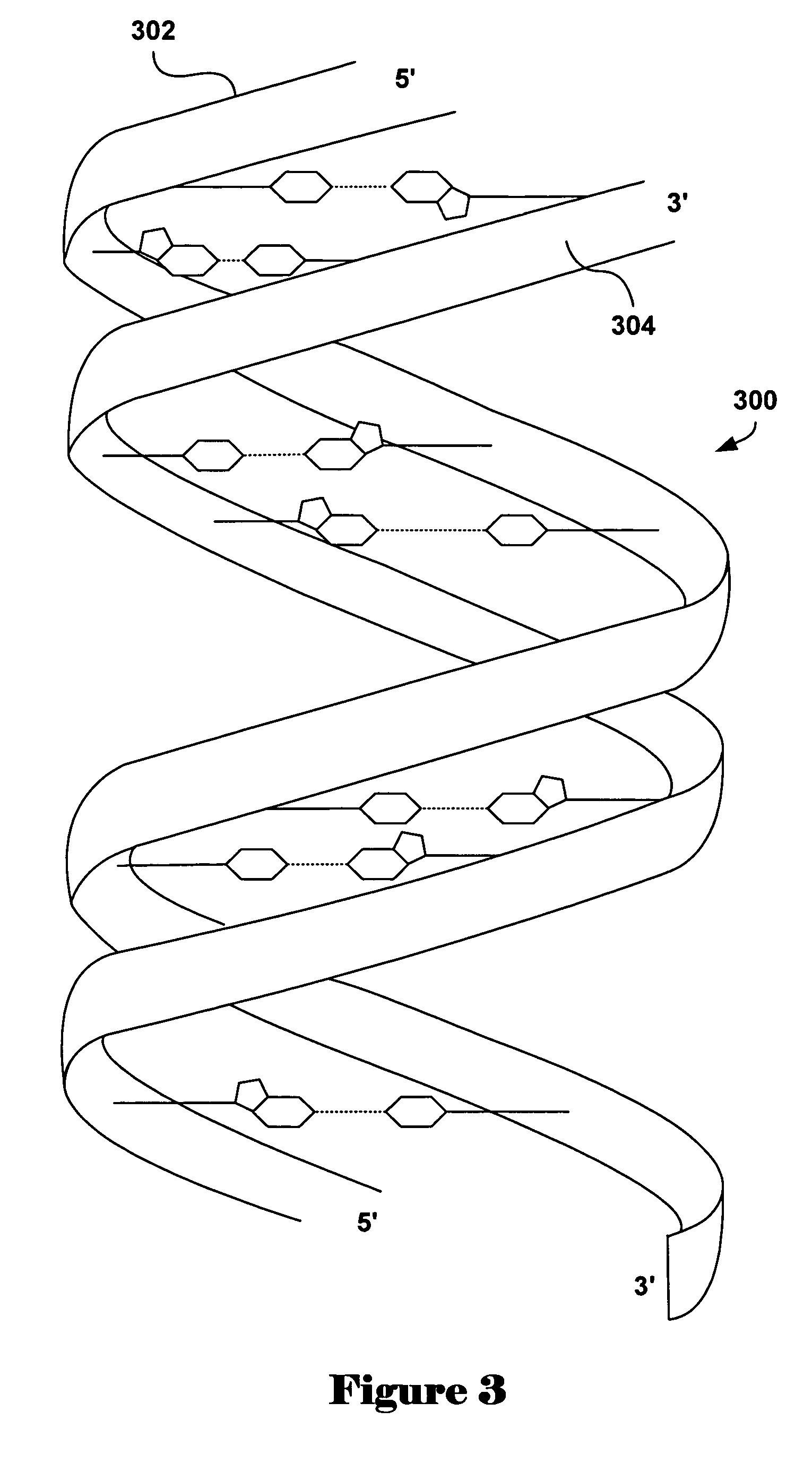
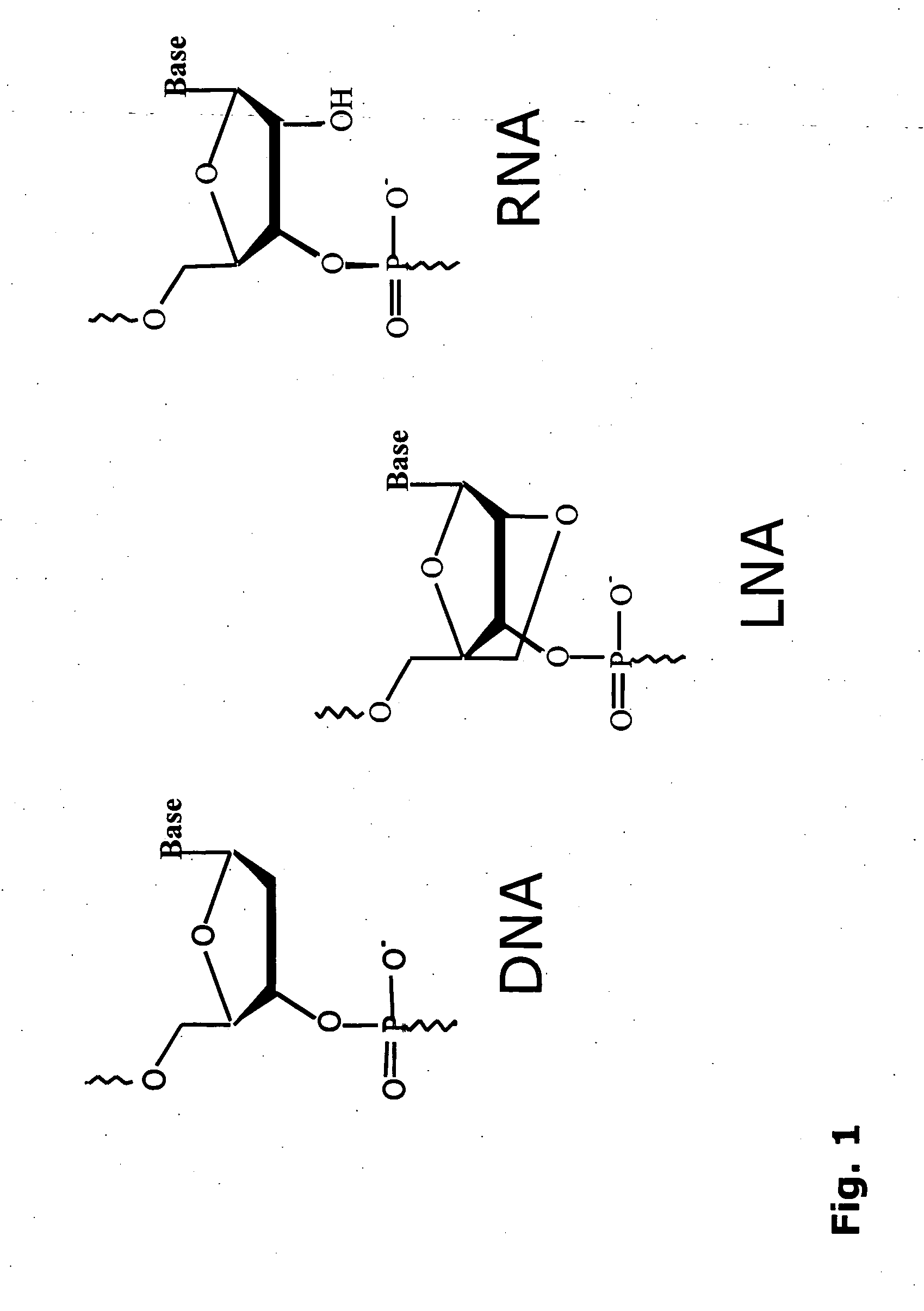
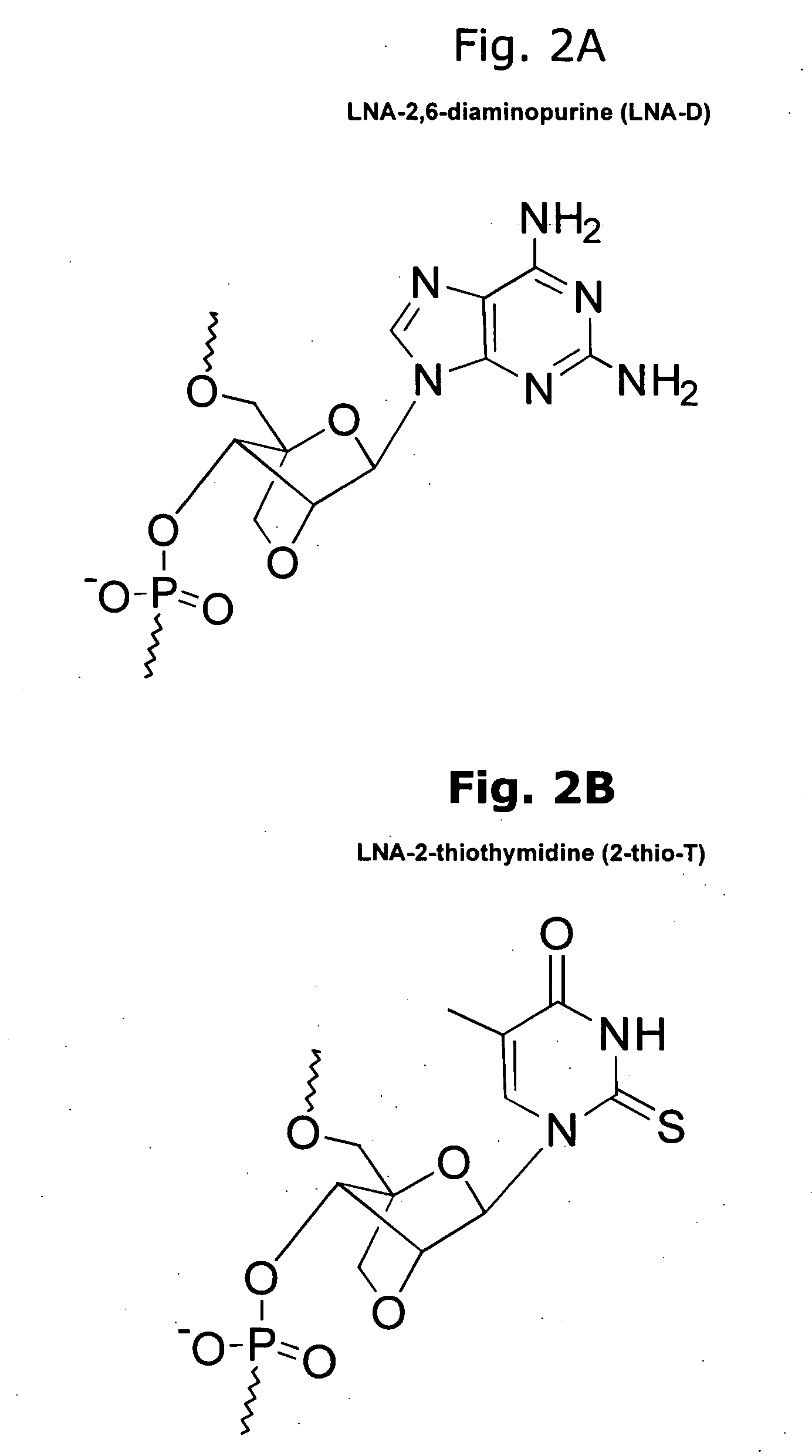
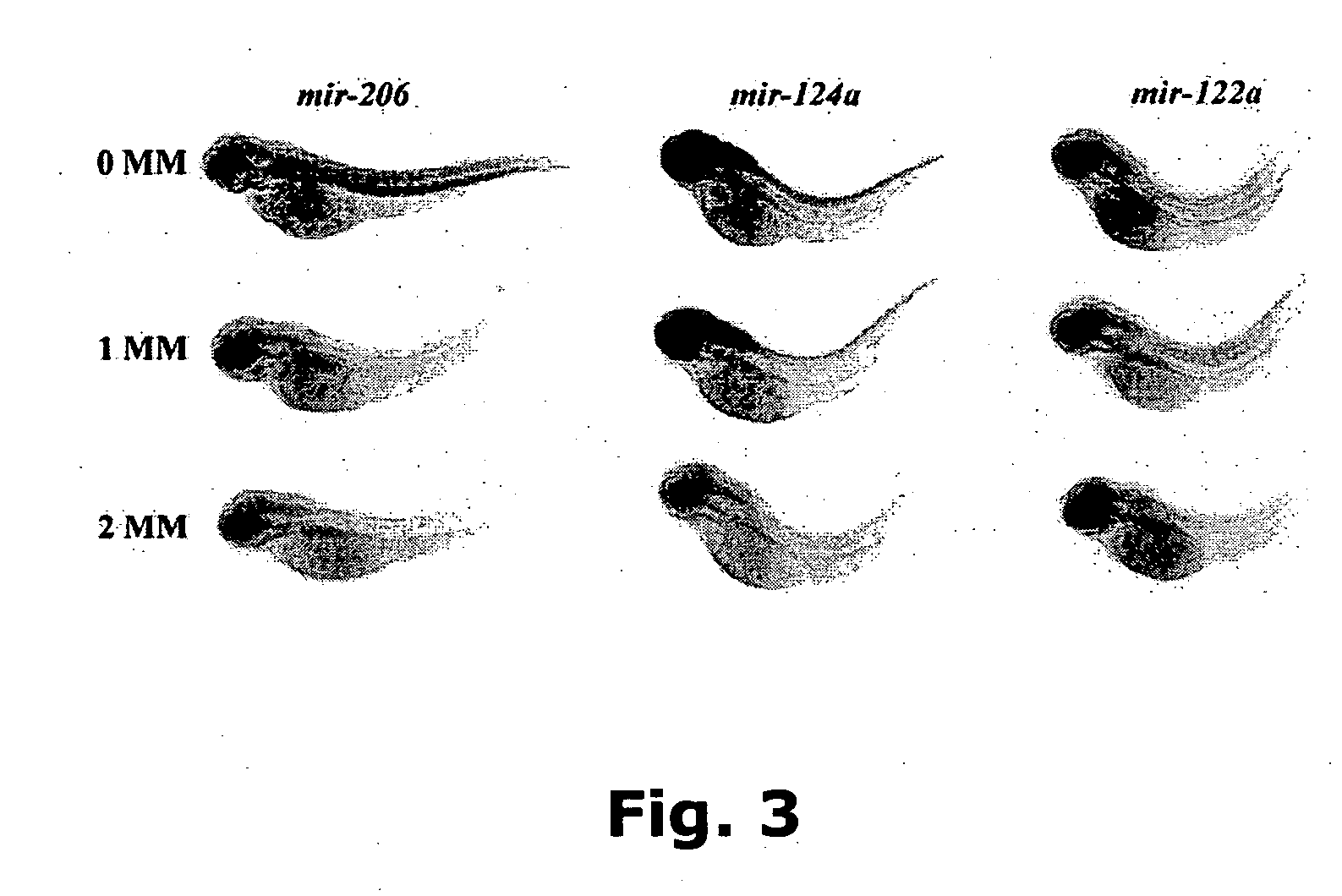
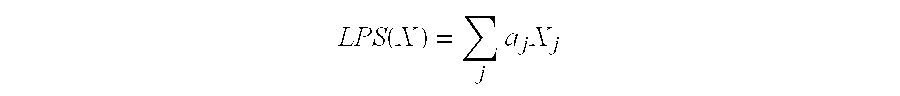Patents
Literature
3241results about "Hybridisation" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
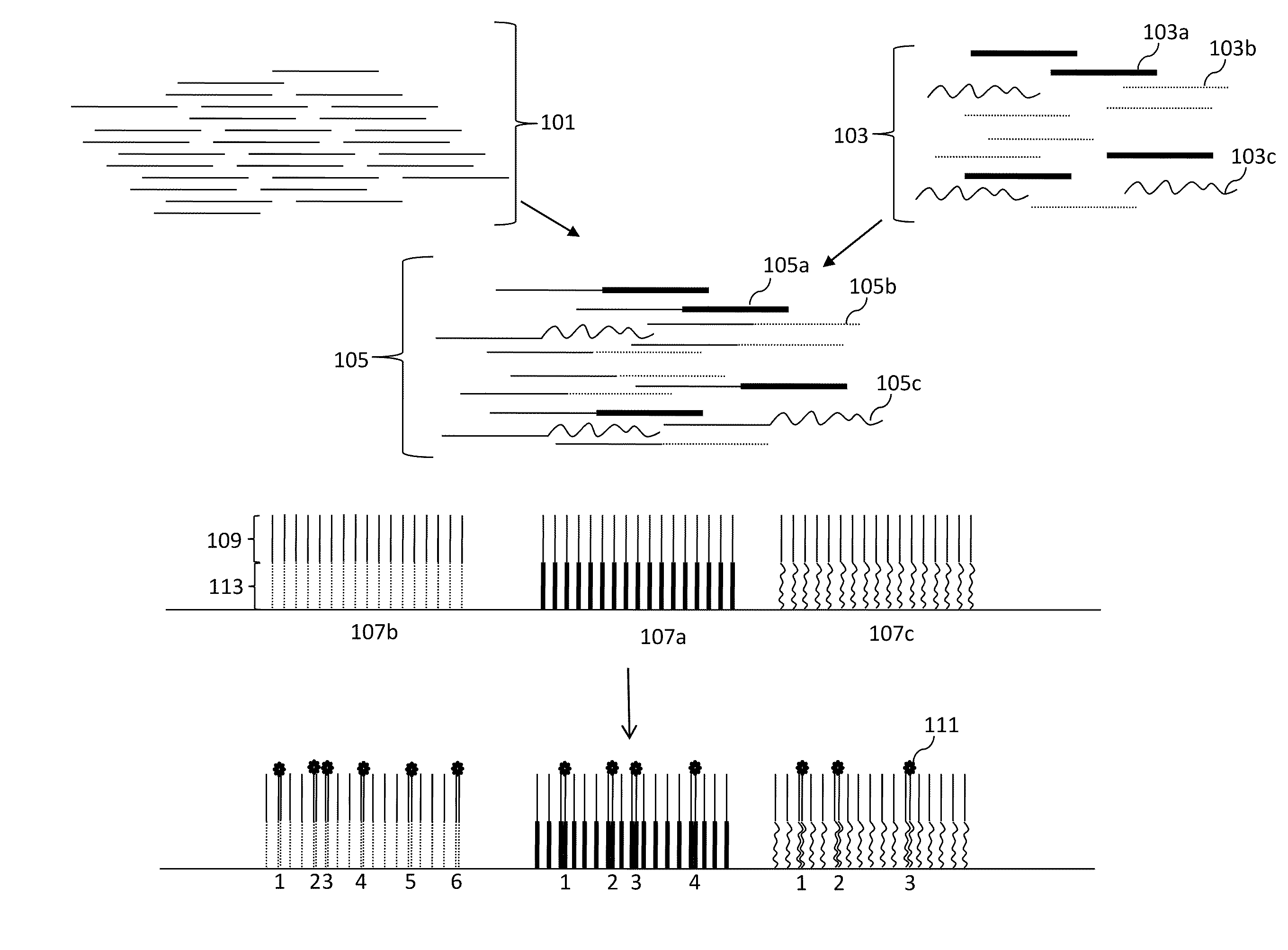
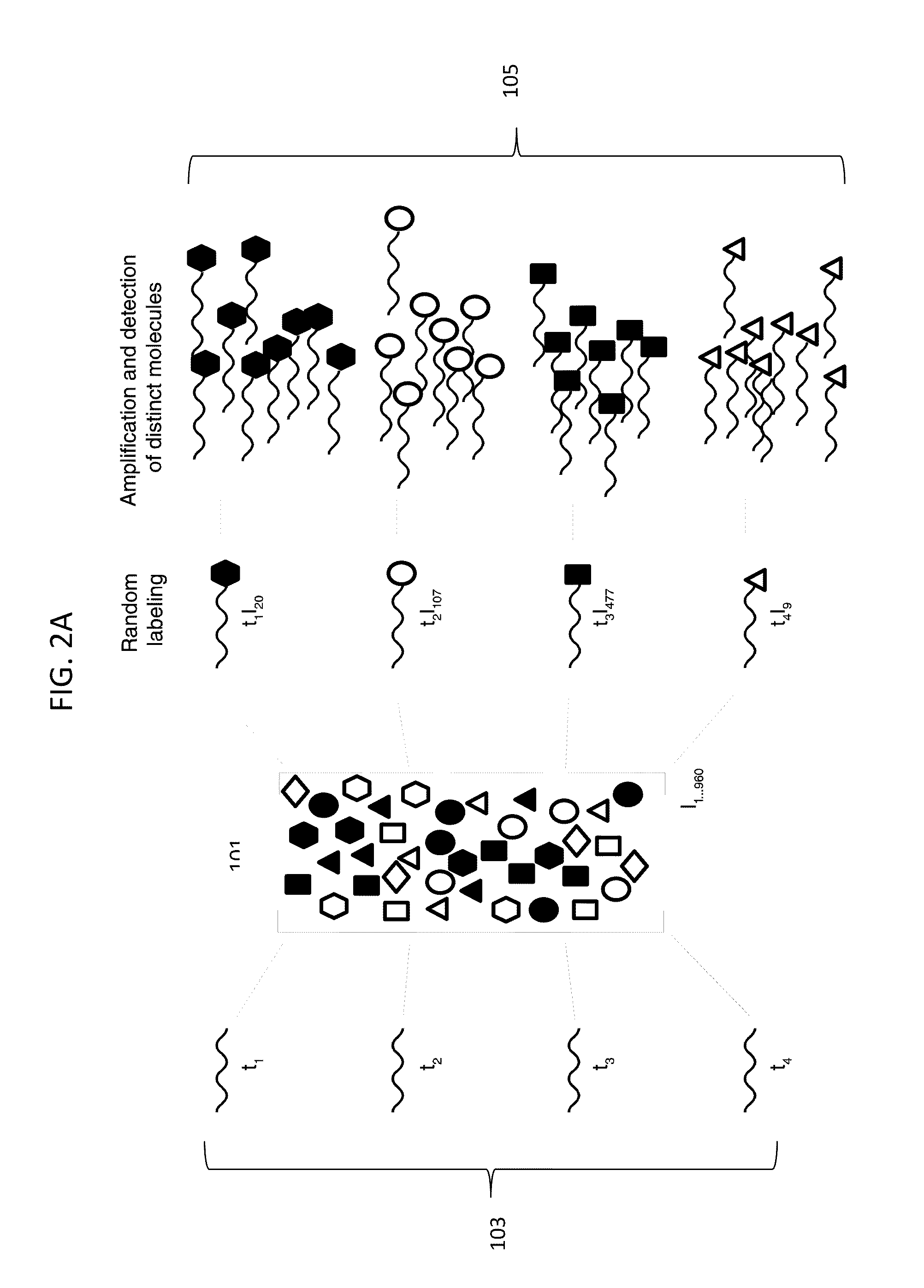
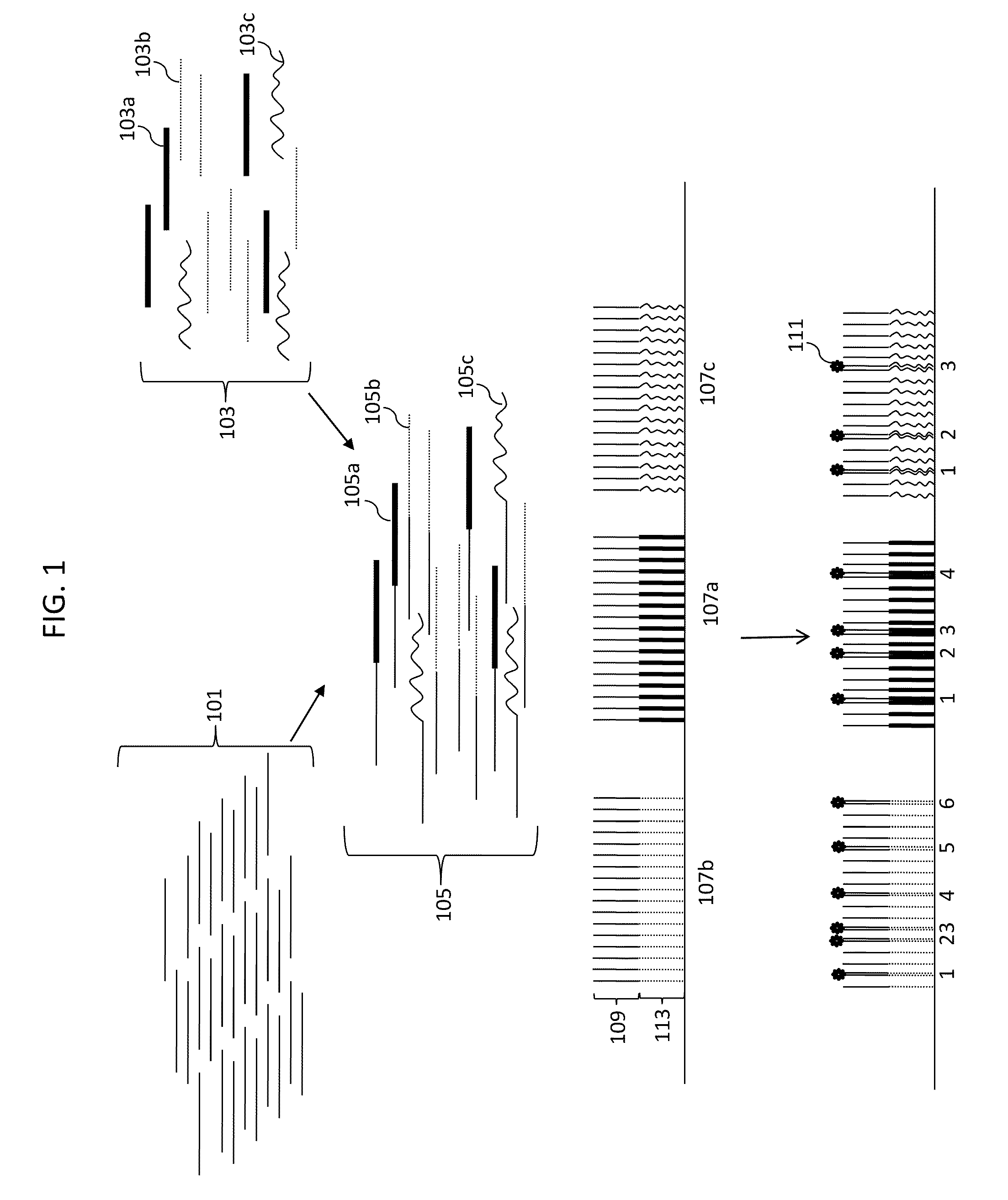
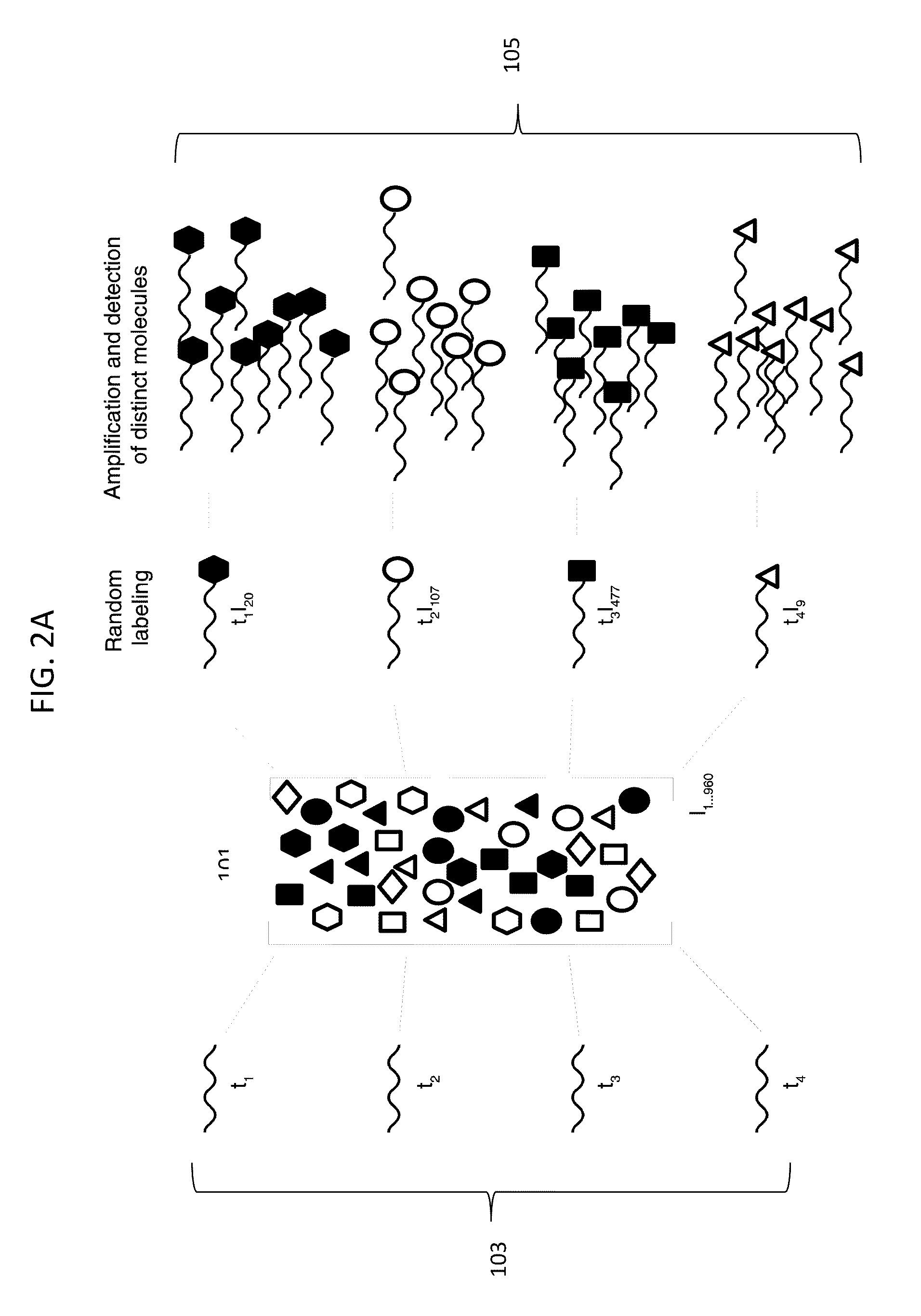
Digital Counting of Individual Molecules by Stochastic Attachment of Diverse Labels
ActiveUS20110160078A1Highly precise relative and absolute counting statisticRaise countNucleotide librariesMicrobiological testing/measurementBiologyMultiple methods
Owner:BECTON DICKINSON & CO
Digital counting of individual molecules by stochastic attachment of diverse labels
Owner:BECTON DICKINSON & CO
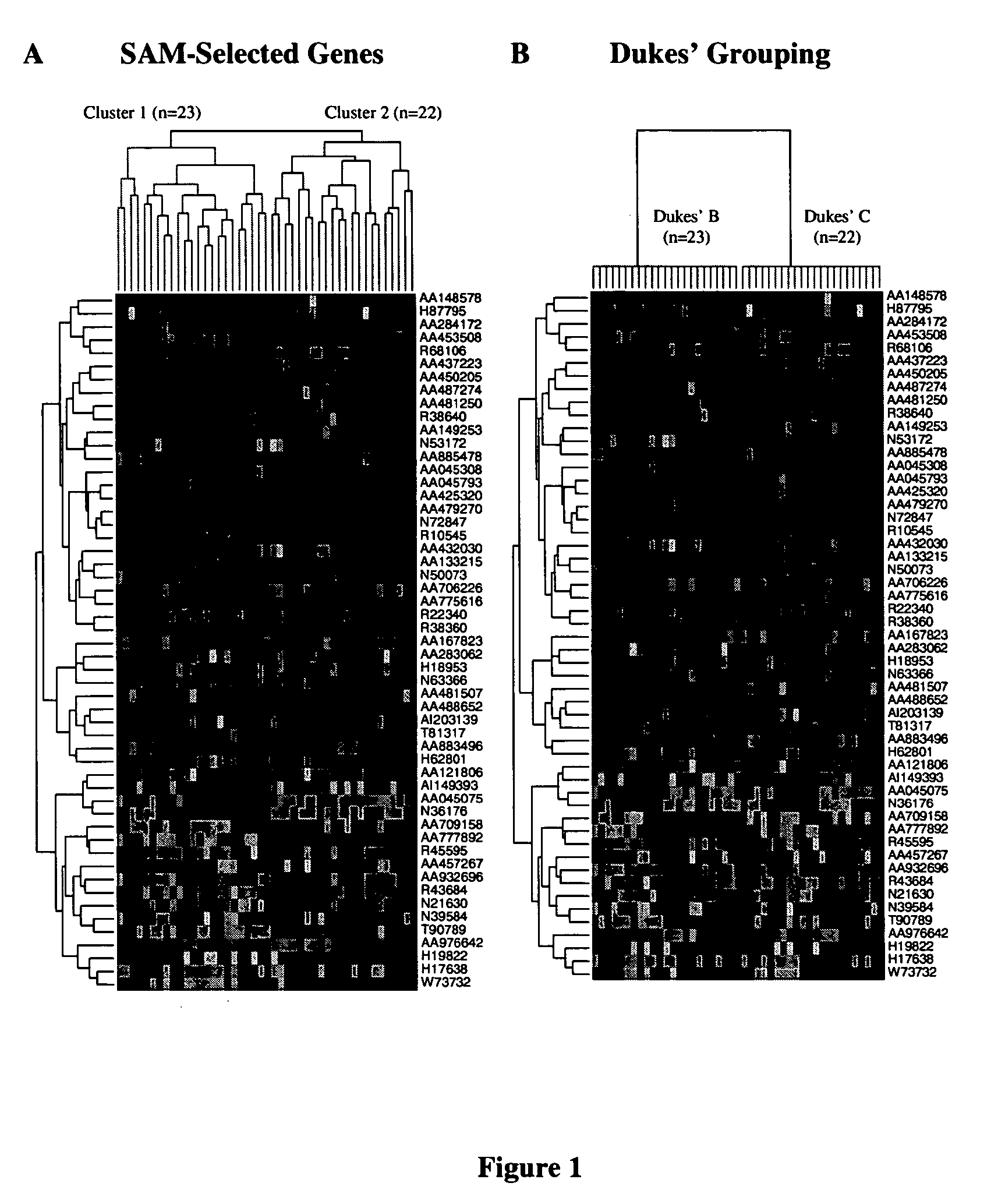
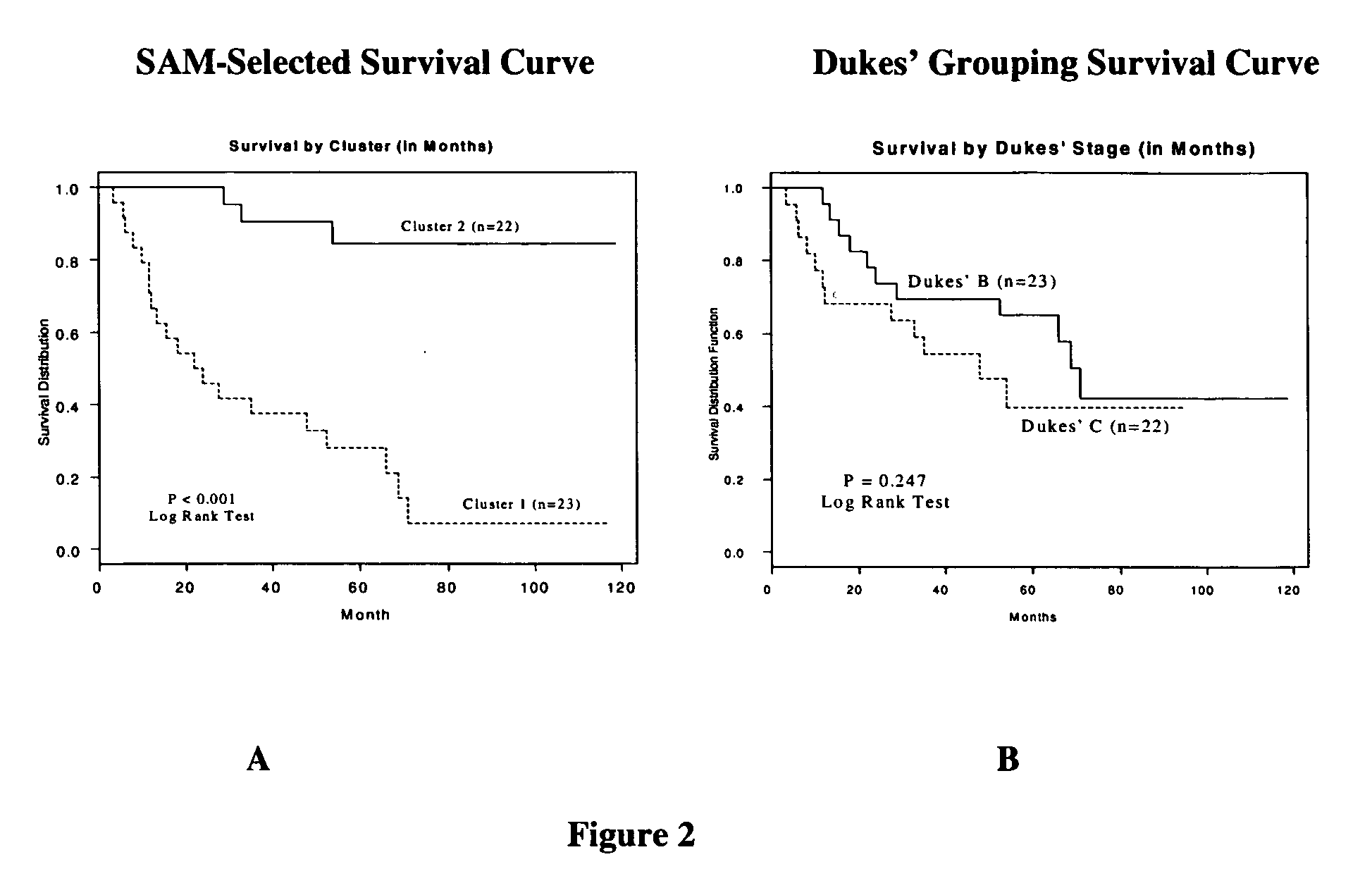
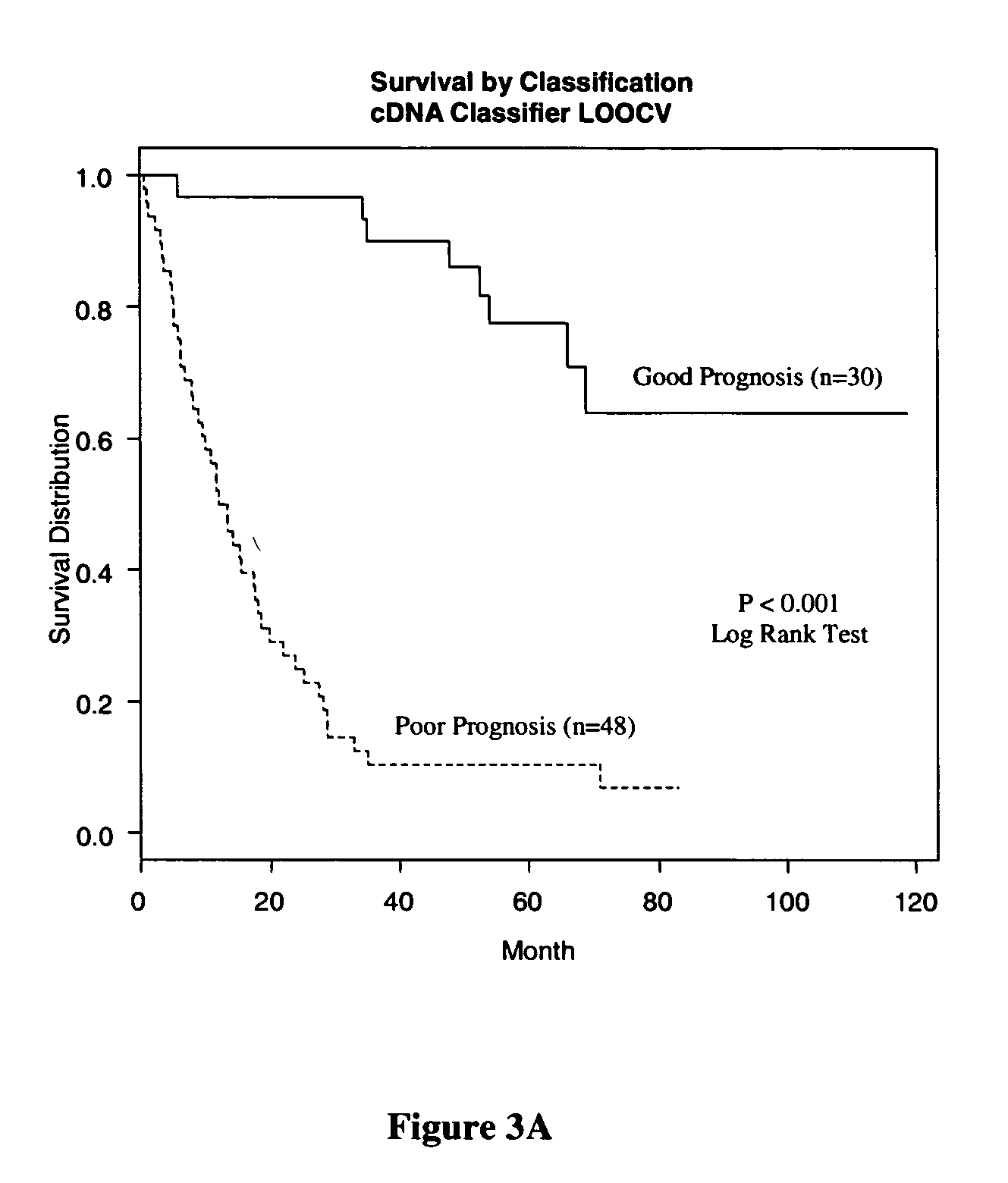
Methods and systems for predicting cancer outcome
The invention provides a molecular marker set that can be used for prognosis of colorectal cancer in a colorectal cancer patient. The invention also provides methods and computer systems for evaluating prognosis of colorectal cancer in a colorectal cancer patient based on the molecular marker set. The invention also provides methods and computer systems for determining chemotherapy for a colorectal cancer patient and for enrolling patients in clinical trials.
Owner:H LEE MOFFITT CANCER CENT & RES INST INC +1
Methods for using co-regulated genesets to enhance detection and classification of gene expression patterns
InactiveUS6203987B1Peptide/protein ingredientsMicrobiological testing/measurementCo-regulationIndividual gene
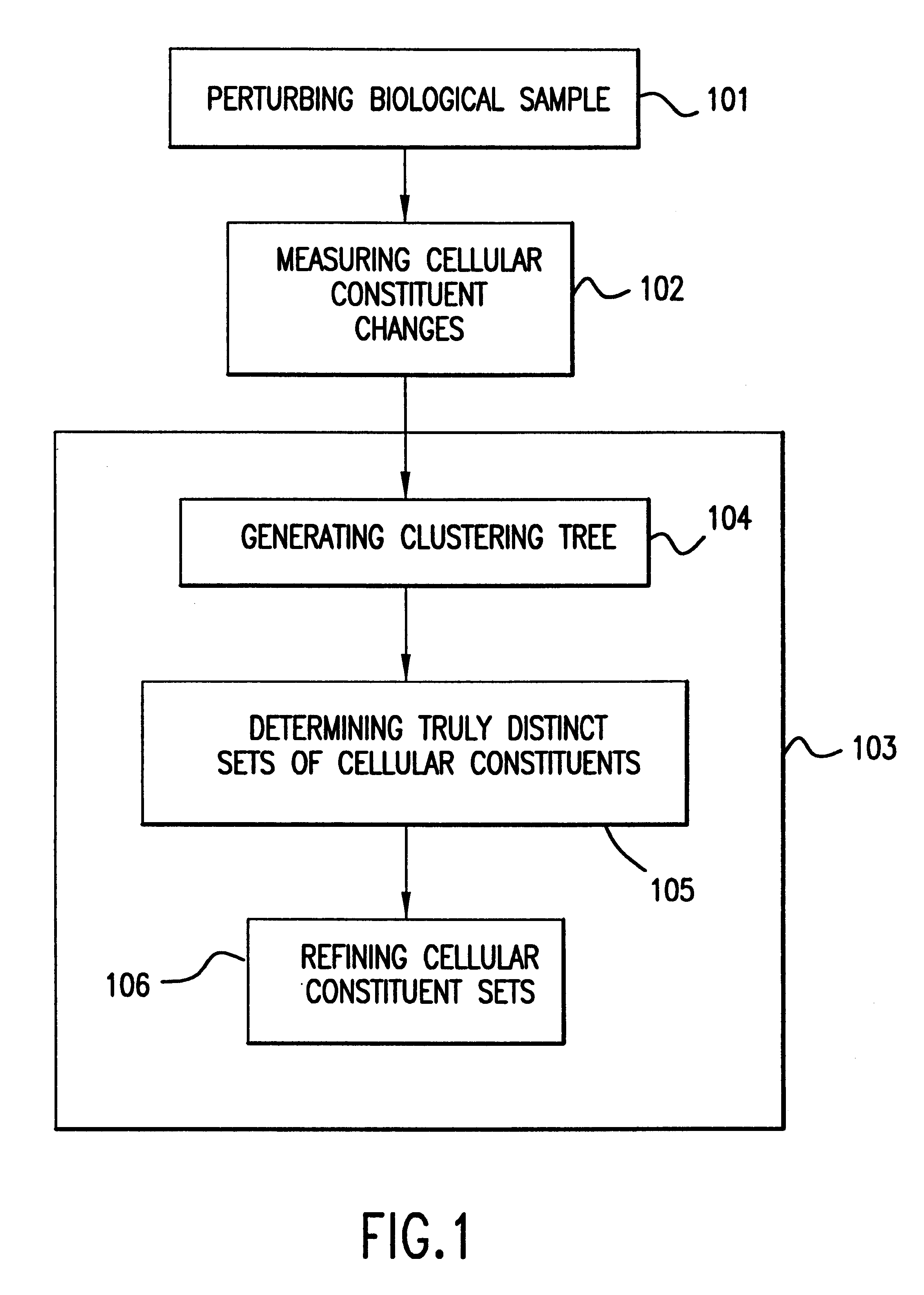
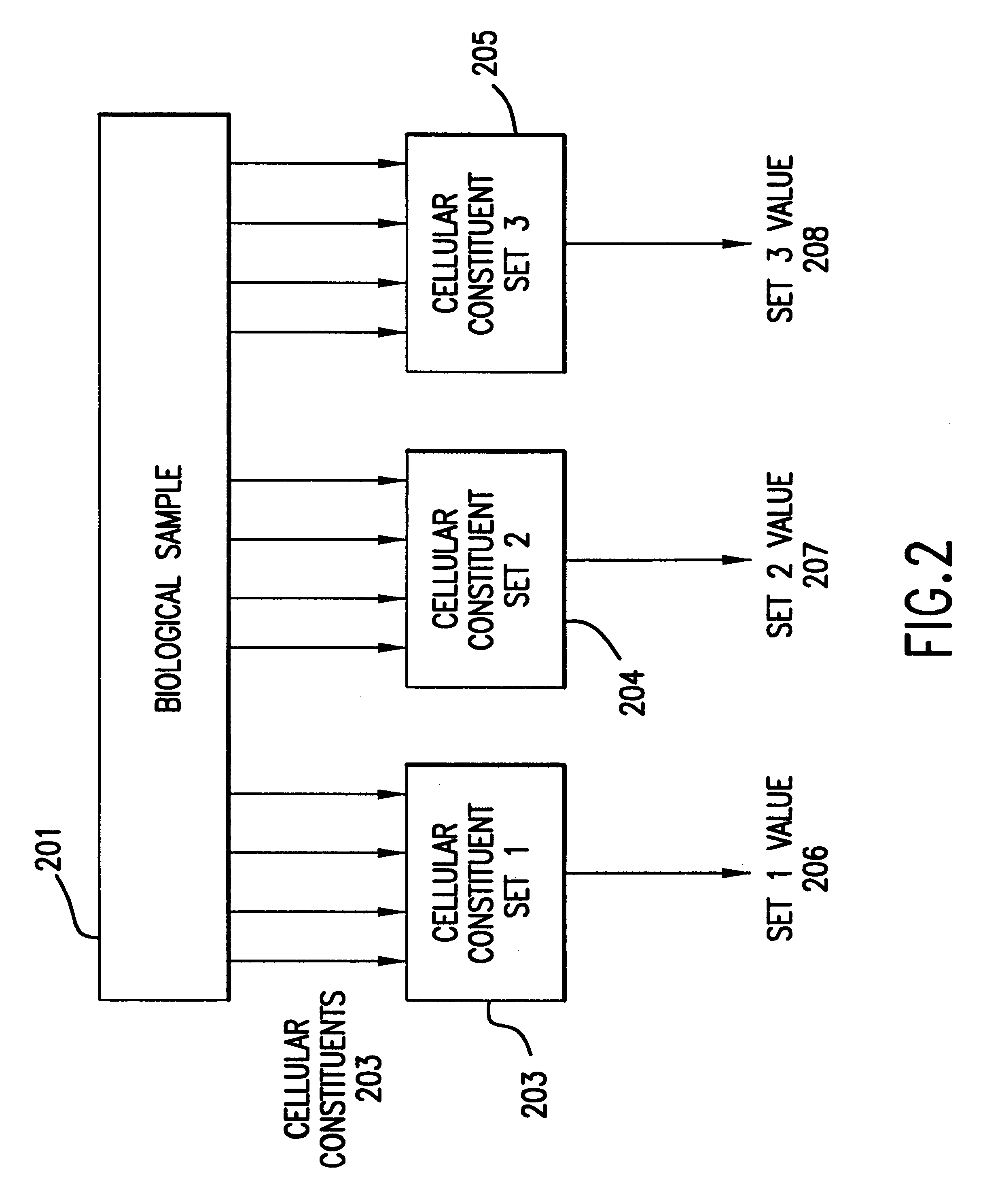
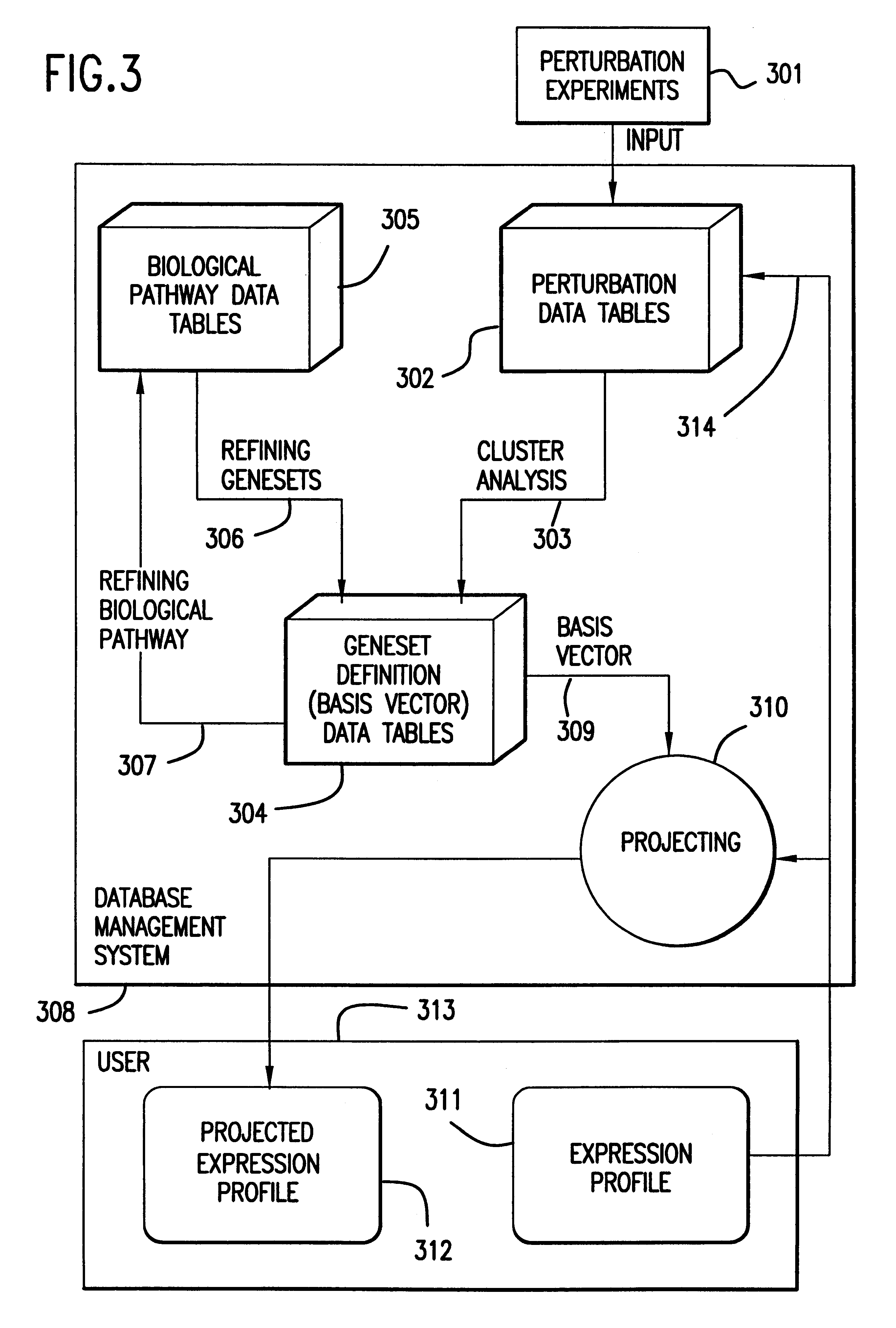
The present invention provides methods for enhanced detection of biological response patterns. In one embodiment of the invention, genes are grouped into basis genesets according to the co-regulation of their expression. Expression of individual genes within a geneset is indicated with a single gene expression value for the geneset by a projection process. The expression values of genesets, rather than the expression of individual genes are then used as the basis for comparison and detection of biological responses with greatly enhanced sensitivity.
Owner:MICROSOFT TECH LICENSING LLC
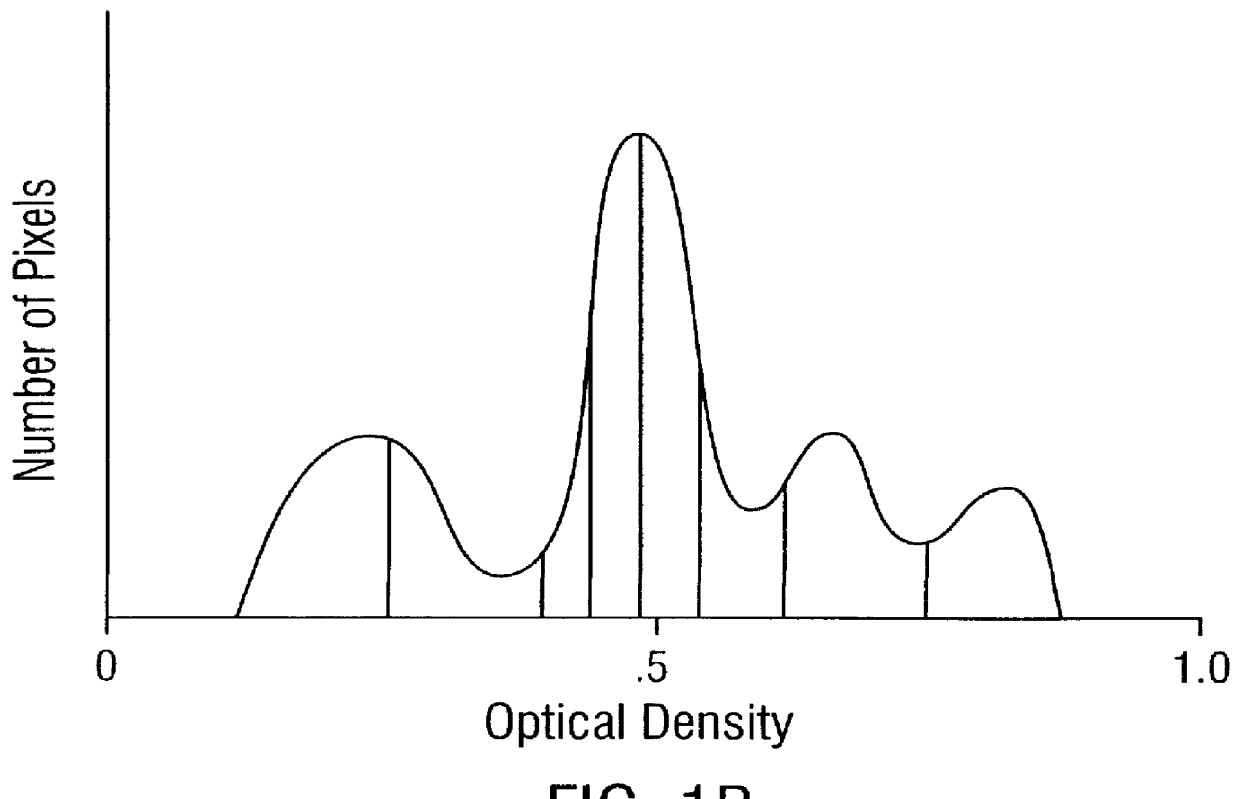
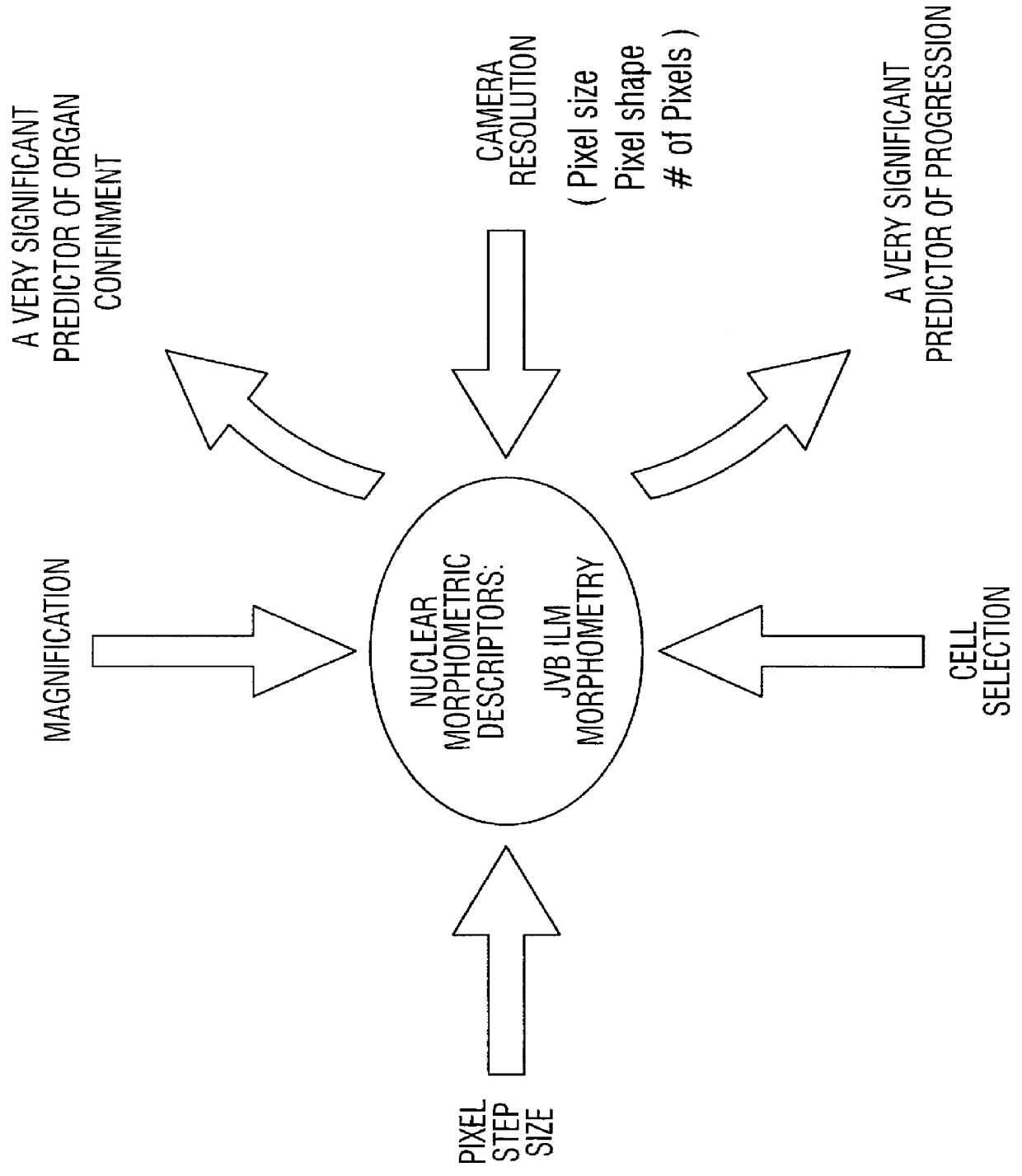
Prediction of prostate cancer progression by analysis of selected predictive parameters
InactiveUS6025128AImprove performanceMicrobiological testing/measurementBiological testingStatistical analysisFactor ii
A method for screening individuals at risk for prostate cancer progression is disclosed. The method is useful for evaluating cells from patients at risk for recurrence of prostate cancer following surgery for prostate cancer. Specifically, the method uses specific Markovian nuclear texture factors, alone or in combination with other biomarkers, to determine whether the cancer will progress or lose organ confinement. In addition, methods of predicting the development of fatal metastatic disease by statistical analysis of selected biomarkers is also disclosed. The invention also contemplates a method that uses a neural network to analyze and interpret cell morphology data. Utilizing Markovian factors and other biomarkers as parameters, the network is first trained with a sets of cell data from known progressors and known non-progressors. The trained network is then used to predict prostate cancer progression in patient samples.
Owner:CYTODIAGNOSTICS +3
Process for discriminating between biological states based on hidden patterns from biological data
The invention describes a process for determining a biological state through the discovery and analysis of hidden or non-obvious, discriminatory biological data patterns. The biological data can be from health data, clinical data, or from a biological sample, (e.g., a biological sample from a human, e.g., serum, blood, saliva, plasma, nipple aspirants, synovial fluids, cerebrospinal fluids, sweat, urine, fecal matter, tears, bronchial lavage, swabbings, needle aspirantas, semen, vaginal fluids, pre-ejaculate.), etc. which is analyzed to determine the biological state of the donor. The biological state can be a pathologic diagnosis, toxicity state, efficacy of a drug, prognosis of a disease, etc. Specifically, the invention concerns processes that discover hidden discriminatory biological data patterns (e.g., patterns of protein expression in a serum sample that classify the biological state of an organ) that describe biological states.
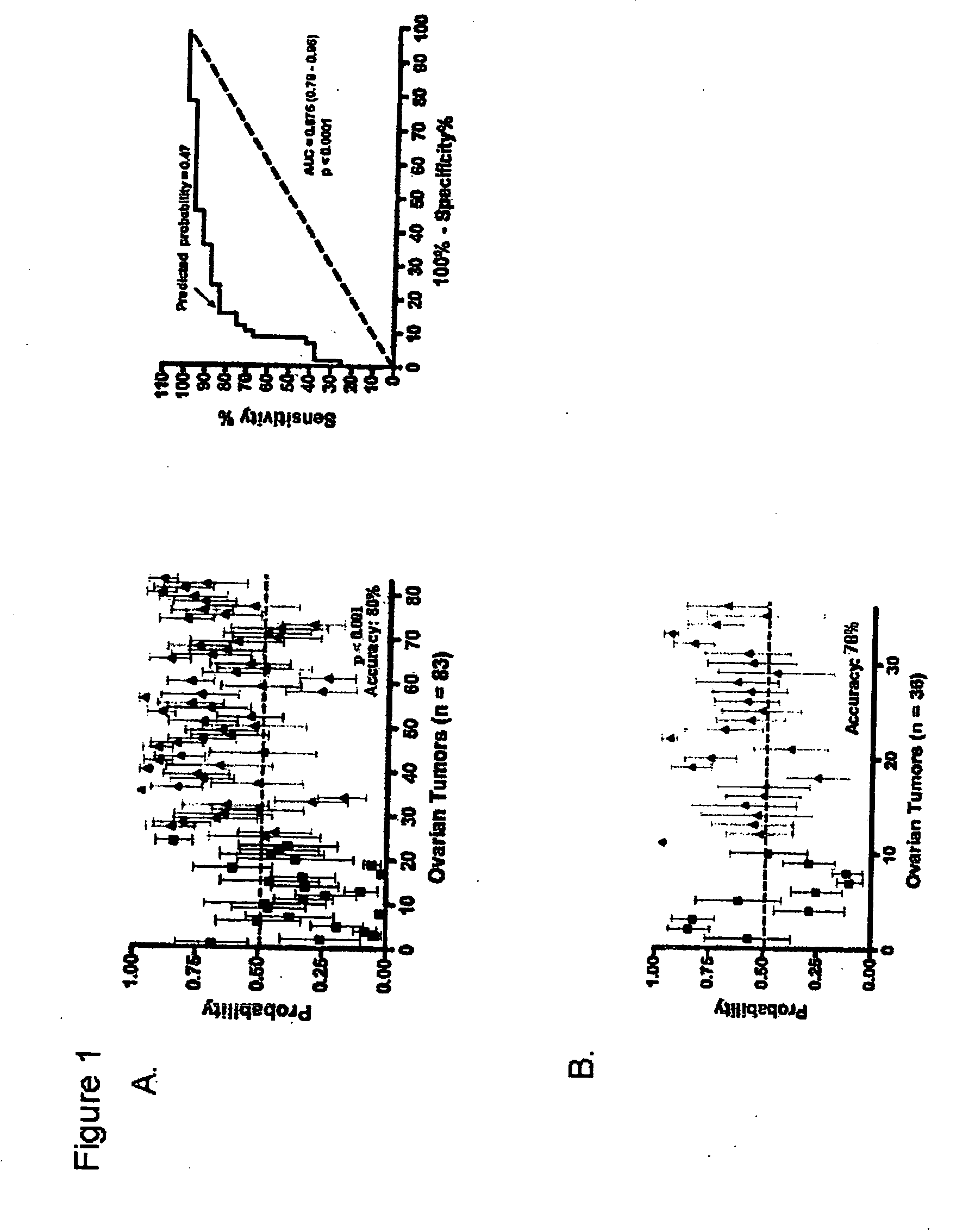
Owner:ASPIRA WOMENS HEALTH INC +1
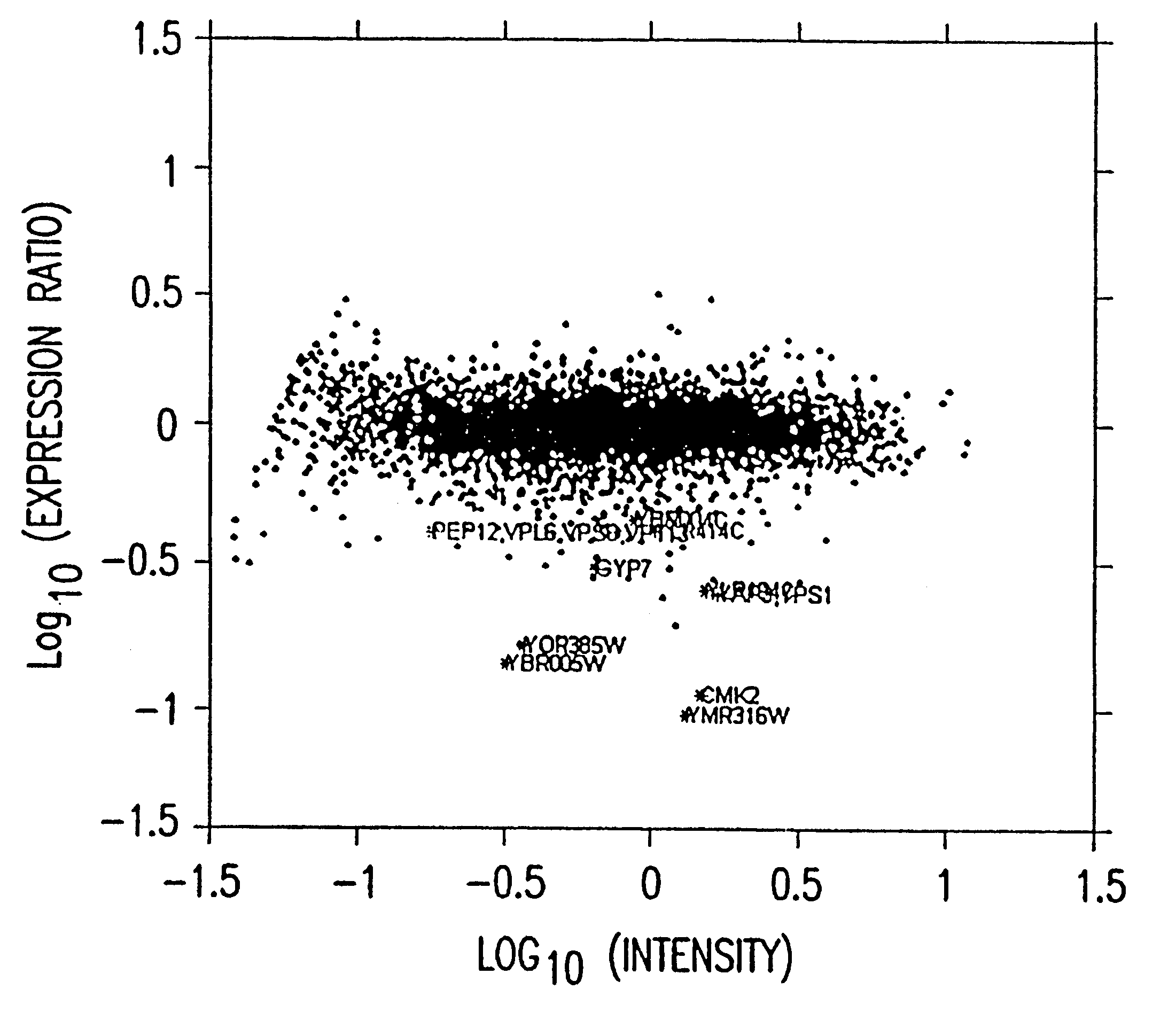
Statistical combining of cell expression profiles
InactiveUS6351712B1Minimize repetitionEliminate errorsMicrobiological testing/measurementRecombinant DNA-technologyFluorophoreErrors and residuals
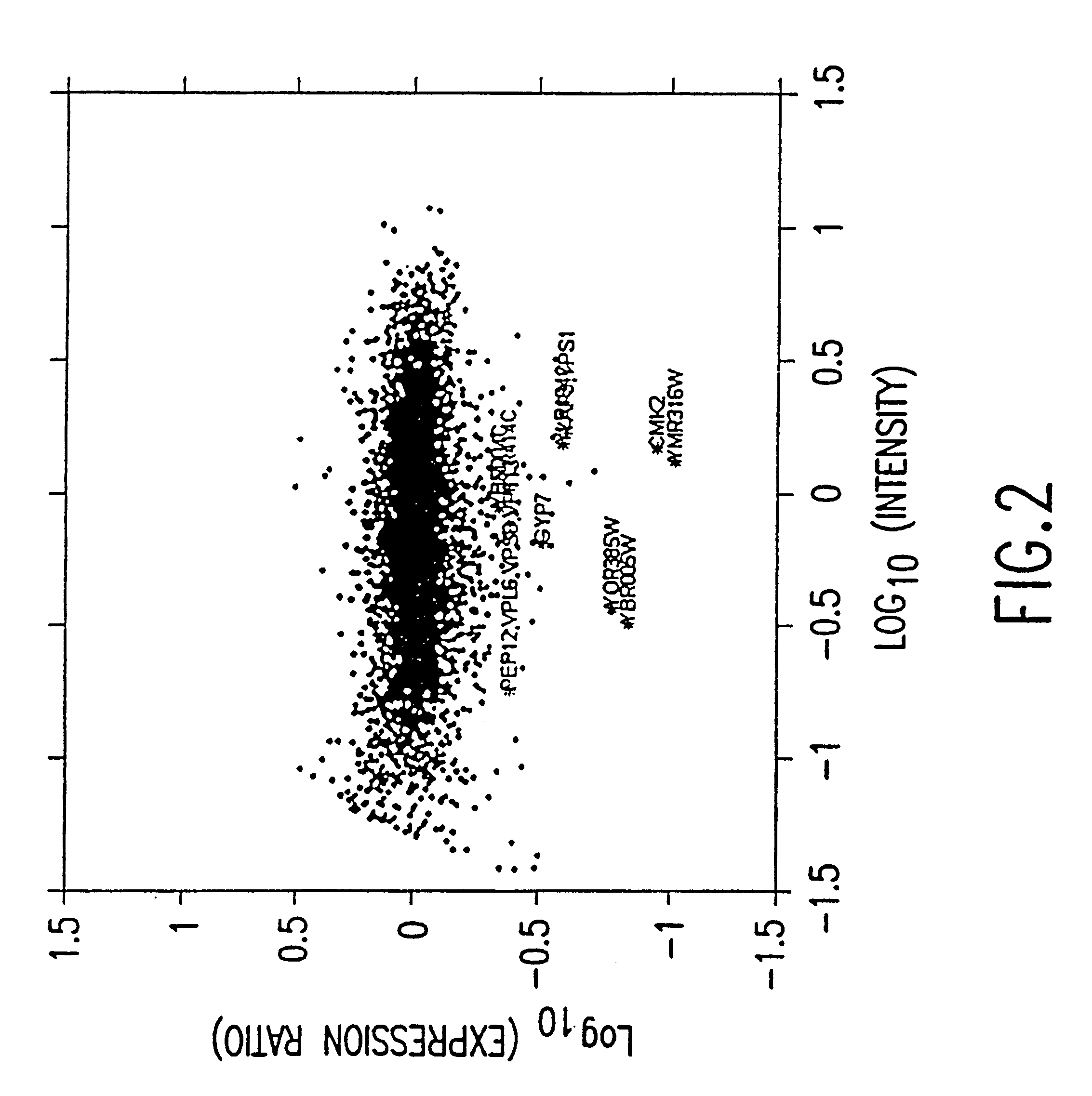
A method for fluorophore bias removal in microarray experiments in which the fluorophores used in microarray experiment pairs are reversed. Further, a method for calculating the individual errors associated with each measurement made in nominally repeated microarray experiments. This error measurement is optionally coupled with rank based methods in order to determine a probability that a cellular constituent is up or down regulated in response to a perturbation. Finally, a method for determining the confidence in the weighted average of the expression level of a cellular constituent in nominally repeated microarray experiments.
Owner:MICROSOFT TECH LICENSING LLC
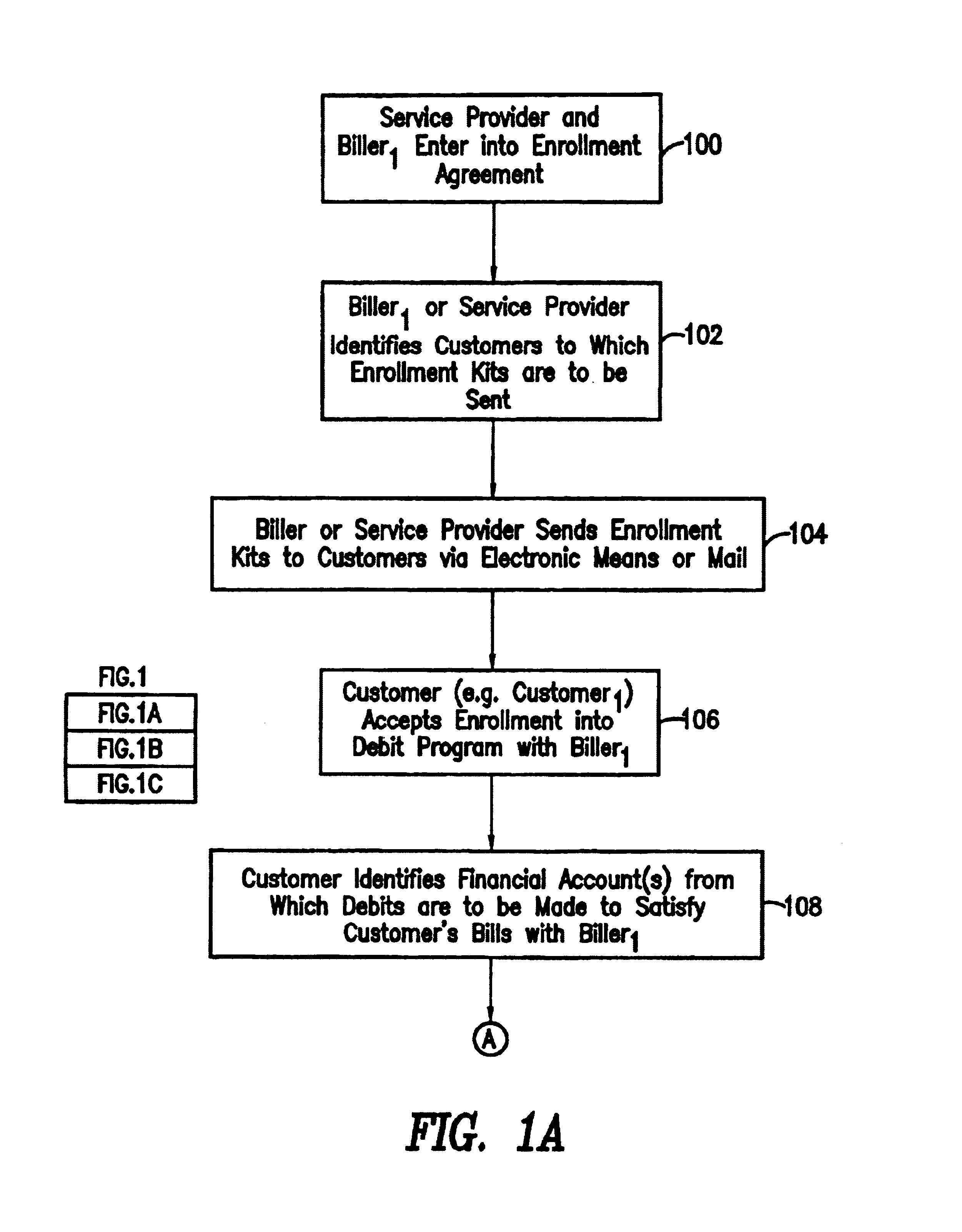
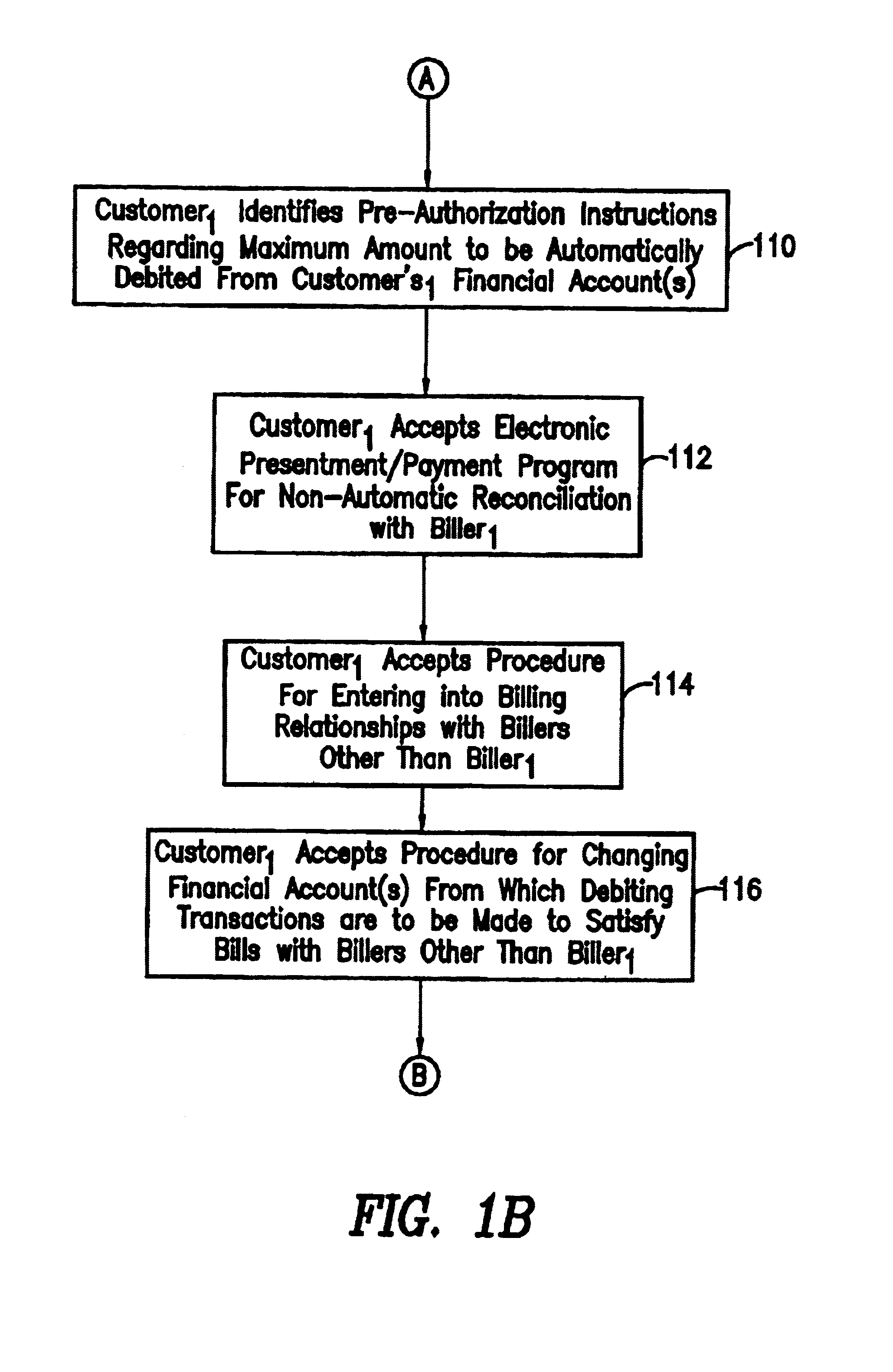
System and method for back office processing of banking transactions using electronic files
InactiveUS7062456B1Reduce equipmentReduce laborFinanceAutomatic teller machinesBank tellerRelevant information
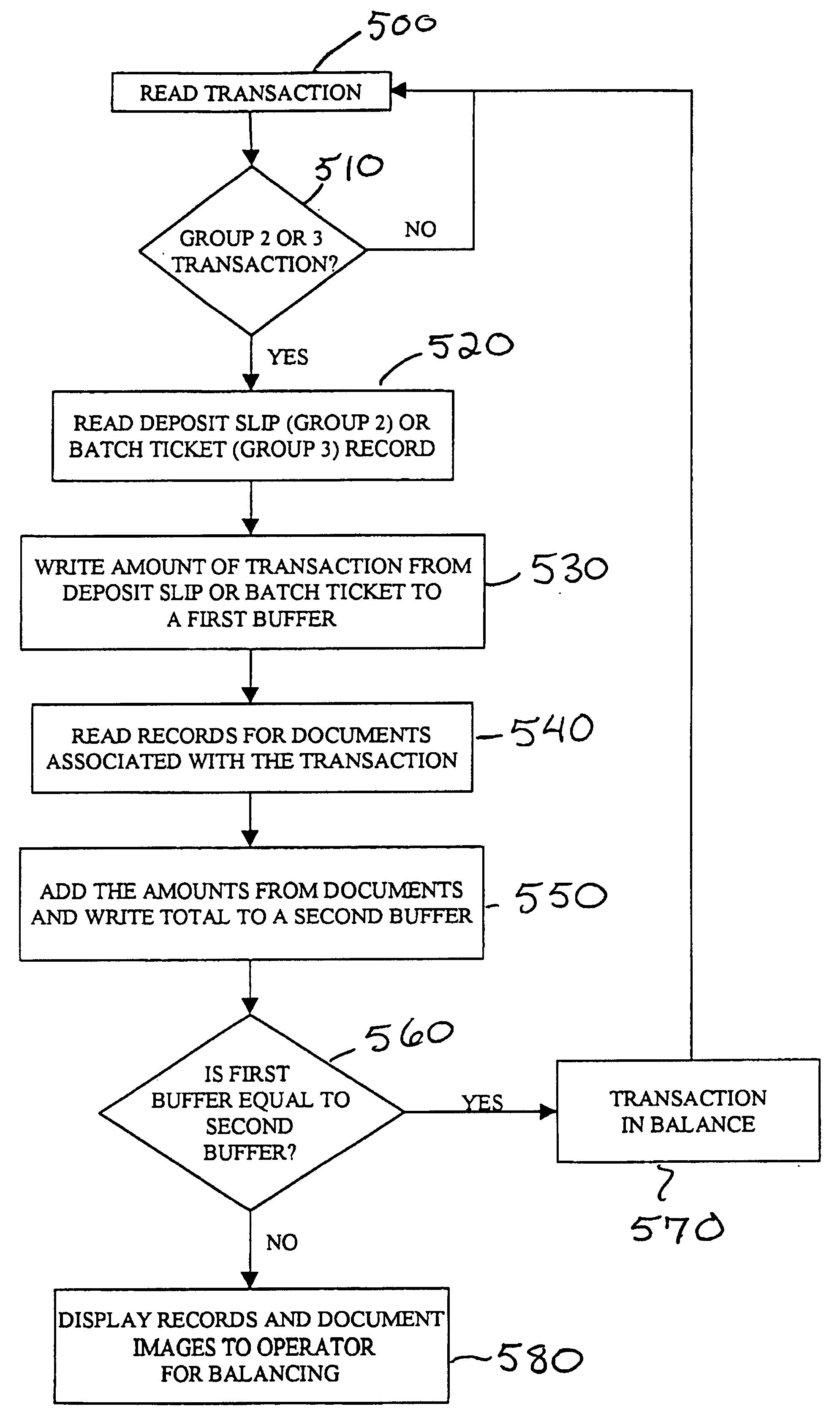
As banking transactions are processed by a bank teller, all of the relevant information with respect to the transaction (e.g., dollar amount) is captured in an electronic file. Each of the electronic files from the various branches of the bank are forwarded to a central back office processing center where the electronic files are combined into a single Transaction Repository. At the end of the branch day, all of the paper associated with the transactions is forwarded from the branches to the back office processing center. The paper transactions are imaged in the conventional manner and the Magnetic Ink Character Recognition (MICR) data is read from the paper. The present invention then automatically correlates the images and MICR data captured from the paper with the complete transaction record contained in the Transaction Repository. Most of the conventional back office processing can now be performed without the need to perform character recognition and without the need for excess human intervention.
Owner:JPMORGAN CHASE BANK NA
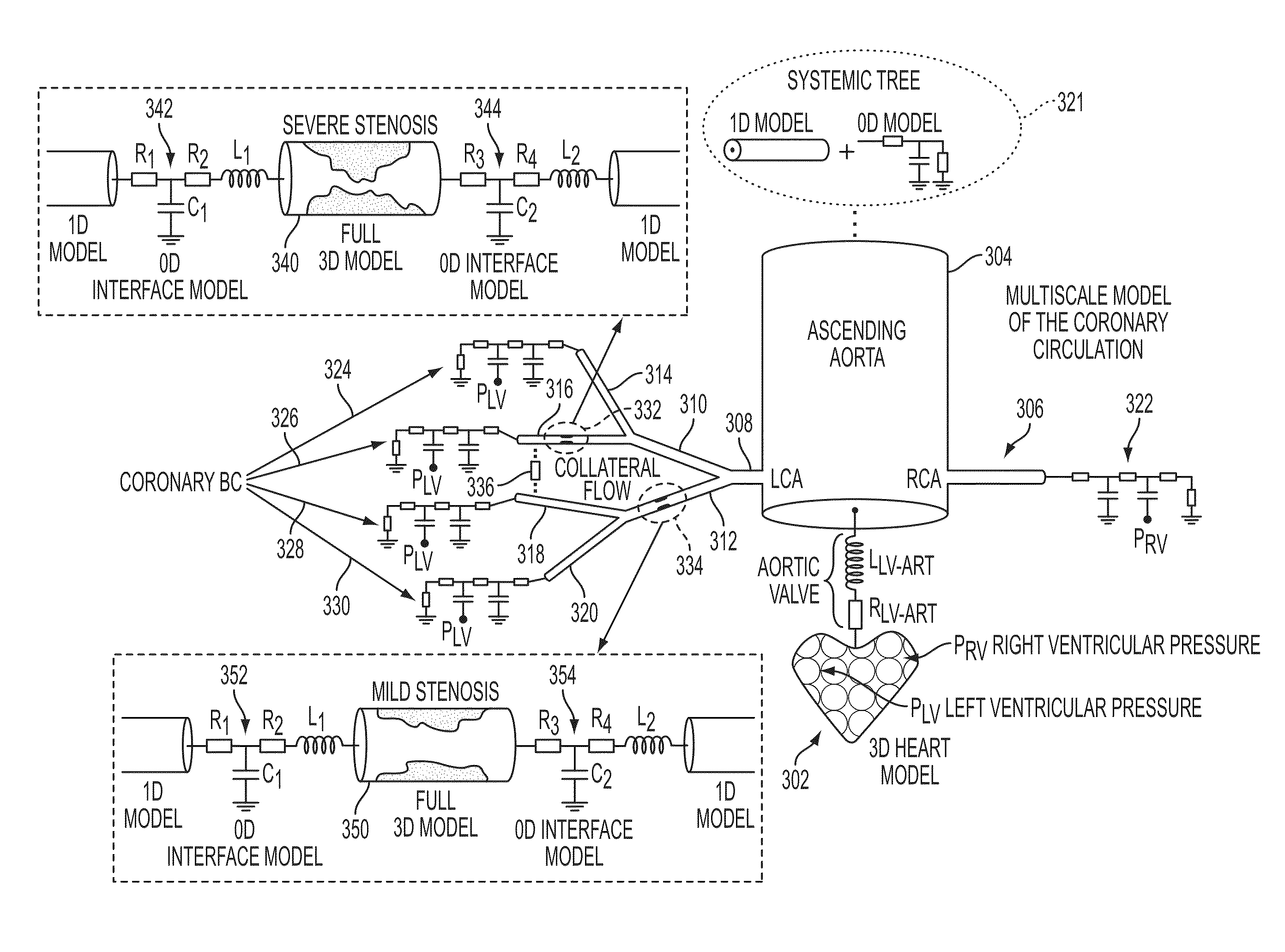
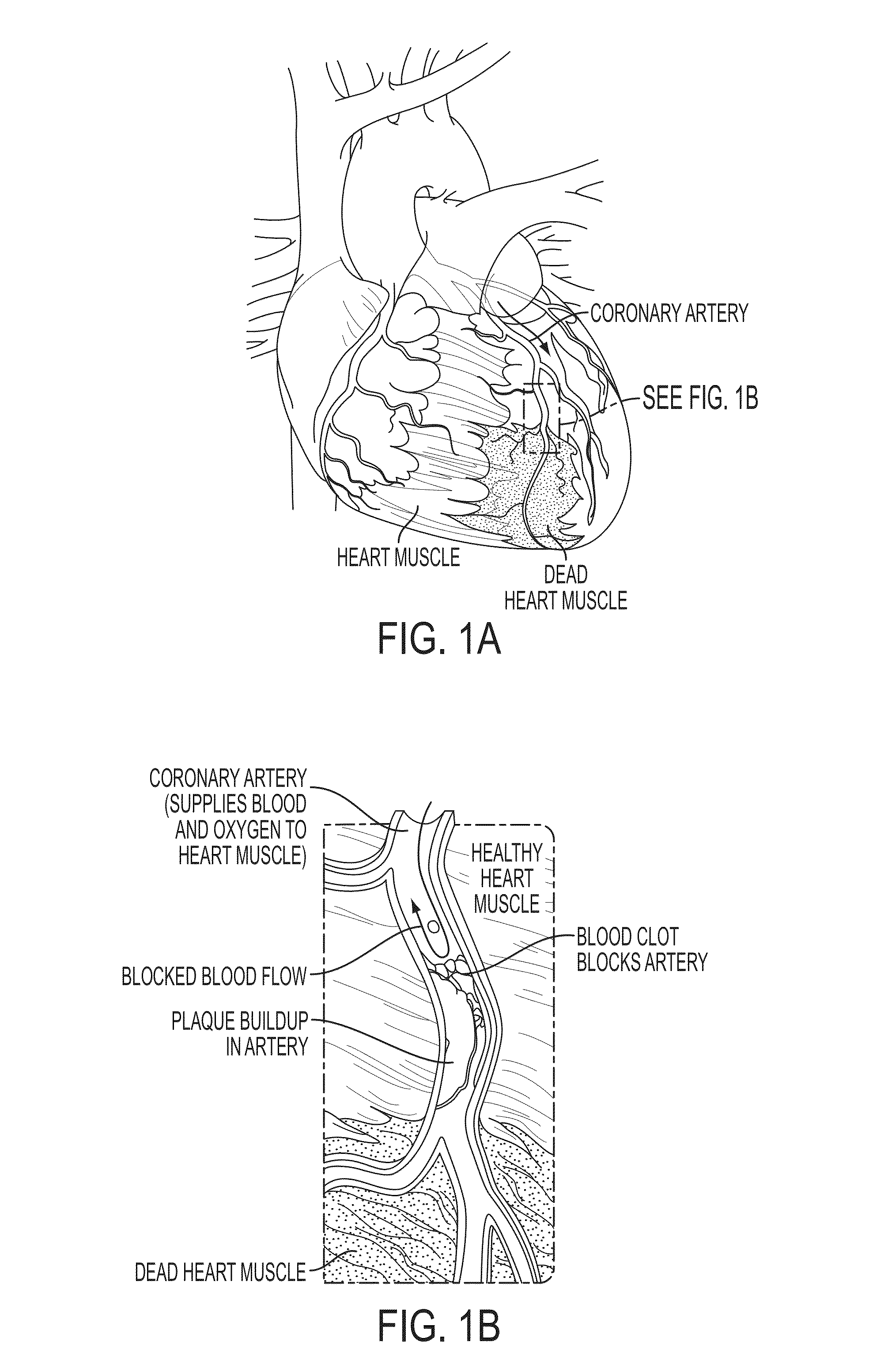
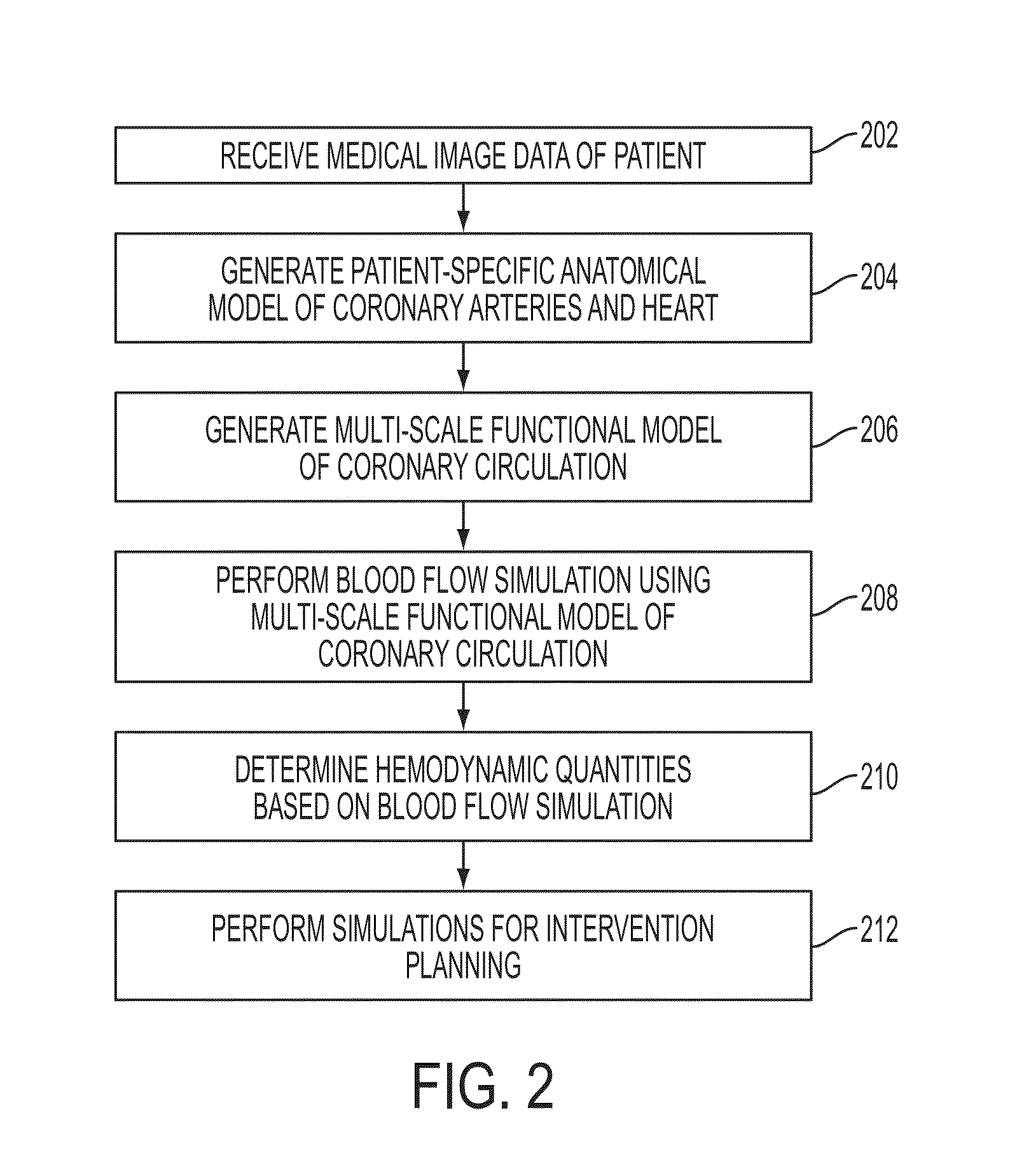
Method and System for Multi-Scale Anatomical and Functional Modeling of Coronary Circulation
ActiveUS20130132054A1Improve predictive performanceImprove clinical managementChemical property predictionChemical structure searchCoronary arteriesIntervention planning
A method and system for multi-scale anatomical and functional modeling of coronary circulation is disclosed. A patient-specific anatomical model of coronary arteries and the heart is generated from medical image data of a patient. A multi-scale functional model of coronary circulation is generated based on the patient-specific anatomical model. Blood flow is simulated in at least one stenosis region of at least one coronary artery using the multi-scale function model of coronary circulation. Hemodynamic quantities, such as fractional flow reserve (FFR), are computed to determine a functional assessment of the stenosis, and virtual intervention simulations are performed using the multi-scale function model of coronary circulation for decision support and intervention planning.
Owner:SIEMENS HEALTHCARE GMBH +1
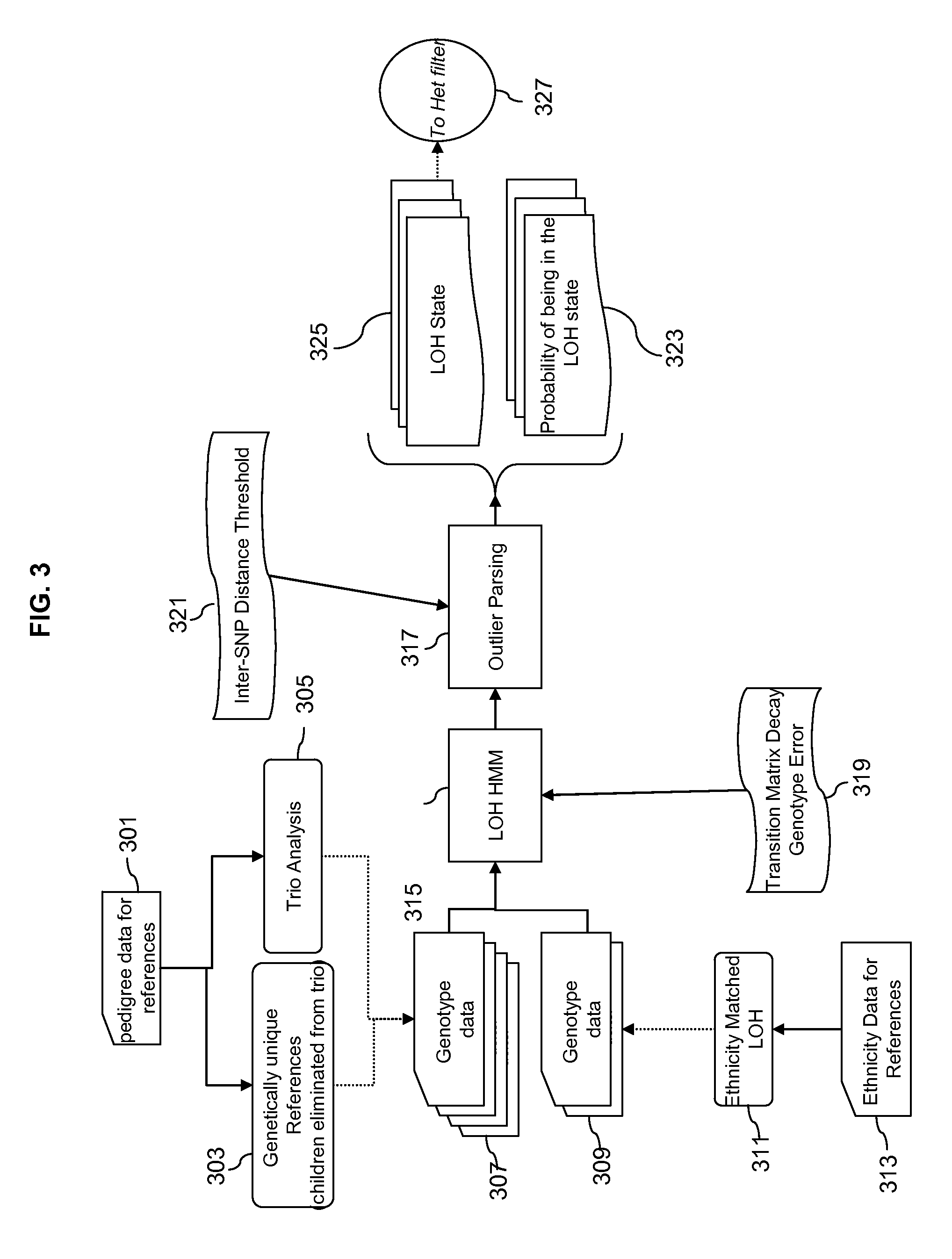
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
ActiveUS20070184467A1Significant resultMicrobiological testing/measurementProteomicsUniparental disomyEmbryo
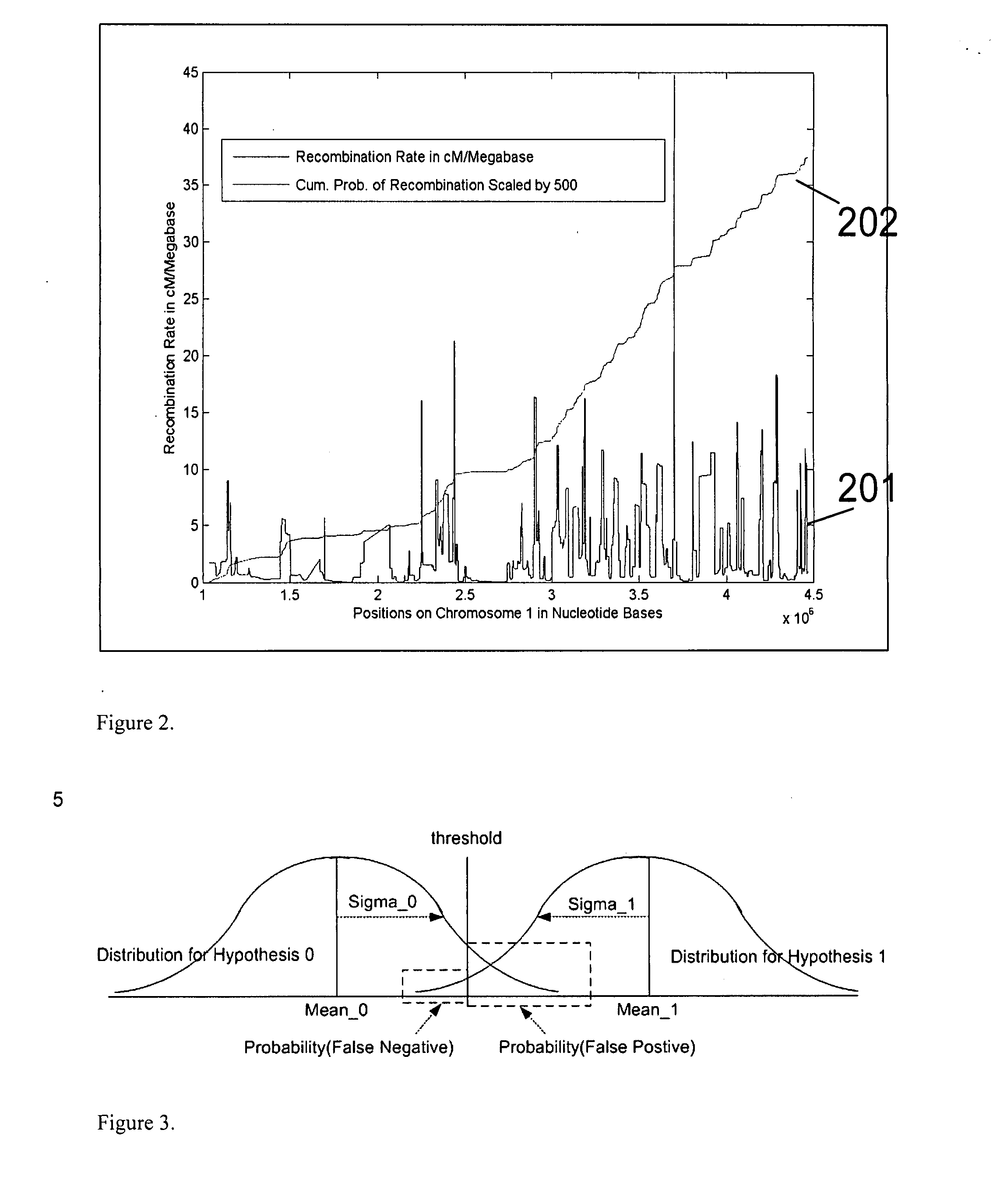
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell is reconstructed using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In accordance with another embodiment of the invention, incomplete genetic data from a fetus is acquired from fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In one embodiment, the genetic data can be reconstructed for the purposes of making phenotypic predictions. In another embodiment, the genetic data can be used to detect for aneuploides and uniparental disomy.
Owner:NATERA
Individualized cancer treatments
InactiveUS20070172844A1Increase probabilityLow toxicityBioreactor/fermenter combinationsBiological substance pretreatmentsDNA microarrayPlatinum
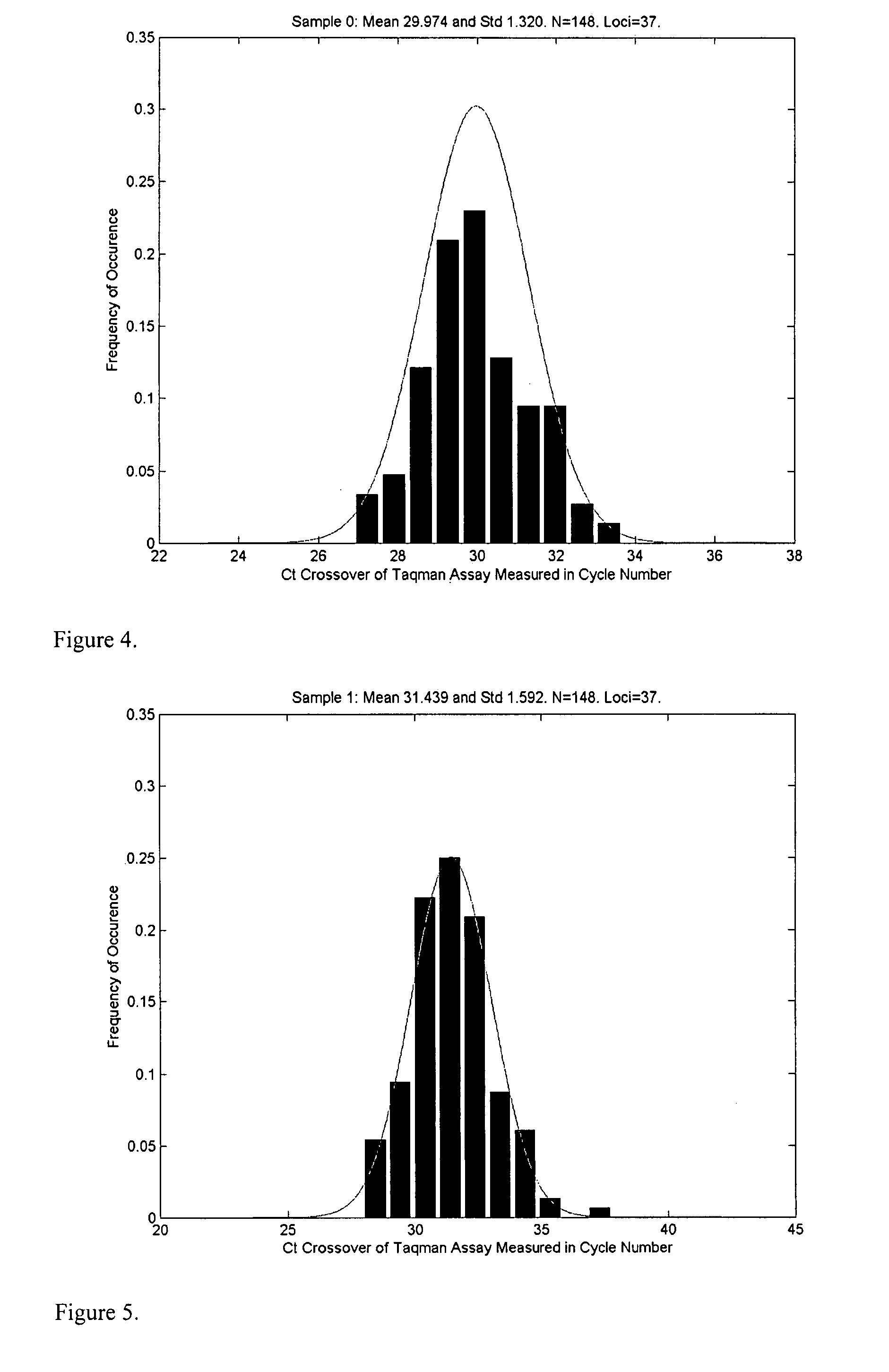
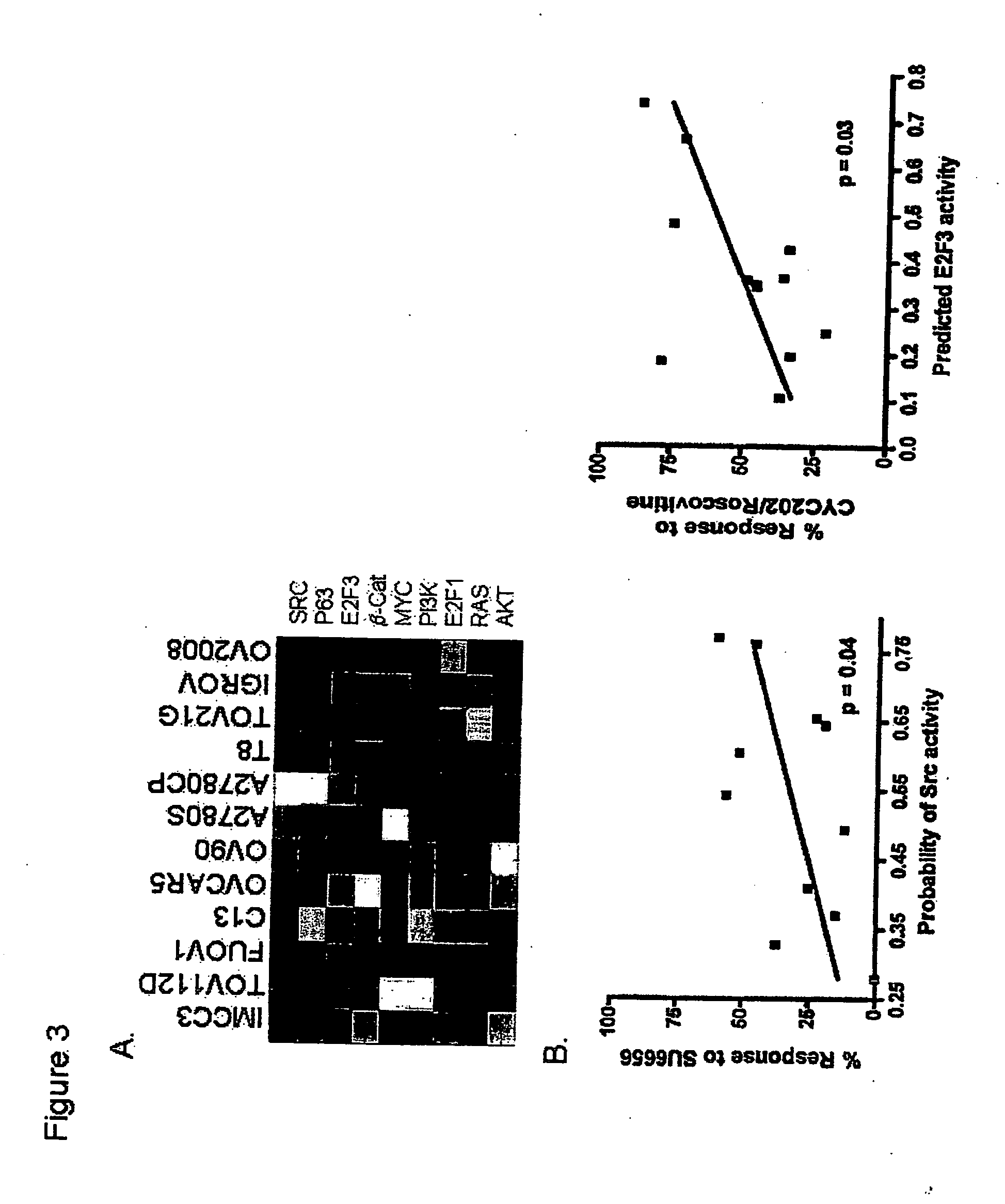
The invention provides for compositions and methods for predicting an individual's responsitivity to cancer treatments and methods of treating cancer. In certain embodiments, the invention provides compositions and methods for predicting an individual's responsitivity to chemotherapeutics, including platinum-based chemotherapeutics, to treat cancers such as ovarian cancer. Furthermore, the invention provides for compositions and methods for predicting an individual's responsivity to salvage therapeutic agents. By predicting if an individual will or will not respond to platinum-based chemotherapeutics, a physician can reduce side effects and toxicity by administering a particular additional salvage therapeutic agent. This type of personalized medical treatment for ovarian cancer allows for more efficient treatment of individuals suffering from ovarian cancer. The invention also provides reagents, such as DNA microarrays, software and computer systems useful for personalizing cancer treatments, and provides methods of conducting a diagnostic business for personalizing cancer treatments.
Owner:UNIV OF SOUTH FLORIDA +1
Random array of micro-spheres for the analysis of nucleic acids
InactiveUS7011945B2Easy to analyzeMore cost-effectivelyBioreactor/fermenter combinationsSequential/parallel process reactionsRandom arrayBarcode
A method of identifying nucleic acid samples comprising: providing a mircoarray including a substrate coated with a composition including a population of micro-spheres dispersed in a fluid containing a gelling agent or a precursor to a gelling agent and immobilized at random positions on the substrate, at least one sub-population of said population micro-spheres containing an optical barcode generated from at least one colorant associated with the micro-spheres and including a nucleic acid probe sequence; contacting said array with a target nucleic acid sequence; and detecting the color barcode of said sub-population of micro-spheres due to the interaction of said probe nucleic acid sequence and said target nucleic acid sequence.
Owner:CARESTREAM HEALTH INC
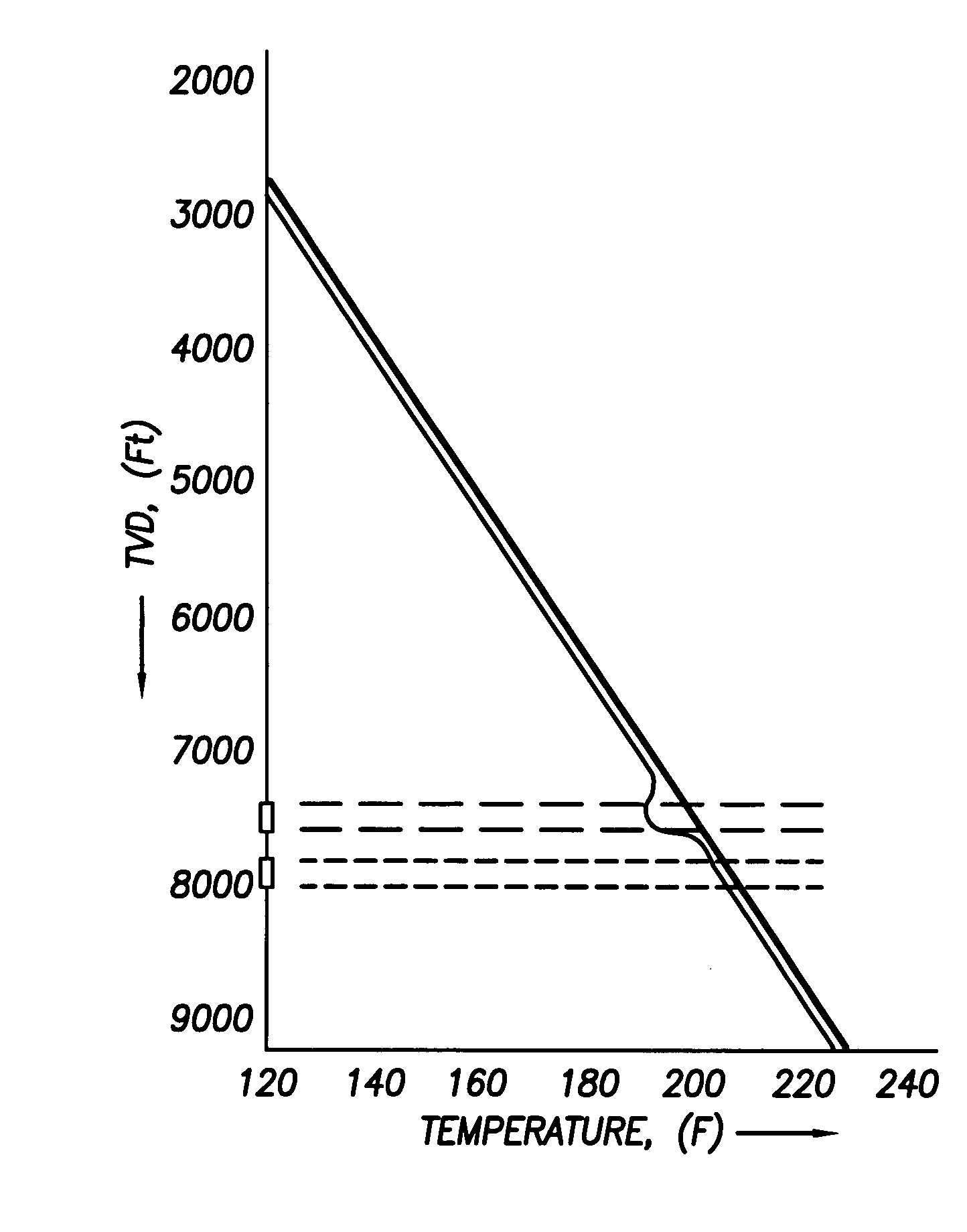
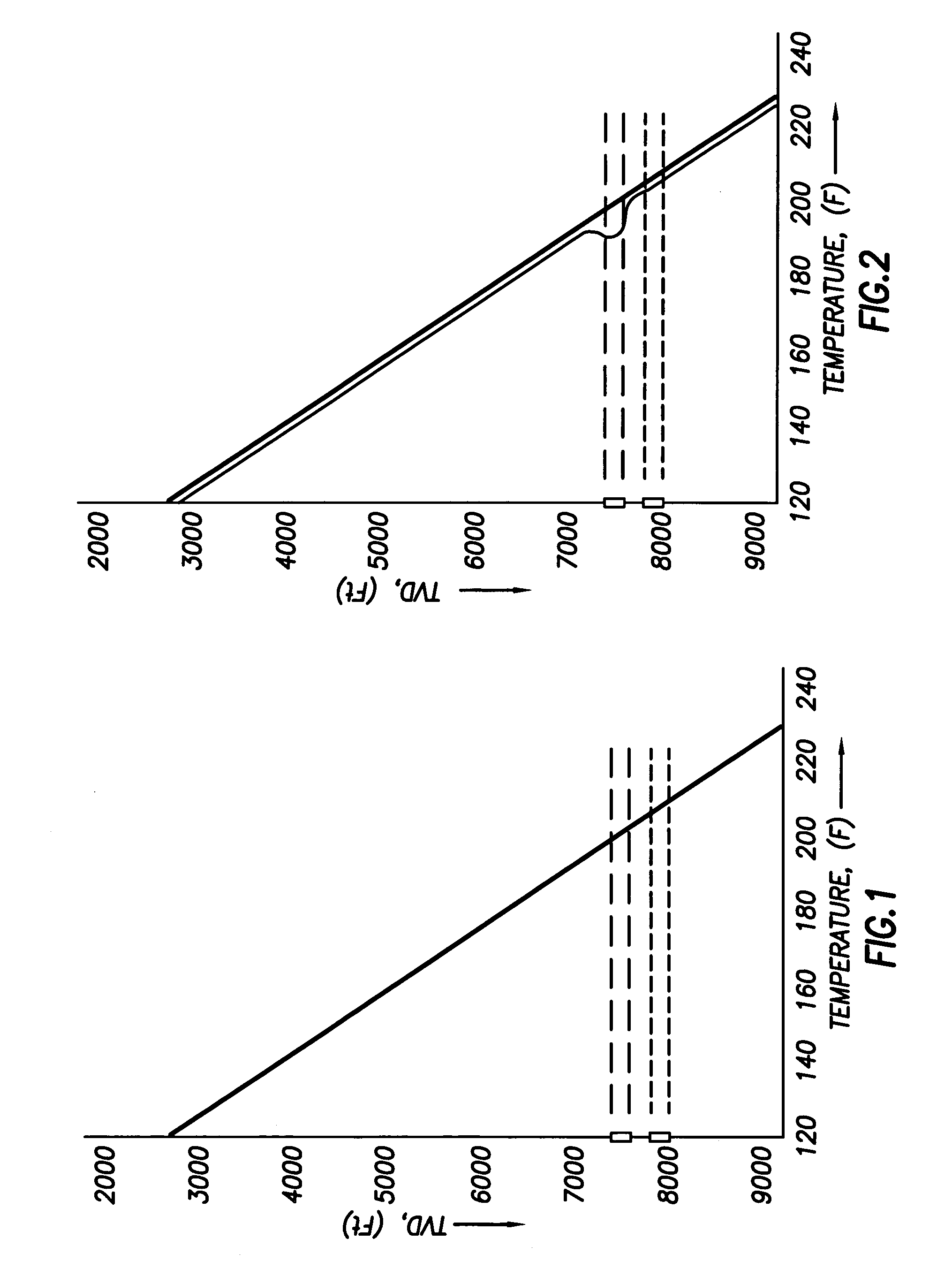
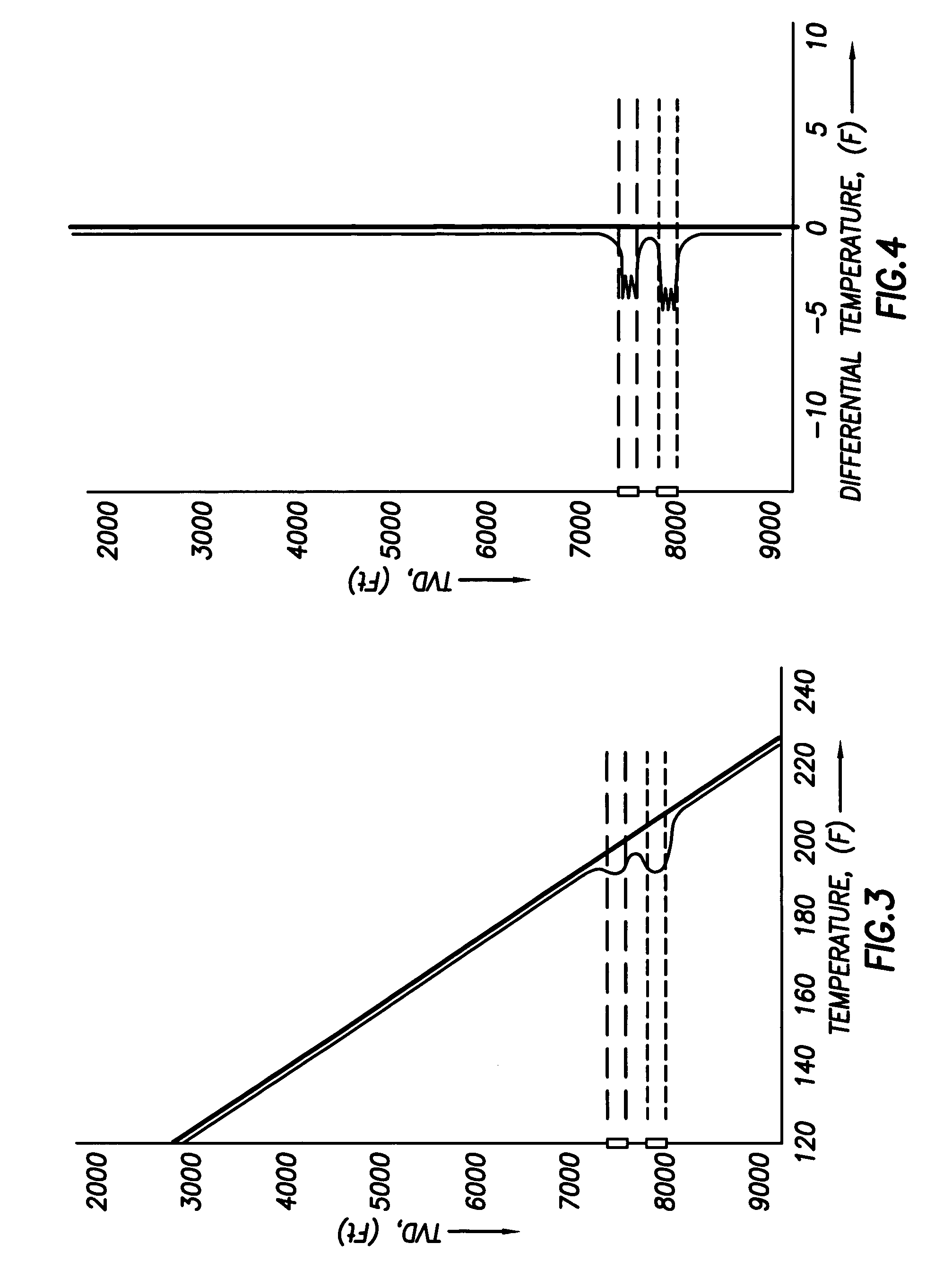
Use of distributed temperature sensors during wellbore treatments
The invention relates to a method for treating subterranean formation comprising providing distributed temperature sensors, injecting a treatment fluid and monitoring the temperature across the treatment interval during the injection process.
Owner:SCHLUMBERGER TECH CORP
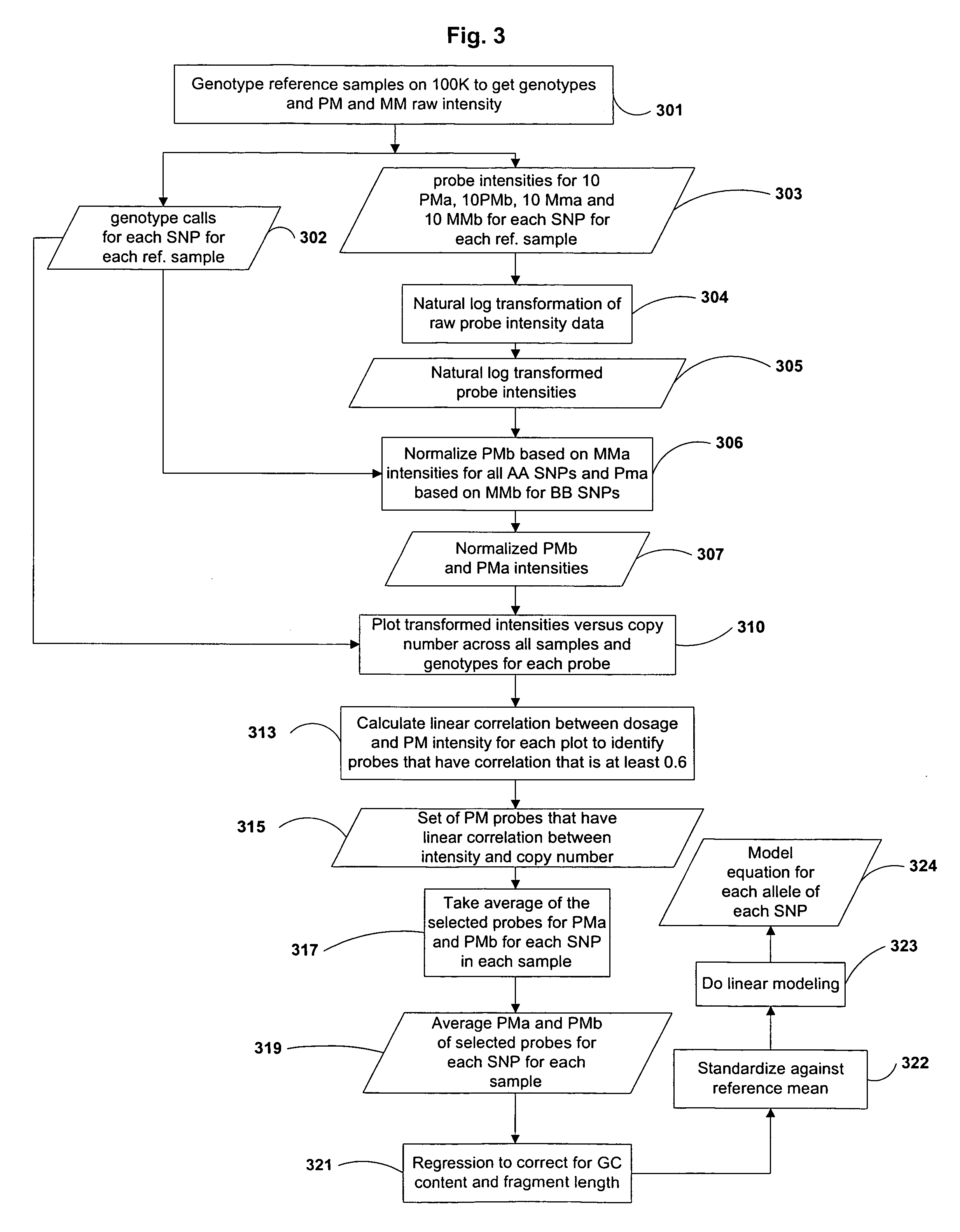
Varietal counting of nucleic acids for obtaining genomic copy number information
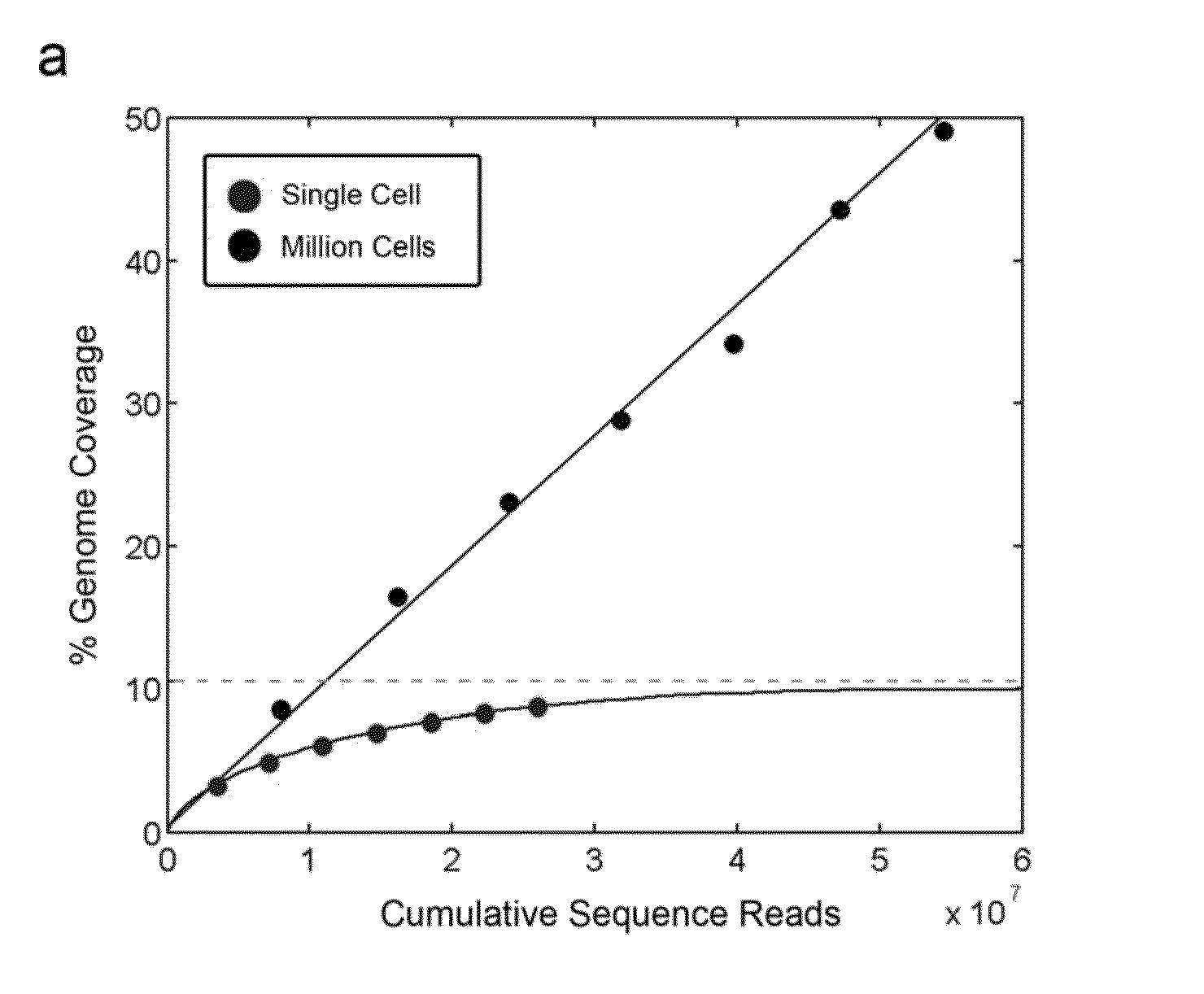
A method for obtaining from genomic material genomic copy number information unaffected by amplification distortion, comprising obtaining segments of the genomic material, tagging the segments with substantially unique tags to generate tagged nucleic acid molecules, such that each tagged nucleic acid molecule comprises one segment of the genomic material and a tag, subjecting the tagged nucleic acid molecules to polymerase chain reaction (PCR) amplification, generating tag associated sequence reads by sequencing the product of the PCR reaction, assigning each tagged nucleic acid molecule to a location on a genome associated with the genomic material by mapping the subsequence of each tag associated sequence read corresponding to a segment of the genomic material to a location on the genome, and counting the number of tagged nucleic acid molecules assigned to the same location on the genome having a different tag, thereby obtaining genomic copy number information unaffected by amplification distortion.
Owner:COLD SPRING HARBOR LAB INC
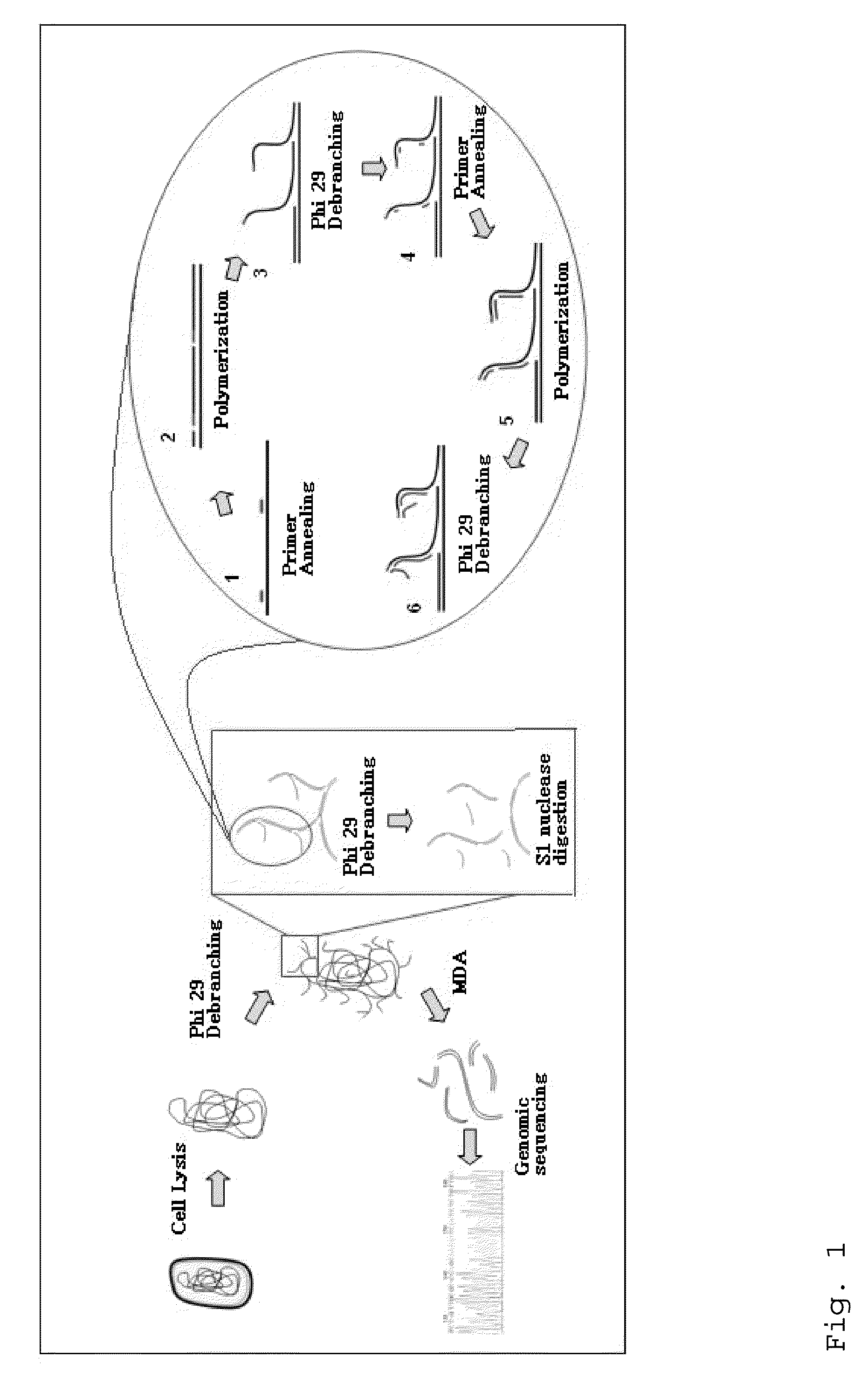
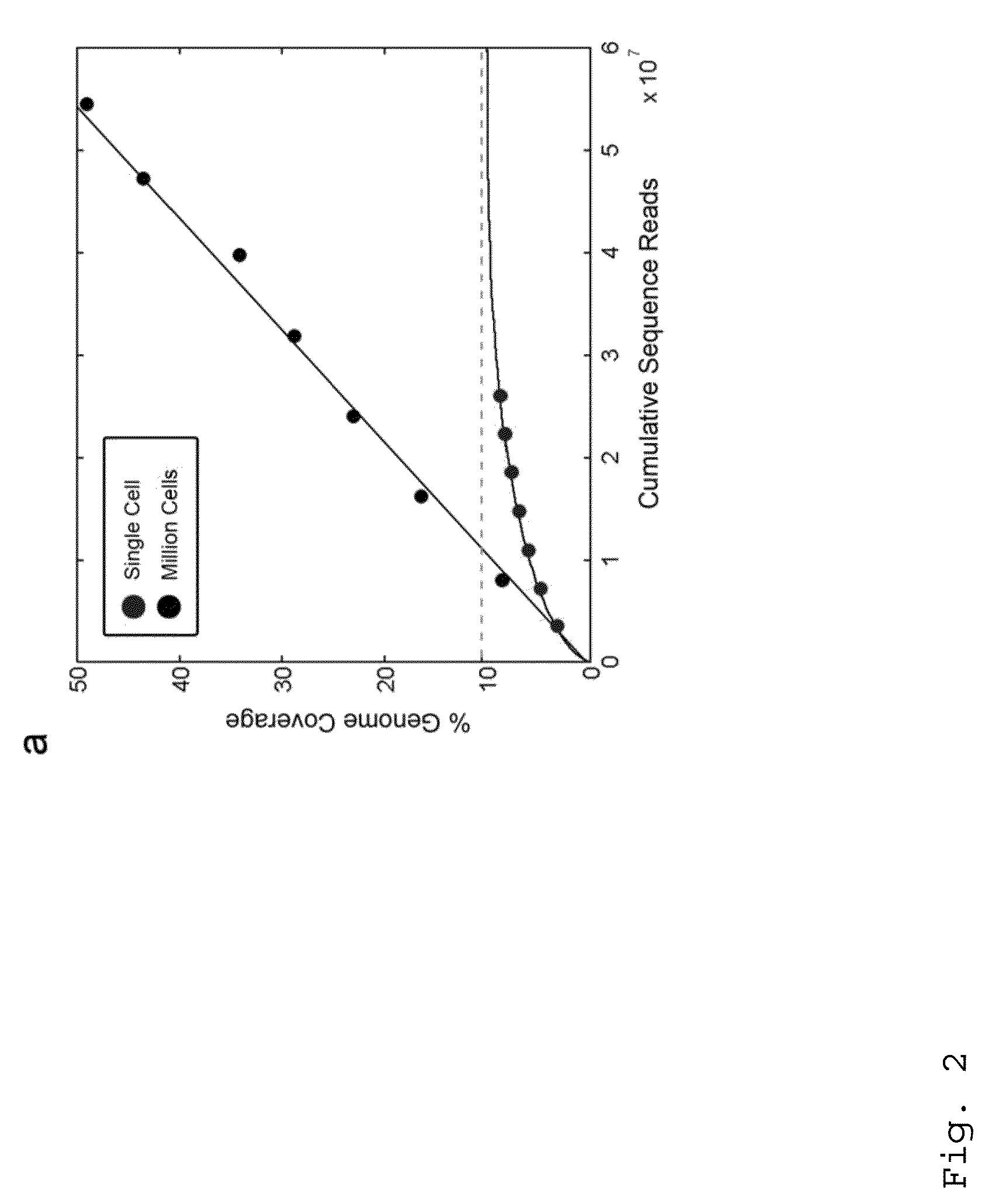
Lung cancer prognostics
InactiveUS20060252057A1Bioreactor/fermenter combinationsBiological substance pretreatmentsOncologyLung cancer
Owner:VERIDEX LCC
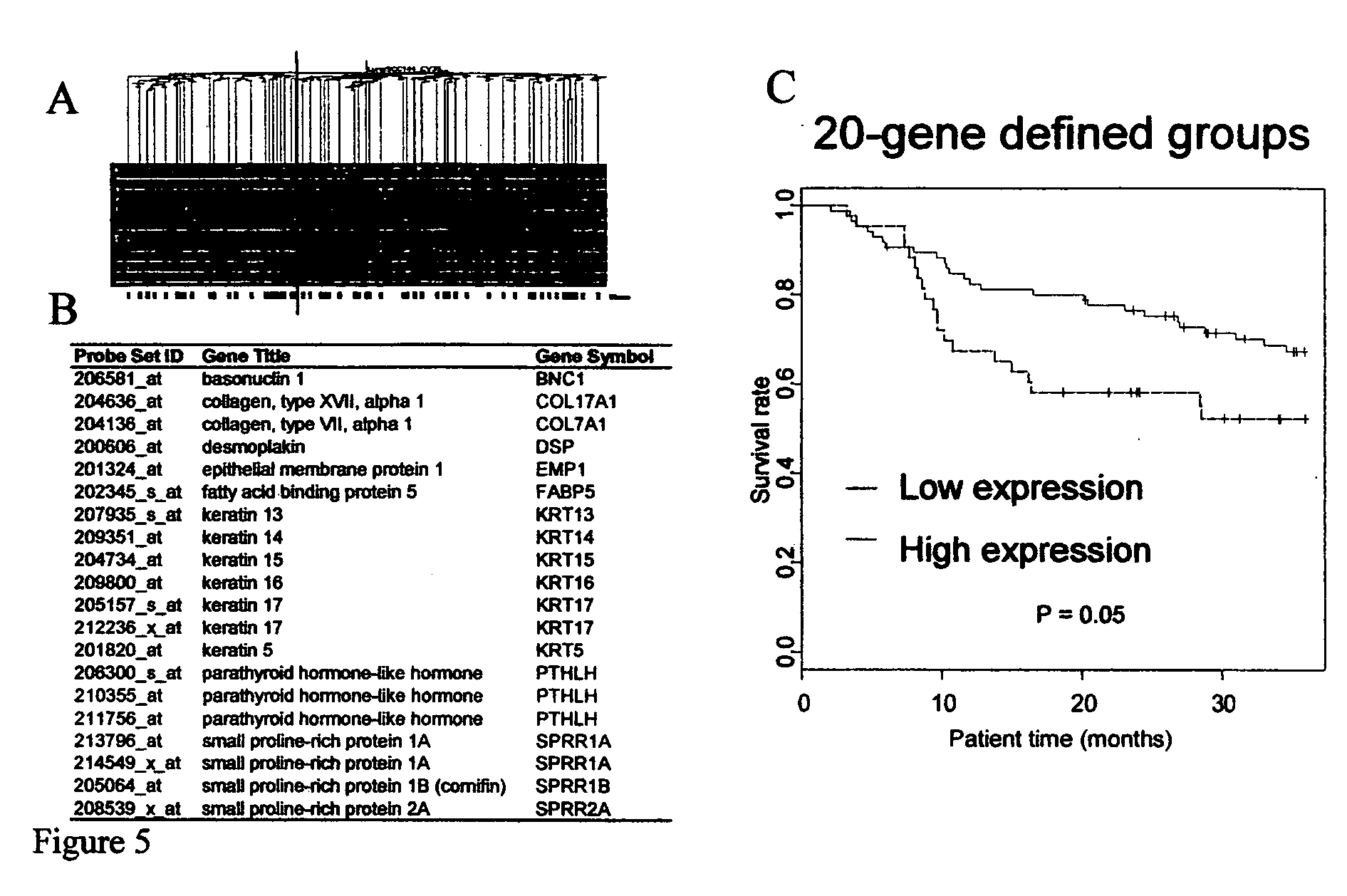
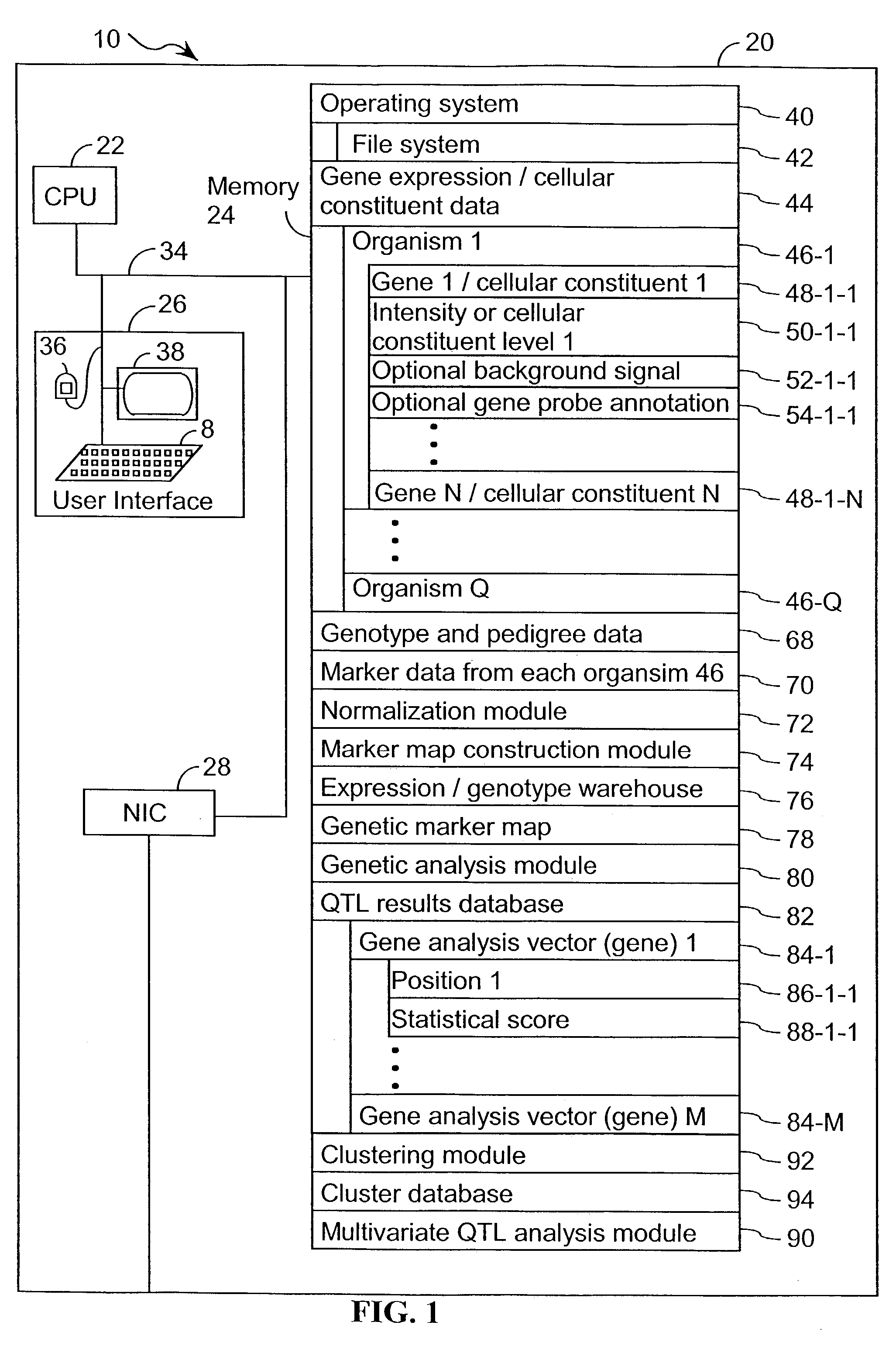
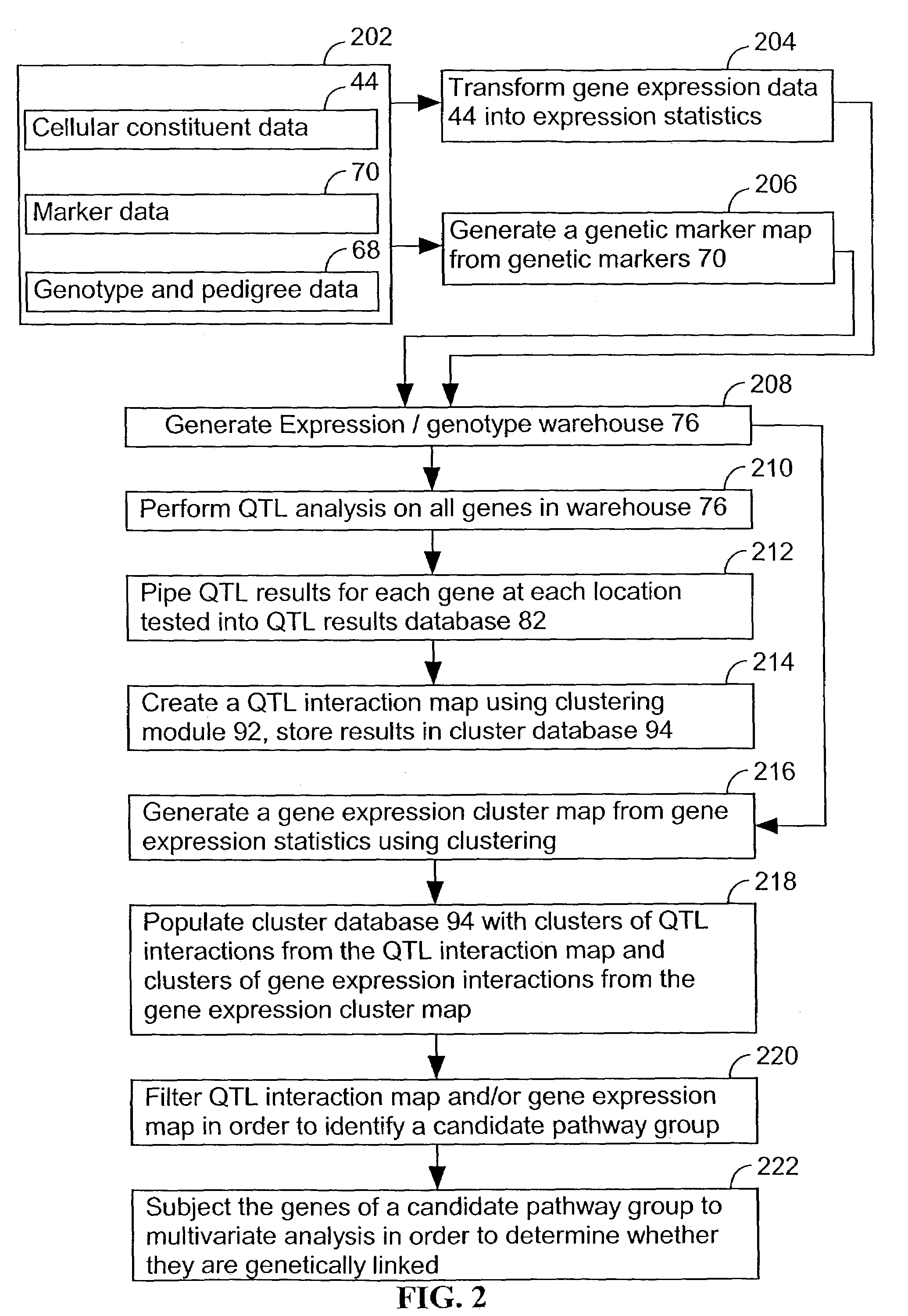
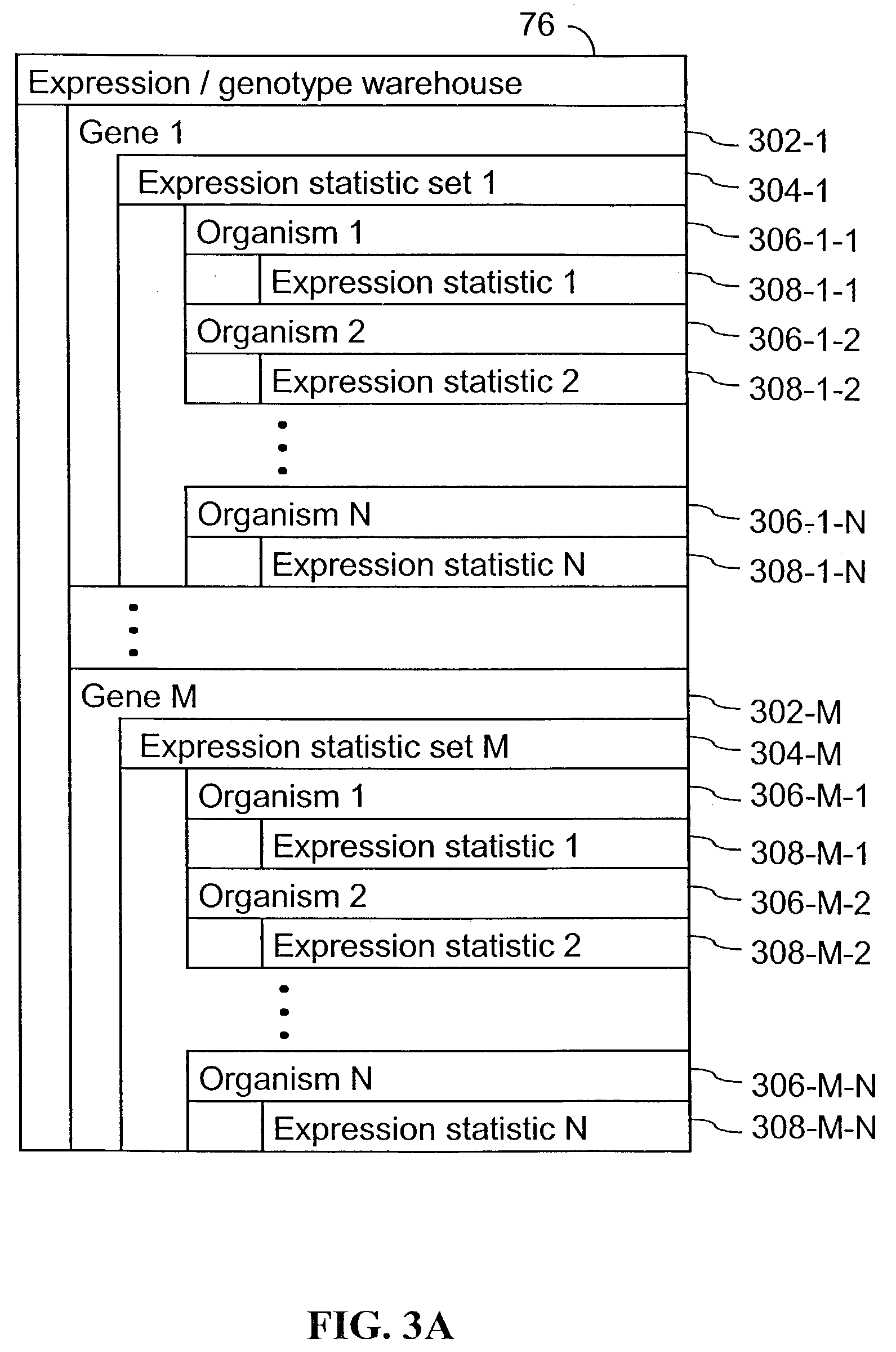
Computer systems and methods for identifying genes and determining pathways associated with traits
ActiveUS7035739B2Bioreactor/fermenter combinationsBiological substance pretreatmentsBiological bodyQuantitative trait locus
Owner:ROSETTA INPHARMATICS LLC
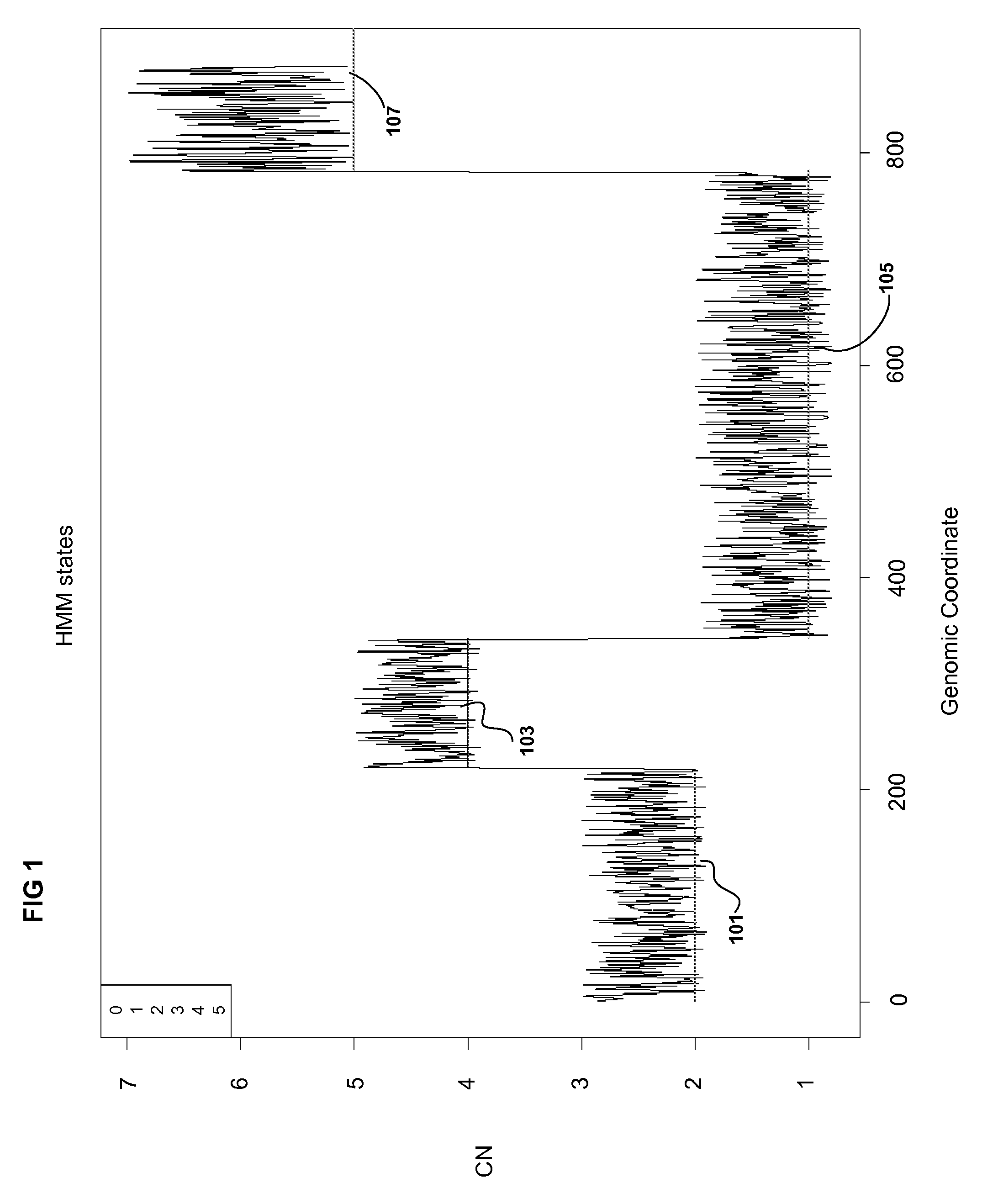
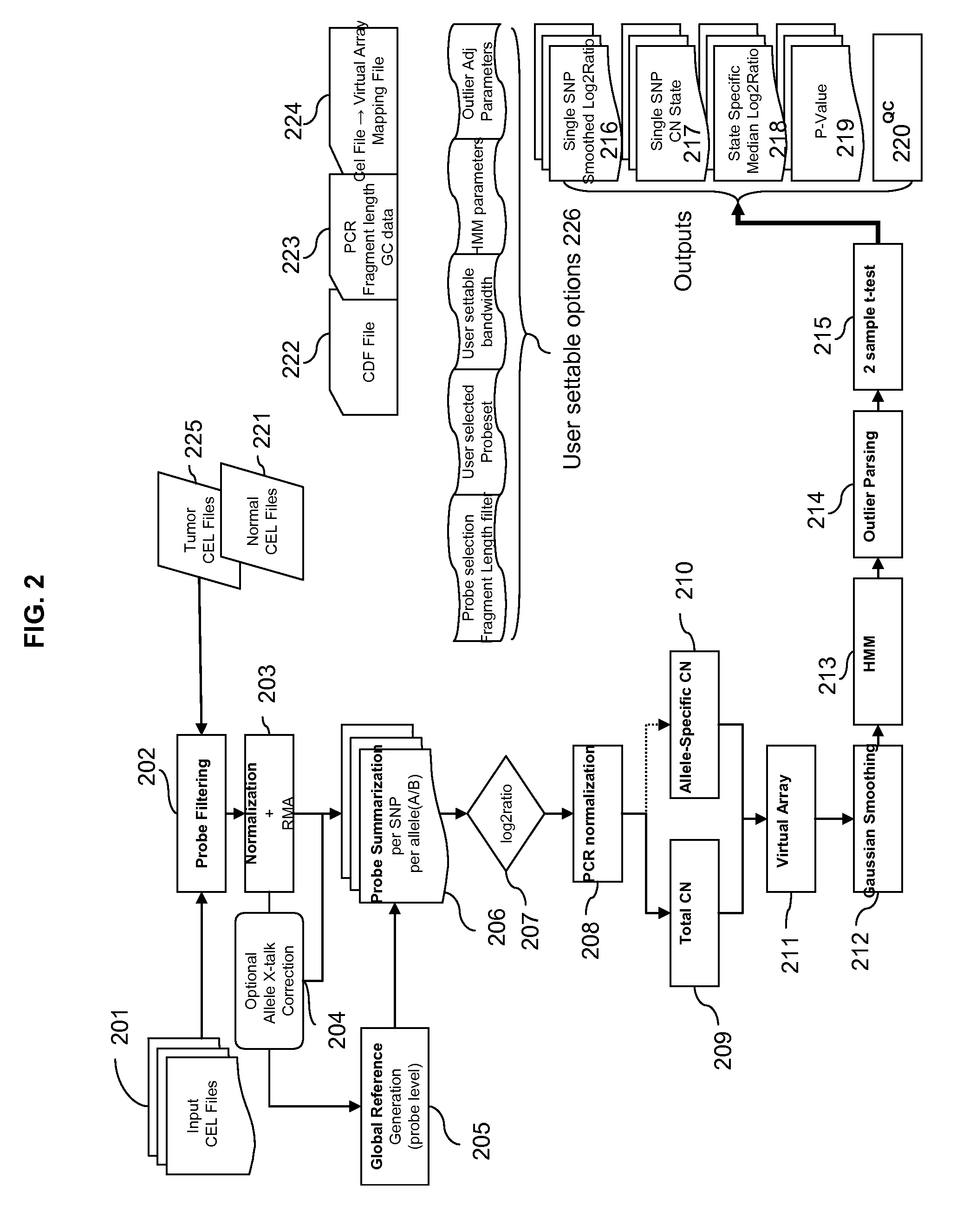
Methods for Identifying DNA Copy Number Changes Using Hidden Markov Model Based Estimations
Methods for estimating genomic copy number and loss of heterozygosity using Hidden Markov Model based estimation are disclosed.
Owner:AFFYMETRIX INC
Methods for the identification, assessment, and treatment of patients with cancer therapy
ActiveUS20060281122A1Reduced growth rateEliminate ineffective or inappropriate therapeutic agentsMechanical/radiation/invasive therapiesMicrobiological testing/measurementAbnormal tissue growthRegimen
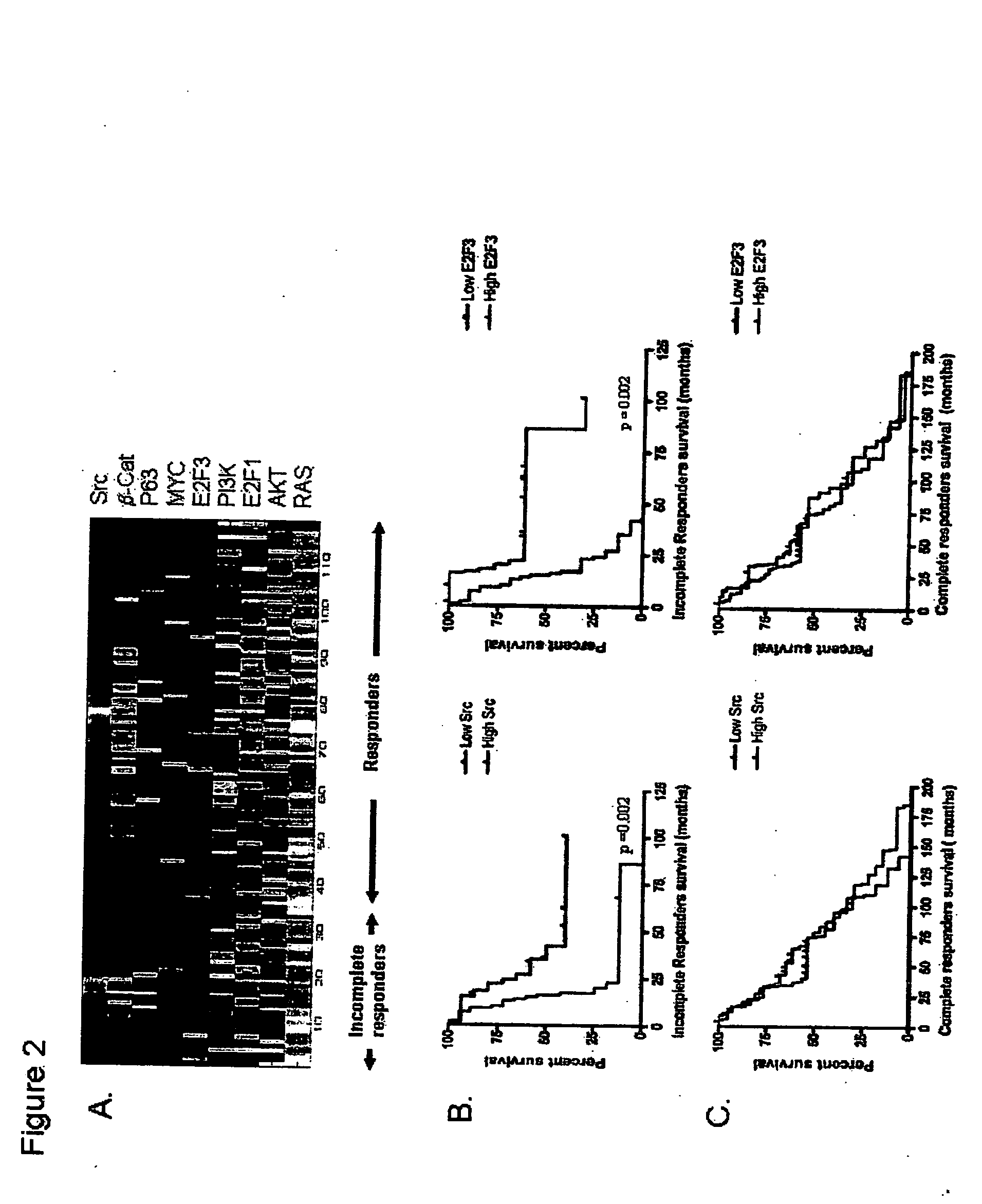
The present invention is directed to the identification of predictive markers that can be used to determine whether patients with cancer are clinically responsive or non-responsive to a therapeutic regimen prior to treatment. In particular, the present invention is directed to the use of certain individual and / or combinations of predictive markers, wherein the expression of the predictive markers correlates with responsiveness or non-responsiveness to a therapeutic regimen. Thus, by examining the expression levels of individual predictive markers and / or predictive markers comprising a marker set, it is possible to determine whether a therapeutic agent, or combination of agents, will be most likely to reduce the growth rate of tumors in a clinical setting.
Owner:MILLENNIUM PHARMA INC
Combinational array for nucleic acid analysis
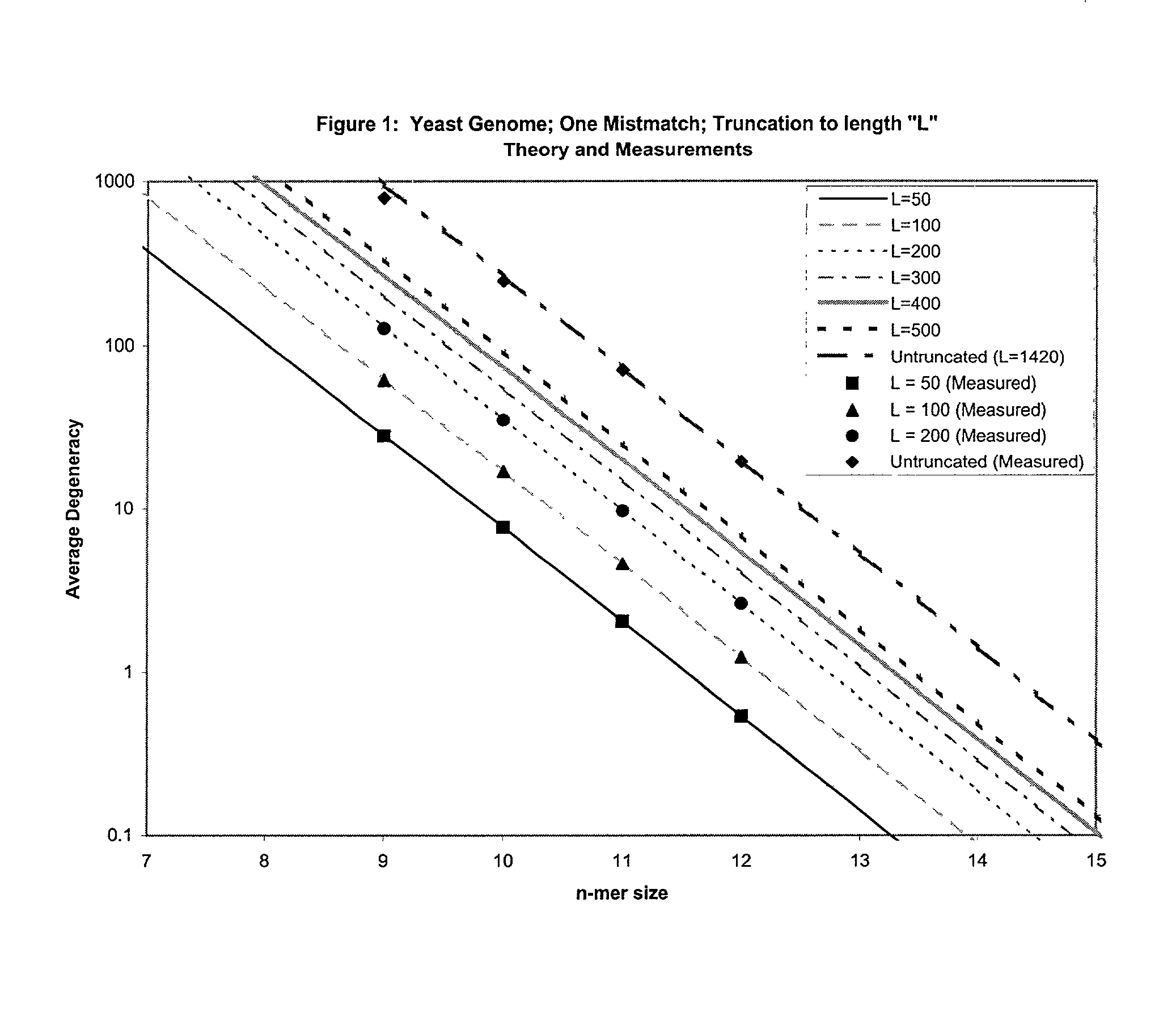
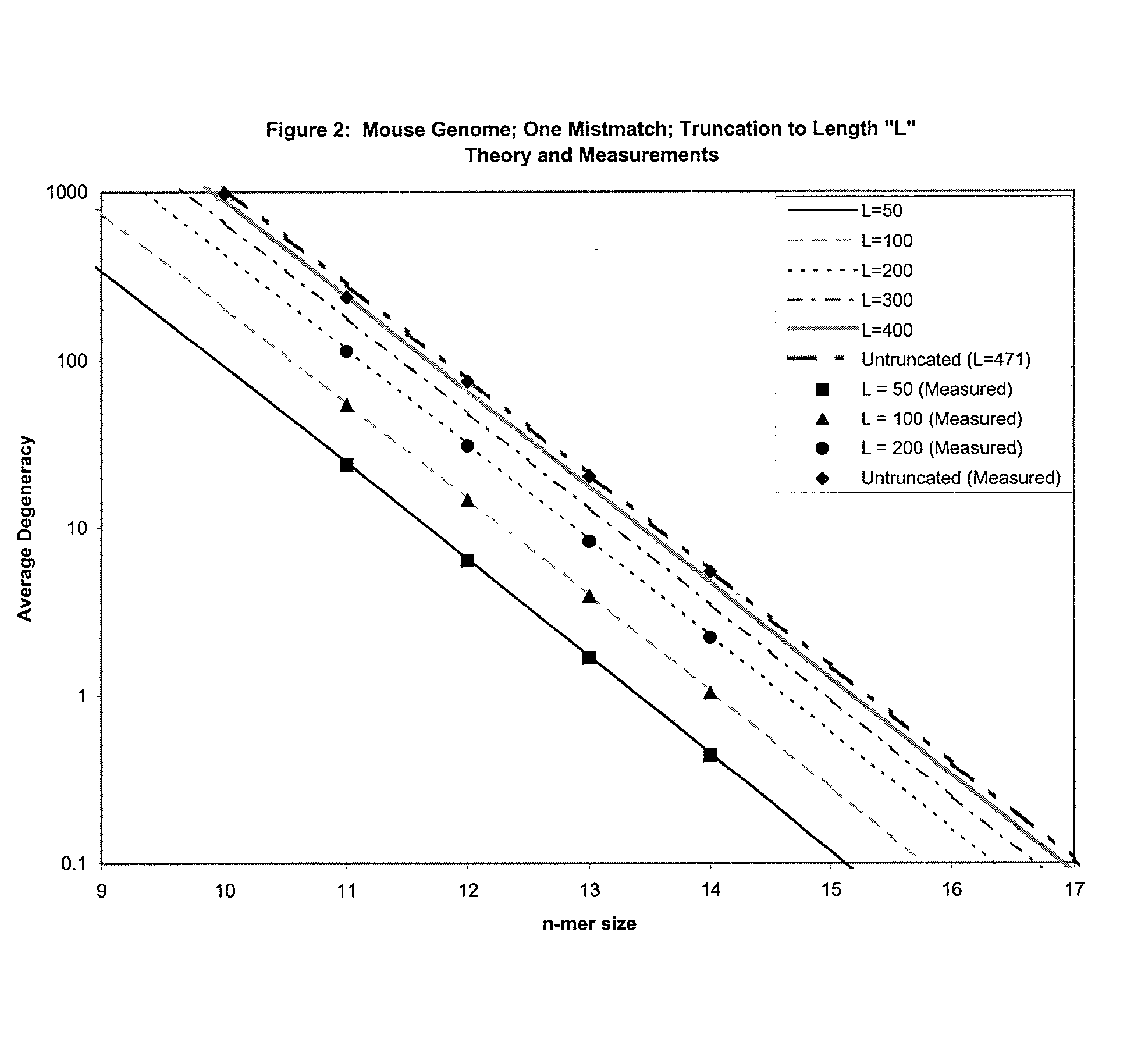
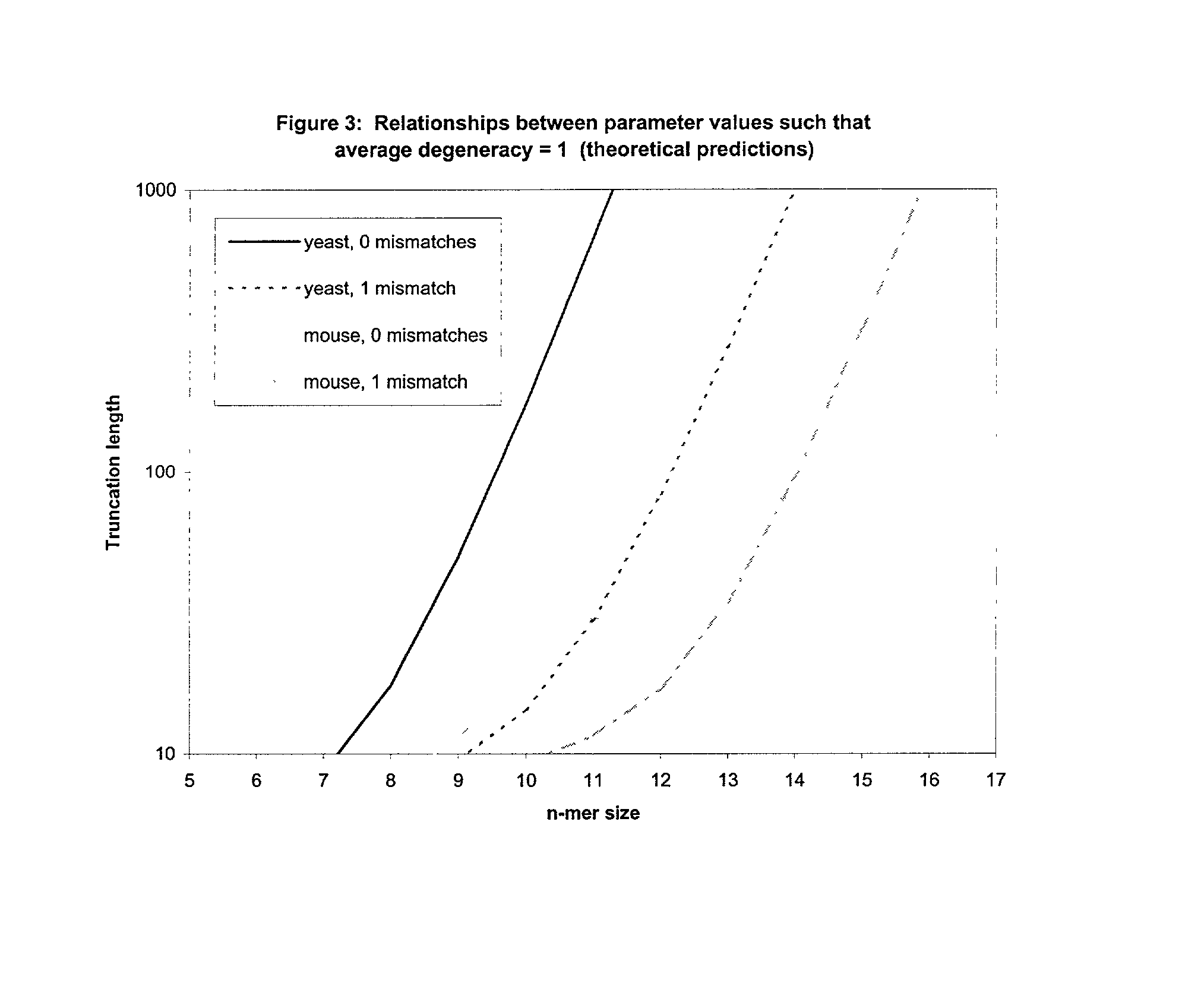
This invention relates to an array, including a universal micro-array, for the analysis of nucleic acids, such as DNA. The devices and methods of the invention can be used for identifying gene expression patterns in any organism. More specifically, all possible oligonucleotides (n-mers) necessary for the identification of gene expression patterns are synthesized. According to the invention, n is large enough to give the specificity to uniquely identify the expression pattern of each gene in an organism of interest, and is small enough that the method and device can be easily and efficiently practiced and made. The invention provides a method of analyzing molecules, such as polynucleotides (e.g., DNA), by measuring the signal of an optically-detectable (e.g., fluorescent, ultraviolet, radioactive or color change) reporter associated with the molecules. In a polynucleotide analysis device according to the invention, levels of gene expression are correlated to a signal from an optically-detectable (e.g. fluorescent) reporter associated with a hybridized polynucleotide. The invention includes an algorithm and method to interpret data derived from a micro-array or other device, including techniques to decode or deconvolve potentially ambiguous signals into unambiguous or reliable gene expression data.
Owner:CALIFORNIA INST OF TECH
Comprehensive Analysis Pipeline for Discovery of Human Genetic Variation
ActiveUS20130311106A1Shorten analysis timeQuick identificationMicrobiological testing/measurementProteomicsData setProcessing core
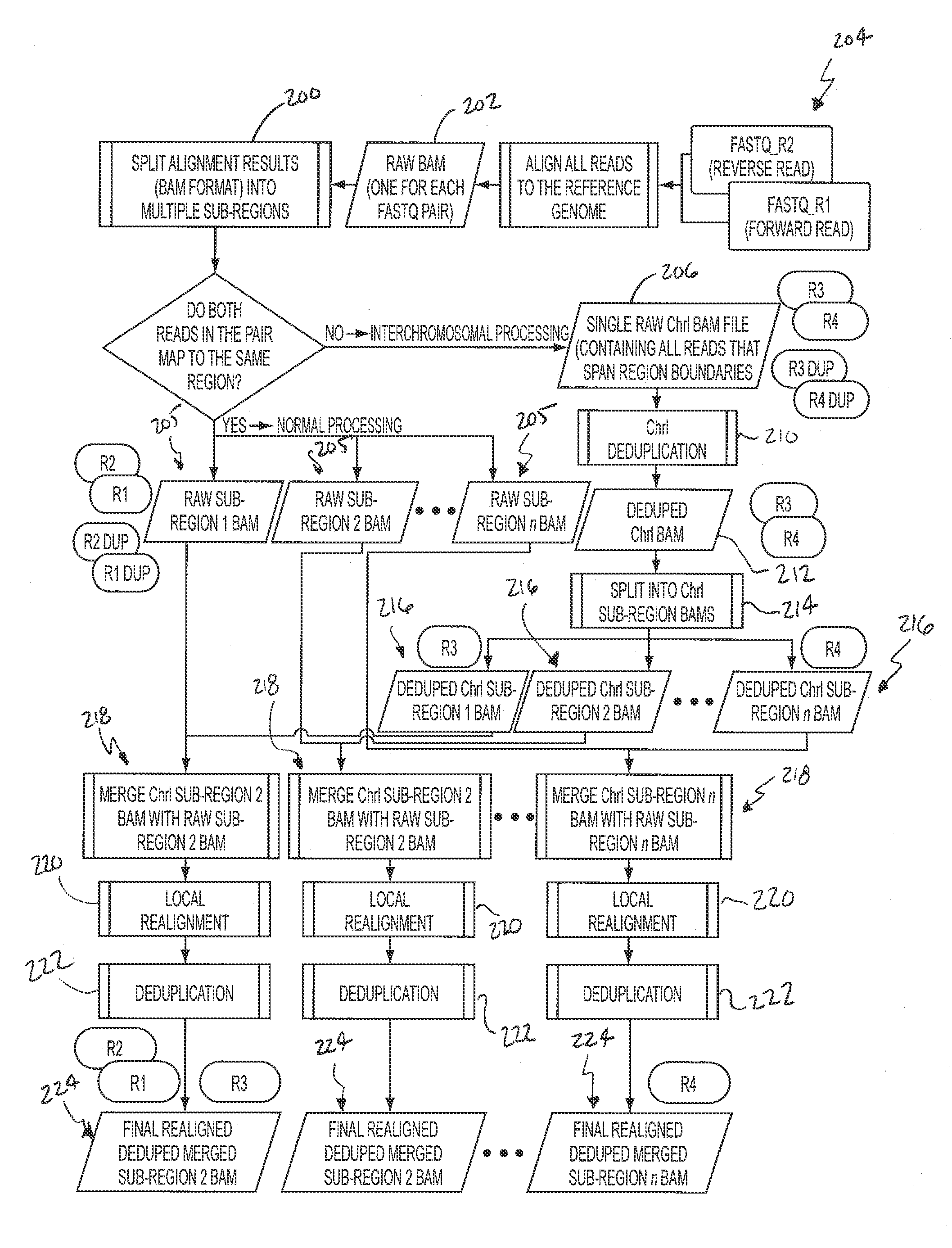
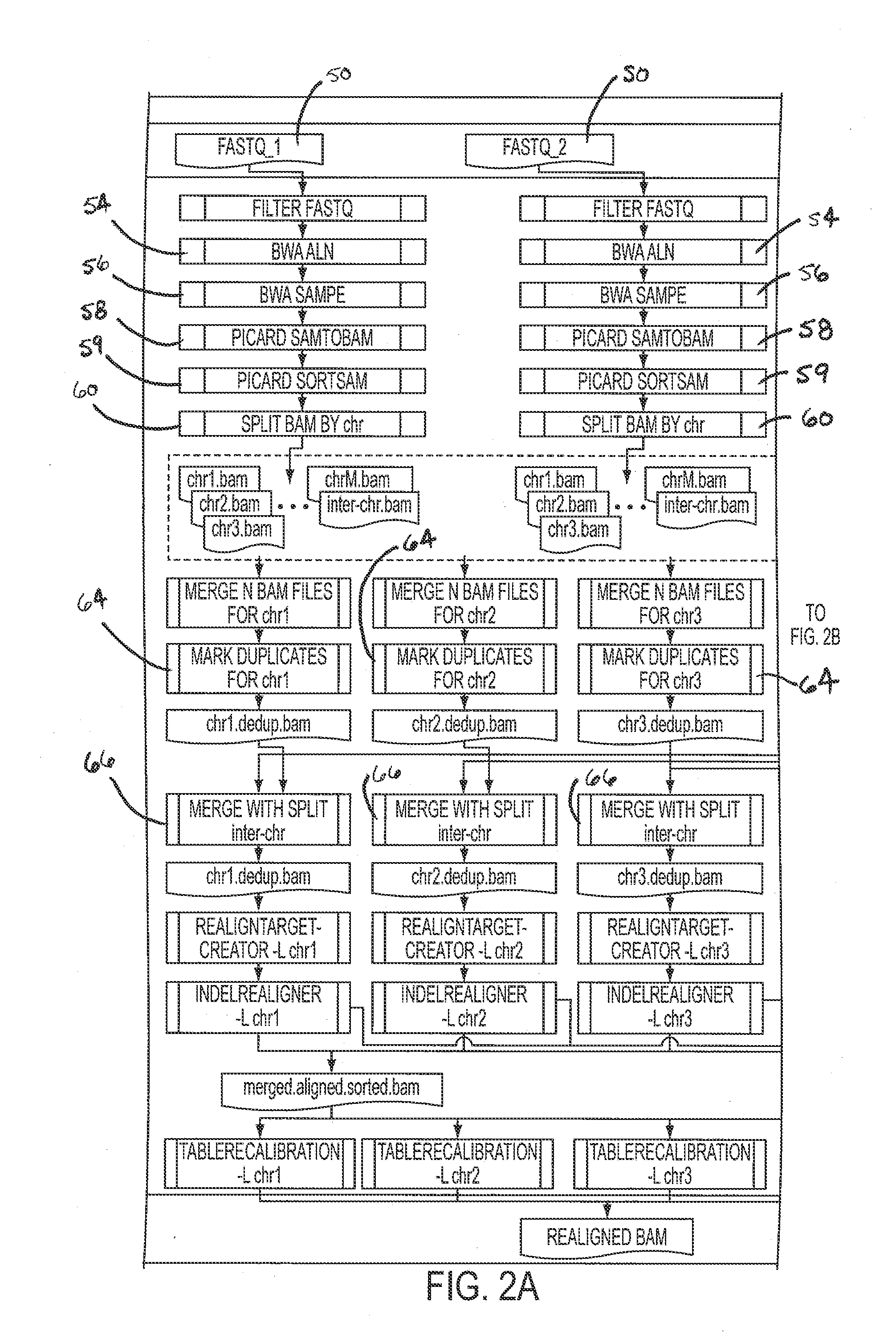
Systems and methods for analyzing genetic sequence data involve: (a) obtaining, by a computer system, genetic sequencing data pertaining to a subject; (b) splitting the genetic sequencing data into a plurality of segments; (c) processing the genetic sequencing data such that intra-segment reads, read pairs with both mates mapped to the same data set, are saved to a respective plurality of individual binary alignment map (BAM) files corresponding to that respective segment; (d) processing the genetic sequencing data such that inter-segment reads, read pairs with both mates mapped to different segments, are saved into at least a second BAM file; and (e) processing at least the first plurality of BAM files along parallel processing paths. The plurality of segments may correspond to any given number of genomic subregions and may be selected based upon the number of processing cores used in the parallel processing.
Owner:RES INST AT NATIONWIDE CHILDRENS HOSPITAL
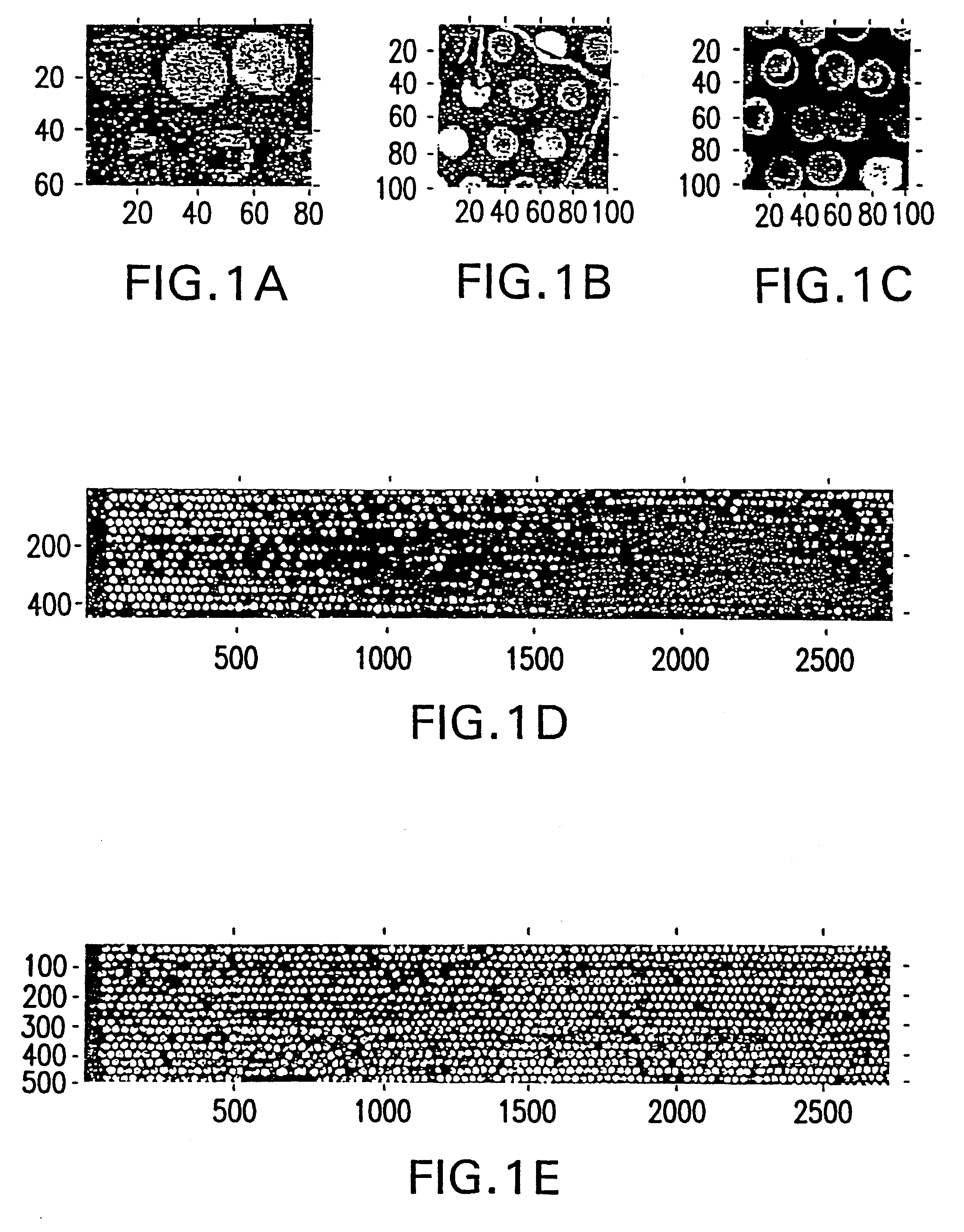
Systems and methods for treating, diagnosing and predicting the occurrence of a medical condition
ActiveUS20100184093A1Reliable and accurate image segmentationMedical simulationBioreactor/fermenter combinationsImmunofluorescenceClinical information
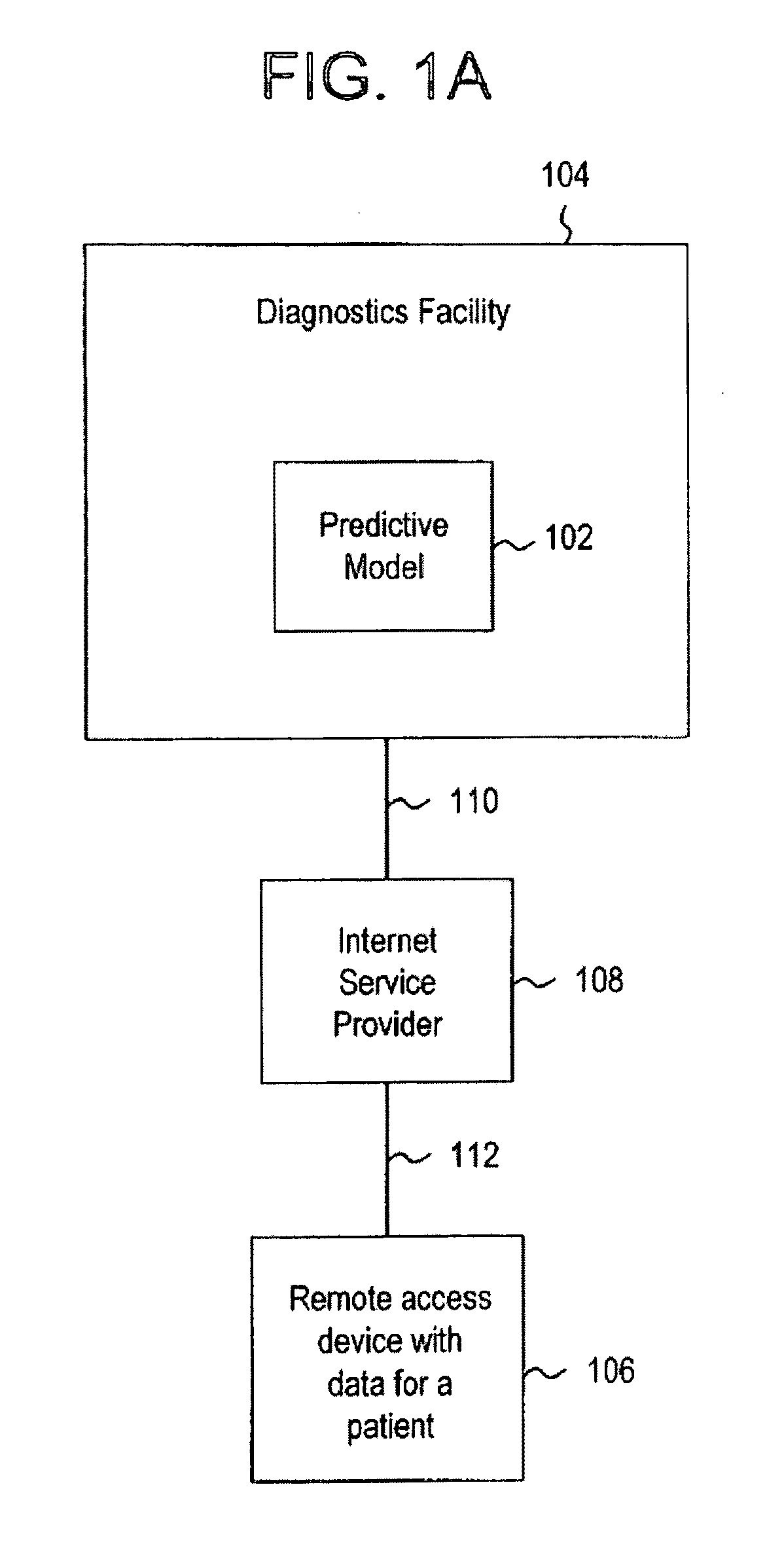
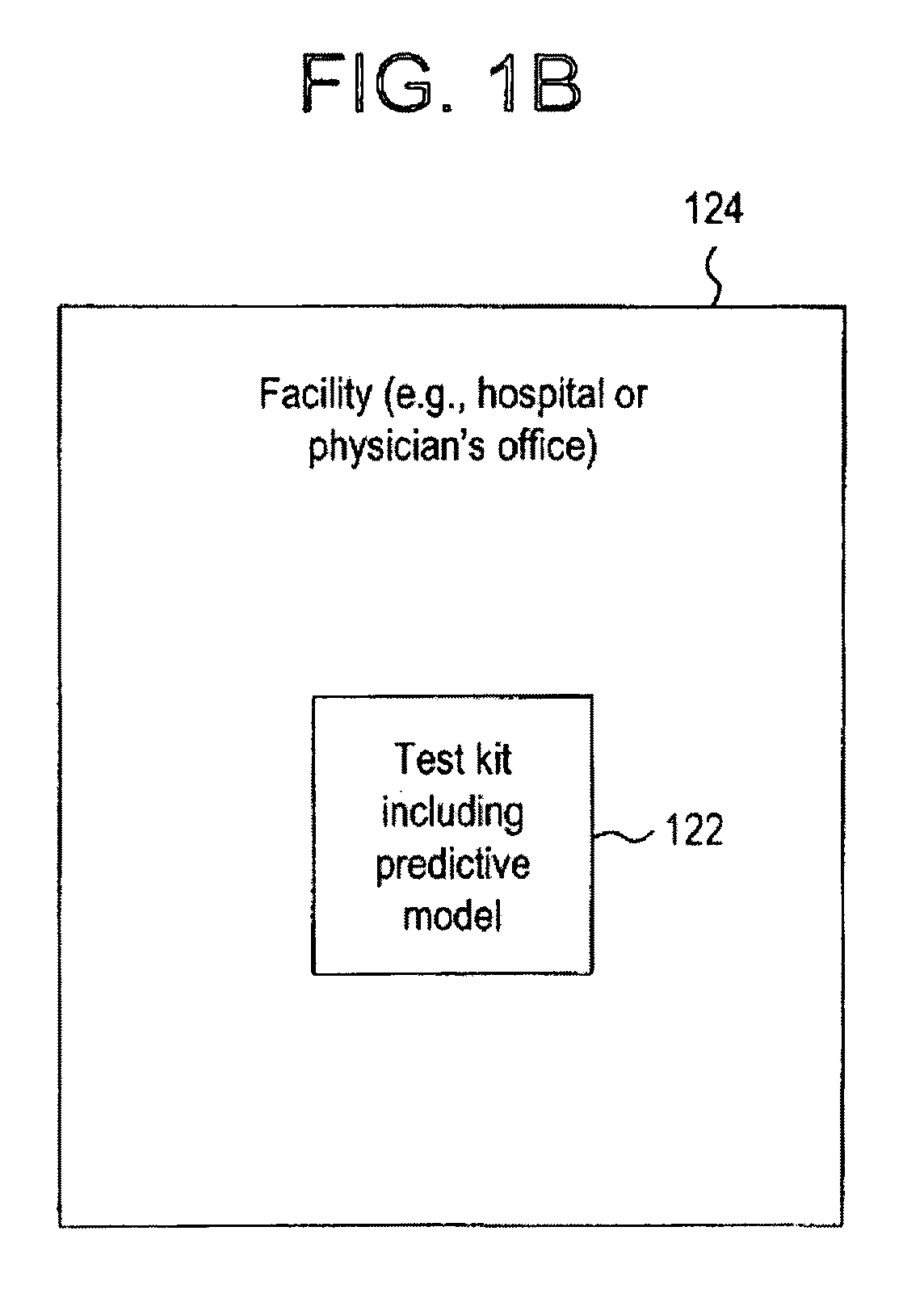
Clinical information, molecular information and / or computer-generated morphometric information is used in a predictive model for predicting the occurrence of a medical condition. In an embodiment, a model predicts whether a patient is likely to have a favorable pathological stage of prostate cancer, where the model is based on features including one or more (e.g., all) of preoperative PSA, Gleason Score, a measurement of expression of androgen receptor (AR) in epithelial and stromal nuclei and / or a measurement of expression of Ki67-positive epithelial nuclei, a morphometric measurement of a ratio of area of epithelial nuclei outside gland units to area of epithelial nuclei within gland units, and a morphometric measurement of area of epithelial nuclei distributed away from gland units. In some embodiments, quantitative measurements of protein expression in cell lines are utilized to objectively assess assay (e.g., multiplex immunofluorescence (IF)) performance and / or to normalize features for use within a predictive model.
Owner:AUREON LAB INC +1
Method and system for quantifying and removing spatial-intensity trends in microarray data
A method and system for quantifying and correcting spatial-intensity trends for each channel of a microarray data set having one or more channels. The method and system of one embodiment of the present invention selects a set of features from each channel of the microarray data set. Based on the selected set of features, a surface is used to determine the intensities for all features in each channel of the microarray data set. Spatial-intensity trends within the microarray data set are quantified, based on the surface to the intensities for each channel of the microarray data set. After the surface has been determined, the spatial-intensity trend can be removed from the microarray data set.
Owner:AGILENT TECH INC
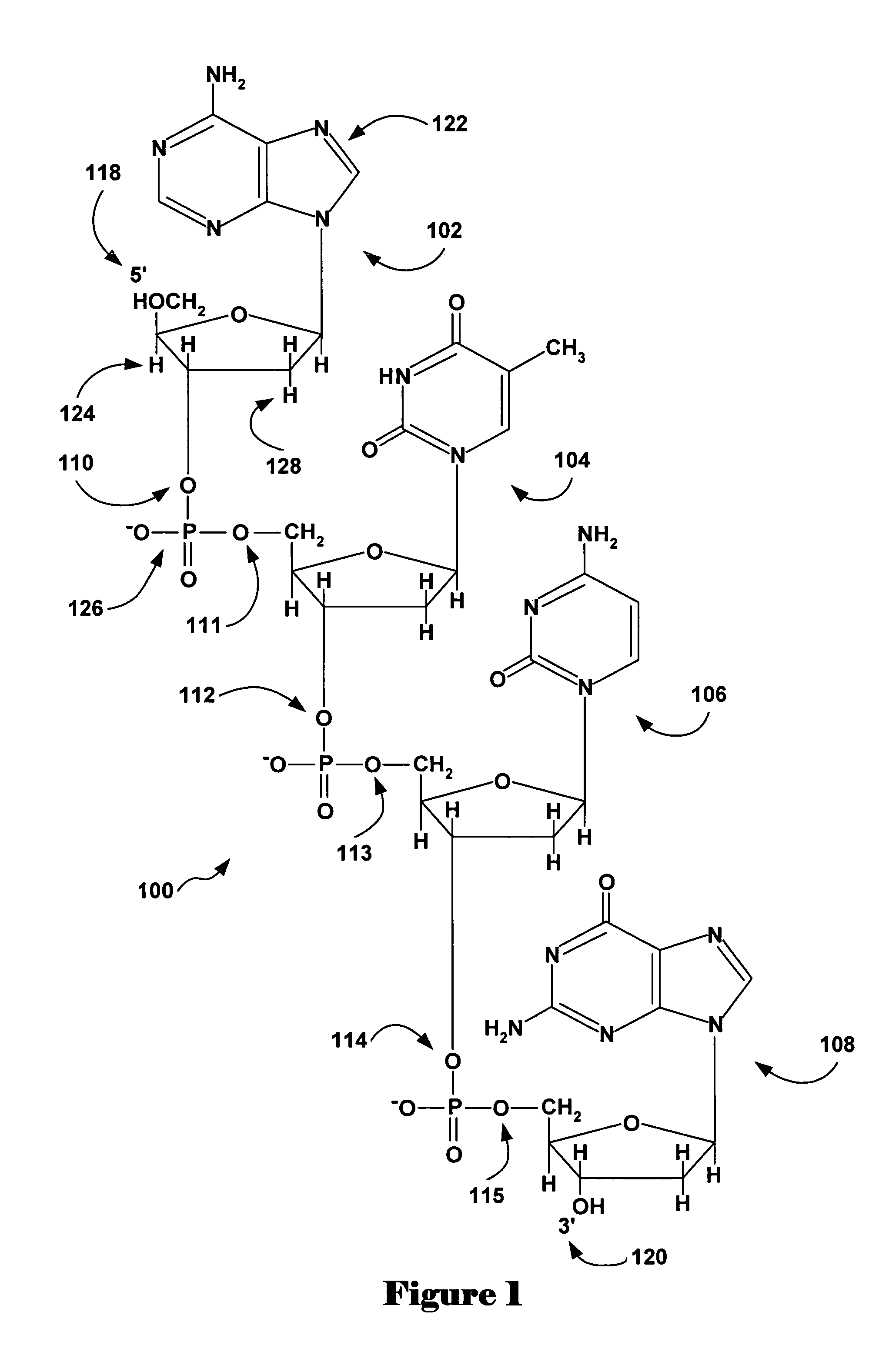
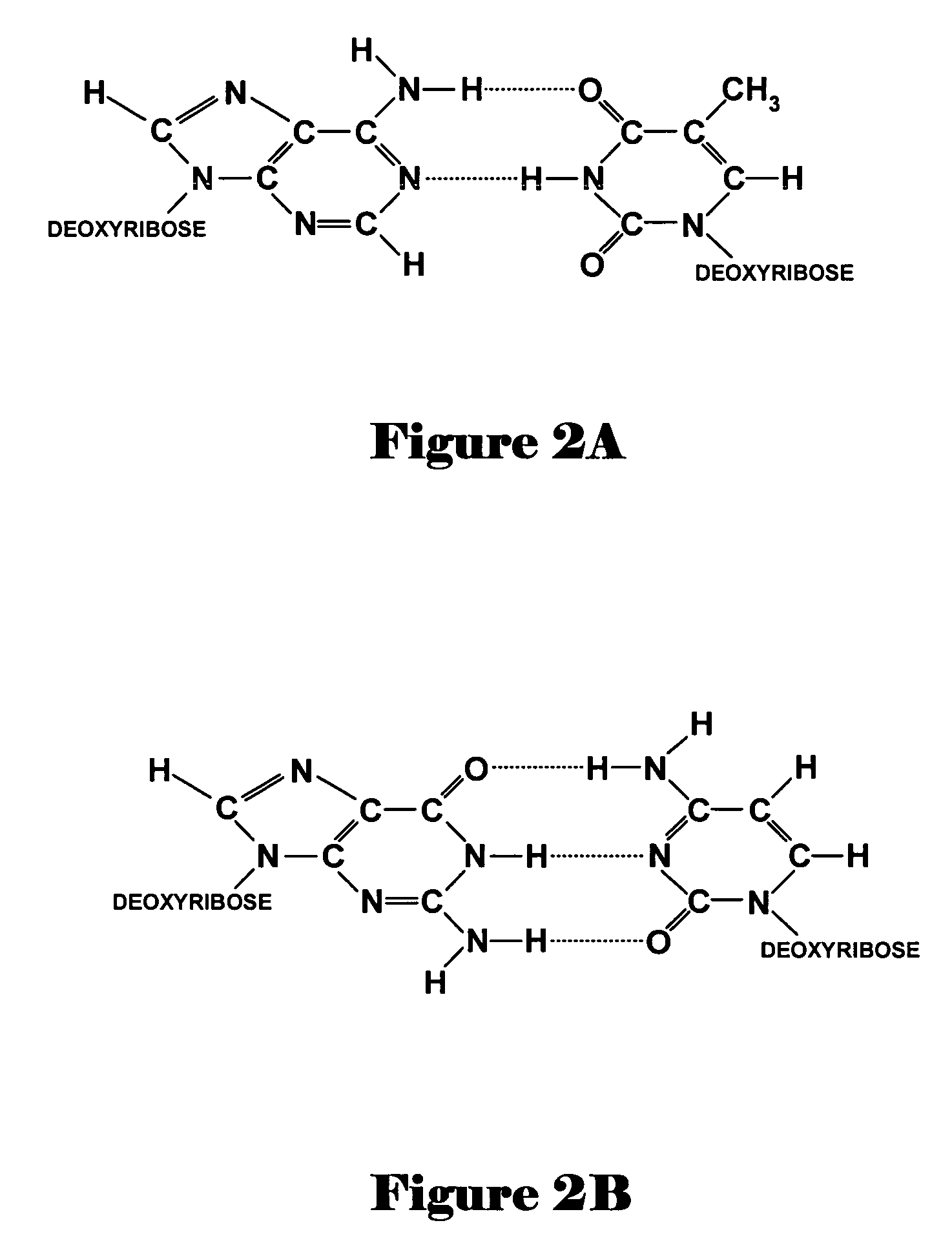
Novel oligonucleotide compositions and probe sequences useful for detection and analysis of micrornas and their target mRNAs
InactiveUS20070099196A1Solve low usageSugar derivativesMicrobiological testing/measurementGeneticsTarget mrna
The invention relates relates to ribonucleic acids and oligonucleotide probes useful for detection and analysis of microRNAs and their target mRNAs, as well as small interfering RNAs (siRNAs).
Owner:EXIQON AS
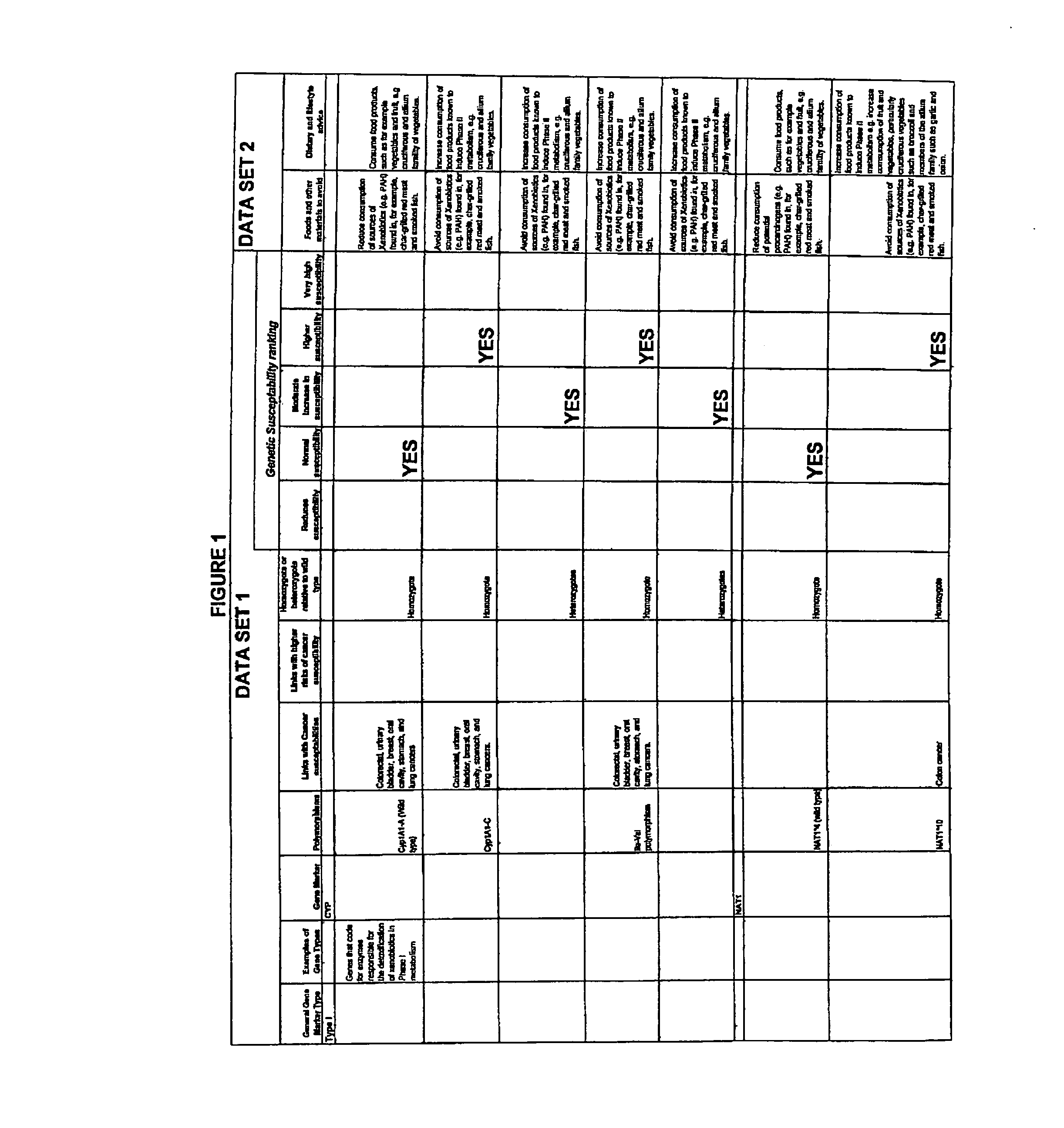
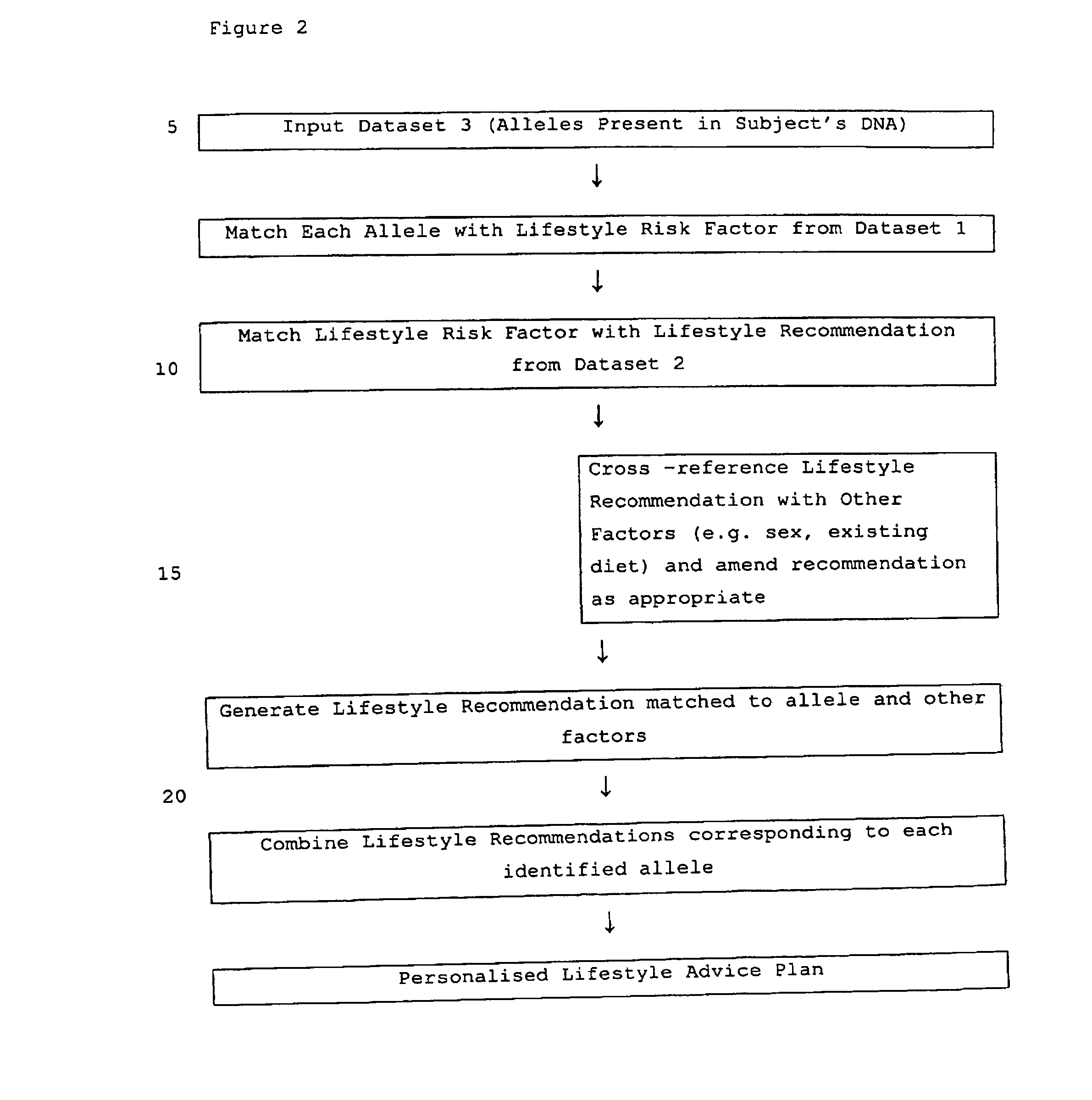
Computer-assisted means for assessing lifestyle risk factors
InactiveUS7054758B2Reduce disease riskProtect healthDigital data processing detailsBiostatisticsPersonalizationData set
The present invention relates to methods of assessing disease susceptibility associated with dietary and lifestyle risk factors. The invention provides for analysis of alleles at loci of genes associated with lifestyle risk factors, and the disease susceptibility profile of an individual is determined by reference to datasets which further match the risk factor with lifestyle recommendations in order to produce a personalized lifestyle advice plan.
Owner:BODYSYNC
Methods for identifying DNA copy number changes
Owner:AFFYMETRIX INC
Methods of identifying biological patterns using multiple data sets
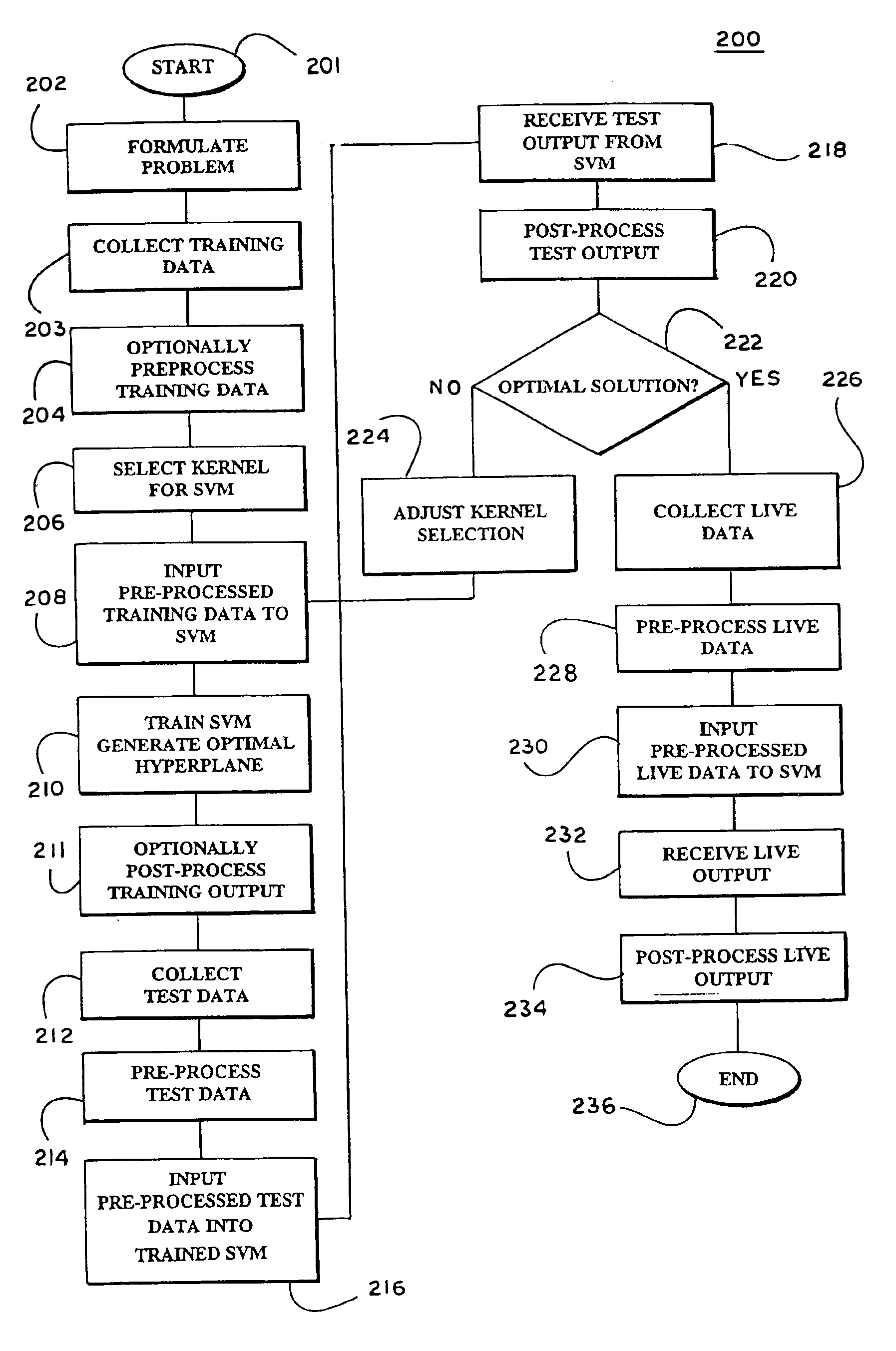
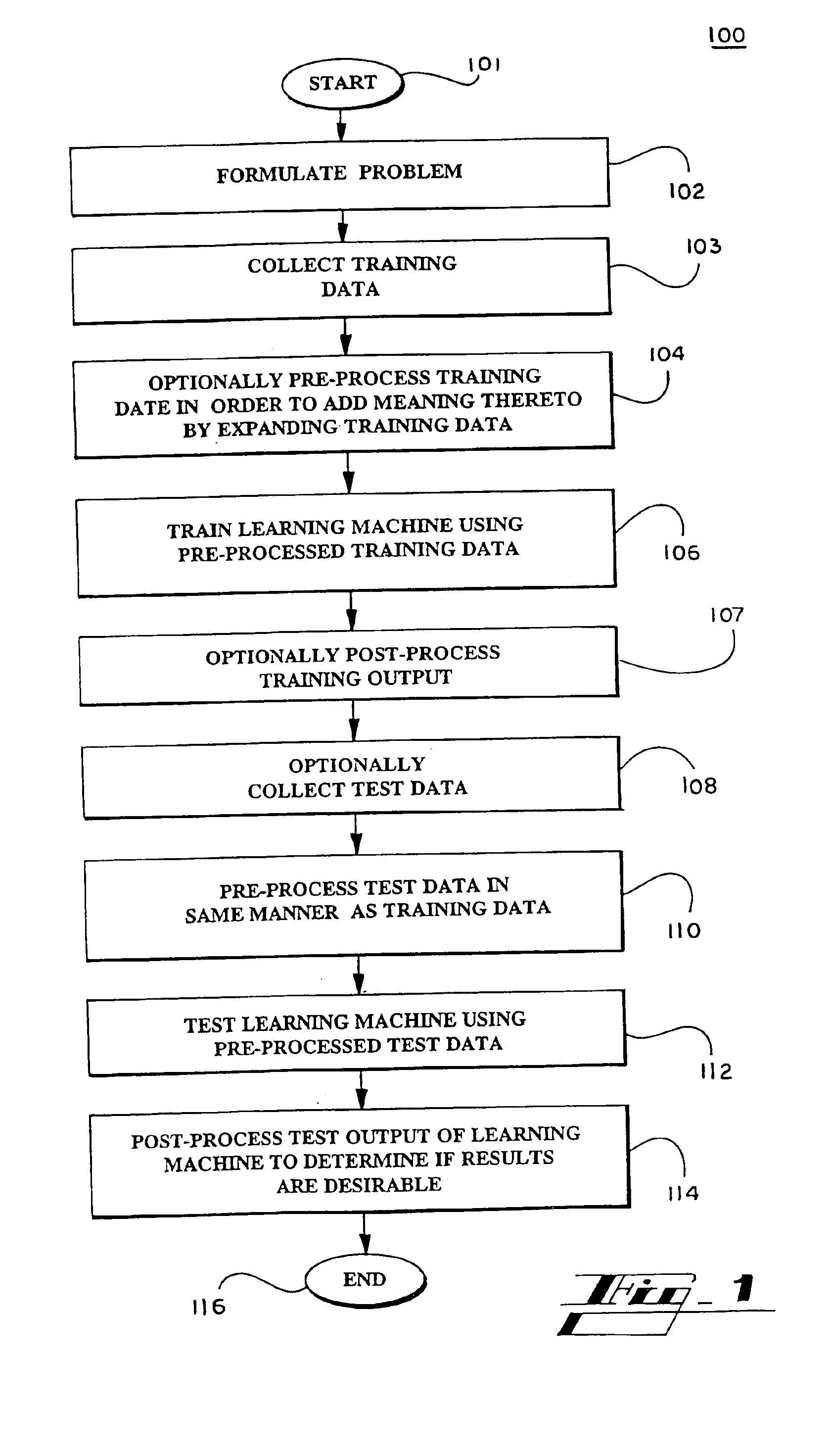
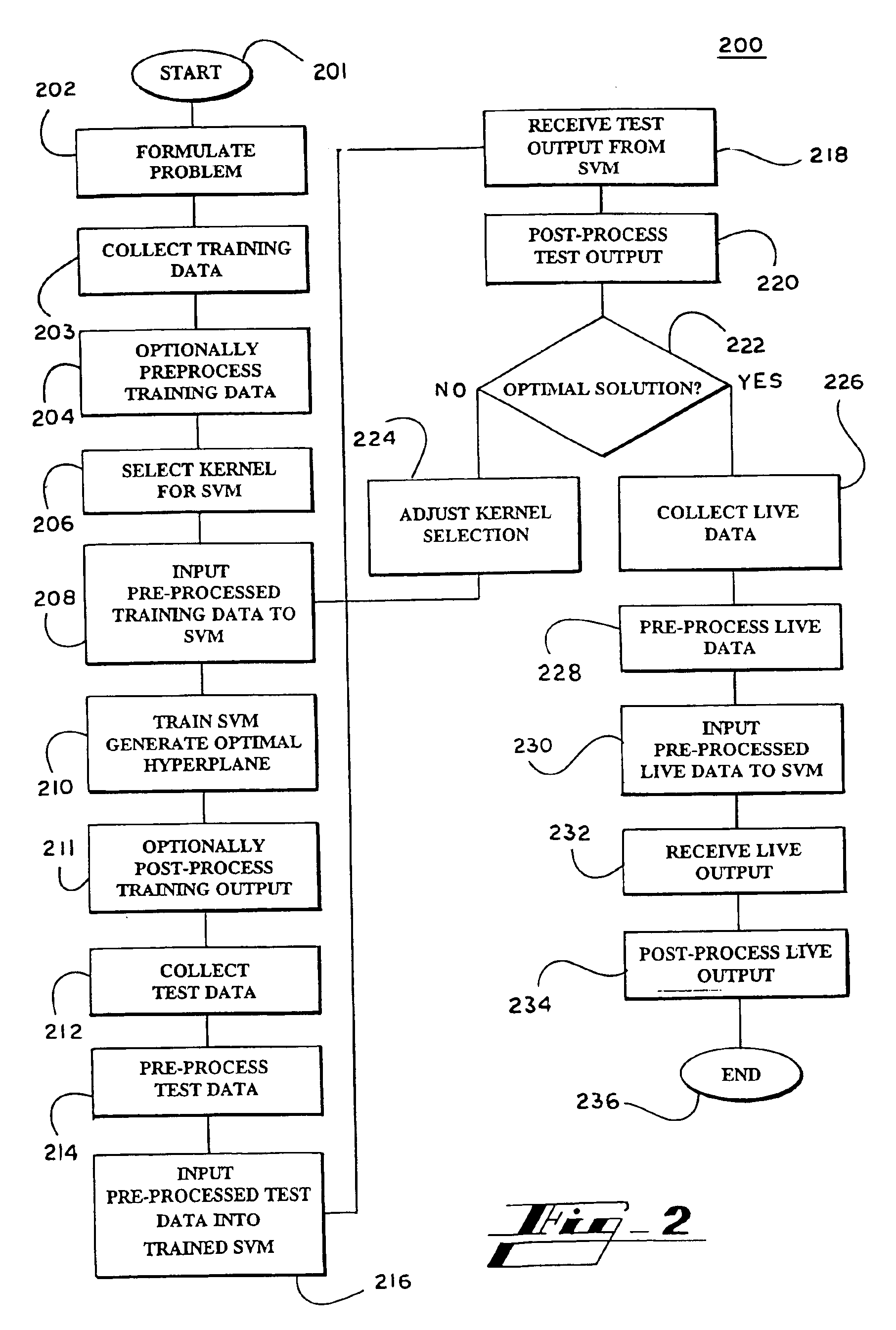
InactiveUS6882990B1Improve abilitiesGreat dimensionalityImage analysisDigital data processing detailsSupport vector machineLearning machine
Systems and methods for enhancing knowledge discovery from data using multiple learning machines in general and multiple support vector machines in particular. Training data for a learning machine is pre-processed in order to add meaning thereto. Multiple support vector machines, each comprising distinct kernels, are trained with the pre-processed training data and are tested with test data that is pre-processed in the same manner. The test outputs from multiple support vector machines are compared in order to determine which of the test outputs if any represents a optimal solution. Selection of one or more kernels may be adjusted and one or more support vector machines may be retrained and retested. Optimal solutions based on distinct input data sets may be combined to form a new input data set to be input into one or more additional support vector machine. The methods, systems and devices of the present invention comprise use of Support Vector Machines for the identification of patterns that are important for medical diagnosis, prognosis and treatment. Such patterns may be found in many different datasets. The present invention also comprises methods and compositions for the treatment and diagnosis of medical conditions.
Owner:HEALTH DISCOVERY CORP +1
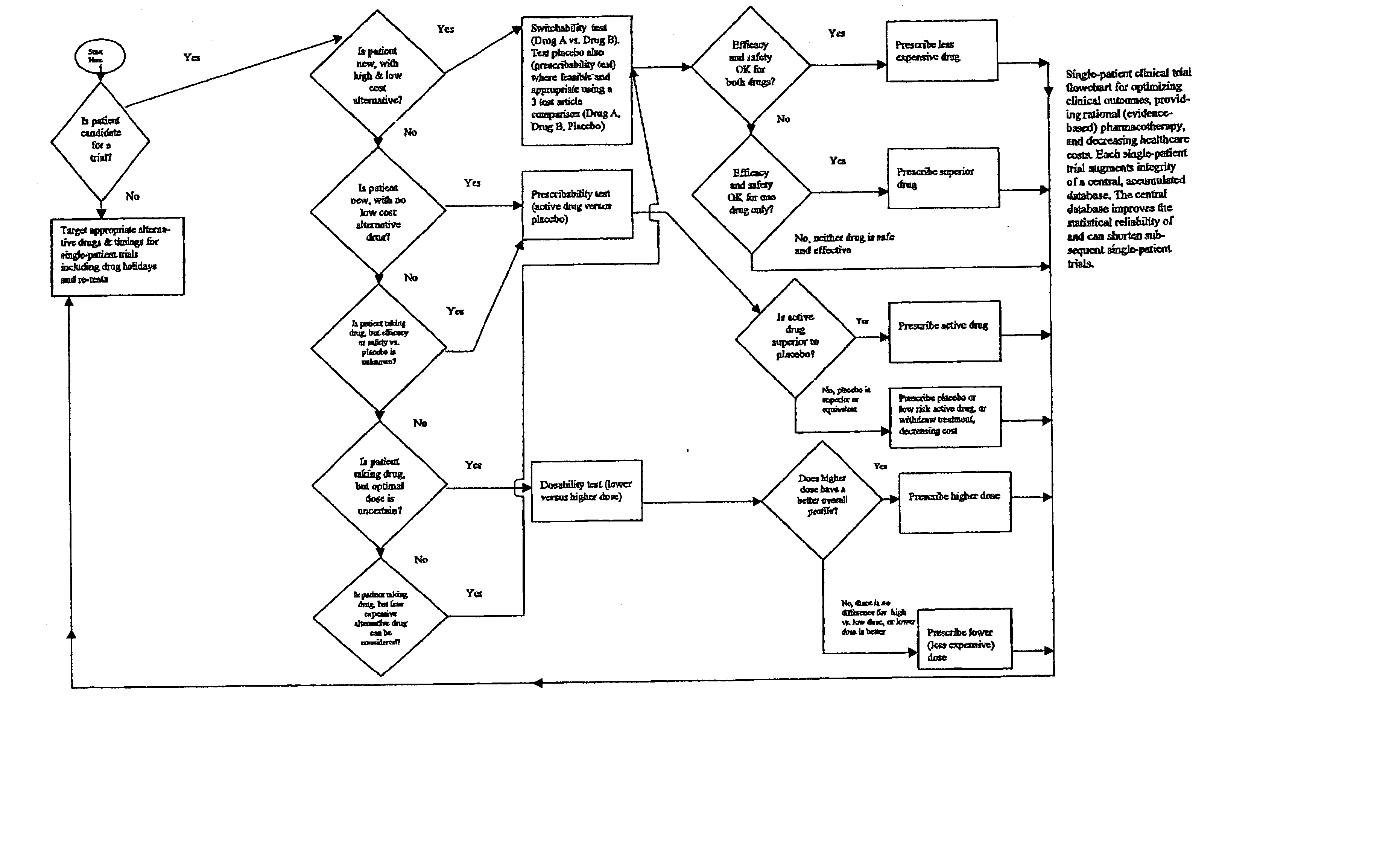
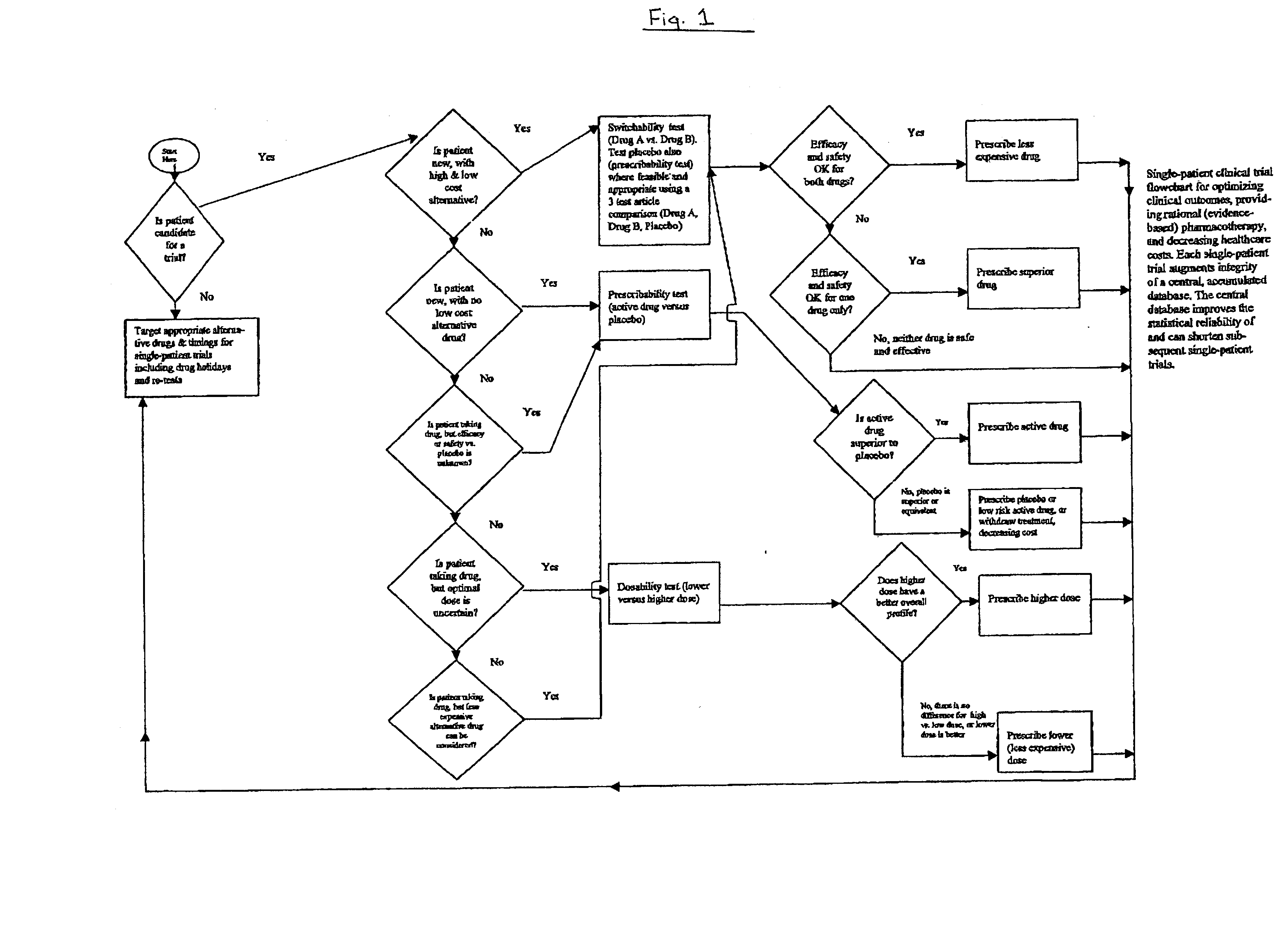
Single-patient drug trials used with accumulated database: risk of habituation
InactiveUS20020032581A1AppropriatenessImprove statistics performancePhysical therapies and activitiesDrug and medicationsHabituationDrug trial
A method of evaluating and / or optimizing clinical outcomes and providing rational pharmacotherapy in an individual or animal requiring chronic drug therapy is provided.
Owner:OPT E SCRIP
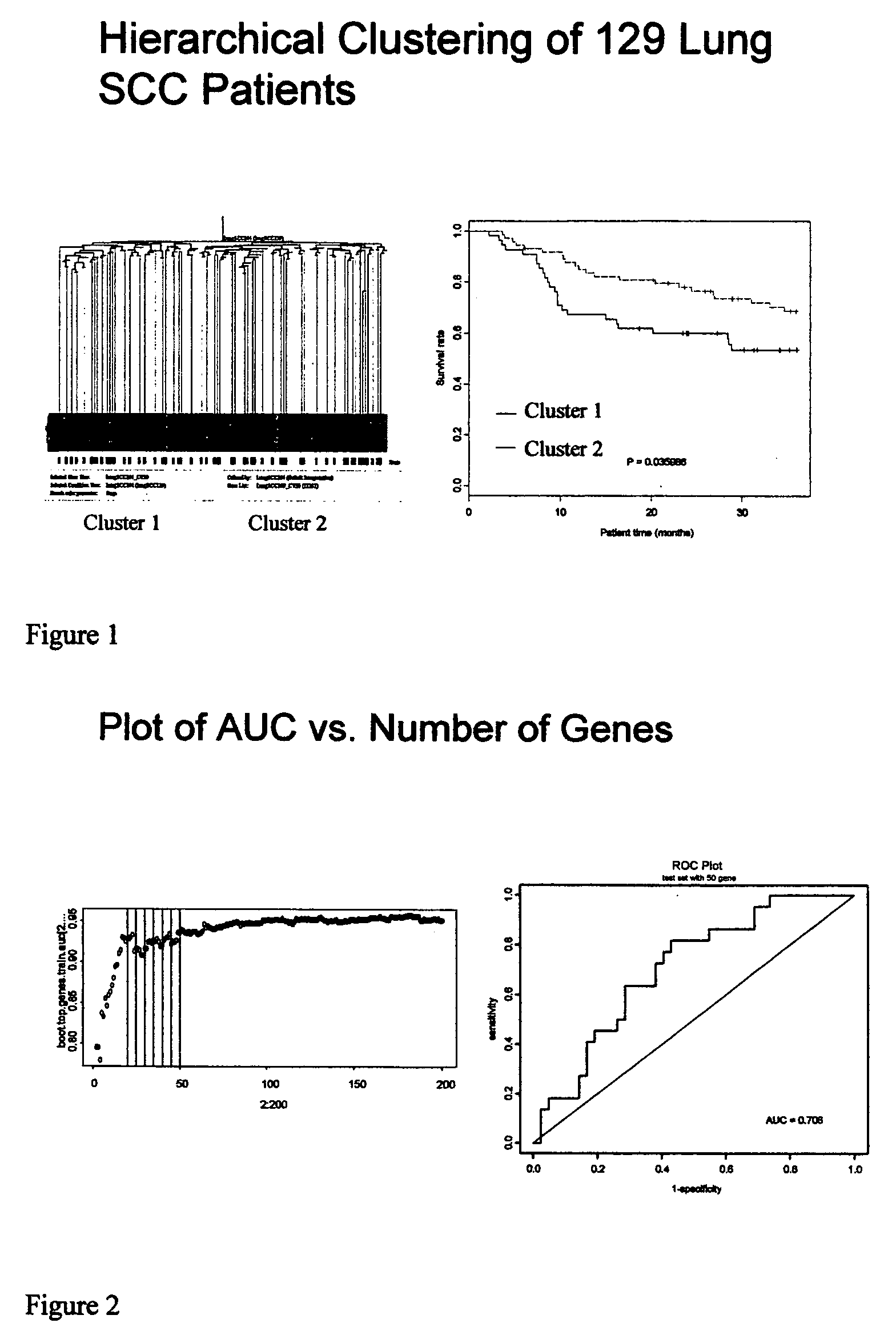
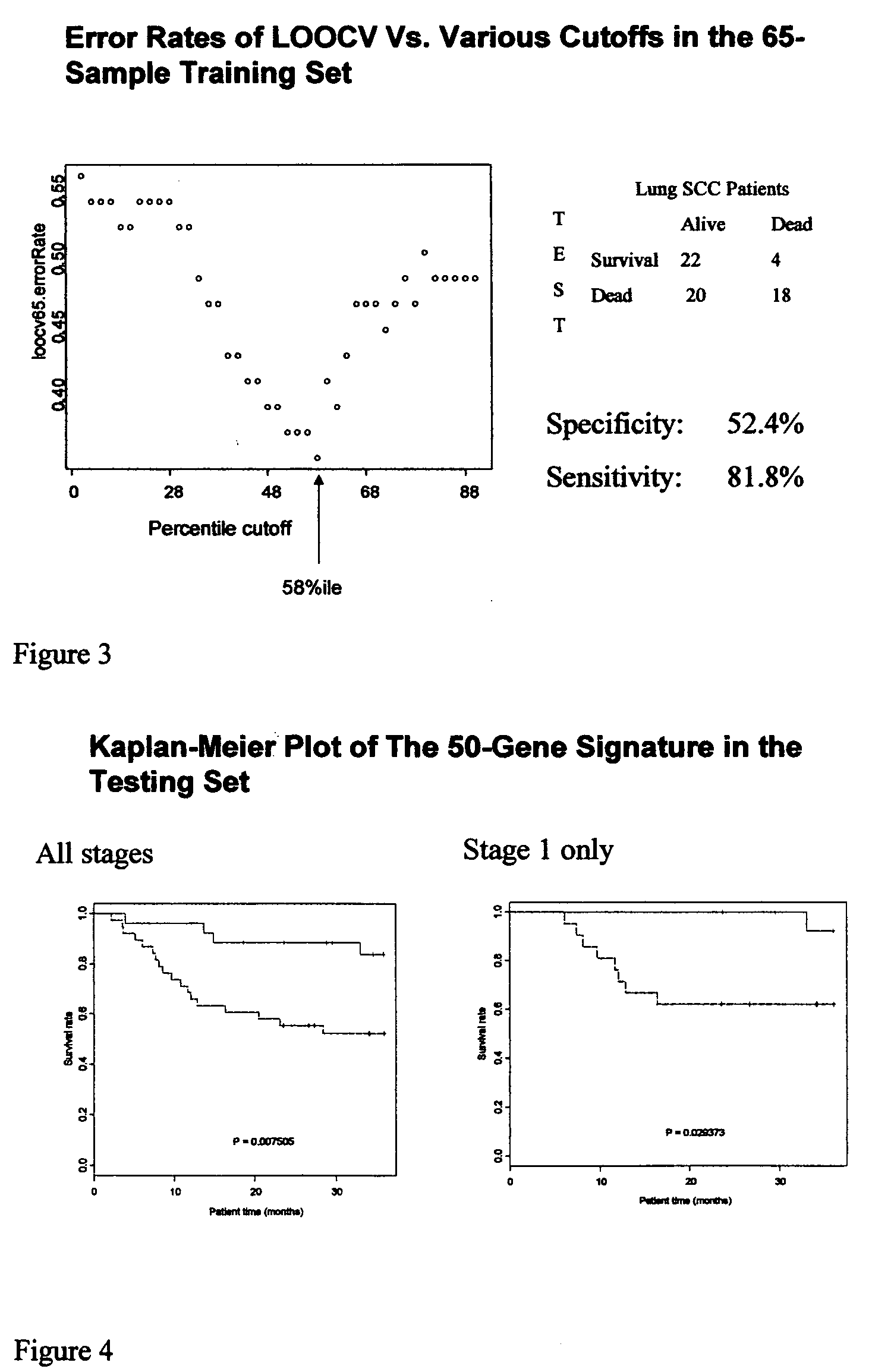
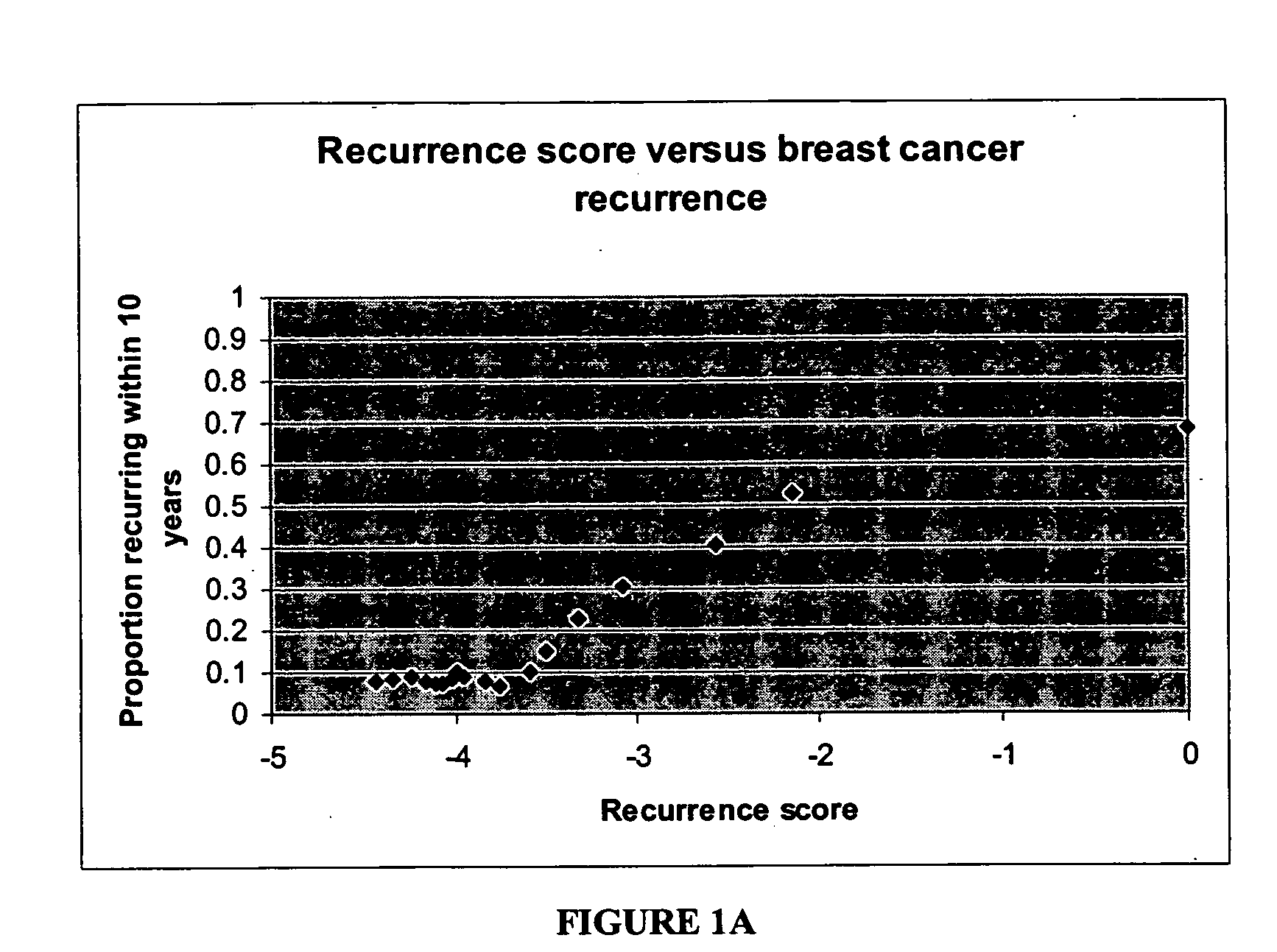
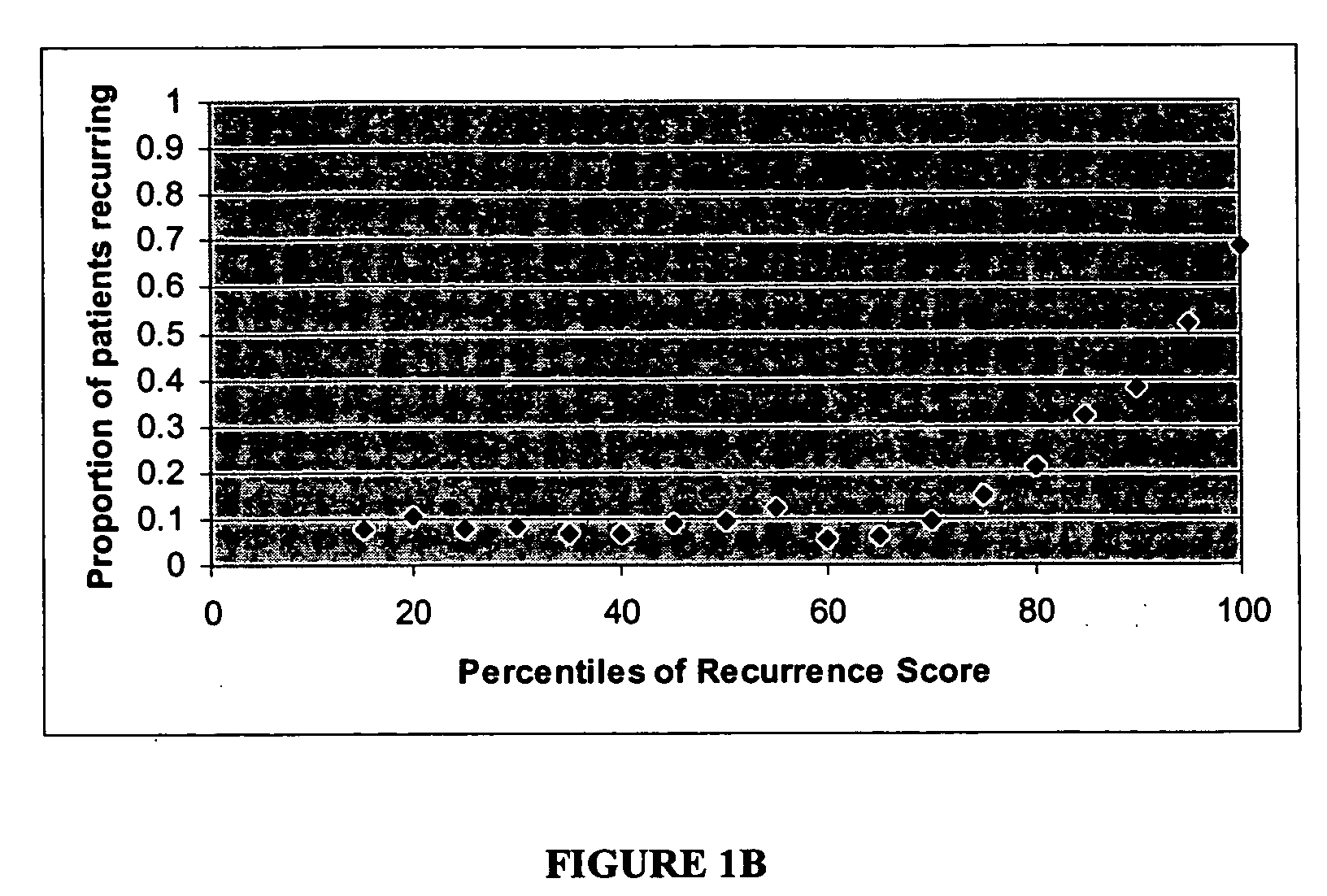
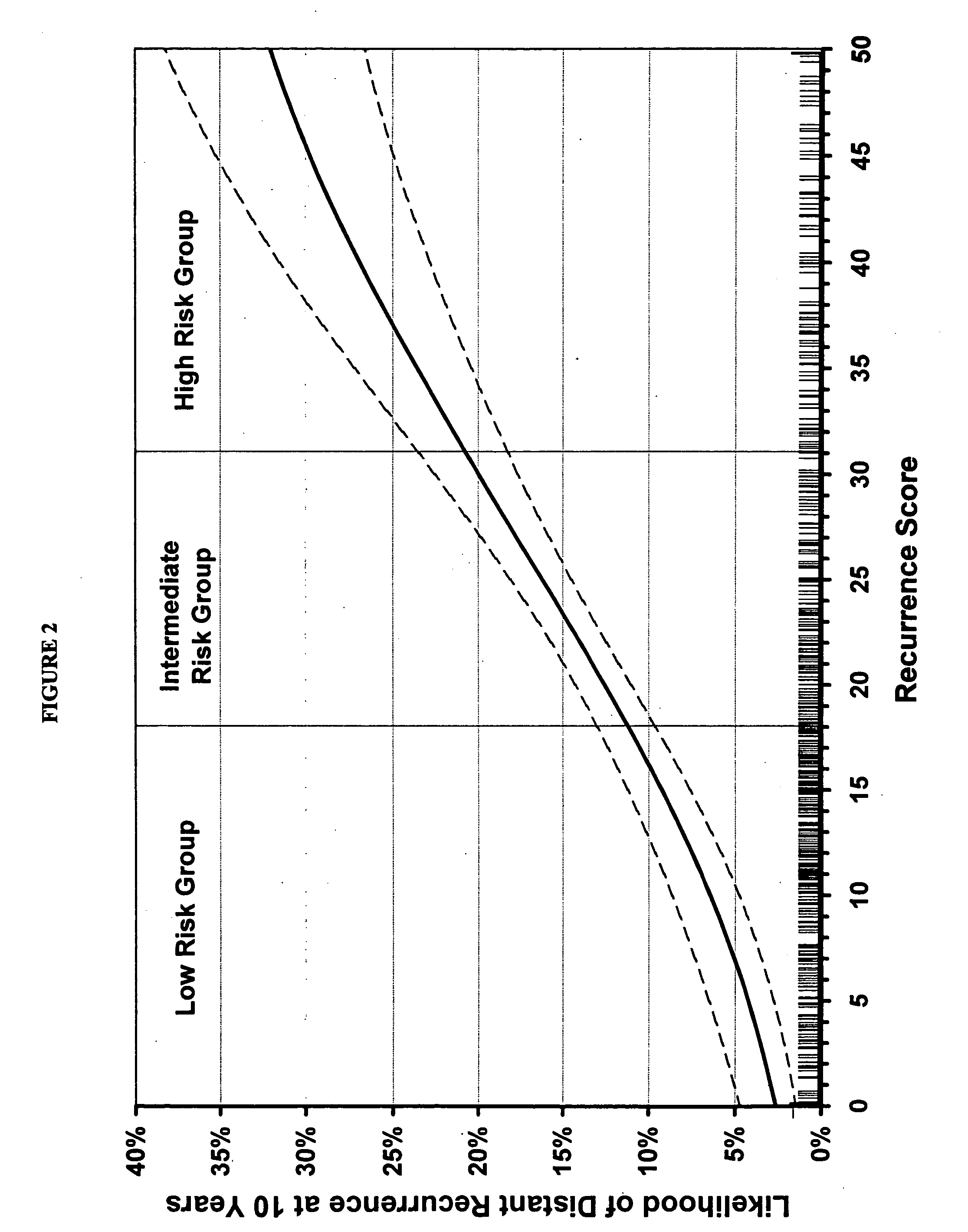
Expression profile algorithm and test for cancer prognosis
The present invention provides a noninvasive, quantitative test for prognosis determination in cancer patients. The test relies on measurements of the tumor levels of certain messenger RNAs (mRNAs). These mRNA levels are inserted into a polynomial formula (algorithm) that yields a numerical recurrence score, which indicates recurrence risk.
Owner:GENOMIC HEALTH INC
Combinatorial array for nucleic acid analysis
InactiveUS20020012926A1Microbiological testing/measurementBiological testingBiological bodyFluorescence
This invention relates to an array, including a universal micro-array, for the analysis of nucleic acids, such as DNA. The devices and methods of the invention can be used for identifying gene expression patterns in any organism. More specifically, all possible oligonucleotides (n-mers) necessary for the identification of gene expression patterns are synthesized. According to the invention, n is large enough to give the specificity to uniquely identify the expression pattern of each gene in an organism of interest, and is small enough that the method and device can be easily and efficiently practiced and made. The invention provides a method of analyzing molecules, such as polynucleotides (e.g., DNA), by measuring the signal of an optically-detectable (e.g., fluorescent, ultraviolet, radioactive or color change) reporter associated with the molecules. In a polynucleotide analysis device according to the invention, levels of gene expression are correlated to a signal from an optically-detectable (e.g. fluorescent) reporter associated with a hybridized polynucleotide. The invention includes an algorithm and method to interpret data derived from a micro-array or other device, including techniques to decode or deconvolve potentially ambiguous signals into unambiguous or reliable gene expression data.
Owner:CALIFORNIA INST OF TECH
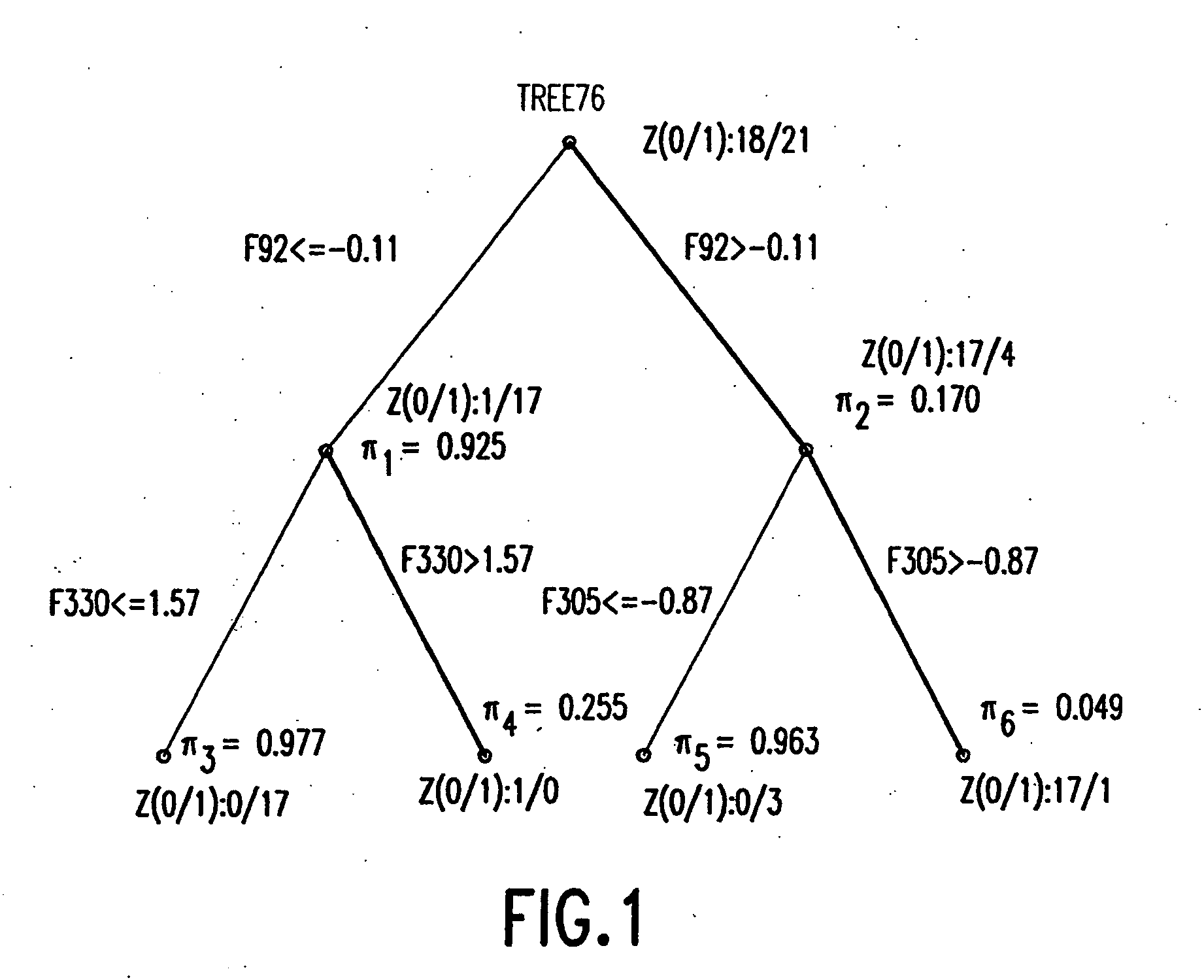
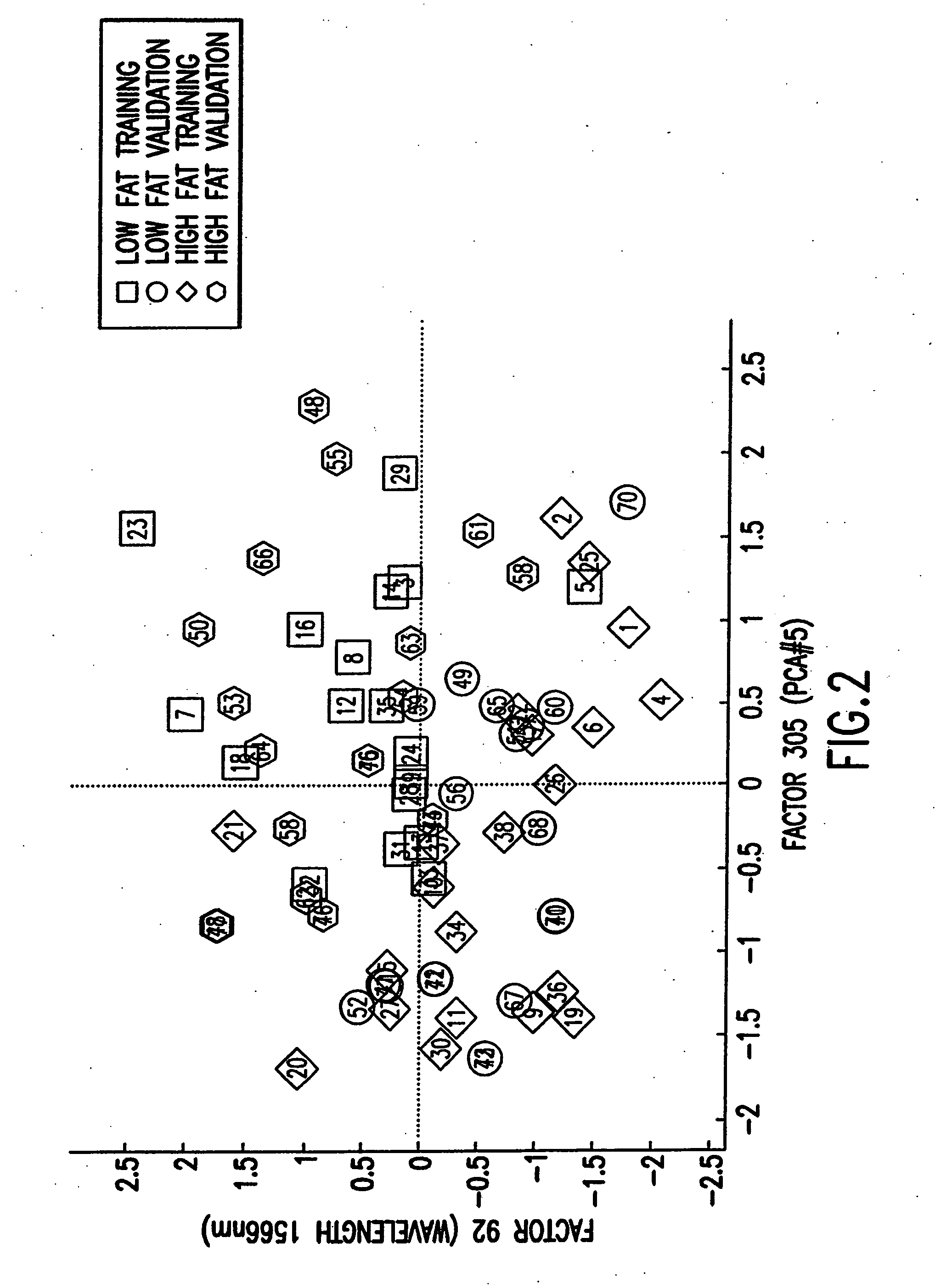
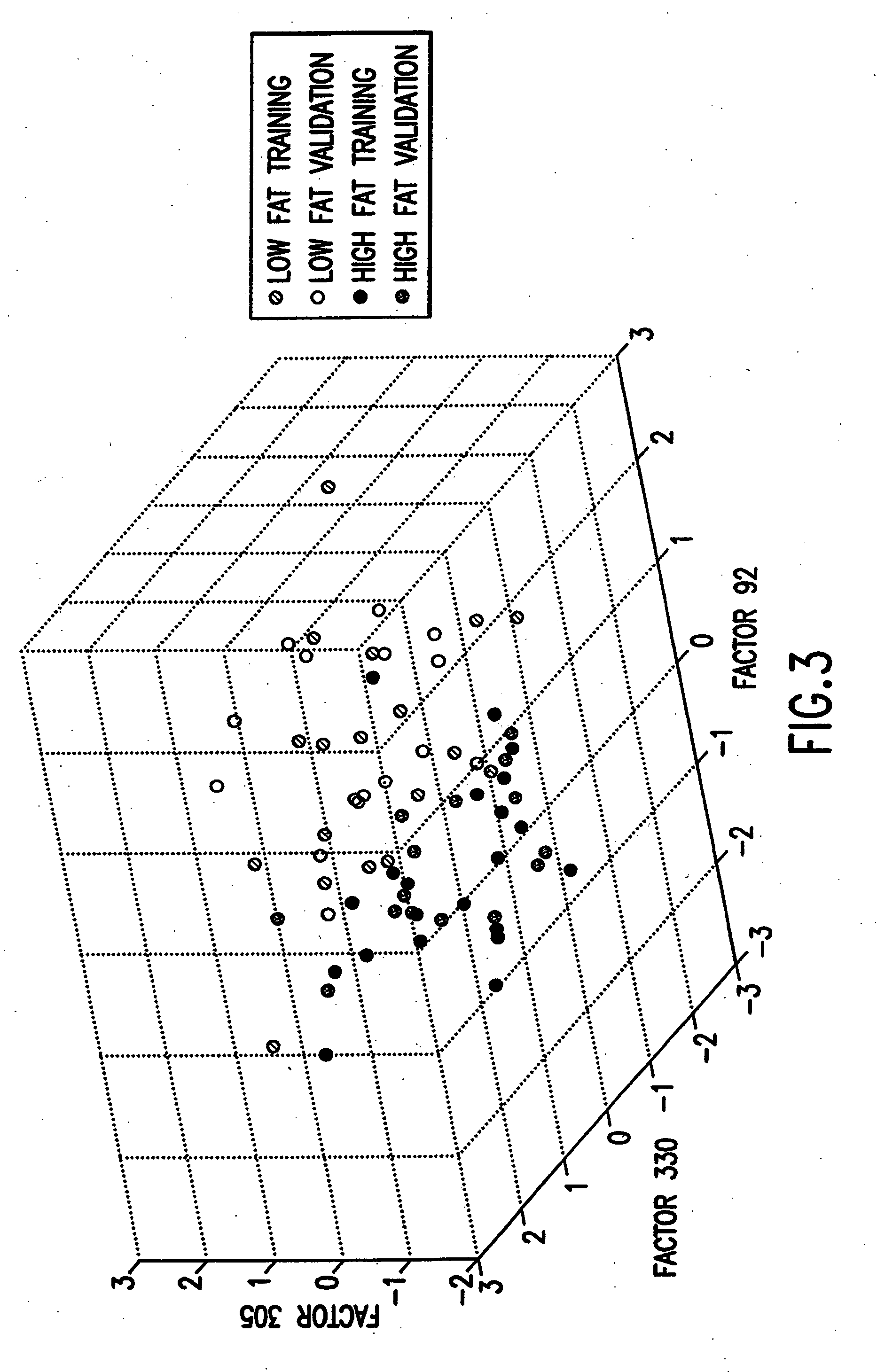
Binary prediction tree modeling with many predictors and its uses in clinical and genomic applications
The statistical analysis described and claimed is a predictive statistical tree model that overcomes several problems observed in prior statistical models and regression analyses, while ensuring greater accuracy and predictive capabilities. Although the claimed use of the predictive statistical tree model described herein is directed to the prediction of a disease in individuals, the claimed model can be used for a variety of applications including the prediction of disease states, susceptibility of disease states or any other biological state of interest, as well as other applicable non-biological states of interest. This model first screens genes to reduce noise, applies k-means correlation-based clustering targeting a large number of clusters, and then uses singular value decompositions (SVD) to extract the single dominant factor (principal component) from each cluster. This generates a statistically significant number of cluster-derived singular factors, that we refer to as metagenes, that characterize multiple patterns of expression of the genes across samples. The strategy aims to extract multiple such patterns while reducing dimension and smoothing out gene-specific noise through the aggregation within clusters. Formal predictive analysis then uses these metagenes in a Bayesian classification tree analysis. This generates multiple recursive partitions of the sample into subgroups (the “leaves” of the classification tree), and associates Bayesian predictive probabilities of outcomes with each subgroup. Overall predictions for an individual sample are then generated by averaging predictions, with appropriate weights, across many such tree models. The model includes the use of iterative out-of-sample, cross-validation predictions leaving each sample out of the data set one at a time, refitting the model from the remaining samples and using it to predict the hold-out case. This rigorously tests the predictive value of a model and mirrors the real-world prognostic context where prediction of new cases as they arise is the major goal.
Owner:DUKE UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com