Methods for identifying DNA copy number changes
a technology of dna copy number and dna copy number, which is applied in the field of methods of estimating the number of copies of a genomic region, can solve the problems of limiting the resolution to 10-20 mb, preventing the detection of small gains and losses, and the hybridization kinetics may not be as uniform, so as to reduce the complexity of the genomic dna in the sampl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
Cell Lines and Nucleic Acid Isolation
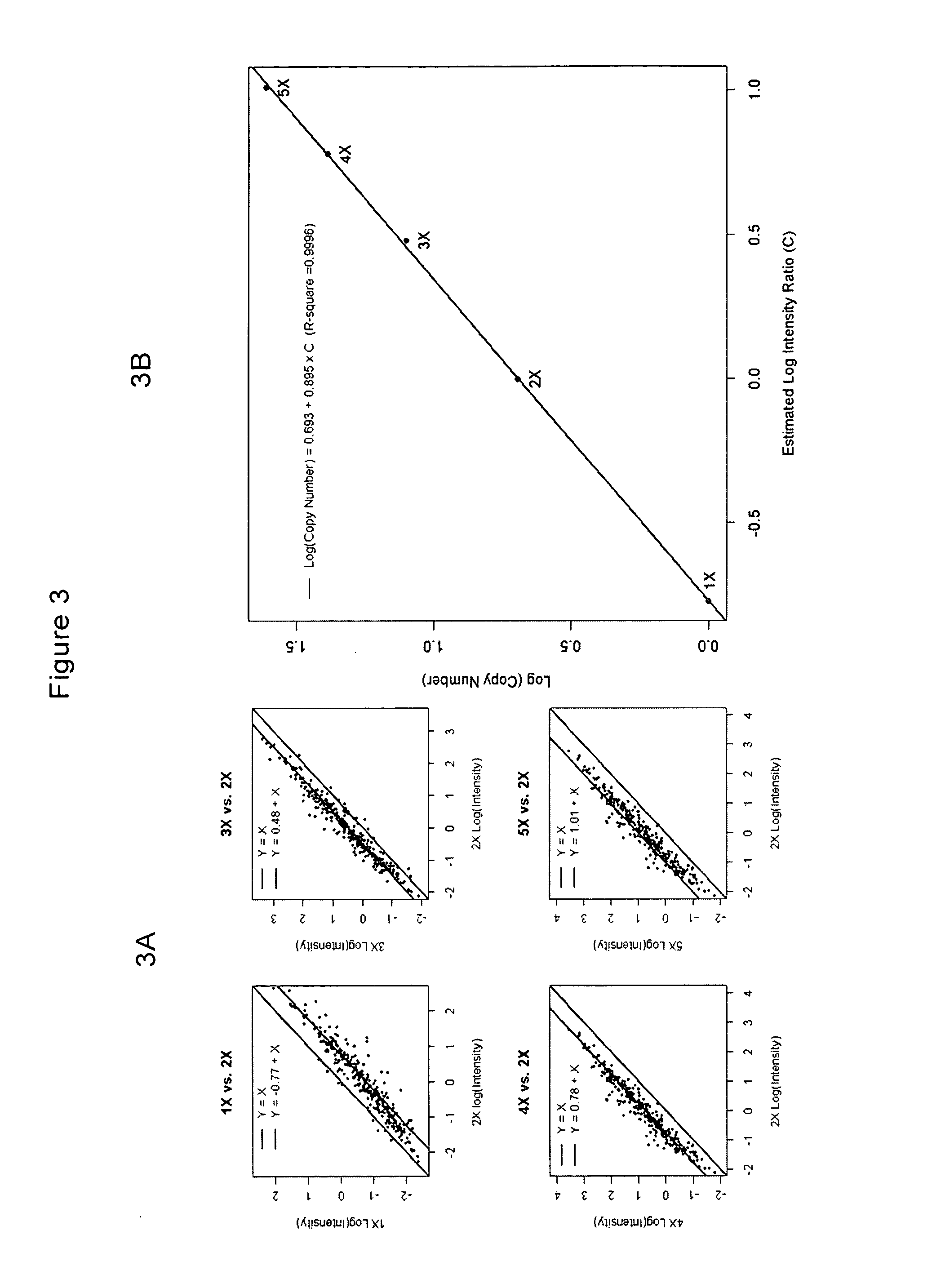
Nine human breast cancer cell lines (BT-20, MCF-7, MCF-12A, MDA-MB-157, MDA-MB-436, MDA-MB-468, SK-BR-3, ZR-75-1, and ZR-75-30) and two syngeneic human breast cancer cell lines (Hs-578T and Hs-578Bst) (Hackett et al. (1977) J Natl Cancer Inst, Vol. 58, pp.1795-806) were obtained from American Type Culture Collection (ATCC). A normal human mammary epithelial cell line (HMEC) was obtained from Clonetics. All cells were grown under recommended culture conditions. Genomic DNA was isolated using QIAGEN QIAamp DNA Blood Mini Kit. DNAs from cell lines containing 3×(NA04626), 4×(NA01416), and 5×(NA06061) chromosomes and DNAs for the normal reference set of 110 individuals (48 males and 62 females) were purchased from NIGMS Human Genetic Cell Repository, Coriell Institute for Medical Research (Camden, N.J.).
The WGSA assay was performed as described in Kennedy et al. (2003) except for modifications to the target amplification and DNA labeling steps. D...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com