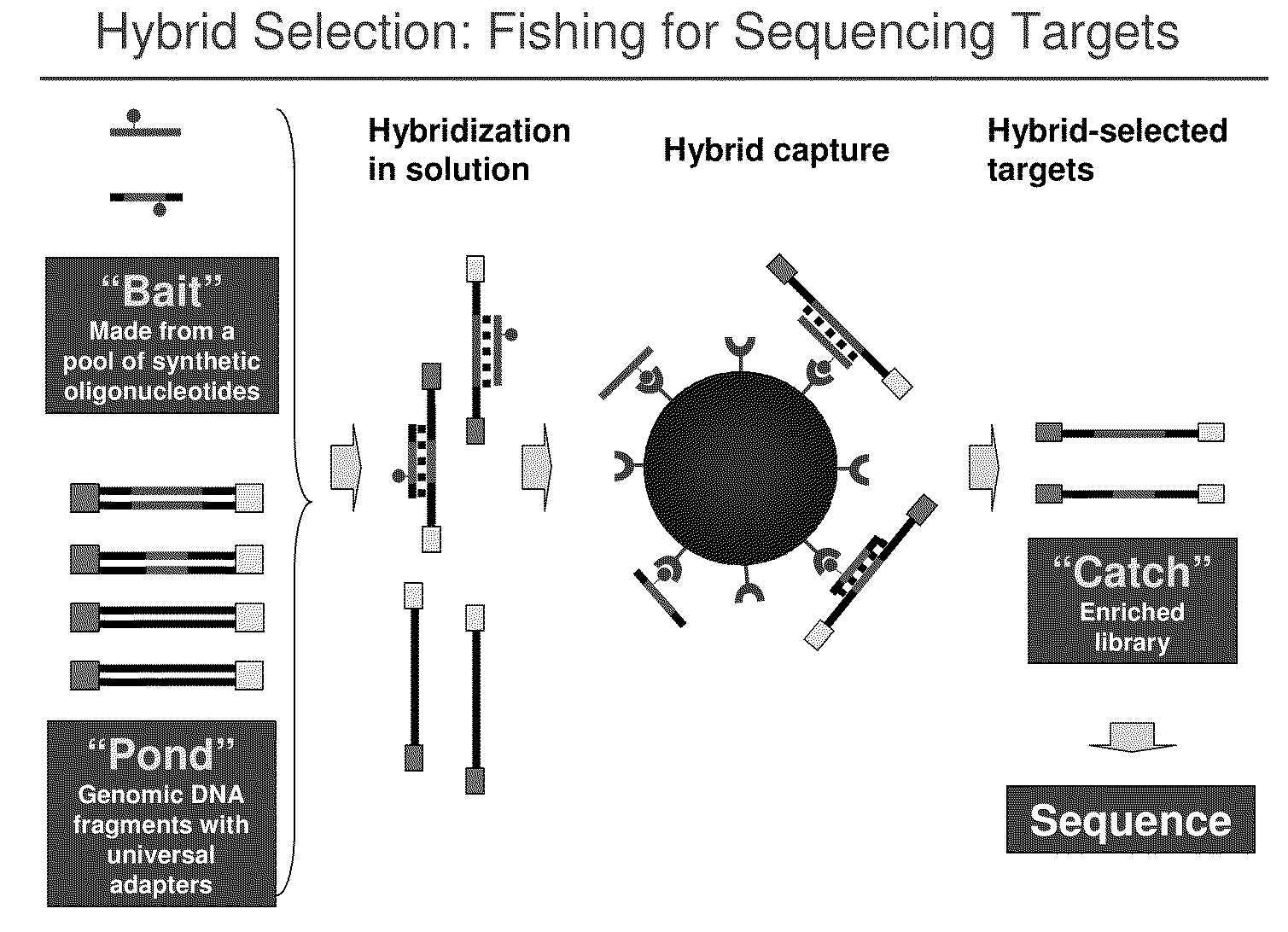
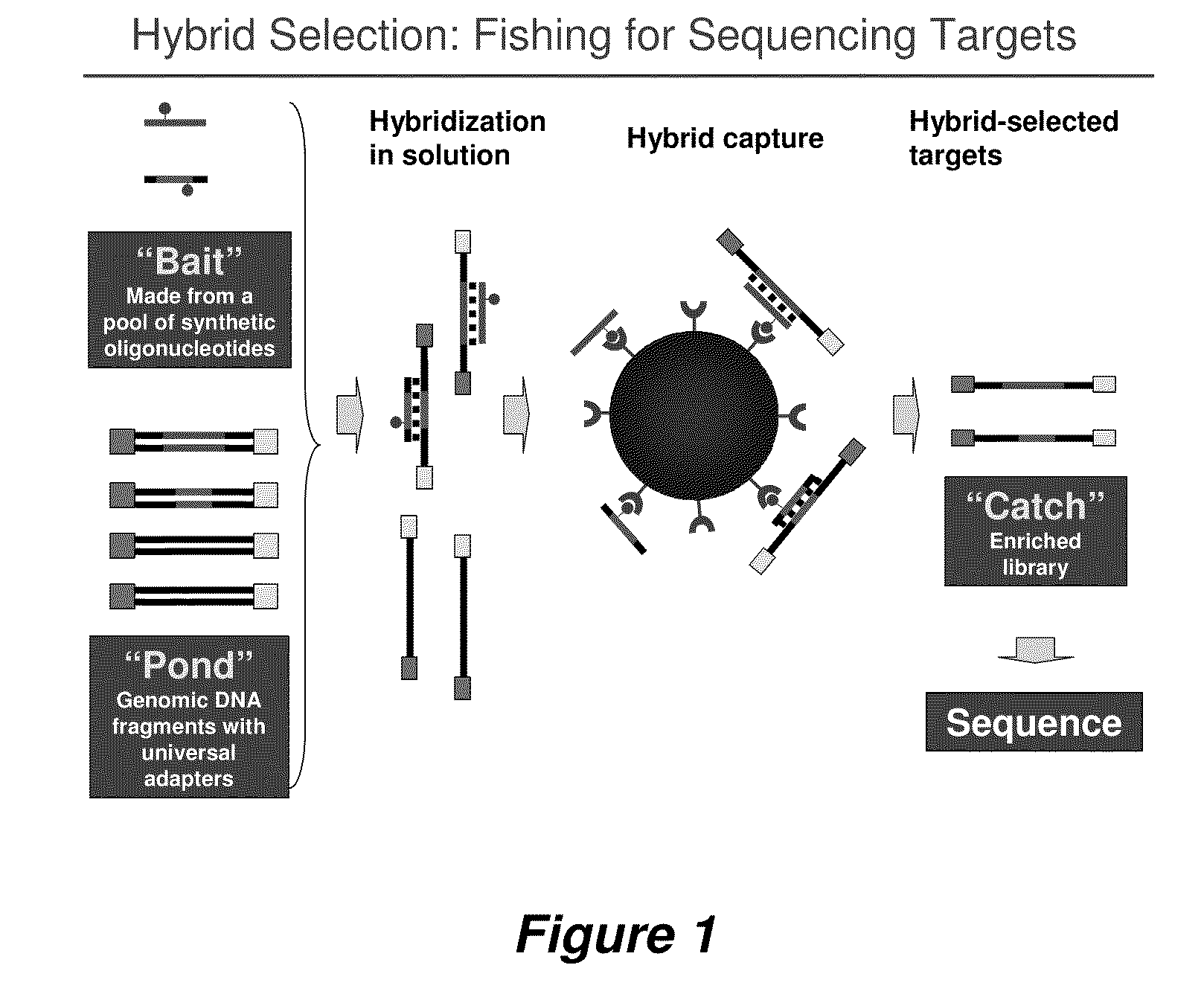
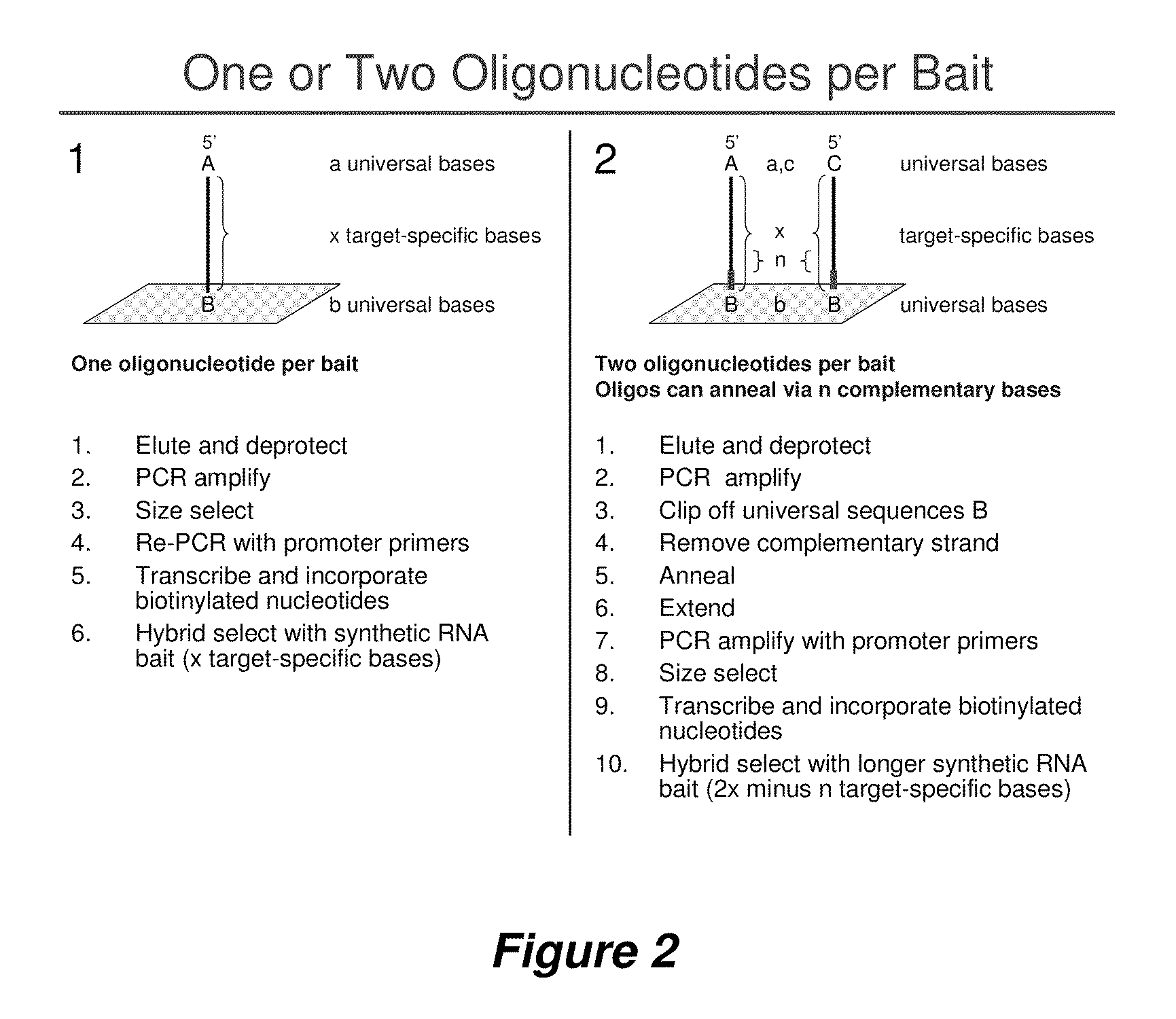
Selection of nucleic acids by solution hybridization to oligonucleotide baits
InactiveUS20100029498A1Minimize the differenceGood reproducibilityMicrobiological testing/measurementLibrary screeningSolution hybridizationOligonucleotide
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +2
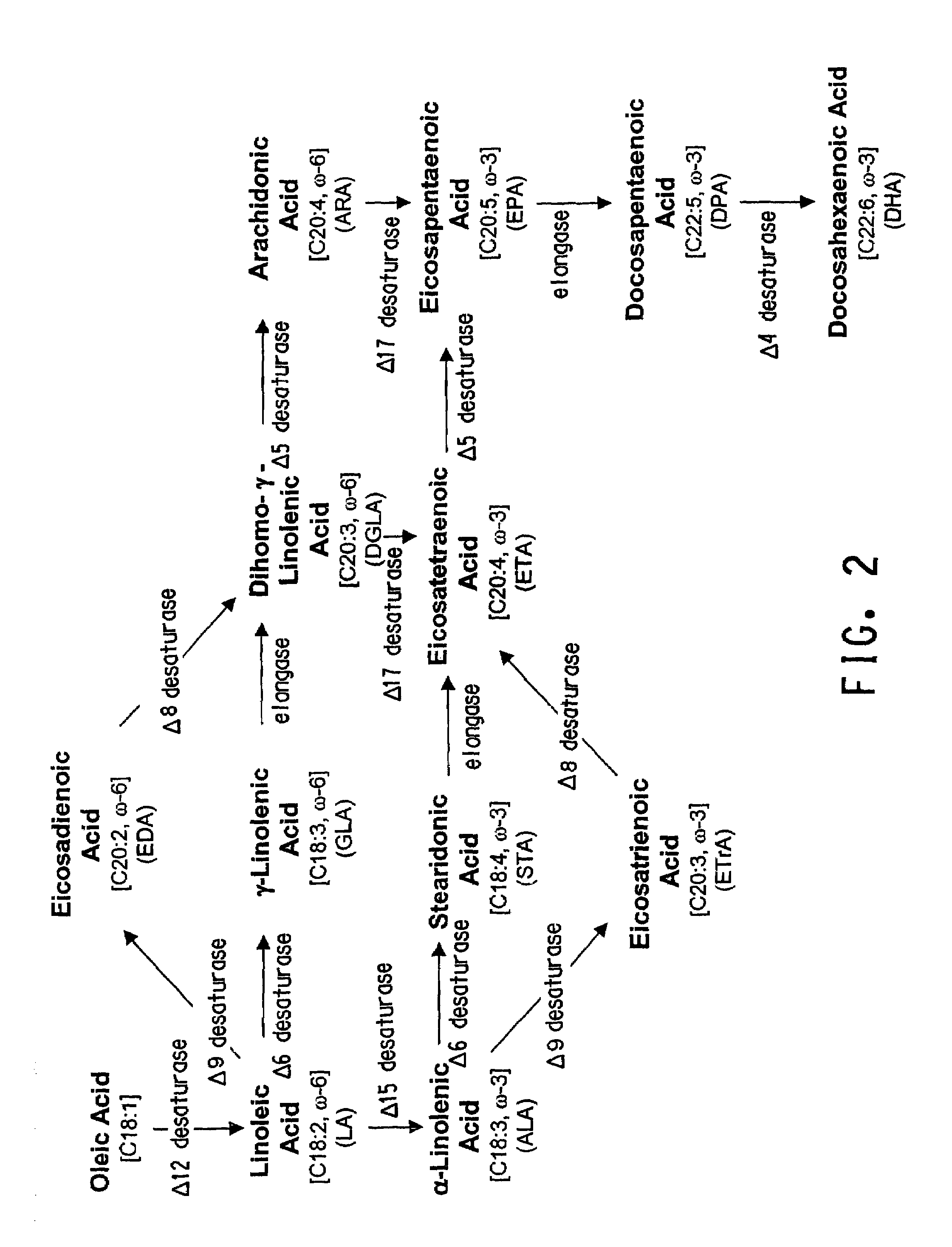
Delta12 desaturases suitable for altering levels of polyunsaturated fatty acids in oleaginous yeast
The present invention relates to fungal Δ12 fatty acid desaturases that are able to catalyze the conversion of oleic acid to linoleic acid (LA; 18:2). Nucleic acid sequences encoding the desaturases, nucleic acid sequences which hybridize thereto, DNA constructs comprising the desaturase genes, and recombinant host microorganisms expressing increased levels of the desaturases are described. Methods of increasing production of specific ω-3 and ω-6 fatty acids by over-expression of the Δ12 fatty acid desaturases are also described herein.
Owner:EI DU PONT DE NEMOURS & CO
Single nucleotide detection using degradation of a fluorescent sequence
Methods and compositions are provided for detecting single nucleotide polymorphisms using a pair of oligonucleotides, a primer and a snp detection sequence, where the snp detection sequence hybridizes to the target DNA downstream from the primer and in the direction of primer extension. The snp detection sequence is characterized by having a nucleotide complementary to the snp and adjacent nucleotide complementary to adjacent nucleotides in the target and an electophoretic tag bonded to the 5'-nucleotide. The pair of oligonucleotides is combined with the target DNA under primer extension conditions, where the polymerase has 5'-3' exonuclease activity. When the snp is present, the electophoretic tag is released from the snp detection sequence, and can be detected by electrophoresis as indicative of the presence of the snp in the target DNA.
Owner:MONOGRAM BIOSCIENCES
Probe-based analysis of heterozygous mutations using two-color labelling
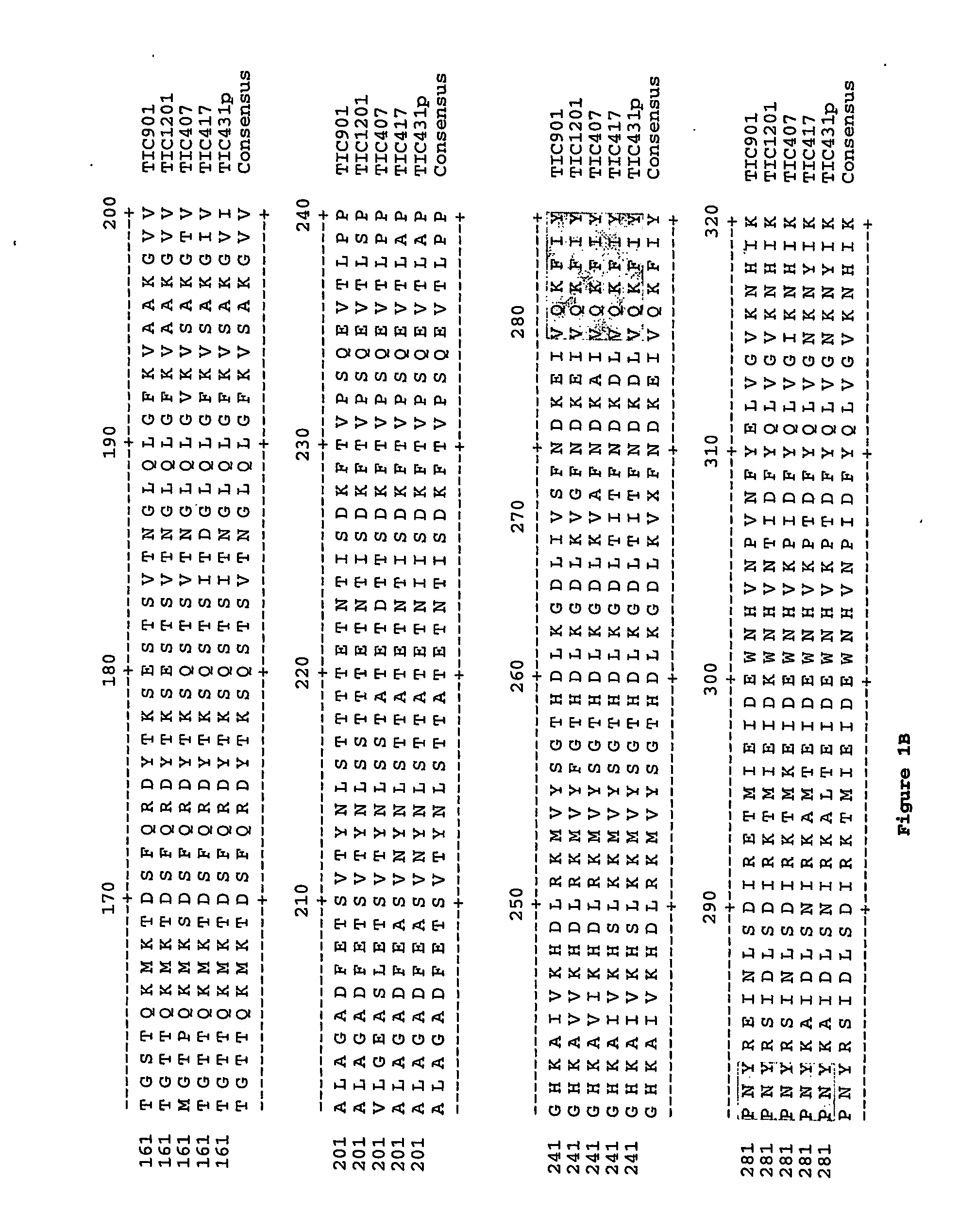
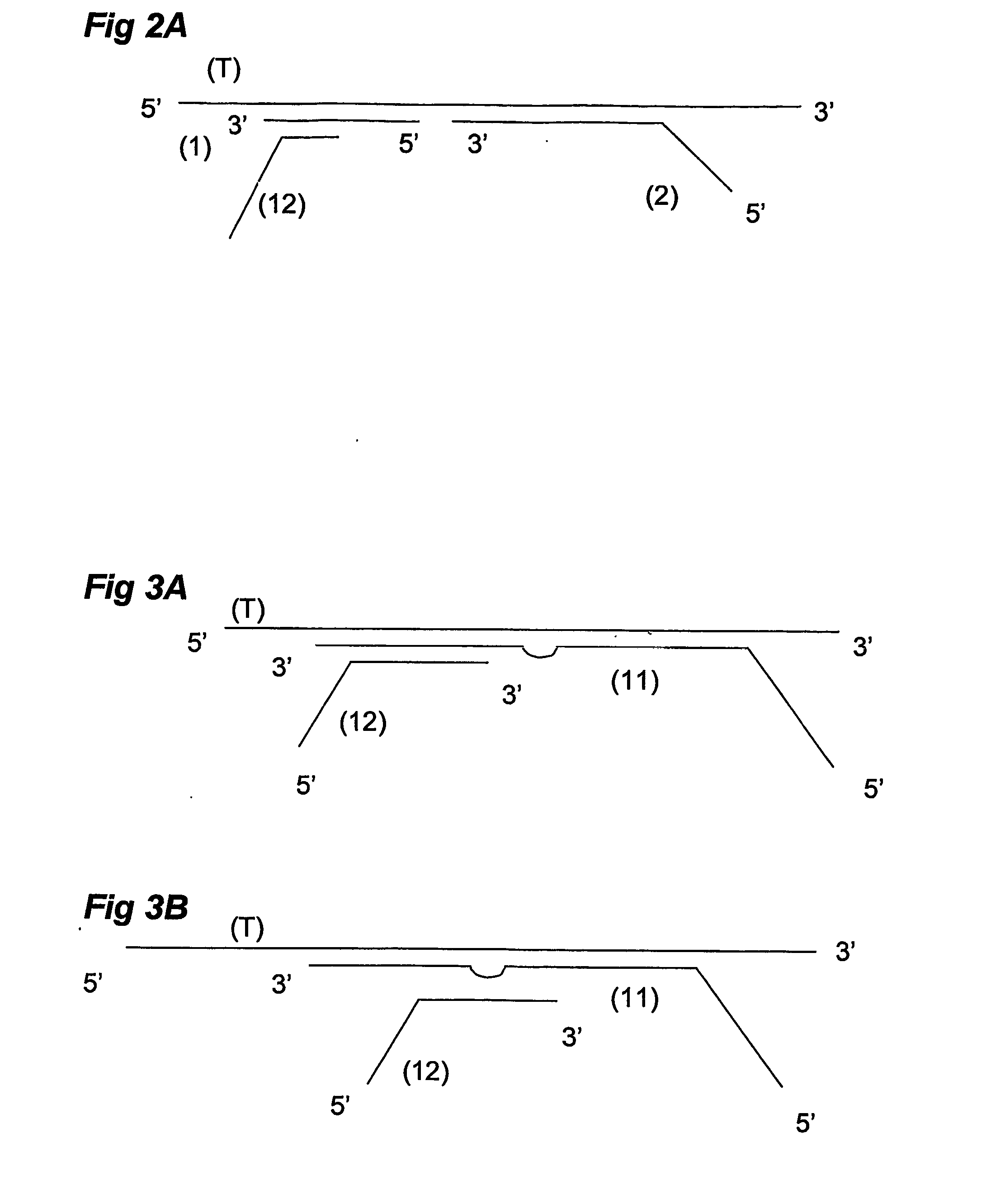
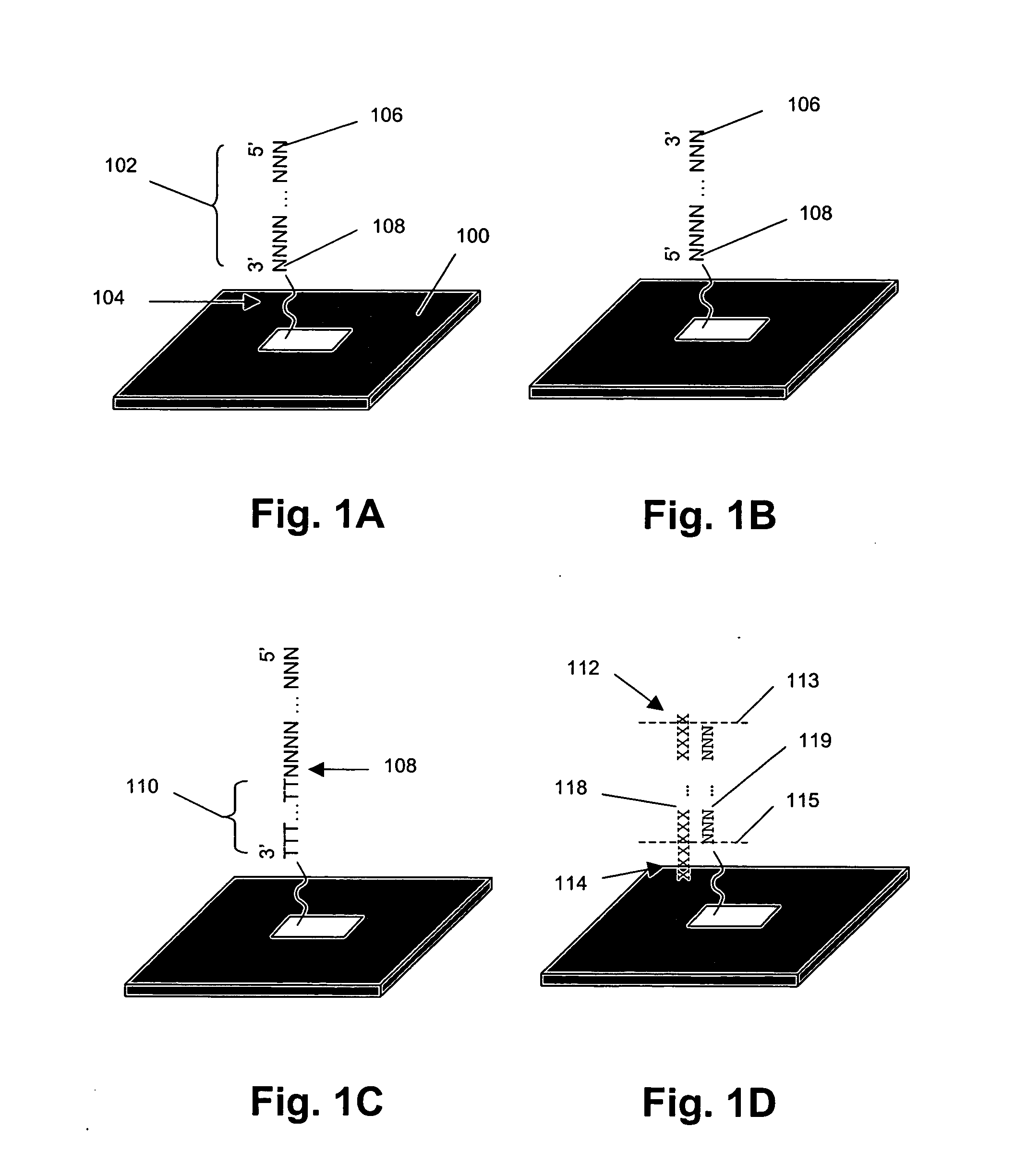
InactiveUS6342355B1Bioreactor/fermenter combinationsBiological substance pretreatmentsVariant alleleGenomic clone
The invention provides methods of analyzing a nucleic acid in a target sample for variant alleles. In such methods, a first-labelled control sample and a second-labelled target sample are hybridized to at least one set of probes. The control sample comprises a homozygous reference allele. The target sample comprises the homozygous reference allele, or variant alleles differing from the reference allele at a locus, or one variant allele differing from the reference allele at the locus and one reference allele. The probes in the probe set span the locus and are complementary to the reference allele. After hybridization the intensity of first and second label bound to each probe in the set is measured. This information is then used to indicate the presence of one variant allele and one reference allele, or the presence of two variant alleles in the target sample.
Owner:UNITED STATES OF AMERICA +1
System and method for inhibiting the decryption of a nucleic acid probe sequence used for the detection of a specific nucleic acid
InactiveUS20060073487A1Reduce or impede a person's ability to designAvoid identificationSugar derivativesMicrobiological testing/measurementGenetic AnomalyNucleic Acid Probes
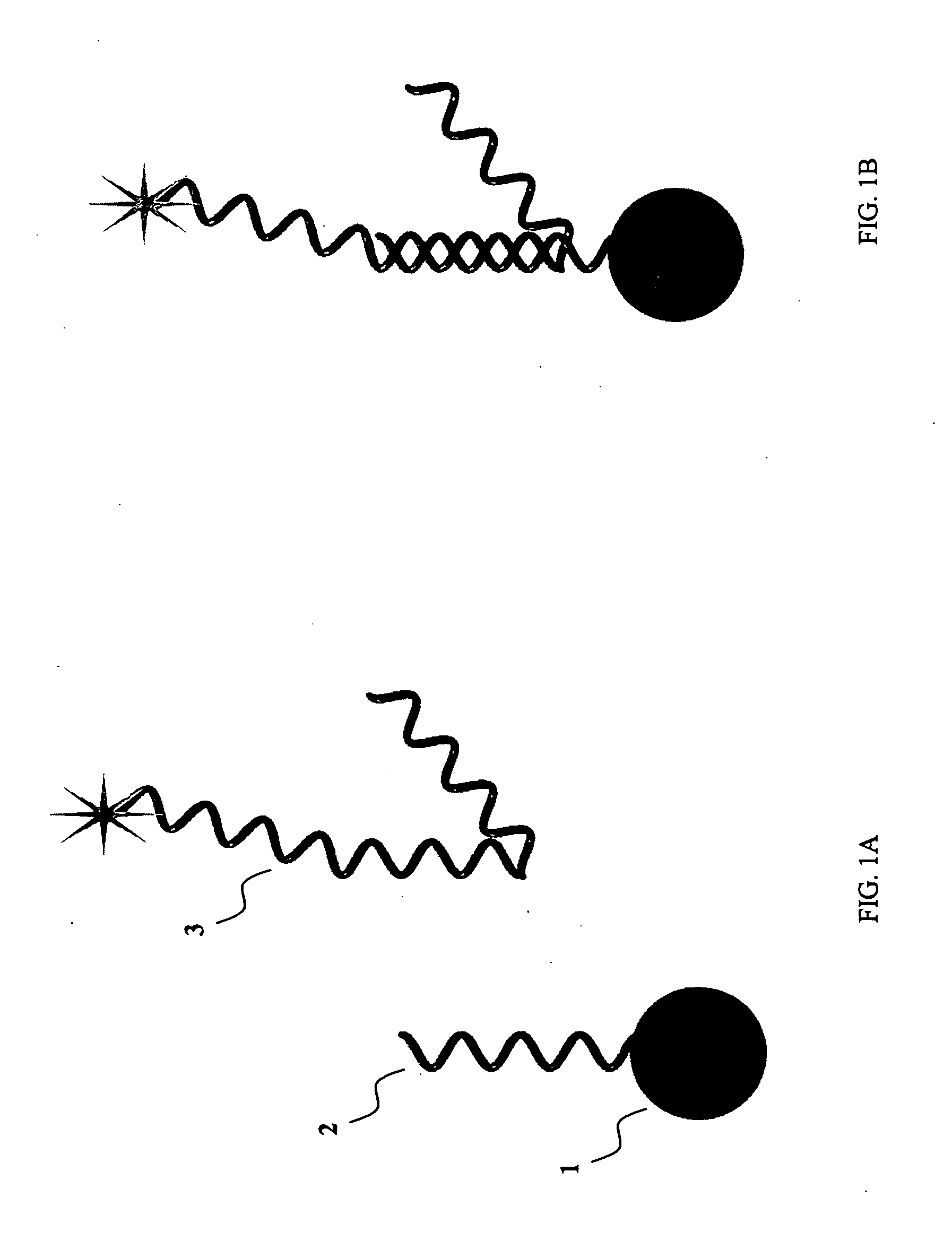
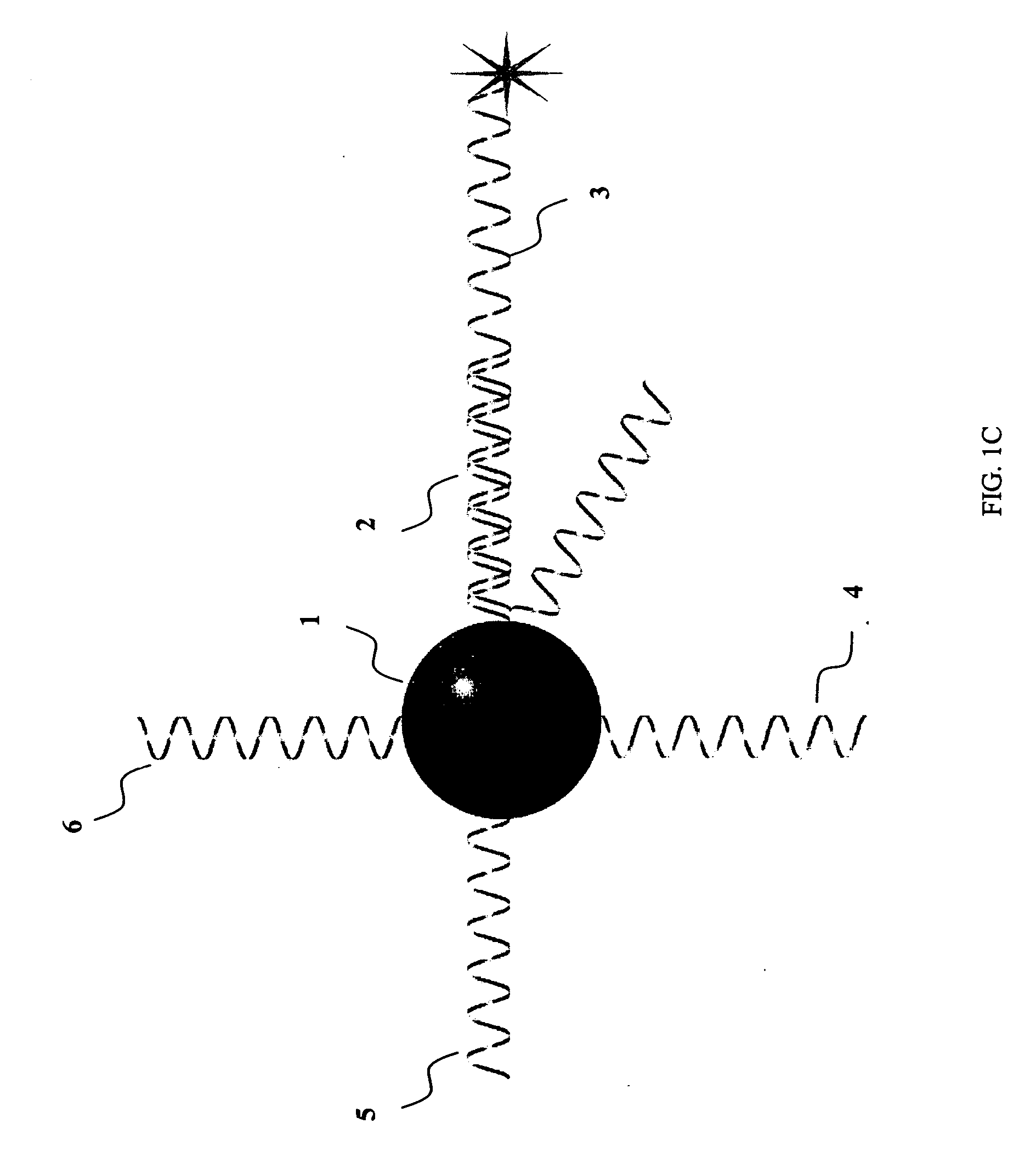
Sequence-specific nucleic acid hybridization assays are used for the detection of specific genetic sequences as indicators of genetic anomalies, mutations, and disease propensity. In addition, they are used for the detection of various biological agents and infectious pathogens. Because a complementary probe or nucleic acid sequence is required to detect a sequence of interest in a hybridization-based assay, nucleic acid sequencing techniques can rapidly determine the specific probe sequence being used for detection. This allows reverse engineered assays to be produced rapidly. In addition, it enables the circumvention of hybridization-based assays for biological agent or infectious pathogen detection by providing the information necessary to create or alter nucleic acid sequences to produce false positives or false negatives. The present invention provides methods and compositions for inhibiting the identification of specific detection sequences. More specifically, the invention provides masking sequences that mask the identity of specific detection sequences.
Owner:RADIX BIOSOLUTIONS
Methods and compositions for analyzing nucleic acids
InactiveUS20060078894A1Minimizes problemReduce capacityMicrobiological testing/measurementFermentationHybridization probeNucleic acid sequencing
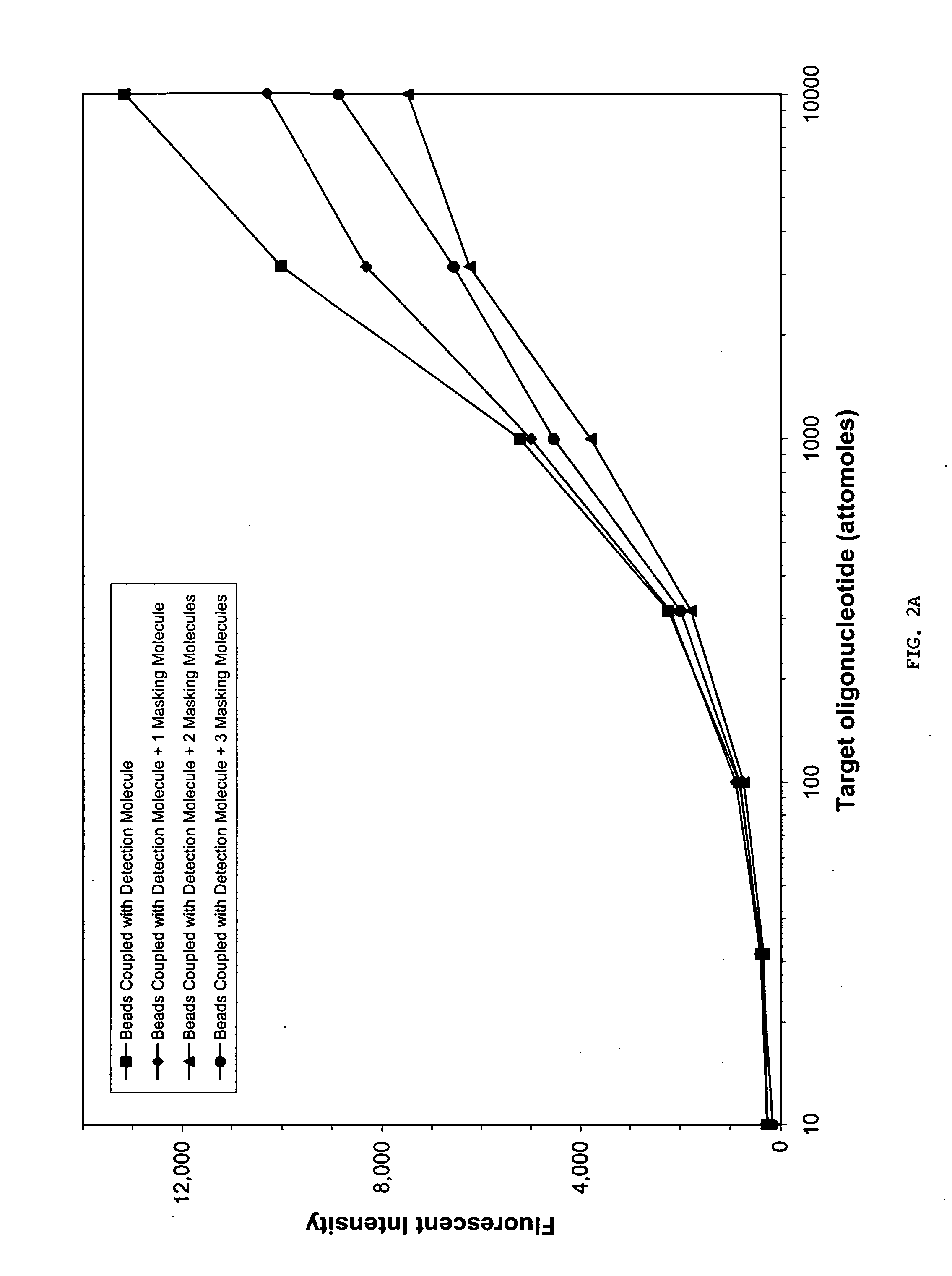
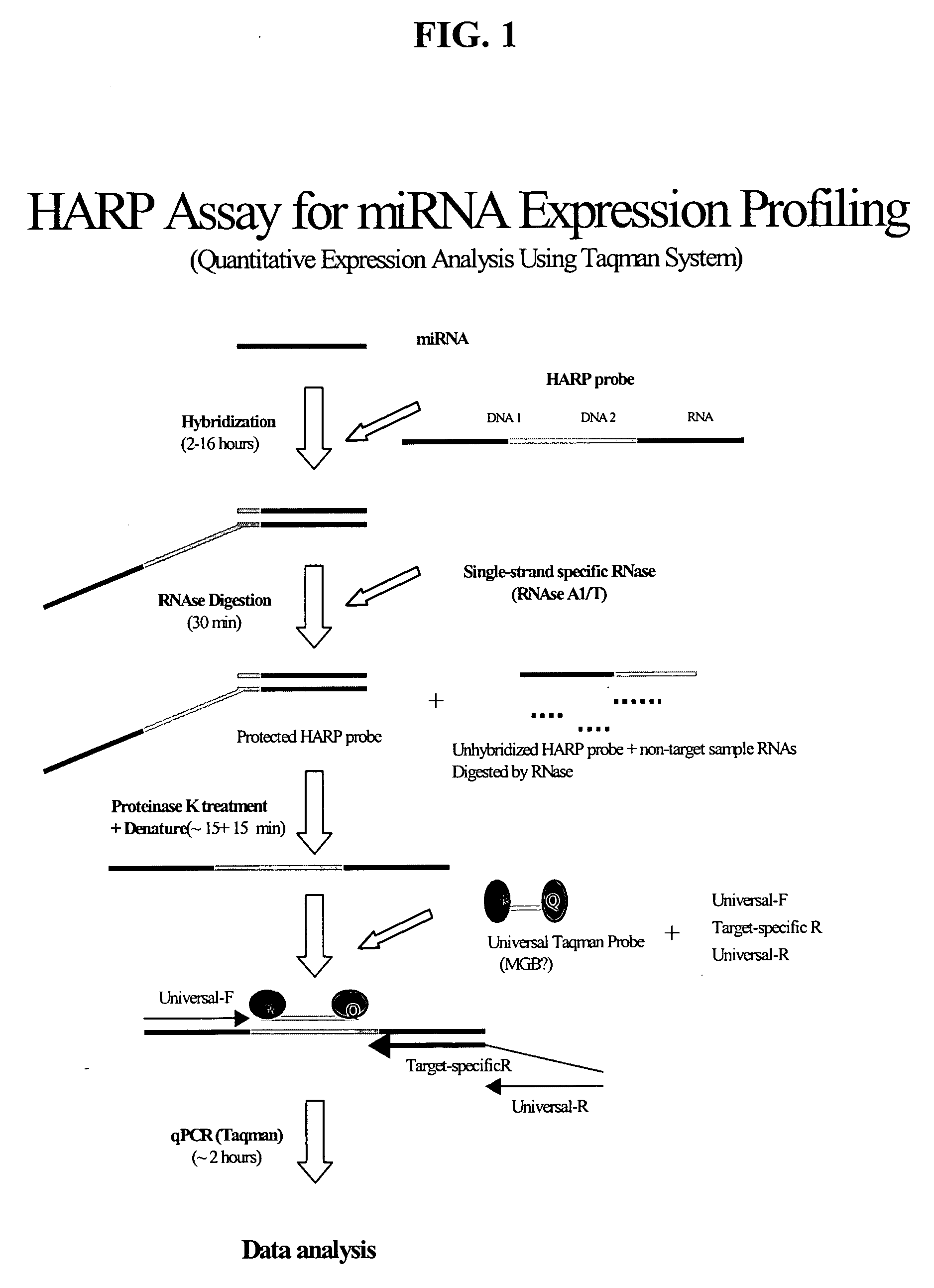
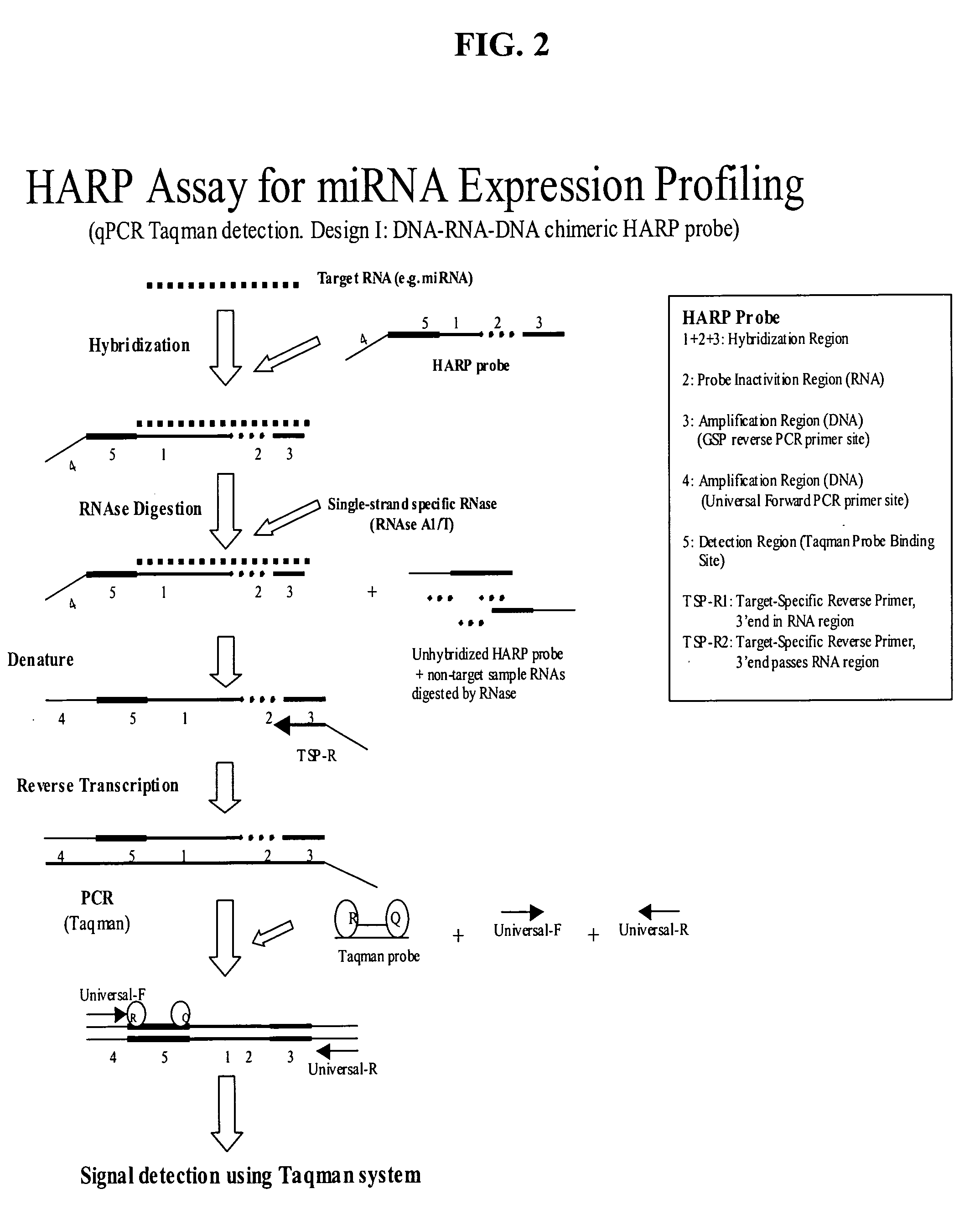
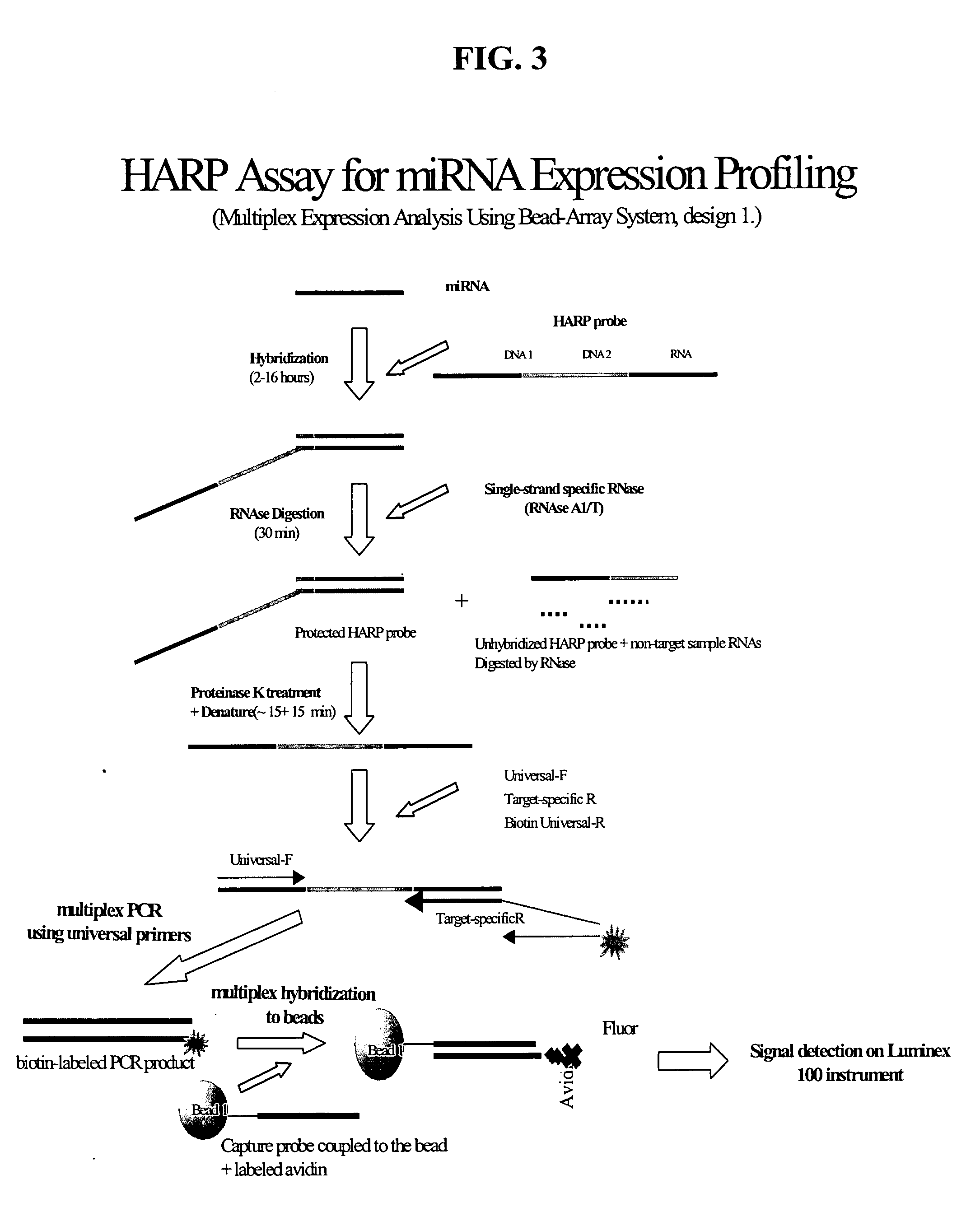
Methods and compositions for probe amplification to detect, identify, quantitate, and / or analyze a targeted nucleic acid sequence. After hybridization between a probe and the targeted nucleic acid, the probe is modified to distinguish hybridized probe from unhybridized probe. Thereafter, the probe is amplified. Moreover, in specific embodiments, the present invention involves a chimeric probe that is particularly effective when the targeted nucleic acid sequence is short and / or has a relatively low concentration, such as with an miRNA molecule.
Owner:ASURAGEN
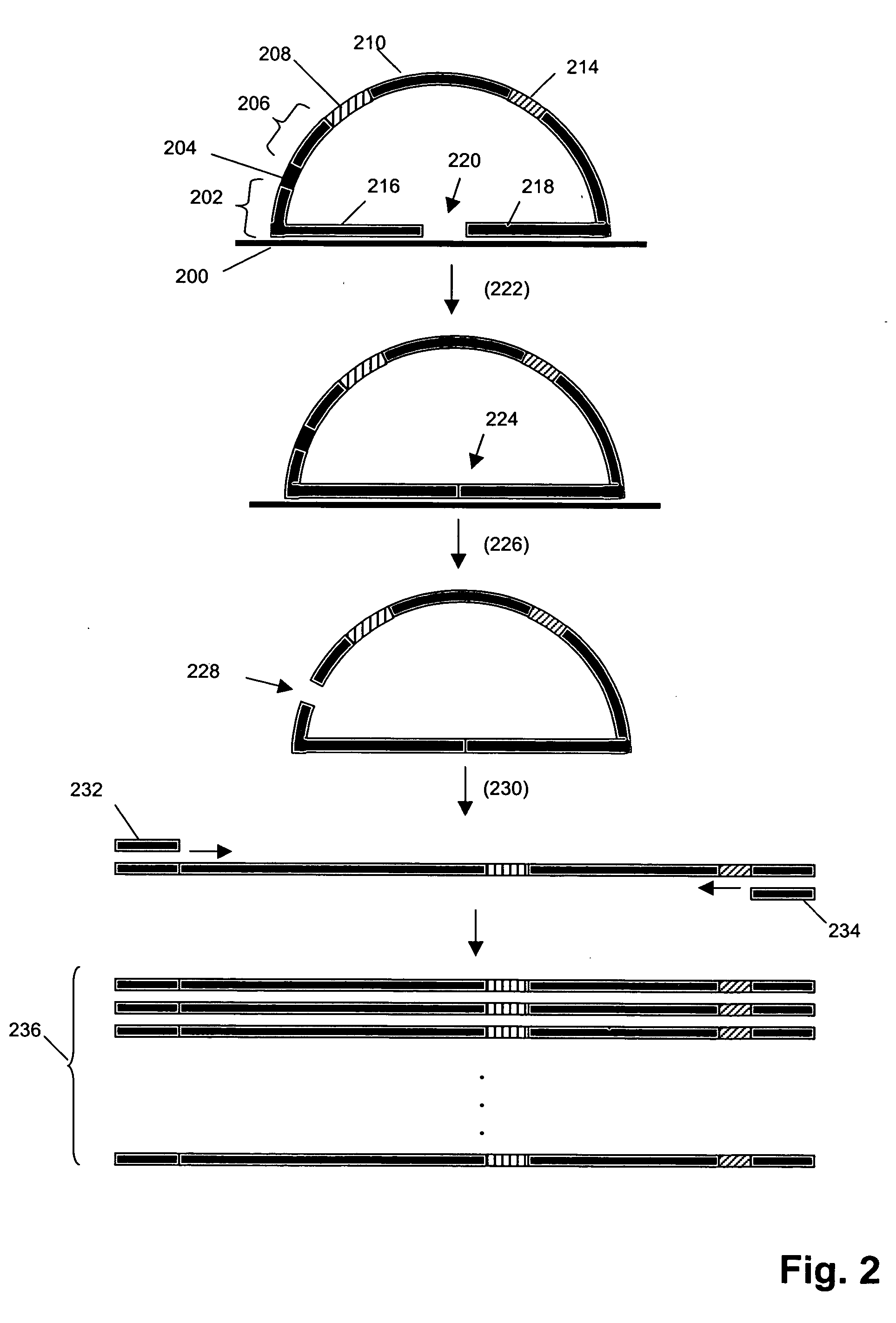
Ola-Based Methods for the Detection of Target Nucleic Avid Sequences
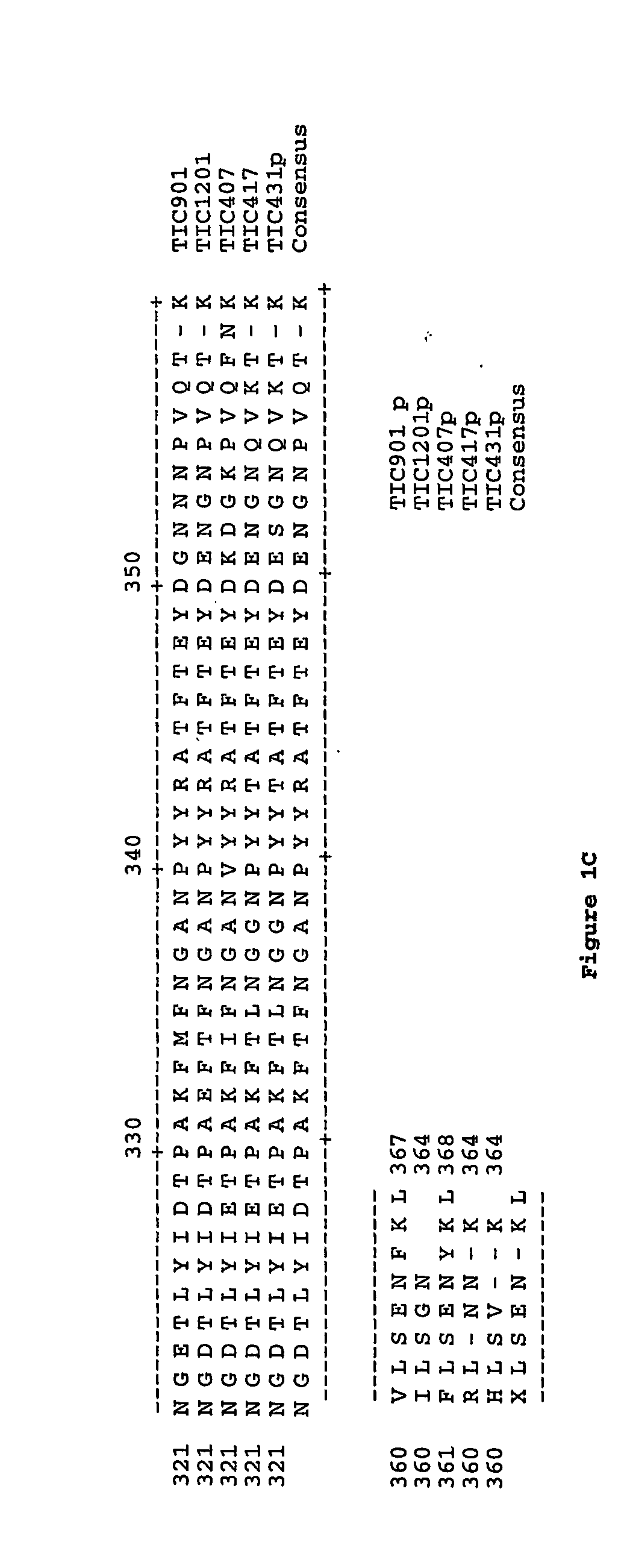
InactiveUS20070269805A1Low costSugar derivativesMicrobiological testing/measurementChemical compoundGenomic clone
Owner:KEYGENE NV
Methods for gene array analysis of nuclear runoff transcripts
InactiveUS6617112B2Rapid and efficient and extensive analysisAccurate predictionSugar derivativesMicrobiological testing/measurementStructure functionOrganism
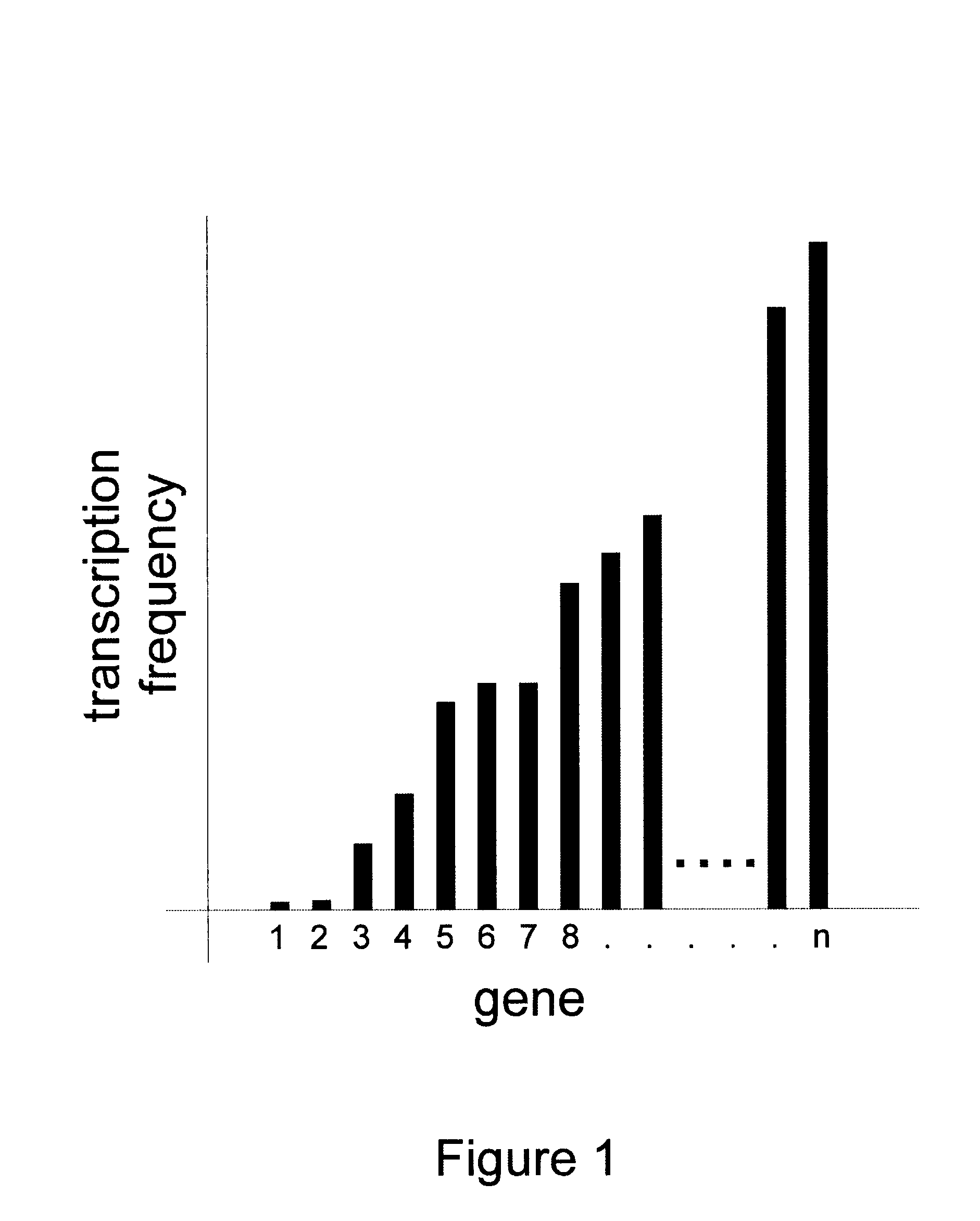
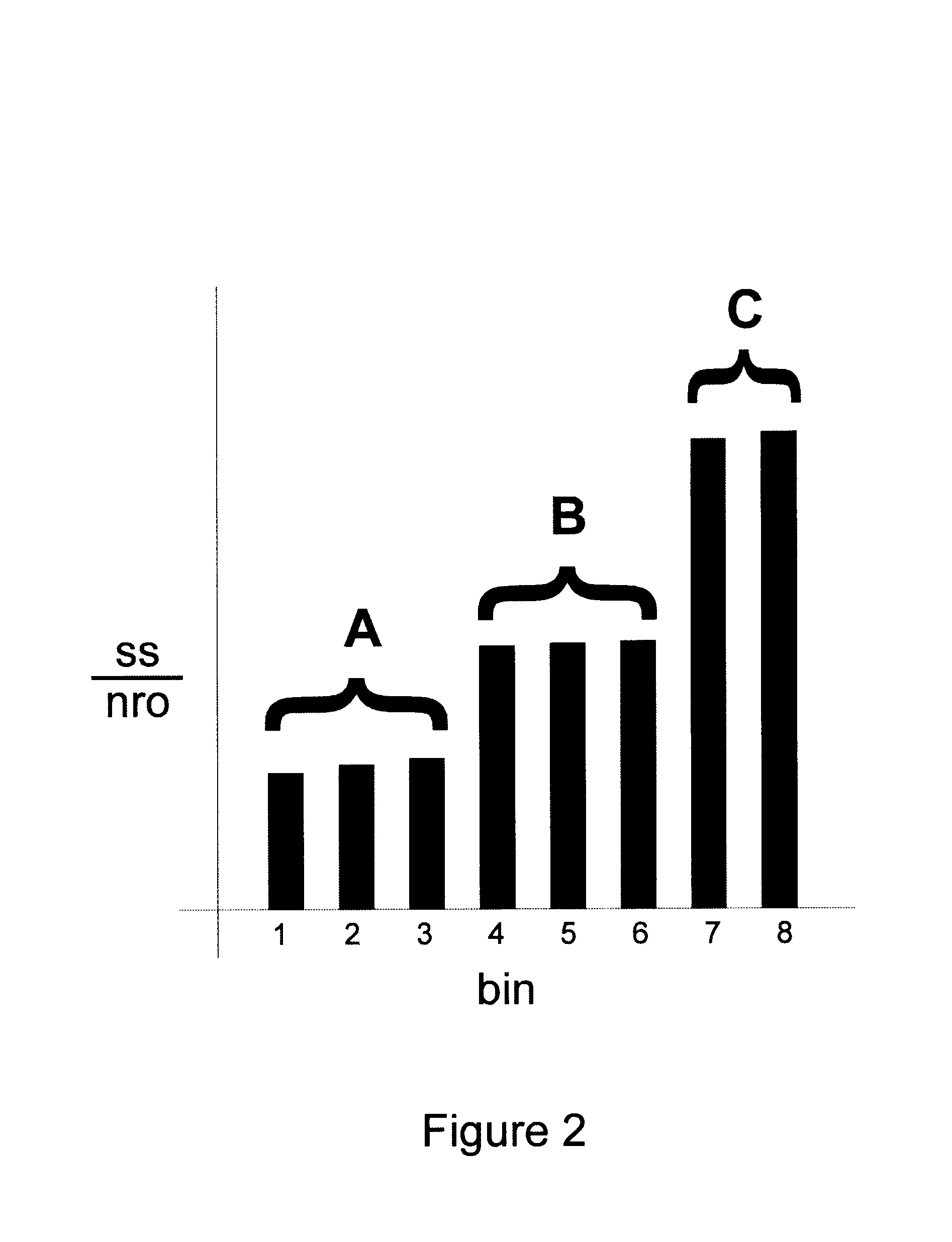
Methods for determining transcription rate of mRNA in eukaryotic cells using nuclear runoff transcription where labeled RNA molecules are hybridized against an array of at least 500 nucleic acid molecule probes representing at least part of the genome of the native eukaryotic organism to identify the quantity of nascent mRNA transcripts in said cells. The method can be used to simultaneously identify the quantity of a large number of mRNA transcripts. A rate of degradation for distinct mRNA in a eukaryotic cell rate is determined by comparing a steady state mRNA with nuclear runoff mRNA. Steady state to nuclear runoff ratios are used to determine gene and mRNA structure function relations that leads to gene expression and mRNA stability, predict structural determinants for mRNA stability and predict regulatory motifs for transcription rates. Methods of constructing recombinant organisms with enhanced stability for mRNA expressed from a gene of interest comprise introducing into the genome of an organism a gene containing one or more sequence elements that confer structural stability on mRNA transcribed from said gene.
Owner:MONSANTO TECH LLC
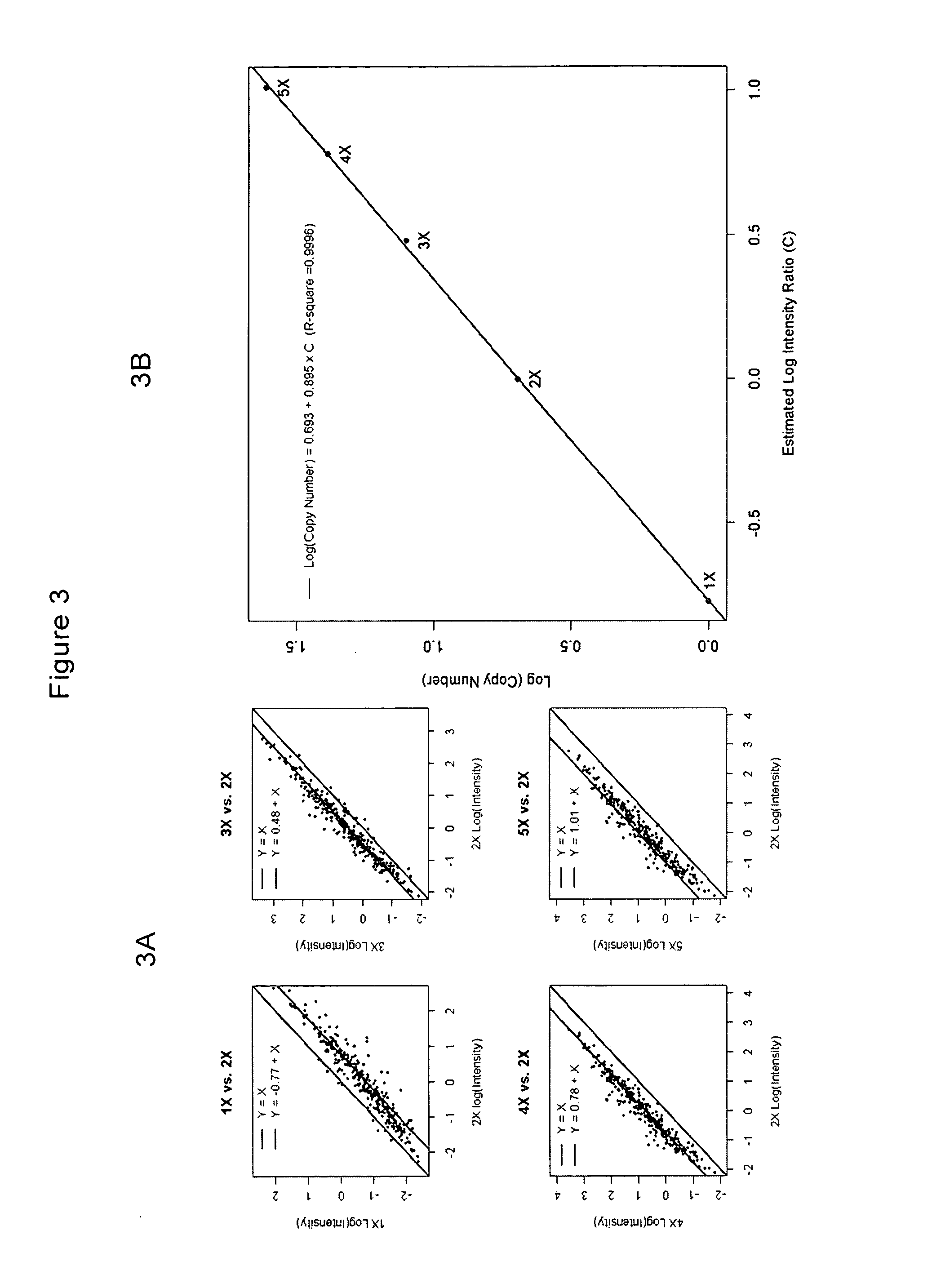
Methods for identifying DNA copy number changes
InactiveUS20050064476A1Reduce complexityMicrobiological testing/measurementProteomicsGenomic DNANormal tissue
Methods of identifying changes in genomic DNA copy number are disclosed. Methods for identifying homozygous deletions and genetic amplifications are disclosed. An array of probes designed to detect presence or absence of a plurality of different sequences is also disclosed. The probes are designed to hybridize to sequences that are predicted to be present in a reduced complexity sample. The methods may be used to detect copy number changes in cancerous tissue compared to normal tissue. The methods may be used to diagnose cancer and other diseases associated with chromosomal anomalies.
Owner:AFFYMETRIX INC
System and methods for enhancing signal-to-noise ratios of microarray-based measurements
InactiveUS20050100939A1Improve signal-to-noise ratioMicrobiological testing/measurementBiological testingCross hybridizationPolynucleotide
The present invention provides systems and methods for large-scale genetic measurements by generating from a sample labeled target sequences whose length, orientation, label, and degree of overlap and complementarity are tailored to corresponding end-attached probes of a solid support so that signal-to-noise ratios of measurement from specifically hybridized labeled target sequences are maximized. Systems for implementing methods of the invention include a set of sample-interacting probes to produce amplicons that either each contain a segment of a target polynucleotide or an oligonucleotide tag that corresponds to a segment of a target polynucleotide, one or more solid phase supports that contain a plurality of end-attached probes, and methods of generating from sample-interacting probe amplicons from which labeled target sequences are tailored for hybridization to the solid phase supports, such as microarrays. In one aspect, labeled target sequences and end-attached probe of the solid phase supports comprise oligonucleotide tags and tag complements, respectively, selected from a minimally cross-hybridizing set.
Owner:PARALLELE BIOSCI
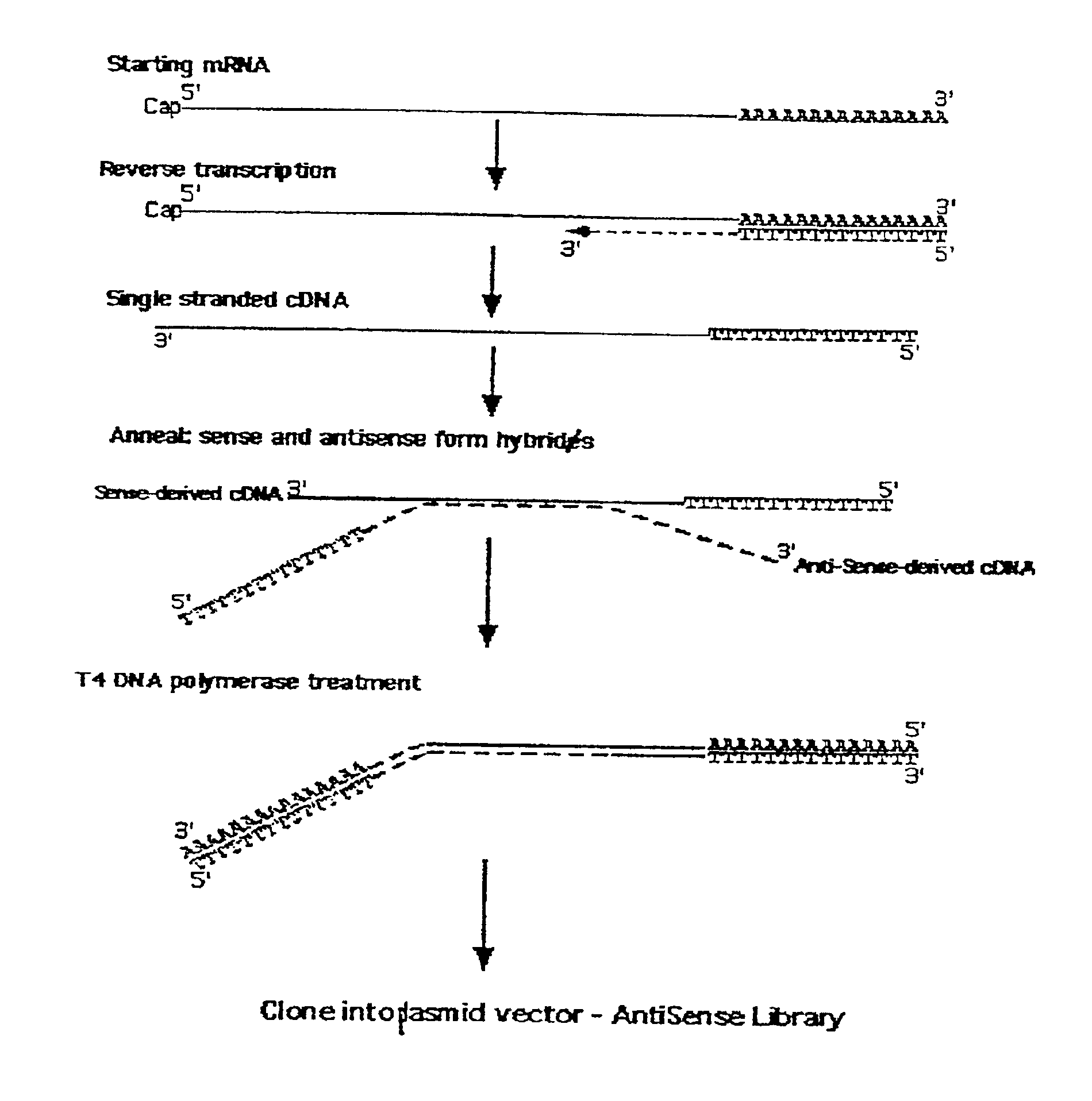
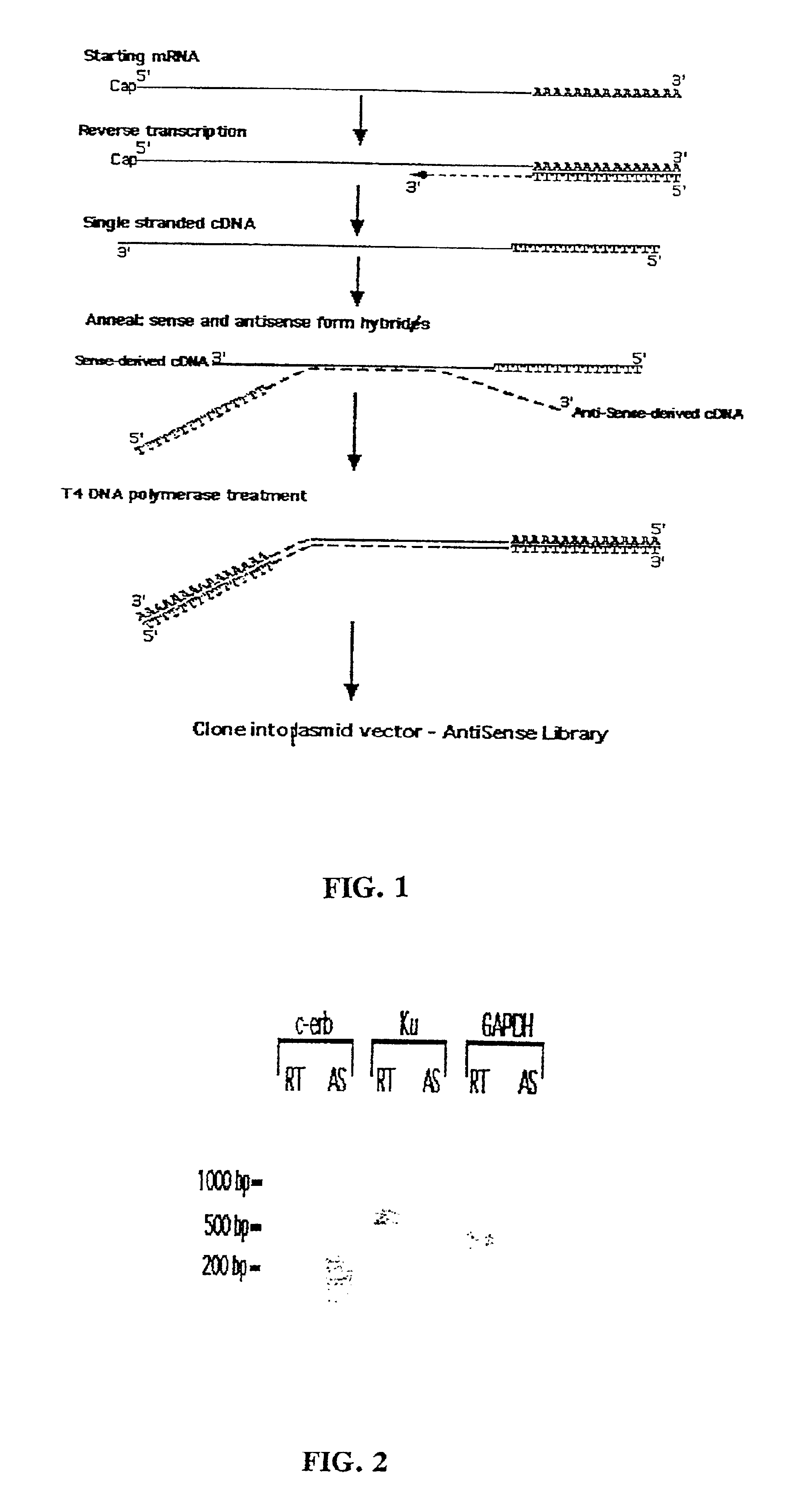
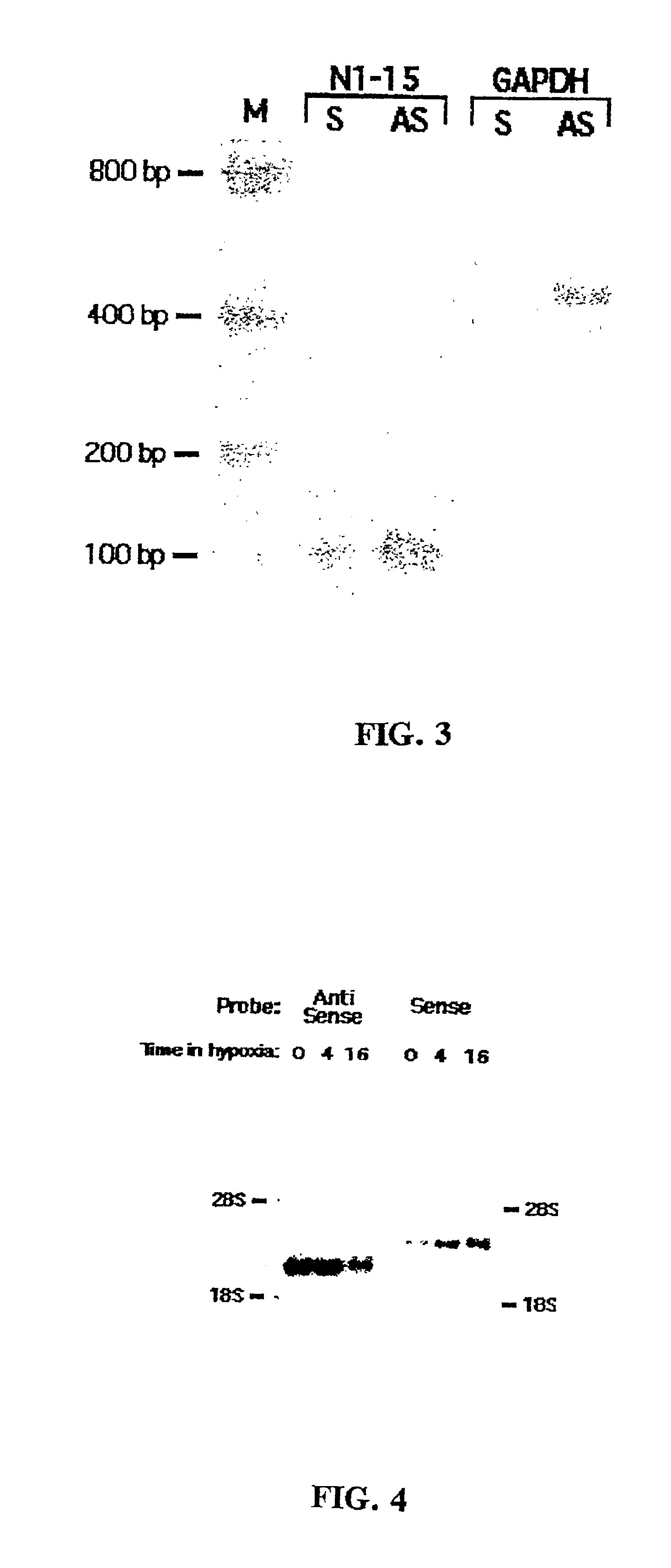
Method for enrichment of natural antisense messenger RNA
A method for enrichment of natural antisense mRNA which involves hybridization of cDNA obtained from sense RNA with cDNA obtained from antisense RNA, followed by DNA polymerase treatment of the sense-antisense hybrid DNA molecule. A natural antisense library can be generated by cloning of sense-antisense hybrid DNA molecules in a vector.
Owner:QUARK FARMACUITIKALS INC
Methods for producing hybrid seed
InactiveUS20070199095A1Suppress expression of proteinFemale sterilityOther foreign material introduction processesFermentationRecombinant DNAHybrid seed
This invention provides methods for producing a non-natural hybrid seed. Also disclosed are specific miRNAs and miRNA recognition sites useful for conferring inducible sterility on a crop plant, and recombinant DNA construct including such exogenous miRNA recognition sites.
Owner:MONSANTO TECH LLC
Methods for detection and quantification of analytes in complex mixtures
InactiveUS20070166708A1Sugar derivativesMaterial analysis by observing effect on chemical indicatorAnalyteNucleic Acid Probes
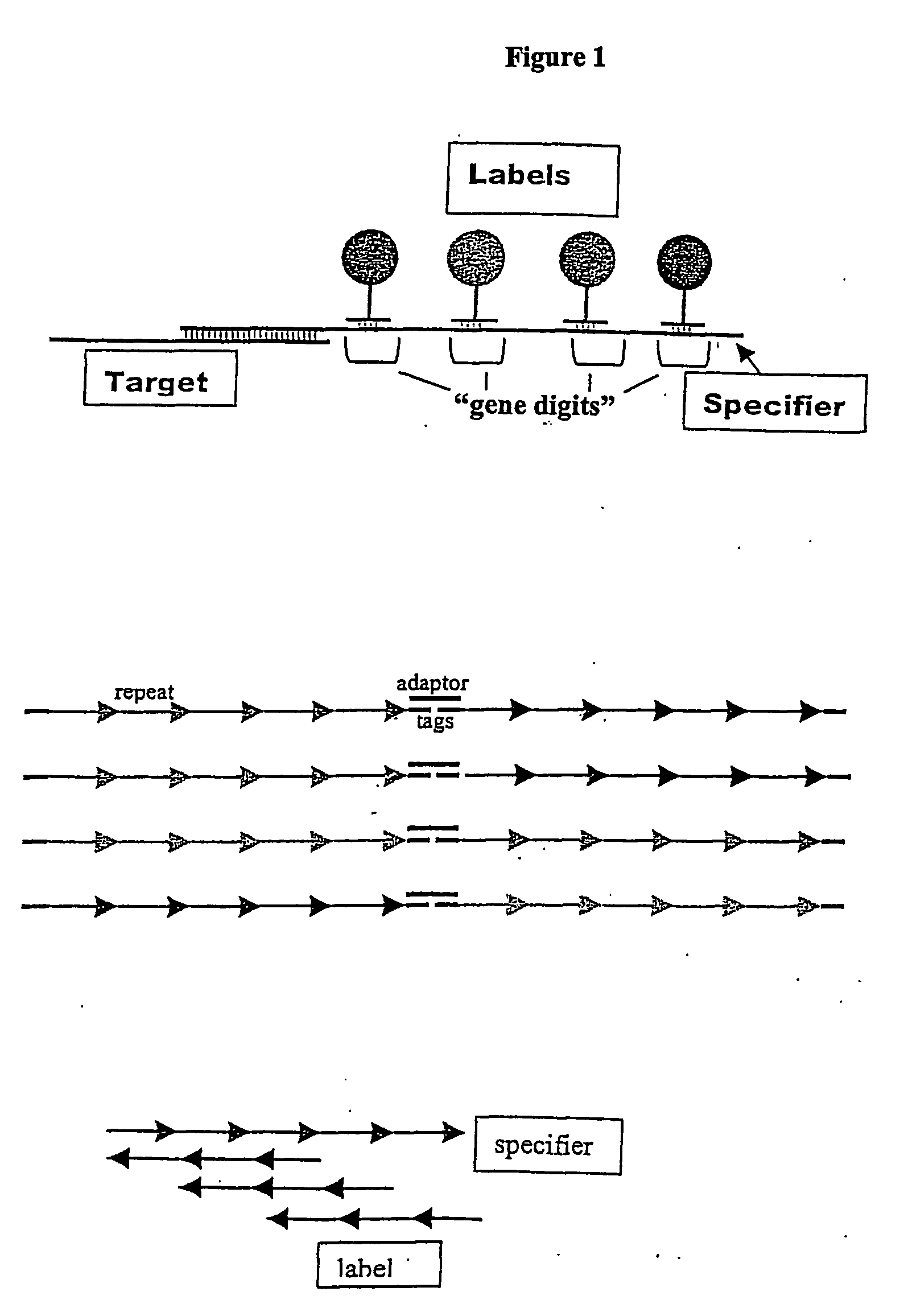
The invention provides a diverse population of uniquely labeled probes, containing about thirty or more target specific nucleic acid probes each attached to a unique label bound to a nucleic acid. Also provided is a method of producing a population of uniquely labeled nucleic acid probes. The method consists of (a) synthesizing a population of target specific nucleic acid probes each having a different specifier; (b) synthesizing a corresponding population of anti-genedigits each having a unique label, the population having a diversity sufficient to uniquely hybridize to genedigits within the specifiers, and (c) hybridizing the populations of target nucleic acid probes to the anti-genedigits, to produce a population in which each of the target specific probes is uniquely labeled. Also provided is a method of detecting a nucleic acid analyte. The method consists of (a) contacting a mixture of nucleic acid analytes under conditions sufficient for hybridization with a plurality of target specific nucleic acid probes each having a different specifier; (b) contacting the mixture under conditions sufficient for hybridization with a corresponding plurality of anti-genedigits each having a unique label, the plurality of anti-genedigits having a diversity sufficient to uniquely hybridize to genedigits within the specifiers, and (c) uniquely detecting a hybridized complex between one or more analytes in the mixture, a target specific probe, and an anti-genedigit.
Owner:INSTITUTE FOR SYSTEMS BIOLOGY
Plants and seeds of corn variety I325350
ActiveUS7157625B1Improve nutritional qualityOther foreign material introduction processesFermentationIsoenzyme typingTissue culture
According to the invention, there is provided seed and plants of the corn variety designated I325350. This invention thus relates to the plants, seeds and tissue cultures of the variety I325350, and to methods for producing a corn plant produced by crossing a corn plant of variety I325350 with itself or with another corn plant, such as a plant of another variety. This invention further relates to corn seeds and plants produced by crossing plants of variety I325350 with plants of another variety, such as another inbred line, and to crosses with related species. This invention further relates to the inbred and hybrid genetic complements of plants of variety I325350, and also to the SSR and isozyme typing profiles of corn variety I325350.
Owner:MONSANTO TECH LLC
Multipotent adult stem cells, sources thereof, methods of obtaining and maintaining same, methods of differentiation thereof, methods of use thereof and cells derived thereof
InactiveUS20050283844A1Improve angiogenesisAntibacterial agentsSenses disorderOligonucleotideBioinformatics
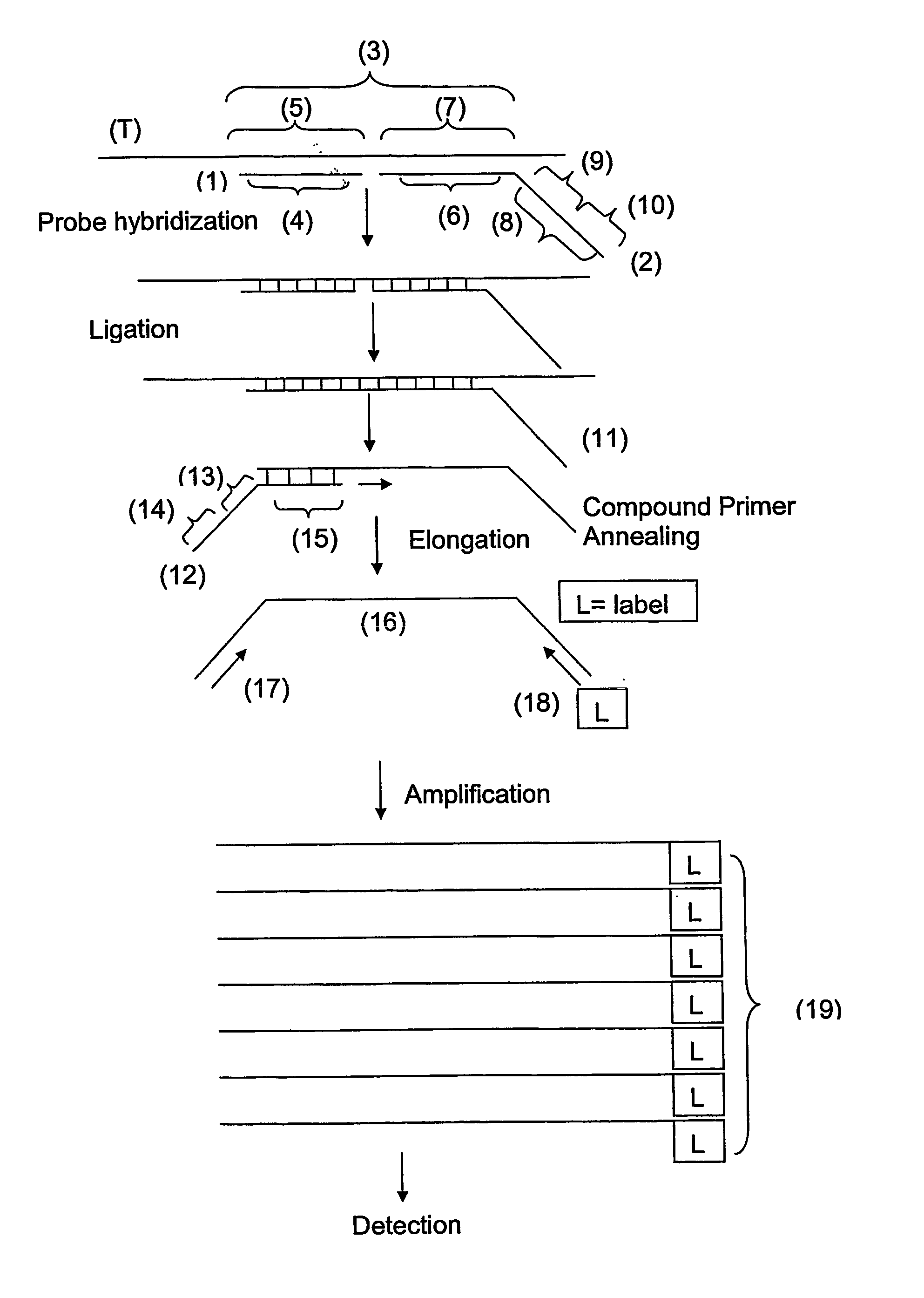
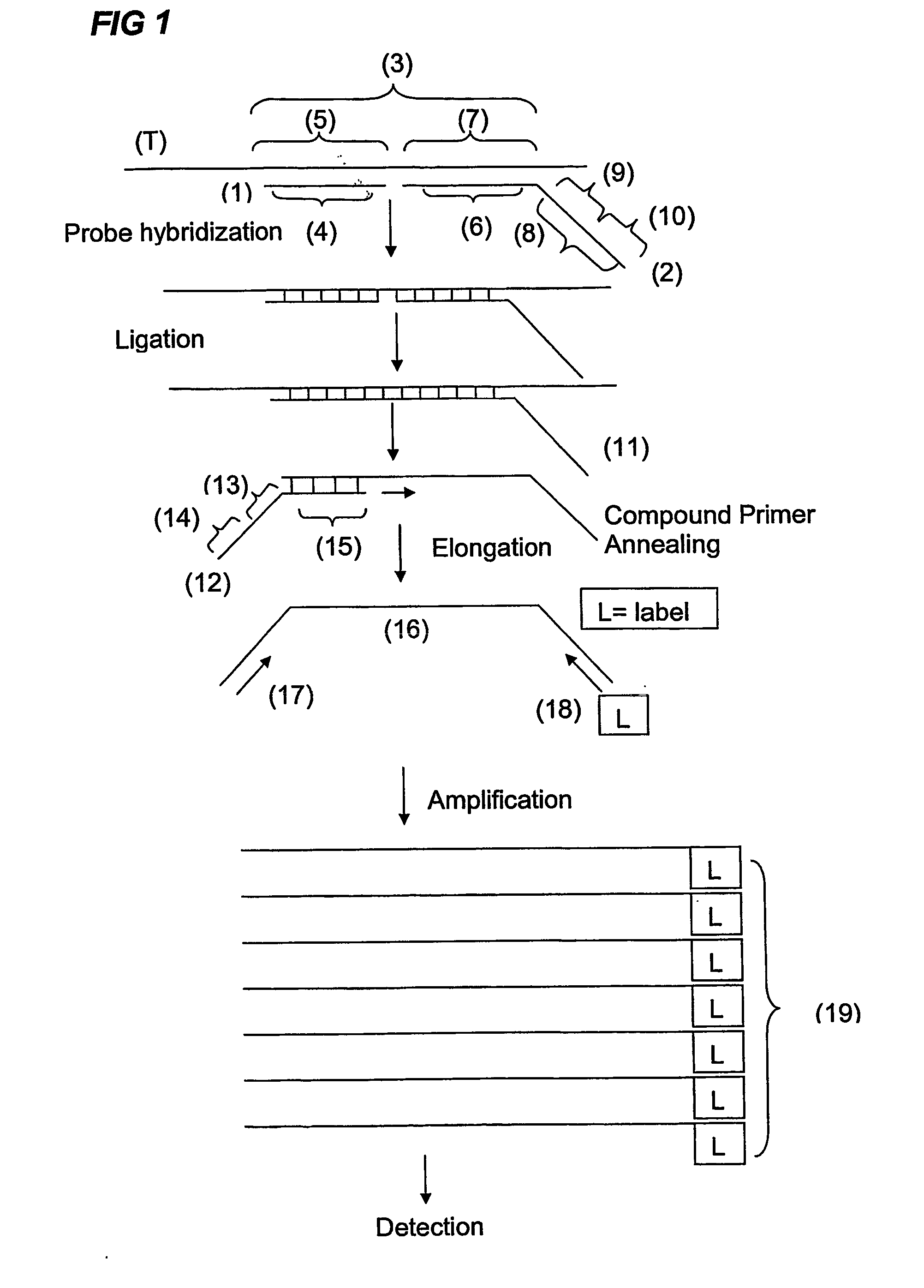
Methods and compositions are provided for circularizing target sequences in a sample. In particular, ligation oligonucleotides are employed to selectively hybridize with the target such that the target can be ligated into a closed circular target. Rolling circle amplification can then be performed directly on the target sequence for subsequent detection and analysis.
Owner:FURCHT LEO +2
Detection of nucleic acids by target-specific hybrid capture method
InactiveUS20060051809A1Rapid and sensitive and accurateFunction increaseSugar derivativesMicrobiological testing/measurementNucleic acid detectionDNA
Target-specific hybrid capture (TSHC) provides a nucleic acid detection method that is not only rapid and sensitive, but is also highly specific and capable of discriminating highly homologous nucleic acid target sequences. The method produces DNA:RNA hybrids which can be detected by a variety of methods.
Owner:QIAGEN GAITHERSBURG
Methods and devices for measuring differential gene expression
InactiveUS6355423B1Efficient and accurateImprove search efficiencySugar derivativesMicrobiological testing/measurementSequence databaseNucleic Acid Sequence Databases
This invention includes methods for identifying nucleic acids in a sample of nucleic acids by observing sequence sets present in the nucleic acids of the sample and then identifying those sequences in a nucleic acid sequence database having the sequence sets observed. In a preferred embodiment, a sequence set consists of two primary subsequences and an additional subsequence having determined mutual relationships. The methods include those for observing the sequence sets and those for performing sequence database searches. This invention also includes devices for recognizing in parallel the additional subsequences in a sample of as well as methods for the use of these devices. In a preferred embodiment, the devices include probes bound to a planar surface that recognize additional subsequence by hybridization, and the methods of use include features to improve the specificity and reproducibility of this hybridization.
Owner:CURAGEN CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com