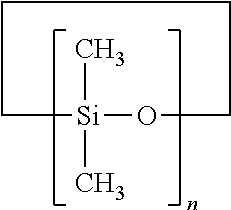
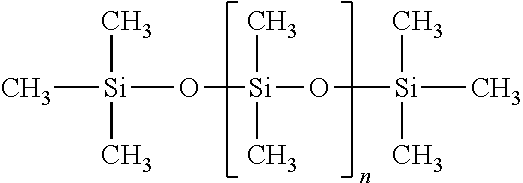
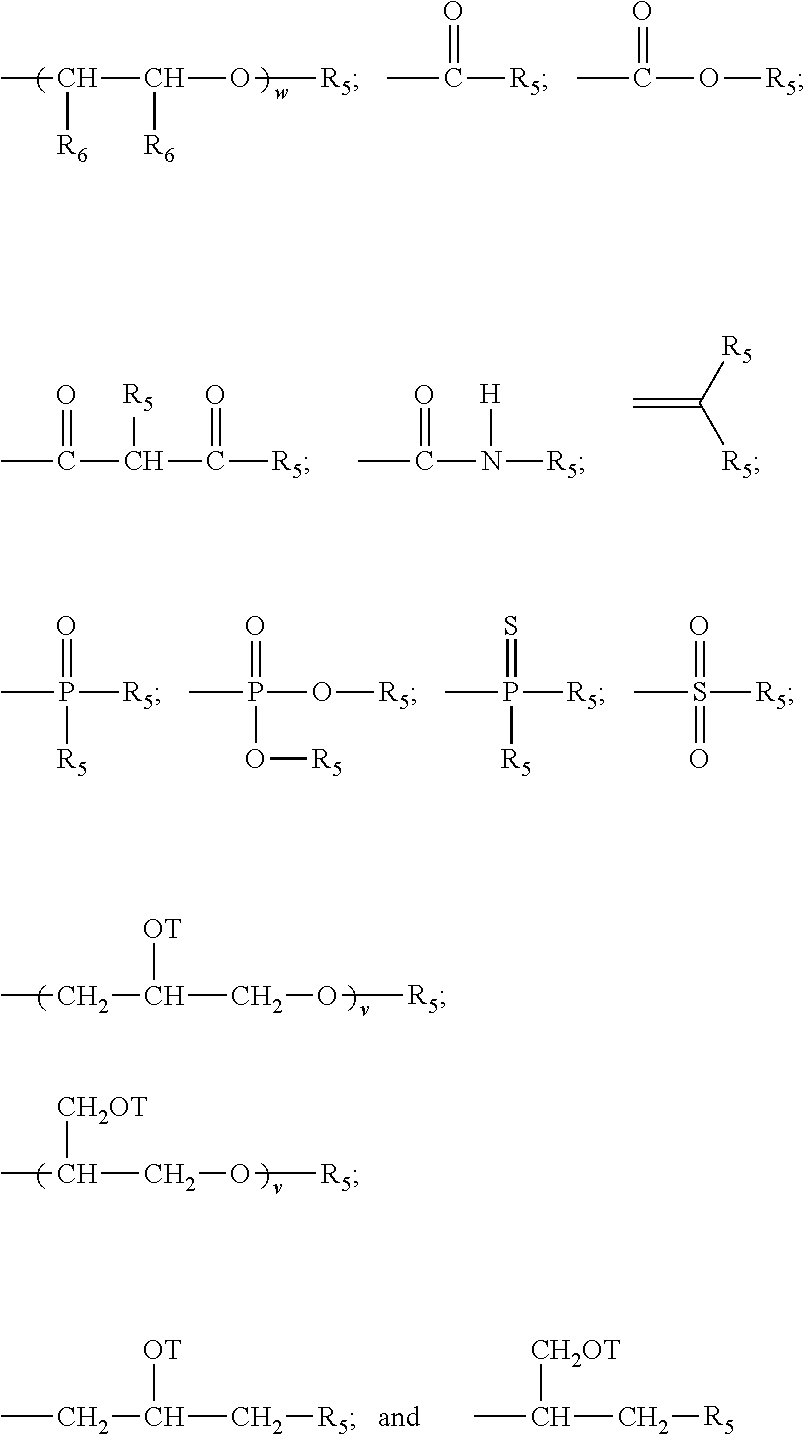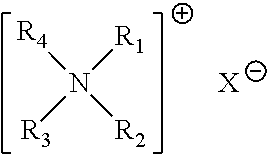Patents
Literature
269results about "Chemical structure search" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
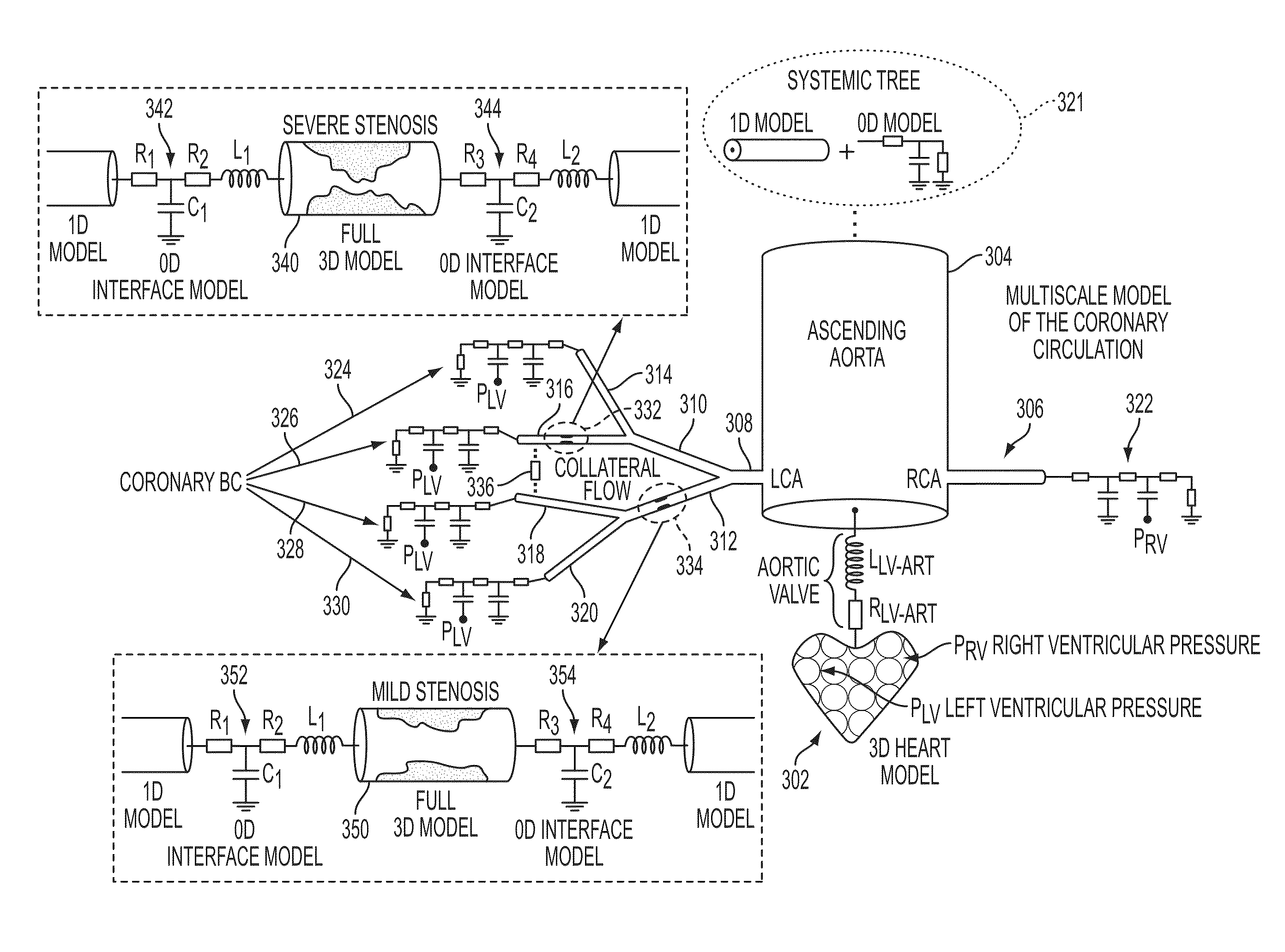
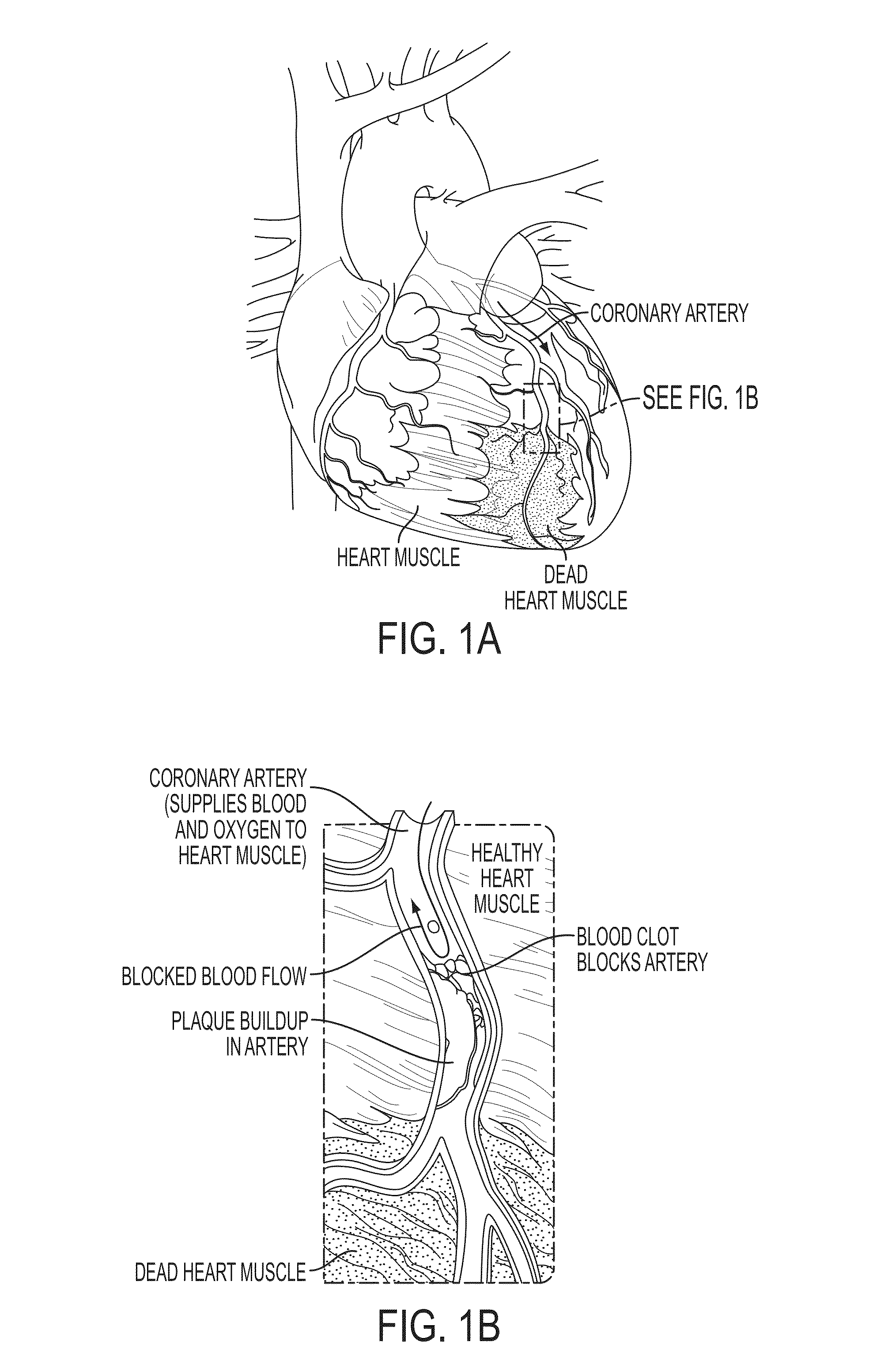
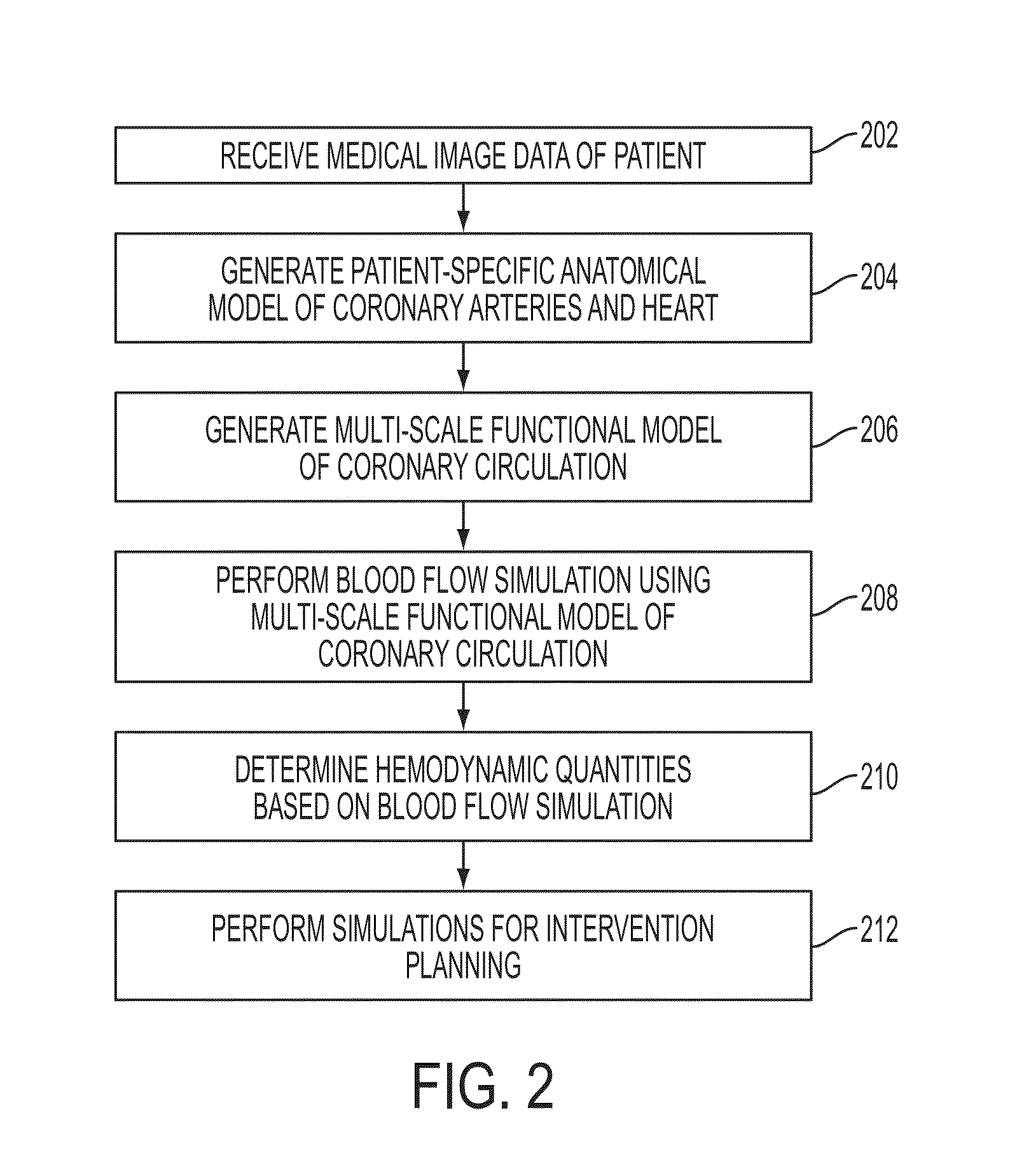
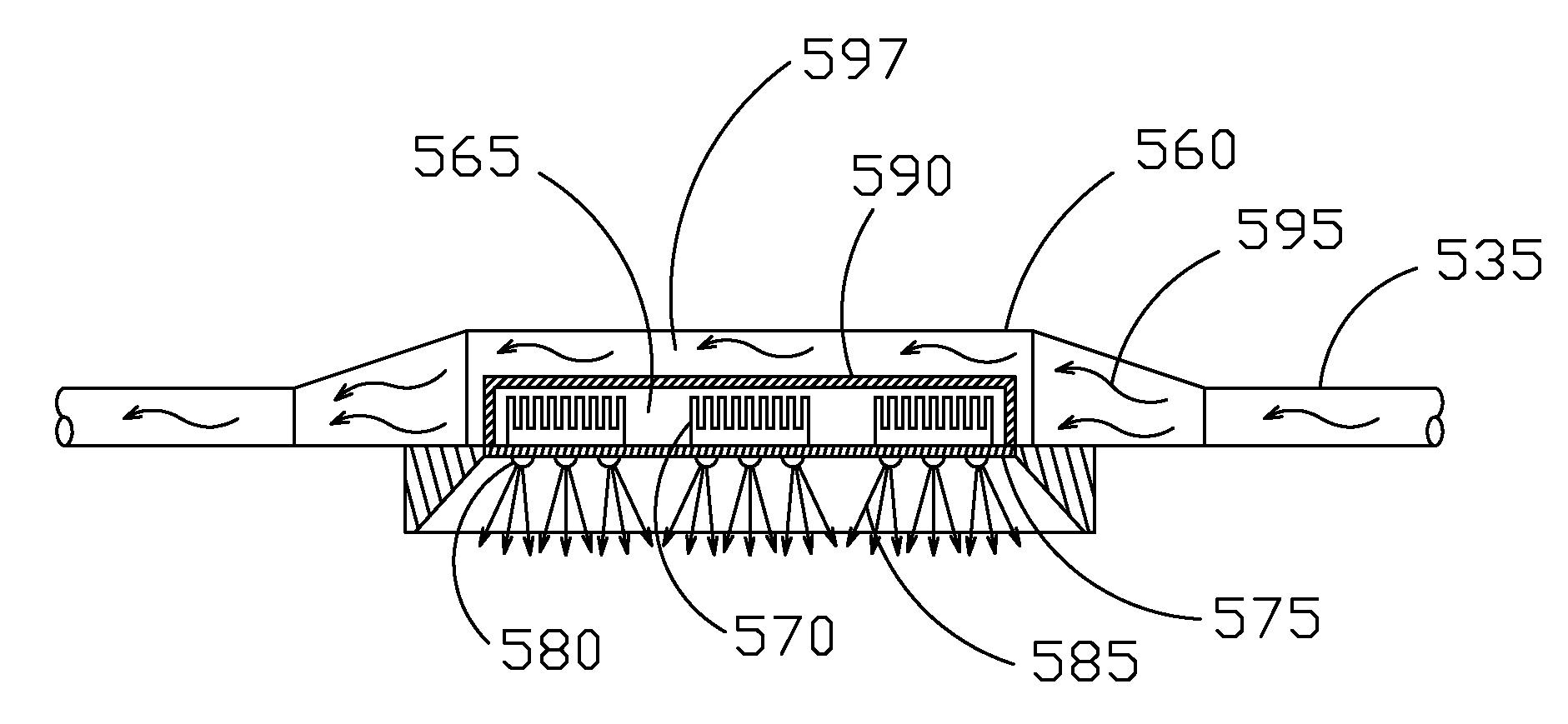
Method and System for Multi-Scale Anatomical and Functional Modeling of Coronary Circulation
ActiveUS20130132054A1Improve predictive performanceImprove clinical managementChemical property predictionChemical structure searchCoronary arteriesIntervention planning
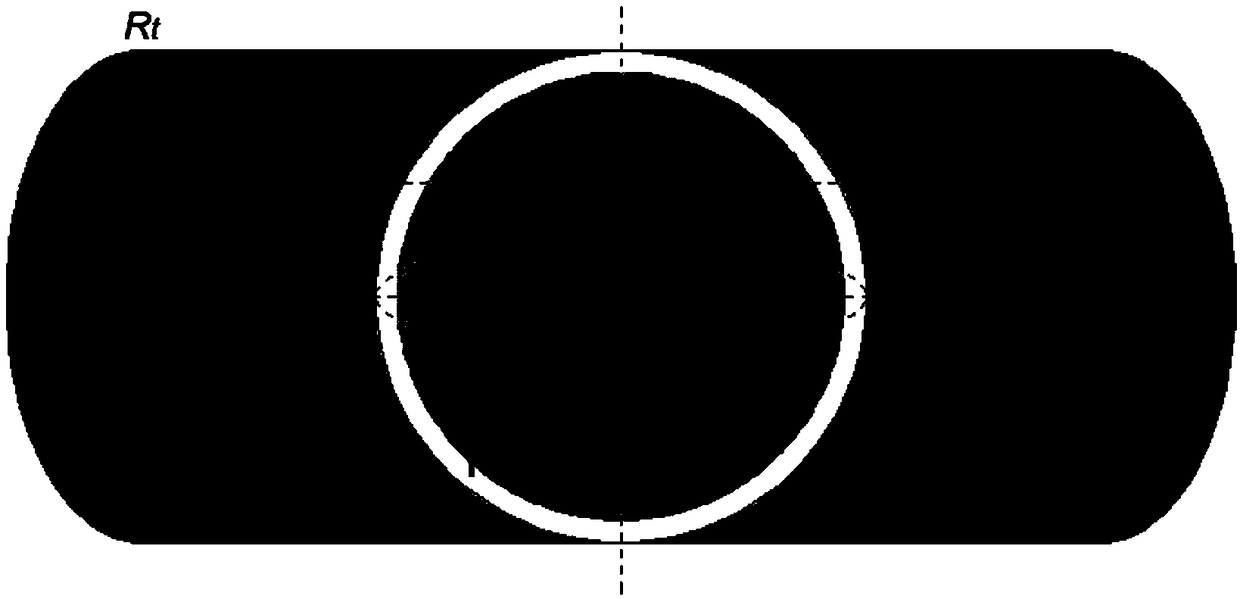
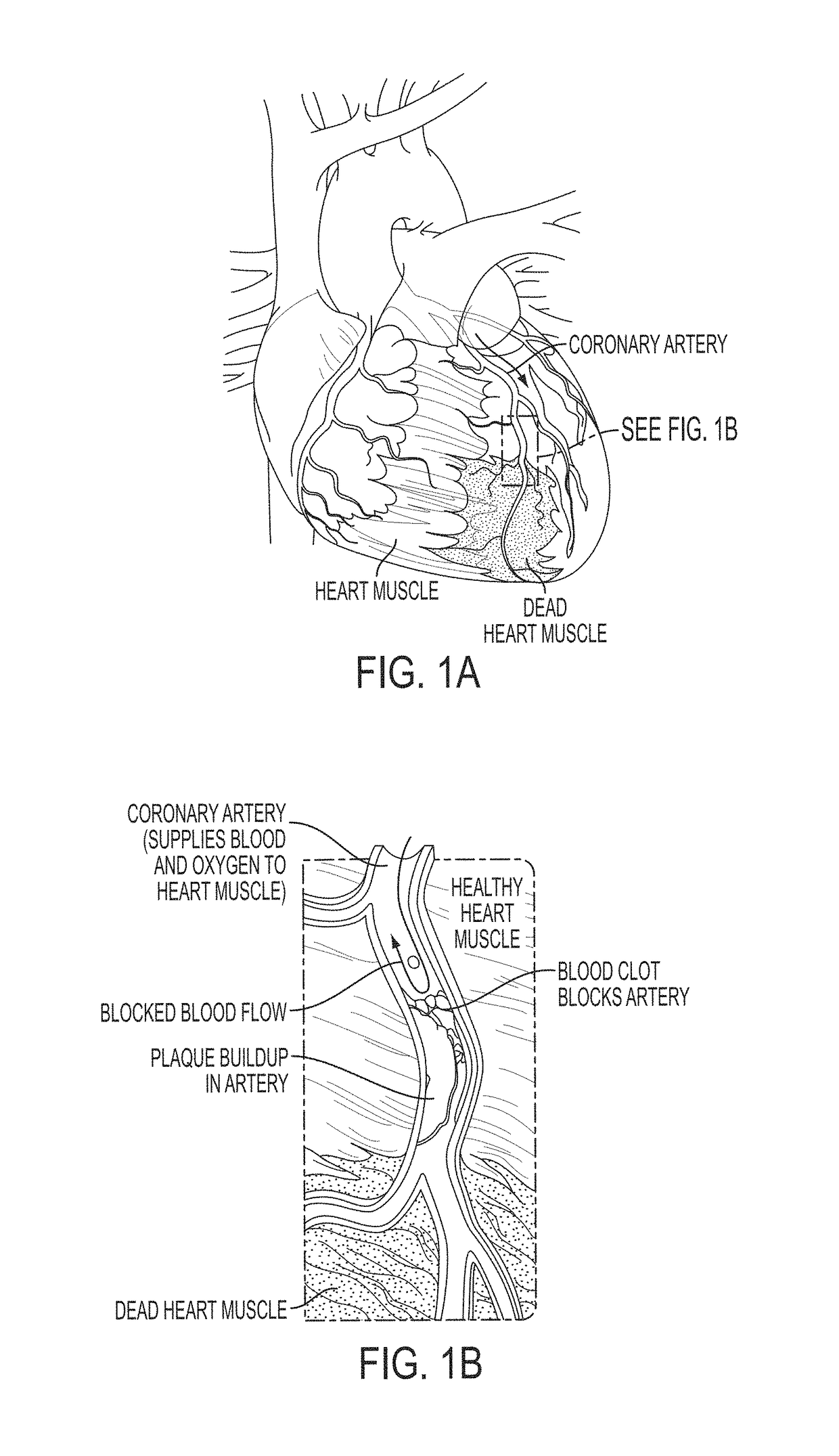
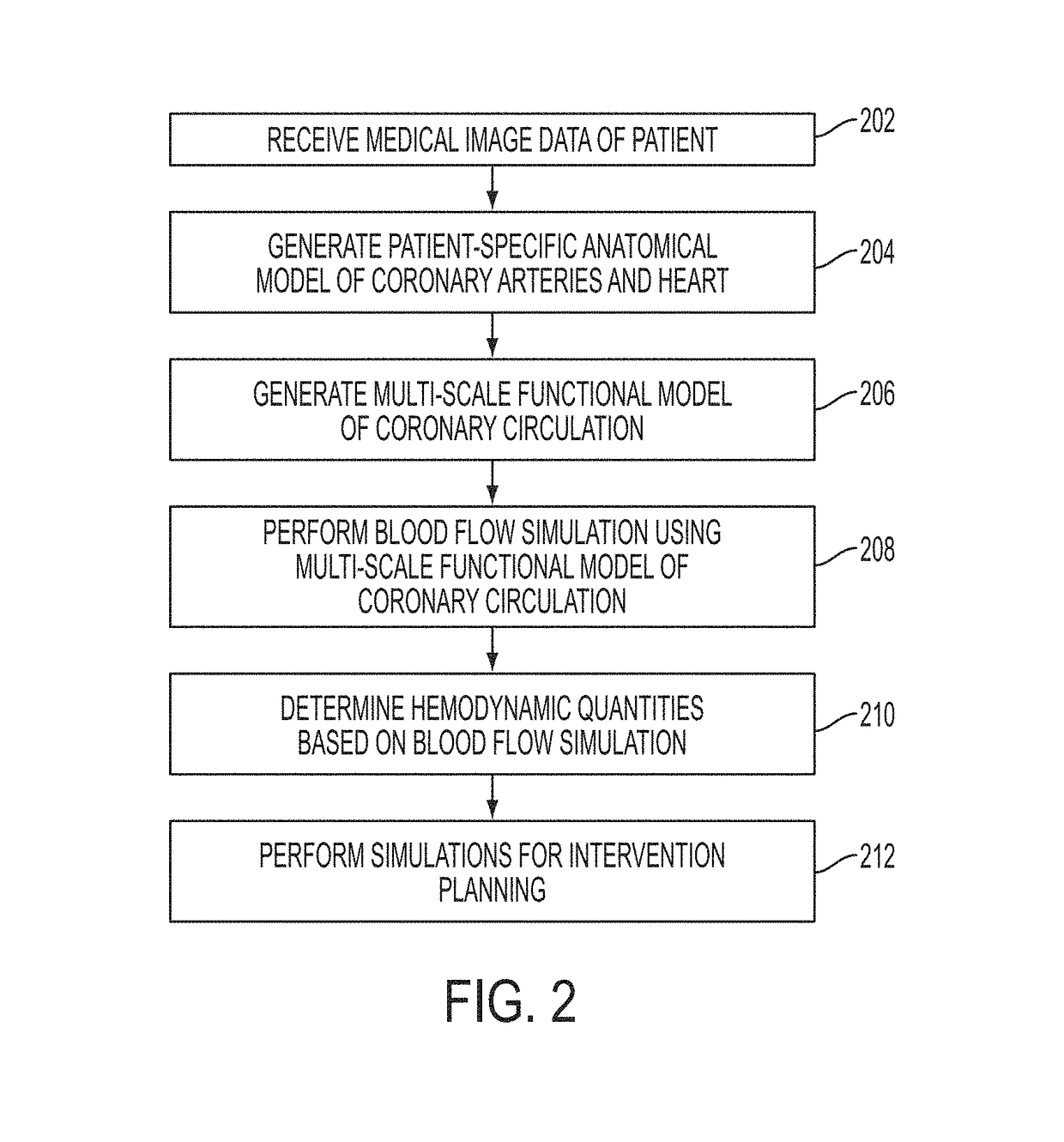
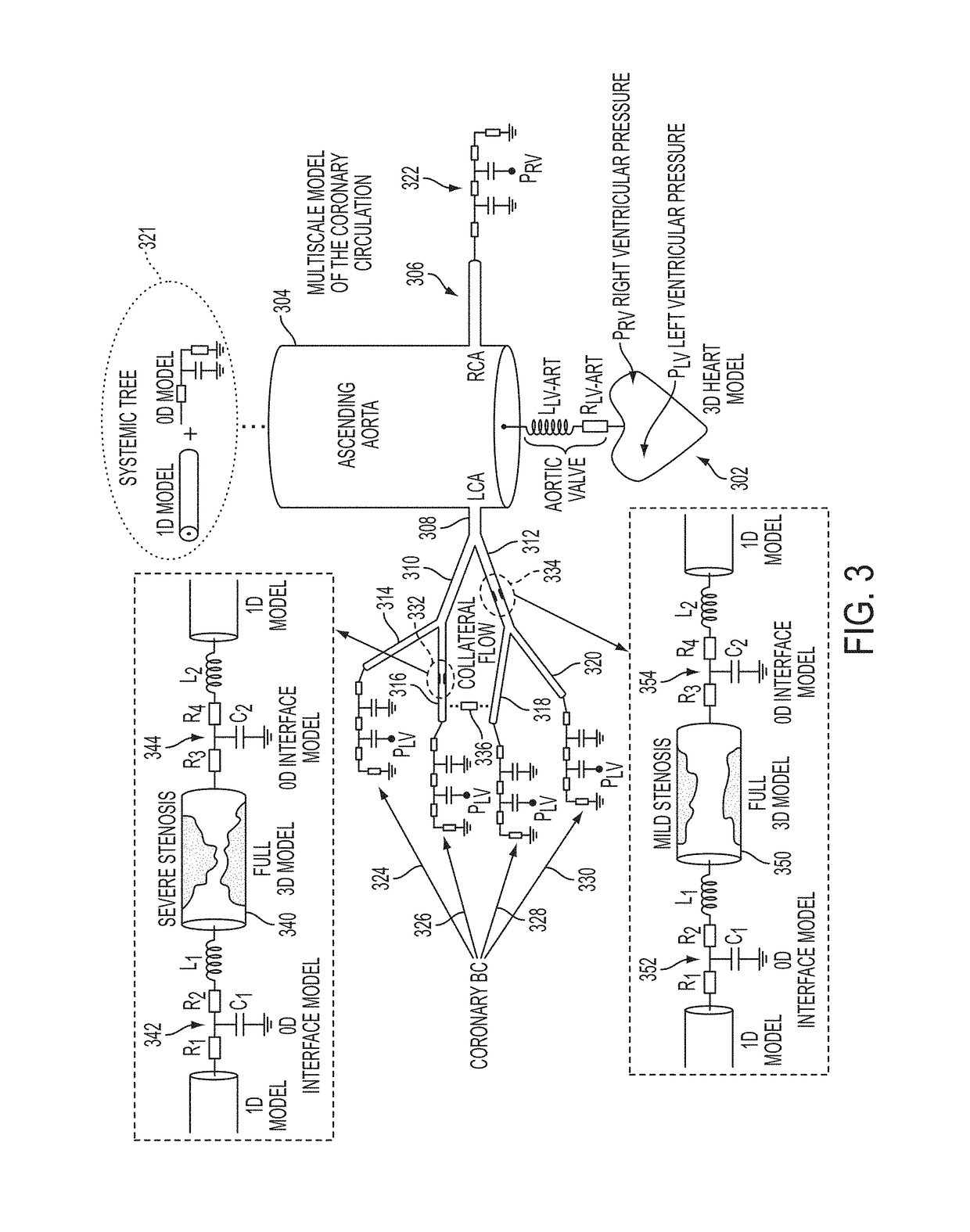
A method and system for multi-scale anatomical and functional modeling of coronary circulation is disclosed. A patient-specific anatomical model of coronary arteries and the heart is generated from medical image data of a patient. A multi-scale functional model of coronary circulation is generated based on the patient-specific anatomical model. Blood flow is simulated in at least one stenosis region of at least one coronary artery using the multi-scale function model of coronary circulation. Hemodynamic quantities, such as fractional flow reserve (FFR), are computed to determine a functional assessment of the stenosis, and virtual intervention simulations are performed using the multi-scale function model of coronary circulation for decision support and intervention planning.
Owner:SIEMENS HEALTHCARE GMBH +1
Text influenced molecular indexing system and computer-implemented and/or computer-assisted method for same
InactiveUS20020087508A1Chemical property predictionMathematical modelsSingular value decompositionAlgorithm
Owner:AXONTOLOGIC
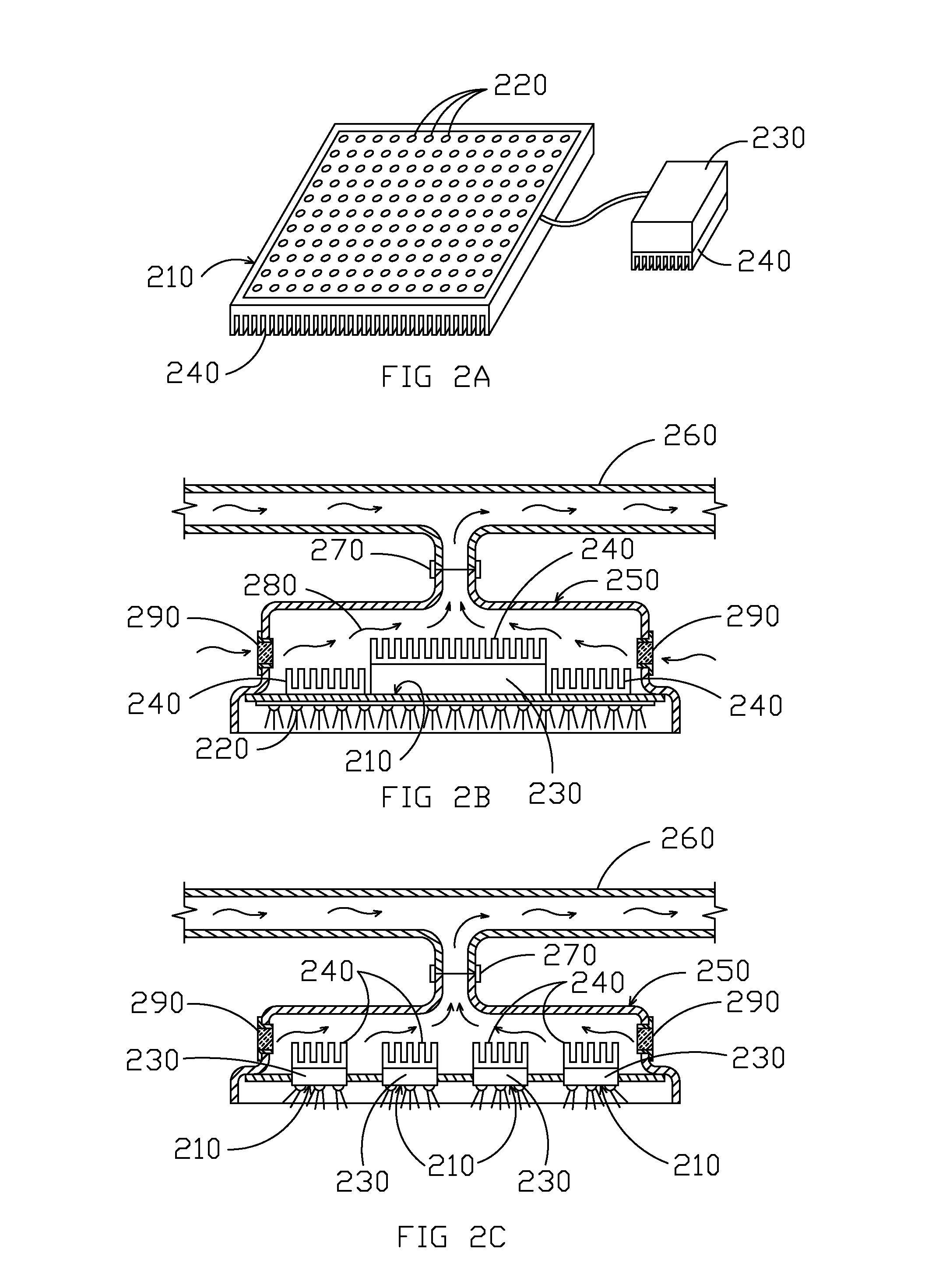
Solid state light fixture with cooling system with heat rejection management
ActiveUS8651704B1Increase operating powerReduce in quantityPlanar light sourcesMechanical apparatusSemiconductor materialsElectrical battery
Systems and methods for solid-state light source heat management are provided. A solid-state light source, e.g. having one or more LEDs, can be mounted within an enclosure. In some embodiments, a heat sink associated with semiconductor material and / or power electronics can be placed on a separate side of the enclosure that is thermally insulated or separated from the state-light source. The enclosure can include a first side or portion that allows light from the solid-state-light source (e.g., LEDs) to pass through to a target area. A fluid transfer conduit may be connected to the second side or portion of the enclosure for passing a fluid (e.g., air, water, coolant, etc.) across the heat sink to transport heat generated by the power electronics and / or semiconductor material away. The heated fluid can then be routed to, as examples, an HVAC system, a vent, water heating systems, heat exchanger systems, and / or storage systems (e.g., batteries).
Owner:MUSCO
System, apparatus, and method for user tunable and selectable searching of a database using a weigthted quantized feature vector
InactiveUS20040006559A1Simplify the search processData processing applicationsDigital data processing detailsFeature vectorChemical compound
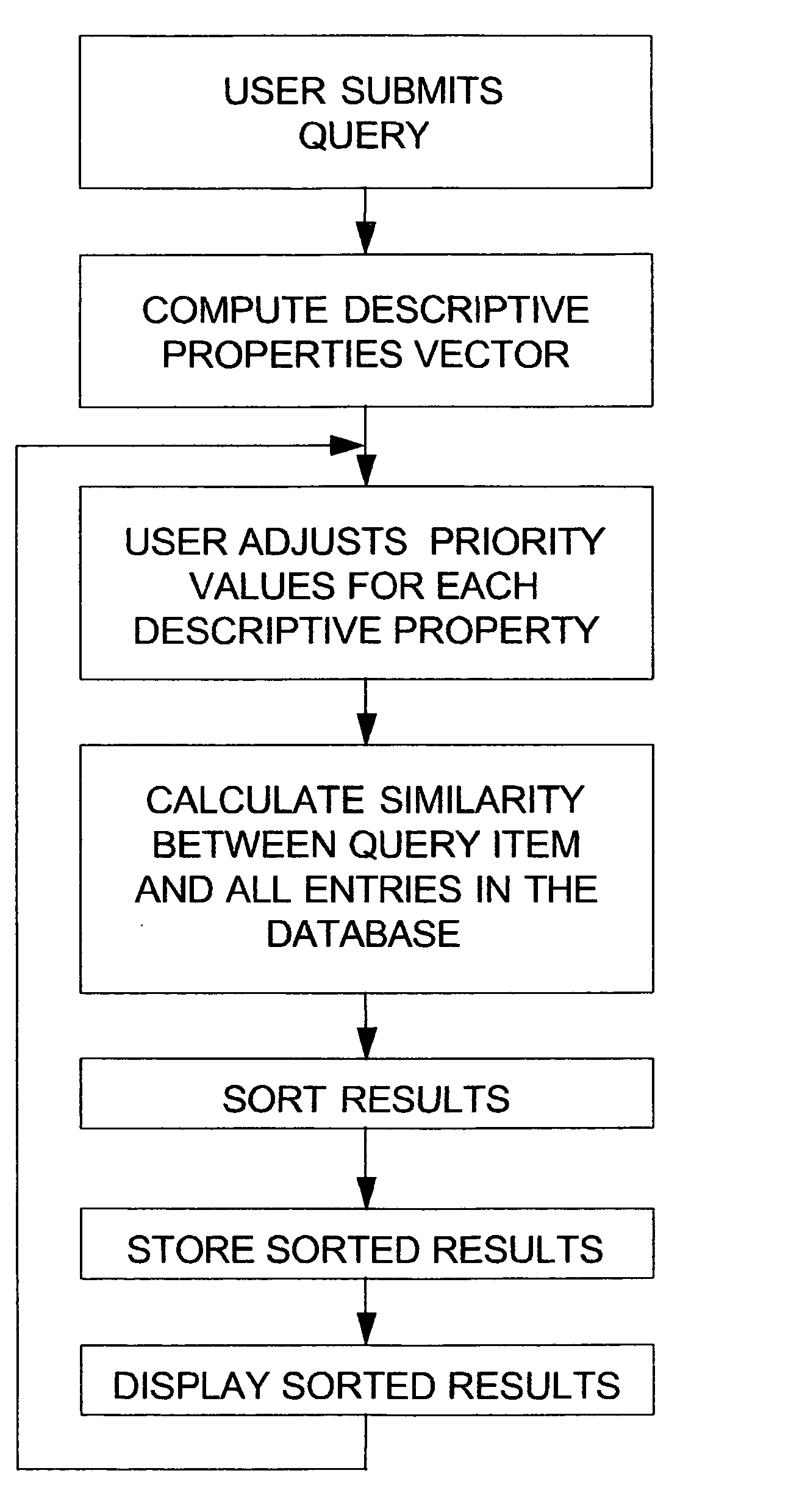
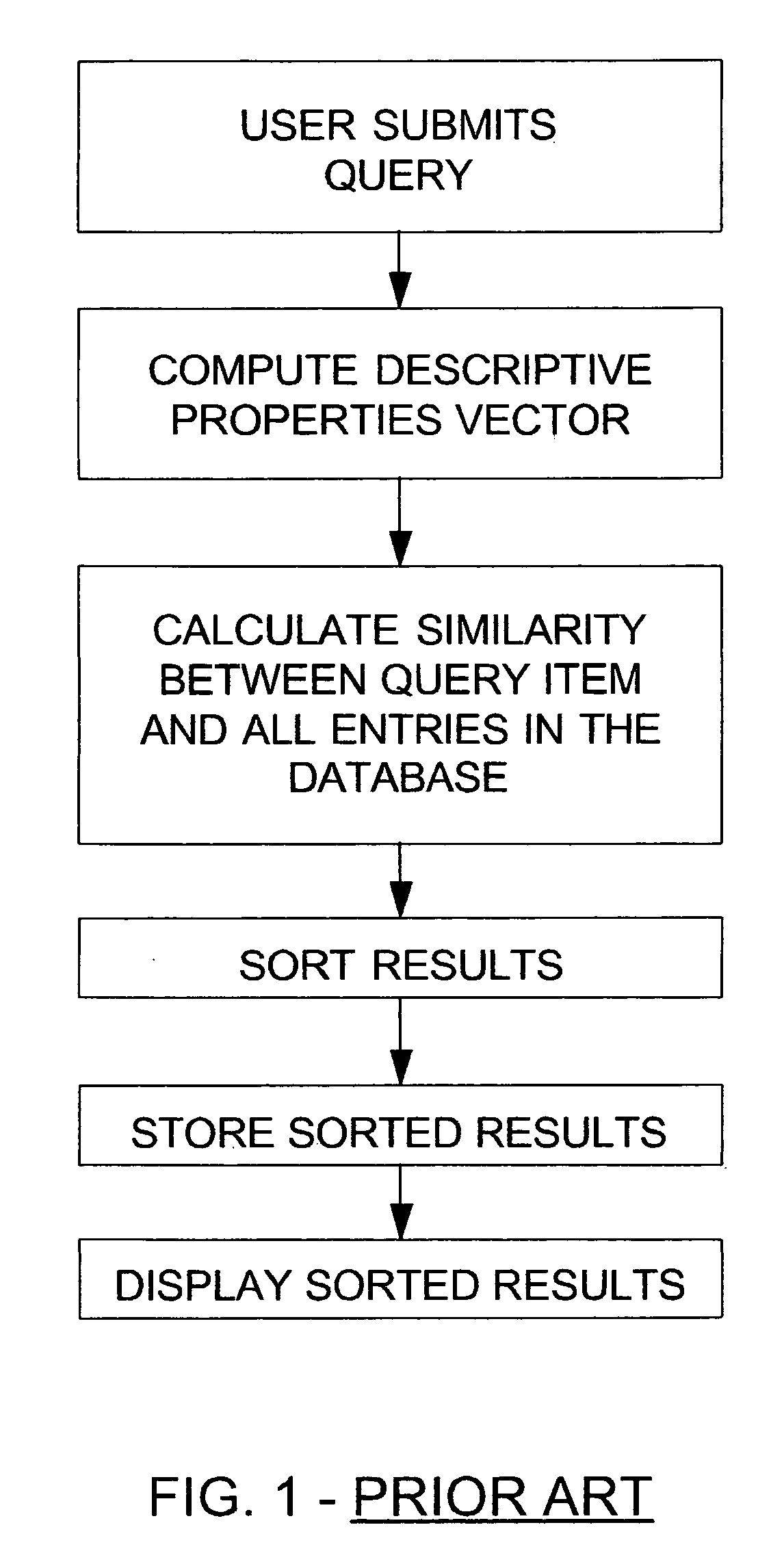
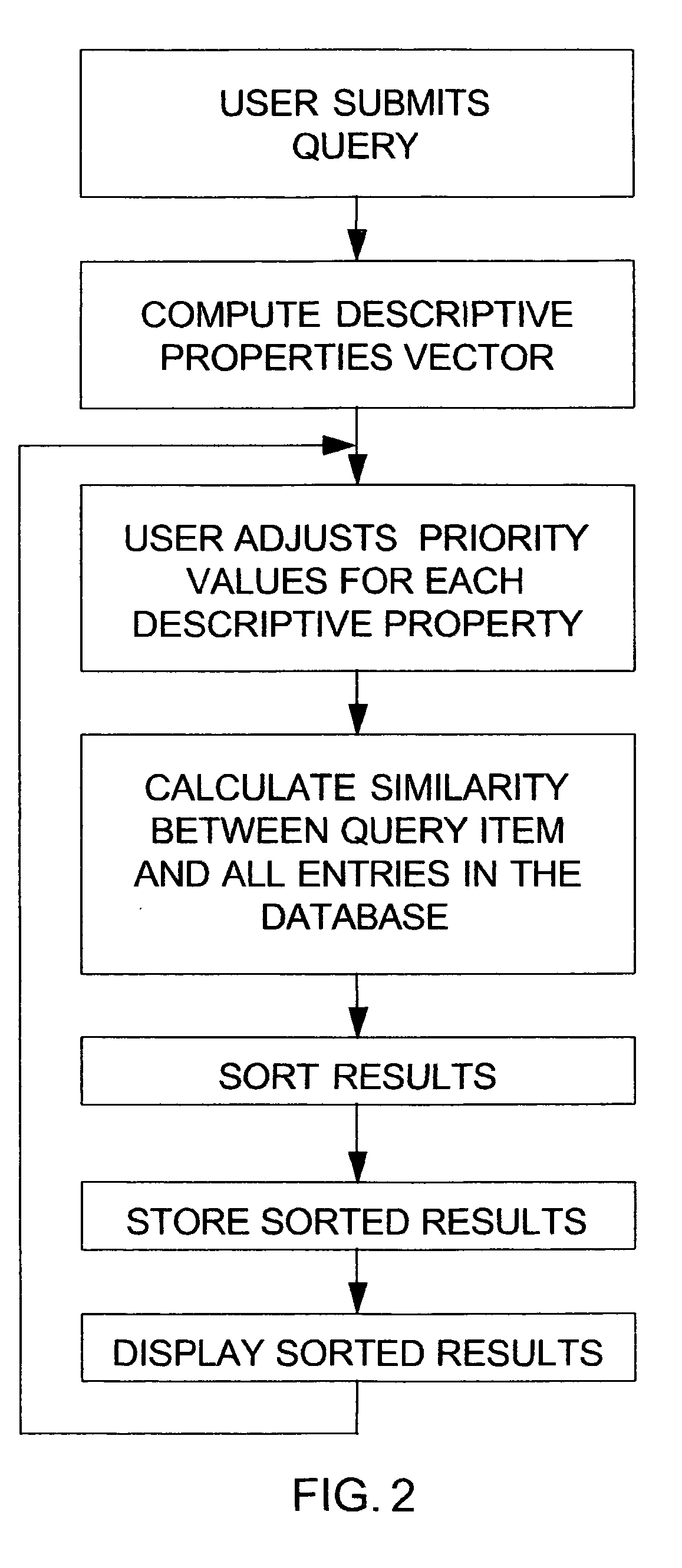
The invention disclosed herein concerns a data processing means for user tunable and selectable searching of a database wherein the data contained therein have associated descriptive properties capable of being expressed in numeric form. A quantized vector representative of the descriptive properties is created for each item in the database. This quantized vector becomes the fingerprint for each data item. The user submits a query item to be matched against the database for similarity. A fingerprint is calculated for the query item. The user may then assign weights to the individual descriptive properties based upon perceived importance. A newly weighted fingerprint for the query item is then compared with the weighted fingerprints for all the data in the database. A list of results sorted in order of decreasing similarity is presented to the user. The user may then change the previously assigned weights and then re-run the similarity search. This may be done as often as necessary to achieve the desired results. The invention describes similarity searching in a generic database. However, this invention is particularly desirable in databases containing chemical compound structure data or biological response screening result data. The process described herein may be run stand alone or as a preliminary screening search in a large database. If used for screening, it can greatly reduce the amount of data required for exactly matching a query item to the data in the database.
Owner:ROW2 TECH
Text influenced molecular indexing system and computer-implemented and/or computer-assisted method for same
InactiveUS6332138B1Chemical property predictionMathematical modelsSingular value decompositionComputer-aided
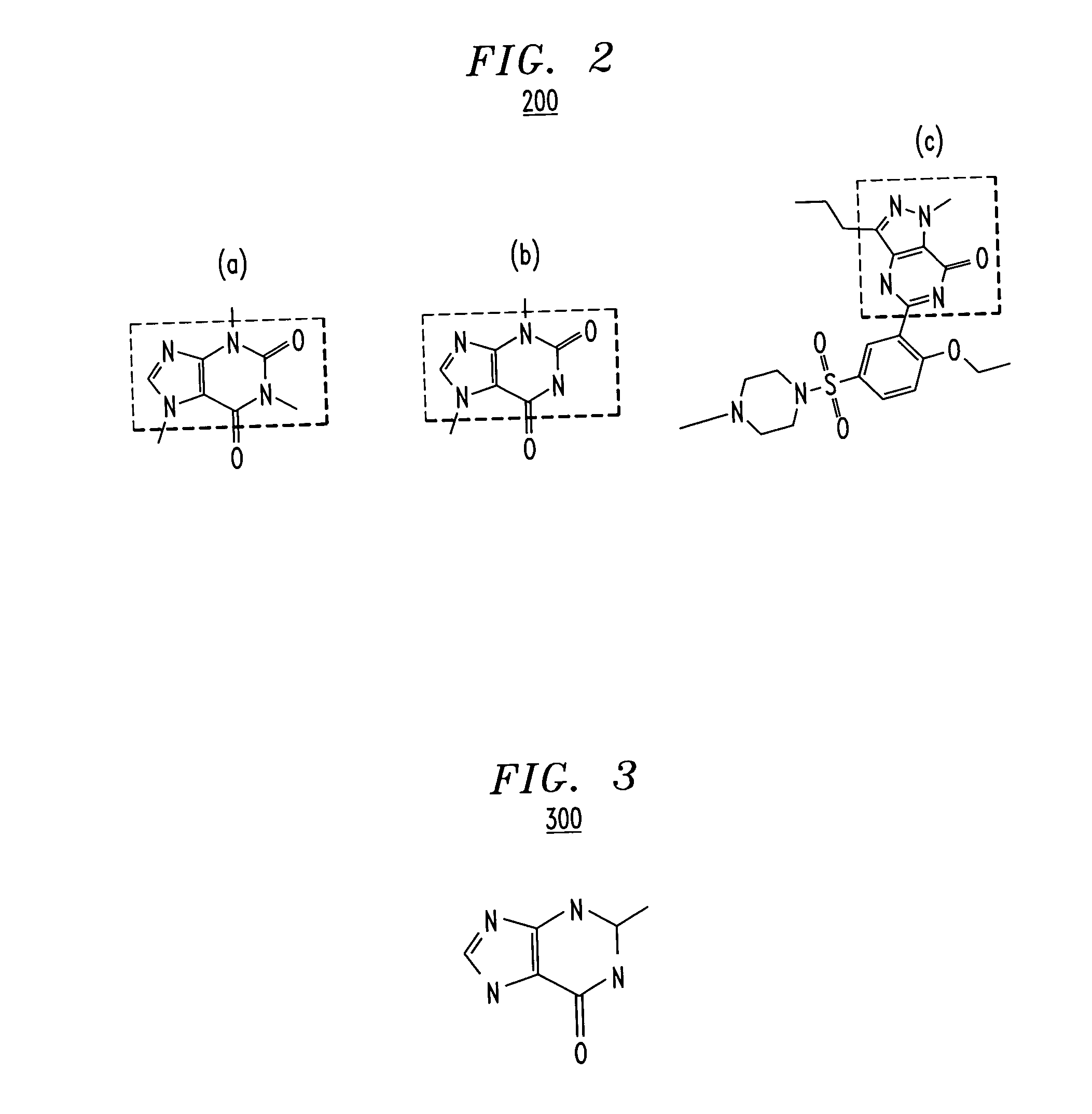
An extension of the vector space model for computing chemical similarity using textual and chemical descriptors is described. The method uses a chemical and / or textual description of a molecule / chemical and a decomposes a molecule / chemical descriptor matrix by a suitable technique such as singular value decomposition to create a low dimensional representation of the original descriptor space. Similarities between a user probe and the textual and / or chemical descriptors are then computed and ranked.
Owner:AXONTOLOGIC
System and method for efficiently performing similarity searches of structural data
InactiveUS20060224562A1Digital data information retrievalChemical structure searchDegree of similarityData mining
Owner:IBM CORP
System, apparatus, and method for user tunable and selectable searching of a database using a weighted quantized feature vector
InactiveUS20060074859A1Simplify the search processData processing applicationsDigital data processing detailsFeature vectorChemical compound
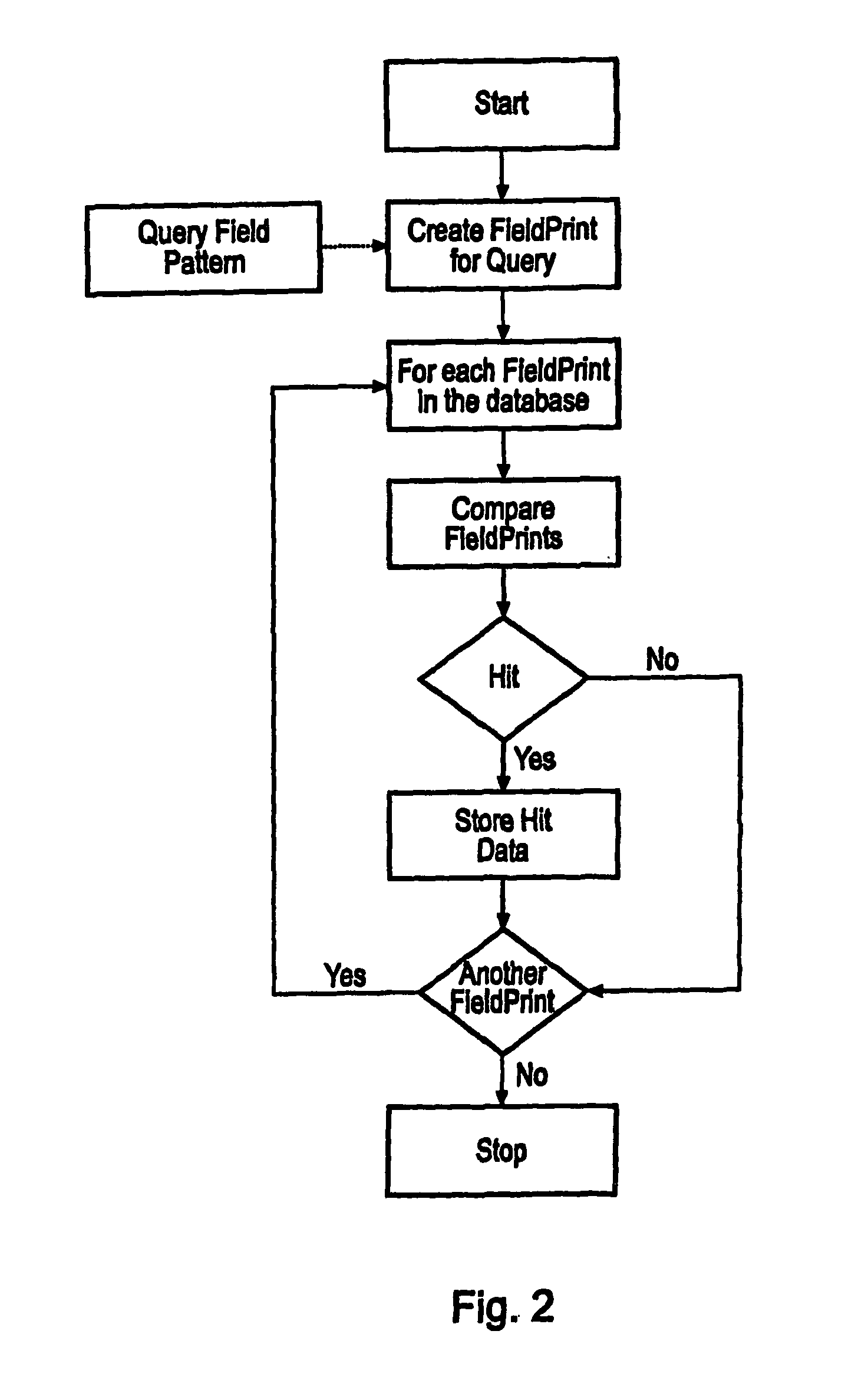
A data processing means for user tunable and selectable (FIG. 2) of a database wherein the data contained therein have associated descriptive properties (FIG. 2) capable of being expressed in numeric form is described. Descriptive property values (FIG. 2) may be standardized numerically to eliminate property value overweighting. A quantized vector (FIG. 2) representative of the descriptive properties is created for each item in the database. This quantized vector becomes the fingerprint for each data item. The user submits a query item to be matched against the database for similarity. A fingerprint is calculated for the query item. The user may then assign weights to the individual descriptive properties based upon perceived importance (FIG. 2). A newly weighted fingerprint for the query item is then compared with the fingerprints for all the data in the database. A list of results is presented to the user (FIG. 2). The user may then change the previously assigned weights and then re-run the similarity search. This may be done as often as necessary to achieve the desired results. Similarity searching in a generic database is described. However, particulary the method is desirable in databases containing chemical compound structure data or biological response screening result data.
Owner:ROW2 TECH INC
Cleaning and/or treatment compositions comprising malodor reduction compositions
Owner:THE PROCTER & GAMBLE COMPANY
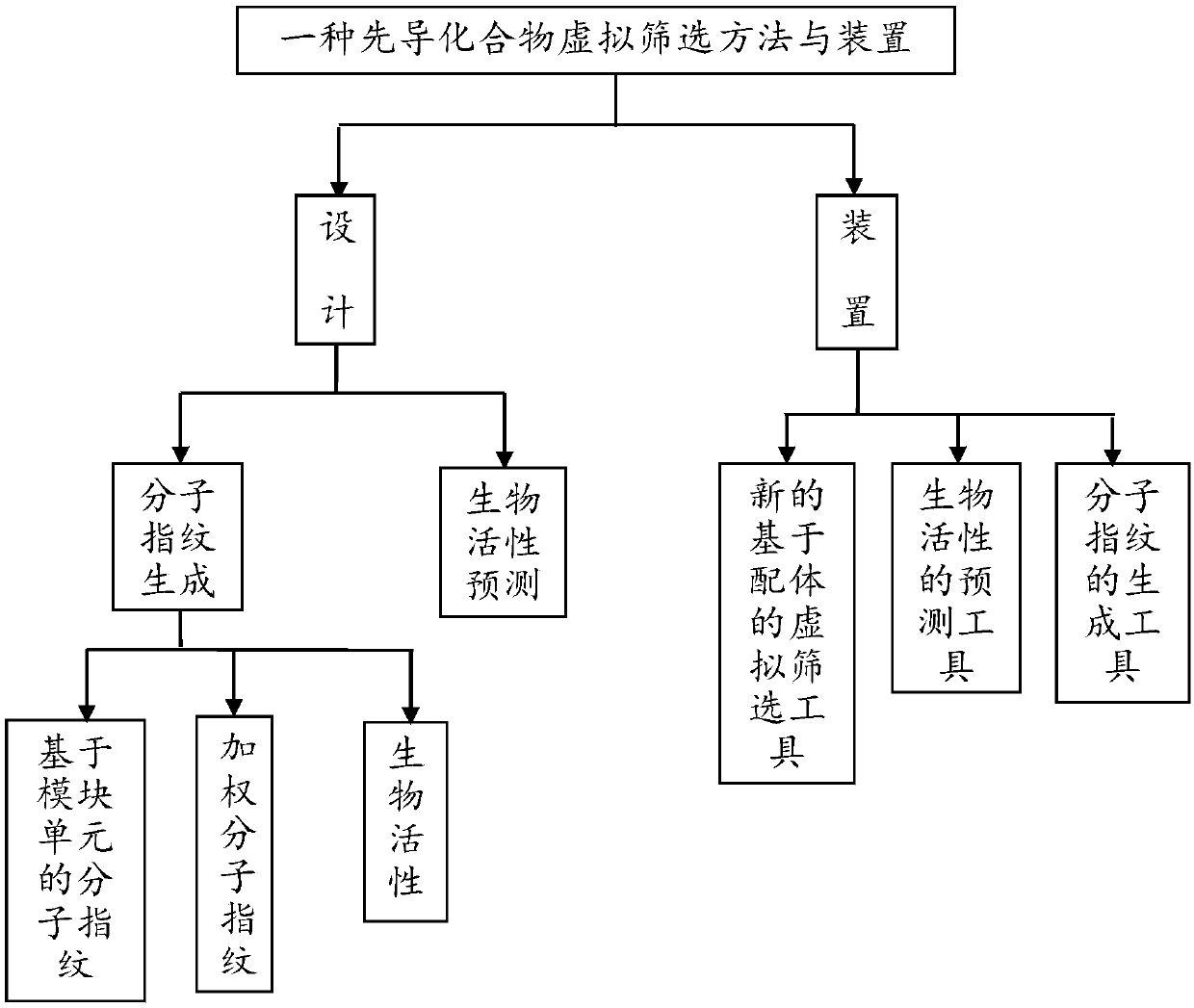
Lead compound virtual screening method and device
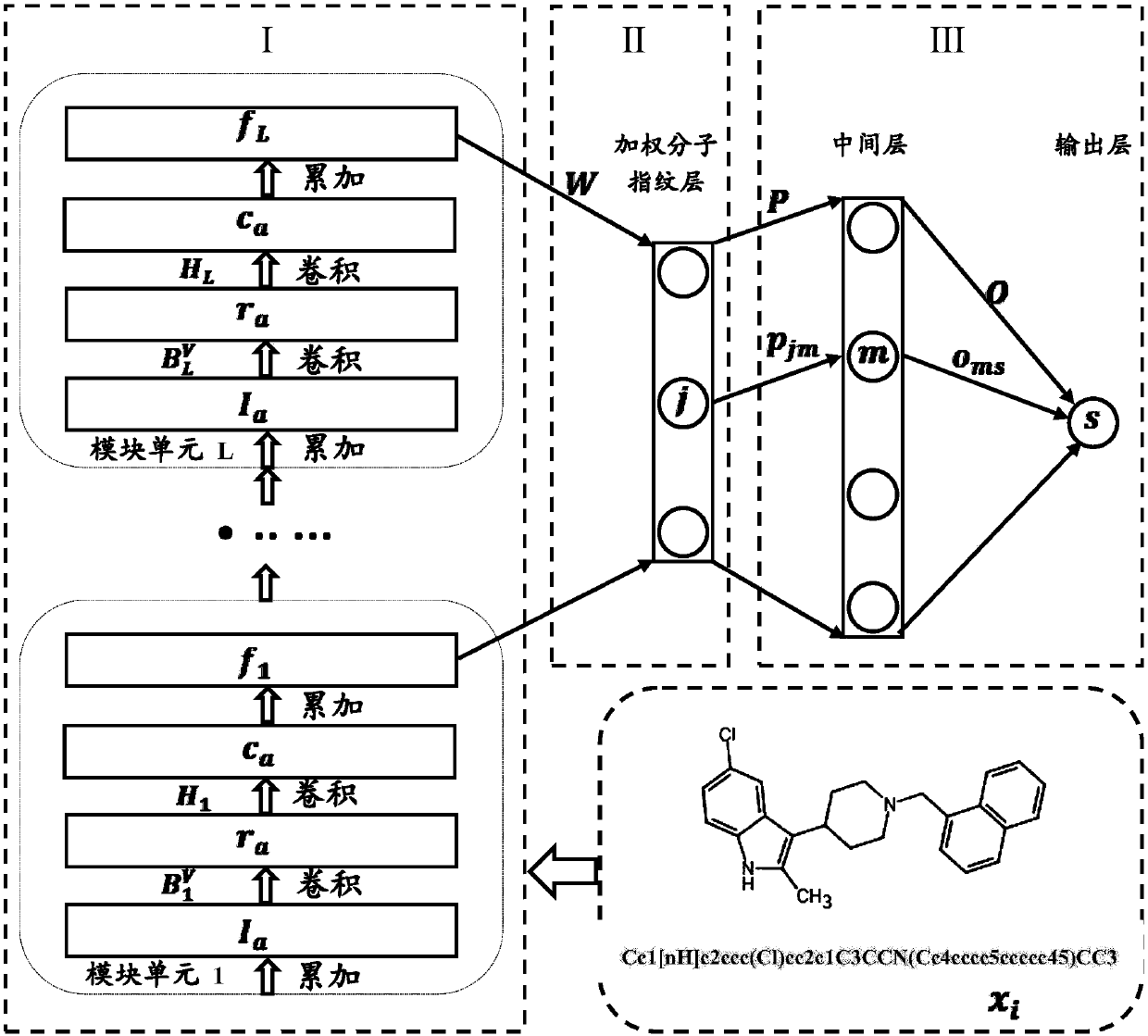
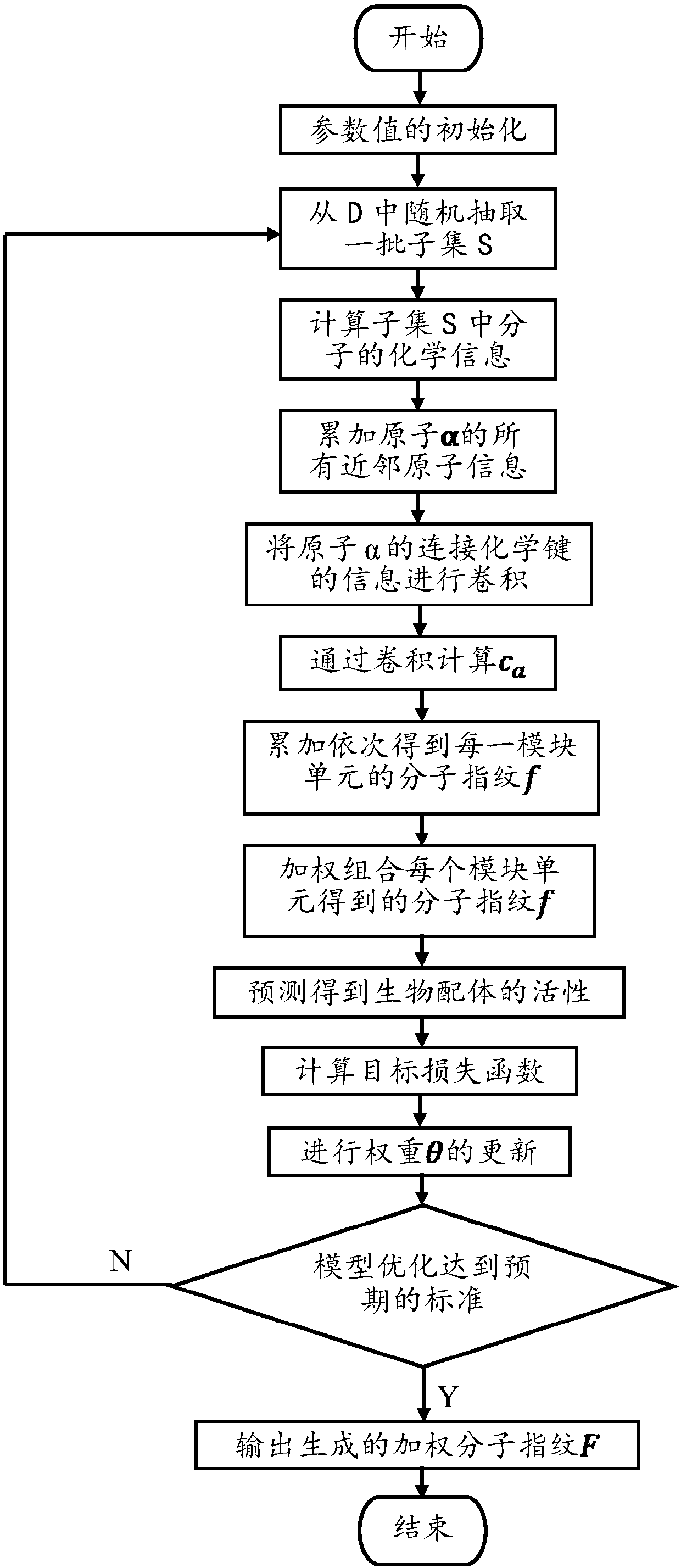
ActiveCN107862173AImprove performanceChemical property predictionChemical structure searchVirtual screeningMedicine
The invention discloses a lead compound virtual screening method and device. The method includes the steps of generation of molecular fingerprints of lead compounds on drug targets and bioactivity prediction of interaction between the lead compounds and the drug targets. The generation of the molecular fingerprints includes a molecular fingerprint part based on a module unit, a weighting molecularfingerprint part and a bioactivity part. During the bioactivity prediction, ligand molecular fingerprints and bioactivity values are utilized to serve as input of a random forest regression model, and a prediction model is constructed. Additionally, the device includes a universal tool for virtual screening on the basis of ligands, a prediction tool for the bioactivity generated when the lead compounds take effects on the drug targets, and a generation tool of the molecular fingerprints of the lead compounds on the drug targets. At present, molecular fingerprints which are excellent in performance and are used for the bioactivity prediction are often greater in length, and however, by adopting a designed deep learning algorithm, molecular fingerprints which are excellent in performance and smaller in length can be generated so that the best bioactivity prediction model of drug target ligands can be obtained.
Owner:NANJING UNIV OF POSTS & TELECOMM
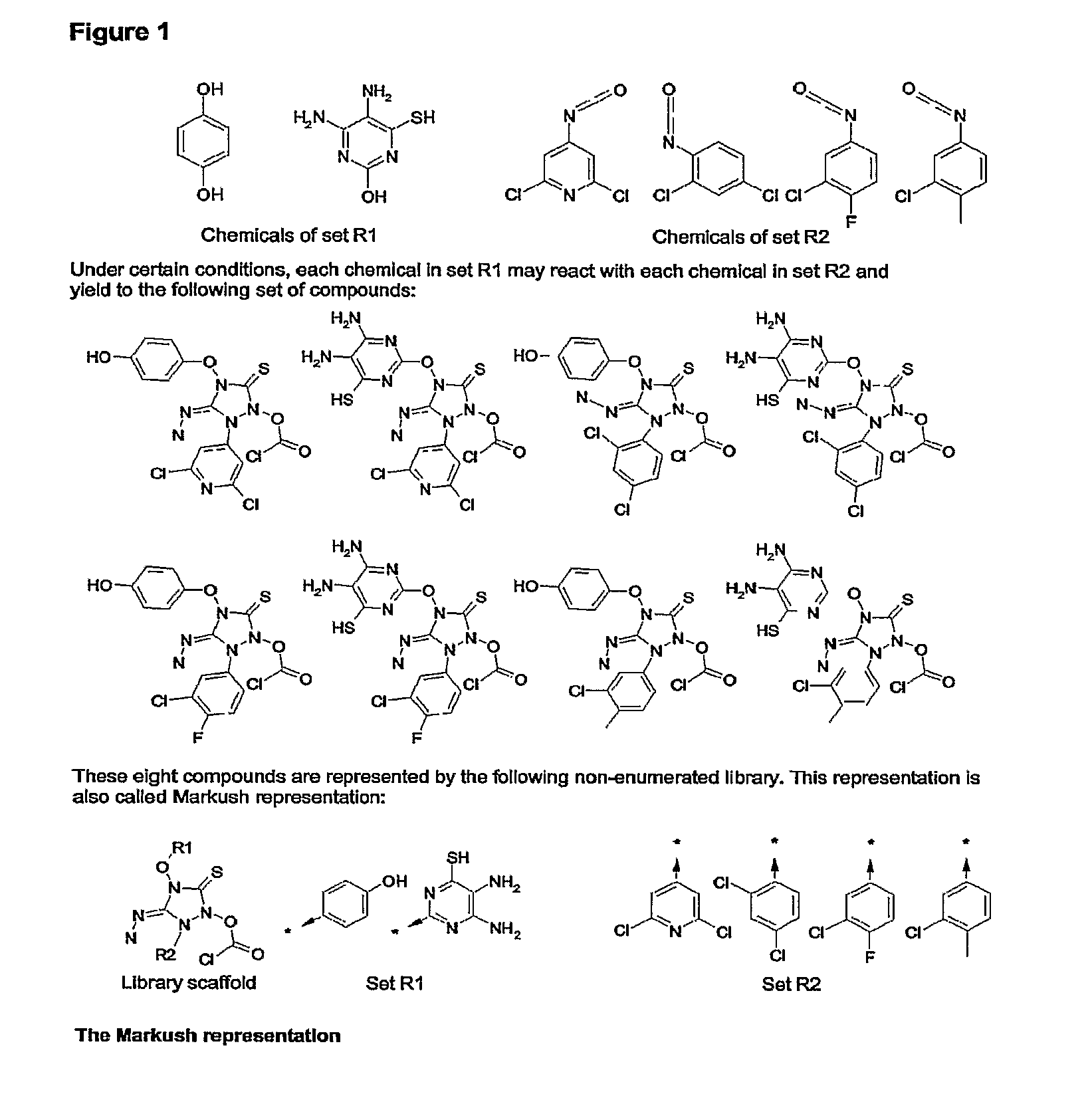
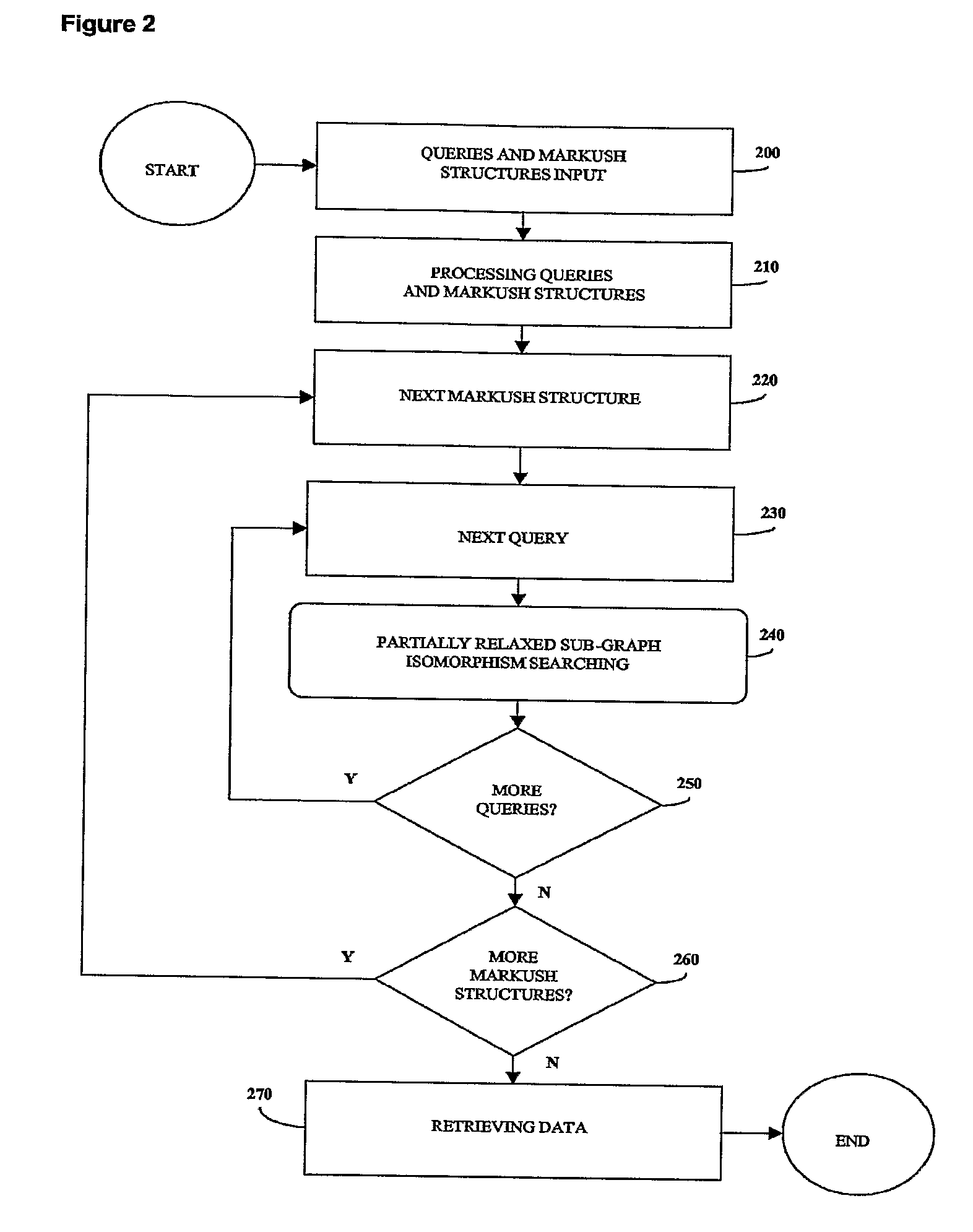
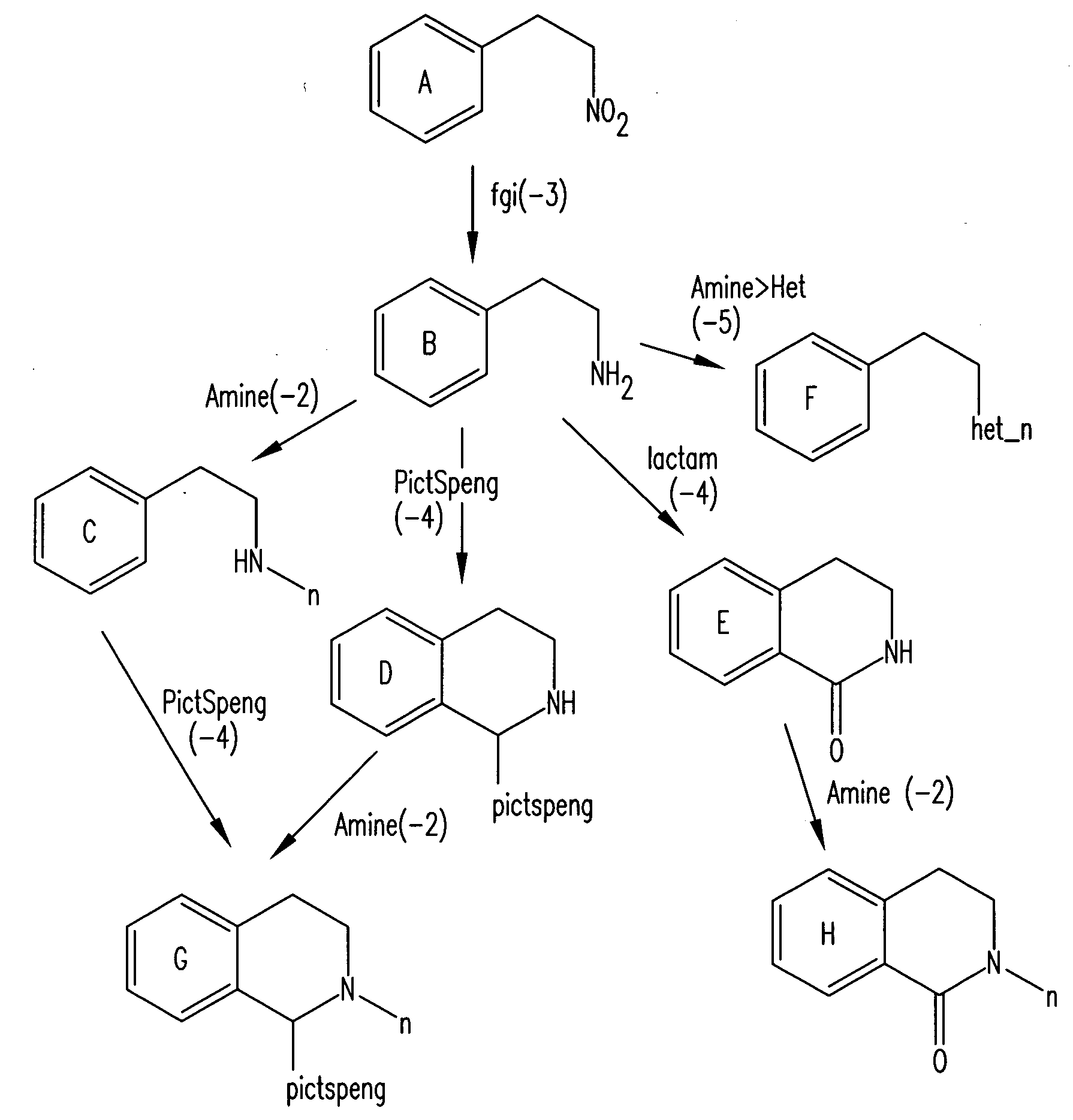
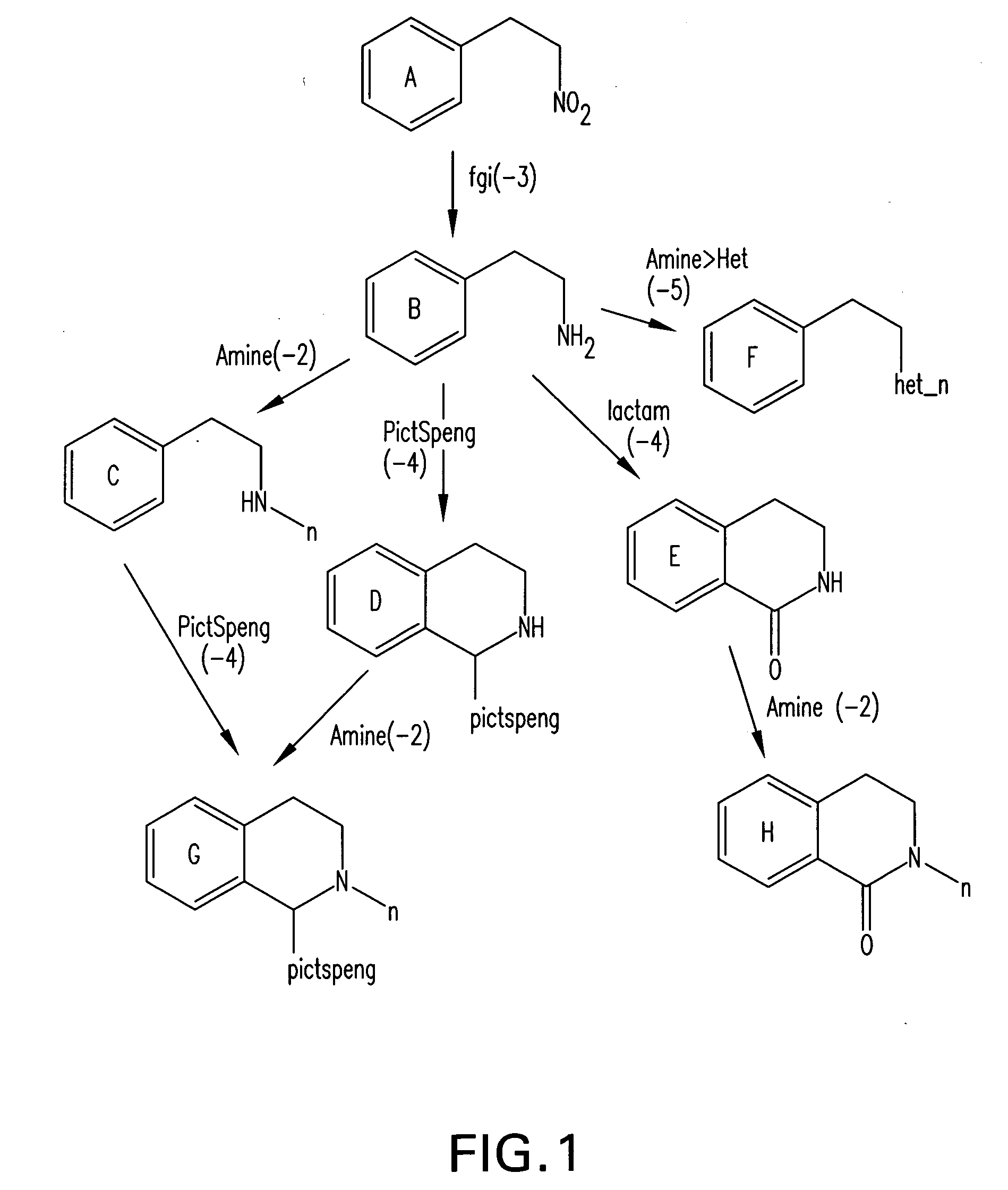
Method for fast substructure searching in non-enumerated chemical libraries
InactiveUS20070260583A1Reduce the required powerRetrieves hits very fastlyChemical structure searchLibrary screeningChemical libraryComputer science
Owner:LAB SERONO SA
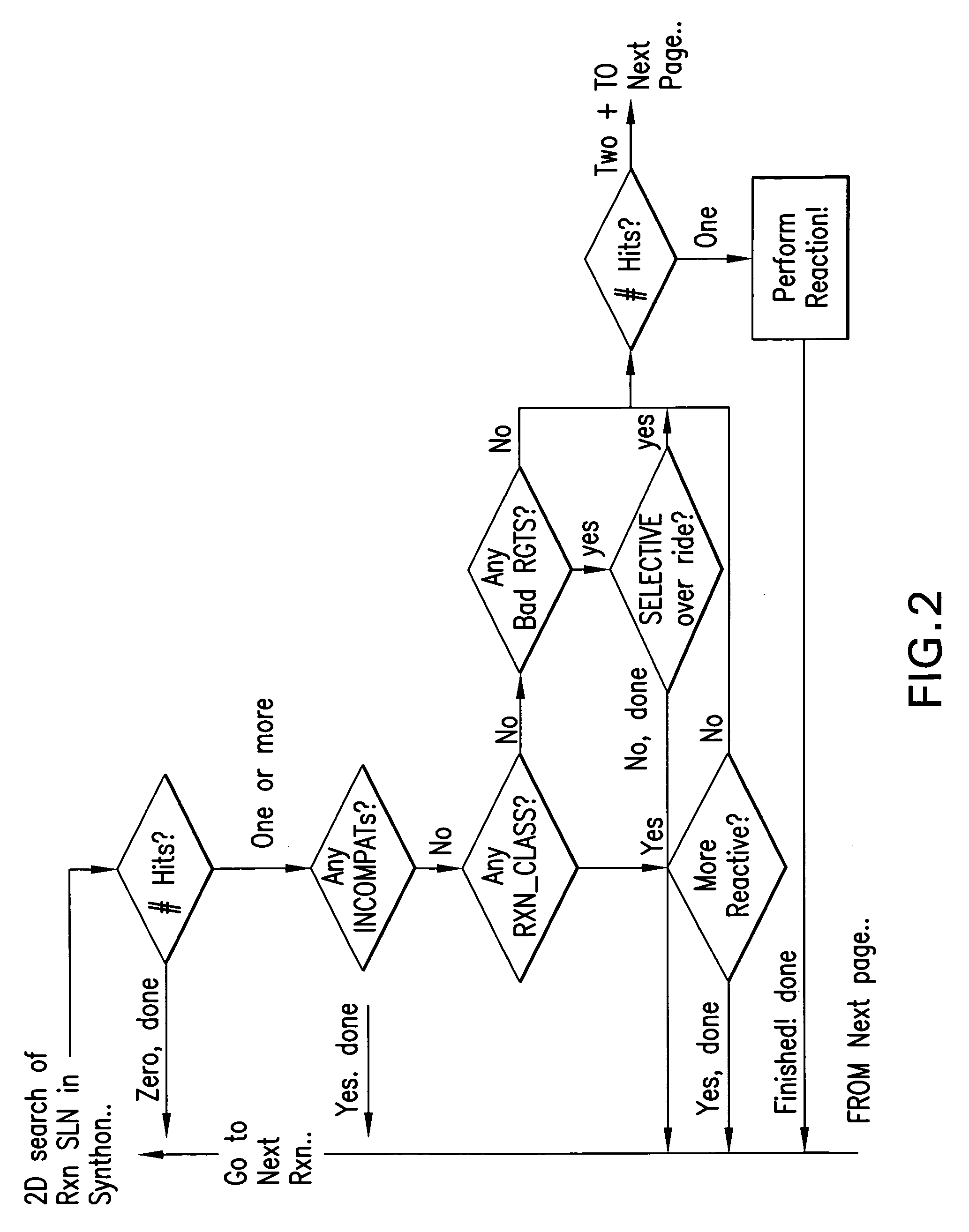
Forward synthetic synthon generation and its useto identify molecules similar in 3 dimensional shape to pharmaceutical lead compounds
ActiveUS20080172216A1Increase richnessIncrease diversityMolecular designChemical structure searchChemical reactionSynthesis methods
A forward synthetic method is described that utilizes recursive application of established organic chemical reactions to derive more complex synthons from available reagents than are available from the reagent synthons themselves. The product of each reaction serves as the starting point for further reactions thereby permitting the generation of multiple complex molecular structures. This synthon generation procedure typically yields 20 ? 30 new structures within the limits of easily accessible syntheses based upon each starting reagent. More complex syntheses yield even more structures. The generated synthons are characterized with a molecular structural descriptor possessing a neighborhood property and can be further characterized with features. The synthons are searched for three dimensional shape and feature similarity to molecular fragments derived from query molecules, typically pharmacological molecules of interest. Identified synthons can be assembled into molecules possessing the same three dimensional shape and likely activity as the molecule of interest.
Owner:CRAMER RICHARD D +1
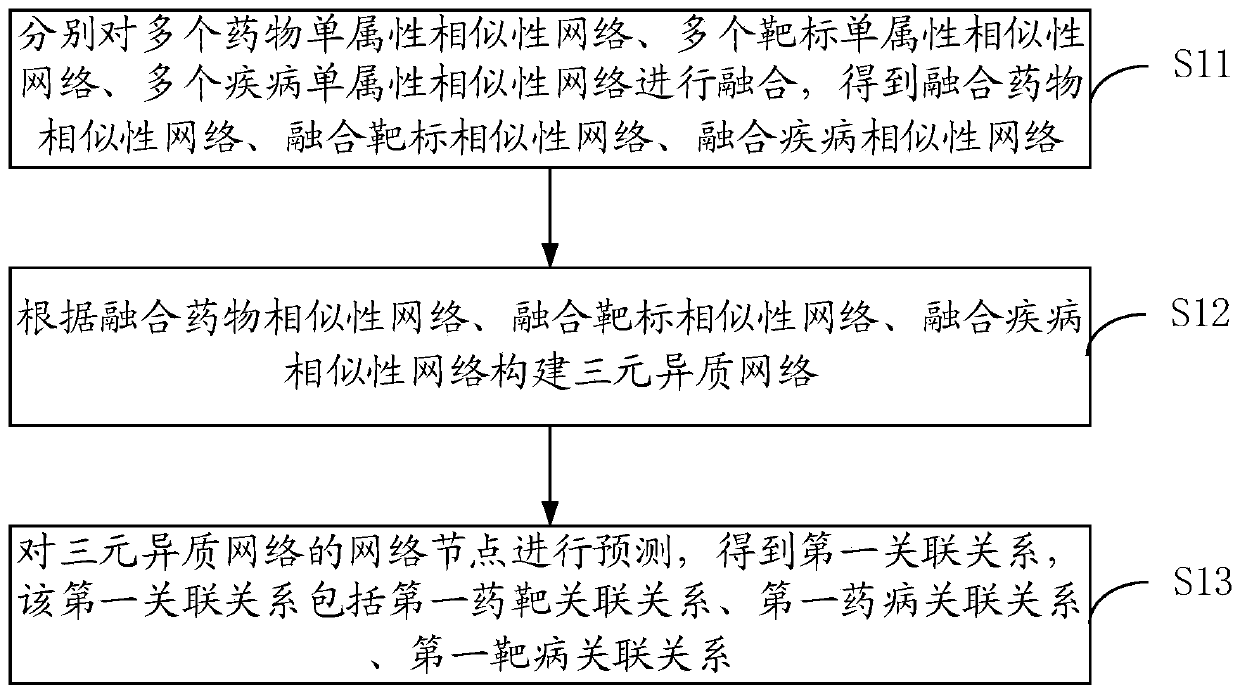
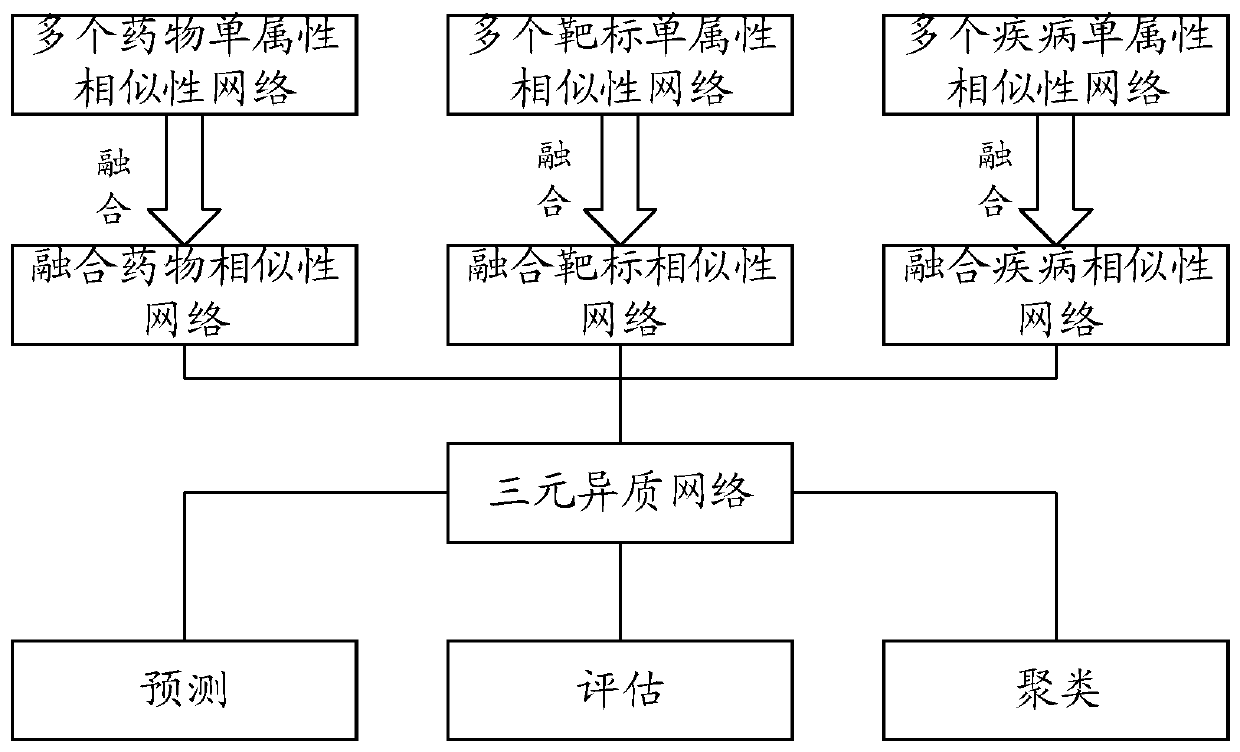
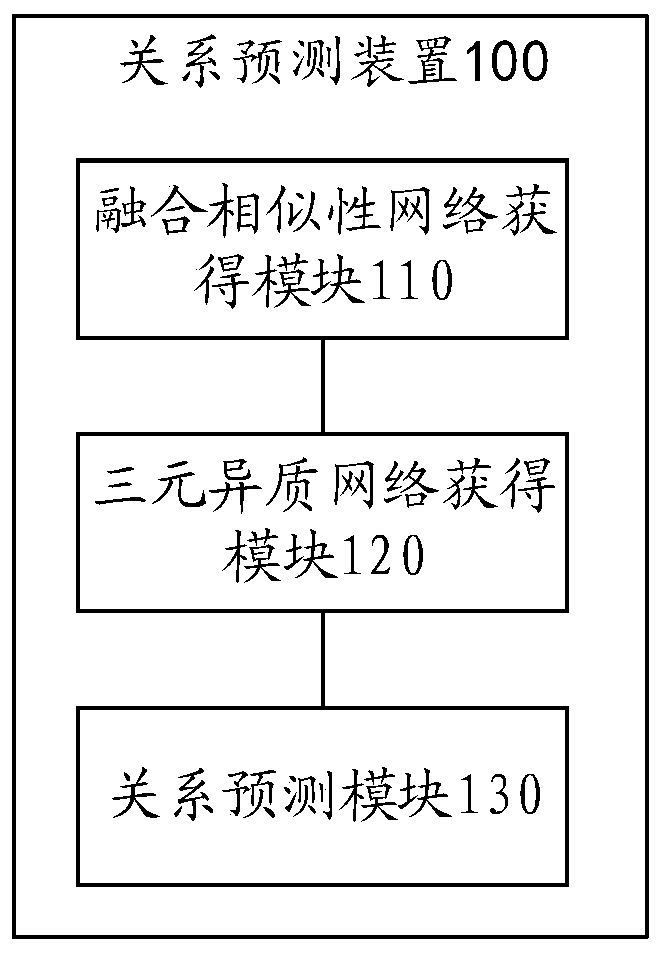
Relation predicating method, device and electronic equipment
ActiveCN109712678AAffect accuracyDigMolecular designChemical structure searchHeterogeneous networkData mining
An embodiment of the invention provides a relation predicating method, a device and electronic equipment. The relation predicating method comprises the steps of fusing a plurality of medicament single-attribute similarity networks, a plurality of target single-attribute similarity network and a plurality of disease single-attribute similarity networks, thereby obtaining a fused medicament similarity network, a fused target similarity network and a fused disease similarity network; constructing a three-element heterogeneous network according to the fused medicament similarity network, the fusedtarget similarity network and the fused disease similarity network; predicting the network node of the three-element heterogeneous network, and obtaining a first association relation which comprisesa first medicament-target association relation, a second medicament-disease association relation and a first target-disease association relation. Therefore medicament-target-disease three-element association relation information can be sufficiently mined. The similarity networks of multiple attributes of medicament, disease and target are fused. Accuracy reduction of the predication result causedby eccentricity caused by a single attribute is prevented.
Owner:ACADEMY OF MILITARY MEDICAL SCI
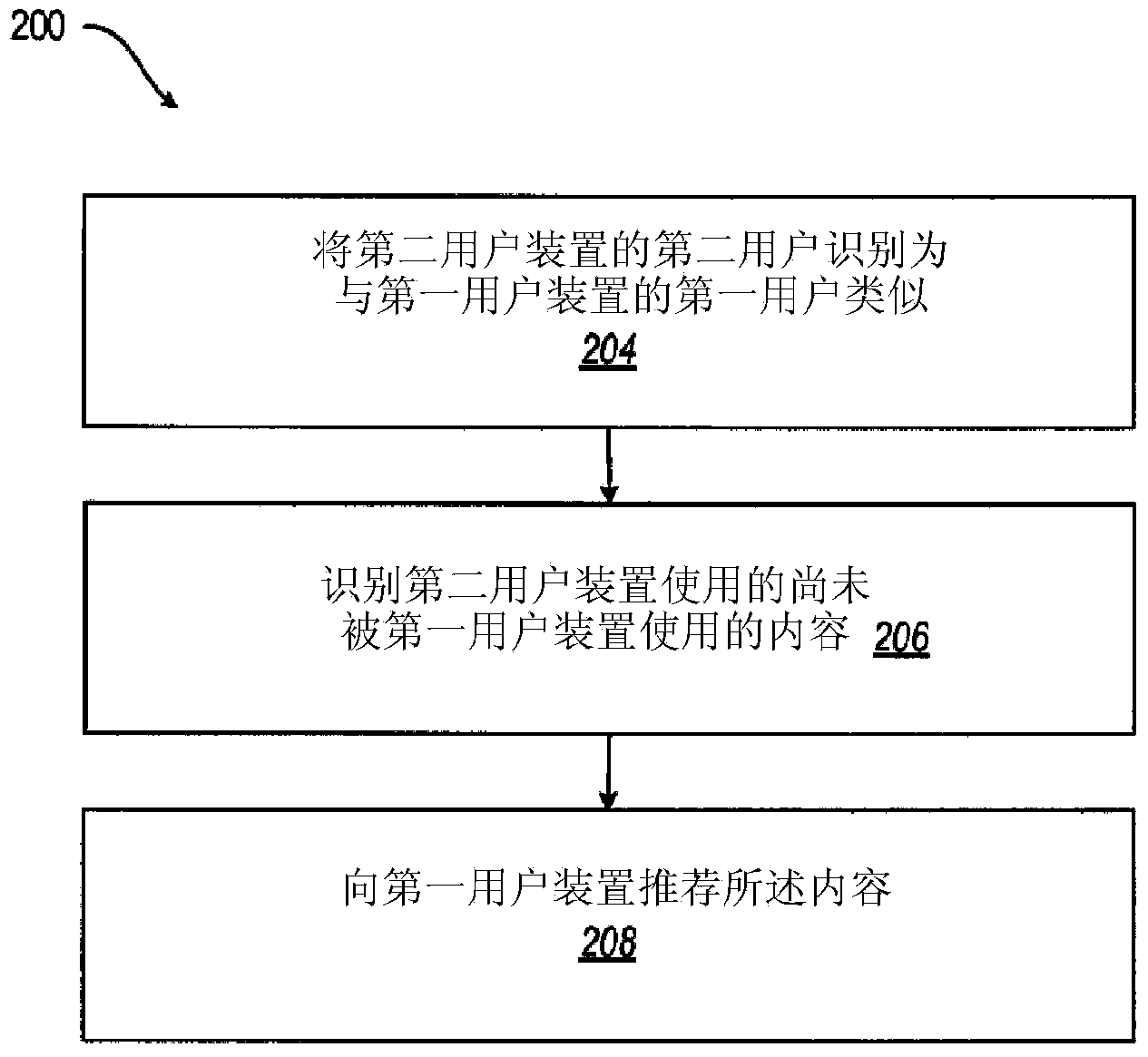
Recommending mobile content by matching similar users
A content recommendation apparatus and method gives users a peer-to-peer, highly personal way to discover new content by being introduced to one or more similar users. A similar user is a user that has a correlation to another user based on their preferences and behaviors (e.g., downloaded content, frequency of use, content currently have on device, browsing behaviors, content organizational habits, etc.). A catalog of content, or a designated portion thereof, can be used at least in part for finding matches and for identifying content to suggest to another. Data about user behavior can serve to connect the user to similar individuals for the purpose of discovering new content.
Owner:QUALCOMM INC
Method for determining a shape space for a set of molecules using minimal metric distances
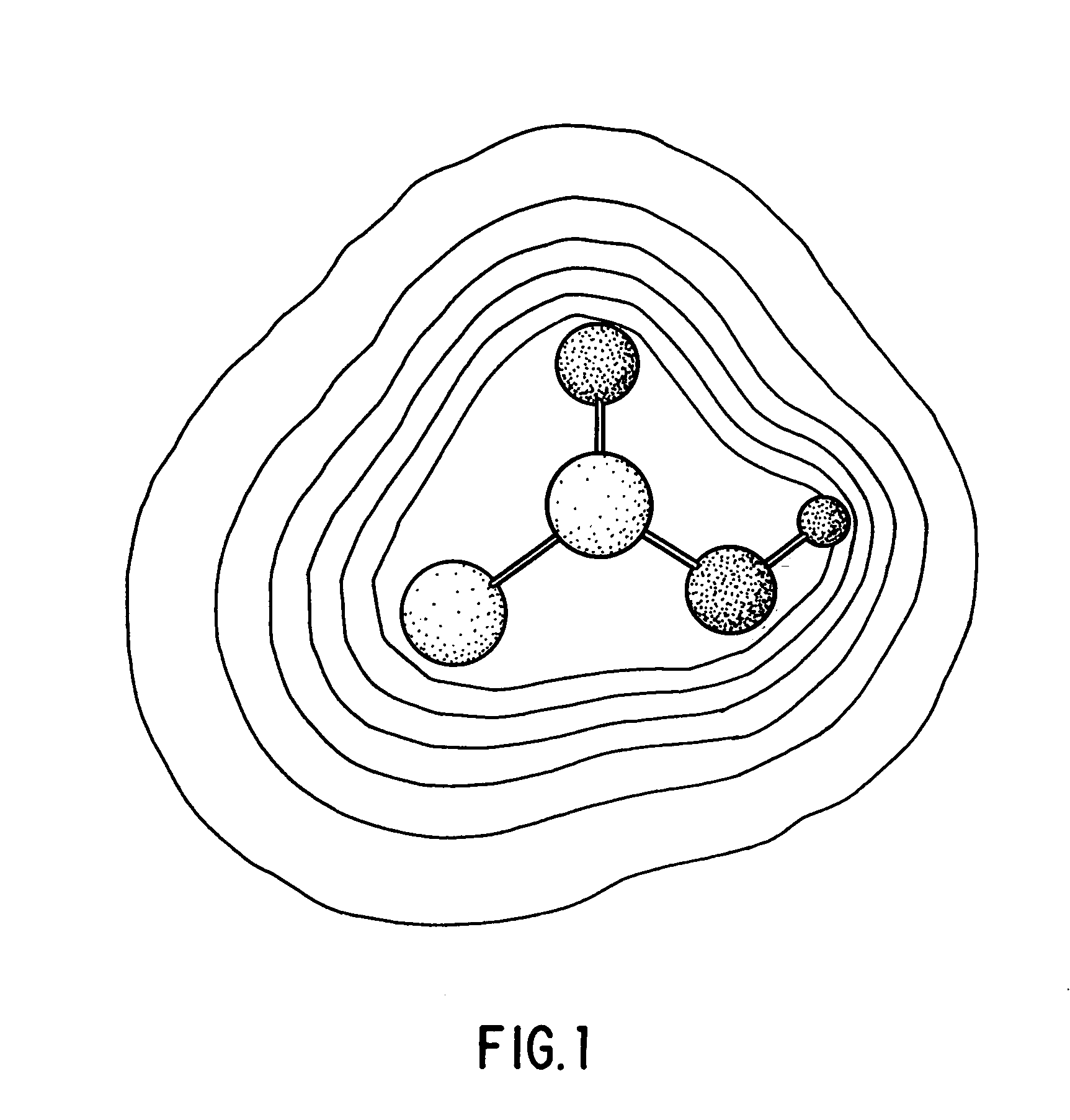
I describe several techniques for characterizing molecules based on the shapes of their fields. The minimal distance between two molecular fields is used as a shape-based metric, independent of the underlying chemical structure, and a high-dimensional shape space description of the molecules is generated. I then show how these attributes can be used in creating, characterizing, and searching databases of molecules based on field similarity. In particular, they allow searches of a database in sublinear time. Next, I extend the utility of this approach by describing a way to automatically break molecules into a series of fragments by using an ellipsoidal Gaussian decomposition. Not only can these fragments then be analyzed by the shape metric technique described above, but the parameters of the decomposition themselves can also be used to further organize and search databases. The most immediate application of these techniques is to pharmaceutical drug discovery and design.
Owner:OPENEYE SCIENTIFIC SOFTWARE
Malodor reduction compositions
InactiveUS20160089464A1Suitable for useCosmetic preparationsOrganic detergent compounding agentsMedicinePlastic film
The present invention relates to malodor reduction compositions and methods of making and using same. The malodor reduction compositions are suitable for use in a variety of applications, including use in consumer products, for example, air freshening compositions, laundry detergents, fabric enhancers, surface cleaners, beauty care products, dish care products, diapers, feminine protection articles, and plastic films for garbage bags. Such malodor control technologies do not unduely interfere with the scent of the perfumed or unperfumed situs that is treated with the malodor control technology.
Owner:THE PROCTER & GAMBLE COMPANY
Mass spectrometry-cleavable cross-linking agents to facilitate structural analysis of proteins and protein complexes, and method of using same
ActiveUS20130144541A1Silicon organic compoundsMolecular designStructure analysisMass Spectrometry-Mass Spectrometry
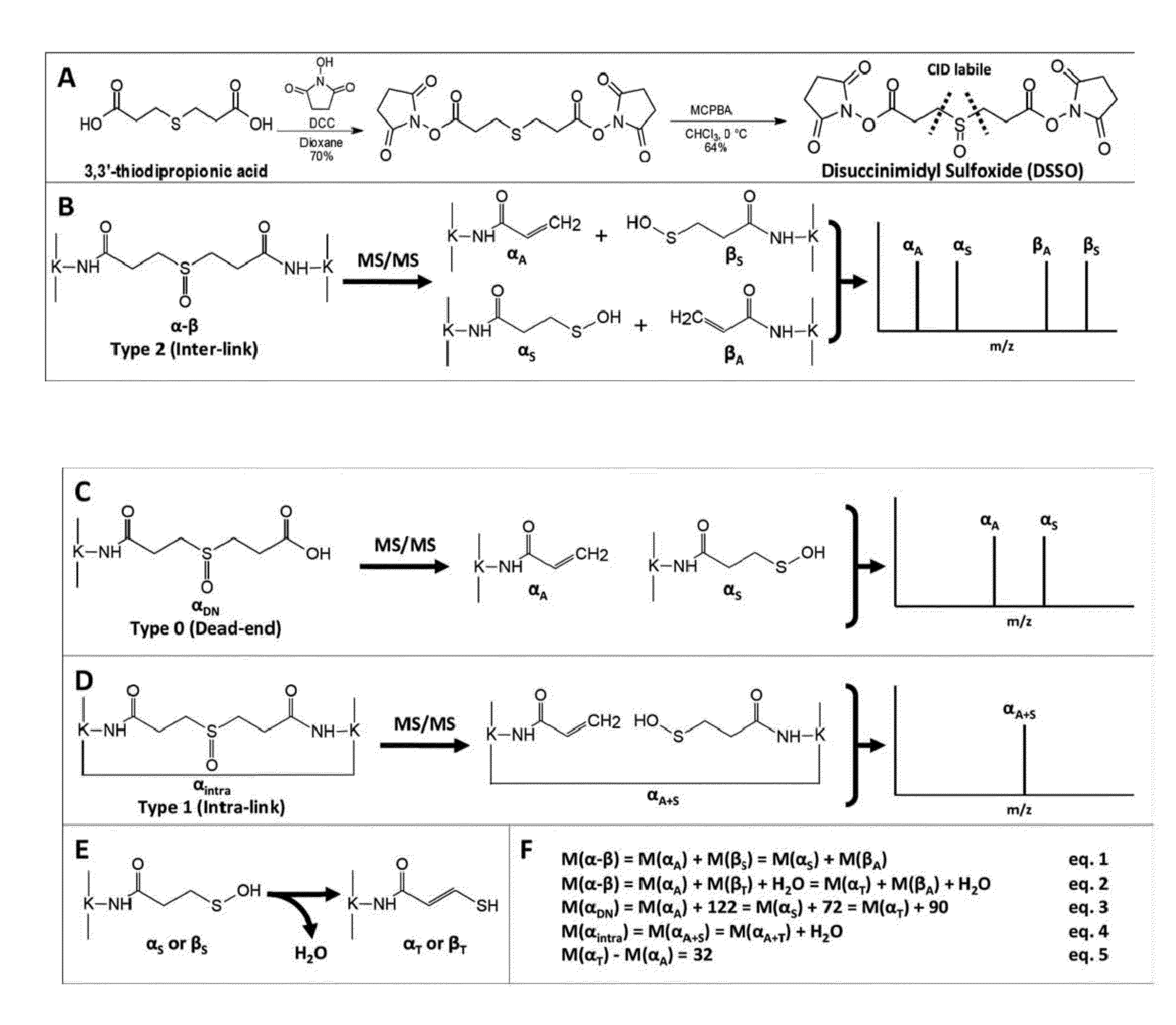
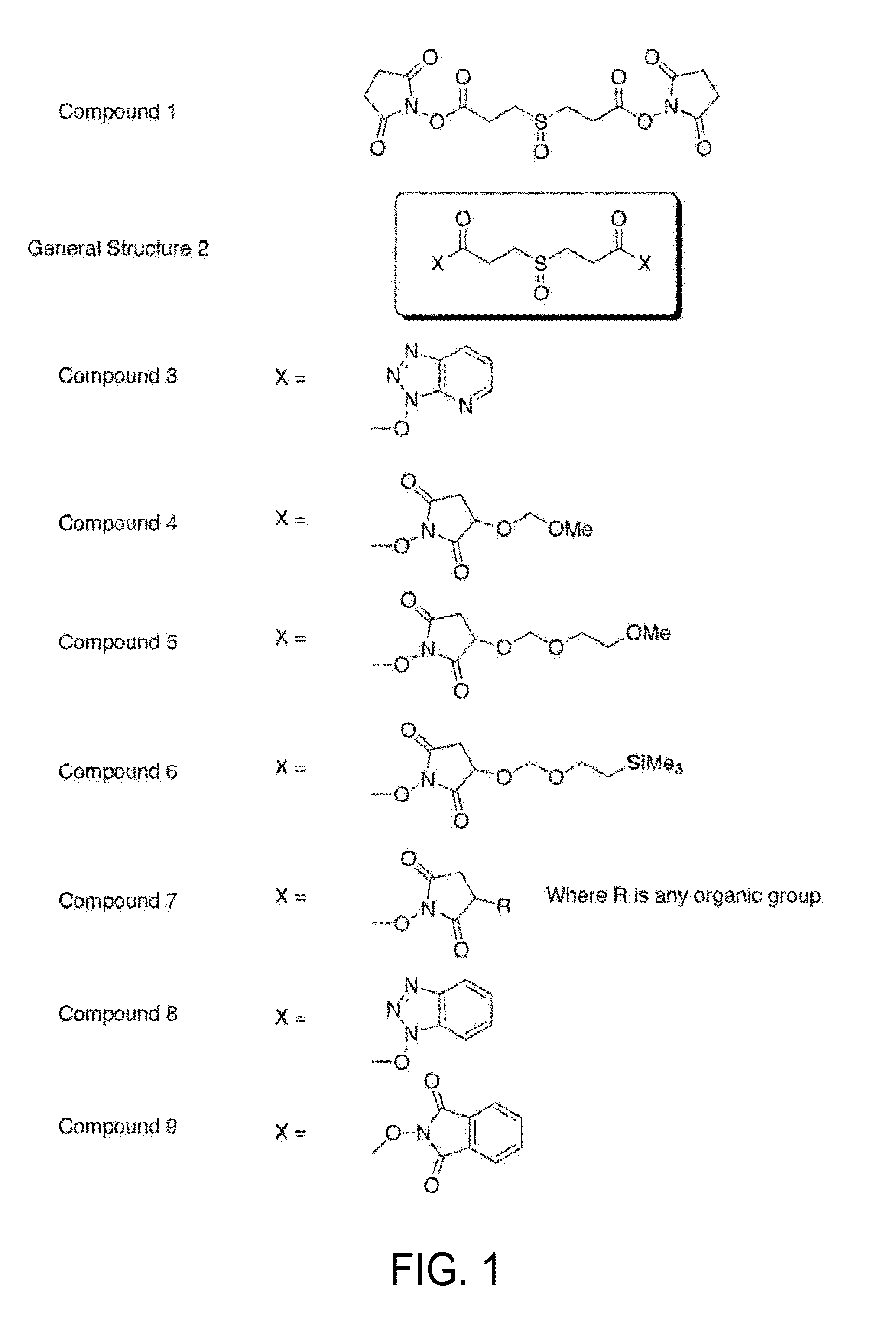
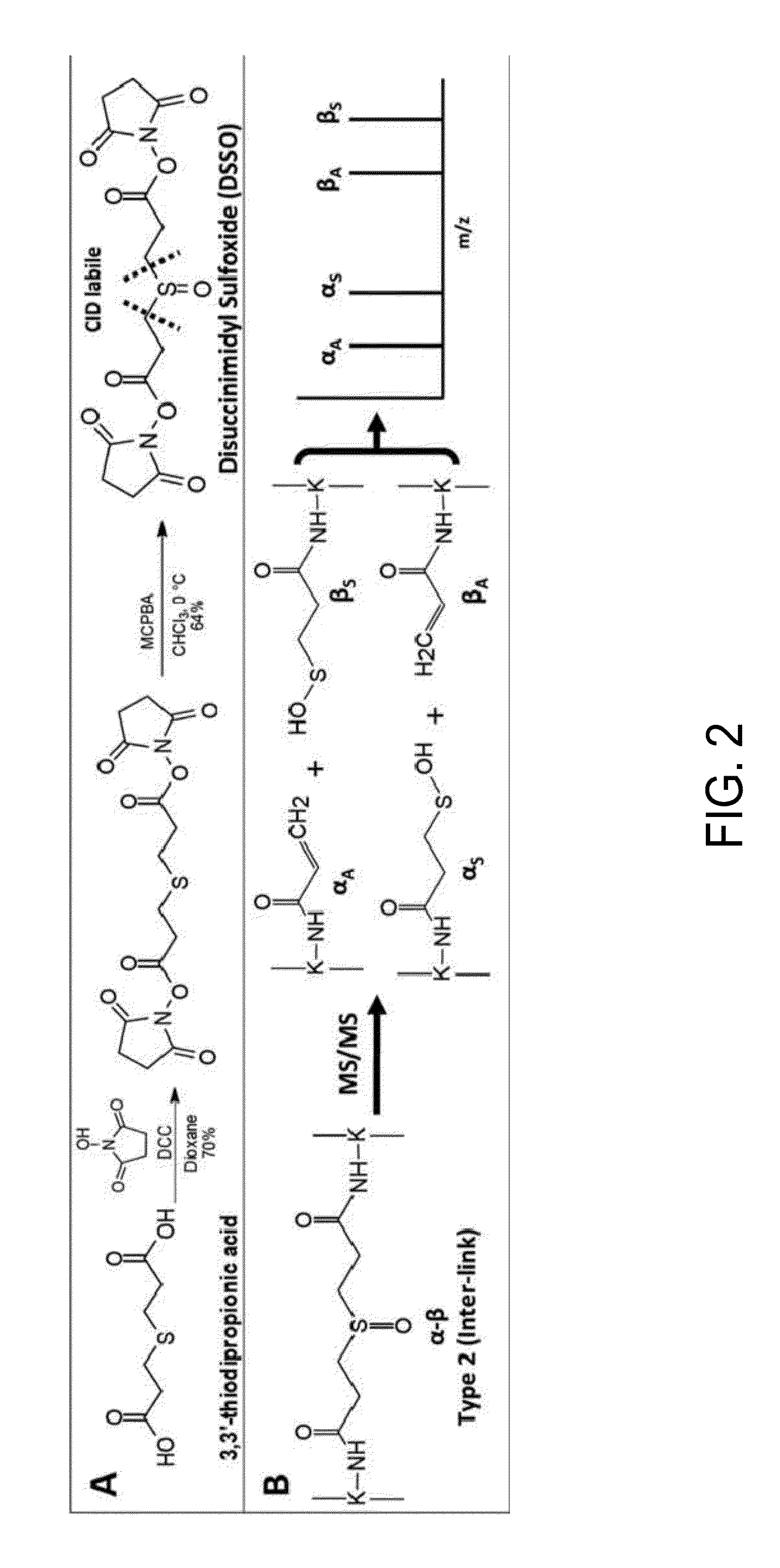
Novel cross-linking compounds that can be used in mass spectrometry, tandem mass spectrometry, and multi-stage tandem mass spectrometry to facilitate structural analysis of proteins and protein complexes are provided and have the formula:where X is an N-hydroxy-succinimidyl or similar heterocyclic group. Also provided is a method of mapping protein-protein interactions of protein complexes using various mass spectrometry techniques.
Owner:RGT UNIV OF CALIFORNIA
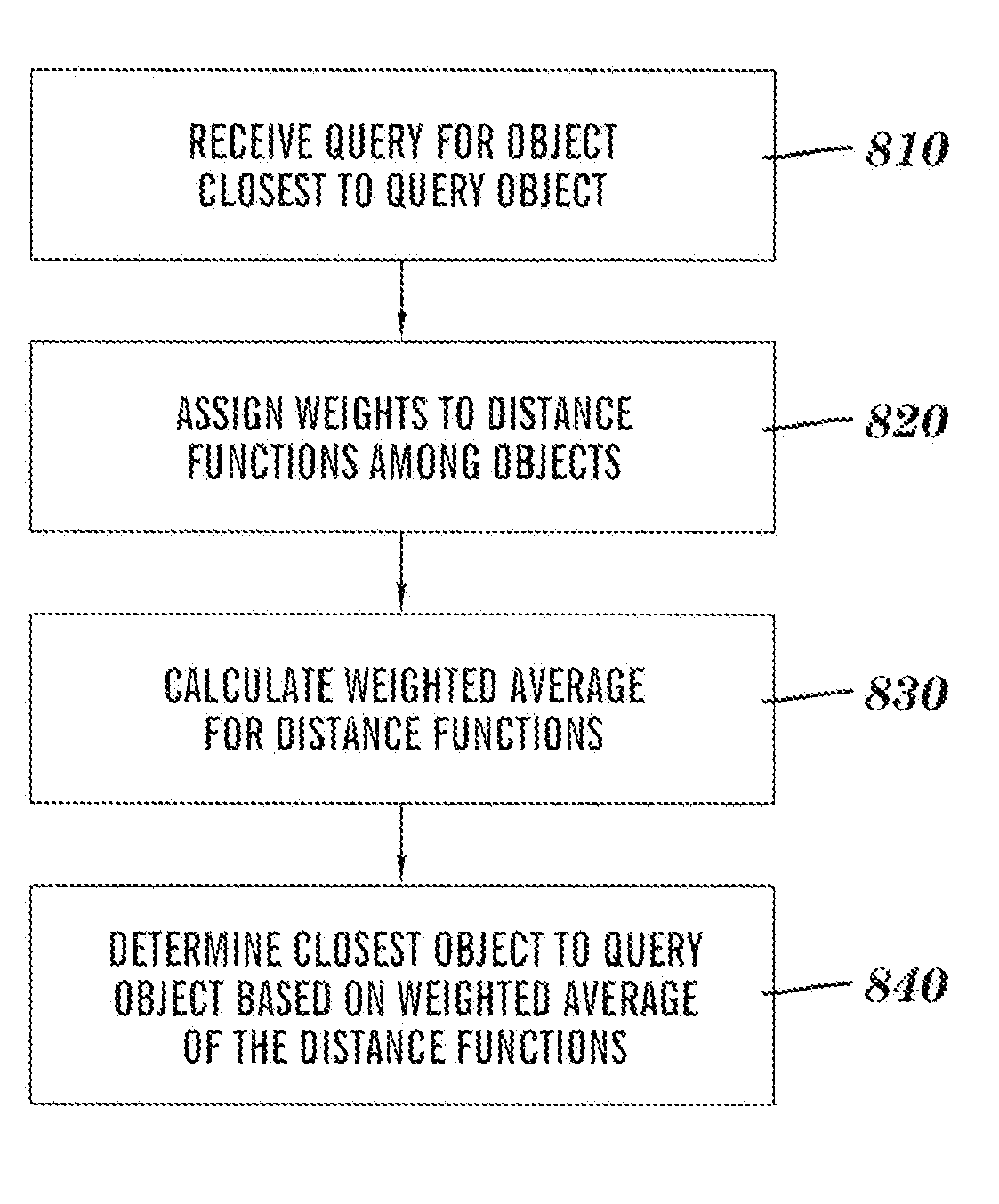
Method, computer program product, and device for conducting a multi-criteria similarity search
Similarities among multiple near-neighbor objects are searched for based on multiple criteria. A query is received for an object closest to an object provided by a user, and weights are assigned by a user to distance functions among the multiple objects at the time of the query. Each distance function represents a different criterion. The weighted average is calculated for the distance functions, and the closest object to the query object based on the weighted average for the distance functions.
Owner:IBM CORP
Molecule attribute prediction method based on artificial neural network
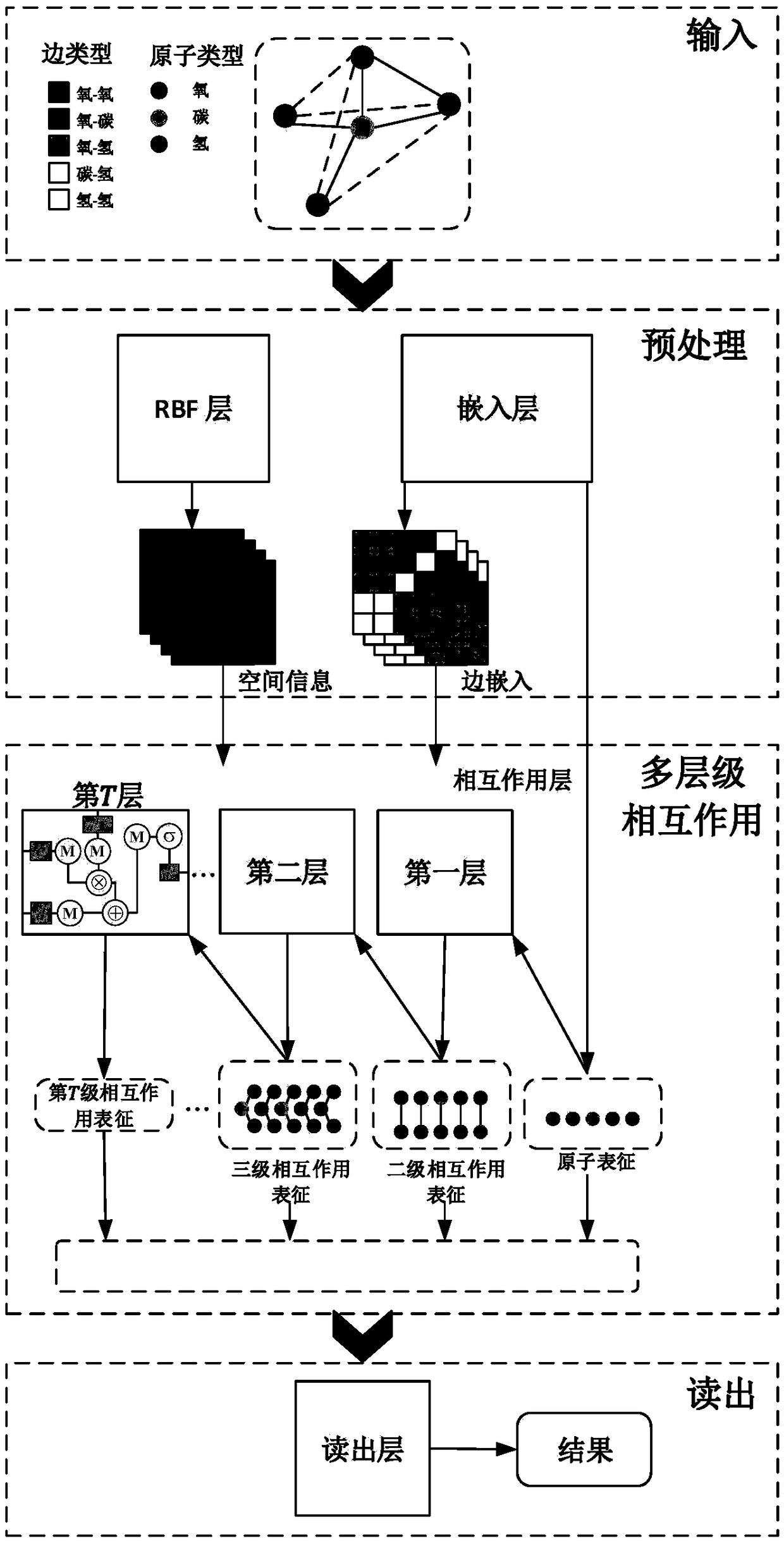
ActiveCN109461475AHigh speedHigh precisionChemical property predictionMolecular designNerve networkPredictive methods
The invention provides a molecule attribute prediction method based on an artificial neural network. The molecule attribute prediction method based on the artificial neural network comprises the following steps: S1) preprocessing of molecule data: acquiring atomic space representation and atomic composition representation through a method for data structure representation of graphs; S2) model establishment: acquiring representation of different stages of molecules from the atomic space representation and the atomic composition representation through a multilayered convolutional neural network,and combining the representations of the different stages of the molecules to obtain a model; and S3) predicting the molecule attribute according to the model. Compared with the prior art, the molecule attribute prediction method based on the artificial neural network has the advantages that the multistage convolutional neural network is used, the relation between the molecule attribute and spacecomposition can be learnt from information of known data and the multistage structure of the molecules, and is used for predicting the related attributes of unknown molecules, and therefore, the speed and the precision are good.
Owner:UNIV OF SCI & TECH OF CHINA
System and method for improved computer drug design
InactiveUS20060041414A1Rapid minimizationWeaken energyMolecular designChemical structure searchComputer-aidedComputer aid
A system and method for computer-aided drug design employs adaptive sampling and iterative fitting of mult-atomic subunits. The iterative fitting is performed by successive perturbation of the location of the multi-atomic subunits in directions that reduces potential energy.
Owner:DRUG DESIGN METHODOLOGIES
System and method for improved computer drug design
InactiveUS7801685B2Rapid minimizationWeaken energyMolecular designChemical structure searchComputer-aidedEngineering
A system and method for computer-aided drug design employs adaptive sampling and iterative fitting of multi-atomic subunits. The iterative fitting is performed by successive perturbation of the location of the multi-atomic subunits in directions that reduces potential energy.
Owner:DRUG DESIGN METHODOLOGIES
FCM (full ceramic microcapsulated) fuel effective multigroup cross section acquiring method
InactiveCN109493924AFast implementation of design calculationsCalculation speedChemical structure searchSelf screeningGroup method
Disclosed is an FCM fuel effective multigroup cross section acquiring method. The FCM fuel effective multigroup cross section acquiring method comprises, in a resonant energy section and based on a superfine group method, solving a TRISO (tristructure isotropic) particle and matrix material containing one-dimensional sphere model to obtain superfine group defect factors; through the superfine group defect factors, correcting the superfine group cross section of all nuclides to homogenize particles and matrix; through Dancoff factor equivalence, acquiring the equivalent one-dimensional rod model of every fuel rod of an FCM fuel, solving the superfine group slowing-down equation of the one-dimensional rod models to acquire the effective self-shielding cross section of the resonant energy section; in a heat energy section, through penetration probability and collision probability equivalence, acquiring multigroup defect factors, correcting the multigroup cross sections of all the nuclidesthrough the multigroup defect factors to homogenize fuel and matrix and to acquire the effective multigroup cross sections of the heat energy section. The FCM fuel effective multigroup cross sectionacquiring method can help effectively process the dual heterogenous effects of the FCM fuel to acquire a precise effective self-screening cross section.
Owner:XI AN JIAOTONG UNIV
Method and system for multi-scale anatomical and functional modeling of coronary circulation
ActiveUS10162932B2Improve predictive performanceEasy to manageMedical simulationChemical property predictionCoronary arteriesIntervention planning
A method and system for multi-scale anatomical and functional modeling of coronary circulation is disclosed. A patient-specific anatomical model of coronary arteries and the heart is generated from medical image data of a patient. A multi-scale functional model of coronary circulation is generated based on the patient-specific anatomical model. Blood flow is simulated in at least one stenosis region of at least one coronary artery using the multi-scale function model of coronary circulation. Hemodynamic quantities, such as fractional flow reserve (FFR), are computed to determine a functional assessment of the stenosis, and virtual intervention simulations are performed using the multi-scale function model of coronary circulation for decision support and intervention planning.
Owner:SIEMENS HEALTHCARE GMBH +1
System and Method for Identifying Similar Molecules
ActiveUS20080004810A1Improve computing efficiencyChemical structure searchBiological testingComputer science
A vectorization process is employed in which chemical identifier strings are converted into respective vectors. These vectors may then be searched to identify molecules that are identical or similar to each other. The dimensions of the vector space can be defined by sequences of symbols that make up the chemical identifier strings. The International Chemical Identifier (InChI) string defined by the International Union of Pure and Applied Chemistry (IUPAC) is particularly well suited for these methods.
Owner:GLOBALFOUNDRIES US INC
Freshening compositions and devices comprising same
ActiveUS20160089462A1Reduce and eliminate malodorousReduce odorCosmetic preparationsOrganic detergent compounding agentsViscosityChemistry
The present invention relates to freshening compositions and devices comprising same that comprise a composition having a viscosity of from about 1 mPa·s to about 50,000 mPa·s comprising malodor reduction compositions and methods of making and using such compositions. The disclosed malodor reduction compositions do not unduely interfere with the scent of the freshening compositions and devices that comprise such technologies and the perfumed or unperfumed situs that is treated with such freshening compositions and devices.
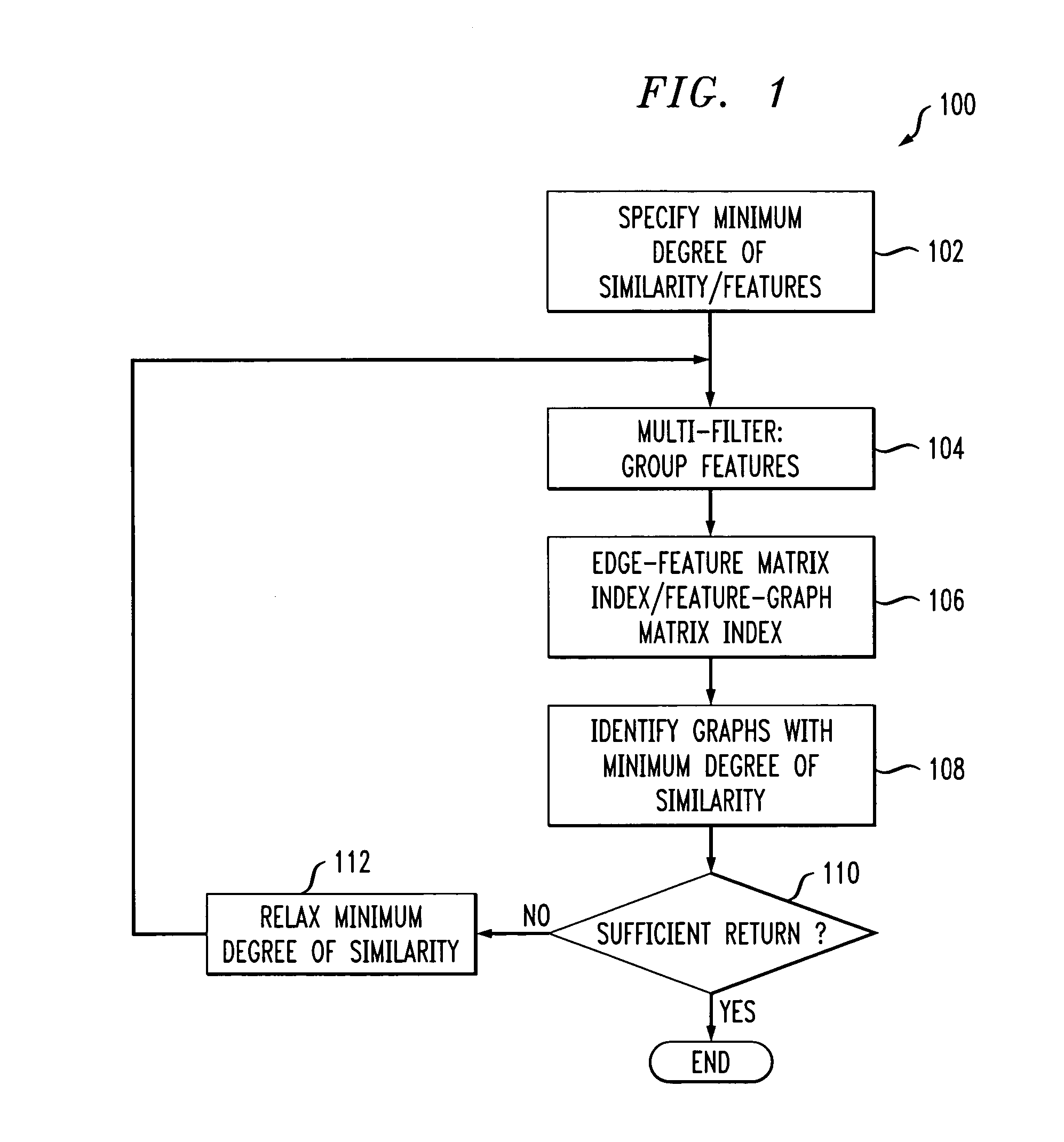
Owner:THE PROCTER & GAMBLE COMPANY
Searchable molecular database
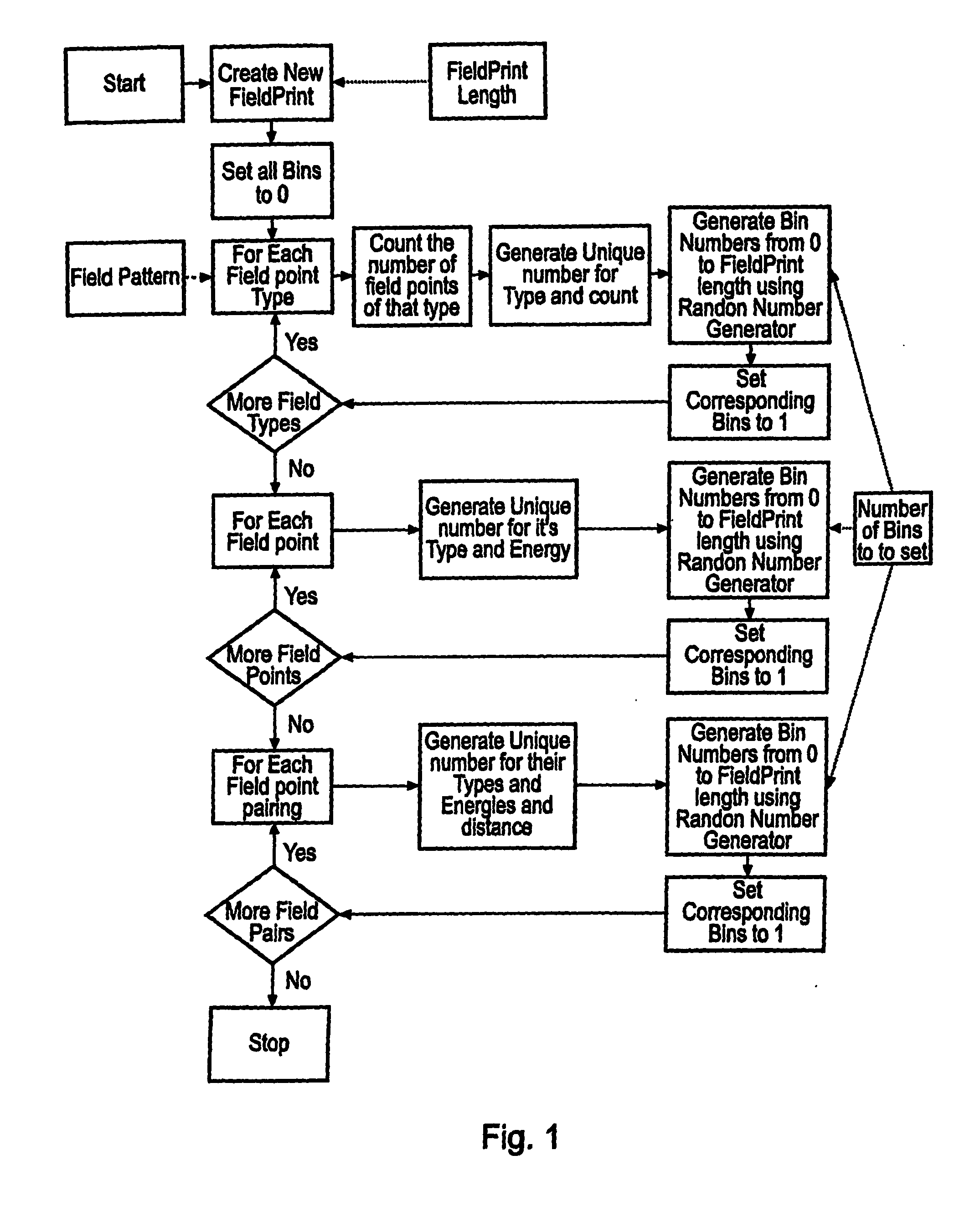
InactiveUS20060116974A1Level be differentRaise the possibilityChemical library matterChemical structure searchChemical structureGraphical user interface
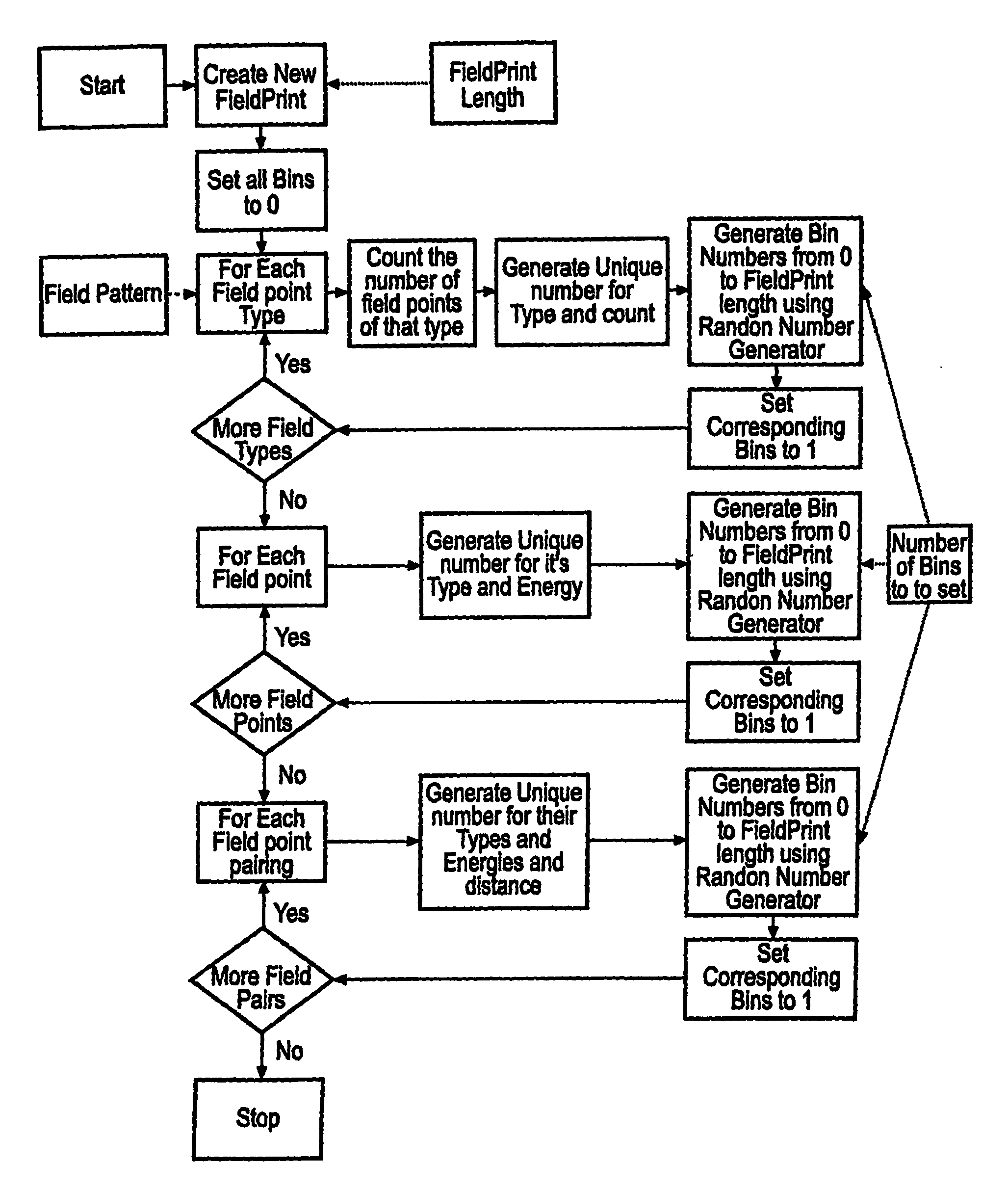
A computer system comprising a database (100) having a plurality of records, is provided. Each record comprises a filed point representation representing field extrema for a conformation of a chemical structure. The database may include records for multiple conformations of the same chemical structure. Each record can have a searchable index of the filed point representation. In one embodiment the index is bit string. An indexing mechanism for generating an index, a searching mechanism for searching the database and a graphical user interface to enable a user to interface with the database (100) are also provided.
Owner:CRESSET BIOMOLECULAR DISCOVERY
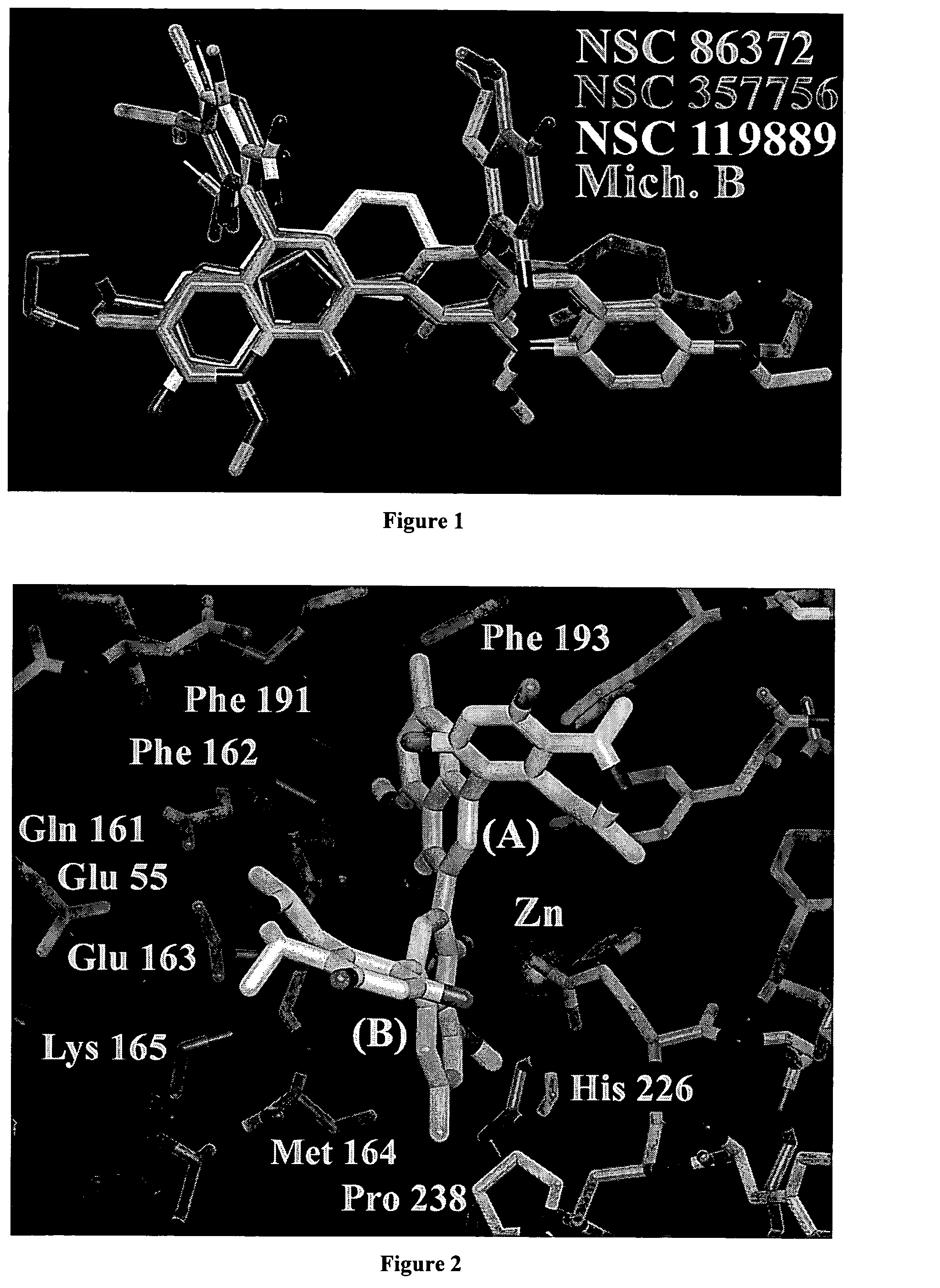
Small molecules and a pharmacophore model for inhibition of botulinum toxin and methods of making and using thereof
Owner:UNITED STATES OF AMERICA THE AS REPRESENTED BY THE SEC OF THE ARMY
Antiperspirant and deodorant compositions comprising malodor reduction compositions
The present invention relates to antiperspirant and deodorant compositions comprising malodor reduction compositions and methods of making and using such antiperspirant and deodorant compositions. Such antiperspirant and deodorant compositions comprising the malodor control technologies disclosed herein provide malodor control without leaving an undesirable scent and when perfume is used to scent such compositions, such scent is not unduely altered by the malodor control technology.
Owner:THE PROCTER & GAMBLE COMPANY
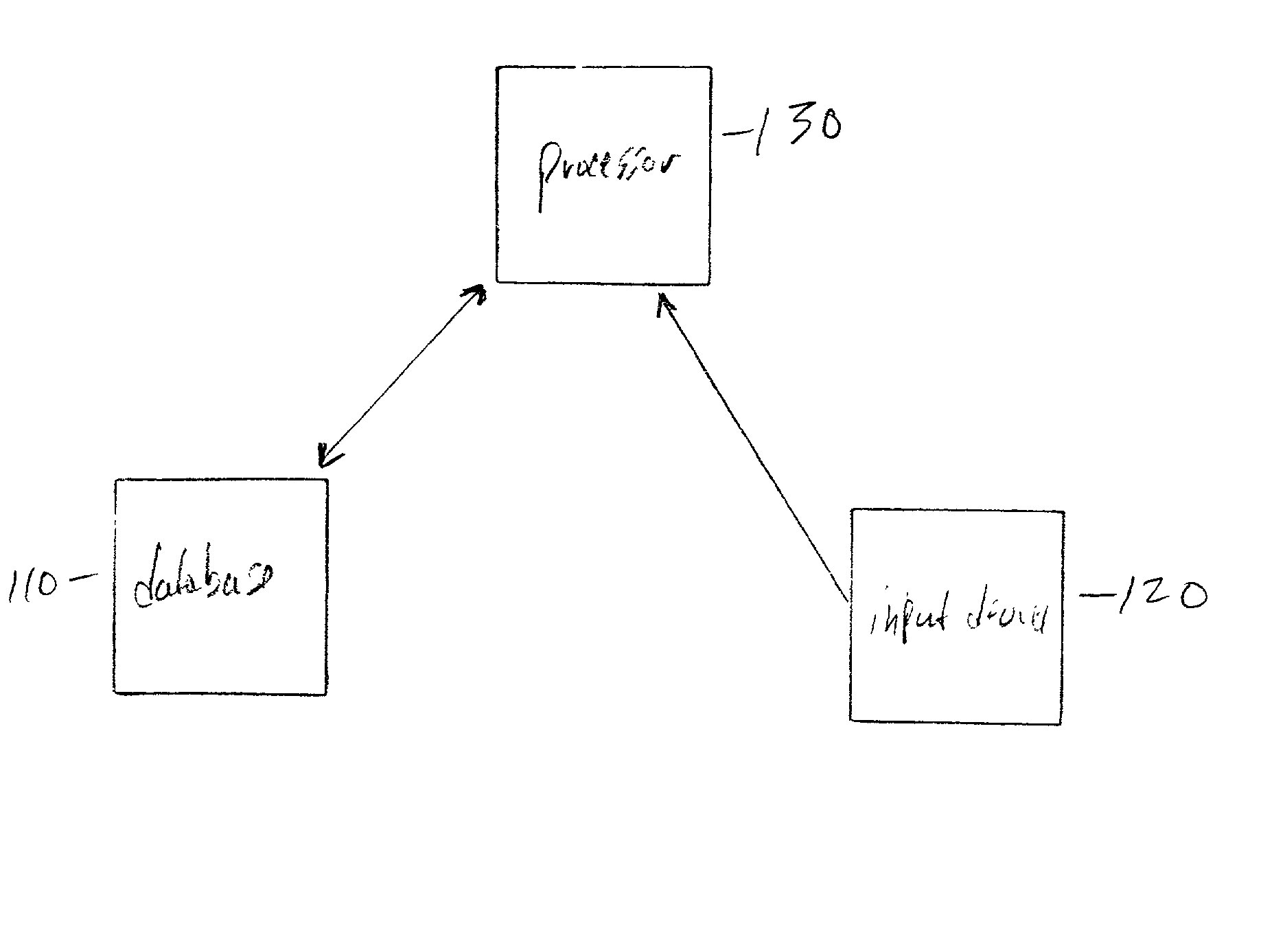
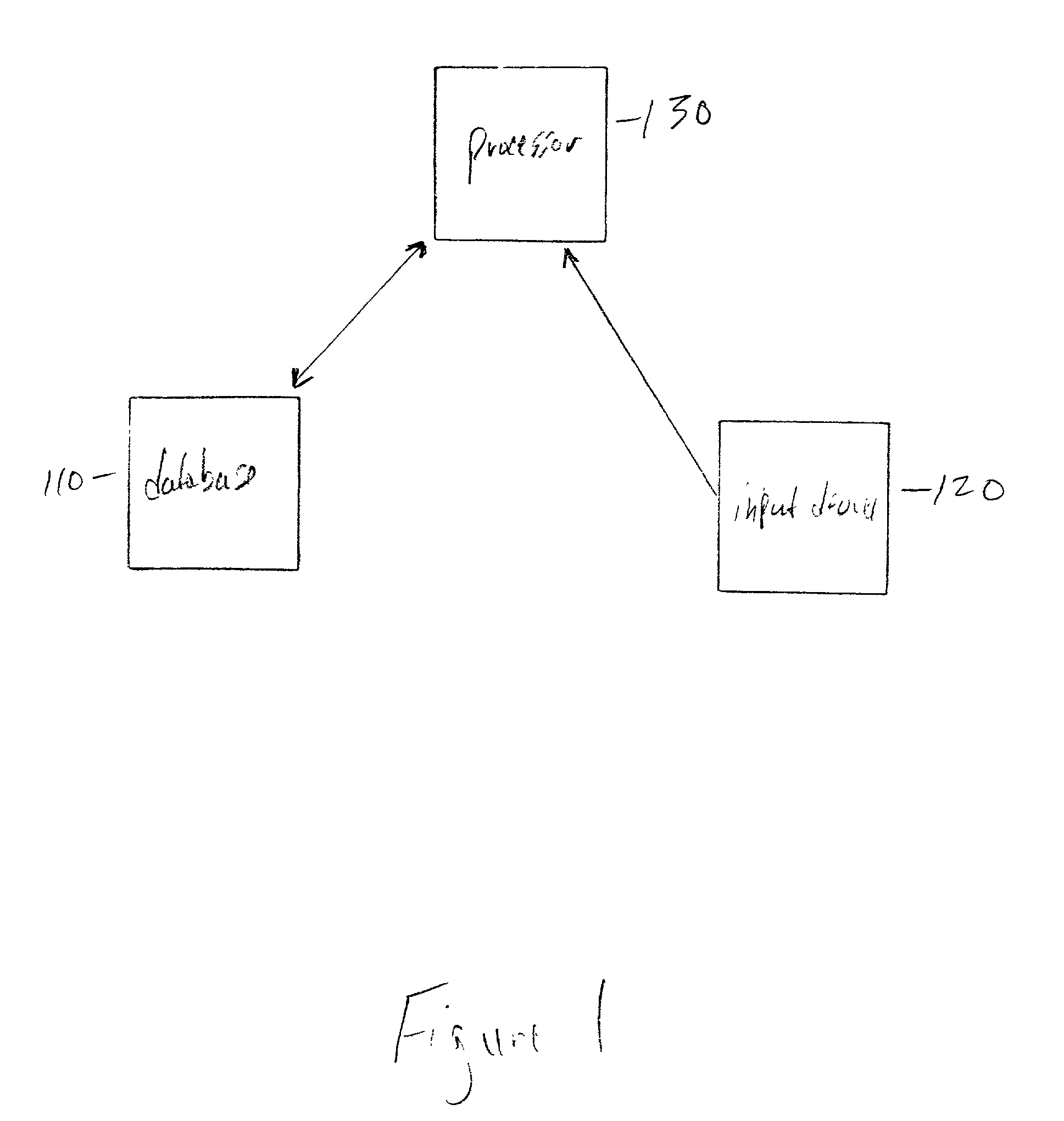
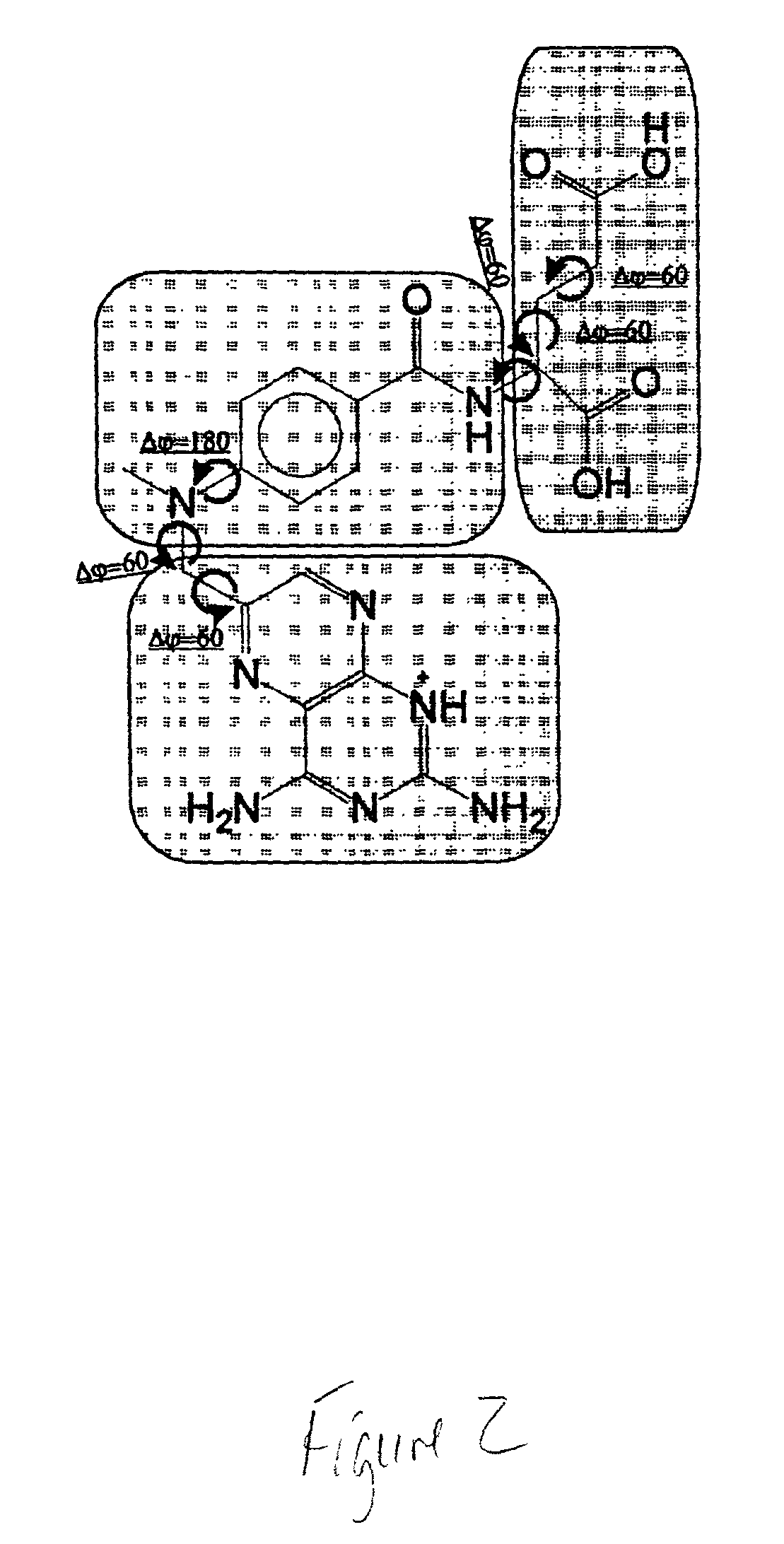
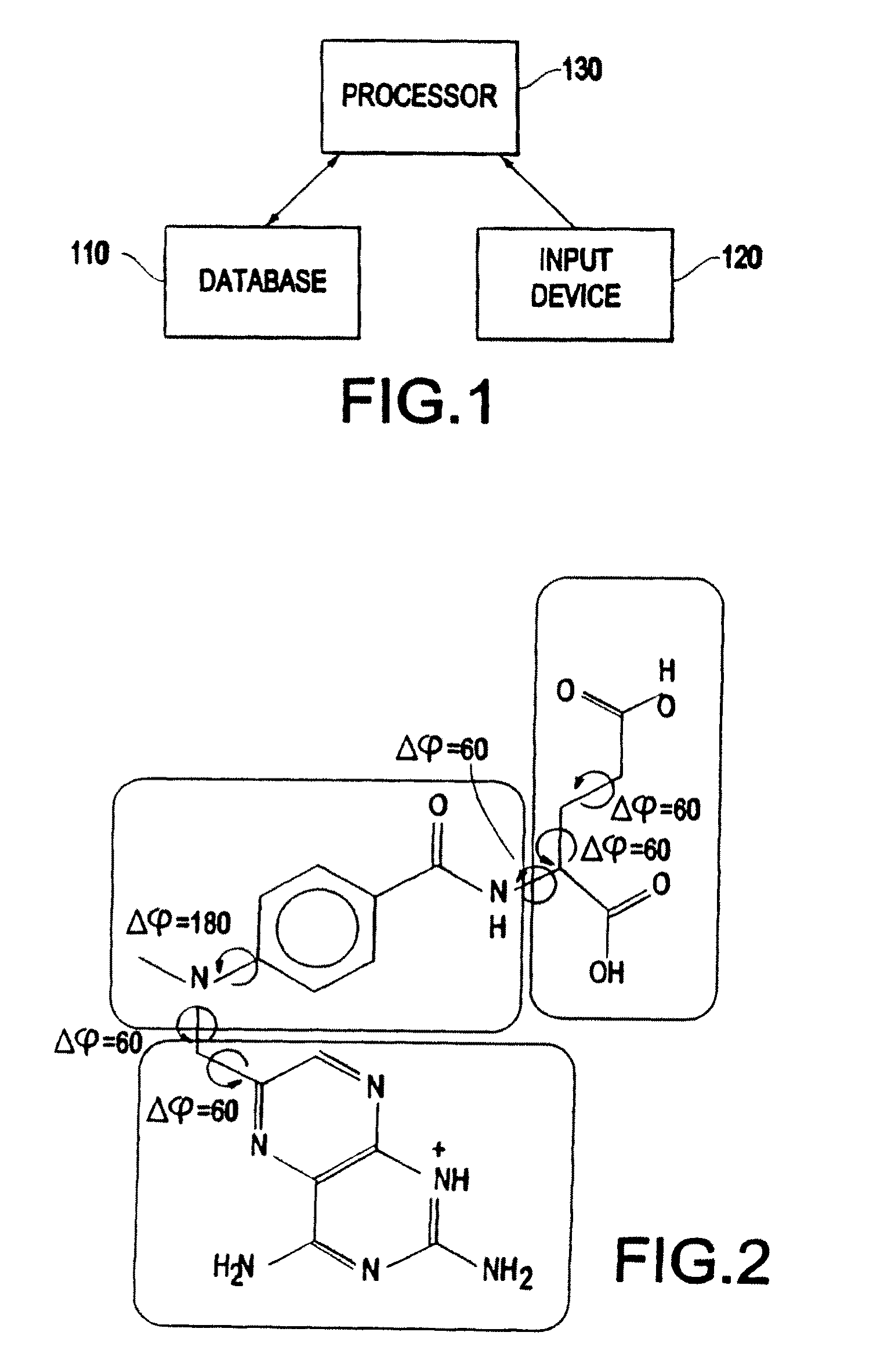
Field-based similarity search system and method
A field-based similarity search system and method includes a database for storing at least one candidate molecule, an input device for inputting a query molecule, and a processor for identifying a candidate molecule which is similar to the query molecule based on a similarity of fragment pair features.
Owner:GLOBALFOUNDRIES INC
Delivery systems comprising malodor reduction compositions
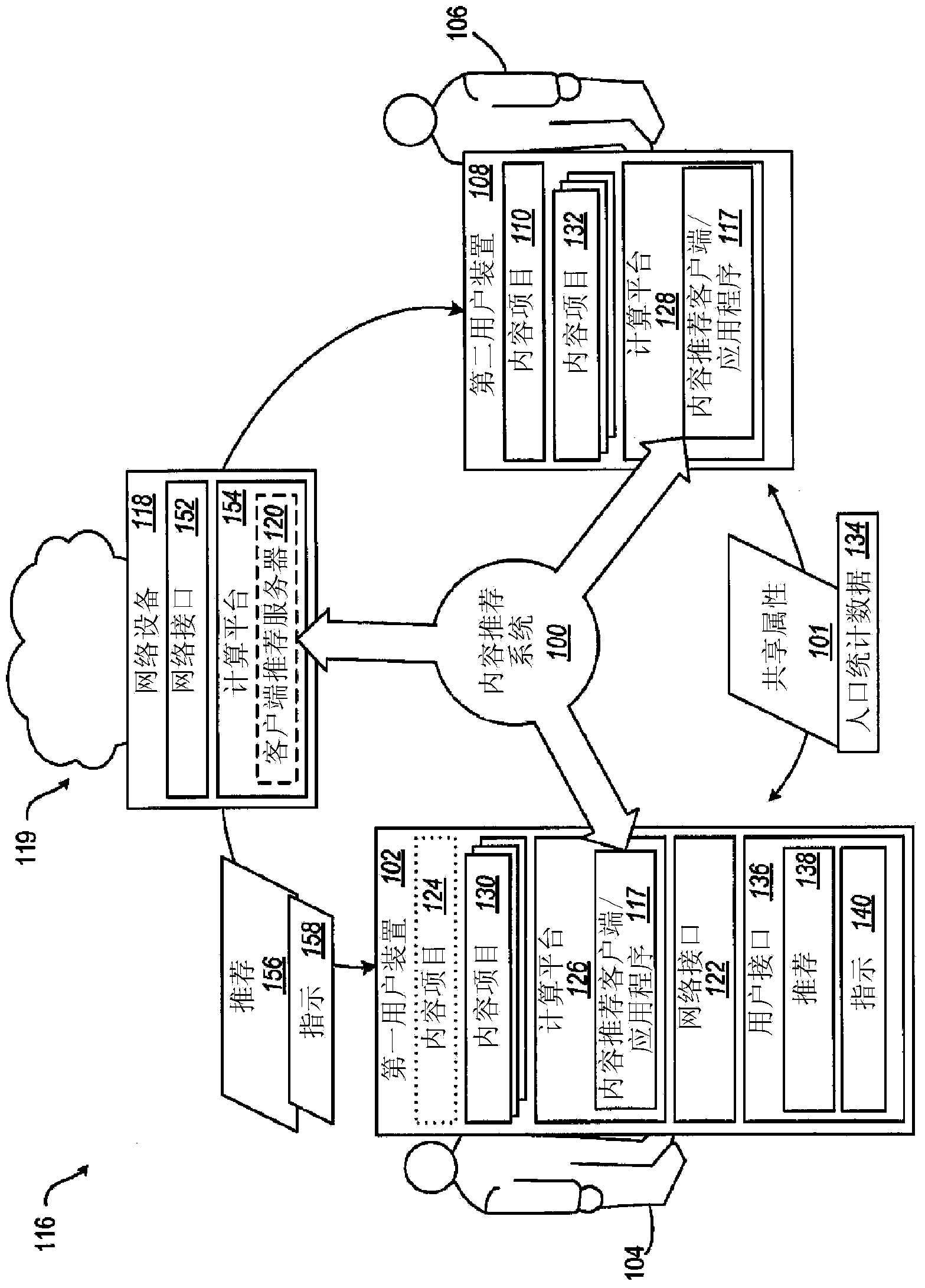
InactiveUS20160090558A1Interference be notReducing malodor trimethylamineCosmetic preparationsOrganic detergent compounding agentsMedicineDelivery system
The present invention relates to delivery systems comprising malodor reduction compositions, methods of making such delivery systems and consumer products made with such delivery systems. Such delivery systems do not unduely interfere with the scent of the perfumed or unperfumed delivery system, perfumed or unperfumed products comprising such delivery systems and any perfumed or unperfumed situs that is treated with such products.
Owner:THE PROCTER & GAMBLE COMPANY
Field-based similarity search system and method
InactiveUS20100057797A1Improve versatilityFast and efficient field-based similarity searchChemical structure searchDigital data processing detailsSimilarity measureField based
A field-based similarity search system and method includes a database for storing at least one candidate molecule, an input device for inputting a query molecule, and a processor for identifying a candidate molecule which is similar to the query molecule based on a similarity of fragment pair features.
Owner:GLOBALFOUNDRIES INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com