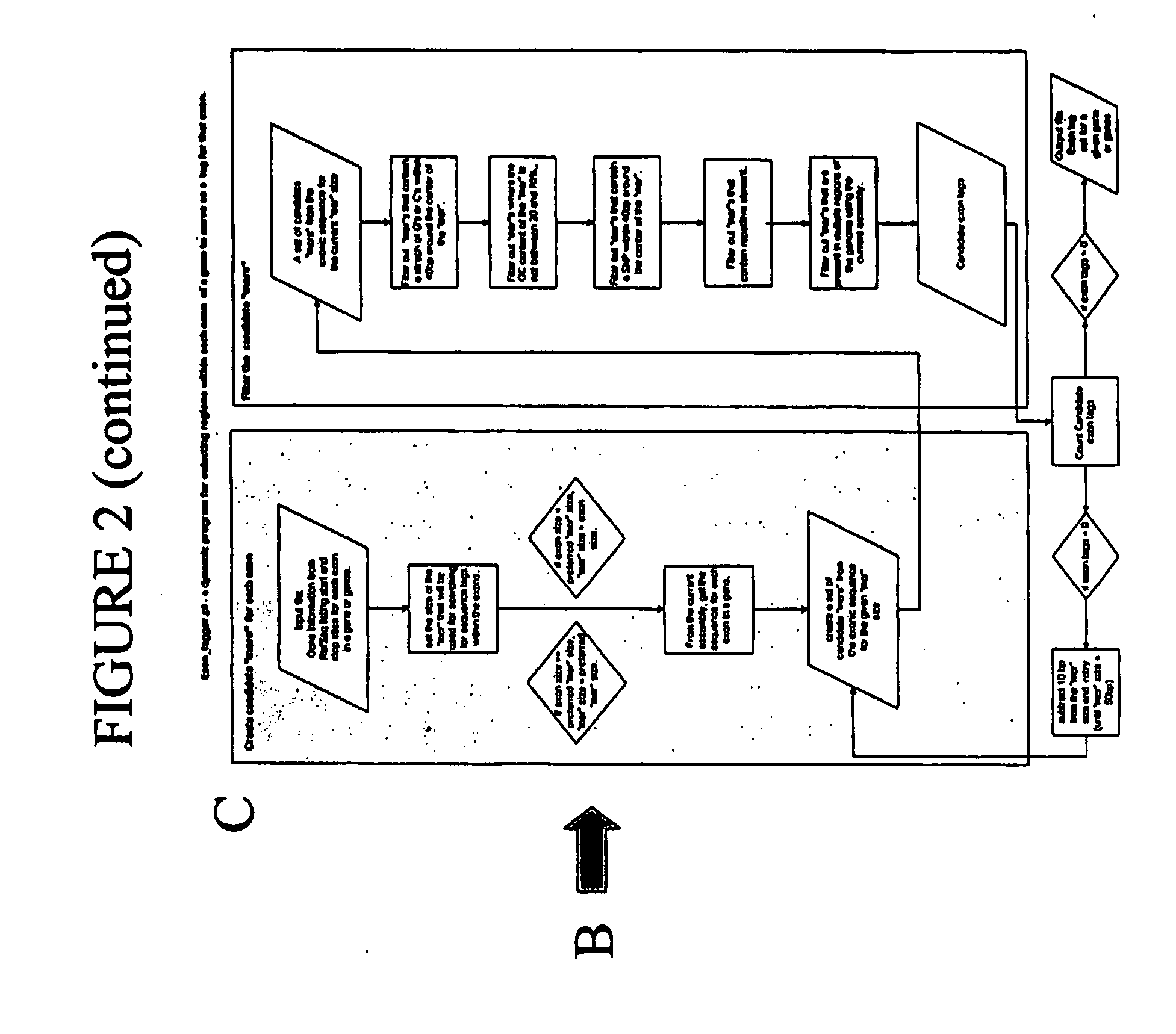
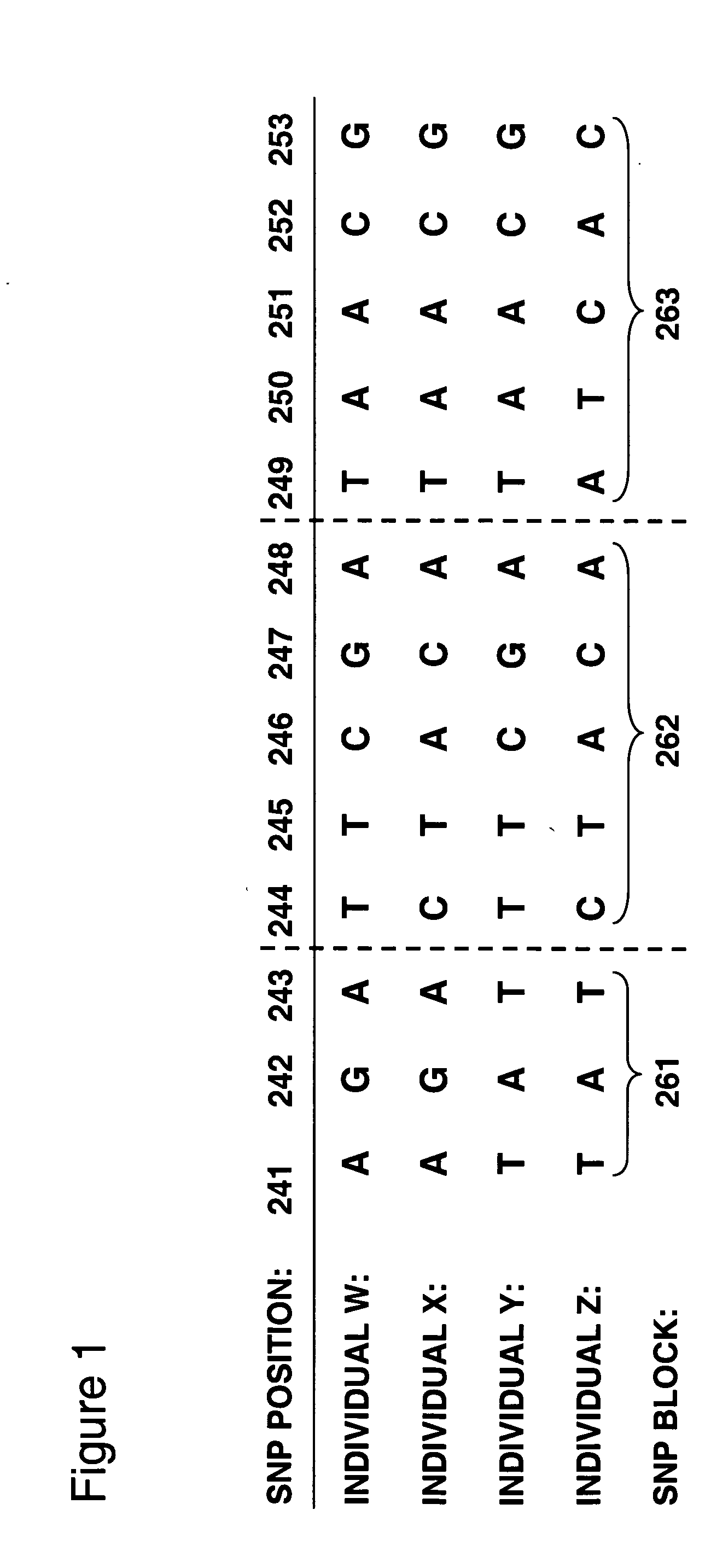
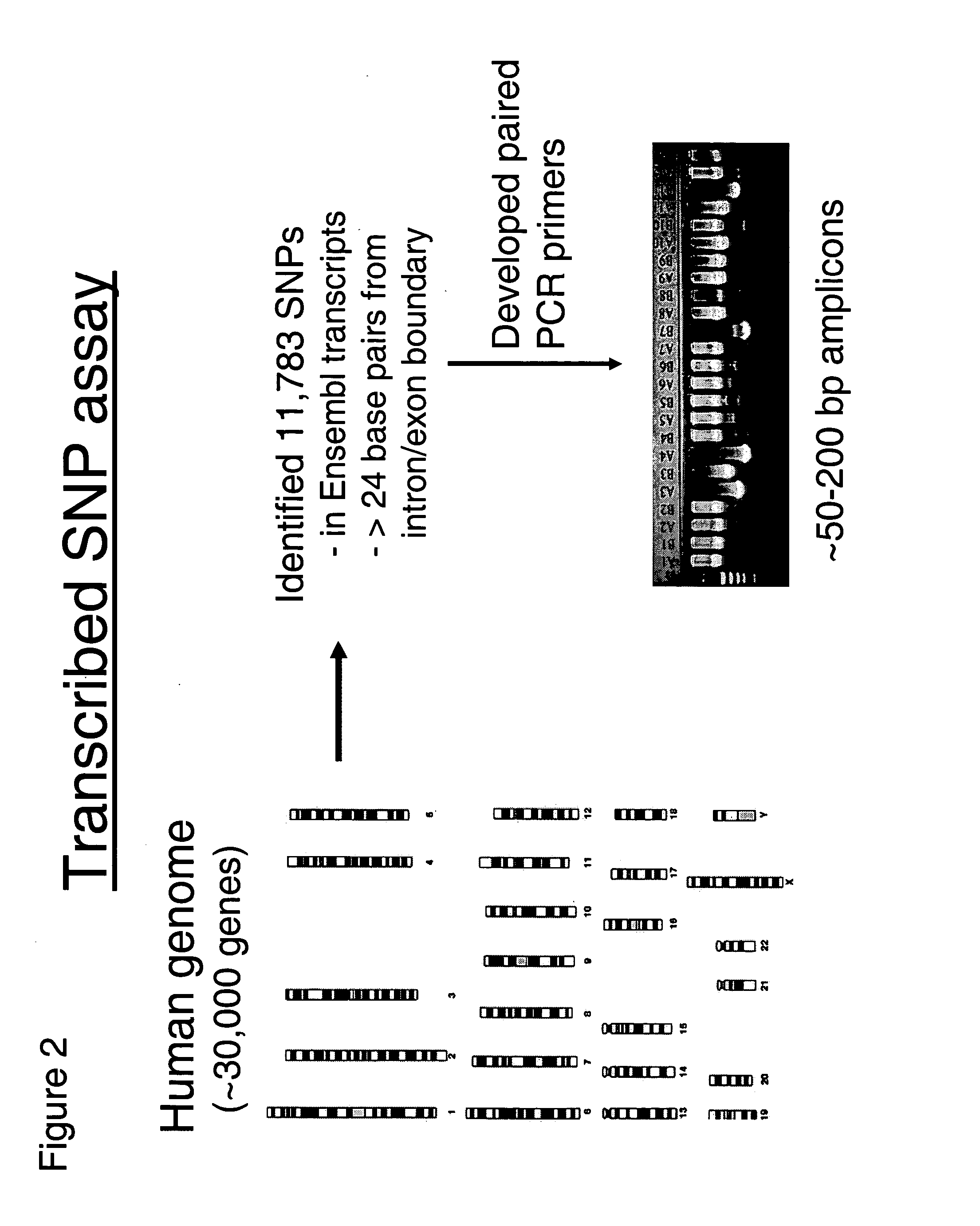
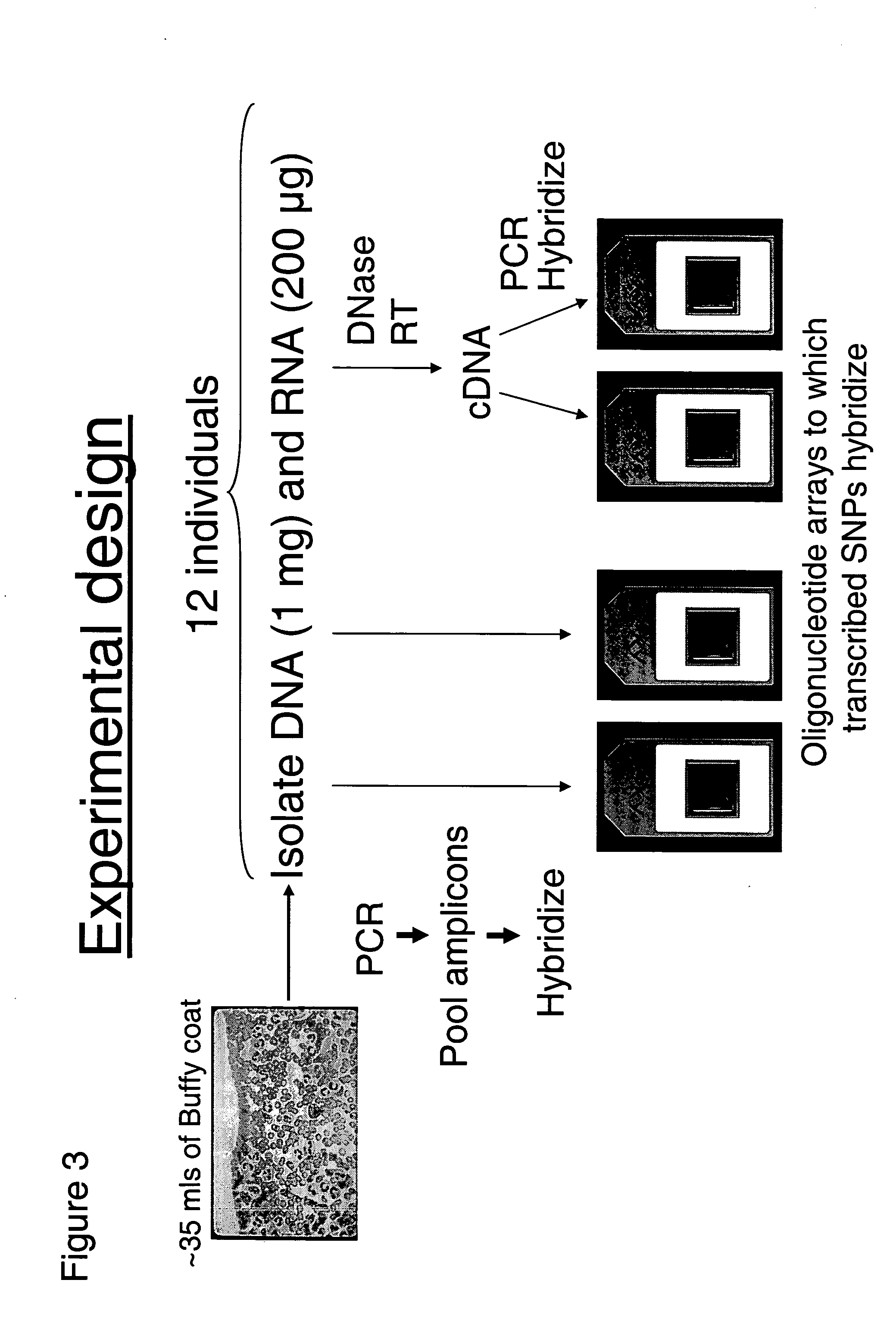
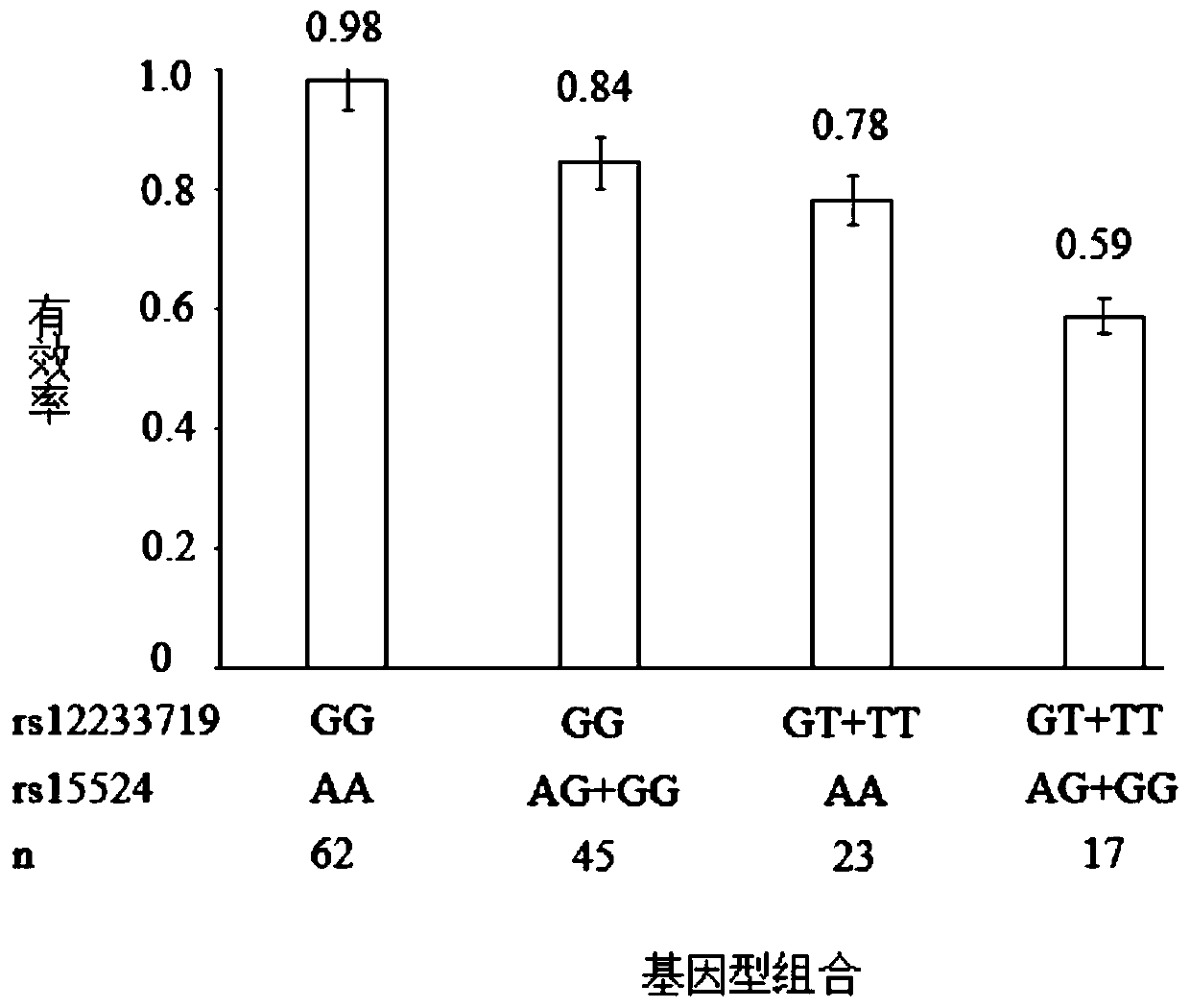
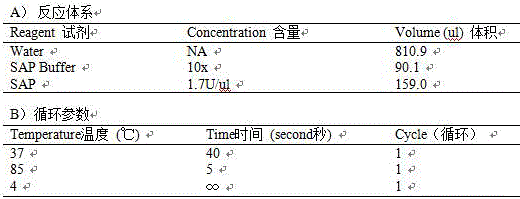
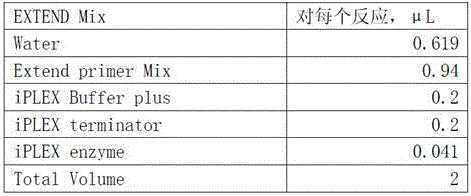
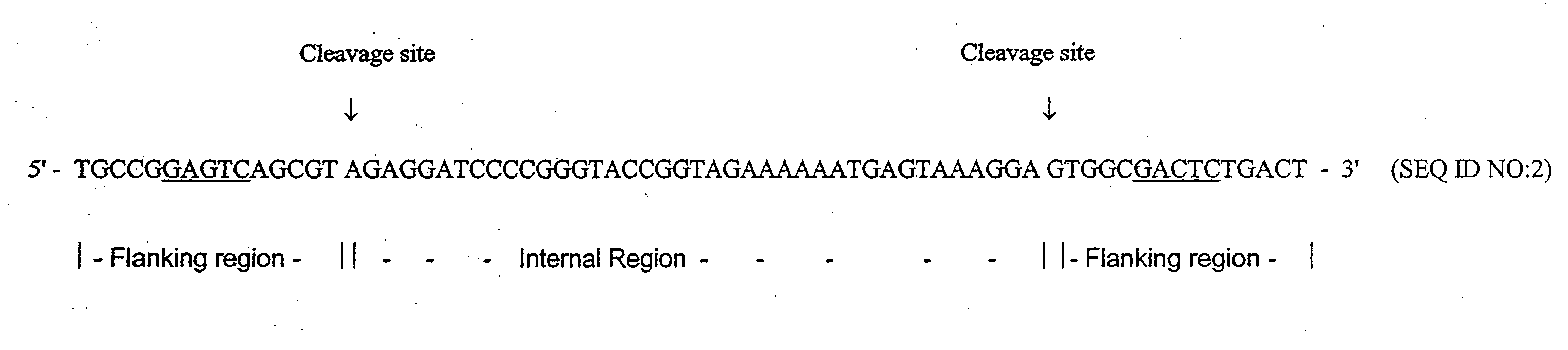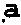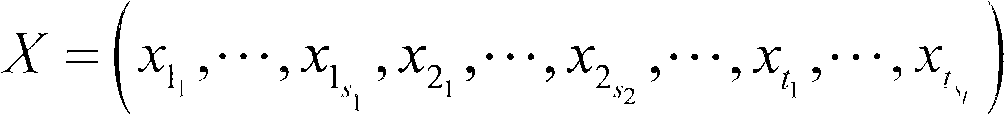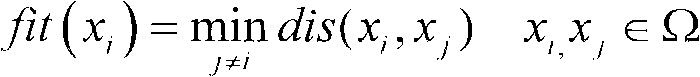Patents
Literature
192 results about "Individual gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
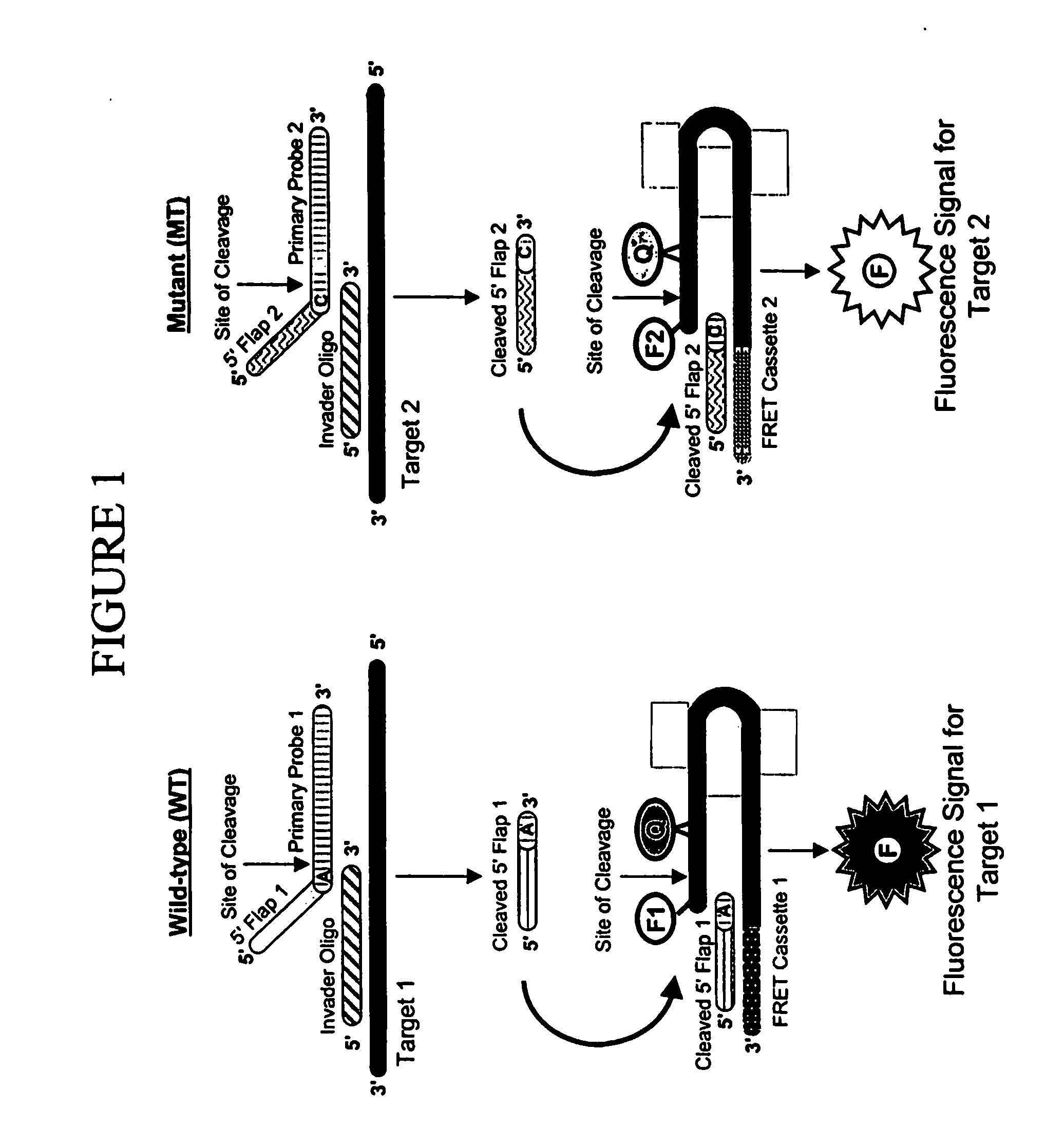
Method and compositions for detection and enumeration of genetic variations
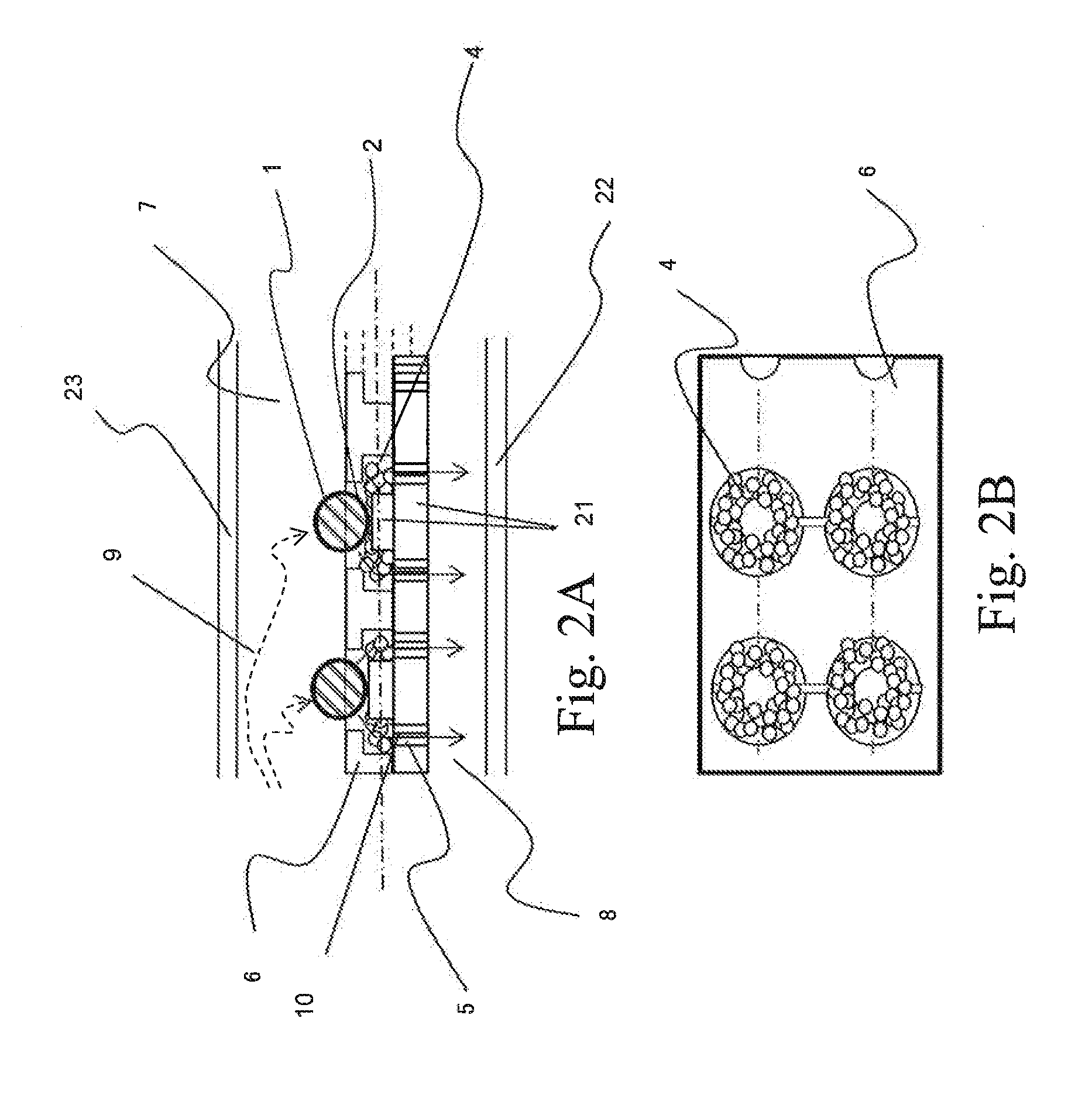
ActiveUS20070065823A1Bioreactor/fermenter combinationsBiological substance pretreatmentsSpecific populationFluorescence
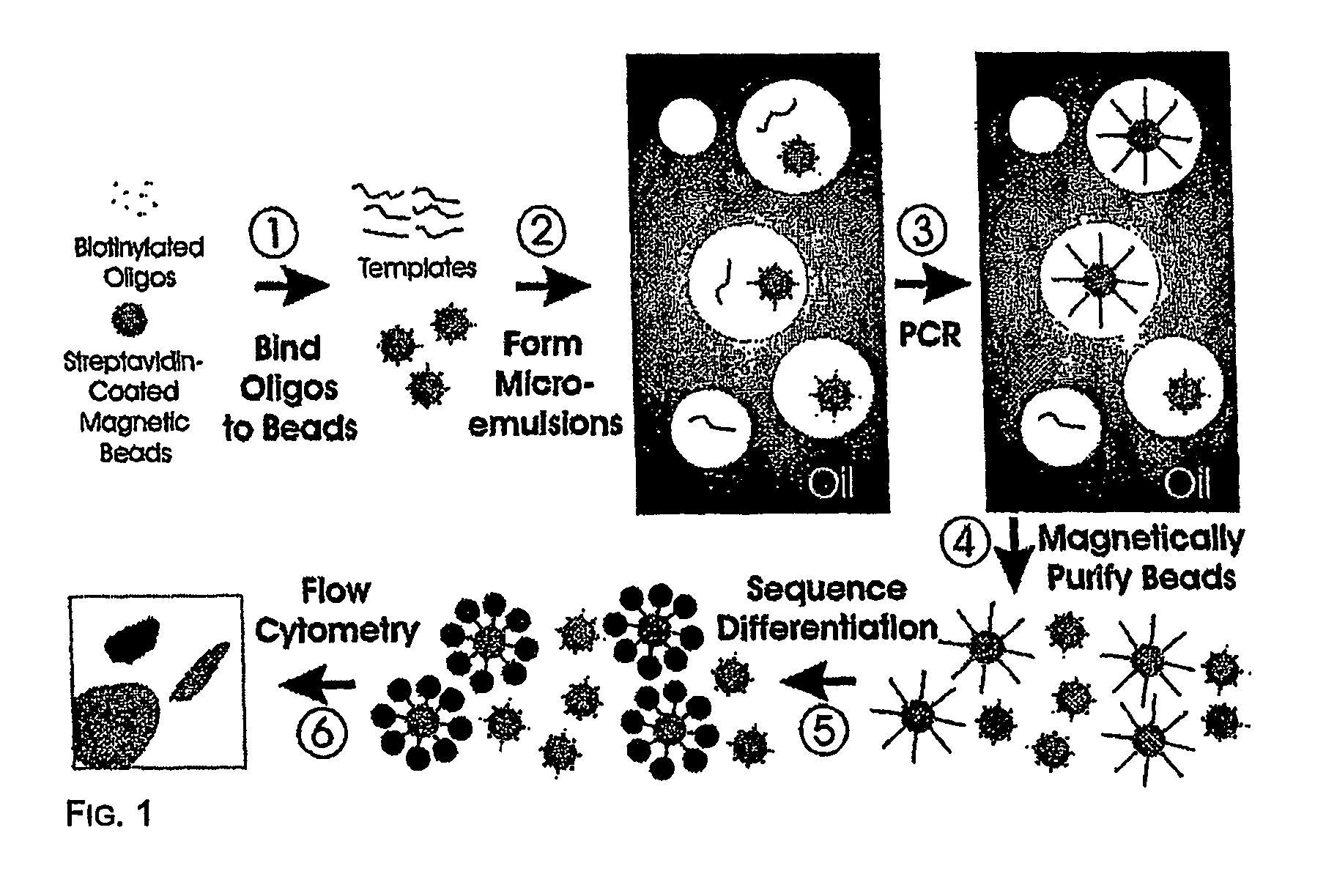
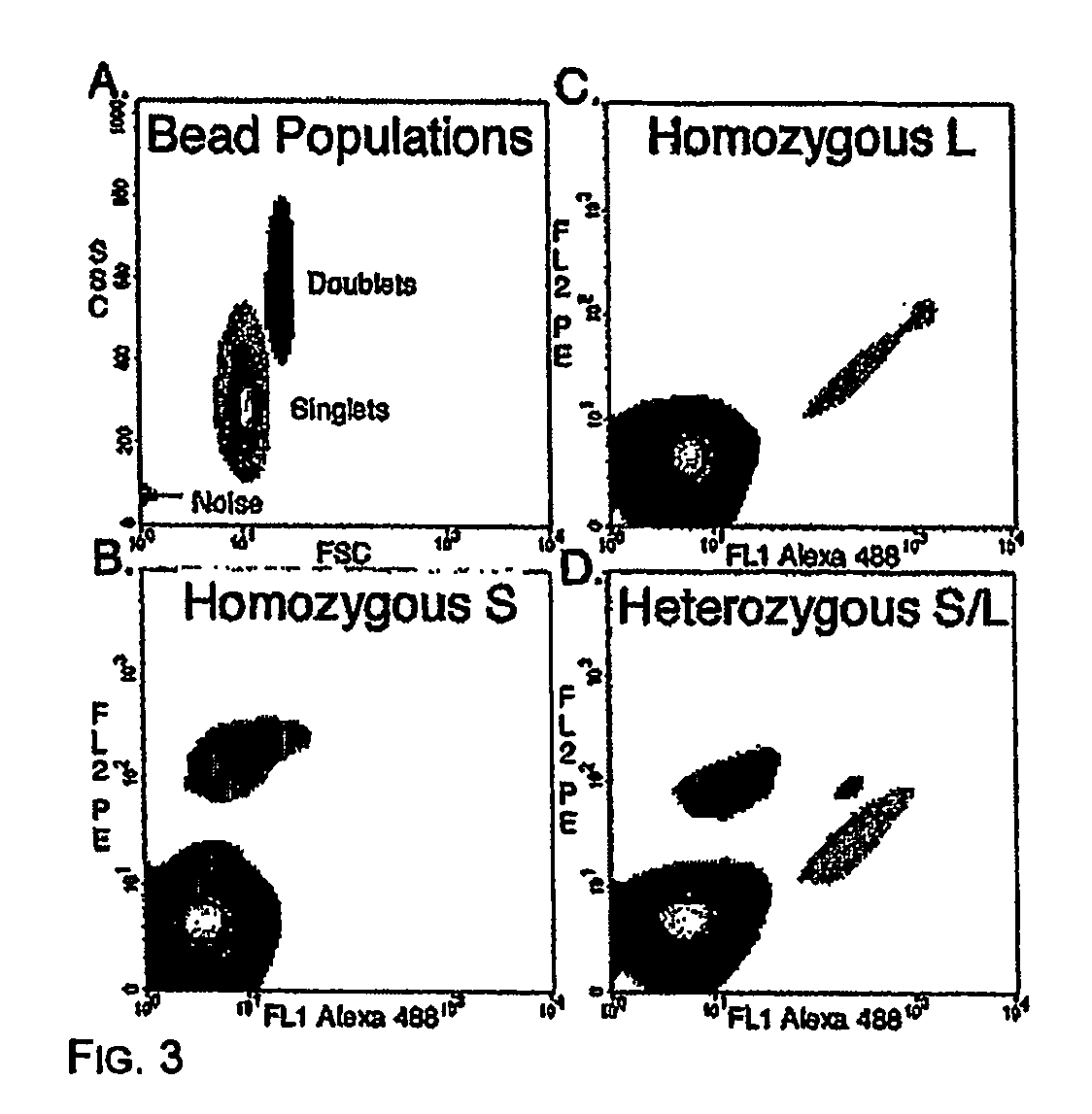
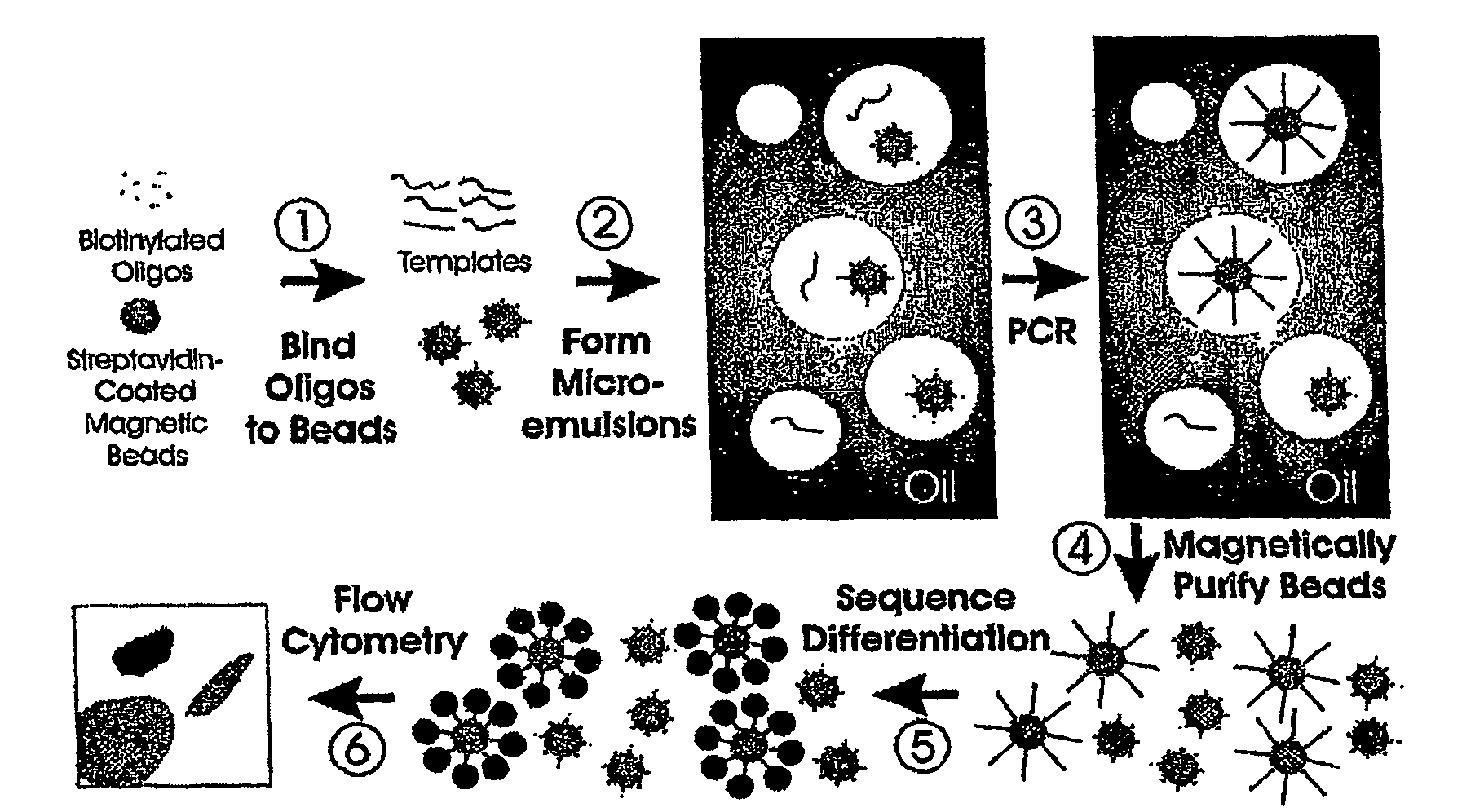
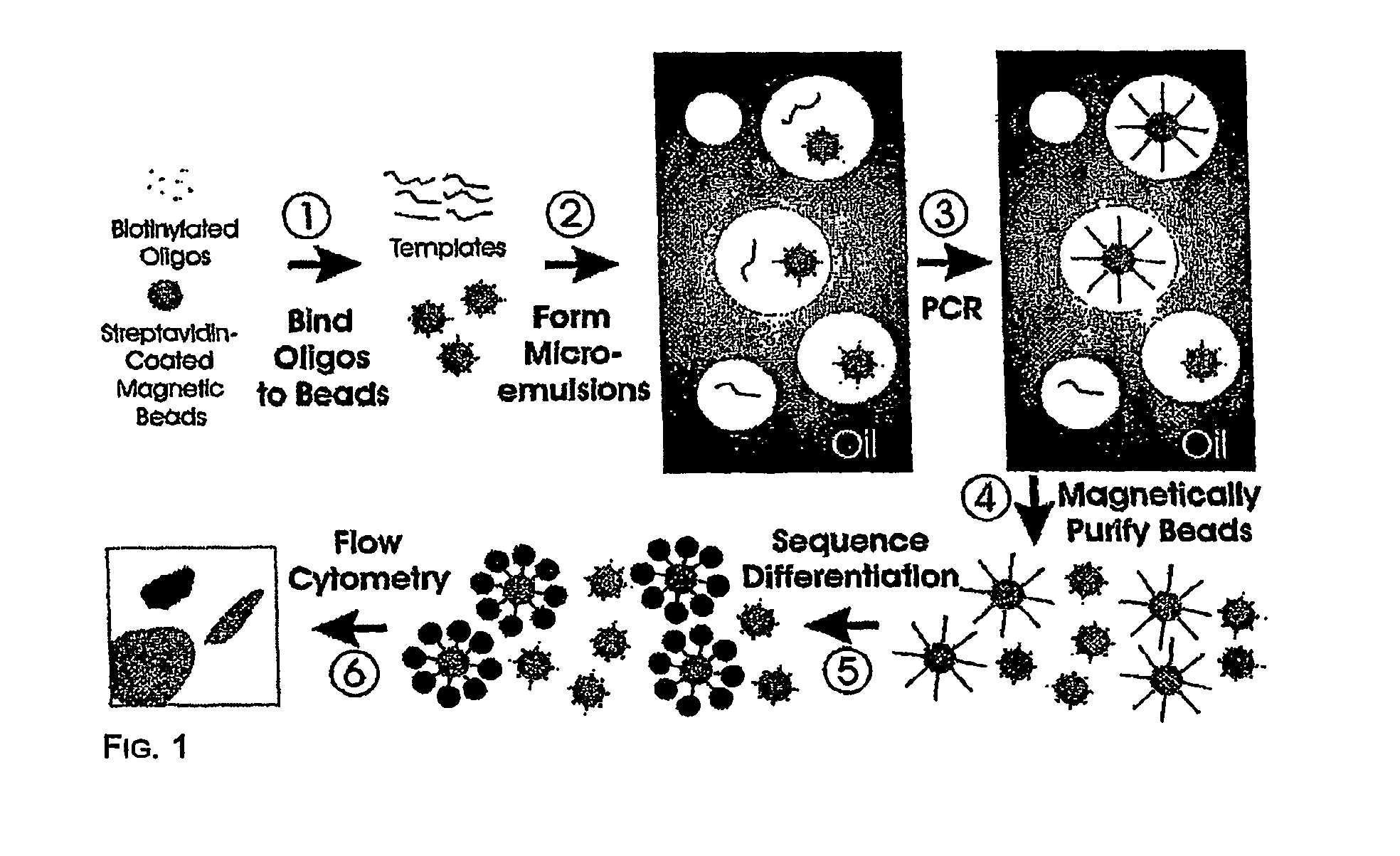
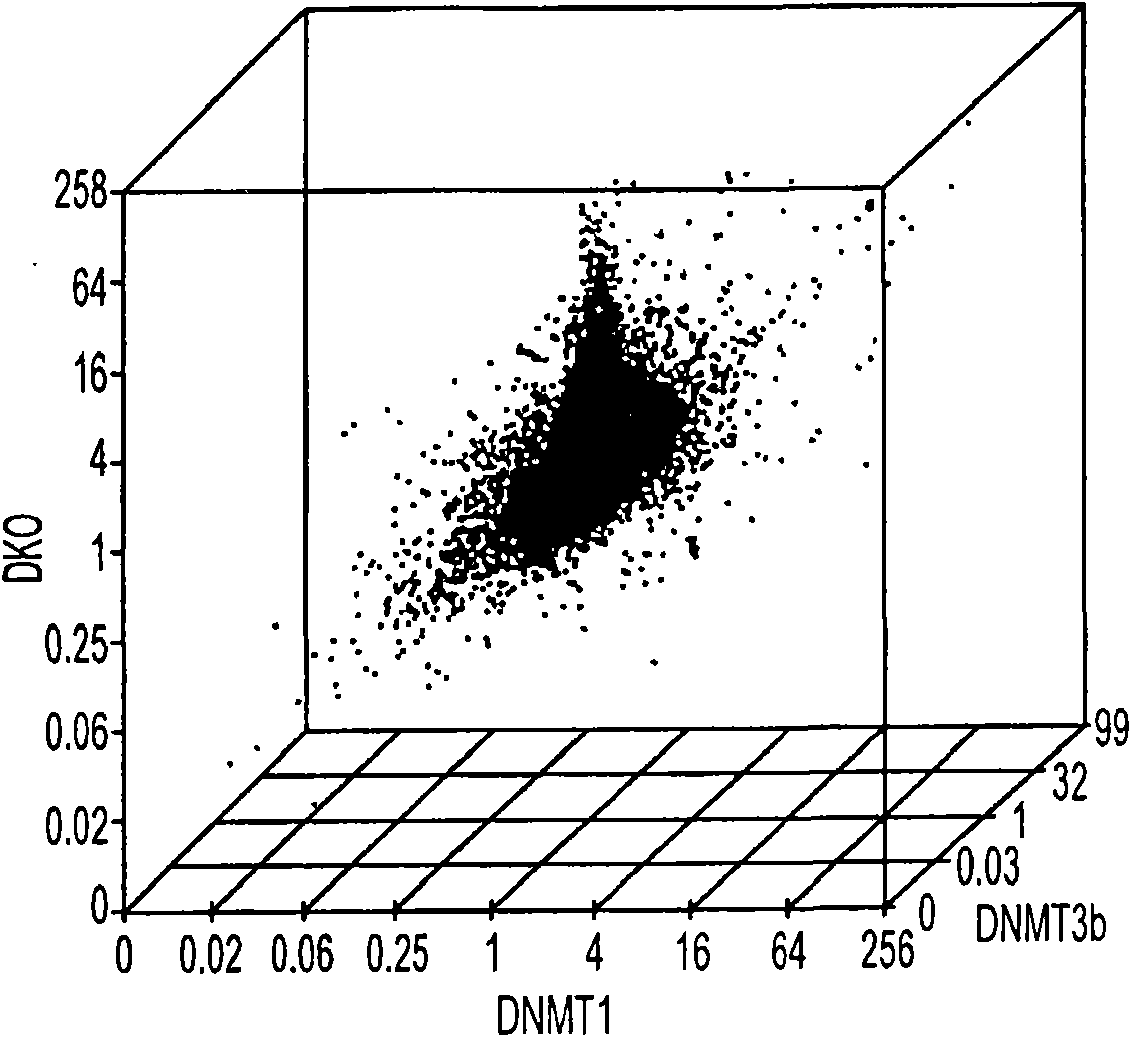
Many areas of biomedical research depend on the analysis of uncommon variations in individual genes or transcripts. Here we describe a method that can quantify such variation at a scale and ease heretofore unattainable. Each DNA molecule in a collection of such molecules is converted into a single particle to which thousands of copies of DNA identical in sequence to the original are bound. This population of beads then corresponds to a one-to-one representation of the starting DNA molecules. Variation within the original population of DNA molecules can then be simply assessed by counting fluorescently-labeled particles via flow cytometry. Millions of individual DNA molecules can be assessed in this fashion with standard laboratory equipment. Moreover, specific variants can be isolated by flow sorting and employed for further experimentation. This approach can be used for the identification and quantification of rare mutations as well as to study variations in gene sequences or transcripts in specific populations or tissues.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
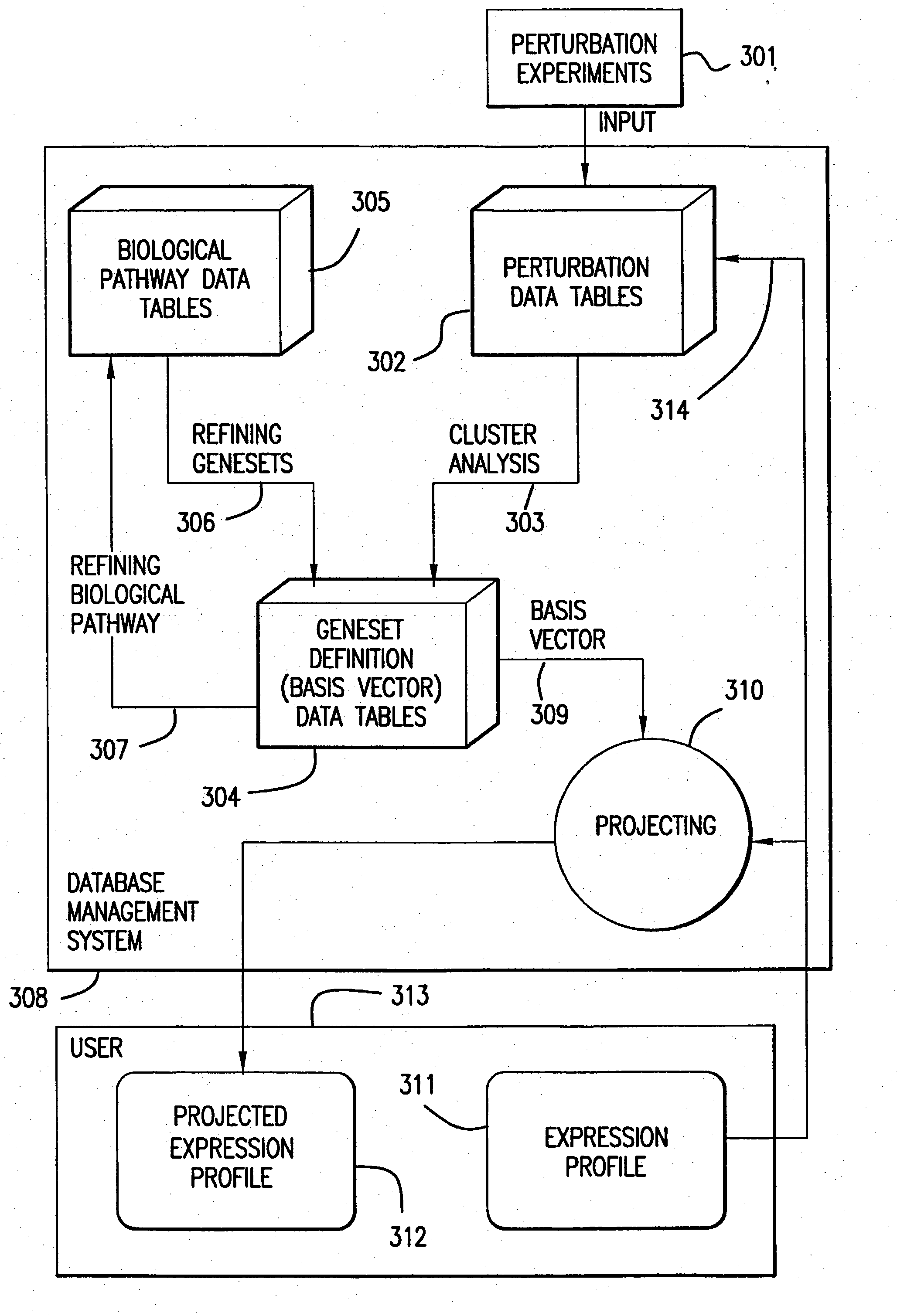
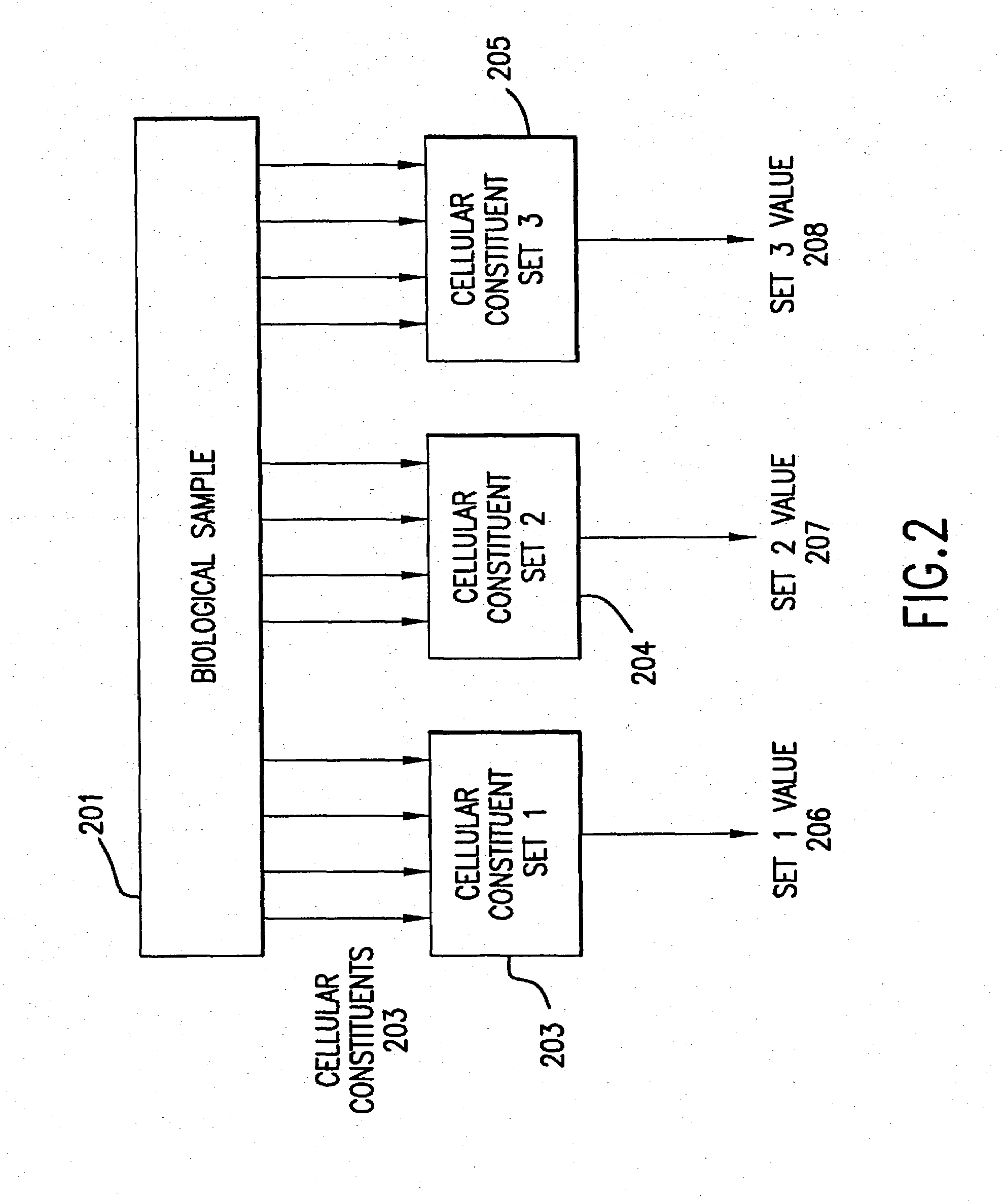
Methods for using co-regulated genesets to enhance detection and classification of gene expression patterns
InactiveUS6203987B1Peptide/protein ingredientsMicrobiological testing/measurementCo-regulationIndividual gene
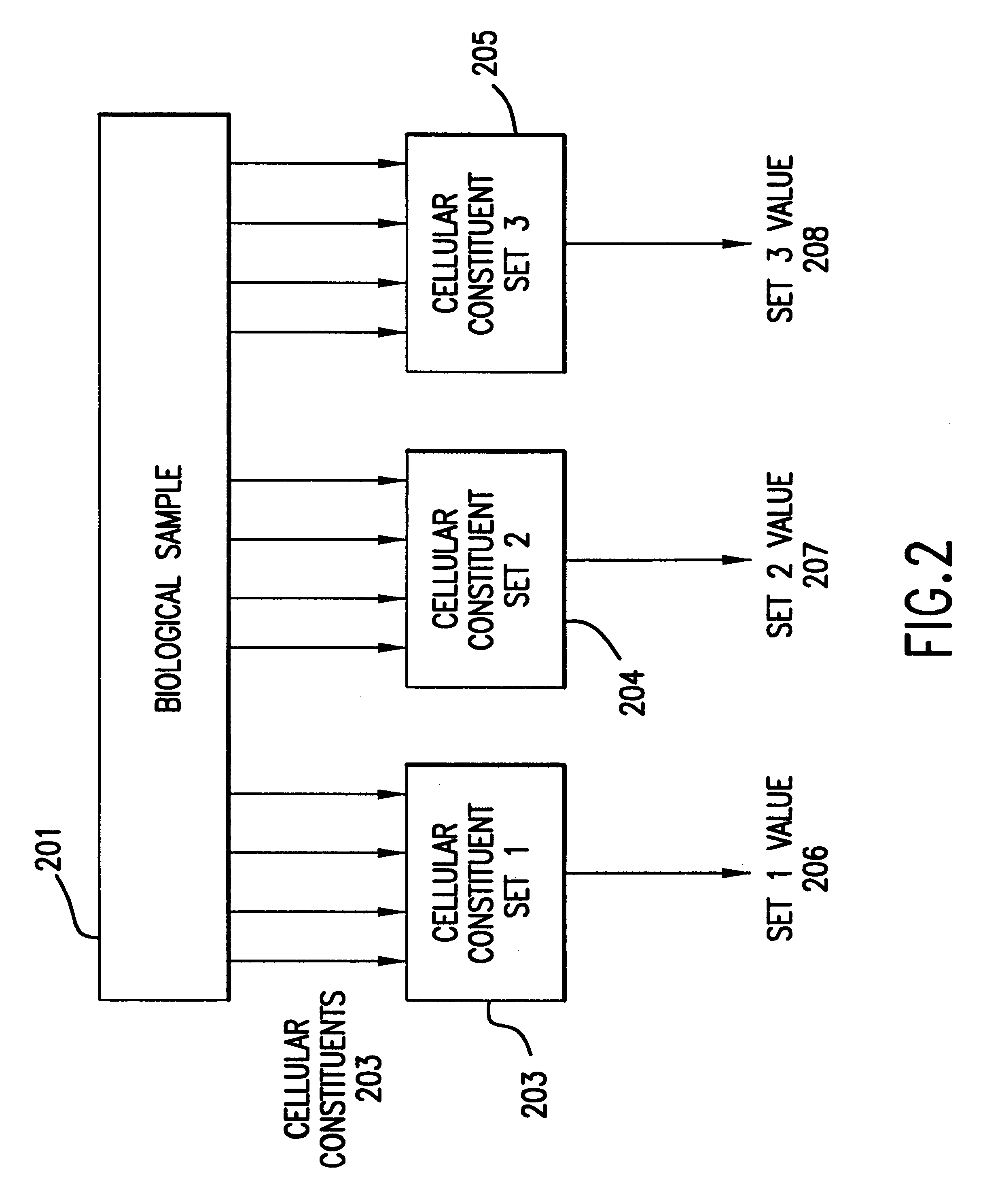
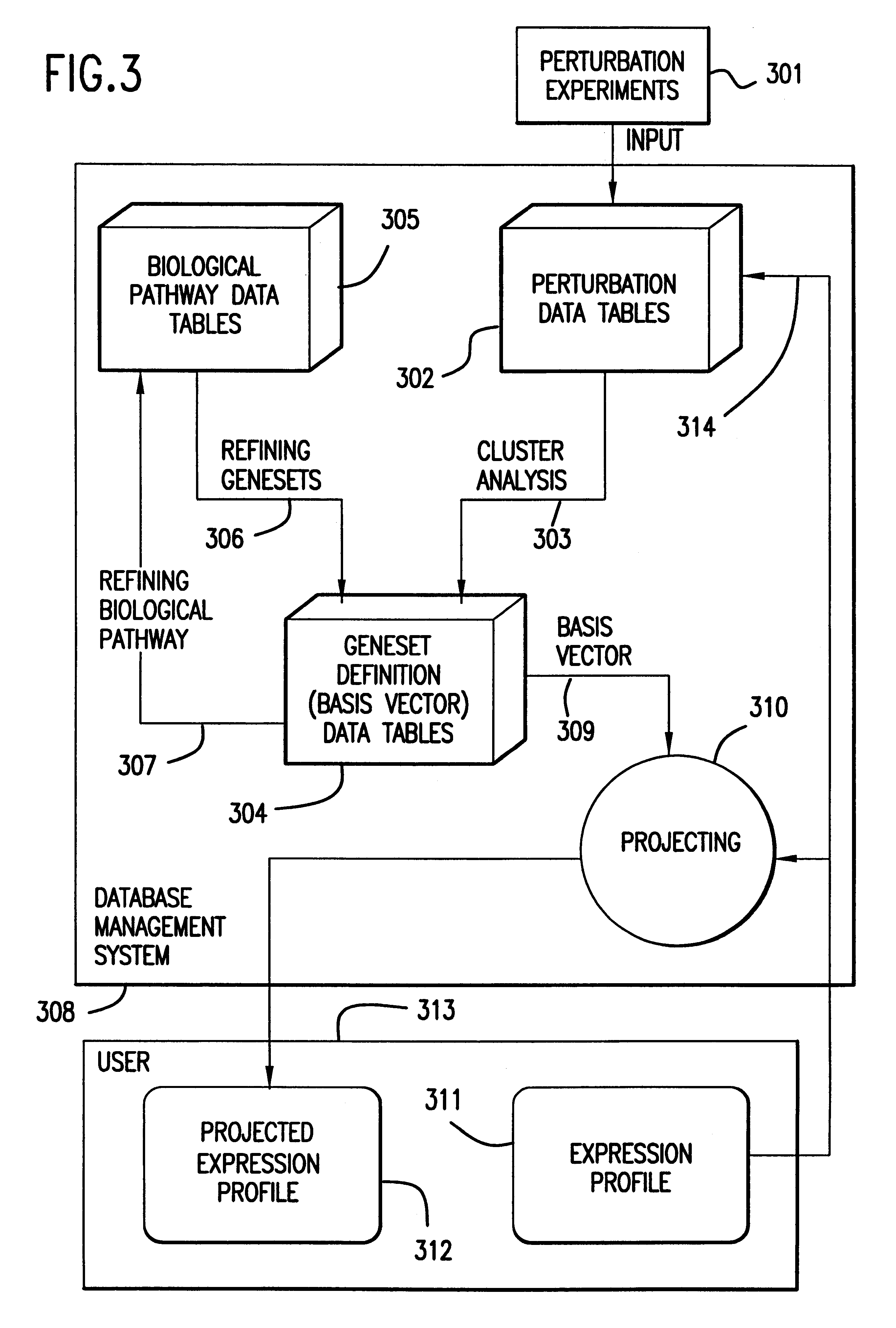
The present invention provides methods for enhanced detection of biological response patterns. In one embodiment of the invention, genes are grouped into basis genesets according to the co-regulation of their expression. Expression of individual genes within a geneset is indicated with a single gene expression value for the geneset by a projection process. The expression values of genesets, rather than the expression of individual genes are then used as the basis for comparison and detection of biological responses with greatly enhanced sensitivity.
Owner:MICROSOFT TECH LICENSING LLC
Methods and compositions for cellular and metabolic engineering
InactiveUS7105297B2Improve abilitiesExcellent catalytic performanceImmunoglobulinsFermentationIndividual geneOrganism
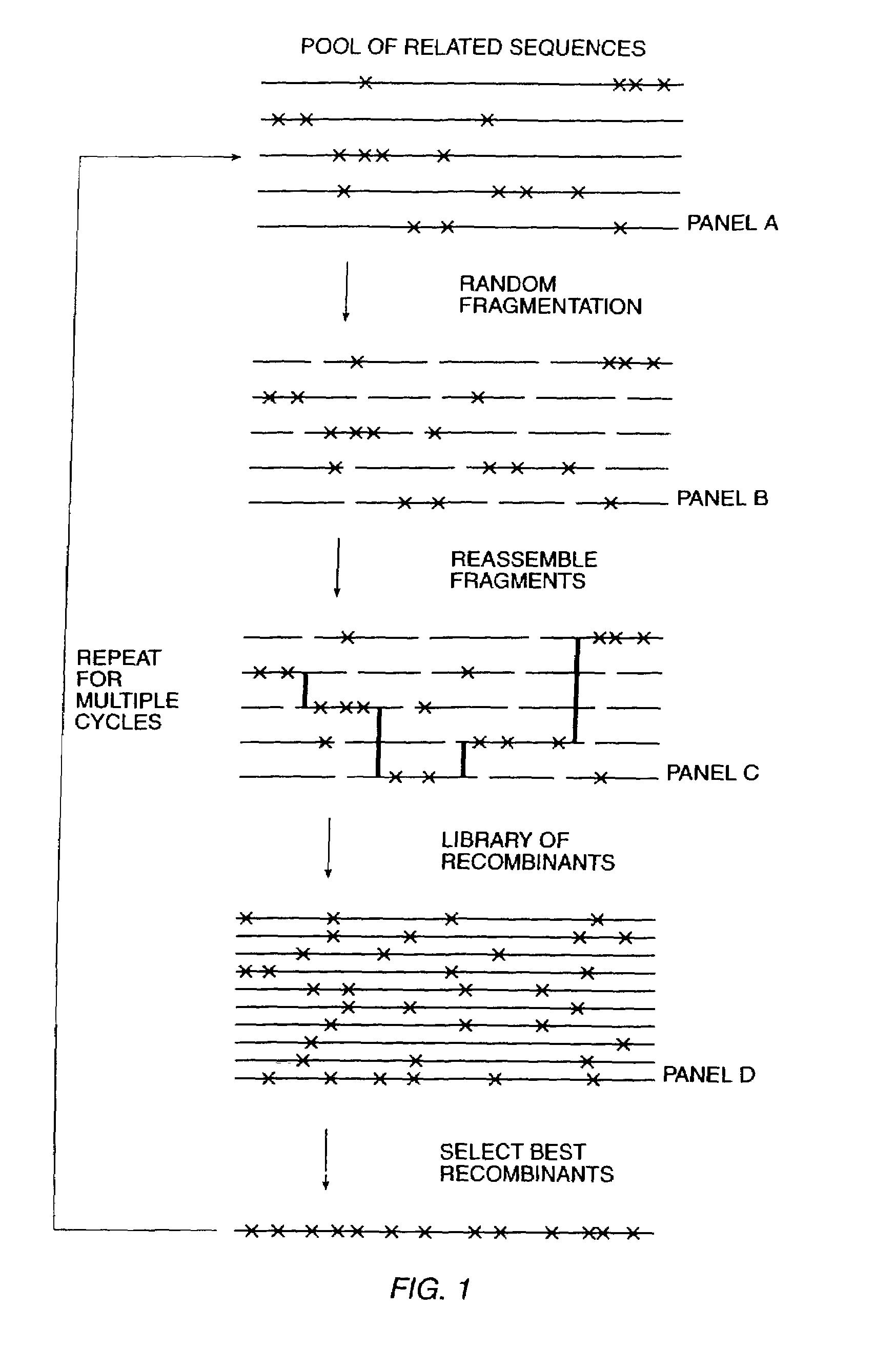
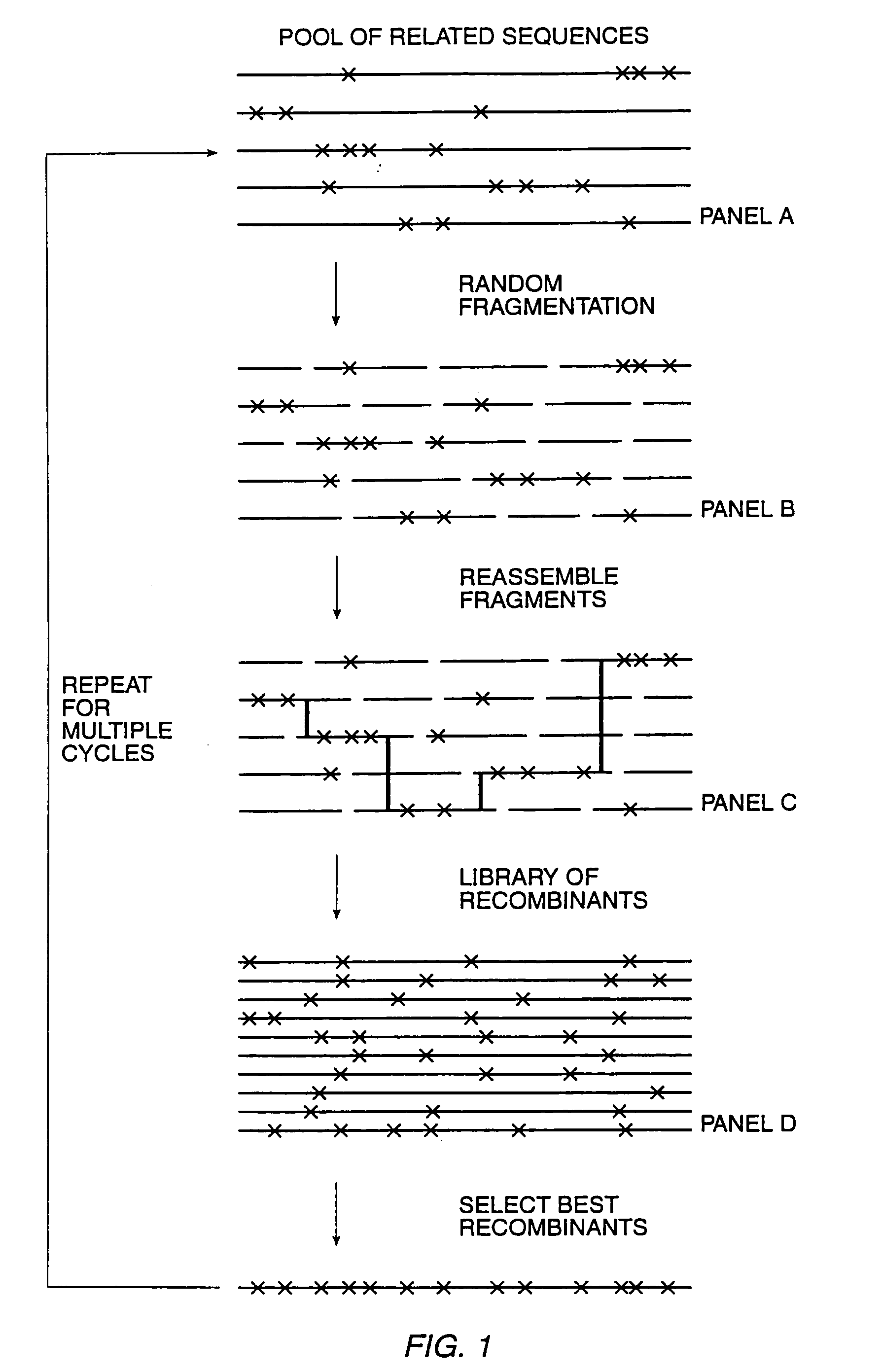
The present invention is generally directed to the evolution of new metabolic pathways and the enhancement of bioprocessing through a process herein termed recursive sequence recombination. Recursive sequence recombination entails performing iterative cycles of recombination and screening or selection to “evolve” individual genes, whole plasmids or viruses, multigene clusters, or even whole genomes. Such techniques do not require the extensive analysis and computation required by conventional methods for metabolic engineering.
Owner:CODEXIS MAYFLOWER HLDG LLC
Method and Compositions for Detection and Enumeration of Genetic Variations
InactiveUS20090286687A1Sugar derivativesMicrobiological testing/measurementSpecific populationIndividual gene
Many areas of biomedical research depend on the analysis of uncommon variations in individual genes or transcripts. Here we describe a method that can quantify such variation at a scale and ease heretofore unattainable. Each DNA molecule in a collection of such molecules is converted into a single particle to which thousands of copies of DNA identical in sequence to the original are bound. This population of beads then corresponds to a one-to-one representation of the starting DNA molecules. Variation within the original population of DNA molecules can then be simply assessed by counting fluorescently-labeled particles via flow cytometry. Millions of individual DNA molecules can be assessed in this fashion with standard laboratory equipment. Moreover, specific variants can be isolated by flow sorting and employed for further experimentation. This approach can be used for the identification and quantification of rare mutations as well as to study variations in gene sequences or transcripts in specific populations or tissues.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Methods for obtaining and using haplotype data
InactiveUS7058517B1Shorten the timeReduce effortMicrobiological testing/measurementProteomicsPopulationS genotyping
Methods, computer program(s) and database(s) to analyze and make use of gene haplotype information. These include methods, program, and database to find and measure the frequency of haplotypes in the general population; methods, program, and database to find correlation's between an individual's haplotypes or genotypes and a clinical outcome; methods, program, and database to predict an individual's haplotypes from the individual's genotype for a gene; and methods, program, and database to predict an individual's clinical response to a treatment based on the individual's genotype or haplotype.
Owner:GENAISSANCE PHARMA INC
Methods for obtaining and using haplotype data
InactiveUS6931326B1Accurate predictionReduce costs and risksMicrobiological testing/measurementProteomicsHaplotypeS genotyping
Methods, computer program(s) and database(s) to analyze and make use of gene haplotype information. These include methods, program, and database to find and measure the frequency of haplotypes in the general population; methods, program, and database to find correlation's between an individual's haplotypes or genotypes and a clinical outcome; methods, program, and database to predict an individual's haplotypes from the individual's genotype for a gene; and methods, program, and database to predict an individual's clinical response to a treatment based on the individual's genotype or haplotype.
Owner:PGXHEALTH
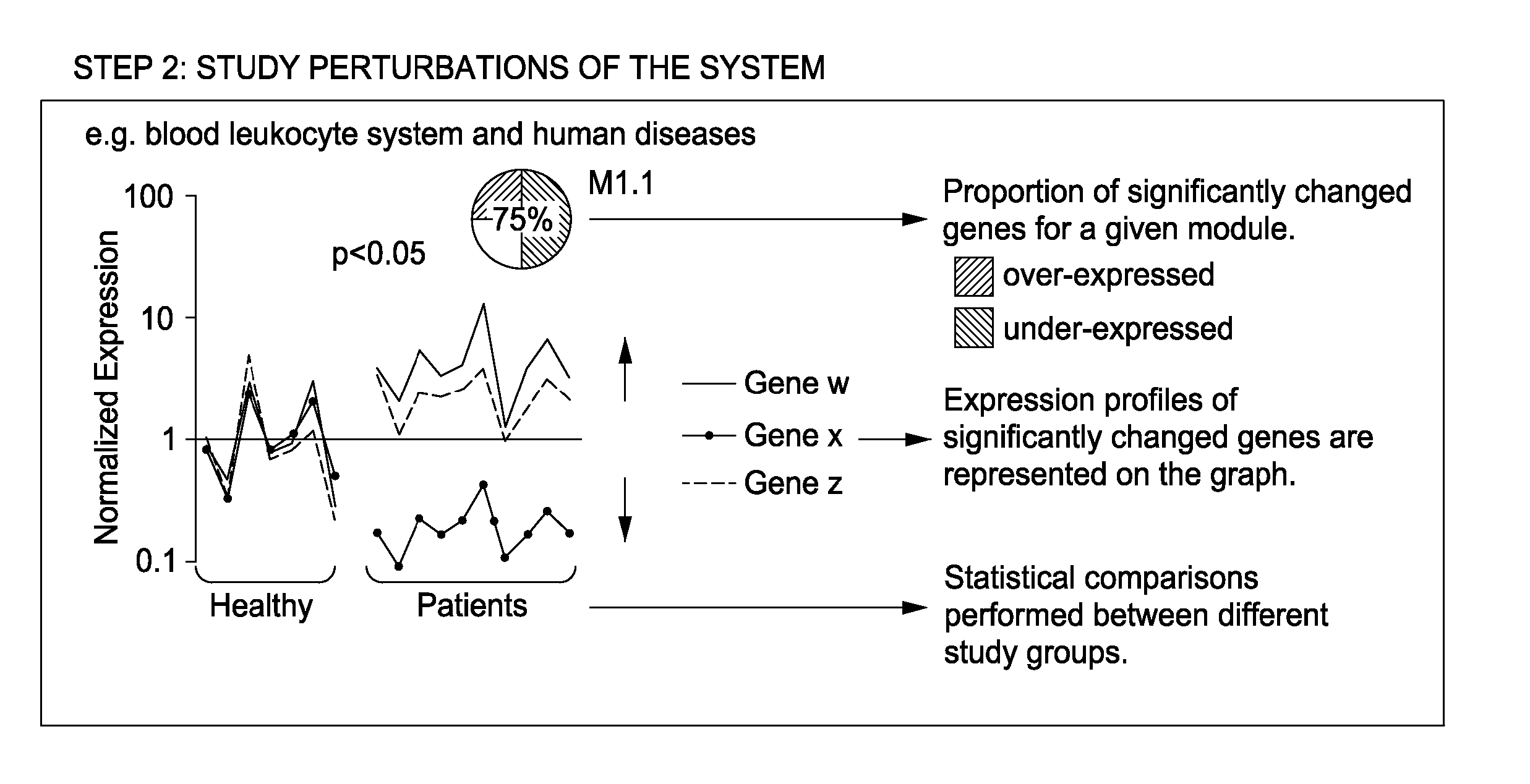
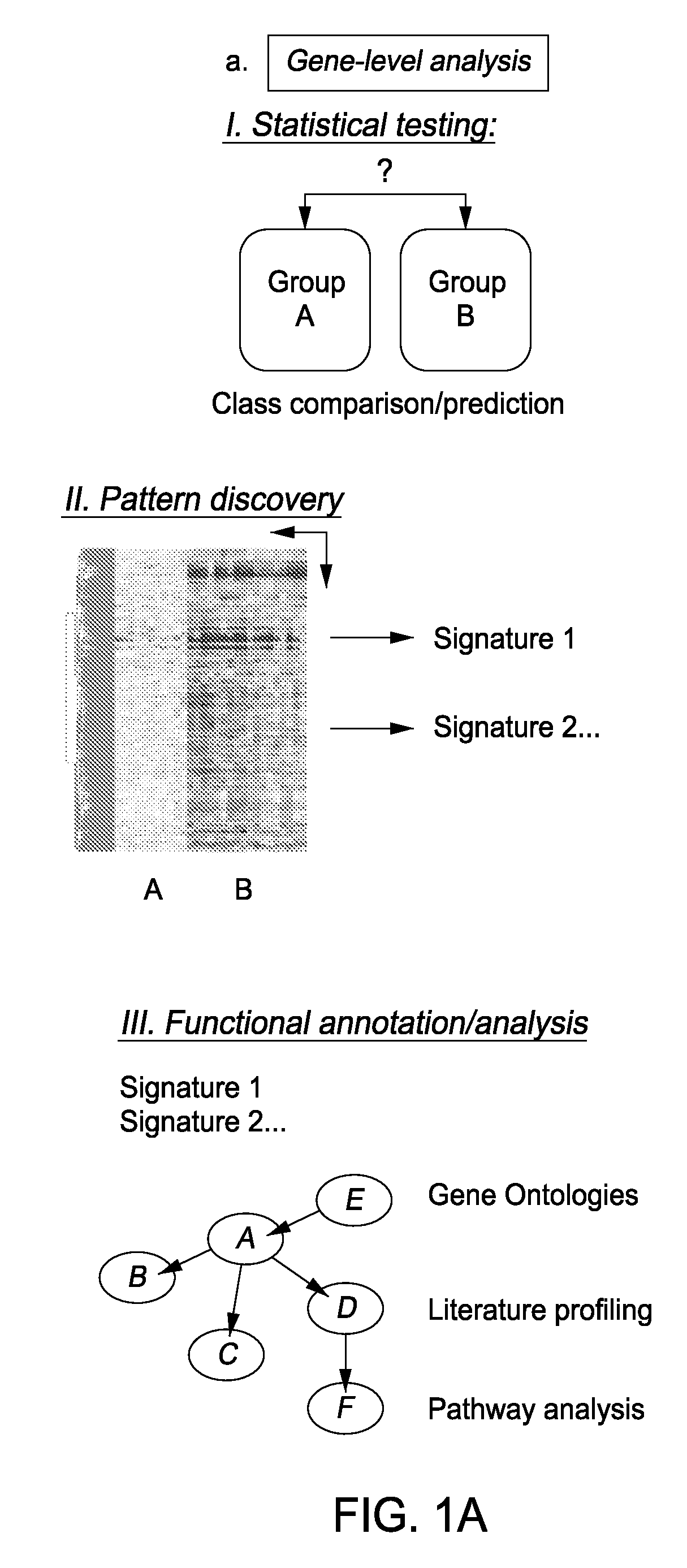
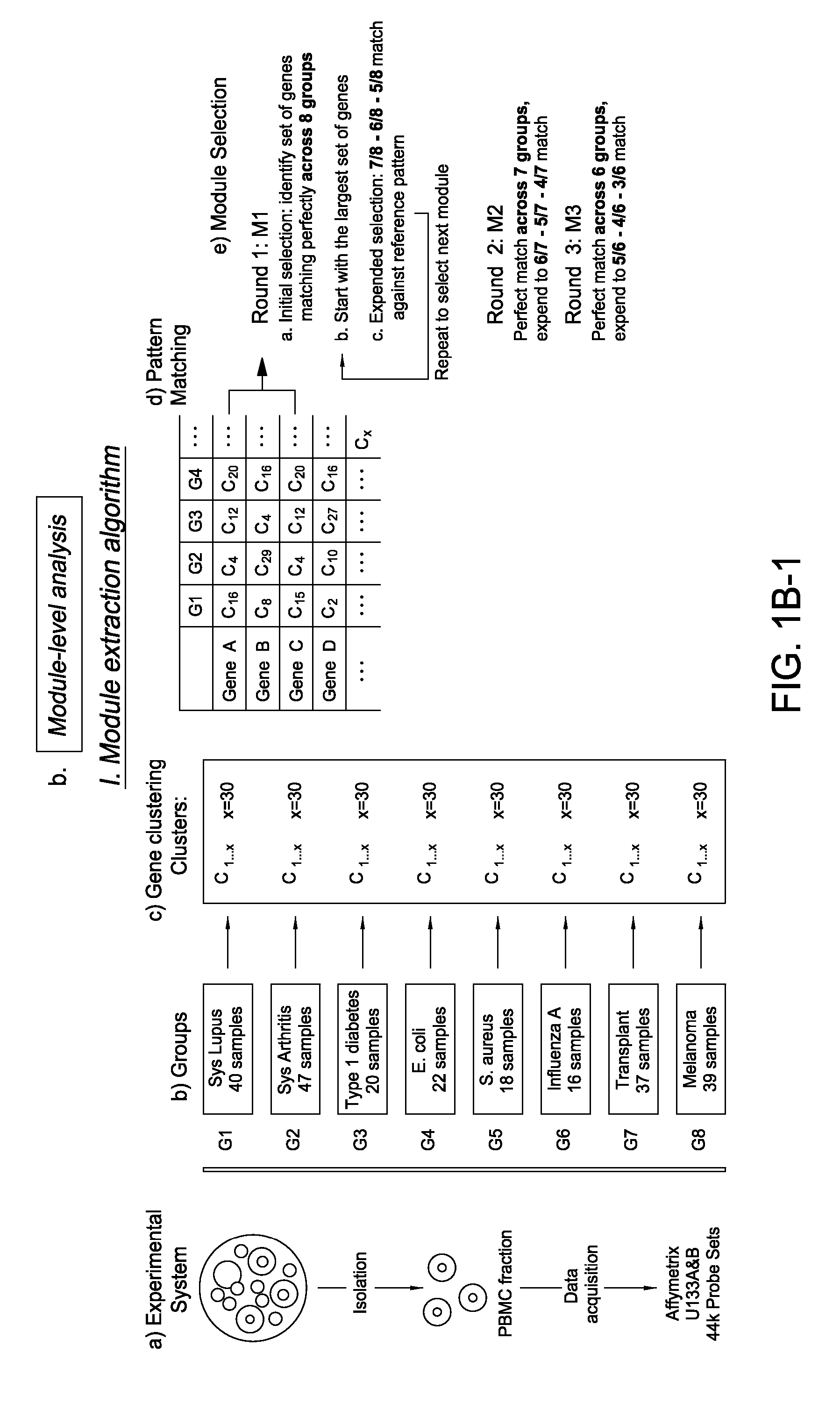
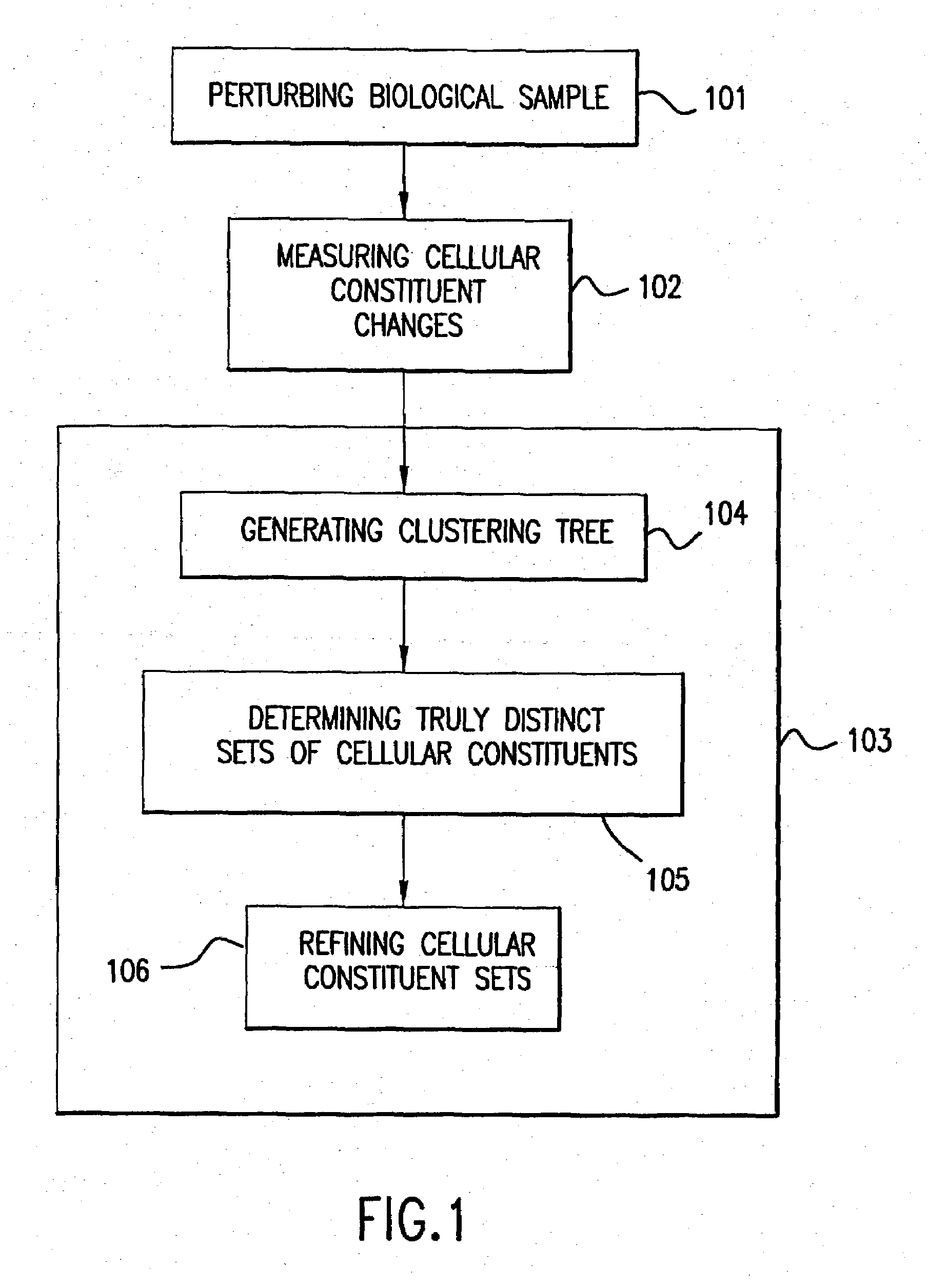
Module-Level Analysis of Peripheral Blood Leukocyte Transcriptional Profiles
InactiveUS20070231816A1Easy to explainDifficult to interpretSugar derivativesMicrobiological testing/measurementIndividual geneBioinformatics
The present invention includes an apparatus, system and method for the development and use of transcriptional modules by obtaining individual gene expression levels from cells obtained from one or more patients with a disease or condition; recording the expression value for each gene in a table that is divided into clusters; iteratively selecting gene expression values for one or more transcriptional modules by: selecting for the module the genes from each cluster that match in every disease or condition; removing the selected genes from the analysis; and repeating the process of gene expression value selection for genes that cluster in a sub-fraction of the diseases or conditions; and iteratively repeating the generation of modules.
Owner:BAYLOR RES INST
Methods for obtaining and using haplotype data
InactiveUS20050191731A1Accurate predictionReduce costs and risksData visualisationBiostatisticsHaplotypeCrowds
Methods, computer program(s) and database(s) to analyze and make use of gene haplotype information. These include methods, program, and database to find and measure the frequency of haplotypes in the general population; methods, program, and database to find correlation's between an individual's haplotypes or genotypes and a clinical outcome; methods, program, and database to predict an individual's haplotypes from the individual's gen type for a gene; and methods, program, and database to predict an individual's clinical response to a treatment based on the individual's genotype or haplotype.
Owner:JUDSON RICHARD S +3
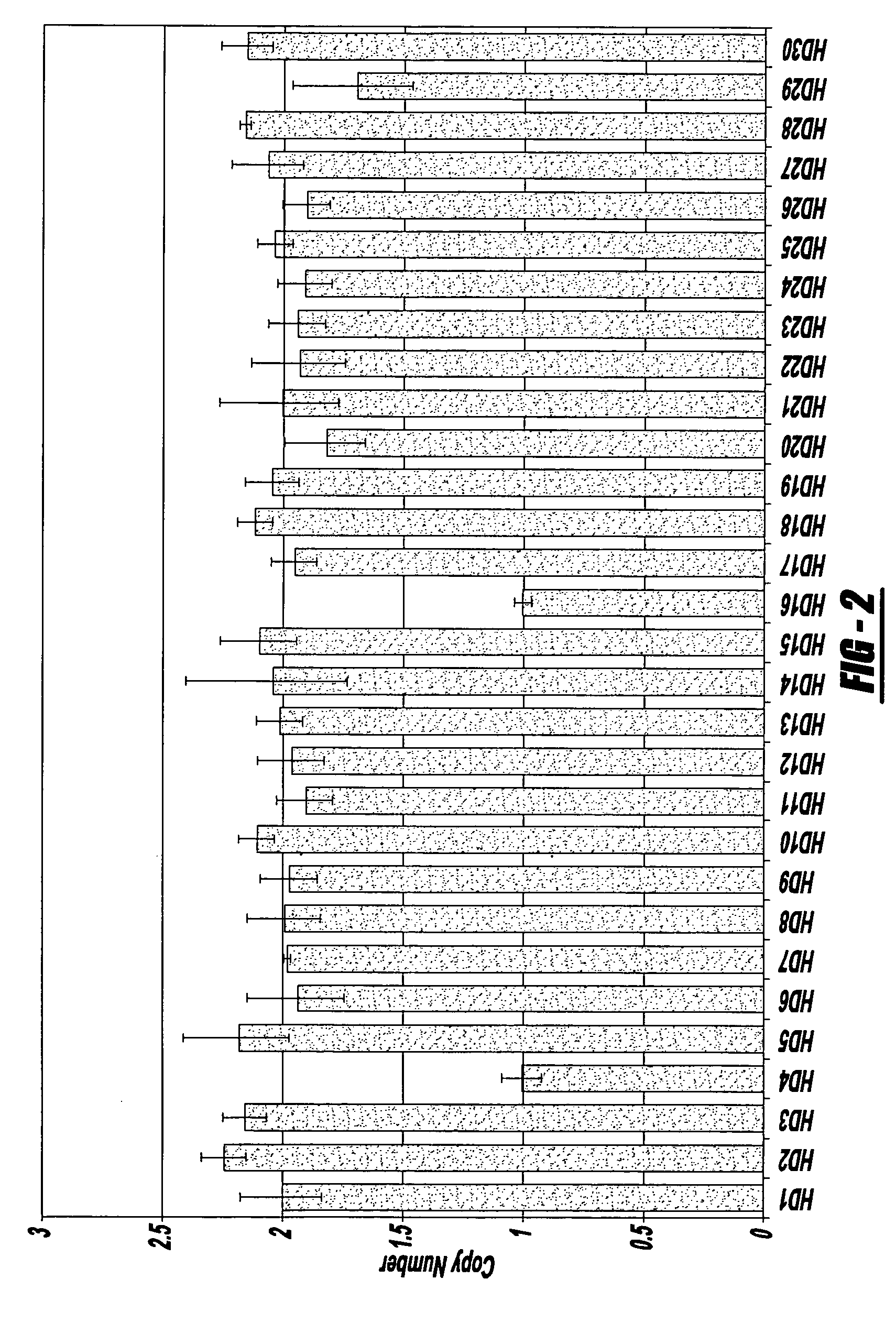
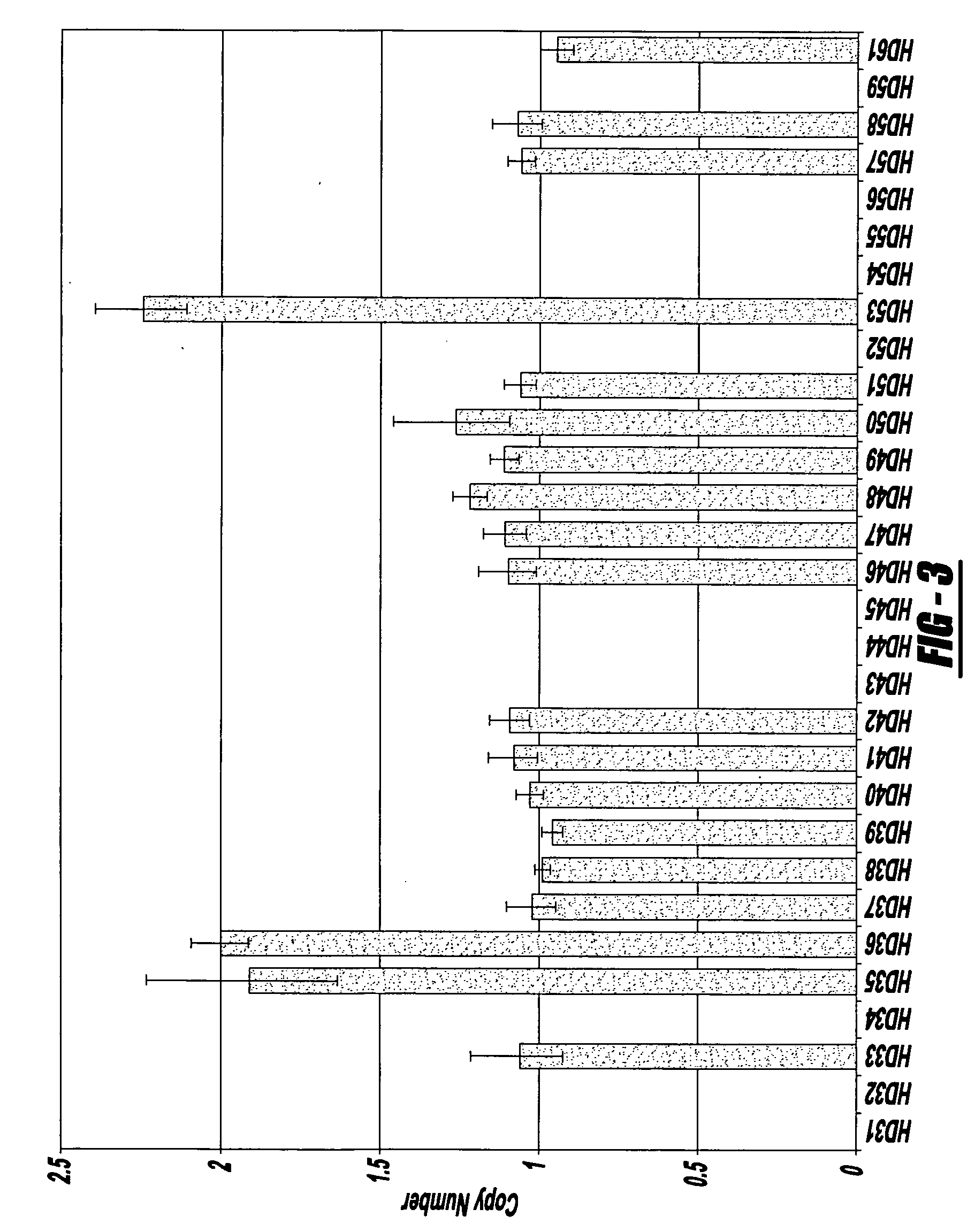
Assays for the direct measurement of gene dosage
InactiveUS20070087345A1Reducing agentMicrobiological testing/measurementIndividual geneImage resolution
The present invention relates to compositions, methods, and kits for quantifying variations in gene copy number, e.g., of individual genes or of chromosomes or portions of chromosomes in an homogeneous reaction, without the need for target amplification, fragment size resolution, or microscopy.
Owner:THIRD WAVE TECH
Allele-specific expression patterns
The invention provides methods of analyzing genes for differential relative allelic expression patterns. Haplotype blocks throughout the genomes of individuals are analyzed to identify haplotype patterns that are associated with specific differential relative allelic expression patterns. Haplotype blocks that contain associated haplotype patterns may be further investigated to identify genes or variants of genes involved in differential relative allelic expression patterns.
Owner:PERLEGEN SCIENCES INC
Methods for using co-regulated genesets to enhance detection and classification of gene expression patterns
InactiveUS20090204333A1Less vulnerableReduced dimensionBioreactor/fermenter combinationsBiological substance pretreatmentsBiologic responseMethod comparison
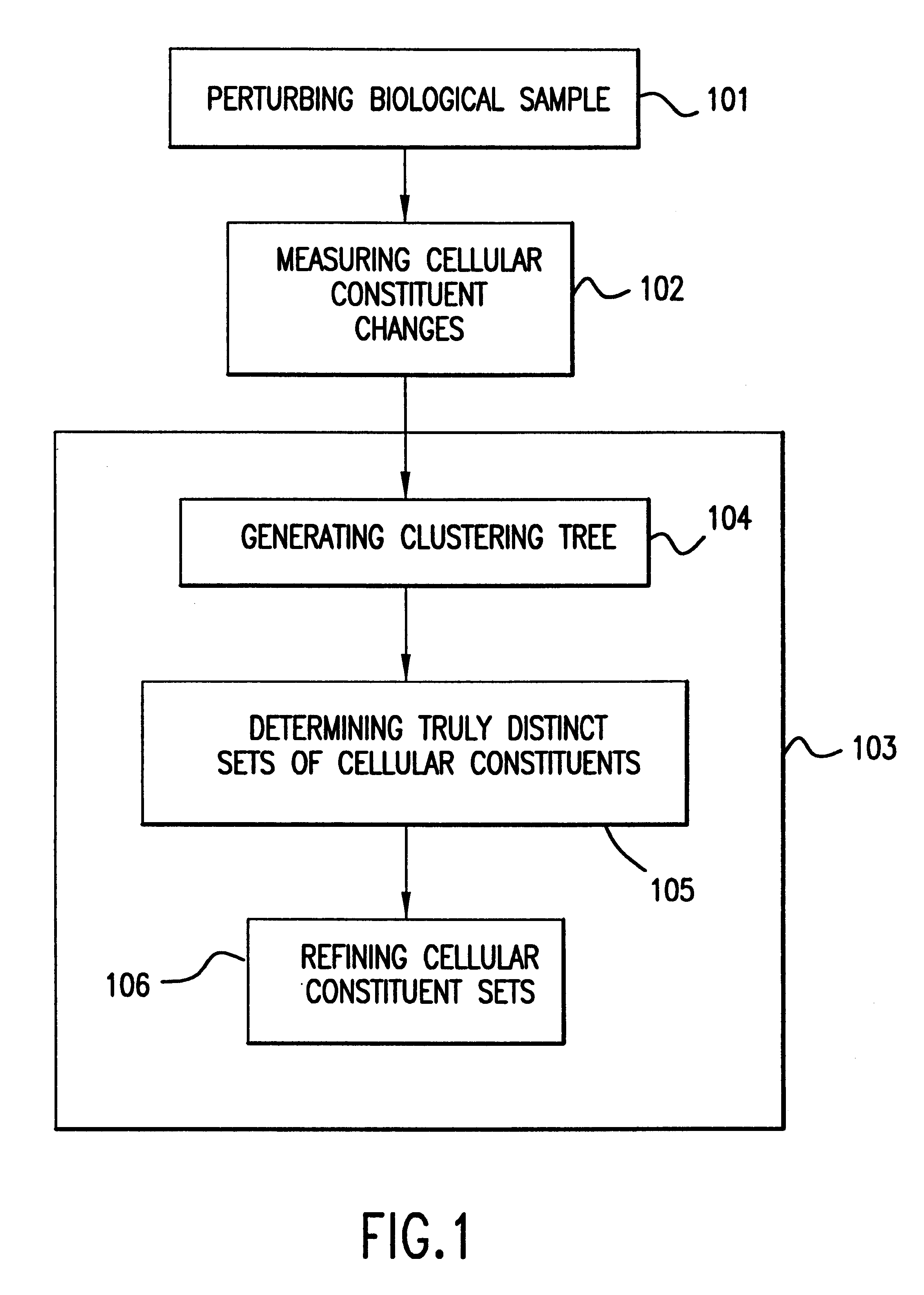
The present invention provides methods for enhanced detection of biological response patterns. In one embodiment of the invention, genes are grouped into basis genesets according to the co-regulation of their expression. Expression of individual genes within a geneset is indicated with a single gene expression value for the geneset by a projection process. The expression values of genesets, rather than the expression of individual genes, are then used as the basis for comparison and detection of biological response with greatly enhanced sensitivity. In another embodiment of the invention, biological responses are grouped according to the similarity of their biological profile.The methods of the invention have many useful applications, particularly in the fields of drug development and discovery. For example, the methods of the invention may be used to compare biological responses with greatly enhanced sensitivity. The biological responses that may be compared according to these methods include responses to single perturbations, such as a biological response to a mutation or temperature change, as well as graded perturbations such as titration with a particular drug. The methods are also useful to identify cellular constituents, particularly genes, associated with a particular type of biological response. Further, the methods may also be used to identify perturbations, such as novel drugs or mutations, which effect one or more particular genesets. The methods may still further be used to remove experimental artifacts in biological response data.
Owner:MICROSOFT TECH LICENSING LLC
Early detection and prognosis of colon cancers
InactiveCN101688239ASugar derivativesMicrobiological testing/measurementGenes mutationTumor specific
We have developed a transcriptome-wide approach to identify genes affected by promoter CpG island hypermethylation and transcriptional silencing in colorectal cancer (CRC). By screening cell lines andvalidating tumor specific hypermethylation in a panel of primary human CRC samples, we estimate that nearly 5% of all known genes may be promoter methylated in an individual tumor. When directly compared to gene mutations, we find a much larger number of genes hypermethylated in individual tumors, and much higher frequency of hypermethylation within individual genes harboring either genetic or epigenetic changes. Thus, to enumerate the full spectrum of alterations in the human cancer genome, and facilitate the most efficacious grouping of tumors to identify cancer biomarkers and tailor therapeutic approaches, both genetic and epigenetic screens should be undertaken. The genes we identified can be used inter alia diagnostically to detect cancer, pre-cancer, and likelihood of developing cancer.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE +1
Analysis of gene family expression
InactiveUS6232065B1Eliminates the cumbersome cloning and sequencing strategiesOvercomes shortcomingSugar derivativesMicrobiological testing/measurementIndividual geneGene expression profiling
The present invention relates to methods and compositions for characterizing the expression patterns of genes and gene families. Specifically, the present invention provides means to generate and monitor gene expression profiles resulting from cellular and physiological changes such that the expression patterns of individual genes or groups of genes can be readily identified and characterized.
Owner:CASE WESTERN RESERVE UNIV
Methods for rapid production of target double-stranded DNA sequences
InactiveUS20100016178A1Increase synthesisIncrease volumeMicrobiological testing/measurementFermentationChemical synthesisIndividual gene
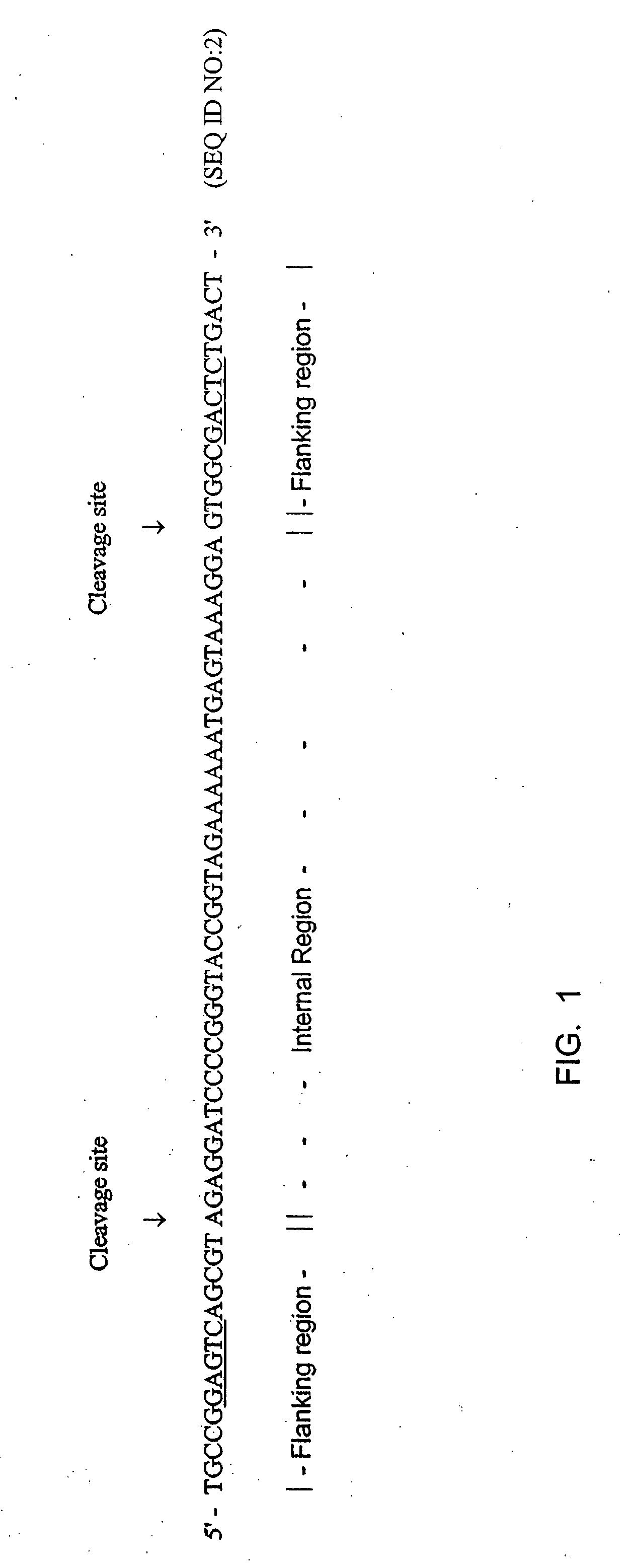
It has been previously disclosed that DNA segments can be made in massively parallel chemical synthesis operations on a common substrate followed by release of the segments from the substrate and assembly of the segments into target DNA molecules. Here it is taught that if the DNA primary constructs are sufficiently long and properly designed, that the copy numbers of the primary constructs can be multiplied as needed by a PCR process using as a template regions at the ends of the primary constructs. The end regions, called flanking regions, can also be designed so that they may be cleaved easily from the amplification products. The target double-stranded DNA can then be assembled from the cleaved fragments. Hundreds of thousands of oligonucleotides can be synthesized and assembled into many different individual genes by this process in a relatively quick and efficient process.
Owner:WISCONSIN ALUMNI RES FOUND
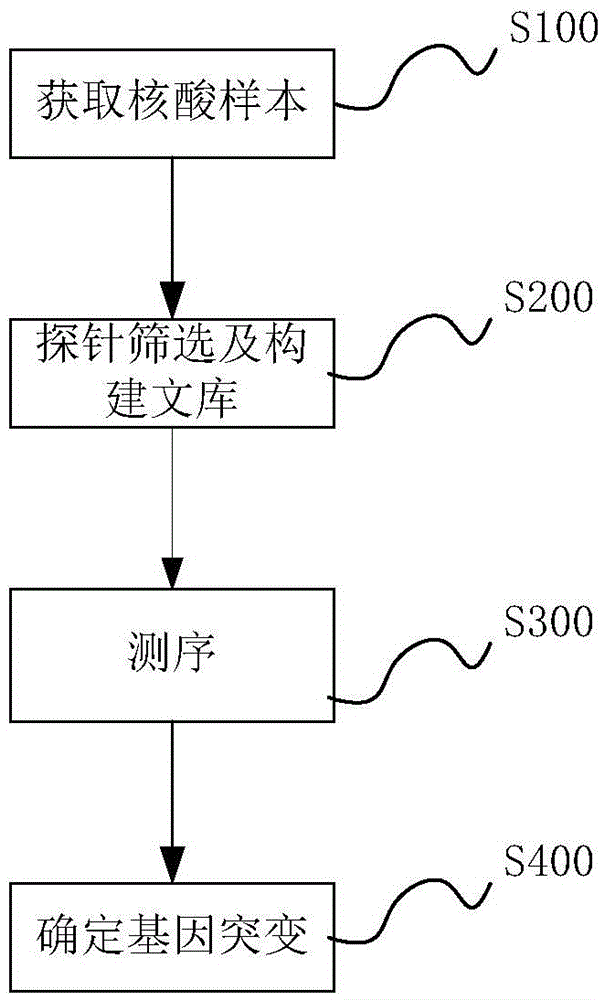
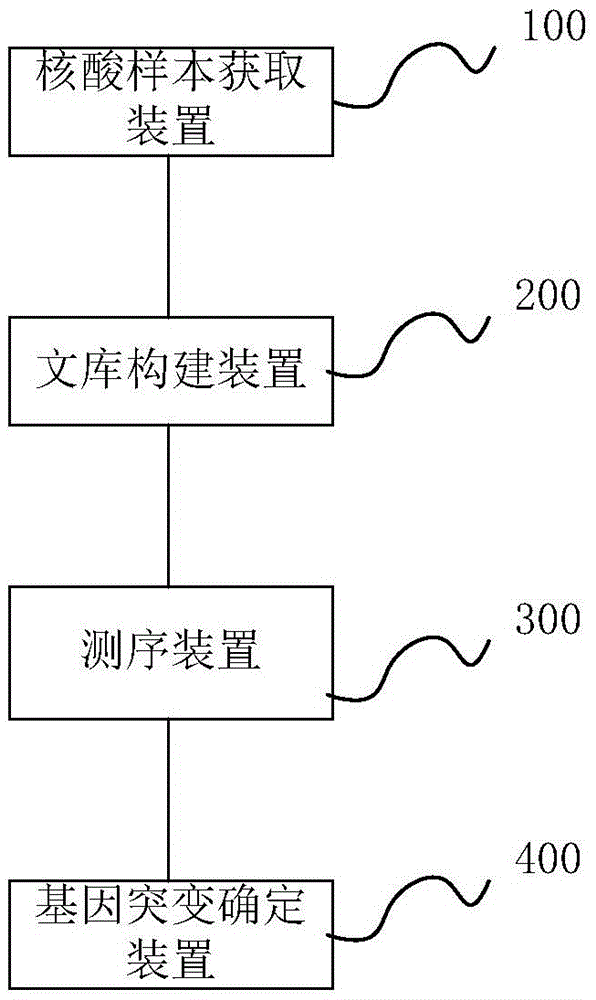
Method and system for determining individual gene mutation
InactiveCN105420351AReduce sample volumeLow costBioreactor/fermenter combinationsBiological substance pretreatmentsDiseaseIndividual gene
Owner:SHENZHEN HUADA GENE INST
Methods and compositions for cellular and metabolic engineering
InactiveUS20060257890A1Improve abilitiesExcellent catalytic performanceMicrobiological testing/measurementLibrary screeningIndividual geneGene cluster
The present invention is generally directed to the evolution of new metabolic pathways and the enhancement of bioprocessing through a process herein termed recursive sequence recombination. Recursive sequence recombination entails performing iterative cycles of recombination and screening or selection to “evolve” individual genes, whole plasmids or viruses, multigene clusters, or even whole genomes. Such techniques do not require the extensive analysis and computation required by conventional methods for metabolic engineering.
Owner:MAXYGEN
Methods and Apparatus for Assigning a Meaningful Numeric Value to Genomic Variants, and Searching and Assessing Same
InactiveUS20120191366A1Easily parseReadily parsableBiological testingSequence analysisNumeric ValueReference genome
The present invention relates to methods, apparatus and computer systems for assigning a numerical value to a genotype at a single- or multi-base segment in an individual's genome to denote the presence of a match or a mismatch of a nucleic acid base sequence of one or more chromosomal copies of the segment, as compared to the nucleic acid base sequence at a reference genome segment that corresponds to the segment of the individual's genome. The methods involve assigning a single digit numerical value to the match or the mismatch of each chromosomal copy of the segment in the genome, so that the numerical value assigned to a mismatch is greater than the numerical value of the match. A null symbol is assigned to a no call determination. The assigned numerical values are summed and a total numerical value which is a single digit or a fixed number of digits is obtained. The steps are repeated to create a vector of total numerical values for the segment among the set of genomes, to thereby obtain a segment-specific pattern of genotype match / mismatch between a set of genomes and the nucleic acid base sequence at the reference genome segment. The segment-specific pattern, also referred to as a “diff pattern” can be used to filter or uncover specific trends or sub-patterns across a set of genomes, and more quickly identify genotypic / phenotypic relationships by identifying sites where the distribution of genotypes in the set of genomes relates in a distinctive, causal way to the distribution of a given phenotype among the individuals whose genomes are under study.
Owner:KNOME
Efficient high-accuracy whole-genome selection method capable of performing parallel operation
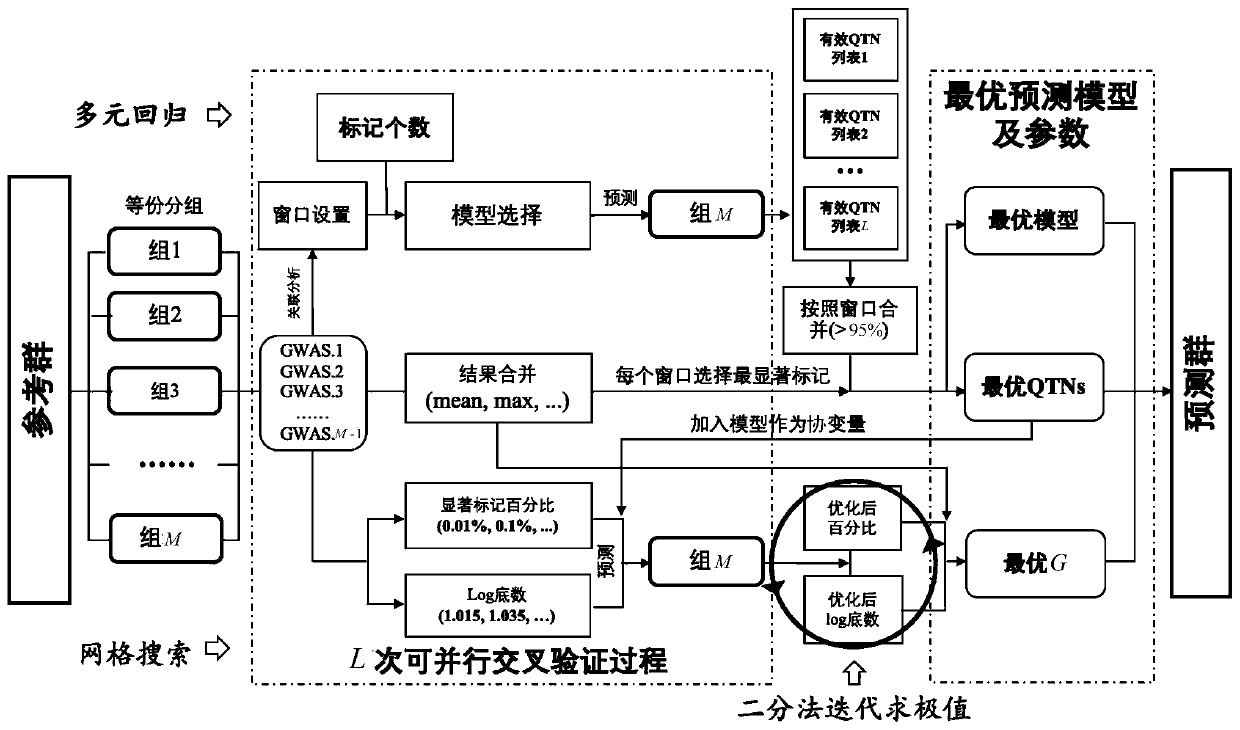
ActiveCN110610744AImprove accuracyImprove computing efficiencyProteomicsGenomicsWhole Genome Association AnalysisAlgorithm
The invention relates to the technical field of animal and plant breeding and human disease prediction, and provides an efficient high-accuracy whole-genome selection method capable of performing parallel operation. The method comprises the following steps: firstly, reading an original genotype file and a phenotype file, constructing a new genotype file and a new phenotype file, and calculating agenetic relationship matrix of all individuals; then, extracting all individuals in the new phenotypic file as a reference group, and extracting all individuals without phenotypic data in the originalgenotypic file as a prediction group; carrying out whole genome association analysis by utilizing the reference group data, and extracting result characteristics of the whole genome association analysis; constructing a model library with specific characters, sequentially optimizing an optimal fixed effect and an optimal random effect by adopting a cross validation strategy, and selecting an optimal prediction model from the model library; and finally, calculating genome estimated breeding values of the prediction group by utilizing the optimal prediction model. The method can quickly, accurately and stably predict individual genome breeding values, and thus the accuracy and efficiency of whole genome selection are improved.
Owner:武汉影子基因科技有限公司
Two-Dimensional Cell Array Device and Apparatus for Gene Quantification and Sequence Analysis
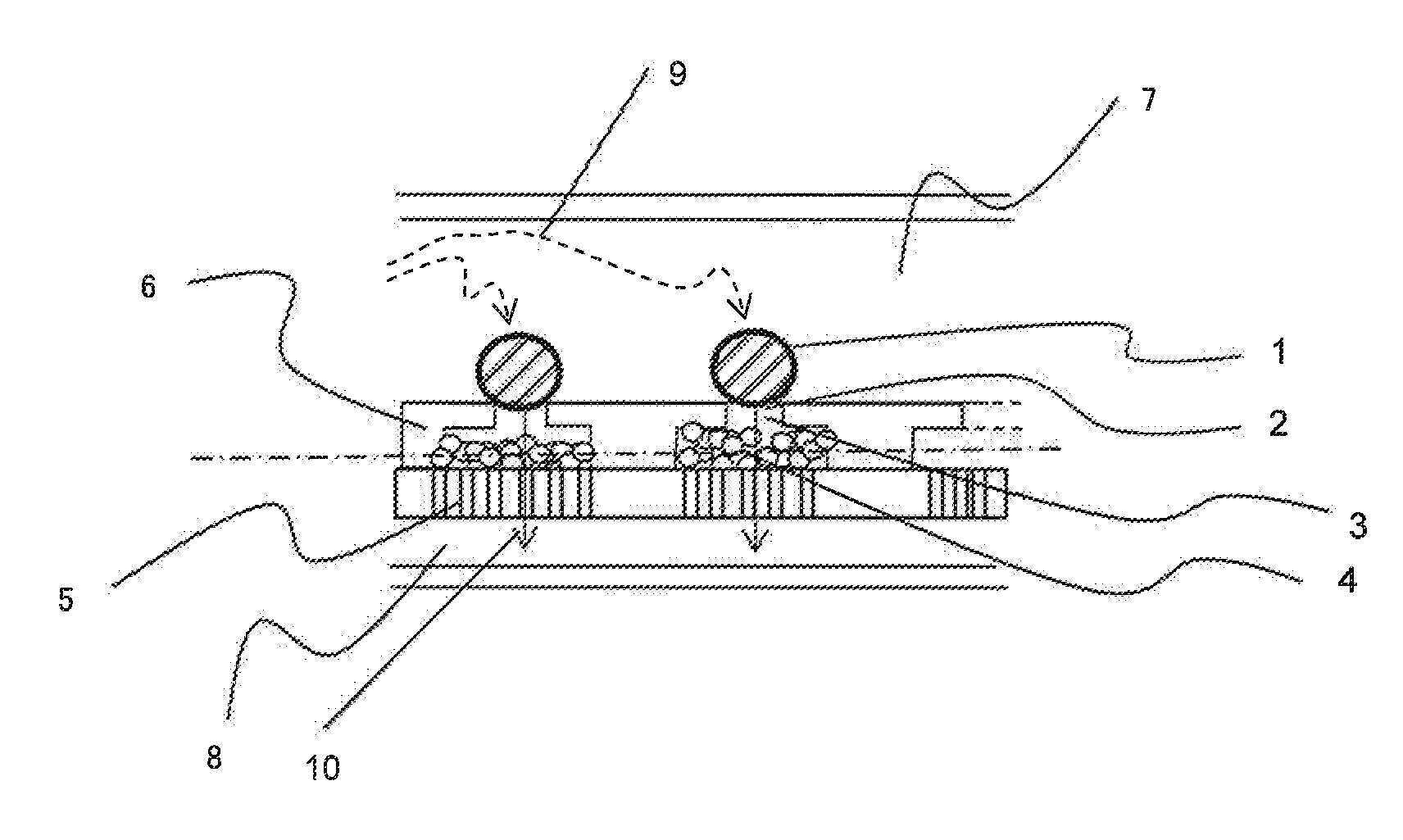
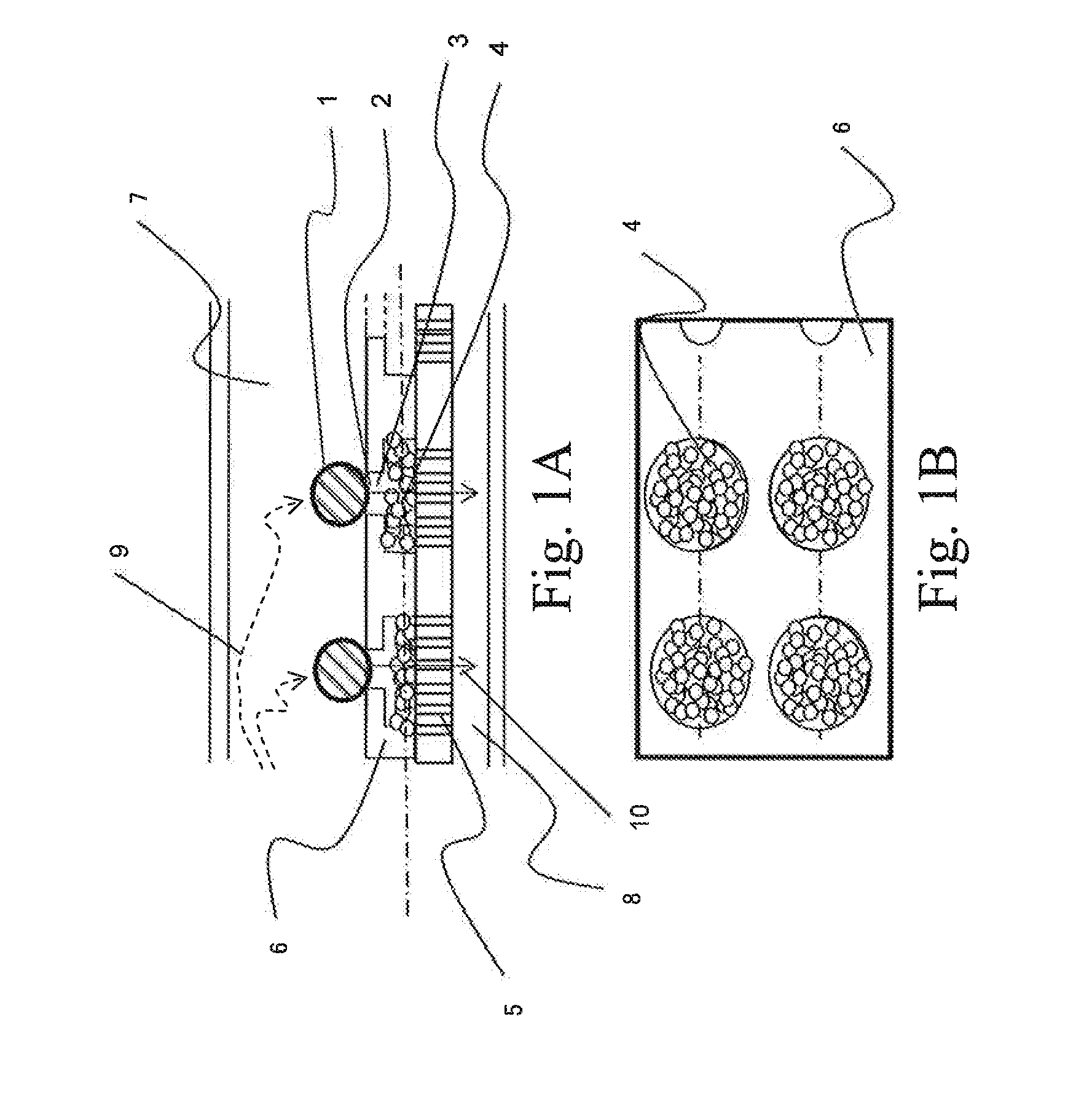
ActiveUS20160010078A1Increase the number of cellsEfficiently treating moleculesLibrary creationLibraries apparatusSequence analysisIndividual gene
In order to conduct gene expression analysis of a number of genes in a number of cells, it has been necessary to separate cells, extract genes therefrom, amplify nucleic acids, and perform sequence analysis. However, separation of cells imposes damages on the cells, and it requires the use of an expensive system. Gene expression analysis in each cell can be carried out with high accuracy by arranging a pair of structures comprising a cell trapping section and a nucleic acid trapping section in a vertical direction to extract individual genes in relevant cells, synthesizing cDNA in the nucleic acid trapping section, amplifying nucleic acids, and analyzing the sequences using a next-generation sequencer.
Owner:HITACHI LTD
Method for extending three-dimensional face database
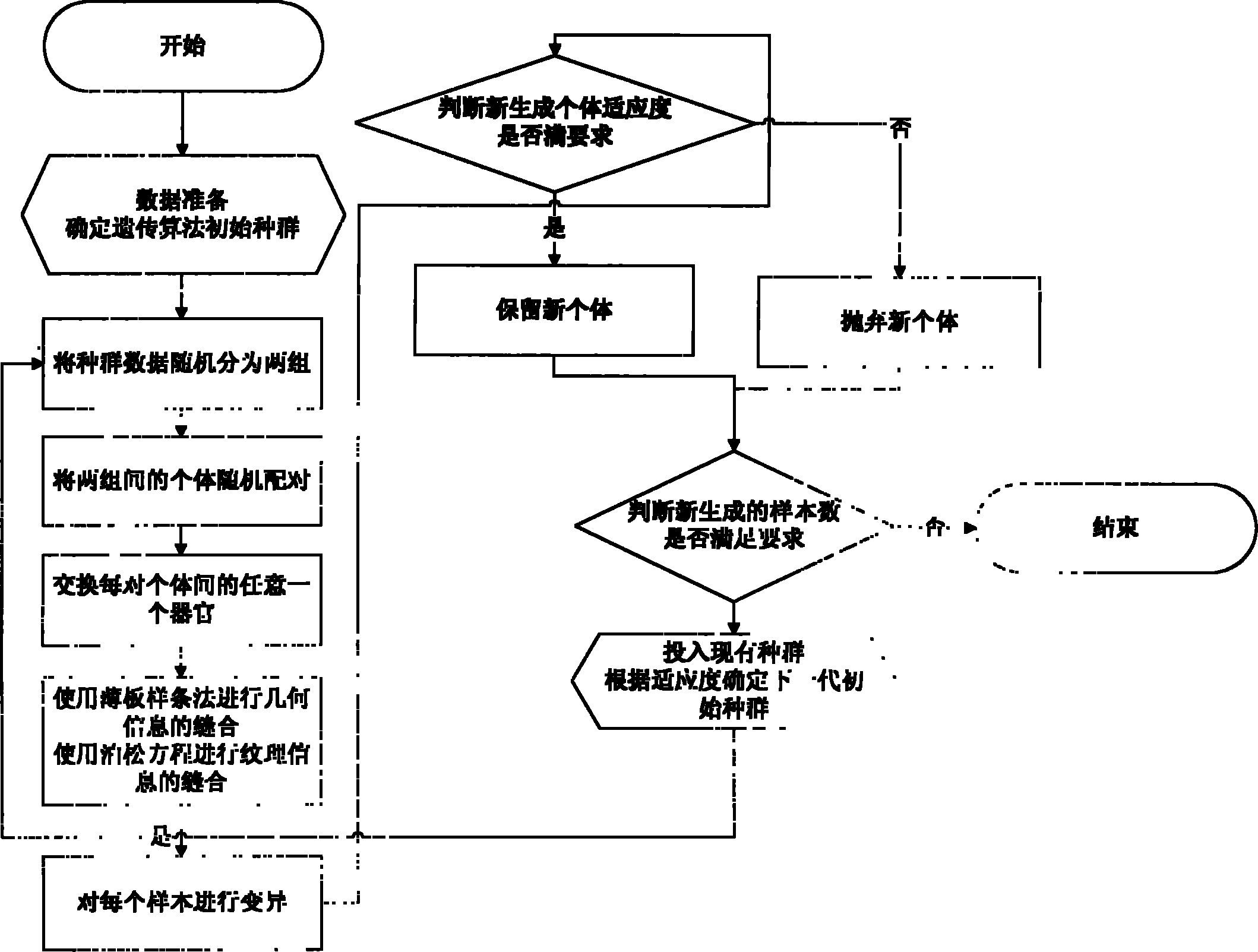
The invention discloses a method for extending a three-dimensional face database, which comprises the following steps of: selecting M samples as an initial database from an existing three-dimensional face database; taking the initial database as an initial population of a genetic algorithm, and taking each sample in the initial database as an individual in the initial population; randomly grouping the individuals in the initial population, randomly pairing the individuals in different two groups, and randomly exchanging organs of each pair of individuals; seaming the exchanged organs and a target face; varying genic values on each pair of individual gene sites; selecting reservation or discard of new individuals according to the similarity of the newly generated individuals and the individuals in the existing population; merging the existing population and the evolved new individual population, and extending the primary three-dimensional face database; and repeating the steps till the extended face database meets the requirement. The method can realize large-scale diverse data extension, and has the advantages of simple realization, good data extending effect, high operating speed and hardware cost saving.
Owner:BEIJING UNIV OF TECH
Early Detection and Prognosis of Colon Cancers
InactiveUS20080221056A1Organic active ingredientsGenetic material ingredientsGenes mutationBiomarker (petroleum)
We have developed a transcriptome-wide approach to identify genes affected by promoter CpG island hypermethylation and transcriptional silencing in colorectal cancer (CRC). By screening cell lines and validating tumor specific hypermethylation in a panel of primary human CRC samples, we estimate that nearly 5% of all known genes may be promoter methylated in an individual tumor. When directly compared to gene mutations, we find a much larger number of genes hypermethylated in individual tumors, and much higher frequency of hypermethylation within individual genes harboring either genetic or epigenetic changes. Thus, to enumerate the full spectrum of alterations in the human cancer genome, and facilitate the most efficacious grouping of tumors to identify cancer biomarkers and tailor therapeutic approaches, both genetic and epigenetic screens should be undertaken. The genes we identified can be used inter alia diagnostically to detect cancer, pre-cancer, and likelihood of developing cancer.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE +1
Detection of gene duplications
InactiveUS20050255485A1Easy to useMicrobiological testing/measurementProteomicsFluorescenceGenetic Anomaly
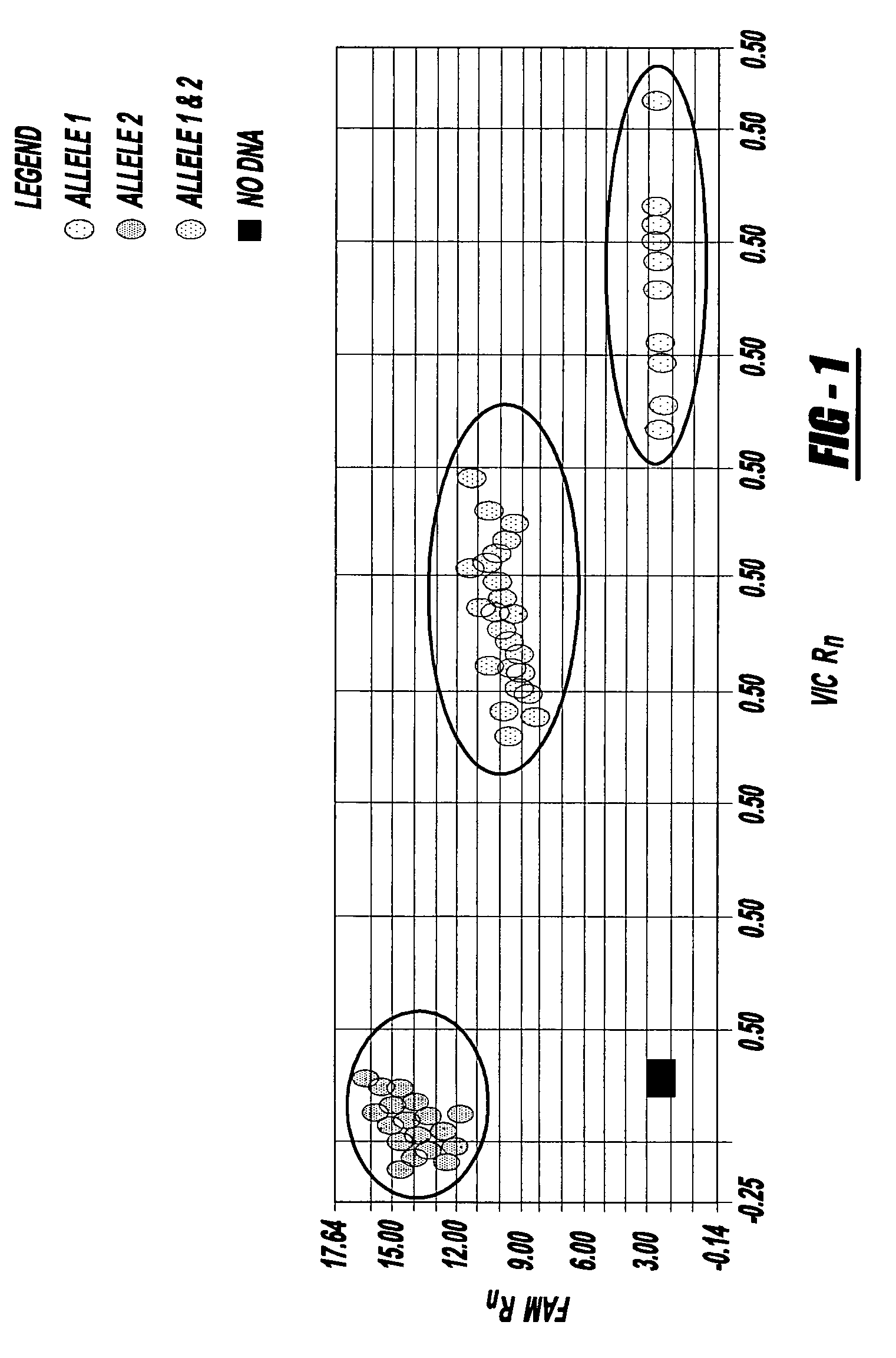
Methods of detecting a candidate genetic anomaly such as a candidate duplication in a genome are disclosed. The methods comprise quantifying fluorogenic assays for alleles of a genetic locus from a plurality of individual genomes, identifying ranges of fluorescent intensities indicative of individual genomes homozygous for a first allele, homozygous for a second allele, or heterozygous for both alleles, and identifying individual genomes in which the fluorescence intensities are outside the range of intensities indicative of homozygosity or heterozygosity for the genetic locus.
Owner:APPL BIOSYSTEMS INC
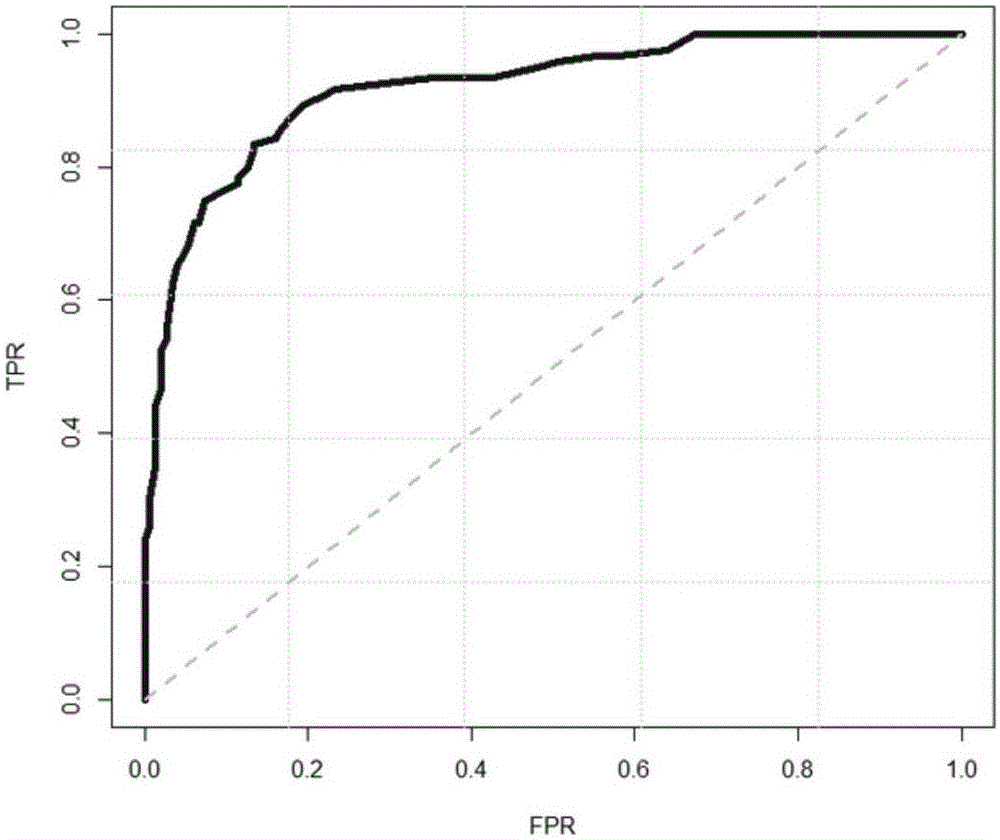
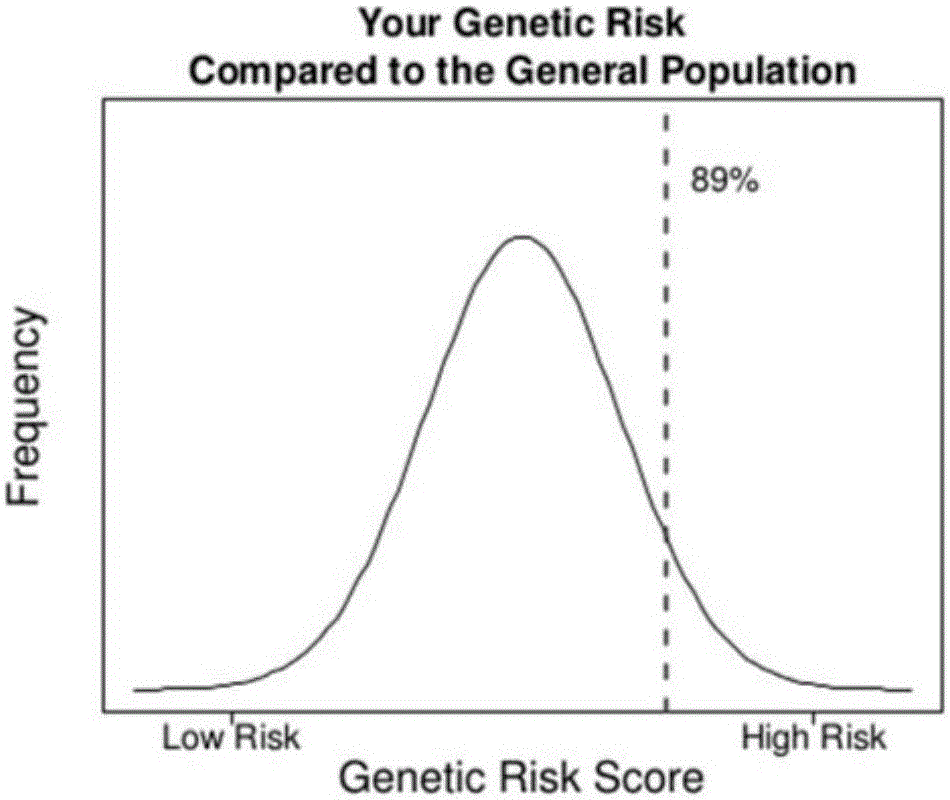
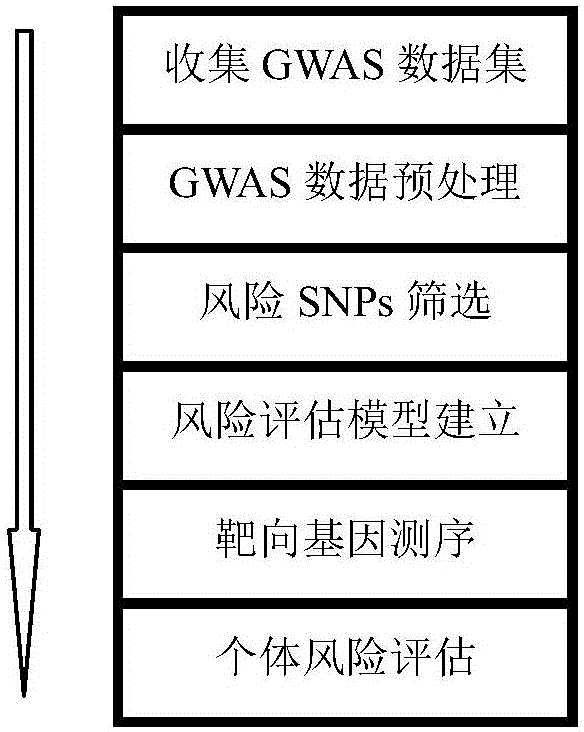
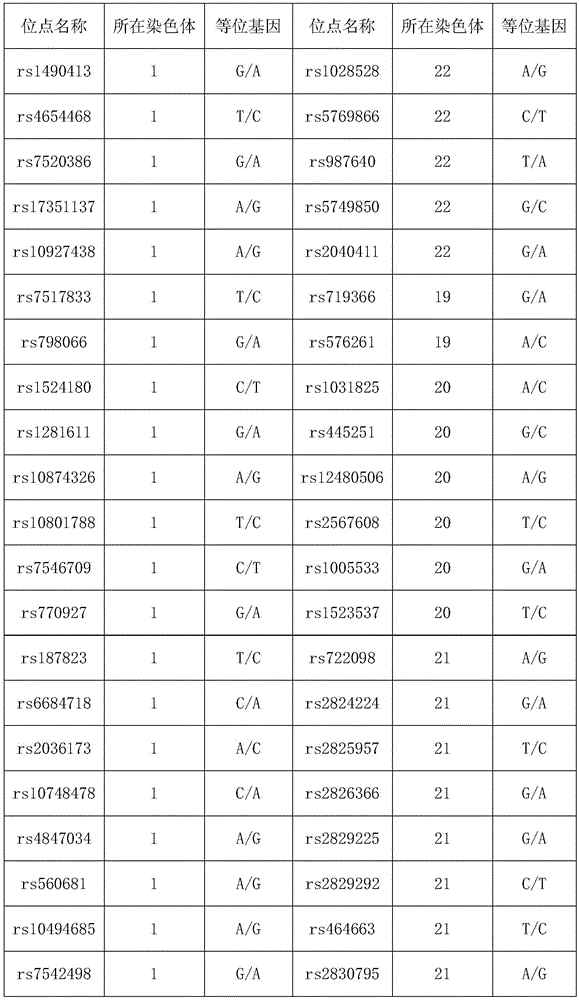
Next generation sequencing based coronary heart disease genetic risk evaluation method
InactiveCN105279369AMethod for assessing genetic risk of coronary heart disease effectiveConducive to interventionsSpecial data processing applicationsGenetic riskMedicine
The invention discloses a next generation sequencing based coronary heart disease genetic risk evaluation method. Particularly, the invention relates to screening of coronary heart disease genetic risk loci and establishment of a coronary heart disease genetic risk evaluation model based on the risk loci. Depending on the model, an individual genome is subjected to targeted sequencing to finally obtain coronary heart disease genetic risk of an individual.
Owner:SUZHOU GENEWORKS TECH CO LTD
Compound typing system used for personal identification
ActiveCN106520982AThe result of identification is reliableThe effect of family identification is excellentMicrobiological testing/measurementDNA/RNA fragmentationHalf sibsTyping
The invention aims at providing a compound typing system used for personal identification, which is used for personal identification, especially the identification for relative relations such as grandparents and grandchild, uncles and nephews, or half sibs, and has the powerful typing efficiency and wide application range. On one aspect, the invention provides a set of genetic marker assembly, and the genetic marker assembly comprises 482 SNP sites positioned on the autosome; on another aspect, the invention provides a typing kit, and the typing kit comprises a reagent for typing the 482 SNP sites in the genetic marker assembly provided by the invention; on still another aspect, the invention provides an individual genotype identity card, and the individual genotype identity card comprises a chip, the genotype information of the genetic marker sites of the individuals is stored in the chip, and the genetic marker sites refer to the 482 SNP sites in the genetic marker assembly provided by the invention.
Owner:INST OF RADIATION MEDICINE ACAD OF MILITARY MEDICAL SCI OF THE PLA +1
Method for high-salt screening of saline-alkali resistant rosa chinensis plant
InactiveCN103004610ABreak growth boundariesHorticulture methodsPlant tissue cultureVascular bundleIndividual gene
The invention relates to a method for high-salt screening of a saline-alkali resistant rosa chinensis plant. In particular, rosa chinensis which is planted in Tibet plateau is obtained by the method for high-salt screening of saline-alkali resistant rosa chinensis. The method comprises the steps of inducing obtained rosa chinensis explant culture medium suitable for rosa chinensis explants out of a sprout texture and then irrigating and screening by a high-salt culture medium to obtain the saline-alkali resistant rosa chinensis; cutting a fibroid branch with thick vascular bundle into 10cm stem sections, sterilizing the culture mediums by 0.5% potassium permanganate according to the ratio of vermiculite to perlite to nutrient soil being 10:2:1, and then spraying the medium by 0.3% rooting liquid, wherein high-salt screening is diluting and irrigating by sodium chloride liquid culture mediums B with different concentrations, and selecting the plant which can normally grow. By adopting the method, the growth boundaries of fresh flowers are broken through; five saline-alkali resistant rosa chinensises can be obtained from genetic modification and individual gene screening, and a blank no fresh flower is available in golmud is filled.
Owner:王昱蓉 +1
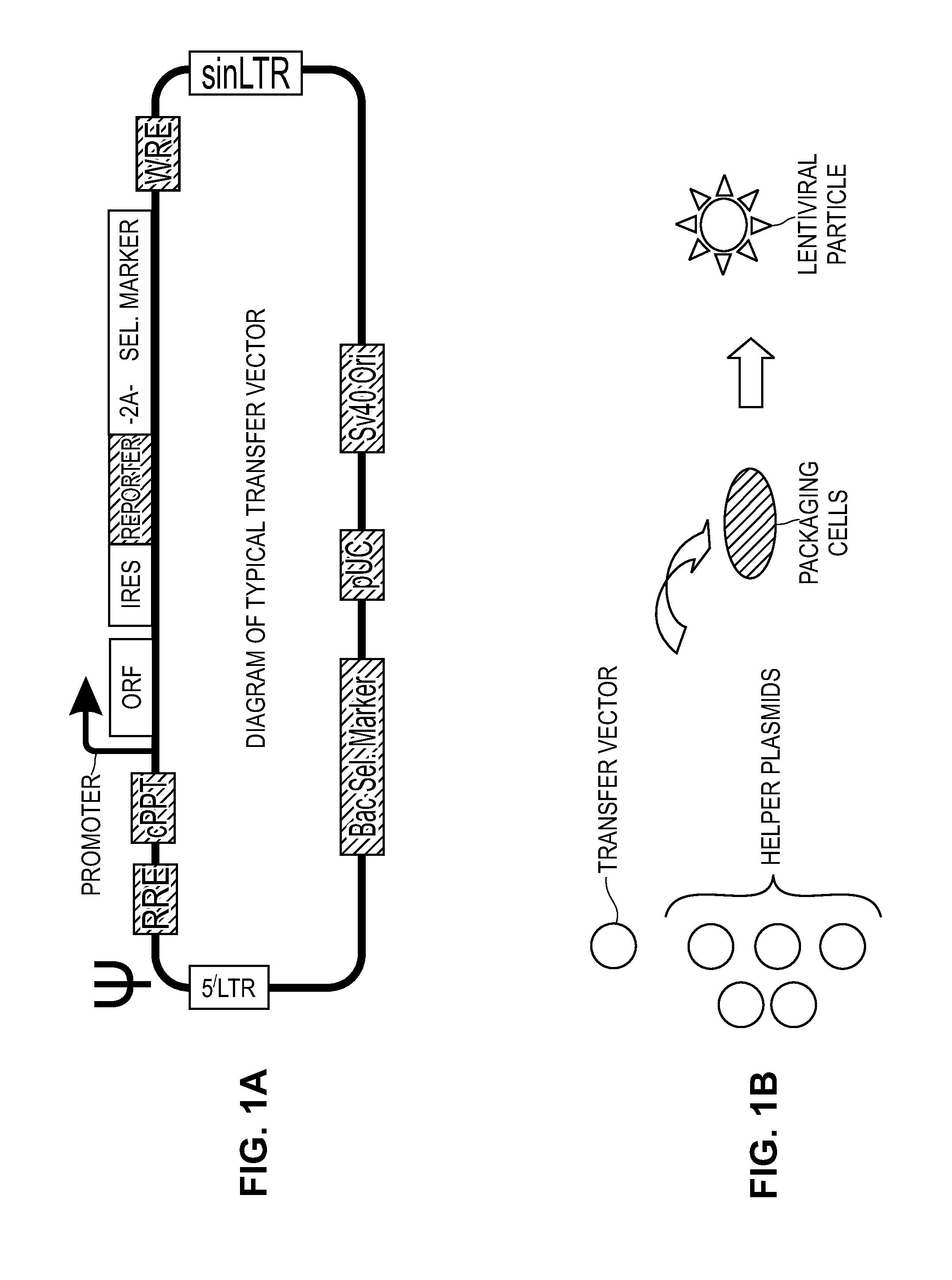
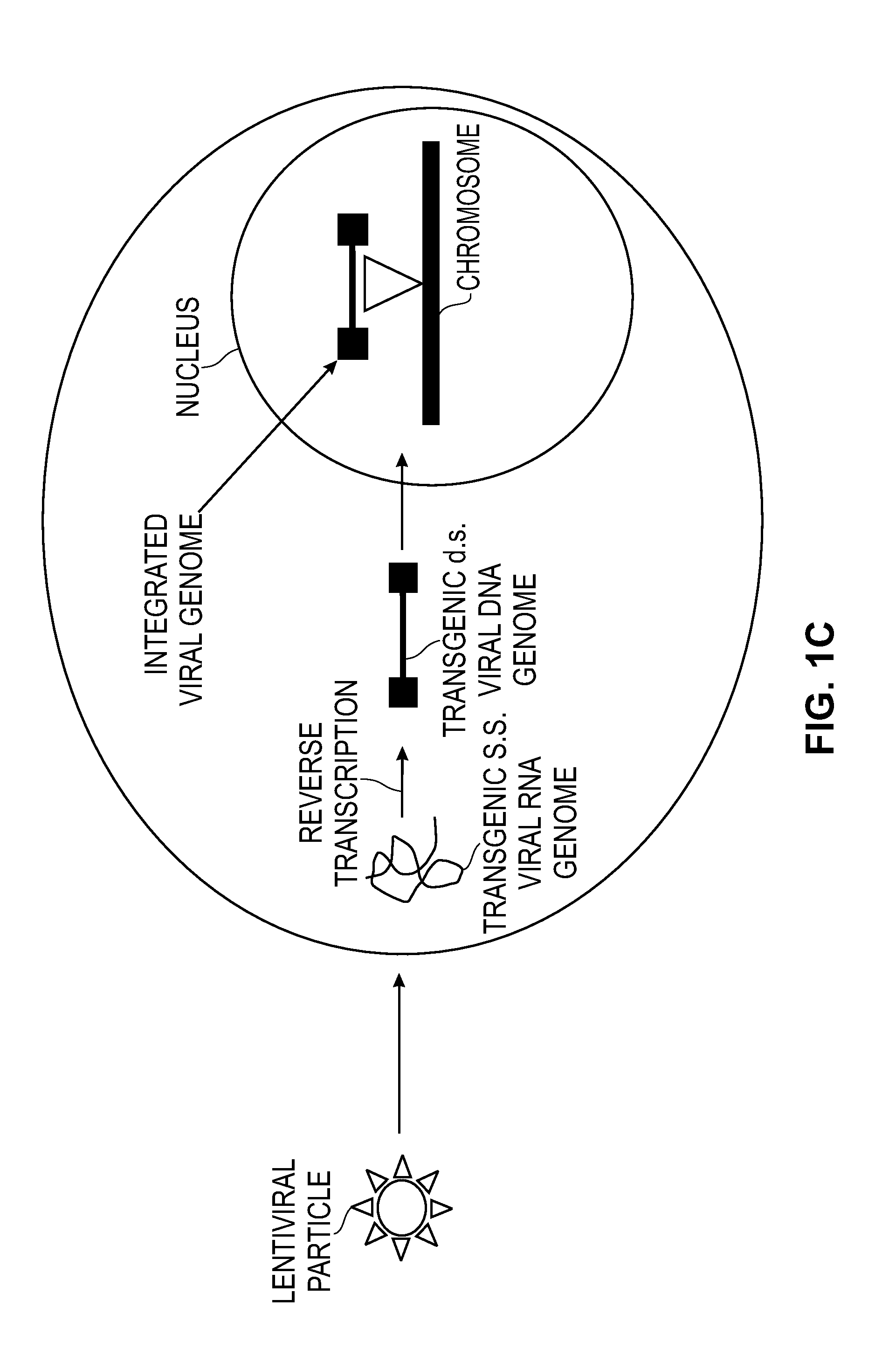
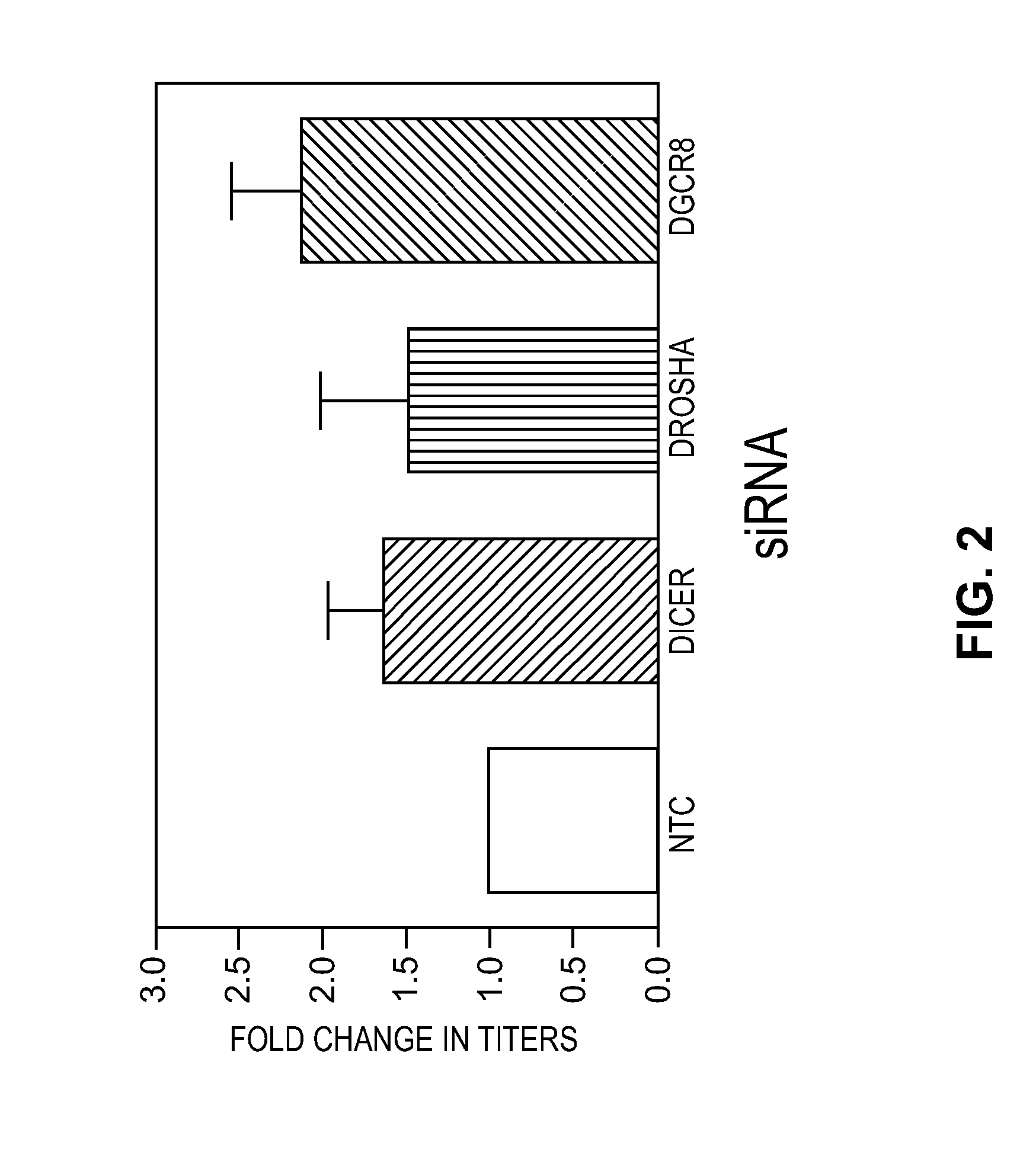
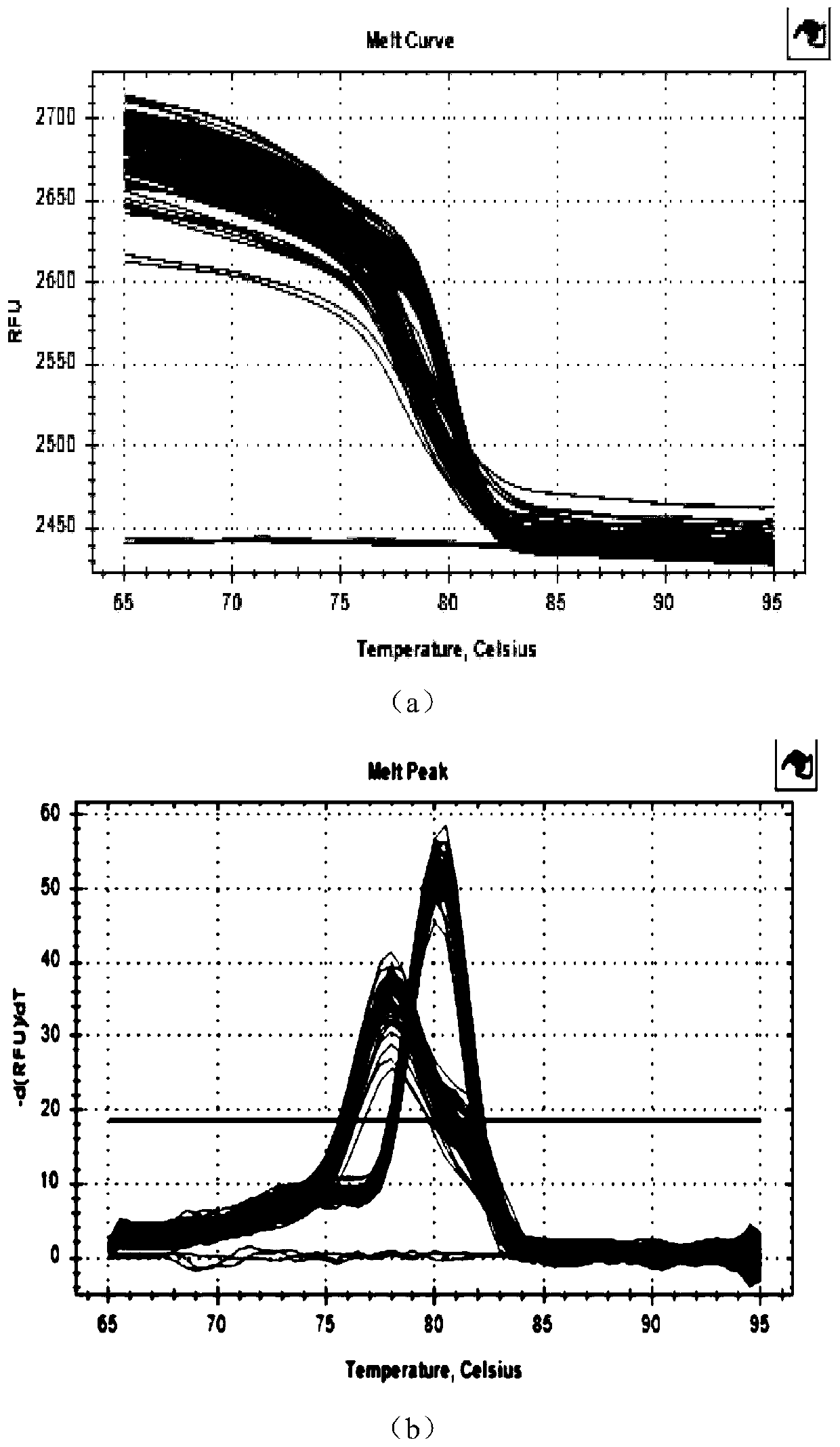
Modified Cell Lines for Increasing Lentiviral Titers
InactiveUS20120083035A1Facilitate viral particle formationFacilitated releaseArtificial cell constructsEmbryonic cellsTiterIndividual gene
The present disclosure provides a method of modifying cells in order to enhance lentiviral titers, cell lines that are modified and modifying reagents. By mediating individual genes and combination thereof, lentiviral titers may be increased.
Owner:DHARMACON INC
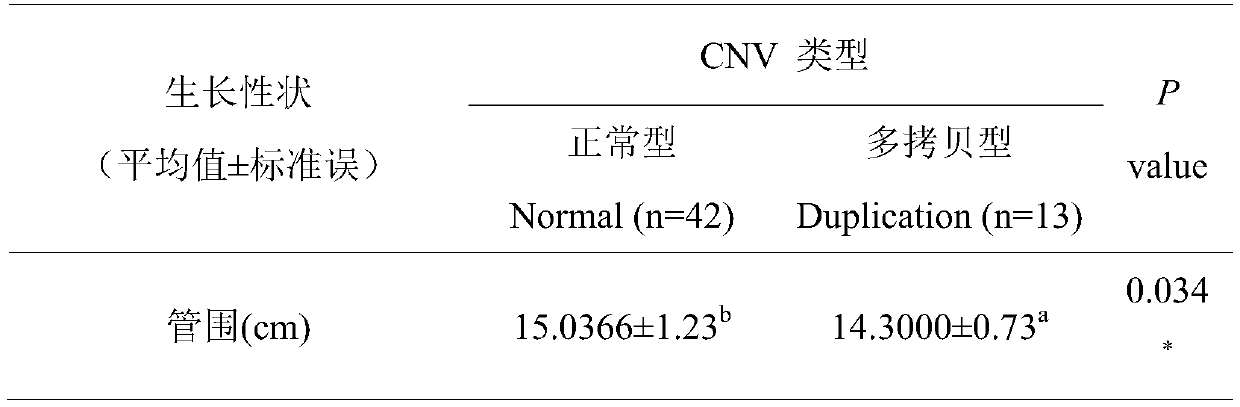
Method for assisting in detecting growth characters by using CNV molecular marker of cattle PLAG1 gene and application thereof
ActiveCN109943646AAccurate identificationLow costMicrobiological testing/measurementClimate change adaptationReference genesMarker-assisted selection
The invention discloses a method for assisting in detecting growth characters by using a CNV molecular marker of cattle PLAG1 genes and an application thereof. The method for assisting in detecting the growth characters by using the CNV molecular marker of the cattle PLAG1 genes comprises the following steps of: respectively amplifying partial fragments of a PLAG1 gene copy number variation regionand a reference gene BTF3 by using a cattle genome DNA as a template through a real-time fluorescence quantitative PCR method, and dividing the quantitative results into multiple copy type and normaltype by 2-delta delta Ct method, thereby identifying copy number variation type of individual PLAG1 gene of a cattle. The copy number variation closely related to the growth characters of the cattleis detected on a DNA level, and the copy number variation can be used as an important candidate molecular marker for marker-assisted selection of the growth characters of Chinese cattles and is used for accelerating the breeding work of the marker-assisted selection of the cattle.
Owner:NORTHWEST A & F UNIV
Epilepsy medication recommendation method and system
PendingCN111462921AHigh control rateShorten the timeBiostatisticsProteomicsAntiepileptic drugMetabolic enzymes
The invention provides an epilepsy medication recommendation method and system, and belongs to the technical field of biomedicine and data processing. According to the invention, deep sequencing is carried out on an antiepileptic drug metabolic enzyme gene, a transporter gene and a drug action target gene, related coding gene polymorphic sites are screened, a Bayesian accumulation regression treemodel and random forest regression are utilized to obtain a personalized medication scheme depending on individual genes, and a subset with high treatment effect is identified, so that clinical medication is guided in a targeted manner, the epilepsy control rate is improved, adverse reactions are reduced, the time for doctors and patients to explore the optimal treatment scheme is shortened, the diagnosis and treatment efficiency is improved, and the method has good practical application value.
Owner:SHANDONG UNIV
Molecular marker identification method related to pig birth litter weight and fetal weight and application of molecular marker identification method
ActiveCN106011245AMicrobiological testing/measurementDNA/RNA fragmentationAllele frequencyFetal weight
The invention discloses a molecular marker identification method related to pig birth litter weight and fetal weight and an application of the molecular marker identification method. The identification method comprises the following steps: collecting a sample; extracting and detecting pig genome DNA; then, conducting SNP (single nucleotide polymorphism) chip genotype judgment and genotype data quality control: adding a PCR amplification primer pair and conducting a PCR amplification reaction by taking DNA of the sample as a template, purifying a reaction product and co-crystallizing the purified reaction product with a mass spectrum chip, conducting detection and typing by virtue of a flight mass spectrum method, detecting an SNP site of a target gene, computing an SNP genotype and an allele frequency by virtue of PopGene3.2, and conducting association analysis on the SNP and characters by virtue of SAS9.2 software; and finally, conducting data collating and analysis. The identification method disclosed by the invention, by determining the genotype of a pig individual, can assist identification of multiparous litter weight and fetal weight.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI +1
SNP molecular marker related to skin color character of black-bone chicken and application
ActiveCN105603094AImprove seed selection efficiencyReduce breeding costsMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionAgricultural science
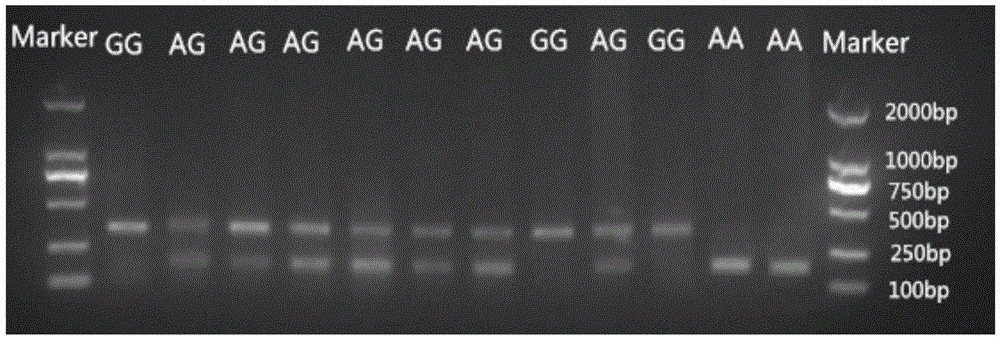
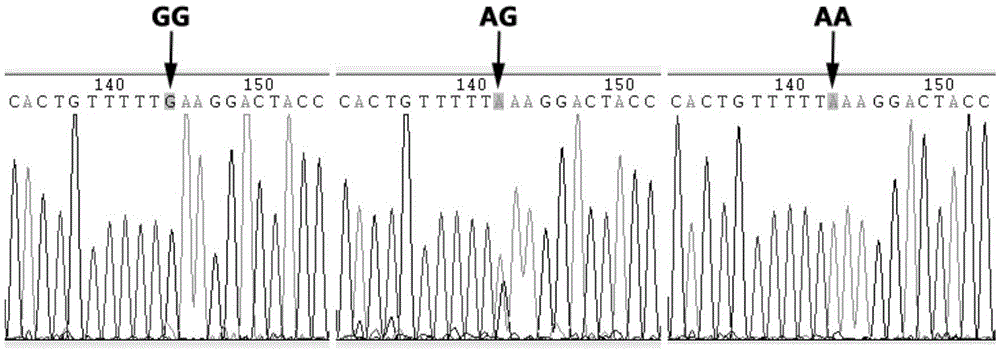
The invention provides application of an SNP molecular marker in detecting the skin color character of a black-bone chicken. The nucleotide sequence of the molecular marker is shown as SEQ ID No.1, and the base mutation type at the 171st bp is A / G. The invention further provides a primer sequence for detecting an SNP locus and a PCR-RFLP method. The genotype of the locus can be distinguished based on the difference of an electrophoretic band after DraI enzyme digestion, and then the homozygosis and heterozygosis of a sample Fm gene can be judged. If the 342 bp band appears, the sample Fm gene is a GG homozygosis-based gene; if the two bands of 171 bp and 342 bp appear, the sample Fm gene is an AG heterozygosis-based gene; if the 171 bp band appears, the sample Fm gene is an AA recessive-homozygosis-based gene. By means of the SNP molecular marker and the detection method, the individual genotype of the black-bone chicken can be judged in the chickling stage, early selection and early elimination of the skin color character can be achieved, and thus significant practical value can be achieved on improving selection efficiency and saving breeding cost.
Owner:HANGZHOU ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com